- 1Department of Molecular and Cell Biology, University of Connecticut, Storrs, CT, United States
- 2Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA, United States
Most mitochondrial proteins are targeted to the organelle by N-terminal mitochondrial targeting sequences (MTSs, or “presequences”) that are recognized by the import machinery and subsequently cleaved to yield the mature protein. MTSs do not have conserved amino acid compositions, but share common physicochemical properties, including the ability to form amphipathic α-helical structures enriched with basic and hydrophobic residues on alternating faces. The lack of strict sequence conservation implies that some polypeptides can be mistargeted to mitochondria, especially under cellular stress. The pathogenic accumulation of proteins within mitochondria is implicated in many aging-related neurodegenerative diseases, including Alzheimer’s, Parkinson’s, and Huntington’s diseases. Mechanistically, these diseases may originate in part from mitochondrial interactions with amyloid-β precursor protein (APP) or its cleavage product amyloid-β (Aβ), α-synuclein (α-syn), and mutant forms of huntingtin (mHtt), respectively, that are mediated in part through their associations with the mitochondrial protein import machinery. Emerging evidence suggests that these amyloidogenic proteins may present cryptic targeting signals that act as MTS mimetics and can be recognized by mitochondrial import receptors and transported into different mitochondrial compartments. Accumulation of these mistargeted proteins could overwhelm the import machinery and its associated quality control mechanisms, thereby contributing to neurological disease progression. Alternatively, the uptake of amyloidogenic proteins into mitochondria may be part of a protein quality control mechanism for clearance of cytotoxic proteins. Here we review the pathomechanisms of these diseases as they relate to mitochondrial protein import and effects on mitochondrial function, what features of APP/Aβ, α-syn and mHtt make them suitable substrates for the import machinery, and how this information can be leveraged for the development of therapeutic interventions.
1 Introduction
Alzheimer’s Disease (AD), Parkinson’s Disease (PD) and Huntington’s Disease (HD) are distinct neurodegenerative disorders that involve the progressive loss of neuronal structure and function. They are collectively classified as proteopathies, which involve severe disruption in cellular protein homeostasis (proteostasis) associated with protein misfolding as well as disruptions in protein processing and localization (Chowhan et al., 2015). Although the mechanistic causes of these diseases are incompletely understood, each is associated with the pathogenic accumulation of specific proteins. Amyloid-β (Aβ), the proteolytic product of amyloid-β precursor protein (APP), is associated with AD (Rocchi et al., 2003; Tcw and Goate, 2017); α-synuclein (α-syn) is associated with PD and other synucleinopathies (Stefanis, 2012); and expanded polyglutamine (poly-Q) repeats underpin a range of neurodegenerative disorders, including HD which is caused by mutant forms of huntingtin (mHtt) (Li et al., 1993; Schilling et al., 1995). Elucidating the mechanisms by which these proteins precipitate their respective pathogenic cascades will be essential in developing new treatments for their associated neurodegenerative diseases.
As protein deposition diseases, AD, PD and HD are associated with the conversion of soluble monomers or oligomers into highly organized insoluble fibrillar aggregates, called amyloid fibrils, that serve as their primary histopathological markers. Each of these neurodegenerative diseases is associated with a specific composition of protein deposits that target certain neuronal subpopulations within the central nervous system (CNS). For decades the dominant and unifying model to explain the etiology of these cerebral proteopathies has focused on aggregates of amyloid fibrils as the causative agents (Hardy and Higgins, 1992). However, in recent years this model has been challenged on two main fronts. First, there is accumulating evidence that monomers or small oligomers of Aβ, α-syn, and mHtt, rather than the large fibrils themselves, may in fact be the cytotoxic species; and second, the extent of fibrillization is not necessarily correlated with disease progression (Uddin et al., 2020; Wells et al., 2021).
These changes in perspective have been accompanied by an increasing recognition that mitochondrial dysfunction plays a key role in neurodegenerative diseases (Swerdlow et al., 2014). More specifically, APP/Aβ, α-syn, and mHtt have all been shown to negatively impact different mitochondrial functions and to accumulate within different mitochondrial subcompartments. Such observations raise the intriguing mechanistic question of how these amyloids target to mitochondria, given that mitochondria contain a network of machineries that are ostensibly designed to selectively import only polypeptides with defined functions in the organelle. Here we review emerging evidence that APP/Aβ, α-syn, and mHtt may contain “cryptic” sequences that mimic the classic N-terminal targeting information of mitochondrial proteins to interact with mitochondrial import complexes, the implications this may have for the role of mitochondrial dysfunction in neurodegeneration, and how such insights could inform the development of novel therapeutic strategies for AD, PD, and HD.
2 Mitochondria of the central nervous system
The CNS is composed of neurons and glial cells (Figure 1A). Neurons are morphologically complex cells that transmit information by receiving a stimulus at dendrites that is transferred to the cell body and then propagated as an action potential (an electrochemical impulse) along the axon. Glial cells provide physical and metabolic support to neurons and include several subtypes including astrocytes, which serve mainly to support neural function and signaling; oligodendrocytes, which form myelin sheaths around axons; and microglia, the immune cells of the CNS. Neurodegenerative disorders predominantly affect neurons in particular anatomical regions of the brain (Dugger and Dickson, 2017; Fu et al., 2018); however, glial cells are purported to also play a direct role in the pathomechanisms of these diseases (Gleichman and Carmichael, 2020).
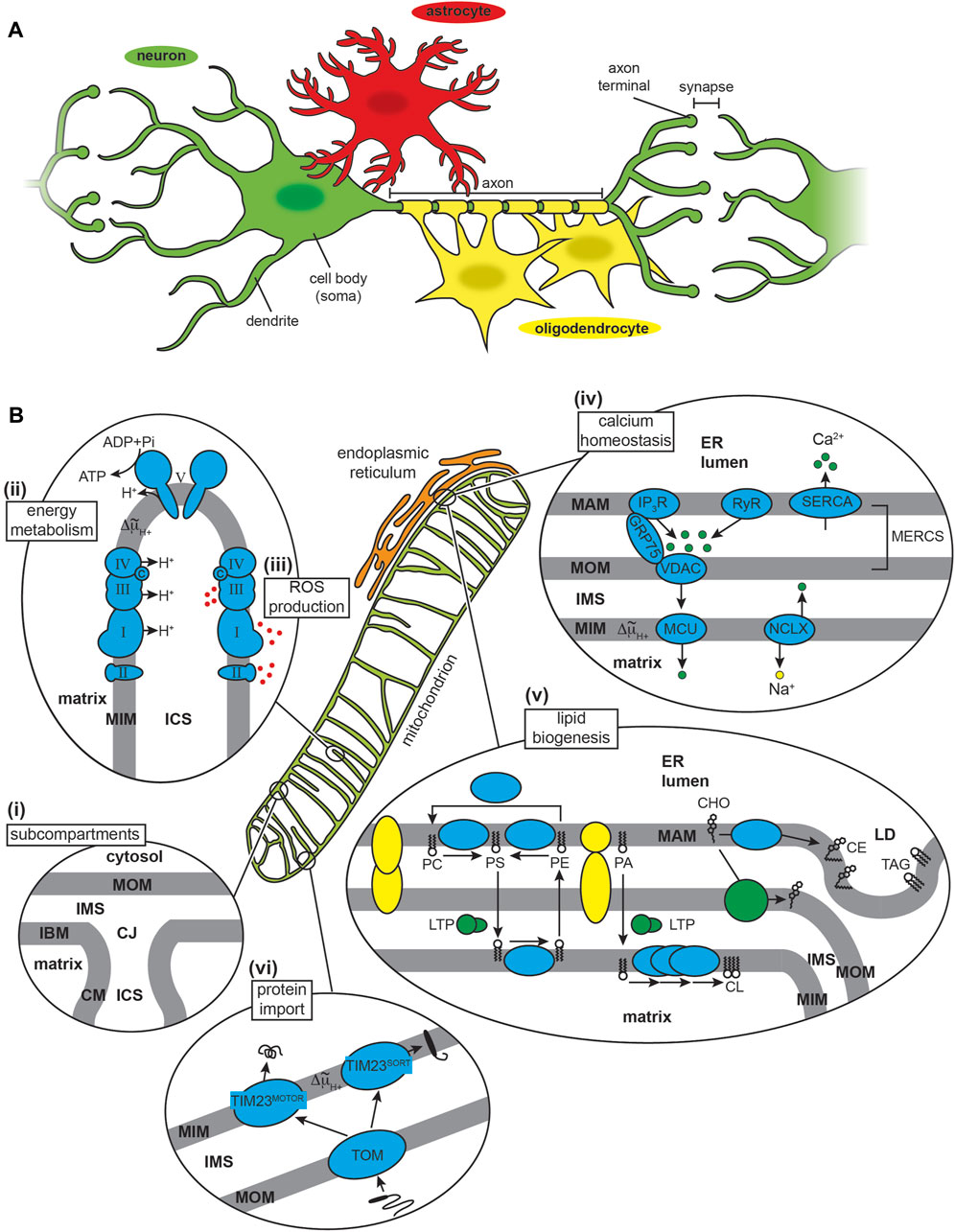
FIGURE 1. The CNS and mitochondria. (A) CNS cell types. The neuron (nerve cell, green) typically consists of a cell body (soma), multiple branching dendrites (afferent processes) that receive signals and transmit them to the cell body, and a single axon (efferent process) that forms an extended cable-like structure ending with axon terminals. Glial cells include astrocytes (red) and oligodendrocytes (yellow), amongst other cell types. (B) Mitochondria structure and major functions. (i) Subcompartments. Mitochondria have a two-membrane organization with a mitochondrial outer membrane (MOM) and inner membrane (MIM), the latter subdivided into an inner boundary membrane (IBM) closely appressed to the MOM, and the cristae membrane (CM). These membranes delineate the innermost matrix compartment from the intermembrane space (IMS) and intracristal space (ICS) that connect at the functional boundary of the crista junction (CJ). (ii) Energy metabolism. The OXPHOS machinery is parsed into complexes that generate an electrochemical proton potential (Δ∼μH+) across the CM (including respiratory complexes I, II, III and IV of the electron transport chain) and the F1FO ATP synthase (complex V) that uses the energy of the proton gradient to drive ATP synthesis. (iii) ROS production. Mitochondrial complexes I, II and III generate superoxide (O2.−) from the one-electron reduction of O2, which can subsequently be catalytically dismutated to H2O2 (a potent signaling molecule) or converted to the cytotoxic hydroxyl radical (OH·). (iv) Calcium homeostasis. The sarco/endoplasmic reticulum acts as a Ca2+ repository based largely on Ca2+ influx from ATP-dependent SERCA pumps and Ca2+ efflux from channels including the inositol (1,4,5)-triphosphate (IP3R) receptor (tethered to the MOM by GRP75) and ryanodine receptor (RyR). Mitochondrial Ca2+ uptake across the MOM occurs through the voltage-dependent anion channel (VDAC) and across the MIM by the mitochondrial calcium uniporter (MCU), with efflux occurring primarily by the Na+/Ca2+ exchanger (NCLX). (v) Lipid biogenesis and trafficking. Lipid biosynthesis complexes (cyan) and lipid transport proteins (LTPs) mediate the production of phospholipids (PC, phosphatidylcholine; PS, phosphatidylserine; PE, phosphatidylethanolamine; PA, phosphatidic acid; and CL, cardiolipin) and cholesterol (CHO). The MAM also mediates formation of lipid droplet (LD), enriched in cholesterol esters (CE) and triacylglycerol (TAG). MAM-MOM tethering complexes are depicted in yellow. (vi) Protein import. Most mitochondrial proteins are nuclear encoded and imported from the cytosol. Nearly all proteins enter mitochondria through the TOM complex, and most are imported to the final destination by modular assemblies of the TIM complex (TIM23MOTOR and TIM23SORT).
Mitochondria are morphologically complex, bound by a mitochondrial outer (MOM) and inner (MIM) membrane that enclose the intermembrane space (IMS), intracristal space (ICS) and matrix aqueous compartments (Figure 1Bi). Mitochondria assume a particularly important role in the physiology of neurons and neuroglia, given the high metabolic and signaling activity of these cells (Kann and Kovacs, 2007). First, mitochondria of the CNS must ensure efficient energy metabolism (Figure 1Bii). The brain comprises only 2%–3% of the human body mass yet accounts for up to 20% of total energy expenditure (Rolfe and Brown, 1997). This high energy demand is due to processes that include the maintenance of ion gradients such as Na+/K+-ATPases and Ca2+-ATPases for neuronal excitability, as well as the synthesis, packaging, and cycling of neurotransmitters (Attwell and Laughlin, 2001). To meet this demand, most neuronal ATP is generated by mitochondrial oxidative phosphorylation (OXPHOS) fueled by glycolysis-derived pyruvate, although some glycolysis-derived ATP is utilized directly at nerve terminals during stress to sustain synaptic transmission (Tsacopoulos and Magistretti, 1996; Murali Mahadevan et al., 2021). The astrocyte-neuron lactatae shuttle model proposes that lactate produced by astrocytes is subsequently taken up by surrounding neurons to support high OXPHOS activity; however, the accuracy and relevance of this model has been questioned (Dienel, 2017). Second, mitochondria are the primary source of reactive oxygen species (ROS) in cells (Figure 1Biii), most of which are generated from the partial reduction of dioxygen (O2) by different enzyme complexes of the electron transport chian (ETC) (Halliwell, 1992). As in other tissues, ROS serve dual roles in CNS cells. On one hand, ROS are critical for signaling processes required for neuronal plasticity and network tuning, and on the other, mitochondrial dysfunction can lead to ROS overproduction and oxidative damage, which is positively associated with neurodegenerative disorders (Milton and Sweeney, 2012; Oswald et al., 2018). Other central functions of mitochondria involve their interactions with the endoplasmic reticulum (ER), specifically at specialized regions of the ER called the mitochondrial-associated membrane (MAM) that is tethered to the MOM at sites termed mitochondria-ER contact sites (MERCSs) (Aoyama-Ishiwatari and Hirabayashi, 2021; Sassano et al., 2022). One function of MERCSs is the regulation of Ca2+ homeostasis (Figure 1Biv). The ER serves as the primary Ca2+ storage organelle, balancing ion uptake by the sarco/endoplasmic reticulum Ca2+-ATPase (SERCA) pumps with transient release from channels that include the inositol 1,4,5-triphosphate receptor (IP3R) and the ryanodine receptor (RyR). Mitochondria serve as temporary stores of cellular Ca2+ (e.g., during Ca2+ transients that occur with action potentials in neurons), taking up ions through the β-barrel voltage-dependent anion channel (VDAC) in the MOM and the mitochondrial calcium uniporter (MCU) of the MIM. Ca2+ dyshomeostasis is a central feature of neurodegenerative diseases (Kolobkova et al., 2017; McDaid et al., 2020; Xu et al., 2022a). MERCSs also regulate lipid biosynthesis (Figure 1Bv) by serving as platforms for the non-vesicular trafficking of phospholipids and cholesterol and lipid droplet formation (Giordano, 2018; Benador et al., 2019). Defects in lipid metabolism are also a central feature of neurodegeneration, much of it attributable to alterations at the MAM-mitochondria interface (Block et al., 2010; Alecu and Bennett, 2019; Yin, 2023). Finally, mitochondria contain protein import machinery for the biogenesis of nuclear-encoded proteins (Figure 1Bvi). The role of the import machinery in mediating mitochondrial interactions with amyloidogenic proteins is the focus of this review.
Mitochondria are highly dynamic organelles, constantly undergoing growth, fission into fragments balanced by fusion into interconnected networks, and selective degradation of dysfunctional organelles by mitophagy (Yapa et al., 2021). Furthermore, neuronal mitochondria are distributed to match the local metabolic and signaling requirements of the somatic, dendritic, axonal, and synaptic regions, a process governed by anterograde and retrograde trafficking (Mandal and Drerup, 2019). Mitochondrial biogenesis requires the regulated import of proteins into the organelle to accommodate growth and replacement of damaged proteins to maintain an adequate population of healthy mitochondria.
Mitochondria also have a specialized lipid composition (Claypool and Koehler, 2012; Poulaki and Giannouli, 2022). The glycerophospholipid cardiolipin is of particular relevance because it is unique to mitochondria. Cardiolipin has an unusual structure, with a two-phosphate headgroup that imparts a strong negative charge to the membrane surface and four acyl tails, creating a molecular geometry that affects lipid packing and stabilizes local membrane curvature (Ikon and Ryan, 2017). Cardiolipin is primarily localized to the MIM, where it accounts for approximately 20 mol% of total phospholipid content, and is less abundant in the MOM, where it makes up less than 5 mol% of phospholipids. Externalization of cardiolipin from the MIM to the MOM can occur with cellular stress, which can serve as a signal for selective mitochondrial autophagy (mitophagy) or programmed cell death (apoptosis) (Li et al., 2015).
A key question surrounding neurodegeneration is why neurons are particularly vulnerable to proteostatic imbalance. The answer is manifold: because neurons are terminally differentiated and non-proliferative cells, they cannot rely on asymmetric mitosis to purge aggregated proteins and must therefore rely on robust proteostatic quality control machinery that can fail with age (Heydari et al., 1994; Conconi et al., 1996; Taylor and Dillin, 2011); because neurons are structurally polarized with long processes, the clearance of protein aggregates from distal parts of the cell is energetically costly and prone to dysregulation (Guo et al., 2020); and because neurons have such specialized functional and metabolic demands (e.g., maintenance of ion gradients, calcium regulation, and neurotransmitter cycling), there is a small energetic margin of error to spare in the face of proteostatic stress, particularly with age-related decreases in energy metabolism (Blaszczyk, 2020).
3 Mitochondrial protein import and the proteostatic network
3.1 The mitochondrial protein import and quality control machinery
The biogenesis and steady-state function of mitochondria require a highly regulated system of protein import, sorting, assembly, and quality control (Figure 2; Supplementary Table S1). The human mitochondrial proteome consists of approximately 1200–1500 individual proteins (Morgenstern et al., 2021; Rath et al., 2021). Being semi-autonomous organelles, mitochondria have the genome (mitochondrial DNA) and the biosynthetic machinery (e.g., mitochondrial ribosomes, RNA/DNA polymerases and tRNAs) to synthesize a handful of their resident proteins, which in humans includes 13 subunits of OXPHOS complexes I, III, IV and V. All other mitochondrial proteins are encoded in nuclear DNA, synthesized on cytosolic ribosomes, and subsequently imported into mitochondria. Multiple pathways exist for the targeting and sorting of nuclear-encoded proteins to the proper mitochondrial membrane or aqueous subcompartment (Busch et al., 2023) (Figure 2A). These proteins are synthesized with mitochondria targeting information encoded in the polypeptide sequence itself, which must be recognized by the dedicated import complexes that direct them to their correct destinations. The majority (about two-thirds) of mitochondria-targeted proteins are imported via the translocase of the mitochondrial inner membrane 23 (TIM23) pathway, which mediates the translocation of soluble proteins into the matrix as well as the integration of membrane proteins into the MIM (Sinha et al., 2014). In this section, we summarize our current understanding of TIM23-based protein biogenesis and the associated mitochondrial proteostasis machinery.
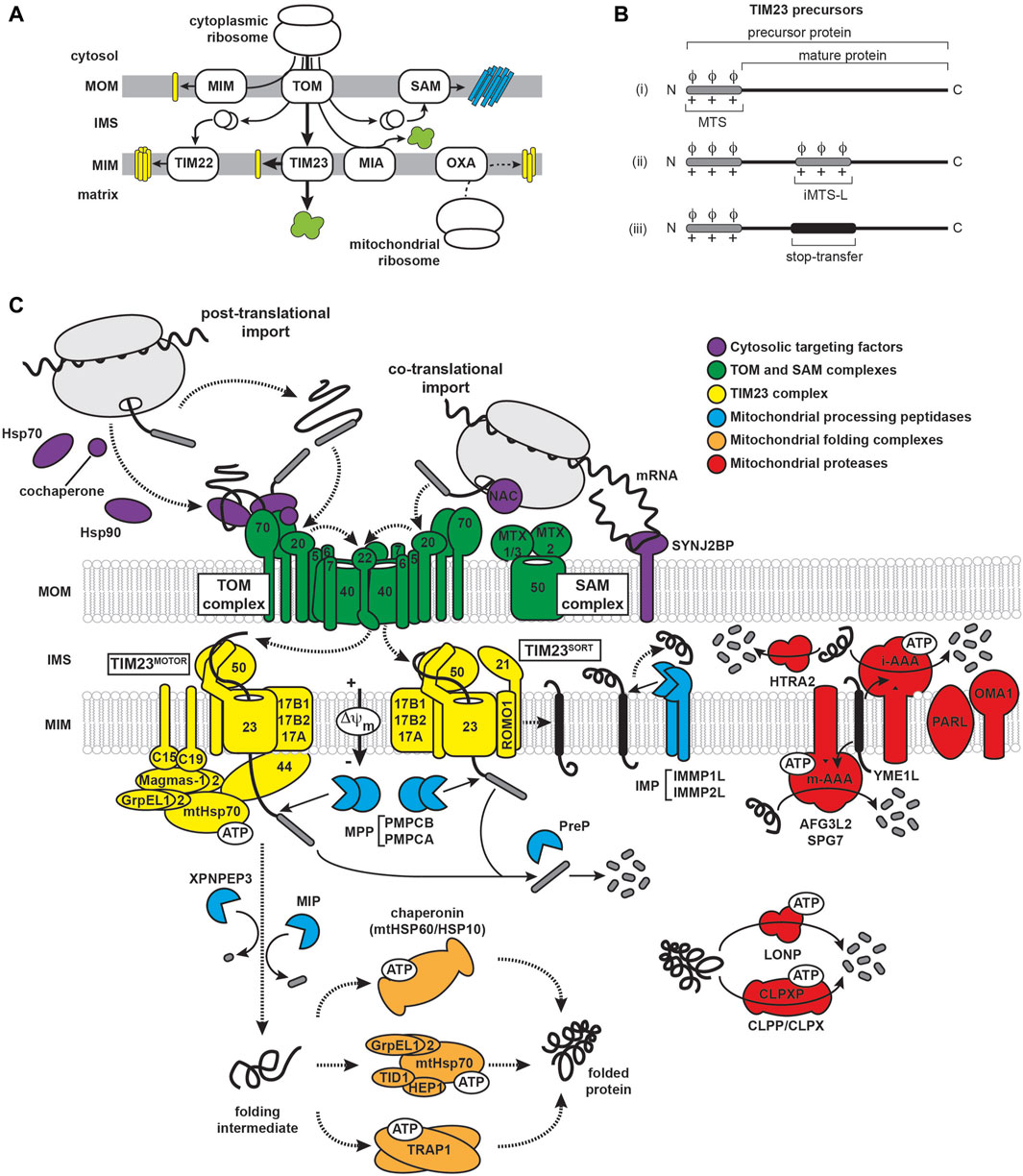
FIGURE 2. Mitochondrial import and proteostatic control. (A) Trafficking routes of mitochondrial proteins. Mitochondrial proteins include soluble proteins of the IMS/ICS and matrix (green), α-helical membrane proteins of the MOM and MIM (yellow) and β-barrel proteins of the MOM (cyan). Trafficking routes are depicted for nuclear-encoded proteins (solid lines) and mitochondrial-encoded proteins (dashed line). The major protein transport complexes include the translocase of the outer membrane (TOM) complex, the mitochondrial import (MIM) complex, the sorting and assembly machinery (SAM) complex, translocases of the inner membrane 22 and 23 (TIM22 and TIM23, respectively), the mitochondrial IMS import and assembly (MIA) complex, and the oxidase assembly (OXA) insertase. The trafficking of MTS-containing precursors via the TIM23 pathway is denoted by the thick arrows. (B) Targeting and topogenic sequences of TIM23 substrates. The MTSs are depicted in gray as containing basic (+) and hydrophobic (ϕ) residues. The stop-transfer segment is depicted in black. (C) The mammalian TIM23 import, processing, and quality control machinery. Components of the biogenesis machinery are categorized as: (i) chaperones that regulate the targeting of precursors from the cytosol to the mitochondrion (violet), (ii) outer membrane (TOM and SAM) complexes (green), (iii) TIM23MOTOR and TIM23SORT complexes (yellow), (iv) proteases that process TIM23 substrates (cyan), (v) matrix chaperones that mediate the folding of TIM23 substrates (orange), and (vi) proteases that mediate the degradation of mitochondrial proteins (red). See Supplementary Table S1 for a listing of all relevant proteins.
3.1.1 TIM23 presequences
A key defining feature of TIM23 substrates is an amino-terminal targeting signal termed the mitochondrial targeting sequence (MTS), or “presequence”. These proteins are synthesized as so-called precursors that are recognized by receptors of the mitochondrial import machinery and typically have their respective MTSs cleaved to yield the mature form of the protein upon reaching their destination. The MTS signals lack sequence conservation, are highly variable in length (ranging from about 15 to 60 amino acids), and are unstructured in aqueous solution. However, they all share a capacity to form amphipathic α-helices with one face enriched in hydrophobic residues and the other enriched in basic residues (mostly Arg). This is coupled with a near absence of acidic residues, resulting in a net charge between +3 and +6 (Roise et al., 1986; Roise et al., 1988; Roise and Schatz, 1988; Pak and Weiner, 1990; Vogtle et al., 2009; Calvo et al., 2017). TIM23-targeted precursors have a diversity of targeting elements (Figure 2B): (i) those containing only the N-terminal MTS followed by a soluble mature protein, (ii) those with MTS-like structures (iMTS-Ls) in the mature protein that bind mitochondrial receptors and enhance import efficiency and kinetics (Backes et al., 2018; Hansen et al., 2018), and (iii) those with a bipartite signal sequence that, in addition to the MTS, contain a topogenic (membrane-active) hydrophobic stop-transfer sequence that partitions into the MIM as a transmembrane segment (Glick et al., 1992). It should also be noted that MTS processing is not a strict feature of TIM23-mediated import, as some TIM23 precursors are not processed following translocation and some retain import competence even with their MTSs deleted (Longen et al., 2014; Woellhaf et al., 2014; Weill et al., 2018). Importantly, the variability of the sequence and the locations of MTS and MTS-like sequences may explain the ability of some amyloidogenic polypeptides to present cryptic TIM23 targeting signals, as we discuss later.
3.1.2 Cytosolic trafficking of TIM23 substrates
The biogenesis of TIM23 substrates begins on cytosolic ribosomes and is regulated by several molecular chaperones and piloting factors (Figure 2C, violet). The targeting of these proteins to mitochondria mostly proceeds post-translationally, whereby the polypeptide is completely synthesized and released from the ribosome before engaging the mitochondrial import machinery. In this case, ATP-dependent cytosolic heat shock proteins (HSPs) of the HSP70 (Deshaies et al., 1988; Murakami et al., 1988; Terada et al., 1995; Endo et al., 1996) and HSP90 families (Young et al., 2003; Fan et al., 2006) may bind precursors at different stages to prevent their aggregation and maintain them in partially unfolded states (Becker et al., 2019; Avendano-Monsalve et al., 2020), which is particularly important for precursor proteins with transmembrane segments (Claros et al., 1995). The HSP70/90 chaperones undergo ATPase cycles that are allosterically coupled to substrate binding and release (Rutledge et al., 2022). HSP70s inhibit folding of their client substrates by extensively interacting with low specificity at binding motifs of short hydrophobic segments flanked by charged residues (Rudiger et al., 1997). By comparison, HSP90s have more extended binding sites that recognize later-folding intermediates (Karagoz and Rudiger, 2015). The activity of HSP70s is modulated by co-chaperones that regulate ATP turnover and substrate specificity (Moran Luengo et al., 2019). HSP70 co-chaperones include J domain (HSP40) proteins, which stimulate ATP hydrolysis, and nucleotide exchange factors (NEFs), which promote exchange of bound ADP for ATP (Kampinga and Craig, 2010). Multiple J domain co-chaperones, including DNAJA1, 2, and 4 in mammals and Djp1, Ydj1 and Sis1 in yeast, have been implicated in precursor protein targeting to mammalian mitochondria (Bhangoo et al., 2007; Papic et al., 2013; Jores et al., 2018).
Alternatively, the targeting of TIM23 substrates can proceed co-translationally, wherein the polypeptide engages the mitochondrial import machinery while it is still being translated on the ribosome (Avendano-Monsalve et al., 2020; Lenkiewicz et al., 2021). The existence of cotranslational import in both yeast and mammalian mitochondria is supported by evidence of mitochondria-bound ribosomes and polysomes that is promoted, for example, following treatment with the translation elongation inhibitor cycloheximide (Crowley and Payne, 1998; Williams et al., 2014; Gold et al., 2017). Other studies similarly support cotranslational targeting of different TIM23 substrates (Ahmed et al., 2006; Yogev et al., 2007). Although mitochondria do not appear to have a dedicated cotranslational targeting route (for instance, analogous to the signal recognition particle-mediated pathway of the ER (Akopian et al., 2013)), there are systems in place for promoting cotranslational mitochondrial import under certain conditions. For example, mRNAs encoding mitochondria-targeted proteins are enriched at the mitochondrial surface (Egea et al., 1997; Matsumoto et al., 2012; Williams et al., 2014; Fazal et al., 2019; Kuzniewska et al., 2020), and stabilized by MOM-localized RNA binding proteins Puf3 in yeast (Wang et al., 2018) and perhaps SYNJ2BP in mammals (Qin et al., 2021). Additionally, a translation stimulator at the mitochondrial surface, the MDI-Larp complex, was shown to enhance protein synthesis in the vicinity of import complexes in Drosophila (Zhang et al., 2016). Finally, mitochondria-targeted proteins can be recognized by the heterodimeric nascent polypeptide-associated complex (NAC) (Beatrix et al., 2000), which simultaneously binds ribosomes and emerging nascent chains and may promote cotranslational targeting to mitochondria (Wiedmann et al., 1994; George et al., 1998; George et al., 2002; del Alamo et al., 2011; Gamerdinger et al., 2019). In yeast, NAC binds to the SAM complex subunit Sam37 (Ponce-Rojas et al., 2017; Avendano-Monsalve et al., 2022) and the MOM protein OM14 (Lesnik et al., 2014); whether NAC engages homologous proteins in mammalian mitochondria remains an open question. NAC may play a special role in amyloidogenic diseases, as it has recently been shown to suppress aggregation of poly-Q expanded proteins (Shen K. et al., 2019). The critical point is that regardless of whether precursor substrates are imported co- or post-translationally, the targeting system is designed to maintain the substrate in an unfolded state in order to preserve its import competence.
3.1.3 Structure and function of the TOM complex
The translocase of the outer mitochondrial membrane (TOM) complex of the MOM serves as the entry site for all TIM23 substrates into mitochondria (Figure 2C, green). This complex contains seven different subunits: the Tom40 β-barrel channel that serves as the aqueous transmembrane conduit for precursors; small TOM proteins (Tom5, Tom6, Tom7) that regulate TOM complex assembly; and several receptors, including Tom22, Tom20 and Tom70 (Pitt and Buchanan, 2021; Araiso and Endo, 2022). The core complex appears to form as equi-stoichiometric assemblies of Tom40/22/5/6/7 that can arrange as dimeric or higher order structures (Kunkele et al., 1998; Model et al., 2008; Mager et al., 2010; Shiota et al., 2015; Bausewein et al., 2017; Sakaue et al., 2019); in contrast, the Tom20 and Tom70 receptors appear to be more loosely bound. Thus, these receptors may instead assemble with the TOM complex in an on-demand basis depending on the presence of substrate (Dekker et al., 1998; Bhagawati et al., 2021). Recent cryo-EM structures of the TOM complex in yeast (Araiso et al., 2019; Tucker and Park, 2019) and human (Wang et al., 2020a; Guan et al., 2021; Su et al., 2022) have shed light on the structural interactions among TOM subunits and how precursor proteins are recognized and translocated. For example, the TOM receptors play complementary and partially overlapping roles in the recognition of MTS-containing proteins. Tom20, Tom70, and Tom22 all have receptor domains containing tetratricopeptide repeat (TPR) motifs that mediate protein interactions (Zeytuni and Zarivach, 2012) and appear to have general protein chaperone function in addition to acting as precursor receptors (Yano et al., 2004; Yamamoto et al., 2009). Tom20 serves as the general receptor for preproteins and is paradigmatic for MTS-receptor interactions because it is the only mitochondrial receptor for which high-resolution structural information is available in the MTS peptide-bound state (Abe et al., 2000; Saitoh et al., 2007; Saitoh et al., 2011). The Tom20 cytosolic C-terminal receptor domain contains two helix-turn-helix motifs that define a single prototypical TPR motif with an embedded nonpolar patch flanked by two acidic regions and a region rich in Gln residues (Figure 3A). The binding groove of Tom20 is shallow and short, accommodating only about eight residues of the MTS. The Tom20 recognition motif within MTSs is ϕχχϕϕ, where ϕ is a nonpolar residue and χ is any residue. This relatively nonspecific recognition motif enables a dynamic, weak-affinity and multi-mode interaction with the substrate, dominated by nonpolar contacts (Muto et al., 2001; Obita et al., 2003). Tom70, by comparison, has a much larger receptor domain containing 11 TPR motifs that are divided into N- and C-terminal parts (Wu and Sha, 2006) (Figure 3B). The N-terminal region of Tom70 contains a homodimerization interface and forms a clamp-like region (TPR motifs 1–3) that binds Hsp70 and Hsp90 chaperones, perhaps serving as a co-chaperone for the transfer of Hsp70/90-bound precursors to the TOM complex (Young et al., 2003; Wu and Sha, 2006). This function underscores its recently discovered role in recruiting chaperones to the mitochondrial surface (Backes et al., 2021). The C-terminal region of Tom70 (TPR motifs 4–11) forms a large pocket that likely binds polytopic membrane precursor proteins destined for the TIM22 pathway (Wiedemann et al., 2001; Rehling et al., 2003), and may bind targeting signals of TIM23 substrates as well (Hines et al., 1990; Hines and Schatz, 1993; Wu and Sha, 2006; Melin et al., 2015). The central receptor Tom22 regulates TOM complex assembly, and unlike the primary Tom20 and Tom70 receptors, it is tightly bound to the TOM complex. Additionally, Tom22 features receptor domains on both the cytosol-facing (Figure 3C) and IMS-facing sides of the MOM (Bolliger et al., 1995; Honlinger et al., 1995; Dekker et al., 1998; van Wilpe et al., 1999; Yamano et al., 2008). While Tom22 and Tom20 have similar substrate profiles (Mayer et al., 1995; Yamano et al., 2008), the Tom20 receptor mediates hydrophobic interactions with the MTS, whereas Tom22 interactions are dominated by electrostatic attraction between the basic face of the MTS and the partially disordered acidic Tom22 binding pocket (Kiebler et al., 1993). However, the strict requirement of these negatively charged residues has been questioned (Nargang et al., 1998). The dominant model describing how precursor proteins traverse the membrane through the TOM complex is by an “acid chain” of negatively charged patches on TOM subunits that guide the positively charged MTS. By this model, Tom5 and Tom22 make up an acidic pathway on the cytosolic face of the MOM (the cis site) (Bolliger et al., 1995; Dietmeier et al., 1997; Schatz, 1997; Komiya et al., 1998), the precursor moves through the Tom40 pore guided by acidic residues on the interior wall of the β-barrel (Suzuki et al., 2000; Gabriel et al., 2003; Shiota et al., 2015), and the precursor then binds sites on the IMS side of the MOM comprised of Tom22, Tom40 and Tom7 (the trans site) (Court et al., 1996; Moczko et al., 1997; Rapaport et al., 1997; Kanamori et al., 1999; Esaki et al., 2004). The selective positioning of acidic binding sites with increasing affinity for the MTS from the cytosolic to the IMS sites drives the vectorial movement of the precursor through the TOM complex.
