- 1National Institute of Plant Genome Research, New Delhi, India
- 2Indian Institute of Toxicology Research, Lucknow, India
The phenomenon of RNA interference (RNAi) which involves sequence-specific gene regulation by small non-coding RNAs, i.e., small interfering RNA (siRNA) and microRNA (miRNA) has emerged as one of most powerful approaches for crop improvement. RNAi based on siRNA is one of the widely used tools of reverse genetics which aid in revealing gene functions in many species. This technology has been extensively applied to alter the gene expression in plants with an aim to achieve desirable traits. RNAi has been used for enhancing the crop yield and productivity by manipulating the gene involved in biomass, grain yield and enhanced shelf life of fruits and vegetables. It has also been applied for developing resistance against various biotic (bacteria, fungi, viruses, nematodes, insects) and abiotic stresses (drought, salinity, cold, etc.). Nutritional improvements of crops have also been achieved by enriching the crops with essential amino acids, fatty acids, antioxidants and other nutrients beneficial for human health or by reducing allergens or anti-nutrients. microRNAs are key regulators of important plant processes like growth, development, and response to various stresses. In spite of similarity in size (20–24 nt), miRNA differ from siRNA in precursor structures, pathway of biogenesis, and modes of action. This review also highlights the miRNA based genetic modification technology where various miRNAs/artificial miRNAs and their targets can be utilized for improving several desirable plant traits. microRNA based strategies are much efficient than siRNA-based RNAi strategies due to its specificity and less undesirable off target effects. As per the FDA guidelines, small RNA (sRNA) based transgenics are much safer for consumption than those over-expressing proteins. This review thereby summarizes the emerging advances and achievement in the field of sRNAs and its application for crop improvement.
Introduction
RNA interference (RNAi) is a gene silencing phenomenon that involves sequence-specific gene regulation induced by double stranded RNA (dsRNA) resulting in inhibition of translation or transcription. During the 1990s, post-transcriptional gene silencing (PTGS) was reported in many organisms including fungi, animals, ciliates, and plants (Baulcombe, 2000; Matzke et al., 2001). The phenomenon of gene silencing was discovered accidentally in petunia flowers where expression of chalcone synthase (pigment producing gene), resulted in variegated flowers instead of expected deep purple color. Since, the expression of both the transgene and the homologous endogenous gene was suppressed, the phenomenon was termed co-suppression (Napoli et al., 1990; Campbell, 2005). A similar phenomenon reported in the fungus Neurospora crassa was named quelling (Romano and Macino, 1992; Cogoni et al., 1996; Fire et al., 1998), and a related mechanism was identified in animals known as RNAi. The phenomenon of RNAi involves small non-coding RNAs called small interfering RNA (siRNA) and microRNA (miRNA) that are the cleavage product of dsRNA. A ribonuclease called DICER or a Dicer-like enzyme (DCL; Pare and Hobman, 2007) carry out this cleavage. The phenomenon of RNAi, in addition to small non-coding RNAs involves a RNA-induced silencing complex (RISC; Redfern et al., 2013; Wilson and Doudna, 2013) and Argonaute proteins (AGOs; Ender and Meister, 2010; Riley et al., 2012).
In plants, miRNAs were identified about 10 years later after discovery of animal miRNAs. In early, Gunnery and Datta (1987) for the first time reported small inhibitory RNA in plant system involved in translational control. They isolated a small molecular weight RNA from barley embryos that specifically inhibits translation initiation. In mid-2002, small RNAs (sRNAs) with miRNA characteristics were reported by different groups to be present in Arabidopsis (Llave et al., 2002b; Mette et al., 2002; Park et al., 2002; Reinhart et al., 2002).
In recent times, RNAi technology has evolved as an important tool of genetic engineering and functional genomics aimed for crop improvement.
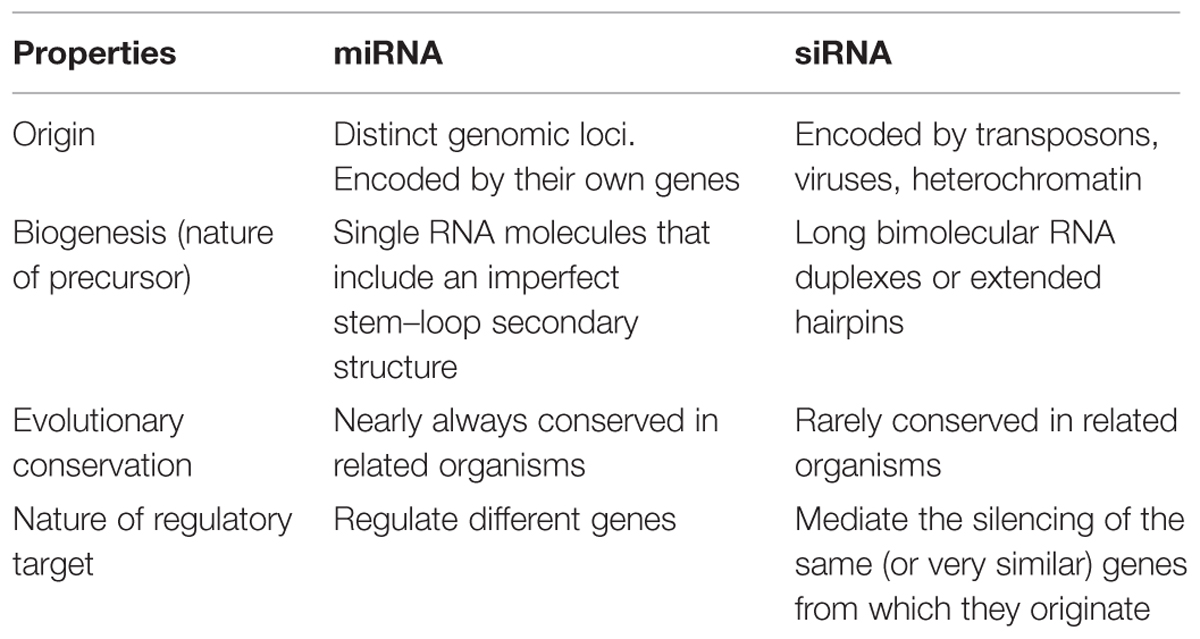
miRNA vs. siRNA
microRNAs (miRNA) and short-interfering RNAs are known to be important regulators of gene expression (Bartel, 2004; Zamore and Haley, 2005; Vazquez, 2006). Though, they show similarity in size (20–24 nt), but differ in precursor structures, pathway of biogenesis, and modes of action (Axtell, 2013; Table 1). Both are processed from long RNA precursors by Dicer-like ribonucleases (Lee et al., 1993; Hamilton and Baulcombe, 1999; Hammond et al., 2000; Zamore et al., 2000; Bernstein et al., 2001; Hutvagner et al., 2001). Both regulate the target gene repression (Lee et al., 1993; Hammond et al., 2000; Zamore et al., 2000) through ribonucleoprotein silencing complexes.
Short-Interfering RNAs (siRNAs)
Long dsRNA or short-hairpin RNA (shRNA) precursors, which are homologous in sequence to the target gene to be silenced (Fire et al., 1998; Tuschl, 2001), initiate the process of RNAi. The entry of a virus, a genetic element like transposons or an introduced transgene provide long dsRNA can triggers the RNAi pathway. The dicer enzyme is recruited in the cell (Bernstein et al., 2001) leading to the cleavage of the dsRNA into siRNA (Hamilton and Baulcombe, 1999) that are short, 5′-phosphorylated dsRNAs (21–25 nt) with two nucleotide overhangs at the 3′ end (Bernstein et al., 2001; Elbashir et al., 2001). The recruitment of siRNA-induced silencing complex (siRISC) leads to degradation of sense strand of siRNA (having the same sequence as the target gene) The siRISC is then incorporated into the antisense strand of siRNA which in association with AGO and other effector proteins brings about cleavage of the target mRNA in sequence-specific manner. The activated RISC can repeatedly participate in mRNA degradation and protein synthesis inhibition resulting into PTGS.
On the basis of origin and mechanism of biogenesis, sRNAs in plants exhibit a range of size classes, predominantly from 20 to 24 nt. Four Dicer-like nucleases (DCLs) have been reported in Arabidopsis (Henderson et al., 2006). Depending on their origin and structure, dsRNA triggers of RNAi are cleaved by one or more DCLs (Zamore et al., 2000; Bernstein et al., 2001). DCL4 and DCL2 are involved in processing of dsRNA with perfectly complementary to produce 20- to 22-nt long short-interfering RNA (Hamilton and Baulcombe, 1999; Hamilton et al., 2002; Hannon, 2002; Tang et al., 2003; Bouche et al., 2006). MiRNAs that are 21-nt long are produced by cleaving of partially paired dsRNA precursors by DCL1 (Denli and Hannon, 2003; Bartel, 2004). DCL3 is involved in biogenesis of 24-nt long repeat-associated sRNAs known as heterochromatic siRNAs (hcsiRNAs; Xie et al., 2005). In a complex called RISC, siRNAs and miRNAs (20- to 21-nt long) in association with AGO proteins, direct sequence-specific cleavage of complementary target RNAs or interfere with their translation (Hammond et al., 2000; Hannon, 2002; Brodersen et al., 2008). Traditionally, sRNA species that are 24-nt long (derived from transposable elements and other repetitive sequences) are linked to transcriptional gene silencing via DNA methylation and repressive chromatin modification (Hamilton et al., 2002; Zilberman et al., 2003; Liu et al., 2004; Fagegaltier et al., 2009; Law and Jacobsen, 2010; Burkhart et al., 2011). SiRNA recruits several DNA- and histone-modifying proteins including the cytosine methyltransferase, chromomethylase3 (Ossowski et al., 2008) which in turn mediate the formation of a transcriptionally inactive silent chromatin state. However, 21 nt long siRNAs may also participate in siRNA-dependent methylation of genomic loci (Pontier et al., 2012). AGO is an important protein that complexes with sRNA to form the core of the RISC. In Arabidopsis thaliana, 10 different AGO proteins are known to mediate the effects of several distinct types of sRNAs (Vaucheret, 2008). Endonucleolytic cleavage (slicing) activity catalyzed by the AGO protein (Huntzinger and Izaurralde, 2011) is an important step in post-transcriptional silencing. Guide RNA confers sequence specificity to any RNA silencing reaction whereas the precise nature of silencing is determined by the properties of the associated AGO protein, including its ribonuclease activity, interacting proteins, and subcellular localization.
Movement of plant sRNAs falls into two main categories: cell-to-cell (short-range) and systemic (long-range) movement (Melnyk et al., 2011). Local movement is symplastic, i.e., from the site of initiation to neighboring cells through channels called plasmodesmata (Lough and Lucas, 2006). RNA silencing also spreads systemically over long distances through the vascular phloem tissue. This long distance or systemic movement of the silencing signal takes place over days (Voinnet et al., 1998) and it is generally from photosynthetic sources (i.e., leaves) to sucrose sinks (i.e., roots and growing points) through a bulk flow process. The systemic RNA silencing signal has been identified in plants by direct sampling of phloem sap (Yoo et al., 2004; Buhtz et al., 2008) and detection of RNAs in stocks and scions of grafted plants (Palauqui et al., 1997; Schwach et al., 2005; Brosnan et al., 2007; Dunoyer et al., 2010a; Molnar et al., 2010). Mobile silencing operates in a nucleotide-sequence-specific manner and its components include sRNA molecules (21–24 nt). Molnar et al. (2010) have reported mobile sRNAs of the 24-nt size class that are associated with DNA methylation of targeted loci. It was consistent with the analysis of viral suppressors of systemic silencing in Nicotiana benthamiana (Hamilton et al., 2002) and the presence of 24-nt sRNA in the phloem sap of oilseed rape (Buhtz et al., 2008) and pumpkin (Yoo et al., 2004). Dunoyer et al. (2010a,b) showed that the mechanically delivered, fluorescently labeled 21- and 24-nt siRNAs move from cell to cell and over long distances. Artificial miRNAs (amiRNAs) were shown to move short distances in leaves (Schwab et al., 2006) or between the pollen vegetative cell cytoplasm and the sperm cells (Slotkin et al., 2009). Endogenous 21-nt miRNAs (miR399) could also be mobile between shoots and roots (Pant et al., 2008) and within the roots (miR165a and miR166b; Carlsbecker et al., 2010).
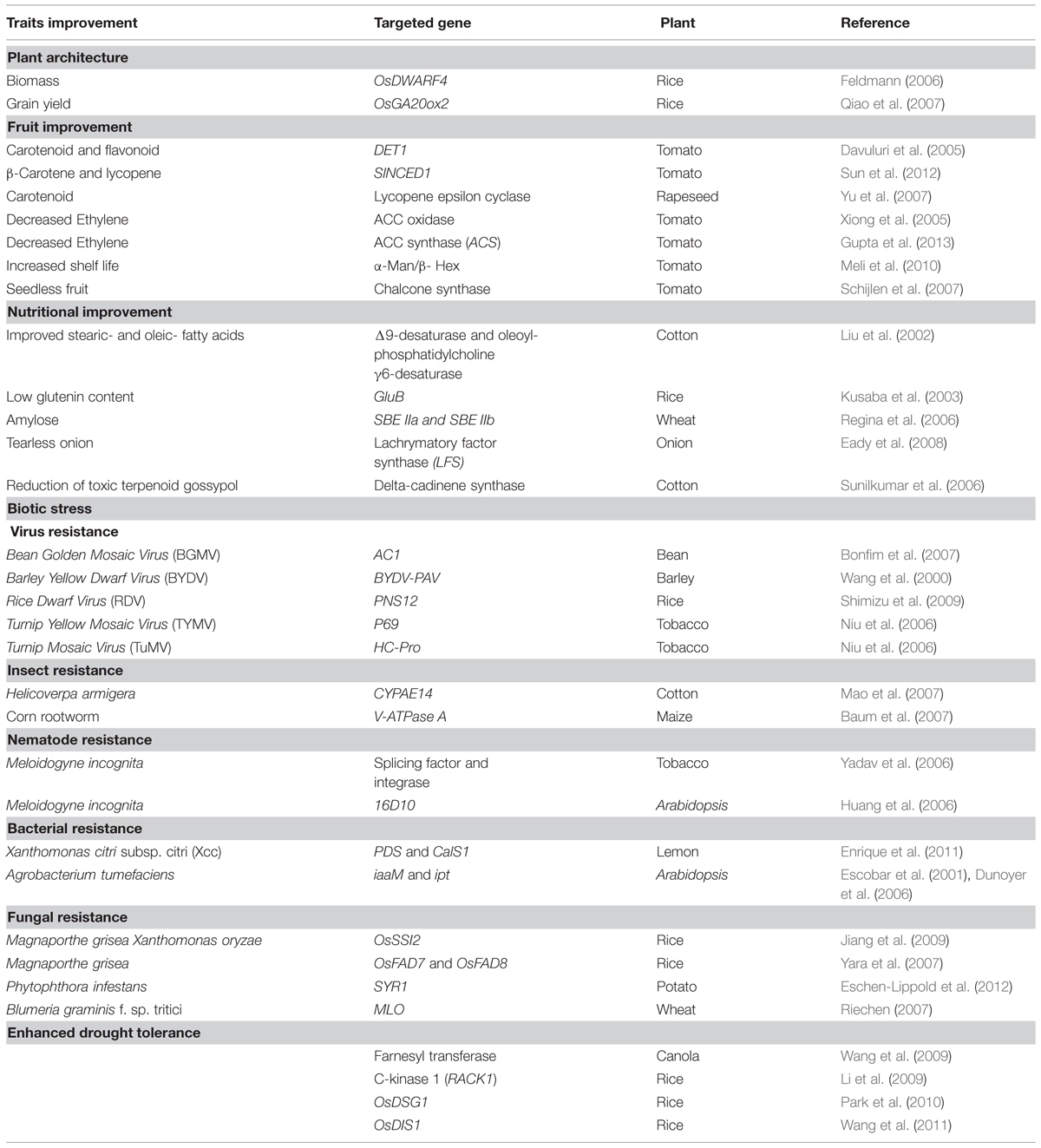
RNA interference can be used to achieve desirable traits in crops by manipulating the gene expression (Table 2). RNAi mediated gene silencing technique mainly involves identification of the target gene(s) followed by generating RNAi construct with hairpin cassette (gene cloned in sense and antisense orientation flanking a spacer or intron), plant transformation and finally screening and evaluating the traits.
microRNA (miRNA)
Plant miRNAs are a class of small regulatory RNAs (20–22 nt) that are encoded by endogenous miRNA genes and transcribed by RNA polymerase II into primary miRNAs (pri-miRNA) having partially double- stranded stem–loop structures (Jones-Rhoades et al., 2006). Following transcription, pri-miRNA is cleaved by a DCL1 enzyme in a two step process resulting in production of a pre-miRNA and a mature miRNA duplex (miRNA/miRNA∗). The mature miRNA is incorporated into RISC and mediates the degradation of mRNA target. miRNA can down-regulate the level of protein of their target genes through either translational repression (Aukerman and Sakai, 2003; Chen, 2004; Brodersen et al., 2008), through cleavage of transcript (Llave et al., 2002a; Xie et al., 2003), or transcriptional inhibition (Bao et al., 2004; Khraiwesh et al., 2010).
