- 1 Department of Plant Biology, Cornell University, Ithaca, NY, USA
- 2 Great Lakes Bioenergy Research Center, Michigan State University, East Lansing, MI, USA
- 3 DOE Plant Research Laboratory, Michigan State University, East Lansing, MI, USA
N-Glycosylation is a common form of eukaryotic protein post-translational modification, and one that is particularly prevalent in plant cell wall proteins. Large scale and detailed characterization of N-glycoproteins therefore has considerable potential in better understanding the composition and functions of the cell wall proteome, as well as those proteins that reside in other compartments of the secretory pathway. While there have been numerous studies of mammalian and yeast N-glycoproteins, less is known about the population complexity, biosynthesis, structural variation, and trafficking of their plant counterparts. However, technical developments in the analysis of glycoproteins and the structures the glycans that they bear, as well as valuable comparative analyses with non-plant systems, are providing new insights into features that are common among eukaryotes and those that are specific to plants, some of which may reflect the unique nature of the plant cell wall. In this review we present an overview of the current knowledge of plant N-glycoprotein synthesis and trafficking, with particular reference to those that are cell wall localized.
Introduction
A common feature of plant proteins that are resident in the cell wall and other compartments of the secretory pathway is glycosylation; a complex form of post-translational modification (PTM) that has been detected in a spectrum of taxonomic groups, including eubacteria and archaea (Lechner and Wieland, 1989; Messner, 1997). Decades of research, primarily focused on yeast and mammalian glycoproteins, have demonstrated that glycosylation is important in numerous biological processes and affects protein characteristics such as folding, enzyme activity, trafficking, localization and ligand interactions (Spiro, 2002; Helenius and Aebi, 2004). However, while glycosylation is particularly prevalent and often extensive in secreted plant proteins, the detailed structures, dynamics, and in most cases functional significance, of their glycans are generally obscure.
Protein glycosylation may be divided into two principal types. The most studied is N-glycosylation, which involves the attachment of the N-acetylglucosamine (GlcNAc) of an oligosaccharide moiety by an amide bond to an asparagine residue (Asn) that is generally referred to as belonging to a consensus sequence N-X-(S/T; also written as N-!P-[S/T]), where X can be any amino acid except proline (Pless and Lennarz, 1977). Alternative non-canonical consensus sequences, such as N-X-C, have been suggested for small subset of N-glycosylation events (Zielinska et al., 2010). The second kind of glycosylation is O-glycosylation, which in plants occurs on serine (S or Ser), threonine (T or Thr), and hydroxyproline (Hyp) residues (Showalter, 2001; Gomord et al., 2010; Velasquez et al., 2011), with no apparent single common core structure or consensus protein sequence. Plant protein O-glycosylation will not be specifically discussed here, but has been the subject of several recent reviews (Gomord et al., 2010; Mohnen and Tierney, 2011; Taylor et al., 2011); rather this article will focus on plant N-glycoproteins.
The glycan structures that are attached to nascent proteins typically reflect the actions of an array of often competing glycosidases and glycosyl transferases in the secretory pathway. While the presence of the consensus sequence is generally required for N-linked glycosylation, the occupation of a potential site is not mandatory. Therefore, a glycoprotein may contain a number of potentially N-glycosylated sites, each of which may or may not be glycosylated (An et al., 2009). This results in a population of decorated proteins with a high degree of microheterogeneity at specific sites, making structural characterization extremely difficult. This challenge is compounded by the large number of possible stereo- and regio-isomers (An et al., 2009; Marino et al., 2010). Moreover, the glycan structures and frequencies cannot be reliably predicted at the level of primary DNA or protein sequences and there is not yet a single analytical platform for their accurate or high-throughput identification (Ruiz-May et al., this issue).
Despite these major obstacles, recent reports have described the characterization of hundreds of glycoproteins from several bacterial (Nothaft and Szymanski, 2010) and animal species (Bunkenborg et al., 2004; Liu et al., 2005b; Kaji et al., 2007; Gundry et al., 2009; Lee et al., 2009; Wollscheid et al., 2009; Zielinska et al., 2010). In contrast, there have been no similar published studies of plant glycoproteomes on this scale. Rather, the emphasis to date has typically been on characterizing the N-glycosylation of individual proteins (Jamet et al., 2008), often in the context of using plants as a factories to produce recombinant proteins for human therapy and diagnostics (Chrispeels and Faye, 1996; Gomord et al., 2010). The purpose of this review is to provide a summary of advances in understanding plant N-glycoprotein synthesis and trafficking, with a specific focus on secreted (i.e., cell wall resident) proteins. Such studies are laying the foundation for a more advanced understanding of the plant N-glycoproteome, and consequently the identity, function and targeting of cell wall proteins.
Biosynthesis of N-Glycoproteins
The yeast Saccharomyces cerevisiae has served as a model for the elucidation of N-glycan biosynthesis in eukaryotes and substantial numbers of the associated genes have been annotated (Burda and Aebi, 1999; Kelleher and Gilmore, 2006; O’Reilly et al., 2006). Considerably fewer have been identified and characterized in plants, but based on existing examples it appears that there is general conservation of the glycosylation machinery (see examples listed in Table 1). There follows an overview of recent developments and questions in the field of plant glycoprotein biosynthesis and trafficking in the context of other experimental model systems.
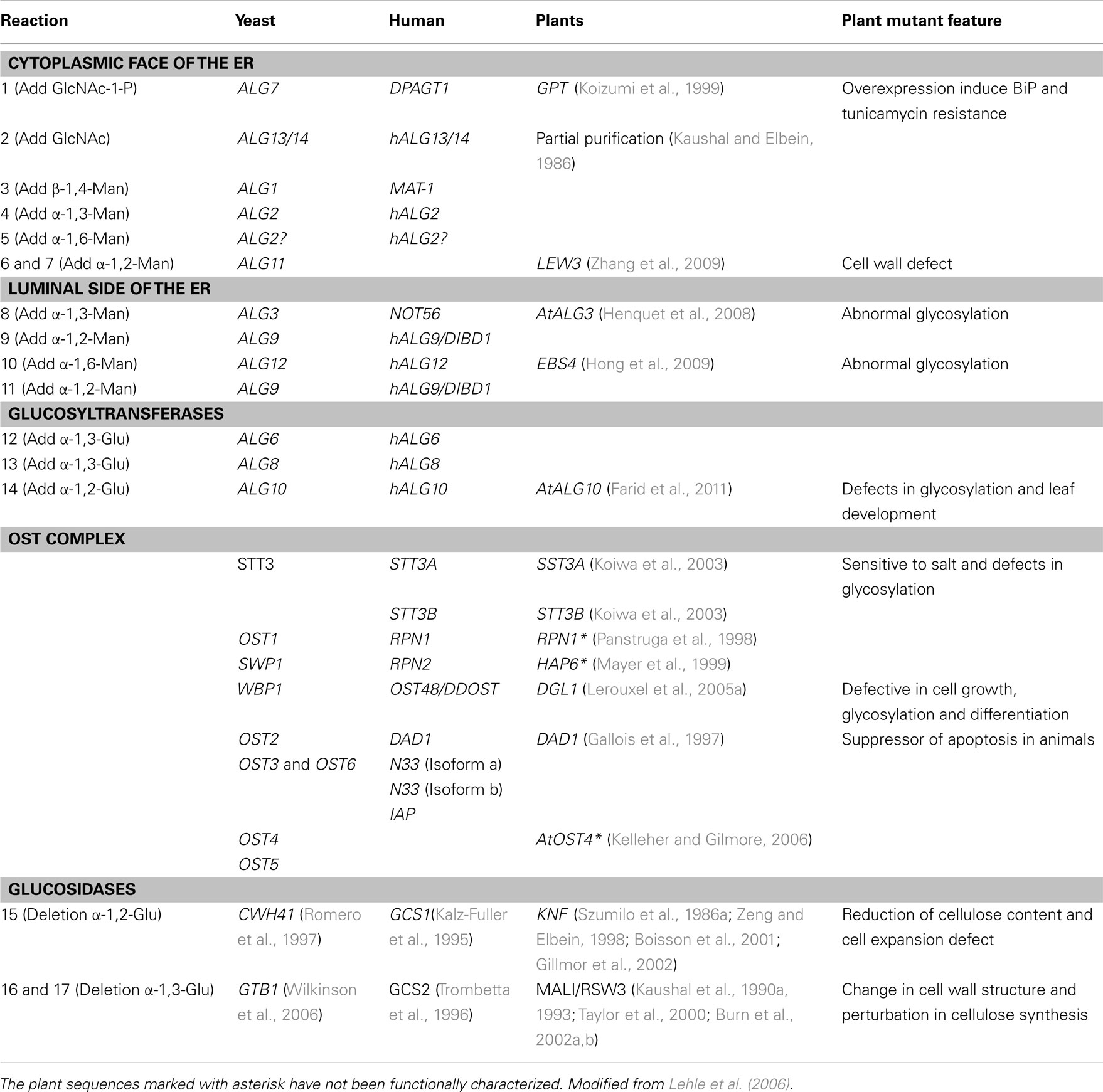
Table 1. Homologous yeast, human and plants genes associated with N-glycosylation and phenotypes of related plant mutants.
Cytoplasmic Face
As with all eukaryotes, protein glycosylation in plants is initiated in the endoplasmic reticulum (ER). However, the biosynthesis of the N-glycan precursor begins on the cytosolic side of the ER (Figure 1; Table 1), starting with the transfer of N-acetylglucosamine-phosphate from soluble UDP-GlcNAc to the lipid membrane bound dolichyl monophosphate (Dol-P), forming N-acetylglucosamine-pyrophosphatidyldolichol (GlcNAc-PP-Dol). Dol-P is one of the rate-limiting factors in N-linked protein glycosylation in yeast and mammalian cells (Burda and Aebi, 1999; Jones et al., 2005) and while this is likely also to be the case in plants, it has not yet been demonstrated. However, mutational defects in the biosynthesis of Dol-P in Arabidopsis have been shown cause multiple physiological effects, such as impaired plasma membrane integrity resulting in electrolyte leakage, reduced cellular turgor and stomatal conductance, and increased drought resistance (Zhang et al., 2008). One GlcNAc and five mannose (Man) residues are subsequently transferred from UDP-GlcNAc, or GDP-Man, respectively, in a controlled, stepwise manner to GlcNAc-PP-Dol, producing the Man5GlcNAc2-PP-Dol branched heptasaccharide intermediate (Helenius and Aebi, 2002).
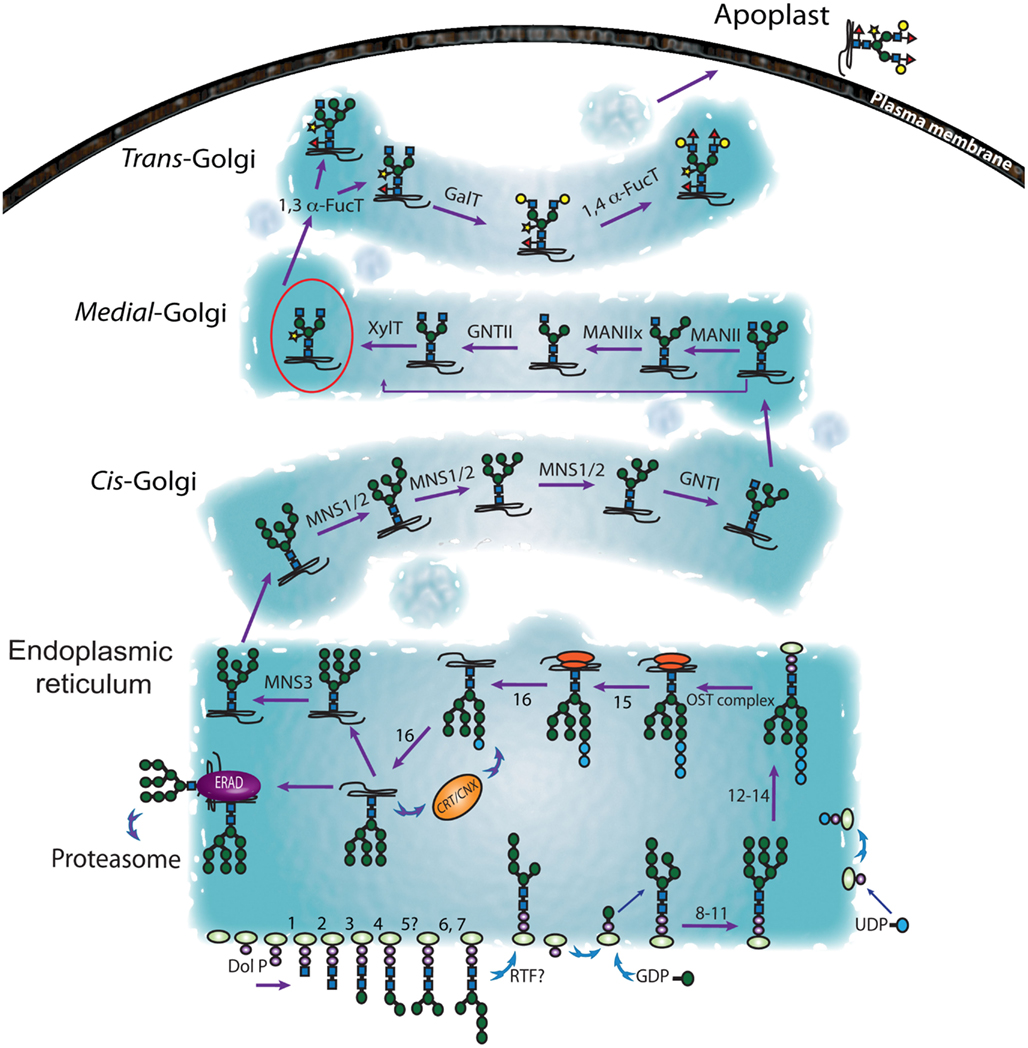
Figure 1. Representation of the secretory pathway followed by the N-glycoproteins. The biosynthesis of the N-glycans is initiated on the cytosolic face of the ER. The resulting Man5GlcNAc-PP-Dol precursor is then flipped onto the luminal side ER lumen where further maturation of the sugar precursor occurs (Glc3Man9GlcNAc-PP-Dol). At this point the N-glycan structure is transferred to the nascent polypeptide. After removal of three Glc residues the N-glycoproteins enter the calnexin-calreticulin cycle (CNX/CRT; Hebert et al., 1995). The alternate action of glucosidase II and UDP-glucose:glycoprotein glucosyltransferase drives the glycoprotein through this cycle until it is correctly folded and exported from the ER to the GA. Misfolded proteins are directed from the ER to the cytosol by the ER-associated degradation (ERAD) machinery for proteasomal hydrolysis (Hebert et al., 1995; Crofts et al., 1998; Helenius and Aebi, 2004; Jin et al., 2007; Lederkremer, 2009). Glucosyl transferase and glucosidases implicated the in the sugar trimming in the ER and identified in plants so far, are enlisted in Table 1. Subsequent modifications of the N-glycans occur in the GA and potentially in other cellular compartments, such as the chloroplast and vacuole (based on a model presented in Gomord et al. (2010). The first indirect insights into the location and orientation of plant glycosyltransferases were provided by immunolocalization of the enzyme products (Laine et al., 1991; Fitchette et al., 1994, 1999). Such studies indicated that β-1,2-xylose is added to plant N-glycans mainly in the medial Golgi while the α-1,3-fucosylation occurs predominantly in the trans-Golgi (Fitchette et al., 1994).
