
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci., 18 January 2012
Sec. Plant Membrane Traffic and Transport
Volume 3 - 2012 | https://doi.org/10.3389/fpls.2012.00002
This article is part of the Research TopicEvolution of transporters in plants (Frontiers in Plant Traffic and Transport)View all 7 articles
Mitochondria play a key role in respiration and energy production and are involved in multiple eukaryotic but also in several plant specific metabolic pathways. Solute carriers in the inner mitochondrial membrane connect the internal metabolism with that of the surrounding cell. Because of their common basic structure, these transport proteins affiliate to the mitochondrial carrier family (MCF). Generally, MCF proteins consist of six membrane spanning helices, exhibit typical conserved domains and appear as homodimers in the native membrane. Although structurally related, MCF proteins catalyze the specific transport of various substrates, such as nucleotides, amino acids, dicarboxylates, cofactors, phosphate or H+. Recent investigations identified MCF proteins also in several other cellular compartments and therefore their localization and physiological function is not only restricted to mitochondria. MCF proteins are a characteristic feature of eukaryotes and bacterial genomes lack corresponding sequences. Therefore, the evolutionary origin of MCF proteins is most likely associated with the establishment of mitochondria. It is not clear whether the host cell, the symbiont, or the chimerical organism invented the ancient MCF sequence. Here, we try to explain the establishment of different MCF proteins and focus on the characteristics of members from plants, in particular from Arabidopsis thaliana.
Already in 1883, the botanist A. Schimper observed that the division process of chloroplasts resembles that of bacteria and he stated briefly that plants represent a consortium of two different organisms (Schimper, 1883; Ward, 1883). Mereschkowsky (1905) focused on plastid evolution in his publication “On the nature and origin of the chromatophores in the plant kingdom” and again proposed that chloroplasts are “semi-autonomous,” endosymbiotic entities of plant cells. About 20 years later Wallin (1923, 1927) postulated that also mitochondria represent a kind of foreign/bacterial cell that thrives in the eukaryotic host. Many striking observations supported a relatedness of bacteria and mitochondria, and in particular of cyanobacteria and chloroplasts however, the pioneering idea of an endosymbiotic origin of these organelles was disregarded for a long time. The endosymbiotic theory was reinvented 1967 by Lynn Margulis (Sagan, 1967) and gained broader scientific attention in the 1970s and 1980s. Detailed electron microscopical data, microbiological investigations, and most importantly the identification of the mitochondrial and the plastidial genome unequivocally clarified that these organelles originated from formerly free-living bacteria that were taken up by another cell (Gray, 1992; Delwiche, 1999). Deciphering organellar and nuclear genomes demonstrated that mitochondria and plastids each evolved by a single endosymbiotic event. Mitochondria were derived from an alpha-proteobacterium whereas chloroplasts originated from a photosynthetic cyanobacterium (Lang et al., 1997; Delwiche, 1999; Martin et al., 2002; Raven and Allen, 2003; Gray et al., 2004; Rodriguez-Ezpeleta et al., 2005; de Duve, 2007). In contrast to the origin of mitochondria, plastid evolution is rather complicated due to additional endosymbioses (secondary, tertiary endosymbioses), due to secondary loss of the plastid, or due to replacement of existing plastids by new ones (Delwiche, 1999; Keeling, 2010).
Although mitochondria and chloroplasts arose by independent evolutionary processes, both share important similarities concerning reduction, modification, and integration of the symbiont into the context of the host cell and have relict bacterial features in common. Establishment of these organelles was accompanied by massive gene transfer from the symbiont to the host nucleus (Martin and Herrmann, 1998; Kurland and Andersson, 2000). Accordingly, the symbiont became metabolically highly impaired due to the loss of genes (and their corresponding functions) and had to be genetically and physiologically integrated within the host cell. Moreover, plastids and mitochondria are surrounded by two membranes. This feature is traced back to their endosymbiotic ancestry from Gram-negative bacteria. The inner organelle membrane represents the former bacterial plasma membrane and the outer organelle membrane descended from the outer bacterial membrane (Cavalier-Smith, 2000; Gross and Bhattacharya, 2009a). Similar to the situation in Gram-negative bacteria, the outer membrane of mitochondria and plastids harbors mainly β-barrel proteins and allows a rather unselective passage of several, small molecules. The inner organellar or bacterial membrane constitutes the permeability barrier and harbors carriers mediating a highly specific and selective solute transfer (Cavalier-Smith, 2000; Gross and Bhattacharya, 2009a). Accordingly, bacterial transport systems were already present in the ancient organelle. Because of the genetic reduction the symbiont lost its independence and relied on additional protein and metabolite provision from the host (Martin and Herrmann, 1998; Kurland and Andersson, 2000). Specific transport systems for solute and protein import into the symbiont were required to compensate for its genetic deficits and to functionally embed the organelle into the host. Pre-existing transport systems in the two surrounding membranes had to be adapted or new systems had to be established prior to or at least synchronously with the transfer of symbiotic genes to the nucleus (Gross and Bhattacharya, 2009a,b; Alcock et al., 2010; Bohnsack and Schleiff, 2010). Moreover, targeting of nuclear encoded proteins to the mitochondrion or plastid and their passage through the two membranes had to be established, guaranteed, and controlled. Protein import systems of mitochondria and plastids consist of specialized translocase complexes in the inner and in the outer membrane (Gross and Bhattacharya, 2009a,b; Alcock et al., 2010; Bohnsack and Schleiff, 2010; Lithgow and Schneider, 2010). An N-terminal extension generally characterizes nuclear encoded proteins with plastidial destination and also several mitochondrial proteins are initially synthesized with mitochondrial presequences that serve as matrix-targeting signals (Lithgow, 2000; Bruce, 2001; Patron and Waller, 2007).
Mitochondria are the main sites of cellular respiration and ATP supply and thus represent an important and characteristic component of eukaryotes. To date no lineage has been shown to have lost or replaced this organelle and also additional endosymbioses apparently did not occur during mitochondrial evolution. However, mitochondria are not homogenous at all; they exhibit substantial genetic, functional, and morphological differences. Interestingly, certain anaerobic or microaerophilic protists possess “anaerobic mitochondria” or mitochondria-related organelles, called hydrogenosomes or mitosomes (Embley and Martin, 2006; Hackstein et al., 2006). Similar to the aerobic “powerhouses,” also “anaerobic” mitochondria and hydrogenosomes are involved in energy production. In the respiratory chain of mitochondria oxygen acts as electron acceptor and water is produced, whereas in hydrogenosomes electrons are transferred to protons resulting in hydrogen generation (Embley and Martin, 2006; Hackstein et al., 2006). “Anaerobic” mitochondria use other electron acceptors than oxygen or protons, like nitrate or fumarate (Embley and Martin, 2006; Hackstein et al., 2006). Mitosomes represent extremely reduced mitochondrial relatives not capable for ATP generation. Accordingly, these mitochondrial remnants rely on ATP import to fuel interior processes (at least iron–sulfur cluster biosynthesis) with energy (Chan et al., 2005; Tsaousis et al., 2008; Williams et al., 2008).
There is ample evidence that mitochondria, hydrogenosomes, and mitosomes evolved from one common facultative anaerobic ancestor (van der Giezen et al., 2002; Voncken et al., 2002; Embley et al., 2003; Embley and Martin, 2006; Hackstein et al., 2006; Williams et al., 2008). However, the nature of the host is uncertain and still debated (de Duve, 2007; Gross and Bhattacharya, 2009a). The hydrogen hypothesis describes metabolic interaction and tight association as a possible reason for the fusion of a facultative aerobic alpha-proteobacterium with an anaerobic methane-producing Archaean (Martin and Müller, 1998). In presence of O2 alpha-proteobacteria respire organic compounds and produce carbon dioxide and water, whereas under anaerobic conditions they perform fermentation and hydrogen instead of water is delivered. Archaeal bacteria can use the H2 and CO2 as sole energy and carbon sources. It is imaginable that ancient methanogenic archaea – due to tight interaction with alpha-proteobacteria – became independent on abiotic H2 and thus were able to colonize new niches. Gain of H2 and CO2 was probably enhanced by enlargement of the contact area between the two species and led to complete surrounding and engulfment of the proteobacterium (Martin and Müller, 1998). During evolution the archaebacterium converted from autotrophy (using H2 and CO2) to heterotrophy (using organic molecules from the environment), gained typical eukaryotic features and the alpha-proteobacterium was reduced to an organelle. Absence of O2 supported the establishment of “early” hydrogenosomes and its presence caused the generation of mitochondria. The conversion of typical aerobic mitochondria into anaerobic organelles and massive reduction to mitochondrial remnants apparently happened several times and might explain why “anaerobic mitochondria,” hydrogenosomes, and mitosomes occur in phylogenetically diverse lineages (Tjaden et al., 2004; Embley and Martin, 2006; Hackstein et al., 2006).
The hydrogen hypothesis tightly connects the establishment of the first eukaryote with the establishment of the first mitochondrion. A second hypothesis bases on a more separated, successive evolution of eukaryotes and mitochondria (Cavalier-Smith, 1983). This hypothesis suggests that primitive early branching eukaryotes (“Archaezoa”) evolved in a pre-mitochondrial era. These “Archaezoa” harbored an internal membrane system and were able to perform phagocytosis and thus possessed important prerequisites for the capture of the symbiont.
The mitochondrial carrier family (MCF) is a large family of proteins with about 30 members in yeast and more than 50 in humans and plants (Palmieri et al., 2000b; Picault et al., 2004; Wohlrab, 2006). MCF proteins are highly heterogenous in terms of substrate specificity and transport mode but all possess a molecular mass of about 30–35 kDa, and exhibit an identical basic structure with six transmembrane domains. MCF proteins are composed of three repetitive modules that take a tilted position in the membrane (Kuan and Saier, 1993; Pebay-Peyroula et al., 2003; Nury et al., 2006). The three repeated homologous regions are of about 100 amino acids in length and each repeat comprises two membrane spanning domains (Saraste and Walker, 1982). Furthermore, the odd-numbered transmembrane domains are kinked and characterized by a conserved MCF motif at the kink (Jezek and Jezek, 2003; Nury et al., 2006). Three loops connect the six transmembrane spanning regions at the matrix side and each loop contains a short amphipathic helical domain with a second conserved MCF motif (Figure 1; Nury et al., 2006). The N- and the C-terminal parts as well as the hydrophilic loops between transmembrane helix 2 and 3 and helix 4 and 5 are exposed to the intermembrane space.
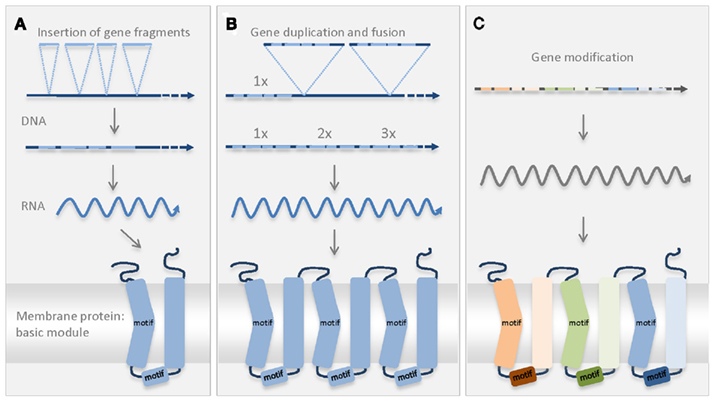
Figure 1. Schematic diagram of the possible scenario explaining the establishment of the first MCF protein. Conserved MCF motives in the secondary structure of the protein are indicated (motif). The odd-numbered transmembrane regions contain one conserved MCF motif (PxD/ExxK/RxK/R) and the proline residue induces the kinked form (Nury et al., 2006). A second MCF motif (D/EGxxxxaK/RG) is located in the helical region of the connecting loop (Nury et al., 2006). (A) In the genome of the host or engulfed bacterium a basic MCF sequence originated by fusion of different smaller gene fragments. The new sequence encodes the postulated basic module of the MCF consisting of two transmembrane helices and of a connecting loop with a helical region. (B) Twofold duplication of the basic sequence and insertion in the genome resulted in the coding sequence of the pre-MCF protein. (C) Gene modification resulted in the establishment of further/different MCF carriers that still exhibit the conserved structure and the MCF motives.
Although MCF proteins are described to exhibit a dimeric structure in their native membrane, more recent analyses suggest that the monomer might be one functional entity (Kunji and Crichton, 2010). The crystal structure of the monomeric ADP/ATP carrier (AAC) in complex with the inhibitor carboxyatractyloside represents the overexpanded form of the cytosolic-state (c-state; Pebay-Peyroula et al., 2003; Klingenberg, 2008). It resembles a cup or a basket with the closed bottom directed to the matrix and a wide opening facing the intermembrane space and possesses a channel-like interior structure. Two salt-bridge networks are suggested to act as gates of the carrier (Nury et al., 2006). In the c-state, one network closes the carrier at the matrix side and the open entry faces the cytosol whereas in the matrix-state (m-state), the other network closes the cytosolic part of the carrier and the internal cavity becomes accessible from the matrix. The mitochondria-specific anionic phospholipid cardiolipin resides between two monomers and was shown to play an import role in the biogenesis and stability of MCF proteins (Jiang et al., 2000; Klingenberg, 2009). Furthermore, transport studies demonstrated that cardiolipin addition enhances the activity of the recombinant mitochondrial AAC and also of other MCF proteins (Jiang et al., 2000; Heimpel et al., 2001). It can be assumed that dimer formation occurred early in MCF evolution because different present-day MCF proteins appear as dimers in their native membranes.
