- 1Laboratory of Marine Biology Protein Engineering, Marine Science and Technical College, Zhejiang Ocean University, Zhoushan, Zhejiang, China
- 2Italian Institute for Environmental Protection and Research (ISPRA), Livorno, Italy
Mytilus coruscus is an economically important marine calcifier living in the Yangtze River estuary sea area, where seasonal fluctuations in natural pH occur owing to freshwater input, resulting in a rapid reduction in seawater pH. In addition, Mytilus constantly suffers from shell fracture or injury in the natural environment, and the shell repair mechanisms in mussels have evolved to counteract shell injury. Therefore, we utilized shell-complete and shell-damaged Mytilus coruscus in this study and performed transcriptomic analysis of the mantle to investigate whether the expression of mantle-specific genes can be induced by acute seawater acidification and how the mantle responds to acute acidification during the shell repair process. We found that acute acidification induced more differentially expressed genes than shell damage in the mantle, and the biomineralization-related Gene Ontology terms and KEGG pathways were significantly enriched by these DEGs. Most DEGs were upregulated in enriched pathways, indicating the activation of biomineralization-related processes in the mussel mantle under acute acidification. The expression levels of some shell matrix proteins and antimicrobial peptides increased under acute acidification and/or shell damage, suggesting the molecular modulation of the mantle for the preparation and activation of the shell repairing and anti-infection under adverse environmental conditions. In addition, morphological and microstructural analyses were performed for the mantle edge and shell cross-section, and changes in the mantle secretory capacity and shell inner film system induced by the two stressors were observed. Our findings highlight the adaptation of M. coruscus in estuarine areas with dramatic fluctuations in pH and may prove instrumental in its ability to survive ocean acidification.
1 Introduction
As carbon dioxide (CO2) is continuously being released into the atmosphere from various anthropogenic sources and absorbed into surface oceans, it forms carbonic acid in seawater, lowering ambient pH and decreasing the abundance of carbonate ions (CO32−) (Sabine et al., 2004; Doney et al., 2009). This process, known as ocean acidification (OA), has far-reaching implications for ocean bio-systems. Thus, OA has emerged as a global environmental issue, and accumulating research has highlighted vulnerabilities in calcification-dependent marine metazoans that are sensitive to changes in carbonate chemistry (Melzner et al., 2020). OA has profound effects on marine organisms and ecosystems, with negative impacts on physiological processes, such as calcification, growth, reproduction, and survival (Strader et al., 2020), in calcifying marine species. Interestingly, some marine species exhibit robust physiological changes relevant to OA (Melzner et al., 2009; Sunday et al., 2014), whereas others are highly sensitive (Teixidó et al., 2018), indicating that the response of bivalves to rising pCO2 is species-specific. However, there is still limited mechanistic understanding of the physiological traits responsible for the differential sensitivities.
The genus Mytilus represents not only a species with significant economic importance in aquaculture worldwide but also shows strong tolerance to a wide range of environmental factors (Beyer et al., 2017). Previous studies have demonstrated the effects of OA on this species. For instance, the embryonic growth of Mytilus edulis appears to be affected by acidified seawater (Bechmann et al., 2011), and the structures of the gills and digestive glands are also affected by seawater acidification, thus inhibiting the energy intake of mussels (Xu et al., 2020). In addition, the calcified shell of Mytilus is a crucial protective exoskeleton for this species, and the structural integrity of Mytilus shell can be affected by high pCO2 (1,000 µatm) even though the biomineralization continues (Fitzer et al., 2014). OA is generally considered to be detrimental to marine shellfishes (Byrne and Fitzer, 2019). The stunting effects of OA on the larval and adult stages of shellfish have been observed in several studies. Reduced seawater CO32− concentration hinders calcification of shellfish, and the acidified seawater can also accelerate the dissolution of CaCO3 minerals of calcified shell (Doney et al., 2009; Harv et al., 2018). However, some studies have reported only minor effects of OA on Mytilus. For example, in a coupled field and laboratory study, Mytilus showed remarkable tolerance to high ambient pCO2 when the food supply was abundant (Thomsen et al., 2013). In addition, high rates of Mytilus calcifying capacity have been observed even in seawater with highly undersaturated CaCO3 (Thomsen and Melzner, 2010), indicating a special biomineralization mechanism of Mytilus to combat OA. In Mytilus, the mantle is the tissue that produces the shell, and the mature shell is composed of different structural layers, including the nacre and prismatic layers (Gao et al., 2015; Liao et al., 2015). Although Mytilus is a globally distributed species with great economic value and ecosystem functions, studies on the effects of OA on Mytilus are limited. Several studies have revealed the effects of OA on Mytilus physiological functions (Broszeit et al., 2016), larval development (Gazeau et al., 2010; Kelly et al., 2016; Ventura et al., 2016), the byssus (Zhao et al., 2017), and mantle gene expression patterns (Hüning et al., 2013). However, the molecular response, especially the biomineralization-related response of Mytilus to OA, remains largely unknown.
As reviewed by Strader et al. (2020), transcriptomics is an effective method and transcriptome profiling is a robust and informative method for examining and elucidating the molecular-level response of marine metazoans to OA. However, the research on the molecular response of Mytilus to OA is still limited. Most experiments have been performed with middle- or long-term exposure to OA, ranging from 1 week to over 3 months (Strader et al., 2020). Previous studies have highlighted the long-term adaptation of Mytilus to OA (Thomsen et al., 2017), and the ability to continue calcification and maintain Mytilus shell integrity under predicted low-pH scenarios has been studied in several key physiological functions of M. edulis under OA (Kroeker et al., 2013). Notably, Kapsenberg et al recently revealed that some genes involved in the shell development of Mytilus larvae exhibited shifts in allele frequencies under low pH conditions, indicating the potential for rapid adaptation to OA in this species (Kapsenberg et al., 2022). To evaluate the molecular responses of Mytilus to acute acidification, the mantle transcriptome was sequenced and analyzed for the mussel with a 48 h of exposure time in acidified seawater, and the transcriptomic data of the mantle revealed a rapid gene expressional change. On the other hand, mussels frequently suffer shell fractures or injuries in their natural environment, and therefore shell repair mechanisms in mussels have evolved to counteract shell injury (Hüning et al., 2016). Recently, Yarra et al. (2021) reported a shell repair experimental model for Mytilus to explore gene expression patterns during the shell damage repair process, and more than 600 differentially expressed genes were identified under these conditions. However, the shell repair process in mussels with OA remains unclear. Therefore, we performed a transcriptomic analysis of Mytilus coruscus mantle with shell damage in normal and acidified seawater. Our data revealed the genes and their functions involved in the rapid response of mussels to OA and provided clues for exploring the biomineralization mechanisms of the mussel mantle under OA and shell damage-repair processes.
2 Materials and methods
2.1 Mussel sampling and treatment
Adult mussels (70–80 mm in length) were collected from a mussel farm located on the Gouqi Islands of the East China Sea in May 2022. Mussels were acclimated at 22°C for 7 days in clean seawater (pH 8.1 and salinity 19‰). The collected mussels were divided into four groups: mussels with complete shell raised in normal seawater and acidified seawater (designated as CN and CA, respectively), and shell-drilled mussels raised in normal seawater and acidified seawater (designated as DN and DA, respectively). Shell damage model was prepared using the shell drilling method described by Yarra et al. (2021). Shell drilled and undrilled mussels were mixed and raised in normal seawater (pH 8.1) and acidified seawater (pH 7.4), respectively, for 48 h. The pH of the seawater was monitored using a pH meter and controlled by a Seawater acidifier (Starfish SF0S02, Qingdao, China) with a CO2 pump. All mussels were fed daily with planktonic food (Chaetoceros mulleri and Isochrysis zhangjiangensis).
2.2 RNA extraction and sequencing
Total RNA was extracted according to the methods used in our previous study (He et al., 2022) from the mantle tissues of the four mussel groups (CN, CA, DN, and DA). In total, there were twelve samples, including three biological replicates for each group, and each sample was collected from six individuals. Complementary DNA (cDNA) from the 12 samples was synthesized using a SuperScript double-stranded cDNA synthesis kit (Invitrogen, Carlsbad, CA, United States of America) with random hexamer primers (Illumina). The cDNA library was then prepared and amplified on cBot (Truseq PE Cluster Kit v3-cBot-HS, Illumina) to generate clusters on the flow cell, which was then paired-end sequenced using a Novaseq 6,000 System (Illumina).
2.3 Transcriptomic data processing and bioinformatics analysis
Using the protocol provided in our previous work (He et al., 2022), the raw sequenced reads were trimmed using SeqPrep and Sickle software, quality control was processed using RSeQC (v2.3.6), and the clean reads were separately aligned to the M. coruscus genome (release number: PRJEB33342) in the orientation mode using HISAT2 software. The mapped reads were further assembled based on the genome data of M. coruscus, and clustered using the Chrysalis clusters software. The “unigenes” were designated by the longest sequences in each cluster. Unigene annotation was performed using BLASTX alignment against NCBI non-redundant protein sequences (NR), Gene Ontology (GO), Eukaryotic Orthologous Groups (KOG), Kyoto Encyclopedia of Genes and Genomes (KEGG), Clusters of Orthologous Groups of proteins (COG), and Pfam databases, with an E-value of <1.0e-5 and the default parameters. The alignment results with the highest homology were used to determine the sequence directions of the unigenes.
To identify differentially expressed genes (DEGs) between the various groups, the expression level of each transcript was calculated according to the transcripts per million reads (TPM) method, and DESeq2 with |log2FC|>1 and Q value ≤0.05 were considered as the threshold for DEGs. In addition, GO and KEGG functional-enrichment analysis were performed at Bonferroni-corrected p-value ≤ 0.05 compared with the whole-transcriptome background. GO functional enrichment and KEGG pathway analyses were performed using GoATools and KOBAS online tools, respectively.
2.4 Quantitative PCR (qPCR) validation of the RNA-Seq analysis
Using the same RNA samples used in the transcriptomic analysis of the mantle, validation of the RNA-seq data was performed based on the randomly selected 22 DEGs from RNASeq data, following the protocol described in our previous work (He et al., 2022). Briefly, a QuantStudio 1 (Applied Biosystems, Thermo Fisher Scientific) was used for qPCR analysis, and the qPCR cycling conditions were as follows: 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60°C for 30 s. EF-1α gene was used as an internal control, and the gene expression level was evaluated using the 2−ΔΔCT method. The primers used for qPCR are listed in Supplementary Table S1. A correlation analysis for the expression variation trend of these randomly selected DEGs was performed using Pearson methods.
2.5 AB-PAS staining of the mantle edge
The mantle edges from four mussel groups (CN, CA, DN, and DA) were collected and immediately fixed in 4% paraformaldehyde. The tissue samples were dehydrated with a gradient series of ethanol and then embedded in paraffin, and sectioned at 5 μm using a microtome (HistoCore BIOCUT, LEICA). Before histological staining, tissue sections were treated using the protocol described in a previous study (Liu et al., 2019), and the sections were stained with Alcian blue using the periodic acid-Schiff (AB-PAS) method (McMANUS, 1946). Stained sections were examined under a light microscope (ECLIPSE E100; NIKON, Japan).
2.6 Transmission electron microscopy (TEM) analysis for the outer fold (OF) in mantle edge and scanning electron microscopy (SEM) analysis for the shell
TEM analysis was performed on the mantle edge according to a previously described protocol (Myers et al., 2007). Briefly, the mantle edges from four mussel groups (CN, CA, DN, and DA) were collected and immediately fixed in a fixative solution containing 4% paraformaldehyde and 2% glutaraldehyde, post-fixed in osmium vapor, and embedded in resin. Sections with thickness of 70–90 nm were cut, floated on copper grids, stained with 2.5% uranyl acetate for 15 min in the dark, and counterstained with lead citrate for 3 min. The sections were viewed and photographed using a Hitachi 600AB transmission microscope (Hitachi, Tokyo, Japan) at an accelerating voltage of 80 kV.
Shells drilled from the mussels under acute acidification were collected and fragmented. The cross-section located at the drilling site was sputter-coated with gold and examined with a SU8010 SEM (Hitachi, Japan) at an accelerating voltage of 3.0 kV.
3 Results
3.1 Transcriptome sequencing, assembly, and functional annotation
Mantle samples were processed, and 12 cDNA libraries were prepared for the four groups (CN, CA, DN, and DA). The sequencing data after sequencing on a NovaSeq 6,000 System (Illumina) are listed in Supplementary Table S2. Briefly, 72.84 Gb clean data for the 12 libraries were obtained, with an average of 6.07 GB and Q30 > 91% for each library. Of these, more than 97% passed the quality standards and were subjected to further analyses. The transcriptomic data were submitted to the NCBI SRA database (Accession No. of PRJNA918971, and the BioSample names TCN, TCS, TAN, and TAS are in correspondence with CN, DN, CA, and DA, respectively, in this study).
