- 1 Department of Biochemistry and Molecular Biology, Michigan State University, East Lansing, MI, USA
- 2 Plant Science Department, Rothamsted Research, Harpenden, UK
- 3 Graduate School of Pharmaceutical Sciences, Chiba University, Chiba, Japan
- 4 Department of Plant Biology, Michigan State University, East Lansing, MI, USA
Sulfate is an essential nutrient cycled in nature. Ion transporters that specifically facilitate the transport of sulfate across the membranes are found ubiquitously in living organisms. The phylogenetic analysis of known sulfate transporters and their homologous proteins from eukaryotic organisms indicate two evolutionarily distinct groups of sulfate transport systems. One major group named Tribe 1 represents yeast and fungal SUL, plant SULTR, and animal SLC26 families. The evolutionary origin of SULTR family members in land plants and green algae is suggested to be common with yeast and fungal SUL and animal anion exchangers (SLC26). The lineage of plant SULTR family is expanded into four subfamilies (SULTR1–SULTR4) in land plant species. By contrast, the putative SULTR homologs from Chlorophyte green algae are in two separate lineages; one with the subfamily of plant tonoplast-localized sulfate transporters (SULTR4), and the other diverged before the appearance of lineages for SUL, SULTR, and SLC26. There also was a group of yet undefined members of putative sulfate transporters in yeast and fungi divergent from these major lineages in Tribe 1. The other distinct group is Tribe 2, primarily composed of animal sodium-dependent sulfate/carboxylate transporters (SLC13) and plant tonoplast-localized dicarboxylate transporters (TDT). The putative sulfur-sensing protein (SAC1) and SAC1-like transporters (SLT) of Chlorophyte green algae, bryophyte, and lycophyte show low degrees of sequence similarities with SLC13 and TDT. However, the phylogenetic relationship between SAC1/SLT and the other two families, SLC13 and TDT in Tribe 2, is not clearly supported. In addition, the SAC1/SLT family is absent in the angiosperm species analyzed. The present study suggests distinct evolutionary trajectories of sulfate transport systems for land plants and green algae.
Introduction
Sulfate is an essential nutrient and the initial substrate for biosynthesis of sulfur-containing metabolites in plants, algae, and microorganisms (Leustek et al., 2000; Saito, 2004; Takahashi et al., 2011). The organic sulfur metabolites synthesized in these autotrophic organisms are the sulfur nutritional resource for animals. However, animals are not devoid of sulfate transport proteins as they play significant roles in reabsorbing sulfate in renal systems to maintain ion homeostasis (Markovich and Murer, 2004; Mount and Romero, 2004). A sulfate transporter is also known to be essential for cartilage formation as it may contribute to supplying sulfate for synthesis of sulfated proteoglycans (Hästbacka et al., 1994). Sulfate transport proteins are found therefore across diverse organisms, although they may facilitate transport of sulfate for different purposes. Apart from the ubiquitous presence among organisms, the expansion of the family members in multi-cellular organisms is most likely an evolutionary development to provide sulfate transporters which are specifically functional in different organs or tissues. The expansion led to distinct spatial distribution and organization of biochemically diversified forms of sulfate transporters, which is necessary for coordinating the overall transport of sulfate within complex biological systems. In addition, the ionic environmental factors are highly variable that may have contributed to develop distinct types of transport systems and regulatory mechanisms.
Previous studies have indicated that sulfate transport proteins can be classified to four different types according to the mechanisms mediating transport of sulfate across the membranes (Figure 1). The influx of sulfate can be coupled with co-transport of positively charged counter ions such as proton (H+) and sodium (Na+). For these mechanisms, the concentration gradients of counter ions serve as driving force for the influx of sulfate across the membranes. The proton gradient is suggested to be the driving force for sulfate uptake systems in yeast and plants (Roomans et al., 1979; Lass and Ullrich-Eberius, 1984; Hawkesford et al., 1993). SUL1 and SUL2 in yeast (Smith et al., 1995a; Cherest et al., 1997) and SULTR family members in plants (Smith et al., 1995b, 1997; Buchner et al., 2004; Takahashi, 2010) are the suggested components of the proton/sulfate co-transport systems. In contrast, animals may use different mechanisms. The sulfate transport activities of SLC13 family proteins are known to be dependent on sodium (Markovich and Murer, 2004; Pajor, 2006). An alternative mechanism is the anion exchange systems facilitating the counter transport of sulfate and other negatively charged ions, such as chloride (Cl−), iodide (I−), and bicarbonate (HCO3−). The SLC26 family proteins facilitate sulfate/anion exchanges in animals (Mount and Romero, 2004). Transport of sulfate can be also driven by an ATP-binding cassette (ABC) transporter complex in bacteria and algal chloroplasts (Sirko et al., 1990; Laudenbach and Grossman, 1991; Lindberg and Melis, 2008). These mechanisms are suggested to have evolved in various ancestral species depending on the ionic environments where those transporters were to be operated.
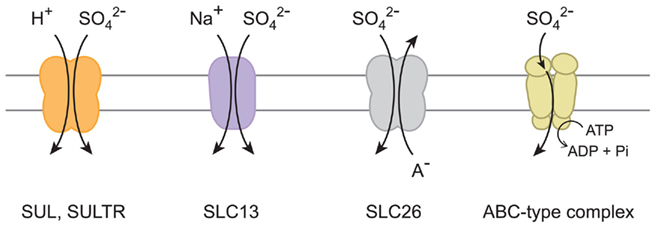
Figure 1. Sulfate transport mechanisms. Proton/sulfate co-transporter, sodium/sulfate co-transporter, sulfate/anion (A−) exchanger, and ABC-type sulfate transporter complex are illustrated. The names of sulfate transport proteins are indicated below the suggested mechanisms.
The various sulfate transport systems are intimately linked with the subsequent metabolism of their transported molecule, sulfate. Once sulfate is delivered to the cell, it serves as a substrate for the sulfur assimilatory enzyme, ATP sulfurylase, both in the cytoplasm and the plastids in plants (Takahashi et al., 2011). The metabolic flux of ATP sulfurylase and subsequent reduction steps in plastids defines the primary requirement of sulfate in metabolism (Vauclare et al., 2002; Kopriva, 2006). Before entering the steps of metabolic conversion, sulfate in the cytoplasm can be sequestered to vacuoles (Buchner et al., 2004; Takahashi, 2010). The export of sulfate to the extra-cellular space would be another factor affecting the rate of sulfate uptake across the plasma membrane. Unknown passive transport systems are suggested for those mechanisms as the membrane potentials are positive at extra-cellular and vacuolar lumen sides (Buchner et al., 2004; Takahashi, 2010). In addition, a steep upward concentration gradient of sulfate may be generated across the plasma membrane under sulfate-starved conditions; active transport systems are necessary to drive the influx of sulfate efficiently under such circumstances. The systems should contain selective mechanisms either coupled with transport of counter ions, or energized by ATP, as catalyzed by ABC transporters (Figure 1).