In animals, miRNA mostly act by translational inhibition, where they often bind motifs in the 3′ UTRs of their targets, which show several mismatches to the miRNA. However, in plants target motifs of miRNA have few mismatches which are most often found in the coding sequences and act mostly by transcript cleavage through a mechanism closely related to RNAi. In animals, miRNA usually consist of 20–22 nucleotides where as in plants it is 20–24 nucleotides (Reinhart et al., 2002; Bartel, 2004). Plant miRNAs are less conserved compared to animal miRNAs. In plants, mostly the mature miRNAs are conserved where as miRNA precursors are usually conserved in animals (Bartel, 2004). Similar to animlas, in Arabidopsis, a RNAse III-like enzymes carry out processing of miRNAs from primary precursors followed by incorporation into a protein complex named RISC. However, an additional step is included in the biogenesis of plant miRNAs, i.e., the methylation on the ribose of the last nucleotide of miRNA by the methyltransferase Hen1 (Park et al., 2002). Unlike animals, plant miRNAs show high degree of sequence complementarity to their target mRNAs (Rhoades et al., 2002). Because of this fact, bioinformatics prediction of plant miRNA targets is much easier and has facilitated prediction and subsequent validation of many plant miRNA targets.
A majority of plant miRNA targets includes genes encoding for transcription factors (Rhoades et al., 2002; Mitsuda and Ohme-Takagi, 2009), few code for hormone receptors (Navarro et al., 2006), some encode enzymes (Xie et al., 2003; Fujii et al., 2005). Many plant miRNAs are known to regulate post-transcriptional gene expression and play critical roles in many developmental processes, abiotic stresses and pathogen responses (Xin et al., 2010), in nutrient homeostasis etc. Functional characterization of several plant miRNA and their target need high-throughput sequencing at global genome-level (Wang et al., 2011). MiRNAs have been extensively studied in model plants such as rice, A. thaliana etc.
Schwab et al. (2006) have designed a series of amiRNAs targeting different endogenous mRNAs and compared their effects to those of natural miRNAs. amiRNAs can be generated by replacing the sequences of miRNA/miRNA*within the miRNA precursor without disrupting its structural features. Like natural miRNAs, amiRNAs had varying number of target mismatches and could silence both single and multiple target genes with high specificity as determined by genome-wide expression profiling. The direct targets of amiRNAs can be accurately predicted by parameters of target selection already determined for natural miRNAs (Schwab et al., 2005).
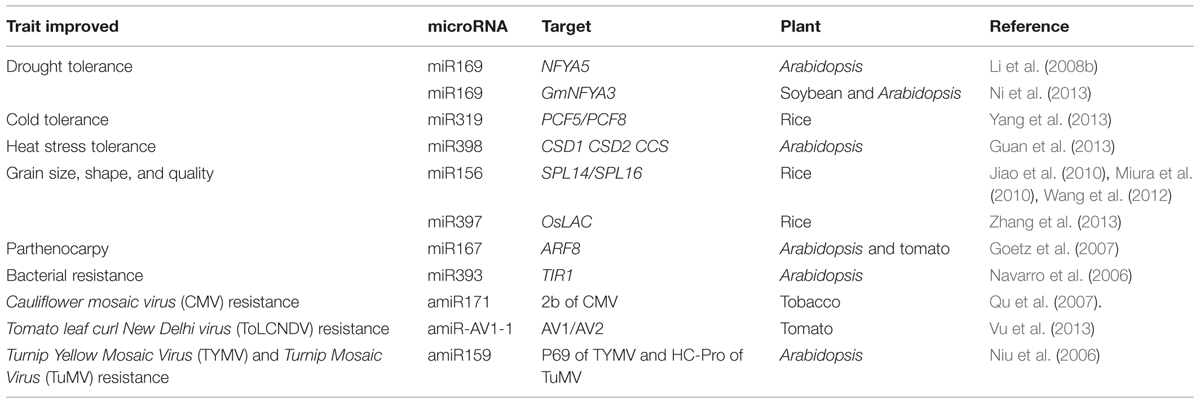
Thus, extensive base pairing with targets is required for plant miRNA function. amiRNAs can be efficiently used for gene silencing, especially when there is need to down regulate several related, but not identical, target gene. Transgenic plants harboring amiRNAs under constitutive and inducible promoters have shown specific and efficient down-regulation of target genes of interest with limited non-autonomous effect. Thus, amiRNAs have great potential for crop improvement (Table 3).
RNAi and its Role in Crop Improvement
Plant Architecture
Biomass
RNA interference can be employed successfully to improve yield of crop and fruit plants by manipulating the basic agronomic traits of plant such as height, inflorescence, branching and size. RNAi mediated knockdown of a gene OsDWARF4 in rice resulted in shorter plants with erect leaf architecture leading to increased photosynthesis in the lower leaves. Such plant has potential for improved yields under dense planting conditions (Feldmann, 2006).
Some plant materials show recalcitrance to a process of saccharification which causes major limitation for conversion of lignocellulosic biomass to ethanol. The problem of recalcitrance of plant cell wall to bioconversion can be overcome by genetic reduction of lignin content (Chen and Dixon, 2007). Independent down-regulation of each of six lignin biosynthesis enzymes in transgenic alfalfa lines, yielded nearly twice as much sugar from stem’s cell walls as compared to wild-type plants (Reddy et al., 2005). Down-regulation of lignin genes like cinnamate 4-hydroxylase (C3H), shikimate hydroxycinnamoyl transferase (HCT) and 4-coumarate-coA ligase (4CL) in plants reduced total lignin content, improved accessibility of cellulases for cellulose degradation and increased dry matter degradability (Hisano et al., 2009). This lignin modification also facilitated bypassing the need for acid pretreatment (Chen and Dixon, 2007). Over-expression of the maize Corngrass1 (Cg1) miRNA that belongs to the MIR156 family caused prolonged vegetative phase and delayed flowering (Chuck et al., 2011) resulting in increased biomass. The transgenic plants showed up to 250% more starch and improved digestibility (Chuck et al., 2011). It was reported that the degrees of morphological changes and biomass yields were related to the expression level of an exogenous rice miRNA Osa-miR156b (Fu et al., 2012). Transgenic switch grass plants with relatively high levels of miR156 showed highly stunted growth, whereas those with moderate and low levels of miR156 expression had 58–101% more biomass production than wild-type controls as a result of increases in tiller numbers. It was also observed that over-expression of rice miR156 could improve the yield of solubilised sugar as well as forage digestibility. Over-expression of miR156 led to increased biomass in other plant species including Arabidopsis and rice (Schwab et al., 2005; Xie et al., 2006, 2012). Cg1 over-expressing, poplar transgenics plants showed significant increase in the growth of axillary meristem, shorterning of internode length, and reduction in stem lignin content (up to 30%) compared to that of the wild-type (Rubinelli et al., 2013).
Grain Yield
Plant architecture can be manipulated in order to achieve increased grain yield like in rice plants (Jiao et al., 2010; Miura et al., 2010; Springer, 2010; Wang et al., 2012). RNAi-mediated suppression of GA 20-oxidase (OsGA20ox2) gene resulted in semi-dwarf plants from a taller rice variety QX1. This RNAi transgenic exhibited significant increase in panicle length, increased number of seeds per panicle and higher test (1000 grain) weight (Qiao et al., 2007). OsSPL14 (Souamosa promoter binding protein-like 14) was reported to be the target of Osa-miR156 in rice (Jiao et al., 2010; Miura et al., 2010) and was shown to positively regulate the rice plant architecture leading to enhanced yield of rice grain (Jiao et al., 2010). Higher expression of OsSPL14 could modify the rice plant architecture resulting in decreased tiller number and increased grain yield (Jiao et al., 2010; Miura et al., 2010; Wang et al., 2012). Wang et al. (2012) showed OsSPL16 to be a positive regulator of cell proliferation with increase in grain width and yield in over-expressing rice plants. However, elevated expression of OsSPL16 resulted in decrease grain appearance quality. This was expected as grain quality and yield are usually negatively correlated. But, decreased expression of OsSPL16 resulted in slender grains with better quality. Thus, miR156 and its targets OsSPL14 or OsSPL16 can be used for modifying plant architecture to create superior rice cultivars with greater yield. Recently, Guo et al. (2013) showed interactions of OsmiR444a regulated OsMADS57, OsTB1 (TEO-SINTE BRANCHED1) and D14 (Dwarf14) in controlling rice tillering. Thus, manupulating OsmiR444a and its targets by genetic engineering can prove to be an important strategy to achieve high grain yield (Guo et al., 2013). Zhang et al. (2013) reported that over-expression of OsmiR397, which is expressed naturally in young panicles and grains of rice promotes panicle branching and enlargement of grain size, causing an increase in overall grain yield of up to 25% in a field trial. OsmiR397 increases grain yield by down-regulating its target, OsLAC coding for a laccase-like protein that is involved in the sensitivity of plants to brassinosteroids. Since, miR397 is highly conserved across different species; this strategy can be extended to other cereal crops for increasing grain yield.
Fruit Improvement
Enhanced Nutritional Value and Edibility
Tomato (Lycopersicon esculentum) is one of the most economically important fruit crop across the world rich in antioxidants, minerals, fibers, and vitamins (Rajam et al., 2007). RNAi has been utilized in development of tomato fruit with enhanced level of carotenoids and flavonoids which are highly beneficial for human health. RNAi in combination with fruit specific promoter has been used to suppress an endogenous DET1 gene in tomato, a photomorphogenesis regulatory gene involved in repression of several light controlled signaling pathways (Davuluri et al., 2005). DET1 was specifically degraded in transgenic tomatoes with suppressed DET1, accompanied by an increase in the level of flavonoid and carotenoid. Abscisic acid (ABA) plays important roles during tomato fruit ripening. The SlNCED1 gene of tomato encoding 9-cis-epoxycarotenoid dioxygenase), an important enzyme in the ABA biosynthesis, was suppressed by RNAi. The fruits from RNAi lines showed enhanced accumulation of upstream compounds in the pathway, chiefly lycopene and β carotene. Similarly, RNAi has been utilized to increase the carotenoid content of rapeseed (Brassica napus) by down-regulating the expression of lycopene epsilon cyclase (ε-CYC) gene. The seeds obtained from transgenic Brassica showed increased in zeaxanthin, violaxanthin, β-carotene, and lutein content (Yu et al., 2007). RNAi approach has also been used in apple to improve the fruit quality by enhanced self-life (Dandekar et al., 2004), reducing the amount of a major apple allergen (Gilisen et al., 2005) and silencing the leaf sorbitol synthesis which affects fruit quality such as starch accumulation and sugar-acid balance (Teo et al., 2006).
Enhanced Shelf Life
The post-harvest deterioration and spoilage of vegetables and fruits are major causes of economic loss. Therefore, increase in shelf life of vegetables and fruits by delayed ripening are another essential agronomic trait that is being addressed through RNAi/microRNA technology. Initiation of ripening in climacteric fruits like tomato is characterized by a climacteric burst of ethylene in, resulting in the regulation of the expression of ripening-specific genes (Osorio et al., 2011). Thus, delayed ripening can be achieved in climacteric fruits by manipulating the gene involved in ethylene production and perception by employing either sense or antisense technology (Hamilton et al., 1990; Oeller et al., 1991; Theologis et al., 1993; Ye et al., 1996; Wilkinson et al., 1997; Xiong et al., 2003, 2005). However, RNAi was shown to be more effective and specific in nature than strategies involving either sense or an antisense RNA (Fire et al., 1998). Xiong et al. (2005) introduced dsRNA targeting a single unit of 1-aminocyclopropane-1-carboxylate (ACC) oxidase, a gene of ethylene biosynthesis pathway in tomato and suppressed the expression of its gene. The rate of ethylene production was significantly inhibited in the ripened fruits of transgenic plants leading to prolonged shelf life. However, a RNAi strategy targeting suppression of more than one homolog would be certainly much effective than knockdown of a single homolog. Gupta et al. (2013) used RNAi technology to develop tomatoes with delayed ripening by targeting three homologs of 1-aminocyclopropane-1-carboxylate (ACC) synthase (ACS) gene during the course of ripening. Delayed ripening leading to extended shelf life for ≈45 days was achieved by effective repression of ethylene production in tomato fruits by using chimeric RNAi-ACS construct designed to target ACS homologs. Two ripening-specific ethylene induced N-glycoprotein modifying enzymes, α-mannosidase (α-Man) and β-D-N-acetylhexosaminidase (β-Hex) were identified and targeted by Meli et al. (2010) through RNAi technique. Enhanced shelf life of ≈30 days and ≈2–2.5 fold firmer fruits were observed after texture analysis of these transgenic tomatoes as rate of fruit softening was significantly reduced.
microRNAs are involved in tomato fruit development and ripening (Pilcher et al., 2007; Itaya et al., 2008; Moxon et al., 2008; Zhang et al., 2011b; Molesini et al., 2012; Karlova et al., 2013). MiR156 targets a important gene in fruit ripening, colorless never ripe (CNR; Moxon et al., 2008; Molesini et al., 2012). Transgenic tomato plants over-expressing sly-miR156 exhibited slightly lighter color than wild-type controls, but could completely ripen eventually (just later; Zhang et al., 2011b). Furthermore, over-expression of miR156 in tomato also resulted in smaller fruit, increased leaf numbers, but reduced height and decreased leaf size (Zhang et al., 2011b), suggesting that higher fruit yield and shorter ripening time in tomato could be expected through either reducing miR156 expression or releasing one or several miR156 target genes. This needs to be tested by transgenic studies. The most recent study shows that CNR is also negatively regulated by APETALA2a (AP2a), a target of miR172, which positively regulates fruit ripening and negatively regulates ethylene synthesis. At the same time, CNR positively regulates AP2a, suggesting a feedback loop regulation of fruit development and ripening (Karlova et al., 2013).
