
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Plant Sci. , 19 January 2012
Sec. Plant Genetics and Genomics
Volume 2 - 2011 | https://doi.org/10.3389/fpls.2011.00116
This article is part of the Research Topic Regulatory Elements in RNA View all 12 articles
 Kentaro Sasaki1,2
Kentaro Sasaki1,2 Ryozo Imai1,2*
Ryozo Imai1,2*The cold shock domain (CSD) is a nucleic acid binding domain that is widely conserved from bacteria to higher plants and animals. In Escherichia coli, cold shock proteins (CSPs) are composed solely of a CSD and function as RNA chaperones that destabilize RNA secondary structures. Cellular RNAs tend to be folded into unfavorable structures under low temperature conditions, and RNA chaperones resolve these structures, recovering functionality of the RNAs. CSP functions are associated mainly with cold adaptation, but they are also involved in other biological processes under normal growth conditions. Eukaryotic CSD proteins contain auxiliary domains in addition to the CSD and regulate many biological processes such as development and stress tolerance. In plants, it has been demonstrated that CSD proteins play essential roles in acquiring freezing tolerance. In addition, it has been suggested that some plant CSD proteins regulate embryo development, flowering time, and fruit development. In this review, we summarize the pleiotropic biological functions of CSP proteins in plants and discuss possible mechanisms by which plant CSD proteins regulate the functions of RNA molecules.
Low temperature is a critical environmental factor that affects growth and survival of many plant species and limits their geographical distribution. Overwintering plants are able to increase their freezing tolerance when exposed to low but non-freezing temperatures, a process known as cold acclimation. During cold acclimation, several cellular and physiological changes occur, including alterations in gene expression. Cold-regulated (COR) genes are highly induced during cold acclimation and are involved in the acquisition of freezing tolerance (Thomashow, 1998). Expression of COR genes is activated by C-repeat binding factors (CBFs), the best-characterized transcription factors related to acquiring freezing tolerance (Jaglo-Ottosen et al., 1998; Liu et al., 1998). Since CBFs are plant specific (Riechmann and Meyerowitz, 1998; Riechmann et al., 2000; Sakuma et al., 2002), it is thought that the CBF signal transduction pathway is conserved only among plants. However, low temperature affects growth and development in diverse organisms. Therefore, it is reasonable to speculate that there are mechanisms for adaptation to low temperature that are conserved throughout evolution, as is known for heat-shock responses. To date, it is not well understood whether there are conserved responses to low temperature within prokaryotes and eukaryotes.
Cold acclimation is also observed in bacteria and has been extensively studied in Escherichia coli. The major cold shock protein (CSP) in E. coli, CspA, is predominantly induced after exposure to low temperature and accumulates to up to 10% of the total protein in the cell (Goldstein et al., 1990). E. coli contains nine cspA family genes (cspA to cspI) and four of them (cspA, cspB, cspG, and cspI) are induced by cold shock (Yamanaka et al., 1998; Wang et al., 1999). The cspC and cspE genes are expressed constitutively and involved in the chromosome partitioning (Yamanaka et al., 1994). Expression of cspD is induced at the stationary phase or upon nutrient starvation. Overexpression of cspD results in a lethal phenotype in E. coli (Yamanaka and Inouye, 1997; Xia et al., 2001). The E. coli cspA, cspB, cspG, and cspE quadruple deletion mutant shows a cold-sensitive phenotype, which can be complemented by overexpression of any one of the CSPs except CspD (Xia et al., 2001). CSPs unwind nucleic acid duplexes in vitro and in vivo (Jiang et al., 1997; Bae et al., 2000; Phadtare et al., 2002). RNA molecules tend to form stable secondary structures at low temperatures, which may impede RNA function such as in translation and transcription. Therefore, it has been suggested that CSPs act as RNA chaperones to destabilize RNA secondary structures, enabling efficient translation at low temperature (Jiang et al., 1997; Phadtare et al., 2002). In addition, CspA, CspC, and CspE are transcription antiterminators, which regulate expression of a set of cold-inducible genes (Bae et al., 2000). Bacterial CSPs are composed of a single nucleic acid binding domain (of about 70 amino acid residues) called the cold shock domain (CSD). The CSD consists of a five-stranded β-barrel containing two consensus RNA binding motifs (RNP-1 and RNP-2), which contribute to single-stranded DNA/RNA binding (Schroder et al., 1995; Hillier et al., 1998; Wang et al., 2000).
