- 1 Department of Plant Biology, Cornell University, Ithaca, NY, USA
- 2 Institute of Biology, University of Kassel, Kassel, Germany
Plant morphogenesis (the process whereby form develops) requires signal cross-talking among all levels of organization to coordinate the operation of metabolic and genomic subsystems operating in a larger network of subsystems. Each subsystem can be rendered as a logic circuit supervising the operation of one or more signal-activated system. This approach simplifies complex morphogenetic phenomena and allows for their aggregation into diagrams of progressively larger networks. This technique is illustrated here by rendering two logic circuits and signal-activated subsystems, one for auxin (IAA) polar/lateral intercellular transport and another for IAA-mediated cell wall loosening. For each of these phenomena, a circuit/subsystem diagram highlights missing components (either in the logic circuit or in the subsystem it supervises) that must be identified experimentally if each of these basic plant phenomena is to be fully understood. We also illustrate the “subsystem incompleteness theorem,” which states that no subsystem is operationally self-sufficient. Indeed, a whole-organism perspective is required to understand even the most simple morphogenetic process, because, when isolated, every biological signal-activated subsystem is morphogenetically ineffective.
Introduction
Three decades ago, the genotype-phenotype relationship was described as follows: “Concerning our understanding of morphological evolution, the consensus view emerged that present knowledge about genome function is not sufficient to make a large direct contribution. We do not know the mechanisms by which gene activity affects the development of an individual animal. Therefore, we cannot come to useful specific conclusions regarding genomic correlates of evolutionary change at the morphological level” (Dawid et al., 1982, pp. 19–20). This perspective highlights a substantial gap between the then current understanding of how the eukaryotic genome functions and how these functions translate into developmental events at the cellular, organ, or organismic levels of organization (i.e., the genotype-to-phenotype relationship). The focus of this paper is to evaluate the extent to which this gap has been shortened in our understanding of plant development, which has experienced tremendous progress in identifying the dedicated genes (e.g., transcription factors) and regulatory systems (e.g., hormone signal perception and transport mechanisms) that play critical roles in embryogenesis and subsequent growth and development.
Despite considerable progress at both ends of the “genome-to-morphology” pathway, we believe that the primary literature justifies the perception that the gap between understanding how the genome functions and how this functionality translates into morphogenesis remains large. For example, despite the elegant work that has revealed the genomics and physiology of perhaps the most important and extensively researched plant hormone auxin (i.e., indole-3-acetic acid, IAA), not a single sequence of events has been discovered that links IAA-response genes directly to a discrete morphogenetic event. Even as basic a phenomenon as cell wall loosening and expansion remains poorly understood despite the identification of the signal transduction pathways IAA-response genes/proteins, and the phenomenology of cell wall expansion (Liepman et al., 2010; Deng et al., 2012).
Here, we review the genotype-to-phenotype relationship using IAA as a model plant hormone in the general context of a “system biology approach” (Pu and Brady, 2010; Yin and Struik, 2010) and in the particular context of the mathematics of logic circuits and the signal-activated subsystems they operate (Stein, 1998; Niklas, 2003). Although other plant hormones are of similar great importance (Kim and Wang, 2010), IAA is selected because it is involved in virtually every phase of plant development (Woodward and Bartel, 2005; Kutschera and Niklas, 2007, 2009) and because it has been extensively studied for over 100 years, notably in grass coleoptiles (Figure 1A). A “systems biology approach” is adopted because it theoretically permits us to integrate, model, and analyze the interactions among all of the components of a complex living system – particularly in the case of feedback loops, which figure prominently in the biology of auxin (as noted by Benjamins and Scheres, 2008).
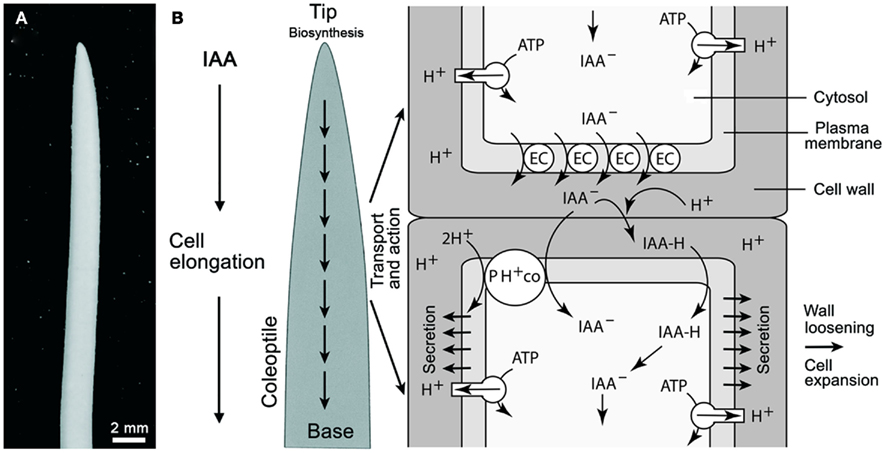
Figure 1. Oat (Avena sativa) coleoptile (A) and schematic rendering of polar auxin (IAA) transport according to the chemiosmotic model (B). Auxin influx can result from the passive diffusion of the protonated form of IAA (IAAH across the plasma membrane) or by active transport of the dissociated form (IAA−) by a 2H+–IAA− permease symporter (denoted as PH+ co). The passive diffusion of IAA is dependent on the pH of the cell wall. Plasma membrane H+-ATPases normally maintain the cell wall at approximately pH 5.0 via protonation. Polar transport is governed by the efflux of IAA at the base of cells via auxin anion efflux carriers (EC) that transport IAA− out of the cell (driven by a negative membrane potential). The PIN family of proteins have been identified as the EC. See Figure 5 for a logic circuit rendering of IAA polar transport. In addition, the mode of IAA action is illustrated.
This paper is organized into three main sections. In the first section, we present IAA as a model signaling molecule and we review our current understanding of its role in transcription regulation, how it is transported from one cell to another, and finally how it participates in cell wall loosening and cell expansion. In the second section, we construct logic circuits and system-activated systems for the transport of IAA and for IAA-mediated cell wall loosening. In the third and last section, we present the “subsystem incompleteness theorem,” which states that the operation of any signal-activated system cannot be truly understood unless its operation is fully integrated with the operation of all other signal-activated subsystems. We also discuss why the transcription factor paradigm is necessary but insufficient to fully explicate morphogenesis and development, even at the cellular level.