The recent expansion of genome sequencing information has enabled the identification of a number of sulfate transporters and homologous proteins from higher plants (Takahashi, 2010). This study focuses on the molecular evolution of the families of sulfate transporters in the green lineage (i.e., land plants and green algae). Phylogenetic analysis was conducted using a diverse set of relevant protein sequences from yeast, fungi, algae, bryophyte, lycophyte, seed plants, and animals to reinterpret their biochemical diversification with respect to the evolution of eukaryotic organisms and to assess the lineage-specific expansion of the family members. The present study aims to provide information of family classifications of sulfate transporters and related proteins based on their phylogenetic relationships and evolution.
Family Classifications
The protein sequences of sulfate transporters were identified from the following organisms: Saccharomyces cerevisiae, Aspergillus niger, Aspergillus nidulans, Chlamydomonas reinhardtii, Volvox carteri, Physcomitrella patens, Selaginella moellendorffii, Arabidopsis thaliana, Glycine max, Populus trichocarpa, Oryza sativa, Brachypodium distachyon, Sorghum bicolor, Caenorhabditis elegans, Drosophila melanogaster, Danio rerio, and Homo sapiens. Metazoan, plant, budding yeast, and Aspergillus protein sequences were obtained from Ensembl (release 621), Phytozome (ver. 52), SGD (Feb 0, 20113), and BROAD4, respectively. To identify sulfate transporter, a three step analysis pipeline was used. First, annotated protein sequences from the 17 representative species were searched against known sulfate transporter from plants and human with a low Expect value threshold of one to include as many candidates as possible. Second, presence of transmembrane regions in these candidate sequences was identified with TMHMM (Krogh et al., 2001). Two types of candidates were analyzed further: (1) the sequence has an Expect value > 1e–5 but has ≥7 transmembrane regions and (2) the sequence has an Expect value ≤ 1e–5 and has ≥1 transmembrane regions. The first criterion is to ensure that divergent transporters are captured. The second is to include partial sequences of true sulfate transporters given many genomes analyzed were not heavily annotated.
In the final step, candidates passing the Expect value and transmembrane region criteria were aligned with annotated sulfate transporters for phylogenetic reconstruction. Due to the sheer number of sequences analyzed, the phylogenetic analysis was done in three iterations starting with computationally straightforward neighbor-joining algorithm with bootstrap as implemented in MEGA (Tamura et al., 2011). After dividing candidates into “tribes” based on neighbor-joining trees, bootstrapped maximum likelihood (ML) trees were generated with RAxML (Stamatakis, 2006). This program has advantages for computation of large phylogenetic trees as in this study. We chose these methods considering accuracy and computational performance. Based on ML tree topology, candidates were subdivided into families and sequences were excluded if they do not resides in the same well supported (>50%) clades with known sulfate transporters. Finally, bootstrapped ML trees were generated for each family.
The members of chloroplast-localized sulfate transporter from Chlamydomonas (Melis and Chen, 2005; Lindberg and Melis, 2008) correspond to bacterial ABC-type sulfate transporter complex, which is composed of a sulfate binding protein, periplasmic membrane-bound proteins, and an ABC protein that hydrolyzes ATP and provides the energy for transport (Sirko et al., 1990; Laudenbach and Grossman, 1991; Figure 1). These components are present only in bacteria and algae but not in any land plant species analyzed (Takahashi, 2010; Takahashi et al., 2011). Horizontal gene transfer of ABC-type sulfate transporter complex may have occurred from bacteria to algae, but it is possible that the complex has been lost in land plants when their ancestor diverged from green algae approximately one billion years ago. Based on phylogenetic analysis, there is no evidence suggesting that the proteins for ABC-type sulfate transporter complex are homologous to other groups of sulfate transporters focused in this study. Although both the eukaryotic sulfate transporters and the ABC-type complexes may transport sulfate, they are structurally and mechanistically different (Figure 1). Plant SULTR, metazoan SLC26, and yeast SUL proteins are predicted to contain 10–14 hydrophobic transmembrane regions (Smith et al., 1995a,b; Cherest et al., 1997; Hawkesford, 2003; Mount and Romero, 2004). SLC13 is predicted to have 8–13 transmembrane regions (Markovich and Murer, 2004; Pajor, 2006). By contrast, the bacterial/algal ABC-type complexes are composed of multiple subunit proteins sharing individual roles in facilitating transport of sulfate across the membranes (Sirko et al., 1990; Laudenbach and Grossman, 1991; Melis and Chen, 2005; Lindberg and Melis, 2008). It is apparent that ABC-type sulfate transporter complex radiated in prokaryotes and algae, and their evolutionary trajectories were completely different from those for the eukaryotic-type sulfate transporters focused in this article.
Two major groups of eukaryotic sulfate transporters were identified and designated Tribe 1 and Tribe 2 (Figures 2–4). Tribe 1 is composed of three major lineages, Family P, Family A1, and Family A2, respectively (Figure 2). The plant SULTR family members are found exclusively in Family P (Figures 2 and 3). The animal SLC26 family members are found in Family A1 and A2. The yeast SUL1/SUL2 and their fungal homologs are found in Family A1, although a few additional homologs including yeast YPR003C and YGR125W exist in groups that diverged earlier than the emergence of plant and animal lineages. The members of algal SULTR also split into two groups; one present in Family P and the other in clades regarded as out-groups. In Tribe 1, the evolutionary origin of plant SULTR family may be tracked back to the fungal–animal–plant common ancestor based on the relationships of plant SULTR (Takahashi, 2010) to yeast and fungal SUL (Smith et al., 1995a; Cherest et al., 1997) and to animal sulfate/anion exchangers (SLC26; Mount and Romero, 2004; Figure 2). The Family P lineage appears to be associated with the Family A1 lineage. Although there still remains ambiguity regarding the exact origin of Family P, the results may well suggest that the ancestral forms of the existing yeast SUL1/SUL2 were the founders of the three major lineages, Family P, A1, and A2 in Tribe 1 (Figure 2). In contrast, the two other yeast SUL homologs, YPR003C and YGR125W, are suggested to have diverged prior to the emergence of those major lineages (Figure 2). Tribe 2 is also composed of three distinct lineages (Figure 4). These lineages represent the families of animal sodium-dependent sulfate/carboxylate transporters (SLC13; Markovich and Murer, 2004; Pajor, 2006), plant tonoplast-localized dicarboxylate transporters (TDT; Emmerlich et al., 2003), and algal putative sulfur-sensing proteins (SAC1; Davies et al., 1996), and SAC1-like transporters (SLT; Pootakham et al., 2010), respectively. The phylogenetic relationships of the family and subfamily members in these two tribes will be discussed in the following sections.
