- 1Fish Genetics Unit, Institute of Genome Biology, Research Institute for Farm Animal Biology (FBN), Dummerstorf, Germany
- 2Department for Marine Aquaculture, Institute of Animal Breeding and Husbandry, Kiel University, Kiel, Germany
- 3Fraunhofer Research Institution for Individualized and Cell-Based Medical Engineering IMTE, Aquaculture and Aquatic Resources, Büsum, Germany
- 4Institute of Toxicology and Pharmacology for Natural Scientists, University Medical School Schleswig-Holstein, Kiel, Germany
- 5Faculty of Agriculture and Environmental Sciences, University of Rostock, Rostock, Germany
Smoltification was found to impact both immune and stress responses of farmed Atlantic salmon (Salmo salar), but little is known about how salinity change affects salmon months after completed smoltification. Here, we examined (1) the effect of salinity change from brackish water to seawater on the stress and immune responses in Atlantic salmon and (2) evaluated if functional diets enriched with microalgae can mitigate stress- and immune-related changes. Groups of Atlantic salmon were fed for 8 weeks with different microalgae-enriched diets in brackish water and were then transferred into seawater. Samples of the head kidney, gill, liver and plasma were taken before seawater transfer (SWT), 20 h after SWT, and 2 weeks after SWT for gene-expression analysis, plasma biochemistry and protein quantification. The salmon showed full osmoregulatory ability upon transfer to seawater reflected by high nkaα1b levels in the gill and tight plasma ion regulation. In the gill, one-third of 44 investigated genes were reduced at either 20 h or 2 weeks in seawater, including genes involved in cytokine signaling (il1b) and antiviral defense (isg15, rsad2, ifit5). In contrast, an acute response after 20 h in SW was apparent in the head kidney reflected by increased plasma stress indicators and induced expression of genes involved in acute-phase response (drtp1), antimicrobial defense (camp) and stress response (hspa5). However, after 2 weeks in seawater, the expression of antiviral genes (isg15, rsad2, znfx1) was reduced in the head kidney. Few genes (camp, clra, c1ql2) in the gill were downregulated by a diet with 8% inclusion of Athrospira platensis. The results of the present study indicate that salinity change months after smoltification evokes molecular stress- and immune responses in Atlantic salmon. However, microalgae-enriched functional diets seem to have only limited potential to mitigate the related changes.
1 Introduction
Atlantic salmon (Salmo salar) is an anadromous salmonid of high economic value (Forseth et al., 2017). Global salmon production of approximately ∼2.6 million tons has stagnated over the last years (Glover et al., 2020) as high mortalities and increasing sea lice infestations of salmon as well as negative environmental impacts caused by nutrient pollution and escapes have hampered further growth of the industry (McGinnity et al., 2003; Quiñones et al., 2019). Traditionally a large part of the production cycle of Atlantic salmon takes place in sea cages, while more recently many producers turn towards using land-based recirculating aquaculture systems (RAS; Dalsgaard et al., 2013). Before Atlantic salmon can be transferred into seawater they have to undergo different physiological and morphological changes known as smoltification to prepare for life in seawater (Hoar, 1988; McCormick et al., 1998). Smoltification not only affects the physiology, but also the immune system of salmon, which manifests in decreased levels of total serum protein and immunoglobulin M (IgM) (Melingen et al., 1995) as well as reduced expression of multiple immune genes in smolts (Johansson et al., 2016). The persistence of these effects after transfer to seawater, coinciding with an increased abundance of pathogens in the marine environment, likely explain some of the high disease-related mortalities seen during the first weeks in seawater (Soares et al., 2011; Bang Jensen et al., 2020). Nowadays, salmon are often reared as larger smolts or post-smolts in RAS on land to shorten the overall time they spend in the sea. As the RAS environment can be fully controlled, many environmental factors such as salinity and water velocity can be adapted for optimal growth and welfare (Ytrestøyl et al., 2020; Mortensen et al., 2022). After their smoltification in freshwater, an acclimation period in brackish water could alleviate the salmon’s transition to salt water and improve their overall health and performance (Ytrestøyl et al., 2020; Ytrestøyl et al., 2022).
Besides changes in the rearing environment, the use of functional feeds can improve the fish’s ability to cope with environmental stressors, stimulate immunity and eventually improve production performance (Dawood et al., 2018; Marimuthu et al., 2022). Microalgae, which have known immune-stimulatory properties in different fish species (Morris et al., 2007; Abdel-Tawwab and Ahmad, 2009; Cerezuela et al., 2012; Zhang et al., 2014; An et al., 2016; Luo et al., 2018), are promising candidates for functional feeds for Atlantic salmon. For example, the addition of Chlorella sp. to diets increased the levels of IgM and IgD antibodies in blood, and the cytokine levels of interleukin-22 (Il-22) and C-C motif chemokine ligand 5 (Ccl-5) in the kidney and liver of Gibel carp Carassius gibelio (Zhang et al., 2014). Supplementation with the microalgae Phaeodactylum tricornutum increased phagocytic activity and Nannochloropsis gaditana enhanced the expression of β-defensin in the head kidney of gilthead seabream Sparus aurata (Cerezuela et al., 2012). Furthermore, it has been shown that diets containing Tetraselmis sp. lowered cortisol levels after confinement stress in gilthead seabream (Pereira et al., 2020) and diets containing Arthrospira platensis counteracted the increasing glucose concentrations after air exposure and enhanced antioxidant activity in gilthead seabream (de Mattos et al., 2019).
A variety of phenotypic and molecular markers are used to assess the health and welfare of farmed fish (Barreto et al., 2022). Gene expression profiling allows the detection of both systemic and local immune responses in key immune organs. Recent studies on Atlantic salmon have revealed a variety of genes that can be used to assess the salmon’s immune status (Krasnov et al., 2020; Lund et al., 2022). Accordingly, expression analysis of stress- and immune-related genes can complement traditional indicators such as growth performance, organ health and plasma indicators. In this study, three key immune organs with varying functions in stress and immune response were analyzed. The head kidney is a major lymphoid and myeloid organ that controls the systemic stress and immune response of fish (Press et al., 1994; Press and Evensen, 1999). The gill is part of the mucosa-associated lymphoid tissue and plays a crucial role in the local immune response, as it is in direct contact with the environment (Haugarvoll et al., 2008; Rességuier et al., 2020). Lastly, the liver was included as the main metabolizing organ to gain insight into the fish’s oxidative status (Birnie-Gauvin et al., 2017; Hussein et al., 2023).
Given the lack of knowledge on how salinity change affects stress physiology and immunity of Atlantic salmon and its potential dietary mitigation, the aim of our study was twofold: (1) assessing the effect of salinity change to seawater following acclimation of Atlantic salmon to brackish water on the stress and immune response and (2) evaluating whether microalgae enriched functional diets can mitigate stress and immune-related changes following salinity change.
2 Material and methods
2.1 Experiment and sampling
The experimental setup has been described in detail in Mueller et al. (2023). Briefly, two identical RAS (7.6 m3, turnover rate 4 times h−1) at the facilities of the Fraunhofer IMTE, Büsum, Germany were used for the trial, where one was operated on brackish water (BW; 13.0 ± 0.8 psu, 13.5 ± 0.4°C, 7.3 ± 0.1 pH, 10.3 ± 0.2 mg/L O2, 0.12 ± 0.07 mg N/L total ammonia 0.06 ± 0.01 mg N/L NO2−) while the other was operated on full marine conditions (SW; 31.8 ± 0.5 psu, 13.4 ± 0.3°C, 7.2 ± 0.1 pH, 10.4 ± 0.3 mg/L O2, 0.12 ± 0.07 mg N/L total ammonia 0.07 ± 0.02 mg N/L NO2−). Atlantic salmon smolts, raised in freshwater, were obtained from Jurassic Salmon, Karnice, Poland and acclimated in brackish water for 2 months in the RAS prior to the start of the experiment. Following acclimation, the fish were fed six different functional diets for 8 weeks in brackish water until 2 weeks after transfer to seawater. This time-interval was chosen as one of the most critical phases during the production of Atlantic salmon (Bang Jensen et al., 2020; Sommerset et al., 2020). The fish were hand-fed twice daily (8 a.m. and 2 p.m.) until apparent satiation. The functional diets were designed to be isonitrogenous and isoenergetic (dry matter basis) and were enriched with one of the following microalgae: Chlorella vulgaris (either intact cell wall, CVI or broken cell wall, CVB), Tetraselmis chuii (TC), Athrospira platensis (AP) or Schizochytrium limacinum (SL) at an inclusion level of 8%. A diet without added microalgae served as a control (CD; Table 1). The microalgae were chosen based on their production volume, allowing for industrial upscaling, as well as containing a diversity of functional compounds. The inclusion level of 8% reflected a compromise of formulating diets without making major adjustments to a variety of ingredients within the feed formulation, as well as making sure sufficient amounts of functional components are available for the fish. At the start of the experiment 28 fish were stocked into each tank (with three tanks assigned to each treatment), but due to serial samplings, 19 fish per tank were transferred to seawater.
