
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Plant Sci. , 16 August 2013
Sec. Plant Physiology
Volume 4 - 2013 | https://doi.org/10.3389/fpls.2013.00314
This article is part of the Research Topic Nitric Oxide Signalling in Plants View all 22 articles
In plants, programed cell death (PCD) is an important mechanism to regulate multiple aspects of growth and development, as well as to remove damaged or infected cells during responses to environmental stresses and pathogen attacks. Under biotic and abiotic stresses, plant cells exhibit a rapid synthesis of nitric oxide (NO) and a parallel accumulation of reactive oxygen species (ROS). Frequently, these responses trigger a PCD process leading to an intrinsic execution of plant cells. The accumulating evidence suggests that both NO and ROS play key roles in PCD. These redox active small molecules can trigger cell death either independently or synergistically. Here we summarize the recent progress on the cross-talk of NO and ROS signals in the hypersensitive response, leaf senescence, and other kinds of plant PCD caused by diverse cues.
Nitric oxide (NO) is a gaseous free radical which was first found to play a crucial role in plant and mediating defense reactions against bacterial pathogens (Noritake et al., 1996; Delledonne et al., 1998). Increasing evidence suggests that NO, as a signal mediator, plays a key role in many physiological and developmental processes, such as germination, leaf expansion, lateral root development, flowering, stomatal closure, crosstalk with plant hormones, defenses against biotic and abiotic stresses (He et al., 2004; Hong et al., 2008; Leitner et al., 2009; Wilkins et al., 2011; Liu et al., 2013; Yadav et al., 2013). In plants, mitochondria and chloroplasts are organelles that are thought to contribute to NO generation in vivo (Galatro et al., 2013; Vanlerberghe, 2013). Although a long standing search for an NO synthase (NOS) in plants similar to NOS enzymes found in mammals has thus far been unsuccessful, suppression of NO signaling in the presence of NOS inhibitors has been reported by several groups, indicating the potential existence of a NOS-like enzyme in plants (Tewari et al., 2013; Figure 1).
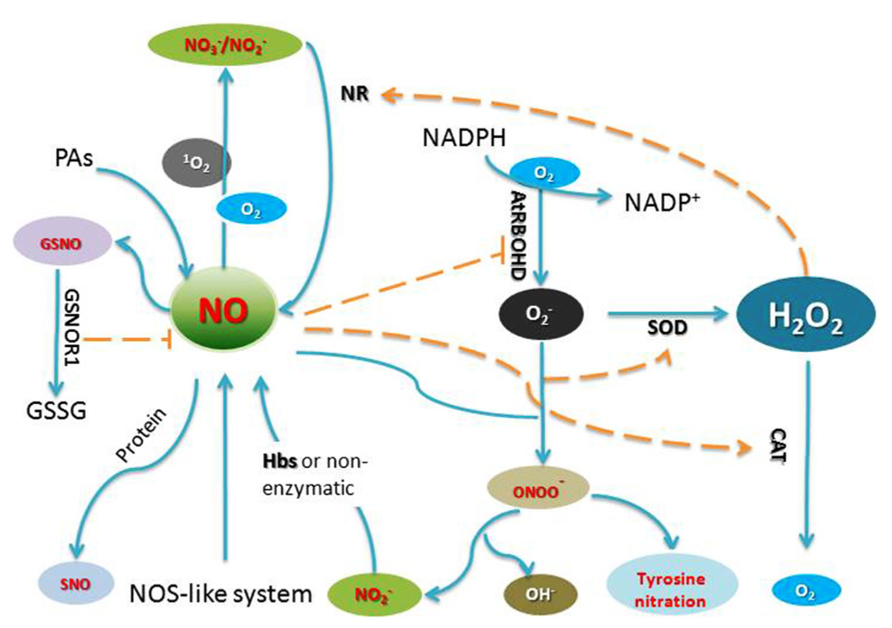
FIGURE 1. Generation of and crosstalk by RNS and ROS in plant cells. AtRBOHD, an NADPH oxidase; GSNO, S-nitrosoglutathione; GSNOR1, S-nitrosoglutathione reductase 1; GSSG, glutathione disulfide; NR, nitrate reductase; SOD, superoxide dismutase; Hbs, Hemoglobin; PAs, polyamines; CAT, catalase.
As a free radical, NO could also react with various intracellular/extracellular targets and form a series of molecules, such as NO radicals (NO-), nitrosonium ions (NO+), peroxynitrite (ONOO-), S-nitrosothiols (SNOs), higher oxides of nitrogen (NOx) and dinitrosyl-iron complexes among others, collectively these NO derivatives are termed reactive nitrogen species (RNS; Di Stasi et al., 2002). The functions of RNS, in plant cells are complex because they are implicated in many different physiological processes. S-nitrosylation, the covalent attachment of an NO moiety to a reactive cysteine thiol to form an SNO, has emerged as a prototypic redox-mediated modification in plants. For example, S-nitrosylation of methionine adenosyltransferase 1 (MAT1; Lindermayr et al., 2006), the Arabidopsis type-II metacaspase AtMC9 (Belenghi et al., 2007), PrxII E, a member of the peroxiredoxin family (Romero-Puertas et al., 2007b), non-expression of pathogenesis-related protein 1 (NPR1; Tada et al., 2008), Arabidopsis thaliana salicylic acid (SA) binding protein 3 (AtSABP3; Wang et al., 2009), TGACG motif binding factor 1 (TGA1) family (Lindermayr et al., 2010), nicotinamide adenine dinucleotide phosphate (NADPH) oxidase AtRBOHD (Yun et al., 2011), cytoskeletal proteins (Yemets et al., 2011), auxin receptor-transport inhibitor response 1/auxin signaling F-box (TIR1/AFB; Terrile et al., 2011), glyceraldehyde-3-phosphate dehydrogenase (GAPDH; Lin et al., 2012) and also Arabidopsis histidine phosphotransfer protein (AHP1; Feng et al., 2013) have been reported. These data implies that protein S-nitrosylation is a key redox-based modification in plants and a pivotal mechanism to convey NO bioactivity. Peroxynitrite (ONOO-), formed from and NO, is also capable of reacting with many classes of biomolecules such as antioxidants and proteins, triggers defense responses in animals and plants (Rubbo et al., 1994a,b). In Arabidopsis, ONOO- could induce hypersensitive response (HR) and defense-related gene expression (Alamillo and Garcia-Olmedo, 2001). Very recently, protein tyrosine nitration, addition of an nitro group (NO2) to one of the two equivalent ortho carbons of the aromatic ring of Tyr residues and metal nitrosylation, was reported as a new important RNS-mediated post-translational modification (Saito et al., 2006; Astier and Lindermayr, 2012; Tanou et al., 2012; Begara-Morales et al., 2013; Chaki et al., 2013). These findings not only deepen our understanding of NO signaling and function in plants, but also indicate the existence of RNS cross-talk with other signaling pathways, such as those orchestrated by auxin, cytokinin, SA, jasmonic acid (JA), ethylene (ET), and reactive oxygen species (ROS).
