- 1 National Engineering Technology Research Center of Oil-tea Camellia, Hunan Academy of Forestry, Changsha, China
- 2 Environmental Horticulture Department and Mid-Florida Research and Education Center, University of Florida, Apopka, FL, USA
Tea oil derived from seeds of Camellia oleifera Abel. is high-quality edible oil in China. This study isolated full-length cDNAs of Rubisco subunits rbcL and rbcS from C. oleifera. The rbcL has 1,522 bp with a 1,425 bp coding region, encoding 475 amino acids; and the rbcS has 615 bp containing a 528 bp coding region, encoding 176 amino acids. The expression level of the two genes, designated as Co-rbcL and Co-rbcS, was determined in three C. oleifera cultivars: Hengchong 89, Xianglin 1, and Xianglin 14 whose annual oil yields were 546.9, 591.4, and 657.7 kg ha-1, respectively. The Co-rbcL expression in ‘Xianglin 14’ was significantly higher than ‘Xianglin 1’, and ‘Xianglin 1’ was greater than ‘Hengchong 89’. The expression levels of Co-rbcS in ‘Xianglin 1’ and ‘Xianglin 14’ were similar but were significantly greater than in ‘Hengchong 89’. The net photosynthetic rate of ‘Xianglin 14’ was significantly higher than ‘Xianglin 1’, and ‘Xianglin 1’ was higher than ‘Hengchong 89’. Pearson’s correlation analysis showed that seed yields and oil yields were highly correlated with the expression level of Co-rbcL at P < 0.001 level; and the expression of Co-rbcS was correlated with oil yield at P < 0.01 level. Net photosynthetic rate was also correlated with oil yields and seed yields at P < 0.001 and P < 0.01 levels, respectively. Our results suggest that Co-rbcS and Co-rbcL in particular could potentially be molecular markers for early selection of high oil yield cultivars. In combination with the measurement of net photosynthetic rates, the early identification of potential high oil production cultivars would significantly shorten plant breeding time and increase breeding efficiency.
Introduction
Camellia oleifera Abel. is a shrub or small tree native to China (Mondal, 2011). It has been cultivated in south-central and southern China for more than 2,000 years primarily for edible oil extracted from seeds (Wei et al., 2012; Figure 1). Commonly known as tea oil or camellia oil, it is comprised of 67.7–76.7% oleic acid, 82–84% unsaturated fatty acids, 68–77% monounsaturated fatty acids, and 7–14% polyunsaturated acid, which is similar to the composition of olive oil (Ma et al., 2011). Additionally, tea oil has been used for a wide range of cosmetic and medicinal purposes (Lee and Yen, 2006; Cheng et al., 2014). There has been increasing demand for producing camellia oil in China (Wei et al., 2012; Liao et al., 2014).

FIGURE 1. Camellia oleifera “Xianglin 14’ produced abundant fruit. Fruit diameters were up to 4 cm (A); seed appearance at seed shell opening (B); a mix of seeds, nutlets, and fruit (C); and tea oil derived from seeds (D).
The production of C. oleifera increased to more than 3.7 million hectares in China (Kong et al., 2013), however, the increased production area still did not meet the demand for tea oil. This is largely due to conventional cultivars having low oil yield ranging from 105 to 225 kg ha-1 (Zhuang, 2008). To improve tea oil yield, the National Engineering and Technology Research Center of Camellia Oil at the Hunan Academy of Forestry in Changsha, Hunan, China initiated a breeding program focusing on germplasm evaluation and new cultivar development of C. oleifera. To date, more than 20 new cultivars have been developed. Tea oil yield of the new cultivars ranged from 450 to 750 kg ha-1 (Zhuang, 2008).
Camellia oleifera is self-sterile, which is caused by prezygotic late-acting self-incompatibility (Liao et al., 2014). Thus, all cultivars are genetically highly heterozygous. Methods currently used for C. oleifera breeding include conventional hybridization and selection of individuals from progenies with high oil content. Because C. oleifera will neither flower and nor bear fruit in the first 4–5 years, full bloom and seed production occurs in 5–7 years. As a result, breeding of C. oleifera is a long and time-consuming process, requiring 7–10 years to develop a new cultivar. After multiple regional trials, new cultivars are propagated vegetatively through grafting or rooting of cuttings. The propagated plants require additional 2–4 years to reach the fruiting stage; therefore, there is a critical need for improving breeding efficiency of C. oleifera.
Marker-assisted selection is an indirect selection method in which a trait of interest is selected based not on the trait itself, but on a marker related to it (Collard and Mackill, 2008). The assumption is that the marker used for trait selection associates at high frequency with the genes of interest. Due to the genetic linkage, marker-assisted selection is particularly useful for traits that are expressed late in development. Recently, a C. oleifera EST library was constructed (Tan et al., 2006), and microsatellite markers for the genus Camellia were also reported (Qiang et al., 2013). However, there are still no morphological, cytological, or molecular markers related to oil yield in C. oleifera, and no any markers have been used for its breeding.
Ribulose 1, 5-bisphosphate carboxylase/oxygenase (EC 4.1. 1.39, Rubisco) should be an important biochemical marker as it can comprise up to 50% of the total soluble protein in plant leaves (Ellis, 1979). Rubisco is the key regulatory enzyme catalyzing CO2 fixation and ribulose diphosphate oxygenase reaction dual function, which determines net photosynthesis (Andersson and Backlund, 2008). The holoenzyme of Rubisco is composed of eight small subunits (SSU, 14–15-kDa) encoded by a nuclear multigene family (rbcS), and eight large subunits (LSUs, 52-kDa) encoded by a single gene (rbcL) in the multicopy chloroplast genome. SSU precursors are processed during entry into the chloroplast and then assembled with the LSUs to yield the Rubisco holoenzyme (Ichikawa et al., 2008). The rbcL gene is highly conserved, while rbcS is less conserved (Manzara and Gruissem, 1988). Recent molecular phylogenetic analysis, however, revealed that positive selection in the rbcL gene of terrestrial land plants is quite a common phenomenon (Kapralov and Filatov, 2007; Galmes et al., 2014). Since plant breeding is considered a man-made evolution (Jauhar, 2006; Ghai, 2009), we assumed that long-term selection of C. oleifera cultivars for high oil yield could lead to enrichment in the abundance of transcript of Rubisco genes for enhancing photosynthetic efficiency and increasing oil biosynthesis. This is based on the notion that the reducing power, ATP, and a range of metabolites derived from photosynthesis support high rates of fatty acid synthesis (Rawsthorne, 2002; Bates et al., 2013). Thus, Rubisco subunit genes could be molecular markers for improving breeding efficiency. Rubisco rbcL and rbcS genes, however, have not been isolated from C. oleifera. Furthermore, since Rubisco activity is directly related to photosynthesis, it is possible that net photosynthetic rate could be a physiological marker for detecting seed yield and increased oil production.
The objectives of this study were to (1) clone and identify rbcL and rbcS genes from C. oleifera; (2) analyze rbcL and rbcS expression in three cultivars that differed in seed and oil yields; (3) examine net photosynthetic rates of the three cultivars; and (4) determine if there were any relationships among rbcL and rbcS gene expressions, net photosynthetic rates, and oil yield.
Materials and Methods
Plant Materials and Growth Conditions
Three cultivars Hengchong 89, Xianglin 1, and Xianglin 14 were planted in 2006 in field plots of the Experimental Station of the National Engineering and Technology Research Center of Oil-Tea Camellia, Hunan Academy of Forestry, Changsha, Hunan, China, which is located between 111° 53′–114° 05′ E longitude and 27° 51′–28° 40′ N latitude. Annual average temperature is 17.03°C, with a mean of 4.6°C in January and 29.0°C in July. Average annual precipitation is 1,331 mm, and a maximum photosynthetic photon flux density of 2,000 mmol m-2 s-1. Hengchong 89 is a popular cultivar in Hunan and other south-central provinces, and Xianglin 1 and Xianglin 14 were developed by the Center of Oil-Tea Camellia. Hengchong 89, a well-established cultivar, was a control. Xianglin 1 and Xianglin 14, due to their adaptation to the climate of south-central China and drought resistance, have been increasingly grown in this region. Three plants of each cultivar were randomly planted in experimental plots that were 54 m2 each, and plants were spaced at 2.0 m × 3.0 m. There were three plots, and each plot represented a block. Plants produced fruit in 2010. Starting from 2011, fruits were harvested and weighed annually.
Extraction of DNA and RNA and Synthesis of first Strand cDNA
The second leaf from the apical meristem was harvested, frozen in liquid nitrogen, and stored in a freezer (-80°C). The total DNA was extracted from the leaves using plant genomic DNA extraction kit (Tiangen Biotech Co., Beijing, China) for isolation of rbcL.
Two sets of total RNA were isolated: one from three cultivars Hengchong 89, Xianglin 1, and Xianglin 14 and the other from Xianglin 14. Frozen tissues were ground in liquid nitrogen and lysed with 600 μl 3x CTAB plus 1% 2-mercaptoethanol, followed by E.Z.N.A.TM Plant RNA Kit Reagent (OMEGA, USA). The RNase-free DNase (OMEGA, USA) was used to remove any remaining contaminating DNA from the total RNA extractions. An aliquot of each sample was checked on an agarose gel. The concentration and purity of each RNA sample was determined using Nanodrop2000 (Thermo scientific, USA).