In animals, the homologous genes to bacterial CSPs have been identified, and have been shown to contain an N-terminal CSD and C-terminal auxiliary domains. The structure of the auxiliary domains varies significantly in different organisms. The vertebrate Y-box binding (YB) proteins have a highly conserved CSD, which shares about 40% amino acid sequence identity with bacterial CSPs (Wistow, 1990; Wolffe et al., 1992). YB proteins contain an N-terminal Ala and Pro rich domain (A/P domain) followed by a CSD and C-terminal alternating clusters of basic and acidic amino acid residues (B/A repeat; Wolffe, 1994; Graumann and Marahiel, 1998). The C-terminal B/A repeat was proposed to have DNA and RNA binding activity and the ability to bind other proteins (Evdokimova and Ovchinnikov, 1999). YB-1, the best-characterized YB protein, is a multifunctional protein that is involved in the regulation of transcription and translation, drug resistance, cell proliferation, and stress adaptation (Kohno et al., 2003). After a temperature downshift, no increase in cell numbers was observed for YB-1-depleted chicken cells, indicating an essential function for YB-1 in cell proliferation under cold conditions (Matsumoto et al., 2005). YB-1 binds to DNA and RNA and shows concentration-dependent melting and annealing activities (Matsumoto and Wolffe, 1998; Skabkin et al., 2001). These activities are probably necessary for the pleiotropic functions of YB-1. Another example of a eukaryotic CSD protein is LIN-28, which was originally identified as an essential regulator of larval development in C. elegans (Moss et al., 1997). LIN-28 comprises a single CSD and two CCHC retroviral-like zinc fingers at the C-terminus. Mammalian LIN-28 is a translational enhancer of IGF-2, which is essential for the growth and differentiation of muscle tissue (Polesskaya et al., 2007). In addition, it has been demonstrated that LIN-28 plays an important role in reprogramming human somatic cells into pluripotent stem cells (Yu et al., 2007; Liao et al., 2008). LIN-28 post-transcriptionally inhibits the biogenesis of let-7 miRNA, which is a regulator of cell growth and differentiation in embryonic cells (Roush and Slack, 2008; Viswanathan et al., 2008). Collectively, these studies suggest that animal CSD proteins play important roles in a variety of biological processes, not only stress adaptation.
Cold shock domain proteins are widespread among lower and higher plants including monocots and dicots (Karlson and Imai, 2003). Plant CSD proteins so far characterized typically contain a large glycine-rich region interspersed with CCHC zinc fingers at the C-terminus (Karlson et al., 2002), a domain structure, and arrangement resembling that of LIN-28. We thus conducted a database search using Plant GDB (http://www.plantgdb.org/) to identify CSD proteins in plant species whose genome sequences are available (Figure 1). All known genome sequences contain at least two CSD proteins that are composed of a CSD, glycine-rich regions and CCHC zinc fingers. The sequence information also indicates that genomic sequences for CSD proteins commonly contain no introns. The number of gene family members in each species ranges from two (Oryza sativa, Zea mays, Sorghum bicolor, and Vitis vinifera) to seven (Glycine max). In addition, individual species have both long and short versions of CSD proteins. In monocots, CSD proteins contain two or four CCHC zinc fingers, whereas the number of CCHC zinc fingers is one, two, four, five, or seven in dicots. The glycine-rich and CCHC zinc finger regions are probably involved in binding of nucleic acids and other proteins (Karlson et al., 2002; Nakaminami et al., 2006; Sasaki et al., 2007). Since all plant CSD proteins contain a highly conserved CSD, it is speculated that the diverse combinations of CCHC zinc fingers are necessary for binding to specific nucleic acids and proteins.