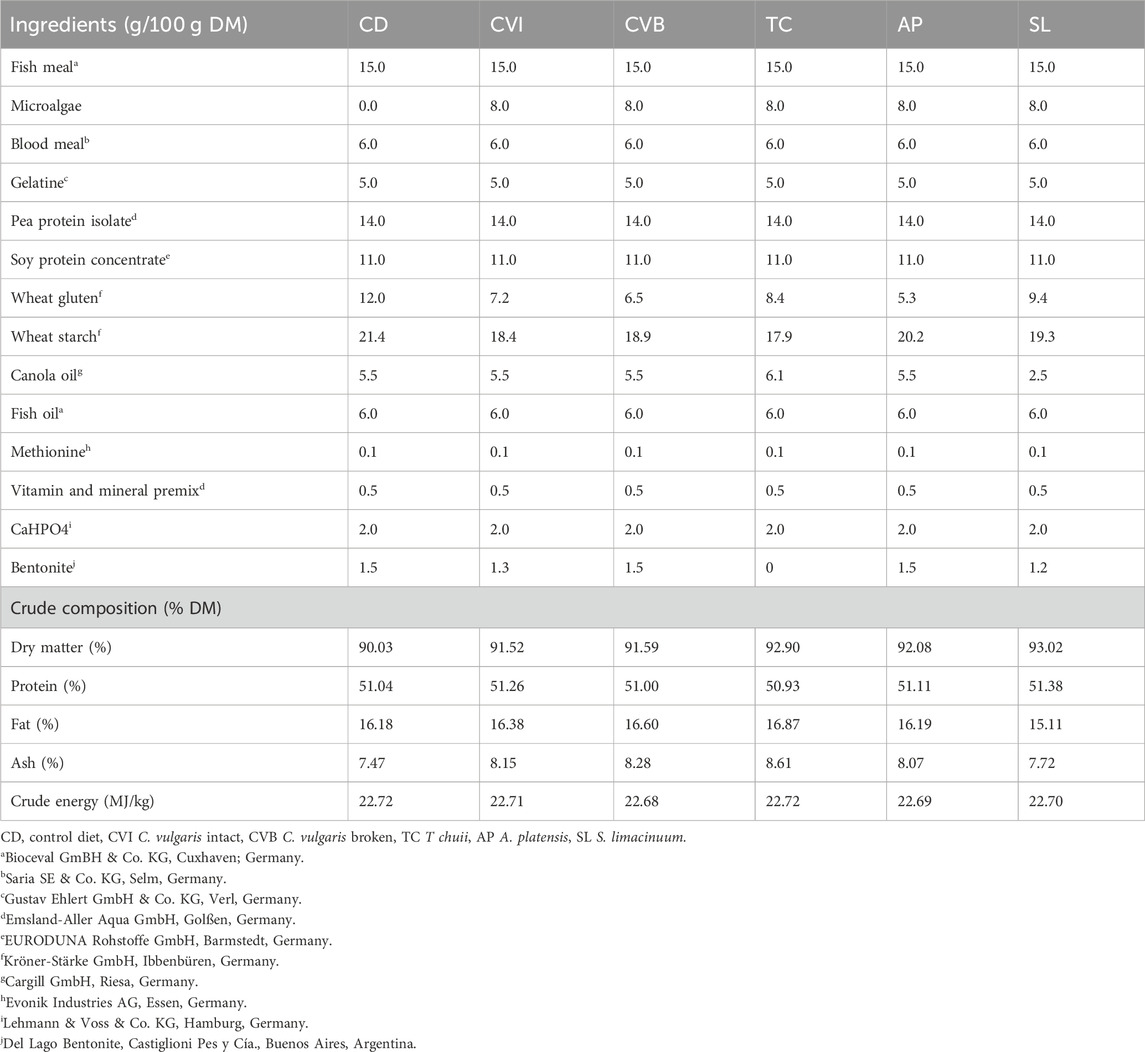
TABLE 1. Formulation of experimental diets in g/100 g dry matter (DM) as well as crude composition in % DM.
Samples were collected after 8 weeks of feeding the experimental diets in brackish water (T1), 20 h following transfer to seawater (T2) and 2 weeks in seawater (T3). These timepoints include both the acute stress and immune response (T2) as well as adaptive immune response and further adaptive responses (T3) to salinity change. Each time nine fish per diet (three per tank) were randomly sampled. The salmon were netted from the tanks and euthanized with MS-222 (0.3 mg/L). Length and weight were recorded and a 2 mL blood sample was withdrawn in heparinized syringes by caudal vein puncture. Blood samples were immediately centrifuged at 4000 g for 8 min and plasma aliquots flash-frozen at −80°C, to be used for the determination of plasma metabolites. The liver and spleen were removed, weighed and a small piece of the liver was flash-frozen on dry ice for determination of protein concentrations. The first left gill arch and the head kidney were sampled for gene-expression analysis and flash-frozen in a RNase-free tube in liquid nitrogen. Somatic indices were calculated as follows:
CF (Fulton´s condition factor) = weight/fish length3 *100.
HSI (hepatosomatic index) = liver weight /fish weight * 100.
SSI (spleen somatic index) = spleen weight /fish weight * 100.
2.2 Plasma enzymes and metabolites
Plasma glucose, Na+ and Cl−-ions in plasma samples were measured on a Fuji Dry Chem NX500i (Fujifilm, Tokyo, Japan) using kits of the manufacturer. Plasma cortisol was determined by an enzyme-linked immunosorbent assay (ELISA) Kit (Demeditec Diagnostics GmbH, Kiel, Germany) following the manufacturer’s instructions.
2.3 Western blots
Liver samples (pooled per tank) were processed to extract total protein based on radioimmunoprecipitation (RIPA) following the manufacturer’s instructions (RIPA Lysis Buffer System, Santa Cruz Biotechnology, Dallas, Texas, United States). Cu, Zn superoxide dismutase (Sod1) and myeloperoxidase (Mpo) proteins were analyzed using SDS-PAGE and Western blot techniques. The protein ß-actin (Actb) was used as a loading control. A no template control and a positive control per antibody were included, with Danio rerio liver used for Mpo, bovine liver for Sod1, and HEK-293 cells for ß-actin. To process 20 µg of total protein, reducing conditions were applied with SDS sample and reducing buffer (both TruPAGE, Sigma-Aldrich, Schnelldorf, Germany) for 10 min at 70°C. SDS-PAGE was performed with precast 4%–12% gradient gels, TruPAGE running buffer, and antioxidant (Sigma-Aldrich) in an Xcell SureLock Mini-Cell (Thermo Fisher Scientific, Waltham, Massachusetts, United States). The proteins were then electro-transferred to a PVDF membrane. The membrane was cut horizontally for parallel protein detection of Mpo and Sod1. Primary antibody incubation was carried out overnight at 4°C using Mpo antibody (ab210563, Abcam, 1:5000 dilution in PBS-T containing 2.5% skim milk) and Sod1 antibody (NBP2-24915, Novus Biologicals, 1:500 dilution in PBS-T containing 2.5% skim milk). Using a dilution of 1:5000, the secondary antibody anti-rabbit IgG conjugated HRP (sc-2357, Santa Cruz Biotechnology) was incubated at room temperature for 90 min. ECL detection reagents and chemiluminescence film (both GE Healthcare, Amersham, United Kingdom) were used for detection, with 40 s of exposure time for both Mpo and Sod1. To detect Actb, the antibodies were stripped in 100 mM glycin buffer (pH 2.5). The membrane was then incubated overnight at 4°C in a 1:5000 dilution of Actb antibody (NB600-503, Novus Biologicals, Wiesbaden, Germany). Protein abundance was quantified by densitometric analysis of the protein bands using GIMP and normalized to the concentration of the reference protein Actb.
2.4 RNA isolation and multiplex gene-expression profiling
TRIzol (Thermo Fisher Scientific) was used to extract total RNA, which was then purified with the ISOLATE II RNA Micro Kit (Meridian Bioscience Inc., Cincinnati, Ohio, United States). NanoDrop One (Thermo Fisher Scientific) was used to determine the concentration of the isolated RNA. High-quality RNA was then reverse-transcribed into cDNA utilizing the Reverse Transcription Master Mix (Standard BioTools, South San Francisco, California, United States). cDNA samples were preamplified with the Fluidigm PreAmp Master Mix before purification with exonuclease I (New England BioLabs, Frankfurt/Main, Germany). All procedures were conducted according to the manufacturer’s instructions.
Exon-skipping oligonucleotide primers for the smoltification markers nkaα1a (homologous to mammalian atp1a1) and nkaα1b were designed using the Pyrosequencing Assay Design software v.1.0.6 (Biotage, Uppsala, Sweden; Supplementary Table S1). To evaluate salmon immunocompetence, Krasnov et al. (2020) designed a set of genes involved in important immunological and stress-relevant pathways and functions, such as antigen presentation, T cell activity, oxidative stress, antiviral and antibacterial defense as well as stress-related pathways such as nf-κb pathway, acute phase response and heat shock protein (HSP) signaling pathway. In addition to 41 of those target genes and three reference genes (Supplementary Table S1), three more immune-specific genes were included (Pyrosequencing Assay Design software v.1.0.6) for hamp, saa5, and sod1 (Supplementary Table S1). The LightCycler-96 system (Roche Diagnostics International AG, Rotkreuz, Switzerland) was utilized to assess the smoltification status using the nkaα1a and nkaα1b primers (1 μL) together with 6 μL SensiFAST SYBR No-ROX Mix (Meridian Bioscience Inc.) and 5 μL cDNA in a 12-μL-reaction volume. The transcripts were amplified according to following program: initial denaturation at 95°C for 300 s, then 40 cycles including a denaturation step at 95°C for 30 s, primer annealing at 60°C for 15 s and at last, elongation step at 72°C for 15 s followed by fluorescence measurement at 72°C for 10 s. Afterwards, gel electrophoresis and melting-curve analysis were conducted to check the quality of the amplicons. Finally, the qPCR results were obtained using the LightCycler-96 analysis software v. 1.1.0.1320 (Roche Diagnostics International AG) and normalized using the geometric mean of two reference genes ribosomal protein L4 (rpl4) and ribosomal protein S20 (rps20). Standard curves of nkaα1a and nkaα1b oligonucleotides were generated (108–103 copies per 5 μL; R2 > 0.99) to calculate the individual copy numbers. Multiplex immunogene-expression profiling was conducted on 48.48 Gene Expression biochips. The pre-amplified cDNA samples and primers were transferred into the sample and assay inlets of these biochips that were first primed in the MX IFC Controller (Standard BioTools). Subsequently, the concentrations of the target transcripts were quantified using the Biomark HD (Standard BioTools) according to the manufacturer’s thermal protocol “GE Fast 48 × 48 PCR + Melt v2.pcl” (application type: gene expression; passive reference: ROX; assay: single probe). By use of Fluidigm real-time PCR analysis software v3.0.2 (Standard BioTools), the raw Cq values were gathered and used for calculation of the relative expression of the target genes based on the ΔCt method. Three reference genes actb, rpl4 and rps20 were employed as internal normalizers. Throughout the calculation, the mean Cq per gene for all samples per organ served as a calibrator. The data collected for four individuals were excluded from the dataset because they exhibited signs of sickness (for example, liver cysts and/or irregular spleen) and subsequently showed abnormal immune-gene expression.