Using HISAT2, the acquired clean reads were mapped to the M. coruscus chromosome-level genome (Yang et al., 2021), with an average alignment ratio of 60.75% and an average unique alignment ratio of 57.23% (Supplementary Table S3). Quality Control (QC) of the mapped reads was assessed using RSeQC (v2.3.6), and the coverage of sequencing and the percentage of reads mapped to genome regions are summarized in Supplementary Figure S1 and Supplementary Table S4, respectively. The uniform distribution of reads in the gene body and the high CDS coverage of the reads indicated the high quality of the transcriptomic data. The filtered high-quality reads were assembled into 86, 924 transcripts using Trinity software. The size distribution of all assembled transcripts is shown in Supplementary Figure S2. Most assembled transcripts had a length of >1,800 bp.
All transcripts acquired in this study were annotated against GO, KEGG, COG, NR, SWISS-PROTEIN, and Pfam databases, and a total of 71,437 (82.18%) transcripts were annotated with an E-value threshold of 1 × 10−5 (Supplementary Figure S3).
3.2 Expression quantity analysis and DEGs
The transcriptomes were quantitatively analyzed using RSEM software, and the TPM value was used to assess expression quantity. The expression distribution of all the transcripts is shown in Figure 1. Principal component analysis (PCA) was further performed for the samples, and the three-dimensional PCA score plots (Figure 2) showed that most samples clustered closely and there was an obvious separation between the treated samples (CA, DN, and DA group) and the control samples (CN group), demonstrating the stability and reproducibility of Illumina sequencing. In addition, the correlation between various samples is summarized in Supplementary Figure S4, and the Venn map showing the conserved and unique transcripts with TPM >1 in all samples is summarized in Supplementary Figure S5, showing 17,521 (40.82%) transcripts shared by all the samples.
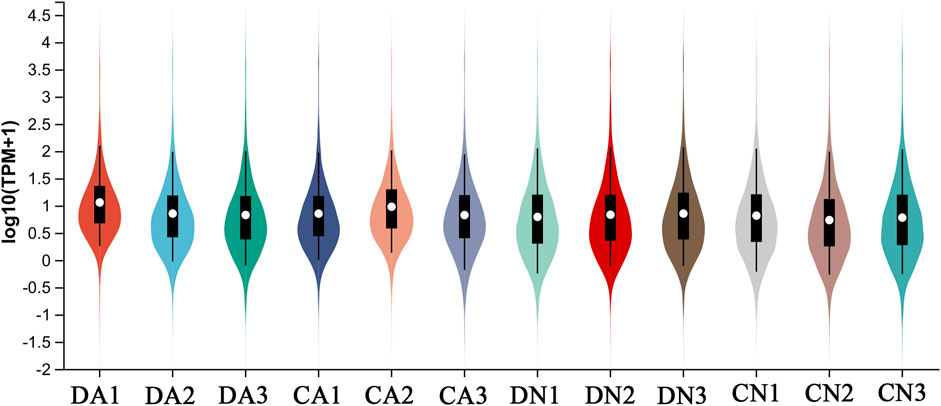
FIGURE 1. Distribution of expression level (Log10TPM+1) of all transcripts from four groups of mantle samples with triplicate for each group. DA, the mussel with drilled shell and raised in acidified sea water (pH 7.4) with exposure time of 48 h; CA, the mussel with complete shell and raised in acidified sea water (pH 7.4) with exposure time of 48 h; DN, the mussel with drilled shell and raised in normal sea water (pH 8.1); CN, the mussel with complete shell and raised in normal sea water (pH 8.1).
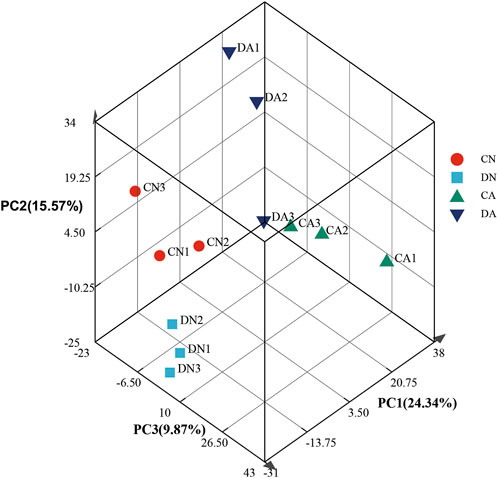
FIGURE 2. Principle component analysis of gene expression profiles at different phases for the twelve mantle samples. DA, the mussel with drilled shell and raised in acidified sea water (pH 7.4) with exposure time of 48 h; CA, the mussel with complete shell and raised in acidified sea water (pH 7.4) with exposure time of 48 h; DN, the mussel with drilled shell and raised in normal sea water (pH 8.1); CN, the mussel with complete shell and raised in normal sea water (pH 8.1).
Using DESeq2 software and Benjamini–Hochberg multiple test calibration method, DEGs with fold change >2 and Q value ≤0.05 were detected from the pair-wise comparisons. Overall, 15,180 DEGs were identified. The majority of DEGs (3,924) were observed in the pairwise comparison of the DA and DN groups, and the minimal DEGs (227) were observed in the DA vs. CA comparison (Figure 3). Venn and volcano maps of these DEGs for the six pairwise comparisons among the four groups are shown in Figure 4 and Supplementary Figure S6, respectively. These results indicated that acidified seawater and shell damage induced completely different responses in the mussel mantle.
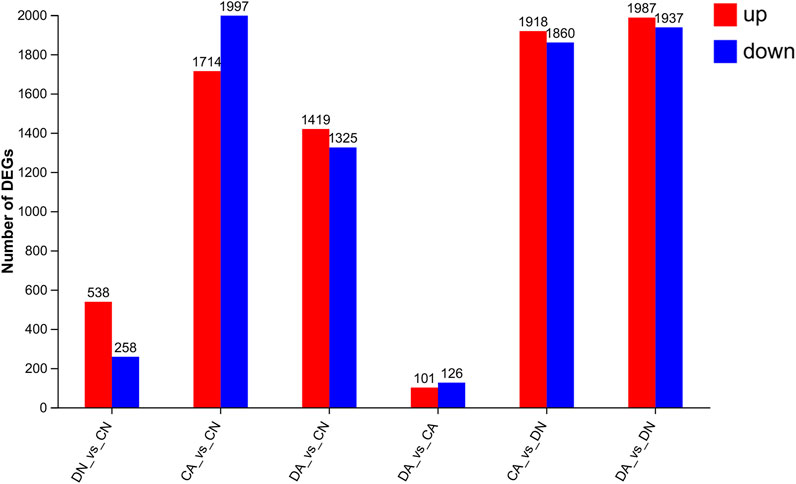
FIGURE 3. Number of DEGs (fold change > 2, p-value < 0.05) from six pairwise comparisons among the mantle samples. DA, the mussel with drilled shell and raised in acidified sea water (pH 7.4) with exposure time of 48 h; CA, the mussel with complete shell and raised in acidified sea water (pH 7.4) with exposure time of 48 h; DN, the mussel with drilled shell and raised in normal sea water (pH 8.1); CN, the mussel with complete shell and raised in normal sea water (pH 8.1).
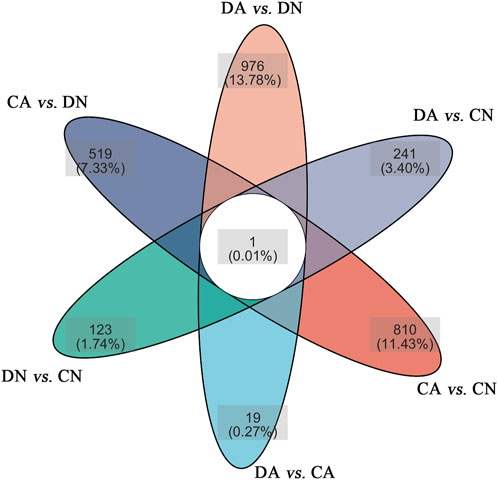
FIGURE 4. Venn maps of the DEGs in six pairwise comparisons of the mantle samples. DA, the mussel with drilled shell and raised in acidified sea water (pH 7.4) with exposure time of 48 h; CA, the mussel with complete shell and raised in acidified sea water (pH 7.4) with exposure time of 48 h; DN, the mussel with drilled shell and raised in normal sea water (pH 8.1); CN, the mussel with complete shell and raised in normal sea water (pH 8.1).
3.3 Functional annotation and enrichment of DEGs
The results of GO annotation for DEGs of the six pairwise compared groups are summarized in Supplementary Figure S7 with the top 20 GO terms, and detailed information is listed in Supplementary Table S5. The DEGs from the six pairwise comparisons were assigned to 47 GO terms in three categories (molecular function, cellular components, and biological processes). The most dominant GO terms presented in the three categories were “Cellular process,” “Membrane part,” and “Binding,” In addition, DEGs were annotated against the COG database, and the results are shown in Supplementary Figure S8. Most DEGs were annotated as “Function unknown”, followed by “Posttranslational modification, protein turnover, chaperones”, “Intracellular trafficking, secretion, and vesicular transport”,” “Transcription”, and “Signal transduction mechanisms.”
Annotated DEGs were subsequently subjected to functional enrichment analysis using Fisher’s exact test (P-adjust <0.05), and significantly enriched GO terms and KEGG pathways were identified and summarized as bubble diagrams for the DEGs from six pairwise comparisons (Figures 5, 6). We observed that acute acidification and shell damage induced different GO responses in mussel mantles. As shown in Figure 5, the cellular component, cellular anatomical entity, and integral/intrinsic component of the membrane appeared to be the most enriched DEGs for DN vs. CN, while heme binding, ligase activity, and microtubule-based processes were the most enriched DEGs for CA vs. CN. In addition, the GO terms of small molecule metabolic processes, localization, and transport were the most enriched DEGs in DA vs. DN, indicating the main responses of shell-damaged mussels under acute acidification (Figure 5). Surprisingly, under the same acidified sea water, the comparison of DA vs. CA showed no enriched GO terms with P-adjust <0.05 for the DEGs, and a p-value < 0.05 was set as threshold and only six GO terms were thus enriched (Figure 5).
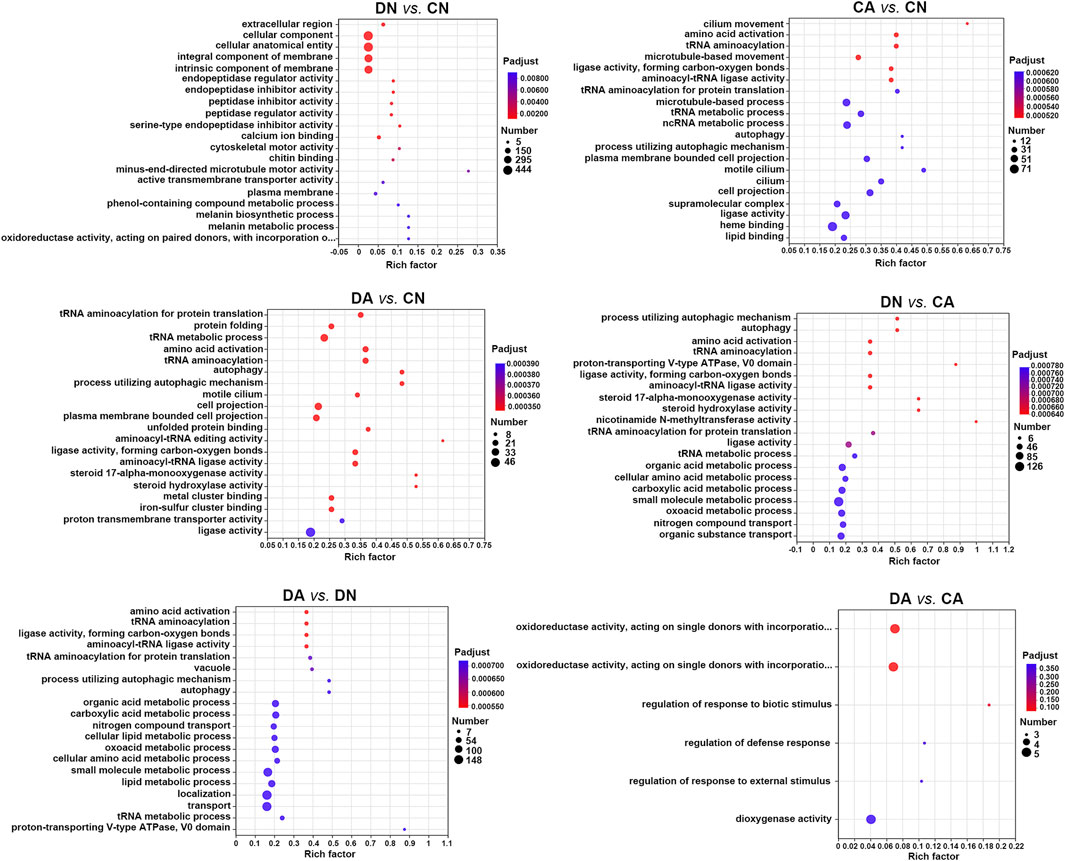
FIGURE 5. Bubble diagram of top 20 GO terms enriched (with P-adjust < 0.05) by DEGs in six pairwise comparisons of mantle samples. DA, the mussel with drilled shell and raised in acidified sea water (pH 7.4) with exposure time of 48 h; CA, the mussel with complete shell and raised in acidified sea water (pH 7.4) with exposure time of 48 h; DN, the mussel with drilled shell and raised in normal sea water (pH 8.1); CN, the mussel with complete shell and raised in normal sea water (pH 8.1). Note that, a p-value <0.05 instead of P-adjust <0.05 was set as the threshold for the enrichment in DA vs. CA.