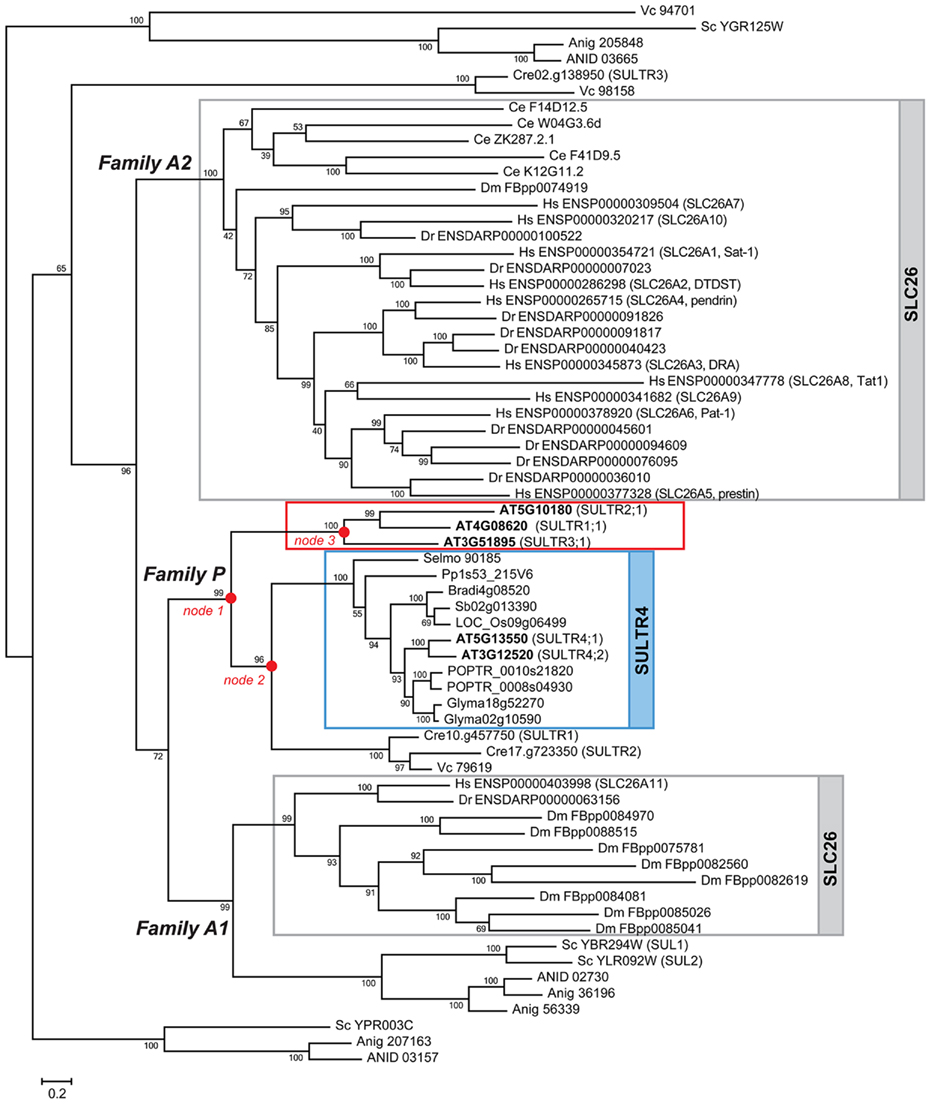
Figure 2. Phylogenetic relationships of SUL, SULTR, and SLC26 in Tribe 1. The lineage that splits to SULTR1, SULTR2, and SULTR3 subfamilies (Figure 3) are boxed in red. SULTR1;1, SULTR2;1, and SULTR3;1 from Arabidopsis were selected as representatives of these subfamilies to construct the phylogenetic tree. The SULTR4 subfamily in plants and SLC26 in animals are highlighted in blue and gray, respectively. The nodes 1–3 where the plant SULTR lineage splits into subfamilies are indicated by red dots. The bootstrap values are shown at the nodes. The locus numbers or gene IDs are indicated according to Ensembl (http://www.ensembl.org), Phytozome (http://www.phytozome.net/), SGD (http://yeastgenome.org/), JGI (http://www.jgi.doe.gov/), and BROAD (http://www.broadinstitute.org). Prefix abbreviations of locus numbers or gene IDs indicate genus and species names: AT, Arabidopsis thaliana; ANID, Aspergillus nidulans; Anig, Aspergillus niger; Bradi, Brachypodium distachyon; Ce, Caenorhabditis elegans; Cre, Chlamydomonas reinhardtti; Dr, Danio rerio; Dm, Drosophila melanogaster; Glyma, Glycine max; Hs, Homo sapiens; Os, Oryza sativa; Pp, Physcomitrella patens; POPTR, Populus trichocarpa; Selmo, Selaginella moellendorffii; Sc, Saccharomyces cerevisiae; Sb, Sorghum bicolor; Vc, Volvox carteri. The locus numbers of Arabidopsis SULTR are highlighted in bold letters. The names of Arabidopsis SULTR, Chlamydomonas SULTR, human SLC26A, and yeast SUL family members are shown in the parentheses next to the locus numbers.
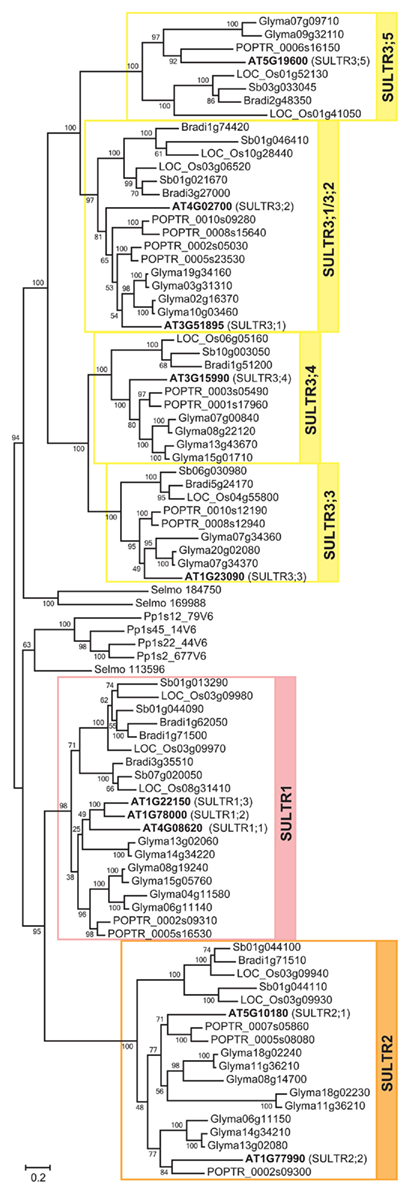
Figure 3. Expansion of SULTR1–SULTR3 subfamilies in plants. The phylogenetic tree indicates expansion of the SULTR1/2/3 lineage (Figure 2) in seed plants. The SULTR1, SULTR2, and SULTR3 subfamilies are highlighted in pink, orange, and yellow, respectively. The bootstrap values are shown at the nodes. The locus numbers or gene IDs are described the same as in Figure 2. The names of Arabidopsis sulfate transporters are shown in the parentheses next to the locus numbers.