For isolation of rbcS, total RNA (500 ng) from the leaf of Xianglin 14 was used to generate a single-stranded cDNAs using an oligo (dT)18 primer and M-MLV reverse transcriptase in the presence of an RNase inhibitor (Thermo Scientific, USA) in a volume of 20 μl.
Cloning and Identification of rbcL and rbcS from C. oleifera
To clone rbcL, forward and reverse primers 5′-GGGAGGGACTTATGTCACCA-3′ and 5′-TGTATTCGGCTCAATCCTTT-3′ were designed in reference to the rbcL nucleotide sequences from Arabidopsis thaliana (GenBank access number: ATU91966), Brassica napus (AF267640), and Glycine max (EU717256 and Z95552) using Primer Premier 5.0. Polymerase chain reaction (PCR) was carried out in a 20 μl reaction containing 12.5 μl PrimeSTAR buffer, 0.5 mM dNTPs, 1.5 mM MgCl2, 0.5 mM of each primer, 1 U PrimeSTAR HS DNA Polymerase (Takara, Dalian, China), and 20 ng of the total genomic DNA as a template. Cycling conditions consisted of pre-cycling at 98°C for 2 min, and 36 cycles of denaturation at 98°C for 40 s, annealing at 54.5 °C for 40 s, elongation at 72°C for 2 min, and an elongation phase at 72°C for 7 min. The PCR products were analyzed on 1.2% (w/v) agarose gel, purified using a gel extraction mini kit, and ligated into pMD18-T vector (Takara, Dalian, China). The mixtures were transformed into Escherichia coli strain DH5α, and the positive clones were sequenced (data not shown).
For cloning of rbcS, a partial cDNA fragment was cloned by degenerate PCR using reverse transcription polymerase chain reaction (RT-PCR). Degenerate primers of rbcS 5′-AAGAAGTTYGAGACSCTSTC-3′ (forward) and 5′-GGCWTGWAGGCGATGAAYCTG-3′ (reverse) were designed using Primer Premier 5.0. PCR was performed in a 20 μl reaction containing 1× PCR buffer, 0.5 mM dNTPs, 1.5 mM MgCl2, 0.5 mM of each degenerate primer of rbcS, 1 U Taq polymerase, and 2.0 μl of the cDNA first strand. Cycling conditions consisted of pre-cycling at 94°C for 5 min followed by 35 cycles at 94°C for 30 s, 53°C for 30 s, and 72°C for 2 min. The reaction was terminated by a 7-min incubation step at 72°C. Rapid amplification of cDNA ends (RACE) was performed by using reagents of 5′ and 3′ RACE purchased from Invitrogen (Carlsbad, CA, USA), and conducted according to their instruction manual.
Gene specific oligonucleotide primers (GSPs) for rbcS 5′ RACE and 3′ RACE were designed as: rbcS 5′ RACE GSP1: 5′-CCTCAACCTCCTTCAACA-3′; rbcS 5′ RACE GSP2: 5′-TCCAGTATCGCCCATCGTAG-3′; and rbcS 5′ RACE GSP3: 5′-CACAAATCCTCCCACAACAG-3′. GSPs for rbcS 3′ RACE were as follows: rbcS 3′ RACE GSP1: 5′-ATGGGCGATACTGGACAATG-3′; rbcS 3’RACE GSP2: 5′-GTGTTGAAGGAGGTTGAGGA-3′; and rbcS 3′RACE GSP3: 5′-AGAAGGAATACCCACAAGCA-3′. rbcS 5’RACE was obtained in a 25 μl reaction containing 12.5 μl 2 × MightyAmp Buffer including Mg2+ 4 mM (2x), dNTP 800 μM (2x); UPM (long); rbcS 5′ RACE GSP 0.75 μl (0.3 μM); the synthesized cDNA as template; and Mighty Amp DNA polymerase 0.5 U (1.25 U/μl). Cycling conditions consisted of pre-cycling at 98°C for 2 min, and 35 cycles of denaturation at 98°C for 40 s, annealing at 54°C for 40 s, and elongation at 68°C for 2 min, and final extension at 72°C for 7 min. For the full-length rbcS gene, the PCRs were performed with the forward and reverse primers 5′-ATGGTTGCCTCCATAC-3′ and 5′-CACAGCCATTGATCTAACGA-3′ in a 20 μl reaction containing PrimeSTAR buffer 12.5 μl, 0.5 mM dNTPs, 1.5 mM MgCl2, 0.5 mM of each primer, 1 U PrimeSTAR HS DNA Polymerase, and 2.0 μl cDNA first strand as template. Cycling conditions consisted of pre-cycling at 98°C for 2 min, and then 35 cycles of denaturation at 98°C for 40 s, annealing at 51°C for 40 s, elongation at 72°C for 2 min, and an elongation phase at 72°C for 7 min. The PCR products were separated, purified, ligated, transformed, and sequenced as described above.
Analysis of Sequence Properties
Database was retrieved using the NCBI server1, and the protein properties were analyzed using ProtParam2, TMpred3, and SignalP 3.0 Server4. The secondary structures were predicted by SOPMA5. The homolog relationships between deduced amino acid sequences of rbcL and rbcS from C. oleifera and other higher plants were analyzed by Vector NTI and displayed by Gendoc. The subcellular location and cleavage site analysis were conducted using the ChloroP 1.1 prediction server6.
Transcript Abundance Analysis by Quantitative RT-PCR
Equal volumes of RNA solution isolated from leaves of three cultivars were used to synthesize single-stranded cDNA in a 20 μl volume using Transcriptor First Strand cDNA Synthesis Kit (Thermo Scientific, USA) with anchored oligo(dT)18 primers. Aliquots of the single-stranded cDNA of the three cultivars were used as templates for qRT-PCR analysis on CFX96 (Bio-Rad, Hercules, CA, USA). PCR amplifications were performed in a total volume of 20 μl using the Maxima SYBR Green qPCR Master Mix (2x) (Thermo Scientific, USA) and the specific primers designed for rbcL of C. oleifera were 5′-TGTACTACAGTTCGGCGGAG-3′ (forward), and 5′-TCCATACCTCACAAGCAGCA-3′ (reverse); for rbcS of C. oleifera, were 5′-TGGGCGATACTGGACAATGT-3′ (forward), and 5′-CAGGCGATGAAACTGATGCA-3′ (reverse). The internal control was GAPDH (glyceraldehyde-3-phosphate-dehydrogenase) gene. The primers of GAPDHF: 5′-GAAGGGTGGTGCAAAGAAGG-3′ and GAPDHR 5′-GACCCTCAACAATGCCAAACT-3′ were designed using Primer 37. The sizes of these amplicons were 194, 165, and 185 bp, respectively.
Photosynthesis Measurement
Net photosynthetic rates (PN) of the three cultivars were determined using a Li-6400 portable photosynthesis meter (Li-COR Bioscience, Lincoln, NE, USA). Other parameters including stomatal conductance (gs), intercellular CO2 concentration (Ci), and transpiration rate (E) were also recorded. The measurements took place on the newest mature leaves at 11:00 am–12:00 pm, four leaves per plant and three readings per leaf. Since each block had nine plants, three plants per cultivar and there were three blocks; a total of nine plants were measured per cultivar in every September from 2011 to 2013. The environmental conditions for the measurements were set as follows: leaf temperature at 25°C, photon flux density at 1,000 μmol m-2 s-1, relative humidity at 65%, and CO2 concentration at 0.04%.
Data Analysis
The experiment for oil production of the three cultivars was arranged as a randomized complete block design with three replications. Fresh fruit was harvested annually from 2011 to 2013; fresh seeds and dry seeds per plant were recorded. Fresh and dry seed yields were calculated based on 1,650 plants per hectare. Tea oil was extracted using the Soxhlet extraction method described by Zeng et al. (2014). Oil yield was calculated based on the amount of oil extracted from seeds.
Data of seed yields, oil yields, and photosynthetic parameters collected in 2011, 2012, and 2013 were analyzed annually (data not shown). The 3-year data were combined based on parameters and analyzed by analysis of variance (SAS GLM, SAS Institute, Cary, NC, USA); mean separations were performed using Duncan’s Multiple Range Test at the 5% level.
The quantitative RT-PCR data were analyzed using the Bio-Rad CFX 2.0 data analysis software. The expression levels of Co-rbcL and Co-rbcS from C. oleifera were normalized based on the internal control of GAPDH gene.
Pearson’s correlation analysis was performed to assess the relationship among seed and oil yields, Co-rbcL and Co-rbcS expressions, and PN. The significance of the correlations was tested using the critical value table (Taylor and Bates, 2013).
Results
Seed and Oil Yields of Three Cultivars
Dry seed yields of three cultivars from 2011 to 2013 averaged 1,540.7, 1,520.3, and 1,806.9 kg ha-1 annually for Hengchong 89, Xianglin 1, and Xianglin 14, respectively (Figure 2A). The seed yield of Xianglin 14 was significantly higher than that of Hengchong 89 and Xianglin 1. Oil extracted from the seeds or oil yield of Xianglin 14 was 657.7 kg ha-1, which was significantly greater than 591.4 kg ha-1 produced by Xianglin 1 (Figure 2B), and oil yield of Xianglin 1 was higher than Hengchong 89 at 546.9 kg ha-1.
