
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Physiol. , 11 January 2024
Sec. Aquatic Physiology
Volume 14 - 2023 | https://doi.org/10.3389/fphys.2023.1332446
This article is part of the Research Topic The Cellular Stress Response and Physiological Adaptations of Corals Subjected to Environmental Stressors and Pollutants, volume II View all 10 articles
 Michael B. Morgan1,2*
Michael B. Morgan1,2* Jacob Williams1
Jacob Williams1 Barrett Breeze1,2
Barrett Breeze1,2 Nicholas English1
Nicholas English1 Nathaniel Higdon1
Nathaniel Higdon1 Kirt Onthank3
Kirt Onthank3 Dominic F. Qualley2
Dominic F. Qualley2Cnidarians face significant threats from ocean acidification (OA) and anthropogenic pollutants such as oxybenzone (BP-3). The convergence of threats from multiple stressors is an important area to investigate because of potential significant synergistic or antagonistic interactions. Real-time quantitative PCR was performed to characterize the expression profiles of twenty-two genes of interest (GOI) in sea anemones (Exaiptasia diaphana) exposed to one of four treatments: 1) 96 h of OA conditions followed by a 4 h exposure to 20 ppb BP-3; 2) Exposure to 4 h 20 ppb BP-3 without 96 h of OA; 3) Exposure to 96 h of OA alone; or 4) laboratory conditions with no exposure to BP-3 and/or OA. These 22 GOIs represent cellular processes associated with proton-dependent transport, sodium-dependent transport, metal cation binding/transport, extracellular matrix, amino acid metabolism/transport, immunity, and/or steroidogenesis. These 22 GOIs provide new insight into vulnerable cellular processes in non-calcifying anthozoans exposed to OA and BP-3. Expression profiles were categorized as synergistic, antagonistic, or additive of BP-3 in the presence of OA. Two GOIs were synergistic. Fifteen GOIs were antagonistic and the remaining five GOIs were additive in response to BP-3 in acidified seawater. A subset of these GOIs appear to be candidate biomarkers for future in situ investigations. In human health, proton-dependent monocarboxylate transporters (MCTs) are promising pharmacological targets and recognized as potential biomarkers. By comparison, these same MCTs appear to be targets of xenobiotic chemical pollutants in cnidarian physiology. In the presence of BP-3, a network of collagen synthesis genes are upregulated and antagonistic in their expression profiles. Cytochrome b561 is a critical protein required for collagen synthesis and in silico modeling demonstrates BP-3 binds in the pocket of cytochrome b561. Understanding the underlying molecular mechanisms of “drug-like” compounds such as BP-3 may lead to a more comprehensive interpretation of transcriptional expression profiles. The collective antagonistic responses of GOIs associated with collagen synthesis strongly suggests these GOIs should be considered candidate biomarkers of effect. GOIs with synergistic and additive responses represent candidate biomarkers of exposure. Results show the effects of OA and BP-3 are interactive with respect to their impact on cnidarians. This investigation offers mechanistic data that supports the expression profiles and underpins higher order physiological responses.
Ocean acidification is a global concern characterized by the increase in the acidity of the world’s oceans due to the uptake of anthropogenic carbon dioxide (CO2) from the atmosphere. Atmospheric CO2 concentrations have risen from pre-industrial levels of approximately 275 ppm (Macfarling Meure et al., 2006) to over 400 ppm (Dunn et al., 2020), with nearly a third of all annual production of anthropogenic production of CO2 being absorbed by the world oceans (Doney et al., 2009). The resulting drop in pH of marine environments poses significant threats to a broad range of marine organisms (Fabry et al., 2008). Meta-analysis of extant taxa responding to OA reveals similar trends in the negative responses of diverse phyla including cnidarians, mollusks, echinoderms, crustaceans, and bony fish (Kroeker et al., 2013; Wittmann and Pörtner, 2013; Medeiros and Souza, 2023). For calcifying organisms, OA leads to the reduction in the saturation state of carbonate ions, which hinders the calcification process. For corals, OA critically impacts growth and survival (Hoegh-Guldberg et al., 2007). A majority of previous cnidarian investigations have focused on calcification, skeletal density, and/or early life stages (Moya et al., 2012; Putnam et al., 2013; Ramos-Silva et al., 2013; Traylor-Knowles and Connelly, 2017). Some phyla such as Cnidaria and Mollusca also have taxa which are not calcifiers, so investigations not directly linked to calcification are also needed for a broader understanding of OA impact. Sea anemones in the genus Exaiptasia are ideal representatives of non-calcifying anthozoans that are easily cultured in the laboratory and utilized for ecotoxicological studies. Representatives of the genus are found worldwide in tropical marine ecosystems, have the same body plan as scleractinian corals, and are capable of establishing/maintaining symbiotic relationships with zooxanthellae (Sunagawa et al., 2009; Main et al., 2010; Schlesinger et al., 2010; Howe et al., 2012; Siddiqui et al., 2015; Duckworth et al., 2017).
Tropical coastal marine ecosystems are particularly notable for a number of diverse features. Coastal upwellings are locations that increase the probability of OA conditions (Manzello, 2010; Murray et al., 2015). Concurrently, anthropogenic activity along coastlines displays a concentration gradient from highest to lowest (nearshore to offshore, respectively) and is routinely driven by seasonal climatic activity (Dachs et al., 1999; Gray et al., 2002; Yamashita and Tanoue, 2003; Fabry et al., 2008; Gavio et al., 2010; Linsmayer et al., 2020; Zhu et al., 2022). As a consequence, coastal marine ecosystems are exposed to a diversity of anthropogenic stressors and these environments are increasingly becoming “sinks” for complex mixtures of anthropogenic chemicals originating from diverse sources (Peters et al., 1997; Atkinson et al., 2003; Wurl and Obbard, 2004; Fent et al., 2006; Schiedek et al., 2007; Halpern et al., 2008; Jones, 2010; Burns and Brinkman, 2011; Dafforn et al., 2011; Vidal-Dorsch et al., 2012; French et al., 2015; Pie et al., 2015; Wear and Thurber, 2015; Downs et al., 2016; Archer et al., 2017; Law, 2017; Worm et al., 2017; Lebreton et al., 2018; Mitchelmore et al., 2019). As a consequence, marine organisms in coastal marine ecosystems face significant threats from diverse anthropogenic chemical pollutants.
Oxybenzone (BP-3), the active ingredient in sunscreen, is just one of many xenobiotics present in the marine environment (Vidal-Dorsch et al., 2012). It is estimated that 14,000 tons of sunscreens enter the oceans annually from beachgoers and domestic sewage effluent, and there is a growing body of evidence of BP-3 acts as an endocrine disruptor on cnidarians (Downs et al., 2016; Mitchelmore et al., 2019; Mitchelmore et al., 2021; Morgan et al., 2022).
The convergence of threats from OA and chemical pollution is an important area of investigation because of potential significant interactions between anthropogenic pollutants and OA conditions (Nikinmaa, 2013). The natural environment is a mixture of multiple stressors causing mixed effects and challenging organisms to acclimatize and/or adapt to physiologically demanding conditions (Evans and Hofmann, 2012; Stillman et al., 2015; Deidda et al., 2021). For example, heavy metal toxicity is known to be exacerbated in sea anemones experiencing OA (Siddiqui et al., 2015; Duckworth et al., 2017). Currently, there are knowledge gaps in cnidarian transcriptional responses to BP-3 within an environment of OA. This study seeks to characterize patterns of transcription in genes of interest (GOIs) in sea anemones responding to BP-3 and acidic seawater.
Two 38 L glass aquaria were filled with raw seawater obtained from the Rosario Beach Marine Laboratory seawater system, which is in turn pumped from Rosario Bay, Fidalgo Island, Washington, US. Seawater was allowed to equilibrate to room temperature, approximately 21°C, before being added to the aquaria. Temperature and pH of each aquarium was monitored using an Open Acidification Tank Controller unit (Onthank et al., 2023). The pH of the treatment aquarium was adjusted lower by bubbling pure CO2 into a bubble stone from a solenoid-equipped CO2 regulator on a 2.3 kg high pressure CO2 cylinder and controlled by the Open Acidification Tank Controller. A pH of 7.5 was the target pH for the treatment aquarium for two reasons: 1) the pH of 7.5 is approximately 0.3 units lower than present day pH of seawater found in Rosario Bay (Onthank et al., 2021), and 2) the pH of 7.5 is below BP-3′s pKa of 7.6 (Fontanals et al., 2010). The pH of the control aquarium was not controlled. The temperature of the aquaria was not controlled and allowed to match the room temperature. Independent measurements of pH were taken of each aquarium daily using the m-cresol purple spectrophotometric method (Dickson et al., 2007) as modified by Culler-Juarez and Onthank (2021). Total pH values were calculated from absorbance values in R using the “specpH” function found the in OTools package version 22.08.01 (https://github.com/KirtOnthank/Otools). Total alkalinity of each aquarium was measured on day 1 and day 4 using the open-cell titration method (Dickson et al., 2007) and alkalinity was calculated from titration data in R using the “at” function in the seacarb package version 3.2.14 (Gattuso et al., 2015). Salinity was measured daily using a Vernier salinity probe (SAL-BTA). Temperature was measured daily using a VWR precision liquid-in-glass thermometer. Partial pressure of carbon dioxide in each aquarium was calculated each day from daily pH, salinity, and temperature measurements, and from average of total alkalinity measurements for each aquarium in R using the “carb” function from in the “seacarb” function version 3.2.14 (Gattuso et al., 2015). All raw data and code to calculate derived values are available in a repository on Zenodo (https://doi.org/10.5281/zenodo.8011603).
Sea anemones (Exaiptasia diaphana) were purchased from a supplier (Carolina Biological Supply, Burlington, NC, United States) and acclimated to laboratory conditions (recirculating natural seawater at 21°C, ambient laboratory lighting, and 29 ppt salinity) for 1 month prior to beginning the experiment. All BP-3 treatments were nominal concentrations for 4 h in 1L seawater under ambient laboratory lighting during the mid-summer season. The 20 ppb concentration was chosen to induce responses and does not necessarily reflect environmental conditions. Previous studies have demonstrated a toxicant concentration of 20 ppb will induce detectable differential gene expression after 4 h of exposure (Morgan et al., 2001; Morgan and Snell, 2002; Morgan and Snell, 2006; Morgan et al., 2012; Morgan et al., 2022). Variations in environmental conditions can influence the toxicity of xenobiotics (Xin et al., 2021). A 4 h exposure was arbitrarily chosen as an initial timeframe since environmental BP-3 is associated with anthropogenic beach activity. The 96 h timeframe of OA exposure was chosen since some coastal upwellings are known to have periodicities of fluctuation (Murray et al., 2015; Rodriguez-Ruano et al., 2023). Dynamic environments such as an acute 96 h exposure of OA followed by a 4 h pulse of BP-3 will challenge organisms to adapt to physiologically demanding conditions (Evans and Hofmann, 2012). BP-3 was solubilized in acetone at 100 μL/L prior to dilution in seawater. Anemones were divided into four treatments: 1) 96 h of OA conditions followed by a 4 h exposure to 20 ppb oxybenzone (BP-3); 2) Exposure to 4 h 20 ppb BP-3 without 96 h of OA; 3) Exposure to 96 h of OA alone; or 4) laboratory conditions without exposure to BP-3 and/or OA conditions. The 96 h timeframe was chosen because there is evidence of different patterns of responses between short- and long-term CO2 exposures (Inbakandan, 2016; Medeiros and Souza, 2023).
A plethora of studies have investigated OA effects on calcifying marine organisms. Meta-analysis of extant taxa responding to OA reveals similar trends in the negative responses of diverse phyla including cnidarias, mollusks, echinoderms, crustaceans, and bony fish (Kroeker et al., 2013; Wittmann and Pörtner, 2013). Some phyla (i.e., Cnidaria and Mollusca) also have taxa which are not calcifiers (i.e., anemones and octopus, respectively). Hence, there is value in investigating responses to OA in non-calcifying taxa within calcifying phyla. In this investigation, genes on a cnidarian stress EST library were compared to an O. rubescens transcriptome from octopus exposed to OA conditions (data unpublished). Previous studies on model eukaryotes have shown that only 5%–10% of their genes respond to stressed conditions (Adams et al., 2000; Gasch et al., 2000; Hill et al., 2000; Goldstone, 2008). Since both phyla contain calcifying and non-calcifying taxa, this comparison offers the opportunity to identify genes that are representative of stress responses of non-calcifying organisms responding to xenobiotic chemicals and OA conditions. Candidate genes of interest (GOIs) were identified from homologous proteins (BLASTX analysis: nr database, standard parameters) found in both the cnidarian stress EST library and the O. rubescens transcriptome data. Subsets of ESTs from the cnidarian stress library have previously been used to characterize other cnidarian stress responses (Morgan et al., 2012; Morgan et al. 2015; Morgan et al. 2022)
Six to 10 anemones were pooled and homogenized for each treatment. Total RNA was isolated using Trizol (Invitrogen, United States). RNA concentration and quality were assessed using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, United States). Total RNA from pooled samples for each treatment was DNase I digested (New England BioLabs, United States), purified by chloroform/phenol extraction, and then reverse transcribed. First stand synthesis used SuperScript IV (Invitrogen, United States) along with random hexamers and oligo-dT primers to reverse transcribe 1 μg of total RNA. Reverse transcription conditions were 1 h at 37°C, followed by 1 min at each temperature between 42°C and 50°C.
