- Institute of Biology/Applied Genetics, Dahlem Centre of Plant Sciences, Freie Universität Berlin, Berlin, Germany
The plant hormone cytokinin realizes at least part of its signaling output through the regulation of gene expression. A great part of the early transcriptional regulation is mediated by type-B response regulators, which are transcription factors of the MYB family. Other transcription factors, such as the cytokinin response factors of the AP2/ERF family, have also been shown to be involved in this process. Additional transcription factors mediate distinct parts of the cytokinin response through tissue- and cell-specific downstream transcriptional cascades. In Arabidopsis, only a single cytokinin response element, to which type-B response regulators bind, has been clearly proven so far, which has 5′-GAT(T/C)-3′ as a core sequence. This motif has served to construct a synthetic cytokinin-sensitive two-component system response element, which is useful for monitoring the cellular cytokinin status. Insight into the extent of transcriptional regulation has been gained by genome-wide gene expression analyses following cytokinin treatment and from plants having an altered cytokinin content or signaling. This review presents a meta analysis of such microarray data resulting in a core list of cytokinin response genes. Genes encoding type-A response regulators displayed the most stable response to cytokinin, but a number of cytokinin metabolism genes (CKX4, CKX5, CYP735A2, UGT76C2) also belong to them, indicating homeostatic mechanisms operating at the transcriptional level. The cytokinin core response genes are also the target of other hormones as well as biotic and abiotic stresses, documenting crosstalk of the cytokinin system with other hormonal and environmental signaling pathways. The multiple links of cytokinin to diverse functions, ranging from control of meristem activity, hormonal crosstalk, nutrient acquisition, and various stress responses, are also corroborated by a compilation of genes that have been repeatedly found by independent gene expression profiling studies. Such functions are, at least in part, supported by genetic studies.
Foreword
The phytohormone cytokinin regulates numerous developmental and physiological processes, including the activity of shoot and root meristems, reproductive behavior, leaf senescence, and responses to environmental cues, in particular light and nutrients (Argueso et al., 2009; Werner and Schmülling, 2009). Regulation of gene expression is a crucial part of realizing the different activities in a developmental- and tissue-specific context. The results of early efforts to identify cytokinin-regulated genes were summarized more than a decade ago (Schmülling et al., 1997). Since then, several major steps toward a more comprehensive understanding of the early and late events of gene regulation by cytokinin have been made. Some important milestones have been the discovery of type-A response regulator genes as primary cytokinin response genes (Brandstatter and Kieber, 1998; Sakakibara et al., 1998); the description of type-B response regulators as transcription factors mediating the cytokinin response (Sakai et al., 2000, 2001; Hwang and Sheen, 2001); the identification of a cis-acting cytokinin response element (Sakai et al., 2000; Taniguchi et al., 2007); and the genome-wide analysis of the cytokinin response of the Arabidopsis transcriptome (Kiba et al., 2004; Brenner et al., 2005). Additional genome-wide studies addressed more specifically the effects of altered cytokinin signaling, the identification of targets of specific signaling components, or organ-specific responses (Rashotte et al., 2006; Lee et al., 2007; Taniguchi et al., 2007; Yokoyama et al., 2007; Argyros et al., 2008; Heyl et al., 2008; Brenner and Schmülling, submitted). More recent work has linked specific transcriptional responses to the biological activities of cytokinin (Dello Ioio et al., 2008a; Müller and Sheen, 2008; Argueso et al., 2010; Zhao et al., 2010; Bishopp et al., 2011c; Köllmer et al., 2011). Taken together, it has become clear that fine-tuned gene regulation is of utmost importance to realize the many different biological activities of cytokinin in diverse tissues.
In this review, we will firstly briefly describe the state-of-the-art knowledge about the early events involved in cytokinin-dependent gene regulation and the main players, and address the question how this system evolved. Then we will summarize the results of a number of genome-wide transcript profiling studies focusing on cytokinin as a regulator of gene expression, present the results of a meta analysis of microarray data and, finally, compile some of the frequently found cytokinin-regulated genes and discuss their functional context. The vast majority of findings come from experiments using Arabidopsis thaliana, thus we will focus on results from this model plant in this review. The key facts presented in this review are summarized in Table 1. For recent reviews addressing various other aspects of the cytokinin signaling system, see To and Kieber (2008), Argueso et al. (2009, 2010), Bishopp et al. (2009, 2011a), Moubayidin et al. (2009), Werner and Schmülling (2009), Hellmann et al. (2010), Kudo et al. (2010), Perilli et al. (2010), Domagalska and Leyser (2011), Heyl et al. (2011), Kiba et al. (2011), and Müller (2011).
The Discovery of Primary Response Genes of Cytokinin
The first primary response genes rapidly induced by cytokinin were identified using the method of differential display in Arabidopsis thaliana (Brandstatter and Kieber, 1998) and maize (Sakakibara et al., 1998). In Arabidopsis, two genes designated IBC6 and IBC7 (INDUCED BY CYTOKININ, IBC) – now known as type-A response regulator genes ARR5 and ARR4, respectively – were identified as being induced after 2 h of cytokinin treatment (Brandstatter and Kieber, 1998). These genes were upregulated within minutes after cytokinin treatment (D’Agostino et al., 2000) and the response was insensitive to cycloheximide, both hallmarks of primary response genes. At the same time, the maize gene ZmCip1, also encoding a type-A response regulator, was found to be induced 30 min after cytokinin treatment (Sakakibara et al., 1998). A number of related Arabidopsis type-A response regulator genes – ARR3, ARR6, ARR7, ARR8, ARR9, ARR15, and ARR16, which were mostly identified by sequence homology to bacterial response regulators (Imamura et al., 1998; Urao et al., 1998) – were characterized as cytokinin-induced genes with different response kinetics (Imamura et al., 1999; Kiba et al., 1999; D’Agostino et al., 2000). It was shown later that type-A ARR proteins act as negative regulators of cytokinin signaling (Hwang and Sheen, 2001; To et al., 2004). Today, by virtue of genome-wide expression analyses, numerous other response genes of cytokinin have been discovered (see Genome-Wide Analysis of the Cytokinin-Regulated Transcriptome) but most of them lack a functional link to the hormone at present.
Transcription Factors Regulating the Primary Cytokinin Response
The discovery of type-A response regulator genes as primary cytokinin response genes was a strong indication that cytokinin signaling may occur through a two-component signaling system (TCS). In fact, the cytokinin signal is perceived by sensor histidine kinases (Inoue et al., 2001; Suzuki et al., 2001; Heyl et al., 2011), named AHK2, AHK3, and CRE1/AHK4 in Arabidopsis, and the following primary signal transduction pathway of cytokinin is a multistep trans-phosphorylation chain similar to the bacterial TCS. Following the transfer of the signal to the nucleus via histidine phosphotransmitter proteins (named AHPs in Arabidopsis; Hwang and Sheen, 2001; Punwani et al., 2010), ultimately a family of transcription factors, the type-B response regulators, is activated and these mediate the transcriptional response to cytokinin (Hwang and Sheen, 2001).
The Arabidopsis genome encodes 11 type-B ARRs. They belong to the MYB family of transcriptions factors and all contain an N-terminal response regulator domain. This domain is thought to negatively regulate the activity of the type-B ARRs as a deletion of this domain leads to a constitutively active form of the protein (Sakai et al., 2000; Imamura et al., 2003; Tajima et al., 2004; Taniguchi et al., 2007). This observation led to a model in which the phosphorylation of the response regulator domain causes a conformational change of the type-B ARRs, thereby activating the proteins. Phosphomimics, in which the canonical Asp of the response regulator domain is exchanged with a Glu leading to a constitutive activation, are supposed to have an altered structure and seem to support this model (Hass et al., 2004). However, structural information confirming the proposed mode of action is still missing. This is in contrast to the DNA-binding MYB-domain. NMR spectroscopy has revealed how the DNA-binding domain of ARR10 is in direct contact with the target DNA, 5′-(A/T)GATT-3′ (Hosoda et al., 2002). The C-terminal part of the type-B ARRs is often enriched by P/Q-rich acidic domains, which have been shown to be important for their transactivation capacity (Sakai et al., 2000; Imamura et al., 2003). The nuclear localization of the type-B ARRs is consistent with their role as transcription factors (Hwang and Sheen, 2001; Lohrmann et al., 2001; Mason et al., 2004; Dortay et al., 2006).
Phylogenetic analyses based on the receiver domains or DNA-binding domains of type-B ARRs revealed a division into three sub-classes (Tajima et al., 2004; To et al., 2004; Mason et al., 2005; Heyl et al., 2008). The largest sub-class consists of seven members (ARR1, ARR2, ARR10, ARR12, ARR11, ARR14, and ARR18) and two other sub-classes containing two members each (ARR13/ARR21 and ARR19/ARR20). The expression patterns of type-B ARR genes reflect this subdivision revealed by phylogenetic analysis. While the members of the largest sub-class are expressed in almost all tissues, members of the other two sub-classes are expressed specifically in reproductive organs (Mason et al., 2004; Tajima et al., 2004; Heyl et al., 2006). The region of expression for ARR19 and ARR21 was narrowed down to the chalazal region of the endosperm (Tiwari et al., 2006; Day et al., 2008).
Analyses have shown the functional redundancy of type-B ARRs (Mason et al., 2005): for example, single knockout mutants of arr1 and arr21 displayed no major alteration of their phenotypes (Sakai et al., 2001; Horák et al., 2003). Only the arr2 knockout mutant showed a slight insensitivity toward cytokinin and ethylene in a hypocotyl elongation assay (Hass et al., 2004). ARR2 has also been implicated in the regulation of leaf senescence (Kim et al., 2006). In contrast, analysis of multiple knockout mutant lines of type-B ARR genes revealed that ARR1, ARR10, and ARR12 mediate the majority of cytokinin action in different processes, such as vascular development, light sensitivity, chlorophyll production, and cell division in the root and shoot (Yokoyama et al., 2007; Argyros et al., 2008; Ishida et al., 2008). Overexpression of full length ARR1 and ARR2 genes did not cause notable phenotypic changes (Sakai et al., 2001; Hass et al., 2004). However, overexpression of only the C-terminal part of the protein, including the DNA-binding domain of ARR1, ARR11, ARR14, ARR20, and ARR21, respectively, caused distinct abnormalities of plant phenotypes during the vegetative stage (ARR1/ARR11/ARR14) or the reproductive stage (ARR20/ARR21; Sakai et al., 2001; Imamura et al., 2003; Tajima et al., 2004). A compilation of the known phenotypes of single and multiple type-B ARR mutants (and other genes of the TCS) has been published recently (Müller, 2011).
Chimeric repressor technology (Hiratsu et al., 2003) was used in order to investigate the functions of type-B ARRs more generally. The addition of the SRDX peptide (LDLDLELRLGFA) to the C-terminal end of ARR1 turned this transcriptional activator into a trans-dominant transcriptional repressor. Protoplast transactivation assays confirmed that ARR1-SRDX represses not only the transactivation activity of ARR1, but also of all other type-B ARRs tested, thus demonstrating its trans-dominant function (Heyl et al., 2008). Plants expressing the ARR1-SRDX construct under the control of the 35S promoter displayed a strong morphological phenotype, reminiscent of the cytokinin deficiency syndrome seen in triple cytokinin receptor mutants or cytokinin oxidase/dehydrogenase (CKX) overexpressors (Werner et al., 2003; Riefler et al., 2006). This phenotype is most likely due to the concerted repression of the activities of the type-B ARR (Heyl et al., 2008). Transcriptional profiling indicated attenuation of the early transcriptional response to cytokinin and suggested that ARR1 and other type-B ARRs use, at least in part, the same DNA-binding sites. Together, the results showed that most, if not all, of the transcriptional response to cytokinin is mediated, at least partly, by the type-B ARRs (Heyl et al., 2008).
Another group of transcription factors regulating the transcriptional response to cytokinin are the cytokinin response factors (CRF). Rashotte and colleagues showed that members of this subfamily of AP2/ERF transcription factors, which is defined by the presence of a common CRF domain, are involved in the control of gene expression of many primary cytokinin response genes (Rashotte et al., 2006; Rashotte and Goertzen, 2010). CRF2 and CRF5 were themselves recognized as primary response genes due to their transcriptional regulation by cytokinin in the presence of cycloheximide (Rashotte et al., 2006). Protoplast experiments showed that all CRFs localize from the cytosol into the nucleus upon cytokinin treatment. This translocation was dependent on the presence of the cytokinin receptors and the AHPs, but not type-A or type-B ARRs (Rashotte et al., 2006). Yeast two-hybrid and Split-YFP experiments showed homo- and heterodimerization among the CRFs and between the AHPs and the CRFs. Deletion experiments indicated that the CRF domain is both necessary and sufficient to mediate these protein interactions. It has been speculated that homodimerization of the CRFs might be necessary for DNA-binding (Cutcliffe et al., 2011). The phenotypic changes of different multiple crf mutants occurred with low penetrance, but point at a role in the development of embryos, cotyledons, and leaves. The severity and the penetrance of the loss-of-function phenotypes increased in higher order mutants indicating functional redundancy (Rashotte et al., 2006).
Evolution of Transcription Factors Mediating the Cytokinin Response
In order to get an insight into the evolution of the TCS, a phylogenetic analysis has been carried out for all four participating protein families (Pils and Heyl, 2009). The type-B RRs are present in all plants species, including algae. Thus, they predate the appearance of the cytokinin receptors and the type-A RRs, which are only found in land plants (Pils and Heyl, 2009). That raises the questions which functions the type-B RRs have in those species missing the cytokinin receptors and whether those ancient functions are still performed by type-B RRs of higher plant species. The CRF protein family has at least eight members in Arabidopsis (Rashotte and Goertzen, 2010). A phylogenetic study showed that CRF genes are present in the genomes of all land plants, but are absent from those of algae. The late-diverging land plants separated into two clades, which coincided with the presence or the absence of an additional domain, the so-called TEH (Thr, Glu, His) region. However, the functional relevance of this TEH region is not yet clear (Rashotte and Goertzen, 2010).
Taken together, these data suggest that the type-B RRs predate the emergence of the cytokinin signaling pathway as we know it from higher plants and were only recruited to it at a later stage of plant evolution. Once the morphology and the developmental program of land plants became more complex, not only did the number of type-B RRs in the genomes increase (Pils and Heyl, 2009; Schaller et al., 2011), but it was also likely that additional factors, such as the CRFs, were recruited to fine-tune the cytokinin response.
cis-Regulatory Cytokinin Response Elements Required for the Regulation of Primary Response Genes
Considering the high conservation of amino acids in the DNA-binding MYB-domain of type-B ARRs (Heyl et al., 2008), it is not surprising that several of them bind to the same or very similar sequence motifs. It has been demonstrated that the DNA-binding domains of both, ARR1 and ARR2, bind in vitro to DNA sequences containing the 5′-AGAT(T/C)-3′ motif. The actual DNA recognition by the DNA-binding domain appeared to occur mainly at the last four positions of this 5-bp sequence (Sakai et al., 2000). The optimal in vitro binding for ARR10 and ARR11 was observed for a DNA sequence containing the motifs 5′-AGATT-3′ and 5′-GGATT-3′, respectively (Hosoda et al., 2002; Imamura et al., 2003). Later, it was shown that ARR1 requires 2-bp extensions on either side of the core sequence for optimal binding in vitro (Taniguchi et al., 2007). Comparing the bases surrounding the 5-bp long core motif, the sequence 5′-AAGAT(T/C)TT-3′ was identified as an extended version of the ARR1-binding motif (Taniguchi et al., 2007).
A synthetic cytokinin reporter that was generated to visualize the output of the TCS in vivo harbors 24 concatemerized copies of a variant of the above-mentioned type-B ARR binding core motif, 5′-(A/G)GATT-3′, in sense and antisense direction and a minimal 35S promoter binding motif upstream of a reporter gene (Müller and Sheen, 2008). The presumed advantage of this synthetic cytokinin reporter compared to other marker genes, such as the commonly used type-A response regulator genes, is that its activity is largely independent of other signaling or tissue-specific information, and thus shows solely the TCS output (Müller and Sheen, 2008). Using this synthetic reporter, the local and temporal defined antagonistic interaction between auxin and cytokinin during the establishment of the root stem cell niche was identified (Müller and Sheen, 2008). Similarly, this TCS reporter or an improved version of it has been used to study cytokinin activity in the shoot apical meristem (Gordon et al., 2009; Yoshida et al., 2011).
