- 1Guizhou Provincial Key Laboratory for Agricultural Pest Management of the Mountainous Region, Institute of Entomology, College of Agriculture, Guizhou University, Guiyang, China
- 2Department of Entomology, College of Plant Protection, Nanjing Agricultural University, Nanjing, China
- 3College of Tobacco Science, Guizhou University, Guiyang, China
Aphidoletes aphidimyza is a predator that is an important biological agent used to control agricultural and forestry aphids. Although many studies have investigated its biological and ecological characteristics, few molecular studies have been reported. The current study was performed to identify suitable reference genes to facilitate future gene expression and function analyses via quantitative reverse transcription PCR. Eight reference genes glyceraldehyde-3-phosphate dehydrogenase (GAPDH), RPS13, RPL8, RPS3, α-Tub, β-actin, RPL32, and elongation factor 1 alpha (EF1-α) were selected. Their expression levels were determined under four different experimental conditions (developmental stages, adult tissues, sugar treatment, and starvation treatment) using qRT-PCR technology. The stability was evaluated with five methods (Ct value, geNorm, NormFinder, BestKeeper, and RefFinder). The results showed that GAPDH, RPL32, and EF1-α were ranked as the best reference gene combinations for measuring gene expression levels among different developing stages and in various starvation treatments. RPL8 and RPS3 were recommended to normalize the gene expression levels among different adult tissues. RPL32, β-actin, and EF1-α were recommended sugar-feeding conditions. To validate the utility of the selected reference pair, RPL8, and RPS3, we estimated the tissue-biased expression level of a chemosensory protein gene (AaphCSP1). As expected, AaphCSP1 is highly expressed in the antennae and lowly expressed in the abdomen. These findings will lay the foundation for future research on the molecular physiology and biochemistry of A. aphidimyza.
Introduction
The Aphidoletes aphidimyza Rondani (Diptera: Cecidomyiidae) is widely utilized as an efficient predator of aphids in various agricultural systems, and its larvae can prey voraciously on more than 80 aphid species (Boulanger et al., 2019). To date studies on A. aphidimyza have focused on biological characteristics (Choi et al., 2004; Guo et al., 2014; Watanabe et al., 2016; De Azevedo et al., 2018; Madahi et al., 2019; Fratoni et al., 2020; Higashida et al., 2022), biological control (Lin et al., 2017; Shang et al., 2019), and the mitochondrial genome (Shen et al., 2022), but little is known about molecular mechanisms. Gene expression-level analysis is fundamental in the study of regulatory mechanisms of genes related to host selection, predation, drug resistance, stagnation, and neuromodulation. However, to date, no study investigating the aphid-eating A. aphidimyza gene expression profile has been published. To support further research on this economically valuable biological control species, basic research such as screening for A. aphidimyza reference genes is necessary.
Quantitative reverse transcription PCR (qRT-PCR) has become one of the most popular methods for studying quantitative gene expression because of its high sensitivity, specificity, and reproducibility (Derveaux et al., 2010; Bustin, 2000; Bustin et al., 2009). The accuracy of qRT-PCR analyses is influenced by many biological and technical factors (Bustin et al., 2009). Stable reference genes should be selected for a wide range of conditions, tissues or organs, and developmental stages (Xie et al., 2021; Xu et al., 2021; Yan et al., 2021). Ribosomal protein genes are often used as reference genes to normalize qRT-PCR data, as are glyceraldehyde-3-phosphate dehydrogenase (GAPDH), β-actin, and elongation factor 1 alpha (EF1-α) (Van Hiel et al., 2009; Ponton et al., 2011; Silva et al., 2012; Fu et al., 2013; Liu et al., 2016). However, organisms do not possess a universal reference gene (Vandesompele et al., 2002). Each candidate reference gene must be validated under specific experimental conditions in order to facilitate the acquisition of accurate results (Pfaffl et al., 2004).
In the current study A. aphidimyza reference genes were evaluated and selected based on stability under a wide range of conditions. Eight potential reference genes of glyceraldehyde-3-phosphate dehydrogenase (GAPDH), RPS13, RPL8, RPS3, α-Tub, β-actin, RPL32, and elongation factor 1 alpha (EF1-α) were assessed for expression stability by ΔCt, geNorm, NormFinder, and BestKeeper during different developmental stages, in different adult tissues, under sugar treatment conditions, and under starvation treatment conditions. Based on rankings produced by the aforementioned four statistical algorithms, an overall ranking for each experimental condition was then generated using RefFinder. The results will function as a foundation for subsequent studies investigating A. aphidimyza gene expression and gene function.
Materials and methods
Insect rearing
The A. aphidimyza strain used in this study was collected at the tobacco station in Leshan Town, Zunyi City, Guizhou Province, China, in May 2017. They were raised in an artificial climate chamber at 24°C ± 1°C, a 16:8 light-dark cycle, and 70% relative humidity. Megara japonica Matsumura on broad bean plants was utilized to feed the larvae, while honey was fed to adults.
Experimental conditions and sample collection
Developmental stage
A. aphidimyza individuals at different development stages, including 10–20 first–third larvae, 10 pupae, and 8 adults (4 females and 4 males) were collected representing different developmental stages. Each experimental treatment included three biological duplicates. All experiments were performed with three independent biological replicates. Individuals of each sample were collected and kept in 1.5 mL centrifuge tubes and then were rapidly frozen in liquid nitrogen, and stored at −80°C before the total RNA extraction.
Adult tissues
A.aphidimyza adults were stunned by keeping them in a −20°C refrigerator for 3 min. Multiple tissue samples, including antennae (500 adults each), head (without antennae, 300 adults), thorax (without wings and legs, 200 adults), abdomen (100 adults), and legs (300 adults) were dissected on ice using a sterilized scalpel and forceps.
Sugar treatment
A. aphidimyza adults emerging 12 h after eclosion were divided into four feeding treatments: no other food was provided, aphid honeydew, 10% sugar solution, and purified water. Each treatment was conducted for 24 h. A total of 15 adults consisting of males and females were collected.
Starvation treatment
The third larvae of A. aphidimyza were collected and starved in 7.5-cm Petri dishes for 0, 1, 3, and 5 days (n = 10 per group).
Total RNA extraction and cDNA synthesis
Total RNA was extracted from each sample using Trizol Reagent (Invitrogen, USA) in accordance with the manufacturer’s instructions. NanoDrop 2000 (NanoDrop Technology, United States) was used to assess the concentration and and purity of RNA in each sample, and the RNA samples with absorbance ratios of A260/A280 around 2.0 were selected for further analysis. RNA integrity was verified via 1.2% agarose gel electrophoresis. Lastly, the extracted RNA was digested by DNase I (Takara, Japan) to remove genomic DNA contamination. 1 μg of total RNA was used to synthesize cDNA using the Prime Script TMRT Reagent kit (Takara, Japan). The cDNA from each sample was stored at −20°C prior to the use of both PCR and RT-qPCR.
