- Department of Biomedical Sciences, Texas A&M University School of Dentistry, Dallas, TX, United States
Upon endoplasmic reticulum (ER) stress, inositol-requiring enzyme 1 (IRE1) is activated, which subsequently converts an unspliced X-box binding protein 1 (XBP1U) mRNA to a spliced mRNA that encodes a potent XBP1S transcription factor. XBP1S is essential for relieving ER stress and secretory cell differentiation. We previously established Twist2-Cre;Xbp1CS/+ mice that constitutively expressed XBP1S in the Twist2-expressing cells as well as in the cells derived from the Twist2-expressing cells. In this study, we analyzed the dental phenotype of Twist2-Cre;Xbp1CS/+ mice. We first generated a mutant Xbp1s minigene that corresponds to the recombinant Xbp1Δ26 allele (the Xbp1CS allele that has undergone Cre-mediated recombination) and confirmed that the Xbp1s minigene expressed XBP1S that does not require IRE1α activation in vitro. Consistently, immunohistochemistry showed that XBP1S was constitutively expressed in the odontoblasts and other dental pulp cells in Twist2-Cre;Xbp1CS/+ mice. Plain X-ray radiography and µCT analysis revealed that constitutive expression of XBP1S altered the dental pulp chamber roof- and floor-dentin formation, resulting in a significant reduction in dentin/cementum formation in Twist2-Cre;Xbp1CS/+ mice, compared to age-matched Xbp1CS/+ control mice. However, there is no significant difference in the density of dentin/cementum between these two groups of mice. Histologically, persistent expression of XBP1S caused a morphological change in odontoblasts in Twist2-Cre;Xbp1CS/+ mice. Nevertheless, in situ hybridization and immunohistochemistry analyses showed that continuous expression of XBP1S had no apparent effects on the expression of the Dspp and Dmp1 genes. In conclusion, these results support that sustained production of XBP1S adversely affected odontoblast function and dentin formation.
1 Introduction
X-box binding protein 1 (XBP1) is a member of the family of basic leucine zipper (bZIP) transcription factors (Liou et al., 1990). Xbp1 is highly expressed in the secretory cells including odontoblasts in tooth and osteoblasts in bone as well as in exocrine glands including pancreas and salivary glands during mouse embryonic development (Clauss et al., 1993). The Xbp1 gene expresses an unspliced XBP1 (XBP1U) mRNA that encodes a transcription factor XBP1U that is subject to rapid degradation (Yoshida et al., 2001; Tirosh et al., 2006; Yoshida et al., 2006; Navon et al., 2010; Luo et al., 2022). XBP1U contains a DNA-binding domain in its amino-terminal region but does not have a transcriptional activation domain, thereby it cannot activate transcription of a gene (Yoshida et al., 2001; Calfon et al., 2002; Tirosh et al., 2006; Navon et al., 2010; Luo et al., 2022). The primary function of XBP1U is to regulate the stability of other transcription factors at the post-translational level in a variety of biological processes (Yoshida et al., 2006; Zhao et al., 2013; Martin et al., 2014; Huang et al., 2017; Zhao et al., 2017; Yang et al., 2022).
Inositol-requiring enzyme 1α (IRE1α) is a Type I transmembrane protein in the endoplasmic reticulum (ER) that is highly conserved across species (Tirasophon et al., 1998; Wang et al., 1998). It consists of an N-terminal ER luminal domain, a transmembrane domain, and a cytosolic domain with serine/threonine kinase and endoribonuclease (RNase) activities (Cox et al., 1993; Shamu and Walter, 1996; Tirasophon et al., 1998). When an accumulation of misfolded/unfolded proteins occurs in the ER, a condition known as “ER stress”, IRE1α is oligomerized and autophosphorylated, resulting in activation of its RNase domain (Shamu and Walter, 1996; Welihinda and Kaufman, 1996; Tirasophon et al., 1998; Lee et al., 2008b; Korennykh et al., 2009; Oikawa et al., 2009; Li et al., 2010; Korennykh et al., 2011a; Korennykh et al., 2011b). The activated IRE1α RNase catalyzes an unconventional splicing of 26 nucleotides from the unspliced XBP1U mRNA to give rise to a spliced XBP1 (XBP1S) mRNA that is translated into a highly active transcription factor XBP1S (Yoshida et al., 2001; Calfon et al., 2002; Lee et al., 2002). XBP1S shares the same N-terminal DNA-binding domain as XBP1U, but it also has a transcriptional activation domain in its carboxy-terminal region, due to the reading frameshift in XBP1S mRNA caused by the splicing (Yoshida et al., 2001; Calfon et al., 2002; Lee et al., 2002). XBP1S enters the nucleus and activates the transcription of the genes encoding the proteins involved in protein folding, ER-associated degradation (ERAD) and lipid biosynthesis, which together help alleviate ER stress (Lee et al., 2003; Yoshida et al., 2003; Shaffer et al., 2004; Sriburi et al., 2004; Oda et al., 2006; Acosta-Alvear et al., 2007; Lee et al., 2008a). Thereby, XBP1S plays an important role in promoting cell survival and adaptive response to ER stress.
In addition, XBP1S is essential for the normal development and function of many secretory organs/cells, such as pancreas (Lee et al., 2005), salivary glands (Lee et al., 2005), plasma cells (Reimold et al., 2000; Reimold et al., 2001; Gass et al., 2002; Iwakoshi et al., 2003; Shaffer et al., 2004), hepatocytes (Reimold et al., 2000; Lee et al., 2005) and osteoblasts (Tohmonda et al., 2011). XBP1 is required for an expansion of the ER to accommodate a high level of nascent secretory proteins as well as for the expression of the genes that code for ER chaperones to facilitate protein folding and subsequent protein trafficking through the secretory pathway during the differentiation of secretory cells (Reimold et al., 2001; Iwakoshi et al., 2003; Shaffer et al., 2004; Lee et al., 2005). Additionally, XBP1S directly promotes the transcription of the osterix (Osx) gene that encodes a transcription factor indispensable for osteoblast differentiation (Nakashima et al., 2002; Tohmonda et al., 2011). Like osteoblasts, odontoblasts are secretory cells that produce a large amount of secretory proteins, which form the organic matrix of dentin. In addition to osteoblasts, OSX is also involved in odontoblast differentiation and function (Kim et al., 2015; Zhang et al., 2015; Yang et al., 2017). However, the roles of XBP1S in odontoblast differentiation and function are largely unknown.
We previously generated a novel mouse model (referred to as “Xbp1CS/+“) with a modified Xbp1 allele that constitutively expressed XBP1S following Cre-recombinase (Cre)-mediated recombination, and showed that Twist2-Cre;Xbp1CS/+ mice constitutively expressed XBP1S in a variety of tissues/organs (Xu et al., 2021). In this study, we examined the role of XBP1S in odontoblast differentiation and dentin formation in Twist2-Cre;Xbp1CS/+ mice. We confirmed that XBP1S was constitutively expressed in the odontoblasts in Twist2-Cre;Xbp1CS/+ mice. We found that persistent expression of XBP1S in mice significantly reduced dentin formation and changed odontoblast morphology, but appeared to have no obvious effects on the expression of the odontoblast differentiation markers. These findings indicate that sustained expression of XBP1S in the odontoblasts inhibited dentin formation.
2 Materials and methods
2.1 Animals
All mice were maintained on a C57BL/6 background and were bred and maintained in community housing (≤4 mice/cage, 22°C) on a 12 h light/dark cycle with free access to water and standard pelleted food. All animal procedures were approved by the Institutional Animal Care and Use Committee (IACUC) of Texas A&M University (Dallas, TX).
