- 1Department of Biology, Queen's University, Kingston, ON, Canada
- 2Hawkesbury Institute for the Environment, University of Western Sydney, Richmond, NSW, Australia
- 3Atlantic Forestry Centre, Canadian Forest Service, Natural Resources Canada, Fredericton, NB, Canada
Plant height is an important agronomic and horticultural trait that impacts plant productivity, durability and esthetic appeal. A number of the plant hormones such as gibberellic acid (GA), auxin and ethylene have been linked to control of plant architecture and size. Reduction in GA synthesis and auxin transport result in dwarfism while ethylene may have a permissive or repressive effect on tissue growth depending upon the age of plant tissues or the environmental conditions considered. We describe here an activation-tagged mutant of Populus tremula x P. alba clone 717-1B4 identified from 2000 independent transgenic lines due to its significantly reduced growth rate and smaller leaf size. Named dwarfy, the phenotype is due to increased expression of PtaACC SYNTHASE8, which codes for an enzyme in the first committed step in the biosynthesis of ethylene. Stems of dwarfy contain fiber and vessel elements that are reduced in length while leaves contain fewer cells. These morphological differences are linked to PtaACS8 inducing different transcriptomic programs in the stem and leaf, with genes related to auxin diffusion and sensing being repressed in the stem and genes related to cell division found to be repressed in the leaves. Altogether, our study gives mechanistic insight into the genetics underpinning ethylene-induced dwarfism in a perennial model organism.
Introduction
Reduced plant height, or dwarfism, is an important agronomic trait linked to higher yields (Huang et al., 1996; Yang and Hwa, 2008), easier harvesting (Adkins et al., 2010) and reduced nutrient demand on soils (Sieling and Kage, 2008). Leaf size, meanwhile, is linked to productivity, predation (Faeth, 1991) and the water status of the plant (Scoffoni et al., 2011). While both height and leaf size are complex traits, they appear to be genetically regulated by a similar panel of plant hormones (Valdovinos et al., 1967; Ephritikhine et al., 1999; Qi et al., 2011; Luo et al., 2013) and cytochrome P450s (Zhang et al., 2014), as well as abiotic factors such as temperature (Yang et al., 2014) and photoperiod (Li et al., 2014). Reductions in organ size are a result of two different physiological phenomena: smaller cells and impeded cellular division (Beemster et al., 2003). These two factors may work independently or synergistically to affect plant stature and organ size (Beemster et al., 2005; Skirycz et al., 2010). Newly produced plant tissues first exhibit growth due to rapid cellular division, a phase that is replaced in a distal-proximal manner by cellular expansion in progenitor cells (Donnelly et al., 1999). Due to the integrated control between these two processes, genetic mutations to single genes can have a drastic impact on plant stature as a whole or at the level of a specific tissue. Altered expression of genes such as ARABIDOPSIS VACUOLAR H+-PYROPHOSPHATASE1 (AVP1; Li et al., 2005), CYTOKININ RESISTANT1 (CNR1; Guo et al., 2010), and ISOPENTENYL TRANSFERASE3 (IPT3; Nobusawa et al., 2013) impact tissue size due to a difference in the total number of cells produced, while EXPANSIN10 (EXP10; Cho and Cosgrove, 2000), ARGOS-LIKE (Hu et al., 2006), and RETINOBLASTOMA-RELATED PROTEIN1 (RBR1; Sabelli et al., 2013) change the final size of plant tissues as a function of altered cell expansion.
The best studied genetic influences on dwarfism are genes and signaling pathways related to hormone production and sensitivity. Within these studies, ethylene, gibberellic acid (GA), auxin, and brassinosteroids (BR) have all been implicated with a role in cell division, cellular growth and overall plant architecture. Blocked BR synthesis (Nakaya et al., 2002) and reduced GA biosynthesis (Tong et al., 2007; Li et al., 2011) or increased GA catabolism (Busov et al., 2003; Schomburg et al., 2003; Curtis et al., 2005; Lee and Zeevaart, 2005; Dijkstra et al., 2008; Zawaski et al., 2011) induce dwarfism in a wide range of model plant systems. Auxin transport, meanwhile, is a critical component of proper plant stem elongation. In rice, auxin transport inhibition has been correlated to slower stem elongation (Yamamoto et al., 2007; Domingo et al., 2009) while reduced basipetal auxin transport in maize and Arabidopsis thaliana results in stunted plant development (Lantican and Muir, 1969; Geisler et al., 2003, 2005; Multani et al., 2003; Geisler and Murphy, 2006). Treatment of plant tissues with ethylene, a gaseous plant hormone, results in stunting (Vahala et al., 2013), a phenotype that has been linked to the induced expression of certain ETHYLENE RESPONSE FACTORs (ERFs; Dubois et al., 2013; Vahala et al., 2013). There also appears to be extensive cross-talk between the different hormone pathways with components of the ethylene pathway controlling GA biosynthesis (Qi et al., 2011) and the activity of DELLA proteins (Luo et al., 2013). Ethylene can also regulate auxin diffusion and biosynthesis (Valdovinos et al., 1967; Stepanova et al., 2005; Ruzicka et al., 2007; Swarup et al., 2007).
Here we characterize an activation tagged mutant of Populus tremula x P. alba clone 717, named “dwarfy,” exhibiting severe dwarfism with both reduced stature and smaller leaves. We show that the gene responsible for this phenotype is annotated as the poplar 1-aminocyclopropane-1-carboxylate synthase (ACS) gene PtaACS8. Ethylene is synthesized in two enzymatic steps from the substrate S-adenosyl-methionine (SAM). The first step is the conversion of SAM into 1-aminocyclopropane-1-carboxylic acid (ACC) by the activity of ACSs followed by the conversion of ACC to ethylene catalyzed by ACC OXIDASEs (ACOs). Ethylene is then perceived by a family of membrane bound receptors that induce the transcription of ETHYLENE RESPONSE FACTORs (ERFs) which, in turn, controls transcription and, ultimately, plant development. We demonstrate that increased expression of PtaACS8 in the dwarfy line results in significantly higher levels of ethylene in all aerial tissues of the plant. Morphologically, the increased expression of PtaACS8 in the stem results in shorter vessels and fibers in secondary growth while endogenous over-expression of the PtaACS8 gene in the leaves results in the production of fewer cells. The reduced growth of stem cells is accompanied by a repression of auxin transport and signaling genes while reduction in cell number in leaves is concurrent with a large reduction in the transcript abundance of a number of cell-cycle genes. Therefore, we conclude that increased expression of PtaACS8 induces stem dwarfism and reduced leaf size through separate genetic pathways.
Materials and Methods
Plant Material
All plants used in Figures 1, 4, 6 were grown under greenhouse conditions at the Canadian Forest Service (CFS), Fredericton, New Brunswick, Canada, while plants used for data analysis in Figures 2, 3, 5 were grown under greenhouse conditions at Queen's University, Kingston, Ontario, Canada. In the former situation, plants were grown under natural daylight and temperature, while in the latter situation, photoperiod was maintained at 16 h per day and temperature at 25°C. The dwarfy mutant was generated as described by Harrison et al. (2007) in a P. tremula x P. alba clone 717-1B4 background and all comparisons of dwarfy were made with this hybrid (wildtype). The dwarfy mutant was initially identified based on the dwarf characteristics such as plant height and leaf size among others in the mutant.
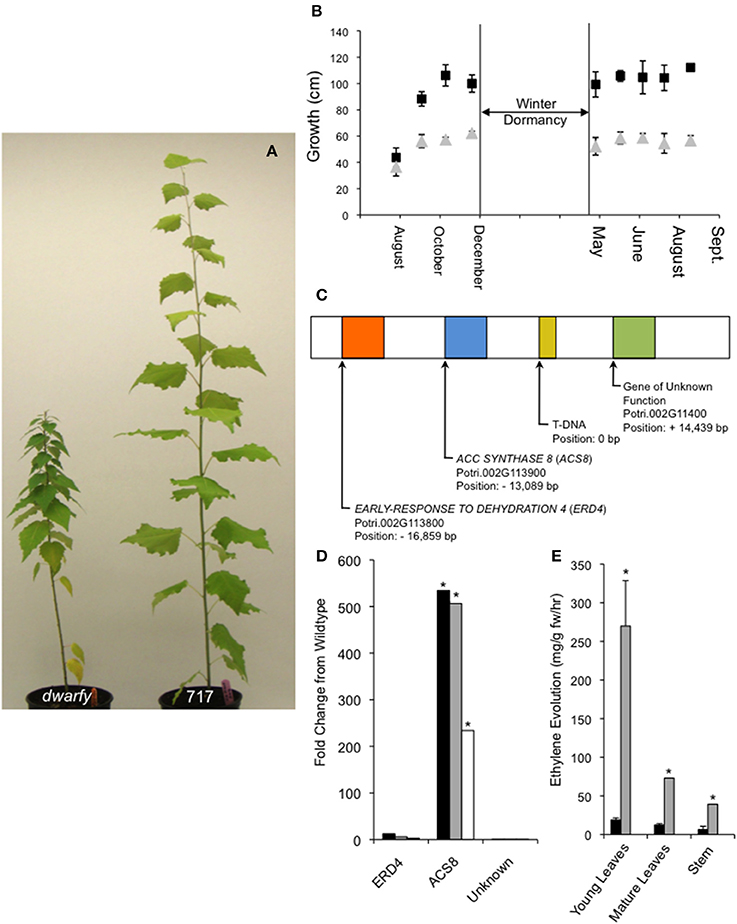
Figure 1. Endogenous over-expression of PtaACS8 results in a dwarfed growth phenotype in P. tremula x P. alba clone 717-1B4. (A) A representative image of the dwarfy mutant P. tremula x P. alba clone as compared to wildtype (717) after 4 months of growth. (B) Graphical mean height growth of dwarfy (gray data points) and wildtype (black data points) over two growth seasons. (C) Graphical representation of the genes found within a 30 kb window around the insertion of the activation tagging T-DNA in the dwarfy mutant. (D) Fold change in the genes found in the genomic vicinity of the T-DNA insertion in the dwarfy mutant as compared to wildtype in immature leaves (black bars), mature leaves (gray bars), stem (white bars). (E) Comparison of ethylene evolution in three different tissues of dwarfy (gray bars) and wildtype (black bars). All values ±Standard Error (SE). In (D,E), *significantly different from wildtype (p < 0.05).