A QuantStudio 7 Flex Real-Time PCR system (Applied Biosystems, United States) used a SYBR Green-based assay to perform qPCR. A 1/100 dilution of first-strand synthesis reactions were used as templates for all qPCR reactions. Primers for each GOI were created using Primer3 (https://primer3.org). Components for each 20 µL qPCR reaction included: 10 µL Luna® universal qPCR Mix (New England BioLabs, United States), 2.5 µL Forward primer (10 µM stock), 2.5 µL Reverse primer (10 µM stock), 2.5 µL dH2O, 2.5 µL sample. Thermocycling conditions were 1 cycle at 95°C for 1 min; 40 cycles of 95°C for 15 s and then 60°C for 30 s; and concluding with 1 cycle of melt curve analysis. Three to four technical replicate reactions were used for analyzing the relative expression of each GOI. Because of a consistent expression pattern, RPL11 was used as the qPCR reference gene (Kenkel et al., 2011). Melt-curve analysis, primer efficiencies, and gel electrophoresis confirmed specificity of priming. Replicate Cq values were averaged to determine ∆Cq and ∆∆Cq for each treatment and GOI. All ∆Cq and ∆∆Cq values are based on the consistent expression of the endogenous reference control gene (RPL11) across all treatments. The ∆∆Cq method was used to determine the differences between targeted GOIs and a single reference gene (Bustin et al., 2009). Two-way ANOVA was performed on Log 2 transformed relative quantity data to determine the significant of the main effects and interactions. All p-values were adjusted using the Benjamini–Hochberg FDR correction.
Human protein homologs have previously been used as substitutes for cnidarian proteins in molecular modelling (Khalturin et al., 2018). Some cnidarian proteins have such significant homology to human proteins that they are even capable of stimulating human responses (Dunn et al., 2006; Wood-Charlson and Weis, 2009; Duffy and Frank, 2011; Dani et al., 2014; Brennan et al., 2017; Mansfield et al., 2017). The most similar vertebrate structures were used for selecting the Protein Databank (PDB) files to be used for initial docking experiments. In the case of MCT1, cryo-EM structures of the homologous human protein in both inward-facing and outward-facing conformations are available in the PDB (PDB IDs 7cko and 7ckr, respectively). These structures were used for docking after removal of water and ligand molecules. The same treatment was applied to cytochrome b561, using 4o79 for the construct facing inward (cytosolic side) and 4o7g for the construct facing outward (non-cytosolic side). For MCTs 2 and 3, models were constructed using H. sapiens sequences corresponding to MCT2 (GenBank: AF049608.1) and MCT3 (GenBank: U81800.1) due to the lack of information on the cnidarian sequences. The cnidarian sequence was used to model MCT10 (GenBank: KXJ27932.1). All models of MCT2, 3, and 10 were constructed with the SWISS-MODEL web server (https://swissmodel.expasy.org) (Bertoni et al., 2017; Bienert et al., 2017; Waterhouse et al., 2018; Studer et al., 2020; Studer et al., 2021) using 7cko for inward-facing constructs and 7ckr for outward-facing constructs as templates. Ligands were created in Avogadro (Hanwell et al., 2012) and energy-minimized prior to docking. Docking was performed using AutoDock Vina (Trott and Olson, 2010); the search space was restricted to the known ligand binding site. Docking included protonated and deprotonated ligands. After docking, the results were analyzed in UCSF Chimera (Pettersen et al., 2004).
After assumptions were tested by Levene’s test for homogeneity of variance across groups and Shapiro-Wilk Normality test, a two-way ANOVA was conducted for each gene to determine the significance of the main effects of OA and BP-3, as well as their potential interactions. The p-values obtained from these ANOVAs underwent an adjustment using the Benjamini–Hochberg False Discovery Rate (FDR) correction method to correct for multiple comparisons.
Genes were classified as “additive” where the interaction p-value exceeded 0.05, suggesting a non-significant interaction effect on gene expression between OA and BP-3. In this scenario, the combined effect of OA and BP-3 on the slope of gene expression can be interpreted as the summation of their independent effects. Genes were categorized as “synergistic” when both the slopes of interaction plots were either both positive or both negative with control OA being less steep than experimental OA exposure. In the context of this study, synergistic effects denote a scenario where the combined influence of OA and BP-3 on gene expression is greater than what would be anticipated from the sum of their individual effects. Lastly, “antagonistic” effects were identified for genes showing opposing slopes in the interaction plots, implying a counteracting influence between OA and BP-3 exposures on the anemone’s gene expression. Data and code underlying these analyses is available at https://zenodo.org/doi/10.5281/zenodo.10056756.
Acclimated anemones were separated into two groups (control and CO2-acidified treatment). At the beginning of the 96 h of exposure to acidified seawater the starting pH was 8.25. At the end of 96 h of exposure to acidified seawater, pCO2, pH, alkalinity, salinity, and temperature was recorded (see Table 1).
This investigation quantified expression of 22 GOIs: 2-oxoglutarate and iron-dependent oxygenase domain-containing protein 2 (2OG-Fe (II)), 17β-hydroxysteroid dehydrogenase type 14 (17βHSD14), 17β-hydroxysteroid dehydrogenase type 12 (17βHSD12), Collagen alpha-2 (I) chain (COL1A2), Collagen alpha-1 (IV) chain (COL4A1), complement component C3 (C3), Cytochrome b561(CYB561), Glutamine synthetase (GLUL), Hephaestin (Heph), Integrin beta-PS (MYS), low affinity copper uptake protein 2 (SLC31A2-1), Monocarboxylate Transporter 2 (MCT2, SLC16A7), Monocarboxylate Transporter 3 (MCT3, SLC16A3), Monocarboxylate Transporter 10 (MCT10, SLC16A10), ornithine aminotransferase, mitochondrial-like (OAT), Serotransferrin-like (TF), Sodium/calcium exchanger 3 (SLC8A3), Soma ferritin-like (FTH1), Tauropine dehydrogenase (TaDH), Tenascin-R (TNR) has a FReD (containing a fibrinogen domain) and is also known as ryncolin-4), Transient receptor potential cation channel subfamily A member 1-like (TRPA1), Zinc transporter ZIP14 (SLC39A14). These GOIs are associated with proton-dependent transport, sodium-dependent transport, metal cation binding/transport, extracellular matrix, amino acid metabolism/transport, immunity, and/or steroidogenesis (see Table 2 for primers; Table 3 for accession numbers).
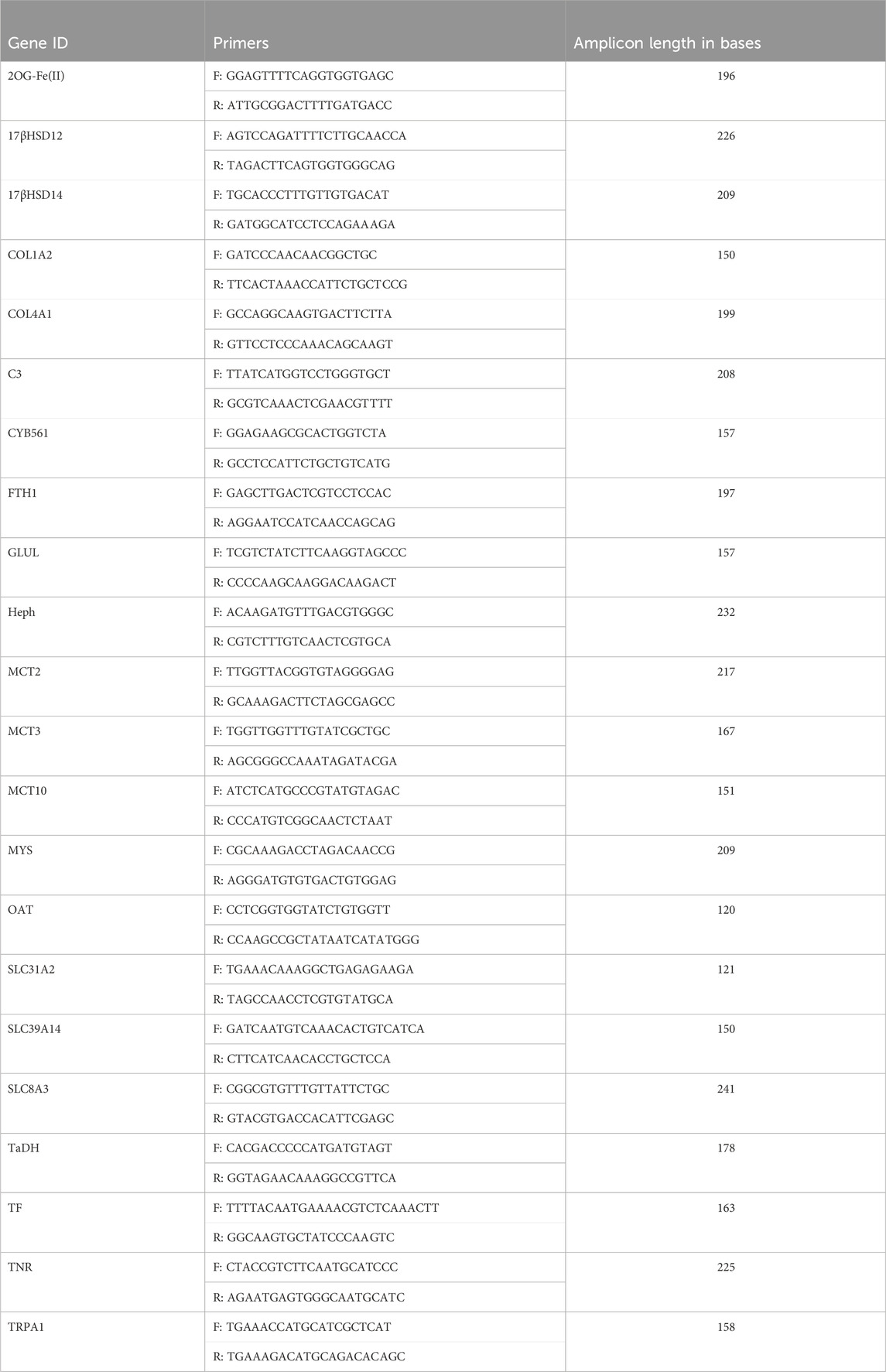
TABLE 2. Genes of Interest and their corresponding primers used in qPCR reaction. Annealing temperature for all primers was 60°C.
All 22 GOIs exhibited significant changes in transcription to OA conditions, and/or BP-3 exposure, and/or the interaction of OA and BP-3 (Figure 1: heatmap Table 4: p-values of significant responses). Twenty GOIs had significant responses to OA and two GOIs (TF, TNR) were not significantly responsive to OA. Fourteen GOIs were responsive to BP-3 exposure while the remaining eight GOIs were not responsive to BP-3 (2OG Fe(II), SLC39A14, MCT2, MCT3, SLC8A3, 17βHSD14, TRPA1, SLC31A2). Five GOIs did not have significant interactions between OA conditions and BP-3 exposure (MCT2, MCT10, GLUL, TF, TNR).
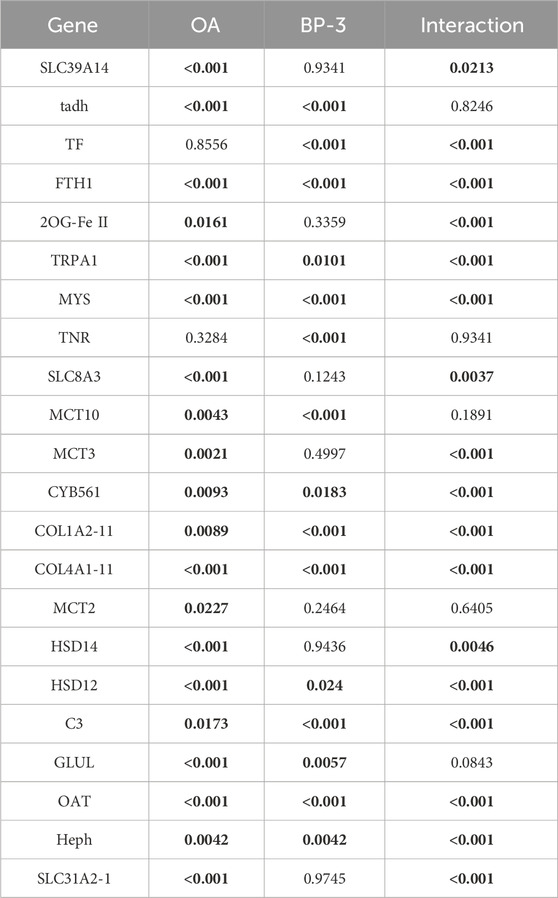
TABLE 4. p-values for each GOI in the presence of OA, BP, or any significant interaction between OA and BP-3. Values in bold are significant.
In silico modeling was performed on MCT2, MCT3, MCT10, and CYB561. MCT2 and MCT3 docking simulations included binding affinities for lactate which is the recognized molecule of transport for proton-dependent transporters. MCT1 was modeled as a comparison since it is the most-well characterized proton-dependent transporter with known inhibitors (BAY8002 and 7ACC2). Docking simulations with MCT10 included amino acids with aromatic side chains which are the recognized molecules of transport (Table 5). Docking simulations of CYB561 reveal there are seven amino acid side chains that interact with BP-3 (Figure 2). By comparison, CYB561 has 4 amino acids interacting with ascorbate (see Figure 2). Table 5 provides a comparison of the binding affinities for BP-3 and ascorbate.
