- 1 Department of Plant Physiology and Biochemistry, University Konstanz, Konstanz, Germany
- 2 Department of Plant Biology, Carnegie Institution for Science, Stanford, CA, USA
- 3 School of Biological Sciences, Washington State University, Pullman, WA, USA
Mechanisms for sensing and regulating metabolic processes at the cellular level are critical for the general physiology and development of living organisms. In higher plants, sugar signaling is crucial for adequate regulation of carbon and energy metabolism and affects virtually every aspect of development. Although many genes are regulated by sugar levels, little is known on how sugar levels are measured by plants. Several components of the sugar signaling network have been unraveled and demonstrated to have extensive overlap with hormone signaling networks. Here we describe the reduced sugar response1-1 (rsr1-1) mutant as a new early flowering mutant that displays decreased sensitivity to abscisic acid. Both hexokinase1 (HXK1)-dependent and glucose phosphorylation-independent signaling is reduced in rsr1-1. Map-based identification of the affected locus demonstrated that rsr1-1 carries a premature stop codon in the gene for a CstF64-like putative RNA processing factor, ESP1, which is involved in mRNA 3′-end formation. The identification of RSR1/ESP1 as a nuclear protein with a potential threonine phosphorylation site may explain the impact of protein phosphorylation cascades on sugar-dependent signal transduction. Additionally, RSR1/ESP1 may be a crucial factor in linking sugar signaling to the control of flowering time.
Introduction
The life cycle of Arabidopsis can be divided into five major phases: seed germination, seedling establishment, vegetative growth phase in which nutrients and biomass are accumulated, flowering, and finally senescence of the rosette to reallocate resources to the seeds during maturation (Pujar et al., 2006). Transition from one phase to another requires the coordinated induction of specific genetic programs. Under constant environmental conditions, the duration of each phase is genetically determined. However, environmental cues and the resulting alterations of the physiological status of the plant can delay or accelerate phase transitions to optimize reproductive success.
Initiation and completion of the individual growth phases can be directly controlled by environmental factors: for example, germination depends on the availability of water as well as stored nutrients. Light is a critical factor that influences the transition from vegetative to reproductive development. In addition, environmental factors are reflected by endogenous physiological parameters, which also act as signals in the regulation of development. Carbohydrate levels especially have been found to play a crucial role in all growth phases. Despite the large number of genes and processes that are known to be regulated by carbohydrates, the complete sensing and signaling cascades are established in only a few cases (Baena-Gonzalez et al., 2007; Smeekens et al., 2010).
During the earliest steps of development, external application of sugars in high concentration inhibits germination and seedling establishment by inducing genetic programs normally active during seed maturation (Lopez-Molina et al., 2002; Dekkers et al., 2008). Inhibition of de-etiolation by glucose (Glc) is dependent on hexokinase1 (HXK1) as a sugar sensor and is discussed to be mediated by elevated abscisic acid (ABA) levels and ABA-dependent signaling. Components of the underlying signaling pathway were revealed by screening for mutants with Glc-insensitive de-etiolation (gin) or similar sugar-dependent phenotypes (Rolland et al., 2002; Rognoni et al., 2007). However, it is so far not known how HXK1 is involved in generating elevated ABA levels (Eveland and Jackson, 2011). Besides seedling establishment, HXK1 also contributes to numerous other Glc-regulated processes. Interestingly, the signaling function of Arabidopsis HXK1 was separable from its catalytic activity (Moore et al., 2003; Cho et al., 2006). Evidence for metabolism-independent Glc sensing and signaling was also derived from effects that were triggered by Glc analogs that are not readily phosphorylated by plant hexokinases or are not further metabolized, such as 3-O-methylglucose (3OmeG) and 6-deoxyglucose (6DG). However, microarray analysis of the full Arabidopsis transcriptome failed to identify genes that were regulated by Glc and such analogs as 3OmeG or 6DG (Villadsen and Smith, 2004; Baena-Gonzalez et al., 2007).
In the adult life of Arabidopsis, the decision to initiate flowering is of crucial importance for reproductive success. Accordingly, this transition is regulated by environmental factors in combination with signals derived from the nutritional status of the plant (Ausin et al., 2005; Srikanth and Schmid, 2011). Day length, temperature, hormones, and autonomous endogenous mechanisms each trigger signaling in partially overlapping cascades that converge on the level of the so called floral pathway integrators, which in turn activate genes that induce and maintain the transition from a vegetative to a floral apical meristem (Srikanth and Schmid, 2011). The output of these signaling events is determined by a combination of classical induction and repression of transcription with posttranscriptional regulation. Alternative pre-mRNA processing, miRNA-mediated mRNA degradation, and regulated changes in mRNA stability all contribute to control the appropriate level of floral pathway integrators and floral identity effectors (Quesada et al., 2005; Kuhn et al., 2007). The nutrient-dependent regulation of flowering seems to depend on the rate of sucrose (Suc) export from source leaves as well as on the availability of proline (Pro), which is found in high concentrations in floral organs (Corbesier et al., 1998; Sivitz et al., 2007; Mattioli et al., 2009).
In the final stage of the life cycle of an Arabidopsis plant, the rosette is sacrificed to recycle resources contained in the leaves for the promotion of seed development and maturation. Leaf senescence is a tightly regulated, specialized form of programmed cell death (PCD) in which events at the cellular and tissue level are highly coordinated (Guiboileau et al., 2010). Other, more localized forms of PCD occur in response to environmental factors, especially in the defense against pathogens. Pathogen-induced PCD is often accompanied by a hypersensitive response (HR), a rapid induction of reactive oxygen species (ROS)-generating processes that kill both the host cells and the intruder (Greenberg and Yao, 2004). At sub-lethal levels, ROS are also used as signaling molecules that trigger acclimation or defense responses, including the induction of PCD (Gechev et al., 2006; Karuppanapandian et al., 2011). In addition, alternative mRNA processing was identified as an additional mechanism in the signaling cascades regulating HR and PCD (Zhang and Gassmann, 2007).
In the present study we describe the influence of the RSR1 locus on multiple developmental transitions, including the spontaneous induction of cell death. The rsr1-1 mutant was identified in a screen for Arabidopsis mutants with altered sugar signaling using the patatin (B33) promoter from potato fused to a Gus reporter gene (Martin et al., 1997). Suc, Glc, and the Glc analog 3OmeG induced expression from the Pat(B33)-Gus promoter-reporter construct, indicating the involvement of HXK-independent sugar signaling. The rsr1-1 mutation blocked sugar induction of the Pat(B33)-promoter almost completely and additionally resulted in an altered regulation of Pro catabolism and hypersensitivity to Pro-induced cell death (Hellmann et al., 2000). In addition to altered cell death regulation, we demonstrate in this study that RSR1 is also a critical factor for sugar signaling during germination and early seedling development. Early flowering of the rsr1-1 mutant indicated that RSR1 is also involved in the regulation of the transition from the vegetative to the generative phase. Map-based cloning revealed that rsr1-1 represents a novel mutant allele of enhanced silencing phenotype 1 (ESP1), encoding a putative component of the mRNA 3′ processing machinery (Herr et al., 2006). The current work establishes RSR1/ESP1 as a mediator in carbohydrate-dependent regulation of developmental processes, potentially by affecting transcript stability.
Results
rsr1-1 is an Early Flowering Mutant that Develops Spontaneous Lesions
Our initial characterization defined rsr1-1 as a mutant with impaired HXK-independent sugar induction of the patatin class I promoter from potato and altered regulation of proline metabolism and sensitivity (Martin et al., 1997; Hellmann et al., 2000). In addition to these phenotypes, rsr1-1 mutants displayed a variety of differences in appearance to the parental line Pat(B33)-Gus when grown on soil: First, the chlorophyll content of rosette leaves was reduced by approximately 25%, which caused a yellowish appearance of the leaves (Figure 1A). Second, the mutant also displayed spontaneous lesions on all leaves under long-day conditions in the greenhouse (Figure 1B). Staining of leaves with 3,3′-diaminobenzidine revealed increased H2O2 production in these lesions (Figure 1C). Furthermore, rsr1-1 mutants developed the first inflorescence about 10 days earlier than control plants (Figures 2A,C), which correlated with a reduced leaf number at the onset of bolting (Figure 2B), classifying rsr1-1 as a novel early flowering mutant. Additionally, changes in root morphology were observed when rsr1-1 mutants were cultured on MS-medium supplemented with 2% Suc: under such conditions roots of rsr1-1 seedlings were almost twice as long as observed for Pat(B33)-Gus plants (Figure 2C). In contrast, root hairs of rsr1-1 seedlings were slightly deformed and shorter in comparison to control plants (Figures 2D,E).