Auxin: A Model Signaling Molecule
The capacity to synthesize phytohormones (auxins, gibberellins, cytokinines, etylene, abscisic acid, and brassinosteroids) is phylogenetically widely distributed. Auxins have been identified in the apical cells of the filamentous brown alga Ectocarpus siliculosis (Le Bail et al., 2010), whereas all of the first five of the principal plant hormones are reported to occur in unicellular photoautotrophs, fungi, and bacteria, including the cyanobacteria (Spaepen et al., 2007; Ross and Reid, 2010). A tryptophan-dependent pathway using indole-3-acetamide (IAM) as an intermediate to synthesize the most common auxin, indole-3-acetic acid (IAA), has been identified in methylobacteria (Hornschuh et al., 2006) and in the pathogenic bacterium Agrobacterium tumefaciens. The IAA synthesized by this and other pathogenic bacteria results in abnormal morphological changes in host plants (Spaepen et al., 2007). Auxin produced by epiphytic bacteria appears to be of importance for symbiotic plant–microbe interactions (Kutschera, 2007; Spaepen et al., 2007), but more work is required to further support this hypothesis. It is unclear however whether or not all of these organisms require endogenous levels of IAA for their normal development.
The molecular biology and physiology of IAA produced endogenously by land plants (embryophytes) has received the most attention. No viable land plant mutant lacking this hormone entirely has been found, suggesting that it is required continuously at some concentration (Zhao, 2010). For this reason, our current state of knowledge about IAA will be used to illustrate the progress made in understanding plant morphogenesis. This emphasis is justified for three reasons: (1) endogenous IAA is involved in a vast range of developmental processes (e.g., cell wall loosening and expansion, phototropism and gravitropism, shoot apical meristem dominance, lateral and adventitious root formation, leaf abscission, floral bud and fruit development, and the differentiation of the vascular tissues), (2) IAA biosynthesis, directional transport, signal perception, and transduction, and cell- or tissue-specific responses triggered by auxin have been extensively studied at the molecular, cellular, and organismic levels, and (3) the interrelations among these hierarchical levels involve numerous protein complexes operating at different developmental levels that serve as a paradigm for plant development and evolution in general (Woodward and Bartel, 2005; Tao et al., 2008).
Auxin Polar Transport
A general scheme of IAA transport and action in the grass coleoptile is shown in Figure 1 (for a more detailed rendering of IAA flux, see Figure 5). A number of reports have shown that the rate of IAA transport from the tip to the base of the organ is in the order of ca. 10 mm/h. There is no evidence for a transfer of auxin from the base to the tip, i.e., the transport occurs only one direction (Kutschera, 2003). Auxin influx occurs either by the passive diffusion of the protonated form (IAAH) across the cell membrane, or by the active transport of the dissociated, anionic form (IAA−) by a permease 2H+-IAA co-transporter, as for example AUX1, which appears to be distributed uniformly in the cell membranes of Arabidopsis root cells (Bennett et al., 1996). Once within a cell, IAA is transported in a polar or in a non-polar manner (Figure 1). Auxin polar transport involves an IAA-influx carrier protein encoded by the AUX1 gene, whereas IAA efflux involves the activity of at least two membrane-bound and -associated proteins (Muday and DeLong, 2001). One of these is a trans-membrane transport protein encoded by members of the PIN gene family that encode for PIN proteins with ten trans-membrane segments and a large hydrophilic loop (Palme and Gälweiler, 1999; Friml et al., 2002). The second category of proteins called IAA-inhibitor-binding proteins perform a regulatory function in response to endogenous, naturally occurring substances, such as flavonoids. These proteins, which were originally described as NPA-binding proteins, are ATP-dependent transporters belonging to the phosphoglycoprotein B subclass of the large superfamily of ATP binding cassette (ABC) integral trans-membrane transporter proteins. The ABCB proteins function as ATP-dependent amphipathic anion carriers involved in auxin efflux (see Geisler and Murphy, 2006). One class of PGPs represented by AtPGP1 catalyze auxin export, while another class with at least one member, AtPGP4, appears to function in auxin import (Luschnig, 2002; Geisler and Murphy, 2006).
Evidence that dephosphorylation and phosphorylation regulate the activity of IAA efflux carrier proteins comes from a variety of sources and suggests that these mechanisms can be used to regulate how and where IAA can be transported to various plant cell- or tissue-types. For example, the Arabidopsis roots curl in NPA1 gene (RCN1) encodes a regulatory subunit of protein phosphatase 2A (PP2A) that is expressed in seedling root tips, lateral root primordia, the pericycle, and the stele. The rcn1 mutant exhibits reduced PP2A activity and defects in IAA-mediated responses involving anisotropic cell expansion (Muday and DeLong, 2001). Phosphorylation mediated by PID kinase and dephosphorylation regulated by RCN1 are known to antagonistically regulate auxin transport and gravity responses in roots. However, PID kinase (which is controlled by IAA as are PIN and ABCB) appears to play a limited role in root development, since loss of PID activity has been shown to alter auxin transport and gravitropism without causing any discernable change on cell polarity in roots (Sukumar et al., 2009). The rcn1 mutant also exhibits a near twofold increase in IAA basipetal transport. Treatment of control plants with phosphatase inhibitors, such as cantharidin, produces the rcn1 phenotype. These lines of evidence indicate that PP2A likely regulates the IAA-polar-efflux carrier protein complex but that the regulatory effects of PP2A on basipetal and acropetal IAA transport are different. The activities of one or more protein kinases may counterbalance the effects of PP2A. Indeed, the results of inhibitor studies suggest that genes encoding several kinases may play a key role in regulating polar IAA transport, e.g., the treatment of tobacco cells with staurosporine or K252a, which are both broad spectrum kinase inhibitors, rapidly reduces IAA efflux without affecting IAA-influx. It must be noted, however, that the non-enzymatic, pH-driven influx of neutral IAA may account for the latter.
Finally, it must be noted that auxin transport generates transcriptional auxin response gradients, but these gradients do not invariably translate into corresponding patterns of auxin response gene expression. For example, the essential root meristem growth regulator gene BREVIS RADIX (BRX; Scacchi et al., 2010) is not spatially expressed in the same way as the IAA gradient in Arabidopsis roots. Santuari et al. (2011) combined a cell-level root meristem model with an overlapping differential endocytosis pattern with a positive auto-regulatory feedback loop via plasma membrane-to-nuclear transfer of BRX. This model successfully mimicked normal root development. Since BRX is required for the expression of some auxin responses genes, Santuari et al. (2011) suggest that organ-specific patterns of endocytosis are required to provide specific positional information to modulate auxin responses.