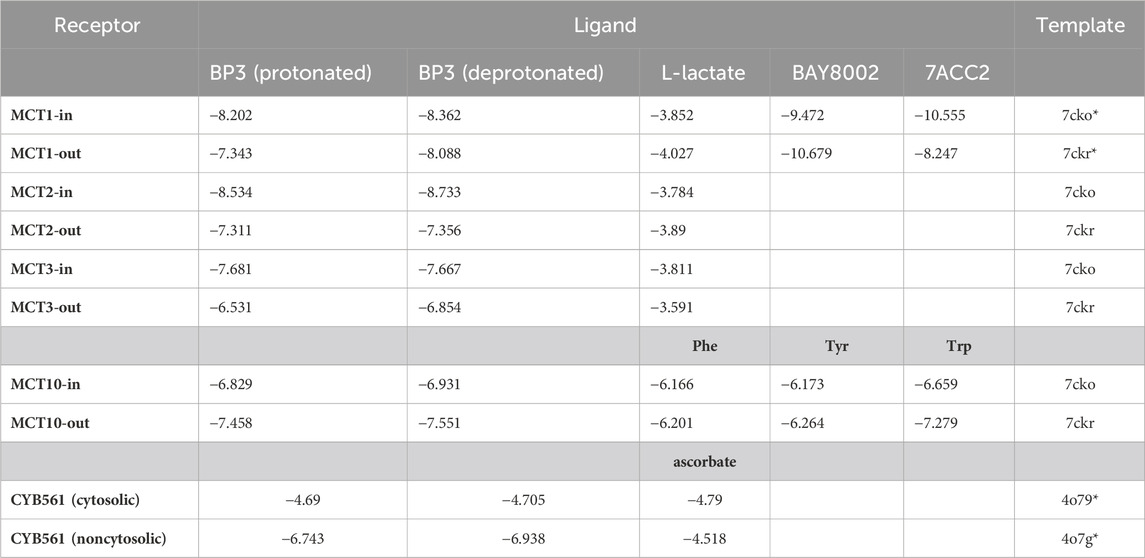
TABLE 5. In silico binding affinities (kcal/mol). BY8002 and 7ACC2 are recognized inhibitors of MCT1. Phe, Tyr, and Trp are the aromatic amino acids normally transported by MCT10. *actual structure used instead of homology model. Transporter orientation towards the cytosolic side of the plasma membrane are labeled “in”. Transporter orientation towards the extracellular side of the plasma membrane are labeled “out”. Values in bold are significant.
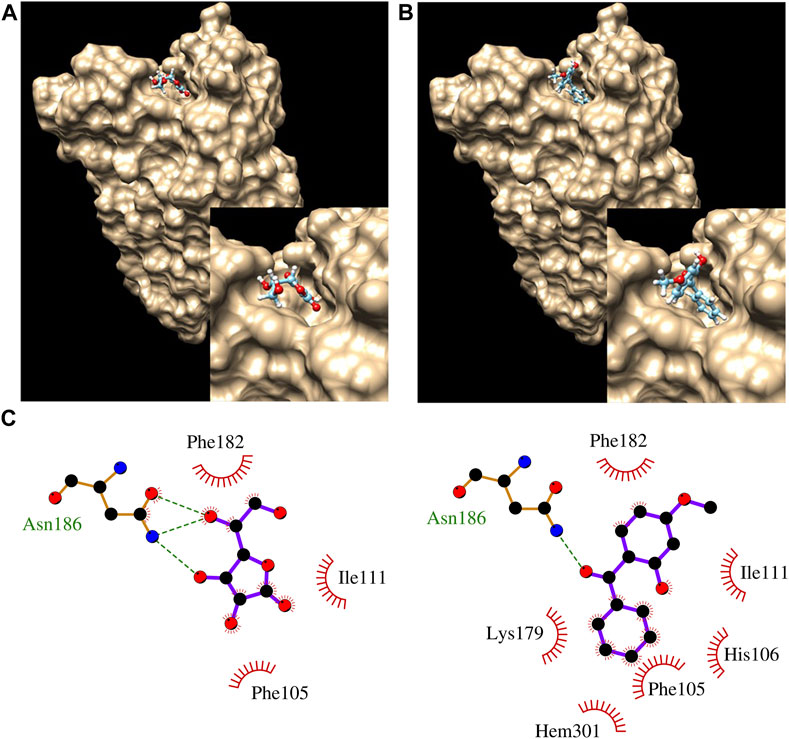
FIGURE 2. Docking results showing ascorbate (A) and BP3 (B) bound to CYB561. The inset pic at bottom right corner of A and B show closeup of ascorbate and BP3, respectively in binding pocket. (C) LigPlot analysis of ascorbate (left) and BP3 (right), showing interactions with nearby residues. Hem301 is a nearby heme group.
Figure 3 identifies which GOIs had synergistic, antagonistic, or additive effects. Table 4 identifies which GOIs had significant interactions. Two GOIs (TF and OAT) had significant synergistic transcriptional interactions on their respective gene networks (see Table 4 and Figure 3). Fifteen GOIs had significant transcriptional interactions with antagonistic effects (see Table 4 and Figure 3). Five GOIs (GLUL, MCT10, MCT2, TaDH, TNR) exhibit additive responses when BP-3 is present in acidified seawater (see Figure 3).
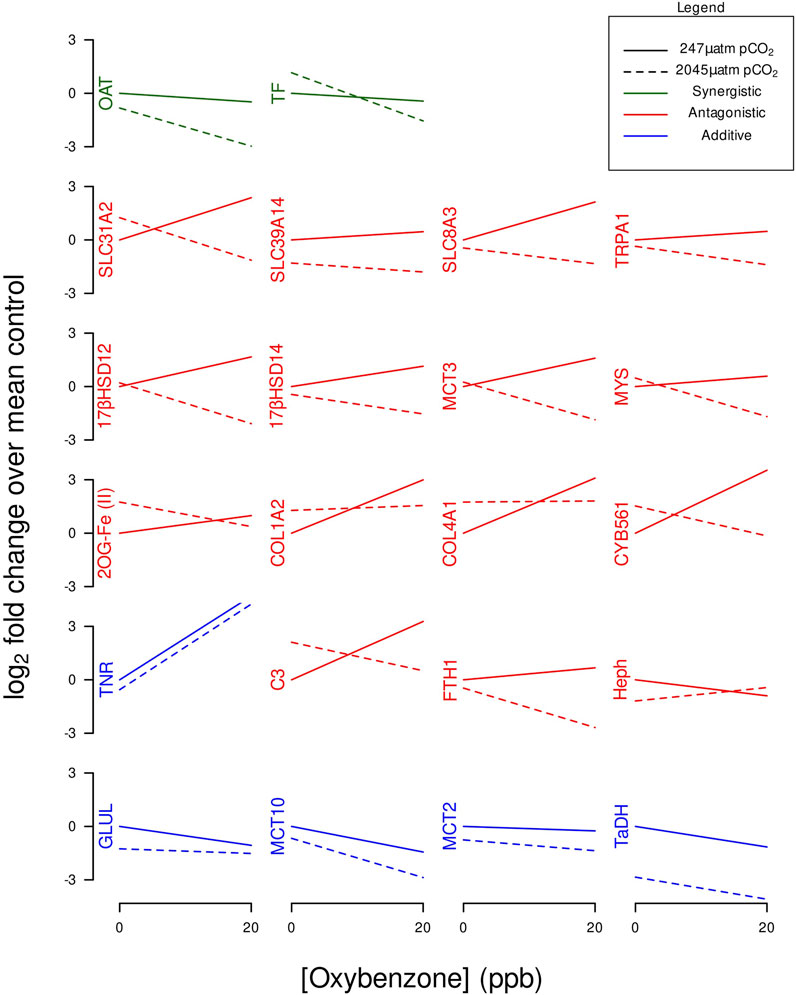
FIGURE 3. Interaction plots of GOI expression profiles categorized by synergistic, antagonistic, or additive effects.
At the beginning of 1 month of acclimation, the pH was not measured. After 1 month of acclimation, the pH of 8.25 was recorded at the beginning of the experiment. The pH of the local seawater is typically between 7.8–8.0 (Murray et al., 2015; Onthank et al., 2021). Differences between the normative pH range of local waters and the pH measured at the beginning of the treatments is likely due to 1) anemone’s algal symbionts favoring carbon fixation (removal of CO2) under the laboratory conditions during the 1 month of acclimation; 2) bubbling of air into the control tank helped drive off excess pCO2 from the local waters. Coral Reefs are known to experience fluctuations in pH of 0.5 units (Birkeland et al., 2008).
The pH range used in this investigation was important because it spanned the pKa of BP-3. Speciation of a compound is determined by pH and the compound’s pKa. If a compound’s pKa is the same as the pH, its ionized and unionized molecules are at equal concentrations (Xin et al., 2021). BP-3′s pKa is 7.6 (Fontanals et al., 2010). The laboratory-controlled acidic conditions of pH 7.5 favored a shift towards a protonated form of BP-3 but there was not a significant difference in the binding affinities of protonated versus deprotonated forms of BP-3 (see Table 5).
While intracellular pH was not directly measured in this investigation, some expression profiles suggest intracellular pH was lowered by exposure to OA. MCT expression is known to be influenced by intracellular pH due to extracellular pH (Gopal et al., 2004). MCTs play a role in the maintenance of intracellular pH which is critical for physiological function (Casey et al., 2010; Halestrap, 2013). Monocarboxylate transporters function as sensors and regulators of pH (Casey et al., 2010; McBrian et al., 2013). MCT 1,2,3, and 4 are symporters of monocarboxylates and protons (1:1 ratio) and the direction of transport across plasma membrane is determined by the relative intra- and extracellular pH (Iwanaga and Kishimoto, 2015). Proton-dependent MCTs are stimulated by decreasing the pH from 8 to 6 and with lowered pH, the Km value (Michaelis constant) for MCTs decreases thereby improving efficiency for binding and transport (Halestrap and Meredith, 2004; Halestrap, 2013). MCT 1&2 pump compounds and protons in one direction while MCT 3&4 pump the same compounds and protons in the opposite direction (Yoshida, 2021). The process of different MCTs coordinating transport of the same compounds in opposite directions is commonly described as metabolic symbiosis (Payen et al., 2020). The expression profiles of MCT2 (SLC16A7) and MCT3 (SLC16A3) in the OA treatment is consistent with the concept of metabolic symbiosis as each would be pumping in opposite direction (see Figure 4A). Data herein is consistent previously published studies demonstrating downregulated transcription of proton-dependent MCTs under acidic conditions (Casey et al., 2010) (see Figure 4A). The gastrovascular pH is known to be more acidic than surrounding seawater (Cai et al., 2016; Bove et al., 2020). Since whole animal homogenate was used in this investigation, future studies should investigate in situ hybridization of MCT2 vs. MCT3 to determine which tissues are most likely to utilize which MCT. Calcium ATPase have previously been the focus of identifying potential proton exchangers that help regulate pH in stony corals (Zoccola et al., 2004; Tambutté et al., 2011). The responsiveness of the proton-dependent MCTs presented herein offers another potential regulator of intracellular pH to consider in future studies. Coupled with the responsiveness of SLC8A3 (Na+/Ca2+ exchanger), these MCTs may provide an alternative mechanistic option for modulating Ca2+ and intracellular pH in stony corals.
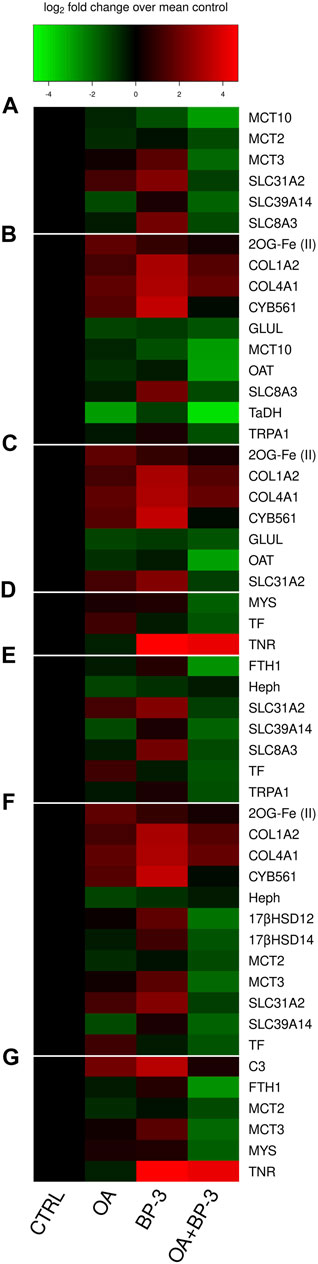
FIGURE 4. Categorized heatmaps of GOIs exhibiting significant differences in expression in response to OA and/or BP-3 exposure. Group (A) Soluble carrier proteins GOIs: SLC31A2, SLC39A14, SLC8A3, MCT3 (SLC16A3), MCT2 (SLC16A7), MCT10 (SLC16A10). Group (B) GOIs associated with amino acid synthesis, reservoirs, and metabolism. Group (C) GOIs associated with Collagen synthesis. Group (D) Extracellular matrix GOIs. Group (E) ion transport GOIs. Group (F) 12 GOIs with functional links to steroidal hormones. Group (G) Immune-related responses.
The following amino acids are associated with expression profiles generated in the investigation: glutamine, glutamic acid, proline, hydroxyproline, amino acids with aromatic side chains, and the non-proteinogenic amino acids taurine and tauropine.
Glutamine synthetase (GLUL) is required to convert glutamic acid into glutamine. Glutamine synthetase regulation is pH-sensitive (Frieg et al., 2021). The downregulated transcription of glutamine synthetase (Figure 4B) suggests lower cellular demand for glutamine allowing glutamate to be utilized/shifted towards other metabolic demands. Under intracellular acidic conditions, four biochemical reactions occur: 1) Glutamine is deamidated by direct hydrolysis (Li et al., 2010), 2) the interconversion of glutamate and α-ketogluarate by glutamate dehydrogenase (GDH) is driven in the direction of glutamate synthesis. 3) the interconversion between glutamate and glutamate-γ-semialdehyde (GSA) is driven towards GSA synthesis which is an intermediate precursor of proline. 4) the interconversion of ornithine and GSA relies on ornithine-δ-aminotransferase (OAT) and the reaction is driven in the direction of GSA (a proline precursor). Under acidic conditions, these four biochemical reactions all move in the direction of proline synthesis. Glutamate metabolism and proline synthesis are functional links between the TCA and urea cycles. Proline plays an important role in regulation of synthesis of ornithine, arginine, polyamines, glutamate and collagen (Karna et al., 2020). In this investigation, there is downregulated transcription of OAT because the cellular demand for converting ornithine into GSA is lessened because glutamate abundance is already elevated in the acidic environment (see Figure 4B). When OAT demand is upregulated, there is increased proline catabolism (Albaugh et al., 2017).
