
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Plant Sci. , 30 March 2012
Sec. Plant Genetics and Genomics
Volume 3 - 2012 | https://doi.org/10.3389/fpls.2012.00058
This article is part of the Research Topic Regulatory Elements in RNA View all 12 articles
 Lukasz Sobkowiak1
Lukasz Sobkowiak1 Dawid Bielewicz1
Dawid Bielewicz1 Ewelina M. Malecka1
Ewelina M. Malecka1 Iver Jakobsen2
Iver Jakobsen2 Merete Albrechtsen3
Merete Albrechtsen3 Zofia Szweykowska-Kulinska1
Zofia Szweykowska-Kulinska1 Andrzej Pacak1*
Andrzej Pacak1*Inorganic phosphate (Pi) is an easily accessible form of phosphorus for plants. Plant Pi uptake is usually limited however by slow Pi diffusion through the soil which strongly adsorps phosphate species. Plants have developed mechanisms to increase Pi availability. There are also abiotic (phosphate level) and biotic (e.g., mycorrhizal) factors regulating the expression of Pi-responsive genes. Transcription factors binding to the promoters of Pi-responsive genes activate different pathways of Pi transport, distribution, and homeostasis maintenance. Pi metabolism involves not only functional proteins but also microRNAs and other non-coding RNAs.
Phosphorus (P) is one of the most important macronutrients in the plant lifecycle. It is involved in the synthesis of nucleic acids and phospholipids, in reactions of phosphorylation, and also in energy delivery (ATP). Lack of phosphate (inorganic phosphate, Pi) inhibits plant growth, whereas excessive phosphate levels are toxic to plants and stimulate algal bloom in water reservoirs (reviewed by Scott, 2008). Here we describe genes and their products involved in Pi transport and maintenance of Pi homeostasis (Figure 1A). Phosphate and its role in plant life have both scientific and economic importance. Figure 1B shows the effect of Pi-deprivation on plant growth: the barley plants grown without Pi in the absence (−Pi) or presence (−Pi, Myc) of mycorrhizal fungi had shoot weights 35.2 and 33% lower, respectively, than the plant grown in Pi replete soil (+Pi) at 23 days post-sowing.