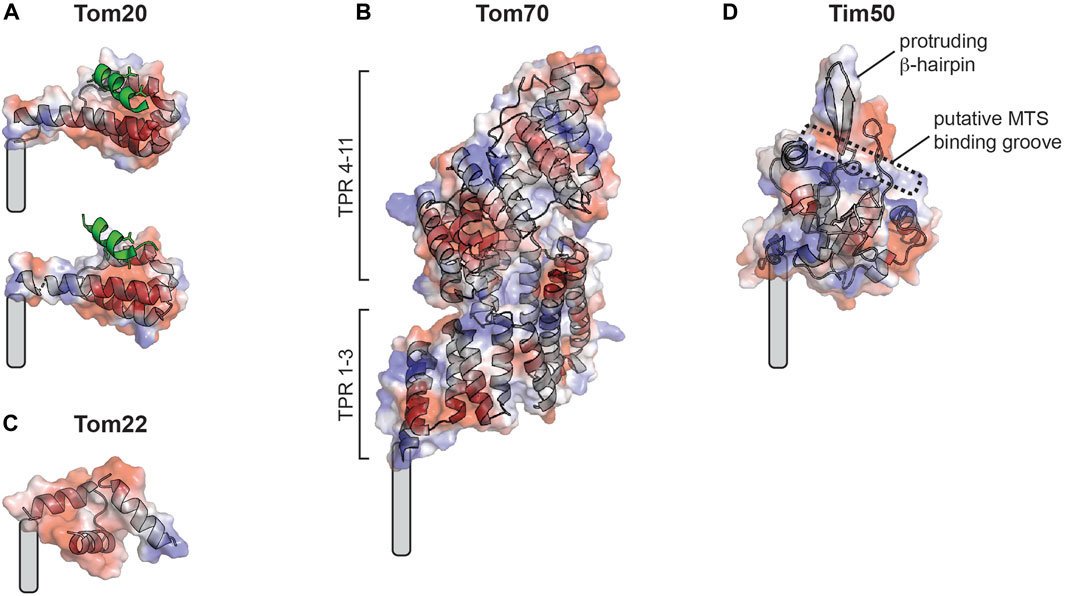
FIGURE 3. Mitochondrial protein import receptors of known structure. Soluble receptor domains are shown as cartoon traces superimposed with electrostatic surfaces (red, acidic; blue, basic; white, neutral). Grey curved rectangles show approximate positions of transmembrane segment attachment. (A) Rat Tom20 receptor from crystal structure with disulfide-bound MTS (green) showing two different binding states resolved by distinct MTS-receptor chemical tethering strategies (the so-called A-linker [PDB 2V1T], above; and the Y-linker [PDB 2V1S], below). (B) Yeast Tom70 receptor from crystal structure (PDB 2GW1). (C) Human Tom22 receptor from cryo-EM structure (PDB 7VDD). (D) Tim50 receptor (homology model of human receptor based on yeast Tim50 core domain) from crystal structure (PDB 3QLE), showing the predicted MTS binding pocket and protruding β-hairpin.
3.1.4 Structure and function of the TIM23 complex
The TIM23 complex is the dedicated machinery of the MIM that mediates the import and sorting of all MTS-containing precursor proteins (Genge and Mokranjac, 2021) (Figure 2, yellow). The general organization of TIM23 is evolutionarily conserved; however, in comparison with the more fully understood yeast complex, human TIM23 forms multiple functionally distinct complexes containing alternate subunit isoforms (Sinha et al., 2014; Pfanner et al., 2019). The core TIM23 complex in humans contains the channel forming Tim23 and Tim17A/B1/B2 isoforms that assemble to make an aqueous conduit across the MIM for the passage of preproteins (Bauer et al., 1999; Moro et al., 1999; Martinez-Caballero et al., 2007; Demishtein-Zohary et al., 2017; Matta et al., 2017). It also contains Tim50, which serves as the main receptor for MTS-bearing precursors (Geissler et al., 2002; Yamamoto et al., 2002; Mokranjac et al., 2003a) and, in mammals, acts as a broad specificity phosphatase (Guo et al., 2004; Chaudhuri et al., 2021). The conserved core domain of the Tim50 receptor contains a putative MTS binding groove lined with negatively charged and hydrophobic residues as well as a prominent β-hairpin (Qian et al., 2011) (Figure 3D). The N-terminal extension of the Tim23 channel is an intrinsically disordered region that specifically interacts with Tim50 near the β-hairpin (Geissler et al., 2002; Yamamoto et al., 2002; Mokranjac et al., 2003a; Meinecke et al., 2006; Alder et al., 2008a; Gevorkyan-Airapetov et al., 2009; Mokranjac et al., 2009; Tamura et al., 2009; Qian et al., 2011; Schulz et al., 2011; Lytovchenko et al., 2013; Malhotra et al., 2017; Dayan et al., 2019; Gunsel et al., 2020). This interaction between the disordered N-terminal extension of Tim23 and Tim50 maintains the Tim23 channel in a quiescent, dimeric state that preserves the transmembrane potential (Δψm) across the MIM (Bauer et al., 1996; Meinecke et al., 2006; Alder et al., 2008a).
To deliver precursors to their correct destination, the compositionally dynamic TIM23 complex forms two different assemblies adapted to the Tim23/17/50 core. The TIM23MOTOR complex mediates the translocation of soluble precursors into the matrix (Mokranjac, 2020), a process that requires the recruitment of a matrix-localized molecular motor system (the presequence translocase-associated motor, or PAM complex) that includes: the central subunit mitochondrial Hsp70 (mtHsp70, also called mortalin), which serves as the ATP-driven molecular motor (Goswami et al., 2010; Esfahanian et al., 2023); Tim44, which anchors mtHsp70 to the TIM23MOTOR complex (Kronidou et al., 1994; Schneider et al., 1994; Silva et al., 2004); and co-chaperones that modulate the ATPase activity of mtHsp70. The latter include the GrpE-Like 1 and 2 (GrpEL1 and GrpEL2) NEFs (Schneider et al., 1996; Naylor et al., 1998; Srivastava et al., 2017), the paralogous DnaJC15 (C15) and DnaJC19 (C19) that are homologous to the yeast J-protein Pam18/Tim14 (Silva et al., 2003; Mokranjac et al., 2003b; Richter-Dennerlein et al., 2014), and isoforms of the mitochondria-associated granulocyte-macrophage colony stimulating factor (GM-CSF) signaling molecule (Magmas-1 and 2) that are orthologs of the yeast J-like protein Pam16/Tim16 (Elsner et al., 2009; Sinha et al., 2010; Waingankar and Silva, 2021). By contrast, the TIM23SORT complex mediates the lateral sorting of membrane-directed precursor proteins into the MIM. This sorting complex lacks the PAM motor and recruits additional membrane subunits. These include Tim21, which makes specific contacts with Tim23 and Tim50 (Tamura et al., 2009; Lytovchenko et al., 2013; Bajaj et al., 2014) and mediates the assembly of membrane-bound subunits of respiratory complex IV (Mick et al., 2012; Richter-Dennerlein et al., 2016), and ROMO1, a subunit homologous to yeast Mgr2 that promotes the interaction of Tim21 with the TIM23SORT complex and is specifically required for the import of mitochondrial proteases (Ieva et al., 2014; Richter et al., 2019; Matta et al., 2020). Apropos of these proposed discrete TIM23SORT and TIM23MOTOR models for MIM integration and matrix import, it should be noted that other experimental results support a model in which TIM23 is a single structural entity that is actively remodeled to support translocation or integration depending on substrate availability instead of existing in two disparate states (Popov-Celeketic et al., 2008).
The import of MTS-containing substrates by the TIM23 complex is a multistep, energy-requiring process. The transfer of precursor proteins to the TIM23 complex is facilitated by the formation of a TOM-TIM23 supercomplex that is stabilized by interactions of the IMS-facing trans site of the TOM complex and the IMS-facing regions of Tim21, Tim50, and Tim23 subunits of the TIM23 complex (Dekker et al., 1997; Chacinska et al., 2003; Mokranjac et al., 2005; Albrecht et al., 2006; van der Laan et al., 2006; Chacinska et al., 2010; Shiota et al., 2011; Gold et al., 2014), which have been verified primarily by crosslinking experiments done with yeast models and with purified proteins (Chacinska et al., 2005; Mokranjac et al., 2005; Albrecht et al., 2006; Tamura et al., 2009; Shiota et al., 2011; Bajaj et al., 2014; Waegemann et al., 2015; Araiso et al., 2019; Gunsel et al., 2020). Upon emerging from the TOM complex, the MTS first binds the Tim50 receptor, thereby displacing receptor interactions with the Tim23 N-terminus and Tim21 and altering the TOM-TIM23 association (Geissler et al., 2002; Yamamoto et al., 2002; Mokranjac et al., 2003a; Mokranjac et al., 2009; Marom et al., 2011a; Shiota et al., 2011; Lytovchenko et al., 2013; Waegemann et al., 2015). The MTS then binds the now-exposed Tim23 N-terminus (Bauer et al., 1996; de la Cruz et al., 2010; Marom et al., 2011a; Lytovchenko et al., 2013) and is directed to the Tim23 channel which, like Tom40, has specific residues along the channel lumen that interact with substrates (Alder et al., 2008b; Denkert et al., 2017). The dynamic Tim23 channel undergoes conformational alterations in response to substrate and changes in the Δψm (Popov-Celeketic et al., 2008; Malhotra et al., 2013) and the presence of the MTS activates the Tim23 channel gating (Bauer et al., 1996; Truscott et al., 2001). The basic MTS is electrophoretically pulled toward the negatively charged matrix through the activated Tim23 channel (Martin et al., 1991), with unidirectional movement imparted by increasing binding affinity between the MTS and Tim23, Tim50, and Tim44 (Marom et al., 2011b). Soluble precursor proteins are then translocated completely into the matrix by the TIM23MOTOR complex, whereby the ATPase activity of mtHsp70, modulated by co-chaperones DnaJC15/19 and Magmas-1/2, pulls the substrate by a Brownian ratchet or active pulling mechanism (Mokranjac et al., 2003a; Mokranjac, 2020; Silva et al., 2003; Mokranjac et al., 2003b; Truscott et al., 2003). By contrast, when a hydrophobic stop-transfer sequence is detected on the substrate, translocation stalls, the complex recruits subunits of the TIM23SORT complex, and the nonpolar segment partitions laterally into the MIM as an α-helical transmembrane segment in a manner driven by the Δψm (Gartner et al., 1995; Gruhler et al., 1997) and mediated by the ROMO1 and Tim21 gatekeepers (van der Laan et al., 2006; Mick et al., 2012; Ieva et al., 2014; Richter-Dennerlein et al., 2016; Richter et al., 2019; Lee et al., 2020). Notably, an alternative structure-based model of TIM23 function suggests that instead of forming an aqueous channel, Tim23 and Tim17 together form lipid-exposed cavities that provide a protein translocation pathway (Sim et al., 2023), consistent with evidence that TIM23 precursors are translocated across the MIM at the Tim17-bilayer interface rather than via a channel defined by Tim23 (Fielden et al., 2023).
3.1.5 Processing and quality control of TIM23 substrates
During import, precursor proteins are selectively processed to their mature forms by a set of mitochondria-localized processing proteases (Gomez-Fabra Gala and Vogtle, 2021; Kunova et al., 2022) (Figure 2C, cyan). The vast majority of TIM23 complex substrates are processed in a way that removes the N-terminal targeting sequences (Vogtle et al., 2009). The main protease is the matrix-localized mitochondrial processing peptidase (MPP), a metalloendopeptidase that forms a dimeric complex (PMPCA and PMPCB subunits in human) (Taylor et al., 2001). MPP cleaves at defined recognition sites (predominantly with the scissile bond two or three residues C-terminal to an Arg residue (Calvo et al., 2017)), thereby releasing the MTS which is subsequently degraded by the presequence protease (PreP) (Alikhani et al., 2011a; Kucukkose et al., 2021). Following MTS cleavage, some matrix-targeted precursors require additional maturation steps at the new N-terminus that involve the removal of either a single residue (mediated by the XPNPEP3 protease (Singh et al., 2017)) or an octapeptide (mediated by the MIP protease (Vogtle et al., 2011)), both of which remove destabilizing N-terminal residues to increase protein half-life (Vogtle et al., 2009; Varshavsky, 2011). Additionally, some TIM23-targeted substrates integrated into the MIM are processed by the inner membrane peptidase IMP (IMMP1L and IMMP2L in human), which releases a soluble IMS-facing domain of the imported protein as the mature, functional form (Nunnari et al., 1993).
To ensure the proper folding of newly-imported proteins, mitochondria contain two main chaperone systems in the matrix (Figure 2C, orange). The mitochondrial Hsp60/Hsp10 chaperonin complex, a homolog of the bacterial GroEL/GroES chaperonin, sequesters unfolded or kinetically-trapped folding intermediates inside an Anfinsen cage-like cavity and undergoes ATPase-driven structural changes to release properly folded proteins (Cheng et al., 1989; Ellis, 1996; Nielsen and Cowan, 1998; Apetri and Horwich, 2008; Chakraborty et al., 2010; Nisemblat et al., 2015). In addition, a soluble complex of mtHsp70 (mortalin) resides in the matrix (Horst et al., 1997; Havalova et al., 2021), where it performs its protein folding functions with three co-chaperones that have been identified in human: the Hsp70-escort protein 1 (HEP1) and J-domain protein tumorous imaginal disc protein 1 (TID-1), which regulate ATPase activity of mtHsp70, and the NEFs GrpEL1/2 (Sichting et al., 2005; Zhai et al., 2008; Iosefson et al., 2012; Dores-Silva et al., 2013; Havalova et al., 2021). These main matrix chaperone systems are supplemented in mammals by the HSP90 paralog TRAP1, which performs diverse functions including acting as a late-stage folding chaperone for mitochondrial matrix proteins (Joshi et al., 2022).
In addition, mitochondria contain several proteases for the degradation of misfolded and damaged proteins (Gomez-Fabra Gala and Vogtle, 2021) (Figure 2C, red). In human mitochondria, four main ATP-fueled proteases of the AAA+ (ATPases associated with diverse cellular activities) superfamily are responsible for the surveillance and clearance of proteins. These include the MIM-bound metalloproteases m-AAA (homo-oligomers of AFG3L2 or hetero-oligomers of AFG3L2 and SPG7 with catalytic domains facing the matrix) and i-AAA (composed of YME1L1 with catalytic domains facing the IMS), both of which can extract and break down MIM proteins (Opalinska and Janska, 2018). Additionally, the matrix contains soluble AAA+ serine proteases. LONP1 is a homohexameric assembly that serves as the central quality control protease in the matrix, degrading misfolded and damaged proteins (Szczepanowska and Trifunovic, 2022) in addition to promoting protein folding by cooperating with mtHsp70 (Shin et al., 2021). The CLPXP complex, on the other hand, is a heterooligomeric protease assembly involved in diverse functions including mitoribosome and OXPHOS maintenance (Szczepanowska and Trifunovic, 2022). The HTRA2 (high temperature requirement) soluble serine protease of the IMS is involved in caspase-dependent apoptosis and has been implicated in PD progression (Vande Walle et al., 2008). Finally, there are additional MIM-bound proteases, including PARL and OMA1, that have a more specific set of substrate proteins that regulate mitochondrial dynamics, mitophagy, and stress responses (Pellegrini and Scorrano, 2007; Jiang et al., 2014). As discussed below, the quality control machinery for newly imported proteins may also be involved in stress responses involving amyloidogenic proteins.
3.2 Amyloid misfolding and cellular proteostasis
Cellular proteostasis involves the regulation of all stages of the protein life cycle (Labbadia and Morimoto, 2015; Klaips et al., 2018): molecular chaperones guide cotranslational folding of nascent chains during ribosomal synthesis and promote protein folding and assembly of oligomeric complexes, degradation mechanisms such as the ubiquitin-proteasome system (UPS) and the autophagy-lysosome pathway (ALP) remove misfolded and aggregated proteins, and stress response pathways respond to protein folding stress, such as the ER-based unfolded protein response (UPRER) (Figure 4, black arrows). All three of these protein quality control processes are implicated in neurodegenerative diseases (Scheper and Hoozemans, 2015; Park et al., 2020; Schmidt et al., 2021).
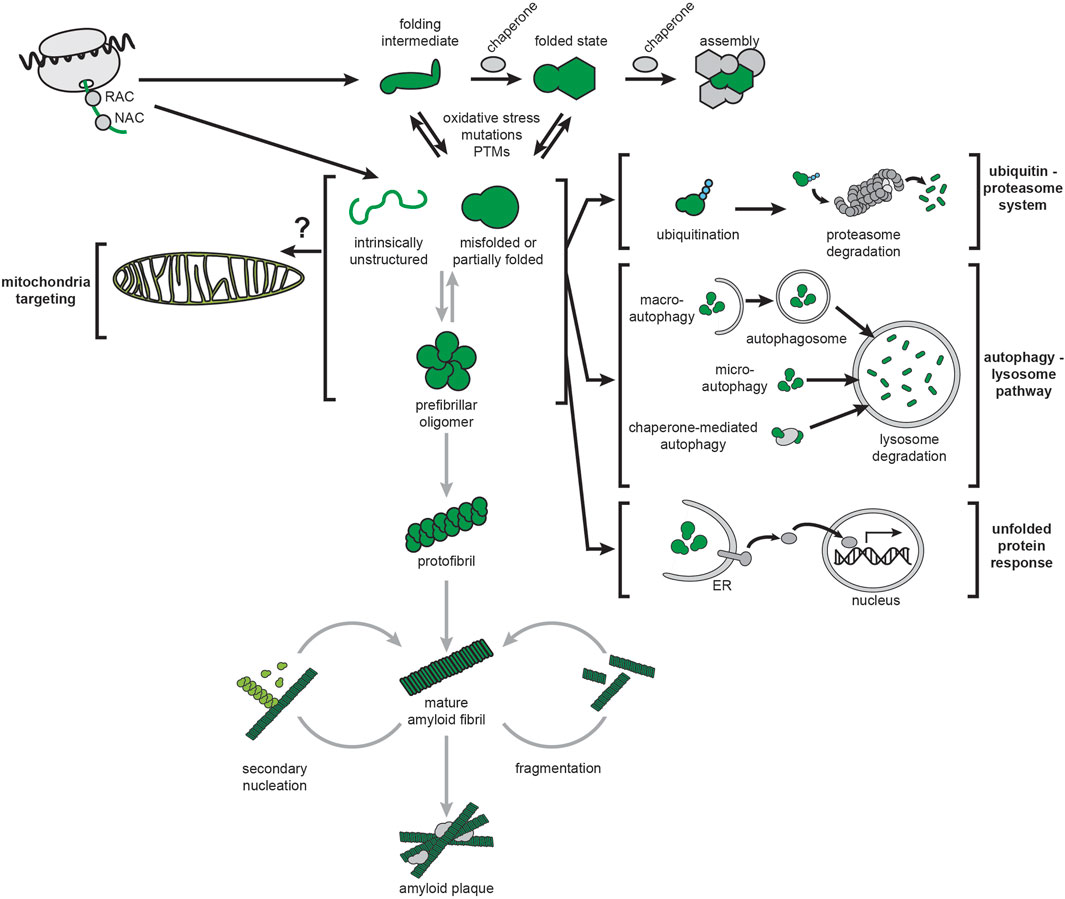
FIGURE 4. Amyloids and cellular proteostasis. The proteostasis network of mammalian cells is depicted by black arrows. Chaperones that guide cotranslational folding on the ribosome include the nascent polypeptide-associated complex (NAC) and the ribosome-associated complex (RAC). Additional chaperones guide monomer folding and assembly into complexes. The ubiquitin-proteasome system (UPS) targets misfolded proteins, or proteins that are no longer needed, by ubiquitination and digestion by the proteasome. The autophagy-lysosome pathway (ALP) targets misfolded proteins (and larger structures such as large aggregates and organelles) by delivering them to the lysosome via distinct pathways (macroautophagy, microautophagy and chaperone-mediated autophagy). Protein unfolding stress can activate complex signal transduction pathways leading to gene expression changes for restoring protein homeostasis, most notably the ER-based unfolded protein response (UPRER). Amyloidogenesis, depicted by gray arrows, can initiate from intrinsically disordered or misfolded structures and proceed by stepwise formation of oligomers, protofibrils, and finally mature fibrils. It can also be accelerated by secondary nucleation and fragmentation of existing fibrils.
AD, PD, and HD are proteostatic diseases associated with the misfolding of Aβ, α-syn, and mHtt, respectively, in a process that leads to the formation of amyloid fibrils (Figure 4, grey arrows). This process can begin with precursors that include intrinsically disordered peptides such as Aβ and α-syn, or with partially and misfolded globular proteins. The kinetics of amyloid fibrillization have a marked dependence on protein concentration, as well as factors that affect the tendency for polypeptides to self-associate, including mutations, post-translational modifications (PTMs), cofactors, and oxidative stress (Alexandrescu, 2005; Rezaei-Ghaleh et al., 2016; Hu et al., 2019; Lontay et al., 2020; Chiki et al., 2021; Gottlieb et al., 2021; He et al., 2021; McGlinchey et al., 2021). Fibrillization follows nucleation kinetics, accounting for the “seeding” properties of amyloids (Chiti and Dobson, 2017; Almeida and Brito, 2020). Nucleation can occur through primary or de novo processes, or secondary mechanisms whereby mature fibril surfaces act as templates for new fibril growth (Ferrone et al., 1985; Padrick and Miranker, 2002; Patil et al., 2011; Tornquist et al., 2018). Amyloid fibrils share a common structural property of protein association through a cross-β spine motif, stabilized by intermolecular hydrogen-bonded parallel β-sheet layers arranged perpendicular to the long axis of the fibril (Nelson et al., 2005; Chiti and Dobson, 2017; Almeida and Brito, 2020). They also share similar morphologies, often being microns long with widths of ∼10 nm and a twisting repeat of ∼100 nm (Dobson, 2003; Qiang et al., 2017).
Amyloidogenic aggregates have historically provided the primary histopathological markers of AD, PD, and HD. These aggregates are generally termed “amyloid fibrils” when formed extracellularly and ‘inclusions’ when formed intracellularly. The hallmarks of AD are extracellular plaques composed primarily of Aβ peptides and intracellular neurofibrillary tangles enriched in hyperphosphorylated variants of the protein tau (Zheng and Koo, 2011). PD is characterized by cytoplasmic aggregates called Lewy bodies and inclusions called Lewy neurites, of which α-syn is the main component (Braak et al., 1999). Intracellular accumulation of mHtt into amyloid-like inclusion bodies is a primary feature of HD (Yamamoto et al., 2000; Arrasate et al., 2004).
The concept that insoluble fibrils, whether extra- or intra-cellular, are the primary cytotoxic factors that initiate amyloidogenic disease is the core tenet of the “amyloid cascade hypothesis” (Figure 5, left) first proposed by Hardy and Higgins to describe the role of Aβ amyloids in AD pathogenesis (Hardy and Higgins, 1992). Subsequently it was proposed that α-syn fibrils are the primary cytotoxic factors in PD (Stefanis, 2012) and that amyloid fibril-like inclusions of mHtt are the primary cytotoxic factors in HD (Scherzinger et al., 1999). There is considerable controversy, however, on whether amyloid fibrils are the only, or even the principal culprits in pathology. A confounding factor is the heterogeneity of species associated with amyloid formation. Amyloidogenesis involves a hierarchy of structures that starts from the functional, soluble form of a protein or peptide and proceeds to oligomers, nuclei, β-sheets, protofilaments, protofilament bundles, and finally mature fibrils (Serpell, 2000) (Figure 4). Within the fibrils themselves, there are structural polymorphs that can differ depending on whether they are grown in vitro or isolated from patients. In fact, increasing evidence suggests distinct structural polymorphs are associated with specific disease subtypes. Therefore, considerable uncertainty surrounds which species represent the culprits in pathogenicity (Carulla et al., 2005; Lansbury and Lashuel, 2006; Haass and Selkoe, 2007; Zraika et al., 2010), with some recent proposals advancing that soluble oligomers (Larson and Lesné, 2012; Arbor et al., 2016; Cline et al., 2018) or even individual proteins/peptides (Hoffner and Djian, 2014; Hillen, 2019) could initiate the phenotypic cascade of amyloidogenic diseases. Because mature fibrils are insoluble and extremely stable, many view soluble oligomeric precursors as the most likely candidates for cytotoxicity. However, studies on amyloid oligomers have been hampered by the low concentrations and transitory nature of these intermediates (Engel, 2009), thereby contributing to a lack of evidence for this model. Finally, it unclear whether cytotoxicity is associated with intracellular or extracellular forms of amyloidogenic proteins (Cuello, 2005; Gurlo et al., 2010; Aston-Mourney et al., 2011).
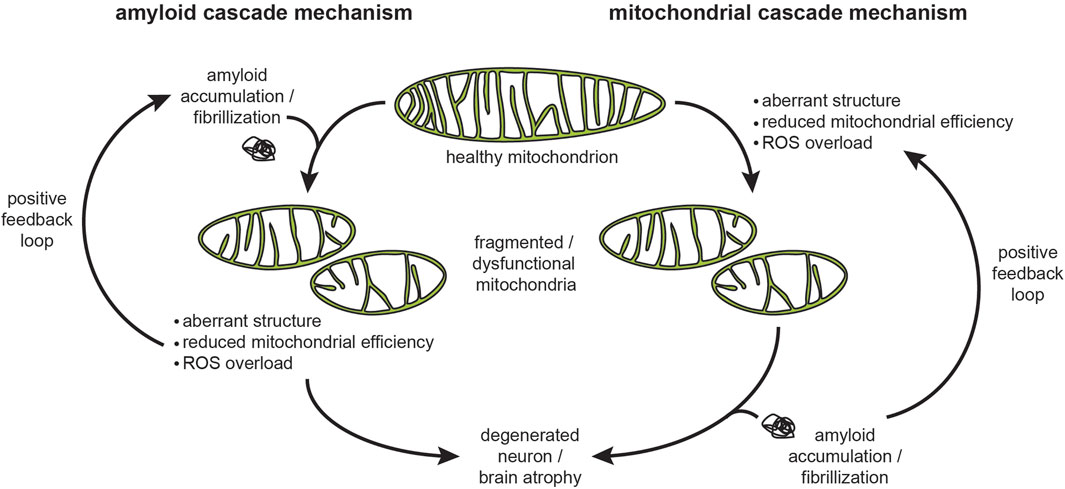
FIGURE 5. The amyloid and mitochondria cascade hypotheses. These two mechanisms describe the cause-and-effect relationships in the progression of neurodegenerative diseases, proposed originally for AD but also relevant for other proteinopathies. The amyloid cascade hypothesis proposes that amyloid accumulation, fibrillization, and plaque formation trigger the pathogenic events, including mitochondrial damage, that lead to neuronal damage. As a corollary to this hypothesis, the most effective interventions would target the accumulation, clearance, and fibril formation of amyloids. The mitochondria cascade hypothesis proposes that mitochondrial dysfunction is the primary driver of neurodegeneration that leads to cellular damage, including amyloid aggregation. As a corollary to this hypothesis, the most effective interventions would be those aimed toward improving mitochondrial physiology. In both mechanisms, amyloid aggregation and mitochondrial dysfunction can be part of a vicious cycle that accelerates disease progression. Note that the interaction of amyloidogenic proteins with the mitochondrial import machinery likely plays a central role in both mechanisms.
Much of the current research on the mechanisms of amyloidogenic diseases has shifted focus towards processes preceding mature fibril formation. As a result, it has now been widely demonstrated that mitochondrial dysfunction occurs early in the pathogenesis of AD (Wang et al., 2020b), PD (Malpartida et al., 2021), and HD (Carmo et al., 2018). The “mitochondrial cascade hypothesis”, originally proposed by Swerdlow and colleagues, proposes that it is in fact the progressive decline in mitochondrial function that promotes AD pathology and influences the progression of the disease (Swerdlow et al., 2014) (Figure 5, right). Therefore, exactly how amyloidogenic proteins interact with mitochondria, and specifically how they may engage the mitochondrial protein import machinery, is a key mechanistic question in understanding the pathology of neurodegenerative disorders.
4 Neurodegenerative diseases and pathogenic mechanisms of amyloids
Aβ, α-syn, and Htt/mHtt are among the approximately 50 amyloidogenic proteins associated with human diseases (Chiti and Dobson, 2017). In this section, we review the roles of these proteins in AD, PD, and HD, respectively, with a special emphasis on their interactions with mitochondria.
4.1 Alzheimer’s disease and the roles of APP and Aβ
AD is the leading cause of senile dementia, affecting several regions of the brain involved in memory and cognition, including the hippocampus, the neocortex, and the basal forebrain (Auld et al., 2002). While the exact etiology of AD is unknown, it is biologically characterized by the aggregation of two misfolded proteins: Aβ, the primary component of extracellular plaques, and hyperphosphorylated variants of the microtubule-associated protein tau, which form intracellular inclusions known as neurofibrillary tangles (NFTs) (Knopman et al., 2021). Aβ is produced through sequential cleavage of APP, a membrane glycoprotein with isoforms ranging from 100 to 140 kDa that predominantly reside at neuronal synapses (Zhou et al., 2011; Hoe et al., 2012) (Figure 6A). APP plays a vital role in neural development and synaptic plasticity, as it is implicated as a receptor involved in kinesin 1 cargo recognition (Lazarov et al., 2005), the Wnt signaling pathway (Liu T. et al., 2021) and other functions including cell adhesion, synaptogenesis (Baumkötter et al., 2014), and iron export (Duce et al., 2010). As a type I integral membrane protein, APP has a large ectodomain at its N-terminus, a single α-helical transmembrane segment, and a small C-terminal intracellular domain. APP belongs to a highly conserved superfamily of genes (Coulson et al., 2000; Jacobsen and Iverfeldt, 2009). In mammals, alternative splicing generates eight APP isoforms, the three most common being APP695, APP751, and APP770, among which APP695 is most highly expressed in neurons (Sandbrink et al., 1996; Belyaev et al., 2010). Like other plasma membrane proteins, the life cycle of APP following synthesis consists of membrane trafficking via the secretory pathway and degradation and recycling by the endocytic system (Lin et al., 2021). The Aβ peptides that form amyloid plaques in AD are derived from the processing of APP through sequential actions of β- and γ-secretases (Selkoe and Hardy, 2016; Zhao et al., 2020) (Figure 6B). The generated Aβ peptides range in size, with the 42-residue Aβ42 being much more aggregation-prone than the more abundant 40-residue Aβ40 (Seubert et al., 1992; Haass and Selkoe, 2007; Chow et al., 2010). Monomeric Aβ40 and Aβ42 are intrinsically disordered in solution (Roche et al., 2016), with Aβ42 having a transient population of β-sheet structure (Kakeshpour et al., 2021).
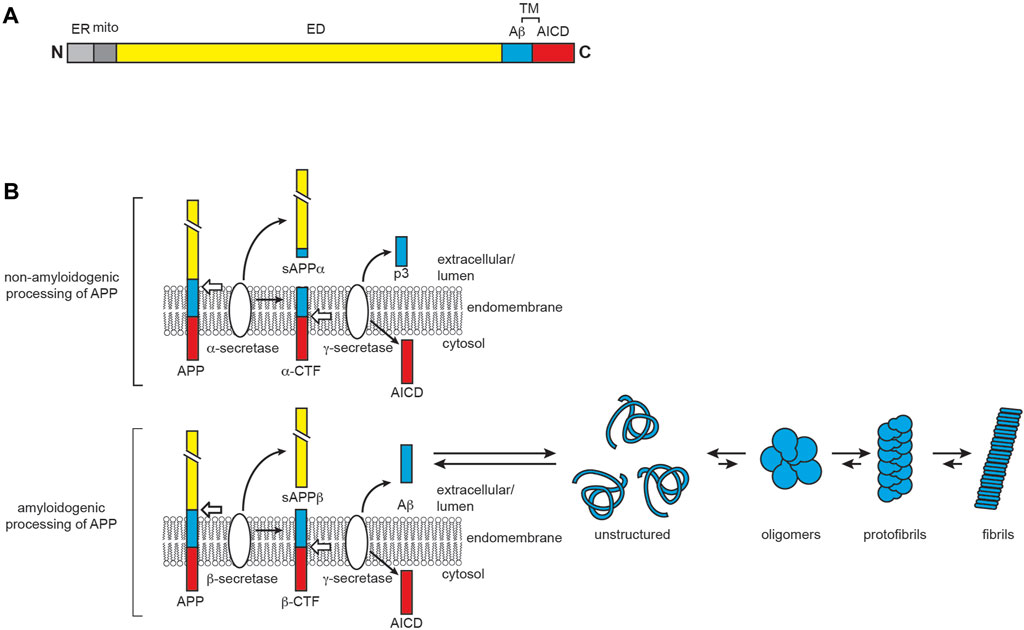
FIGURE 6. AD and the role of APP and Aβ. (A) APP domain organization. Full-length APP contains an extracellular domain (ED, yellow), a transmembrane region (TM), and the APP intracellular domain (AICD, red). The proteolysis product corresponding to Aβ is shown in cyan. The N-terminal ER- and mitochondria-targeting sequences are shown in light and dark gray, respectively. (B) APP processing and Aβ aggregation. APP has two main proteolytic fates. In the non-amyloidogenic pathway (top), APP is first cleaved by α-secretase to yield soluble APPα (sAPPα) and the α-C-terminal fragment (α-CTF), the latter of which is then cleaved by γ-secretase to produce AICD and the non-amyloidogenic extracellular peptide p3. In the amyloidogenic pathway (bottom), β-secretase proteolyzes full-length APP to yield soluble APPβ (sAPPβ) and the β-C-terminal fragment (β-CTF), the latter of which is then cleaved by γ-secretase to produce AICD and the amyloid beta peptide Aβ. The Aβ peptide, particularly Aβ42, is disordered in solution and prone to aggregation and fibrillization.
The neuropathology of AD has historically been described by the amyloid cascade hypothesis, whereby Aβ deposition is the causative event (Hardy and Higgins, 1992). In support of this hypothesis, early-onset familial AD (FAD) is caused by autosomal dominant inheritance mutations in the genes encoding APP or APP processing enzymes (Bekris et al., 2010; Ricciarelli and Fedele, 2017). However, the vast majority of cases are sporadic, late onset AD (LOAD) occurring without mutations in the genes encoding APP or APP processing enzymes (Bekris et al., 2010). While the correlation between Aβ and AD remains strongly supported (Herrup, 2015), intracellular Aβ in the form of soluble monomers and oligomers are now considered to be the primary toxic species rather than insoluble, fibrillar assemblies of Aβ that form extracellular plaques (Goure et al., 2014; Gallego Villarejo et al., 2022). Despite this, soluble Aβ species precede the formation of plaques, which are found in patient brains prior to the onset of clinical symptoms, and it has been shown that Aβ alone is not sufficient to induce disease pathogenesis (Herrup, 2015). This suggests a more complex pathological mechanism at play that led to the development of a variety of alternative hypotheses surrounding AD etiology.
