- 1Department of Plant Biology, University of Vermont, Burlington, VT, USA
- 2Norwegian University of Life Sciences, Ås, Norway
Flowering plants initially diversified during the Mesozoic era at least 140 million years ago in regions of the world where temperate seasonal environments were not encountered. Since then several cooling events resulted in the contraction of warm and wet environments and the establishment of novel temperate zones in both hemispheres. In response, less than half of modern angiosperm families have members that evolved specific adaptations to cold seasonal climates, including cold acclimation, freezing tolerance, endodormancy, and vernalization responsiveness. Despite compelling evidence for multiple independent origins, the level of genetic constraint on the evolution of adaptations to seasonal cold is not well understood. However, the recent increase in molecular genetic studies examining the response of model and crop species to seasonal cold offers new insight into the evolutionary lability of these traits. This insight has major implications for our understanding of complex trait evolution, and the potential role of local adaptation in response to past and future climate change. In this review, we discuss the biochemical, morphological, and developmental basis of adaptations to seasonal cold, and synthesize recent literature on the genetic basis of these traits in a phylogenomic context. We find evidence for multiple genetic links between distinct physiological responses to cold, possibly reinforcing the coordinated expression of these traits. Furthermore, repeated recruitment of the same or similar ancestral pathways suggests that land plants might be somewhat pre-adapted to dealing with temperature stress, perhaps making inducible cold traits relatively easy to evolve.
Introduction
Since the late Eocene and Oligocene around 47.5 to 26 million years ago (mya) the Earth has experienced dramatic cooling events, resulting in an overall contraction of the tropics, and establishment of novel temperate zones in both northern and southern hemispheres (Zachos et al., 2001; Stickley et al., 2009). In response to this cooling, several ancestrally tropical lineages have successfully diversified outside their ecological zone of origin, becoming adapted to cooler and often more seasonal environments (Latham and Ricklefs, 1993; Sandve and Fjellheim, 2010). However, the fact that less than half the families of modern angiosperms are represented in the temperate zones suggests that adaptations to cold seasonal climates might be difficult to evolve (Ricklefs and Renner, 1994; Donoghue, 2008).
Support for the hypothesis that adaptations to low or freezing seasonal temperatures are relatively hard and/or slow to evolve comes from the fact that climate cooling during the Eocene–Oligocene boundary was associated with large-scale extinctions of both animals and plants (Ivany et al., 2000), and by the apparent complexity of physiological and morphological adaptations to cold (see later sections). However, as an alternative hypothesis, it has been postulated that, since climate cooling has been an ongoing process throughout the Cenozoic, the relatively recent expansion of cold temperate zones has meant that only a minority of plant families have been historically party to selection by cold winters (Fine and Ree, 2006). Thus, there is still much debate about whether different adaptations to extended periods of cold can evolve quickly enough to allow range expansions and/or local adaptation under gradual or rapid climate change conditions (Franks et al., 2007; Cook et al., 2012). The focus of this review is to highlight the major ways in which plants have adapted physiologically to cold seasonal environments, and to synthesize some of the current available data on the genetic basis of these adaptations, with the general goal of understanding the evolutionary lability of cold-season traits.
Physiological and Morphological Adaptations to Seasonal Cold
Plant species in both the cold temperate zones and the tropical highlands can experience periods of cold or freezing temperatures that are potentially detrimental to growth and development. However, a key difference between temperate and tropical highland species is the timing and duration of cold, either occurring on a diurnal cycle (tropical highland species) or an annual cycle (temperate species). Despite evidence for overlap in the genetic response to cold (Dhillion et al., 2010; Greenup et al., 2011), the extent to which cold occurs diurnally or seasonally has major implications for the life history strategies that are adopted by plant populations in a given environment (e.g., Teutonico and Osborn, 1995). The focus of this review is physiological adaptation to seasonal cold, i.e., adaptation to the coldest season of the year in temperate climates.
Unlike animals, individual plants are immobile. Thus, in order to reduce the negative effects of winter cold, many temperate plants must synchronize their sensitive reproductive output with favorable environmental conditions of the spring and summer (Bradshaw, 1972; King and Heide, 2009). In the case of spring annuals, germination, reproduction, and senescence occur during the warm seasons. In order to avoid winter growth, spring annual seeds remain dormant during the winter, only to germinate in response to inductive temperatures in the spring (Hemming and Trevaskis, 2011). By contrast, winter annuals set seed and germinate in the fall, overwinter in a vegetative growth state, and flower in the spring. The ability of plants to respond to an extended period of cold to rapidly attain flowering competency is termed vernalization responsiveness (Chouard, 1960). Furthermore, since the aerial vegetative structures (i.e., stems and leaves) of winter annuals are subject to cold, vernalization responsive plants are often, but not always (Rapacz and Markowski, 1999), induced into a state of cold and/or freezing tolerance.
Similar to annuals, temperate perennials vary in their life history strategies for dealing with seasonal cold. In the case of herbaceous perennials, germination, reproduction, and senescence occur during the warm seasons. However, rather than relying only on seed to produce the next generation, herbaceous perennials are capable of secondary rounds of vegetative growth from dormant underground meristems (e.g., rhizomes), which occurs at the conclusion of winter. By contrast, woody perennials such as trees, often delay their flowering for several years until a critical biomass is achieved (Rohde and Bhalerao, 2007). As in the case of winter annuals, temperate herbaceous perennials are often responsive to vernalization, and can tolerate chilling and frost. Furthermore, in addition to cold tolerance, many temperate and boreal trees are able to protect new growth from harsh winter conditions by becoming dormant prior to winter (endodormancy; Lang et al., 1987; Howe et al., 2003; Campoy and Egea, 2011). Recent studies on the genetic basis of these varied adaptations to winter cold offer exciting opportunities to understand constraints on plant transitions from the tropical to the temperate zone, and vice versa. This is particularly relevant in the face of current and projected changes to our climate. The following sections will focus on the evolution and genetic basis of three important physiological cold adaptations: cold acclimation (i.e., the seasonal acquisition of cold and freezing tolerance), endodormancy, and vernalization responsiveness. However, first we will consider what is currently known about the phylogenetic pattern of these traits across seed plants, and their relationships to climate.
Phenotypic Correlations and the Phylogenetic Distribution of Cold Adaptive Traits
In temperate plants that experience prolonged cold to sub-zero winter temperatures, above ground tissues are susceptible to delayed growth and damage by frost. Thus, high latitude plants that undergo endodormancy (woody perennials), or are responsive to vernalization (herbaceous annuals and perennials), are often also able to induce cold tolerance through a process known as cold acclimation (Howe et al., 2003; see Cold Acclimation and Cold Tolerance). For example, non-vernalization responsive (spring) wheat varieties generally have lower freezing tolerance than vernalization responsive (winter) wheat, and in winter wheat length of vernalization requirement is positively correlated with speed of cold acclimation (Prasil et al., 2004). Furthermore, in temperate trees such as Pinus contorta and Douglas-fir (Pseudotsuga menziesii), elevation and distance from warmer ocean climates are strongly associated with the timing of growth cessation and endodormancy and the temperature required to induce cold acclimation and subsequent freezing tolerance (Campbell and Sugano, 1979; Howe et al., 2003). A similar trend has been found between length of vernalization needed to elicit flowering and continental-oceanic gradients in A. thaliana (Lewandowska-Sabat et al., 2012).
Despite the correlation between cold adaptive traits, there are examples of endodormant and vernalization responsive plants that cannot induce cold tolerance, and vice versa. For example, Thuja plicata and Tsuga heterophylla both acclimate to cold, but do not experience endodormancy (Silim and Lavender, 1994), several trees undergo endodormancy without acclimating to cold (Kramer and Kozlowski, 1979), and several vernalization-responsiveness cereal cultivars are considered cold-sensitive (Tester and Bacic, 2005). The lack of a strict association between endodormancy/vernalization responsiveness and cold acclimation might be explained by the fact that low non-freezing temperatures can be detrimental to young bud development without affecting growth of other plant structures. For example, in plants adapted to, or derived from, subtropical climates, synchronization of bud development with warm conditions might be enough to escape the negative effects of occasional low winter temperatures, without the additional need for cold/freezing tolerance. Alternatively, boreal plants that experience sub-zero temperatures for large parts of the year might benefit from constitutive freezing tolerance, negating the importance of cold acclimation.
As outlined in the introduction, angiosperm families containing temperate species are less common than families confined to the tropics (Ricklefs and Renner, 1994; Donoghue, 2008). However, temperate taxa are distributed throughout the seed plant phylogeny (Ricklefs, 2005; Figure 1). Thus, the timing of angiosperm diversification relative to global Eocene cooling events suggests numerous independent origins of temperate seed plants (Wang et al., 2009b). How many of these major lineages evolved physiological adaptations to seasonal cold? With the exception of early-diverging angiosperms and several tropical gymnosperms for which there are no or limited experimental data, a broad literature search suggests that cold acclimation, endodormancy, and vernalization responsiveness have evolved in all major seed plant clades (Figure 1). Thus, at a broad phylogenetic scale, adaptations to cold might be relatively easy to evolve. In the following sections, we review available genetic data to determine whether multiple independent origins of cold traits can be explained by the modification of pre-adapted pathways (e.g., exaptations, sensu Gould and Vrba, 1982), and/or represent novel evolutionary innovations. We also suggest future studies that could be carried out to determine the potential for evolution of seasonal cold traits on shorter timescales and in response to current global change.
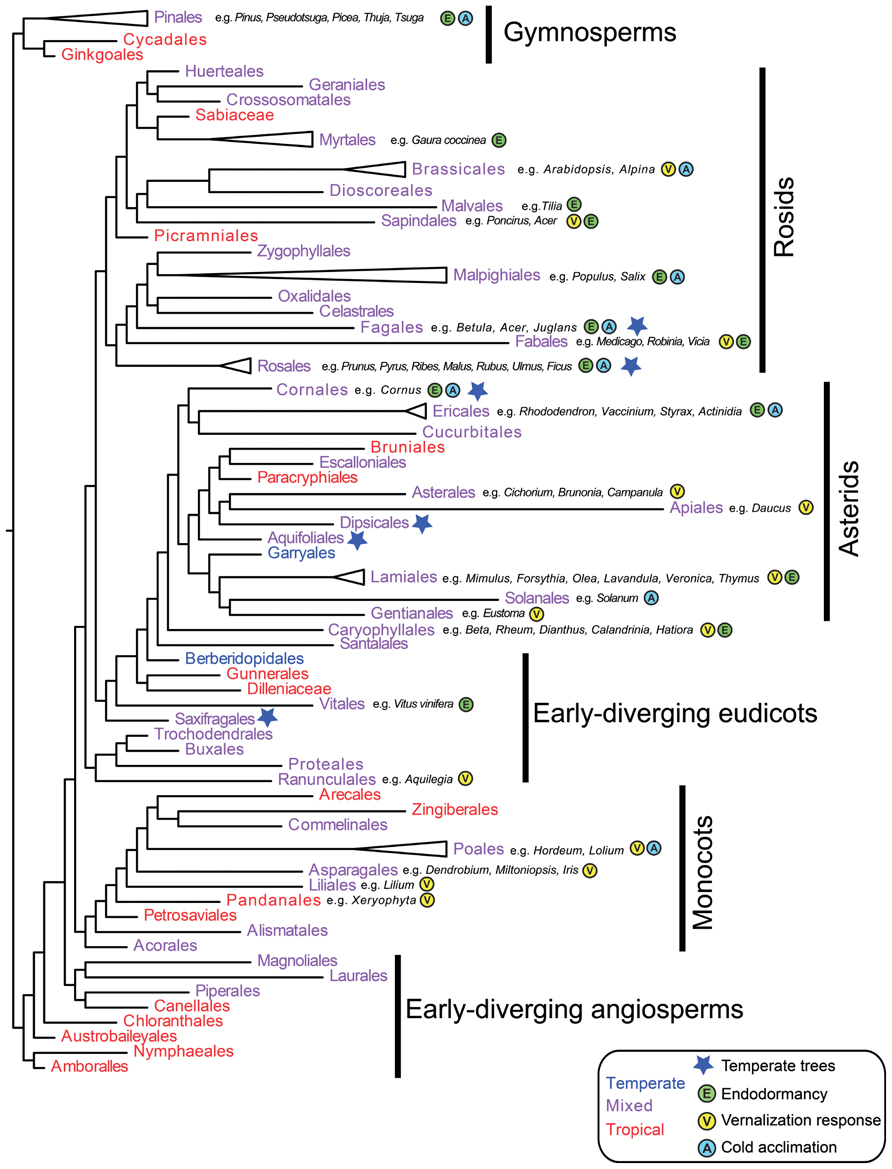
FIGURE 1. Evolution of cold adaptive traits in seed plants. Relationships among major seed plant orders are inferred using representative taxa from Smith et al. (2011) in phylomatic (Webb and Donoghue, 2005). Orders are color-coded as primarily temperate (blue), broadly distributed, or primarily tropical (red) based on the APG website (Stevens, 2001 onward). Blue stars indicate orders where trees are primarily temperate based on Ricklefs (2005). Evidence for endodormancy (E), cold acclimation/freezing tolerance (A), and vernalization responsiveness (V) are denoted for each order with example species. Since most species have not been tested for cold adaptations, absence of data does not necessarily indicate absence of traits. However, since cold climates arose after major radiations in seed plants, presence data (based on Krug, 1991; De la Rosa et al., 2000; Kawamata et al., 2002; Wilson et al., 2002; Karlson et al., 2004; Streck and Schuh, 2005; Lopez and Runkle, 2006; Fausey and Cameron, 2007; Kalberer et al., 2007; Mewes and Pank, 2007; Rohwer and Heins, 2007; Svendsen et al., 2007; Padhye and Cameron, 2008, 2009; Pietsch et al., 2009; Zlesak and Anderson, 2009; Biasi et al., 2010; Byard et al., 2010; Ghelardini et al., 2010; Kaymak and Guvenc, 2010; Kubota et al., 2010; Lenahan et al., 2010; Rantasen and Palonen, 2010; Caffarra et al., 2011; Cave et al., 2011; Charrier et al., 2011; Dogramaci et al., 2011; Lin et al., 2011; Adhikari et al., 2012; Andreini et al., 2012; Bilavcik et al., 2012; Diaz-Riquelme et al., 2012; Nishitani et al., 2012; Sanchez-Perez et al., 2012; Whitman and Runkle, 2012; Alessandro et al., 2013; Guzy-Wrobelska et al., 2013; Jones et al., 2013; Mojtahedi et al., 2013) indicates multiple origins of cold adaptive traits across the phylogeny.
Cold Acclimation and Cold Tolerance
Cold tolerance is a highly complex trait that encompasses both the ability to tolerate the direct effects of low temperatures on plant function and the indirect effects of ice formation in and surrounding the plant. Direct effects of cold conditions change biological thermodynamic processes, biomolecule stability and function, and alter normal cellular processes such as photosynthesis, inter- and intracellular transport, and the balance between production and neutralization of toxic reactive oxygen species (ROS; Thomashow, 1998; Figure 2). By contrast, indirect effects of ice formation that occur during sub-zero temperatures create a different kind of cellular stress. First, extracellular ice formation depletes available water in and around cells, affecting normal water dependent processes and inducing freezing dehydration with associated cell membrane disruption (Steponkus et al., 1998). Second, large ice crystals grow at the expense of the formation of small new crystals, a process referred to as ice recrystallization (Capicciotti et al., 2012). This process generates large and potentially damaging expanding crystals in extracellular spaces. Although most species have some innate tolerance to a sudden exposure to cold, many temperate species have evolved the ability to gradually increase their freezing tolerance during extended periods of cold, but non-freezing, temperatures and changing photoperiod during autumn (Thomashow, 1999; Catalá et al., 2011). This inducible process is referred to as cold acclimation, and ultimately leads to healthy plants that can successfully reproduce the following spring (Figure 2).