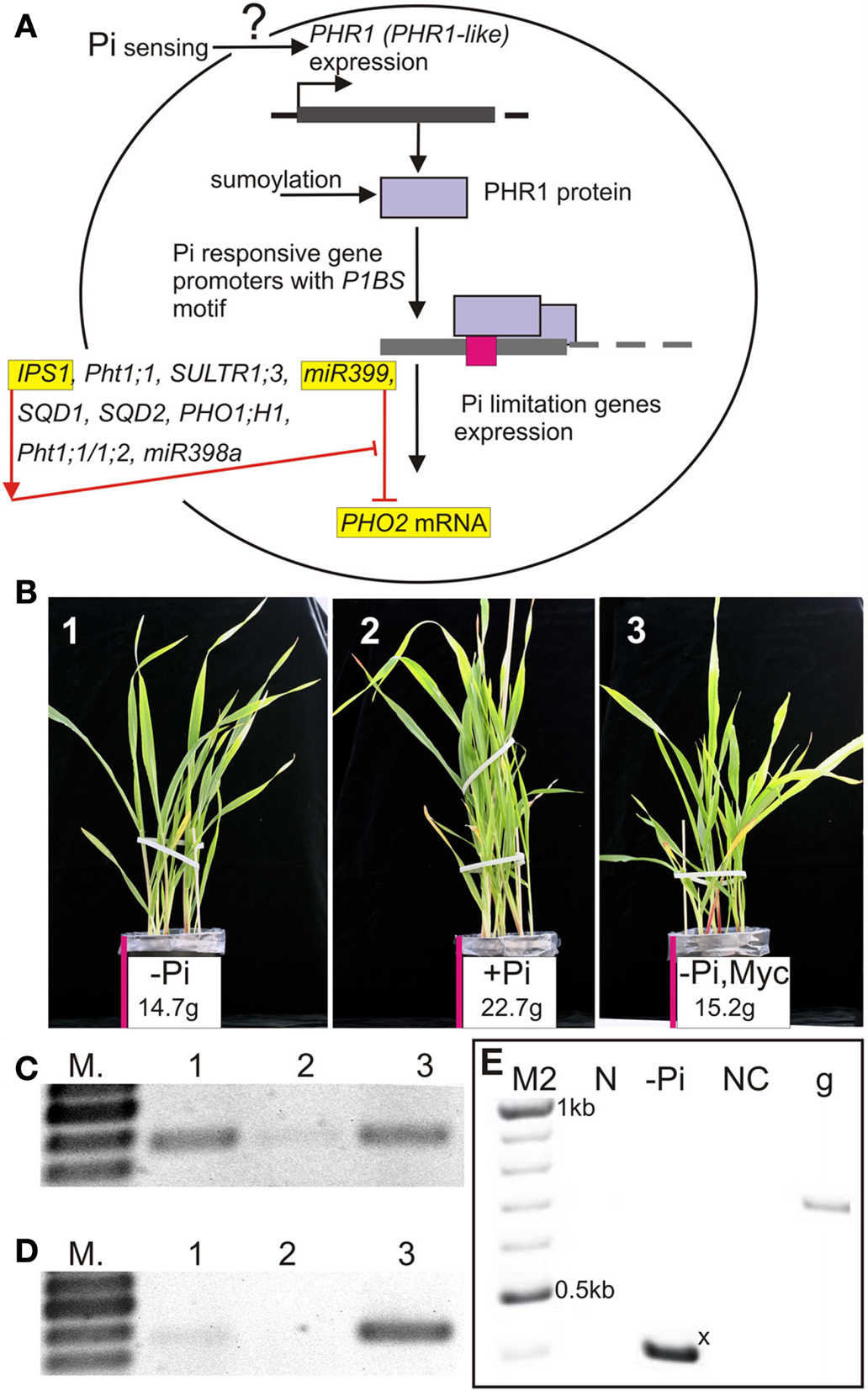
Figure 1. (A) Schematic representation of the relation between Arabidopsis PHR1 or its orthologs and examples of Pi-responsive genes possessing PHR1 binding sites in their promoters. Arabidopsis: IPS1 and Pht1;1 genes have been described by Martin et al. (2000) and Muchhal et al. (1996), respectively; SULTR1;3 by Rouached et al. (2011); miR399 by Pant et al. (2009) and Kuo and Chiou (2011); SQD1, SQD2 (involved in sulfolipid biosynthesis) by Franco-Zorrilla et al. (2004); PHO1;H1 (involved in Pi loading to the root xylem) by Stefanovic et al. (2007); barley Pht1;1/1;2 by Schünmann et al. (2004a); rice miR398a by Gu et al. (2010); (B) Barley grown without or with inorganic phosphate (−Pi, +Pi) or without Pi but in the presence of mycorrhizal fungi (−Pi Myc); (C) Barley HvPht1;1/1;2 expression in roots under different soil Pi and mycorrhiza conditions; (D) Barley HvPht1;8 expression in roots under different soil Pi and mycorrhiza conditions; (E) Arabidopsis pri-miR399a isoform expression under Pi sufficient growing conditions (N) and during Pi starvation (−Pi). The short pri-microRNA399a isoform is marked with an x (see Figure 2B). Lanes/pictures 1, 3 (−Pi), lane/picture 2 (+Pi), lane/picture 3 depicts roots colonized by the mycorrhizal fungus Glomus intraradices (28A). N, sufficient Pi conditions; −Pi, phosphate starvation; NC, negative control; g, genomic DNA positive control. M – GeneRuler 100 bp DNA ladder, M2 – GeneRuler 100 bp Plus DNA ladder (Thermo Scientific, formerly Fermentas). Barley – Hordeum vulgare cv. Black Hulless. Presented data: (B–D) Pacak et al., unpublished data, (E) Sobkowiak, unpublished data.
