- Donald Danforth Plant Science Center, St. Louis, MO, USA
Multicellularity has independently evolved numerous times throughout the major lineages of life. Often, multicellularity can enable complex, macroscopic organismal architectures but it is not required for the elaboration of morphology. Several alternative cellular strategies have arisen as solutions permitting exquisite forms. The green algae class Ulvophyceae, for example, contains truly multicellular organisms, as well as macroscopic siphonous cells harboring one or multiple nuclei, and siphonocladous species, which are multinucleate and multicellular. These diverse cellular organizations raise a number of questions about the evolutionary and molecular mechanisms underlying complex organismal morphology in the green plants. Importantly, how does morphological patterning arise in giant coenocytes, and do nuclei, analogous to cells in multicellular organisms, take on distinct somatic and germline identities? Here, we comparatively explore examples of patterning and differentiation in diverse coenocytic and single-cell organisms and discuss possible mechanisms of development and nuclear differentiation in the siphonous algae.
Introduction
Among the green plants (Viridiplantae), green algae (Chlorophyta) show remarkable diversity in morphology and cellular organization. Members of class Ulvophyceae include truly multicellular organisms, such as sea lettuce (Ulva) and various terrestrial green algae (e.g., Trentepohlia), macroscopic uninucleate cells, such as Acetabularia, giant siphonous coenocytes harboring multiple nuclei, such as Caulerpa, and siphonocladous species such as Cladophora, which are multicellular and multinucleate (Figure 1). Phylogenetic reconstruction suggests that the common ancestor of these species was a unicellular, uninucleate organism (Cocquyt et al., 2010), demonstrating that in the green plants macroscopic growth forms independently arose from diverse cellular strategies.
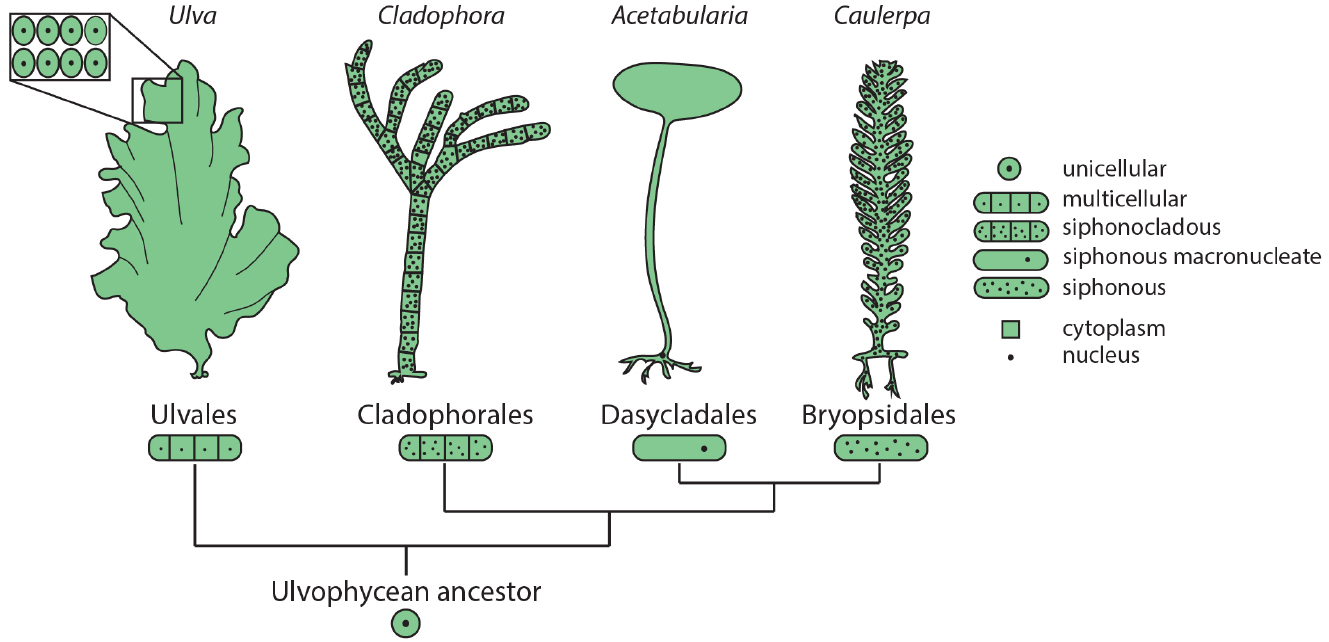
Figure 1. Diverse morphologies and cellular organization in the green algae. Orders within class Ulvophyceae contain examples of multicellular organisms (Ulva), siphonocladous species with multinucleate, multicellular organization (Cladophora), giant uninucleate cells (Acetabularia), and multinucleate siphonous algae (Caulerpa). The relationships among Ulvophycean classes (not drawn to scale) are based on the molecular phylogeny of Cocquyt et al. (2010).