Auxin Signaling and Transcriptional Regulation
Once auxin enters a cell it is capable of modulating the transcription of a large number of genes (Key, 1989). The most intensively characterized auxin response cascade involves proteins transcribed by three genes: auxin/indole-3-acetic acid (AUX–IAA), auxin response factor (ARF), and transport inhibitor response1/auxin signaling f-box protein 1–3 (TIR1–AFB1–3). At low IAA concentrations, AUX–IAA proteins form dimers with ARF transcription factors bond to consensus TGTCTC auxin-repressive promoter elements (Lau et al., 2009). These AUX–IAA/ARF protein complexes block the transcription of early auxin response genes. At higher concentrations, auxin interacts with TIR1–AFB1–3 receptors, which are an integral part of the SKP1-CULLIN-F-BOX PROTEIN (SCF) complex, and targets bound and ubiquitinated AUX–IAA for degradation by the 26S proteosome, thereby activating ARF to activate early auxin gene expression. Early auxin gene expression obtains a variety of cell- or tissue-specific auxin responses, along with a negative feedback loop due to the expression of AUX–IAA (Figure 2). Likewise, rapid auxin effects related to the Auxin binding protein 1 (ABP 1) Golgi-secretion have been discovered at the level of the proteome in epidermal cells of coleoptile sections (Sauer and Kleine-Vehn, 2011; Deng et al., 2012).

Figure 2. Schematic view of the auxin response cascade. This model involves proteins transcribed by AUX–IAA, ARF, and TIR1–AFB1–3. At high concentrations, auxin interacts with TIR1–AFB1–3 receptors, which are part of the SCF complex, and targets bound and ubiquitinated AUX–IAA for degradation by the 26S proteosome, thereby activating ARF to de-repress early auxin gene expression. (At low IAA concentrations, AUX–IAA proteins form dimers with ARF transcription factors bond to consensus TGTCTC auxin-repressive promoter elements. These AUX–IAA/ARF protein complexes block the transcription of early auxin response genes.) Along with a negative feedback loop due to the expression of AUX–IAA, early auxin gene expression obtains a variety of cell- or tissue-specific auxin responses one among which is the production of V-type H+-ATPases that can promote the Golgi-mediated secretion of wall proteins and instigate cell wall loosing when activated by IAA and auxin binding proteins (ABP; see Figure 6 for a logic circuit rendering of auxin-mediated cell wall loosening).
Given the large number of developmental processes in which auxin participates, it is reasonable to suppose that auxin-specific responses require mechanisms that evoke specific control of transcript or protein levels and the differential subsequent developmental interpretation of auxin concentrations and gradients by means of specific combinations of auxin signaling components. Unfortunately, very little is known about the transcriptional regulation of auxin-induced genes (Paponov et al., 2008). For example, the uncharacterized locus PARTIAL SUPPRESSOR OF AXR3–1 (PAX1) has been shown to positively regulate the expression of AUXIN RESISTANT3 (AXR3)–IAA17 and the expression of other AUX–IAAs (Tanimoto et al., 2007). However, it is uncertain whether PAX1 transcriptional regulation is the result of a direct mechanism or by means of a feedback loop involving AXR3–IAA17. Targeted ARF degradation could provide a mechanism for specific auxin responses in specific cells or tissues. For example, Salmon et al. (2008) have shown that the proteosome-dependent degradation of ARF1 does not invariably involve the SCF complex (Figure 2). Another possible mechanism for generating cell- or tissue-specific auxin responses is the recruitment of other proteins into the core AUX–IAA/ARF complex. Shin et al. (2007) have shown that MYB DOMAIN PROTEIN77 (MYB77) is a co-activator of ARF (via the C terminus) and interacts with IAA19 to promote auxin response gene expression in vitro. These authors have also shown that ARF7 and MYB77 interact synergistically based on the behavior of the lateral root phenotype of the arf7 and myb77 double mutant in vivo (Shin et al., 2007). It is possible therefore that ARF and MYB interact as co-promotors that can combinatorially and perhaps differentially regulate gene expression in different cells and tissues.
In summary, our mechanistic understanding of how PIN polarization is affected by developmental status and ambient environmental conditions is comparatively meager, albeit increasing rapidly. For example, recent computer and theoretic modeling of polar auxin transport consider ever more complex scenarios for extracellular (apoplastic) transport and feedback loops between auxin transport gradients and PIN protein localization. These models continue to provided deeper understanding into the transport of auxin in the multicellular plant body (reviewed by Wabnik et al., 2011).
Cell Wall Loosening and Cell Expansion
A permanent increase in cell size requires the plastification of the cell wall, which is mediated by IAA. Among monocots, this role has been extensively experimentally studied in terms of the coleoptile of the grass seedling. This sheath-like organ protects the enclosed primary leaf during growth of the shoot through the soil (Kutschera and Niklas, 2009). Figure 3A shows an etiolated maize seedling (Zea mays), and an excised coleoptile segment, 10 mm in length, from the sub-apical region of the shoot. Numerous studies have shown that, in etiolated maize coleoptiles, cell elongation is controlled by IAA supplied from the tip of the organ, and that the endogenous level of auxin corresponds to ca. 50% of the concentration that causes a maximum growth response in the excised segment (i.e., organ growth in vitro, Figure 3B; Kutschera, 1994, 2003).
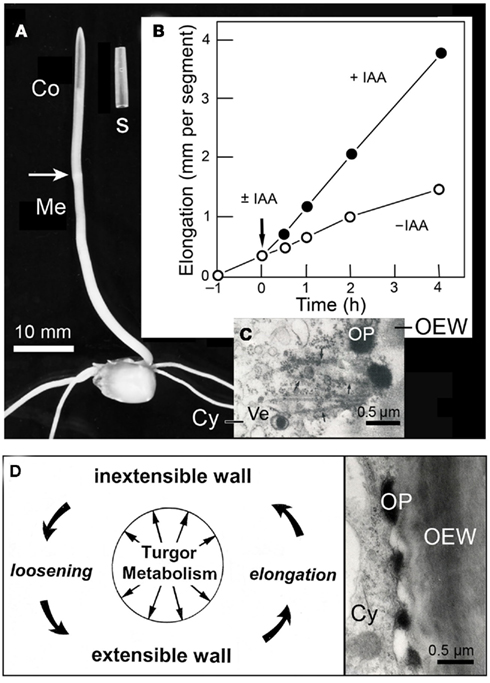
Figure 3. Auxin action in the maize (Zea mays) coleoptile. Photograph of a 4-day-old etiolated seedling and an excised segment, 10 mm in length (A). Growth response of maize coleoptile segments in the presence or absence of auxin (±IAA, 10 μM) (B). Transmission electron micrograph of the peripheral cytoplasm of an IAA-treated segment (after 1 h of auxin treatment) (C), and scheme of IAA-mediated loosening of the growth-controlling outer epidermal wall (D). Co, coleoptile, Cy, cytoplasm, OEW, outer epidermal wall, OP, osmiophilic nano-particle, Ve, Golgi-vesicle.
