
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Plant Sci. , 22 December 2011
Sec. Plant Genetics and Genomics
Volume 2 - 2011 | https://doi.org/10.3389/fpls.2011.00099
This article is part of the Research Topic Regulatory Elements in RNA View all 12 articles
Initiation of RNA polymerase II transcription signals the beginning of a series of physically and functionally coupled pre-mRNA processing events that transform an RNA transcript into a highly structured, mature ribonucleoprotein complex. With such a complexity of co-transcriptional processes comes the need to identify and degrade improperly processed transcripts. Quality control of mRNA expression primarily involves exonucleolytic degradation of aberrant RNAs. RNA silencing, on the other hand, tends to be viewed separately as a pathway that primarily functions in regulating endogenous gene expression and in genome defense against transposons and viruses. Here, we review current knowledge of these pathways as they exist in plants and draw parallels to similar pathways in other eukaryotes. We then highlight some unexplored overlaps that exist between the RNA silencing and RNA decay pathways of plants, as evidenced by their shared RNA substrates and shared genetic requirements.
Mature mRNA formation involves a series of inter-dependent and physically coupled co-transcriptional processing events. With such a complexity of co-transcriptional processes comes the need to identify and degrade improperly processed transcripts. Aberrant transcripts are not just a misuse of cellular resources, but could potentially be very damaging if translated into atypical proteins.
Exonucleolytic RNA turnover generally begins with the removal of the poly(A) tail, and proceeds from both the 5′ and 3′ ends of the transcript. In one pathway, the body of the RNA is degraded in a 3′–5′ direction by a multi-subunit complex of exoribonucleases called the exosome. The diversity of proteins that compose a functional exosome varies depending both on cellular conditions, and the nature of the RNA substrate (Lange et al., 2009). This diversity allows for controlled degradation of RNA, for example in the case of 5.8S rRNA synthesis, where limited, but precise 3′–5′ processing is required (Belostotsky, 2009). More commonly, however, the exosome carries out complete RNA degradation, such as for the turnover of tRNAs, small nuclear RNAs (snRNAs), small nucleolar RNAs (snoRNAs), and mRNAs (Allmang et al., 1999, 2000; Bousquet-Antonelli et al., 2000; Libri et al., 2002). The second pathway of RNA turnover involves removal of the 5′ 7-methylguanosine cap, followed by processive digestion by a 5′–3′ exoribonuclease. In Arabidopsis, XRN4 is the predominant cytoplasmic exoribonuclease, while XRN2 and XRN3 have additional roles in degrading transcripts within the nucleus (Kastenmayer and Green, 2000; Gy et al., 2007).
While polyadenylation by canonical poly(A) polymerases typically stabilizes transcripts, in some cases, polyadenylation by non-canonical poly(A) polymerases can flag transcripts as targets for 5′ or 3′ exonucleolytic degradation (Anderson and Wang, 2009; Lange et al., 2009).
The nonsense-mediated decay (NMD) pathway is responsible for degrading transcripts with premature termination codons (PTCs) that normally arise through improper splicing (Rebbapragada and Lykke-Andersen, 2009). In doing so, NMD prevents the formation of potentially deleterious C-terminally truncated proteins.
The UP-FRAMESHIFT gene family, consisting of three members (UPF1–3), is conserved between yeast, plants, and humans. The first member, UPF1, is the most conserved and in metazoans at least, ultimately directs transcript degradation following phosphorylation by SMG1 with assistance from UPF2 and UPF3 (Page et al., 1999; Yamashita et al., 2001; Kashima et al., 2006). In accordance with this role, Arabidopsis upf1 mutants over-accumulate transcripts containing PTCs (Yoine et al., 2006; Hayden and Jorgensen, 2007). In plants, as in mammals, tethering of UPF1 to transcripts targets them for NMD (Lykke-Andersen et al., 2000; Kertesz et al., 2006). How termination codons are recognized as premature is a topic of intense study, and as yet remains unresolved. In plants, NMD degrades transcripts with either a long 3′ UTR or with a termination codon in close proximity to an exon junction (Kertesz et al., 2006; Kerenyi et al., 2008).
In mammals, early studies showed that the presence of an exon junction complex (EJC) greater than 50 nucleotides (nt) downstream of a termination codon was capable of triggering NMD (Le Hir and Séraphin, 2008). A model for PTC definition was thus proposed to involve the position of the termination codon relative to EJCs during early rounds of translation (Kashima et al., 2006). UPF2 and UPF3 are bound to the core EJC, while a complex of proteins including SMG1 and UPF1 are bound to the terminating ribosome. The EJC (including UPF2 and UPF3) is normally removed during these early rounds of translation, except if the EJC is greater than 50 nt from the termination codon. In this case, the EJC remains present following translation termination, allowing UPF2 to bind to UPF1 and trigger SMG1 to phosphorylate UPF1, which ultimately leads to RNA decay (Kashima et al., 2006).
However, this model does not account for NMD in Saccharomyces cerevisiae, which has few intron-containing transcripts. Furthermore, subsequent studies on intronless mammalian and plant genes have shown that transcripts with long 3′ UTRs can be targeted for NMD despite the absence of a downstream EJC (Chan and Yu, 1998; Rajavel and Neufeld, 2001; Buhler et al., 2004; LeBlanc and Beemon, 2004). An alternative model, termed the faux 3′ UTR model, posits that the EJC is not a requisite for NMD, but rather that it is the length of the 3′ UTR that marks the transcript for NMD (Amrani et al., 2004). In the case of a proper termination codon, stimulatory factors present in the 3′ UTR may promote efficient termination of translation. However, when the distance between these 3′ UTR factors and the terminating ribosome becomes too great, termination is stalled, and NMD is triggered.