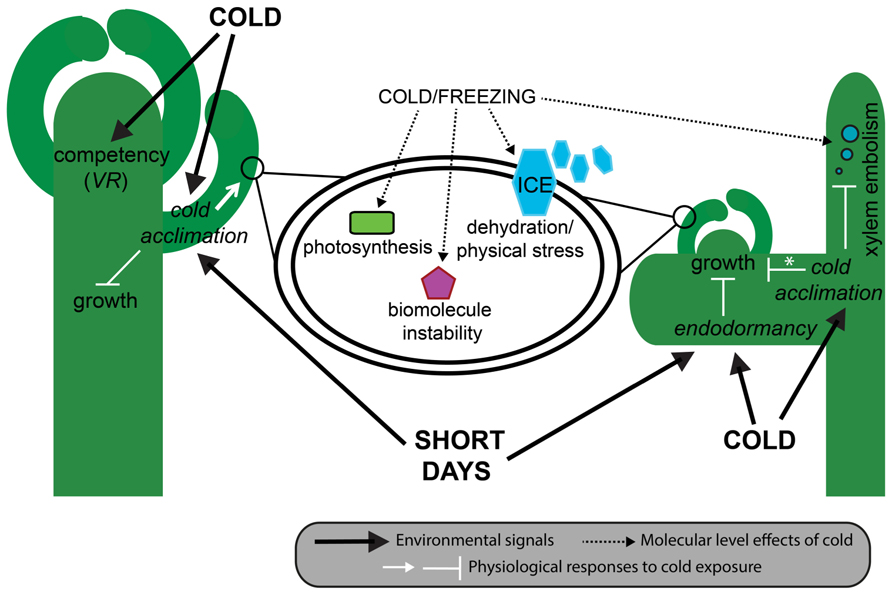
FIGURE 2. Effect of cold on plant phenotype. In herbaceous plants (left) such as A. thaliana and wheat, cool autumnal temperatures induce cold acclimation mostly in young leaves, and the acquisition of floral competency in the shoot apical meristem. In A. thaliana, cold acclimation is also induced by short days, and results in a decrease in growth rate through the gibberellic acid pathway, and an increase in freezing tolerance within cells (middle). In woody plants (right) such as pines and birch, endodormancy can be induced as early as late summer and results in the complete cessation of meristematic mitosis. As in herbaceous plants, cool temperatures lead to cold acclimation and a gradual increase in freezing tolerance, sometimes (e.g., deciduous trees) but not always (e.g., evergreen trees) resulting in a reduction in whole-plant growth. At the cellular level, freezing tolerance results from the ability of cells to deal with dehydration, ice crystal formation, biomolecule instability, and disruption to photosynthesis. At the whole-plant level, some freezing tolerant woody plants are able to deal with the increased likelihood of embolism resulting from bubbles formed when dissolved gases are released from frozen xylem sap. Asterisk denotes species-level differences in growth responses to cold acclimation. VR, vernalization responsiveness.
Molecular and Physiological Changes Associated with Cold Acclimation and Cold Tolerance
Cold acclimation involves major changes in the biochemical and physiological state of the plant, improving low temperature stress tolerance. In general, proteins and compounds with various protective functions are increased, while photosynthesis and several other metabolism-related biochemical pathways are suppressed (e.g., Fowler and Thomashow, 2002; Lee et al., 2005; Rudi et al., 2010; Winfield et al., 2010; Table 1). Although cold acclimation genes and genetic pathways can vary widely between species, some molecular and physiological changes seem to be similar across major angiosperm clades (reviewed in Sandve et al., 2011).
The ability to manipulate ice formation has arisen multiple times throughout angiosperm evolution, and is achieved either by decreasing the freezing point (thermal hysteresis) or by inhibiting ice recrystallization (Griffith and Yaish, 2004; Byard et al., 2010; Figure 2). Although thermal hysteresis is commonly found in plants, it is not affected much by cold acclimation (Urrutia et al., 1992); ice recrystallization inhibition is believed to be more important for plant cold acclimation (Griffith and Yaish, 2004; Table 1). Ice recrystallization inhibition is found in species of many plant lineages (Doucet et al., 2000) and is caused by a range of diverse proteins, including beta-1,3-glucanases, WRKY proteins, chitinases, and thaumatin-like proteins (Griffith and Yaish, 2004 and references therein). For example, in eudicots the carrot (Daucus carota) polygalacturonase inhibitor protein inhibits ice recrystallization and decreases the freezing point; its expression in tobacco (Solanum tabacum) and A. thaliana results in inhibition of ice recrystallization and increased freezing tolerance (Worrall et al., 1998; Meyer et al., 1999). In monocots, a different Pooideae (Poaceae)-specific inhibitor of ice recrystallization-protein (IRIP) family shows strong inhibition of ice recrystallization (Sidebottom et al., 2000), which has been shown to increase freezing tolerance in planta (Zhang et al., 2010).
Freezing induced cell dehydration is another factor plants have to deal with during winter (Uemura et al., 1995; Figure 2). During cold acclimation plants gain the ability to modify membrane stability, which involves both changes to the cell membrane lipid content (Li et al., 2004; Moellering et al., 2010) and production of membrane-interacting protective compounds, such as proline and a diversity of carbohydrates (Fujikawa et al., 1999; Hisano et al., 2004; Valluru and Van den Ende, 2008; Table 1). Manipulation of lipid metabolism and membrane lipid composition in transgenic plants has improved freezing and chilling tolerance in tobacco (Khodakovskaya et al., 2006), poplar (Populus sp.; Zhou et al., 2009), and tomato (S. lycopersicum; Domínguez et al., 2010).
In addition to proteins and compounds with direct protective action in cold, modulation of photosynthetic processes is a common cold acclimation response among angiosperms (Figure 2). During photosynthesis, light energy is absorbed and converted to chemical energy in thylakoid membranes of chloroplasts, and then used for CO2-fixation in the Calvin cycle. Absorption of light by photosystem II (PSII) normally leads to light-induced damage of the PSII, a process referred to as photoinhibition, which is counteracted by a PSII damage repair mechanism that restores PSII function (Hetherington et al., 1989; Aro et al., 1993, 2005; Melis, 1999; Bascuñán-Godoy et al., 2012). However, if the level of absorbed light energy greatly exceeds that of the consumed chemical energy this will impair PSII damage repair and accelerate photoinhibition (Takahashi and Murata, 2008), with potential detrimental consequences for plant growth (Melis, 1999). Low temperatures promote increased photoinhibition (Hetherington et al., 1989), and to minimize photoinhibition-associated damage, higher plants undergo photosynthetic acclimation during cold acclimation either by increasing the energy demand through increased carbon assimilation and carbon metabolism (Huner et al., 1993), dissipation of excess excitation energy as heat (Dall’Osto et al., 2005), or improving the PSII repair machinery (Bascuñán-Godoy et al., 2012; Table 1). Photoinhibition is also detrimental to the entire cell due to the associated increase of ROS (Krause, 1988). Photoinhibition has been shown to regulate the expression of genes in cold acclimation (Gray et al., 1997), hence, a plants’ freezing tolerance is inherently linked to temperature-induced photoinhibition. Variation in the capacity for photosynthetic acclimation during cold acclimation is correlated with genotypic differences in winter survival and freezing tolerance of grasses (Rapacz et al., 2004). Moreover, the C-REPEAT binding factor (CBF) pathway has been shown to alleviate photoinhibition in autumn conditions (Yang et al., 2010).
In woody species, freezing temperatures can disrupt whole-plant functioning by limiting long-distance water transport in the xylem (Sperry et al., 1994; Améglio et al., 2001). This is particularly true following xylem embolisms (Figure 2), which are often induced by freeze–thaw cycles in regions that experience freezing winter nights and above freezing winter days, such as North Africa, the Mediterranean region of Europe, and southern parts of North America (Cavendar-Bares et al., 2005; Meideros and Pockman, 2011). Although variation in resistance to embolism can be explained by differences in vessel diameter and architecture (Sperry et al., 1994; Améglio et al., 2001), cold acclimation has been found to reduce xylem embolisms in oak (Quercus) and several conifers (Hammel, 1967; Sperry et al., 1994; Cavendar-Bares et al., 2005). Furthermore, it is hypothesized that some woody plants can repair winter embolism; the mechanisms underlying repair and induced resistance are largely unknown (Cavendar-Bares et al., 2005).
Sensing and Signaling Cold
How low temperature is sensed and then signaled to the cell nucleus is generally not well understood. The best-studied temperature sensing mechanism is membrane fluidity. Membranes surrounding cells, mitochondria, and chloroplasts consist of a lipid bilayer, and low temperatures causes lipid membranes to become more rigid (Alonso et al., 1997). Chemically induced membrane rigidity results in cytoskeleton changes, increased Ca2+ influx to the cell, and changes in activity of certain protein kinases, which ultimately result in transcription of cold-induced genes that artificially mimic the process of cold acclimation and improve freezing tolerance of plants (Örvar et al., 2000; Sangwan et al., 2001). In addition to cell membrane changes, abscisic acid (ABA; Llorente et al., 2000; Xiong et al., 2001) and ROS (Lee et al., 2002) accumulation in warm conditions has been shown to initiate processes similar to cold acclimation, resulting in increased freezing tolerance.
Recently, a molecular model of cold signaling, linking cold sensing and transcription, has been put forward (Doherty et al., 2009). The model includes four components; Ca2+, calcium modulated proteins (calmodulins), calmodulin binding transcriptional activators (CAMTAs), and cold responsive transcription factors. Low temperatures increase the Ca2+ influx (probably as an indirect response to more rigid membranes) and activate calmodulin proteins, which subsequently activate other Ca2+-unresponsive CAMTA proteins essential for cold acclimation. The A. thaliana genome contains six CAMTA genes with calmodulin binding- and DNA binding CG-1 (CGCG) domains (Bouché et al., 2002). Absence of the CG-1 element in promoters of cold responsive transcription factors (both CBF and non-CBF), leads to a decrease in their transcript levels of up to 40–50% (Doherty et al., 2009). Loss-of-function CAMTA gene mutants are unable to acclimate to cold and drought, and so are sensitive to freezing (Doherty et al., 2009; Pandey et al., 2013). It should be noted that the validity of this model with respect to the role of calmodulin as Ca2+ signal sensors has not been experimentally tested.
Transcriptional Regulation of the Cold Acclimation Process
At least 50–60 transcription factors are known to be important in initiation of cold acclimation in A. thaliana, including members of the AP2, MYB, MYC, bZIP, and Zn-FINGER transcription factor families, but relatively little is known about their downstream targets (Fowler and Thomashow, 2002; Lee et al., 2005; Vogel et al., 2005; Knight et al., 2009). The only well characterized cold acclimation pathway in plants is the INDUCER OF CBF EXPRESSION (ICE)-CBF-COLD-RESPONSIVE (COR) cold response pathway (Gilmour et al., 1988; Chinnusamy et al., 2003; Stockinger et al., 2007). During plant chilling, expression of ICE genes triggers rapid (~15 min) transient up-regulation of CBFs (Chinnusamy et al., 2003; Dong et al., 2006), which together directly or indirectly regulate approximately 30% of all cold-induced transcriptional changes (Vogel et al., 2005; Svensson et al., 2006; Van Buskirk and Thomashow, 2006; Table 1). The initial ICE-CBF regulatory switch was described in A. thaliana, but data suggest it is functionally conserved in apple (Malus domestica, Rosaceae), and (at least partially) in grasses, subfamily Pooideae (Chinnusamy et al., 2003; Badawi et al., 2008; Feng et al., 2012). Cold-induced CBFs are found in species across all land plant lineages (e.g., Skinner et al., 2005; Xiong and Fei, 2006; Pennycooke et al., 2008), and their protein products bind to CCGAC core motifs in the promoters of diverse cold and dehydration responsive genes, including themselves (Liu et al., 1998; Novillo et al., 2004; Stockinger et al., 2007).
In addition to cold, CBF transcription is affected by photoperiod. Expression of A. thaliana CBFs fluctuates on a diurnal basis, peaking around 8 h after the dawn zeitgeber (Fowler et al., 2005). Under long day conditions, CBF gene expression is repressed by the action of PHYTOCHROME B (PHYB), PHYTOCHROME-INTERACTING FACTOR 4 (PIF4) and PIF7 (Lee and Thomashow, 2012). By contrast, under short days, reduced mRNA levels and stability of PHYB, PIF4, PIF7 and their protein products, respectively, result in the derepression of CBF transcription (Lee and Thomashow, 2012). Although the circadian regulation of CBFs is not well understood, the differential regulation of CBFs under short versus long days provides a secondary mechanism by which freezing tolerance can be timed to coincide with winter.
The plant hormone ABA is a major player in regulating genes involved in plant stress response through the transcriptional activation of ABA-dependent transcription factors (Shinozaki and Yamaguchi-Shinozaki, 2000). The importance of ABA in freezing tolerance is debated (Gusta et al., 2005), but much evidence supports a role for ABA in cold acclimation under natural conditions. First, endogenous ABA-levels increase in A. thaliana and wheat (Triticum aestivum) during low temperature exposure (Cuevas et al., 2008; Shakirova et al., 2009). Second, application of exogenous ABA enhances freezing tolerance in whole plants (Chen and Gusta, 1983; Mantyla et al., 1995) and calli (Dallaire et al., 1994). Third, many genes expressed during cold acclimation are regulated by ABA, including the CBF genes (Hoth et al., 2002; Knight et al., 2004; Cuevas et al., 2008; Kobayashi et al., 2008; Shakirova et al., 2009; Agarwal and Jha, 2010). Involvement of ABA in cold transcriptional regulation is observed in bryophytes (Bhyan et al., 2012), monocots, and eudicots (reviewed in Gusta et al., 2005).
Conservation and Diversification of Cold Acclimation and Cold Tolerance
Despite independent evolution over hundreds of millions of years, some pathways and mechanisms involved in cold acclimation are similar between species of bryophytes, monocots, and eudicots (see above). This could be interpreted as evidence for conservation of ancestral cold response pathways from the earliest land plant through the diversification of all major land plant lineages. An alternative interpretation is that similarities in cold responses across land plant lineages are due to genetic parallelisms. The latter could occur if an ancient stress response pathway was recruited to cold acclimation and cold/freezing tolerance multiple times independently. Predictions of this hypothesis include substantial overlap between the cold acclimation/tolerance and other stress pathways.
One potential pathway that might have been recruited for cold acclimation and cold/freezing tolerance is the drought tolerance pathway. All land plants are constantly battling to minimize water loss at the atmosphere–plant boundary and prevent cellular dehydration. Adaptations to withstand dehydration were probably some of the main evolutionary innovations when plants moved from aquatic to terrestrial life ~500 mya; hence basic molecular responses to dehydration can be assumed to have a common ancestry in all land plants. Interestingly, many key cold acclimation responses are tightly linked to dehydration. For example, in A. thaliana gene expression correlations of 0.15–0.30 are found between response to dehydration stresses (drought, salt, and osmotic stress) and cold stress (Swindell, 2006). This strong molecular connection between dehydration-like stress responses and cold can only be explained by the control of these responses through non-specific stress triggers, perhaps by cellular redox states, or some other shared signaling mechanisms.
Following this logic, distantly related cold or freezing tolerant species are predicted to have adapted to cold through changes in partially overlapping molecular pathways, resulting in the recurring recruitment of (a few) similar pathways. If this hypothesis holds, we would expect relatively similar initial transcriptional responses and more diverse downstream molecular changes between species with independent adaptations to cold. Comparative transcriptome analyses have shown conserved expression profiles during cold acclimation in potato (S. tuberosum) and A. thaliana, despite over 100 mya of independent evolution (Carvallo et al., 2011). However, more comprehensive comparative studies of stress responses among species are needed to better understand the patterns of transcriptional conservation over macro-evolutionary time scales.