So far, the functionality of binding motifs identified for ARR1 and other type-B ARRs have been supported mainly by in vitro experiments (Sakai et al., 2001; Imamura et al., 2003; Taniguchi et al., 2007) and their use for the TCS reporter (Müller and Sheen, 2008). There are only a very few loss-of-function studies confirming the functionality of these sequences in planta. One example is the transcriptional regulation of the non-symbiotic hemoglobin (NSHB) genes (Hunt et al., 2001; Ross et al., 2004). It was shown by using transient bombardment assays in tobacco leaf disks that cytokinin activates the Oryza sativa NSHB2 (OsNSHB2) promoter and that this regulation can be mediated by ARR1 of Arabidopsis (Ross et al., 2004). Deletion and site-directed mutational analyses of the predicted ARR1-binding cis-element 5′-AGATT-3′ in the OsNSHB2 promoter confirmed the functionality of this element in the cytokinin response. Deletion led to a dramatic reduction in cytokinin-inducibility of this promoter (Ross et al., 2004). These experiments were not fully conclusive, as the cytokinin-dependent activation of a rice promoter by an Arabidopsis transcription factor was tested in tobacco cells. However, it has been shown recently that mutation of this core motif in the ARR6 promoter strongly reduces its response toward cytokinin in its original host (Ramireddy, 2009).
The afore-mentioned work confirmed the functional relevance of the core motif in planta. However, it should be noted that because of its statistically frequent occurrence, this short motif is not diagnostic for the cytokinin responsiveness of a given promoter. It is not significantly enriched in the top 20 cytokinin-responsive promoters within the 1-kb 5′ of the translational start site (Figure 1), where it occurs at a frequency of 0.68% in comparison to the promoters of the 20 genes least likely to be regulated by cytokinin, where it has a frequency of 0.57%. The extended motif, 5′-AAGAT(T/C)TT-3′, which was identified for ARR1 (Taniguchi et al., 2007), is enriched in the top 20 cytokinin-responsive promoters, where it occurs at a frequency of 0.16% compared to 0.01% in the promoters of the 20 genes least likely to be regulated by cytokinin. However, it is missing in 5 of the top 20 genes (Figure 1). This could indicate that ARR1 is not involved in the transcriptional regulation of all these cytokinin response genes, or that other variants of the extended motif allow for the binding of and transcriptional activation by type-B RRs.
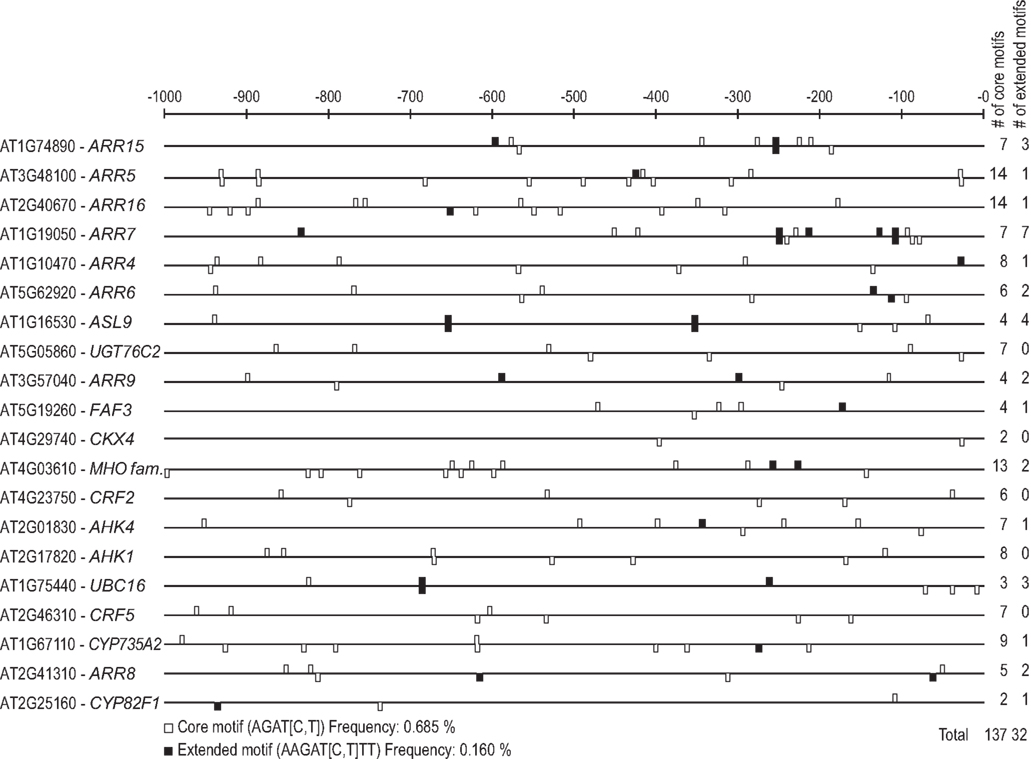
Figure 1. Motif mapping of type-B response regulator binding motifs in the promoters of the top 20 cytokinin-regulated genes. The core motif (Sakai et al., 2000) and the extended motif determined for ARR1 (Taniguchi et al., 2007) were mapped onto the promoter regions 1000 bp upstream of the translational start sites of the 20 highest ranking genes of the meta analysis (Table 3) using the MotifMapper of the TOUCAN 2 workbench (Aerts et al., 2005). The motifs were counted, and their frequency of occurrence (in percent) calculated using the formula (number of motifs * 100)/(20 promoters * 1000 bp).
Another putative cytokinin response element has been identified in cucumber. Expression analysis showed that cytokinin and light activate the transcription of the NADPH–protochlorophyllide reductase (POR) gene which has a key role in chlorophyll synthesis (Kuroda et al., 2000). Gel shift experiments demonstrated that the promoter of the POR gene contains a cis-element (5′-TATTAG-3′) which binds nuclear extracts in a cytokinin-dependent manner (Fusada et al., 2005). Mutation of this sequence in the POR promoter caused a reduced cytokinin-dependent binding of nuclear proteins and caused a reduced cytokinin responsiveness in transient expression assays (Fusada et al., 2005). However, the protein factor(s) binding to this motif are not known. This new element, mediating part of the cytokinin response, is present in only 6 of the top 20 cytokinin-regulated genes of Arabidopsis in the meta analysis, indicating that this motif also is not absolutely required for cytokinin responsiveness or might be species-specific (data not shown).
Genome-Wide Analysis of the Cytokinin-Regulated Transcriptome
With the dawn of the transcriptomics age, enabled by the wide availability of microarrays, several attempts have been made to identify more cytokinin-responsive genes. Over time, the themes of the experimental approaches have changed. The first transcriptome studies were purely discovery-based approaches in order to find as many cytokinin-regulated genes as possible for a number of different time points. The focus then shifted toward the identification of genes being targeted directly by single or multiple transcription factors of the type-B response regulator and CRF families. The latest investigations have shown two trends: firstly, the attempt to investigate the transcriptome of smaller expression domains; secondly, an enhanced focus on physiological and developmental downstream processes influenced by cytokinin.
Published Studies on the Cytokinin-Regulated Transcriptome
A first study investigating cytokinin effects on the transcript level of many genes in 2002 used a microarray containing 8297 probe sets from Arabidopsis and assessed gene expression during shoot induction from root callus (Che et al., 2002). As the samples were taken days after transferring the explants to cytokinin-containing shoot induction medium, early changes in gene expression were not identified and only the expression data of known cytokinin and auxin responsive genes were shown. The results confirmed the cytokinin induction of the type-A ARR genes and CRE1/AHK4. Another early study focusing on long-term transcriptomic changes used the method of massively parallel signature sequencing and analyzed seedlings harboring an inducible cytokinin synthesizing bacterial IPT gene (Hoth et al., 2003). This work also confirmed a number of known cytokinin-regulated genes and it was noted that subsets of genes are regulated independently of tissue type or developmental stage. Both studies also identified numerous novel genes regulated or potentially regulated by cytokinin.
The first investigation devoted to the immediate–early cytokinin response also used the microarray with 8297 probe sets (Rashotte et al., 2003). Samples were obtained from seedlings treated with different cytokinins (benzyladenine, trans-zeatin) in different concentrations (1–5 μM) and for different time periods (15 min to 24 h). The wol and cre1 cytokinin receptor mutants were analyzed, too. More than 70 novel cytokinin-regulated genes were identified. Two cytokinin-responsive “putative AP2 genes” (AT2G46310 and AT4G23750) were discovered, which were classified as CRFs in later research (Rashotte et al., 2006). Homeostatic mechanisms were revealed as two of the cytokinin-induced genes encoded cytokinin metabolic enzymes: the cytokinin oxidase/dehydrogenase CKX4 (Werner et al., 2001) and a cytochrome P450 enzyme (CYP735A2) with cytokinin trans-hydroxylase activity (Takei et al., 2004). Another potential pattern of cytokinin-regulated genes indicated an effect on the redox state (Rashotte et al., 2003).
One year later, a first analysis using a whole-genome microarray featuring 22,810 probe sets (ATH1 array) was published (Kiba et al., 2004). Although the work focused on the type-C (by then still classified as type-A) response regulator ARR22, it also contained information on cytokinin-induced genes in wild-type Arabidopsis. There, a large number of additional genes were found to be cytokinin-regulated, including several heat shock-related genes and the histidine kinase gene AHK1. This study was extended to plants overexpressing the C-terminal half of the type-B response regulator ARR21 (ARR21-C-ox) causing a constitutive activation of the cytokinin response, in order to contrast ARR22 overexpressors (ARR22-ox) attenuating the cytokinin response (Kiba et al., 2005). A set of 68 robustly cytokinin-responsive genes was generated by comparing transcript levels in wild type, cytokinin-treated wild type, ARR22-ox, and ARR21-C-ox.
Another study using the ATH1 array also appeared in 2005 comparing cytokinin-induced and cytokinin-deficient seedlings (35S:CKX1), with the uninduced wild type as the reference (Brenner et al., 2005). An extensive evaluation of the functional context of the regulated genes identified transcriptional regulation, signal transduction, interaction with other hormones (particularly gibberellin), light signaling, primary metabolism, and transport as cytokinin target processes. It was found that transcription factor genes were largely overrepresented among the cytokinin-regulated genes in comparison to their proportion in the whole genome. Many of these genes were only temporarily induced, while others were upregulated at a late time point, suggesting that transcriptional cascades are triggered during the course of cytokinin action. The MapMan software (Thimm et al., 2004) was used to identify cytokinin-regulated physiological processes. Among all the genes of cytokinin metabolism and signaling, only the type-A response regulators showed a clearly opposite regulation in cytokinin-induced and cytokinin-deficient seedlings. In addition, two processes were identified with an opposite regulation of their genes depending on the cytokinin status, namely metabolism of the regulator of carbon utilization, trehalose-6-phosphate, and valine and leucine metabolism. Finally, the upregulation of several plastidal transcripts was noted. The data generated in this study spawned further research on four cytokinin-regulated transcription factor genes, namely GATA22 (also known as CGA1/GNL), two genes coding for members of the homeodomain zip (HD zip) class II transcription factor family (HAT4, HAT22), and bHLH64 (Köllmer et al., 2011).
Transcript profiling of cytokinin-treated arr1 arr12, crf1 crf2 crf5, and crf2 crf3 crf6 mutants was conducted in order to identify the target genes of the CRFs. The results led to the conclusion that both, type-B ARRs and CRFs, are necessary for cytokinin induction of many of the early cytokinin response genes, because their regulation was attenuated in higher order mutants of either transcription factor family. Interestingly, while the expression of the type-A ARR genes in response to cytokinin treatment was reduced in the arr1 arr12 mutant, it was unaffected or even slightly upregulated in the crf mutants tested (Rashotte et al., 2006).
The reduced induction of cytokinin-regulated genes in ARR7 overexpressing plants showed clearly that ARR7 is a negative regulator of cytokinin signaling (Lee et al., 2007). However, the regulation of only 30% to 50% of all cytokinin-regulated genes was attenuated by ARR7 overexpression, indicating that the protein acts only on part of the cytokinin signaling pathway. The regulation of transcription factor genes, hormone-related and stress-related genes, as well as the finding that the responsiveness of most cytokinin-induced expansin genes and several LATERAL ORGAN BOUNDARIES DOMAIN (LBD) genes was attenuated by ARR7, were specifically addressed.
Direct target genes of ARR1 were identified using plants expressing a dexamethasone-inducible variant of ARR1 (Sakai et al., 2001) and an AFLP-based expression profiling method called “high-coverage expression profiling” (HiCEP; Fukumura et al., 2003) as the analytical tool (Taniguchi et al., 2007). An improved sequence motif for ARR1-binding was characterized in the promoters of the 23 ARR1 target genes identified (see cis-Regulatory Cytokinin Response Elements Required for the Regulation Of Primary Response Genes).
The roots of an arr10 arr12 mutant were studied using the ATH1 microarray (Yokoyama et al., 2007). This mutant combination was selected because it causes a strong root phenotype, suggesting that ARR10 and ARR12 (together with ARR1) transduce most of the cytokinin signal in the root. It was found that the transcriptional response to cytokinin in this mutant was attenuated for most of the known cytokinin response genes, as well as for several genes newly discovered in this study. The induction of the type-A response regulator genes ARR15 and ARR16 was particularly strongly diminished, leading the authors to suggest a functional link between ARR10 and ARR12, on the one hand, and ARR15 and ARR16, on the other hand.
Following a similar approach, a transcription profiling of the shoots of seedlings was part of the characterization of an arr1 arr10 arr12 triple mutant (Argyros et al., 2008). In these plants, the cytokinin-induced increase in transcript levels of 71 genes, including many of the then well-known primary response genes, was severely attenuated. In addition, the basal transcript levels of almost 90% of these genes were reduced more than twofold, indicating a reduced response to endogenous cytokinin. The cytokinin-induced repression of the transcript levels of a number of genes was also diminished in the mutant plants. These results added convincing evidence that most of the general cytokinin response during the seedling stage is signaled through the three type-B response regulators ARR1, ARR10, and ARR12 (Argyros et al., 2008).
A study looking more broadly into the action of type-B response regulators was carried out using transgenic plants overexpressing a trans-dominant chimeric repressor of type-B response regulator action, ARR1-SRDX (see Transcription Factors Regulating the Primary Cytokinin Response; Heyl et al., 2008). Transcriptome comparison between ARR1-SRDX transgenic seedlings and wild-type seedlings identified transcripts regulated by type-B response regulators. About three-quarters of the genes induced by cytokinin in the wild type were not induced in ARR1-SRDX plants, and a total of 658 genes were found whose transcripts were less abundant in the ARR1-SRDX overexpressing plants. The functional context of these genes indicated that type-B ARRs regulate a number of processes, such as nitrate transport and metabolism, red light signaling, and crosstalk with other hormones (auxin, ethylene, and gibberellin). A possible negative feedback on ARR1 expression, as well as on cytokinin metabolism, was also detected. It was proposed that type-B ARRs are possible nodes for the integration of different signaling pathways (Heyl et al., 2008).
The latest cytokinin-related microarray study so far investigated similarities and differences of the transcriptomic response to cytokinin induction and cytokinin deficiency in roots and shoots using the CATMA microarray (Brenner and Schmülling, submitted). The rationale for this study were the marked differences of the cytokinin effects on roots and shoots which could be reflected by differences in the set of responsive genes. It was found by principal component analysis that the early pattern of the transcriptomic cytokinin response was largely similar in roots and shoots while the later response showed some notable differences: Cytokinin-induced roots had a more shoot-like global transcript profile and, similarly, cytokinin-deficient shoots had a global transcript profile shifted toward a more root-like fashion. Interestingly, nuclear genes encoding plastid-localized proteins were strongly overrepresented among these differentially regulated genes. These observations suggest that cytokinin promotes shoot identity on the transcript level at least partially independently of morphological organ identity. Several genes with organ-specific responses were identified, including root-specificity for the cytokinin hydroxylase gene CYP735A2 and shoot specificity for the cell cycle regulator gene CDKA;1. However, contrasting with the fundamental difference of the growth response of roots and shoots to the hormone, the vast majority of the cytokinin-regulated transcriptome showed similar response patterns in roots and shoots (Brenner and Schmülling, submitted).
In summary, the microarray studies have shown that the multitude of processes influenced by cytokinin (Werner and Schmülling, 2009) are reflected by changes of the abundance of transcripts encoding proteins of diverse function. There is, in part, little overlap between the genes identified by different studies, indicating their context-dependent regulation. The transcriptomic results can serve as a springboard to the investigation of downstream cytokinin signaling events, as will be discussed further below.