Seedless Fruit Development (Parthenocarpy)
Parthenocarpy or seedlessness is a process involving production of seedless fruits, developed from the ovary without pollination and fertilization. Parthenocarpy or seedlessness is a highly desirable agronomic trait for fruit which enhances its marketing values (Molesini et al., 2012) because high yield can be achieved even under environmental conditions unfavorable for pollination and fertilization. In tomato, seedless fruits have been achieved by down-regulating a chalcone synthase, a gene involved in first step of flavonoid biosynthesis (Schijlen et al., 2007). Phytohormones such as auxin and gibberellins are closely associated with the trait of parthenocarpy (Molesini et al., 2012) which in turn are regulated by many miRNAs. Thus, manipulating the level of phytohormones by controlling activities of miRNAs or their targets can prove to be an important approach to achieve parthenocarpy. For example, expression of an aberrant form of auxin response factor 8 (ARF8), a target of miR167 (Ru et al., 2006; Wu et al., 2006; Molesini et al., 2012), resulted in parthenocarpic fruit in both Arabidopsis and tomato (Goetz et al., 2007). Seedless fruits were observed in tomato plants when ARF7 function was suppressed using RNAi (De Jong et al., 2009). Parthenocarpic fruits were also observed in tomato in which genes of the AUCSIA family coding for 53-amino-acid-long (protein or peptide) were functionally suppressed by RNAi (Molesini et al., 2009).
Biotic Stress Resistance
Phytopathogens are the cause many plant diseases leading to substantial damage to crop production and thereby heavy economic loss. Several RNAi strategies have been employed to improve the defense mechanism in crop plants against various biotic stresses including attack by virus, bacteria, fungi, nematodes, and insects.
Virus Resistance
Virus-induced gene silencing (VIGS) is a RNA-mediated PTGS mechanism that protects plants against foreign gene invasion (Beclin et al., 2002; Ding, 2010). VIGS has emerged as an extremely powerful functional genomics tool for knocking out gene expression of target plant genes in some plants. The concept of pathogen derived resistance (PDR) has promoted research aimed at achieving plants resistant to virus through genetic engineering (Simon-Mateo and García, 2011). PDR is either protein mediated involving protein encoded by the transgene or RNA-mediated, i.e., by the transcript produced from the transgene. In order to achieve PDR, hairpin dsRNA including small hairpin RNA (shRNA), self-complementary hpRNA, and intron-spliced hpRNA were formed in vivo using inverse repeat sequences from viral genomes. Among these, PTGS with the highest efficiency was elicited by the method involving self-complementary hairpin RNAs separated by an intron (Smith et al., 2000; Wesley et al., 2001). High resistance against viruses has been observed in plants even in the presence of inverted repeats of dsRNA-induced PTGS (IR-PTGS; Beclin et al., 2002; Pandolfini et al., 2003; Zrachya et al., 2007). Resistance to Potato Spindle Tuber Viroid (PSTVd) infection was achieved in transgenic tomato plants producing dsRNA against PSTVd sequences (Nora et al., 2009). Similar strategy was used to successfully engineer resistance in cassava plants against African Cassava Mosaic Virus (ACMV; Vanderschuren et al., 2009). Virus resistance has been engineered successfully in plants by targeting the coat protein (CP) gene through RNAi. Powell-Abel et al. (1986) showed that transgenic tobacco expressing the CP gene of Tobacco Mosaic Virus (TMV) was resistant to TMV and that the resistance was due to the expressed CP. Later, this strategy was extended to generate resistance against many different viruses as potato resistant to Potato Virus Y (PVY; Missiou et al., 2004), tobacco resistant to Beet Necrotic Yellow Vein Virus (BNYVV; Andika et al., 2005), Cucumis cv. melo resistant to Papaya Ring Spot Virus type W(PRSV-W; Krubphachaya et al., 2007), N. benthamiana resistant to Cucumber Green Mottle Mosaic Virus (CGMMV; Kamachi et al., 2007), N. benthamiana and Prunus domestica resistant to Plum Pox virus (PPV; Hily et al., 2007).
The use of RNA silencing strategy to engineer resistance is not limited to RNA viruses but can successfully be applied to DNA viruses. For example blackgram plants recovered efficiently from geminivirus Vigna mungo yellow mosaic virus (VMYMV) infection when inoculated with hpRNA construct containing the promoter sequence of VMYMV under the control of the 35S promoter (Pooggin et al., 2003). RNAi method has been used to generate common bean resistant to geminivirus Beans Golden Mosaic Virus (BGMV; Bonfim et al., 2007). A broad-spectrum resistance has been developed against tospoviruses in tomato plants by targeting multiple regions of a viral gene (Bucher et al., 2006). Zhou et al. (2012) have used sequences from disease-specific protein gene and CP gene from Rice Stripe Virus to develop resistance against Rice Stripe Disease.
Many viruses express viral silencing repressors (VSRs) proteins to counter host antiviral RNA silencing (Burgyan and Havelda, 2011). One of the strategies to improve virus resistance involves targeting the miRNA against these VSR. Niu et al. (2006) developed Arabidopsis plants with specific resistance against Turnip Yellow Mosaic Virus (TYMV) and Turnip Mosaic Virus (TuMV) by expressing amiRNAs based on miR159 precursor of A. thaliana. These amiRNAs target the sequence of two VSRs, P69 of TYMV and HC-Pro of TuMV. Transgenic tobacco resistant to Cucumber Mosaic Virus (CMV) was generated by targeting a VSR, 2b of CMV through expression of an amiRNA based on an A. thaliana miR171 precursor (Qu et al., 2007). TGBp1/p25 of Potato Virus X (PVX) was targeted in tobacco by expression of amiRNAs based on an A. thaliana miR159a, miR167b, and miR171a precursors (Ai et al., 2011). Singh et al. (2014) generated transgenic tobacco plants resistant to Tomato leaf curl New Delhi virus (ToLCNDV) by transformation with trans-acting siRNA generating constructs against RNAi suppressor proteins (AC2 and AC4) of the geminivirus. Another promising strategy to reduce the multiplication and spread of virus in the plant includes the use of amiRNAs targeting the viral genes involved in replication, transmission, and symptom development after viral infection. Vu et al. (2013) used two amiRNA targeting the middle region of the AV1 (coat protein) transcript (amiR-AV1-3) and the overlapping region of the AV1 and AV2 (pre-coat protein) transcripts (amiR-AV1-1) of a geminivirus, Tomato leaf curl virus (ToLCV). Transgenic tomato plants expressing amiR-AV1-1, were highly tolerant to ToLCNDV and could successfully propagate until the T2 generation.
Bacterial Resistance
Bacterial diseases are extremely difficult to control due to rapid rate of spreading. RNAi mediated supression of two genes of Agrobacterium tumefaciens involved in crown gall tumor formation (iaaM and ipt) could significantly reduce the production of tumors in Arabidopsis (Escobar et al., 2001; Dunoyer et al., 2006). This strategy could be further extended to other plants. Fatty acids and their derivatives are important signaling molecule reported to negatively regulate plant’s resistance to bacterial disease (Li et al., 2008a; Jiang et al., 2009). Arabidopsis and soybean plants showing enhanced resistance to multiple pathogens were generated by RNAi mediated suppression of SACPD gene encoding a fatty acid desaturase (Jiang et al., 2009).
In Arabidopsis, miR393 was reported to repress auxin signaling by negatively regulating the F-box auxin receptors like TIR1, thereby restricting the infection by bacteria Pseudomonas syringae (Navarro et al., 2006). Transgenic Arabidopsis plants over-expressing miR393 had enhanced bacterial resistance but with some developmental alterations (Navarro et al., 2006). Two different miRNAs, miR398 (Jagadeeswaran et al., 2009) and miR825 (Fahlgren et al., 2007) were reported to be down-regulated by bacterial infections. In the case of miR398, the expression of miR398 targets coding for two Cu/Zn superoxide dismutases (CSD1 and CSD2) was analyzed, and CSD1 was up-regulated upon bacterial infection in accordance with the down-regulation of miR398 under biotic stress (Jagadeeswaran et al., 2009). Very recently, new insight into miRNA function was gained with the discovery, that several miRNA families target genes of plant innate immune receptors (NBS-LRR) in Legumes (Zhai et al., 2011) and Solanaceae (Li et al., 2012). MiR482/2118 family of miRNAs were shown to target numerous NBS-LRR mRNAs encoding disease resistance proteins in tomato (Solanum lycopersicum) and other members of the Solanaceae (Shivaprasad et al., 2012). Viral and bacterial infection suppresses miR482-mediated silencing of R genes. Thus, these pathogen responsive miRNA are either up- or down-regulated in response to bacterial invasion and effect gene expression by suppressing negative regulators and inducing positive regulators of immune responses. Identification and characterization of the targets of these miRNAs would help decipher new players in the pathways of host defense. If these pathogen-regulated miRNAs serve as positive regulator of bacterial resistance, strategy to generate transgenics with enhanced bacterial resistance involves constitutive over-expression of miRNA or amiRNA. When miRNAs act as negative regulators, transgenic plants over-expressing these miRNAs become more sensitive to bacteria. In these cases, up-regulation of their target genes might be an effective strategy for improving plant stress tolerance, which can be achieved by over-expressing a miRNA-resistant form of its target or using amiRNA target mimic (Franco-Zorrilla et al., 2007).
Fungal Resistance
Targeting the genes of fatty acid metabolism through RNAi has proved to be a important strategy to generate diseases tolerance in various crop plants. RNAi-mediated suppression of a rice gene OsSSI2 led to enhanced resistance to blast fungus Magnaporthe grisea and leaf blight bacterium Xanthomonas oryzae (Jiang et al., 2009). Besides, enhanced disease resistance against M. grisea in rice was achieved by suppression of two genes namely OsFAD7 and OsFAD8 which are Ω-3 fatty acid desaturase (Yara et al., 2007). RNAi mediated targeting of genes involved in lignin production, led to enhanced resistance of soybean to phytopathogen Sclerotinia sclerotiorum due to reduced lignin content (Peltier et al., 2009).
Very recently, 24 miRNAs were shown to be involved in responses to attack by the fungus Blumeria graminis f. sp. tritici (Bgt) in wheat which cause a devastating diseases of wheat-powdery mildew (Xin et al., 2010). A rice miRNA osa-miR7695 was found to negatively regulate a natural resistance-associated macrophage protein 6 (OsNramp6) in response to the blast fungus Magnaporthe oryzae. Improved resistance to rice blast infection was achieved by over-expression of Osa-miR7696 (Campo et al., 2013).
Insect and Nematode Resistance
RNA interference has also been applied to control insect pests which lead to substantial crop loss (Huvenne and Smagghe, 2010). The development of a new generation of insect-resistant crops involves feeding of dsRNA as a diet component to insect which was shown to efficiently down-regulate the targeted genes in insect (Price and Gatehouse, 2008). The strategy was used in corn plants by expression of dsRNAs for tubulin or vacuolar ATPase genes to develop western corn rootworm (WCR) resistant transgenic corn plants (Baum et al., 2007). Cotton bollworm (Helicoverpa armigera) larvae showed reduced growth when fed on plant material expressing dsRNA specific to cytochrome P450 gene (CYP6AE14; Mao et al., 2007). Silencing of genes involved in parasitism or housekeeping genes in the root-knot nematode by expression of dsRNA in host plant resulted in enhanced resistance to the nematode (Gheysen and Vanholme, 2007). Sindhu et al. (2009) has targeted four genes involved in parasitism of sugar beet cyst nematode (Heterodera schachtii), having host A. thaliana by RNAi. Though complete resistance was not achieved but the number of mature nematode females in different RNAi lines were reduced to 23–64%. Ibrahim et al. (2011) could successfully reduce the gall formation by Meloidogyne incognita in soybean roots by suppressing the genes encoding tyrosine phosphatase (TP) and mitochondrial stress-70 protein precursor (MSP).
microRNAs were also reported to be involved in plant nematode interactions. In response to infection by the nematode, H. schachtii, miR161, miR164, miR167a, miR172c, miR396c, miR396a,b, and miR398a were down-regulated in Arabidopsis (Hewezi et al., 2008; Khraiwesh et al., 2012). Comparative analysis of miRNA profiling in soybean indicated that 101 miRNAs belonging to 40 families were responsive to the infection of the soybean cyst nematode (SCN; Heterodera glycines), the most devastating pathogen in soybean. It was further revealed that 20 miRNAs were differentially expressed between SCN resistant and susceptible soybean cultivars (Li et al., 2012). Moreover, it has been suggested that nematode induced miRNAs and sRNAs likely to be involved in feeding site establishment and parasitism, respectively (Hewezi et al., 2008). Over-expression of these nematode induced candidate miRNAs, and/or silencing of their corresponding targets, may provide clear insight about plant-nematode parasitism, and also may provide nematode resistance to crop plants. Nematode resistance can also be achieved by expressing amiRNA, containing known miRNA genes with the replaced seed region of the vital gene (parasitism or housekeeping) of plant parasitic nematodes.
Abiotic Stress Tolerance
Drought and Salinity Tolerance
Water deficit or drought is one the major environmental stresses limiting crop productivity. RNAi has been applied successfully to develop drought tolerant crops. RNAi mediated down-regulation of farnesyl transferase in canola using the AtHPR1 promoter showed more resistance to seed abortion during flowering induced by water deficiency without affecting yield during drought stress (Wang et al., 2009). Transgenic rice plants tolerant to drought stress were developed by silencing of receptor for activated C-kinase 1 (RACK1; Li et al., 2009). The ubiquitin ligase gene has been targeted for RNAi in rice to enhance drought tolerance. Rice knockdown of a RING finger E3 ligase gene-OsDSG1 leads to enhanced drought tolerance (Park et al., 2010). Similarly, silencing of OsDIS1 (for Oryza sativa drought-induced SINA protein 1), a C3HC4 RING finger E3 ligase by RNAi enhanced drought tolerance.
