
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Plant Sci., 26 February 2014
Sec. Plant Cell Biology
Volume 5 - 2014 | https://doi.org/10.3389/fpls.2014.00069
This article is part of the Research TopicEndoplasmic reticulum – shape and function in stress translationView all 12 articles
Stress caused by environmental conditions or physiological growth can lead to an accumulation of unfolded proteins in the endoplasmic reticulum (ER) causing ER stress, which in turn triggers a cytoprotective mechanism termed the unfolded protein response (UPR). Under mild-short stress conditions the UPR can restore ER functioning and cell growth, such as reducing the load of unfolded proteins through the upregulation of genes involved in protein folding and in degrading mis-folded proteins, and through autophagy activation, but it can also lead to cell death under prolonged and severe stress conditions. A diversified suite of sensors has been evolved in the eukaryotic lineages to orchestrate the UPR most likely to suit the cell’s necessity to respond to the different kinds of stress in a conserved as well as species-specific manner. In plants three UPR sensors cooperate with non-identical signaling pathways: the protein kinase inositol-requiring enzyme (IRE1), the ER-membrane-associated transcription factor bZIP28, and the GTP-binding protein β1 (AGB1). In this mini-review, we show how plants differ from the better characterized metazoans and fungi, providing an overview of the signaling pathways of the UPR, and highlighting the overlapping and the peculiar roles of the different UPR branches in light of evolutionary divergences in eukaryotic kingdoms.
Environmental or physiological conditions that interfere with the proper protein folding in the endoplasmic reticulum (ER) lead to an accumulation of potentially toxic mis-folded proteins, a condition generally termed as “ER stress”. To restore ER homeostasis, a network of inter-organelle signaling pathways mediates the “unfolded protein response” (UPR), leading to an increase of protein folding capacity in the ER (Walter and Ron, 2011). If these mechanisms of adaptation and survival to ER stress fail, the UPR signaling leads the cells toward cell death (Hetz, 2012). Even if several aspects of the UPR are conserved across eukaryotes, the mechanisms to counteract the ER stress can vary across plants, metazoans and yeast (Kimata and Kohno, 2011; Chen and Brandizzi, 2013a; Howell, 2013). This review focuses on the current understanding of how the UPR signaling pathways initiate and progress in response to the severity and duration of ER stress and addresses the overlapping and unique roles of the UPR response in eukaryotes with emphasis on multicellular eukaryotes.
In the yeast Saccharomyces cerevisiae, the UPR is mediated by inositol-requiring enzyme1 (IRE1; Ire1p; Cox et al., 1993; Mori et al., 1993), an ER-resident protein largely conserved in eukaryotes. IRE1 is a type I transmembrane protein, with an N-terminal ER luminal stress-sensing domain, and a Ser/Thr kinase domain and an endoribonuclease domain in the cytosol (Figure 1A). Metazoans and plants have expanded their UPR signaling pathways with additional ER stress sensors (Figures 1B,C). Indeed, in metazoans, at least three ER transmembrane sensors initiate the UPR: IRE1 (IRE1α and IRE1β isoforms), the activating transcription factor 6 proteins (ATF6α and ATF6β isoforms), and the protein kinase RNA-like ER kinase (PERK) (Hetz, 2012). ATF6 is a type II transmembrane protein, characterized by a C-terminal ER lumen domain and an N-terminal cytosolic domain containing a bZIP (basic leucine zipper) transcriptional factor domain, while PERK is a type I transmembrane protein, with an ER-luminal stress-sensing domain and a cytosolic Ser/Thr kinase domain (Figure 1A). In plants, the UPR regulators so far identified are two IRE1 homologs (IRE1A and IRE1B; Koizumi et al., 2001), a functional homolog of ATF6 (bZIP28; Liu et al., 2007a), and a component of the G protein complex (AGB1; Wang et al., 2007). Intriguingly, the involvement of G protein complex in UPR has not been observed in other eukaryotes possibly because of a redundancy of the multiple isoforms of the heterotrimeric GTP-binding proteins in metazoans. Moreover, a functional PERK ortholog has not been identified in plants.
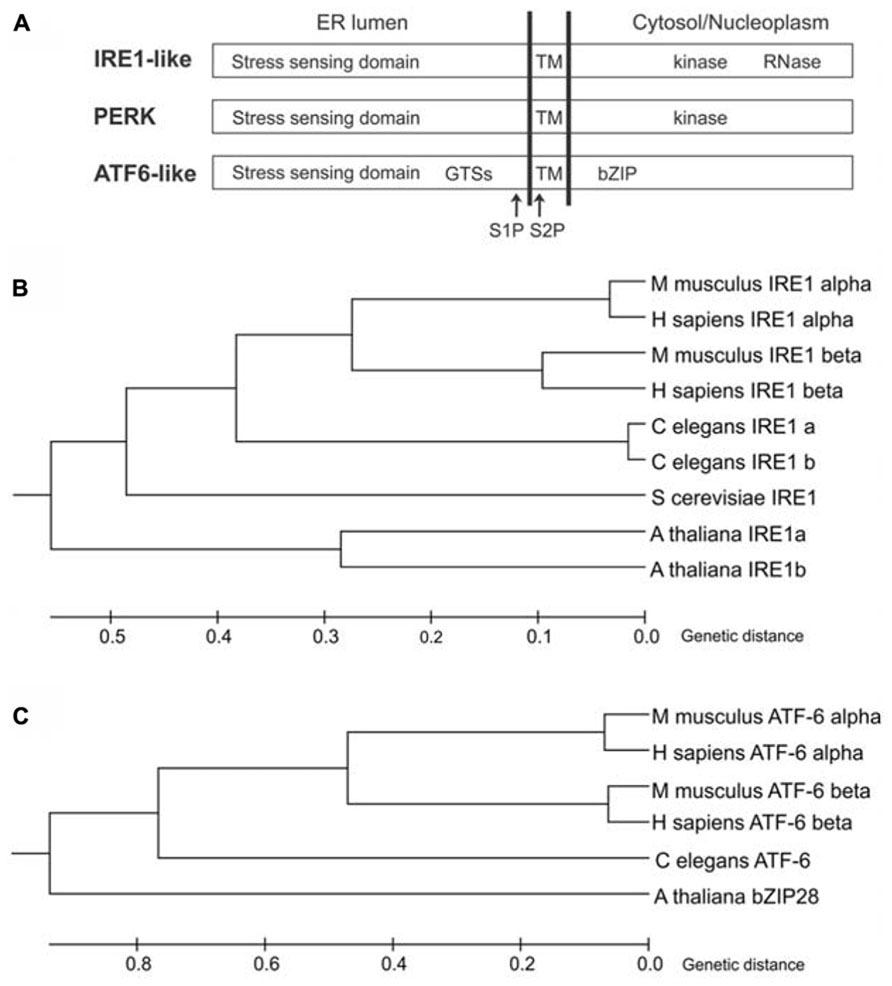
FIGURE 1. ER stress-sensing proteins in eukaryotes. (A) Schematic diagram of IRE1-like, PERK and ATF6-like proteins (not to scale). GTSs, Golgi trafficking signals; TM, transmembrane domain; S1P, site 1 protease cleavage site; S2P, site 2 protease cleavage site; bZIP, basic leucine zipper domain. (B) Phylogenetic tree analysis of IRE1-like proteins from Animalia (Homo sapiens, Mus musculus and Caenorhabditis elegans), Fungi (Saccharomyces cerevisiae) and Plantae (Arabidopsis thaliana) was constructed by the Unweighted Pair Group Method with Arithmetic Mean (UPGMA) method using MEGA6 software. NCBI reference sequences: IRE1 alpha (H.s)-NP_001424, IRE1 beta (H.s)-NP_150296, IRE1 alpha (M.m)-NP_076402, IRE1 beta (M.m)-NP_036146, IRE1a (C.e)-NP_001254135, IRE1b (C.e)-NP_001254136, IRE1 (S.c)-NP_011946, IRE1a (A.t)-NP_565419, IRE1b (A.t)-NP_568444. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Poisson correction method. (C) Phylogenetic tree analysis of ATF6-like proteins from Animalia (H. sapiens, M. musculus and C. elegans) and Plantae (A. thaliana) was constructed by the UPGMA method using MEGA6 software. NCBI reference sequences: ATF6 alpha (H.s)-NP_031374, ATF6 beta (H.s)-NP_001129625, ATF6 alpha (M.m)-NP_001074773, ATF6 beta (M.m)-NP_059102, ATF6 (C.e)-NP_510094, bZIP28 (A.t)-NP_187691. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Poisson correction method.
