- 1 College of Life Sciences, Northwest A&F University, Yangling, China
- 2 National Key Laboratory of Crop Genetic Improvement, National Center of Plant Gene Research (Wuhan), Huazhong Agricultural University, Wuhan, China
The phytohormone auxin has been known to be a regulator of plant growth and development ever since its discovery. Recent studies on plant–pathogen interactions identify auxin as a key character in pathogenesis and plant defense. Like plants, diverse pathogens possess the capacity to synthesize indole-3-acetic acid (IAA), the major form of auxin in plants. The emerging knowledge on auxin-signaling components, auxin metabolic processes, and indole-derived phytoalexins in plant responses to pathogen invasion has provided putative mechanisms of IAA in plant susceptibility and resistance to non-gall- or tumor-inducing pathogens.
Introduction
Many physiologic processes, including tropisms, root and shoot morphologic architecture, flower and fruit development, vascular formation, tissue differentiation, and cell elongation, are directed primarily by auxin (Davies, 2004). Almost every event in auxin signaling and metabolism, including auxin perception, signaling derepression, biosynthesis, and inactivation is intimately correlated with plant defense or disease development. Indole-3-acetic acid (IAA) is the generally occurring form of auxin in plants, and the compound can also be produced by microbes. Many studies have elucidated how and why phytopathogens like tumor- and gall-inducing species and plant growth-promoting rhizobacteria produce IAA (Liu et al., 1982; Glick, 1995; Patten and Glick, 2002; Barash and Manulis-Sasson, 2007; Reineke et al., 2008; Lee et al., 2009; Apine and Jadhav, 2011). Non-gall- or tumor-inducing phytopathogens occupy a special position among IAA-producing microbes (Fett et al., 1987; Glickmann et al., 1998; Hasan, 2002; Maor et al., 2004; Yang et al., 2007; Fu et al., 2011); however, the details of their IAA-related functional determinants remain ambiguous. This review focuses on the phenomena and the possible mechanisms of auxin in the interactions of plants and non-gall- or tumor-inducing pathogens.
Auxin Performance in Plant–Pathogen Interactions
Auxin behavior in plant–pathogen interactions is complicated. Thus far, most studies are related to its role during pathogenesis. Auxin can promote disease symptoms in many plants. Treatments with the auxin analogs 2,4-dichlorophenoxyacetic acid (2,4-D) or 1-naphthalacetic acid (NAA) enhance disease symptoms in Arabidopsis infected by Pseudomonas syringae pv. tomato (Pto) DC3000 (Navarro et al., 2006; Chen et al., 2007). Pseudomonas syringae pv. maculicola inoculation supplemented with NAA not only increases susceptibility of Arabidopsis to this pathogen but also accelerates its growth (Wang et al., 2007). Exogenous application of IAA or 2,4-D increases the lesion areas of resistance rice lines inoculated with Xanthomonas oryzae pv. oryzae (Xoo; Ding et al., 2008). External IAA treatment aggravates the disease progress caused by Xanthomonas oryzae pv. oryzicola (Xoc) and Magnaporthe oryzae in rice (Fu et al., 2011). Heterologous expression of a fungal polygalacturonase in tobacco or exogenous treatment of tobacco with oligogalacturonide (OG) enhances plant resistance to the necrotrophic fungal pathogen Botrytis cinerea, but pretreatment with IAA recovers the susceptibility (Ferrari et al., 2008). Recently, IAA was found to antagonize OG-induced resistance against B. cinerea in Arabidopsis (Savatin et al., 2011). Auxin transport inhibitor naphthylphthalamic acid can attenuate canker development of sweet orange infected by Xanthomonas axonopodis pv. citri, but NAA can provoke more serious disease symptoms (Cernadas and Benedetti, 2009).
Elevated levels of endogenous plant IAA have been observed during pathogen infection. Arabidopsis showed increased IAA concentration from 48 to 96 h post inoculation with PstDC3000 in comparison with mock-inoculated control plants (Schmelz et al., 2004). A salicylic acid (SA)-deficient NahG line and ethylene-insensitive mutants etr1 and etr2 were used to examine the phytohormone interactions corresponding with Xanthomonas campestris pv. campestris (Xcc) invasion; the results suggested that all three tested materials accumulate IAA independent of either SA or ethylene action (O’Donnell et al., 2003). Susceptible rice plants accumulated more IAA than resistant plants during infection with different pathogens (Ding et al., 2008; Fu et al., 2011).
A few studies have revealed that auxin can enhance disease resistance in Arabidopsis against some necrotrophic pathogens, which will be discussed in the next part of this review.
Auxin Signaling in Plant–Pathogen Interactions
Auxin can be perceived by a family of F-box proteins including transport inhibitor response 1 (TIR1) and auxin signaling F-box proteins 1, 2, and 3 (AFB1, AFB2, and AFB3; Dharmasiri et al., 2005; Kepinski and Leyser, 2005). A bacterial peptide flg22, a pathogen-associated molecular pattern, at a high concentration (10 μM), induces the microRNA miR393 leading to degradation of the mRNAs of auxin receptors TIR1, AFB2, and AFB3, but transcriptional repression of AFB1 is microRNA independent (Navarro et al., 2006). TIR1 is the core member of the ubiquitin–ligase complex SCFTIR1 that degrades transcriptional repressor Aux/IAA and thus releases auxin response factor (ARF) to regulate the transcription of primary auxin-responsive genes after the interaction of TIR1 and auxin (Woodward and Bartel, 2005). The flg22-elicited down-regulation of TIR1 and stabilization of an Aux/IAA protein happen simultaneously, and some primary auxin-responsive genes are suppressed (Navarro et al., 2006). Overexpression of AFB1 in a tir1 mutant background results in enhanced disease sensitivity to virulent Pto DC3000, and constitutive expression of miR393 triggers decreased TIR1 transcript and increased resistance to P. syringae in Arabidopsis (Navarro et al., 2006). According to the results from above-mentioned studies, it could be hypothesized that suppressing auxin signaling at the onset of auxin perception would contribute to disease resistance.
The study on OG–IAA antagonism in Arabidopsis discovered that, when the elicitor flg22 was applied at a lower concentration (1 μM), miR393 expression was unchanged, and the mRNAs of auxin receptors were not decreased; when flg22 was applied at a high concentration (10 μM), it did not stabilize Aux/IAA repressor in the presence or absence of exogenous IAA (Savatin et al., 2011). Thus, it needs to be deliberated whether miR393 is directly responsible for the repressed auxin signaling by flg22.