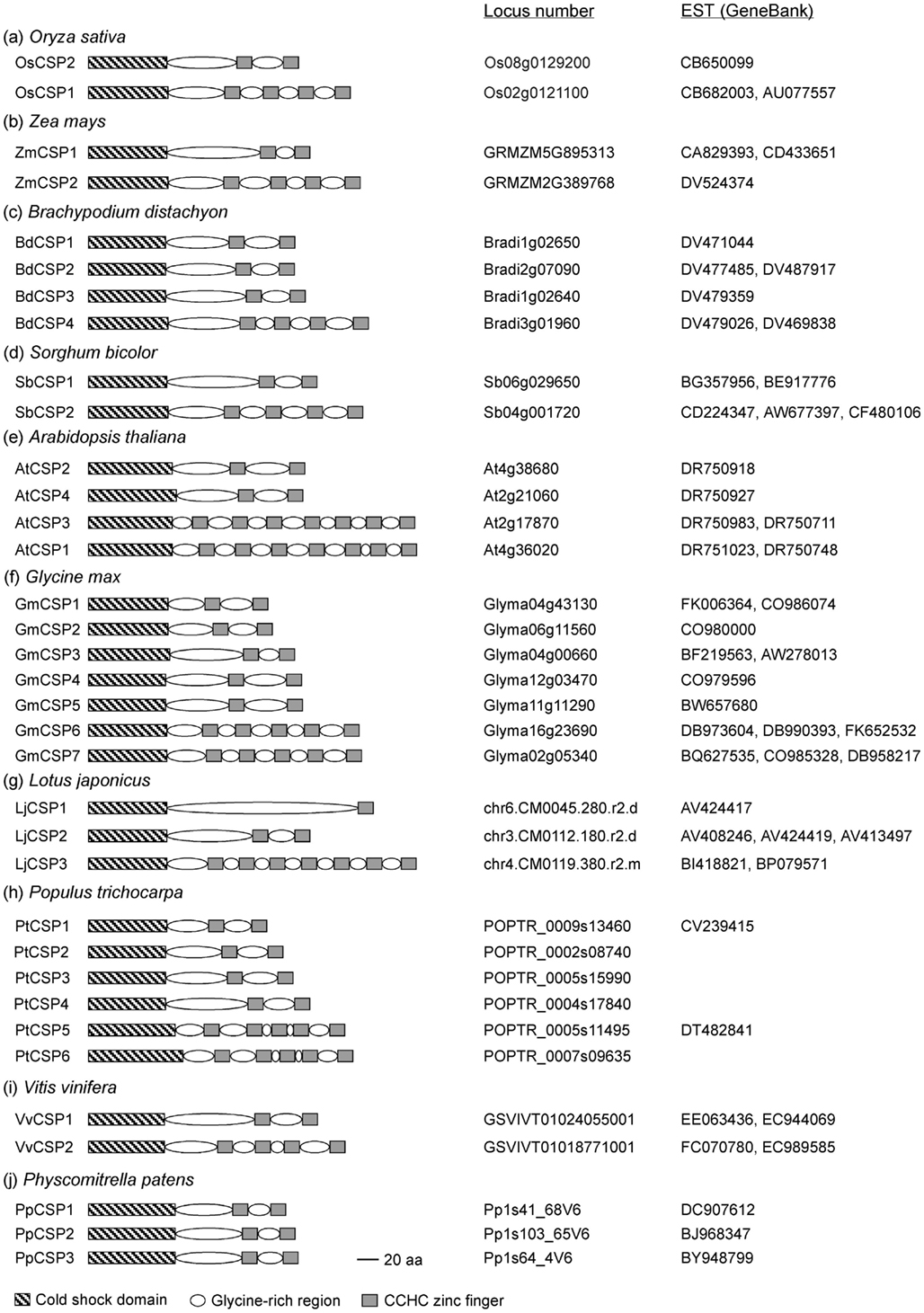
Figure 1. Schematic representation of the domain organization of cold shock domain proteins in plant species whose genome sequences are available. Locus numbers and corresponding EST accession numbers are shown. Only very short EST sequences, or none at all, are available for LjCSP1 (Lotus japonicas) and PtCSPs (Populus trichocarpa).
Whereas a considerable amount of research has been carried out to characterize CSD proteins in bacteria and animals, little is known about their functions in plants. The first functionally characterized plant CSD protein was the wheat CSP (WCSP1; Karlson et al., 2002; Table 1). WCSP1 contains a glycine-rich region interspersed with three C-terminal CCHC zinc fingers. WCSP1 mRNA is up-regulated in response to cold and the corresponding protein is substantially accumulated in crown tissue during prolonged cold acclimation. Transcript levels of WCSP1 are not modulated by other environmental stresses such as salt, drought and heat, or treatment with abscisic acid (Karlson et al., 2002), suggesting that the function of WCSP1 is specific to cold adaptation. WCSP1 binds to DNA and RNA and melts double-stranded nucleic acids in vitro and in vivo (Karlson et al., 2002; Nakaminami et al., 2005, 2006). In addition, WCSP1 complements a cold-sensitive phenotype of the E. coli csp mutant (Nakaminami et al., 2006). These data suggest that WCSP1 shares a conserved function with E. coli CSPs and is involved in the regulation during cold acclimation.