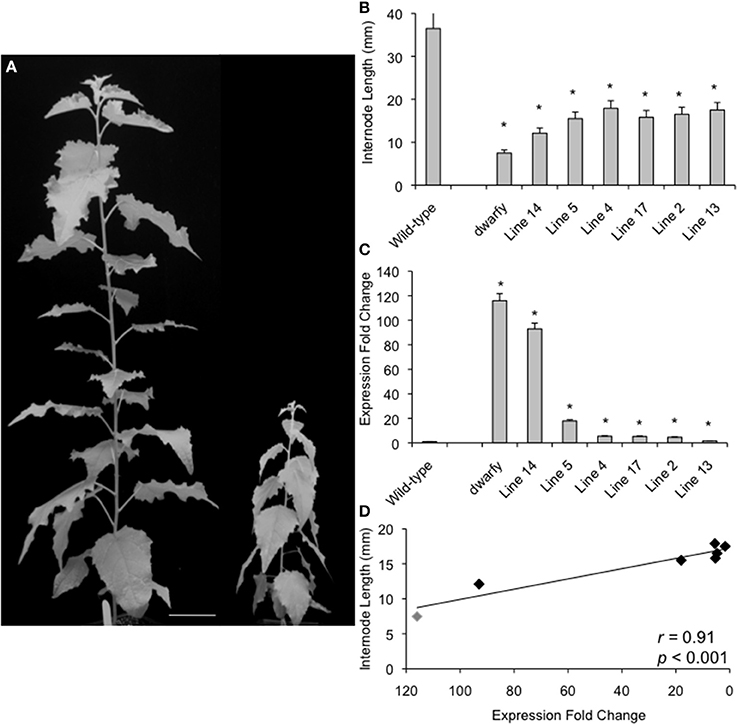
Figure 2. Plant growth retardation in dwarfy mutant is significantly correlated to the expression level of PtaACS8. (A) A representative image of one independent line of the 35S::PtaACS8 mutant P. tremula x P. alba clone as compared to wildtype (717) after 2 months of growth. Scale bar = 8 cm. (B) Relative expression of PtaACS8 in wildtype, dwarfy and 6 independent transgenic lines containing the 35S::PtaACS8 construct. (C) Internode length of wildtype, dwarfy and 6 independent transgenic lines containing the 35S::PtaACS8 construct. (D) Correlation between PtaACS8 expression levels and internode length in wildtype, dwarfy and 6 independent transgenic lines containing the 35S::PtaACS8 construct. All values ±Standard Error (SE). In (B,C), *significantly different from wildtype (p < 0.05).
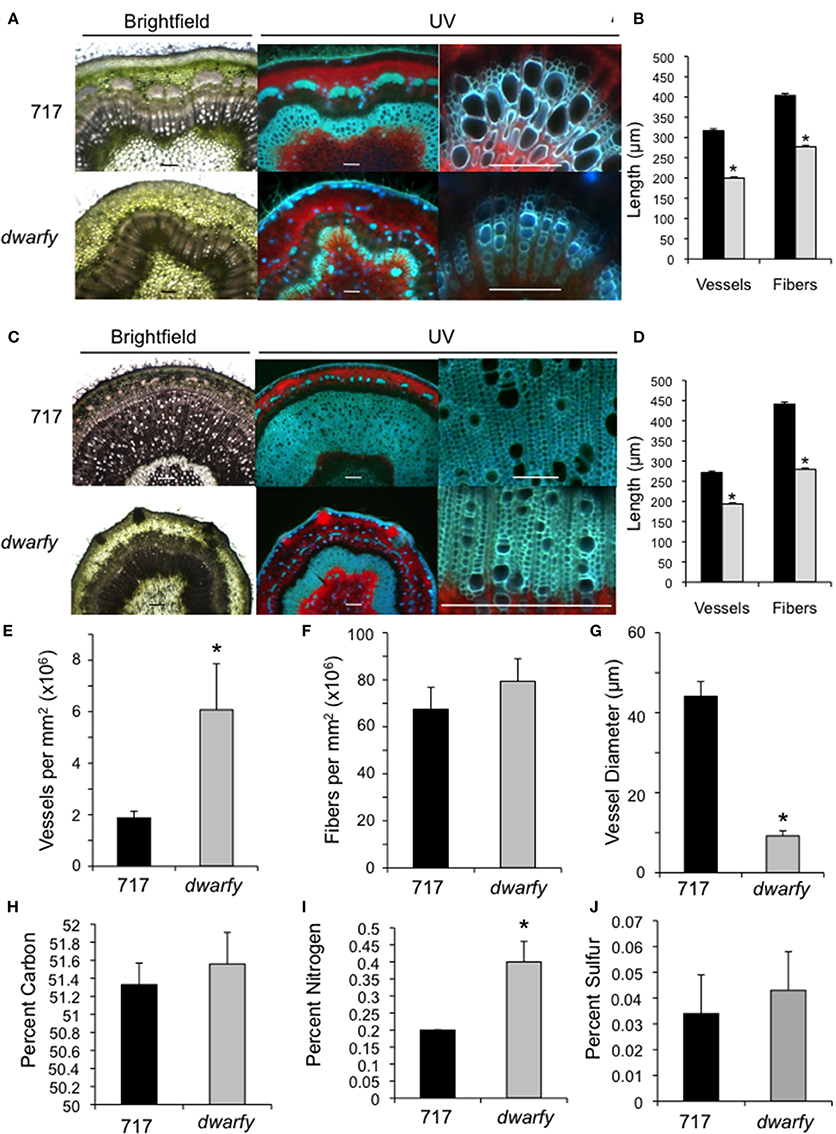
Figure 3. Elevated expression levels of PtaACS8 result in significant changes in stem architecture and physical characteristics. (A) Transverse cross section of wild-type and dwarfy stems at the leaf 10–11 internode as observed under brightfield and UV autofluorescence. Scale bar = 1 mm for first two images and 100 μm for the third panel (B) Vessel and fiber lengths of wild-type (black bars) and dwarfy (gray bars) stems between the leaf 10–11 internode. (C) Transverse cross section of wildtype and dwarfy stems at the leaf 20–21 internode as observed under brightfield and UV autofluorescence. Scale bar = 0.5 mm for first two images and 100 μm for the third panel. (D) Vessel and fiber lengths of wild-type (black bars) and dwarfy (gray bars) stems between the leaf 20–21 internode. Vessel (E) and fiber (F) density in wild-type and dwarfy stems between the leaf 20–21 internode. (G) Vessel diameter in wild-type and dwarfy stems between the leaf 20–21 internode. Relative percentage of carbon (H), nitrogen (I) and sulfur (J) in the stems of wildtype and dwarfy. All values ±Standard Error. *Significantly different from wildtype (p < 0.05).
T-DNA Insertion Analysis
Southern analysis of the dwarfy poplar mutant line was done and confirmed the presence of one T-DNA insertion event (Harrison et al., 2007). Genomic DNA (gDNA) was extracted from CFS greenhouse dwarfy mutant leaves using the cetyltrimethylammonium bromide (CTAB) method and gDNA was quantified using an Nanodrop1000 spectrophotometer and quality was checked on 0.8% (w/v) agarose Tris-acetate EDTA ethidium bromide gel. To identify the site of T-DNA insertion, the Genome Walker™ universal kit (Clontech, http://www.clontech.com) was used according to the manufacturer's protocol to create 4 restriction digested gDNA libraries. Each library was analyzed by primary and nested PCR using T-DNA vector specific primers (VSP 1 and VSP 2) designed from the T-DNA sequence and adapter primers AP1 & AP2 provided in the kit (Table S1). Primary PCRs were done on 1 μl of each library except using High Fidelity Platinum Taq (Invitrogen) for the PCR reaction mix. Primary PCR reactions from each library were diluted 50 times in H2O and 1 μl of the each dilution was used for nested PCR analysis using the same PCR reaction components except primers VSP2 & AP2 were used. Cycling parameters for both primary & nested PCRs were the same ones stated in the kit except that the elongation time was increased to 5 min. Primary and nested PCRs were analyzed by gel electrophoresis and bands from the nested PCR reaction that were over 1 kb in size were subcloned in pCR4-topo vector using the TOPO TA cloning kit (Invitrogen, http://www.invitrogen.com) and fully sequenced at the McGill University and Genome Quebec Innovation Centre (http://gqinnovationcenter.com). Based on flanking sequence information, a flanking gDNA primer FSP1 was designed and PCR was done on dwarfy gDNA using FSP1 and VSP 2 primers to confirm the location of the T-DNA insertion site. This amplicon was TOPO TA cloned and fully sequenced. Localisation of insertion site was determined by BLASTn using flanking sequence as query against the P. trichocarpa genome (Tuskan et al., 2006) (Populus trichocarpa v3.0, DOE-JGI, http://www.phytozome.net/poplar).
Affymetrix Array Analysis
For gene expression analysis, total RNA was extracted from 0.5 g of different tissues of wildtype and dwarfy taken from CFS greenhouse grown plants using a modification of Chang et al., 1993 and the RNeasy kit (QIAGEN). Total RNA quality and quantity was determined by Nanodrop1000 and by electrophoresis. Triplicate samples of dwarfy and wild-type leaf and stem total RNA were sent to the Microarray Centre (University Health Network, Toronto (UHN)) for sample processing and analysis. Sample quality was verified with the Agilent 2100 Bioanalyser before analysis with the GeneChip® Poplar Genome Array. Data was also analyzed by the Microarray Center (UHN) using Gene Spring software.
Isolation of ACS8 Candidate Gene Coding Sequence
The ACS8 cDNA was isolated from wild-type leaf tissues using the Smart RACE kit (Clontech, http://www.clontech.com) according to manufacturer's protocol. One microgram of total RNA was used to produce the 5′&3′RACE ready products and ACS8 gene specific primers ACS8RACE.fwd and ACS8RACE.rev were used along with the Universal Primer (UP) provided in the kit (Table S1). 5′&3′ RACE products were subcloned in pCR4-topo vector using the TOPO TA cloning kit and sequenced. Gene specific primers; ACS8-ATG.fwd and ACS8-Stop.rev primers were designed and used to isolate the full ACS8 CDS using the 3′RACE ready product previously generated and the amplicon was cloned in pBluescript II (+) (Fermentas, http://www.fermentas.com) using HindIII-XbaI restriction sites. The resulting construct carrying the full ACS8 CDS was fully sequenced.
Agrobacterium Tumefaciens Mediated Tranformation of P. Tremula x P. Alba Clone 717-1B4
In order to generate a binary plant transformation vector, ACS8/pBluescript II (+) construct was digested with EcoRI and subcloned in pART7 (Gleave, 1992) and verified by restriction digest for correct orientation between the CaMV promoter and ocs 3′ region. The ACS8/pART7 construct was subsequently digested with NotI and the whole cassette was ligated in the binary vector pART 27 (Gleave, 1992) prior to Agrobacterium tumefaciens transformation in line 717-1-B4 (Harrison et al., 2007). Out of 17 independent transgenic lines generated, 6 lines survived the transfer to greenhouse conditions and these lines were analyzed. Total RNA from leaf tissue was extracted from newly transformed lines and RT-qPCRs were done for gene expression analysis of lines generated using the procedures as stated below. Each line was analyzed in duplicate technical replicates. Internode lengths were measured after 3 months of growth.
