
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci., 25 June 2014
Sec. Plant Development and EvoDevo
Volume 5 - 2014 | https://doi.org/10.3389/fpls.2014.00284
This article is part of the Research TopicMolecular basis of fruit developmentView all 12 articles
Plant evolution is largely driven by adaptations in seed protection and dispersal strategies that allow diversification into new niches. This is evident by the tremendous variation in flowering and fruiting structures present both across and within different plant lineages. Within a single plant family a staggering variety of fruit types can be found such as fleshy fruits including berries, pomes, and drupes and dry fruit structures like achenes, capsules, and follicles. What are the evolutionary mechanisms that enable such dramatic shifts to occur in a relatively short period of time? This remains a fundamental question of plant biology today. On the surface it seems that these extreme differences in form and function must be the consequence of very different developmental programs that require unique sets of genes. Yet as we begin to decipher the molecular and genetic basis underlying fruit form it is becoming apparent that simple genetic changes in key developmental regulatory genes can have profound anatomical effects. In this review, we discuss recent advances in understanding the molecular mechanisms of fruit endocarp tissue differentiation that have contributed to species diversification within three plant lineages.
In general, fruits can be divided into two classes; dry fruits and fleshy fruits. Dry fruits are thought to predate their fleshy counterparts and are typically dispersed by physical forces (Scutt et al., 2006). Once the seeds mature, they are ejected by pod shattering, swept up by the wind, or adhere to animal surfaces for transport (epizoochory). In contrast, seed dispersal in fleshy fruits most often depends on animals consuming the fruit and dispersing the seeds after ingesting or discarding them. Whether it is a dry or fleshy fruit, all fruits contain tissue layers derived from the carpel ovary which are collectively called the pericarp (Figure 1). The pericarp can often be further differentiated into additional layers called endocarp (innermost layer), mesocarp (intermediate layer), and exocarp (skin or surface layer). Pericarp differentiation in dry fruits is often difficult to discern as each layer sometimes only contains a few rows of cells. In most fleshy fruits, the mesocarp comprises the soft edible portion of the fruit but in some exceptions the fleshy portion is formed from tissues other than the ovary (Figure 1). These are sometimes known as false fruits. For example, apple produces a pome fruit in which the core represents the true ovary derived fruit and the edible portion originates from the hypanthium; formed from the fused base of petals and sepals. In contrast, the fleshy portion of the strawberry is formed from the flower receptacle.
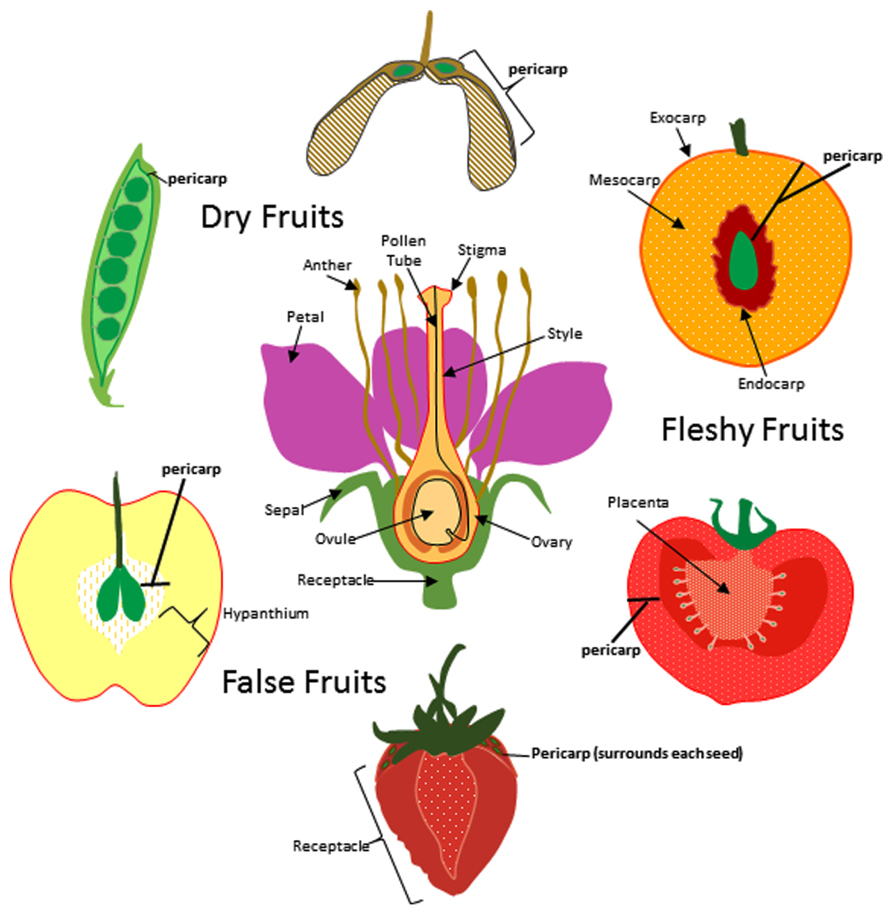
FIGURE 1. Origin of fruit tissue layers in dry, fleshy, and false fruits. For simplicity a flower with a single carpel is shown at center though it is important to note that many of the flowers that give rise to the fruits depicted here produce multiple carpels. The ovary and other floral tissues are indicated and the carpel is outlined in red. Pericarp (bold) is indicated for pea, maple, peach, tomato, strawberry and apple fruits. Exocarp, mesocarp, and endocarp are indicated for peach.
The endocarp is differentiated from the inner layer of the ovary and is the tissue layer immediately adjacent to the seed. It plays diverse roles in fruit function and can be fleshy as found in watermelon, fibrous like in mango, or extremely hard and durable as in a peach. Fruits with a hardened endocarp are called drupes. Drupes include a number of economically important crops such as peach, cherry, plum, almond, coffee, mango, olive, coconut, pistachio, date, raspberry, oil palm, and walnuts (Figure 2). The hardened endocarp provides a physical barrier around the seed protecting it from disease and herbivory (Doster and Michailides, 1999). The seeds of drupes are dispersed by animals either after consumption (blackberries) or upon being discarded (peaches). Once dispersed the seeds escape their woody enclosure via cracking and splitting of the endocarp shell due to environmental exposure.

FIGURE 2. Examples of lignified endocarps in drupes after removal of exocarp and mesocarp. Seeds are contained inside and not shown (lower row is magnified for visibility). (1) Coconut, (2) mango, (3) Walnut, (4) Peach, (5) Apricot, (6) Olive, (7) Date, (8) Pistachio, (9) Blackberry, and (10) Cherry.
In dry fruits the endocarp plays a primary role in seed dispersal. Dry fruits are generally categorized as either dehiscent or indehiscent depending on whether or not the pericarp splits open at maturity. Dehiscence is a mechanism of seed dispersal whereby the pod is forcibly opened by internal physical tension which builds during fruit maturation, causing the seeds to be suddenly discharged. Wisteria represents an extreme case in which the pods are explosive, ejecting the seeds very long distances. Other examples of dehiscent fruits include sweet pea, soybean, alfalfa, milkweed, mustard, cabbage, and poppy. Dry indehiscent fruits do not undergo this process and include a number of nuts, sunflowers, and windborn seed types such as the winged seeds found in maple and ash or cypsela-type structures produced by dandelions.
In dehiscent fruits, differentiation of the endocarp and specialized adjoining tissue layers from the mesocarp regulates pod shatter. This process has been extensively studied in Arabidopsis thaliana which is in the family Brassicaceae (reviewed by Ferrándiz, 2002; Dinneny and Yanofsky, 2004; Dinneny et al., 2005; Lewis et al., 2006). Arabidopsis fruits form as a bivalved silique containing regularly arranged seeds (Figure 3). The pericarp in each silique forms two valves that sandwich a thin papery tissue called the septum onto which the seeds are attached. The valves are connected to the septum on two sides by an external part of the septum called the replum. The endocarp is sub-divided into two layers; endocarp A (ena) and endocarp B (enb) that line the inner surface of the valves. A distinct tissue layer referred to as the valve margin forms a hinge on either side of the replum at the tip of the silique. Upon maturation, cells within the ena layer secrete cell wall degrading enzymes while cells within the enb layer, vascular bundles within the replum, the valve margins, and patches of neighboring mesocarp lignify and harden (Ferrándiz, 2002; Liljegren et al., 2004; Ogawa et al., 2009). This simultaneous separation and hardening of the enb, valve margin, inner replum, and adjoining mesocarp tissues creates tension forces that eventually cause the pod to split open along a distinct separation layer that divides the valve margins from the replum. Silique dehiscence is a highly coordinated process that is tightly coupled to complex differential pericarp tissue patterning of the mesocarp, endocarp, valve, valve margins, separation layer, and replum.