The common basic structure of all present-day MCF proteins suggests that all carriers arose from one single ancestral sequence that was duplicated and differentiated in several rounds. So far, the evolutionary origin of the first MCF protein is not clarified. The threefold repetitive structure of MCF carriers (Kuan and Saier, 1993) most likely reflects also the structure of the first MCF protein. During its evolution, pre-existing gene fragments were probably fused to a basic module encoding a protein with two transmembrane helices (Figure 1). Two subsequent gene duplications of this basic module and fusion of the three identical copies probably gave rise to the ancestor of all MCF proteins (pre-MCF protein, Figure 1). It is imaginable that the postulated single basic module itself also arose from gene duplication of a fragment encoding a single transmembrane domain (Palmieri et al., 2011). The ancestral MCF protein might have originated prior to, during or after establishment of the mitochondrial organelle. It might have been introduced by the proteobacterial symbiont or by the host (archaebacteria or “Archaezoa”) or was newly invented in the consortium. The fact that MCF proteins or sequences that substantially resemble the postulated basic module have never been identified in bacterial species argues against a bacterial origin. Whereas, the presence of MCF proteins in almost all eukaryotes (very few exceptions exist) tightly connects the MCF ancestry with the eukaryotic cell.
Taken the “Archaezoa” hypothesis as a possible evolutionary basis, the first MCF sequence might have been established in the early amitochondriate eukaryote to fulfill a function in a different organelle. In fact, MCF proteins are not only restricted to mitochondria but also present in many other cellular compartments (Bedhomme et al., 2005; Bouvier et al., 2006; Thuswaldner et al., 2007; Kirchberger et al., 2008; Leroch et al., 2008; Linka et al., 2008; Palmieri et al., 2009; Bernhardt et al., 2011; Rieder and Neuhaus, 2011). However, the majority of MCF proteins reside in the inner mitochondrial membrane. Accordingly, their main metabolic role and thus probably also their origin is rather connected with mitochondria. Moreover, organisms that harbor relicts of mitochondria contain only few or no MCF proteins (Chan et al., 2005; Tsaousis et al., 2008; Williams et al., 2008). The important relevance of MCF proteins with respect to mitochondria is further supported by the fact that these proteins represent the only solute carriers in the inner mitochondrial membrane whereas membranes from other organelles harbor a mixture of different transport proteins which belong to different families and apparently originated due to different events. For example, carriers of the inner envelope of the chloroplast were derived from proteins of other organelles, some have clearly cyanobacterial ancestors (endosymbiotic gene transfer), others were acquired by horizontal gene transfer (from other bacteria, such as chlamydial species; Tyra et al., 2007).
It is tempting to speculate that the basic module of MCF proteins was newly created from the scratch by fusion of smaller DNA segments (Figure 1) shortly before or during endosymbiosis and that establishment of the first MCF protein drove or at least supported mitochondrial genesis. Gene duplications and modifications were apparently required to obtain diverse MCF proteins with different characteristics. Carriers of the MCF mediating the exchange of adenine nucleotides are a characteristic feature of all present-day mitochondria, of mitochondria-derived hydrogenosomes and were also detected in mitosomes (Chan et al., 2005; Tsaousis et al., 2008; Williams et al., 2008; Klingenberg, 2009). Because of this wide distribution and because of the important role of mitochondria in energy production it can be hypothesized that the first MCF protein was also involved in energy passage (ADP/ATP exchange). However, due to the fact that present-day MCF proteins exhibit a broad range of different substrate spectra (Figure 2) one might also assume that the first MCF protein was not a highly specific carrier but rather an “all-rounder” mediating the translocation of various solutes. A supposed relatively broad substrate spectrum of the first MCF protein might have provided an advantageous basis for fast establishment of a subset of different, more specific carriers. Some present-day MCF carriers exhibit relatively broad substrate specificities; although certain substrates are often preferred and others are transported with lower affinity or velocity. This suggests that the carriers binding pocket is not always highly selective and restrictive and that several substrates loosely fit in (Nury et al., 2006; Klingenberg, 2008). Interestingly, recent studies of metazoan MCF carriers showed that the ratio of hydrophobic amino acids at specific helix–helix interfaces increased during their evolution which indicates a trend toward a higher selectivity and to smaller substrates (Gong et al., 2010).
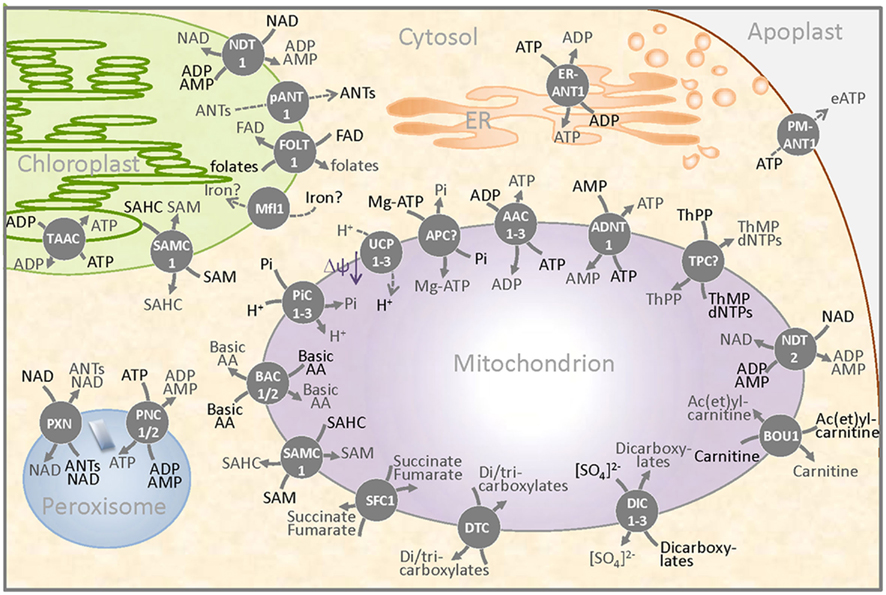
Figure 2. Schematic overview of a plant cell detail presenting the occurrence of MCF carriers in membranes of different compartments. MCF proteins were identified in the inner membrane of mitochondria and plastids, in the thylakoids, in the endoplasmic reticulum (ER), in peroxisomes, and in the plasma membrane (separating the cytosol and the apoplast). Three ADP/ATP carriers (AAC1–3), the ATP/AMP carrier (ADNT1), putative ATP–Mg/Pi carriers (APC?), and the NAD/adenine nucleotide exchanger mediate the passage of ATP, ADP, AMP, or of the adenylated cofactor NAD across the inner mitochondrial membrane. The mitochondrial transport of carboxylates is mediated by three dicarboxylate carriers (DIC1–3), by a dicarboxylate/tricarboxylate carrier (DTC), and by a carrier probably specific for succinate/fumarate exchange. The controlled flux of protons reduces the mitochondrial membrane potential (ΔΨ) and is catalyzed by two proven and one predicted uncoupling protein (UCP1–3). The transport of the cofactor thiamine pyrophosphate in exchange with thiamine monophosphate or (d)NTPs is probably catalyzed by a postulated mitochondrial thiamine pyrophosphate carrier (TPC?). Basic amino acids leave and enter the mitochondrion via the carriers BAC1 and BAC2 (BAC1–2) and the carnitine/ac(et)ylcarnitine carrier and the S-adenosylmethionine carrier (SAMC1) provide carbon moieties to the organelle. The three carriers PiC1–3 allow the net import of phosphate that can act as substrate of ATP synthesis and as counter exchange substrate of other MCF proteins. Plastids also harbor MCF proteins. An adenine nucleotide exporter (pANT1), a NAD/adenine nucleotide exchanger (NDT1), a possible folate carrier (FOLT1), and a Mitoferrin-like protein reside in the inner envelope membrane and provide important substrates and cofactors to the plastid. In addition to its localization in mitochondria, the carrier SAMC1 was also identified in the plastid. ATP provision to the thylakoid and to the ER is probably facilitated by the respective ATP/ADP exchangers – TAAC and ER-ANT1. Peroxisomes contain specific adenine nucleotide carriers (PNC1/2) as well as the NAD transporter PXN. In the plasma membrane, the protein PM-ANT1 might be involved in the provision of extracellular ATP.
Most eukaryotic organisms possess a large set of MCF proteins and phylogenetic analyses of the corresponding amino acid sequences demonstrate that the various MCF carriers from plants (Arabidopsis thaliana), animals (Homo sapiens), and fungi (Saccharomyces cerevisiae) build independent clades (Figure 3; Figures A1–A5 in Appendix; Palmieri et al., 2011). The branching pattern of these main clades generally does not reflect the affiliation to the different organismic groups but is rather caused by differences in functional properties (Palmieri et al., 2011). For example the cluster containing basic amino acid and carnitine/acylcarnitine carriers is clearly separated from the clusters comprising phosphate carriers (PiC), iron transporters, adenine nucleotide carriers, etc. (Figure 3). Almost every functional cluster comprises carriers from animals, yeast, and plants. This fact suggests that functionally different carriers probably evolved by multiple events of gene duplication, sequence alterations, and protein specification before the separation of the three eukaryotic kingdoms. A basic set of MCF carriers with different transport properties, substrate specificities, and localization might have already existed in ancient protists and these carriers were the evolutionary basis for the functional groups that are still required and retained in present-day eukaryotes (Figure 3). Multi-gene families are often arranged in physical clusters in the respective genomes. A collective grouping of the respective members is indicative for their establishment by recent gene duplications (Sappl et al., 2004). However, sequences encoding different eukaryotic MCF carriers are generally widely distributed across the different chromosomes and most are separated by a high number of other genes. This pattern also argues against recent duplication events as basis of many MCF proteins.
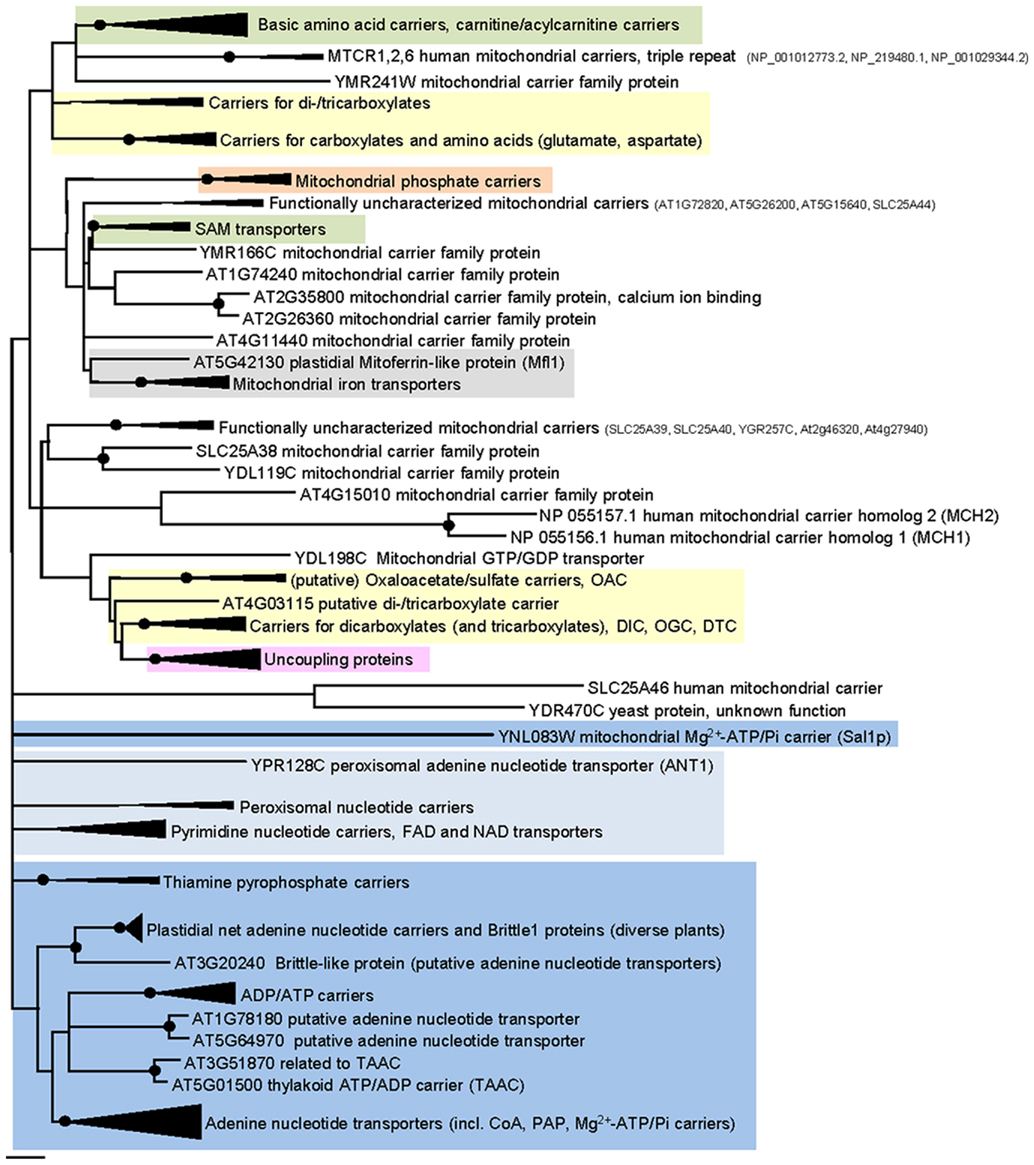
Figure 3. Phylogenetic relationships of MCF proteins. A phylogenetic tree based on amino acid sequences of the MCF proteins from Homo sapiens, Saccharomyces cerevisiae, and Arabidopsis thaliana is shown. The tree was calculated with MEGA5 using the maximum likelihood treeing method and the JTT model of amino acid substitution. The analysis involved 164 amino acid sequences (Table S1 in Supplementary Material). All positions containing gaps and missing data were eliminated. There were a total of 147 positions in the final dataset. Nodes that are supported by bootstrap values (maximum likelihood and maximum parsimony, 1000 replications) higher than 70 are indicated by black dots. Different functional groups (clusters or sub-clusters) are shaded: basic amino acid carriers, carnitine/acylcarnitine carriers, and SAM transporters in green, carriers for carboxylates, dicarboxylates, tricarboxylates, glutamate, and aspartate in yellow, mitochondrial phosphate carriers in orange, iron transporters in gray, uncoupling proteins in purple, peroxisomal ADP/ATP carriers, pyrimidine nucleotide, FAD, and NAD carriers in very pale blue, adenine nucleotide and thiamine pyrophosphate transporters transporters in blue). The shaded subsets are displayed in detail in Figures A1–A5 in Appendix.