Quantitative Gene Expression Analysis
For in gene expression analysis in transgenic 35S::PtaACS8 lines, total RNA was extracted from 100 mg of shoot apical tissue using the RNeasy kit (QIAGEN). Total RNA quality and quantity was determined by Nanodrop1000 and by electrophoresis. Two to four micrograms of total RNA was treated with Turbo DNaseI (Ambion, http://www.ambion.com) and RT-qPCR was done with 50 ng of total RNA/reaction using the one step Quantitect SYBR Green RT-PCR kit (QIAGEN, http://www.qiagen.com). RT-qPCR cycling conditions were: 30 min at 50°C for reverse transcriptase reaction and 15 min at 95°C for enzyme inactivation followed by 40 cycles of 15 s at 94°C, 15 s denaturation at 55°C (annealing) and 30 s at 72°C elongation followed by fluorescence measurement. The relative expression of PtaACS8 was compared to the UBQ10 reference gene (Plett et al., 2010).
The amplification efficiencies of each gene primer set were determined by E = 10[−1/slope] and were calculated using the slopes of n-fold serial dilution standard curves. Fold change ratios were determined using the comparative Ct method (ΔΔCt method) since amplification efficiencies were approximately equal in all target and reference genes measured in the study. Samples were analyzed in triplicates of each wildtype and the dwarfy mutant plants. Each total RNA sample was analyzed in duplicate. A No Reverse Transcriptase (NoRT) for each sample was included and a No Template Control (NTC) was included for each primer pair to make sure no contamination was present in the experiments. Amplicon specificity was confirmed by electrophoresis (single band at the right size), by melt curve analysis (single peak and Tm) and by sequencing.
Ethylene Determination
Leaf and stem samples were removed from wild-type and dwarfy poplar plants between 10 AM and 12 PM, and incubated in 20 mL headspace vials for 4 h at ambient temperature. Fresh weight was recorded and time between vial seal and sample injection were noted to have an exact incubation time. Ethylene content within this headspace was determined by gas chromatography coupled to a flame ionization detector (Gas Chromatography- Flame Ionization Detector GC-FID, Agilent 7890A) with an injector (splitless mode) temperature of 240°C and oven temperature at 60°C (isothermal) using helium as carrier gas (3 mL/min). A30 m × 0.53 mm ID (30 μm average thickness) Carboxen 1006 PLOT column (Supelco, Sigma Aldrich, Ontario Canada) was used to separate ethylene from the mixture. The FID (heated to 240°C) hydrogen:air:makeup flows were 30:400:25 (mL/min). Two measurements of 0.1 mL gas aliquot was taken from a headspace vial using a gas tight syringe (Hamilton 1700 series) and immediately injected for each sample. At these conditions, the observed retention time of ethylene was 3.14 min. A calibration curve was generated to cover the range of 0.1–20 mg ethylene. Ethylene standard gas mixture was made by drawing a volume of 99.5% ethylene (Praxair) and injecting it in a previously vacuum-purged sealed headspace vial (volume determined by water capacity), then breaking the vacuum with a syringe needle and filling the vial with ambient air to atmospheric pressure. The diluted ethylene was allowed to stand for 1 h to reach dispersal equilibrium. Increasing volumes were injected to cover the desired ethylene range and each injection was repeated in triplicate.
Physical Characteristics Analysis
Cuttings of wildtype and dwarfy were established by cutting 4–5 cm shoot explants from stock plants. Cuttings were planted in Jiffy 42 mm peat plugs grown under greenhouse conditions June to August (natural lighting, watered twice daily) for 7 weeks. After 7 weeks, cuttings were transferred to greenhouse pots (15 cm diameter, 19 cm long). After a total of 9 weeks, 3 trees were randomly selected every month and height was measured and data analyzed using basic statistical tools in Excel (Microsoft Office). Leaf cell size, trichome density and cell density were performed as per Plett et al. (2010). Fiber and vessel isolation and measurements were performed as per Chaffey et al. (2002).
GA and Ethylene Biosynthetic Inhibitor Growth Effect Analysis
Plants used for GA effect on growth were grown under normal greenhouse conditions as mentioned above. A triplicate (for GA) or a duplicate (for AVG) of wild-type and dwarfy plants of similar heights, grown for 5 weeks from cuttings, were used for each treatment for the experiment. Plant height was measured before the experiment and measured prior to each new application of GA or AVG. A total of 3 applications of 10 μl of 3 mM GA/water or ETOH (for GA analysis) or of water or 100 μM AVG were added every fourth day to the shoot apex of each plant and the total length of the experiment was 12 days. Data was analyzed using the height difference between the first measurement (before first application) and before 3rd application (3rd measurement), since 2 of the dwarfy/GA treated shoot apex samples had dried up and were dead before the final measurement.
Percent Carbon, Nitrogen and Sulfur Analysis
Dried leaf, stem, and roots samples from both wildtype and dwarfy were ground with a bead mill, and kept under vacuum in order to keep moisture out of the samples prior to carbon (C), nitrogen (N) and sulfur (S) analysis (CNS) by the CFS analytical laboratory according to the method of Kalra and Maynard (1991). A triplicate of each clone for each tissue types was measured and data was analyzed using basic statistical tools in Excel (Microsoft Office). Results presented are the measure of C, N and S from healthy mature leaves and internode tissues harvested in June of the growing season.
Results
Endogenous Over-Expression of PtaACS8 Induces Dwarfism in Populus
From a large population of activation-tagged P. tremula x P. alba clone 717-1B4 (2000 independent transgenic lines; Harrison et al., 2007), we identified one line with a consistent reduction in growth rate over multiple growing seasons (Figures 1A,B). This mutant was named dwarfy. Using Southern blotting only one T-DNA insert in dwarfy and located this insert on chromosome 2 using a modified TAIL PCR was identified. Within a window of ±20 Kb around the T-DNA, 3 genes annotated in Phytozome (Figure 1C) were found as follows: a gene of unknown function (Potri.002G11400; +14.4 Kb up-stream), PtaACC SYNTHASE8 (PtaACS8; Potri.002G113900; 13.1 Kb down-stream) and PtaEARLY-RESPONSE TO DEHYDRATION 4 (ERD4; Potri.002G113800; 16.9 Kb down-stream). A quantification of the expression of these genes in the dwarfy mutant line relative to wild-type P. tremula x P. alba clone 717-1B4 demonstrated that only PtaACS8 exhibited increased gene expression in all aerial tissues of the plant (Figure 1D). As ACC synthases are involved in the first step in the biosynthesis of the plant hormone ethylene, ethylene production was measured in the same three compartments as used for gene expression analysis in wild-type and mutant plants (i.e., young and mature leaves and stem tissues). Compared to wildtype, the mutant line produced 14× higher levels of ethylene in younger leaves and 6× higher levels of ethylene in mature leaves and the stems (Figure 1E).
To verify that increased transcript abundance of PtaACS8 was indeed responsible for the dwarfism phenotype of the mutant, the Potri.002G113900 gene was cloned and expressed ectopically in the P. tremula x P. alba clone 717-1B4 genetic background under the control of the 35S-cauliflower mosaic virus promoter. We were able to regenerate six independent transgenic lines from callus culture which, when grown alongside age-equivalent wildtype (i.e., propagated at the same time and treated in the same manner as the 35S::PtaACS8 lines), displayed a dwarf phenotype (Figure 2A). This reduction in growth and internode length was significant as compared to wildtype in all lines tested although the plants were consistently bigger than dwarfy (Figures 2A,B). The discrepancy in height difference is likely due to the fact that none of the 35S::PtaACS8 transgenic lines displayed the same level of PtaACS8 transcript accumulation as dwarfy (Figure 2C). As there was a significant correlation between the transcript abundance of PtaACS8 and the dwarf phenotype in the transgenic lines (Figure 2D; r = 0.91; p < 0.001), we conclude that increased transcript abundance of PtaACS8 in the original dwarfy transgenic line is responsible for the reduction in plant stature.
Increased Transcript Abundance of PtaACS8 Leads to Altered Stem Characteristics
The dwarfy mutant line exhibited alterations to the morphology of all aerial parts of the plant. While the internode length of the dwarfy line was significantly reduced (Figure 2B), there were also significant alterations to the microscopic anatomy of the stem (Figure 3). Due to the great difference in height of the two plants being compared, we used a plastochron index to identify and compare the same internode between the mutant line and wildtype. We used different microscopy techniques to observe different stem properties: brightfield to gain a general over-view of the stem architecture, UV excitation to differentiate chlorophyll autofluorescence (red signal) from secondary cell wall fluorescence (blue-green signal; Figures 3A,C). In young stems (internode between leaves 10 and 11), there was a reduction in the amount of secondary xylem formed in dwarfy as compared to wildtype (Figure 3A) as well as a significant reduction in the length of xylem fibers and vessels (Figure 3B; p < 0.05). In older stem tissues (internode between leaves 20 and 21), the reduction in secondary xylem formation (Figure 3C) and fiber/vessel lengths were still observed (Figure 3D). Detailed analysis of wood formation in these older tissues also revealed a difference in cell density: dwarfy had a higher density of xylem vessels per square millimeter with a significantly smaller outer diameter as compared to wild-type stems (Figures 3A,C,E–G) while there was no significant difference in the density of fibers. As alterations to the cell make-up of the stem and alteration in growth rate may influence nutrient deposition in the stem, we analyzed the percentage of carbon, nitrogen and sulfur in these mature internodes of both wildtype and dwarfy. No significant difference in percent accumulation of carbon and sulfur in the stems of dwarfy and wildtype were observed while the stems of the former accumulated a significantly higher concentration of nitrogen-containing compounds (Figures 3H–J; p < 0.05).
Hormone- and Nutrient-Related Genes Display Altered Abundance in Dwarfy Stems
In order to understand the transcriptomic profile of dwarfy stems, we performed a whole genome oligo-array transcriptomic analysis of whole stem tissues. We found 223 genes differentially expressed (≥2-fold; p < 0.05) as compared to wild-type P. tremula x P. alba clone 717-1B4 stems of the same age (Table S2). Within these genes we found that there were a number of ethylene and auxin related genes and genes coding for proteins involved in nutrient transport and biosynthesis (Table 1). Genes related to the ethylene pathway included PtaACS8 (>230-fold increase) a number of ETHYLENE RESPONSE FACTOR (ERF) genes, two serine-threonine receptor kinases (PtaCTR3, PtaCTR4) and two ethylene receptor genes (PtaETR1, PtaETR5). The majority of genes associated with the auxin pathway, meanwhile, were repressed in the stems of dwarfy while a gene encoding an IAA-amido-synthetase glycosyl-hydrolase (GH) family protein displayed increased abundance. Nutrient transport and synthesis was also affected with two sugar transporters and an amino acid transporter being repressed while the transcript accumulation of a glutamine synthase was increased (Table 1).