In yeast, it has been shown that ribosomes do not dissociate from mRNA when terminating at a PTC, possibly because they cannot receive the termination-stimulating factor (Amrani et al., 2004). The nature of the termination-stimulating factor is not fully known, but studies by Behm-Ansmant et al. (2007) have shown that artificial tethering of poly(A)-binding protein (PABP) close to a PTC efficiently suppresses NMD, suggesting that PABP may be one possible termination-stimulating factor. To account for the mammalian involvement of the EJC, it has been suggested that UPF1 phosphorylation by SMG1 may be made more efficient by the binding of an EJC that remains intact following the early rounds of translation (Stalder and Muhlemann, 2008).
Although the issue of PTC definition remains contentious, it is generally accepted to depend on an early pioneer round of translation, whereby a nuclear-localized ribosome scans the transcript for PTCs. Support for the pioneer round of translation was provided by the discovery that most mammalian nonsense transcripts are down-regulated in the nuclear fractions of cells (Urlaub et al., 1989; Belgrader et al., 1994; Kugler et al., 1995; Carter et al., 1996). Furthermore, various forms of evidence have demonstrated that NMD acts primarily on transcripts bound at their 5′ ends by the cap-binding complex (CBC; Ishigaki et al., 2001; Lejeune et al., 2002; Hosoda et al., 2005). The CBC is replaced by eukaryotic translation initiation factor 4E (eIF4E) soon after nuclear export, in order to support the bulk of translation. That PTC transcripts might be detected in the nucleus has led many to believe that translation occurs there also (Wilkinson, 2003). Others believe that scanning occurs in both the nucleus and the cytoplasm, or that translation occurs during nuclear export, when mRNAs are still associated with the nucleus (Chang et al., 2007). Following recognition of a PTC, transcripts are generally sent down the typical deadenylation and decapping decay pathways. In Drosophila, degradation is usually initiated by endonucleolytic cleavage followed by 5′–3′ and 3′–5′ exonucleolytic degradation of the cleavage fragments (Gatfield and Izaurralde, 2004). In humans, there is also evidence for the existence of endonucleolytic cleavage of nonsense transcripts (Eberle et al., 2009).
Unspliced or mis-spliced transcripts may be subject to several different RNA decay pathways, with the specific pathway taken depending on the nature of the mis-spliced or unspliced transcript. Transcripts containing introns that fail to be recognized by the spliceosome are exported to the cytoplasm, just as an intronless transcript would be, where they are likely to be translated and subject to NMD (He et al., 1993; Jaillon et al., 2008; Sayani et al., 2008; Kawashima et al., 2009). Evidence for the widespread use of NMD in degrading unspliced transcripts has come from studies by Jaillon et al. (2008), who found that unspliced eukaryotic introns rarely maintain the translation reading frame, and thus result in premature termination and NMD. In yeast and animal systems, intron-containing transcripts are recognized and committed to splicing by formation of the commitment complex (Legrain and Rosbash, 1989; Rain and Legrain, 1997; Galy et al., 2004). Improper splicing will lead predominantly to 3′–5′ degradation by the nuclear exosome, with a contribution from the 5′–3′ exonuclease-mediated decay pathway (Bousquet-Antonelli et al., 2000; Hilleren et al., 2001).
Most studies tracing the fate of unspliced pre-mRNAs have been conducted in yeast and animal systems, and little is known about the processes that regulate the fate of unspliced transcript levels in plants. Interestingly however, it has been shown that unspliced pre-mRNAs accumulate in the nucleoli of Arabidopsis, together with components of the NMD (Kim et al., 2009) and gene silencing (Pontes and Pikaard, 2008) pathways. How these aberrant RNAs are recognized is not known, but they are thought to induce gene silencing or are exported from the nucleus into specialized processing bodies (P-bodies), where the degradation phase of NMD takes place (Kim et al., 2009).
The gene silencing pathways of plants are characterized by duplicated genes with unique, redundant, and/or overlapping functions. They are crucial to regulating endogenous gene expression and protecting plants from transposons and viruses. RNA-mediated gene silencing pathways vary in their processing steps and their target loci, but all share three features: (i) production of double-stranded RNA (dsRNA), (ii) dicing of dsRNA by Dicer-like (DCL) enzymes to generate small interfering RNA (siRNA) or microRNA (miRNA), ~21–24 nt in length, and (iii) siRNA- or miRNA-directed execution of gene silencing. These small regulatory RNAs, together with Argonaute (AGO) proteins, can direct either post-transcriptional gene silencing (PTGS) by mRNA cleavage and/or translational repression, or transcriptional gene silencing (TGS) through DNA methylation and chromatin modifications. The production of dsRNA can be brought about either by direct transcription of an inverted repeat locus (Waterhouse et al., 1998; Smith et al., 2000) or through the activity of an RNA-dependent RNA polymerase (RDR) on a single-stranded RNA template (Dalmay et al., 2000; Mourrain et al., 2000).