One prevailing proposal is the ApoE cascade hypothesis (Martens et al., 2022), based on the genetic association of LOAD with the apolipoprotein ε4 allele (APOE4) (Corder et al., 1993). APOE4 carriers have a vastly increased risk of developing LOAD and a reduced age of onset compared to carriers of the APOE3 and APOE2 isoforms (Corder et al., 1993; Sando et al., 2008). ApoE is a 34 kDa lipid-binding protein that primarily functions in lipid transport and metabolism (Mahley, 2016), with differences among the ApoE2, E3, and E4 isoforms confined to two sites (residues 112 and 158) (Najm et al., 2019; Martens et al., 2022). The primary functional difference in ApoE4 is its binding preference for very-low density lipoprotein (VLDL) over high-density lipoprotein (HDL), whereas the ApoE2 and ApoE3 isoforms preferentially bind HDL (Raffai et al., 2001; Belloy et al., 2019). While ApoE contributes to Aβ synthesis, accumulation, and clearance in an isoform-dependent manner, there is conflicting evidence on the isoform-specific roles of ApoE in Aβ pathology (Koistinaho et al., 2004; Castellano et al., 2011; Huang and Mahley, 2014; Najm et al., 2019). The ApoE cascade hypothesis does not include Aβ as a contributing factor in disease pathogenesis, stating that the biophysical and structural properties dependent of the ApoE isoform initiate a cascade of events driving AD and aging-related pathogenic condition (Martens et al., 2022). ApoE4 has been shown to alter lipid homeostasis and metabolism, leading to changes in lipid droplet formation, cholesterol turnover, and the PC/PE ratio (Farmer et al., 2019; Lazar et al., 2022; Yang et al., 2023). Consistent with MAM alterations, several lines of work support the involvement of ApoE in mitochondrial dysfunction as a contributing component to disease pathogenesis, including recent findings suggesting mitochondrial dysfunction influences ApoE expression and secretion (Dose et al., 2016; Martens et al., 2022; Wynne et al., 2023).
AD is associated with pronounced defects in mitochondrial function (Cenini and Voos, 2019; Wang et al., 2020c). These include alterations in mitochondrial biogenesis and morphology, and decreases in mitochondrial number (Hirai et al., 2001; Qin et al., 2009; Oka et al., 2016; Brustovetsky et al., 2023); compromised energy metabolism, including glucose hypometabolism (Kumar et al., 2022) and deterioration of the TCA and OXPHOS systems (particularly CIV) (Bubber et al., 2005; Liang et al., 2008; Zhang et al., 2015; Mastroeni et al., 2017; Sorrentino et al., 2017; Adav et al., 2019; Ryu et al., 2021); oxidative stress (Misrani et al., 2021); effects on mitochondrial dynamics, most notably a general increase in fragmentation (Wang et al., 2008a; Wang et al., 2009); impairment of mitochondrial trafficking (Calkins et al., 2011); and altered ER-mitochondria apposition with associated cellular Ca2+ dyshomeostasis (Leal et al., 2020; Fernandes et al., 2021; Li et al., 2023). While these effects could be associated with the interaction of APP/Aβ with mitochondria (see below), other AD-related factors could also be involved.
4.2 Parkinson’s disease and the role of α-syn
PD involves the selective degeneration of nigrostriatal dopaminergic neurons, manifesting as progressive effects on movement that include rigidity, resting tremor, and bradykinesia (Zhai et al., 2018). The main histological features of PD are cytoplasmic inclusions called Lewy bodies (LBs) and neuritic inclusions called Lewy neurites (LNs) whose core component is α-syn (Baba et al., 1998; Spillantini et al., 1998) but also contain vesicular structures, membranes and fragmented organelles (Shahmoradian et al., 2019; Ericsson et al., 2021). These α-syn aggregates are characteristic of a general group of neurodegenerative disorders termed synucleinopathies. α-syn is a 140 residue protein encoded by the synuclein alpha (SNCA) gene that contains three domains: (i) an N-terminal amphipathic region, (ii) a highly hydrophobic region called the non-amyloid-β component (NAβC), and (iii) a C-terminal acidic region (Bisi et al., 2021) (Figure 7A). Being enriched in the axon terminals of presynaptic neurons, α-syn is a membrane-interactive protein that plays a role in synaptic function, including vesicle trafficking and exocytosis by assembly of SNARE complexes, synaptic membrane remodeling, and maintenance of neurotransmitter vesicle pools (Bendor et al., 2013; Burre, 2015). α-syn also localizes to the nucleus, where it regulates gene expression (Somayaji et al., 2021).
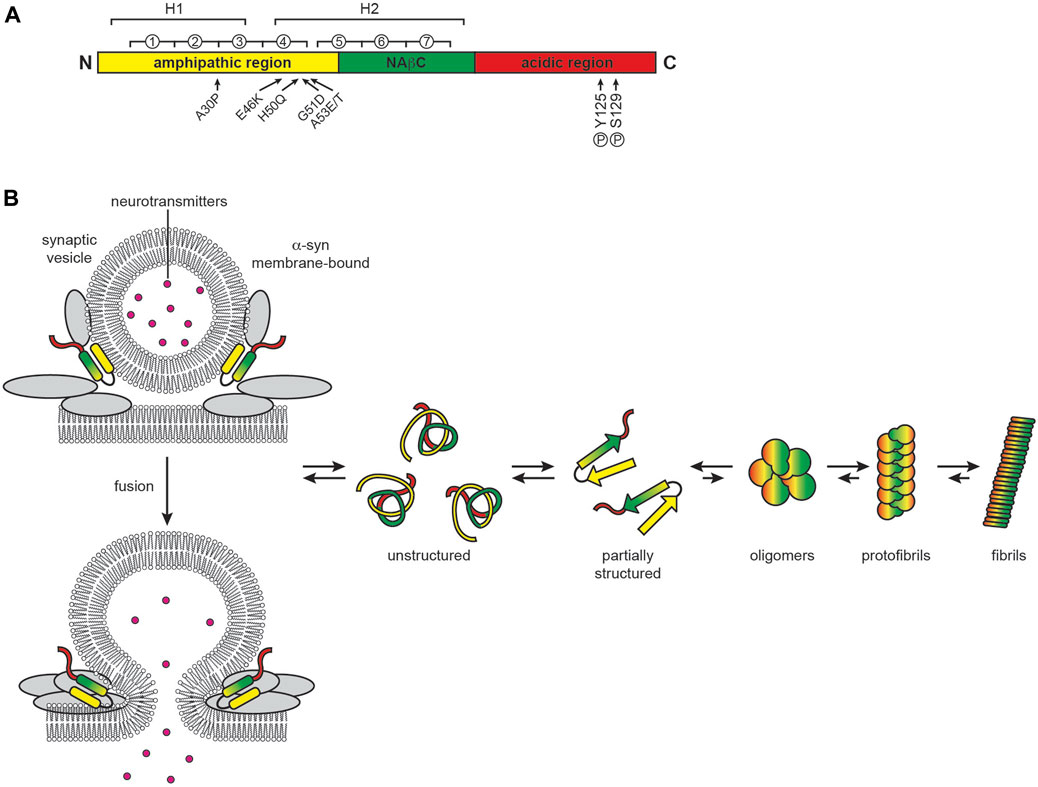
FIGURE 7. PD and the role of α-syn. (A) α-syn domain organization. α-syn contains an N-terminal amphipathic region involved in membrane interactions (yellow, residues 1–60), a central non-amyloid β component (NAβC) responsible for protein fibrillization (green, residues 61–95) and a highly acidic C-terminal region (red, residues 96–140). Circled numbers indicate the seven 11-residue repeats containing the KTKEGV motif; H1 and H2 indicate the regions of helical propensity that form two helices in the micelle-bound state or can form an elongated helix when bound to lower-curvature membranes. Arrows indicate sites of point mutations and phosphorylation, as indicated, that enhance α-syn misfolding and aggregation (B) α-syn function and aggregation. When membrane bound, α-syn folds into a single elongated helix or forms a two-helix structure. In solution, α-syn is largely disordered and prone to aggregation and fibrillization.
Monomeric α-syn is intrinsically disordered in solution, dynamically interconverting among many different conformations that differ with respect to short- and long-range electrostatic interactions and transient secondary structures (Weinreb et al., 1996). The N-terminus of α-syn in solution has helical propensity with a low fractional population assuming α-helix conformation; however, when bound to membranes, it adopts a stable helical structure that is either a continuous helix or two separate helices (Davidson et al., 1998; Bussell and Eliezer, 2003; Chandra et al., 2003; Ulmer et al., 2005; Croke et al., 2008; Georgieva et al., 2008; Jao et al., 2008; Stockl et al., 2008; Cho et al., 2009; Bodner et al., 2010; Rao et al., 2010; Croke et al., 2011; Ullman et al., 2011; Adao et al., 2020; Musteikyte et al., 2021). Membrane binding is facilitated by seven imperfect repeats of the 11-residue pattern xKTKEGVxxxx that extend from the amphipathic domain into the NAβC domain, a motif also found in apolipoproteins A2 (Bussell and Eliezer, 2003). This membrane-interactive region imparts classic amphitropic behavior to α-syn, which consequently promotes its interaction with curved bilayers, particularly with those enriched in negatively charged phospholipids (Jo et al., 2002) and cholesterol (Fantini et al., 2011). This membrane-binding activity is critical for its function in synaptic vesicle interaction (Fortin et al., 2005; Sarchione et al., 2021). The hydrophobic NAβC domain nucleates α-syn aggregation, forming the canonical β-structure of amyloid fibrils along with parts of the N-terminal domain (Rodriguez et al., 2015). The fraction of α-syn that natively exists in neurons in a membrane-bound state, as an unfolded monomer (Fauvet et al., 2012; Theillet et al., 2016), or as aggregation-resistant folded tetramers (Bartels et al., 2011; Wang et al., 2011) remains an open question.
Familial autosomal dominant PD is linked to defects in several genes, most of which have a connection to mitochondrial physiology (Cieri et al., 2017; Billingsley et al., 2018). Among them, the six known heritable mutations in the SNCA gene (A30P, E46K, H50Q, G51D, and A53E/T) all localize to the amphipathic N-terminal region of α-syn and promote aggregation (Flagmeier et al., 2016). α-syn aggregation can also be potentiated by increases in its copy number (Srinivasan et al., 2021). However, PD is primarily an idiopathic disease in which aging-related increases in α-syn aggregation and/or certain post-translational modifications of α-syn are commonly involved (Anderson et al., 2006; Manzanza et al., 2021; Roshanbin et al., 2021). The balance between α-syn function and pathogenicity is depicted in Figure 7B.
Mitochondrial dysfunction plays a key role in the pathogenesis of PD (Vicario et al., 2018). The enhanced expression of α-syn or particular mutants of α-syn known to promote PD cause the disruption of mitochondrial dynamics (Xie and Chung, 2012; Thorne and Tumbarello, 2022) and promote fragmentation (Kamp et al., 2010; Nakamura et al., 2011; Gui et al., 2012; Pozo Devoto et al., 2017), impair mitochondrial trafficking (Pozo Devoto et al., 2017) and autophagic clearance (Gao et al., 2017), impact mitochondrial energetics (Banerjee et al., 2010), and alter Ca2+ flux by the enhancement (Cali et al., 2012) or disruption (Guardia-Laguarta et al., 2014; Guardia-Laguarta et al., 2015; Paillusson et al., 2017) of specific ER-mitochondria tethering sites. One of the most prominent effects of PD is the impairment of respiratory Complex I associated with reduced energetic output and an overproduction of ROS (Devi et al., 2008; Marella et al., 2009). This multitude of effects is in line with the many mitochondrial subcompartments α-syn has been found to target, including the MOM (Li et al., 2007; Cole et al., 2008; McFarland et al., 2008; Nakamura et al., 2008; Zhang et al., 2008; Kamp et al., 2010; Nakamura et al., 2011; Di Maio et al., 2016a; Pozo Devoto et al., 2017), the MIM/IMS (McFarland et al., 2008; Nakamura et al., 2011; Zhu et al., 2011; Lu et al., 2013; Robotta et al., 2014; Amorim et al., 2017), the matrix (McFarland et al., 2008; Ludtmann et al., 2016; Ludtmann et al., 2018) and the MAM (Cali et al., 2012; Guardia-Laguarta et al., 2014; Paillusson et al., 2017). Importantly, this extensive interaction of α-syn with mitochondria is mediated in part through lipid bilayer interactions, particularly with regions enriched with the mitochondrial lipid cardiolipin (Ramakrishnan et al., 2003; Cole et al., 2008; Nakamura et al., 2008; Grey et al., 2011; Zigoneanu et al., 2012; Robotta et al., 2014; Ryan et al., 2018) that provides the negative surface, high curvature, and acyl packing defects necessary to promote α-syn binding (Sharon et al., 2003; Nuscher et al., 2004; Middleton and Rhoades, 2010; Pfefferkorn et al., 2012; Ouberai et al., 2013; Gilmozzi et al., 2020).
4.3 Huntington’s disease and the role of mHtt
HD is associated with neurodegeneration of the basal ganglion, with preferential deterioration of striatal medium spiny GABAergic neurons, that clinically presents as progressive loss of motor control and cognition (Morigaki and Goto, 2017). This monogenic disease is caused by heritable alterations in Htt (Jurcau and Jurcau, 2022). Structurally, Htt is a 348 kDa protein that contains two domains of HEAT tandem repeats that form α-solenoid structures connected by a bridge domain, and an N-terminal region with a tripartite organization that contains a highly conserved sequence of 17 N-terminal amino acids (Nt17), a stretch of glutamine residues (poly-Q), and a proline-rich domain (PRD) (Saudou and Humbert, 2016; Guo et al., 2018) (Figure 8A). Htt is conformationally dynamic, with an extensive list of interaction partners likely owing to the protein-interaction functions of its HEAT repeats (Shirasaki et al., 2012). As such, it serves as a multivalent molecular scaffold, a function that likely supports its many interactions in synapses including axonal transport, vesicle recycling, autophagy, transcriptional regulation, and endocytosis (Barron et al., 2021).
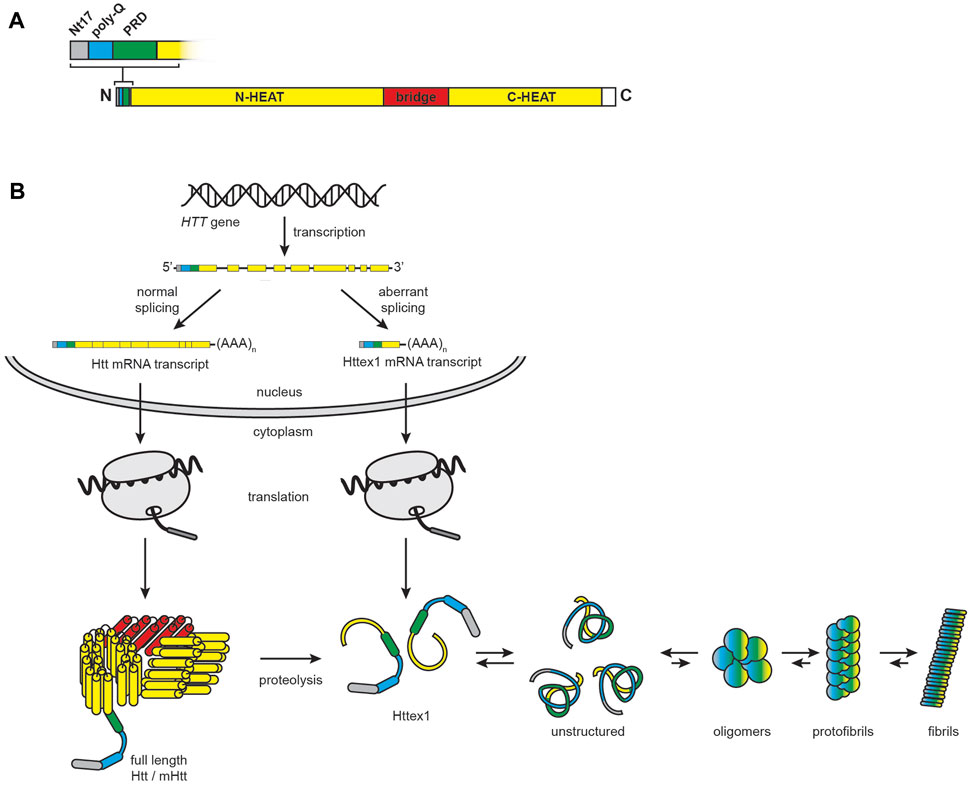
FIGURE 8. HD and the role of mHtt. (A) Htt domain organization. Full-length huntingtin is a 3144-residue protein that consists of N-terminal (residues 91–1683) and C-terminal (residues 2092–3057) α-helical solenoid domains containing multiple HEAT (huntingtin, elongation factor 3, protein phosphatase 2A, lipid kinase TOR) repeats (yellow) connected by a helical linker region (red, residues 1684–2091). The full-length protein also features an N-terminal region consisting of the Nt17 sequence (grey, residues 1–17), the poly-Q tract, which is expanded in HD (cyan, residues 18–40), and a proline-rich domain (green, residues 41–90). (B) Htt biogenesis, processing, and misfolding. In the nucleus, normal splicing of the HTT gene results in a full-length transcript, whereas alternative splicing and premature polyadenylation results in a transcript encoding Httex1. Following translation in the cytosol, full-length Htt is subject to extensive proteolysis, which also contributes to the pool of Httex1 protein. Httex1 is unstructured in solution and prone to aggregation and fibrillization.
HD is an autosomal dominant disorder caused by an extension of the poly-Q tracts resulting from polymorphic CAG trinucleotide repeat expansion in exon 1 of the HTT gene (MacDonald et al., 1993). HD is part of a larger family of neurodegenerative diseases associated with polyglutamine expansion of proteins (Lieberman et al., 2019). Whereas Htt in healthy individuals contains fewer than 36 CAG repeats, the pathogenically expanded poly-Q region can contain between 42 and 250 CAG repeats, with disease severity and earlier age of onset directly related to the extent of expansion (Rubinsztein et al., 1996; Penney et al., 1997). Because Htt is ubiquitously expressed, HD affects not only the brain but also peripheral tissues with high metabolic activity (Chuang and Demontis, 2021). Poly-Q expansion causes the resulting mHtt protein to have altered folding (Vijayvargia et al., 2016) and interactions (Ratovitski et al., 2012; Greco et al., 2022). This makes mHtt prone to aggregation, forming inclusion bodies in the cytoplasm and nucleus (Davies et al., 1997) with diverse aggregate structures (Tanaka et al., 2001; Poirier et al., 2002) and a complex composition that includes lipids, proteins, and membrane-bound organelles (Kegel-Gleason, 2013; Riguet et al., 2021). Notably, the primary cytotoxic, aggregation-prone species in HD may not be full-length mHtt, but rather heterogeneous populations of N-terminal fragments of mHtt exon 1 (Httex1) (Mangiarini et al., 1996; Cooper et al., 1998; Hackam et al., 1998; Martindale et al., 1998; Hoffner et al., 2005; Barbaro et al., 2015) that can arise by cleavage of the mature protein by caspase, calpain, or other proteases (Goldberg et al., 1996; Wellington et al., 1998; Kim et al., 2001; Gafni and Ellerby, 2002; Lunkes et al., 2002; Graham et al., 2006; Schilling et al., 2007; Landles et al., 2010; Tebbenkamp et al., 2012; Martin et al., 2019), or by aberrant splicing and premature polyadenylation of pathogenic HTT exon 1 with expanded CAG repeats (HTTex1 transcripts) (Sathasivam et al., 2013; Neueder et al., 2017; Neueder et al., 2018) (Figure 8B).
Mitochondrial dysfunction plays a central role in HD pathogenesis (Bossy-Wetzel et al., 2008; Carmo et al., 2018; Jurcau and Jurcau, 2023). HD is associated with reduced mitochondrial biogenesis and quality control (Steffan et al., 2000; Cui et al., 2006; Weydt et al., 2006; Jin and Johnson, 2010; Wong and Holzbaur, 2014; Khalil et al., 2015; Guo et al., 2016; Dubois et al., 2021; Sonsky et al., 2021), mtDNA heteroplasmy (Wang et al., 2021), defective energy metabolism and OXPHOS activity (Sawa et al., 1999; Klivenyi et al., 2004; Seong et al., 2005; Benchoua et al., 2006; Pandey et al., 2008; Mochel et al., 2012; Silva et al., 2013; Naia et al., 2015; Burtscher et al., 2020), altered mitochondrial Ca2+ handling and sensitivity to the mitochondrial permeability transition pore (mPTP) (Panov et al., 2002; Choo et al., 2004; Milakovic et al., 2006; Giacomello et al., 2013), increased mitochondrial oxidative stress (Sorolla et al., 2008; Chae et al., 2012; Ribeiro et al., 2012; Ribeiro et al., 2014; Chen et al., 2017; Moretti et al., 2021; Lopes et al., 2022; Egidio et al., 2023), defective mitochondrial dynamics and hyper-fission (Costa et al., 2010; Song et al., 2011; Shirendeb et al., 2012; Manczak and Reddy, 2015; Cherubini et al., 2020), defective mitochondrial trafficking (Trushina et al., 2004; Chang et al., 2006; Orr et al., 2008; Shirendeb et al., 2012; Berth and Lloyd, 2023), and altered cytochrome c release with apoptosis (Chen et al., 2000; Kiechle et al., 2002; Zhang et al., 2006; Wang et al., 2008b). Although many of these effects are related to spurious interactions of mHtt with cytosolic proteins, there are several reports that full-length and N-terminal fragments of Htt/mHtt interact directly with mitochondria (Panov et al., 2002; Choo et al., 2004; Petrasch-Parwez et al., 2007; Orr et al., 2008; Song et al., 2011; Guo et al., 2016). Some of these studies specifically implicate interactions with the MOM (Hamilton et al., 2020) and some show interactions with internal mitochondrial compartments (Yano et al., 2014; Yablonska et al., 2019). In general, accumulation of N-terminal mHtt fragments with mitochondria increases with age (Orr et al., 2008). Potential interactions between Htt/mHtt and the mitochondrial import machinery are discussed below.
5 Interactions of amyloidogenic proteins with the mitochondrial protein import machinery
5.1 Noncanonical and multi-specific targeting signals
Most proteins that are targeted to cellular compartments other than their site of ribosomal synthesis contain unambiguous targeting sequences that faithfully direct them to a specific location. For example, as described above, the MTSs of TIM23 substrates form an N-terminal amphipathic α-helix that serves as the primary recognition element of the TIM23 import machinery. Three examples of MTSs shown in Figure 9A illustrate their amphipathic character: they have an appreciable number of nonpolar residues quantified as the mean hydrophobicity (<H>) (Fauchere and Pliska, 1983), a net charge (z) that reflects a preponderance of basic residues, and an asymmetric distribution of nonpolar and basic residues quantified by the hydrophobic moment (<μH>) (Eisenberg et al., 1982). Yet it is also notable that MTSs vary significantly with respect to sequence, length, and extent of amphipathicity. Thus, while canonical MTSs encompass a range of physicochemical properties, they are sufficiently well-defined enough to be recognized by a single import pathway.
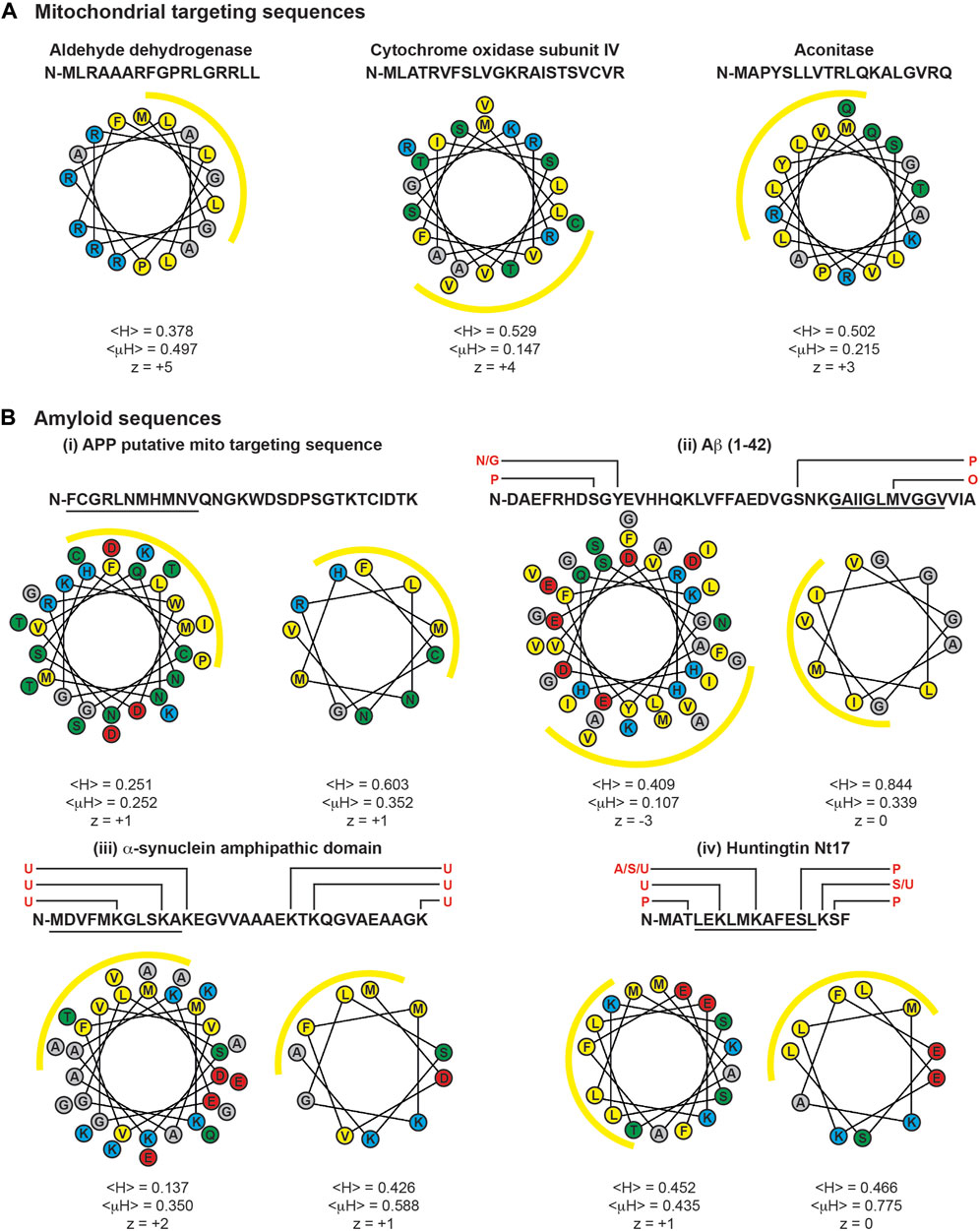
FIGURE 9. MTSs and cryptic targeting sequences of amyloids. Sequences and helical wheel diagrams of MTSs and select regions of amyloidogenic proteins are depicted. Helical projections and parameters were determined using the HeliQuest server (Gautier et al., 2008). Mean hydrophobicity is denoted as <H>, hydrophobic moment as <μH>, and net charge as z. For all projections, the N-terminal residue is oriented due North (0o), and yellow arcs indicate the hydrophobic face of the amphipathic helices determined by <μH> values. (A) Example MTSs. Human variants of select TIM23 substrates aldehyde dehydrogenase, subunit IV of Cytochrome c oxidase, and aconitase are shown. (B) Cryptic targeting signals of amyloidogenic proteins. These include the putative MTS of: (i) APP, (ii) Aβ (1-42), (iii) α-syn (1-32), and (iv) huntingtin (1-17). Helical projections to the left show the entire sequence. Helical projections to the right show the 11-residue (∼3 helical turn) section with the strongest amphipathic character, indicated by the underlined residues. Known sites of post-translational modification are indicated above the primary sequences in red (A, acetylation; G, glycosylation; N, nitration; O, oxidation; P, phosphorylation; S, SUMOylation; U, ubiquitinylation). Single letter amino acid codes colored by side chain functionality: yellow, hydrophobic; red, acidic; cyan, basic; green, polar uncharged.
For some proteins, however, the relationship between the targeting sequence and the organellar import pathway is not straightforward and some targeting sequences can direct passenger proteins to multiple cellular compartments by non-mutually exclusive mechanisms (Karniely and Pines, 2005). Indeed, targeting fidelity requires multiple levels of regulation and precise recognition because mitochondrial MTSs, ER signal peptides, chloroplast transit peptides, and even some peroxisomal targeting signals share similarity in that they all form N-terminal amphipathic α-helices with a hydrophobic face and a basic (or polar) face (Kunze and Berger, 2015).
Some proteins contain an ambiguous (or cryptic) targeting sequence that can be recognized by multiple import machineries (Lu et al., 2004; Ventura et al., 2004; Chatre et al., 2009). Other proteins contain well-defined targeting signals whose accessibility or targeting efficiency can be modified by folding, protein binding, or post-translational modifications (Karniely and Pines, 2005). Still other proteins contain multiple targeting signals that direct them to different compartments (Petrova et al., 2004; Pino et al., 2007). For example, some cytochromes P450 (CYPs) expressed in hepatocytes contain chimeric signals with amino-terminal ER-targeting sequences in tandem with a mitochondrial-targeting sequence (Addya et al., 1997; Anandatheerthavarada et al., 1999; Robin et al., 2001; Robin et al., 2002). The bipartite targeting sequence of these cytochromes contain a cryptic mitochondria localization signal that remains idle until activation. In the case of CYP2E1, the mitochondrial targeting signal is activated by cyclic-AMP-dependent phosphorylation of a Ser residue in the cryptic sequence (Robin et al., 2002). Moreover, an inducible endoprotease has been identified that activates mitochondrial import by cleaving bimodal targeting signals to expose the cryptic mitochondrial targeting signal (Boopathi et al., 2008). It is unclear whether such cleavage of chimeric signals is a general requirement for mitochondrial targeting.
By allowing greater diversity of potential cellular locations, noncanonical targeting imparts flexibility to the functional range of individual proteins that can be subject to regulation by the cell in response to different physiological demands. However, cryptic targeting sequences can also cause the pathogenic accumulation of proteins in organelles. In the following sections, we review the cryptic mitochondria targeting sequences present in Aβ/APP, α-syn, and Htt/mHtt that may facilitate their interactions with the mitochondrial import machinery.
5.2 Interactions of Aβ and APP with the mitochondrial import machinery
Several lines of evidence indicate that full-length APP engages the TIM23 import pathway. The canonical biogenesis route of APP entails targeting to the ER via the SRP/Sec61 pathway, translocation across the ER membrane attendant with glycosylation of the ectodomain, integration as a type I membrane protein, and trafficking by the secretory, endocytic and recycling routes (Muresan and Ladescu Muresan, 2015). APP contains a classic N-terminal Sec-targeting signal with a predicted Signal Peptidase cleavage site (AxA) between positions 17 and 18 (Auclair et al., 2012; Almagro Armenteros et al., 2019). Following this ER-targeting signal is a potential cryptic MTS spanning residues 37 to 67, segments of which could form an amphipathic helix (Figure 9Bi). Thus, together they form a chimeric ER-mitochondria targeting signal (Devi and Anandatheerthavarada, 2010). Indeed, it has been shown using human cortical neuronal cells that a non-glycosylated form of APP695 can stall in the mitochondrial import pathway with a predicted Nmito/Ccyto topology. This stalled intermediate made crosslinking-detected interactions with Tom40, Tim23, and Tim44, in a manner that required the Δψm and the positive residues Arg40, His44, and Lys51, all three of which reside on a common face of the putative MTS (Anandatheerthavarada et al., 2003) (Figure 9Bi). Similar translocation intermediates of APP were observed in mitochondria of transgenic AD mouse models (Anandatheerthavarada et al., 2003) and in postmortem samples of AD brains (Devi et al., 2006), the latter showing stable association of APP with ∼480 kDa complexes containing the TOM machinery and ∼620 kDa complexes containing the TIM23 machinery. The stalling of APP in the import machinery appeared to be due to the tight folding of the APP acidic domain (residues 220–290) blocking transport along the TOM complex, as deletion of this domain facilitated complete translocation (Anandatheerthavarada et al., 2003). This observation is consistent with the formation of stalled translocation intermediates observed with native mitochondria-targeted substrates with tightly folded (cofactor-bound) C-terminal domains and fusion constructs with MTSs fused to cofactor-stabilized domains like DHFR (Schneider, 2018). In agreement with the known effects of AD on mitochondria function, APP translocation intermediates were shown to interfere with the import of native mitochondrial proteins (Devi et al., 2006). The extent to which mitochondrial targeting of APP may be facilitated by endoprotease cleavage of the ER targeting signal to expose the cryptic MTS (Boopathi et al., 2008) is an open question. Figure 10A (left) summarizes the interactions of APP with mitochondria.
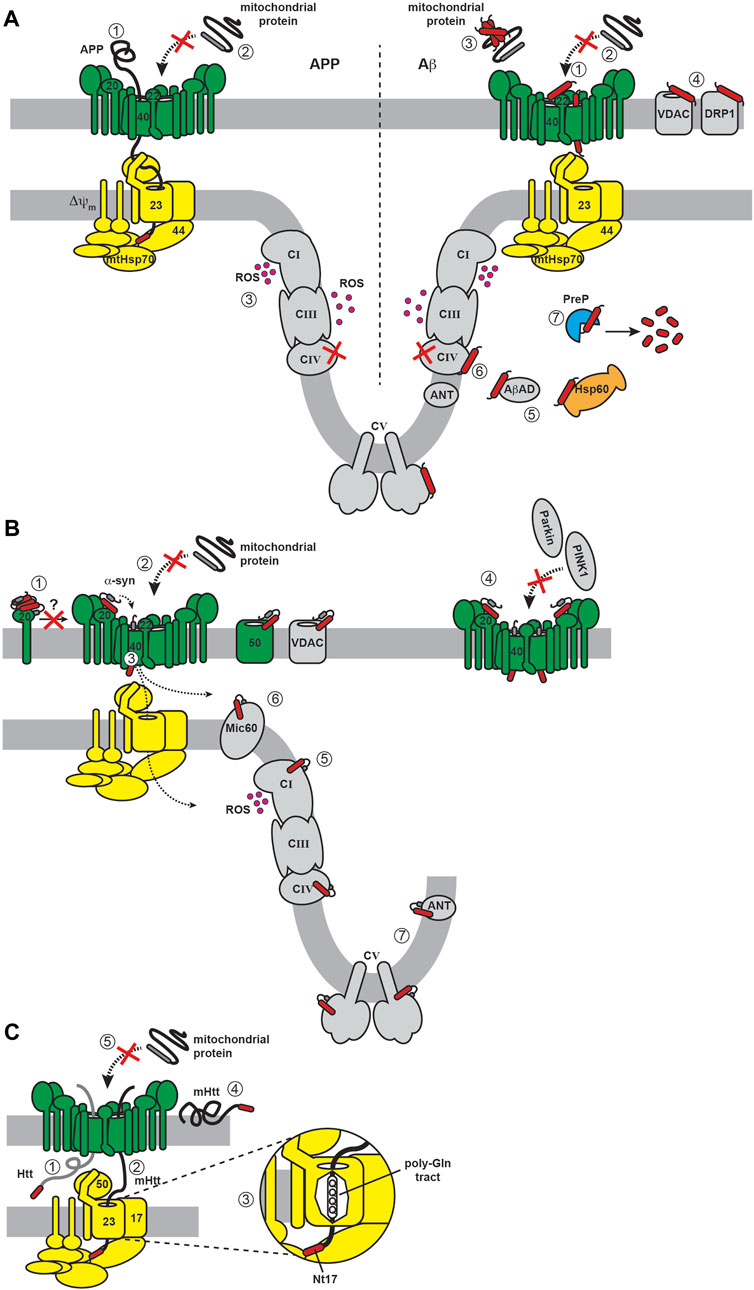
FIGURE 10. Interactions of amyloidogenic proteins with the TIM23 complex. Models show import complex color scheme used in Figure 2 (TOM/SAM complex, green; TIM23 complex, yellow) with other proteins depicted in grey. Subunits known to interact with amyloids are explicitly indicated by their names. Putative cryptic targeting signals of Aβ/APP, α-syn, and Htt/mHtt are shown in red. [(A), left] APP. (1) APP forms a two membrane-spanning intermediate, engaging both TOM and TIM23 complexes in a Δψm-dependent manner, with complete translocation blocked by the tight folding of the C-terminal AICD domain. (2) Stalled APP intermediates block the import of native mitochondrial precursor proteins. (A, right) Aβ. (1) Aβ interacts with the TOM complex (subunits Tom22 and Tom40), which (2) disrupts the import of native mitochondrial precursors. (3) Aβ forms co-aggregates with mitochondrial precursors in the cytosol. Aβ interacts with proteins of the (4) MOM, (5) matrix, and (6) MIM, most notably CIV and CV. (7) Aβ is degraded by the matrix PreP protease. A general feature of APP/Aβ stress is dysfunctional CIV and excess ROS production by the OXPHOS machinery. (B) α-syn. (1) Multimers of α-syn interact with free Tom20, blocking its assembly with the TOM complex. The engagement of α-syn with TOM complex receptors and channel may (2) hinder import of native mitochondrial proteins, (3) reduce Tom40 expression, and/or (4) disrupt PINK1/Parkin-mediated mitophagy; (5) Interaction of α-syn with CI or CIV may disrupt ETC activity and cause ROS overproduction; (6) Interaction of α-syn with MICOS subunit Mic60 may affect cristae morphogenesis; (7) Interaction of α-syn with ATP synthase (CV) or the adenine nucleotide translocase (ANT) may disrupt adenine nucleotide flux and/or ATP production. (C) Htt/mHtt. (1) Htt may engage the TOM machinery with a tendency to accumulate in the IMS. (2) mHtt engages the TIM23 complex, (3) possibly facilitated by the unstructured poly-Q segment C-terminal to Nt17, which provides a flexible linker for Nt17 to engage TIM23 subunits. (4) mHtt may alternatively accumulate on the cytosolic side of the MOM. (5) Blockage of the import machinery by Htt/mHtt hinders import of mitochondrial precursors.
