- Department of Plant and Environmental Sciences, University of Copenhagen, Frederiksberg, Denmark
Plants are sessile organisms that are under constant attack from microbes. They rely on both preformed defenses, and their innate immune system to ward of the microbial pathogens. Preformed defences include for example the cell wall and cuticle, which act as physical barriers to microbial colonization. The plant immune system is composed of surveillance systems that perceive several general microbe elicitors, which allow plants to switch from growth and development into a defense mode, rejecting most potentially harmful microbes. The elicitors are essential structures for pathogen survival and are conserved among pathogens. The conserved microbe-specific molecules, referred to as microbe- or pathogen-associated molecular patterns (MAMPs or PAMPs), are recognized by the plant innate immune systems pattern recognition receptors (PRRs). General elicitors like flagellin (Flg), elongation factor Tu (EF-Tu), peptidoglycan (PGN), lipopolysaccharides (LPS), Ax21 (Activator of XA21-mediated immunity in rice), fungal chitin, and β-glucans from oomycetes are recognized by plant surface localized PRRs. Several of the MAMPs and their corresponding PRRs have, in recent years, been identified. This review focuses on the current knowledge regarding important MAMPs from bacteria, fungi, and oomycetes, their structure, the plant PRRs that recognizes them, and how they induce MAMP-triggered immunity (MTI) in plants.
Introduction
Bacteria, fungi, oomycetes, and viruses attack plants in an attempt to gain nutrients from them. During the course of evolution both plants and pathogens have evolved features to combat each other; the plant is equipped with sophisticated and rapidly mounted defense mechanisms, while their cognate pathogens have developed counterstrategies to overcome those defenses, the so called “arms race” between plant and pathogens (Bent and MacKey, 2007). The interplay between the plant defense systems and its suppression by pathogens has been portrayed as a “zigzag model” by Jones and Dangl (2006). This model proposes that the plants' immune responses consist of two branches. The first line of defense in plants is the recognition of conserved molecules characteristic of many microbes. These elicitors are also known as microbe- or pathogen-associated molecular patterns (MAMPs or PAMPs) (Table 1). MAMPs are essential structures for the microbes and are for that reason conserved both among pathogens, non-pathogenic and saprophytic microorganisms. MAMPs are recognized by pattern recognition receptors (PRRs), which are localized on the surface of plant cells; this first phase of defense induction is called MAMP-triggered immunity (MTI) (Ausubel, 2005; Jones and Dangl, 2006). All known plant PRRs are plasma membrane-localized receptor-like kinases (RLKs) or receptor-like proteins (RLPs) with modular functional domains. RLKs contain an extracellular domain (ECD), a single-pass transmembrane (TM) domain, and an intracellular kinase domain. RLPs contain an ECD and a TM but have only a short cytosolic domain without an obvious signaling domain (Table 1). Notably, in contrast to mammals, no intracellular nucleotide-binding-leucine-rich repeat (NB-LRR) protein recognizing a MAMP has yet been identified in plants (Maekawa et al., 2011). Bacterial effector proteins, injected directly into the host plants' cytoplasm via the pathogens type III secretion system (TTSS), have been demonstrated to suppress MTI (Jamir et al., 2004; He et al., 2006; Nomura et al., 2006), resulting in effector-triggered susceptibility (ETS). The second line of the plants' defense is direct or indirect recognition of a given effector through a set of plant resistance (R) gene products resulting in effector-triggered immunity (ETI) (Jones and Dangl, 2006); also named the gene-for-gene interaction as early as 1942 by Flor. ETI is generally an accelerated and amplified MTI response, and as such it is an effective defense response (resistance) that in most cases leads to a localized cell death, known as the hypersensitive response (HR). The majority of the R proteins are intracellular receptor proteins of the NB-LRR type. In most cases the interaction between NB-LRRs and the effectors are indirect (van der Biezen and Jones, 1998).
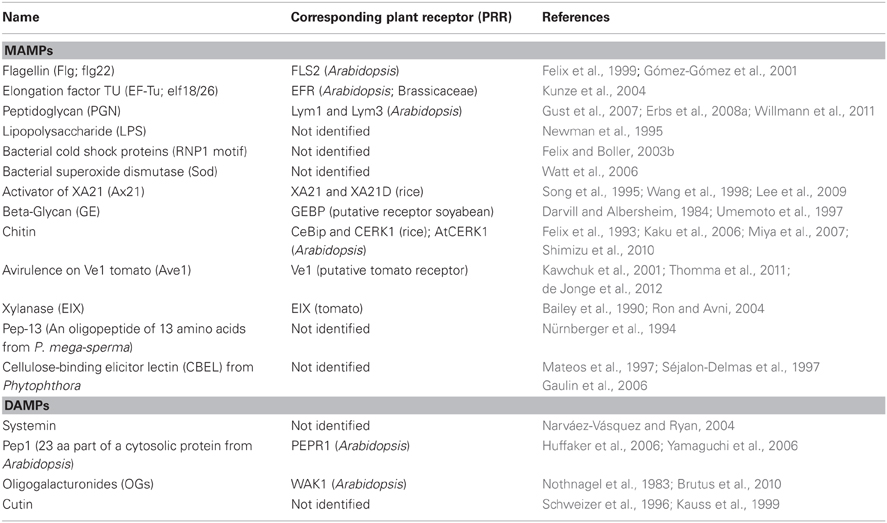
Table 1. Microbe-associated molecular patterns (MAMPs) and Damage-associated molecular patterns (DAMPs).
MAMP-induced defense responses include the production of reactive oxygen species (ROS, also called the oxidative burst), production of reactive nitrogen species such as nitric oxide (NO), alterations in the plant cell wall, induction of antimicrobial compounds and the synthesis of pathogenesis-related (PR) proteins. ROS and NO can act in signaling and have direct antimicrobial effects. ROS can also drive oxidative cross-linking of polymers in the plant cell wall to strengthen it against degradation, which may restrict pathogen spread. Other alterations in the plant wall include the deposition of the β-(1–3) linked glucan callose. PR proteins comprise a number of families that include enzymes, such as β-(1–3) glucanase and chitinase, which can directly attack pathogen structures, antimicrobial peptides and small proteins, and PR1, which is of unknown function [for reviews, see Hammond-Kosack and Jones (1996); Greenberg (1997); Lamb and Dixon (1997); Nürnberger and Kemmerling (2006)]. The induction of MTI in plants has been most extensively studied using the small peptides flg22 and elf18 derived from the bacterial MAMPs flagellin (Flg) and the translation elongation factor Tu (EF-Tu), respectively (Felix and Boller, 2003a; Zipfel et al., 2006). Bacterial glycoconjugates, such as the peptidoglycan (PGN), which provides rigidity and structure to the cell envelopes of both Gram-negative and Gram-positive bacteria (Erbs et al., 2008a; Willmann et al., 2011), and lipopolysaccharides (LPS) from the outer membrane of Gram-negative bacteria have been found to act as elicitors of plant innate immunity (Silipo et al., 2005; Erbs and Newman, 2012). Oligosaccharides derived from cell wall polymers of fungi and oomycetes also act as MAMPs. Fungal chitin and its degradation products N-acetyl-chito-oligosaccharides, i.e., chitin oligomers induce various defense responses in both monocot and dicot plants (Kaku et al., 2006; Miya et al., 2007). In the oomycetes, the cell walls are composed of β-glucans and cellulose, rather than chitin, as in the fungi. Some of the earliest work on the role of glycosylated compounds in triggering plant defenses has examined the effects of β-(1→3/1→6)-linked glucans from the cell walls of Phytophthora megasperma f. sp. glycinea on the induction of phytoalexin accumulation in soybean [reviewed in Cheong and Hahn (1991)]. In the plant-virus interactions no conserved viral MAMP has been identified so far, and the primary plant defense is thought to be based mainly on RNA silencing (RNAi). By analogy with the zigzag model, viral-derived double-stranded RNA (dsRNA) is regarded as the MAMP inducing RNAi, a general plant defense mechanism or the MTI. To counteract this defense, plant viruses express RNA silencing suppressors (RSSs), many of which bind to dsRNA and attenuate RNAi (Csorba et al., 2009; Ruiz-Ferrer and Voinnet, 2009).
This review will focus on some of the important MAMPs from bacteria, fungi, and oomycetes and review the current knowledge of their structure, how they are recognized and how they induce MTI in plants. We include the slightly more unusual MAMP Ax21 from the rice pathogenic bacteria Xanthomonas oryzae pv. oryzae (Xoo). MAMPs in general activate MTI directly via their respective plant receptors, whereas Ax21 is secreted out of the bacterium via the type-I secretion system (TOSS), where it is then recognized by the rice receptor XA21, and induction of MTI follows. Finally, we will briefly describe damage-associated molecular patterns (DAMPs) of plant origin, which also induce MTI in the plant.
Bacterial MAMPs
Flagellin (Flg)
Flagella are essential structures for the pathogenic bacteria as they provide motility and often increase adhesion of the bacteria to its host. Flg, the main building block of bacterial flagella, is well-established as a major activator of innate immunity in animals [reviewed by Ramos et al. (2004)]. Some of the first MAMP recognition studies in plants were carried using Flg. Studies in mammals have shown that at least one of the conserved domains in the N-terminal and C-terminal part of the bacterial Flg, found to be involved in bacterial motility as well, is recognized by Toll-like receptor 5 (TLR5) (Hayashi et al., 2001; Smith et al., 2003). Studies in Arabidopsis, tomato, and other plants, revealed that plants respond to a highly conserved domain in the N-terminal part of the bacterial Flg, a 22 amino acid (aa) peptide, flg22 (Felix et al., 1999). In order to identify the gene involved in recognition and transduction of the flg22 elicitor signal, Gomez-Gomez and Boller (2000), used a genetic approach to screen Arabidopsis mutants after flg22 treatment and isolated several Flg sensing 2 (FLS2) mutants, which mapped to the FLS2 locus on chromosome 5. FLS2 belongs to the RLK family and has an ECD with 28 LRRs, a TM domain, and an intracellular serine/threonine kinase domain. No high-affinity binding site was found, after treatment with a radiolabeled derivative of flg22, in the Flg insensitive Arabidopsis ecotype Ws-0 and in plants carrying mutations in the LRR domain of the FLS2 gene, indicating a role for LRR in Flg binding (Gomez-Gomez and Boller, 2000; Bauer et al., 2001). Later work revealed that both an extracellular LRR domain and kinase activity of FLS2 were necessary for high affinity binding and binding specificity for Flg (Gómez-Gómez et al., 2001). Chinchilla et al. (2006) showed the specific interaction of flg22 with FLS2 in Arabidopsis. The recognized domain within Flg is not the same for all plant species. For example, flg15, an N-terminally shortened version of flg22, was shown to be highly active in tomato, while it only elicits immune responses at higher concentrations in Arabidopsis. Rice is able to recognize flg22, but its defense response is greater to the full length Flg (Takai et al., 2008). The functionality of the FLS2 receptor was tested by heterologous expression of the Arabidopsis FLS2 receptor in tomato cells. In these expression studies, tomato cells gained the Flg perception system characteristic for Arabidopsis, demonstrating that FLS2 represents the PRR that determines the specificity of Flg perception (Meindl et al., 2000; Bauer et al., 2001; Chinchilla et al., 2006). The difference in recognition of the Flg epitope is not restricted to different plant families; variations have also been found between species in the same family. A 15 aa peptide derived from E. coli Flg was shown only to be highly active in tomato (Solanum lycopersicum previously called Lycopersicon esculentum), but not in tobacco. Furthermore, the tomato Flg receptor, SlFLS2, an ortholog of the Arabidopsis FLS2 receptor, has now been identified and used in expression studies with Nicotiana benthamiana, where N. benthamiana expressing SlFLS2 gained the Flg perception system specific for tomato (Robatzek et al., 2007). In addition to this, studies focusing on host recognition of Xanthomonas campestris pv. campestris (Xcc) Flg have revealed within species and within pathovar variations for defense eliciting activity of Flgs among Xcc strains (Sun et al., 2006). Confirmation of FLS2 as a surface receptor came with studies using transgenic Arabidopsis Ws-0, expressing FLS2 fused to the green fluorescent protein (GFP), which revealed a cell membrane localization of FLS2. Additionally, FLS2 was found to undergo ligand-induced endocytosis; it is thought that this subcellular redistribution of FLS2, or any other surface receptor, from the plasma membrane to cytoplasmic vesicles may be a central point in signaling during immune responses (McCoy et al., 2004; Robatzek et al., 2006). Flg-induced activation of FLS2 in Arabidopsis, involves a complex formation with the Brassinosteroid-Insensitive 1 (BRI1)-associated receptor kinase 1 (BAK1) (Chinchilla et al., 2007). BAK1 has also been reported to be involved in BRI1 endocytosis (Russinova et al., 2004). Furthermore, BAK1 is required for the immune responses triggered by multiple MAMPs other than Flg, including the bacterial elongation factor EF-Tu (see below) (Roux et al., 2011). The activities of MAP kinases (MAPK) were delayed and reduced or even absent in response to flg22 or elf18, a fully active EF-Tu derivative, in bak1 mutants, compared to wild type plants. This indicates that BAK1 acts as a positive regulator of MAMP signaling in Arabidopsis. In addition, it was revealed that FLS2, after flg22 stimulation, interacts with BAK1 in a ligand-dependent manner (Chinchilla et al., 2007). This interaction allows phosphorylation and activation of the receptor complex (Schulze et al., 2010). Downstream of the FLS2-BAK1 receptor complex is a cytoplasmic receptor kinase Botrytis-induced kinase 1 (BIK1), which constitutively associate with FLS2. After FLS2-BAK1 dimerization, BIK1 dissociate from FLS2, possibly allowing BIK1 to phosphorylate downstream components, and thus linking the MAMP receptor complex to downstream intracellular signaling leading to MTI (Lu et al., 2010). However, the substrates of FLS2 and BAK1 kinases have yet to be identified, and how the MAMP signal is transmitted from the BAK1-associated receptor complexes at the plasma membrane to intracellular events is largely unknown.