Selection and identification of candidate reference genes
Eight candidate genes, namely, GAPDH, RPL32, RPS13, β-actin, RPS3, EF1-α,
Quantitative real-time PCR
Primer sequences were designed using NCBI Primer-BLAST in Supplementary Table S2. qRT-PCR reactions were conducted in accordance with the manufacturer’s instructions using TB Green® Premix Ex Taq™ Tli RNaseH Plus (Takara, Japan) and the QuantStudio™ 7Pro Real-Time PCR System (Applied Biosystems, Life Technologies, Carlsbad, CA, United States). The reaction mixture consisted of a 20 μL mixture containing 10 µL of 2 × SYBR Green qPCR Master Mix, 0.4 µL of forwarding primer (10 Mmol L−1), 0.4 µL of reverse primer (10 Mmol L−1), 0.4 µL of ROX Reference Dye II (50X), 1 µL of cDNA template, and 7.8 µL of double-distilled water. The qRT-PCR reaction conditions were as follows: initial denaturation at 95°C for 30 s, followed by 40 cycles at 95°C for 3 s, and ended with an annealing step at 60°C for 30 s. A melting curve analysis was conducted in the 60°C–95°C temperature range to ensure the specificity of the primers. Three independent biological replicates were set. A standard curve was generated from the five-fold dilution series of cDNA, the slopes were analyzed, and the corresponding amplification efficiencies were calculated. RT-qPCR efficiency (E) was determined via the following equation:
Expression stability of candidate reference genes under different treatments
Evaluations of the stability of selected reference genes were conducted using geNorm (Andersen et al., 2004), BestKeeper (Silver et al., 2006), NormFinder (Szabo et al., 2004), and the ΔCt method (Yang et al., 2015). Lastly, a comprehensive ranking of under different conditions was performed using the web-based tool “RefFinder” (Livak and Schmittgen, 2001; Shi and Zhang, 2016). P The optimal number of reference genes for accurate normalization of the target gene was determined by the variation value (Vn/Vn+1) calculated by geNorm. Pairwise variation (Vn/n+1 = 0.15) Vn/n + 1 < 0.15 indicates that the optimal number of reference genes is n, and Vn/n+1 > 0.15 indicates that the optimal number is n + 1 (Vandesompele et al., 2002).
Stability verification of candidate reference genes
The chemosensory protein (CSP) of A. aphidimyza was selected as the target gene to verify the stability of candidate reference genes (GenBank: OP321094). The primer sequence of the target gene was as follows:
Forward: AACGCTTTTGTTGGACAGCTAC.
Reverse: CAATGAATCGAAGCACACGA.
Based on the stability (RPL8 and RPS13) and instability (β-actin) of primary reference genes The average relative expressions of AaphCSP1 in different female tissues were computed based on the 2−ΔΔCT method (Livak and Schmittgen, 2001) and three independent biological replicates.
Results
Selection of candidate reference gene
Eight candidate genes of A. aphidimyza with a complete open reading frame (ORF) were identified by RT-PCR. The GAPDH, RPL32, RPS13, β-actin, RPS3, EF1-α,
Expression variations of candidate reference gene
Eight candidate reference genes were analyzed by qRT-PCR using cycle threshold values (Ct) reflecting their expression under different conditions.The gene expression analysis of the eight candidate reference genes in all samples under four conditions showed a range of Ct means of 13.34 (β-actin) −30.75 (RPL32) (Figure 1). At different developmental stages, GAPDH and RPPL8 had the smaller gene expression variation, whereas β-actin and EF1-α had the higher expression difference (Figure 1A). Among various tissues, except for GAPDH, the expression fluctuations were higher in selected reference genes (Figure 1B). Adults feeding on different sugar varieties, RPPL8 and RPS3 had smaller gene expression variation (Figure 1C). Larvae starvation treatment conditions, GAPDH had a smaller gene variation (Figure 1D).
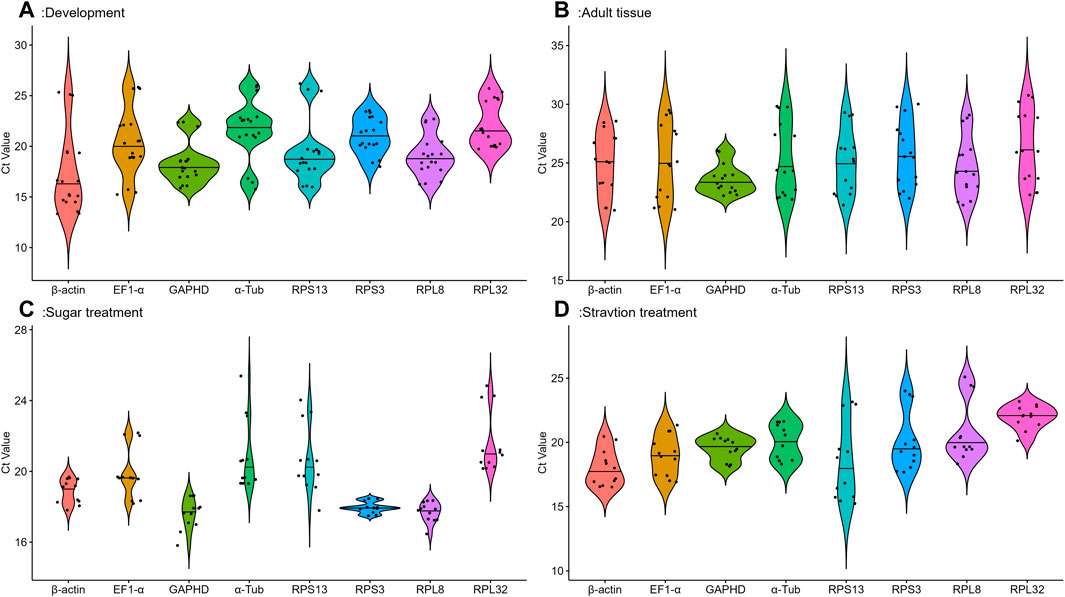
FIGURE 1. Expression levels of eight housekeeping genes in A. aphidimyza were investigated in four different experiments; (A) (developmental stages), (B) (adult tissues), (C) (sugar treatments), and (D) (starvation treatments). Mean Ct values for the eight candidate reference genes are presented in box plots, where each box indicates the 25th and 75th percentiles, and the line across the box represents the median. The eight genes analyzed were GAPDH, RPL32, RPS13, β-actin, RPS3, EF1-α, α-Tub, and RPL8.