A Meta Analysis of Publicly Available Microarray Data Sets
In addition to the published and commented microarray results described in Section “Published Studies on the Cytokinin-Regulated Transcriptome,” a number of unpublished datasets are available in the microarray databases. As of July 2011, there were 28 sets of cytokinin-related microarray-derived data stored in the ArrayExpress database (www.ebi.ac.uk/arrayexpress), and a lot more are probably archived in individual labs inaccessible to the community. Arabidopsis thaliana is the experimental system of 25 of the publicly available datasets; one dataset comes from Medicago truncatula, one dataset from Solanum tuberosum and one from hybrid poplar (Populus tremula × Populus alba; Table 2). Despite great efforts to get the data into a uniform format and high-quality annotation, such as the MIAME standard for the description of a microarray experiment (Brazma et al., 2001), the data are not readily comparable for several reasons. Firstly, different array platforms were used; data from single-color microarrays are not directly comparable with data from two-color arrays. Secondly, the annotation standards are different because data originally submitted to different databases were combined. Thirdly, the experiments submitted had been designed for distinct research purposes and different growth conditions or developmental stages were used. Last but not least, each laboratory has its individual plant growth system resulting in differences in global gene expression (Massonnet et al., 2010) and, presumably, also the cytokinin response. For these reasons, we have chosen eight microarray experiments which have used the same array platform, the same organism (i.e., Arabidopsis thaliana), a roughly comparable experimental layout and a sufficient number of replicates, included wild-type data, and had a good overall quality of the data and traceable sample annotations for the meta analysis (Table 2).
The meta analysis based on these selected microarray datasets summarizes the genes most robustly regulated by cytokinin (Table 3). At the top of the list, there are 6 of the 10 type-A response regulator genes and 2 more are found among the top 20 cytokinin response genes. ARR3 ranked in position 66 of the list, while ARR17 did not have a significant meta-P-value and thus does not appear to be regulated by cytokinin. Some other genes of cytokinin metabolism and signaling are also part of this list, including CKX4 and CKX5, which encode cytokinin oxidases/dehydrogenases (Werner et al., 2001), the UDP-glucosyltransferase gene UGT76C2 encoding a cytokinin-O-glycosyltransferase (Hou et al., 2004), the cytochrome P450 gene CYP735A2 encoding a cytokinin trans-hydroxylase involved in trans-zeatin synthesis (Takei et al., 2004), and the cytokinin receptor gene CRE1/AHK4 (Inoue et al., 2001; Suzuki et al., 2001). The other histidine kinase gene in the list, AHK1, encodes a sensor of osmotic stress (Urao et al., 1999; Tran et al., 2007; Wohlbach et al., 2008). Other well-characterized genes are also part of this list, such as the cytokinin response factors CRF2 and CRF5 (Rashotte et al., 2006). Interestingly, in a co-expression analysis, almost all the genes of the top 25 list that have a known function in cytokinin metabolism and signaling clustered together (Figure 2A). Expression of CYP735A2 clustered separately with ASL9 (Figure 2A). ASL9 encodes a LOB-domain transcription factor whose overexpression results in a dwarf phenotype and alters the cytokinin response of seedlings, but a loss-of-function phenotype is not known (Kiba et al., 2005; Naito et al., 2007). One other gene that has not yet been functionally linked to cytokinin, FAF3, which is a member of the newly established FANTASTIC FOUR (FAF) gene family (Wahl et al., 2010), is part of the central cluster (Figure 2A). FAF3 is most strongly expressed in the vasculature, where it might have a function in differentiation, which is regulated by cytokinin (Mähönen et al., 2006; Bishopp et al., 2011b). No functional information exists for the genes encoding the ubiquitin-conjugating enzyme 16 (UBC16), the metallo-hydroxylase/oxidoreductase superfamily protein (MHO), and cytochrome P450 CYP82F1. These three genes show, together with ARR16, the less related (most distinct) expression profile compared to the known cytokinin-related genes (Figure 2B).
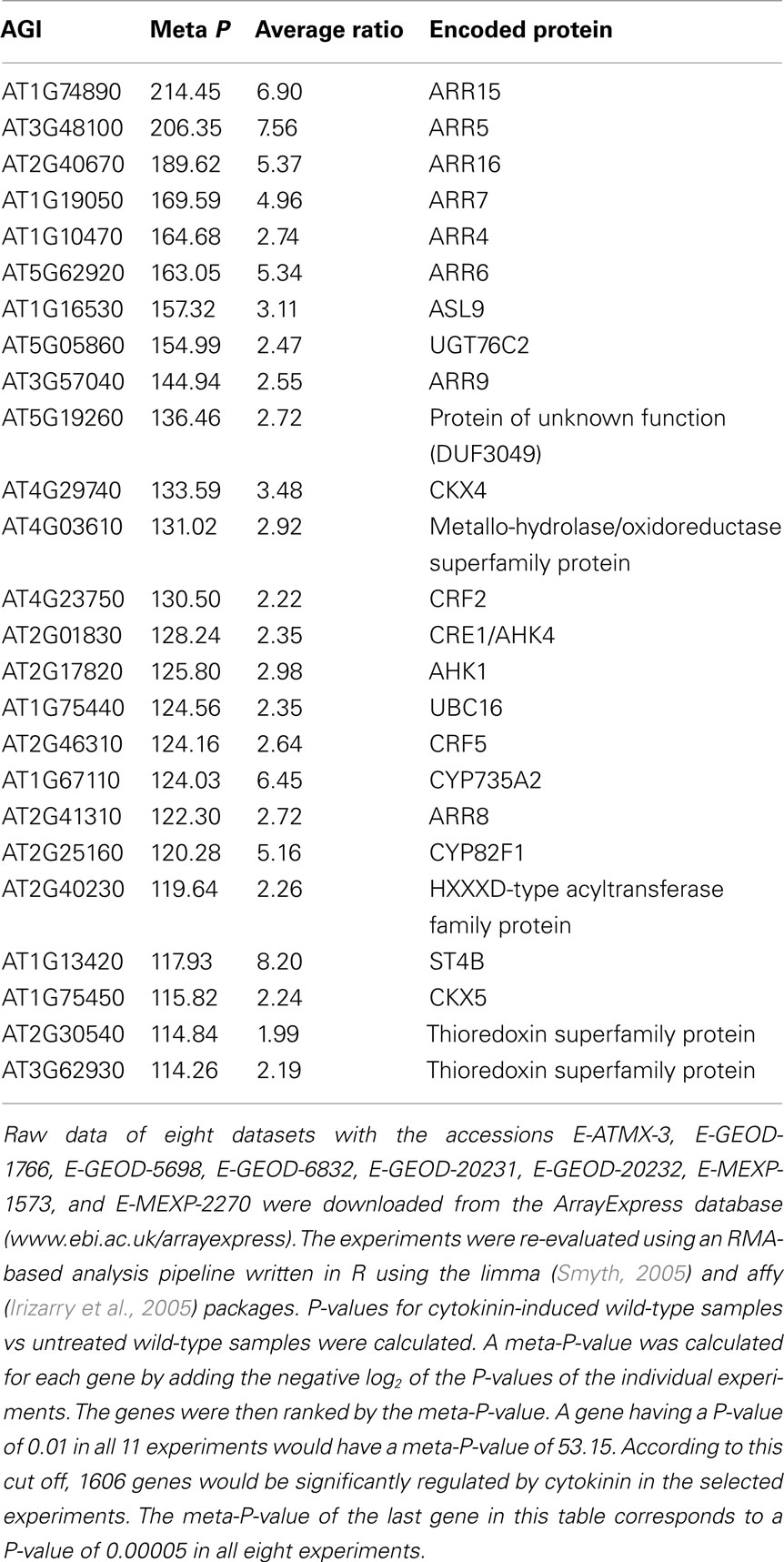
Table 3. Top 25 genes of a meta analysis across selected transcript profiling experiments of Arabidopsis thaliana performed with Affymetrix ATH1 arrays.
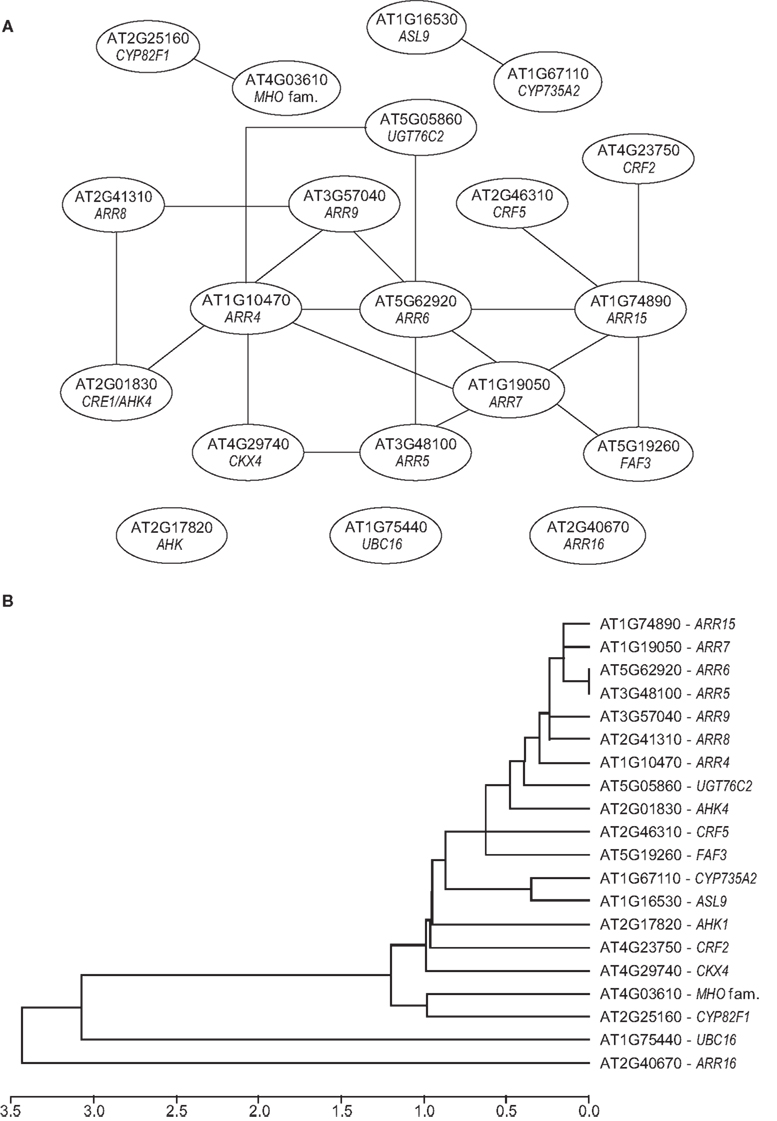
Figure 2. Co-expression and clustering analysis of cytokinin response genes. (A) The relationships among the top 20 cytokinin response genes (see Table 2) is visualized by co-expression analysis. For this analysis, expression profiles of the top 20 gene were obtained from 1388 array experiments from the ATTED-II database (Obayashi et al., 2011). Among these were 236 tissue-specific arrays, 534 abiotic stress arrays, 200 biotic stress arrays, 200 hormone arrays, and 48 light arrays. The expression profiles were analyzed using the ATTED-II database Mutual Rank (MR) calculations were used to quantify the similarity of the gene expression profiles (Obayashi et al., 2009). The co-expression network was drawn with the Network Drawer (http://atted.jp/top_draw.shtml#NetworkDrawer) and modified. (B) Hierarchical clustering of cytokinin response genes. The clustering was performed by using the single linkage method (ATTED-II database; Obayashi et al., 2011). For each query gene, the Mutual Rank was averaged from its co-expressed gene list and their distances were plotted as log10 of the Mutual Rank value.
We were interested whether the top 20 cytokinin-responsive genes were also targets of other signaling pathways. We, therefore, analyzed their response to different hormones and environmental cues using the data sets available from Genevestigator (Hruz et al., 2008). We selected hormone experiments that employed conditions similar to those of the cytokinin experiments (Goda et al., 2008). The heatmap, shown in Figure 3, reveals that several of the top 20 cytokinin response genes are the subject of rapid regulation by other hormones. In most cases, this regulation appears to cause a reduction of the transcript levels rather than an induction, and most genes respond at a later time point to these other hormones than to cytokinin. Treatment with auxin, and partly also with ABA, for example, causes reduced transcript levels of numerous cytokinin-responsive genes after 1–3 h (Figure 3). Surprisingly, the strongest and earliest reactions were seen with brassinolide (BL) and methyl jasmonate. Expression of CKX4 is upregulated and expression of CRE1/AHK4, CRF2, and ARR9 is downregulated only 30 min after BL treatment. Stronger and broader effects of the different hormones on the transcript levels of cytokinin-regulated genes are seen at later time points (data not shown). Taken together, this supports the idea that the antagonistic and agonistic action of cytokinin and different hormones are, at least partly, realized through transcriptional regulation of the same target genes.
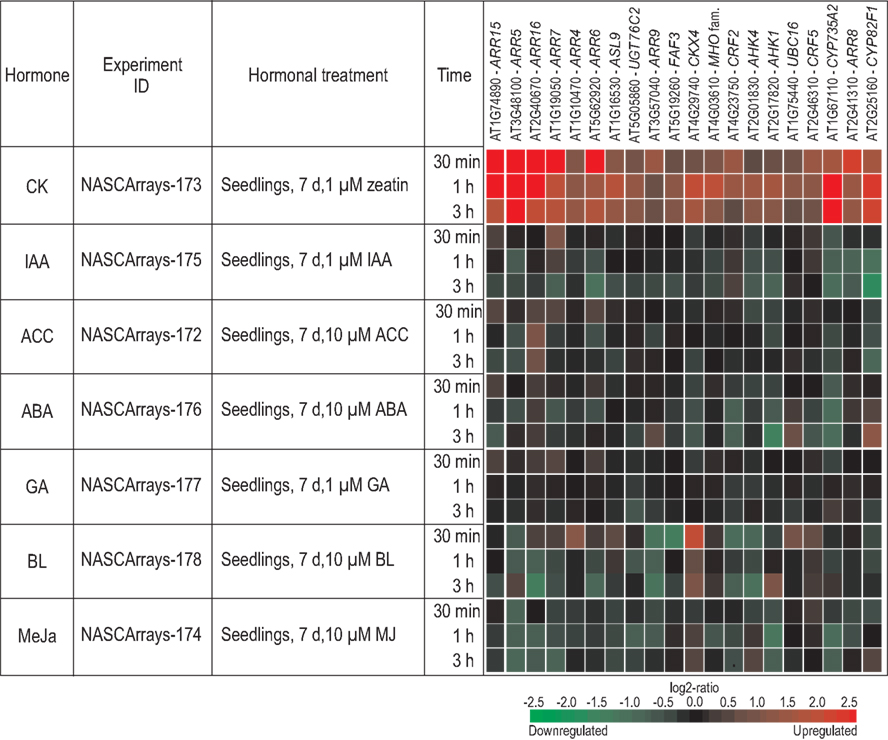
Figure 3. Regulation of cytokinin response genes by different hormones. The hormonal meta-profile analysis of the top 20 cytokinin response genes (see Table 2) was established using the Genevestigator database (http://www.genevestigator.com/gv/index.jsp; Hruz et al., 2008) and modified. Only the hormone experiments described by Goda et al. (2008) were considered as tissues (i.e., seedlings); time points and other experimental conditions were most similar to those used for the cytokinin treatments. The gene expression responses are calculated as ratios between hormone-treated and mock-treated samples. The resulting heatmap is color coded as indicated and thus reflects the up- (red color) or downregulation (green color) of genes. Each experiment can be retrieved from the NASCArrays database (Craigon et al., 2004) by using their unique ID. Abbreviations used in the figure: d, days; h, hours; min, minutes; μM, micromolar; CK, cytokinin; IAA, indole-3-acetic acid; ACC, 1-aminocyclopropane-1-carboxylic acid; ABA, abscisic acid; GA, gibberellic acid; BL, brassinolide; MeJa, methyl jasmonate.