Intriguingly, cold acclimation has also been demonstrated in green algae (Nagao et al., 2008), but the alga process does not respond to ABA as many land plants do, suggesting involvement of different pathways. Algae do contain AP2 domain encoding genes like the CBF transcription factors, but it is not clear if these transcription factors are involved in cold acclimation. More detailed studies in algae will enable us to understand if the basic molecular modules of plant cold acclimation evolved as early as in an aquatic land plant ancestor.
Endodormancy
Meristem dormancy is a common phenomenon in plants and has been linked to variation in a number of developmental genes (reviewed in Doust, 2007; Domagalska and Leyser, 2011). The most common type of meristem dormancy derives from the ability of the main axis to suppress the outgrowth of axillary meristems (apical dominance or paradormancy), and is a major determinant of architecture and growth habit across higher plants (Lang et al., 1987). By contrast, endodormancy is specific to temperate woody perennials, being shaped by both internal factors and seasonal fluctuations in both temperature and photoperiod (Chuine et al., 2001; reviewed in Campoy and Egea, 2011; Figures 2 and 3). Although closely linked to cold acclimation, endodormancy is a distinct physiological process that is sensitive to, but does not require, cold or other external factors to be induced (Faust et al., 1995). Endodormancy can be defined as dormancy under conditions that are conducive to growth (Dogramaci et al., 2011). By contrast, bud flush, which is induced after endodormancy is broken and mitotic division reinitiated, relies on a particular regime of cold followed by warm temperatures, the duration and timing of which is cultivar/species specific, and is often correlated with latitude (Saure, 1985; Erez and Couvillon, 1987; Naor et al., 2003; Campoy et al., 2011).
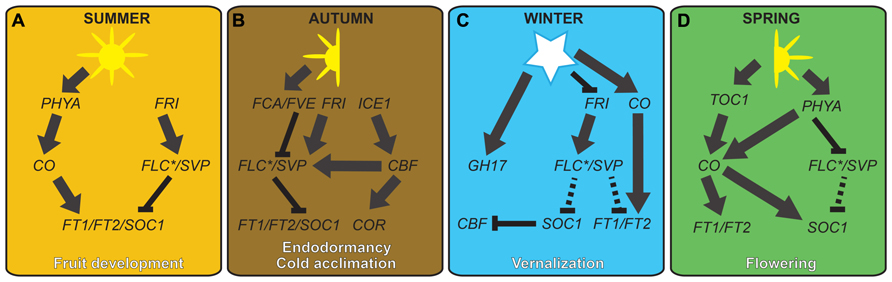
FIGURE 3. Hypothetical model showing possible genetic links between cold-regulated growth and reproduction in temperate plants. Expression levels of FT- and SOC1-like flowering integrator genes are maintained at moderate levels in summer by the antagonistic action of long day and warm temperature-regulated genes (A). During cooler temperatures of the autumn, the ICE-CBF-COR cold acclimation pathway is initiated, resulting in a high level of flowering repressors, such as FLC- and SVP-like genes. Although SVP-like gene transcription is dampened by the negative action of FCA- and FVE-like proteins, levels are high enough to work with FLC to repress FT-like and SOC1-like genes, resulting in endodormancy in woody perennials (B). Freezing winter temperatures negatively regulate FRI- and FLC-like genes, or functional equivalents, resulting in the derepression of FT- and SOC1-like genes, and the subsequent negative regulation of cold acclimation genes (C). However, despite up-regulation of FT-like genes in leaves, callus plugs in the shoot apices hinder the floral transition. During warm conditions of the spring, the combination of callus plug decay and long day regulation of FT-like genes results in bud flush and the induction of flowering (D). Unbroken and broken lines represent strong and weak interactions, respectively.
Despite inherent difficulties in working with woody species, research on both temperate gymnosperms and angiosperms, such as apple (M. domestica), apricot (Prunus armeniaca), poplar (Populus trichocarpa), hybrid aspen (Populus tremula × Populus tremuloides), and woody spurge (Euphorbia esula), have revealed many genes and gene networks underlying endodormancy and bud break (Dogramaci et al., 2011; Hsu et al., 2011; Karlberg et al., 2011; reviewed in Campoy and Egea, 2011; Figure 3). Available data suggest a complex interplay between the circadian clock and ABA (autonomous), gibberellic acid, photoperiod, and temperature pathways (reviewed in Campoy and Egea, 2011). In species such as poplar and grape (Vitis vinifera), photoperiod and temperature appear to be major determinants of the timing of bud set (defined as bud formation and growth cessation) and endodormancy (Howe et al., 2003; Rohde et al., 2011). By contrast, species such as Pinus contorta that can become endodormant as early as late summer use alternative cues such as drought and/or node number (Chuine et al., 2001; Howe et al., 2003). The endodormancy model presented below takes into account the former; future work is required to determine the genetic basis for summer onset endodormancy.
Genetic Model for Bud Set and Endodormancy
Although not all aspects of autumn endodormancy have been worked out, the most plausible genetic model is that bud set and endodormancy are controlled largely through the differential regulation of FLOWERING LOCUS T (FT) by photoperiod and/or temperature (Figure 3). According to research on poplar and A. thaliana, long summer days stabilize the CONSTANS (CO) protein through the action of the light-absorbing protein complex GIGANTIA (GI)/FLAVIN KELCH F BOX (FKF1; Sawa et al., 2007; Song et al., 2012). Late in the day, GI/FKF1 also acts to degrade CYCLING DOF FACTORs (CDFs), which are transcriptional repressors of CO, and CO protein levels are stabilized through the action of another light-absorbing protein PHYTOCHROME A (PHYA; Yanofsky and Kay, 2002; Fornara et al., 2009; Andres and Coupland, 2012). In turn CO up-regulates the major flowering pathway integrator FT, resulting in meristem outgrowth and the development of leaf and branch initials (bud set; Samach et al., 2000). It is postulated that FT-regulated summer growth is mediated by the action of AINTEGUMENTALIKE1 (AIL) genes that are the direct targets of FT. Unlike A. thaliana, AIL genes in poplar and hybrid aspen positively control growth regulators such as D-type cyclins, suggesting that these genes have been recruited to the endodormancy pathway (Karlberg et al., 2011).
In contrast to the summer growth model, the poplar endodormancy model posits that as temperatures become cooler with the onset of autumn, expression of circadian clock genes, such as TIMING OF CAB EXPRESSION1 (TOC1) and PSEUDO-RESPONSE REGULATORs (PRRs), is repressed so that CO expression is no longer induced (Mas et al., 2003; reviewed in Imaizumi, 2010; Figure 3). The ability of temperature to regulate circadian clock gene expression is still being investigated. However, evidence suggests that diurnal hot/cold cycles can replace photoperiod cycles for entrainment of the circadian clock (Salomé and McClung, 2005; Yamashino et al., 2008; Zuther et al., 2012). Furthermore, the reduced day length of autumn means that PHYA is no longer able to repress the expression of dark-regulated genes that reduce CO protein stability, such as CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1) and SUPPRESSOR OF PHYTOCHROME A 1 (SPA1; Jang et al., 2008; Zuo et al., 2011). In addition to CO-dependent reduction of FT levels in the autumn, it is speculated that poplar FT is further repressed by the action of the SHORT VEGETATIVE PHASE (SVP)-like genes DORMANCY ASSOCIATED MADS-BOX 5 (DAM5) and DAM6; reviewed in Campoy and Egea, 2011). Thus, since poplar FT potentially mediates signals from the leaves to shoot apical meristems, SVP-like mediated AIL gene repression might cause the autumnal cessation of bud growth, which marks the start of leaf fall and the beginning of cold acclimation (Karlberg et al., 2011; Figure 3). Further testing of the genetic interactions between these genes and their protein products will be required to validate this model.
Genetic Model for Endodormancy Release and Bud Flush
Endodormancy occurs either in the absence of cold or with intermittent cold, whereas release of endodormancy occurs in response to continuous above-freezing temperatures of the late autumn and winter (Vegis, 1964). As outlined above, levels of the growth activator/floral pathway integrator protein FT are reduced under short days from autumn onward due to low levels of CO (Yoo et al., 2007; Joon Seo et al., 2013). However, during chilling conditions of the late autumn and early winter, repression of the FT inhibitor SVP is also reduced, resulting in a potentially moderate increase of FT during winter (Lee et al., 2007; Figure 3). In addition to A. thaliana SVP, repression in response to prolonged cold was recently demonstrated for the SVP-like DAM6 genes in Japanese apricot (Prunus mume; Sasaki et al., 2011). Moreover, SVP-like genes have been implicated in induction of endodormancy in leafy spurge (Euphorbia esula; Horvath et al., 2008). Thus, temperature regulation of SVP-like genes could be a common mechanism for endodormancy release during the winter through the negative regulation of FT-like genes.
Rinne et al. (2011) recently suggested a potential complicating factor regarding the involvement of FT in endodormancy release in temperate and boreal trees. During cool autumnal temperatures, mobile signals are potentially blocked from entering hybrid aspen shoot apices due to the presence of callus plugs (Rinne et al., 2001). However, during winter freezing temperatures, callus plugs are gradually removed through the activation of gibberellic acid responsive genes that regulate GH17 proteins, the latter being associated with lipid bodies that help to breakdown callose (Rinne et al., 2011). Since FT in A. thaliana is a mobile protein that travels from leaves to shoot apices, the formation of callus plugs is one mechanism that might delay FT signaling in the shoot apex during autumn. An explicit test of this hypothesis will be required to determine whether FT is mobile in hybrid aspen and other temperate trees. Furthermore, if FT levels do gradually increase in the shoot apex during the winter it will be interesting to determine whether this influences the timing of endodormancy release, bud flush, or both.
Bud Flush
Endodormancy affects both vegetative and reproductive meristems through the negative regulation of FT. However, since FT is a positive regulator of inflorescence meristem genes, it has been unclear how vegetative meristems maintain their identity during bud flush. As a possible solution to this puzzle, Hsu et al. (2011) recently demonstrated that poplar contains two FT paralogs, FT1 and FT2, that are differentially expressed both spatially and temporally in response to temperature (both genes) and daylength (FT2; Figure 3). Whereas expression of FT1 peaks during winter in leaves, shoots, vegetative buds, and reproductive buds, FT2 expression is highest during spring, and is confined to leaves and reproductive buds (Hsu et al., 2011). The sequential action of FT1 and FT2 on axillary meristems in winter and spring, respectively, results in discrete zones of floral growth and vegetative growth along lateral shoots. A similar partitioning of FT-like gene function has been described for the vernalization response of sugar beet (Beta vulgaris; Pin et al., 2010; see later section).
Evolution of Endodormancy
Like tree habit, endodormancy has multiple independent evolutionary origins, being found in diverse lineages of both gymnosperms and angiosperms (Figure 1). Recent studies in poplar suggest that duplication and diversification of FT-like genes has been important for the periodic growth of vegetative and inflorescence structures along the shoot axis (Hsu et al., 2011). However, it is unclear whether diversification of FT-like genes contributed to the evolution of endodormancy per se. Intriguingly, functional analyses of the closest FT homologs in spruces (Picea sp.) and pines (Pinus sp.) suggest that the positive role of FT in flowering time evolved after the split of gymnosperms and angiosperms (Klintenäs et al., 2012). Constitutive expression of spruce FT-like genes in A. thaliana results in late flowering phenotypes, suggesting that the gymnosperm FT-like genes repress flowering similar to the A. thaliana FT paralog TFL1 (Klintenäs et al., 2012). In Norway spruce (Picea abies), PaFTL1 and PaFTL2 are up-regulated under short days and spring conditions, respectively (Gyllenstrand et al., 2007; Asante et al., 2011; Karlgren et al., 2011). Together these studies suggest independent recruitment of FT-like genes in angiosperm and gymnosperm bud set and bud burst. Similar studies in other plants will be required to determine the prevalence of FT-like gene involvement in the evolution of endodormancy. Furthermore, future research is needed to determine the nature of regulatory evolution in the FT gene family.
Vernalization Responsiveness
Vernalization is the process by which an extended period of cold makes plants competent to flower (Chouard, 1960). In other words, vernalization responsive individuals will flower earlier under inductive conditions (long days and warm temperatures) when those conditions are preceded by a prolonged exposure to cold. This allows plants to synchronize flowering with favorable conditions of the spring (Amasino, 2010). Unlike endodormancy, shoot apices of vernalized plants continue to undergo some level of mitotic division, so that vegetative growth is maintained. In addition, vernalization is distinct from seed stratification, the latter being the release of seed dormancy through chilling (reviewed in Finch-Savage and Leubner-Metzger, 2006).
Extensive variation for vernalization responsiveness is found within many lineages of angiosperms, and is associated with both latitudinal clines and temperature/precipitation variables (Briggs and Walters, 1997; Stinchcombe et al., 2004; Franks et al., 2007; Samis et al., 2008; Kim et al., 2009; Méndez-Vigo et al., 2011; Figure 1). Thus, vernalization responsiveness appears to have evolved multiple times in response to selection by cold seasonal climates, and is hypothesized to have allowed expansion of clades within temperate zones (Preston and Kellogg, 2008; Kim et al., 2009; Edwards and Smith, 2010). For example, at least 3 out of 17 rosid, 5 out of 16 asterid, 1 out of 9 early-diverging eudicot, and 4 out of 10 monocot orders contain species that respond to vernalization. This conservative estimate suggests that vernalization responsiveness is a relatively evolvable trait at higher taxonomic levels. Whether this can be explained by relatively simple changes to pre-existing pathways is discussed below.
Genetic Models of Vernalization Responsiveness
In A. thaliana, vernalization responsiveness is mediated through epigenetic silencing of the flowering repressor gene FLC, and possibly its five MADS AFFECTING FLOWERING (MAF) paralogs, by the Plant-HomeoDomain-Polycomb Repressive Complex 2- (PHD-PRC2) complex (Ratcliffe et al., 2003; Kim et al., 2009). The PHD-PRC2 complex initiates trimethylation of histone 2 lysine 27 (H3K27me3) and becomes progressively localized to the first intron of FLC during exposure to cold (Shindo et al., 2006; Angel et al., 2011; Strange et al., 2011). Recent evidence suggests that the mechanism for PHD-PRC2 recruitment to FLC is associated with the FLC locus itself (Heo and Sung, 2010). Two non-coding transcripts that initiate from the first intron [COOL ASSISTED INTRONIC NONCODING RNA (COLDAIR)] and 3′-UTR (COOLAIR) of FLC are up-regulated in response to cold, and negatively regulate the transcription of FLC through recruitment of PRC2 (Heo and Sung, 2010; reviewed in Ietswaart et al., 2012). Reduction of FLC transcription in response to vernalization results in the release of FT and SOC1 from negative regulation, permitting the shoot apex to respond to inductive flowering signals (Searle et al., 2006). Following cold treatment, warm temperatures and long days promote the expression of FT through the photoperiod, temperature-, and age-dependent pathways. This results in an FT-mediated morphological shift in the shoot apex from vegetative to inflorescence identity, via induction of MADS-box genes such as FRUITFULL (FUL) and APETALA1 (AP1), and the eventual production of flowers, fruits (siliques), and seeds (reviewed in Adrian et al., 2009; Amasino, 2010).
Several members of the temperate grass subfamily Pooideae also respond to vernalization. However, since the ancestor of grasses was likely tropical, vernalization responsiveness in pooids is inferred to have evolved independently from vernalization responsiveness in the Brassicaceae (Clayton and Renvoize, 1986; Davis and Soreng, 2008; Preston and Kellogg, 2008; Edwards and Smith, 2010). In the closely related crop species wheat and barley (Pooideae), differences in vernalization responsiveness are largely a result of variation at three major loci: VERNALIZATION1 (VRN1), VRN2, and VRN3 (reviewed in Trevaskis et al., 2007a; Distelfeld et al., 2009). However several other genes are implicated in the pathway (Greenup et al., 2011). VRN1 is homologous to the flower development genes AP1, CAULIFLOWER (CAL), and FUL in A. thaliana, and its expression is progressively induced during long durations of cold in response to vernalization-induced changes to chromatin at the VRN1 locus (Oliver et al., 2009; Alonso-Peral et al., 2011). In wheat and barley cultivars that respond to vernalization, VRN1 expression is repressed prior to winter by chromatin modifications mediated by proteins that interact with regulatory sites within the promoter or first intron. Simultaneously, the long day induction of VRN1 is repressed by the zinc-finger CO-like gene VRN2 (Hemming et al., 2009). Independent of VRN1 expression, another MADS-box genes, ODDSOC2, is repressed during exposure to cold, resulting in the loss of transcriptional inhibition of downstream flowering genes (Greenup et al., 2010). Expression of VRN1 is required for long-term repression of ODDSOC2 and VRN2 (Trevaskis et al., 2006; Hemming et al., 2008), ODDSOC2 negatively regulates the flower development gene FLOWERING PROMOTING FACTOR 1 (FPF1; Greenup et al., 2010), and VRN2 negatively regulates the temperate cereal FT ortholog VRN3 above a certain threshold (Yan et al., 2004, 2006).