Some functional groups are not present in all eukaryotic lineages (Palmieri et al., 2011). Succinate/fumarate carriers are apparently absent in animals or yeasts lack uncoupling proteins (UCPs; Figure A1 in Appendix). The presence of a functional group in two of the three lineages suggests that the remaining lineage lost the respective carrier function because it was compensated by other transporters and/or no more required. The presence of a carrier function in solely one lineage however, might be indicative for its new invention after the separation of the eukaryotes. A mitochondrial GTP/GDP carrier exists only in yeasts and the subgroup of plastidial adenine nucleotide carriers and Brittle1 proteins is restricted to plants (Figure 3). In the functional clades, paralogs/isoforms of one species often form a highly related subgroup (Figures A1–A5 in Appendix). The relation in between these paralogs is more pronounced than that to the corresponding orthologs from other species. The three AACs from Arabidopsis for example, form a small sub-cluster distinct from that of the yeast isoforms and from the human orthologs (Figure A2 in Appendix). This phylogenetic positioning suggests that the additional isoforms evolved by gene duplications independently in the different eukaryotic lineages (Palmieri et al., 2011).
In certain functional clades, plants exhibit a higher number of proven or predicted MCF paralogs. This might indicate that gene duplication and/or gene retention occurred more often in plants. Immotile plants might exhibit additional mcf genes that are differentially regulated (expressed) to react more flexible to changing environmental conditions, biotic or abiotic stresses. However, also mobile algae possess a higher number of MCF paralogs than human or yeast, at least in some functional clades (Palmieri et al., 2011). It is tempting to assume that plants require additional MCF proteins because they possess an additional organelle, the plastid. MCF proteins that are directly or indirectly associated with plastid function might have originated by gene duplication of the mitochondrial pendant and rerouting to the plastid, like the plastidial NAD transporter (NDT1) which is a close homolog to the mitochondrial NAD transporter (NDT2; Figure A3 in Appendix; Palmieri et al., 2009). Furthermore, plastids might have gained MCF proteins also due to dual targeting of previously mitochondrial localized carriers. Recent studies revealed that several MCF proteins exhibit dual targeting and thus might fulfill functions in different organelles, like mitochondria as well as plastids (Agrimi et al., 2004; Bouvier et al., 2006; Palmieri et al., 2006a; Bahaji et al., 2011a,b). The existence of certain plastid-specific MCF proteins suggests that the respective gene duplications and modifications occurred after separation of the plant lineage. It is important to mention that plant specific MCF proteins (e.g., the adenine nucleotide carriers ER-ANT1 and PM-ANT1) do not only comprise plastidial carriers (Leroch et al., 2008; Rieder and Neuhaus, 2011).
Plant and algal genomes contain a high number of MCF coding sequences. In the past decades, the function of more and more carriers was identified; however, the physiological role of the majority of plant MCF proteins is still unclear. Not even one-half of the 58 MCF proteins of the frequently analyzed model plant Arabidopsis thaliana are biochemically characterized in detail. In rare cases, the physiological impact of carriers was deduced from metabolic defects of corresponding loss-of-function mutants. Import studies performed with intact yeast or E. coli cells expressing the respective carriers, or with the purified, recombinant proteins reconstituted into lipid vesicles, allowed the identification of specific inhibitors and the determination of detailed biochemical properties, like substrate specificities, transport kinetics, modes, and electrogenicity (Palmieri et al., 2000a; Picault et al., 2004). According to the transport characteristics, four functional subfamilies of the MCF from human, yeast, and Arabidopsis were recently proposed (Palmieri et al., 2011). The first subfamily comprises nucleotide and nucleotide derivate transporters. MCF proteins mediating the passage of di- and tricarboxylates or keto acids constitute the second subfamily. Amino acid carriers and carnitine/acylcarnitine carriers are affiliated to a third functional group and transporters with other substrates like UCPs (H+ passage) or PiCs represent the fourth subdivision. The MCF subfamilies can be further sub-divided into functionally related groups (for example AACs, ATP–Mg/Pi transporters, and NAD transporters, etc., constitute the subfamily of nucleotide and dinucleotide carriers; Palmieri et al., 2011). Each of these MCF groups is characterized by typical triplets in conserved important domains (such as regions extending into the translocation cavity). Subgroups that share some substrates often also share some of these characteristic triplets (Palmieri et al., 2011). The affiliation of a biochemically yet uncharacterized MCF protein to one functional group seems to be a proper basis allowing the prediction of its putative transport substrates and thus of a possible physiological function. In the following, we focus on functional groups and subgroups of our phylogenetic analysis (Figure 3; Table S1 in Supplementary Material) and describe the properties of biochemically characterized members from Arabidopsis.
Arabidopsis possesses three typical AACs (AAC1: At3g08580, AAC2: At5g13490, and AAC3: At4g28390; Figure 2) involved in mitochondrial energy export (Haferkamp et al., 2002). These carriers are structurally and functionally highly related to their orthologs from animals or yeast, they exhibit identical substrate specificities and inhibitor sensitivities and their transport characteristics are similarly influenced by the membrane potential (Haferkamp et al., 2002). However, in contrast to the mitochondrial AACs from other organisms, plant AACs are characterized by an N-terminal extension (approximately 60 amino acid residues in length) that is not essential for but might support correct targeting to the mitochondrion. In phylogenetic analyzes of MCF carriers AACs cluster together (Palmieri et al., 2011). However, the plant paralogs are separated from fungal and animal AACs (Figure A2 in Appendix). Interestingly, the Arabidopsis genome encodes two additional AAC-related proteins (At5g17400, At5g56450) that share significant homology to the AACs (Figure A2 in Appendix). Recently, the physiological function of these carriers was clarified.
The first AAC-related carrier (At5g17400) lacks the mitochondrial targeting sequence of plant AACs. This carrier was shown to reside in the plant ER (Leroch et al., 2008) and therefore was called ER-located adenine nucleotide transporter 1 (ER-ANT1; Figure 2). When heterologously expressed in E. coli, the recombinant protein was able to mediate ATP/ADP exchange and thus ER-ANT1 might fuel ATP dependent processes in the ER lumen, like protein translocation, folding, or maturation. Interestingly, yeast and animals do not possess a definite ER-ANT1 ortholog or AAC-related proteins (with comparably high sequence similarities to mitochondrial AACs) and apparently use another MCF member or a completely different carrier-type for energy provision to the ER. The existence of other/additional ER-located ATP importers is also supported by the fact that ER-ANT1 Arabidopsis knock out mutants (although impaired in growth) are still able to survive and to produce fertile seeds (Leroch et al., 2008).
The second functionally investigated AAC-related carrier (PM-ANT1: At5g56450) is N-terminally slightly longer (plus 20 amino acids) than ER-ANT1 but a sequence extension comparable to that of the plant mitochondrial AACs is missing (Rieder and Neuhaus, 2011). The GFP-fusion-protein was targeted to the plant plasma membrane, the recombinant carrier (PM-ANT1) showed ATP and ADP uptake into E. coli and probably also accepts a broad range of (desoxy) nucleotides as substrates. Until now the transport mode of PM-ANT1 is not clarified, however, its relatedness to AAC proteins might suggest a function in nucleotide exchange. In animal systems but also in plant cells extracellular ATP (eATP) represents an important signal molecule and the localization of PM-ANT1 in the plasma membrane indicates a possible role in eATP supply and metabolism (Figure 2; Rieder and Neuhaus, 2011). PM-ANT1 from Arabidopsis is highly expressed in developing pollen and mutant plants reduced in PM-ANT1 activity are impaired in flower development, in particular in anther dehiscence. Therefore, PM-ANT1 was suggested to mediate ATP export specifically from pollen cells and that enhanced eATP acts as a signal received by the anther stomium cells. In addition to the PM-ANT1 also other proteins or mechanisms have to be involved in eATP release, because solely the flowers of the mutant plants show developmental or metabolic defects whereas other plant organs seem to be unaffected (Rieder and Neuhaus, 2011). In fact, several other transport processes were identified to be involved in ATP release from animal but also plant cells. The lack of clear orthologs of ER-ANT1 and PM-ANT1 in yeast or animals (Figure A2 in Appendix) suggests that these proteins were newly invented or retained to fulfill specific functions in plants.
Several MCF carriers from Arabidopsis (At1g78180, At5g64970, At3g51870 At5g01500, At1g14560, At4g26180, At4g01100, At3g53940, At3g55640, At2g37890, At5g61810, At5g07320, and At5g51050) show lower but still important amino acid similarities to the AACs and the AAC-related proteins and thus can be supposed to mediate adenine nucleotide transport (Figure 3; Figure A2 in Appendix). Two of these carriers were characterized and proven to catalyze the exchange of adenine nucleotides or related compounds.
The first one (At4g01100) is affiliated to a phylogenetic subgroup containing mitochondrial CoA and phospho-adenosyl-phosphate (PAP) transporters from human (including the Graves’ disease carrier) and yeast (Fiermonte et al., 2009; Palmieri et al., 2011; Figure A2 in Appendix). This carrier was named adenine nucleotide transporter (ADNT1: At4g01100) because the reconstituted recombinant protein transported several adenylated (desoxy) nucleotides and adenosine 5-sulfophosphate (APS) in a counter exchange mode, whereas CoA transport was not measurable (Palmieri et al., 2008b). Whether also PAP represents a further substrate of ADNT1 was not investigated. Localization studies revealed that the ADNT1–GFP-fusion-protein is targeted to mitochondria. Several facts supported the idea that ADNT1 might stimulate mitochondrial energy provision of heterotrophic tissues by facilitating the import of cytosolic AMP in exchange with mitochondrial ATP (Figure 2). First, the carrier favors AMP instead of ADP as counter exchange substrate of ATP. Second, the adnt1 gene is highly expressed in non-photosynthetic as well as in fast growing tissues. Third, roots of ADNT1 loss-of-function mutants are shorter in size than that of wild type plants and are impaired in mitochondrial respiration (Palmieri et al., 2008b).
The second characterized protein with moderate similarities to the AAC proteins (At5g01500) does not clearly affiliate to the AACs or to the CoA and PAP carrier subgroup (Figure 3). This carrier was shown to mediate ATP/ADP exchange across the bacterial plasma membrane when heterologously expressed in E. coli and localization studies revealed a presence in the thylakoid membrane (Figure 2; Thuswaldner et al., 2007). Investigations of TAAC loss-of-function mutants led to the assumption that TAAC mediated ATP provision to the thylakoid lumen plays a role in photoinhibition and photoprotection of photosystem II and in the regulation of the electrochemical H+ gradient across the thylakoid membrane (Yin et al., 2010). However, it is important to mention that TAAC is not only located in thylakoids but also in the plastid envelope and thus a dual targeting of the carrier is likely (Thuswaldner et al., 2007). Furthermore, increased transcript and protein abundance in growing and heterotrophic tissues points to an alternative/additional function of TAAC in developing and “thylakoid free” plastids. Due to certain important structural similarities to CoA/PAP transporters and ADNT1 it was recently suggested that TAAC accepts further substrates than solely ATP and ADP (Palmieri et al., 2011). The high sequence similarity of one functionally uncharacterized carrier (At3g51870) and TAAC indicates a similar biochemical function (Figure 3).
In human and yeast, Mg2+–ATP/Pi carriers (APC, Sal1p) are involved in the replenishment of the mitochondrial purine nucleotide pool in accordance to organellar demands (Fiermonte et al., 2004; Traba et al., 2008, 2009). These carriers catalyze an electroneutral Mg2+–ATP/phosphate exchange and hence, allow net provision of ATP. Three of the remaining putative adenine nucleotide carriers (At5g07320, At5g51050, At5g61810) form a well supported sub-cluster with the human APCs (Figure A2 in Appendix). Accordingly, these proteins are supposed to catalyze net nucleotide provision however, their biochemical and physiological characteristics have not been analyzed yet.
Two further proteins from Arabidopsis (At4g32400, At3g20240) exhibit at least moderate similarities to the different groups of adenine nucleotide carriers. These carriers are absent in yeast and human and thus probably originated after the establishment of the plant kingdom (Leroch et al., 2005; Comparot-Moss and Denyer, 2009). One of these carriers (pANT1: At4g32400) was shown to reside in plastids and to mediate a net transport of adenylated nucleotides (Kirchberger et al., 2008). In yeast and human the de novo synthesis of nucleotides takes place in the cytosol whereas in plants these pathways are located in the plastid (Zrenner et al., 2006). Accordingly, pANT1 might possibly supply newly synthesized adenine nucleotides to the cytosol (Figure 2). The potential Arabidopsis APC proteins are supposed to mediate net uptake of these molecules into mitochondria. Recent investigations demonstrated that pANT1 exhibits also a mitochondrial localization (Bahaji et al., 2011b). The N-terminal extension alone possesses the capacity to guide GFP to the plastid whereas the N-terminally truncated (mature carrier) is targeted to mitochondria. Until now, the physiological role of such a net adenine nucleotide transporter in mitochondria is unclear (Bahaji et al., 2011a). In the context of a possible evolution of this protein by gene duplication of a mitochondrial adenine nucleotide carrier sequence and by addition of a plastidial targeting sequence one might speculate that the interior targeting information represents a relict of an ancient mitochondrial localization.
Both carriers (the plastidial adenine nucleotide transporter pANT1 and At3g20240) form a well supported subgroup with functionally related carriers from other plants including, ADP glucose carriers (Brittle1, BT1) from cereals (Figure 3; Kirchberger et al., 2007; Comparot-Moss and Denyer, 2009). In most plants, ADP glucose, the carbohydrate precursor for starch production, is synthesized in the plastid. However, culture forms of cereals with high starch levels possess a cytosolic variant of the ADP glucose pyrophosphorylase (AGPase; Denyer et al., 1996; Beckles et al., 2001). Accordingly, in these plants ADP glucose has to be transported from the cytosol into the plastids. The protein BT1 is highly abundant in plastids of maize endosperm (Cao et al., 1995). Furthermore, a maize mutant with a defect in the brittle1 locus is impaired in starch synthesis and possesses enhanced cytosolic ADP glucose levels in immature kernels (Shannon et al., 1998). BT1 shows important similarities to pANT1. Actually, they exhibit 79% identity and 89% similarity in the transport domain and identical substrate contact points (Traba et al., 2011). However, BT1 facilitates the exchange of ADP glucose with ADP or AMP (Kirchberger et al., 2007) whereas pANT1 catalyzes a unidirectional transport of AMP, ADP, and ATP but not of ADP glucose. Accordingly, modification of the substrate spectrum and the change from uniporter to antiporter apparently requires only minor modifications. In fact, the transport mode of some MCF carriers can be changed solely by treatment with reducing agents and hence modifications of thiol groups are sufficient to convert the transport properties (Dierks et al., 1990).