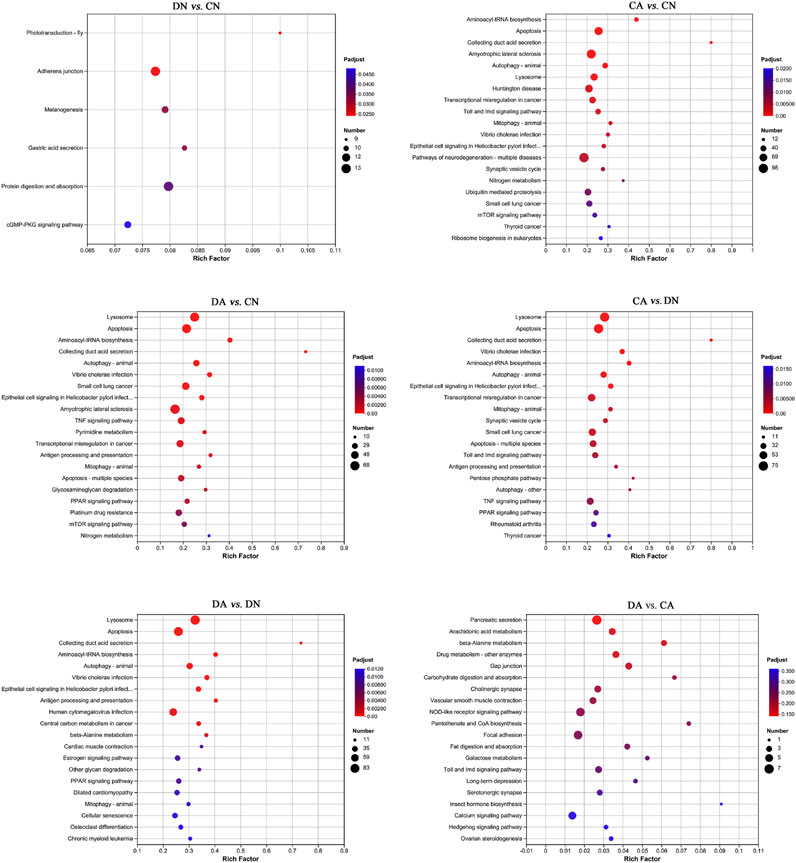
FIGURE 6. Bubble diagram of top 20 KEGG pathways enriched (with P-adjust < 0.05) by DEGs in six pairwise comparisons of mantle samples. DA, the mussel with drilled shell and raised in acidified sea water (pH 7.4) with exposure time of 48 h; CA, the mussel with complete shell and raised in acidified sea water (pH 7.4) with exposure time of 48 h; DN, the mussel with drilled shell and raised in normal sea water (pH 8.1); CN, the mussel with complete shell and raised in normal sea water (pH 8.1). Note that, a p-value <0.05 instead of P-adjust <0.05 was set as the threshold for the enrichment in DA vs. CA.
The top 20 enriched KEGG pathways for the DEGs of the six paired comparisons are summarized in Figure 6. KEGG enrichment results revealed strong responses in the pathways of adherens junction and protein digestion and absorption in DN vs. CN, pathways of neurodegeneration and amyotrophic lateral sclerosis in CA vs. CN, pathways of lysosomes and apoptosis in DA vs. DN, and pathways of pancreatic secretion and NOD-like receptor signaling in DA vs. CA. No KEGG enrichments with P-adjust <0.05 can were acquired for DA vs. CA, and a p-value <0.05 was used for the enrichment analysis in this comparison. In addition, the number of upregulated and downregulated DEGs for each enriched KEGG pathway with P-adjust <0.05 or p-value <0.05 are listed in Table 1. Table 6 shows the 29, 38, 24, 26, and 30 enriched KEGG pathways for DN vs. CN, CA vs. CN, DA vs. CN, DA vs. CA, CA vs. DN, and DA vs. DN, respectively. Most KEGG pathways showed more enriched upregulated DEGs than downregulated DEGs.
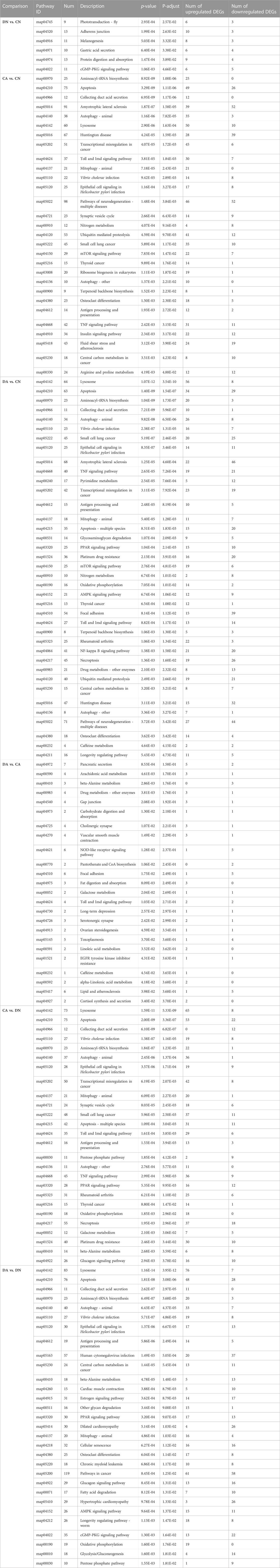
TABLE 1. KEGG pathways enriched by DEGs in six pairwise comparisons of mantle samples. DA, the mussel with drilled shell and raised in acidified sea water (pH 7.4) with exposure time of 48 h; CA, the mussel with complete shell and raised in acidified sea water (pH 7.4) with exposure time of 48 h; DN, the mussel with drilled shell and raised in normal sea water (pH 8.1); CN, the mussel with complete shell and raised in normal sea water (pH 8.1).
3.4 Expression levels of representative biomineralization-related and immune-related transcripts
To further explore the biomineralization-related responses of the mussel mantle to different stresses, representative shell matrix proteins (SMPs) were selected based on the shell proteome data of Mytilus (Liao et al., 2015). A total of 80 transcripts corresponding to the 12 SMP families were acquired, most of which presented significant changes in TPM value among the four groups. The heatmaps of the expression level for these transcripts are summarized in Figure 7. Detailed information on these transcripts is provided in Supplementary Table S6. The TPM values of the biomineralization-related transcripts showed different expression patterns depending on shell damage, acute acidification, or a combination of the two stresses.
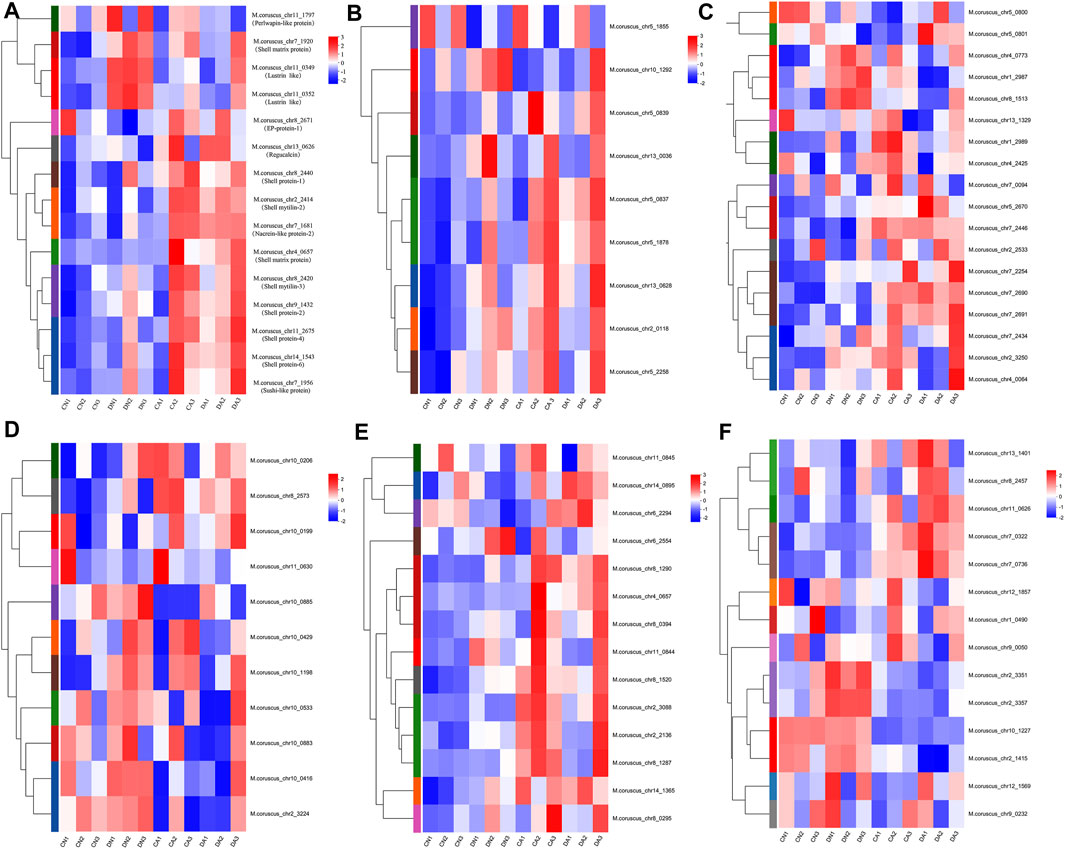
FIGURE 7. Heatmap of the expression level of transcripts corresponding to the biomineralization-related proteins in each mantle sample. (A), the expressional heatmap for the transcripts of shell proteins, shell matrix proteins, lustrins, nacreins, EP-protein, regucalcin, and perlwapin; (B), the expressional heatmap for the transcripts corresponding to Tyrosinase; (C); the expressional heatmap for the transcripts corresponding to Carbonic anhydrase; (D), the expressional heatmap for the transcripts corresponding to Chitinase; (E), the expressional heatmap for the transcripts corresponding to Chitin synthase; (F), the expressional heatmap for the transcripts corresponding to Perlucin. DA, the mussel with drilled shell and raised in acidified sea water (pH 7.4) with exposure time of 48 h; CA, the mussel with complete shell and raised in acidified sea water (pH 7.4) with exposure time of 48 h; DN, the mussel with drilled shell and raised in normal sea water (pH 8.1); CN, the mussel with complete shell and raised in normal sea water (pH 8.1).
In addition, transcripts of antimicrobial peptide (AMP) and peptidoglycan recognition protein (PGRP) were selected with TPM>0.01 from the mantle transcriptome, and the averaged TPM values of these transcripts in each group are listed in Table 2. Most AMP transcripts showed decreased TPM value under shell damage (DN group) and increased TPM value under acute acidification (CA and DA groups) compared to the TPM value in the CN group. For the PGRP transcripts, three of the five transcripts showed a higher expression level under shell damage (DN group) and acute acidification (CA group) than the TPM value in the CN group. Combined shell damage and acute acidification decreased the expression levels of these PGRP transcripts, with the TPM value being significantly downregulated in the DA group compared to that in the CA group (Table 2).
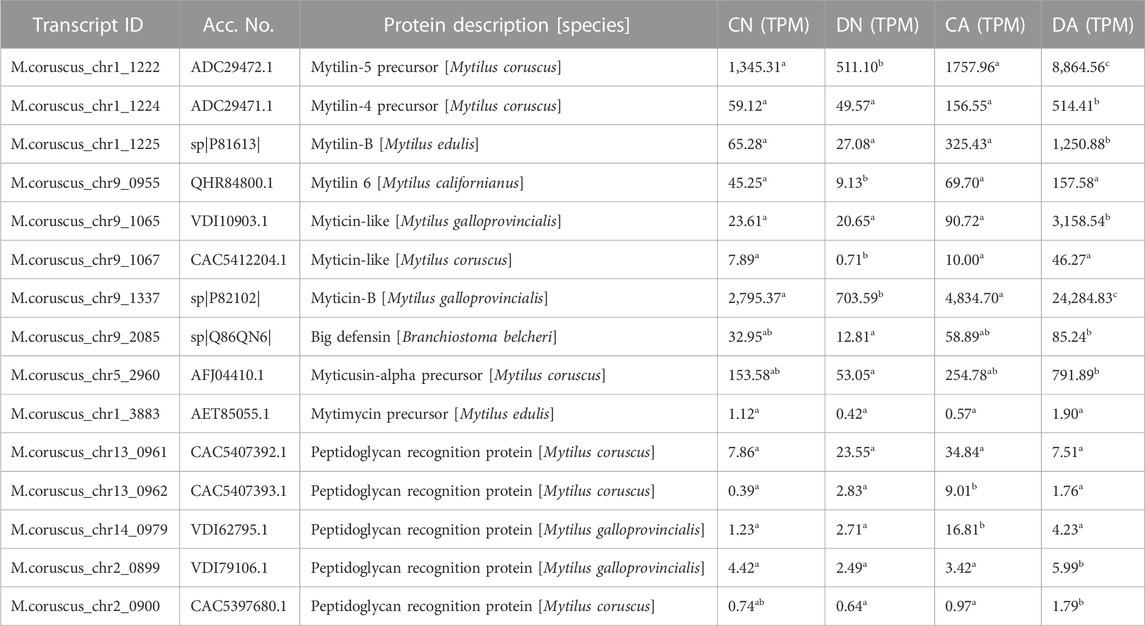
TABLE 2. The averaged expression levels of transcripts of antimicrobial peptides and peptidoglycan recognition proteins in four groups of mantle sample. DA, the mussel with drilled shell and raised in acidified sea water (pH 7.4) with exposure time of 48 h; CA, the mussel with complete shell and raised in acidified sea water (pH 7.4) with exposure time of 48 h; DN, the mussel with drilled shell and raised in normal sea water (pH 8.1); CN, the mussel with complete shell and raised in normal sea water (pH 8.1). The data were statistically analyzed using F test for variance homogeneity and one-way analysis of variance (ANOVA). Significant difference (p < 0.05) between the groups was denoted by different letters.