However, recent studies have also shown that intact auxin signaling is indispensable for resistance against necrotrophic pathogens. Mutations in AXR1, which encodes protein related to auxin signal-mediated ubiquitination of repressor Aux/IAA, cause susceptibility to necrotrophic oomycete Pythium irregulare (Tiryaki and Staswick, 2002). Arabidopsis mutants defective in auxin-stimulated ubiquitination pathway display increased susceptibility to necrotrophic fungi Plectosphaerella cucumerina and B. cinerea; AXR3 is an auxin transcriptional repressor that undergoes degradation by ubiquitination machinery, and heat shock-induced AXR3-GUS protein becomes more stable upon P. cucumerina infection (Llorente et al., 2008). These results suggest that the global down-regulation of auxin response genes observed after virulent necrotrophic B. cinerea or P. cucumerina infections may be caused by reduced degradation of auxin-signaling repressors (Llorente et al., 2008).
Auxin Conjugation in Plant–Pathogen Interactions
Additional evidence for the disease resistance conferred by suppressing auxin signaling comes from auxin conjugation machinery that is mostly controlled by GH3 family proteins (Woodward and Bartel, 2005).
GH3 belongs to a multi-gene family that encodes proteins having acyl acid amido synthetase function (Woodward and Bartel, 2005; Westfall et al., 2010). The first GH3 gene was identified as a rapid early auxin-responsive gene in soybean (Hagen and Guilfoyle, 1985). The transcription of GH3 genes can be regulated by ARFs (Ulmasov et al., 1999). However, not all GH3 genes are auxin-responsive (Woodward and Bartel, 2005).
GH3 proteins are divided into three groups (I to III). The representative of GH3 family group I orthologs, AtGH3.11/FIN219/JAR1, regulates jasmonic acid (JA)-signaling responses by catalyzing the formation of a bioactive JA-isoleucine conjugate in Arabidopsis (Staswick et al., 2002). Mutant jar1 causes susceptibility to the soil fungus P. irregulare (Staswick et al., 1998). Phylogenetic analyses show that the group III members of the GH3 family have only been discovered in the genomes of some dicots such as Arabidopsis, Brassica napus, and Gossypium hirsutum so far (Terol et al., 2006). The only biochemically characterized GH3 family group III protein is Arabidopsis AtGH3.12, which preferentially catalyzes the formation of 4-substituted benzoates-amido conjugates (Okrent et al., 2009).
So far, all the reported GH3 proteins that have IAA-amido synthetase function for catalyzing the formation of IAA-amido conjugates and thus inactivating IAA belong to group II, with the exception of AtGH3.12, which possibly also promotes the formation of IAA-glutamate conjugates (Staswick et al., 2002, 2005; Ding et al., 2008; Chen et al., 2009, 2010; Zhang et al., 2009; Böttcher et al., 2010; Westfall et al., 2010; Fu et al., 2011). ATP–PPi isotope exchange assays carried out on thin-layer chromatography showed that Arabidopsis AtGH3.2, AtGH3.3, AtGH3.4, AtGH3.6, AtGH3.9, and AtGH3.17 and grape berry VvGH3.1 have adenylation activity that enables conjugating amino acids to IAA, and AtGH3.5 shows adenylation activity for both IAA and SA (Staswick et al., 2002, 2005; Böttcher et al., 2010). Enzyme activity analyses using liquid chromatography–tandem mass spectrometry assays prove that rice OsGH3.2, OsGH3.8, and OsGH3.13 and Arabidopsis AtGH3.17 all have IAA-amido synthetase function (Ding et al., 2008; Chen et al., 2009, 2010; Zhang et al., 2009; Westfall et al., 2010; Fu et al., 2011). The group II GH3 proteins use various amino acids as substrates to form IAA-amido conjugates (Westfall et al., 2010). Some of these conjugates, like IAA-alanine and IAA-leucine, serve to temporarily store IAA and can be hydrolyzed to release free IAA when needed, whereas IAA-aspartic acid and IAA-glutamate sequester IAA for degradation (Staswick et al., 2005). Taken together, synthesis of these IAA conjugates is important to maintain IAA homeostasis.
Some group II GH3-overexpressing transgenic plants or activation-tagged mutants show auxin-deficient phenotypes, among which Arabidopsis AtGH3.5 and rice OsGH3.1, OsGH3.2, and OsGH3.8 mediate plant disease resistance associated with suppression of auxin signaling (Nakazawa et al., 2001; Takase et al., 2004; Park et al., 2007a,b; Zhang et al., 2007, 2009; Ding et al., 2008; Domingo et al., 2009; Fu et al., 2011). Arabidopsis mutant wes1-D exhibits growth retardation and a reduced endogenous IAA level due to activation of AtGH3.5, and wes1-D plants showed enhanced resistance to P. syringae accompanied by activation of defense-responsive gene PR-1 (Park et al., 2007a). The IAA content and plant height negatively correlate with the expression level of OsGH3.1 in rice. OsGH3.1-overexpressing plants have enhanced resistance to fungal pathogen M. oryzae, which may be associated with activation of defense-responsive genes (Domingo et al., 2009). Activation of OsGH3.2 or OsGH3.8 leads to diminished free IAA contents but elevated IAA-aspartic acid content in rice, and the plants show enhanced broad-spectrum resistance to M. oryzae, Xoc, and Xoo, which is closely associated with the inhibition of cell wall-loosening protein expansins (Ding et al., 2008; Fu et al., 2011).
Indole Derivatives in Plant–Pathogen Interactions
There are many roads leading to the synthesis of IAA in plant and microbe. Based on whether the precursor tryptophan (Trp) is required, IAA biosynthesis is divided into Trp-dependent and Trp-independent routes. The Trp-dependent IAA biosynthesis route contains at least six different pathways: indole-3-acetamide (IAM), indole-3-pyruvate (IPA), tryptamine (TAM), indole-3-acetaldoxime (IAOx), indole-3-acetonitrile (IAN), and Trp side-chain oxidase (TSO; Figure 1; Woodward and Bartel, 2005; Spaepen et al., 2007). In the IAM pathway, Trp is changed to IAM by a Trp monooxygenase (IaaM) first, and then IAM is transformed to IAA by an amidohydrolase (Patten and Glick, 1996; Pollmann et al., 2003). In the IPA pathway, Trp undergoes first transamination, then decarboxylation, and finally oxidation to become IAA (Koga et al., 1994; Koga, 1995; Basse et al., 1996; Seo et al., 1998; Stepanova et al., 2008). In the TAM pathway, Trp is decarboxylated to TAM at the beginning by Trp decarboxylase and TAM goes through two steps of oxidation to form IAA in microbes, but in plants, TAM is converted to IAOx by YUCCA first and then follows the IAOx pathway to form IAA (Perley and Stowe, 1966; Hartmann et al., 1983; Facchini et al., 2000; Zhao et al., 2001). In the IAOx pathway, two plant P450 monooxygenases, CYP79B2 and CYP79B3, oxidize Trp to IAOx, which is either metabolized to IAN by another P450 monooxygenase, CYP71A13, or forms indole-3-acetaldehyde (IAAld) by some unidentified enzymes (Barlier et al., 2000; Hull et al., 2000; Mikkelsen et al., 2000; Nafisi et al., 2007). In the IAN pathway, IAN is generated through the IAOx pathway in plants, but the IAN source in microbes is still unclear; however, nitrilase is responsible for modifying IAN to IAA in both plants and microbes (Nagasawa et al., 1990; Bartling et al., 1992; Kobayashi et al., 1993). The TSO pathway is unique to microbes. In this pathway, Trp is directly converted to IAAld, which is then turned into IAA by IAAld dehydrogenase (Oberhänsli et al., 1991). Both plants and microbes can use the Trp-independent route to synthesize IAA, but the enzymes and intermediates involved still need to be determined (Woodward and Bartel, 2005; Spaepen et al., 2007).