The Aβ peptide has also been widely reported to accumulate in mitochondria through the import machinery. The presence of Aβ within mitochondria is well documented by its direct interactions with many mitochondrial proteins, including MOM proteins VDAC (Manczak and Reddy, 2012) and Drp1 (Manczak et al., 2011), matrix proteins Hsp60 (Caspersen et al., 2005), Aβ-binding alcohol dehydrogenase (ABAD) (Lustbader et al., 2004) and cyclophilin D (Du et al., 2008), and OXPHOS complexes including Complex IV (Hernandez-Zimbron et al., 2012) and ATP synthase (Beck SJ. et al., 2016). Furthermore, a pronounced feature of AD pathogenesis is the mitochondrial accumulation of Aβ (Caspersen et al., 2005; Manczak et al., 2006; Cha et al., 2012). Aβ42 has been shown to enter mitochondria through the TOM complex, associating with the MIM fraction in a Δψm-independent manner and localizing to cristae in both in vitro import systems and in mitochondria from human brain biopsies (Hansson Petersen et al., 2008). The specific interactions of Aβ with the TOM complex were explored using a yeast mitochondria model system, indicating that Aβ binds directly to the Tom22 receptor, but not Tom20 or Tom70, and that Aβ residues 25–42 were indispensable for this interaction (Hu et al., 2018). The accumulation of Aβ within the import machinery is functionally relevant because it inhibits the import of native nuclear-encoded proteins (Sirk et al., 2007). However, it should also be noted that cytosolic Aβ may inhibit the import of mitochondrial proteins by coaggregation in the cytosol that does not involve blocking the mitochondrial import machinery (Cenini et al., 2016). A common theme in these studies is that the more aggregation-prone Aβ42 has a stronger interaction with the import machinery than the Aβ40 variant (Sirk et al., 2007; Cenini et al., 2016; Hu et al., 2018). Figure 10A (right) summarizes the interactions of Aβ with mitochondria.
As mentioned, ApoE4, the primary genetic risk factor for AD, induces increased connectivity between MAMs and mitochondria in different AD models (Area-Gomez et al., 2012; Tambini et al., 2016). MAMs have lipid raft-like membranes, enriched in sphingolipids and cholesterol, which promote the interaction between APP and its processing enzymes (β-secretase and γ-secretase) that have been found to localize at the MAM. The biophysical properties of these microdomains may enhance amyloidogenic processing of APP (Diaz et al., 2015; Del Prete et al., 2017; Area-Gomez et al., 2018). Additionally, the protein interactome of MAMs in cells with mutant APP is enriched in mitochondrial import components Tom22, Tim17b, and Sam50 (Del Prete et al., 2017). Enhanced proximity of APP to the mitochondrial import machinery could contribute to spurious import of APP.
There are some potentially confounding factors regarding the import of the Aβ peptide through the mitochondrial import machinery. The first is that Aβ produced from APP proteolysis is released into exocytoplasmic compartments (ER lumen or the extracellular space), whereas engagement with the mitochondrial import machinery occurs from the cytosol. However, there is evidence that proteolytically produced or externally added Aβ can be taken up by cells via clathrin-dependent or -independent endocytosis, followed by endosomal escape, which could allow it to localize to mitochondria (Saavedra et al., 2007; Hansson Petersen et al., 2008; Berridge, 2010; Friedrich et al., 2010). Furthermore, Aβ produced in the vicinity of mitochondria by MAM-localized secretases could directly access mitochondria (Area-Gomez et al., 2009). The second is that the presence of Aβ in the matrix would necessitate TIM23-mediated translocation; however, to date there is no evidence of a direct interaction between Aβ and the TIM23 complex. One explanation could be that the association between the short Aβ peptide and the TIM23 receptors and channel is too transient to be captured by techniques like crosslinking or immunoprecipitation. Lastly, the Aβ sequence does not contain any segments with strong amphipathicity (opposing basic and nonpolar faces) that are present in canonical MTSs (Figure 9Bii). This feature may hinder transport along the “acid chain” of the TIM23 pathway, possibly explaining how Aβ could stall nonproductively at the TOM complex.
It is noteworthy that AD is associated with a general disruption of the mitochondrial import machinery. Analysis of RNA-seq datasets from brains of AD patients versus age-matched healthy individuals showed that expression of mitochondrial import genes was decreased with AD (Sorrentino et al., 2017), consistent with the observed decrease in expression of Tom20 and Tom70 in postmortem neocortex samples of AD patients that correlate with higher Aβ42/Aβ40 ratios (Chai et al., 2018). It has also been proposed that the length of a poly-T polymorphism in the TOMM40 gene correlates with LOAD, supporting the involvement of the TOM complex channel in AD pathogenesis (Roses et al., 2010); however, this finding has been questioned (Chiba-Falek et al., 2018). It is possible that altered expression of TOM complex subunits could modulate mitochondrial interactions of APP/Aβ; for instance, decreased Tom20/70 expression relative to Tom22 could promote Aβ binding to the TOM complex (Hu et al., 2018).
The most compelling evidence that the mitochondrial import and processing machinery serves as a mechanism for amyloid clearance comes from the interaction between Aβ and PreP, the matrix-localized protease that digests MTSs (Pinho et al., 2014). This degradation step is critical because MTSs, being amphipathic and membrane-interactive, can disrupt MIM integrity and their buildup can lead to Δψm collapse and OXPHOS uncoupling (Teixeira and Glaser, 2013). Importantly, PreP was shown to degrade Aβ variants in vitro (Falkevall et al., 2006) and PreP activity is lower in the temporal lobes of AD patients and transgenic AD mice compared with controls, which may be further attributed to disease-related oxidative modifications (Alikhani et al., 2011b). Given the functional coupling between precursor processing and MTS turnover (Kucukkose et al., 2021), the accumulation of Aβ could overcome the MTS-clearing capacity of PreP and cause feedback inhibition of MIP and MPP, thereby inhibiting import by the toxic buildup of precursors and MTSs in the matrix (Mossmann et al., 2014). Conversely, overexpression of PreP in AD mouse models decreases mitochondrial Aβ concentrations and improves organellar function (Fang et al., 2015; Brunetti et al., 2016; Du et al., 2021). Given the key role of PreP in proteostasis (Brunetti et al., 2021), mitochondrial import of Aβ and its subsequent degradation may be essential for balancing cellular Aβ concentrations.
5.3 Interactions of α-syn with the mitochondrial import machinery
Several studies support the entry of α-syn into mitochondria through the import complexes. α-syn was first shown to accumulate in mitochondria in a manner that required the Δψm across the MIM and an accessible Tom40 channel (Devi et al., 2008). Furthermore, the possibility that the N-terminus of α-syn targets mitochondria is supported by the indispensable role of residues 1–32 in mitochondrial uptake (Devi et al., 2008) and of residues 1–11 in association of α-syn with isolated mitochondria (Robotta et al., 2014). When folded as an α-helix (stabilized by membrane interactions), the N-terminal 32 residues of α-syn form an amphipathic structure (Figure 9Biii), made up in part by the first two imperfect KTKEGV repeats that produce a Lys-rich basic face and a Val-rich nonpolar face. The N-terminus of α-syn therefore has strong potential to act as a cryptic MTS (Devi and Anandatheerthavarada, 2010).
TIM23-mediated uptake of α-syn is also supported by its physical and genetic interactions with subunits of the import machinery. Based on proteomics analysis of the interactome of the α-syn C-terminal peptide and its variants phosphorylated at Tyr125 and Ser129 that are known to promote aggregation (Samuel et al., 2016; Fayyad et al., 2020), the unmodified and phosphorylated peptides were found to preferentially interact with mitochondrial and cytosolic proteins, respectively (McFarland et al., 2008). In this study, the unmodified α-syn C-terminal peptide bound Tom40, Sam50, and Tom22, with the interaction to MOM proteins Tom40 and Sam50 strongly reduced by phosphorylation, suggesting the preferential import of unmodified α-syn (McFarland et al., 2008). This connection between α-syn aggregation propensity and mitochondrial import is further supported by the fact that all known PD-related missense mutations reside in the N-terminal region (Flagmeier et al., 2016) near the cryptic targeting sequence (Figure 7A). Perhaps more compelling are the specific ways in which N-terminal mutants of α-syn feature altered cellular interactions; namely, the A53T mutant shows increased affinity for mitochondria and the MAM, whereas the A30P mutant shows weaker affinity (Cole et al., 2008; Devi et al., 2008; Guardia-Laguarta et al., 2014; Pozo Devoto et al., 2017). This may be related to the decreased binding affinity of A30P mutant α-syn for lipid membranes (Jo et al., 2002; Fortin et al., 2004). Therefore, these PTMs and site mutations likely alter the aggregation propensity and membrane interactions of α-syn that subsequently affect its interaction with the mitochondrial import machinery in complex ways.
The involvement of the TOM complex in α-syn import was further investigated in two other studies. In one report, brain tissue taken from postmortem PD patients and transgenic mice overexpressing α-syn showed decreased expression of Tom40 concurrent with mtDNA damage, oxidative stress, and reduced bioenergetic efficiency (Bender et al., 2013). Another report used a rotenone-induced Complex I dysfunction model of PD, as well as postmortem brain tissue of PD patients, to show that α-syn (specifically, the S129 phosphomimetic and soluble oligomers) demonstrated strong but reversible binding to Tom20 and reduced Tom20 expression. This binding thereby prevented interactions between the Tom20/Tom22 receptors, inhibited the import of mitochondrial proteins, and inhibited respiration (Di Maio et al., 2016b). In these studies, the overexpression of either Tom40 (Bender et al., 2013) or Tom20 (Di Maio et al., 2016b; De Miranda et al., 2020) reversed α-syn aggregation and its associated mitochondrial defects. Taken together, these results show a direct interaction between α-syn and the TOM complex, although exactly how the oligomeric state of α-syn or its PTMs may modify such interactions remain open questions. It should be noted that to date, there is no strong evidence for the specific interaction between α-syn and the TIM23 import complex of the MIM. However, transport and sorting of α-syn via the TIM23 machinery is highly likely given the submitochondrial distribution of α-syn, which includes the MIM and matrix. Figure 10B summarizes the interactions of α-syn with mitochondria.
Another feature supporting the mitochondria-targeting capacity of the α-syn N-terminus is its membrane-interactive nature. In its functional role of fusing synaptic vesicles with the presynaptic membrane, α-syn binds anionic phospholipid bilayers through its N-terminus, which then adopts a stable α-helical structure and displaces the unstructured C-terminal end resulting in an elongated conformation (Bartels et al., 2010). Similar membrane interactions have long been known for bona fide mitochondrial MTSs, which can undergo a coil-to-helix transition upon anchoring to lipid bilayers without perturbing membrane integrity (Skerjanc et al., 1987; Hoyt et al., 1991; Wieprecht et al., 2000). Functionally, this could allow the mitochondrial precursor to undergo a two-dimensional random walk on the membrane surface to seek out its cognate import receptor more efficiently. It is possible that α-syn adopts a similar mechanism to accumulate on membrane surfaces to increase its local concentration in the vicinity of import complexes.
The interaction of α-syn with the mitochondrial import machinery has important mechanistic implications for mitochondrial stress responses and quality control related to PD (Thorne and Tumbarello, 2022). One of the main pathways for mitophagy-based removal of damaged mitochondria is the PINK1/Parkin system. Under non-stressed conditions, the serine/threonine kinase PINK1 engages mitochondrial import complexes and becomes proteolyzed and efficiently degraded; however, under stress conditions that lower the Δψm of the MIM, PINK1 accumulates at the MOM and recruits the E3 ubiquitin ligase Parkin, which ubiquitinylates mitochondrial proteins. This signals for autophagic degradation of dysfunctional organelles and consequently blunts cellular expansion of the damage (Ge et al., 2020). Importantly, defects in PINK1 and Parkin (encoded by PINK1 and PRKN genes, respectively) cause the deregulation of mitochondrial quality control and together they represent the preeminent monogenic forms of heritable PD (Klein and Westenberger, 2012).
Moreover, the specific affinity of the α-syn N-terminal region for cardiolipin (Ramakrishnan et al., 2003; Cole et al., 2008; Nakamura et al., 2008; Grey et al., 2011; Robotta et al., 2014) may explain its interaction with mitochondria during stress. The translocation of cardiolipin from the MIM to the MOM is an early signaling event in mitophagy and apoptosis (Li et al., 2015), and it has been shown using α-syn mutant models of PD that cardiolipin becomes externalized to the cytosol, where it then binds α-syn and promotes the refolding of α-syn monomers from aggregated fibrils (Ryan et al., 2018). This may represent a feed-forward process whereby the stress of α-syn burden causes cardiolipin externalization, thereby resulting in the recruitment of more α-syn to the mitochondrial surface. Mitophagy then results when α-syn burden outmatches the refolding ability of cardiolipin.
It should be emphasized that α-syn may play a physiological role in mitochondria (Faustini et al., 2017). For example, consistent with the general role that α-syn appears to play in cellular lipid metabolism and signaling (Jenco et al., 1998; Golovko et al., 2005; Narayanan et al., 2005; Golovko et al., 2006), the lack of α-syn in SNCA-/- mice causes mitochondria to have reduced cardiolipin with altered acyl compositions (Ellis et al., 2005; Barcelo-Coblijn et al., 2007). This effect on cardiolipin in turn alters the physical properties of the MIM coupled with reduced Complex I and III activity (Ellis et al., 2005) and is accompanied by increases in neutral lipids including cholesterol and cholesterol esters (Barcelo-Coblijn et al., 2007). Other studies indicate that α-syn may play critical roles in mitochondrial dynamics, quality control and transport (reviewed in (Pozo Devoto and Falzone, 2017)). While these roles of α-syn could in principle be exerted within or outside the mitochondrion, some reports indicate a role of α-syn within the organelle. For example, α-syn appears to interact directly with the matrix-facing catalytic domain of mitochondrial ATP synthase, with monomers positively regulating its catalytic activity (Ludtmann et al., 2016) and oligomers promoting the mitochondria permeability transition involved in cell death (Ludtmann et al., 2018). This functional duality indicates that, as with cytoplasmic α-syn, it may be the abundance and/or aggregation of α-syn within the mitochondrion and not the mere presence of α-syn itself, that dictates pathogenicity inside the organelle. As such, the TIM23 machinery must then play a key role in regulating this balance of mitochondrial α-syn.
5.4 Interactions of Htt/mHtt with the mitochondrial import machinery
In Htt, the site of pathogenic poly-Q expansion is flanked by the first 17 residues of the N-terminus (Nt17) and the proline-rich domain (PRD) (Figure 8A). The Nt17 sequence has the hallmarks of a moderately amphipathic MTS (Figure 9Biv). Many structural and computational studies have addressed the conformational dynamics of the Nt17 sequence in the context of the tripartite structure of Htt. Nt17 itself does not adopt a stable secondary structure but has the characteristics of a compact coil (Thakur et al., 2009), which is similar to other IDPs (Uversky, 2002), and has the propensity to form α-helical structures, particularly in the presence of membranes (Davies et al., 1997; Tam et al., 2009). Indeed, Nt17 forms α-helices in the context of oligomers or fibrils of Httex1 fragments (Kim et al., 2009; Sivanandam et al., 2011; Michalek et al., 2013). The α-helical Nt17 structure promotes aggregation of Httex1 by promoting helical structure within the poly-Q tract (Thakur et al., 2009). By contrast, the PRD forms a polyproline II structure (PPII), which reduces aggregation propensity (Bhattacharyya et al., 2006). Importantly, Htt constructs lacking the Nt17 fail to localize to mitochondria, supporting a role of this segment in mitochondrial targeting (Rockabrand et al., 2007).
Using a combination of biochemical and microscopy-based analyses with striatum-derived cell lines and murine models of HD, Friedlander and colleagues found that Htt and mHtt (and N-terminal fragments thereof) interact directly with the mitochondrial TIM23 complex and are localized to the IMS (Yano et al., 2014; Yablonska et al., 2019) (Figure 10C). In these studies, immunoprecipitation/mass spectrometry and surface plasmon resonance spectroscopy were used to show interactions with TIM23 complex subunits Tim23, Tim50 and Tim17a. They revealed that mHtt had higher affinity interactions with TIM23 subunits and inhibited the TIM23-mediated import of native mitochondrial proteins to a greater extent than wild type Htt counterparts. Importantly, these studies showed that both the Nt17 segment and the expanded poly-Q tract are crucial for interactions with the TIM23 complex. The authors concluded that this inhibition of mitochondrial import by mHtt is an early event in HD pathology (detected pre-symptomatically) and that this effect alone can result in neuronal cell death. Figure 10C summarizes the interactions of mHtt with the mitochondrial import machinery.
These observations provide potential clues as to how Htt, specifically N-terminal segments of mHtt, may engage and disrupt the TIM23 machinery during HD-related proteostatic stress. First, the requirement of Nt17 for TIM23 interactions suggests that this sequence acts like a MTS, assuming an amphipathic α-helical structure to engage the TOM/TIM23 receptors and accumulate Htt/mHtt at mitochondria during HD progression. Second, because TIM23 substrates must be unfolded to traverse the import pathway, N-terminal fragments of Htt/mHtt may be more likely to engage TIM23 than full-length Htt/mHtt, as this would require significant unfolding for import. Finally, the selective interaction of mHtt fragments with TIM23 suggests that the poly-Q expansion provides a sufficiently long, unstructured tether for the Nt17 to access the TIM23 binding sites. Indeed, it has been shown that a sufficiently long presequence is required for TIM23 precursors to span both mitochondrial membranes and engage matrix mtHsp70, which translocates substrates by an active power stroke or Brownian ratchet mechanism (Matouschek et al., 1997; Okamoto et al., 2002). The localization of Htt/mHtt to the IMS suggests incomplete translocation of these polypeptides into the matrix, creating stalled translocation intermediates that could explain the inhibition of TIM23 import. It should be noted that the findings of the Friedlander group were subsequently questioned in a study that found mitochondria-localized mHtt to reside only on the cytosolic side of the MOM and to have no measurable effect on the import of TIM23 substrates (Hamilton et al., 2020). Future work will be required to reconcile these contradictory findings.
6 Discussion
We have reviewed current evidence that APP/Aβ, α-syn, and Htt/mHtt interact with the mitochondrial import machinery through their cryptic N-terminal targeting sequences. These interactions have implications for the physiological roles of these proteins as well as their pathogenic interactions in AD, PD, and HD, respectively. They also set the stage for addressing how mitochondrial import may serve as a clearance mechanism for amyloids during proteostatic stress, as well as presenting new directions for developing interventions for neurodegenerative diseases. These two questions are addressed below.
6.1 Proteostatic mechanisms that may involve mitochondrial import
Mitochondrial proteostasis involves many stress response pathways that either directly or indirectly involve the protein import machinery (Figure 11), and any combination of them could be marshalled in response to amyloid burden at the TIM23 complex. First, some responses involve the degradation of toxic peptides or proteins in the matrix subcompartment. As noted above, the MTS-degrading enzyme PreP (Figure 11A) has an established role in the breakdown of Aβ (Falkevall et al., 2006) and may assume a more general role in the clearance of other amyloidogenic peptides. Other pathways have been described whereby mitochondria import unfolded or aggregated proteins into the matrix for degradation, providing a proteostatic mechanism for the clearance of cytotoxic proteins. These mechanisms, termed Mitochondria as Guardian in Cytosol (MAGIC) (Ruan et al., 2017) and the FUNDC1/HSC70 pathway (Li Y. et al., 2019), both entail the uptake of aggregation-prone proteins into mitochondria with subsequent degradation by matrix proteases. Thus, these pathways may explain why some amyloidogenic proteins are ectopically imported into mitochondria (Figure 11B). Alternative quality control pathways involve feedback mechanisms between the mitochondria and the nucleus to signal defects in mitochondrial protein biogenesis. Among them, the best characterized is the mitochondrial unfolded protein response (UPRmt), which senses the accumulation of unfolded proteins and ROS in mitochondrial compartments and activates transcription factors to express nuclear genes encoding mitochondrial chaperones, proteases, and antioxidant systems (Wodrich et al., 2022) (Figure 11C). Indeed, UPRmt activation is implicated in neurodegeneration (Zhu et al., 2021), and particularly in AD (Beck JS. et al., 2016; Sorrentino et al., 2017; Shen Y. et al., 2019), PD (Cooper et al., 2017), and HD (Berendzen et al., 2016; Fu et al., 2019). Other stress responses serve to clear the cytosol of mitochondrial precursor proteins when import is compromised. Namely, the UPR activated by the mistargeting of proteins (UPRam) (Wrobel et al., 2015) and the associated mitochondrial precursor overaccumulation stress (mPOS) response (Wang and Chen, 2015) (Figure 11D) respond to increased cytosolic concentrations of precursors, which are normally very low, by activating the proteasome and downregulating cytosolic protein synthesis, respectively. Other similar responses are designed to extract stalled translocation intermediates from the TOM complex and direct them to the proteasome for degradation. These include the mitochondrial compromised import response (MitoCPR) system (Weidberg and Amon, 2018) and the mitochondrial protein translocation-associated degradation (MitoTAD) system (Martensson et al., 2019) (Figure 11E). Although many of these stress responses have been resolved in lower eukaryotes, there is evidence that they exist in mammalian systems as well, although the mechanistic details have yet to be elucidated.
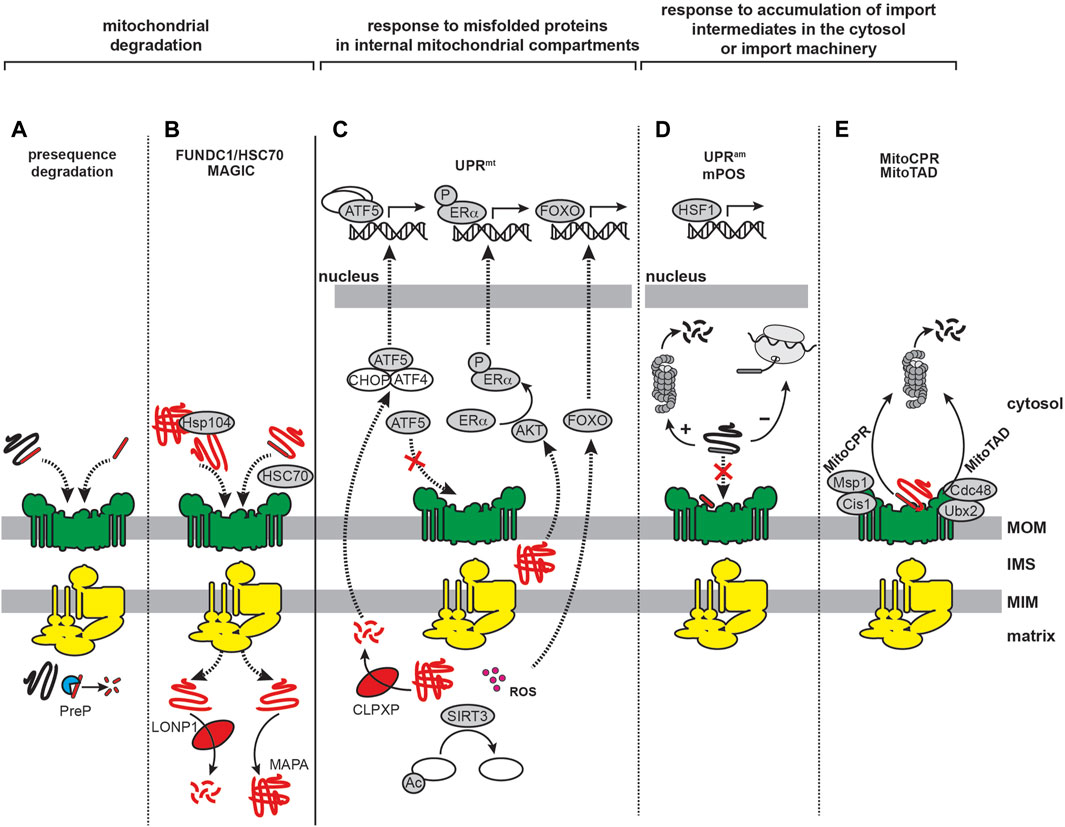
FIGURE 11. Mitochondrial proteostatic stress responses involving the import machinery. Depicted are complementary stress responses of mammalian systems that involve the mitochondrial import machinery and may be upregulated with amyloid burden. (A) PreP degradation. The PreP peptidase that cleaves MTSs also degrades Aβ in the matrix and may play a broader role in clearing imported peptides, including amyloids. (B) MAGIC and FUNDC1/HSC70. By these processes, cytosolic proteins are imported into mitochondria for sequestration and proteolysis. In the MAGIC pathway, cytosolic Hsp104 dissociates aggregated proteins, which are imported via the TOM complex into the matrix, where they are degraded by the Pim1 (yeast)/LONP1 (human) protease. In the FUNDC1/HSC70 axis, the MOM protein FUNDC1 interacts with cytosolic HSC70 to import unfolded proteins that are either degraded by LONP1 or assembled into non-aggresomal mitochondrion-associated protein aggregates (MAPAs) that can be subsequently degraded by autophagy. (C) UPRmt. This process involves transcriptional reprogramming based on mitochondria-nuclear communication in response to protein aggregates in the matrix. The UPRmt involves multiple pathways (1). The ATF5 transcription factor is imported into mitochondria under non-stressed conditions, but when its import is hindered it traffics to the nucleus with other transcription factors (ATF4/CHOP) to activate genes encoding mitochondrial chaperones and proteases. Peptides from the degradation of misfolded mitochondrial proteins, likely produced by CLPXP, may then be exported to facilitate ATF5 translocation (2). Protein aggregates in the IMS activate the AKT kinase, which phosphorylates ERα, serving as a transcription factor to induce expression of IMS-specific proteases (3). Matrix aggregates and ROS activate the sirtuin SIRT3 to deacetylate matrix proteins and activate the FOXO transcription factor that activates mitochondrial antioxidant genes. (D) UPRam and mPOS. These reactions respond to cytotoxic accumulation of mitochondrial precursor proteins in the cytosol resulting from defective import. The response of UPRam includes activation of the proteasome, whereas that of the mPOS pathway involves global reduction in cytosolic protein synthesis. Upregulation of the HSP1 transcription factor increases expression of chaperones and other stress response factors. (E) MitoTAD and MitoCPR. These systems remove stalled, nonproductive intermediates from the import machinery by continual monitoring of the TOM complex. TOM complex-associated adaptor proteins Cis1/Ubx2 recruit AAA ATPases Msp1/Cdc48 to extract stalled precursors and direct them to the proteasome for degradation.
One confounding factor in these mitochondria-based proteostasis mechanisms is the question of how aggregated proteins in the cytosol could access the TOM/TIM23 machinery, given that the import requires an unfolded precursor. A potential solution may come from the observation that a cytosolic HSP70/co-chaperone system is capable of disaggregating amyloid structures (Wentink et al., 2020). Such a system could in principle dislodge amyloid proteins and deliver them as monomers in a post-translational, chaperone-bound fashion to the mitochondrial import machinery. Indeed, several amyloids are found as distinct disease-associated polymorph structures (Qiang et al., 2017; Yang et al., 2022) that could differ in their stabilities and proclivities to dissociate soluble species. A second possibility is that since amyloid fibril structures feature rigid cores surrounded by disordered “fuzzy coats”, the solvent exposure of these flexible regions could promote PTMs and/or dissociation of monomers (Lin et al., 2017). Thus, shed monomers or proteolytic fragments of amyloidogenic proteins from fibrillar aggregates could then be free to interact with the mitochondrial protein machinery.
In summary, the degree to which PreP, MAGIC, and FUNDC1/HSC70 may assist as general clearance systems for amyloidogenic proteins like Aβ, α-syn, or mHtt requires further investigation, as do the pathways by which these amyloids may trigger mitochondrial import-related stress responses.
6.2 Prospects for drug development
The interactions between amyloidogenic proteins and the mitochondrial import machinery may inform strategies for developing novel therapeutic interventions for neurodegenerative diseases. Clinical studies and drug discovery efforts have led researchers to question the role of amyloid fibrils themselves as the primary causative agents in AD, PD, and HD. For example, the extent of fibril formation does not always correlate with disease severity (Breydo et al., 2012; Ow and Dunstan, 2014), fibrils can be present even in the absence of disease symptoms (Ke et al., 2020), and therapeutic strategies designed to disrupt fibril formation have been met with mixed success (Pardridge, 2016; Barker and Mason, 2019; Paolini Paoletti et al., 2020; Pardridge, 2020; Yiannopoulou and Papageorgiou, 2020). Hence, alternative intervention approaches, including targeting mitochondrial dysfunction, are being developed for neurodegenerative disorders (Xu et al., 2022b). Here we review some promising interventions that directly or indirectly involve the amyloid-TIM23 pathway interaction.
From a broad perspective, given that AD, PD, and HD are all diseases of aging (Blumenthal, 2004; Taylor and Dillin, 2011), targeting general aging-related mitochondrial dysfunction may be an effective strategy. For example, the efficiency of the UPRmt declines with age (Bennett and Kaeberlein, 2014; Munoz-Carvajal and Sanhueza, 2020) making it a potential target for neurodegenerative diseases (Suarez-Rivero et al., 2022). Indeed, compounds that increase cellular NAD+ levels, including nicotinamide riboside and olaparib, improved longevity in C. elegans models of Aβ proteotoxicity by activation of UPRmt and mitophagy (Sorrentino et al., 2017). Because the UPRmt involves upregulation of the TIM23 machinery (Xin et al., 2022), mitochondrial import may either be an important target of UPRmt-modulating compounds, or serve as a marker for pharmacological UPRmt activation. Another mitochondrial parameter that declines with age is the Δψm (Sugrue and Tatton, 2001; Berry and Kaeberlein, 2021; Berry et al., 2023). Caloric restriction (CR) and rapamycin, which extend lifespan in several wild type organisms (Nakagawa et al., 2012; Bitto et al., 2016; Shindyapina et al., 2022), can mediate their effects by regulating Δψm (Paglin et al., 2005; Cheema et al., 2021; Berry et al., 2023). These treatments could therefore operate in part by rescuing TIM23-mediated protein import, which functions in a Δψm-dependent manner. However, thorough pre-clinical testing of these interventions in AD, PD, and HD models would be warranted as CR and UPRmt activation can shorten or extend lifespan depending on the context in which the treatments are applied (Liao et al., 2010; Bennett and Kaeberlein, 2014; Angeli et al., 2021; Xin et al., 2022). Notably, a recent study with rapamycin in a mouse model of AD showed increased amyloid plaque formation upon treatment (Shi et al., 2022).
Given the potential role of mitochondria-resident proteases in amyloid degradation, these may represent pharmacological targets. For instance, as mentioned, the mitochondrial PreP enzyme degrades MTSs and has been implicated in degradation of Aβ in the matrix (Falkevall et al., 2006; Alikhani et al., 2011b; Mossmann et al., 2014; Fang et al., 2015; Du et al., 2021); thus, PreP may represent a potential drug target for neurodegeneration (Brunetti et al., 2021). In fact, researchers using the Senescence Accelerated Mouse Prone 8 (SAMP8) AD model found that restoring global mitochondrial function with metabolic modulators (essential and branched-chain amino acids) was accompanied by a reversal of aging-related declines in PreP levels (Brunetti et al., 2020). Similarly, researchers using the APPswe/PS1dE9 AD model, associated with FAD mutations in APP and γ-secretase, found that treatment with the neuroprotective compound Ligustilide reduced disease progression with a concurrent increase in PreP levels (Xu et al., 2018). Very recently, Pioglitazone, an antagonist of the PPAR-γ transcription factor that regulates mitochondria structure and function, was shown to upregulate PreP and the insulin degrading enzyme IDE, thereby restoring the peptide processing machinery (Di Donfrancesco et al., 2023). Original efforts toward developing agonists of PreP were focused on small molecule benzimidiazole derivatives (Vangavaragu et al., 2014), but these results were called into question (Li NS. et al., 2019). Therefore, the potential efficacy of specific effectors of PreP requires further validation.
Finally, there are several therapeutic compounds effective in treating neurodegenerative diseases that improve mitochondrial function as part of their mechanism of action. Three such compounds, described here, could have direct or indirect effects on mitochondrial protein import. First, the synthetic tetrapeptide SS-31 (elamipretide) targets mitochondria by interacting with the cardiolipin-rich MIM (Szeto, 2014). SS-31 is protective against mitochondrial dysfunction associated with a range of diseases, including myopathy, cardiac, retinal, and kidney diseases, and has demonstrated neuroprotective function (Zhu et al., 2018; Zhao et al., 2019; Liu Y. et al., 2021; Nhu et al., 2021), including efficacy in models of AD (Manczak et al., 2010; Calkins et al., 2011; Reddy et al., 2017; Reddy et al., 2018), PD (Yang et al., 2009), and HD (Yin et al., 2016). In these studies, SS-31 has been shown to restore mitochondrial biogenesis, dynamics and energetic output, reduce oxidative stress, and preserve mitochondria structure, with a notable effect on upregulating TOM complex receptor expression (Reddy et al., 2018). SS-31 and its side chain variants may act by modulating membrane electrostatics (Mitchell et al., 2020; Mitchell et al., 2022), mitigating Ca2+ stress at the MIM (Mitchell et al., 2020), and/or preserving the Δψm (Zhang et al., 2020). Furthermore, SS-31 has an extensive interactome in mitochondria (Chavez et al., 2020) and may therefore attenuate pathogenic interactions of peptides such as Aβ and α-syn with different mitochondrial proteins. Second, the curcumin derivative J147 is a neuroprotective compound that has been explored as an effective treatment in AD models (Prior et al., 2013; Goldberg et al., 2018; Currais et al., 2019; Goldberg et al., 2020; Kepchia et al., 2021; Kepchia et al., 2022). Based on these studies, several molecular mechanisms have been ascribed to J147, including improved energy metabolism, modulation of Ca2+ flux and activation of the AMPK/mTOR pathway, reduction of plasma free fatty acid levels, and regulation of acetyl-CoA metabolism. Third, polyphenols constitute a broad class of phytochemicals, many of which show neuroprotective properties (Naoi et al., 2019). Among them, urolithin A has demonstrated efficacy in models of AD (Ballesteros-Alvarez et al., 2023) and PD (Liu et al., 2022). Mechanistically, urolithin A activates mitophagy (Ryu et al., 2016; Amico et al., 2021), which may reverse defects in PINK1/Parkin mitophagy that are implicated in both familial and sporadic PD (Dawson and Dawson, 2010). Because the PINK1/Parkin mechanism is based on interactions with the TIM23 pathway, urolithin A may help reduce proteostatic stress caused by interactions of α-syn with the import machinery. Notably, SS-31, J147, and polyphenolics are all known to interact with mitochondrial F1FO ATP synthase (Ahmad and Laughlin, 2010; Goldberg et al., 2018; Chavez et al., 2020), an enzyme whose dysfunction is implicated in neurodegeneration, particularly AD (Ebanks et al., 2020). Increasing evidence supports that dysfunction of ATP synthase coincides with AD progression (Liang et al., 2008; Terni et al., 2010; Beck SJ. et al., 2016), which may mechanistically occur by interactions of Aβ with the OSCP subunit (Beck SJ. et al., 2016). It has also been shown that loss of OSCP drives the opening of the mPTP and activates the UPRmt (Angeli et al., 2021). Hence, compounds that bind ATP synthase may help maintain its function during amyloidogenic stress and/or inhibit mPTP pore opening induced by the pore initiator cyclophilin D, producing a result similar to the action of cyclosporin A (Connern and Halestrap, 1994; Gauba et al., 2019). Such interventions could increase ATP synthase activity and prevent membrane depolarization, thereby improving available ATP levels to enhance proteostatic clearance and matrix-directed import through the TIM23 pathway.