3.5 Validation of DEGs via qRT-PCR
To validate the reliability of the DEG results, 22 transcripts were randomly selected from the DEGs in the mantle, and their expression levels were validated based on their differential expression profiles under the same conditions described in Section 2.1. qPCR was performed for validation, and the relative expressional levels and the correlation between the expression values from qPCR and RNA-seq is shown in Supplementary Figure S9. In most cases, the qPCR results matched the RNA-seq results, and the coefficient of association (r value) is 0.656, which means a medium correlation between RNA-Seq and qPCR data. These results indicate that the RNA-seq data reliably identified potential genes under different conditions.
3.6 AB-PAS staining of the mantle edge
As shown in Figure 8, three folds can be observed in the mantle edge area: inner fold (IF), middle fold (MF), and OF, representing three different regions of the mantle edge. Many invaginations along the mantle epithelium cover the morphogenetic zones of the three folds, and a columnar epithelial layer can be observed on the surface of the three folds (Figure 8). The OF was similar to the MF in terms of length and shape, whereas the IFs were different. As reviewed previously (Kocot et al., 2016; Clark et al., 2020), most bivalve mantles have three folds at the edges. In addition, histological observations revealed an epidermal layer at the surface of the mantle folds, a basement layer under the epidermal layer, and a connective layer under the basement layer (Figure 8). The epidermal layer has been suggested to be a biological regulator of shell formation (Mao et al., 2019; Zhang et al., 2021).
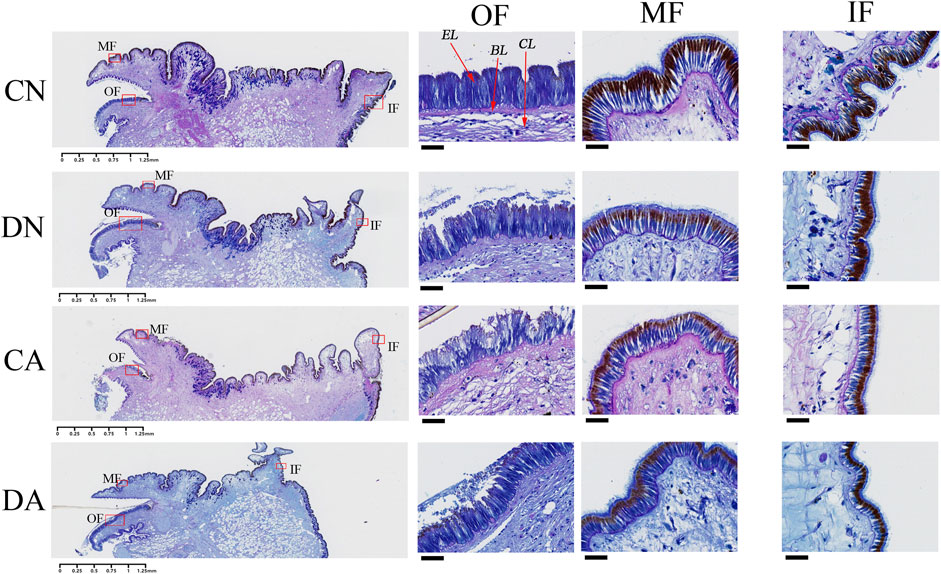
FIGURE 8. Histological observation of the mantle edge with AB-PAS staining. CN, the mussel with complete shell and raised in normal sea water (pH 8.1); DN, the mussel with drilled shell and raised in normal sea water (pH 8.1); CA, the mussel with complete shell and raised in acidified sea water (pH 7.4) with exposure time of 48 h; DA, the mussel with drilled shell and raised in acidified sea water (pH 7.4) with exposure time of 48 h; OF, outer fold; MF, middle fold; IF, inner fold. EL, epidermis layer; BL, basement layer; CL, connective layer. The left panel represents the whole mantle edge area and the enlarged photos corresponding to the red frames are respectively listed in right three panels. The scar bar is 1.25 mm for the left panel, and 25 μm for the right three panels.
AB-PAS staining also revealed that sulfated and sialic mucosubstances (stained blue) were present mainly in the epidermal layer of the three mantle folds (Figure 8). In addition, neutral mucosubstances (red or purple) were observed mainly in the connective and basement layers of the mantle edge and the epidermis layer of the MFs and IFs. In the OF, both shell damage and acute acidification induced invagination in the epidermal layer and cell morphological changes (Figure 8). Cell damage in the epidermal layer of this fold was observed under both shell damage and acute acidification. Interestingly, the epidermal layers of the MFs and IFs were not affected by either shell damage or acute acidification, as no morphological changes were observed in these two folds (Figure 8). These observations highlight an alteration of the mantle OF epithelium in response to shell damage and acute acidification.
3.7 TEM observation of the mantle edge OF
TEM analysis revealed the presence of different secretory vesicles in mantle epithelial cells. As shown in Figures 9A–C, at least three types of secretory vesicle can be discriminated from the mantle of the control group (CN group), including the vesicle with a diameter of 1–2 μm, electron-lucent core (Figure 9A) with a diameter of 1–2 μm, one or two electron-dense cores surrounded by electron-lucent area (Figure 9B), and vesicles with a diameter of 0.1–0.5 μm and high electron-density (Figure 9C). In the mantle of DN group, vesicles with diameter of 0.2–0.5 μm and high electron-density can be observed as packed tightly in the cells and surrounded by a cytoplasm (Figures 9D–F). In the mantle of the CA group, the number of the observed vesicles was lower than that of other groups, and only some small vesicles with diameter of 0.1–0.4 μm can be observed as packed loosely with or without membrane (Figures 9G–I). While in the mantle of DA group, vesicles with diameter of 0.1–0.8 μm and different electron density can be observed in the mantle epithelia cells (Figures 9J–L). Our results revealed that compared to the CN group, the observed vesicles in the DN, CA, and DA groups were somewhat different in terms of size, electron density, and number, indicating different secretion behaviors of the mantle cells in response to shell damage and/or acute acidification.
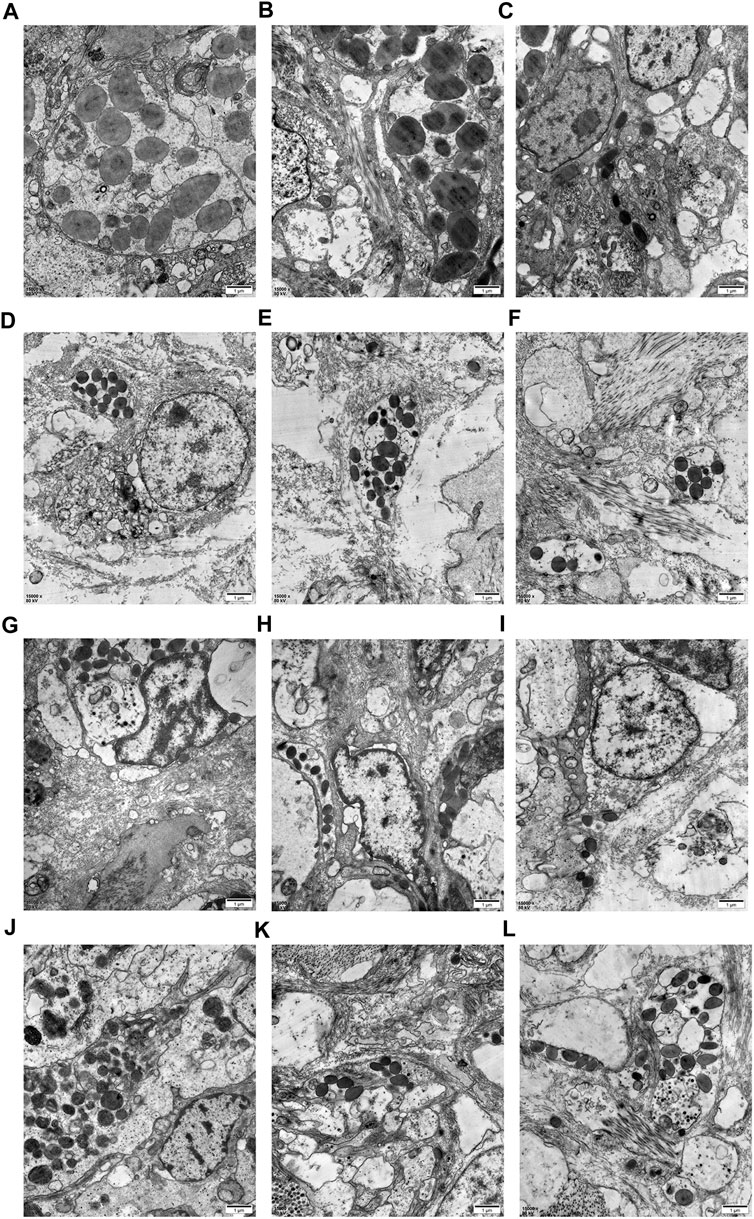
FIGURE 9. TEM analysis for the secretory cells containing different vesicles or granules in the mantle edge epithelia from the outer fold. (A–C), three duplicate samples from the mussel with complete shell and raised in normal sea water (pH 8.1); (D–F), three duplicate samples from the mussel with drilled shell and raised in normal sea water (pH 8.1); (G–I), three duplicate samples from the mussel with complete shell and raised in acidified sea water (pH 7.4); (J–L), three duplicate samples from the mussel with drilled shell and raised in acidified sea water (pH 7.4).
3.8 SEM observation of the mussel shell under different stresses
The cross sections of the shell drilling site and the neighboring area were analyzed via SEM, and the microstructures are presented in Figure 10. Compared to the shell-drilled mussels under normal sea water, acute acidification induced slight textural changes at the shell-drilling site and the neighboring area. At the drilling site, a film was observed on the surface of the cross-section of the shell under normal seawater (Figure 10C), and this film disappeared partially at the drilling site after 48 h of acute acidification (Figure 10D). In the area near the drilling site, a smooth film was observed for the shell under normal seawater (Figure 10E), and thickening of this film with wavy textures was observed for the mussel under acute acidification (Figure 10F). These observations indicate a microstructural change in the inner shell film caused by acidification of the mussel shell structure.
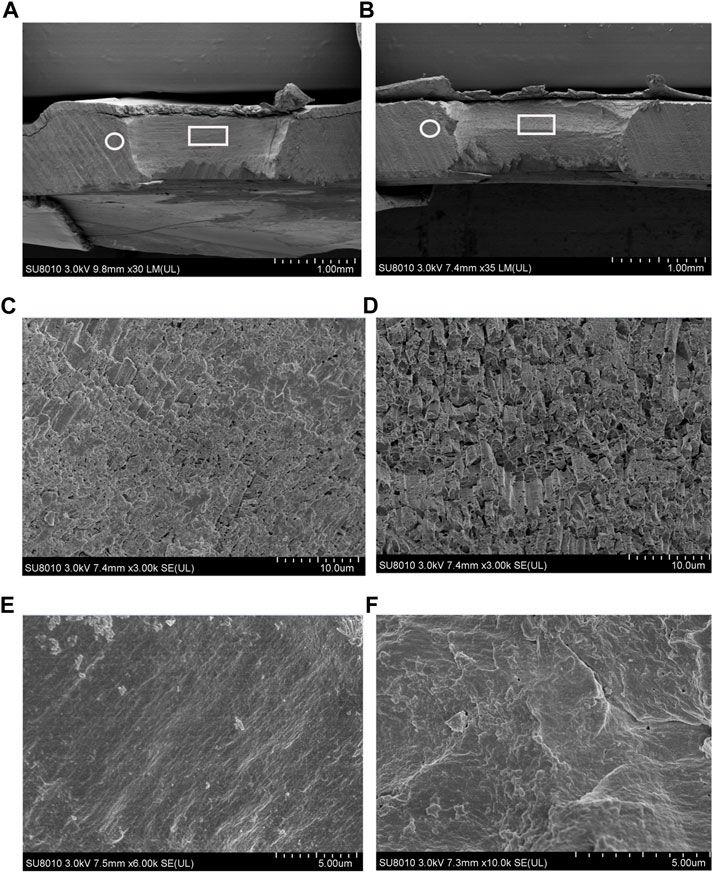
FIGURE 10. SEM observation of shell cross section located at the drilling site (denoted by a white frame) and the adjacent area (denoted by a white circle). (A), the shell cross section from the mussel under normal sea water (pH 8.1).; (B), the shell cross section from the mussel under acidified sea water (pH 7.4) with exposure time of 48 h; (C), enlargement of the area denoted by a white frame in A; (D), enlargement of the area denoted by a white frame in B; (E), enlargement of the area denoted by a white circle in A; (F), enlargement of the area denoted by a white circle in B.
