- 1Saclay Plant Science, Institut de Biologie des Plantes, Université Paris-Sud 11, Orsay, France
- 2UMR 8618, CNRS, Orsay, France
- 3Interdisciplinary Scientific Research Institute, Jianghan University, Wuhan, China
Histone H3 lysine 4 trimethylation (H3K4me3) has been shown to be involved in stress-responsive gene expression and gene priming in plants. However, the role of H3K4me3 resetting in the processes is not clear. In this work we studied the expression and function of Arabidopsis H3K4 demethylase gene JMJ15. We show that the expression of JMJ15 was relatively low and was limited to a number of tissues during vegetative growth but was higher in young floral organs. Over-expression of the gene in gain-of-function mutants reduced the plant height with accumulation of lignin in stems, while the loss-of-function mutation did not produce any visible phenotype. The gain-of-function mutants showed enhanced salt tolerance, whereas the loss-of-function mutant was more sensitive to salt compared to the wild type. Transcriptomic analysis revealed that over-expression of JMJ15 down-regulated many genes which are preferentially marked by H3K4me3 and H3K4me2. Many of the down-regulated genes encode transcription regulators involved in stress responses. The data suggest that increased JMJ15 levels may regulate the gene expression program that enhances stress tolerance.
Introduction
Histone lysine methylation is an important epigenetic modification for gene expression in eukaryotic cells (Martin and Zhang, 2005; Kouzarides, 2007). Genome-wide analysis has revealed that about two-thirds of Arabidopsis genes are marked by mono-, di- or tri-methylation of histone H3 at residue lysine 4 (H3K4me1/2/3) (Zhang et al., 2009). H3K4me3 is predominantly found at the promoter and 5′ end of genes and is strongly associated with transcriptional activation in plants. H3K4me3 level was found to be increased on responsive genes upon stress treatment, but in some cases the increase was found to be lagged behind gene activation (Kim et al., 2008; Hu et al., 2011), suggesting that H3K4me3 may have a function to mark the active gene state. In addition, H3K4me3 in gene body has been suggested to play a role in transcriptional memory of stress-responsive genes in Arabidopsis (Alvarez-Venegas et al., 2007; Jaskiewicz et al., 2011). Recent results indicated that the H3K4me3 level in gene body was decreased after stress recovery but remained higher than basal state, suggesting that a regulated resetting mechanism is involved in partial removal of H3K4me3 and that remaining H3K4me3 may contribute to the transcriptional memory in Arabidopsis (Ding et al., 2012; Kim et al., 2012).
Histone methylation marks are established by evolutionarily conserved SET-domain proteins (named after 3 Drosophila genes: Su(var)3–9, Enhancer of zeste and Trithorax). H3K4 methylation is mediated by the Trithorax group proteins (TRX). Arabidopsis Trithorax ATX1 and ATX2 respectively trimethylate and dimethylate H3K4 (Saleh et al., 2008). ATX1 was found to be necessary for stress-induced gene expression (Alvarez-Venegas and Avramova, 2005; Alvarez-Venegas et al., 2007; Ding et al., 2011). Other SET-domain genes (SDG) such as SDG4 and SDG2 are also involved in H3K4 methylation and control of many aspects of plant development (Cartagena et al., 2008; Berr et al., 2010; Guo et al., 2010).
Histone methylation is reversed by histone demethylases. Lysine Specific Demethylase 1 (LSD1) is the first identified histone demethylase to remove mono- and di-methyl groups from H3K4 (Shi et al., 2004). In Arabidopsis there are 4 LSD1-like genes including FLOWERING LOCUS D (FLD), LSD1-LIKE 1 (LDL1), and LSD1-LIKE 2 (LDL2) that are shown to be involved in flowering time control (Jiang et al., 2007). The second class of histone demethylases that contain the jumonji C (JmjC) domain catalyze histone lysine demethylation through a ferrous ion (Fe(II)) and α-ketoglutaric acid (α-KG)-dependent oxidative reaction (Tsukada et al., 2006). Multiple JmjC domain-containing histone demethylases are identified in animal cells, which are divided into distinct groups including JARID/KDM5, JMJD1/JHDM2/KDM3, JMJD2/KDM4, JMJD3/UTX/KDM6, JHDM1/FBX/KDM2 and the “JmjC domain-only” group. Members of each group target to specific histone lysine residues at different methylation states (Klose et al., 2006). About 20 JmjC domain-containing protein genes are found in Arabidopsis (Lu et al., 2008; Sun and Zhou, 2008; Chen et al., 2011). Most animal and plant JmjC proteins are conserved, while some animal proteins, such as the JMJD3/UTX/KDM6 group that has the H3K27 demethylase activity is not found in plants. Recent data have shown that plant JMJD2/KDM4 homologs can demethylate H3K27 (Lu et al., 2011; Li et al., 2013).
The JARID/KDM5 group catalyzes H3K4me2/3 demethylation in mammalian cells. Arabidopsis genome has one JARID/KDM5-like gene (JMJ17), whose function is presently unknown. There is a specific group in plants which includes Arabidopsis JMJ14, JMJ15, JMJ16, JMJ18, and JMJ19. The JmjC domains of this group are more closely related to that of the JARID, but structurally similar to that of JMJD2/KDM4 (Chen et al., 2013). JMJ14, JMJ15 and JMJ18 have been reported to have the H3K4me2/3 demethylase activity and to regulate diverse aspects of chromatin function and plant development (Deleris et al., 2010; Lu et al., 2010; Searle et al., 2010; Le Masson et al., 2012; Yang et al., 2012a,b; Cui et al., 2013). However, the function of these H3K4 demethylases in plant stress tolerance has not been evaluated. In this work we provide evidence that increased expression of JMJ15 preferentially down-regulates H3K4me2/3-marked stress-related genes and enhances salt stress tolerance.
Materials and Methods
Plant Growth
The Arabidopsis thaliana ecotype Columbia (Col-0) was used throughout this study. T-DNA mutant lines jmj15-1 (GABI_257F10), jmj15-2 (GABI_876B01) and jmj15-3 (GABI_663C11) were obtained from the Nottingham Arabidopsis Stock Center (NASC) and confirmed by PCR. Seeds were surface-sterilized and plants were grown on 0.5 x Murashige and Skoog (MS) medium after stratification at 4°C for 2 days. Plants were analyzed on plates under long-day (LD, 16 h light/8 h dark) or short-day (SD, 8 h light/16 h dark) photoperiods at 20°C. Ten days after germination, plants were transferred to soil and kept in growth rooms under LD conditions.
To test gene expression in response to salt, experiments were carried out with 8 day-old plants, treated with 0.5 x MS supplemented with or without 100 to 150 mM NaCl for 1-5 h. For germination tests, seeds of wild type and jmj15 mutants were sown on medium containing 130-150 mM NaCl. Images of the Petri dishes were taken 10 days after germination.