Indeed, although land plants (Embryophyta) are truly multicellular organisms, they possess siphonous-like properties at the cellular level (Kaplan and Hagemann, 1991; Niklas et al., 2013). In contrast to animal cell division, cytokinesis in land plants involves phragmoplast formation, which leaves cytoplasmic connections between daughter cells. This symplastic connectivity is further maintained via the formation of plasmodesmata. Beyond cytoplasmic connectivity, several lines of evidence suggest that land plant morphology is specified at an organismal level rather than through emergent properties of cells, lessening the contribution of multicellularity to plant architecture compared to animal morphology. For example, irradiation studies and clonal analyses have demonstrated that organ initiation and elaboration are largely independent of cell lineage patterns (Haber, 1962; Foard et al., 1965; Foard, 1971; Poethig, 1987). Furthermore, the phenomenon of compensation ensures that drastic changes in the cell cycle have remarkably little effect on overall organ shape in land plants (Tsukaya, 2006). Finally, morphological convergence over large evolutionary distances, between land plants, which exhibit true three-dimensional parenchymatous tissue patterning, red algae (Rhodophyta), which grow by two-dimensional filament elaboration, and unicellular siphonous growth in the green algae (Chlorophyta) highlights the relative independence of organismal morphology from cell division patterns (Kaplan and Hagemann, 1991). Thus, siphonous algae allow an opportunity to explore basic mechanisms enabling the elaboration of complex morphologies independent of histogenesis, and provide insights into the evolution of diverse plant architectures.
Two green algae, Acetabularia acetabulum (Dasycladales), which is a uninucleate single-celled organism throughout most of its lifecycle, and Caulerpa taxifolia (Bryopsidales), a multinucleate siphonous coenocyte, show striking patterns of local transcript accumulation superimposed on morphological structures convergent with the roots, stems, and leaves of land plants (Vogel et al., 2002; Ranjan et al., 2015). How siphonous algae achieve the feat of establishing compartmentalized transcript accumulation without cellularization of the cytoplasmic space remains largely unexplored. Although transcript partitioning is likely necessary for the elaboration of complex morphology in Caulerpa the fact that this organism consists of a single cell poses fundamental questions: (1) How widespread is mRNA movement throughout the Caulerpa coenocyte?; (2) Are nuclei in the shared cytoplasm functionally—or even genetically—equivalent?; and (3) How is the division between germline and soma achieved within a single cell, if at all? Here, we explore evolutionary solutions to these challenges in diverse organisms and outline hypotheses to examine the mechanisms underlying the complex architectures found in the siphonous algae.
Patterning in a Single Cell: Morphology via mRNA Movement
Morphogenesis in multicellular organisms involves the establishment and maintenance of spatiotemporal transcript accumulation patterns. In the absence of cellular compartmentalization, localized gene expression or regulated transcript movement must act to create the mRNA patterns observed in Caulerpa and Acetabularia (Vogel et al., 2002; Ranjan et al., 2015), while processes such as tethering and regulation of protein translation may be in place to maintain them.