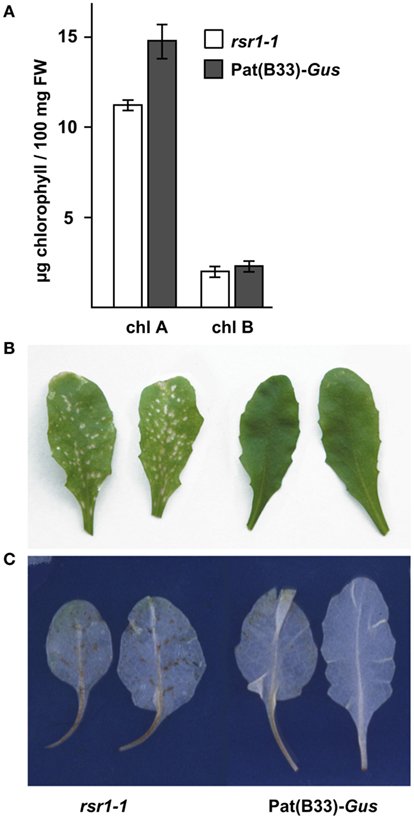
Figure 1. rsr1-1 has reduced chlorophyll and forms spontaneous lesions. (A) Chlorophyll was extracted from 30-day-old soil grown plants. Chlorophyll A and B content was significantly reduced in the rsr1-1 mutant compared to the parental line Pat(B33)-Gus. Values presented are the mean ± SD from 10 plants. (B) Under long-day conditions in the greenhouse, rsr1-1 mutants displayed spontaneous lesion formation on all leaves. (C) Infiltration with 3,3′-diaminobenzidine demonstrated ROS accumulation in lesions of rsr1-1 leaves.
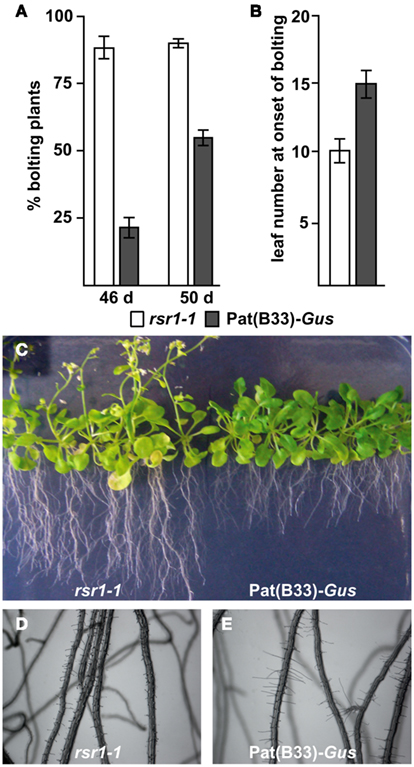
Figure 2. rsr1-1 is early flowering and has an altered root morphology. (A) Under short-day conditions rsr1-1 plants started flowering much earlier than the parental line Pat(B33)-Gus. (B) Under long-day conditions, rsr1-1 plants formed fewer primary rosette leaves than Pat(B33)-Gus. In (A,B) the values presented are the mean ± SD of ≥15 plants. (C) In sterile culture on MS-medium with 60 mM Suc, rsr1-1 mutants maintained the early flowering phenotype and produced roots nearly twice as long as those of Pat(B33)-Gus. Under the same conditions, root hairs were shorter in rsr1-1 (D) than in Pat(B33)-Gus (E) and slightly irregular in shape.
rsr1-1 is Tolerant to High Glc and ABA Levels at Early Developmental Stages
To determine if altered sugar sensitivity of rsr1-1 was manifested already at the embryonic stage, germination of Pat(B33)-Gus and rsr1-1 seeds was assayed on MS-plates containing either 30 mM Glc, 330 mM Glc, or 30 mM Glc plus 300 mM 3Omeg. As an osmotic control, a combination of 30 mM Glc and 300 mM sorbitol was applied. Germination was defined as the time point when the radicle breaks through the seed coat. At 30 mM Glc, more than 95% of the seeds germinated within 3 days after plating, and no difference was detected between rsr1-1 and Pat(B33)-Gus seeds (Figures 3A,B). All other treatments delayed germination of both lines markedly. Interestingly, high concentrations of 3Omeg or Glc slowed germination to a greater extent than sorbitol in Pat(B33)-Gus seeds (Figure 3A). A sugar-specific effect that is mimicked by 3Omeg indicates that the underlying signaling cascade is independent of Glc phosphorylation and further metabolism. In rsr1-1 seeds, the effects of Glc and 3Omeg did not differ from the osmotic control, indicating that the sugar-specific signal was not correctly transmitted (Figure 3B). The effect of 3Omeg was specific for germination, as seedlings of both lines, Pat(B33)-Gus and rsr1-1, did not display any developmental arrest on medium containing 30 mM Glc plus 300 mM 3Omeg (data not shown).
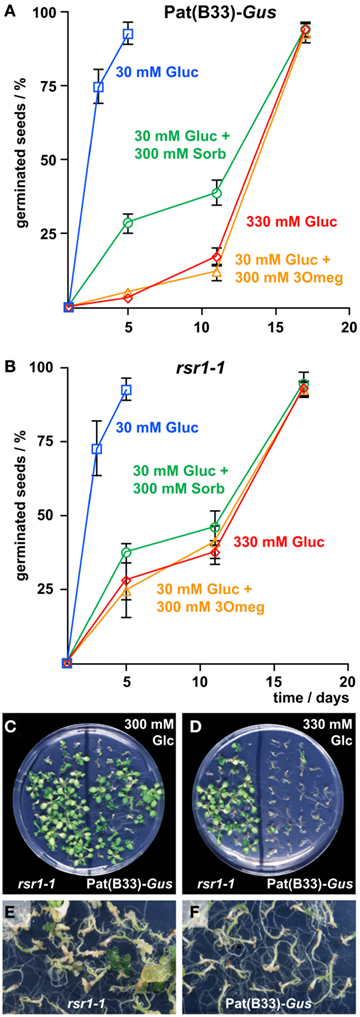
Figure 3. Sugar insensitivity of germination and establishment in rsr1-1. (A,B) Germination rates of rsr1-1 and Pat(B33)-Gus seeds on MS-medium supplemented with different concentrations of Glc, 3Omeg, or sorbitol. Only on 330 mM Glc and on 30 mM Glc + 300 mM 3Omeg significant differences were observed between rsr1-1 and Pat(B33)-Gus. Values represent the mean ± SD of three replica of 70 seeds each. (C) On plates containing 300 mM Glc both rsr1-1 and Pat(B33)-Gus were able to initiate chlorophyll synthesis and the formation of true leaves. (D) 330 mM D-Glc inhibited establishment of autotrophic growth of Pat(B33)-Gus whereas rsr1-1 could still develop normally. (E) After 2 months on MS-medium supplemented with 360 mM D-Glc, rsr1-1 developed callus-like enlargements of the apical meristem, which were not observed in Pat(B33)-Gus(F).
At later developmental stages, it is known that seedling establishment is inhibited by high sugar concentrations, and a variety of mutants have been described that are insensitive to this inhibitory effect (Pego et al., 2000; Rolland et al., 2002). To investigate whether rsr1-1 is also a Glc-insensitive mutant, Pat(B33)-Gus and rsr1-1 seedlings were cultured on growth medium supplemented with increasing D-Glc concentrations. While Pat(B33)-Gus plants were arrested in development at the cotyledon stage, and chlorophyll synthesis was blocked at concentrations higher than 300 mM (Figures 3C,D), rsr1-1 seedlings still developed green cotyledons followed by true leaves on medium containing up to 330 mM Glc. However, further increase in Glc (360–380 mM) also resulted in arrest of rsr1-1 development. After 6–8 weeks on 360 mM Glc, rsr1-1 plants developed callus-like tissue at the apical meristem, which was not observed in Pat(B33)-Gus (Figures 3E,F). These experiments demonstrate that mutation of RSR1 shifted the upper limit of Glc-tolerance, but did not result in complete Glc insensitivity.
Earlier studies had demonstrated that the inhibitory effect of high Glc concentrations on seedling establishment involves ABA signaling (Dekkers et al., 2008; Wingler and Roitsch, 2008), while low Glc concentrations were shown to reduce the inhibitory effect of ABA on germination and development (Finkelstein and Lynch, 2000). Addition of 3 μM ABA to the growth medium strongly delayed germination seeds from both, rsr1-1 mutants and the parental line Pat(B33)-Gus, but still allowed germination of nearly all seeds. Seedling establishment in both lines was completely blocked by 3 μM ABA on sugar-free MS-medium (Figure 4A). Addition of 30 mM Glc attenuated the delay in germination in both lines. While rsr1-1 plants accumulated chlorophyll and later on developed true leaves under these conditions, Pat(B33)-Gus seedlings remained white and cotyledons did not expand (Figure 4B). However, combinations of 5 μM ABA with 30 mM Glc or 3 μM ABA with more than 90 mM Glc inhibited development of rsr1-1 seedlings equal to Pat(B33)-Gus (data not shown).