ADP glucose transporters are restricted to monocotyledonous cereals with starch-rich tissues (Comparot-Moss and Denyer, 2009) and form a phylogenetic clade closely related to two clades containing proven or suggested plastidial adenine nucleotide transporters (clade pANT1 and clade pANT2; Figure A2 in Appendix). Clade one consists of pANT1 proteins from diverse plant lineages whereas the pANT2s clade comprises solely members from cereals and grasses. The monocot-specific genes of pANT2 and the ADP glucose carriers are suggested to have originated by segmental or whole-genome duplication (Comparot-Moss and Denyer, 2009). The first pANT was probably established before the separation of monocotyledonous and dicotyledonous plants and pant gene duplication in the monocotyledonous line probably gave rise to the homologs pANT1 and pANT2. In ancestral grasses an additional gene duplication of a pANT gene (maybe pANT2) occurred and provided the basis for the establishment of the ADP glucose transporter (Comparot-Moss and Denyer, 2009).
Thiamine pyrophosphate is a coenzyme involved in diverse metabolic pathways. In insects, human, and yeast MCF proteins specific for this cofactor (thiamine pyrophosphate carriers, TPC) have been identified (Marobbio et al., 2002; Lindhurst et al., 2006; Iacopetta et al., 2010). Generally, nucleotides and deoxynucleotides represent additional substrates of the TPCs and in fact, the human TPC was initially described to act as a deoxynucleotide carrier (Iacobazzi et al., 2001). TPCs from the different eukaryotic lineages exhibit overlapping substrate spectra, however, they also differ in certain aspects (like in the preference of different nucleotides). The proteins from human and Drosophila melanogaster act in a strict antiport mode (Iacopetta et al., 2010) whereas the yeast pendant is also capable for substrate uniport (Marobbio et al., 2002). In contrast to the other characterized TPC the insect homolog does not accept thiamine monophosphate (ThMP) as substrate (Marobbio et al., 2002). The Arabidopsis genome encodes two homologous sequences (At3g21390 and At5g48970) similar and related to the TPCs and thus, one or both of the corresponding proteins might act as a transporter of this cofactor in plant mitochondria (Figure 2; Palmieri et al., 2011). The TPCs clearly constitute a phylogenetic group distantly related to the groups of adenine nucleotide carriers (Figure 3).
Further phylogenetic groups of MCF carriers also rather distantly related to the mitochondrial adenine nucleotide carriers comprise nucleotide transporters from peroxisomes, mitochondrial pyrimidine nucleotide carriers (PyrNC), NAD/ADP exchangers, and FAD transporters (Palmieri et al., 2011; Figure 3; Figure A3 in Appendix). Until now, in Arabidopsis, three MCF proteins with peroxisomal localization (At3g05290, At5g27520, and At2g39970) have been identified. The peroxisomal nucleotide carriers PNC1 (At3g05290) and PNC2 (At5g27520) are functionally similar to their peroxisomal counterpart from yeast (ANT1; Palmieri et al., 2002; Linka et al., 2008). They catalyze the specific exchange of ATP with ADP or AMP (Figure 2) and are able to complement a yeast mutant (Δant) deficient in peroxisomal ATP uptake (Linka et al., 2008). Investigations of transgenic Arabidopsis lines demonstrated that PNC1 and PNC2 play an essential role in energy provision to plant peroxisomes and that lack of these proteins significantly impairs fatty acid breakdown and other peroxisomal reactions (Linka et al., 2008).
Nicotinamide adenine dinucleotide (NAD) and its phosphorylated derivative NADP are characteristic factors of redox homeostasis. In addition to their basic role in cellular metabolism these pyridine nucleotides were shown to control intracellular processes, like Ca2+ signaling or transcriptional regulation. NAD is synthesized in the cytosol but is required in several compartments and thus has to be delivered to the respective organelles. Recently, two plant homologs (NDT1: At2g47490 and NDT2: At1g25380) of the mitochondrial NAD transporters from yeast (NDT1: YIL006W and NDT2: YEL006W) were identified (Todisco et al., 2006; Palmieri et al., 2009). Complementation of yeast double mutants lacking its mitochondrial NDTs suggested that also the plant MCF homologs mediate NAD import into mitochondria. Import studies with reconstituted plant NDTs demonstrated that both proteins perform a fast NAD counter exchange with various nucleotides – preferentially with ADP or AMP as well as a slow unidirectional NAD transport. However, GFP-based targeting analyzes revealed a mitochondrial localization only for plant NDT2, whereas plant NDT1 apparently resides in plastids (Figure 2; Palmieri et al., 2009). Plastids and mitochondria of fast growing tissues require high amounts of NAD. Significant net NAD uptake via the plant NDTs is probably facilitated by interaction with a net adenine nucleotide transporter that provides the counter exchange adenylates to the respective organelle.
Just recently the function of the third peroxisomal MCF protein (At2g399970) was identified. Because of certain similarities and to the peroxisomal adenine nucleotide carriers (PNCs from Arabidopsis and ANT1 from yeast) this protein initially was assumed to catalyze ATP transport. However, it was not able to restore ATP import into peroxisomes of the yeast Δant mutant (Linka et al., 2008; Bernhardt et al., 2011). Biochemical studies demonstrated that this peroxisomal protein (called PXN) catalyzes NAD uptake in exchange with mainly AMP but also with NADH, ADP, and with nicotinate adenine dinucleotide, an intermediate of NAD synthesis (Figure 2). The absence of a functional peroxisomal NAD carrier in Arabidopsis mutants led to an impaired NAD-dependent β-oxidation and to the accumulation of oil bodies in the seedlings (Bernhardt et al., 2011). Apart from its possible role as a peroxisomal redox shuttle (NAD/NADH exchange) this carrier might also be (directly or indirectly) involved in net uptake of NAD and in the establishment of the peroxisomal NAD pool.
Similar to NAD also FAD operates as an important cofactor in multiple cellular pathways and in different organelles. Moreover, FAD synthesis involves cytosolic reactions, mitochondrial, and plastidial enzymes and thus precursors and intermediates must enter and leave the respective compartments. Complementation studies with a yeast mutant deficient for its mitochondrial folic acid carrier suggested that the Arabidopsis MCF protein FOLT1 (At5g66380) acts as a transporter for folic acid and folate derivates (Tzagoloff et al., 1996; Bedhomme et al., 2005). FOLT1 was targeted to plastids when fused to GFP (Figure 2). At first glance Arabidopsis does not encode a clearly identifiable second FOLT isoform. The absence of a visible phenotype in Arabidopsis mutant plants lacking functional FOLT1 however is indicative for the existence of an alternative plastidial folate transport system (Bedhomme et al., 2005). Interestingly, a transporter of the major facilitator superfamily was shown to mediate FAD transport into plastids (Klaus et al., 2005).
Generally, the plastidial NAD and FAD carriers form a phylogenetic sub-cluster together with their functional mitochondrial counterparts as well as with mitochondrial PyrNC from yeast and human (Figure 3; Figure A3 in Appendix). The peroxisomal NAD and adenine nucleotide transporters from different species exhibits low (plant and human) or rather insignificant support (yeast; Figure 3; Figure A3 in Appendix). Nevertheless, the phylogenetic relation of functional homologs from one species is well supported and argues for their establishment by recent gene duplications.
Di- and tricarboxylates are important substrates and intermediates of the mitochondrial Krebs cycle and they are required in several other metabolic processes, for example in the de novo synthesis of amino acids and nucleotides, in gluconeogenesis or in the glyoxylate cycle.
In Arabidopsis, three mitochondrial dicarbonic acid carriers (DIC1: At2g22500, DIC2: At4g24570, and DIC3: At5g09470; Figure 2) were recently identified (Palmieri et al., 2008a). DIC1 and DIC2 are highly expressed in different tissues, whereas dic3-RNA is hardly detectable (restricted to flower buds and siliques). Similar to their counterparts from yeast or animals (Palmieri et al., 1996; Fiermonte et al., 1998), DIC1–3 are able to transport malate, oxaloacetate, succinate, maleate, malonate, phosphate, sulfate, and thiosulfate; however they exhibit slightly different substrate preferences than their non-plant orthologs (Palmieri et al., 2008a). DIC mediated transport of dicarboxylates in exchange with phosphate or sulfate allows the provision of substrates for mitochondrial reactions/respiration. Whereas malate/oxaloacetate exchange by the DIC proteins might be an important factor of the mitochondrial redox shuttle (metabolic interaction with cytosolic and mitochondrial NAD-dependent malate dehydrogenases).
The dicarboxylate/tricarboxylate carrier from Arabidopsis (DTC: At5g19760; Figure 2) exhibits high similarities to the DICs and was shown to mediate an electroneutral transport of different single protonated tricarboxylates (citrate, isocitrate, and aconitate) in exchange with a broad variety of unprotonated dicarboxylates (2-oxoglutarate, oxaloacetate, malate, maleate, succinate, and malonate; Picault et al., 2002). Due to these exchange capacities DTC is able to adapt and control the amount of Krebs cycle intermediates according to the cellular metabolism. In fact, in every plant tissue dtc transcripts were detectable, however at different levels (Picault et al., 2002). Apart from a proposed basic metabolic role in the Krebs cycle DTC was additionally supposed to be involved in the mitochondrial provision of organic acids for ammonium assimilation (in the plastids) because the dtc gene expression was significantly increased by nitrate application (after a nitrate starvation phase; Picault et al., 2002). Interestingly, plants seem to contain “hybrid” carriers when compared to animals and yeast. The DTC from Arabidopsis possesses characteristics overlapping with that of the mammalian oxoglutarate and citrate carriers and the DIC proteins from Arabidopsis exhibit combined properties of the DICs and oxaloacetate/sulfate carrier (OAC) from yeast (Catoni et al., 2003a,b).
One MCF sequence (At5g01340: SFC1) from Arabidopsis forms a subgroup with the succinate/fumarate carrier from yeast (ACR1) but also exhibits moderate similarities to the citrate and oxodicarboxylate carriers from yeast and human (Fiermonte et al., 2001; Palmieri et al., 2001a, 2011; Catoni et al., 2003b). The yeast carrier ACR1 accepts a broad spectrum of di- and tricarboxylates with preference of succinate and fumarate (Palmieri et al., 1997). The plant carrier SFC1 was suggested to act in a similar way (Figure 2) and to exhibit overlapping biochemical properties as ACR1 because it complemented the growth phenotype of a yeast mutant lacking this mitochondrial protein (Catoni et al., 2003b). The expression pattern of the sfc1 gene indicates a possible involvement of the carrier in gluconeogenesis, in particular of young (germinating) seedlings and pollen, as well as in ethanolic fermentation (Catoni et al., 2003b).
Phylogenetic analyses of members of the MCF showed that carriers transporting di- and tricarboxylates form two distinct clusters (Figure 3; Figure A1 in Appendix; Catoni et al., 2003b). One cluster comprises the diverse DIC proteins, the DTC from plant and the human oxoglutarate carrier (OGC). Moreover, plant DIC isoforms are highly homologous and clearly separated from the DICs of yeast and animals and therefore, most likely arose by gene duplication of one common ancestral plant dic sequence (Figure A1 in Appendix; Catoni et al., 2003b). UCPs from different organisms as well as a group consisting of an OAC from yeast, two putative homologs from human and a so far biochemically uncharacterized transporter from Arabidopsis (At4g03115) are more distantly related to the DIC proteins but also affiliated to the first cluster (Catoni et al., 2003b; Palmieri et al., 2011). The succinate/fumarate carriers from plant and yeast (SFC1, ACR1) are clearly separated from the cluster containing the DIC proteins and are rather related to citrate carriers and different carbonic acid and amino acid transporters from yeast and human (citrate, oxodicarboxylate oxoglutarate, glutamate, aspartate carriers; Catoni et al., 2003b; Palmieri et al., 2011).
Uncoupling proteins are generally known to mediate the passage of protons across the inner mitochondrial membrane (Figure 2) in a nucleotide-sensitive and fatty-acid-dependent manner (Klingenberg, 1990; Garlid et al., 1996). UCP activity reduces the proton gradient and hence ATP production, it stimulates respiratory (enzyme) activity and finally leads to heat generation. Accordingly, UCPs were thought to play an exclusive role in thermogenesis of newborn, cold-acclimated, and hibernating animals and to be absent in yeast or plants. However, UCP proteins were identified in diverse plant lineages and UCP1 from Arabidopsis (also called PUMP1: At3g54110) and related proteins from other plants were demonstrated to play a role in proton transport at low temperatures or in thermogenesis (Vercesi et al., 1995; Borecky et al., 2001; Ito et al., 2003, 2006). The expression of the remaining two Arabidopsis UCPs (UCP2: At5g58970 and UCP3: At1g14140) indicates a different physiological function (Watanabe et al., 1999; Borecky et al., 2006). The activation of plant UCPs by reactive oxygen species and their expression pattern in non-thermogenic plants suggests that the primary role of most plant UCPs is not the organ specific production of heat but rather the regulation of mitochondrial energy synthesis in response to stress factors (Laloi, 1999; Ito et al., 2003, 2006; Borecky et al., 2006; Vercesi et al., 2006). Plant UCP1 and UCP2 form a subgroup related to human UCP1–3 whereas the third UCP from Arabidopsis clusters with human UCP4 and UCP4-related proteins and thus was supposed to represent a rather ancient UCP form (Figure A1 in Appendix; Hanak and Jezek, 2001). As mentioned in the previous chapter UCPs and DICs exhibit important amino acid sequence similarities and phylogenetic relation (Figure 3; Borecky et al., 2006). And in fact, prior to their biochemical investigation it was controversially discussed whether the DIC proteins from plants represent dicarbonic acids carriers or UCPs (Millar and Heazlewood, 2003; Picault et al., 2004; Borecky et al., 2006).