Interestingly, the C. taxifolia transcript accumulation profile shows a marked enrichment of RNA polymerase II transcripts in the holdfast, a pseudo-organ analogous to roots in land plants (Ranjan et al., 2015). As RNA polymerase II is a necessary component of the transcription machinery, a tempting hypothesis is that transcription either exclusively or preferentially occurs in holdfast nuclei, while transcripts observed throughout the rest of the cell have moved. Examples of regulated intracellular mRNA movement are well documented and often mechanistically dependent on the cytoskeleton. Actin filaments are employed for unidirectional mRNA translocation, such as polar localization of transcripts regulating asymmetric mating type switching in Saccharomyces cerevisiae (Paquin et al., 2007) and hyphal tip growth of filamentous ascomycete fungi, while microtubule-associated transcript movement enables long-distance bi-directional localization in the coenocyte of the basidiomycete fungus Ustilago maydis (Zarnack and Feldbrügge, 2010). Classical Acetabularia grafting experiments by Hämmerling (1953) provided the first clear demonstration that the single basal nucleus, through its implied molecular action at a distance, specifies cap morphology in this unicellular macroalga. Recent studies have demonstrated that actin-dependent mRNA movement is at least one component of this long-distance patterning signal during vegetative growth (Mine et al., 2001). Similarly, localized transcription and subsequent transcript movement may generate spatio-temporal transcript partitioning in the Caulerpa coenocyte (Ranjan et al., 2015). Nuclear run-on assays and targeted disruption of actin polymerization can determine if indeed transcription is restricted solely to the holdfast. In addition to cytoskeleton-associated transcript movement, the prevalence of cytoplasmic streaming in Caulerpa suggests that bulk cytoplasmic flow may aid the transport of patterning machinery (Sabnis and Jacobs, 1967; Herth et al., 1972; Kuroda and Manabe, 1983; Menzel, 1987). Live imaging approaches to simultaneously track microinjected labeled mRNAs and to visualize distinct cellular components can address the extent of transcript movement in Caulerpa and the contributions of cytoplasmic streaming and the cytoskeleton to this process.
Many Nuclei, One Cytoplasm: Heterogeneity and Local Subfunctionalization
Local gene expression differences within a single multinucleate cell may arise due to the existence of genetically distinct nuclear populations harboring varying cis-regulatory alleles. Some filamentous fungi, for example, actively maintain a diverse nuclear population within a single cell (Roper et al., 2011). An important implication of nuclear heterogeneity is that it necessitates coordination of nuclear activities to decrease intra-organismal genetic conflict (Michod and Roze, 2001; Roper et al., 2011; Niklas, 2014). During vegetative growth of filamentous ascomycetes, for example, heterokaryosis through hyphal fusion between distinct genotypes is regulated by an elaborate somatic incompatibility checking system (Roper et al., 2011). In discussing the possibility of nuclear heterogeneity in Caulerpa, the fact that these species are capable of regenerating the entire coenocyte from any vegetative fragment emerges as an important consideration. Moreover, in C. taxifolia sexual reproduction is limited, and there are reports that the invasive strain may only be male (Phillips, 2009). Since vegetative propagation predominates, nuclear inheritance is mainly cytoplasmic and the accumulation of spontaneous mutations defining multiple intracellular nuclear lineages within a single Caulerpa coenocyte is likely. This would create a high genetic burden and conflict between distinct nuclear lineages, and blur the distinction between individual and the population. As we discuss subsequently, a soma germline divide may maintain transgenerational genomic integrity. Additionally, mechanisms to rein in genomic diversity within a single coenocyte may also exist. Indeed, genome-wide comparisons of individual nuclei in the asexually reproducing endomycorrhizal fungus Rhizophagus irregularis show remarkable inter-nuclear homogeneity (Lin et al., 2014). Cycles of genome rearrangements or parasexual nuclear recombination and gene conversion may contribute to diversification and homogenization in the absence of sex in these fungi and other asexual organisms (Lin et al., 2014; Seidl and Thomma, 2014). Currently, there is little evidence of the precise nuclear make-up of a single Caulerpa species (see Varela-Álvarez et al., 2012, for a comparison of genome size and ploidy across species). Thus, the extent of intracellular nuclear genetic diversity in C. taxifolia needs to be resolved in order to address outstanding questions, including its impact on organismal morphology, reproduction, and population genetics. A consensus Caulerpa genome and genomic resequencing of single nuclei will be instrumental in understanding nuclear population dynamics in the Caulerpa coenocyte.
