
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 19 March 2012
Sec. Plant Genetics and Genomics
Volume 3 - 2012 | https://doi.org/10.3389/fpls.2012.00046
This article is part of the Research Topic Regulatory Elements in RNA View all 12 articles
Arabidopsis miR319a/b/c primary transcripts are unusual due to the presence of a long stem and loop structure containing functional miR319a/b/c molecules. In our experiments carried out using high throughput sequencing (HTS), we have shown that additional microRNAs (miRNAs), miR319a.2/b.2/c.2 are generated from the upper part of the same hairpin structure. We have also found cognate miRNAa.2*/b.2*/c.2* to be present in the HTS results with a considerably lower number of reads. Northern hybridization revealed that miR319b.2 is mainly expressed in 35-day-old plant rosette leaves, as well as in stem and inflorescences of 42- and 53-day-old plants. Moreover, it carries multiple signatures of a functional miRNA, including as follows: (i) its biogenesis is HYL1-dependent; (ii) it is incorporated in a substantial amount into RISC complexes containing AGO1, AGO2, or AGO4 protein; (iii) 24 nt-long species of miR319b.2 have been found in inflorescences to be more abundant than 21 nt miR319b.2 species; (iv) it is present in various ratios to miR319b during plant development, which suggests the existence of a regulatory mechanism responsible for its biogenesis/processing; (v) there is an observed cross-species conservation of the miR319a/b/c stem nucleotide sequence extending beyond mature miRNA region; and (vi) all evidence suggests that intron-containing RAP2.12 mRNA isoform is the target for miR319b.2. All these features prompt us to claim miR319b.2 as a functional miRNA molecule.
MicroRNAs (miRNAs) represent a class of endogenous, 21–24 nt-long regulatory RNA molecules (Llave et al., 2002; Reinhart et al., 2002; Palatnik et al., 2003). They are involved in the regulation of gene expression by targeting the cognate mRNA molecules for cleavage, or by inhibiting their translation. In Arabidopsis thaliana plants, the mature miRNAs are generated from the fold-back hairpin-like structure of nuclear-localized MIR gene transcripts termed pri-miRNAs. At least eight proteins are involved in plant miRNA precursor’s maturation process: type-III ribonuclease DICER-LIKE 1 (DCL1), dsRNA binding protein HYPONASTIC LEAVES 1 (HYL1, DRB1), zinc-finger protein SERRATE (SE), forkhead-associated (FHA) domain-containing protein DAWDLE (DDL), CAP BINDING PROTEIN 20 (CBP20), CAP BINDING PROTEIN 80 (CBP80/ABH1), methyltransferase HUA ENHANCER 1 (HEN1), and HASTY – an ortholog of animal Exportin 5 (Park et al., 2002, 2005; Han et al., 2004; Kurihara and Watanabe, 2004; Vazquez et al., 2004; Lobbes et al., 2006; Yang et al., 2006b,a; Kim et al., 2008; Laubinger et al., 2008; Yu et al., 2008; Huang et al., 2009).
Plant miR159 and miR319 genes are highly conserved and their representatives can be found in a variety of plants from mosses to flowering plants (Kozomara and Griffiths-Jones, 2011). It is postulated that miR159 and miR319 evolved from a common ancestor and share sequence identity in 17 out of 21 nt (Kozomara and Griffiths-Jones, 2011; Li et al., 2011). However, due to sequence specificity and temporal and special expression patterns in Arabidopsis plants, they have distinct targets; miR159 downregulates the expression of MYB, while miR319 downregulates the expression of TCP transcription factor, respectively. The miR159 is involved in flowering, male fertility, and ABA-dependent seed germination processes, while miR319 plays a role in leaf and flower development; thus, both are critical for plant development, growth, morphogenesis, and reproduction (Palatnik et al., 2007; Reyes and Chua, 2007; Nag et al., 2009). Both MIR159 and MIR319 families contain three genes: MIR159a/b/c, and MIR319a/b/c, respectively. A unique future of MIR159 and MIR319 genes is the presence of a long stem and loop structure. The base-proximal segment of miR159/319 stem and loop precursors is conserved, while the loop-proximal part is conserved to a lesser extent (Bologna et al., 2009). The biogenesis pathway of both miRNAs is unusual since it is a loop-to-base processing mechanism that begins with the cleavage of the loop, instead of the usual cut at the base of the stem-loop structure. Moreover, in several cases, additional small RNAs generated from miRNA 319a/b/c precursors were observed. It is possible that a non-canonical mechanism of the miRNA159/319 maturation potentially generates additional sRNAs from pri-miRNA159/319 hairpins. These additional sRNAs may be non-functional intermediates generated during the miRNA biogenesis pathway or functional, regulatory RNA molecules (Addo-Quaye et al., 2009; Bologna et al., 2009). It was shown by Zhang et al. (2010) that additional sRNAs generated from miR159a, miR319a, and miR319b precursors are stable in A. thaliana plants infected by various Pseudomonas syringae strains. They have shown that miR319b.2 is the main sRNA molecule derived from pri-miRNA319b. In this paper, we show that miR319b.2 sRNA is also present at a relatively high level in Arabidopsis stems and inflorescences in non-infected plants. Moreover, our analysis shows that these additional small RNAs exhibit many futures similar to functional miRNA molecules, namely, they are associated with AGO proteins, and our results point to an intron-containing RAP2.12 mRNA isoform as a target of these miRNAs.
Arabidopsis thaliana (L.) Heynh, Col-0 wild type plants, homozygous se-1 plants (NASC, N3257), homozygous T-DNA insertion line hyl1-2 (SALK_064863), homozygous line ΔmiR319b (SALK_037093), and miR319boe (SALK_059451) were grown in conditions as described in (Szarzynska et al., 2009). Plants for developmental and organ specific northern blot analyses and developmental quantitative real-time PCR profiling of pri-miRNAs from MIR319 family were grown in the hydroponic growing system for A. thaliana (Araponics SA, Liege, Belgium).
For northern blot analyses, total RNA was isolated from 14-day-old seedlings, 20- day-old plants, and from organs (roots, rosette leaves, stems, and inflorescences) collected in 35, 42, and 53 days after sowing (DAS), using the method described in (Pant et al., 2009). For quantitative real-time PCR analyses, total RNA was isolated as described in (Szarzynska et al., 2009).