Evolution of Vernalization Responsiveness
Vernalization responsiveness has evolved independently in several plant lineages, presumably in response to climate cooling events over the past 47.5 million years. Comparative genetic studies in a range of different angiosperm species suggest that vernalization responsiveness has evolved primarily through the neo-functionalization of ancient photoperiod pathway genes, including CO-like (i.e., VRN2), FT-like, FUL-like (i.e., grass VRN1), and SOC1-like (i.e., FLC), following their duplication (Figure 4). For example, phylogenetic analyses suggest that the FLC-like gene clade of A. thaliana is restricted to the Brassicaceae (Becker and Theissen, 2003; Ballerini and Kramer, 2011; Figure 4). Furthermore, although FLC/MAF-like genes are found in other core eudicots, gene expression tends to be positively rather than negatively regulated by cold. This is the case for Arabidopsis MAF5, the Texas bluebell (Eustoma grandiflorum, Gentianaceae) EgFLCL, and trifoliate orange (Poncirus trifoliate, Rutaceae) PtFLC (Zhang et al., 2009; Nakano et al., 2011). In A. thaliana, natural variation in vernalization responsiveness has been linked to variation in the promoter, first exon, and first intron of FLC, and within the positive regulator of FLC, FRIGIDA (FRI; Michaels and Amasino, 1999, 2001; El-Din El-Assal et al., 2001; Werner et al., 2005; Balasubramanian et al., 2006; Angel et al., 2011; Bond et al., 2011; Coustham et al., 2012; Wollenberg and Amasino, 2012). A similar role has been afforded to the FLC ortholog PERPETUAL FLOWERING 1 (PEP1) in the Brassicaceae species Arabis alpina. However, since Arabis alpina is a perennial species, cold-induced chromatin modification of PEP1 is only transient, being reset every growing season (Wang et al., 2009a).
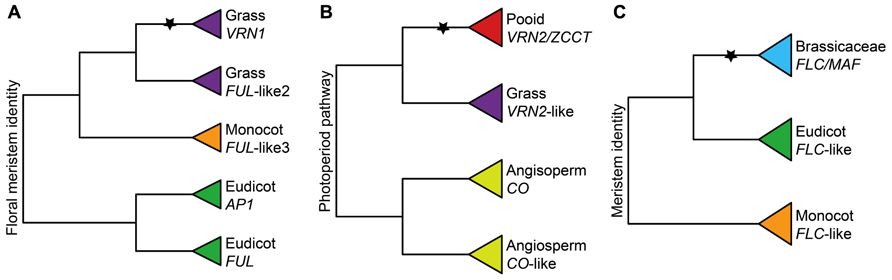
FIGURE 4. Importance of gene duplications for the functional evolution of vernalization responsive genes. (A) Simplified phylogeny of the APETALA1/FRUITFULL (AP1/FUL) gene family showing the grass-specific duplication that gave rise to VRN1 based on Litt and Irish (2003) and Preston and Kellogg (2006). (B) Simplified phylogeny of the CCT zinc-finger gene family showing the pooid-specific duplication that gave rise to ZCCT1 and ZCCT2 based on Yan et al. (2004). (C) Simplified phylogeny of the FLOWERING LOCUS C (FLC) gene family showing the Brassicaceae-specific duplication that gave rise to FLC and MAF genes based on Becker and Theissen (2003). Inferred ancestral functions are stated at the base of each tree. Stars denote important duplication events for the evolution of VR. Pooid clades are red, grass clades are purple, monocot clades are orange, Brassicaceae clades are blue, eudicot clades are green, and angiosperm clades are yellow.
In sugar beet (B. vulgaris, Amaranthaceae, Caryophyllales) recruitment of a lineage-specific FT-like gene duplication has been implicated in the independent origin of vernalization responsiveness (Pin et al., 2010; Mutasa-Gottgens et al., 2012). Under warm conditions, the FT homolog BvFT1 represses its paralog BvFT2, resulting in a block to flowering. By contrast, under cold conditions, BvFT1 is down-regulated, causing derepression of BvFT2 and promoting flowering (Pin et al., 2010). It was recently discovered that down-regulation of BvFT1 during winter is due to the repressive action of BOLTING TIME CONTROL 1 (BvBTC1), which is related to the circadian clock PRR genes in A. thaliana (Pin et al., 2012). Moreover, variation in the response of BvBTC1 alleles to vernalization has been linked to growth habit differences in domesticated sugar beet (Pin et al., 2012). Together with the fact that VRN1 and VRN2 are inferred to have evolved somewhere at the base of (Preston and Kellogg, 2006) or within (Yan et al., 2004) the Poaceae, respectively, these data suggest that lineage-specific flowering gene duplications have been important for independent origins of vernalization responsiveness, either through subtle switches from flowering inducers to repressors (e.g., VRN2, FLC, and BvFT1) or more dramatic changes in regulation and downstream targeting (e.g., VRN1 and BvBTC1; Figure 3).
Genetic Links between Seasonal Adaptations to Cold
Given the temporal overlap between endodormancy and cold acclimation, an interesting question is whether these two processes are linked at the genetic level. Presently, such links can be tentatively formed by combining data from cold acclimating but not endodormant A. thaliana, and endodormant trees. In the case of A. thaliana, CO is a regulator of both FT and SOC1. Similar to FT, SOC1 positively regulates the expression of meristem identity genes, such as FUL and LFY. However, SOC1 also negatively regulates the cold responsive CBF genes (Seo et al., 2009; Figure 3). Thus, although it needs to be explicitly tested in endodormant species, these data suggest a mechanism by which the break of endodormancy can influence the loss of cold acclimation.
Similar genetic associations can also be postulated for cold acclimation and vernalization responsiveness. Recent studies have shown that wheat VRN1 has CBF-binding sites in its promoter, and that VRN1 negatively regulates CBF genes (Alonso-Peral et al., 2011). This suggests a negative feedback loop between cold acclimation and vernalization. However, it remains to be tested whether CBF proteins actually bind to the VRN1 promoter, and if so, whether this interaction is positive or negative. Interestingly, in A. thaliana genetic evidence suggests that CBF proteins positively regulate FLC in the autumn, accentuating the repression of flowering over winter (Seo et al., 2009; Figures 2 and 3). If this connection also exists in pooid grasses we would predict that CBF proteins positively regulate repressors of flowering such as VRN2 and homologs of SVP. Consistent with this, it has been demonstrated that the barley SVP-like genes Barley MADS1 (BM1), BM10, and Vegetative to Reproductive Transition gene 2 (VRT2), and wheat VRT2 are up-regulated by cold (Trevaskis et al., 2007b; Sutton et al., 2009; but see Kane et al., 2005). Alternatively, CBF proteins could directly repress flowering by negatively regulating VRN1 or VRN3.
Conclusions and Future Prospects
Low to freezing temperatures are major determinants of latitudinal and altitudinal ranges of plants (Cavendar-Bares et al., 2005), and less than half of angiosperm plant families are distributed in regions with seasonally low temperatures (Ricklefs and Renner, 1994; Larcher, 2005). In the next 80 years it is predicted that global temperatures will increase by 1.1–6.4°C, and that there will be an increase in the frequency and/or severity of warm spells during winter months (USGCRP, 2009; National Research Council, 2010). Both these escalating intermittent temperature fluctuations and long-term climate changes have the potential to affect the phenology of cold temperate adapted species that rely on an extended period of cold for timely flowering (Loik et al., 2004; Offord, 2011). However, flowering time will ultimately result from the interaction of different genetic pathways in response to environmental factors such as photoperiod, cold, heat, water-stress, and developmental age (Cook et al., 2012). These interactions are only starting to be worked out.
Contrary to the hypothesis that cold-induced traits are hard to evolve, phylogenetic analyses in combination with past climate and trait data suggest multiple independent origins of cold acclimation, endodormancy, and vernalization responsiveness, at least in angiosperms. Furthermore, although often correlated, cold acclimation and endodormancy/vernalization responsiveness can be uncoupled at the physiological level, potentially allowing increased flexibility in species-specific responses to seasonal cold. Does this imply parallel evolution of the same ancestral genes and/or pre-adapted genetic pathways?
Available data support the hypothesis that cold-induced traits have evolved multiple times independently through the modification of the same genetic pathways. This suggests that these pathways are somewhat pre-adapted to providing avoidance of or tolerance to cold stress. However, the exact genes and proteins that have been recruited to cold adaptive traits differ from clade to clade. Many of the known key regulators of cold-induced physiological traits are members of large gene families that have broadly conserved roles in stress responses (e.g., dehydrin proteins) and/or developmental transitions (e.g., VRN1/FUL-like genes). In Pooideae, Brassicaceae, and poplar several genes involved in cold acclimation (e.g., CBF/DRE genes and LRR-containing genes), vernalization responsiveness (CO-like and FLC), and endodormancy (FT-like genes) have evolved from lineage-specific duplications (Becker and Theissen, 2003; Yan et al., 2004; Sandve et al., 2008; Sandve and Fjellheim, 2010; Figure 4). Loss of vernalization responsiveness has been documented for multiple cultivars of wheat and barley under artificial selection either through the loss of VRN2, or the loss of negative-cis-regulatory elements in VRN1 and VRN3 (Yan et al., 2003, 2004; Fu et al., 2005; Andersen et al., 2006; Szucs et al., 2007; Schwartz et al., 2010; Alonso-Peral et al., 2011). Whether these changes can happen rapidly enough in natural populations to combat human-induced climate change is a hot topic of debate.
Despite an unfolding picture at the broad phylogenetic scale that suggests multiple evolutionary origins of cold adaptive traits (Figure 1), relatively little physiological, developmental, or genomic data are available for understanding the evolutionary lability of seasonal cold adaptations at the family level and below. This is particularly true for species that might have retained cold adaptive traits following secondary shifts to the tropics, thus hampering our understanding of adaptation on relatively short timescales. Nonetheless, exciting recent and ongoing experimental and phylogenomic studies in both tropical and temperate taxa of the Brassicaceae (Brassicales), Poaceae (Poales), Pinaceae (Pinales), and Phrymaceae (Lamiales) are providing novel insights into the tempo, ancestral selection pressures, and potential constraints related to the evolution of cold acclimation/tolerance, endodormancy, and vernalization responsiveness (e.g., Lin et al., 2005; Preston and Kellogg, 2008; Sandve and Fjellheim, 2010; Méndez-Vigo et al., 2011; Sandve et al., 2011; Friedman and Willis, 2013; Humphreys and Linder, 2013). Successful studies will need to combine physiological observations with ancestral state reconstruction and genetic/genomic analyses to determine the direction of trait shifts, and any prerequisites for their evolution. Finally, population-level studies will continue to provide insight into evolutionary responses to more subtle (both temporal and quantitative) seasonal variation in temperature across species’ ranges.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Toby Kellogg and three anonymous reviewers for helpful suggestions on an earlier version of the manuscript. This work was funded by a United States Department of Agriculture (USDA) Hatch grant to Jill C. Preston.
References
Adhikari, K. N., Buirchell, B. J., and Sweetingham, M. W. (2012). Length of vernalization period affects flowering time in three lupin species. Plant Breed. 131, 631–636. doi: 10.1111/j.1439-0523.2012.01996.x
Adrian, J., Torti, S., and Turck, F. (2009). From decision to commitment: the molecular memory of flowering. Mol. Plant 2, 628–642. doi: 10.1093/mp/ssp031
Agarwal, P. K., and Jha, B. (2010). Transcription factors in plants and ABA dependent and independent abiotic stress signaling. Biol. Plant. 54, 201–212. doi: 10.1007/s10535-010-0038-7
Alessandro, M. S., Galmarini, C. R., Iorizzo, M., and Simon, P. W. (2013). Molecular mapping of vernalization requirement and fertility restoration genes in carrot. Theor. Appl. Genet. 126, 415–423. doi: 10.1007/s00122-012-1989-1
Alonso, A., Queiroz, C. S., and Magalhaes, A. C. (1997). Chilling stress leads to increased cell membrane rigidity in roots of coffee (Coffea arabica L.) seedlings. Biochim. Biophys. Acta 1323, 75–84. doi: 10.1016/S0005-2736(96)00177-0
Alonso-Peral, M., Oliver, S. N., Casao, M. C., Greenup, A. A., and Trevaskis, B. (2011). The promoter of the cereal VERNALIZATION1 gene is sufficient for transcriptional induction by prolonged cold. PLoS ONE 6:e29456. doi: 10.1371/journal.pone.0029456
Amasino, R. (2010). Arabidopsis: a rich harvest 10 years after completion of the genome sequence. Plant J. 61, 1001–1013. doi: 10.1111/j.1365-313X.2010.04148.x
Améglio, T., Cochard, H., and Ewers, F. W. (2001). Stem diameter variations and cold hardiness in walnut trees. J. Exp. Bot. 52, 2135–2142. doi: 10.1093/jexbot/52.364.2135
Andersen, J. R., Jensen, L. B., Asp, T., and Lubberstedt, T. (2006). Vernalization response in perennial ryegrass (Lolium perenne L.) and wheat (Triticum monococcum) VRN1 and rice (Oryza sativa) Hd1. Plant Mol. Biol. 60, 481–494. doi: 10.1007/s11103-005-4815-1
Andreini, L., Viti, R., Bartolini, S., Ruiz, D., Egea, J., and Campoy, J. A. (2012). The relationship between xylem differentiation and dormancy evolution in apricot flower buds (Prunus armeniaca L.): the influence of environmental conditions in two Mediterranean areas. Trees Struct. Funct. 26, 919–928. doi: 10.1007/s00468-011-0668-1
Andres, F., and Coupland, G. (2012). The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 13, 627–639. doi: 10.1038/nrg3291
Angel, A., Song, J., Dean, C., and Howard, M. (2011). A polycomb-based switch underlying quantitative epigenetic memory. Nature 476, 105–109. doi: 10.1038/nature10241
Aro, E. M., Suorsa, M., Rokka, A., Allahverdiyeva, Y., Paakkarinen, V., Saleem, A., et al. (2005). Dynamics of photosystem II: a proteomic approach to thylakoid protein complexes. J. Exp. Bot. 56, 347–356. doi: 10.1093/jxb/eri041
Aro, E. M., Virgin, I., and Andersson, B. (1993). Photoinhibition of photosystem II. Inactivation, protein damage, and turnover. Biochim. Biophys. Acta 1143, 113–134. doi: 10.1016/0005-2728(93)90134-2
Asante, D. K. A., Yakovlev, I. A., Fossdal, C. G., Holefors, A., Opseth, L., Olsen, J. E., et al. (2011). Gene expression changes during short day induced terminal bud formation in Norway spruce. Plant Cell Environ. 34, 332–346. doi: 10.1111/j.1365-3040.2010.02247.x
Badawi, M., Reddy, Y. V., Agharbaoui, Z., Tominaga, Y., Danyluk, J., Sarhan, F., et al. (2008). Structure and functional analysis of wheat ICE (inducer of CBF expression) Genes. Plant Cell Physiol. 49, 1237–1249. doi: 10.1093/pcp/pcn100
Balasubramanian, S., Sureshkumar, S., Agawal, M., Michael, T. P., Wessinger, C., Maloof, J. N., et al. (2006). The PHYTOCHROME C photoreceptor mediates natural variation in flowering and growth responsiveness in Arabidopsis thaliana. Nat. Genet. 38, 71–715. doi: 10.1038/ng1818
Ballerini, E. S., and Kramer, E. M. (2011). Environmental and molecular analysis of the floral transition in the lower eudicot Aquilegia formosa. Evodevo 2, 4. doi: 10.1186/2041-9139-2-4
Bascuñán-Godoy, L., Sanhueza, C., Cuba, M., Zuniga, G. E., Corcuera, L. J., and Bravo, L. A. (2012). Cold-acclimation limits low temperature induced photoinhibition by promoting a higher photochemical quantum yield and a more effective PSII restoration in darkness in the Antarctic rather than the Andean ecotype of Colobanthus quitensis Kunt Bartl (Caryophyllaceae). BMC Plant Biol. 12:114. doi: 10.1186/1471-2229-12-114
Becker, A., and Theissen, G. (2003). The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Mol. Phylogenet. Evol. 29, 464–489. doi: 10.1016/S1055-7903(03)00207-0
Bhyan, S. B., Minami, A., Kaneko, Y., Suzuki, S., Arakawa, K., Sakata, Y., et al. (2012). Cold acclimation in the moss Physcomitrella patens involves abscisic acid-dependent signaling. J. Plant Physiol. 169, 137–145. doi: 10.1016/j.jplph.2011.08.004
Biasi, L. A., de Carvalho, R. I. N., and Zannette, F. (2010). Dormancy dynamics of grape and kiwi tree buds in a region of low chilling occurrence. Rev. Bras. Frutic. 32, 1244–1249. doi: 10.1590/S0100- 29452010005000118
Bilavcik, A., Zamecnik, J., Grospietsch, M., Faltus, M., and Jadrna, P. (2012). Dormancy development during cold hardening of in vitro cultured Malus domestica Borkh. plants in relation to their frost resistance and cryotolerance. Trees Struct. Funct. 26, 1181–1192. doi: 10.1007/s00468-012-0694-7
Bond, D. M., Dennis, E. S., and Finnegan, E. J. (2011). The low temperature response pathways for cold acclimation and vernalization are independent. Plant Cell Environ. 34, 1737–1748. doi: 10.1111/j.1365-3040.2011.02370.x
Bouché, N., Scharlat, A., Snedden, W., Bouchez, D., and Fromm, H. (2002). A novel family of calmodulin-binding transcription activators in multicellular organisms. J. Biol. Chem. 277, 21851–21861. doi: 10.1074/jbc.M200268200
Bradshaw, A. D. (1972). Some evolutionary consequences of being a plant. Evol. Biol. 5, 25–47. doi: 10.1007/978-1-4757-0256-9_2
Briggs, D., and Walters, S. M. (1997). Plant Variation and Evolution. Cambridge: Cambridge University Press.