Expression stability of candidate reference gene
Developmental stages
The geNorm algorithm evaluates the candidate reference genes based on their expression stability values (M-values) and pairwise variations (Vn/Vn+1). The expression stability values revealed that α-Tub and EF1-α were the better reference genes during different developmental stages, with M-values below 1 (Table 1; Supplementary Figure S1). Pair-wise variation analysis of reference genes showed that V2/3 was less than 0.15 (Supplementary Figure S2), indicating that gene expression analysis required two different reference genes in the developmental stage. Based on the above comprehensive ranking, we recommended the following two genes as reference genes in developmental stages: GAPDH and RPS13 (Supplementary Figure S2). According to the NormFinder, the stable gene was GAPDH, with a p-value less than 0.2. The most unstable gene was β-actin, with a p-value of 0.314 (Table 1; Supplementary Figure S1). Based on the BestKeeper analysis, GAPDH was the most stable gene (Table 1; Supplementary Figure S1). The stability of the eight A. aphidimyza candidate reference genes were ranked by RefFinder at various developmental stages from high to low: GAPDH > RPL32 > EF1-α > RPS3 > α-Tub > RPS13 > RPL8 > β-actin (Figure 2A). Therefore, GAPDH and RPS13 are ranked as the best reference gene combinations for measuring target genes among different developing stages (Figure 2).
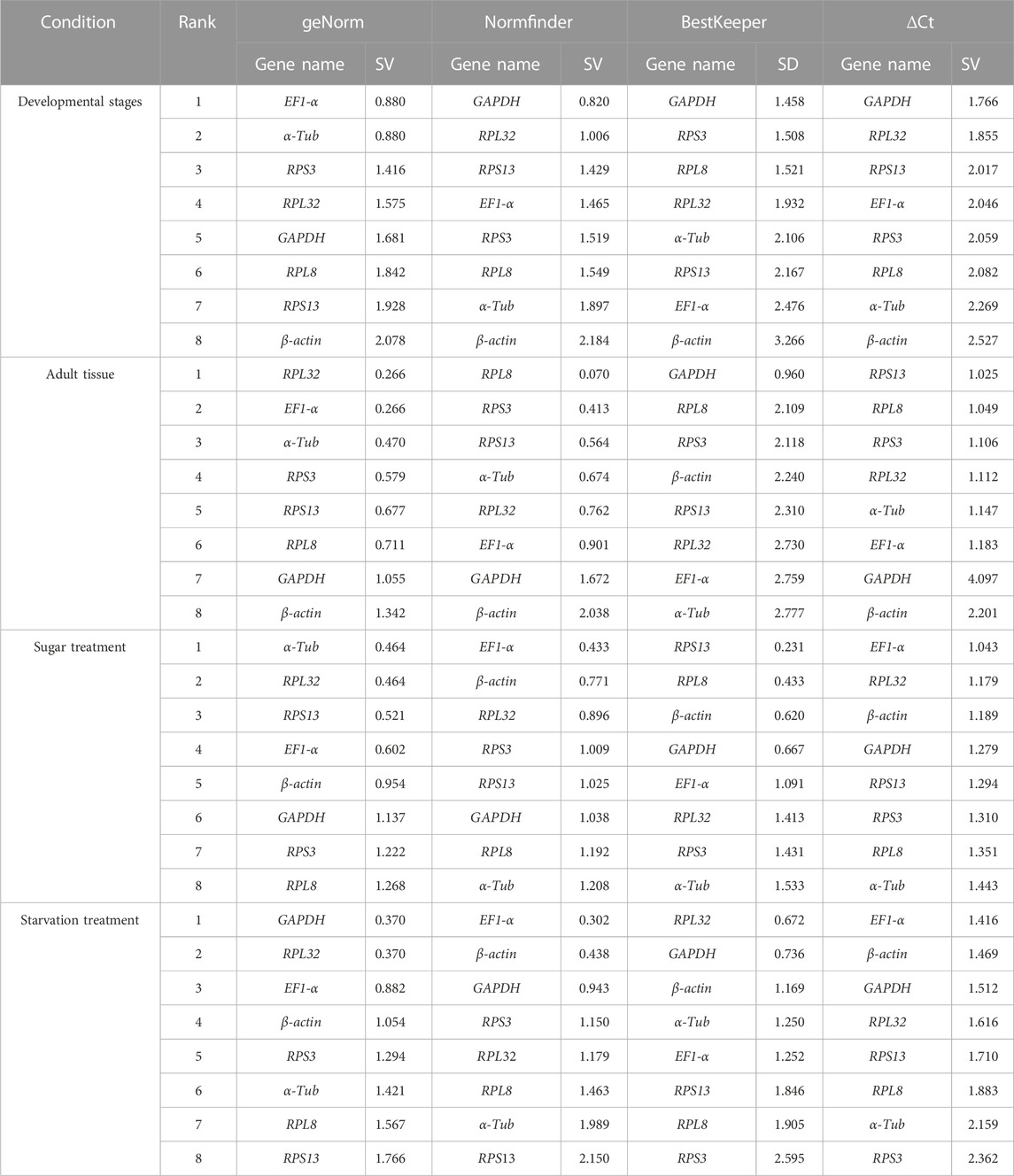
TABLE 1. Stability of the expression of eight candidate reference genes under different experimental conditions.
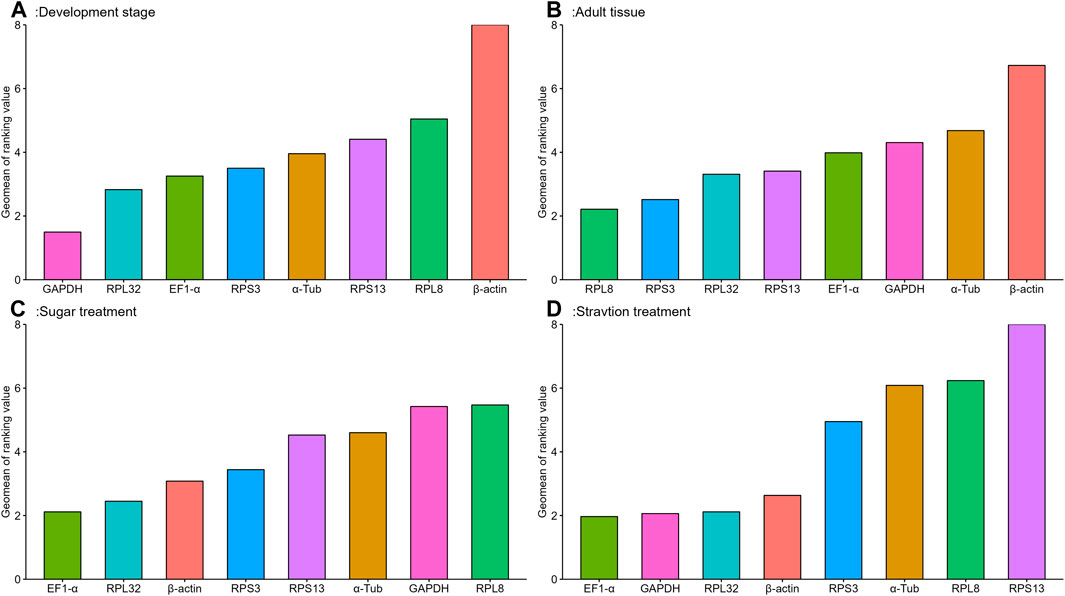
FIGURE 2. Stability of expression analysis of eight candidate reference genes of A.aphidimyza in four different types of experiments calculated by RefFinder.