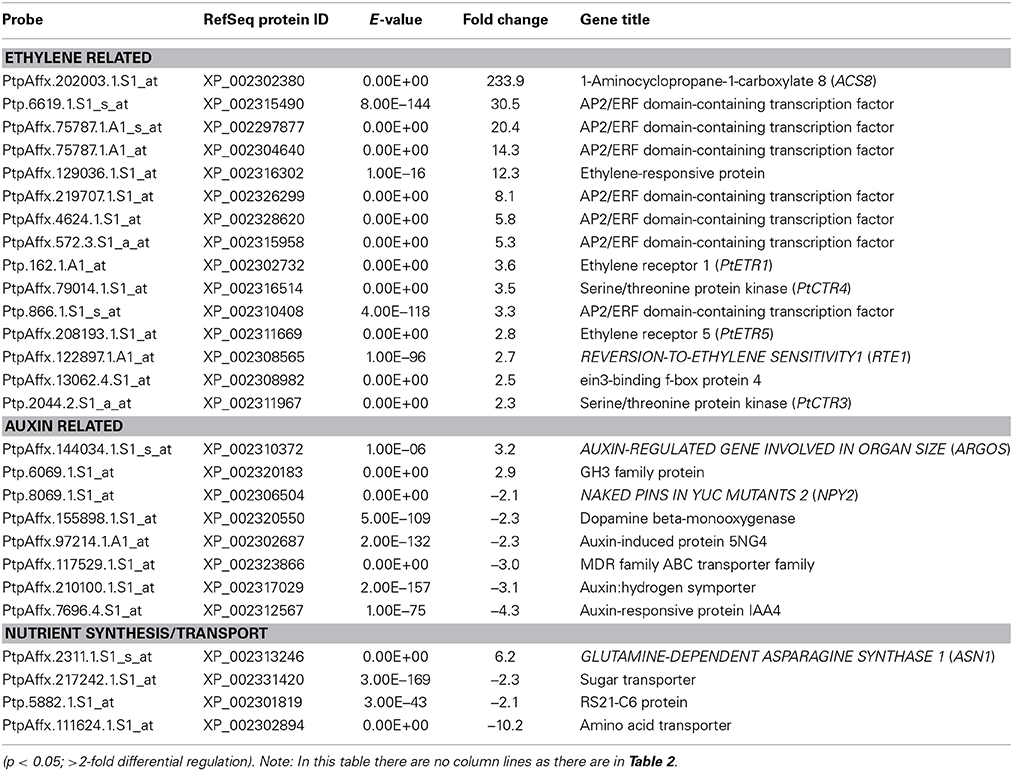
Table 1. Genes found to have significantly different abundance in the stems of dwarfy as compared to wildtype.
GA has been linked to enhanced growth phenotypes through the induction of auxin biosynthesis and polar transportation (Björklund et al., 2007). Therefore, as our transcriptional analysis of the dwarfy mutant indicated that auxin transport and signaling was affected, we tested whether GA application to the growing apex of dwarfy would be able to rescue the growth phenotype of the mutant. We found that the growth rate of dwarfy was significantly increased by treatment with GA (Figures 4A,B). Therefore GA is able to rescue the dwarfy phenotype. We also treated dwarfy with the ethylene biosynthetic inhibitor AVG. This treatment resulted in an increase in internode length (Figures 4C–E), demonstrating that blocking ethylene synthesis also rescues the dwarfy phenotype.
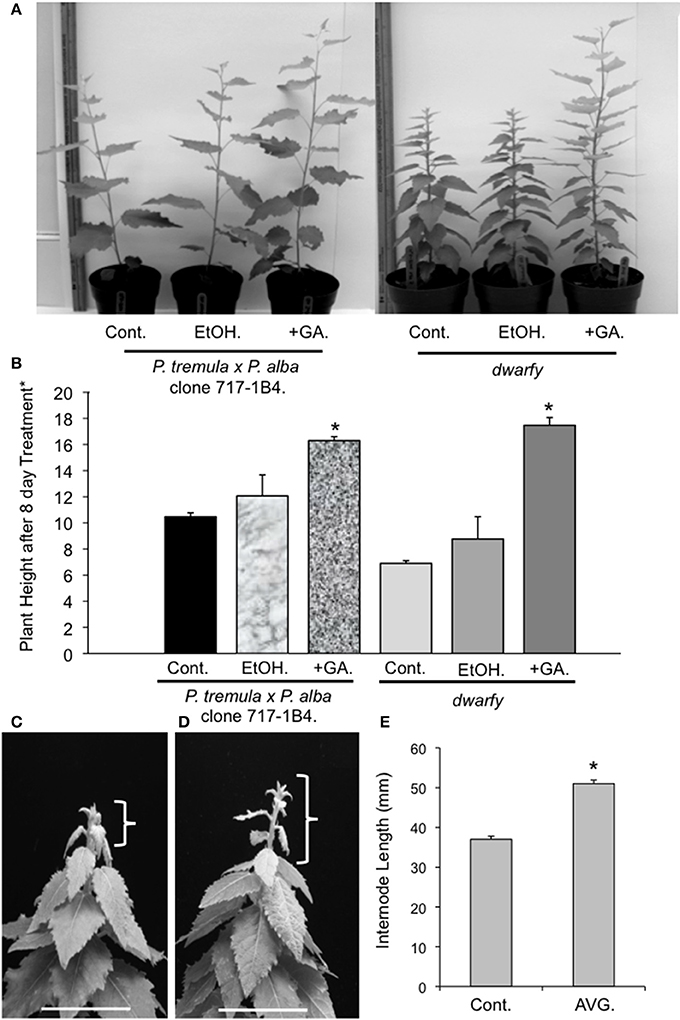
Figure 4. Application of GA and AVG to dwarfy apexes induces faster growth rate. (A) Representative image of GA influence on the growth rates of dwarfy and wildtype (+GA) as compared to ethanol control (+EtOH) and untreated control (Cont.). (B) Mean heights of wild-type and dwarfy saplings treated with GA (+GA) as compared to ethanol control (+EtOH) and untreated control (Cont.). (C) Representative image of water and (D) AVG influence on the growth rates of dwarfy and wild-type. Parentheses indicate growth of main stem for the treatment period. Scale bar = 3 cm. (E) Mean internode lengths wild-type and dwarfy saplings treated with AVG as compared to water control (Cont.). All values ±SE. *Significantly different from wildtype (p < 0.05).
Increased Transcript Abundance of PtaACS8 Leads to Altered Leaf Characteristics
Mature leaves in the dwarfy mutant also showed altered size when compared to wild-type leaves (Figure 5A). The leaves of dwarfy were much smaller than those of wildtype (Figure 1A). Despite the alterations in leaf size, the epidermal cell size of dwarfy was not significantly altered compared to wildtype (Figure 5B). Trichome density was also not affected, but stomate density was significantly higher in the dwarfy mutant (Figures 5C,D). Only nitrogen content was significantly higher in dwarfy stems, as compared to wildtype (Figure 3I). Unlike stems, the total percentage of carbon in leaves was significantly reduced in dwarfy as compared to wildtype (Figure 5E), while nitrogen levels were not altered (Figure 5F). Sulfur levels in mutant leaves showed a tendency toward a lower accumulation compared to wildtype, but this difference was not found to be significant (Figure 5G; p < 0.05). The date at which leaves became chlorotic and dropped off the stem in dwarfy as compared to wild-type plants was assessed as increased ethylene levels have been correlated to early leaf senescence (Breeze et al., 2011; Koyama et al., 2013). When grown under natural conditions, chlorosis of 1-year-old dwarfy leaves happens earlier as compared to wild-type plants (Figures 6A–D) and significant leaf drop occurred in dwarfy plants in the month of November while there was no significant leaf drop in the same period in wild-type trees (Figures 6E,F). It is interesting to speculate that the reduced C and S observed in the dwarfy leaves may be related to the shorter growing season for these leaves.
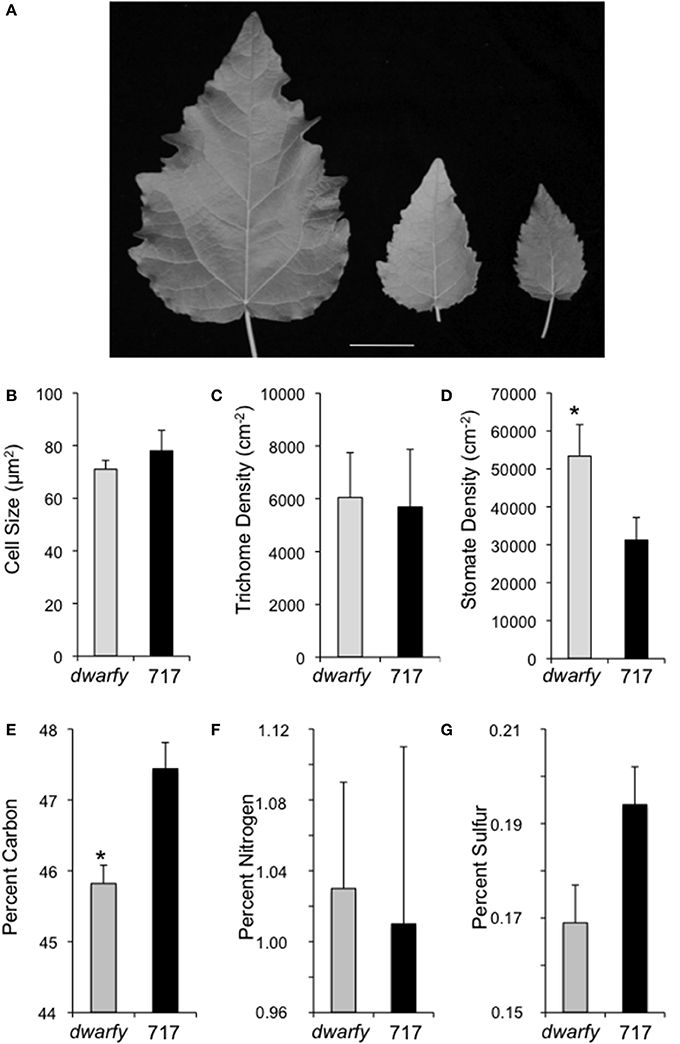
Figure 5. Elevated expression levels of PtaACS8 result in significant changes in leaf architecture and physical characteristics. (A) A representative image of fully expanded leaves of wildtype, one independent line of the 35S::PtaACS8 mutant P. tremula x P. alba clone 717-1B4 and dwarfy, respectively, after 2 months of growth. Scale bar = 2 cm. Epidermal cell density (B), trichome density (C) and stomate density (D) in fully expanded leaves of dwarfy (gray bars) as compared to wild-type leaves (black bars). Relative percentage of carbon (E), nitrogen (F), and sulfur (G) in mature leaves of wildtype (black bars) and dwarfy (gray bars). All values ±SE. *Significantly different from wildtype (p < 0.05).