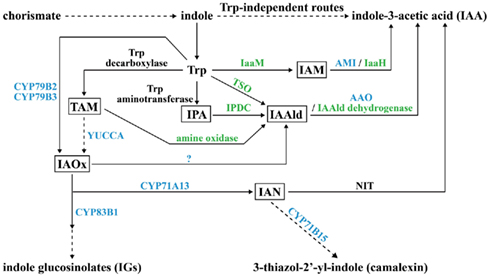
Figure 1. Potential pathways for biosynthesis of IAA and secondary metabolites IGs and camalexin in plant and microbe. The key intermediate product for each tryptophan (Trp)-dependent IAA biosynthesis pathway is boxed. Enzymes for which plant genes are identified are in blue; enzymes for which microbial genes are identified are in green; enzymes for which both plant and microbe genes are identified are in black. A question mark indicates that the enzyme is unknown. Dashed arrows indicate that the conversion needs to be accomplished in more than one step. IAM, indoleacetamide; IaaM, Trp-2-monooxygenase; AMI, amidohydrolase; IaaH, IAM hydrolase; IPA, indole-3-pyruvic acid; IPDC, indole-3-pyruvate decarboxylase; IAAld, indole-3-acetaldehyde; AAO, aldehyde oxidase; TAM, tryptamine; YUCCA, flavin monooxygenase (FMO)-like enzyme; IAOx, indole-3-acetaldoxime; IAN, indole-3-acetonitrile; NIT, nitrilase; TSO, Trp side-chain oxidase.
Some plant-produced secondary metabolites, known as phytoalexins, serve as antibiotics in response to pathogen invasion. Phytoalexins camalexin (3-thiazol-2′-yl-indole), which is toxic to necrotrophic fungi, and indole glucosinolates (IGs), which are broad-spectrum defense compounds, are indole derivatives that share overlapping biosynthesis steps with Trp-dependent IAA synthesis pathways in plants (Figure 1). IAOx is the key branching point for the formation of camalexin and IGs. Through the P450 monooxygenase CYP83B1, IAOx enters a secondary metabolism pathway leading to IGs, which has been identified by direct biochemical evidence (Bednarek et al., 2009). When IAOx is converted to IAN by CYP71A13, IAN can alternatively be hydrolyzed to IAA by NIT or feed into the camalexin synthesis pathway with the final step catalyzed by one more P450 monooxygenase, CYP71B15 (Figure 1; Bartling et al., 1992; Schuhegger et al., 2006). Thus, there is balanced regulation between the biosynthesis of IAA and indole-derived phytoalexins in plant–pathogen interactions.
The biosynthesis of camalexin and IGs is also under the control of auxin signaling. In Arabidopsis miR393-elicited suppression of auxin signaling, ARF is sequestered by stable heterodimerization with Aux/IAA (Woodward and Bartel, 2005). It was recently reported that ARF1 and ARF9 are negative regulators for IG accumulation and that ARF9 functions positively in camalexin accumulation in Arabidopsis (Robert-Seilaniantz et al., 2011).
Emerging evidence shows that camalexin and IGs are important for plants to establish resistance against pathogens. The flg22-triggered callose deposition enhances plant cell wall strength, which contributes to restricting Pto DC3000 growth. This kind of callose deposition requires IG biosynthesis, and mutants defective in the IG biosynthesis pathway or transcriptional activation of IG biosynthesis fail to form callose deposition upon the flg22 trigger (Clay et al., 2009). A reduction in the levels of IGs and camalexin in auxin polar transport mutant aux1 may be the reason for the completely abolished systemic acquired resistance in this mutant (Truman et al., 2010). Similarly, camalexin and IGs are found to participate in mlo-conditioned powdery mildew immunity in Arabidopsis (Consonni et al., 2010). Overexpression of miR393 confers Arabidopsis resistance to biotrophic pathogens but susceptibility to necrotrophic pathogens. The different responses of Arabidopsis to different types of pathogens are associated with transcriptional induction of IG synthesis-related genes but suppression of camalexin synthesis-related genes (Robert-Seilaniantz et al., 2011). The opposite reaction to different pathogens can be indirectly clarified by the general rule that miR393-overexpressing plants accumulate more IGs, but less camalexin after flg22 treatment (Robert-Seilaniantz et al., 2011).
Presumed Mechanisms of Auxin in Plant–Pathogen Interactions
The involvement of auxin in disease development appears to include the following aspects. First, auxin can weaken the plant cell wall, a natural barrier against pathogens. Exogenous auxin can promote rapid elongation of plant tissues by increasing the extensibility of cell wall (Cosgrove, 1993). The plant cell wall is composed of cellulose microfibrils and polysaccharides, and the rigidity of the plant cell wall is sustained by some structural proteins (Darley et al., 2001). Three types of cell wall structural proteins take part in acid-induced wall extension: the endo-β-1,4-glucanases (EGases), xyloglucan endotransglycosylases (XETs), and expansins. The expression of EGases and XETs can be induced by auxin, and they function during cell wall loosening in association with depolymerization of wall polysaccharides (Fry et al., 1992; Brummell et al., 1994; Catalá et al., 1997). At the initial stage of infection, pathogens generate IAA to stimulate EGases and XETs to hydrolyze plant cell wall polysaccharides, and the cell wall saccharides that are released can serve as an ideal nutrition supply to pathogens for survival and later proliferation (Lindow and Brandl, 2003). Auxin also promotes the expression of expansins. Tomato expansin gene LeExp2 shows a high expression level at hypocotyl and fruit growth stages, and auxin up-regulates the expression of LeExp2 remarkably (Catalá et al., 2000). Some expansin genes of soybean and rice can be induced by auxin as well (Downes et al., 2001; Ding et al., 2008). Expansins mediate long-term extension of the cell wall by breaking the hydrogen bonds between cellulose microfibrils and polysaccharides in the wall (McQueen-Mason and Cosgrove, 1995). This process can make the plant cell wall vulnerable and create an opportunity for pathogen attack. Rice plants overexpressing expansin genes are more susceptible to Xoo, Xoc, and M. oryzae infections compared with susceptible wild-type plants; resistant rice plants overexpressing OsGH3.2 or OsGH3.8 to suppress auxin signaling show decreased expression levels of expansin genes before and after infection by any of these three pathogens (Ding et al., 2008; Fu et al., 2011). Microarray analysis demonstrates that several genes encoding proteins concerning cell wall loosening are down-regulated in rice plants overexpressing OsGH3.1, which suppresses auxin signaling (Domingo et al., 2009).