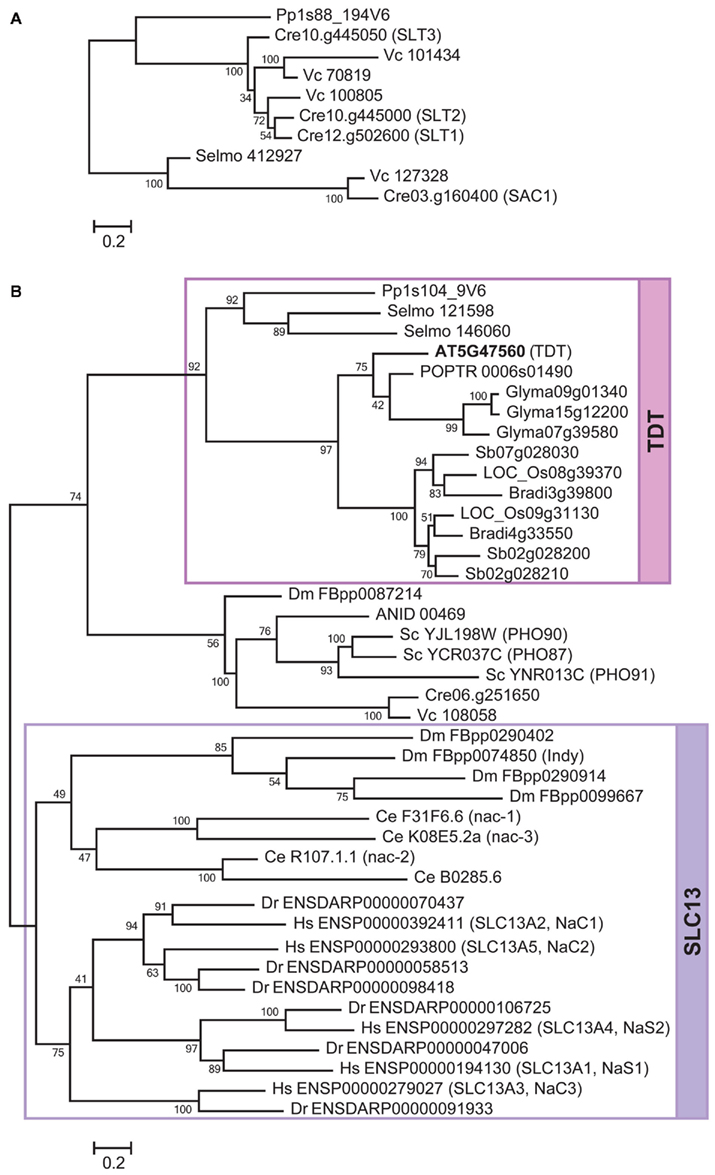
Figure 4. Phylogenetic relationships of SAC1/SLT, SLT13, and TDT in Tribe 2. The phylogenetic trees of SAC1/SLT (A), and SLC 13 and TDT (B), were constructed separately. The lineages for SLC13 and TDT are highlighted in violet and magenta, respectively. The bootstrap values are shown at the nodes. The locus numbers or gene IDs are described the same as in Figure 2. The locus number of Arabidopsis TDT is highlighted in bold letters. The names of Chlamydomonas SAC1 and SLT, Arabidopsis TDT, and animal SLC13 family members are shown in the parentheses next to the locus numbers.
Plant Sulfate Transporters (SULTR)
The plant specific lineage of Tribe 1 is composed of four distinct subfamilies of SULTR-type sulfate transporters, SULTR1, SULTR2, SULTR3, and tonoplast-localized sulfate transporters (SULTR4; Figure 2, Family P; Figure 3). These four subfamilies correspond with the nomenclatures of sulfate transporters identified from Arabidopsis and other vascular plant species (Hawkesford, 2003; Buchner et al., 2004; Takahashi, 2010). Family P was fairly specific to land plant species, although part of algal SULTR members (Pootakham et al., 2010) was present in an out-group diverged from SULTR4 subfamily (Figure 2, node 2). The first gene duplication event in Family P was the split of the land plant SULTR1/2/3 and SULTR4 forms (Figure 2, node 1). Two distinct types of SULTR were subsequently generated. The lineage of SULTR4 (Kataoka et al., 2004a) was diverged by subsequent gene duplication (Figure 2, node 2). The vacuole localization of the ancestral form of SULTR4 was probably defined after this gene duplication event, because a plasma membrane localized sulfate transporter from Chlamydomonas (SULTR2; Pootakham et al., 2010) is found in the clade diverged from this lineage. The branching of SULTR4 subfamily members in bryophyte and lycophyte is somewhat irregular as Selaginella SULTR4 appears to have diverged earlier than Physcomitrella SULTR4. This may be due to the ambiguity of branch position of Physcomitrella SULTR4 with relatively low bootstrap support.