Constructs and Transformation
For the histochemical GUS assay, the 2 kb promoter of JMJ15 was amplified from wild type genomic DNA using the following primers: 5′-GGATCCAGAGCTTGGCCATTTCTTGA-3′ (forward) and 5′-GGTACCGCACTGAAAGGCTCCATTG-3′ (reverse). BamHI and KpnI (underlined) were used for digestions. The JMJ15 promoter fragment was inserted as translational fusion with the uidA gene into the pPR97 vector. To generate the 35S-JMJ15-FLAG-HA construct, the full length cDNA without the stop codon was amplified from total cDNA isolated from Col-0 plants using primers: TCTAGACCTTTGGGTTTTGTGGAGTG (forward) and TCTAGACCAATTCAAATCAACCCCAAA (reverse). Using XbaI site, JMJ15 cDNA was inserted into the binary vector pFA121, which was modified based on pBI121 and contained 2 × FLAG-HA tag. The pJMJ15-GUS and 35S-JMJ15-FLAG-HA constructs were transformed into Agrobacterium tumefaciens strain GV3101 and then transformed the plants using floral dip method.
Microarray Analysis
Total RNA was extracted from 12 day-old seedling using Trizol (Invitrogen) and cleaned using the RNeasy isolation kit (Qiagen). Hybridization with Affymetrix GeneChip Arabidopsis ATH1 Genome Array was performed at CapitalBio Corporation. Wild type and both jmj15 over-expression alleles were performed in two biological repeats. Gene expression changes between the samples were analyzed by the AffylmGUI package from R software. GO annotation was carried out with the GO terms of the TAIR database (http://arabidopsis.org/tools/bulk/go/index.jsp). The percentage of significantly gene enrichment in each TAIR annotated category was calculated as follows: the number of enriched genes divided by N × 100, where N represents the total number of genes annotated in each category. Significantly enriched genes were subsequently analyzed for their H3K4 methylation levels at epigenomics database (http://epigenomics.mcdb.ucla.edu/H3K4m1m2m3/).
Real-Time PCR
For gene expression analysis, two micrograms of total RNA were reverse transcribed into cDNA by ImPromII reverse transcriptase (Promega). Real-time PCR was performed with the LightCycler® 480 SYBR Green I Master (Roche) on a LightCycler 480 (Roche). At least two biological replicates and two technical repeats for every biological replicate were tested. The primers used in this study are listed in Supplementary Table 1.
Histochemical GUS and Lignin Staining
GUS staining was performed as previously described (Bertrand et al., 2003). Briefly, plant samples were fixed with 90% acetone on ice for 20 min and were washed with staining buffer (0.2% Triton X-100, 5 mM potassium ferrocyanide, 5 mM potassium ferricyanide, 100 mM NaH2PO4 and 100 mM Na2HPO4 pH 7.2). Then the samples were immersed in GUS staining solution with 1 mM X-Gluc and placed under vacuum for 20 min. After incubation at 37°C overnight, the staining solution was removed and samples were cleared by sequential changes of 70% (v/v) ethanol and stored at 4°C.
The histological comparative analysis of inflorescence stems between Col-0 and jmj15 mutants was done at the stage of newly formed green siliques, about 2 weeks after bolting, when the inflorescence stems of wild type reach 20 cm in height. Cross-sections of the inflorescence stems at the basal end were stained for 3 min in phloroglucinol-HCl reagent (Prolabo, VMR International, France) and then observed in ethanol 100%: HCl 37% (9/1, v/v) using a light microscope (Nikon, MULTIZOOM AZ 100).
Results
Expression Levels of H3K4 Demethylase Genes
To investigate whether H3K4 demethylase genes are involved in plant stress responses, we analyzed the mRNA levels of JMJ14 (At4g20400), JMJ15 (At2g34880), JMJ16 (At1g08620), JMJ17 (At1g63490), JMJ18 (At3g30810), and JMJ19 (At2g38950) genes in 8 day-old seedlings grown in ½MS media under continuous light, then transferred to 100 mM NaCl or to ½MS solution for 5 hours. In untreated (½MS) seedlings, the expression levels of the 6 genes varied considerably. The relative expression levels of JMJ17, JMJ18, and JMJ19 were much higher (>102) than that of JMJ14, JMJ15, and JMJ16 (Supplementary Figure 1). NaCl treatment did not dramatically affect the expression of these genes, although some decrease of JMJ14 and JMJ18 and some increase of JMJ15 transcript levels were detected.
JMJ15 Displayed a Highly Tissue-Specific Expression Pattern
The relatively low expression level of JMJ15 was in agreement with previous data showing that the 1.5 kb promoter region of JMJ15 is weak in vegetative tissues (Hong et al., 2009). To study the temporal and spatial expression pattern of JMJ15, we used a larger promoter region of JMJ15 (−2051 to +14 bp relative to ATG) to make a GUS reporter translational fusion construct and transformed Arabidopsis Col-0 plants. Three independent GUS reporter lines were characterized. All showed a similar pattern of GUS expression. In seedlings, GUS activity was detected only at the base of rosette leaves and root vascular tissues (Figures 1A–C). Interestingly, a higher accumulation of GUS activity was detected in pericycle cells that initiated to lateral root meristem (Figures 1B–D). The GUS activity remained to be detected at the base of the growing lateral roots (Figures 1E–G), but not in the root tip (Figure 1H). In the inflorescence, GUS activity was strongly detected in young anthers and was detectable in carpels, but the activities became weaker in the mature flower (Figures 1I–L). This temporal and tissue-specific expression pattern suggested that JMJ15 may have a function in plant development.
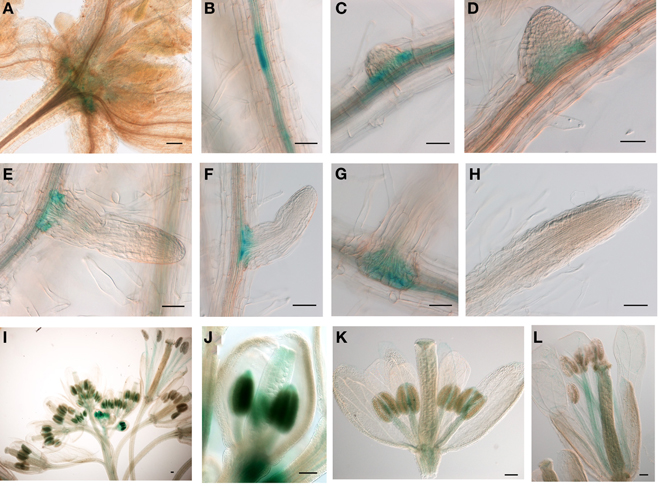
Figure 1. The 2.0 kb promoter activity of JMJ15 in transgenic plants. In seedlings, the GUS activity was detected in the base of rosette leaves (A), root vascular tissues (B), and bases of growing lateral roots (C–G), but not in root tip (H). In flower buds, the GUS activity was detected in anthers and in carpels (I,J), which become weak in opened flowers, but remained in stamen filaments (K,L). Bar = 0.1 mm.