If, indeed, nuclear genetic diversity is limited within the Caulerpa coenocyte, it is interesting to speculate that groups of nuclei may somehow become locally committed to different expression patterns. Nuclear subfunctionalization is supported by the observation of “nucleo-cytoplasmic domains”—regularly spaced nuclei hypothesized to represent local zones of influence—in many syncytia and coenocytes (Goff and Coleman, 1987; McNaughton and Goff, 1990; Bruusgaard et al., 2003; Anderson et al., 2013). The demonstration of exclusive associations between individual nuclei and the plasma membrane during syncytial stages of Drosophila melanogaster embryogenesis is further evidence that the establishment of functional nuclear domains may be a means of spatial subfunctionalization in a shared cytoplasm (Mavrakis et al., 2009). Moreover, the formation and maintenance of the morphogenic bicoid gradient in Drosophila (Driever and Nüsslein-Volhard, 1988) is intricately intertwined with local nuclear dynamics (Gregor et al., 2007). These examples illustrate that, analogous to cells in multicellular organisms, the delineation of nuclear zones of influence may be a hallmark of coenocytic and syncytial growth. An intriguing implication of this observation is that in the absence of physical cell boundaries, nucleo-cytoplasmic domains, instead of cells, may form an elemental, subcellular functional unit of eukaryotic life (Baluška et al., 2004), consistent with the organismal theory proposed by Kaplan and Hagemann (1991).
Reconciling Nuclear Activity and Potential: A Possible Role for Small RNAs
As discussed in the previous section, the prevalence of vegetative propagation in C. taxifolia may necessitate mechanisms to rein in the effects of spontaneous mutations. Moreover, regeneration implies that fragments derived from any portion of the coenocyte can give rise to the entire organism. Thus, similar to the ability of differentiated cells of land plants to revert to a totipotent state, mechanisms to preserve nuclear potential may be in place in Caulerpa. However, recent evidence that transcripts with nuclear-related functions (such as transcription, DNA replication, chromatin, and RNAi) are highly enriched in the stolon and holdfast regions (Ranjan et al., 2015) suggests that nuclear populations in Caulerpa may be functionally distinct. To reconcile these observations, it is tantalizing to speculate of a nuclear division of labor into transcriptionally active and inert nuclei, analogous to the specification of somatic and germline cellular fates.
The setting aside of specialized germ cells, which act as propagules of a “true” copy of an organism’s genome, provides a buffer from the variance to which somatic genomes are subject. The use of such a divide is amplified with increasing organismal size and life span, which correlate with increased propensity for the accumulation of deleterious somatic variants (Michod and Roze, 2001; Umen, 2014). In C. taxifolia, which as discussed, may be a long-lived clonal lineage, the need for a mechanism to reduce the transgenerational effects of deleterious somatic variants is particularly acute given that genetically distinct nuclear populations may be present in a single cell.
Germline fidelity is intimately connected with small RNAs and their mobility, leading to speculation about the significance of the observed restriction of transcripts related to RNAi biogenesis to Caulerpa stolons (Ranjan et al., 2015). A conceptual basis for how small RNA-mediated nuclear germline identity may be accomplished within a shared cytoplasm is provided by the angiosperm microgametophyte. During pollen formation in Arabidopsis thaliana, a vegetative and a generative nucleus share a common cytoplasm in the microspore. The generative nucleus gives rise to sperm nuclei, which ultimately contribute their genomes to the next generation. Transposable elements are activated in the vegetative nucleus allowing for the generation of small interfering RNAs (siRNAs). siRNAs accumulate in sperm nuclei and direct transposon silencing ensuring transgenerational genomic fidelity (Slotkin et al., 2009). Although mechanistically distinct, a small RNA-dependent phenomenon is also involved in inter-nuclear communication during conjugation in ciliated protozoans, which exhibit nuclear dimorphism with a transcriptionally inert germline micronucleus and an active somatic macronucleus. Ciliates undergo conjugative sexual reproduction during which micronuclei recombine and are transmitted to the next generation, while somatic macronuclei are degraded. A special feature of macronuclei, which likely contributes to their role as transcriptionally active entities, is that site-specific DNA rearrangements result in the removal of ∼15% of the micronucleus genome from which the zygotic macronucleus is derived. Interestingly, the site-specificity of these macronuclear genome rearrangements is guided by a specific class of small RNAs, termed scnRNAs (Mochizuki et al., 2002; Duharcourt et al., 2009; Fang et al., 2012) illustrating another context in which small RNAs act to coordinate inter-nuclear division of labor and to preserve germline integrity.