Silencing of transgenes is a problem plaguing plant molecular biologists aiming to achieve high and stable transgene expression. Initial studies of transgene silencing in plants noted a correlation between high expression levels and the induction of silencing (Napoli et al., 1990; Smith et al., 1994; Elmayan and Vaucheret, 1996). It was proposed that gene silencing would be induced following a “quantitative aberration” in gene expression (Smith et al., 1994; Elmayan and Vaucheret, 1996). While this model held true in most cases, a report showing that equivalent transcription rates could exist in both silenced and expressing plants, meant that refinements to the model were needed (English et al., 1996). It was proposed that specific RNA molecules may have qualitative aberrations that lead to the induction of RNA silencing (English et al., 1996), perhaps by acting as a template for an RDR and leading to the synthesis of dsRNA. Supporting this idea, it has been shown that de-capped and improperly terminated read-through transcripts are direct substrates for RDR6, the key RDR required for transgene silencing in Arabidopsis (Gazzani et al., 2004; Luo and Chen, 2007). Under high rates of transcription, these aberrant RNA molecules would be more likely to accumulate as a result of transcriptional errors, and would therefore become increasingly likely to trigger the RNA silencing cascade. This so-called “threshold model” accounts for both qualitative and quantitative aspects of gene expression, and posits that excessive transcription can lead to an accumulation of aberrant transcripts. These abnormal transcripts would in turn increase the likelihood of being recognized by an RDR and ultimately trigger RNA silencing.
As more is learnt about the mechanisms of mRNA decay and RNA silencing, it is becoming increasingly apparent that spatial and functional overlaps exist between them (Figure 1). Similar to siRNA-directed gene silencing that targets transposons and viruses, miRNAs target endogenous transcripts as a way of negatively regulating gene expression. In animals, some miRNA targets are degraded in a similar way as NMD targets; via the deadenylation and decapping pathways (Bagga et al., 2005; Rehwinkel et al., 2005; Behm-Ansmant et al., 2006; Eulalio et al., 2009). Accordingly, the 5′–3′ exonuclease XRN1 and the decapping DCP1:DCP2 complex are required for both NMD as well as miRNA-mediated gene silencing of many metazoan mRNAs (Souret et al., 2004; Orban and Izaurralde, 2005; Rehwinkel et al., 2005). In humans, UPF1 is involved in miRNA-directed transcript down-regulation, and co-localizes with AGO1 and AGO2 in P-bodies (Jin et al., 2009). The C. elegans UPF1 ortholog, SMG2, has also been shown to be required for the maintenance of RNA silencing (Domeier et al., 2000). In Arabidopsis, UPF1 is primarily involved in NMD but has also been shown to play a role in inverted repeat-induced PTGS (Arciga-Reyes et al., 2006).
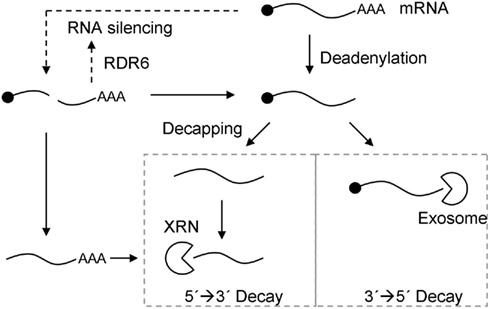
Figure 1. Model for the relationship between RNA silencing and RNA decay pathways. Exonucleolytic mRNA decay can be initiated by deadenylation, followed by 3′ → 5′ degradation by the exosome, or decapping, followed by 5′ → 3′ degradation by the XRN family of exoribonucleases (closed arrows). Alternatively, RNA silencing can be induced by miRNA- or siRNA-guided cleavage of mRNA (dotted arrows). This generates uncapped and unpolyadenylated RNA products, which can either be substrates for RDR6 and secondary siRNA biogenesis to reinforce RNA silencing (dotted arrow), or substrates for the exonucleolytic RNA decay pathways (closed arrows).
Interestingly, a genome-wide analysis of UPF1 targets in Arabidopsis found that in addition to many PTC-containing transcripts, several non-coding RNAs are upregulated in upf1 mutants, which, given the translational requirement of NMD, implicates UPF1 in other additional RNA decay pathways (Kurihara et al., 2009). Moreover, UPF1 and AGO1, as well as SDE3, an RNA helicase with an undetermined role in gene silencing, co-localize to Arabidopsis P-bodies (Zhang et al., 2006; Pomeranz et al., 2010; Xu and Chua, 2011). Plant P-bodies are largely defined by the accumulation of the decapping enzymes DCP1, DCP2, DCP5 and VARICOSE (VCS), and the 5′–3′ exoribonuclease XRN4, suggesting that, as in animals, plant P-bodies are sites of mRNA decapping and decay (Xu and Chua, 2011).
Whereas translational repression has long been observed in animals, only recently, the widespread existence of miRNA- and siRNA-guided translational repression has been reported in Arabidopsis (Brodersen et al., 2008). The importance of translational repression in gene silencing however has been questioned by the Bartel group, who found that mRNA decay accounts for most of the miRNA-mediated translational repression observed in mammals (Guo et al., 2010). Similar small RNA-directed decay pathways have not yet been observed in plants. However, the Arabidopsis decapping protein, VCS, has been shown to contribute to translational repression (Brodersen et al., 2008), warranting further investigations into this possibility.