The upregulated transcription profiles of collagen chains I and IV (see Figure 4B) are consistent with an increased metabolic demand for proline synthesis since proline is a major component of collagen. Newly synthesized proline normally accumulates in cytoplasm where it helps buffer cytosolic pH (Meena et al., 2019). If cells are experiencing an intracellular acidic pH, then upregulated synthesis of proline won’t offset intracellular acidic pH because newly synthesized proline is utilized in collagen synthesis. The upregulated transcription of collagen chains (COL1A2; COL4A1) (see Figure 4B) suggests increased cellular demand for synthesizing proline which must then be post-translationally modified to produce the hydroxyproline residues needed for collagen. Both proline and hydroxyproline have important metabolic and physiological roles (Li and Wu, 2018).
Tauropine dehydrogenase (TaDH) is responsible for the interconversion of two non-proteinogenic amino acids Tauropine and Taurine. The abundance of Tauropine typically correlates with the concentration of taurine (derivative of methionine or cysteine) (Linsmayer et al., 2020). The expression profile of TaDH (see Figure 4B) reveals Tauropine and Taurine are relevant in the context of OA where the interconversion reaction is driven towards Taurine accumulation. Tauropine is produced under hypoxic (fermentation) conditions. There is growing recognition of the importance of fermentative metabolism for cnidarian biology (Linsmayer et al., 2020). Fermentation will increase demand for proton-dependent MCTs to pump lactate out of cells. Taurine levels increase in pathological conditions where its antioxidant capabilities help to downregulate of inflammatory mediators (Voskanyan et al., 2014). Taurine’s buffering capacity reduces the negative impact of lactate by modulating intracellular free Ca2+ during fermentation conditions. Taurine lowers intracellular Ca2+ levels by inhibiting calcium influx from calcium channels including the reverse mode of Na+/Ca2+ exchanger (SLC8A3) (Li et al., 2005). Calcium influx can also occur through TRPA1 which is responsive to taurine (Redmond et al., 2014; Meents et al., 2019). Under acidic conditions, glutamate synthesis is elevated which elevates the intracellular Ca2+ concentration. Taurine reduces glutamate-induced elevated Ca2+ concentration (Ripps and Shen, 2012). Expression profiles for both SLC8A3 and TRPA1 are downregulated which reaffirms a reduced cellular demand for Ca2+ influx (see Figure 4B).
MCT10 transports amino acids with aromatic side chains (Imagawa et al., 2004; Yoshida, 2021). Cnidarians import essential amino acids (including phenylalanine and tyrosine) directly from symbiotic algae (Wang and Douglas, 1999) and tyrosine is known to be can be transferred to marine invertebrate larvae via the food chain (Heyland and Moroz, 2005). MCT10’s recognized function offers a potential mechanistic explanation for how cnidarians import the amino acids referenced in these previous studies. It has also recently been discovered that another monocarboxylate transporter MCT7 will transport taurine, a non-proteinogenic amino acid (Higuchi et al., 2022).
The four major classes of proteins associated with the ECM are structural proteins, glycoproteins, glycosaminoglycans and proteoglycans, and matricellular proteins (Jones et al., 2015). Seven GOIs (COL1A2, COL4A1, CYB561, 2OG-Fe(II), GLUL, OAT, SLC31A2) are associated the collagen synthesis (Figure 4C). Collagen belongs to the structural class of the ECM proteins. The upregulated transcription of collagen chains I & 4 (COL1A2; COL4A1, respectively) suggests increased cellular demand for proline which must be post-translationally modified to produce hydroxyproline for collagen. Proline synthesis occurs through the glutamate cycle (requiring GOI: GLUL) and/or through modifications of ornithine (requiring GOI: OAT). Ascorbate is needed to hydroxylate proline residues in collagen processing which also requires 2-oxoglutarate oxygenase with Fe2+ as cofactor (GOI: 2OG-Fe(II)) and cytochrome b561 (GOI: CYB561) to bind ascorbate (Peterkofsky, 1991; Loenarz and Schofield, 2008; Berczi and Zimanyi, 2014). Cu2+ uptake is stimulated by ascorbate, which acts as a Cu2+ reductant (Schweigel-Röntgen, 2014). The upregulation of SLC31A2 (aka: CTR2) is consistent with expression profiles of genes associated the collagen synthesis and an increased metabolic demand for ascorbate (see Figure 4C).
Collagen is upregulated in wound healing (Albaugh et al., 2017). Under acidic conditions, collagen synthesis is upregulated and the proline incorporated into collagen removes it from the metabolic pool (Chalecka et al., 2021). Mechanical properties of collagen formed under alkaline conditions are better than collagen formed under acidic conditions (Zhang et al., 2022). Data herein cannot discern whether upregulated collagen synthesis is due to demand for additional collagen or replacement of existing collagen.
Three GOIs (Mys, TNR, TF) have functional associations with the extracellular matrix (ECM) (Figure 4D). Integrins are trans-membrane proteins that facilitate interactions between cells and ECM (Gotwals et al., 1994; White et al., 2004). Tenascin (TNR) (aka: ryncolin-4) has a FReD (fibrinogen domain), associated with wound healing, and known to be responsive to acidified seawater (Tran et al., 2004; Moya et al., 2012). TNR belongs to the matricellular class of ECM proteins. Serotransferrin (TF) is an iron-binding glycoprotein delivering iron to cells (Gkouvatsos et al., 2012). TF belongs to the glycoprotein class of ECM proteins. MCT 1-4 require and bind directly to basigin or embigin, single-pass transmembrane glycoproteins that are necessary for MCT activity (Halestrap, 2013; Muramatsu, 2016). Integrins strongly interact with glycoproteins (basigin or embigin) to facilitate integrin signaling inside of cell as well as the glycoprotein’s activity (Muramatsu, 2016). During an acute phase of wound healing, a drop in pH can cause hypoxia and increased production of lactic acid (Jones et al., 2015). Expression profiles of the GOIs associated with collagen synthesis and the other ECM transcripts are consistent with previous cnidarian research demonstrating CO2-driven acidification enhances synthesis of the extracellular matrix (Moya et al., 2012).
Seven GOIs (FTH1, Heph, SLC31A2, SLC39A14, SLC8A3, TF, TRPA1,) function in cation transport (Figure 4E). Two GOIs (SLC8A3, TRPA1) transport Ca2+. The Na+/Ca2+ exchanger (SLC8A3) exchanges one Ca2+ for three Na+. The Na+/Ca2+ exchanger functions in both import and export of Ca2+ for maintenance of intracellular Ca2+ homeostasis, however the predominant mode is to export Ca2+ (Khananshvili, 2013; Barron et al., 2018). Transient receptor potential cation channel subfamily A member 1 (TRPA1) is a nonselective cation channel permeable to Ca2+, Na+, and K+. The TRPA1 channel is strongly regulated by both extracellular and intracellular calcium (Meents et al., 2019). While SLC8A2 and TRPA1 are linked to calcium transport, they are not exclusively linked to calcification since anemones are not calcifiers.
Three GOIs (Heph, FTH1, TF) function in iron binding/transport. Hephaestin, a transmembrane ceruloplasmin homolog involved with iron efflux and considered an important link between copper and iron metabolism (Vulpe et al., 1999; Lang et al., 2012). Serotransferrin (TF) is an iron-binding glycoprotein involved with iron influx to cells (Gkouvatsos et al., 2012). Ceruloplasmin and TF are known to interact as positive and negative controls in an acute phase response to perturbations of copper and iron metabolism (Golizeh et al., 2017). Hephaestin, a ceruloplasmin homolog has previously demonstrated acute responsiveness to exogenous copper (Morgan and Snell, 2006). Somaferratin (FTH1) functions in iron storage, detoxification, and innate immunity (Voolstra et al., 2009). Expression profiles for Heph, FTH1, and TF reveal the cellular demand for iron influx/eflux (TF expression) varied significantly with the presence/absence of BP-3 in OA conditions (see Figure 4E).
Intracellular concentrations of Cu2+ and Zn2+ are altered in response to OA. As a member of the SLC39 family, SLC39A14 is plasma membrane protein involved with Zn2+ influx and increasing the availability of cytosolic Zn2+ (Schweigel-Röntgen, 2014). Members of the SLC31 family act to regulate the intracellular Cu2+ concentration. SLC31A2 is mainly found in intracellular membranes of the late endosome and lysosome and is known to regulate intracellular Cu2+ concentration (Wee et al., 2013; Schweigel-Röntgen, 2014). Dysregulation of Zn2+ and Cu2+ are known to be associated with diseased conditions (Lynes et al., 2007; Schweigel-Röntgen, 2014).
Twelve GOIs (2OG-Fe (II), 17βHSD14, 17βHSD12, COL1A2, COL4A1, CYB561, Heph, MCT2, MCT3, SLC31A2, SLC39A14, TF) have functional links to steroidal or tyrosine-based hormones (Figure 4F). BP-3 is recognized as an endocrine disruptor (Downs et al., 2016; LaPlante et al., 2018; Matouskova et al., 2020). 17βHSD14 and 17βHSD12 are associated with steroidogenesis. 17βHSD14 converts estradiol (E2) into estrone (E1); and testosterone (T) into androstenedione (A4), while 17βHSD12 performs the reverse reactions, E1 into E2, and A4 into T (Tarrant et al., 2003; Luu-The et al., 2006; Shikina and Chang, 2016; Salah et al., 2019). Proton-dependent MCTs are regulated by sex hormones (Felmlee et al., 2020). Solute carrier transporters SLC39A and SLC31A families are known to be regulated by estrogen (Schweigel-Röntgen, 2014). Estrogen can transcriptionally regulate synthesis of TF and ceruloplasmin (Golizeh et al., 2017), therefore it is highly probable Hephaestin (a homolog of ceruloplasmin) will respond in a similar manner to estrogen. Biosynthesis of collagen is subject to steroidal hormone regulation (Karna et al., 2020).
The variability in expression of these 12 GOIs reveals how BP-3 exposure influences their expression profiles in the presence/absence of OA conditions. Expression profiles of 17βHSD14 and 17βHSD12 in the presence of BP-3 are consistent with a previous investigation where there was upregulated transcription of both 17βHSD14 and 17βHSD12 in the presence of BP-3 (Morgan et al., 2022). However, upregulated expression of 17βHSD14 in response to BP-3 in this investigation was not statistically significant (see Table 4) suggesting the cellular demand for converting E2 into E3 was not significantly different from the control condition. Future studies investigating the reproductive status of anemones at the time of BP-3 exposure can be important to ascertaining the sensitivity of the anemone’s response to BP-3 alone versus in an environment of acidified seawater and BP-3. This will be particularly important in the abilities of these GOIs to detect endocrine disruption in a multi-stressor environment.
Six GOIs (FTH1, C3, MCT2, MCT3, MYS, TNR) have functional links to immune responses (Figure 4G). Cnidarian integrins are involved in multiple immunological cellular processes such as cell migration, differentiation, signal transduction, and wound repair (Miller et al., 2007; Knack et al., 2008; Shearer et al., 2012). Ferritins such as somaferritin (FTH1) are associated with innate immunity (Voolstra et al., 2009). C3 compliment is a recognized component of cnidarian innate immune system (Miller et al., 2007; Schwarz et al., 2008; Kvennefors et al., 2010; Palmer and Traylor-Knowles, 2012; Ocampo et al., 2015). Tenascin-R (TNR) is also known as ryncolin-4 is considered part of the innate immune system, involved with activating the Compliment pathway, and is upregulated in acidic conditions (OmPraba et al., 2010; Zúñiga-Soto et al., 2023). To be activated, proton-dependent MCTs need to bind immuno-globulin transmembrane glycoproteins (Muramatsu, 2016).
In silico modeling demonstrates BP-3 favorably binds to MCT2, MCT3 with affinities much closer to recognized inhibitors (see Table 5). MCT10′s binding affinity to BP-3 is most similar to tryptophan (see Table 5) which suggests a plausible mechanism for importing BP-3 into a cell. BP-3 binding into the pocket of CYB561 facing the extracellular side of the plasma membrane suggests exogenous BP-3 may act as competitive inhibitor of ascorbate (see Table 5 and Figure 2). This conclusion is supported by the upregulated transcription of CYB561 (see Figures 1, 3). Structurally, Asn 186 functions by hydrogen bonding to the ligand; there are three putative hydrogen bonds to ascorbate and one hydrogen bond to BP-3. However, BP-3 has a greater surface area and is more hydrophobic compared to ascorbate, and the increase in van der Waals interactions between CYB561 and BP-3 explains the stronger binding affinity predicted by docking experiments. His 106, Lys 179, and the nearby heme group (Hem 301) are all involved in BP-3 binding, while these interactions are absent when ascorbate is bound. His 106 is highly conserved and known to participate in binding the two heme-b centers. In a previous study, a laboratory-induced F105W/H106E double mutation completely abolished the ascorbate-dependent redox activity of CYB561, and thus demonstrated His 106 is the single most critical residue for binding to the non-cytosolic side of the protein (Berczi and Zimanyi, 2014).
Results herein reveal how synergistic, antagonistic, and/or additive responses potentially work in concert to produce cellular responses that might be described as homeostatic or adaptive.
While Serotransferrin (TF) had a significant synergistic interaction and Heph had a significant antagonistic interaction, both GOIs contribute to modulating the intracellular transcriptional demands for iron. TF is an iron-binding glycoprotein involved with iron influx (Castellano et al., 1994; Gkouvatsos et al., 2012). Hephaestin (Heph) is a transmembrane-bound homolog of ceruloplasmin involved with iron efflux and considered an important link between copper and iron metabolism (Vulpe et al., 1999; Lang et al., 2012). Ceruloplasmin and TF are known to interact as positive and negative controls in an acute phase response to perturbations of iron metabolism (Golizeh et al., 2017). These GOIs (TF and Heph) with significant but opposite interactions (see Figure 3) appear to represent the molecular underpinnings of iron homeostasis.
Under acidic conditions, the synergistic transcriptional response of OAT (reduce the biosynthesis of ornithine) coupled with the additive transcriptional response of GLUL (reduce the synthesis of glutamine) drive both pathways in the direction of proline synthesis. The biosynthesis of glutamate and proline can be linked to ornithine modifications (Majumdar et al., 2016).
