
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Plant Sci., 02 June 2015
Sec. Plant Physiology
Volume 6 - 2015 | https://doi.org/10.3389/fpls.2015.00405
Reactive oxygen species (ROS) are engaged in several processes essential for normal cell functioning, such as differentiation, anti-microbial defense, stimulus sensing and signaling. Interestingly, recent studies imply that cellular signal transduction and gene regulation are mediated not only directly by ROS but also by the molecules derived from ROS-mediated oxidation. Lipid peroxidation leads to non-enzymatic formation of oxylipins. These molecules were shown to modulate expression of signaling associated genes including genes encoding phosphatases, kinases and transcription factors. Oxidized peptides derived from protein oxidation might be engaged in organelle-specific ROS signaling. In turn, oxidation of particular mRNAs leads to decrease in the level of encoded proteins and thus, contributes to the post-transcriptional regulation of gene expression. Present mini review summarizes latest findings concerning involvement of products of lipid, protein and RNA oxidation in signal transduction and gene regulation.
It is well established that reactive oxygen species (ROS) are engaged in the cellular signal transduction network (Neill et al., 2002; Ślesak et al., 2007; Wrzaczek et al., 2013). In the case of plants they have been shown to mediate morphogenesis, stomatal closure, gravitropism, programmed cell death (PCD), and responses to stress factors, including systematic acquired resistance (SAR) and systematic acquired acclimation (SAA; Neill et al., 2002; Ślesak et al., 2007). In recent years progress has been made in understanding the molecular language associated with ROS-mediated signal transduction. Changes in ROS level affect oxidative status of redox-sensitive signal regulators such as ascorbate, glutathione and thioredoxins, leading to modulation of metabolism and gene expression. For example, shift of ascorbate/dehydroascorbate ratio toward more oxidized state was shown to inhibit cell cycle (Kopczewski and Kuźniak, 2013). In turn, glutathione and thioredoxins are major players in protein thiol switching—reversible modification of proteins causing alterations in their conformation and functioning. Thiol switches were shown to regulate various cellular processes including translation, transcription and signaling (Brandes et al., 2009; Dietz and Hell, 2015). Changes in cellular redox status affects disulfide bonds in proteins associated with transcription, leading to the alterations in their polymerization and DNA binding capacity (Chi et al., 2013). Proteins subjected to the described oxidative-dependent regulation include NPR1 transcription co-activator and TGA9, RAP2.4a, and R2R3 transcription factors (Wrzaczek et al., 2013). ROS influence signal transduction pathways also indirectly through interaction with other signaling elements, such as nitric oxide, calcium ions and plant hormones (Neill et al., 2002; Chi et al., 2013). Direct and indirect ROS action modulates gene expression. The global Arabidopsis transcriptome analysis has revealed that application of H2O2 causes changes in the expression of approximately 1% of genes (Desikan et al., 2001).
Lately, new players have emerged on the ROS signaling arena. It has been suggested that oxidation of cellular compounds leads to the formation of new signaling molecules: non-enzymatically formed oxylipins, peptides derived from ROS-dependent protein degradation and oxidatively modified mRNAs. The present mini review summarizes their role in cellular signaling and gene regulation (illustrated on Figure 1).