2.5 Statistical analysis
All statistical analyses were performed using R (v4.1.2; R Core Team 2021; Vienna, Austria) in the environment R studio. The relative gene expression values obtained from the multigene expression assays as well as the actual copy numbers from the qPCR evaluation of the smoltification markers (nkaα1a/b) were log2-transformed before statistical analysis. However, the copy numbers of the smoltification markers were displayed in a boxplot on a log10 scale to guide visual interpretation. Mixed effect models were defined for the response variable expression of gene of interest, plasma parameter or organ health index. For testing the expression of the smoltification markers, the model included isoform (nkaα1a/b) and timepoint as fixed factors and tank was included as a random factor. For the other gene expression data as well as plasma parameter the model included diet, timepoint and their interaction as fixed factors and the tank as a random factor. In case of testing the organ health index, as well as plasma ion concentrations among the diet groups, timepoint was not included in the model. Model residuals were inspected graphically and found to be normally distributed and heteroscedastic. Analysis of variance (ANOVA) was conducted followed by multiple contrast tests for heteroscedastic data (Hasler and Hothorn, 2008). This was done to compare the microalgae-enriched diets to the control within a given time point as well as within each diet for effects over time. Comparisons were considered significant at p < 0.05. In case an interaction effect of diet and timepoint was absent, subsequent contrasts were pooled over the remaining factor. For the analysis of liver protein concentrations (pooled on tank level), a model based on generalized least squares (GLS) was defined, which included only the fixed factors diet and timepoint and subsequently model evaluation followed the procedure described above.
To evaluate the overall effect of seawater transfer on gene expression data, principal component analysis (PCA) with time point as grouping was conducted on log2-transformed relative expression values for the 44 target genes investigated per tissue, as implemented in the ade4 package in R (Bougeard and Dray, 2018). The individual contribution of each gene to the overall separation of the time points in the PCA was visualized using factor maps implemented in the facto extra package in R (Kassambara and Mundt, 2020). Fold changes of each gene across the factor time, relative to the timepoint in brackish water (T2), were calculated from relative gene expression values. Subsequently fold changes were visualized in a hierarchically clustered heatmap for both organs separately using the ppheatmap package (Kolde, 2022). A range of 0.33 to 3-fold change based on the relative gene expression values was considered as basal levels. The relative expression levels of three differentially expressed genes in at least one microalgae group (clra, c1ql2, and camp) in the gill were visualized as log2-transformed relative expression in boxplots.
3 Results
3.1 Smoltification status and osmoregulation
Salmon acclimated to BW and transferred to full strength SW had a slight but significant increase in plasma Na+ and Cl− concentrations (Figures 1A,B) following 20 h in SW (p < 0.05) in the control group. However, after 2 weeks in SW, plasma ion levels returned to levels prior to transfer. This was furthermore reflected in the expression of the two NKA isoforms in the gill. The transcripts of the “seawater isoform” nkaα1b were 5.1 times more abundant in the gill by the end of the brackish water phase than the “freshwater isoform” nkaα1a (p < 0.001) and this ratio increased even more 20 h and 2 weeks in SW where nkaα1a transcript abundance was significantly reduced further (p < 0.05) while nkaα1b transcript abundance increased (p < 0.001; Figure 1C).
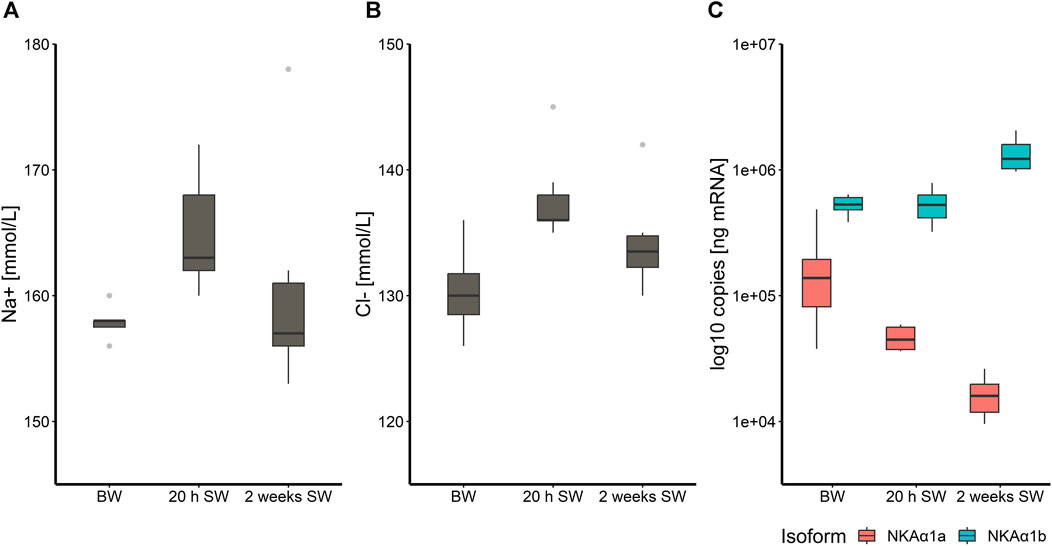
FIGURE 1. Boxplot with median and 1.5 x interquartile range whiskers showing sodium-ion (A) and chloride-ion (B) concentration in plasma of Atlantic salmon fed the control diet (CD) after 8 weeks experimental period in brackish water (T1), 20 h in seawater (T2) and 2 weeks in seawater (T3), as well as log-10- transformed copy numbers of nkaα1a/b transcripts for the three timepoints (C); n = 6–8. Note that the y-axis in panel (A), (B) and (C) does not start at zero.
3.2 Effects of salinity change on expression of immune and stress-related genes
In the head kidney, when salinity changed from BW to SW, levels of immune- and stress-related genes were significantly modulated in a time-dependent manner (Figure 2A). Principal component analysis (PCA) revealed a 21% share of variability of the first principal component for the conditions timepoint/salinity. The overall gene-expression pattern of salmon from brackish water (T1) grouped between early (T2) and the later (T3) timepoint in seawater. This pattern was driven by the differential transcript concentrations of ifit5 (interferon-induced protein with tetratricopeptide repeats 5), isg15 (interferon-stimulated gene 15), rsad2 (viperin alias radical S-adenosyl methionine domain containing 2), znfx1 (NFX1-type zinc finger containing protein), cat (catalase), mpo (myeloperoxidase) and il18 (interleukin 18; Figure 2B). Similarly to the head kidney, the overall expression pattern in the gill was modulated in a time-dependent manner (Figure 3A). In this case, the first principal component explained 29.2% of the variance and this effect was primarily driven by cd40 (cluster of differentiation 40), irf1 (interferon regulatory factor 1) and, like in the head kidney, isg15, rsad2, mpo and il18 (Figure 3B).
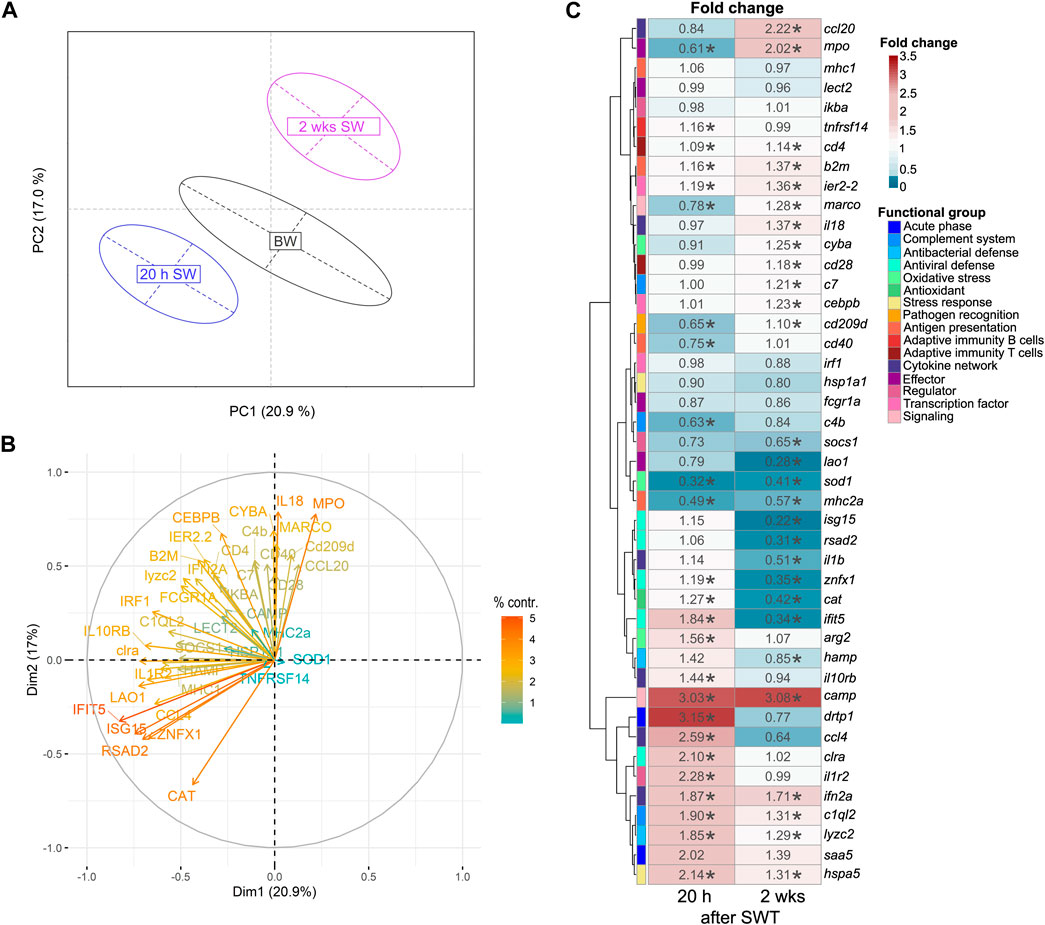
FIGURE 2. Principal component analysis (A) of 44 stress-and immune-regulated genes in the head kidney of Atlantic salmon after 8 weeks experimental period in brackish water (BW, T1), 20 h in seawater (T2) and 2 weeks in seawater (T3). Factor map (B) explaining the contribution of individual genes to the overall separation of the PCA. Length and color of the arrows indicate percent contribution of each gene. Heatmap (C) of fold changes for the genes investigated in relation to timepoint T1 (BW) based on relative gene expression values. Statistical significance compared to timepoint T1 was assessed using multiple contrast tests (*, p < 0.05). The rows represent different genes categorized into functional groups as illustrated in the legend on the right. The columns display the time points 20 h in seawater (T2) and 2 weeks in seawater (T3). Each cell is colorized based on the fold change of that gene, as visualized in the legend on the right. For each figure panel (A–C) n = 35–50 per timepoint.