In Arabidopsis, RNA silencing and RNA decay pathways share similar substrates, suggesting that competition or antagonism may exist between these pathways (Figure 1). De-capped RNAs, such as those arising from siRNA-directed cleavage, are substrates for both RDR6 as well as XRN2, 3, and 4 (Gazzani et al., 2004; Gy et al., 2007). Accordingly, XRN2, 3, and 4 are endogenous suppressors of RNA silencing (Gazzani et al., 2004; Gy et al., 2007). Improperly terminated transcripts are another form of aberrant RNA demonstrated to be a template for RDR6 (Herr et al., 2006; Luo and Chen, 2007). The first indication that improperly terminated transcripts might be substrates for RDR6 came from Herr et al. (2006), who isolated a series of mutants that were defective in 3′ end processing and also hyper-sensitive to RNA silencing. Later, this hypothesis was tested empirically with a GUS reporter, which displayed a progressive reduction in transgene silencing when cloned with either zero, one, or multiple terminators (Luo and Chen, 2007). In theory, read-through transcripts possess long 3′ UTRs and might therefore also be targeted for NMD (Kertesz et al., 2006). Indeed, a genome-wide analysis of exosome targets in Arabidopsis found that mRNAs extending beyond their annotated 3′ end, indicative of defective 3′ end processing, are targets for exosome-mediated decay (Chekanova et al., 2007).
Given the involvement of the poly(A) tail in nuclear mRNA export (Eckner et al., 1991), it is likely that at least some forms of aberrant RNA would become concentrated in the nucleus, and may be recognized by either the RNA silencing or NMD machinery. Indeed, as mentioned earlier, components of both RNA silencing and NMD pathways have been localized to sub-nuclear compartments, as have their substrates (Pontes and Pikaard, 2008; Kim et al., 2009; Hoffer et al., 2011). The question remains then as to how aberrant RNAs are sorted between the NMD and RNA silencing pathways. With RNA silencing acting in trans to degrade all homologous transcripts and NMD degrading individual nonsense transcripts in cis, the division of RNA substrates between these pathways is likely to have major consequences for the genome-wide regulation of gene expression.
One possible scenario is that NMD acts before RNA silencing, but has a limited capacity for RNA substrates. Normally, in the event of a spontaneous defect in 3′ end formation, a defective transcript will be recognized and degraded by the NMD machinery. When aberrant mRNA formation becomes more frequent, for example following a mutation in critical motifs within the terminator sequence, these NMD degradation pathways may become inadequate, leading to a build up of aberrant RNAs in the cell. This, according to the threshold model, would trigger RNA silencing, leading to the degradation of all transcripts homologous to the defective transcripts. This model posits that RNA decay is the front line RNA quality control pathway, and that RNA silencing may be induced only when the capacity of the NMD pathways become saturated.
For plant biotechnologists, this could present an opportunity to limit the undesirable effects of transgene silencing. According to the threshold model of silencing induction, by intentionally targeting NMD against transcripts expressed from a transgenic locus, the incidence of RNA silencing could be reduced (Figure 2). Partial triggering of NMD may be achieved by exploiting the nature of translational initiation, which in eukaryotes, occurs mainly at the 5′ end of an mRNA, with ribosomes generally finding the start codon (i.e., AUG) via a scanning mechanism (Kozak, 1986, 2002). Inclusion of PTCs in small upstream open reading frames (uORFs) can decrease but does not prevent translation of the main open reading frame (Wang and Rothnagel, 2004). Re-initiation can still occur when the ribosome has completed translation of the uORF but has not completely disassociated from the transcript and continues to translate the major downstream open reading frame (Wang and Rothnagel, 2004). Inclusion of uORFs, terminating with a PTC upstream of the main AUG, could therefore induce NMD but still allow sufficient translation re-initiation on the main coding sequence of the transgene. Interestingly, ~27% of endogenous Arabidopsis mRNAs contain one or more uORFs within their 5′ UTRs (Kawaguchi and Bailey-Serres, 2005). In plant viruses, leaky ribosome scanning associated with uORFs and translation re-initiation are used for the production of multiple separately initiated proteins from a single mRNA (Ryabova et al., 2006). Another possible role for these uORFs in viral transcripts, and indeed one that has not been investigated, is to trigger NMD and consequently limit viral RNA levels below the threshold limits of the host’s RNA silencing machinery. The use of such decoy PTCs could enhance NMD and permit the expression of transgenes at levels below the threshold that would induce gene silencing.
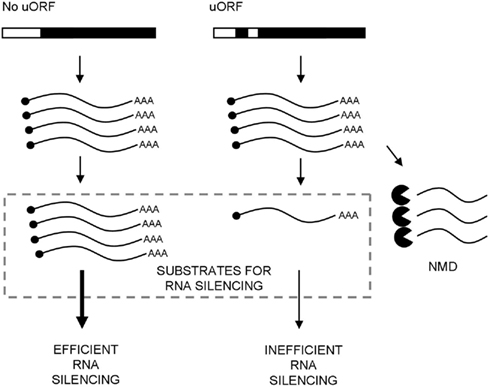
Figure 2. Model for the relationship between nonsense-mediated decay (NMD) and induction of RNA silencing in plants. The gene on the left hand side expresses a single open reading frame (filled), while the gene on the right hand side has an additional small upstream ORF (uORF; filled) in the 5′ UTR (open) that causes premature termination by the scanning ribosome. Transcripts containing uORFs may therefore be partially subject to NMD and reduce the pool of RNA molecules that would otherwise contribute the threshold-dependent induction of RNA silencing.