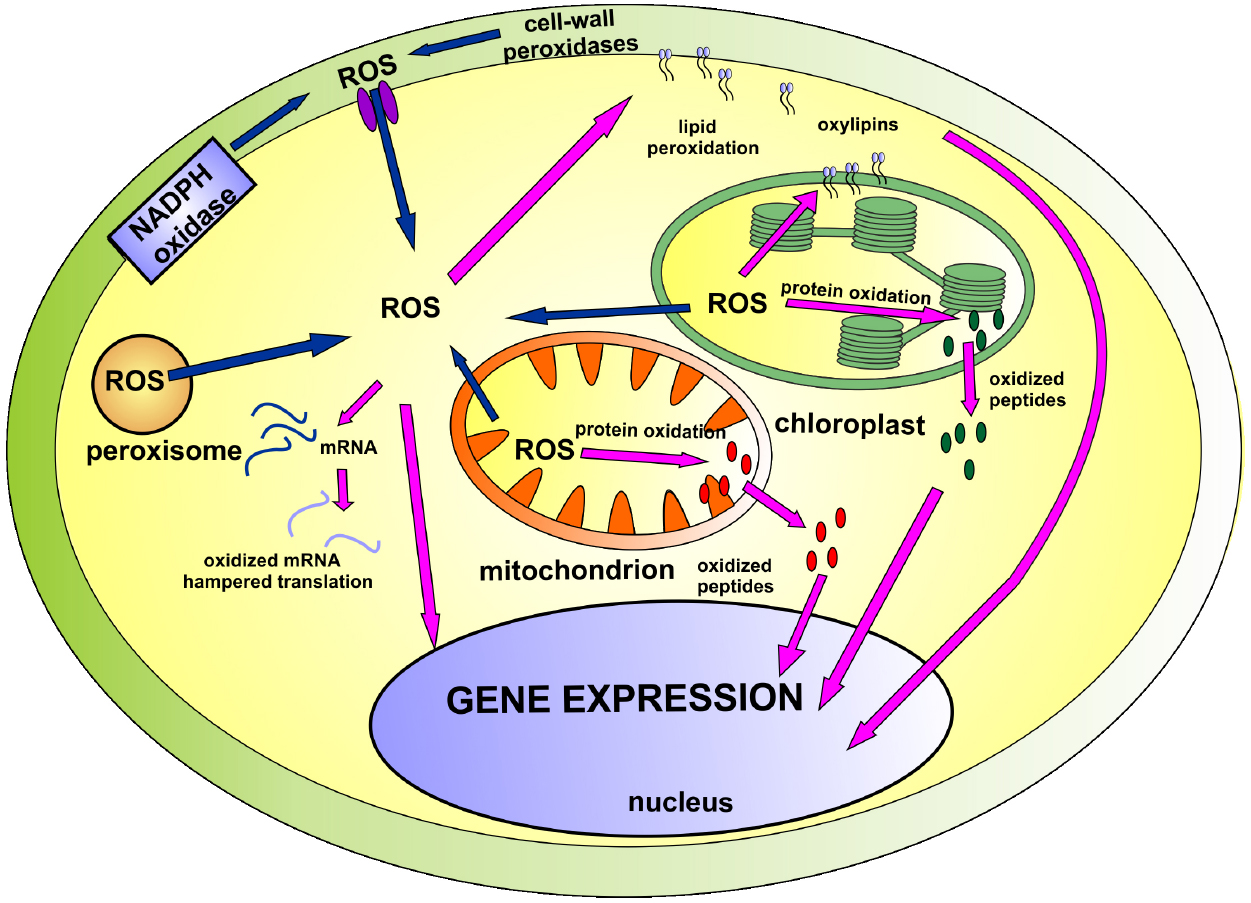
Figure 1. The involvement of ROS and products of ROS-dependent oxidation in cellular signaling network. The main sources of ROS (marked with blue arrows) include mitochondria, chloroplast, NADPH oxidase and cell-wall peroxidases. Accumulated ROS participate in cellular signaling (marked with pink arrows) directly or indirectly through derivatives of lipid, protein and mRNA oxidation. Detailed description can be found in the text.
Oxygenation of polyunsaturated fatty acids leads to the production of oxylipins via enzymatic or non-enzymatic pathways. The study of plant lipid peroxidation products, thus far has been focused mainly on the enzymatically synthesized jasmonate family of signaling molecules, and their roles in the regulation of developmental and defense-related processes. Importantly, it should be emphasized that the enzymatic process, which is generally seen as beneficial for survival, generates relatively small numbers of lipid regulators. Recent evidence indicates that a number of biological active oxylipins are generated in a non-enzymatic pathway, initiated by enhanced formation of ROS, mainly hydroxyl radical and superoxide anion. Moreover, a growing body of evidence suggests that some non-enzymatically formed oxylipins, such as phytoprostanes (PP), may also function as endogenous mediators that regulate various signal transduction pathways (Thoma et al., 2003; Loeffler et al., 2005; Grun et al., 2007). Transcriptome analysis have revealed that in Arabidopsis and tomato plants A1- and B1-type phytoprostanes (PPA1 and PPB1) induce a variety of genes associated with secondary metabolism, stress responses and cellular detoxification. Importantly, the induced enzymes included glutathione-S-transferase (GST) and cytochrome P450 enzymes are engaged in inactivation of toxic electrophiles, which are also formed in a result of lipid peroxidation. Therefore, it has been suggested that phytoprostanes are not only markers of oxidative injury, but may also act as ROS scavengers (Thoma et al., 2003; Loeffler et al., 2005; Mueller et al., 2008). Moreover, it has been shown that 24% of the genes in Arabidopsis up-regulated by PP are related to signal transduction, including transcription factors, phosphatases and kinases (Mueller et al., 2008). Interestingly, Mueller et al. (2008) also reported that approximately 50% of the PPA1-induced genes in both Arabidopsis cell culture and whole-plant experiments contain a TGA motif (TGACG) in their promoters. These are putative binding sites for basic/leucine zipper transcription factors of the TGA family. Subsequent analysis revealed that 60% of all genes induced by PPA1 could not be induced in Arabidopsis mutants deficient in class II TGA transcriptions factors—tga2, tga5, and tga6 mutants (Mueller et al., 2008). This class of transcription factors are known to be involved in plant xenobiotic signaling (Behringer et al., 2011). In response to a variety of xenobiotics, they recruit the SCARECROW-LIKE 14 (SCL14) transcriptions coactivator to promoters with TGA motifs and induce the expression of detoxification genes. The fact that Cyp81D11 (At3g28740) gene encoding a putative cytochrome P450 monooxygenase, is induced by both PPA1 and TGA/SCL14 complex indicates that there is a link between oxylipin signaling and TGA-mediated gene regulation in response to xenobiotic metabolism (Fode et al., 2008; Mueller et al., 2008). Although the substrate of CYP81D11 is not known, its presumed function as a monooxygenase and its expression pattern suggest a role in plant detoxification processes (Fode et al., 2008; Mueller et al., 2008).