The mechanisms of how ER stress is sensed have been partially defined: the UPR sensors may detect ER stress (1) through the dissociation of their ER-luminal stress-sensing domain from the ER chaperones, which would be induced by the binding of ER chaperones to unfolded proteins, as shown for IRE1α, PERK, and ATF6 in metazoans (Kimata and Kohno, 2011); (2) through the direct binding of the ER-luminal domain of the UPR sensors to the unfolded proteins, as shown for IRE1 in yeast (Gardner and Walter, 2011); and (3) through post-translational modifications within the luminal domain, as observed in mammals for ATF6, where for example the hypoglycosylation is a monitoring mechanism for ER homeostasis to sense the glucose starvation or N-linked glycosylation impairment (Hong et al., 2004).
Upon ER stress induction, the UPR sensors are activated as follows: (1) the RNase domain of IRE1 is activated through its oligomerization and trans-autophosphorylation via its own kinase domain (Korennykh et al., 2009; Ali et al., 2011); (2) ATF6-orthologs are transported to the Golgi likely via specific trafficking signals (Shen et al., 2002; Srivastava et al., 2012) with the COPII vesicles (Schindler and Schekman, 2009; Srivastava et al., 2012), where the ATF6-orthologs are cleaved by the sequential action of the site 1 and site 2 proteases (S1P and S2P), thus releasing their N-terminal cytosolic transcription factor for translocation to the nucleus (Ye et al., 2000; Liu et al., 2007a); and (3) after PERK dimerization, one PERK homodimer likely inserts its flexible activation loop into the catalytic site of the adjacent homodimer, resulting in an interdimer trans-phosphorylation (Marciniak et al., 2006; Cui et al., 2011), that activates the kinase domain of PERKs. The mechanisms that lead to a role of AGB1 in the UPR are unknown (Wang et al., 2007; Chen and Brandizzi, 2012). In subcellular fractionation experiments, AGB1 has been found to be largely associated with the ER (Wang et al., 2007). Because AGB1 lacks a transmembrane domain, it is possible that post-translational modifications may modulate its UPR signaling function.
The different arms of the UPR have evolved to activate overlapping but non-identical pathways in order to restore homeostasis, or, if the ER stress persists, to trigger programed cell death (PCD) in plants and yeasts, and apoptosis in metazoans (Figure 2). In the early phases of ER stress responses in metazoans, the overload of newly synthesized proteins into the protein-overloaded ER is reduced through the selective degradation of many mRNAs encoding ER-translocating proteins by the IRE1 endonuclease activity, a process termed “regulated IRE1 dependent decay” (RIDD), and through the transient attenuation of global protein translation via PERK. RIDD promotes the rapid mRNA decay of genes encoding secretory proteins (Hollien and Weissman, 2006; Han et al., 2009) and a similar mechanism most likely operates in plants (Mishiba et al., 2013). Interestingly, RIDD-mediated decrease in ER protein overload was also demonstrated in fission yeast Schizosaccharomyces pombe, where it functions as the exclusive UPR mechanism (Kimmig et al., 2012), but not in the budding yeast Saccharomyces cerevisiae, where, indeed, the UPR does not appear to attenuate protein translation (Spear and Ng, 2003).