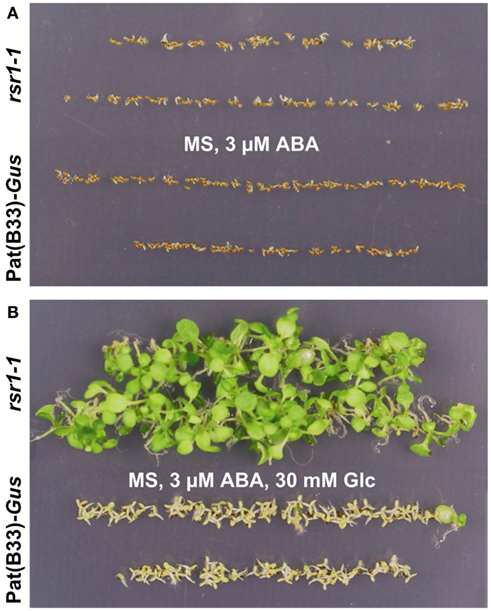
Figure 4. Abscisic acid (ABA)-insensitivity of rsr1-1 is Glc-dependent. Seeds of rsr1-1 (upper lanes) and Pat(B33)-Gus (lower lanes) were plated on MS-medium supplemented either with (A) 3 μM ABA or (B) 3 μM ABA + 30 mM Glc. In the presence of both Glc and ABA, rsr1-1 seedlings had developed chlorophyll and primary leaves 4 weeks after plating, whereas 3 μM ABA alone delayed germination and completely inhibited development in both rsr1-1 and Pat(B33)-Gus.
rsr1-1 is a Novel Mutant Allele of ESP1, a Putative CstF64-like RNA Processing Factor
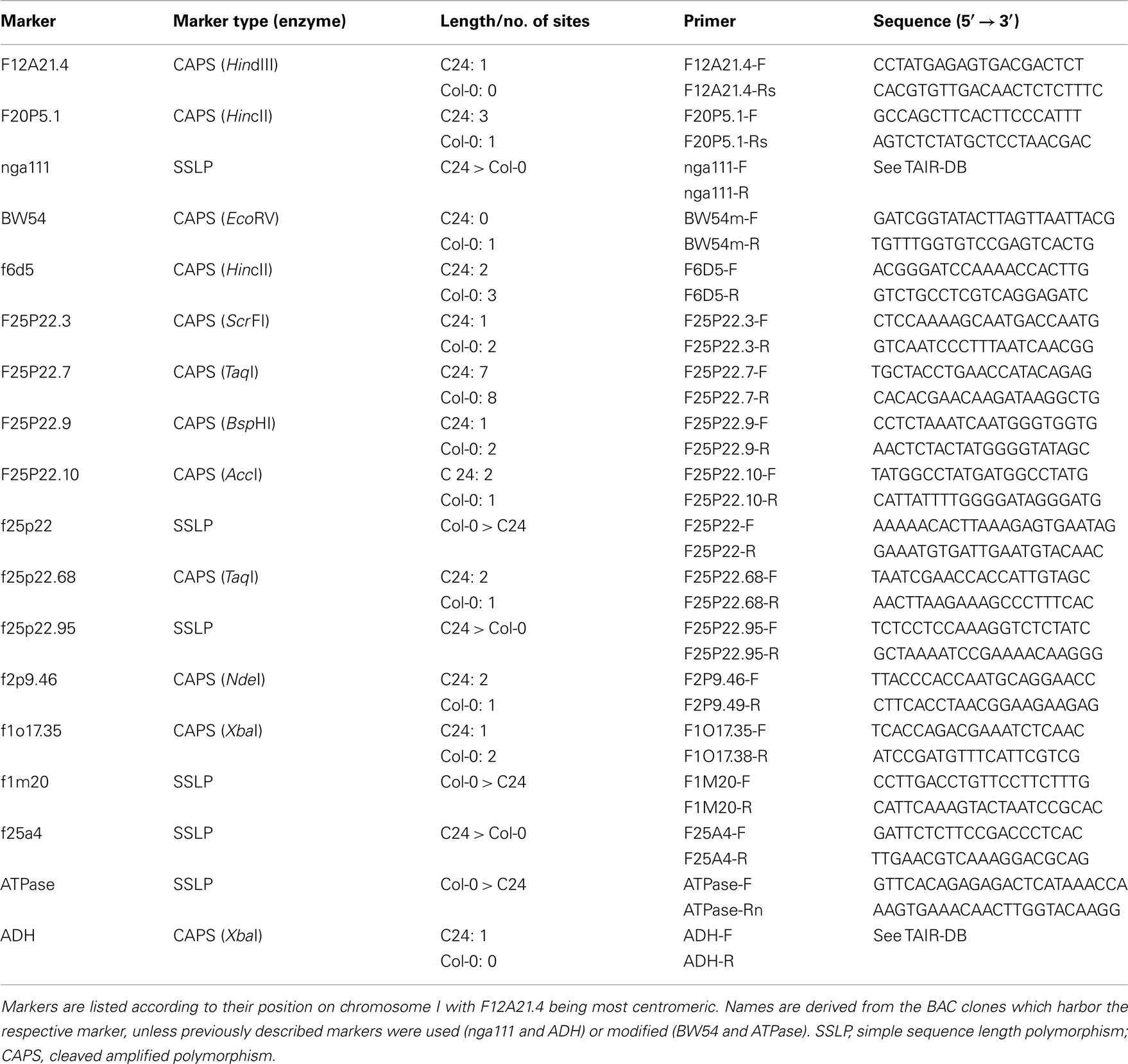
The mutation in rsr1-1 plants had previously been mapped to a region between nga111 and ADH on the long arm of chromosome I (Martin et al., 1997), and was therefore not allelic to any gin or abi mutants that have been characterized at the molecular level (Zhou et al., 1998; Arenas-Huertero et al., 2000; Huijser et al., 2000; Laby et al., 2000; Rook et al., 2001). To identify the RSR1 locus, 18 markers polymorphic between C24 and Col-0 were analyzed in a population of 1315 F2 plants from a cross between rsr1-1 and Col-0. A total of 20 informative recombination events between markers nga111 and f1m20 were detected that placed RSR1 on a 130-kbp fragment between markers f25p22.3 and f2p9.46 (Figure 5A; Table A1 in Appendix). This region contains 40 predicted protein-coding genes, one micro-RNA and one pre-tRNA (TAIR10 genome annotation)1. Double recombination events within this region indicated a higher probability for a position of RSR1 on the distal half of BAC clone F25P22.
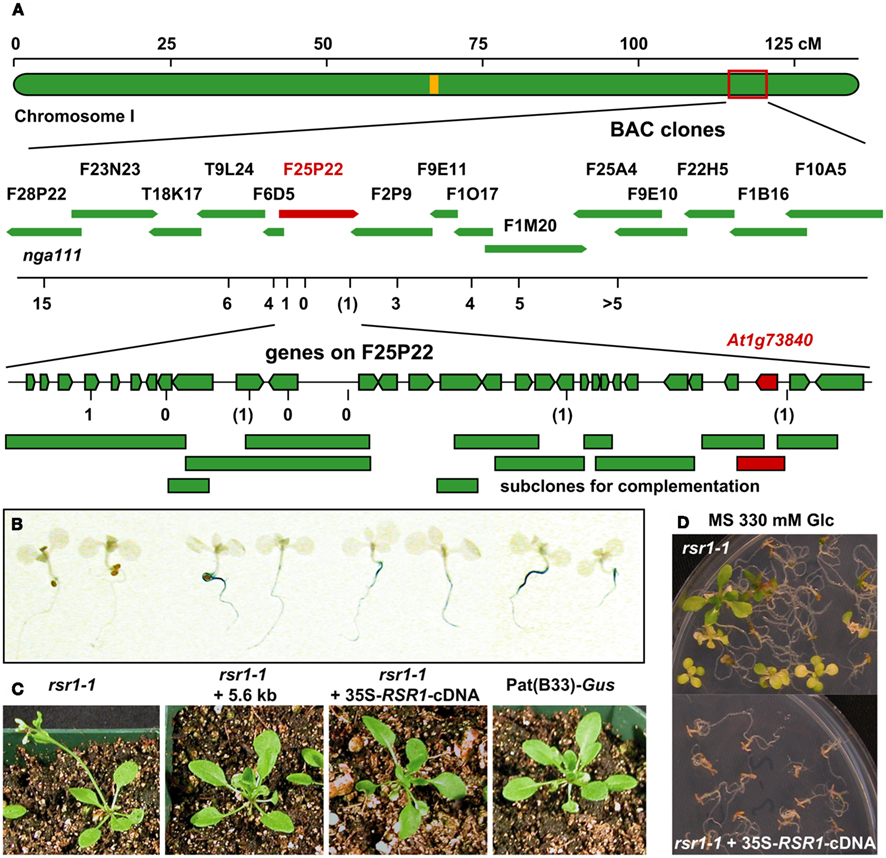
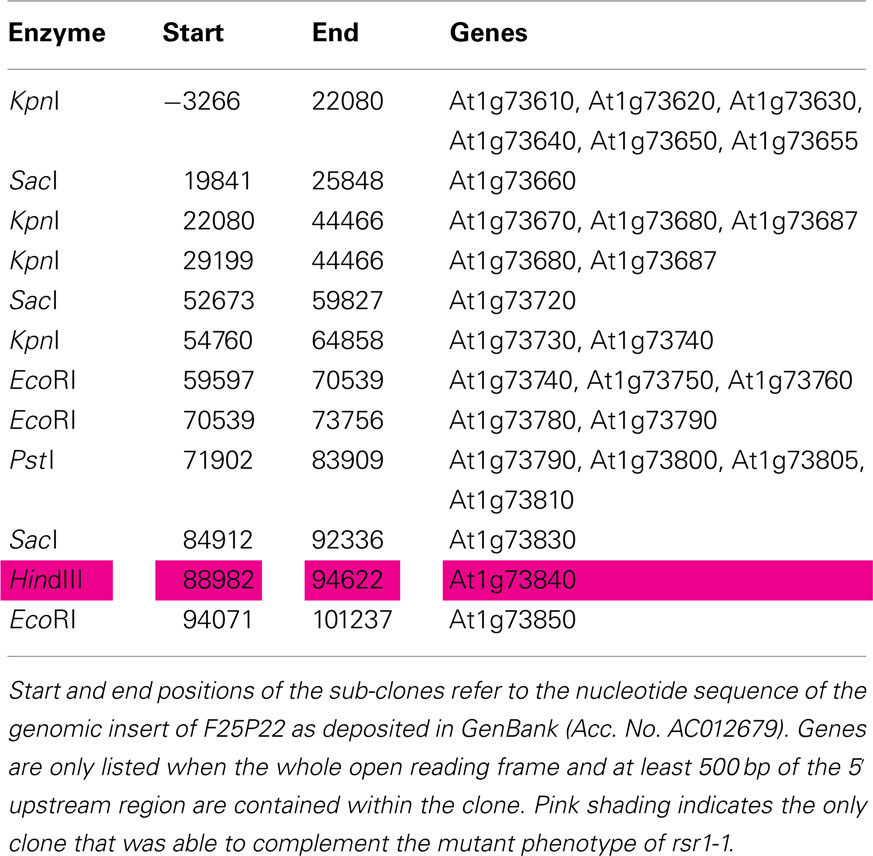
Figure 5. Identification of RSR1 and complementation of mutant phenotypes. (A) Schematic view of chromosome I with two subsequent enlargements of the region around RSR1. BAC clones of the region between nga111 and ADH were screened for markers polymorphic between C24 and Col-0. The second enlargement shows the position and orientation of annotated genes on BAC clone F25P22. Vertical black lines indicate the positions of markers used for the mapping of rsr1-1. The numbers below the markers are the numbers of recombination events between RSR1 and the respective marker, observed in a population of 1315 F2 plants from a cross between rsr1-1 and Col-8. Numbers in brackets indicate that a double recombination event to the left and right of the respective marker was observed. Green bars below BAC F25P22 indicate genomic fragments that could not complement rsr1-1. The single complementing fragment in which At1g73840 is the only intact gene is shown in red. (B) GUS-staining in roots of seedlings grown in the presence of 90 mM sucrose demonstrated that sugar induction of the patatin class II promoter was lost in rsr1-1 plants, but restored by transformation with either the 5.6-kb genomic fragment shown above or the cDNA of At1g73840 under control of the CaMV 35S promoter. (C) Normal flowering was restored in rsr1-1 plants expressing a wildtype copy of At1g73840. (D) Overexpression of the At1g73840 cDNA in rsr1-1 reverted the gin-phenotype.
