- Center for Plant Molecular Biology, University of Tübingen, Tübingen, Germany
14-3-3 Dimers are well known to interact with diverse target proteins throughout eukaryotes. Most notably, association of 14-3-3s commonly requires phosphorylation of a serine or threonine residue within a specific sequence motif of the client protein. Studies with a focus on individual target proteins have unequivocally demonstrated 14-3-3s to be the crucial factors modifying the client’s activity state upon phosphorylation and, thus, finishing the job initiated by a kinase. In this respect, a recent in-depth analysis of the rice transcription factor FLOWERING LOCUS D1 (OsFD1) revealed 14-3-3s to be essential players in floral induction. Such fascinating discoveries, however, can often be ascribed to the random identification of 14-3-3 as an interaction partner of the favorite protein. In contrast, our understanding of 14-3-3 function in higher organisms is frustratingly limited, mainly due to an overwhelming spectrum of putative targets in combination with the existence of a multigene 14-3-3 family. In this review we will discuss our current understanding of the function of plant 14-3-3 proteins, taking into account recent surveys of the Arabidopsis 14-3-3 interactome.
14-3-3 Proteins mediate the effects of certain protein kinases through their ability to bind well-defined phosphoserine or phosphothreonine containing peptide motifs (Muslin et al., 1996; Yaffe et al., 1997; Johnson et al., 2010), the coordination of which finally results in modification of the activity, stability, subcellular localization or interaction capability of the client protein (reviewed in Chevalier et al., 2009; Gökirmak et al., 2010). The fact that 14-3-3s form clamp-like dimers with each monomer capable of binding a phosphopeptide within an amphipathic groove (see Figure 1) immediately suggests that 14-3-3s function as an intermolecular bridge linking two different phosphoproteins. However, evidence that 14-3-3s act as adapters is surprisingly limited. More commonly, a 14-3-3 dimer seems to engage with two tandem phosphorylated sites in the same protein (Johnson et al., 2010).
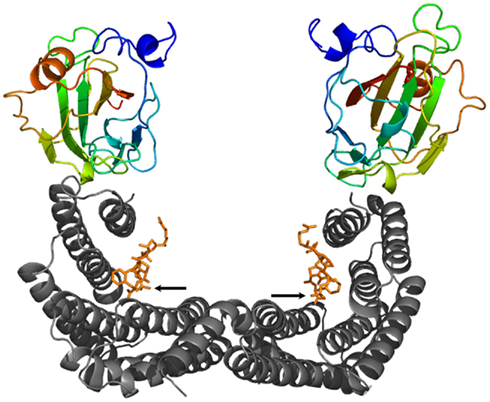
Figure 1. Ribbon plot of a 14-3-3 dimer (gray) in complex with two phosphopeptides derived from the transcription factor OsFD1 (orange, the phosphorylated residue is indicated by an arrow, respectively) and two Hd3a proteins [color coded from the N-terminus (blue) to the C-terminus (red)] (Taoka et al., 2011). While binding of the OsFD1 phosphopeptides occurs in an extended conformation within the amphipathic groove of each 14-3-3 monomer, the “florigen” Hd3a attaches to the “outside” of the groove, respectively.
Research in recent years has revealed an impressive list of putative 14-3-3 targets in both animals and plants (reviewed in MacKintosh, 2004; Chevalier et al., 2009; Oecking and Jaspert, 2009; Gökirmak et al., 2010; Denison et al., 2011; Johnson et al., 2011). The emerging picture is that 14-3-3s may function as dynamic coordinators of cellular responses to distinct signaling cues, due to their concerted regulation of cellular signaling, organization, and metabolism. In other words, depending on the environmental and cellular stimuli perceived by a cell, a specific subset of 14-3-3 clients become phosphorylated and – upon 14-3-3 association – brings about an integrated cellular response. Since 14-3-3 association normally relies on phosphorylation, such a scenario can in principle be achieved by 14-3-3 dependent regulation of almost all the substrates of particular kinases/phosphatases and/or of the kinases/phosphatases themselves.
Historically, plant 14-3-3s were thought to play a prominent role in primary metabolism (reviewed in Huber et al., 2002; Comparot et al., 2003; Denison et al., 2011) and ion homeostasis. Several recent high-throughput proteomic studies, however, have suggested them to also be key regulatory components in signaling cascades, in particular phytohormone mediated processes. Since some of these 14-3-3 interactions have already been reviewed (Chevalier et al., 2009; Oecking and Jaspert, 2009; Gökirmak et al., 2010; Denison et al., 2011) we will focus on recently identified putative interactors in Arabidopsis that represent the “historical” and “novel” 14-3-3 targets, namely transporters and proteins involved in hormonal signaling.
Do 14-3-3s Fine-Tune Membrane Transport Processes?