Total RNA isolated from the wild type A. thaliana Col-0 35-day-old rosette leaves (plants were grown in soil) was used for a small RNA library construction. The small RNA libraries were generated and sequenced by Illumina (Fasteris SA, Plan-les-Ouates, Switzerland and British Columbia Cancer Agency, Vancouver, BC, Canada). Adaptor sequences were identified and trimmed from each read using a customized Perl script. Reads in which the adaptor could not be identified were discarded. Two independent rounds of sequencing resulted in a total of more than five million unique, quality-filtered, and adaptor-trimmed reads. As expected for a sRNA sequencing procedure, the size distribution of the short sequences revealed the presence of dominating classes of 24 and 21 nt reads. The maximum read count in our sample was 657050. A BWA program was used to align the trimmed reads to the set of pri-miRNAs and miRNA genes (Li and Durbin, 2010). For each library, we counted the number of trimmed reads within the 18–24 nt range. Reads with counts of less than 5 were discarded giving final number of 41829 and 167616 sequences for two separate replicas.
Counts for the sequences which were mapping to the pri-miRNA were normalized by the total number of 18–24 nt trimmed reads in the library and presented in the form of “reads per million (RPM)”. Trimmed reads that were <18 nt or >24 nt were not considered in this analysis. Three libraries were downloaded from the GEO database: GSM253622, GSM253623, and GSM253624, representing the sequences immunopurified with AGO1, AGO2, and AGO4 complexes, respectively. The reads were mapped to pri-miRNA with the BWA program.
The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE353351.
RNA (30–50 μg) was fractionated on a 15–17% denaturing (7 M urea, Sigma, Deisenhofen, Germany) polyacrylamide gel (PAGE), transferred to a Hybond-NX membrane (Amersham Biosciences-GE Healthcare, Little Chalfont, UK) by capillary transfer, using 20xSSC buffer, and fixed by UV-crosslinking. Pre-hybridization was carried out at 42°C for 2 × 30 min, using PerfectHyb Hybridization Buffer (Sigma, Deisenhofen, Germany). Probes were labeled with γ32P ATP (6000 Ci/mmol; NEN-PerkinElmer Life and Analytical Sciences, Waltham, MA, USA), using T4 polynucleotide kinase (Roche, Mannheim, Germany) and purified on Illustra Micro Spin G-25 Columns (GE Healthcare). Hybridization was performed overnight at 42°C. Lengths of RNA molecules were estimated using 32P-labeled Decade Marker System (Ambion). Oligonucleotides complementary to miR159/319 and miR319b.2 (see Table A1 in Appendix) were used as probes. A probe complementary to U6 snRNA was used as a loading control.
The concentration of cDNA from the wild type, miR319boe and ΔmiR319b mutant plants was normalized against the β-actin as described in (Szarzynska et al., 2009). The PCR products were electrophoresed on 1.2% agarose gels in 1 × TBE buffer (for primer sequences see Table A1 in Appendix).
Real-time PCR analyses were carried out using methodology described in (Szarzynska et al., 2009). Log10 were calculated from the fold change of particular pri-miRNAs or RAP2.12 mRNA to PP2A transcript (At1g69960).
5′-RACE experiments were performed using SMARTer RACE cDNA Amplification Kit (Clontech, Mountain View, CA, USA), according to the manufacturer’s protocol. All primers used in the experiments are listed in Table A1.
We carried out SOLEXA sequencing reactions of RNA enriched with small RNAs isolated from 35-day-old A. thaliana rosette leaves. As already reported by Bologna et al. (2009) and Zhang et al. (2010), we also found extra sRNA species generated from the same stem and loop structure where miRNA 319a/b/c and miRNAa*/b*/c* are embedded. They are located in the loop-proximal part of the stem and loop precursor structure (Figure 1). These most abundant species were named miRNA 319a.2/319b.2/319c.2, respectively. We also found sRNA species that can be regarded as miRNA* to miRNA 319a.2/b.2. Table 1 shows normalized counts for the reads of the miRNAs derived from 319a/b/c precursors found in SOLEXA deep sequencing results. File A1 in Appendix contains the full list of all matching sRNAs, which originate from pri-miR319a/b/c. Analysis of these data clearly shows that the number of reads for miR319b.2 is the highest, even exceeding the number of the reads obtained for miR319a/b, miR319a, and miR319b are identical in sequence; therefore, it is not feasible to distinguish the number of reads derived from each locus separately.
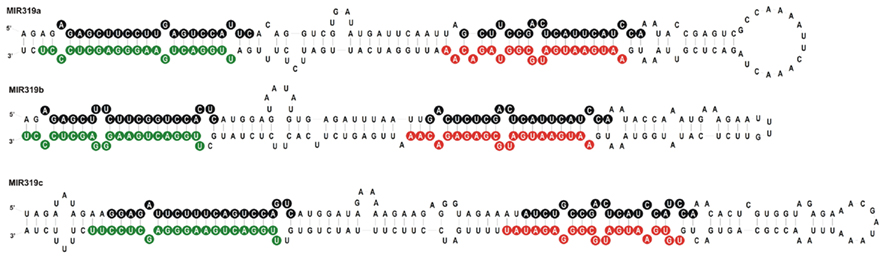
Figure 1. Stem and loop structures of Arabidopsis pre-miR319 precursors. Sequences of miR319a, b, and c are in green, sequences representing miRNA319a.2, b.2, and c.2 are in red, and star sequences are in black circles, respectively.
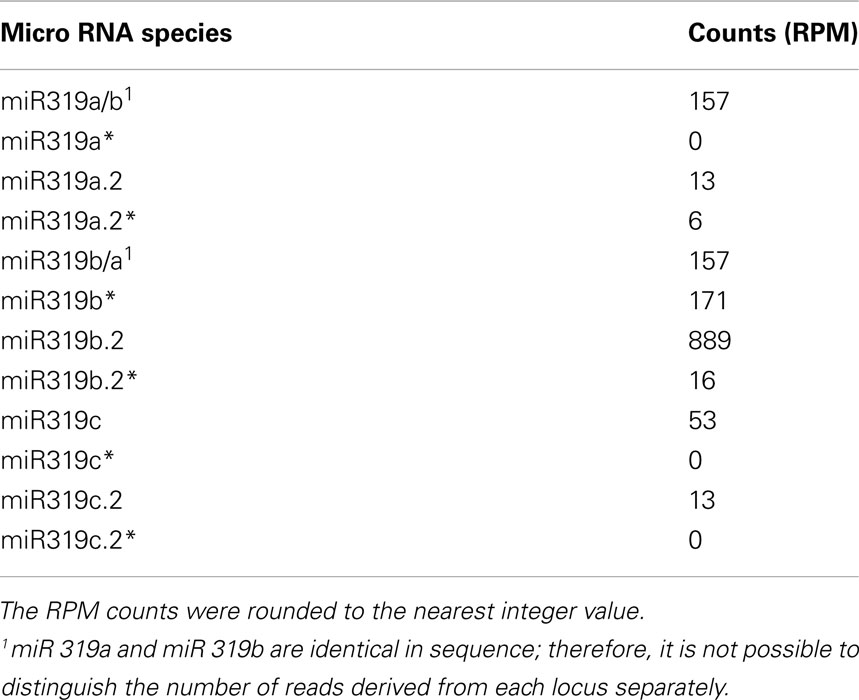
Table 1. Number of representative reads for specific miR319 species and additional miRNAs generated from their precursors.