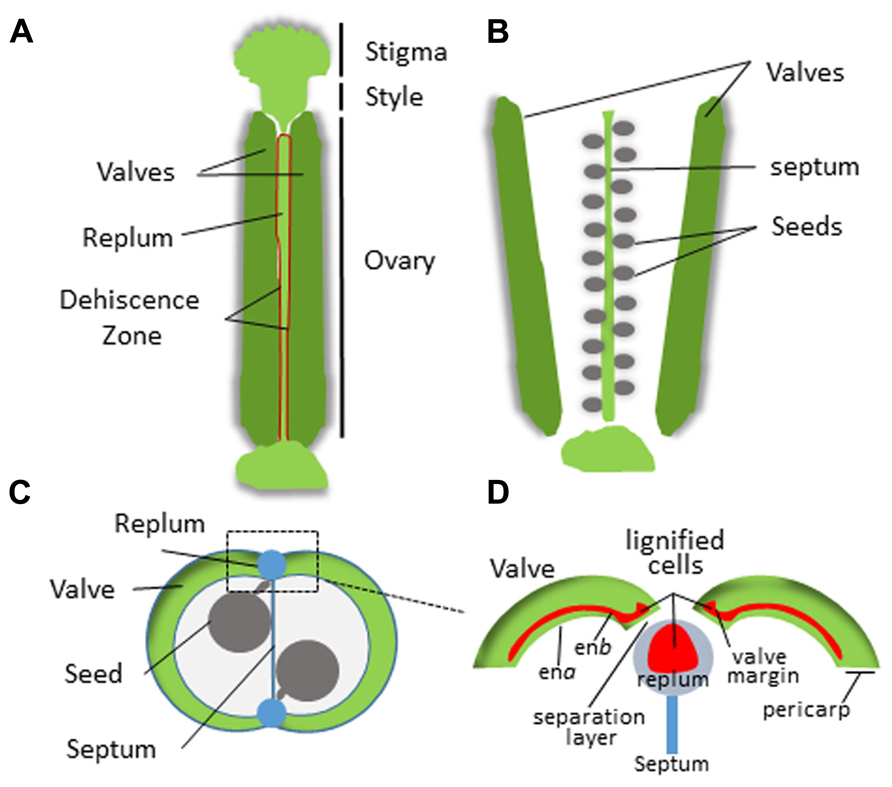
FIGURE 3. Structure of the Arabidopsis silique. (A) Intact silique prior to dehiscence. Dehiscence zone is highlighted in red. (B) Valve separation after dehiscence revealing the seeds attached to the septum. (C) Cross section of the silique. (D) Magnified view of the replum region. The pericarp tissue is indicated and lignification zones are shown in red.
In addition to seed protection and dispersal, the endocarp also plays an important role in sustaining and communicating with developing seeds. Seeds are connected to the maternal fruit tissue via an umbilical structure called the funiculus. The funiculus initiates from the seed coat and attaches to the placenta on the ovary wall. As the fruit matures, the placental layer of the ovary often becomes part of, or is fused to the endocarp.
A hallmark of both drupes and dehiscent fruits is the hardening of the endocarp as the fruit matures. Hardening occurs via secondary cell wall formation and lignification. The process of secondary wall formation in fruit tissues has not been studied to any great extent. However, based on the structural similarities between endocarp tissue and wood, information about this process can be inferred from studies on wood formation. In plant stems, xylem cells undergo a series of changes as they transform from fleshy to woody tissue. These include cell elongation, cell expansion, secondary cell wall deposition, programmed cell death, and finally heartwood formation (Dejardin et al., 2010). Secondary walls are comprised of multiple layers made up of cellulose, hemi-cellulose, and lignin with smaller amounts of pectin and proteins.
Lignin provides a matrix within secondary cell walls for polymerization of cellulosic and hemi-cellulosic polymers which together contribute to providing tissue rigidity and tensile strength (Novaes et al., 2010). Most of the genes for the major enzymes in the pathway and the potential regulatory points have been identified (Figure 4; Boerjan et al., 2003). Lignin is formed from the phenylpropanoid pathway, the end products of which are coniferyl and sinapyl alcohols. These lignin monomers serve as the basis for lignification which is the process of producing the lignin polymer via oxidative reactions aided by peroxidases and laccases. Radical coupling of the monomers, particularly cross-coupling with the growing polymer, is a multi-step process that produces the complex lignin polymer.
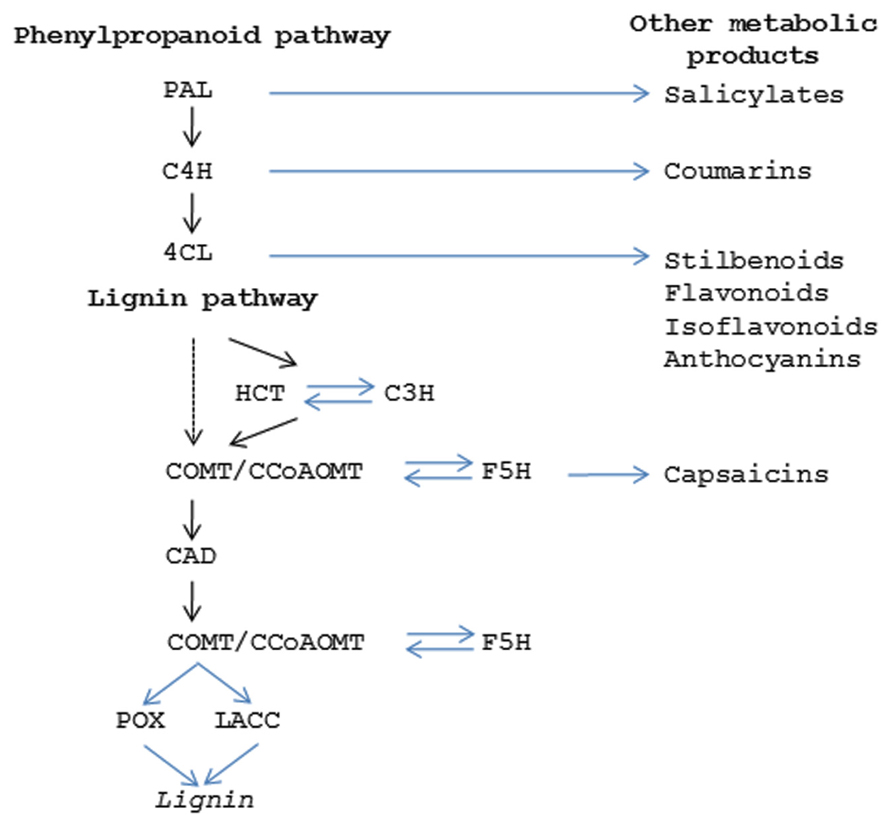
FIGURE 4. Secondary metabolic pathways. Diagram showing the enzymes in the phenylpropanoid pathway (PAL, phenylalanine ammonia lyase; C4H, cinnamic acid 4-hydroxylase; 4CL,4-coumarate:coenzyme A ligase) which produces the precursor products for lignin (HCT, hydroxycinnamoyl-CoA shikimate/quinate hydroxycinnamoyl transferase; C3H, 4-coumarate 3 –hydroxylase; COMT, caffeic acid O-methyltransferase; CCoAOMT, Caffeoyl CoA O-methyltransferase; F5H, ferulate 5-hydroxylase; CAD, cinnamyl alcohol dehydrogenase; POX, peroxidase; LACC, laccase). Steps in phenylpropanoid and ligin pathways that give rise to other secondary metabolism products are indicated.
The mechanism of endocarp hardening in peach has been investigated to a limited extent examining only one or two components or enzymes in the composition and formation of the stone tissue (Ryugo, 1963; Abeles and Biles, 1991; Alba et al., 2000; Hayama et al., 2006) Ryugo documented in the early 1960s that peach stones are rich in lignin, the seasonal pattern of lignin accumulation, and the presence of lignin biosynthesis intermediates (Ryugo, 1961, 1963). These studies and others have shown an increase in stone dry weight and lignification that begins in the second stage of fruit development until maturity (Ryugo, 1961; Nakano and Nakamura, 2002). More recently, biochemical analysis of drupes including olive, black walnut, peach, and coconut indicate they contain nearly twice as much lignin as wood, suggesting that the process of secondary wall formation can occur to a relatively extreme degree in fruit endocarp tissues (Mendu et al., 2011).