Elongation Factor Tu (EF-Tu)
In protein biosynthesis, the ribosomes translate the sequence of nucleotides in mRNA into the sequence of aa's in a protein. During the phase of elongation the ribosome is associated with elongation factors. One such elongation factor is EF-Tu, the most abundant protein in the bacterial cell (Jeppesen et al., 2005). The elicitor activity is attributed to a highly conserved part of the N-terminus of EF-Tu, either a 26 or 18 aa peptide named elf26 or elf18. The perception of EF-Tu by the EF-Tu Receptor (EFR) is independent of Flg perception, as EF-Tu is active in plants carrying mutations in FLS2 (Kunze et al., 2004). Although many of the signaling components downstream of EFR and FLS2 are shared between them (see above). EF-Tu recognition has only been found to elicit innate immunity in members of the family Brassicaceae (Zipfel et al., 2006). Studies using crosslinking assays in Arabidopsis cells, confirmed that elf18 and flg22 bind to different high-affinity binding receptors. Nevertheless, elf18 and flg22 were found by microarray analysis to induce the same pool of genes, and also a common set of responses in Arabidopsis. In addition to this, a combined treatment with both MAMPs, elf26/18 and flg22, was shown to induce the same kinases without an additive effect (Zipfel et al., 2006). An EF-Tu insensitive efr-1 mutant did not respond with an oxidative burst, increased ethylene biosynthesis or induced resistance to Pseudomonas syringae pv. tomato (Pst) DC3000 in response to EF-Tu-derived elicitors, whereas Arabidopsis Col-0 and the fls2 mutant did respond to EF-Tu elicitors. Heterologous expression studies of EFR in N. benthamiana, a plant lacking a perception system for EF-Tu, resulted in N. benthamiana with a perception system for EF-Tu, confirming the role for EFR as a functional receptor for EF-Tu (Zipfel et al., 2006). In addition to this, efr mutants were found to be more susceptible to Agrobacterium tumefaciens (At)-mediated transformation than wild type plants, indicating that EF-Tu recognition and the subsequent defense responses reduce Agrobacterium-mediated plant transformation (Zipfel et al., 2006).
Similar to FLS2, EFR belongs to the RLK family and has an ECD with 24 LRRs, a single TM domain and an intracellular serine/threonine kinase domain (Zipfel et al., 2006). Both FLS2 and EFR are members of the subfamily LRR-XII of RLKs (Shiu and Bleecker, 2003). Besides FLS2 and EFR from Arabidopsis, the rice pathogen recognition receptor, XA21 (see below), which confers resistance to Xoo strains is also a member of the LRR-XII subfamily (Song et al., 1995; Shiu et al., 2004; Lee et al., 2006). In contrast to FLS2, but like XA21, N-glycosylation is required for EFR functionality. Mutation of a single predicted glycosylation site compromised elf18 binding despite correct localization of the mutated protein to the plasma membrane (Häweker et al., 2010).
Peptidoglycan (PGN)
PGN, a molecule never found in eukaryotes, is an essential and unique membrane envelope component of all bacteria, making it an excellent target for the eukaryotic innate immune system [reviewed by McDonald et al. (2005); Dziarski and Gupta (2006)]. PGN, which provides rigidity and structure to the cell envelope of both Gram-positive and Gram-negative bacteria, is a complex molecule consisting of numerous glycan chains that are cross-linked by oligo-peptides. These glycan chains are composed of altering N-acetylglucosamine (GlcNAc) and N-acetylmuramic acid (MurNAc), with short peptides attached by an amide linkage to the lactyl group of MurNAc. Several types of PGN, classified by the nature of the third residue of the stem peptide are commonly found. Typically, this is m-diaminopimelic acid (mDAP) PGN in Gram-negative bacteria and in some Gram-positive bacilli (genus Bacillus and Clostridium), whereas most other Gram-positive bacteria have L-lysine (LYS) PGN. In a recent study in tomato Nguyen et al. (2010) showed that pre-inoculation into tomato with Staphylococcus aureus PGN reduced the growth of a subsequent bacterial infection in PGN-treated tissue. This priming of defense with a MAMP is similar to that previously described for LPS (Newman et al., 2002). Early experiments with plant cells showed that the Gram-positive human pathogen S. aureus PGN was active as an elicitor in inducing extracellular alkalization of cultured tobacco cells, while no response was observed in cultured tomato cells, suggesting a different perception system for PGN within the Solanaceae (Felix and Boller, 2003a). PGN from both Gram-positive and Gram-negative bacteria was later found to act as elicitors of plant innate immunity in Arabidopsis (Gust et al., 2007; Erbs et al., 2008a). Gust et al. (2007) showed that it was the sugar backbone of the Gram-positive S. aureus PGN that was responsible for triggering immune responses and not the breakdown product of PGN, the muramyl dipeptide (MDP) or the muropeptide dimer, which is known to be the minimal chemical structure required for triggering the innate immune system in vertebrates and insects [reviewed by Traub et al. (2006)].
Erbs et al. (2008a), on the contrary, using PGN from two Gram-negative bacterial plant pathogens, Xcc and At found that both Xcc and At PGN and its constituents functioned as MAMPs in Arabidopsis and induced immune responses such as generation of ROS, extracellular pH increase, PR1 gene expression, and callose deposition. Furthermore, they showed that the muropeptides were significantly more effective at inducing defense responses than the intact PGN molecule. These observations could be indicative of different perception systems for PGN from Gram-positive and Gram-negative bacteria or differences in structures, and therefore recognition sites, of the muropetides of human vs. plant pathogens. So far, the full structure of PGN from a Gram-positive plant pathogen has not been elucidated. In a study from 2009, Gimenez-Ibanez et al. showed that PGN from the bacterial pathogen Pst DC3000 induced the generation of ROS in Arabidopsis cerk1 (chitin elicitor receptor kinase1) mutant plants, which indicated that Pst DC3000 PGN perception is independent of CERK1. In contrast, Willmann et al. (2011) reported that two of three Arabidopsis chitin oligosaccharide elicitor-binding proteins (AtCEBiP), LYM1, and LYM3, are involved in the perception/signaling of PGN (from various sources) together with AtCERK1, indicating the presence of a two-component receptor system similar to the rice chitin receptor OsCEBiP and OsCERK1 (Shimizu et al., 2010) (also see text below). All three proteins are required for PGN perception in vivo and for resistance to bacterial pathogens. Willmann et al.'s findings also showed that AtCERK1 is involved in the perception of at least two MAMP molecules, chitin and PGN in Arabidopsis. Interestingly, Shinya et al. (2012) showed that AtCERK1 serves both for chitin and PGN signaling, but AtCERK1 seems to contribute differently to the signaling. In the case of PGN signaling, the binding proteins LYM1 and LYM3 not only bind the ligand, but also contribute to the activation of AtCERK1 and downstream signaling, similarly to the function of OsCEBiP in rice. On the other hand, in the case of chitin signaling, AtCERK1 seems to function for both ligand perception and signaling (also see text below). Structurally, CEBiP is a receptor protein that contains extracellular LysM domains that are ~40 aa in length lacking a recognizable intracellular signaling domain. The LysM domains are considered to generally mediate binding to GlcNAc-containing glycans, like chitin and the backbone of PGN [reviewed by Gust et al. (2012)]. Also in monocots two LysM-containing PRRs have been shown to recognize PGN. Liu et al. (2012) reported two homologous rice lysine-motif containing proteins, LYP4 and LYP6. Both proteins bound PGN and chitin, but not LPS in vitro. Silencing either of the two proteins impaired the PGN or chitin-induced defense responses, and compromised the resistance against Xoo or the fungal pathogen Magnaporthe oryzae. These results suggest that PGN and chitin have overlapping perception components in rice.
In mammals, the recognition of PGN is complex, e.g., different receptors are found for PGN (extracellularly) and muropeptides (intracellularly). The cytosolic protein Nod2 can recognize MDP, from both Gram-positive and Gram-negative bacteria, and a lysine-containing muramyl tripeptide, but not a DAP-containing muramyl tripeptide (Girardin et al., 2003). In contrast, Nod1 only detects DAP-containing muropeptides. For instance, the human Nod1 recognizes the DAP-containing GlcNAc and MurNAc tripeptide (Chamaillard et al., 2003). The structure of the mammalian NOD proteins is similar to that of the plant R proteins, which are intracellular receptor proteins of the NB-LRR type (Inohara et al., 2005). In plants these proteins are involved in the recognition of specialized pathogen effectors leading to ETI (see above). However, in animals they seem to be involved in MAMP recognition rather than recognition of pathogen effectors. The future will show if a system similar to the Nod system, detecting intracellular microbial molecules, could be a possibility in plants.
Lipopolysaccharides (LPS)
LPS, the major component of the outer membrane of Gram-negative bacteria, have been shown to have multiple roles in plant microbe interactions; it is thought to contribute to the restrictive Gram-negative membrane permeability, allowing bacterial growth in unfavorable environments. LPS and its derivatives act as MAMPs and induce innate immune responses in plants (Newman et al., 1995; Dow et al., 2000; Bedini et al., 2005; Silipo et al., 2005). Earlier studies in plants have shown that LPS can prevent the HR induced by bacteria. Pre-treatment of Arabidopsis leaves with LPS and its derivatives was found to prevent the HR caused by strains of Pst carrying the avrRpm1 or the avrRps4 genes, a phenomenon referred to as localized induced response (LIR; Newman et al., 2002; Silipo et al., 2005). The mechanisms behind HR prevention are still unknown, but the effects of LPS pre-treatment are considered to be associated with enhanced resistance of the plant tissue to pathogenic bacteria, which is thought to occur through an LPS-dependent potentiation of plant defense responses (Newman et al., 2002). LPS consists of a lipid, a core oligosaccharide, and an O-polysaccharide part. The lipid, referred to as lipid A, is embedded in the outer part of the phospholipid bilayer in the bacterial membrane. Lipid A and the core oligosaccharide are linked, usually by the sugar 3-deoxy-d-manno-2-octulosonate (KDO). The core oligosaccharide consists of a short series of sugars and ends in the O-antigen, which is composed of repeating oligosaccharide units (Raetz and Whitfield, 2002). The O-antigen of the LPS from many phytopathogenic bacteria has shown to consist of oligorhamnans (Bedini et al., 2002).