Lumen of the Endoplasmic Reticulum
The second phase of synthesis involves the translocation of the Man5GlcNAc2-PP-Dol moiety across the ER membrane to the luminal leaflet (Figure 1; Table 1). This is thought to be catalyzed by a flippase, although the identity of this protein has not yet been confirmed in eukaryotic cells (Bugg and Brandish, 1994; Higgins, 1994; Sprong et al., 2001). Following transfer to the luminal side of the ER membrane, the oligosaccharide moiety is extended by the progressive addition of four mannose (Man) and three glucose (Glc) residues by several luminal glycosyltransferases to form GlcMan9GlcNAc2-PP-Dol (Gomord et al., 2010). Genes encoding a variety of plant glycosyltransferases have been identified (Ross et al., 2001) and several enzymes involved in N-glycan biosynthesis have been characterized in recombinant forms (Leonard et al., 2004). However, the functions of many plant glycosyltransferase homologs remain to be elucidated (Table 1). The basic step of the biosynthesis of the N-glycan precursor is conserved among eukaryotic cells but the regulation mechanism and the number of intermediates vary among species, and even between tissues (Pattison and Amtmann, 2009).
The third phase of N-linked protein glycosylation (Figure 1) is the transfer of a mature Glc3Man9GlcNAc2 oligosaccharide to select Asn residues that are components of the N-!P-[S/T] consensus sequence (Pless and Lennarz, 1977). This step is catalyzed by the oligomeric oligosaccharyltransferase (OST, Table 1) complex (Yan and Lennarz, 1999), which in mammals consists of seven or eight non-identical subunits, but nine protein subunits in yeast (Knauer and Lehle, 1999). The active site subunits of the eukaryotic OST are termed STT3 proteins (Nilsson and von Heijne, 1993; Yan and Lennarz, 2002) and in mammalian systems it has been shown that two STT3 isoforms (STT3A and STT3B) are responsible for co- and post-translational N-polypeptide glycosylation (Ruiz-Canada et al., 2009). Sequence homology searches of the Arabidopsis genome identified five genes encoding putative OST subunits and two STT3 isoforms (Gallois et al., 1997), but only three of these have been functionally characterized (Koiwa et al., 2003; Lerouxel et al., 2005b).
Modification in the Endoplasmic Reticulum
After transfer from dolichol to the nascent glycoprotein, the N-glycan is trimmed (Figure 1; Table 1), involving hydrolytic removal of the distal α-1,2-linked Glc by glucosidase I (Grinna and Robbins, 1979; Hubbard and Ivatt, 1981), two adjacent α-1,3-linked Glc residues by glucosidase II (Michael and Kornfeld, 1980; Kilker et al., 1981) and a single specific Man residue. Glucosidases I and II were first purified from mung bean (Szumilo et al., 1986a; Kaushal et al., 1990a, 1993; Zeng and Elbein, 1998) and the characterization of the Arabidopsis mutants gsc1-1 and gsc1-2 resulted in the cloning of a glucosidase I gene. The encoded protein has homology to animal and yeast α-glucosidase I, which is involved in the first step of N-glycan trimming (Boisson et al., 2001). The first plant glucosidase II gene was identified from potato (Taylor et al., 2000) and the gene encoding the catalytic subunit was discovered through a study of an Arabidopsis temperature sensitive mutant (rsw3) with perturbed cellulose synthesis and a swollen root phenotype (Burn et al., 2002a).
With regard to subsequent trimming and modification of the Man component by mannosidases, studies in mammalians systems have resulted in the definition of three subgroups of the class 1 α-mannosidase family, based on sequence similarity and proposed function: ER-α1, 2, mannosidases I (ER-MNSIs), Golgi-α-mannosidases I (Golgi-MNSIs), and ER degradation-α-enhancing mannosidase (EDEM) like proteins (Mast and Moremen, 2006). In humans, only one ER-MNS1 cleaves one terminal Man from Man9GlcNAc2 (referred to as b-branch in Liebminger et al., 2009) to generate Man8GlcNAc2. In mammals, some N-glycoproteins bearing Glc1Man9GlcNAc2 (i.e., retaining an extra Glc due to incomplete processing in the ER) have been shown to traffic to the cis-Golgi. In such cases, a Golgi resident endo-α-D-mannosidase can then cleave this molecule internally between two Man residues, resulting in a Man8GlcNAc2 glycan (Lubas and Spiro, 1987). However, phylogenetic surveys and enzymatic assays suggests the absence of such Golgi endo-α-D-mannosidase in higher plants (Dairaku and Spiro, 1997). More recently, an ER-MNSI (MNS3) from Arabidopsis was identified and biochemically characterized (Liebminger et al., 2009). Arabidopsis MNS3 showed 47% of identity to human ER-MNSI and is required for the efficient trimming of Man9GlcNAc2 to Man8GlcNAc2 (Liebminger et al., 2009). The apparent absence of Golgi endo-α-D-mannosidases in higher plants (Dairaku and Spiro, 1997) and the fact that ER resident plant glycoproteins predominantly bear Man8GlcNAc2 and minimal amount of Man9GlcNAc2, might suggest that MNS3 resides in the ER. However, transient expression of MNS3-GFP in leaf epidermal cells of Nicotiana benthamiana showed overlapping expression with the Golgi marker GnTI-CTS-mRFP (Liebminger et al., 2009). In mammalian cells the ER-MNS1 has observed to be located in the ER-derived quality control compartment (Avezov et al., 2008), which is adjacent to, but not overlapping with the Golgi and the ER-to-Golgi intermediate compartment (Kamhi-Nesher et al., 2001). One hypothesis then is that MNS3 is localized in a similar, but as yet unconfirmed subcellular compartment.
Quality Control CNX/CRT Cycle
During translation and glycosylation in the ER, N-glycoproteins undergo a quality control process that prevents the arrival of incorrectly folded proteins at their final destinations and involves their redirection to the protein degradation machinery (Figure 1, and for more information see Liu and Howell, 2010). The N-glycan structure assembled in the ER serve as a recognition tags for the ER quality control and reflect the folding status of proteins (Helenius and Aebi, 2004). The membrane bound calnexin (CNX) and its soluble luminal homolog calreticulin (CRT) are chaperones that facilitate maturation and detection of defective folding (Lederkremer, 2009). The cleavage of the terminal glucose residues of the N-glycans can be crucial to protein folding (Lupattelli et al., 1997). Indeed, the mono-glycosylated core glycan (Figure 1, GlcMan9GlcNAc2) generates the interaction with the CNX/CRT complex (Ruddock and Molinari, 2006). The dissociation of the CNX/CRT complex from the glycoproteins is made possible by the removal of the third glucose linked mannose residue by glucosidase II. However, the enzyme UDP-glucose: glycoprotein glucosyltransferase (GT) can then re-glucosylate the N-glycan chain and recreate a ligand for CNX/CRT (Hebert et al., 1995). An Arabidopsis GT has been identified that plays such a role (Jin et al., 2007). It is not clear how this cycle of glycoprotein binding and glycan modification promotes protein folding or oligomerization, but one suggestion is that CNX/CRT facilitates ER retention once the GT has recognized and signaled the unfolded, or partially folded, state of a protein (Crofts et al., 1998). This cycle continues until proper folding is achieved, which prevents further recognition by the GT folding sensor (Jin et al., 2007). Another contributor to this process is the luminal binding protein (BiP). It is thought that BiP binds to translocation intermediates, misfolded proteins and peptides with exposed hydrophobic regions (Blond-Elguindi et al., 1993; Gething, 1999), preventing aggregation that could lead to permanent misfolding (Gaut and Hendershot, 1993; Hendershot et al., 1996). However, the nature and extent of any interaction between CNX/CRT and BiP that allows the proper folding of the glycoprotein intermediates through the ER is unclear at present. Similarly, it is not known whether other ER resident proteins or some other interacting molecules are also involved. Misfolded proteins released from the CNX/CRT cycle are redirected from the ER to the cytosol for proteasomal degradation; a poorly understood process in plants, referred to as ER-associated protein degradation (ERAD; Di Cola et al., 2001, 2005; Lederkremer, 2009; Liebminger et al., 2010; Liu and Howell, 2010).
ER Export of Glycosylated Proteins
After the initial glycosylation event involving the addition of Man and Glc residues with transfer of the N-glycan donor on the protein and final deletion of three glucose and one MAN residues, N-glycoproteins carrying Man8GlcNAc2 are delivered from the ER to the cis-Golgi generally via the COPII machinery (Figure 2) using cargo receptors or bulk flow transport (Kuehn et al., 1998; Phillipson et al., 2001). Several soluble cargo receptors have been characterized in mammals and yeasts by mutant analysis. For example, ERGIC-53 and Emp46p/47p are soluble ER resident receptors that interact with the glycosylation motif of soluble cargos, while Erv29p is a cargo receptor that interacts with the ILV motif of glycosylated proteins (Appenzeller et al., 1999; Belden and Barlowe, 2001; Otte and Barlowe, 2004). Both in vitro and in vivo interaction experiments have demonstrated that the absence of these functional cargo receptors leads to defective secretion, suggesting that they are essential for packing soluble cargo into COPII vesicles prior to transport from the ER to the Golgi. Considering their binding specificity for the glycosylation motif, ERGIC-53, and Emp46p/47p could be regarded as a glycosylation checkpoint (Appenzeller et al., 1999; Otte and Barlowe, 2004). Although receptor mediated cargo recruitment by the COPII machinery has not been characterized in plants, delivery of soluble glycoproteins by bulk flow via COPII machinery has been shown in tobacco, using calreticulin without the ER retention signal HDEL (calreticulin ΔHDEL) and α-amylase fused with HDEL (Phillipson et al., 2001). Calreticulin binds to glycosylated proteins for quality control and has the ER retention signal (HDEL) that mediates retrieval from the Golgi to ER. It has been reported that over-expressed calreticulin ΔHDEL is secreted by the default secretory pathway; however, secretion of calreticulin ΔHDEL decreases when COPII machinery is partially inhibited (Phillipson et al., 2001). These results demonstrate the existence of COPII-mediated bulk flow of glycosylated proteins in tobacco. Interestingly, no close homologs of soluble cargo receptors have been identified in plants, although given the conservation of the COPII machinery among eukaryotes, it is reasonable to hypothesize that plants may also have such receptors.
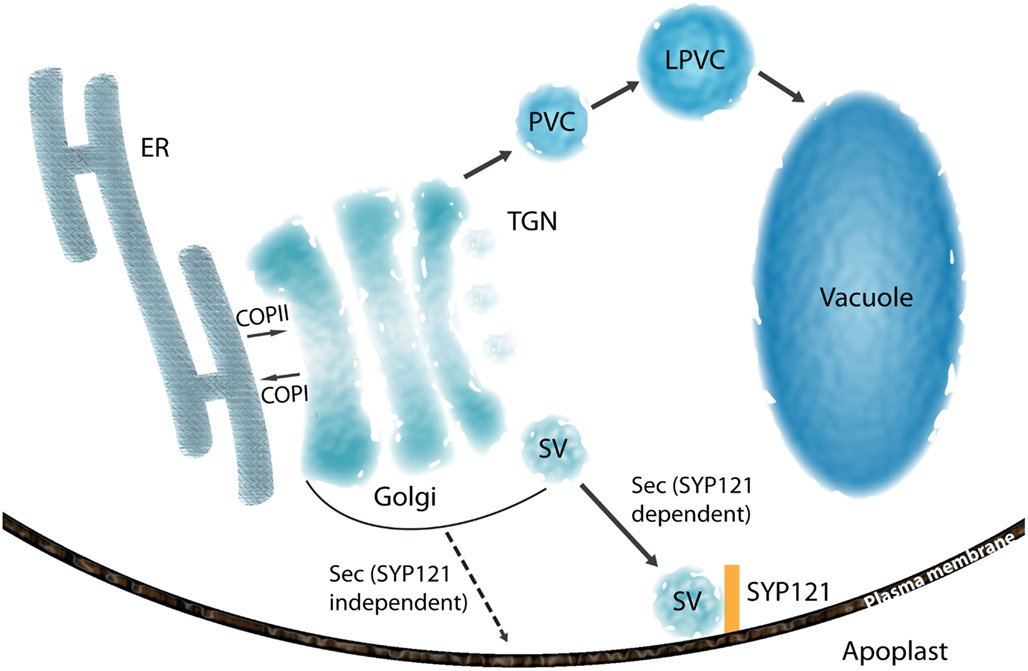
Figure 2. Overview of the secretory pathway of glycosylated proteins. Glycosylated proteins are transported from ER to cis-Golgi by either bulk flow transport or receptor mediated transport. COPII and COPI proteins are involved in anterograde and retrograde trafficking between ER and Golgi, respectively. Secretory and vacuolar proteins are sorted at TGN. Vacuolar proteins in TGN are transported to vacuole via prevacuolar compartment (PVC) formed from maturation of TGN as well as late PVC (LPVC). Secretory proteins are accumulated in the secretory vesicles (SV) and delivered to the cell surface. A transport route indicated by dashed arrows represents a hypothetical pathway for GPI-anchored proteins (PMEI1 and PGIP2).