Sub-clones of F25P22 containing 16 out of the 20 predicted genes in this region were tested for complementation of the sugar signaling deficiency of rsr1-1 mutant plants. Complementation was exclusively observed with a 5.6-kb HindIII fragment spanning nucleotides 88982–94626 of the genomic insert of BAC clone F25P22 (Figure 5; Table A2 in Appendix). This fragment contained a single complete gene, At1g73840, encoding for a predicted proline-rich protein of 388 amino acids. Complementation of the rsr phenotype was observed in two independently transformed lines as well as in rsr1-1 plants expressing the cDNA of At1g73840 under the control of a CaMV 35S promoter. In the presence of 90 mM sucrose, strong GUS-staining demonstrated induction of the patatin class I promoter in Pat(B33)-Gus plants, while no GUS-staining was observed in rsr1-1 plants (Figure 5B; Martin et al., 1997). Seedlings from all three complementation lines with either the genomic fragment or with CaMV 35S-driven expression of the cDNA of At1g73840 showed equally strong GUS-staining as the original line Pat(B33)-Gus. Moreover, the genomic fragment and the cDNA also complemented the gin, early flowering and spontaneous lesion formation phenotypes of rsr1-1 (Figures 5C,D and data not shown).
The ORF of At1g73840 contains a single base exchange at position 442 (C to T transition, typical for EMS mutagenesis) in rsr1-1, resulting in a premature stop codon in the fourth out of seven exons (Figure 6A). The Baulcombe lab had previously identified a different mutation in At1g73840 in a screen for genes involved in restricting RNAi-mediated gene silencing of a phytoene desaturase gene, and named this mutant enhanced silencing phenotype 1-1 (esp1-1; Herr et al., 2006). The esp1-1 mutant carries a point mutation 25 codons upstream of the base exchange in rsr1-1, which also leads to a premature stop codon.
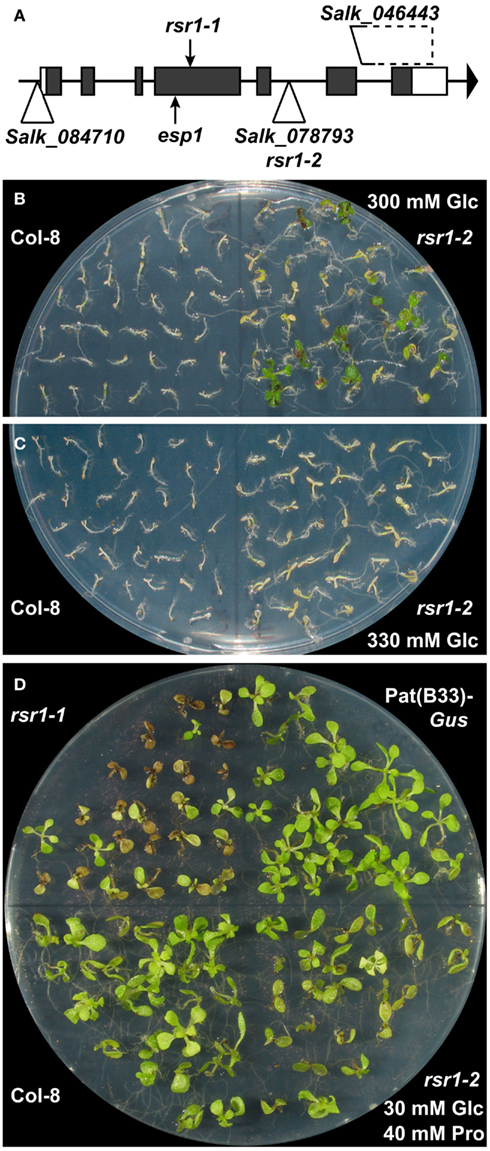
Figure 6. A T-DNA insertion line of At1g73840 mimics rsr1-1 phenotypes. (A) Schematic drawing of the exon/intron structure of At1g73840 with the positions of the premature stop codons in rsr1-1 and esp1-1 alleles and insertion sites of T-DNA lines analyzed in this study. (B) rsr1-2 mutants displayed increased tolerance to 300 mM Glc compared to Col-8. (C) At 330 mM Glc, also rsr1-2 plants were no longer able to establish autotrophic growth. (D) Like rsr1-1, rsr1-2 mutants were hypersensitive to 40 mM Pro in the culture medium.
T-DNA Insertion in RSR1 Causes Glucose Tolerance and Proline Hypersensitivity
To further confirm that the phenotypic changes in rsr1-1 plants are caused by the mutation in At1g73840, several T-DNA insertion lines from the Salk collection (Alonso et al., 2003) were characterized (Figure 6A). The Salk_084710 line carried a T-DNA insertion 54 bp upstream of the ATG of At1g73840 and did not show any alterations in phenotype or RSR1/ESP1 expression in comparison to wildtype plants (data not shown). The Salk_046443 line carried a larger deletion downstream of RSR1/ESP1, which also affects an unknown gene that is essential for embryo development (data not shown). Of the analyzed lines, only the Salk_078793 line, with an insertion in the fifth intron, specifically affected the expression of RSR1/ESP1. Homozygous plants of this line contained a much larger RSR1/ESP1-containing transcript and had a slightly increased organ initiation rate, while they were otherwise phenotypically very similar to wildtype plants under greenhouse conditions (Figure A1 in Appendix). In sterile culture, the Salk_078793 line displayed a decreased Glc sensitivity compared to the parental line Col-8 when grown on MS-medium containing 300 mM Glc, and was therefore named rsr1-2 (Figure 6B). At a Glc concentration of 330 mM, neither rsr1-2 nor Col-8 were able to de-etiolate and establish autotrophic growth (Figure 6C). rsr1-2 plants did not flower earlier than Col-8, which generally flowers earlier than C24 and other late flowering accessions due to allelic variations at the Frigida (Fri) locus (Gazzani et al., 2003; Shindo et al., 2005). Spontaneous lesion formation was not observed in rsr1-2, but this T-DNA insertion mutant was hypersensitive to Pro, similar to rsr1-1 (Figure 6D). In summary, our observations of early flowering, glucose-insensitivity and spontaneous lesion formation in rsr1-1 strongly support that this mutant is affected in At1g73840 and is therefore allelic to esp1-1.