7 Conclusion
Aβ/APP, α-syn, and Htt/mHtt all contain MTS-like sequences that promote their interactions with the mitochondrial TIM23 import machinery. These interactions can directly impact mitochondrial protein import function, as well as allow incorporation of amyloids into mitochondrial subcompartments, which could contribute to pathogenicity in AD, PD, and HD, respectively. The engagement of Aβ/APP, α-syn, and Htt/mHtt with the import complexes may be part of the normal physiological function of these proteins, may represent an amyloid clearance mechanism, or may be purely pathogenic. Addressing these questions will be critical in understanding the role of mitochondria in neurodegeneration. We note that evidence for the engagement of amyloids with the import machinery does not necessarily favor either the amyloid or mitochondrial cascade hypotheses, as these interactions could occur under both scenarios. However, determining whether the amyloid-import machinery interaction is an upstream cause, or a downstream effect will help resolve the role of mitochondrial dysfunction in the sequence of events associated with neurodegeneration. Further investigation of the interactions of amyloidogenic proteins with the mitochondrial import machinery will add to our understanding of the role of mitochondria in proteostatic stress and facilitate the development of therapeutic interventions.
Author contributions
AR: Writing–original draft, Writing–review and editing. WM: Writing–original draft, Writing–review and editing. AA: Writing–original draft, Writing–review and editing. NA: Funding acquisition, Writing–original draft, Writing–review and editing.
Funding
The authors declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by a grant from the NIH (R01AG065879) to NA (PI) and AA (co-I).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2023.1263420/full#supplementary-material
References
Abe Y., Shodai T., Muto T., Mihara K., Torii H., Nishikawa S., et al. (2000). Structural basis of presequence recognition by the mitochondrial protein import receptor Tom20. Cell. 100 (5), 551–560. doi:10.1016/s0092-8674(00)80691-1
Adao R., Cruz P. F., Vaz D. C., Fonseca F., Pedersen J. N., Ferreira-da-Silva F., et al. (2020). DIBMA nanodiscs keep alpha-synuclein folded. Biochim. Biophys. Acta Biomembr. 1862 (9), 183314. doi:10.1016/j.bbamem.2020.183314
Adav S. S., Park J. E., Sze S. K. (2019). Quantitative profiling brain proteomes revealed mitochondrial dysfunction in Alzheimer's disease. Mol. Brain 12 (1), 8. doi:10.1186/s13041-019-0430-y
Addya S., Anandatheerthavarada H. K., Biswas G., Bhagwat S. V., Mullick J., Avadhani N. G. (1997). Targeting of NH2-terminal-processed microsomal protein to mitochondria: a novel pathway for the biogenesis of hepatic mitochondrial P450MT2. J. Cell. Biol. 139 (3), 589–599. doi:10.1083/jcb.139.3.589
Ahmad Z., Laughlin T. F. (2010). Medicinal chemistry of ATP synthase: a potential drug target of dietary polyphenols and amphibian antimicrobial peptides. Curr. Med. Chem. 17 (25), 2822–2836. doi:10.2174/092986710791859270
Ahmed A. U., Beech P. L., Lay S. T., Gilson P. R., Fisher P. R. (2006). Import-associated translational inhibition: novel in vivo evidence for cotranslational protein import into Dictyostelium discoideum mitochondria. Eukaryot. Cell. 5 (8), 1314–1327. doi:10.1128/EC.00386-05
Akopian D., Shen K., Zhang X., Shan S. O. (2013). Signal recognition particle: an essential protein-targeting machine. Annu. Rev. Biochem. 82, 693–721. doi:10.1146/annurev-biochem-072711-164732
Albrecht R., Rehling P., Chacinska A., Brix J., Cadamuro S. A., Volkmer R., et al. (2006). The Tim21 binding domain connects the preprotein translocases of both mitochondrial membranes. EMBO Rep. 7 (12), 1233–1238. doi:10.1038/sj.embor.7400828
Alder N. N., Jensen R. E., Johnson A. E. (2008b). Fluorescence mapping of mitochondrial TIM23 complex reveals a water-facing, substrate-interacting helix surface. Cell. 134 (3), 439–450. doi:10.1016/j.cell.2008.06.007
Alder N. N., Sutherland J., Buhring A. I., Jensen R. E., Johnson A. E. (2008a). Quaternary structure of the mitochondrial TIM23 complex reveals dynamic association between Tim23p and other subunits. Mol. Biol. Cell. 19 (1), 159–170. doi:10.1091/mbc.e07-07-0669
Alecu I., Bennett S. A. L. (2019). Dysregulated lipid metabolism and its role in alpha-synucleinopathy in Parkinson's disease. Front. Neurosci. 13, 328. doi:10.3389/fnins.2019.00328
Alexandrescu A. T. (2005). Amyloid accomplices and enforcers. Protein Sci. 14 (1), 1–12. doi:10.1110/ps.04887005
Alikhani N., Berglund A. K., Engmann T., Spanning E., Vogtle F. N., Pavlov P., et al. (2011a). Targeting capacity and conservation of PreP homologues localization in mitochondria of different species. J. Mol. Biol. 410 (3), 400–410. doi:10.1016/j.jmb.2011.05.009
Alikhani N., Guo L., Yan S., Du H., Pinho C. M., Chen J. X., et al. (2011b). Decreased proteolytic activity of the mitochondrial amyloid-beta degrading enzyme, PreP peptidasome, in Alzheimer's disease brain mitochondria. J. Alzheimers Dis. 27 (1), 75–87. doi:10.3233/JAD-2011-101716
Almagro Armenteros J. J., Tsirigos K. D., Sonderby C. K., Petersen T. N., Winther O., Brunak S., et al. (2019). SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 37 (4), 420–423. doi:10.1038/s41587-019-0036-z
Almeida Z. L., Brito R. M. M. (2020). Structure and aggregation mechanisms in amyloids. Molecules 25 (5), 1195. doi:10.3390/molecules25051195
Amico D., Andreux P. A., Valdes P., Singh A., Rinsch C., Auwerx J. (2021). Impact of the natural compound urolithin A on health, disease, and aging. Trends Mol. Med. 27 (7), 687–699. doi:10.1016/j.molmed.2021.04.009
Amorim I. S., Graham L. C., Carter R. N., Morton N. M., Hammachi F., Kunath T., et al. (2017). Sideroflexin 3 is an alpha-synuclein-dependent mitochondrial protein that regulates synaptic morphology. J. Cell. Sci. 130 (2), 325–331. doi:10.1242/jcs.194241
Anandatheerthavarada H. K., Biswas G., Mullick J., Sepuri N. B., Otvos L., Pain D., et al. (1999). Dual targeting of cytochrome P4502B1 to endoplasmic reticulum and mitochondria involves a novel signal activation by cyclic AMP-dependent phosphorylation at ser128. EMBO J. 18 (20), 5494–5504. doi:10.1093/emboj/18.20.5494
Anandatheerthavarada H. K., Biswas G., Robin M. A., Avadhani N. G. (2003). Mitochondrial targeting and a novel transmembrane arrest of Alzheimer's amyloid precursor protein impairs mitochondrial function in neuronal cells. J. Cell. Biol. 161 (1), 41–54. doi:10.1083/jcb.200207030
Anderson J. P., Walker D. E., Goldstein J. M., de Laat R., Banducci K., Caccavello R. J., et al. (2006). Phosphorylation of Ser-129 is the dominant pathological modification of alpha-synuclein in familial and sporadic Lewy body disease. J. Biol. Chem. 281 (40), 29739–29752. doi:10.1074/jbc.M600933200
Angeli S., Foulger A., Chamoli M., Peiris T. H., Gerencser A., Shahmirzadi A. A., et al. (2021). The mitochondrial permeability transition pore activates the mitochondrial unfolded protein response and promotes aging. Elife 10, 10. doi:10.7554/elife.63453
Aoyama-Ishiwatari S., Hirabayashi Y. (2021). Endoplasmic reticulum-mitochondria contact sites-emerging intracellular signaling hubs. Front. Cell. Dev. Biol. 9, 653828. doi:10.3389/fcell.2021.653828
Apetri A. C., Horwich A. L. (2008). Chaperonin chamber accelerates protein folding through passive action of preventing aggregation. Proc. Natl. Acad. Sci. U. S. A. 105 (45), 17351–17355. doi:10.1073/pnas.0809794105
Araiso Y., Endo T. (2022). Structural overview of the translocase of the mitochondrial outer membrane complex. Biophys. Physicobiol 19, e190022. doi:10.2142/biophysico.bppb-v19.0022
Araiso Y., Tsutsumi A., Qiu J., Imai K., Shiota T., Song J., et al. (2019). Structure of the mitochondrial import gate reveals distinct preprotein paths. Nature 575 (7782), 395–401. doi:10.1038/s41586-019-1680-7
Arbor S. C., LaFontaine M., Cumbay M. (2016). Amyloid-beta Alzheimer targets - protein processing, lipid rafts, and amyloid-beta pores. Yale J. Biol. Med. 89 (1), 5–21.
Area-Gomez E., de Groof A., Bonilla E., Montesinos J., Tanji K., Boldogh I., et al. (2018). A key role for MAM in mediating mitochondrial dysfunction in Alzheimer disease. Cell. Death Dis. 9 (3), 335. doi:10.1038/s41419-017-0215-0
Area-Gomez E., de Groof A. J., Boldogh I., Bird T. D., Gibson G. E., Koehler C. M., et al. (2009). Presenilins are enriched in endoplasmic reticulum membranes associated with mitochondria. Am. J. Pathol. 175 (5), 1810–1816. doi:10.2353/ajpath.2009.090219
Area-Gomez E., Del Carmen Lara Castillo M., Tambini M. D., Guardia-Laguarta C., de Groof A. J., Madra M., et al. (2012). Upregulated function of mitochondria-associated ER membranes in Alzheimer disease. EMBO J. 31 (21), 4106–4123. doi:10.1038/emboj.2012.202
Arrasate M., Mitra S., Schweitzer E. S., Segal M. R., Finkbeiner S. (2004). Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature 431 (7010), 805–810. doi:10.1038/nature02998
Aston-Mourney K., Hull R. L., Zraika S., Udayasankar J., Subramanian S. L., Kahn S. E. (2011). Exendin-4 increases islet amyloid deposition but offsets the resultant beta cell toxicity in human islet amyloid polypeptide transgenic mouse islets. Diabetologia 54 (7), 1756–1765. doi:10.1007/s00125-011-2143-3
Attwell D., Laughlin S. B. (2001). An energy budget for signaling in the grey matter of the brain. J. Cereb. Blood Flow. Metab. 21 (10), 1133–1145. doi:10.1097/00004647-200110000-00001
Auclair S. M., Bhanu M. K., Kendall D. A. (2012). Signal peptidase I: cleaving the way to mature proteins. Protein Sci. 21 (1), 13–25. doi:10.1002/pro.757
Auld D. S., Kornecook T. J., Bastianetto S., Quirion R. (2002). Alzheimer's disease and the basal forebrain cholinergic system: relations to beta-amyloid peptides, cognition, and treatment strategies. Prog. Neurobiol. 68 (3), 209–245. doi:10.1016/s0301-0082(02)00079-5
Avendano-Monsalve M. C., Mendoza-Martinez A. E., Ponce-Rojas J. C., Poot-Hernandez A. C., Rincon-Heredia R., Funes S. (2022). Positively charged amino acids at the N terminus of select mitochondrial proteins mediate early recognition by import proteins αβ'-NAC and Sam37. J. Biol. Chem. 298 (6), 101984. doi:10.1016/j.jbc.2022.101984
Avendano-Monsalve M. C., Ponce-Rojas J. C., Funes S. (2020). From cytosol to mitochondria: the beginning of a protein journey. Biol. Chem. 401 (6-7), 645–661. doi:10.1515/hsz-2020-0110
Baba M., Nakajo S., Tu P. H., Tomita T., Nakaya K., Lee V. M., et al. (1998). Aggregation of alpha-synuclein in Lewy bodies of sporadic Parkinson's disease and dementia with Lewy bodies. Am. J. Pathol. 152 (4), 879–884.
Backes S., Bykov Y. S., Flohr T., Raschle M., Zhou J., Lenhard S., et al. (2021). The chaperone-binding activity of the mitochondrial surface receptor Tom70 protects the cytosol against mitoprotein-induced stress. Cell. Rep. 35 (1), 108936. doi:10.1016/j.celrep.2021.108936
Backes S., Hess S., Boos F., Woellhaf M. W., Godel S., Jung M., et al. (2018). Tom70 enhances mitochondrial preprotein import efficiency by binding to internal targeting sequences. J. Cell. Biol. 217 (4), 1369–1382. doi:10.1083/jcb.201708044
Bajaj R., Jaremko L., Jaremko M., Becker S., Zweckstetter M. (2014). Molecular basis of the dynamic structure of the TIM23 complex in the mitochondrial intermembrane space. Structure 22 (10), 1501–1511. doi:10.1016/j.str.2014.07.015
Ballesteros-Alvarez J., Nguyen W., Sivapatham R., Rane A., Andersen J. K. (2023). Urolithin A reduces amyloid-beta load and improves cognitive deficits uncorrelated with plaque burden in a mouse model of Alzheimer's disease. Geroscience 45 (2), 1095–1113. doi:10.1007/s11357-022-00708-y
Banerjee K., Sinha M., Pham Cle L., Jana S., Chanda D., Cappai R., et al. (2010). Alpha-synuclein induced membrane depolarization and loss of phosphorylation capacity of isolated rat brain mitochondria: implications in Parkinson's disease. FEBS Lett. 584 (8), 1571–1576. doi:10.1016/j.febslet.2010.03.012
Barbaro B. A., Lukacsovich T., Agrawal N., Burke J., Bornemann D. J., Purcell J. M., et al. (2015). Comparative study of naturally occurring huntingtin fragments in Drosophila points to exon 1 as the most pathogenic species in Huntington's disease. Hum. Mol. Genet. 24 (4), 913–925. doi:10.1093/hmg/ddu504
Barcelo-Coblijn G., Golovko M. Y., Weinhofer I., Berger J., Murphy E. J. (2007). Brain neutral lipids mass is increased in alpha-synuclein gene-ablated mice. J. Neurochem. 101 (1), 132–141. doi:10.1111/j.1471-4159.2006.04348.x
Barker R., Mason S. L. (2019). The hunt for better treatments for Huntington's disease. Lancet Neurol. 18 (2), 131–133. doi:10.1016/S1474-4422(18)30448-4
Barron J. C., Hurley E. P., Parsons M. P. (2021). Huntingtin and the synapse. Front. Cell. Neurosci. 15, 689332. doi:10.3389/fncel.2021.689332
Bartels T., Ahlstrom L. S., Leftin A., Kamp F., Haass C., Brown M. F., et al. (2010). The N-terminus of the intrinsically disordered protein α-synuclein triggers membrane binding and helix folding. Biophys. J. 99 (7), 2116–2124. doi:10.1016/j.bpj.2010.06.035
Bartels T., Choi J. G., Selkoe D. J. (2011). α-Synuclein occurs physiologically as a helically folded tetramer that resists aggregation. Nature 477 (7362), 107–110. doi:10.1038/nature10324
Bauer M. F., Gempel K., Reichert A. S., Rappold G. A., Lichtner P., Gerbitz K. D., et al. (1999). Genetic and structural characterization of the human mitochondrial inner membrane translocase. J. Mol. Biol. 289 (1), 69–82. doi:10.1006/jmbi.1999.2751
Bauer M. F., Sirrenberg C., Neupert W., Brunner M. (1996). Role of Tim23 as voltage sensor and presequence receptor in protein import into mitochondria. Cell. 87 (1), 33–41. doi:10.1016/s0092-8674(00)81320-3
Baumkötter F., Schmidt N., Vargas C., Schilling S., Weber R., Wagner K., et al. (2014). Amyloid precursor protein dimerization and synaptogenic function depend on copper binding to the growth factor-like domain. J. Neurosci. 34 (33), 11159–11172. doi:10.1523/JNEUROSCI.0180-14.2014
Bausewein T., Mills D. J., Langer J. D., Nitschke B., Nussberger S., Kuhlbrandt W. (2017). Cryo-EM structure of the TOM core complex from neurospora crassa. Cell. 170 (4), 693–700. doi:10.1016/j.cell.2017.07.012
Beatrix B., Sakai H., Wiedmann M. (2000). The alpha and beta subunit of the nascent polypeptide-associated complex have distinct functions. J. Biol. Chem. 275 (48), 37838–37845. doi:10.1074/jbc.M006368200
Beck J. S., Mufson E. J., Counts S. E. (2016b). Evidence for mitochondrial UPR gene activation in familial and sporadic alzheimer's disease. Curr. Alzheimer Res. 13 (6), 610–614. doi:10.2174/1567205013666151221145445
Beck S. J., Guo L., Phensy A., Tian J., Wang L., Tandon N., et al. (2016a). Deregulation of mitochondrial F1FO-ATP synthase via OSCP in Alzheimer's disease. Nat. Commun. 7, 11483. doi:10.1038/ncomms11483
Becker T., Song J., Pfanner N. (2019). Versatility of preprotein transfer from the cytosol to mitochondria. Trends Cell. Biol. 29 (7), 534–548. doi:10.1016/j.tcb.2019.03.007
Bekris L. M., Yu C. E., Bird T. D., Tsuang D. W. (2010). Genetics of alzheimer disease. J. Geriatr. Psychiatry Neurol. 23 (4), 213–227. doi:10.1177/0891988710383571
Belloy M. E., Napolioni V., Greicius M. D. (2019). A quarter century of APOE and alzheimer's disease: progress to date and the path forward. Neuron 101 (5), 820–838. doi:10.1016/j.neuron.2019.01.056
Belyaev N. D., Kellett K. A., Beckett C., Makova N. Z., Revett T. J., Nalivaeva N. N., et al. (2010). The transcriptionally active amyloid precursor protein (APP) intracellular domain is preferentially produced from the 695 isoform of APP in a {beta}-secretase-dependent pathway. J. Biol. Chem. 285 (53), 41443–41454. doi:10.1074/jbc.M110.141390
Benador I. Y., Veliova M., Liesa M., Shirihai O. S. (2019). Mitochondria bound to lipid droplets: where mitochondrial dynamics regulate lipid storage and utilization. Cell. Metab. 29 (4), 827–835. doi:10.1016/j.cmet.2019.02.011
Benchoua A., Trioulier Y., Zala D., Gaillard M. C., Lefort N., Dufour N., et al. (2006). Involvement of mitochondrial complex II defects in neuronal death produced by N-terminus fragment of mutated huntingtin. Mol. Biol. Cell. 17 (4), 1652–1663. doi:10.1091/mbc.e05-07-0607
Bender A., Desplats P., Spencer B., Rockenstein E., Adame A., Elstner M., et al. (2013). TOM40 mediates mitochondrial dysfunction induced by α-synuclein accumulation in Parkinson's disease. PLoS One 8 (4), e62277. doi:10.1371/journal.pone.0062277
Bendor J. T., Logan T. P., Edwards R. H. (2013). The function of alpha-synuclein. Neuron 79 (6), 1044–1066. doi:10.1016/j.neuron.2013.09.004
Bennett C. F., Kaeberlein M. (2014). The mitochondrial unfolded protein response and increased longevity: cause, consequence, or correlation? Exp. Gerontol. 56, 142–146. doi:10.1016/j.exger.2014.02.002
Berendzen K. M., Durieux J., Shao L. W., Tian Y., Kim H. E., Wolff S., et al. (2016). Neuroendocrine coordination of mitochondrial stress signaling and proteostasis. Cell. 166 (6), 1553–1563. doi:10.1016/j.cell.2016.08.042
Berridge M. J. (2010). Calcium hypothesis of Alzheimer's disease. Pflugers Arch. 459 (3), 441–449. doi:10.1007/s00424-009-0736-1
Berry B. J., Kaeberlein M. (2021). An energetics perspective on geroscience: mitochondrial protonmotive force and aging. Geroscience 43 (4), 1591–1604. doi:10.1007/s11357-021-00365-7
Berry B. J., Mjelde E., Carreno F., Gilham K., Hanson E. J., Na E., et al. (2023). Preservation of mitochondrial membrane potential is necessary for lifespan extension from dietary restriction. Geroscience 45, 1573–1581. doi:10.1007/s11357-023-00766-w
Berth S. H., Lloyd T. E. (2023). Disruption of axonal transport in neurodegeneration. J. Clin. Investig. 133 (11), e168554. doi:10.1172/JCI168554
Bhagawati M., Arroum T., Webeling N., Montoro A. G., Mootz H. D., Busch K. B. (2021). The receptor subunit Tom20 is dynamically associated with the TOM complex in mitochondria of human cells. Mol. Biol. Cell. 32 (20), br1. doi:10.1091/mbc.E21-01-0042
Bhangoo M. K., Tzankov S., Fan A. C., Dejgaard K., Thomas D. Y., Young J. C. (2007). Multiple 40-kDa heat-shock protein chaperones function in Tom70-dependent mitochondrial import. Mol. Biol. Cell. 18 (9), 3414–3428. doi:10.1091/mbc.e07-01-0088
Bhattacharyya A., Thakur A. K., Chellgren V. M., Thiagarajan G., Williams A. D., Chellgren B. W., et al. (2006). Oligoproline effects on polyglutamine conformation and aggregation. J. Mol. Biol. 355 (3), 524–535. doi:10.1016/j.jmb.2005.10.053
Billingsley K. J., Bandres-Ciga S., Saez-Atienzar S., Singleton A. B. (2018). Genetic risk factors in Parkinson's disease. Cell. Tissue Res. 373 (1), 9–20. doi:10.1007/s00441-018-2817-y
Bisi N., Feni L., Peqini K., Perez-Pena H., Ongeri S., Pieraccini S., et al. (2021). α-Synuclein: an all-inclusive trip around its structure, influencing factors and applied techniques. Front. Chem. 9, 666585. doi:10.3389/fchem.2021.666585
Bitto A., Ito T. K., Pineda V. V., LeTexier N. J., Huang H. Z., Sutlief E., et al. (2016). Transient rapamycin treatment can increase lifespan and healthspan in middle-aged mice. Elife 5, e16351. doi:10.7554/eLife.16351
Blaszczyk J. W. (2020). Energy metabolism decline in the aging brain-pathogenesis of neurodegenerative disorders. Metabolites 10 (11), 450. doi:10.3390/metabo10110450
Block R. C., Dorsey E. R., Beck C. A., Brenna J. T., Shoulson I. (2010). Altered cholesterol and fatty acid metabolism in Huntington disease. J. Clin. Lipidol. 4 (1), 17–23. doi:10.1016/j.jacl.2009.11.003
Blumenthal H. T. (2004). Amyloidosis: a universal disease of aging? J. Gerontol. A Biol. Sci. Med. Sci. 59 (4), 361–369. doi:10.1093/gerona/59.4.m361
Bodner C. R., Maltsev A. S., Dobson C. M., Bax A. (2010). Differential phospholipid binding of alpha-synuclein variants implicated in Parkinson's disease revealed by solution NMR spectroscopy. Biochemistry 49 (5), 862–871. doi:10.1021/bi901723p
Bolliger L., Junne T., Schatz G., Lithgow T. (1995). Acidic receptor domains on both sides of the outer membrane mediate translocation of precursor proteins into yeast mitochondria. EMBO J. 14 (24), 6318–6326. doi:10.1002/j.1460-2075.1995.tb00322.x
Boopathi E., Srinivasan S., Fang J. K., Avadhani N. G. (2008). Bimodal protein targeting through activation of cryptic mitochondrial targeting signals by an inducible cytosolic endoprotease. Mol. Cell. 32 (1), 32–42. doi:10.1016/j.molcel.2008.09.008
Bossy-Wetzel E., Petrilli A., Knott A. B. (2008). Mutant huntingtin and mitochondrial dysfunction. Trends Neurosci. 31 (12), 609–616. doi:10.1016/j.tins.2008.09.004
Braak H., Sandmann-Keil D., Gai W., Braak E. (1999). Extensive axonal Lewy neurites in Parkinson's disease: a novel pathological feature revealed by alpha-synuclein immunocytochemistry. Neurosci. Lett. 265 (1), 67–69. doi:10.1016/s0304-3940(99)00208-6
Breydo L., Wu J. W., Uversky V. N. (2012). Α-synuclein misfolding and Parkinson's disease. Biochimica Biophysica Acta (BBA) - Mol. Basis Dis. 1822 (2), 261–285. doi:10.1016/j.bbadis.2011.10.002
Brunetti D., Bottani E., Segala A., Marchet S., Rossi F., Orlando F., et al. (2020). Targeting multiple mitochondrial processes by a metabolic modulator prevents sarcopenia and cognitive decline in SAMP8 mice. Front. Pharmacol. 11, 1171. doi:10.3389/fphar.2020.01171
Brunetti D., Catania A., Viscomi C., Deleidi M., Bindoff L. A., Ghezzi D., et al. (2021). Role of PITRM1 in mitochondrial dysfunction and neurodegeneration. Biomedicines 9 (7), 833. doi:10.3390/biomedicines9070833
Brunetti D., Torsvik J., Dallabona C., Teixeira P., Sztromwasser P., Fernandez-Vizarra E., et al. (2016). Defective PITRM1 mitochondrial peptidase is associated with Aβ amyloidotic neurodegeneration. EMBO Mol. Med. 8 (3), 176–190. doi:10.15252/emmm.201505894
Brustovetsky T., Khanna R., Brustovetsky N. (2023). CRMP2 participates in regulating mitochondrial morphology and motility in alzheimer's disease. Cells 12 (9), 1287. doi:10.3390/cells12091287
Bubber P., Haroutunian V., Fisch G., Blass J. P., Gibson G. E. (2005). Mitochondrial abnormalities in Alzheimer brain: mechanistic implications. Ann. Neurol. 57 (5), 695–703. doi:10.1002/ana.20474
Burre J. (2015). The synaptic function of alpha-synuclein. J. Park. Dis. 5 (4), 699–713. doi:10.3233/JPD-150642
Burtscher J., Di Pardo A., Maglione V., Schwarzer C., Squitieri F. (2020). Mitochondrial respiration changes in R6/2 huntington's disease model mice during aging in a brain region specific manner. Int. J. Mol. Sci. 21 (15), 5412. doi:10.3390/ijms21155412
Busch J. D., Fielden L. F., Pfanner N., Wiedemann N. (2023). Mitochondrial protein transport: versatility of translocases and mechanisms. Mol. Cell. 83 (6), 890–910. doi:10.1016/j.molcel.2023.02.020
Bussell R., Eliezer D. (2003). A structural and functional role for 11-mer repeats in alpha-synuclein and other exchangeable lipid binding proteins. J. Mol. Biol. 329 (4), 763–778. doi:10.1016/s0022-2836(03)00520-5
Cali T., Ottolini D., Negro A., Brini M. (2012). α-Synuclein controls mitochondrial calcium homeostasis by enhancing endoplasmic reticulum-mitochondria interactions. J. Biol. Chem. 287 (22), 17914–17929. doi:10.1074/jbc.M111.302794
Calkins M. J., Manczak M., Mao P., Shirendeb U., Reddy P. H. (2011). Impaired mitochondrial biogenesis, defective axonal transport of mitochondria, abnormal mitochondrial dynamics and synaptic degeneration in a mouse model of Alzheimer's disease. Hum. Mol. Genet. 20 (23), 4515–4529. doi:10.1093/hmg/ddr381
Calvo S. E., Julien O., Clauser K. R., Shen H., Kamer K. J., Wells J. A., et al. (2017). Comparative analysis of mitochondrial N-termini from mouse, human, and yeast. Mol. Cell. Proteomics 16 (4), 512–523. doi:10.1074/mcp.M116.063818
Carmo C., Naia L., Lopes C., Rego A. C. (2018). Mitochondrial dysfunction in huntington's disease. Adv. Exp. Med. Biol. 1049, 59–83. doi:10.1007/978-3-319-71779-1_3
Carulla N., Caddy G. L., Hall D. R., Zurdo J., Gairi M., Feliz M., et al. (2005). Molecular recycling within amyloid fibrils. Nature 436 (7050), 554–558. doi:10.1038/nature03986
Caspersen C., Wang N., Yao J., Sosunov A., Chen X., Lustbader J. W., et al. (2005). Mitochondrial Abeta: a potential focal point for neuronal metabolic dysfunction in Alzheimer's disease. FASEB J. 19 (14), 2040–2041. doi:10.1096/fj.05-3735fje
Castellano J. M., Kim J., Stewart F. R., Jiang H., DeMattos R. B., Patterson B. W., et al. (2011). Human apoE isoforms differentially regulate brain amyloid-beta peptide clearance. Sci. Transl. Med. 3 (89), 89ra57. 89ra57. doi:10.1126/scitranslmed.3002156
Cenini G., Rüb C., Bruderek M., Voos W. (2016). Amyloid β-peptides interfere with mitochondrial preprotein import competence by a coaggregation process. Mol. Biol. Cell. 27 (21), 3257–3272. doi:10.1091/mbc.E16-05-0313
Cenini G., Voos W. (2019). Mitochondria as potential targets in alzheimer disease therapy: an update. Front. Pharmacol. 10, 902. doi:10.3389/fphar.2019.00902
Cha M. Y., Han S. H., Son S. M., Hong H. S., Choi Y. J., Byun J., et al. (2012). Mitochondria-specific accumulation of amyloid beta induces mitochondrial dysfunction leading to apoptotic cell death. PLoS One 7 (4), e34929. doi:10.1371/journal.pone.0034929
Chacinska A., Lind M., Frazier A. E., Dudek J., Meisinger C., Geissler A., et al. (2005). Mitochondrial presequence translocase: switching between TOM tethering and motor recruitment involves Tim21 and Tim17. Cell. 120 (6), 817–829. doi:10.1016/j.cell.2005.01.011
Chacinska A., Rehling P., Guiard B., Frazier A. E., Schulze-Specking A., Pfanner N., et al. (2003). Mitochondrial translocation contact sites: separation of dynamic and stabilizing elements in formation of a TOM-TIM-preprotein supercomplex. EMBO J. 22 (20), 5370–5381. doi:10.1093/emboj/cdg532
Chacinska A., van der Laan M., Mehnert C. S., Guiard B., Mick D. U., Hutu D. P., et al. (2010). Distinct forms of mitochondrial TOM-TIM supercomplexes define signal-dependent states of preprotein sorting. Mol. Cell. Biol. 30 (1), 307–318. doi:10.1128/MCB.00749-09
Chae J. I., Kim D. W., Lee N., Jeon Y. J., Jeon I., Kwon J., et al. (2012). Quantitative proteomic analysis of induced pluripotent stem cells derived from a human Huntington's disease patient. Biochem. J. 446 (3), 359–371. doi:10.1042/BJ20111495
Chai Y. L., Xing H., Chong J. R., Francis P. T., Ballard C. G., Chen C. P., et al. (2018). Mitochondrial translocase of the outer membrane alterations may underlie dysfunctional oxidative phosphorylation in alzheimer's disease. J. Alzheimers Dis. 61 (2), 793–801. doi:10.3233/JAD-170613
Chakraborty K., Chatila M., Sinha J., Shi Q., Poschner B. C., Sikor M., et al. (2010). Chaperonin-catalyzed rescue of kinetically trapped states in protein folding. Cell. 142 (1), 112–122. doi:10.1016/j.cell.2010.05.027
Chandra S., Chen X., Rizo J., Jahn R., Sudhof T. C. (2003). A broken alpha -helix in folded alpha -Synuclein. J. Biol. Chem. 278 (17), 15313–15318. doi:10.1074/jbc.M213128200
Chang D. T., Rintoul G. L., Pandipati S., Reynolds I. J. (2006). Mutant huntingtin aggregates impair mitochondrial movement and trafficking in cortical neurons. Neurobiol. Dis. 22 (2), 388–400. doi:10.1016/j.nbd.2005.12.007
Chatre L., Matheson L. A., Jack A. S., Hanton S. L., Brandizzi F. (2009). Efficient mitochondrial targeting relies on co-operation of multiple protein signals in plants. J. Exp. Bot. 60 (3), 741–749. doi:10.1093/jxb/ern319
Chaudhuri M., Tripathi A., Gonzalez F. S. (2021). Diverse functions of Tim50, a component of the mitochondrial inner membrane protein translocase. Int. J. Mol. Sci. 22 (15), 7779. doi:10.3390/ijms22157779
Chavez J. D., Tang X., Campbell M. D., Reyes G., Kramer P. A., Stuppard R., et al. (2020). Mitochondrial protein interaction landscape of SS-31. Proc. Natl. Acad. Sci. U. S. A. 117 (26), 15363–15373. doi:10.1073/pnas.2002250117
Cheema N. J., Cameron J. M., Hood D. A. (2021). Effect of rapamycin on mitochondria and lysosomes in fibroblasts from patients with mtDNA mutations. Am. J. Physiol. Cell. Physiol. 321 (1), C176–C186. doi:10.1152/ajpcell.00471.2020
Chen C. M., Wu Y. R., Chang K. H. (2017). Altered aconitase 2 activity in huntington's disease peripheral blood cells and mouse model striatum. Int. J. Mol. Sci. 18 (11), 2480. doi:10.3390/ijms18112480
Chen M., Ona V. O., Li M., Ferrante R. J., Fink K. B., Zhu S., et al. (2000). Minocycline inhibits caspase-1 and caspase-3 expression and delays mortality in a transgenic mouse model of Huntington disease. Nat. Med. 6 (7), 797–801. doi:10.1038/77528
Cheng M. Y., Hartl F. U., Martin J., Pollock R. A., Kalousek F., Neupert W., et al. (1989). Mitochondrial heat-shock protein hsp60 is essential for assembly of proteins imported into yeast mitochondria. Nature 337 (6208), 620–625. doi:10.1038/337620a0
Cherubini M., Lopez-Molina L., Gines S. (2020). Mitochondrial fission in Huntington's disease mouse striatum disrupts ER-mitochondria contacts leading to disturbances in Ca(2+) efflux and Reactive Oxygen Species (ROS) homeostasis. Neurobiol. Dis. 136, 104741. doi:10.1016/j.nbd.2020.104741
Chiba-Falek O., Gottschalk W. K., Lutz M. W. (2018). The effects of the TOMM40 poly-T alleles on Alzheimer's disease phenotypes. Alzheimers Dement. 14 (5), 692–698. doi:10.1016/j.jalz.2018.01.015
Chiki A., Zhang Z., Rajasekhar K., Abriata L. A., Rostami I., Krapp L. F., et al. (2021). Investigating crosstalk among PTMs provides novel insight into the structural basis underlying the differential effects of Nt17 PTMs on mutant Httex1 aggregation. Front. Mol. Biosci. 8, 686086. doi:10.3389/fmolb.2021.686086
Chiti F., Dobson C. M. (2017). Protein misfolding, amyloid formation, and human disease: a summary of progress over the last decade. Annu. Rev. Biochem. 86, 27–68. doi:10.1146/annurev-biochem-061516-045115
Cho M. K., Nodet G., Kim H. Y., Jensen M. R., Bernado P., Fernandez C. O., et al. (2009). Structural characterization of alpha-synuclein in an aggregation prone state. Protein Sci. 18 (9), 1840–1846. doi:10.1002/pro.194
Choo Y. S., Johnson G. V., MacDonald M., Detloff P. J., Lesort M. (2004). Mutant huntingtin directly increases susceptibility of mitochondria to the calcium-induced permeability transition and cytochrome c release. Hum. Mol. Genet. 13 (14), 1407–1420. doi:10.1093/hmg/ddh162
Chow V. W., Mattson M. P., Wong P. C., Gleichmann M. (2010). An overview of APP processing enzymes and products. Neuromolecular Med. 12 (1), 1–12. doi:10.1007/s12017-009-8104-z
Chowhan R. K., Dar T. A., Singh L. R. (2015). “Proteopathies: biological, molecular and clinical perspectives,” in Proteostasis and chaperone surveillance. Editors L. R. Singh, T. A. Dar, and P. Ahmad (New Delhi: Springer India), 139–169.