Peptide sequences in the cytosolic domain of glycoproteins have been reported to be important for their transport from the ER to the cis-Golgi. For example, a GDP-mannose transporter (GONST1) and Arabidopsis AtCASP have diacidic DXE motifs in their cytosolic domains that are required for efficient ER export (Hanton et al., 2005). Similarly, the diacidic motifs are also important for efficient ER export in yeast, where the interaction between the diacidic motif and a Sec23p/24p complex has been studied (Nishimura and Balch, 1997; Votsmeier and Gallwitz, 2001). Physical association between the diacidic motif of a K+-channel (KAT1) and the COPII coat component Sec24 has been demonstrated in vivo in guard cells using a fluorescence resonance energy transfer (FRET) approach (Sieben et al., 2008). Sec24 isoforms from yeast and mammals have been reported to have their own specific binding domains for cargo selection (Miller et al., 2002; Mancias and Goldberg, 2008) and studies of the three Arabidopsis Sec24 isoforms (Faso et al., 2009) suggest that they may also have a specific binding motif (Wendeler et al., 2007). Further characterization of sorting signals other than a diacidic motif for the interaction with Sec24 isoforms in plants is an important future goal.
Considering the proposed specific membrane-anchored cargo selection by Sec24 isoforms, it is possible that membrane-anchored cargos are sequestered at different ER domains, dependent upon the isoform. This hypothesis finds further support in light of the recent result that a partial loss of function of Arabidopsis Sec24A led to the deformation of the ER into skein-like structures at the perinuclear area, suggesting that Sec24A may control ER export of cargo that is important for ER morphology in specific regions of the cell (Faso et al., 2009). This hypothesis has yet to be tested since it is still not know whether the COPII coat at the sites of ER protein export, the so-called ER export/exit sites (ERES), may contain different Sec24 isoforms, or only one isoform. A diversification of ERES within the cell might facilitate ER export of cargo to specific regions of the cell and allow efficient communication with the surrounding environment. A detailed analysis using fluorescent protein tagged Sec24 isoforms under the control of endogenous promoters could provide important insights into the composition of ER export sites and test the potential importance of ERES diversification.
Maturation of Plant N-glycoprotein Glycans in the Golgi Apparatus
Once N-glycosylated proteins reach the cis-Golgi, a number of enzymes then contribute to further the N-glycan processing and maturation (Figure 1; Lerouge et al., 1998). After the generation of Man5GlcNAc2 by the MNS1/2 2 (Liebminger et al., 2009; Schoberer and Strasser, 2011), the N-acetylglucosaminyltransferase I (GNT I) catalyze the addition of GlcNAc yielding GlcNAcMan5GlcNAc2 (Johnson and Chrispeels, 1987; Tezuka et al., 1992). The cDNA encoding GNT I was isolated from tobacco and its identity confirmed by heterologous expression and activity assays (Strasser et al., 1999) and several other cDNA clones encoding GNT I from potato and Arabidopsis have been characterized (Wenderoth and von Schaewen, 2000). Subsequently, α-mannosidase II (MAN II) acts to sequentially remove two Man residues (Kaushal et al., 1990b; Strasser et al., 2006) and N-acetylglucosaminyltransferase II (GNT) adds a single GlcNAc (Johnson and Chrispeels, 1987; Tezuka et al., 1992; Strasser et al., 1999) to generate GlcNAc2Man3GlcNAc2. MAN II has been purified from mung bean (Szumilo et al., 1986b; Kaushal et al., 1990b) and the corresponding gene was identified from Arabidopsis based on homology with human and Drosophila sequences (Strasser et al., 2006). At this point additional structural features are introduced that are apparently unique to plants (Fitchette et al., 1994), insects (Kubelka et al., 1993; Altmann et al., 1999), certain invertebrates (van Kuik et al., 1985), and parasitic nematodes (Haslam et al., 2001; Faveeuw et al., 2003). In plants, these comprise the addition of an β-1,2-xylose (Xyl) by β-1, 2-xylosyltransferases (Zeng et al., 1997; Strasser et al., 2000; Pagny et al., 2003) and α-1,3-fucose (Fuc) to the 3-position of the innermost GlcNAc residue by an α-1,3-fucosyltransferase (Leiter et al., 1999; Wilson et al., 2001a; Figure 1). The resulting N-glycans can be further decorated by the consecutive addition of β-1,3-galactose and α-1,4-fucose residues to the outermost GlcNAc residue to yield the so-called Lewis (Lea) epitope (Fitchette-Laine et al., 1997; Melo et al., 1997). The first reaction is catalyzed by a β-1,3-galactosyltransferase (1,3 β-GalT), which has been identified and characterized in Arabidopsis (Strasser et al., 2007). The second reaction is catalyzed by an α-1,4-fucosyltransferase (1,4 α-FucT). In humans, α-1,4-fucosylation is primarily mediated by the Lewis fucosyltransferase, Fuc-TIII, a member of the α-1,3-fucosyltransferase family, which can catalyze the formation of both α-1,3- and α-1,4-fucose linkages (Palma et al., 2001). Genes encoding 1,4 α-FucT have been cloned from several plant species and the corresponding enzymatic activity shown to have strict acceptor substrate specificity for type 1 chain-based glycan structures (Galβ1-3GlcNAc), but not type 2 chains (Galβ1-4GlcNAc) that are typical of mammalian N-linked glycans (Fitchette-Laine et al., 1997; Oriol et al., 1999; Bakker et al., 2001; Palma et al., 2001; Wilson, 2001; Wilson et al., 2001b; Leonard et al., 2002, 2005).
These plant-associated differences in N-glycan structures have important implications for human health as plant glycoproteins are often extremely immunogenic (Garcia-Casado et al., 1996) and there are a number of reports that ascribe IgE cross reaction with pollen and dietary components to the carbohydrate component of glycoproteins (Wilson et al., 2001b). Indeed, it has been claimed that the major food allergens are typically water soluble glycoproteins (van Ree et al., 2000). Plant N-glycan structural differences are also significant in terms of experimental procedures that can be used for their analysis. For example, GlcNAc substituted with α-1, 3-fucose is not recognized by the enzyme PNGase-F, which is commonly used to deglycosylate mammalian, fungal and yeast proteins. PNGase-A can be used as an alternative, but this enzyme is only active on shorter peptides (Ytterberg and Jensen, 2010), which is an obstacle to the systematic study of plant glycoproteins.
Golgi Distribution of Glycan Modifying Enzymes
It has been proposed that the N-glycan modifying enzymes described above are spatially separated in the different Golgi stacks consistent with the sequence of glycan processing and maturation (Lerouge et al., 1998). Thus, their localization must be highly regulated in the Golgi. Previous efforts to explain this regulation have shown several Golgi localization or retention determinants in type-II membrane enzymes for N-glycosylation and type-I membrane proteins. The transmembrane domain (TMD) itself has been found to be important for the ER exit and Golgi localization (Brandizzi et al., 2002; Saint-Jore-Dupas et al., 2006). The membrane thickness is different among endomembrane systems: the ER and the Golgi membranes are 4–5 nm thick, while the post-Golgi membranes are thicker than those of the ER, or the Golgi membranes of fungi and mammals (Grove et al., 1968), suggesting that the length of the TMD domain is important for insertion into the correct membrane. Studies of the human lysosomal protein LAMP1 demonstrated that TMD lengths of 17, 20, and 23 amino acids are required for the insertion into the membranes of the ER, Golgi or the plasma membrane, respectively. A similar distribution dependency on TMD length has been shown for the TMD of pea vacuolar sorting receptor BP-80, a type-I membrane protein (Brandizzi et al., 2002), and the TMD of soybean α-1,2 mannosidase I (ManI) appears to be sufficient for the Golgi targeting (Saint-Jore-Dupas et al., 2006). While TMD length is important for membrane insertion, it has also been proposed that the composition of amino acids in the TMD, together with membrane lipid composition, is important for the Golgi localization in mammalian cells. Cis-, medial-, and trans-Golgi membranes have been found to have different sphingolipid:glycerophospholipid ratios and the nature of the amino acids that comprise the TMD may influence the affinities or partitioning with the membrane structural variants, and thus localization in different Golgi compartments (Patterson et al., 2008; Jackson, 2009). It is not yet known whether this model also applies to plants.
In addition to the TMD, the cytosolic tail (CT) domain of ER or Golgi membrane bound enzymes contains sorting signals. The CT domain has been investigated for ER retention in plant and mammalian cells, as well as intra-Golgi trafficking in yeast and mammalian cells. It has been suggested that there are several types of CT domain, which have a role in determining the localization of the proteins. The di-lysine motif in the CT of type-I membrane proteins and di-arginine motif in the CT of type-II membrane proteins are necessary for the COPI-mediated ER retrieval and are conserved in mammals, yeasts, and plants (Benghezal et al., 2000; McCartney et al., 2004; Kabuss et al., 2005; Okamoto et al., 2008; Boulaflous et al., 2009; Uemura et al., 2009). A number of yeast proteins have been found to have Vps74p binding motifs in their CT domain (Tu and Banfield, 2010). Vps74p forms a tetramer complex and functions as a bridge between COPI coat protein and CT domain (Schmitz et al., 2008). The localization of Vps74p is dependent on phosphatidylinositol 4-phosphate (PtdIns4P), synthesized by the enzyme phosphatidylinositol 4-kinase, and lack of PtdIns4P causes mislocalization of a Vps74p interacting Golgi resident protein, α-1,2-mannosyltransferase, to the vacuole (Tu et al., 2008; Wood et al., 2009). In addition, knock-down of a gene encoding the lipid phosphatase Suppressor of actin mutations 1 (SAC1) disrupts the restricted distribution of PtdIns4P in the trans-Golgi network (TGN), resulting abnormal Golgi morphology and mislocalization of Golgi enzymes in mammals. This result suggests that enrichment of PtdIns4P in the TGN is required for maintaining Golgi structure and proper distribution of Golgi proteins (Cheong et al., 2010).
In addition to Vps74p, COG complexes have been shown to be involved in the distribution of Golgi proteins in yeast and Arabidopsis (Bruinsma et al., 2004; Ishikawa et al., 2008). The COG complex is considered to be a tethering factor and is required for the typical SNARE- and COPI-mediated vesicle transport (Brown and Pfeffer, 2010). Mutation in one subunit in the Arabidopsis COG complex causes mislocalization of Golgi proteins and abnormal cell growth and organization, presumably by affecting localization of glycosylation enzymes (Ishikawa et al., 2008). These results suggest that the COPI machinery contributes to steady-state Golgi distribution of enzymes with specific CT domains, which may be required for the interaction with trafficking components in COPI vesicles. Considering the suggested role of the COPI vesicles in retrograde trafficking from the cis-Golgi to the ER, or from the trans-Golgi to cis-/medial-Golgi (Donohoe et al., 2007), and the conserved role of the COG complex in Golgi distribution (Ishikawa et al., 2008), the COPI machinery could contribute to steady-state Golgi distribution of enzymes with specific CT domain, which is necessary for the interaction with trafficking components in COPI vesicles in plants. Although little is known about PtdIns4P in plants, it is known to be enriched in the trans-Golgi and plasma membrane in BY-2 cells, cowpea protoplasts and Arabidopsis (Vermeer et al., 2009). Receptors such as Vps74p and cognate interacting motif in Golgi proteins are not present in plants (Schoberer and Strasser, 2011), but a conserved role for the COG complex and high accumulation of PtdIns4P in the plant Golgi apparatus suggests that plants may utilize distinct cargo receptors and interacting motifs that utilize COPI machinery.
N-Glycoprotein Sorting and Trafficking to the Cell Surface and Apoplast
Following maturation in the ER and the GA (Figure 1), plant N-glycans can be further modified during the transit of glycoproteins to their final destinations, which can include the chloroplast, vacuole, and apoplast (Rose and Lee, 2010). Although cargo sorting can occur in earlier secretory compartments and also continue beyond the TGN, it may reach a particularly high level of complexity and sophistication in the TGN, where the sorting machinery controls multiple divergent pathways directed to spatially segregated acceptor compartments (De Matteis and Luini, 2008). No specific sorting determinants have been reported in secreted proteins, which are normally sorted at the TGN and transported to the plasma membrane/apoplast by the default pathway. This has been demonstrated by the secretion of non-plant soluble proteins fused with the signal peptide for ER translocation (Denecke et al., 1990; Batoko et al., 2000). At the TGN, vacuolar proteins are transported to the vacuole via the prevacuolar compartment (PVC), which originates from the TGN, and late PVC (Figure 2; Scheuring et al., 2011; Bottanelli et al., 2012). However, secretory vesicles have been reported to have no coat proteins and form secretory vesicle clusters at the TGN (Toyooka et al., 2007). The secretory carrier membrane protein 2 (SCAMP2) has been established as a marker of secretory vesicles containing pectin and secretory soluble GFP fusion marker (secGFP), and mammalian SCAMP2 has been reported to be involved in vesicle fusion at the PM (Liu et al., 2002, 2005a; Toyooka et al., 2007). Considering the localization and high degree of sequence conservation of plant SCAMP2 with its mammalian homolog, it could also be involved in membrane fusion at PM.