Byard, S., Wisniewski, M., Li, J., and Karlson, D. (2010). Interspecific analysis of xylem freezing responses in Acer and Betula. HortScience 45, 165–168.
Caffarra, A., Donnelly, A., Chuine, I., and Jones, M. B. (2011). Modelling the timing of Betula pubescens budburst. I. Temperature and photoperiod: a conceptual model. Clim. Res. 46, 147–157. doi: 10.3354/BR00980
Campbell, R. K., and Sugano, A. I. (1979). Genecology of bud-burst phenology in Douglas-fir: response to flushing temperature and chilling. Bot. Gaz. 140, 223–231. doi: 10.1086/337079
Campoy, J. A., and Egea, D. R. J. (2011). Dormancy in temperate fruit trees in a global warming context. Sci. Hortic. (Amsterdam) 130, 357–372. doi: 10.1016/j.scienta.2011.07.011
Campoy, J. A., Ruiz, D., Cook, N. G., Allderman, L., and Egea, J. (2011). High temperatures and time to budbreak in low chill apricot ‘Palsteyn’. Towards a better understanding of chill and heat requirements fulfillment. Sci. Hortic. (Amsterdam) 129, 649–655. doi: 10.1016/j.scienta.2011.05.008
Capicciotti, C. J., Leclere, M., Perras, F. A., Bryce, D. L., Paulin, H., Harden, J., et al. (2012). Potent inhibition of ice recrystallization by low molecular weight carbohydrate-based surfactants and hydrogelators. Chem. Sci. 3, 1408–1416. doi: 10.1039/c2sc00885h
Carvallo, M. A., Pino, M. T., Jeknic, Z., Zou, C., Doherty, C. J., Shiu, S. H., et al. (2011). A comparison of the low temperature transcriptomes and CBF regulons of three plant species that differ in freezing tolerance: Solanum commersonii, Solanum tuberosum, and Arabidopsis thaliana. J. Exp. Bot. 62, 3807–3819. doi: 10.1093/jxb/err066
Catalá, R., Medina, J., and Salina, J. (2011). Integration of low temperature and light signaling during cold acclimation response in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108, 16475–16480. doi: 10.1073/pnas.1107161108
Cave, R. L., Birch, C. J., Hammer, G. L., Erwin, G. E., and Johnston, M. E. (2011). Juvenility and flowering of Brunonia australis (Goodeniaceae) and Calandrinia sp. (Portulacaceae) in relation to vernalization and daylength. Ann. Bot. 108, 215–220. doi: 10.1093/aob/mBR116
Cavendar-Bares, J., Cortes, P., Rambal, S., Joffre, R., Miles, B., and Rocheteau, A. (2005). Summer and winter sensitivity of leaves and xylem to minimum freezing temperatures: a comparison of co-occurring Mediterranean oaks that differ in leaf lifespan. New Phytol. 168, 597–612. doi: 10.1111/j.1469-8137.2005.01555.x
Charrier, G., Bonhomme, M., Lacointe, A., and Ameglio, T. (2011). Are budburst dates, dormancy and cold acclimation in walnut trees (Juglans regia L.) under mainly genotypic or environmental control? Int. J. Biometeorol. 55, 763–774. doi: 10.1007/s00484-011-0470-1
Chen, T. H., and Gusta, L. V. (1983). Abscisic acid-induced freezing resistance in cultured plant cells. Plant Physiol. 73, 71–75. doi: 10.1104/pp.73.1.71
Chinnusamy, V., Ohta, M., Kanrar, S., Lee, B. H., Hong, X., Agarwal, M., et al. (2003). ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 17, 1043–1054. doi: 10.1101/gad.1077503
Chouard, P. (1960). Vernalization and its relations to dormancy. Annu. Rev. Plant Physiol. 11, 191–238. doi: 10.1146/annurev.pp.11.060160.001203
Chuine, I., Aitken, S. N., and Ying, C. C. (2001). Temperature thresholds of shoot elongation in provenances of Pinus contorta. Can. J. For. Res. 31, 1444–1455.doi: 10.1139/x01-072
Clayton, W. D., and Renvoize, S. A. (1986). Genera Graminum: Grasses of the World. Kew Bulletin Additional Series XIII. Kew: Royal Botanic Gardens.
Cook, B. I., Wolkovich, E. M., and Parmesan, C. (2012). Divergent responses to spring and winter warming drive community level flowering trends. Proc. Natl. Acad. Sci. U.S.A. 109, 9000–9005. doi: 10.1073/pnas.1118364109
Coustham, V., Li, P., Strange, A., Lister, C., Song, J., and Dean, C. (2012). Quantitative modulation of polycomb silencing underlies natural variation in vernalization. Science 337, 584–587. doi: 10.1126/science.1221881
Cuevas, J. C., Lopez-Cobollo, R., Alcazar, R., Zarza, X., Koncz, C., Altabella, T., et al. (2008). Putrescine is involved in Arabidopsis freezing tolerance and cold acclimation by regulating abscisic acid levels in response to low temperature. Plant Physiol. 148, 1094–1105. doi: 10.1104/pp.108.122945
Dallaire, S., Houde, M., Gagne, Y., Saini, H. S., Boileau, S., Chevrier, N., et al. (1994). ABA and low temperature induce freezing tolerance via distinct regulatory pathways in wheat. Plant Cell Physiol. 35, 1–9.
Dall’Osto, L., Caffarri, S., and Bassi, R. (2005). A mechanism of nonphotochemical energy dissipation, independent from PsbS, revealed by a conformational change in the antenna protein CP26. Plant Cell 17, 1217–1232. doi: 10.1105/ tpc.104.030601
Davis, J. I., and Soreng, R. J. (2008). A phylogenetic analysis of the grasses (Poaceae), with attention to subfamily Pooideae and structural features of the plastid and nuclear genomes, including an intron loss in GBSSI. Aliso 23, 335–348.
De la Rosa, R., Rallo, L., and Rapoport, H. F. (2000). Olive floral bud growth and starch content during winter rest and spring budbreak. HortScience 35, 1223–1227.
Dhillion, T., Pearce, S. P., Stockinger, E. J., Distelfeld, A., Li, C., Knox, A. K., et al. (2010). Regulation of freezing tolerance and flowering in temperate cereals: the VRN-1 connection. Plant Physiol. 153, 1846–1858. doi: 10.1104/pp.110.159079
Diaz-Riquelme, J., Grimplet, J., Martinez-Zapater, J. M., and Carmona, M. J. (2012). Transcriptome variation along bud development in grapevine (Vitis vinifera L.). BMC Plant Biol. 12:181. doi: 10.1186/1471-2229-12-181 doi: 10.1186/1471-2229-12-181
Distelfeld, A., Li, C., and Dubcovsky, J. (2009). Regulation of flowering in temperate cereals. Curr. Opin. Plant Biol. 12, 178–184. doi: 10.1016/j.pbi.2008.12.010
Dogramaci, M., Horvath, D. P., Christoffers, M. J., and Anderson, J. V. (2011). Dehydration and vernalization treatments identify overlapping molecular networks impacting endodormancy maintenance in leafy spurge crown buds. Funct. Integr. Genomics 11, 611–626. doi: 10.1007/s10142-011-0239-y
Doherty, C. J., Van Buskirk, H. A., Myers, S. J., and Thomashow, M. F. (2009). Roles for Arabidopsis CAMTA transcription factors in cold-regulated gene expression and freezing tolerance. Plant Cell 21, 972–984. doi: 10.1105/ tpc.108.063958
Domagalska, M. A., and Leyser, O. (2011). Signal integration in the control of shoot branching. Nat. Rev. Mol. Cell Biol. 12, 211–221. doi: 10.1038/nrm3088
Domínguez, T., Hernández, M. L., Pennycooke, J. C., Jiménez, P., Martínez-Rivas, J. M., Sanz, C., et al. (2010). Increasing omega-3 desaturase expression in tomato results in altered aroma profile and enhanced resistance to cold stress. Plant Physiol. 153, 655–665. doi: 10.1104/pp.110.154815
Dong, C. H., Hu, X., Tang, W., Zheng, X., Kim, Y. S., Lee, B. H., et al. (2006). A putative Arabidopsis nucleoporin, AtNUP160, is critical for RNA export and required for plant tolerance to cold stress. Mol. Cell. Biol. 26, 9533–9543. doi: 10.1128/MCB.01063-06
Donoghue, M. J. (2008). A phylogenetic perspective on the distribution of plant diversity. Proc. Natl. Acad. Sci. U.S.A. 105, 11549–11555. doi: 10.1073/pnas.0801962105
Doucet, C. J., Byass, L., Elias, L., Worrall, D., Smallwood, M., and Bowles, D. J. (2000). Distribution and characterization of recrystallization inhibitor activity in plant and lichen species from the UK and maritime Antarctic. Cryobiology 40, 218–227. doi: 10.1006/cryo.2000.2241
Doust, A. N. (2007). Grass architecture: genetic and environmental control of branching. Curr. Opin. Plant Biol. 10, 1–5. doi: 10.1016/j.pbi.2006.11.015
Edwards, E. J., and Smith, S. A. (2010). Phylogenetic analyses reveal the shady history of C4 grasses. Proc. Natl. Acad. Sci. U.S.A. 107, 2532–2537. doi: 10.1073/pnas.0909672107
El-Din El-Assal, S., Alonso-Blanco, C., Peeters, A. J., Raz, V., and Koorneef, M. (2001). A QTL for flowering time in Arabidopsis reveals a novel allele of CRY2. Nat. Genet. 29, 435–440. doi: 10.1038/ng767
Erez, A., and Couvillon, G. A. (1987). Characterization of the influence of moderate temperatures on rest completion in peach. J. Am. Soc. Hortic. Sci. 112, 677–680.
Fausey, B. A., and Cameron, A. C. (2007). Differing vernalization responses of Veronica spicata ‘Red Fox’ and Laurentia axillaris. J. Am. Soc. Hortic. Sci. 132, 751–757.
Faust, M., Liu, D., Wang, S. Y., and Stutte, G. W. (1995). Involvement of apical dormancy in winter dormancy of apple buds. Acta Hortic. 395, 47–56.
Feng, X. M., Zhao, Q., Zhao, L. L., Qiao, Y., Xie, X. B., Li, H. F., et al. (2012). The cold-induced basic helix-loop-helix transcription factor gene MdClbHLH1 encodes and ICE-like protein in apple. BMC Plant Biol. 12:22. doi: 10.1186/1471-2229-12-22
Finch-Savage, W. E., and Leubner-Metzger, G. (2006). Seed dormancy and the control of germination. New Phytol. 171, 501–523. doi: 10.1111/j.1469-8137.2006.01787.x
Fine, P. V. A., and Ree, R. H. (2006). Evidence for a time-integrated species-area effect on the latitudinal gradient in tree diversity. Am. Nat. 168, 796–804. doi: 10.1086/508635
Fornara, F., Panigrahi, K. C., Gissot, L., Sauerbrunn, N., Rühl, M., Jarillo, J. A., et al. (2009). Arabidopsis DOF transcription factors act redundantly with CONSTANS protein in photoperiodic flowering. Science 303, 1003–1006. doi: 10.1016/j.devcel.2009.06.015
Fowler, S., and Thomashow, M. F. (2002). Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 14, 1675–1690. doi: 10.1105/tpc.003483
Fowler, S. G., Cook, D., and Thomashow, M. F. (2005). Low temperature induction of Arabidopsis CBF1, 2, and 3 is gated by the circadian clock. Plant Physiol. 137, 961–968. doi: 10.1104/pp.104.058354
Franks, S. J., Sim, S., and Weis, A. E. (2007). Rapid evolution of flowering time by an annual plant in response to a climate fluctuation. Proc. Natl. Acad. Sci. U.S.A. 104, 1278–1282. doi: 10.1073/pnas.0608379104
Friedman, J., and Willis, J. H. (2013). Major QTLs for critical photoperiod and vernalization underlie extensive variation in flowering time in the Mimulus guttatus species complex. New Phytol. doi: 10.1111/nph.12260 [Epub ahead of print].
Fu, D., Szucs, P., Yan, L., Helguera, M., Skinner, J. S., von Zitzewitz, J., et al. (2005). Large deletions within the first intron in VRN-1 are associated with spring growth habit in barley and wheat. Mol. Genet. Genomics 273, 54–65. doi: 10.1007/s00438-004-1095-4
Fujikawa, S., Jitsuyama, Y., and Kuroda, K. (1999). Determination of the role of cold acclimation-induced diverse changes in plant cells from the viewpoint of avoidance of freezing injury. J. Plant Res. 112, 237–244. doi: 10.1007/PL00013880
Ghelardini, L., Santini, A., Black-Samuelsson, S., Myking, T., and Falusi, M. (2010). Bud dormancy release in elm (Ulmus spp.) clones – a case study of photoperiod and temperature responses. Tree Physiol. 30, 264–274. doi: 10.1093/treephys/tpp110
Gilmour, S. J., Hajela, R. K., and Thomashow, M. F. (1988). Cold acclimation in Arabidopsis thaliana. Plant Physiol. 87, 745–750. doi: 10.1104/pp.87.3.745
Gould, S. J., and Vrba, E. S. (1982). Exaptation: a missing term in the science of form. Paleobiology 8, 4–15.