While defined K+-channels (Sottocornola et al., 2006; Latz et al., 2007; Voelker et al., 2010) and the plasma membrane localized P-type H+-ATPase are among the “old players” in the Arabidopsis 14-3-3 field (reviewed in Oecking and Jaspert, 2009; Denison et al., 2011), recent studies have suggested a much wider diversity of target proteins involved in membrane transport processes than had previously been realized (Chang et al., 2009; Shin et al., 2011). Table 1 summarizes these analyses, including data obtained via a yeast two-hybrid screen in our lab (Oecking et al., unpublished, see Table A1 in Appendix). There are several exciting aspects that should be highlighted. The (putative) 14-3-3 clients include:
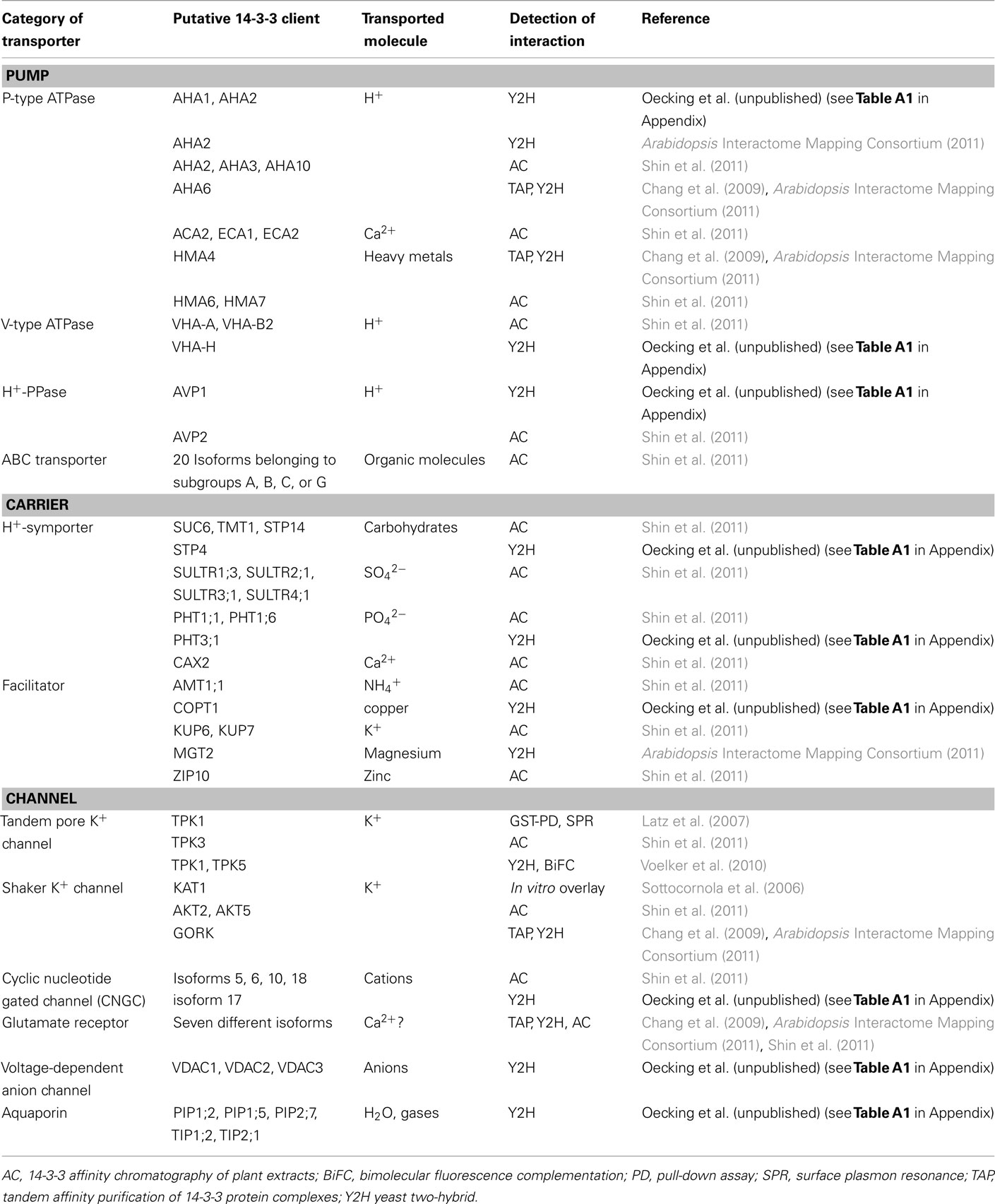
Table 1. A summary of prominent membrane transporters recently identified as putative Arabidopsis 14-3-3 interactors.
(i) All three proton-pumps found in plant cells [i.e., P-type Arabidopsis H+-ATPase (AHA), V-type H+-ATPase (VHA), and the H+-pyrophosphatase (H+-PPase)]. These primary active transporters establish proton gradients across plasma and endosomal membranes, thus enabling secondary active transport processes and/or endosomal acidification (Gaxiola et al., 2007). This suggests 14-3-3s to be of central importance for the interdependent posttranslational regulation of the cellular complement of proton-pumps. However, with the exception of the P-type H+-ATPase which is activated by 14-3-3s (reviewed in Duby and Boutry, 2009), the functional consequences of their interaction with the remaining proton-pumps are not yet known. Furthermore, the major role of the H+-PPase AVP1 has recently been shown to rely on removal of inhibitory cytosolic PPi rather than acidification of the vacuole (Ferjani et al., 2011).