Adult tissue
Based on the geNorm algorithm, the M-values of EF1-α and RPL32 were below 0.4 (Table 1; Supplementary Figure S1). The pairwise variation analysis displayed that the V2/3 values were close to 0.15 (Supplementary Figure S2), indicating that two reference genes should be selected as reference genes in subsequent studies on other genes in adult tissues. We recommended RPS13 and RPL8 as reference genes (Supplementary Table S3). Based on the BestKeeper, GAPDH was the most stable gene. According to the NormFinder, RPL8 was the most stable gene (Table 1; Supplementary Figure S1). Combining the four algorithms, the comprehensive ranking by RefFinder was as follows: RPL8 > RPS3 > RPL32 > RPS13 > EF1-α > GAPDH > α-Tub > β-actin (Figure 2B). Therefore, RPL8 and RPS3 are ranked as the best reference gene combinations for measuring target genes among different adult tissues (Figure 2).
Sugar treatment
In this study, the geNorm algorithm results showed that the comprehensive reference gene rankings from the best to the least stable were α-Tub, RPL32, RPS13, EF1-α, β-actin, GAPDH, RPS3, and RPL8 (Table 1; Supplementary Figure S1). Except for GAPDH, RPS3, and RPL8, the other genes in the selected reference genes showed values below 1, indicating their stabilities were similar. Moreover, the pairwise variation analysis showed that the V2/3 value exceeded 0.15, indicating three different reference genes are needed for gene expression analysis in sugar treatment (Supplementary Figure S2). The NormFinder analysis revealed that the stability of the selected reference genes was EF1-α, β-actin, and RPL32, with p-values of 0.433, 0.771, and 0.896, respectively, indicating their similar stability (Table 1; Supplementary Figure S1). The BestKeeper data showed that RPS13, RPL8, β-actin, and GAPDH were the most stable because they showed Cp values of 0.231, 0.433, 0.620, and 0.667, respectively (Table 1; Supplementary Figure S1). The stability of reference genes was evaluated comprehensively using RefFinder, which ranked them as EF1-α > RPL32 > β-actin > RPS3 > RPS13 > α-Tub > GAPDH > RPL8 (Figure 2C). Thus, the three reference genes (EF1-α, RPL32, and β-actin) are recommended to be used to test the target gene expression levels in sugar treatment.
Starvation treatment
Based on the geNorm algorithm, the M-values of GAPDH, EF1-α, and RPL32 were below 0.4 (Table 1; Supplementary Figure S1). The pairwise variation analysis displayed that the V2/3 values exceeded 0.15, suggesting three reference genes are enough for gene expression determination within starvation treatment (Supplementary Figure S2). The NormFinder analysis revealed that the stability of the selected reference genes was EF1-α > β-actin > GAPDH > RPS3 > RPL32 > RPL8 > α-Tub > RPS13, with the p-value of 0.302, 0.438, 0.943, 1.150, 1.179, 1.463, 1.989, and 2.150, respectively. Again, the p values of EF1-α and β-actin were below 0.5 (Table 1; Supplementary Figure S1), indicating their similar stability. The BestKeeper data revealed that RPL32 and GAPDH were the most stable because they showed Cp values of 0.672 and 0.736, respectively. The Cp values of β-actin, α-Tub, EF1-α, RPS13, and RPL8 were more than 1.0, and the Cp value of RPS3 was more than 2.0 (Table 1; Supplementary Figure S1). The RefFinder showed a comprehensive ranking order from the most to the least stable: EF1-α > GAPDH > RPL32 > β-actin > RPS3 > α-Tub > RPL8 > RPS13 (Figure 2D). Thus, we considered EF1-α, GAPDH, and RPL32 as the most appropriate reference gene combinations.
Verification of candidate reference genes
To evaluate the stability of the selected reference genes, we selected AaphCSP1 as the target gene and analyzed the expression level of AaphCSP1 in the antenna, head, leg, thorax, and abdomen. The following reference genes were used to normalize: RPL8, RPS3, RPL8 + RPS3 (the most stable reference gene), and β-actin (the least stable reference gene). The highest accumulated mRNA level of AaphCSP1 was found in the antenna, followed by those in the head, the lowest level was detected in the leg, thorax, and abdomen. However, β-actin was used as a reference gene, and AaphCSP1 was highly expressed in the thorax exceeded (p < 0.05) (Figure 3).
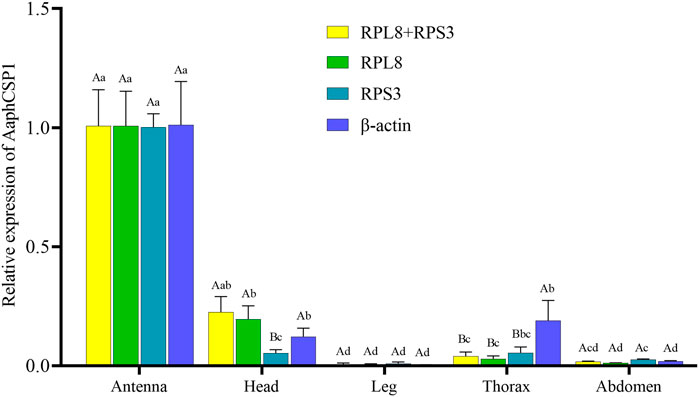
FIGURE 3. Expression of AaphCSP1 gene under different female tissue. Three reference gene combinations (RPL8 + RPS3, RPS3, RPL8, β-actin) were used for the normalization. The values were calculated using the 2−ΔΔCT method. The relative transcripts are the ratios of copy numbers in different tissue relative to the antenna, which is set as 1. The data in the figure were the mean ± standard error. Different lowercase letters indicate that, after normalization with the same reference gene, there was a significant difference in the expression level of AaphCSP1 in female adult A. aphidimyza (Tukey’s HSD-p < 0.05). Different uppercase letters indicate that there were significant differences in the normalization results of each reference gene (Tukey’s HSD-p < 0.05).