Reactive oxygen species including hydrogen peroxide (H2O2), superoxide anion , hydroxyl radicals (.OH) and singlet oxygen (1O2) have all been implicated in the control of biological processes in plants. Mitochondria as an “energy factory” are believed to be a major site of ROS production. Alternative oxidase (AOX) has an important influence on both ROS and RNS generation by the respiratory chain in mitochondria (Vanlerberghe, 2013). Peroxisomes are subcellular organelles with an essentially oxidative type of metabolism and produce superoxide radicals as a consequence of their normal metabolism. Chloroplasts are also a major site of ROS generation in plants (Hideg et al., 2006). The superoxide radicals and singlet oxygen (1O2) are produced in chloroplasts by photo-reduction of oxygen and energy transfer from triplet excited chlorophyll to oxygen, respectively (Figure 1).
Hydrogen peroxide, a ROS of major biological significance, can form as a result of the reaction of superoxide and also can be generated by specific enzymes (Noctor et al., 2000; Gechev et al., 2006). An oxidative burst, with rapid synthesis and its subsequent dismutation to H2O2 in the apoplast, is a common response to pathogens, elicitors, wounding, heat, ultra-violet light, and ozone (Orozco-Cardenas et al., 2001; Rao and Davis, 2001). Besides its directly oxidative activity, it is now clear that H2O2 has a key signaling role in plants (Gechev et al., 2006; Jiang et al., 2011). H2O2 can induce gene expression and modulates signaling proteins, such as protein phosphatases (PP), protein kinases (PK), transcription factors and calcium channels that are located in the plasma membrane or elsewhere (Neill et al., 2002; Lin et al., 2012).
A well-documented form of plant programed cell death (PCD) is the HR, characterized by the rapid cell death surrounding infection sites. The HR shows some similarity to the characteristics of animal apoptosis, such as membrane dysfunction, vacuolization of the cytoplasm, chromatin condensation, and endonucleolytic cleavage of DNA (Greenberg and Yao, 2004; Choi et al., 2013; Iakimova et al., 2013). Both NO and ROS have been implicated in controlling the HR process. One of the key determinants for the HR is the balance between intracellular NO and ROS levels (Delledonne et al., 2001; Zaninotto et al., 2006). Following pathogen recognition, NO accumulation occurs concomitant with an oxidative burst, which consists of a biphasic production of apoplastic ROS at the site of attempted invasion (Romero-Puertas et al., 2004). In this context, NO and H2O2 are thought to function in combination to promote HR cell death. For example, either of them could cause the release of cytochrome c from mitochondria, and affect the caspase-like signaling cascade, leading to the HR (Mur et al., 2006; Tan et al., 2013). Some key components of the defense signaling cascade that are known to be affected by ROS and NO activity include mitogen-activated protein kinases (MAPKs) and phosphatases (Figure 2). Thus, modulation of a central MAPK cascade may converge both H2O2 and NO signaling pathways activated in response to pathogen infection. In tomato cell suspensions, upon xylanase perception, cells activate a protein kinase pathway required for NO formation and S-nitrosylation-dependent mechanisms which are involved in downstream signaling, leading to production of polyamine and ROS production (Lanteri et al., 2011).
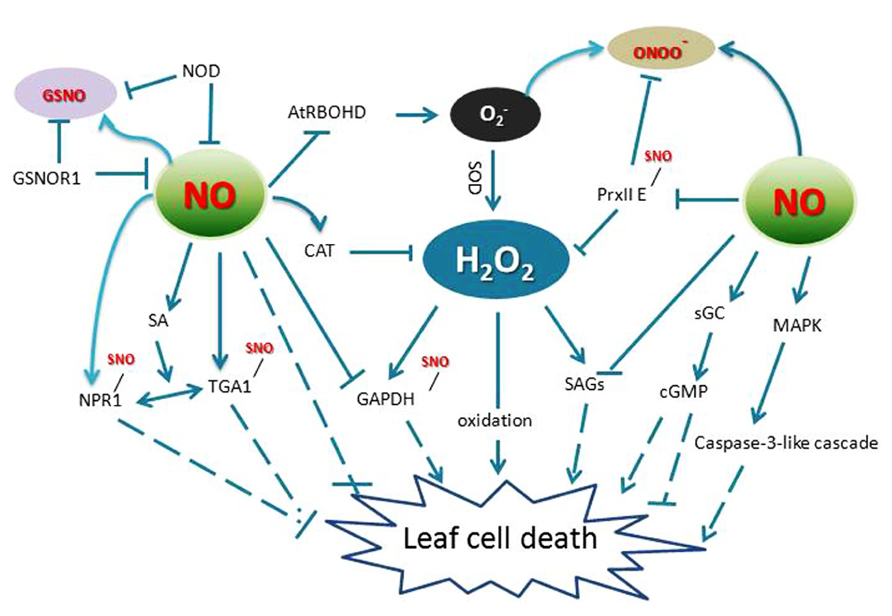
FIGURE 2. Crosstalk of RNS and ROS in leaf cell death. AtRBOHD, an NADPH oxidase; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; GSNO, S-nitrosoglutathione; GSNOR1, S-nitrosoglutathione reductase 1; NPR1, non-expression of pathogenesis related protein 1; TGA1, TGACG motif binding factor 1; NR, nitrate reductase; SAG, senescence-associated genes; PrxII E, peroxiredoxin II E; NOD, NO degrading dioxygenase; sGC, soluble guanylate cyclase. MAPK, mitogen-activated protein kinase; SOD, superoxide dismutase; CAT, catalase; cGMP, cyclic guanosine monophosphate; sGC, soluble guanylate cyclase.