Biophysical studies reveal that IAA acts primarily by mechanically loosening the thick, growth-controlling outer wall of the outer epidermis, an “organ-sheath” that is maintained under tension via the turgor pressure of the thin-walled inner cells of the coleoptile (Kutschera et al., 1987; Kutschera and Niklas, 2007). Cytological analyses document that, in coleoptiles of maize, rye (Secale cereale), and oats (Avena sativa), IAA rapidly causes the appearance of osmiophilic nano-particles that are secreted via the Golgi system and are incorporated into the outer epidermal wall (Kutschera et al., 1987; Bergfeld et al., 1988; Kutschera and Edelmann, 2005; Deng et al., 2012). These conspicuous IAA-induced particles, which are composed of glycoproteins and fuse with the growing wall, are shown in Figures 3C,D. It has been postulated that these granules act as “wall loosening factors” in the coleoptile (Kutschera, 1994, 2003), but more work is required to validate the “protein secretion hypothesis of IAA action,” which has been supported by recent proteomic analyses (Kutschera et al., 2010; Deng et al., 2012).
In addition to IAA, the fungal phytotoxin fusicoccin (Fc) has been extensively used to elucidate the mechanism of cell elongation. In contrast to auxin, Fc causes wall loosening and organ elongation in coleoptile segments via the rapid secretion of protons (H+-ions) according to the “acid-growth theory of Fc action” (see Kutschera, 1994, 2006). This Fc (acid)-induced growth response is attributable to a non-enzymatic (or expansin-mediated) wall loosening process that has been analyzed in detail. The role of the much weaker and slower proton secretion response elicited by IAA is unclear, but it is likely that it acts to establish a pH-gradient between the cytoplasm (pH ∼ 6.0) and the wall space (pH ∼ 5.0 in IAA-treated segments). Upon addition of Fc, a wall pH of 3.5–4.0 is rapidly established that triggers acid-mediated, turgor-driven organ elongation (Kutschera, 1994, 2003, 2006).
A general scheme of auxin-regulated, turgor- and metabolism-dependent wall expansion resulting in organ growth is shown in Figure 3D (for a more detailed rendering of wall loosening, see Figure 6). It is known that turgor pressure and oxygen-dependent cellular respiration are pre-requisites for IAA action and growth. However, auxin does not promote cell elongation via an enhancement of turgor- or osmotic pressure. Likewise, in coleoptiles, the rate of oxygen uptake per gram of fresh mass (i.e., organ-specific cellular respiration) is not enhanced by IAA, but by Fc (Kutschera, 2003). Although we know many details about IAA action in grass coleoptiles, fewer data are available for the auxin response in the axial organs of dicotyledonous plants, such as sunflower hypocotyls (Kutschera and Niklas, 2007). We therefore have restricted our discussion of auxin action to the coleoptile of the Poaceae, which is a classic model system in this area of plant research.
Logic Circuits, Signal-Activation Systems, and Auxin
A synoptic treatment of logic circuits is beyond the scope of this paper (for classic references, see Halmos, 1963; Harrison, 1965; for an application to plant development, see Stein, 1998; for an application to gene networks, see Yant, 2012). However, at a very basic level, there are only two kinds: combinatorial circuits, in which the output signal depends exclusively on the near instantaneous value of the input signal, and sequential circuits, in which the output signal also depends on the history of previous inputs. Both types conceptualize a signaling pathway as an electrical circuit containing one or more switches. The “logic” of a circuit is the formal algorithm that describes the conditions (logical propositions) that dictate whether a signal passes through a circuit. Parallel and serial circuits exist. The former provides manifold responses to the same signal depending on instantaneous conditions, because parallel circuits allow an initial input signal to flow through two or more pathways, permitting two or more output signals at each terminus. Consider a parallel circuit constructed with three switches, A, B, and C. This simple signal pathway has eight possible combinatorial responses. More generally, the number of responses is given by 2N, where N is the number of switches. Responses coordinated by parallel logic circuits can achieve seemingly continuous variation in response to the passage of a single input signal if they contain even a modest number of switches (i.e., 2N = 10 = 1024), if some switches activate or suppress other switches in the circuit, if the circuit has two or more input signals, or if the output signals interact combinatorially. Also, if switches respond to more than one signal, the number of possible output signals is 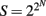 .
.
Although a complex logic circuit can be simplified mathematically, four caveats are self-evident when biological systems are approached in this way (Niklas, 2003). First, there is no a priori method to determine which among logically equivalent circuits is biologically real, i.e., over-simplification can produce false circuit diagrams. Second, incomplete signaling pathways may appear to “work” when diagrammed, i.e., missing components are not invariably obvious. Third, parallel logic circuits may obtain invariant output signals that give the appearance that input signals pass through serial switches, i.e., a bifurcating signal transduction pathway is more readily misdiagnosed than a serial pathway. Fourth, nothing in a logic circuit per se indicates when and how long a switch is turned on or off or how long a genomic or metabolic product lasts, i.e., the temporal components of signaling can be lost.
To be useful, logic circuits must be wedded to the subsystems they supervise. There are a variety of signal-activated subsystem configurations. However, the simplest is error-activated, that is, the output signal is used to modulate the input signal. This configuration has four essential components (see Harrison, 1965; Hill and Peterson, 1968): (1) a comparator to measure the difference (error) between the actual and the desired output of the subsystem, (2) an actuator/suppressor to convert the error-signal into an internal signal, (3) the actual machinery or assemblage that is controlled (the subsystem assembly), and (4) a feedback element to direct the immediate output signal of the assemblage back to the comparator (Figure 4A).
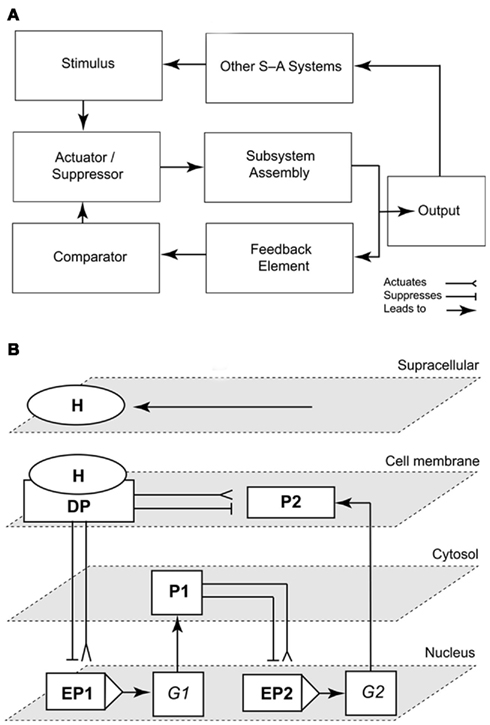
Figure 4. Representations of a simple signal-activated subsystem and the emergence of the “subsystem incompleteness theorem.” Scheme of a signal-activated (S-A) subsystem with four basic components (an actuator/suppressor, a subsystem assembly, a feedback element, and a comparator). The output signal (response to an input stimulus) feeds into other signal-activated (S-A) systems (A). Schematic illustrating the “Incompleteness theorem” for a circuit/subsystem consisting of a docking protein DP activated by a hormone H that mediates signaling to a cell membrane-bound protein P2, a cytoplasmic protein P1, and the enhancers/promoters (EP) of two genes G1–2(B). The network is structurally self-contained, but its operation depends on the delivery of the external signal (stimulus, H; adapted from Niklas, 2003).