Discussion
Quantitative real-time PCR is a crucial technique for studying gene expression analysis. This technique uses a quantitative method to detect gene expression, with the internal reference gene serving as a standard to calibrate the expression of the target gene. It is highly sensitive and has good repeatability, making it a standard technique for detecting or comparing mRNA levels in gene expression studies. The expression level of any single reference gene does not remain constant under all experimental treatments however, so appropriate reference genes must be chosen before RT-qPCR is performed (Shi and Zhang, 2016). Reference genes have been screened in a variety of insects, such as Bombyx mori (Peng et al., 2012), Podoptera frugiperda (Zhou et al., 2021), and Anastatus japonicus (Liu et al., 2022).
The stability evaluation of candidate reference genes is often performed by multiple methods based on a comprehensive analysis of relevant parameters, and the candidate reference genes are ranked according to their stability. In the present study five methods were used individually to rank stability at various developmental stages and in different tissues of A. aphidimyza adults, and the most stable reference genes and their rankings were not entirely consistent. For example, BestKeeper and ΔCt determined that GAPDH was the most stable gene during the various development stages of A. aphidimyza development, whereas GeNorm determined that EF1-α = α-Tub was the most stable gene, and NormFinder determined that GAPDH was the most stable gene (Table 1; Supplementary Figure S1). Similar discrepancies were observed among different adults tissues, sugar treatment conditions, and starvation treatment conditions. To lessen disparities in outcomes between software, RefFinder was used to synthetically rate the stability of each possible reference gene (Figure 2). Similar situations have been observed in the endogenous screening of other insects (Yang et al., 2017).
In different adult tissues, the reliable reference genes were RPS3 and RPL8 (Table 1; Figure 2). Ribosomal proteins are essential housekeeping components that regulate the synthesis of cellular ribosomes, and participate in cellular translation processes. Members of the ribosomal gene family have been used as reference genes in many studies. For example, RPS18 had the highest expression stability across tissues in Tetranychus cinnabarinus (Sun et al., 2010), RPS11 had the highest expression stability in Nilaparvata lugens (Yuan et al., 2014), and RPS13 had the highest expression stability in Sesamia inferens (Sun et al., 2015). RPS15 and RPL13 were also reported as a stable pair of reference genes in different tissues of larvae of the cotton bollworm Helicoverpa armigera (Zhang et al., 2015). In Mythimna separata larvae RPL12 was the most stable gene in different tissues (Li et al., 2018). RPS13 had the highest expression stability in different adult tissues of Tuta absoluta (Yang et al., 2021), whereas RPL13 exhibited higher stability in specific larval tissues of Phthorimaea operculella (Shen et al., 2022).
GAPDH remained stable during developmental stages and under sugar treatment conditions in adult A. aphidimyza (Table 1; Figure 2). The GAPDH enzyme regulates glycolysis, gluconeogenesis, and other mechanisms of energy metabolism. For example, GAPDH was the most stable reference gene during various developmental stages in Schistocerca gregaria (Van Hiel et al., 2009), Spodoptera litura (Lu et al., 2013), Spodoptera exigua (Zhu et al., 2014), and Sesamia inferens (Lu et al., 2015). However, numerous investigations in different insects have revealed that GAPDH expression can occasionally be erratic, such as in Harmonia axyridis (Qu et al., 2018), Tetranychus cinnabarinus (Sun et al., 2010), Sogatella furcifera (An et al., 2016), Bombus terrestris, and Bombus lucorum (Horňáková et al., 2010). EF1-α has been identified as a highly stable reference gene in various insect species. In both Thitarodes armoricanus larvae (Liu et al., 2016) and Danaus plexippus (Pan et al., 2015) EF1-α is evidently insensitive to changes in tissue type or developmental stage. EF1-α has exhibited good expression stability in Phthorimaea operculella during several developmental stages and at different temperatures (Shen et al., 2022). Notably, under numerous sugar treatment conditions in the current study EF1-α was the most reliable reference gene in adult A. aphidimyza (Table 1; Figure 2).
β-actin is a highly conserved cytoskeletal protein that plays a crucial role in various eukaryotic physiological processes such as cell division, chromosome movement, organelle movement, and cytoplasmic flow. It has been identified as one of the most stable reference genes in several insect species, including Liriomyza trifolii (Chang et al., 2017), and Drosophila melanogaster (Ponton et al., 2011). However, in the present study, it was the worst reference gene in different tissues and under different sugar treatment conditions in adult A. aphidimyza (Table 1; Figure 2). This observation is consistent with previous reports that β-actin was less stable across temperature and photoperiod treatments in Helicoverpa armigera (Shakeel et al., 2015), and in Coleopteran insect species such as Coccinella septempunctata (Yang et al., 2016), Henosepilachna vigintioctomaculata (Lü et al., 2018), Phaedon brassicae (Ma et al., 2021), and Phthorimaea operculella (Shen et al., 2022). α-Tub is a gene that encodes microtubule proteins involved in a number of physiological processes such as the construction and maintenance of cell morphology, intracellular transport, chromosome movement, and cell division. It is widely considered to be a reliable reference gene in several insect species, including Chilo suppressalis larvae (Xu et al., 2019) and different developmental stages of Lymantria dispar (Yin et al., 2020). In the present study however, in different tissues and under different starvation treatment conditions in A. aphidimyza, α-Tub was the worst reference gene (Table 1; Figure 2).