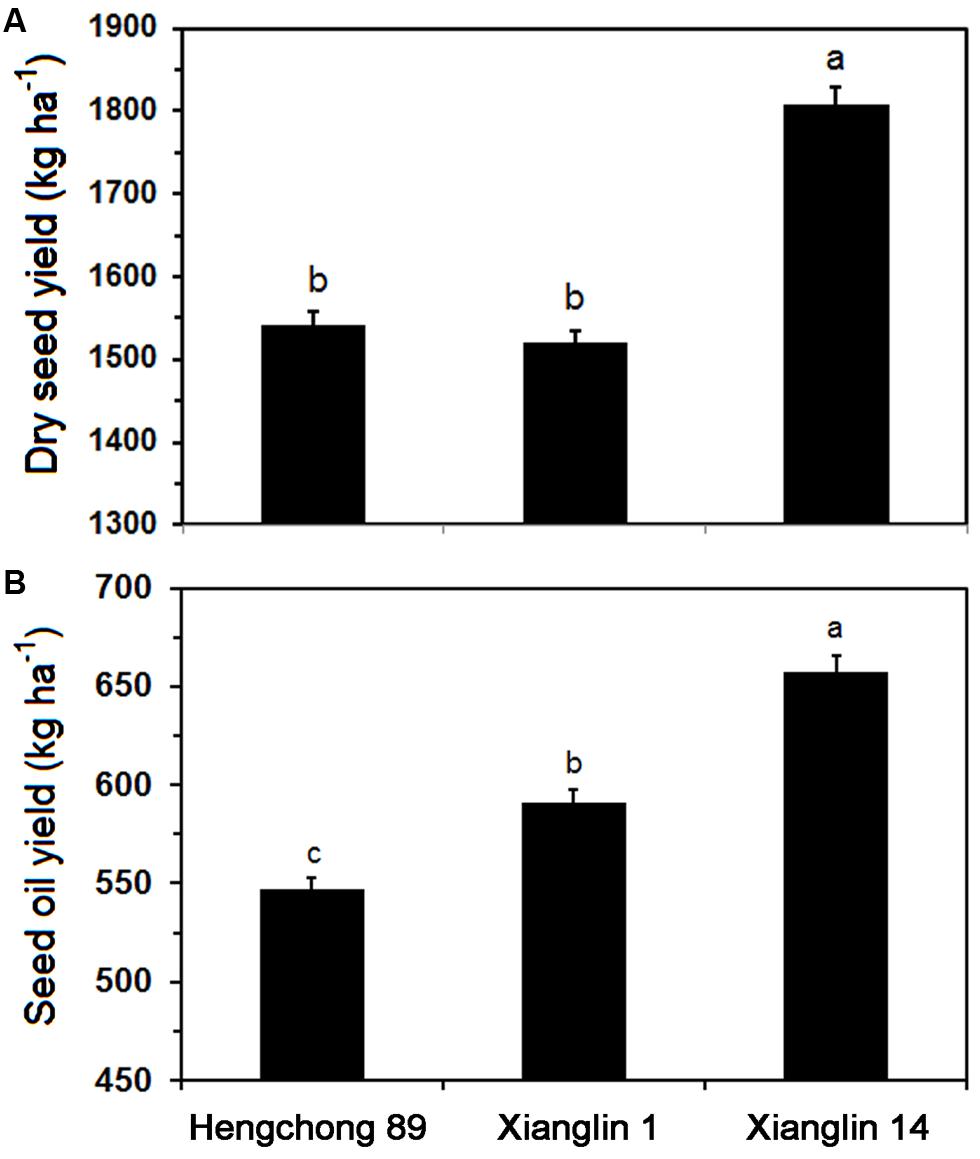
FIGURE 2. Mean dry seed (A) and oil (B) yields of three cultivars Hengchong 89, Xianglin 1, and Xianglin 14 grown at the Experimental Station of the National Engineering and Technology Research Center of Oil-Tea Camellia, Changsha, Hunan, China from 2011 to 2013. Bars represent standard error with n = 9. Different letters above the bar represent significant difference at P < 0.05 level by Duncan’s Multiple Range Test.
Cloning and Identification of rbcL and rbcS cDNAs from C. oleifera
Polymerase chain reaction amplification generated a product about 1,500 bp (Figure 3). After sequencing, the product was confirmed to be 1,522 bp containing a 1,425 bp coding region and encoding 475 amino acids with a molecular mass of 52.63 kDa. The nucleotide sequence was 99% homologous with that from other Camellia species, such as Camellia sinensis (GenBank accession number: KC143082), Camellia japonica (L12602), and Camellia granthamiana (AF380034). Such a high homology suggested that this fragment from C. oleifera was rbcL gene, which was designated as Co-rbcL. The sequence has been deposited in GenBank with an accession no. KJ721197.
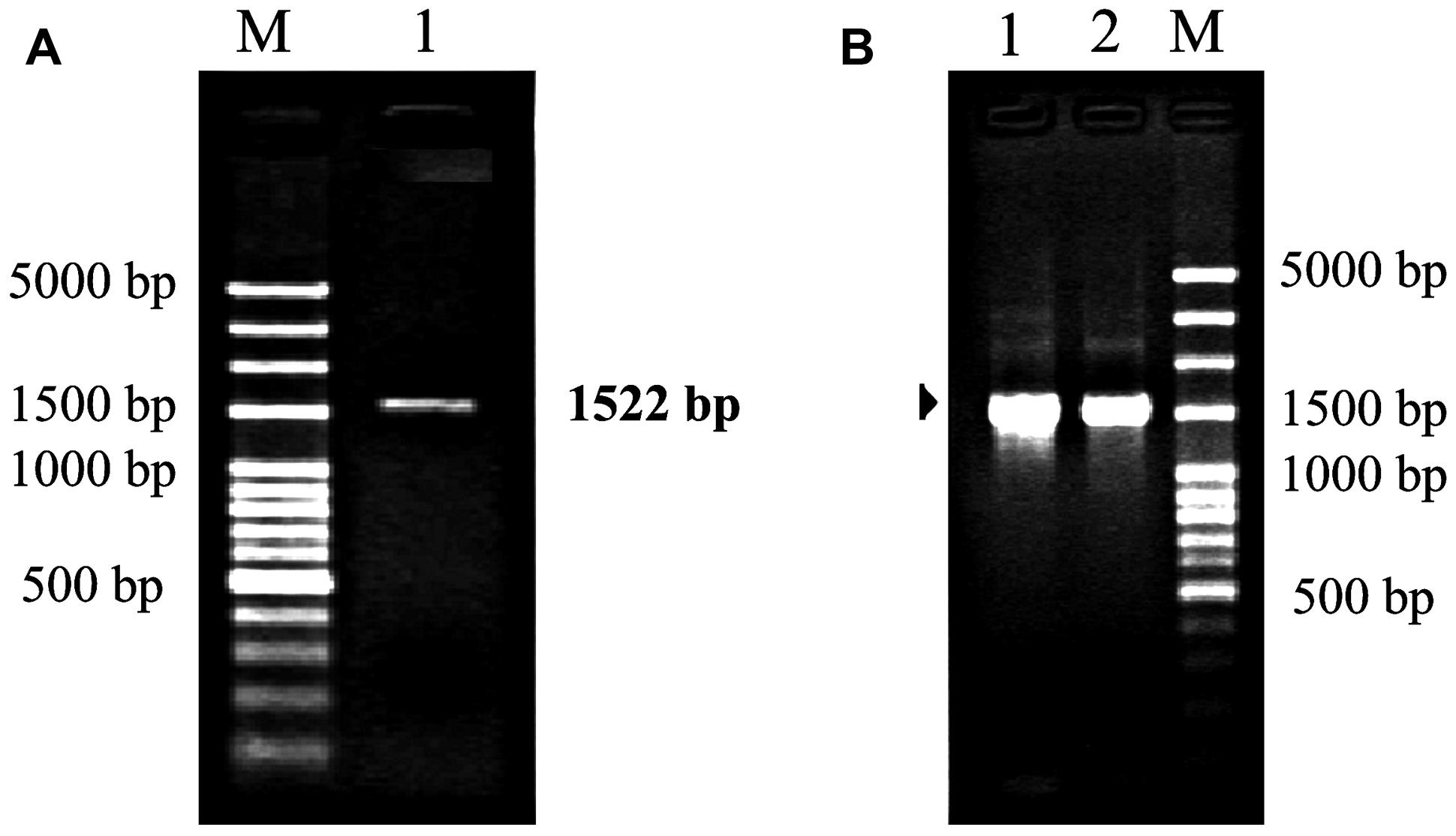
FIGURE 3. Polymerase chain reaction cloning of rbcL from leaves of C. oleifera and its recombinant strains. (A) Lane 1: the PCR amplification fragment of rbcL with an expected band at 1,522 bp for rbcL amplified by PCR. (B) Lanes 1 and 2 represent the positive recombinant strains. M represents 100 bp plus DNA ladder.
A partial sequence of rbcS gene was first obtained from degenerate RT-PCR reactions. Sequence analysis showed that this fragment had 324 bp (Figures 4A,B) encoding a deduced peptide sequence of 122 amino acids. This sequence shared 94% homology with rbcS of C. sinensis (GenBank accession no. EF011075), suggesting that it was a fragment of rbcS gene. Based on this fragment, a set of gene specific primers were designed in order to obtain the 5′ and 3′ ends (Figures 4C–F). The resultant full-length rbcS gene was found to be 615 bp. This sequence had an open reading frame of 528 bp starting with an initiation codon ATG at position of the 2nd and 4th nucleotides and ending with a termination codon TAA at position of the 530th and 532nd nucleotides, encoding 176 amino acid residues with a molecular mass of 19.69 kDa. The GC content of this sequence was about 49.15%. The predicted nucleotide sequence of the coding region shared more than 95% homology with that of rbcS mRNA from C. sinensis (GenBank accession no. EF011075). This sequence was designated as Co-rbcS, and has been deposited in GenBank with an accession no. KJ721196.