The mechanisms of Pi acquisition include secretion of carbon dioxide, and plant enzymes, e.g., acid phosphatases and organic acids (citric and malic acids), which target organic and inorganic soil P species, respectively, and release Pi from the soil particles (Scott, 2008; Richardson et al., 2011). Phosphate starvation induces expression of transcription factors (TFs) like phosphorus starvation response 1 (Psr1), which shows at least 10-fold increase of expression during Pi deficiency in Chlamydomonas reinhardtii (AF174532; Wykoff et al., 1999). In contrast, the related Arabidopsis thaliana ortholog phosphate starvation response 1 (AtPHR1, At4g28610) is only weakly responsive to Pi starvation (Rubio et al., 2001). The key regulatory functions of AtPHR1 and other TFs involved in Pi response have been reviewed by Nilsson et al. (2010) and Rouached et al. (2010). Signaling networks including TFs, Pi-responsive microRNAs, hormones, and sugars implicated in Pi sensing have been recently extensively reviewed by Chiou and Lin (2011). Located in the nucleus AtPHR1 can bind as a dimer via its MYB domain to the PHR1 binding site (P1BS, sequence GNATATNC), a motif present in the promoters of crucial Pi-responsive genes (Rubio et al., 2001; Schünmann et al., 2004a; Bustos et al., 2010; Nilsson et al., 2010; Oropeza-Aburto et al., 2012). A set of Pi-responsive genes with P1BS elements in their promoters is outlined in Figure 1A. This motif is frequent and considerably enriched in the promoters of Pi-responsive genes of Arabidopsis compared with the entire genome (Müller et al., 2007; Bustos et al., 2010). AtPHR1 also regulates genes not directly involved in phosphate metabolism but possessing the P1BS element: e.g., P1BS occurs in the promoter of the Arabidopsis sulfate transporter 1;3 gene (AtSULTR1;3, At1g22150), which is up-regulated during Pi-deficient conditions in wild-type plants but much less in phr1 mutant plants (Rouached et al., 2011). Bustos et al. (2010) showed that expression of Pi-responsive genes in Arabidopsis also requires the PHR1-LIKE1 TF (AtPHL1, At5g29000), which also contains a MYB domain. The double mutant phr1 phl1 exhibited lower expression levels of Pi transporter AtPht1;1 (U62330, At5g43350, three P1BS elements) compared with either of the single mutants phr1 and phl1 or wild-type plants (Franco-Zorrilla et al., 2004; Bustos et al., 2010). Since the expression of neither AtPHR1 nor AtPHL1 is strongly induced by Pi starvation, it is still unclear how Pi-limitation influences Pi-responsive genes. It has been shown in Arabidopsis that there are at least four MYB-CC (CC – coiled coil domain) proteins highly similar to both AtPHR1 and AtPHL1, i.e., At2g20400, At3g04450, At3g13040, and At5g06800 (Bustos et al., 2010). It is possible that these proteins are also involved in regulating the Pi-deprivation response. Furthermore, the AtPHR1 protein is a target for sumoylation by the SUMO E3 ligase, AtSIZ1 (At5g60410), which is also needed for Pi starvation-dependent responses (Miura et al., 2005). The expression of AtSIZ1 is not strongly induced by Pi starvation, however (Miura et al., 2005).
There are four Pi transporter classes, Pht1, Pht2, Pht3, and Pht4, which are responsible for phosphate transport across membranes of cells, chloroplasts, mitochondria, and Golgi, respectively (Karandashov and Bucher, 2005; Rouached et al., 2010). Analysis of six barley (Hordeum vulgare) Pht1 gene promoters revealed that all analyzed promoters possess P1BS elements (Schünmann et al., 2004a). These genes encode proteins belonging to the Pht1 family of phosphate transporters, which represents plasma membrane phosphate-H+ symporters, containing 12 trans-membrane helices connected by a hydrophilic loop (reviewed by Karandashov and Bucher, 2005). Expression of the AtPht1;1, HvPht1;1, HvPht1;2 (almost identical CDS to HvPht1;1), Triticum aestivum Pht1;2 (AJ344241) gene members of the Pht1 family, is largely restricted to roots (Muchhal et al., 1996; Davies et al., 2002; Schünmann et al., 2004a). The HvPht1;1 (AF543197) and HvPht1;2 (AY187019) promoters possess three and two P1BS-like motifs, respectively, and are Pi-limitation induced genes (Schünmann et al., 2004a,b; Glassop et al., 2005; Pacak et al., unpublished data; Figure 1C). Mutations in two out of three P1BS elements present in the HvPht1;1 rearranged promoter completely abolished promoter completely abolished low-Pi induction (Schünmann et al., 2004b). Expression of the TaPht1;2 gene is induced by Pi starvation in the wheat cultivar Dalcahue, although the full P1BS motif is not present in the promoter. Instead, four other conserved motifs have been identified. Three of them are also present in the promoter of HvPht1;1, whereas the fourth one is similar to the P1BS element (motif4; ATATRCA sequence; Tittarelli et al., 2007). Some phosphate transporter genes are expressed only in the presence of mycorrhizal fungi (reviewed by Javot et al., 2007; Smith et al., 2011). Mycorrhizal fungi do not colonize the roots of Arabidopsis, and no mycorrhiza-specific Pi transporter genes have been discovered in this species. They are, however, present in barley (HvPht1;8, AY187023; Glassop et al., 2005; Pacak et al., unpublished data; Figure 1D). Chen et al. (2011) described a set of mycorrhiza-activated phosphate transporters from eudicots. Apart from a mycorrhiza transcription factor binding sequence (MYCS), P1BS motifs were also found in the promoters of these genes. Mutation or deletion of either of these motifs resulted in a remarkable decrease or even the complete absence of gene expression (Chen et al., 2011).