2.2 DNA constructs
Two DNA constructs, Xbp1 minigene and Xbp1s minigene constructs, were generated (Figure 1). The former construct expressed an unspliced XBP1 mRNA that could undergo an unconventional splicing to form a spliced XBP1S mRNA by activated IRE1α RNase, whereas the latter only produced a spliced XBP1S mRNA. The Xbp1 minigene was generated from three Xbp1 gene fragments, A, B and C. Fragment A is a 4.6-kb fragment containing promoter, exon 1, intron 1, exon 2, intron 2, exon 3 and 5′ part of intron 3. It was released from the targeting construct used to generate Xbp1CS/+ mice (Xu et al., 2021) by restriction endonuclease AscI and NsiI. Fragment B is a 950-bp fragment containing 3′ part of intron 3, exon 4, intron 4, and 5′ part of exon 5. It was released by enzymes NsiI and EcoRV from the 1.3 kb PCR product amplified from the genomic DNA extracted from a wild-type C57/BL6 mouse using the following primers, Xbp1-F and Xbp1-R1, as previously described (Xu et al., 2021). Fragment C is a 1.3-kb fragment containing most of exon 5. It was first released from the targeting construct (Xu et al., 2021) by restriction enzymes EcoRV and Hind III, and subcloned into the EcoRV and HindIII sites of pBluescript SK(−) vector to generate an intermediate construct which appended a SalI cut site to the 3′ terminus of fragment C; fragment C was subsequently released from the intermediate construct by restriction enzymes EcoRV and SalI. Fragments A, B and C were then ligated into the MluI and SalI sites of a pGL3-basic vector (Promega) to replace the luciferase gene and generate the Xbp1 minigene (Figure 1A). The Xbp1s minigene was also generated from three DNA fragments A′, B′ and C’. Fragment A′ is a 4.7-kb fragment containing promoter, exon 1, intron 1, exon 2, intron 2, exon 3 and 5′ part of intron 3. It was released from the targeting construct by restriction endonucleases AscI and SspI. Fragment B′ is a 700-bp fragment containing 3′ part of intron 3, a modified exon 4 lacking the 26 intronic sequence (Δ26), and 5′ part of intron 4. It was released by enzymes SspI and AflII from the 1.3 kb PCR products amplified from the recombinant Xbp1 allele of a Twist2-Cre;Xbp1CS/CS mouse using the following primers, Xbp1-F and Xbp1-R1, as previously described (Xu et al., 2021). Fragment C′ is a 1.5-kb fragment containing 3′ part of intron 4 and exon 5. It was released from the Xbp1 minigene by restriction endonucleases AflII and SalI. Fragments A′, B′ and C’ were then ligated into the MluI and SalI sites of the pGL3-basic vector to replace the luciferase gene and generate the Xbp1s minigene (Figure 1B). The Xbp1s minigene ended up with a loxP site in intron 3. All restriction endonucleases were purchased from New England Biolabs. In addition, a plasmid expressing human IRE1α was a gift from Dr. Randal Kaufman (Addgene plasmid # 21892). The plasmid encoding a kinase-defective IRE1α-K599A (Tirasophon et al., 1998) or RNase-defective IRE1α-N906A variant (Han et al., 2009) was generated using QuikChange II XL site-directed mutagenesis kit (Agilent Technologies, Texas, USA). All DNA constructs were confirmed by restriction enzyme digestion and/or and DNA sequencing.
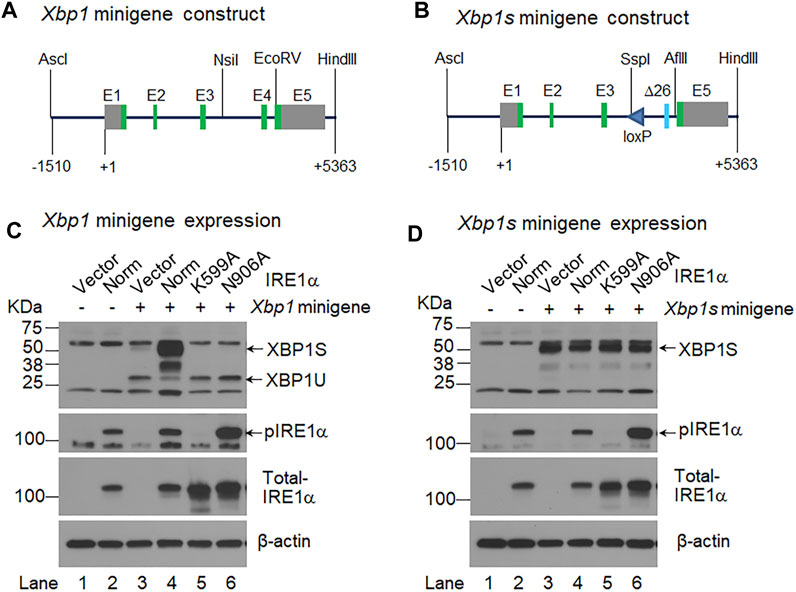
FIGURE 1. IRE1α activation-independent expression of XBP1S. (A, B). Shown are the schematic representations of the Xbp1 minigene (A) and Xbp1s minigene (B) constructs. The Xbp1s minigene contains a modified exon 4 (Δ26) that lack the 26 intronic sequence, and it also contains a loxP site in intron 3. +1, the transcription start site; E, exon; green boxes, coding exons; and grey boxes, non-coding exons. AscI, NsiI, EcoRV, HindIII, SspI and AflII are restriction endonucleases. (C, D). HEK 293 cells were transiently transfected with the Xbp1 minigene (C) or Xbp1s minigene (D) together with the pCDNA3 empty vector (Vector) (lane 3) or a construct expressing normal (Norm) IRE1α (lane 4) or two IRE1α variants, K599A (lane 5) and N906A (lane 6). HEK 293 cells were transfected with the pCDNA3 empty vector alone (lane 1) or the construct expressing normal IRE1α alone (lane 2) as controls. Total cell lysate was harvested 48 h after transfection and analyzed by Western-blotting with an antibody that recognizes both XBP1U and XBP1S, the blot was then stripped and sequentially probed with an antibody against phosphorylated IRE1α and total IRE1α. The blot was probed with mouse monoclonal β-actin antibody as the loading control.
2.3 Cell culture and DNA transfection
HEK293 EBNA cells were grown and maintained in Dulbecco’s modified Eagle’s medium (DMEM) (Corning, USA) supplemented with 10% heat inactivated fetal bovine serum (FBS), GlutaMAX™ and penicillin/streptomycin (Gibco, USA) at 37°C with 5% CO2 and 95% humidity, as described previously (Liang et al., 2019). DNA transfections were performed using X-tremeGENE™ 9 transfection reagent (Roche, Mannheim, Germany), according to the manufacturer’s instruction. Briefly, HEK293 cells were plated into a 6-well plate at a density of 8 × 105 cells per well; on the next day, the cells were transiently transfected with a total of 3 µg of the Xbp1 or Xbp1s minigene construct together with the pCDNA3 empty vector or a construct expressing normal IRE1α or two IRE1α variants, K599A and N906A. HEK293 cells were transfected with the empty vector alone or the construct expressing normal IRE1α together with the empty vector as controls. Total cell lysates were harvested 48 h after transfection and analyzed by Western-blotting analysis as described below.
2.4 Western-blotting analysis
Western-blotting analysis was performed as previously described (Liang et al., 2019). Briefly, 40 µg of the total cell lysates were electrophoresed on a 4%–15% gradient SDS-PAGE (sodium dodecyl sulfate-polyacrylamide gel electrophoresis) gel (BioRad, Hercules, CA), and the proteins were subsequently transferred onto a PVDF membrane (EMD Millipore, Billerica, MA). The membrane was sequentially immunoblotted with rabbit anti-XBP1 polyclonal antibody that recognizes both XBP1U and XBP1S (1:4000, Abcam, Cambridge, MA), horseradish peroxidase (HRP)-conjugated rabbit polyclonal anti-phosphorylated IRE1α (pSer724) (1:5000; Novus Biologicals) and mouse monoclonal anti-IRE1α antibody (1:1000; Santa Cruz Biotechnology, Inc.). The secondary antibodies used included HRP-conjugated goat anti-rabbit IgG antibody (1:2000; Santa Cruz Biotechnology, Inc.) and HRP-conjugated goat anti-mouse IgG antibody (1:2000; Santa Cruz Biotechnology), β-actin was immunoblotted with peroxidase-conjugated mouse monoclonal anti-β-actin antibody (1:50,000; Sigma). The immunostained protein bands were detected with ECL™ Chemiluminescent detection reagents (Pierce Biotechnology, Inc., Rockford, IL), and imaged by using a CL-XPosure film (Pierce Biotechnology).
2.5 Generation of Twist2-cre;Xbp1CS/+ mice
The Xbp1CS/+ female mice were mated with Twist2-Cre knock-in male mice (Twist2Cre/+; Stock No. 008712, the Jackson Laboratory) to generate Twist2-Cre;Xbp1CS/+ mice, as previously described (Xu et al., 2021). Twist2-Cre expresses Cre recombinase in the dental mesenchyme that later gives rise to odontoblasts (Yu et al., 2003; Meng et al., 2015). The Xbp1CS/+ mice carries a modified Xbp1 allele that constitutively express spliced XBP1S following Cre-recombinase (Cre)-mediated recombination event. Thus, XBP1S was constitutively expressed in the odontoblasts and their progenitors in Twist2-Cre;Xbp1CS/+ mice. The dental phenotype of Twist2-Cre;Xbp1CS/+ mice were analyzed, in comparison with age-matched Xbp1CS/+ control mice. Both male and female mice were analyzed as no phenotypic differences were noted between different sexes. PCR genotyping was performed using genomic DNA extracted from mouse tail biopsies, as previously described (Xu et al., 2021).