Second, auxin appears to be involved in the opening of stomata, which provide a route of invasion into plants for some phytopathogens. Several pathogens, such as Fusicoccum amygdali, Plasmopara viticola, Xanthomonas axonopodis pv. citri (Xac), Xcc, and Xoc, use stomata as gateways to colonize the interior of plant tissues (Turner and Graniti, 1969; Niño-Liu et al., 2006; Allègre et al., 2007; Gottig et al., 2009; Gudesblat et al., 2009). Auxin might take charge of stomatal opening and thereby assist pathogens to conquer stoma-conferred plant innate immunity (Schroeder et al., 2001; Melotto et al., 2006). However, this hypothesis needs to be validated by more experimental evidence.
Finally, IAA signaling may be antagonistic to the SA-mediated disease resistance pathway (Figure 2). The P. syringae type III effector AvrRpt2 promotes virulence in Arabidopsis by suppressing pathogenesis-related (PR) gene expression that could be activated in SA-dependent defense (Chen et al., 2004). This process is notably paralleled by an increased IAA content (Chen et al., 2007). SA-mediated plant immunity usually accompanies the repression of the auxin-signaling pathway, including down-regulation of small auxin-up RNA (SAUR) family genes, Aux/IAA family genes, polar auxin transport genes, and auxin receptor genes, and up-regulation of some IAA-amido synthase genes (GH3 family; Wang et al., 2007). However, the enhanced resistance to Xoo by suppressing auxin signaling does not require activation of SA signaling in rice (Ding et al., 2008; Fu et al., 2011). In addition, microarray data clearly indicate that some defense-responsive genes are down-regulated in IAA-treated Arabidopsis or IAA-overproduction Arabidopsis mutants (Zhao et al., 2002; Redman et al., 2004).
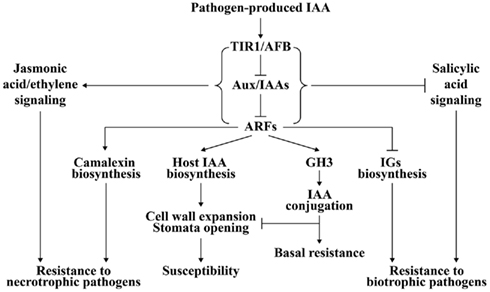
Figure 2. A model of auxin signaling in plant–pathogen interaction. After pathogen-induced IAA accumulation is perceived by auxin receptor TIR1/AFB, TIR1/AFB is activated for degradation of auxin-signaling repressor Aux/IAAs and subsequent derepression of ARFs. Plant IAA biosynthesis may be up-regulated by some ARFs. The elevated IAA level will cause plant cell wall expansion and stomatal opening, which promotes disease development. Some members of ARFs positively regulate camalexin biosynthesis but negatively regulate biosynthesis of indole glucosinolates (IGs), which results in resistance to necrotrophic pathogens and susceptibility to biotrophic pathogens. The expression of some GH3 genes is also activated by ARFs. GH3 sequesters IAA into a conjugated form that inhibits the function of IAA on cell wall and stomata, and thus confers plant resistance. Additionally, auxin-signaling acts antagonistically toward salicylic acid signaling essential for biotrophic resistance, but synergistically with jasmonic acid/ethylene-signaling required for necrotrophic resistance.
The reason underlying the auxin-signaling pathway in disease resistance against necrotrophic pathogens might be allied with JA- and ethylene-signaling pathways that have positive effects on defense against necrotrophic pathogens (Figure 2; Glazebrook, 2005). The induction of JA synthesis-related genes and JA-regulated defense genes, including AOS (allene oxide synthase), LOX2 (lipoxygenase 2), and AtVSP (vegetative storage protein), by IAA is suppressed in axr1-24, which might explain the increased susceptibility of this auxin-signaling mutant to the necrotrophic pathogen P. irregulare (Tiryaki and Staswick, 2002). However, during susceptible interactions between Arabidopsis auxin-signaling mutants aux1-12 and axr2-1 and the necrotrophic pathogen P. cucumerina, activation of SA- and JA/ethylene-signaling pathways is not affected, which means the auxin-signaling pathway is significant in defense against necrotrophic pathogens (Llorente et al., 2008).
Virulence Mechanism of Pathogen-Generated IAA during Disease Development
The IAA synthesized and secreted by pathogens may act as a virulence factor during disease development. A spontaneous mutant of Pseudomonas fluorescens, HP72, produces only low amounts of IAA and has a weakened ability to colonize bentgrass root (Suzuki et al., 2003). Knockout of the IAA synthesis-related gene iaaM (Trp-2-monooxygenase) in Erwinia chrysanthemi reduces bacterial IAA production, diminishes pectate lyase production, and impairs local maceration of pathogenicity on the African violet Saintpaulia ionantha (Yang et al., 2007).
Although the exact role of pathogen-generated IAA in plant–pathogen interactions is still unclear, two models may provide an explanation. The first model describes how pathogen-generated IAA may have a direct virulence effect on plants by loosening the cell wall, opening stomata, and inhibiting SA-dependent defense signaling as discussed above (Chen et al., 2004, 2007; Ding et al., 2008; Domingo et al., 2009; Fu et al., 2011). The second model outlines how IAA secreted by the pathogen may evoke plant endogenous IAA biosynthesis resulting in amplification of the virulence effect caused by pathogenic IAA (Figure 2).