Among the Pi-responsive microRNAs which have been described in the latest review published by Kuo and Chiou (2011), miR399 is especially interesting owing to its interaction with an RNA molecule named induced by phosphate starvation 1 (IPS1). The miR399 promoters possess the P1BS motif (Kuo and Chiou, 2011) and expression of their primary transcripts (pri-microRNAs) is activated by Pi starvation (Pant et al., 2009; Sobkowiak, unpublished data; Figure 1E). Two P1BS elements were found in the promoter of the Arabidopsis gene AtIPS1 (AF236376, At3g09922; Martin et al., 2000; Rubio et al., 2001; Bustos et al., 2010). Originally AtIPS1 was described as a highly Pi starvation inducible gene belonging to the Mt4/TPSI1 family, which possesses a conserved nucleotide motif and encodes only short open reading frames (Martin et al., 2000). Excellent work done by Franco-Zorrilla et al. (2007) showed that AtIPS1 sequesters miR399, which is bound to the mentioned conserved region, thereby preventing complete degradation of miR399 targets such as the mRNA for the phosphate2 protein (PHO2, At2g33770). AtIPS1 itself is not cleaved by miR399 owing to the sequence mismatches (Figure 2A). As we showed in our previous report about miR gene structures and the processing of Arabidopsis HYL1-dependent pri-microRNAs, alternative splicing events and alternative polyadenylation of microRNA precursors are often observed (Szarzynska et al., 2009). Interestingly, the Arabidopsis pri-microRNA399a transcript undergoes alternative splicing, with 3′ alternative splice site selection resulting in completely different sequences of the 3′ exons in the two pri-miR399a isoforms as well as different polyadenylation sites (Figure 2B; Sobkowiak, unpublished data). The role of these processes in miR399a expression regulation is still elusive.
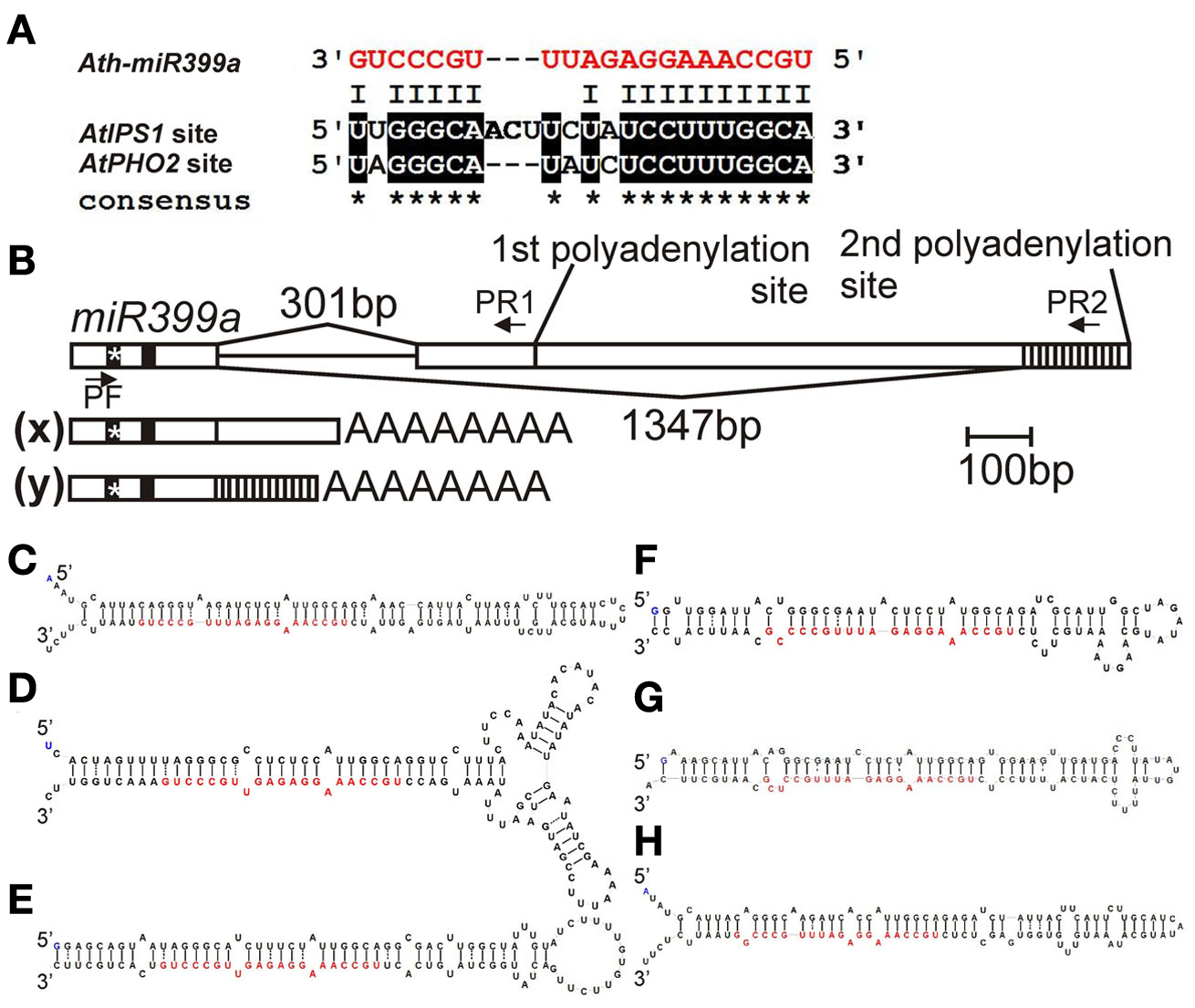
Figure 2. (A) Alignment of mature Arabidopsis miR399a with miR399 binding site present in AtIPS1 sequence and in 5′ UTR of AtPHO2 mRNA (binding site no. 2 according to Lin et al., 2008); (B) Arabidopsis miR399a gene structure and its pri-miRNA species. The position of primers used for amplification of both pri-miR399a isoforms is marked by arrows. (x) pri-microRNA399a isoform marked in Figure 1E with an x (primers: PF and PR1), (y) second pri-microRNA399a isoform (primers: PF and PR2), after alternative 3′ splice site recognition. The position of mature microRNA is depicted by a vertical black line, the position of the microRNA* is depicted by * symbol on a vertical black line; (C–H) Stem-loop structures of Arabidopsis microRNA399 precursors with Minimum folding Free Energy (MFE); mature microRNA399 is marked by red fonts; (C) pre-miR399a (dG = −51.60 kcal/mol, one of two structures with the same energy, MI0001020 – accession number in the miRBase, www.mirbase.org); (D) pre-miR399b (dG = −61.70 kcal/mol, MI0001021); (E) pre-miR399c (dG = −57.89 kcal/mol, MI0001022); (F) pre-miR399d (dG = −45.40 kcal/mol, MI0001023); (G) pre-miR399e (dG = −48.70, one of six structures with the same energy, MI0001024); (H) pre-miR399f (dG = −49.60, one of two structures with the same energy, MI0001025); structures were designed with Folder v. 1.11 beta software, algorithm RNAfold (Hansen, 2007).
The six microRNA399 species (a–f) in Arabidopsis are considered as the immediate mediators of PHO2 mRNA silencing. They are derived from six pre-microRNAs that differ in structure and sequence (Figures 2C–H). Mature miR399s can bind to five predicted target sites (differing in sequence from each other) present in the 5′ UTR of the PHO2 mRNA (Allen et al., 2005; Bari et al., 2006). Only miR399a and miR399f have full complementarity (including GU base pairings) with target sequences – sites 1 and 3 in the 5′ UTR of PHO2 mRNA, respectively (Lin et al., 2008). In Arabidopsis, an overexpression of miR399f leads to overaccumulation of Pi in shoots resembling the pho2 mutant phenotype (Chiou et al., 2006). Suppression of PHO2 by overexpressed miR399b/c is less efficient (Lin et al., 2008). The differences in microRNA affinity for the PHO2 mRNA are probably owed to a nucleotide substitution at position 13 in the miR399b/c sequence, which reduces base pairing between microRNA399b/c and the target sites (Lin et al., 2008). Interestingly, the same nucleotide substitution improves base pairing of miR399b/c with the conserved region of the At4/IPS1 family. A similar variation in miR399 sequences has been found in rice, Medicago, and Populus, where certain miR399 variants show reduced base pairing with target sequences in PHO2 but improved base pairing with the IPS1 homolog of the respective species (Lin et al., 2008).