Gadjev et al. (2006) conducted analysis of data on changes in Arabidopsis transcriptome in response to oxidative stress of various origins. Four out of nine analyzed microarray studies were conducted on plants treated with ROS-generating agents: ozone leading to the formation of H2O2 and O2– in apoplast, methyl viologen (MV)—redox-cycling herbicide generating O2– in chloroplasts and mitochondria, 3-aminotriazole (AT)—herbicide causing H2O2 accumulation in peroxisomes and AAL—toxin leading to drastic increase of cellular H2O2 level. The rest of the experiments were preformed on either flu mutant characterized by rapid accumulation of 1O2 in chloroplasts after transfer from dark to light conditions or transgenic plants with alerted antioxidant systems in specific cell compartments including chloroplasts, mitochondria, cytoplasm and peroxisomes (Gadjev et al., 2006; and references therein). The results of the study indicated that, in addition to hallmark transcripts induced by general oxidative stress, a number of genes respond specifically only to a particular ROS specie or even to ROS originating from a specific cell compartment. It is worth mentioning that these specific ROS signatures can be now identified in Arabidopsis transcriptome by a bioinformatic tool—ROSMETER. The software estimates correlation between microarray data of interest and indices compiled from available transcriptomic data on Arabidopsis plants undergoing oxidative stress of different type and origin (Rosenwasser et al., 2013). Although, it seems that plants possess a sensory system which enables them to distinguish between ROS produced in specific organelles, the exact mechanism of such system is still unknown. It has been proposed that the required information is transmitted through H2O2 waves differing in frequency and amplitude, as has been observed in the case of Ca2+ (Mittler et al., 2011). However, Ca2+-signaling requires the maintenance of low Ca2+ basal level by efficient removal and storing in relaying stations, such as vacuoles and endoplasmic reticulum (ER). In contrast, the level of H2O2 in plant cells is relatively high. The steady state concentration of H2O2 has been estimated in the range between 60 μM and 2 mM depending on plant species. However, it is possible that such wide range might result from technical difficulties in ROS quantification (Neill et al., 2002; and references therein; Vestergaard et al., 2012). There is also no evidence that ROS can be stored in any relaying station. A study based on extensive mathematical modeling demonstrated that, taking into account the above-mentioned facts, the transmission of a signal by diffusing H2O2 is unlikely (Vestergaard et al., 2012). Another possibility is that the generated ROS activate other, more specific signaling elements (Mittler et al., 2011). Møller and Sweetlove (2010) have proposed that functions of such specific signaling elements could be performed by peptides derived from ROS-dependent degradation of proteins. The peptides derived from protein oxidation are capable of transmitting several pieces of information. Firstly, the origin of degraded protein gives information about the localization of oxidative stress. Secondly, the number of derived peptides reflects the intensity of oxidative stress. Lastly, the modification of determined amino acid residues is a signature of the type of over-produced ROS specie (Møller and Sweetlove, 2010). Interestingly, it has been already demonstrated that peptides derived from protein degradation exhibit signaling functions. In C. elegans response to unfolded protein stress in mitochondria, involving induction of chaperone genes, requires re-localization of the ZC376.7 protein from cytoplasm to nucleus. Translocation of this transcription factor is dependent on the activity of two mitochondria-localized proteins—Clp protease and HAF-1 transporter. The authors propose that unfolded/misfolded proteins are degraded by Clp protease, and the derived peptides are transported by HAF-1 to cytoplasm, where they interact with ZC376.7, leading to its translocation to the nucleus (Haynes et al., 2010). However, to our best knowledge, so far there are no studies fully supporting the idea that also peptides formed as a result of protein oxidation act as signaling elements. Providing experimental evidence for involvement of such peptides in organelle-specific ROS signaling is an interesting future research challenge.