Another characterized protein in the secretory pathway is SYP121 (Figure 2), a soluble NSF (N-ethylmaleimide-sensitive factor) Attachment Protein Receptors (SNARE) at the plasma membrane. It has been reported that SYP121 is involved in many aspects of the response of plants to their environment, such as non-host resistance phenomena and cellular processes including the regulation of ion channels and membrane fusion at the plasma membrane (Geelen et al., 2002; Sutter et al., 2006; Zhang et al., 2007). The Sp2 cytosolic domain of SYP121 was reported to inhibit secretion of the fluorescent secreted marker protein secGFP, presumably by preventing SNARE-mediated vesicle fusion at the plasma membrane (Tyrrell et al., 2007). This dominant-negative feature of the Sp2 domain suggests that SYP121 is a SNARE that controls bulk flow secretory pathways. Recently, new secretory pathways for cell wall proteins, a pectin methylesterase inhibitor protein (PMEI1) and a polygalacturonase inhibitor protein (PGIP2) have been proposed (De Caroli et al., 2011). The secretion of these two proteins was not inhibited by the Sp2 domain, and a glycosylphosphatidylinositol (GPI) anchoring of PMEI1 was found to be essential for the secretion of PMEI1. The new proposed secretory pathway for PMEI1 and PGIP2 suggests that the SYP121 independent vesicles are responsible for the transport of the GPI-anchored proteins. For the characterization of the new type of vesicles, the characterization of SNAREs for membrane fusion and the visualization of vesicles containing PMEI1 or PGIP2 using electron microscopy will provide new insights into the future of the alternative secretory pathway for GPI-anchored proteins.
In addition to the above examples of features of cell wall protein associated trafficking pathways, the microtubule-dependent secretion pathway of the cellulose synthase complex (CSC) has also been established. The CSC, which is responsible for cellulose biosynthesis, is formed in the trans-Golgi, and microtubules and kinesins are involved in the distribution of CSC and cellulose deposition (Haigler and Brown, 1986; Zhong et al., 2002; Lloyd, 2006; Wightman and Turner, 2008). Recently, it was reported that CSC is transported from the Golgi to the plasma membrane along cortical microtubules (Crowell et al., 2009; Gutierrez et al., 2009), consistent with previous results. Additionally, CSC was found to be internalized under abiotic stress or cellulose synthesis inhibitor treatment, indicating the putative regulation of the CSC by endocytosis (Crowell et al., 2009; Gutierrez et al., 2009). Endocytosis in plants is important for maintaining cellular functions and transmitting signals resulting from developmental or environmental cues. Other examples include the polar distribution of auxin efflux carriers and the shuttling of the receptor-like kinase, BRI1 from the plasma membrane to endosomes for signaling (Geldner et al., 2003, 2007; Abas et al., 2006). Endocytosis of the CSC would presumably allow cellulose synthesis to be directed to new sites by redistribution to the PM, or rapidly inhibit cellulose synthesis by compartmentalizing CSC in endosomes, although a detailed mechanism has not yet been determined. Thus, investigation of the endocytic regulation of the CSC, as well as related trafficking components and signaling pathways by endocytosis is likely to provide important information on the regulation of cellulose synthesis during development or under various environmental conditions.
Functional Significance of Cell Wall Protein N-Glycosylation
There are numerous reports, spanning several decades, describing the secretion of plant glycoproteins both in vitro and in vivo (Jamet et al., 2008; Agrawal et al., 2010; Rose and Lee, 2010; Ruiz-May et al., 2010). In vitro culture systems, such as suspension cell cultures (SCCs), have provided a useful platform to study secreted glycoproteins through characterization of protein populations in the cell culture medium (Kieliszewski and Lamport, 1994; Fitchette-Laine et al., 1997; Fitchette et al., 1999; Kieliszewski, 2001; Misaki et al., 2001; Luczak et al., 2008; Agrawal et al., 2010; Ruiz-May et al., 2010). While there are several reports describing the analysis of secreted plant protein populations (Rose and Lee, 2010), there have been few systematic surveys that specifically target plant glycoproteins from complex tissues. One of the first efforts involved an analysis of N-glycoproteins from Arabidopsis stems using affinity chromatography with the carbohydrate binding lectin Concanavalin A (Con A; Minic et al., 2007). This resulted in the identification of 102 glycoproteins, 94% of which were predicted to be targeted to the secretory pathway, while 87% had putative cell wall or PM localization (Minic et al., 2007). Zhang et al. (2010) identified 127 putative Arabidopsis glycoproteins using a combination of multidimensional lectin chromatography and boric acid chromatography. N-glycopeptides for 20 proteins were also predicted using ProTerNyc software (Albenne et al., 2009). Recently, Catala et al. (2011) used affinity chromatography with Con A to identify a population of proteins that are predicted to be resident in the cell wall in ripe tomato (Solanum lycopersicum) fruit. However, in these studies no glycosylation sites or N-glycan identification and characterization were described.
While the functions of large numbers of plant cell wall proteins have been determined (Lee et al., 2004; Jamet et al., 2008; Agrawal et al., 2010; Rose and Lee, 2010), it is rare that the associated glycan structures or their functional significance are addressed. One early report described proteins that are secreted into the medium of carrot cell cultures coincident with the formation of embryogenic cells and somatic embryos (van Engelen et al., 1991). These transitions were inhibited by treatment with tunicamycin at an early preglobular stage, leading to the suggestion that N-glycoproteins secreted to the extracellular environment contribute to cell development: the active glycoprotein was subsequently identified as a secreted cationic peroxidase (Cordewener et al., 1991). However, it is important to note that such cell cultures are highly artificial experimental systems. In a related study, it was shown that a somatic embryogenesis defect in a temperature sensitive carrot cell mutant was effectively rescued by the addition to the culture medium of a mixture of proteins secreted by wild type embryo cultures (Lo Schiavo et al., 1990). The active component was purified and shown to be an acidic endochitinase bearing complex type N-glycans (De Jong et al., 1992).
In addition to the glycans that are attached to proteins, a variety of free N-glycans have been found in plant tissues as precursors of glycosylation or resulting from glycoprotein proteolysis (Priem and Gross, 1992; Nakamura et al., 2008; Meli et al., 2010). Particular attention has been paid to N-glycans that are present in the pericarp tissue of tomato fruit and that both accumulate during ripening (Priem et al., 1993) and that have been reported to promote ripening when infiltrated into fruit tissues. Blocking N-glycosylation with tunicamycin slows the rate of ripening (Handa et al., 1985), but it is not clear whether this is a consequence of altering levels of free N-glycans, or modifying the properties of the many ripening related N-glycoproteins. Recently, Meli et al. (2010) reported that suppression of α-mannosidase or β-N-acetyl hexosaminidase genes reduced rates of softening in transgenic tomato fruits. α-mannosidase cleaves the terminal α-mannosidic linkages from both the high mannose type and plant complex type N-glycans (Kimura et al., 2002), while β-N-acetyl hexosaminidase removes terminal N-acetyl-D-hexosamine residues and generates the paucimannosidic N-glycans present in most plant glycoproteins (Takahashi and Nishibe, 1978; Nakamura et al., 2009). Both enzymes are present at high levels in ripening fruit from a range of species, including tomato (Suvarnalatha and Prabha, 1999; Jagadeesh et al., 2004). The authors also reported that the α-mannosidase and β-N-acetyl hexosaminidase suppressed fruits showed reduced levels of the transcripts expression encoding a range of glycoproteins that are associated with cell wall degradation in (Meli et al., 2010). In this study, impaired glycosylation in the transgenic fruit would clearly have affected many glycoproteins and no new insights were provided into the specific proteins that contribute to fruit softening, but the results do suggest the existence of some currently unknown mechanism of feedback regulation.
Conclusion and Future Perspectives
Glycoproteins have long been of interest to biochemists and biologists from a wide range of fields. It is now clear that that the glycans on glycoproteins play numerous important roles, such as influencing protein folding, regulation of protein function by differential glycan processing (Moloney et al., 2000; Lee et al., 2005; Lauc, 2006; Lauc et al., 2010), providing protection from proteases, acting as recognition motifs for specific lectins and mediating cell-cell interactions (Lee and Lee, 1995), as well as enabling intracellular protein transport (Lauc and Heffer-Lauc, 2006) and many other functions, some of which are still poorly understood (Varki, 1993).
Most studies of glycoproteins focus on their protein components (Lauc, 2006). However, in order to have a comprehensive knowledge of their functions and properties there can be great value in adopting an integrative approach to study both the sugar and protein moieties as a functional unit. For example, Con A, one of the most studied plant lectins, is synthesized as an inactive glycoprotein precursor and various post-translational events, such as endoproteolytic cleavages and deglycosylation, are required to generate the mature lectin. Importantly, deglycosylation appears to be a key step and N-glycanase action is sufficient for activation of the lectin precursor (Sheldon and Bowles, 1992; Ramis et al., 2001), suggesting that de-N-glycosylation is a control key for glycoprotein activation. Another example is a prion protein that has two variably occupied glycosylation sites and generally a GPI anchor (Stahl et al., 1987). The implications of glycosylation for the structure and function of this glycoprotein are far reaching and the regulation of glycan modifications and their effects on the subsequent structure and function continue to be an extremely important and active area of current research into prion diseases (Rudd et al., 2002). In such cases critical aspects of protein function would be obscure if the research focus was solely directed to the polypeptide.
The same generic conclusions should likely be applied to the glycans that decorate plant proteins, but studies of the plant glycoproteome are still in their infancy and are lagging behind equivalent analyses of their microbial, yeast and animal counterparts. Indeed, as far as we are aware, there exist no reports of systematic screening of plant glycoproteins: a remarkable deficiency in the “omics” compendium that will doubtless change in the near future as plant scientists exploit the current explosion of gene sequence information and new mass spectrometry technologies.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Funding to Jocelyn K. C. Rose for research in this area is provided by the NSF Plant Genome Research Program (DBI-0606595) and the New York State Office of Science, Technology and Academic Research (NYSTAR). Funding to Federica Brandizzi is provided by the Great Lake Bioenergy Center (GLBRC) under the grant (DE-FC02-07ER64494).
References
Abas, L., Benjamins, R., Malenica, N., Paciorek, T., Wisniewska, J., Moulinier-Anzola, J. C., Sieberer, T., Friml, J., and Luschnig, C. (2006). Intracellular trafficking and proteolysis of the Arabidopsis auxin-efflux facilitator PIN2 are involved in root gravitropism. Nat. Cell Biol. 8, 249–256.
Agrawal, G. K., Jwa, N. S., Lebrun, M. H., Job, D., and Rakwal, R. (2010). Plant secretome: unlocking secrets of the secreted proteins. Proteomics 10, 799–827.
Albenne, C., Canut, H., Boudart, G., Zhang, Y., San Clemente, H., Pont-Lezica, R., and Jamet, E. (2009). Plant cell wall proteomics: mass spectrometry data, a trove for research on protein structure/function relationships. Mol. Plant 2, 977–989.
Altmann, F., Staudacher, E., Wilson, I. B., and Marz, L. (1999). Insect cells as hosts for the expression of recombinant glycoproteins. Glycoconj. J. 16, 109–123.
An, H. J., Froehlich, J. W., and Lebrilla, C. B. (2009). Determination of glycosylation sites and site-specific heterogeneity in glycoproteins. Curr. Opin. Chem. Biol. 13, 421–426.
Appenzeller, C., Andersson, H., Kappeler, F., and Hauri, H. P. (1999). The lectin ERGIC-53 is a cargo transport receptor for glycoproteins. Nat. Cell Biol. 1, 330–334.
Avezov, E., Frenkel, Z., Ehrlich, M., Herscovics, A., and Lederkremer, G. Z. (2008). Endoplasmic reticulum (ER) mannosidase I is compartmentalized and required for N-glycan trimming to Man5-6GlcNAc2 in glycoprotein ER-associated degradation. Mol. Biol. Cell 19, 216–225.
Bakker, H., Schijlen, E., De Vries, T., Schiphorst, W. E., Jordi, W., Lommen, A., Bosch, D., and van Die, I. (2001). Plant members of the alpha1→3/4-fucosyltransferase gene family encode an alpha1→4-fucosyltransferase, potentially involved in Lewis(a) biosynthesis, and two core alpha1→3-fucosyltransferases. FEBS Lett. 507, 307–312.
Batoko, H., Zheng, H. Q., Hawes, C., and Moore, I. (2000). A rab1 GTPase is required for transport between the endoplasmic reticulum and golgi apparatus and for normal Golgi movement in plants. Plant Cell 12, 2201–2218.
Belden, W. J., and Barlowe, C. (2001). Role of Erv29p in collecting soluble secretory proteins into ER-derived transport vesicles. Science 294, 1528–1531.
Benghezal, M., Wasteneys, G. O., and Jones, D. A. (2000). The C-terminal dilysine motif confers endoplasmic reticulum localization to type I membrane proteins in plants. Plant Cell 12, 1179–1201.
Blond-Elguindi, S., Cwirla, S. E., Dower, W. J., Lipshutz, R. J., Sprang, S. R., Sambrook, J. F., and Gething, M. J. (1993). Affinity panning of a library of peptides displayed on bacteriophages reveals the binding specificity of BiP. Cell 75, 717–728.
Boisson, M., Gomord, V., Audran, C., Berger, N., Dubreucq, B., Granier, F., Lerouge, P., Faye, L., Caboche, M., and Lepiniec, L. (2001). Arabidopsis glucosidase I mutants reveal a critical role of N-glycan trimming in seed development. EMBO J. 20, 1010–1019.