The Arabidopsis PHO2 protein is responsible for the Pi level decrease in shoots and Pi remobilization. pho2 mutants grown under Pi replete conditions accumulate Pi in shoots, but not in roots, and show induction of some phosphate starvation-induced genes, e.g., acid phosphatase 5 (AtACP5, At3g17790) and AtPht1;4 – At2g38940 (Delhaize and Randall, 1995; Bari et al., 2006). Down-regulation of PHO2 expression in barley produced a similar effect of increased Pi levels in shoots and decreased Pi levels in roots (Pacak et al., 2010). PHO2 possesses the UBiquitin-Conjugating E2 enzyme catalytic domain (UBCc domain). Since other UBC domain-containing proteins are usually smaller, 20 vs. 100 kDa (Bari et al., 2006), PHO2 may contain unidentified additional domains. Such domains may interact with phosphate-related proteins, e.g., targeting them for degradation. Degradation of these proteins may affect the phosphate homeostasis. Bioinformatic analysis showed that, apart from PHO2 mRNA, other transcripts derived from the following genes, At3g11130 (encoding Clathrin, heavy chain), At3g25905 (clavata3/ESR-related27 protein), At3g54700 (AtPht1;7), At4g00170 (vesicle-associated membrane protein, VAMP), and At4g09730 (DEAD-box protein), are potential targets for miR399 (Pant et al., 2009; Kuo and Chiou, 2011). These complex connections between the miR399 family members and their targets can explain the fact that miR399b-overexpressing plants and a pho2 mutant exhibited not only Pi-related changes, but also an early flowering phenotype observed only at normal temperature (23°C; Kim et al., 2011).
The data presented above show that the presence of one or more P1BS elements in a gene promoter is associated with low-Pi induction. Other factors, however, can modulate the response. In the promoter of the Arabidopsis phospholipase DZ2 gene (PLDZ2, At3g05630), apart from five P1BS copies, elements such as SRE (sugar-repressive element) have been found. A 65-bp promoter fragment spanning two of the P1BS motifs (the EZ2 region) has been identified as particularly important for the Pi-limitation response but strong induction also required sucrose and was negatively affected by cytokinins (Oropeza-Aburto et al., 2012). Exactly how the information carried by the P1BS elements and by the various sugar and hormone responsive promoter elements is integrated by the MYB-CC TFs PHR1 and PHL1 and possibly others is still unknown. Furthermore, additional studies are necessary to find out how Pi-limitation directly affects the function and/or expression of Pi-related TFs. Finally, the influence of other factors which can modulate the Pi response of genes containing P1BS elements should be further investigated.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to thank Anne Olsen (DTU) for collaboration in barley cultivation in different phosphate conditions. Andrzej Pacak and Zofia Szweykowska-Kulinska were supported by the European Regional Development Fund through the Innovative Economy for Poland 2007–2013, project WND-POIG.01.03.01-00-101/08 POLAPGEN-BD “Biotechnological tools for breeding cereals with increased resistance to drought.” Andrzej Pacak and Merete Albrechtsen were supported by Marie Curie Intra-European Fellowships – BARPHO, 025110. Dawid Bielewicz was supported by the Foundation for Polish Science (FNP) – MPD 2010/3. Lukasz Sobkowiak was supported by a Ph.D. grant awarded by the Polish Ministry of Higher Education and Sciences no. NN301035839, a grant for scientific research from the Dean of Biology Faculty, Adam Mickiewicz University, Poznan, and the European Fund “Scholarship support for Ph.D. students specializing in majors strategic for Wielkopolska’s development,” Sub-measure 8.2.2, Human Capital Operational Programme. The work was also supported by grant MNiSW No. 3011/B/P01/2009/37.
Allen, E., Xie, Z., Gustafson, A. M., and Carrington, J. C. (2005). MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121, 207–221.
Bari, R., Pant, B. D., Stitt, M., and Scheible, W. R. (2006). PHO2, microRNA, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol. 141, 988–999.
Bustos, R., Castrillo, G., Linhares, F., Puga, M. I., Rubio, V., Pérez-Pérez, J., Solano, R., Leyva, A., and Paz-Ares, J. (2010). A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PLoS Genet. 6, e1001102.
Chen, A., Gu, M., Sun, S., Zhu, L., Hong, S., and Xu, G. (2011). Identification of two conserved cis-acting elements, MYCS and P1BS, involved in the regulation of mycorrhiza-activated phosphate transporters in eudicot species. New Phytol. 189, 1157–1169.
Chiou, T. J., Aung, K., Lin, S. I., Wu, C. C., Chiang, S. F., and Su, C. I. (2006). Regulation of phosphate homeostasis by microRNA in Arabidopsis. Plant Cell 18, 412–421.
Chiou, T. J., and Lin, S. I. (2011). Signaling network in sensing phosphate availability in plants. Annu. Rev. Plant Biol. 62, 185–206.