Chuang C. L., Demontis F. (2021). Systemic manifestation and contribution of peripheral tissues to Huntington's disease pathogenesis. Ageing Res. Rev. 69, 101358. doi:10.1016/j.arr.2021.101358
Cieri D., Brini M., Cali T. (2017). Emerging (and converging) pathways in Parkinson's disease: keeping mitochondrial wellness. Biochem. Biophys. Res. Commun. 483 (4), 1020–1030. doi:10.1016/j.bbrc.2016.08.153
Claros M. G., Perea J., Shu Y., Samatey F. A., Popot J. L., Jacq C. (1995). Limitations to in vivo import of hydrophobic proteins into yeast mitochondria. The case of a cytoplasmically synthesized apocytochrome b. Eur. J. Biochem. 228 (3), 762–771. doi:10.1111/j.1432-1033.1995.tb20321.x
Claypool S. M., Koehler C. M. (2012). The complexity of cardiolipin in health and disease. Trends Biochem. Sci. 37 (1), 32–41. doi:10.1016/j.tibs.2011.09.003
Cline E. N., Bicca M. A., Viola K. L., Klein W. L. (2018). The amyloid-beta oligomer hypothesis: beginning of the third decade. J. Alzheimers Dis. 64 (1), S567-S610–S610. doi:10.3233/JAD-179941
Cole N. B., Dieuliis D., Leo P., Mitchell D. C., Nussbaum R. L. (2008). Mitochondrial translocation of alpha-synuclein is promoted by intracellular acidification. Exp. Cell. Res. 314 (10), 2076–2089. doi:10.1016/j.yexcr.2008.03.012
Conconi M., Szweda L. I., Levine R. L., Stadtman E. R., Friguet B. (1996). Age-related decline of rat liver multicatalytic proteinase activity and protection from oxidative inactivation by heat-shock protein 90. Arch. Biochem. Biophys. 331 (2), 232–240. doi:10.1006/abbi.1996.0303
Connern C. P., Halestrap A. P. (1994). Recruitment of mitochondrial cyclophilin to the mitochondrial inner membrane under conditions of oxidative stress that enhance the opening of a calcium-sensitive non-specific channel. Biochem. J. 302 (2), 321–324. doi:10.1042/bj3020321
Cooper J. F., Machiela E., Dues D. J., Spielbauer K. K., Senchuk M. M., Van Raamsdonk J. M. (2017). Activation of the mitochondrial unfolded protein response promotes longevity and dopamine neuron survival in Parkinson's disease models. Sci. Rep. 7 (1), 16441. doi:10.1038/s41598-017-16637-2
Cooper J. K., Schilling G., Peters M. F., Herring W. J., Sharp A. H., Kaminsky Z., et al. (1998). Truncated N-terminal fragments of huntingtin with expanded glutamine repeats form nuclear and cytoplasmic aggregates in cell culture. Hum. Mol. Genet. 7 (5), 783–790. doi:10.1093/hmg/7.5.783
Corder E. H., Saunders A. M., Strittmatter W. J., Schmechel D. E., Gaskell P. C., Small G. W., et al. (1993). Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science 261 (5123), 921–923. doi:10.1126/science.8346443
Costa V., Giacomello M., Hudec R., Lopreiato R., Ermak G., Lim D., et al. (2010). Mitochondrial fission and cristae disruption increase the response of cell models of Huntington's disease to apoptotic stimuli. EMBO Mol. Med. 2 (12), 490–503. doi:10.1002/emmm.201000102
Coulson E. J., Paliga K., Beyreuther K., Masters C. L. (2000). What the evolution of the amyloid protein precursor supergene family tells us about its function. Neurochem. Int. 36 (3), 175–184. doi:10.1016/s0197-0186(99)00125-4
Court D. A., Nargang F. E., Steiner H., Hodges R. S., Neupert W., Lill R. (1996). Role of the intermembrane-space domain of the preprotein receptor Tom22 in protein import into mitochondria. Mol. Cell. Biol. 16 (8), 4035–4042. doi:10.1128/mcb.16.8.4035
Croke R. L., Patil S. M., Quevreaux J., Kendall D. A., Alexandrescu A. T. (2011). NMR determination of pKa values in α-synuclein. Protein Sci. 20 (2), 256–269. doi:10.1002/pro.556
Croke R. L., Sallum C. O., Watson E., Watt E. D., Alexandrescu A. T. (2008). Hydrogen exchange of monomeric alpha-synuclein shows unfolded structure persists at physiological temperature and is independent of molecular crowding in Escherichia coli. Protein Sci. 17 (8), 1434–1445. doi:10.1110/ps.033803.107
Crowley K. S., Payne R. M. (1998). Ribosome binding to mitochondria is regulated by GTP and the transit peptide. J. Biol. Chem. 273 (27), 17278–17285. doi:10.1074/jbc.273.27.17278
Cuello A. C. (2005). Intracellular and extracellular Abeta, a tale of two neuropathologies. Brain Pathol. 15 (1), 66–71. doi:10.1111/j.1750-3639.2005.tb00101.x
Cui L., Jeong H., Borovecki F., Parkhurst C. N., Tanese N., Krainc D. (2006). Transcriptional repression of PGC-1alpha by mutant huntingtin leads to mitochondrial dysfunction and neurodegeneration. Cell. 127 (1), 59–69. doi:10.1016/j.cell.2006.09.015
Currais A., Huang L., Goldberg J., Petrascheck M., Ates G., Pinto-Duarte A., et al. (2019). Elevating acetyl-CoA levels reduces aspects of brain aging. Elife 8, e47866. doi:10.7554/eLife.47866
Davidson W. S., Jonas A., Clayton D. F., George J. M. (1998). Stabilization of alpha-synuclein secondary structure upon binding to synthetic membranes. J. Biol. Chem. 273 (16), 9443–9449. doi:10.1074/jbc.273.16.9443
Davies S. W., Turmaine M., Cozens B. A., DiFiglia M., Sharp A. H., Ross C. A., et al. (1997). Formation of neuronal intranuclear inclusions underlies the neurological dysfunction in mice transgenic for the HD mutation. Cell. 90 (3), 537–548. doi:10.1016/s0092-8674(00)80513-9
Dawson T. M., Dawson V. L. (2010). The role of parkin in familial and sporadic Parkinson's disease. Mov. Disord. 25(1), S32–S39. doi:10.1002/mds.22798
Dayan D., Bandel M., Gunsel U., Nussbaum I., Prag G., Mokranjac D., et al. (2019). A mutagenesis analysis of Tim50, the major receptor of the TIM23 complex, identifies regions that affect its interaction with Tim23. Sci. Rep. 9 (1), 2012. doi:10.1038/s41598-018-38353-1
de la Cruz L., Bajaj R., Becker S., Zweckstetter M. (2010). The intermembrane space domain of Tim23 is intrinsically disordered with a distinct binding region for presequences. Protein Sci. 19 (11), 2045–2054. doi:10.1002/pro.482
De Miranda B. R., Rocha E. M., Castro S. L., Greenamyre J. T. (2020). Protection from α-Synuclein induced dopaminergic neurodegeneration by overexpression of the mitochondrial import receptor TOM20. NPJ Park. Dis. 6 (1), 38. doi:10.1038/s41531-020-00139-6
Dekker P. J., Martin F., Maarse A. C., Bomer U., Muller H., Guiard B., et al. (1997). The Tim core complex defines the number of mitochondrial translocation contact sites and can hold arrested preproteins in the absence of matrix Hsp70-Tim44. EMBO J. 16 (17), 5408–5419. doi:10.1093/emboj/16.17.5408
Dekker P. J., Ryan M. T., Brix J., Muller H., Honlinger A., Pfanner N. (1998). Preprotein translocase of the outer mitochondrial membrane: molecular dissection and assembly of the general import pore complex. Mol. Cell. Biol. 18 (11), 6515–6524. doi:10.1128/mcb.18.11.6515
del Alamo M., Hogan D. J., Pechmann S., Albanese V., Brown P. O., Frydman J. (2011). Defining the specificity of cotranslationally acting chaperones by systematic analysis of mRNAs associated with ribosome-nascent chain complexes. PLoS Biol. 9 (7), e1001100. doi:10.1371/journal.pbio.1001100
Del Prete D., Suski J. M., Oules B., Debayle D., Gay A. S., Lacas-Gervais S., et al. (2017). Localization and processing of the amyloid-beta protein precursor in mitochondria-associated membranes. J. Alzheimers Dis. 55 (4), 1549–1570. doi:10.3233/JAD-160953
Demishtein-Zohary K., Gunsel U., Marom M., Banerjee R., Neupert W., Azem A., et al. (2017). Role of Tim17 in coupling the import motor to the translocation channel of the mitochondrial presequence translocase. Elife 6, e22696. doi:10.7554/eLife.22696
Denkert N., Schendzielorz A. B., Barbot M., Versemann L., Richter F., Rehling P., et al. (2017). Cation selectivity of the presequence translocase channel Tim23 is crucial for efficient protein import. Elife 6, e28324. doi:10.7554/eLife.28324
Deshaies R. J., Koch B. D., Werner-Washburne M., Craig E. A., Schekman R. (1988). A subfamily of stress proteins facilitates translocation of secretory and mitochondrial precursor polypeptides. Nature 332 (6167), 800–805. doi:10.1038/332800a0
Devi L., Anandatheerthavarada H. K. (2010). Mitochondrial trafficking of APP and alpha synuclein: relevance to mitochondrial dysfunction in Alzheimer's and Parkinson's diseases. Biochimica Biophysica Acta (BBA) - Mol. Basis Dis. 1802 (1), 11–19. doi:10.1016/j.bbadis.2009.07.007
Devi L., Prabhu B. M., Galati D. F., Avadhani N. G., Anandatheerthavarada H. K. (2006). Accumulation of amyloid precursor protein in the mitochondrial import channels of human Alzheimer's disease brain is associated with mitochondrial dysfunction. J. Neurosci. 26 (35), 9057–9068. doi:10.1523/JNEUROSCI.1469-06.2006
Devi L., Raghavendran V., Prabhu B. M., Avadhani N. G., Anandatheerthavarada H. K. (2008). Mitochondrial import and accumulation of alpha-synuclein impair complex I in human dopaminergic neuronal cultures and Parkinson disease brain. J. Biol. Chem. 283 (14), 9089–9100. doi:10.1074/jbc.M710012200
Di Donfrancesco A., Berlingieri C., Giacomello M., Frascarelli C., Magalhaes Rebelo A. P., Bindoff L. A., et al. (2023). PPAR-gamma agonist pioglitazone recovers mitochondrial quality control in fibroblasts from PITRM1-deficient patients. Front. Pharmacol. 14, 1220620. doi:10.3389/fphar.2023.1220620
Di Maio R., Barrett P. J., Hoffman E. K., Barrett C. W., Zharikov A., Borah A., et al. (2016a). α-Synuclein binds to TOM20 and inhibits mitochondrial protein import in Parkinson's disease. Sci. Transl. Med. 8 (342), 342ra78. doi:10.1126/scitranslmed.aaf3634
Di Maio R., Barrett P. J., Hoffman E. K., Barrett C. W., Zharikov A., Borah A., et al. (2016b). α-Synuclein binds to TOM20 and inhibits mitochondrial protein import in Parkinson's disease. Sci. Transl. Med. 8 (342), 342ra78. doi:10.1126/scitranslmed.aaf3634
Diaz M., Fabelo N., Martin V., Ferrer I., Gomez T., Marin R. (2015). Biophysical alterations in lipid rafts from human cerebral cortex associate with increased BACE1/AβPP interaction in early stages of Alzheimer's disease. J. Alzheimers Dis. 43 (4), 1185–1198. doi:10.3233/JAD-141146
Dienel G. A. (2017). Lack of appropriate stoichiometry: strong evidence against an energetically important astrocyte-neuron lactate shuttle in brain. J. Neurosci. Res. 95 (11), 2103–2125. doi:10.1002/jnr.24015
Dietmeier K., Honlinger A., Bomer U., Dekker P. J., Eckerskorn C., Lottspeich F., et al. (1997). Tom5 functionally links mitochondrial preprotein receptors to the general import pore. Nature 388 (6638), 195–200. doi:10.1038/40663
Dobson C. M. (2003). Protein folding and misfolding. Nature 426 (6968), 884–890. doi:10.1038/nature02261
Dores-Silva P. R., Minari K., Ramos C. H., Barbosa L. R., Borges J. C. (2013). Structural and stability studies of the human mtHsp70-escort protein 1: an essential mortalin co-chaperone. Int. J. Biol. Macromol. 56, 140–148. doi:10.1016/j.ijbiomac.2013.02.009
Dose J., Huebbe P., Nebel A., Rimbach G. (2016). APOE genotype and stress response - a mini review. Lipids Health Dis. 15, 121. doi:10.1186/s12944-016-0288-2
Du F., Yu Q., Yan S., Zhang Z., Vangavaragu J. R., Chen D., et al. (2021). Gain of PITRM1 peptidase in cortical neurons affords protection of mitochondrial and synaptic function in an advanced age mouse model of Alzheimer's disease. Aging Cell. 20 (5), e13368. doi:10.1111/acel.13368
Du H., Guo L., Fang F., Chen D., Sosunov A. A., McKhann G. M., et al. (2008). Cyclophilin D deficiency attenuates mitochondrial and neuronal perturbation and ameliorates learning and memory in Alzheimer's disease. Nat. Med. 14 (10), 1097–1105. doi:10.1038/nm.1868
Dubois C., Kong G., Tran H., Li S., Pang T. Y., Hannan A. J., et al. (2021). Small non-coding RNAs are dysregulated in huntington's disease transgenic mice independently of the therapeutic effects of an environmental intervention. Mol. Neurobiol. 58 (7), 3308–3318. doi:10.1007/s12035-021-02342-9
Duce J. A., Tsatsanis A., Cater M. A., James S. A., Robb E., Wikhe K., et al. (2010). Iron-export ferroxidase activity of beta-amyloid precursor protein is inhibited by zinc in Alzheimer's disease. Cell. 142 (6), 857–867. doi:10.1016/j.cell.2010.08.014
Dugger B. N., Dickson D. W. (2017). Pathology of neurodegenerative diseases. Cold Spring Harb. Perspect. Biol. 9 (7), a028035. doi:10.1101/cshperspect.a028035
Ebanks B., Ingram T. L., Chakrabarti L. (2020). ATP synthase and Alzheimer's disease: putting a spin on the mitochondrial hypothesis. Aging (Albany NY) 12 (16), 16647–16662. doi:10.18632/aging.103867
Egea G., Izquierdo J. M., Ricart J., San Martin C., Cuezva J. M. (1997). mRNA encoding the beta-subunit of the mitochondrial F1-ATPase complex is a localized mRNA in rat hepatocytes. Biochem. J. 322(2), 557–565. doi:10.1042/bj3220557
Egidio F., Castelli V., Cimini A., d'Angelo M. (2023). Cell rearrangement and oxidant/antioxidant imbalance in huntington's disease. Antioxidants (Basel) 12 (3), 571. doi:10.3390/antiox12030571
Eisenberg D., Weiss R. M., Terwilliger T. C. (1982). The helical hydrophobic moment: a measure of the amphiphilicity of a helix. Nature 299 (5881), 371–374. doi:10.1038/299371a0
Ellis C. E., Murphy E. J., Mitchell D. C., Golovko M. Y., Scaglia F., Barcelo-Coblijn G. C., et al. (2005). Mitochondrial lipid abnormality and electron transport chain impairment in mice lacking alpha-synuclein. Mol. Cell. Biol. 25 (22), 10190–10201. doi:10.1128/MCB.25.22.10190-10201.2005
Ellis R. J. (1996). Revisiting the anfinsen cage. Fold. Des. 1 (1), R9–R15. doi:10.1016/S1359-0278(96)00004-1
Elsner S., Simian D., Iosefson O., Marom M., Azem A. (2009). The mitochondrial protein translocation motor: structural conservation between the human and yeast Tim14/Pam18-Tim16/Pam16 co-chaperones. Int. J. Mol. Sci. 10 (5), 2041–2053. doi:10.3390/ijms10052041
Endo T., Mitsui S., Nakai M., Roise D. (1996). Binding of mitochondrial presequences to yeast cytosolic heat shock protein 70 depends on the amphiphilicity of the presequence. J. Biol. Chem. 271 (8), 4161–4167. doi:10.1074/jbc.271.8.4161
Engel M. F. (2009). Membrane permeabilization by islet amyloid polypeptide. Chem. Phys. Lipids 160 (1), 1–10. doi:10.1016/j.chemphyslip.2009.03.008
Ericsson M., von Saucken V., Newman A. J., Doehr L., Hoesch C., Kim T. E., et al. (2021). Crowded organelles, lipid accumulation, and abnormal membrane tubulation in cellular models of enhanced α-synuclein membrane interaction. Brain Res. 1758, 147349. doi:10.1016/j.brainres.2021.147349
Esaki M., Shimizu H., Ono T., Yamamoto H., Kanamori T., Nishikawa S., et al. (2004). Mitochondrial protein import. Requirement of presequence elements and tom components for precursor binding to the TOM complex. J. Biol. Chem. 279 (44), 45701–45707. doi:10.1074/jbc.M404591200
Esfahanian N., Knoblich C. D., Bowman G. A., Rezvani K. (2023). Mortalin: protein partners, biological impacts, pathological roles, and therapeutic opportunities. Front. Cell. Dev. Biol. 11, 1028519. doi:10.3389/fcell.2023.1028519
Falkevall A., Alikhani N., Bhushan S., Pavlov P. F., Busch K., Johnson K. A., et al. (2006). Degradation of the amyloid beta-protein by the novel mitochondrial peptidasome, PreP. J. Biol. Chem. 281 (39), 29096–29104. doi:10.1074/jbc.M602532200
Fan A. C., Bhangoo M. K., Young J. C. (2006). Hsp90 functions in the targeting and outer membrane translocation steps of Tom70-mediated mitochondrial import. J. Biol. Chem. 281 (44), 33313–33324. doi:10.1074/jbc.M605250200
Fang D., Wang Y., Zhang Z., Du H., Yan S., Sun Q., et al. (2015). Increased neuronal PreP activity reduces Aβ accumulation, attenuates neuroinflammation and improves mitochondrial and synaptic function in Alzheimer disease's mouse model. Hum. Mol. Genet. 24 (18), 5198–5210. doi:10.1093/hmg/ddv241
Fantini J., Carlus D., Yahi N. (2011). The fusogenic tilted peptide (67-78) of alpha-synuclein is a cholesterol binding domain. Biochim. Biophys. Acta 1808 (10), 2343–2351. doi:10.1016/j.bbamem.2011.06.017
Farmer B. C., Kluemper J., Johnson L. A. (2019). Apolipoprotein E4 alters astrocyte fatty acid metabolism and lipid droplet formation. Cells 8 (2), 182. doi:10.3390/cells8020182
Fauchere J., Pliska V. (1983). Hydrophobic parameters II of amino acid side-chains from the partitioning of N-acetyl-amino acid amides. Eur. J. Med. Chem. 18, 369–375.
Faustini G., Bono F., Valerio A., Pizzi M., Spano P., Bellucci A. (2017). Mitochondria and alpha-synuclein: friends or foes in the pathogenesis of Parkinson's disease? Genes. (Basel) 8 (12), 377. doi:10.3390/genes8120377
Fauvet B., Mbefo M. K., Fares M. B., Desobry C., Michael S., Ardah M. T., et al. (2012). α-Synuclein in central nervous system and from erythrocytes, mammalian cells, and Escherichia coli exists predominantly as disordered monomer. J. Biol. Chem. 287 (19), 15345–15364. doi:10.1074/jbc.M111.318949
Fayyad M., Erskine D., Majbour N. K., Vaikath N. N., Ghanem S. S., Sudhakaran I. P., et al. (2020). Investigating the presence of doubly phosphorylated alpha-synuclein at tyrosine 125 and serine 129 in idiopathic Lewy body diseases. Brain Pathol. 30 (4), 831–843. doi:10.1111/bpa.12845
Fazal F. M., Han S., Parker K. R., Kaewsapsak P., Xu J., Boettiger A. N., et al. (2019). Atlas of subcellular RNA localization revealed by APEX-seq. Cell. 178 (2), 473–490. doi:10.1016/j.cell.2019.05.027
Fernandes T., Resende R., Silva D. F., Marques A. P., Santos A. E., Cardoso S. M., et al. (2021). Structural and functional alterations in mitochondria-associated membranes (MAMs) and in mitochondria activate stress response mechanisms in an in vitro model of alzheimer's disease. Biomedicines 9 (8), 881. doi:10.3390/biomedicines9080881
Ferrone F. A., Hofrichter J., Eaton W. A. (1985). Kinetics of sickle hemoglobin polymerization. II. A double nucleation mechanism. J. Mol. Biol. 183 (4), 611–631. doi:10.1016/0022-2836(85)90175-5
Fielden L. F., Busch J. D., Merkt S. G., Ganesan I., Steiert C., Hasselblatt H. B., et al. (2023). Central role of Tim17 in mitochondrial presequence protein translocation. Nature 621, 627–634. doi:10.1038/s41586-023-06477-8
Flagmeier P., Meisl G., Vendruscolo M., Knowles T. P., Dobson C. M., Buell A. K., et al. (2016). Mutations associated with familial Parkinson's disease alter the initiation and amplification steps of alpha-synuclein aggregation. Proc. Natl. Acad. Sci. U. S. A. 113 (37), 10328–10333. doi:10.1073/pnas.1604645113
Fortin D. L., Nemani V. M., Voglmaier S. M., Anthony M. D., Ryan T. A., Edwards R. H. (2005). Neural activity controls the synaptic accumulation of alpha-synuclein. J. Neurosci. 25 (47), 10913–10921. doi:10.1523/JNEUROSCI.2922-05.2005
Fortin D. L., Troyer M. D., Nakamura K., Kubo S., Anthony M. D., Edwards R. H. (2004). Lipid rafts mediate the synaptic localization of alpha-synuclein. J. Neurosci. 24 (30), 6715–6723. doi:10.1523/JNEUROSCI.1594-04.2004
Friedrich R. P., Tepper K., Ronicke R., Soom M., Westermann M., Reymann K., et al. (2010). Mechanism of amyloid plaque formation suggests an intracellular basis of Abeta pathogenicity. Proc. Natl. Acad. Sci. U. S. A. 107 (5), 1942–1947. doi:10.1073/pnas.0904532106
Fu H., Hardy J., Duff K. E. (2018). Selective vulnerability in neurodegenerative diseases. Nat. Neurosci. 21 (10), 1350–1358. doi:10.1038/s41593-018-0221-2
Fu Z., Liu F., Liu C., Jin B., Jiang Y., Tang M., et al. (2019). Mutant huntingtin inhibits the mitochondrial unfolded protein response by impairing ABCB10 mRNA stability. Biochim. Biophys. Acta Mol. Basis Dis. 1865 (6), 1428–1435. doi:10.1016/j.bbadis.2019.02.015
Gabriel K., Egan B., Lithgow T. (2003). Tom40, the import channel of the mitochondrial outer membrane, plays an active role in sorting imported proteins. EMBO J. 22 (10), 2380–2386. doi:10.1093/emboj/cdg229
Gafni J., Ellerby L. M. (2002). Calpain activation in Huntington's disease. J. Neurosci. 22 (12), 4842–4849. doi:10.1523/JNEUROSCI.22-12-04842.2002
Gallego Villarejo L., Bachmann L., Marks D., Brachthauser M., Geidies A., Muller T. (2022). Role of intracellular amyloid beta as pathway modulator, biomarker, and therapy target. Int. J. Mol. Sci. 23 (9), 4656. doi:10.3390/ijms23094656
Gamerdinger M., Kobayashi K., Wallisch A., Kreft S. G., Sailer C., Schlomer R., et al. (2019). Early scanning of nascent polypeptides inside the ribosomal tunnel by NAC. Mol. Cell. 75 (5), 996–1006. doi:10.1016/j.molcel.2019.06.030
Gao F., Yang J., Wang D., Li C., Fu Y., Wang H., et al. (2017). Mitophagy in Parkinson's disease: pathogenic and therapeutic implications. Front. Neurol. 8, 527. doi:10.3389/fneur.2017.00527
Gartner F., Bomer U., Guiard B., Pfanner N. (1995). The sorting signal of cytochrome b2 promotes early divergence from the general mitochondrial import pathway and restricts the unfoldase activity of matrix Hsp70. EMBO J. 14 (23), 6043–6057. doi:10.1002/j.1460-2075.1995.tb00293.x
Gauba E., Chen H., Guo L., Du H. (2019). Cyclophilin D deficiency attenuates mitochondrial F1Fo ATP synthase dysfunction via OSCP in Alzheimer's disease. Neurobiol. Dis. 121, 138–147. doi:10.1016/j.nbd.2018.09.020
Gautier R., Douguet D., Antonny B., Drin G. (2008). HELIQUEST: a web server to screen sequences with specific alpha-helical properties. Bioinformatics 24 (18), 2101–2102. doi:10.1093/bioinformatics/btn392
Ge P., Dawson V. L., Dawson T. M. (2020). PINK1 and Parkin mitochondrial quality control: a source of regional vulnerability in Parkinson's disease. Mol. Neurodegener. 15 (1), 20. doi:10.1186/s13024-020-00367-7
Geissler A., Chacinska A., Truscott K. N., Wiedemann N., Brandner K., Sickmann A., et al. (2002). The mitochondrial presequence translocase: an essential role of Tim50 in directing preproteins to the import channel. Cell. 111 (4), 507–518. doi:10.1016/s0092-8674(02)01073-5
Genge M. G., Mokranjac D. (2021). Coordinated translocation of presequence-containing precursor proteins across two mitochondrial membranes: knowns and unknowns of how TOM and TIM23 complexes cooperate with each other. Front. Physiol. 12, 806426. doi:10.3389/fphys.2021.806426
George R., Beddoe T., Landl K., Lithgow T. (1998). The yeast nascent polypeptide-associated complex initiates protein targeting to mitochondria in vivo. Proc. Natl. Acad. Sci. U. S. A. 95 (5), 2296–2301. doi:10.1073/pnas.95.5.2296
George R., Walsh P., Beddoe T., Lithgow T. (2002). The nascent polypeptide-associated complex (NAC) promotes interaction of ribosomes with the mitochondrial surface in vivo. FEBS Lett. 516 (1-3), 213–216. doi:10.1016/s0014-5793(02)02528-0
Georgieva E. R., Ramlall T. F., Borbat P. P., Freed J. H., Eliezer D. (2008). Membrane-bound alpha-synuclein forms an extended helix: long-distance pulsed ESR measurements using vesicles, bicelles, and rodlike micelles. J. Am. Chem. Soc. 130 (39), 12856–12857. doi:10.1021/ja804517m
Gevorkyan-Airapetov L., Zohary K., Popov-Celeketic D., Mapa K., Hell K., Neupert W., et al. (2009). Interaction of Tim23 with Tim50 Is essential for protein translocation by the mitochondrial TIM23 complex. J. Biol. Chem. 284 (8), 4865–4872. doi:10.1074/jbc.M807041200
Giacomello M., Oliveros J. C., Naranjo J. R., Carafoli E. (2013). Neuronal Ca(2+) dyshomeostasis in Huntington disease. Prion 7 (1), 76–84. doi:10.4161/pri.23581
Gilmozzi V., Gentile G., Castelo Rueda M. P., Hicks A. A., Pramstaller P. P., Zanon A., et al. (2020). Interaction of alpha-synuclein with lipids: mitochondrial cardiolipin as a critical player in the pathogenesis of Parkinson's disease. Front. Neurosci. 14, 578993. doi:10.3389/fnins.2020.578993
Giordano F. (2018). Non-vesicular lipid trafficking at the endoplasmic reticulum-mitochondria interface. Biochem. Soc. Trans. 46 (2), 437–452. doi:10.1042/BST20160185
Gleichman A. J., Carmichael S. T. (2020). Glia in neurodegeneration: drivers of disease or along for the ride? Neurobiol. Dis. 142, 104957. doi:10.1016/j.nbd.2020.104957
Glick B. S., Brandt A., Cunningham K., Muller S., Hallberg R. L., Schatz G. (1992). Cytochromes c1 and b2 are sorted to the intermembrane space of yeast mitochondria by a stop-transfer mechanism. Cell. 69 (5), 809–822. doi:10.1016/0092-8674(92)90292-k
Gold V. A., Chroscicki P., Bragoszewski P., Chacinska A. (2017). Visualization of cytosolic ribosomes on the surface of mitochondria by electron cryo-tomography. EMBO Rep. 18 (10), 1786–1800. doi:10.15252/embr.201744261
Gold V. A., Ieva R., Walter A., Pfanner N., van der Laan M., Kuhlbrandt W. (2014). Visualizing active membrane protein complexes by electron cryotomography. Nat. Commun. 5, 4129. doi:10.1038/ncomms5129
Goldberg J., Currais A., Ates G., Huang L., Shokhirev M., Maher P., et al. (2020). Targeting of intracellular Ca(2+) stores as a therapeutic strategy against age-related neurotoxicities. NPJ Aging Mech. Dis. 6, 10. doi:10.1038/s41514-020-00048-1
Goldberg J., Currais A., Prior M., Fischer W., Chiruta C., Ratliff E., et al. (2018). The mitochondrial ATP synthase is a shared drug target for aging and dementia. Aging Cell. 17 (2), e12715. doi:10.1111/acel.12715
Goldberg Y. P., Nicholson D. W., Rasper D. M., Kalchman M. A., Koide H. B., Graham R. K., et al. (1996). Cleavage of huntingtin by apopain, a proapoptotic cysteine protease, is modulated by the polyglutamine tract. Nat. Genet. 13 (4), 442–449. doi:10.1038/ng0896-442
Golovko M. Y., Faergeman N. J., Cole N. B., Castagnet P. I., Nussbaum R. L., Murphy E. J. (2005). Alpha-synuclein gene deletion decreases brain palmitate uptake and alters the palmitate metabolism in the absence of alpha-synuclein palmitate binding. Biochemistry 44 (23), 8251–8259. doi:10.1021/bi0502137
Golovko M. Y., Rosenberger T. A., Faergeman N. J., Feddersen S., Cole N. B., Pribill I., et al. (2006). Acyl-CoA synthetase activity links wild-type but not mutant alpha-synuclein to brain arachidonate metabolism. Biochemistry 45 (22), 6956–6966. doi:10.1021/bi0600289
Gomez-Fabra Gala M., Vogtle F. N. (2021). Mitochondrial proteases in human diseases. FEBS Lett. 595 (8), 1205–1222. doi:10.1002/1873-3468.14039
Goswami A. V., Chittoor B., D'Silva P. (2010). Understanding the functional interplay between mammalian mitochondrial Hsp70 chaperone machine components. J. Biol. Chem. 285 (25), 19472–19482. doi:10.1074/jbc.M110.105957
Gottlieb L., Guo L., Shorter J., Marmorstein R. (2021). N-alpha-acetylation of Huntingtin protein increases its propensity to aggregate. J. Biol. Chem. 297 (6), 101363. doi:10.1016/j.jbc.2021.101363
Goure W. F., Krafft G. A., Jerecic J., Hefti F. (2014). Targeting the proper amyloid-beta neuronal toxins: a path forward for Alzheimer's disease immunotherapeutics. Alzheimers Res. Ther. 6 (4), 42. doi:10.1186/alzrt272
Graham R. K., Deng Y., Slow E. J., Haigh B., Bissada N., Lu G., et al. (2006). Cleavage at the caspase-6 site is required for neuronal dysfunction and degeneration due to mutant huntingtin. Cell. 125 (6), 1179–1191. doi:10.1016/j.cell.2006.04.026
Greco T. M., Secker C., Ramos E. S., Federspiel J. D., Liu J. P., Perez A. M., et al. (2022). Dynamics of huntingtin protein interactions in the striatum identifies candidate modifiers of Huntington disease. Cell. Syst. 13 (4), 304–320.e5. doi:10.1016/j.cels.2022.01.005
Grey M., Linse S., Nilsson H., Brundin P., Sparr E. (2011). Membrane interaction of α-synuclein in different aggregation states. J. Park. Dis. 1 (4), 359–371. doi:10.3233/JPD-2011-11067
Gruhler A., Arnold I., Seytter T., Guiard B., Schwarz E., Neupert W., et al. (1997). N-terminal hydrophobic sorting signals of preproteins confer mitochondrial hsp70 independence for import into mitochondria. J. Biol. Chem. 272 (28), 17410–17415. doi:10.1074/jbc.272.28.17410
Guan Z., Yan L., Wang Q., Qi L., Hong S., Gong Z., et al. (2021). Structural insights into assembly of human mitochondrial translocase TOM complex. Cell. Discov. 7 (1), 22. doi:10.1038/s41421-021-00252-7
Guardia-Laguarta C., Area-Gomez E., Rub C., Liu Y., Magrane J., Becker D., et al. (2014). α-Synuclein is localized to mitochondria-associated ER membranes. J. Neurosci. 34 (1), 249–259. doi:10.1523/JNEUROSCI.2507-13.2014
Guardia-Laguarta C., Area-Gomez E., Schon E. A., Przedborski S. (2015). A new role for alpha-synuclein in Parkinson's disease: alteration of ER-mitochondrial communication. Mov. Disord. 30 (8), 1026–1033. doi:10.1002/mds.26239
Gui Y. X., Wang X. Y., Kang W. Y., Zhang Y. J., Zhang Y., Zhou Y., et al. (2012). Extracellular signal-regulated kinase is involved in alpha-synuclein-induced mitochondrial dynamic disorders by regulating dynamin-like protein 1. Neurobiol. Aging 33 (12), 2841–2854. doi:10.1016/j.neurobiolaging.2012.02.001
Gunsel U., Paz E., Gupta R., Mathes I., Azem A., Mokranjac D. (2020). InVivo dissection of the intrinsically disordered receptor domain of Tim23. J. Mol. Biol. 432 (10), 3326–3337. doi:10.1016/j.jmb.2020.03.031
Guo Q., Bin H., Cheng J., Seefelder M., Engler T., Pfeifer G., et al. (2018). The cryo-electron microscopy structure of huntingtin. Nature 555 (7694), 117–120. doi:10.1038/nature25502
Guo W., Stoklund Dittlau K., Van Den Bosch L. (2020). Axonal transport defects and neurodegeneration: molecular mechanisms and therapeutic implications. Semin. Cell. Dev. Biol. 99, 133–150. doi:10.1016/j.semcdb.2019.07.010
Guo X., Sun X., Hu D., Wang Y. J., Fujioka H., Vyas R., et al. (2016). VCP recruitment to mitochondria causes mitophagy impairment and neurodegeneration in models of Huntington's disease. Nat. Commun. 7, 12646. doi:10.1038/ncomms12646
Guo Y., Cheong N., Zhang Z., De Rose R., Deng Y., Farber S. A., et al. (2004). Tim50, a component of the mitochondrial translocator, regulates mitochondrial integrity and cell death. J. Biol. Chem. 279 (23), 24813–24825. doi:10.1074/jbc.M402049200