Feedback is defined as that property of a closed-loop system that permits the comparison of the output signal (or some other variable controlled by the subsystem) to the input of the subsystem (or an input to some other internal component) so that the control action is some function of the input-to-output ratio. In many ways, the feedback element is the most important of the four components because it confers four characteristics: (1) an increased range of input signals over which the subsystem responds satisfactorily, (2) reduced sensitivity to variations in the output to input signal ratio, (3) reduced effects of non-linear distortions, and (4) a tendency toward initial oscillatory behavior. Negative and positive feedback loops exist and a single loop can serve in both capacities, especially in the case of a subsystem “hot-wired” by a sequential (history-dependent) logic circuit (for an interesting example, see Bhalla et al., 2002).
A Logic Circuit for IAA Polar Transport
A logic circuit for the polar and lateral transport of IAA is shown in Figure 5. This rendering indirectly addresses the manner in which carrier protein-membrane asymmetries for IAA efflux are established and maintained. Among asymmetric animal cell types (e.g., nerve cells), proteins characterizing different membrane domains are sorted by means of targeting Golgi-vesicle delivery. Attachment to the actin cytoskeleton, either directly or by means of protein complexes, maintains membrane asymmetry once achieved. This model may hold true for plants. PIN gene family proteins typically show asymmetric plasmalemma localization, whereas inhibitors of Golgi-vesicle secretion impede IAA transport. There is also some evidence that IAA-influx and -efflux carrier proteins are attached to the actin cytoskeleton. Lateral membrane asymmetries are suggested by recent genetic studies and implied by the Cholodny–Went hypothesis, i.e., lateral IAA transport across light- or gravity-stimulated tissues drives differential cell growth in volume. For example, mutations in genes encoding for IAA transport proteins (e.g., aux 1 and pin 2) obtain agravitropic phenotypes. Changes in the gravity vector produce asymmetric patterns of IAA-induced gene expression in the shoots and roots of transgenic plants expressing the GUS reporter under the control of IAA-response promoters. Also, some IAA transport inhibitors block gravitropic bending, have no evident affect on polar IAA transport, and evoke differential IAA-regulated gene expression. The existence of numerous PIN genes with distinct patterns of expression and PIN proteins with different patterns of membrane location indicate that polar and lateral IAA efflux involves different efflux carriers (Teale et al., 2006; Figure 5).
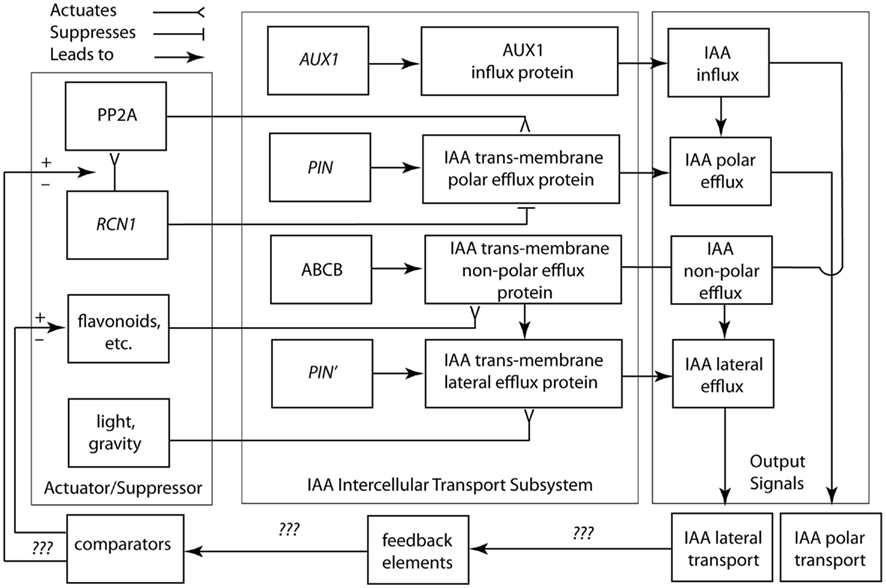
Figure 5. Circuit/subsystem diagram for IAA polar and lateral transport. This diagram is based on studies showing that IAA polar transport involves an IAA-influx protein carrier encoded by the AUX1 gene, whereas IAA efflux involves the activity of at least two membrane-bound and -associated proteins. One of these is a trans-membrane transport protein, which is encoded by members of the PIN gene family. The second are ABCB transporter proteins. See text for additional details. PP2A, protein phosphatase 2A; RCN1, Arabidopsis roots curl in NPA1 gene.
Based on this limited information, the suppressor/activator in the IAA polar transport system consists de minimis of IAA efflux protein inhibitors (e.g., flavonoids) and the RCN1 gene encoding for the regulatory subunit of PP2A. Details regarding the feedback loop and comparator in this system are largely unknown, although it appears that IAA is involved in its own regulation. Viable candidates for missing components include intracellular IAA concentrations, which may indirectly or directly regulate RCN1 gene transcription. Nevertheless, we are woefully ignorant of how the important morphogenetic system operates and we lack a clear understanding of the manner in which carrier protein-membrane asymmetries for IAA efflux are established and maintained. Among asymmetric animal cell types (e.g., nerve cells), proteins characterizing different membrane domains are sorted by means of targeting Golgi-vesicle delivery. Attachment to the actin cytoskeleton, either directly or by means of protein complexes, maintains membrane asymmetry once achieved. This model may hold true for plants. PIN gene family proteins typically show asymmetric plasma membrane localization, whereas inhibitors of Golgi-vesicle secretion impede IAA transport and auxin-mediated cell elongation (Kutschera, 2003). There is also evidence that IAA-influx and -efflux carrier proteins are attached to the actin cytoskeleton (Cox and Muday, 1994).
Lateral membrane asymmetries are suggested by recent genetic studies and implied by the Cholodny–Went hypothesis, i.e., lateral IAA transport across light- or gravity-stimulated tissues drives differential cell growth in volume. For example, mutations in genes encoding for IAA transport proteins (e.g., aux 1 and pin 2) obtain agravitropic phenotypes. Changes in the gravity vector produce asymmetric patterns of IAA-induced gene expression in the shoots and roots of transgenic plants expressing the GUS reporter under the control of IAA-response promoters. Also, some IAA transport inhibitors that block gravitropic bending, have no evident affect on polar IAA transport, and evoke differential IAA-regulated gene expression. The existence of numerous PIN genes with distinct patterns of expression and PIN proteins with different membrane locations indicate that polar and lateral IAA efflux involves a variety of efflux carriers.