Wang et al. (2011) studied legume model plant, Medicago truncatula and identified several drought-responsive miRNAs. The already known and predicted targets of these miRNAs were found to be involved in wide variety of processes in plant cell including development, transcription, protein degradation, detoxification, etc. It was observed that miR393 was strongly up-regulated by dehydration, cold, high salinity, and ABA treatments (Sunkar and Zhu, 2004). On exposing Arabidopsis to varying degrees of different abiotic stress, up regulation was observed in miR402, miR319c, miR389a, and miR397b. Xin et al. (2010) identified and reported 12 miRNAs responsive to heat stress in wheat (Triticum aestivum L.). MiRNA responsive to various stresses (drought, cold, salinity, and ABA) were identified in rice seedlings (Jian et al., 2010). Among many stress responsive miRNAs discovered in rice, only two miR393 and miR169 were found to be responsive to abiotic stress like dehydration (Zhao et al., 2007). In Arabidopsis, miR169 was down-regulated by drought stress, and its target nuclear factor YA5 (NF-YA5) was significantly induced upon drought stress. Transgenic plants over-expressing miR169a were more sensitive to drought stress with increased leaf water loss as compared to wild-type controls, and over-expression of NF-YA5 led to enhanced drought tolerance in transgenic plants (Li et al., 2008a). Similarly, a newly characterized Soybean (Glycine max L.) gene GmNFYA3, a target of miR169, was demonstrated to positively modulate drought stress tolerance in transgenic Arabidopsis plants (Ni et al., 2013). However, over-expression of GmNFYA3 in Arabidopsis resulted in increased sensitivity to salinity stress and exogenous ABA (Ni et al., 2013). These findings were contrary to the observation in tomato (S. lycopersicum), in which miR169 was induced by drought stress and its four targets NF-YA1/2/3 and multidrug resistance-associated protein gene 1 (MRP1) were all down-regulated by drought stress. Constitutive over-expression of tomato miR169c led to reduced stomatal openings, transpiration rate, and leaf water loss, thus enhanced drought tolerance in transgenic plants in comparison to wild-type controls (Zhang et al., 2011a). It is notable that transgenic tomato plants had no noticeable morphological and developmental alterations under field conditions, indicating that miR169 or its targets could be potential candidate genes for genetic engineering to achieve enhanced abiotic stress tolerance in transgenic plants (Zhang et al., 2011a). However, as mentioned above, miR169 positively regulates plant response to drought stress in tomato, but negatively in Arabidopsis. The opposite roles miR169 play in different plant species suggest that even conserved miRNAs might function in a species-specific manner and this should be taken into careful consideration when tailoring GM strategies for specific target species.
In a recent study, transgenic creeping bentgrass (Agrostis stolonifera) plants over-expressing a rice miR319 gene (Osa-miR319) exhibited enhanced tolerance to drought and salinity that was associated with increased leaf wax content and water retention but reduced sodium uptake. Gene expression analysis indicated that the enhanced abiotic stress tolerance can be attributed to a significant down-regulation of at least four putative turf miR319 target genes teosinte branched/cycloidea/proliferating factors (TCP)—AsPCF5, AsPCF6, AsPCF8, and AsTCP14 and a homolog of a rice NAC domain gene AsNAC60 (Zhou et al., 2013). It is noticeable that over-expression of Osa-miR319 caused pleiotropic phenotypes including increased leaf size, enlarged stems and decreased tiller number, which are undesirable traits for creeping bent grass. To better utilize the miR319-based GM strategy, further functional characterization of miR319 targets or downstream genes in the related biological pathways needs to be undertaken. However, the mechanism of miR319-mediated plant abiotic stress response revealed from this study provides important information to develop novel strategies for plant genetic engineering and has the potential to be applied in other important crop species. MiR159 was reported to respond to hormone signaling and dehydration responses in Arabidopsis (Achard et al., 2004; Reyes and Chua, 2007).
Hu et al. (2009) showed that most of the rice histone deacetylases genes are responsive to drought or salt stresses with specific patterns of expression and divergent developmental functions compared to closely related homologs in Arabidopsis. Trindade et al. (2010) identified several conserved miRNAs in M. truncatula plants which are expressed differentially in water-deficit condition. MiR169 was shown to be down-regulated in roots, whereas miR398a/b and miR408 showed very high expression in shoots as well as roots. Hwang et al. (2011) reported a drought stress-responsive miR171 family in potato plants, Solanum tuberosum which miR171a, miR171b, and miR171c. The RNAi-mediated silencing of farnesyl transferase genes (FTA or FTB) in canola resulted in reduced transpiration rate due to decreased stomatal conductance, thereby promoting yield (Wang et al., 2005, 2009).
Cold and Heat Stress Tolerance
The expression of miR319 was reported to change in response to cold stress in Arabidopsis (Sunkar and Zhu, 2004; Liu et al., 2008), rice (Lv et al., 2010), and sugarcane (Thiebaut et al., 2011). Further transgenic studies using wild-type plants as controls indicated that over-expression of Osa-miR319 gene led to increased cold stress tolerance (4°C) after chilling acclimation(12°C) of plants (Yang et al., 2013). However, miR319 transgenic rice plants displayed severe developmental delay. To avoid the pleiotropic effect of miR319, two RNAi lines for the miR319 targets, OsPCF5 and OsTCP21, were generated. These RNAi lines, upon chilling acclimation, also exhibited better cold tolerance than wild-type controls, but were phenotypically normal. It should be noted that improvement of plant cold tolerance in the miR319 over-expression lines was more significant than that in the OsPCF5 and OsTCP21 RNAi lines (Yang et al., 2013), most likely due to target function redundancy in the latter case. This is also a challenge when manipulating miRNA targets instead of miRNAs themselves for plant trait modification, because in some cases multiple targets have to be simultaneously down-regulated to achieve the same level of effect as over-expression of an individual miRNA gene. Guan et al. (2013) discovered a novel plant thermotolerance mechanism, especially for the protection of reproductive organs. It involves induction of miR398 to downregulate its targets CSD (copper/zinc superoxide dismutase) genes, CSD1 and CSD2 as well as CCS (a gene encoding copper chaperone for both CSD1 and CSD2; Guan et al., 2013). They found that csd1, csd2, and ccs mutants displayed higher heat stress tolerance than wild-type plants associated with increased accumulation of heat stress transcription factors and heat shock proteins and reduced damage to flowers (Guan et al., 2013). These results strongly suggest that manipulating miR398 or its targets can be an applicable strategy to increase heat tolerance in crop species, especially in corn, which suffers damage to its reproductive tissues by prolonged periods of high summer temperatures.
Oxidative Stress Tolerance
Plants tend to accumulate reactive oxygen species (ROS) in response to environmental stimuli such as high intensity light, extreme temperatures, UV radiation, heavy metals, salinity, drought stresses, and mechanical stresses (Khraiwesh et al., 2012). Superoxide dismutases (SODs) in plants can detoxify superoxide radicals by converting them into molecular oxygen and hydrogen peroxide. Several studies have been conducted to improve plant stress tolerance by over-expression of superoxide dismutase (Cu/Zn-SODs) in transgenic plants which detoxify superoxide radicals (Tepperman and Dunsmuir, 1990; Pitcher et al., 1991; Gupta et al., 1993; Perl et al., 1993; Sunkar et al., 2006). However, in some of the studies, the transgenic plants exhibited minimal or no increase in stress tolerance (Tepperman and Dunsmuir, 1990; Pitcher et al., 1991). Sunkar et al. (2006) offered an improved strategy to solve this problem by over-expressing miR398-resistant form of CSD2, which led to increased tolerance to high intensity light, heavy metals and other oxidative stresses (Table 1; Sunkar et al., 2006). The exploration of interactions between miR398 and its targets (CSD1 and CSD2) also provided a possible explanation to the previously failed attempts. The expression of the introduced SOD transgenes containing the miR398 target sites were negatively impacted by miR398-mediated gene regulation.
Nutritional Improvement
Biofortification
Essential fatty acids have important role in maintaining heart health in human. RNAi technology has also been used successfully or modification of fatty acid composition of oil. Two key fatty acid desaturase genes encoding stearoyl-acyl-carrier protein Δ9-desaturase and oleoyl-phosphatidylcholine γ6-desaturase (Liu et al., 2002) targeted by RNAi for nutritional enhancement of cotton seed oil with high-oleic (HO) and high-stearic (HS) contents. Soybean oil flavor and stability can be improved and process of hydrogenation can be avoided by reducing the content of alpha-linolenic acid (18:3). Flores et al. (2008) have used hairpin RNA mediated RNAi strategy (glycinin promoterwas used for seed-specific silencing) to down-regulate omega-3 fatty acid desaturase [an enzyme converting linoleic acid (18:2) to alpha-linolenic acid (18:3)]. Transgenic soybean seed were reported to significant reduction in alpha-linoleic acid content (1–3%) compared to non-transgenic soybean seed (7–10%).
Seed-specific RNAi approaches have also successfully been used to generate dominant high lysine corn by suppressing the expression of 22-kDa maize zein storage proteins, a group of abundant proteins in maize seed but poor in lysine content (Segal et al., 2003). RNAi was also used to down-regulate the starch-branching enzyme resulting in high-amylose wheat, which has a great potential to improve human health (Regina et al., 2006). RNAi technology can be applied to increase the starch content in the leaves. The process of phosphorylation and dephosphorylation of starch are crucial steps of leaf starch degradation. Based on this fact, starch content has been increased by manipulation of phosphate metabolism genes in Arabidopsis and maize (Weise et al., 2012). RNAi-mediated gene silencing has been utilized to reduce glutenin content in rice which is difficult to digest by kidney patients. To achieve low glutenin content GluB hairpin RNA was employed by Kusaba et al. (2003) to produce a rice variety called LGC-1 (low glutenin content 1).
The two main enzymatic steps involved in sucrose biosynthesis involve synthesis of sucrose-6-phosphate (Suc6P) by sucrose-phosphate synthase (SPS) which is hydrolysed to sucrose and inorganic phosphate (Pi) by an another enzyme sucrose phosphatase (SPP). At low temperature (4°C), potato tubers undergo an unfavorable phenomenon of, i.e., ‘cold sweetening’ which involves conversion of starch to reducing sugars glucose and fructose. To address this problem of cold induced sweetening, Chen et al. (2008) have targeted NtSPP2 gene of tobacco for RNAi mediated suppression of SPP in transgenic potato tubers. The transgenic potato tubers showed accumulation of Suc6P when exposed to cold storage. In SPP-silenced tubers, the process of sucrose-to-hexose conversion was reduced as cold induced expression of vacuolar invertase (VI) was found to be blocked.
RNA interference technology was used by Gil-Humanes et al. (2008) to significantly reduce the level of γ-gliadins in different wheat cultivars by silencing the expression of specific γ-gliadins. Gil-Humanes et al. (2012) further, reported slight increase in the protein content of transgenic lines due to compensatory effect produced by down-regulation of γ-gliadins on the rest of gluten proteins. The glutenin content was found to increase whereas no significant changes were observed in the total gliadins content.
Allergen and Anti-Nutrient Elimination
Certain food like peanuts, apple, etc. consists of certain allergens that lead to allergic response when consumed. Besides many commonly consumed plants consist of natural toxin which is harmful to human health so, reduction or elimination of these allergens or toxins from food by RNAi will enhance its edibility and food quality. Expression of one of the major apple allergen Mald1 was reduced significantly by Gilisen et al. (2005) through RNAi-based gene silencing. RNAi was used by Le et al. (2006) to suppress the expression of a tomato allergen-Lyce3 which is a non-specific lipid transfer protein from tomato peel. Dodo et al. (2008) reduced the content of Arah2, a major peanut allergen by 25% in crude peanut extract. In onion, Eady et al. (2008) using hp RNAi suppressed the tear inducing lachrymatory factor synthase (LFS) gene which is involved in conversion of 1-propenyl sulfenic acid to propanthial S-oxide [tear-inducing, lachrymatory factor (LF)] resulting in the production of tearless onion. The RNAi technology has also been used to reduce the level of, a neurotoxin β-N-oxalyl-amino-alanine-L-alanine (BOAA) in the Grass pea. (Lathyrus sativus) or chickling pea which can cause paralytic disease called lathyrism (Angaji et al., 2010). Though cotton seeds are rich source of dietary protein but consist of a toxic terpenoid-gossypol which plays an important role in plant defense mechanism. RNAi has been used to lower gossypol content in cotton seeds by reducing seed specific expression of delta-cadinene synthase, an enzyme in gossypol biosynthesis pathway (Sunilkumar et al., 2006). Recently, Rathore et al. (2012) reported that RNAi-mediated silencing of delta-cadinene synthase gene resulted in cotton plants that produced ultra-low gossypol cottonseed (ULGCS). Lewis et al. (2008) used RNAi for silencing the nicotine demethylase gene to suppress the conversion of nicotine to nor-nicotine by six fold which is precursor of a carcinogenic compound in tobacco (N. tabacum).
Conclusion and Future Prospects
RNA interference technology involving siRNA and miRNA have emerged as an attractive tool used by plant biologists not only to decipher the plant function but also to develop plants with improved and novel traits by manipulation of both desirable and undesirable genes. It is a powerful tool to understand the functions of individual genes and also proved useful to molecular breeders in producing improved crop varieties. sRNAs have been utilized either for silencing a gene of interest by RNAi or altering the expression of a gene by over-expressing a miRNA or introduction of artificially synthesized miRNA targeting it. The technology has well been utilized in crops not only to improve the food productivity (biomass and grain yield) but its nutritional value (cereals, fruits, and vegetables enriched with essential minerals, vitamins, fatty acids, and amino acids). RNAi technology has also been exploited to develop plants with improved resistance against various environmental stresses (especially drought) and biotic stresses like pathogen attack (virus, bacteria, insects, and nematodes).
Thus applications of research related with small non-coding RNAs has the potential to contribute to food safety by generating crop cultivars with improved agronomic traits leading to increasing yield and nutritional value. Research focussing on interaction of miRNAs and their targets can provides valuable insight into mechanisms of post-transcriptional gene regulation and multiple molecular pathways controlling plant stress responses. After completion of several recent genome sequencing projects, the study of RNAi has become increasingly more expanding and challenging. Researchers can manipulate several newly explored RNAi pathways in order to amend gene expression in crops, which can result in new and better quality traits.
Although RNAi technology can serve as potential tools for crop improvement but certain limitations are also associated with it. Altering the expression of a target gene or its miRNA might lead to undesirable pleiotropic changes in plant morphology and development. Therefore transgenic strategies should be designed only after completely understanding the mechanism of miRNA regulation. This will minimize undesirable trade-off in transgenic products. MiRNA strategy useful for one plant species may not work for another species, since regulations of evolutionarily conserved miRNAs may vary in different species (Li et al., 2008a; Zhang et al., 2011a). A family of related genes can be silenced with only one RNAi construct due to the sequence heterogeneity of siRNAs, but can cause off-target effects leading to unintentional silencing of genes with regions of homology to the intended target. Besides, post-transcriptional silencing in plants is mobile that can be induced locally and then spreads throughout the plant. Thus, siRNA-based RNAi strategies might not be suitable for some applications requiring tissue-specific silencing of genes. Atificial miRNAs can be used to overcome these limitations as it brings about much more specific silencing of genes. There is a reduced chance of off-target effects with miRNA-based RNAi, since miRNAs use only a single 21- or 22-nt sequence to identify the target, whereas siRNAs comprise a population of sequences. Thus, with miRNA-based RNAi, it is easier to target individual genes, even in a closely related gene family. amiRNAs are also suitable for tissue-specific silencing because miRNA-directed silencing does not spread throughout the plant. However, silencing might not be very durable with amiRNA because only a single 21- or 22-nt specificity determinant is involved. Mutation of the target can lead to easier escape from miRNA-directed silencing compared to siRNA-directed silencing which targets much larger sequence. This problem can be overcome by using two (or more) different amiRNAs against the target.