In plants, basic amino acids serve as an important nitrogen source and are mobilized from storage proteins during seed germination. Following protein degradation the basic amino acid arginine enters plant mitochondria where it is converted by arginase to urea and ornithine (Goldraij and Polacco, 2000). Ornithine acts as precursor for the synthesis of glutamine, proline, polyamines and alkaloids (and also arginine) and accordingly has to leave the mitochondrion.
In Arabidopsis two carriers for basic amino acids (BAC1: At2g33820 and BAC2: At1g79900; Figure 2) were identified by complementation of the yeast mutant arg11 defective in mitochondrial ornithine/arginine transport (carrier: ORT1; Catoni et al., 2003a; Hoyos et al., 2003; Palmieri et al., 2006b). Transport studies with reconstituted recombinant proteins revealed that BAC1 and BAC2 mediate the exchange of the basic amino acids arginine, lysine, ornithine, and histidine (Hoyos et al., 2003; Palmieri et al., 2006b). BAC2 has a broader substrate spectrum than BAC1; it additionally transports citrulline and is less stereospecific because it also allows the passage of D-amino acids at high rates. BAC1 is most likely preferentially involved in the mobilization of storage proteins, whereas BAC2 was shown to contribute to proline accumulation in response to hyperosmotic stress (Hoyos et al., 2003; Toka et al., 2010).
In phylogenetic analyzes, the Arabidopsis carriers BAC1, BAC2, and BOU1 (“A BOUT DE SOUFFLE”; At5g46800) are positioned in an MCF subgroup consisting of basic amino acid carriers and carnitine carriers as well as of so far biochemically non-characterized MCF proteins from animals or yeast (Figure 3; Figure A4 in Appendix). The BOU1 protein exhibits important structural similarities to BAC1 and to the ornithine carriers from yeast and human and was suggested to fulfill a role in the degradation of storage lipids during seedling germination. In peroxisomes fatty acids are mobilized and oxidized and subsequently delivered to mitochondria to undergo respiration. BOU1 was hypothesized to catalyze the exchange of ac(et)ylcarnitine and carnitine at the mitochondrial membrane and by this to supply carbons to the mitochondrion (Figure 2; Lawand et al., 2002). Because plant peroxisomes export the majority of carbons in form of citrate and not in form of ac(et)ylcarnitine and because an acyl-CoA carnitine acetyltransferase has not yet been identified in plants the primarily supposed function is questionable (Pracharoenwattana et al., 2005). Accordingly, a detailed investigation of the functional characteristics of BOU1 is required to further clarify its physiological role. Just recently, the mammalian carnitine–acylcarnitine carrier-like protein (SLC25A29) with palmitoylcarnitine transporting activity (Sekoguchi et al., 2003) was shown to additionally transport basic amino acids, because it was able to rescue impaired ornithine transport when overexpressed in fibroblasts lacking the ornithine carrier (ORT1; hyperornithinemia–hyperammonemia–homocitrullinuria syndrome; Camacho and Rioseco-Camacho, 2009).
It is important to mention that MCF carriers for the transport of non-basic amino acids are known in animals and fungi but not in plants. Carriers specific for glutamate were identified in human (Fiermonte et al., 2002) and for glutamate and aspartate in human and yeast (Palmieri et al., 2001b; Cavero et al., 2003). These carriers are phylogenetically related to carboxylate carriers from different organisms (Figure A1 in Appendix).
In nearly all cellular methylation-processes the amino acid derivative S-adenosylmethionine (SAM) acts as a C1-group donor, however, in specific reactions it also operates as a provider of amino, ribosyl, or aminopropyl groups. SAM is exclusively synthesized in the plant cytosol and has to be imported into mitochondria as well as into plastids where it is required as substrate for, e.g., methylation of organellar DNA, RNA, and proteins (Hanson and Roje, 2001). For re-methylation the resulting product S-adenosylhomocysteine (SAHC) has to be exported from the respective organelle into the cytosol. Arabidopsis possesses two homologs (SAMC1: At4g39460 and SAMC2: At1g34065) to the SAM transporters from yeast and mammalia (Marobbio et al., 2003; Agrimi et al., 2004). The SAM transporters form a (sub)-cluster clearly separated from other amino acid carriers (Figure 3).
The amino acid sequence of both plant proteins possesses an N-terminal extension. The N-terminal sequence of SAMC1 targets GFP to the plastid, whereas the full-length SAMC1 resides in both, chloroplasts and mitochondria (Figure 2; Bouvier et al., 2006; Palmieri et al., 2006a). SAMC1 was shown to mediate the counter exchange of SAM and SAHC, to be of higher abundance than SAMC2 and to be expressed in various plant tissues (Bouvier et al., 2006; Palmieri et al., 2006a). Furthermore, Arabidopsis plants lacking functional SAMC1 exhibit a dwarf phenotype and are impaired prenyl-lipid metabolism (Bouvier et al., 2006). In comparison to SAMC1 the paralog SAMC2 is rather poorly investigated; its biochemical properties, subcellular localization, and physiological function are still unclear. However, due to high sequence similarities to SAMC1 and phylogenetic relation to SAM carriers from other organisms, a function of SAMC2 in SAM transport seems very likely (Figure A4 in Appendix).
Apart from ADP also phosphate has to be imported to fuel energy regeneration in mitochondria with substrates. Accordingly, mitochondrial ATP/ADP exchange requires a concerted phosphate transport. PiCs in the inner mitochondrial membrane physiologically interact with AAC proteins, they catalyze a Pi/H+ symport (or Pi/OH− antiport) and by this supply phosphate for ATP synthesis (Pratt et al., 1991; Stappen and Krämer, 1994). Arabidopsis encodes three proteins (At5g14040: PiC1, At3g48850: PiC2, and At2g17270: PiC3; Figure 2) related to mitochondrial PiC from human and yeast. In phylogenetic analyses all PiCs proteins form a (sub)-cluster separated from functionally different MCF proteins (Figure 3; Palmieri et al., 2011).
Complementation studies with a yeast mutant deficient in mitochondrial phosphate import confirmed that PiC1 and PiC2 from Arabidopsis act as PiCs (Hamel et al., 2004). The function of the third putative Arabidopsis PiC (more distantly related to the PiC1 and 2 plant isoforms, Figure A5 in Appendix) remains to be experimentally proven. AAC1 and PiC1 are supposed to be the predominant carriers providing the substrates for ATP synthesis because they are expressed in several tissues and show highest abundance in the mitochondrial membrane (Millar and Heazlewood, 2003).
Two so far uncharacterized carriers from Arabidopsis (At1g07030 and At2g30160) exhibit high similarities and well supported phylogenetic relation to mitochondrial iron transporters from animals (Mitoferrin 1 and 2) and yeast (MRS3 and 4; Mühlenhoff et al., 2003; Shaw et al., 2006; Froschauer et al., 2009; Paradkar et al., 2009). Therefore, these carriers might represent the plant homologs (Figure A5 in Appendix). The functional cluster of iron transporters is separated from functionally different MCF carriers but contains an additional member that exhibits lower similarities and a more distant relationship (with low bootstrap support) to the Mitoferrins (Figure 3). The corresponding protein is called Mitoferrin-like transporter (AtMfl1: At5g42130), is a component of the inner plastid envelope and investigations of mutant plants suggested its involvement in iron metabolism (Tarantino et al., 2011). Accordingly, AtMfl1 was supposed to mediate iron uptake into the chloroplast (Figure 2).
Interestingly, just recently the PyrNC from yeast (Marobbio et al., 2006) that clusters with the NAD and FAD carriers (Figure 3; Figure A3 in Appendix) was also suggested to be involved in mitochondrial iron transport (Yoon et al., 2011).
Until now the biochemical properties of many MCF members from plants are not or only poorly investigated. The vast majority of biochemically characterized plant MCF proteins comprises carriers from Arabidopsis, whereas carriers from other plant species or algae are clearly underrepresented. However, the investigation of MCF proteins that are restricted to plants and algae allows elucidating plant specific functions and properties. Phylogenetic analyses demonstrate that the various MCF carriers from Arabidopsis, human, and yeast build independent clades. Generally, the branching pattern of these main clades reflects differences in their biochemical and physiological properties and MCF proteins with similar functions often form distinct clusters. Several functional MCF groups can be deduced from phylogenetic analysis (Table S1 in Supplementary Material). These functional groups are generally in line with the groups recently proposed by Palmieri et al. (2011). The affiliation to a phylogenetic cluster or sub-cluster (containing functionally characterized carriers from other organisms) might help to get an idea about putative transport substrates of functionally uncharacterized pendants. Proteins of unknown function that possess functionally characterized paralogs or orthologs apparently are the most promising candidates for fast elucidation of their biochemical properties. However, proteins are not always clearly affiliated to a functional MCF cluster or form a cluster with solely biochemically uncharacterized MCF proteins. The identification of the functional properties of these proteins (of no distinct cluster, or of an uncharacterized cluster) is a particular challenge and of significant importance. It will extend our knowledge about the capacities of MCF proteins; it will provide new and probably unexpected insights into the general capacities of MCF proteins and represents an important basis for the characterization of related carriers.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at http://www.frontiersin.org/Plant_Traffic_and_Transport/10.3389/fpls.2012.00002/abstract
Agrimi, G., Di Noia, M. A., Marobbio, C. M., Fiermonte, G., Lasorsa, F. M., and Palmieri, F. (2004). Identification of the human mitochondrial S-adenosylmethionine transporter: bacterial expression, reconstitution, functional characterization and tissue distribution. Biochem. J. 379, 183–190.
Alcock, F., Clements, A., Webb, C., and Lithgow, T. (2010). Evolution. Tinkering inside the organelle. Science 327, 649–650.
Bahaji, A., Munoz, F. J., Ovecka, M., Baroja-Fernandez, E., Montero, M., Li, J., Hidalgo, M., Almagro, G., Sesma, M. T., Ezquer, I., and Pozueta-Romero, J. (2011a). Specific delivery to mitochondria of AtBT1 complements the aberrant growth and sterility phenotype of homozygous Atbt1 Arabidopsis mutants. Plant J. 68, 1115–1121.
Bahaji, A., Ovecka, M., Barany, I., Risueno, M. C., Munoz, F. J., Baroja-Fernandez, E., Montero, M., Li, J., Hidalgo, M., Sesma, M. T., Ezquer, I., Testillano, P. S., and Pozueta-Romero, J. (2011b). Dual targeting to mitochondria and plastids of AtBT1 and ZmBT1, two members of the mitochondrial carrier family. Plant Cell Physiol. 52, 597–609.
Beckles, D. M., Smith, A. M., and ap Rees, T. (2001). A cytosolic ADP-glucose pyrophosphorylase is a feature of graminaceous endosperms, but not of other starch-storing organs. Plant Physiol. 125, 818–827.
Bedhomme, M., Hoffmann, M., McCarthy, E. A., Gambonnet, B., Moran, R. G., Rebeille, F., and Ravanel, S. (2005). Folate metabolism in plants: an Arabidopsis homolog of the mammalian mitochondrial folate transporter mediates folate import into chloroplasts. J. Biol. Chem. 280, 34823–34831.
Bernhardt, K., Wilkinson, S., Weber, A. P., and Linka, N. (2011). A peroxisomal carrier delivers NAD+ and contributes to optimal fatty acid degradation during storage oil mobilisation. Plant J. 69, 1–13.
Bohnsack, M. T., and Schleiff, E. (2010). The evolution of protein targeting and translocation systems. Biochim. Biophys. Acta 1803, 1115–1130.
Borecky, J., Maia, I. G., Costa, A. D., Jezek, P., Chaimovich, H., de Andrade, P. B., Vercesi, A. E., and Arruda, P. (2001). Functional reconstitution of Arabidopsis thaliana plant uncoupling mitochondrial protein (AtPUMP1) expressed in Escherichia coli. FEBS Lett. 505, 240–244.
Borecky, J., Nogueira, F. T., de Oliveira, K. A., Maia, I. G., Vercesi, A. E., and Arruda, P. (2006). The plant energy-dissipating mitochondrial systems: depicting the genomic structure and the expression profiles of the gene families of uncoupling protein and alternative oxidase in monocots and dicots. J. Exp. Bot. 57, 849–864.
Bouvier, F., Linka, N., Isner, J. C., Mutterer, J., Weber, A. P., and Camara, B. (2006). Arabidopsis SAMT1 defines a plastid transporter regulating plastid biogenesis and plant development. Plant Cell 18, 3088–3105.
Bruce, B. D. (2001). The paradox of plastid transit peptides: conservation of function despite divergence in primary structure. Biochim. Biophys. Acta 1541, 2–21.
Camacho, J. A., and Rioseco-Camacho, N. (2009). The human and mouse SLC25A29 mitochondrial transporters rescue the deficient ornithine metabolism in fibroblasts of patients with the hyperornithinemia-hyperammonemia-homocitrullinuria (HHH) syndrome. Pediatr. Res. 66, 35–41.
Cao, H., Sullivan, T. D., Boyer, C. D., and Shannon, J. C. (1995). Bt1, a structural gene for the major 39-44 kDa amyloplast membrane polypeptides. Physiol. Plant 95, 176–186.
Catoni, E., Desimone, M., Hilpert, M., Wipf, D., Kunze, R., Schneider, A., Flügge, U. I., Schumacher, K., and Frommer, W. B. (2003a). Expression pattern of a nuclear encoded mitochondrial arginine-ornithine translocator gene from Arabidopsis. BMC Plant Biol. 3, 1.
Catoni, E., Schwab, R., Hilpert, M., Desimone, M., Schwacke, R., Flügge, U. I., Schumacher, K., and Frommer, W. B. (2003b). Identification of an Arabidopsis mitochondrial succinate-fumarate translocator. FEBS Lett. 534, 87–92.
Cavalier-Smith, T. (1983). “A 6 kingdom classification and a unified phylogeny,” in Endocytobiology I1, eds W. Schwemmler, and H. E. A. Schenk (Berlin: De Gruyter), 1027–1034.
Cavalier-Smith, T. (2000). Membrane heredity and early chloroplast evolution. Trends Plant Sci. 5, 174–182.