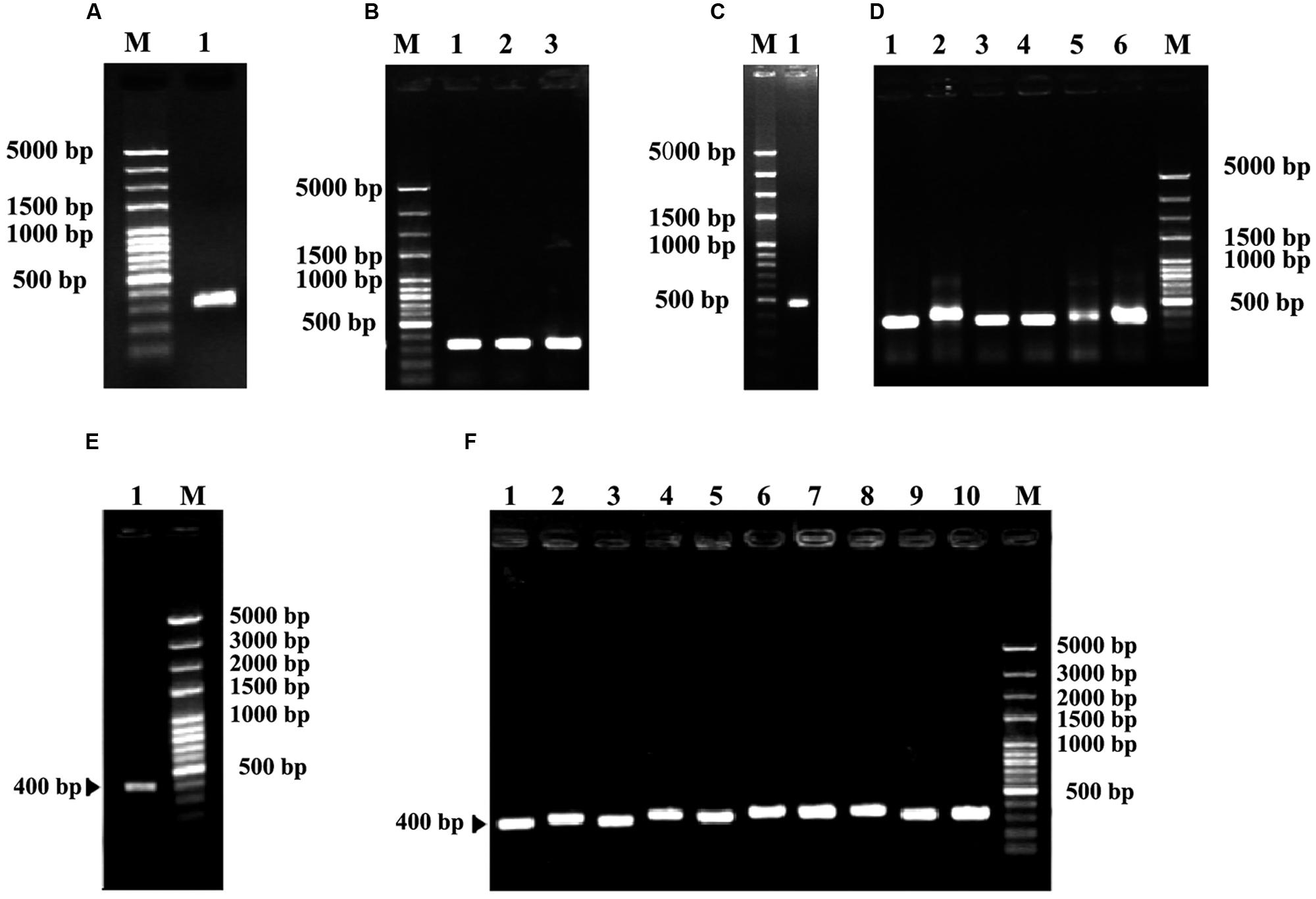
FIGURE 4. Polymerase chain reaction cloning of rbcS from leaves of C. oleifera and its recombinant strains. (A) The degenerate PCR amplification fragment of rbcS in lane 1. (B) Lanes 1, 2, and 3 are positive recombinants of rbcS. (C) Lane 1 indicates the 5′ RACER result of Co-rbcS. (D) Lanes 1–6 were positive recombinants of the 5′ RACER of Co-rbcS. (E) Lane 1 is the 3′ RACER result of Co-rbcS. (F) Lanes 1–10 are positive clones of Co-rbcS. M represents 100 bp plus DNA ladder.
Characteristics of Co-rbcL and Co-rbcS Proteins
The putative Co-rbcL amino acid sequence shared an overall homology of 100, 99, 98, 98, 94, and 92% with that of Camellia taliensis (Yang et al., 2013), C. sinensis (GenBank accession no. YP_007317256), peach (Prunus persica, GenBank accession no. YP_004021673), Pyrus pyrifolia (GenBank accession no. YP_004842247), tobacco (Nicotiana tabacum; Shinozaki and Sugiura, 1982), and pine (Pinus thunbergii; Mukai et al., 1991), respectively (Figure 5). Noticeably, there were active sites residing in these aligned sequences, including 165YGRPLLGCTIKPK177, K-177, 321SGGDHI/VHAGTVVGKLEGER339, K-334, K-201, and C-459 (Mukai et al., 1991). The Co-rbcL processing site was postulated to occur between the Lys(K)51 and Arg(A)9 of the protein sequence using the ChloroP 1.1 Prediction Server6.
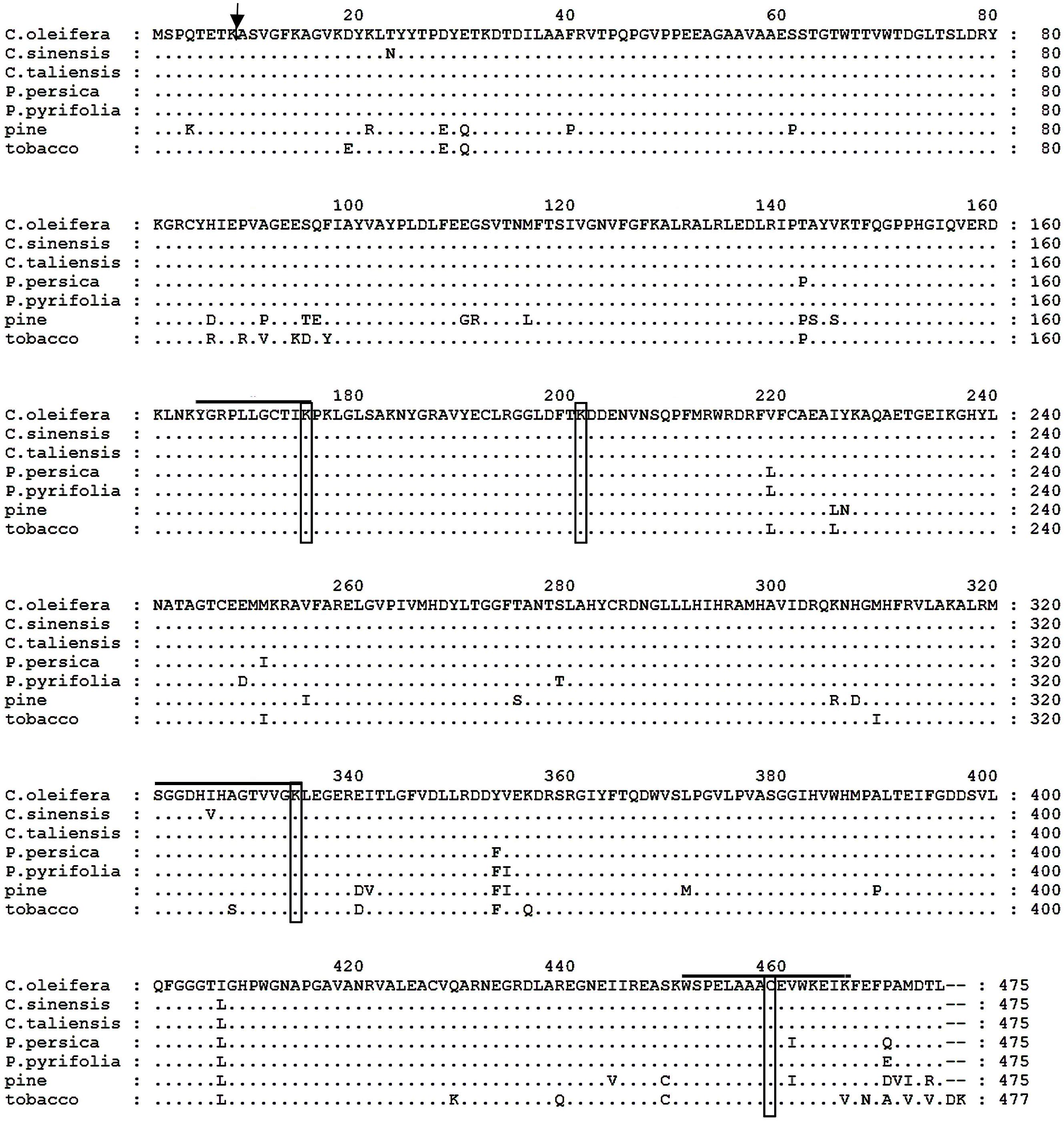
FIGURE 5. Comparison of the deduced amino acid sequences of the rbcL protein from Camellia oleifera, Camellia sinensis, Camellia taliensis, Prunus persica, Pyrus pyrifolia, pine (Pinus thunbergii), and tobacco (Nicotiana tabacum). Amino acid residues identical to those of C. oleifera rbcL protein are indicated by dots. Hyphens indicate gaps. The arrow indicates the possible processing site of mature protein in C. oleifera; and active site residues are overlined and boxed.