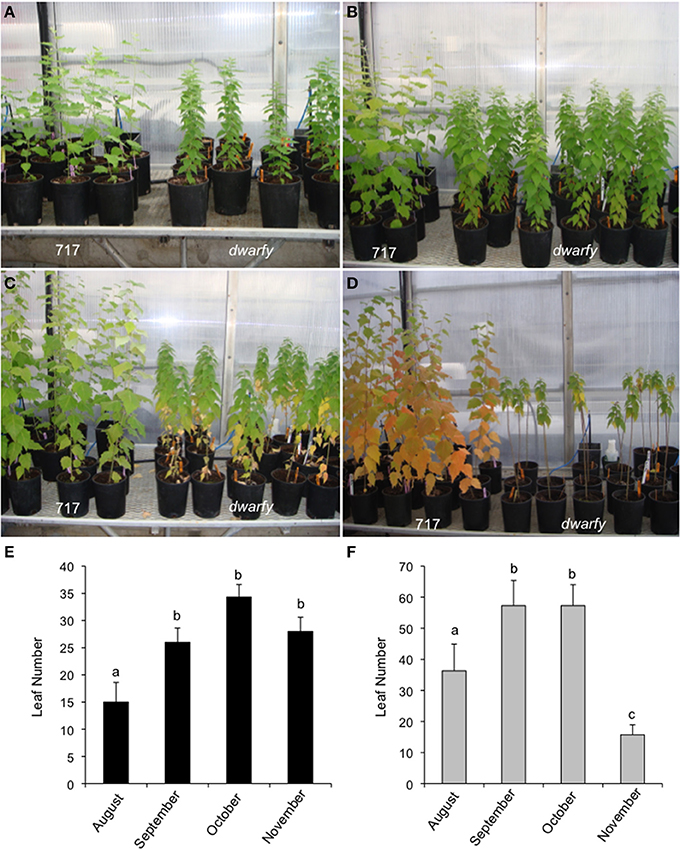
Figure 6. dwarfy mutants exhibit pre-mature leaf senescence. Comparison of leaf senescence rates in wildtype and dwarfy mutant clones within their first year of growth in August (A), September (B), October (C), and November (D). Leaf numbers on wild-type trees (E) and dwarfy trees (F) over the same time period are presented. ±SE; superscript letters indicate significant differences between treatments as determined by One-Way analysis of variance (ANOVA) followed by a Tukey HSD (honestly significant difference) multiple comparison test (p < 0.05).
Senescence- and Cell Cycle-Related Genes Exhibit Altered Abundance in Dwarfy Leaves
We found that 183 genes were significantly regulated in fully expanded leaves of dwarfy as compared to wild-type P. tremula x P. alba clone 717-1B4. A large number of hormone-related genes with altered transcription were observed in the stems of dwarfy, while only two of these genes (PtaACS8 and a GH3 family protein) were significantly differentially regulated in mature dwarfy leaves (≥2-fold; p < 0.05; Table 2; Table S3). A number of nutrient transporters displayed altered transcript abundance, although they were different from those identified in dwarfy stems (Table 1). Three other classes of genes were differentially regulated in mature dwarfy leaves that were not observed in the stems: defense-, senescence- and cell cycle/expansion-related genes (Table 2). The majority of the defense-related genes were associated with pathogen attack, including a chitinase, a lipase and a glyoxal oxidase. As the leaf tissues were healthy at the time of harvest and displayed no infection structures, the activation of these genes is likely constitutive in the dwarfy background. Three genes associated with leaf senescence were also up-regulated. One group that only showed reduced levels of transcript abundance was that of genes associated with cell cycle and cellular growth (Table 2). Within this group of genes were a number of cyclins, calmodulin-like proteins and one expansin.
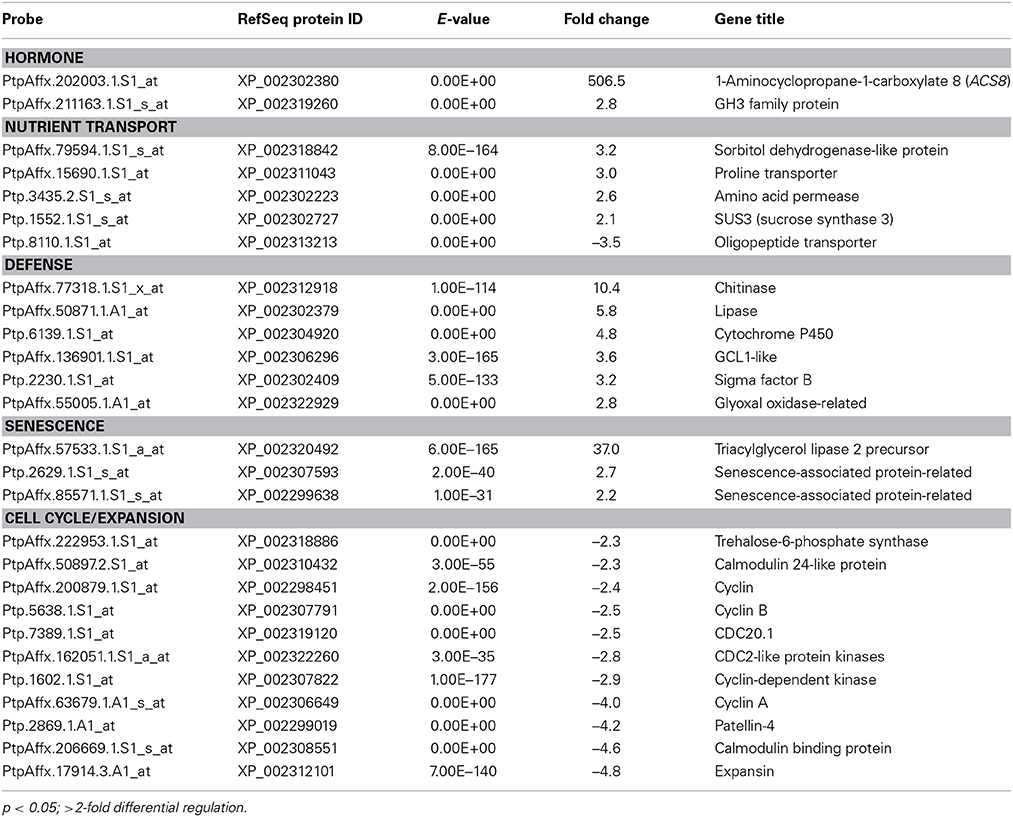
Table 2. Genes found to have significantly different abundance in the leaves of dwarfy as compared to wildtype.
Discussion
Due to ornamental value and to wide-ranging applications within agriculture, the genetic traits that control cell size and dwarfism in plants have been widely studied (Valdovinos et al., 1967; Ephritikhine et al., 1999; Busov et al., 2003; Qi et al., 2011; Luo et al., 2013; Li et al., 2014; Yang et al., 2014; Zhang et al., 2014). Through breeding practices and applications of growth regulators, dozens of different dwarf plant varieties have been produced over the past few decades (Parker, 2012; Jiang et al., 2013; Wang et al., 2014a,b). Largely studied in annual models such as Arabidopsis, Zea and Oryza, control of plant stature has been linked most readily to plant hormones. For example, Arabidopsis mutants with increased ethylene production (e.g., eto; Woeste et al., 1999) exhibit thickening of the hypocotyl while increased ethylene signaling (e.g., ctr1-1; ACS6DDD; Liu and Zhang, 2004) has been found to result in reduced stature and smaller leaf size. Due to advancements in insertional mutagenesis and other transgenic technologies, it is now becoming feasible to also screen perennial plants for the genes that control dwarfism (Busov et al., 2003; Harrison et al., 2007; Vahala et al., 2013). Here we characterize a mutant line of P. tremula x P. alba clone 717-1B4 that exhibits higher transcript accumulation of PtaACS8 and that produces a significantly higher level of ethylene in all aerial tissues as compared to wild-type trees. Increased transcript abundance of PtaACS8 is correlated to reduced plant stature and smaller leaves while treatment of dwarfy shoots induces increases in internode length. The transcriptional cascade induced by altered levels of PtaACS8 are very different in stem and leaf tissue with a transcriptional reduction in genes associated with auxin transport and signaling evident in stems and repressed cell cycle genes in the leaves. These results place PtaACS8, and likely ethylene, as regulators that control two major morphological traits associated with dwarfism and reduced tissue size.
Using transformation technologies such as activation tagging is a very useful approach to identifying and characterizing the role of genes in a physiologically relevant manner. Rather than ectopic over-expression of a gene, the inserted enhancer used in activation tagging only enhances expression in its native expression pattern. This mutagenesis approach has been used in a number of model plant systems including Arabidopsis (Weigel et al., 2000), tomato (Mathews et al., 2003), rice (Jeong et al., 2006), and poplar (Harrison et al., 2007). Using this approach Busov et al. (2003) were able to identify a poplar GA2-OXIDASE that resulted in a plant with a very similar phenotype to that described here for dwarfy; reduced stature and smaller leaves. Since their publication, dwarfism in a native dwarf plum tree cultivar has also been linked to a GA2-OXIDASE (El-Sharkawy et al., 2012) demonstrating that findings from activation tagging studies can be extended to natural plant populations.
As opposed to a strictly GA-dependent phenotype, our results support the hypothesis that growth retardation in dwarfy is driven largely by ethylene, the endpoint of the biochemical pathway in which PtaACS8 operates. This is based on the evidence that increased PtaACS8 transcripts are correlated to significant increases in ethylene production in the stem (Figure 1E), whose signal is being relayed by the activation of several ERF genes (Table 1). Our results also demonstrate that blocking of ethylene biosynthesis resuces the dwarfy phenotype (Figures 4C–E). Further, the reduction in xylem fiber and vessel length described here-in has also previously been observed after ethylene treatment of poplar stems (Junghans et al., 2004; Love et al., 2009; Vahala et al., 2013). While we cannot rule out the possibility that the reduced stature in dwarfy is a result of increased ACC accumulation, our results support the hypothesis that stunting of the dwarfy stem is controlled in an ethylene-dependent manner. Increased ethylene, however, is likely not the only causative factor in explaining the stature of dwarfy. Rather, ethylene appears to be influencing another pathway associated with plant stature: the auxin pathway. We found evidence for a repression of auxin-homeostasis and transport genes in the stem of dwarfy (Table 1). Ethylene has long been tied to a negative effect on auxin diffusion (von Guttenberg and Steinmetz, 1947; Morgan and Gausman, 1966; Valdovinos et al., 1967; Suttle, 1988; Andersson-Gunneras et al., 2003; Ruzicka et al., 2007; Stepanova et al., 2007; Swarup et al., 2007). As inhibition of auxin diffusion has been correlated to a reduction in stem cell elongation of poplar (Junghans et al., 2004), pea (Lantican and Muir, 1969), tomato (Higashide et al., 2014), tulip (Okubo and Uemoto, 1985), Arabidopsis (Franklin et al., 2011; Chae et al., 2012), gourds (Wang et al., 2014a) amongst many other systems. Our results give a molecular framework by which ethylene affects dwarfy height where increased expression of PtaACS8 results in greater production of ethylene which, upon perception in plant stem tissue, represses genes related to auxin diffusion and synthesis which would then curtail cell elongation in the stem. GA treatment of growth apexes can also rescue the dwarfy phenotype, although we cannot conclude from present data if GA generates this phenotype by acting downstream of the ethylene signal in the dwarfy mutant or in a separate pathway.