Cavero, S., Vozza, A., del Arco, A., Palmieri, L., Villa, A., Blanco, E., Runswick, M. J., Walker, J. E., Cerdan, S., Palmieri, F., and Satrustegui, J. (2003). Identification and metabolic role of the mitochondrial aspartate-glutamate transporter in Saccharomyces cerevisiae. Mol. Microbiol. 50, 1257–1269.
Chan, K. W., Slotboom, D. J., Cox, S., Embley, T. M., Fabre, O., van der Giezen, M., Harding, M., Horner, D. S., Kunji, E. R., Leon-Avila, G., and Tovar, J. (2005). A novel ADP/ATP transporter in the mitosome of the microaerophilic human parasite Entamoeba histolytica. Curr. Biol. 15, 737–742.
Comparot-Moss, S., and Denyer, K. (2009). The evolution of the starch biosynthetic pathway in cereals and other grasses. J. Exp. Bot. 60, 2481–2492.
Delwiche, C. F. (1999). Tracing the thread of plastid diversity through the tapestry of life. Am. Nat. 154, 164–177.
Denyer, K., Dunlap, F., Thorbjørnsen, T., Keeling, P., and Smith, A. E. (1996). The major form of ADP-glucose pyrophosphorylase in maize endosperm is extra-plastidial. Plant Physiol. 112, 779–785.
Dierks, T., Salentin, A., Heberger, C., and Krämer, R. (1990). The mitochondrial asparate/glutamate and ADT/ATP carrier switch from obligate counterexchange to unidirectional transport after modification by SH-reagents. Biochim. Biophys. Acta 1028, 268–280.
Embley, T. M., and Martin, W. (2006). Eukaryotic evolution, changes and challenges. Nature 440, 623–630.
Embley, T. M., van der Giezen, M., Horner, D. S., Dyal, P. L., Bell, S., and Foster, P. G. (2003). Hydrogenosomes, mitochondria and early eukaryotic evolution. IUBMB Life 55, 387–395.
Fiermonte, G., De Leonardis, F., Todisco, S., Palmieri, L., Lasorsa, F. M., and Palmieri, F. (2004). Identification of the mitochondrial ATP-Mg/Pi transporter. Bacterial expression, reconstitution, functional characterization, and tissue distribution. J. Biol. Chem. 279, 30722–30730.
Fiermonte, G., Dolce, V., Palmieri, L., Ventura, M., Runswick, M. J., Palmieri, F., and Walker, J. E. (2001). Identification of the human mitochondrial oxodicarboxylate carrier. Bacterial expression, reconstitution, functional characterization, tissue distribution, and chromosomal location. J. Biol. Chem. 276, 8225–8230.
Fiermonte, G., Palmieri, L., Dolce, V., Lasorsa, F. M., Palmieri, F., Runswick, M. J., and Walker, J. E. (1998). The sequence, bacterial expression, and functional reconstitution of the rat mitochondrial dicarboxylate transporter cloned via distant homologs in yeast and Caenorhabditis elegans. J. Biol. Chem. 273, 24754–24759.
Fiermonte, G., Palmieri, L., Todisco, S., Agrimi, G., Palmieri, F., and Walker, J. E. (2002). Identification of the mitochondrial glutamate transporter. Bacterial expression, reconstitution, functional characterization, and tissue distribution of two human isoforms. J. Biol. Chem. 277, 19289–19294.
Fiermonte, G., Paradies, E., Todisco, S., Marobbio, C. M., and Palmieri, F. (2009). A novel member of solute carrier family 25 (SLC25A42) is a transporter of coenzyme A and adenosine 3′,5′-diphosphate in human mitochondria. J. Biol. Chem. 284, 18152–18159.
Froschauer, E. M., Schweyen, R. J., and Wiesenberger, G. (2009). The yeast mitochondrial carrier proteins Mrs3p/Mrs4p mediate iron transport across the inner mitochondrial membrane. Biochim. Biophys. Acta 1788, 1044–1050.
Garlid, K. D., Orosz, D. E., Modriansky, M., Vassanelli, S., and Jezek, P. (1996). On the mechanism of fatty acid-induced proton transport by mitochondrial uncoupling protein. J. Biol. Chem. 271, 2615–2620.
Goldraij, A., and Polacco, J. C. (2000). Arginine degradation by arginase in mitochondria of soybean seedling cotyledons. Planta 210, 652–658.
Gong, M., Li, J., Wang, M., Wang, J., Zen, K., and Zhang, C. Y. (2010). The evolutionary trajectory of mitochondrial carrier family during metazoan evolution. BMC Evol. Biol. 10, 282.
Gray, M. W., Lang, B. F., and Burger, G. (2004). Mitochondria of protists. Annu. Rev. Genet. 38, 477–524.
Gross, J., and Bhattacharya, D. (2009a). Mitochondrial and plastid evolution in eukaryotes: an outsiders’ perspective. Nat. Rev. Genet. 10, 495–505.
Gross, J., and Bhattacharya, D. (2009b). Revaluating the evolution of the Toc and Tic protein translocons. Trends Plant Sci. 14, 13–20.
Hackstein, J. H., Tjaden, J., and Huynen, M. (2006). Mitochondria, hydrogenosomes and mitosomes: products of evolutionary tinkering! Curr. Genet. 50, 225–245.
Haferkamp, I., Hackstein, J. H., Voncken, F. G., Schmit, G., and Tjaden, J. (2002). Functional integration of mitochondrial and hydrogenosomal ADP/ATP carriers in the Escherichia coli membrane reveals different biochemical characteristics for plants, mammals and anaerobic chytrids. Eur. J. Biochem. 269, 3172–3181.
Hamel, P., Saint-Georges, Y., de Pinto, B., Lachacinski, N., Altamura, N., and Dujardin, G. (2004). Redundancy in the function of mitochondrial phosphate transport in Saccharomyces cerevisiae and Arabidopsis thaliana. Mol. Microbiol. 51, 307–317.
Hanak, P., and Jezek, P. (2001). Mitochondrial uncoupling proteins and phylogenesis – UCP4 as the ancestral uncoupling protein. FEBS Lett. 495, 137–141.
Hanson, A. D., and Roje, S. (2001). One-carbon metabolism in higher plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 119–137.
Heimpel, S., Basset, G., Odoy, S., and Klingenberg, M. (2001). Expression of the mitochondrial ADP/ATP carrier in Escherichia coli. Renaturation, reconstitution, and the effect of mutations on 10 positive residues. J. Biol. Chem. 276, 11499–11506.
Hoyos, M. E., Palmieri, L., Wertin, T., Arrigoni, R., Polacco, J. C., and Palmieri, F. (2003). Identification of a mitochondrial transporter for basic amino acids in Arabidopsis thaliana by functional reconstitution into liposomes and complementation in yeast. Plant J. 33, 1027–1035.
Iacobazzi, V., Ventura, M., Fiermonte, G., Prezioso, G., Rocchi, M., and Palmieri, F. (2001). Genomic organization and mapping of the gene (SLC25A19) encoding the human mitochondrial deoxynucleotide carrier (DNC). Cytogenet. Cell Genet. 93, 40–42.
Iacopetta, D., Carrisi, C., De Filippis, G., Calcagnile, V. M., Cappello, A. R., Chimento, A., Curcio, R., Santoro, A., Vozza, A., Dolce, V., Palmieri, F., and Capobianco, L. (2010). The biochemical properties of the mitochondrial thiamine pyrophosphate carrier from Drosophila melanogaster. FEBS J. 277, 1172–1181.
Ito, K., Abe, Y., Johnston, S. D., and Seymour, R. S. (2003). Ubiquitous expression of a gene encoding for uncoupling protein isolated from the thermogenic inflorescence of the dead horse arum Helicodiceros muscivorus. J. Exp. Bot. 54, 1113–1114.
Ito, K., Matsukawa, K., and Kato, Y. (2006). Functional analysis of skunk cabbage SfUCPB, a unique uncoupling protein lacking the fifth transmembrane domain, in yeast cells. Biochem. Biophys. Res. Commun. 349, 383–390.
Jezek, P., and Jezek, J. (2003). Sequence anatomy of mitochondrial anion carriers. FEBS Lett. 534, 15–25.
Jiang, F., Ryan, M. T., Schlame, M., Zhao, M., Gu, Z., Klingenberg, M., Pfanner, N., and Greenberg, M. L. (2000). Absence of cardiolipin in the crd1 null mutant results in decreased mitochondrial membrane potential and reduced mitochondrial function. J. Biol. Chem. 275, 22387–22394.
Keeling, P. J. (2010). The endosymbiotic origin, diversification and fate of plastids. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365, 729–748.
Kirchberger, S., Leroch, M., Huynen, M. A., Wahl, M., Neuhaus, H. E., and Tjaden, J. (2007). Molecular and biochemical analysis of the plastidic ADP-glucose transporter (ZmBT1) from Zea mays. J. Biol. Chem. 282, 22481–22491.
Kirchberger, S., Tjaden, J., and Neuhaus, H. E. (2008). Characterization of the Arabidopsis Brittle1 transport protein and impact of reduced activity on plant metabolism. Plant J. 56, 51–63.
Klaus, S. M., Kunji, E. R., Bozzo, G. G., Noiriel, A., de la Garza, R. D., Basset, G. J., Ravanel, S., Rébeillé, F., Gregory, J. F. III, and Hanson, A. D. (2005). Higher plant plastids and cyanobacteria have folate carriers related to those of trypanosomatids. J. Biol. Chem. 280, 38457–38463.
Klingenberg, M. (1990). Mechanism and evolution of the uncoupling protein of brown adipose tissue. Trends Biochem. Sci. 15, 108–112.
Klingenberg, M. (2008). The ADP and ATP transport in mitochondria and its carrier. Biochim. Biophys. Acta. 1778, 1978–2021.
Klingenberg, M. (2009). Cardiolipin and mitochondrial carriers. Biochim. Biophys. Acta 1788, 2048–2058.
Kuan, J., and Saier, M. H. Jr. (1993). The mitochondrial carrier family of transport proteins: structural, functional, and evolutionary relationships. Crit. Rev. Biochem. Mol. Biol. 28, 209–233.
Kunji, E. R., and Crichton, P. G. (2010). Mitochondrial carriers function as monomers. Biochim. Biophys. Acta 1797, 817–831.
Kurland, C. G., and Andersson, S. G. (2000). Origin and evolution of the mitochondrial proteome. Microbiol. Mol. Biol. Rev. 64, 786–820.
Lang, B. F., Burger, G., O’Kelly, C. J., Cedergren, R., Golding, G. B., Lemieux, C., Sankoff, D., Turmel, M., and Gray, M. W. (1997). An ancestral mitochondrial DNA resembling a eubacterial genome in miniature. Nature 387, 493–497.
Lawand, S., Dorne, A. J., Long, D., Coupland, G., Mache, R., and Carol, P. (2002). Arabidopsis A BOUT DE SOUFFLE, which is homologous with mammalian carnitine acyl carrier, is required for postembryonic growth in the light. Plant Cell 14, 2161–2173.
Leroch, M., Kirchberger, S., Haferkamp, I., Wahl, M., Neuhaus, H. E., and Tjaden, J. (2005). Identification and characterization of a novel plastidic adenine nucleotide uniporter from Solanum tuberosum. J. Biol. Chem. 280, 17992–18000.
Leroch, M., Neuhaus, H. E., Kirchberger, S., Zimmermann, S., Melzer, M., Gerhold, J., and Tjaden, J. (2008). Identification of a novel adenine nucleotide transporter in the endoplasmic reticulum of Arabidopsis. Plant Cell 20, 438–451.
Lindhurst, M. J., Fiermonte, G., Song, S., Struys, E., De Leonardis, F., Schwartzberg, P. L., Chen, A., Castegna, A., Verhoeven, N., Mathews, C. K., Palmieri, F., and Biesecker, L. G. (2006). Knockout of Slc25a19 causes mitochondrial thiamine pyrophosphate depletion, embryonic lethality, CNS malformations, and anemia. Proc. Natl. Acad. Sci. U.S.A. 103, 15927–15932.
Linka, N., Theodoulou, F. L., Haslam, R. P., Linka, M., Napier, J. A., Neuhaus, H. E., and Weber, A. P. (2008). Peroxisomal ATP import is essential for seedling development in Arabidopsis thaliana. Plant Cell 20, 3241–3257.
Lithgow, T., and Schneider, A. (2010). Evolution of macromolecular import pathways in mitochondria, hydrogenosomes and mitosomes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365, 799–817.
Marobbio, C. M., Agrimi, G., Lasorsa, F. M., and Palmieri, F. (2003). Identification and functional reconstitution of yeast mitochondrial carrier for S-adenosylmethionine. EMBO J. 22, 5975–5982.
Marobbio, C. M., Di Noia, M. A., and Palmieri, F. (2006). Identification of a mitochondrial transporter for pyrimidine nucleotides in Saccharomyces cerevisiae: bacterial expression, reconstitution and functional characterization. Biochem. J. 393, 441–446.
Marobbio, C. M., Vozza, A., Harding, M., Bisaccia, F., Palmieri, F., and Walker, J. E. (2002). Identification and reconstitution of the yeast mitochondrial transporter for thiamine pyrophosphate. EMBO J. 21, 5653–5661.
Martin, W., and Herrmann, R. G. (1998). Gene transfer from organelles to the nucleus: how much, what happens, and why? Plant Physiol. 118, 9–17.
Martin, W., and Müller, M. (1998). The hydrogen hypothesis for the first eucaryote. Nature 392, 37–41.
Martin, W., Rujan, T., Richly, E., Hansen, A., Cornelsen, S., Lins, T., Leister, D., Stoebe, B., Hasegawa, M., and Penny, D. (2002). Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc. Natl. Acad. Sci. U.S.A. 99, 12246–12251.
Mereschkowsky, C. (1905). Über Natur und Ursprung der Chromatophoren im Pflanzenreiche. Biol. Centralbl. 25, 593–596. [English translation in Martin, W., and Kowallik, K. V. (1999). Annotated English translation of Mereschkowsky’s 1905 paper Über Natur und Ursprung der Chromatophoren im Pflanzenreiche. Eur. J. Phycol. 34, 287–295].