2.6 Plain X-ray radiography and micro-computed tomography (μCT)
The mandibles were dissected from 3- and 7-week-old Xbp1CS/+ and Twist2-Cre;Xbp1CS/+ mice and fixed in 4% paraformaldehyde (PFA) in diethylpyrocarbonate (DEPC) - treated 0.1 M phosphate-buffered saline (PBS) overnight. The left halves of the mandibles were then stored in 70% ethanol for plain x-ray radiography and µCT analyses, and the right halves were processed for histological analysis (see below). For plain x-ray radiography analysis, the left halves of the mandibles were analyzed by a high-resolution Faxitron X-Ray MX-20 Specimen Radiography System (Faxitron X-Ray Corp., Tucson, AZ) at 6s/26 kV for 3-week-old mice and at 10.6s/26 kV for 7-week-old mice. For µCT analysis, the first molars of the left halves of the mandibles were scanned with a high-resolution Scanco μCT35 imaging system (Scanco Medical, Brüttisellen, Switzerland) in 6-μm slice increment at 70 kV, 116 μA, as previously described (Zhang et al., 2018; Chavez et al., 2021). For three-dimensional (3D) structure construction and morphometric analysis of the mandibular first molars, the whole teeth were outlined. Thresholds were determined for each age based on visual comparisons that could distinguish the tissue of interest from the surrounding tissues (Christiansen, 2016). For dentin and cementum, a threshold of 250 was used for 3-week-old mice, whereas a threshold of 270 was used for 7-week-old mice. For measuring roof and floor dentin thickness, the lowest point at the upper border of the roof dentin concave and the highest point at the lower border of the floor dentin convex were taken as reference points. The roof dentin thickness and floor dentin thickness were defined as the thickness of dentin on the line determined by the two reference points on the sagittal plane that transverses the center of the mandibular first molars. The center of the mandibular first molar was defined as the sagittal (mesial to distal) section crossing both the most proximal and distal pulp horns, which usually bring two more pulp horns between them, and with the largest openings of both proximal and distal root apexes. The central 10 slices were measured for the roof dentin thickness and floor dentin thickness for each mouse. An average of 10 measurements were taken as the thickness of roof dentin and floor dentin, respectively, for each mouse. The morphometric parameters, including the volume and density, were evaluated using the μCT built-in software. The data obtained from 3 to 5 independent mice for each genotype were used for quantitative analysis.
2.7 Histological analysis
The right halves of the mandibles from the 3- and 7-week-old Xbp1CS/+ and Twist2-Cre;Xbp1CS/+ mice were used for histological analysis. Following fixation, the mandibles were decalcified in 15% ethylenediaminetetraacetic acid (EDTA) solution (pH 7.4) at 4°C for 7 days to 2 weeks, depending on the age of the mice. The decalcified mandibles were then dehydrated in a series of gradient ethanol (50% ethanol for 1 h, 70% for 1 h, 95% for 2 h and 100% for 1 h twice and 100% overnight), followed by incubation in xylene for 1 h twice. The mandibles were subsequently embedded in paraffin, and were cut into serial mesio-distal sections at a thickness of 5 μm for Hematoxylin and Eosin (H&E) staining and other histological analyses. Images were taken using Leica DM4 B upright microscope equipped with a Flexacam C1 camera (Leica Biosystems, Wetzlar, Germany).
2.8 In situ hybridization
In situ hybridization (ISH) was performed as previously described (Gibson et al., 2013; Liang et al., 2019). Briefly, the 5-µm tissue sections were processed in xylene and gradient ethanol for dewax and rehydration, followed by antigen retrieval with 10 μg/mL protease K (Ambion, Austin, TX) for 5 min at room temperature. The sections were then hybridized with 1 μg/mL antisense complementary RNA (cRNA) probe at 65 °C for 14–16 h. The probes used include 1.1 kb digoxigenin (DIG)-labeled DSPP cRNA probe and 0.8 kb DIG-labeled DMP1 cRNA probe. The sections were blocked and immunostained with an anti-DIG antibody conjugated to alkaline phosphatase (1:2000, Roche, Mannheim, Germany). The signals were developed with an NBT/BCIP (nitro blue tetrazolium/5-bromo-4-chloro-3-indolyl-phosphate) chromogenic substrate system (Roche ). The sections were counterstained with nuclear fast red (Sigma, Saint Louis, MO) and mounted with Permount mounting medium (Fisher Scientific, Waltham, MA). Images were taken using Leica DM4 B upright microscope (Leica Biosystems).
2.9 Immunohistochemistry
Immunohistochemistry (IHC) was performed to detect total XBP1 (XBP1U and XBP1S), spliced XBP1S, DSPP and DMP1, as previously described (Gibson et al., 2013; Liang et al., 2021; Xu et al., 2021). The 5-µm tissue sections were processed in xylene and gradient ethanol for dewax and rehydration, and were then incubated in sodium citrate buffer (pH 6.0) for antigen retrieval and 3% hydrogen peroxidase (H2O2) in PBS to quench endogenous peroxidase. The sections were blocked with 3% bovine serum albumin (BSA) and 10% normal goat serum (NGS) in 0.1 M PBST (0.1M PBS with 0.1% Tween-20), and incubated with primary then secondary antibodies diluted in 2% NGS. The primary antibodies used include rabbit anti-XBP1 polyclonal antibody that recognizes both XBP1U and XBP1S (1:200, Abcam, Cambridge, MA), rabbit anti-XBP1S monoclonal antibody (E9V3E) that specifically recognizes XBP1S (1:50, Cell Signaling Technology, Danvers, MA), rabbit anti-DSPP polyclonal antibody (1:1000) (Gibson et al., 2013; Liang et al., 2019), and rabbit anti-DMP1 polyclonal antibody (1:600, #857-3) (Gibson et al., 2013). The secondary antibody used was the biotinylated goat anti-rabbit IgG (H + L) antibody (1:200, Vector Laboratories, Burlingame, CA). The immunostaining signals were visualized using DAB (3.3′-diaminobenzidine) kit (Vector Laboratories, Burlingame, CA), according to the manufacturer’s instructions. The sections were counterstained with methyl green (Sigma, Saint Louis, MO) for better visualization of tissue morphology. The sections were then mounted with Permount mounting medium (Fisher Scientific, Waltham, MA). Pictures were taken using Leica DM4 B upright microscope (Leica Biosystems).
2.10 Statistical analysis
Statistical analysis was conducted using the GraphPad Prism 9.0 software package (GraphPad Software, San Diego, CA). Student’s t test was employed to compare the difference between Xbp1CS/+ and Twist2-Cre;Xbp1CS/+ mice. The quantified results were expressed as mean ± standard deviation (SD). p < 0.05 was considered statistically significant.
3 Results
3.1 The Xbp1s minigene constitutively expressed spliced XBP1S only
We previously demonstrated that Twist2-Cre;Xbp1CS/+ mice (bearing a recombinant Xbp1Δ26 allele, i.e., the Xbp1CS allele that has undergone Cre-recombinase mediated recombination) constitutively expressed XBP1S (Xu et al., 2021). Here we generated two DNA constructs, Xbp1 minigene and Xbp1s minigene. The Xbp1 minigene corresponds to the wild-type Xbp1 gene (Figure 1A), whereas the Xbp1s minigene carries a modified version of exon 4 (E4Δ26, encoding XBP1S) and a loxP site in intron 3 (Figure 1B), which is equivalent to the recombinant Xbp1Δ26 allele in Twist2-Cre;Xbp1CS/+ mice (Xu et al., 2021). We then co-transfected the Xbp1 or Xbp1s minigene construct along with the pCDNA3 empty vector or a construct expressing normal IRE1α or two IRE1α variants, K599A and N906A into HEK293 cells, and analyzed XBP1U and XBP1S by Western-blotting analyses. The IRE1α K599A variant has a defective kinase domain (Tirasophon et al., 1998), whereas the IRE1α N906A variant retains kinase activity but is defective in RNase activity (Han et al., 2009). As expected, we found that the Xbp1 minigene produced a high level of unspliced XBP1U but a relatively low level of spliced XBP1S. We also found that co-transfection of the construct expressing normal IRE1α, but not the construct expressing either of the two IRE1α variants, dramatically increased the level of spliced XBP1S produced by the Xbp1 minigene (Figure 1C). In contrast, the Xbp1s minigene only generated spliced XBP1S, and the level of XBP1S was not altered by the presence of either normal IRE1α or its two variants (Figure 1D). Moreover, consistent with previous studies showing that IRE1α overexpression alone leads to the autophosphorylation and activation of its RNase domain (Shamu and Walter, 1996; Han et al., 2009), we found that overexpression of normal IRE1α as well as the RNase-defective N906A variant, but not the Kinase-defective K599A variant, led to the phosphorylation of IRE1α (Figures 1C, D). It is also of note that no XBP1 protein was detected in the cells transfected with either the empty vector or the construct expressing normal IRE1α alone (Figures 1C, D). These in vitro results further confirmed that the mutant Xbp1Δ26 allele would constitutively express spliced XBP1S mRNA and protein, regardless of whether or not IRE1α was activated in Twist2-Cre;Xbp1CS/+ mice.