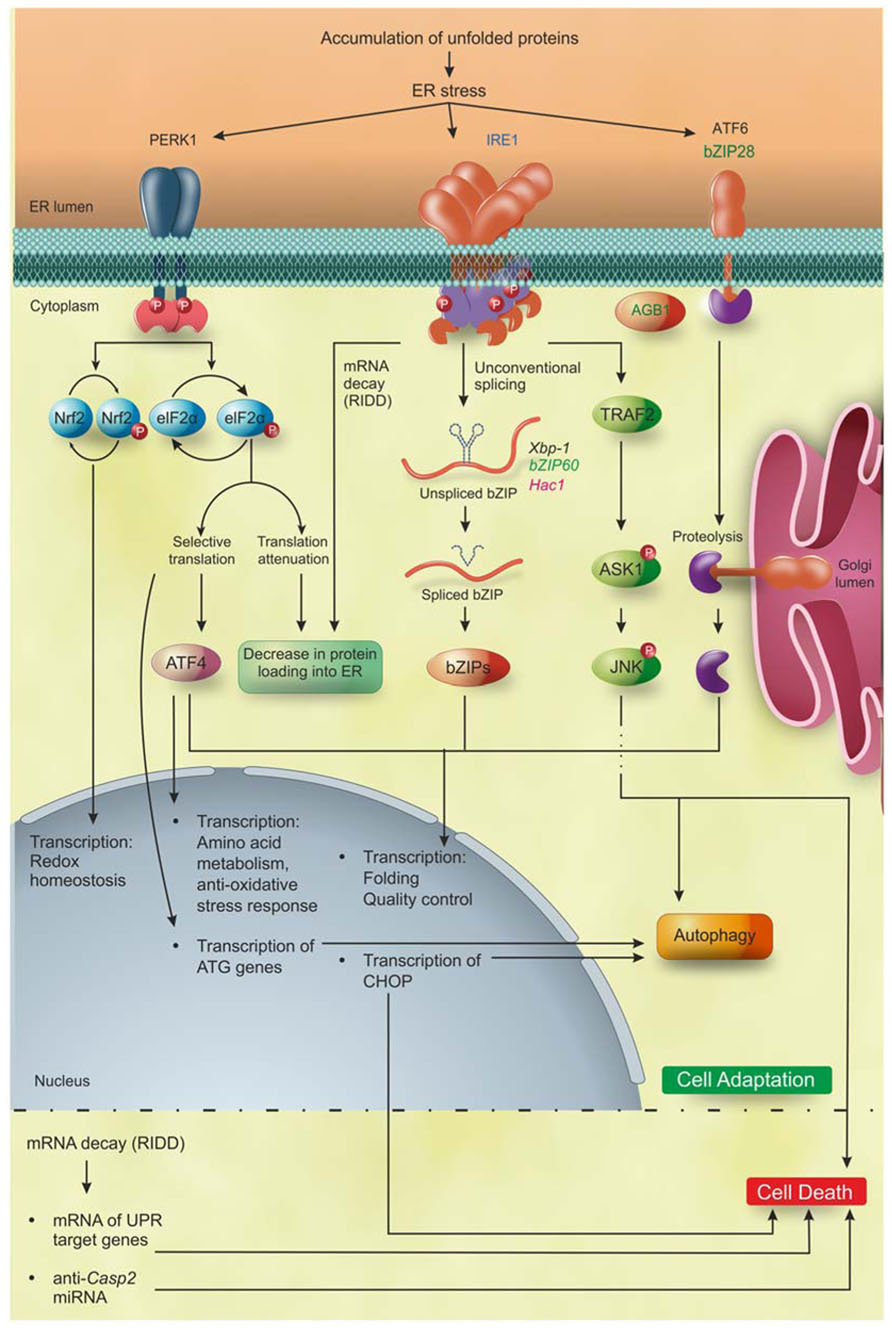
FIGURE 2. PR pathways in eukaryotes. Accumulation of unfolded proteins inside the ER lumen triggers to UPR that, during the first layer of response, leads the cell to adaptation. IRE1 is the only identified ER stress sensor in yeast and it is widely conserved in metazoan and plants. Moreover, additional ER stress sensors so far identified are bZIP28 and AGB1 in plants, and ATF6 and PERK1 in metazoan. Activation of PERK leads to phosphorylation of two proteins: Nrf2 that activates the transcription of genes involved in redox homeostasis; and eIF2α that decreases the overload of newly synthetized proteins into the ER and increases the specific translation of the transcription factor ATF4. Subsequently, ATF4 induces the expression of genes involved in amino acid metabolism, anti-oxidative stress response, folding, quality control and autophagy. The activation of IRE1 leads (1) to the selective degradation of mRNAs (RIDD) encoding ER-translocating proteins, decreasing the protein loading into the ER, and (2) to the translation of the bZIP transcription factors Xbp1/bZIP60/Hac1 mRNA in metazoan, plants and yeast, respectively, that upregulate the UPR target genes mainly involved in folding, quality control and autophagy. ATF6 in metazoans and bZIP28 in Arabidopsis are translocated into the Golgi apparatus and cleaved by Site1 and Site2 proteases, releasing the ATF6/bZIP28 transcription factors domain, which then translocates to the nucleus where it increases the expression of genes involved in folding, quality control and autophagy. Upon excessive and prolonged ER stress, all the UPR pathways lead to cell death in yeast, metazoan, and plants. In plants, the cell death executioners are still largely unknown, while in metazoan they have been partially defined. In detail, IRE1 regulates the cell death through the mRNA decay of UPR target genes and of anti-apoptotic protein (anti-Casp2). In addition, IRE1 interacting with TRAF2 triggers the ASK1/JNK pathway promoting apoptosis. Moreover, PERK/eIF2α/ATF4, IRE1, and ATF6 induce the expression of the transcription factor CHOP, involved in the induction of apoptosis. Blue, eukaryotes; black, metazoan; green, plants; pink, yeast. Figure adapted from Chen and Brandizzi (2012).
To attenuate ER stress in metazoans in addition to RIDD, PERK phosphorylates the α subunit of the eukaryotic translation initiation factor 2 (eIF2α), inhibiting the 80s ribosome assembly and down-regulating protein synthesis (Harding et al., 1999). In mammals, the phosphorylation of eIF2α is a conserved mechanism to block general protein translation not solely restricted to ER stress, and it is carried out by different kinases activated by diverse cellular stresses. The only eIF2 α-kinase conserved among eukaryotes is the GCN2 protein: GCN2/eIF2 α pathway attenuates protein translation under nutrient limitation in yeasts and mammals (Berlanga et al., 1999; Harding et al., 2000), and under amino acid starvation, abiotic and biotic stresses and plants (Lageix et al., 2008; Zhang et al., 2008). Whether the plant GCN2 may function as the metazoan PERK in the UPR is yet unknown.
Once the first layer of response to ER stress conditions is completed, in plants, metazoans and yeasts all the UPR stress sensors promote a coordinated adaptive response to protect the cell against oxidative stress, to augment protein-folding and secretory capacity in order to ensure that protein exit the ER productively, and to degrade potentially toxic unfolded proteins, by up-regulating the genes encoding oxido-reductases, ER chaperones, vesicle trafficking proteins, ER-associated degradation (ERAD) and ER-quality control (ERQC) components (Travers et al., 2000; Martínez and Chrispeels, 2003; Kamauchi et al., 2005).