JMJ15 Gain-of-Function Mutations Showed a Reduced Plant Height Phenotype
The JMJ15 gene contains 10 exons and encodes a polypeptide of 806 amino acids with distinct domains, including the JmjN domain, JmjC domain, a C5HC2 zinc finger, and the FY-rich N-terminus (FYRN)/FY-rich C-terminus (FYRC) domains, which are conserved in JMJ14, JMJ16, and JMJ18 (Supplementary Figure 2) (Lu et al., 2008). To study the function of JMJ15 in gene expression and plant development, we characterized 3 T-DNA insertion mutants: jmj15-1(GK-257F10), jmj15-2 (GK-876B01) and jmj15-3 (GK-663C11). In jmj15-1 and jmj15-2, the T-DNA was inserted in the 5′ end, and in jmj15-3 the T-DNA was inserted in the seventh exon of the gene (Figure 2). RT-PCR analysis with 4 pairs of primers that covered the whole coding region of the gene, revealed that the transcript level of JMJ15 was dramatically increased in jmj15-1 and jmj15-2, but the transcript was interrupted in jmj15-3 compared to wild-type (Figure 2). The insertion in jmj15-1 and jmj15-2 did not alter the 5′ end of the coding region, as the primer set F1 (that cover the 5′end of the coding region) and R1 successfully amplify the transcripts from the mutants. The data suggested that jmj15-1 and jmj15-2 were gain-of-function mutants that overexpressed the gene and that jmj15-3 was a loss-of-function mutant.
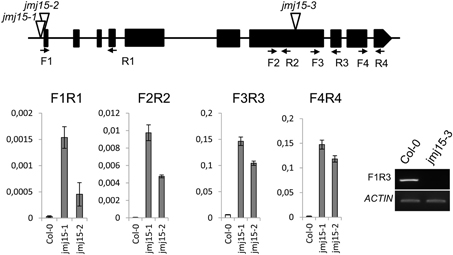
Figure 2. Characterization of JMJ15 T-DNA insertion mutants. The insertion positions of the 3 alleles are indicated by open arrows. The exons are represented by black boxes. The positions of forward (F) and reverse (R) primers are indicated by arrows. The expression levels (relative to ACTIN2 mRNA) in jmj15-1 and jmj15-2 compared to wild type were tested by qRT-PCR using the 4 indicated primer sets. The transcript in jmj15-3 compared to wild type was analyzed by RT-PCR using the indicated primers.
The jmj15-3 loss-of-function mutation did not display any visible phenotype in normal growth conditions, confirming previous observations (Yang et al., 2012a). However, in short day (8 h light/16 h dark)-grown seedlings, jmj15-1 and jmj15-2 mutants produced slightly shorter hypocotyls compared to wild type (Figure 3A). At the mature stage, the plant height of jmj15-1 and jmj15-2 were clearly reduced compared to wild type (Figure 3B). To study whether the plant height phenotype of jmj15-1 and jmj15-2 was due to increased expression of the gene, we made 35S-JMJ15-FLAG-HA construct and obtained JMJ15 over-expression transgenic plants. The transgenic plants also displayed the reduced plant height phenotype at mature stage (Figures 3B,C).
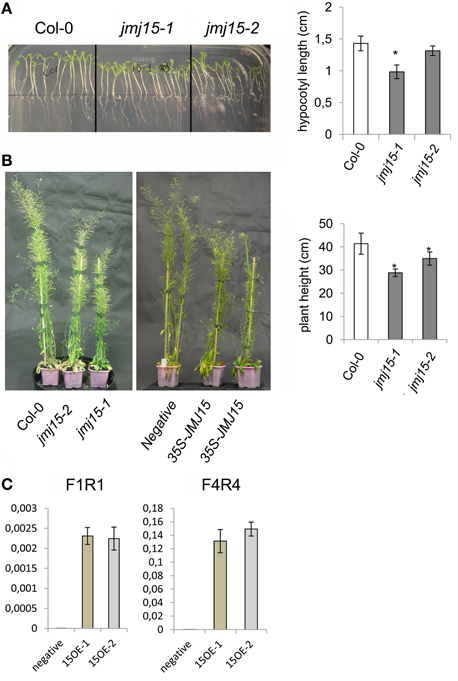
Figure 3. Jmj15 gain-of-function mutations reduced plant height. (A) Phenotype of hypocotyls of short day-grown (in ½MS medium) seedlings of jmj15-1 and jmj15-2 compared to wild type col-0 (left). Lengths of hypocotyls were measured from 30 plants per genotype (right). (B) Plant height at mature stage of jmj15-1 and jmj15-2 compared to wild type (left) and of 35S-JMJ15-HA over-expression plants compared to negative transgenic plants (middle) grown in soil in greenhouse under long day conditions. Plant heights were measured from 10 plants per genotype (right). Bar = means ± SD. Asterisks indicate the significance of difference from wild type by student t-tests (P < 0.01). (C) JMJ15 transcript levels (relative to ACTIN2 transcripts) in over-expression transgenic positive (15OE-1 and 15OE-2) and negative plants.
The plant height phenotype of the jmj15 gain-of-function mutants prompted us to further investigate the stem structure by using histochemical method. Sections of the basal part of the inflorescence stem of 5 week-old plants (grown in long day in greenhouse) were stained with phloroglucinol and examined by light microscopy. Phloroglucinol reacts with coniferaldehyde groups in lignin, and the color intensity reflects the total lignin content. The analysis revealed that jmj15-1 and jmj15-2 exhibited a significantly deeper red staining in the stem vascular system and interfascicular fibers compared to that in wild type and jmj15-3 (Figure 4). This observation suggested that over-expression of JMJ15 resulted in an increase of the total lignin content in the stems concurrently with stem growth reduction.
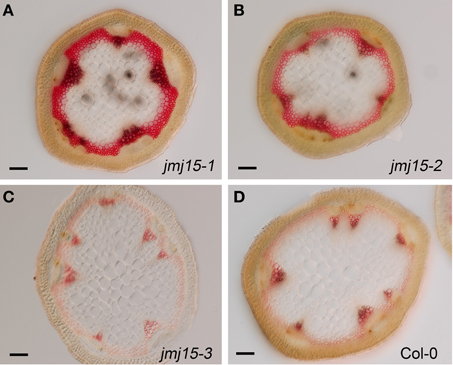
Figure 4. Lignin accumulation in jmj15-1, jmj15-2, and jmj15-3 mutants compared to wild type. Sections of the basal part of the inflorescence stem of 5 week-old plants (grown in long day in greenhouse) were stained with phloroglucinol and examined by light microscopy. Phloroglucinol reacts with coniferaldehyde groups in lignin, and the color intensity reflects the total lignin content. Bar = 0.1 mm.
