
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Physiol., 27 November 2023
Sec. Invertebrate Physiology
Volume 14 - 2023 | https://doi.org/10.3389/fphys.2023.1274416
The eusocial pest, red imported fire ant (Solenopsis invicta), is a highly invasive species that poses significant threats to public safety, agriculture, and the ecological environment. Cycloxaprid, a newly identified effective, slow-acting, and non-repellent insecticide against S. invicta, allows contaminated individuals to transfer the insecticide among nestmates through body contact. However, the molecular-level changes occurring in S. invicta post cycloxaprid exposure and any molecular alterations contributing to the slow demise or decreased sensitivity remain unclear. In this study, transcriptomic and metabolomic techniques were used to investigate the molecular mechanisms of S. invicta exposed to cycloxaprid. Differential analysis results revealed 275, 323, and 536 differentially expressed genes at 12, 24, and 48 h, respectively. Genes involved in lipid and energy metabolism, DNA integration, and hormone synthesis were largely upregulated at 12 h, suggesting S. invicta might actively resist cycloxaprid impacts, and predominantly downregulated at 48 h, indicating further functional impairment and impending death. Also, we observed an imbalance in olfactory perception pathways at 12 h, which may indicate a disruption in the olfactory system of S. invicta. Metabolomic results showed that the regulation of most differential metabolites (DMs) was consistent with the expression changes of their related DEGs at different time points. Our study provides insights into the mechanism underlying slow-acting and non-repellent properties of cycloxaprid against S. invicta.
The red imported fire ant, Solenopsis invicta, is a highly destructive eusocial pest that has invaded more than 20 countries and regions worldwide, including the United States, Australia, and China (Ascunce et al., 2011; Wetterer, 2013). This pest poses significant threats to public safety, agriculture, and the ecological environment (Gutrich et al., 2007; Vinson, 2013; Wang et al., 2020). Medically, S. invicta stings can cause intense pain, blistering, pustules, allergic reactions, and, in extreme cases, anaphylactic shock or fatal outcomes (Haddad Junior and Larsson, 2015; Bernaba et al., 2019). Also, S. invicta can cause damage to the roots and seeds of various crops, leading to reduced yields and economic losses in agricultural systems (Chan and Guénard, 2020). Additionally, the invasion of S. invicta significantly deceased the abundance and diversity of native ants and other arthropods (Morrison, 2002). Given these issues, S. invicta resulted in massive global economic costs for damage and management (Angulo et al., 2022). For example, S. invicta was estimated to lead to annual costs of USD 6 billion in the United States and AUD 1.65 billion in Australia (Pereira et al., 2002; Wylie and Janssen-May, 2017).
Eusocial insects exhibit complex behaviors and biological features (e.g., highly organized social structures, hierarchical communication systems, task specialization, and unique food-sharing behaviors) that differentiate them from solitary insects (Robinson, 1992; Gordon, 2019). These characteristics not only ensure the high adaptability and flexibility of eusocial pests to invade diverse habitats but also pose challenges to their control. Traditional insecticides can rapidly eliminate individuals of eusocial pests outside the nest, but they cannot easily reach the reproducers and larvae concealed deep within the nest, resulting in inadequate control at a colony level (Drees and Gold, 2003; Silverman and Brightwell, 2008; Rust and Su, 2012). Therefore, insecticides to control eusocial pests such as S. invicta should be slow-acting and non-repellent to ensure the horizontal transfer of insecticides among nestmates through physical contact and eventually eradicate the whole colony. Currently, insecticides such as imidacloprid, fipronil, rotenone, and spinosad have been widely applied for S. invicta management (Chen and Oi, 2020).
Cycloxaprid is a novel oxabridged cis-nitromethylene neonicotinoid insecticide characterized by a nitro (-NO2) group in a cis-configuration (Shao et al., 2013a). This insecticide appears to activate a different site compared to traditional neonicotinoid insecticide on the nicotinic acetylcholine receptors (nAChRs) and exhibits high insecticidal activity (Shao et al., 2013b; Zhang et al., 2019). For now, it has been applied to control various pests, including A. gossypii, Bemisia tabaci, and Sitobion avenae (Cui et al., 2016; Wang et al., 2018a; Cui et al., 2020). For example, Cui et al. (2012) reported that cycloxaprid exhibited intense contact and root-systemic activity against S. avenae, negatively affecting its feeding behavior and growth rate. Also, exposure to sublethal levels of cycloxaprid significantly shortened the pre-oviposition period and life expectancy of Aphis gossypii and inhibited the activity of various detoxification enzymes (Cui et al., 2018).
Our previous studies showed that cycloxaprid is a slow-acting and non-repellent insecticide against S. invicta workers at certain concentrations (Zhang et al., 2022). For example, surfaces treated with cycloxaprid (0.3 μg/cm2) led to high mortality in S. invicta workers. However, cycloxaprid caused a much slower speed of kill than other fire ant control agents such as bifenthrin. In addition, cycloxaprid did not significantly influence the foraging, digging, and nesting behaviors of S. invicta workers. Interestingly, both corpses and live ants previously exposed to cycloxaprid caused horizontal toxicity against untreated ants. However, the molecular-level changes within S. invicta post cycloxaprid exposure remain unclear. In this study, we analyzed changes in gene expression and metabolites in S. invicta workers after exposure to cycloxaprid for 12, 24, and 48 h. By examining the differences in genes and metabolites, we aimed to reveal the mechanism of action of cycloxaprid on S. invicta and discuss the potential reasons underlying its slow-acting and non-repellent properties.
Six S. invicta colonies were collected from Tianlu Lake Forest Park (23°12′ N, 113°8′ E), Guangzhou, China, on 3 November 2021. Mounds were more than 5 m apart at the collection site. Nest materials with immatures (eggs, larvae, and pupae) and adults (workers and queens) of S. invicta were rapidly shoveled into a plastic container (length: 50 cm, width: 37 cm, height: 26 cm; Kangyi Co., Ltd., Hubei, China). The walls of the containers were previously smeared with talcum powder, and therefore S. invicta cannot climb and escape (Ning et al., 2019). The collected S. invicta colonies were brought to the laboratory and reared under relatively constant conditions (a photoperiod of 14 h light: 10 h dark, and room temperature of 21°C–23°C). Ten days before the experiment, S. invicta colonies were extracted from the soil using the water-dropping method provided by Chen (2007). The extracted ants were transferred to another plastic container (the inner walls were smeared with the talcum powder as mentioned above). Frozen crickets, water, and 20% honey water were supplied.
Cycloxaprid was obtained from the School of Pharmacy, East China University of Science and Technology, Shanghai, China. Plastic boxes (length: 44 cm, width: 29 cm, height: 19 cm; Kangyi Co., Ltd., Hubei, China) with talcum powder-coated inner walls were used as experimental arenas. Cycloxaprid was dissolved in acetone (analytical reagent, Fuyu Chemical Co., Ltd., Tianjin, China) to prepare for the aimed concentrations. The required amount of solution was then evenly smeared onto the bottom of the boxes to reach the cycloxaprid concentration of 0.3 μg/cm2 after complete evaporation of the acetone. The same amount of acetone (without cycloxaprid) was added to the control boxes. For each colony, 10 g of S. invicta workers were weighed and transferred to a cycloxaprid-treated or untreated box, and 10-mL tubes containing water or 20% honey water were supplied. Here, we only tested worker ants because the immatures and reproductive castes (i.e., queens) may have different molecular responses that may interfere with the results of workers (Kohlmeier et al., 2017; Negroni et al., 2021). Each box was placed in a larger plastic container (length: 50 cm, width: 37 cm, height: 26 cm), where moistened paper towels were placed on the bottom to maintain humidity (Wen et al., 2022). The bioassays were maintained at room temperature (21°C–23°C). At 12, 24, and 48 h, 500 mg of live S. invicta workers were randomly collected from each experimental box and immediately transferred to liquid nitrogen, then stored in a −80°C freezer.
The transcriptome of S. invicta workers collected from cycloxaprid-treated or untreated boxes was analyzed using RNA-seq. Solenopsis invicta colonies 1-3 were used in this test, with each colony as an independent biological replicate. In the sample preparation process, 20 mg S. invicta workers were pulverized into a fine powder under liquid nitrogen. Subsequently, the total RNA was isolated using TRIzol reagent (Invitrogen, USA), adhering strictly to the manufacturer’s guidelines. The RNA purification, library construction, and sequencing were carried out utilizing the Illumina sequencing platform, with all services provided by Metware Biotechnology Co., Ltd. (Wuhan, China). We employed Fastp (v0.19.3) to filter out low-quality reads and adapter sequences (Chen et al., 2018). We also utilized HISAT (v2.1.0) to construct the index and align clean reads with the reference genome (https://www.ncbi.nlm.nih.gov/assembly/GCF_016802725.1/#/st) (Kim et al., 2015). StringTie (v1.3.4d) was adopted to predict new genes (Pertea et al., 2016), and the transcripts were annotated using Diamond (v2.1.8) (Buchfink et al., 2015). The task of counting aligned reads and calculating the Fragments Per Kilobase of transcript per Million (FPKM) of each gene according to the gene length was performed using FeatureCounts (v1.6.2) (Liao et al., 2014). The differential expression between cycloxaprid-treated and untreated ants was analyzed by utilizing DESeq2 (v1.22.1) (Love et al., 2014) and EdgeR (Robinson et al., 2010), with p values corrected through the Benjamini–Hochberg method (Haynes, 2013). To identify differentially expressed genes (DEGs), a threshold of |log2foldchange| (log2FC) > 1 and a p-value < 0.05 were applied as the markers of significant differential expression.