3.2 Constitutive expression of XBP1S in the odontoblasts in Twist2-Cre;Xbp1CS/+ mice
We next performed IHC to analyze the protein levels of XBP1U and XBP1S in the dental pulps of the mandibular first molars of 3-week-old Xbp1CS/+ and Twist2-Cre;Xbp1CS/+ mice. When an antibody that recognizes both XBP1U and XBP1S was used, we found that immunostaining signals for total XBP1 (XBP1U and XBP1S) were detected in the odontoblasts and other dental pulp cells in both Xbp1CS/+ and Twist2-Cre;Xbp1CS/+ mice (Figure 2A, A1–A3). Further, the intensity of total XBP1 immunostaining signals appeared to be comparable between these two groups of mice. However, when a monoclonal antibody that specifically recognizes XBP1S, but not XBP1U, was used, the immunostaining signals for XBP1S were observed in the odontoblasts and other dental pulp cells in Twist2-Cre;Xbp1CS/+ mice, but were barely detectable in Xbp1CS/+ mice (Figure 2B, B1, B2–B3). These results corroborated that XBP1S was constitutively expressed in the odontoblasts and other dental pulp cells in Twist2-Cre;Xbp1CS/+ mice.
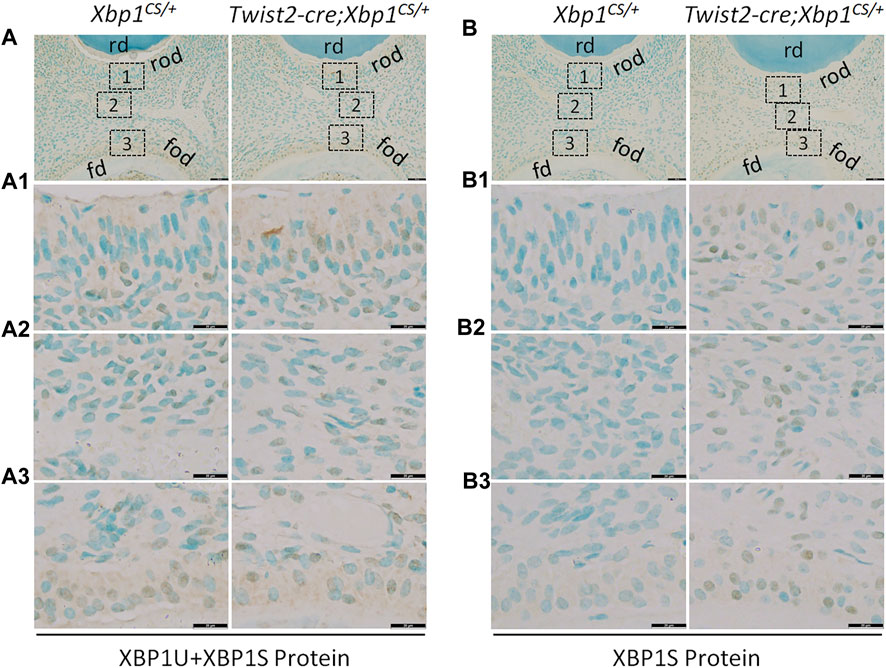
FIGURE 2. Immunohistochemical staining of total XBP1 (XBP1U and XBP1S) and XBP1S. Shown are the representative images of IHC staining of total XBP1 (including XBP1U and XBP1S) (A); signal in brown) and XBP1S (B); signal in brown) in the mandibular first molars of 3-week-old Xbp1CS/+ and Twist2-Cre;Xbp1CS/+ mice. Each image in (A, B) is from the middle region of the crown of a sagittally-sectioned mandibular first molar. (A1-A3, B1-B3) are the higher magnification views of the roof-forming odontoblasts (box1), central dental pulp cells (box 2) and floor-forming odontoblasts (box 3) in (A, B), respectively. rd, roof dentin; fd, floor dentin; rod, roof-forming odontoblasts; fod, floor-forming odontoblasts. Note that the signals for total XBP1 were found in the dental pulps of Xbp1CS/+ and Twist2-Cre;Xbp1CS/+ mice, whereas the signals for XBP1S were strongly detected in the dental pulps of Twist2-Cre;Xbp1CS/+ mice, but were barely detectable in those of Xbp1CS/+ mice. Scale bars: 50 μm in (A, B); 20 μm in (A1-A3, B1-B3).
3.3 Dental defects associated with constitutive expression of XBP1S in the odontoblasts
We then characterized the dental phenotype of 3- and 7-week-old Twist2-Cre;Xbp1CS/+ mice, in comparison with age-matched Xbp1CS/+ control mice, by plain X-ray radiography and μCT analysis. X-ray radiography and 3D reconstructed μCT images showed that Twist2-Cre;Xbp1CS/+ mice developed a thinner pulp chamber roof dentin and a thicker pulp chamber floor dentin by the age of 7 weeks, compared to the age-matched Xbp1CS/+ mice (Figures 3A–D). Moreover, the μCT images also showed that Twist2-Cre;Xbp1CS/+ mice form less cementum than Xbp1CS/+ mice by the age of 7 weeks (Figure 3D), which was further confirmed by H&E staining (Supplementary Figure S1). Consistently, quantitative μCT analyses demonstrated that Twist2-Cre;Xbp1CS/+ mice, by the age of 7 weeks, acquired a significant increase in the pulp floor dentin thickness and pulp volume, but a significant decrease in the pulp roof dentin thickness as well as a significant decrease in total dentin/cementum volume, compared to age-matched Xbp1CS/+ control mice (Figures 4A–D). There is no significant difference in the density of dentin/cementum between these two groups of mice at either age (Figure 4E). These findings indicate that constitutive expression of XBP1S resulted in altered roof and floor dentin formation in Twist2-Cre;Xbp1CS/+ mice.
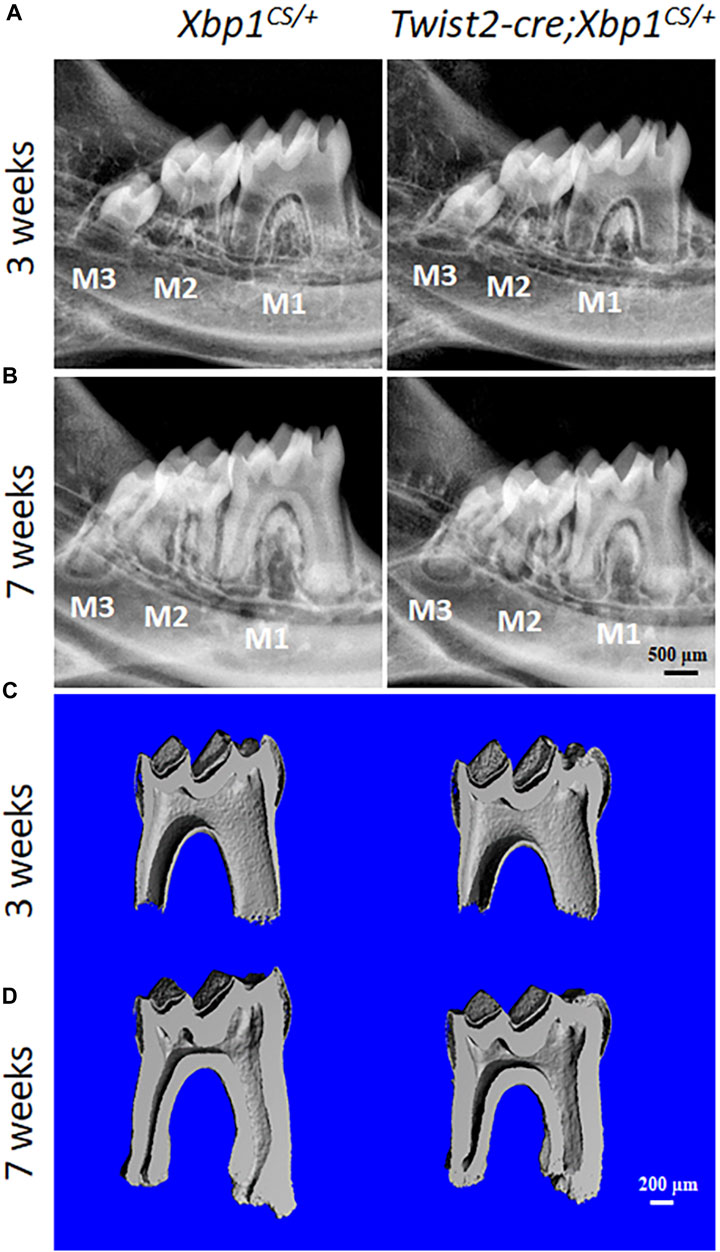
FIGURE 3. Plain X-ray radiography and micro-computed tomography (µCT) analyses of the mandibular molars. (A, B). Representative plain X-ray radiographs of the mandibular molars of 3-week-old and 7-week-old Xbp1CS/+ and Twist2-Cre;Xbp1CS/+ mice. M1, first molar; M2, second molar; M3, third molar. Scale bar: 500 μm. (C, D). Representative 3-dimensional reconstructed μCT images (sagittal sections) of the mandibular first molars of 3-week-old and 7-week-old Xbp1CS/+ and Twist2-Cre;Xbp1CS/+ mice. The mesial side of each molar is on the right, and the distal side is on the left. Scale bar: 200 μm.