Rice has two CSD proteins [OsCSP1 (Os02g0121100) and OsCSP2 (Os08g0129200)], which exhibit nucleic acid binding activity and complement the cold sensitivity of the E. coli csp mutant (Chaikam and Karlson, 2008). Expression of OsCSPs was slightly increased in shoot and root tissues by short term low temperature treatment (Chaikam and Karlson, 2008; Table 1). However, OsCSP protein levels were not increased in crown tissue during 10 days of low temperature treatment (Chaikam and Karlson, 2008). These data are in great contrast to the observed expression characteristics for WCSP1. Tissue specific expression patterns of OsCSPs revealed that OsCSP proteins are highly accumulated in the developing panicle, flower, and seed (Chaikam and Karlson, 2008). Thus, the functions of OsCSPs may be more associated with developmental processes than with cold tolerance.
In Arabidopsis, four CSD proteins (AtCSP1–AtCSP4) were identified and functional analyses of AtCSPs have been performed with overexpression lines and mutants (Table 1). An AtCSP3 (At2g17870) knock-out mutant (atcsp3-2) was more sensitive to freezing than was wild-type under both non-acclimated and cold-acclimated conditions (Kim et al., 2009). Overexpression of AtCSP3 confers increased freezing tolerance in Arabidopsis without obvious developmental defects (Kim et al., 2009). AtCSP3 does not affect the expression of CBFs and COR genes, but it regulates the expression of stress-related genes whose roles in freezing tolerance are unknown (Kim et al., 2009). Interestingly, several genes down-regulated in atcsp3-2 are known to be up-regulated in the ada2b-1 mutant, which is more freezing tolerant than wild-type without up-regulation of COR gene expression (Vlachonasios et al., 2003). Since ADA2b is a component of histone acetyltransferase complexes, it is speculated that there is crosstalk between AtCSP3 and histone modification. It has been demonstrated that AtCSP2 (AtGRP2/CSDP2; At4g38680) transcript is highly expressed in meristematic and developing tissues (Fusaro et al., 2007; Sasaki et al., 2007; Nakaminami et al., 2009). Consistent with its expression patterns, functional analyses using RNAi knock-down transgenic plants indicated that AtCSP2 negatively regulates flowering time, and positively regulates seed/embryo development (Fusaro et al., 2007). Recently, it was demonstrated that AtCSP4 (AtGRP2b; At2g21060), the closest paralog of AtCSP2, also plays an important role in development. Overexpression of AtCSP4 resulted in reduced silique length and embryo lethality (Yang and Karlson, 2011). Expression of several MADS-box and endosperm development genes is altered in the AtCSP4-overexpressing line during floral and silique development. Park et al. (2009) reported that overexpression of AtCSP1 (CSDP1; At4g36020) delays seed germination under dehydration or salt stress conditions, whereas overexpression of AtCSP2 accelerated seed germination under salt stress conditions. Although overexpression of AtCSP1 or AtCSP2 did not enhance freezing tolerance in Arabidopsis, they each complement the freezing-sensitive phenotype of grp7, which is a mutant of glycine-rich RNA binding protein 7 (GRP7) with RNA chaperone activity (Kim et al., 2007, 2008). These functional studies of plant CSD proteins reveal that the expressions of multiple CSD proteins are differentially regulated by developmental and stress cues (Table 1). Furthermore, plant CSD proteins commonly exhibit RNA chaperone activity and function as regulatory proteins.