4 Discussion
The M. coruscus used in this study was collected from the Yangtze River estuary sea area, where seasonal fluctuation of natural pH occurs due to freshwater input; thus, the seawater pH value is reduced rapidly in summer (Wei et al., 2021). This suggests that M. coruscus would be impacted if they cannot recover from short-term exposure to coastal acidification. Therefore, short-term exposure of M. coruscus to acidification was performed in this study to explore the molecular mechanisms underlying the rapid adaptation of this mussel to coastal acidification.
4.1 Functional changes of the mantle cell induced by the stressors
We utilized shell-complete and shell-damaged mussels of M. coruscus to investigate whether the expression of mantle-specific genes can be induced by acute seawater acidification and how the mantle responds to acute acidification at the beginning of the shell repair process. According to the number of DEGs between various comparisons, we observed that in the mantle, low pH induced more DEGs than that of shell damage, as 796 and 227 DEGs could be detected in DN vs. CN and DA vs. CA, respectively, whereas more than 3,000 DEGs could be detected in comparisons with changed pH, such as CA vs. CN and DA vs. DN (Figure 3). These results indicate that the mussel mantle is sensitive to acute acidification. GO enrichment analysis further revealed different response patterns of DEGs induced by acute acidification and shell damage. As shown in Figure 5, some reported biomineralization-related GO terms, such as calcium ion binding (Smeets et al., 2015), chitin binding (Furuhashi et al., 2010), and melanin-related processes (Huang et al., 2021), were enriched for DN vs. CN, suggesting rapid activation of the shell repair process, whereas acidification alone induced DEGs to be mainly enriched in metabolic-related processes, such as organic acids, amino acids, and small molecules. Similar results were observed for DA vs. DN and DA vs. CA. Interestingly, no enrichment GO term could be detected with P-adjust <0.05 in DA vs. CA, and only five GO terms related to redox reaction and stimuli response regulation were found with a p-value <0.35 in this comparison (Figure 5).
KEGG enrichment analysis revealed the responses of mussel mantle to acute acidification and/or shell damage. Under acute acidification, both shell-complete and shell-damaged mussels presented similar responses in the KEGG enrichment analysis, and DEGs from the mussel mantle were mainly enriched in pathways related to cell processes, signal transduction, and metabolism. Apoptosis, lysosomes, and autophagy were apparently the primary enriched KEGG pathways, with the number of upregulated DEGs being much higher than that of downregulated DEGs (Figure 6; Table 1), indicating the activation of these pathways under acute acidification. In Mytilus, autophagy, lysosomal function, and mTOR signaling are intrinsically interlinked in response to environmental stress (Sforzini et al., 2018). In marine metazoans, apoptosis is a well-characterized short-term reaction of physiological tolerance to abiotic stress in which damaged cells are removed (Kültz, 2005; Wang et al., 2021). In addition, autophagy is a highly dynamic and conserved pathway that maintains cellular homeostasis via the sequestration and delivery of damaged proteins and organelles into lysosomes for degradation (Parzych and Klionsky, 2014), which is consistent with the activation of the ubiquitin-mediated proteolytic pathway in the comparison of CA vs. CN in this study (Figure 6; Table 1). Lysosomes and mitochondria are decisive controllers of cell death or apoptosis; acidification of lysosomes activates autophagy and triggers apoptosis (Li et al., 2013). Our findings indicate that the pathways of apoptosis, lysosomes, and autophagy in the mussel mantle are induced by acute acidification, and the activation of these pathways may mitigate potential cell damage from reduced pH through the maintenance of cellular homeostasis in the mussel mantle. We also observed that the mTOR signaling pathway, a highly conserved signaling network regulating cell growth in response to nutrients, hormones, and stresses (Wang and Zhang, 2019), was significantly enriched in CA vs. CN, with more upregulated DEGs than downregulated DEGs (Figure 6; Table 1). mTOR has been reported to regulate autophagy, and activation of the mTOR signaling pathway can inhibit autophagy (Al-Bari and Xu, 2020). Surprisingly, in our study, acute acidification induced both the mTOR signaling pathway and autophagy in the mussel mantle. This suggests that in Mytilus, the regulation of autophagy may be mTOR-independent and that other signals, such as Ca2+ (Al-Bari and Xu, 2020), may play a key role in the activation of autophagy in mantle cells during acute acidification. However, this hypothesis must be verified in future studies.
Shell damage also induced DEGs (DN vs. CN) enriched in the adherens junction, melanogenesis, gastric acid secretion, and cGMP-PKG signaling pathway, with the number of upregulated DEGs being greater than that of downregulated DEGs (Figure 6; Table 1). Adherens junctions play a key role in mediating osteogenesis (Guntur et al., 2012). Melanogenesis is related to the shell pigmentation process, and thus to the formation of the shell periostracum layer (Malachowicz and Wenne, 2019). Gastric acid secretion is an important process for calcium uptake and bone growth in humans (Kopic et al., 2013), and the cGMP-PKG signaling pathway can activate osteoblast function and bone formation in mice (Kalyanaraman et al., 2018). Although the biomineralization-related functions of these KEGG pathways have mainly been reported in the human body or in mice, we cannot exclude the roles of these pathways in shell formation and reparation of Mytilus. In addition, other potential biomineralization-related KEGG pathways were detected in the comparisons of CA vs. CN and DA vs. DN, indicating that acute acidification induces a shell formation response in the mussel mantle. For example, collecting duct acid secretion regulates acid–base transport (Wagner et al., 2009) and renal calcification (Moochhala et al., 2008). Amyotrophic lateral sclerosis is another potential biomineralization-related pathway with reported functions in bone metabolism in mice and humans (Joyce et al., 2012; Mizuno et al., 2020). Based on the abovementioned findings, we speculate that different shell repair mechanisms may be adopted by Mytilus mantle under normal conditions and in acidified seawater, and the different patterns of enriched biomineralization-related KEGG pathways presented in this study provide clues for exploring specific mechanisms during the shell formation and shell repair processes of mussels under OA.
4.2 Morphology and microstructure of the mantle tissue and the shell
Histological observations of the mantle edge highlighted the effects of shell damage and acute acidification on the morphology of the columnar epithelial layer, particularly the OF (Figure 8). Previous studies have revealed that the three folds of the mantle edge are associated with different functions in the bivalve mantle: the IF is involved in water inflow, the MF is involved in sensory functions, and the OF is involved in shell secretion and formation of the periostracum layer (i.e., the shell skin) (Clark, 2020; Clark et al., 2020; Salas et al., 2022). In addition, the OF has been suggested to play a critical role in promoting calcium carbonate crystal formation, as many shell matrix proteins have been identified in this fold (Wang et al., 2011; Nakayama et al., 2013). The shell formation of mussel involves the transformation of HCO3− to CO32-, and a hydrogen ion (H+) is thus released (Sillanpää et al., 2018; Sillanpää et al., 2020). Together with the increased H+ generated by OA, the acid–base homeostasis of mantle edge cells may consequently be altered (Ramesh et al., 2020). Therefore, cell morphological changes are understandable for the mantle edge as a cellular response and secretory activity employed by mantle cells to cope with shell damage and/or acute acidification. In this study, we found that epithelial cells covering the mantle edge-stained blue using the AB-PAS technique, indicating that they contain acidic mucosubstances. These acidic mucosubstances may play active roles in the mineralization of mollusk shells by aiding the absorption or binding of calcium ions (Á et al., 2015; Lee et al., 2014). In the mussels under acute acidification, the epithelium of the mantle OF showed fewer AB-PAS-positive cells than the mussels under normal seawater (Figure 8), suggesting a decrease in the secretory activity in the mantle OF and highlighting the negative effect of acute acidification on the shell formation of mussels. TEM observations of the mantle edge presented similar results for the vesicles of mantle secretory cells. The size, electron density, and number of vesicles significantly changed with shell damage and/or acute acidification (Figure 9). However, we found that the vesicles in the shell-damaged mussel mantle were not affected by acute acidification, and even more vesicles could be observed in the mantle cells of the mussel under combined shell damage and acute acidification (Figure 9J-L), suggesting that the mussel tried to start a shell repair process even at a low pH. In addition to the responses of cellular and secretory activity in the mussel mantle under different stressors, further microstructural observation also demonstrated the morphological change of the cross-section at the shell drilling site and the adjacent area, as shown in Figure 10. In this study, we observed that the film covering the prism layer of the shell-drilling site changed, and some erosive holes present on the calcite crystals could be observed in the mussel under acute acidification (Figure 10D). In addition, the smooth film covering the shell underling site became thicker and rougher with a wavy texture presented for the mussel under acute acidification (Figure 10F). These results indicate that acute acidification impairs the shell repair process by affecting the inner shell film and calcite integrity of the prism layer. Disturbed shell ultrastructures in acidified seawater have been previously observed in other Mytilus species (Fitzer et al., 2014; Fitzer et al., 2015) and oysters (Fitzer et al., 2018). Although increasing the pCO2 had no significant effect on calcite growth, the structural integrity of the shell was disturbed owing to the structural disorientation of the calcite crystals. The inner-shell film system has been reported to be a key component for shell formation and repair in mollusks (Watabe, 1965; Hüning et al., 2016). Morphological changes in the inner-shell film indicate the effects of shell damage and/or acidification on shell biomineralization of Mytilus, and the mantle cellular function may thus change in response to stressors.
4.3 Expression of shell matrix protein-related transcripts
In M. coruscus, a set of 63 SMPs was identified from mussel shells using proteomic techniques (Liao et al., 2015). The genes corresponding to these SMPs were selected from the RNA-Seq data in this study, and the genes with an expression level (TPM value) of >0.01 in each group and are listed in Table 2. A heat map based on this table is summarized in Figure 7. Eighty transcripts corresponding to 12 SMP families were identified, and their expression levels showed complex responses to acute acidification and/or shell damage. Shell proteins, such as shell proteins −1, −2, −4, and −6, were induced with increasing TPM values by acute acidification or shell damage (Figure 7A). Similar results were observed for nacrein, sushi-like protein, EP protein, and regucalcin (Figure 7A), which are all members of the proposed universal molluscan biomineralization-related proteins (Arivalagan et al., 2017). We also noted that nacrein and lustrin, both SMPs involved in shell nacre layer formation (She et al., 1997; Song et al., 2014), presented much higher expression levels under acute acidification than those under shell damage alone, suggesting a compensatory response of these two SMPs to counter OA. In addition, most transcripts of tyrosinase also presented similar changes (Figure 7B), as a high TPM value for this SMP was observed under acute acidification alone or combined with shell damage, which implied vital roles of tyrosinase in the formation and repair of mussel shells under OA. Similar results were observed for Crassostrea angulata larvae. The mRNA level of tyrosinase was significantly upregulated in early d-veliger larvae in the 3,000 ppm CO2 treatment group (Yang et al., 2017). However, in another case, the expression of the tyrosinase gene was significantly downregulated in Pacific oyster Crassostrea gigas larvae in acidified seawater (Liu et al., 2020). This may be due to the different sensitivities of these two closely related Crassostrea species to low pH stress, as described in a previous study (Moreira et al., 2016). Carbonic anhydrase (CA) is another SMP involving in biomineralization, playing a key role in CO2/HCO3− conversion for CaCO3 precipitation of Bivalvia shell (Roleda et al., 2012). The crystallographic control and CaCO3 mineral growth may be disrupted if OA inhibits CA (Fitzer et al., 2014). In this study, 18 transcripts of CA were identified, most of which were upregulated under acute acidification and/or shell damage (Figure 7C), suggesting rapid activation of CA to counter OA and shell damage in the mussel mantle. Similar results were reported in the case of Crassostrea species, and increasing CA activity has been proposed as a high tolerance capacity of the oyster to the tested conditions (Moreira et al., 2016). However, we also noted that several CA transcripts, such as M. coruscus _chr13_1329 and M. coruscus _chr5_0800, were downregulated under low pH and/or shell damage conditions (Figure 7C; Table 2), suggesting that the role of CA in mussel shell biomineralization is complex. Considering the role of chitin in the shell composition of Mytilus and snails (Schönitzer and Weiss, 2007; Yonezawa et al., 2016), chitinase and chitin synthase are the two key enzymes that regulate shell chitin metabolism and provide a chitin layer inside the shell during growth. Both chitinases and chitin synthases have been identified in M. coruscus (Liao et al., 2015). In this study, most transcripts of both chitinase and chitin synthase were upregulated under shell-damage stress (Figures 7D,E), suggesting critical roles for these two enzymes in the shell-repair process. Similar results were observed in a previous study (Yarra et al., 2021). Interestingly, acute acidification inhibited the expression of most chitinase (Figure 7D; Table 2) and upregulated the expression of most chitin synthase transcripts (Figure 7E; Table 2). This indicates that Mytilus may simultaneously promote chitin synthesis and inhibit chitin degradation to maintain the integrity of the chitin shell, thus mitigating the impairments caused by OA. Perlucin is a lectin C-type domain-containing SMP with a reported function in calcium carbonate crystallization in mussel shells (Blank et al., 2003). In this study, the expression level of most perlucin transcripts in the mussel mantle showed a complex response, as some perlucin transcripts showed inhibition by shell damage and activation by combined shell damage and acute acidification, whereas others showed a reverse trend (Figure 7F; Table 2). This indicates different functions of different perlucin transcripts under different stressors. Perlucin has been reported to be involved in the innate immune defense in shrimp and abalones (Bi et al., 2020; Choi et al., 2022). Considering the multiple functions of perlucin in both biomineralization and innate immunity, we speculate that some perlucins may participate in innate immune defense, while others are involved in shell formation in Mytilus, and different responses of different perlucin transcripts to acute acidification and shell damage may reflect the complex roles of this molecule.