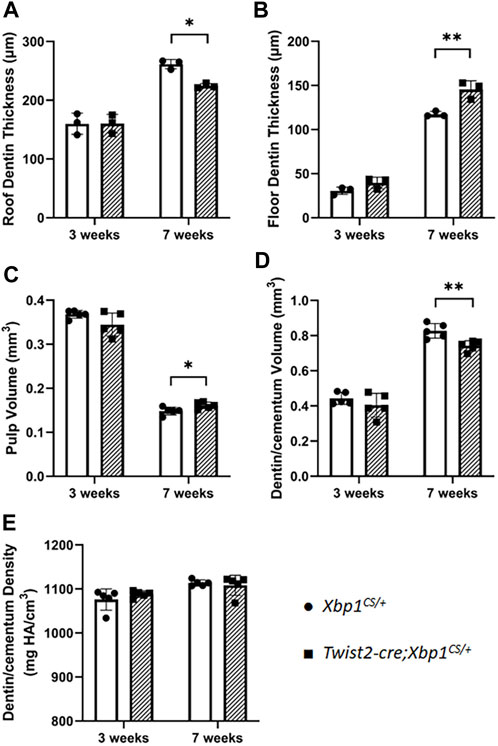
FIGURE 4. Quantitative µCT analysis of mandibular first molars. µCT analysis was performed to quantify the roof dentin thickness (A), floor dentin thickness (B), pulp volume (C), dentin/cementum volume (D) and dentin/cementum density (E) of mandibular first molars of 3- and 7-week-old mice. Student’s t test was used to compare the difference between Xbp1CS/+ and Twist2-Cre;Xbp1CS/+ mice. All values are mean ± SD. n = 3 for each group in A-B; n = 5 for each group in C-E; * p < 0.05; ** p < 0.01.
3.4 Changes in odontoblast morphology in Twist2-Cre;Xbp1CS/+ mice
Histologically, Hematoxylin and Eosin (H&E) staining showed that the mandibular first molars of Xbp1CS/+ mice had completely erupted by the age of 3 weeks, whereas those in age-matched Twist2-Cre;Xbp1CS/+ mice were still covered by reduced enamel epithelium, indicating delayed eruption of the mandibular first molars (Figure 5A). Moreover, the roof-forming odontoblasts in the mandibular first molars of the 3-week-old Xbp1CS/+ mice were aligned as a single layer of tall columnar and highly polarized cells (Figures 5A,A1). However, the roof-forming odontoblasts in Twist2-Cre;Xbp1CS/+ mice were dramatically shorter and irregular (Figures 5A,A1). The morphological differences were less apparent in the floor-forming odontoblasts between these two groups of mice (Figures 5A,A2). Moreover, the predentin of the pulp chamber roof in Twist2-Cre;Xbp1CS/+ mice was thinner than that of Xbp1CS/+ mice (Figure 5A1). These results demonstrate that persistent expression of XBP1S altered odontoblast morphology in Twist2-Cre;Xbp1CS/+ mice.
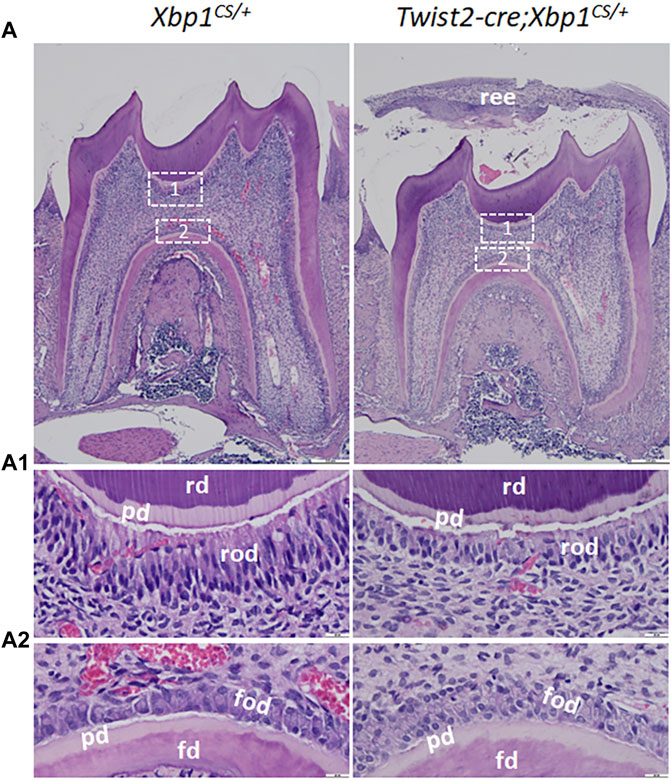
FIGURE 5. H&E staining of the mandibular first molars. (A). Shown are the representative images of H&E staining of a sagittally-sectioned mandibular first molars of 3-week-old Xbp1CS/+ and Twist2-Cre;Xbp1CS/+ mice. (A1, A2) are the higher magnification views of the roof-forming odontoblasts (box1) and floor-forming odontoblasts (box 2) in A. Note that the roof-forming odontoblasts were long, columnar-shaped and highly polarized in Xbp1CS/+ mice, but the roof-forming odontoblasts became shorter and irregular in Twist2-Cre;Xbp1CS/+ mice. ree, reduced enamel epithelium; pd, predentin, rd, roof dentin; fd, floor dentin; rod, roof-forming odontoblasts; fod, floor-forming odontoblasts. Scale bars: 200 μm in A; 20 μm in (A1, A2).
3.5 Odontoblast differentiation in Twist2-Cre;Xbp1CS/+ mice
We further analyzed the expression of the odontoblast differentiation markers, including Dspp and Dmp1, in Twist2-Cre;Xbp1CS/+ mice by in situ hybridization and immunohistochemistry. In situ hybridization showed that DSPP mRNAs were highly expressed in both roof- and floor-forming odontoblasts in Xbp1CS/+ mice and the level of DSPP mRNAs in the odontoblasts in Twist2-Cre;Xbp1CS/+ mice was comparable to that of Xbp1CS/+ mice (Figure 6A, A1, A2). Immunohistochemistry demonstrated that DSP/DSPP immunostaining signals were strongly detected in the roof dentin matrix, with a relatively low level in the floor dentin matrix; and the levels of DSP/DSPP immunostaining signals in Twist2-Cre;Xbp1CS/+ mice were similar to those in Xbp1CS/+ mice (Figure 7A, A1, A2). In situ hybridization also showed that a low level of DMP1 mRNAs was expressed in both roof- and floor-forming odontoblasts in Xbp1CS/+ mice; and the level of DMP1 mRNAs in Twist2-Cre;Xbp1CS/+ mice was similar to that in Xbp1CS/+ mice (Figure 6B, B1, B2). Consistently, immunohistochemistry indicated that the intensity of DMP1 immunostaining signals were comparable in the roof and floor dentin matrices in Xbp1CS/+ mice and Twist2-Cre;Xbp1CS/+ mice (Figure 7B, B1, B2). These results suggest that sustained expression of XBP1S had no obvious effects on the expression of Dspp and Dmp1 in the odontoblasts in Twist2-Cre;Xbp1CS/+ mice.