Davies, T. G. E., Ying, J., Xu, Q., Li, Z. S., Li, J., and Gordon-Weeks, R. (2002). Expression analysis of putative high-affinity phosphate transporters in Chinese winter wheats. Plant Cell Environ. 25, 1325–1339.
Delhaize, E., and Randall, P. J. (1995). Characterization of a phosphate-accumulator mutant of Arabidopsis thaliana. Plant Physiol. 107, 207–213.
Franco-Zorrilla, J. M., González, E., Bustos, R., Linhares, F., Leyva, A., and Paz-Ares, J. (2004). The transcriptional control of plant responses to phosphate limitation. J. Exp. Bot. 55, 285–293.
Franco-Zorrilla, J. M., Valli, A., Todesco, M., Mateos, I., Puga, M. I., Rubio-Somoza, I., Leyva, A., Weigel, D., Garcia, J. A., and Paz-Ares, J. (2007). Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 39, 1033–1037.
Glassop, D., Smith, S. E., and Smith, F. W. (2005). Cereal phosphate transporters associated with the mycorrhizal pathway of phosphate uptake into roots. Planta 222, 688–698.
Gu, M., Xu, K., Chen, A., Zhu, Y., Tang, G., and Xu, G. (2010). Expression analysis suggests potential roles of microRNAs for phosphate and arbuscular mycorrhizal signaling in Solanum lycopersicum. Physiol. Plant. 138, 226–237.
Hansen, T. (2007). Folder Version 1.11 Beta. Denmark: MBI, University of Aarhus. Available at: http://www.ncrnalab.dk/rnafolder/
Javot, H., Pumplin, N., and Harrison, M. J. (2007). Phosphate in the arbuscular mycorrhizal symbiosis: transport properties and regulatory roles. Plant Cell Environ. 30, 310–322.
Karandashov, V., and Bucher, M. (2005). Symbiotic phosphate transport in arbuscular mycorrhizas. Trends Plant Sci. 10, 22–29.
Kim, W., Ahn, H. J., Chiou, T. J., and Ahn, J. H. (2011). The role of the miR399-PHO2 module in the regulation of flowering time in response to different ambient temperatures in Arabidopsis thaliana. Mol. Cells 32, 83–88.
Kuo, H. F., and Chiou, T. J. (2011). The role of microRNAs in phosphorus deficiency signaling. Plant Physiol. 156, 1016–1024.
Lin, S. I., Chiang, S. F., Lin, W. Y., Chen, J. W., Tseng, C. Y., Wu, P. C., and Chiou, T. J. (2008). Regulatory network of microRNA399 and PHO2 by systemic signaling. Plant Physiol. 147, 732–746.
Martin, A. C., del Pozo, J. C., Iglesias, J., Rubio, V., Solano, R., de La Peña, A., Leyva, A., and Paz-Ares, J. (2000). Influence of cytokinins on the expression of phosphate starvation responsive genes in Arabidopsis. Plant J. 24, 559–567.
Miura, K., Rus, A., Sharkhuu, A., Yokoi, S., Karthikeyan, A. S., Raghothama, K. G., Baek, D., Koo, Y. D., Jin, J. B., Bressan, R. A., Yun, D. J., and Hasegawa, P. M. (2005). The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proc. Natl. Acad. Sci. U.S.A. 102, 7760–7765.
Muchhal, U. S., Pardo, J. M., and Raghothama, K. G. (1996). Phosphate transporters from the higher plant Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 93, 10519–10523.
Müller, R., Morant, M., Jarmer, H., Nilsson, L., and Nielsen, T. H. (2007). Genome-wide analysis of the Arabidopsis leaf transcriptome reveals interaction of phosphate and sugar metabolism. Plant Physiol. 143, 156–171.
Nilsson, L., Müller, R., and Nielsen, T. H. (2010). Dissecting the plant transcriptome and the regulatory responses to phosphate deprivation. Physiol. Plant. 139, 129–143.
Oropeza-Aburto, A., Cruz-Ramírez, A., Acevedo-Hernández, G. J., Pérez-Torres, C. A., Caballero-Pérez, J., and Herrera-Estrella, L. (2012). Functional analysis of the Arabidopsis PLDZ2 promoter reveals an evolutionarily conserved low-Pi-responsive transcriptional enhancer element. J. Exp. Bot. 63, 2189–2202.
Pacak, A., Geisler, K., Jørgensen, B., Barciszewska-Pacak, M., Nilsson, L., Nielsen, T. H., Johansen, E., Grønlund, M., Jakobsen, I., and Albrechtsen, M. (2010). Investigations of barley stripe mosaic virus as a gene silencing vector in barley roots and in Brachypodium distachyon and oat. Plant Methods 6, 26.