A second analysis for the changes in transcript abundance of the top 20 cytokinin response genes was made in a similar fashion for their responses to different stresses or environmental cues. An arbitrary selection of the resulting heatmaps is shown in Figure 4. Most stress conditions (heat, drought, osmotic and biotic stress, high light irradiation) caused a reduced expression of cytokinin response genes. An exception is cold stress, which induced an increase of transcript levels in most cases. Short treatments (45 min) with blue or red light did not cause changes of the transcript abundance of the genes analyzed, while UV-B light was repressive, at least after 6 h treatment. As most stress treatments inhibit growth, it could be that the downregulation of cytokinin-responsive genes is an early indication of such growth inhibitory activity. We hypothesize that modulation (i.e., mostly a reduction) of the cytokinin status is a common part of the general stress response, and that this modulation is required to redirect the use of plant resources for the stress response and no longer to maintain growth. Consistently, there is a tendency that stress responses are accompanied by a reduction of the cytokinin content (Hare et al., 1997).
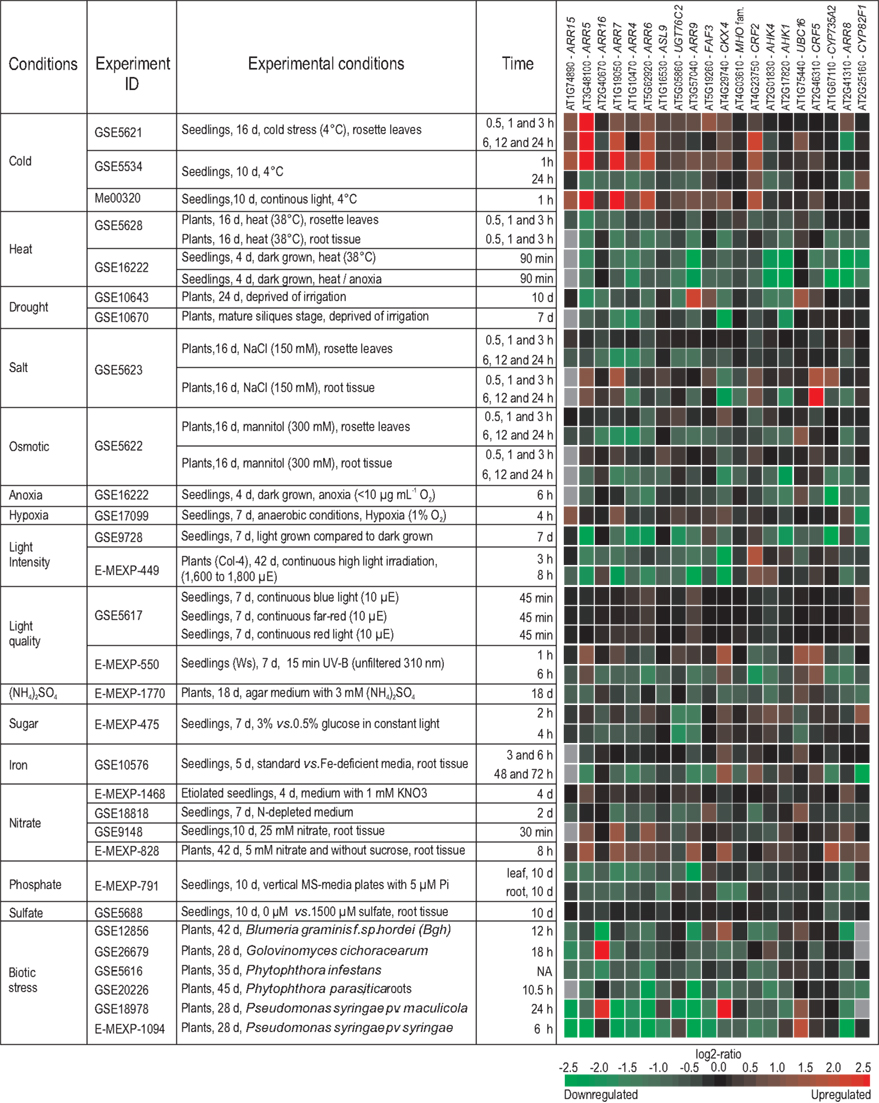
Figure 4. Regulation of cytokinin response genes by different environmental cues. The meta-profile analysis for the changes in transcript abundance of the top 20 cytokinin response genes (Table 2) in response to different stresses or environmental cues was established with the Genevestigator database (http://www.genevestigator.com/gv/index.jsp; Hruz et al., 2008) and modified. The figure shows an arbitrary selection of experiments which was selected based on one of the following criteria: (i) Stress (abiotic and biotic) treatment of wild-type plants; (ii) nutrient changes deprivation) and other treatments of wild-type plants; and (iii) only experiments using Col-0 were considered unless otherwise specified. If data for several time points after the treatments were available, data showing early time points were preferred. The gene expression responses are calculated as log2-ratios between the signal intensities from different stress or nutritional treatments compared to control or mock-treated samples. The resulting heatmap is color coded as indicated, and thus reflects the up- (red color) or downregulation (green color) of genes. A gray color indicates that gene expression was not detectable in both treatment and control conditions. The individual experiments are available from various repositories, such as gene expression omnibus (GEO; Edgar et al., 2002), ArrayExpress (Rocca-Serra et al., 2003), and TAIR (Swarbreck et al., 2008), and can be retrieved by using their unique ID. Abbreviations used in the figure: d, days; vs., versus; Ws, Wassileskija; Col-4, Columbia-4; NA, not available.
The Functional Context of Cytokinin-Regulated Genes
Regulation of the transcript abundance of a given gene can be an indication for its functional relevance in a cytokinin-regulated process. The gene expression profiling studies described a plethora of candidate genes. More than 3000 Arabidopsis genes in total have been described as being regulated by cytokinin. We were interested to know which genes related to which processes would be found independently by different laboratories. We have screened, therefore, the existing publications and show in Table 4 an arbitrary and non-exhaustive compilation of genes that were identified at least three times in the 13 existing studies (see Published Studies on the Cytokinin-Regulated Transcriptome; Table 2). Some of the genes were identified up to 10 times, indicating that they are highly sensitive to a change in the cytokinin status (Table 4). Interestingly, the same cytokinin metabolism genes as in the meta analysis (Table 3), namely CKX4, CYP735A2, and UGT76C2 (see previous chapter), were found most often by different laboratories. This overlap indicates that similar relevant genes might be found by both approaches supporting the validity of the meta analysis (Table 3).
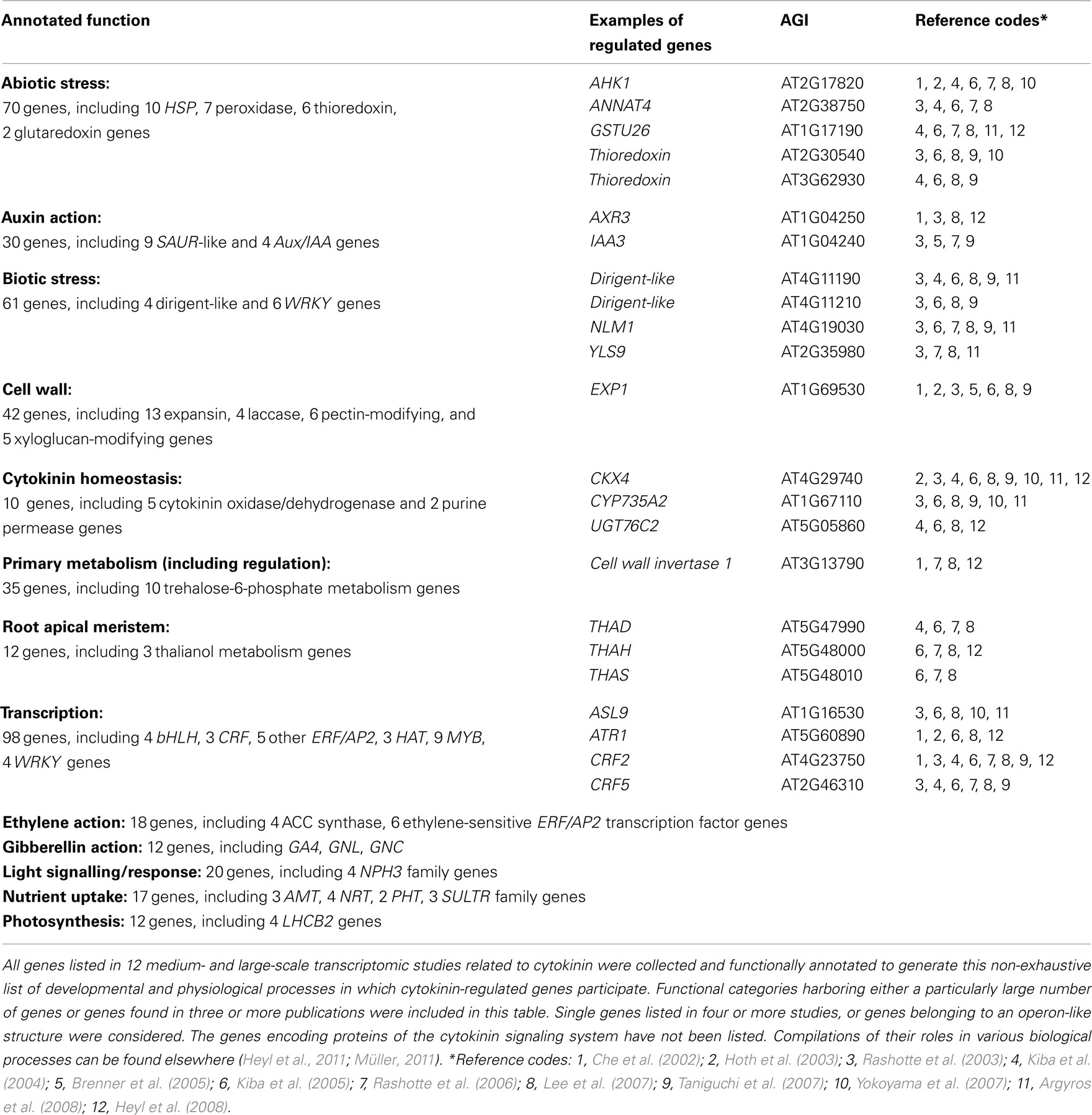
Table 4. Selection of developmental and physiological processes in which cytokinin-regulated genes participate.
A closer inspection of the functional context of the cytokinin-regulated genes has revealed a number of common themes which provide insights into the great variety of cytokinin actions. The processes that were identified range from crosstalk with other hormones, to different events related to growth (e.g., cell expansion, meristem activity), the redox state, and various biotic and abiotic stresses (Table 4). In the following sections, we discuss several of these genes. Detailed discussions of the functional context of some other cytokinin-regulated genes can also be found in several of the transcriptomic studies (Rashotte et al., 2003; Kiba et al., 2004, 2005; Brenner et al., 2005; Brenner and Schmülling, submitted). Importantly, genetic studies have provided evidence in some cases that the indications of a putative functional relevance concluded from cytokinin-sensitive changes of transcript abundance are indeed reflected by a role of the hormone in the respective process.
Crosstalk between cytokinin and other phytohormones
Among the more than 20 auxin-related genes that were found to be regulated by cytokinin are many that are also regulated by auxin, indicating that the auxin–cytokinin crosstalk may often be realized through transcriptional regulation. The list comprises five AUX/IAA genes and eight SAUR-like genes, including SAUR-AC1 (Table 4). However, only two of these, SHY2/IAA3 and AXR3/IAA17, were found in four independent studies. The AUX/IAA genes encode negative regulators of auxin signaling rapidly degraded upon an auxin stimulus through the ubiquitin–proteasome pathway (Dharmasiri et al., 2005; Kepinski and Leyser, 2005). Transcription of SHY2/IAA3 is directly activated by the type-B response regulator ARR1 in the root meristem (Dello Ioio et al., 2008b). Its activation leads to cell differentiation by auxin redistribution caused by its negative effect on genes encoding PIN auxin efflux carriers. Regulation of PIN genes by cytokinin has also been reported by others (Laplaze et al., 2007; Růžicka et al., 2009; Bishopp et al., 2011b,c; Yoshida et al., 2011). The function of AXR3 in auxin signaling is of a more general type. It was reported to have functions in leaf development, apical dominance, several aspects of root development, and crosstalk with brassinosteroids (Leyser et al., 1996; Pérez-Pérez et al., 2010). The role of the SAUR-like genes has remained unclear thus far (Jain et al., 2006), apart from reports about one rice SAUR gene, which acts as a negative regulator of auxin synthesis and transport (Kant and Rothstein, 2009; Kant et al., 2009). No specific role related to cytokinin has been revealed for these genes.
Auxin–cytokinin interaction also plays a role in shoot apical meristem control (Zhao et al., 2010). The absence of meristem control genes, which are known to be regulated by cytokinin, such as CLV1 (Lindsay et al., 2006; Gordon et al., 2009; Bartrina et al., 2011), from the microarray data is probably due to the fact that either their regulation was not detected, or that the transcripts were too low in abundance, both due to the small size of their respective expression domains in comparison to the tissues which were sampled for the transcriptome studies. Similarly, cytokinin-regulated auxin genes involved in the patterning of the root vasculature, where auxin and cytokinin act in a signaling loop causing the mutual restriction of the active domains of the respective other hormone (Bishopp et al., 2011b,c), were missed, probably for the same reasons. A possible exception is FAF3, a novel gene which was discussed in Section “A Meta Analysis of Publicly Available Microarray Data Sets.”
Cytokinin also interacts with ethylene. It causes the upregulation of four aminocyclopropane-1-carboxylate synthase (ACS) genes involved in ethylene biosynthesis, as well as six ethylene-sensitive transcription factor genes of the ERF/AP2 family. However, none of these genes has been identified in more than two different studies. Whether the upregulation of the transcription factors is directly caused by cytokinin, or whether it is indirectly mediated by increased ethylene levels due to the induction of ACS genes, cannot be decided based on the array experiments summarized here. It is known that part of the cytokinin response is mediated through ethylene as a second messenger, such as the regulation of root architecture (Cary et al., 1995; Laplaze et al., 2007; Růžicka et al., 2009).
The interaction of cytokinin with gibberellins (GA) is reflected on the molecular level by the regulation of two GATA transcription factor genes, GNL/CGA1/GATA22 and GNC/GATA21. It was shown in a recent genetic study that both GNL and GNC are direct targets of the GA signaling pathway, and that both negatively regulate germination, greening, leaf expansion, and flowering time (Richter et al., 2010). Additionally, GNL seems to have a role in chloroplast development, as GNL overexpressors developed chloroplasts in illuminated roots, while a gnl mutant formed fewer chloroplasts in the hypocotyl of seedlings. It has been proposed that GNL integrates signaling pathways of light, GA, and cytokinin (Köllmer et al., 2011). Cytokinin also appears to interfere with GA activity by regulating the GA synthesis gene GA4 and the GA signaling genes RGA and GAI, both encoding DELLA repressor proteins. Together, cytokinin action caused a reduction of GA activity (Brenner et al., 2005).
Cytokinin affects different parts of the plant cell
One of the most frequently listed genes in the present microarray studies encodes the expansin EXP1. Twelve other expansin genes were found in at least two microarray studies, along with 18 other cell wall-related genes (Table 4). Expansins are thought to be involved in weakening the structure of the cell wall, which is required for cell expansion, and thus growth. Links between cytokinin and cell wall-related genes have been found previously (Downes and Crowell, 1998). A cytokinin-induced change of wall extensibility has also been directly measured (Thomas et al., 1981), and a negative influence of cytokinin signaling on cell wall thickness has been reported (Jung et al., 2008). However, detailed investigations on the role of EXP1 and other cell wall-related genes in growth regulation by cytokinin have not yet been published.
Cytokinin has been known to have a positive effect on chloroplast development, performance, and longevity for a long time (Staetler and Laetsch, 1965; Parthier, 1979; Chory et al., 1994; Yaronskaya et al., 2006; Okazaki et al., 2009; Brenner and Schmülling, submitted), and may thus influence photosynthesis. It affects the transcript abundance of genes encoding plastid-localized proteins (Zubo et al., 2008), and a proteomic study identified about 50% of the cytokinin-regulated proteins to be localized in the chloroplast (Cerný et al., 2011). Furthermore, cytokinin appears to exert a protective activity on biochemical processes associated with photosynthesis (Rivero et al., 2009). Four of the six genes found to be induced by cytokinin in at least two publications belong to the LHCB2 family, encoding components of the light harvesting complexes. This is in accordance with the influence of cytokinin on the relative amount of light harvesting complexes (Šiffel et al., 1992; Catský et al., 1993) and on the accumulation of light harvesting complex mRNAs and proteins (Marziani Longo et al., 1990).