The logic circuit presented in Figure 5 does not address in detail non-polar auxin transport, which is the principal translocation mechanism in the root, via the phloem (Aloni, 1995; Swarup et al., 2001). This mode of hormone transfer is passive and therefore does not require the direct expenditure of energy via active transport mechanisms. More important, this logic circuit does not entertain recent insights gained from empirical studies and theoretical models that consider PIN polarization both with and against auxin concentration gradients in which cells with low auxin concentrations manifest an “up-the-gradient” pattern of behavior, while cells with high auxin concentrations follow a “down-the-flow pattern” of behavior (see Wabnik et al., 2011).
A Logic Circuit for Cell Wall Loosening
A logic circuit for the regulation of cell wall loosening as mediated by IAA is shown in Figure 6. In this rendering, the plasmalemma-bound ABP 1–IAA conjugate is diagrammed as the actuator/suppressor switch for V-type ATPases. Once activated, the cell wall is loosened via the secretion of glycoproteins, and turgor pressure drives cell expansion. The “ABP 1–IAA switch” (Sauer and Kleine-Vehn, 2011) is also diagrammed to trigger delayed cytoplasmic and genomic responses involving the synthesis and delivery of other, not growth-limiting cell wall components, such as cellulose. This logic circuit diagram shows that sustained turgor maintenance via osmoregulation and cell wall loosening are required for continued cell expansion. The diagram also illustrates that the feedback loop and comparator for the output signal of the cell expansion machinery are unknown and must be sought experimentally. The IAA degradation, the down-regulation of solute concentrations, the synthesis of new cell wall-binding polymers, the re-orientation of cellulose microfibrils, the deposition of secondary wall layers, and the degradation of wall loosening enzymes are among the many viable candidates for these missing network components. However, it is clear that cell wall loosening involves numerous other suppressor/actuator subsystems, many of which remain poorly understood (Liepman et al., 2010; Deng et al., 2012).
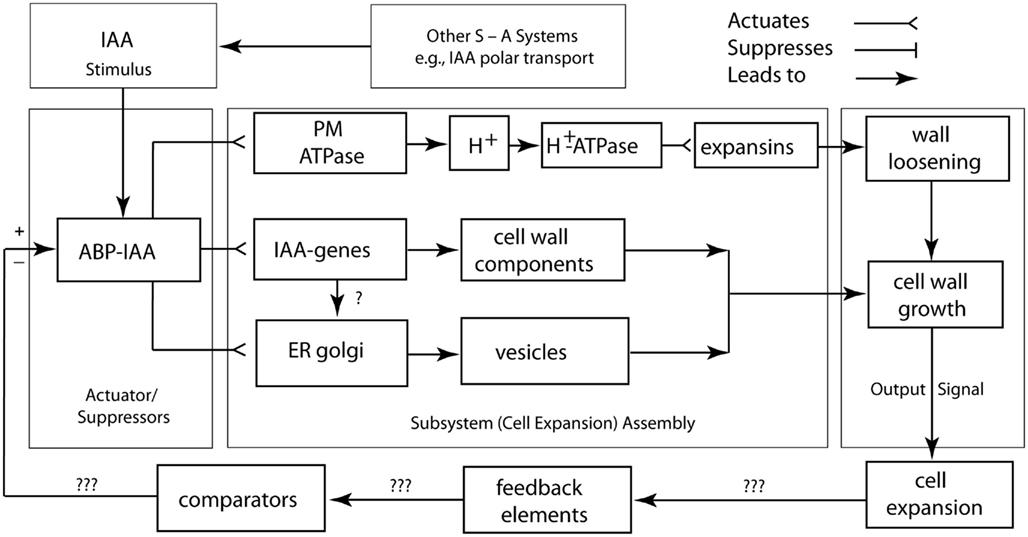
Figure 6. Circuit/subsystem diagram for cell expansion mediated by auxin. The IAA binding protein ABP 1 is implicated as the central plasma membrane-associated receptor and Golgi-mediated secretion of cell wall components are assumed to be responsible for the loosening of the growth-controlling outer epidermal wall. PM, plasma membrane, ER, endoplasmic reticulum, CWC, cell wall components. See text and Figure 3 for additional details.
The Subsystem Incompleteness Theoreom and the Transcription Factor Paradigm
The Subsystem Incompleteness Theorem
By definition, logic circuits and the biological subsystems they regulate are networked to other logic circuits and subsystems by shared circuits. This feature reduces the erratic behavior of the system as a whole. It allows the organism as a whole to achieve global stability by the synergistic feedback signaling of numerous components. Systems engineers call this “recursive combinatorial regulation.” Biologists sometimes refer to this as “homeostasis.” In systems engineering, recursive combinatorial regulation permits a network to repeatedly cycle through a programmed series of transformations. In biology, this kind of regulation permits “normal development.”
Importantly, no single actuator/suppressor switch exists in isolation because each subsystem circuit requires an activation or suppression signal. One subsystem must receive a signal and temporarily function as an epistatic actuator. When switched on or off, this subsystem sparks the operation of the entire network, suppressing or activating one or more of the networked subsystems. However, once the entire system is set into operation, no “master” switch exists.
This feature governs the working of any networked system and obtains what has been called the “subsystem incompleteness theorem,” which states that the operation of any biological subsystem cannot be fully diagnosed in isolation of the operations of all of the other subsystems to which it is networked (Niklas, 2003). The subsystem incompleteness theorem may be thought of a biological analog to Gödel’s incompleteness principles. Loosely, these principles argue that no algorithmic system can be used to prove its own self-consistency (Gödel, 1931). In an analogous manner, no developmental subsystem can be used to demonstrate how the subsystem fully operates because each subsystem is “wired” to all other subsystems.
To illustrate this conclusion, consider two nuclear genes (G1, G2), their enhancer-promoters (EP1, EP2), a cell membrane docking protein (DP) activated or suppressed by a hormone (H), and two regulatory proteins (P1, P2). For convenience, all the elements in this network may be diagrammed as suppression- or activation-switches (Figure 4B). At the structural level, this network is a self-contained system regardless of the presence of H. However, in terms of its operation, this network is suppressed or activated by H, which is delivered from an external source that is regulated by one or more other network systems. This simple subsystem mirrors auxin polar transport (the “delivery system”), auxin signaling and transcriptional regulation (in the role of H), and auxin-mediated cell wall loosening (as the subsystem that is activated).