Interestingly, many proteins are targets of both NO and H2O2 (Figure 2). For example, GAPDH that plays a role in mediating ROS signaling in plants is a direct target of H2O2 and it is also a target of NO-mediated S-nitrosylation, which blunts its activity (Lindermayr et al., 2005). Also, MAT in mammals is inactivated by H2O2 through a reversible and covalent oxidation of a Cys residue. The same Cys residue is also a target for NO, which similarly causes enzyme inactivation (Hancock et al., 2005). Further, PrxII E not only reduces H2O2 and alkyl hydroperoxides (Dietz, 2003a,b; Horling et al., 2003), but also functions in detoxifying peroxynitrite. S-nitrosylation of PrxII E during the defense response regulates the antioxidant function of this key enzyme and might contribute to the HR (Romero-Puertas et al., 2007a,b; Figure 2). As a useful tool to elicit ROS-activated responses, ozone (O3) has been shown to induce HR-like cell death. During this process, NO accumulation preceded accumulation of ET, JA, SA, and leaf injury, implies that NO is an important signaling molecule in response to O3 exposure (Rao and Davis, 2001; Ahlfors et al., 2009).
Contrary to its program cell death functions in the HR, NO can also scavenge H2O2 and protects plant cells from damage under certain circumstances (Beligni et al., 2002; Crawford and Guo, 2005). NO donors affect both wounding-induced H2O2 synthesis and wounding- or JA-induced expression of defense genes (Grun et al., 2006). In Arabidopsis, S-nitrosoglutathione reductase 1 (GSNOR1) is a key regulator that indirectly controls the global levels of protein S-nitrosylation (SNO). Loss-of-function mutations in GSNOR1 increased total cellular NO and SNO content and compromised both non-host and resistance (R) gene-mediated protection and also disabled basal defense responses (Feechan et al., 2005; Wang et al., 2010; Figure 2). Further, the mutant atgsnor1–3 was also perturbed in thermotolerance and resistance to paraquat (1,1′-dimethyl-4,4′-bipyridinium dichloride), which induces the production of superoxide and H2O2 in wild type leaves (Lee et al., 2008; Chen et al., 2009). Consistent with these results, wild-type plants treated with an NO donor displayed resistance to paraquat (Chen et al., 2009). These studies showed that the Arabidopsis GSNOR1/HOT5/PAR-2 gene not only regulates SA signaling and thermotolerance by modulating the intracellular SNO level, but also acts downstream of superoxide to regulate cell death.
Interestingly, the increased levels of SNOs in atgsnor1–3 plants potentiated the HR even in the absence of the cell death agonist SA and apoplastic ROS synthesis. Surprisingly, NO S-nitrosylates the NADPH oxidase, AtRBOHD, at Cys890, diminishes its ability to synthesize ROS. This cysteine is also evolutionarily conserved and specifically S-nitrosylated in both human and fly NADPH oxidases, suggesting that this mechanism may govern immune responses in both plants and animals (Yun et al., 2011). Thus, NO may control ROS production through protein S-nitrosylation to further control the development of cell death processes. Collectively, these findings have provided significant insights into the understanding of the mechanisms underpinning ROS and RNS function in plants, revealing that the ROS/RNS pathway in plant PCD is highly complex and is mediated at least in part by crosstalk with several phytohormone signaling networks.
Leaf senescence, thought to be another form of plant PCD, is the final stage of leaf development, which is not only controlled by organ age but also triggered by adverse environmental factors (Pourtau et al., 2004; Munns, 2005; Masclaux-Daubresse et al., 2007; Jing et al., 2008; Wu et al., 2012). Additionally, phytohormones such as ET, SA, JA, auxin, ABA, and cytokinins all affect leaf senescence (Lim et al., 2007). In Arabidopsis, the level of H2O2 increases dramatically in leaf tissue during senescence. In addition to its role in oxidizing macromolecules such as proteins and lipids, H2O2 has also been proposed to function as a signal to induce the expression of genes involved in the senescence process (Cui et al., 2013). In agreement with its lower antioxidant capacity, senescent leaf tissue was found to contain elevated levels of ROS. In this context, a number of senescence-associated genes (SAGs) characterized from Arabidopsis could be induced by ozone (Miller et al., 1999) and the expression of many other SAGs were also induced by ROS (Navabpour et al., 2003), indicating that ROS might function as a signal to promote senescence. Interestingly, senescence-associated NAC genes (senNACs), key regulators of leaf senescence, were also found to be rapidly and strongly induced by H2O2 treatment in both leaves and roots (Balazadeh et al., 2010, 2011). Thus, ROS has a dual role in leaf senescence: to promote the cell death process by directly oxidizing target macromolecules and to drive the expression of senescence-related genes.
Distinct from the positive role of ROS in senescence, NO can both provoke and impede this process, dependent upon its concentration and subcellular location. NO may alleviate the toxicity of ROS and has thus acted as a leaf senescence delaying factor in plants. The NO-deficient mutant nos1/noa1 showed early leaf senescence (Niu and Guo, 2012) and similarly Arabidopsis expressing an NO degrading dioxygenase (NOD) displayed a senescence-like phenotype (Mishina et al., 2007; Figure 2). Furthermore, the level of NO is related with the senescence process and is thought to be an essential component involved in plant senescence signaling cascades. In Arabidopsis mutant dnd1, which lacks a plasma membrane-localized cation channel (CNGC2), early senescence-associated phenotypes (such as loss of chlorophyll, expression level of senescence associated genes, H2O2 generation, lipid peroxidation, tissue necrosis, and SA levels) were all elevated relative to wild type. Basal levels of NO in dnd1 leaves were lower than wild type, suggesting that the function of CNGC2 may impact downstream “basal” NO production in addition to its role linked to NO signaling (Ma et al., 2010). NO generation is therefore thought to act as a negative regulator during plant leaf senescence signaling. The protective effect of NO against ROS induced cell death can also be linked to the enhanced activity of antioxidant enzymes, as negative regulator of the chlorophyll catabolic pathway and as drivers for positively maintaining the stability of thylakoid membranes during leaf senescence (Liu and Guo, 2013).