Over-Expression of JMJ15 Preferentially Repressed Genes Marked by H3K4 Methylation
To determine the effect of JMJ15 over-expression on gene expression, the transcriptomes of jmj15-1, jmj15-2 and wild type seedlings (12 day-old, grown in ½MS media) were analyzed by using the Affymetrix Arabidopsis ATH1 Genome Array. Two biological replicates for each sample were analyzed. Pair-wise plots of the microarray data revealed a good correlation of the hybridization signals between the biological replicates of each sample and between the 2 mutant alleles (Figure 5A). The average hybridization signals of the replicates of both mutants were normalized and compared with the wild type signals. Up- and down-regulated genes in both jmj15-1 and jmj15-2 were filtrated with the threshold >2 fold changes (p-value < 0.01) compared to wild type. The analysis revealed 23 up-regulated and 164 down-regulated genes in the mutant lines (Supplementary Dataset 1). In addition, the analysis revealed a much high expression level of JMJ15 itself (>7-8 folds) in the mutants compared to wild type (Supplementary Dataset 1), confirming the over-expression of the gene in the mutants. The higher number of down-regulated genes compared to up-regulated ones suggested that elevated JMJ15 expression mainly repressed genes and that JMJ15 acted as a transcriptional repressor, consistent with its H3K4 demethylase activity (Liu et al., 2010; Yang et al., 2012a). To validate the microarray data, we checked 5 down-regulated and 4 up-regulated (including JMJ15) genes by RT-qPCR. The relative transcript level changes in the mutants compared to wild type detected by RT-qPCR were in agreement with that from the microarray analysis (Figure 5B).
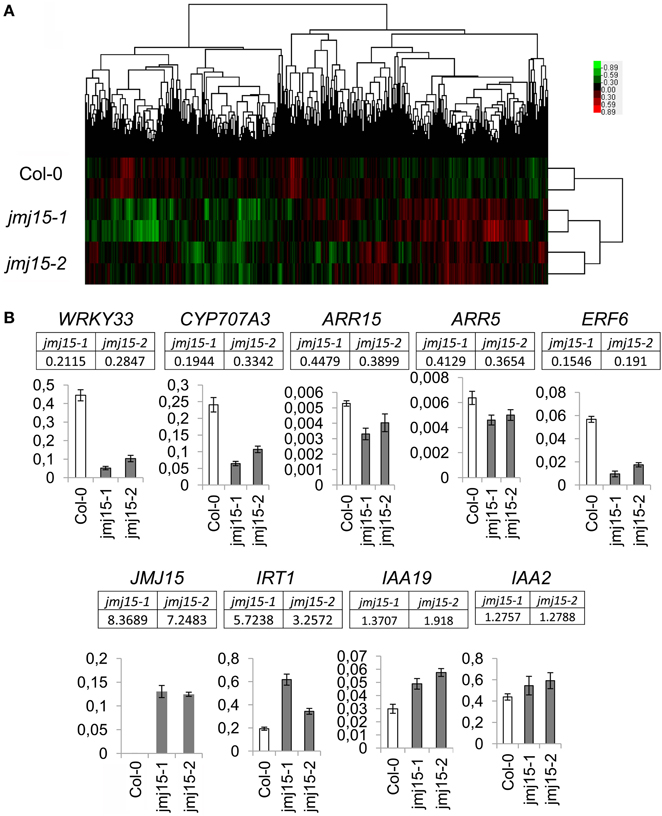
Figure 5. Transcriptomic analysis of jmj15-1 and jmj15-2 (12 day-old) seedlings compared to wild type grown in ½MS medium. (A) Pair-wise plots of the microarray hybridization signals between the biological replicates of each sample and the between wild type and two 2 mutant alleles. Green: down-regulated genes; red: up-regulated genes. (B) Five down-regulated and 4 up-regulated genes in the two mutants (microarray signals relative to wild type are indicated below the respective genes) were validated by qRT-PCR. Bar = means ± SD from 3 replicates.
To evaluate whether there was any enrichment of the deregulated genes for H3K4me2/3, we compared the deregulated genes with the genome-wide H3K4me2/3/1 data obtained from wild type seedlings (Zhang et al., 2009). The analysis revealed that about 83% of the down-regulated genes were marked by the H3K4 methylation (mostly by H3K4me2, H3K4me3, or both) in the gene bodies, compared to about 52% of up-regulated genes (Figure 6, Supplementary Dataset 1). About 58% of the down-regulated genes were marked by H3K4me3, H3K4me2, or H3K4me2/3 in the promoter region (in the −500 bp region relative to TSS), compared to about 32% of up-regulated genes. Compared to up-regulated genes, the down-regulated ones were clearly enriched for the H3K4me2/3 double methylation marks. This analysis suggested that JMJ15-mediated gene repression might be achieved by demethylating H3K4 and indicated that JMJ15 preferentially repressed genes that have the H3K4me2/3 double methylation marks.
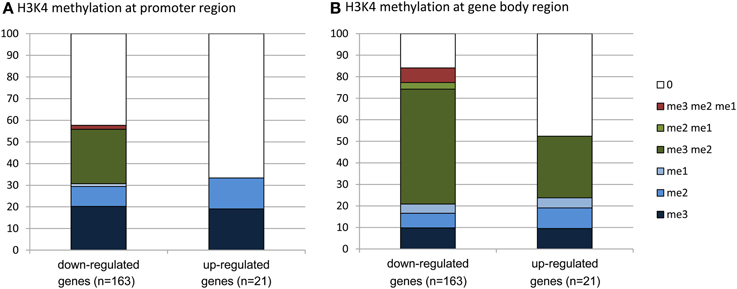
Figure 6. Down-regulated genes are highly enriched for H3K4me2/3. Up-regulated (21) and down-regulated (163) genes in jmj15-1 or jmj15-2 mutants (changes >2 folds) were compared with genome-wide H3K4 methylation data. Percentages of H3K4me3, H3K4me2, H3K4me1 and their combinations on the promoter (A) and the gene body (B) regions are presented.