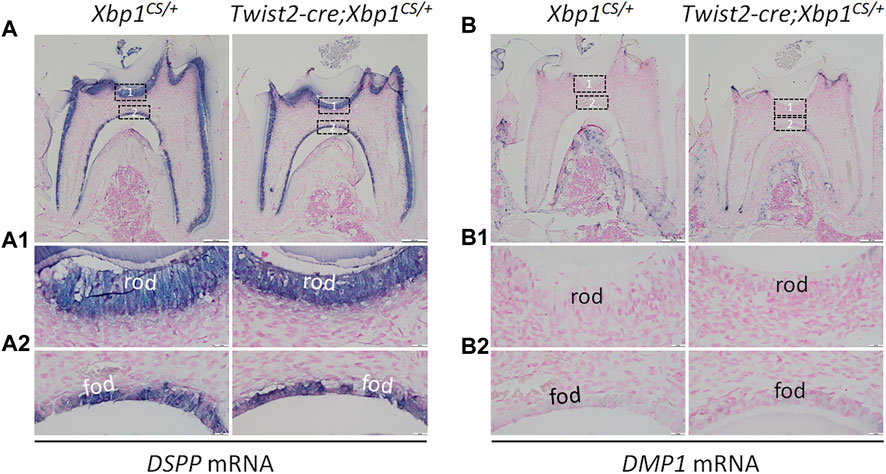
FIGURE 6. In situ hybridization analyses of DSPP and DMP1 mRNA. Shown are the representative in situ hybridization analyses of DSPP mRNA (A); signal in purple) and DMP1 mRNA (B); signal in purple) in the mandibular first molars of 3-week-old Xbp1CS/+ and Twist2-Cre;Xbp1CS/+ mice. Each image in (A, B) is from the middle region of the crown of a sagittally-sectioned mandibular first molar. (A1-A2, B1-B2) are the higher magnification views of the roof-forming odontoblasts (box1) and floor-forming odontoblasts (box 2) in (A, B), respectively. rod, roof-forming odontoblasts; fod, floor-forming odontoblasts. Scale bars: 200 μm in (A, B); 20 μm in (A1-A2, B1-B2).
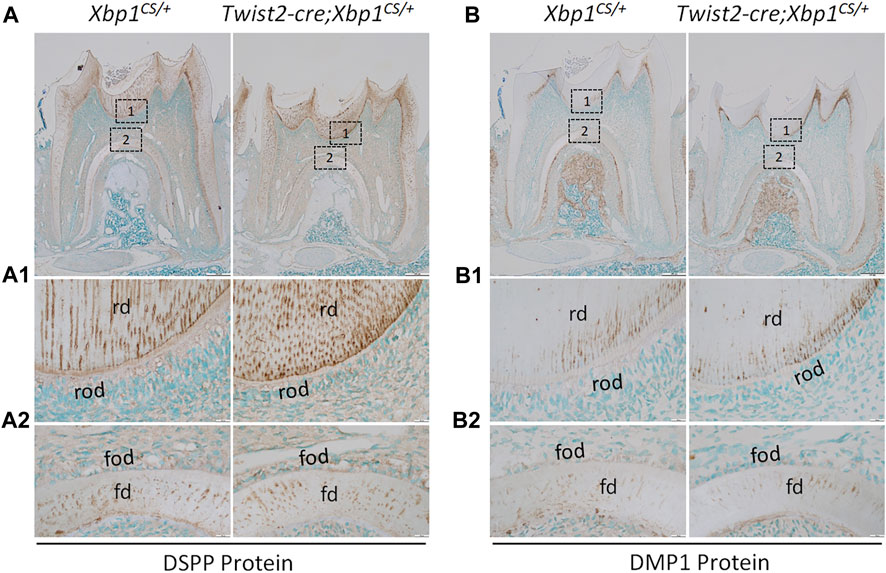
FIGURE 7. Immunohistochemical staining of DSP/DSPP and DMP1 protein. Shown are the representative images of IHC staining of DSP/DSPP (A); signal in brown) and DMP1 (B); signal in brown) in the mandibular first molars of 3-week-old Xbp1CS/+ and Twist2-Cre;Xbp1CS/+ mice. Each image in (A, B) is from the middle region of the crown of a sagittally-sectioned mandibular first molar. (A1-A2, B1-B2) are the higher magnification views of box 1 and box 2 in (A, B), respectively. rd, roof dentin; fd, floor dentin; rod, roof-forming odontoblasts; fod, floor-forming odontoblasts. Scale bars: 200 μm in (A, B); 20 μm in (A1-A2, B1-B2).
4 Discussion
XBP1S plays important roles in both ER stress and cell differentiation. Here we examined the roles of XBP1S in odontoblast differentiation during mouse tooth development by gain of function approach. We found that persistent expression of XBP1S in mice led to a significant reduction in dentin formation as well as a morphological change in odontoblasts, but had no apparent effects on the expression of the odontoblast differentiation markers.
Previous studies have demonstrated that XBP1S production depends on an unconventional splicing of unspliced XBP1U mRNA by activated IRE1α RNase (Yoshida et al., 2001; Calfon et al., 2002; Lee et al., 2002). We have shown that Twist2-Cre;Xbp1CS/+ mice expressed a high level of XBP1S following Cre-mediated recombination by immunohistochemistry (Xu et al., 2021). To further confirm that XBP1S production was independent of IRE1α activation in these mice, we generated a wild-type Xbp1 minigene and a mutant Xbp1s minigene, which corresponds to the wild-type Xbp1 and recombinant Xbp1Δ26 allele, respectively. As expected, we found that the Xbp1 minigene expressed XBP1S in a way that primarily depends on IRE1α activation, when transfected into cells in vitro. In contrast, the Xbp1s minigene constitutively expressed XBP1S, regardless of whether IRE1α was activated or not. Consistent with the in vitro results, immunohistochemistry demonstrated that both XBP1U and XBP1S were detected in the odontoblasts and other dental pulp cells in Xbp1CS/+ and Twist2-Cre;Xbp1CS/+ mice, when an antibody that recognizes both XBP1U and XBP1S was used. However, when an antibody that only reacts with XBP1S was used, XBP1S immunostaining signals were readily found in Twist2-Cre;Xbp1CS/+ mice, yet were barely detectable in Xbp1CS/+ mice. These results confirmed that XBP1S was constitutively produced in the odontoblasts and other pulp cells in Twist2-Cre;Xbp1CS/+ mice.
Constant production of XBP1S affected dentin formation in Twist2-Cre;Xbp1CS/+ mice. Plain X-ray radiography and μCT analysis demonstrated that Twist2-Cre;Xbp1CS/+ mice had enlarged dental pulp chambers, altered roof and floor dentin formation and a significant decrease in dentin/cementum formation by the age of 7 weeks. Histologically, the roof-forming odontoblasts were dramatically shorter and irregular in Twist2-Cre;Xbp1CS/+ mice, compared to those in age-matched Xbp1CS/+ control mice. Nevertheless, in situ hybridization and immunohistochemistry showed that increased XBP1S appeared to have no apparent effects on the expression of the odontoblast differentiation markers including Dspp and Dmp1. OSX is a transcription factor that is essential for osteoblast and odontoblast differentiation (Nakashima et al., 2002; Kim et al., 2015; Zhang et al., 2015; Yang et al., 2017). Moreover, it has been shown that XBP1S stimulates the expression of Osx during osteoblast differentiation (Tohmonda et al., 2011). However, immunohistochemistry demonstrated that the OSX immunostaining signals in Twist2-Cre;Xbp1CS/+ mice were comparable to those in Xbp1CS/+ mice (Supplementary Figure S2). Taken together, these findings suggest that constitutive expression of XBP1S negatively affected odontoblast function and dentin formation in mice. Further studies are needed to determine how sustained XBP1S caused a negative impact on odontoblast function in the future.
Accumulating evidence supports that the level and transcriptional activity of XBP1S is tightly regulated at the translational and post-translational levels during ER stress. First, XBP1S is only translated when the unspliced XBP1U mRNA is converted to the spliced mRNA by IRE1α RNase that is activated upon ER stress (Yoshida et al., 2001; Calfon et al., 2002). Second, XBP1S is subject to ubiquitin-mediated proteasomal degradation (Lee et al., 2018; Wang et al., 2019; Sun et al., 2020). Third, XBP1U also binds to XBP1S and enhances its degradation by proteasome (Tirosh et al., 2006; Yoshida et al., 2006). Lastly, it has been shown that other posttranslational modifications, such as phosphorylation, acetylation/deacetylation and sumoylation/desumoylation, can affect XBP1S protein stability and transcriptional activity (Lee et al., 2011; Wang et al., 2011; Jiang et al., 2012; Liu et al., 2016; Wang et al., 2016). Overall, these mechanisms ensure that XBP1S is only produced and functional when it is needed during ER stress, and that it is degraded once ER stress is relieved.