The functional analyses with gene annotation and enrichment analyses were conducted to investigate the functions of DEGs at different time points. DEGs were annotated using the Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), Non-Redundant Protein Sequence Database (NR), Kentucky Online Gateway (KOG), Swiss-Prot, and Pfam databases. We executed enrichment analysis using the hypergeometric test. In the case of KEGG, the test was carried out at the pathway level, whereas for GO, the testing was based on the specific GO term.
To ascertain the precision and dependability of the transcriptome, we conducted an analysis using qRT-PCR on the total RNA of samples previously treated or untreated with cycloxaprid. Solenopsis invicta colonies 1-3 were tested, with each colony as an independent biological replicate. Eight selected genes (AASS, ACO, ORAI, SCD, CYP450-1, CYP450-2, CYP450-3, GST) were chosen based on their significant differential expression (see results) as well as their important roles in key biological processes (i.e., fatty acid synthesis and metabolism [AASS, ACO, SCD], energy conversion [ORAI], and detoxification [CYP450, GST]). We employed Ribosomal protein S3 (RPS3; XM_011172385) as the internal reference gene for normalization (Chen et al., 2022). Specific gene primer pairs were crafted utilizing the NCBI Primer-BLAST tool (http://www.ncbi.nlm.nih.gov/tools/primer-blast/). The primers used in this study are detailed in Supplementary Table S1. Each sample’s total RNA (2 µg) was transcribed in reverse to cDNA utilizing HiScript III RT SuperMix for qPCR (+gDNA wiper) (Vazyme, Nanjing, China), adhering strictly to the manufacturer’s guidelines. Subsequently, the qRT-PCR process was executed employing the Option Real-Time PCR System CFX96 (Bio-Rad, CA). The procedure conditions comprised an initial pre-denaturation phase at 95°C lasting for 2 min, followed by 39 cycles of denaturation at 95°C for 5 s and annealing at 60°C for 30 s. Following this, a melting curve was created by gradually escalating the temperature from 65°C to 95°C. Each qPCR operation was carried out in triplicate as three technical replicates to assess the variability introduced by the experimental procedure. The 2−ΔΔCT method (Livak and Schmittgen, 2001) was employed to determine the relative expression levels of the target genes. Statistical scrutiny was undertaken using a paired t-test, with a p-value < 0.05 indicating the statistical significance.
The metabolome of S. invicta workers collected from cycloxaprid-treated or untreated boxes was analyzed using Widely-targeted metabolomic analysis (Chen et al., 2013). Six S. invicta colonies were tested, each as an independent biological replicate. Samples kept at −80°C were defrosted on ice and then pulverized using liquid nitrogen. A solution of 400 μL, comprising methanol and water in a ratio of 7:3, was blended with an internal standard and added to the 20 mg of the crushed sample. This composite was then agitated at 1,500 rpm for 5 min. Following a 15-min icing period, the samples were centrifuged at 12,000 rpm for 10 min at 4°C. A portion of the supernatant (300 μL) was set aside and cooled at −20°C for 30 min, then centrifuged at 12,000 rpm for 3 min at 4°C. For Liquid chromatography-mass spectrometry (LC-MS) analysis, a 200 μL aliquot of the supernatant was processed using the Ultra-Performance Liquid Chromatography and Tandem Mass Spectrometry (UPLC-MS/MS) systems provided by Wuhan Metware Biotechnology Co., Ltd. (China).
To identify Differential metabolites (DMs), a threshold of |log2foldchange| (log2FC) > 1 and a VIP >1 were applied as the markers of significant difference. VIP values were derived from Partial least squares discriminant analysis (PLS-DA) outcomes using the MetaboAnalystR package in R. Before PLS-DA, the data underwent log-transformation and mean-centering. A permutation test involving 200 permutations was implemented to prevent overfitting. The R function ‘plsr’ was employed to perform unsupervised PLS-DA, following a unit variance-scaling of the data. Annotation of DMs was accomplished through the KEGG Compound database, with mapping to the KEGG Pathway database. Statistically significant enrichment pathways were identified using the p-value from a hypergeometric test for a specified list of metabolites.
A correlation analysis was carried out between transcriptome and metabolome to gain deeper insight into the regulatory network of S. invicta workers responding to cycloxaprid. The transcriptomic and metabolomic analysis data from samples of colonies 1-3 were used for correlation analysis. The Pearson Correlation Coefficient (PCC) for differentially expressed genes and metabolites was computed employing the ‘cor’ function within the R package. To showcase the fold changes in genes and metabolites across different groups, a nine-quadrant plot was generated with a PCC >0.6 and p-value <0.05, using the ggplot2 and getopt packages in R. For generating a correlation network diagram, genes and metabolites with a PCC >0.85 and a p-value <0.05 were chosen, using Cytoscape (v3.9.1) as per Shannon et al. (2003).
Each sample yielded 42,596,782 to 56,609,672 clean reads, with Q30 percentages ranging from 90.38% to 92.81% and GC content ranging from 44.29% to 48.15% (Supplementary Table S2), reflecting the high quality of the transcriptome sequencing data. Following the alignment of clean reads to the reference genome of S. invicta, we identified 15,673 to 15,967 expressed genes for each sample. A total of 275, 323, and 536 DEGs at 12, 24, and 48 h were identified, respectively, using a threshold of |log2FC| > 1 and p-value < 0.05 (Figure 1A). Venn diagram (Figure 1B) revealed 23 shared DEGs between 12 and 24 h, 24 shared DEGs between 12 and 48 h, 56 shared DEGs between 24 and 48 h, and 9 shared DEGs across all three time points.
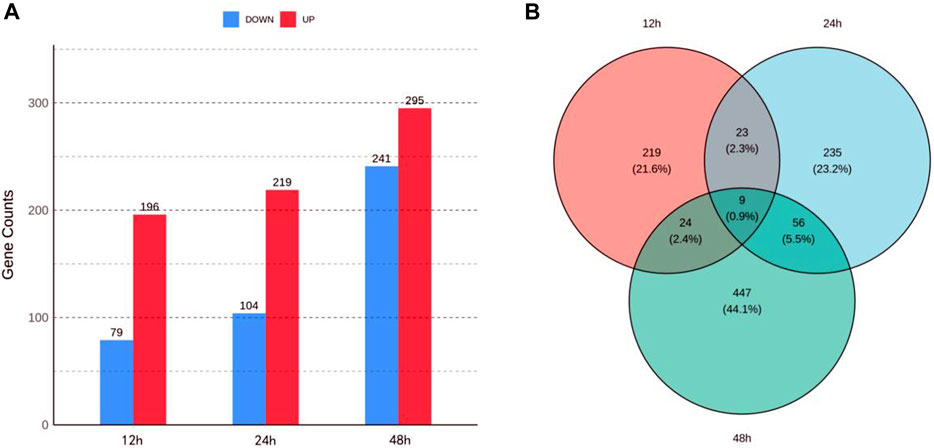
FIGURE 1. Transcriptomic analysis of Solenopsis invicta at three time points post cycloxaprid exposure. (A) The number of upregulated and downregulated differentially expressed genes (DEGs) at three time points; (B) Venn diagram illustrating the overlap of DEGs at the three time points, highlighting the shared and unique responses to cycloxaprid exposure.
Functional analyses provided insights into significantly enriched pathways (Figure 2; Figure 3) and the involvement of upregulated and downregulated DEGs in these pathways (Supplementary Figure S1, S2). At 12 h, the significantly enriched pathways primarily consisted of upregulated DEGs associated with fatty acid metabolism, fatty acid biosynthesis, AMPK signaling, insulin signaling, acyltransferase activity, DNA integration, and insect hormone biosynthesis. At 24 h, the significantly enriched pathways also primarily comprised upregulated DEGs, which were involved in protein digestion and absorption, pancreatic secretion, proteolysis, extracellular region, and amino sugar and nucleotide sugar metabolism. At 48 h, the significantly enriched pathways included both upregulated and downregulated DEGs. Specifically, DEGs involved in fatty acid metabolism, fatty acid biosynthesis, AMPK signaling pathway, insulin signaling pathway, iron ion binding, and heme binding were downregulated. In contrast, some DEGs involved with transmembrane transport, extracellular region, and cAMP signaling were upregulated. In addition, 22 DEGs involved in sensory perception of smell, odorant binding, and olfactory receptor activity displayed an inconsistent distribution at 12 h, with 13 upregulated and 9 downregulated.