Plants might perceive pathogenic IAA in the manner of auxin receptors such as TIR1. TIR1 bound with IAA facilitates the interaction of TIR1 and auxin-signaling repressor protein Aux/IAA, which leads to degradation of Aux/IAA and derepression of ARF-controlled auxin-regulated gene expression (Woodward and Bartel, 2005; Tan et al., 2007). A series of downstream events in the auxin-signaling pathway are activated (Woodward and Bartel, 2005) and may promote auxin biosynthesis in plants (Figure 2). This hypothesis is supported by the evidence that IAA or pathogen infection can induce plant IAA synthesis-related genes. Exogenous IAA can induce the expression of some paralogs of the YUCCA family, which is important for plant auxin biosynthesis in Arabidopsis, in the black cottonwood Populus trichocarpa (Figure 1; Zhao et al., 2001; Xia et al., 2009). Indole-3-acetaldehyde oxidase (AAO) and nitrilase (NIT) are two protein families involved in the two Trp-dependent pathways for IAA biosynthesis in plants, respectively (Woodward and Bartel, 2005). Some paralogs of AAO and NIT families are induced by exogenous IAA treatment in rice (Fu et al., 2011). The expression of an NIT gene is increased in Xcc-infected Arabidopsis (O’Donnell et al., 2003). AAO and NIT genes are markedly induced in susceptible rice plants after infection with M. oryzae, Xoc, or Xoo, which is accompanied by accumulation of IAA in the infected tissues (Ding et al., 2008; Fu et al., 2011). Furthermore, IG and camalexin accumulation is negatively and positively controlled by some members of ARFs, respectively (Robert-Seilaniantz et al., 2011), which endows the plant with greater susceptibility to biotrophic pathogens and more resistance to necrotrophic pathogens (Figure 2). However, both models may be active in one plant–pathogen interaction.
Conclusion
Plants and pathogens communicate with each other in a conversation through auxin. Further studies on the physiologic and pathogenetic effects of auxin, the bilateral performance of auxin to different kinds of pathogens, and the fine-tuned interplay between auxin signaling and secondary metabolism and other hormone signaling will lead to a better understanding of auxin signaling in plant–pathogen interactions. Besides these theoretical inquiries, manipulating auxin signaling to endow plants with enhanced disease resistance will attract increasing attention and be put into practice in the future.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (30930063, 30921091).
References
Allègre, M., Daire, X., Héloir, M. C., Trouvelot, S., Mercier, L., Adrian, M., and Pugin, A. (2007). Stomatal deregulation in Plasmopara viticola-infected grapevine leaves. New Phytol. 173, 832–840.
Apine, O. A., and Jadhav, J. P. (2011). Optimization of medium for indole-3-acetic acid production using Pantoea agglomerans strain PVM. J. Appl. Microbiol. 110, 1235–1244.
Barash, I., and Manulis-Sasson, S. (2007). Virulence mechanisms and host specificity of gall-forming Pantoea agglomerans. Trends Microbiol. 15, 538–545.
Barlier, I., Kowalczyk, M., Marchant, A., Ljung, K., Bhalerao, R., Bennett, M., Sandberg, G., and Bellini, C. (2000). The SUR2 gene of Arabidopsis thaliana encodes the cytochrome P450 CYP83B1, a modulator of auxin homeostasis. Proc. Natl. Acad. Sci. U.S.A. 97, 14819–14824.
Bartling, D., Seedorf, M., Mithöfer, A., and Weiler, E. W. (1992). Cloning and expression of an Arabidopsis nitrilase which can convert indole-3-acetonitrile to the plant hormone, indole-3-acetic acid. Eur. J. Biochem. 205, 417–424.
Basse, C. W., Lottspeich, F., Steglich, W., and Kahmann, R. (1996). Two potential indole-3-acetaldehyde dehydrogenases in the phytopathogenic fungus Ustilago maydis. Eur. J. Biochem. 242, 648–656.
Bednarek, P., Pislewska-Bednarek, M., Svatoš, A., Schneider, B., Doubsky, J., Mansurova, M., Humphry, M., Consonni, C., Panstruga, R., Sanchez-Vallet, A., Molina, A., and Schulze-Lefert, P. (2009). A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science 323, 101–106.
Böttcher, C., Keyzers, R. A., Boss, P. K., and Davies, C. (2010). Sequestration of auxin by the indole-3-acetic acid-amido synthetase GH3-1 in grape berry (Vitis vinifera L.) and the proposed role of auxin conjugation during ripening. J. Exp. Bot. 61, 3615–3625.
Brummell, D. A., Lashbrook, C. C., and Bennett, A. B. (1994). Plant endo-1,4-β-glucanases: structure, properties and physiological function. ACS Symp. Ser. 566, 100–129.
Catalá, C., Rose, J. K., and Bennett, A. B. (1997). Auxin regulation and spatial localization of an endo-1,4-β-D-glucanase and a xyloglucan endotransglycosylase in expanding tomato hypocotyls. Plant J. 12, 417–426.
Catalá, C., Rose, J. K., and Bennett, A. B. (2000). Auxin-regulated genes encoding cell wall-modifying proteins are expressed during early tomato fruit growth. Plant Physiol. 122, 527–534.
Cernadas, R. A., and Benedetti, C. E. (2009). Role of auxin and gibberellin in citrus canker development and in the transcriptional control of cell-wall remodeling genes modulated by Xanthomonas axonopodis pv. citri. Plant Sci. 177, 190–195.
Chen, Q., Westfall, C. S., Hicks, L. M., Wang, S., and Jez, J. M. (2010). Kinetic basis for the conjugation of auxin by a GH3 family indole-acetic acid-amido synthetase. J. Biol. Chem. 285, 29780–29786.
Chen, Q., Zhang, B., Hicks, L. M., Wang, S., and Jez, J. M. (2009). A liquid chromatography-tandem mass spectrometry-based assay for indole-3-acetic acid-amido synthetase. Anal. Biochem. 390, 149–154.
Chen, Z., Agnew, J. L., Cohen, J. D., He, P., Shan, L., Sheen, J., and Kunkel, B. N. (2007). Pseudomonas syringae type III effector AvrRpt2 alters Arabidopsis thaliana auxin physiology. Proc. Natl. Acad. Sci. U.S.A. 104, 20131–20136.
Chen, Z., Kloek, A. P., Cuzick, A., Moeder, W., Tang, D., Innes, R. W., Klessig, D. F., McDowell, J. M., and Kunkel, B. N. (2004). The Pseudomonas syringae type III effector AvrRpt2 functions downstream or independently of SA to promote virulence on Arabidopsis thaliana. Plant J. 37, 494–504.
Clay, N. K., Adio, A. M., Denoux, C., Jander, G., and Ausubel, F. M. (2009). Glucosinolate metabolites required for an Arabidopsis innate immune response. Science 323, 95–101.