In order to know more about the structures within LPS that trigger immune responses in plants, synthetic O-antigen polysaccharides, oligorhamnans of increasing chain lengths, were tested in Arabidopsis. Tri-, hexa-, and nonasaccharides were synthesized and found to suppress the HR, as well as act as MAMPs and elicit the induction of the PR genes PR1and PR2 in Arabidopsis. The efficiency of HR suppression and PR gene induction improved with increasing chain lengths of sugars in the synthetic O-antigen. In addition, a coiled structure was observed with the increasing chain length, indicating a role for this structure as a MAMP and by correlation a role for the O-antigen from phytopathogenic bacteria in plant innate immunity (Bedini et al., 2005). Studies in mammalian cells have shown that LPS is recognized through their lipid A moiety and this recognition was shown to govern the interactions with the innate immune system (Loppnow et al., 1989). In addition to this, the molecular shape of lipid A was found to directly correlate with its activity as a conical shape of lipid A was associated with endotoxicity and a cylindrical shape with antagonistic activity. A net negative charge of lipid A was found to influence its molecular conformation, and with that, its biological activity (Schromm et al., 1998, 2000). To study if the innate immune system from the mammalian system has parallels in the plant system, the role and mechanisms of action of LPS and its derivatives, the core oligosaccharide and the lipid A moiety, in plant-bacteria interactions were investigated in Arabidopsis. Initially, the complete structure of purified Xcc lipo-oligosaccharides (LOS), LPS without the O-chain, was determined. Xcc LOS was found to be a unique molecule with a high negative charge density and a phosphoramide group never found in such molecules before (Silipo et al., 2005). Xcc LOS and derivatives have been shown to elicit induction of the PR genes in Arabidopsis. LOS was found to induce the defense-related PR1 and PR2 genes in two temporal phases: the core oligosaccharide induced only the early phase and the lipid A moiety only the later phase, which suggests that both the core oligosaccharide and the lipid A are recognized by plant cells, e.g., both act as elicitors. These findings support the role of Xcc lipid A and the Xcc core oligosaccharide as MAMPs of innate immunity in plants. Silipo et al. (2005) speculated that the different LPS fragments are recognized by different plant receptors. This elicitor activity of Xcc lipid A correlates with earlier studies by Zeidler et al. (2004), who showed that lipid A preparations from various bacteria induced a rapid burst of NO production that was associated with the induction of defense-related genes in Arabidopsis. In a recent study by Madala et al. (2011) where the structure of Burkholderia cepacia strain ASP B 2D lipid A was determined, the role of lipid A as a MAMP in Arabidopsis was confirmed, and it was found to induce transcriptional changes associated with plant defense responses. Contrary to this, studies in tobacco cells, have shown that neither the lipid A nor the O-chain of the Xcc LPS molecule could induce the oxidative burst alone, but rather it was the inner core part of the LPS molecule that was responsible (Braun et al., 2005). The conflict in results could reflect the different defense responses measured after treatment with LPS and its derivatives in different plants.
In correlation to studies in the mammalian system, where it is well-established that the phosphorylation pattern of lipid A affects its biological activity [reviewed by Gutsmann et al. (2007)], it was tested whether de-phosphorylated Xcc LOS could be recognized in plants. After de-phosphorylation of Xcc LOS the molecule maintained only the negative charge of the KDO residue, and rendered the molecule unable to induce LIR, suggesting that the charged groups present in LOS play a key role in inducing defense responses in Arabidopsis (Silipo et al., 2005). Furthermore, from these experiments it could be concluded that the electrostatic interactions involving the phosphate groups seem to have a crucial function in binding not only lipid A, but also the core oligosaccharide, to putative receptors in plants (Silipo et al., 2005). LPS has been found, not only to induce defense responses, but also to prime expression of plant defense responses upon subsequent bacterial inoculation, e.g., promote an early triggering of the synthesis of the antimicrobial compounds feruloyl tyramine (FT) and p-coumaroyl tyramine (CT) (Newman et al., 2002, 2007; Prime-A-Plant Group, 2006). The O-antigen part of the LPS molecule is thought to be responsible for induced systemic resistance (ISR) in Arabidopsis. Early studies showed that LPS from the rhizobacterium Pseudomonas fluorescens, as well as the live bacteria, induced ISR in carnation and radish, whereas mutant bacteria, lacking the O-antigen side chain could not induce ISR (Leeman et al., 1995; van Loon et al., 1998). In contrast to the rhizobacteria-mediated ISR, systemic activation of defense-related responses in plants upon local necrotizing pathogen infection is referred to as systemic acquired resistance (SAR). SAR is accompanied by a systemic increase in salicylic acid (SA), and SA is required for SAR signaling (Ryals et al., 1996; Schneider et al., 1996). However, recent studies suggest that recognition of the MAMPs, LPS, or Flg, and not necrotic lesion formations contribute to the bacterial induction of SAR in Arabidopsis. Treatment of Arabidopsis with P. aeruginosa LPS, Flg or non-host bacteria were shown to be associated with accumulation of SA, expression of the PR genes and expression of the SAR marker gene Flavin-dependent monooxygenase 1 in treated as well as in distant leaves (Mishina and Zeier, 2006, 2007). The signaling cascades underlying SAR and NO production after perception of LPS by plant cells have not yet been resolved. Sun et al. (2012) investigated the biosynthetic origin of NO and the role of Non-expressor of Pathogenesis-Related Genes (NPR1) to gain insight into the mechanism involved in LPS-induced resistance of Arabidopsis. NPR1 is a key regulator of SAR, and is essential for SA signal transduction (Rockel et al., 2002). Analysis of inhibitors and mutants showed that LPS-induced NO synthesis was mainly mediated by an arginine-utilizing source of NO generation. LPS (Sigma)-activated defense responses, including callose deposition and defense-related gene expression, were found to be regulated through an NPR1-dependent pathway. In contrast, Xcc LPS can induce defense responses in pepper without triggering the oxidative burst or SA synthesis (Newman et al., 2002).
The activity of LPS in plants has mostly been described in dicots, but studies in rice cells have revealed that LPS, from various pathogenic and non-pathogenic bacteria, induce a generation of ROS and defense-related gene expression in monocots, indicating that the machinery recognizing LPS is evolutionary conserved in monocots and dicots (Desaki et al., 2006, 2012). Furthermore, the two MAMPs, LPS and chitin oligosaccharide, induced a close correlation of genes in rice cells, indicating a convergence in signaling cascades downstream of recognition. In addition, the effect of LPS from various bacteria was shown to be associated with a programmed cell death (PCD) in rice cells. In contrast, LPS has never been shown to elicit PCD in dicots (Desaki et al., 2006). The mechanism by which LPS is perceived by plants is still not understood. Recent studies with fluorescein-labeled Xcc LPS in cultured N. tabacum cells revealed that LPS was rapidly bound to the cell wall and then internalized into the cell, and eventually, LPS was found exclusively inside the vacuole. These findings suggest endocytosis, comparable to the mammalian system, of Xcc LPS in tobacco cells (Gross et al., 2005). However, no PRRs for LPS and its derivatives have been characterized in plants. In the mammalian immune system, LPS form complexes with LPS-binding proteins (LBP), and this LPS-LBP complex is recognized by the membrane-bound CD14 receptor, glycosylphosphatidyl inositol (GPI)-anchored glycoprotein (Wright et al., 1990), which again is thought to associate with TLR4-MD2 to participate in LPS-induced signaling (Jiang et al., 2000; Miyake, 2004). The concentrations of LPS required to elicit most of the effects described above are in the 5–100 μg per ml range, suggesting that plants do not have the sensitivity to LPS shown by mammalian cells, which can respond at concentrations in the pg to ng per ml range. These considerations have led to suggestions that plants possess only low affinity systems to detect LPS (Zeidler et al., 2004), although plants can detect other bacterial MAMPs such as the peptides derived from Flg and EF-Tu elongation factor at sub nM levels. One complicating factor is the aggregation of LPS molecules within the purified preparations, which may affect the ability of LPS to cross the matrix of the plant cell wall to reach presumed membrane-associated receptors (Aslam et al., 2009).
Many groups have attempted to identify plant components involved in LPS recognition and perception often with conflicting results. Livaja et al. (2008) found that in Arabidopsis cells, B. cepacia LPS induced a leucine-rich repeat RLK At5g45840 by nearly 17-fold after 30 min. Furthermore, in a proteomic analysis of the changes following perception of LPS from an endophytic strain of B. cepacia in N. tabacum BY-2 cells, 88 LPS induced/regulated proteins, and phosphoproteins were identified, many of which were found to be involved in metabolism and energy-related processes. Moreover, proteins were found that are known to be involved in protein synthesis, protein folding, vesicle trafficking, and secretion (Gerber et al., 2006, 2008). In a transcription profiling of A. thaliana cells treated with LPS from B. cepacia, Livaja et al. (2008) surprisingly did not find any genes involved in callose synthesis. Furthermore, genes involved in ROS production were found to be upregulated at a very low level by B. cepacia LPS. In addition, Livaja et al. (2008) found that B. cepacia LPS only induced the PR genes PR3 and PR4, whereas studies in B. cepacia LPS treated Arabidopsis leaves revealed induction of several PR genes (Zeidler et al., 2004). Other LPS preparations, from P. aeruginosa and Escherichia coli, respectively, induce PR1 and PR5 in Arabidopsis leaves (Mishina and Zeier, 2007). The variation in results both reflects the different plant systems (Arabidopsis cell cultures contra the whole plant) and the origin of the LPS. All the above very specific effects show the ability of particular plants to recognize structural features within LPS that are not necessarily widely conserved.
Recognition of LPS/LOS in mammals is rather complex; how complex this recognition is in plants is still not known, and the mechanism of this recognition and consequent transduction steps in plants remains obscure. Gross et al. (2005) showed that, in tobacco cells, Xcc LPS was internalized 2 h after its introduction to a cell suspension, where it co-localized with Ara6, a plant homolog of Rab5 which is known to regulate early endosomal functions in mammals. It was speculated that this endocytosis in tobacco cells was, in correlation with the mammalian system, part of a down regulation of defense responses. In a recent study by Zeidler et al. (2010) localization and mobilization of fluorescein-labeled Salmonella minnesota LPS was studied in Arabidopsis. Leaves were pressure infiltrated with fluorescein-labeled S. minnesota LPS and the mobility of LPS was studied over time by fluorescence microscopy. After 1 h a fluorescent signal was observed in the intercellular space of the infiltrated leaf. The labeled LPS were visible in the midrib of the leaves after 4 h, whereas this fluorescence had spread to the smaller leaf veins near the midrib after 6 h. After 24 h it was detectable in the lateral veins. Moreover, cross-sections of the midrib 3 h after supplementation with fluorescein-labeled LPS revealed a fluorescent signal in the xylem. Using capillary zone electrophoresis a distribution of fluorescein-labeled S. minnesota LPS was found in the treated as well as in systemic leaves of the plant (Zeidler et al., 2010). In contrast to the results reported by Gross et al. (2005), no intracellular accumulation of the labeled LPS was observed in Arabidopsis. This disparity in outcomes might again be a reflection of the use of different plants, the difference in the age of the plants used (plant cell cultures vs. seedlings vs. fully developed plants) and the different defense responses measured after treatment with LPS and its derivatives.
Alterations in lipid A or other structures within LPS are known to occur during symbiotic interactions with plants (Kannenberg and Carlson, 2001) and in response to compounds in plant root exudates (Fischer et al., 2003) and may occur during plant pathogenesis. These alterations may serve both to increase the resistance of the bacteria against host defenses and to attenuate the activity of lipid A or LPS in triggering those defenses. Characterization of the structure and function of LOS from a non-pathogenic Xcc mutant strain 8530, which carries a Tn5 insertion in a gene of unknown function (Dow et al., 1995), revealed that this mutant had a truncated core region. The fact that Xcc strain 8530 was defective in core completion led to significant modifications in the acylation and phosphorylation patterns of its lipid A, and these changes had influence on its ability to trigger innate immune responses in Arabidopsis (Silipo et al., 2008). The core sugars provide protection against antimicrobial compounds and attenuate the endotoxic properties of lipid A, similar to lipid A modifications seen in mammalian pathogens (Raetz et al., 2007). These findings indicate that Xcc has the capacity to modify the structure of lipid A and thus reduce its activity as a MAMP in plants (Silipo et al., 2008). The acyl chains of lipid A can vary, as can the number and length of them depending on growth conditions and bacterial species. Studies in mammalian cells have shown that LPS from Shigella flexneri elicit a weaker TLR4-mediated response than E. coli LPS due to differences in the acylation status of their lipid A moieties (Rallabhandi et al., 2008).
Lipid A from Halomonas magadiensis, an extremophilic and alkaliphilic Gram-negative bacteria, isolated from a soda lake in an East African Rift Valley has been found to act as an LPS antagonist in human cells (Silipo et al., 2004). H. magadiensis lipid A, characterized by an unusual and very low degree of acylation, was verified to inhibit E. coli lipid A-induced immune responses in human cells (Ialenti et al., 2006). E. coli lipid A, which is an effective agonistic structure of immune responses in mammalian cells, is composed of a bis-phosphorylated hexa-acylated disaccharide backbone with an asymmetric distribution of the acyl residues. Studies have revealed that structural differences on the lipid A skeleton, for example, acylation can affect its agonist/antagonist activity (Munford and Varley, 2006). In consonant with the ability in blocking enteric LPS-induced human monocyte activation, our laboratory found that H. magadiensis lipid A was able to antagonize the action of E. coli lipid A when inducing PR1 gene expression in Arabidopsis. Even though the mode of perception of LPS in plants is far less-understood than in mammals and insects, these results indicate that Arabidopsis is sensitive to the same structures of lipid A that determine biological activity in humans (Erbs et al., 2008b).