In addition to lignin, the phenylpropanoid pathway produces other secondary metabolic products that play important roles in fruit function (Figure 4). In some cases these compounds are critical for conferring seed protection and specifying seed dispersal. Coumarins, stilbenes, flavonols, and isoflavonoids have anti-microbial properties that limit bacterial and fungal disease (Dixon and Paiva, 1995). Other compounds contribute to fruit flavor and aroma; either attracting or deterring herbivores (Smith, 1982; Biggs and Northover, 1988; Peters and Constabel, 2002; Vom Endt et al., 2002). Herbivores are also strongly influenced by fruit coloration which is often attributable to anthocyanins and confer red or purple colorations. While this topic will not be extensively covered here, the fact that many of these functions arise from modifications of the same core enzymatic pathway highlights how relatively small changes in the control of secondary metabolic products can have large impacts on fruit phenotypes.
Advances in genetics and genomics technologies are speeding identification of the underlying genes and signaling pathways that control differentiation of ovarian tissues into endocarp, mesocarp, and exocarp. Arabidopsis is leading the way and the information gained is now being translated to numerous other crops. While our current knowledge is still limited, it is becoming apparent that the same or very similar cellular programs contribute to pericarp tissue differentiation in a variety of species. Here, we review and discuss the developments regarding this emerging field of study in the Brassicaceae, Rosaceae, and Solanaceae families.
Brassicaceae includes a number of economically important plants such as mustard, cabbage, radish, broccoli, and turnips. The model plant Arabidopsis thaliana is also a member of this family. Most Brassica species have a dehiscent pod-like fruit called a silique (long and narrow) or silicle (short and wide) and contain a distinctive replum tissue that separates the two valve margins. Mutagenesis screens in Arabidopsis have generated a large number of fruit morphology mutants. Some of these were found to contain defects in the dehiscence process and were named according to their phenotypes including indehiscent (ind), shatterproof (shp), alcatraz (alc), spatula (spt), fruitfull (ful), and replumless (rpl; reviewed by Dinneny and Yanofsky, 2004; Dinneny et al., 2005; Lewis et al., 2006). The identification and cloning of the underlying genes has provided insight into the molecular mechanisms of dehiscence and how pericarp tissues differentiate and lignify during development.
Specific zones within the pericarp are controlled by a coordinated set of transcription factors (TF) that specify tissue fate (Figure 5). The lignified valve margin layer responsible for pod shatter is determined by two partially redundant genes, SHP1 and SHP2, which encode MADS-BOX TFs. SHP is closely related to the class C gene Agamous (AG) that regulates flower carpel and stamen identity (Liljegren et al., 2000). Siliques in shp1/shp2 double mutants do not lignify within the valve margin layer and fail to dehisce. Specification of valve cell fate by SHP1 and SHP2 is delimited by another MADS-BOX TF called FUL. FUL is expressed throughout the valves and negatively regulates SHP1 and SHP2, restricting their activity to the valve margin. Arabidopsis ful mutants produce siliques where the entire valve mesocarp lignifies while FUL over-expression leads to conversion of valve margins and outer replum into non-lignified valve tissue; resulting in indehiscent siliques (Ferrándiz et al., 2000). SHP1 and SHP2 positively regulate a basic helix loop helix (bHLH) TF called IND (Liljegren et al., 2004). IND is also negatively regulated by FUL and has been shown to prevent valve margin cells from adopting a valve identity (Girin et al., 2010). It does this by coordinating an auxin gradient in the separation layer cells resulting in the formation of lignified valve margin tissues required for valve separation (Sorefan et al., 2009).
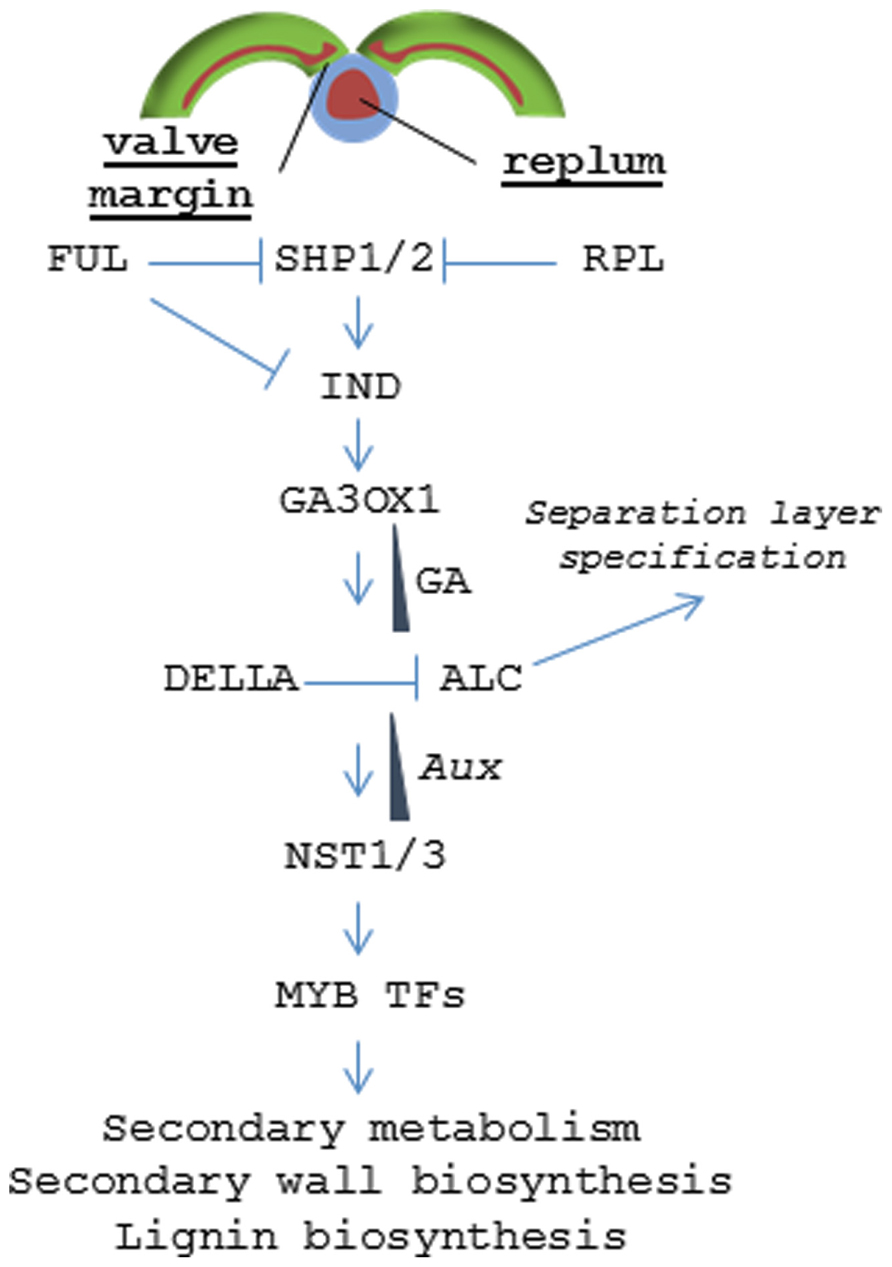
FIGURE 5. Signaling pathway regulating dehiscence in Arabidopsis. Model depicting the hierarchy of transcription factors that control tissue patterning in the valve margin and replum.
The non-lignified valve margin layer is determined by ALC and SPT, two partially redundant bHLH class TFs (Rajani and Sundaresan, 2001; Groszmann et al., 2011). alc mutants develop relatively normal siliques but lack the non-lignified layer (separation layer) that separates the lignified valve margin from the replum lignification zone. This blocks valve separation resulting in an indehiscent phenotype. ALC is negatively regulated by gibberellic acid (GA) through the DELLA repressor. IND induces expression of a gene encoding a GA activating enzyme (GA3OX1) resulting in GA accumulation in the separation layer and subsequent dissociation of the DELLA protein from ALC (Arnaud et al., 2010).
In the replum, RPL serves a similar function as FUL and prevents those non-lignifying cells from adopting a valve margin cell fate by inhibiting SHP2 expression in the replum (Roeder et al., 2003). In the more severe rpl mutant phenotypes, the lignified valve margin layer intrudes into the replum lignification layer resulting in a partially indehiscent silique. RPL encodes a BELL1 family homeodomain TF (Roeder et al., 2003). BELL1 had been previously known to negatively regulate AG (Yanofsky et al., 1990; Western and Haughn, 1999).