RSR1/ESP1 Localizes to the Nucleus and is Subject to Threonine Phosphorylation
A survey of publicly available microarray data showed that RSR1/ESP1 is expressed at similar levels in all plant tissues and expression does not respond strongly to developmental or environmental stimuli (Genevestigator V.3, Zimmermann et al., 2005). However, a post-translational modification of the RSR1/ESP1 protein was detected by high-resolution proteomics analysis, namely phosphorylation of the threonine residue at position 33 of the predicted protein sequence (PhosPhAt database, release 3.0; Durek et al., 2010). Computational analysis of the RSR1/ESP1 protein sequence did not reveal any well-defined targeting signals. However, SubLoc v1.0 and WolFPSort predicted a slight preference for a nuclear localization of RSR1/ESP1, which is in agreement with its proposed function in mRNA processing (Hua and Sun, 2001; Horton et al., 2007). To verify the computational predictions, the cDNA of RSR1/ESP1 was translationally fused with GFP at its C-terminus and expressed under the control of the CaMV 35S promoter. In both transiently transformed Nicotiana benthamiana leaves and in stable Arabidopsis transformants, GFP fluorescence was most prominent in the nucleus but also detectable in the cytosol (Figures 7A,B).
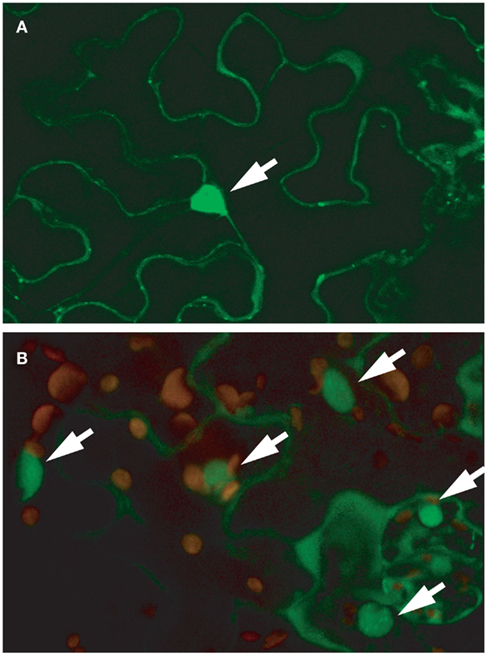
Figure 7. RSR1–GFP localizes predominantly to the nucleus. Confocal microscopy images of leaf epidermal cells expressing an RSR1–GFP fusion protein under the control of a CaMV 35S promoter. (A) Single focal plane in the epidermis of a tobacco (Nicotiana benthamiana) leaf imaged 48 h after infiltration with Agrobacterium tumefaciens suspension. (B) Leaf epidermis and parenchyma cells of a stably transformed Arabidopsis plant. The image shows a projection of 15 optical planes along the z-axis and an overlay of the GFP signal (green) with chlorophyll autofluorescence (red). GFP fluorescence was strongest in nuclei but also present in the cytoplasm. Nuclei are marked with arrows.
Protein Phosphorylation Regulates RSR1/ESP1-Dependent Sugar Signaling
It had previously been demonstrated that sugar and ABA signaling is mediated by protein phosphorylation events. For example, the Glc-insensitive mutant gin4 affects the protein kinase CTR1 (Zhou et al., 1998), and SNF1-related protein kinases are critical factors in mediating ABA responses (Fujii et al., 2011). To investigate whether protein phosphorylation plays a role in the sugar- and RSR1/ESP1-dependent regulation of the patatin promoter, we applied protein kinase and phosphatase inhibitors alone or in combination with Suc.
After incubation with 200 mM Suc for 30 h, rsr1-1 mutants showed a 1.5-fold increase in GUS-activity, compared to a threefold induction in Pat(B33)-Gus (Figure 8A). Staurosporine (ST), a broad range inhibitor of protein kinases, induced GUS-activity in roots of Pat(B33)-Gus plants in the absence of Suc, whereas okadaic acid (OA), an inhibitor of protein phosphatases, reduced the sugar-dependent response. A combination of ST and Suc led to an additional increase in GUS-activity. In contrast, these responses to altered protein phosphorylation were severely reduced in rsr1-1 demonstrating that phosphorylation of either RSR1/ESP1 or upstream components in the same signaling cascade mediate sugar-dependent regulation of the Pat(B33)-Gus construct (Figure 8B).
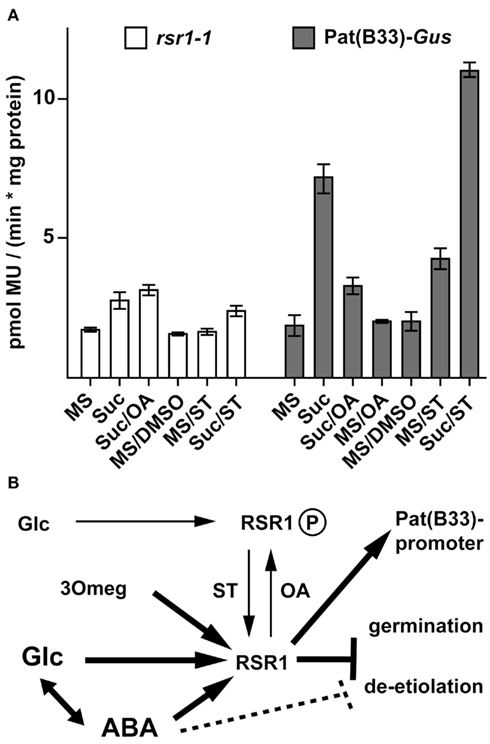
Figure 8. Protein phosphatases and kinases regulate RSR1-mediated effects. (A) To determine expression levels of the Pat(B33)-Gus transgene, GUS-activity was determined in roots after 30 h incubation with liquid MS-medium supplemented with either 200 mM Suc, 0.4 mM okadaic acid (OA), 0.001% DMSO, 0.4 mM staurosporine (ST, in 0.001% DMSO) or combinations of inhibitors with 200 mM Suc. For details see Section “Materials and Methods.” OA suppressed Suc-dependent induction of GUS expression in Pat(B33)-Gus whereas rsr1-1 mutants were insensitive to ST-dependent stimulation of GUS-activity. The values presented are the mean ± SD of ≥4 biological replicates. (B) Tentative model for the regulation of Pat(B33) promoter activity and seedling development by RSR1/ESP1. Low levels of Glc render RSR1/ESP1 inactive, potentially via phosphorylation. This effect can be mimicked by OA and overcome by ST. Intermediate levels of Glc or the Glc analog 3Omeg activate RSR1/ESP1, resulting in activation of the Pat(B33) promoter and inhibition of germination. High levels of ABA and/or Glc but not 3Omeg inhibit de-etiolation of seedlings, probably by a combinatorial effect of several pathways.
Discussion
In this study, we describe the detailed physiological and molecular characterization of the rsr1-1 mutant that we identified earlier as defective in sugar-dependent induction of a transgenic patatin class I promoter from potato. We found that RSR1/ESP1 also contributes to the sugar-dependent regulation of endogenous processes, since rsr1-1 mutants were insensitive to Glc-dependent repression of germination and seedling establishment.
Interestingly, germination of our control line, Pat(B33)-Gus, was inhibited by high concentrations of Glc as well as the poorly metabolized sugar analog 3Omeg, whereas sorbitol had a weaker effect. Like the Glc and 3Omeg driven induction of the patatin promoter, this inhibitory effect on germination was lost in rsr1-1 mutant seedlings (Martin et al., 1997). We conclude that 3Omeg can mimic a subset of metabolism-independent Glc signals, although global transcript profiling has so far failed to identify genes that respond to 3Omeg or 6-deoxyglucose (Villadsen and Smith, 2004). The loss of Glc- and 3Omeg-specific repression of germination in rsr1-1 mutant seeds provides a novel approach to unravel the underlying signaling cascade. Previous studies with mutants that were insensitive to Glc- and HXK1-dependent repression of seedling establishment had demonstrated that Glc-inhibition of germination is controlled by a separate pathway, probably targeting the degradation rate of ABA (Price et al., 2003). Accordingly, neither Pat(B33)-Gus nor rsr1-1 seedlings displayed a block in de-etiolation in response to high concentrations of 3Omeg.
The signaling cascade leading to Glc-dependent inhibition of seedling establishment was unraveled by forward genetic screens for Glc-insensitive (gin) mutants. Identification of genes affected by gin mutations demonstrated that Glc signaling in this case depends on HXK1, which acts directly as a part of a nuclear protein complex in the regulation of gene expression (Moore et al., 2003; Cho et al., 2006). Other sugar insensitive mutants identified in similar screens were affected in either ABA synthesis or signaling, which led to the hypothesis that HXK1-dependent sugar signaling may lead to an accumulation of ABA (Rolland et al., 2006). This hypothesis was further supported by sugar insensitive phenotypes of additional mutants that were isolated in screens for defects in ABA synthesis or signaling (Dekkers et al., 2008). Identification of rsr1-1 as a Glc and ABA insensitive mutant likely places RSR1 downstream of HXK1 and ABA accumulation in this sugar signaling pathway (Figure 8B). Since the mutation in rsr1-1 disrupts HXK-independent as well as HXK-dependent pathways, RSR1 might be an integrator of multiple sugar sensing pathways, or it could act independently in several signaling cascades.
The identification of rsr1-1 as a novel mutant allele of the enhanced silencing phenotype 1 (ESP1) gene (At1g73840) suggests that RSR1/ESP1 acts upon mRNA processing. RSR1/ESP1 has homology to the mammalian mRNA 3′-end Cleavage Stimulation Factor CstF64 but lacks a conserved RNA binding motive (Herr et al., 2006). ESP1 was previously characterized by a single mutant allele, esp1-1, that, like rsr1-1, was identified in a screen of EMS mutagenized populations of transgenic lines derived from the ecotype C24. Both mutations introduced premature stop codons in the fourth exon of RSR1/ESP1, after the first of two domains that are homologous to CstF64.