The small number of reads for miR319c, miR319c.2, and miR319a.2 obtained in the SOLEXA experiments induced us to evaluate the expression profiles of all MIR319 family members in various developmental stages and organs studied.
We designed primers to specifically amplify each of the particular MIR319 gene family transcripts, and using real-time PCR, we carried out pri-miR319a/b/c expression profiling. Figure 2 shows the obtained results. All family members show differences at the transcript level during plant development. For instance, pri-miR319c expression decreases, while pri-miR319a and pri-miR319b increase in roots of 42-day-old plants as compared to their level in 35- and 53-day-old plants, respectively. We were not able to detect pri-miR319a in rosette leaves of 42- and 53-day-old plants, while at the same time, we observed the expression of two other family members with the highest level of pri-miR319c. The levels of all pri-miRs319 differ in rosette leaves of 35-day-old plants. Pri-miR319b and pri-miR319c are almost to the same level. However, our high throughput sequencing (HTS) results do not show the same amount of mature miR319a/b and miR319c in rosette leaves of the 35-day-old plants. It indicates that the level of pri-miR does not necessarily mirror the level of mature miRNA. Generally, the highest expression of all MIR319 family genes occurs in stems and inflorescences. Our real-time PCR data shows that pri-miR319b from the MIR319 family is the dominating transcript present in the majority of tissues studied.
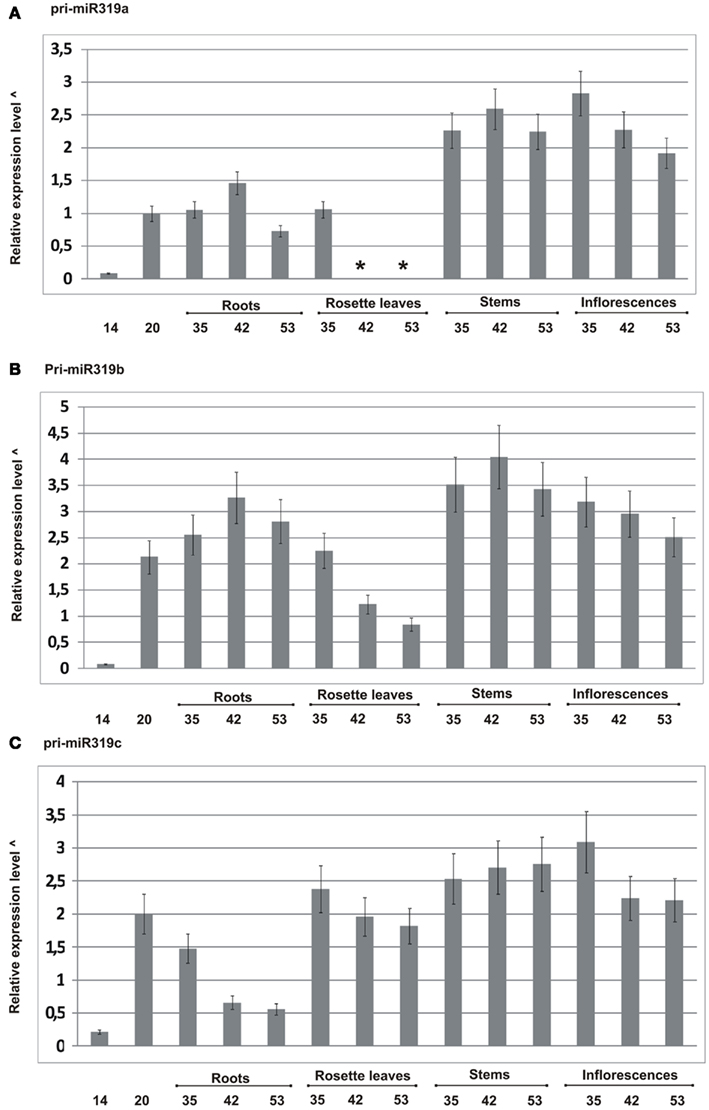
Figure 2. Quantitative real-time PCR profiling of pri-miRNAs from MIR319 family in different developmental stages in wild type A. thaliana plants (14-, 20-, 35-, 42-, and 53-day-old plants). (A–C) Expression patterns of pri-miR319a, pri-miR319b, and pri-miR319c, respectively. *Not detected, ^log10 calculated from the fold change of particular pri-miRNAs standardized to the PP2A transcript level (At1g69960). Because the expression of pri-miR319a/b/c is in most cases lower than the level of PP2A transcript, the graph was rescaled according to the following formula: abs(xmax-x), where abs denotes absolute value, xmax represents the lowest integer value of relative expression level in the original graph and x represents actual expression level for a given pri-miRNA in the particular developmental stage. Real-time PCR for all pri-miR319 was carried out in three biological replicates. Thin black lines represent SD.
We identified a null-insertional T-DNA Arabidopsis mutant in the MIR319b locus (SALK_037093; ΔmiR319b), and a MIR319b overexpression mutant (Salk_059451; miR319boe) within SALK collection. Both of them showed phenotypic differences when compared to wild type plants (Figure 3A). To our surprise, both mutants exhibited growth retardation. However, the retardation is more profound in the case of the overexpression mutant. Also, the shape of rosette leaves is changed when compared to wild type plants. Leaves of the null mutant plant are narrower, their margins are rolled inwards, while the morphological changes in the case of the plant overexpressing MIR319b are more profound, showing a more compact rosette of arrow-shaped leaves, leaf serrated margin, its blade sharply pointed at the tip, and a strong leaf curvature when compared to wt plants. We did not observe any differences in flower shape and structure. In general, the phenotype of the miR319boe mutant resembles strongly the phenotype of MIR-JAW (MIR319a) overexpressing mutant described by Weigel et al. (2000). RT-PCR reaction carried out using RNA isolated from 42-day-old stems for MIR319b transcript detection revealed the increased level of primary transcript in the miR319boe mutant and the lack of transcript in the case of ΔmiR319b mutant plants (Figure 3B). Northern hybridization shows the lack of miR319b.2 in the null mutant plants and increased levels of miR319a/b/c and miR319b.2 in the miR319boe mutant, as compared to wild plants. The very faint hybridization signal observed in the case of the ΔmiR319b mutant, when the radioactive probe for miR319b.2 was used, derives probably from the weak expression of miR319a.2 that may weakly cross-hybridize (there are two mismatches between miR319a.2 and miR319b.2, Figures 3C,D). Since there are 4 nt mismatches between miR319b.2/miR319c.2 and five between miR319a.2/miR319c.2, we anticipate it to be unlikely that the probe for miR319b.2 detection is cross-hybridizing with the miR319c.2. Northern hybridization results are in agreement with the expression profile of pri-miR319a/b/c described in previous chapter (see Figure 2). All these observations, together with the deep sequencing data, suggest that miR319b.2 represents a stable molecule being a product of pre-miR319b processing.