Gurlo T., Ryazantsev S., Huang C. J., Yeh M. W., Reber H. A., Hines O. J., et al. (2010). Evidence for proteotoxicity in beta cells in type 2 diabetes: toxic islet amyloid polypeptide oligomers form intracellularly in the secretory pathway. Am. J. Pathol. 176 (2), 861–869. doi:10.2353/ajpath.2010.090532
Haass C., Selkoe D. J. (2007). Soluble protein oligomers in neurodegeneration: lessons from the Alzheimer's amyloid beta-peptide. Nat. Rev. Mol. Cell. Biol. 8 (2), 101–112. doi:10.1038/nrm2101
Hackam A. S., Singaraja R., Wellington C. L., Metzler M., McCutcheon K., Zhang T., et al. (1998). The influence of huntingtin protein size on nuclear localization and cellular toxicity. J. Cell. Biol. 141 (5), 1097–1105. doi:10.1083/jcb.141.5.1097
Halliwell B. (1992). Reactive oxygen species and the central nervous system. J. Neurochem. 59 (5), 1609–1623. doi:10.1111/j.1471-4159.1992.tb10990.x
Hamilton J., Brustovetsky T., Khanna R., Brustovetsky N. (2020). Mutant huntingtin does not cross the mitochondrial outer membrane. Hum. Mol. Genet. 29 (17), 2962–2975. doi:10.1093/hmg/ddaa185
Hansen K. G., Boos F., Herrmann J. M. (2018). Accessory signals in protein translocation. Aging (Albany NY) 10 (4), 530–531. doi:10.18632/aging.101435
Hansson Petersen C. A., Alikhani N., Behbahani H., Wiehager B., Pavlov P. F., Alafuzoff I., et al. (2008). The amyloid beta-peptide is imported into mitochondria via the TOM import machinery and localized to mitochondrial cristae. Proc. Natl. Acad. Sci. U. S. A. 105 (35), 13145–13150. doi:10.1073/pnas.0806192105
Hardy J. A., Higgins G. A. (1992). Alzheimer's disease: the amyloid cascade hypothesis. Science 256 (5054), 184–185. doi:10.1126/science.1566067
Havalova H., Ondrovicova G., Keresztesova B., Bauer J. A., Pevala V., Kutejova E., et al. (2021). Mitochondrial HSP70 chaperone system-the influence of post-translational modifications and involvement in human diseases. Int. J. Mol. Sci. 22 (15), 8077. doi:10.3390/ijms22158077
He S., Wang F., Yung K. K. L., Zhang S., Qu S. (2021). Effects of α-synuclein-associated post-translational modifications in Parkinson's disease. ACS Chem. Neurosci. 12 (7), 1061–1071. doi:10.1021/acschemneuro.1c00028
Hernandez-Zimbron L. F., Luna-Munoz J., Mena R., Vazquez-Ramirez R., Kubli-Garfias C., Cribbs D. H., et al. (2012). Amyloid-beta peptide binds to cytochrome C oxidase subunit 1. PLoS One 7 (8), e42344. doi:10.1371/journal.pone.0042344
Herrup K. (2015). The case for rejecting the amyloid cascade hypothesis. Nat. Neurosci. 18 (6), 794–799. doi:10.1038/nn.4017
Heydari A. R., Takahashi R., Gutsmann A., You S., Richardson A. (1994). Hsp70 and aging. Experientia 50 (11-12), 1092–1098. doi:10.1007/BF01923466
Hillen H. (2019). The beta amyloid dysfunction (BAD) hypothesis for Alzheimer’s disease. Front. Neurosci. 13, 1154. doi:10.3389/fnins.2019.01154
Hines V., Brandt A., Griffiths G., Horstmann H., Brutsch H., Schatz G. (1990). Protein import into yeast mitochondria is accelerated by the outer membrane protein MAS70. EMBO J. 9 (10), 3191–3200. doi:10.1002/j.1460-2075.1990.tb07517.x
Hines V., Schatz G. (1993). Precursor binding to yeast mitochondria. A general role for the outer membrane protein Mas70p. J. Biol. Chem. 268 (1), 449–454. doi:10.1016/s0021-9258(18)54172-7
Hirai K., Aliev G., Nunomura A., Fujioka H., Russell R. L., Atwood C. S., et al. (2001). Mitochondrial abnormalities in Alzheimer's disease. J. Neurosci. 21 (9), 3017–3023. doi:10.1523/JNEUROSCI.21-09-03017.2001
Hoe H. S., Lee H. K., Pak D. T. (2012). The upside of APP at synapses. CNS Neurosci. Ther. 18 (1), 47–56. doi:10.1111/j.1755-5949.2010.00221.x
Hoffner G., Djian P. (2014). Monomeric, oligomeric and polymeric proteins in huntington disease and other diseases of polyglutamine expansion. Brain Sci. 4 (1), 91–122. doi:10.3390/brainsci4010091
Hoffner G., Island M. L., Djian P. (2005). Purification of neuronal inclusions of patients with Huntington's disease reveals a broad range of N-terminal fragments of expanded huntingtin and insoluble polymers. J. Neurochem. 95 (1), 125–136. doi:10.1111/j.1471-4159.2005.03348.x
Honlinger A., Kubrich M., Moczko M., Gartner F., Mallet L., Bussereau F., et al. (1995). The mitochondrial receptor complex: mom22 is essential for cell viability and directly interacts with preproteins. Mol. Cell. Biol. 15 (6), 3382–3389. doi:10.1128/mcb.15.6.3382
Horst M., Oppliger W., Rospert S., Schonfeld H. J., Schatz G., Azem A. (1997). Sequential action of two hsp70 complexes during protein import into mitochondria. EMBO J. 16 (8), 1842–1849. doi:10.1093/emboj/16.8.1842
Hoyt D. W., Cyr D. M., Gierasch L. M., Douglas M. G. (1991). Interaction of peptides corresponding to mitochondrial presequences with membranes. J. Biol. Chem. 266 (32), 21693–21699. doi:10.1016/s0021-9258(18)54692-5
Hu W., Wang Z., Zheng H. (2018). Mitochondrial accumulation of amyloid β (Aβ) peptides requires TOMM22 as a main Aβ receptor in yeast. J. Biol. Chem. 293 (33), 12681–12689. doi:10.1074/jbc.RA118.002713
Hu Z. W., Vugmeyster L., Au D. F., Ostrovsky D., Sun Y., Qiang W. (2019). Molecular structure of an N-terminal phosphorylated beta-amyloid fibril. Proc. Natl. Acad. Sci. U. S. A. 116 (23), 11253–11258. doi:10.1073/pnas.1818530116
Huang Y., Mahley R. W. (2014). Apolipoprotein E: structure and function in lipid metabolism, neurobiology, and Alzheimer's diseases. Neurobiol. Dis. 72 Pt, 3–12. doi:10.1016/j.nbd.2014.08.025
Ieva R., Schrempp S. G., Opalinski L., Wollweber F., Hoss P., Heisswolf A. K., et al. (2014). Mgr2 functions as lateral gatekeeper for preprotein sorting in the mitochondrial inner membrane. Mol. Cell. 56 (5), 641–652. doi:10.1016/j.molcel.2014.10.010
Ikon N., Ryan R. O. (2017). Cardiolipin and mitochondrial cristae organization. Biochim. Biophys. Acta Biomembr. 1859 (6), 1156–1163. doi:10.1016/j.bbamem.2017.03.013
Iosefson O., Sharon S., Goloubinoff P., Azem A. (2012). Reactivation of protein aggregates by mortalin and Tid1-the human mitochondrial Hsp70 chaperone system. Cell. Stress Chaperones 17 (1), 57–66. doi:10.1007/s12192-011-0285-3
Jacobsen K. T., Iverfeldt K. (2009). Amyloid precursor protein and its homologues: a family of proteolysis-dependent receptors. Cell. Mol. Life Sci. 66 (14), 2299–2318. doi:10.1007/s00018-009-0020-8
Jao C. C., Hegde B. G., Chen J., Haworth I. S., Langen R. (2008). Structure of membrane-bound alpha-synuclein from site-directed spin labeling and computational refinement. Proc. Natl. Acad. Sci. U. S. A. 105 (50), 19666–19671. doi:10.1073/pnas.0807826105
Jenco J. M., Rawlingson A., Daniels B., Morris A. J. (1998). Regulation of phospholipase D2: selective inhibition of mammalian phospholipase D isoenzymes by alpha- and beta-synucleins. Biochemistry 37 (14), 4901–4909. doi:10.1021/bi972776r
Jiang X., Jiang H., Shen Z., Wang X. (2014). Activation of mitochondrial protease OMA1 by Bax and Bak promotes cytochrome c release during apoptosis. Proc. Natl. Acad. Sci. U. S. A. 111 (41), 14782–14787. doi:10.1073/pnas.1417253111
Jin Y. N., Johnson G. V. (2010). The interrelationship between mitochondrial dysfunction and transcriptional dysregulation in Huntington disease. J. Bioenerg. Biomembr. 42 (3), 199–205. doi:10.1007/s10863-010-9286-7
Jo E., Fuller N., Rand R. P., St George-Hyslop P., Fraser P. E. (2002). Defective membrane interactions of familial Parkinson's disease mutant A30P alpha-synuclein. J. Mol. Biol. 315 (4), 799–807. doi:10.1006/jmbi.2001.5269
Jores T., Lawatscheck J., Beke V., Franz-Wachtel M., Yunoki K., Fitzgerald J. C., et al. (2018). Cytosolic Hsp70 and Hsp40 chaperones enable the biogenesis of mitochondrial beta-barrel proteins. J. Cell. Biol. 217 (9), 3091–3108. doi:10.1083/jcb.201712029
Joshi A., Ito T., Picard D., Neckers L. (2022). The mitochondrial HSP90 paralog TRAP1: structural dynamics, interactome, role in metabolic regulation, and inhibitors. Biomolecules 12 (7), 880. doi:10.3390/biom12070880
Jurcau A., Jurcau C. M. (2023). Mitochondria in Huntington's disease: implications in pathogenesis and mitochondrial-targeted therapeutic strategies. Neural Regen. Res. 18 (7), 1472–1477. doi:10.4103/1673-5374.360289
Jurcau A., Jurcau M. C. (2022). Therapeutic strategies in huntington's disease: from genetic defect to gene therapy. Biomedicines 10 (6), 1895. doi:10.3390/biomedicines10081895
Kakeshpour T., Ramanujam V., Barnes C. A., Shen Y., Ying J., Bax A. (2021). A lowly populated, transient β-sheet structure in monomeric Aβ1-42 identified by multinuclear NMR of chemical denaturation. Biophys. Chem. 270, 106531. doi:10.1016/j.bpc.2020.106531
Kamp F., Exner N., Lutz A. K., Wender N., Hegermann J., Brunner B., et al. (2010). Inhibition of mitochondrial fusion by alpha-synuclein is rescued by PINK1, Parkin and DJ-1. EMBO J. 29 (20), 3571–3589. doi:10.1038/emboj.2010.223
Kampinga H. H., Craig E. A. (2010). The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat. Rev. Mol. Cell. Biol. 11 (8), 579–592. doi:10.1038/nrm2941
Kanamori T., Nishikawa S., Nakai M., Shin I., Schultz P. G., Endo T. (1999). Uncoupling of transfer of the presequence and unfolding of the mature domain in precursor translocation across the mitochondrial outer membrane. Proc. Natl. Acad. Sci. U. S. A. 96 (7), 3634–3639. doi:10.1073/pnas.96.7.3634
Kann O., Kovacs R. (2007). Mitochondria and neuronal activity. Am. J. Physiol. Cell. Physiol. 292 (2), C641–C657. doi:10.1152/ajpcell.00222.2006
Karagoz G. E., Rudiger S. G. (2015). Hsp90 interaction with clients. Trends Biochem. Sci. 40 (2), 117–125. doi:10.1016/j.tibs.2014.12.002
Karniely S., Pines O. (2005). Single translation-dual destination: mechanisms of dual protein targeting in eukaryotes. EMBO Rep. 6 (5), 420–425. doi:10.1038/sj.embor.7400394
Ke P. C., Zhou R., Serpell L. C., Riek R., Knowles T. P. J., Lashuel H. A., et al. (2020). Half a century of amyloids: past, present and future. Chem. Soc. Rev. 49 (15), 5473–5509. doi:10.1039/c9cs00199a
Kegel-Gleason K. B. (2013). Huntingtin interactions with membrane phospholipids: strategic targets for therapeutic intervention? J. Huntingt. Dis. 2 (3), 239–250. doi:10.3233/JHD-130068
Kepchia D., Currais A., Dargusch R., Finley K., Schubert D., Maher P. (2021). Geroprotective effects of Alzheimer's disease drug candidates. Aging (Albany NY) 13 (3), 3269–3289. doi:10.18632/aging.202631
Kepchia D., Huang L., Currais A., Liang Z., Fischer W., Maher P. (2022). The Alzheimer's disease drug candidate J147 decreases blood plasma fatty acid levels via modulation of AMPK/ACC1 signaling in the liver. Biomed. Pharmacother. 147, 112648. doi:10.1016/j.biopha.2022.112648
Khalil B., El Fissi N., Aouane A., Cabirol-Pol M. J., Rival T., Lievens J. C. (2015). PINK1-induced mitophagy promotes neuroprotection in Huntington's disease. Cell. Death Dis. 6 (1), e1617. doi:10.1038/cddis.2014.581
Kiebler M., Keil P., Schneider H., van der Klei I. J., Pfanner N., Neupert W. (1993). The mitochondrial receptor complex: a central role of MOM22 in mediating preprotein transfer from receptors to the general insertion pore. Cell. 74 (3), 483–492. doi:10.1016/0092-8674(93)80050-o
Kiechle T., Dedeoglu A., Kubilus J., Kowall N. W., Beal M. F., Friedlander R. M., et al. (2002). Cytochrome C and caspase-9 expression in Huntington's disease. Neuromolecular Med. 1 (3), 183–195. doi:10.1385/NMM:1:3:183
Kim M. W., Chelliah Y., Kim S. W., Otwinowski Z., Bezprozvanny I. (2009). Secondary structure of Huntingtin amino-terminal region. Structure 17 (9), 1205–1212. doi:10.1016/j.str.2009.08.002
Kim Y. J., Yi Y., Sapp E., Wang Y., Cuiffo B., Kegel K. B., et al. (2001). Caspase 3-cleaved N-terminal fragments of wild-type and mutant huntingtin are present in normal and Huntington's disease brains, associate with membranes, and undergo calpain-dependent proteolysis. Proc. Natl. Acad. Sci. U. S. A. 98 (22), 12784–12789. doi:10.1073/pnas.221451398
Klaips C. L., Jayaraj G. G., Hartl F. U. (2018). Pathways of cellular proteostasis in aging and disease. J. Cell. Biol. 217 (1), 51–63. doi:10.1083/jcb.201709072
Klein C., Westenberger A. (2012). Genetics of Parkinson's disease. Cold Spring Harb. Perspect. Med. 2 (1), a008888. doi:10.1101/cshperspect.a008888
Klivenyi P., Starkov A. A., Calingasan N. Y., Gardian G., Browne S. E., Yang L., et al. (2004). Mice deficient in dihydrolipoamide dehydrogenase show increased vulnerability to MPTP, malonate and 3-nitropropionic acid neurotoxicity. J. Neurochem. 88 (6), 1352–1360. doi:10.1046/j.1471-4159.2003.02263.x
Knopman D. S., Amieva H., Petersen R. C., Chetelat G., Holtzman D. M., Hyman B. T., et al. (2021). Alzheimer disease. Nat. Rev. Dis. Prim. 7 (1), 33. doi:10.1038/s41572-021-00269-y
Koistinaho M., Lin S., Wu X., Esterman M., Koger D., Hanson J., et al. (2004). Apolipoprotein E promotes astrocyte colocalization and degradation of deposited amyloid-beta peptides. Nat. Med. 10 (7), 719–726. doi:10.1038/nm1058
Kolobkova Y. A., Vigont V. A., Shalygin A. V., Kaznacheyeva E. V. (2017). Huntington's disease: calcium dyshomeostasis and pathology models. Acta Naturae 9 (2), 34–46. doi:10.32607/20758251-2017-9-2-34-46
Komiya T., Rospert S., Koehler C., Looser R., Schatz G., Mihara K. (1998). Interaction of mitochondrial targeting signals with acidic receptor domains along the protein import pathway: evidence for the 'acid chain' hypothesis. EMBO J. 17 (14), 3886–3898. doi:10.1093/emboj/17.14.3886
Kronidou N. G., Oppliger W., Bolliger L., Hannavy K., Glick B. S., Schatz G., et al. (1994). Dynamic interaction between Isp45 and mitochondrial hsp70 in the protein import system of the yeast mitochondrial inner membrane. Proc. Natl. Acad. Sci. U. S. A. 91 (26), 12818–12822. doi:10.1073/pnas.91.26.12818
Kucukkose C., Taskin A. A., Marada A., Brummer T., Dennerlein S., Vogtle F. N. (2021). Functional coupling of presequence processing and degradation in human mitochondria. FEBS J. 288 (2), 600–613. doi:10.1111/febs.15358
Kumar V., Kim S. H., Bishayee K. (2022). Dysfunctional glucose metabolism in alzheimer's disease onset and potential pharmacological interventions. Int. J. Mol. Sci. 23 (17), 9540. doi:10.3390/ijms23179540
Kunkele K. P., Heins S., Dembowski M., Nargang F. E., Benz R., Thieffry M., et al. (1998). The preprotein translocation channel of the outer membrane of mitochondria. Cell. 93 (6), 1009–1019. doi:10.1016/s0092-8674(00)81206-4
Kunova N., Havalova H., Ondrovicova G., Stojkovicova B., Bauer J. A., Bauerova-Hlinkova V., et al. (2022). Mitochondrial processing peptidases-structure, function and the role in human diseases. Int. J. Mol. Sci. 23 (3), 1297. doi:10.3390/ijms23031297
Kunze M., Berger J. (2015). The similarity between N-terminal targeting signals for protein import into different organelles and its evolutionary relevance. Front. Physiol. 6, 259. doi:10.3389/fphys.2015.00259
Kuzniewska B., Cysewski D., Wasilewski M., Sakowska P., Milek J., Kulinski T. M., et al. (2020). Mitochondrial protein biogenesis in the synapse is supported by local translation. EMBO Rep. 21 (8), e48882. doi:10.15252/embr.201948882
Labbadia J., Morimoto R. I. (2015). The biology of proteostasis in aging and disease. Annu. Rev. Biochem. 84, 435–464. doi:10.1146/annurev-biochem-060614-033955
Landles C., Sathasivam K., Weiss A., Woodman B., Moffitt H., Finkbeiner S., et al. (2010). Proteolysis of mutant huntingtin produces an exon 1 fragment that accumulates as an aggregated protein in neuronal nuclei in Huntington disease. J. Biol. Chem. 285 (12), 8808–8823. doi:10.1074/jbc.M109.075028
Lansbury P. T., Lashuel H. A. (2006). A century-old debate on protein aggregation and neurodegeneration enters the clinic. Nature 443 (7113), 774–779. doi:10.1038/nature05290
Larson M. E., Lesné S. E. (2012). Soluble Aβ oligomer production and toxicity. J. Neurochem. 120(1), 125–139. doi:10.1111/j.1471-4159.2011.07478.x
Lazar A. N., Hanbouch L., Boussicaut L., Fourmaux B., Daira P., Millan M. J., et al. (2022). Lipid dys-homeostasis contributes to APOE4-associated AD pathology. Cells 11 (22), 3616. doi:10.3390/cells11223616
Lazarov O., Morfini G. A., Lee E. B., Farah M. H., Szodorai A., DeBoer S. R., et al. (2005). Axonal transport, amyloid precursor protein, kinesin-1, and the processing apparatus: revisited. J. Neurosci. 25 (9), 2386–2395. doi:10.1523/JNEUROSCI.3089-04.2005
Leal N. S., Dentoni G., Schreiner B., Naia L., Piras A., Graff C., et al. (2020). Amyloid beta-peptide increases mitochondria-endoplasmic reticulum contact altering mitochondrial function and autophagosome formation in alzheimer's disease-related models. Cells 9 (12), 2552. doi:10.3390/cells9122552
Lee S., Lee H., Yoo S., Ieva R., van der Laan M., von Heijne G., et al. (2020). The Mgr2 subunit of the TIM23 complex regulates membrane insertion of marginal stop-transfer signals in the mitochondrial inner membrane. FEBS Lett. 594 (6), 1081–1087. doi:10.1002/1873-3468.13692
Lenkiewicz A. M., Krakowczyk M., Bragoszewski P. (2021). Cytosolic quality control of mitochondrial protein precursors-the early stages of the organelle biogenesis. Int. J. Mol. Sci. 23 (1), 7. doi:10.3390/ijms23010007
Lesnik C., Cohen Y., Atir-Lande A., Schuldiner M., Arava Y. (2014). OM14 is a mitochondrial receptor for cytosolic ribosomes that supports co-translational import into mitochondria. Nat. Commun. 5, 5711. doi:10.1038/ncomms6711
Li N. S., Liang W., Piccirilli J. A., Tang W. J. (2019b). Reinvestigating the synthesis and efficacy of small benzimidazole derivatives as presequence protease enhancers. Eur. J. Med. Chem. 184, 111746. doi:10.1016/j.ejmech.2019.111746
Li S. H., Schilling G., Young W. S., Li X. J., Margolis R. L., Stine O. C., et al. (1993). Huntington's disease gene (IT15) is widely expressed in human and rat tissues. Neuron 11 (5), 985–993. doi:10.1016/0896-6273(93)90127-d
Li W. W., Yang R., Guo J. C., Ren H. M., Zha X. L., Cheng J. S., et al. (2007). Localization of alpha-synuclein to mitochondria within midbrain of mice. Neuroreport 18 (15), 1543–1546. doi:10.1097/WNR.0b013e3282f03db4
Li X. X., Tsoi B., Li Y. F., Kurihara H., He R. R. (2015). Cardiolipin and its different properties in mitophagy and apoptosis. J. Histochem Cytochem 63 (5), 301–311. doi:10.1369/0022155415574818
Li Y., Xue Y., Xu X., Wang G., Liu Y., Wu H., et al. (2019a). A mitochondrial FUNDC1/HSC70 interaction organizes the proteostatic stress response at the risk of cell morbidity. EMBO J. 38 (3), e98786. doi:10.15252/embj.201798786
Li Z., Cao Y., Pei H., Ma L., Yang Y., Li H. (2023). The contribution of mitochondria-associated endoplasmic reticulum membranes (MAMs) dysfunction in Alzheimer's disease and the potential countermeasure. Front. Neurosci. 17, 1158204. doi:10.3389/fnins.2023.1158204
Liang W. S., Reiman E. M., Valla J., Dunckley T., Beach T. G., Grover A., et al. (2008). Alzheimer's disease is associated with reduced expression of energy metabolism genes in posterior cingulate neurons. Proc. Natl. Acad. Sci. U. S. A. 105 (11), 4441–4446. doi:10.1073/pnas.0709259105
Liao C. Y., Rikke B. A., Johnson T. E., Diaz V., Nelson J. F. (2010). Genetic variation in the murine lifespan response to dietary restriction: from life extension to life shortening. Aging Cell. 9 (1), 92–95. doi:10.1111/j.1474-9726.2009.00533.x
Lieberman A. P., Shakkottai V. G., Albin R. L. (2019). Polyglutamine repeats in neurodegenerative diseases. Annu. Rev. Pathol. 14, 1–27. doi:10.1146/annurev-pathmechdis-012418-012857
Lin H. K., Boatz J. C., Krabbendam I. E., Kodali R., Hou Z., Wetzel R., et al. (2017). Fibril polymorphism affects immobilized non-amyloid flanking domains of huntingtin exon1 rather than its polyglutamine core. Nat. Commun. 8, 15462. doi:10.1038/ncomms15462
Lin T., Tjernberg L. O., Schedin-Weiss S. (2021). Neuronal trafficking of the amyloid precursor protein—what do we really know? Biomed 9 (7), 801. doi:10.3390/biomedicines9070801
Liu J., Jiang J., Qiu J., Wang L., Zhuo J., Wang B., et al. (2022). Urolithin A protects dopaminergic neurons in experimental models of Parkinson's disease by promoting mitochondrial biogenesis through the SIRT1/PGC-1α signaling pathway. Food Funct. 13 (1), 375–385. doi:10.1039/d1fo02534a
Liu T., Zhang T., Nicolas M., Boussicault L., Rice H., Soldano A., et al. (2021a). The amyloid precursor protein is a conserved Wnt receptor. eLife 10, e69199. doi:10.7554/eLife.69199
Liu Y., Fu H., Wu Y., Nie B., Liu F., Wang T., et al. (2021b). Elamipretide (SS-31) improves functional connectivity in Hippocampus and other related regions following prolonged neuroinflammation induced by lipopolysaccharide in aged rats. Front. Aging Neurosci. 13, 600484. doi:10.3389/fnagi.2021.600484
Longen S., Woellhaf M. W., Petrungaro C., Riemer J., Herrmann J. M. (2014). The disulfide relay of the intermembrane space oxidizes the ribosomal subunit mrp10 on its transit into the mitochondrial matrix. Dev. Cell. 28 (1), 30–42. doi:10.1016/j.devcel.2013.11.007
Lontay B., Kiss A., Virag L., Tar K. (2020). How do post-translational modifications influence the pathomechanistic landscape of huntington's disease? A comprehensive review. Int. J. Mol. Sci. 21 (12), 4282. doi:10.3390/ijms21124282
Lopes C., Ferreira I. L., Maranga C., Beatriz M., Mota S. I., Sereno J., et al. (2022). Mitochondrial and redox modifications in early stages of Huntington's disease. Redox Biol. 56, 102424. doi:10.1016/j.redox.2022.102424
Lu L., Zhang C., Cai Q., Lu Q., Duan C., Zhu Y., et al. (2013). Voltage-dependent anion channel involved in the alpha-synuclein-induced dopaminergic neuron toxicity in rats. Acta Biochim. Biophys. Sin. (Shanghai). 45 (3), 170–178. doi:10.1093/abbs/gms114
Lu Z. H., Chakraborty G., Ledeen R. W., Yahya D., Wu G. (2004). N-Acetylaspartate synthase is bimodally expressed in microsomes and mitochondria of brain. Brain Res. Mol. Brain Res. 122 (1), 71–78. doi:10.1016/j.molbrainres.2003.12.002
Ludtmann M. H., Angelova P. R., Ninkina N. N., Gandhi S., Buchman V. L., Abramov A. Y. (2016). Monomeric alpha-synuclein exerts a physiological role on brain ATP synthase. J. Neurosci. 36 (41), 10510–10521. doi:10.1523/JNEUROSCI.1659-16.2016
Ludtmann M. H. R., Angelova P. R., Horrocks M. H., Choi M. L., Rodrigues M., Baev A. Y., et al. (2018). α-synuclein oligomers interact with ATP synthase and open the permeability transition pore in Parkinson's disease. Nat. Commun. 9 (1), 2293. doi:10.1038/s41467-018-04422-2
Lunkes A., Lindenberg K. S., Ben-Haiem L., Weber C., Devys D., Landwehrmeyer G. B., et al. (2002). Proteases acting on mutant huntingtin generate cleaved products that differentially build up cytoplasmic and nuclear inclusions. Mol. Cell. 10 (2), 259–269. doi:10.1016/s1097-2765(02)00602-0
Lustbader J. W., Cirilli M., Lin C., Xu H. W., Takuma K., Wang N., et al. (2004). ABAD directly links Abeta to mitochondrial toxicity in Alzheimer's disease. Science 304 (5669), 448–452. doi:10.1126/science.1091230
Lytovchenko O., Melin J., Schulz C., Kilisch M., Hutu D. P., Rehling P. (2013). Signal recognition initiates reorganization of the presequence translocase during protein import. EMBO J. 32 (6), 886–898. doi:10.1038/emboj.2013.23
MacDonald M. E., Ambrose C. M., Duyao M. P., Myers R. H., Lin C., Srinidhi L., et al. (1993). A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. The Huntington's Disease Collaborative Research Group. Cell. 72 (6), 971–983. doi:10.1016/0092-8674(93)90585-e
Mager F., Sokolova L., Lintzel J., Brutschy B., Nussberger S. (2010). LILBID-mass spectrometry of the mitochondrial preprotein translocase TOM. J. Phys. Condens Matter 22 (45), 454132. doi:10.1088/0953-8984/22/45/454132
Mahley R. W. (2016). Central nervous system lipoproteins: apoE and regulation of cholesterol metabolism. Arterioscler. Thromb. Vasc. Biol. 36 (7), 1305–1315. doi:10.1161/ATVBAHA.116.307023
Malhotra K., Modak A., Nangia S., Daman T. H., Gunsel U., Robinson V. L., et al. (2017). Cardiolipin mediates membrane and channel interactions of the mitochondrial TIM23 protein import complex receptor Tim50. Sci. Adv. 3 (9), e1700532. doi:10.1126/sciadv.1700532
Malhotra K., Sathappa M., Landin J. S., Johnson A. E., Alder N. N. (2013). Structural changes in the mitochondrial Tim23 channel are coupled to the proton-motive force. Nat. Struct. Mol. Biol. 20 (8), 965–972. doi:10.1038/nsmb.2613
Malpartida A. B., Williamson M., Narendra D. P., Wade-Martins R., Ryan B. J. (2021). Mitochondrial dysfunction and mitophagy in Parkinson's disease: from mechanism to therapy. Trends Biochem. Sci. 46 (4), 329–343. doi:10.1016/j.tibs.2020.11.007
Manczak M., Anekonda T. S., Henson E., Park B. S., Quinn J., Reddy P. H. (2006). Mitochondria are a direct site of A beta accumulation in Alzheimer's disease neurons: implications for free radical generation and oxidative damage in disease progression. Hum. Mol. Genet. 15 (9), 1437–1449. doi:10.1093/hmg/ddl066
Manczak M., Calkins M. J., Reddy P. H. (2011). Impaired mitochondrial dynamics and abnormal interaction of amyloid beta with mitochondrial protein Drp1 in neurons from patients with Alzheimer's disease: implications for neuronal damage. Hum. Mol. Genet. 20 (13), 2495–2509. doi:10.1093/hmg/ddr139
Manczak M., Mao P., Calkins M. J., Cornea A., Reddy A. P., Murphy M. P., et al. (2010). Mitochondria-targeted antioxidants protect against amyloid-beta toxicity in Alzheimer's disease neurons. J. Alzheimers Dis. 20(2), S609–S631. doi:10.3233/JAD-2010-100564
Manczak M., Reddy P. H. (2012). Abnormal interaction between the mitochondrial fission protein Drp1 and hyperphosphorylated tau in Alzheimer's disease neurons: implications for mitochondrial dysfunction and neuronal damage. Hum. Mol. Genet. 21 (11), 2538–2547. doi:10.1093/hmg/dds072
Manczak M., Reddy P. H. (2015). Mitochondrial division inhibitor 1 protects against mutant huntingtin-induced abnormal mitochondrial dynamics and neuronal damage in Huntington's disease. Hum. Mol. Genet. 24 (25), 7308–7325. doi:10.1093/hmg/ddv429
Mandal A., Drerup C. M. (2019). Axonal transport and mitochondrial function in neurons. Front. Cell. Neurosci. 13, 373. doi:10.3389/fncel.2019.00373
Mangiarini L., Sathasivam K., Seller M., Cozens B., Harper A., Hetherington C., et al. (1996). Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell. 87 (3), 493–506. doi:10.1016/s0092-8674(00)81369-0
Manzanza N. O., Sedlackova L., Kalaria R. N. (2021). Alpha-synuclein post-translational modifications: implications for pathogenesis of lewy body disorders. Front. Aging Neurosci. 13, 690293. doi:10.3389/fnagi.2021.690293
Marella M., Seo B. B., Yagi T., Matsuno-Yagi A. (2009). Parkinson's disease and mitochondrial complex I: a perspective on the Ndi1 therapy. J. Bioenerg. Biomembr. 41 (6), 493–497. doi:10.1007/s10863-009-9249-z
Marom M., Azem A., Mokranjac D. (2011a). Understanding the molecular mechanism of protein translocation across the mitochondrial inner membrane: still a long way to go. Biochimica Biophysica Acta (BBA) - Biomembr. 1808 (3), 990–1001. doi:10.1016/j.bbamem.2010.07.011
Marom M., Dayan D., Demishtein-Zohary K., Mokranjac D., Neupert W., Azem A. (2011b). Direct interaction of mitochondrial targeting presequences with purified components of the TIM23 protein complex. J. Biol. Chem. 286 (51), 43809–43815. doi:10.1074/jbc.M111.261040
Martens Y. A., Zhao N., Liu C. C., Kanekiyo T., Yang A. J., Goate A. M., et al. (2022). ApoE Cascade Hypothesis in the pathogenesis of Alzheimer's disease and related dementias. Neuron 110 (8), 1304–1317. doi:10.1016/j.neuron.2022.03.004
Martensson C. U., Priesnitz C., Song J., Ellenrieder L., Doan K. N., Boos F., et al. (2019). Mitochondrial protein translocation-associated degradation. Nature 569 (7758), 679–683. doi:10.1038/s41586-019-1227-y
Martin D. D. O., Schmidt M. E., Nguyen Y. T., Lazic N., Hayden M. R. (2019). Identification of a novel caspase cleavage site in huntingtin that regulates mutant huntingtin clearance. FASEB J. 33 (3), 3190–3197. doi:10.1096/fj.201701510RRR
Martin J., Mahlke K., Pfanner N. (1991). Role of an energized inner membrane in mitochondrial protein import. Delta psi drives the movement of presequences. J. Biol. Chem. 266 (27), 18051–18057. doi:10.1016/s0021-9258(18)55235-2
Martindale D., Hackam A., Wieczorek A., Ellerby L., Wellington C., McCutcheon K., et al. (1998). Length of huntingtin and its polyglutamine tract influences localization and frequency of intracellular aggregates. Nat. Genet. 18 (2), 150–154. doi:10.1038/ng0298-150
Martinez-Caballero S., Grigoriev S. M., Herrmann J. M., Campo M. L., Kinnally K. W. (2007). Tim17p regulates the twin pore structure and voltage gating of the mitochondrial protein import complex TIM23. J. Biol. Chem. 282 (6), 3584–3593. doi:10.1074/jbc.M607551200
Mastroeni D., Khdour O. M., Delvaux E., Nolz J., Olsen G., Berchtold N., et al. (2017). Nuclear but not mitochondrial-encoded oxidative phosphorylation genes are altered in aging, mild cognitive impairment, and Alzheimer's disease. Alzheimers Dement. 13 (5), 510–519. doi:10.1016/j.jalz.2016.09.003
Matouschek A., Azem A., Ratliff K., Glick B. S., Schmid K., Schatz G. (1997). Active unfolding of precursor proteins during mitochondrial protein import. EMBO J. 16 (22), 6727–6736. doi:10.1093/emboj/16.22.6727
Matsumoto S., Uchiumi T., Saito T., Yagi M., Takazaki S., Kanki T., et al. (2012). Localization of mRNAs encoding human mitochondrial oxidative phosphorylation proteins. Mitochondrion 12 (3), 391–398. doi:10.1016/j.mito.2012.02.004