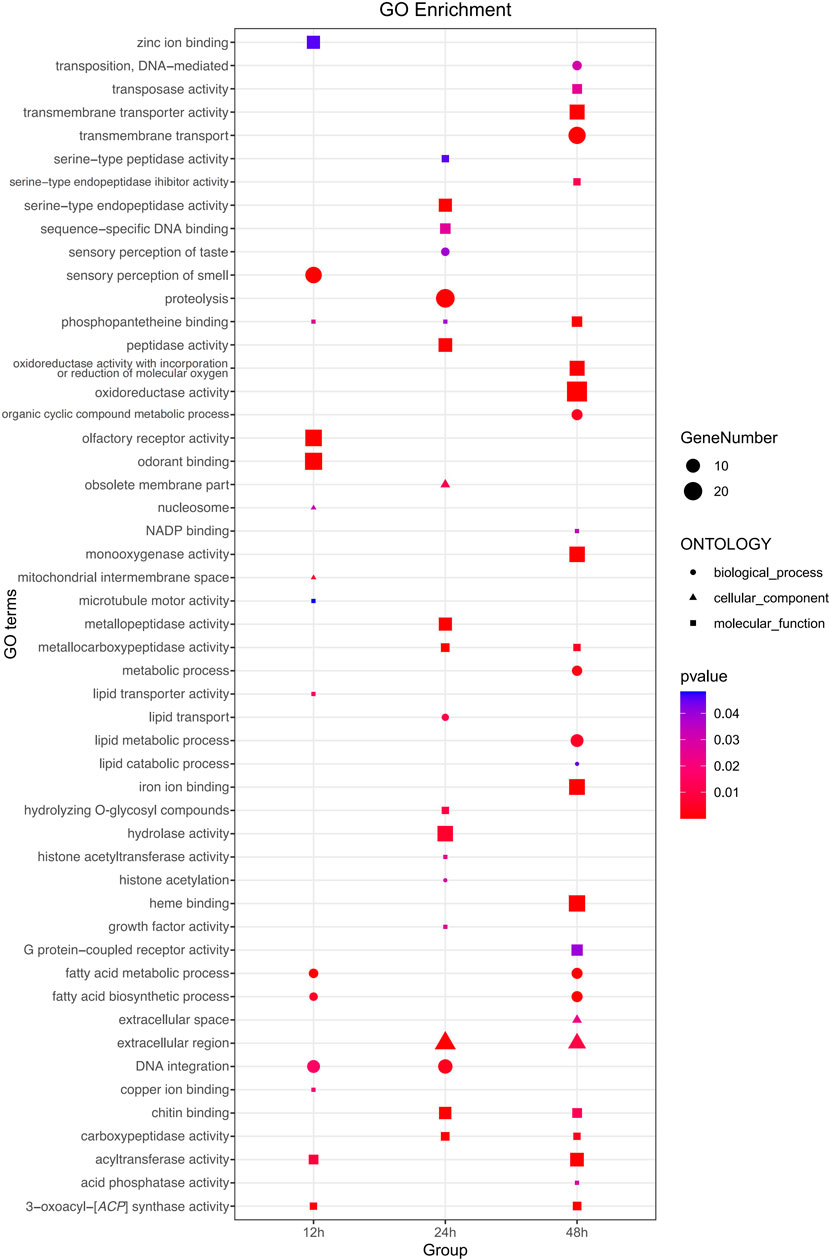
FIGURE 2. Comprehensive Gene Ontology (GO) enrichment analysis of DEGs at three distinct time points post cycloxaprid exposure. The size of each dot represents the number of genes involved in the corresponding GO term, while the shape symbolizes the specific functional classification of the respective GO term. p-values, calculated using hypergeometric tests, demonstrate the statistical significance of the enrichment results.
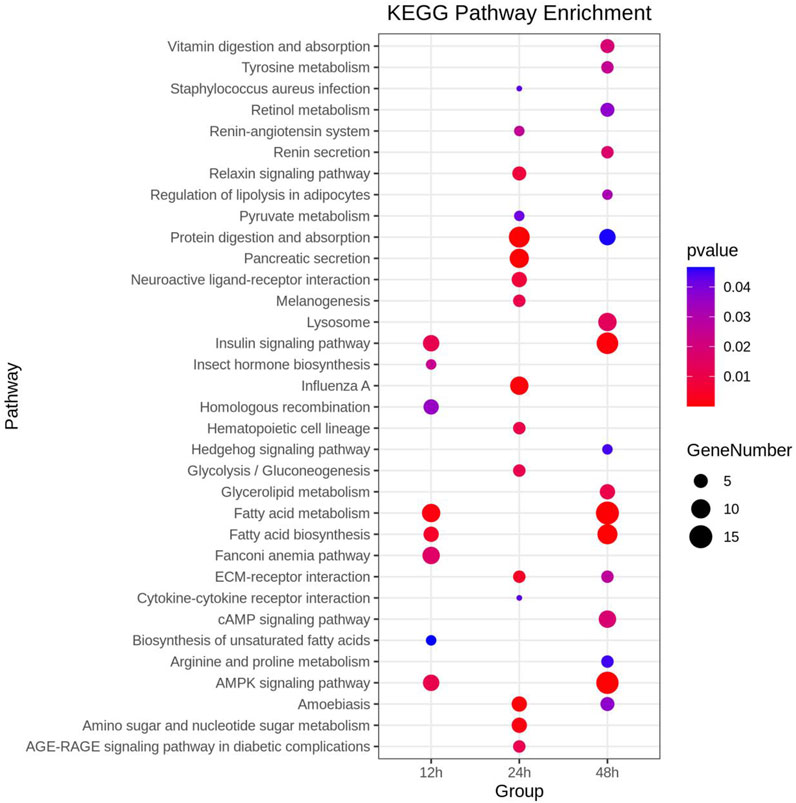
FIGURE 3. Comprehensive Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of DEGs at three distinct time points post cycloxaprid exposure. The size of each dot represents the number of genes involved in the corresponding KEGG pathways. p-values, calculated using hypergeometric tests, demonstrate the statistical significance of the enrichment results.
Accordance was observed between the qRT-PCR expression patterns and the FPKM values valued from RNA-seq data (Figure 4). At specific time points, statistical analysis revealed significant differences (p-value < 0.05) in the expression levels of selected genes between cycloxaprid-treated and untreated ants. Specifically, AASS and ORAI were significantly upregulated at 48 h; CYP450-2 was significantly downregulated at 24 h; ACO, CYP450-1, and CYP450-3 were significantly downregulated at 48 h; SCD and GST were significantly downregulated at 24 h and 48 h. The consistency of the RNA-seq and qRT-PCR data underscored the precision and dependability of our transcriptomic analysis.
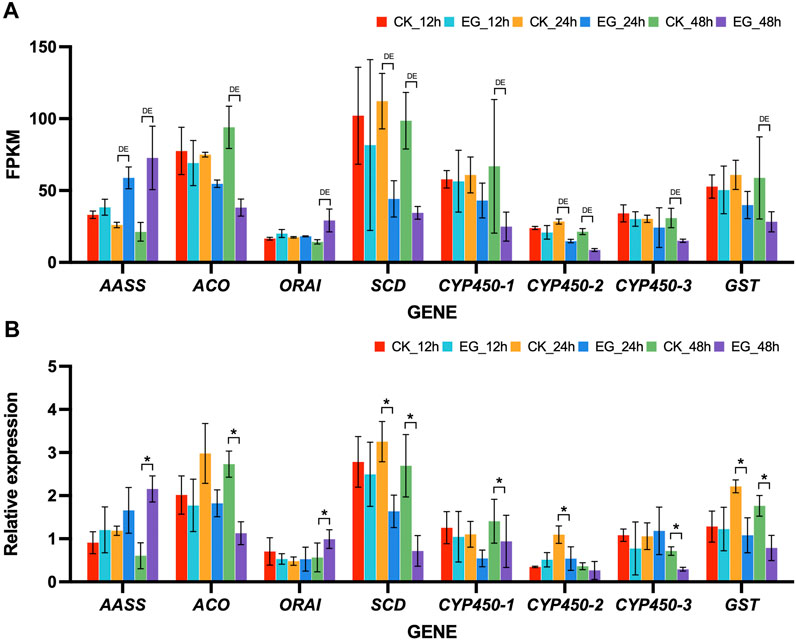
FIGURE 4. Expression patterns of 8 selected DEGs in Solenopsis invicta across three time points post cycloxaprid exposure. (A) Expression levels of the selected genes based on Fragments Per Kilobase of transcript per Million (FPKM) values obtained from RNA-seq data. “DE” denotes differential expression, which has been validated through the use of “DESeq2” or “EdgeR” tests, indicating a statistically significant difference in gene expression between the control and experimental groups at the corresponding time point. (B) Relative expression levels of the selected genes were obtained via quantitative real-time PCR (qRT-PCR) analysis. Error bars represent the standard error of the mean (SEM) from three replicates. “*” marks instances where the paired t-test yielded a p-value <0.05, suggesting a significant difference in gene expression between the control and experimental groups at the corresponding time point. CK and, EG indicate the control group and the experimental group (fire ants treated with cycloxaprid), respectively, followed by the time point of sampling. The consistency between the RNA-seq and qRT-PCR data highlights the reliability of our transcriptomic analysis.
UPLC-MS/MS detected 24 classes and 1,135 metabolites (Figure 5A). The PLS-DA analysis results demonstrated a progressive separation between the experimental and control groups across the three time points (12, 24, and 48 h). The separation gradually became more evident as the exposure time increased, and by 48 h, the clustering was distinctly clear (Figure 5B). A total of 24, 23, and 68 DMs at 12, 24, and 48 h were identified, respectively, using a threshold of |log2FC| > 2 and VIP >0.05 (Figure 5C). Venn diagram (Figure 5D) revealed 5 shared DEGs between 12 and 24 h, 2 shared DEGs between 12 and 48 h, 4 shared DEGs between 24 and 48 h, and 3 shared DMs across all three time points.