Like rsr1-1, esp1-1 mutant plants displayed spontaneous lesion formation on leaves and early flowering time. Other esp and gin mutants were also affected in timing of the transition to flowering. The gin1/aba2 mutant flowered normal on soil, but unlike the wildtype, flowering was not delayed by high Glc concentrations (Zhou et al., 1998). On the contrary, deletion of the vacuolar Glc transporter VGT1 caused a late flowering phenotype (Aluri and Buttner, 2007). While ESP4 and ESP5 have been identified as core components of the Cleavage Polyadenylation Specificity Complex (CSPF), which affects mRNA processing of the flowering regulator FCA, the endogenous target of RSR1/ESP1 in flowering regulation remains unknown (Herr et al., 2006).
In this context it is interesting to note that, unlike the two chemically induced point mutation alleles in the C24 background, the T-DNA insertion allele rsr1-2, in the early flowering Col-8 background, did not show an additional acceleration of flowering. Vernalization-independent early flowering of Col-8 and other early flowering ecotypes was correlated to allelic variations at the Fri locus often resulting in low levels of FRI protein (Shindo et al., 2005). Thus early flowering of rsr1-1 and esp1-1, but not rsr1-2, plants suggests that RSR1/ESP1 might affect flowering via FRI, although additional factors were identified that contribute to the variation of flowering time between different Arabidopsis accessions (Werner et al., 2005). It remains possible that the T-DNA insertion in rsr1-2 plants affects the RSR1/ESP1 protein in a different way than the premature stop codons in rsr1-1 and esp1-1 plants.
A molecular function of RSR1/ESP1 in mRNA processing and gene silencing may also explain the Pro hypersensitive phenotype of the rsr1-1 and rsr1-2 mutants (Hellmann et al., 2000). Pro hypersensitivity was attributed to defects in Pro degradation, since mutants in Pro dehydrogenase (ProDH) and pyrroline-5-carboxylate dehydrogenase (P5CDH) were also hypersensitive to external Pro (Mani et al., 2002; Nanjo et al., 2003; Deuschle et al., 2004). Expression of P5CDH was found to be regulated by gene silencing via the overlapping gene Sro5 on the complementary DNA strand, but so far, our analyses did not clearly demonstrate an altered regulation of P5CDH in the rsr1-1 mutant (Funck, 2001; Borsani et al., 2005). A recent study on transcriptome changes in a splicing factor mutant suggests that RSR1/ESP1 itself may be regulated by alternative mRNA processing (Yoshimura et al., 2011).
Lesion formation in leaves and ROS accumulation were prominent features of damages induced by external Pro application (Deuschle et al., 2004). Spontaneous lesion formation and H2O2 accumulation in rsr1-1 and esp1-1 mutants indicate an imbalance in ROS homeostasis independent of Pro treatment. This disturbance in ROS metabolism may also account for the decreased length of root hairs in rsr1-1 mutant seedlings. Directional tip growth of root hairs is a highly regulated process, in which ROS production and signaling play an important role (Cardenas, 2009). It remains to be analyzed whether the enhanced root growth of rsr1-1 seedlings is a compensatory response to the shorter root hairs or if both phenotypes are regulated independently.
Interestingly, the experimental evidence for threonine phosphorylation of RSR1/ESP1 was obtained in an experiment in which nitrogen-starved seedlings were re-supplied with ammonium (Engelsberger and Schulze, 2012). Phosphorylation of RSR1/ESP1 in response to a nitrogen stimulus indicates that RSR1/ESP1 may contribute to nutrient signaling for both carbon and nitrogen. Involvement of the yeast Suc Non-Fermenting 1 (SNF1) protein kinase and the Arabidopsis SNF1-Related Kinases (SnRKs) in Glc signaling are well established, but the exact links between SnRKs and Glc signaling are still unclear (Smeekens et al., 2010). RSR1/ESP1 could either be a target of SnRK-dependent phosphorylation or it could contribute to the regulation of SnRK activity. Supporting these scenarios, signaling via SnRKs was independent of HXK1 and therefore similar to the regulation of the patatin promoter by Glc analogs that are not phosphorylated by HXK1 (Martin et al., 1997; Baena-Gonzalez et al., 2007).
Our experiments with protein kinase and phosphatase inhibitors demonstrate that Glc-dependent induction of the patatin promoter is negatively regulated by protein phosphorylation. In rsr1-1 mutants, Glc-dependent and staurosporine-dependent induction of GUS expression were equally suppressed, indicating that RSR1/ESP1 acts downstream of the phosphorylation events or may itself be the target for inhibitory phosphorylation (Figure 8B).
Taken together, the experimental and bioinformatics data available for RSR1/ESP1 suggest that regulated mRNA processing is a novel part of nutrient sensing and signaling in plants. The absence of a predicted RNA binding domain in RSR1/ESP1 indicates that RSR1/ESP1 has to act in concert with other proteins or protein complexes to regulate a specific subset of mRNAs. The multiple mutant phenotypes of rsr1-1 and esp1-1 mainly affect processes that are already known to be regulated by the nutrient status, such as germination, root growth, chlorophyll synthesis and flowering. Many key regulators of these processes are already known and targeted searches for alternative mRNA processing may help to bridge the gaps between nutrient sensing, signaling, and effector protein expression.
Materials and Methods
Plant Growth
Arabidopsis (A. thaliana (L.) Heynh.) lines Pat(B33)-Gus and rsr1-1 (descendent from ecotype C24; Martin et al., 1997) were grown in sterile culture under short-day conditions on MS-media (Murashige and Skoog, 1965) supplemented with different concentrations of Suc, Glc, 3Omeg, sorbitol, Pro, or ABA. Respective concentrations are given in the text or figure legends. Seeds were stratified for 24 h at 4°C in 0.1% agarose. Transformation by floral dip was performed according to (Clough and Bent, 1998).
Three lines (Salk lines 084710, 046443, and 078793) predicted to carry a T-DNA insertion in the RSR1 gene (At1g73840) were obtained from the Nottingham Arabidopsis Stock Center. The insertion sites for all three lines were confirmed by PCR with LB primer (gccctttgacgttggaggccac) and RSR1-specific primers (for 084710: ccacacggattgcagatttag, for 078793: atgttcgagctctctgattgg, and for 046443: cccagcaacctccttttcatc). The insertion sites were further confirmed by sequencing of the PCR products in the case of lines 084710 and 078793 (rsr1-2). All insertion lines were backcrossed three times to the parental line (ABRC accession Col-8) to eliminate secondary mutations.
Phenotypic Analyses
Glc-tolerance and ABA-insensitivity were tested by germinating seedlings on MS-media supplemented with various concentrations of Glc, 3Omeg, sorbitol, or ABA. Seeds were scored as germinated when the radicle had emerged through the seed coat. Seedling establishment was scored after 6–28 days. For analysis of root growth and root hairs, the plates were placed vertically in a growth cabinet. To test for hypersensitivity to Pro, plants were cultivated on MS-medium with 30 mM Glc and 40 mM Pro (Hellmann et al., 2000). For bolting time analysis, transformation and pigment quantification plants were grown in the greenhouse (≥16 h light; 20°C). Pigments were extracted twice with 80% acetone from fully expanded rosette leaves ground in liquid nitrogen. Chlorophyll A and B were quantified photometrically under dimmed light according to (Lichtenthaler, 1987). For ROS detection, detached leaves were infiltrated by transpiration with 1 mM 3,3′-diaminobenzidine at pH 5.5 in dim light. After 4 h, the leaves were destained in 80% ethanol.
Fine Mapping of RSR1
For mapping of the RSR1 locus, F2 plants from a cross between rsr1-1 (C24 ecotype) and Col-0 were assayed for sugar induction of the Pat(B33)-Gus construct using in vivo GUS-staining (see below). Genomic DNA was isolated from 1315 kanamycin-resistant plants and used for PCR-based genotyping in the proximity of marker nga111. Eighteen previously described or novel SSLP or CAPS markers were found to be polymorphic between C24 and Col-0 in the region of interest (see Table A1 in Appendix).
Complementation and Subcellular Localization
For complementation of the rsr1-1 mutant with genomic fragments, the BAC clone F25P22 (Medline AC012679) was digested with different restriction enzymes and fragments containing single or multiple genes were sub-cloned into pPZP221 (Hajdukiewicz et al., 1994; Table A2 in Appendix). For complementation assays using the RSR1 cDNA, the full length cDNA (RIKEN, clone RAFL 09-93-F21) was cloned between the CaMV 35S promoter and terminator in the EcoRI/BamHI sites of pRT101 (Töpfer et al., 1987). The cassette was excised with HindIII and transferred into a pCAMBIA-derived binary vector conferring phosphinothricin resistance2.