SHP1 and SHP2 have retained some class C function and are marginally redundant with AG in ovule and floral organ differentiation and appear to be regulated by AG (Pinyopich et al., 2003). Ectopic expression of SHP resulted in conversion of sepals to carpel-like structures even in the absence of AG (Favaro et al., 2003). Thus, sub-functionalization of AG genes has resulted in overlapping and partially redundant pathways controlling different stages of flower and fruit development (Savidge et al., 1995; Colombo et al., 2010). This is also evident for STK, another AG-like MADS-BOX gene that resides in a distinct clade. STK controls funiculus development and seed release and shares partial redundancy with AG, SHP1 and SHP2 in specifying ovule cell fate but does not have a class C function (Pinyopich et al., 2003; Zahn et al., 2006). Still, STK has retained the capacity for class C function as ectopic STK expression can complement carpel formation in an agamous mutant (Favaro et al., 2003). All four members of the AG lineage are known to function in multi-meric MADS-BOX protein complexes with proteins encoded by members of the SEPALLATA (SEP) floral organ identity genes that together modulate downstream transcriptional activation (Davies et al., 1996; Pelaz et al., 2000; Favaro et al., 2003). The finding that AG-like genes independently control both dehiscence and seed release imply that this closely related family of transcriptional regulators has evolved to control distinct fruit development processes (Pinyopich et al., 2003).
While the mechanisms regarding valve margin and replum specification are known in Arabidopsis, signaling associated with enb determination is less clear. Each of the dehiscence mutants shp1, shp2, ind, alc, ful, and rpl show relatively normal endocarp development with the exception of the quintuple mutant ind alc shp1 shp2 ful that displays a complete loss of pericarp lignification (Liljegren et al., 2004). This finding suggests that enb cell fate requires this same pathway but there may be significant redundancy and/or signaling feedback loops that are not fully understood. But once pericarp tissue identity has been established, at least one downstream pathway leading to tissue differentiation and lignification is known. Two NAC (NO APICAL MERISTEM) family TFs called NST1 and NST3 (NO SECONDARY WALL THICKENING) [also known as SND (SECONDARY WALL-ASSOCIATED NAC DOMAIN)] were found to regulate secondary wall formation and lignification within the endocarp layers (Mitsuda and Ohme-Takagia, 2008). nst1 nst3 double mutants show little or no lignin accumulation and were found to be required for expression of genes involved in cell wall biosynthesis and secondary metabolism (Mitsuda and Ohme-Takagia, 2008). NST1 was also shown to regulate anther dehiscence and lignification of woody and vascular tissues (Mitsuda et al., 2005; Zhong et al., 2007). NST1 acts upstream of a series of MYB (myeloblastosis) TFs that, in turn, directly regulate the expression of genes encoding key enzymes in the phenylpropanoid pathway that drives lignin biosynthesis (Mitsuda et al., 2007; Zhong et al., 2007, 2008). Orthologs of NST1 have similar functions in Medicago and poplar vascular tissues; suggesting that lignification in endocarp tissues occurs via the same pathway as that in vegetative tissues and wood (Zhao et al., 2010; Zhong et al., 2010). Still, there is a gap in our understanding of how NST1 and NST3 become activated in a tissue specific fashion. The finding that IND controls auxin patterning may hold the key as wood formation is also known to be regulated by the establishment of local auxin gradients (Nilsson et al., 2008; Sorefan et al., 2009).
Based on the knowledge gained in Arabidopsis, a number of researchers have evaluated whether these same genetic pathways are conserved in other Brassica species. The valve margins of dehiscent fruit in Lepidium campestre are very similar to that of Arabidopsis and expression of ALC, IND, SHP1, and SHP2 was likewise found to be limited to the valve margins (Mummenhoff et al., 2009). Lenser and Theißen (2013) showed that RNAi knock-down or over-expression of IND, ALC, SHP, or FUL resulted in the anticipated indehiscent phenotypes and mimicked those observed in Arabidopsis with only minor differences. The regulatory interactions among these genes in FUL or IND lines were also conserved as ALC was found to be a negative regulator of IND in both L. campestre and Arabidopsis (Lenser and Theißen, 2013). In contrast, gene expression of ALC, IND, SHP1, and SHP2 was found to be abolished in the tissue corresponding to the valve margins in Lepidium appelianum, a Brassica species that produces an indehiscent fruit lacking the separation layer (Mummenhoff et al., 2009). The authors concluded that the evolution of indehiscence in this species likely involved changes in an upstream regulator of the pathway. Expression and functional studies ruled out known regulators including orthologs of FUL, RPL, and APETALA2 (AP2; Mühlhausen et al., 2013). Some dehiscent Brassica species vary with respect to the development of valve margins. Erucaria erucarioides and Cakile lanceolata produce heteroarthrocarpic fruits where only the proximal segment of the silique dehisces while the distal portion remains indehiscent (Avino et al., 2012). Expression of the valve margin identify genes ALC, FUL, IND, RPL, SHP1, and SHP2 was largely conserved in the proximal dehiscent part of the fruit but absent in the distal indehiscent portion. Collectively, these studies indicate that evolutionary adaptations in Brassica siliques are, in part, driven by changes in the expression of a single coordinated developmental pathway that helps define the valve, valve margin, separation layer, and replum lignification zones.
In contrast to the Brassicaceae, plants in the family Rosaceae encompass an extremely wide range of fruiting types including drupes, pomes, achenes, as well as a number of dry dehiscent and indehiscent fruits. The genus Prunus exclusively contains drupes including peaches, plums, apricots, almonds, and cherries which produce a large lignified endocarp that surrounds the seed; commonly called the stone. These fruits grow in a sigmoidal pattern and display a pause in growth that coincides with endocarp hardening. This may be a consequence of the increased carbon and energy demands associated with lignification (Callahan et al., 2009). Recent studies on the pattern and timing of endocarp lignification reveal it is a highly coordinated process that occurs over a 2- to 3-week period (Tani et al., 2007; Dardick et al., 2010; Hu et al., 2011; Lombardo et al., 2011; Figure 6). While the timing can vary between cultivars, lignin is often first detectable approximately 35-45 days after bloom in a thin endocarp layer along the fruit suture and in the funiculus. But after several days the entire endocarp begins to lignify. Hardening appears to follow the same pattern as lignin accumulation since the tissue in which lignin is first detectable is also the first to harden.
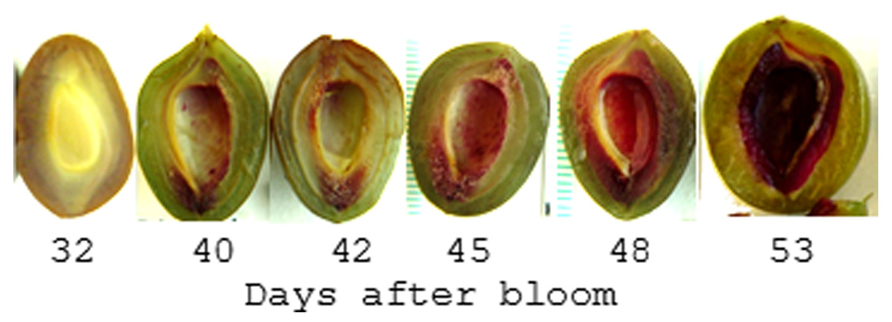
FIGURE 6. Pattern of lignin production in plum endocarp. Shown is a plum fruit series sectioned parallel to the suture line and stained with Phloroglucinol-HCL which turns red in the presence of lignin. After 53 days the endocarp begins to harden such that it can no longer be cut with a scalpel.
While functional studies are still lacking, expression profiling data suggests that many of the same genes that control dehiscence in Brassica species also control endocarp development in peach (Prunus persica; Dardick et al., 2010; Figure 7). The peach homologs of SHP and STK were found to be up-regulated in the endocarp shortly after pollination. SHP and STK expression were restricted to the endocarp and seed but gradually decline near the onset of lignin accumulation. Likewise, FUL expression remained higher in the mesocarp and exocarp but was constitutively low in the endocarp. This is consistent with a possible role in delimiting endocarp lignification margins. Upon the decline of SHP and STK, the expression of a peach NST1 homolog rapidly accumulated along with secondary metabolism and cell wall biosynthesis genes. While clear homologs of ALC and IND were not found in peach, the two most similar genes were not endocarp specific (Dardick et al., 2010). ALC was previously shown to be specific to Brassica species and evolved as a recent duplication of another bHLH TF called SPATULA (SPT; Groszmann et al., 2008). Tani et al., 2011 showed that the expression patterns of peach SPT were consistent with a role in specifying endocarp margins. Collectively, these data imply that highly similar pathways likely control pericarp development in both Prunus and Brassica fruits.