A different genetic pathway is likely responsible for the observed reduction in leaf size in dwarfy. While increases in PtaACS8 transcripts and ethylene evolution in the stem coincided with stunted fiber and vessel growth, no change in leaf epidermal cell size is observed despite higher levels of PtaACS8 transcripts and higher ethylene evolution in the leaves. This would indicate that the leaf is smaller due to the absolute number of cells making up the tissue rather than the size of cell generated. It is interesting in the leaves of dwarfy that we see no evidence of compensation by leaf epidermal cells to maintain a larger leaf area. “Compensation” occurs when upstream inhibition of cell division initiates a secondary signaling pathway that increases cell size to maintain proper tissue growth (Hemerly et al., 1995; DeVeylder et al., 2001; Tsukaya, 2002; Horiguchi et al., 2006). Ethylene treatment has been associated with both stimulation of cell division (Love et al., 2009) and inhibition of cellular division (Edwards and Miller, 1972; Lee and LaRue, 1992; Heidstra et al., 1997; Dubois et al., 2013; Luo et al., 2013). In Arabidopsis, ethylene has been associated with reduced petal and leaf size in mutants with constitutive ethylene signaling (Kieber et al., 1993; Roman and Ecker, 1995; Luo et al., 2013) and under water limiting conditions due to the activity of ERF6 through its control of GA2-OXIDASE (Dubois et al., 2013). In the transcriptomic analysis of dwarfy leaves we do not see evidence of either ERF or GA2-OXIDASE genes accumulating at altered abundances. Rather, within the group of genes regulated in dwarfy leaves, we observed the repression of a large class of cell cycle genes including CYCLIN-DEPENDENT KINASE1 (CDK1), CYCLIN A, and CYCLIN B1 (Table 2). In eukaryotic cells, CYCLIN A initiates the cellular transition from G2 to prophase after which CYCLIN B1 enters the nucleus and, together with CDK1, induces mitosis by phosphorylation and activation of enzymes regulating chromatin condensation, nuclear membrane breakdown and mitosis-specific microtubule and microfilament re-orientation (Nigg, 2001; Smits and Medema, 2001; Gavet and Pines, 2010; Suryadinata et al., 2010; Rattani et al., 2014). As this whole suite of proteins is necessary for cellular division, repression of their transcription in the leaves of dwarfy, as compared to wild-type leaves, is likely the key pathway by which leaf size is being affected. These results are reminiscent of earlier observations that ethylene in Pisum sativum stopped cell division prior to entry into prophase (Apelbaum and Burg, 1972).
The leaf drop date of natural grown-year old dwarfy and wild-type plants was assessed as increased ethylene levels have been correlated to early leaf senescence (Breeze et al., 2011; Koyama et al., 2013). Leaf yellowing, considered to be the first visible senescent event (Quirino et al., 2000) was present in only the dwarfy basal leaves in October (Figure 6C), while in the wild-type, senescence related-color changes were prevalent in November basal leaves (Figure 6D) by which time dwarfy basal leaves had dehisced. Buchanan-Wollaston et al. (2003) noted that plants exposed to exogenous ethylene do exhibit premature senescence with the older leaves yellowing first; similar to the results here-in. November dwarfy and wild-type plants both had green apical leaves suggesting the onset of senescence and leaf dehiscence in all but the apical leaves in the dwarfy mutant were altered.
Our results support the hypothesis that there are two different developmental programs regulating tissue size in the dwarfy mutant. In the stem, we observe alterations to ethylene response factors and an inhibition of auxin homeostasis genes suggesting that ethylene inhibits stem elongation as previously observed in model organisms such as Arabidopsis (Guzman and Ecker, 1990), poplar (Junghans et al., 2004; Love et al., 2009; Vahala et al., 2013), tobacco (Romano et al., 1993), and tomato (Huang and Lin, 2003) through its influence on the auxin pathway. In the leaves, we find that the leaves of dwarfy produce fewer cells and are, thereby, smaller. This phenotype is likely tied to the differential expression of the protein group responsible for the induction of mitosis. Altogether, our study of the dwarfy mutant poplar has given insight into the genetics underpinning ethylene-induced dwarfism.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fpls.2014.00514/abstract
Table S1. Primers used in this study.
Table S2. Genes differentially regulated in the stems of dwarfy as compared to wildtype stems.
Table S3. Genes differentially regulated in the leaves of dwarfy as compared to wild-type leaves.
References
Adkins, S., Foale, M., and Harries, H. (2010). “Growth and production of coconut. Soils, plant growth and crop production,” in Encyclopedia of Life Support Systems (EOLSS), Developed Under the Auspices of UNESCO (Oxford, UK: Eolss Publishers). Available online at: http://greenplanet.eolss.net/EolssLogn/mss/C10/E1-05A/E1-05A-40/E1-05A-40-TXT.aspx
Andersson-Gunneras, S., Hellgren, J. M., Bjorklund, S., Regan, S., Moritz, T., and Sundberg, B. (2003). Asymmetric expression of a poplar ACC oxidase controls ethylene production during gravitational induction of tension wood. Plant J. 34, 339–349. doi: 10.1046/j.1365-313X.2003.01727.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Apelbaum, A., and Burg, S. P. (1972). Effect of ethylene on cell division and deoxyribonucleic acid synthesis in Pisum sativum. Plant Physiol. 50, 117–124. doi: 10.1104/pp.50.1.117
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Beemster, G. T., de Veylder, L., Vercruysse, S., West, G., Rombault, D., van Hummelen, P., et al. (2005). Genome-wide analysis of gene expression profiles associated with cell cycle transitions in growing organs of Arabidopsis. Plant Physiol. 138, 734–743. doi: 10.1104/pp.104.053884
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Beemster, G. T., Fiorani, F., and Inze, D. (2003). Cell cycle: the key to plant growth control? Trends Plant Sci. 8, 154–158. doi: 10.1016/S1360-1385(03)00046-3
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Björklund, S., Antti, H., Uddestrand, I., Moritz, T., and Sundberg, B. (2007). Cross-talk between gibberellin and auxin in development of Populus wood: gibberellin stimulates polar auxin transport and has a common transcriptome with auxin. Plant J. 52, 499–511. doi: 10.1111/j.1365-313X.2007.03250.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Breeze, E., Harrison, E., McHattie, S., Hughes, L., Hickman, R., Hill, C., et al. (2011). High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell 23, 873–894. doi: 10.1105/tpc.111.083345
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Buchanan-Wollaston, V., Earl, S., Harrison, E., Mathas, E., Navabpour, S., Page, T., et al. (2003). The molecular analysis of leaf senescence - a genomic approach. Plant Biotech. J. 1, 3–22. doi: 10.1046/j.1467-7652.2003.00004.x
Busov, V. B., Meilan, R., Pearce, D. W., Ma, C., Rood, S. B., and Strauss, S. H. (2003). Activation tagging of a dominant gibberellin catabolism gene (GA 2-OXIDASE) from poplar that regulates tree stature. Plant Physiol. 132, 1283–1291. doi: 10.1104/pp.103.020354
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chae, K., Isaacs, C. G., Reeves, P. H., Maloney, G. S., Muday, G. K., Nagpal, P., et al. (2012). Arabidopsis SMALL AUXIN UP RNA63 promotes hypocotyl and stamen filament elongation. Plant J. 71, 684–697. doi: 10.1111/j.1365-313X.2012.05024.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chaffey, N., Cholewa, E., Regan, S., and Sundberg, B. (2002). Secondary xylem development in Arabidopsis: a model for wood formation. Physiol. Plant. 114, 594–600. doi: 10.1034/j.1399-3054.2002.1140413.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chang, S., Puryear, J., and Carney, J. (1993). A simple and efficient method for isolating RNA from pine trees. Plant Mol. Biol. Rep. 11, 113–116. doi: 10.1007/BF02670468
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cho, H. T., and Cosgrove, D. J. (2000). Altered expression of EXPANSIN modulates leaf growth and pedicel abscission in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 97, 9783–9788. doi: 10.1073/pnas.160276997
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Curtis, I. S., Hanada, A., Yamaguchi, S., and Kamiya, Y. (2005). Modification of plant architecture through the expression of GA 2-OXIDASE under the control of an estrogen inducible promoter in Arabidopsis thaliana. Planta 222, 957–967. doi: 10.1007/s00425-005-0037-7
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
DeVeylder, L., Beeckman, T., Beemster, G. T. S., Krols, L., Terras, F., Landrieu, I., et al. (2001). Functional analysis of cyclin-dependent kinase inhibitors of Arabidopsis. Plant Cell 13, 1653–1667. doi: 10.1105/tpc.13.7.1653
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Dijkstra, C., Adams, E., Bhattacharya, A., Page, A. F., Anthony, P., Kourmpetli, S., et al. (2008). Over-expression of a GIBBERELLIN 2-OXIDASE gene from Phaseolus coccineus L. enhances gibberellin inactivation and induces dwarfism in Solanum species. Plant Cell Rep. 27, 463–470. doi: 10.1007/s00299-007-0471-z
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Domingo, C., Andres, F., Tharreau, D., Iglesias, D. J., and Talon, M. (2009). Constitutive expression of OsGH3.1 reduces auxin content and enhances defense response and resistance to a fungal pathogen in rice. Mol. Plant Microbe Interact. 22, 201–210. doi: 10.1094/MPMI-22-2-0201
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Donnelly, P. M., Bonetta, D., Tsukaya, H., Dengler, R. E., and Dengler, N. G. (1999). Cell cycling and cell enlargement in developing leaves of Arabidopsis. Dev. Biol. 215, 407–419. doi: 10.1006/dbio.1999.9443
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Dubois, M., Skirycz, A., Claeys, H., Maleux, K., Dhondt, S., De Bodt, S., et al. (2013). ETHYLENE RESPONSE FACTOR6 acts as a central regulator of leaf growth under water-limiting conditions in Arabidopsis. Plant Physiol. 162, 1319–1332. doi: 10.1104/pp.113.216341
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Edwards, M. E., and Miller, J. H. (1972). Growth regulation by ethylene in fern gametophytes. II. Inhibition of cell division. Am. J. Bot. 59, 450–457.