Cytokinin as a regulator of metabolism genes
Several genes annotated as linked to root growth but rarely considered in cytokinin-related studies were identified. The THAD, THAH, and THAS genes are required for the synthesis of thalianol, which is a triterpene of unknown function (Field and Osbourn, 2008). The genes are organized in an operon-like fashion (Field and Osbourn, 2008). The operon includes another gene encoding an acyltransferase of unknown function (At5g47980) which has also been found in numerous microarray studies to be regulated by cytokinin (Kiba et al., 2004, 2005; Rashotte et al., 2006; Lee et al., 2007; Argyros et al., 2008; Heyl et al., 2008). Interestingly, plants with a higher thalianol content show a stunted shoot growth but have longer roots (Field and Osbourn, 2008), both phenotypic features reminiscent of cytokinin-deficient plants (Werner et al., 2003). However, the exact role of thalianol in regulating root and shoot growth and its possible interplay with cytokinin is unknown.
A function of cytokinin in regulating uptake and metabolism of different nutrients – nitrogen, sulfate, phosphate, and iron – has been known for some time (Sakakibara et al., 2006; Ribot et al., 2008; Argueso et al., 2009; Igarashi et al., 2009; Rubio et al., 2009). This is supported by the regulation of nitrogen (AMT, NRT), sulfate (SULTR), and phosphate (PHT) transporter genes by the hormone (Table 4). In the case of nitrate, genes encoding both high-affinity (NRT1) and low-affinity (NRT2) transporters (Crawford and Glass, 1998) are regulated by cytokinin. NRT1.1 – also cytokinin-regulated – functions as a nitrate sensor (Ho et al., 2009), suggesting an influence of cytokinin on nitrate sensing. The reciprocal signaling relations between nitrogen and cytokinin were reviewed recently (Kiba et al., 2011). One of the three ammonium transporters, AMT1;3, has a major role in ammonium-triggered lateral root branching (Lima et al., 2010). The sulfate transporter SULTR1;2 is responsible for sulfate uptake into the roots, while the exact role of the other two sulfate transporters, SULTR3;1 and SULTR3;4, is not yet clear. SULTR1;2 and SULTR1;1 were shown to be negatively regulated by cytokinin in a partially CRE1/AHK4-dependent manner, associated by a decrease in sulfate uptake (Maruyama-Nakashita et al., 2004). The functions of the phosphate transporters PHT1;2 and PHT1;4 are not restricted to phosphate uptake from the soil into the root, but extend to broader roles in phosphate remobilization and distribution throughout the whole plant (Mudge et al., 2002).
Nineteen genes involved in primary metabolism and its regulation were identified, however, only the one encoding cell wall invertase 1 was found repeatedly (Table 4). Cell wall invertases provide hexose sugars to cells by cleavage of sucrose in the cell wall. The provision of either hexoses or sucrose has a regulatory potential for source–sink relationships and carbon utilization, as well as for development (Sherson et al., 2003). Total cell wall invertase activity was shown to be reduced in the source leaves of cytokinin-deficient plants (Werner et al., 2008), and cell wall invertase 1 expression was consistently found to be increased by cytokinin in the transcriptomics studies summarized here. Ten of the 19 genes assigned to the category primary metabolism are involved in trehalose-6-phosphate metabolism. Trehalose-6-phosphate is also regulated by source–sink relationships and carbon utilization, and plants with a disturbed trehalose-6-phosphate homeostasis show severe developmental and metabolic defects (Schluepmann et al., 2003, 2004; Dijken et al., 2004). It was shown that trehalose synthesis genes are regulated by cytokinin in an opposite fashion than genes encoding trehalose-degrading enzymes, supporting the idea that trehalose-6-phosphate levels are regulated, at least partly, by cytokinin (Brenner et al., 2005). In summary, the gene regulatory data indicate that cytokinin action on source–sink relationships and carbon utilization could be accomplished through the regulation of cell wall invertase and trehalose-6-phosphate metabolism genes.
Cytokinin and light
Cytokinin regulates the transcript levels of four NPH3-like genes, which have an annotated function in light signaling. However, NPH3 proteins are involved in various signaling pathways. Its founding member interacts with the blue light receptor NPH1/PHOT1 and with the E3 ubiquitin ligase component CUL3; the resulting degradation contributes to photomorphogenesis and leaf positioning (Motchoulski and Liscum, 1999; Pedmale and Liscum, 2007; Inoue et al., 2008). Five other NPH3-like genes have been linked with auxin-regulated organogenesis acting through influencing the polar localization of PIN proteins (Treml et al., 2005; Cheng et al., 2007; Furutani et al., 2007, 2011). The four NPH3-like genes found repeatedly to be regulated by cytokinin, two of which are closely related phylogenetically, have not yet been functionally described.
The transcript level of three genes encoding members of the phytochrome signaling pathway is regulated by cytokinin in the combined microarray dataset: PHYA, the phytochrome kinase substrate PKS1, and the GRAS transcription factor gene PAT1. PHYA has functions in far-red-dependent de-etiolation, day-length sensing, internode and leaf growth, and shade avoidance (Schepens et al., 2004). PKS1 is a negative regulator of phytochrome signaling (Fankhauser et al., 1999; Lariguet et al., 2003) and is also necessary for blue light-induced phototropism in a phyA-dependent manner (Lariguet et al., 2006; Boccalandro et al., 2008). PAT1 is a member of the transcription factor GRAS family and specifically involved in an early stage of phyA signaling (Bolle et al., 2000). The transcriptional regulation of genes involved in phytochrome signaling pathways is consistent with known links between cytokinin and light responses (Argueso et al., 2009; Werner and Schmülling, 2009; Yoshida et al., 2011).
Cytokinin regulation of transcription factor genes
Gene expression studies have revealed very early that cytokinin acts downstream of the TCS through activating transcriptional cascades (Rashotte et al., 2003; Brenner et al., 2005), and several studies have identified functionally relevant cytokinin-regulated transcription factor genes (Argueso et al., 2010). Other cytokinin-regulated transcription factor genes are still looking for a function (CRF2, CRF5; see above), or have, so far, only be studied by overexpression, such as ASL9 (Naito et al., 2007), bHLH64, HAT4, HAT22, and GATA22/CGA1/GNL (Köllmer et al., 2011). Cytokinin regulates the transcript levels of many other transcription factor genes, and seven of them have been detected in at least four publications describing transcriptomic studies (Table 4). No function is yet known for most of them, such as the zinc finger transcription factor At1g68360, which was found to be also regulated by auxin (Pufky et al., 2003) and cold stress (Vergnolle et al., 2005), or the B-box zinc finger transcription factor STH7 (Datta et al., 2008), which responded to molecules in wildfire smoke that stimulate seed germination (Nelson et al., 2010). The bZIP transcription factor BZIP11 (formerly known as ATB2) is interesting because it has been proposed to be a powerful regulator of carbohydrate metabolism that functions in a growth regulatory network including trehalose-6-phosphate (see Cytokinin as a Regulator of Metabolism Genes; Ma et al., 2011). BZIP11 controls the sink strength by regulating the expression of genes in young sink tissue (Delatte et al., 2011). In addition, it has been shown to control two genes encoding enzymes in amino acid metabolism, ASN1 and ProDH2. ATR1 encodes a transcription factor of the MYB family, which is the key regulator of genes involved in the biosynthesis of glucosinolates and auxin from tryptophane (Celenza et al., 2005). Its regulation by cytokinin provides another facet to the cytokinin–auxin crosstalk. TZF1 encodes a zinc finger protein which has been identified as a regulated gene in studies on the carbon status (Contento et al., 2004; Lin et al., 2011), disease and insect defense (Cartieaux et al., 2003; Ascencio-Ibáñez et al., 2008), stress tolerance (Kasukabe et al., 2004; Lin et al., 2011), development of male and female reproductive organs (Alves-Ferreira et al., 2007; Skinner and Gasser, 2009), and ABA and GA signaling (Lin et al., 2011). Its mode of action is currently under investigation (Pomeranz et al., 2011)
A role for cytokinin in the stress response
Many cytokinin-responsive genes are also regulated by different abiotic and biotic stresses (Figure 4). One important type of abiotic stress influenced by cytokinin appears to be oxidative stress, as many genes encoding enzymes involved in relieving oxidative stress – peroxidases, thioredoxins, and glutaredoxins – were found to be regulated by the hormone in different microarray studies (Table 4). A beneficial effect of cytokinin in coping with oxidative stress has been described (Zavaleta-Mancera et al., 2007). Two thioredoxin genes, At2g30540 and At3g62930, were particularly often named in the publications listed in Table 4. They were consistently found among the most strongly cytokinin-regulated genes in the meta analysis, namely at position 24 and 25, respectively (Table 3). No precise function has yet been assigned to these genes. Generally, thioredoxins regulate the redox status of disulfide bonds of their target proteins, thereby supplying reducing power for reductases, which detoxify lipid hydroperoxides or repair damaged proteins. In addition to these functions, they may have roles in oxidative stress signaling or as regulators of scavenging mechanisms (Vieira Dos Santos and Rey, 2006). Three other genes related to abiotic stress, AHK1, ANNAT4, and GSTU26, were listed in 6 or 7 of the 13 cytokinin-related transcriptomics studies, suggesting a relevant influence of cytokinin on their related field of action. AHK1 encodes a histidine kinase receptor structurally related to the cytokinin receptors and functions as a positive regulator of drought and salt stress responses and ABA signaling (Tran et al., 2007). ANNAT4 encodes an annexin, a calcium-dependent membrane-binding protein that mediates osmotic stress and abscisic acid signal transduction in Arabidopsis (Lee et al., 2004). The glutathione S-transferase gene GSTU26 is also induced by low temperature (Nutricati et al., 2006).
A link between cytokinin and biotic stress has been suggested because altered cytokinin levels have been observed after pathogen infection (Clarke et al., 1999; Maksimov et al., 2002) and an altered susceptibility to pathogens due to an altered cytokinin status has been observed (Clarke et al., 1998; Pertry et al., 2009). Therefore, it was not surprising that several defense-related genes were found to be regulated by cytokinin in multiple transcriptomic studies, among them three dirigent-like proteins. They are known to be transcriptionally induced by biotic stress (Fristensky et al., 1985; Görlach et al., 1996; Lee et al., 1996; Ralph et al., 2006) and participate as mediators of stereospecificity in the formation of lignans (Davin et al., 1997; Burlat et al., 2001), which possess antiviral, antimicrobial, and cytotoxic properties (MacRae and Towers, 1984; Mendonça Pauletti et al., 2000; Rahman and Gray, 2002). NLM1, also known as NIP1;1, encodes an aquaporin homolog that determines antimonite and arsenite sensitivity (Kamiya and Fujiwara, 2009; Kamiya et al., 2009) and also responds to auxin and osmotic stress (Kreps et al., 2002; Goda et al., 2004). YLS9 is induced in response to various biotic triggers, including flagellin and the oviposition of butterfly eggs (Navarro et al., 2004; Michel et al., 2006; Little et al., 2007; Zhao et al., 2007).
Future Prospects
A wealth of information has been generated by transcriptomic studies creating new hypotheses about the functions of cytokinin and functional mechanisms of its action. Numerous interesting areas deserve further investigation, such as the regulation of ribosomal genes by cytokinin suggesting translational control of cytokinin-induced changes (Brenner et al., 2005; Kiba et al., 2005; Rashotte and Goertzen, 2010; Brenner and Schmülling, submitted). Several developmental processes are affected by mutations in ribosomal genes, including embryogenesis, root growth, and leaf development. This points out the functional relevance of the specific assembly of ribosomal proteins in addition to their fundamental role in translation, presumably because they have an effect on the translation of specific proteins (Weijers et al., 2001; Byrne, 2009; Horiguchi et al., 2011). With the dawn of the era of next-generation sequencing-based transcriptomics, more data can be expected from species whose genomes have not been sequenced thus far. Data from crop species will be of special interest for applied research given the potential of manipulating the cytokinin status to achieve yield increase (Ashikari et al., 2005; Bartrina et al., 2011) or to ameliorate other plant traits relevant for agricultural performance (Werner et al., 2010; Zalewski et al., 2010). In addition, experimental limitations of the hybridization-based microarray platform, especially regarding annotation dependency, sensitivity, dynamic range, and reproducibility, will be overcome. With the new technologies, the transcriptome of smaller samples can be evaluated, detecting smaller changes in gene expression with higher confidence. Therefore, we are looking forward to an exciting expansion of high-fidelity transcriptomic knowledge across many species. New tools to master the challenge of making proper use of the huge body of data generated will certainly facilitate an understanding of the functions of the networks discovered.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was funded by the DFG grant Schm 814/22-1 within the framework of the Arabidopsis Functional Genomics Network (AFGN).
References
Aerts, S., Van Loo, P., Thijs, G., Mayer, H., de Martin, R., Moreau, Y., and De Moor, B. (2005). TOUCAN 2: the all-inclusive open source workbench for regulatory sequence analysis. Nucleic Acids Res. 33, W393–W396.
Alves-Ferreira, M., Wellmer, F., Banhara, A., Kumar, V., Riechmann, J. L., and Meyerowitz, E. M. (2007). Global expression profiling applied to the analysis of Arabidopsis stamen development. Plant Physiol. 145, 747–762.
Argueso, C. T., Ferreira, F. J., and Kieber, J. J. (2009). Environmental perception avenues: the interaction of cytokinin and environmental response pathways. Plant Cell Environ. 32, 1147–1160.
Argueso, C. T., Raines, T., and Kieber, J. J. (2010). Cytokinin signaling and transcriptional networks. Curr. Opin. Plant Biol. 13, 533–539.
Argyros, R. D., Mathews, D. E., Chiang, Y.-H., Palmer, C. M., Thibault, D. M., Etheridge, N., Argyros, D. A., Mason, M. G., Kieber, J. J., and Schaller, G. E. (2008). Type B response regulators of Arabidopsis play key roles in cytokinin signaling and plant development. Plant Cell 20, 2102–2116.
Ascencio-Ibáñez, J. T., Sozzani, R., Lee, T.-J., Chu, T.-M., Wolfinger, R. D., Cella, R., and Hanley-Bowdoin, L. (2008). Global analysis of Arabidopsis gene expression uncovers a complex array of changes impacting pathogen response and cell cycle during geminivirus infection. Plant Physiol. 148, 436–454.
Ashikari, M., Sakakibara, H., Lin, S., Yamamoto, T., Takashi, T., Nishimura, A., Angeles, E. R., Qian, Q., Kitano, H., and Matsuoka, M. (2005). Cytokinin oxidase regulates rice grain production. Science 309, 741–745.
Bartrina, I., Otto, E., Strnad, M., Werner, T., and Schmülling, T. (2011). Cytokinin regulates the activity of reproductive meristems, flower organ size, ovule formation, and thus seed yield in Arabidopsis thaliana. Plant Cell 23, 69–80.
Bishopp, A., Benková, E., and Helariutta, Y. (2011a). Sending mixed messages: auxin-cytokinin crosstalk in roots. Curr. Opin. Plant Biol. 14, 10–16.
Bishopp, A., Help, H., El-Showk, S., Weijers, D., Scheres, B., Friml, J., Benková, E., Mähönen, A. P., and Helariutta, Y. (2011b). A mutually inhibitory interaction between auxin and cytokinin specifies vascular pattern in roots. Curr. Biol. 21, 917–926.
Bishopp, A., Lehesranta, S., Vatén, A., Help, H., El-Showk, S., Scheres, B., Helariutta, K., Mähönen, A. P., Sakakibara, H., and Helariutta, Y. (2011c). Phloem-transported cytokinin regulates polar auxin transport and maintains vascular pattern in the root meristem. Curr. Biol. 21, 927–932.
Bishopp, A., Help, H., Helariutta, Y., and Kwang, W. J. (2009). “Cytokinin signaling during root development,” in International Review of Cell and Molecular Biology, ed. K. Jeon (San Diego: Academic Press), 1–48.
Boccalandro, H. E., De Simone, S. N., Bergmann-Honsberger, A., Schepens, I., Fankhauser, C., and Casal, J. J. (2008). PHYTOCHROME KINASE SUBSTRATE1 regulates root phototropism and gravitropism. Plant Physiol. 146, 108–115.
Bolle, C., Koncz, C., and Chua, N.-H. (2000). PAT1, a new member of the GRAS family, is involved in phytochrome A signal transduction. Genes Dev. 14, 1269–1278.
Brandstatter, I., and Kieber, J. J. (1998). Two genes with similarity to bacterial response regulators are rapidly and specifically induced by cytokinin in Arabidopsis. Plant Cell 10, 1009–1020.