Figure 3. The phenotypes of 42-day-old A. thaliana wild type, ΔmiR319b (Salk_037093), and miR319boe (Salk_059451) mutant plant (A). RT-PCR profiling of the pri-miR319b level. (B) The upper panel shows the increased level of primary transcript in the miR319boe mutant and the lower panel – the lack of transcript in the case of ΔmiR319b mutant plants. Northern blot hybridization (C,D) of miR319 and miR319b.2 in the wild type, miR319boe and ΔmiR319b mutant plants. RNA was isolated from stems [left panel (C)] and inflorescenses [right panel (D)]. For better visualization of miR319b.2 hybridization signals, these blots were longer exposed than in the case of miR319b.
The inspection of the GEO database of the HTS results for AGO1, AGO2, and AGO4 immunoprecipitates for miR319a/b, miR319c, and miR319a.2/b.2/c.2 revealed the highest number of reads for miRNA 319a/b and miR319b.2 as RNA species incorporated into RISC complexes. This suggests a functional role for miR319b.22. Table 2 shows the numbers of particular miRNAs and miRNAs from the miR319 family found in AGO1, AGO2, and AGO4 immunoprecipitates. Zhang et al. (2010) have recently shown the presence of miR319a.2, as well as miR319b.2, in small RNA libraries from Arabidopsis infected with various P. syringae pv. tomato (Pst) DC3000 strains, which again strengthens the idea that these miRNAs may be functional.
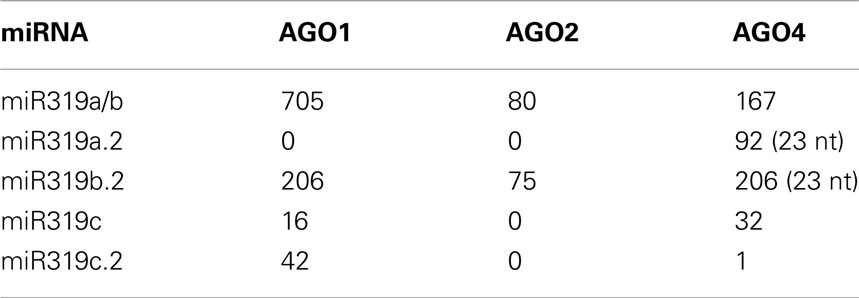
Table 2. Detection of miR319 a/b/c and additional sRNAs from miR319a/b/c precursors within AGO1, AGO2, and AGO4 immunoprecipitates.
We studied the presence of miR319b.2 in plants by carrying out northern hybridization for miR159/319 and miR319b.2 in selected developmental stages and organs (see Figure 4). Because of high similarity between miR159 and miR319 families that share sequence identity of 17 out 21 nt, both species hybridize to miR319a/b probe. Since in the ΔmiR319b mutant background only a very faint hybridization signal for miR319a.2 was observed (Figure 3), we omitted MIR319a and MIR319c expression in miR319b/miR319b.2 ratio calculations. Analysis of hybridization signals shows that miR159/319b and miR319b.2 are detectable predominantly in rosette leaves, stems, and inflorescences (Figures 4C–E). The ratio of hybridization signals for miR319b to miR319b.2 in stems shows a non-equimolar abundance of both micro RNAs, with three times higher abundance of miR319b in 35-day-old and 42-day-old plants. However, in 53-day-old stems this ratio is changed and shows only 1.7 higher expression of miR319b in comparison to miR319b.2. Interestingly, the amount of miR319b.2 in 35-day-old rosette leaves exceeds the amount of miR319b (Figure 4C). It is in agreement with our HTS data that shows a substantially higher number of miR319b.2 reads than that of miR 319a/b. Changes in stechiometry between miR319b and miR319b.2, and the higher amount of the miR319b.2 in comparison to miR319b in 35-day-old rosette leaves supports our idea of a possible functional and regulatory role of miR319b.2.
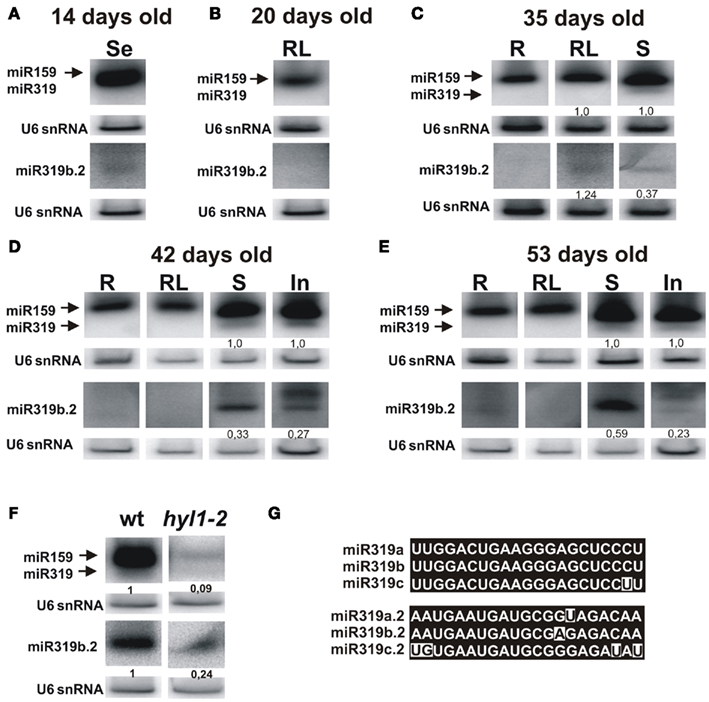
Figure 4. Detection of the mature miR319 and miR319b.2 in the wild type A. thaliana plants. Northern hybridization was performed with total RNA enriched for small RNAs from 14-day-old seedlings (A), 20-day-old rosette leaves (B), 35-day-old plants (roots, rosette leaves, and stems) (C), 42-day-old (D), and 53-day-old plants (E) (roots, rosette leaves, stems, and inflorescences) using the probe for miR319a/b and miR319b.2. Biogenesis of miR319b.2 is HYL1-dependent (F). In (F), hybridization was carried out using RNA isolated from the stems of 42-day-old plants. The ratio between 319b and 319b.2 signals is indicated in (C–E), and between miR319b or miR319b.2 in wild type and hyl1-2 mutant plants in (F). For better visualization of miR319b.2 hybridization signals, these blots were longer exposed than in the case of miR319b. The probe for U6 snRNA was used as RNA loading control. (G) Shows nucleotide sequences of miR319a/b/c, and miR319a.2/b.2/c.2, respectively.