An increasing number of studies are demonstrating a strong integration between RNA processing and RNA silencing pathways in plants, suggesting that an unforeseen role for RNA silencing may be in mRNA quality control (Chen, 2008). Additionally, a growing overlap is being uncovered between RNA silencing and RNA decay pathways. Perhaps the most exciting possibility not yet reported in plants, is the existence of small RNA-guided exonucleolytic removal of targeted RNAs. siRNA- or miRNA-directed exonucleolytic decay would avoid the generation of 5′ unpolydenylated and 3′ de-capped fragments that would otherwise become substrates to RDR-dependent gene silencing. As such, this form of silencing might limit the activity of RDRs, and therefore allow for subtle regulation of endogenous mRNA levels. miRNA-directed decay has already been demonstrated to account for most of the translational repression observed in mammals (Guo et al., 2010), but similar experiments have not yet been performed to determine whether the same is true for the translational repression observed in plants (Brodersen et al., 2008; Lanet et al., 2009). The involvement of VCS, a protein demonstrated to have decapping activity, in translational repression hints of this possibility, especially when one considers that decapping usually takes place following deadenylation (Brodersen et al., 2008). The existence of both translational repression and RNA decay may be resolved by a kinetic model in which the formation of translationally repressed RNAs precedes mRNA decapping and subsequent degradation (Franks and Lykke-Andersen, 2008; Xu and Chua, 2009). From the work presented in this review, it is clear that while much is known about the RNA decay and RNA silencing pathways of plants, there is a distinct possibility of further overlap between the two pathways that remains to be uncovered. This area of investigation could provide more important discoveries into the mechanisms of small RNA-directed gene regulation in plants.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Allmang, C., Kufel, J., Chanfreau, G., Mitchell, P., Petfalski, E., and Tollervey, D. (1999). Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J. 18, 5399–5410.
Allmang, C., Mitchell, P., Petfalski, E., and Tollervey, D. (2000). Degradation of ribosomal RNA precursors by the exosome. Nucleic Acids Res. 28, 1684–1691.
Amrani, N., Ganesan, R., Kervestin, S., Mangus, D. A., Ghosh, S., and Jacobson, A. (2004). A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature 432, 112–118.
Anderson, J. T., and Wang, X. (2009). Nuclear RNA surveillance: no sign of substrates tailing off. Crit. Rev. Biochem. Mol. Biol. 44, 16–24.
Arciga-Reyes, L., Wootton, L., Kieffer, M., and Davies, B. (2006). UPF1 is required for nonsense-mediated mRNA decay (NMD) and RNAi in Arabidopsis. Plant J. 47, 480–489.
Bagga, S., Bracht, J., Hunter, S., Massirer, K., Holtz, J., Eachus, R., and Pasquinelli, A. E. (2005). Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell 122, 553–563.
Behm-Ansmant, I., Gatfield, D., Rehwinkel, J., Hilgers, V., and Izaurralde, E. (2007). A conserved role for cytoplasmic poly(A)-binding protein 1 (PABPC1) in nonsense-mediated mRNA decay. EMBO J. 26, 1591–1601.
Behm-Ansmant, I., Rehwinkel, J., Doerks, T., Stark, A., Bork, P., and Izaurralde, E. (2006). MRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 20, 1885–1898.
Belgrader, P., Cheng, J., Zhou, X. B., Stephenson, L. S., and Maquat, L. E. (1994). Mammalian nonsense codons can be cis effectors of nuclear mRNA half-life. Mol. Cell. Biol. 14, 8219–8228.
Belostotsky, D. (2009). Exosome complex and pervasive transcription in eukaryotic genomes. Curr. Opin. Cell Biol. 21, 352–358.
Bousquet-Antonelli, C., Presutti, C., and Tollervey, D. (2000). Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell 102, 765–775.
Brodersen, P., Sakvarelidze-Achard, L., Bruun-Rasmussen, M., Dunoyer, P., Yamamoto, Y. Y., Sieburth, L., and Voinnet, O. (2008). Widespread translational inhibition by plant miRNAs and siRNAs. Science 320, 1185–1190.
Buhler, M., Paillusson, A., and Muhlemann, O. (2004). Efficient downregulation of immunoglobulin mu mRNA with premature translation-termination codons requires the 5′-half of the VDJ exon. Nucleic Acids Res. 32, 3304–3315.
Carter, M. S., Li, S. L., and Wilkinson, M. F. (1996). A splicing-dependent regulatory mechanism that detects translation signals. EMBO J. 15, 5965–5975.
Chan, M. T., and Yu, S. M. (1998). The 3′ untranslated region of a rice alpha-amylase gene functions as a sugar-dependent mRNA stability determinant. Proc. Natl. Acad. Sci. U.S.A. 95, 6543–6547.
Chang, Y. F., Imam, J. S., and Wilkinson, M. E. (2007). The nonsense-mediated decay RNA surveillance pathway. Annu. Rev. Biochem. 76, 51–74.
Chekanova, J. A., Gregory, B. D., Reverdatto, S. V., Chen, H., Kumar, R., Hooker, T., Yazaki, J., Li, P., Skiba, N., Peng, Q., Alonso, J., Brukin, V., Grossniklaus, U., Ecker, J., and Belostotsky, D. A. (2007). Genome-wide high-resolution mapping of exosome substrates reveals hidden features in the Arabidopsis transcriptome. Cell 131, 1340–1353.