Millar, A. H., and Heazlewood, J. L. (2003). Genomic and proteomic analysis of mitochondrial carrier proteins in Arabidopsis. Plant Physiol. 131, 443–453.
Mühlenhoff, U., Stadler, J. A., Richhardt, N., Seubert, A., Eickhorst, T., Schweyen, R. J., Lill, R., and Wiesenberger, G. (2003). A specific role of the yeast mitochondrial carriers MRS3/4p in mitochondrial iron acquisition under iron-limiting conditions. J. Biol. Chem. 278, 40612–40620.
Nury, H., Dahout-Gonzalez, C., Trezeguet, V., Lauquin, G. J., Brandolin, G., and Pebay-Peyroula, E. (2006). Relations between structure and function of the mitochondrial ADP/ATP carrier. Annu. Rev. Biochem. 75, 713–741.
Palmieri, F., Pierri, C. L., De Grassi, A., Nunes-Nesi, A., and Fernie, A. R. (2011). Evolution, structure and function of mitochondrial carriers: a review with new insights. Plant J. 66, 161–181.
Palmieri, F., Rieder, B., Ventrella, A., Blanco, E., Do, P. T., Nunes-Nesi, A., Trauth, A. U., Fiermonte, G., Tjaden, J., Agrimi, G., Kirchberger, S., Paradies, E., Fernie, A. R., and Neuhaus, H. E. (2009). Molecular identification and functional characterization of Arabidopsis thaliana mitochondrial and chloroplastic NAD+ carrier proteins. J. Biol. Chem. 284, 31249–31259.
Palmieri, L., Agrimi, G., Runswick, M. J., Fearnley, I. M., Palmieri, F., and Walker, J. E. (2001a). Identification in Saccharomyces cerevisiae of two isoforms of a novel mitochondrial transporter for 2-oxoadipate and 2-oxoglutarate. J. Biol. Chem. 276, 1916–1922.
Palmieri, L., Pardo, B., Lasorsa, F. M., del Arco, A., Kobayashi, K., Iijima, M., Runswick, M. J., Walker, J. E., Saheki, T., Satrustegui, J., and Palmieri, F. (2001b). Citrin and aralar1 are Ca2+-stimulated aspartate/glutamate transporters in mitochondria. EMBO J. 20, 5060–5069.
Palmieri, L., Arrigoni, R., Blanco, E., Carrari, F., Zanor, M. I., Studart-Guimaraes, C., Fernie, A. R., and Palmieri, F. (2006a). Molecular identification of an Arabidopsis S-adenosylmethionine transporter. Analysis of organ distribution, bacterial expression, reconstitution into liposomes, and functional characterization. Plant Physiol. 142, 855–865.
Palmieri, L., Todd, C. D., Arrigoni, R., Hoyos, M. E., Santoro, A., Polacco, J. C., and Palmieri, F. (2006b). Arabidopsis mitochondria have two basic amino acid transporters with partially overlapping specificities and differential expression in seedling development. Biochim. Biophys. Acta 1757, 1277–1283.
Palmieri, L., Lasorsa, F. M., de Palma, A., Palmieri, F., Runswick, M. J., and Walker, J. E. (1997). Identification of the yeast ACR1 gene product as a succinate-fumarate transporter essential for growth on ethanol or acetate. FEBS Lett. 417, 114–118.
Palmieri, L., Lasorsa, F. M., Vozza, A., Agrimi, G., Fiermonte, G., Runswick, M. J., Walker, J. E., and Palmieri, F. (2000a). Identification and functions of new transporters in yeast mitochondria. Biochim. Biophys. Acta 1459, 363–369.
Palmieri, L., Runswick, M. J., Fiermonte, G., Walker, J. E., and Palmieri, F. (2000b). Yeast mitochondrial carriers: bacterial expression, biochemical identification and metabolic significance. J. Bioenerg. Biomembr. 32, 67–77.
Palmieri, L., Palmieri, F., Runswick, M. J., and Walker, J. E. (1996). Identification by bacterial expression and functional reconstitution of the yeast genomic sequence encoding the mitochondrial dicarboxylate carrier protein. FEBS Lett. 399, 299–302.
Palmieri, L., Picault, N., Arrigoni, R., Besin, E., Palmieri, F., and Hodges, M. (2008a). Molecular identification of three Arabidopsis thaliana mitochondrial dicarboxylate carrier isoforms: organ distribution, bacterial expression, reconstitution into liposomes and functional characterization. Biochem. J. 410, 621–629.
Palmieri, L., Santoro, A., Carrari, F., Blanco, E., Nunes-Nesi, A., Arrigoni, R., Genchi, F., Fernie, A. R., and Palmieri, F. (2008b). Identification and characterization of ADNT1, a novel mitochondrial adenine nucleotide transporter from Arabidopsis. Plant Physiol. 148, 1797–1808.
Palmieri, L., Rottensteiner, H., Girzalsky, W., Scarcia, P., Palmieri, F., and Erdmann, R. (2002). Identification and functional reconstitution of the yeast peroxisomal adenine nucleotide transporter. EMBO J. 20, 5049–5059.
Paradkar, P. N., Zumbrennen, K. B., Paw, B. H., Ward, D. M., and Kaplan, J. (2009). Regulation of mitochondrial iron import through differential turnover of mitoferrin 1 and mitoferrin 2. Mol. Cell. Biol. 29, 1007–1016.
Patron, N. J., and Waller, R. F. (2007). Transit peptide diversity and divergence: a global analysis of plastid targeting signals. Bioessays 29, 1048–1058.
Pebay-Peyroula, E., hout-Gonzalez, C., Kahn, R., Trezeguet, V., Lauquin, G. J., and Brandolin, G. (2003). Structure of mitochondrial ADP/ATP carrier in complex with carboxyatractyloside. Nature 426, 39–44.
Picault, N., Hodges, M., Palmieri, L., and Palmieri, F. (2004). The growing family of mitochondrial carriers in Arabidopsis. Trends Plant Sci. 9, 138–146.
Picault, N., Palmieri, L., Pisano, I., Hodges, M., and Palmieri, F. (2002). Identification of a novel transporter for dicarboxylates and tricarboxylates in plant mitochondria. Bacterial expression, reconstitution, functional characterization, and tissue distribution. J. Biol. Chem. 277, 24204–24211.
Pracharoenwattana, I., Cornah, J. E., and Smith, S. M. (2005). Arabidopsis peroxisomal citrate synthase is required for fatty acid respiration and seed germination. Plant Cell 17, 2037–2048.
Pratt, R. D., Ferreira, G. C., and Pedersen, P. L. (1991). Mitochondrial phosphate transport. Import of the H+/Pi symporter and role of the presequence. J. Biol. Chem. 266, 1276–1280.
Raven, J. A., and Allen, J. F. (2003). Genomics and chloroplast evolution: what did cyanobacteria do for plants? Genome Biol. 4, 209.
Rieder, B., and Neuhaus, H. E. (2011). Identification of an Arabidopsis plasma membrane-located ATP transporter important for anther development. Plant Cell 23, 1932–1944.
Rodriguez-Ezpeleta, N., Brinkmann, H., Burey, S. C., Roure, B., Burger, G., Löffelhardt, W., Bohnert, H. J., Philippe, H., and Lang, B. F. (2005). Monophyly of primary photosynthetic eukaryotes: green plants, red algae, and glaucophytes. Curr. Biol. 15, 1325–1330.
Sappl, P. G., Heazlewood, J. L., and Millar, A. H. (2004). Untangling multi-gene families in plants by integrating proteomics into functional genomics. Phytochemistry 65, 1517–1530.
Saraste, M., and Walker, J. E. (1982). Internal sequence repeats and the path of polypeptide in mitochondrial ADP/ATP translocase. FEBS Lett. 144, 250–254.
Schimper, A. F. W. (1883). Über die Entwicklung der Chlorophyllkörner und Farbkörper. Bot. Zeitung 41, 105–162. [For an English publication concerning this topic see Ward, H. M. (1883) ].
Sekoguchi, E., Sato, N., Yasui, A., Fukada, S., Nimura, Y., Aburatani, H., Ikeda, K., and Matsuura, A. (2003). A novel mitochondrial carnitine-acylcarnitine translocase induced by partial hepatectomy and fasting. J. Biol. Chem. 278, 38796–38802.
Shannon, J. C., Pien, F. M., Cao, H., and Liu, K. C. (1998). Brittle-1, an adenylate translocator, facilitates transfer of extraplastidial synthesized ADP – glucose into amyloplasts of maize endosperms. Plant Physiol. 117, 1235–1252.
Shaw, G. C., Cope, J. J., Li, L., Corson, K., Hersey, C., Ackermann, G. E., Gwynn, B., Lambert, A. J., Wingert, R. A., Traver, D., Trede, N. S., Barut, B. A., Zhou, Y., Minet, E., Donovan, A., Brownlie, A., Balzan, R., Weiss, M. J., Peters, L. L., Kaplan, J., Zon, L. I., and Paw, B. H. (2006). Mitoferrin is essential for erythroid iron assimilation. Nature 440, 96–100.
Stappen, R., and Krämer, R. (1994). Kinetic mechanism of phosphate/phosphate and phosphate/OH−antiports catalyzed by reconstituted phosphate carrier from beef heart mitochondria. J. Biol. Chem. 269, 11240–11246.
Tarantino, D., Morandini, P., Ramirez, L., Soave, C., and Murgia, I. (2011). Identification of an Arabidopsis mitoferrinlike carrier protein involved in Fe metabolism. Plant Physiol. Biochem. 49, 520–529.
Thuswaldner, S., Lagerstedt, J. O., Rojas-Stutz, M., Bouhidel, K., Der, C., Leborgne-Castel, N., Mishra, A., Marty, F., Schoefs, B., Adamska, I., Persson, B. L., and Spetea, C. (2007). Identification, expression, and functional analyses of a thylakoid ATP/ADP carrier from Arabidopsis. J. Biol. Chem. 282, 8848–8859.
Tjaden, J., Haferkamp, I., Boxma, B., Tielens, A. G., Huynen, M., and Hackstein, J. H. (2004). A divergent ADP/ATP carrier in the hydrogenosomes of Trichomonas gallinae argues for an independent origin of these organelles. Mol. Microbiol. 51, 1439–1446.
Todisco, S., Agrimi, G., Castegna, A., and Palmieri, F. (2006). Identification of the mitochondrial NAD+ transporter in Saccharomyces cerevisiae. J. Biol. Chem. 281, 1524–1531.
Toka, I., Planchais, S., Cabassa, C., Justin, A. M., De Vos, D., Richard, L., Savoure, A., and Carol, P. (2010). Mutations in the hyperosmotic stress-responsive mitochondrial BASIC AMINO ACID CARRIER2 enhance proline accumulation in Arabidopsis. Plant Physiol. 152, 1851–1862.
Traba, J., Froschauer, E. M., Wiesenberger, G., Satrustegui, J., and del Arco, A. (2008). Yeast mitochondria import ATP through the calcium-dependent ATP-Mg/Pi carrier Sal1p, and are ATP consumers during aerobic growth in glucose. Mol. Microbiol. 69, 570–585.
Traba, J., Satrustegui, J., and del Arco, A. (2009). Characterization of SCaMC-3-like/slc25a41, a novel calcium-independent mitochondrial ATP-Mg/Pi carrier. Biochem. J. 418, 125–133.
Traba, J., Satrustegui, J., and del Arco, A. (2011). Adenine nucleotide transporters in organelles: novel genes and functions. Cell. Mol. Life Sci. 68, 1183–1206.
Tsaousis, A. D., Kunji, E. R., Goldberg, A. V., Lucocq, J. M., Hirt, R. P., and Embley, T. M. (2008). A novel route for ATP acquisition by the remnant mitochondria of Encephalitozoon cuniculi. Nature 453, 553–556.
Tyra, H. M., Linka, M., Weber, A. P. M., and Bhattacharya, D. (2007). Host origin of plastid solute transporters in the first photosynthetic eukaryotes. Genome Biol. 8, R212.
Tzagoloff, A., Jang, J., Glerum, D. M., and Wu, M. (1996). FLX1 codes for a carrier protein involved in maintaining a proper balance of flavin nucleotides in yeast mitochondria. J. Biol. Chem. 271, 7392–7397.
van der Giezen, M., Slotboom, D. J., Horner, D. S., Dyal, P. L., Harding, M., Xue, G. P., Embley, T. M., and Kunji, E. R. (2002). Conserved properties of hydrogenosomal and mitochondrial ADP/ATP carriers: a common origin for both organelles. EMBO J. 21, 572–579.
Vercesi, A. E., Borecky, J., Maia, I. G., Arruda, P., Cuccovia, I. M., and Chaimovich, H. (2006). Plant uncoupling mitochondrial proteins. Annu. Rev. Plant Biol. 57, 383–404.
Vercesi, A. E., Martins, I. S., Silva, M. A. P., Leite, H. M. F., Cuccovia, I. M., and Chaimovich, H. (1995). Pumping plants. Nature 375, 24.
Voncken, F., Boxma, B., Tjaden, J., Akhmanova, A., Huynen, M., Verbek, F., Tielens, A. G. M., Haferkamp, I., Neuhaus, H. E., Vogels, G., Veenhuis, M., and Hackstein, J. H. P. (2002). Multiple origins of hydrogenosomes: functional and phylogenetic evidence from the ADP/ATP carrier of the anaerobic chytrid Neocallimastix sp. Mol. Microbiol. 44, 1441–1454.
Watanabe, A., Nakazono, M., Tsutsumi, N., and Hirai, A. (1999). AtUCP2: a novel isoform of the mitochondrial uncoupling protein of Arabidopsis thaliana. Plant Cell Physiol. 40, 1160–1166.
Williams, B. A., Haferkamp, I., and Keeling, P. J. (2008). An ADP/ATP-specific mitochondrial carrier protein in the microsporidian Antonospora locustae. J. Mol. Biol. 375, 1249–1257.
Wohlrab, H. (2006). The human mitochondrial transport/carrier protein family. Nonsynonymous single nucleotide polymorphisms (nsSNPs) and mutations that lead to human diseases. Biochim. Biophys. Acta 1757, 1263–1270.