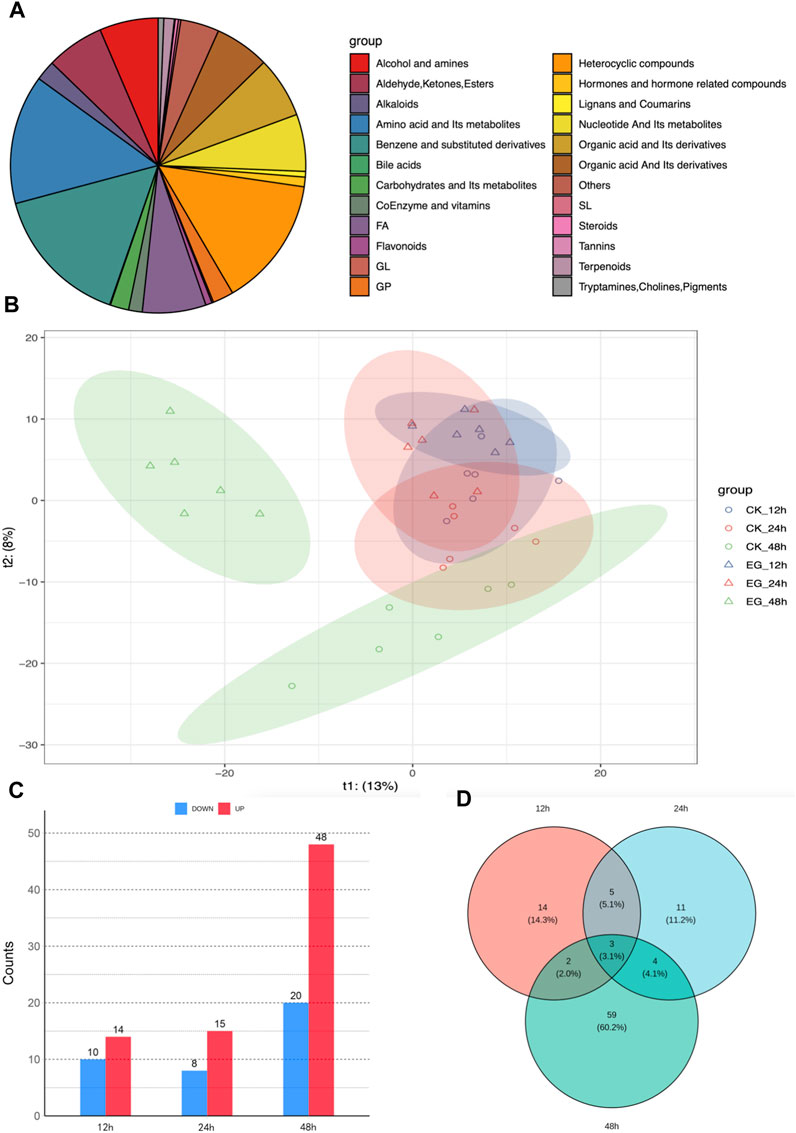
FIGURE 5. Metabolomic analysis of Solenopsis invicta at three time points post cycloxaprid exposure. (A) Classification of detected metabolites; (B) Partial Least Squares Discriminant Analysis (PLS-DA) results for six sample groups, illustrating the progressive separation between the experimental and control groups across the three time points; (C) The number of upregulated and downregulated differentially expressed metabolites (DMs) at three time points; (D) Venn diagram illustrating the overlap of DMs at the three time points, highlighting the shared and unique responses to cycloxaprid exposure.
Utilizing the KEGG database, 346 out of 1,135 metabolites were annotated. Additionally, KEGG enrichment analysis revealed that DMs were enriched in 11, 13, and 15 pathways at 12, 24, and 48 h, respectively (Figure 6). Pathways, including metabolic pathways, neuroactive ligand-receptor interaction, biosynthesis of cofactors, and tyrosine metabolism, were significantly enriched at all three time points. Differential abundance score plots illustrated the changing trends of enriched pathways (Supplementary Figure S3–S5). At 12 h, DMs in the fructose and mannose metabolism and antifolate resistance were upregulated, whereas DMs in the caffeine metabolism were downregulated. At 24 h, DMs in the pertussis and Fc epsilon RI signaling pathway were upregulated, whereas DMs in the caffeine metabolism were downregulated. At 48 h, DMs in the pertussis and riboflavin metabolism were upregulated, whereas DMs in the sphingolipid metabolism and D-Glutamine and D-glutamate metabolism were downregulated.
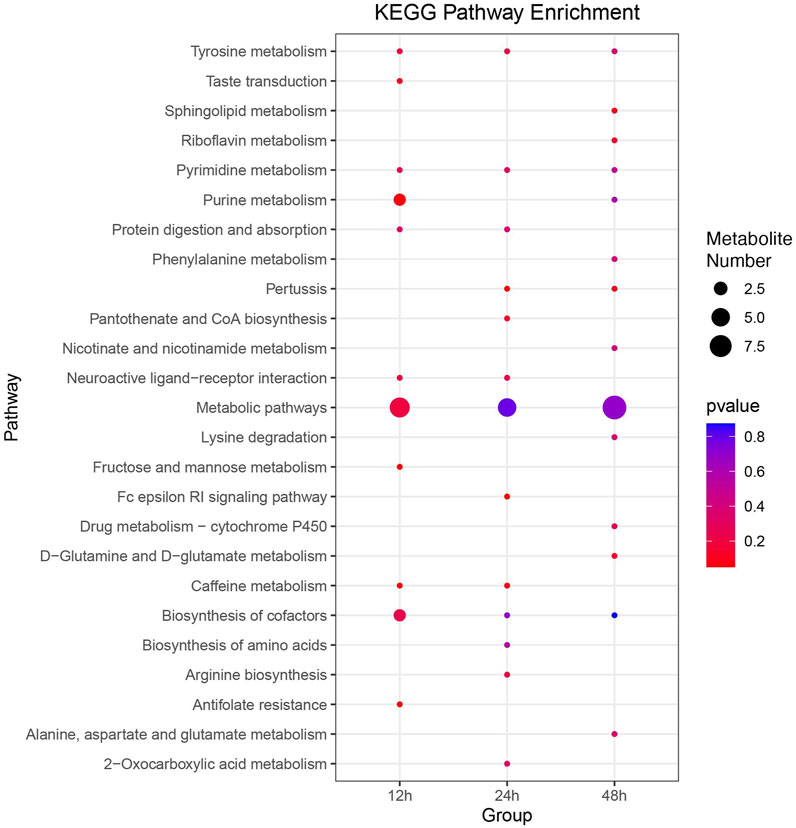
FIGURE 6. Comprehensive KEGG enrichment analysis of DMs at three distinct time points post cycloxaprid exposure. The size of each dot represents the number of genes involved in the corresponding KEGG pathways. p-values, calculated using hypergeometric tests, demonstrate the statistical significance of the enrichment results.
Correlation analysis revealed a significant association between 226 DEGs and 54 DMs (|R| > 0.8, p < 0.05). The nine-quadrant diagram (Figure 7) showed that the regulatory relationships between DEGs and DMs changed over time post cycloxaprid exposure. There was a notably higher number of points in Quadrants 1 and 3 at 48 h (Figure 7C) compared with 12 and 24 h (Figures 7A,B), indicating a significant shift in the gene expression patterns that caused an increased number of metabolites influenced by both upregulated and downregulated DEGs. Meanwhile, the number of points in Quadrant 7 remained relatively stable across the time points, suggesting a persistent association between downregulated DEGs and decreased DMs throughout the experiment. In addition, we generated a series of correlation network diagrams (|R| > 0.85, p < 0.05) to provide a more extensive perspective on the interaction between DEGs and DMs in S. invicta post cycloxaprid exposure (Figure 8), which highlights the intricate relationships between the transcriptomic and metabolomic changes in response to cycloxaprid exposure.
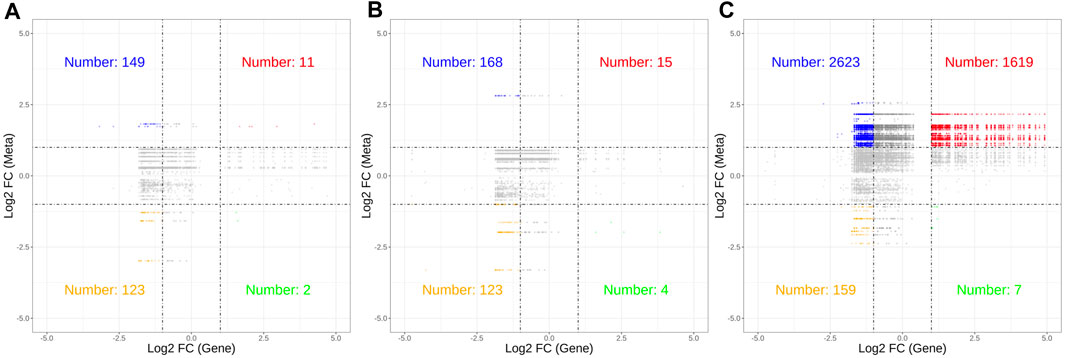
FIGURE 7. Nine-quadrant diagrams showed correlations between genes and metabolites in Solenopsis invicta at 12 (A), 24 (B), and 48 h (C) post cycloxaprid exposure. These diagrams display dynamic relationships between gene expression and metabolic changes in response to cycloxaprid exposure over time.