(ii) Other members of the P-type ATPase family (reviewed in Axelsen and Palmgren, 2001) that transport calcium (ACA or ECA, reviewed in Geisler et al., 2000; Bonza and De Michelis, 2011) or metals (HMA, reviewed in Krämer et al., 2007) and are, thus, essential for cellular ion homeostasis.
(iii) Almost 20 pumps belonging to the ATP-binding cassette (ABC) family (reviewed in Martinoia et al., 2002), the members of which can carry large organic molecules, including the phytohormone auxin.
(iv) Several carriers – some of which act as proton symporters [see (i)] – that permit the uptake of nutrients, such as phosphate, sulfate, carbohydrates, and ammonium.
(v) A multitude of ion channels. Besides the “old players” mediating potassium transport, some poorly selective cation channels (cyclic nucleotide gated channel, CNGC), glutamate receptors which are implicated in cellular Ca2+ homeostasis [see (ii)] as well as voltage-dependent anion-selective channels (VDAC) may be subject to 14-3-3 regulation.
(vi) Finally some aquaporins (reviewed in Maurel et al., 2008) localized either in the plasma membrane (PIP) or the tonoplast (TIP) and known to facilitate the transport of water and/or gases, amongst others.
Taken together, this suggests that 14-3-3 proteins exert an impressively widespread influence on membrane transport processes in plants. Since plants are experts in maintaining optimal metabolic conditions under numerous environmental constraints, 14-3-3s may be the key to a tight and coordinated regulation of transporters that are directly or indirectly involved in ion and nutrient transport. Nevertheless, the number and diversity of these putative 14-3-3 clients are also somewhat intimidating and give rise to numerous questions. Which signals regulate which targets and which kinases are involved? Which mechanisms could elicit a concerted regulation of transporters whose interplay would make sense, such as proton-pumps and secondary active transporters? As the list of 14-3-3 clients is continually growing, the question whether the cellular “14-3-3 pool” is limiting under certain circumstances is becoming more important. However, it should be noted that, in the majority of cases, a mere handful of transporters has been identified. Indeed, the bulk of the above-mentioned clients was discovered by one study (Shin et al., 2011), suggesting that the given developmental stage and growth conditions of the plant material used favor 14-3-3 mediated regulation of membrane transporters.
Are 14-3-3s General Regulators of Phytohormone Mediated Processes?
Meanwhile, Arabidopsis 14-3-3 proteins have been proved to be important mediators in signaling cascades (reviewed in Chevalier et al., 2009; Oecking and Jaspert, 2009; Gökirmak et al., 2010; Denison et al., 2011). This breakthrough was achieved by several elegant studies demonstrating 14-3-3s to be essential players in brassinosteroid (BR) signaling due to their modification of the subcellular localization of key transcription factors, such as brassinazole resistant1 (BZR1; Gampala et al., 2007; Ryu et al., 2007) and BRI1 EMS suppressor1 (BES1; Ryu et al., 2010). Currently available Arabidopsis 14-3-3 interactome data moreover suggest 14-3-3s to participate in processes mediated by almost any phytohormone (Chang et al., 2009; Paul et al., 2009; Shin et al., 2011; Swatek et al., 2011; summarized in Table 2 which includes data obtained via a yeast two-hybrid screen in our lab; Oecking et al., unpublished, see Table A1 in Appendix). First of all, not only a multitude of further proteins likewise involved in BR signaling but also polypeptides critical for its biosynthesis have been identified as potential 14-3-3 clients. A comparable scenario emerges with respect to the gaseous hormone ethylene (Table 2). At present, we can only speculate as to why multiple 14-3-3 interactions might be required to control a defined cellular response. One conceivable scenario is that 14-3-3 function as molecular gauges, thereby forcing the cell to react to a given phytohormone according to the signal strength. Taking into account that clients involved in several cellular processes might compete for binding to 14-3-3s, numerous targets within one defined pathway are assumed to channel the overall cellular response.