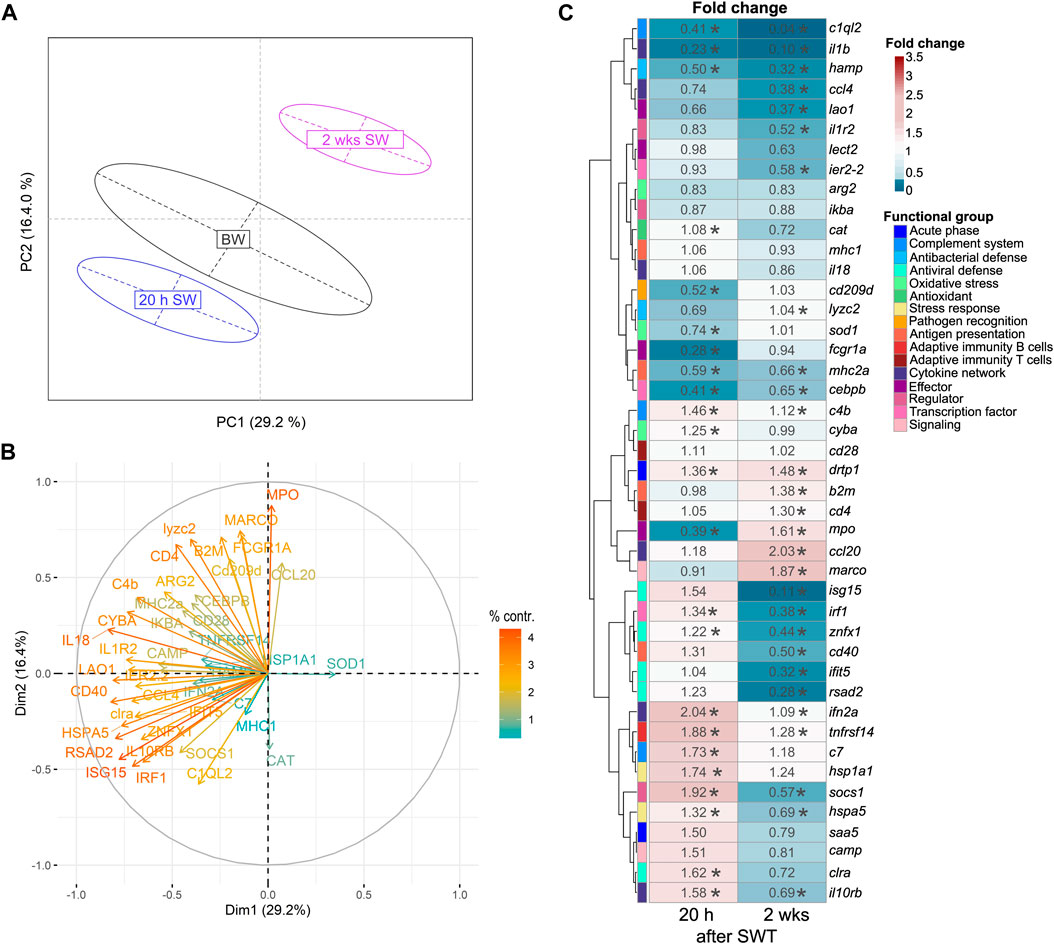
FIGURE 3. Principal component analysis (A) of 44 stress-and immune-regulated genes in the gill of Atlantic salmon after 8 weeks experimental period in brackish water (BW, T1), 20 h in seawater (T2) and 2 weeks in seawater (T3). Factor map (B) explaining the contribution of individual genes to the overall separation of the PCA. Length and color of the arrows indicate percent contribution of each gene. Heatmap (C) of fold changes for the genes investigated in relation to timepoint T1 (BW) based on relative gene expression values. Statistical significance compared to timepoint T1 was assessed using multiple contrast tests (*, p < 0.05). The rows represent different genes categorized into functional groups as illustrated in the legend on the right. The columns display the time points 20 h in seawater (T2) and 2 weeks in seawater (T3). Each cell is colorized based on the fold change of that gene, as visualized in the legend on the right. For each figure panel (A–C) n = 36–50 per timepoint.
After seawater transfer, none of the selected genes was increased in the gill above the defined threshold for basal levels (fc > 3). In contrast, the levels of several genes were significantly reduced (fc < 0.33) in the gill after SWT, such as the reduced (0.28-fold; p < 0.001) transcript level of fcgr1a (Fc gamma receptor Ia) shortly after SWT (Figure 3C). Several genes were reduced at both 20 h and 2 weeks after SWT (Figure 3C), including c1ql2 (complement C1q-like 2; 0.41- and 0.04-fold; p < 0.001); il1b (interleukin 1β; 0.23- and 0.10-fold; p < 0.001); and hamp (hepcidin; 0.5- and 0.32-fold; p < 0.001). The antiviral genes isg15, rsad2 and ifit5 were only found significantly reduced (up to 0.11-fold; p < 0.001) at 2 weeks after salinity change.
The transcript profile in the head kidney differed from that in the gill. Most notably, the transcript levels of camp (cathelicidin) were enhanced by > 3-fold 20 h and 2 weeks after seawater transfer (p < 0.001, Figure 2C). Furthermore, drtp1 (differentially regulated trout protein 1) and saa5 (serum amyloid A-5 protein) were 2.01- to 3.15-fold induced 20 h after seawater transfer (p < 0.001). After salinity change, also reduced levels of genes involved in different immune pathways were observed in the head kidney. Sod1 (superoxide dismutase 1) was reduced 20 h (0.32-fold) and 2 weeks (0.41-fold) after seawater transfer (p < 0.001). Furthermore, lao1 was reduced (0.28-fold; p < 0.001) 2 weeks after seawater transfer. Noteworthy, the genes isg15, rsad2, znfx1 and ifit5 which are all involved in antiviral defense, were reduced (0.22- to 0.35-fold; p < 0.001) 2 weeks after seawater transfer.
3.3 Mitigation potential of microalgae on immune and stress responses
At the end of the trial in seawater, the condition of the salmon, their hepatosomatic index (HSI) and spleen somatic index (SSI) across all diet groups was similar (Table 2). Moreover, no diet-dependent effects were observed in plasma concentrations of sodium and chloride after 20 h in seawater (Figure 4). Plasma cortisol levels decreased significantly (p < 0.001) between timepoints in seawater for fish fed CD but remained unchanged in SW for the other diet groups (Figure 5A). Furthermore, cortisol levels after 20 h in seawater were elevated as compared to control fish in brackish water (Supplementary Figure S1). Glucose levels increased over time in seawater, which was significant for fish fed AP (p < 0.01) and SL (p < 0.001; Figure 5B). The protein levels of myeloperoxidase increased over time in seawater for fish fed CD (p < 0.05), CVB (p < 0.001) and AP (p < 0.01; Figure 6A). At both timepoints, the concentration of Sod1 was not significantly different in the microalgae-fed groups as compared to the control group (Figure 6B). However, for fish fed CVI and TC levels decreased over time in seawater (p < 0.05; Figure 6B).
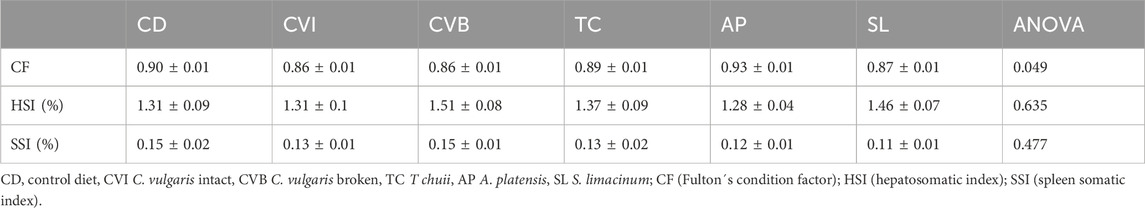
TABLE 2. Organ indices (n = 9, mean ± sem) of Atlantic salmon fed with six microalgae enriched diets sampled 2 weeks in seawater (T3).
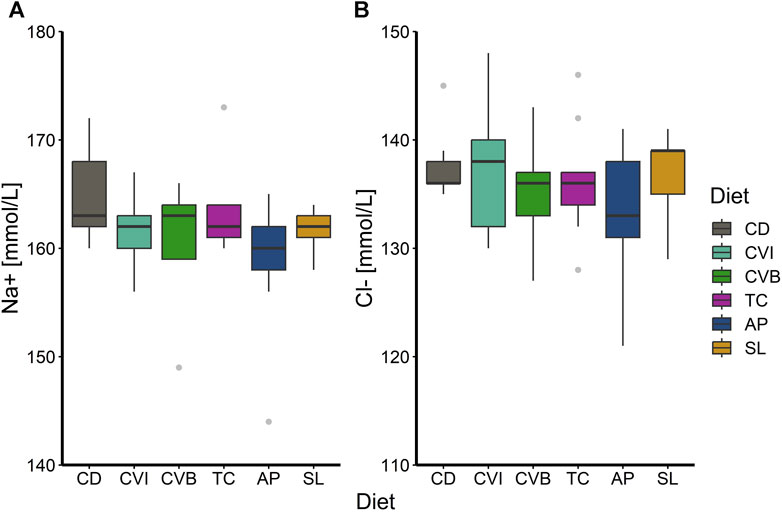
FIGURE 4. Boxplot with median and 1.5 x interquartile range whiskers showing sodium-ion (A) and chloride-ion (B) concentration in plasma of Atlantic salmon 20 h in seawater (T2) fed with the microalgae enriched diets C. vulgaris intact (CVI), C. vulgaris broken (CVB), T. chuii (TC), A. platensis (AP) or S. limacinum (SL) at 8% inclusion or a control diet (CD); n = 9. Note that the y-axis in panel (A) and (B) does not start at zero.
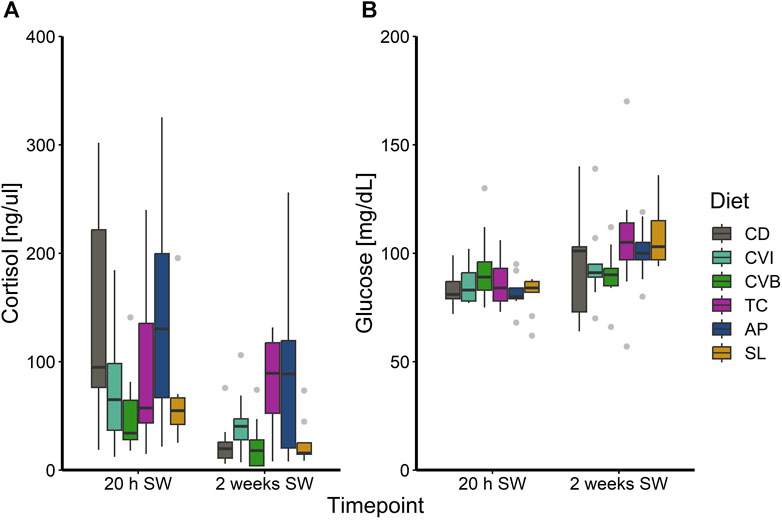
FIGURE 5. Boxplot with median and 1.5 x interquartile range whiskers showing plasma cortisol (A) and glucose (B) of Atlantic salmon 20 h in seawater (T2) and 2 weeks in seawater (T3) fed with the microalgae enriched diets C. vulgaris intact (CVI), C. vulgaris broken (CVB), T. chuii (TC), A. platensis (AP) or S. limacinum (SL) at 8% inclusion or a control diet (CD); n = 9.