Over-Expression of JMJ15 Preferentially Repressed Stress Regulatory Genes
Gene ontology (GO) analysis of the deregulated genes using the GO Slim Classification (http://www.arabidopsis.org/help/helppages/go_slim_help.jsp) revealed that a large proportion of the deregulated genes (23.73%) in the jmj15 mutants had a function in the nucleus (Supplementary Figure 3A). Interestingly, 36 out of the 164 down-regulated genes were transcriptional regulators (Supplementary Dataset 1). Remarkably, about 50% (82/164) of the down-regulated genes belonged to the stress-responsive categories (Supplementary Figure 3B, Supplementary Dataset 1), suggesting that JMJ15 may have a function in stress-responsive gene expression. We noticed that among the greater than 5 fold down-regulated genes (31), about 50% (15) encode transcription factors (Supplementary Dataset 1). These included the stress-responsive zinc finger protein STZ/Zat10 (At1g27730) (Sakamoto et al., 2000), the stress-responsive WRKY proteins WRKY40 (At1g80840) and WRKY33 (At2g38470) (Jiang and Deyholos, 2009), the cold-responsive factor CBF2 (At4g25470) (Vogel et al., 2005), the ethylene-responsive-element binding proteins ATERF6 (At4g17490) and ATERF11 (At1g28370) (Li et al., 2011; Dubois et al., 2013) (Supplementary Dataset 1). Among them, the decreased expression of WRKY33 and ERF6 was validated by RT-qPCR (Figure 5B).
JMJ15 Gain-of-Function Mutations Enhanced Salt Stress Tolerance
To study whether jmj15 mutations affected plant tolerance to stress, we germinated seeds of wild type, jmj15-1, jmj15-2 and jmj15-3 mutants on ½MS media containing 130 mM or 150 mM NaCl. The seedling growth phenotype shown in Figure 7A indicated that the gain-of-function mutations (jmj15-1 and jmj15-2) enhanced plant tolerance to salt stress, whereas the loss-of-function mutation (jmj15-3) reduced the stress resistance. To study whether JMJ15 over-expression affected stress-responsive gene expression, we analyzed the transcript levels of several stress-responsive marker genes (i.e., RD29A, RD29B, RD22, COR15A, COR47, P5CS1, and P5CS2) in the gain-of-function mutants grown in normal conditions then treated with or without 150 mM NaCl for 1 h. Without treatment, the expression of these genes was not clearly affected by JMJ15 over-expression. After 1h treatment with 150 mM NaCl, the expression of the marker genes was induced in both wild type and the gain-of-function mutants, but the induction of RD29A, RD22, and COR15 was clearly higher in the mutants (Figure 7B). The higher induction of the stress-responsive genes might be associated with the enhanced salt tolerance phenotype of the mutants.
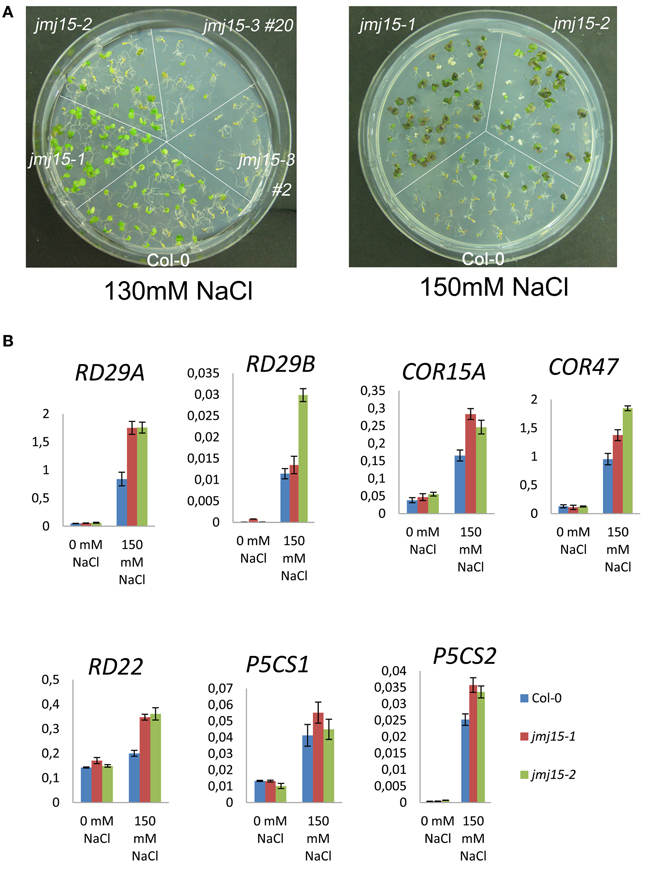
Figure 7. Comparison of sensitivity to NaCl and stress-responsive gene expression between jmj15 mutants with wild type. (A) Wild type, jmj15-1, jmj15-2, and jmj15-3 seeds were germinated for 12 days on ½MS supplemented with indicated concentrations of NaCl. Images of the Petri dishes were taken 10 days after germination. (B) Transcript levels (relative to ACTIN2) of indicated stress-responsive marker genes in wild type and jmj15-1 and jmj15-2 mutant seedlings treated with or without 150 mM NaCl for 1 h.
Discussion
Function of JMJ15 in Stress Tolerance
In this work we have shown that JMJ15 displayed a temporal and tissue-specific expression pattern. Constitutive or over-expression of the gene, as a result of T-DNA insertions in the 5′ region, preferentially repressed genes marked by H3K4me2/3, suggesting that JMJ15-mediated gene repression may be achieved mainly through demethylation of H3K4me2/3. The observation that about a third of the down-regulated genes are related to stress implies that JMJ15-dependent H3K4me2/3 levels are important for the expression of this category of genes. This is consistent with numerous observations that H3K4me3 is associated with the induction of biotic and abiotic stress-responsive genes (van Dijk et al., 2010; Hu et al., 2011; Jaskiewicz et al., 2011; Zong et al., 2013; To and Kim, 2014), and that ATX1 that trimethylates H3K4 in the genic region is required for stress-responsive gene expression (Ding et al., 2009, 2011). Microarray analysis of atx1 mutant seedlings revealed that 424 genes were up-regulated and 328 genes were down-regulated more than 2 folds compared to wild type (Alvarez-Venegas et al., 2006). Similarly, 271 genes were found to be up-regulated and 321 genes down-regulated in mutant seedlings of another H3K4 methyltransferase gene SDG2 (Guo et al., 2010). However there was no clear correlation between transcription changes in sdg2 and atx1 (Guo et al., 2010), suggesting that the two enzymes may regulate different targets. Comparison of the deregulated genes did not reveal a clear overlap, although there was a relatively higher number of overlapped genes between jmj15 and atx1 than between jmj15 and sdg2 (Supplementary Figure 4). These proteins may targets to different loci. In addition, the present data showing that the mostly down-regulated genes encode transcription factors involved in stress responses raise the hypothesis that JMJ15 might be a higher hierarchical regulator primarily to regulate stress-responsive gene transcription programs in Arabidopsis. Since JMJ15 is closely related to JMJ14, JMJ16, and JMJ18 (Lu et al., 2008; Sun and Zhou, 2008), the ectopically expressed JMJ15 may also regulate the targets of the other related demethylases.