Chen, X. (2008). A silencing safeguard: links between RNA silencing and mRNA processing in Arabidopsis. Dev. Cell 14, 811–812.
Dalmay, T., Hamilton, A., Rudd, S., Angell, S., and Baulcombe, D. C. (2000). An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell 101, 543–553.
Domeier, M. E., Morse, D. P., Knight, S. W., Portereiko, M., Bass, B. L., and Mango, S. E. (2000). A link between RNA interference and nonsense-mediated decay in Caenorhabditis elegans. Science 289, 1928–1930.
Eberle, A. B., Lykke-Andersen, S., Muhlemann, O., and Jensen, T. H. (2009). SMG6 promotes endonucleolytic cleavage of nonsense mRNA in human cells. Nat. Struct. Mol. Biol. 16, 49–55.
Eckner, R., Ellmeier, W., and Birnstiel, M. L. (1991). Mature mRNA 3′ end formation stimulates RNA export from the nucleus. EMBO J. 10, 3513–3522.
Elmayan, T., and Vaucheret, H. (1996). Expression of single copies of a strongly expressed 35S transgene can be silenced post-transcriptionally. Plant J. 9, 787–797.
English, J. J., Mueller, E., and Baulcombe, D. C. (1996). Suppression of virus accumulation in transgenic plants exhibiting silencing of nuclear genes. Plant Cell 8, 179–188.
Eulalio, A., Huntzinger, E., Nishihara, T., Rehwinkel, J., Fauser, M., and Izaurralde, E. (2009). Deadenylation is a widespread effect of miRNA regulation. RNA 15, 21–32.
Franks, T. M., and Lykke-Andersen, J. (2008). The control of mRNA decapping and P-body formation. Mol. Cell 32, 605–615.
Galy, V., Gadal, O., Fromont-Racine, M., Romano, A., Jacquier, A., and Nehrbass, U. (2004). Nuclear retention of unspliced mRNAs in yeast is mediated by perinuclear Mlp1. Cell 116, 63–73.
Gatfield, D., and Izaurralde, E. (2004). Nonsense-mediated messenger RNA decay is initiated by endonucleolytic cleavage in Drosophila. Nature 429, 575–578.
Gazzani, S., Lawrenson, T., Woodward, C., Headon, D., and Sablowski, R. (2004). A link between rnRNA turnover and RNA interference in Arabidopsis. Science 306, 1046–1048.
Guo, H. L., Ingolia, N. T., Weissman, J. S., and Bartel, D. P. (2010). Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466, U835–U866.
Gy, I., Gasciolli, V., Lauressergues, D., Morel, J. B., Gombert, J., Proux, F., Proux, C., Vaucheret, H., and Mallory, A. C. (2007). Arabidopsis FIERY1, XRN2, and XRN3 are endogenous RNA silencing suppressors. Plant Cell 19, 3451–3461.
Hayden, C. A., and Jorgensen, R. A. (2007). Identification of novel conserved peptide uORF homology groups in Arabidopsis and rice reveals ancient eukaryotic origin of select groups and preferential association with transcription factor-encoding genes. BMC Biol. 5, 32. doi:10.1186/1741-7007-5-32
He, F., Peltz, S. W., Donahue, J. L., Rosbash, M., and Jacobson, A. (1993). Stabilization and ribosome association of unspliced pre-mRNAs in a yeast upfl mutant. Proc. Natl. Acad. Sci. U.S.A. 90, 7034–7038.
Herr, A. J., Molnar, A., Jones, A., and Baulcombe, D. C. (2006). Defective RNA processing enhances RNA silencing and influences flowering of Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 103, 14994–15001.
Hilleren, P., McCarthy, T., Rosbash, M., Parker, R., and Jensen, T. H. (2001). Quality control of mRNA 3′-end processing is linked to the nuclear exosome. Nature 413, 538–542.
Hoffer, P., Ivashuta, S., Pontes, O., Vitins, A., Pikaard, C., Mroczka, A., Wagner, N., and Voelker, T. (2011). Posttranscriptional gene silencing in nuclei. Proc. Natl. Acad. Sci. U.S.A. 108, 409–414.
Hosoda, N., Kim, Y. K., Lejeune, F., and Maquat, L. E. (2005). CBP80 promotes interaction of Upf1 with Upf2 during nonsense-mediated mRNA decay in mammalian cells. Nat. Struct. Mol. Biol. 12, 893–901.
Ishigaki, Y., Li, X. J., Serin, G., and Maquat, L. E. (2001). Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell 106, 607–617.
Jaillon, O., Bouhouche, K., Gout, J. F., Aury, J. M., Noel, B., Saudemont, B., Nowacki, M., Serrano, V., Porcel, B. M., Segurens, B., Le Mouël, A., Lepère, G., Schächter, V., Bétermier, M., Cohen, J., Wincker, P., Sperling, L., Duret, L., and Meyer, E. (2008). Translational control of intron splicing in eukaryotes. Nature 451, 359–362.
Jin, H., Suh, M. R., Han, J., Yeom, K. H., Lee, Y., Heo, I., Ha, M., Hyun, S., and Kim, V. N. (2009). Human UPF1 participates in small RNA-induced mRNA downregulation. Mol. Cell. Biol. 29, 5789–5799.