Thus far, LPS preparations used for the analysis of plant responses and for structural studies have been derived from bacteria grown in culture. We know nothing about the alterations in LPS that might occur when bacteria are within plants, although this may be highly relevant for recognition and signaling. Changes could occur in both the size distribution of LPS (alteration in the ratio of LOS to LPS) and/or in decoration of LPS with saccharide, fatty acid, phosphate, or other constituents. Increases in the sensitivity of mass spectrometric methodologies may allow development of micro-methods to analyse such changes in bacteria isolated from plants. Transcriptome or proteome profiling of bacteria isolated from plants may also give clues as to possible LPS modifications.
Intriguingly, although lipid A-like molecules have not been reported in plants, many plants, including Arabidopsis, encode full-length nuclear orthologs of six of the nine enzymes of the E. coli biosynthetic genes for lipid A. Arabidopsis mutants generated by knock-out of these genes are viable under laboratory conditions. However, they accumulate (wild type) or lose (mutant) the expected lipid A precursors (Li et al., 2011). The lipid A biosynthetic genes of higher plants may have been acquired from Gram-negative bacteria with the endosymbiosis of mitochondria. Plant lipid A may therefore play a structural role in mitochondrial or perhaps chloroplast membranes. Alternatively, lipid A-like molecules in Arabidopsis may be involved in signal transduction of plant defense responses. Although the mechanisms by which plants detect LPS remain unknown, lipid A-like molecules in plants might serve as signals to regulate cellular responses during plant pathogen invasion.
Activator of XA21-Mediated Immunity (Ax21)
Even though the rice receptor XA21 has been known for a long time, the corresponding ligand Ax21 (previously known as avrXa21) was identified only recently (Lee et al., 2009). The conservation of Ax21 in all sequenced Xanthomonas spp., Xylella fastidiosa, and the human pathogen Stenotrophomonas maltophila suggests that it plays a key role in a biological function. Ax21 encodes a 194 aa protein (Bogdanove et al., 2011). The minimal recognized epitope mimicking Ax21 activity is a 17 aa sulfated peptide, called axYs22, which has been shown to be 100% identical among six different Xanthomonas spp. (Lee et al., 2009). XA21, is, together with FLS2 and EFR, among the best studied PRRs, they all belong to subfamily LRR XII of the non-RD class of receptor kinases (Shiu and Bleecker, 2003; Shiu et al., 2004; Dardick and Ronald, 2006). Xa21 was originally identified as a dominant-resistant locus conferring resistance to multiple Xoo races in the wild rice species O. longistaminata (Khush et al., 1990). Xa21 maps to chromosome 11, and already upon its discovery it was speculated to encode a gene product recognizing a determinant present in all Xoo races (Ronald et al., 1992). Later, the resistance of locus Xa21 was linked to a single gene, also named Xa21, encoding a receptor kinase-like protein with predicted LRR, TM, juxtamembrane (JM), and intracellular kinase domains and that this single gene was sufficient to confer resistance to a number of Xoo isolates (Song et al., 1995; Wang et al., 1996). Xa21 is a member of a gene family with at least seven members in rice. The closest relative to Xa21 is Xa21D and the spectrum of resistance is identical between the two genes, but the level of resistance differs as XA21D only confers partial resistance. The LRR domains of Xa21 and Xa21D are more than 99% identical, but Xa21D lacks the TM and kinase domains and it may therefore have an extracellular function, but the mode of action is unknown (Song et al., 1997; Wang et al., 1998).
Several proteins have been shown to interact with XA21. The ATPase XB24 (XA21-binding protein 24) promotes autophosphorylation of XA21, which is thereby kept in an inactive state. When Ax21 binds to XA21, the XB24/XA21 protein complex probably dissociates, and XA21 is activated (Chen et al., 2010). After activation, the phosphatase XB15 (XA21-binding protein 15) dephosphorylates XA21 in order to deactivate it again (Park et al., 2008). A recent study has shown that upon Ax21 recognitions by XA21, the intercellular kinase domain is released and translocated to the cell nucleus, a translocation that is necessary for the XA21-mediated immune response (Park and Ronald, 2012). The rice transcription factor OsWKKY62 (also called XB10), which has previously been shown to be a negative regulator of XA21 activity, is needed for this translocation (Peng et al., 2008; Park and Ronald, 2012). The E3 ubiquitin kinase XB3 (XA21-binding protein 3) is important for XA21-mediated resistance, as rice lines silenced in XB3 both has a decreased level of XA21 and display reduced resistance to Xoo (Wang et al., 2006). Less thoroughly studied are the genes Rox1, 2, and 3 (Regulator of XA21-mediated immunity 1, 2, and 3 encoding a thiamine phosphokinase, a NOL1/NOL2/sun gene family member and a nuclear migration protein, respectively), which have also been shown to affect Xoo resistance in XA21-containing rice plants (Lee et al., 2011).
Interestingly, the Arabidopsis FLS2 and EFR receptors bind the artificial Ax21 derived peptide axYs22-A1, this binding trigger responses similar to the ones triggered by Flg. axYs22-A1 is identical to axYs22, except that the first aa in the peptide has been changed from Ala to Glu (Danna et al., 2011). It was previously thought that FLS2 was specific to Flg. Even though the authors analyzed their axYs22-A1 peptide stocks for the presence of flg22 by mass spectrometry, a question was later raised whether the observations were caused by flg22 contamination (Danna et al., 2011; Mueller et al., 2012). Mueller et al. (2012) had observed incidences of commercially produced peptides contaminated with flg22 and even minute amounts (in the range of 1 ppm) will activate FLS2 responses. Furthermore, Arabidopsis cell cultures did not respond to treatment with axYs22 in their laboratory, therefore they concluded that the FLS2 binding observed by Danna et al. (2011) could be caused by contamination (Mueller et al., 2012). These concerns were dismissed by Danna et al. (2012), who believe that the difference in peptides used (axYs22-A1 vs. axYs22) and the differences in their experimental set-ups, could explain different results.
Ax21 is secreted from the bacterial cell through the TOSS—a fact that has been known longer than the identity of Ax21 (da Silva et al., 2004). TOSS is a relatively simple system consisting of only three protein subunits: a membrane fusion protein (MFP), an adenosine triphosphate-binding cassette (ABC) transporter, and an outer membrane protein (OMP). Three Xoo genes with homology to the TOSS components have been shown to be required for Ax21 activity (the so called rax genes). The genes raxA, raxB, and raxC are identified as coding for a MFP, an ABC transporter, and an OMP, respectively (da Silva et al., 2004). raxA and raxB are arranged in a putative operon (called raxSTAB) together with the gene raxST, which is not a part of the TOSS (da Silva et al., 2004). Instead raxST is a sulfotransferase (Shuguo et al., 2012), which catalyze the transfer of sulfate from PAPS (3′-phosphoadenosine 5′-phosphosulfate) to a tyrosine residue. Sulfation of secreted peptides is often important for their biological function. This is also the case for the Ax21-derived peptide axYs22, as a non-sulfated version of the peptide, axY22 is not recognized by XA21 (Lee et al., 2009). Based on genetic similarity and complementation studies, the two genes raxP and raxQ have been suggested to be responsible for the PAPS synthesis as they encode proteins with ATP sulfohydrolase and APS kinase activities (Shen et al., 2002). Downstream of raxSTAB another putative operon, comprised of the two genes raxR and raxH, has been found. Xoo strains mutated in these two genes do not express full Ax21 activity. These two genes are probably encoding proteins involved in a bacterial two component regulatory system (a response regulator and a histidine protein kinase), and they could be involved in the regulation of a number of genes (Burdman et al., 2004). The two component system composed of RaxR and RaxH have also been found to regulate the expression of another two component system composed of PhoP and PhoQ. The PhoPQ system also seems to control the TTSS, important for delivering bacterial effector molecules to the host cell, through regulation of the hrpG gene (Lee et al., 2008).
As Ax21 was shown to be a secreted molecule (Lee et al., 2009) and the finding that the expression of raxST, raxP, raxR, and raxC are density-dependent it was suggested that Ax21 is a quorum sensing (QS) molecule (Lee et al., 2006). This was supported by a finding in S. maltophila, showing that mutants lacking Ax21 display reduced motility and biofilm formation. Also in this organism it appears that RaxH and RaxR are part of a two-component system (McCarthy et al., 2011). Han et al. (2011) published the evidence for this hypothesis, thereby making Ax21 the first QS factor also functioning as a MAMP. But unfortunately critical errors of a central Xoo strain used in the study has been found and the authors of the original paper are now in the process of repeating the experiments with a new validated strain in order to confirm the results (Comment on the PLoS homepage since January 2013). The outcome of these experiments should be followed with great interest. Knowledge of detection of small proteins, like QS in rice and other species, can be used to develop reagents to disrupt QS-mediated virulence activities.
Fungal and Oomycete MAMPs
Chitin and β-Glucan
Examples of MAMPs from fungi and oomycetes include the fungal chitin and β-glucan from P. megasperma. However, data describing how these MAMPs are recognized and how the following signal transduction is mediated has only in a few cases been accomplished.
In fungal cell walls branched β-glucan is cross-linked to chitin and in oomycetes to cellulose. In soybean the PRR recognizing P. megasperma β-glucan was identified as the β-glucan binding protein (GBP) (Umemoto et al., 1997). This MAMP and its corresponding PRR has not been studied further. On the other hand chitin and its fragments chitin oligosaccharides have been shown to trigger defense responses in both monocots and dicots. Together with CEBiP, CERK1 recognizes fungal chitin (Kaku et al., 2006; Shimizu et al., 2010). In rice, the RLP CEBiP binds chitin oligosaccharides at the cell surface and interacts with the LysM-RLK OsCERK1 for signaling. In Arabidopsis, only the LysM-containing RLK CERK1 was found to be essential for chitin elicitor signaling (Miya et al., 2007). Three CEBiP-like proteins, LYM1-3, have been identified in Arabidopsis. Using heterologous expression of these three Arabidopsis CEBiP homologous in tobacco BY-2 cells Shinya et al. (2012) tested for their ability to bind chitin oligosaccharides and found that only LYM2, also referred to as AtCEBiP, showed high affinity binding to chitin oligosaccharides. Even though affinity labeling with biotinylated (GlcNAc)8 indicated that AtCEBiP represent a cell surface chitin-binding protein, knockout (KO) of AtCEBiP, LYM1, or LYM3, single or triple KO, together with AtCEBiP overexpression studies suggested that AtCEBiP does not contribute to chitin signaling in Arabidopsis (Shinya et al., 2012). These studies reveal that Arabidopsis and rice exploit different chitin receptor systems. Similar results were obtained by Wan et al. (2012) who showed that mutations in each of the three Arabidopsis CEBiP-like proteins 1, 2, or 3, or in a combination resulting in a triple mutant, had no effect on the plant response to chitin. Arabidopsis has five LysM RLKs1-5 (Lyk1, 2, 3, 4, and 5) one of them, Lyk1, is also known as CERK1. Wan et al. (2012) tested the Arabidopsis lyk2, 3, 4, and 5 KO mutants, respectively, to see if they were involved in chitin signaling. They found that the plant immune response to chitin was reduced only in the lyk4 mutant suggesting Lyk4 to be involved in a chitin recognition receptor complex (Wan et al., 2012). Furthermore, the lyk4 plants were more susceptible to the fungal pathogen Alternaria brassicicola and the bacterial pathogen Pst DC3000 than wild type plant (Wan et al., 2012). In addition to this it has been reported that two rice lysine-motif containing proteins, LYP4 and LYP6, could bind both PGN and chitin acting as dual functional PRRs in rice innate immunity (Liu et al., 2012). Their results further suggest that overlapping perception systems exist for bacterial PGN and fungal chitin in rice. In contrast, LYM1 and LYM3 the orthologs of LYP4 and LYP6 in Arabidopsis were only able to bind PGN and not chitin (Willmann et al., 2011). Further details on PGN recognition can be found in the text above.