Two strong transmembrane helices, signal peptides or transmembrane regions were found in the putative polypeptide of Co-rbcS by SignalP 3.0 Server using neural networks (NNs) and hidden Markov models (HMMs) trained on eukaryotes (Figure 6). The most likely cleavage site appeared to be between positions 16 and 17. Acidic residues and basic residues account for 25.5 and 12.25%, respectively. Hydrophobic residues and charged residues are 28.90 and 18.89%, respectively. Alignment of the putative Co-rbcS amino acid sequence with rbcS from the other plants showed that Co-rbcS shared 96% homology with that of C. sinensis (ABK15574), 78% homology with that of Gossypium hirsutum (AFS41732) and P. pyrifolia var. culta (BAA00450) rbcS, and 77% homology with that of B. napus rbcS (ABB51649) and A. thaliana small chain 3B (NP_198657) as well as 75 and 76% homology with A. thaliana ribulose bisphosphate carboxylase 2B small chain (NP_198658) and A. thaliana ribulose bisphosphate carboxylase small chain 1B (NP_198659), respectively.
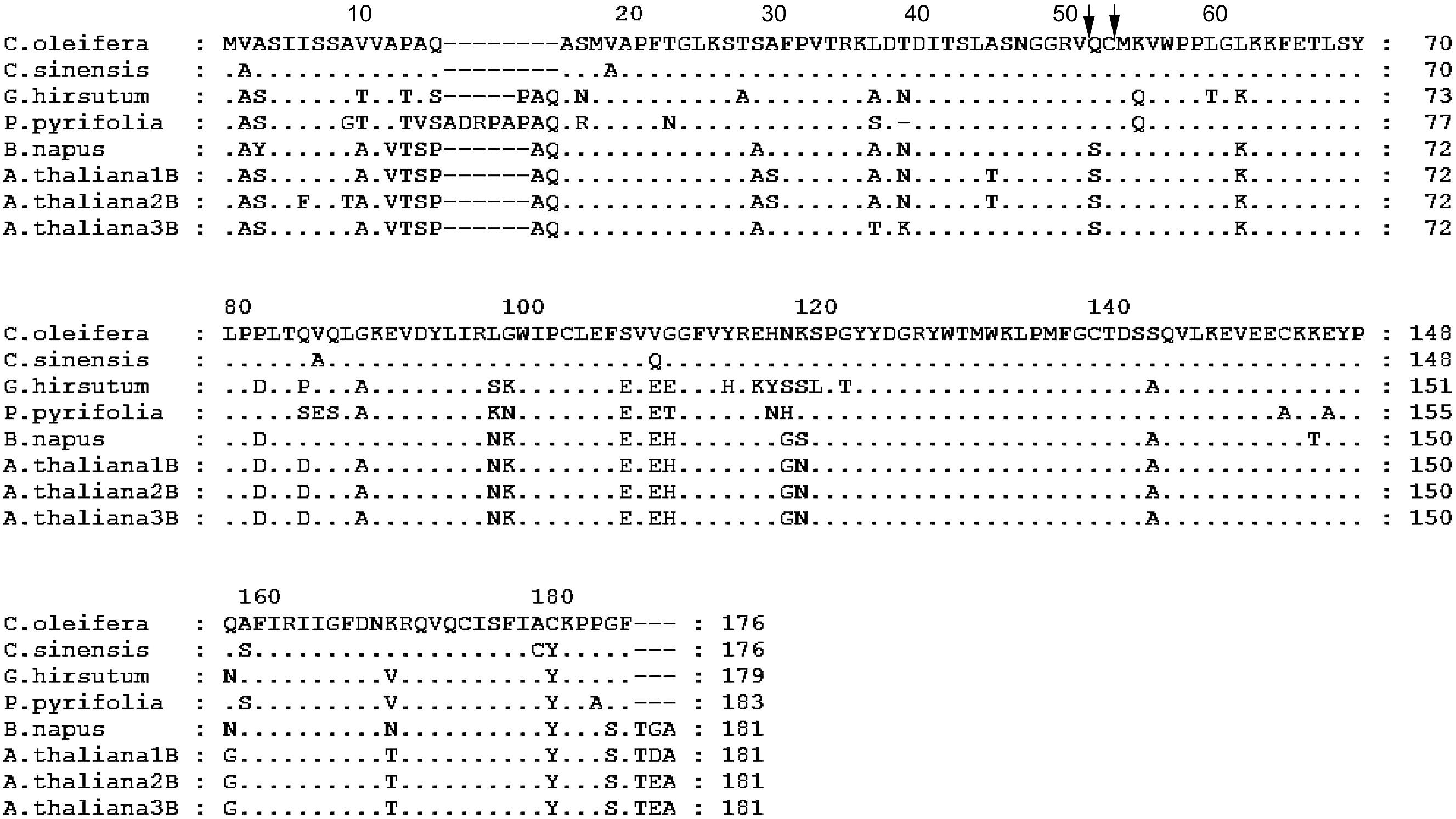
FIGURE 6. Alignment of the amino acid sequence of rbcS from Camellia oleifera with the sequence from Camellia sinensis, Gossypium hirsutum, Pyrus pyrifolia, Brassica napus, and Arabidopsis thaliana. A. thaliana 1B, 2B, and 3B represent A. thaliana ribulose bisphosphate carboxylase small chain 1B (NP_198659), A. thaliana ribulose bisphosphate carboxylase 2B small chain (NP_198658), and A. thaliana small chain 3B (NP_198657), respectively. Amino acid residues identical to those of C. oleifera rbcS protein are indicated by dots. Hyphens indicate gaps. The arrows indicate the possible processing site of mature protein in C. oleifera.
Expression of CO-rbcL and CO-rbcS in C. oleifera Cultivars
Real-time quantitative PCR analysis showed that Co-rbcL expression in cultivar Xianglin 14 was 42% higher than Xianglin 1 and 103% higher than Hengchong 89; Co-rbcL expression in Xianglin 1 was 41% higher than Hengchong 89 (Figure 7A). The expression levels of Co-rbcS in Xianglin 1 and Xianglin 14 were similar (Figure 7B) but were more than 2.5 fold higher than Hengchong 89.
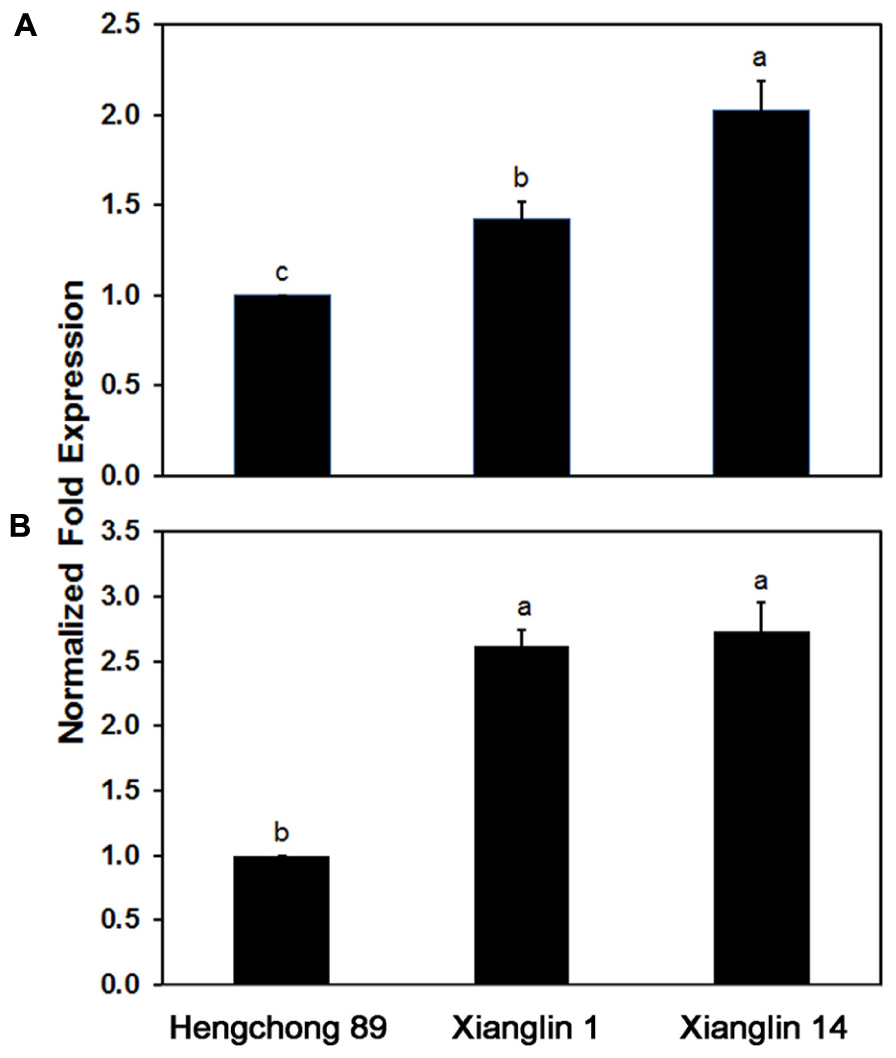
FIGURE 7. Expression of Co-rbcL (A) and Co-rbcS (B) in leaves of three cultivars Hengchong 89, Xianglin 1, and Xianglin 14 analyzed by real-time quantitative PCR, and the expression levels were normalized based on internal control of GAPDH gene. Bars represent standard error with n = 3. Different letters above the bar represent significant difference at P < 0.05 level by Duncan’s Multiple Range Test.