In order to demonstrate the utility of RPL8 and RPS3 inaccurate gene expression analysis in A. aphidimyza, we evaluated the relative gene expression level of AaphCSP1 in the antenna, head, leg, thorax, and abdomen. Our results showed that AaphCSP1 was abundantly expressed in the antenna, moderately transcribed in the head, and lowly expressed in the thorax and abdomen (Figure 3). Our expression data are consistent with the fact that CSPs are highly expressed in pest and parasite antennae, their main function is thought to be chemoreception (Liu et al., 2012). Including pheromone detection (Angeli et al., 1999), reproduction (Zeng et al., 2020), and host hunting (Peng et al., 2021). Thus, the tissue-biased expression pattern of AaphCSP1 demonstrates that RPL8 and RPS3 can be used as endogenous controls to assess gene expression in A. aphidimyza.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
X-XS: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Software, Validation, Visualization, Writing–original draft, Writing–review and editing. G-QZ: Conceptualization, Data curation, Formal Analysis, Software, Validation, Writing–original draft, Writing–review and editing. Y-XZ: Conceptualization, Data curation, Investigation, Software, Validation, Visualization, Writing–original draft, Writing–review and editing. X-XZ: Data curation, Investigation, Software, Validation, Visualization, Writing–original draft, Writing–review and editing. X-FY: Conceptualization, Data curation, Methodology, Project administration, Software, Writing–original draft, Writing–review and editing. M-FY: Funding acquisition, Project administration, Resources, Supervision, Visualization, Writing–original draft, Writing–review and editing. FZ: Funding acquisition, Project administration, Resources, Supervision, Visualization, Writing–original draft, Writing–review and editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was funded by the Technology Innovation Talent Team Project [Qian Ke He Pingtai Rencai-CXTD (2021)004] and the Guizhou Province Science and Zunyi Branch Tobacco Company (Project: 2022520300270188).
Acknowledgments
We sincerely thank the editors and reviewers for their valuable suggestions and comments on this study. We greatly thank Dr. Chen-Hui Shen and Ms. Meng-Jiao Lin (both from Nanjing Agricultural University) for their molecular experiments and data analysis. We also sincerely thank Associate Professor Lin Jin (Nanjing Agricultural University) for her guidance on this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2023.1277942/full#supplementary-material
SUPPLEMENTARY FIGURE S1 | Stability of expression of eight candidate reference genes in A. aphidimyza calculated by three different software programs (geNorm, NormFinder, and BestKeeper). (A) Different developmental stages, (B) adult tissues, (C) sugar treatment conditions, and (D) starvation treatment conditions.
SUPPLEMENTARY FIGURE S2 | Evaluation of the optimal number of reference genes for normalization of A. aphidimyza in different treatments, determined via geNorm. The dashed line indicates that the pairwise variation is 0.15. Vn/n + 1 < 0.15 indicates that the optimal number of reference genes for normalization is n, and the Vn/n + 1 > 0.15 indicates that the optimal number is n + 1.
References
An X. K., Hou M. L., Liu Y. D. (2016). Reference gene selection and evaluation for gene expression studies using qRT-PCR in the white-backed planthopper, Sogatella furcifera (Hemiptera: delphacidae). J. Econ. Entomol. 109 (2), 879–886. doi:10.1093/jee/tov333
Andersen C. L., Jensen J. L., Ørntoft T. F. (2004). Normalization of real-time quantitative reverse transcription–PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 64 (15), 5245–5250. doi:10.1158/0008–5472.CAN–04–0496
Angeli S., Ceron F., Scaloni A., Monti M., Monteforti G., Minnocci A., et al. (1999). Purification, structural characterization, cloning and immunocytochemical localization of chemoreception proteins from Schistocerca gregaria. Eur. J. Biochem. 262 (3), 745–754. doi:10.1046/j.1432–1327.1999.00438.x
Boulanger F. X., Jandricic S., Bolckmans K., Wäckers F. L., Pekas A. (2019). Optimizing aphid biocontrol with the predator Aphidoletes aphidimyza, based on biology and ecology. Pest Manag. Sci. 75 (6), 1479–1493. doi:10.1002/ps.5270
Bustin S. A. (2000). Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol. 25 (2), 169–193. doi:10.1677/jme.0.0250169
Bustin S. A., Benes V., Garson J. A., Hellemans J., Huggett J., Kubista M., et al. (2009). The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55 (4), 611–622. doi:10.1373/clinchem.2008.112797
Chang Y. W., Chen J. Y., Lu M. X., Gao Y., Tian Z. H., Gong W. R., et al. (2017). Selection and validation of reference genes for quantitative real-time PCR analysis under different experimental conditions in the leafminer Liriomyza trifolii (Diptera: agromyzidae). PloS One 12 (7), e0181862. doi:10.1371/journal.pone.0181862
Choi M. Y., Roitberg B. D., Shani A., Raworth D. A., Lee G. H. (2004). Olfactory response by the aphidophagous gall midge, Aphidoletes aphidimyza to honeydew from green peach aphid, Myzus persicae. Entomol. Exp. Appl. 111 (1), 37–45. doi:10.1111/j.0013–8703.2004.00151.x
De Azevedo A. G. C., Stuart R. M., Sigsgaard L. (2018). Presence of a generalist entomopathogenic fungus influences the oviposition behaviour of an aphid-specific predator. Bio Control 63 (5), 655–664. doi:10.1007/s10526–018–9889–1
Derveaux S., Vandesompele J., Hellemans J. (2010). How to do successful gene expression analysis using real-time PCR. Methods 50 (4), 227–230. doi:10.1016/j.ymeth.2009.11.001
Fratoni S., Duarte M. V., Vangansbeke D., Wäckers F. L., Dicke M., Pekas A. (2020). A bittersweet meal: the impact of sugar solutions and honeydew on the fitness of two predatory gall midges. Biol. Control 140, 104098. doi:10.1016/j.biocontrol.2019.104098
Fu W., Xie W., Zhang Z., Wang S., Wu Q., Liu Y., et al. (2013). Exploring valid reference genes for quantitative real-time PCR analysis in Plutella xylostella (Lepidoptera: plutellidae). Int. J. Biol. Sci. 9 (8), 792–802. doi:10.7150/ijbs.5862