On the other hand, NO can also promote the leaf senescence. Arabidopsis AtFer1, one of the best characterized plant ferritin isoforms to date, strongly accumulates upon treatment with excess iron, via an NO-mediated pathway. The AtFer1 isoform is functionally involved in events leading to the onset of age-dependent senescence in Arabidopsis and its iron-detoxification function during senescence is required when ROS accumulates (Murgia et al., 2007). Recently identification of an NO accrual mutant noe1 (nitric oxide excess 1) in rice revealed that NOE1 encoded a rice catalase (CAT) OsCATC. Interestingly, noe1 plants exhibited an increase of H2O2 in their leaves, which consequently promoted NO production via activation of nitrate reductase. Removal of excess NO reduced cell death in both leaves and suspension cultures derived from noe1 plants, implicating that NO acts as an important endogenous mediator of H2O2-induced leaf cell death. Reduction of intracellular SNO levels, generated by over-expression of OsGSNOR alleviated leaf cell death in noe1 plants. Thus, S-nitrosylation was also involved in light-dependent leaf cell death in noe1. Collectively, these data suggest that both NO and SNOs are important mediators in the process of H2O2-induced leaf cell death in rice (Lin et al., 2012; Wang et al., 2013). OsGSNOR in noe1 plants reduced SNO levels, consistent with a key role for this enzyme in SNO homeostasis. Moreover, the results show that no change in H2O2 content occurred in either GSNOR-overexpressing or GSNOR-RNAi transgenic lines in the context of noe1 background, suggesting that NO might function downstream of H2O2 in a light-driven leaf cell death in rice. It was found that NO treatment led to rapid cell death and induced H2O2 accumulation in maize leaves, and pharmacological studies also suggested that NO-induced cell death is in part mediated via H2O2, therefore H2O2 may be involved in NO-induced cell death in maize leaves (Kong et al., 2013). These discrepancies for the role of NO in cell death might be due to the differences in plant species, redox state, and growth conditions. Both NO and H2O2 could induce leaf cell death during which they could crosstalk with each other through different pathways.
Some reports also describe the cross-talk of NO and ROS in other kinds of cell death in plants. Gibberellin (GA)-induced PCD in barley (Hordeum vulgare cv. Himalaya) aleurone layers is mediated by ROS and NO is a protective antioxidant. NO donors delay this PCD process, but do not inhibit metabolism in general, or the GA-induced synthesis and secretion of alpha-amylase. The amounts of CAT and superoxide dismutase (SOD) are greatly reduced in aleurone layers treated with GA. Treatment with GA in the presence of NO donors delays the loss of CAT and SOD. Thus, NO may be an endogenous modulator of PCD in barley aleurone cells (Beligni et al., 2002). Furthermore, the exogenous application of NO rendered the plants more tolerant to arsenic (As)-induced oxidative damage by enhancing their antioxidant defense and glyoxalase system (Hasanuzzaman and Fujita, 2013). Previous work has also shown that NO acts as a pivotal positive mediator in cadmium (Cd)-induced PCD in suspension cell cultures. NO strongly counteracts Cd-induced ROS mediated cytotoxicity in Brassica juncea by controlling antioxidant metabolism (De Michele et al., 2009; Verma et al., 2013). Similarly, a role for NO as an antioxidant during heavy metal mediated toxicity has been highlighted recently by Saxena and Shekhawat (2013).
On the other hand, NO could also aid ROS-induced PCD. In pollen-pistil interactions, self-incompatibility (SI) induces relatively rapid and transient increases in ROS and NO. As ROS/NO scavengers alleviated both the formation of SI-induced actin punctate foci and also activation of a DEVDase/caspase-3-like activity (Wilkins et al., 2011). In tobacco BY-2 cells, sphinganine or dihydrosphingosine (d18:0, DHS) induce a calcium dependent PCD and trigger H2O2 production via the activation of NADPH oxidase(s). They also promote NO production, which is required for cell death induction (Da Silva et al., 2011). NO accumulated in Cd-induced PCD and promoted Cd-induced Arabidopsis PCD by promoting MPK6-mediated caspase-3-like activation (Ye et al., 2013). So the different roles of RNS in PCD and their crosstalk with ROS depend on the plant species, growth conditions and redox status.
In plants, RNS and ROS synthesis is a routine requirement for cells to undergo PCD, these small molecules can act either synergistically or independently (Clarke et al., 2000; Orozco-Cardenas and Ryan, 2002; Bright et al., 2006). The accumulating data suggests significant cross-talk occurs between RNS and ROS (Figure 1), although the clear relationship of RNS and ROS in the process of cell death remains elusive. NO and ROS could regulate the synthesis each other. During HR, NO can affect ROS synthesis through S-nitrosylating NADPH oxidase AtRBOHD (Yun et al., 2011). On the other hand, in rice noe1 mutant, in the absence of OsNOE1/OsCATC function, the accumulation of H2O2 induces NO production through elevating nitrate reductase expression, which is further integral to H2O2 induced leaf cell death through S-nitrosylation of GAPDH and thioredoxin (Lin et al., 2012; Wang et al., 2013). Cross-talk of NO and H2O2 is a prominent feature in the activities of these small molecules. RNS and ROS also play important roles in modulating the activity of target proteins. A complete list of signaling pathways regulated by ROS or RNS still awaits identification, the data presented in this review are therefore far from offering a comprehensive picture of the function of NO and ROS during plant PCD. Thus, further work is needed to understand how these key molecules trigger the onset and development of plant cell death.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We apologize for not being able to cite many relevant original papers, replaced by reviews, due to space limitation. This work was supported by grants from National Natural Science Foundation of China (Grant No. 31171514) and an international exchange grant provided by the National Natural Science Foundation of China and the Royal Society of Edinburgh (Grant No. 30811130222).