Although most of the research concerning nucleic acid oxidation is concentrated on DNA, several reports imply that RNA is more susceptible to this process (Hofer et al., 2006; Shan et al., 2007; Liu et al., 2012). Importantly, the accumulation of oxidized RNA has been shown to be associated with several disorders, such as Parkinson’s and Alzheimer’s diseases, amyotrophic lateral sclerosis (ALS), dementia with Lewy bodies, schizophrenia, diabetes and hemochromatosis (Nunomura et al., 2004; Poulsen et al., 2012; Jorgensen et al., 2013 and reference therein). Immunoprecipitation with monoclonal antibody 15A3 recognizing markers of nucleic acid oxidation, 8-oxo-7,8-hydroguanosine (8OHG) and 8-oxo-7,8-dihydroguanine (8OHdG), enabled identification of mRNAs highly oxidized in postmortem isolated brain tissues of patients suffering from Alzheimer’s disease. Interestingly, oxidation did not occur in abundant transcripts encoding for example β-actin. Instead the most prominent oxidation was observed in the case of a specific set of mRNAs associated with signaling, protein transport, regulation of gene expression, response to oxidative stress and metabolism. Thus, the authors conclude that mRNA oxidation is not a random process but targets specific transcripts (Shan et al., 2003). Similar results were obtained in motor neurons of transgenic mice expressing familial ALS-linked mutation in superoxide dismutase gene. In three independent microarray analysis performed in identical experimental conditions oxidation affected same RNA species. Interestingly some of the highly oxidized mRNAs were associated with ASL pathogenesis (e.g., superoxide dismutase, amyloid beta precursor-like protein, prion protein) and cellular signaling (e.g., calmodulin, mitogen-activated protein kinases, transcription factors; Chang et al., 2008). Targeted mRNA oxidation has been also observed in plants. Increased levels of 8-oxo-7,8-dihydroguanosine (8-OHG), a marker of RNA oxidation, were noted in sunflowers seeds. The observed oxidation was not a random process—reproducible microarray analysis showed that only a specific set of mRNAs has been oxidatively modified (Bazin et al., 2011). Thus far it is not clear what determines the susceptibility of mRNA to oxidation. However, the possibilities that its sensitivity is dependent on the abundance of specific mRNA species, the frequency of guanine in its sequence, or the occurrence of any specific motif have been excluded (Chang et al., 2008; Bazin et al., 2011). Importantly accumulation of oxidized transcripts causes ribosome stalling leading to the slowing down of translation (Shan et al., 2007; Simms et al., 2014), which can explain frequently observed reduction in the level of encoded proteins (Shan et al., 2003, 2007; Tanaka et al., 2006; Chang et al., 2008). Thus, targeted RNA oxidation leading to the diminished expression of definite proteins might constitute a newly discovered mechanism of post-transcriptional gene expression regulation. Moreover, as highly oxidized transcripts encode inter alia signaling elements, this process likely affects cellular signaling network (Shan et al., 2003; Chang et al., 2008; Bazin et al., 2011).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Research concerning the role of oxidized RNA in plants response to cadmium stress is conducted in the Department of Plant Ecophysiology with financial support of National Science Centre, Poland, granted on the basis of decision number DEC-2014/13/D/NZ9/04812.