Proline is a fundamental structural component of collagen. COL1A2, COL4A1, CYB561, and 2OG-Fe(II) are all involved with collagen synthesis and exhibit similar expression profiles under OA or BP-3 exposure (see Figures 1, 3). Multiple GOIs with similar significant interactions (i.e., antagonistic effects) involving the same process (i.e., collagen synthesis) appear to be in synchrony. Transcription of multiple GOIs needed for collagen synthesis represent the molecular underpinnings for the physiological response of adaptation to an acidic environment.
Antagonistic effects are representative of gene networks where the impacts of OA and BP-3 are working in opposition. Fifteen GOIs exhibit antagonistic effects when exposed to OA and BP-3 (see Figure 3). Twelve of these GOIs were downregulated when exposed to BP-3 in presence of acidified seawater reveals, whereas 14 GOIs were upregulated in the presence of BP-3 alone (see Figure 3). Under normal conditions, 17βHSD14 and 17βHSD12 perform functions that are opposite of each other. The expression profiles of these GOIs are consistent with a previous investigation where these GOIs exhibited transcriptional responses to an endocrine disruptive 20 ppb BP-3 exposure (Morgan et al., 2022). Future studies investigating longer exposures to BP-3 in an environment of OA will generate more comprehensive dose-response curves that will contribute to better characterization of the responsiveness of these GOIs and the other antagonistic responses.
Influx and efflux transporters can act with intracellular metabolizing enzymes to regulate drug effects (Giacomini and Sugiyama, 2006; Goldstone, 2008). There is growing evidence that MCTs and solute carrier transporters (SLC8A3, SLC31A2) are responsive to various drugs (Halestrap and Wilson, 2012; Njiaju et al., 2012; Halestrap, 2013; Khananshvili, 2013; Iwanaga and Kishimoto, 2015). MCT 1&2 are drug targets for compounds acting as selective inhibitors (Payen et al., 2020; Wang et al., 2021). Anthropogenic chemicals can upregulate expression MCTs (Genter et al., 2002). In human health, MCTs are promising pharmacological targets and in cnidarian physiology, MCTs interact with xenobiotic chemical pollutants (Goldstone, 2008). Structures that are similar to ascorbate can inhibit enzymes usually working on ascorbate (Majamaa et al., 1986). TRPA1 is a promiscuous chemical sensor capable of responding to an unusually wide variety of chemically diverse compounds including many natural products and irritants, environmental chemicals, and common pharmaceuticals (Bautista et al., 2006; Jagadeesan, 2016; Meents et al., 2019). Tauropine dehydrogenase (TaDH) belongs to the ornithine cyclodeaminase (OCD)/mu-crystallin family. Substrate utilization of opine dehydrogenases is notoriously promiscuous (Linsmayer et al., 2020). Understanding the underlying molecular mechanisms of “drug-like” compounds such as BP-3 may lead to a more comprehensive interpretation of the expression profiles herein.
Developing transcriptional biomarkers in the laboratory will help to establish links between laboratory-based testing and adverse effects in situ (Depledge and Fossi, 1994). Development of biomarkers for cnidarian health are far behind biomarkers of human health. Using a reductionist approach, candidate biomarkers identified in this investigation can be valuable preliminary tools to future, more complex studies involving in situ monitoring. A first step biomarker development is to identify GOIs responsive to laboratory-controlled conditions on a single stressor (i.e., exposure to OA or BP-3). Eight GOIs representing collagen synthesis, zinc transport, transient receptor cation channel, and proteins containing a fibrinogen domain have previously demonstrated cnidarian responsiveness to OA (Moya et al., 2012; Wu et al., 2022). Three GOIs (C3, 17βHSD14, 17βHSD12) previously demonstrated responsiveness to BP-3 (Morgan et al., 2022). The only GOIs with known responsiveness to other anthropogenic chemicals include TNR, Heph, and select members of the solute carrier protein family (Morgan and Snell, 2006; Goldstone, 2008; Morgan et al., 2012).
A second step in biomarker development is to use the same GOIs to investigate responses under conditions of two stressors such as OA and BP-3 exposure. MCT 1-4 are recognized as potential biomarkers of human diseases (Felmlee et al., 2020). This investigation offers evidence (expression of proton-dependent MCTs in an acidic environment and in silico modeling of BP-3 binding to proton-dependent MCTs) which may represent a mechanism not previously recognized for modulating changes in intracellular pH in cnidarians. Both MCT2 and MCT3 discern differences in OA vs. normal pCO2 conditions which is consistent with MCT 2 and MCT 3 as pH regulators, but these MCTs cannot discern when BP-3 is present (see Table 4). MCT3 has a significant interaction between OA and BP-3 while MCT2 and MCT10 show no significant interactions between BP-3 and OA (see Table 4). MCT10 is downregulated in the presence of BP-3 (see Table 4) which could be environmentally relevant since increasing concentrations of free amino acids (including those with aromatic side chains) are known to occur in locations closer to anthropogenic sources (Yamashita and Tanoue, 2003). The value of CYB561 as a biomarker can be established by the OA/BP-3 and BP-3 expression profiles and confirmed by in silico docking of BP-3 which binds directly in the ascorbate binding pocket of CYB561 (see Figure 2).
A third step in the development of GOIs as biomarkers would be to investigate how they respond to multiple stressors such as OA, BP-3 exposure, and another stressor of global importance, thermal stress. GOIs associated with collagen synthesis and iron transport have previously demonstrated cnidarian responsiveness to elevated temperatures (Voolstra et al., 2009; DeSalvo et al., 2010; Kenkel et al., 2011). Critics cite multiple stressors causing mixed effects as being a vulnerable weakness for interpreting complex real-world conditions (Deidda et al., 2021). Anthropogenic activities represent complex mixtures of multiple stressors (Jones, 2010; Wear and Thurber, 2015; Mitchelmore et al., 2019; Mitchelmore et al., 2021). Cnidarian biomarkers for sensing changes in intracellular pH or pesticide exposure are emerging (Morgan and Snell, 2002; Barott et al., 2017). A biomarker developed in the lab subsequently demonstrated detectable responses in field sampling (Morgan and Snell, 2002). Characterizing the GOIs as synergistic, antagonistic, and additive responses provides a systematic process for interpreting the effects of multiple stressors. GOIs with synergistic and additive responses represent candidate biomarkers of exposure. The collective antagonistic responses of COL4A1, COL1A2, CYB561, and 2OG-Fe (II) strongly suggests these GOIs should be considered candidate biomarkers of effect. The observed changes in expression profiles of GOIs associated with collagen synthesis coupled with in silico binding of BP-3 to CYB561 offers a mechanistic explanation that underpins higher order physiological responses of organisms restructuring their extracellular matrix in response to an acidic environment.
Candidate biomarkers responsive to different stressed conditions can also be sensitive to different timeframes of stressor exposure. Results herein correlate well with previous transcriptional studies of cnidarian responses to short-term acidified seawater exposures or an acute exposure to 20ppb BP-3 (Moya et al., 2012; Morgan et al., 2022; Woo and Yum, 2022). The GOIs in this investigation represent responses not detected in study of >8000 genes in adult A. millepora after a 28-day exposure to acidic seawater (similar to pH in this investigation) that stimulated >600 differentially expressed transcripts (Kaniewska et al., 2012). Future studies establishing dose-response curves for these GOIs should be productive in elucidating differences in short-term vs. long-term responses to OA, exposure to BP-3, and/or the interaction between OA conditions and exposure to BP-3.
Transcriptional responses are the foundational framework for identifying candidate biomarkers. Gene expression profiling can be used to detect sub-lethal stress responses, differentiate the contribution of different anthropogenic stressors, and access an organism’s condition in natural environments (Snell et al., 2003; Evans and Hofmann, 2012). Gene expression profiling provides a portal into responsive cellular processes that converge and culminate into altered physiology. Gene expression profiles reveal responses that precede translational activity. In silico modeling provides molecular details about the precise location of impact an exogenous chemical can have on a specific protein. Transcriptional responses coupled with in silico modeling offer new insight into how environmentally relevant exogenous anthropogenic chemicals impact specific proteins. Results herein are consistent with responses of SLCs known to be associated with transport of H+, Na+, Ca2+, small chain aliphatic hydrocarbons, and amino acids (both proteinogenic and nonproteinogenic). The solute carrier family of proteins deserves further exploration in the context of xenobiotic chemical pollution in an environment of OA. Venn et al. (2013) stated “It is only with a better mechanistic perspective of the response of coral physiology to environmental change that forecasts of the future of coral reefs under ocean acidification and climate change can be improved”. Results from this investigation offer seven distinctive elements: 1) The expression profiles of these 22 GOIs are representative of important cellular processes impacting non-calcifying cnidarians and 17 of the 22 GOIs have significant interactions between OA and BP-3. 2) Fifteen GOIs exhibit antagonistic interactions to the presence of BP-3 in an environment of acidified seawater. The compensatory actions to one stressor versus both stressors depend on the cellular process impacted. 3) Data herein provides a plausible mechanistic explanation for how BP-3 binds to CYB561 and stimulates upregulation of genes associated with collagen synthesis in an environment of OA; 4) Data herein offers a plausible mechanism for how cnidarians in an environment of OA can modulate intracellular pH through proton-dependent monocarboxylate transporters; 5) Identification of 5 GOIs that should be considered “drug targets” for binding exogenous BP-3; 6) Identifying 12 GOIs with recognized functional links to hormones that are also responding to BP-3 and OA represent new avenues to pursue in the context of endocrine disruption; 7) By characterizing the expression profiles of 22 GOIs in a laboratory-controlled multiple stressor environment, these GOIs are one step closer to being candidate biomarkers for in situ field testing. Servetto et al. (2021) succinctly articulated how an understanding the molecular mechanisms underpinning the physiological processes is critical to properly interpreting cnidarian responses to acidic pH and their ability cope in the Anthropocene. Interactive effects of global stressors (e.g., ocean warming and/or acidification) and local stressors (e.g., pollution) can be advanced with a better understanding of the transcriptional responsiveness of these GOIs in the presence of multiple stressors. These 22 GOIs offer new directions for future studies on the interactions of ocean acidification and xenobiotic chemicals in cnidarians as well as other phyla containing calcifying and non-calcifying taxa.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
The manuscript presents research on animals that do not require ethical approval for their study.