Guo H., Meng L., Wang Y., Zheng L., Li B. (2014). Oviposition behavior of the predatory midge Aphidoletes aphidimyza in response to aphid patch quality. J. Insect Behav. 27, 816–825. doi:10.1007/s10905–014–9473–1
Higashida K., Yano E., Takabayashi J., Ozawa R., Yoneya K. (2022). Volatiles from eggplants infested by Aphis gossypii induce oviposition behavior in the aphidophagous gall midge Aphidoletes aphidimyza. Arthropod-Plant Inte. 16 (1), 45–52. doi:10.1007/s11829–021–09882–w
Hornáková D., Matousková P., Kindl J., Valterová I., Pichová I. (2010). Selection of reference genes for real-time polymerase chain reaction analysis in tissues from Bombus terrestris and Bombus lucorum of different ages. Anal. Biochem. 397 (1), 118–120. doi:10.1016/j.ab.2009.09.019
Li K., Xu N., Yang Y. J., Zhang J. H., Yin H. (2018). Identification and validation of reference genes for RT-qPCR normalization in Mythimna separata (Lepidoptera: noctuidae). Biomed. Res. Int. 2018, 1828253. doi:10.1155/2018/1828253
Lin Q., Zhai Y., Chen H., Yin Y., Sun M., Yu Y., et al. (2017). Predatory capacity of Aphidoletes aphidimyza (Rondani). Chin. J. Biol. Control 33 (2/3), 171. doi:10.16409/j.cnki.2095–039x.2017.02.004
Liu G., Qiu X., Cao L., Zhang Y., Zhan Z., Han R. (2016). Evaluation of reference genes for reverse transcription quantitative PCR studies of physiological responses in the ghost moth, Thitarodes armoricanus (Lepidoptera, Hepialidae). PloS One 11 (7), e0159060. doi:10.1371/journal.pone.0159060
Liu R., He X., Lehane S., Lehane M., Hertz-Fowler C., Berriman M., et al. (2012). Expression of chemosensory proteins in the tsetse fly Glossina morsitans is related to female host–seeking behaviour. Insect Mol. Biol. 21 (1), 41–48. doi:10.1111/j.1365–2583.2011.01114.x
Liu Z., Xiao J., Xia Y., Wu Q., Zhao C., Li D. (2022). Selection and validation of reference genes for RT-qPCR–based analyses of Anastatus japonicus Ashmead (Hymenoptera: helicopteridae). Front. Physiol. 13, 1046204. doi:10.3389/fphys.2022.1046204
Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2- ΔΔCT method. Methods 25 (4), 402–408. doi:10.1006/meth.2001.1262
Lü J., Chen S., Guo M., Ye C., Qiu B., Wu , et al. (2018). Selection and validation of reference genes for RT-qPCR analysis of the ladybird beetle Henosepilachna vigintioctomaculata. Front. Physiol. 9, 1614. doi:10.3389/fphys.2018.01614
Lu Y., Yuan M., Gao X., Kang T., Zhan S., Wan H., et al. (2013). Identification and validation of reference genes for gene expression analysis using quantitative PCR in Spodoptera litura (Lepidoptera: noctuidae). PloS One 8 (7), e68059. doi:10.1371/journal.pone.0068059
Lu Y., Zheng X., Liang Q., Xu H., Yang Y., Tian J., et al. (2015). Evaluation and validation of reference genes for SYBR Green qRT-PCR normalization in Sesamia inferens (Lepidoptera: noctuidae). J. Asia Pac Entomol. 18 (4), 669–675. doi:10.1016/j.aspen.2015.08.002
Ma L., Jiang T., Liu X., Xiao H., Peng Y., Zhang W. (2021). Evaluation of candidate reference genes for gene expression analysis in the brassica leaf beetle, Phaedon brassicae (Coleoptera: chrysomelidae). PloS One 16 (6), e0251920. doi:10.1371/journal.pone.0251920
Madahi K., Sahragard A., Hosseini R., Baniameri V. (2019). Bottom-up effect of host plants on life-history characteristics of Aphidoletes aphidimyza feeding on Aphis gossypii. Entomol. Exp. Appl. 167 (9), 835–847. doi:10.1111/eea.12824
Pan H., Yang X., Bidne K., Hellmich R. L., Siegfried B. D., Zhou X. (2015). Selection of reference genes for RT-qPCR analysis in the monarch butterfly, Danaus plexippus (L.), a migrating bio–indicator. PloS One 10 (6), e0129482. doi:10.1371/journal.pone.0129482
Peng R., Zhai Y., Ding H., Di T., Zhang T., Li B., et al. (2012). Analysis of reference gene expression for real-time PCR based on relative quantitation and dual spike–in strategy in the silkworm Bombyx mori. Acta Biochim. Biophys. Sin. 44 (7), 614–622. doi:10.1093/abbs/gms040
Peng X., Liu L., Huang Y. X., Wang S. J., Li D. X., Chen S. T., et al. (2021). Involvement of chemosensory proteins in host plant searching in the bird cherry–oat aphid. Insect Sci. 28 (5), 1338–1353. doi:10.1111/1744–7917.12865
Pfaffl M. W., Tichopad A., Prgomet C., Neuvians T. P. (2004). Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: bestKeeper–Excel–based tool using pair–wise correlations. Biotechnol. Lett. 26, 509–515. doi:10.1023/b:bile.0000019559.84305.47
Ponton F., Chapuis M. P., Pernice M., Sword G. A., Simpson S. J. (2011). Evaluation of potential reference genes for reverse transcription–qPCR studies of physiological responses in Drosophila melanogaster. J. Insect Physiol. 57 (6), 840–850. doi:10.1016/j.jinsphys.2011.03.014
Qu C., Wang R., Che W., Zhu X., Li F., Luo C. (2018). Selection and evaluation of reference genes for expression analysis using quantitative real-time PCR in the Asian Ladybird Harmonia axyridis (Coleoptera: coccinellidae). PloS One 13 (6), e0192521. doi:10.1371/journal.pone.0192521
Shakeel M., Zhu X., Kang T., Wan H., Li J. (2015). Selection and evaluation of reference genes for quantitative gene expression studies in cotton bollworm, Helicoverpa armigera (Lepidoptera: noctuidae). J. Asia-Pac. Entomol. 18 (2), 123–130. doi:10.1016/j.aspen.2015.01.001
Shang S., Huang C., Shen X., Yu X., Cao Y., Liu M., et al. (2019). Field control effect of Aphidoletes aphidimyza on tobacco Myzus persicae. Guizhou Agric. Sci. 47, 41–44. doi:10.3969/j.issn.1001–3601.2019.06.010
Shen C. H., Peng L. J., Zhang Y. X., Zeng H. R., Yu H. F., Jin L., et al. (2022). Reference genes for expression analyses by qRT-PCR in Phthorimaea operculella (Lepidoptera: gelechiidae). Insects 13 (2), 140. doi:10.3390/insects13020140
Shi C., Zhang Y. (2016). Advances in reference gene for real-time quantitative reverse transcription PCR (qRT-PCR) of insects research. Chin. J. Appl. Entomology 53 (2), 237–246. doi:10.7679/j.issn.2095–1353.2016.031