Brazma, A., Hingamp, P., Quackenbush, J., Sherlock, G., Spellman, P., Stoeckert, C., Aach, J., Ansorge, W., Ball, C. A., Causton, H. C., Gaasterland, T., Glenisson, P., Holstege, F. C. P., Kim, I. F., Markowitz, V., Matese, J. C., Parkinson, H., Robinson, A., Sarkans, U., Steffen, S.-K., Stewart, J., Taylor, R., Vilo, J., and Vingron, M. (2001). Minimum information about a microarray experiment (MIAME): toward standards for microarray data. Nat. Genet. 29, 365–371.
Brenner, W. G., Romanov, G. A., Köllmer, I., Bürkle, L., and Schmülling, T. (2005). Immediate-early and delayed cytokinin response genes of Arabidopsis thaliana identified by genome-wide expression profiling reveal novel cytokinin-sensitive processes and suggest cytokinin action through transcriptional cascades. Plant J. 44, 314–333.
Burlat, V., Kwon, M., Davin, L. B., and Lewis, N. G. (2001). Dirigent proteins and dirigent sites in lignifying tissues. Phytochemistry 57, 883–897.
Cartieaux, F., Thibaud, M.-C., Zimmerli, L., Lessard, P., Sarrobert, C., David, P., Gerbaud, A., Robaglia, C., Somerville, S., and Nussaume, L. (2003). Transcriptome analysis of Arabidopsis colonized by a plant-growth promoting rhizobacterium reveals a general effect on disease resistance. Plant J. 36, 177–188.
Cary, A. J., Liu, W., and Howell, S. H. (1995). Cytokinin action is coupled to ethylene in its effects on the inhibition of root and hypocotyl elongation in Arabidopsis thaliana seedlings. Plant Physiol. 107, 1075–1082.
Celenza, J. L., Quiel, J. A., Smolen, G. A., Merrikh, H., Silvestro, A. R., Normanly, J., and Bender, J. (2005). The Arabidopsis ATR1 Myb transcription factor controls indolic glucosinolate homeostasis. Plant Physiol. 137, 253–262.
Che, P., Gingerich, D. J., Lall, S., and Howell, S. H. (2002). Global and hormone-induced gene expression changes during shoot development in Arabidopsis. Plant Cell 14, 2771–2785.
Cheng, Y., Qin, G., Dai, X., and Zhao, Y. (2007). NPY1, a BTB-NPH3-like protein, plays a critical role in auxin-regulated organogenesis in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 104, 18825–18829.
Chory, J., Reinecke, D., Sim, S., Washburn, T., and Brenner, M. (1994). A role for cytokinins in de-etiolation in Arabidopsis. Plant Physiol. 104, 339–347.
Clarke, S. F., Burritt, D. J., Jameson, P. E., and Guy, P. L. (1998). Influence of plant hormones on virus replication and pathogenesis-related proteins in Phaseolus vulgaris L. infected with white clover mosaic potexvirus. Physiol. Mol. Plant Pathol. 53, 195–207.
Clarke, S. F., McKenzie, M. J., Burritt, D. J., Guy, P. L., and Jameson, P. E. (1999). Influence of white clover mosaic potexvirus infection on the endogenous cytokinin content of bean. Plant Physiol. 120, 547–552.
Contento, A. L., Kim, S.-J., and Bassham, D. C. (2004). Transcriptome profiling of the response of Arabidopsis suspension culture cells to suc starvation. Plant Physiol. 135, 2330–2347.
Craigon, D. J., James, N., Okyere, J., Higgins, J., Jotham, J., and May, S. (2004). NASCArrays: a repository for microarray data generated by NASC’s transcriptomics service. Nucleic Acids Res. 32, D575–D577.
Crawford, N. M., and Glass, A. D. M. (1998). Molecular and physiological aspects of nitrate uptake in plants. Trends Plant Sci. 3, 389–395.
Cutcliffe, J. W., Hellmann, E., Heyl, A., and Rashotte, A. M. (2011). CRFs form protein–protein interactions with each other and with members of the cytokinin signalling pathway in Arabidopsis via the CRF domain. J. Exp. Bot. 62, 4995–5002.
D’Agostino, I. B., Deruere, J., and Kieber, J. J. (2000). Characterization of the response of the Arabidopsis response regulator gene family to cytokinin. Plant Physiol. 124, 1706–1717.
Datta, S., Johansson, H., Hettiarachchi, C., Irigoyen, M. L., Desai, M., Rubio, V., and Holm, M. (2008). LZF1/SALT TOLERANCE HOMOLOG3, an Arabidopsis B-box protein involved in light-dependent development and gene expression, undergoes cop1-mediated ubiquitination. Plant Cell 20, 2324–2338.
Davin, L. B., Wang, H.-B., Crowell, A. L., Bedgar, D. L., Martin, D. M., Sarkanen, S., and Lewis, N. G. (1997). Stereoselective bimolecular phenoxy radical coupling by an auxiliary (dirigent) protein without an active center. Science 275, 362–367.
Day, R. C., Herridge, R. P., Ambrose, B. A., and Macknight, R. C. (2008). Transcriptome analysis of proliferating Arabidopsis endosperm reveals biological implications for the control of syncytial division, cytokinin signaling, and gene expression regulation. Plant Physiol. 148, 1964–1984.
Delatte, T. L., Sedijani, P., Kondou, Y., Matsui, M., de Jong, G. J., Somsen, G. W., Wiese-Klinkenberg, A., Primavesi, L. F., Paul, M. J., and Schluepmann, H. (2011). Growth arrest by trehalose-6-phosphate: an astonishing case of primary metabolite control over growth by way of the SnRK1 signaling pathway. Plant Physiol. 157, 160–174.
Dello Ioio, R., Linhares, F. S., and Sabatini, S. (2008a). Emerging role of cytokinin as a regulator of cellular differentiation. Curr. Opin. Plant Biol. 11, 23–27.
Dello Ioio, R., Nakamura, K., Moubayidin, L., Perilli, S., Taniguchi, M., Morita, M. T., Aoyama, T., Costantino, P., and Sabatini, S. (2008b). A genetic framework for the control of cell division and differentiation in the root meristem. Science 322, 1380–1384.
Dharmasiri, N., Dharmasiri, S., and Estelle, M. (2005). The F-box protein TIR1 is an auxin receptor. Nature 435, 441–445.
Dijken, A. J. H. V., Schluepmann, H., and Smeekens, S. C. M. (2004). Arabidopsis trehalose-6-phosphate synthase 1 is essential for normal vegetative growth and transition to flowering. Plant Physiol. 135, 969–977.
Domagalska, M. A., and Leyser, O. (2011). Signal integration in the control of shoot branching. Nat. Rev. Mol. Cell Biol. 12, 211–221.
Dortay, H., Mehnert, N., Bürkle, L., Schmülling, T., and Heyl, A. (2006). Analysis of protein interactions within the cytokinin-signaling pathway of Arabidopsis thaliana. FEBS J. 273, 4631–4644.
Downes, B. P., and Crowell, D. N. (1998). Cytokinin regulates the expression of a soybean β-expansin gene by a post-transcriptional mechanism. Plant Mol. Biol. 37, 437–444.
Edgar, R., Domrachev, M., and Lash, A. E. (2002). Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210.
Fankhauser, C., Yeh, K. C., Lagarias, J. C., Zhang, H., Elich, T. D., and Chory, J. (1999). PKS1, a substrate phosphorylated by phytochrome that modulates light signaling in Arabidopsis. Science 284, 1539–1541.
Field, B., and Osbourn, A. E. (2008). Metabolic diversification –independent assembly of operon-like gene clusters in different plants. Science 320, 543–547.
Fristensky, B., Riggleman, R. C., Wagoner, W., and Hadwiger, L. A. (1985). Gene expression in susceptible and disease resistant interactions of peas induced with Fusarium solani pathogens and chitosan. Physiol. Mol. Plant Pathol. 27, 15–28.
Fukumura, R., Takahashi, H., Saito, T., Tsutsumi, Y., Fujimori, A., Sato, S., Tatsumi, K., Araki, R., and Abe, M. (2003). A sensitive transcriptome analysis method that can detect unknown transcripts. Nucleic Acids Res. 31, e94.
Furutani, M., Kajiwara, T., Kato, T., Treml, B. S., Stockum, C., Torres-Ruiz, R. N. A., and Tasaka, M. (2007). The gene MACCHI-BOU 4/ENHANCER OF PINOID encodes a NPH3-like protein and reveals similarities between organogenesis and phototropism at the molecular level. Development 134, 3849–3859.
Furutani, M., Sakamoto, N., Yoshida, S., Kajiwara, T., Robert, H. S., Friml, J., and Tasaka, M. (2011). Polar-localized NPH3-like proteins regulate polarity and endocytosis of PIN-FORMED auxin efflux carriers. Development 138, 2069–2078.
Fusada, N., Masuda, T., Kuroda, H., Shimada, H., Ohta, H., and Takamiya, K.-I. (2005). Identification of a novel cis-element exhibiting cytokinin-dependent protein binding in vitro; in the 5’-region of NADPH-protochlorophyllide oxidoreductase gene in cucumber. Plant Mol. Biol. 59, 631–645.
Goda, H., Sasaki, E., Akiyama, K., Maruyama-Nakashita, A., Nakabayashi, K., Li, W., Ogawa, M., Yamauchi, Y., Preston, J., Aoki, K., Kiba, T., Takatsuto, S., Fujioka, S., Asami, T., Nakano, T., Kato, H., Mizuno, T., Sakakibara, H., Yamaguchi, S., Nambara, E., Kamiya, Y., Takahashi, H., Hirai, M. Y., Sakurai, T., Shinozaki, K., Saito, K., Yoshida, S., and Shimada, Y. (2008). The AtGenExpress hormone and chemical treatment data set: experimental design, data evaluation, model data analysis and data access. Plant J. 55, 526–542.
Goda, H., Sawa, S., Asami, T., Fujioka, S., Shimada, Y., and Yoshida, S. (2004). Comprehensive comparison of auxin-regulated and brassinosteroid-regulated genes in Arabidopsis. Plant Physiol. 134, 1555–1573.
Gordon, S. P., Chickarmane, V. S., Ohno, C., and Meyerowitz, E. M. (2009). Multiple feedback loops through cytokinin signaling control stem cell number within the Arabidopsis shoot meristem. Proc. Natl. Acad. Sci. U.S.A. 106, 16529–16534.
Görlach, J., Volrath, S., Knauf-Beiter, G., Hengy, G., Beckhove, U., Kogel, K. H., Oostendorp, M., Staub, T., Ward, E., Kessmann, H., and Ryals, J. (1996). Benzothiadiazole, a novel class of inducers of systemic acquired resistance, activates gene expression and disease resistance in wheat. Plant Cell 8, 629–643.
Hare, P. D., Cress, W. A., and van Staden, J. (1997). The involvement of cytokinins in plant responses to environmental stress. Plant Growth Regul. 23, 79–103.
Hass, C., Lohrmann, J., Albrecht, V., Sweere, U., Hummel, F., Yoo, S. D., Hwang, I., Zhu, T., Schäfer, E., Kudla, J., and Harter, K. (2004). The response regulator 2 mediates ethylene signalling and hormone signal integration in Arabidopsis. EMBO J. 23, 3290–3302.
Hellmann, E., Gruhn, N., and Heyl, A. (2010). The more, the merrier: cytokinin signaling beyond Arabidopsis. Plant Signal. Behav. 5, 1384–1390.
Heyl, A., Ramireddy, E., Brenner, W. G., Riefler, M., Allemeersch, J., and Schmülling, T. (2008). The transcriptional repressor ARR1-SRDX suppresses pleiotropic cytokinin activities in Arabidopsis. Plant Physiol. 147, 1380–1395.
Heyl, A., Riefler, M., Romanov, G. A., and Schmülling, T. (2011). Properties, functions and evolution of cytokinin receptors. Eur. J. Cell Biol. (in press). doi: 10.1016/j.ejcb.2011.1002.1009.
Heyl, A., Werner, T., and Schmülling, T. (eds). (2006). “Cytokinin metabolism and signal transduction,” in Plant Hormone Signaling, Annual Plant Reviews, eds P. Hedden, and S. Thomas (Oxford: Blackwell Publishing), 93–123.
Hiratsu, K., Matsui, K., Koyama, T., and Ohme-Takagi, M. (2003). Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 34, 733–739.
Ho, C.-H., Lin, S.-H., Hu, H.-C., and Tsay, Y.-F. (2009). CHL1 functions as a nitrate sensor in plants. Cell 138, 1184–1194.
Horák, J., Brzobohatý, B., and Lexa, M. (2003). Molecular and physiological characterisation of an insertion mutant in the ARR21 putative response regulator gene from Arabidopsis thaliana. Plant Biol. 5, 245–254.
Horiguchi, G., Mollá-Morales, A., Pérez-Pérez, J. M., Kojima, K., Robles, P., Ponce, M. R., Micol, J. L., and Tsukaya, H. (2011). Differential contributions of ribosomal protein genes to Arabidopsis thaliana leaf development. Plant J. 65, 724–736.
Hosoda, K., Imamura, A., Katoh, E., Hatta, T., Tachiki, M., Yamada, H., Mizuno, T., and Yamazaki, T. (2002). Molecular structure of the GARP family of plant Myb-related DNA binding motifs of the Arabidopsis response regulators. Plant Cell 14, 2015–2029.
Hoth, S., Ikeda, Y., Morgante, M., Wang, X., Zuo, J., Hanafey, M. K., Gaasterland, T., Tingey, S. V., and Chua, N. H. (2003). Monitoring genome-wide changes in gene expression in response to endogenous cytokinin reveals targets in Arabidopsis thaliana. FEBS Lett. 554, 373–380.
Hou, B., Lim, E. K., Higgins, G. S., and Bowles, D. J. (2004). N-glucosylation of cytokinins by glycosyltransferases of Arabidopsis thaliana. J. Biol. Chem. 279, 47822–47832.
Hruz, T., Laule, O., Szabo, G., Wessendorp, F., Bleuler, S., Oertle, L., Widmayer, P., Gruissem, W., and Zimmermann, P. (2008). Genevestigator V3: a reference expression database for the meta-analysis of transcriptomes. Adv. Bioinformatics 2008, Article ID 420747.
Hunt, P. W., Watts, R. A., Trevaskis, B., Llewelyn, D. J., Burnell, J., Dennis, E. S., and Peacock, W. J. (2001). Expression and evolution of functionally distinct haemoglobin genes in plants. Plant Mol. Biol. 47, 677–692.
Hwang, I., and Sheen, J. (2001). Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature 413, 383–389.
Igarashi, D., Izumi, Y., Dokiya, Y., Totsuka, K., Fukusaki, E., and Ohsumi, C. (2009). Reproductive organs regulate leaf nitrogen metabolism mediated by cytokinin signal. Planta 229, 633–644.
Imamura, A., Hanaki, N., Nakamura, A., Suzuki, T., Taniguchi, M., Kiba, T., Ueguchi, C., Sugiyama, T., and Mizuno, T. (1999). Compilation and characterization of Arabidopsis thaliana response regulators implicated in His-Asp phosphorelay signal transduction. Plant Cell Physiol. 40, 733–742.
Imamura, A., Hanaki, N., Umeda, H., Nakamura, A., Suzuki, T., Ueguchi, C., and Mizuno, T. (1998). Response regulators implicated in His-to-Asp phosphotransfer signaling in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 95, 2691–2696.
Imamura, A., Kiba, T., Tajima, Y., Yamashino, T., and Mizuno, T. (2003). In vivo and in vitro characterization of the ARR11 response regulator implicated in the His-to-Asp phosphorelay signal transduction in Arabidopsis thaliana. Plant Cell Physiol. 44, 122–131.
Inoue, S.-I., Kinoshita, T., Takemiya, A., Doi, M., and Shimazaki, K.-I. (2008). Leaf positioning of Arabidopsis in response to blue light. Mol. Plant 1, 15–26.
Inoue, T., Higuchi, M., Hashimoto, Y., Seki, M., Kobayashi, M., Kato, T., Tabata, S., Shinozaki, K., and Kakimoto, T. (2001). Identification of CRE1 as a cytokinin receptor from Arabidopsis. Nature 409, 1060–1063.
Irizarry, R. A., Gautier, L., Bolstad, B. M., Miller, C., Astrand, M., Cope, L. M., Gentleman, R., Gentry, J., Halling, C., Huber, W., MacDonald, J., Rubinstein, B. I. P., Workman, C., and Zhang, J. (2005). affy: Methods for Affymetrix Oligonucleotide Arrays. R Package Version 1.8.1.