Bazin, J., Langlade, N., Vincourt, P., Arribat, S., Balzergue, S., El-Maarouf-Bouteau, H., et al. (2011). Targeted mRNA oxidation regulates sunflower seed dormancy alleviation during dry after-ripening. Plant Cell 23, 2196–2208. doi: 10.1105/tpc.111.086694
Behringer, C., Bartsch, K., and Schaller, A. (2011). Safeners recruit multiple signalling pathways for the orchestrated induction of the cellular xenobiotic detoxification machinery in Arabidopsis. Plant Cell Environ. 34, 1970–1985. doi: 10.1111/j.1365-3040.2011.02392.x
Brandes, N., Schmitt, S., and Jakob, U. (2009). Thiol-based redox switches in eukaryotic proteins. Antioxid. Redox Sign. 11, 997–1014. doi: 10.1089/ars.2008.2285
Chang, Y., Kong, Q., Shan, X., Tian, G., Llieva, H., Cleveland, D. W., et al. (2008). Messenger RNA oxidation occurs early in disease pathogenesis and promotes motor neuron degeneration in ALS. PLoS ONE 3:e2849. doi: 10.1371/journal.pone.0002849
Chi, Y. H., Paeng, S. K., Kim, M. J., Hwang, G. Y., Melencion, S. M. B., Oh, H. T., et al. (2013). Redox-dependent functional switching of plant proteins accompanying with their structural changes. Front. Plant Sci. 4:277. doi: 10.3389/fpls.2013.00277
Desikan, R., A-H-Mackerness, S., Hancock, J. T., and Neill, S. J. (2001). Regulation of the Arabidopsis transcriptome by oxidative stress. Plant Physiol. 127, 159–172. doi: 10.1104/pp.127.1.159
Dietz, K. J., and Hell, R. (2015). Thiol switches in redox regulation of chloroplasts: balancing redox state, metabolism and oxidative stress. Biol. Chem. 396, 483–494. doi: 10.1515/hsz-2014-0281
Fode, B., Siemsen, T., Thurow, C., Weigel, R., and Gatz, C. (2008). The Arabidopsis GRAS protein SCL14 interacts with class II TGA transcription factors and is essential for the activation of stress-inducible promoters. Plant Cell 20, 3122–3135. doi: 10.1105/tpc.108.058974
Gadjev, I., Vanderauwera, S., Gechev, T. S., Laloi, C., Minkov, I. N., Shulev, V., et al. (2006). Transcriptomic footprints disclose specificity of reactive oxygen species signaling in Arabidopsis. Plant Physiol. 141, 436–445. doi: 10.1104/pp.106.078717
Grun, G., Berger, S., Matthes, D., and Mueller, M. J. (2007). Early accumulation of non-enzymatically synthesized oxylipins in Arabidopsis thaliana after infection with Pseudomonas syringae. Funct. Plant Biol. 34, 65–71. doi: 10.1071/FP06205
Haynes, C. M., Yang, Y., Blais, S. P., Neubert, T. A., and Ron, D. (2010). The matrix peptide exporter HAF-1 signals mitochondrial UPR by activating the transcript factor ZC376.7 in C. elegans. Mol. Cell 37, 529–540. doi: 10.1016/j.molcel.2010.01.015
Hofer, T., Seo, A. Y., Prudencio, M., and Leeuwenburgh, C. (2006). A method to determine RNA and DNA oxidation stimutaneously by HPLC-ECD: greater RNA than DNA oxidation in rat liver after doxorubicin. Biol. Chem. 287, 103–111. doi: 10.1515/BC.2006.014
Jorgensen, A., Broedbaek, K., Jensen-Fink, A., Knorr, U., Greisen Soendergaard, M., Henriksen, T., et al. (2013). Increased systematic oxidatively generated DNA and RNA damage in schizophrenia. Psychiatry Res. 209, 417–423. doi: 10.1016/j.psychres.2013.01.033
Kopczewski, T., and Kuźniak, E. (2013). Redox signals as a language of interorganellar communication in plant cells. Centr. Eur. J. Biol. 8, 1153–1163. doi: 10.2478/s11535-013-0243-4
Liu, M., Gong, X., Alluri, R. K., Wu, J., Sablo, T., and Li, Z. (2012). Characterization of RNA damage under oxidative stress in Escherichia coli. Biol. Chem. 393, 123–132. doi: 10.1515/hsz-2011-0247
Loeffler, C., Berger, S., Guy, A., Durand, T., Bringmann, G., Dreyer, M., et al. (2005). B1-phytoprostanes trigger plant defense and detoxification responses. Plant Physiol. 137, 328–340. doi: 10.1104/pp.104.051714
Mittler, R., Vanderauwera, S., Suzuki, N., Miller, G., Tognetti, V. B., Vandepoele, K., et al. (2011). ROS signaling: the new wave? Trends Plant Sci. 16, 300–309. doi: 10.1016/j.tplants.2011.03.007