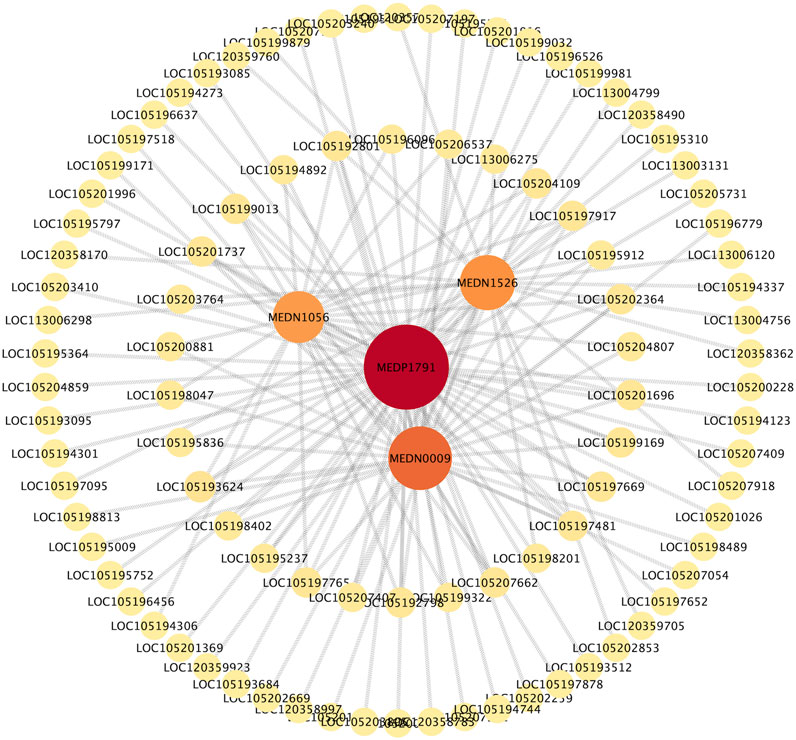
FIGURE 8. The correlation network showed connections between genes and metabolites in Solenopsis invicta post cycloxaprid exposure. The network indicated the complex relationships and potential interactions between the transcriptomic and metabolomic changes post cycloxaprid exposure.
A few recent studies focused on the molecular effect of insecticides on S. invica. For example, Siddiqui et al. (2022a) utilized transcriptomic analysis to investigate the detoxification mechanisms of S. invicta in response to beta-cypermethrin and fipronil, revealing significant alterations in metabolic pathways, insulin signaling pathways, and fatty acid metabolism. This study also detected changes in the expression of detoxification genes within the families of acetylcholinesterase (AChE), carboxylesterase (CarE), glutathione S-transferase (GST), and cytochrome P450 (CYP450). Similarly, Siddiqui et al. (2022b) reported that indoxacarb treatment also affected these pathways and genes in S. invicta. In addition, Xiong et al. (2022) utilized qRT-PCR and RNA interference (RNAi) technologies to investigate the function of specific CYP450 family genes of S. invicta treated with fluralaner, and they found that several genes, such as CYP9AS16, CYP6AS161, CYP6SQ20, and CYP336A45, were closely related to the detoxification process of fluralaner. These findings align with the present study, providing a consistent view of the insecticide response at the molecular level in S. invicta.
Cycloxaprid has recently garnered substantial attention as a promising alternative to traditional insecticides, owing to its potential to counter resistance issues for agricultural pests (Wang et al., 2018a; Cui et al., 2020). For example, Zhang et al. (2018) found a slower rate of resistance development to cycloxaprid in Laodelphax striatellus compared to chlorpyrifos and buprofezin. Remarkably, only a 10-fold increase in resistance was observed in an L. striatellus variant with cycloxaprid resistance, contrasting with the increases of 106-fold and 332-fold for buprofezin and chlorpyrifos, respectively, under equivalent selection pressures. Dong et al. (2022) revealed that an A. gossypii variant with cycloxaprid resistance demonstrated minimal cross-resistance to six of other insecticides. The fold increases in resistance were 4.3 for imidacloprid, 2.9 for acetamiprid, 3.7 for thiamethoxam, 6.1 for nitenpyram, 2.2 for flupyradifurone, and 4.5 for sulfoxaflor. Similarly, Cui et al. (2016) and Zhang et al. (2019) suggested no cross-resistance between cycloxaprid and imidacloprid in A. gossypii and Nilaparvata lugens. Some recent research has investigated the molecular effects of cycloxaprid on various sucking insect pests (Yang et al., 2016; Jin et al., 2017). For example, Jin et al. (2021) identified 22 genes (e.g., genes in the families of CYP450 and GST) potentially associated with Sogatella furcifera’s resistance to cycloxaprid using transcriptomic analysis. Zhang et al. (2018) found that the resistance to cycloxaprid in L. striatellus was linked to the modified expression of nicotinic acetylcholine receptor subunits. Moreover, they reported that a knockdown of the Lsa1 gene significantly amplified the susceptibility of the cycloxaprid-resistant strain to cycloxaprid. While these insights shed light on the resistance mechanisms of sucking insects, it is crucial to note that successful control of eusocial pests, such as S. invicta, primarily depends on the slow-action and non-repellency of cycloxaprid.
In our study, transcriptomic analyses revealed alterations in various physiological processes of S. invicta post cycloxaprid exposure over different durations. At 12 h, most DEGs were upregulated and enriched pathways involved in fatty acid metabolism, fatty acid biosynthesis, AMPK signaling pathway, insulin signaling pathway, and acyltransferase activity. Fatty acids are essential for energy storage and release, cell membrane assembly, and the production of signaling molecules in insects (Canavoso et al., 2001; Arrese and Soulages, 2010). The insulin signaling pathway assumes a crucial role in sustaining energy homeostasis through regulating carbohydrate, lipid, protein, and energy metabolism in insects (Wu and Brown, 2006; Géminard et al., 2009; Nässel and Broeck, 2016), which promotes glucose uptake and glycolysis, stimulates lipogenesis, and suppresses lipolysis and gluconeogenesis (Broughton et al., 2005). Acyltransferases are critical in synthesizing and degrading various lipids, including fatty acids, glycerolipids, and phospholipids (Van der Horst, 2003; Arrese and Soulages, 2010). In addition, the AMPK is activated under energy-deprived conditions (e.g., low ATP levels) and can initiate various downstream processes that help maintain energy balance by regulating fatty acid synthesis, oxidation, and other related processes (Pan and Hardie, 2002; Hardie, 2007; Sinnett and Brenman, 2016). Changes in these pathways collectively point to an enhancement in lipid and energy metabolism in S. invicta. Neonicotinoid insecticides can bind to nAChRs located in the central nervous systems, leading to excitation, convulsions, and death of insects, accompanied by increased energy consumption (Matsuda et al., 2001; Tomizawa and Casida, 2003; Jeschke et al., 2011). It is probable that S. invicta actively enhanced its lipid metabolism to generate more energy to compensate for the increased energy expenditure after cycloxaprid exposure, thereby maintaining survival and recovery.
At 24 h, most DEGs were upregulated and enriched in pathways involved with protein digestion and absorption, pancreatic secretion, proteolysis, and amino sugar and nucleotide sugar metabolism. The upregulation of genes involved in protein digestion and absorption may suggest increased efficiency in breaking down dietary proteins and assimilating amino acids (Holtof et al., 2019). This process could be essential for maintaining energy balance because amino acids can be an energy source when required (MacFarlane, 2018). Similarly, the upregulation of genes involved in pancreatic secretion and proteolysis indicates the enhanced capacity to produce and secrete digestive enzymes, which are crucial for breaking down proteins into smaller peptides and amino acids (van Hoef et al., 2011). These results strongly correlate with findings at 12 h, indicating that S. invicta may enhance its protein and carbohydrate metabolism to obtain more energy to support the heightened energy consumption post cycloxaprid exposure.
At 48 h, most DEGs were downregulated and enriched in pathways of fatty acid metabolism, fatty acid biosynthesis, AMPK signaling, and insulin signaling. This may indicate a significant reduction in lipid and energy metabolism of S. invicta at the later stage after cycloxaprid exposure. Furthermore, the downregulation of genes involved in iron ion and heme binding suggests a decreased ability to bind and utilize iron and heme. This could impact various biological processes such as oxygen transport, energy production, and DNA synthesis (Nichol et al., 2002; Walter-Nuno et al., 2013). Upregulated pathways mainly include oxidoreductase activity, transmembrane transport, extracellular region, and cAMP signaling, implying the increased oxidative stress response that may alter membrane transport, enhance extracellular communication, and modulate intracellular signaling cascades (Kunieda et al., 2006; Holtof et al., 2019; Wu et al., 2019; Liu et al., 2021).