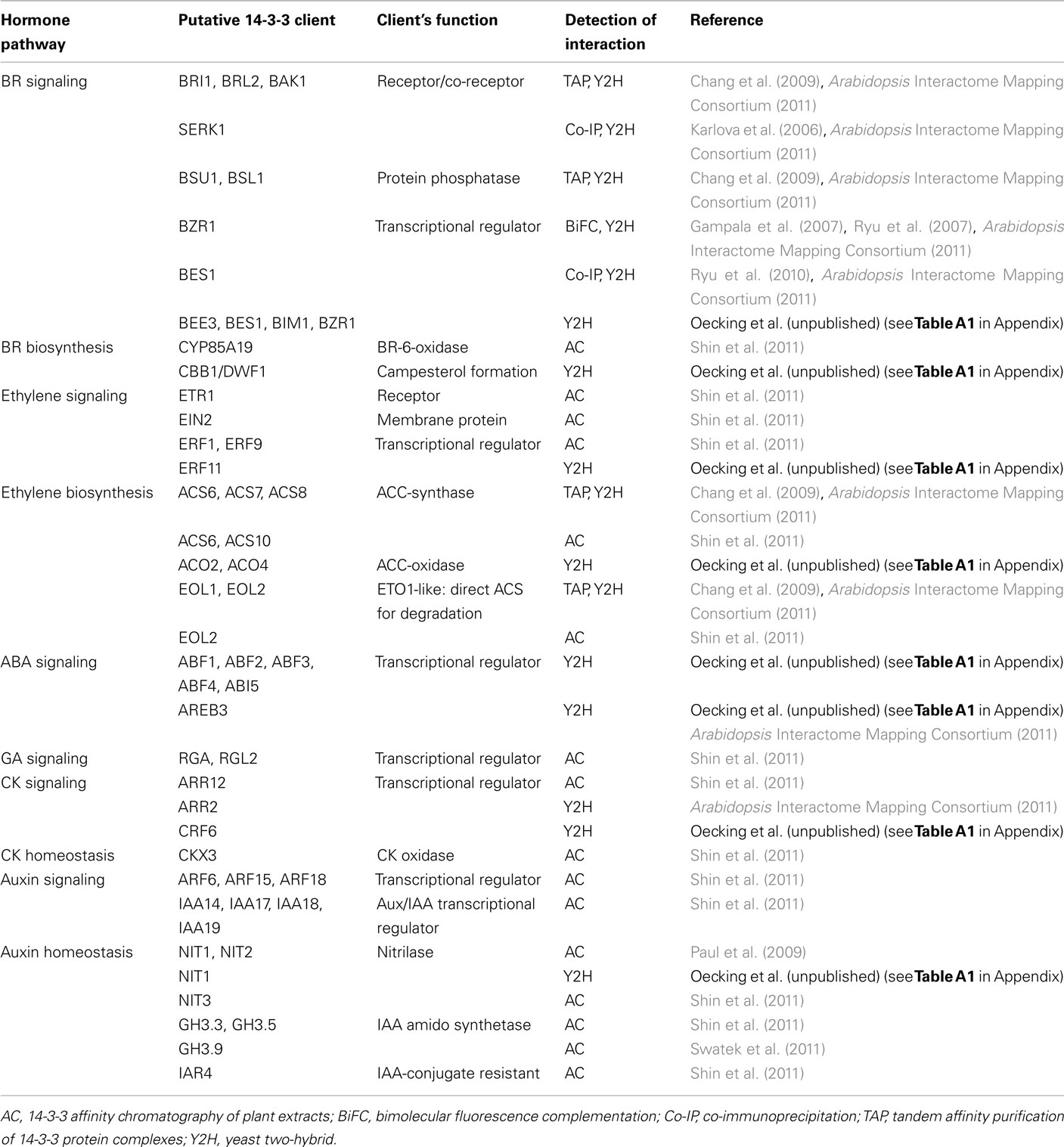
Table 2. A summary of recently identified putative Arabidopsis 14-3-3 interactors involved in phytohormone signaling or biosynthesis.
Remarkably, the modification of transcriptional regulators involved in hormonal signaling seems to be another hotspot in Arabidopsis 14-3-3 biology. In this regard, several prominent proteins, which act as either repressors or activators of gene expression in BR (BZR1, BES1, BIM1), ethylene [ethylene responsive factor (ERF)], auxin [auxin/indole-3-acetic acid proteins (Aux/IAA), auxin response transcription factor (ARF)], gibberellin (GA) [repressor of GA1-3 (RGA), RGA-like (RGL)], abscisic acid (ABA) [ABA response element binding factor (ABF)], and cytokinin (CK) [Arabidopsis response regulator (ARR), cytokinin response factor (CRF)] responses, are among the putative 14-3-3 clients in Arabidopsis (Table 2). In contrast to BZR1/BES1, which are retained in the cytosol upon 14-3-3 association (Gampala et al., 2007; Ryu et al., 2007, 2010), the members of the ABF subfamily of BASIC REGION/LEUCINE ZIPPER (bZIP) transcription factors constitutively localize to the nucleus, as shown for ABA INSENSITIVE 5 (ABI5; Lopez-Molina et al., 2002), ABF3 (Sirichandra et al., 2010) and the rice homolog OREB1 (Hong et al., 2011).
In this respect, 14-3-3s have been proposed to be critical for the stability of the ABF3 protein, the proteasomal turnover rate of which is high in the absence of ABA and 14-3-3s (Sirichandra et al., 2010). Even though 14-3-3 dependent modification of transcriptional regulators mediating ethylene, auxin, GA, and CK responses has yet to be verified, 14-3-3s might in summary be far more important for assuring the developmental plasticity of plants than has yet been assumed. The fact that more and more proteins are identified as putative targets moreover implies that 14-3-3 interactions are highly dynamic which would enable the cellular 14-3-3 “pool” to immediately and precisely react to altered signaling cues.