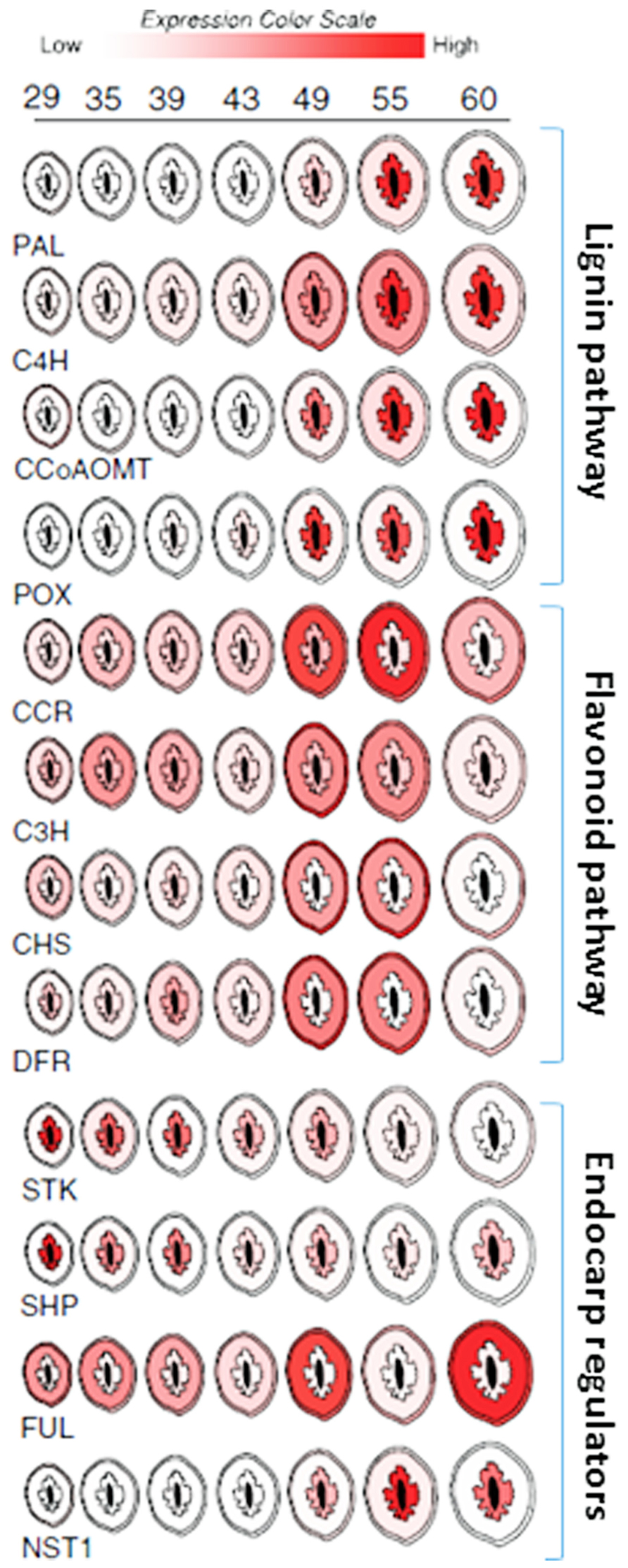
FIGURE 7. Spatial/temporal pattern of gene expression in developing peach fruit. Cross sections of peach fruits from 29 to 60 days after bloom are depicted. Relative gene expression levels within the exocarp (outer skin), mesocarp (fleshy middle), and endocarp (inner stone) sections are color coded (scale bar at top). Expression in seed (black center) is not shown. Target gene abbreviations are listed below each series and the relevant pathways are delimited by brackets and labeled (right).
Peach mesocarp and exocarp tissues accumulate other secondary metabolic compounds including flavonoids. Flavonoids are an important class of compounds found in nearly all fruit. They provide resistance against disease and pests and contribute to fruit flavor and color. Well known examples include the anthocyanins which are commonly responsible for the orange, red, and purple colorations found in many fruits. Like lignin, flavonoids are also synthesized via secondary metabolism pathways which are thought to be competitive with lignin since both draw on the same precursors of the phenylpropanoid pathway. Peach fruit showed simultaneous activation of the lignin and flavonoid pathways during early fruit development (Dardick et al., 2010; Hu et al., 2011). These events were spatially coordinated such that phenylpropanoid pathway genes were induced in all three pericarp layers; endocarp, mesocarp, and exocarp (though to a much greater degree in endocarp). But in the endocarp this upregulation was accompanied by lignin pathway induction and concomitant flavonoid pathway repression while in the mesocarp and exocarp flavonoid pathway genes were induced and lignin genes were repressed (Figure 7). Presumably this coordination allows the fruit to accumulate defense compounds, flavor, and color development in the mesocarp and exocarp while simultaneously enabling endocarp lignification. Thus, seed protection via endocarp lignification appears to be coordinated with the production of compounds necessary for defense, herbivore attraction, and seed dispersal.
There is tremendous variation in Prunus endocarp phenotypes which have been selected through breeding. For example, almond shells vary with respect to endocarp thickness, hardness, and brittleness. These agronomic qualities are critical for processing almonds and other types of nuts. Some peach varieties suffer from a phenotype called “split pit” where the endocarp does not seal along the suture leaving the seed vulnerable to pests and disease. Peach cultivars that resume rapid fruit growth before the stone has completely hardened are more likely to have split pits. Tani et al. (2007) found that SHP expression in a split pit resistant variety was lower during the lignification stage while FUL expression was significantly elevated in the sensitive variety during later stages of fruit growth.
“Stoneless” is a naturally occurring phenotype first found in a wild-type plum (Prunus domestica) species from France, Sans Noyau (Callahan et al., 2009). “Stoneless” does not completely develop the endocarp layer resulting in a partially naked seed that sits within an empty fruit cavity (Figure 8). We have observed that the “Stoneless” phenotype is strongly influenced by the environment since in years with hot spring temperatures fruit tend to contain a more complete stone while in cooler years very little stone is present (Callahan et al., 2009). The hardened tissue that remains in “Stoneless” appears to coincide with the funiculus and a portion of the plancental endocarp wall (Callahan et al., 2009). Expression studies show that the lignification process likely functions normally in “Stoneless” since secondary metabolism genes are still induced. The lack of endocarp tissue suggests that this mutant does not contain a complete endocarp layer.

FIGURE 8. Naturally occurring prunus mutant phenotype. (A) Transverse section of a wild-type plum (left) and “Stoneless” (right). The endocarp is visible as a whitish tissue layer surrounding the seed. In “Stoneless” a cavity is visible where endocarp tissue is normally found. (B) Same fruit stained with phloroglucinol-HCL to visualize lignin production (red color). Little or no lignin staining is found in “Stoneless.”
A handful of studies have also been carried out in other Rosaceous genera. Hawthorn (Crataegus spp.) produces a pome fruit that often contains a hardened endocarp like a drupe, however, some species are known to produce soft, edible endocarps. Expression profiling studies revealed that unlike that observed in those with a hardened endocarp, the lignin pathway was not upregulated in the endocarp of soft hawthorns (Dai et al., 2013). In another pome fruit, japanese pears (Pyrus pyrifolia), examination of the gene expression patterns of SHP and FUL among numerous other MADS-box genes showed that SHP expression was limited to the fruit core during early fruit development and was largely absent in the fruit cortex and skin, consistent with the ovarian origin of the core (Ubi et al., 2013). In contrast, FUL expression was more uniform and was present in skin, cortex, and core regions. While the pear core itself does not lignify, the adjoining layer is lined by disorganized stone cells or schlereids that can be found scattered throughout the hypanthium resulting in a gritty flesh texture (Tao et al., 2009). In strawberry (Fragaria ananassa), Daminato et al. (2013) found that silencing or over-expression of SHP did not appreciably alter fruit form. This may be expected since the flesh of strawberry is derived from the flower receptacle and not the pericarp. However, SHP transgenic lines did show significant changes in ripening time. These results are consistent with similar experiments in tomato where SHP was also shown to be a key regulator of fruit ripening (Itkin et al., 2009). Due to availability of whole genome sequences for a number of Rosaceous species including strawberry, apple, and peach along with established transformation systems, this family offers an excellent opportunity to further study the diversification of fruit development (Velasco et al., 2010; Shulaev et al., 2011; Verde et al., 2013).
The family Solanaceae also contains a wide variety of both dry and fleshy fruit types which have repeatedly undergone a number of berry-to-capsule and capsule-to-berry transitions. A detailed developmental analysis by Pabón-Mora and Litt (2011) showed that early developmental stages are similar among capsular and berry type fruits. Later developmental stages were marked by differentiation of endocarp including changes in cell number, cell expansion, and sclerification.