Ishida, K., Yamashino, T., Yokoyama, A., and Mizuno, T. (2008). Three type-B response regulators, ARR1, ARR10 and ARR12, play essential but redundant roles in cytokinin signal transduction throughout the life cycle of Arabidopsis thaliana. Plant Cell Physiol. 49, 47–57.
Jain, M., Tyagi, A. K., and Khurana, J. P. (2006). Genome-wide analysis, evolutionary expansion, and expression of early auxin-responsive SAUR gene family in rice (Oryza sativa). Genomics 88, 360–371.
Jung, K. W., Oh, S. I., Kim, Y. Y., Yoo, K. S., Cui, M. H., and Shin, J. S. (2008). Arabidopsis histidine-containing phosphotransfer factor 4 (AHP4) negatively regulates secondary wall thickening of the anther endothecium during flowering. Mol. Cells 25, 294–300.
Kamiya, T., and Fujiwara, T. (2009). Arabidopsis NIP1;1 transports antimonite and determines antimonite sensitivity. Plant Cell Physiol. 50, 1977–1981.
Kamiya, T., Tanaka, M., Mitani, N., Ma, J. F., Maeshima, M., and Fujiwara, T. (2009). NIP1;1, an aquaporin homolog, determines the arsenite sensitivity of Arabidopsis thaliana. J. Biol. Chem. 284, 2114–2120.
Kant, S., Bi, Y.-M., Zhu, T., and Rothstein, S. J. (2009). SAUR39, a small auxin-up RNA gene, acts as a negative regulator of auxin synthesis and transport in rice. Plant Physiol. 151, 691–701.
Kant, S., and Rothstein, S. (2009). Auxin-responsive SAUR39 gene modulates auxin level in rice. Plant Signal. Behav. 4, 1174–1175.
Kasukabe, Y., He, L., Nada, K., Misawa, S., Ihara, I., and Tachibana, S. (2004). Overexpression of spermidine synthase enhances tolerance to multiple environmental stresses and up-regulates the expression of various stress-regulated genes in transgenic Arabidopsis thaliana. Plant Cell Physiol. 45, 712–722.
Kepinski, S., and Leyser, O. (2005). The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435, 446–451.
Kiba, T., Aoki, K., Sakakibara, H., and Mizuno, T. (2004). Arabidopsis response regulator, ARR22, ectopic expression of which results in phenotypes similar to the wol cytokinin-receptor mutant. Plant Cell Physiol. 45, 1063–1077.
Kiba, T., Kudo, T., Kojima, M., and Sakakibara, H. (2011). Hormonal control of nitrogen acquisition: roles of auxin, abscisic acid, and cytokinin. J. Exp. Bot. 62, 1399–1409.
Kiba, T., Naitou, T., Koizumi, N., Yamashino, T., Sakakibara, H., and Mizuno, T. (2005). Combinatorial microarray analysis revealing Arabidopsis genes implicated in cytokinin responses through the His?Asp phosphorelay circuitry. Plant Cell Physiol. 46, 339–355.
Kiba, T., Taniguchi, M., Imamura, A., Ueguchi, C., Mizuno, T., and Sugiyama, T. (1999). Differential expression of genes for response regulators in response to cytokinins and nitrate in Arabidopsis thaliana. Plant Cell Physiol. 40, 767–771.
Kim, H. J., Ryu, H., Hong, S. H., Woo, H. R., Lim, P. O., Lee, I. C., Sheen, J., Nam, H. G., and Hwang, I. (2006). Cytokinin-mediated control of leaf longevity by AHK3 through phosphorylation of ARR2 in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 103, 814–819.
Köllmer, I., Werner, T., and Schmülling, T. (2011). Ectopic expression of different cytokinin-regulated transcription factor genes of Arabidopsis thaliana alters plant growth and development. J. Plant Physiol. 168, 1320–1327.
Kreps, J. A., Wu, Y., Chang, H.-S., Zhu, T., Wang, X., and Harper, J. F. (2002). Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol. 130, 2129–2141.
Kudo, T., Kiba, T., and Sakakibara, H. (2010). Metabolism and long-distance translocation of cytokinins. J. Integr. Plant Biol. 52, 53–60.
Kuroda, H., Masuda, T., Fusada, N., Ohta, H., and Takamiya, K. (2000). Expression of NADPH-protochlorophyllide oxidoreductase gene in fully green leaves of cucumber. Plant Cell Physiol. 41, 226–229.
Laplaze, L., Benkova, E., Casimiro, I., Maes, L., Vanneste, S., Swarup, R., Weijers, D., Calvo, V., Parizot, B., Herrera-Rodriguez, M. B., Offringa, R., Graham, N., Doumas, P., Friml, J., Bogusz, D., Beeckman, T., and Bennett, M. (2007). Cytokinins act directly on lateral root founder cells to inhibit root Initiation. Plant Cell 19, 3889–3900.
Lariguet, P., Boccalandro, H. E., Alonso, J. M., Ecker, J. R., Chory, J., Casal, J. J., and Fankhauser, C. (2003). A growth regulatory loop that provides homeostasis to phytochrome A signaling. Plant Cell 15, 2966–2978.
Lariguet, P., Schepens, I., Hodgson, D., Pedmale, U. V., Trevisan, M., Kami, C., de Carbonnel, M., Alonso, J. M., Ecker, J. R., Liscum, E., and Fankhauser, C. (2006). PHYTOCHROME KINASE SUBSTRATE 1 is a phototropin 1 binding protein required for phototropism. Proc. Natl. Acad. Sci. U.S.A. 103, 10134–10139.
Lee, D. J., Park, J. Y., Ku, S. J., Ha, Y. M., Kim, S., Kim, M. D., Oh, M. H., and Kim, J. (2007). Genome-wide expression profiling of ARABIDOPSIS RESPONSE REGULATOR 7(ARR7) overexpression in cytokinin response. Mol. Genet. Genomics 277, 115–137.
Lee, J., Parthier, B., and Löbler, M. (1996). Jasmonate signalling can be uncoupled from abscisic acid signalling in barley: identification of jasmonate-regulated transcripts which are not induced by abscisic acid. Planta 199, 625–632.
Lee, S., Lee, E. J., Yang, E. J., Lee, J. E., Park, A. R., Song, W. H., and Park, O. K. (2004). Proteomic identification of annexins, calcium-dependent membrane binding proteins that mediate osmotic stress and abscisic acid signal transduction in Arabidopsis. Plant Cell 16, 1378–1391.
Leyser, H. M. O., Pickett, F. B., Dharmasiri, S., and Estelle, M. (1996). Mutations in the AXR3 gene of Arabidopsis result in altered auxin response including ectopic expression from the SAUR-AC1 promoter. Plant J. 10, 403–413.
Lima, J. E., Kojima, S., Takahashi, H., and von Wirén, N. (2010). Ammonium triggers lateral root branching in Arabidopsis in an AMMONIUM TRANSPORTER1;3-dependent manner. Plant Cell 22, 3621–3633.
Lin, P.-C., Pomeranz, M. C., Jikumaru, Y., Kang, S. G., Hah, C., Fujioka, S., Kamiya, Y., and Jang, J.-C. (2011). The Arabidopsis tandem zinc finger protein AtTZF1 affects ABA- and GA-mediated growth, stress and gene expression responses. Plant J. 65, 253–268.
Lindsay, D. L., Sawhney, V. K., and Bonham-Smith, P. C. (2006). Cytokinin-induced changes in CLAVATA1 and WUSCHEL expression temporally coincide with altered floral development in Arabidopsis. Plant Sci. 170, 1111–1117.
Little, D., Gouhier-Darimont, C., Bruessow, F., and Reymond, P. (2007). Oviposition by pierid butterflies triggers defense responses in Arabidopsis. Plant Physiol. 143, 784–800.
Lohrmann, J., Sweere, U., Zabaleta, E., Bäurle, I., Keitel, C., Kozma, B. L., Brennicke, A., Schäfer, E., Kudla, J., and Harter, K. (2001). The response regulator ARR2: a pollen-specific transcription factor involved in the expression of nuclear genes for components of mitochondrial complex I in Arabidopsis. Mol. Genet. Genomics 265, 2–13.
Ma, J., Hanssen, M., Lundgren, K., Hernández, L., Delatte, T., Ehlert, A., Liu, C.-M., Schluepmann, H., Dröge-Laser, W., Moritz, T., Smeekens, S., and Hanson, J. (2011). The sucrose-regulated Arabidopsis transcription factor bZIP11 reprograms metabolism and regulates trehalose metabolism. New Phytol. 191, 733–745.
MacRae, W. D., and Towers, G. H. N. (1984). Biological activities of lignans. Phytochemistry 23, 1207–1220.
Mähönen, A. P., Bishopp, A., Higuchi, M., Nieminen, K. M., Kinoshita, K., Tormakangas, K., Ikeda, Y., Oka, A., Kakimoto, T., and Helariutta, Y. (2006). Cytokinin signaling and its inhibitor AHP6 regulate cell fate during vascular development. Science 311, 94–98.
Maksimov, I. V., Ganiev, R. M., and Khairullin, R. M. (2002). Changes in the levels of IAA, ABA, and cytokinins in wheat seedlings infected with Tilletia caries. Russ. J. Plant Physiol. 49, 221–224.
Maruyama-Nakashita, A., Nakamura, Y., Yamaya, T., and Takahashi, H. (2004). A novel regulatory pathway of sulfate uptake in Arabidopsis roots: implication of CRE1/WOL/AHK4-mediated cytokinin-dependent regulation. Plant J. 38, 779–789.
Marziani Longo, G. P., Bracale, M., Rossi, G., and Longo, C. P. (1990). Benzyladenine induces the appearance of LHCP-mRNA and of the relevant protein in dark-grown excised watermelon cotyledons. Plant Mol. Biol. 14, 569–573.
Mason, M. G., Li, J., Mathews, D. E., Kieber, J. J., and Schaller, G. E. (2004). Type-B response regulators display overlapping expression patterns in Arabidopsis. Plant Physiol. 135, 927–937.
Mason, M. G., Mathews, D. E., Argyros, D. A., Maxwell, B. B., Kieber, J. J., Alonso, J. M., Ecker, J. R., and Schaller, G. E. (2005). Multiple type-B response regulators mediate cytokinin signal transduction in Arabidopsis. Plant Cell 17, 3007–3018.
Massonnet, C., Vile, D., Fabre, J., Hannah, M. A., Caldana, C., Lisec, J., Beemster, G. T. S., Meyer, R. C., Messerli, G., Gronlund, J. T., Perkovic, J., Wigmore, E., May, S., Bevan, M. W., Meyer, C., Rubio-Díaz, S., Weigel, D., Micol, J. L., Buchanan-Wollaston, V., Fiorani, F., Walsh, S., Rinn, B., Gruissem, W., Hilson, P., Hennig, L., Willmitzer, L., and Granier, C. (2010). Probing the reproducibility of leaf growth and molecular phenotypes: a comparison of three Arabidopsis accessions cultivated in ten laboratories. Plant Physiol. 152, 2142–2157.
Mendonça Pauletti, P., Araújo, A. R., Young, M. C. M., Giesbrecht, A. M., and da Silva Bolzani, V. (2000). nor-Lignans from the leaves of Styrax ferrugineus (Styracaceae) with antibacterial and antifungal activity. Phytochemistry 55, 597–601.
Michel, K., Abderhalden, O., Bruggmann, R. M., and Dudler, R. (2006). Transcriptional changes in powdery mildew infected wheat and Arabidopsis leaves undergoing syringolin-triggered hypersensitive cell death at infection sites. Plant Mol. Biol. 62, 561–578.
Motchoulski, A., and Liscum, E. (1999). Arabidopsis NPH3: a NPH1 photoreceptor-interacting protein essential for phototropism. Science 286, 961–964.
Moubayidin, L., Di Mambro, R., and Sabatini, S. (2009). Cytokinin-auxin crosstalk. Trends Plant Sci. 14, 557–562.
Mudge, S. R., Rae, A. L., Diatloff, E., and Smith, F. W. (2002). Expression analysis suggests novel roles for members of the Pht1 family of phosphate transporters in Arabidopsis. Plant J. 31, 341–353.
Müller, B. (2011). Generic signal-specific responses: cytokinin and context-dependent cellular responses. J. Exp. Bot. 62, 3273–3288.
Müller, B., and Sheen, J. (2008). Cytokinin and auxin interaction in root stem-cell specification during early embryogenesis. Nature 453, 1094–1097.
Naito, T., Yamashino, T., Kiba, T., Koizumi, N., Kojima, M., Sakakibara, H., and Mizuno, T. (2007). A link between cytokinin and ASL9 (ASYMMETRIC LEAVES 2 LIKE 9) that belongs to the AS2/LOB (LATERAL ORGAN BOUNDARIES) family genes in Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 71, 1269–1278.
Navarro, L., Zipfel, C., Rowland, O., Keller, I., Robatzek, S., Boller, T., and Jones, J. D. G. (2004). The transcriptional innate immune response to flg22. Interplay and overlap with Avr gene-dependent defense responses and bacterial pathogenesis. Plant Physiol. 135, 1113–1128.
Nelson, D. C., Flematti, G. R., Riseborough, J.-A., Ghisalberti, E. L., Dixon, K. W., and Smith, S. M. (2010). Karrikins enhance light responses during germination and seedling development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 107, 7095–7100.
Nutricati, E., Miceli, A., Blando, F., and De Bellis, L. (2006). Characterization of two Arabidopsis thaliana glutathione S-transferases. Plant Cell Rep. 25, 997–1005.
Obayashi, T., Hayashi, S., Saeki, M., Ohta, H., and Kinoshita, K. (2009). ATTED-II provides coexpressed gene networks for Arabidopsis. Nucleic Acids Res. 37, D987–D991.
Obayashi, T., Nishida, K., Kasahara, K., and Kinoshita, K. (2011). ATTED-II updates: condition-specific gene coexpression to extend coexpression analyses and applications to a broad range of flowering plants. Plant Cell Physiol. 52, 213–219.
Okazaki, K., Kabeya, Y., Suzuki, K., Mori, T., Ichikawa, T., Matsui, M., Nakanishi, H., and Miyagishima, S.-Y. (2009). The PLASTID DIVISION1 and 2 components of the chloroplast division machinery determine the rate of chloroplast division in land plant cell differentiation. Plant Cell 21, 1769–1780.
Parthier, B. (1979). The role of phytohormones (cytokinin) in chloroplasts development. Biochem. Physiol. Pflanz. 174, 173–214.
Pedmale, U. V., and Liscum, E. (2007). Regulation of phototropic signaling in Arabidopsis via phosphorylation state changes in the phototropin 1-interacting protein NPH3. J. Biol. Chem. 282, 19992–20001.
Pérez-Pérez, J. M., Candela, H., Robles, P., López-Torrejón, G., del Pozo, J. C., and Micol, J. L. (2010). A role for AUXIN RESISTANT3 in the coordination of leaf growth. Plant Cell Physiol. 51, 1661–1673.
Perilli, S., Moubayidin, L., and Sabatini, S. (2010). The molecular basis of cytokinin function. Curr. Opin. Plant Biol. 13, 21–26.
Pertry, I., Václavíková, K., Depuydt, S., Galuszka, P., Spíchal, L., Temmerman, W., Stes, E., Schmülling, T., Kakimoto, T., Van Montagu, M. C. E., Strnad, M., Holsters, M., Tarkowski, P., and Vereecke, D. (2009). Identification of Rhodococcus fascians cytokinins and their modus operandi to reshape the plant. Proc. Natl. Acad. Sci. U.S.A. 106, 929–934.
Pils, B., and Heyl, A. (2009). Unraveling the evolution of cytokinin signaling. Plant Physiol. 151, 782–791.
Pomeranz, M., Zhang, L., Finer, J., and Jang, J. C. (2011). Can AtTZF1 act as a transcriptional activator or repressor in plants? Plant Signal. Behav. 6, 719–722.
Pufky, J., Qiu, Y., Rao, M., Hurban, P., and Jones, A. (2003). The auxin-induced transcriptome for etiolated Arabidopsis seedlings using a structure/function approach. Funct. Integr. Genomics 3, 135–143.
Punwani, J. A., Hutchison, C. E., Schaller, G. E., and Kieber, J. J. (2010). The subcellular distribution of the Arabidopsis histidine phosphotransfer proteins is independent of cytokinin signaling. Plant J. 62, 473–482.