In summary, we have shown that persistent production of XBP1S adversely affected odontoblast function and dentin formation. These findings further highlight the importance of controlling the level and transcriptional activity of XBP1S within a cell. Loss of function study is warranted to determine if XBP1S is essential for odontoblast differentiation and function in the future.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Ethics statement
The animal study was approved by the Institutional Animal Care and Use Committee (IACUC) of Texas A&M University. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
QX: Writing–original draft, Data curation, Formal Analysis, Investigation, Methodology. JL: Data curation, Formal Analysis, Investigation, Writing–original draft. HZ: Investigation, Methodology, Writing–original draft. SW: Data curation, Formal Analysis, Investigation, Writing–original draft. CQ: Conceptualization, Funding acquisition, Writing–original draft. YL: Conceptualization, Funding acquisition, Methodology, Project administration, Supervision, Validation, Visualization, Writing–original draft.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by National Institute of Dental & Craniofacial Research (NIDCR) grant DE027345.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2023.1319954/full#supplementary-material
References
Acosta-Alvear D., Zhou Y., Blais A., Tsikitis M., Lents N. H., Arias C., et al. (2007). XBP1 controls diverse cell type- and condition-specific transcriptional regulatory networks. Mol. Cell. 27, 53–66. doi:10.1016/j.molcel.2007.06.011
Calfon M., Zeng H., Urano F., Till J. H., Hubbard S. R., Harding H. P., et al. (2002). IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 415, 92–96. doi:10.1038/415092a
Chavez M. B., Chu E. Y., Kram V., De Castro L. F., Somerman M. J., Foster B. L. (2021). Guidelines for micro-computed tomography analysis of rodent dentoalveolar tissues. JBMR Plus 5, e10474. doi:10.1002/jbm4.10474
Christiansen B. A. (2016). Effect of micro-computed tomography voxel size and segmentation method on trabecular bone microstructure measures in mice. Bone Rep. 5, 136–140. doi:10.1016/j.bonr.2016.05.006
Clauss I. M., Gravallese E. M., Darling J. M., Shapiro F., Glimcher M. J., Glimcher L. H. (1993). In situ hybridization studies suggest a role for the basic region-leucine zipper protein hXBP-1 in exocrine gland and skeletal development during mouse embryogenesis. Dev. Dyn. 197, 146–156. doi:10.1002/aja.1001970207
Cox J. S., Shamu C. E., Walter P. (1993). Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell. 73, 1197–1206. doi:10.1016/0092-8674(93)90648-a
Gass J. N., Gifford N. M., Brewer J. W. (2002). Activation of an unfolded protein response during differentiation of antibody-secreting B cells. J. Biol. Chem. 277, 49047–49054. doi:10.1074/jbc.M205011200
Gibson M. P., Zhu Q., Wang S., Liu Q., Liu Y., Wang X., et al. (2013). The rescue of dentin matrix protein 1 (DMP1)-deficient tooth defects by the transgenic expression of dentin sialophosphoprotein (DSPP) indicates that DSPP is a downstream effector molecule of DMP1 in dentinogenesis. J. Biol. Chem. 288, 7204–7214. doi:10.1074/jbc.M112.445775
Han D., Lerner A. G., Vande Walle L., Upton J. P., Xu W., Hagen A., et al. (2009). IRE1alpha kinase activation modes control alternate endoribonuclease outputs to determine divergent cell fates. Cell. 138, 562–575. doi:10.1016/j.cell.2009.07.017
Huang C., Wu S., Ji H., Yan X., Xie Y., Murai S., et al. (2017). Identification of XBP1-u as a novel regulator of the MDM2/p53 axis using an shRNA library. Sci. Adv. 3, e1701383. doi:10.1126/sciadv.1701383
Iwakoshi N. N., Lee A. H., Vallabhajosyula P., Otipoby K. L., Rajewsky K., Glimcher L. H. (2003). Plasma cell differentiation and the unfolded protein response intersect at the transcription factor XBP-1. Nat. Immunol. 4, 321–329. doi:10.1038/ni907
Jiang Z., Fan Q., Zhang Z., Zou Y., Cai R., Wang Q., et al. (2012). SENP1 deficiency promotes ER stress-induced apoptosis by increasing XBP1 SUMOylation. Cell. Cycle 11, 1118–1122. doi:10.4161/cc.11.6.19529
Kim T. H., Bae C. H., Lee J. C., Kim J. E., Yang X., De Crombrugghe B., et al. (2015). Osterix regulates tooth root formation in a site-specific manner. J. Dent. Res. 94, 430–438. doi:10.1177/0022034514565647
Korennykh A. V., Egea P. F., Korostelev A. A., Finer-Moore J., Stroud R. M., Zhang C., et al. (2011a). Cofactor-mediated conformational control in the bifunctional kinase/RNase Ire1. BMC Biol. 9, 48. doi:10.1186/1741-7007-9-48
Korennykh A. V., Egea P. F., Korostelev A. A., Finer-Moore J., Zhang C., Shokat K. M., et al. (2009). The unfolded protein response signals through high-order assembly of Ire1. Nature 457, 687–693. doi:10.1038/nature07661
Korennykh A. V., Korostelev A. A., Egea P. F., Finer-Moore J., Stroud R. M., Zhang C., et al. (2011b). Structural and functional basis for RNA cleavage by Ire1. BMC Biol. 9, 47. doi:10.1186/1741-7007-9-47
Lee A. H., Chu G. C., Iwakoshi N. N., Glimcher L. H. (2005). XBP-1 is required for biogenesis of cellular secretory machinery of exocrine glands. EMBO J. 24, 4368–4380. doi:10.1038/sj.emboj.7600903
Lee A. H., Iwakoshi N. N., Glimcher L. H. (2003). XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol. Cell. Biol. 23, 7448–7459. doi:10.1128/mcb.23.21.7448-7459.2003
Lee A. H., Scapa E. F., Cohen D. E., Glimcher L. H. (2008a). Regulation of hepatic lipogenesis by the transcription factor XBP1. Science 320, 1492–1496. doi:10.1126/science.1158042
Lee J., Salazar Hernandez M. A., Auen T., Mucka P., Lee J., Ozcan U. (2018). PGC-1α functions as a co-suppressor of XBP1s to regulate glucose metabolism. Mol. Metab. 7, 119–131. doi:10.1016/j.molmet.2017.10.010
Lee J., Sun C., Zhou Y., Lee J., Gokalp D., Herrema H., et al. (2011). p38 MAPK-mediated regulation of Xbp1s is crucial for glucose homeostasis. Nat. Med. 17, 1251–1260. doi:10.1038/nm.2449
Lee K., Tirasophon W., Shen X., Michalak M., Prywes R., Okada T., et al. (2002). IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes. & Dev. 16, 452–466. doi:10.1101/gad.964702
Lee K. P., Dey M., Neculai D., Cao C., Dever T. E., Sicheri F. (2008b). Structure of the dual enzyme Ire1 reveals the basis for catalysis and regulation in nonconventional RNA splicing. Cell. 132, 89–100. doi:10.1016/j.cell.2007.10.057
Li H., Korennykh A. V., Behrman S. L., Walter P. (2010). Mammalian endoplasmic reticulum stress sensor IRE1 signals by dynamic clustering. Proc. Natl. Acad. Sci. U. S. A. 107, 16113–16118. doi:10.1073/pnas.1010580107
Liang T., Xu Q., Zhang H., Wang S., Diekwisch T. G. H., Qin C., et al. (2021). Enamel defects associated with dentin sialophosphoprotein mutation in mice. Front. Physiol. 12, 724098. doi:10.3389/fphys.2021.724098
Liang T., Zhang H., Xu Q., Wang S., Qin C., Lu Y. (2019). Mutant dentin sialophosphoprotein causes dentinogenesis imperfecta. J. Dent. Res. 22034519854029, 912–919. doi:10.1177/0022034519854029
Liou H. C., Boothby M. R., Finn P. W., Davidon R., Nabavi N., Zeleznik-Le N. J., et al. (1990). A new member of the leucine zipper class of proteins that binds to the HLA DR alpha promoter. Science 247, 1581–1584. doi:10.1126/science.2321018
Liu J., Ibi D., Taniguchi K., Lee J., Herrema H., Akosman B., et al. (2016). Inflammation improves glucose homeostasis through ikkβ-XBP1s interaction. Cell. 167, 1052–1066. doi:10.1016/j.cell.2016.10.015
Luo X., Alfason L., Wei M., Wu S., Kasim V. (2022). Spliced or unspliced, that is the question: the biological roles of XBP1 isoforms in pathophysiology. Int. J. Mol. Sci. 23, 2746. doi:10.3390/ijms23052746