Interestingly, 22 DEGs were enriched in pathways related to odorant perception at 12 h. Solenopsis invicta typically detects odor molecules through olfactory receptors located on their antennae. The alterations in these genes suggest a disturbance of olfactory perception in S. invicta, which may ultimately cause reduced sensitivity to cycloxaprid. Additional researches are required in the future to examine the potential involvement of these genes in the non-repellent effect of cycloxaprid on S. invicta. We also investigated the expression of detoxification-related genes, and identified 22 CYP450 family genes and 2 GST family genes. At 12 h, LOC105207882 and LOC120359407 were upregulated, suggesting that ants may actively adjust their detoxification mechanisms to counteract the effects of cycloxaprid during early exposure. At 24 h, the detoxification response seems to weaken because only one gene (LOC105206318) was upregulated, whereas four genes (LOC105198272, LOC105203688, LOC120357488, and LOC120359317) were downregulated. This shift indicates that S. invicta may experience increased stress due to continued exposure to cycloxaprid, thereby hindering their ability to maintain effective detoxification processes. At 48 h, 18 detoxification DEGs were downregulated, and only two genes (LOC105205661 and LOC105206584) were upregulated, indicating that S. invicta may be unable to sustain its detoxification capacity after prolonged exposure, which may lead to impaired physiological function and eventually death. It is worth noting that the expression changes in the CYP450 family and GST family genes could have interconnected impacts on other physiological processes, such as lipid metabolism, energy metabolism, and immune function. For instance, at 12 h, the activation of these genes may necessitate energy, thus promoting lipid and energy metabolism. However, at 48 h, the decline in lipid and energy metabolism may contribute to the inability of these genes to maintain the function of detoxification.
Metabolomic analyses showed the diversity and functions of DMs in S. invicta following exposure to cycloxaprid over different durations. These DMs are various fatty acids, nucleotides, hormones, and amines, which participate in numerous physiological processes, including lipid and energy metabolism, DNA synthesis, hormone signaling, neurotransmission, and overall metabolic activity (Weyel and Wegener, 1996; Gäde et al., 2003; Kaczmarek and Boguś, 2021; Barbero et al., 2023). Correlation analysis showed that 226 DEGs exhibited significant correlations with 54 DMs (|R| > 0.8, p < 0.05), indicating the tight interplay between metabolites and genes in S. invicta treated with cycloxaprid. The correlation network (Figure 8) showed that the nicotinamide riboside (NR, MEDP1791) was strongly interconnected with numerous metabolites and genes. NR is a pyridine-nucleoside and a form of vitamin B3 and serves as an essential precursor to nicotinamide adenine dinucleotide (NAD+) (Bogan and Brenner, 2008), which is a cofactor for enzymes involved in energy metabolism and various metabolic pathways such as glycolysis and fatty acid oxidation (Xie et al., 2020). Therefore, NR might contribute to the adaptive responses of S. invicta to cycloxaprid, and future studies are needed to verify this.
Notably, we found that changes in fatty acids were inversely correlated with the expression levels of genes implicated in fatty acid synthesis and metabolism pathways. Transcriptomic data revealed upregulation of DEGs associated with fatty acid pathways at 12 h and downregulation at 48 h. In contrast, metabolomic analyses detected only two upregulated fatty acids at 12 h, and eight carnitines were still upregulated at 48 h. Carnitine is an endogenous compound carrier molecule in lipid metabolism and energy homeostasis (Bremer, 1983; Bieber, 1988; Pande and Murthy, 1989). The diminished lipid and energy metabolism in S. invicta may disrupt fatty acid cycling, resulting in carnitine accumulation at 48 h. Numerous studies involving humans and animals have demonstrated the beneficial effects of carnitine accumulation in treating diverse health conditions, such as arrhythmia, gastrointestinal discomfort, and epilepsy (Longo et al., 2006; Fukuda et al., 2015; Wang et al., 2018b). Concurrently, recent research suggests that the regulation of specific fatty acids might play a significant role in determining the potential of Leptinotarsa decemlineata to develop resistance to neonicotinoid insecticide (Clements et al., 2020). Therefore, the alterations in fatty acid (including carnitine) might also enhance the capacity of S. invicta to adapt to cycloxaprid. It is important to note that the present study was conducted using S. invicta workers, and future studies would be valuable to assess the impacts of cycloxaprid on different developmental stages of S. invicta.
In conclusion, our study provides a comprehensive understanding of the slow-acting and non-repellent properties of cycloxaprid against S. invicta. The slow-acting nature of cycloxaprid can be ascribed to the dynamic physiological responses observed in S. invicta post exposure, encompassing the enhancement of lipid and energy metabolism, alterations in immune function, and adjustments in detoxification mechanisms. These enhancements enable S. invicta to maintain their survival and recovery during the initial stages of exposure to cycloxaprid. However, they are insufficient to completely escape the lethal threat posed by the insecticide. As exposure duration increases, cycloxaprid progressively impairs physiological function, ultimately leading to the ants’ death. Furthermore, the non-repellent characteristic of cycloxaprid is likely associated with changes in olfactory perception-related genes, which may induce a disorder in S. invicta’s olfactory perception, resulting in reduced sensitivity to cycloxaprid. Consequently, the ants do not avoid their counterparts contaminated with cycloxaprid, thereby enhancing the effectiveness of this insecticide in controlling S. invicta at colony levels.
The original contributions presented in the study are included in the article/Supplementary Material. The datasets presented in this study can be found in online repositories. This data can be found in National Center for Biotechnology Information (NCBI) BioProject database under the accession number PRJNA1005227.
CD: Data curation, Formal Analysis, Investigation, Methodology, Software, Visualization, Writing–original draft. KJ: Data curation, Methodology, Visualization, Writing–review and editing. ZX: Methodology, Writing–review and editing. LW: Methodology, Writing–review and editing. JC: Data curation, Methodology, Project administration, Supervision, Writing–review and editing. CW: Conceptualization, Funding acquisition, Investigation, Methodology, Project administration, Supervision, Writing–review and editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Key R&D Program of China (No. 2021YFD1000500).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2023.1274416/full#supplementary-material
Angulo E., Hoffmann B. D., Ballesteros-Mejia L., Taheri A., Balzani P., Bang A., et al. (2022). Economic costs of invasive alien ants worldwide. Biol. Invasions 24 (7), 2041–2060. doi:10.1007/s10530-022-02791-w
Arrese E. L., Soulages J. L. (2010). Insect fat body: energy, metabolism, and regulation. Annu. Rev. Entomol. 55, 207–225. doi:10.1146/annurev-ento-112408-085356
Ascunce M. S., Yang C.-C., Oakey J., Calcaterra L., Wu W.-J., Shih C.-J., et al. (2011). Global invasion history of the fire ant Solenopsis invicta. Science 331 (6020), 1066–1068. doi:10.1126/science.1198734
Barbero F., Mannino G., Casacci L. P. (2023). The role of biogenic amines in social insects: with a special focus on ants. Insects 14, 386. doi:10.3390/insects14040386
Bernaba M., Power E., Campion J., Gotzek D., Schmidt J. O., Klotz S. A. (2019). Unconscious woman in shock and covered with ants pulled from an abandoned automobile. Am. J. Med. 132 (10), 1239–1241. doi:10.1016/j.amjmed.2019.02.053
Bieber L. L. (1988). Carnitine. Annu. Rev. Biochem. 57 (1), 261–283. doi:10.1146/annurev.bi.57.070188.001401
Bogan K. L., Brenner C. (2008). Nicotinic acid, nicotinamide, and nicotinamide riboside: a molecular evaluation of NAD+ precursor vitamins in human nutrition. Annu. Rev. Nutr. 28, 115–130. doi:10.1146/annurev.nutr.28.061807.155443