Ave1 Peptide and Ethylene-Inducing Xylanase (EIX)
In tomato a Verticillium resistance locus Ve was identified that mediates resistance against race 1 strains of Verticillium dahlia and V. albo-atrium, respectively (Kawchuk et al., 2001). The characterization of the Ve locus revealed two genes Ve1 and Ve2 that encode cell-surface receptors belonging to the LRR class of RLP. Only Ve1 was found to confer resistance in tomato. Moreover, tomato plants silenced in BAK1, showed higher susceptibility to infection with Verticillium indicating that BAK1 is involved in Ve1-induced defense responses in tomato (Fradin et al., 2009). A putative ligand for the LRR-RLP Ve1 is the Ave1 (avirulence on Ve1 tomato) peptide. Ave1, a conserved peptide identified in several fungi and in the plant pathogenic bacteria Xanthomonas axonopodis pv. citri, has been found to have homology to plant natriuretic peptides (PNPs). PNPs are extracellular signaling molecules that have been shown to have a role in regulation of homeostasis under several stress conditions (Wang et al., 2011). Ave1 acts as an elicitor of disease resistance mediated by the LRR-RLP Ve1 in tomato (de Jonge et al., 2012), but a direct binding between Ave1 and Ve1 remains to be shown. Ve1 has been referred to as a PRR or an R protein accompanied by speculations that the Ave1 peptide could be an effector acting as a MAMP (Thomma et al., 2011). Future results will reveal, if it is possible to differentiate as strictly, as we do today, between MAMPs and effectors, as between PRRs and R proteins.
Two other PRRs in tomato, the LRR-RLPs SlEix1 and SlEix2, which have been shown to have homology to the tomato Ve and Cf PRRs, recognize the fungal ethylene-inducing xylanase (EIX) (Ron and Avni, 2004). EIX is a 22-kD fungal protein (β-1-4-endoxylanase) from Trichoderma viride that independent of its endoxylanase activity can act as an elicitor of defense responses in tomato and tobacco plants (Furman-Matarasso et al., 1999; Ron and Avni, 2004). The aa sequence of SlEix1 and SlEix2 are 81.4% identical, SlEix1 and SlEix2 both bind EIX, but their functions differ. The SlEix2 receptor has been shown to be internalized upon EIX application (Bar and Avni, 2009) and only SlEix2 transmits the signal mediated by EIX leading to plant immune responses (Ron and Avni, 2004). SlEix1, on the other hand, block the EIX signaling and the authors suggested that SlEix1 functions in inhibition of plant defense signaling and plant cell death in response to EIX (Bar et al., 2010). Using BAK1-silenced tobacco plants Bar et al. (2010) further showed that BAK1 was required for this inhibitory activity of SlEix1 on SlEix2 signaling and endocytosis.
Damage-Associated Molecular Patterns (DAMPs)
The plant defense system is not only recognizing microbial elicitors, some plant-derived molecules also induce plant defense responses. This sensing of infectious-self or modified-self is mediated by DAMPs (Seong and Matzinger, 2004; Boller and Felix, 2009), also referred to as microbe-induced molecular patterns (MIMPs, Mackey and McFall, 2006). Similarly the mammalian immune system detects “danger” through a series of DAMPs, now also in in this system named damage-associated. The mammalian DAMPs are derived from other tissues activating intracellular cascades that lead to an inflammatory response (Lotze et al., 2007).
In Plants the 18 aa tomato peptide systemin is an endogenous elicitor of plant defense (Pearce et al., 1991; McGurl et al., 1992). The systemin precursor prosystemin is a cytoplasmic protein and upon cell damage the released systemin acts as a DAMP on surrounding cells (Narváez-Vásquez and Ryan, 2004). Early reports showed that the RLK SR160 (the tomato ortholog of BRI1) was the receptor for systemin (Scheer and Ryan, 1999, 2002), but later reports found that null mutants were sensitive to systemin (Holton et al., 2007; Lanfermeijer et al., 2008). Also in Arabidopsis a system with a putative cytosolic peptide (Pep1) activates transcription of defense-related genes and induces alkalization in cell cultures. The 23 aa Pep1 and the seven homologous in Arabidopsis (Pep 1–7) are derived from the C-terminal part of their precursor proteins PROPEP1–7 (Huffaker et al., 2006). The Pep1 receptor, PEPR1, was found to be a LRR receptor belonging to the LRR XI subfamily (Yamaguchi et al., 2006). Based on sequence similarity a second receptor of Pep-peptides, PEPR2, has been identified. Transcription of both PEPR1 and 2 is activated by wounding, Methyl-Jasmonate (MeJA), Pep-peptides, and specific MAMPs. A level of redundancy is found regarding ligand specificity of PEPR1 and 2 as they both bind Pep1 and 2, and in addition PEPR1 binds Pep3–6 (Yamaguchi et al., 2010).
Oligogalacturonides (OG) and cutin released from plant cell walls also function as DAMPs (Schweizer et al., 1996; Denoux et al., 2008). Using a domain swap approach Brutus et al. (2010) proved that WAK1 (Wall-Associated Kinase 1) function as an OG receptor whereas a receptor for cutin still remains to be found.
Concluding Remarks
Even though MAMPs are much conserved, they are under selective pressure in adapted pathogens to evade recognition. For example in the case of bacterial Flg, a potent inducer of MTI in most plants, mutations in key residues of the flg22 epitope that abolish recognition by the receptor FLS2 have been selected for in several plant pathogens and symbionts (Boller and Felix, 2009; Lopez-Gomez et al., 2012). Notably, this positive selection seems to be more rapid than previously thought, as modern natural isolates of Pst adapt to their tomato host through non-synonymous mutations in the Flg-encoding gene fliC (Cai et al., 2011). The best example so far of a glycosylated MAMP not being recognized in plants, is the LPS molecule from the nitrogen-fixing soil bacterium Bradyrhizobium sp. BTAi1, this LPS does not trigger the innate immune response in different plant families. Aeschynomene indica (the natural host of Bradyrhizobium), Lotus japonicus, and Arabidopsis were tested for perception of Bradyrhizobium LPS. Defense responses were not induced in any of the tested plants. The authors determined the structure of Bradyrhizobium LPS and found a unique LPS with an, in nature, unprecedented chemical structure of the monosaccharide forming the polymer, this “different” structure probably prevents recognition by the LPS receptor complex in plants (Silipo et al., 2011). MAMPs are necessary for microbial life and therefore under strong negative selection, but their immunogenic epitopes are under positive selection to evade host immune detection. These opposing evolutionary forces were recently used to identify novel candidate MAMPs from Pseudomonas and Xanthomonas species through an innovative bioinformatics approach. Identifying new MAMPs may prove to be a source of new antimicrobial agents (McCann et al., 2012).
Although plant receptors for bacterial PGN and the proteinaceous MAMPs Flg and EF-Tu elongation factor have been identified, those involved in perception of LPS remain obscure. In conclusion we expect that in the next few years we will see a substantial increase in our understanding of the processes of MAMPs perception and signal transduction in plants through the deployment of cross disciplinary approaches and ever expanding ranges of molecular experimental tools. Despite their critical role in immunity, we know remarkably little about the range and diversity of MAMPs. Most studies have focused on a limited number of MAMPs as described in this review. The identification of new MAMPs will give insight into the molecular and evolutionary mechanisms underlying host-pathogen interactions, and greater understanding of the mechanisms by which MAMPs elicits defense responses may have considerable impact on the improvement of plant health and disease resistance.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Gitte Erbs and Mari-Anne Newman acknowledge funding by The Danish Council for Independent Research, Technology, and Production Sciences (FTP) and VILLUM FONDEN, Denmark.
References
Aslam, S. N., Erbs, G., Morrissey, K. L., Newman, M.-A., Chinchilla, D., Boller, T., et al. (2009). MAMPs signatures, synergy, size and charge: influences on perception or mobility and host defense responses. Mol. Plant Pathol. 10, 375–387.
Ausubel, F. (2005). Are innate immune signalling pathways in plants and animals conserved? Nat. Immunol. 6, 973–979.
Bailey, B. A., Dean, J. F. D., and Anderson, J. D. (1990). An ethylene biosynthesis inducing endoxylanase elicits electrolyte leakage and necrosis in Nicotiana tabacum cv xanthi Leaves. Plant Physiol. 94, 1849–1854.
Bar, M., and Avni, A. (2009). EHD2 inhibits ligand-induced endocytosis and signaling of the leucine-rich repeat receptor-like protein LeEix2. Plant J. 59, 600–611.
Bar, M., Sharfman, M., Ron, M., and Avni, A. (2010). BAK1 is required for the attenuation of ethylene-inducing xylanase (Eix)-induced defense responses by the decoy receptor LeEix1. Plant J. 63, 791–800.
Bauer, Z., Gomez-Gomez, L., Boller, T., and Felix, G. (2001). Sensitivity of different ecotypes and mutants of Arabidopsis thaliana toward the bacterial elicitor flagellin correlates with the presence of receptorbinding sites. J. Biol. Chem. 276, 45669–45676.
Bedini, E., De Castro, C., Erbs, G., Mangoni, L., Dow, J. M., Newman, M.-A., et al. (2005). Structure-dependent modulation of a pathogen response in plants by synthetic O-antigen polysaccharides. J. Am. Chem. Soc. 127, 2414–2416.
Bedini, E., Parrilli, M., and Unverzagt, C. (2002). Oligomerization of a rhamnanic trisaccharide repeating unit of O-chain polysaccharides from phytopathogenic bacteria. Tetrahedron Lett. 43, 8879–8882.
Bent, A. F., and MacKey, D. (2007). Elicitors, effectors, and R genes: the new paradigm and a lifetime supply of questions. Annu. Rev. Phytopathol. 45, 399–436.
Bogdanove, A. J., Koebnik, R., Lu, H., Furutani, A., Angiuoli, S. V., Patil, P. B., et al. (2011). Two new complete genome sequences offer insight into host and tissue specificity of plant pathogenic Xanthomonas spp. J. Bacteriol. 193, 5450–5464.
Boller, T., and Felix, G. (2009). A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60, 379–406.
Braun, S. G., Meyer, A., Holst, O., Pühler, A., and Niehaus, K. (2005). Characterization of the Xanthomonas campestris pv. campestris lipopolysaccharide substructures essential for elicitation of an oxidative burst in tobacco cells. Mol. Plant Microbe Interact. 18, 674–681.
Brutus, A., Sicilia, F., Macone, A., Cervone, F., and De Lorenzo, G. (2010). A domain swap approach reveals a role of the plant wall-associated kinase 1 (WAK1) as a receptor of oligogalacturonides. Proc. Natl. Acad. Sci. U.S.A. 107, 9452–9457.
Burdman, S., Shen, Y., Lee, S. W., Xue, Q., and Ronald, P. C. (2004). RaxH/RaxR: a two-component regulatory system in Xanthomonas oryzae pv. oryzae required for AvrXa21 activity. Mol. Plant-Microbe Interact. 17, 602–612.
Cai, R., Lewis, J., Yan, S., Liu, H., Clarke, C. R., Campanile, F., et al. (2011). The plant pathogen Pseudomonas syringae pv. tomato is genetically monomorphic and under strong selection to evade tomato immunity. PLoS Pathog. 7:e1002130. doi: 10.1371/journal.ppat.1002130
Chamaillard, M., Hashimoto, M., Horie, Y., Masumoto, J., Qiu, S., Saab, L., et al. (2003). An essential role for NOD1 in host recognition of bacterial peptidoglycan containing diaminopimelic acid. Nat. Immunol. 4, 702–707.
Chen, X., Chern, M., Canlas, P. E., Ruan, D., Jiang, C., and Ronald, P. C. (2010). An ATPase promotes autophosphorylation of the pattern recognition receptor XA21 and inhibits XA21-mediated immunity. Proc. Natl. Acad. Sci. U.S.A. 107, 8029–8034.
Cheong, J.-J., and Hahn, M. G. (1991). A specific, high-affinity binding site for the hepta-β-glucoside elicitor exists in soybean membranes. Plant Cell 3, 137–147.
Chinchilla, D., Bauer, Z., Regenass, M., Boller, T., and Felix, G. (2006). The Arabidopsis receptor kinase FLS2 binds flg22 and determines the specificity of flagellin perception. Plant Cell 18, 465–476.
Chinchilla, D., Zipfel, C., Robatzek, S., Kemmerling, B., Nurnberger, T., Jones, D. G. J., et al. (2007). A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defense. Nature 448, 497–500.