Consonni, C., Bednarek, P., Humphry, M., Francocci, F., Ferrari, S., Harzen, A., Ver Loren van Themaat, E., and Panstruga, R. (2010). Tryptophan-derived metabolites are required for antifungal defense in the Arabidopsis mlo2 mutant. Plant Physiol. 152, 1544–1561.
Cosgrove, D. J. (1993). Wall extensibility: its nature, measurement, and relationship to plant cell growth. New Phytol. 124, 1–23.
Darley, C. P., Forrester, A. M., and McQueen-Mason, S. J. (2001). The molecular basis of plant cell wall extension. Plant Mol. Biol. 47, 179–195.
Davies, P. J. (2004). “The plant hormones: their nature, occurrence, and function,” in Plant Hormones: Biosynthesis, Signal Transduction, Action, ed. P. J. Davies (Dordrecht: Kluwer Academic Publishers), 1–15.
Dharmasiri, N., Dharmasiri, S., and Estelle, M. (2005). The F-box protein TIR1 is an auxin receptor. Nature 435, 441–445.
Ding, X., Cao, Y., Huang, L., Zhao, J., Xu, C., Li, X., and Wang, S. (2008). Activation of the indole-3-acetic acid-amido synthetase GH3-8 suppresses expansin expression and promotes salicylate- and jasmonate-independent basal immunity in rice. Plant Cell 20, 228–240.
Domingo, C., Andrés, F., Tharreau, D., Iglesias, D. J., and Talón, M. (2009). Constitutive expression of OsGH3.1 reduces auxin content and enhances defense response and resistance to a fungal pathogen in rice. Mol. Plant Microbe Interact. 22, 201–210.
Downes, B. P., Steinbaker, C. R., and Crowell, D. N. (2001). Expression and processing of a hormonally regulated β-expansin from soybean. Plant Physiol. 126, 244–252.
Facchini, P. J., Huber-Allanach, K. L., and Tari, L. W. (2000). Plant aromatic L-amino acid decarboxylases: evolution, biochemistry, regulation, and metabolic engineering applications. Phytochemistry 54, 121–138.
Ferrari, S., Galletti, R., Pontiggia, D., Manfredini, C., Lionetti, V., Bellincampi, D., Cervone, F., and De Lorenzo, G. (2008). Transgenic expression of a fungal endo-polygalacturonase increases plant resistance to pathogens and reduces auxin sensitivity. Plant Physiol. 146, 669–681.
Fett, W. F., Osman, S. F., and Dunn, M. F. (1987). Auxin production by plant-pathogenic Pseudomonads and Xanthomonads. Appl. Environ. Microbiol. 53, 1839–1845.
Fry, S. C., Smith, R. C., Renwick, K. F., Martin, D. J., Hodge, S. K., and Matthews, K. J. (1992). Xyloglucan endotransglycosylase, a new wall-loosening enzyme activity from plants. Biochem. J. 282, 821–828.
Fu, J., Liu, H., Li, Y., Yu, H., Li, X., Xiao, J., and Wang, S. (2011). Manipulating broad-spectrum disease resistance by suppressing pathogen-induced auxin accumulation in rice. Plant Physiol. 155, 589–602.
Glazebrook, J. (2005). Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43, 205–227.
Glick, B. R. (1995). The enhancement of plant growth by free-living bacteria. Can. J. Microbiol. 41, 109–117.
Glickmann, E., Gardan, L., Jacquet, S., Hussain, S., Elasri, M., Petit, A., and Dessaux, Y. (1998). Auxin production is a common feature of most pathovars of Pseudomonas syringae. Mol. Plant Microbe Interact. 11, 156–162.
Gottig, N., Garavaglia, B. S., Garofalo, C. G., Orellano, E. G., and Ottado, J. (2009). A filamentous hemagglutinin-like protein of Xanthomonas axonopodis pv. citri, the phytopathogen responsible for citrus canker, is involved in bacterial virulence. PLoS ONE 4, e4358. doi:10.1371/journal.pone.0004358
Gudesblat, G. E., Torres, P. S., and Vojnov, A. A. (2009). Xanthomonas campestris overcomes Arabidopsis stomatal innate immunity through a DSF cell-to-cell signal-regulated virulence factor. Plant Physiol. 149, 1017–1027.
Hagen, G., and Guilfoyle, T. J. (1985). Rapid induction of selective transcription by auxins. Mol. Cell Biol. 5, 1197–1203.
Hartmann, A., Singh, M., and Klingmuller, W. (1983). Isolation and characterization of Azospirillum mutants excreting high amounts of indoleacetic acid. Can. J. Microbiol. 29, 916–923.
Hasan, H. A. (2002). Gibberellin and auxin-indole production by plant root-fungi and their biosynthesis under salinity-calcium interaction. Acta Microbiol. Immunol. Hung. 49, 105–118.
Hull, A. K., Vij, R., and Celenza, J. L. (2000). Arabidopsis cytochrome P450s that catalyze the first step of tryptophan-dependent indole-3-acetic acid biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 97, 2379–2384.
Kepinski, S., and Leyser, O. (2005). The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435, 446–451.
Kobayashi, M., Izui, H., Nagasawa, T., and Yamada, H. (1993). Nitrilase in biosynthesis of the plant hormone indole-3-acetic acid from indole-3-acetonitrile-cloning of the Alcaligenes gene and site-directed mutagenesis of cysteine residues. Proc. Natl. Acad. Sci. U.S.A. 90, 247–251.
Koga, J. (1995). Structure and function of indolepyruvate decarboxylase, a key enzyme in indole-3-acetic acid biosynthesis. Biochim. Biophys. Acta 1249, 1–13.
Koga, J., Syono, K., Ichikawa, T., and Adachi, T. (1994). Involvement of L-tryptophan aminotransferase in indole-3-acetic acid biosynthesis in Enterobacter cloacae. Biochim. Biophys. Acta 1209, 241–247.
Lee, C. W., Efetova, M., Engelmann, J. C., Kramell, R., Wasternack, C., Ludwig-Müller, J., Hedrich, R., and Deeken, R. (2009). Agrobacterium tumefaciens promotes tumor induction by modulating pathogen defense in Arabidopsis thaliana. Plant Cell 21, 2948–2962.
Lindow, S. E., and Brandl, M. T. (2003). Microbiology of the phyllosphere. Appl. Environ. Microbiol. 69, 1875–1883.
Liu, S. T., Perry, K. L., Schardl, C. L., and Kado, C. I. (1982). Agrobacterium Ti plasmid indoleacetic acid gene is required for crown gall oncogenesis. Proc. Natl. Acad. Sci. U.S.A. 79, 2812–2816.