Gray, G. R., Chauvin, L. P., Sarhan, F., and Huner, N. P. A. (1997). Cold acclimation and freezing tolerance (a complex interaction of light and temperature). Plant Physiol. 114, 467–474.
Greenup, A. G., Sasani, S., Oliver, S. N., Talbot, M. J., Dennis, E. S., Hemming, M. N., et al. (2010). ODDSOC2 is a MADS-box floral repressor that is down-regulated by vernalization in temperate cereals. Plant Physiol. 153, 1062–1073. doi: 10.1104/pp.109.152488
Greenup, A. G., Sasani, S., Oliver, S. N., Walford, S. A., Millar, A. A., and Trevaskis, B. (2011). Transcriptome analysis of the vernalization response in barley (Hordeum vulgare) seedlings. PLoS ONE 6:e17900. doi: 10.1371/journal.pone.0017900
Griffith, M., and Yaish, M. W. F. (2004). Antifreeze proteins in overwintering plants: a tale of two activities. Trends Plant Sci. 9, 399–405. doi: 10.1016/j.tplants.2004.06.007
Gusta, L., Trischuk, R., and Weiser, C. J. (2005). Plant cold acclimation: the role of abscisic acid. J. Plant Growth Regul. 24, 308–318. doi: 10.1007/s00344-005-0079-x
Guzy-Wrobelska, J., Filek, M., Kaliciak, A., Szarejko, I., Machackova, I., Krekule, J., et al. (2013). Vernalization and photoperiod-related changes in the DNA methylation state in winter and spring rapeseed. Acta Physiol. Plant. 35, 817–827. doi: 10.1007/s11738-012-1126-4
Gyllenstrand, N., Clapham, D., Kallman, T., and Lagercrantz, U. (2007). A Norway spruce FLOWERING LOCUS T homolog is implicated in control of growth rhythm in conifers. Plant Physiol. 144, 248–257. doi: 10.1104/pp.107.095802
Hammel, H. T. (1967). Freezing of xylem sap without cavitation. Plant Physiol. 42, 55–66. doi: 10.1104/pp.42.1.55
Hanin, M., Brini, F., Ebel, C., Toda, Y., Takeda, S., and Masmoudi, K. (2011). Plant dehydrins and stress tolerance: versatile proteins for complex mechanisms. Plant Signal. Behav. 6, 1503–1509. doi: 10.4161/psb.6.10.17088
Hemming, M. N., Fieg, S., Peacock, J., Dennis, E. A., and Trevaskis, B. (2009). Regions associated with repression of the barley (Hordeum vulgare) VERNALIZATION1 gene are not required for cold induction. Mol. Genet. Genomics 282, 107–117. doi: 10.1007/s00438-009-0449-3
Hemming, M. N., Peacock, W. J., Dennis, E. S., and Trevaskis, B. (2008). Low-temperature and daylength cue are integrated to regulated FLOWERING LOCUS T in barley. Plant Physiol. 147, 1–12. doi: 10.1104/pp.108.116418
Hemming, M. N., and Trevaskis, B. (2011). Make hay when the sun shines: the role of MADS-box genes in temperature-dependant seasonal flowering responses. Plant Sci. 180, 447–453. doi: 10.1016/j.plantsci.2010.12.001
Heo, J. B., and Sung, S. (2010). Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 331, 76–79. doi: 10.1126/science.1197349
Hetherington, S. E., He, J., and Smillie, R. M. (1989). Photoinhibition at low temperature in chilling-sensitive and -resistant plants. Plant Physiol. 90, 1609–1615. doi: 10.1104/pp.90.4.1609
Hisano, H., Kanazawa, A., Kawakami, A., Yoshida, M., Shimamoto, Y., and Yamada, T. (2004). Transgenic perennial ryegrass plants expressing wheat fructosyltransferase genes accumulate increased amounts of fructan and acquire increased tolerance on a cellular level to freezing. Plant Sci. 167, 861–868. doi: 10.1016/j.plantsci.2004.05.037
Horvath, D. P., Chao, W. S., Suttle, J. C., Thimmapuram, J., and Anderson, J. V. (2008). Transcriptome analysis identifies novel responses and potential regulatory genes involved in seasonal dormancy transitions in leafy spurge (Euphorbia esula L.). BMC Genomics 9:536. doi: 10.1186/1471-2164-9-536
Hoth, S., Morgante, M., Sanchez, J. P., Hanafey, M. K., Tingey, S. V., and Chua, N. H. (2002). Genome-wide gene expression profiling in Arabidopsis thaliana reveals new targets of abscisic acid and largely impaired gene regulation in the abi1-1 mutant. J. Cell Sci. 115, 4891–4900. doi: 10.1242/jcs.00175
Howe, G. T., Aitken, S. N., Neale, D. B., Jermstad, K. D., Wheeler, N. C., and Chen, T. H. H. (2003). From genotype to phenotype: unraveling the complexities of cold adaptation in forest trees. Can. J. Bot. 81, 1247–1266. doi: 10.1139/b03-141
Hsu, C.-Y., Adams, J. P., Kim, H., No, K., Ma, C., Strauss, S. H., et al. (2011). FLOWERING LOCUS T duplication coordinates reproductive and vegetative growth in perennial poplar. Proc. Natl. Acad. Sci. U.S.A. 108, 10756–10771. doi: 10.1073/pnas.1104713108
Humphreys, A. M., and Linder, H. P. (2013). Evidence for recent evolution of cold tolerance in grasses suggests current distribution is not limited by (low) temperature. New Phytol. 198, 1261–1273. doi: 10.1111/ nph.12244.
Huner, N., Oguist, G., Hurry, V., Krol, M., Falk, S., and Griffith, M. (1993). Photosynthesis, photoinhibition and low temperature acclimation in cold tolerant plants. Photosynth. Res. 37, 19–39. doi: 10.1007/ BF02185436
Ietswaart, R., Wu, Z., and Dean, C. (2012). Flowering time control: another window to the connection between antisense RNA and chromatin. Trends Genet. 28, 445–453. doi: 10.1016/j.tig.2012.06.002
Imaizumi, T. (2010). Arabidopsis circadian clock and photoperiodism: time to think about location. Curr. Opin. Plant Biol. 13, 83–89. doi: 10.1016/j.pbi.2009.09.007
Ivany, L. C., Patterson, W. P., and Lohmann, K. C. (2000). Cooler winters as a possible cause of mass extinctions at the Eocene/Oligocene boundary. Nature 407, 887–890. doi: 10.1038/35038044
Jang, S., Marchal, V., Panigrahi, K. C., Wenkel, S., Soppe, W., Deng, X. W., et al. (2008). Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiod flowering response. EMBO J. 27, 1277–1288. doi: 10.1038/emboj.2008.68
Janská, A., Aprile, A., Zámečnik, J., Cattivelli, L., and Ovesná, J. (2011). Transcriptional responses of winter barley to cold indicate nucleosome remodelling as a specific feature of crown tissues. Funct. Integr. Genomics 11, 307–325. doi: 10.1007/s10142-011-0213-8
Jones, H. G., Hillis, R. M., Gordon, S. L., and Brennan, R. M. (2013). An approach to the determination of winter chill requirements for different Ribes cultivars. Plant Biol. 15, 18–27. doi: 10.1111/j.1438-8677.2012.00590.x
Joon Seo, P., Jung, J. H., Park, M. J., Lee, K., and Park, C. M. (2013). Controlled turnover of CONSTANS protein by the HOS1 E3 ligase regulates floral transition at low temperatures. Plant Signal. Behav. 8, e23780. doi: 10.4161/psb.23780
Kalberer, S. R., Arora, R., Leyva-Estrada, N., and Krebs, S. L. (2007). Cold hardiness of floral buds of deciduous azaleas: dehardening, rehardening, and endodormancy in late winter. J. Am. Soc. Hortic. Sci. 132, 73–79.
Kane, N. A., Danyluk, J., Tardif, G., Ouellet, F., Laliberte, J.-F., Limin, A. E., et al. (2005). TaVRT-2, a member of the StMADS-11 clade of flowering repressors, is regulated by vernalization and photoperiod in wheat. Plant Physiol. 138, 2354–2363. doi: 10.1104/pp.105.061762
Karlberg, A., Bako, L., and Bhalerao, R. P. (2011). Short day-mediated cessation of growth requires the downregulation of AINTEGUMENTALIKE1 transcription factor in hybrid aspen. PLoS Genet. 7:e1002361. doi: 10.1371/journal.pgen.1002361
Karlgren, A., Gyllenstrand, N., Kallman, T., Sundstrom, J. F., Moore, D., Lascoux, M., et al. (2011). Evolution of the PEBP gene family in plants: functional diversification of seed plant evolution. Plant Physiol. 156, 1967–1977. doi: 10.1104/pp.111.176206
Karlson, D. T., Xiang, Q. Y., Stirm, V. E., Shirazi, A. M., and Ashworth, E. N. (2004). Phylogenetic analyses in cornus substantiate ancestry of xylem supercooling freezing behavior and reveal lineage of desiccation related proteins. Plant Physiol. 135, 1654–1665. doi: 10.1104/pp.103.037473
Kawamata, M., Nishida, E., Ohara, H., Ohkawa, K., and Matsui, H. (2002). Changes in the intensity of bud dormancy and internal compositions of current shoot in fig. J. Jpn. Soc. Hortic. Sci. 71, 177–182. doi: 10.2503/jjshs.71.177
Kaymak, H. C., and Guvenc, I. (2010). The influence of vernalization time and day length on flower induction of radish (Raphanus sativus L.) under controlled and field conditions. Turk. J. Agric. For. 34, 401–413. doi: 10.3906/tar-0901-14
Khodakovskaya, M., McAvoy, R., Peters, J., Wu, H., and Li, Y. (2006). Enhanced cold tolerance in transgenic tobacco expressing a chloroplast ω-3 fatty acid desaturase gene under the control of a cold-inducible promoter. Planta 223, 1090–1100. doi: 10.1007/s00425-005-0161-4
Kim, D.-H., Doyle, M. R., Sung, S., and Amasino, R. M. (2009). Vernalization: winter and the timing of flowering in plants. Annu. Rev. Cell Dev. Biol. 25, 277–299. doi: 10.1146/annurev.cellbio. 042308.113411
King, R. W., and Heide, O. M. (2009). Seasonal flowering and evolution: the heritage from Charles Darwin. Funct. Plant Biol. 36, 1027–1036. doi: 10.1071/FP09170
Klintenäs, M., Pin, P. A., Benlloch, R., Ingvarsson, P. K., and Nilsson, O. (2012). Analysis of conifer FLOWERING LOCUS T/TERMINAL-FLOWER1-like genes provides evidence for dramatic biochemical evolution in the angiosperm FT lineage. New Phytol. 196, 1260–1273. doi: 10.1111/j.1469-8137.2012.04332.x
Knight, H., Mugford, S. G., Ulker, B., Gao, D., Thorlby, G., and Knight, M. R. (2009). Identification of SFR6, a key component in cold acclimation acting post-translationally on CBF function. Plant J. 58, 97–108. doi: 10.1111/j.1365-313X. 2008.03763.x
Knight, H., Zarka, D. G., Okamoto, H., Thomashow, M. F., and Knight, M. R. (2004). Abscisic acid induces CBF gene transcription and subsequent induction of cold-regulated genes via the CRT promoter element. Plant Physiol. 135, 1710–1707. doi: 10.1104/pp.104.043562
Kobayashi, F., Takumi, S., and Handa, H. (2008). “Characterization of ABA sensitivity in mutants with altered abiotic stress tolerance in common wheat,” in Proceedings of the 11th International Wheat Genetics Symposium, Vol. 3 (Sydney: Sydney University Press), 907–909.
Krause, G. H. (1988). Photoinhibition of photosynthesis. an evaluation of damaging and protective mechanisms. Physiol. Plant. 74, 566–574. doi: 10.1111/j.1399-3054.1988.tb02020.x
Krug, H. (1991). Investigations of changes in growth-activity of rhubarb (Rheum-rhaponticum × rhabarbarum) and consequences for production and forcing. Gartenbauwissenschaft 56, 93–98.
Kubota, S., Momose, H., Yoneda, K., and Koshioka, M. (2010). Lavandula × intermedia is a vernalization type plant. Jpn. Agric. Res. Q. 44, 67–72. doi: 10.6090/jarq.44.67
Lang, G. A., Early, J. D., Martin, G. C., and Darnell, R. L. (1987). Endodormancy, paradormancy, and ecodormancy – physiological terminology and classification for dormancy research. HortScience 22, 371–377.
Larcher, W. (2005). Climatic constraints drive the evolution of low temperature resistance in woody plants. J. Agric. Meterol. 61, 189–202. doi: 10.2480/agrmet.61.189
Latham, R. E., and Ricklefs, R. E. (1993). Global patterns of tree species richness in moist forests: energy-diversity theory does not account for variation in species richness. Oikos 67, 325–333. doi: 10.2307/3545479
Lee, B.-H., Henderson, D. A., and Zhu, J.-K. (2005). The Arabidopsis cold-responsive transcriptome and its regulation by ICE1. Plant Cell 17, 3155–3175. doi: 10.1105/tpc.105.035568
Lee, B.-H., Lee, H., Xiong, L., and Zhu, J.-K. (2002). A mitochondrial complex I defect impairs cold-regulated nuclear gene expression. Plant Cell 14, 1235–1251. doi: 10.1105/tpc.010433
Lee, C. M., and Thomashow, M. F. (2012). Photoperiodic regulation of the C-repeat binding factor (CBF) cold acclimation pathway and freezing tolerance in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 109, 15054–15059. doi: 10.1073/pnas.1211295109
Lee, J. H., Yoo, S. J., Park, S. H., Hwang, I., Lee, J. S., and Ahn, J. H. (2007). Role of SVP in the control of flowering time by ambient temperatures in Arabidopsis. Genes Dev. 21, 397–402. doi: 10.1101/gad.1518407
Lenahan, O. M., Graves, W. R., and Arora, R. (2010). Cold-hardiness and deacclimation of Styrax americanus from three provenances. HortScience 45, 1819–1823.
Lewandowska-Sabat, A. M., Winge, P., Fjellheim, S., Dorum, G., Bones, A. M., and Rognli, O. A. (2012). Genome wide transcriptional profiling of acclimation to photoperiod in high-latitude accessions of Arabidopsis thaliana. Plant Sci. 186, 143–155. doi: 10.1016/j.plantsci.2011.10.009
Li, W., Li, M., Zhang, W., Welti, R., and Wang, X. (2004). The plasma membrane-bound phospholipase D[delta] enhances freezing tolerance in Arabidopsis thaliana. Nat. Biotechnol. 22, 427–433. doi: 10.1038/nbt949
Lin, M., Starman, T. W., Wang, Y. T., Niu, G. H., and Cothren, J. T. (2011). Deferring flowering of nobile dendrobium hybrids by holding plants under low temperature after vernalization. Sci. Hortic. 130, 869–873. doi: 10.1016/j.scienta.2011.08.027
Lin, S.-I., Wang, J.-G., Poon, S.-Y., Su, C.-L., Wang, S.-S., and Chiou, T.-J. (2005). Differential regulation of FLOWERING LOCUS C expression by vernalization in cabbage and Arabidopsis. Plant Physiol. 137, 1037–1048. doi: 10.1104/pp. 104.058974
Litt, A., and Irish, V. F. (2003). Duplication and diversification in the APETALA1/FRUITFULL floral homeotic gene lineage: implications for evolution of floral development. Genetics 165, 821–833.
Liu, Q., Kasuga, M., Sakuma, Y., Abe, H., Miura, S., Yamaguchi-Shinozaki, K., et al. (1998). Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10, 1391–1406.