Csorba, T., Pantaleo, V., and Burgyán, J. (2009). RNA silencing: an antiviral mechanism. Adv. Virus Res. 75, 35–71.
da Silva, F. G., Shen, Y., Dardick, C., Burdman, S., Yadav, R. C., de Leon, A. L., et al. (2004). Bacterial genes involved in type I secretion and sulfation are required to elicit the rice Xa21-mediated innate immune response. Mol. Plant-Microbe Interact. 17, 593–601.
Danna, C. H., Millet, Y. A., Koller, T., Han, S. W., Bent, A. F., Ronald, P. C., et al. (2011). The Arabidopsis flagellin receptor FLS2 mediates the perception of Xanthomonas Ax21 secreted peptides. Proc. Natl. Acad. Sci. U.S.A. 108, 9286–9291.
Danna, C. H., Zhang, X. C., Khatri, A., Bent, A. F., Ronald, P. C., and Ausubel, F. M. (2012). FLS2-mediated responses to Ax21-derived peptides: response to the Mueller et al. commentary. Plant Cell 24, 3174–3176.
Dardick, C., and Ronald, P. (2006). Plant and animal pathogen recognition receptors signal through non-RD kinases. PLoS Pathog. 2:e2. doi: 10.1371/journal.ppat.0020002
Darvill, A. G., and Albersheim, P. (1984). Phytoalexins and their elicitors-a defense against microbial infection in plants. Annu. Rev. Plant Physiol. 35, 243–275.
de Jonge, R., Peter van Esse, H., Maruthachalam, K., Bolton, M. D., Santhanam, P., Saber, M. K., et al. (2012). Tomato immune receptor Ve1 recognizes effector of multiple fungal pathogens uncovered by genome and RNA sequencing. Proc. Natl. Acad. Sci. U.S.A. 109, 5110–5115.
Denoux, C., Galletti, R., Mammarella, N., Gopalan, S., Werck, D., De Lorenzo, G., et al. (2008). Activation of defense response pathways by OGs and Flg22 elicitors in Arabidopsis seedlings. Mol. Plant 1, 423–445.
Desaki, Y., Miya, A., Venkatesh, B., Tsuyumu, S., Yamane, H., Kaku, H., et al. (2006). Bacterial lipopolysaccharides induce defense responses associated with programmed cell death in rice cells. Plant Cell Physiol. 47, 1530–1540.
Desaki, Y., Otomo, I., Kobayashi, D., Jikumaru, Y., Kamiya, Y., Venkatesh, B., et al. (2012). Positive crosstalk of MAMP signaling pathways in rice cells. PLoS ONE 7:e51953. doi: 10.1371/journal.pone.0051953
Dow, M., Newman, M.-A., and von Roepenack, E. (2000). The induction and modulation of plant defense responses by bacterial lipopolysaccharides. Annu. Rev. Phytopathol. 38, 241–261.
Dow, J. M., Osbourn, A. E., Wilson, T. J. G., and Daniels, M. J. (1995). A locus determining pathogenicity of Xanthomonas-campestris is involved in lipopolysaccharide biosynthesis. Mol. Plant Microbe Interact. 8, 768–777.
Dziarski, R., and Gupta, D. (2006). The peptidoglycan recognition proteins (PGRPs). Genome Biol. 7, 232–245.
Erbs, G., and Newman, M.-A. (2012). The role of lipopolysaccharide and peptidoglycan, two glycosylated bacterial microbe-associated molecular patterns (MAMPs), in plant innate immunity. Mol. Plant Pathol. 13, 95–104.
Erbs, G., Silipo, A., Aslam, S., De Castro, C., Liparoti, V., Flagiello, A., et al. (2008a). Peptidoglycan and muropeptides from pathogens Agrobacterium and Xanthomanas elicit innate immunity: structure and activity. Chem. Biol. 15, 438–448.
Erbs, G., Jensen, T. T., Silipo, A., Grant, W., Dow, J. M., Molinaro, A., et al. (2008b). An antagonist of lipid A action in mammals has complex effects on lipid A induction of defense responses in the model plant Arabidopsis thaliana. Microbes Infect. 10, 571–574.
Felix, G., and Boller, T. (2003a). Molecular sensing of bacteria in plants. J. Biol. Chem. 278, 6201–6208.
Felix, G., and Boller, T. (2003b). Molecular sensing of bacteria in plants - the highly conserved RNA-binding motif RNP-1 of bacterial cold shock proteins is recognized as an elicitor signal in tobacco. J. Biol. Chem. 278, 6201–6208.
Felix, G., Duran, J. D., Volko, S., and Boller, T. (1999). Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 18, 265–276.
Felix, G., Regenass, M., and Boller, T. (1993). Specific perception of subnanomolar concentrations of chitin fragments by tomato cells—induction of extracellular alkalinization, changes in protein phosphorylation, and establishment of a refractory state. Plant J. 4, 307–316.
Fischer, S. E., Miguel, M. J., and Mori, G. B. (2003). Effect of root exudates on the exopolysaccharide composition and the lipopolysaccharide profile of Azospirillum brasilense Cd under saline stress. FEMS Microbiol. Lett. 219, 53–62.
Fradin, E. F., Zhang, Z., Ayala, J. C. J., Castroverde, C. D. M., Nazar, R. N., Robb, J., et al. (2009). Genetic dissection of Verticillium wilt resistance mediated by tomato Ve1. Plant Physiol. 150, 320–332.
Furman-Matarasso, N., Cohen, E., Du, Q., Chejanovsky, N., Hanania, U., and Avni, A. (1999). A point mutation in the ethylene-inducing xylanase elicitor inhibits the β-1-4-endoxylanase activity but not the elicitation activity. Plant Physiol. 121, 345–351.
Gaulin, E., Dramé, N., Lafitte, C., Torto-Alalibo, T., Martinez, Y., Ameline-Torregrosa, C., et al. (2006). Cellulose binding domains of a Phytophthora cell wall protein are novel pathogen-associated molecular patterns. Plant Cell 18, 1766–1777.
Gerber, I. B., Laukens, K., De Vijlder, T., Witters, E., and Dubery, I. A. (2008). Proteomic profiling of cellular targets of lipopolysaccharide-induced signaling in Nicotiana tabacum BY-2 cells. Biochim. Biophys. Acta 1784, 1750–1762.
Gerber, I. B., Laukens, K., Witters, E., and Dubery, I. A. (2006). Lipopolysaccharide-responsive phosphoproteins in Nicotiana tabacum cells. Plant Physiol. Biochem. 44, 369–379.
Gimenez-Ibanez, S., Ntoukakis, V., and Rathjen, J. P. (2009). The LysM receptor kinase CERK1 mediates bacterial perception in Arabidopsis. Plant Signal. Behav. 4, 539–541.
Girardin, S. E., Travassos, L. H., Herve, M., Blanot, D., Boneca, I. G., Philpott, D. J., et al. (2003). Peptidoglycan molecular requirements allowing detection by Nod1 and Nod2. J. Biol. Chem. 278, 41702–41708.
Gómez-Gómez, L., Bauer, Z., and Boller, T. (2001). Both the extracellular leucine-rich repeat domain and the kinase activity of FLS2 are required for flagellin binding and signalling in Arabidopsis. Plant Cell 13, 1155–1163.
Gomez-Gomez, L., and Boller, T. (2000). FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 5, 1003–1011.
Greenberg, J. T. (1997). Programmed cell death in plant-pathogen interactions. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 525–545.
Gross, A., Kapp, D., Nielsen, T., and Niehaus, K. (2005). Endocytosis of Xathomonas campestris pathovar campestris lipopolysaccharides in non-host plant cells of N. benthamiana. New Phytol. 165, 215–226.
Gust, A., Biswas, R., Lenz, H. D., Rauhut, T., Ranf, S., Kemmerling, B., et al. (2007). Bacteria-derived peptidoglycans constitute pathogen-associated molecular patterns triggering innate immunity in Arabidopsis. J. Biol. Chem. 282, 32338–32348.
Gust, A. A., Willmann, R., Desaki, Y., Grabherr, H. M., and Nürnberger, T. (2012). Plant LysM proteins: modules mediating symbiosis and immunity. Trends Plant Sci. 17, 495–502.
Gutsmann, T., Schromm, A. B., and Brandenburg, K. (2007). The physiochemistry of endotoxins in relation to bioactivity. J. Med. Microbiol. 297, 341–352.
Hammond-Kosack, K. E., and Jones, J. D. (1996). Resistance gene-dependent plant defense responses. Plant Cell 8, 1773–1791.
Han, S. W., Sriariyanum, M., Lee, S. W., Sharma, M., Bahar, O., Bower, Z., et al. (2011). Small protein-mediated quorum sensing in a gram negative bacterium. PLoS ONE 6:e29192. doi: 10.1371/journal.pone.0029192
Häweker, H., Rips, S., Koiwa, H., Salomon, S., Saijo, Y., Chinchilla, D., et al. (2010). Pattern recognition receptors require N-glycosylation to mediate plant immunity. J. Biol.Chem. 285, 4629–4636.
Hayashi, F., Smith, K. D., Ozinsky, A., Hawn, T. R., Yi, E. C., Goodlett, D. R., et al. (2001). The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature 410, 1099–1103.
He, P., Shan, L., Lin, N.-C., Martin, G. B., Kemmerling, B., Nürnberger, T., et al. (2006). Specific bacterial suppressors of MAMP signalling upstream of MAPKKK in Arabidopsis innate immunity. Cell 125, 563–575.
Holton, N., Caño-Delgado, A., Harrison, K., Montoya, T., Chory, J., and Bishop, G. J. (2007). Tomato BRASSINOSTEROID INTENSITIVE1 is required for systemin-induced root elongation in Solanum pimpinellifolium but is not essential for wound signaling. Plant Cell 19, 1709–1717.
Huffaker, A., Pearce, G., and Ryan, C. A. (2006). An endogenous peptide signal in Arabidopsis activates components of the innate immune response. Proc. Natl. Acad. Sci. U.S.A. 103, 10098–10103.
Ialenti, A., Di Meglio, P., Grassia, G., Maffia, P., Di Rosa, M., Lanzetta, R., et al. (2006). A novel lipid A from Halomonas magadiensis inhibits enteric LPS-induced human monocyte activation. Eur. J. Immunol. 36, 354–360.
Inohara, N., Chamaillard, M., McDonald, C., and Nunez, G. (2005). NOD-LRR proteins: role in host-microbial interactions and inflammatory disease. Annu. Rev. Biochem. 74, 355–383.
Jamir, Y., Guo, M., Oh, H.-S., Petnicki-Ocwieja, T., Chen, S., Tang, X., et al. (2004). Identification of Pseudomonas syringae type III effectors that can suppress programmed cell death in plants and yeast. Plant J. 37, 554–565.
Jeppesen, M. G., Navratil, T., Spremulli, L. L., and Nyborg, J. (2005). Crystal structure of the bovine mitochondrial elongation factor Tu.Ts complex. J. Biol. Chem. 280, 5071–5081.
Jiang, S., Naito, M., Kaizu, C., Kuwata, K., Hasegawa, G., Mukaida, N., et al. (2000). Lipopolysaccharide-induced cytokine and receptor expression and neutrophil infiltration in the liver of osteopetrosis (op/op) mutant mice. Liver 20, 465–474.
Kaku, H., Nishizawa, Y., Ishll-Minami, N., Akimoto-Tomlyama, C., Dohmae, N., Taklo, K., et al. (2006). Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc. Natl. Acad. Sci. U.S.A. 103, 11086–11091.
Kannenberg, E. L., and Carlson, R. W. (2001). Lipid A and O-chain modifications cause rhizobium lipopolysaccharides to become hydrophobic during bacteroid development. Mol. Microbiol. 39, 379–391.
Kauss, H., Fauth, M., Merten, A., and Jeblick, W. (1999). Cucumber hypocotyls respond to cutin monomers via both an inducible and a constitutive H2O2-generating system. Plant Physiol. 120, 1175–1182.
Kawchuk, L., Hachey, J., Lynch, D. R., Klcsar, F., van Rooijen, G., Waterer, D. R., et al. (2001). Tomato Ve disease resistance genes encode cell surface-like receptors. Proc. Natl Acad. Sci. U.S.A. 98, 6511–6515.
Khush, G. S., Bacalangco, E., and Ogawa, T. (1990). A new resistance gene to bacterial blight derived from O. longistaminata. Rice Genet. 7, 121–122.
Kunze, G., Zipfel, C., Robatzek, S., Niehaus, K., Boller, T., and Felix, G. (2004). The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cell 16, 3496–3507.
Lamb, C., and Dixon, R. A. (1997). The oxidative burst in plant disease resistance. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 251–275.