The presence of core machinery for small RNA-mediated silencing in the Ulvophycean lineage (Cerutti et al., 2011) supports the possibility that small RNA-directed pathways may be used by siphonous green algae such as Caulerpa. Moreover, miRNAs in Chlamydomonas reinhardtii peak in abundance during gametogenesis (Zhao et al., 2007), suggesting a role for small RNA pathways as not only regulators of development and morphology, but perhaps guardians of the germline in this Chlorophyte. Thus, generating a small RNA atlas of the Caulerpa taxifolia coenocyte will be instrumental in evaluating the possibility that small RNAs may pattern nuclei with somatic and germline characteristics and maintain genomic fidelity. While enrichment of RNAi-related transcripts in the Caulerpa stolon suggests that small RNA production may be localized to this pseudo-organ (Ranjan et al., 2015), the extent and mechanisms of small RNA mobility and its local accumulation in the shared cytoplasm remain open questions. Active nuclei in the stolon, acting analogously to somatic cells, might produce small RNAs that travel to silence nuclei in the pinnules, where gametogenesis is localized (Figure 2).
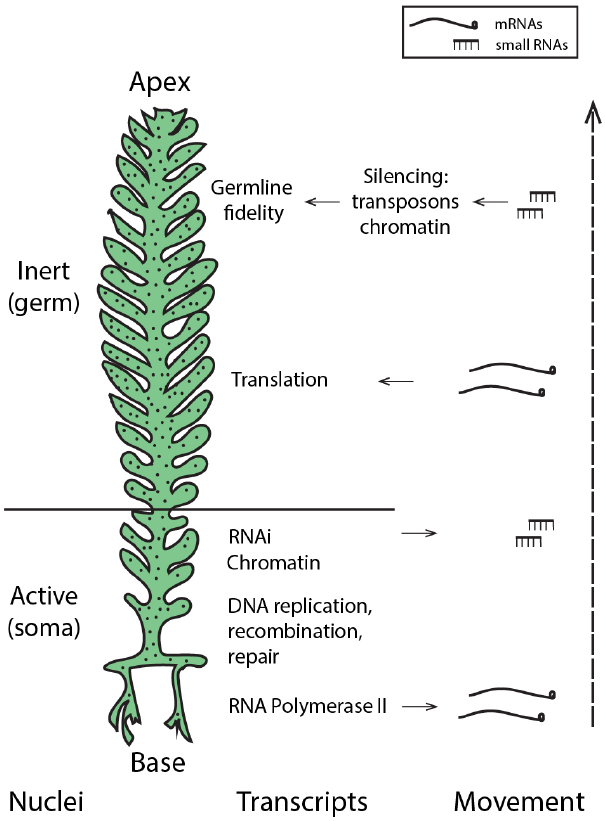
Figure 2. Hypothetical model: localized transcription and regulated movement of mRNAs and small RNAs may pattern the Caulerpa coenocyte. Transcriptionally active nuclei in the stolon and frond base produce transcripts and small RNAs. mRNA movement, coupled with local tethering and translation outside the region of transcription, establish spatio-temporal patterning. Small RNAs move from the stolon apically to induce silencing in the frond pinnules, creating germline-like, transcriptionally inert nuclei at the site of gametogenesis. “Nuclei” denotes hypothesized nuclear states; localized accumulation of transcript classes (as described in Ranjan et al., 2015) is shown by “Transcripts” and “Movement” shows the acropetal transport of mRNAs and small RNAs.