Crops engineered with constructs encoding heterologous proteins or over-expressing protein are fundamentally different from those consisting of RNA-mediated gene suppression cassettes because these constructs do not encode any protein(s) and are intended to express only non-coding RNAs. Transgenic proteins are recommended to undergo digestibility studies, to evaluate their potential for digestion, as allergenic and toxic proteins may be refractory to digestion (Astwood et al., 1996). RNA is not known to produce oral toxicity in humans but proteins in some rare cases can produce oral toxicity. According to the US FDA, “Introduced nucleic acids [in biotech crops], in and of themselves, do not raise safety concerns (FDA, 1992).” Furthermore, with regards to RNA-mediated gene regulation in genetically engineered crops, the US FDA states, “thus, for example, the introduction of a gene encoding an anti-sense ribonucleic acid (RNA) would not raise concerns about either the gene or the anti-sense RNA. Any safety considerations would focus on the intended effects of the anti-sense RNA.” Thus, transgenic crops with introduced RNA based traits are much safe for human consumptions than crops over-expressing proteins and need not require acute oral toxicity studies and evaluation of digestibility of the introduced RNA component. Certain bio safety concerns do arise on use of RNAi transgenic as transcriptional gene silencing by chromatin modification might lead to hereditary changes associated with adverse effects. However, crop plants developed with thoughtfully designed RNAi strategy and proper assessment of risk related to food safety may help overcome the biosafety concerns. In spite of few limitations, crop improvement strategies based on small non-coding RNAs have enormous potential to increase productivity as well as nutritional value.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank the reviewers for their comments and suggestions which helped to improve the manuscript. We sincerely apologize to those authors whose work we have inadvertently omitted, or could not review at length due to space limitations.
References
Achard, P., Herr, A., Baulcombe, D. C., and Harberd, N. P. (2004). Modulation of floral development by gibberellin-regulated microRNA. Development 131, 3357–3365. doi: 10.1242/dev.01206
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ai, T., Zhang, L., Gao, Z., Zhu, C. X., and Guo, X. (2011). Highly efficient virus resistance mediated by artificial microRNAs that target the suppressor of PVX and PVY in plants. Plant Biol. 13, 304–316. doi: 10.1111/j.1438-8677.2010.00374.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Andika, I. B., Kondo, H., and Tamada, T. (2005). Evidence that RNA silencing-mediated resistance to beet necrotic yellow vein virus is less effective in roots than in leaves. Mol. Plant Microbe Interact. 18, 194–204. doi: 10.1094/MPMI-18-0194
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Angaji, S. A., Hedayati, S. S., Hosein poor, R., Samad poor, S., Shiravi, S., and Madani, S. (2010). Application of RNA interference in plants. Plant Omics J. 3, 77–84.
Astwood, J. D., Leach, J. N., and Fuchs, R. L. (1996). Stability of food allergens to digestion in vitro. Nat. Biotechnol. 14, 1269–1273. doi: 10.1038/nbt1096-1269
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Aukerman, M. J., and Sakai, H. (2003). Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 15, 2730–2741. doi: 10.1105/tpc.016238
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Axtell, M. J. (2013). Classification and comparison of small RNAs from plants. Annu. Rev. Plant Biol. 64, 137–159. doi: 10.1146/annurev-arplant-050312-120043
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Bao, N., Lye, K. W., and Barton, M. K. (2004). MicroRNA binding sites in Arabidopsis class III HD-ZIP mRNAs are required for methylation of the template chromosome. Dev. Cell 7, 653–662. doi: 10.1016/j.devcel.2004.10.003
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Bartel, D. P. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297. doi: 10.1016/S0092-8674(04)00045-5
Baulcombe, D. (2000). Unwinding RNA silencing. Science 290, 1108–1109. doi: 10.1126/science.290.5494.1108
Baum, J. A., Bogaert, T., Clinton, W., Heck, G. R., Feldmann, P., Ilagan, O., et al. (2007). Control of coleopteran insect pests through RNA interference. Nat. Biotechnol. 25, 1322–1326. doi: 10.1038/nbt1359
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Beclin, C., Boutet, S., Waterhouse, P., and Vaucheret, H. (2002). A branched pathway for transgene-induced RNA silencing in plants. Curr. Biol. 12, 684–688. doi: 10.1016/S0960-9822(02)00792-3
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Bernstein, E., Caudy, A. A., Hammond, S. M., and Hannon, G. J. (2001). Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409, 363–366. doi: 10.1038/35053110
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Bonfim, K., Faria, J. C., Nogueira, E. O., Mendes, E. A., and Aragao, F. J. (2007). RNAi-mediated resistance to bean golden mosaic virus in genetically engineered common bean (Phaseolus vulgaris). Mol. Plant Microbe Interact. 20, 717–726. doi: 10.1094/MPMI-20-6-0717
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Bouche, N., Lauressergues, D., Gasciolli, V., and Vaucheret, H. (2006). An antagonistic function for Arabidopsis DCL2 in development and a new function for DCL4 in generating viral siRNAs. EMBO J. 25, 3347–3356. doi: 10.1038/sj.emboj.7601217
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Brodersen, P., Sakvarelidze-Achard, L., Bruun-Rasmussen, M., Dunoyer, P., Yamamoto, Y. Y., Sieburth, L., et al. (2008). Widespread translational inhibition by plant miRNAs and siRNAs. Science 320, 1185–1190. doi: 10.1126/science.1159151
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Brosnan, C. A., Mitter, N., Christie, M., Smith, N. A., Waterhouse, P. M., and Carroll, B. J. (2007). Nuclear gene silencing directs reception of long-distance mRNA silencing in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 104, 14741–14746. doi: 10.1073/pnas.0706701104
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Bucher, E., Lohius, D., van Poppel, P. M., Geerts-Dimitriadou, C., Gold-bach, R., and Prins, M. (2006). Multiple virus resistance at a high frequency using a single transgene construct. J. Gen. Virol. 87, 697–701. doi: 10.1099/vir.0.82276-0
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Buhtz, A., Springer, F., Chappell, L., Baulcombe, D. C., and Kehr, J. (2008). Identification and characterization of small RNAs from the phloem of Brassica napus. Plant J. 53, 739–749. doi: 10.1111/j.1365-313X.2007.03368.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Burgyan, J., and Havelda, Z. (2011). Viral suppressors of RNA silencing. Trends Plant Sci. 16, 265–272. doi: 10.1016/j.tplants.2011.02.010
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Burkhart, K. B., Guang, S., Buckley, B. A., Wong, L., Bochner, A. F., and Kennedy, S. (2011). A pre-mRNA-associating factor links endogenous siRNAs to chromatin regulation. PLoS Genet. 7:e1002249. doi: 10.1371/journal.pgen.1002249
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Campbell, T. N. (2005). Choy FYM, RNA interference: past, present and future. Curr. Issues Mol. Biol. 7, 1–6.
Campo, S., Peris-Peris, C., Sire, C., Moreno, A. B., Donaire, L., Zytnicki, M., et al. (2013). Identification of a novel microRNA (miRNA) from rice that targets an alternatively spliced transcript of the Nramp6 (Natural resistance-associated macrophage protein 6) gene involved in pathogen resistance. New Phytol. 199, 212–227. doi: 10.1111/nph.12292
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Carlsbecker, A., Lee, J. Y., Roberts, C. J., Dettmer, J., Lehesranta, S., Zhou, J., et al. (2010). Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature 465, 316–321. doi: 10.1038/nature08977
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Chen, F., and Dixon, R. A. (2007). Lignin modification improves fermentable sugar yields for biofuel production. Nat. Biotechnol. 25, 759–761. doi: 10.1038/nbt1316
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Chen, S., Hajirezaei, M. R., Zanor, M. I., Hornyik, C., Debast, S., Lacomme, C., et al. (2008). RNA interference-mediated repression of sucrose-phosphatase in transgenic potato tubers (Solanum tuberosum) strongly affects the hexose-to-sucrose ratio upon cold storage with only minor effects on total soluble carbohydrate accumulation. Plant Cell Environ. 31, 165–176.
Chen, X. (2004). A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303, 2022–2025. doi: 10.1126/science.1088060
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Chuck, G. S., Tobias, C., Sun, L., Kraemer, F., Li, C., Dibble, D., et al. (2011). Overexpression of the maize corngrass1 microRNA prevents flowering, improves digestibility, and increases starch content of switchgrass. Proc. Natl. Acad. Sci. U.S.A. 108, 17550–17555. doi: 10.1073/pnas.1113971108
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Cogoni, C., Irelan, J. T., Schumacher, M., Schmidhauser, T., Selker, E. U., and Macino, G. (1996). Transgene silencing of the al-1 gene in vegetative cells of Neurospora is mediated by a cytoplasmic effector and does not depend on DNA-DNA interactions or DNA methylation. EMBO J. 15, 3153–3163.
Dandekar, A. M., Teo, G., Defilippi, B. G., Uratsu, S. L., Passey, A. J., Kader, A. A., et al. (2004). Effect of down-regulation of ethylene biosynthesis on fruit flavor complex in apple fruit. Transgenic Res. 13, 373–384. doi: 10.1023/B:TRAG.0000040037.90435.45
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Davuluri, G. R., Tuinen, A., Fraser, P. D., Manfredonia, A., Newman, R., Burgess, D., et al. (2005). Fruit-specific RNAi-mediated suppression of DET1 enhances carotenoid and flavonoid content in tomatoes. Nat. Biotechnol. 23, 890–895. doi: 10.1038/nbt1108
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
De Jong, M., Wolters-Arts, M., Feron, R., Mariani, C., and Vriezen, W. H. (2009) The Solanum lycopersicum auxin response factor 7 (SlARF7) regulates auxin signaling during tomato fruit set and development. Plant J. 5, 160–170. doi: 10.1111/j.1365-313X.2008.03671.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Denli, A., and Hannon, G. (2003) RNAi: an ever-growing puzzle. Trends Biochem. Sci. 28, 196–201. doi: 10.1016/S0968-0004(03)00058-6
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ding, S. W. (2010) RNA-based antiviral immunity. Nat. Rev. Immunol. 10, 632–644. doi: 10.1038/nri2824
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Dodo, H. W., Konan, K. N., Chen, F. C., Egnin, M., and Viquez, O. M. (2008) Alle viating peanut allergy using genetic engineering: the silencing of the immunodominant allergen Ara h 2 leads to its significant reduction and a decrease in peanut allergenicity. Plant Biotechnol. J. 6, 135–145. doi: 10.1111/j.1467-7652.2007.00292.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Dunoyer, P., Himber, C., and Voinnet, O. (2006) Induction, suppression and requirement of RNA silencing pathways in virulent Agrobacterium tumefaciens infections. Nat. Genet. 38, 258–263. doi: 10.1038/ng1722
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Dunoyer, P., Schott, G., Himber, C., Meyer, D., Takeda, A., Carrington, J. C., et al. (2010a) Small RNA duplexes function as mobile silencing signals between plant cells. Science 328, 912–916. doi: 10.1126/science.1185880
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Dunoyer, P., Brosnan, C. A., Schott, G., Wang, Y., Jay, F., Alioua, A., et al. (2010b) An endogenous, systemic RNAi pathway in plants. EMBO J. 29, 1699–1712. doi: 10.1038/emboj.2010.65
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Eady, C. C., Kamoi, T., Kato, M., Porter, N. G., Davis, S., Shaw, M., et al. (2008) Silencing onion lachrymatory factor synthase causes a significant change in the sulfur secondary metabolite profile. Plant Physiol. 147, 2096–2106. doi: 10.1104/pp.108.123273
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Elbashir, S. M., Lendeckel, W., and Tuschl, T. (2001) RNA interference is mediated by 21-and 22-nucleotide RNAs. Genes Dev. 15, 188–200. doi: 10.1101/gad.862301
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ender, C., and Meister, G. (2010) Argonaute proteins at a glance. J. Cell Sci. 123, 1819–1823. doi: 10.1242/jcs.055210
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Enrique, R., Siciliano, F., Favaro, M. A., Gerhardt, N., Roeschlin, R., Rigano, L., et al. (2011) Novel demonstration of RNAi in citrus reveals importance of citrus callose synthase in defence against Xanthomonas citrisubsp.Citri. Plant Biotechnol. J. 9, 394–407. doi: 10.1111/j.1467-7652.2010.00555.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Eschen-Lippold, L., Landgraf, R., Smolka, U., Schulze, S., Heilmann, M., Heilmann, I., et al. (2012) Activation of defense against Phytophthora infestans in potato by down-regulation of syntaxin gene expression. New Phytol. 193, 985–996. doi: 10.1111/j.1469-8137.2011.04024.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Escobar, M. A., Civerolo, E. L., Summerfelt, K. R., and Dandekar, A. M. (2001) RNAi-mediated oncogene silencing confers resistance to crown gall tumorigenesis. Proc. Natl. Acad. Sci. U.S.A. 98, 13437–13442. doi: 10.1073/pnas.241276898
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Fagegaltier, D., Bouge, A. L., Berry, B., Poisot, E., Sismeiro, O., Coppee, J. Y., et al. (2009) The endogenous siRNA pathway is involved in heterochromatin formation in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 106, 21258–21263. doi: 10.1073/pnas.0809208105
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Fahlgren, N., Howell, M. D., Kasschau, K. D., Chapman, E. J., Sullivan, C. M., Cumbie, J. S., et al. (2007) High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA Genes. PLoS ONE 2:e219. doi: 10.1371/journal.pone.0000219
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
FDA. (1992) Statement of policy: foods derived from new plant varieties. Fed. Regist. 57, 22984–23005.