Photosynthesis
The mean net photosynthetic rate (PN) of cultivar Xianglin 14 from 3-year data (2011–2013) was 12.55 μmol CO2 m-2 s-1, which was the highest (Table 1). The PN of Xianglin 1 was significantly higher than Hengchong 89. Stomatal conductance (gs) of Xianglin 14 was also significantly higher than Xianglin 1, and Xianglin 1 was greater than Hengchong 89. On the other hand, intercellular CO2 concentration (Ci) was significantly higher in ‘Hengchong 89’ than both ‘Xianglin 14’ and ‘Xianglin 1’. Transpiration rates (E) of ‘Xianglin 14’ and ‘Xianglin 1’ were similar but both were significantly higher than‘Hengchong 89’.
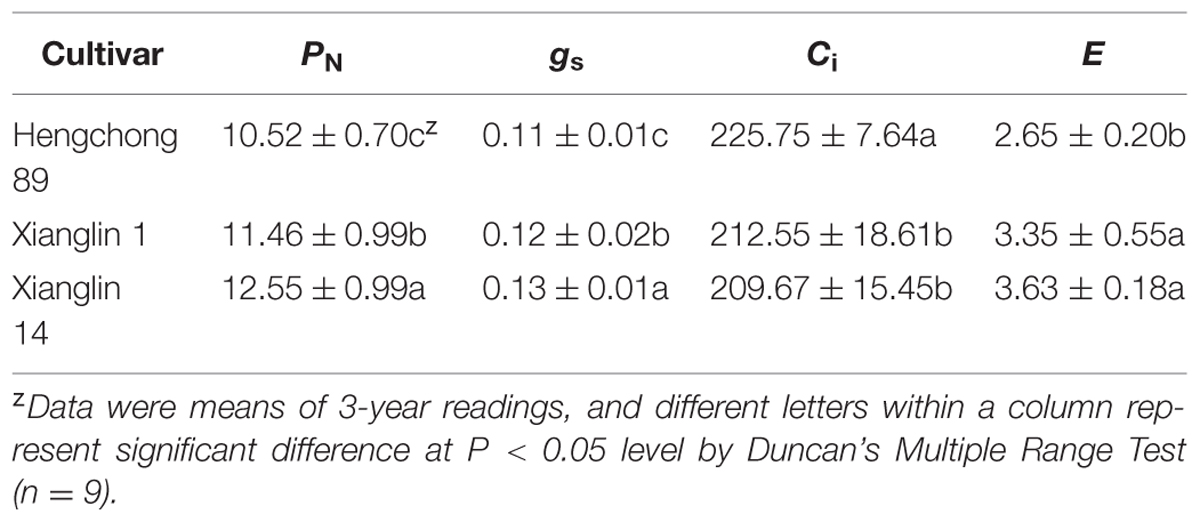
TABLE 1. Net photosynthetic rate, PN [μmol(CO2) m-2 s-1]; stomatal conductance, gs [mol(H2O) m-2 s-1]; intercellular CO2 concentration, Ci [cm3 m-3]; and transpiration rate, E [mmol(H2O) m-2 s-1] of three C. oleifera cultivars determined from 2011 to 2013 when grown in Hunan, China.
Relationships among Oil Yield, PN, and CO-rbcL and CO-rbcS Expression
Pearson’s correlation analysis showed that oil yield was significantly correlated with the expression levels of Co-rbcL and Co-rbcS at the P < 0.001 level (r = 0.94) and P < 0.01 level (r = 0.78), respectively (Table 2). The oil yield was also highly correlated with PN at the P < 0.001 level (r = 0.97). Furthermore, the expression level of Co-rbcL was closely correlated with dry seed yield at P < 0.001 level (r = 0.94) and fresh seed yield P < 0.01 level (r = 0.86); however, there was no significant correlation between Co-rbcS expression and fresh and dry seed yields. Additionally, PN was significantly correlated with dry seed yield but was not significant with fresh seed yield.
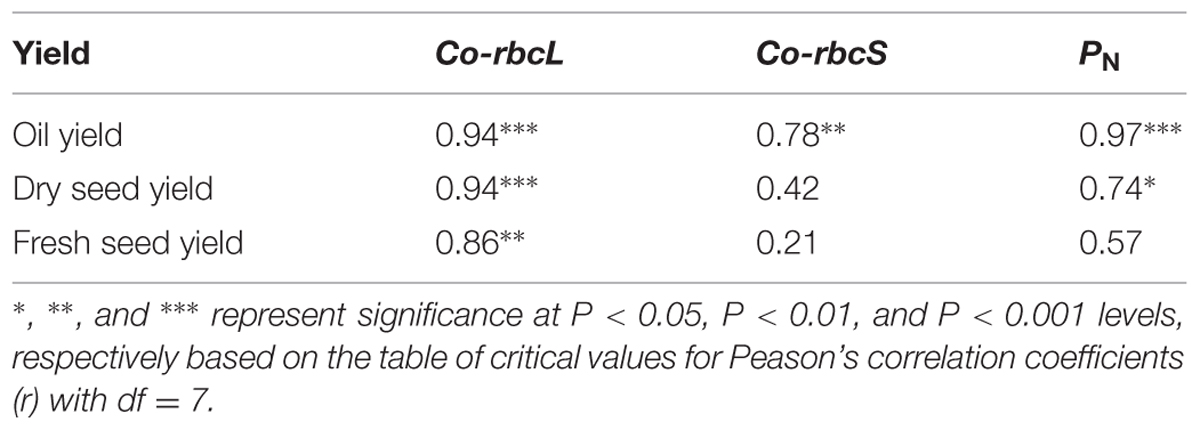
TABLE 2. Person’s correlation coefficients (r) of oil yield, dry seed and fresh seed yields in relation to the normalized fold expression of Co-rbcL and Co-rbcS and net photosynthetic rate (PN) in three cultivars of C. oleifera.
Discussion
The primary photosynthetic CO2 reduction reaction, the binding of CO2 to the acceptor-molecule ribulose-1,5-bisphosphate (RuBP) to form two molecules of 3-phosphoglycerate, is catalyzed by the enzyme Rubisco (Andersson and Backlund, 2008). Rubisco is the most abundant soluble protein in the plant leaf (Ellis, 1979). Increasing photosynthesis has been shown to increase crop yield (Ainsworth and Long, 2005; Raines, 2011). Photosynthesis provides the source of carbon and the reducing power and ATP for de novo fatty acid synthesis, and increased carbons enhance the rate of fatty acid synthesis (Rawsthorne, 2002; Bates et al., 2013). In the search for potential molecular markers that might help improve C. oleifera breeding efficiency for high oil yields, Rubisco genes could be a subject of interest.
The present study isolated Rubisco subunits rbcL and rbcS genes from C. oleifera. As illustrated in Figure 5, rbcL amino acid sequence of C. oleifera is 100 and 99% homologous to its relatives C. taliensis (Yang et al., 2013) and C. sinensis (GenBank accession number: YP_007317256), respectively and is more than 90% homologous to other higher plant species, which supports the conclusion that the rbcL gene is highly conserved (Manzara and Gruissem, 1988). In addition to the identified active sites and transcription processing site in the Co-rbcL, the Co-rbcL protein also included eight cysteine residues at positions of 84, 172, 192, 221, 247, 284, 427, and 459. The Cys pairs 172 to 192 and 449 to 459 can create disulfide bridges within each LSU, while Cys 247 is thought to bridge between two adjacent subunits (Cohen et al., 2005). Residue Asp473 was proposed to serve as a latch responsible for placing the large-subunit carboxy-terminus over loop 6 and stabilizing the closed conformation required for catalysis (Duff et al., 2000); however, it is not essential for catalysis but for CO2/O2 specificity (Satagopan and Spreitzer, 2004). Lysine 334 is thought to play a specific role in stabilizing the transition state intermediates of both the carboxylation and oxygenation reactions, thereby facilitating the reaction of the enefiolate with the gaseous substrate (Parry et al., 2003). The amino acid sequence of rbcS of C. oleifera is only slightly above 75% homologous to the other plant species (Figure 6), indicating that rbcS is less conserved than rbcL (Manzara and Gruissem, 1988). Based on SOPMA prediction (Combet et al., 2000), the protein includes 22.16% of α helix, 24.43% of β sheet, 6.25% of β turn, and 47.16% of random coil. Moreover, the Co-rbcS precursors appeared to be processed to mature protein during entry into the chloroplast, and the processing site was postulated to occur between residues Gln(Q)52 and Cys(C)53 using ChloroP 1.1 Prediction Server6.