In 42-day-old and 53-day-old inflorescences we observed (in addition to 21 nt long miR319b.2) 24 nt-long species, which constitute the main hybridization band (Figures 4D,E). The 24 nt-long miRNA species have already been reported and represent the products of DCL3 activity (Vazquez et al., 2008; Zhou et al., 2010). Moreover, it was shown by Vazquez et al. (2008), that 24 nt-long miRNA species are present predominantly in the inflorescences. The observed 24 nt length of miR319b.2 in inflorescences emphasizes its miRNA-like biogenesis and characteristics specific of miRNA species.
It was shown that the biogenesis of the miR319b is HYL1-dependent (Feijie and Yuke, 2007; Szarzynska et al., 2009). Therefore, we tested whether the amount of mature miR319b.2 also depended on the activity of the HYL1 protein. Figure 4F shows that, indeed, the accumulation of miR319b.2 is decreased in the hyl1-2 mutant, and the same was observed for the miR159/319. Thus we concluded that the biogenesis of both miR319b.2 and miR319b is HYL1-dependent.
A bioinformatic search of putative miR319b.2 targets pointed to five mRNAs (see Table A2 in Appendix). One of them was the At1g53910 gene encoding RAP2.12, a member of the ERF (ethylene response factor) subfamily B-2 of ERF/AP2 transcription factors family. Figure 5 shows the structure of RAP2.12 gene and its two mRNA isoforms. There are two alternatively spliced mRNA isoforms. One of them is fully spliced, while the other retains a second intron, which is located within the 3′-UTR. As shown in Figures 6A,B, real-time PCR measurements revealed an approximately ninefold higher amount of RAP2.12 mRNA isoform containing intron in comparison to the one that is fully spliced; both in 42-day-old wild type plant stems and inflorescences (Figures 6A,B). Only RAP2.12 mRNA isoform containing intron can be targeted by miR319b.2 within 3′-UTR region (Figure 5B). In the NEOMORPH database3, there are RAP2.12 mRNA fragments which are cut exactly at the putative slicing site, which is located in the middle of the intron. However, using the 5′-RACE approach we were not able to prove experimentally that RAP2.12 mRNA intron-containing isoform is cleaved by miR319b.2-guided RISC complex at the exactly predicted site. Our 5′-RACE results show mRNA fragments cut close, and always downstream from the putative slicing site (Figure 5B). We decided to compare the amount of putative 3′-intron-containing RAP2.12 mRNA isoform cleavage product in wt plants, miR319boe, and ΔmiR319b mutant plants. Agarose gel electrophoresis of 5′-RACE product revealed almost no product in the ΔmiR319b mutant background, while it was present in higher amounts in the miR319boe mutant background as compared with the wild type plants (Figure 5C). Apart from the 5′-RACE slicing product of the expected length, we also observed degradation products (marked with a star in Figure 5C) that are present only in wt and miR139boe mutant lines.
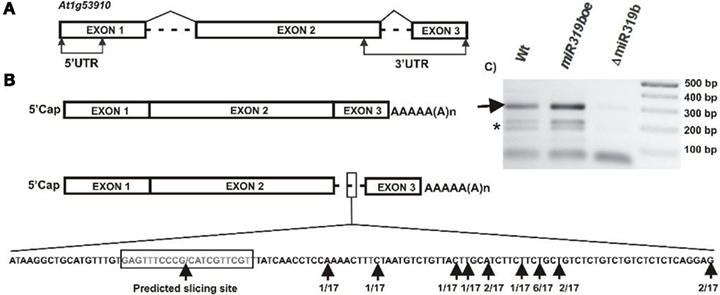
Figure 5. The structure of RAP2.12 gene (A) and its two mRNA isoforms (B). The predicted slicing site within intron-retaining mRNA isoform is marked and enlarged. Black arrows point to predicted slicing site and identified products of 5′-RACE carried out for RAP2.12 slicing products. (C) The agarose gel showing the 3′-RAP2.12 intron-containing mRNA isoform cleavage products in wt plants, miR319boe, and ΔmiR319b mutant plants. Arrow points to the expected length of 5′-RACE product, star depicts degradation products obtained during 5′-RACE. In each line, five 5′-RACE reaction products were pooled together and loaded on gel.
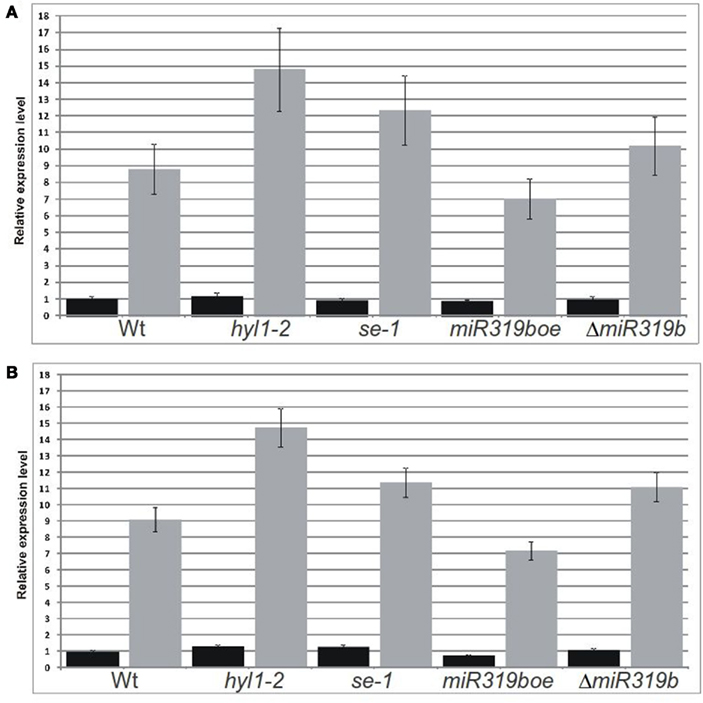
Figure 6. Real-time PCR analyses performed for two RAP2.12 mRNA isoforms in 42-day-old plants. Fully spliced isoform (black bars) was amplified using primers flanking exon–exon junction and the intron-retaining isoform (gray bars) was amplified with primers flanking predicted slicing site (A, B). Relative expression level of both mRNA isoforms of wild type and mutant hyl1-2, se-1, miR319boe, and ΔmiR319b plants, were analyzed in stems and inflorescences, respectively. Ct values for all mRNA transcripts were normalized against the PP2A (At1g69960) Ct value.