Silva A. X., Jander G., Samaniego H., Ramsey J. S., Figueroa C. C. (2012). Insecticide resistance mechanisms in the green peach aphid Myzus persicae (Hemiptera: aphididae) I: a transcriptomic survey. PloS One 7 (6), e36366. doi:10.1371/journal.pone.0036366
Silver N., Best S., Jiang J., Thein S. L. (2006). Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 7 (1), 1–9. doi:10.1186/1471–2199–7–33
Sun M., Lu M. X., Tang X. T., Du Y. Z. (2015). Exploring valid reference genes for quantitative real-time PCR analysis in Sesamia inferens (Lepidoptera: noctuidae). PloS One 10 (1), e0115979. doi:10.1371/journal.pone.0115979
Sun W., Jin Y., He L., Lu W. C., Li M. (2010). Suitable reference gene selection for different strains and developmental stages of the carmine spider mite, Tetranychus cinnabarinus, using quantitative real-time PCR. J. Insect Sci. 10, 208. doi:10.1673/031.010.20801
Szabo A., Perou C. M., Karaca M., Perreard L., Quackenbush J. F., Bernard P. S. (2004). Statistical modeling for selecting housekeeper genes. Genome Biol. 5, 1–10. doi:10.1186/gb–2004–5–8–r59
Van Hiel M. B., Van Wielendaele P., Temmerman L., Van Soest S., Vuerinckx K., Huybrechts R., et al. (2009). Identification and validation of housekeeping genes in brains of the desert locust Schistocerca gregaria under different developmental conditions. BMC Mol. Biol. 10, 1–10. doi:10.1186/1471–2199–10–56
Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., et al. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3 (7), 1–12. doi:10.1186/gb–2002–3–7–research0034
Watanabe H., Yano E., Higashida K., Hasegawa S., Takabayashi J., Ozawa R. (2016). An attractant of the aphidophagous gall midge Aphidoletes aphidimyza from honeydew of Aphis gossypii. J. Chem. Ecol. 42, 149–155. doi:10.1007/s10886–016–0666–2
Xie L. C., Tian J. C., Lu Y. H., Xu H. X., Zang L. S., Lu Z. X., et al. (2021). Selection of reference genes for RT-qPCR analysis in Trichogramma chilonis (Hymenoptera: trichogrammatidae). J. Asia–Pac. Entomol. 24 (3), 679–687. doi:10.1016/j.aspen.2021.05.016
Xu H., Wang G., Lu Y., Yang Y., Zheng X., Tian J. (2019). Screening reference genes and evaluating of their expression stability for qRT-PCR normalization in Chilo suppressalis (Lepidoptera: pyralididae). Chin. J. Rice Sci. 33 (1), 75–84. doi:10.16819/j.1001–7216.2019.8035
Xu J., Welker D. L., James R. R. (2021). Variation in expression of reference genes across life stages of a bee. Megachile Rotundata. Insects. 12 (1), 36. doi:10.3390/insects12010036
Yan X., Zhang Y., Xu K., Wang Y., Yang W. (2021). Selection and validation of reference genes for gene expression analysis in Tuta absoluta Meyrick (Lepidoptera: gelechiidae). Insects 12 (7), 589. doi:10.3390/insects12070589
Yang A. P., Wang Y. S., Huang C., Lv Z. C., Liu W. X., Bi S. Y., et al. (2021). Screening potential reference genes in Tuta absoluta with real-time quantitative PCR analysis under different experimental conditions. Genes 12 (8), 1253. doi:10.3390/genes12081253
Yang C., Pan H., Liu Y., Zhou X. (2015). Stably expressed housekeeping genes across developmental stages in the two–spotted spider mite, Tetranychus urticae. PloS One 10 (3), e0120833. doi:10.1371/journal.pone.0120833
Yang C., Preisser E. L., Zhang H., Liu Y., Dai L., Pan H., et al. (2016). Selection of reference genes for RT-qPCR analysis in Coccinella septempunctata to assess un–intended effects of RNAi transgenic plants. Front. Plant Sci. 7, 1672. doi:10.3389/fpls.2016.01672
Yang Y., Li Z., Cao J., Li Y., Li H., Yang Q., et al. (2017). Identification and evaluation of suitable reference genes for normalization of MicroRNA expression in Helicoverpa armigera (Lepidoptera: noctuidae) using quantitative real-time PCR. J. Insect Sci. 17 (2), 33. doi:10.1093/jisesa/iex007
Ye J., Coulouris G., Zaretskaya I., Cutcutache I., Rozen S., Madden T. L. (2012). Primer–BLAST: a tool to design target–specific primers for polymerase chain reaction. BMC Bioinforma. 13, 134. doi:10.1186/1471–2105–13–134
Yin J., Sun L., Zhang Q., Cao C. (2020). Screening and evaluation of the stability of expression of reference genes in Lymantria dispar (Lepidoptera: erebidae) using qRT-PCR. Gene 749, 144712. doi:10.1016/j.gene.2020.144712
Yuan M., Lu Y., Zhu X., Wan H., Shakeel M., Zhan S., et al. (2014). Selection and evaluation of potential reference genes for gene expression analysis in the brown planthopper, Nilaparvata lugens (Hemiptera: delphacidae) using reverse–transcription quantitative PCR. PloS One 9 (1), e86503. doi:10.1371/journal.pone.0086503
Zeng Y., Merchant A., Wu Q., Wang S., Kong L., Zhou X., et al. (2020). A chemosensory protein BtabCSP11 mediates reproduction in Bemisia tabaci. Front. Physiol. 11, 709. doi:10.3389/fphys.2020.00709
Zhang S., An S., Li Z., Wu F., Yang Q., Liu Y., et al. (2015). Identification and validation of reference genes for normalization of gene expression analysis using qRT-PCR in Helicoverpa armigera (Lepidoptera: noctuidae). Gene 555 (2), 393–402. doi:10.1016/j.gene.2014.11.038
Zhou L., Meng J. Y., Ruan H. Y., Yang C. L., Zhang C. Y. (2021). Expression stability of candidate RT-qPCR housekeeping genes in Spodoptera frugiperda (Lepidoptera: noctuidae). Arch. Insect Biochem. Physiol. 108 (1), e21831. doi:10.1002/arch.21831
Keywords: Aphidoletes aphidimyza, chemosensory protein, expression stability, housekeeping genes, real-time quantitative PCR
Citation: Shen X-X, Zhang G-Q, Zhao Y-X, Zhu X-X, Yu X-F, Yang M-F and Zhang F (2023) Selection and validation of optimal reference genes for RT-qPCR analyses in Aphidoletes aphidimyza Rondani (Diptera: Cecidomyiidae). Front. Physiol. 14:1277942. doi: 10.3389/fphys.2023.1277942
Received: 15 August 2023; Accepted: 06 October 2023;
Published: 25 October 2023.
Edited by:
Zhijun Yu, Hebei Normal University, ChinaCopyright © 2023 Shen, Zhang, Zhao, Zhu, Yu, Yang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mao-Fa Yang, Z2RnZGx5QDEyNi5jb20=; Feng Zhang, ZnpoYW5nQG5qYXUuZWR1LmNu
 Xiu-Xian Shen
Xiu-Xian Shen Guo-Qiang Zhang2
Guo-Qiang Zhang2 Mao-Fa Yang
Mao-Fa Yang Feng Zhang
Feng Zhang