Ahlfors, R., Brosche, M., Kollist, H., and Kangasjarvi, J. (2009). Nitric oxide modulates ozone-induced cell death, hormone biosynthesis and gene expression in Arabidopsis thaliana. Plant J. 58, 1–12. doi: 10.1111/j.1365-313X.2008.03756.x
Alamillo, J. M., and Garcia-Olmedo, F. (2001). Effects of urate, a natural inhibitor of peroxynitrite-mediated toxicity, in the response of Arabidopsis thaliana to the bacterial pathogen Pseudomonas syringae. Plant J. 25, 529–540. doi: 10.1046/j.1365-313x.2001.00984.x
Astier, J., and Lindermayr, C. (2012). Nitric oxide-dependent posttranslational modification in plants: an update. Int. J. Mol. Sci. 13, 15193–15208. doi: 10.3390/ijms131115193
Balazadeh, S., Kwasniewski, M., Caldana, C., Mehrnia, M., Zanor, M. I., Xue, G. P., et al. (2011). ORS1, an H2O2-responsive NAC transcription factor, controls senescence in Arabidopsis thaliana. Mol. Plant 4, 346–360. doi: 10.1093/mp/ssq080
Balazadeh, S., Siddiqui, H., Allu, A. D., Matallana-Ramirez, L. P., Caldana, C., Mehrnia, M., et al. (2010). A gene regulatory network controlled by the NAC transcription factor ANAC092/AtNAC2/ORE1 during salt-promoted senescence. Plant J. 62, 250–264. doi: 10.1111/j.1365-313X.2010.04151.x
Begara-Morales, J. C., Chaki, M., Sanchez-Calvo, B., Mata-Perez, C., Leterrier, M., Palma, J. M., et al. (2013). Protein tyrosine nitration in pea roots during development and senescence. J. Exp. Bot. 64, 1121–1134. doi: 10.1093/jxb/ert006
Belenghi, B., Romero-Puertas, M. C., Vercammen, D., Brackenier, A., Inze, D., Delledonne, M., et al. (2007). Metacaspase activity of Arabidopsis thaliana is regulated by S-nitrosylation of a critical cysteine residue. J. Biol. Chem. 282, 1352–1358. doi: 10.1074/jbc.M608931200
Beligni, M. V., Fath, A., Bethke, P. C., Lamattina, L., and Jones, R. L. (2002). Nitric oxide acts as an antioxidant and delays programmed cell death in barley aleurone layers. Plant Physiol. 129, 1642–1650. doi: 10.1104/pp.002337
Bright, J., Desikan, R., Hancock, J. T., Weir, I. S., and Neill, S. J. (2006). ABA-induced NO generation and stomatal closure in Arabidopsis are dependent on H2O2 synthesis. Plant J. 45, 113–122. doi: 10.1111/j.1365-313X.2005.02615.x
Chaki, M., Carreras, A., Lopez-Jaramillo, J., Begara-Morales, J. C., Sanchez-Calvo, B., Valderrama, R., et al. (2013). Tyrosine nitration provokes inhibition of sunflower carbonic anhydrase (beta-CA) activity under high temperature stress. Nitric Oxide 29, 30–33. doi: 10.1016/j.niox.2012.12.003
Chen, R., Sun, S., Wang, C., Li, Y., Liang, Y., An, F., et al. (2009). The Arabidopsis PARAQUAT RESISTANT2 gene encodes an S-nitrosoglutathione reductase that is a key regulator of cell death. Cell Res. 19, 1377–1387. doi: 10.1038/cr.2009.117
Choi, H. W., Kim, N. H., Lee, Y. K., and Hwang, B. K. (2013). The pepper extracellular xyloglucan-specific endo-beta-1,4-glucanase inhibitor protein gene, CaXEGIP1, is required for plant cell death and defense responses. Plant Physiol. 161, 384–396. doi: 10.1104/pp.112.203828
Clarke, A., Desikan, R., Hurst, R. D., Hancock, J. T., and Neill, S. J. (2000). NO way back: nitric oxide and programmed cell death in Arabidopsis thaliana suspension cultures. Plant J. 24, 667–677. doi: 10.1046/j.1365-313x.2000.00911.x
Crawford, N. M., and Guo, F. Q. (2005). New insights into nitric oxide metabolism and regulatory functions. Trends Plant Sci. 10, 195–200. doi: 10.1016/j.tplants.2005.02.008
Cui, M. H., Ok, S. H., Yoo, K. S., Jung, K. W., Yoo, S. D., and Shin, J. S. (2013). An Arabidopsis cell growth defect factor-related protein, CRS, promotes plant senescence by increasing the production of hydrogen peroxide. Plant Cell Physiol. 54, 155–167. doi: 10.1093/pcp/pcs161
Da Silva, D., Lachaud, C., Cotelle, V., Briere, C., Grat, S., Mazars, C., et al. (2011). Nitric oxide production is not required for dihydrosphingosine-induced cell death in tobacco BY-2 cells. Plant Signal. Behav. 6, 736–739. doi: 10.4161/psb.6.5.15126
De Michele, R., Vurro, E., Rigo, C., Costa, A., Elviri, L., Di Valentin, M., et al. (2009). Nitric oxide is involved in cadmium-induced programmed cell death in Arabidopsis suspension cultures. Plant Physiol. 150, 217–228. doi: 10.1104/pp.108.133397
Delledonne, M., Xia, Y., Dixon, R. A., and Lamb, C. (1998). Nitric oxide functions as a signal in plant disease resistance. Nature 394, 585–588. doi: 10.1038/29087
Delledonne, M., Zeier, J., Marocco, A., and Lamb, C. (2001). Signal interactions between nitric oxide and reactive oxygen intermediates in the plant hypersensitive disease resistance response. Proc. Natl. Acad. Sci. U.S.A. 98, 13454–13459. doi: 10.1073/pnas.231178298
Dietz, K. J. (2003a). Plant peroxiredoxins. Annu. Rev. Plant Biol. 54, 93–107. doi: 10.1146/annurev.arplant.54.031902.134934
Dietz, K. J. (2003b). Redox control, redox signaling, and redox homeostasis in plant cells. Int. Rev. Cytol. 228, 141–193. doi: 10.1016/S0074-7696(03)28004-9
Di Stasi, A. M., Mallozzi, C., Macchia, G., Maura, G., Petrucci, T. C., and Minetti, M. (2002). Peroxynitrite affects exocytosis and SNARE complex formation and induces tyrosine nitration of synaptic proteins. J. Neurochem. 82, 420–429. doi: 10.1046/j.1471-4159.2002.00980.x