The observations that the jmj15 gain-of-function mutants showed enhanced salt tolerance at seedling stage and that the loss-of-function mutant was more sensitive to salt stress than wild type, suggest that JMJ15 is required for salt tolerance. The data showing that the gain-of-function mutants displayed reduced growth and increased stem lignification, which are suggested to be associated with stress responses (Moura et al., 2010; Golldack et al., 2013), support the hypothesis that increased JMJ15 levels may regulate the gene expression program that integrates plant growth to stress tolerance. Among the mostly repressed genes in jmj15 gain-of-function mutants was STZ/Zat10 that encodes a C2H2-zinc finger protein associated with the ERF amphiphilic repression (EAR) domain (Supplementary Dataset 1). STZ/Zat10 has been shown to be a transcriptional suppressor of stress-responsive genes (Sakamoto et al., 2004). Knockout and RNAi of the gene could enhance plant tolerance to abiotic stress (Mittler et al., 2006). As the STZ locus displays a high level of H3K4me3 (Supplementary Figure 5), it is possible that the repression of STZ/Zat10 by JMJ15 through H3K4me2/3 demethylation is associated with the enhanced salt tolerance phenotype of the mutants. STZ/Zat10 may be a major player in JMJ15-mediated regulatory network of stress tolerance. However, the observations that several tested stress-responsive marker genes were not changed in the over-expression plants under normal conditions but showed a greater induction during salt stress (Figure 7B), suggest that they might be among the target genes of the transcription repressors ST/Zat10 and AtERF11 under salt stress. However, among the repressed transcription factor genes, some are likely activators (WRKY33). The mechanism of JMJ15-mediated salt tolerance is complex, which may be resulted from a combination of different functions of JMJ15 in gene regulation.
Developmental Function of JMJ15
Consistent with previous results (Yang et al., 2012a), the loss-of-function mutation identified in this study (jmj15-3) did not produce any visible phenotype. JMJ15 was first identified as Maternal Effect Embryo Arrest 27 (MEE27) in a genetic screen for mutants defective in female gametophyte development (Pagnussat et al., 2005). However, no embryonic defect was observed in jmj15 loss-of-function mutants (Yang et al., 2012a). Either the mutation was compensated by highly expressed homologs (e.g., JMJ18, Hong et al., 2009) or JMJ15-dependent H3K4 demethylation is not sufficient to lead to any morphological change. In addition, another study has identified JMJ15 as a maternally imprinted gene (Hsieh et al., 2011), however, our data showing the high promoter activity of JMJ15 in anthers do not support that observation.
It is reported that JMJ14 demethylates H3K4me2/3 at the Flowering Locus T (FT) locus and represses expression of the gene and that jmj14 loss-of-function mutants display an early flowering phenotype (Jeong et al., 2009; Lu et al., 2010). Conversely, JMJ18 directly binds to and represses the flowering repressor gene, Flowering Locus C (FLC), through H3K4me2/3 demethylation. Consequently, loss-of-function mutations of JMJ18 result in a weak late-flowering phenotype, while JMJ18 overexpressors exhibit an early flowering phenotype (Yang et al., 2012b). These observations support the notion that members of this H3K4 demethylase group target to different loci and have distinct functions in plant development. However, Yang et al have shown that, like JMJ18, JMJ15 over-expression plants showed repressed FLC expression and produced an early flower phenotype (Yang et al., 2012a). But unlike jmj18 mutants (Yang et al., 2012a), the jmj15-3 loss-of-function mutation did not alter the flowering phenotype. Possibly, JMJ15 at elevated levels may demethylate and repress genes that normally targeted by JMJ18 in wild type plants. However, the jmj15-1 and jmj15-2 gain-of-mutation mutants did not show any clear flowering phenotype. This discrepancy may be due to difference in expression levels or tissue-specificity of JMJ15 in the over-expression plants and the mutant alleles.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the French Agence Nationale de Recherche grant ANR-10-BLAN-1238 (CERES) and ANR-12-BSV6-0010 (NERTHPATH). Yuan Shen was supported by a PhD Fellowship from the Chinese Scholar Council.
Supplementary Material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fpls.2014.00290/abstract
References
Alvarez-Venegas, R., Al Abdallat, A., Guo, M., Alfano, J. R., and Avramova, Z. (2007). Epigenetic control of a transcription factor at the cross section of two antagonistic pathways. Epigenetics 2, 106–113. doi: 10.4161/epi.2.2.4404
Alvarez-Venegas, R., and Avramova, Z. (2005). Methylation patterns of histone H3 Lys 4, Lys 9 and Lys 27 in transcriptionally active and inactive Arabidopsis genes and in atx1 mutants. Nucleic Acids Res. 33, 5199–5207. doi: 10.1093/nar/gki830
Alvarez-Venegas, R., Sadder, M., Hlavacka, A., Baluska, F., Xia, Y. N., Lu, G. Q., et al. (2006). The Arabidopsis homolog of trithorax, ATX1, binds phosphatidylinositol 5-phosphate, and the two regulate a common set of target genes. Proc. Natl. Acad. Sci. U.S.A. 103, 6049–6054. doi: 10.1073/pnas.0600944103
Berr, A., McCallum, E. J., Menard, R., Meyer, D., Fuchs, J., Dong, A., et al. (2010). Arabidopsis SET DOMAIN GROUP2 is required for H3K4 trimethylation and is crucial for both sporophyte and gametophyte development. Plant Cell 22, 3232–3248. doi: 10.1105/tpc.110.079962
Bertrand, C., Bergounioux, C., Domenichini, S., Delarue, M., and Zhou, D. X. (2003). Arabidopsis histone acetyltransferase AtGCN5 regulates the floral meristem activity through the WUSCHEL/AGAMOUS pathway. J. Biol. Chem. 278, 28246–28251. doi: 10.1074/jbc.M302787200