The IRE1 endonuclease domain catalyzes the non-conventional cytoplasmic splicing of the mRNA encoding bZIP60/XBP1/HAC1 (in Arabidopsis, metazoans and budding yeast respectively), leading to the translation of a transcription factor (bZIP60s/XBP1s/HAC1s) that mainly upregulates the expression of ERQC and ERAD-related genes (Yoshida et al., 2001; Iwata et al., 2008; Deng et al., 2011). Among the species, the amino acid sequences of these transcription factors are not highly conserved; however, a two stem–loop structure accompanied by a consensus sequence in each loop of the IRE1-splicing mRNA substrates is remarkably conserved and associated with IRE1-mediated cleavage (Oikawa et al., 2010; Deng et al., 2011; Nagashima et al., 2011). In yeast and metazoans, the spliced substrate becomes a potent transcriptional activator, since it gains a transcriptional activation domain in the new C-terminal tail (Mori et al., 2000; Yoshida et al., 2001). It is noteworthy that in the absence of induced ER stress in budding yeasts, the unspliced HAC1 (HAC1u) mRNA is not translated, since an intron in the HAC1u mRNA blocks its translation (Chapman and Walter, 1997), while in mammalian cells the unspliced XBP1 (XBP1u) mRNA is translated. XBP1u protein along with the XBP1u mRNA is associated peripherally with the ER membrane through an amphipathic region, where it facilitates the targeting of XBP1u mRNA to IRE1 presumably to increase the cytoplasmic splicing efficiency providing a rapid response to ER stress (Yanagitani et al., 2009). Upon prolonged ER stress, XBP1u forms a complex with XBP1s, leading it to be exported from the nucleus to the cytoplasm and rapidly degraded by the proteasome, presumably shutting down the transcription of the XBP1-target genes (Yoshida et al., 2006). Intriguingly, the IRE1-spliced XBP1s mRNA loses the ER membrane-anchor domain and it is released in the cytosol, indicating a different translational place for XBP1u and XBP1s mRNAs that presumably prevents the excess of degradation of the XBP1s by XBP1u protein during ER stress (Yanagitani et al., 2009). In plants, experiments based on the expression of in-frames fluorescent protein fusion with unspliced bZIP60u, have shown an association of bZIP60u with the ER through its putative C-terminal transmembrane domain (Deng et al., 2011). However, the biological roles of unspliced bZIP60 (bZIP60u) on the ER membrane are currently unknown. Unlike yeast and metazoans, spliced bZIP60 (bZIP60s) does not gain a transcriptional activation activity, since the transcriptional activation domain is located in the N-terminal tail along with a nuclear localization signal. The IRE1-splicing produces instead a new protein deprived of the transmembrane domain (Deng et al., 2011; Nagashima et al., 2011) and characterized by an improved fine regulatory modulation of its transcription activity as recently reported in rice (Lu et al., 2012). However, how this tuning is achieved is still not clear.
Under ER stress, the transcription factor domain of bZIP28/ATF6 increases the expression of genes involved in protein folding and of other ER-stress related transcription factors such as bZIP60/XBP-1, providing a positive feedback for augmenting the UPR (Yoshida et al., 2001; Liu and Howell, 2010). Moreover, in metazoans, ATF6 enhances the expression of genes involved in ERAD, lipid biosynthesis and ER expansion, which are required to improve the capacity of the secretory pathway (Bommiasamy et al., 2009).
The UPR genes are induced through the recognition of cis-acting elements on their promoter regions by the UPR transcription factors. In budding yeast, HAC1s binds and activates the UPR element-I (UPRE-I, consensus region CAGNGTG; Mori et al., 1996) and UPRE-II (TACGTG; Fordyce et al., 2012). In mammalians, XBP1 efficiently binds the UPRE-I (GA-TGACGT-G[G/A]; Wang et al., 2000; Yamamoto et al., 2004) and the ER stress responsive element-II (ERSE-II, ATTGG–N–CCACG; Kokame et al., 2001; Yamamoto et al., 2004), while ATF6 binds the ERSE-I (ERSE-I, CCAAT–N9–CCACG; Roy and Lee, 1999) and the ERSE-II (ATTGG–N–CCACG; Kokame et al., 2001) only in the presence of the nuclear transcription factor NF-Y (Yamamoto et al., 2004). Interestingly, the mammalian ERSE-I and UPRE-I elements are conserved in plants (Oh et al., 2003; Iwata et al., 2008; Liu and Howell, 2010), where other cis-elements have also been found, such as the pUPRE-II (GATGACGCGTAC; Hayashi et al., 2013) and the pUPRE-III (TCATCG; Sun et al., 2013). Similar to ATF6 in mammals, bZIP28 binds the ERSE-I element with assistance from the transcription factors NF-Y (Liu and Howell, 2010), while bZIP60s directly binds the pUPRE-III (Sun et al., 2013) and it regulates also promoters containing the cis-element ERSE-I and UPRE-I (Iwata and Koizumi, 2005). The multiple cis-elements involved in the ER stress response and their differential binding affinities for the UPR transcription factors presumably fine temporal modulate the UPR signaling and the ER stress response. Moreover, plants have evolved an additional layer of UPR regulation. The plant-specific nuclear transcription factor NAC103 is indeed induced by ER stress presumably through bZIP60s, and the encoded protein NAC103 in turn regulates the UPR downstream genes (Sun et al., 2013). However, the precise mechanisms of the gene regulation networks are largely unknown.
In metazoans, although PERK induces general protein translation attenuation, it also favors selective protein translation. Specifically, the PERK-phosphorylated eIF2α activates the translation of mRNAs with uORF (upstream open reading frame) within their 5’ untranslated region (UTR), like the activating transcription factor 4 (ATF4; Harding et al., 2000). ATF4 protects cells against oxidative stress and ensures the supply of reducing substances (i.e., glutathione) by enhancing the metabolism of their precursors (i.e., sulfur-containing amino acids; Harding et al., 2003). Also PERK phosphorylates the transcription factor Nrf2 (nuclear factor erythroid2-releated factor 2), which translocates to the nucleus, heterodimerizes with the small Maf proteins and activates the transcription of genes involved in the redox homeostasis by binding to the antioxidant response elements on the target gene promoters (Cullinan et al., 2003).
Autophagy is an evolutionarily conserved process of bulk degradation, whereby large portions of cytoplasmic and organellar components are engulfed by double membrane vesicles (termed as “autophagosome”) and delivered to the lysosome in metazoan, or to the vacuole in yeast and plants, for degradation and recycling of macromolecules (Liu and Bassham, 2012). During ER stress, autophagy is activated in yeast, mammals and plants, and it is involved in clearing unfolded protein from the ER by supplementing the ERAD pathway and, in turn, alleviating stress (Ding et al., 2007; Liu et al., 2012). In yeast, IRE1 regulates autophagy through the splicing of HAC1, which induces the production of Atg8p, an ubiquitin-like protein required for autophagosome formation (Yorimitsu et al., 2006). Unlike yeasts, in metazoans, autophagy is triggered by the kinase activity of IRE1 and PERK. IRE1, indeed, recruits the adaptor protein TNFR-associated factor2 (TRAF2) on the ER membranes, thus triggering the activation of the apoptosis signal regulating kinase1 (ASK1), which in turn regulates the activation of the c-Jun-N-terminal kinase (JNK), whose pathway induces the autophagosome formation (Pattingre et al., 2009). Moreover, PERK promotes autophagy through the phosphorylation of eIF2α, which induces the expression of ATF4, recently considered a key signal for autophagy activation (Matsumoto et al., 2013). ATF4 in turn activates the expression of autophagy-related (ATG) genes and of C/EBP-homologous protein (CHOP) transcription factor. CHOP and ATF4 together promote and modulate the induction of genes implicated in the formation, elongation and function of the autophagosome (B’chir et al., 2013). Similar to metazoans, in plants, it has been recently found that IRE1, specifically the IRE1b isoform, activates autophagy upon ER stress response independently from the IRE1-mediated bZIP60 mRNA splicing (Liu et al., 2012). However, the mechanistic features of plant autophagy under ER stress are mainly unknown, in terms of the upstream regulator/s of IRE1b as well as its downstream targets.