Møller, I. M., and Sweetlove, L. J. (2010). ROS signalling -specificity is required. Trends Plant Sci. 15, 370–374. doi: 10.1016/j.tplants.2010.04.008
Mueller, S., Hilbert, B., Dueckershoff, K., Roitsch, T., Krischke, M., Mueller, J. M., et al. (2008). General detoxification and stress responses are mediated by oxidized lipids through TGA transcription factors in Arabidopsis. Plant Cell 20, 768–785. doi: 10.1105/tpc.107.054809
Neill, S., Desikan, R., and Hancock, J. (2002). Hydrogen peroxide signaling. Curr. Opin. Plant Biol. 5, 388–395. doi: 10.1016/S1369-5266(02)00282-0
Nunomura, A., Chiba, S., Lippa, C. F., Cras, P., Kalaria, R. N., Takeda, A., et al. (2004). Neuronal RNA oxidation is a prominent feature of familial Alzheimer’s disease. Neurobiol. Dis. 17, 108–113. doi: 10.1016/j.nbd.2004.06.003
Poulsen, H. E., Specht, E., Broedbaek, K., Henriksen, T., Ellervik, C., Mandrup-Poulsen, T., et al. (2012). RNA modifications by oxidation: a novel disease mechanism?. Free Radical. Bio. Med. 52, 1353–1361. doi: 10.1016/j.freeradbiomed.2012.01.009
Rosenwasser, S., Fluhr, R., Joshi, J. R., Leviatan, N., Sela, N., Hetzroni, A., et al. (2013). ROSMETER: a bioinformatic tool for the identification of transcriptomic imprints related to reactive oxygen species type and origin provides new insights into stress responses. Plant. Physiol. 163, 1071–1083. doi: 10.1104/pp.113.218206
Shan, X., Chang, Y., and Lin, C. L. (2007). Messenger RNA oxidation is an early event preceding cell death and causes reduced protein expression. FASB. J. 21, 2753–2764. doi: 10.1096/fj.07-8200com
Shan, X., Tashiro, H., and Lin, C. G. (2003). The identification and characterization of oxidized RNAs in Alzheimer’s disease. J. Neurosci. 23, 4913–4921.
Simms, C. L., Hudson, B. H., Mosior, J. W., Rangwala, A. S., and Zaher, H. S. (2014). An active role for the ribosome in determining the fate of oxidized mRNA. Cell Rep. 9, 1256–1264. doi: 10.1016/j.celrep.2014.10.042
Ślesak, I., Libik, M., Karpinska, B., Karpinksi, S., and Miszalski, Z. (2007). The role of hydrogen peroxide in regulation of plant metabolism and cellular signalling in response to environmental stresses. Acta Biochim. Pol. 54, 39–50.
Tanaka, M., Chock, P. B., and Stadtman, E. R. (2006). Oxidized Messenger RNA induces translation errors. Proc. Natl. Acad. Sci. U.S.A. 104, 66–71. doi: 10.1073/pnas.0609737104
Thoma, I., Loeffler, C., Sinha, A. K., Gupta, M., Krischke, M., Steffan, B., et al. (2003). isoprostanes induced by reactive oxygen species trigger defense gene activation and phytoalexin accumulation in plants. Plant J. 34, 363–375. doi: 10.1046/j.1365-313X.2003.01730.x
Vestergaard, C. L., Flyvbjerg, H., and Møller, I. M. (2012). Intracellular signal by diffusion: can waves of hydrogen peroxide transmit intracellular information in plant cells? Front. Plant Sci. 3:295. doi: 10.3389/fpls.2012.00295
Keywords: reactive oxygen species, signaling, oxidation, oxylipins, oxidized proteins, oxidized RNA
Citation: Chmielowska-Bąk J, Izbiańska K and Deckert J (2015) Products of lipid, protein and RNA oxidation as signals and regulators of gene expression in plants. Front. Plant Sci. 6:405. doi: 10.3389/fpls.2015.00405
Received: 04 March 2015; Accepted: 19 May 2015;
Published: 02 June 2015.
Edited by:
Richard Sayre, New Mexico Consortium at Los Alamos National Laboratory, USAReviewed by:
Karl-Josef Dietz, Universität Bielefeld, GermanyCopyright © 2015 Chmielowska-Bąk, Izbiańska and Deckert. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Joanna Deckert, Department of Plant Ecophysiology, Institute of Experimental Biology, Faculty of Biology, Adam Mickiewicz University, ul.Umultowska 89, 61-614 Poznań, Poland,am9hbm5hLmRlY2tlcnRAYW11LmVkdS5wbA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.