MM: Conceptualization, Project Administration, Resources, Writing–original draft, Writing–editing and review. JW: Formal analysis, Funding acquisition, Validation, Writing–original draft. BB: Data curation, Investigation, Validation. NE: Funding acquisition, Investigation, Validation. NH: Funding acquisition, Investigation, Validation, Writing–original draft. KO: Conceptualization, Data curation, Formal analysis, Investigation, Resources, Visualization, Writing–original draft, Writing–editing and review. DQ: Formal analysis, Investigation, Resources, Visualization, Writing–original draft.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. Financial support was provided by Berry College for one Richards Scholarship (JW), and two Richards Undergraduate Support Grants (NE; NH), Funding for publishing from the Berry College School of MNS Dean (MM; DQ). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Adams M., Celniker S., Holt R., Evans C., Gocayne J., Amanatides P., et al. (2000). The genome sequence of Drosophila melanogaster. Science 287 (5461), 2185–2195. doi:10.1126/science.287.5461.2185
Albaugh V. L., Mukherjee K., Barbul A. (2017). Proline precursors and collagen synthesis: biochemical challenges of nutrient supplementation and wound healing. J. Nutr. 147 (11), 2011–2017. doi:10.3945/jn.117.256404
Archer E., Petrie B., Kasprzyk-Hordern B., Wolfaardt G. M. (2017). The fate of pharmaceuticals and personal care products (PPCPs), endocrine disrupting contaminants (EDCs), metabolites and illicit drugs in a WWTW and environmental waters. Chemosphere 174, 437–446. doi:10.1016/j.chemosphere.2017.01.101
Atkinson S., Atkinson M. J., Tarrant A. M. (2003). Estrogens from sewage in coastal marine environments. Environ. Health Perspec 111 (4), 531–535. doi:10.1289/ehp.5233
Barott K. L., Barron M. E., Tresguerres M. (2017). Identification of a molecular pH sensor in coral. Proc. R. Soc. B Biol. Sci. 284 (1866), 20171769. doi:10.1098/rspb.2017.1769
Barron M. E., Thies A. B., Espinoza J. A., Barott K. L., Hamdoun A., Tresguerres M. (2018). A vesicular Na+/Ca2+ exchanger in coral calcifying cells. PLoS One 13 (10), e0205367. doi:10.1371/journal.pone.0205367
Bautista D. M., Jordt S.-E., Nikai T., Tsuruda P. R., Read A. J., Poblete J., et al. (2006). TRPA1 mediates the inflammatory actions of environmental irritants and proalgesic agents. Cell. 124 (6), 1269–1282. doi:10.1016/j.cell.2006.02.023
Berczi A., Zimanyi L. (2014). The trans-membrane cytochrome b561 proteins: structural information and biological function. Curr. Protein Pept. S. C. 15 (8), 745–760. doi:10.2174/1389203715666140828100351
Bertoni M., Kiefer F., Biasini M., Bordoli L., Schwede T. (2017). Modeling protein quaternary structure of homo-and hetero-oligomers beyond binary interactions by homology. Sci. Rep. 7 (1), 10480. doi:10.1038/s41598-017-09654-8
Bienert S., Waterhouse A., de Beer T. A. P., Tauriello G., Studer G., Bordoli L., et al. (2017). The SWISS-MODEL Repository—new features and functionality. Nuc Acids Res. 45 (D1), D313–D319. doi:10.1093/nar/gkw1132
Birkeland C., Craig P., Fenner D., Smith L., Kiene W. E., Riegl B. M. (2008). Geologic setting and ecological functioning of coral reefs in American Samoa. Coral Reefs U. S. A., 741–765. doi:10.1007/978-1-4020-6847-8_20
Bove C. B., Whitehead R. F., Szmant A. M. (2020). Responses of coral gastrovascular cavity pH during light and dark incubations to reduced seawater pH suggest species-specific responses to the effects of ocean acidification on calcification. Coral Reefs 39 (6), 1675–1691. doi:10.1007/s00338-020-01995-7
Brennan J. J., Messerschmidt J. L., Williams L. M., Matthews B. J., Reynoso M., Gilmore T. D. (2017). Sea anemone model has a single Toll-like receptor that can function in pathogen detection, NF-κB signal transduction, and development. P Natl. Acad. Sci. 114 (47), E10122–E10131. doi:10.1073/pnas.1711530114
Burns K., Brinkman D. (2011). Organic biomarkers to describe the major carbon inputs and cycling of organic matter in the central Great Barrier Reef region. Estuar. Coast. Shelf Sci. 93 (2), 132–141. doi:10.1016/j.ecss.2011.04.001
Bustin S. A., Benes V., Garson J. A., Hellemans J., Huggett J., Kubista M., et al. (2009). The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55 (4), 611–622. doi:10.1373/clinchem.2008.112797
Cai W.-J., Ma Y., Hopkinson B. M., Grottoli A. G., Warner M. E., Ding Q., et al. (2016). Microelectrode characterization of coral daytime interior pH and carbonate chemistry. Nat. Commun. 7 (1), 11144. doi:10.1038/ncomms11144
Casey J. R., Grinstein S., Orlowski J. (2010). Sensors and regulators of intracellular pH. Nat. Rev. Mol. Cell. Biol. 11 (1), 50–61. doi:10.1038/nrm2820
Castellano A. C., Barteri M., Castagnola M., Bianconi A., Borghi E., Dellalonga S. (1994). Structure-function relationship in the serotransferrin: the role of the pH on the conformational change and the metal ions release. Biochem. Biophys. Res. Commun. 198 (2), 646–652. doi:10.1006/bbrc.1994.1094
Chalecka M., Kazberuk A., Palka J., Surazynski A. (2021). P5C as an interface of proline interconvertible amino acids and its role in regulation of cell survival and apoptosis. Int. J. Mol. Sci. 22 (21), 11763. doi:10.3390/ijms222111763
Culler-Juarez M. E., Onthank K. L. (2021). Elevated immune response in Octopus rubescens under ocean acidification and warming conditions. Mar. Biol. 168 (9), 137. doi:10.1007/s00227-021-03913-z
Dachs J., Bayona J. M., Ittekkot V., Albaigés J. (1999). Monsoon-driven vertical fluxes of organic pollutants in the western arabian sea. Environ. Sci. Technol. 33 (22), 3949–3956. doi:10.1021/es990200e
Dafforn K. A., Lewis J. A., Johnston E. L. (2011). Antifouling strategies: history and regulation, ecological impacts and mitigation. Mar. Poll. Bull. 62 (3), 453–465. doi:10.1016/j.marpolbul.2011.01.012
Dani V., Ganot P., Priouzeau F., Furla P., Sabourault C. (2014). Are Niemann-Pick type C proteins key players in cnidarian–dinoflagellate endosymbioses? Mol. Ecol. 23 (18), 4527–4540. doi:10.1111/mec.12876
Deidda I., Russo R., Bonaventura R., Costa C., Zito F., Lampiasi N. (2021). Neurotoxicity in marine invertebrates: an update. Biology 10 (2), 161. doi:10.3390/biology10020161
Depledge M. H., Fossi M. C. (1994). The role of biomarkers in environmental assessment (2). Invertebrates. Ecotoxicology 3 (3), 161–172. doi:10.1007/bf00117081
DeSalvo M. K., Sunagawa S., Voolstra C. R., Medina M. (2010). Transcriptomic responses to heat stress and bleaching in the elkhorn coral Acropora palmata. Mar. Ecol. Prog. Ser. 402, 97–113. doi:10.3354/meps08372
Dickson A. G., Sabine C. L., Christian J. R. (2007). Guide to best practices for ocean CO2 measurements. New York: North Pacific Marine Science Organization.
Doney S. C., Fabry V. J., Feely R. A., Kleypas J. A. (2009). Ocean acidification: the other CO2 problem. Ann. Rev. Mar. Sci. 1, 169–192. doi:10.1146/annurev.marine.010908.163834
Downs C. A., Kramarsky-Winter E., Segal R., Fauth J., Knutson S., Bronstein O., et al. (2016). Toxicopathological effects of the sunscreen UV filter, oxybenzone (Benzophenone-3), on coral planulae and cultured primary cells and its environmental contamination in Hawaii and the U.S. Virgin islands. Arch. Environ. Contam. Tox 70 (2), 265–288. doi:10.1007/s00244-015-0227-7
Duckworth C. G., Picariello C. R., Thomason R. K., Patel K. S., Bielmyer-Fraser G. K. (2017). Responses of the sea anemone, Exaiptasia pallida, to ocean acidification conditions and zinc or nickel exposure. Aqua Tox 182, 120–128. doi:10.1016/j.aquatox.2016.11.014
Duffy D. J., Frank U. (2011). Modulation of COUP-TF expression in a Cnidarian by ectopic wnt signalling and allorecognition. PLoS One 6 (4), e19443. doi:10.1371/journal.pone.0019443
Dunn R. J., Stanitski D. M., Gobron N., Willett K. M., Ades M., Adler R., et al. (2020). "Global climate." Bull. Am. Meteorological Soc. 101(101 S9–S128. doi:10.1175/BAMS-D-20-0104.1
Dunn S. R., Phillips W. S., Spatafora J. W., Green D. R., Weis V. M. (2006). Highly conserved caspase and bcl-2 homologues from the sea anemone Aiptasia pallida: lower metazoans as models for the study of apoptosis evolution. J. Mol. Evol. 63 (1), 95–107. doi:10.1007/s00239-005-0236-7
Evans T. G., Hofmann G. E. (2012). Defining the limits of physiological plasticity: how gene expression can assess and predict the consequences of ocean change. Philos. Trans. R. Soc. Lond B Biol. Sci. 367 (1596), 1733–1745. doi:10.1098/rstb.2012.0019
Fabry V. J., Seibel B. A., Feely R. A., Orr J. C. (2008). Impacts of ocean acidification on marine fauna and ecosystem processes. ICES J. Mar. Sci. 65 (3), 414–432. doi:10.1093/icesjms/fsn048
Felmlee M. A., Jones R. S., Rodriguez-Cruz V., Follman K. E., Morris M. E. (2020). Monocarboxylate transporters (SLC16): function, regulation, and role in health and disease. Pharmacol. Rev. 72 (2), 466–485. doi:10.1124/pr.119.018762
Fent K., Weston A. A., Caminada D. (2006). Ecotoxicology of human pharmaceuticals. Aqua Tox 76 (2), 122–159. doi:10.1016/j.aquatox.2005.09.009
Fontanals N., Cormack P. A. G., Sherrington D. C., Marcé R. M., Borrull F. (2010). Weak anion-exchange hypercrosslinked sorbent in on-line solid-phase extraction–liquid chromatography coupling to achieve automated determination with an effective clean-up. J. Chromatogr. A 1217 (17), 2855–2861. doi:10.1016/j.chroma.2010.02.064
French V. A., King S. C., Kumar A., Northcott G., McGuinness K., Parry D. (2015). Characterisation of microcontaminants in Darwin Harbour, a tropical estuary of northern Australia undergoing rapid development. Sci. Total Environ. 536, 639–647. doi:10.1016/j.scitotenv.2015.07.114
Frieg B., Görg B., Gohlke H., Häussinger D. (2021). Glutamine synthetase as a central element in hepatic glutamine and ammonia metabolism: novel aspects. Biol. Chem. 402 (9), 1063–1072. doi:10.1515/hsz-2021-0166
Gasch A. P., Spellman P. T., Kao C. M., Carmel-Harel O., Eisen M. B., Storz G., et al. (2000). Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell. 11, 4241–4257. doi:10.1091/mbc.11.12.4241
Gattuso J.-P., Epitalon J.-M., Lavigne H., Orr J., Gentili B., Hagens M., et al. (2015). Package seacarb. Available at http://cran.r-project.org/package=seacarb.
Gavio B., Palmer-Cantillo S., Mancera E. J. (2010). Historical analysis (2000–2005) of the coastal water quality in san andrés Island, SeaFlower biosphere reserve, caribbean Colombia. Mar. Poll. Bull. 60 (7), 1018–1030. doi:10.1016/j.marpolbul.2010.01.025
Genter M. B., Burman D. M., Vijayakumar S., Ebert C. L., Aronow B. J. (2002). Genomic analysis of alachlor-induced oncogenesis in rat olfactory mucosa. Physiol. Genomics 12 (1), 35–45. doi:10.1152/physiolgenomics.00120.2002
Giacomini K. M., Sugiyama Y. (2006). Membrane transporters and drug response. Goodman Gilman’s Pharmacol. basis Ther. 11, 41–70.
Gkouvatsos K., Papanikolaou G., Pantopoulos K. (2012). Regulation of iron transport and the role of transferrin. Biochim. Biophys. Acta (BBA) - Gen. Subj. 1820 (3), 188–202. doi:10.1016/j.bbagen.2011.10.013
Goldstone J. V. (2008). Environmental sensing and response genes in cnidaria: the chemical defensome in the sea anemone Nematostella vectensis. Cell. Biol. Toxicol. 24 (6), 483–502. doi:10.1007/s10565-008-9107-5
Golizeh M., Lee K., Ilchenko S., Ösme A., Bena J., Sadygov R. G., et al. (2017). Increased serotransferrin and ceruloplasmin turnover in diet-controlled patients with type 2 diabetes. Free Radic. Biol. Med. 113, 461–469. doi:10.1016/j.freeradbiomed.2017.10.373
Gopal E., Fei Y.-J., Sugawara M., Miyauchi S., Zhuang L., Martin P., et al. (2004). Expression of slc5a8 in kidney and its role in Na+-coupled transport of lactate. J. Biol. Chem. 279 (43), 44522–44532. doi:10.1074/jbc.M405365200
Gotwals P. J., Fessler L. I., Wehrli M., Hynes R. O. (1994). Drosophila PS1 integrin is a laminin receptor and differs in ligand specificity from PS2. P Natl. Acad. Sci. 91 (24), 11447–11451. doi:10.1073/pnas.91.24.11447
Gray J. S., Wu R. S., Or Y. Y. (2002). Effects of hypoxia and organic enrichment on the coastal marine environment. Mar. Ecol. Prog. Ser. 238, 249–279. doi:10.3354/meps238249
Halestrap A. P. (2013). Monocarboxylic acid transport. Compr. Physiol. 3 (4), 1611–1643. doi:10.1002/cphy.c130008
Halestrap A. P., Meredith D. (2004). The SLC16 gene family—from monocarboxylate transporters (MCTs) to aromatic amino acid transporters and beyond. Pflügers Arch. 447, 619–628. doi:10.1007/s00424-003-1067-2
Halestrap A. P., Wilson M. C. (2012). The monocarboxylate transporter family—role and regulation. IUBMB Life 64 (2), 109–119. doi:10.1002/iub.572
Halpern B. S., Walbridge S., Selkoe K. A., Kappel C. V., Micheli F., D'Agrosa C., et al. (2008). A global map of human impact on marine ecosystems. Science 319 (5865), 948–952. doi:10.1126/science.1149345
Hanwell M. D., Curtis D. E., Lonie D. C., Vandermeersch T., Zurek E., Hutchison G. R. (2012). Avogadro: an advanced semantic chemical editor, visualization, and analysis platform. J. Cheminformatics 4 (1), 17. doi:10.1186/1758-2946-4-17
Heyland A., Moroz L. L. (2005). Cross-kingdom hormonal signaling: an insight from thyroid hormone functions in marine larvae. J. Exp. Biol. 208 (23), 4355–4361. doi:10.1242/jeb.01877
Higuchi K., Sugiyama K., Tomabechi R., Kishimoto H., Inoue K. (2022). Mammalian monocarboxylate transporter 7 (MCT7/Slc16a6) is a novel facilitative taurine transporter. J. Biol. Chem. 298 (4), 101800. doi:10.1016/j.jbc.2022.101800
Hill A. A., Hunter C. P., Tsung B. T., Tucker-Kellogg G., Brown E. L. (2000). Genomic analysis of gene expression in C. elegans. Science 290, 809–812. doi:10.1126/science.290.5492.809
Hoegh-Guldberg O., Mumby P. J., Hooten A. J., Steneck R. S., Greenfield P., Gomez E., et al. (2007). Coral reefs under rapid climate change and ocean acidification. Science 318 (5857), 1737–1742. doi:10.1126/science.1152509
Howe P. L., Reichelt-Brushett A. J., Clark M. W. (2012). Aiptasia pulchella: a tropical cnidarian representative for laboratory ecotoxicological research. Environ. Tox Chem. 31 (11), 2653–2662. doi:10.1002/etc.1993
Imagawa S., Nakano Y., Watanabe T. (2004). Molecular analysis of a major soluble egg protein in the scleractinian coral Favites chinensis. Comp. Biochem. Physiol. B 137 (1), 11–19. doi:10.1016/j.cbpc.2003.09.011
Inbakandan D. (2016). “Predicting the impacts of ocean acidifcation: approaches from an omics perspective,” in Marine omics (USA: C. Press), 559–592.
Iwanaga T., Kishimoto A. (2015). Cellular distributions of monocarboxylate transporters: a review. Biomed. Res. 36 (5), 279–301. doi:10.2220/biomedres.36.279
Jagadeesan S. K. (2016). Experimental approach for drug profiling of calcitriol in yeast. China: Université d'Ottawa/University of Ottawa.