Conclusion and Perspectives
Siphonous green algae exhibit remarkable morphological complexity, convergent with the architecture of land plants. The elaboration of form in a single cell highlights the necessity to address the mechanisms by which morphology arises in a supra-cellular, organismal context. At the very least, morphology requires the establishment and maintenance of spatiotemporal gene expression patterns. In multicellular organisms, developmentally regulated variation in gene expression defines cellular, tissue, and organ levels of organization. Evidence suggests that transcript partitioning contributes to morphological complexity in coenocytic organisms as well. How such patterns are generated within a shared cytoplasm remains speculative. Additionally, the implications of cytoplasmic inheritance of the nuclear genome on intracellular genetic diversity and conflict, organismal morphology, and the maintenance of genomic fidelity are all open questions. Precedent from studies in diverse multinucleate organisms helps to outline a conceptual basis of development and inheritance in siphonous algae. Small RNA movement may locally specify nuclear activity, while transcript movement and tethering may contribute to differential gene expression and morphological patterning. The generation of genomic resources, a small RNA expression atlas, as well as development of live imaging and cytoplasmic manipulation techniques in giant coenocytes such as Caulerpa will provide a strong foundation to address these hypotheses. Ultimately, understanding the mechanisms by which complex morphology arises in the absence of multicellularity has fundamental implications for our understanding of the evolution of plant architecture.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Margaret Frank for helpful discussions and insightful comments on the manuscript.
References
Anderson, C. A., Ester, U., Korndorf, T., Borsuk, M. E., Skotheim, J. M., and Gladfelter, A. S. (2013). Nuclear repulsion enables division autonomy in a single cytoplasm. Curr. Biol. 23, 1999–2010. doi: 10.1016/j.cub.2013.07.076
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Baluška, F., Volkmann, D., and Barlow, P. (2004). Eukaryotic cells and their cell bodies: cell theory revisited. Annu. Bot. 94, 9–32. doi: 10.1093/aob/mch109
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Bruusgaard, J. C., Liestøl, K., Ekmark, M., Kollstad, K., and Gundersen, K. (2003). Number and spatial distribution of nuclei in the muscle fibres of normal mice studied in vivo. J. Physiol. 551, 467–478. doi: 10.1113/jphysiol.2003.045328
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Cerutti, H., Ma, X., Msanne, J., and Repas, T. (2011). RNA-mediated silencing in algae: biological roles and tools for analysis of gene function. Eukaryot. Cell 10, 1164–1172. doi: 10.1128/EC.05106-11
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Cocquyt, E., Verbruggen, H., Leliaert, F., and De Clerck, O. (2010). Evolution and cytological diversification of the green seaweeds (Ulvophyceae). Mol. Biol. Evol. 27, 2052–2061. doi: 10.1093/molbev/msq091
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Driever, W., and Nüsslein-Volhard, C. (1988). A gradient of bicoid protein in Drosophila embryos. Cell 54, 83–89. doi: 10.1016/0092-8674(88)90182-1
Duharcourt, S., Lepère, G., and Meyer, E. (2009). Developmental genome rearrangements in ciliates: a natural genomic subtraction mediated by non-coding transcripts. Trends Genet. 25, 344–350. doi: 10.1016/j.tig.2009.05.007
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Fang, W., Wang, X., Bracht, J. R., Nowacki, M., and Landweber, L. F. (2012). Piwi-interacting RNAs protect DNA against loss during Oxytricha genome rearrangement. Cell 151, 1243–1255. doi: 10.1016/j.cell.2012.10.045
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Foard, D. E. (1971). The initial protrusion of a leaf primordium can form without concurrent periclinal cell divisions. Can. J. Bot. 49, 1601–1603. doi: 10.1139/b71-223
Foard, D. E., Haber, A. H., and Fishman, T. N. (1965). Initiation of lateral root primordia without completion of mitosis and without cytokinesis in uniseriate pericycle. Am. J. Bot. 52, 580–590. doi: 10.2307/2440119
Goff, L. J., and Coleman, A. W. (1987). The solution to the cytological paradox of isomorphy. J. Cell Biol. 104, 739–748. doi: 10.1083/jcb.104.3.739