Lanfermeijer, F. C., Staal, M., Malinowski, R., Stratmann, J. W., and Elzenga, T. M. (2008). Micro-electrode flux estimation confirms that the Solanum pimpinellifolium cu3 mutant still responds to systemin. Plant Physiol. 146, 129–139.
Lee, I., Seo, Y. S., Coltrane, D., Hwang, S., Oh, T., Marcotte, E. M., et al. (2011). Genetic dissection of the biotic stress response using a genome-scale gene network for rice. Proc. Natl. Acad. Sci. U.S.A. 108, 18548–18553.
Lee, S. W., Han, S. W., Sririyanum, M., Park, C. J., Seo, Y. S., and Ronald, P. C. (2009). A type I-secreted, sulfated peptide triggers XA21-mediated innate immunity. Science 326, 850–853.
Lee, S.-W., Han, S.-W., Bartley, L. E., and Ronald, P. C. (2006). Unique characteristics of Xanthomonas oryzae pv. oryzae AvrXa21 and implications for plant innate immunity. Proc. Natl. Acad. Sci. U.S.A. 103, 18395–18400.
Lee, S. W., Jeong, K. S., Han, S. W., Lee, S. E., Phee, B. K., Hahn, T. R., et al. (2008). The Xanthomonas oryzae pv. oryzae PhoPQ two-component system is required for AvrXA21 activity, hrpG expression, and virulence. J. Bacteriol. 190, 2183–2196.
Leeman, M., Vanpelt, J. A., Denouden, F. M., Heinsbroek, M., Pahm, B., and Schippers, B. (1995). Induction of systemic resistance against fusarium-wilt of radish by lipopolysaccharides of Pseudomonas fluorescens. Phytopathol 85, 1021–1027.
Li, C., Guan, Z., Liu, D., and Raetz, C. R. H. (2011). Pathway for lipid A biosynthesis in Arabidopsis thaliana resembling that of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A 108, 11397–11392.
Liu, B., Li, J.-F., Ao, Y., Qu, J., Li, Z., Su, J., et al. (2012). Lysin motif-containing proteins LYP4 and LYP6 play dual roles in peptidoglycan and chitin perception in rice innate immunity. Plant Cell 24, 3406–3419.
Livaja, M., Zeidler, D., von Rad, U., and Durner, J. (2008). Transcriptional responses of Arabidopsis thaliana to the bacteria-derived PAMPs harpin and lipopolysaccharide. Immunobiology 213, 161–171.
Lopez-Gomez, M., Sandal, N., Stougaard, J., and Boller, T. (2012). Interplay of flg22-induced defense responses and nodulation in Lotus japonicas. J. Exp. Bot. 63, 393–401.
Loppnow, H., Brade, H., Durrbaum, I., Dinarello, C. A., Kusumoto, S., Rietschel, E. T., et al. (1989). IL-1 induction-capacity of defined lipopolysaccharide partial structures. J. Immunol. 142, 3229–3238.
Lotze, M. T., Zeh, H. J., Rubartelli, A., Sparvero, L. J., Amoscato, A. A., Washburn, N. R., et al. (2007). The grateful dead: damage associated molecular pattern molecules and reduction/oxidation regulates immunity. Immunol. Rev. 220, 60–81.
Lu, D., Wu, S., Gao, X., Zhang, Y., Shan, L., and He, P. (2010). A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc. Natl. Acad. Sci. U.S.A. 107, 496–501.
Mackey, D., and McFall, A. J. (2006). MAMPs and MIMPs: proposed classifications for inducers of innate immunity. Mol. Microbiol. 61, 1365–1371.
Madala, N. E., Leone, M. R., Molinaro, A., and Dubery, I. A. (2011). Deciphering the structural and biological properties of the lipid A moiety of lipopolysaccharides from Burkholderia cepacia strain ASP B 2D, in Arabidopsis thaliana. Glycobiology 21, 184–194.
Maekawa, T., Kufer, T. A., and Schulze-Lefert, P. (2011). NLR functions in plant and animal immune systems: so far and yet so close. Nat. Immunol. 12, 817–826.
Mateos, F. V., Rickauer, M., and Esquerré-Tugayé, M. T. (1997). Cloning and characterization of a cDNA encoding an elicitor of Phytophthora parasitica var. nicotianae that shows cellulose-binding lectin-like activities. Mol. Plant Microbe Interact. 10, 1045–1053.
McCann, H. C., Nahal, H., Thakur, S., and Guttman, D. S. (2012). Identification of innate immunity elicitors using molecular signatures of natural selection. Proc. Natl. Acad. Sci. U.S.A. 109, 4215–4220.
McCarthy, Y., Dow, J. M., and Ryan, R. P. (2011). The Ax21 protein is a cell-cell signal that regulates virulence in the nosocomial pathogen Stenotrophomonas maltophilia. J. Bacteriol. 193, 6375–6378.
McCoy, S. L., Kurtz, S. E., Hausman, F. A., Trune, D. R., Bennett, R. M., and Hefeneider, S. H. (2004). Activation of RAW264.7 macrophages by bacterial DNA and lipopolysaccharides increases cell surface DNA binding and internalization. J. Biol. Chem. 279, 17217–17223.
McDonald, C., Inohara, N., and Nuñez, G. (2005). Peptidoglycan signalling in innate immunity and inflammatory disease. J. Biol. Chem. 280, 20177–20180.
McGurl, B., Pearce, G., Orozco-Cardemas, M., and Ryan, C. A. (1992). Structure, expression, and antisese inhibition of the systemin precursor gene. Science 255, 1570–1573.
Meindl, T., Boller, T., and Felix, G. (2000). The bacterial elicitor flagellin activates its receptor in tomato cells according to the address-message concept. Plant Cell 12, 1783–1794.
Miyake, K. (2004). Innate recognition of lipopolysaccharide by Toll-like receptor 4-MD-2. Trends Microbiol. 12, 186–192.
Mishina, T. E., and Zeier, J. (2006). The Arabidopsis flavin-dependent monooxygenase FMO1 is an essential component of biologically induced systemic acquired resistance. Plant Physiol. 141, 1666–1675.
Mishina, T. E., and Zeier, J. (2007). Pathogen-associated molecular pattern recognition rather than development of tissue necrosis contributes to bacterial induction of systemic acquired resistance in Arabidopsis. Plant J. 50, 500–513.
Miya, A., Albert, P., Shinya, T., Desaki, Y., Ichimura, K., Shirasu, K., et al. (2007). CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 104, 19613–19618.
Mueller, K., Chinchilla, D., Albert, M., Jehle, A. K., Kalbacher, H., Boller, T., et al. (2012). Contamination risks in work with synthetic peptides: flg22 as an example of a pirate in commercial peptide preparations. Plant Cell 24, 3193–3197.
Munford, R. S., and Varley, A. W. (2006). Shield as signal: lipopolysaccharide and the evolution of immunity to Gram-negative bacteria. PLoS Pathog. 2:e67. doi: 10.1371/journal.ppat.0020067
Narváez-Vásquez, J., and Ryan, C. A. (2004). The cellular localization of prosystemin: a functional role for phloem parenchyma in systemic wound signaling. Planta 218, 360–369.
Newman, M.-A., Daniels, M. J., and Dow, J. M. (1995). Lipopolysaccharide from Xanthomonas campestris induces defense-related gene expression in Brassica campestris. Mol. Plant Microbe Interact. 8, 778–780.
Newman, M.-A., Dow, J. M., Molinaro, A., and Parrilli, M. (2007). Priming, induction and modulation of plant defense responses by bacterial lipopolysaccharides. J. Endotoxin Res. 13, 68–79.
Newman, M. A., von Roepenack-Lahaye, E., Parr, A., Daniels, M. J., and Dow, J. M. (2002). Prior exposure to lipopolysaccharide potentiates expression of plant defenses in response to bacteria. Plant J. 29, 487–495.
Nguyen, H. P., Chakravarthy, S., Velásquez, A. C., McLane, H. L., Zeng, L., Nakayashiki, H., et al. (2010). Methods to Study PAMP-triggered immunity using tomato and Nicotiana benthamiana. Mol. Plant Microbe Interact 23, 991–999.
Nomura, K., DebRoy, S., Lee, Y. H., Pumplin, N., Jones, J., and He, S. Y. (2006). A bacterial virulence protein suppresses host innate immunity to cause plant disease. Science 313, 220–223.
Nothnagel, E. A., McNeil, M., Albersheim, P., and Dell, A. (1983). Host-pathogen interactions: XXII. A galacturonic acid oligosaccharide from plant cell walls elicits phytoalexins. Plant Physiol. 71, 916–926.
Nürnberger, T., and Kemmerling, B. (2006). Receptor protein kinases – pattern recognition receptors in plant immunity. Trends Plant Sci. 11, 519–522.
Nürnberger, T., Nennstiel, D., Jabs, T., Sacks, W. R., Hahlbrock, K., and Scheel, D. (1994). High affinity binding of a fungal oligopeptide elicitor to parsley plasma membranes triggers multiple defense responses. Cell 78, 449–460.
Park, C. J., Peng, Y., Chen, X., Dardick, C., Ruan, D., Bart, R., et al. (2008). Rice XB15, a protein phosphatase 2C negatively regulates cell death and XA21-mediated innate immunity. PLoS Biol. 6:e231. doi: 10.1371/journal.pbio.0060231
Park, C. J., and Ronald, P. C. (2012). Cleavage and nuclear localization of the rice XA21 immune receptor. Nat. Commun. 3, 920.
Pearce, G., Strydom, D., Johnson, S., and Ryan, C. A. (1991). A polypeptide from tomato leaves induces wound-inducible proteinase inhibitor proteins. Science 253, 895–898.
Peng, Y., Bartley, L. E., Chen, X., Dardick, C., Chern, M., Ruan, R., et al. (2008). OsWRKY62 is a negative regulator of basal and Xa21-mediated defense against Xanthomonas oryzae pv. oryzae in rice. Mol. Plant 1, 446–458.
Prime-A-Plant Group: Conrath, U., Beckers, G. J. M., Flors, V., García-Agustín, P., Jakab, G., Mauch, F., et al. (2006). Priming: getting ready for battle. Mol. Plant-Microbe Interact. 19, 1062–107.
Raetz, C. R. H., Reynolds, C. M., Trent, M. S., and Bishop, R. E. (2007). Lipid A modification systems in Gram-negative bacteria. Annu. Rev. Biochem. 76, 295–329.
Raetz, C. R. H., and Whitfield, C. (2002). Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71, 635–700.
Rallabhandi, P., Awomoyi, A., Thomas, K. E., Phalipon, A., Fujimoto, Y., Fukase, K., et al. (2008). Differential activation of human TLR4 by Escherichia coli and Shigella flexneri 2a lipopolysaccharide: combined effects of lipid A acylation state and TLR4 polymorphisms on signaling. J. Immunol. 180, 1139–1147.
Ramos, H. C., Rumbo, M., and Sirard, J.-C. (2004). Bacterial flagellins: mediators of pathogenicity and host immune responses in mucosa. Trends Microbiol. 12, 509–517.
Robatzek, S., Bittel, P., Chinchilla, D., Kochner, P., Felix, G., Shiu, S. H., et al. (2007). Molecular identification and characterization of the tomato flagellin LeFLS2, an orthologue of Arabidopsis FLS2 exhibiting characteristically different perception specificities. Plant Mol. Biol. 64, 539–547.
Robatzek, S., Chinchilla, D., and Boller, T. (2006). Ligand-induced endocytosis of the pattern recognition receptor FLS2 in Arabidopsis. Genes Dev. 20, 537–542.
Rockel, P., Strube, F., Rockel, A., Wildt, J., and Kaiser, W. M. (2002). Regulation of nitric oxide (NO) production by plant nitrate reductase in vivo and in vitro. J. Exp. Bot. 53, 103–110.
Ron, M., and Avni, A. (2004). The receptor for the fungal elicitor ethylene-inducing xylanase is a member of a resistance-like gene family in tomato. Plant Cell 16, 1604–1615.
Ronald, P. C., Albano, B., Tabien, R., Abenes, L., Wu, K. S., McCouch, S., et al. (1992). Genetic and physical analysis of the rice bacterial blight disease resistance locus, Xa21. Mol. Gen. Genet. 236, 113–120.
Roux, M., Schwessinger, B., Albrecht, C., Chinchilla, D., Jones, A., Holton, N., et al. (2011). The Arabidopsis leucine-rich repeat receptor-like kinases BAK1/SERK3 and BKK1/SERK4 are required for innate immunity to hemibiotrophic and biotrophic pathogens. Plant Cell 23, 2440–2455.