4.4 Expression of immune-related and energy-related transcripts
Notably, the mantle is not only a tissue for shell formation but also an immune-related tissue in the mussel innate immune system (Huang et al., 2022; Li et al., 2022). Previous studies have demonstrated that elevated pCO2 has a significant effect on the immune system of Pacific oysters, with the mRNA expression patterns of several immune-related genes varying depending on the exposure time and tissues (Wang et al., 2016). In addition, the upregulation of C-type lectin was observed in Mytilus under long-term exposure to high pCO2, indicating that pCO2-driven OA may trigger specific immune-related genes (Castillo et al., 2017). In the present study, immune-related pathways in the mantle were significantly affected by acute acidification, particularly in mussels with shell damage. We noted that the pathways of Toll and Imd signaling, V. cholerae infection, and ubiquitin-mediated proteolysis were significantly enriched in the CA vs. CN comparison (Figure 6). In Mytilus, Toll signal transduction pathway is an important immune-related pathway that can be triggered by either Gram-positive or Gram-negative bacteria (Castillo et al., 2017). The Vibrio cholerae infection pathway is enriched in the mussel gills and gonads under bacterial stress (He et al., 2022), indicating an immune-related function of this pathway in Mytilus. Interestingly, for the shell-damaged mussels, extra immune-related pathways were detected under acute acidification, as enriched TNF signaling and antigen processing and presentation were observed in the DA vs. CN comparison. The function of the TNF signaling pathway in Mytilus immunity has been confirmed in previous studies (Betti et al., 2006; Toubiana et al., 2014; Qi et al., 2018), and enrichment of this pathway in bacteria-induced mussels has also been detected in the gills and gonads of Mytilus (He et al., 2022). Interestingly, no immune-related KEGG pathway was enriched in the comparison of DN vs. CN, suggesting that shell damage stress alone did not induce a strong immune response in the mussel mantle. In addition, most AMP-related transcripts were downregulated under shell-damaged conditions, indicating immune suppression in shell-damaged mussels. Interestingly, acute acidification induced a rapid upregulation of these AMP transcripts, including mytilin, myticin, big defensin, and myticusin, and the highest TPM value was observed in mussels with combined acute acidification and shell damage (Table 2), suggesting immunomodulation following acute acidification exposure, and a preparation of the mantle for possible subsequent infection under OA. The expression pattern of PGRPs showed a slightly different response to that of AMPs. Of the five PGRP transcripts, three were upregulated and two were downregulated in response to shell damage alone, and acute acidification induced high TPM values in these PGRPs. These results are partially consistent with those reported previously, as pCO2-driven OA promotes the transcription of several AMPs and PGRP in mussel gills (Castillo et al., 2017).
Previous studies have revealed the effects of OA on energy-related metabolism in marine metazoans, and metabolic depression is commonly observed with the downregulation of genes associated with ATP production in response to OA (Hüning et al., 2013). However, some species with high tolerance to OA have adapted to constitutively express genes that enable high ATP production (Evans et al., 2017). These studies highlighted that the core response of marine metazoans to a decrease in pH involves reallocation and/or changes in the production of ATP, often as a trade-off in maintaining ion homeostasis, calcification, and control of internal pH levels (Strader et al., 2020). In the present study, we observed that only a few energy-related pathways in the mussel mantle were affected by low pH stress. For example, oxidative phosphorylation, fat digestion and absorption, and glycolysis/gluconeogenesis were enriched in the comparisons of DA vs. CN, DA vs. CA, and DA vs. DN (Table 1). Notably, the most enriched DEGs in the oxidative phosphorylation pathway were upregulated under acute acidification, indicating increased ATP production capacity in the mussel mantle in response to acute acidification. This finding suggests that mussels invest more energy in acclimatizing to acute acidification via the activation of oxidative phosphorylation. Similar results have been reported for other species with a high tolerance to OA (Davies et al., 2016; Evans et al., 2017). In addition, a previous study revealed that the concentrations of ATP and ADP in the adductor muscle of M. edulis showed no significant changes in response to high pCO2, indicating the tolerance of Mytilus to OA (Matoo et al., 2021). Our results partially confirm this view, and the elevated oxidative phosphorylation and upregulated gene expression of AMPs and biomineralization-related genes may represent positive responses of M. coruscus adapted to OA resistance. Taken together, our study provides important data on the extant levels of plasticity in the mantle transcriptome of mussels, as well as insights into the potential adaptation of mussels to future global change.
5 Conclusion
In the present study, we conducted a transcriptomic analysis of the bivalve M. coruscus under various conditions to ascertain the molecular responses of the mussel mantle to different stressors and comment on the potential adaptation strategy of M. coruscus in estuarine conditions that naturally experience low pH and frequent shell breakage. Here, we report the different gene expression patterns of the mussel mantle under different stresses (shell damage, acute acidification, and a combination of these two stressors), showing a strong response in the mussel mantle to acute acidification. Functional analysis of the DEGs from various comparisons in this study revealed that M. coruscus appear to improve its ability to continue biomineralization under OA by activating biomineralization-related pathways and upregulating the expression levels of some SMPs and AMPs, which suggests tolerance to acute acidification and a capacity to maintain the shell-repair process even under high pCO2. It should be noted that other genes with reported potential function in shell-repairing are not significantly modulated in this study, such as genes with function in proteases and protease inhibitors, methyl translation, cell signalling, and ion balance (Yarra et al., 2021). Considering the differences of our study and other studies in the species and the exposure time of mussel to acidification and shell-damage, our study revealed the earlier gene modulation of mussel mantle more than that of whole shell-repairing process under normal or acidified seawater. Morphological analysis further highlighted the importance of the OF at the mantle edge in the shell repair process. Microstructural analysis revealed changes in M. coruscus mantle cells and the shell cross-section under shell damage and/or acute acidification, implying increased secretory activity and a cost to the structural integrity of the shell for maintaining shell formation and repair under acute acidification. Our findings highlight the adaptation of M. coruscus in estuarine areas with dramatic fluctuations in pH and may prove instrumental in its ability to survive OA. However, to project the future implications of OA on the biomineralization mechanisms of Mytilus and elucidate how it can potentially adapt to predicted global climate changes. Further long-term and possibly multi-generational experimental studies are warranted to quantify the adaptation of M. coruscus to predict future climate change.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: [https://www.ncbi.nlm.nih.gov/bioproject/ and PRJNA918971].
Ethics statement
The animal study was approved by Animal Experimental Ethical Inspection, Institutional Animals Care and Use Committee of Zhejiang Ocean University. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
ZL: Conceptualization, Writing–original draft, Writing–review and editing. FL: Formal Analysis, Writing–original draft. YW: Validation, Writing–original draft. XF: Investigation, Validation, Writing–original draft. YL: Formal Analysis, Investigation, Writing–original draft. JH: Investigation, Visualization, Writing–original draft. IB: Writing–review and editing. XY: Resources, Writing–review and editing. XZ: Conceptualization, Resources, Writing–review and editing. GS: Conceptualization, Investigation, Validation, Writing–review and editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by the National Natural Science Foundation of China (Grant Nos. 32271580, 42020104009, and 32200083). Zhejiang Provincial Natural Science Foundation (Grant Nos. LQ23D060002 and LTGS23C010001).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2023.1289655/full#supplementary-material
SUPPLEMENTARY FIGURE S1 | The coverage of reads mapped to M. coruscus genome regions. DA, the mussel with drilled shell and raised in acidified sea water (pH 7.4) with exposure time of 48 h; CA, the mussel with complete shell and raised in acidified sea water (pH 7.4) with exposure time of 48 h; DN, the mussel with drilled shell and raised in normal sea water (pH 8.1); CN, the mussel with complete shell and raised in normal sea water (pH 8.1).
SUPPLEMENTARY FIGURE S2 | The length distribution of all the assembled transcripts.
SUPPLEMENTARY FIGURE S3 | The number of all the assembled transcripts annotated in different databases.
SUPPLEMENTARY FIGURE S4 | heatmap of correlation relationship among the 12 mantle samples. DA, the mussel with drilled shell and raised in acidified sea water (pH 7.4) with exposure time of 48 h; CA, the mussel with complete shell and raised in acidified sea water (pH 7.4) with exposure time of 48 h; DN, the mussel with drilled shell and raised in normal sea water (pH 8.1); CN, the mussel with complete shell and raised in normal sea water (pH 8.1).
SUPPLEMENTARY FIGURE S5 | Venn map of transcripts from four groups of mantle sample. DA, the mussel with drilled shell and raised in acidified sea water (pH 7.4) with exposure time of 48 h; CA, the mussel with complete shell and raised in acidified sea water (pH 7.4) with exposure time of 48 h; DN, the mussel with drilled shell and raised in normal sea water (pH 8.1); CN, the mussel with complete shell and raised in normal sea water (pH 8.1).
SUPPLEMENTARY FIGURE S6 | The volcano maps of DEGs in six pairwise compared samples from mantle. DA, the mussel with drilled shell and raised in acidified sea water (pH 7.4) with exposure time of 48 h; CA, the mussel with complete shell and raised in acidified sea water (pH 7.4) with exposure time of 48 h; DN, the mussel with drilled shell and raised in normal sea water (pH 8.1); CN, the mussel with complete shell and raised in normal sea water (pH 8.1).
SUPPLEMENTARY FIGURE S7 | GO annotation of DEGs from six pairwise comparisons of mantle sample. DA, the mussel with drilled shell and raised in acidified sea water (pH 7.4) with exposure time of 48 h; CA, the mussel with complete shell and raised in acidified sea water (pH 7.4) with exposure time of 48 h; DN, the mussel with drilled shell and raised in normal sea water (pH 8.1); CN, the mussel with complete shell and raised in normal sea water (pH 8.1).
SUPPLEMENTARY FIGURE S8 | COG annotation of DEGs in six pairwise comparisons of mantle sample. DA, the mussel with drilled shell and raised in acidified sea water (pH 7.4) with exposure time of 48 h; CA, the mussel with complete shell and raised in acidified sea water (pH 7.4) with exposure time of 48 h; DN, the mussel with drilled shell and raised in normal sea water (pH 8.1); CN, the mussel with complete shell and raised in normal sea water (pH 8.1).
SUPPLEMENTARY FIGURE S9 | Validation of expression levels of 22 randomly selected transcripts by qPCR comparing with RNA-seq in mantle samples. Correlation analysis was performed using Pearson methods. DA, the mussel with drilled shell and raised in acidified sea water (pH 7.4) with exposure time of 48 h; CA, the mussel with complete shell and raised in acidified sea water (pH 7.4) with exposure time of 48 h; DN, the mussel with drilled shell and raised in normal sea water (pH 8.1); CN, the mussel with complete shell and raised in normal sea water (pH 8.1). The relative expression level was represented by 2–ΔΔCT for qPCR data, and fold change values for RNA-seq data, comparing to the values in CN group.
SUPPLEMENTARY TABLE S1 | The primers used for qPCR validation of transcriptome data.
SUPPLEMENTARY TABLE S2 | The sequencing statistics of the 12 libraries from mantle samples.
SUPPLEMENTARY TABLE S3 | The statistics of clean reads mapped to the M. coruscus genome.
SUPPLEMENTARY TABLE S4 | The percentage of reads mapped to different genome regions.
SUPPLEMENTARY TABLE S5 | Statistics of GO annotated DEGs from six pairwise comparisons of mantle samples. DA, the mussel with drilled shell and raised in acidified sea water (pH 7.4) with exposure time of 48 h; CA, the mussel with complete shell and raised in acidified sea water (pH 7.4) with exposure time of 48 h; DN, the mussel with drilled shell and raised in normal sea water (pH 8.1); CN, the mussel with complete shell and raised in normal sea water (pH 8.1).