14-3-3s are Critical for Floral Transition in Rice
While global studies are essential to get an impression of the extent of the plant 14-3-3 interactome, the necessity to address the biological significance of individual interactions is more pressing than ever. In this respect, a recent study elegantly combining biochemistry, cell biology, and genetics has impressively proved 14-3-3s to be essential components of the florigen activation complex (FCA) that promotes flowering in the short day plant rice (Taoka et al., 2011). The term “florigen” was created in 1936 and refers to a molecule that is generated in leaves under inductive photoperiodic conditions and subsequently transported to the shoot apex (Chailakhyan, 1936). Evidence could only recently be provided that the protein flowering locus T (FT) represents such a long-distance signal in the facultative long day plant Arabidopsis (Mathieu et al., 2007). In the shoot apex, a complex of FT and the bZIP transcription factor flowering locus D (FD) initiates floral development through transcriptional activation of floral identity genes (Abe et al., 2005; Wigge et al., 2005).
Using the rice FT homolog Hd3a as a bait in a yeast two-hybrid screen, (Taoka et al., 2011) identified rice 14-3-3 isoforms as well as OsFD1, a rice homolog of the Arabidopsis FD, as putative binding partners. The initially astonishing observation that Hd3a is unable to interact directly with OsFD1 gave rise to subsequent in-depth analyses demonstrating 14-3-3s to mediate this interaction. Remarkably, the binding sites in 14-3-3 for Hd3a and OsFD1 are separated, indicating that the two partners bind in different manner. OsFD1 represents the typical 14-3-3 target, in that its coordination within the typical groove of a 14-3-3 monomer depends on phosphorylation. However, in contrast with most physiological targets, Hd3a does not have to be phosphorylated in order to associate with 14-3-3s. The obtained crystal structure of the Hd3a:14-3-3 complex revealed a fascinating difference as compared to the canonical 14-3-3 interactions: an unphosphorylated Hd3a monomer binds “on top” of each 14-3-3 monomer, thus extending the W-shaped structure of the 14-3-3 dimer (Taoka et al., 2011; Figure 1). Crystal soaking in the presence of a phosphorylated OsFD1 peptide representing the 14-3-3 binding site finally allowed the modeling of the FCA holocomplex composed of 14-3-3, OsFD1 and Hd3a. Notably, even though 14-3-3s mediate the Hd3a:OsFD1 interaction, the two proteins do not come in contact with each other, suggesting 14-3-3s to function as a platform enabling spatial proximity but not direct linkage (Taoka et al., 2011). On the basis of cell biological data Taoka et al. (2011) proposed a model according to which 14-3-3s act as cytoplasmic receptors for the “florigen” Hd3a in the shoot apex. Once this protein couple enters the nucleus, a ternary complex including phosphorylated OsFD1 is built, which in turn is retained in the nucleus and activates transcription of genes crucial for floral induction. Finally, several experiments performed with mutant versions of either Hd3a or OsFD1 that had lost their ability to interact with 14-3-3s have impressively proved the in vivo significance of 14-3-3 association for flowering in rice.
However, based upon the current structural knowledge nearly each 14-3-3 client should be able to associate with the Hd3a:14-3-3 complex. Thus, one crucial question is still unanswered: what determines specificity of the Hd3a:14-3-3 complex to interact exclusively with OsFD?
Minor Knowledge about the Biological Role of Distinct 14-3-3 Isoforms
The above-mentioned studies, demonstrating that 14-3-3s are of the utmost importance for BR signaling and timing of floral transition, were focused on particular 14-3-3 client proteins and have generated substantial insight into the function of plant 14-3-3s. Considering the enormous quantity and functional complexity of the recently identified putative 14-3-3 targets, the subset of which may vary considerably as a function of the developmental and physiological stage, the question as to whether Arabidopsis 14-3-3 mutants can broaden our understanding of 14-3-3 function becomes more and more critical. Arabidopsis expresses thirteen 14-3-3 isoforms that can be divided into two major phylogenetic groups, the epsilon and the non-epsilon group, the latter consisting of three organizational subgroups. Since the epsilon group is considered to harbor living fossil isoforms which may fulfill fundamental eukaryotic functions, non-epsilon members may be responsible for organism-specific regulatory aspects.
The current knowledge about plants characterized by reduced or absent expression of specific 14-3-3 isoforms is limited (see Oecking and Jaspert, 2009; Denison et al., 2011). We identified T-DNA induced loss-of-function alleles of several individual non-epsilon 14-3-3 isoforms, which collectively do not show a statistically significant phenotype under normal growth conditions. The same applies to the simultaneous loss-of-function of two 14-3-3 isoforms (kappa/lambda) constituting a phylogenetic non-epsilon subgroup, suggesting functional redundancy among members of the non-epsilon group at least with respect to fundamental functions (data not shown). Beyond that, although lambda and kappa were the most frequently 14-3-3 isoforms identified in a yeast two-hybrid screen to interact with BZR1, the corresponding double knockout mutants did not display altered BR responses (Gampala et al., 2007). Taken together, redundancy of all non-epsilon isoforms independent of their belonging to different phylogenetic subgroups is likely. Thus, higher order loss-of-function mutants seem to be required to produce a phenotype, which in turn is expected to be pleiotropic and is thus difficult to interpret. Nevertheless, focusing on well-defined developmental stages could provide hints for the dominant and major 14-3-3 targets under the given circumstances.