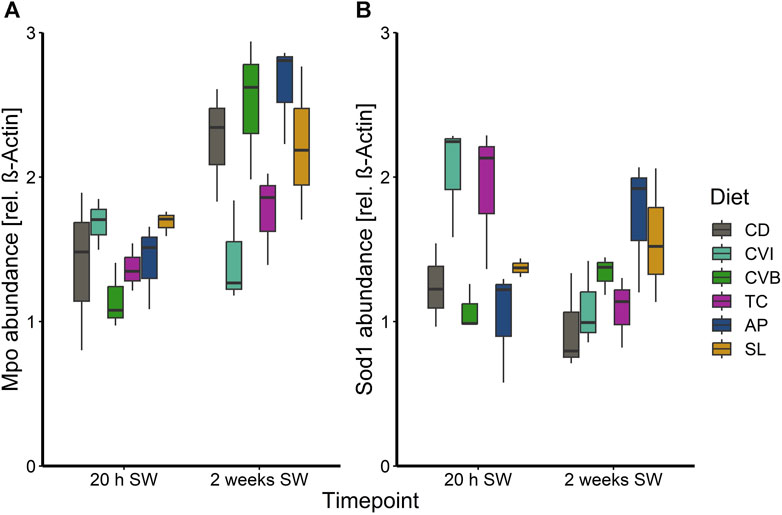
FIGURE 6. Boxplot with median and 1.5 x interquartile range whiskers showing protein abundance of myeloperoxidase (A) and superoxide dismutase 1 (B) in liver tissue of Atlantic salmon 20 h in seawater (T2) and 2 weeks in seawater (T3) fed with the microalgae enriched diets C. vulgaris intact (CVI), C. vulgaris broken (CVB), T. chuii (TC), A. platensis (AP) or S. limacinum (SL) at 8% inclusion or a control diet (CD); n = 3 pool of 3 fish per tank.
The different diet groups followed a similar trend of gene expression over the three timepoints corresponding to brackish water phase, acute seawater transfer and 2 weeks in seawater (Supplementary Figure S2, S3). None of the investigated genes in the head kidney were differentially expressed comparing microalgae fed fish to control fish, although a diet effect in the main model was detected (Supplementary Table S2). A few genes were found to be differentially regulated in the gill of salmon fed particular microalgae as compared to the control (Supplementary Table S3; Figure 7). Clra, c1ql2 and camp were reduced in fish fed AP 20 h after seawater transfer (Figures 7A–C). While expression of clra was reduced only modest as compared to the control (0.43-fold, p < 0.05), c1ql2 (0.10-fold, p < 0.01) and camp (0.23-fold, p = 0.077) were more strongly reduced.
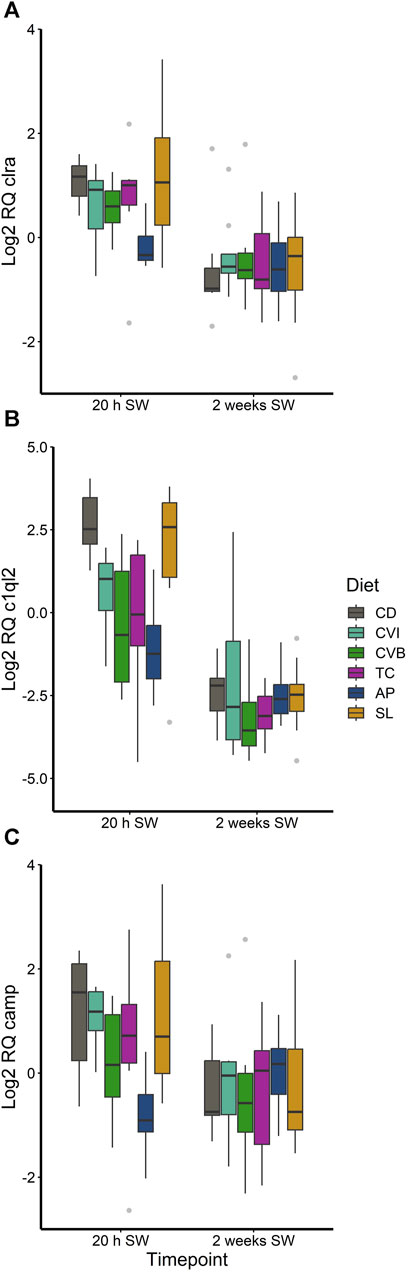
FIGURE 7. Boxplot with median and 1.5 x interquartile range whiskers showing gene expression of clra (A), c1ql2 (B) and camp (C) in the gill of Atlantic salmon 20 h in seawater (SW) and 2 weeks in seawater fed with the microalgae enriched diets C. vulgaris intact (CVI), C. vulgaris broken (CVB), T. chuii (TC), A. platensis (AP) or S. limacinum (SL) at 8% inclusion or a control diet (CD), n = 6-9.
4 Discussion
4.1 Salinity change following BW acclimation does not impair the osmoregulatory ability
Upon completed smoltification salmon smolts show hypo-osmoregulatory capacity and are ready for entering seawater. However, it is generally accepted that holding salmon smolts for longer periods after smoltification in freshwater or brackish water can induce a process known as desmoltification, where the physiological ability to tolerate seawater is transiently lost (Mortensen and Damsgård, 1998; Stefansson et al., 1998). Although the salmon in the present study were reared in brackish water for several months, the fish were able to regulate their ion balance well within physiological limits upon transfer to seawater. The slight increase in plasma Na+ and Cl− concentrations 20 h after SWT returned to basal levels after 2 weeks in SW, similar to findings from McCormick et al. (2013). In the gill, the sodium-potassium pump Na+/K+-ATPase (NKA), plays a crucial role in regulating osmolality and ion homeostasis: the NKA regulates the absorption of ions in freshwater and their secretion in seawater (Zaugg and Wagner, 1973; Evans et al., 2005). While the NKA isoform NKA α1a dominates in pre-smolts in freshwater, levels of the seawater isoform NKA α1b increase during smoltification and dominate in seawater (McCormick et al., 2009). The ability to regulate osmolality properly in seawater was also reflected by a greatly increased nkaα1b/nkaα1a ratio in the gill. In line with previous findings (Nilsen et al., 2007; McCormick et al., 2013) nkaα1b expression increased further in SW, while expression of nkaα1a decreased strongly.
4.2 Acute and prolonged salinity exposure induce divergent responses in the head kidney and gill of Atlantic salmon
Seawater transfer is a critical process during the life-cycle of Atlantic salmon in the wild, as well as under production. To cope with the associated altered environmental conditions, a stress response is triggered that leads to the activation of the hypothalamic-pituitary-interrenal (HPI) axis (Wendelaar Bonga, 1997). Hereby, stress hormones (such as cortisol and adrenaline) are released that induce a variety of physiological changes, allowing the fish to adapt to new the environment and return to internal homeostasis (Wendelaar Bonga, 1997). In addition to changes in, for example, oxygen uptake, metabolism or hydromineral balance, the immune system is also affected, mainly with regard to lymphocyte activities and the levels of circulating cytokines (Tort, 2011). Due to the wide range of stressors and the duration of their exposure, the stress response and also the interlinked immune response adapts accordingly, at the systemic but also at the local tissue level.
In this study, we examined how salinity change affects the expression of immune- and stress-related genes in the head kidney and gill, to get an understanding of the systemic and local immune and stress response after seawater transfer. We found that in the head kidney, the genes dominating the overall gene-expression pattern were mainly involved in antiviral defense (ifit5, isg15, rsad2, znfx1), and additionally in anti-oxidant activity (cat), antimicrobial defense (mpo) and Th1 immunity (il18). Transcriptional suppression of immune genes, particularly related to antiviral defense, were found during smoltification, and these effects continued after seawater transfer (Johansson et al., 2016). Interestingly, our data indicates that salinity change per se has an effect on antiviral immunity. This might be an evolutionary conserved response resulting from an energy-allocation trade-off upon acclimation to seawater. Concomitantly, the modulated expression of antiviral genes may be directly connected to the abundance of different viruses in freshwater and marine habitats (Maranger and Bird, 1995; Clasen et al., 2008) which requires further investigation.
Similar to the head kidney, the overall expression pattern in the gill was dominated by the antimicrobial gene mpo and antiviral-related genes isg15 and rsad2. In addition, the antiviral gene irf1 and the transmembrane protein-encoding gene cd40 (alias tnfrsf5) majorly influenced the expression pattern of the gill. CD40 is a marker for antigen-presenting cells including neutrophilic granulocytes, dendritic cells and macrophages (Lagos et al., 2012) and could thus indicate the remodeling of the cellular immunity in the gill after salinity change, which has been found to occur in Atlantic salmon during smoltification and following seawater transfer from freshwater (West et al., 2021).