SUPPLEMENTARY TABLE S6 | TPM values and annotations of biomineralization-related transcripts screened from the transcriptome data. DA, the mussel with drilled shell and raised in acidified sea water (pH 7.4) with exposure time of 48 h; CA, the mussel with complete shell and raised in acidified sea water (pH 7.4) with exposure time of 48 h; DN, the mussel with drilled shell and raised in normal sea water (pH 8.1); CN, the mussel with complete shell and raised in normal sea water (pH 8.1). Only those ranscripts with TPM value >0.01 in each group were listed here.
References
Álvarez N. R., Molist G. (2015). The outer mantle epithelium of Haliotis tuberculata (Gastropoda Haliotidae): an ultrastructural and histochemical study using lectins. Acta Zool. 96 (4), 452–459. doi:10.1111/azo.12090
Al-Bari M. A. A., Xu P. (2020). Molecular regulation of autophagy machinery by mTOR-dependent and -independent pathways. Ann. N. Y. Acad. Sci. 1467 (1), 3–20. doi:10.1111/nyas.14305
Arivalagan J., Yarra T., Marie B., Sleight V. A., Duvernois-Berthet E., Clark M. S., et al. (2017). Insights from the shell proteome: biomineralization to adaptation. Mol. Biol. Evol. 34 (1), 66–77. doi:10.1093/molbev/msw219
Bechmann R. K., Taban I. C., Westerlund S., Godal B. F., Arnberg M., Vingen S., et al. (2011). Effects of ocean acidification on early life stages of shrimp (Pandalus borealis) and mussel (Mytilus edulis). J. Toxicol. Environ. Health A 74 (7-9), 424–438. doi:10.1080/15287394.2011.550460
Betti M., Ciacci C., Lorusso L. C., Canonico B., Falcioni T., Gallo G., et al. (2006). Effects of tumour necrosis factor alpha (TNFalpha) on Mytilus haemocytes: role of stress-activated mitogen-activated protein kinases (MAPKs). Biol. Cell 98 (4), 233–244. doi:10.1042/BC20050049
Beyer J., Green N. W., Brooks S., Allan I. J., Ruus A., Gomes T., et al. (2017). Blue mussels (Mytilus edulis spp.) as sentinel organisms in coastal pollution monitoring: a review. Mar. Environ. Res. 130, 338–365. doi:10.1016/j.marenvres.2017.07.024
Bi J., Ning M., Xie X., Fan W., Huang Y., Gu W., et al. (2020). A typical C-type lectin, perlucin-like protein, is involved in the innate immune defense of whiteleg shrimp Litopenaeus vannamei. Fish. Shellfish Immunol. 103, 293–301. doi:10.1016/j.fsi.2020.05.046
Blank S., Arnoldi M., Khoshnavaz S., Treccani L., Kuntz M., Mann K., et al. (2003). The nacre protein perlucin nucleates growth of calcium carbonate crystals. J. Microsc. 212 (3), 280–291. doi:10.1111/j.1365-2818.2003.01263.x
Broszeit S., Hattam C., Beaumont N. (2016). Bioremediation of waste under ocean acidification: reviewing the role of Mytilus edulis. Mar. Pollut. Bull. 103 (1-2), 5–14. doi:10.1016/j.marpolbul.2015.12.040
Byrne M., Fitzer S. (2019). The impact of environmental acidification on the microstructure and mechanical integrity of marine invertebrate skeletons. Conserv. Physiol. 7 (1), coz062. doi:10.1093/conphys/coz062
Castillo N., Saavedra L. M., Vargas C. A., Gallardo-Escárate C., Détrée C. (2017). Ocean acidification and pathogen exposure modulate the immune response of the edible mussel Mytilus chilensis. Fish. Shellfish Immunol. 70, 149–155. doi:10.1016/j.fsi.2017.08.047
Choi M. J., Kim Y. R., Park N. G., Kim C. H., Oh Y. D., Lim H. K., et al. (2022). Characterization of a C-Type lectin domain-containing protein with antibacterial activity from Pacific abalone (Haliotis discus hannai). Int. J. Mol. Sci. 23 (2), 698. doi:10.3390/ijms23020698
Clark M. S. (2020). Molecular mechanisms of biomineralization in marine invertebrates. J. Exp. Biol. 223 (11), jeb206961. doi:10.1242/jeb.206961
Clark M. S., Peck L. S., Arivalagan J., Backeljau T., Berland S., Cardoso J. C. R., et al. (2020). Deciphering mollusc shell production: the roles of genetic mechanisms through to ecology, aquaculture and biomimetics. Biol. Rev. Camb Philos. Soc. 95 (6), 1812–1837. doi:10.1111/brv.12640
Davies S. W., Marchetti A., Ries J. B., Castillo K. D. (2016). Thermal and pCO2 stress elicit divergent transcriptomic responses in a resilient coral. Front. Mar. Sci. 3, 1–15. doi:10.3389/fmars.2016.00112
Doney S. C., Fabry V. J., Feely R. A., Kleypas J. A. (2009). Ocean acidification: the other CO2 problem. Ann. Rev. Mar. Sci. 1, 169–192. doi:10.1146/annurev.marine.010908.163834
Evans T. G., Pespeni M. H., Hofmann G. E., Palumbi S. R., Sanford E. (2017). Transcriptomic responses to seawater acidification among sea urchin populations inhabiting a natural pH mosaic. Mol. Ecol. 26 (8), 2257–2275. doi:10.1111/mec.14038
Fitzer S. C., Phoenix V. R., Cusack M., Kamenos N. A. (2014). Ocean acidification impacts mussel control on biomineralisation. Sci. Rep. 4, 6218. doi:10.1038/srep06218
Fitzer S. C., Torres Gabarda S., Daly L., Hughes B., Dove M., O'Connor W., et al. (2018). Coastal acidification impacts on shell mineral structure of bivalve mollusks. Ecol. Evol. 8 (17), 8973–8984. doi:10.1002/ece3.4416
Fitzer S. C., Zhu W., Tanner K. E., Phoenix V. R., Kamenos N. A., Cusack M. (2015). Ocean acidification alters the material properties of Mytilus edulis shells. J. R. Soc. Interface 12 (103), 20141227. doi:10.1098/rsif.2014.1227
Furuhashi T., Miksik I., Smrz M., Germann B., Nebija D., Lachmann B., et al. (2010). Comparison of aragonitic molluscan shell proteins. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 155 (2), 195–200. doi:10.1016/j.cbpb.2009.11.007
Gao P., Liao Z., Wang X. X., Bao L. F., Fan M. H., Li X. M., et al. (2015). Layer-by-Layer proteomic analysis of Mytilus galloprovincialis shell. PLoS One 10 (7), e0133913. doi:10.1371/journal.pone.0133913
Gazeau F., Gattuso J. P., Dawber C., Pronker A. E., Peene F., Peene J., et al. (2010). Effect of ocean acidification on the early life stages of the blue mussel Mytilus edulis. Biogeosciences 7, 2051–2060. doi:10.5194/bg-7-2051-2010
Guntur A. R., Rosen C. J., Naski M. C. (2012). N-cadherin adherens junctions mediate osteogenesis through PI3K signaling. Bone 50 (1), 54–62. doi:10.1016/j.bone.2011.09.036
Harvey B. P., Agostini S., Wada S., Inaba K., Hall-Spencer J. M. (2018). Dissolution: the Achilles’ heel of the triton shell in an acidifying ocean. Front. Mar. Sci. 5, 371. doi:10.3389/fmars.2018.00371
He Z., He J., Wang J., Zhang X., Fan M., Butino I., et al. (2022). Comparative transcriptomic analysis of gill and gonad from mytilus under antibiotics treatment followed by different bacteria challenge. Aquaculture 547 (5), 737457. doi:10.1016/j.aquaculture.2021.737457
Huang J., Li L., Jiang T., Xie L., Zhang R. (2022). Mantle tissue in the pearl oyster Pinctada fucata secretes immune components via vesicle transportation. Fish. Shellfish Immunol. 121, 116–123. doi:10.1016/j.fsi.2022.01.001
Huang S., Jiang H., Zhang L., Gu Q., Wang W., Wen Y., et al. (2021). Integrated proteomic and transcriptomic analysis reveals that polymorphic shell colors vary with melanin synthesis in Bellamya purificata snail. J. Proteomics 230, 103950. doi:10.1016/j.jprot.2020.103950
Hüning A. K., Lange S. M., Ramesh K., Jacob D. E., Jackson D. J., Panknin U., et al. (2016). A shell regeneration assay to identify biomineralization candidate genes in mytilid mussels. Mar. Genomics 27, 57–67. doi:10.1016/j.margen.2016.03.011
Hüning A. K., Melzner F., Thomsen J., Gutowska M. A., Krämer L., Frickenhaus S., et al. (2013). Impacts of seawater acidification on mantle gene expression patterns of the Baltic Sea blue mussel: implications for shell formation and energy metabolism. Mar. Biol. 160, 1845–1861. doi:10.1007/s00227-012-1930-9
Joyce N. C., Hache L. P., Clemens P. R. (2012). Bone health and associated metabolic complications in neuromuscular diseases. Phys. Med. Rehabil. Clin. N. Am. 23 (4), 773–799. doi:10.1016/j.pmr.2012.08.005
Kalyanaraman H., Schwaerzer G., Ramdani G., Castillo F., Scott B. T., Dillmann W., et al. (2018). Protein kinase G activation reverses oxidative stress and restores osteoblast function and bone formation in male mice with type 1 diabetes. Diabetes 67 (4), 607–623. doi:10.2337/db17-0965
Kapsenberg L., Bitter M. C., Miglioli A., Aparicio-Estalella C., Pelejero C., Gattuso J. P., et al. (2022). Molecular basis of ocean acidification sensitivity and adaptation in Mytilus galloprovincialis. iScience 25 (8), 104677. doi:10.1016/j.isci.2022.104677
Kelly M. W., Padilla-Gamiño J. L., Hofmann G. E. (2016). High pCO2 affects body size, but not gene expression in larvae of the California mussel (Mytilus californianus). ICES J. Mar. Sci. 73, 962–969. doi:10.1093/icesjms/fsv184
Kocot K. M., Aguilera F., Mcdougall C., Jackson D. J., Degnan B. M. (2016). Sea shell diversity and rapidly evolving secretomes: insights into the evolution of biomineralization. Front. Zool. 13, 23. doi:10.1186/s12983-016-0155-z
Kopic S., Geibel J. P. (2013). Gastric acid, calcium absorption, and their impact on bone health. Physiol. Rev. 93 (1), 189–268. doi:10.1152/physrev.00015.2012
Kroeker K. J., Kordas R. L., Crim R., Hendriks I. E., Ramajo L., Singh G. S., et al. (2013). Impacts of ocean acidification on marine organisms: quantifying sensitivities and interaction with warming. Glob. Chang. Biol. 19 (6), 1884–1896. doi:10.1111/gcb.12179
Kültz D. (2005). Molecular and evolutionary basis of the cellular stress response. Annu. Rev. Physiol. 67, 225–257. doi:10.1146/annurev.physiol.67.040403.103635
Lee D. G., Park M. W., Kim B. H., Kim H., Jeon M. A., Lee J. S. (2014). Microanatomy and ultrastructure of outer mantle epidermis of the cuttlefish, Sepia esculenta (Cephalopoda: sepiidae). Micron 58, 38–46. doi:10.1016/j.micron.2013.11.004
Li M., Khambu B., Zhang H., Kang J. H., Chen X., Chen D., et al. (2013). Suppression of lysosome function induces autophagy via a feedback down-regulation of MTOR complex 1 (MTORC1) activity. J. Biol. Chem. 288 (50), 35769–35780. doi:10.1074/jbc.M113.511212
Li X., Feng S., Xuan X., Wang H., Shen X., Chen Y., et al. (2022). A proteomic approach reveals biomineralization and immune response for mantle to pearl sac in the freshwater pearl mussel (Hyriopsis cumingii). Fish. Shellfish Immunol. 127, 788–796. doi:10.1016/j.fsi.2022.06.057
Liao Z., Bao L. F., Fan M. H., Gao P., Wang X. X., Qin C. L., et al. (2015). In-depth proteomic analysis of nacre, prism, and myostracum of Mytilus shell. J. Proteomics 122, 26–40. doi:10.1016/j.jprot.2015.03.027
Liu H. H., Sun Q., Jiang Y. T., Fan M. H., Wang J. X., Liao Z. (2019). In-depth proteomic analysis of Boleophthalmus pectinirostris skin mucus. J. Proteomics 200, 74–89. doi:10.1016/j.jprot.2019.03.013
Liu Z., Zhou Z., Zhang Y., Wang L., Song X., Wang W., et al. (2020). Ocean acidification inhibits initial shell formation of oyster larvae by suppressing the biosynthesis of serotonin and dopamine. Sci. Total Environ. 735, 139469. doi:10.1016/j.scitotenv.2020.139469
Malachowicz M., Wenne R. (2019). Mantle transcriptome sequencing of Mytilus spp. and identification of putative biomineralization genes. Peer J. 6, e6245. doi:10.7717/peerj.6245