Upon excessive and prolonged ER stress, in metazoans, all the three UPR signaling pathways lead to cell death through apoptosis via the intrinsic mitochondrial pathway. Indeed, UPR regulates the activity of the pro-apoptotic members of the Bcl-2 family via transcriptional and post-transcriptional mechanisms, leading the BAX/BAK-mediated pore formation in the mitochondrial outer membrane, release of cytochrome c from the mitochondria, and subsequent activation of caspases, which are critical regulators of apoptosis via their role in propagating apoptotic signaling cascades (Rodriguez et al., 2011). However, it is not yet clear whether other types of cell death occur to eliminate terminally compromised cells under irreversible ER stress. Intriguingly in metazoans, under prolonged ER stress autophagy may switch from a pro-survival process to apoptosis. Several regulators of autophagy machinery are indeed involved in the apoptosis, such as JNK and CHOP (Rodriguez et al., 2011).
In mammals RIDD activity mediated by IRE1α enhances (1) ER stress intensity through the decay of mRNA encoding UPR target genes during the transition phase between the adaptive and apoptotic response (Han et al., 2009), and (2) expression of pro-apoptotic proteases, like the Caspase 2 (Upton et al., 2008), through the decay of selected antiapoptotic pre-miRNAs during the apoptotic response (Upton et al., 2012). In plants, the biological significance of RIDD activity in cell fate determination is still unknown.
In yeast, ER stress can induce PCD with apoptotic phenotypes (Hauptmann et al., 2006), as well as in a non-apoptotic process, where vacuole fragmentation and leaking of vacuolar materials are cell death features (Kim et al., 2012). Plant cell death executioners in the UPR are instead largely unknown. Unlike in metazoans, the plant IRE1 does not seem to have a pro-apoptotic role, given that the Arabidopsis ire1 double mutants display compromised ER stress tolerance, instead of a greater survival rate (Nagashima et al., 2011; Chen and Brandizzi, 2012). Although neither homologs of Bcl-2 family proteins nor components of the PERK-CHOP pathways have been identified in plants yet, some regulators of ER-PCD seem to be conserved across kingdoms. These include the Bax inhibitor1 (BI-1)-like protein, an ER transmembrane protein that protects cells against ER-stress induced-cell death (Chae et al., 2003; Watanabe and Lam, 2008), and the chaperone BiP, that has a protective function against ER stress induced-cell death in both mammalian and plant cells (Kishi et al., 2010; Reis et al., 2011).
Plants have developed UPR roles for evolutionarily conserved gene family, like for the Bcl-2-associated athanogene7 (BAG7) protein, and AGB1, the Gβ subunit of heterotrimeric GTP-binding protein family. In detail, BAG7 a plant ER-localized protein involved in the UPR aids chaperones like BiP in the protection of cells via a co-chaperone activity, while in yeast and mammals BAGs have a nuclear/cytoplasmic localization and are not involved in the maintenance of the UPR, but rather in other processes ranging from proliferation to growth arrest and cell death (Williams et al., 2010). Furthermore, in plants, AGB1 and IRE1 have antagonistic roles in the UPR gene induction and they regulate essential and independent UPR signaling arms (Wang et al., 2007; Chen and Brandizzi, 2012, 2013b), but the underlying mechanisms are unclear.
Environmental conditions, such as heat and salt stresses (Liu et al., 2007b; Deng et al., 2011), as well as physiological events, like growth and developmental processes (Chen and Brandizzi, 2012; Deng et al., 2013) evoked ER stress. Moreover, UPR has been recently linked to the phytohormone auxin, a master regulator of plant physiology, revealing a plant-specific strategy to maintain balance between stress adaptation and growth regulation (Chen et al., 2014). Additional studies are required to elucidate the plant UPR signaling and its molecular components, and how it is fine regulated during physiological events and environmental stresses. Also further work is needed to clarify the mechanisms leading the UPR to switch from cell survival to cell death and to identify the precise steps downstream of each UPR arm across different kingdoms.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We apologize to colleagues whose work was not discussed owing to space constraints. We thank Drs. Yani Chen and Francesco D’Onofrio for assistance in figure illustration. This work was primarily supported by National Institutes of Health (R01 GM101038) with contributing support from the Chemical Sciences, Geosciences and Biosciences Division, Office of Basic Energy Sciences, Office of Science, U.S. Department of Energy (award number DE-FG02-91ER20021).
Ali, M. M., Bagratuni, T., Davenport, E. L., Nowak, P. R., Silva-Santisteban, M. C., Hardcastle, A., et al. (2011). Structure of the Ire1 autophosphorylation complex and implications for the unfolded protein response. EMBO J. 30, 894–905. doi: 10.1038/emboj.2011.18
B’chir, W., Maurin, A. C., Carraro, V., Averous, J., Jousse, C., Muranishi, Y., et al. (2013). The eIF2α/ATF4 pathway is essential for stress-induced autophagy gene expression. Nucleic Acids Res. 41, 7683–7699. doi: 10.1093/nar/gkt563
Berlanga, J. J., Santoyo, J., and De Haro, C. (1999). Characterization of a mammalian homolog of the GCN2 eukaryotic initiation factor 2alpha kinase. Eur. J. Biochem. 265, 754–762. doi: 10.1046/j.1432-1327.1999.00780.x
Bommiasamy, H., Back, S. H., Fagone, P., Lee, K., Meshinchi, S., Vink, E., et al. (2009). ATF6alpha induces XBP1-independent expansion of the endoplasmic reticulum. J. Cell Sci. 122, 1626–1636. doi: 10.1242/jcs.045625