Feldmann, K. A. (2006) Steroid regulation improves crop yield. Nat. Biotechnol. 24, 46–47. doi: 10.1038/nbt0106-46
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Fire, A., Xu, S., Montgomery, M. K., Kostas, S. A., Driver, S. E., and Mello, C. C. (1998) Potent and specific genetic inteference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811. doi: 10.1038/35888
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Flores, T., Karpova, O., Su, X., Zeng, P., Bilyeu, K., Sleper, D. A., et al. (2008) Silencing of GmFAD3 gene by siRNA leads to low alpha-linolenic acids (18:3) of fad3-mutant phenotype in soybean [Glycine max (Merr.)]. Transgenic Res. 17, 839–850. doi: 10.1007/s11248-008-9167-6
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Franco-Zorrilla, J. M., Valli, A., Todesco, M., Mateos, I., Puga, M. I., Rubio-Somoza, I., et al. (2007) Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 39, 1033–1037. doi: 10.1038/ng2079
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Fu, C., Sunkar, R., Zhou, C., Shen, H., Zhang, J. Y., Matts, J., et al. (2012) Overexpression of miR156 in switchgrass (Panicum virgatum L.) results in various morphological alterations and leads to improved biomass production. Plant Biotechnol. J. 10, 443–452. doi: 10.1111/j.1467-7652.2011.00677.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Fujii, H., Chiou, T. J., Lin, S. I., Aung, K., and Zhu, J. K. (2005) A miRNA involved in phosphate-starvation response in Arabidopsis. Curr. Biol. 15, 2038–2043. doi: 10.1016/j.cub.2005.10.016
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Gheysen, G., and Vanholme, B. (2007) RNAi from plants to nematodes. Trends Biotechnol. 25, 89–92. doi: 10.1016/j.tibtech.2007.01.007
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Gil-Humanes, J., Piston, F., Hernando, A., Alvarez, J. B., Shewry, P. R., and Barro, F. (2008) Silencing of γ-gliadins by RNA interference (RNAi) in bread wheat. J. Cereal Sci. 48, 565–568. doi: 10.1016/j.jcs.2008.03.005
Gil-Humanes, J., Pistón, F., Rosell, C. M., and Barro, F. (2012) Significant down-regulation of γ-gliadins has minor effect on gluten and starch properties of bread wheat. J. Cereal Sci. 56, 161–170. doi: 10.1016/j.jcs.2012.02.009
Gilisen, L. J., Bolhaar, S. T., Matos, C. I., Rouwendal, G. J., Boone, M. J., Krens, F. A., et al. (2005) Silencing of major apple allergen Mal d 1 by using the RNA interference approach. J. Allergy Clin. Immunol. 115, 364–369. doi: 10.1016/j.jaci.2004.10.014
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Goetz, M., Hooper, L. C., Johnson, S. D., Rodrigues, J. C., Vivian-Smith, A., and Koltunow, A. M. (2007) Expression of aberrant forms of auxin response factor8 stimulates parthenocarpy in Arabidopsis and tomato. Plant Physiol. 145, 351–366. doi: 10.1104/pp.107.104174
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Guan, Q., Lu, X., Zeng, H., Zhang, Y., and Zhu, J. (2013) Heat stress induction of miR398 triggers a regulatory loop that is critical for thermo tolerance in Arabidopsis. Plant J. 74, 840–851. doi: 10.1111/tpj.12169
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Gunnery, S., and Datta, A. (1987) An inhibitor RNA of translation from Barley embryo. Biochem. Biophys. Res. Comm. 142, 383–388. doi: 10.1016/0006-291X(87)90285-3
Guo, S., Xu, Y., Liu, H., Mao, Z., Zhang, C., Ma, Y., et al. (2013) The interaction between OsMADS57 and OsTB1 modulates rice tillering via DWARF14. Nat. Comm. 4:1566. doi: 10.1038/ncomms2542
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Gupta, A. S., Heinen, J. L., Holaday, A. S., Burke, J. J., and Allen, R. D. (1993) Increased resistance to oxidative stress in transgenic plants that overexpress chloroplastic Cu/Zn superoxide dismutase. Proc. Natl. Acad. Sci. U.S.A. 90, 1629–1633. doi: 10.1073/pnas.90.4.1629
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Gupta, A., Pal, R. K., and Rajama, M. V. (2013) Delayed ripening and improved fruit processing quality in tomato by RNAi-mediated silencing of three homologs of 1-aminopropane-1-carboxylate synthase gene. J. Plant Physiol. 170, 987–995. doi: 10.1016/j.jplph.2013.02.003
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Hamilton, A. J., and Baulcombe, D. C. (1999) A novel species of small antisense RNA in posttranscriptional gene silencing. Science 286, 950–952. doi: 10.1126/science.286.5441.950
Hamilton, A. J., Lycett, G. W., and Grierson, D. (1990) Antisense gene that inhibits synthesis of the hormone ethylene in transgenic plants. Nature 346, 284–287. doi: 10.1038/346284a0
Hamilton, A., Voinnet, O., Chappell, L., and Baulcombe, D. C. (2002) Two classes of short interfering RNA in RNA silencing. EMBO J. 21, 4671–4679. doi: 10.1093/emboj/cdf464
Hammond, S. M., Bernstein, E., Beach, D., and Hannon, G. J. (2000) An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404, 293–296. doi: 10.1038/35005107
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Hannon, G. J. (2002) RNA interference. Nature 418, 244–251. doi: 10.1038/418244a
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Henderson, I. R., Zhang, X., Lu, C., Johnson, L., Meyers, B. C., Green, P. J., et al. (2006) Dissecting Arabidopsis thaliana DICER function in small RNA processing, gene silencing and DNA methylation patterning. Nat. Genet. 38, 721–725. doi: 10.1038/ng1804
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Hewezi, T., Howe, P., Maier, T. R., and Baum, T. J. (2008) Arabidopsis small RNAs and their targets during cyst nematode parasitism. Mol. Plant Microbe Interact. 21, 1622–1634. doi: 10.1094/MPMI-21-12-1622
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Hily, J. M., Ravelonandro, M., Damsteegt, V., Basset, C., Petri, C., Liu, Z., et al. (2007) Plum pox virus coat protein gene intron-hair pin-RNA (ihpRNA) constructs provide resistance to Plum pox virus in Nicotiana bethamiana and Prunus domestica. J. Am. Soc. Hort. Sci. 132, 850–858.
Hisano, H., Nandakumar, R., and Wang, Z. Y. (2009) Genetic modification of lignin biosynthesis for improved biofuel production. Vitro Cell. Dev. Biol. Plant 45, 306–313. doi: 10.1007/s11627-009-9219-5
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Hu, Y., Qin, F., Huang, L., Sun, Q., Li, C., Zhao, Y., et al. (2009) Rice histone deacetylase genes display specific expression patterns and developmental functions. Biochem. Biophys. Res. Commun. 388, 266–271. doi: 10.1016/j.bbrc.2009.07.162
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Huang, G., Allen, R., Davis, E. L., Baum, T. J., and Hussey, R. S. (2006) Engineering broad root-knot resistance in transgenic plants by RNAi silencing of a conserved and essential root-knot nematode parasitism gene. Proc. Natl. Acad. Sci. U.S.A. 103, 14302–14306. doi: 10.1073/pnas.0604698103
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Huntzinger, E., and Izaurralde, E. (2011) Gene silencing by micro-RNAs: contributions of translational repression and mRNA decay. Nat. Rev. Genet. 12, 99–110. doi: 10.1038/nrg2936
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Hutvagner, G., McLachlan, J., Pasquinelli, A. E., Balint, E., Tuschl, T., and Zamore, P. D. (2001) A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293, 834–838. doi: 10.1126/science.1062961
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Huvenne, H., and Smagghe, G. (2010) Mechanisms of dsRNA uptake in insects and potential of RNAi for pest control: a review. J. Insect. Physiol. 56, 227–235. doi: 10.1016/j.jinsphys.2009.10.004
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Hwang, E. W., Shin, S. J., Yu, B. K., Byun, M. O., and Kwon, H. B. (2011) miR171 family members are involved in drought response in Solanum tuberosum. J. Plant Biol. 54, 43–48. doi: 10.1007/s12374-010-9141-8
Ibrahim, H. M., Alkharouf, N. W., Meyer, S. L., Aly, M. A., El-Din, A. K., Hussein, E. H., et al. (2011) Post-transcriptional gene silencing of root-knot nematode in transformed soybean roots. Exp. Parasitol. 127, 90–99. doi: 10.1016/j.exppara.2010.06.037
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Itaya, A., Bundschuh, R., Archual, A. J., Joung, J. G., Fei, Z., Dai, X., et al. (2008) Small RNAs in tomato fruit and leaf development. Biochim. Biophys. Acta 1779, 99–107. doi: 10.1016/j.bbagrm.2007.09.003
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Jagadeeswaran, G., Saini, A., and Sunkar, R. (2009) Biotic and abiotic stress down-regulate miR398 expression in Arabidopsis. Planta 229, 1009–1014. doi: 10.1007/s00425-009-0889-3
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Jian, X., Zhang, L., Li, G., Zhang, L., Wang, X., Cao, X., et al. (2010) Identification of novel stress-regulated microRNAs from Oryza sativa L. Genomics 95, 47–50. doi: 10.1016/j.ygeno.2009.08.017
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Jiang, C. J., Shimono, M., Maeda, S., Inoue, H., Mori, M., Hasegawa, M., et al. (2009) Suppression of the rice fatty-acid desaturase gene OsSSI2 enhances resistance to blast and leaf blight diseases in rice. Mol. Plant Microbe Int. 22, 820–829. doi: 10.1094/MPMI-22-7-0820
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Jiao, Y., Wang, Y., Xue, D., Wang, J., Yan, M., Liu, G., et al. (2010) Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 42, 541–544. doi: 10.1038/ng.591
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Jones-Rhoades, M. W., Bartel, D. P., and Bartel, B. (2006) MicroRNAs and their regulatory roles in plants. Annu. Rev. Plant Biol. 57, 19–53. doi: 10.1146/annurev.arplant.57.032905.105218
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kamachi, S., Mochizuki, A., Nishiguchi, M., and Tabei, Y. (2007) Transgenic Nicotiana benthamiana plants resistant to cucumber green mottle mosaic virus based on RNA silencing. Plant Cell Rep. 26, 1283–1288. doi: 10.1007/s00299-007-0358-z
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Karlova, R., van Haarst, J. C., Maliepaard, C., van de Geest, H., Bovy, A. G., Lammers, M., et al. (2013) Identification of microRNA targets in tomato fruit development using high-throughput sequencing and degradome analysis. J. Exp. Bot. 64, 1863–1878. doi: 10.1093/jxb/ert049
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Khraiwesh, B., Arif, M. A., Seumel, G. I., Ossowski, S., Weigel, D., Reski, R., et al. (2010) Transcriptional control of gene expression by microRNAs. Cell 140, 111–122. doi: 10.1016/j.cell.2009.12.023
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Khraiwesh, B., Zhu, J. K., and Zhu, J. (2012) Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochim. Biophys. Acta 1819, 137–148. doi: 10.1016/j.bbagrm.2011.05.001
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Krubphachaya, P., Jurícek, M., and Kertbundit, S. (2007) Induction of RNA mediated resistance to papaya ring spot virus type W. J. Biochem. Mol. Biol. 40, 401–411. doi: 10.5483/BMBRep.2007.40.3.404
Kusaba, M., Miyahara, K., Iida, S., Fukuoka, H., Takano, T., Sassa, H., et al. (2003) Low glutelin content 1: a dominant mutation that suppresses the glutelin multigene family via RNA silencing in rice. Plant Cell 15, 1455–1467. doi: 10.1105/tpc.011452
Law, J. A., and Jacobsen, S. E. (2010) Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 11, 204–220. doi: 10.1038/nrg2719
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Le, L., Lorenz, Y., Scheurer, S., Fotisch, K., Enrique, E., Bartra, J., et al. (2006) Design of tomato fruits with reduced allergenicity by dsRNAi-mediated inhibition of ns-LTP (Lyc e 3) expression. Plant Biotechnol. J. 4, 231–242. doi: 10.1111/j.1467-7652.2005.00175.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Lee, R. C., Feinbaum, R. L., and Ambros, V. (1993) The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843–854. doi: 10.1016/0092-8674(93)90529-Y
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Lewis, R. S., Jack, A. M., Morris, J. W., Robert, V. J., Gavilano, L. B., Siminszky, B., et al. (2008) RNA interference (RNAi)-induced suppression of nicotine demethylase activity reduces levels of a key carcinogen in cured tobacco leaves. Plant Biotechnol. J. 6, 346–354. doi: 10.1111/j.1467-7652.2008.00324.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Li, D. H., Liu, H., Yang, Y. l., Zhen, P. P., and Liang, J. S. (2009) Down-regulated expression of RACK1 gene by RNA interference enhances drought tolerance in rice. Rice Sci. 16, 14–20. doi: 10.1016/S1672-6308(08)60051-7
Li, K., Yuh-Shuh, W., Srinivasa Rao, U., Keri, W., Yuhong, T., Vatsala, V., et al. (2008a) Overexpression of a fatty acid amide hydrolase compromises innate immunity in Arabidopsis. Plant J. 56, 336–349. doi: 10.1111/j.1365-313X.2008.03603.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Li, W. X., Oono, Y., Zhu, J., He, X. J., Wu, J. M., Iida, K., et al. (2008b) The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and post transcriptionally to promote drought resistance. Plant Cell 20, 2238–2251. doi: 10.1105/tpc.108.059444
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Li, X., Wang, X., Zhang, S., Liu, D., Duan, Y., and Dong, W. (2012) Identification of soybean microRNAs involved in soybean cyst nematode infection by deep sequencing. PLoS ONE 7:e39650. doi: 10.1371/journal.pone.0039650
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Liu, H. H., Tian, X., Li, Y. J., Wu, C. A., and Zheng, C. C. (2008) Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. RNA 14, 836–843. doi: 10.1261/rna.895308
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Liu, J., He, Y., Amasino, R., and Chen, X. (2004) siRNA s targeting an intronic transposon in the regulation of natural flowering behavior in Arabidopsis. Genes Dev. 18, 2873–2878. doi: 10.1101/gad.1217304
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Liu, Q., Singh, P. S., and Green, A. G. (2002) High-stearic and high-oleic cotton seed oils produced by hairpin RNA-mediated post-transcriptional gene silencing. Plant Physiol. 129, 1732–1743. doi: 10.1104/pp.001933
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Llave, C., Kasschau, K. D., Rector, M. A., and Carrington, J. C. (2002a) Endogenous and silencing associated small RNAs in plants. Plant Cell 14, 1605–1619. doi: 10.1105/tpc.003210
Llave, C., Xie, Z., Kasschau, K. D., and Carrington, J. C. (2002b) Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297, 2053–2056. doi: 10.1126/science.1076311
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Lough, T. J., and Lucas, W. J. (2006) Integrative plant biology: role of phloem long-distance macromolecular trafficking. Annu. Rev. Plant Biol. 57, 203–232. doi: 10.1146/annurev.arplant.56.032604.144145
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Lv, D. K., Bai, X., Li, Y., Ding, X. D., Ge, Y., Cai, H., et al. (2010) Profiling of cold-stress-responsive miRNAs in rice by microarrays. Gene 459, 39–47. doi: 10.1016/j.gene.2010.03.011
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Mao, Y. B., Cai, W. J., Wang, J. W., Hong, G. J., Tao, X. Y., Wang, L. J., et al. (2007) Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi impairs larval tolerance of gossypol. Nat. Biotechnol. 25, 1307–1313. doi: 10.1038/nbt1352
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Matzke, M., Matzke, A., Pruss, G., and Vance, V. (2001) RNA-based silencing strategies in plants. Curr. Opin. Genet. Dev. 11, 221–227. doi: 10.1016/S0959-437X(00)00183-0