Llorente, F., Oliveros, J. C., Martinez-Zapater, J. M., and Salinas, J. (2000). A freezing-sensitive mutant of Arabidopsis, frs1, is a new aba3 allele. Planta 211, 648–655. doi: 10.1007/s004250000340
Loik, M. E., Still, C. J., Huxman, T. E., and Harte, J. (2004). In situ photosynthetic freezing tolerance for plants exposed to a global warming manipulation in the Rocky Mountains, Colorado, USA. New Phytol. 162, 331–341. doi: 10.1111/j.1469-8137.2004.01002.x
Lopez, R. G., and Runkle, E. S. (2006). Temperature and photoperiod regulate flowering of potted Miltoniopsis orchids. Hortic. Sci. 41, 593–597.
Mantyla, E., Lang, V., and Palva, E. T. (1995). Role of abscisic acid in drought-induced freezing tolerance, cold acclimation, and accumulation of LT178 and RAB18 proteins in Arabidopsis thaliana. Plant Physiol. 107, 141–148.
Maruyama, K., Takeda, M., Kidokoro, S., Yamada, K., Sakuma, Y., Urano, K., et al. (2009). Metabolic pathways involved in cold acclimation identified by integrated analysis of metabolites and transcripts regulated by DREB1A and DREB2A. Plant Physiol. 150, 1972–1980. doi: 10.1104/pp.109.135327
Mas, P., Kim, W. Y., Somers, D. E., and Kay, S. A. (2003). Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature 426, 567–570. doi: 10.1038/nature02163
Meideros, J. A. S., and Pockman, W. T. (2011). Drought increases freezing tolerance of both leaves and xylem of Larrea tridentata. Plant Cell Environ. 34, 43–51. doi: 10.1111/j.1365-3040.2010.02224.x
Melis, A. (1999). Photosystem II damage and repair cycle in chloroplasts: what modulates the rate of photodamage in vivo? Trends Plant Sci. 4, 130–135. doi: 10.1016/S1360-1385(99)01387-4
Méndez-Vigo, B., Picó, F. X., Ramiro, M., Martínez-Zapater, J. M., and Alonso-Blanco, C. (2011). Altitudinal and climatic adaptation is mediated by flowering traits and FRI, FLC, and PHYC genes in Arabidopsis. Plant Physiol. 157, 1942–1955. doi: 10.1104/pp.111.183426
Mewes, S., and Pank, F. (2007). Influence of cold temperature and light on the flower induction of thyme (Thymus vulgaris L.). Eur. J. Hortic. Sci. 72, 80–84.
Meyer, K., Keil, M., and Naldrett, M. J. (1999). A leucine-rich repeat protein of carrot that exhibits antifreeze activity. FEBS Lett. 447, 171–178. doi: 10.1016/S0014- 5793(99)00280-X
Michaels, S. D., and Amasino, R. M. (1999). FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11, 949–956. doi: 10.1105/tpc.11.5.949
Michaels, S. D., and Amasino, R. (2001). Loss of FLOWERING LOCUS C activity eliminates the late-flowering phenotype of FRIGIDA and autonomous-pathway mutations, but not responsiveness to vernalization. Plant Cell 13, 935–941. doi: 10.1105/tpc.13.4.935
Moellering, E. R., Muthan, B., and Benning, C. (2010). Freezing tolerance in plants requires lipid remodeling at the outer chloroplast membrane. Science 330, 226–228. doi: 10.1126/science.1191803
Mojtahedi, N., Masuda, J., Hiramatstu, M., Nguyen, T. L. H., and Okubo, H. (2013). Role of temperature in dormancy induction and release in one-year-old seedlings of Lilium longiflorum populations. J. Jpn. Soc. Hortic. Sci. 82, 63–68. doi: 10.2503/jjshs1.82.63
Mutasa-Gottgens, E. S., Joshi, A., Holmes, H. F., Hedden, P., and Gottgens, B. (2012). A new RNAseq-based reference transcriptome for sugar beet and its application in transcriptome-scale analysis of vernalization and gibberellin responses. BMC Genomics 13:99. doi: 10.1186/1471-2164-13-99
Nagao, M., Matsui, K., and Uemura, M. (2008). Klebsormidium flaccidum, a charophycean green alga, exhibits cold acclimation that is closely associated with compatible solute accumulation and ultrastructural changes. Plant Cell Environ. 31, 872–885. doi: 10.1111/j.1365-3040. 2008.01804.x
Nakano, Y., Kawashima, H., Kinoshita, T., Yoshikawa, H., and Hisamatsu, T. (2011). Characterization of FLC, SOC1 and FT homologs in Eustoma grandiflorum: effects of vernalization and post-vernalization conditions on flowering and gene expression. Physiol. Plant. 141, 383–393. doi: 10.1111/j.1399-3054.2011.01447.x
Naor, A., Flaishman, M., Stern, R., Moshe, A., and Erez, A. (2003). Temperature effects on dormancy completion of vegetative buds in apple. J Am. Soc. Hortic. Sci. 128, 636–641.
National Research Council. (2010). Advancing the Science of Climate Change. Washington, DC: The National Academies Press.
Nishitani, C., Saito, T., Ubi, B. E., Moriguchi, T., Saito, T., Yamamoto, T., et al. (2012). Transcriptome analysis of Pyrus pyrifolia leaf buds during transition from endodormancy to ecodormancy. Sci. Hortic. 147, 49–55. doi: 10.1016/j.scienta.2012.09.001
Novillo, F., Alonso, J. M., Ecker, J. R., and Salinas, J. (2004). CBF2/DREB1C is a negative regulator of CBF1/DREB1B and CBF3/DREB1A expression and plays a central role in stress tolerance in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 101, 3985–3990. doi: 10.1073/pnas.0303029101
Offord, C. A. (2011). Pushed to the limit: consequences of climate change for the Araucariaceae: a relictual rain forest family. Ann. Bot. 108, 347–357. doi: 10.1093/aob/mBR135
Oliver, S. N., Finnegan, E. J., Dennis, E. S., Peacock, W. J., and Trevaskis, B. (2009). Vernalization-induced flowering in cereals is associated with changes in histone methylation at the VERNALIZATION1 gene. Proc. Natl. Acad. Sci. U.S.A. 106, 8336–8391. doi: 10.1073/pnas.0903566106
Örvar, B. L., Sangwan, V., Omann, F., and Dhindsa, R. S. (2000). Early steps in cold sensing by plant cells: the role of actin cytoskeleton and membrane fluidity. Plant J. 23, 785–794. doi: 10.1046/j.1365-313x.2000.00845.x
Padhye, S. R., and Cameron, A. C. (2008). Dianthus gratianopolitanus Vill. ‘Bath’s Pink’ has a near-obligate vernalization requirement. HortScience 43, 346–349.
Padhye, S. R., and Cameron, A. C. (2009). Vernalization responses of campanula ‘Birch Hybrid’. J. Am. Soc. Hortic. Sci. 134, 497–504.
Pandey, N., Ranjan, A., Pant, P., Tripathi, R. K., Ateek, F., Pandley, H. P., et al. (2013). CAMTA 1 regulates drought responses in Arabidopsis thaliana. BMC Genomics 14, 216. doi: 10.1186/1471-2164-14-216
Pennycooke, J., Cheng, H., Roberts, S., Yang, Q., Rhee, S., and Stockinger, E. J. (2008). The low temperature-responsive, Solanum CBF1 genes maintain high identity in their upstream regions in a genomic environment undergoing gene duplications, deletions, and rearrangements. Plant Mol. Biol. 67, 483–497. doi: 10.1007/s11103-008-9333-5
Pietsch, G. M., Anderson, N. O., and Li, P. H. (2009). Cold tolerance and short day acclimation in perennial Gaura coccinea and G. Drummondii. Sci. Hortic. 120, 418–425. doi: 10.1016/j.scienta.2008.11.030
Pin, P. A., Benlloch, R., Bonnet, D., Wremerth-Weich, E., Kraft, T., Gielen, J. J. L., et al. (2010). An antagonistic pair of FT homologs mediates the control of flowering in sugar beet. Science 330, 1397–1400. doi: 10.1126/science.1197004
Pin, P. A., Zhang, W., Vogt, S. H., Dally, N., Buttner, B., Schulze-Buxloh, G., et al. (2012). The role of a pseudo-response regulator gene in life cycle adaptation and domestication of beet. Curr. Biol. 22, 1095–1101. doi: 10.1016/j.cub.2012.04.007
Prasil, I. T., Prasilova, P., and Pankova, K. (2004). Relationship among vernalization, shoot apex development and frost tolerance in wheat. Ann. Bot. 94, 413–418. doi: 10.1093/aob/mch158
Preston, J. C., and Kellogg, E. A. (2006). Reconstructing the evolutionary history of paralogous APETALA1/FRUITFULL-like genes in grasses (Poaceae). Genetics 174, 421–437. doi: 10.1534/genetics.106.057125
Preston, J. C., and Kellogg, E. A. (2008). Discrete developmental roles for temperate cereal grass VERNALIZATION 1/FRUITFUL-like genes in flowering competency and the transition to flowering. Plant Physiol. 146, 265–276. doi: 10.1104/pp.107.109561
Rantasen, M., and Palonen, P. (2010). Hot water treatment released endodormancy but reduced number of flowers in potted red raspberry plants. HortScience 45, 894–898.
Rapacz, M., Gasior, D., Zwierzyowski, Z., Lesniewska-Bocianowska, A., Humphreys, M. W., and Gay, A. P. (2004). Changes in cold tolerance and the mechanisms of acclimation of photosystem II to cold hardening generated by anther culture of Festuca pratensis × Lolium multiflorum cultivars. New Phytol. 162, 105–114. doi: 10.1111/j.1469-8137.2004.01024.x
Rapacz, M., and Markowski, A. (1999). Winter hardiness, frost resistance and vernalization requirement of European winter oilseed rape (Brassica napus var. oleifera) cultivars within the last 20 years. J. Agron. Crop Sci. 183, 243–253. doi: 10.1046/j.1439-037x.1999.00346.x
Ratcliffe, O. J., Kumimoto, R. W., Wong, B. J., and Riechmann, J. L. (2003). Analysis of the Arabidopsis MADS AFFECTING FLOWERING gene family: MAF2 prevents vernalization by short periods of cold. Plant Cell 15, 1159–1169. doi: 10.1105/tpc.009506
Ricklefs, R. E. (2005). Historical and ecological dimensions of global patterns in plant diversity. Biol. Skr. 55, 583–603.
Ricklefs, R. E., and Renner, S. S. (1994). Species richness within families of flowering plants. Evolution 48, 1619–1636. doi: 10.2307/2410252
Rinne, P. L. H., Kaikuranta, P. M., and van der Schoot, C. (2001). The shoot apical meristem restores its symplasmic organization during chilling-induced release from dormancy. Plant J. 26, 249–264. doi: 10.1046/j.1365-313X.2001.01022.x
Rinne, P. L. H., Welling, A., Vahala, J., Ripel, L., Ruonala, R., Kangasjarvi, J., et al. (2011). Chilling of dormant buds hyperinduces FLOWERING LOCUS T and recruits GA-inducible 1,3-beta-glucanases to reopen signal conduits and release dormancy in Populus. Plant Cell 23, 130–146. doi: 10.1105/tpc.110.081307
Rohde, A., and Bhalerao, R. P. (2007). Plant dormancy in the perennial context. Trends Plant Sci. 12, 217–223. doi: 10.1016/j.tplants.2007.03.012
Rohde, A., Storme, V., Jorge, V., Gaudet, M., Vitacolonna, N., Fabbrini, F., et al. (2011). Bud set in poplar – genetic dissection of a complex trait in natural and hybrid populations. New Phytol. 189, 106–121. doi: 10.1111/j.1469-8137.2010.03469.x
Rohwer, C. L., and Heins, R. A. (2007). Daily light integral, prevernalization photoperiod, and vernalization temperature and duration control flowering of easter cactus. HortScience 42, 1596–1604.
Rudi, H., Sandve, S. R., Opseth, L., and Rognli, O. A. (2010). Identification of candidate genes important for frost tolerance in Festuca pratensis Huds. by transcriptional profiling. Plant Sci. 180, 78–85. doi: 10.1016/j.plantsci.2010.07.014
Salomé, P. A., and McClung, C. R. (2005). PSEUDO-RESPONSE REGULATOR 7 and 9 are partially redundant genes essential for the temperature responsiveness of the Arabidopsis circadian clock. Plant Cell 17, 791–803. doi: 10.1105/tpc.104.029504
Samach, A., Onouchi, H., Gold, S. E., Ditta, G. S., Schwarz-Sommer, Z., Yanofsky, M. F., et al. (2000). Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288, 1613–1616. doi: 10.1126/science.288.5471.1613
Samis, K. E., Heath, K. D., and Stinchcombe, J. R. (2008). Discordant longitudinal clines in flowering time and phytochrome C in Arabidopsis thaliana. Evolution 62, 2971–2983. doi: 10.1111/j.1558-5646.2008.00484.x
Sanchez-Perez, R., Dicenta, F., and Martinez-Gomez, P. (2012). Inheritance of chilling and heat requirements for flowering in almond and QTL analysis. Trees Genet. Genom. 8, 379–389. doi: 10.1007/s11295-011-0448-5
Sandve, S. R., and Fjellheim, S. (2010). Did gene family expansions during the Eocene–Oligocene boundary climate cooling play a role in Pooideae adaptation to cool climates? Mol. Ecol. 19, 2075–2088. doi: 10.1111/j.1365-294X.2010.04629.x
Sandve, S. R., Kosmala, A., Rudi, H., Fjellheim, S., Rapacz, M., Yamada, T., et al. (2011). Molecular mechanisms underlying frost tolerance in perennial grasses adapted to cold climates. Plant Sci. 180, 69–77. doi: 10.1016/j.plantsci.2010.07.011
Sandve, S. R., Rudi, H., Asp, T., and Rognli, O. A. (2008). Tracking the evolution of a cold stress associated gene family in cold tolerant grasses. BMC Evol. Biol. 8:245. doi: 10.1186/1471-2148-8-245
Sangwan, V., Foulds, I., Singh, J., and Dhindsa, R. S. (2001). Cold-activation of Brassica napus BN115 promoter is mediated by structural changes in membranes and cytoskeleton, and requires Ca2+ influx. Plant J. 27, 1–12. doi: 10.1046/j.1365-313x.2001.01052.x
Sasaki, R., Yamane, H., Ooka, T., Jotatsu, H., Kitamura, Y., Akagi, T., et al. (2011). Functional and expressional analyses of PmDAM genes associated with endodormancy in Japanese apricot. Plant Physiol. 157, 485–497. doi: 10.1104/pp.111.181982
Saure, M. C. (1985). Dormancy release in deciduous fruit trees. Hortic. Rev. 7, 239–300. doi: 10.1002/9781118060735.ch6
Sawa, M., Nusinow, D. A., Kay, S. A., and Imaizumi, T. (2007). FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 318, 261–265. doi: 10.1126/ science.1146994
Schwartz, C. J., Doyle, M. R., Manzandeda, A. J., Rey, P. J., Mitchell-Olds, T., and Amasino, R. M. (2010). Natural variation of flowering time and vernalization responsiveness in Brachypodium distachyon. Bioenergy Res. 3, 38–46. doi: 10.1007/s12155-009-9069-3
Searle, I., He, Y., Turck, F., Vincent, C., Fornara, F., Krober, S., et al. (2006). The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev. 20, 898–912. doi: 10.1101/gad.373506
Seo, E., Lee, H., Jeon, J., Park, H., Kim, J., Noh, Y.-S., et al. (2009). Crosstalk between cold response and flowering time in Arabidopsis is mediated through the flowering-time gene SOC1 and its upstream negative regulator FLC. Plant Cell 21, 3185–3197. doi: 10.1105/tpc.108.063883
Shakirova, F., Allagulova, C., Bezrukova, M., Aval’Baev, A., and Gimalov, F. (2009). The role of endogenous ABA in cold-induced expression of the TADHN dehydrin gene in wheat seedlings. Russ. J. Plant Physiol. 56, 720–723. doi: 10.1134/S1021443709050203
Shindo, C., Lister, C., Crevillen, P., Nordborg, M., and Dean, C. (2006). Variation in the epigenetic silencing of FLC contributes to natural variation in Arabidopsis vernalization response. Genes Dev. 20, 3079–3083. doi: 10.1101/gad.405306
Shinozaki, K., and Yamaguchi-Shinozaki, K. (2000). Molecular responses to dehydration and low temperatures: differences and cross-talk between two stress signaling pathways. Curr. Opin. Plant Biol. 3, 217–223.