What about epsilon group members? While many of the recent proteome wide approaches in Arabidopsis have focused on the identification of putative non-epsilon 14-3-3 interactors, only one study compared two phylogenetically distinct isoforms (Swatek et al., 2011). The results suggest not only isoform specificity of several target proteins but also preference for 14-3-3 dimer formation between phylogenetically similar 14-3-3 isoforms. Hence, functional specialization may exist, at least between members of the non-epsilon and epsilon group.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Felicity de Courcy for critical reading of the manuscript. The research in our lab is supported by grants from the German Research Foundation (AFGN).
References
Abe, M., Kobayashi, Y., Yamamoto, S., Daimon, Y., Yamaguchi, A., Ikeda, Y., Ichinoki, H., Notaguchi, M., Goto, K., and Araki, T. (2005). FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309, 1052–1056.
Arabidopsis Interactome Mapping Consortium. (2011). Evidence for network evolution in an Arabidopsis interactome map. Science 333, 601–607.
Axelsen, K. B., and Palmgren, M. G. (2001). Inventory of the superfamily of P-type ion pumps in Arabidopsis. Plant Physiol. 126, 696–706.
Bonza, M. C., and De Michelis, M. I. (2011). The plant Ca2+-ATPase repertoire: biochemical features and physiological functions. Plant Biol. (Stuttg.) 13, 421–430.
Chailakhyan, M. K. (1936). New facts in support of the hormonal theory of plant development. C. R. Acad. Sci. URSS 13, 79–83.
Chang, I. F., Curran, A., Woolsey, R., Quilici, D., Cushman, J. C., Mittler, R., Harmon, A., and Harper, J. F. (2009). Proteomic profiling of tandem affinity purified 14-3-3 protein complexes in Arabidopsis thaliana. Proteomics 9, 2967–2985.
Chevalier, D., Morris, E. R., and Walker, J. C. (2009). 14-3-3 and FHA domains mediate phosphoprotein interactions. Annu. Rev. Plant Biol. 60, 67–91.
Comparot, S., Lingiah, G., and Martin, T. (2003). Function and specificity of 14-3-3 proteins in the regulation of carbohydrate and nitrogen metabolism. J. Exp. Bot. 54, 595–604.
Denison, F. C., Paul, A. L., Zupanska, A. K., and Ferl, R. J. (2011). 14-3-3 proteins in plant physiology. Semin. Cell Dev. Biol. 22, 720–727.
Duby, G., and Boutry, M. (2009). The plant plasma membrane proton pump ATPase: a highly regulated P-type ATPase with multiple physiological roles. Pflugers Arch. 457, 645–655.
Ferjani, A., Segami, S., Horiguchi, G., Muto, Y., Maeshima, M., and Tsukaya, H. (2011). Keep an eye on PPi: the vacuolar-type H+-pyrophosphatase regulates postgerminative development in Arabidopsis. Plant Cell 23, 2895–2908.
Gampala, S. S., Kim, T. W., He, J. X., Tang, W., Deng, Z., Bai, M. Y., Guan, S., Lalonde, S., Sun, Y., Gendron, J. M., Chen, H., Shibagaki, N., Ferl, R. J., Ehrhardt, D., Chong, K., Burlingame, A. L., and Wang, Z. Y. (2007). An essential role for 14-3-3 proteins in brassinosteroid signal transduction in Arabidopsis. Dev. Cell 13, 177–189.
Gaxiola, R. A., Palmgren, M. G., and Schumacher, K. (2007). Plant proton pumps. FEBS Lett. 581, 2204–2214.
Geisler, M., Axelsen, K. B., Harper, J. F., and Palmgren, M. G. (2000). Molecular aspects of higher plant P-type Ca(2+)-ATPases. Biochim. Biophys. Acta 1465, 52–78.
Gökirmak, T., Paul, A. L., and Ferl, R. J. (2010). Plant phosphopeptide-binding proteins as signaling mediators. Curr. Opin. Plant Biol. 13, 527–532.
Hong, J. Y., Chae, M. J., Lee, I. S., Lee, Y. N., Nam, M. H., Kim, D. Y., Byun, M. O., and Yoon, I. S. (2011). Phosphorylation-mediated regulation of a rice ABA responsive element binding factor. Phytochemistry 72, 27–36.
Huber, S. C., Mackintosh, C., and Kaiser, W. M. (2002). Metabolic enzymes as targets for 14-3-3 proteins. Plant Mol. Biol. 50, 1053–1063.
Johnson, C., Crowther, S., Stafford, M. J., Campbell, D. G., Toth, R., and MacKintosh, C. (2010). Bioinformatic and experimental survey of 14-3-3-binding sites. Biochem. J. 427, 69–78.