Bottanelli, F., Gershlick, D. C., and Denecke, J. (2012). Evidence for sequential action of Rab5 and Rab7 GTPases in prevacuolar organelle partitioning. Traffic 13, 338–354.
Boulaflous, A., Saint-Jore-Dupas, C., Herranz-Gordo, M. C., Pagny-Salehabadi, S., Plasson, C., Garidou, F., Kiefer-Meyer, M. C., Ritzenthaler, C., Faye, L., and Gomord, V. (2009). Cytosolic N-terminal arginine-based signals together with a luminal signal target a type II membrane protein to the plant ER. BMC Plant Biol. 9, 144. doi:10.1186/1471-2229-9-144
Brandizzi, F., Frangne, N., Marc-Martin, S., Hawes, C., Neuhaus, J. M., and Paris, N. (2002). The destination for single-pass membrane proteins is influenced markedly by the length of the hydrophobic domain. Plant Cell 14, 1077–1092.
Brown, F. C., and Pfeffer, S. R. (2010). An update on transport vesicle tethering. Mol. Membr. Biol. 27, 457–461.
Bruinsma, P., Spelbrink, R. G., and Nothwehr, S. F. (2004). Retrograde transport of the mannosyltransferase Och1p to the early Golgi requires a component of the COG transport complex. J. Biol. Chem. 279, 39814–39823.
Bugg, T. D., and Brandish, P. E. (1994). From peptidoglycan to glycoproteins: common features of lipid-linked oligosaccharide biosynthesis. FEMS Microbiol. Lett. 119, 255–262.
Bunkenborg, J., Pilch, B. J., Podtelejnikov, A. V., and Wisniewski, J. R. (2004). Screening for N-glycosylated proteins by liquid chromatography mass spectrometry. Proteomics 4, 454–465.
Burda, P., and Aebi, M. (1999). The dolichol pathway of N-linked glycosylation. Biochim. Biophys. Acta 1426, 239–257.
Burn, J. E., Hocart, C. H., Birch, R. J., Cork, A. C., and Williamson, R. E. (2002a). Functional analysis of the cellulose synthase genes CesA1, CesA2, and CesA3 in Arabidopsis. Plant Physiol. 129, 797–807.
Burn, J. E., Hurley, U. A., Birch, R. J., Arioli, T., Cork, A., and Williamson, R. E. (2002b). The cellulose-deficient Arabidopsis mutant rsw3 is defective in a gene encoding a putative glucosidase II, an enzyme processing N-glycans during ER quality control. Plant J. 32, 949–960.
Catala, C., Howe, K. J., Hucko, S., Rose, J. K., and Thannhauser, T. W. (2011). Towards characterization of the glycoproteome of tomato (Solanum lycopersicum) fruit using Concanavalin A lectin affinity chromatography and LC-MALDI-MS/MS analysis. Proteomics 11, 1530–1544.
Cheong, F. Y., Sharma, V., Blagoveshchenskaya, A., Oorschot, V. M., Brankatschk, B., Klumperman, J., Freeze, H. H., and Mayinger, P. (2010). Spatial regulation of Golgi phosphatidylinositol-4-phosphate is required for enzyme localization and glycosylation fidelity. Traffic 11, 1180–1190.
Chrispeels, M. J., and Faye, L. (1996). “Production of recombinant glycoproteins with define no-immunogenic glycans,” in Transgenic Plants: A Production of Recombinant System for Industrial and Pharmaceutical Proteins, eds M. R. L. Owen and J. Pen (Chinchester: John Wiley), 99–113.
Cordewener, J., Booij, H., Vanderzandt, H., Vanengelen, F., Vankammen, A., and Devries, S. (1991). Tunicamycin-inhibited carrot somatic embryogenesis can be restored by secreted cationic peroxidase isoenzymes. Planta 184, 478–486.
Crofts, A. J., Leborgne-Castel, N., Pesca, M., Vitale, A., and Denecke, J. (1998). BiP and calreticulin form an abundant complex that is independent of endoplasmic reticulum stress. Plant Cell 10, 813–824.
Crowell, E. F., Bischoff, V., Desprez, T., Rolland, A., Stierhof, Y. D., Schumacher, K., Gonneau, M., Hofte, H., and Vernhettes, S. (2009). Pausing of Golgi bodies on microtubules regulates secretion of cellulose synthase complexes in Arabidopsis. Plant Cell 21, 1141–1154.
Dairaku, K., and Spiro, R. G. (1997). Phylogenetic survey of endomannosidase indicates late evolutionary appearance of this N-linked oligosaccharide processing enzyme. Glycobiology 7, 579–586.
De Caroli, M., Lenucci, M. S., Di Sansebastiano, G. P., Dalessandro, G., De Lorenzo, G., and Piro, G. (2011). Protein trafficking to the cell wall occurs through mechanisms distinguishable from default sorting in tobacco. Plant J. 65, 295–308.
De Jong, A. J., Cordewener, J., Lo Schiavo, F., Terzi, M., Vandekerckhove, J., van Kammen, A., and De Vries, S. C. (1992). A carrot somatic embryo mutant is rescued by chitinase. Plant Cell 4, 425–433.
De Matteis, M. A., and Luini, A. (2008). Exiting the Golgi complex. Nat. Rev. Mol. Cell Biol. 9, 273–284.
Denecke, J., Botterman, J., and Deblaere, R. (1990). Protein secretion in plant cells can occur via a default pathway. Plant Cell 2, 51–59.
Di Cola, A., Frigerio, L., Lord, J. M., Ceriotti, A., and Roberts, L. M. (2001). Ricin A chain without its partner B chain is degraded after retrotranslocation from the endoplasmic reticulum to the cytosol in plant cells. Proc. Natl. Acad. Sci. U.S.A. 98, 14726–14731.
Di Cola, A., Frigerio, L., Lord, J. M., Roberts, L. M., and Ceriotti, A. (2005). Endoplasmic reticulum-associated degradation of ricin A chain has unique and plant-specific features. Plant Physiol. 137, 287–296.
Donohoe, B. S., Kang, B. H., and Staehelin, L. A. (2007). Identification and characterization of COPIa- and COPIb-type vesicle classes associated with plant and algal Golgi. Proc. Natl. Acad. Sci. U.S.A. 104, 163–168.
Farid, A., Pabst, M., Schoberer, J., Altmann, F., Glossl, J., and Strasser, R. (2011). Arabidopsis thaliana alpha1,2-glucosyltransferase (ALG10) is required for efficient N-glycosylation and leaf growth. Plant J. 68, 314–325.
Faso, C., Chen, Y. N., Tamura, K., Held, M., Zemelis, S., Marti, L., Saravanan, R., Hummel, E., Kung, L., Miller, E., Hawes, C., and Brandizzi, F. (2009). A missense mutation in the Arabidopsis COPII coat protein Sec24A induces the formation of clusters of the endoplasmic reticulum and Golgi apparatus. Plant Cell 21, 3655–3671.
Faveeuw, C., Mallevaey, T., Paschinger, K., Wilson, I. B., Fontaine, J., Mollicone, R., Oriol, R., Altmann, F., Lerouge, P., Capron, M., and Trottein, F. (2003). Schistosome N-glycans containing core alpha 3-fucose and core beta 2-xylose epitopes are strong inducers of Th2 responses in mice. Eur. J. Immunol. 33, 1271–1281.
Fitchette, A. C., Cabanes-Macheteau, M., Marvin, L., Martin, B., Satiat-Jeunemaitre, B., Gomord, V., Crooks, K., Lerouge, P., Faye, L., and Hawes, C. (1999). Biosynthesis and immunolocalization of Lewis a-containing N-glycans in the plant cell. Plant Physiol. 121, 333–344.
Fitchette, A. C., Gomord, V., Chekkafi, A., and Faye, L. (1994). Distribution of xylosylation and fucosylation in the plant Golgi apparatus. Plant J. 5, 673–682.
Fitchette-Laine, A. C., Gomord, V., Cabanes, M., Michalski, J. C., Saint Macary, M., Foucher, B., Cavelier, B., Hawes, C., Lerouge, P., and Faye, L. (1997). N-glycans harboring the Lewis a epitope are expressed at the surface of plant cells. Plant J. 12, 1411–1417.
Gallois, P., Makishima, T., Hecht, V., Despres, B., Laudie, M., Nishimoto, T., and Cooke, R. (1997). An Arabidopsis thaliana cDNA complementing a hamster apoptosis suppressor mutant. Plant J. 11, 1325–1331.
Garcia-Casado, G., Sanchez-Monge, R., Chrispeels, M. J., Armentia, A., Salcedo, G., and Gomez, L. (1996). Role of complex asparagine-linked glycans in the allergenicity of plant glycoproteins. Glycobiology 6, 471–477.
Gaut, J. R., and Hendershot, L. M. (1993). The modification and assembly of proteins in the endoplasmic reticulum. Curr. Opin. Cell Biol. 5, 589–595.
Geelen, D., Leyman, B., Batoko, H., Di Sansabastiano, G. P., Moore, I., and Blatt, M. R. (2002). The abscisic acid-related SNARE homolog NtSyr1 contributes to secretion and growth: evidence from competition with its cytosolic domain. Plant Cell 14, 387–406.
Geldner, N., Anders, N., Wolters, H., Keicher, J., Kornberger, W., Muller, P., Delbarre, A., Ueda, T., Nakano, A., and Jurgens, G. (2003). The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell 112, 219–230.
Geldner, N., Hyman, D. L., Wang, X. L., Schumacher, K., and Chory, J. (2007). Endosomal signaling of plant steroid receptor kinase BRI1. Genes Dev. 21, 1598–1602.
Gething, M. J. (1999). Role and regulation of the ER chaperone BiP. Semin. Cell Dev. Biol. 10, 465–472.
Gillmor, C. S., Poindexter, P., Lorieau, J., Palcic, M. M., and Somerville, C. (2002). Alpha-glucosidase I is required for cellulose biosynthesis and morphogenesis in Arabidopsis. J. Cell Biol. 156, 1003–1013.
Gomord, V., Fitchette, A. C., Menu-Bouaouiche, L., Saint-Jore-Dupas, C., Plasson, C., Michaud, D., and Faye, L. (2010). Plant-specific glycosylation patterns in the context of therapeutic protein production. Plant Biotechnol. J. 8, 564–587.
Grinna, L. S., and Robbins, P. W. (1979). Glycoprotein biosynthesis. Rat liver microsomal glucosidases which process oligosaccharides. J. Biol. Chem. 254, 8814–8818.
Grove, S. N., Bracker, C. E., and Morre, D. J. (1968). Cytomembrane differentiation in endoplasmic reticulum-Golgi apparatus-vesicle complex. Science 161, 171–173.
Gundry, R. L., Raginski, K., Tarasova, Y., Tchernyshyov, I., Bausch-Fluck, D., Elliott, S. T., Boheler, K. R., van Eyk, J. E., and Wollscheid, B. (2009). The mouse C2C12 myoblast cell surface N-linked glycoproteome: identification, glycosite occupancy, and membrane orientation. Mol. Cell. Proteomics 8, 2555–2569.
Gutierrez, R., Lindeboom, J. J., Paredez, A. R., Emons, A. M., and Ehrhardt, D. W. (2009). Arabidopsis cortical microtubules position cellulose synthase delivery to the plasma membrane and interact with cellulose synthase trafficking compartments. Nat. Cell Biol. 11, 797–806.
Haigler, C. H., and Brown, R. M. (1986). Transport of rosettes from the golgi-apparatus to the plasma-membrane in isolated mesophyll-cells of zinnia-elegans during differentiation to tracheary elements in suspension-culture. Protoplasma 134, 111–120.
Handa, A. K., Singh, N. K., and Biggs, M. S. (1985). Effect of tunicamycin on invitro ripening of tomato pericarp tissue. Physiol. Plant 63, 417–424.
Hanton, S. L., Renna, L., Bortolotti, L. E., Chatre, L., Stefano, G., and Brandizzi, F. (2005). Diacidic motifs influence the export of transmembrane proteins from the endoplasmic reticulum in plant cells. Plant Cell 17, 3081–3093.
Haslam, S. M., Morris, H. R., and Dell, A. (2001). Mass spectrometric strategies: providing structural clues for helminth glycoproteins. Trends Parasitol. 17, 231–235.
Hebert, D. N., Foellmer, B., and Helenius, A. (1995). Glucose trimming and reglucosylation determine glycoprotein association with calnexin in the endoplasmic reticulum. Cell 81, 425–433.
Helenius, A., and Aebi, M. (2004). Roles of N-linked glycans in the endoplasmic reticulum. Annu. Rev. Biochem. 73, 1019–1049.
Helenius, J., and Aebi, M. (2002). Transmembrane movement of dolichol linked carbohydrates during N-glycoprotein biosynthesis in the endoplasmic reticulum. Semin. Cell Dev. Biol. 13, 171–178.
Hendershot, L., Wei, J., Gaut, J., Melnick, J., Aviel, S., and Argon, Y. (1996). Inhibition of immunoglobulin folding and secretion by dominant negative BiP ATPase mutants. Proc. Natl. Acad. Sci. U.S.A. 93, 5269–5274.
Henquet, M., Lehle, L., Schreuder, M., Rouwendal, G., Molthoff, J., Helsper, J., van der Krol, S., and Bosch, D. (2008). Identification of the gene encoding the alpha1,3-mannosyltransferase (ALG3) in Arabidopsis and characterization of downstream n-glycan processing. Plant Cell 20, 1652–1664.
Hong, Z., Jin, H., Fitchette, A. C., Xia, Y., Monk, A. M., Faye, L., and Li, J. (2009). Mutations of an alpha1,6 mannosyltransferase inhibit endoplasmic reticulum-associated degradation of defective brassinosteroid receptors in Arabidopsis. Plant Cell 21, 3792–3802.