Three-year field studies showed that the highest seed yield was produced by the C. oleifera cultivar Xianglin 14 (Figure 2A), which also produced the highest amount of oil (Figure 2B). Hengchong 89 had the lowest seed yield and was also the lowest in oil production. Xianglin 1 had a similar seed yield as Hengchong 89; however, its oil yield was significantly greater than Hengchong 89, suggesting that long-term selection for oil yield may result in cultivars also differing in fatty acid biosynthesis. Subsequent real-time quantitative PCR analysis showed that the level of Co-rbcL expression in Xianglin 14 was substantially higher than Xianglin 1; and Xianglin 1 was significantly greater than Hengchong 89 (Figure 7A). The expression levels of rbcS in Xianglin 14 and Xianglin 1 were similar, but their expressions were much greater than Hengchong 89 (Figure 7B). Examining leaf photosynthesis showed that PN were significantly higher in Xianglin 14 than Xianglin 1; and Xianglin 1 was higher than Hengchong 89 (Table 1). Pearson’s correlation analysis demonstrated that Co-rbcL expression levels were significantly correlated with oil and dry seed yields at P < 0.001 level as well as with fresh yield at P < 0.01 level; while Co-rbcS expression was only correlated with oil yield at P < 0.01 level. The PN was correlated with oil yields at P < 0.001 level and dry seed yields at P < 0.05 level (Table 2). Our results suggest that Co-rbcL and Co-rbcS, particularly Co-rbcL, could potentially be molecular markers for early selection of higher oil producing cultivars. Furthermore, the selection should also take the net photosynthetic rate into consideration. The incorporation of Co-rbcL and Co-rbcS markers into breeding programs could allow early recognition of potential high oil production cultivars and could significantly shorten tea oil plant breeding time and increase breeding efficiency.
The interest in the relationships of Rubisco activity, Rubisco proteins, and/or their mRNAs with crop yield dates back to the 1970s (Frey and Moss, 1976; Murthy and Singh, 1979). Some contradictory reports on the relationship of photosynthesis and crop yield in company with suggestions that the crop is a sink and not photosynthesis limited (Kuo et al., 1980; Loza-Tavera et al., 1990; Evans, 1998; Borras et al., 2004) have led to the view that improving photosynthesis is unlikely to increase crop yield (Borras et al., 2004; Sinclair et al., 2004). Information regarding improving photosynthesis to enhance crop yield became limited, even though traditional breeders have been continuously working on the improvement of photosynthesis and other economically important traits for yield increase. The reappraisal of the importance of photosynthesis for yield enhancement has been a more recent event (Long et al., 2006; Fischer and Edmeades, 2010; von Caemmerer and Evans, 2010), which is based on at least three lines of evidence: first, leaf photosynthesis increase resulted from CO2 enrichment generally leads to increased crop yield. Second, C4 plants have greater rates of photosynthesis and produce more biomass per unit of intercepted sunlight than C3 plants. Third, the introduction of dwarf genes into rice and wheat for effectively capturing light significantly increased the fraction of biomass in grain. Several recent publications have documented photosynthesis improvement in enhancing corn, rice, soybean, and wheat yields (Long et al., 2006; Fischer and Edmeades, 2010; Zhu et al., 2010; Parry et al., 2011; Ainsworth et al., 2012). Phylogenetic analysis also showed that positive selection in the rbcL gene of higher plants is common (Kapralov and Filatov, 2007), and Rubisco kinetics is quite variable among plant species (Flood et al., 2011; Galmes et al., 2014). Furthermore, two transplastomic tobacco lines with functional Rubisco from the cyanobacterium Synechococcus elongatus had higher rates of CO2 fixation per unit of enzyme than the control plant (Lin et al., 2014), suggesting that Rubisco activity and regulation should be targets for improving plant productivity.
We consider our study as a part of the effort on the recognition of the importance of photosynthesis for yield improvement. The continuing and long-term selection of high seed and high oil yields in C. oleifera could lead to enrich the abundances of transcripts of Rubisco genes that enhance photosynthetic properties, photosynthetic efficiency or capacity. Since the foremost breeding goal has been selecting cultivars with high oil production, the selective action may also enhance fatty acid biosynthesis, thus improving crop oil yield (Long et al., 2006; Zhu et al., 2010; Parry et al., 2011). This could be due to the fact that Rubisco determines photosynthetic efficiency, and photosynthesis is the source of carbon and reducing power and ATP for de novo fatty acid synthesis. The present study may also suggest that genes related the biosynthesis of oil could be enriched by the selection (Zeng et al., 2014). In addition to investigating mechanisms underlying the enhanced Co-rbcL and Co-rbcS expression, further studies are warranted to examine if some oil biosynthesis genes are also highly expressed in ‘Xianglin 14’. Nevertheless, the identified Rubisco genes, particularly Co-rbcL could potentially be molecular markers for improving breeding efficiency in C. oleifera.
Conclusion
The present study is the first report of Co-rbcL and Co-rbcS in C. oleifera, a tree species producing high-quality edible oil. Analyzing the gene expression and net photosynthetic rates of three cultivars differing in oil production showed that oil yields were highly correlated with the expression levels of Co-rbcS and Co-rbcL in particular as well as net photosynthetic rates. These results suggest that Co-rbcL and Co-rbcS could potentially be molecular markers for early selection of high oil production cultivars of C. oleifera.
Author Contributions
YC, BW, and JC conceived and designed the experiments. BW, XW, RW, SP, LC, LM, and JL conducted the experiments. All the authors participated in the analysis and interpretation of the results. YC, BW, and JC wrote this manuscript, which was approved by all authors.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was supported in part by National Natural Science Foundation of China (Grant No. 31370677). The authors wish to thank Ms. Terri A. Mellich and Mrs. Barbara Henny for critical reading of this manuscript.
Footnotes
- ^http://www.ncbi.nlm.nih.gov
- ^http://www.expasy.ch/tools/protparam.html
- ^http://www.ch.embnet.org/software/TMPRED_form.html
- ^http://www.cbs.dtu.dk/services/SignalP/
- ^http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_sopma.html
- ^http://www.cbs.dtu.dk/services/ChloroP/
- ^http://primer3.ut.ee/
References
Ainsworth, E. A., and Long, S. P. (2005). What have we learned from 15 years of free-air CO2 enrichment (FACE)? A meta-analytic review of the responses of photosynthesis, canopy properties and plant production to rising CO2. New Phytol. 165, 351–371. doi: 10.1111/j.1469-8137.2004.01224.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ainsworth, E. A., Yendrek, C. R., Skoeczka, J. A., and Long, S. P. (2012). Accelerating yield potential in soybean: potential targets for biotechnological improvement. Plant Cell Environ. 35, 38–52. doi: 10.1111/j.1365-3040.2011.02378.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Andersson, I., and Backlund, A. (2008). Structure and function of rubisco. Plant Physiol. Biochem. 46, 275–291. doi: 10.1016/j.plaphy.2008.01.001
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Bates, P. D., Stymne, S., and Ohlrogge, J. (2013). Biochemical pathways in seed oil synthesis. Curr. Opin. Plant Biol. 16, 358–364. doi: 10.1016/j.pbi.2013.02.015
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Borras, L., Slafer, G. A., and Otegui, M. E. (2004). Seed dry weight response to source-sink manipulations in wheat, maize and soybean: a quantitative reappraisal. Field Crop Res. 86, 131–146. doi: 10.1016/j.fcr.2003.08.002
Cheng, Y., Wu, S., Ho, C., Huang, S., Cheng, C., and Yen, G. (2014). Beneficial effects of Camellia oil (Camellia oleifera Abel.) on ketoprofen-induced gastrointestinal mucosal damage through upregulation of HO-1 and VEGF. J. Agri. Food Chem. 62, 642–650. doi: 10.1021/jf404614k
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Cohen, I., Knopf, J. A., Irihimovitch, V., and Shapira, M. (2005). A proposed mechanism for the inhibitory effects of oxidative stress on Rubisco assembly and its subunit expression. Plant Physiol. 137, 738–746. doi: 10.1104/pp.104.056341
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Collard, B. C. Y., and Mackill, D. J. (2008). Marker-assisted selection: an approach for precision plant breeding in the twenty-first century. Philos. Trans. R. Soc. Lond. B Biol. Sci. 363, 557–572. doi: 10.1098/rstb.2007.2170
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Combet, C., Blanchet, C., Geourjon, C., and Deléage, G. (2000). NPS@: Network Protein Sequence Analysis. Trends Biochem. Sci. 25, 147–150. doi: 10.1016/S0968-0004(99)01540-6
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Duff, A. P., Andrews, T. J., and Curmi, P. M. G. (2000). The transition between the open and closed states of Rubisco is triggered by the inter-phosphate distance of the bound bisphosphate. J. Mol. Biol. 298, 903–916. doi: 10.1006/jmbi.2000.3724
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ellis, R. J. (1979). Most abundant protein in the world. Trends Biochem. Sci. 4, 241–244. doi: 10.1016/0968-0004(79)90212-3
Evans, L. T. (1998). “Greater crop production: whence and whither?” in Feeding a World Population of More Than Eight Billion People- A Challenge to Science, eds J. C. Waterlow, D. G. Armstrong, L. Fowdenand, and R. Riley (Cary: Oxford University Press), 89–97.