Feechan, A., Kwon, E., Yun, B. W., Wang, Y., Pallas, J. A., and Loake, G. J. (2005). A central role for S-nitrosothiols in plant disease resistance. Proc. Natl. Acad. Sci. U.S.A. 102, 8054–8059. doi: 10.1073/pnas.0501456102
Feng, J., Wang, C., Chen, Q., Chen, H., Ren, B., Li, X., et al. (2013). S-nitrosylation of phosphotransfer proteins represses cytokinin signaling. Nat. Commun. 4, 1529. doi: 10.1038/ncomms2541
Galatro, A., Puntarulo, S., Guiamet, J. J., and Simontacchi, M. (2013). Chloroplast functionality has a positive effect on nitric oxide level in soybean cotyledons. Plant Physiol. Biochem. 66, 26–33. doi: 10.1016/j.plaphy.2013.01.019
Gechev, T. S., Van Breusegem, F., Stone, J. M., Denev, I., and Laloi, C. (2006). Reactive oxygen species as signals that modulate plant stress responses and programmed cell death. Bioessays 28, 1091–1101. doi: 10.1002/bies.20493
Greenberg, J. T., and Yao, N. (2004). The role and regulation of programmed cell death in plant-pathogen interactions. Cell. Microbiol. 6, 201–211. doi: 10.1111/j.1462-5822.2004.00361.x
Grun, S., Lindermayr, C., Sell, S., and Durner, J. (2006). Nitric oxide and gene regulation in plants. J. Exp. Bot. 57, 507–516. doi: 10.1093/jxb/erj053
Hancock, J. T., Henson, D., Nyirenda, M., Desikan, R., Harrison, J., Lewis, M., et al. (2005). Proteomic identification of glyceraldehyde 3-phosphate dehydrogenase as an inhibitory target of hydrogen peroxide in Arabidopsis. Plant Physiol. Biochem. 43, 828–835. doi: 10.1016/j.plaphy.2005.07.012
Hasanuzzaman, M., and Fujita, M. (2013). Exogenous sodium nitroprusside alleviates arsenic-induced oxidative stress in wheat (Triticum aestivum L.) seedlings by enhancing antioxidant defense and glyoxalase system. Ecotoxicology 22, 584–596. doi: 10.1007/s10646-013-1050-4
He, Y., Tang, R. H., Hao, Y., Stevens, R. D., Cook, C. W., Ahn, S. M., et al. (2004). Nitric oxide represses the Arabidopsis floral transition. Science 305, 1968–1971. doi: 10.1126/science.1098837
Hideg, E., Kalai, T., Kos, P. B., Asada, K., and Hideg, K. (2006). Singlet oxygen in plants-its significance and possible detection with double (fluorescent and spin) indicator reagents. Photochem. Photobiol. 82, 1211–1218. doi: 10.1562/2006-02-06-RA-797
Hong, J. K., Yun, B. W., Kang, J. G., Raja, M. U., Kwon, E., Sorhagen, K., et al. (2008). Nitric oxide function and signalling in plant disease resistance. J. Exp. Bot. 59, 147–154. doi: 10.1093/jxb/erm244
Horling, F., Lamkemeyer, P., Konig, J., Finkemeier, I., Kandlbinder, A., Baier, M., et al. (2003). Divergent light-, ascorbate-, and oxidative stress-dependent regulation of expression of the peroxiredoxin gene family in Arabidopsis. Plant Physiol. 131, 317–325. doi: 10.1104/pp.010017
Iakimova, E. T., Sobiczewski, P., Michalczuk, L., Wegrzynowicz-Lesiak, E., Mikicinski, A., and Woltering, E. J. (2013). Morphological and biochemical characterization of Erwinia amylovora-induced hypersensitive cell death in apple leaves. Plant Physiol. Biochem. 63, 292–305. doi: 10.1016/j.plaphy.2012.12.006
Jiang, H., Chen, Y., Li, M., Xu, X., and Wu, G. (2011). Overexpression of SGR results in oxidative stress and lesion-mimic cell death in rice seedlings. J. Integr. Plant Biol. 53, 375–387. doi: 10.1111/j.1744-7909.2011.01037.x
Jing, H. C., Hebeler, R., Oeljeklaus, S., Sitek, B., Stuhler, K., Meyer, H. E., et al. (2008). Early leaf senescence is associated with an altered cellular redox balance in Arabidopsis cpr5/old1 mutants. Plant Biol. (Stuttg.) 10(Suppl. 1), 85–98. doi: 10.1111/j.1438-8677.2008.00087.x
Kong, X., Zhang, D., Pan, J., Zhou, Y., and Li, D. (2013). Hydrogen peroxide is involved in nitric oxide-induced cell death in maize leaves. Plant Biol. (Stuttg.) 15, 53–59. doi: 10.1111/j.1438-8677.2012.00598.x
Lanteri, M. L., Lamattina, L., and Laxalt, A. M. (2011). Mechanisms of xylanase-induced nitric oxide and phosphatidic acid production in tomato cells. Planta 234, 845–855. doi: 10.1007/s00425-011-1446-4
Lee, U., Wie, C., Fernandez, B. O., Feelisch, M., and Vierling, E. (2008). Modulation of nitrosative stress by S-nitrosoglutathione reductase is critical for thermotolerance and plant growth in Arabidopsis. Plant Cell 20, 786–802. doi: 10.1105/tpc.107.052647
Leitner, M., Vandelle, E., Gaupels, F., Bellin, D., and Delledonne, M. (2009). NO signals in the haze: nitric oxide signalling in plant defence. Curr. Opin. Plant Biol. 12, 451–458. doi: 10.1016/j.pbi.2009.05.012
Lim, P. O., Kim, H. J., and Nam, H. G. (2007). Leaf senescence. Annu. Rev. Plant Biol. 58, 115–136. doi: 10.1146/annurev.arplant.57.032905.105316
Lin, A., Wang, Y., Tang, J., Xue, P., Li, C., Liu, L., et al. (2012). Nitric oxide and protein S-nitrosylation are integral to hydrogen peroxide-induced leaf cell death in rice. Plant Physiol. 158, 451–464. doi: 10.1104/pp.111.184531
Lindermayr, C., Saalbach, G., Bahnweg, G., and Durner, J. (2006). Differential inhibition of Arabidopsis methionine adenosyltransferases by protein S-nitrosylation. J. Biol. Chem. 281, 4285–4291. doi: 10.1074/jbc.M511635200
Lindermayr, C., Saalbach, G., and Durner, J. (2005). Proteomic identification of S-nitrosylated proteins in Arabidopsis. Plant Physiol. 137, 921–930. doi: 10.1104/pp.104.058719