Bremer J. (1983). Carnitine--metabolism and functions. Physiol. Rev. 63 (4), 1420–1480. doi:10.1152/physrev.1983.63.4.1420
Broughton S. J., Piper M. D. W., Ikeya T., Bass T. M., Jacobson J., Driege Y., et al. (2005). Longer lifespan, altered metabolism, and stress resistance in Drosophila from ablation of cells making insulin-like ligands. P. Natl. Acad. Sci. U. S. A. 102 (8), 3105–3110. doi:10.1073/pnas.0405775102
Buchfink B., Xie C., Huson D. (2015). Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60. doi:10.1038/nmeth.3176
Canavoso L. E., Jouni Z. E., Karnas K. J., Pennington J. E., Wells M. A. (2001). Fat metabolism in insects. Annu. Rev. Nutr. 21 (1), 23–46. doi:10.1146/annurev.nutr.21.1.23
Chan K. H., Guénard B. (2020). Ecological and socio-economic impacts of the red import fire ant, Solenopsis invicta (Hymenoptera: Formicidae), on urban agricultural ecosystems. Urban Ecosyst. 23, 1–12. doi:10.1007/s11252-019-00893-3
Chen J. (2007). Advancement on techniques for the separation and maintenance of the red imported fire ant colonies. Insect Sci. 14 (1), 1–4. doi:10.1111/j.1744-7917.2007.00120.x
Chen J., Oi D. H. (2020). Naturally occurring compounds/materials as alternatives to synthetic chemical insecticides for use in fire ant management. Insects 11, 758. doi:10.3390/insects11110758
Chen J., Zhou Y., Lei Y., Shi Q., Qi G., He Y., et al. (2022). Role of the foraging gene in worker behavioral transition in the red imported fire ant, Solenopsis invicta (Hymenoptera: Formicidae). Pest Manag. Sci. 78 (7), 2964–2975. doi:10.1002/ps.6921
Chen S., Zhou Y., Chen Y., Gu J. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi:10.1093/bioinformatics/bty560
Chen W., Gong L., Guo Z., Wang W., Zhang H., Liu X., et al. (2013). A novel integrated method for large-scale detection, identification, and quantification of widely targeted metabolites: application in the study of rice metabolomics. Mol. Plant 6 (6), 1769–1780. doi:10.1093/mp/sst080
Clements J., Olson J. M., Sanchez-Sedillo B., Bradford B., Groves R. L. (2020). Changes in emergence phenology, fatty acid composition, and xenobiotic-metabolizing enzyme expression is associated with increased insecticide resistance in the Colorado potato beetle. Arch. Insect Biochem. 103 (3), e21630. doi:10.1002/arch.21630
Cui L., Qi H., Yang D., Yuan H., Rui C. (2016). Cycloxaprid: a novel cis-nitromethylene neonicotinoid insecticide to control imidacloprid-resistant cotton aphid (Aphis gossypii). Pestic. Biochem. Phys. 132, 96–101. doi:10.1016/j.pestbp.2016.02.005
Cui L., Sun L., Yang D., Yan X., Yuan H. (2012). Effects of cycloxaprid, a novel cis-nitromethylene neonicotinoid insecticide, on the feeding behaviour of Sitobion avenae. Pest Manag. Sci. 68, 1484–1491. doi:10.1002/ps.3333
Cui L., Wang Q., Wang Q., Wang L., Yuan H., Rui C. (2020). Cycloxaprid: a novel cis-nitromethylene neonicotinoid insecticide to control Bemisia tabaci. Pest Manag. Sci. 76 (5), 1705–1712. doi:10.1002/ps.5693
Cui L., Yuan H., Wang Q., Wang Q., Rui C. (2018). Sublethal effects of the novel cis-nitromethylene neonicotinoid cycloxaprid on the cotton aphid Aphis gossypii Glover (Hemiptera: aphididae). Sci. Rep. 8, 8915. doi:10.1038/s41598-018-27035-7
Dong W., Yang H., Wang C., Li H., Shang J., Chen Z., et al. (2022). Cross-resistance and fitness costs of the cis-nitromethylene neonicotinoid cycloxaprid resistance in melon aphid, Aphis gossypii (Hemiptera: aphididae). J. Econ. Entomol. 115 (5), 1668–1675. doi:10.1093/jee/toac112
Drees B. M., Gold R. E. (2003). Development of integrated pest management programs for the red imported fire ant (Hymenoptera: Formicidae). J. Entomol. Sci. 38 (2), 170–180. doi:10.18474/0749-8004-38.2.170
Fukuda M., Kawabe M., Takehara M., Iwano S., Kuwabara K., Kikuchi C., et al. (2015). Carnitine deficiency: risk factors and incidence in children with epilepsy. Brain Dev. 37 (8), 790–796. doi:10.1016/j.braindev.2014.12.004
Gäde G., Goldsworthy G. J., Gäde G., Goldsworthy G. J., Gäde G., Goldsworthy G. J. (2003). Insect peptide hormones: a selective review of their physiology and potential application for pest control. Pest Manag. Sci. 59, 1063–1075. doi:10.1002/ps.755
Géminard C., Rulifson E. J., Léopold P. (2009). Remote control of insulin secretion by fat cells in Drosophila. Cell Metab. 10 (3), 199–207. doi:10.1016/j.cmet.2009.08.002
Gordon D. M. (2019). The ecology of collective behavior in ants. Annu. Rev. Entomol. 64, 35–50. doi:10.1146/annurev-ento-011118-111923
Gutrich J. J., VanGelder E., Loope L. (2007). Potential economic impact of introduction and spread of the red imported fire ant, Solenopsis invicta, in Hawaii. Environ. Sci. Policy 10 (7-8), 685–696. doi:10.1016/j.envsci.2007.03.007
Haddad Junior V., Larsson C. E. (2015). Anaphylaxis caused by stings from the Solenopsis invicta, lava-pés ant or red imported fire ant. An. Bras. Dermatol. 90, 22–25. doi:10.1590/abd1806-4841.20153420
Hardie D. G. (2007). AMP-activated protein kinase as a drug target. Annu. Rev. Pharmacol. 47, 185–210. doi:10.1146/annurev.pharmtox.47.120505.105304
Haynes W. (2013). “Benjamini–hochberg method,” in Encyclopedia of systems biology. Editors W. Dubitzky, O. Wolkenhauer, K. H. Cho, and H. Yokota (New York, NY: Springer). doi:10.1007/978-1-4419-9863-7_1215
Holtof M., Lenaerts C., Cullen D., Vanden Broeck J. (2019). Extracellular nutrient digestion and absorption in the insect gut. Cell Tissue Res. 377, 397–414. doi:10.1007/s00441-019-03031-9
Jeschke P., Nauen R., Schindler and M., Elbert A. (2011). Overview of the status and global strategy for neonicotinoids. J. Agr. Food Chem. 59 (7), 2897–2908. doi:10.1021/jf101303g
Jin J.-X., Jin D.-C., Li F.-L., Cheng Y., Li W.-H., Ye Z.-C., et al. (2017). Expression differences of resistance-related genes induced by cycloxaprid using qRT-PCR in the female adult of Sogatella furcifera (Hemiptera: Delphacidae). J. Econ. Entomol. 110 (4), 1785–1793. doi:10.1093/jee/tox155
Jin J.-X., Ye Z.-C., Jin D.-C., Li F.-L., Li W.-H., Cheng Y., et al. (2021). Changes in transcriptome and gene expression in Sogatella furcifera (Hemiptera: Delphacidae) in response to cycloxaprid. J. Econ. Entomol. 114 (1), 284–297. doi:10.1093/jee/toaa238
Kaczmarek A., Boguś M. (2021). The metabolism and role of free fatty acids in key physiological processes in insects of medical, veterinary and forensic importance. PeerJ 9, e12563. doi:10.7717/peerj.12563
Kim D., Langmead B., Salzberg S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12 (4), 357–360. doi:10.1038/nmeth.3317
Kohlmeier P., Negroni M. A., Kever M., Emmling S., Stypa H., Feldmeyer B., et al. (2017). Intrinsic worker mortality depends on behavioral caste and the queens’ presence in a social insect. Sci. Nat. 104, 34. doi:10.1007/s00114-017-1452-x
Kunieda T., Fujiyuki T., Kucharski R., Foret S., Ament S. A., Toth A. L., et al. (2006). Carbohydrate metabolism genes and pathways in insects: insights from the honey bee genome. Insect Mol. Biol. 15 (5), 563–576. doi:10.1111/j.1365-2583.2006.00677.x
Liao Y., Smyth G. K., Shi W. (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30 (7), 923–930. doi:10.1093/bioinformatics/btt656
Liu N., Wang Y., Li T., Feng X. (2021). G-protein coupled receptors (GPCRs): signaling pathways, characterization, and functions in insect physiology and toxicology. Int. J. Mol. Sci. 22 (10), 5260. doi:10.3390/ijms22105260
Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 25 (4), 402–408. doi:10.1006/meth.2001.1262
Longo N., Amat di San Filippo C., Pasquali M. (2006). Disorders of carnitine transport and the carnitine cycle. Am. J. Med. Genet. 142C (2), 77–85. doi:10.1002/ajmg.c.30087
Love M. I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15 (12), 550. doi:10.1186/s13059-014-0550-8
MacFarlane N. G. (2018). Digestion and absorption. Anaesth. Intensive Care Med. 19 (3), 125–127. doi:10.1016/j.mpaic.2018.01.001
Matsuda K., Buckingham S. D., Kleier D., Rauh J. J., Grauso M., Sattelle D. B. (2001). Neonicotinoids: insecticides acting on insect nicotinic acetylcholine receptors. Trends Pharmacol. Sci. 22 (11), 573–580. doi:10.1016/S0165-6147(00)01820-4
Morrison L. W. (2002). Long-term impacts of an arthropod-community invasion by the imported fire ant, Solenopsis invicta. Ecology 83, 2337–2345. doi:10.1890/0012-9658(2002)083[2337:ltioaa]2.0.co;2