Cartagena, J. A., Matsunaga, S., Seki, M., Kurihara, D., Yokoyama, M., Shinozaki, K., et al. (2008). The Arabidopsis SDG4 contributes to the regulation of pollen tube growth by methylation of histone H3 lysines 4 and 36 in mature pollen. Dev. Biol. 315, 355–368. doi: 10.1016/j.ydbio.2007.12.016
Chen, Q. F., Chen, X. S., Wang, Q., Zhang, F. B., Lou, Z. Y., Zhang, Q. F., et al. (2013). Structural basis of a histone H3 lysine 4 demethylase required for stem elongation in rice. PLoS Genet. 9:e1003239. doi: 10.1371/journal.pgen.1003239
Chen, X. S., Hu, Y. F., and Zhou, D. X. (2011). Epigenetic gene regulation by plant Jumonji group of histone demethylase. Bba Gene. Regul. Mech. 1809, 421–426. doi: 10.1016/j.bbagrm.2011.03.004
Cui, X., Jin, P., Gu, L., Lu, Z., Xue, Y., Wei, L., et al. (2013). Control of transposon activity by a histone H3K4 demethylase in rice. Proc. Natl. Acad. Sci. U.S.A. 110, 1953–1958. doi: 10.1073/pnas.1217020110
Deleris, A., Greenberg, M. V., Ausin, I., Law, R. W., Moissiard, G., Schubert, D., et al. (2010). Involvement of a Jumonji-C domain-containing histone demethylase in DRM2-mediated maintenance of DNA methylation. EMBO Rep. 11, 950–955. doi: 10.1038/embor.2010.158
Ding, Y., Avramova, Z., and Fromm, M. (2011). The Arabidopsis trithorax-like factor ATX1 functions in dehydration stress responses via ABA-dependent and ABA-independent pathways. Plant J. 66, 735–744. doi: 10.1111/j.1365-313X.2011.04534.x
Ding, Y., Lapko, H., Ndamukong, I., Xia, Y., Al-Abdallat, A., Lalithambika, S., et al. (2009). The Arabidopsis chromatin modifier ATX1, the myotubularin-like AtMTM and the response to drought. Plant Signal. Behav. 4, 1049–1058. doi: 10.4161/psb.4.11.10103
Ding, Y., Ndamukong, I., Xu, Z., Lapko, H., Fromm, M., and Avramova, Z. (2012). ATX1-generated H3K4me3 is required for efficient elongation of transcription, not initiation, at ATX1-regulated genes. PLoS Genet. 8:e1003111. doi: 10.1371/journal.pgen.1003111
Dubois, M., Skirycz, A., Claeys, H., Maleux, K., Dhondt, S., De Bodt, S., et al. (2013). Ethylene Response Factor6 acts as a central regulator of leaf growth under water-limiting conditions in Arabidopsis. Plant Physiol. 162, 319–332. doi: 10.1104/pp.113.216341
Golldack, D., Li, C., Mohan, H., and Probst, N. (2013). Gibberellins and abscisic acid signal crosstalk: living and developing under unfavorable conditions. Plant Cell Rep. 32, 1007–1016. doi: 10.1007/s00299-013-1409-2
Guo, L., Yu, Y. C., Law, J. A., and Zhang, X. Y. (2010). SET DOMAIN GROUP2 is the major histone H3 lysie 4 trimethyltransferase in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 107, 18557–18562. doi: 10.1073/pnas.1010478107
Hong, E. H., Jeong, Y. M., Ryu, J. Y., Amasino, R. M., Noh, B., and Noh, Y. S. (2009). Temporal and spatial expression patterns of nine Arabidopsis genes encoding Jumonji C-domain proteins. Mol. Cells 27, 481–490. doi: 10.1007/s10059-009-0054-7
Hsieh, T. F., Shin, J., Uzawa, R., Silva, P., Cohen, S., Bauer, M. J., et al. (2011). Regulation of imprinted gene expression in Arabidopsis endosperm. Proc. Natl. Acad. Sci. U.S.A. 108, 1755–1762. doi: 10.1073/pnas.1019273108
Hu, Y. F., Shen, Y., Silva, N. C. E., and Zhou, D. X. (2011). The role of histone methylation and H2A.Z occupancy during rapid activation of ethylene responsive genes. PLoS ONE 6:e28224. doi: 10.1371/journal.pone.0028224
Jaskiewicz, M., Conrath, U., and Peterhansel, C. (2011). Chromatin modification acts as a memory for systemic acquired resistance in the plant stress response. EMBO Rep. 12, 50–55. doi: 10.1038/embor.2010.186
Jeong, J. H., Song, H. R., Ko, J. H., Jeong, Y. M., Kwon, Y. E., Seol, J. H., et al. (2009). Repression of FLOWERING LOCUS T chromatin by functionally redundant histone H3 lysine 4 demethylases in Arabidopsis. PLoS ONE 4:e8033. doi: 10.1371/journal.pone.0008033
Jiang, D., Yang, W., He, Y., and Amasino, R. M. (2007). Arabidopsis relatives of the human lysine-specific Demethylase1 repress the expression of FWA and FLOWERING LOCUS C and thus promote the floral transition. Plant Cell 19, 2975–2987. doi: 10.1105/tpc.107.052373
Jiang, Y., and Deyholos, M. K. (2009). Functional characterization of Arabidopsis NaCl-inducible WRKY25 and WRKY33 transcription factors in abiotic stresses. Plant Mol. Biol. 69, 91–105. doi: 10.1007/s11103-008-9408-3
Kim, J. M., To, T. K., Ishida, J., Matsui, A., Kimura, H., and Seki, M. (2012). Transition of chromatin status during the process of recovery from drought stress in Arabidopsis thaliana. Plant Cell Physiol. 53, 847–856. doi: 10.1093/pcp/pcs053
Kim, J. M., To, T. K., Ishida, J., Morosawa, T., Kawashima, M., Matsui, A., et al. (2008). Alterations of lysine modifications on the histone H3 N-tail under drought stress conditions in Arabidopsis thaliana. Plant Cell Physiol. 49, 1580–1588. doi: 10.1093/pcp/pcn133
Klose, R. J., Kallin, E. M., and Zhang, Y. (2006). JmjC-domain-containing proteins and histone demethylation. Nat. Rev. Genet. 7, 715–727. doi: 10.1038/nrg1945
Kouzarides, T. (2007). Chromatin modifications and their function. Cell 128, 693–705. doi: 10.1016/j.cell.2007.02.005
Le Masson, I., Jauvion, V., Bouteiller, N., Rivard, M., Elmayan, T., and Vaucheret, H. (2012). Mutations in the Arabidopsis H3K4me2/3 Demethylase JMJ14 suppress posttranscriptional gene silencing by decreasing transgene transcription. Plant Cell 24, 3603–3612. doi: 10.1105/tpc.112.103119
Li, T. T., Chen, X. S., Zhong, X. C., Zhao, Y., Liu, X. Y., Zhou, S. L., et al. (2013). Jumonji C domain protein JMJ705-mediated removal of histone H3 lysine 27 trimethylation is involved in defense-related gene activation in rice. Plant Cell 25, 4725–4736. doi: 10.1105/tpc.113.118802
Li, Z. F., Zhang, L. X., Yu, Y. W., Quan, R. D., Zhang, Z. J., Zhang, H. W., et al. (2011). The ethylene response factor AtERF11 that is transcriptionally modulated by the bZIP transcription factor HY5 is a crucial repressor for ethylene biosynthesis in Arabidopsis. Plant J. 68, 88–99. doi: 10.1111/j.1365-313X.2011.04670.x