Hubbard, S. C., and Ivatt, R. J. (1981). Synthesis and processing of asparagine-linked oligosaccharides. Annu. Rev. Biochem. 50, 555–583.
Ishikawa, T., Machida, C., Yoshioka, Y., Ueda, T., Nakano, A., and Machida, Y. (2008). EMBRYO YELLOW gene, encoding a subunit of the conserved oligomeric Golgi complex, is required for appropriate cell expansion and meristem organization in Arabidopsis thaliana. Genes Cells 13, 521–535.
Jackson, C. L. (2009). Mechanisms of transport through the Golgi complex. J. Cell Sci. 122, 443–452.
Jagadeesh, B. H., Prabha, T. N., and Srinivasan, K. (2004). Activities of glycosidases during fruit development and ripening of tomato (Lycopersicum esculantum L.): implication in fruit ripening. Plant Sci. 166, 1451–1459.
Jamet, E., Albenne, C., Boudart, G., Irshad, M., Canut, H., and Pont-Lezica, R. (2008). Recent advances in plant cell wall proteomics. Proteomics 8, 893–908.
Jin, H., Yan, Z., Nam, K. H., and Li, J. (2007). Allele-specific suppression of a defective brassinosteroid receptor reveals a physiological role of UGGT in ER quality control. Mol. Cell 26, 821–830.
Johnson, K. D., and Chrispeels, M. J. (1987). Substrate Specificities of N-acetylglucosaminyl-, fucosyl-, and xylosyltransferases that modify glycoproteins in the Golgi apparatus of bean cotyledons. Plant Physiol. 84, 1301–1308.
Jones, J., Viswanathan, K., Krag, S. S., and Betenbaugh, M. J. (2005). Polyprenyl lipid synthesis in mammalian cells expressing human cis-prenyl transferase. Biochem. Biophys. Res. Commun. 331, 379–383.
Kabuss, R., Ashikov, A., Oelmann, S., Gerardy-Schahn, R., and Bakker, H. (2005). Endoplasmic reticulum retention of the large splice variant of the UDP-galactose transporter is caused by a dilysine motif. Glycobiology 15, 905–911.
Kaji, H., Kamiie, J., Kawakami, H., Kido, K., Yamauchi, Y., Shinkawa, T., Taoka, M., Takahashi, N., and Isobe, T. (2007). Proteomics reveals N-linked glycoprotein diversity in Caenorhabditis elegans and suggests an atypical translocation mechanism for integral membrane proteins. Mol. Cell Proteomics 6, 2100–2109.
Kalz-Fuller, B., Bieberich, E., and Bause, E. (1995). Cloning and expression of glucosidase I from human hippocampus. Eur. J. Biochem. 231, 344–351.
Kamhi-Nesher, S., Shenkman, M., Tolchinsky, S., Fromm, S. V., Ehrlich, R., and Lederkremer, G. Z. (2001). A novel quality control compartment derived from the endoplasmic reticulum. Mol. Biol. Cell 12, 1711–1723.
Kaushal, G. P., and Elbein, A. D. (1986). Purification and Properties of UDP-GlcNAc:dolichyl-pyrophosphoryl-GlcNAc GlcNAc transferase from mung bean seedling. Plant Physiol. 81, 1086–1091.
Kaushal, G. P., Pastuszak, I., Hatanaka, K., and Elbein, A. D. (1990a). Purification to homogeneity and properties of glucosidase II from mung bean seedlings and suspension-cultured soybean cells. J. Biol. Chem. 265, 16271–16279.
Kaushal, G. P., Szumilo, T., Pastuszak, I., and Elbein, A. D. (1990b). Purification to homogeneity and properties of mannosidase II from mung bean seedlings. Biochemistry 29, 2168–2176.
Kaushal, G. P., Zeng, Y., and Elbein, A. D. (1993). Biosynthesis of glucosidase II in suspension-cultured soybean cells. J. Biol. Chem. 268, 14536–14542.
Kelleher, D. J., and Gilmore, R. (2006). An evolving view of the eukaryotic oligosaccharyltransferase. Glycobiology 16, 47R–62R.
Kieliszewski, M. J. (2001). The latest hype on Hyp-O-glycosylation codes. Phytochemistry 57, 319–323.
Kieliszewski, M. J., and Lamport, D. T. (1994). Extensin: repetitive motifs, functional sites, post-translational codes, and phylogeny. Plant J. 5, 157–172.
Kilker, R. D. Jr., Saunier, B., Tkacz, J. S., and Herscovics, A. (1981). Partial purification from Saccharomyces cerevisiae of a soluble glucosidase which removes the terminal glucose from the oligosaccharide Glc3Man9GlcNAc2. J. Biol. Chem. 256, 5299–5603.
Kimura, Y., Matsuo, S., Tsurusaki, S., Kimura, M., Hara-Nishimura, I., and Nishimura, M. (2002). Subcellular localization of endo-beta-N-acetylglucosaminidase and high-mannose type free N-glycans in plant cell. Biochim. Biophys. Acta 1570, 38–46.
Knauer, R., and Lehle, L. (1999). The oligosaccharyltransferase complex from Saccharomyces cerevisiae. Isolation of the OST6 gene, its synthetic interaction with OST3, and analysis of the native complex. J. Biol. Chem. 274, 17249–17256.
Koiwa, H., Li, F., Mccully, M. G., Mendoza, I., Koizumi, N., Manabe, Y., Nakagawa, Y., Zhu, J., Rus, A., Pardo, J. M., Bressan, R. A., and Hasegawa, P. M. (2003). The STT3a subunit isoform of the Arabidopsis oligosaccharyltransferase controls adaptive responses to salt/osmotic stress. Plant Cell 15, 2273–2284.
Koizumi, N., Ujino, T., Sano, H., and Chrispeels, M. J. (1999). Overexpression of a gene that encodes the first enzyme in the biosynthesis of asparagine-linked glycans makes plants resistant to tunicamycin and obviates the tunicamycin-induced unfolded protein response. Plant Physiol. 121, 353–361.
Kubelka, V., Altmann, F., Staudacher, E., Tretter, V., Marz, L., Hard, K., Kamerling, J. P., and Vliegenthart, J. F. (1993). Primary structures of the N-linked carbohydrate chains from honeybee venom phospholipase A2. Eur. J. Biochem. 213, 1193–1204.
Kuehn, M. J., Herrmann, J. M., and Schekman, R. (1998). COPII-cargo interactions direct protein sorting into ER-derived transport vesicles. Nature 391, 187–190.
Laine, A. C., Gomord, V., and Faye, L. (1991). Xylose-specific antibodies as markers of subcompartmentation of terminal glycosylation in the Golgi apparatus of sycamore cells. FEBS Lett. 295, 179–184.
Lauc, G., and Heffer-Lauc, M. (2006). Shedding and uptake of gangliosides and glycosylphosphatidylinositol-anchored proteins. Biochim. Biophys. Acta 1760, 584–602.
Lauc, G., Rudan, I., Campbell, H., and Rudd, P. M. (2010). Complex genetic regulation of protein glycosylation. Mol. Biosyst. 6, 329–335.
Lechner, J., and Wieland, F. (1989). Structure and biosynthesis of prokaryotic glycoproteins. Annu. Rev. Biochem. 58, 173–194.
Lederkremer, G. Z. (2009). Glycoprotein folding, quality control and ER-associated degradation. Curr. Opin. Struct. Biol. 19, 515–523.
Lee, A., Kolarich, D., Haynes, P. A., Jensen, P. H., Baker, M. S., and Packer, N. H. (2009). Rat liver membrane glycoproteome: enrichment by phase partitioning and glycoprotein capture. J. Proteome Res. 8, 770–781.
Lee, R. T., Lauc, G., and Lee, Y. C. (2005). Glycoproteomics: protein modifications for versatile functions. Meeting on glycoproteomics. EMBO Rep. 6, 1018–1022.
Lee, S. J., Saravanan, R. S., Damasceno, C. M., Yamane, H., Kim, B. D., and Rose, J. K. (2004). Digging deeper into the plant cell wall proteome. Plant Physiol. Biochem. 42, 979–988.
Lee, Y. C., and Lee, R. T. (1995). Carbohydrate-Protein Interactions – basis of glycobiology. Acc. Chem. Res. 28, 321–327.
Lehle, L., Strahl, S., and Tanner, W. (2006). Protein glycosylation, conserved from yeast to man: a model organism helps elucidate congenital human diseases. Angew. Chem. Int. Ed. Engl. 45, 6802–6818.
Leiter, H., Mucha, J., Staudacher, E., Grimm, R., Glossl, J., and Altmann, F. (1999). Purification, cDNA cloning, and expression of GDP-L-Fuc:Asn-linked GlcNAc alpha1,3-fucosyltransferase from mung beans. J. Biol. Chem. 274, 21830–21839.
Leonard, R., Costa, G., Darrambide, E., Lhernould, S., Fleurat-Lessard, P., Carlue, M., Gomord, V., Faye, L., and Maftah, A. (2002). The presence of Lewis a epitopes in Arabidopsis thaliana glycoconjugates depends on an active alpha4-fucosyltransferase gene. Glycobiology 12, 299–306.
Leonard, R., Kolarich, D., Paschinger, K., Altmann, F., and Wilson, I. B. (2004). A genetic and structural analysis of the N-glycosylation capabilities. Plant Mol. Biol. 55, 631–644.
Leonard, R., Lhernould, S., Carlue, M., Fleurat, P., Maftah, A., and Costa, G. (2005). Biochemical characterization of Silene alba alpha4-fucosyltransferase and Lewis a products. Glycoconj. J. 22, 71–78.
Lerouge, P., Cabanes-Macheteau, M., Rayon, C., Fischette-Laine, A. C., Gomord, V., and Faye, L. (1998). N-glycoprotein biosynthesis in plants: recent developments and future trends. Plant Mol. Biol. 38, 31–48.
Lerouxel, O., Mouille, G., Andeme-Onzighi, C., Bruyant, M. P., Seveno, M., Loutelier-Bourhis, C., Driouich, A., Hofte, H., and Lerouge, P. (2005a). Mutants in DEFECTIVE GLYCOSYLATION, an Arabidopsis homolog of an oligosaccharyltransferase complex subunit, show protein underglycosylation and defects in cell differentiation and growth. Plant J. 42, 455–468.
Lerouxel, O., Mouille, G., Andeme-Onzighi, C., Bruyant, M. P., Seveno, M., Loutelier-Bourhis, C., Driouich, A., Hofte, H., and Lerouge, P. (2005b). Mutants in DEFECTIVE GLYCOSYLATION, an Arabidopsis homolog of an oligosaccharyltransferase complex subunit, show protein underglycosylation and defects in cell differentiation and growth. Plant J. 42, 455–468.
Liebminger, E., Huttner, S., Vavra, U., Fischl, R., Schoberer, J., Grass, J., Blaukopf, C., Seifert, G. J., Altmann, F., Mach, L., and Strasser, R. (2009). Class I alpha-mannosidases are required for N-glycan processing and root development in Arabidopsis thaliana. Plant Cell 21, 3850–3867.
Liebminger, E., Veit, C., Mach, L., and Strasser, R. (2010). Mannose trimming reactions in the early stages of the N-glycan processing pathway. Plant Signal. Behav. 5, 476–478.
Liu, J. X., and Howell, S. H. (2010). Endoplasmic reticulum protein quality control and its relationship to environmental stress responses in plants. Plant Cell 22, 2930–2942.
Liu, L. X., Guo, Z. H., Tieu, Q. Y., Castle, A., and Castle, D. (2002). Role of secretory carrier membrane protein SCAMP2 in granule exocytosis. Mol. Biol. Cell 13, 4266–4278.
Liu, L. X., Liao, H. N., Castle, A., Zhang, J., Casanova, J., Szabo, G., and Castle, D. (2005a). SCAMP2 interacts with Arf6 and phospholipase D1 and links their function to exocytotic fusion pore formation in PC12 cells. Mol. Biol. Cell 16, 4463–4472.
Liu, T., Qian, W. J., Gritsenko, M. A., Camp, D. G. II, Monroe, M. E., Moore, R. J., and Smith, R. D. (2005b). Human plasma N-glycoproteome analysis by immunoaffinity subtraction, hydrazide chemistry, and mass spectrometry. J. Proteome Res. 4, 2070–2080.
Lo Schiavo, F., Giuliano, G., De Vries, S. C., Genga, A., Bollini, R., Pitto, L., Cozzani, F., Nuti-Ronchi, V., and Terzi, M. (1990). A carrot cell variant temperature sensitive for somatic embryogenesis reveals a defect in the glycosylation of extracellular proteins. Mol. Gen. Genet. 223, 385–393.
Lubas, W. A., and Spiro, R. G. (1987). Golgi endo-alpha-D-mannosidase from rat liver, a novel N-linked carbohydrate unit processing enzyme. J. Biol. Chem. 262, 3775–3781.
Luczak, M., Bugajewska, A., and Wojtaszek, P. (2008). Inhibitors of protein glycosylation or secretion change the pattern of extracellular proteins in suspension-cultured cells of Arabidopsis thaliana. Plant Physiol. Biochem. 46, 962–969.
Lupattelli, F., Pedrazzini, E., Bollini, R., Vitale, A., and Ceriotti, A. (1997). The rate of phaseolin assembly is controlled by the glucosylation state of its N-linked oligosaccharide chains. Plant Cell 9, 597–609.
Mancias, J. D., and Goldberg, J. (2008). Structural basis of cargo membrane protein discrimination by the human COPII coat machinery. EMBO J. 27, 2918–2928.
Marino, K., Bones, J., Kattla, J. J., and Rudd, P. M. (2010). A systematic approach to protein glycosylation analysis: a path through the maze. Nat. Chem. Biol. 6, 713–723.