Jones E. M., Cochrane C. A., Percival S. L. (2015). The effect of pH on the extracellular matrix and biofilms. Adv. Wound Care 4 (7), 431–439. doi:10.1089/wound.2014.0538
Jones R. (2010). Environmental contamination associated with a marine landfill (‘seafill’) beside a coral reef. Mar. Poll. Bull. 60 (11), 1993–2006. doi:10.1016/j.marpolbul.2010.07.028
Kaniewska P., Campbell P. R., Kline D. I., Rodriguez-Lanetty M., Miller D. J., Dove S., et al. (2012). Major cellular and physiological impacts of ocean acidification on a reef building coral. PLoS One 7 (4), e34659. doi:10.1371/journal.pone.0034659
Karna E., Szoka L., Huynh T. Y. L., Palka J. A. (2020). Proline-dependent regulation of collagen metabolism. Cell. Mol. Life Sci. 77 (10), 1911–1918. doi:10.1007/s00018-019-03363-3
Kenkel C. D., Aglyamova G., Alamaru A., Bhagooli R., Capper R., Cunning R., et al. (2011). Development of gene expression markers of acute heat-light stress in reef-building corals of the genus porites. PLoS One 6 (10), e26914. doi:10.1371/journal.pone.0026914
Khalturin K., Billas I., Chebaro Y., Reitzel A. M., Tarrant A. M., Laudet V., et al. (2018). NR3E receptors in cnidarians: a new family of steroid receptor relatives extends the possible mechanisms for ligand binding. J. Steroid Biochem. Mol. Biol. 184, 11–19. doi:10.1016/j.jsbmb.2018.06.014
Khananshvili D. (2013). The SLC8 gene family of sodium–calcium exchangers (NCX)–Structure, function, and regulation in health and disease. Mol. aspects Med. 34 (2-3), 220–235. doi:10.1016/j.mam.2012.07.003
Knack B. A., Iguchi A., Shinzato C., Hayward D. C., Ball E. E., Miller D. J. (2008). Unexpected diversity of cnidarian integrins: expression during coral gastrulation. BMC Evol. Biol. 8, 136. doi:10.1186/1471-2148-8-136
Kroeker K. J., Kordas R. L., Crim R., Hendriks I. E., Ramajo L., Singh G. S., et al. (2013). Impacts of ocean acidification on marine organisms: quantifying sensitivities and interaction with warming. Glob. Change Biol. 19 (6), 1884–1896. doi:10.1111/gcb.12179
Kvennefors E. C. E., Leggat W., Kerr C. C., Ainsworth T. D., Hoegh-Guldberg O., Barnes A. C. (2010). Analysis of evolutionarily conserved innate immune components in coral links immunity and symbiosis. Dev. Comp. Immunol. 34 (11), 1219–1229. doi:10.1016/j.dci.2010.06.016
Lang M. L., Braun C. L., Kanost M. R., Gorman M. J. (2012). Multicopper oxidase-1 is a ferroxidase essential for iron homeostasis in Drosophila melanogaster. Proc. Natl. Acad. Sci. U. S. A. 109 (33), 13337–13342. doi:10.1073/pnas.1208703109
LaPlante C. D., Bansal R., Dunphy K. A., Jerry D. J., Vandenberg L. N. (2018). Oxybenzone alters mammary gland morphology in mice exposed during pregnancy and lactation. J. Endocr. Soc. 2 (8), 903–921. doi:10.1210/js.2018-00024
Law K. (2017). Plastics in the marine environment. Ann. Rev. Mar. Sci. 9 (1), 205–229. doi:10.1146/annurev-marine-010816-060409
Lebreton L., Slat B., Ferrari F., Sainte-Rose B., Aitken J., Marthouse R., et al. (2018). Evidence that the great pacific garbage patch is rapidly accumulating plastic. Sci. Rep. 8 (1), 4666. doi:10.1038/s41598-018-22939-w
Li F., Obrosova I. G., Abatan O., Tian D., Larkin D., Stuenkel E. L., et al. (2005). Taurine replacement attenuates hyperalgesia and abnormal calcium signaling in sensory neurons of STZ-D rats. Am. J. Physiol.-Endocrinol. Metab. 288 (1), E29–E36. doi:10.1152/ajpendo.00168.2004
Li P., Wu G. (2018). Roles of dietary glycine, proline, and hydroxyproline in collagen synthesis and animal growth. Amino Acids 50 (1), 29–38. doi:10.1007/s00726-017-2490-6
Li X., Lin C., O’Connor P. B. (2010). Glutamine deamidation: differentiation of glutamic acid and γ-glutamic acid in peptides by electron capture dissociation. Anal. Chem. 82 (9), 3606–3615. doi:10.1021/ac9028467
Linsmayer L. B., Deheyn D. D., Tomanek L., Tresguerres M. (2020). Dynamic regulation of coral energy metabolism throughout the diel cycle. Sci. Rep. 10 (1), 19881. doi:10.1038/s41598-020-76828-2
Loenarz C., Schofield C. J.(2008). Expanding chemical biology of 2-oxoglutarate oxygenases. Nat. Chem. Biol. 4, 152–156. doi:10.1038/nchembio0308-152
Luu-The V., Tremblay P., Labrie F. (2006). Characterization of type 12 17beta-hydroxysteroid dehydrogenase, an isoform of type 3 17beta-hydroxysteroid dehydrogenase responsible for estradiol formation in women. Mol. Endocrinol. 20 (2), 437–443. doi:10.1210/me.2005-0058
Lynes M. A., Kang J. Y., Sensi S. L., Perdrizet G. A., Hightower L. E. (2007). Heavy metal ions in normal physiology, toxic stress, and cytoprotection. Ann. N. Y. Acad. Sci. 1113 (1), 159–172. doi:10.1196/annals.1391.010
Macfarling Meure C., Etheridge D., Trudinger C., Steele P., Langenfelds R., Van Ommen T., et al. (2006). Law Dome CO2, CH4 and N2O ice core records extended to 2000 years BP. Geophys. Res. Lett. 33 (14). doi:10.1029/2006GL026152
Main W. P. L., Ross C., Bielmyer G. K. (2010). Copper accumulation and oxidative stress in the sea anemone, Aiptasia pallida, after waterborne copper exposure. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 151 (2), 216–221. doi:10.1016/j.cbpc.2009.10.008
Majamaa K., Günzler V., Hanauske-Abel H. M., Myllylä R., Kivirikko K. I. (1986). Partial identity of the 2-oxoglutarate and ascorbate binding sites of prolyl 4-hydroxylase. J. Biol. Chem. 261 (17), 7819–7823. doi:10.1016/s0021-9258(19)57475-0
Majumdar R., Barchi B., Turlapati S. A., Gagne M., Minocha R., Long S., et al. (2016). Glutamate, ornithine, arginine, proline, and polyamine metabolic interactions: the pathway is regulated at the post-transcriptional level. Front. Plant Sci. 7, 78. doi:10.3389/fpls.2016.00078
Mansfield K. M., Carter N. M., Nguyen L., Cleves P. A., Alshanbayeva A., Williams L. M., et al. (2017). Transcription factor NF-κB is modulated by symbiotic status in a sea anemone model of cnidarian bleaching. Sci. Rep. 7 (1), 16025. doi:10.1038/s41598-017-16168-w
Manzello D. P. (2010). Ocean acidification hotspots: spatiotemporal dynamics of the seawater CO2 system of eastern Pacific coral reefs. Limnol. Ocean. 55 (1), 239–248. doi:10.4319/lo.2010.55.1.0239
Matouskova K., Jerry D. J., Vandenberg L. N. (2020). Exposure to low doses of oxybenzone during perinatal development alters mammary gland morphology in male and female mice. Reprod. Toxicol. 92, 66–77. doi:10.1016/j.reprotox.2019.08.002
McBrian M. A., Behbahan S., Ferrari R., Su T., Huang T.-W., Li K., et al. (2013). Histone acetylation regulates intracellular pH. Mol. Cell. 49 (2), 310–321. doi:10.1016/j.molcel.2012.10.025
Medeiros I. P. M., Souza M. M. (2023). Acid times in physiology: a systematic review of the effects of ocean acidification on calcifying invertebrates. Environ. Res. 231 (Pt 1), 116019. doi:10.1016/j.envres.2023.116019
Meena M., Divyanshu K., Kumar S., Swapnil P., Zehra A., Shukla V., et al. (2019). Regulation of L-proline biosynthesis, signal transduction, transport, accumulation and its vital role in plants during variable environmental conditions. Heliyon 5 (12), e02952. doi:10.1016/j.heliyon.2019.e02952
Meents J. E., Ciotu C. I., Fischer M. J. M. (2019). TRPA1: a molecular view. J. Neurophysiol. 121 (2), 427–443. doi:10.1152/jn.00524.2018
Miller D. J., Hemmrich G., Ball E. E., Hayward D. C., Khalturin K., Funayama N., et al. (2007). The innate immune repertoire in cnidaria--ancestral complexity and stochastic gene loss. Genome Biol. 8 (4), R59. doi:10.1186/gb-2007-8-4-r59
Mitchelmore C. L., Burns E. E., Conway A., Heyes A., Davies I. A. (2021). A critical review of organic ultraviolet filter exposure, hazard, and risk to corals. Environ. Tox Chem. 40, 967–988. doi:10.1002/etc.4948
Mitchelmore C. L., He K., Gonsior M., Hain E., Heyes A., Clark C., et al. (2019). Occurrence and distribution of UV-filters and other anthropogenic contaminants in coastal surface water, sediment, and coral tissue from Hawaii. Sci. Total Environ. 670, 398–410. doi:10.1016/j.scitotenv.2019.03.034
Morgan M., Goodner K., Ross J., Poole A. Z., Stepp E., Stuart C. H., et al. (2015). Development and application of molecular biomarkers for characterizing caribbean yellow band disease in Orbicella faveolata. PeerJ 3, e1371. doi:10.7717/peerj.1371
Morgan M. B., Parker C. C., Robinson J. W., Pierce E. M. (2012). Using Representational Difference Analysis to detect changes in transcript expression of Aiptasia genes after laboratory exposure to lindane. Aqua Tox 110, 66–73. doi:10.1016/j.aquatox.2012.01.001
Morgan M. B., Ross J., Ellwanger J., Phrommala R. M., Youngblood H., Qualley D., et al. (2022). Sea anemones responding to sex hormones, oxybenzone, and benzyl butyl phthalate: transcriptional profiling and in silico modelling provide clues to decipher endocrine disruption in Cnidarians. Front. Genet. 12, 793306. doi:10.3389/fgene.2021.793306
Morgan M. B., Snell T. W. (2002). Characterizing stress gene expression in reef-building corals exposed to the mosquitoside dibrom. Mar. Poll. Bull. 44 (11), 1206–1218. doi:10.1016/S0025-326X(02)00177-7
Morgan M. B., Snell T. W. (2006). Expression of a ceruloplasmin homolog in corals: an informative biomarker of stress. Okinawa, Japan: The 10th International Coral Reef Symposium, 822–830. doi:10.13140/2.1.5077.3762
Morgan M. B., Vogelien D. L., Snell T. W. (2001). Assessing coral stress responses using molecular biomarkers of gene transcription. Environ. Tox Chem. 20 (3), 537–543. doi:10.1002/etc.5620200312
Moya A., Huisman L., Ball E. E., Hayward D. C., Grasso L. C., Chua C. M., et al. (2012). Whole transcriptome analysis of the coral Acropora millepora reveals complex responses to CO₂-driven acidification during the initiation of calcification. Mol. Ecol. 21 (10), 2440–2454. doi:10.1111/j.1365-294X.2012.05554.x
Muramatsu T. (2016). Basigin (CD147), a multifunctional transmembrane glycoprotein with various binding partners. J. Biochem. 159 (5), 481–490. doi:10.1093/jb/mvv127
Murray J. W., Roberts E., Howard E., O'Donnell M., Bantam C., Carrington E., et al. (2015). An inland sea high nitrate-low chlorophyll (HNLC) region with naturally high pCO2. Limnol. Ocean. 60 (3), 957–966. doi:10.1002/lno.10062
Nikinmaa M. (2013). Climate change and ocean acidification—interactions with aquatic toxicology. Aqua Tox 126, 365–372. doi:10.1016/j.aquatox.2012.09.006
Njiaju U. O., Gamazon E. R., Gorsic L. K., Delaney S. M., Wheeler H. E., Im H. K., et al. (2012). Whole-genome studies identify solute carrier transporters in cellular susceptibility to paclitaxel. Pharmacogenet. Genom 22 (7), 498–507. doi:10.1097/fpc.0b013e328352f436
Ocampo I. D., Zárate-Potes A., Pizarro V., Rojas C. A., Vera N. E., Cadavid L. F. (2015). The immunotranscriptome of the Caribbean reef-building coral Pseudodiploria strigosa. Immunogenetics 67 (9), 515–530. doi:10.1007/s00251-015-0854-1
OmPraba G., Chapeaurouge A., Doley R., Devi K. R., Padmanaban P., Venkatraman C., et al. (2010). Identification of a novel family of snake venom proteins veficolins from cerberus rynchops using a venom gland transcriptomics and proteomics approach. J. Proteome Res. 9 (4), 1882–1893. doi:10.1021/pr901044x
Onthank K. L., Foster J., Carman E. P., Foster J. E., Culler-Juarez M., Calvo E., et al. (2023). The Open Acidification Tank Controller: an open-source device for the control of pH and temperature in ocean acidification experiments. HardwareX 14, e00435. doi:10.1016/j.ohx.2023.e00435
Onthank K. L., Trueblood L. A., Schrock-Duff T., Kore L. G. (2021). Impact of short-and long-term exposure to elevated seawater Pco2 on metabolic rate and hypoxia tolerance in Octopus rubescens. Physio Biochem. Zoo. 94 (1), 1–11. doi:10.1086/712207
Palmer C. V., Traylor-Knowles N. (2012). Towards an integrated network of coral immune mechanisms. Proc. R. Soc. B-Biol. Sci. 279 (1745), 4106–4114. doi:10.1098/rspb.2012.1477
Payen V. L., Mina E., Hée V. F. V., Porporato P. E., Sonveaux P. (2020). Monocarboxylate transporters in cancer. Mol. Metab. 33, 48–66. doi:10.1016/j.molmet.2019.07.006