SULTR1/2/3 subsequently split into SULTR1/2 and SULTR3 clades (Figure 2, node 3; Figure 3). SULTR1 and SULTR2 in Arabidopsis are functional sulfate transporters that can restore the sulfate uptake activity of the yeast sul1 sul2 mutant (Takahashi et al., 1997, 2000; Shibagaki et al., 2002; Yoshimoto et al., 2002, 2007). The phylogenetic relationships between SULTR1 and SULTR2 support their functional similarities as being sulfate transporters. Within the clade of SULTR1/2, there is a sister group of sulfate transporters for Physcomitrella and Selaginella. The ancestor of this group likely emerged prior to the division of the SULTR1 and SULTR2 subfamilies. Their phylogenetic relationships with SULTR1/2 lineage may well suggest that these bryophyte and lycophyte SULTR homologs would have sulfate transport activities, although their functional identities are not confirmed. With respect to the substrate specificities of the SULTR1 and SULTR2 subfamilies, the results suggest that the differences of their kinetic properties are evolutionarily derived. Consistent with previous findings, SULTR1 and SULTR2 can be defined as subfamilies of high- and low-affinity sulfate transporters, respectively (Takahashi et al., 2000; Yoshimoto et al., 2002). Within each subfamily, SULTR1 first splits to dicotyledonous and monocotyledonous groups and subsequently duplicates to have the subfamily members in different flowering plant lineages. SULTR2 also splits to dicotyledonous and monocotyledonous groups, followed by specialization of SULTR2;1 and SULTR2;2. Supported by the experimental evidence for the kinetic properties of Arabidopsis SULTR2;1 and SULTR2;2 (Takahashi et al., 2000), the divergence of these two forms at least in dicots may indicate the difference in their affinities to sulfate.
SULTR3 is principally composed of subfamily members from angiosperms. Before the expansion in the angiosperms, the ancestral lineage of SULTR3 gave rise to Selaginella SULTR (Figure 3). Since there is no Physcomitrella SULTR in this subfamily, the lineage of SULTR3 appears to be specific to vascular plant species. The SULTR3 subfamily subsequently divided to four classes. They were designated SULTR3;5, SULTR3;1/3;2, SULTR3;3, and SULTR3;4 according to the names of the subfamily members from Arabidopsis (Takahashi, 2010). As with the SULTR1 subfamily, SULTR3;3, SULTR3;4, and SULTR3;5 first split to dicotyledonous and monocotyledonous groups and then diverge to have the subfamily members in individual plant species. With respect to the expansion of the SULTR3;1/3;2 subfamily, SULTR3;1 and SULTR3;2 are founded after the division of dicotyledonous and monocotyledonous plants as described for the evolution of SULTR2;1/2;2. The SULTR3 family members in Arabidopsis are suggested to be involved in internal transport of sulfate in vasculature and developing seeds (Kataoka et al., 2004b; Zuber et al., 2010). In addition, a SULTR3;5 homolog in Lotus japonicus appears to mediate intracellular transport of sulfate to symbiosomes (Krusell et al., 2005). In spite of these indications from the physiological characterizations of plant mutant lines, the exact biochemical features of SULTR3 subfamily members are yet unverified. At this point, their sulfate uptake activities are suggested to be very low or barely detectable (Kataoka et al., 2004b) except for the case in L. japonicus SST1 (Krusell et al., 2005). In contrast to the SULTR1/2 subfamilies, the lack of biochemical information hampers us to interpret the evolutionary diversification of the individual classes of SULTR3 subfamily based on their molecular functions.
Some earlier studies have annotated an additional family of putative sulfate transporter (group 5) based on its partial sequence similarities with plant SULTR (Hawkesford, 2003; Buchner et al., 2004). However, the transmembrane structured proteins in group 5 contain no sulfate transporter motif (Leves et al., 2008) and STAS domain (Aravind and Koonin, 2000) which is the typical signature for SUL, SULTR, and SLC26 proteins. Later identification of its role as a molybdate transporter (MOT) may explain its considerable divergence from sulfate transporters (Tejada-Jiménez et al., 2007; Tomatsu et al., 2007; Baxter et al., 2008; Gasber et al., 2011). Consistent with functional studies, we did not find clear support for an evolutionary relationship between MOT and families of sulfate transporters in both prokaryotes and eukaryotes.
SUL and SLC26
The yeast and fungal SUL and animal SLC26 sulfate/anion exchangers are present in two distinct lineages of Tribe 1 (Figure 2, Family A1, and Family A2). Family A1 contains SUL1/SUL2 sulfate transporters from yeast (Smith et al., 1995a; Cherest et al., 1997) and their closest homologs in Aspergillus. An animal-specific lineage is present in this family. It was composed of SLC26A11 from human and zebra fish, and eight homologous proteins from Drosophila. It is likely that the majority of the existing SLC26 members in Drosophila occurred through gene duplications in this animal-specific lineage in Family A1. Only one SLC26 member from Drosophila is found in the other animal clade (Family A2). By contrast, SLC26A11 is the only member from human and zebra fish present in Family A1, while the rest of the members (SLC26A1–A10) are found in Family A2. Family A2 is a lineage of SLC26 from C. elegans, human, and zebra fish (Figure 2). This animal-specific lineage eventually expands to human and zebra fish SLC26A1–A10 members. The ancestor of this group appears to have first duplicated to form a subfamily of C. elegans SLC26, and subsequently expanded to the vertebrate SLC26A1–A10 members (Mount and Romero, 2004).
The high bootstrap value provides supports that the existing members of sulfate transporters in Family P, A1, and A2 are originated from the same ancestral form. The results from phylogenetic analysis further suggested additional members of putative sulfate transporters which could have diverged prior to the emergence of these three major lineages (Figure 2). The SULTR homologs from Chlamydomonas (SULTR3; Pootakham et al., 2010) and Volvox (Vc 98158) in the out-group may derive from earlier evolutionary events. Yeast YPR003C and YGR125W and their homologs in Aspergillus form additional clades, suggesting their ancestral forms may have first diverged from the major lineages of existing sulfate transporters. The yeast YPR003C and YGR125W are putative sulfate transporters yet to be characterized. They may contribute to the residual sulfate transport activities in yeast sul1 and sul1 sul2 mutants (Smith et al., 1995a; Cherest et al., 1997), although the biochemical function of these putative sulfate transporters awaits further investigation. It is notable that a SULTR homolog from Volvox is present in the clade of YGR125W (Figure 2; Vc 94701). The phylogenetic relationship between algal SULTR and putative sulfate transporters from yeast and fungi suggests that they have shared a common ancestor prior to the divergence of the green algal and fungal lineages.
Putative Sulfur-Sensing Protein (SAC1) and SAC1-Like Transporters (SLT)
Among the three lineages in Tribe 2, the SAC1/SLT is relatively independent of the other two lineages, SLC13 and TDT (Figure 4). Considering the overall similarities of protein sequences among these family members, SAC1/SLT (Davies et al., 1996; Pootakham et al., 2010), SLC13 (Markovich and Murer, 2004; Pajor, 2006), and TDT (Emmerlich et al., 2003) may have originated from a common ancestor. However, the phylogenetic relationships of SAC1/SLT with SLC13 and TDT were not clearly supported according to the ML tree (Figure 4). SAC1/SLT is a family specific to Chlorophyte algae Chlamydomonas and Volvox, a bryophyte Physcomitrella and a lycophyte Selaginella (Figure 4A). No seed plant homologs have been identified. Within this family, a SAC1 homolog is identified from Selaginella, suggesting that SAC1/SLT is present at least in the ancestor of vascular plants but has been lost when seed plant ancestors diverged from non-seed plants approximately 400 million years ago.