Fischer, R. A., and Edmeades, G. O. (2010). Breeding and cereal yield progress. Crop Sci. 50, S85–S98. doi: 10.2135/cropsci2009.10.0564
Flood, P. J., Harbinson, J., and Aarts, M. G. M. (2011). Natural genetic variation in plant photosynthesis. Trends Plant Sci. 16, 327–335. doi: 10.1016/j.tplants.2011.02.005
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Frey, N. M., and Moss, D. N. (1976). Variation in RuDPCase activity in barley. Crop Sci. 16, 209–213. doi: 10.2135/cropsci1976.0011183X001600020011x
Galmes, J., Lapralov, M. V., Andralojc, P. J. Conesa, M. A., Keys, A. J., Parry, M.A. J., et al. (2014). Expanding knowledge of the Rubisco kinetics variability in plant species: environmental and evolutionary trends. Plant Cell Environ. 37, 1989–2001. doi: 10.1111/pce.12335
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ichikawa, K., Miyake, C., Iwano, M., Sekine, M., Shinmyo, A., and Kato, K. (2008). Ribulose 1,5-bisphosphate carboxylase/oxygenase large subunit translation is regulated in a small subunit-independent manner in the expanded leaves of tobacco. Plant Cell Physiol. 49, 214–225. doi: 10.1093/pcp/pcm179
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Jauhar, P. P. (2006). Modern biotechnology as an integral supplement to conventional plant breeding: the prospects and challenges. Crop Sci. 46, 1841–1859. doi: 10.2135/cropsci2005.07-0223
Kapralov, M. V., and Filatov, D. A. (2007). Widespread positive selection in the photosynthetic Rubisco enzyme. BMC Evol. Biol. 7:73. doi: 10.1186/1471-2148-7-73
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kong, W., Liu, X., Yao, X., Wang, K., Ren, H., and Cao, Y. (2013). Research on photosynthetic characteristics of four oil-tea (Camellia) species. J. Southwest Univ. (Natl. Sci. Ed.) 35, 1–7.
Kuo, C. G., Hsu, F. H., Tsay, J. S., and Park, H. G. (1980). Variation in specific leaf weight and RuDPCase activity in mungbean. Can. J. Plant Sci. 60, 1059–1062. doi: 10.4141/cjps80-155
Lee, C., and Yen, G. (2006). Antioxidant activity and bioactive compounds of tea seed (Camellia oleifera Abel.) oil. J. Agric. Food Chem. 54, 779–784. doi: 10.1021/jf052325a
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Liao, T., Yuan, D., Zou, F., Gao, C., Yang, Y., Zhang, L., et al. (2014). Self-sterility in Camellia oleifera may be due to the prezygotic late-acting self-incompatibility. PLoS ONE 9:e99639. doi: 10.1371/journal.pone.0099639
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Lin, M. T., Occhialini, A., Andralojc, P. J., Parry, M. A. J., and Hanson, M. R. (2014). A faster Rubisco with potential to increase photosynthesis in crops. Nature 513, 547–550. doi: 10.1038/nature13776
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Long, S. P., Zhu, X., Naidu, S. L., and Ort, D. R. (2006). Can improvement in photosynthesis increase crop yields? Plant Cell Environ. 29, 315–330. doi: 10.1111/j.1365-3040.2005.01493.x
Loza-Tavera, H., Martinez-Barajas, E., and Sanchez-de-Jimenez, E. (1990). Regulation of ribulose-1,5-bisphosphate carboxylase expression in second leaves of maize seedlings from low and high yield populations. J. Plant Physiol. 93, 541–548. doi: 10.1104/pp.93.2.541
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ma, J., Ye, H., Rui, Y., Chen, G., and Zhang, N. (2011). Fatty acid composition of Camellia oleifera oil. J. Verbrauch. Lebensm. 6, 9–12. doi: 10.1007/s00003-010-0581-3
Manzara, T., and Gruissem, W. (1988). Organization and expression of the genes encoding ribulose-l,5-bisphosphate carboxylase in higher plants. Photosynth. Res. 16, 117–139. doi: 10.1007/BF00039489
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Mondal, T. K. (2011). “Camellia,” in Wild Crop Relatives: Genomics and Breeding Resources, ed. C. Kole (Berlin: Springer-Verlag), 15–39. doi: 10.1007/978-3-642-21201-7_2
Mukai, Y., Yamamoto, N., and Odani, K. (1991). Structure and expression of a gene for the large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase from pine. Plant Cell Physiol. 32, 273–282.
Murthy, K. K., and Singh, M. (1979). Photosynthesis, chlorophyll content and ribulose diphosphate carboxylase activity in relation to yield in wheat genotypes. J. Agri. Sci. 93, 7–11. doi: 10.1017/S0021859600086068
Parry, M. A. J., Andralojc, P. J., Mitchell, R. A. C., Madgwick, P., and Keys, A. J. (2003). Manipulation of Rubisco: the amount, activity, function, and regulation. J. Exp. Bot. 54, 1321–1333. doi: 10.1093/jxb/erg141
Parry, M. A. J., Reynolds, M., Salvucci, M. E., Raines, C., Andralojc, P. J., Zhu, X., et al. (2011). Raising yield potential of wheat. II. Increasing photosynthetic capacity and efficiency. J. Exp. Bot. 62, 453–467. doi: 10.1093/jxb/erq304
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Qiang, W., Xu, L., Jiang, X., Li, J., Gu, Y., Xu, L., et al. (2013). Survey and analysis of microsatellites from DNA sequences in Camellia species using 454 pyrosequencing. Sci. Silvae Sin. 49, 43–50.
Raines, C. A. (2011). Increasing photosynthetic carbon assimilation in C3 plants to improve crop yield: current and future strategies. Plant Physiol. 155, 36–42. doi: 10.1104/pp.110.168559
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Rawsthorne, S. (2002). Carbon flux and fatty acid synthesis in plants. Prog. Lipid Res. 41, 182–196. doi: 10.1016/S0163-7827(01)00023-6
Satagopan, S., and Spreitzer, R. J. (2004). Substitutions at the Asp-473 latch residue of Chlamydomonas ribulosebisphosphate carboxylase/oxygenase cause decreases in carboxylation efficiency and CO2/O2 specificity. J. Biol. Chem. 279, 14240–14244. doi: 10.1074/jbc.M313215200
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Sinclair, T. R., Purcell, L. C., and Sneller, C. H. (2004). Crop transformation and the challenge to increase yield potential. Trends Plant Sci. 9, 70–75. doi: 10.1016/j.tplants.2003.12.008
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Shinozaki, K., and Sugiura, M. (1982). The nucleotide sequence of the tobacco chloroplast gene for the large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase. Gene 20, 91–102. doi: 10.1016/0378-1119(82)90090-7
Tan, X., Hu, F., Xie, L., Shi, M., Zhang, D., and Wuyun, T. (2006). Construction of EST library and analysis of main expressed genes of Camellia oleifera seeds. Scientia Silvae Sinicae 42, 43–48.
Taylor, J. A., and Bates, T. R. (2013). A discussion on the significance associated with Pearson’s correlation in precision agriculture studies. Precision Agric. 14, 558–564. doi: 10.1007/s11119-013-9314-9
von Caemmerer, S., and Evans, J. R. (2010). Enhancing C3 photosynthesis. Plant Physiol. 154, 589–592. doi: 10.1104/pp.110.160952
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Wei, J., Chen, X., Sun, H., and Shen, G. (2012). Research progress on breeding and utilization of oil-used Camellia L. Acta. Agric. Zhejiang. 24, 533–540.
Yang, J. B., Yang, S. X., Li, H. T., Yang, J., and Li, D. Z. (2013). Comparative chloroplast genomes of Camellia species. PLoS ONE 8:e73053. doi: 10.1371/journal.pone.0073053
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zeng, Y., Tan, X., Zhang, L., Jiang, N., and Cao, H. (2014). Identification and expression of fructose-1,6-bisphosphate aldolase genes and their relations to oil content in developing seeds of tea oil tree (Camellia oleifera). PLoS ONE 9:e107422. doi: 10.1371/journal.pone.0107422
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zhuang, R. L. (2008), Oil-Tea Camellia in China, 2nd Edn. Beijing: Chinese Forestry Publishing House.
Zhu, X., Long, S. P., and Ort, D. R. (2010). Improving photosynthetic efficiency for greater yield. Annu. Rev. Plant Biol. 61, 235–261. doi: 10.1146/annurev-arplant-042809-112206
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Keywords: Camellia oleifera, camellia oil, photosynthesis, Rubisco, rbcL, rbcS, tea oil
Citation:Chen Y, Wang B, Chen J, Wang X, Wang R, Peng S, Chen L, Ma L and Luo J (2015) Identification of Rubisco rbcL and rbcS in Camellia oleifera and their potential as molecular markers for selection of high tea oil cultivars. Front. Plant Sci. 6:189. doi: 10.3389/fpls.2015.00189
Received: 20 December 2014; Accepted: 09 March 2015;
Published online: 31 March 2015
Edited by:
Jaime Prohens, Universitat Politècnica de València, SpainReviewed by:
Grama Nanjappa Dhanapal, University of Agricultural Sciences, Bengaluru, IndiaSandhya Mehrotra, Birla Institute of Technology and Science, India
Copyright © 2015 Chen, Wang, Chen, Wang, Wang, Peng, Chen, Ma and Luo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yongzhong Chen, National Engineering Technology Research Center of Oil-tea Camellia, Hunan Academy of Forestry, Changsha, Hunan 410004, ChinaY2hlbnlvbmd6aG9uZzA2QDE2My5jb20=Jianjun Chen, Environmental Horticulture Department and Mid-Florida Research and Education Center, University of Florida, 2725 South Binion Road, Apopka, FL 32703, USAampjaGVuQHVmbC5lZHU=
 Yongzhong Chen
Yongzhong Chen Baoming Wang
Baoming Wang Jianjun Chen
Jianjun Chen Xiangnan Wang
Xiangnan Wang Rui Wang
Rui Wang Shaofeng Peng
Shaofeng Peng Longsheng Chen
Longsheng Chen Li Ma
Li Ma Jian Luo
Jian Luo