Pant, B. D., Musialak-Lange, M., Nuc, P., May, P., Buhtz, A., Kehr, J., Walther, D., and Scheible, W. R. (2009). Identification of nutrient-responsive Arabidopsis and rapeseed microRNAs by comprehensive real-time polymerase chain reaction profiling and small RNA sequencing. Plant Physiol. 150, 1541–1555.
Richardson, A. E., Lynch, J. P., Ryan, P. R., Delhaize, E., Smith, F. A., Smith, S. E., Harvey, P. R., Ryan, M. H., Veneklaas, E. J., Lambers, H., Oberson, A., Culvenor, R. A., and Simpson, R. J. (2011). Plant and microbial strategies to improve the phosphorus efficiency of agriculture. Plant Soil 349, 121–156.
Rouached, H., Arpat, A. B., and Poirier, Y. (2010). Regulation of phosphate starvation responses in plants: signaling players and cross-talks. Mol. Plant 3, 288–299.
Rouached, H., Secco, D., Bulak, A., and Poirier, Y. (2011). The transcription factor PHR1 plays a key role in the regulation of sulfate shoot-to-root flux upon phosphate starvation in Arabidopsis. BMC Plant Biol. 11, 19.
Rubio, V., Linhares, F., Solano, R., Martín, A. C., Iglesias, J., Leyva, A., and Paz-Ares, J. (2001). A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev. 15, 2122–2133.
Schünmann, P. H. D., Richardson, A. E., Smith, F. W., and Delhaize, E. (2004a). Characterization of promoter expression patterns derived from the Pht1 phosphate transporter genes of barley (Hordeum vulgare L.). J. Exp. Bot. 55, 855–865.
Schünmann, P. H. D., Richardson, A. E., Vickers, C. E., and Delhaize, E. (2004b). Promoter analysis of the barley Pht1;1 phosphate transporter gene identifies regions controlling root expression and responsiveness to phosphate deprivation. Plant Physiol. 136, 4205–4214.
Smith, S. E., Jakobsen, I., Grønlund, M., and Smith, F. A. (2011). Roles of arbuscular mycorrhizas in plant phosphorus nutrition: interactions between pathways of phosphorus uptake in arbuscular mycorrhizal roots have important implications for understanding and manipulating plant phosphorus acquisition. Plant Physiol. 156, 1050–1057.
Stefanovic, A., Ribot, C., Rouached, H., Wang, Y., Chong, J., Belbahri, L., Delessert, S., and Poirier, Y. (2007). Members of the PHO1 gene family show limited functional redundancy in phosphate transfer to the shoot, and are regulated by phosphate deficiency via distinct pathways. Plant J. 50, 982–994.
Szarzynska, B., Sobkowiak, L., Pant, B. D., Balazadeh, S., Scheible, W. R., Mueller-Roeber, B., Jarmolowski, A., and Szweykowska-Kulinska, Z. (2009). Gene structures and processing of Arabidopsis thaliana HYL1-dependent pri-miRNAs. Nucleic Acids Res. 37, 3083–3093.
Tittarelli, A., Milla, L., Vargas, F., Morales, A., Neupert, C., Meisel, L. A., Salvo, G. H., Penaloza, E., Munoz, G., Corcuera, L. J., and Silva, H. (2007). Isolation and comparative analysis of the wheat TaPT2 promoter: identification in silico of new putative regulatory motifs conserved between monocots and dicots. J. Exp. Bot. 58, 2573–2582.
Keywords: phosphate, Pi-responsiveness, P1BS, microRNA
Citation: Sobkowiak L, Bielewicz D, Malecka EM, Jakobsen I, Albrechtsen M, Szweykowska-Kulinska Z and Pacak A (2012) The role of the P1BS element containing promoter-driven genes in Pi transport and homeostasis in plants. Front. Plant Sci. 3:58. doi: 10.3389/fpls.2012.00058
Received: 30 November 2011; Accepted: 07 March 2012;
Published online: 30 March 2012.
Edited by:
Bernie Carroll, The University of Queensland, AustraliaReviewed by:
Chris Helliwell, Commonwealth Scientific and Industrial Research Organisation, AustraliaCopyright: © 2012 Sobkowiak, Bielewicz, Malecka, Jakobsen, Albrechtsen, Szweykowska-Kulinska and Pacak. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Andrzej Pacak, Department of Gene Expression, Adam Mickiewicz University, Umultowska 89, Poznan 61-614, Poland. e-mail:YXBhY2FrQGFtdS5lZHUucGw=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.