The Transcription Factor Paradigm and Rules of the Game
The transcription factor paradigm is pervasive in the literature discussing developmental biology. It has implicated at least six molecular mechanisms for phenotypic evolution: (1) gene array duplication and subsequent sub-functionalization, (2) changes in the spatial expression patterns of pre-existing arrays, (3) homeo-domain protein sequence alterations, (4) modifications of DNA binding domains, (5) alterations in downstream regulated gene networks, and (6) changes in upstream regulatory genes (Niklas and Kutschera, 2009, 2010). Even when the mode of action and the spatial domain of gene expression remain unchanged, this paradigm suggests that modifications in the interactions between regulatory and downstream target genes participate to evoke significant phenotypic evolutionary change.
By the same token, a few general plant developmental “rules” have emerged largely using maize (Zea mays) and Thale cress (Arabidopsis thaliana) as model organisms (see Feuillet and Eversole, 2009; Koornneef and Meinke, 2010): (1) dedicated gene expression determines cell, tissue, and organ identity, (2) cell-fate is determined by position and not by clonal history, (3) developmental and signal transduction pathways are often controlled by very large gene networks, and (4) development is modulated by cell-to-cell signaling. Likewise, molecular phylogenetic analyses across a broad spectrum of plant lineages and clades reveal that a limited number of genomic/developmental subsystems or “modules” (many of which are extremely ancient) have been re-deployed in ways that are responsible for major morphological transformations (Niklas and Kutschera, 2009, 2010).
Concluding Remarks
Our review of the available literature leads us to conclude that the perspective offered by the transcription factor paradigm is incomplete. We find substantial evidence that, in complex multicellular systems such as seed plants (Figures 1A and 3A), a “system biology approach” is necessary to understand the genotype-to-phenotype relationship. It is becoming increasingly evident that we must understand how developmental modules communicate with one another and become operationally integrated as well as understanding how these modules operate internally. This depth of comprehension remains a challenging and exciting endeavor – one that has been a focus in the literature devoted to theoretical developmental biology, but one that has been, in our opinion, largely absent in most attempts to synthesize the empirical data. In the final analysis, the transcription factor paradigm must, at the very least, be modified to appreciate that development is not the simple result of “gene-to-phenotype mapping” but rather a manifestation of very complex signal cross-talking among numerous subsystems that collectively operate as a single, integrated living system (i.e., the organism).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank three anonymous reviewers for their helpful and insightful comments. Part of this work was carried out in the Department of Plant Biology, Carnegie Institution for Science, Stanford, CA 94305, USA. We thank Prof. Winslow R. Briggs for the provision of lab space and consultation. The collaboration of the authors was supported by the Alexander von Humboldt-Stiftung (Bonn, Germany; AvH-Fellowship Stanford 2010/2011 to UK).
References
Aloni, R. (1995). “The induction of vascular tissue by auxin and cytokinins,” in Plant Hormones and Their Role in Plant Development, 2nd Edn, ed. P. J. Davies (Dordrecht: Kluwer), 531–546.
Benjamins, R., and Scheres, B. (2008). Auxin: the looping star in plant development. Annu. Rev. Plant Biol. 59, 443–465.
Bennett, M. J., Marchand, A., Green, H. G., May, S. T., Ward, S. P., Millner, P. A., Walker, A. S., Schultz, B., and Feldmann, K. A. (1996). Arabidopsis AUX1 gene: a permease-like regulator of root gravitropism. Science 273, 948–950.
Bergfeld, R., Speth, V., and Schopfer, P. (1988). Reorientation of microfibrils and microtubules at the outer epidermal wall of maize coleoptiles during auxin-mediated growth. Bot. Acta 101, 57–67.
Bhalla, U. S., Ram, P. T., and Iyengar, R. (2002). MAP kinase phosphatase as a locus of flexibility in a mitogen-activated protein kinase signaling network. Science 297, 1018–1023.
Cox, D. N., and Muday, G. K. (1994). NPA binding activity is peripheral to the plasma membrane and is associated with the cytoskeleton. Plant Cell 6, 1941–1953.
Dawid, I., Britten, R. J., Davidson, E. H., Dover, G. A., Gallwitz, D. F., Garcia-Bellido, A., Kafatos, F. C., Kauffman, S. A., Moritz, K., Ohno, S., Schmidtke, J., and Schütz, G. (1982). “Adaptive aspects of development group report,” in Evolution and Development, ed. J. Bonner (Berlin: Springer-Verlag), 19–39.
Deng, Z., Xu, S., Chalkley, R. J., Oses-Prieto, J. A., Burlingame, A. L., Wang, Z.-Y., and Kutschera, U. (2012). Rapid auxin-mediated changes in the proteome of the epidermal cells in rye coleoptiles: implications for the initiation of growth. Plant Biol. doi: 10.111/j.1438-8677.2011.00513.x
Friml, J., Wi′sniewska, J., Benková, E., Mendgen, K., and Palme, K. (2002). Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature 415, 806–809.
Geisler, M., and Murphy, A. S. (2006). The ABC of auxin transport: the role of p-glycoproteins in plant development. FEBS Lett. 580, 1094–1102.
Gödel, K. (1931). Über formal unentscheidbare Sätze der Principia Mathematica und verwandter Systeme, I. Monatshefte für Mathematik und Physik 38, 173–198.
Hill, F. J., and Peterson, G. R. (1968). Introduction to Switching Theory and Logical Design. New York, NY: John Wiley.
Hornschuh, M., Grotha, R., and Kutschera, U. (2006). Moss-associated methylobacteria as phytosymbionts: an experimental study. Naturwissenschaften 93, 480–486.
Kim, T. W., and Wang, Z. Y. (2010). Brassinosteroid signal transduction from receptor kinase to transcription factors. Annu. Rev. Plant Biol. 61, 681–704.
Koornneef, M., and Meinke, D. (2010). The development of Arabidopsis as a model plant. Plant J. 61, 909–921.
Kutschera, U. (2003). Auxin-induced cell elongation in grass coleoptiles: a phytohormone in action. Curr. Top. Plant Biol. 4, 27–46.
Kutschera, U. (2007). Plant-associated methylobacteria as co-evolved phytosymbionts: a hypothesis. Plant Signal. Behav. 2, 74–78.
Kutschera, U., Bergfeld, R., and Schopfer, P. (1987). Cooperation of epidermis and inner tissues in auxin-mediated growth of maize coleoptiles. Planta 170, 168–180.
Kutschera, U., Deng, Z., Oses-Prieto, J. A., Burlingame, A. L., and Wang, Z.-Y. (2010). Cessation of coleoptile elongation and loss of auxin sensitivity in developing rye seedlings. A quantitative proteomic analysis. Plant Signal. Behav. 5, 509–517.
Kutschera, U., and Edelmann, H. G. (2005). Osmiophilic nanoparticles in epidermal cells of grass coleoptiles: implications for growth and gravitropism. Recent Res. Dev. Plant Sci. 3, 1–14.