Martin D., Li Y., Yang J., Wang G., Margariti A., Jiang Z., et al. (2014). Unspliced X-box-binding protein 1 (XBP1) protects endothelial cells from oxidative stress through interaction with histone deacetylase 3. J. Biol. Chem. 289, 30625–30634. doi:10.1074/jbc.M114.571984
Meng T., Huang Y., Wang S., Zhang H., Dechow P. C., Wang X., et al. (2015). Twist1 is essential for tooth morphogenesis and odontoblast differentiation. J. Biol. Chem. 290, 29593–29602. doi:10.1074/jbc.M115.680546
Nakashima K., Zhou X., Kunkel G., Zhang Z., Deng J. M., Behringer R. R., et al. (2002). The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell. 108, 17–29. doi:10.1016/s0092-8674(01)00622-5
Navon A., Gatushkin A., Zelcbuch L., Shteingart S., Farago M., Hadar R., et al. (2010). Direct proteasome binding and subsequent degradation of unspliced XBP-1 prevent its intracellular aggregation. FEBS Lett. 584, 67–73. doi:10.1016/j.febslet.2009.11.069
Oda Y., Okada T., Yoshida H., Kaufman R. J., Nagata K., Mori K. (2006). Derlin-2 and Derlin-3 are regulated by the mammalian unfolded protein response and are required for ER-associated degradation. J. Cell. Biol. 172, 383–393. doi:10.1083/jcb.200507057
Oikawa D., Kimata Y., Kohno K., Iwawaki T. (2009). Activation of mammalian IRE1alpha upon ER stress depends on dissociation of BiP rather than on direct interaction with unfolded proteins. Exp. Cell. Res. 315, 2496–2504. doi:10.1016/j.yexcr.2009.06.009
Reimold A. M., Etkin A., Clauss I., Perkins A., Friend D. S., Zhang J., et al. (2000). An essential role in liver development for transcription factor XBP-1. Genes. & Dev. 14, 152–157. doi:10.1101/gad.14.2.152
Reimold A. M., Iwakoshi N. N., Manis J., Vallabhajosyula P., Szomolanyi-Tsuda E., Gravallese E. M., et al. (2001). Plasma cell differentiation requires the transcription factor XBP-1. Nature 412, 300–307. doi:10.1038/35085509
Shaffer A. L., Shapiro-Shelef M., Iwakoshi N. N., Lee A. H., Qian S. B., Zhao H., et al. (2004). XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity 21, 81–93. doi:10.1016/j.immuni.2004.06.010
Shamu C. E., Walter P. (1996). Oligomerization and phosphorylation of the Ire1p kinase during intracellular signaling from the endoplasmic reticulum to the nucleus. EMBO J. 15, 3028–3039. doi:10.1002/j.1460-2075.1996.tb00666.x
Sriburi R., Jackowski S., Mori K., Brewer J. W. (2004). XBP1: a link between the unfolded protein response, lipid biosynthesis, and biogenesis of the endoplasmic reticulum. J. Cell. Biol. 167, 35–41. doi:10.1083/jcb.200406136
Sun H., Wei G., Liu H., Xiao D., Huang J., Lu J., et al. (2020). Inhibition of XBP1s ubiquitination enhances its protein stability and improves glucose homeostasis. Metabolism 105, 154046. doi:10.1016/j.metabol.2019.154046
Tirasophon W., Welihinda A. A., Kaufman R. J. (1998). A stress response pathway from the endoplasmic reticulum to the nucleus requires a novel bifunctional protein kinase/endoribonuclease (Ire1p) in mammalian cells. Genes. Dev. 12, 1812–1824. doi:10.1101/gad.12.12.1812
Tirosh B., Iwakoshi N. N., Glimcher L. H., Ploegh H. L. (2006). Rapid turnover of unspliced Xbp-1 as a factor that modulates the unfolded protein response. J. Biol. Chem. 281, 5852–5860. doi:10.1074/jbc.M509061200
Tohmonda T., Miyauchi Y., Ghosh R., Yoda M., Uchikawa S., Takito J., et al. (2011). The IRE1α-XBP1 pathway is essential for osteoblast differentiation through promoting transcription of Osterix. EMBO Rep. 12, 451–457. doi:10.1038/embor.2011.34
Wang F. F., Liu M. Z., Sui Y., Cao Q., Yan B., Jin M. L., et al. (2016). Deficiency of SUMO-specific protease 1 induces arsenic trioxide-mediated apoptosis by regulating XBP1 activity in human acute promyelocytic leukemia. Oncol. Lett. 12, 3755–3762. doi:10.3892/ol.2016.5162
Wang F. M., Chen Y. J., Ouyang H. J. (2011). Regulation of unfolded protein response modulator XBP1s by acetylation and deacetylation. Biochem. J. 433, 245–252. doi:10.1042/BJ20101293
Wang X. Z., Harding H. P., Zhang Y., Jolicoeur E. M., Kuroda M., Ron D. (1998). Cloning of mammalian Ire1 reveals diversity in the ER stress responses. EMBO J. 17, 5708–5717. doi:10.1093/emboj/17.19.5708
Wang Y., Zhang Y., Yi P., Dong W., Nalin A. P., Zhang J., et al. (2019). The IL-15-AKT-XBP1s signaling pathway contributes to effector functions and survival in human NK cells. Nat. Immunol. 20, 10–17. doi:10.1038/s41590-018-0265-1
Welihinda A. A., Kaufman R. J. (1996). The unfolded protein response pathway in Saccharomyces cerevisiae. Oligomerization and trans-phosphorylation of Ire1p (Ern1p) are required for kinase activation. J. Biol. Chem. 271, 18181–18187. doi:10.1074/jbc.271.30.18181
Xu Q., Zhang H., Wang S., Qin C., Lu Y. (2021). Constitutive expression of spliced XBP1 causes perinatal lethality in mice. Genesis 59, e23420. doi:10.1002/dvg.23420
Yang G., Yuan G., Macdougall M., Zhi C., Chen S. (2017). BMP-2 induced Dspp transcription is mediated by Dlx3/Osx signaling pathway in odontoblasts. Sci. Rep. 7, 10775. doi:10.1038/s41598-017-10908-8
Yang L., Dai R., Wu H., Cai Z., Xie N., Zhang X., et al. (2022). Unspliced XBP1 counteracts β-catenin to inhibit vascular calcification. Circ. Res. 130, 213–229. doi:10.1161/CIRCRESAHA.121.319745
Yoshida H., Matsui T., Hosokawa N., Kaufman R. J., Nagata K., Mori K. (2003). A time-dependent phase shift in the mammalian unfolded protein response. Dev. Cell. 4, 265–271. doi:10.1016/s1534-5807(03)00022-4
Yoshida H., Matsui T., Yamamoto A., Okada T., Mori K. (2001). XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 107, 881–891. doi:10.1016/s0092-8674(01)00611-0
Yoshida H., Oku M., Suzuki M., Mori K. (2006). pXBP1(U) encoded in XBP1 pre-mRNA negatively regulates unfolded protein response activator pXBP1(S) in mammalian ER stress response. J. Cell. Biol. 172, 565–575. doi:10.1083/jcb.200508145
Yu K., Xu J., Liu Z., Sosic D., Shao J., Olson E. N., et al. (2003). Conditional inactivation of FGF receptor 2 reveals an essential role for FGF signaling in the regulation of osteoblast function and bone growth. Development 130, 3063–3074. doi:10.1242/dev.00491
Zhang H., Jiang Y., Qin C., Liu Y., Ho S. P., Feng J. Q. (2015). Essential role of osterix for tooth root but not crown dentin formation. J. bone mineral Res. official J. Am. Soc. Bone Mineral Res. 30, 742–746. doi:10.1002/jbmr.2391
Zhang H., Xie X., Liu P., Liang T., Lu Y., Qin C. (2018). Transgenic expression of dentin phosphoprotein (DPP) partially rescued the dentin defects of DSPP-null mice. PLoS One 13, e0195854. doi:10.1371/journal.pone.0195854
Zhao G., Fu Y., Cai Z., Yu F., Gong Z., Dai R., et al. (2017). Unspliced XBP1 confers VSMC homeostasis and prevents aortic aneurysm formation via FoxO4 interaction. Circ. Res. 121, 1331–1345. doi:10.1161/CIRCRESAHA.117.311450
Keywords: X-box binding protein 1 (XBP1), tooth development, odontoblast, cell differentiation, dentin, dentinogenesis
Citation: Xu Q, Li J, Zhang H, Wang S, Qin C and Lu Y (2024) Constitutive expression of spliced X-box binding protein 1 inhibits dentin formation in mice. Front. Physiol. 14:1319954. doi: 10.3389/fphys.2023.1319954
Received: 11 October 2023; Accepted: 26 December 2023;
Published: 10 January 2024.
Edited by:
Guohua Yuan, Wuhan University, ChinaReviewed by:
Brian L. Foster, The Ohio State University, United StatesYong-Hee Patricia Chun, The University of Texas Health Science Center at San Antonio, United States
Copyright © 2024 Xu, Li, Zhang, Wang, Qin and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yongbo Lu, eWx1QHRhbXUuZWR1
 Qian Xu
Qian Xu Jiahe Li
Jiahe Li Hua Zhang
Hua Zhang Chunlin Qin
Chunlin Qin Yongbo Lu
Yongbo Lu