Tomato (Solanum lycopersicum) has long served as a model for fleshy fruit development and ripening. The role of a tomato SHP homolog called TOMATO AGAMOUS-LIKE 1 (TAGL1) has been extensively studied (Vrebalov et al., 2009; Itkin et al., 2009). Silencing of TAGL1 resulted in both a thinner pericarp layer and impaired ripening. Pericarp thickness was reduced by approximately 50% in TAGL1 silenced lines compared to wild-type which was attributed to fewer numbers of cell layers (Vrebalov et al., 2009). This same effect on pericarp thickness was not observed in the small fruited MicroTom tomato variety which has a naturally thin pericarp (Pan et al., 2010). Ripening in TAGL1 silenced lines or lines expressing a chimeric dominant TAGL1 repressor displayed reduced carotenoids (the pigments responsible for fruit coloration in tomato), lower levels of ethylene, and repression of ripening associated genes including those associated with ethylene biosynthesis and signaling (Itkin et al., 2009; Vrebalov et al., 2009; Pan et al., 2010). The role of TAGL1 in ripening was distinct from the previously described MADS-BOX gene RIPENING INHIBITOR (RIN), however, these two MADS-BOX genes may overlap in their ability to induce ethylene as TAGL1 protein was shown to bind the ACC synthase2 (ASC2) promoter in a transient assay (Itkin et al., 2009). In contrast, TAGL1 over-expression led to increased fruit fleshiness, fruit-like sepals that ripened, and increased accumulation of carotenoids (Itkin et al., 2009; Vrebalov et al., 2009). When transformed into an Arabidopsis shp1 shp2 double mutant, TAGL1 did not rescue the indehiscent phenotype suggesting that TAGL1 and SHP1 may have functionally diverged. In contrast, over-expression of the peach SHP homolog (also called PpPLENA) in tomato gave rise to a phenotype reminiscent of that observed for TAGL1 (Tadiello et al., 2009). Experiments to test whether peach SHP can complement Arabidopsis mutants have not yet been reported. It was shown, however, that ectopic expression of SHP derived from the Rosaceous species Taihangia rupestris led to conversion of sepals to carpelloid structures and promoted premature pod shatter (Lü et al., 2007). Collectively, these findings suggest that SHP-like genes have conserved functions but may have differentiated during the evolution of new fruiting structures and seed dispersal strategies.
Tobacco species are also members of the Solanaceae and produce dry capsular fruits that dehisce upon maturation. Over-expression of a Nicotiana tobacum homolog of FUL led to indehiscent phenotypes in both N. tobacum and N. sylvestris that was attributed to reduced lignification along the carpel midrib (Smykal et al., 2007). Knock down of SHP through virus induced gene silencing (VIGS) led to a complete loss of dehiscence and lack of lignified layers lining the dehiscence zones (Fourquin and Ferrándiz, 2012). In addition, SHP silencing caused significant alterations in flower development marked by incomplete carpel fusion and shortened styles. Meristem specification was also altered leading to additional carpel and stamen abnormalities. Simultaneous silencing of SHP and AG led to further loss of stamen and carpel identity as did silencing AG alone, suggesting that SHP plays only a minor role in C-function (Fourquin and Ferrándiz, 2012). The data imply a limited sub-functionalization of SHP from the progenitor C-class TF AG in N. benthamiana. Similar C-function overlap between AG homologs in snapdragon (called FARINELLI and PLENA, respectively) and in Petunia (called PETUNIA MADS-BOX GENE 3 (PMADS3) and FLORAL BINDING PROTEIN 6 (FBP6)) was also observed (Causier et al., 2005; Heijmans et al., 2012).
As our knowledge of fruit development expands beyond model crops, some of the genes responsible for natural variation in fruit forms are beginning to emerge. A recent report on Medicago showed that the coiled pod morphology unique to some members of that genus was likely the result of amino acid changes with a SHP homolog that promotes increased valve margin lignification (Fourquin et al., 2013). The loss of the hardened endocarp in commercial oil palm varieties was recently traced to mutations in the DNA binding domain of SHELL, a STK homolog, that were shown to prevent association with SEP (Singh et al., 2013). This stands in contrast to Arabidopsis where STK does not appear to play a role in endocarp differentiation (Pinyopich et al., 2003; Zahn et al., 2006). Findings such as these provide our first glimpse into how plants have evolved such a dizzying array of fruiting structures and seed dispersal strategies. It is now becoming clear that rapid conversions of fruit form and function are possible through changes in the expression patterns and/or activity of sub-functionalized AG-like genes or their associated regulators. These changes can lead to spatial/temporal shifts in cell fate determination accompanied by modifications in secondary metabolic activities that mediate downstream events such as lignification, coloration, and/or generation of herbivore attractants/repellents.
The current emphasis on Arabidopsis as a model system has undoubtedly introduced some level of bias into our current level of knowledge and there is a clear need for plant biologists to expand molecular developmental studies to other crops. For example, the degree to which AG-like genes and their known partners have played a role in natural selection of plant species remains to be seen. New sequencing technologies that enable gene mapping through genome-wide association studies (GWAS) along with a growing genomic toolkit promise to address these questions. Ongoing experiments to unveil the specific changes that have allowed different fruit forms to emerge within the same plant lineage will help shed light on the identity of key developmental pathways, the degree of plasticity of these regulatory systems, and how specific plants have adapted to occupy new niches.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We sincerely thank Dr. Zhongchi Liu (University of Maryland, College Park) and Dr. Courtney Hollender for their critical reading of the manuscript. We also acknowledge all of those colleagues who have made important contributions to this field but whose work was not cited.
Abeles, F., and Biles, C. (1991). Characterization of peroxidases in lignifying peach fruit endocarp. Plant Physiol. 95, 269–273. doi: 10.1104/pp.95.1.269
Alba, C. M., de Forchetti, S. M., and Tigier, H. A. (2000). Phenoloxidase of peach (Prunus persica) endocarp: Its relationship with peroxidases and lignification. Physiol. Plant. 109, 382–387. doi: 10.1034/j.1399-3054.2000.100403.x
Arnaud, N., Girin, T., Sorefan, K., Fuentes, S., Wood, T. A., Lawrenson, T., et al. (2010). Gibberellins control fruit patterning in Arabidopsis thaliana. Genes Dev. 24, 2127–2132. doi: 10.1101/gad.593410
Avino, M., Kramer, E. M., Donohue, K., Hammel, A. J., and Hall, J. C. (2012). Understanding the basis of a novel fruit type in Brassicaceae, conservation and deviation in expression patterns of six genes. Evo devo 3, 20. doi: 10.1186/2041-9139-3-20
Biggs, R., and Northover, J. (1988). Early and late-season susceptibility of peach fruits to Monilinia fructicola. Plant Dis. 72, 1070–1074. doi: 10.1094/PD-72-1070
Boerjan, W., Ralph, J., and Baucher, M. (2003). Lignin biosynthesis. Annu. Rev. Plant Biol. 54, 519–546. doi: 10.1146/annurev.arplant.54.031902.134938
Callahan, A. M., Dardick, C., and Scorza, R. (2009). Characterization of ‘Stoneless’, a naturally occurring, partially stoneless plum cultivar. J. Am. Soc. Hort. Sci. 134, 120–125.
Causier, B., Castillo, R., Zhou, J., Ingram, R., Xue, Y., Schwarz-Sommer, Z., et al. (2005). Evolution in action: following function in duplicated floral homeotic genes. Curr. Biol. 15, 1508–1512. doi: 10.1016/j.cub.2005.07.063
Colombo, M., Brambilla, V., Marcheselli, R., Caporali, E., Kater, M. M., and Colombo, L. (2010). A new role for the SHATTERPROOF genes during Arabidopsis gynoecium development. Dev. Biol. 337, 294–302. doi: 10.1016/j.ydbio.2009.10.043
Dai, H., Han, G., Yan, Y., Zhang, F., Liu, Z., Li, X., et al. (2013). Transcript assembly and quantification by RNA-Seq reveals differentially expressed genes between soft-endocarp and hard-endocarp hawthorns. PLoS ONE 8:e72910. doi: 10.1371/journal.pone.0072910
Daminato, M., Guzzo, F., and Casadoro, G. (2013). A SHATTERPROOF-like gene controls ripening in non-climacteric strawberries, and auxin and abscisic acid antagonistically affect its expression. J. Exp. Bot. 64, 3775–3786. doi: 10.1093/jxb/ert214
Dardick, C. D., Callahan, A. M., Chiozzotto, R., Schaffer, R. J., Piagnani, M. C., and Scorza, R. (2010). Stone formation in peach fruit exhibits spatial coordination of the lignin and flavonoid pathways and similarity to Arabidopsis dehiscence. BMC Biol. 9:13. doi: 10.1186/1741-7007-8-13
Davies, B., Egea-Cortines, M., de Andrade Silva, E., Saedler, H., and Sommer, H. (1996). Multiple interactions amongst floral homeotic MADS box proteins. EMBO J. 15, 4330–4343.