The real-time PCR was performed using primers flanking the predicted slicing site for five putative miR319b.2 targets in wt plants and hyl1-2, se-1, miR319boe, and ΔmiR319b mutants, respectively. The experiments were done for 42-day-old plants and RNA was isolated from stems and inflorescences. Only in the case of RAP2.12 we observed the expected results. For the intron-retaining mRNA isoform, the level in hyl1-2, se-1, and ΔmiR319b mutants was increased, while in the miR319boe mutant – decreased, when compared to Arabidopsis wt plants (Figures 6A,B). In parallel experiments, no significant changes were observed in case of the fully spliced RAP2.12 mRNA isoform (Figures 6A,B). All these results indicate that RAP2.12 may be a target for miR319b.2 in stem and inflorescences of A. thaliana plants.
Recent bioinformatic analyses of eukaryotic transcriptome sequences generated using new generation sequencing (NGS) approaches revealed the presence of multiple types of small RNA species with known and unknown functions (Sobala and Hutvagner, 2011). Studies carried out on Arabidopsis transcriptome revealed the presence of a new class 19 nt-long small RNAs corresponding to the 5′ end of the specific tRNA species. These small RNAs accumulate to high levels in phosphate-starved roots. However, their biological function is still not clear (Hsieh et al., 2010). In addition, it was found that small RNAs with evolutionary conservation of size and position are derived from the vast majority of snoRNA loci in Arabidopsis, as well as in other species. These sno-derived RNAs (sdRNAs) are associated with Arabidopsis AGO7. It is postulated that there is an interplay between the RNA silencing and snoRNA-mediated RNA processing and RNA-directed regulatory system (Taft et al., 2009). In humans, many small RNAs derived mainly from 3′-ends of intron sequences and 3′-UTRs were reported. They were found to be associated with AGO1/2. Valen et al. (2011) revealed that these sRNAs are the products of non-canonical miRNA biogenesis pathways. Finally, it was shown that viral infection by rice stripe virus (RSV) induces expression of novel-phased miRNAs and the accumulation of miRNA*s from rice conserved cellular miRNA precursors (Du et al., 2011). All of these data show that we experience the genesis of the still-expanding realm of small regulatory RNAs deriving from the known, canonical RNA species.
Our studies regarding miR319b.2 indicate that new, functional sRNA molecules are generated from already known, conservative Arabidopsis miRNA genes. As mentioned previously, Zhang et al. (2010) have shown the presence of miR319a.2 and miR319b.2 in small RNA libraries from Arabidopsis infected with various P. syringae pv. tomato (Pst) DC3000 strains. However, our results show that it is mainly miR319b.2, which is expressed during plant development in non-stressed conditions. We cannot rule out the possibility that miR319a.2 and miR319c.2 are generated at higher levels in other developmental stages than the ones we studied, or in a plants response to stress conditions. In this paper, we show that miR319b.2 derived from pri-miR319b has properties of many functional miRNAs, such as: (i) its biogenesis is HYL1-dependent; (ii) it is incorporated in a substantial amount into RISC complexes containing AGO1, AGO2, or AGO4 proteins; (iii) 24 nt-long species of miR319b.2 have been found in inflorescences, where they are more abundant than 21 nt-long miR319b.2 species (Vazquez et al., 2008; Hu et al., 2011); (iv) it is present in various ratios to miR319b during plant development, which suggests the existence of a regulatory mechanism responsible for its biogenesis/processing; (v) there is cross-species conservation of the miR319a/b/c stem nucleotide sequence extending beyond the mature miRNA region. This conservation is clearly visible in the regions where miR319a.2/b.2/c.2 are located. Although this conservation is not as pronounced as in the case of the mature miR319a/b/c sequences, one can observe a clustering of these sequences within the main taxonomic groups (dicots, monocots, lower plants; Li et al., 2011).
Previously, using in silico approach, Zhang et al. (2010) predicted eight putative targets for miR319b.2. Our bioinformatic analyses found four additional presumed targets. The five best predictions were evaluated experimentally. Among these, RAP2.12 mRNA is one of the most likely. Target sequences matching the miR319b.2, miR319a.2, and putative miR319c.2 sequences was found in the RAP2.12 intron that is retained in the mature mRNA in the course of alternative splicing. Analyses of NEOMORPH database show that the RAP2.12 mRNA fragments are cleaved exactly at the predicted site in at least two independent experiments. However, the 5′-RACE for RAP2.12 cleavage site was not conclusive and we were not able to confirm experimentally that the 3′-mRNA slice product cleaved exactly in the predicted slicing site. Instead, the 3′-products located 20–60 nt downstream from the predicted slice site were observed. It is likely that sliced mRNA 3′-product is unstable and undergoes rapid degradation in tissues and organs studied. This supposition is in agreement with our results of slicing site detection using the 5′-RACE approach shown in Figure 5C. In addition to the 5′-RACE product of the expected length, we also observed shorter products derived from the intron region.
Assuming that the intron-containing RAP2.12 isoform is a target of miR319b.2, we predicted that its expression would change in the miRNA biogenesis mutants, miR319boe, and ΔmiR319b mutant plants when compared to wt plants. In the case of fully spliced RAP2.12 isoform, we did not expect any changes. qPCR analyses of the RAP2.12 two isoforms in the wt, hyl1-2, se-1, miR319boe, and ΔmiR319b mutant plants confirmed in general our predictions. No fluctuations were noted in the fully spliced RAP2.12 mRNA isoform level in any mutant background studied, while the level of the intron-containing RAP2.12 isoform was higher in the miRNA biogenesis mutant plants and in the ΔmiR319b mutant, and lower in the miR319boe mutant plants when compared to wt plants. However, we expected the expression of intron-retained RAP2.12 isoform in the ΔmiR319b mutant plants to be higher than in the wild type plants and at least the same as is in the case of hyl1-2 or se-1 mutants. The level of the RAP2.12 intron-retained isoform is higher than that in the wild type plants, but it does not reach the level of this isoform observed for miRNA biogenesis mutants. We anticipate that the low levels of miR319a.2 and miR319c.2 still present in the ΔmiR319b mutant plants may be responsible for targeting the RAP2.12 intron-retained isoform and down-regulating its expression, in comparison to hyl1-2 or se-1 mutants in which the biogenesis of all of these miRNAs is affected. This prediction is also supported by the very weak presence of the slicing product in the ΔmiR319b mutant plants that can be the result of miR319a.2 and/or miR319c.2 activities.