Nässel D. R., Broeck J. V. (2016). Insulin/IGF signaling in Drosophila and other insects: factors that regulate production, release and post-release action of the insulin-like peptides. Cell Mol. Life Sci. 73, 271–290. doi:10.1007/s00018-015-2063-3
Negroni M. A., Macit M. N., Stoldt M., Feldmeyer B., Foitzik S. (2021). Molecular regulation of lifespan extension in fertile ant workers. Phil. Trans. R. Soc. B 376, 20190736. doi:10.1098/rstb.2019.0736
Nichol H., Law J. H., Winzerling J. J. (2002). Iron metabolism in insects. Annu. Rev. Entomol. 47 (1), 535–559. doi:10.1146/annurev.ento.47.091201.145237
Ning D., Yang F., Xiao Q., Ran H., Xu Y. (2019). A simple and efficient method for preventing ant escape (Hymenoptera: Formicidae). Myrmecol. News 29, 57–65. doi:10.25849/myrmecol.news_029:057
Pan D. A., Hardie D. G. (2002). A homologue of AMP-activated protein kinase in Drosophila melanogaster is sensitive to AMP and is activated by ATP depletion. Biochem. J. 367 (1), 179–186. doi:10.1042/bj20020703
Pande S. V., Murthy M. S. R. (1989). Carnitine: vitamin for an insect, vital for man. Biochem. Cell Biol. 67 (10), 671–673. doi:10.1139/o89-100
Pereira R. M., Williams D. F., Becnel J. J., Oi D. H. (2002). Yellow-head disease caused by a newly discovered Mattesia sp. in populations of the red imported fire ant, Solenopsis invicta. J. Invertebr. Pathol. 81, 45–48. doi:10.1016/S0022-2011(02)00116-7
Pertea M., Kim D., Pertea G. M., Leek J. T., Salzberg S. L. (2016). Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 11 (9), 1650–1667. doi:10.1038/nprot.2016.095
Robinson G. E. (1992). Regulation of division of labor in insect societies. Annu. Rev. Entomol. 37 (1), 637–665. doi:10.1146/annurev.en.37.010192.003225
Robinson M. D., McCarthy D. J., Smyth G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26 (1), 139–140. doi:10.1093/bioinformatics/btp616
Rust M. K., Su N.-Y. (2012). Managing social insects of urban importance. Annu. Rev. Entomol. 57, 355–375. doi:10.1146/annurev-ento-120710-100634
Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13 (11), 2498–2504. doi:10.1101/gr.1239303
Shao X., Liu Z., Xu X., Li Z., Qian X. (2013a). Overall status of neonicotinoid insecticides in China: production, application and innovation. J. Pestic. Sci. 38, 1–9. doi:10.1584/jpestics.D12-037
Shao X., Swenson T. L., Casida J. E. (2013b). Cycloxaprid insecticide: nicotinic acetylcholine receptor binding site and metabolism. J. Agric. Food Chem. 61, 7883–7888. doi:10.1021/jf4030695
Siddiqui J. A., Luo Y., Sheikh U. A. A., Bamisile B. S., Khan M. M., Imran M., et al. (2022a). Transcriptome analysis reveals differential effects of beta-cypermethrin and fipronil insecticides on detoxification mechanisms in Solenopsis invicta. Front. Physiol. 13, 1018731. doi:10.3389/fphys.2022.1018731
Siddiqui J. A., Zhang Y., Luo Y., Bamisile B. S., Rehman N. U., Islam W., et al. (2022b). Comprehensive detoxification mechanism assessment of red Imported fire ant (Solenopsis invicta) against Indoxacarb. Molecules 27, 870. doi:10.3390/molecules27030870
Silverman J., Brightwell R. J. (2008). The argentine ant: challenges in managing an invasive unicolonial pest. Annu. Rev. Entomol. 53, 231–252. doi:10.1146/annurev.ento.53.103106.093450
Sinnett S. E., Brenman J. E. (2016). The role of AMPK in Drosophila melanogaster. AMP-Activated protein kinase 107, 389–401. doi:10.1007/978-3-319-43589-3_16
Tomizawa M., Casida J. E. (2003). Selective toxicity of neonicotinoids attributable to specificity of insect and mammalian nicotinic receptors. Annu. Rev. Entomol. 48 (1), 339–364. doi:10.1146/annurev.ento.48.091801.112731
Van der Horst D. J. (2003). Insect adipokinetic hormones: release and integration of flight energy metabolism. Comp. Biochem. Physiology Part B 136 (2), 217–226. doi:10.1016/S1096-4959(03)00151-9
Van Hoef V., Breugelmans B., Spit J., Simonet G., Zels S., Billen J., et al. (2011). Functional analysis of a pancreatic secretory trypsin inhibitor-like protein in insects: silencing effects resemble the human pancreatic autodigestion phenotype. Insect biochem. molec. 41 (9), 688–695. doi:10.1016/j.ibmb.2011.04.012
Vinson S. B. (2013). Impact of the invasion of the imported fire ant. Insect Sci. 20 (4), 439–455. doi:10.1111/j.1744-7917.2012.01572.x
Walter-Nuno A. B., Oliveira M. P., Oliveira M. F., Gonçalves R. L., Ramos I. B., Koerich L. B., et al. (2013). Silencing of maternal heme-binding protein causes embryonic mitochondrial dysfunction and impairs embryogenesis in the blood sucking insect Rhodnius prolixus. J. Biol. Chem. 288 (41), 29323–29332. doi:10.1074/jbc.M113.504985
Wang L., Zeng L., Xu Y., Lu Y. (2020). Prevalence and management of Solenopsis invicta in China. Neobiota 54, 89–124. doi:10.3897/neobiota.54.38584
Wang R., Fang Y., Mu C., Qu C., Li F., Wang Z., et al. (2018a). Baseline susceptibility and cross-resistance of cycloxaprid, a novel cis-nitromethylene neonicotinoid insecticide, in Bemisia tabaci MED from China. Crop Prot. 110, 283–287. doi:10.1016/j.cropro.2017.02.012
Wang R., Liu Y.-Y., Liu G.-H., Lu H.-B., Mao C.-Y. (2018b). l-Carnitine and heart disease. Life Sci. 194, 88–97. doi:10.1016/j.lfs.2017.12.015
Wen C., Shen L., Chen J., Zhang J., Feng Y., Wang Z., et al. (2022). Red imported fire ants cover the insecticide-treated surfaces with particles to reduce contact toxicity. J. Pest Sci. 95 (3), 1135–1150. doi:10.1007/s10340-021-01474-0
Wetterer J. K. (2013). Exotic spread of Solenopsis invicta buren (hymenoptera: Formicidae) beyond north America. Sociobiology 60 (1), 50–55. doi:10.13102/sociobiology.v60i1.50-55
Weyel W., Wegener G. (1996). Adenine nucleotide metabolism during anoxia and postanoxic recovery in insects. Experientia 52, 474–480. doi:10.1007/BF01919319
Wu C., Chakrabarty S., Jin M., Liu K., Xiao Y. (2019). Insect ATP-binding cassette (ABC) transporters: roles in xenobiotic detoxification and bt insecticidal activity. Int. J. Mol. Sci. 20 (11), 2829. doi:10.3390/ijms20112829
Wu Q., Brown M. R. (2006). Signaling and function of insulin-like peptides in insects. Annu. Rev. Entomol. 51, 1–24. doi:10.1146/annurev.ento.51.110104.151011
Wylie F. R., Janssen-May S. (2017). Red imported fire ant in Australia: what if we lose the war? Ecol. Manag. Restor. 18, 32–44. doi:10.1111/emr.12238
Xie N., Zhang L., Gao W., Huang C., Huber P. E., Zhou X., et al. (2020). NAD+ metabolism: pathophysiologic mechanisms and therapeutic potential. Sig. Transduct. Target. Ther. 5, 227–237. doi:10.1038/s41392-020-00311-7
Xiong T., Ling S., Liu J., Zeng X. (2022). Insecticidal and P450 mediate metabolism of fluralaner against red imported fire ant, Solenopsis invicta (Hymenoptera: Formicidae). Pestic. Biochem. Physiol. 187, 105184. doi:10.1016/j.pestbp.2022.105184
Yang Y., Zhang Y., Yang B., Fang J., Liu Z. (2016). Transcriptomic responses to different doses of cycloxaprid involved in detoxification and stress response in the whitebacked planthopper, Sogatella furcifera. Entomol. Exp. Appl. 158 (3), 248–257. doi:10.1111/eea.12406
Zhang L., Wang L., Chen J., Zhang J., He Y., Lu Y., et al. (2022). Toxicity, horizontal transfer, and physiological and behavioral effects of cycloxaprid against Solenopsis invicta (Hymenoptera: Formicidae). Pest Manag. Sci. 78 (6), 2228–2239. doi:10.1002/ps.6847
Zhang Y., Han Y., Yang Q., Wang L., He P., Liu Z., et al. (2018). Resistance to cycloxaprid in Laodelphax striatellus is associated with altered expression of nicotinic acetylcholine receptor subunits. Pest Manag. Sci. 74 (4), 837–843. doi:10.1002/ps.4757
Keywords: eusocial insect, formicidae, red imported fire ant, neonicotinoid insecticide, cycloxaprid
Citation: Du C, Jiang K, Xu Z, Wang L, Chen J and Wang C (2023) Transcriptome and metabolome comprehensive analysis reveal the molecular basis of slow-action and non-repellency of cycloxaprid against an eusocial pest, Solenopsis invicta. Front. Physiol. 14:1274416. doi: 10.3389/fphys.2023.1274416
Received: 01 September 2023; Accepted: 13 November 2023;
Published: 27 November 2023.
Edited by:
Natraj Krishnan, Mississippi State University, United StatesReviewed by:
Yifan Zhai, Shandong Academy of Agricultural Sciences, ChinaCopyright © 2023 Du, Jiang, Xu, Wang, Chen and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jie Chen, Y2hlbmpAZ2RwcHJpLmNvbQ==; Cai Wang, d2FuZ2NhaUBzY2F1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.