Ruiz-Ferrer, V., and Voinnet, O. (2009). Roles of plant small RNAs in biotic stress responses. Annu. Rev. Plant Biol. 60, 485–510.
Russinova, E., Borst, J.-W., Kwaaitaal, M., Cano-Delgado, A., Yin, Y., Chory, J., et al. (2004). Heterodimerization and endocytosis of Arabidopsis brassinosteroid receptors BRI1 and AtSERK3 (BAK1). Plant Cell 16, 3216–3229.
Ryals, J. A., Neuenschwander, U. H., Willits, M. G., Molina, A., Steiner, H.-Y., and Hunt, M. D. (1996). Systemic acquired resistance. Plant Cell 8, 1809–1819.
Scheer, J. M., and Ryan, C. A. (1999). A 160-kD systemin receptor on the Surface of Lycopersicon peruvianum suspension-cultured cells. Plant Cell 11, 1525–1535.
Scheer, J. M., and Ryan, C. A. (2002). The systemin receptor SR160 from Lycopersicon peruvianum is a member of the LRR receptor kinase family. Proc. Natl. Acad. Sci. U.S.A. 99, 9585–9590.
Schneider, M., Schweizer, P., Meuwly, P., and Metraux, J. P. (1996). Systemic acquired resistance in plants. Int. Rev. Cytol. 168, 303–340.
Schromm, A., Brandenburg, K., Loppnow, H., Moran, A., Koch, M., Rietschel, E., et al. (2000). Biological activities of lipopolysaccharides are determined by the shape of their lipid A portion. Eur. J. Biochem. 267, 2008–2013.
Schromm, A., Brandenburg, K., Loppnow, H., Zahringer, U., Rietschel, E., Carroll, S., et al. (1998). The charge of endotoxin molecules influences their conformation and IL-6-inducing capacity. J. Immunol. 161, 5464–5471.
Schulze, B., Mentzel, T., Jehle, A. K., Mueller, K., Beeler, S., Boller, T., et al. (2010). Rapid heteromerization and phosphorylation of ligand-activated plant transmembrane receptors and their associated kinase BAK1. J. Biol. Chem. 285, 9444–9451.
Schweizer, P., Felix, G., Buchala, A., Müller, C., and Métraux, J. P. (1996). Perception of free cutin monomers by plant cells. Plant J. 10, 331–341.
Séjalon-Delmas, N., Mateos, F. V., Bottin, A., Rickauer, M., Dargent, R., and Esquerré-Tugayé, M. T. (1997). Purification, elicitor activity, and cell wall localization of a glycoprotein from phytophthora parasitica var. nicotianae, a fungal pathogen of tobacco. Phytopathology 9, 899–909.
Seong, S. Y., and Matzinger, P. (2004). Hydrophobicity: an ancient damage-associated molecular pattern that initiates innate immune responses. Nat. Immunol. 4, 469–478.
Shen, Y., Sharma, P., da Silva, F. G., and Ronald, P. (2002). The Xanthomonas oryzae pv. oryzae raxP and raxQ genes encode an ATP sulphurylase and adenosine-5'-phosphosulphate kinase that are required for AvrXa21 avirulence activity. Mol. Microbiol. 44, 37–48.
Shimizu, T., Nakano, T., Takamizawa, D., Desaki, Y., Ishii-Minami, N., Nishizawa, Y., et al. (2010). Two Lys M receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J. 64, 204–214.
Shinya, T., Motoyama, N., Ikeda, A., Wada, M., Kamiya, K., Hayafune, M., et al. (2012). Functional characterization of CEBiP and CERK1 homologs in Arabidopsis and rice reveals the presence of different chitin receptor systems in plants. Plant Cell Physiol. 53, 1696–1706.
Shiu, S. H., and Bleecker, A. B. (2003). Expansion of the receptor-like kinase/pelle gene family and receptor-like proteins in Arabidopsis. Plant Physiol. 132, 530–543.
Shiu, S.-H., Karlowski, W. M., Pan, R., Tzeng, Y.-H., Mayer, K. F. X., and Li, W.-H. (2004). Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 16, 1220–1234.
Shuguo, H., Wei, Z., Chao, Z., and Daoji, W. (2012). One-step expression and tyrosine O-sulfonation of Ax21 in Escherichia coli. Appl. Biochem. Biotechnol. 166, 1368–1379.
Silipo, A., Leone, M. R., Erbs, G., Lanzetta, R., Parrilli, M., Chang, W.-S., et al. (2011). A unique bicyclic monosaccharide from the Bradyrhizobium lipopolysaccharide and its role in the molecular interaction with plants. Angew. Chem. 50, 12610–12612.
Silipo, A., Molinaro, A., Sturiale, L., Dow, J. M., Erbs, G., Lanzetta, R., et al. (2005). The elicitation of plant innate immunity by lipooligosaccharide of Xanthomonas campestris. J. Biol. Chem. 280, 33660–33668.
Silipo, A., Sturiale, L., Garozzo, D., de Castro, C., Lanzetta, R., Parrilli, M., et al. (2004). Structure elucidation of the highly heterogeneous lipid A from the lipopolysaccharide of the Gram-negative extremophile bacterium Halomonas magadiensis strain 21 M1. Eur. J. Org. Chem. 2004, 2263–2271.
Silipo, A., Sturiale, L., Garozzo, D., Erbs, G., Tandrup Jensen, T., Lanzetta, R., et al. (2008). The acylation and phosphorylation pattern of lipid A from Xanthomonas campestris strongly influence its ability to trigger the innate immune response in Arabidopsis. Chembiochem 9, 896–904.
Smith, K. D., Andersen-Nissen, E., Hayashi, F., Strobe, K., Bergman, M. A., Rassoulian Barrett, S. L., et al. (2003). Toll-like receptor 5 recognizes a conserved site on flagellin required for protofilament formation and bacterial mobility. Nat. Immunol. 4, 1247–1253.
Song, W. Y., Pi, L. Y., Wang, G. L., Gardner, J., Holsten, T., and Ronald, P. (1997). Evolution of the rice Xa21 disease resistance gene family. Plant Cell 9, 1279–1287.
Song, W. Y., Wang, G. L., Chen, L. L., Kim, H. S., Pi, L. Y., Holsten, T., et al. (1995). A receptor kinase-like protein encoded by the rice disease resistance gene Xa21. Science 270, 1804–1806.
Sun, A. Z., Nie, S. J., and Xing, D. (2012). Nitric oxide-mediated maintenance of redox homeostasis contributes to NPR1-dependent plant innate immunity triggered by lipopolysaccharides. Plant Physiol. 160, 1081–1096.
Sun, W., Dunning, F. M., Pfund, C., Weingarten, R., and Bent, A. F. (2006). Within-species flagellin polymorphism in Xanthomonas campestris pv campestris and its impact on elicitation of Arabidopsis FLAGELLIN SENSING2-dependent defenses. Plant Cell 18, 764–779.
Takai, R., Isogai, A., Takayama, S., and Che, F. S. (2008). Analysis of flagellin perception mediated by flg22 receptor OsFLS2 in rice. Mol. Plant Microbe Interact. 21, 1635–1642.
Thomma, B. P. H. J., Nürnberger, T., and Joosten, M. H. A. J. (2011). Of PAMPs and effectors: the blurred PTI-ETI dichotomy. Plant Cell 23, 4–15.
Traub, S., von Aulock, S., Hartung, T., and Herman, C. (2006). MDP and other muropeptides -direct and synergistic effects on the immune system. J. Endotoxin Res. 12, 69–85.
Umemoto, N., Kakitani, M., Iwamatsu, A., Yoshikawa, M., Yamaoka, N., and Ishida, I. (1997). The structure and function of a soybean β-glucan-elicitor-binding protein. Proc. Natl. Acad. Sci. U.S.A. 94, 1029–1034.
van der Biezen, E. A., and Jones, J. D. (1998). Plant disease-resistance proteins and the gene-for-gene concept. Trends Biochem. Sci. 23, 454–456.
van Loon, L. C., Bakker, P. A., and Pieterse, C. M. (1998). Systemic resistance induced by rhizosphere bacteria. Annu. Rev. Phytopathol. 36, 453–483.
Wan, J., Tanaka, K., Zhang, X.-C., Son, G. H., Brechenmacher, L., Nguyen, T. H. N., et al. (2012). LYK4, a LysM receptor-like kinase, is important for chitin signaling and plant innate immunity in Arabidopsis. Plant Physiol. 160, 396–406.
Wang, Y. H., Gehring, C., and Irving, H. R. (2011). Plant natriuretic peptides are apoplastic and paracrine stress response molecules. Plant Cell Physiol. 52, 837–850.
Wang, Y. S., Pi, L. Y., Chen, X., Chakrabarty, P. K., Jiang, J., De Leon, A. L., et al. (2006). Rice XA21 binding protein 3 is a ubiquitin ligase required for full Xa21-mediated disease resistance. Plant Cell 18, 3635–3646.
Wang, G. L., Ruan, D. L., Song, W. Y., Sideris, S., Chen, L. L., Pi, L. Y., et al. (1998). Xa21D Encodes a receptor-like molecule with a leucine-rich repeat domain that determines race-specific recognition and is subject to adaptive evolution. Plant Cell 10, 765–779.
Wang, G. L., Song, W. Y., Ruan, D. L., Sideris, S., and Ronald, P. C. (1996). The cloned gene, Xa21, confers resistance to multiple Xanthomonas oryzae pv. oryzae isolates in transgenic plants. Mol. Plant Microbe Interact. 9, 850–855.
Watt, S. A., Tellstrom, V., Patschkowski, T., and Niehaus, K. (2006). Identification of the bacterial superoxide dismutase (SodM) as plant-inducible elicitor of an oxidative burst reaction in tobacco cell suspension cultures. J. Biotechnol. 126, 78–86.
Willmann, R., Lajunen, H. M., Erbs, G., Newman, M.-A., Kolb, D., Tsuda, K., et al. (2011). Arabidopsis lysin-motif proteins LYM1 LYM3 CERK1 mediate bacterial peptidoglycan sensing and immunity to bacterial infection. Proc. Natl. Acad. Sci. U.S.A. 108, 19824–19829.
Wright, S. D., Ramos, R. A., Tobias, P. S., Ulevitch, R. J., and Mathison, J. C. (1990). CD14 serves as the cellular receptor for complexes of lipopolysaccharides with lipopolysaccharide binding protein. Science 249, 1431–1433.
Yamaguchi, Y., Huffaker, A., Bryan, A. C., Tax, F. E., and Ryan, C. A. (2010). PEPR2 is a second receptor for the Pep1 and Pep2 peptides and contributes to defense responses in Arabidopsis. Plant Cell 22, 508–522.
Yamaguchi, Y., Pearce, G., and Ryan, C. A. (2006). The cell surface leucine-rich repeat receptor for AtPep1, an endogenous peptide elicitor in Arabidopsis, is functional in transgenic tobacco cells. Proc. Natl. Acad. Sci. U.S.A. 103, 10104–10109.
Zeidler, D., Dubery, I. A., Schmitt-Kopplin, P., Von Rad, U., and Durner, J. (2010). Lipopolysaccharide mobility in leaf tissue of Arabidopsis thaliana. Mol. Plant Pathol. 11, 747–755.
Zeidler, D., Zähringer, U., Gerber, I., Dubery, I., Hertung, T., Bors, W., et al. (2004). Innate immunity in Arabidopsis thaliana lipopolysaccharides activate nitric oxide synthase NOS and induce defense genes. Proc. Natl. Acad. Sci. U.S.A. 101, 15811–15816.
Keywords: innate immunity, MAMPs, Flg22, EF-Tu, Ax21, PGN, LPS, Chitin
Citation: Newman M-A, Sundelin T, Nielsen JT and Erbs G (2013) MAMP (microbe-associated molecular pattern) triggered immunity in plants. Front. Plant Sci. 4:139. doi: 10.3389/fpls.2013.00139
Received: 15 March 2013; Accepted: 23 April 2013;
Published online: 16 May 2013.
Edited by:
Saskia C. Van Wees, Utrecht University, NetherlandsCopyright © 2013 Newman, Sundelin, Nielsen and Erbs. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and subject to any copyright notices concerning any third-party graphics etc.
*Correspondence: Mari-Anne Newman, Department of Plant and Environmental Sciences, Faculty of Science, University of Copenhagen, Thorvaldsensvej 40, 1871 Frederiksberg C, Denmark. e-mail:bWFyaUBsaWZlLmt1LmRr