The gill is in direct contact with the environment and is therefore responsible for a local immune response. In our study, salinity change caused a suppression of several genes in the gill. Shortly after SWT, fcgr1a encoding an immunoglobulin Fc receptor (FcRs) was reduced. These receptors are present on the surface of phagocytes (Stafford et al., 2006). The reduction of fcgr1a likely indicate a decreased number of phagocytes in the gill maybe associated with an impaired antigen recognition capability. This effect is probably related to the acute stress resulting from acute salinity change, as 2 weeks later fcgr1a returned to the level similar to before seawater transfer. Furthermore, we observed decreased transcript levels of c1ql2 20 h and 2 weeks after salinity change. C1ql2 encodes a protein which is structurally related to C1q (Köbis et al., 2017) and although its precise role is not fully defined in salmonids yet, c1ql2 might serve as a carbohydrate-binding protein (known as lectins) and bridge between innate and adaptive immunity (Köbis et al., 2017; Hillestad et al., 2020). Johansson et al. (2016) described a similar pattern in the gill of Atlantic salmon one and 3 weeks after seawater transfer, with c1ql2 transcript numbers in gill half those relative to pre-smolts. Additionally, two genes (il1b, hamp) involved in early immune-related pathways were reduced 20 h after SWT, and still 2 weeks later. Il1b encodes the pro-inflammatory cytokine il-1β (Secombes et al., 2011) and hamp encodes for the antimicrobial peptide hepcidin (Douglas et al., 2003). Lastly, the transcript levels of the antiviral defense genes isg15, rsad2 and ifit5 were reduced 2 weeks after SWT and this observation may suggest that alternative defense strategies may be more beneficial in salmon exposed to salinity change. In this context it may be noted that isg15 and rsad2 were downregulated both in the head kidney and gill during acclimation to seawater and could thus indicate a systemic reaction.
Several genes in the head kidney were activated in our study after salinity change. Especially camp was greatly enhanced at both timepoints in seawater. Camp encodes an antimicrobial peptide with immunomodulatory activities (Scocchi et al., 2009). Furthermore, two biomarker genes (drtp1, saa5) of the acute-phase and stress response (Talbot et al., 2009; Lee et al., 2017) in fish were induced shortly after SWT. In parallel, several genes were suppressed in the head kidney after salinity change, similar as observed in the gill. Sod1 is one of the genes, which was downregulated 20 h and 2 weeks after SWT. Since this biomarker gene of oxidative stress encodes a potent antioxidant, the reduction of sod1 therefore suggests a diminished antioxidant defense or decreased amount of reactive oxygen species (ROS) in the gill after salinity change. In previous research, a change in salinity has been found to induce oxidative stress in fish (Liu et al., 2007; Lushchak, 2011). Blood samples of the freshwater fish Nile tilapia Oreochromus niloticus had a lower superoxide dismutase and catalase activity when fish were held in higher salinity (4 or 12 psu, respectively) than in the control group for 2 weeks (Elarabany, 2017). In contrast, in liver of anadromous chum salmon Oncorhynchus keta, superoxide dismutase and catalase activity was significantly enhanced in fish held for 42 days in 8, 16 and 24 psu compared to fish kept in 0 psu (Li et al., 2022). Moreover, in addition to antiviral genes (isg15, rsad2, znfx1, ifit5), lao1 was also reduced merely 2 weeks after SWT. Lao1 encodes for L-amino acid oxidases with antibacterial and antiparasitic activity (Kitani and Nagashima, 2020). The downregulation of these genes with important roles in immune defense may be due to the low pathogen pressure in RAS and therefore, energy could be rather directed to cope with salinity change than to maintain increased immune barriers.
In this study, expression of several immune-genes (c1ql2, fcgr1a, hamp, ifit5, il1b, isg15, rsad2) was reduced in the gill while in the head kidney both an induction (camp, drtp1, saa5) and reduction (ifit5, isg15, lao1, sod1, znfx1) of genes was observed after salinity change. However, these alterations in transcript levels were rather modest, possibly due to the transfer within the protected RAS environment with a lower pathogen pressure than the open marine environment. Due to different experimental setups, it is difficult to compare our results with other studies investigating freshwater to seawater transfer (Johansson et al., 2016; Karlsen et al., 2018; Lund et al., 2022), but the general pattern of gene expression is similar.
4.3 Microalgae have limited potential for modulating the response to changing salinity
Microalgae could contribute to improve the fish’s ability to cope with salinity, as their immune-stimulatory and stress-modulatory properties have previously been demonstrated (Cerezuela et al., 2012; Zhang et al., 2014; Raji et al., 2018). Since each of the microalgae species have their own beneficial characteristics (based on various functional compounds), this study evaluated the effects of different microalgae-enriched functional diets on the stress and immune status of salmon after salinity change. The overall growth performance among the different diet groups was not different over the 10-week trial period (Mueller et al., 2023) and furthermore similar condition and organ indices among the diet groups indicate that the incorporated microalgae did not impair growth nor energy reserves of the fish.
Cortisol is the primary stress hormone in fish and rapidly elevated in response to acute stress (Wendelaar Bonga, 1997; Barton, 2002) and alongside with glucose used as the primary stress indicator in fishes (Sopinka et al., 2016). Moreover, high cortisol levels induce hypo-osmoregulatory ability during smoltification (Langhorne and Simpson, 1986). We also observed increased levels of cortisol in post-smolts after 20 h in seawater, probably resulting from a response to salinity change rather than to handling, as cortisol in similarly handled fish that had been transferred back into brackish water, remained on a basal level. After 2 weeks in seawater, the levels of cortisol decreased, most notable for the control group and likely due to adaptation to the new environment of the fish. Notably, the functional diets did not influence this primary stress response. Interestingly glucose levels showed an opposite trend and we can only speculate why glucose levels were not elevated initially but increased slightly over time in seawater. The slightly lower glucose levels at 20 h in SW might have resulted from the usage of glucose as an energy substrate.
In addition to the transcriptional profiles in the head kidney and gills, we determined the protein levels of myeloperoxidase in the liver. Myeloperoxidase is an antimicrobial enzyme characteristic for neutrophilic granulocytes (Castro et al., 2008; Aratani, 2018; Chen et al., 2019). Fish fed with the control diet, or diets enriched with C. vulgaris broken and A. platensis showed an increase of Mpo levels over time in seawater, suggesting an increase in antimicrobial activity in the liver of these fish. The hepatic abundance of Sod1 can give insights into the antioxidant activity in the liver of the fish. The high Sod1 concentration in liver of fish fed C. vulgaris intact and T. chuii acutely after salinity change (20 h in seawater) may provide a better oxidative state during this time period but protein levels potentially decreased 2 weeks in seawater because lower ROS levels were subsequently present. In line with these results, previous research on salmon fed with a pre-extruded Tetraselmis diet also revealed significantly induced sod1 expression in the liver (Sørensen et al., 2021).
Overall, microalgae enriched diets only induced subtle changes in immune-gene expression. This can be likely explained by the fact that feed intake in seawater was drastically reduced (Mueller et al., 2023), which has limited the uptake of functional compounds and their immune modulatory effects. The functional diet supplemented with A. platensis caused a reduction of genes in the gill involved in antimicrobial defense (camp) and pathogen recognition (clra, c1ql2) and we can only speculate if these modest reductions in gene expressions are reflected at the protein level. However, extracts of A. platensis showed strong antimicrobial activity (Metekia and Ulusoy, 2023) and this could have reduced the need to produce antimicrobial peptides as well as pathogen recognition receptors, explaining its reduced expression.
Altogether, the transfer from brackish water to seawater had a more pronounced effect on the immune status of the salmon than the diet, which caused subtle changes in immune-gene expression. This is an important finding for the development of future stress and immune-mitigation strategies in aquaculture.
5 Conclusion
The current study evaluated the effect of salinity change from brackish water to seawater on Atlantic salmon. Although the fish showed full osmoregulatory ability after rearing in brackish water, salinity change caused a reduction of immune genes in the head kidney and gill even several months after smoltification. The herein-investigated microalgae had only marginal potential to modulate the observed responses. Future research should explore whether other functional feed additives, e.g., beta-glucans can stimulate the immune system of salmon during this critical period. Furthermore, different salinity adaptation strategies and their effect on the stress and immune status of Atlantic salmon need to be evaluated experimentally.
Data availability statement
The raw data supporting the conclusion of this article will be made available by the authors, without undue reservation.
Ethics statement
The animal study was approved by the animal welfare officer of the “Fraunhofer IMTE Büsum” and the local authority of Schleswig-Holstein, according to the German animal welfare law (NTP—ID 00043858-1-0). The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
DM: Data curation, Visualization, Writing–original draft, Investigation, Formal Analysis. JM: Writing–original draft, Data curation, Investigation, Methodology, Visualization, Conceptualization, Formal Analysis. JL: Data curation, Writing–review and editing. TS: Data curation, Writing–review and editing. EM: Funding acquisition, Writing–review and editing. HS: Supervision, Writing–review and editing, Project administration. AR: Supervision, Writing–review and editing. CS: Conceptualization, Funding acquisition, Writing–review and editing, Project administration. TG: Conceptualization, Funding acquisition, Writing–review and editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was funded by the BMBF within the project BioFiA (Project numbers 031B09151E1, 031B09151E2, 031B09151E8) and the project AQUATOR (Project number 031B0915H3).