Liu, C. Y., Lu, F. L., Cui, X., and Cao, X. F. (2010). Histone methylation in higher plants. Annu. Rev. Plant Biol. 61, 395–420. doi: 10.1146/annurev.arplant.043008.091939
Lu, F., Cui, X., Zhang, S., Jenuwein, T., and Cao, X. (2011). Arabidopsis REF6 is a histone H3 lysine 27 demethylase. Nat. Genet. 43, 715–719. doi: 10.1038/ng.854
Lu, F. L., Cui, X., Zhang, S. B., Liu, C. Y., and Cao, X. F. (2010). JMJ14 is an H3K4 demethylase regulating flowering time in Arabidopsis. Cell Res. 20, 387–390. doi: 10.1038/cr.2010.27
Lu, F. L., Li, G. L., Cui, X., Liu, C. Y., Wang, X. J., and Cao, X. F. (2008). Comparative analysis of JmjC domain-containing proteins reveals the potential histone demethylases in Arabidopsis and rice. J. Integr. Plant Biol. 50, 886–896. doi: 10.1111/j.1744-7909.2008.00692.x
Martin, C., and Zhang, Y. (2005). The diverse functions of histone lysine methylation. Nat. Rev. Mol. Cell Biol. 6, 838–849. doi: 10.1038/nrm1761
Mittler, R., Kim, Y., Song, L. H., Coutu, J., Coutu, A., Ciftci-Yilmaz, S., et al. (2006). Gain- and loss-of-function mutations in Zat10 enhance the tolerance of plants to ablotic stress. FEBS Lett. 580, 6537–6542. doi: 10.1016/j.febslet.2006.11.002
Moura, J. C. M. S., Bonine, C. A. V., Viana, J. D. F., Dornelas, M. C., and Mazzafera, P. (2010). Abiotic and biotic stresses and changes in the lignin content and composition in plants. J. Integr. Plant Biol. 52, 360–376. doi: 10.1111/j.1744-7909.2010.00892.x
Pagnussat, G. C., Yu, H. J., Ngo, Q. A., Rajani, S., Mayalagu, S., Johnson, C. S., et al. (2005). Genetic and molecular identification of genes required for female gametophyte development and function in Arabidopsis. Development 132, 1161–1161. doi: 10.1242/dev.01595
Sakamoto, H., Araki, T., Meshi, T., and Iwabuchi, M. (2000). Expression of a subset of the Arabidopsis Cys(2)/His(2)-type zinc-finger protein gene family under water stress. Gene 248, 23–32. doi: 10.1016/S0378-1119(00)00133-5
Sakamoto, H., Maruyama, K., Sakuma, Y., Meshi, T., Iwabuchi, M., Shinozaki, K., et al. (2004). Arabidopsis Cys2/His2-type zinc-finger proteins function as transcription repressors under drought, cold, and high-salinity stress conditions. Plant Physiol. 136, 2734–2746. doi: 10.1104/pp.104.046599
Saleh, A., Alvarez-Venegas, R., Yilmaz, M., Le, O., Hou, G., Sadder, M., et al. (2008). The highly similar Arabidopsis homologs of trithorax ATX1 and ATX2 encode proteins with divergent biochemical functions. Plant Cell 20, 568–579. doi: 10.1105/tpc.107.056614
Searle, I. R., Pontes, O., Melnyk, C. W., Smith, L. M., and Baulcombe, D. C. (2010). JMJ14, a JmjC domain protein, is required for RNA silencing and cell-to-cell movement of an RNA silencing signal in Arabidopsis. Genes Dev. 24, 986–991. doi: 10.1101/gad.579910
Shi, Y., Lan, F., Matson, C., Mulligan, P., Whetstine, J. R., Cole, P. A., et al. (2004). Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119, 941–953. doi: 10.1016/j.cell.2004.12.012
Sun, Q. W., and Zhou, D. X. (2008). Rice jmjC domain-containing gene JMJ706 encodes H3K9 demethylase required for floral organ development. Proc. Natl. Acad. Sci. U.S.A. 105, 13679–13684. doi: 10.1073/pnas.0805901105
To, T. K., and Kim, J. M. (2014). Epigenetic regulation of gene responsiveness in Arabidopsis. Front. Plant Sci. 4:548. doi: 10.3389/fpls.2013.00548
Tsukada, Y., Fang, J., Erdjument-Bromage, H., Warren, M. E., Borchers, C. H., Tempst, P., et al. (2006). Histone demethylation by a family of JmjC domain-containing proteins. Nature 439, 811–816. doi: 10.1038/nature04433
van Dijk, K., Ding, Y., Malkaram, S., Riethoven, J. J. M., Liu, R., Yang, J. Y., et al. (2010). Dynamic changes in genome-wide histone H3 lysine 4 methylation patterns in response to dehydration stress in Arabidopsis thaliana. BMC Plant Biol. 10:238. doi: 10.1186/1471-2229-10-238
Vogel, J. T., Zarka, D. G., Van Buskirk, H. A., Fowler, S. G., and Thomashow, M. F. (2005). Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis. Plant J. 41, 195–211. doi: 10.1111/j.1365-313X.2004.02288.x
Yang, H., Mo, H., Fan, D., Cao, Y., Cui, S., and Ma, L. (2012a). Overexpression of a histone H3K4 demethylase, JMJ15, accelerates flowering time in Arabidopsis. Plant Cell Rep. 31, 1297–1308. doi: 10.1007/s00299-012-1249-5
Yang, H. C., Han, Z. F., Cao, Y., Fan, D., Li, H., Mo, H. X., et al. (2012b). A companion cell-dominant and developmentally regulated H3K4 demethylase controls flowering time in arabidopsis via the repression of FLC expression. PLoS Genet. 8:e1002664. doi: 10.1371/journal.pgen.1002664
Zhang, X., Bernatavichute, Y. V., Cokus, S., Pellegrini, M., and Jacobsen, S. E. (2009). Genome-wide analysis of mono-, di- and trimethylation of histone H3 lysine 4 in Arabidopsis thaliana. Genome Biol. 10, R62. doi: 10.1186/gb-2009-10-6-r62
Keywords: histone methylation, jumonji demethylase, JMJ15, abiotic stress tolerance gene, epigenetic regulation, H3K4me3, chromatin modification
Citation: Shen Y, Conde e Silva N, Audonnet L, Servet C, Wei W and Zhou D-X (2014) Over-expression of histone H3K4 demethylase gene JMJ15 enhances salt tolerance in Arabidopsis. Front. Plant Sci. 5:290. doi: 10.3389/fpls.2014.00290
Received: 08 April 2014; Accepted: 04 June 2014;
Published online: 24 June 2014.
Edited by:
Keqiang Wu, National Taiwan University, TaiwanReviewed by:
Judy Brusslan, California State University, USAXuncheng Liu, Chinese Academy of Sciences, China
Copyright © 2014 Shen, Conde e Silva, Audonnet, Servet, Wei and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dao-Xiu Zhou, Institut de Biologie des Plantes, Université Paris-Sud 11, B630, 91405 Orsay, France e-mail:ZGFvLXhpdS56aG91QHUtcHN1ZC5mcg==
 Yuan Shen
Yuan Shen Natalia Conde e Silva
Natalia Conde e Silva Laure Audonnet1
Laure Audonnet1 Wei Wei
Wei Wei Dao-Xiu Zhou
Dao-Xiu Zhou