The phylogenetic tree provides further support that SLT1–SLT3 (Pootakham et al., 2010) are well conserved in Chlorophyte green algae as they are found in Chlamydomonas and Volvox (Figure 4A). The closest family member from Physcomitrella is suggested to have diverged from this algal SLT group, although the phylogenetic relationship is not strongly supported. The branch organization apparently suggests that Physcomitrella and Selaginella may have lost SAC1 and SLT, respectively. Since the position of a Physcomitrella homolog is not supported (Figure 4A), it may be also associated with the SAC1 clade. Accordingly, the clade of SLT1–SLT3 will become distinguishable as an algal specific lineage. Although the biological functions of SAC1/SLT from Physcomitrella and Selaginella are yet to be verified, this alternative interpretation may simply explain the divergence of algal SLT sulfate transporters from SAC1. It is hypothesized that Chlamydomonas or green algae in general may have the flexibility to utilize proton/sulfate transporter (SULTR) or sodium/sulfate transporter (SLT) depending on the environmental conditions such as pH and sodium concentrations which they need to acclimate (Pootakham et al., 2010). The sodium-dependency of sulfate transport activity of SLT needs to be verified to support this model.
SLC13 and TDT
The lineage of SLC13 is composed of animal SLC13 (Markovich and Murer, 2004; Pajor, 2006; Figure 4B). No plant or algal proteins are associated with this family. Based on the phylogeny, the expansion pattern of this SLC13 family can be interpreted in a straightforward way. The SLC13 ancestor first diverged to form two lineages; one specific to vertebrates and the other that further duplicates to form clades specific to Drosophila and C. elegans (Figure 4B). A Drosophila lifespan determinant protein, Indy, is known to function as a sodium-independent electro-neutral citrate transporter (Rogina et al., 2000; Inoue et al., 2002). By contrast, NaC family members of C. elegans co-transport sodium and dicarboxylate (Fei et al., 2003). These lines of evidence suggest that sodium-dependency is not always conserved among SLC13. Among the five SLC13 proteins in human, two are sodium/sulfate transporters (NaS1 and NaS2) and the rest three are sodium/carboxylate transporters (NaC1, NaC2, and NaC3; Markovich and Murer, 2004; Pajor, 2006). The phylogenetic tree indicates that both NaS1/NaS2 and NaC1/NaC2 originally come from the ancestor of NaC3. It is suggested that the substrate specificities of NaS1 and NaS2 for sulfate may have been developed later when they diverged from the lineage leading to NaC1 and NaC2.
The TDT family is specific to land plants (Figure 4B). The tonoplast-localized dicarboxylate transporter from Arabidopsis provides the biochemical evidence for this family (Emmerlich et al., 2003). The Arabidopsis TDT is capable of transporting malate and fumarate to vacuoles (Emmerlich et al., 2003; Hurth et al., 2005). There is no direct experimental evidence showing a sulfate transport activity, although a certain degree of sequence similarity with SLC13 has been detected. In addition, it is reported that sodium does not stimulate the dicarboxylate transport activity of TDT (Emmerlich et al., 2003), suggesting it is not functionally equivalent to SLC13. The phylogenetic tree indicates that the ancestral protein of TDT was first split to give a clade specific to seed plants and a separate clade for Selaginella and Physcomitrella (Figure 4B). The family members in dicotyledonous and monocotyledonous plant species are suggested to have expanded through subsequent gene duplications. There appears to be two separate forms for monocotyledonous TDT, although their functional differences are not known.
Intriguingly, a group of phosphate transporters (PHO) that may share a common ancestry with TDT was identified. This group is composed of low-affinity PHO from yeast (Wykoff and O’Shea, 2001; Hürlimann et al., 2007). Yeast PHO87, PHO90, and PHO91, and homologs from Aspergillus, Chlamydomonas, Volvox, and Drosophila are present in this clade. It may be hypothesized that the substrate specificity could have been low during the ancient period and was defined when the ancestor split to TDT and PHO. The low substrate affinity of the existing PHO87 and PHO90 (Wykoff and O’Shea, 2001) may be considered as a remnant of the ancestral trait.
Conclusion
Sulfate transporters are essential biological components. The occurrence of types of sulfate transporters may vary depending on genotype, the environment, as well as location at the organ or subcellular compartment level. A number of studies have described the biochemical and physiological functions of sulfate transporters from various organisms. The present study is intended to provide a framework to reinterpret the biological information in the context of their evolutionary relationships. Using protein sequences of sulfate transporters from a diverse set of organisms, distinct evolutionary origins of sulfate transporters were identified. The results suggest that they subsequently underwent gene duplications and eventually expanded to have multiple subfamily members playing potentially specialized roles in sulfate transport processes.
The phylogenetic analysis indicates the evolutionary trajectories of two distinct families of sulfate transporters in green algae and land plant species: (i) Chlorophyte green algae contain both SULTR and SAC1/SLT family members; (ii) SULTR family in chlorophytes is associated with the lineage of plant SULTR4 subfamily but also contains divergent members likely originating from earlier evolutionary events; (iii) Angiosperms has multiple SULTR1–SULTR4 subfamily members but are devoid of SAC1/SLT; (iv) Selaginella and Physcomitrella are at intermediate positions between algae and angiosperms, as they seem to have partially developed SULTR subfamilies, and have SAC1/SLT homologs as well that are absent in angiosperms. These lines of evidence suggest that plants and algae individually may have developed mechanisms for sulfate transport and sensing to adapt to their natural environments. The upstream sulfur-sensing system may be different between plants and algae, as suggested by the absence of SAC1 in plants. In addition, their sulfate transport systems appear to have different biochemical characteristics and physiological roles.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Shin-Han Shiu is partially supported by NSF grants IOS-1126998 and MCB-0929100. Rothamsted Research receives grant aided support from the Biotechnology and Biological Sciences Research Council (BBSRC), UK.
Footnotes
References
Aravind, L., and Koonin, E. V. (2000). The STAS domain – a link between anion transporters and antisigma-factor antagonists. Curr. Biol. 10, R53–R55.
Baxter, I., Muthukumar, B., Park, H. C., Buchner, P., Lahner, B., Danku, J., Zhao, K., Lee, J., Hawkesford, M. J., Guerinot, M. L., and Salt, D. E. (2008). Variation in molybdenum content across broadly distributed populations of Arabidopsis thaliana is controlled by a mitochondrial molybdenum transporter (MOT1). PLoS Genet. 4, e1000004. doi: 10.1371/journal.pgen.1000004
Buchner, P., Takahashi, H., and Hawkesford, M. J. (2004). Plant sulphate transporters: co-ordination of uptake, intracellular and long-distance transport. J. Exp. Bot. 55, 1765–1773.