Kutschera, U., and Niklas, K. J. (2007). The epidermal-growth-control theory of stem elongation: an old and a new perspective. J. Plant Physiol. 164, 1395–1409.
Kutschera, U., and Niklas, K. J. (2009). Evolutionary plant physiology: Charles Darwin’s forgotten synthesis. Naturwissenschaften 96, 1339–1354.
Lau, S., Shao, N., Bock, R., Jürgens, G., and De Smet, I. (2009). Auxin signaling in algal lineages: fact or myth? Trends Plant Sci. 14, 1360–1385.
Le Bail, A., Billoud, B., Kowalczyk, N., Kowalczyk, M., Gicquel, M., Le Panse, S., Stewart, S., Scornet, D., Cock, J. M., Ljung, K., and Charrier, B. (2010). Auxin metabolism and function in the multicellular brown alga Ectocarpus siliculosus. Plant Physiol. 153, 128–144.
Liepman, A. H., Wightman, R., Geshi, N., Turner, S. R., and Scheller, H. V. (2010). Arabidopsis a powerful model system for plant cell wall research. Plant J. 61, 1107–1121.
Muday, G. K., and DeLong, A. (2001). Polar auxin transport: controlling where and how much. Trends Plant Sci. 6, 535–542.
Niklas, K. J., and Kutschera, U. (2009). The evolutionary development of plant body plans. Funct. Plant Biol. 36, 682–695.
Niklas, K. J., and Kutschera, U. (2010). The evolution of the land plant life cycle. New Phytol. 185, 27–41.
Palme, K., and Gälweiler, L. (1999). PIN-pointing the molecular basis of auxin transport. Curr. Opin. Plant Biol. 2, 375–381.
Paponov, I. A., Paponov, M. T., Teale, W., Menges, M., Chakrabortee, S., Murray, J. A. H., and Palme, K. (2008). Comprehensive transcriptome analysis of auxin responses in Arabidopsis. Mol. Plant 1, 321–337.
Pu, L., and Brady, S. (2010). Systems biology update: cell type-specific transcriptional regulatory networks. Plant Physiol. 152, 411–419.
Ross, J. J., and Reid, J. B. (2010). Evolution of growth-promoting plant hormones. Funct. Plant Biol. 37, 795–805.
Salmon, J., Ramos, J., and Callis, J. (2008). Degradation of the auxin response factor ARF1. Plant J. 54, 118–128.
Santuari, L., Scacchi, E., Rodriguez-Villalon, A., Salinas, P., Dohmann, E. M. N., Brunoud, G., Vernoux, T., Smith, R. S., and Hardtke, C. S. (2011). Positional information by differential endocytosis splits auxin response to drive Arabidopsis root meristem growth. Curr. Biol. 21, 1918–1923.
Sauer, M., and Kleine-Vehn, J. (2011). Auxin binding protein 1: the outsider. Plant Cell 23, 2033–2043.
Scacchi, E., Salinas, P., Gujas, B., Santuari, L., Krogan, N., Ragni, L., Berleth, T., and Hardtke, C. S. (2010). Spatio-temporal sequence of cross-regulatory events in root meristem growth. Proc. Natl. Acad. Sci. U.S.A. 107, 22734–22739.
Shin, R., Burch, A. Y., Huppert, K. A., Tiwari, S. B., Murphy, A. S., Guilfoyle, T. J., and Schachtman, D. P. (2007). The Arabidopsis transcription factor MYB77 modulates auxin signal transcription. Plant Cell 19, 2440–2453.
Spaepen, S., Vanderleyden, J., and Remans, R. (2007). Indole-3-acetic acid in micobial and microorganism-plant signaling. FEMS Microbiol. Rev. 31, 425–448.
Stein, W. E. (1998). Developmental logic: establishing a relationship between developmental process and phylogenetic pattern in primitive vascular plants. Rev. Palaeobot. Palynol. 102, 15–42.
Sukumar, P., Edwards, K. S., Rahman, A., DeLong, A., and Muday, G. K. (2009). PINOID kinase regulates root gravitropism through modulation of PIN2-dependent basipetal auxin transport in Arabidopsis. Plant Physiol. 150, 722–735.
Swarup, R., Friml, J., Marchant, A., Ljung, K., Sandberg, G., Palme, K., and Bennett, M. (2001). Localization of the auxin permease AUX1 suggests two functionally distinct hormone pathways operate in the Arabidopsis root apex. Genes Dev. 15, 2648–2653.
Tanimoto, M., Jowett, J., Stirnberg, P., Rouse, D., and Leyser, O. (2007). pax1-1 Partially suppresses gain-of-function mutations in Arabidopsis AXR3/IAA17. BMC Plant Biol. 7, 20. doi:10.1186/1471-2229-7-20
Tao, Y., Ferrer, J.-L., Ljung, K., Pojer, F., Hong, F., Long, J. A., Li, L., Moreno, J. E., Bowman, M. E., Ivans, L. J., Cheng, Y., Lim, J., Zhao, Y., Ballaré, C. L., Sandberg, G., Noel, J. P., and Chory, J. (2008). Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133, 164–176.
Teale, W. D., Paponov, I. A., and Palme, K. (2006). Auxin in action: signalling, transport and the control of plant growth and development. Nat. Rev. Mol. Cell Biol. 7, 847–859.
Wabnik, K., Govaerts, W., Friml, F., and Klein-Vehn, J. (2011). Feedback models for polarized auxin transport: an emerging trend. Mol. Biosyst. 7, 2352–2359.
Woodward, A. W., and Bartel, B. (2005). Auxin: regulation, action, and interaction. Ann. Bot. 95, 707–735.
Yant, L. (2012). Genome-wide mapping of transcription factor binding reveals developmental process integration and a fresh look at evolutionary dynamics. Am. J. Bot. 99, 277–290.
Yin, X., and Struik, P. C. (2010). Modeling the crop: from systems dynamic to systems biology. J. Exp. Bot. 61, 2171–2183.
Keywords: auxin, gene networks, IAA, plant evolution, transcription factors
Citation: Niklas KJ and Kutschera U (2012) Plant development, auxin, and the subsystem incompleteness theorem. Front. Plant Sci. 3:37. doi: 10.3389/fpls.2012.00037
Received: 21 November 2011;
Accepted: 13 February 2012;
Published online: 01 March 2012.
Edited by:
Mariana Benítez, Masaryk University, Czech RepublicReviewed by:
Mark E. Olson, Universidad Nacional Autónoma de México, MexicoMarkus Geisler, University of Fribourg, Switzerland
Copyright: © 2012 Niklas and Kutschera. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Karl J. Niklas, Department of Plant Biology, Cornell University, Ithaca, NY 14853, USA. e-mail:a2puMkBjb3JuZWxsLmVkdQ==