Lindermayr, C., Sell, S., Muller, B., Leister, D., and Durner, J. (2010). Redox regulation of the NPR1-TGA1 system of Arabidopsis thaliana by nitric oxide. Plant Cell 22, 2894–2907. doi: 10.1105/tpc.109.066464
Liu, F., and Guo, F. Q. (2013). Nitric oxide deficiency accelerates chlorophyll breakdown and stability loss of thylakoid membranes during dark-induced leaf senescence in Arabidopsis. PLoS ONE 8:e56345. doi: 10.1371/journal.pone.0056345
Liu, W. Z., Kong, D. D., Gu, X. X., Gao, H. B., Wang, J. Z., Xia, M., et al. (2013). Cytokinins can act as suppressors of nitric oxide in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 110, 1548–1553. doi: 10.1073/pnas.1213235110
Ma, W., Smigel, A., Walker, R. K., Moeder, W., Yoshioka, K., and Berkowitz, G. A. (2010). Leaf senescence signaling: the Ca2+-conducting Arabidopsis cyclic nucleotide gated channel2 acts through nitric oxide to repress senescence programming. Plant Physiol. 54, 733–743. doi: 10.1104/pp.110.161356
Masclaux-Daubresse, C., Purdy, S., Lemaitre, T., Pourtau, N., Taconnat, L., Renou, J. P., et al. (2007). Genetic variation suggests interaction between cold acclimation and metabolic regulation of leaf senescence. Plant Physiol. 143, 434–446. doi: 10.1104/pp.106.091355
Miller, J. D., Arteca, R. N., and Pell, E. J. (1999). Senescence-associated gene expression during ozone-induced leaf senescence in Arabidopsis. Plant Physiol. 120, 1015–1024. doi: 10.1104/pp.120.4.1015
Mishina, T. E., Lamb, C., and Zeier, J. (2007). Expression of a nitric oxide degrading enzyme induces a senescence programme in Arabidopsis. Plant Cell Environ. 30, 39–52. doi: 10.1111/j.1365-3040.2006.01604.x
Munns, R. (2005). Genes and salt tolerance: bringing them together. New Phytol. 167, 645–663. doi: 10.1111/j.1469-8137.2005.01487.x
Mur, L. A., Carver, T. L., and Prats, E. (2006). NO way to live; the various roles of nitric oxide in plant-pathogen interactions. J. Exp. Bot. 57, 489–505. doi: 10.1093/jxb/erj052
Murgia, I., Vazzola, V., Tarantino, D., Cellier, F., Ravet, K., Briat, J. F., et al. (2007). Knock-out of ferritin AtFer1 causes earlier onset of age-dependent leaf senescence in Arabidopsis. Plant Physiol. Biochem. 45, 898–907. doi: 10.1016/j.plaphy.2007.09.007
Navabpour, S., Morris, K., Allen, R., Harrison, E., A-H-Mackerness, S., and Buchanan-Wollaston, V. (2003). Expression of senescence-enhanced genes in response to oxidative stress. J. Exp. Bot. 54, 2285–2292. doi: 10.1093/jxb/erg267
Neill, S., Desikan, R., and Hancock, J. (2002). Hydrogen peroxide signalling. Curr. Opin. Plant Biol. 5, 388–395. doi: 10.1016/S1369-5266(02)00282-0
Niu, Y. H., and Guo, F. Q. (2012). Nitric oxide regulates dark-induced leaf senescence through EIN2 in Arabidopsis. J. Integr. Plant Biol. 54, 516–525. doi: 10.1111/j.1744-7909.2012.01140.x
Noctor, G., Veljovic-Jovanovic, S., and Foyer, C. H. (2000). Peroxide processing in photosynthesis: antioxidant coupling and redox signalling. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355, 1465–1475. doi: 10.1098/rstb.2000.0707
Noritake, T., Kawakita, K., and Doke, N. (1996). Nitric oxide induces phytoalexin accumulation in potato tuber tissues. Plant Cell Physiol. 37, 113–116. doi: 10.1093/oxfordjournals.pcp.a028908
Orozco-Cardenas, M. L., Narvaez-Vasquez, J., and Ryan, C. A. (2001). Hydrogen peroxide acts as a second messenger for the induction of defense genes in tomato plants in response to wounding, systemin, and methyl jasmonate. Plant Cell 13, 179–191. doi: 10.1105/tpc.13.1.179
Orozco-Cardenas, M. L., and Ryan, C. A. (2002). Nitric oxide negatively modulates wound signaling in tomato plants. Plant Physiol. 130, 487–493. doi: 10.1104/pp.008375
Pourtau, N., Mares, M., Purdy, S., Quentin, N., Ruel, A., and Wingler, A. (2004). Interactions of abscisic acid and sugar signalling in the regulation of leaf senescence. Planta 219, 765–772. doi: 10.1007/s00425-004-1279-5
Rao, M. V., and Davis, K. R. (2001). The physiology of ozone induced cell death. Planta 213, 682–690. doi: 10.1007/s004250100618
Romero-Puertas, M. C., Corpas, F. J., Rodriguez-Serrano, M., Gomez, M., Del Rio, L. A., and Sandalio, L. M. (2007a). Differential expression and regulation of antioxidative enzymes by cadmium in pea plants. J. Plant Physiol. 164, 1346–1357. doi: 10.1016/j.jplph.2006.06.018
Romero-Puertas, M. C., Laxa, M., Matte, A., Zaninotto, F., Finkemeier, I., Jones, A. M., et al. (2007b). S-nitrosylation of peroxiredoxin II E promotes peroxynitrite-mediated tyrosine nitration. Plant Cell 19, 4120–4130. doi: 10.1105/tpc.107.055061
Romero-Puertas, M. C., Perazzolli, M., Zago, E. D., and Delledonne, M. (2004). Nitric oxide signalling functions in plant-pathogen interactions. Cell. Microbiol. 6, 795–803. doi: 10.1111/j.1462-5822.2004.00428.x
Rubbo, H., Denicola, A., and Radi, R. (1994a). Peroxynitrite inactivates thiol-containing enzymes of Trypanosoma cruzi energetic metabolism and inhibits cell respiration. Arch. Biochem. Biophys. 308, 96–102. doi: 10.1006/abbi.1994.1014
Rubbo, H., Radi, R., Trujillo, M., Telleri, R., Kalyanaraman, B., Barnes, S., et al. (1994b). Nitric oxide regulation of superoxide and peroxynitrite-dependent lipid peroxidation. Formation of novel nitrogen-containing oxidized lipid derivatives. J. Biol. Chem. 269, 26066–26075.