Our understanding of plant CSD proteins has progressed significantly in recent years. The evolutionarily conserved structures and biochemical activities of CSD proteins suggest that these proteins are indispensable for cold adaptation in both prokaryotes and eukaryotes. In addition, regulatory functions of CSD proteins extend to developmental processes in both animals and plants. Whereas information regarding the biological functions of plant CSD proteins is accumulating, the cellular function of plant CSD proteins still remains to be elucidated. Whether or not CSD proteins have specific target mRNAs in plant cells needs to be addressed. In Chlamydomonas reinhardtii, the NAB1 CSD protein stabilizes the mRNA of LHCBM (major light-harvesting complex of photosynthesis II) and represses its translation at the pre-initiation stage (Mussgnug et al., 2005). NAB1-like proteins have not been identified in higher plants. However, RNA stabilization and translational repression have been described for animal CSD proteins such as YB-1 and FRGY2 (Matsumoto et al., 1996; Evdokimova et al., 2001). Conserved post-transcriptional regulation mechanisms may exist for NAB1 and animal CSD proteins. Further studies are necessary to investigate possible CSD-target mRNAs in higher plants. These investigations could reveal novel mechanisms of gene regulation through CSD proteins.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work was supported by a grant from NARO (Development of innovative crops through the molecular analysis of useful genes) to Kentaro Sasaki and Grants-in-aid for Scientific Research from the Japan Society of Promotion of Science (KAKENHI Scientific Research B nos. 22380063 and 19380063) to Ryozo Imai.
Bae, W., Xia, B., Inouye, M., and Severinov, K. (2000). Escherichia coli CspA-family RNA chaperones are transcription antiterminators. Proc. Natl. Acad. Sci. U.S.A. 97, 7784–7789.
Chaikam, V., and Karlson, D. (2008). Functional characterization of two cold shock domain proteins from Oryza sativa. Plant Cell Environ. 31, 995–1006.
Evdokimova, V., Ruzanov, P., Imataka, H., Raught, B., Svitkin, Y., Ovchinnikov, L. P., and Sonenberg, N. (2001). The major mRNA-associated protein YB-1 is a potent 5′ cap-dependent mRNA stabilizer. EMBO J. 20, 5491–5502.
Evdokimova, V. M., and Ovchinnikov, L. P. (1999). Translational regulation by Y-box transcription factor: involvement of the major mRNA-associated protein, p50. Int. J. Biochem. Cell Biol. 31, 139–149.
Fusaro, A. F., Bocca, S. N., Ramos, R. L., Barroco, R. M., Magioli, C., Jorge, V. C., Coutinho, T. C., Rangel-Lima, C. M., De Rycke, R., Inze, D., Engler, G., and Sachetto-Martins, G. (2007). AtGRP2, a cold-induced nucleo-cytoplasmic RNA-binding protein, has a role in flower and seed development. Planta 225, 1339–1351.
Goldstein, J., Pollitt, N. S., and Inouye, M. (1990). Major cold shock protein of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 87, 283–287.
Graumann, P. L., and Marahiel, M. A. (1998). A superfamily of proteins that contain the cold-shock domain. Trends Biochem. Sci. 23, 286–290.
Hillier, B. J., Rodriguez, H. M., and Gregoret, L. M. (1998). Coupling protein stability and protein function in Escherichia coli CspA. Fold. Des. 3, 87–93.
Jaglo-Ottosen, K. R., Gilmour, S. J., Zarka, D. G., Schabenberger, O., and Thomashow, M. F. (1998). Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 280, 104–106.
Jiang, W., Hou, Y., and Inouye, M. (1997). CspA, the major cold-shock protein of Escherichia coli, is an RNA chaperone. J. Biol. Chem. 272, 196–202.
Karlson, D., and Imai, R. (2003). Conservation of the cold shock domain protein family in plants. Plant Physiol. 131, 12–15.
Karlson, D., Nakaminami, K., Toyomasu, T., and Imai, R. (2002). A cold-regulated nucleic acid-binding protein of winter wheat shares a domain with bacterial cold shock proteins. J. Biol. Chem. 277, 35248–35256.
Kim, J. S., Jung, H. J., Lee, H. J., Kim, K. A., Goh, C. H., Woo, Y., Oh, S. H., Han, Y. S., and Kang, H. (2008). Glycine-rich RNA-binding protein 7 affects abiotic stress responses by regulating stomata opening and closing in Arabidopsis thaliana. Plant J. 55, 455–466.
Kim, J. S., Park, S. J., Kwak, K. J., Kim, Y. O., Kim, J. Y., Song, J., Jang, B., Jung, C. H., and Kang, H. (2007). Cold shock domain proteins and glycine-rich RNA-binding proteins from Arabidopsis thaliana can promote the cold adaptation process in Escherichia coli. Nucleic Acids Res. 35, 506–516.