Mast, S. W., and Moremen, K. W. (2006). Family 47 alpha-mannosidases in N-glycan processing. Meth. Enzymol. 415, 31–46.
Mayer, K., Schuller, C., Wambutt, R., Murphy, G., Volckaert, G., Pohl, T., Dusterhoft, A., Stiekema, W., Entian, K. D., Terryn, N., Harris, B., Ansorge, W., Brandt, P., Grivell, L., Rieger, M., Weichselgartner, M., De Simone, V., Obermaier, B., Mache, R., Muller, M., Kreis, M., Delseny, M., Puigdomenech, P., Watson, M., Schmidtheini, T., Reichert, B., Portatelle, D., Perez-Alonso, M., Boutry, M., Bancroft, I., Vos, P., Hoheisel, J., Zimmermann, W., Wedler, H., Ridley, P., Langham, S. A., Mccullagh, B., Bilham, L., Robben, J., van der Schueren, J., Grymonprez, B., Chuang, Y. J., Vandenbussche, F., Braeken, M., Weltjens, I., Voet, M., Bastiaens, I., Aert, R., Defoor, E., Weitzenegger, T., Bothe, G., Ramsperger, U., Hilbert, H., Braun, M., Holzer, E., Brandt, A., Peters, S., van Staveren, M., Dirkse, W., Mooijman, P., Lankhorst, R. K., Rose, M., Hauf, J., Kotter, P., Berneiser, S., Hempel, S., Feldpausch, M., Lamberth, S., van den Daele, H., De Keyser, A., Buysshaert, C., Gielen, J., Villarroel, R., De Clercq, R., van Montagu, M., Rogers, J., Cronin, A., Quail, M., Bray-Allen, S., Clark, L., Doggett, J., Hall, S., Kay, M., Lennard, N., Mclay, K., Mayes, R., Pettett, A., Rajandream, M. A., Lyne, M., Benes, V., Rechmann, S., Borkova, D., Blocker, H., Scharfe, M., Grimm, M., Lohnert, T. H., Dose, S., De Haan, M., Maarse, A., Schafer, M., Müller-Auer, S., Gabel, C., Fuchs, M., Fartmann, B., Granderath, K., Dauner, D., Herzl, A., Neumann, S., Argiriou, A., Vitale, D., Liguori, R., Piravandi, E., Massenet, O., Quigley, F., Clabauld, G., Mündlein, A., Felber, R., Schnabl, S., Hiller, R., Schmidt, W., Lecharny, A., Aubourg, S., Chefdor, F., Cooke, R., Berger, C., Montfort, A., Casacuberta, E., Gibbons, T., Weber, N., Vandenbol, M., Bargues, M., Terol, J., Torres, A., Perez-Perez, A., Purnelle, B., Bent, E., Johnson, S., Tacon, D., Jesse, T., Heijnen, L., Schwarz, S., Scholler, P., Heber, S., Francs, P., Bielke, C., Frishman, D., Haase, D., Lemcke, K., Mewes, H. W., Stocker, S., Zaccaria, P., Bevan, M., Wilson, R. K., de la Bastide, M., Habermann, K., Parnell, L., Dedhia, N., Gnoj, L., Schutz, K., Huang, E., Spiegel, L., Sehkon, M., Murray, J., Sheet, P., Cordes, M., Abu-Threideh, J., Stoneking, T., Kalicki, J., Graves, T., Harmon, G., Edwards, J., Latreille, P., Courtney, L., Cloud, J., Abbott, A., Scott, K., Johnson, D., Minx, P., Bentley, D., Fulton, B., Miller, N., Greco, T., Kemp, K., Kramer, J., Fulton, L., Mardis, E., Dante, M., Pepin, K., Hillier, L., Nelson, J., Spieth, J., Ryan, E., Andrews, S., Geisel, C., Layman, D., Du, H., Ali, J., Berghoff, A., Jones, K., Drone, K., Cotton, M., Joshu, C., Antonoiu, B., Zidanic, M., Strong, C., Sun, H., Lamar, B., Yordan, C., Ma, P., Zhong, J., Preston, R., Vil, D., Shekher, M., Matero, A., Shah, R., Swaby, I K., O’Shaughnessy, A., Rodriguez, M., Hoffmann, J., Till, S., Granat, S., Shohdy, N., Hasegawa, A., Hameed, A., Lodhi, M., Johnson, A., Chen, E., Marra, M., Martienssen, R., and McCombie, W. R. (1999). Sequence and analysis of chromosome 4 of the plant Arabidopsis thaliana. Nature 402, 769–777.
McCartney, A. W., Dyer, J. M., Dhanoa, P. K., Kim, P. K., Andrews, D. W., Mcnew, J. A., and Mullen, R. T. (2004). Membrane-bound fatty acid desaturases are inserted co-translationally into the ER and contain different ER retrieval motifs at their carboxy termini. Plant J. 37, 156–173.
Meli, V. S., Ghosh, S., Prabha, T. N., Chakraborty, N., Chakraborty, S., and Datta, A. (2010). Enhancement of fruit shelf life by suppressing N-glycan processing enzymes. Proc. Natl. Acad. Sci. U.S. A. 107, 2413–2418.
Melo, N. S., Nimtz, M., Conradt, H. S., Fevereiro, P. S., and Costa, J. (1997). Identification of the human Lewis(a) carbohydrate motif in a secretory peroxidase from a plant cell suspension culture (Vaccinium myrtillus L.). FEBS Lett. 415, 186–191.
Michael, J. M., and Kornfeld, S. (1980). Partial purification and characterization of the glucosidases involved in the processing of asparagine-linked oligosaccharides. Arch. Biochem. Biophys. 199, 249–258.
Miller, E., Antonny, B., Hamamoto, S., and Schekman, R. (2002). Cargo selection into COPII vesicles is driven by the Sec24p subunit. EMBO J. 21, 6105–6113.
Minic, Z., Jamet, E., Negroni, L., Arsene Der Garabedian, P., Zivy, M., and Jouanin, L. (2007). A sub-proteome of Arabidopsis thaliana mature stems trapped on Concanavalin A is enriched in cell wall glycoside hydrolases. J. Exp. Bot. 58, 2503–2512.
Misaki, R., Kimura, Y., Fujiyama, K., and Seki, T. (2001). Glycoproteins secreted from suspension-cultured tobacco BY2 cells have distinct glycan structures from intracellular glycoproteins. Biosci. Biotechnol. Biochem. 65, 2482–2488.
Mohnen, D., and Tierney, M. L. (2011). Plant science. Plants get Hyp to O-glycosylation. Science 332, 1393–1394.
Moloney, D. J., Shair, L. H., Lu, F. M., Xia, J., Locke, R., Matta, K. L., and Haltiwanger, R. S. (2000). Mammalian Notch1 is modified with two unusual forms of O-linked glycosylation found on epidermal growth factor-like modules. J. Biol. Chem. 275, 9604–9611.
Nakamura, K., Inoue, M., Maeda, M., Nakano, R., Hosoi, K., Fujiyama, K., and Kimura, Y. (2009). Molecular cloning and gene expression analysis of tomato endo-beta-N-acetylglucosaminidase, an endoglycosidase involved in the production of high-mannose type free N-glycans during tomato fruit ripening. Biosci. Biotechnol. Biochem. 73, 461–464.
Nakamura, K., Inoue, M., Yoshiie, T., Hosoi, K., and Kimura, Y. (2008). Changes in structural features of free N-glycan and endoglycosidase activity during tomato fruit ripening. Biosci. Biotechnol. Biochem. 72, 2936–2945.
Nilsson, I. M., and von Heijne, G. (1993). Determination of the distance between the oligosaccharyltransferase active site and the endoplasmic reticulum membrane. J. Biol. Chem. 268, 5798–5801.
Nishimura, N., and Balch, W. E. (1997). A di-acidic signal required for selective export from the endoplasmic reticulum. Science 277, 556–558.
Nothaft, H., and Szymanski, C. M. (2010). Protein glycosylation in bacteria: sweeter than ever. Nat. Rev. Microbiol. 11, 765–778.
Okamoto, M., Yoko-O, T., Miyakawa, T., and Jigami, Y. (2008). The cytoplasmic region of alpha-1,6-mannosyltransferase Mnn9p is crucial for retrograde transport from the Golgi apparatus to the endoplasmic reticulum in Saccharomyces cerevisiae. Eukaryotic Cell 7, 310–318.
O’Reilly, M. K., Zhang, G., and Imperiali, B. (2006). In vitro evidence for the dual function of Alg2 and Alg11: essential mannosyltransferases in N-linked glycoprotein biosynthesis. Biochemistry 45, 9593–9603.
Oriol, R., Mollicone, R., Cailleau, A., Balanzino, L., and Breton, C. (1999). Divergent evolution of fucosyltransferase genes from vertebrates, invertebrates, and bacteria. Glycobiology 9, 323–334.
Otte, S., and Barlowe, C. (2004). Sorting signals can direct receptor-mediated export of soluble proteins into COPII vesicles. Nat. Cell Biol. 6, 1189–1194.
Pagny, S., Bouissonnie, F., Sarkar, M., Follet-Gueye, M. L., Driouich, A., Schachter, H., Faye, L., and Gomord, V. (2003). Structural requirements for Arabidopsis beta1,2-xylosyltransferase activity and targeting to the Golgi. Plant J. 33, 189–203.
Palma, A. S., Vila-Verde, C., Pires, A. S., Fevereiro, P. S., and Costa, J. (2001). A novel plant alpha4-fucosyltransferase (Vaccinium myrtillus L.) synthesises the Lewis(a) adhesion determinant. FEBS Lett. 499, 235–238.
Panstruga, R., Buschges, R., Piffanelli, P., and Schulze-Lefert, P. (1998). A contiguous 60 kb genomic stretch from barley reveals molecular evidence for gene islands in a monocot genome. Nucleic Acids Res. 26, 1056–1062.
Patterson, G. H., Hirschberg, K., Polishchuk, R. S., Gerlich, D., Phair, R. D., and Lippincott-Schwartz, J. (2008). Transport through the Golgi apparatus by rapid partitioning within a two-phase membrane system. Cell 133, 1055–1067.
Pattison, R. J., and Amtmann, A. (2009). N-glycan production in the endoplasmic reticulum of plants. Trends Plant Sci. 14, 92–99.
Phillipson, B. A., Pimpl, P., Dasilva, L. L. P., Crofts, A. J., Taylor, J. P., Movafeghi, A., Robinson, D. G., and Denecke, J. (2001). Secretory bulk flow of soluble proteins is efficient and COPII dependent. Plant Cell 13, 2005–2020.
Pless, D. D., and Lennarz, W. J. (1977). Enzymatic conversion of proteins to glycoproteins. Proc. Natl. Acad. Sci. U.S.A. 74, 134–138.
Priem, B., Gitti, R., Bush, C. A., and Gross, K. C. (1993). Structure of 10 free N-glycans in ripening tomato fruit – arabinose is a constituent of a plant N-glycan. Plant Physiol. 102, 445–458.
Priem, B., and Gross, K. C. (1992). Mannosyl-containing and xylosyl-containing glycans promote tomato (Lycopersicon-esculentum Mill) fruit ripening. Plant Physiol. 98, 399–401.
Ramis, C., Gomord, V., Lerouge, P., and Faye, L. (2001). Deglycosylation is necessary but not sufficient for activation of proconcanavalin A. J. Exp. Bot. 52, 911–917.
Romero, P. A., Dijkgraaf, G. J., Shahinian, S., Herscovics, A., and Bussey, H. (1997). The yeast CWH41 gene encodes glucosidase I. Glycobiology 7, 997–1004.
Rose, J. K. C., and Lee, S. J. (2010). Straying off the highway: trafficking of secreted plant proteins and complexity in the plant cell wall proteome. Plant Physiol. 153, 433–436.
Ross, J., Li, Y., Lim, E., and Bowles, D. J. (2001). Higher plant glycosyltransferases. Genome Biol. 2, reviews3004.
Rudd, P. M., Merry, A. H., Wormald, M. R., and Dwek, R. A. (2002). Glycosylation and prion protein. Curr. Opin. Struct. Biol. 12, 578–586.
Ruddock, L. W., and Molinari, M. (2006). N-glycan processing in ER quality control. J. Cell. Sci. 119, 4373–4380.
Ruiz-Canada, C., Kelleher, D. J., and Gilmore, R. (2009). Cotranslational and posttranslational N-glycosylation of polypeptides by distinct mammalian OST isoforms. Cell 136, 272–283.
Ruiz-May, E., De La Peña, C., Ayil-Gutiérrez, B. A., Nic-Can, G. I., Mukul-López, H. G., Galaz-Ávalos, R. M., and Loyola-Vargas, V. M. (2010). Protein secretion by cell cultures: an essential biological issue. Acta Hortic. 849, 213–222.
Saint-Jore-Dupas, C., Nebenfuhr, A., Boulaflous, A., Follet-Gueye, M. L., Plasson, C., Hawes, C., Driouich, A., Faye, L., and Gomord, V. (2006). Plant N-glycan processing enzymes employ different targeting mechanisms for their spatial arrangement along the secretory pathway. Plant Cell 18, 3182–3200.
Scheuring, D., Viotti, C., Kruger, F., Kunzl, F., Sturm, S., Bubeck, J., Hillmer, S., Frigerio, L., Robinson, D. G., Pimpl, P., and Schumacher, K. (2011). Multivesicular bodies mature from the trans-golgi network/early endosome in Arabidopsis. Plant Cell 23, 3463–3481.
Schmitz, K. R., Liu, J., Li, S., Setty, T. G., Wood, C. S., Burd, C. G., and Ferguson, K. M. (2008). Golgi localization of glycosyltransferases requires a Vps74p oligomer. Dev. Cell 14, 523–534.