Meli, V. S., Ghosh, S., Prabha, T. N., Chakraborty, N., Chakraborty, S., and Datta, A. (2010) Enhancement of fruit shelf life by suppressing N-glycan processing enzymes. Proc. Natl. Acad. Sci. U.S.A. 107, 2413–2418. doi: 10.1073/pnas.0909329107
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Melnyk, C. W., Molnar, A., and Baulcombe, D. C. (2011) Intercellular and systemic movement of RNA silencing. EMBO J. 30, 3553–3563 doi: 10.1038/emboj.2011.274
Mette, M. F., van der Winden, J., Matzke, M., and Matzke, A. J. (2002) Short RNAs can identify new candidate transposable element families in Arabidopsis. Plant Physiol. 130, 6–9. doi: 10.1104/pp.007047
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Missiou, A., Kalantidis, K., Boutla, A., Tzortzakaki, S., Tabler, M., and Tsagris, M. (2004) Generation of transgenic potato plants highly resistant to potato virus Y (PVY) through RNA silencing. Mol. Breed. 14, 185–197. doi: 10.1023/B:MOLB.0000038006.32812.52
Mitsuda, N., and Ohme-Takagi, M. (2009) Functional analysis of transcription factors in Arabidopsis. Plant Cell Physiol. 50, 1232–1248. doi: 10.1093/pcp/pcp075
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Miura, K., Ikeda, M., Matsubara, A., Song, X. J., Ito, M., Asano, K., et al. (2010) OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat. Genet. 42, 545–549. doi: 10.1038/ng.592
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Molesini, B., Pandolfini, T., Rotino, G. L., Dani, V., and Spena, A. (2009) Aucsia gene silencing causes parthenocarpic fruit development in tomato. Plant Physiol. 149, 534–548. doi: 10.1104/pp.108.131367
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Molesini, B., Pii, Y., and Pandolfini, T. (2012) Fruit improvement using intragenesis and artificial microRNA. Trends Biotechnol. 30, 80–88. doi: 10.1016/j.tibtech.2011.07.005
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Molnar, A., Melnyk, C. W., Bassett, A., Hardcastle, T. J., Dunn, R., and Baulcombe, D. C. (2010) Small silencing RNAs in plants are mobile and direct epigenetic modification in recipient cells. Science 328, 872–875. doi: 10.1126/science.1187959
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Moxon, S., Jing, R., Szittya, G., Schwach, F., Pilcher, R. L. R., Moulton, V., et al. (2008) Deep sequencing of tomato short RNAs identifies microRNAs targeting genes involved in fruit ripening. Genome Res. 18, 1602–1609. doi: 10.1101/gr.080127.108
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Napoli, C., Lemieux, C., and Jorgensen, R. (1990) Introduction of chimeric chalcone synthase gene into Petunia results in reversible cosuppression of homologous genes in trans. Plant Cell 2, 279–289. doi: 10.1105/tpc.2.4.279
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Navarro, L., Dunoyer, P., Jay, F., Arnold, B., Dharmasiri, N., Estelle, M., et al. (2006) A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312, 436–439. doi: 10.1126/science.1126088
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ni, Z., Hu, Z., Jiang, Q., and Zhang, H. (2013) GmNFYA3, a target gene of miR169, is a positive regulator of plant tolerance to drought stress. Plant Mol. Biol. 82, 113–129. doi: 10.1007/s11103-013-0040-5
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Niu, Q. W., Lin, S. S., Reyes, J. L., Chen, K. C., Wu, H. W., Yeh, S. D., et al. (2006) Expression of artificial microRNAs in transgenic Arabidopsis thaliana confers virus resistance. Nat. Biotechnol. 24, 1420–1428. doi: 10.1038/nbt1255
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Nora, S., MichÈLe, Z., Asuka, I., Biao, D., Ming-Bo, W., Gabi, K., et al. (2009) RNAi-mediated resistance to Potato spindle tuber viroid in transgenic tomato expressing a viroid hairpin RNA construct. Mol. Plant Pathol. 10, 459–469. doi: 10.1111/j.1364-3703.2009.00546.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Oeller, P. W., Lu, M. W., Taylor, L. P., Pike, D. A., and Theologis, A. (1991) Reversible inhibition of tomato fruit senescence by antisense RNA. Science 254, 437–439. doi: 10.1126/science.1925603
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Osorio, S., Alba, R., Damasceno, C. M., Lopez-Casado, G., Lohse, M., Zanor, M. I., et al. (2011) Systems biology of tomato fruit development: combined transcript, protein, and metabolite analysis of tomato transcription factor (nor, rin) and ethylene receptor (Nr) mutants reveals novel regulatory interactions. Plant Physiol. 157, 405–425. doi: 10.1104/pp.111.175463
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ossowski, S., Schwab, R., and Weigel, D. (2008) Gene silencing in plants using artificial microRNAs and other small RNAs. Plant J. 53, 674–690. doi: 10.1111/j.1365-313X.2007.03328.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Palauqui, J.-C., Elmayan, T., Pollien, J.-M., and Vaucheret, H. (1997) Systemic acquired silencing: transgene-specific post-transcriptional silencing is transmitted by grafting from silenced stocks to non-silenced scions. EMBO J. 16, 4738–4745. doi: 10.1093/emboj/16.15.4738
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Pandolfini, T., Molesini, B., Avesani, L., Spena, A., and Polverari, A. (2003) Expression of self-complementary hairpin RNA under the control of the rolC promoter confers systemic disease resistance to plum pox virus without preventing local infection. BMC Biotechnol. 3:7. doi: 10.1186/1472-6750-3-7
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Pant, B. D., Buhtz, A., Kehr, J., and Scheible, W. R. (2008) MicroRNA399 is a long-distance signal for the regulation of plant phosphate homeostasis. Plant J. 53, 731–738. doi: 10.1111/j.1365-313X.2007.03363.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Pare, J. M., and Hobman, T. C. (2007) “Dicer: structure, function and role in RNA-dependent gene-silencing pathways,” in Industrial Enzymes, eds J. Polaina and A. P. MacCabe (Berlin: Springer), 421–438.
Park, G. G., Park, J. J., Yoon, J., Yu, S. N., and An, G. (2010) A Ring finger E3 ligase gene, Oryza sativa delayed seed germination 1 (OsDSG1), controls seed germination and stress responses in rice. Plant Mol. Biol. 74, 467–478. doi: 10.1007/s11103-010-9687-3
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Park, W., Li, J., Song, R., Messing, J., and Chen, X. (2002) Carpel factory, a dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol. 12, 1484–1495. doi: 10.1016/S0960-9822(02)01017-5
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Peltier, A. J., Hatfield, R. D., and Grau, C. R. (2009) Soybean stem lignin concentration relates to resistance to Sclerotinia sclerotiorum. Plant Dis. 93, 149–154. doi: 10.1094/PDIS-93-2-0149
Perl, A., Perl-Treves, R., Galili, S., Aviv, D., Shalgi, E., Malkin, S., et al. (1993) Enhanced oxidative-stress defense in transgenic potato expressing tomato Cu, Zn superoxide dismutases. Theor. Appl. Genet. 85, 568–576. doi: 10.1007/BF00220915
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Pilcher, R. L., Moxon, S., Pakseresht, N., Moulton, V., Manning, K., Seymour, G., et al. (2007) Identification of novel small RNAs in tomato (Solanum lycopersicum). Planta 226, 709–717. doi: 10.1007/s00425-007-0518-y
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Pitcher, L. H., Brennan, E., Hurley, A., Dunsmuir, P., Tepperman, J. M., and Zilinskas, B. A. (1991) Overproduction of petunia chloroplastic copper/zinc superoxide dismutase does not confer ozone tolerance in transgenic tobacco. Plant Physiol. 97, 452–455. doi: 10.1104/pp.97.1.452
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Pontier, D., Picart, C., Roudier, F., Garcia, D., Lahmy, S., Azevedo, J., et al. (2012) NERD, a plant-specific GW protein, defines an additional RNAi-dependent chromatin based pathway in Arabidopsis. Mol. Cell 48, 121–132. doi: 10.1016/j.molcel.2012.07.027
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Pooggin, M., Shivaprasad, P. V., Veluthambi, K., and Hohn, T. (2003) RNAi targeting of DNA virus in plants. Nat. Biotechnol. 21, 131–132. doi: 10.1038/nbt0203-131b
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Powell-Abel, P., Nelson, R. S., De, B., Hoffmann, N., Rogers, S. G., Fraley, R. T., et al. (1986) Delay of disease development in transgenic plants that express the tobacco mosaic virus coat protein gene. Science 232, 738–743. doi: 10.1126/science.3457472
Price, D. R. G., and Gatehouse, J. A. (2008) RNAi-mediated crop protection against insects. Trends Biotechnol. 26, 393–400. doi: 10.1016/j.tibtech.2008.04.004
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Qiao, F., Yang, Q., Wang, C. L., Fan, Y. L., Wu, X. F., and Zhao, K. J. (2007) Modification of plant height via RNAi suppression of OsGA20ox2 gene in rice. Euphytica 158, 35–45. doi: 10.1007/s10681-007-9422-6
Qu, J., Ye, J., and Fang, R. (2007) Artificial microRNA-mediated virus resistance in plants. J. Virol. 81, 6690–6699. doi: 10.1128/JVI.02457-06
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Rajam, M. V., Madhulatha, P., Pandey, R., Hazarika, P. J., and Razdan, M. K. (2007) “Applications of genetic engineering in tomato,” in Genetic Improvement of Solanaceae Crops: Tomato, Vol. 2, eds M. K. Razdan and A. K. Mattoo (Enfield, NH: Science Publishers), 285–311.
Rathore, K. S., Sundaram, S., Sunilkumar, G., Campbell, L. M., Puckhaber, L., Marcel, S., et al. (2012) Ultra-low gossypol cottonseed: generational stability of the seed-specific, RNAi-mediated phenotype and resumption of terpenoid profile following seed germination. Plant Biotechnol. J. 10, 174–183. doi: 10.1111/j.1467-7652.2011.00652.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Reddy, M. S. S., Chen, F., Shadle, G., Jackson, L., Aljoe, H., and Dixon, R. A. (2005) Targeted down-regulation of cytochrome P450 enzymes for forage quality improvement in alfalfa (Medicago sativa L.). Proc. Natl. Acad. Sci. U.S.A. 102, 16573–16578. doi: 10.1073/pnas.0505749102
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Redfern, A. D., Colley, S. M., Beveridge, D. J., Ikeda, N., Epis, M. R., Li, X., et al. (2013) RNA-induced silencing complex (RISC) proteins PACT, TRBP, and Dicer are SRA binding nuclear receptor coregulators. Proc. Natl. Acad. Sci. U.S.A. 110, 6536–6541. doi: 10.1073/pnas.1301620110
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Regina, A., Bird, A., Topping, D., Bowden, S., Freeman, J., Barsby, T., et al. (2006) High amylose wheat generated by RNA interference improves indices of large-bowel health in rats. Proc. Natl. Acad. Sci. U.S.A. 103, 3546–3551. doi: 10.1073/pnas.0510737103
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Reinhart, B. J., Weinstein, E. G., Rhoades, M. W., Bartel, B., and Bartel, D. P. (2002) MicroRNAs in plants. Genes Dev. 16, 1616–1626. doi: 10.1101/gad.1004402
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Reyes, J. L., and Chua, N. H. (2007) ABA induction of miR159 controls transcript levels of two MYB factors during Arabidopsis seed germination. Plant J. 49, 592–606. doi: 10.1111/j.1365-313X.2006.02980.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Rhoades, M. W., Reinhart, B. J., Lim, L. P., Burge, C. B., Bartel, B., and Bartel, D. P. (2002) Prediction of plant microRNA targets. Cell 110, 513–520. doi: 10.1016/S0092-8674(02)00863-2
Riechen, J. (2007) Establishment of broad-spectrum resistance against Blumeria graminis f. sp. tritici in Triticum aestivum by RNAi-mediated knock-down of MLO. J. Verbrauch. Lebensm. 2:120. doi: 10.1007/s00003-007-0282-8
Riley, K. J., Yario, T. A., and Steitz, J. A. (2012) Association of argonaute proteins and microRNAs can occur after cell lysis. RNA 18, 1581–1585. doi: 10.1261/rna.034934.112
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Romano, N., and Macino, G. (1992) Quelling: transient inactivation of gene expression in Neurospora crassa by transformation with homologous sequences. Mol. Microbiol. 6, 3343–3353. doi: 10.1111/j.1365-2958.1992.tb02202.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ru, P., Xu, L., Ma, H., and Huang, H. (2006) Plant fertility defects induced by the enhanced expression of microRNA167. Cell Res. 16, 457–465. doi: 10.1038/sj.cr.7310057
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Rubinelli, P. M., Chuck, G., Li, X., and Meilan, R. (2013) Constitutive expression of the Corn-grass1 microRNA in poplar affects plant architecture and stem lignin content and composition. Biomass Bioenerg. 54, 312–321. doi: 10.1016/j.biombioe.2012.03.001
Schijlen, E. G. W. M., de Vos, R. C. H., Martens, S., Jonker, H. H., Rosin, F. M., Molthoff, J. W., et al. (2007) RNA Interference silencing of chalcone synthase, the first step in the flavonoid biosynthesis pathway, leads to parthenocarpic tomato fruits. Plant Physiol. 144, 1520–1530. doi: 10.1104/pp.107.100305
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Schwab, R., Ossowski, S., Riester, M., Warthmann, M., and Weigel, D. (2006) Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18, 1121–1133. doi: 10.1105/tpc.105.039834
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Schwab, R., Palatnik, J. F., Riester, M., Schommer, C., Schmid, M., and Weigel, D. (2005) Specific effects of microRNAs on the plant transcriptome. Dev. Cell 8, 517–527. doi: 10.1016/j.devcel.2005.01.018
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Schwach, F., Vaistij, F. E., Jones, L., and Baulcombe, D. C. (2005) An RNA dependent RNA-polymerase prevents meristem invasion by Potato virus X and is required for the activity but not the production of a systemic silencing signal. Plant Physiol. 138, 1842–1852. doi: 10.1104/pp.105.063537
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Segal, G., Song, R., and Messing, J. (2003) A new opaque variant of maize by a single dominant RNA-interference inducing transgene. Genetics 165, 387–397.
Shimizu, T., Yoshii, M., Wei, T., Hirochika, H., and Omura, T. (2009) Silencing by RNAi of the gene for Pns12, a viroplasm matrix protein of Rice dwarf virus, results in strong resistance of transgenic rice plants to the virus. Plant Biotechnol. J. 7, 24–32. doi: 10.1111/j.1467-7652.2008.00366.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Shivaprasad, P. V., Chen, H. M., Patel, K., Bond, D. M., Santos, B. A., and Baulcombe, D. C. (2012) A microRNA superfamily regulates nucleotide binding site leucine rich repeats and other mRNAs. Plant Cell 24, 859–874. doi: 10.1105/tpc.111.095380
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Simon-Mateo, C., and García, J. A. (2011) Antiviral strategies in plants based on RNA silencing. Biochim. Biophys. Acta 1809, 722–731. doi: 10.1016/j.bbagrm.2011.05.011
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Sindhu, A. S., Maier, T. R., Mitchum, M. G., Hussey, R. S., Davis, E. L., and Baum, T. J. (2009) Effective and specific in planta RNAi in cyst nematodes: expression interference of four parasitism genes reduces parasitic success. J. Exp. Bot. 60, 315–324. doi: 10.1093/jxb/ern289
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Singh, A., Taneja, J., Dasgupta, I., and Mukherjee, S. K. (2014) Development of plants resistant to tomato geminiviruses using artificial trans-acting small interfering RNA. Mol. Plant Patholol. doi: 10.1111/mpp.12229 [Epub ahead of print].