Kim, M. H., Sasaki, K., and Imai, R. (2009). Cold shock domain protein 3 regulates freezing tolerance in Arabidopsis thaliana. J. Biol. Chem. 284, 23454–23460.
Kohno, K., Izumi, H., Uchiumi, T., Ashizuka, M., and Kuwano, M. (2003). The pleiotropic functions of the Y-box-binding protein, YB-1. Bioessays 25, 691–698.
Liao, J., Wu, Z., Wang, Y., Cheng, L., Cui, C., Gao, Y., Chen, T., Rao, L., Chen, S., Jia, N., Dai, H., Xin, S., Kang, J., Pei, G., and Xiao, L. (2008). Enhanced efficiency of generating induced pluripotent stem (iPS) cells from human somatic cells by a combination of six transcription factors. Cell Res. 18, 600–603.
Liu, Q., Kasuga, M., Sakuma, Y., Abe, H., Miura, S., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1998). Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10, 1391–1406.
Matsumoto, K., Meric, F., and Wolffe, A. P. (1996). Translational repression dependent on the interaction of the Xenopus Y-box protein FRGY2 with mRNA. Role of the cold shock domain, tail domain, and selective RNA sequence recognition. J. Biol. Chem. 271, 22706–22712.
Matsumoto, K., Tanaka, K. J., and Tsujimoto, M. (2005). An acidic protein, YBAP1, mediates the release of YB-1 from mRNA and relieves the translational repression activity of YB-1. Mol. Cell. Biol. 25, 1779–1792.
Matsumoto, K., and Wolffe, A. P. (1998). Gene regulation by Y-box proteins: coupling control of transcription and translation. Trends Cell Biol. 8, 318–323.
Moss, E. G., Lee, R. C., and Ambros, V. (1997). The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell 88, 637–646.
Mussgnug, J. H., Wobbe, L., Elles, I., Claus, C., Hamilton, M., Fink, A., Kahmann, U., Kapazoglou, A., Mullineaux, C. W., Hippler, M., Nickelsen, J., Nixon, P. J., and Kruse, O. (2005). NAB1 is an RNA binding protein involved in the light-regulated differential expression of the light-harvesting antenna of Chlamydomonas reinhardtii. Plant Cell 17, 3409–3421.
Nakaminami, K., Hill, K., Perry, S. E., Sentoku, N., Long, J. A., and Karlson, D. T. (2009). Arabidopsis cold shock domain proteins: relationships to floral and silique development. J. Exp. Bot. 60, 1047–1062.
Nakaminami, K., Karlson, D. T., and Imai, R. (2006). Functional conservation of cold shock domains in bacteria and higher plants. Proc. Natl. Acad. Sci. U.S.A. 103, 10122–10127.
Nakaminami, K., Sasaki, K., Kajita, S., Takeda, H., Karlson, D., Ohgi, K., and Imai, R. (2005). Heat stable ssDNA/RNA-binding activity of a wheat cold shock domain protein. FEBS Lett. 579, 4887–4891.
Park, S. J., Kwak, K. J., Oh, T. R., Kim, Y. O., and Kang, H. (2009). Cold shock domain proteins affect seed germination and growth of Arabidopsis thaliana under abiotic stress conditions. Plant Cell Physiol. 50, 869–878.
Phadtare, S., Inouye, M., and Severinov, K. (2002). The nucleic acid melting activity of Escherichia coli CspE is critical for transcription antitermination and cold acclimation of cells. J. Biol. Chem. 277, 7239–7245.
Polesskaya, A., Cuvellier, S., Naguibneva, I., Duquet, A., Moss, E. G., and Harel-Bellan, A. (2007). Lin-28 binds IGF-2 mRNA and participates in skeletal myogenesis by increasing translation efficiency. Genes Dev. 21, 1125–1138.
Riechmann, J. L., Heard, J., Martin, G., Reuber, L., Jiang, C., Keddie, J., Adam, L., Pineda, O., Ratcliffe, O. J., Samaha, R. R., Creelman, R., Pilgrim, M., Broun, P., Zhang, J. Z., Ghandehari, D., Sherman, B. K., and Yu, G. (2000). Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290, 2105–2110.
Riechmann, J. L., and Meyerowitz, E. M. (1998). The AP2/EREBP family of plant transcription factors. Biol. Chem. 379, 633–646.