Rahman, M. M., and Gray, A. I. (2002). Antimicrobial constituents from the stem bark of Feronia limonia. Phytochemistry 59, 73–77.
Ralph, S., Park, J.-Y., Bohlmann, J. R., and Mansfield, S. (2006). Dirigent proteins in conifer defense: gene discovery, phylogeny, and differential wound- and insect-induced expression of a family of DIR and DIR-like genes in spruce (Picea spp.). Plant Mol. Biol. 60, 21–40.
Ramireddy, E. (2009). Functional Characterization of B-Type Response Regulators of Arabidopsis thaliana. Dissertation, Freie Universität Berlin, Berlin.
Rashotte, A., and Goertzen, L. (2010). The CRF domain defines cytokinin response factor proteins in plants. BMC Plant Biol. 10, 74. doi: 10.1186/1471-2229-10-74
Rashotte, A. M., Carson, S. D., To, J. P., and Kieber, J. J. (2003). Expression profiling of cytokinin action in Arabidopsis. Plant Physiol. 132, 1998–2011.
Rashotte, A. M., Mason, M. G., Hutchison, C. E., Ferreira, F. J., Schaller, G. E., and Kieber, J. J. (2006). A subset of Arabidopsis AP2 transcription factors mediates cytokinin responses in concert with a two-component pathway. Proc. Natl. Acad. Sci. U.S.A. 103, 11081–11085.
Ribot, C. C., Wang, Y., and Poirier, Y. (2008). Expression analyses of three members of the AtPHO1 family reveal differential interactions between signaling pathways involved in phosphate deficiency and the responses to auxin, cytokinin, and abscisic acid. Planta 227, 1025–1036.
Richter, R., Behringer, C., Müller, I. K., and Schwechheimer, C. (2010). The GATA-type transcription factors GNC and GNL/CGA1 repress gibberellin signaling downstream from DELLA proteins and PHYTOCHROME-INTERACTING FACTORS. Genes Dev. 24, 2093–2104.
Riefler, M., Novak, O., Strnad, M., and Schmülling, T. (2006). Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism. Plant Cell 18, 40–54.
Rivero, R. M., Shulaev, V., and Blumwald, E. (2009). Cytokinin-dependent photorespiration and the protection of photosynthesis during water deficit. Plant Physiol. 150, 1530–1540.
Rocca-Serra, P., Brazma, A., Parkinson, H., Sarkans, U., Shojatalab, M., Contrino, S., Vilo, J., Abeygunawardena, N., Mukherjee, G., Holloway, E., Kapushesky, M., Kemmeren, P., Lara, G. G., Oezcimen, A., and Sansone, S. A. (2003). ArrayExpress: a public database of gene expression data at EBI. C. R. Biol. 326, 1075–1078.
Ross, E. J. H., Stone, J. M., Elowsky, C. G., Arredondo-Peter, R., Klucas, R. V., and Sarath, G. (2004). Activation of the Oryza sativa non-symbiotic haemoglobin-2 promoter by the cytokinin-regulated transcription factor, ARR1. J. Exp. Bot. 55, 1721–1731.
Rubio, V., Bustos, R., Irigoyen, M. A., Cardona-López, X., Rojas-Triana, M. N., and Paz-Ares, J. (2009). Plant hormones and nutrient signaling. Plant Mol. Biol. 69, 361–373.
Růžicka, K., Šimášková, M., Duclercq, J., Petrášek, J., Zažímalová, E., Simon, S., Friml, J., Van Montagu, M. C. E., and Benková, E. (2009). Cytokinin regulates root meristem activity via modulation of the polar auxin transport. Proc. Natl. Acad. Sci. U.S.A. 106, 4284–4289.
Sakai, H., Aoyama, T., and Oka, A. (2000). Arabidopsis ARR1 and ARR2 response regulators operate as transcriptional activators. Plant J. 24, 703–711.
Sakai, H., Honma, T., Aoyama, T., Sato, S., Kato, T., Tabata, S., and Oka, A. (2001). ARR1, a transcription factor for genes immediately responsive to cytokinins. Science 294, 1519–1521.
Sakakibara, H., Suzuki, M., Takei, K., Deji, A., Taniguchi, M., and Sugiyama, T. (1998). A response-regulator homologue possibly involved in nitrogen signal transduction mediated by cytokinin in maize. Plant J. 14, 337–344.
Sakakibara, H., Takei, K., and Hirose, N. (2006). Interactions between nitrogen and cytokinin in the regulation of metabolism and development. Trends Plant Sci. 11, 440–448.
Schaller, G. E., Shiu, S.-H., and Armitage, J. P. (2011). Two-component systems and their co-option for eukaryotic signal transduction. Curr. Biol. 21, R320–R330.
Schepens, I., Duek, P., and Fankhauser, C. (2004). Phytochrome-mediated light signalling in Arabidopsis. Curr. Opin. Plant Biol. 7, 564–569.
Schluepmann, H., Pellny, T., Van, D. A., Smeekens, S., and Paul, M. (2003). Trehalose 6-phosphate is indispensable for carbohydrate utilization and growth in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 100, 6849–6854.
Schluepmann, H., van Dijken, A., Aghdasi, M., Wobbes, B., Paul, M., and Smeekens, S. (2004). Trehalose mediated growth inhibition of Arabidopsis seedlings is due to trehalose-6-phosphate accumulation. Plant Physiol. 135, 879–890.
Schmülling, T., Schäfer, S., and Romanov, G. (1997). Cytokinins as regulators of gene expression. Physiol. Plant 100, 505–519.
Sherson, S. M., Alford, H. L., Forbes, S. M., Wallace, G., and Smith, S. M. (2003). Roles of cell-wall invertases and monosaccharide transporters in the growth and development of Arabidopsis. J. Exp. Bot. 54, 525–531.
Šiffel, P., Sindelková, E., Durchan, M., and Zajicová, M. (1992). Photosynthetic characteristics of Solanum tuberosum L. plants transformed by genes for synthesis of phytohormones. Photosynthetica 27, 441–447.
Skinner, D., and Gasser, C. (2009). Expression-based discovery of candidate ovule development regulators through transcriptional profiling of ovule mutants. BMC Plant Biol. 9, 29. doi: 10.1186/1471-2229-9-29
Smyth, G. K. (2005). “Limma: linear models for microarray data,” in Bioinformatics and Computational Biology Solutions Using R and Bioconductor, eds R. Gentleman, V. Carey, S. Dudoit, R. Irizarry, and W. Huber (New York: Springer), 397–420.
Staetler, D. A., and Laetsch, W. M. (1965). Kinetin-induced chloroplast maturation in cultures of tobacco tissue. Science 149, 1387–1388.
Suzuki, T., Miwa, K., Ishikawa, K., Yamada, H., Aiba, H., and Mizuno, T. (2001). The Arabidopsis sensor His-kinase, AHK4, can respond to cytokinins. Plant Cell Physiol. 42, 107–113.
Swarbreck, D., Wilks, C., Lamesch, P., Berardini, T. Z., Garcia-Hernandez, M., Foerster, H., Li, D., Meyer, T., Muller, R., Ploetz, L., Radenbaugh, A., Singh, S., Swing, V., Tissier, C., Zhang, P., and Huala, E. (2008). The Arabidopsis Information Resource (TAIR): gene structure and function annotation. Nucleic Acids Res. 36, D1009–D1014.
Tajima, Y., Imamura, A., Kiba, T., Amano, Y., Yamashino, T., and Mizuno, T. (2004). Comparative studies on the type-B response regulators revealing their distinctive properties in the His-to-Asp phosphorelay signal transduction of Arabidopsis thaliana. Plant Cell Physiol. 45, 28–39.
Takei, K., Yamaya, T., and Sakakibara, H. (2004). Arabidopsis CYP735A1 and CYP735A2 encode cytokinin hydroxylases that catalyze the biosynthesis of trans-zeatin. J. Biol. Chem. 279, 41866–41872.
Taniguchi, M., Sasaki, N., Tsuge, T., Aoyama, T., and Oka, A. (2007). ARR1 directly activates cytokinin response genes that encode proteins with diverse regulatory functions. Plant Cell Physiol. 48, 263–277.
Thimm, O., Blaesing, O., Gibon, Y., Nagel, A., Meyer, S., Krueger, P., Selbig, J., Mueller, L. A., Rhee, S. Y., and Stitt, M. (2004). MAPMAN: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37, 914–939.
Thomas, J., Ross, C. W., Chastain, C. J., Koomanoff, N., Hendrix, J. E., and Van Volkenburgh, E. (1981). Cytokinin-induced wall extensibility in excised cotyledons of radish and cucumber. Plant Physiol. 68, 107–110.
Tiwari, S., Spielman, M., Day, R. C., and Scott, R. J. (2006). Proliferative phase endosperm promoters from Arabidopsis thaliana. Plant Biotechnol. J. 4, 393–407.
To, J. P., Haberer, G., Ferreira, F. J., Deruere, J., Mason, M. G., Schaller, G. E., Alonso, J. M., Ecker, J. R., and Kieber, J. J. (2004). Type-A Arabidopsis response regulators are partially redundant negative regulators of cytokinin signaling. Plant Cell 16, 658–671.
To, J. P. C., and Kieber, J. J. (2008). Cytokinin signaling: two-components and more. Trends Plant Sci. 13, 85–92.
Tran, L.-S. P., Urao, T., Qin, F., Maruyama, K., Kakimoto, T., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2007). Functional analysis of AHK1/ATHK1 and cytokinin receptor histidine kinases in response to abscisic acid, drought, and salt stress in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 104, 20623–20628.
Treml, B. S., Winderl, S., Radykewicz, R., Herz, M., Schweizer, G. N., Hutzler, P., Glawischnig, E., and Ruiz, R. N. A. T. (2005). The gene ENHANCER OF PINOID controls cotyledon development in the Arabidopsis embryo. Development 132, 4063–4074.
Urao, T., Yakubov, B., Satoh, R., Yamaguchi-Shinozaki, K., Seki, M., Hirayama, T., and Shinozaki, K. (1999). A transmembrane hybrid-type histidine kinase in Arabidopsis functions as an osmosensor. Plant Cell 11, 1743–1754.
Urao, T., Yakubov, B., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1998). Stress-responsive expression of genes for two-component response regulator-like proteins in Arabidopsis thaliana. FEBS Lett. 427, 175–178.
Vergnolle, C., Vaultier, M.-N. l., Taconnat, L., Renou, J.-P., Kader, J.-C., Zachowski, A., and Ruelland, E. (2005). The cold-induced early activation of phospholipase C and D pathways determines the response of two distinct clusters of genes in Arabidopsis cell suspensions. Plant Physiol. 139, 1217–1233.
Vieira Dos Santos, C., and Rey, P. (2006). Plant thioredoxins are key actors in the oxidative stress response. Trends Plant Sci. 11, 329–334.
Wahl, V., Brand, L. H., Guo, Y. L., and Schmid, M. (2010). The FANTASTIC FOUR proteins influence shoot meristem size in Arabidopsis thaliana. BMC Plant Biol. 10, 285. doi: 10.1186/1471-2229-10-285
Weijers, D., Franke-van Dijk, M., Vencken, R.-J., Quint, A., Hooykaas, P., and Offringa, R. (2001). An Arabidopsis minute-like phenotype caused by a semi-dominant mutation in a RIBOSOMAL PROTEIN S5 gene. Development 128, 4289–4299.
Werner, T., Holst, K., Pors, Y., Guivarc’h, A., Mustroph, A., Chriqui, D., Grimm, B., and Schmulling, T. (2008). Cytokinin deficiency causes distinct changes of sink and source parameters in tobacco shoots and roots. J. Exp. Bot. 59, 2659–2672.
Werner, T., Motyka, V., Laucou, V., Smets, R., van Onckelen, H., and Schmülling, T. (2003). Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 15, 2532–2550.
Werner, T., Motyka, V., Strnad, M., and Schmülling, T. (2001). Regulation of plant growth by cytokinin. Proc. Natl. Acad. Sci. U.S.A. 98, 10487–10492.
Werner, T., Nehnevajova, E., Köllmer, I., Novak, O., Strnad, M., Krämer, U., and Schmülling, T. (2010). Root-specific reduction of cytokinin causes enhanced root growth, drought tolerance, and leaf mineral enrichment in Arabidopsis and tobacco. Plant Cell 22, 3905–3920.
Werner, T., and Schmülling, T. (2009). Cytokinin action in plant development. Curr. Opin. Plant Biol. 12, 527–538.
Wohlbach, D. J., Quirino, B. F., and Sussman, M. R. (2008). Analysis of the Arabidopsis histidine kinase ATHK1 reveals a connection between vegetative osmotic stress sensing and seed maturation. Plant Cell 20, 1101–1117.
Yaronskaya, E., Vershilovskaya, I., Poers, Y., Alawady, A., Averina, N., and Grimm, B. (2006). Cytokinin effects on tetrapyrrole biosynthesis and photosynthetic activity in barley seedlings. Planta 224, 700–709.
Yokoyama, A., Yamashino, T., Amano, Y., Tajima, Y., Imamura, A., Sakakibara, H., and Mizuno, T. (2007). Type-B ARR transcription factors, ARR10 and ARR12, are implicated in cytokinin-mediated regulation of protoxylem differentiation in roots of Arabidopsis thaliana. Plant Cell Physiol. 48, 84–96.
Yoshida, S., Mandel, T., and Kuhlemeier, C. (2011). Stem cell activation by light guides plant organogenesis. Genes Dev. 25, 1439–1450.
Zalewski, W., Galuszka, P., Gasparis, S., Orczyk, W., and Nadolska-Orczyk, A. (2010). Silencing of the HvCKX1 gene decreases the cytokinin oxidase/dehydrogenase level in barley and leads to higher plant productivity. J. Exp. Bot. 61, 1839–1851.
Zavaleta-Mancera, H. A., López-Delgado, H., Loza-Tavera, H., Mora-Herrera, M., Trevilla-García, C., Vargas-Suárez, M., and Ougham, H. (2007). Cytokinin promotes catalase and ascorbate peroxidase activities and preserves the chloroplast integrity during dark-senescence. J. Plant Physiol. 164, 1572–1582.
Zhao, J., Wang, J., An, L., Doerge, R., Chen, Z., Grau, C., Meng, J., and Osborn, T. (2007). Analysis of gene expression profiles in response to Sclerotinia sclerotiorum in Brassica napus. Planta 227, 13–24.
Zhao, Z., Andersen, S. U., Ljung, K., Doležal, K., Miotk, A., Schultheiss, S. J., and Lohmann, J. U. (2010). Hormonal control of the shoot stem-cell niche. Nature 465, 1089–1092.
Zubo, Y. O., Yamburenko, M. V., Selivankina, S. Y., Shakirova, F. M., Avalbaev, A. M., Kudryakova, N. V., Zubkova, N. K., Liere, K., Kulaeva, O. N., Kusnetsov, V. V., and Börner, T. (2008). Cytokinin stimulates chloroplast transcription in detached barley leaves. Plant Physiol. 148, 1082–1093.
Catský, J., Pospíšilová, J., Machácková, I., Wilhelmová, N., and Šesták, Z. (1993). Photosynthesis and water relations in transgenic tobacco plants with T-DNA carrying gene 4 for cytokinin synthesis. Biol. Plant. 35, 393–399.
Keywords: cytokinin, gene regulation, transcription factor, cis-element, transcriptomics, meta analysis, signal transduction, regulatory network
Citation: Brenner WG, Ramireddy E, Heyl A and Schmülling T (2012) Gene regulation by cytokinin in Arabidopsis. Front. Plant Sci. 3:8. doi: 10.3389/fpls.2012.00008
Received: 30 September 2011; Paper pending published: 21 October 2011;
Accepted: 06 January 2012; Published online: 31 January 2012.
Edited by:
Andreas P. M. Weber, University of Düsseldorf, GermanyCopyright: © 2012 Brenner, Ramireddy, Heyl and Schmülling. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Alexander Heyl and Thomas Schmülling, Institute of Biology/Applied Genetics, Dahlem Centre of Plant Sciences, Freie Universität Berlin, Albrecht-Thaer-Weg 6, D-14195 Berlin, Germany. e-mail:aGV5bEB6ZWRhdC5mdS1iZXJsaW4uZGU=;dGhvbWFzLnNjaG11ZWxsaW5nQGZ1LWJlcmxpbi5kZQ==


 Alexander Heyl*
Alexander Heyl*