Chae, H. J., Ke, N., Kim, H. R., Chen, S., Godzik, A., Dickman, M., et al. (2003). Evolutionarily conserved cytoprotection provided by Bax Inhibitor-1 homologs from animals, plants, and yeast. Gene 323, 101–113. doi: 10.1016/j.gene.2003.09.011
Chapman, R. E., and Walter, P. (1997). Translational attenuation mediated by an mRNA intron. Curr. Biol. 7, 850–859. doi: 10.1016/S0960-9822(06)00373-3
Chen, Y., Aung, K., Rolčík, J., Walicki, K., Friml, J., and Brandizzi, F. (2014). Inter-regulation of the unfolded protein response and auxin signaling. Plant J. 77, 97–107. doi: 10.1111/tpj.12373
Chen, Y., and Brandizzi, F. (2013a). IRE1: ER stress sensor and cell fate executor. Trends Cell Biol. 23, 547–555. doi: 10.1016/j.tcb.2013.06.005
Chen, Y., and Brandizzi, F. (2013b). Analysis of unfolded protein response in Arabidopsis. Methods Mol. Biol. 1043,73–80. doi: 10.1007/978-1-62703-532-3_8
Chen, Y., and Brandizzi, F. (2012). AtIRE1A/AtIRE1B and AGB1 independently control two essential unfolded protein response pathways in Arabidopsis. Plant J. 69, 266–277. doi: 10.1111/j.1365-313X.2011.04788.x
Cox, J. S., Shamu, C. E., and Walter, P. (1993). Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell 73, 1197–1206. doi: 10.1016/0092-8674(93)90648-A
Cui, W., Li, J., Ron, D., and Sha, B. (2011). The structure of the PERK kinase domain suggests the mechanism for its activation. Acta Crystallogr. D Biol. Crystallogr. 67, 423–428. doi: 10.1107/S0907444911006445
Cullinan, S. B., Zhang, D., Hannink, M., Arvisais, E., Kaufman, R. J., and Diehl, J. A. (2003). Nrf2 is a direct PERK substrate and effector of PERK-dependent cell survival. Mol. Cell. Biol. 23, 7198–7209. doi: 10.1128/MCB.23.20.7198-7209.2003
Deng, Y., Humbert, S., Liu, J. X., Srivastava, R., Rothstein, S. J., and Howell, S. H. (2011). Heat induces the splicing by IRE1 of a mRNA encoding a transcription factor involved in the unfolded protein response in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108, 7247–7252. doi: 10.1073/pnas.1102117108
Deng, Y., Srivastava, R., and Howell, S. H. (2013). Protein kinase and ribonuclease domains of IRE1 confer stress tolerance, vegetative growth, and reproductive development in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 110, 19633–19638. doi: 10.1073/pnas.1314749110
Ding, W. X., Ni, H. M., Gao, W., Yoshimori, T., Stolz, D. B., Ron, D., et al. (2007). Linking of autophagy to ubiquitin-proteasome system is important for the regulation of endoplasmic reticulum stress and cell viability. Am. J. Pathol. 171, 513–524. doi: 10.2353/ajpath.2007.070188
Fordyce, P. M., Pincus, D., Kimmig, P., Nelson, C. S., El-Samad, H., Walter, P., et al. (2012). Basic leucine zipper transcription factor Hac1 binds DNA in two distinct modes as revealed by microfluidic analyses. Proc. Natl. Acad. Sci. U.S.A. 109, E3084–E3093. doi: 10.1073/pnas.1212457109
Gardner, B. M., and Walter, P. (2011). Unfolded proteins are Ire1-activating ligands that directly induce the unfolded protein response. Science 333, 1891–1894. doi: 10.1126/science.1209126
Han, D., Lerner, A. G., Vande Walle, L., Upton, J. P., Xu, W., Hagen, A., et al. (2009). IRE1alpha kinase activation modes control alternate endoribonuclease outputs to determine divergent cell fates. Cell 138, 562–575. doi: 10.1016/j.cell.2009.07.017
Harding, H. P., Novoa, I., Zhang, Y., Zeng, H., Wek, R., Schapira, M., et al. (2000). Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell 6, 1099–1108. doi: 10.1016/S1097-2765(00)00108-8
Harding, H. P., Zhang, Y., and Ron, D. (1999). Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature 397, 271–274. doi: 10.1038/16729
Harding, H. P., Zhang, Y., Zeng, H., Novoa, I., Lu, P. D., Calfon, M., et al. (2003). An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell 11, 619–633. doi: 10.1016/S1097-2765(03)00105-9
Hauptmann, P., Riel, C., Kunz-Schughart, L. A., Fröhlich, K. U., Madeo, F., and Lehle, L. (2006). Defects in N-glycosylation induce apoptosis in yeast. Mol. Microbiol. 59, 765–778. doi: 10.1111/j.1365-2958.2005.04981.x
Hayashi, S., Takahashi, H., Wakasa, Y., Kawakatsu, T., and Takaiwa, F. (2013). Identification of a cis-element that mediates multiple pathways of the endoplasmic reticulum stress response in rice. Plant J. 74, 248–257. doi: 10.1111/tpj.12117
Hetz, C. (2012). The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat. Rev. Mol. Cell Biol. 13, 89–102. doi: 10.1038/nrm3270
Hollien, J., and Weissman, J. S. (2006). Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science 313, 104–107. doi: 10.1126/science.1129631
Hong, M., Luo, S., Baumeister, P., Huang, J. M., Gogia, R. K., Li, M., et al. (2004). Underglycosylation of ATF6 as a novel sensing mechanism for activation of the unfolded protein response. J. Biol. Chem. 279, 11354–11363. doi: 10.1074/jbc.M309804200
Howell, S. H. (2013). Endoplasmic reticulum stress responses in plants. Annu. Rev. Plant Biol. 64, 477–499. doi: 10.1146/annurev-arplant-050312-120053
Iwata, Y., Fedoroff, N. V., and Koizumi, N. (2008). Arabidopsis bZIP60 is a proteolysis-activated transcription factor involved in the endoplasmic reticulum stress response. Plant Cell 20, 3107–3121. doi: 10.1105/tpc.108.061002
Iwata, Y., and Koizumi, N. (2005). An Arabidopsis transcription factor, AtbZIP60, regulates the endoplasmic reticulum stress response in a manner unique to plants. Proc. Natl. Acad. Sci. U.S.A. 102, 5280–5285. doi: 10.1073/pnas.0408941102
Kamauchi, S., Nakatani, H., Nakano, C., and Urade, R. (2005). Gene expression in response to endoplasmic reticulum stress in Arabidopsis thaliana. FEBS J. 272, 3461–3476. doi: 10.1111/j.1742-4658.2005.04770.x
Kim, H., Kim, A., and Cunningham, K. W. (2012). Vacuolar H+-ATPase (V-ATPase) promotes vacuolar membrane permeabilization and nonapoptotic death in stressed yeast. J. Biol. Chem. 287, 19029–12039. doi: 10.1074/jbc.M112.363390