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Gregor, T., Wieschaus, E. F., McGregor, A. P., Bialek, W., and Tank, D. W. (2007). Stability and nuclear dynamics of the bicoid morphogen gradient. Cell 130, 141–152. doi: 10.1016/j.cell.2007.05.026
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Haber, A. H. (1962). Nonessentiality of concurrent cell divisions for degree of polarization of leaf growth. I. Studies with radiation-induced mitotic inhibition. Am. J. Bot. 49, 583–589. doi: 10.2307/2439715
Herth, W., Franke, W. W., and Vanderwoude, W. J. (1972). Cytochalasin stops tip growth in plants. Naturswissenschaften 59, 38–39. doi: 10.1007/BF00594629
Hämmerling, J. (1953). Nucleo-cytoplasmic relationship in the development of Acetabularia. J. Intern. Rev. Cytol. 2, 475–498. doi: 10.1016/S0074-7696(08)61042-6
Kaplan, D. R., and Hagemann, W. (1991). The relationship of cell and organism in vascular plants. Bioscience 41, 693–703. doi: 10.2307/1311764
Kuroda, K., and Manabe, E. (1983). Microtubule-associated cytoplasmic streaming in Caulerpa. Proc. Japan Acad. 59, 131–134. doi: 10.2183/pjab.59.131
Lin, K., Limpens, E., Zhang, Z., Ivanov, S., Saunders, D. G. O., Mu, D., et al. (2014). Single nucleus genome sequencing reveals high similarity among nuclei of an endomycorrhizal fungus. PLoS Genet. 10:e1004078. doi: 10.1371/journal.pgen.1004078
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Mavrakis, M., Rikhy, R., and Lippincott-Schwartz, J. (2009). Plasma membrane polarity and compartmentalization are established before cellularization in the fly embryo. Dev. Cell 16, 93–104. doi: 10.1016/j.devcel.2008.11.003
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
McNaughton, E. E., and Goff, L. (1990). The role of microtubules in establishing nuclear spatial patterns in multinucleate green algae. Protoplasma 157, 19–37. doi: 10.1007/BF01322636
Menzel, D. (1987). The cytoskeleton of the giant coenocytic alga Caulerpa visualized by immunocytochemistry. Protoplasma 139, 71–76. doi: 10.1007/BF01282277
Michod, R. E., and Roze, D. (2001). Cooperation and conflict in the evolution of multicellularity. Heredity 86, 1–7. doi: 10.1046/j.1365-2540.2001.00808.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Mine, I., Okuda, K., and Mnzel, D. (2001). Poly(A)+ RNA during vegetative development of Acetabularia peniculus. Protoplasma 216, 56–65. doi: 10.1007/BF02680131
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Mochizuki, K., Fine, N. A., Fujisawa, T., and Gorovsky, M. A. (2002). Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in Tetrahymena. Cell 110, 689–699. doi: 10.1016/S0092-8674(02)00909-1
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Niklas, K. J. (2014). The evolutionary-developmental origins of multicellularity. Am. J. Bot. 101, 6–25. doi: 10.3732/ajb.1300314
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Niklas, K. J., Cobb, E. D., and Crawford, D. R. (2013). The evo-devo of multinucleate cells, tissues, and organisms, and an alternative route to multicellularity. Evol. Dev. 15, 466–474. doi: 10.1111/ede.12055
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Paquin, N., Ménade, M., Poirier, G., Donato, D., Drouet, E., and Chartrand, P. (2007). Local activation of yeast ASH1 mRNA translation through phosphorylation of Khd1p by the casein kinase Yck1p. Mol. Cell 26, 795–809. doi: 10.1016/j.molcel.2007.05.016
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Phillips, J. A. (2009). Reproductive ecology of Caulerpa taxifolia (Caulerpaceae, Bryopsidales) in subtropical eastern Australia. Eur. J. Phycol. 44, 81–88. doi: 10.1080/09670260802343640
Poethig, R. S. (1987). Clonal analysis of cell lineage patterns in plant development. Am. J. Bot. 74, 5581–5594. doi: 10.2307/2443838
Ranjan, A., Townsley, B. T., Ichihashi, Y., Sinha, N. R., and Chitwood, D. H. (2015). An intracellular transcriptomic atlas of the giant coenocyte Caulerpa taxifolia. PLoS Genet. 11:e1004900. doi: 10.1371/journal.pgen.1004900
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Roper, M., Ellison, C., Taylor, J. W., and Glass, N. L. (2011). Nuclear and genome dynamics in multinucleate ascomycete fungi. Curr. Biol. 21, R786–R793. doi: 10.1016/j.cub.2011.06.042
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Sabnis, D. D., and Jacobs, W. P. (1967). Cytoplasmic streaming and microtubules in the coenocytic marine alga, Caulerpa prolifera. J. Cell Sci. 2, 465–472.