Dejardin, A., Laurans, F., Arnaud, D., Breton, C., Pilate, G., and Leple, J. C. (2010). Wood formation in Angiosperms. C. R. Biol. 333, 325–334. doi: 10.1016/j.crvi.2010.01.010
Dinneny, J., Weigel, D., and Yanofsky, M. (2005). A genetic framework for fruit patterning in Arabidopsis thaliana. Development 132, 4687–4696. doi: 10.1242/dev.02062
Dinneny, J., and Yanofsky, M. (2004). Drawing lines and borders: how the dehiscent fruit of Arabidopsis is patterned. Bio Essays 27, 42–49. doi: 10.1002/bies.20165
Dixon, R. A., and Paiva, N. L. (1995). Stress-induced phenylpropanoid metabolism. Plant Cell 7, 1085–1097. doi: 10.1105/tpc.7.7.1085
Doster, M. A., and Michailides, T. J. (1999). Relationship between shell discoloration of pistachio nuts and incidence of fungal decay and insect infestation. Plant Dis. 83, 259–264. doi: 10.1094/PDIS.1999.83.3.259
Favaro, R., Pinyopich, A., Battaglia, R., Kooiker, M., Borghi, L., Ditta, G., et al. (2003). MADS-box protein complexes control carpel and ovule development in Arabidopsis. Plant Cell 15, 2603–2611. doi: 10.1105/tpc.015123
Ferrándiz, C. (2002). Regulation of fruit dehiscence in Arabidopsis. J. Exp. Bot. Plant J. 53, 2031–2038. doi: 10.1093/jxb/erf082
Ferrándiz, C., Liljegren, S. J., and Yanofsky, M. F. (2000). Negative regulation of the SHATTERPROOF genes by FRUITFULL during Arabidopsis fruit development. Science 289, 436–438. doi: 10.1126/science.289.5478.436
Fourquin, C., del Cerro, C., Victoria, F. C., Vialette-Guiraud, A., de Oliveira, A. C., and Ferrándiz, C. (2013). A change in SHATTERPROOF protein lies at the origin of a fruit morphological novelty and a new strategy for seed dispersal in medicago genus. Plant Physiol. 162, 907–917. doi: 10.1104/pp.113.217570
Fourquin, C., and Ferrándiz, C. (2012). Functional analyses of AGAMOUS family members in Nicotiana benthamiana clarify the evolution of early and late roles of C-function genes in eudicots. Plant J. 71, 990–1001. doi: 10.1111/j.1365-313X.2012.05046.x
Girin, T., Stephenson, P., Goldsack, C. M., Kempin, S. A., Perez, A., Pires, N., et al. (2010). Brassicaceae INDEHISCENT genes specify valve margin cell fate and repress replum formation. Plant J. 63, 329–338. doi: 10.1111/j.1365-313X.2010.04244.x
Groszmann, M., Paicu, T., Alvarez, J. P., Swain, S. M., and Smyth, D. R. (2011). SPATULA and ALCATRAZ, are partially redundant, functionally diverging bHLH genes required for Arabidopsis gynoecium and fruit development. Plant J. 68, 816–829. doi: 10.1111/j.1365-313X.2011.04732.x
Groszmann, M., Paicu, T., and Smyth, D. R. (2008). Functional domains of SPATULA, a bHLH transcription factor involved in carpel and fruit development in Arabidopsis. Plant J. 55, 40–52. doi: 10.1111/j.1365-313X.2008.03469.x
Hayama, H., Ito, A., Shimada, T., and Kashimura, Y. (2006). Cellulose synthesis during endocarp hardening of peach fruit. J. Hort. Sci. Biotechnol. 81, 651–655.
Heijmans, K., Ament, K., Rijpkema, A. S., Zethof, J., Wolters-Arts, M., Gerats, T., et al. (2012). Redefining C and D in the petunia ABC. Plant Cell 24, 2305–2317. doi: 10.1105/tpc.112.097030
Hu, H., Liu, Y., Shi, G. L., Liu, Y. P., Wu, R. J., Yang, A. Z., et al. (2011). Proteomic analysis of peach endocarp and mesocarp during early fruit development. Physiol. Plant. 142, 390–406. doi: 10.1111/j.1399-3054.2011.01479.x
Itkin, M., Seybold, H., Breitel, D., Rogachev, I., Meir, S., and Aharoni, A. (2009). TOMATO AGAMOUS-LIKE 1 is a component of the fruit ripening regulatory network. Plant J. 60, 1081–1095. doi: 10.1111/j.1365-313X.2009.04064.x
Lenser T., and Theißen G. (2013). Conservation of fruit dehiscence pathways between Lepidium campestre and Arabidopsis thaliana sheds light on the regulation of INDEHISCENT. Plant J. 76, 545–556. doi: 10.1111/tpj.12321
Lewis, M. W., Leslie, M. E., and Liljegren, S. J. (2006). Plant separation, 50 ways to leave your mother. Curr. Opin. Plant Biol. 9, 59–65. doi: 10.1016/j.pbi.2005.11.009
Liljegren, S. J., Ditta, G. S., Eshed, Y., Savidge, B., Bowman, J. L., and Yanofsky, M. F. (2000). SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature 404, 766–770. doi: 10.1038/35008089
Liljegren, S. J., Roeder, A. H., Kempin, S. A., Gremski, K., Østergaard, L., Guimil, S., et al. (2004). Control of fruit patterning in Arabidopsis by INDEHISCENT. Cell 116, 843–853. doi: 10.1016/S0092-8674(04)00217-X
Lombardo, V. A., Osorio, S., Borsani, J., Lauxmann, M. A., Bustamante, C. A., Budde, C. O., et al. (2011). Metabolic profiling during peach fruit development and ripening reveals the metabolic networks that underpin each developmental stage. Plant Physiol. 157, 1696–1710. doi: 10.1104/pp.111.186064
Lü, S., Du, X., Lu, W., Chong, K., and Meng, Z. (2007). Two AGAMOUS-like MADS-box genes from Taihangia rupestris (Rosaceae) reveal independent trajectories in the evolution of class C and class D floral homeotic functions. Evol. Dev. 9, 92–104. doi: 10.1111/j.1525-142X.2006.00140.x
Mendu, V., Harman-Ware, A. E., Crocker, M., Jae, J., Stork, J., Morton, S. III, et al. (2011). Identification and thermochemical analysis of high-lignin feedstocks for biofuel and biochemical production. Biotechnol. Biofuels 4, 43. doi: 10.1186/1754-6834-4-43
Mitsuda, N., Iwase, A., Yamamoto, H., Yoshida, M., Seki, M., Shinozaki, K., et al. (2007). NAC transcription factors, NST1 and NST3, are key regulators of the formation of secondary walls in woody tissues of Arabidopsis. Plant Cell 19, 270–280. doi: 10.1105/tpc.106.047043
Mitsuda, N., and Ohme-Takagia, M. (2008). NAC transcription factors NST1 and NST3 regulate pod shattering in a partially redundant manner by promoting secondary wall formation after the establishment of tissue identity. Plant J. 56, 768–778. doi: 10.1111/j.1365-313X.2008.03633.x
Mitsuda, N., Seki, M., Shinozaki, K., and Ohme-Takagi, M. (2005). The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. Plant Cell 17, 2993–3006. doi: 10.1105/tpc.105.036004
Mühlhausen, A., Lenser, T., Mummenhoff, K., and Theißen, G. (2013). Evidence that an evolutionary transition from dehiscent to indehiscent fruits in Lepidium (Brassicaceae) was caused by a change in the control of valve margin identity genes. Plant J. 73, 824–835. doi: 10.1111/tpj.12079
Mummenhoff, K., Polster, A., Mühlhausen, A., and Theissen, G. (2009). Lepidium as a model system for studying the evolution of fruit development in Brassicaceae. J. Exp. Bot. 60, 1503–1513. doi: 10.1093/jxb/ern304
Nakano, M., and Nakamura, M. (2002). Cracking and mechanical properties of the stone in peach cultivars after severe thinning. Acta Horticult. 592, 531–536.