The miR319a/b/c-targeting of the TCP transcription factors was shown in jasmonate biosynthesis, which is involved in plant senescence (Schommer et al., 2008). RAP2.12 with one alternatively spliced mRNA isoform is potentially targeted by miR319b.2 and is also involved in plant senescence and plant response to osmotic stress (Papdi et al., 2008). Thus, it is feasible that both types of miRNAs: miR319a/b/c and miR319a.2/b.2/c.2 are involved in a similar physiological processes in plant development.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This study was supported financially with a grant awarded by the Ministry of Higher Education and Sciences, Poland (NN301301137); Ph.D. grant awarded by the Ministry of Higher Education and Sciences, Poland (NN301035839 to Lukasz Sobkowiak and Zofia Szweykowska-Kulinska); a grant for scientific research from the Dean of Biology Faculty, Adam Mickiewicz University, Poznan, Poland, and European Funds, Human Capital Operation Program 8.2.2, Poznan (to Lukasz Sobkowiak). We thank Prof. Ewa Czarnecka-Verner for a critical review of this manuscript and Lance Verner for proof reading.
Addo-Quaye, C., Snyder, J. A., Park, Y. B., Li, Y. F., Sunkar, R., and Axtell, M. J. (2009). Sliced microRNA targets and precise loop-first processing of MIR319 hairpins revealed by analysis of the Physcomitrella patens degradome. RNA 15, 2112–2121.
Bologna, N. G., Mateos, J. L., Bresso, E. G., and Palatnik, J. F. (2009). A loop-to-base processing mechanism undrlies the biogenesis of plant microRNAs miR319 and miR159. EMBO J. 28, 3646–3656.
Du, P., Wu, J., Zhang, J., Zhao, S., Zheng, H., Gao, G., Wei, L., and Li, Y. (2011). Viral infection induces expression of novel phased microRNAs from conserved cellular microRNA precursors. PLoS Pathog. 7, e1002176. doi: 10.1371/journal.ppat.1002176
Feijie, W., and Yuke, H. (2007). The N-terminal double-stranded RNA binding domains of Arabidopsis HYPONASTIC LEAVES 1 are sufficient for pre-microRNA processing. Plant Cell 19, 914–925.
Han, M. H., Goud, S., Song, L., and Fedoroff, N. (2004). The Arabidopsis double-stranded RNA-binding protein HYL1 plays a role in microRNA-mediated gene regulation. Proc. Natl. Acad. Sci. U.S.A. 101, 1093–1098.
Hsieh, L. C., Lin, S. I., Kuo, H. F., and Chiou, T. J. (2010). Abundance of tRNA-derived small RNAs in phosphate-starved Arabidopsis roots. Plant Signal. Behav. 5, 537–539.
Hu, Q., Hollunder, J., Niehl, A., Korner, C. J., Gereige, D., Windels, D., Arnold, A., Kuiper, M., Vazquez, F., Pooggin, M., and Heinlein, M. (2011). Speciefic impact of tobamovirus infection on the Arabidopsis small RNA profile. PLoS ONE 6, e19549. doi: 10.1371/journal.pone.0019549
Huang, Y., Ji, L., Huang, Q., Vassylyev, D. G., Chen, X., and Ma, J. B. (2009). Structural insights into mechanisms of the small RNA methyltransferase HEN1. Nature 461, 823–827.
Kim, S., Yang, J. Y., Xu, J., Jang, I. C., Prigge, M. J., and Chua, N. H. (2008). Two cap-binding proteins CBP20 and CBP80 are involved in processing primary microRNAs. Plant Cell Physiol. 49, 1634–1644.
Kozomara, A., and Griffiths-Jones, S. (2011). miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 39, D152–D157.
Kurihara, Y., and Watanabe, Y. (2004). Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc. Natl. Acad. Sci. U.S.A. 101, 12753–12758.
Laubinger, S., Sachsenberg, T., Zeller, G., Busch, W., Lohmann, J. U., Rätsch, G., and Weigel, D. (2008). Dual roles of the nuclear cap-binding complex and SERRATE in pre-mRNA splicing and microRNA processing in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 105, 8795–8800.
Li, H., and Durbin, R. (2010). Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595.
Li, Y., Li, C., Ding, G., and Jin, Y. (2011). Evolution of MIR159/319 microRNA genes and their post-transcriptional regulatory link to siRNA pathways. BMC Evol. Biol. 11, 122. doi: 10.1186/1471-2148-11-122
Llave, C., Xie, Z., Kasschau, K. D., and Carrington, J. C. (2002). Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297, 2053–2056.
Lobbes, D., Rallapalli, G., Schmidt, D. D., Martin, C., and Clarke, J. (2006). SERRATE: a new player on the plant microRNA scene. EMBO Rep. 7, 1052–1058.
Nag, A., King, S., and Jack, T. (2009). miR319a targeting of TCP4 is critical for petal growth and development in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 106, 22534–22539.
Palatnik, J. F., Allen, E., Wu, X., Schommer, C., Schwab, R., Carrington, J. C., and Weigel, D. (2003). Control of leaf morphogenesis by microRNAs. Nature 18, 257–263.
Palatnik, J. F., Wollmann, H., Schommer, C., Schwab, R., Boisbouvier, J., Rodriguez, R., Warthmann, N., Allen, E., Dezulian, T., Huson, D., Carrington, J. C., and Weigel, D. (2007). Sequence and expression differences underlie functional specialization of Arabidopsis microRNAs miR159 and miR319. Dev. Cell 13, 115–125.
Pant, B. D., Musialak-Lange, M., Nuc, P., May, P., Buhtz, A., Kehr, J., Walther, D., and Scheible, W. R. (2009). Identification of nutrient-responsive Arabidopsis and rapeseed microRNAs by comprehensive real-time polymerase chain reaction profiling and small RNA sequencing. Plant Physiol. 150, 1541–1555.
Papdi, C., Abraham, E., Joseph, M. P., Popescu, C., Koncz, C., and Szabados, L. (2008). Functional identification of Arabidopsis stress regulatory genes using the controlled cDNA overexpression system. Plant Physiol. 147, 528–542.
Park, M. Y., Wu, G., Gonzalez-Sulser, A., Vaucheret, H., and Poethig, R. S. (2005). Nuclear processing and export of microRNAs in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 102, 3691–3696.
Park, W., Li, J., Song, R., Messing, J., and Chen, X. (2002). Carpel factory, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol. 12, 1484–1495.