Acknowledgments
We are grateful to Michael Schlachter for his advice on experimental design and Mario Hasler for statistical advice. The authors would like to thank Matteo Pauly, Anna Simon, Thilo Kock and Frederike Schmachtl for their help during sampling. Julian Krinitskij is greatly acknowledged for his excellent technical assistance.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2024.1338858/full#supplementary-material
References
Abdel-Tawwab M., Ahmad M. H. (2009). Live Spirulina (Arthrospira platensis) as a growth and immunity promoter for Nile tilapia, Oreochromis niloticus (L.), challenged with pathogenic Aeromonas hydrophila. Aquac. Res. 40 (9), 1037–1046. doi:10.1111/j.1365-2109.2009.02195.x
An B. K., Kim K. E., Jeon J. Y., Lee K. W. (2016). Effect of dried Chlorella vulgaris and Chlorella growth factor on growth performance, meat qualities and humoral immune responses in broiler chickens. SpringerPlus 5 (718), 718. doi:10.1186/s40064-016-2373-4
Aratani Y. (2018). Myeloperoxidase: its role for host defense, inflammation, and neutrophil function. Archives Biochem. Biophysics 640 (January), 47–52. doi:10.1016/j.abb.2018.01.004
Bang Jensen B., Qviller L., Toft N. (2020). Spatio-temporal variations in mortality during the seawater production phase of Atlantic salmon (Salmo salar) in Norway. J. Fish Dis. 43 (4), 445–457. doi:10.1111/jfd.13142
Barreto M. O., Rey Planellas S., Yang Y., Phillips C., Descovich K. (2022). Emerging indicators of fish welfare in aquaculture. Rev. Aquac., 14(1), 343–361. doi:10.1111/raq.12601
Barton B. A. (2002). Stress in fishes: a diversity of responses with particular reference to changes in circulating corticosteroids. Integr. Comp. Biol. 42 (3), 517–525. doi:10.1093/icb/42.3.517
Birnie-Gauvin K., Costantini D., Cooke S. J., Willmore W. G. (2017). A comparative and evolutionary approach to oxidative stress in fish: a review. Fish Fish. 18 (5), 928–942. doi:10.1111/faf.12215
Bougeard S., Dray S. (2018). Supervised multiblock analysis in R with the ade4 package. J. Stat. Softw. 86, 1–17. doi:10.18637/jss.v086.i01
Castro R., Piazzon M. C., Noya M., Leiro J. M., Lamas J. (2008). Isolation and molecular cloning of a fish myeloperoxidase. Mol. Immunol. 45 (2), 428–437. doi:10.1016/j.molimm.2007.05.028
Cerezuela R., Guardiola F. A., González P., Meseguer J., Esteban M. Á. (2012). Effects of dietary Bacillus subtilis, Tetraselmis chuii, and Phaeodactylum tricornutum, singularly or in combination, on the immune response and disease resistance of sea bream (Sparus aurata L.). Fish Shellfish Immunol. 33 (2), 342–349. doi:10.1016/j.fsi.2012.05.004
Chen C., Yang B., Abbas Raza S. H., Zhang D., Wu T., Zhang Z., et al. (2019). Role of Myeloperoxidase of northern snakehead (Channa argus) in Aeromonas veronii infection. Microb. Pathog. 135 (July), 103622. doi:10.1016/j.micpath.2019.103622
Clasen J. L., Bridgen S. M., Payet J. P., Suttle C. A. (2008). Evidence that viral abundance across oceans and lakes is driven by different biological factors. Freshw. Biol. 53 (6), 1090–1100. doi:10.1111/j.1365-2427.2008.01992.x
Dalsgaard J., Lund I., Thorarinsdottir R., Drengstig A., Arvonen K., Pedersen P. B. (2013). Farming different species in RAS in Nordic countries: current status and future perspectives. Aquac. Eng. 53, 2–13. doi:10.1016/j.aquaeng.2012.11.008
Dawood M. A. O., Koshio S., Esteban M. Á. (2018). Beneficial roles of feed additives as immunostimulants in aquaculture: a review. Rev. Aquac. 10 (4), 950–974. doi:10.1111/raq.12209
de Mattos B. O., López-Olmeda J. F., Guerra-Santos B., Ruiz C. E., García-Beltrán J. M., Ángeles-Esteban M., et al. (2019). Coping with exposure to hypoxia: modifications in stress parameters in gilthead seabream (Sparus aurata) fed spirulina (Arthrospira platensis) and brewer’s yeast (Saccharomyces cerevisiae). Fish Physiology Biochem. 45 (6), 1801–1812. doi:10.1007/s10695-019-00677-8
Douglas S. E., Gallant J. W., Liebscher R. S., Dacanay A., Tsoi S. C. M. (2003). Identification and expression analysis of hepcidin-like antimicrobial peptides in bony fish. Dev. Comp. Immunol. 27 (6–7), 589–601. doi:10.1016/S0145-305X(03)00036-3
Elarabany N. (2017). Effects of Salinity on Some Haematological and Biochemical Parameters in Nile Tilapia, <i&gt;Oreochromus niloticus&lt;/i&gt;. Agric. For. Fish. 6 (6), 200–205. doi:10.11648/j.aff.20170606.13
Evans D. H., Piermarini P. M., Choe K. P. (2005). The multifunctional fish gill: dominant site of gas exchange, osmoregulation, acid-base regulation, and excretion of nitrogenous waste. Physiol. Rev. 85 (1), 97–177. doi:10.1152/physrev.00050.2003
Forseth T., Barlaup B. T., Finstad B., Fiske P., Gjøsæter H., Falkegård M., et al. (2017). The major threats to Atlantic salmon in Norway. ICES J. Mar. Sci. 74 (6), 1496–1513. doi:10.1093/icesjms/fsx020
Glover K. A., Wennevik V., Hindar K., Skaala Ø., Fiske P., Solberg M. F., et al. (2020). The future looks like the past: introgression of domesticated Atlantic salmon escapees in a risk assessment framework. Fish Fish., 21(6), 1077–1091. doi:10.1111/faf.12478
Hasler M., Hothorn L. A. (2008). Multiple contrast tests in the presence of heteroscedasticity. Biometrical J. 50 (5), 793–800. doi:10.1002/bimj.200710466
Haugarvoll E., Bjerkås I., Nowak B. F., Hordvik I., Koppang E. O. (2008). Identification and characterization of a novel intraepithelial lymphoid tissue in the gills of Atlantic salmon. J. Anat. 213 (2), 202–209. doi:10.1111/j.1469-7580.2008.00943.x
Hillestad B., Makvandi-Nejad S., Krasnov A., Moghadam H. K. (2020). Identification of genetic loci associated with higher resistance to pancreas disease (PD) in Atlantic salmon (Salmo salar L.). BMC Genomics 21 (1), 388–413. doi:10.1186/s12864-020-06788-4
Hoar W. S. (1988). 4 the physiology of smolting salmonids. Fish. Physiol. 11 (PB), 275–343. doi:10.1016/S1546-5098(08)60216-2
Hussein M. M., Sayed R. K. A., Mokhtar D. M. (2023). Structural and immunohistochemical analysis of the cellular compositions of the liver of molly fish (Poecilia sphenops), focusing on its immune role. Zool. Lett. 9 (1), 1–13. doi:10.1186/s40851-022-00200-7
Johansson L. H., Timmerhaus G., Afanasyev S., Jørgensen S. M., Krasnov A. (2016). Smoltification and seawater transfer of Atlantic salmon (Salmo salar L.) is associated with systemic repression of the immune transcriptome. Fish Shellfish Immunol. 58, 33–41. doi:10.1016/j.fsi.2016.09.026
Karlsen C., Ytteborg E., Timmerhaus G., Høst V., Handeland S., Jørgensen S. M., et al. (2018). Atlantic salmon skin barrier functions gradually enhance after seawater transfer. Sci. Rep. 8 (1), 9510–9512. doi:10.1038/s41598-018-27818-y
Kassambara A., Mundt F. (2020). Package “factoextra”. Extract and visualize the results of multivariate data analyses. Documentation R programme.
Kitani Y., Nagashima Y. (2020). L-Amino acid oxidase as a fish host-defense molecule. Fish Shellfish Immunol. 106 (August), 685–690. doi:10.1016/j.fsi.2020.08.028
Köbis J. M., Rebl H., Goldammer T., Rebl A. (2017). Multiple gene and transcript variants encoding trout C-polysaccharide binding proteins are differentially but strongly induced after infection with Aeromonas salmonicida. Fish Shellfish Immunol. 60, 509–519. doi:10.1016/j.fsi.2016.11.021
Krasnov A., Afanasyev S., Nylund S., Rebl A. (2020). Multigene expression assay for assessment of the immune status of Atlantic Salmon. Genes. 11 (11), 1236–1311. doi:10.3390/genes11111236
Lagos L. X., Iliev D. B., Helland R., Rosemblatt M., Jørgensen J. B. (2012). CD40L - a costimulatory molecule involved in the maturation of antigen presenting cells in Atlantic salmon (Salmo salar). Dev. Comp. Immunol. 38 (3), 416–430. doi:10.1016/j.dci.2012.07.011
Langhorne P., Simpson T. H. (1986). The interrelationship of cortisol, Gill (Na + K) ATPase, and homeostasis during the Parr-Smolt transformation of atlantic salmon (Salmo salar L.). General Comp. Endocrinol. 61 (2), 203–213. doi:10.1016/0016-6480(86)90198-X
Lee P. T., Bird S., Zou J., Martin S. A. M. (2017). Phylogeny and expression analysis of C-reactive protein (CRP) and serum amyloid-P (SAP) like genes reveal two distinct groups in fish. Fish Shellfish Immunol. 65, 42–51. doi:10.1016/j.fsi.2017.03.037
Li P., Liu W., Lu W., Wang J. (2022). Biochemical indices, gene expression, and SNPs associated with salinity adaptation in juvenile chum salmon (Oncorhynchus keta) as determined by comparative transcriptome analysis. PeerJ 10, e13585. doi:10.7717/peerj.13585
Liu Y., Wang W. N., Wang A. L., Wang J. M., Sun R. Y. (2007). Effects of dietary vitamin E supplementation on antioxidant enzyme activities in Litopenaeus vannamei (Boone, 1931) exposed to acute salinity changes. Aquaculture 265 (1–4), 351–358. doi:10.1016/j.aquaculture.2007.02.010
Lund H., Bakke A., Boysen P., Afanasyev S., Rebl A., Manji F., et al. (2022). Evaluation of immune status in two cohorts of atlantic salmon raised in different aquaculture systems (case study). Genes. 13 (5), 736. doi:10.3390/genes13050736
Luo Z., Ye H. M., Gao Y., Ling S. C., Wei C. C., Zhu X. (2018). Chlorella additive increased growth performance, improved appetite and immune response of juvenile crucian carp Carassius auratus. Aquac. Res. 49 (10), 3329–3337. doi:10.1111/are.13797
Lushchak V. I. (2011). Environmentally induced oxidative stress in aquatic animals. Aquat. Toxicol. 101 (1), 13–30. doi:10.1016/j.aquatox.2010.10.006
Maranger R., Bird D. F. (1995). Viral abundance in aquatic systems: a comparison between marine and fresh waters. Mar. Ecol. Prog. Ser., 121, 217–226. doi:10.3354/meps121217