El-Sharkawy, I., El-Kayal, W., Prasath, D., Fernandex, H., Bouzayen, M., Svircev, A. M., et al. (2012). Identification and genetic characterization of GIBBERELLIN 2-OXIDASE gene that controls tree stature and reproductive growth in plum. J. Exp. Bot. 63, 1225–1239. doi: 10.1093/jxb/err345
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ephritikhine, G., Gellner, M., Vannini, C., Lapous, D., and Barbier-Brygoo, H. (1999). The sax1 dwarf mutant of Arabidopsis thaliana shows altered sensitivity of growth responses to abscisic acid, auxin, gibberellins and ethylene and is partially rescued by exogenous brassinosteroid. Plant J. 18, 303–314. doi: 10.1046/j.1365-313X.1999.00454.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Faeth, S. H. (1991). Effect of oak leaf size on abundance, dispersion and survival of the leafminer Cameraria sp. (Lepidoptera: Gracillariidae). Environ. Ent. 20, 196–204.
Franklin, K. A., Lee, S. H., Patel, D., Kumar, S. V., Spartz, A. K., Gu, C., et al. (2011). Phytochrome-interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc. Nat. Acad. Sci. U.S.A. 108, 20231–20235. doi: 10.1073/pnas.1110682108
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gavet, O., and Pines, J. (2010). Progressive activation of CyclinB1-Cdk1 co-ordinates entry to mitosis. Develop. Cell 18, 533–543. doi: 10.1016/j.devcel.2010.02.013
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Geisler, M., Blakeslee, J. J., Bouchard, R., Lee, O. R., Vincenzetti, V., Brandyopadhyay, A., et al. (2005). Cellular efflux of auxin mediated by the Arabidopsis MDR/PGP transporter AtPGP1. Plant J. 44, 179–194. doi: 10.1111/j.1365-313X.2005.02519.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Geisler, M., Kolukisaoglu, H. U., Bouchard, R., Billion, K., Berger, J., Saal, B., et al. (2003). TWISTED DWARF1, a unique plasma membrane-anchored immunophilin-like protein, interacts with Arabidopsis multidrug resistance-like transporters AtPGP1 and AtPGP19. Mol. Biol. Cell 14, 4238–4249. doi: 10.1091/mbc.E02-10-0698
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Geisler, M., and Murphy, A. S. (2006). The ABC of auxin transport: the role of p-glycoproteins in plant development. FEBS Lett. 580, 1094–1102. doi: 10.1016/j.febslet.2005.11.054
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gleave, A. P. (1992). A versatile binary vector system with a T-DNA organizational structure conducive to efficient integreation of cloned DNA into the plant genome. Plant Mol. Biol. 20, 1203–1207.
Guo, M., Rupe, M. A., Dieter, J. A., Zou, J., Spielbauer, D., Duncan, K. E., et al. (2010). CELL NUMBER REGULATOR 2 affects plant and organ size in maize: implications for crop yield enhancement and heterosis. Plant Cell 22, 1057–1073. doi: 10.1105/tpc.109.073676
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Guzman, P., and Ecker, J. R. (1990). Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell 2, 513–523.
Harrison, E. J., Bush, M., Plett, J. M., McPhee, D. P., Vitez, R., O'Malley, B., et al. (2007). Diverse developmental mutants revealed in an activation-tagged population of poplar. Can. J. Bot. 85, 1071–1081. doi: 10.1139/B07-063
Heidstra, R., Yang, W. C., Yalcin, Y., Peck, S., Emons, A. M., van Kammen, A., et al. (1997). Ethylene provides positional information on cortical cell division but not involved in Nod factor-induced root hair tip growth in Rhizobium-legume interaction. Development 124, 1781–1787.
Hemerly, A., de Almei da Engler, J., Bergounioux, C., Van Montagu, M., Engler, G., Inze, D., et al. (1995). Dominant negative mutants of the CDC2 kinase uncouple cell division from iterative plant development. EMBO J. 14, 3925–3936.
Higashide, T., Narukawa, M., Shimada, Y., and Soeno, K. (2014). Suppression of elongation and growth of tomato seedlings by auxin biosynthesis inhibitors and modeling of the growth and environmental response. Sci. Rep. 4:4556. doi: 10.1038/srep04556
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Horiguchi, G., Ferjani, A., Fujikura, U., and Tsukaya, H. (2006). Coordination of cell proliferation and cell expansion in the control of leaf size in Arabidopsis thaliana. J. Plant Res. 119, 37–42. doi: 10.1007/s10265-005-0232-4
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hu, Y., Poh, H. M., and Chua, N. H. (2006). The Arabidopsis ARGOS-LIKE gene regulates cell expansion during organ growth. Plant J. 47, 1–9. doi: 10.1111/j.1365-313X.2006.02750.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Huang, J.-Y., and Lin, C.-H. (2003). Cold water treatment promotes ethylene production and dwarfing in tomato seedlings. Plant Physiol. Biochem. 41, 283–288. doi: 10.1016/S0981-9428(03)00020-2
Huang, N., Courtois, B., Khush, G. S., Lin, H., Wang, G., Wu, P., et al. (1996). Association of quantitative trait loci for plant height with major dwarfing genes in rice. Heredity 77, 130–137. doi: 10.1038/hdy.1996.117
Jeong, D.-H., An, S., Park, S., Kang, H.-G., Park, B.-B., Kim, S.-R., et al. (2006). Generation of a flanking sequence-tag database for activation-tagging lines in japonica rice. Plant J. 45, 123–132. doi: 10.1111/j.1365-313X.2005.02610.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jiang, L., Liu, X., Xiong, G., Liu, H., Chen, F., Wang, L., et al. (2013). DWARF 53 act as a repressor of strigolactone signaling in rice. Nature 504, 401–405. doi: 10.1038/nature12870
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Junghans, U., Langenfeld-Heyser, R., Polle, A., and Teichmann, T. (2004). Effects of auxin transport inhibitors and ethylene on the wood anatomy of poplar. Plant Biol. 6, 22–29. doi: 10.1055/s-2003-44712
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kalra, Y. P., and Maynard, D. G. (1991). Methods Manual for Forest Soil and Plant Analysis. Information Report NOR-X-319E. Edmonton, AB: Canadian Forest Service Publications.
Kieber, J. J., Rothenberg, M., Roman, G., Feldmann, K. A., and Ecker, J. R. (1993). CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the Raf family of protein kinases. Cell 72, 427–441.
Koyama, T., Nil, H., Mitsuda, N., Ohta, M., Kitajima, S., Ohme-Takagi, M., et al. (2013). A regulatory cascade involving class II ETHYLENE RESPONSE FACTOR transcriptional repressors operates in the progression of leaf senescence. Plant Physiol. 162, 991–1005. doi: 10.1104/pp.113.218115
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lantican, B. P., and Muir, R. M. (1969). Auxin physiology of dwarfism in Pisum sativum. Physiol. Plant. 22, 412–423. doi: 10.1111/j.1399-3054.1969.tb07393.x
Lee, D. J., and Zeevaart, J. A. (2005). Molecular cloning of GA 2-OXIDASE3 from spinach and its ectopic expression in Nicotiana sylvestris. Plant Physiol. 138, 243–254. doi: 10.1104/pp.104.056499
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lee, K. H., and LaRue, T. A. (1992). Exogenous ethylene inhibits nodulation of Pisum sativum L. cv Sparkle. Plant Physiol. 100, 1759–1763.
Li, J., Jiang, J., Qian, Q., Xu, Y., Zhang, C., Xiao, J., et al. (2011). Mutation of rice BC12/GDD1, which encodes a kinesin-like protein that binds to a GA biosynthesis gene promoter, leads to dwarfism with impaired cell elongation. Plant Cell 23, 628–640. doi: 10.1105/tpc.110.081901
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Li, J., Yang, H., Peer, W. A., Richter, G., Blakeslee, J., Bandyopadhyay, A., et al. (2005). Arabidopsis H+-PPase AVP1 regulates auxin-mediated organ development. Science 310, 121–125. doi: 10.1126/science.1115711
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Li, R., Zia, J., Xu, Y., Zhao, X., Liu, Y.-G., and Chen, Y. (2014). Characterization and genetic mapping of a Photoperiod-sensitive dwarf 1 locus in rice (Oryze sativa L.). Theor. Appl. Genet. 127, 241–250. doi: 10.1007/s00122-013-2213-7
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Liu, Y., and Zhang, S. (2004). Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. Plant Cell 16, 3386–3399. doi: 10.1105/tpc.104.026609
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Love, J., Björklund, S., Vahala, J., Hertzberg, M., Kangasjärvi, J., and Sundberg, B. (2009). Ethylene is an endogenous stimulator of cell division in the cambial meristem of Populus. Proc. Natl. Acad. Sci. U.S.A. 106, 5984–5989. doi: 10.1073/pnas.0811660106
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Luo, J., Ma, N., Pei, H., Chen, J., Li, J., and Gao, J. (2013). A DELLA gene, RhGAI1, is a direct target of EIN3 and mediates ethylene-regulated rose petal expansion via repressing the expression of RhCesA2. J. Exp. Bot. 64, 5075–5084. doi: 10.1093/jxb/ert296
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mathews, H., Clendennen, S. K., Caldwell, C. G., Liu, X. L., Connors, K., Matheis, N., et al. (2003). Activation tagging in tomato identifies a transcriptional regulator of anthocyanin biosynthesis, modification and transport. Plant Cell 15, 1689–1703. doi: 10.1105/tpc.012963
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Morgan, P. W., and Gausman, H. W. (1966). Effects of ethylene on auxin transport. Plant Physiol. 41, 45–52.
Multani, D. S., Briggs, S. P., Chamberlin, M. A., Blakeslee, J. J., Murphy, A. S., and Johal, G. S. (2003). Loss of an MDR transporter in compact stalks of maize br2 and sorgum dw3 mutants. Science 302, 81–84. doi: 10.1126/science.1086072
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nakaya, M., Tsukaya, H., Murakami, N., and Kato, M. (2002). Brassinosteroids control the proliferation in leaf cells of Arabidopsis thaliana. Plant Cell Physiol. 43, 239–244. doi: 10.1093/pcp/pcf024
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nigg, E. A. (2001). Mitotic kinases as regulators of cell division and its checkpoints. Nat. Rev. Mol. Cell Biol. 2, 21–32. doi: 10.1038/35048096
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nobusawa, T., Okushima, Y., Nagata, N., Kojima, M., Sakakibara, H., and Umeda, M. (2013). Synthesis of very-long-chain fatty acids in the epidermis controls plant organ growth by restricting cell proliferation. PLoS Biol. 11:e1001531. doi: 10.1371/journal.pbio.1001531
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Okubo, H., and Uemoto, S. (1985). Changes in endogenous gibberellin and auxin activities during first internode elongation in tulip flower stalk. Plant Cell Physiol. 26, 709–719.