Kashima, I., Yamashita, A., Izumi, N., Kataoka, N., Morishita, R., Hoshino, S., Ohno, M., Dreyfuss, G., and Ohno, S. (2006). Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev. 20, 355–367.
Kastenmayer, J. P., and Green, P. J. (2000). Novel features of the XRN-family in Arabidopsis: evidence that AtXRN4, one of several orthologs of nuclear Xrn2p/Rat1p, functions in the cytoplasm. Proc. Natl. Acad. Sci. U.S.A. 97, 13985–13990.
Kawaguchi, R., and Bailey-Serres, J. (2005). mRNA sequence features that contribute to translational regulation in Arabidopsis. Nucleic Acids Res. 33, 955–965.
Kawashima, T., Pellegrini, M., and Chanfreau, G. F. (2009). Nonsense-mediated mRNA decay mutes the splicing defects of spliceosome component mutations. RNA 15, 2236–2247.
Kerenyi, Z., Merai, Z., Hiripi, L., Benkovics, A., Gyula, P., Lacomme, C., Barta, E., Nagy, F., and Silhavy, D. (2008). Inter-kingdom conservation of mechanism of nonsense-mediated mRNA decay. EMBO J. 27, 1585–1595.
Kertesz, S., Kerenyi, Z., Merai, Z., Bartos, I., Palfy, T., Barta, E., and Silhavy, D. (2006). Both introns and long 3′-UTRs operate as cis-acting elements to trigger nonsense-mediated decay in plants. Nucleic Acids Res. 34, 6147–6157.
Kim, S., Koroleva, O. A., Lewandowska, D., Pendle, A. F., Clark, G. P., Simpson, C. G., Shaw, P. J., and Brown, J. W. S. (2009). Aberrant mRNA transcripts and the nonsense-mediated decay proteins UPF2 and UPF3 are enriched in the Arabidopsis nucleolus. Plant Cell 21, 2045–2057.
Kozak, M. (1986). Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell 44, 283–292.
Kozak, M. (2002). Pushing the limits of the scanning mechanism for initiation of translation. Gene 299, 1–34.
Kugler, W., Enssle, J., Hentze, M. W., and Kulozik, A. E. (1995). Nuclear degradation of nonsense mutated beta-globin mRNA – a posttranscriptional mechanism to protect heterozygotes from severe clinical manifestations of beta-thalassemia. Nucleic Acids Res. 23, 413–418.
Kurihara, Y., Matsui, A., Hanada, K., Kawashima, M., Ishida, J., Morosawa, T., Tanaka, M., Kaminuma, E., Mochizuki, Y., Matsushima, A., Toyoda, T., Shinozaki, K., and Seki, M. (2009). Genome-wide suppression of aberrant mRNA-like noncoding RNAs by NMD in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 106, 2453–2458.
Lanet, E., Delannoy, E., Sormani, R., Floris, M., Brodersen, P., Crete, P., Voinnet, O., and Robaglia, C. (2009). Biochemical evidence for translational repression by Arabidopsis microRNAs. Plant Cell 21, 1762–1768.
Lange, H., Sement, F. M., Canaday, J., and Gagliardi, D. (2009). Polyadenylation-assisted RNA degradation processes in plants. Trends Plant Sci. 14, 497–504.
LeBlanc, J. J., and Beemon, K. L. (2004). Unspliced Rous sarcoma virus genomic RNAs are translated and subjected to nonsense-mediated mRNA decay before packaging. J. Virol. 78, 5139–5146.
Legrain, P., and Rosbash, M. (1989). Some cis-acting and trans-acting mutants for splicing target pre-mRNA to the cytoplasm. Cell 57, 573–583.
Lejeune, F., Ishigaki, Y., Li, X. J., and Maquat, L. E. (2002). The exon junction complex is detected on CBP80-bound but not eIF4E-bound mRNA in mammalian cells: dynamics of mRNP remodeling. EMBO J. 21, 3536–3545.
Libri, D., Dower, K., Boulay, J., Thomsen, R., Rosbash, M., and Jensen, T. H. (2002). Interactions between mRNA export commitment, 3′-end quality control, and nuclear degradation. Mol. Cell. Biol. 22, 8254–8266.
Luo, Z., and Chen, Z. (2007). Improperly terminated, unpolyadenylated mRNA of sense transgenes is targeted by RDR6-mediated RNA silencing in Arabidopsis. Plant Cell 19, 943–958.
Lykke-Andersen, J., Shu, M.-D., and Steitz, J. A. (2000). Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell 103, 1121–1131.
Mourrain, P., Beclin, C., Elmayan, T., Feuerbach, F., Godon, C., Morel, J. B., Jouette, D., Lacombe, A. M., Nikic, S., Picault, N., Rémoué, K., Sanial, M., Vo, T. A., and Vaucheret, H. (2000). Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell 101, 533–542.
Napoli, C., Lemieux, C., and Jorgensen, R. (1990). Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant Cell 2, 279–289.
Orban, T. I., and Izaurralde, E. (2005). Decay of mRNAs targeted by RISC requires XRN1, the Ski complex, and the exosome. RNA 11, 459–469.
Page, M. F., Carr, B., Anders, K. R., Grimson, A., and Anderson, P. (1999). SMG-2 is a phosphorylated protein required for mRNA surveillance in Caenorhabditis elegans and related to Upf1p of yeast. Mol. Cell. Biol. 19, 5943–5951.