Llorente, F., Muskett, P., Sánchez-Vallet, A., López, G., Ramos, B., Sánchez-Rodríguez, C., Jordá, L., Parker, J., and Molina, A. (2008). Repression of the auxin response pathway increases Arabidopsis susceptibility to necrotrophic fungi. Mol. Plant 1, 496–509.
Maor, R., Haskin, S., Levi-Kedmi, H., and Sharon, A. (2004). In planta production of indole-3-acetic acid by Colletotrichum gloeosporioides f. sp. aeschynomene. Appl. Environ. Microbiol. 70, 1852–1854.
McQueen-Mason, S. J., and Cosgrove, D. J. (1995). Expansin mode of action on cell walls. Analysis of wall hydrolysis, stress relaxation, and binding. Plant Physiol. 107, 87–100.
Melotto, M., Underwood, W., Koczan, J., Nomura, K., and He, S. Y. (2006). Plant stomata function in innate immunity against bacterial invasion. Cell 126, 969–980.
Mikkelsen, M. D., Hansen, C. H., Wittstock, U., and Halkier, B. A. (2000). Cytochrome P450 CYP79B2 from Arabidopsis catalyzes the conversion of tryptophan to indole-3-acetaldoxime, a precursor of indole glucosinolates and indole-3-acetic acid. J. Biol. Chem. 275, 33712–33717.
Nafisi, M., Goregaoker, S., Botanga, C. J., Glawischnig, E., Olsen, C. E., Halkier, B. A., and Glazebrook, J. (2007). Arabidopsis cytochrome P450 monooxygenase 71A13 catalyzes the conversion of indole-3-acetaldoxime in camalexin synthesis. Plant Cell 19, 2039–2052.
Nagasawa, T., Mauger, J., and Yamada, H. (1990). A novel nitrilase, arylacetonitrilase, of Alcaligenes faecalis JM3-purification and characterization. Eur. J. Biochem. 194, 765–772.
Nakazawa, M., Yabe, N., Ichikawa, T., Yamamoto, Y. Y., Yoshizumi, T., Hasunuma, K., and Matsui, M. (2001). DFL1, an auxin-responsive GH3 gene homologue, negatively regulates shoot cell elongation and lateral root formation, and positively regulates the light response of hypocotyl length. Plant J. 25, 213–221.
Navarro, L., Dunoyer, P., Jay, F., Arnold, B., Dharmasiri, N., Estelle, M., Voinnet, O., and Jones, J. D. (2006). A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312, 436–439.
Niño-Liu, D. O., Ronald, P. C., and Bogdanove, A. J. (2006). Xanthomonas oryzae pathovars: model pathogens of a model crop. Mol. Plant Pathol. 7, 303–324.
Oberhänsli, T., Défago, G., and Haas, D. (1991). Indole-3-acetic-acid (IAA) synthesis in the biocontrol strain CHA0 of Pseudomonas fluorescens: role of tryptophan side-chain oxidase. J. Gen. Microbiol. 137, 2273–2279.
O’Donnell, P. J., Schmelz, E. A., Moussatche, P., Lund, S. T., Jones, J. B., and Klee, H. J. (2003). Susceptible to intolerance – a range of hormonal actions in a susceptible Arabidopsis pathogen response. Plant J. 33, 245–257.
Okrent, R. A., Brooks, M. D., and Wildermuth, M. C. (2009). Arabidopsis GH3.12 (PBS3) conjugates amino acids to 4-substituted benzoates and is inhibited by salicylate. J. Biol. Chem. 284, 9742–9754.
Park, J. E., Park, J. Y., Kim, Y. S., Staswick, P. E., Jeon, J., Yun, J., Kim, S. Y., Kim, J., Lee, Y. H., and Park, C. M. (2007a). GH3-mediated auxin homeostasis links growth regulation with stress adaptation response in Arabidopsis. J. Biol. Chem. 282, 10036–10046.
Park, J. E., Seo, P. J., Lee, A. K., Jung, J. H., Kim, Y. S., and Park, C. M. (2007b). An Arabidopsis GH3 gene, encoding an auxin-conjugating enzyme, mediates phytochrome B-regulated light signals in hypocotyl growth. Plant Cell Physiol. 48, 1236–1241.
Patten, C. L., and Glick, B. R. (1996). Bacterial biosynthesis of indole-3-acetic acid. Can. J. Microbiol. 42, 207–220.
Patten, C. L., and Glick, B. R. (2002). Role of Pseudomonas putida indoleacetic acid in development of the host plant root system. Appl. Environ. Microbiol. 68, 3795–3801.
Perley, J. E., and Stowe, B. B. (1966). On the ability of Taphrina deformans to produce indoleacetic acid from tryptophan by way of tryptamine. Plant Physiol. 41, 234–237.
Pollmann, S., Neu, D., and Weiler, E. W. (2003). Molecular cloning and characterization of an amidase from Arabidopsis thaliana capable of converting indole-3-acetamide into the plant growth hormone, indole-3-acetic acid. Phytochemistry 62, 293–300.
Redman, J. C., Haas, B. J., Tanimoto, G., and Town, C. D. (2004). Development and evaluation of an Arabidopsis whole genome Affymetrix probe array. Plant J. 38, 545–561.
Reineke, G., Heinze, B., Schirawski, J., Buettner, H., Kahmann, R., and Basse, C. W. (2008). Indole-3-acetic acid (IAA) biosynthesis in the smut fungus Ustilago maydis and its relevance for increased IAA levels in infected tissue and host tumour formation. Mol. Plant Pathol. 9, 339–355.
Robert-Seilaniantz, A., MacLean, D., Jikumaru, Y., Hill, L., Yamaguchi, S., Kamiya, Y., and Jones, J. D. G. (2011). The microRNA miR393 re-directs secondary metabolite biosynthesis away from camalexin and towards glucosinolates. Plant J. 67, 218–231.
Savatin, D. V., Ferrari, S., Sicilia, F., and De Lorenzo, G. (2011). Oligogalacturonide-auxin antagonism does not require post-transcriptional gene silencing or stabilization of auxin response repressors in Arabidopsis thaliana. Plant Physiol. (in press), doi:10.1104/pp.111.184663.
Schmelz, E. A., Engelberth, J., Tumlinson, J. H., Block, A., and Alborn, H. T. (2004). The use of vapor phase extraction in metabolic profiling of phytohormones and other metabolites. Plant J. 39, 790–808.
Schroeder, J. I., Allen, G. J., Hugouvieux, V., Kwak, J. M., and Waner, D. (2001). Guard cell signal transduction. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 627–658.