Cherest, H., Davidian, J. C., Thomas, D., Benes, V., Ansorge, W., and Surdin-Kerjan, Y. (1997). Molecular characterization of two high affinity sulfate transporters in Saccharomyces cerevisiae. Genetics 145, 627–635.
Davies, J. P., Yildiz, F. H., and Grossman, A. (1996). Sac1, a putative regulator that is critical for survival of Chlamydomonas reinhardtii during sulfur deprivation. EMBO J. 15, 2150–2159.
Emmerlich, V., Linka, N., Reinhold, T., Hurth, M. A., Traub, M., Martinoia, E., and Neuhaus, H. E. (2003). The plant homolog to the human sodium/dicarboxylic cotransporter is the vacuolar malate carrier. Proc. Natl. Acad. Sci. U.S.A. 100, 11122–11126.
Fei, Y. J., Inoue, K., and Ganapathy, V. (2003). Structural and functional characteristics of two sodium-coupled dicarboxylate transporters (ceNaDC1 and ceNaDC2) from Caenorhabditis elegans and their relevance to life span. J. Biol. Chem. 278, 6136–6144.
Gasber, A., Klaumann, S., Trentmann, O., Trampczynska, A., Clemens, S., Schneider, S., Sauer, N., Feifer, I., Bittner, F., Mendel, R. R., and Neuhaus, H. E. (2011). Identification of an Arabidopsis solute carrier critical for intracellular transport and inter-organ allocation of molybdate. Plant Biol. 13, 710–718.
Hästbacka, J., de la Chapelle, A., Mahtani, M. M., Clines, G., Reeve-Daly, M. P., Daly, M., Hamilton, B. A., Kusumi, K., Trivedi, B., Weaver, A., Coloma, A., Lovett, M., Buckler, A., Kaitila, I., and Lander, E. S. (1994). The diastrophic dysplasia gene encodes a novel sulfate transporter: positional cloning by fine-structure linkage disequilibrium mapping. Cell 78, 1073–1087.
Hawkesford, M. J. (2003). Transporter gene families in plants: the sulfate transporter gene family – redundancy or specialization? Physiol. Plant 117, 155–163.
Hawkesford, M. J., Davidian, J. C., and Grignon, C. (1993). Sulphate/proton cotransport in plasma-membrane vescicles isolated from roots of Brassica napus L.: increased transport in membranes isolated from sulphur-starved plants. Planta 190, 297–304.
Hürlimann, H. C., Stadler-Waibel, M., Werner, T. P., and Freimoser, F. M. (2007). Pho91 is a vacuolar phosphate transporter that regulates phosphate and polyphosphate metabolism in Saccharomyces cerevisiae. Mol. Biol. Cell 18, 4438–4445.
Hurth, M. A., Suh, S. J., Kretzschmar, T., Geis, T., Bregante, M., Gambale, F., Martinoia, E., and Neuhaus, H. E. (2005). Impaired pH homeostasis in Arabidopsis lacking the vacuolar dicarboxylate transporter and analysis of carboxylic acid transport across the tonoplast. Plant Physiol. 137, 901–910.
Inoue, K., Fei, Y. J., Huang, W., Zhuang, L., Chen, Z., and Ganapathy, V. (2002). Functional identity of Drosophila melanogaster Indy as a cation-independent, electroneutral transporter for tricarboxylic acid-cycle intermediates. Biochem. J. 367, 313–319.
Kataoka, T., Watanabe-Takahashi, A., Hayashi, N., Ohnishi, M., Mimura, T., Buchner, P., Hawkesford, M. J., Yamaya, T., and Takahashi, H. (2004a). Vacuolar sulfate transporters are essential determinants controlling internal distribution of sulfate in Arabidopsis. Plant Cell 16, 2693–2704.
Kataoka, T., Hayashi, N., Yamaya, T., and Takahashi, H. (2004b). Root-to-shoot transport of sulfate in Arabidopsis: evidence for the role of SULTR3;5 as a component of low-affinity sulfate transport system in the root vasculature. Plant Physiol. 136, 4198–4204.
Kopriva, S. (2006). Regulation of sulfate assimilation in Arabidopsis and beyond. Ann. Bot. 97, 479–495.
Krogh, A., Larsson, B., von Heijne, G., and Sonnhammer, E. L. (2001). Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580.
Krusell, L., Krause, K., Ott, T., Desbrosses, G., Krämer, U., Sato, S., Nakamura, Y., Tabata, S., James, E. K., Sandal, N., Stougaard, J., Kawaguchi, M., Miyamoto, A., Suganuma, N., and Udvardi, M. K. (2005). The sulfate transporter SST1 is crucial for symbiotic nitrogen fixation in Lotus japonicus root nodules. Plant Cell 17, 1625–1636.
Lass, B., and Ullrich-Eberius, C. L. (1984). Evidence for proton/sulfate cotransport and its kinetics in Lemna gibba G1. Planta 161, 53–60.
Laudenbach, D. E., and Grossman, A. R. (1991). Characterization and mutagenesis of sulfur-regulated genes in a cyanobacterium: evidence for function in sulfate transport. J. Bacteriol. 173, 2739–2750.
Leustek, T., Martin, M. N., Bick, J. A., and Davies, J. P. (2000). Pathways and regulation of sulfur metabolism revealed through molecular and genetic studies. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51, 141–165.
Leves, F. P., Tierney, M. L., and Howitt, S. M. (2008). Polar residues in a conserved motif spanning helices 1 and 2 are functionally important in the SulP transporter family. Int. J. Biochem. Cell Biol. 40, 2596–2605.
Lindberg, P., and Melis, A. (2008). The chloroplast sulfate transport system in the green alga Chlamydomonas reinhardtii. Planta 228, 951–961.
Markovich, D., and Murer, H. (2004). The SLC13 gene family of sodium sulphate/carboxylate cotransporters. Pflugers Arch. 447, 594–602.
Melis, A., and Chen, H. C. (2005). Chloroplast sulfate transport in green algae – genes, proteins and effects. Photosyn. Res. 86, 299–307.
Mount, D. B., and Romero, M. F. (2004). The SLC26 gene family of multifunctional anion exchangers. Pflugers Arch. 447, 710–721.
Pajor, A. M. (2006). Molecular properties of the SLC13 family of dicarboxylate and sulfate transporters. Pflugers Arch. 451, 597–605.