Mao J., Zhang W., Wang X., Song J., Yin D., Tian Y., et al. (2019). Histological and expression differences among different mantle regions of the yesso scallop (patinopecten yessoensis) provide insights into the molecular mechanisms of biomineralization and pigmentation. Mar. Biotechnol. (NY) 21 (5), 683–696. doi:10.1007/s10126-019-09913-x
Matoo O. B., Lannig G., Bock C., Sokolova I. M. (2021). Temperature but not ocean acidification affects energy metabolism and enzyme activities in the blue mussel, Mytilus edulis. Ecol. Evol. 11 (7), 3366–3379. doi:10.1002/ece3.7289
McMANUS J. F. (1946). Histological demonstration of mucin after periodic acid. Nature 158, 202. doi:10.1038/158202a0
Melzner F., Gutowska M. A., Langenbuch M., Dupont S., Lucassen M., Thorndyke M. C., et al. (2009). Physiological basis for high CO2 tolerance in marine ectothermic animals: pre-adaptation through lifestyle and ontogeny? Biogeosciences 6 (10), 2313–2331. doi:10.5194/bg-6-2313-2009
Melzner F., Mark F. C., Seibel B. A., Tomanek L. (2020). Ocean acidification and coastal marine invertebrates: tracking CO2 effects from seawater to the cell. Ann. Rev. Mar. Sci. 12, 499–523. doi:10.1146/annurev-marine-010419-010658
Mizuno N., Iwata T., Ohsawa R., Ouhara K., Matsuda S., Kajiya M., et al. (2020). Optineurin regulates osteoblastogenesis through STAT1. Biochem. Biophys. Res. Commun. 525 (4), 889–894. doi:10.1016/j.bbrc.2020.03.028
Moochhala S. H., Sayer J. A., Carr G., Simmons N. L. (2008). Renal calcium stones: insights from the control of bone mineralization. Exp. Physiol. 93 (1), 43–49. doi:10.1113/expphysiol.2007.040790
Moreira A., Figueira E., Soares A. M., Freitas R. (2016). The effects of arsenic and seawater acidification on antioxidant and biomineralization responses in two closely related Crassostrea species. Sci. Total Environ. 545-546, 569–581. doi:10.1016/j.scitotenv.2015.12.029
Myers J. M., Johnstone M. B., Mount A. S., Silverman H., Wheeler A. P. (2007). TEM immunocytochemistry of a 48 kDa MW organic matrix phosphoprotein produced in the mantle epithelial cells of the Eastern oyster (Crassostrea virginica). Tissue Cell 39 (4), 247–256. doi:10.1016/j.tice.2007.03.006
Nakayama S., Suzuki M., Endo H., Iimura K., Kinoshita S., Watabe S., et al. (2013). Identification and characterization of a matrix protein (PPP-10) in the periostracum of the pearl oyster, Pinctada fucata. FEBS Open Bio 3, 421–427. doi:10.1016/j.fob.2013.10.001
Parzych K. R., Klionsky D. J. (2014). An overview of autophagy: morphology, mechanism, and regulation. Antioxid. Redox Signal 20 (3), 460–473. doi:10.1089/ars.2013.5371
Qi P., He Y., Liao Z., Dong W., Xia H. (2018). Molecular cloning and functional analysis of tumor necrosis factor receptor-associated factor 6 (TRAF6) in thick shell mussel, Mytilus coruscus. Fish. Shellfish Immunol. 80, 631–640. doi:10.1016/j.fsi.2018.05.053
Ramesh K., Hu M. Y., Melzner F., Bleich M., Himmerkus N. (2020). Intracellular pH regulation in mantle epithelial cells of the Pacific oyster, Crassostrea gigas. J. Comp. Physiol. B 190 (6), 691–700. doi:10.1007/s00360-020-01303-3
Roleda M. Y., Boyd P. W., Hurd C. L. (2012). Before Ocean acidification: calcifier chemistry lessons†. J. Phycol. 48 (4), 840–843. doi:10.1111/j.1529-8817.2012.01195.x
Sabine C. L., Feely R. A., Gruber N., Key R. M., Lee K., Bullister J. L., et al. (2004). The oceanic sink for anthropogenic CO2. Science 305 (5682), 367–371. doi:10.1126/science.1097403
Salas C., Bueno-Pérez J. D., López-Téllez J. F., Checa A. G. (2022). Form and function of the mantle edge in Protobranchia (Mollusca: bivalvia). Zool. (Jena) 153, 126027. doi:10.1016/j.zool.2022.126027
Schönitzer V., Weiss I. M. (2007). The structure of mollusc larval shells formed in the presence of the chitin synthase inhibitor Nikkomycin Z. BMC Struct. Biol. 7, 71. doi:10.1186/1472-6807-7-71
Sforzini S., Moore M. N., Oliveri C., Volta A., Jha A., Banni M., et al. (2018). Role of mTOR in autophagic and lysosomal reactions to environmental stressors in molluscs. Aquat. Toxicol. 195, 114–128. doi:10.1016/j.aquatox.2017.12.014
Shen X., Belcher A. M., Hansma P. K., Stucky G. D., Morse D. E. (1997). Molecular cloning and characterization of lustrin A, a matrix protein from shell and pearl nacre of Haliotis rufescens. J. Biol. Chem. 272 (51), 32472–32481. doi:10.1074/jbc.272.51.32472
Sillanpää J. K., Cardoso JCDR, Félix R. C., Anjos L., Power D. M., Sundell K. (2020). Dilution of seawater affects the Ca2 + transport in the outer mantle epithelium of Crassostrea gigas. Front. Physiol. 11, 1. doi:10.3389/fphys.2020.00001
Sillanpää J. K., Sundh H., Sundell K. S. (2018). Calcium transfer across the outer mantle epithelium in the Pacific oyster, Crassostrea gigas. Proc. Biol. Sci. 285 (1891), 20181676. doi:10.1098/rspb.2018.1676
Smeets P. J., Cho K. R., Kempen R. G., Sommerdijk N. A., De Yoreo J. J. (2015). Calcium carbonate nucleation driven by ion binding in a biomimetic matrix revealed by in situ electron microscopy. Nat. Mater 14 (4), 394–399. doi:10.1038/nmat4193
Song X., Wang X., Li L., Zhang G. (2014). Identification two novel nacrein-like proteins involved in the shell formation of the Pacific oyster Crassostrea gigas. Mol. Biol. Rep. 41 (7), 4273–4278. doi:10.1007/s11033-014-3298-z
Strader M. E., Wong J. M., Hofmann G. E. (2020). Ocean acidification promotes broad transcriptomic responses in marine metazoans: a literature survey. Front. Zool. 17, 7. doi:10.1186/s12983-020-0350-9
Sunday J. M., Calosi P., Dupont S., Munday P. L., Stillman J. H., Reusch T. B. (2014). Evolution in an acidifying ocean. Trends Ecol. Evol. 29 (2), 117–125. doi:10.1016/j.tree.2013.11.001
Teixidó N., Gambi M. C., Parravacini V., Kroeker K., Micheli F., Villéger S., et al. (2018). Functional biodiversity loss along natural CO2 gradients. Nat. Commun. 9 (1), 5149. doi:10.1038/s41467-018-07592-1
Thomsen J., Casties I., Pansch C., Körtzinger A., Melzner F. (2013). Food availability outweighs ocean acidification effects in juvenile Mytilus edulis: laboratory and field experiments. Glob. Chang. Biol. 19 (4), 1017–1027. doi:10.1111/gcb.12109
Thomsen J., Melzner F. (2010). Moderate seawater acidification does not elicit long-term metabolic depression in the blue mussel Mytilus edulis. Mar. Biol. 157, 2667–2676. doi:10.1007/s00227-010-1527-0
Thomsen J., Stapp L. S., Haynert K., Schade H., Danelli M., Lannig G., et al. (2017). Naturally acidified habitat selects for ocean acidification-tolerant mussels. Sci. Adv. 3 (4), e1602411. doi:10.1126/sciadv.1602411
Toubiana M., Rosani U., Giambelluca S., Cammarata M., Gerdol M., Pallavicini A., et al. (2014). Toll signal transduction pathway in bivalves: complete cds of intermediate elements and related gene transcription levels in hemocytes of immune stimulated Mytilus galloprovincialis. Dev. Comp. Immunol. 45 (2), 300–312. doi:10.1016/j.dci.2014.03.021
Ventura A., Schulz S., Dupont S. (2016). Maintained larval growth in mussel larvae exposed to acidified under-saturated seawater. Sci. Rep. 6, 23728. doi:10.1038/srep23728
Wagner C. A., Devuyst O., Bourgeois S., Mohebbi N. (2009). Regulated acid-base transport in the collecting duct. Pflugers Arch. 458 (1), 137–156. doi:10.1007/s00424-009-0657-z
Wang Q., Cao R., Ning X., You L., Mu C., Wang C., et al. (2016). Effects of ocean acidification on immune responses of the Pacific oyster Crassostrea gigas. Fish. Shellfish Immunol. 49, 24–33. doi:10.1016/j.fsi.2015.12.025
Wang X., Li C., Jia Z., Xu T., Wang Y., Sun M., et al. (2021). Regulation of apoptosis by Pacific oyster Crassostrea gigas reveals acclimation strategy to CO2 driven acidification. Ecotoxicol. Environ. Saf. 217, 112235. doi:10.1016/j.ecoenv.2021.112235
Wang X., Liu S., Xie L., Zhang R., Wang Z. (2011). Pinctada fucata mantle gene 3 (PFMG3) promotes differentiation in mouse osteoblasts (MC3T3-E1). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 158 (2), 173–180. doi:10.1016/j.cbpb.2010.11.004
Wang Y., Zhang H. (2019). Regulation of autophagy by mTOR signaling pathway. Adv. Exp. Med. Biol. 1206, 67–83. doi:10.1007/978-981-15-0602-4_3
Watabe N. (1965). Studies on shell formation. XI. Crystal–matrix relationships in the inner layer of mollusk shells. J. Ultrastruct. Res. 12, 351–370. doi:10.1016/S0022-5320(65)80104-6
Wei Q., Wang B., Zhang X., Ran X., Fu M., Sun X., et al. (2021). Contribution of the offshore detached changjiang (Yangtze River) diluted water to the formation of hypoxia in summer. Sci. Total Environ. 764, 142838. doi:10.1016/j.scitotenv.2020.142838
Xu M., Sun T., Tang X., Lu K., Jiang Y., Cao S., et al. (2020). Title: CO2 and HCl-induced seawater acidification impair the ingestion and digestion of blue mussel Mytilus edulis. Chemosphere 240, 124821. doi:10.1016/j.chemosphere.2019.124821
Yang B., Pu F., Li L., You W., Ke C., Feng D. (2017). Functional analysis of a tyrosinase gene involved in early larval shell biogenesis in Crassostrea angulata and its response to ocean acidification. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 206, 8–15. doi:10.1016/j.cbpb.2017.01.006
Yang J. L., Feng D. D., Liu J., Xu J. K., Chen K., Li Y. F., et al. (2021). Chromosome-level genome assembly of the hard-shelled mussel Mytilus coruscus, a widely distributed species from the temperate areas of East Asia. Gigascience 10 (4), giab024. doi:10.1093/gigascience/giab024
Yarra T., Ramesh K., Blaxter M., Hüning A., Melzner F., Clark M. S. (2021). Transcriptomic analysis of shell repair and biomineralization in the blue mussel, Mytilus edulis. BMC Genomics 22 (1), 437. doi:10.1186/s12864-021-07751-7
Yonezawa M., Sakuda S., Yoshimura E., Suzuki M. (2016). Molecular cloning and functional analysis of chitinases in the fresh water snail, Lymnaea stagnalis. J. Struct. Biol. 196 (2), 107–118. doi:10.1016/j.jsb.2016.02.021
Zhang Y., Jiao Y., Li Y., Tian Q., Du X., Deng Y. (2021). Comprehensive analysis of microRNAs in the mantle central and mantle edge provide insights into shell formation in pearl oyster Pinctada fucata martensii. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 252, 110508. doi:10.1016/j.cbpb.2020.110508
Keywords: Mytilus coruscus, acute acidification, shell damage, biomineralization, shell matrix protein
Citation: Liao Z, Liu F, Wang Y, Fan X, Li Y, He J, Buttino I, Yan X, Zhang X and Shi G (2023) Transcriptomic response of Mytilus coruscus mantle to acute sea water acidification and shell damage. Front. Physiol. 14:1289655. doi: 10.3389/fphys.2023.1289655
Received: 06 September 2023; Accepted: 16 October 2023;
Published: 26 October 2023.
Edited by:
Valerio Matozzo, University of Padua, ItalyReviewed by:
Antonio Figueras, Spanish National Research Council (CSIC), SpainMirella Vazzana, University of Palermo, Italy
Copyright © 2023 Liao, Liu, Wang, Fan, Li, He, Buttino, Yan, Zhang and Shi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ge Shi, c2cyNjEwMTA1QDEyNi5jb20=; Xiaolin Zhang, emhhbmd4aWFvbGluQHpqb3UuZWR1LmNu
†These authors have contributed equally to this work
 Zhi Liao
Zhi Liao Fei Liu1†
Fei Liu1† Isabella Buttino
Isabella Buttino Xiaojun Yan
Xiaojun Yan Xiaolin Zhang
Xiaolin Zhang