Sakuma, Y., Liu, Q., Dubouzet, J. G., Abe, H., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2002). DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 290, 998–1009.
Sasaki, K., Kim, M. H., and Imai, R. (2007). Arabidopsis COLD SHOCK DOMAIN PROTEIN2 is a RNA chaperone that is regulated by cold and developmental signals. Biochem. Biophys. Res. Commun. 364, 633–638.
Schroder, K., Graumann, P., Schnuchel, A., Holak, T. A., and Marahiel, M. A. (1995). Mutational analysis of the putative nucleic acid-binding surface of the cold-shock domain, CspB, revealed an essential role of aromatic and basic residues in binding of single-stranded DNA containing the Y-box motif. Mol. Microbiol. 16, 699–708.
Skabkin, M. A., Evdokimova, V., Thomas, A. A., and Ovchinnikov, L. P. (2001). The major messenger ribonucleoprotein particle protein p50 (YB-1) promotes nucleic acid strand annealing. J. Biol. Chem. 276, 44841–44847.
Thomashow, M. F. (1998). Role of cold-responsive genes in plant freezing tolerance. Plant Physiol. 118, 1–8.
Viswanathan, S. R., Daley, G. Q., and Gregory, R. I. (2008). Selective blockade of microRNA processing by Lin28. Science 320, 97–100.
Vlachonasios, K. E., Thomashow, M. F., and Triezenberg, S. J. (2003). Disruption mutations of ADA2b and GCN5 transcriptional adaptor genes dramatically affect Arabidopsis growth, development, and gene expression. Plant Cell 15, 626–638.
Wang, N., Yamanaka, K., and Inouye, M. (1999). CspI, the ninth member of the CspA family of Escherichia coli, is induced upon cold shock. J. Bacteriol. 181, 1603–1609.
Wang, N., Yamanaka, K., and Inouye, M. (2000). Acquisition of double-stranded DNA-binding ability in a hybrid protein between Escherichia coli CspA and the cold shock domain of human YB-1. Mol. Microbiol. 38, 526–534.
Wolffe, A. P. (1994). Structural and functional properties of the evolutionarily ancient Y-box family of nucleic acid binding proteins. Bioessays 16, 245–251.
Wolffe, A. P., Tafuri, S., Ranjan, M., and Familari, M. (1992). The Y-box factors: a family of nucleic acid binding proteins conserved from Escherichia coli to man. New Biol. 4, 290–298.
Xia, B., Ke, H., and Inouye, M. (2001). Acquirement of cold sensitivity by quadruple deletion of the cspA family and its suppression by PNPase S1 domain in Escherichia coli. Mol. Microbiol. 40, 179–188.
Yamanaka, K., Fang, L., and Inouye, M. (1998). The CspA family in Escherichia coli: multiple gene duplication for stress adaptation. Mol. Microbiol. 27, 247–255.
Yamanaka, K., and Inouye, M. (1997). Growth-phase-dependent expression of cspD, encoding a member of the CspA family in Escherichia coli. J. Bacteriol. 179, 5126–5130.
Yamanaka, K., Mitani, T., Ogura, T., Niki, H., and Hiraga, S. (1994). Cloning, sequencing, and characterization of multicopy suppressors of a mukB mutation in Escherichia coli. Mol. Microbiol. 13, 301–312.
Yang, Y., and Karlson, D. T. (2011). Overexpression of AtCSP4 affects late stages of embryo development in Arabidopsis. J. Exp. Bot. 62, 2079–2091.
Keywords: cold shock protein, cold shock domain, cold acclimation, freezing tolerance, development
Citation: Sasaki K and Imai R (2012) Pleiotropic roles of cold shock domain proteins in plants. Front. Plant Sci. 2:116. doi: 10.3389/fpls.2011.00116
Received: 13 December 2011;
Accepted: 27 December 2011;
Published online: 19 January 2012.
Edited by:
Anireddy S. N. Reddy, Colorado State University, USAReviewed by:
Hunseung Kang, Chonnam National University, South KoreaCopyright: © 2012 Sasaki and Imai. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Ryozo Imai, Hokkaido Agriculture Research Center, National Agriculture and Food Research Organization, Hitsujigaoka 1, Toyohira-ku, Sapporo 062-8555, Japan. e-mail:cnppQGFmZnJjLmdvLmpw
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.