Schuhegger, R., Nafisi, M., Mansourova, M., Petersen, B. L., Olsen, C. E., Svatoš, A., Halkier, B. A., and Glawischnig, E. (2006). CYP71B15 (PAD3) catalyzes the final step in camalexin biosynthesis. Plant Physiol. 141, 1248–1254.
Seo, M., Akaba, S., Oritani, T., Delarue, M., Bellini, C., Caboche, M., and Koshiba, T. (1998). Higher activity of an aldehyde oxidase in the auxin-overproducing superroot1 mutant of Arabidopsis thaliana. Plant Physiol. 116, 687–693.
Spaepen, S., Vanderleyden, J., and Remans, R. (2007). Indole-3-acetic acid in microbial and microorganism-plant signaling. FEMS Microbiol. Rev. 31, 425–448.
Staswick, P. E., Serban, B., Rowe, M., Tiryaki, I., Maldonado, M. T., Maldonado, M. C., and Suza, W. (2005). Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell 17, 616–627.
Staswick, P. E., Tiryaki, I., and Rowe, M. (2002). Jasmonate response locus JAR1 and several related Arabidopsis genes encode enzymes of the firefiy luciferase superfamily that show activity on jasmonic, salicylic, and indole-3-acetic acids in an assay for adenylation. Plant Cell 14, 1405–1415.
Staswick, P. E., Yuen, G. Y., and Lehman, C. C. (1998). Jasmonate signaling mutants of Arabidopsis are susceptible to the soil fungus Pythium irregulare. Plant J. 15, 747–754.
Stepanova, A. N., Robertson-Hoyt, J., Yun, J., Benavente, L. M., Xie, D. Y., Dolezal, K., Schlereth, A., Jürgens, G., and Alonso, J. M. (2008). TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133, 177–191.
Suzuki, S., He, Y., and Oyaizu, H. (2003). Indole-3-acetic acid production in Pseudomonas fluorescens HP72 and its association with suppression of creeping bentgrass brown patch. Curr. Microbiol. 47, 138–143.
Takase, T., Nakazawa, M., Ishikawa, A., Kawashima, M., Ichikawa, T., Takahashi, N., Shimada, H., Manabe, K., and Matsui, M. (2004). ydk1-D, an auxin-responsive GH3 mutant that is involved in hypocotyl and root elongation. Plant J. 37, 471–483.
Tan, X., Calderon-Villalobos, L. L., Sharon, M., Zheng, C., Robinson, C. V., Estelle, M., and Zheng, N. (2007). Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446, 640–645.
Terol, J., Domingo, C., and Talón, M. (2006). The GH3 family in plants: genome wide analysis in rice and evolutionary history based on EST analysis. Gene 371, 279–290.
Tiryaki, I., and Staswick, P. E. (2002). An Arabidopsis mutant defective in jasmonate response is allelic to the auxin-signaling mutant axr1. Plant Physiol. 130, 887–894.
Truman, W. M., Bennett, M. H., Turnbull, C. G. N., and Grant, M. R. (2010). Arabidopsis auxin mutants are compromised in systemic acquired resistance and exhibit aberrant accumulation of various indolic compounds. Plant Physiol. 152, 1562–1573.
Turner, N. C., and Graniti, A. (1969). Fusicoccin: a fungal toxin that opens stomata. Nature 223, 1070–1071.
Ulmasov, T., Hagen, G., and Guilfoyle, T. J. (1999). Activation and repression of transcription by auxin-response factors. Proc. Natl. Acad. Sci. U.S.A. 96, 5844–5849.
Wang, D., Pajerowska-Mukhtar, K., Culler, A. H., and Dong, X. (2007). Salicylic acid inhibits pathogen growth in plants through repression of the auxin signaling pathway. Curr. Biol. 17, 1784–1790.
Westfall, C. S., Herrmann, J., Chen, Q., Wang, S., and Jez, J. M. (2010). Modulating plant hormones by enzyme action: the GH3 family of acyl acid amido synthetases. Plant Signal. Behav. 61, 752–766.
Woodward, A. W., and Bartel, B. (2005). Auxin: regulation, action and interaction. Ann. Bot. 95, 707–735.
Xia, Y., Kang, B. G., Osburn, L. D., Li, Y., and Cheng, Z. M. (2009). Identification of the flavin-dependent monooxygenase-encoding YUCCA gene family in Populus trichocarpa and their expression in vegetative tissues and in response to hormone and environmental stresses. Plant Cell Tissue Organ Cult. 97, 271–283.
Yang, S., Zhang, Q., Guo, J., Charkowski, A. O., Glick, B. R., Ibekwe, A. M., Cooksey, D. A., and Yang, C. H. (2007). Global effect of indole-3-acetic acid biosynthesis on multiple virulence factors of Erwinia chrysanthemi 3937. Appl. Environ. Microbiol. 73, 1079–1088.
Zhang, S. W., Li, C. H., Cao, J., Zhang, Y. C., Zhang, S. Q., Xia, Y. F., Sun, D. Y., and Sun, Y. (2009). Altered architecture and enhanced drought tolerance in rice via the down-regulation of indole-3-acetic acid by TLD1/OsGH3.13 activation. Plant Physiol. 151, 1889–1901.
Zhang, Z., Li, Q., Li, Z., Staswick, P. E., Wang, M., Zhu, Y., and He, Z. (2007). Dual regulation role of GH3.5 in salicylic acid and auxin signaling during Arabidopsis-Pseudomonas syringae interaction. Plant Physiol. 145, 450–464.
Zhao, Y., Christensen, S. K., Fankhauser, C., Cashman, J. R., Cohen, J. D., Weigel, D., and Chory, J. (2001). A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291, 306–309.
Keywords: defense, disease, indole-3-acetic acid, microbe, phytoalexin
Citation: Fu J and Wang S (2011) Insights into auxin signaling in plant–pathogen interactions. Front. Plant Sci. 2:74. doi: 10.3389/fpls.2011.00074
Received: 15 June 2011;
Accepted: 17 October 2011;
Published online: 01 November 2011.
Edited by:
Giulia de Lorenzo, Sapienza – Università di Roma, ItalyReviewed by:
Murray Grant, University of Exeter, UKAntonio Molina, Centro de Biotecnología y Genómica de Plantas, Spain
Copyright: © 2011 Fu and Wang. This is an open-access article subject to a non-exclusive license between the authors and Frontiers Media SA, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and other Frontiers conditions are complied with.
*Correspondence: Shiping Wang, National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, Wuhan 430070, China. e-mail:c3dhbmdAbWFpbC5oemF1LmVkdS5jbg==