Yin, L., Lundin, B., Bertrand, M., Nurmi, M., Solymosi, K., Kangasjarvi, S., Aro, E. M., Schoefs, B., and Spetea, C. (2010). Role of thylakoid ATP/ADP carrier in photoinhibition and photoprotection of photosystem II in Arabidopsis. Plant Physiol. 153, 666–677.
Yoon, H., Zhang, Y., Pain, J., Lyver, E. R., Lesuisse, E., Pain, D., and Dancis, A. (2011). Rim2, pyrimidine nucleotide exchanger, is needed for iron utilization in mitochondria. Biochem. J. 440, 137–146.
Zrenner, R., Stitt, M., Sonnewald, U., and Boldt, R. (2006). Pyrimidine and purine biosynthesis and degradation in plants. Annu. Rev. Plant Biol. 57, 805–836.
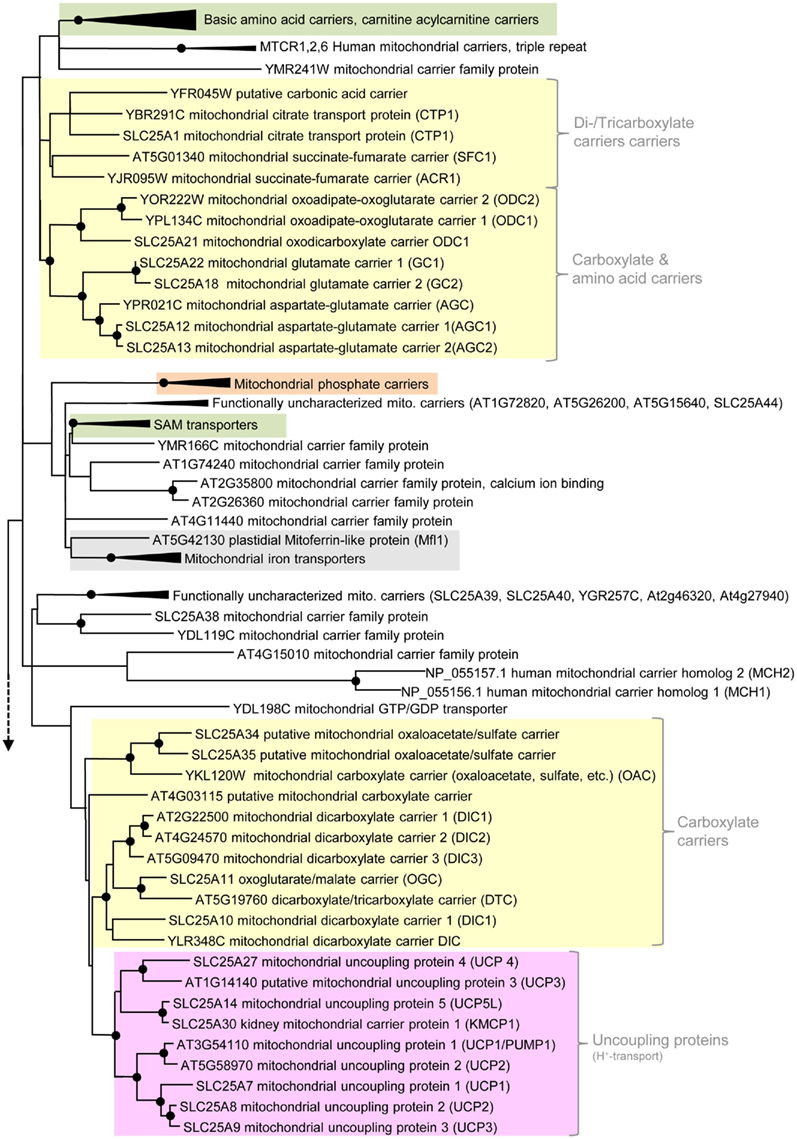
Figure A1. Phylogenetic relationships of putative and proven carbonic acid carriers and uncoupling proteins. Detailed subsets of the phylogenetic tree shown in Figure 3 are displayed (yellow and purple shaded subsets). Nodes that are supported by bootstrap values (maximum likelihood and maximum parsimony, 1000 replications) higher than 70 are indicated by black dots.
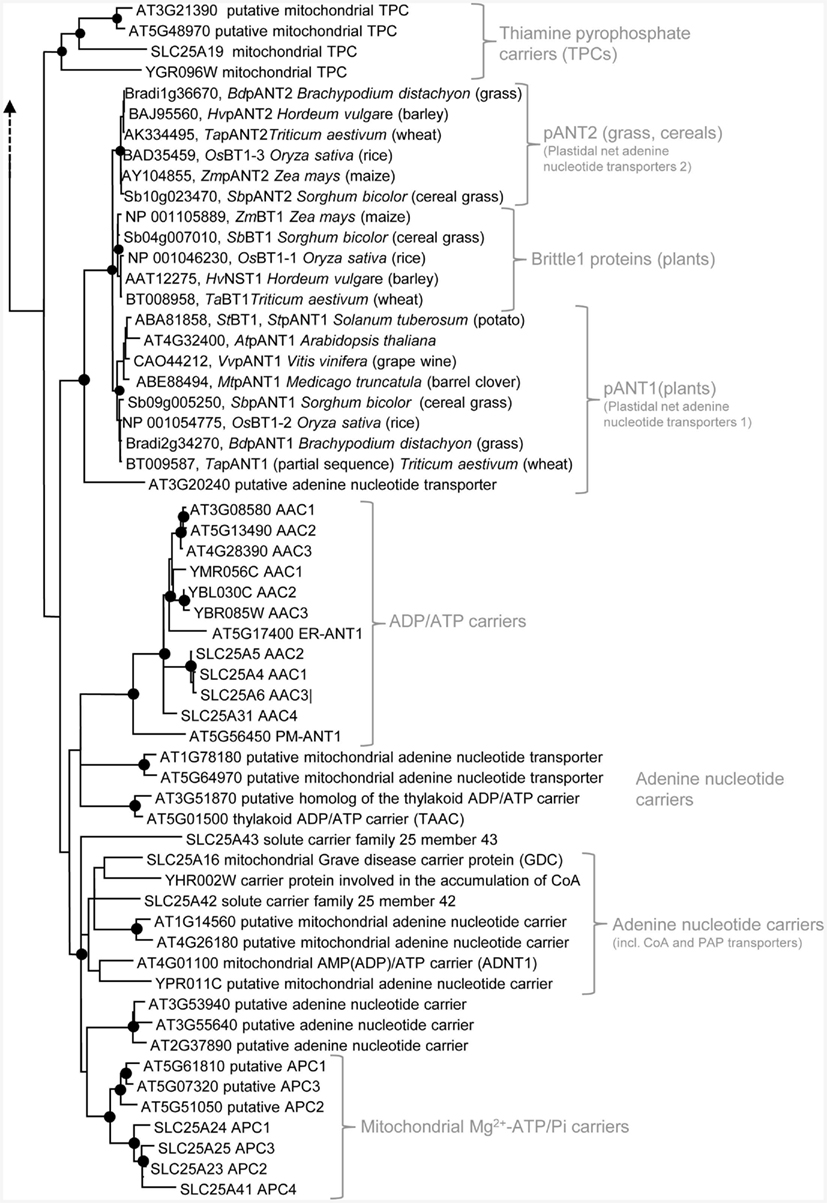
Figure A2. Phylogenetic relationships of putative and proven adenine nucleotide transporters. A subset of the phylogenetic tree shown in Figure 3 is displayed [majority of the blue shaded subset, the mitochondrial Mg2+–ATP/Pi carrier (Sallp) is included in the subset of Figure A3]. Nodes that are supported by bootstrap values (maximum likelihood and maximum parsimony, 1000 replications) higher than 70 are indicated by black dots.
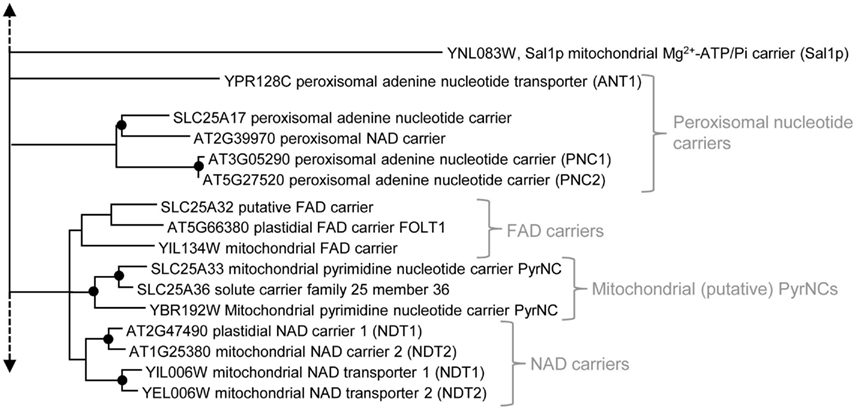
Figure A3. Phylogenetic relationships of peroxisomal adenine nucleotide carriers, pyrimidine nucleotide, FAD, and NAD carriers. A subset of the phylogenetic tree shown in Figure 3 is displayed [pale blue shaded subset and the mitochondrial Mg2+–ATP/Pi carrier (Sallp)]. Nodes that are supported by bootstrap values (maximum likelihood and maximum parsimony, 1000 replications) higher than 70 are indicated by black dots.
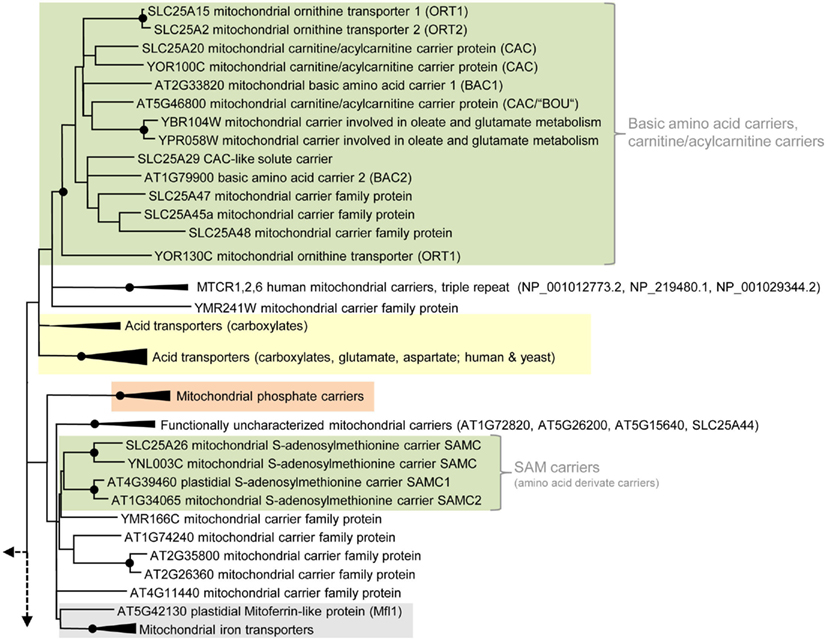
Figure A4. Phylogenetic relationships of putative and proven basic amino acid carriers, carnitine/acylcarnitine carriers, and S-adenosylmethionine carriers. Detailed subsets of the phylogenetic tree shown in Figure 3 are displayed (green shaded subsets). Nodes that are supported by bootstrap values (maximum likelihood and maximum parsimony, 1000 replications) higher than 70 are indicated by black dots.
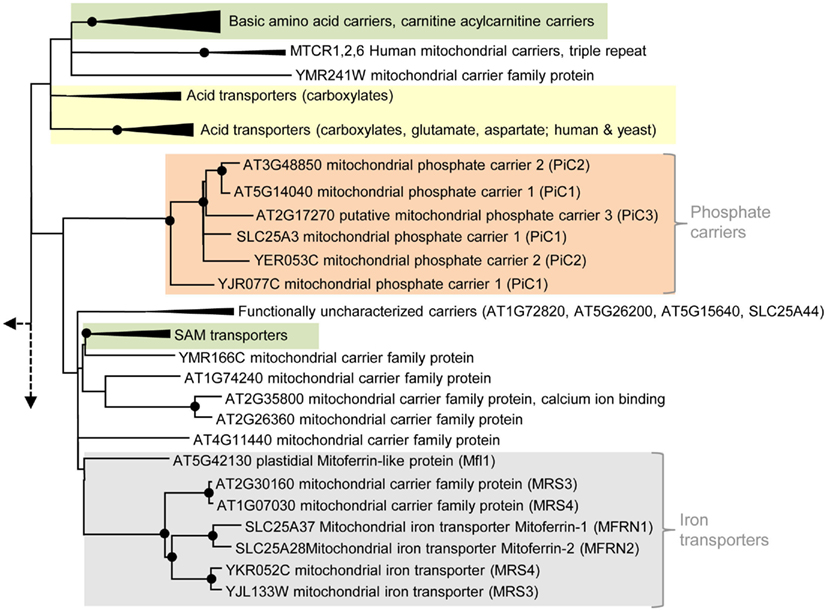
Figure A5. Phylogenetic relationships of putative and proven phosphate and iron carriers. Detailed subsets of the phylogenetic tree shown in Figure 3 are displayed (orange and gray shaded subsets). Nodes that are supported by bootstrap values (maximum likelihood and maximum parsimony, 1000 replications) higher than 70 are indicated by black dots.
Keywords: mitochondrial carrier family, MCF, solute carriers, evolution, function, mitochondria, plant, Arabidopsis thaliana
Citation: Haferkamp I and Schmitz-Esser S (2012) The plant mitochondrial carrier family: functional and evolutionary aspects. Front. Plant Sci. 3:2. doi: 10.3389/fpls.2012.00002
Received: 04 October 2011; Accepted: 03 January 2012;
Published online: 18 January 2012.
Edited by:
Markus Geisler, University of Fribourg, SwitzerlandReviewed by:
Nicole Linka, Heinrich-Heine Universität Düsseldorf, GermanyCopyright: © 2012 Haferkamp and Schmitz-Esser. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Ilka Haferkamp, Biologie, Zelluläre Physiologie/Membrantransport, Technische Universität Kaiserslautern, Erwin-Schrödinger-Str. 22, 67653 Kaiserslautern, Germany. e-mail:aGFmZXJrQHJocmsudW5pLWtsLmRl
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.