Johnson, C., Tinti, M., Wood, N. T., Campbell, D. G., Toth, R., Dubois, F., Geraghty, K. M., Wong, B. H., Brown, L. J., Tyler, J., Gernez, A., Chen, S., Synowsky, S., and MacKintosh, C. (2011). Visualization and biochemical analyses of the emerging mammalian 14-3-3-phosphoproteome. Mol. Cell Proteomics 10. doi: 10.1074/mcp.M110.005751.
Karlova, R., Boeren, S., Russinova, E., Aker, J., Vervoort, J., and de Vries, S. (2006). The Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE1 protein complex includes BRASSINOSTEROID-INSENSITIVE1. Plant Cell 18, 626–638.
Kim, J., Harter, K., and Theologis, A. (1997). Protein-protein interactions among the Aux/IAA proteins. Proc. Natl. Acad. Sci. U.S.A. 94, 11786–11791.
Krämer, U., Talke, I. N., and Hanikenne, M. (2007). Transition metal transport. FEBS Lett. 581, 2263–2272.
Latz, A., Becker, D., Hekman, M., Muller, T., Beyhl, D., Marten, I., Eing, C., Fischer, A., Dunkel, M., Bertl, A., Rapp, U. R., and Hedrich, R. (2007). TPK1, a Ca(2+)-regulated Arabidopsis vacuole two-pore K(+) channel is activated by 14-3-3 proteins. Plant J. 52, 449–459.
Lopez-Molina, L., Mongrand, S., Mclachlin, D. T., Chait, B. T., and Chua, N. H. (2002). ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J. 32, 317–328.
MacKintosh, C. (2004). Dynamic interactions between 14-3-3 proteins and phosphoproteins regulate diverse cellular processes. Biochem. J. 381, 329–342.
Martinoia, E., Klein, M., Geisler, M., Bovet, L., Forestier, C., Kolukisaoglu, U., Muller-Rober, B., and Schulz, B. (2002). Multifunctionality of plant ABC transporters – more than just detoxifiers. Planta 214, 345–355.
Mathieu, J., Warthmann, N., Kuttner, F., and Schmid, M. (2007). Export of FT protein from phloem companion cells is sufficient for floral induction in Arabidopsis. Curr. Biol. 17, 1055–1060.
Maurel, C., Verdoucq, L., Luu, D. T., and Santoni, V. (2008). Plant aquaporins: membrane channels with multiple integrated functions. Annu. Rev. Plant Biol. 59, 595–624.
Muslin, A. J., Tanner, J. W., Allen, P. M., and Shaw, A. S. (1996). Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell 84, 889–897.
Oecking, C., and Jaspert, N. (2009). Plant 14-3-3 proteins catch up with their mammalian orthologs. Curr. Opin. Plant Biol. 12, 760–765.
Paul, A. L., Liu, L., Mcclung, S., Laughner, B., Chen, S., and Ferl, R. J. (2009). Comparative interactomics: analysis of Arabidopsis 14-3-3 complexes reveals highly conserved 14-3-3 interactions between humans and plants. J. Proteome Res. 8, 1913–1924.
Ryu, H., Cho, H., Kim, K., and Hwang, I. (2010). Phosphorylation dependent nucleocytoplasmic shuttling of BES1 is a key regulatory event in brassinosteroid signaling. Mol. Cells 29, 283–290.
Ryu, H., Kim, K., Cho, H., Park, J., Choe, S., and Hwang, I. (2007). Nucleocytoplasmic shuttling of BZR1 mediated by phosphorylation is essential in Arabidopsis brassinosteroid signaling. Plant Cell 19, 2749–2762.
Shin, R., Jez, J. M., Basra, A., Zhang, B., and Schachtman, D. P. (2011). 14-3-3 proteins fine-tune plant nutrient metabolism. FEBS Lett. 585, 143–147.
Sirichandra, C., Davanture, M., Turk, B. E., Zivy, M., Valot, B., Leung, J., and Merlot, S. (2010). The Arabidopsis ABA-activated kinase OST1 phosphorylates the bZIP transcription factor ABF3 and creates a 14-3-3 binding site involved in its turnover. PLoS ONE 5, e13935. doi:10.1371/journal.pone.0013935
Sottocornola, B., Visconti, S., Orsi, S., Gazzarrini, S., Giacometti, S., Olivari, C., Camoni, L., Aducci, P., Marra, M., Abenavoli, A., Thiel, G., and Moroni, A. (2006). The potassium channel KAT1 is activated by plant and animal 14-3-3 proteins. J. Biol. Chem. 281, 35735–35741.
Swatek, K. N., Graham, K., Agrawal, G. K., and Thelen, J. J. (2011). The 14-3-3 isoforms chi and epsilon differentially bind client proteins from developing Arabidopsis seed. J. Proteome Res. 10, 4076–4087.
Taoka, K., Ohki, I., Tsuji, H., Furuita, K., Hayashi, K., Yanase, T., Yamaguchi, M., Nakashima, C., Purwestri, Y. A., Tamaki, S., Ogaki, Y., Shimada, C., Nakagawa, A., Kojima, C., and Shimamoto, K. (2011). 14-3-3 Proteins act as intracellular receptors for rice Hd3a florigen. Nature 476, 332–335.