Pootakham, W., Gonzalez-Ballester, D., and Grossman, A. R. (2010). Identification and regulation of plasma membrane sulfate transporters in Chlamydomonas. Plant Physiol. 153, 1653–1668.
Rogina, B., Reenan, R. A., Nilsen, S. P., and Helfand, S. L. (2000). Extended life-span conferred by cotransporter gene mutations in Drosophila. Science 290, 2137–2140.
Roomans, G. M., Kuypers, G. A. J., Theuvenet, A. P. R., and Borst-Pauwels, G. W. F. H. (1979). Kinetics of sulfate uptake by yeast. Biochim. Biophys. Acta 551, 197–206.
Saito, K. (2004). Sulfur assimilatory metabolism. The long and smelling road. Plant Physiol. 136, 2443–2450.
Shibagaki, N., Rose, A., Mcdermott, J. P., Fujiwara, T., Hayashi, H., Yoneyama, T., and Davies, J. P. (2002). Selenate-resistant mutants of Arabidopsis thaliana identify SULTR1;2, a sulfate transporter required for efficient transport of sulfate into roots. Plant J. 29, 475–486.
Sirko, A., Hryniewicz, M., Hulanicka, D., and Böck, A. (1990). Sulfate and thiosulfate transport in Escherichia coli K-12: nucleotide sequence and expression of the cysTWAM gene cluster. J. Bacteriol. 172, 3351–3357.
Smith, F. W., Hawkesford, M. J., Ealing, P. M., Clarkson, D. T., Vanden Berg, P. J., Belcher, A. R., and Warrilow, A. G. S. (1997). Regulation of expression of a cDNA from barley roots encoding a high affinity sulfate transporter. Plant J. 12, 875–884.
Smith, F. W., Hawkesford, M. J., Prosser, I. M., and Clarkson, D. T. (1995a). Isolation of a cDNA from Saccharomyces cerevisiae that encodes a high affinity sulphate transporter at the plasma membrane. Mol. Gen. Genet. 247, 709–715.
Smith, F. W., Ealing, P. M., Hawkesford, M. J., and Clarkson, D. T. (1995b). Plant members of a family of sulfate transporters reveal functional subtypes. Proc. Natl. Acad. Sci. U.S.A. 92, 9373–9377.
Stamatakis, A. (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690.
Takahashi, H. (2010). Regulation of sulfate transport and assimilation in plants. Int. Rev. Cell Mol. Biol. 281, 129–159.
Takahashi, H., Kopriva, S., Giordano, M., Saito, K., and Hell, R. (2011). Sulfur assimilation in photosynthetic organisms: molecular functions and regulations of transporters and assimilatory enzymes. Annu. Rev. Plant Biol. 62, 157–184.
Takahashi, H., Watanabe-Takahashi, A., Smith, F. W., Blake-Kalff, M., Hawkesford, M. J., and Saito, K. (2000). The role of three functional sulfate transporters involved in uptake and translocation of sulfate in Arabidopsis thaliana. Plant J. 23, 171–182.
Takahashi, H., Yamazaki, M., Sasakura, N., Watanabe, A., Leustek, T., de Almeida Engler, J., Engler, G., Van Montagu, M., and Saito, K. (1997). Regulation of sulfur assimilation in higher plants: a sulfate transporter induced in sulfate starved roots plays a central role in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 94, 11102–11107.
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739.
Tejada-Jiménez, M., Llamas, A., Sanz-Luque, E., Galván, A., and Fernández, E. (2007). A high-affinity molybdate transporter in eukaryotes. Proc. Natl. Acad. Sci. U.S.A. 104, 20126–20130.
Tomatsu, H., Takano, J., Takahashi, H., Watanabe-Takahashi, A., Shibagaki, N., and Fujiwara, T. (2007). An Arabidopsis thaliana high-affinity molybdate transporter required for efficient uptake of molybdate from soil. Proc. Natl. Acad. Sci. U.S.A. 104, 18807–18812.
Vauclare, P., Kopriva, S., Fell, D., Suter, M., Sticher, L., von Ballmoos, P., Krähenbühl, U., den Camp, R. O., and Brunold, C. (2002). Flux control of sulphate assimilation in Arabidopsis thaliana: adenosine 5′-phosphosulphate reductase is more susceptible than ATP sulphurylase to negative control by thiols. Plant J. 31, 729–740.
Wykoff, D. D., and O’Shea, E. K. (2001). Phosphate transport and sensing in Saccharomyces cerevisiae. Genetics 159, 1491–1499.
Yoshimoto, N., Inoue, E., Watanabe-Takahashi, A., Saito, K., and Takahashi, H. (2007). Posttranscriptional regulation of high-affinity sulfate transporters in Arabidopsis by sulfur nutrition. Plant Physiol. 145, 378–388.
Yoshimoto, N., Takahashi, H., Smith, F. W., Yamaya, T., and Saito, K. (2002). Two distinct high-affinity sulfate transporters with different inducibilities mediate uptake of sulfate in Arabidopsis roots. Plant J. 29, 465–473.
Zuber, H., Davidian, J. C., Aubert, G., Aimé, D., Belghazi, M., Lugan, R., Heintz, D., Wirtz, M., Hell, R., Thompson, R., and Gallardo, K. (2010). The seed composition of Arabidopsis mutants for the group 3 sulfate transporters indicates a role in sulfate translocation within developing seeds. Plant Physiol. 154, 913–926.
Keywords: evolution, plant, sulfate, transporter
Citation: Takahashi H, Buchner P, Yoshimoto N, Hawkesford MJ and Shiu S-H (2012) Evolutionary relationships and functional diversity of plant sulfate transporters. Front. Plant Sci. 2:119. doi: 10.3389/fpls.2011.00119
Received: 30 September 2011;
Paper pending published: 10 November 2011;
Accepted: 31 December 2011;
Published online: 19 January 2012.
Edited by:
Heven Sze, University of Maryland, USAReviewed by:
Li-Qing Chen, Carnegie Institution for Science, USAJoseph M. Jez, Washington University in St. Louis, USA
Copyright: © 2012 Takahashi, Buchner, Yoshimoto, Hawkesford and Shiu. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Hideki Takahashi, Department of Biochemistry and Molecular Biology, Michigan State University, 209 Biochemistry Building, East Lansing, MI 48824, USA. e-mail:aHRha2FoYUBtc3UuZWR1; Shin-Han Shiu, Department of Plant Biology, Michigan State University, S308 Plant Biology Building, East Lansing, MI 48824, USA. e-mail:c2hpdXNAbXN1LmVkdQ==

 Peter Buchner2
Peter Buchner2