Nilsson, J., Karlberg, A., Antti, H., Lopez-Vernaza, M., Mellerowicz, E., Perrot-Rechenmann, C., et al. (2008). Dissecting the molecular basis of the regulation of wood formation by auxin in hybrid aspen. Plant Cell 20, 843–855. doi: 10.1105/tpc.107.055798
Novaes, E., Kirst, M., Chiang, V., Winter-Sederoff, H., and Sederoff, R. (2010). Lignin and biomass, a negative correlation for wood formation and lignin content in trees. Plant Physiol. 154, 555–561. doi: 10.1104/pp.110.161281
Ogawa, M., Kay, P., Wilson, S., and Swain, S. (2009). Arabidopsis dehiscence zone polygalacturonase1 (ADPG1), ADPG2, and QUARTET2 are polygalacturonases required for cell separation during reproductive development in Arabidopsis. Plant Cell 21, 216–233. doi: 10.1105/tpc.108.063768
Pabón-Mora, N., and Litt, A. (2011). Comparative anatomical and developmental analysis of dry and fleshy fruits of Solanaceae. Am. J. Bot. 98, 1415–1436. doi: 10.3732/ajb.1100097
Pan, I. L., McQuinn, R., Giovannoni, J. J., and Irish, V. F. (2010). Functional diversification of AGAMOUS lineage genes in regulating tomato flower and fruit development. J. Exp. Bot. 61, 1795–1806. doi: 10.1093/jxb/erq046
Pelaz, S., Ditta, G. S., Baumann, E., Wisman, E., and Yanofsky, M. F. (2000). B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405, 200–203. doi: 10.1038/35012103
Peters, D. J., and Constabel, C. P. (2002). Molecular analysis of herbivore-induced condensed tannin synthesis: cloning and expression of dihydroflavonol reductase from trembling aspen (Populus tremuloides). Plant J. 32, 701–712. doi: 10.1046/j.1365-313X.2002.01458.x
Pinyopich, A., Ditta, G. S., Savidge, B., Liljegren, S. J., Baumann, E., Wisman, E., et al. (2003). Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature 424, 85–88. doi: 10.1038/nature01741
Rajani, S., and Sundaresan, V. (2001). The Arabidopsis myc/bHLH gene ALCATRAZ enables cell separation in fruit dehiscence. Curr. Biol. 11, 1914–1922. doi: 10.1016/S0960-9822(01)00593-0
Roeder, A. H., Ferrándiz, C., and Yanofsky, M. F. (2003). The role of the REPLUMLESS homeodomain protein in patterning the Arabidopsis fruit. Curr. Biol. 13, 1630–1635. doi: 10.1016/j.cub.2003.08.027
Ryugo, K. (1961). The rate of dry weight accumulation by the peach pit during the hardening process. Am. Soc. Horticult. Sci. 78, 132–137.
Ryugo, K. (1963). Changes in methoxyl content in the peach endocarp and some of its soluble phenolic constituents during lignification. Am. Soc. Horticult. Sci. 84, 110–115.
Savidge, B., Rounsley, S. D., and Yanofsky, M. F. (1995). Temporal relationship between the transcription of two Arabidopsis MADS box genes and the floral organ identity genes. Plant Cell 7, 721–733. doi: 10.1105/tpc.7.6.721
Scutt, C. P., Vinauger-Douard, M., Fourquin, C., Finet, C., and Dumas, C. (2006). An evolutionary perspective on the regulation of carpel development. J. Exp. Bot. 57, 2143–2152. doi: 10.1093/jxb/erj188
Shulaev, V., Sargent, D. J., Crowhurst, R. N., Mockler, T. C., Folkerts, O., Delcher, A. L., et al. (2011). The genome of woodland strawberry (Fragaria vesca). Nat. Genet. 43, 109–116. doi: 10.1038/ng.740
Singh, R., Low, E. T., Ooi, L. C., Ong-Abdullah, M., Ting, N. C., Nagappan, J., et al. (2013). The oil palm SHELL gene controls oil yield and encodes a homologue of SEEDSTICK. Nature 500, 340–344. doi: 10.1038/nature12356
Smith, D. A. (1982). “Toxicity of phytoalexins,” in Phytoalexins, eds J. A. Bailey and J. W. Mansfield (New York: John Wiley and Sons), 215-252.
Smykal, P., Gennen, J., De Bodt, S., Ranganath, V., and Melzer, S. (2007). Flowering of strict photoperiodic Nicotiana varieties in non-inducive conditions by transgenic approaches. Plant Mol. Biol. 65, 233–242. doi: 10.1007/s11103-007-9211-6
Sorefan, K., Girin, T., Liljegren, S. J., Ljung, K., Robles, P., Galván-Ampudia, C. S., et al. (2009). A regulated auxin minimum is required for seed dispersal in Arabidopsis. Nature 459, 583–586. doi: 10.1038/nature07875
Tadiello, A., Pavanello, A., Zanin, D., Caporali, E., Colombo, L., Rotino, G. L., et al. (2009). A PLENA-like gene of peach is involved in carpel formation and subsequent transformation into a fleshy fruit. J. Exp. Bot. 60, 651–661. doi: 10.1093/jxb/ern313
Tani, E., Polidoros, A., and Tsaftaris, A. (2007). Characterization and expression analysis of FRUITFULL-and SHATTERPROOF-like genes from peach (Prunus persica) and their role in split-pit formation. Tree Physiol. 27, 649–659. doi: 10.1093/treephys/27.5.649
Tani, E., Tsaballa, A., Stedel, C., Kalloniati, C., Papaefthimiou, D., Polidoros, A., et al. (2011). The study of a SPATULA-like bHLH transcription factor expressed during peach (Prunus persica) fruit development. Plant Physiol. Biochem. 49, 654–663. doi: 10.1016/j.plaphy.2011.01.020
Tao, S., Khanizadeh, S., Zhang, H., and Zhang, S. (2009). Anatomy, ultrastructure and lignin distribution of stone cells in two Pyrus species. Plant Sci. 176, 413–419. doi: 10.1016/j.plantsci.2008.12.011
Ubi, B. E., Saito, T., Bai, S., Nishitani, C., Ban, Y., Ikeda, K., et al. (2013). Characterization of 10 MADS-box genes from Pyrus pyrifolia and their differential expression during fruit development and ripening. Gene 528, 183–194. doi: 10.1016/j.gene.2013.07.018
Velasco, R., Zharkikh, A., Affourtit, J., Dhingra, A., Cestaro, A., Kalyanaraman, A., et al. (2010). The genome of the domesticated apple (Malus × domestica Borkh.). Nat. Genet. 42, 833–839. doi: 10.1038/ng.654
Verde, I., Abbott, A. G., Scalabrin, S., Jung, S., Shu, S., Marroni, F., et al. (2013). The high-quality draft genome of peach (Prunus persica) identifies unique patterns of genetic diversity, domestication and genome evolution. Nat. Genet. 45, 487–494. doi: 10.1038/ng.2586
Vom Endt, D., Kijne, J. W., and Memelink, J. (2002). Transcription factors controlling plant secondary metabolism, what regulates the regulators? Phytochemistry 61, 107–114. doi: 10.1016/S0031-9422(02)00185-1
Vrebalov, J., Pan, I. L., Arroyo, A. J., McQuinn, R., Chung, M., Poole, M., et al. (2009). Fleshy fruit expansion and ripening are regulated by the Tomato SHATTERPROOF gene TAGL1. Plant Cell 21, 3041–3062. doi: 10.1105/tpc.109.066936
Western, T. L., and Haughn, G. W. (1999). BELL1 and AGAMOUS genes promote ovule identity in Arabidopsis thaliana. Plant J. 18, 329–336. doi: 10.1046/j.1365-313X.1999.00448.x
Yanofsky, M. F., Ma, H., Bowman, J. L., Drews, G. N., Feldmann, K. A., and Meyerowitz, E. M. (1990). The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature 346, 35–39. doi: 10.1038/346035a0
Zahn, L. M., Leebens-Mack, J. H., Arrington, J. M., Hu, Y., Landherr, L. L., dePamphilis, C. W., et al. (2006). Conservation and divergence in the AGAMOUS subfamily of MADS-box genes: evidence of independent sub- and neofunctionalization events. Evol. Dev. 8, 30–45. doi: 10.1111/j.1525-142X.2006.05073.x
Zhao, Q., Wang, H., Yin, Y., Xu, Y., Chen, F., and Dixon, R. A. (2010). Syringyl lignin biosynthesis is directly regulated by a secondary cell wall master switch. Proc. Natl. Acad. Sci. U.S.A. 107, 14496–14501. doi: 10.1073/pnas.1009170107
Zhong, R., Lee, C., and Ye, Z. H. (2010). Functional characterization of poplar wood-associated NAC domain transcription factors. Plant Physiol. 152, 1044–1055. doi: 10.1104/pp.109.148270
Zhong, R., Lee, C., Zhou, J., McCarthy, R. L., and Ye, Z. H. (2008). A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell 20, 2763–2782. doi: 10.1105/tpc.108.061325
Keywords: fruit development, endocarp, dehiscence, lignification, fruit evolution
Citation: Dardick C and Callahan AM (2014) Evolution of the fruit endocarp: molecular mechanisms underlying adaptations in seed protection and dispersal strategies. Front. Plant Sci. 5:284. doi: 10.3389/fpls.2014.00284
Received: 07 March 2014; Accepted: 30 May 2014;
Published online: 25 June 2014.
Edited by:
Zhongchi Liu, Harvard University, USAReviewed by:
Angela Hay, University of Oxford, UKCopyright © 2014 Dardick and Callahan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chris Dardick, Appalachian Fruit Research Station, United States Department of Agriculture – Agricultural Research Service, 2217 Wiltshire Road, Kearneysville, WV 25442, USA e-mail:Y2hyaXMuZGFyZGlja0BhcnMudXNkYS5nb3Y=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.