To analyze the subcellular localization of RSR1/ESP1, the RSR1 ORF was amplified by PCR from RALF cDNA clone 09-93-F21 using the following primers: ataggtaccatggcgggtaagcagatcgg and cttggatccgctgtcggagaatctgttgaag. The resulting PCR fragment was cloned into the KpnI/BamHI restriction sites of vector pCF203, a derivative of the pPZP212 vector, creating a CaMV 35S-driven C-terminal GFP fusion construct. Localization of GFP expression was analyzed in N. benthamiana transiently transformed by Agrobacterium-mediated transformation, as well as in stable Arabidopsis Col-0 transformants. Cells expressing the RSR1–GFP fusion were imaged using a Nipkow spinning disk confocal microscope as described (Deuschle et al., 2006).
Inhibitor Assays and Analysis of GUS-Activity
Plants were cultured on solid MS-medium containing 30 mM Glc. Twenty-day-old plants were transferred to glass jars, in which only the roots had contact to liquid medium. Plants were pre-treated for 2 h with 0.4 mM okadaic acid (Calbiochem) or 0.4 mM staurosporine (from a 40 mM stock in DMSO, Sigma) in MS-medium before Suc was added. After 30 h, roots were dissected and frozen in liquid nitrogen. Proteins were extracted from roots in GUS extraction buffer (Jefferson et al., 1987) in the presence of protease inhibitors (Complete, Roche). Fluorimetric GUS-assays were performed with 10 μg total protein by measuring the fluorescence of 4-methyl-umbelliferone as described by (Martin et al., 1997) using a Fluoroskan II fluorimeter (Lab-systems). In vivo GUS-staining and staining of whole seedlings was performed for 18 h at 37°C according to (Martin et al., 1997).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Yvonne Sauermann, Bettina Stadelhofer, and Gabi Fiene for excellent technical assistance. We are grateful to Sutton Mooney for critically reading the manuscript for this article. This work was supported by grants to Wolf B. Frommer from the US Department of Energy (DOE; DE-FG02-04ER15542).
Footnotes
References
Alonso, J. M., Stepanova, A. N., Leisse, T. J., Kim, C. J., Chen, H., Shinn, P., Stevenson, D. K., Zimmerman, J., Barajas, P., Cheuk, R., Gadrinab, C., Heller, C., Jeske, A., Koesema, E., Meyers, C. C., Parker, H., Prednis, L., Ansari, Y., Choy, N., Deen, H., Geralt, M., Hazari, N., Hom, E., Karnes, M., Mulholland, C., Ndubaku, R., Schmidt, I., Guzman, P., Aguilar-Henonin, L., Schmid, M., Weigel, D., Carter, D. E., Marchand, T., Risseeuw, E., Brogden, D., Zeko, A., Crosby, W. L., Berry, C. C., and Ecker, J. R. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657.
Aluri, S., and Buttner, M. (2007). Identification and functional expression of the Arabidopsis thaliana vacuolar glucose transporter 1 and its role in seed germination and flowering. Proc. Natl. Acad. Sci. U.S.A. 104, 2537–2542.
Arenas-Huertero, F., Arroyo, A., Zhou, L., Sheen, J., and Leon, P. (2000). Analysis of Arabidopsis glucose insensitive mutants, gin5 and gin6, reveals a central role of the plant hormone ABA in the regulation of plant vegetative development by sugar. Genes Dev. 14, 2085–2096.
Ausin, I., Alonso-Blanco, C., and Martinez-Zapater, J. M. (2005). Environmental regulation of flowering. Int. J. Dev. Biol. 49, 689–705.
Baena-Gonzalez, E., Rolland, F., Thevelein, J. M., and Sheen, J. (2007). A central integrator of transcription networks in plant stress and energy signalling. Nature 448, 938–942.
Borsani, O., Zhu, J., Verslues, P. E., Sunkar, R., and Zhu, J. K. (2005). Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell 123, 1279–1291.
Cardenas, L. (2009). New findings in the mechanisms regulating polar growth in root hair cells. Plant Signal. Behav. 4, 4–8.
Cho, Y. H., Yoo, S. D., and Sheen, J. (2006). Regulatory functions of nuclear hexokinase1 complex in glucose signaling. Cell 127, 579–589.
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743.
Corbesier, L., Lejeune, P., and Bernier, G. (1998). The role of carbohydrates in the induction of flowering in Arabidopsis thaliana: comparison between the wild type and a starchless mutant. Planta 206, 131–137.
Dekkers, B. J., Schuurmans, J. A., and Smeekens, S. C. (2008). Interaction between sugar and abscisic acid signalling during early seedling development in Arabidopsis. Plant Mol. Biol. 67, 151–167.
Deuschle, K., Chaudhuri, B., Okumoto, S., Lager, I., Lalonde, S., and Frommer, W. B. (2006). Rapid metabolism of glucose detected with FRET glucose nanosensors in epidermal cells and intact roots of Arabidopsis RNA-silencing mutants. Plant Cell 18, 2314–2325.
Deuschle, K., Funck, D., Forlani, G., Stransky, H., Biehl, A., Leister, D., Van Der Graaff, E., Kunze, R., and Frommer, W. B. (2004). The role of Δ1-pyrroline-5-carboxylate dehydrogenase in proline degradation. Plant Cell 16, 3413–3425.
Durek, P., Schmidt, R., Heazlewood, J. L., Jones, A., Maclean, D., Nagel, A., Kersten, B., and Schulze, W. X. (2010). PhosPhAt: the Arabidopsis thaliana phosphorylation site database. An update. Nucleic Acids Res. 38, D828–D834.
Engelsberger, W. R., and Schulze, W. X. (2012). Nitrate and ammonium lead to distinct global dynamic phosphorylation patterns when resupplied to nitrogen starved Arabidopsis seedlings. Plant J. 69, 978–995.
Eveland, A. L., and Jackson, D. P. (2011). Sugars, signalling, and plant development. J. Exp. Bot. doi: 10.1093/jxb/err379. [Epub ahead of print].
Finkelstein, R. R., and Lynch, T. J. (2000). Abscisic acid inhibition of radicle emergence but not seedling growth is suppressed by sugars. Plant Physiol. 122, 1179–1186.
Fujii, H., Verslues, P. E., and Zhu, J. K. (2011). Arabidopsis decuple mutant reveals the importance of SnRK2 kinases in osmotic stress responses in vivo. Proc. Natl. Acad. Sci. U.S.A. 108, 1717–1722.
Funck, D. (2001). Glucose and Proline Dependent Signal Transduction in Arabidopsis: Contributions to Balancing Life and Death. Ph.D. thesis, Eberhard-Karls-Universität, Tübingen.
Gazzani, S., Gendall, A. R., Lister, C., and Dean, C. (2003). Analysis of the molecular basis of flowering time variation in Arabidopsis accessions. Plant Physiol. 132, 1107–1114.
Gechev, T. S., Van Breusegem, F., Stone, J. M., Denev, I., and Laloi, C. (2006). Reactive oxygen species as signals that modulate plant stress responses and programmed cell death. Bioessays 28, 1091–1101.
Greenberg, J. T., and Yao, N. (2004). The role and regulation of programmed cell death in plant-pathogen interactions. Cell. Microbiol. 6, 201–211.
Guiboileau, A., Sormani, R., Meyer, C., and Masclaux-Daubresse, C. (2010). Senescence and death of plant organs: nutrient recycling and developmental regulation. C. R. Biol. 333, 382–391.
Hajdukiewicz, P., Svab, Z., and Maliga, P. (1994). The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25, 989–994.
Hellmann, H., Funck, D., Rentsch, D., and Frommer, W. B. (2000). Hypersensitivity of an Arabidopsis sugar signaling mutant toward exogenous proline application. Plant Physiol. 123, 779–789.
Herr, A. J., Molnar, A., Jones, A., and Baulcombe, D. C. (2006). Defective RNA processing enhances RNA silencing and influences flowering of Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 103, 14994–15001.
Horton, P., Park, K. J., Obayashi, T., Fujita, N., Harada, H., Adams-Collier, C. J., and Nakai, K. (2007). Wolf Psort: protein localization predictor. Nucleic Acids Res. 35, W585–W587.
Hua, S. J., and Sun, Z. R. (2001). Support vector machine approach for protein subcellular localization prediction. Bioinformatics 17, 721–728.
Huijser, C., Kortstee, A., Pego, J., Weisbeek, P., Wisman, E., and Smeekens, S. (2000). The Arabidopsis sucrose uncoupled-6 gene is identical to abscisic acid insensitive-4: involvement of abscisic acid in sugar responses. Plant J. 23, 577–585.
Jefferson, R. A., Kavanagh, T. A., and Bevan, M. W. (1987). GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907.
Karuppanapandian, T., Moon, J. C., Kim, C., Manoharan, K., and Kim, W. (2011). Reactive oxygen species in plants: their generation, signal transduction, and scavenging mechanisms. Aust. J. Crop. Sci. 5, 709–725.
Kuhn, J. M., Breton, G., and Schroeder, J. I. (2007). mRNA metabolism of flowering-time regulators in wild-type Arabidopsis revealed by a nuclear cap binding protein mutant, abh1. Plant J. 50, 1049–1062.
Laby, R. J., Kincaid, M. S., Kim, D., and Gibson, S. I. (2000). The Arabidopsis sugar-insensitive mutants sis4 and sis5 are defective in abscisic acid synthesis and response. Plant J. 23, 587–596.
Lichtenthaler, H. K. (1987). Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Meth. Enzymol. 148, 350–382.