Kimata, Y., and Kohno, K. (2011). Endoplasmic reticulum stress-sensing mechanisms in yeast and mammalian cells. Curr. Opin. Cell Biol. 23, 135–142. doi: 10.1016/j.ceb.2010.10.008
Kimmig, P., Diaz, M., Zheng, J., Williams, C. C., Lang, A., Aragón, T., et al. (2012). The unfolded protein response in fission yeast modulates stability of select mRNAs to maintain protein homeostasis. Elife 1, e00048. doi: 10.7554/eLife.00048
Kishi, S., Shimoke, K., Nakatani, Y., Shimada, T., Okumura, N., Nagai, K., et al. (2010). Nerve growth factor attenuates 2-deoxy-d-glucose-triggered endoplasmic reticulum stress-mediated apoptosis via enhanced expression of GRP78. Neurosci. Res. 66, 14–21. doi: 10.1016/j.neures.2009.09.003
Koizumi, N., Martinez, I. M., Kimata, Y., Kohno, K., Sano, H., and Chrispeels, M. J. (2001). Molecular characterization of two Arabidopsis Ire1 homologs, endoplasmic reticulum-located transmembrane protein kinases. Plant Physiol. 127, 949–962. doi: 10.1104/pp.010636
Kokame, K., Kato, H., and Miyata, T. (2001). Identification of ERSE-II, a new cis-acting element responsible for the ATF6-dependent mammalian unfolded protein response. J. Biol. Chem. 276, 9199–9205. doi: 10.1074/jbc.M010486200
Korennykh, A. V., Egea, P. F., Korostelev, A. A., Finer-Moore, J., Zhang, C., Shokat, K. M., et al. (2009). The unfolded protein response signals through high-order assembly of Ire1. Nature 457, 687–693. doi: 10.1038/nature07661
Lageix, S., Lanet, E., Pouch-Pélissier, M. N., Espagnol, M. C., Robaglia, C., Deragon, J. M., et al. (2008). Arabidopsis eIF2alpha kinase GCN2 is essential for growth in stress conditions and is activated by wounding. BMC Plant Biol. 8:134. doi: 10.1186/1471-2229-8-134
Liu, J. X., and Howell, S. H. (2010). bZIP28 and NF-Y transcription factors are activated by ER stress and assemble into a transcriptional complex to regulate stress response genes in Arabidopsis. Plant Cell 22, 782–796. doi: 10.1105/tpc.109.072173
Liu, J. X., Srivastava, R., Che, P., and Howell, S. H. (2007a). An endoplasmic reticulum stress response in Arabidopsis is mediated by proteolytic processing and nuclear relocation of a membrane-associated transcription factor, bZIP28. Plant Cell 19, 4111–4119. doi: 10.1105/tpc.106.050021
Liu, J. X., Srivastava, R., Che, P., and Howell, S. H. (2007b). Salt stress responses in Arabidopsis utilize a signal transduction pathway related to endoplasmic reticulum stress signaling. Plant J. 51, 897–909. doi: 10.1111/j.1365-313X.2007.03195.x
Liu, Y., and Bassham, D. C. (2012). Autophagy: pathways for self-eating in plant cells. Annu. Rev. Plant Biol. 63, 215–237. doi: 10.1146/annurev-arplant-042811-105441
Liu, Y., Burgos, J. S., Deng, Y., Srivastava, R., Howell, S. H., and Bassham, D. C. (2012). Degradation of the endoplasmic reticulum by autophagy during endoplasmic reticulum stress in Arabidopsis. Plant Cell 24, 4635–4651. doi: 10.1105/tpc.112.101535
Lu, S. J., Yang, Z. T., Sun, L., Sun, L., Song, Z. T., and Liu, J. X. (2012). Conservation of IRE1-regulated bZIP74 mRNA unconventional splicing in rice (Oryza sativa L.) involved in ER stress responses. Mol. Plant. 5, 504–514. doi: 10.1093/mp/ssr115
Marciniak, S. J., Garcia-Bonilla, L., Hu, J., Harding, H. P., and Ron, D. (2006). Activation-dependent substrate recruitment by the eukaryotic translation initiation factor 2 kinase PERK. J. Cell Biol. 172, 201–209. doi: 10.1083/jcb.200508099
Martínez, I. M., and Chrispeels, M. J. (2003). Genomic analysis of the unfolded protein response in Arabidopsis shows its connection to important cellular processes. Plant Cell 15, 561–576. doi: 10.1105/tpc.007609
Matsumoto, H., Miyazaki, S., Matsuyama, S., Takeda, M., Kawano, M., Nakagawa, H., et al. (2013). Selection of autophagy or apoptosis in cells exposed to ER-stress depends on ATF4 expression pattern with or without CHOP expression. Biol. Open 2, 1084–1090. doi: 10.1242/bio.20135033
Mishiba, K., Nagashima, Y., Suzuki, E., Hayashi, N., Ogata, Y., Shimada, Y., et al. (2013). Defects in IRE1 enhance cell death and fail to degrade mRNAs encoding secretory pathway proteins in the Arabidopsis unfolded protein response. Proc. Natl. Acad. Sci. U.S.A. 110, 5713–5718. doi: 10.1073/pnas.1219047110
Mori, K., Kawahara, T., Yoshida, H., Yanagi, H., and Yura, T. (1996). Signalling from endoplasmic reticulum to nucleus: transcription factor with a basic-leucine zipper motif is required for the unfolded protein-response pathway. Genes Cells 1, 803–817. doi: 10.1046/j.1365-2443.1996.d01-274.x
Mori, K., Ogawa, N., Kawahara, T., Yanagi, H., and Yura, T. (2000). mRNA splicing-mediated C-terminal replacement of transcription factor Hac1p is required for efficient activation of the unfolded protein response. Proc. Natl. Acad. Sci. U.S.A. 97, 4660–4665. doi: 10.1073/pnas.050010197
Mori, K., Ma, W., Gething, M. J., and Sambrook, J. (1993). A transmembrane protein with a cdc2+/CDC28-related kinase activity is required for signaling from the ER to the nucleus. Cell 74, 743–756. doi: 10.1016/0092-8674(93)90521-Q
Nagashima, Y., Mishiba, K., Suzuki, E., Shimada, Y., Iwata, Y., and Koizumi, N. (2011). Arabidopsis IRE1 catalyses unconventional splicing of bZIP60 mRNA to produce the active transcription factor. Sci. Rep. 1, 29. doi: 10.1038/srep00029
Oh, D. H., Kwon, C. S., Sano, H., Chung, W. I., and Koizumi, N. (2003). Conservation between animals and plants of the cis-acting element involved in the unfolded protein response. Biochem. Biophys. Res. Commun. 301, 225–230. doi: 10.1016/S0006-291X(02)03019-X
Oikawa, D., Tokuda, M., Hosoda, A., and Iwawaki, T. (2010). Identification of a consensus element recognized and cleaved by IRE1 alpha. Nucleic Acids Res. 38, 6265–6673. doi: 10.1093/nar/gkq452
Pattingre, S., Bauvy, C., Carpentier, S., Levade, T., Levine, B., and Codogno, P. (2009). Role of JNK1-dependent Bcl-2 phosphorylation in ceramide-induced macroautophagy. J. Biol. Chem. 284, 2719–2728. doi: 10.1074/jbc.M805920200