Pomeranz, M. C., Hah, C., Lin, P. C., Kang, S. G., Finer, J. J., Blackshear, P. J., and Jang, J. C. (2010). The Arabidopsis tandem zinc finger protein AtTZF1 traffics between the nucleus and cytoplasmic foci and binds both DNA and RNA. Plant Physiol. 152, 151–165.
Pontes, O., and Pikaard, C. S. (2008). siRNA and miRNA processing: new functions for Cajal bodies. Curr. Opin. Genet. Dev. 18, 197–203.
Rain, J. C., and Legrain, P. (1997). In vivo commitment to splicing in yeast involves the nucleotide upstream from the branch site conserved sequence and the Mud2 protein. EMBO J. 16, 1759–1771.
Rajavel, K. S., and Neufeld, E. F. (2001). Nonsense-mediated decay of human HEXA mRNA. Mol. Cell. Biol. 21, 5512–5519.
Rebbapragada, I., and Lykke-Andersen, J. (2009). Execution of nonsense-mediated mRNA decay: what defines a substrate? Curr. Opin. Cell Biol. 21, 394–402.
Rehwinkel, J., Letunic, I., Raes, J., Bork, P., and Izaurralde, E. (2005). Nonsense-mediated mRNA decay factors act in concert to regulate common mRNA targets. RNA 11, 1530–1544.
Ryabova, L. A., Pooggin, M. M., and Hohn, T. (2006). Translation reinitiation and leaky scanning in plant viruses. Virus Res. 119, 52–62.
Sayani, S., Janis, M., Lee, C. Y., Toesca, I., and Chanfreau, G. F. (2008). Widespread impact of nonsense-mediated mRNA decay on the yeast intronome. Mol. Cell 31, 360–370.
Smith, H. A., Swaney, S. L., Parks, T. D., Wernsman, E. A., and Dougherty, W. G. (1994). Transgenic plant-virus resistance mediated by untranslatable sense RNAs – expression, regulation, and fate of nonessential RNAs. Plant Cell 6, 1441–1453.
Smith, N. A., Singh, S. P., Wang, M. B., Stoutjesdijk, P. A., Green, A. G., and Waterhouse, P. M. (2000). Gene expression – total silencing by intron-spliced hairpin RNAs. Nature 407, 319–320.
Souret, F. F., Kastenmayer, J. P., and Green, P. J. (2004). AtXRN4 degrades mRNA in Arabidopsis and its substrates include selected miRNA targets. Mol. Cell 15, 173–183.
Urlaub, G., Mitchell, P. J., Ciudad, C. J., and Chasin, L. A. (1989). Nonsense mutations in the dihydrofolate-reductase gene affect RNA processing. Mol. Cell. Biol. 9, 2868–2880.
Wang, X. Q., and Rothnagel, J. A. (2004). 5′-Untranslated regions with multiple upstream AUG codons can support low-level translation via leaky scanning and reinitiation. Nucleic Acids Res. 32, 1382–1391.
Waterhouse, P. M., Graham, H. W., and Wang, M. B. (1998). Virus resistance and gene silencing in plants can be induced by simultaneous expression of sense and antisense RNA. Proc. Natl. Acad. Sci. U.S.A. 95, 13959–13964.
Xu, J., and Chua, N. H. (2009). Arabidopsis decapping 5 is required for mRNA decapping, P-body formation, and translational repression during postembryonic development. Plant Cell 21, 3270–3279.
Xu, J., and Chua, N. H. (2011). Processing bodies and plant development. Curr. Opin. Plant Biol. 14, 88–93.
Yamashita, A., Ohnishi, T., Kashima, I., Taya, Y., and Ohno, S. (2001). Human SMG-1, a novel phosphatidylinositol 3-kinase-related protein kinase, associates with components of the mRNA surveillance complex and is involved in the regulation of nonsense-mediated mRNA decay. Genes Dev. 15, 2215–2228.
Yoine, M., Ohto, M. A., Onai, K., Mita, S., and Nakamura, K. (2006). The lba1 mutation of UPF1 RNA helicase involved in nonsense-mediated mRNA decay causes pleiotropic phenotypic changes and altered sugar signalling in Arabidopsis. Plant J. 47, 49–62.
Keywords: RNA silencing, RNA decay, exoribonuclease, aberrant RNA
Citation: Christie M, Brosnan CA, Rothnagel JA and Carroll BJ (2011) RNA decay and RNA silencing in plants: competition or collaboration? Front. Plant Sci. 2:99. doi: 10.3389/fpls.2011.00099
Received: 09 August 2011; Accepted: 30 November 2011;
Published online: 22 December 2011.
Edited by:
Richard Jorgensen, LANGEBIO, MexicoReviewed by:
Celine A. Hayden, Applied Biotechnology Institute, USACopyright: © 2011 Christie, Brosnan, Rothnagel and Carroll. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Bernard J. Carroll, School of Chemistry and Molecular Biosciences, The University of Queensland, Cooper Road, St Lucia, QLD 4072, Australia. e-mail:Yi5jYXJyb2xsQHVxLmVkdS5hdQ==
†Present address: Christopher A. Brosnan, Department of Biology, Swiss Federal Institute of Technology, Zurich, Switzerland.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.