Peterkofsky B. (1991). Ascorbate requirement for hydroxylation and secretion of procollagen: relationship to inhibition of collagen synthesis in scurvy. Am. J. Clin. Nutr. 54 (6), 1135S–1140S. doi:10.1093/ajcn/54.6.1135s
Peters E. C., Gassman N. J., Firman J. C., Richmond R. H., Power E. A. (1997). Ecotoxicology of tropical marine ecosystems. Environ. Tox Chem. 16 (1), 12–40. doi:10.1002/etc.5620160103
Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., et al. (2004). UCSF Chimera?A visualization system for exploratory research and analysis. J. Comp. Chem. 25 (13), 1605–1612. doi:10.1002/jcc.20084
Pie H. V., Heyes A., Mitchelmore C. L. (2015). Investigating the use of oil platform marine fouling invertebrates as monitors of oil exposure in the Northern Gulf of Mexico. Sci. Total Environ. 508, 553–565. doi:10.1016/j.scitotenv.2014.11.050
Putnam H. M., Mayfield A. B., Fan T. Y., Chen C. S., Gates R. D. (2013). The physiological and molecular responses of larvae from the reef-building coral Pocillopora damicornis exposed to near-future increases in temperature and pCO2. Mar. Biol. 160 (8), 2157–2173. doi:10.1007/s00227-012-2129-9
Ramos-Silva P., Kaandorp J., Huisman L., Marie B., Zanella-Cleon I., Guichard N., et al. (2013). The skeletal proteome of the coral Acropora millepora: the evolution of calcification by Co-option and domain shuffling. Mol. Biol. Evol. 30 (9), 2099–2112. doi:10.1093/molbev/mst109
Redmond W. J., Gu L., Camo M., McIntyre P., Connor M. (2014). Ligand determinants of fatty acid activation of the pronociceptive ion channel TRPA1. PeerJ 2, e248. doi:10.7717/peerj.248
Rodriguez-Ruano V., Toth L. T., Enochs I. C., Randall C. J., Aronson R. B. (2023). Upwelling, climate change, and the shifting geography of coral reef development. Sci. Rep. 13 (1), 1770. doi:10.1038/s41598-023-28489-0
Salah M., Abdelsamie A. S., Frotscher M. (2019). Inhibitors of 17β-hydroxysteroid dehydrogenase type 1, 2 and 14: structures, biological activities and future challenges. Mol. Cell. Endocrinol. 489, 66–81. doi:10.1016/j.mce.2018.10.001
Schiedek D., Sundelin B., Readman J. W., Macdonald R. W. (2007). Interactions between climate change and contaminants. Mar. Poll. Bull. 54 (12), 1845–1856. doi:10.1016/j.marpolbul.2007.09.020
Schlesinger A., Kramarsky-Winter E., Rosenfeld H., Armoza-Zvoloni R., Loya Y. (2010). Sexual plasticity and self-fertilization in the sea anemone aiptasia diaphana. PLoS One 5 (7), e11874. doi:10.1371/journal.pone.0011874
Schwarz J. A., Brokstein P. B., Voolstra C., Terry A. Y., Manohar C. F., Miller D. J., et al. (2008). Coral life history and symbiosis: functional genomic resources for two reef building Caribbean corals, Acropora palmata and Montastraea faveolata. BMC Genom 9, 97. doi:10.1186/1471-2164-9-97
Schweigel-Röntgen M. (2014). The families of zinc (SLC30 and SLC39) and copper (SLC31) transporters. Curr. Top. Membr. 73, 321–355. doi:10.1016/b978-0-12-800223-0.00009-8
Servetto N., Aranzamendi M. C. d., Bettencourt R., Held C., Abele D., Movilla J., et al. (2021). Molecular mechanisms underlying responses of the Antarctic coral Malacobelemnon daytoni to ocean acidification. Mar. Environ. Res. 170, 105430. doi:10.1016/j.marenvres.2021.105430
Shearer T. L., Rasher D. B., Snell T. W., Hay M. E. (2012). Gene expression patterns of the coral Acropora millepora in response to contact with macroalgae. Coral Reefs 31 (4), 1177–1192. doi:10.1007/s00338-012-0943-7
Shikina S., Chang C.-F. (2016). “Sexual reproduction in stony corals and insight into the evolution of oogenesis in Cnidaria,” in The Cnidaria, past, present and future (Germany: Springer), 249–268. doi:10.1007/978-3-319-31305-4_16
Siddiqui S., Goddard R. H., Bielmyer-Fraser G. K. (2015). Comparative effects of dissolved copper and copper oxide nanoparticle exposure to the sea anemone, Exaiptasia pallida. Aqua Tox 160, 205–213. doi:10.1016/j.aquatox.2015.01.007
Snell T. W., Brogdon S. E., Morgan M. B. (2003). Gene expression profiling in ecotoxicology. Ecotoxicology 12 (6), 475–483. doi:10.1023/B:ECTX.0000003033.09923.a8
Stillman J. H., Paganini A. W., Podrabsky J. E., Tomanek L. (2015). Biochemical adaptation to ocean acidification. J. Exp. Biol. 218 (12), 1946–1955. doi:10.1242/jeb.115584
Studer G., Rempfer C., Waterhouse A. M., Gumienny R., Haas J., Schwede T. (2020). QMEANDisCo—distance constraints applied on model quality estimation. Bioinformatics 36 (6), 2647–1771. doi:10.1093/bioinformatics/btaa058
Studer G., Tauriello G., Bienert S., Biasini M., Johner N., Schwede T. (2021). ProMod3—a versatile homology modelling toolbox. PLoS Comput. Biol. 17 (1), e1008667. doi:10.1371/journal.pcbi.1008667
Sunagawa S., Wilson E. C., Thaler M., Smith M. L., Caruso C., Pringle J. R., et al. (2009). Generation and analysis of transcriptomic resources for a model system on the rise: the sea anemone Aiptasia pallida and its dinoflagellate endosymbiont. BMC Genom 10, 258. doi:10.1186/1471-2164-10-258
Tambutté S., Holcomb M., Ferrier-Pagès C., Reynaud S., Tambutté É., Zoccola D., et al. (2011). Coral biomineralization: from the gene to the environment. J. Exp. Mar. Biol. Eco 408 (1-2), 58–78. doi:10.1016/j.jembe.2011.07.026
Tarrant A. M., Blomquist C. H., Lima P. H., Atkinson M. J., Atkinson S. (2003). Metabolism of estrogens and androgens by scleractinian corals. Comp. Biochem. Physiol. B 136 (3), 473–485. doi:10.1016/s1096-4959(03)00253-7
Tran K. T., Griffith L., Wells A. (2004). Extracellular matrix signaling through growth factor receptors during wound healing. Wound Repair Regen. 12 (3), 262–268. doi:10.1111/j.1067-1927.2004.012302.x
Traylor-Knowles N., Connelly M. T. (2017). What is currently known about the effects of climate change on the coral immune response. Curr. Clim. Change Rep. 3 (4), 252–260. doi:10.1007/s40641-017-0077-7
Trott O., Olson A. J. (2010). AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comp. Chem. 31 (2), 455–461. doi:10.1002/jcc.21334
Venn A. A., Tambutté E., Holcomb M., Laurent J., Allemand D., Tambutté S. (2013). Impact of seawater acidification on pH at the tissue–skeleton interface and calcification in reef corals. P Natl. Acad. Sci. 110 (5), 1634–1639. doi:10.1073/pnas.1216153110
Vidal-Dorsch D. E., Bay S. M., Maruya K., Snyder S. A., Trenholm R. A., Vanderford B. J. (2012). Contaminants of emerging concern in municipal wastewater effluents and marine receiving water. Environ. Tox Chem. 31 (12), 2674–2682. doi:10.1002/etc.2004
Voolstra C. R., Schnetzer J., Peshkin L., Randall C. J., Szmant A. M., Medina M. (2009). Effects of temperature on gene expression in embryos of the coral Montastraea faveolata. BMC Genom 10 (1), 627. doi:10.1186/1471-2164-10-627
Voskanyan A., Vardapetyan H., Hovhannisyan M., Antonyan M., Darbinyan A. (2014). THE ANTINOCICEPTIVE EFFECT OF TAURINE AGAINST LEVANTINE VIPER (Macrovipera lebetina obtusa) VENOM. J. Exp. Biol. 2, 6.
Vulpe C. D., Kuo Y. M., Murphy T. L., Cowley L., Askwith C., Libina N., et al. (1999). Hephaestin, a ceruloplasmin homologue implicated in intestinal iron transport, is defective in the sla mouse. Nat. Genet. 21, 195–199. doi:10.1038/5979
Wang J. T., Douglas A. E. (1999). Essential amino acid synthesis and nitrogen recycling in an alga–invertebrate symbiosis. Mar. Biol. 135 (2), 219–222. doi:10.1007/s002270050619
Wang N., Jiang X., Zhang S., Zhu A., Yuan Y., Xu H., et al. (2021). Structural basis of human monocarboxylate transporter 1 inhibition by anti-cancer drug candidates. Cell. 184 (2), 370–383.e13. doi:10.1016/j.cell.2020.11.043
Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., et al. (2018). SWISS-MODEL: homology modelling of protein structures and complexes. Nuc Acids Res. 46, W296. Web Server issue). doi:10.1093/nar/gky427
Wear S. L., Thurber R. (2015). Sewage pollution: mitigation is key for coral reef stewardship. Ann. N. Y. Acad. Sci. 1355 (1), 15–30. doi:10.1111/nyas.12785
Wee N. K. Y., Weinstein D. C., Fraser S. T., Assinder S. J. (2013). The mammalian copper transporters CTR1 and CTR2 and their roles in development and disease. Int. J. Biochem. Cell. Biol. 45 (5), 960–963. doi:10.1016/j.biocel.2013.01.018
White D. J., Puranen S., Johnson M. S., Heino J. (2004). The collagen receptor subfamily of the integrins. Int. J. Biochem. Cell. Biol. 36 (8), 1405–1410. doi:10.1016/j.biocel.2003.08.016
Wittmann A. C., Pörtner H.-O. (2013). Sensitivities of extant animal taxa to ocean acidification. Nat. Clim. Chang. 3 (11), 995–1001. doi:10.1038/nclimate1982
Woo S., Yum S. (2022). Transcriptional response of the azooxanthellate octocoral Scleronephthya gracillimum to seawater acidification and thermal stress. Comp. Biochem. Physiology Part D Genomics Proteomics 42, 100978. doi:10.1016/j.cbd.2022.100978
Wood-Charlson E. M., Weis V. M. (2009). The diversity of C-type lectins in the genome of a basal metazoan, Nematostella vectensis. Dev. Comp. Immunol. 33 (8), 881–889. doi:10.1016/j.dci.2009.01.008
Worm B., Lotze H. K., Jubinville I., Wilcox C., Jeambeck J. (2017). Plastic as a persistent marine pollutant. Ann. Rev. Environ. Resour. 42, 1–26. doi:10.1146/annurev-environ-102016-060700
Wu H., Li J., Chai G., Xiao Y., Song Q., Li Z. (2022). Synergistic effects of ocean acidification and warming on coral host, Symbiodinium and nutrients exchange-based symbioses indicated by metatranscriptome. Authorea. doi:10.22541/au.164864741.18158605/v1
Wurl O., Obbard J. (2004). A review of pollutants in the sea-surface microlayer (SML): a unique habitat for marine organisms. Mar. Poll. Bull. 48 (11-12), 1016–1030. doi:10.1016/j.marpolbul.2004.03.016
Xin X., Huang G., Zhang B. (2021). Review of aquatic toxicity of pharmaceuticals and personal care products to algae. J. Hazard Mater 410, 124619. doi:10.1016/j.jhazmat.2020.124619
Yamashita Y., Tanoue E. (2003). Distribution and alteration of amino acids in bulk DOM along a transect from bay to oceanic waters. Mar. Chem. 82 (3-4), 145–160. doi:10.1016/s0304-4203(03)00049-5
Yoshida G. J. (2021). The harmonious interplay of amino acid and monocarboxylate transporters induces the robustness of cancer cells. Metabolites 11 (1), 27. doi:10.3390/metabo11010027
Zhang T., Yu Z., Ma Y., Chiou B.-S., Liu F., Zhong F. (2022). Modulating physicochemical properties of collagen films by cross-linking with glutaraldehyde at varied pH values. Food Hydrocoll. 124, 107270. doi:10.1016/j.foodhyd.2021.107270
Zhu W., Ren Y., Liu X., Huang D., Xia J., Zhu M., et al. (2022). The impact of coastal upwelling on coral reef ecosystem under anthropogenic influence: coral reef community and its response to environmental factors. Front. Mar. Sci. 9, 888888. doi:10.3389/fmars.2022.888888
Zoccola D., Tambutté E., Kulhanek E., Puverel S., Scimeca J.-C., Allemand D., et al. (2004). Molecular cloning and localization of a PMCA P-type calcium ATPase from the coral Stylophora pistillata. Biochim. Biophys. Acta (BBA) - Biomembr. 1663 (1-2), 117–126. doi:10.1016/j.bbamem.2004.02.010
Keywords: ocean acidification, oxybenzone, gene expression profiles, biomarkers, multiple stressors, in silico modeling, synergistic response, antagonistic response
Citation: Morgan MB, Williams J, Breeze B, English N, Higdon N, Onthank K and Qualley DF (2024) Synergistic and antagonistic interactions of oxybenzone and ocean acidification: new insight into vulnerable cellular processes in non-calcifying anthozoans. Front. Physiol. 14:1332446. doi: 10.3389/fphys.2023.1332446
Received: 02 November 2023; Accepted: 28 December 2023;
Published: 11 January 2024.
Edited by:
Ranjeet Bhagooli, University of Mauritius, MauritiusReviewed by:
Amilcare Barca, University of Salento, ItalyCopyright © 2024 Morgan, Williams, Breeze, English, Higdon, Onthank and Qualley. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michael B. Morgan, bWJtb3JnYW5AYmVycnkuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.