Sidebottom, C., Buckley, S., Pudney, P., Twigg, S., Jarman, C., Holt, C., et al. (2000). Phytochemistry: heat-stable antifreeze protein from grass. Nature 406, 256. doi: 10.1038/35018639
Silim, S. N., and Lavender, D. P. (1994). Seasonal patterns of environmental regulation of frost hardiness in shoots of seedlings of Thuja plicata, Chamaecyparis nootkatensis, and Picea glauca. Can. J. Bot. 72, 309–316. doi: 10.1139/b94-040
Skinner, J., Zitzewitz, J., Szucs, P., Marquez-Cedillo, L., Filichkin, T., Amundsen, K., et al. (2005). Structural, functional, and phylogenetic characterization of a large CBF gene family in barley. Plant Mol. Biol. 59, 533–551. doi: 10.1007/s11103-005-2498-2
Smith, S. A., Beaulieu, J., Stamatakis, A., and Donoghue, M. J. (2011). Understanding angiosperm diversification using large and small phylogenies. Am. J. Bot. 98, 404–414. doi: 10.3732/ajb.1000481
Song, Y. H., Smith, R. H., To, B. J., Millar, A. J., and Imaizumi, T. (2012). FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering. Science 336, 1045–1049. doi: 10.1126/science.1219644
Sperry, J. S., Nichols, K. L., Sullivan, J. E., and Eastlack, S. E. (1994). Xylem embolism in ring-porous, diffuse-porous, and coniferous trees of northern Utah and interior Alaska. Ecology 75, 1736–1752. doi: 10.2307/1939633
Steponkus, P. L., Uemura, M., Joseph, R. A., Gilmour, S. J., and Thomashow, M. F. (1998). Mode of action of the COR15a gene on the freezing tolerance of Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 95, 14570–14575. doi: 10.1073/pnas.95.24.14570
Stevens, P. F. (2001 onward). Angiosperm Phylogeny Website, Version 12, July 2012 [and more or less continuously updated since].
Stickley, C. E., St John, K., Koc, N., Jordan, R. W., Passchier, S., Pearce, R. B., et al. (2009). Evidence for middle Eocene Arctic sea ice from diatoms and ice-rafted debris. Nature 460, 376–379. doi: 10.1038/nature08163
Stinchcombe, J. R., Weinig, C., Ungerer, M., Olsen, K. M., Mays, C., Halldorsdottir, S. S., et al. (2004). A latitudinal cline in flowering time in Arabidopsis thaliana modulated by the flowering time gene FRIGIDA. Proc. Natl. Acad. Sci. U.S.A. 101, 4712–4717. doi: 10.1073/pnas.0306401101
Stockinger, E. J., Skinner, J. S., Gardner, K. G., Francia, E., and Pecchioni, N. (2007). Expression levels of barley Cbf genes at the Frost resistance-H2 locus are dependent upon alleles at Fr-H1 and Fr-H2. Plant J. 51, 308–321. doi: 10.1111/j.1365-313X.2007.0141.x
Strange, A., Li, P., Lister, C., Anderson, J., Warthmann, N., Shindo, C., et al. (2011). Major-effect alleles at relatively few loci underlie distinct vernalization and flowering variation in Arabidopsis accessions. PLoS ONE 6:e19949. doi: 10.1371/journal.pone.0019949
Streck, N. A., and Schuh, M. (2005). Simulating the vernalization response of the “snow queen” lily (Lilium longiflorum Thunb.). Sci. Agric. 62, 117–121. doi: 10.1590/S0103-90162005000200004
Sutton, F., Chen, D.-G., Ge, X., and Kenefick, D. (2009). Cbf genes of the Fr-A2 alleles are differentially regulated between long-term cold acclimated crown tissue of freeze-resistant and – susceptible, winter wheat mutant lines. BMC Plant Biol. 9:34. doi: 10.1186/1471-2229-9-34
Svendsen, E., Wilsen, R., Stevenson, R., Liu, R. S., and Tanino, K. K. (2007). A molecular marker associated with low-temperature induction of dormancy in red osier dogwood (Cornus sericea). Tree Physiol. 27, 385–397. doi: 10.1093/treephys/27.3.385
Svensson, J. T., Crosatti, C., Campoli, C., Bassi, R., Stanca, A. M., Close, T. J., et al. (2006). Transcriptome analysis of cold acclimation in barley albina and xantha mutants. Plant Physiol. 141, 257–270. doi: 10.1104/pp.105.072645
Swindell, W. R. (2006). The association among gene expression responses to nine abiotic stress treatments in Arabidopsis thaliana. Genetics 174, 1811–1824. doi: 10.1534/genetics.106.061374
Szucs, P., Skinner, J. S., Karsai, I., Cuesta-Marcos, A., Haggard, K. G., Corey, A. E., et al. (2007). Validation of the VRN-H2/VRN-H1 epistatic model in barley reveals that intron length variation in VRN-H1 may account for a continuum of vernalization sensitivity. Mol. Genet. Genomics 277, 249–261. doi: 10.1007/s00438-006-0195-8
Takahashi, S., and Murata, N. (2008). How do environmental stresses accelerate photoinhibition? Trends Plant Sci. 13, 178–182. doi: 10.1016/j.tplants.2008.01.005
Tester, M., and Bacic, A. (2005). Abiotic stress tolerance in grasses. From model plants to crop plants. Plant Physiol. 137, 791–793. doi: 10.1104/pp.104.900138
Teutonico, R. A., and Osborn, T. C. (1995). Mapping loci controlling vernalization requirement in Brassica rapa. Theor. Appl. Genet. 91, 1279–1283. doi: 10.1007/BF00220941
Thomashow, M. F. (1998). Role of cold-responsive genes in plant freezing tolerance. Plant Physiol. 118, 1–8. doi: 10.1104/pp.118.1.1
Thomashow, M. F. (1999). Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 571–599. doi: 10.1146/annurev.arplant.50.1.571
Trevaskis, B., Hemming, M. N., Dennis, E. S., and Peacock, W. J. (2007a). The molecular basis of vernalization-induced flowering in cereals. Trends Plant Sci. 12, 352–357. doi: 10.1016/j.tplants.2007.06.010
Trevaskis, B., Tadege, M., Hemming, M. N., Peacock, W. J., Dennis, E. S., and Sheldon, C. (2007b). Short vegetative phase-like MADS-box genes inhibit floral meristem identity in barley. Plant Physiol. 143, 225–235. doi: 10.1104/pp.106.090860
Trevaskis, B., Hemming, M. N., Peacock, W. J., and Dennis, E. S. (2006). HvVRN2 responds to daylength, whereas HvVRN1 is regulated by vernalization and developmental status. Plant Physiol. 140, 1397–1405. doi: 10.1104/pp.105.073486
Uemura, M., Joseph, R. A., and Steponkus, P. L. (1995). Cold acclimation of Arabidopsis thaliana (effect on plasma membrane lipid composition and freeze-induced lesions). Plant Physiol. 109, 15–30.
Urrutia, M. E., Duman, J. G., and Knight, C. A. (1992). Plant thermal hysteresis proteins. Biochim. Biophys. Acta 1121, 199–206. doi: 10.1016/0167-4838(92)90355-H
USGCRP. (2009). Global Climate Change Impacts in the United States. New York: Cambridge University Press.
Valluru, R., and Van den Ende, W. (2008). Plant fructans in stress environments: emerging concepts and future prospects. J. Exp. Bot. 59, 2905–2916. doi: 10.1093/jxb/ern164
Van Buskirk, H. A., and Thomashow, M. F. (2006). Arabidopsis transcription factors regulating cold acclimation. Physiol. Plant. 126, 72–80. doi: 10.1111/j.1399-3054. 2006.00625.x
Vegis, A. (1964). Dormancy in higher plants. Annu. Rev. Plant Physiol. 15, 185–224. doi: 10.1146/annurev.pp. 15.060164.001153
Vogel, J. T., Zarka, D. G., Buskirk, H. A. V., Fowler, S. G., and Thomashow, M. F. (2005). Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis. Plant J. 41, 195–211. doi: 10.1111/j.1365-313X.2004.02288.x
Wang, R., Farrona, S., Vincent, C., Joecker, A., Schoof, H., Turck, F., et al. (2009a). PEP1 regulates perennial flowering in Arabis alpina. Nature 459, 423–429. doi: 10.1038/nature07988
Wang, H., Moore, M. J., Soltis, P. S., Bell, C. D., Brockington, S. F., Alexandre, R., et al. (2009b). Rosid radiation and the rapid rise of angiosperm-dominated forests. Proc. Natl. Acad. Sci. U.S.A. 106, 3853–3858. doi: 10.1073/pnas.0813376106
Webb, C. O., and Donoghue, M. J. (2005). Phylomatic: tree assembly for applied phylogenetics. Mol. Ecol. Notes 5, 181–183. doi: 10.1111/j.1471-8286.2004.00829.x
Werner, J. D., Borevitz, J. O., Uhlenhaut, N. H., Ecker, J. R., Chory, J., and Weigel, D. (2005). FRIGIDA-independent variation in flowering time of natural Arabidopsis thaliana accessions. Genetics 170, 1197–1207. doi: 10.1534/genetics.104.036533
Whitman, C. M., and Runkle, E. S. (2012). Determining the flowering requirements of two Aquilegia cultivars. HortScience 47, 1261–1264.
Wilson, B. C., Sibley, J. L., and Atland, J. E. (2002). Chilling duration affects foliar budbreak of linden cultivars. Horttechnology 12, 660–662.
Winfield, M. O., Lu, C., Wilson, I. D., Coghill, J. A., and Edwards, E. J. (2010). Plant responses to cold: transcriptome analysis of wheat. Plant Biotechnol. J. 8, 749–771. doi: 10.1111/j.1467-7652.2010.00536.x
Wollenberg, A. C., and Amasino, R. M. (2012). Natural variation in the temperature range permissive for vernalization in accessions of Arabidopsis thaliana. Plant Cell Environ. 35, 2181–2191. doi: 10.1111/j.1365-3040.2012.02548.x
Worrall, D., Elias, L., Ashford, D., Smallwood, M., Sidebottom, C., Liliford, P., et al. (1998). A carrot leucine-rich-repeat protein that inhibits ice recrystallization. Science 282, 115–117. doi: 10.1126/science.282.5386.115
Xiong, L. M., Ishitani, M., Lee, H., and Zhu, J.-K. (2001). The Arabidopsis LOS5/ABA3 locus encodes a molybdenum cofactor sulfurase and modulates cold stress- and osmotic stress-responsive gene expression. Plant Cell 13, 2063–2083.
Xiong, Y., and Fei, S.-Z. (2006). Functional and phylogenetic analysis of a DREB/CBF-like gene in perennial ryegrass (Lolium perenne L.). Planta 224, 878–888. doi: 10.1007/s00425-006-0273-5
Yamashino, T., Ito, S., Niwa, Y., Kunihiro, A., Nakamichi, N., and Mizuno, T. (2008). Involvement of Arabidopsis clock-associated pseudo-response regulators in diurnal oscillations of gene expression in the presence of environmental time cues. Plant Cell Physiol. 49, 1839–1850. doi: 10.1093/pcp/pcn165
Yan, L., Fu, D., Li, C., Blechl, A., Tranquilli, G., Bonafede, M., et al. (2006). The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proc. Natl. Acad. Sci. U.S.A. 103, 19581–19586. doi: 10.1073/pnas.0607142103
Yan, L., Loukoianov, A., Blechl, A., Tranquilli, G., Ramakrishna, W., SanMiguel, P., et al. (2004). The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science 303, 1640–1644. doi: 10.1126/science.1094305
Yan, L., Loukoianov, A., Tranquilli, G., Helguera, M., Fahima, T., and Dubcovsky, J. (2003). Positional cloning of the wheat vernalization gene VRN1. Proc. Natl. Acad. Sci. U.S.A. 100, 6263–6268.
Yanofsky, M. J., and Kay, S. A. (2002). Molecular basis of seasonal time measurement in Arabidopsis. Nature 419, 308–312. doi: 10.1038/nature00996
Yang, J.-S., Wang, R., Meng, J.-J., Bi, Y.-P., Xu, P.-L., Guo, F., et al. (2010). Overexpression of Arabidopsis CBF1 gene in transgenic tobacco alleviates photoinhibition of PSII and PSI during chilling stress under low irradiance. J. Plant Physiol. 167, 534–539. doi: 10.1016/j.jplph.2009.11.005
Yoo, S. Y., Kim, S. Y., Lee, J. S., and Ahn, J. H. (2007). Control of flowering time and cold response by a NAC-domain protein in Arabidopsis. PLoS ONE 2:e642. doi: 10.1371/journal.pone.0000642
Zachos, J., Pagani, M., Sloan, L., Thomas, E., and Billups, K. (2001). Trends, rhythms, and aberrations in global climate 65 Ma to present. Science 292, 686–693. doi: 10.1126/science.1059412
Zhang, C., Fei, C.-Z., Arora, R., and Hannapel, D. (2010). Ice recrystallization inhibition proteins of perennial ryegrass enhance freezing tolerance. Planta 232, 155–164. doi: 10.1007/s00425-010-1163-4
Zhang, J. Z., Li, Z. M., Mei, L., Yao, J. L., and Hu, C. G. (2009). PtFLC homolog from trifoliate orange (Poncirus trifoliata) is regulated by alternative splicing and experiences seasonal fluctuations in expression level. Planta 229, 847–859. doi: 10.1007/s00425-008-0885-z
Zhou, Z., Wang, M.-J., Zhao, S.-T., Hu, J.-J., and Lu, M.-Z. (2009). Changes in freezing tolerance in hybrid poplar caused by up- and down-regulation of PtFAD2 gene expression. Transgenic Res. 19, 647–654. doi: 10.1007/s11248-009-9349-x
Zlesak, D. C., and Anderson, N. O. (2009). Inheritance of non-obligate vernalization requirement for flowering in Lilium formosanum Wallace. Isr. J. Plant Sci. 57, 315–327. doi: 10.1560/IJPS.57.4.315
Zuo, Z., Liu, H., Liu, B., and Lin, C. (2011). Blue light-dependent interaction of CRY2 with SPA1 regulates COP1 activity and floral initiation in Arabidopsis. Curr. Biol. 21, 841–847. doi: 10.1016/ j.cub.2011.03.048
Keywords: cold acclimation, freezing tolerance, endodormancy, plant adaptation, seasonality, vernalization responsiveness
Citation: Preston JC and Sandve SR (2013) Adaptation to seasonality and the winter freeze. Front. Plant Sci. 4:167. doi: 10.3389/fpls.2013.00167
Received: 04 February 2013; Accepted: 13 May 2013;
Published online: 03 June 2013.
Edited by:
Madelaine Bartlett, Brigham Young University, USAReviewed by:
Rishi Bhalerao, Sveriges Lantbruksuniversitet, SwedenBen Trevaskis, Commonwealth Scientific and Industrial Research Organisation, Australia
Erika Edwards, Brown University, USA
Copyright: © 2013 Preston and Sandve. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and subject to any copyright notices concerning any third-party graphics etc.
*Correspondence: Jill C. Preston, Department of Plant Biology, University of Vermont, 111 Jeffords Hall, 63 Carrigan Drive, Burlington, VT 05405, USA e-mail:amlsbC5wcmVzdG9uQHV2bS5lZHU=