Seidl, M. F., and Thomma, B. P. H. J. (2014). Sex or no sex: evolutionary adaptation occurs regardless. Bioessays 36, 335–345. doi: 10.1002/bies.201300155
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Slotkin, R. K., Vaughn, M., Borges, F., Tanurdzic, M., Becker, J. D., Feijo, J. A., et al. (2009). Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell 136, 461–472. doi: 10.1016/j.cell.2008.12.038
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Tsukaya, H. (2006). Mechanism of leaf-shape determination. Annu. Rev. Plant Biol. 57, 477–496. doi: 10.1146/annurev.arplant.57.032905.105320
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Umen, J. G. (2014). Green algae and the origins of multicellularity in the plant kingdom. CSH Perspect. Biol. 6:a016170. doi: 10.1101/cshperspect.a016170
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Varela-Álvarez, E., Garreta, A. G., Lluch, J. R., Soler, N. S., Serrao, E. A., and Siguán, M. A. R. (2012). Mediterranean species of Caulerpa are polyploid with smaller genomes in the invasive ones. PLoS ONE 7:e47728. doi: 10.1371/journal.pone.0047728
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Vogel, H., Grieninger, G. E., and Zetsche, K. H. (2002). Differential messenger RNA gradients in the unicellular alga Acetabularia acetabulum. Role of the cytoskeleton. Plant Physiol. 129, 1407–1416. doi: 10.1104/pp.010983
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zarnack, K., and Feldbrügge, M. (2010). Microtubule-dependent mRNA transport in fungi. Eukaryot. Cell 9, 982–990. doi: 10.1128/EC.00030-10
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zhao, T., Li, G., Mi, S., Li, S., Hannon, G., Wang, X. J., et al. (2007). A complex system of small RNAs in the unicellular green alga Chlamydomonas reinhardtii. Gene. Dev. 15, 1190–1203. doi: 10.1101/gad.1543507
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Keywords: morphology, multicellularity, coenocyte, green algae, transcript accumulation, patterning, small RNAs
Citation: Coneva V and Chitwood DH (2015) Plant architecture without multicellularity: quandaries over patterning and the soma-germline divide in siphonous algae. Front. Plant Sci. 6:287. doi: 10.3389/fpls.2015.00287
Received: 20 February 2015; Accepted: 09 April 2015;
Published: 24 April 2015.
Edited by:
Bénédicte Charrier, Centre National de la Recherche Scientifique, FranceReviewed by:
Stefan A. Rensing, University of Marburg, GermanyJ. M. Cock, Centre National de la Recherche Scientifique, France
Copyright © 2015 Coneva and Chitwood. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daniel H. Chitwood, Donald Danforth Plant Science Center, 975 North Warson Road, St. Louis, MO 63132, USA dchitwood@danforthcenter.org
 Viktoriya Coneva
Viktoriya Coneva