Saito, S., Yamamoto-Katou, A., Yoshioka, H., Doke, N., and Kawakita, K. (2006). Peroxynitrite generation and tyrosine nitration in defense responses in tobacco BY-2 cells. Plant Cell Physiol. 47, 689–697. doi: 10.1093/pcp/pcj038
Saxena, I., and Shekhawat, G. S. (2013). Nitric oxide (NO) in alleviation of heavy metal induced phytotoxicity and its role in protein nitration. Nitric Oxide 32, 13–20. doi: 10.1016/j.niox.2013.03.004
Tada, Y., Spoel, S. H., Pajerowska-Mukhtar, K., Mou, Z., Song, J., Wang, C., et al. (2008). Plant immunity requires conformational changes [corrected] of NPR1 via S-nitrosylation and thioredoxins. Science 321, 952–956. doi: 10.1126/science.1156970
Tan, J., Zhuo, C., and Guo, Z. (2013). Nitric oxide mediates cold- and dehydration-induced expression of a novel MfHyPRP that confers tolerance to abiotic stress. Physiol. Plant. doi: 10.1111/ppl.12032 [Epub ahead of print].
Tanou, G., Filippou, P., Belghazi, M., Job, D., Diamantidis, G., Fotopoulos, V., et al. (2012). Oxidative and nitrosative-based signaling and associated post-translational modifications orchestrate the acclimation of citrus plants to salinity stress. Plant J. 72, 585–599. doi: 10.1111/j.1365-313X.2012.05100.x
Terrile, M. C., Paris, R., Calderon-Villalobos, L. I., Iglesias, M. J., Lamattina, L., Estelle, M., et al. (2011). Nitric oxide influences auxin signaling through S-nitrosylation of the Arabidopsis transport inhibitor response1 auxin receptor. Plant J. 70, 492–500. doi: 10.1111/j.1365-313X.2011.04885.x
Tewari, R. K., Prommer, J., and Watanabe, M. (2013). Endogenous nitric oxide generation in protoplast chloroplasts. Plant Cell Rep. 32, 31–44. doi: 10.1007/s00299-012-1338-5
Vanlerberghe, G. C. (2013). Alternative oxidase: a mitochondrial respiratory pathway to maintain metabolic and signaling homeostasis during abiotic and biotic stress in plants. Int. J. Mol. Sci. 14, 6805–6847. doi: 10.3390/ijms14046805
Verma, K., Mehta, S. K., and Shekhawat, G. S. (2013). Nitric oxide (NO) counteracts cadmium induced cytotoxic processes mediated by reactive oxygen species (ROS) in Brassica juncea: cross-talk between ROS, NO and antioxidant responses. Biometals 26, 255–269. doi: 10.1007/s10534-013-9608-4
Wang, Y., Chen, C., Loake, G. J., and Chu, C. (2010). Nitric oxide: promoter or suppressor of programmed cell death? Protein Cell 1, 133–142. doi: 10.1007/s13238-010-0018-x
Wang, Y., Lin, A., Loake, G. J., and Chu, C. (2013). H2O2-induced leaf cell death and the crosstalk of reactive nitric/oxygen species. J. Integr. Plant. Biol. 55, 202–208. doi: 10.1111/jipb.12032
Wang, Y. Q., Feechan, A., Yun, B. W., Shafiei, R., Hofmann, A., Taylor, P., et al. (2009). S-nitrosylation of AtSABP3 antagonizes the expression of plant immunity. J. Biol. Chem. 284, 2131–2137. doi: 10.1074/jbc.M806782200
Wilkins, K. A., Bancroft, J., Bosch, M., Ings, J., Smirnoff, N., and Franklin-Tong, V. E. (2011). ROS and NO mediate actin reorganization and programmed cell death in the self-incompatibility response of Papaver. Plant Physiol. 156, 404–416. doi: 10.1104/pp.110.167510
Wu, X. Y., Kuai, B. K., Jia, J. Z., and Jing, H. C. (2012). Regulation of leaf senescence and crop genetic improvement. J. Integr. Plant. Biol. 54, 936–952. doi: 10.1111/jipb.12005
Yadav, S., David, A., Baluska, F., and Bhatla, S. C. (2013). Rapid auxin-induced nitric oxide accumulation and subsequent tyrosine nitration of proteins during adventitious root formation in sunflower hypocotyls. Plant Signal. Behav. 8, pii: e23196 doi: 10.4161/psb.23196 [Epub ahead of print].
Ye, Y., Li, Z., and Xing, D. (2013). Nitric oxide promotes MPK6-mediated caspase-3-like activation in cadmium-induced Arabidopsis thaliana programmed cell death. Plant Cell Environ. 36, 1–15. doi: 10.1111/j.1365-3040.2012.02543.x
Yemets, A. I., Krasylenko, Y. A., Lytvyn, D. I., Sheremet, Y. A., and Blume, Y. B. (2011). Nitric oxide signalling via cytoskeleton in plants. Plant Sci. 181, 545–554. doi: 10.1016/j.plantsci.2011.04.017
Yun, B. W., Feechan, A., Yin, M., Saidi, N. B., Le Bihan, T., Yu, M., et al. (2011). S-nitrosylation of NADPH oxidase regulates cell death in plant immunity. Nature 478, 264–268. doi: 10.1038/nature10427
Keywords: nitric oxide, reactive oxygen species, programed cell death, hypersensitive response, leaf senescence
Citation: Wang Y, Loake GJ and Chu C (2013) Cross-talk of nitric oxide and reactive oxygen species in plant programed cell death. Front. Plant Sci. 4:314. doi: 10.3389/fpls.2013.00314
Received: 30 May 2013; Paper pending published: 17 June 2013;
Accepted: 26 July 2013; Published online: 16 August 2013.
Edited by:
Emmanuel Baudouin, Université Pierre et Marie Curie - Paris 6, FranceReviewed by:
Vasileios Fotopoulos, Cyprus University of Technology, CyprusCopyright © 2013 Wang, Loake and Chu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chengcai Chu, State Key Laboratory of Plant Genomics and National Center for Plant Gene Research (Beijing), Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, No. 1 West Beichen Road, Chaoyang, Beijing 100101, People’s Republic of China e-mail:Y2NjaHVAZ2VuZXRpY3MuYWMuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.