Schoberer, J., and Strasser, R. (2011). Sub-compartmental organization of Golgi-resident N-glycan processing enzymes in plants. Mol. Plant 4, 220–228.
Sheldon, P. S., and Bowles, D. J. (1992). The Glycoprotein precursor of concanavalin-a is converted to an active lectin by deglycosylation. EMBO J. 11, 1297–1301.
Showalter, A. M. (2001). Arabinogalactan-proteins: structure, expression and function. Cell. Mol. Life Sci. 58, 1399–1417.
Sieben, C., Mikosch, M., Brandizzi, F., and Homann, U. (2008). Interaction of the K(+)-channel KAT1 with the coat protein complex II coat component Sec24 depends on a di-acidic endoplasmic reticulum export motif. Plant J. 56, 997–1006.
Spiro, R. G. (2002). Protein glycosylation: nature, distribution, enzymatic formation, and disease implications of glycopeptide bonds. Glycobiology 12, 43R–56R.
Sprong, H., van der Sluijs, P., and van Meer, G. (2001). How proteins move lipids and lipids move proteins. Nat. Rev. Mol. Cell Biol. 2, 504–513.
Stahl, N., Borchelt, D. R., Hsiao, K., and Prusiner, S. B. (1987). Scrapie prion protein contains a phosphatidylinositol glycolipid. Cell 51, 229–240.
Strasser, R., Bondili, J. S., Vavra, U., Schoberer, J., Svoboda, B., Glossl, J., Leonard, R., Stadlmann, J., Altmann, F., Steinkellner, H., and Mach, L. (2007). A unique beta1,3-galactosyltransferase is indispensable for the biosynthesis of N-glycans containing Lewis a structures in Arabidopsis thaliana. Plant Cell 19, 2278–2292.
Strasser, R., Mucha, J., Mach, L., Altmann, F., Wilson, I. B., Glossl, J., and Steinkellner, H. (2000). Molecular cloning and functional expression of beta1, 2-xylosyltransferase cDNA from Arabidopsis thaliana. FEBS Lett. 472, 105–108.
Strasser, R., Mucha, J., Schwihla, H., Altmann, F., Glossl, J., and Steinkellner, H. (1999). Molecular cloning and characterization of cDNA coding for beta1, 2N-acetylglucosaminyltransferase I (GlcNAc-TI) from Nicotiana tabacum. Glycobiology 9, 779–785.
Strasser, R., Schoberer, J., Jin, C., Glossl, J., Mach, L., and Steinkellner, H. (2006). Molecular cloning and characterization of Arabidopsis thaliana Golgi alpha-mannosidase II, a key enzyme in the formation of complex N-glycans in plants. Plant J. 45, 789–803.
Sutter, J. U., Campanoni, P., Tyrrell, M., and Blatt, M. R. (2006). Selective mobility and sensitivity to SNAREs is exhibited by the Arabidopsis KAT1 K+ channel at the plasma membrane. Plant Cell 18, 935–954.
Suvarnalatha, G., and Prabha, T. N. (1999). Alpha-D-mannosidase from Lycopersicon esculentum – II. Phytochemistry 50, 1111–1115.
Szumilo, T., Kaushal, G. P., and Elbein, A. D. (1986a). Purification and properties of glucosidase I from mung bean seedlings. Arch. Biochem. Biophys. 247, 261–271.
Szumilo, T., Kaushal, G. P., Hori, H., and Elbein, A. D. (1986b). Purification and properties of a glycoprotein processing alpha-mannosidase from mung bean seedlings. Plant Physiol. 81, 383–389.
Takahashi, N., and Nishibe, H. (1978). Some characteristics of a new glycopeptidase acting on aspartylglycosylamine linkages. J. Biochem. 84, 1467–1473.
Taylor, C. M., Karunaratne, C. V., and Xie, N. (2011). Glycosides of hydroxyproline: some recent, unusual discoveries. Glycobiology 22, 757–767.
Taylor, M. A., Ross, H. A., Mcrae, D., Stewart, D., Roberts, I., Duncan, G., Wright, F., Millam, S., and Davies, H. V. (2000). A potato alpha-glucosidase gene encodes a glycoprotein-processing alpha-glucosidase II-like activity. Demonstration of enzyme activity and effects of down-regulation in transgenic plants. Plant J. 24, 305–316.
Tezuka, K., Hayashi, M., Ishihara, H., Akazawa, T., and Takahashi, N. (1992). Studies on synthetic pathway of xylose-containing N-linked oligosaccharides deduced from substrate specificities of the processing enzymes in sycamore cells (Acer pseudoplatanus L.). Eur. J. Biochem. 203, 401–413.
Toyooka, K., Goto, Y., Asatsuma, S., and Matsuoka, K. (2007). SCAMP-containing vesicle cluster (SVC), a post-Golgi exocytotic organelle. Plant Cell Physiol. 48, S122–S122.
Trombetta, E. S., Simons, J. F., and Helenius, A. (1996). Endoplasmic reticulum glucosidase II is composed of a catalytic subunit, conserved from yeast to mammals, and a tightly bound noncatalytic HDEL-containing subunit. J. Biol. Chem. 271, 27509–27516.
Tu, L., Tai, W. C. S., Chen, L., and Banfield, D. K. (2008). Signal-mediated dynamic retention of glycosyltransferases in the Golgi. Science 321, 404–407.
Tu, L. N., and Banfield, D. K. (2010). Localization of Golgi-resident glycosyltransferases. Cell. Mol. Life Sci. 67, 29–41.
Tyrrell, M., Campanoni, P., Sutter, J. U., Pratelli, R., Paneque, M., Sokolovski, S., and Blatt, M. R. (2007). Selective targeting of plasma membrane and tonoplast traffic by inhibitory (dominant-negative) SNARE fragments. Plant J. 51, 1099–1115.
Uemura, S., Yoshida, S., Shishido, F., and Inokuchi, J. (2009). The cytoplasmic tail of Gm3 synthase defines its subcellular localization, stability, and in vivo activity. Mol. Biol. Cell 20, 3088–3100.
van Engelen, F. A., Sterk, P., Booij, H., Cordewener, J. H., Rook, W., van Kammen, A., and De Vries, S. C. (1991). Heterogeneity and cell type-specific localization of a cell wall glycoprotein from carrot suspension cells. Plant Physiol. 96, 705–712.
van Kuik, J. A., van Halbeek, H., Kamerling, J. P., and Vliegenthart, J. F. (1985). Primary structure of the low-molecular-weight carbohydrate chains of Helix pomatia alpha-hemocyanin. Xylose as a constituent of N-linked oligosaccharides in an animal glycoprotein. J. Biol. Chem. 260, 13984–13988.
van Ree, R., Cabanes-Macheteau, M., Akkerdaas, J., Milazzo, J. P., Loutelier-Bourhis, C., Rayon, C., Villalba, M., Koppelman, S., Aalberse, R., Rodriguez, R., Faye, L., and Lerouge, P. (2000). Beta(1,2)-xylose and alpha(1,3)-fucose residues have a strong contribution in IgE binding to plant glycoallergens. J. Biol. Chem. 275, 11451–11458.
Varki, A. (1993). Biological roles of oligosaccharides – all of the theories are correct. Glycobiology 3, 97–130.
Velasquez, S. M., Ricardi, M. M., Dorosz, J. G., Fernandez, P. V., Nadra, A. D., Pol-Fachin, L., Egelund, J., Gille, S., Harholt, J., Ciancia, M., Verli, H., Pauly, M., Bacic, A., Olsen, C. E., Ulvskov, P., Petersen, B. L., Somerville, C., Iusem, N. D., and Estevez, J. M. (2011). O-glycosylated cell wall proteins are essential in root hair growth. Science 332, 1401–1403.
Vermeer, J. E., Thole, J. M., Goedhart, J., Nielsen, E., Munnik, T., and Gadella, T. W. Jr. (2009). Imaging phosphatidylinositol 4-phosphate dynamics in living plant cells. Plant J. 57, 356–372.
Votsmeier, C., and Gallwitz, D. (2001). An acidic sequence of a putative yeast Golgi membrane protein binds COPII and facilitates ER export. EMBO J. 20, 6742–6750.
Wendeler, M. W., Paccaud, J. P., and Hauri, H. P. (2007). Role of Sec24 isoforms in selective export of membrane proteins from the endoplasmic reticulum. EMBO Rep. 8, 258–264.
Wenderoth, I., and von Schaewen, A. (2000). Isolation and characterization of plant N-acetyl glucosaminyltransferase I (GntI) cDNA sequences. Functional analyses in the Arabidopsis cgl mutant and in antisense plants. Plant Physiol. 123, 1097–1108.
Wightman, R., and Turner, S. R. (2008). The roles of the cytoskeleton during cellulose deposition at the secondary cell wall. Plant J. 54, 794–805.
Wilkinson, B. M., Purswani, J., and Stirling, C. J. (2006). Yeast GTB1 encodes a subunit of glucosidase II required for glycoprotein processing in the endoplasmic reticulum. J. Biol. Chem. 281, 6325–6333.
Wilson, I. B. (2001). Identification of a cDNA encoding a plant Lewis-type alpha1,4-fucosyltransferase. Glycoconj. J. 18, 439–447.
Wilson, I. B., Rendic, D., Freilinger, A., Dumic, J., Altmann, F., Mucha, J., Muller, S., and Hauser, M. T. (2001a). Cloning and expression of cDNAs encoding alpha1,3-fucosyltransferase homologues from Arabidopsis thaliana. Biochim. Biophys. Acta 1527, 88–96.
Wilson, I. B., Zeleny, R., Kolarich, D., Staudacher, E., Stroop, C. J., Kamerling, J. P., and Altmann, F. (2001b). Analysis of Asn-linked glycans from vegetable foodstuffs: widespread occurrence of Lewis a, core alpha1,3-linked fucose and xylose substitutions. Glycobiology 11, 261–274.
Wollscheid, B., Bausch-Fluck, D., Henderson, C., O’Brien, R., Bibel, M., Schiess, R., Aebersold, R., and Watts, J. D. (2009). Mass-spectrometric identification and relative quantification of N-linked cell surface glycoproteins. Nat. Biotechnol. 27, 378–386.
Wood, C. S., Schmitz, K. R., Bessman, N. J., Setty, T. G., Ferguson, K. M., and Burd, C. G. (2009). PtdIns4P recognition by Vps74/GOLPH3 links PtdIns 4-kinase signaling to retrograde Golgi trafficking. J. Cell Biol. 187, 967–975.
Yan, Q., and Lennarz, W. J. (1999). Oligosaccharyltransferase: a complex multisubunit enzyme of the endoplasmic reticulum. Biochem. Biophys. Res. Commun. 266, 684–689.
Yan, Q., and Lennarz, W. J. (2002). Studies on the function of oligosaccharyl transferase subunits. Stt3p is directly involved in the glycosylation process. J. Biol. Chem. 277, 47692.
Ytterberg, A. J., and Jensen, O. N. (2010). Modification-specific proteomics in plant biology. J. Proteomics 73, 2249–2266.
Zeng, Y., Bannon, G., Thomas, V. H., Rice, K., Drake, R., and Elbein, A. (1997). Purification and specificity of beta1,2-xylosyltransferase, an enzyme that contributes to the allergenicity of some plant proteins. J. Biol. Chem. 272, 31340–31347.
Zeng, Y. C., and Elbein, A. D. (1998). Purification to homogeneity and properties of plant glucosidase I. Arch. Biochem. Biophys. 355, 26–34.
Zhang, H., Ohyama, K., Boudet, J., Chen, Z., Yang, J., Zhang, M., Muranaka, T., Maurel, C., Zhu, J. K., and Gong, Z. (2008). Dolichol biosynthesis and its effects on the unfolded protein response and abiotic stress resistance in Arabidopsis. Plant Cell 20, 1879–1898.
Zhang, M., Henquet, M., Chen, Z., Zhang, H., Zhang, Y., Ren, X., van der Krol, S., Gonneau, M., Bosch, D., and Gong, Z. (2009). LEW3, encoding a putative alpha-1,2-mannosyltransferase (ALG11) in N-linked glycoprotein, plays vital roles in cell-wall biosynthesis and the abiotic stress response in Arabidopsis thaliana. Plant J. 60, 983–999.
Zhang, Y., Giboulot, A., Zivy, M., Valot, B., Jamet, E., and Albenne, C. (2010). Combining various strategies to increase the coverage of the plant cell wall glycoproteome. Phytochemistry 72, 1109–1123.
Zhang, Z., Feechan, A., Pedersen, C., Newman, M. A., Qiu, J. L., Olesen, K. L., and Thordal-Christensen, H. (2007). A SNARE-protein has opposing functions in penetration resistance and defence signalling pathways. Plant J. 49, 302–312.
Zhong, R., Burk, D. H., Morrison, W. H. III, and Ye, Z. H. (2002). A kinesin-like protein is essential for oriented deposition of cellulose microfibrils and cell wall strength. Plant Cell 14, 3101–3117.
Keywords: glycoprotein, glycoproteome, secretory pathway, N-glycan, protein sorting
Citation: Ruiz-May E, Kim S-J, Brandizzi F and Rose JKC (2012) The secreted plant N-glycoproteome and associated secretory pathways. Front. Plant Sci. 3:117. doi: 10.3389/fpls.2012.00117
Received: 30 March 2012; Accepted: 15 May 2012;
Published online: 06 June 2012.
Edited by:
Seth DeBolt, University of Kentucky, USAReviewed by:
Guillaume Pilot, Virginia Tech, USAJoshua L. Heazlewood, Lawrence Berkeley National Laboratory, USA
Copyright: © 2012 Ruiz-May, Kim, Brandizzi and Rose. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Jocelyn K. C. Rose, Department of Plant Biology, Cornell University, 412 Mann Library Building, Ithaca, NY 14853 USA. e-mail: jr286@cornell.edu