Voelker, C., Gomez-Porras, J. L., Becker, D., Hamamoto, S., Uozumi, N., Gambale, F., Mueller-Roeber, B., Czempinski, K., and Dreyer, I. (2010). Roles of tandem-pore K+ channels in plants – a puzzle still to be solved. Plant Biol. (Stuttg.) 12(Suppl. 1), 56–63.
Wigge, P. A., Kim, M. C., Jaeger, K. E., Busch, W., Schmid, M., Lohmann, J. U., and Weigel, D. (2005). Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309, 1056–1059.
Yaffe, M. B., Rittinger, K., Volinia, S., Caron, P. R., Aitken, A., Leffers, H., Gamblin, S. J., Smerdon, S. J., and Cantley, L. C. (1997). The structural basis for 14-3-3:phosphopeptide binding specificity. Cell 91, 961–971.
Appendix
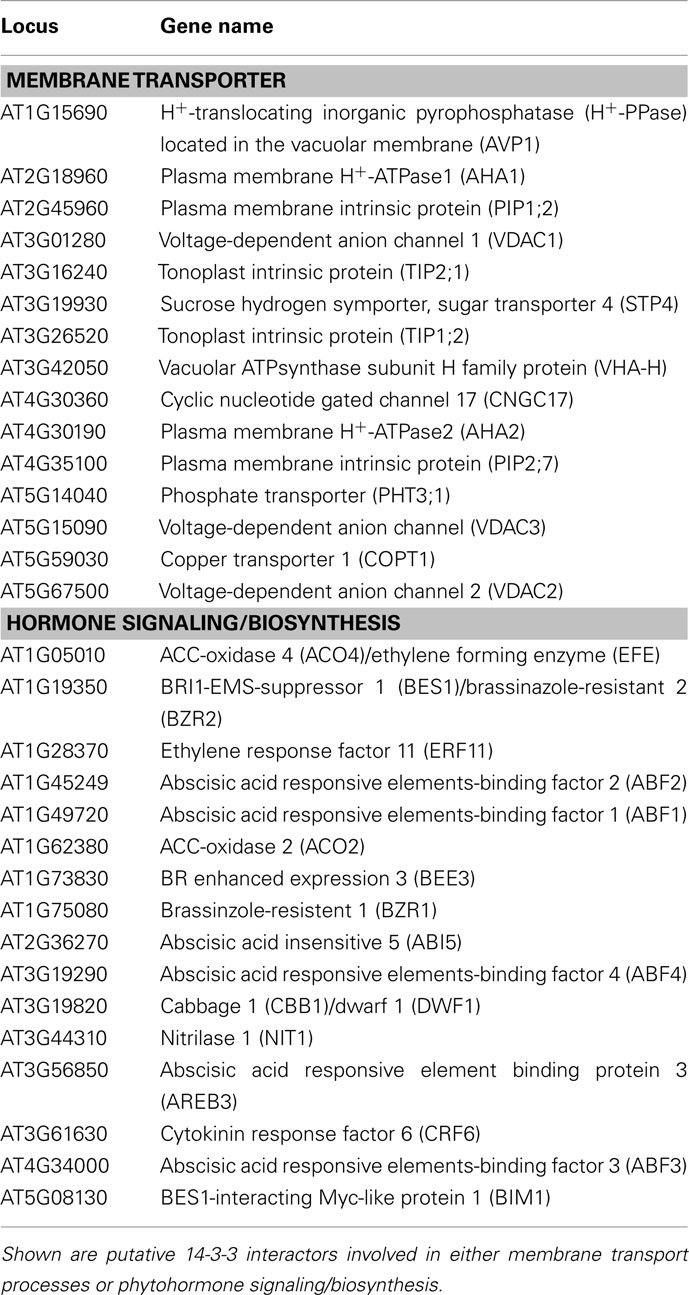
Table A1. Results of a yeast two-hybrid screen using a cDNA library obtained from 3 days old etiolated Arabidopsis seedlings (Kim et al., 1997) and different Arabidopsis 14-3-3 isoforms as a bait.
Keywords: 14-3-3, interactome
Citation: Jaspert N, Throm C and Oecking C (2011) Arabidopsis 14-3-3 proteins: fascinating and less fascinating aspects. Front. Plant Sci. 2:96. doi: 10.3389/fpls.2011.00096
Received: 20 September 2011;
Paper pending published: 21 October 2011;
Accepted: 24 November 2011;
Published online: 09 December 2011.
Edited by:
Andreas P. M. Weber, University of Duesseldorf, GermanyReviewed by:
Jeffrey F. Harper, University of Nevada Reno, USABurkhard Schulz, Purdue University, USA
Copyright: © 2011 Jaspert, Throm and Oecking. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Claudia Oecking, Center for Plant Molecular Biology, University of Tübingen, Auf der Morgenstelle 5, 72076 Tübingen, Germany. e-mail:Y2xhdWRpYS5vZWNraW5nQHptYnAudW5pLXR1ZWJpbmdlbi5kZQ==
 Nina Jaspert
Nina Jaspert