Marimuthu V., Shanmugam S., Sarawagi A. D., Kumar A., Kim I. H., Balasubramanian B. (2022). A glimpse on influences of feed additives in aquaculture. EFood 3 (1–2), 1–10. doi:10.1002/efd2.6
McCormick S. D., Hansen L. P., Quinn T. P., Saunders R. L. (1998). Movement, migration, and smolting of Atlantic salmon (Salmo salar). Can. J. Fish. Aquatic Sci. 55 (Suppl. 1), 77–92. doi:10.1139/d98-011
McCormick S. D., Regish A. M., Christensen A. K. (2009). Distinct freshwater and seawater isoforms of Na+/K +-ATPase in gill chloride cells of Atlantic salmon. J. Exp. Biol. 212 (24), 3994–4001. doi:10.1242/jeb.037275
McCormick S. D., Regish A. M., Christensen A. K., Björnsson B. T. (2013). Differential regulation of sodium-potassium pump isoforms during smolt development and seawater exposure of atlantic salmon. J. Exp. Biol. 216 (7), 1142–1151. doi:10.1242/jeb.080440
McGinnity P., Prodöhl P., Ferguson A., Hynes R., Maoiléidigh N. Ó., Baker N., et al. (2003). Fitness reduction and potential extinction of wild populations of Atlantic salmon, Salmo salar, as a result of interactions with escaped farm salmon. Proc. R. Soc. B Biol. Sci. 270 (1532), 2443–2450. doi:10.1098/rspb.2003.2520
Melingen G. O., Stefansson S. O., Berg A., Wergeland H. I. (1995). Changes in serum protein and IgM concentration duringsmolting and early post-smolt period in vaccinated and unvaccinated Atlantic salmon (Salmo salar L.). Fish Shellfish Immunol. 5 (3), 211–221. doi:10.1016/S1050-4648(05)80015-4
Metekia W. A., Ulusoy B. H. (2023). Antimicrobial activity of Spirulina platensis extract on total mesophilic and psychrophilic bacteria of fresh tilapia fillet. Sci. Rep. 13 (1), 13081. doi:10.1038/s41598-023-40260-z
Morris H. J., Carrillo O., Almarales A., Bermúdez R. C., Lebeque Y., Fontaine R., et al. (2007). Immunostimulant activity of an enzymatic protein hydrolysate from green microalga Chlorella vulgaris on undernourished mice. Enzyme Microb. Technol. 40 (3), 456–460. doi:10.1016/j.enzmictec.2006.07.021
Mortensen A., Damsgård B. (1998). The effect of salinity on desmoltification in Atlantic salmon. Aquaculture, 168(1), 407–411. doi:10.1016/S0044-8486(98)00366-4
Mortensen H. S., Jacobsen E., Kolarevic J., Vang A. (2022). Exposing atlantic salmon post-smolts to fluctuating sublethal nitrite concentrations in a commercial recirculating aquaculture system (RAS) may have negative consequences. Front. Animal Sci. 3 (July), 1–7. doi:10.3389/fanim.2022.886071
Mueller J., Pauly M., Molkentin J., Ostermeyer U., van Muilekom D. R., Rebl A., et al. (2023). Microalgae as functional feed for Atlantic salmon: effects on growth, health, immunity, muscle fatty acid and pigment deposition. Front. Mar. Sci. 10 (October). doi:10.3389/fmars.2023.1273614
Nilsen T. O., Ebbesson L. O. E., Madsen S. S., McCormick S. D., Andersson E., Björnsson B. T., et al. (2007). Differential expression of gill Na+,K+-ATPase alpha- and beta-subunits, Na+,K+,2Cl-cotransporter and CFTR anion channel in juvenile anadromous and landlocked Atlantic salmon Salmo salar. J. Exp. Biol. 210 (16), 2885–2896. doi:10.1242/jeb.002873
Pereira H., Sardinha M., Santos T., Gouveia L., Barreira L., Dias J., et al. (2020). Incorporation of defatted microalgal biomass (Tetraselmis sp. CTP4) at the expense of soybean meal as a feed ingredient for juvenile gilthead seabream (Sparus aurata). Algal Res. 47 (March), 101869. doi:10.1016/j.algal.2020.101869
Press C. M., Dannevig B. H., Landsverk T. (1994). Immune and enzyme histochemical phenotypes of lymphoid and nonlymphoid cells within the spleen and head kidney of atlantic salmon (salmo salar l.). Fish Shellfish Immunol. 4 (2), 79–93. doi:10.1006/fsim.1994.1007
Press C. M. L., Evensen O. (1999). The morphology of the immune system in teleost fishes. Fish Shellfish Immunol. 9 (4), 309–318. doi:10.1006/fsim.1998.0181
Quiñones R. A., Fuentes M., Montes R. M., Soto D., León-Muñoz J. (2019). Environmental issues in Chilean salmon farming: a review. Rev. Aquac. 11 (2), 375–402. doi:10.1111/raq.12337
Raji A. A., Alaba P. A., Yusuf H., Abu Bakar N. H., Mohd Taufek N., Muin H., et al. (2018). Fishmeal replacement with Spirulina Platensis and Chlorella vulgaris in African catfish (Clarias gariepinus) diet: effect on antioxidant enzyme activities and haematological parameters. Res. Veterinary Sci. 119 (April), 67–75. doi:10.1016/j.rvsc.2018.05.013
Rességuier J., Dalum A. S., Du Pasquier L., Zhang Y., Koppang E. O., Boudinot P., et al. (2020). Lymphoid tissue in teleost gills: variations on a theme. Biology 9 (6), 127–214. doi:10.3390/biology9060127
Scocchi M., Pallavicini A., Salgaro R., Bociek K., Gennaro R. (2009). The salmonid cathelicidins: a gene family with highly varied C-terminal antimicrobial domains. Comp. Biochem. Physiology - B Biochem. Mol. Biol. 152 (4), 376–381. doi:10.1016/j.cbpb.2009.01.003
Secombes C. J., Wang T., Bird S. (2011). The interleukins of fish. Dev. Comp. Immunol. 35 (12), 1336–1345. doi:10.1016/j.dci.2011.05.001
Soares S., Green D. M., Turnbull J. F., Crumlish M., Murray A. G. (2011). A baseline method for benchmarking mortality losses in Atlantic salmon (Salmo salar) production. Aquaculture 314 (1–4), 7–12. doi:10.1016/j.aquaculture.2011.01.029
Sommerset I., Walde C. S., Bang Jensen B., Bornø B., Haukaas A., Brun E. (2020). Fiskehelserapporten 2019. Report 5a/2020. China: Norwegian Veterinary Institute.
Sopinka N. M., Donaldson M. R., O’Connor C. M., Suski C. D., Cooke S. J. (2016). 11 - stress indicators in fish. In C. B. Schreck, L. Tort, A. P. Farrell, and C. J. B. T.-F. P. Brauner (Eds.), Biology of stress in fish China, (Vol. 35, pp. 405–462). Academic Press. doi:10.1016/B978-0-12-802728-8.00011-4
Sørensen S. L., Ghirmay A., Gong Y., Dahle D., Vasanth G., Sørensen M., et al. (2021). Growth, chemical composition, histology and antioxidant genes of atlantic salmon (Salmo salar) fed whole or pre-processed nannochloropsis oceanica and tetraselmis sp. Fishes 6 (3), 23. doi:10.3390/fishes6030023
Stafford J. L., Wilson M., Nayak D., Quiniou S. M., Clem L. W., Miller N. W., et al. (2006). Identification and characterization of a FcR homolog in an ectothermic vertebrate, the channel catfish (Ictalurus punctatus). J. Immunol. 177 (4), 2505–2517. doi:10.4049/jimmunol.177.4.2505
Stefansson S. O., Berge Å. I., Gunnarsson G. S. (1998). Changes in seawater tolerance and gill Na+,K+-ATPase activity during desmoltification in Atlantic salmon kept in freshwater at different temperatures. Aquaculture, 168(1), 271–277. doi:10.1016/S0044-8486(98)00354-8
Talbot A. T., Smith T. J., Cairns M. T. (2009). Characterisation of the differentially regulated trout protein 1 (DRTP1) gene in rainbow trout (Oncorhynchus mykiss). Fish Shellfish Immunol. 26 (4), 589–598. doi:10.1016/j.fsi.2008.09.013
Tort L. (2011). Stress and immune modulation in fish. Dev. Comp. Immunol. 35 (12), 1366–1375. doi:10.1016/j.dci.2011.07.002
Wendelaar Bonga S. E. (1997). The stress response in fish. Physiol. Rev. 77 (3), 591–625. doi:10.1152/physrev.1997.77.3.591
West A. C., Mizoro Y., Wood S. H., Ince L. M., Iversen M., Jørgensen E. H., et al. (2021). Immunologic profiling of the Atlantic salmon gill by single nuclei transcriptomics. Front. Immunol. 12, 669889. doi:10.3389/fimmu.2021.669889
Ytrestøyl T., Hjelle E., Kolarevic J., Takle H., Rebl A., Afanasyev S., et al. (2022). Photoperiod in recirculation aquaculture systems and timing of seawater transfer affect seawater growth performance of Atlantic salmon (Salmo salar). J. World Aquac. Soc. March. doi:10.1111/jwas.12880
Ytrestøyl T., Takle H., Kolarevic J., Calabrese S., Timmerhaus G., Rosseland B. O., et al. (2020). Performance and welfare of Atlantic salmon, <i>Salmo salar</i> L. post-smolts in recirculating aquaculture systems: importance of salinity and water velocity. J. World Aquac. Soc. 51 (2), 373–392. doi:10.1111/jwas.12682
Zaugg W. S., Wagner H. H. (1973). Gill atpase activity related to parr-smolt transformation and migration in steelhead trout (Salmo gairdneri): influence of photoperiod and temperature. Comp. Biochem. Physiology -- Part B Biochem. 45 (4), 955–965. doi:10.1016/0305-0491(73)90156-9
Keywords: microalgae, salmon, smoltification, salinity, immunity, functional feed
Citation: van Muilekom DR, Mueller J, Lindemeyer J, Schultheiß T, Maser E, Seibel H, Rebl A, Schulz C and Goldammer T (2024) Salinity change evokes stress and immune responses in Atlantic salmon with microalgae showing limited potential for dietary mitigation. Front. Physiol. 15:1338858. doi: 10.3389/fphys.2024.1338858
Received: 15 November 2023; Accepted: 22 January 2024;
Published: 12 February 2024.
Edited by:
Carlo C. Lazado, Norwegian Institute of Food, Fisheries and Aquaculture Research, NorwayReviewed by:
Benjamin Costas, University of Porto, PortugalBrian D. Ott, United States Department of Agriculture (USDA), United States
Copyright © 2024 van Muilekom, Mueller, Lindemeyer, Schultheiß, Maser, Seibel, Rebl, Schulz and Goldammer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jonas Mueller, am11ZWxsZXJAdGllcnp1Y2h0LnVuaS1raWVsLmRl; Tom Goldammer, dG9tZ29sZGFtbWVyQGZibi1kdW1tZXJzdG9yZi5kZQ==
†These authors have contributed equally to this work and share first authorship
 Doret R. van Muilekom
Doret R. van Muilekom Jonas Mueller
Jonas Mueller Jacqueline Lindemeyer
Jacqueline Lindemeyer Thekla Schultheiß
Thekla Schultheiß Edmund Maser
Edmund Maser Henrike Seibel3
Henrike Seibel3 Alexander Rebl
Alexander Rebl Tom Goldammer
Tom Goldammer