Parker, T. (2012). An apple a day!: new dwarf apple ‘leprechaun’ in the perfect planting partner for ‘pinkbelle.’ Medicus 52, 65. Available online at: http://search.informit.com.au/documentSummary;dn=214566906243183;res=IELHEA
Plett, J. M., Wilkins, O., Campbell, M., Ralph, S., and Regan, S. (2010). Endogenous over-expression of Populus MYB186 increases trichome density, improves insect pest resistance, and impacts plant growth. Plant J. 64, 419–432. doi: 10.1111/j.1365-313X.2010.04343.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Qi, W., Sun, F., Wang, Q., Chen, M., Huang, Y., Feng, Y.-Q., et al. (2011). Rice ethylene-response AP2/ERF factor OsEATB restricts internode elongation by down-regulating a gibberellin biosynthetic gene. Plant Physiol. 157, 216–228. doi: 10.1104/pp.111.179945
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Quirino, B. F., Noh, Y. S., Himelblau, E., and Amasino, R. M. (2000). Molecular aspects of leaf senescence. Trends Plant Sci. 5, 278–282. doi: 10.1016/S1360-1385(00)01655-1
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rattani, A., Vinod, P. K., Godwin, J., Tachibana-Konwalski, K., Wolna, M., Malumbres, M., et al. (2014). Dependency of the spindle assembly checkpoint on Cdk1 renders the anaphase transition irreversible. Curr. Biol. 24, 630–637. doi: 10.1016/j.cub.2014.01.033
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Roman, G., and Ecker, J. R. (1995). Genetic analysis of a seedling stress response to ethylene in Arabidopsis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 350, 75–81.
Romano, C. P., Cooper, M. L., and Klee, H. J. (1993). Uncoupling auxin and ethylene effects in transgenic tobacco and Arabidopsis plants. Plant Cell 5, 181–189.
Ruzicka, K., Ljung, K., Vanneste, S., Podhorska, R., Beechman, T., Frimi, J., et al. (2007). Ethylene regulates root growth through effects on auxin biosynthesis and transport-dependent auxin distribution. Plant Cell 19, 2197–2212. doi: 10.1105/tpc.107.052126
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sabelli, P. A., Liu, Y., Dante, R. A., Lizarraga, L. E., Nguyen, H. N., Brown, S. W., et al. (2013). Control of cell proliferation, endoreduplicaiton, cell size, and cell death by the retinoblastoma-related pathway in maize endosperm. Proc. Natl. Acad. Sci. U.S.A. 110, E1827–E1836. doi: 10.1073/pnas.1304903110
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Schomburg, F. M., Bizzell, C. M., Lee, D. J., Zeevaart, J. A., and Amasino, R. M. (2003). Overexpression of a novel class of gibberellin 2-oxidases decreases gibberellin levels and creates dwarf plants. Plant Cell 15, 151–163. doi: 10.1105/tpc.005975
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Scoffoni, C., Rawis, M., McKown, A., Cochard, H., and Sack, L. (2011). Decline of leaf hydraulic conductance with dehydration: relationship of leaf size and venation architecture. Plant Physiol. 156, 832–843. doi: 10.1104/pp.111.173856
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sieling, K., and Kage, H. (2008). The potential of semi-dwarf oilseed rape genotypes to reduce the risk of N leaching. J. Agric. Sci. 146, 77–84. doi: 10.1017/S0021859607007472
Skirycz, A., de Bodt, S., Obata, T., de Clercq, I., Claeys, H., de Rycke, R., et al. (2010). Developmental stage specificity and the role of mitochondrial metabolism in the response of Arabidopsis leaves to prolonged mild osmotic stress. Plant Physiol. 152, 226–244. doi: 10.1104/pp.109.148965
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Smits, V. A., and Medema, R. H. (2001). Checking out the G2/M transition. Biochim. Biophys. Acta 1519, 1–12. doi: 10.1016/S0167-4781(01)00204-4
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Stepanova, A. N., Hoyt, J. M., Hamilton, A. A., and Alonso, J. M. (2005). A link between ethylene and auxin uncovered by the characterization of two root-specific ethylene-insensitive mutants in Arabidopsis. Plant Cell 17, 2230–2242. doi: 10.1105/tpc.105.033365
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Stepanova, A. N., Yun, J., Likhacheva, A. V., and Alonso, J. M. (2007). Multilevel interactions between ethylene and auxin in Arabidopsis roots. Plant Cell 19, 2169–2185. doi: 10.1105/tpc.107.052068
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Suryadinata, R., Sadowski, M., and Sarcevic, B. (2010). Control of cell cycle progression by phosphorylation of cyclin-dependent kinase (CDK) substrates. Biosci. Rep. 30:bsr0300243. doi: 10.1042/BSR20090171
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Suttle, J. C. (1988). Effect of ethylene treatment on polar IAA transport, net IAA uptake and specific binding of N-1-naphthylphthalamic acid in tissues and microsomes isolated from etiolated pea epicotyls. Plant Physiol. 88, 795–799.
Swarup, R., Perry, P., Hagenbeek, D., van der Straeten, D., Beemster, G. T. S., Sandberg, G., et al. (2007). Ethylene upregulates auxin biosynthesis in Arabidopsis seedlings to enhance inhibition of root cell elongation. Plant Cell 19, 2186–2196. doi: 10.1105/tpc.107.052100
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tong, J.-P., Liu, X.-J., Zhang, S.-Y., Li, S.-Q., Peng, X.-J., Yang, J., et al. (2007). Identification, genetic characterization, GA response and molecular mapping of Sdt97: a dominant mutant gene conferring semi-dwarfism in rice (Oryza sativa L.). Genet. Res. 89, 221–230. doi: 10.1017/S0016672307009020
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tsukaya, H. (2002). Interpretation of mutants in leaf morphology: genetic evidence for a compensatory system in leaf morphogenesis that provides a new link between cell and organismal theories. Int. Rev. Cytol. 217, 1–39. doi: 10.1016/S0074-7696(02)17011-2
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tuskan, G. A., DiFazio, S., Jansson, S., Bohlmann, J., Grigoriev, I., Hellsten, U., et al. (2006). The genome of black cottonwood, Populus trichocarpa (Torr.and Gray). Science 15, 1596–1604. doi: 10.1126/science.1128691
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Vahala, J., Felten, J., Love, J., Gorzsás, A., Gerber, L., Lamminmäki, A., et al. (2013). A genome-wide screen for ethylene-induced Ethylene Response Factors (ERFs) in hybrid aspen stem identifies ERF genes that modify stem growth and wood properties. New Phytol. 200, 511–522. doi: 10.1111/nph.12386
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Valdovinos, J. G., Ernest, L. C., and Henry, E. W. (1967). Effect of ethylene and gibberellic acid on auxin synthesis in plant tissues. Plant Physiol. 42, 1803–1806.
von Guttenberg, H., and Steinmetz, E. (1947). The effects of ethylene on growth hormone and growth. Pharmazie 2, 17–21.
Wang, R., Huang, H., Lin, Y., Chen, Q., Liang, Z., and Wu, T. (2014a). Genetic and gene expression analysis of dm1, a dwarf mutant from Cucurbita maxima Duch. ex Lam, based on the AFLP method. Can. J. Plant Sci. 94, 293–302. doi: 10.4141/cjps2013-016
Wang, W., Li, G., Zhao, J., Chu, H., Lin, W., Zhang, D., et al. (2014b). DWARF TILLER1, a WUSCHEL-related homeobox transcription factor, is required for tiller growth in rice. PLoS Genet. 10:e1004154. doi: 10.1371/journal.pgen.1004154
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Weigel, D., Ahn, J. H., Blazquez, M. A., Borevitz, J. O., Christensen, S. K., Fankhause, C., et al. (2000). Activation tagging in Arabidopsis. Plant Physiol. 122, 1003–1014. doi: 10.1104/pp.122.4.1003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Woeste, K. E., Ye, C., and Kieber, J. J. (1999). Two Arabidopsis mutants that overproduce ethylene are affected in the posttranscriptional regulation of 1-aminocyclopropane-1-carboxylic acid synthase. Plant Physiol. 119, 521–530. doi: 10.1104/pp.119.2.521
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yamamoto, Y., Kamiya, N., Morinaka, Y., Matsuoka, M., and Sazuka, T. (2007). Auxin biosynthesis by the YUCCA genes in rice. Plant Physiol. 143, 1362–1371. doi: 10.1104/pp.106.091561
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yang, X. C., and Hwa, C. M. (2008). Genetic modification of plant architecture and variety improvement in rice. Heredity 101, 296–404. doi: 10.1038/hdy.2008.90
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yang, Y., Massa, G. D., and Mitchell, C. A. (2014). Temperature DIP at the beginning of the photoperiod reduces plant height but not seed yield of maize grown in controlled environments. Ind. Crops Prod. 53, 120–127. doi: 10.1016/j.indcrop.2013.12.002
Zawaski, C., Kadmiel, M., Pickens, J., Ma, C., Strauss, S., and Busov, V. (2011). Repression of gibberellin biosynthesis or signaling produces striking alterations in poplar growth, morphology and flowering. Planta 234, 1285–1298. doi: 10.1007/s00425-011-1485-x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhang, J., Liu, X., Li, S., Cheng, Z., and Li, C. (2014). The rice semi-dwarf mutant sd37, caused by a mutation in CYP96B4, plays an important role in the fine-tuning of plant growth. PLoS ONE 9:e88068. doi: 10.1371/journal.pone.0088068
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: poplar, activation tagging, wood formation, plant stature, plant growth rate
Citation: Plett JM, Williams M, LeClair G, Regan S and Beardmore T (2014) Heterologous over-expression of ACC SYNTHASE8 (ACS8) in Populus tremula x P. alba clone 717-1B4 results in elevated levels of ethylene and induces stem dwarfism and reduced leaf size through separate genetic pathways. Front. Plant Sci. 5:514. doi: 10.3389/fpls.2014.00514
Received: 30 July 2014; Accepted: 13 September 2014;
Published online: 04 November 2014.
Edited by:
Domenico De Martinis, Energy and Sustainable Economic Development, ItalyReviewed by:
Dierk Wanke, Tuebingen University, GermanySara Maldonado, University of Buenos Aires, Argentina
Copyright © 2014 Plett, Williams, LeClair, Regan and Beardmore. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tannis Beardmore, Canadian Forest Service, Natural Resources Canada, Fredericton, NB, E3B 5P7, Canada e-mail:dGFubmlzLmJlYXJkbW9yZUBucmNhbi1ybmNhbi5nYy5jYQ==
†These authors have contributed equally to this work.
 Jonathan M. Plett
Jonathan M. Plett Martin Williams3
Martin Williams3 Sharon Regan
Sharon Regan