Lopez-Molina, L., Mongrand, S., Mclachlin, D. T., Chait, B. T., and Chua, N.-H. (2002). ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J. 32, 317–328.
Mani, S., Van De Cotte, B., Van Montagu, M., and Verbruggen, N. (2002). Altered levels of proline dehydrogenase cause hypersensitivity to proline and its analogs in Arabidopsis. Plant Physiol. 128, 73–83.
Martin, T., Hellmann, H., Schmidt, R., Willmitzer, L., and Frommer, W. B. (1997). Identification of mutants in metabolically regulated gene expression. Plant J. 11, 53–62.
Mattioli, R., Costantino, P., and Trovato, M. (2009). Proline accumulation in plants: not only stress. Plant Signal. Behav. 4, 1016–1018.
Moore, B., Zhou, L., Rolland, F., Hall, Q., Cheng, W. H., Liu, Y. X., Hwang, I., Jones, T., and Sheen, J. (2003). Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science 300, 332–336.
Murashige, T., and Skoog, F. (1965). A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant 15, 473–497.
Nanjo, T., Fujita, M., Seki, M., Kato, T., Tabata, S., and Shinozaki, K. (2003). Toxicity of free proline revealed in an Arabidopsis T-DNA-tagged mutant deficient in proline dehydrogenase. Plant Cell Physiol. 44, 541–548.
Pego, J. V., Kortstee, A. J., Huijser, C., and Smeekens, S. C. (2000). Photosynthesis, sugars and the regulation of gene expression. J. Exp. Bot. 51, 407–416.
Price, J., Li, T. C., Kang, S. G., Na, J. K., and Jang, J. C. (2003). Mechanisms of glucose signaling during germination of Arabidopsis. Plant Physiol. 132, 1424–1438.
Pujar, A., Jaiswal, P., Kellogg, E. A., Ilic, K., Vincent, L., Avraham, S., Stevens, P., Zapata, F., Reiser, L., Rhee, S. Y., Sachs, M. M., Schaeffer, M., Stein, L., Ware, D., and Mccouch, S. (2006). Whole-plant growth stage ontology for angiosperms and its application in plant biology. Plant Physiol. 142, 414–428.
Quesada, V., Dean, C., and Simpson, G. G. (2005). Regulated RNA processing in the control of Arabidopsis flowering. Int. J. Dev. Biol. 49, 773–780.
Rognoni, S., Teng, S., Arru, L., Smeekens, S., and Perata, P. (2007). Sugar effects on early seedling development in Arabidopsis. Plant Growth Regul. 52, 217.
Rolland, F., Baena-Gonzalez, E., and Sheen, J. (2006). Sugar sensing and signaling in plants: conserved and novel mechanisms. Annu. Rev. Plant Biol. 57, 675–709.
Rolland, F., Moore, B., and Sheen, J. (2002). Sugar sensing and signaling in plants. Plant Cell 14(Suppl.), S185–S205.
Rook, F., Corke, F., Card, R., Munz, G., Smith, C., and Bevan, M. W. (2001). Impaired sucrose-induction mutants reveal the modulation of sugar-induced starch biosynthetic gene expression by abscisic acid signalling. Plant J. 26, 421–433.
Shindo, C., Aranzana, M. J., Lister, C., Baxter, C., Nicholls, C., Nordborg, M., and Dean, C. (2005). Role of FRIGIDA and FLOWERING LOCUS C in determining variation in flowering time of Arabidopsis. Plant Physiol. 138, 1163–1173.
Sivitz, A. B., Reinders, A., Johnson, M. E., Krentz, A. D., Grof, C. P., Perroux, J. M., and Ward, J. M. (2007). Arabidopsis sucrose transporter AtSUC9. High-affinity transport activity, intragenic control of expression, and early flowering mutant phenotype. Plant Physiol. 143, 188–198.
Smeekens, S., Ma, J., Hanson, J., and Rolland, F. (2010). Sugar signals and molecular networks controlling plant growth. Curr. Opin. Plant Biol. 13, 274–279.
Srikanth, A., and Schmid, M. (2011). Regulation of flowering time: all roads lead to Rome. Cell. Mol. Life Sci. 68, 2013–2037.
Töpfer, R., Matzeit, V., Gronenborn, B., Schell, J., and Steinbiss, H. H. (1987). A set of plant expression vectors for transcriptional and translational fusions. Nucleic Acids Res. 15, 5890.
Villadsen, D., and Smith, S. M. (2004). Identification of more than 200 glucose-responsive Arabidopsis genes none of which responds to 3-O-methylglucose or 6-deoxyglucose. Plant Mol. Biol. 55, 467–477.
Werner, J. D., Borevitz, J. O., Uhlenhaut, N. H., Ecker, J. R., Chory, J., and Weigel, D. (2005). FRIGIDA-independent variation in flowering time of natural Arabidopsis thaliana accessions. Genetics 170, 1197–1207.
Wingler, A., and Roitsch, T. (2008). Metabolic regulation of leaf senescence: interactions of sugar signalling with biotic and abiotic stress responses. Plant Biol. (Stuttg.) 10(Suppl. 1), 50–62.
Yoshimura, K., Mori, T., Yokoyama, K., Koike, Y., Tanabe, N., Sato, N., Takahashi, H., Maruta, T., and Shigeoka, S. (2011). identification of alternative splicing events regulated by an Arabidopsis serine/arginine-like protein, atSR45a, in response to high-light stress using a tiling array. Plant Cell Physiol. 52, 1786–1805.
Zhang, X. C., and Gassmann, W. (2007). Alternative splicing and mRNA levels of the disease resistance gene RPS4 are induced during defense responses. Plant Physiol. 145, 1577–1587.
Zhou, L., Jang, J. C., Jones, T. L., and Sheen, J. (1998). Glucose and ethylene signal transduction crosstalk revealed by an Arabidopsis glucose-insensitive mutant. Proc. Natl. Acad. Sci. U.S.A. 95, 10294–10299.
Zimmermann, P., Hennig, L., and Gruissem, W. (2005). Gene-expression analysis and network discovery using Genevestigator. Trends Plant Sci. 10, 407–409.
Appendix
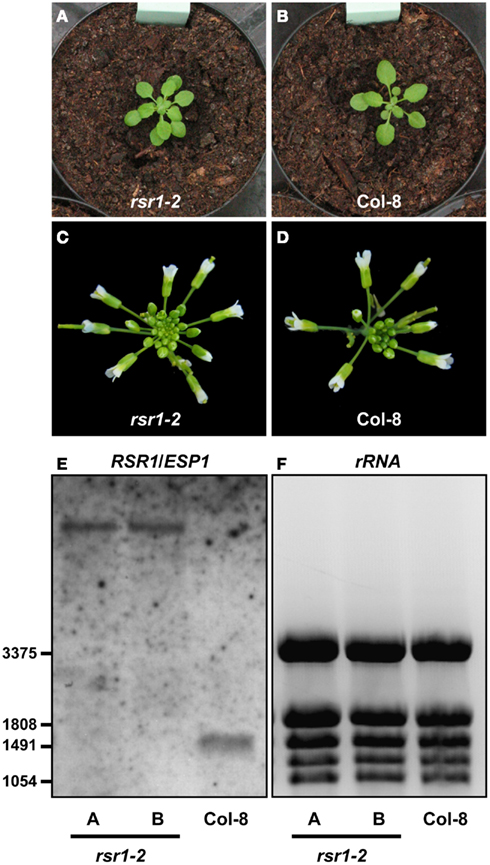
Figure A1. Organ initiation rate is increased in rsr1-2. (A,B) Rosettes of rsr1-2 plants had approximately 1.5 times more leaves than Col-8 plants of the same age. (C,D) Also inflorescences of rsr1-2 contained higher numbers of buds, resulting in more open flowers per day. (E) By northern blot, no native RSR1/ESP1 transcripts (expected size 1455 b) were detected in homozygous rsr1-2 plants. (F) EtBR stained rRNA confirms equal loading of the gel. rRNA bands were also used as size markers.
Keywords: Arabidopsis, 3-O-methylglucose, patatin, mRNA processing, protein phosphorylation, signal transduction, proline-rich protein
Citation: Funck D, Clauß K, Frommer WB and Hellmann HA (2012) The Arabidopsis CstF64-like RSR1/ESP1 protein participates in glucose signaling and flowering time control. Front. Plant Sci. 3:80. doi: 10.3389/fpls.2012.00080
Received: 22 February 2012; Paper pending published: 12 March 2012;
Accepted: 10 April 2012; Published online: 04 May 2012.
Edited by:
Sjef Smeekens, Utrecht University, NetherlandsReviewed by:
Fred Rook, University of Copenhagen, DenmarkSangdong Yoo, Sungkyunkwan University, South Korea
Copyright: © 2012 Funck, Clauß, Frommer and Hellmann. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Wolf B. Frommer, Department of Plant Biology, Carnegie Institution for Science, 260 Panama Street, Stanford, CA 94306, USA. e-mail:d2Zyb21tZXJAc3RhbmZvcmQuZWR1
†Present address: Karen Clauß, Institute of Biophysics, University of Ulm, Ulm, Germany.
‡Dietmar Funck and Karen Clauß have contributed equally to this work.

 Karen Clauß2†‡
Karen Clauß2†‡