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Slotkin, R. K., Vaughn, M., Borges, F., Tanurdzic, M., Becker, J. D., Feijo, J. A., et al. (2009) Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell 136, 461–472. doi: 10.1016/j.cell.2008.12.038
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Smith, N. A., Singh, S. P., Wang, M. B., Stoutjesdijk, P. A., Green, A. G., and Waterhouse, P. M. (2000) Total silencing by intron-spliced hairpin RNAs. Nature 407, 319–320. doi: 10.1038/35036500
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Springer, N. (2010) Shaping a better rice plant. Nat. Genet. 42:475. doi: 10.1038/ng0610-475
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Sun, L., Yuan, B., Zhang, M., Wang, L., Cui, M., Wang, Q., et al. (2012) Fruit-specific RNAi mediated suppression of SlNCED1 increases both lycopene and E-carotene contents in tomato fruit. J. Exp. Bot. 63, 3097–3108. doi: 10.1093/jxb/ers026
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Sunilkumar, G., Campbell, L. M., Puckhaber, L., Stipanovic, R. D., and Rathore, K. S. (2006) Engineering cottonseed for use in human nutrition by tissue-specific reduction of toxic gossypol. Proc. Natl. Acad. Sci. U.S.A. 103, 18054–18059. doi: 10.1073/pnas.0605389103
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Sunkar, R., Kapoor, A., and Zhu, J. K. (2006) Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell 18, 2051–2065. doi: 10.1105/tpc.106.041673
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Sunkar, R., and Zhu, J. K. (2004) Novel and stress-regulated micro RNAs and other small RNAs from Arabidopsis. Plant Cell 16, 2001–2019. doi: 10.1105/tpc.104.022830
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Tang, G., Reinhart, B. J., Bartel, D. P., and Zamore, P. D. (2003) A biochemical framework for RNA silencing in plants. Genes Dev. 17, 49–63. doi: 10.1101/gad.1048103
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Teo, G., Suziki, Y., Uratsu, S. L., Lampien, B., Ormonde, N., Hu, W. K., et al. (2006) Silencing leaf sorbitol synthesis alters long-distance portioning and apple fruit quality. Proc. Natl. Acad. Sci. U.S.A. 103, 18842–18847. doi: 10.1073/pnas.0605873103
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Tepperman, J. M., and Dunsmuir, P. (1990) Transformed plants with elevated levels of chloroplastic SOD are not more resistant to superoxide toxicity. Plant Mol. Biol. 14, 501–511. doi: 10.1007/BF00027496
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Theologis, A., Oeller, P. W., Wong, L. M., Rottmann, W. H., and Gantz, D. M. (1993) Use of a tomato mutant constructed with reverse genetics to study fruit ripening, a complex developmental process. Dev. Gen. 14, 282–295. doi: 10.1002/dvg.1020140406
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Thiebaut, F., Rojas, C. A., Almeida, K. L., Grativol, C., Domiciano, G. C., Lamb, C. R., et al. (2011) Regulation of miR319 during cold stress in sugarcane. Plant Cell Environ. 35, 502–512. doi: 10.1111/j.1365-3040.2011.02430.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Trindade, I., CapitCapitão, C., Dalmay, T., Fevereiro, M. P., and Santos, D. M. (2010) miR398 and miR408 are up-regulated in response to water deficit in Medicago truncatula. Planta 231, 705–716. doi: 10.1007/s00425-009-1078-0
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Tuschl, T. (2001) RNA interference and small interfering RNAs. Chembiochem 2, 239–145. doi: 10.1002/1439-7633(20010401)2:4<239::AID-CBIC239>3.0.CO;2-R
Vanderschuren, H., Alder, A., Zhang, P., and Gruissem, W. (2009) Dose-dependent RNAi-mediated geminivirus resistance in the tropical root crop cassava. Plant Mol. Biol. 70, 265–272. doi: 10.1007/s11103-009-9472-3
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Vaucheret, H. (2008) Plant ARGONAUTES. Trends Plant Sci. 13, 350–358. doi: 10.1016/j.tplants.2008.04.007
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Vazquez, F. (2006) Arabidopsis endogenous small RNAs: highways and byways. Trends Plant Sci. 11, 460–468. doi: 10.1016/j.tplants.2006.07.006
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Voinnet, O., Vain, P., Angell, S., and Baulcombe, D. C. (1998) Systemic spread of sequence-specific transgene RNA degradation is initiated by localised introduction of ectopic promoterless DNA. Cell 95, 177–187. doi: 10.1016/S0092-8674(00)81749-3
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Vu, T. V., Choudhury, N. R., and Mukherjee, S. K. (2013) Transgenic tomato plants expressing artificial microRNAs for silencing the pre-coat and coat proteins of a begomovirus, tomato leaf curl New Delhi virus, show tolerance to virus infection. Virus Res. 172, 35–45. doi: 10.1016/j.virusres.2012.12.008
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Wang, M., Abbott, D. C., and Waterhouse, P. M. (2000) A single copy of a virus-derived transgene encoding hairpin RNA gives immunity to barley yellow dwarf virus. Mol. Plant Pathol. 1, 347–356. doi: 10.1046/j.1364-3703.2000.00038.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Wang, S., Wu, K., Yuan, Q., Liu, X., Liu, Z., Lin, X., et al. (2012) Control of grain size, shape and quality by OsSPL16 in rice. Nat. Genet. 44, 950–954. doi: 10.1038/ng.2327
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Wang, T., Chen, L., Zhao, M., Tian, Q., and Zhang, W. H. (2011) Identification of drought-responsive microRNAs in Medicago truncatula by genome-wide high-throughput sequencing. BMC Genomics 12:367–378. doi: 10.1186/1471-2164-12-367
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Wang, Y., Beaith, M., Chalifoux, M., Ying, J., Uchacz, T., Sarvas, C., et al. (2009) Shoot-specific down-regulation of protein farnesyltransferase (a-subunit) for yield protection against drought in canola. Mol. Plant 2, 191–200. doi: 10.1093/mp/ssn088
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Wang, Y., Ying, J., Kuzma, M., Chalifoux, M., Sample, A., McArthur, C., et al. (2005) Molecular tailoring of farnesylation for plant drought tolerance and yield protection. Plant J. 43, 413–424. doi: 10.1111/j.1365-313X.2005.02463.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Weise, S. E., Aung, K., Jarou, Z. J., Mehrshahi, P., Li, Z., Hardy, A. C., et al. (2012) Engineering starch accumulation by manipulation of phosphate metabolism of starch. Plant Biotechnol. J. 10, 545–554. doi: 10.1111/j.1467-7652.2012.00684.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Wesley, S. V., Helliwell, C. A., Smith, N. A., Wang, M. B., Rouse, D. T., Liu, Q., et al. (2001) Construct design for efficient, effective and high throughput gene silencing in plants. Plant J. 27, 581–590. doi: 10.1046/j.1365-313X.2001.01105.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Wilkinson, J. Q., Lanahan, M. B., Clark, D. G., Bleecker, A. B., Chang, C., Meyerowitz, E. M., et al. (1997) A dominant mutant receptor from Arabidopsis confers ethylene insensitivity in heterologous plants. Nat. Biotechnol. 15, 444–447. doi: 10.1038/nbt0597-444
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Wilson, R. C., and Doudna, J. A. (2013) Molecular mechanisms of RNA interference. Annu. Rev. Biophys. 42, 217–239. doi: 10.1146/annurev-biophys-083012-130404
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Wu, M. F., Tian, Q., and Reed, J. W. (2006) Arabidopsis microRNA167 controls patterns of ARF6 and ARF8 expression, and regulates both female and male reproduction. Development 133, 4211–4218. doi: 10.1242/dev.02602
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Xie, K., Shen, J., Hou, X., Yao, J., Li, X., Xiao, J., et al. (2012) Gradual increase of miR156 regulates temporal expression changes of numerous genes during leaf development in rice. Plant Physiol. 158, 1382–1394. doi: 10.1104/pp.111.190488
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Xie, K., Wu, C., and Xiong, L. (2006) Genomic organization, differential expression, and interaction of Squamosa promoter-binding- like transcription factors and microRNA156 in rice. Plant Physiol. 142, 280–293. doi: 10.1104/pp.106.084475
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Xie, Z., Allen, E., Wilken, A., and Carrington, J. C. (2005) DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 102, 12984–12989. doi: 10.1073/pnas.0506426102
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Xie, Z., Kasschau, K. D., and Carrington, J. C. (2003) Negative feedback regulation of Dicer-Like1 in Arabidopsis by microRNA-guided mRNA degradation. Curr. Biol. 13, 784–789. doi: 10.1016/S0960-9822(03)00281-1
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Xin, M., Wang, Y., Yao, Y., Xie, C., Peng, H., Ni, Z., et al. (2010) Diverse set of microRNAs are responsive to powdery mildew infection and heat stress in wheat (Triticum aestivum L.). BMC Plant Biol. 10:123–134. doi: 10.1186/1471-2229-10-123
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Xiong, A. S., Yao, Q. H., Li, X., Fan, H. Q., and Peng, R. H. (2003) Double antisense ACC oxidase and ACC synthase fusion gene introduced into tomato by Agrobacterium-mediated transformation and analysis the ethylene production of transgenic plants. Acta Biol. Exp. Sin. 36, 35–41.
Xiong, A. S., Yao, Q. H., Peng, R. H., Li, X., Han, P. L., and Fan, H. Q. (2005) Different effects on ACC oxidase genesilencing triggered by RNA interference in transgenic tomato. Plant Cell Rep. 23, 639–646. doi: 10.1007/s00299-004-0887-7
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Yadav, B. C., Veluthambi, K., and Subramaniam, K. (2006) Host-generated double stranded RNA induces RNAi in plant-parasitic nematodes and protects the host from infection. Mol. Biochem. Parasitol. 148, 219–222. doi: 10.1016/j.molbiopara.2006.03.013
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Yang, C., Li, D., Mao, D., Liu, X., Li, C., Li, X., et al. (2013) Overexpression of microRNA319 impacts leaf morphogenesis and leads to enhanced cold tolerance in rice (Oryza sativa L.). Plant Cell Environ. 36, 2207–2218. doi: 10.1111/pce.12130
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Yara, A., Yaeno, T., Hasegawa, M., Seto, H., Montillet, J. L., Kusumi, K., et al. (2007) Disease resistance against Magnaporthe grisea is enhanced in transgenic rice with suppression of o-3 fatty acid desaturases. Plant Cell Physiol. 48, 1263–1274. doi: 10.1093/pcp/pcm107
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ye, Z. B., Li, H. X., Zheng, Y. L., and Liu, H. L. (1996) Inhibition of introducing antisense ACC oxidase gene into tomato genome on expression of its endogeous gene. J. Huazhong Agric. Univ. 15, 305–309.
Yoo, B.-C., Kragler, F., Varkonyi-Gasic, E., Haywood, V., Archer-Evans, S., Lee, Y. M., et al. (2004) A systemic small RNA signaling system in plants. Plant Cell 16, 1979–2000. doi: 10.1105/tpc.104.023614
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Yu, B., Lydiate, D. J., Young, L. W., Schafer, U. A., and Hannoufa, A. (2007) Enhancing the carotenoid content of Brassica napus seeds by downregulating lycopene epsilon cyclase. Transgenic Res. 17, 573–585. doi: 10.1007/s11248-007-9131-x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zamore, P. D., and Haley, B. (2005) Ribo-genome: the big world of small RNAs. Science 309, 1519–1524. doi: 10.1126/science.1111444
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zamore, P. D., Tuschl, T., Sharp, P. A., and Bartel, D. P. (2000) RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 101, 25–33. doi: 10.1016/S0092-8674(00)80620-0
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zhai, J., Jeong, D. H., De Paoli, E., Park, S., Rosen, B. D., Li, Y., et al. (2011) MicroRNAs as master regulators of the plant NBLRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev. 25, 2540–2553. doi: 10.1101/gad.177527.111
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zhang, X., Zou, Z., Gong, P., Zhang, J., Ziaf, K., Li, H., et al. (2011a) Over-expression of microRNA169 confers enhanced drought tolerance to tomato. Biotechnol. Lett. 33, 403–409. doi: 10.1007/s10529-010-0436-0
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zhang, X., Zou, Z., Zhang, J., Zhang, Y., Han, Q., Hu, T., et al. (2011b) Over-expression of sly-miR156a in tomato results in multiple vegetative and reproductive trait alterations and partial phenocopy of the sft mutant. FEBS Lett. 585, 435–439. doi: 10.1016/j.febslet.2010.12.036
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zhang, Y. C., Yu, Y., Wang, C. Y., Li, Z. Y., Liu, Q., Xu, J., et al. (2013) Overexpression of microRNA OsmiR397 improves rice yield by increasing grain size and promoting panicle branching. Nat. Biotechnol. 31, 848–852. doi: 10.1038/nbt.2646
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zhao, B. T., Liang, R. Q., Ge, L. F., Li, W., Xiao, H. S., Lin, H. X., et al. (2007) Identification of drought-induced microRNAs in rice. Biochem. Biophy. Res. Comm. 354, 585–590. doi: 10.1016/j.bbrc.2007.01.022
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zhou, M., Li, D., Li, Z., Hu, Q., Yang, C., Zhu, L., et al. (2013) Constitutive expression of a miR319 gene alters plant development and enhances salt and drought tolerance in transgenic creeping bentgrass. Plant Physiol. 161, 1375–1391. doi: 10.1104/pp.112.208702
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zhou, Y., Yuan, Y., Yuan, F., Wang, M., Zhong, H., Gu, M., et al. (2012) RNAi-directed down-regulation of RSV results in increased resistance in rice (Oryza sativa L.). Biotechnol. Lett. 34, 965–978. doi: 10.1007/s10529-012-0848-0
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zilberman, D., Cao, X., and Jacobsen, S. E. (2003) ARGONAUTE4 control of locus specific siRNA accumulation and DNA and histone methylation. Science 299, 716–719. doi: 10.1126/science.1079695
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zrachya, A., Glick, E., Levy, Y., Arazi, T., Citovsky, V., and Gafni, Y. (2007) Suppressor of RNA silencing encoded by tomato yellow curl virus-Israel. Virology 358, 159–165. doi: 10.1016/j.virol.2006.08.016
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Keywords: miRNA, siRNA, RNAi, gene silencing, crop improvement
Citation: Kamthan A, Chaudhuri A, Kamthan M and Datta A (2015) Small RNAs in plants: recent development and application for crop improvement. Front. Plant Sci. 6:208. doi: 10.3389/fpls.2015.00208
Received: 12 December 2014; Accepted: 16 March 2015;
Published online: 02 April 2015
Edited by:
Thomas Debener, Leibniz University of Hannover, GermanyReviewed by:
Shogo Matsumoto, Nagoya University, JapanSunil Kumar Mukherjee, Biotech Consortium India Limited, India
Copyright © 2015 Kamthan, Chaudhuri, Kamthan and Datta. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Asis Datta, National Institute of Plant Genome Research, Lab No 105 NIPGR, Jawaharlal Nehru University Campus, Aruna Asaf Ali Marg, New Delhi-110067, Delhi, IndiaYXNpc19kYXR0YUByZWRpZmZtYWlsLmNvbQ==
 Ayushi Kamthan
Ayushi Kamthan Abira Chaudhuri
Abira Chaudhuri Mohan Kamthan
Mohan Kamthan Asis Datta
Asis Datta