Reis, P. A., Rosado, G. L., Silva, L. A., Oliveira, L. C., Oliveira, L. B., Costa, M. D., et al. (2011). The binding protein BiP attenuates stress-induced cell death in soybean via modulation of the N-rich protein-mediated signaling pathway. Plant Physiol. 157, 1853–1865. doi: 10.1104/pp.111.179697
Rodriguez, D., Rojas-Rivera, D., and Hetz, C. (2011). Integrating stress signals at the endoplasmic reticulum: the BCL-2 protein family rheostat. Biochim. Biophys. Acta 1813, 564–574. doi: 10.1016/j.bbamcr.2010.11.012
Roy, B., and Lee, A. S. (1999). The mammalian endoplasmic reticulum stress response element consists of an evolutionarily conserved tripartite structure and interacts with a novel stress-inducible complex. Nucleic Acids Res. 27, 1437–1443. doi: 10.1093/nar/27.6.1437
Schindler, A. J., and Schekman, R. (2009). In vitro reconstitution of ER-stress induced ATF6 transport in COPII vesicles. Proc. Natl. Acad. Sci. U.S.A. 106, 17775–17780. doi: 10.1073/pnas.0910342106
Shen, J., Chen, X., Hendershot, L., and Prywes, R. (2002). ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev. Cell 3, 99–111. doi: 10.1016/S1534-5807(02)00203-4
Spear, E. D., and Ng, D. T. W. (2003). Stress tolerance of misfolded carboxypeptidase Y requires maintenance of protein trafficking and degradative pathways. Mol. Biol. Cell 14, 2756–2767. doi: 10.1091/mbc.E02-11-0717
Srivastava, R., Chen, Y., Deng, Y., Brandizzi, F., and Howell, S. H. (2012). Elements proximal to and within the transmembrane domain mediate the organelle-to-organelle movement of bZIP28 under ER stress conditions. Plant J. 70, 1033–1042. doi: 10.1111/j.1365-313X.2012.04943.x
Sun, L., Yang, Z. T., Song, Z. T., Wang, M. J., Sun, L., Lu, S. J., et al. (2013). The plant-specific transcription factor gene NAC103 is induced by bZIP60 through a new cis-regulatory element to modulate the unfolded protein response in Arabidopsis. Plant J. 76, 274–286. doi: 10.1111/tpj.12287
Travers, K. J., Patil, C. K., Wodicka, L., Lockhart, D. J., Weissman, J. S., and Walter, P. (2000). Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 101, 249–258. doi: 10.1016/S0092-8674(00)80835-1
Upton, J. P., Austgen, K., Nishino, M., Coakley, K. M., Hagen, A., Han, D., et al. (2008). Caspase-2 cleavage of BID is a critical apoptotic signal downstream of endoplasmic reticulum stress. Mol. Cell. Biol. 28, 3943–3951. doi: 10.1128/MCB.00013-08
Upton, J. P., Wang, L., Han, D., Wang, E. S., Huskey, N. E., Lim, L., et al. (2012). IRE1α cleaves select microRNAs during ER stress to derepress translation of proapoptotic Caspase-2. Science 338, 818–822. doi: 10.1126/science.1226191
Walter, P., and Ron, D. (2011). The unfolded protein response: from stress pathway to homeostatic regulation. Science 334, 1081–1086. doi: 10.1126/science.1209038
Wang, S., Narendra, S., and Fedoroff, N. (2007). Heterotrimeric G protein signaling in the Arabidopsis unfolded protein response. Proc. Natl. Acad. Sci. U.S.A. 104, 3817–3822. doi: 10.1073/pnas.0611735104
Wang, Y., Shen, J., Arenzana, N., Tirasophon, W., Kaufman, R. J., and Prywes, R. (2000). Activation of ATF6 and an ATF6 DNA binding site by the endoplasmic reticulum stress response. J. Biol. Chem. 275, 27013–27020.
Watanabe, N., and Lam, E. (2008). Arabidopsis Bax inhibitor-1 functions as an attenuator of biotic and abiotic types of cell death. Plant J. 45, 884–894. doi: 10.1111/j.1365-313X.2006.02654.x
Williams, B., Kabbage, M., Britt, R., and Dickman, M. B. (2010). AtBAG7, an Arabidopsis Bcl-2-associated athanogene, resides in the endoplasmic reticulum and is involved in the unfolded protein response. Proc. Natl. Acad. Sci. U.S.A. 107, 6088–6093. doi: 10.1073/pnas.0912670107
Yamamoto, K., Yoshida, H., Kokame, K., Kaufman, R. J., and Mori, K. (2004). Differential contributions of ATF6 and XBP1 to the activation of endoplasmic reticulum stress-responsive cis-acting elements ERSE, UPRE and ERSE-II. J. Biochem. 136, 343–350. doi: 10.1093/jb/mvh122
Yanagitani, K., Imagawa, Y., Iwawaki, T., Hosoda, A., Saito, M., Kimata, Y., et al. (2009). Cotranslational targeting of XBP1 protein to the membrane promotes cytoplasmic splicing of its own mRNA. Mol. Cell 34, 191–200. doi: 10.1016/j.molcel.2009.02.033
Ye, J., Rawson, R. B., Komuro, R., Chen, X., Davé, U. P., Prywes, R., et al. (2000). ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol. Cell 6, 1355–1364. doi: 10.1016/S1097-2765(00)00133-7
Yorimitsu, T., Nair, U., Yang, Z., and Klionsky, D. J. (2006). Endoplasmic reticulum stress triggers autophagy. J. Biol. Chem. 281, 30299–30304. doi: 10.1074/jbc.M607007200
Yoshida, H., Matsui, T., Yamamoto, A., Okada, T., and Mori, K. (2001). XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107, 881–891. doi: 10.1016/S0092-8674(01)00611-0
Yoshida, H., Oku, M., Suzuki, M., and Mori, K. (2006). pXBP1(U) encoded in XBP1 pre-mRNA negatively regulates unfolded protein response activator pXBP1(S) in mammalian ER stress response. J. Cell Biol. 172, 565–575. doi: 10.1083/jcb.200508145
Keywords: unfolded protein response, ER stress, UPR temporal activation, adaptation, autophagy, programed cell death, apoptosis, eukaryotes
Citation: Ruberti C and Brandizzi F (2014) Conserved and plant-unique strategies for overcoming endoplasmic reticulum stress. Front. Plant Sci. 5:69. doi: 10.3389/fpls.2014.00069
Received: 06 January 2014; Accepted: 10 February 2014;
Published online: 26 February 2014.
Edited by:
Lorenzo Frigerio, University of Warwick, UKReviewed by:
Aldo Ceriotti, Consiglio Nazionale delle Ricerche, ItalyCopyright © 2014 Ruberti and Brandizzi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Federica Brandizzi, Plant Research Laboratory, Department of Energy, Michigan State University, 612 Wilson Road, East Lansing, MI 48824, USA e-mail:ZmJAbXN1LmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.