Reinhart, B. J., Weinstein, E. G., Rhoades, M. W., Bartel, B., and Bartel, D. P. (2002). MicroRNAs in plants. Genes Dev. 16, 1616–1626.
Reyes, J. L., and Chua, N. H. (2007). ABA induction of miR159 controls transcripts levels of two MYB factors during Arabidopsis seed germination. Plant J. 49, 592–606.
Schommer, C., Palatnik, J. F., Aggarwal, P., Chételat, A., Cubas, P., Farmer, E. E., Nath, U., and Weigel, D. (2008). Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol. 6, e230. doi: 10.1371/journal.pbio.0060230
Sobala, A., and Hutvagner, G. (2011). Transfer RNA-derived fragments: origins, processing, and functions. Wiley Interdiscip. Rev. RNA 2, 853–862.
Szarzynska, B., Sobkowiak, L., Pant, B. D., Balazadeh, S., Scheible, W. R., Mueller-Roeber, B., Jarmolowski, A., and Szweykowska-Kulinska, Z. (2009). Gene structures and processing of Arabidopsis thaliana HYL1-dependent pri-miRNAs. Nucleic Acids Res. 37, 3083–3093.
Taft, R. J., Glazov, E. A., Lassmann, T., Hayashizaki, Y., Carninci, P., and Mattick, J. S. (2009). Small RNAs derived from snoRNAs. RNA 15, 1233–1240.
Valen, E., Preker, P., Andersen, P. R., Zhao, X., Chen, Y., Ender, C., Dueck, A., Meister, G., Sandelin, A., and Jensen, T. H. (2011). Biogenic mechanisms and utilization of small RNAs derived from human protein-coding genes. Nat. Struct. Mol. Biol. 18, 1075–1082.
Vazquez, F., Blevins, T., Ailhas, J., Boller, T., and Meins, F. Jr. (2008). Evolution of Arabidopsis MIR genes generates novel microRNA classes. Nucleic Acids Res. 36, 6429–6438.
Vazquez, F., Gasciolli, V., Crété, P., and Vaucheret, H. (2004). The nuclear dsRNA binding protein HYL1 is required for microRNA accumulation and plant development, but not posttranscriptional transgene silencing. Curr. Biol. 14, 346–351.
Weigel, D., Ahn, J. F., Blazquez, M. A., Borevitz, J. O., Christensen, S. K., Fankhauser, C., Ferrandiz, C., Karadailsky, I., Malancharuvil, E. J., Neff, M. M., Nguyen, J. T., Sato, S., Wang, Z. Y., Xia, Y., Dixon, R. A., Harrison, M. J., Lamb, C. J., Yanofsky, M. F., and Chory, J. (2000). Activation tagging in Arabidopsis. Plant Physiol. 122, 1003–1014.
Yang, L., Liu, Z., Lu, F., Dong, A., and Huang, H. (2006a). SERRATE is a novel nuclear regulator in primary microRNA processing in Arabidopsis. Plant J. 47, 841–850.
Yang, Z., Ebright, Y. W., Yu, B., and Chen, X. (2006b). HEN1 recognizes 21-24 nt small RNA duplexes and deposits a methyl group onto the 2′ OH of the 3′ terminal nucleotide. Nucleic Acids Res. 34, 667–675.
Yu, B., Bi, L., Zheng, B., Ji, L., Chevalier, D., Agarwal, M., Ramachandran, V., Li, W., Lagrange, T., Walker, J. C., and Chen, X. (2008). The FHA domain proteins DAWDLE in Arabidopsis and SNIP1 in humans act in small RNA biogenesis. Proc. Natl. Acad. Sci. U.S.A. 105, 10073–10078.
Zhang, W., Gao, S., Zhou, X., Xia, J., Chellappan, P., Zhou, X., Zhang, X., and Jin, H. (2010). Multiple distinct small RNAs originate from the same micro RNA precursors. Genome Biol. 11, R81.
Zhou, H., Zhang, Q., Zhang, J., Ni, F., Liu, C., and Qi, Y. (2010). DNA methylation mediated by a micro RNA pathway. Mol. Cell 38, 465–475.
List of all sRNA sequences which match the miR319 primary transcripts
# pri-miR_id start:end_position strand raw_counts miR_id
ath-MIR319a 48:69 + 8 miR319a.2*
ath-MIR319a 110:132 + 16 miR319a.2
ath-MIR319a 152:171 + 6
ath-MIR319a 154:169 + 5
ath-MIR319a 154:174 + 90 miR319a
ath-MIR319a 155:173 + 5
ath-MIR319b 3:21 + 5
ath-MIR319b 3:23 + 213 miR319b*
ath-MIR319b 4:22 + 10
ath-MIR319b 4:23 + 35
ath-MIR319b 4:24 + 6
ath-MIR319b 48:69 + 8 miR319b.2*
ath-MIR319b 108:122 + 8
ath-MIR319b 108:123 + 7
ath-MIR319b 108:124 + 10
ath-MIR319b 108:125 + 22
ath-MIR319b 108:126 + 21
ath-MIR319b 108:127 + 73
ath-MIR319b 108:128 + 538 miR319b.2
ath-MIR319b 108:129 + 132
ath-MIR319b 108:130 + 98
ath-MIR319b 108:131 + 11
ath-MIR319b 109:129 + 8
ath-MIR319b 152:167 + 5
ath-MIR319b 152:172 + 90 miR319b
ath-MIR319b 153:171 + 5
ath-MIR319c 126:146 + 16 miR319c.2
ath-MIR319c 170:185 + 5
ath-MIR319c 170:190 + 31 miR319c
Keywords: miRNA, pri-miRNA, RAP2.12, splicing, gene expression
Citation: Sobkowiak L, Karlowski W, Jarmolowski A and Szweykowska-Kulinska Z (2012) Non-canonical processing of Arabidopsis pri-miR319a/b/c generates additional microRNAs to target one RAP2.12 mRNA isoform. Front. Plant Sci. 3:46. doi: 10.3389/fpls.2012.00046
Received: 21 November 2011; Accepted: 25 February 2012;
Published online: 19 March 2012.
Edited by:
Bernie Carroll, The University of Queensland, AustraliaReviewed by:
Hui Wang, Centre for Ecology and Hydrology, Natural Environmental Research Council, UKCopyright: © 2012 Sobkowiak, Karlowski, Jarmolowski and Szweykowska-Kulinska. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Zofia Szweykowska-Kulinska, Department of Gene Expression, Faculty of Biology, Institute of Molecular Biology and Biotechnology, Adam Mickiewicz University, Umultowska 89, 61-614 Poznan, Poland. e-mail:em9mc3p3ZXlAYW11LmVkdS5wbA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.