Matta S. K., Kumar A., D'Silva P. (2020). Mgr2 regulates mitochondrial preprotein import by associating with channel-forming Tim23 subunit. Mol. Biol. Cell. 31 (11), 1112–1123. doi:10.1091/mbc.E19-12-0677
Matta S. K., Pareek G., Bankapalli K., Oblesha A., D'Silva P. (2017). Role of Tim17 transmembrane regions in regulating the architecture of presequence translocase and mitochondrial DNA stability. Mol. Cell. Biol. 37 (6), e00491-16. doi:10.1128/MCB.00491-16
Mayer A., Nargang F. E., Neupert W., Lill R. (1995). MOM22 is a receptor for mitochondrial targeting sequences and cooperates with MOM19. EMBO J. 14 (17), 4204–4211. doi:10.1002/j.1460-2075.1995.tb00094.x
McDaid J., Mustaly-Kalimi S., Stutzmann G. E. (2020). Ca(2+) dyshomeostasis disrupts neuronal and synaptic function in alzheimer's disease. Cells 9 (12), 2655. doi:10.3390/cells9122655
McFarland M. A., Ellis C. E., Markey S. P., Nussbaum R. L. (2008). Proteomics analysis identifies phosphorylation-dependent alpha-synuclein protein interactions. Mol. Cell. Proteomics 7 (11), 2123–2137. doi:10.1074/mcp.M800116-MCP200
McGlinchey R. P., Ni X., Shadish J. A., Jiang J., Lee J. C. (2021). The N terminus of α-synuclein dictates fibril formation. Proc. Natl. Acad. Sci. U. S. A. 118 (35), e2023487118. doi:10.1073/pnas.2023487118
Meinecke M., Wagner R., Kovermann P., Guiard B., Mick D. U., Hutu D. P., et al. (2006). Tim50 maintains the permeability barrier of the mitochondrial inner membrane. Science 312 (5779), 1523–1526. doi:10.1126/science.1127628
Melin J., Kilisch M., Neumann P., Lytovchenko O., Gomkale R., Schendzielorz A., et al. (2015). A presequence-binding groove in Tom70 supports import of Mdl1 into mitochondria. Biochim. Biophys. Acta 1853 (8), 1850–1859. doi:10.1016/j.bbamcr.2015.04.021
Michalek M., Salnikov E. S., Bechinger B. (2013). Structure and topology of the huntingtin 1-17 membrane anchor by a combined solution and solid-state NMR approach. Biophys. J. 105 (3), 699–710. doi:10.1016/j.bpj.2013.06.030
Mick D. U., Dennerlein S., Wiese H., Reinhold R., Pacheu-Grau D., Lorenzi I., et al. (2012). MITRAC links mitochondrial protein translocation to respiratory-chain assembly and translational regulation. Cell. 151 (7), 1528–1541. doi:10.1016/j.cell.2012.11.053
Middleton E. R., Rhoades E. (2010). Effects of curvature and composition on α-synuclein binding to lipid vesicles. Biophys. J. 99 (7), 2279–2288. doi:10.1016/j.bpj.2010.07.056
Milakovic T., Quintanilla R. A., Johnson G. V. (2006). Mutant huntingtin expression induces mitochondrial calcium handling defects in clonal striatal cells: functional consequences. J. Biol. Chem. 281 (46), 34785–34795. doi:10.1074/jbc.M603845200
Milton V. J., Sweeney S. T. (2012). Oxidative stress in synapse development and function. Dev. Neurobiol. 72 (1), 100–110. doi:10.1002/dneu.20957
Misrani A., Tabassum S., Yang L. (2021). Mitochondrial dysfunction and oxidative stress in alzheimer's disease. Front. Aging Neurosci. 13, 617588. doi:10.3389/fnagi.2021.617588
Mitchell W., Ng E. A., Tamucci J. D., Boyd K. J., Sathappa M., Coscia A., et al. (2020). The mitochondria-targeted peptide SS-31 binds lipid bilayers and modulates surface electrostatics as a key component of its mechanism of action. J. Biol. Chem. 295 (21), 7452–7469. doi:10.1074/jbc.RA119.012094
Mitchell W., Tamucci J. D., Ng E. L., Liu S., Birk A. V., Szeto H. H., et al. (2022). Structure-activity relationships of mitochondria-targeted tetrapeptide pharmacological compounds. Elife 11, e75531. doi:10.7554/eLife.75531
Mochel F., Durant B., Meng X., O'Callaghan J., Yu H., Brouillet E., et al. (2012). Early alterations of brain cellular energy homeostasis in Huntington disease models. J. Biol. Chem. 287 (2), 1361–1370. doi:10.1074/jbc.M111.309849
Moczko M., Bomer U., Kubrich M., Zufall N., Honlinger A., Pfanner N. (1997). The intermembrane space domain of mitochondrial Tom22 functions as a trans binding site for preproteins with N-terminal targeting sequences. Mol. Cell. Biol. 17 (11), 6574–6584. doi:10.1128/mcb.17.11.6574
Model K., Meisinger C., Kuhlbrandt W. (2008). Cryo-electron microscopy structure of a yeast mitochondrial preprotein translocase. J. Mol. Biol. 383 (5), 1049–1057. doi:10.1016/j.jmb.2008.07.087
Mokranjac D. (2020). How to get to the other side of the mitochondrial inner membrane - the protein import motor. Biol. Chem. 401 (6-7), 723–736. doi:10.1515/hsz-2020-0106
Mokranjac D., Paschen S. A., Kozany C., Prokisch H., Hoppins S. C., Nargang F. E., et al. (2003a). Tim50, a novel component of the TIM23 preprotein translocase of mitochondria. EMBO J. 22 (4), 816–825. doi:10.1093/emboj/cdg090
Mokranjac D., Popov-Celeketic D., Hell K., Neupert W. (2005). Role of Tim21 in mitochondrial translocation contact sites. J. Biol. Chem. 280 (25), 23437–23440. doi:10.1074/jbc.C500135200
Mokranjac D., Sichting M., Neupert W., Hell K. (2003b). Tim14, a novel key component of the import motor of the TIM23 protein translocase of mitochondria. EMBO J. 22 (19), 4945–4956. doi:10.1093/emboj/cdg485
Mokranjac D., Sichting M., Popov-Celeketic D., Mapa K., Gevorkyan-Airapetov L., Zohary K., et al. (2009). Role of Tim50 in the transfer of precursor proteins from the outer to the inner membrane of mitochondria. Mol. Biol. Cell. 20 (5), 1400–1407. doi:10.1091/mbc.e08-09-0934
Moran Luengo T., Mayer M. P., Rudiger S. G. D. (2019). The hsp70-hsp90 chaperone cascade in protein folding. Trends Cell. Biol. 29 (2), 164–177. doi:10.1016/j.tcb.2018.10.004
Moretti D., Tambone S., Cerretani M., Fezzardi P., Missineo A., Sherman L. T., et al. (2021). NRF2 activation by reversible KEAP1 binding induces the antioxidant response in primary neurons and astrocytes of a Huntington's disease mouse model. Free Radic. Biol. Med. 162, 243–254. doi:10.1016/j.freeradbiomed.2020.10.022
Morgenstern M., Peikert C. D., Lubbert P., Suppanz I., Klemm C., Alka O., et al. (2021). Quantitative high-confidence human mitochondrial proteome and its dynamics in cellular context. Cell. Metab. 33 (12), 2464–2483.e18. doi:10.1016/j.cmet.2021.11.001
Morigaki R., Goto S. (2017). Striatal vulnerability in huntington's disease: neuroprotection versus neurotoxicity. Brain Sci. 7 (6), 63. doi:10.3390/brainsci7060063
Moro F., Sirrenberg C., Schneider H. C., Neupert W., Brunner M. (1999). The TIM17.23 preprotein translocase of mitochondria: composition and function in protein transport into the matrix. EMBO J. 18 (13), 3667–3675. doi:10.1093/emboj/18.13.3667
Mossmann D., Vogtle F. N., Taskin A. A., Teixeira P. F., Ring J., Burkhart J. M., et al. (2014). Amyloid-beta peptide induces mitochondrial dysfunction by inhibition of preprotein maturation. Cell. Metab. 20 (4), 662–669. doi:10.1016/j.cmet.2014.07.024
Munoz-Carvajal F., Sanhueza M. (2020). The mitochondrial unfolded protein response: a hinge between healthy and pathological aging. Front. Aging Neurosci. 12, 581849. doi:10.3389/fnagi.2020.581849
Murakami H., Pain D., Blobel G. (1988). 70-kD heat shock-related protein is one of at least two distinct cytosolic factors stimulating protein import into mitochondria. J. Cell. Biol. 107 (1), 2051–2057. doi:10.1083/jcb.107.6.2051
Murali Mahadevan H., Hashemiaghdam A., Ashrafi G., Harbauer A. B. (2021). Mitochondria in neuronal health: from energy metabolism to Parkinson's disease. Adv. Biol. (Weinh). 5 (9), e2100663. doi:10.1002/adbi.202100663
Muresan V., Ladescu Muresan Z. (2015). Amyloid-beta precursor protein: multiple fragments, numerous transport routes and mechanisms. Exp. Cell. Res. 334 (1), 45–53. doi:10.1016/j.yexcr.2014.12.014
Musteikyte G., Jayaram A. K., Xu C. K., Vendruscolo M., Krainer G., Knowles T. P. J. (2021). Interactions of α-synuclein oligomers with lipid membranes. Biochim. Biophys. Acta Biomembr. 1863 (4), 183536. doi:10.1016/j.bbamem.2020.183536
Muto T., Obita T., Abe Y., Shodai T., Endo T., Kohda D. (2001). NMR identification of the Tom20 binding segment in mitochondrial presequences. J. Mol. Biol. 306 (2), 137–143. doi:10.1006/jmbi.2000.4397
Naia L., Ferreira I. L., Cunha-Oliveira T., Duarte A. I., Ribeiro M., Rosenstock T. R., et al. (2015). Activation of IGF-1 and insulin signaling pathways ameliorate mitochondrial function and energy metabolism in Huntington's Disease human lymphoblasts. Mol. Neurobiol. 51 (1), 331–348. doi:10.1007/s12035-014-8735-4
Najm R., Jones E. A., Huang Y. (2019). Apolipoprotein E4, inhibitory network dysfunction, and Alzheimer's disease. Mol. Neurodegener. 14 (1), 24. doi:10.1186/s13024-019-0324-6
Nakagawa S., Lagisz M., Hector K. L., Spencer H. G. (2012). Comparative and meta-analytic insights into life extension via dietary restriction. Aging Cell. 11 (3), 401–409. doi:10.1111/j.1474-9726.2012.00798.x
Nakamura K., Nemani V. M., Azarbal F., Skibinski G., Levy J. M., Egami K., et al. (2011). Direct membrane association drives mitochondrial fission by the Parkinson disease-associated protein alpha-synuclein. J. Biol. Chem. 286 (23), 20710–20726. doi:10.1074/jbc.M110.213538
Nakamura K., Nemani V. M., Wallender E. K., Kaehlcke K., Ott M., Edwards R. H. (2008). Optical reporters for the conformation of alpha-synuclein reveal a specific interaction with mitochondria. J. Neurosci. 28 (47), 12305–12317. doi:10.1523/JNEUROSCI.3088-08.2008
Naoi M., Wu Y., Shamoto-Nagai M., Maruyama W. (2019). Mitochondria in neuroprotection by phytochemicals: bioactive polyphenols modulate mitochondrial apoptosis system, function and structure. Int. J. Mol. Sci. 20 (10), 2451. doi:10.3390/ijms20102451
Narayanan V., Guo Y., Scarlata S. (2005). Fluorescence studies suggest a role for alpha-synuclein in the phosphatidylinositol lipid signaling pathway. Biochemistry 44 (2), 462–470. doi:10.1021/bi0487140
Nargang F. E., Rapaport D., Ritzel R. G., Neupert W., Lill R. (1998). Role of the negative charges in the cytosolic domain of TOM22 in the import of precursor proteins into mitochondria. Mol. Cell. Biol. 18 (6), 3173–3181. doi:10.1128/mcb.18.6.3173
Naylor D. J., Stines A. P., Hoogenraad N. J., Hoj P. B. (1998). Evidence for the existence of distinct mammalian cytosolic, microsomal, and two mitochondrial GrpE-like proteins, the Co-chaperones of specific Hsp70 members. J. Biol. Chem. 273 (33), 21169–21177. doi:10.1074/jbc.273.33.21169
Nelson R., Sawaya M. R., Balbirnie M., Madsen A. O., Riekel C., Grothe R., et al. (2005). Structure of the cross-beta spine of amyloid-like fibrils. Nature 435 (7043), 773–778. doi:10.1038/nature03680
Neueder A., Dumas A. A., Benjamin A. C., Bates G. P. (2018). Regulatory mechanisms of incomplete huntingtin mRNA splicing. Nat. Commun. 9 (1), 3955. doi:10.1038/s41467-018-06281-3
Neueder A., Landles C., Ghosh R., Howland D., Myers R. H., Faull R. L. M., et al. (2017). The pathogenic exon 1 HTT protein is produced by incomplete splicing in Huntington's disease patients. Sci. Rep. 7 (1), 1307. doi:10.1038/s41598-017-01510-z
Nhu N. T., Xiao S. Y., Liu Y., Kumar V. B., Cui Z. Y., Lee S. D. (2021). Neuroprotective effects of a small mitochondrially-targeted tetrapeptide elamipretide in neurodegeneration. Front. Integr. Neurosci. 15, 747901. doi:10.3389/fnint.2021.747901
Nielsen K. L., Cowan N. J. (1998). A single ring is sufficient for productive chaperonin-mediated folding in vivo. Mol. Cell. 2 (1), 93–99. doi:10.1016/s1097-2765(00)80117-3
Nisemblat S., Yaniv O., Parnas A., Frolow F., Azem A. (2015). Crystal structure of the human mitochondrial chaperonin symmetrical football complex. Proc. Natl. Acad. Sci. U. S. A. 112 (19), 6044–6049. doi:10.1073/pnas.1411718112
Nunnari J., Fox T. D., Walter P. (1993). A mitochondrial protease with two catalytic subunits of nonoverlapping specificities. Science 262 (5142), 1997–2004. doi:10.1126/science.8266095
Nuscher B., Kamp F., Mehnert T., Odoy S., Haass C., Kahle P. J., et al. (2004). Alpha-synuclein has a high affinity for packing defects in a bilayer membrane: a thermodynamics study. J. Biol. Chem. 279 (21), 21966–21975. doi:10.1074/jbc.M401076200
Obita T., Muto T., Endo T., Kohda D. (2003). Peptide library approach with a disulfide tether to refine the Tom20 recognition motif in mitochondrial presequences. J. Mol. Biol. 328 (2), 495–504. doi:10.1016/s0022-2836(03)00288-2
Oka S., Leon J., Sakumi K., Ide T., Kang D., LaFerla F. M., et al. (2016). Human mitochondrial transcriptional factor A breaks the mitochondria-mediated vicious cycle in Alzheimer's disease. Sci. Rep. 6, 37889. doi:10.1038/srep37889
Okamoto K., Brinker A., Paschen S. A., Moarefi I., Hayer-Hartl M., Neupert W., et al. (2002). The protein import motor of mitochondria: a targeted molecular ratchet driving unfolding and translocation. EMBO J. 21 (14), 3659–3671. doi:10.1093/emboj/cdf358
Opalinska M., Janska H. (2018). AAA proteases: guardians of mitochondrial function and homeostasis. Cells 7 (10), 163. doi:10.3390/cells7100163
Orr A. L., Li S., Wang C. E., Li H., Wang J., Rong J., et al. (2008). N-terminal mutant huntingtin associates with mitochondria and impairs mitochondrial trafficking. J. Neurosci. 28 (11), 2783–2792. doi:10.1523/JNEUROSCI.0106-08.2008
Oswald M. C., Brooks P. S., Zwart M. F., Mukherjee A., West R. J., Giachello C. N., et al. (2018). Reactive oxygen species regulate activity-dependent neuronal plasticity in Drosophila. Elife 7, e39393. doi:10.7554/eLife.39393
Ouberai M. M., Wang J., Swann M. J., Galvagnion C., Guilliams T., Dobson C. M., et al. (2013). α-Synuclein senses lipid packing defects and induces lateral expansion of lipids leading to membrane remodeling. J. Biol. Chem. 288 (29), 20883–20895. doi:10.1074/jbc.M113.478297
Ow S. Y., Dunstan D. E. (2014). A brief overview of amyloids and Alzheimer's disease. Protein Sci. 23 (10), 1315–1331. doi:10.1002/pro.2524
Padrick S. B., Miranker A. D. (2002). Islet amyloid: phase partitioning and secondary nucleation are central to the mechanism of fibrillogenesis. Biochemistry 41 (14), 4694–4703. doi:10.1021/bi0160462
Paglin S., Lee N. Y., Nakar C., Fitzgerald M., Plotkin J., Deuel B., et al. (2005). Rapamycin-sensitive pathway regulates mitochondrial membrane potential, autophagy, and survival in irradiated MCF-7 cells. Cancer Res. 65 (23), 11061–11070. doi:10.1158/0008-5472.CAN-05-1083
Paillusson S., Gomez-Suaga P., Stoica R., Little D., Gissen P., Devine M. J., et al. (2017). α-Synuclein binds to the ER-mitochondria tethering protein VAPB to disrupt Ca2+ homeostasis and mitochondrial ATP production. Acta Neuropathol. 134 (1), 129–149. doi:10.1007/s00401-017-1704-z
Pak Y. K., Weiner H. (1990). Import of chemically synthesized signal peptides into rat liver mitochondria. J. Biol. Chem. 265 (24), 14298–14307. doi:10.1016/s0021-9258(18)77300-6
Pandey M., Varghese M., Sindhu K. M., Sreetama S., Navneet A. K., Mohanakumar K. P., et al. (2008). Mitochondrial NAD+-linked State 3 respiration and complex-I activity are compromised in the cerebral cortex of 3-nitropropionic acid-induced rat model of Huntington's disease. J. Neurochem. 104 (2), 420–434. doi:10.1111/j.1471-4159.2007.04996.x
Panov A. V., Gutekunst C. A., Leavitt B. R., Hayden M. R., Burke J. R., Strittmatter W. J., et al. (2002). Early mitochondrial calcium defects in Huntington's disease are a direct effect of polyglutamines. Nat. Neurosci. 5 (8), 731–736. doi:10.1038/nn884
Paolini Paoletti F., Gaetani L., Parnetti L. (2020). The challenge of disease-modifying therapies in Parkinson's disease: role of CSF biomarkers. Biomolecules 10 (2), 335. doi:10.3390/biom10020335
Papic D., Elbaz-Alon Y., Koerdt S. N., Leopold K., Worm D., Jung M., et al. (2013). The role of Djp1 in import of the mitochondrial protein Mim1 demonstrates specificity between a cochaperone and its substrate protein. Mol. Cell. Biol. 33 (20), 4083–4094. doi:10.1128/MCB.00227-13
Pardridge W. M. (2016). Re-engineering therapeutic antibodies for Alzheimer's disease as blood-brain barrier penetrating bi-specific antibodies. Expert Opin. Biol. Ther. 16 (12), 1455–1468. doi:10.1080/14712598.2016.1230195
Pardridge W. M. (2020). Treatment of alzheimer's disease and blood-brain barrier drug delivery. Pharm. (Basel) 13 (11), 394. doi:10.3390/ph13110394
Park H., Kang J. H., Lee S. (2020). Autophagy in neurodegenerative diseases: a hunter for aggregates. Int. J. Mol. Sci. 21 (9), 3369. doi:10.3390/ijms21093369
Patil S. M., Mehta A., Jha S., Alexandrescu A. T. (2011). Heterogeneous amylin fibril growth mechanisms imaged by total internal reflection fluorescence microscopy. Biochemistry 50 (14), 2808–2819. doi:10.1021/bi101908m
Pellegrini L., Scorrano L. (2007). A cut short to death: parl and Opa1 in the regulation of mitochondrial morphology and apoptosis. Cell. Death Differ. 14 (7), 1275–1284. doi:10.1038/sj.cdd.4402145
Penney J. B., Vonsattel J. P., MacDonald M. E., Gusella J. F., Myers R. H. (1997). CAG repeat number governs the development rate of pathology in Huntington's disease. Ann. Neurol. 41 (5), 689–692. doi:10.1002/ana.410410521
Petrasch-Parwez E., Nguyen H. P., Lobbecke-Schumacher M., Habbes H. W., Wieczorek S., Riess O., et al. (2007). Cellular and subcellular localization of Huntingtin [corrected] aggregates in the brain of a rat transgenic for Huntington disease. J. Comp. Neurol. 501 (5), 716–730. doi:10.1002/cne.21272
Petrova V. Y., Drescher D., Kujumdzieva A. V., Schmitt M. J. (2004). Dual targeting of yeast catalase A to peroxisomes and mitochondria. Biochem. J. 380 (2), 393–400. doi:10.1042/BJ20040042
Pfanner N., Warscheid B., Wiedemann N. (2019). Mitochondrial proteins: from biogenesis to functional networks. Nat. Rev. Mol. Cell. Biol. 20 (5), 267–284. doi:10.1038/s41580-018-0092-0
Pfefferkorn C. M., Jiang Z., Lee J. C. (2012). Biophysics of alpha-synuclein membrane interactions. Biochim. Biophys. Acta 1818 (2), 162–171. doi:10.1016/j.bbamem.2011.07.032
Pinho C. M., Teixeira P. F., Glaser E. (2014). Mitochondrial import and degradation of amyloid-β peptide. Biochim. Biophys. Acta 1837 (7), 1069–1074. doi:10.1016/j.bbabio.2014.02.007
Pino P., Foth B. J., Kwok L. Y., Sheiner L., Schepers R., Soldati T., et al. (2007). Dual targeting of antioxidant and metabolic enzymes to the mitochondrion and the apicoplast of Toxoplasma gondii. PLoS Pathog. 3 (8), e115. doi:10.1371/journal.ppat.0030115
Pitt A. S., Buchanan S. K. (2021). A biochemical and structural understanding of TOM complex interactions and implications for human health and disease. Cells 10 (5), 1164. doi:10.3390/cells10051164
Poirier M. A., Li H., Macosko J., Cai S., Amzel M., Ross C. A. (2002). Huntingtin spheroids and protofibrils as precursors in polyglutamine fibrilization. J. Biol. Chem. 277 (43), 41032–41037. doi:10.1074/jbc.M205809200
Ponce-Rojas J. C., Avendano-Monsalve M. C., Yanez-Falcon A. R., Jaimes-Miranda F., Garay E., Torres-Quiroz F., et al. (2017). αβ'-NAC cooperates with Sam37 to mediate early stages of mitochondrial protein import. FEBS J. 284 (5), 814–830. doi:10.1111/febs.14024
Popov-Celeketic D., Mapa K., Neupert W., Mokranjac D. (2008). Active remodelling of the TIM23 complex during translocation of preproteins into mitochondria. EMBO J. 27 (10), 1469–1480. doi:10.1038/emboj.2008.79
Poulaki A., Giannouli S. (2022). Mitochondrial lipids: from membrane organization to apoptotic facilitation. Int. J. Mol. Sci. 23 (7), 3738. doi:10.3390/ijms23073738
Pozo Devoto V. M., Dimopoulos N., Alloatti M., Pardi M. B., Saez T. M., Otero M. G., et al. (2017). αSynuclein control of mitochondrial homeostasis in human-derived neurons is disrupted by mutations associated with Parkinson's disease. Sci. Rep. 7 (1), 5042. doi:10.1038/s41598-017-05334-9
Pozo Devoto V. M., Falzone T. L. (2017). Mitochondrial dynamics in Parkinson's disease: a role for alpha-synuclein? Dis. Model. Mech. 10 (9), 1075–1087. doi:10.1242/dmm.026294
Prior M., Dargusch R., Ehren J. L., Chiruta C., Schubert D. (2013). The neurotrophic compound J147 reverses cognitive impairment in aged Alzheimer's disease mice. Alzheimers Res. Ther. 5 (3), 25. doi:10.1186/alzrt179
Qian X., Gebert M., Hopker J., Yan M., Li J., Wiedemann N., et al. (2011). Structural basis for the function of Tim50 in the mitochondrial presequence translocase. J. Mol. Biol. 411 (3), 513–519. doi:10.1016/j.jmb.2011.06.020
Qiang W., Yau W. M., Lu J. X., Collinge J., Tycko R. (2017). Structural variation in amyloid-beta fibrils from Alzheimer's disease clinical subtypes. Nature 541 (7636), 217–221. doi:10.1038/nature20814
Qin W., Haroutunian V., Katsel P., Cardozo C. P., Ho L., Buxbaum J. D., et al. (2009). PGC-1alpha expression decreases in the Alzheimer disease brain as a function of dementia. Arch. Neurol. 66 (3), 352–361. doi:10.1001/archneurol.2008.588
Qin W., Myers S. A., Carey D. K., Carr S. A., Ting A. Y. (2021). Spatiotemporally-resolved mapping of RNA binding proteins via functional proximity labeling reveals a mitochondrial mRNA anchor promoting stress recovery. Nat. Commun. 12 (1), 4980. doi:10.1038/s41467-021-25259-2
Raffai R. L., Dong L. M., Farese R. V., Weisgraber K. H. (2001). Introduction of human apolipoprotein E4 "domain interaction" into mouse apolipoprotein E. Proc. Natl. Acad. Sci. U. S. A. 98 (20), 11587–11591. doi:10.1073/pnas.201279298
Ramakrishnan M., Jensen P. H., Marsh D. (2003). Alpha-synuclein association with phosphatidylglycerol probed by lipid spin labels. Biochemistry 42 (44), 12919–12926. doi:10.1021/bi035048e
Rao J. N., Jao C. C., Hegde B. G., Langen R., Ulmer T. S. (2010). A combinatorial NMR and EPR approach for evaluating the structural ensemble of partially folded proteins. J. Am. Chem. Soc. 132 (25), 8657–8668. doi:10.1021/ja100646t
Rapaport D., Neupert W., Lill R. (1997). Mitochondrial protein import. Tom40 plays a major role in targeting and translocation of preproteins by forming a specific binding site for the presequence. J. Biol. Chem. 272 (30), 18725–18731. doi:10.1074/jbc.272.30.18725
Rath S., Sharma R., Gupta R., Ast T., Chan C., Durham T. J., et al. (2021). MitoCarta3.0: an updated mitochondrial proteome now with sub-organelle localization and pathway annotations. Nucleic Acids Res. 49 (D1), D1541–D1547. doi:10.1093/nar/gkaa1011
Ratovitski T., Chighladze E., Arbez N., Boronina T., Herbrich S., Cole R. N., et al. (2012). Huntingtin protein interactions altered by polyglutamine expansion as determined by quantitative proteomic analysis. Cell. Cycle 11 (10), 2006–2021. doi:10.4161/cc.20423
Reddy P. H., Manczak M., Kandimalla R. (2017). Mitochondria-targeted small molecule SS31: a potential candidate for the treatment of Alzheimer's disease. Hum. Mol. Genet. 26 (8), 1483–1496. doi:10.1093/hmg/ddx129
Reddy P. H., Manczak M., Yin X., Reddy A. P. (2018). Synergistic protective effects of mitochondrial division inhibitor 1 and mitochondria-targeted small peptide SS31 in alzheimer's disease. J. Alzheimers Dis. 62 (4), 1549–1565. doi:10.3233/JAD-170988
Rehling P., Model K., Brandner K., Kovermann P., Sickmann A., Meyer H. E., et al. (2003). Protein insertion into the mitochondrial inner membrane by a twin-pore translocase. Science 299 (5613), 1747–1751. doi:10.1126/science.1080945
Rezaei-Ghaleh N., Amininasab M., Kumar S., Walter J., Zweckstetter M. (2016). Phosphorylation modifies the molecular stability of beta-amyloid deposits. Nat. Commun. 7, 11359. doi:10.1038/ncomms11359
Ribeiro M., Rosenstock T. R., Cunha-Oliveira T., Ferreira I. L., Oliveira C. R., Rego A. C. (2012). Glutathione redox cycle dysregulation in Huntington's disease knock-in striatal cells. Free Radic. Biol. Med. 53 (10), 1857–1867. doi:10.1016/j.freeradbiomed.2012.09.004
Ribeiro M., Rosenstock T. R., Oliveira A. M., Oliveira C. R., Rego A. C. (2014). Insulin and IGF-1 improve mitochondrial function in a PI-3K/Akt-dependent manner and reduce mitochondrial generation of reactive oxygen species in Huntington's disease knock-in striatal cells. Free Radic. Biol. Med. 74, 129–144. doi:10.1016/j.freeradbiomed.2014.06.023
Ricciarelli R., Fedele E. (2017). The amyloid cascade hypothesis in alzheimer's disease: it's time to change our mind. Curr. Neuropharmacol. 15 (6), 926–935. doi:10.2174/1570159X15666170116143743
Richter F., Dennerlein S., Nikolov M., Jans D. C., Naumenko N., Aich A., et al. (2019). ROMO1 is a constituent of the human presequence translocase required for YME1L protease import. J. Cell. Biol. 218 (2), 598–614. doi:10.1083/jcb.201806093
Richter-Dennerlein R., Korwitz A., Haag M., Tatsuta T., Dargazanli S., Baker M., et al. (2014). DNAJC19, a mitochondrial cochaperone associated with cardiomyopathy, forms a complex with prohibitins to regulate cardiolipin remodeling. Cell. Metab. 20 (1), 158–171. doi:10.1016/j.cmet.2014.04.016
Richter-Dennerlein R., Oeljeklaus S., Lorenzi I., Ronsor C., Bareth B., Schendzielorz A. B., et al. (2016). Mitochondrial protein synthesis adapts to influx of nuclear-encoded protein. Cell. 167 (2), 471–483. doi:10.1016/j.cell.2016.09.003
Riguet N., Mahul-Mellier A. L., Maharjan N., Burtscher J., Croisier M., Knott G., et al. (2021). Nuclear and cytoplasmic huntingtin inclusions exhibit distinct biochemical composition, interactome and ultrastructural properties. Nat. Commun. 12 (1), 6579. doi:10.1038/s41467-021-26684-z
Robin M. A., Anandatheerthavarada H. K., Biswas G., Sepuri N. B., Gordon D. M., Pain D., et al. (2002). Bimodal targeting of microsomal CYP2E1 to mitochondria through activation of an N-terminal chimeric signal by cAMP-mediated phosphorylation. J. Biol. Chem. 277 (43), 40583–40593. doi:10.1074/jbc.M203292200
Robin M. A., Anandatheerthavarada H. K., Fang J. K., Cudic M., Otvos L., Avadhani N. G. (2001). Mitochondrial targeted cytochrome P450 2E1 (P450 MT5) contains an intact N terminus and requires mitochondrial specific electron transfer proteins for activity. J. Biol. Chem. 276 (27), 24680–24689. doi:10.1074/jbc.M100363200
Robotta M., Gerding H. R., Vogel A., Hauser K., Schildknecht S., Karreman C., et al. (2014). Alpha-synuclein binds to the inner membrane of mitochondria in an α-helical conformation. Chembiochem 15 (17), 2499–2502. doi:10.1002/cbic.201402281
Rocchi A., Pellegrini S., Siciliano G., Murri L. (2003). Causative and susceptibility genes for Alzheimer's disease: a review. Brain Res. Bull. 61 (1), 1–24. doi:10.1016/s0361-9230(03)00067-4
Roche J., Shen Y., Lee J. H., Ying J., Bax A. (2016). Monomeric aβ(1-40) and aβ(1-42) peptides in solution adopt very similar ramachandran map distributions that closely resemble random coil. Biochemistry 55 (5), 762–775. doi:10.1021/acs.biochem.5b01259
Rockabrand E., Slepko N., Pantalone A., Nukala V. N., Kazantsev A., Marsh J. L., et al. (2007). The first 17 amino acids of Huntingtin modulate its sub-cellular localization, aggregation and effects on calcium homeostasis. Hum. Mol. Genet. 16 (1), 61–77. doi:10.1093/hmg/ddl440
Rodriguez J. A., Ivanova M. I., Sawaya M. R., Cascio D., Reyes F. E., Shi D., et al. (2015). Structure of the toxic core of alpha-synuclein from invisible crystals. Nature 525 (7570), 486–490. doi:10.1038/nature15368
Roise D., Horvath S. J., Tomich J. M., Richards J. H., Schatz G. (1986). A chemically synthesized pre-sequence of an imported mitochondrial protein can form an amphiphilic helix and perturb natural and artificial phospholipid bilayers. EMBO J. 5 (6), 1327–1334. doi:10.1002/j.1460-2075.1986.tb04363.x
Roise D., Schatz G. (1988). Mitochondrial presequences. J. Biol. Chem. 263 (10), 4509–4511. doi:10.1016/s0021-9258(18)68809-x
Roise D., Theiler F., Horvath S. J., Tomich J. M., Richards J. H., Allison D. S., et al. (1988). Amphiphilicity is essential for mitochondrial presequence function. EMBO J. 7 (3), 649–653. doi:10.1002/j.1460-2075.1988.tb02859.x
Rolfe D. F., Brown G. C. (1997). Cellular energy utilization and molecular origin of standard metabolic rate in mammals. Physiol. Rev. 77 (3), 731–758. doi:10.1152/physrev.1997.77.3.731
Roses A. D., Lutz M. W., Amrine-Madsen H., Saunders A. M., Crenshaw D. G., Sundseth S. S., et al. (2010). A TOMM40 variable-length polymorphism predicts the age of late-onset Alzheimer's disease. Pharmacogenomics J. 10 (5), 375–384. doi:10.1038/tpj.2009.69
Roshanbin S., Aniszewska A., Gumucio A., Masliah E., Erlandsson A., Bergstrom J., et al. (2021). Age-related increase of alpha-synuclein oligomers is associated with motor disturbances in L61 transgenic mice. Neurobiol. Aging 101, 207–220. doi:10.1016/j.neurobiolaging.2021.01.010
Ruan L., Zhou C., Jin E., Kucharavy A., Zhang Y., Wen Z., et al. (2017). Cytosolic proteostasis through importing of misfolded proteins into mitochondria. Nature 543 (7645), 443–446. doi:10.1038/nature21695
Rubinsztein D. C., Leggo J., Coles R., Almqvist E., Biancalana V., Cassiman J. J., et al. (1996). Phenotypic characterization of individuals with 30-40 CAG repeats in the Huntington disease (HD) gene reveals HD cases with 36 repeats and apparently normal elderly individuals with 36-39 repeats. Am. J. Hum. Genet. 59 (1), 16–22.
Rudiger S., Germeroth L., Schneider-Mergener J., Bukau B. (1997). Substrate specificity of the DnaK chaperone determined by screening cellulose-bound peptide libraries. EMBO J. 16 (7), 1501–1507. doi:10.1093/emboj/16.7.1501
Rutledge B. S., Choy W. Y., Duennwald M. L. (2022). Folding or holding? Hsp70 and Hsp90 chaperoning of misfolded proteins in neurodegenerative disease. J. Biol. Chem. 298 (5), 101905. doi:10.1016/j.jbc.2022.101905
Ryan T., Bamm V. V., Stykel M. G., Coackley C. L., Humphries K. M., Jamieson-Williams R., et al. (2018). Cardiolipin exposure on the outer mitochondrial membrane modulates alpha-synuclein. Nat. Commun. 9 (1), 817. doi:10.1038/s41467-018-03241-9
Ryu D., Mouchiroud L., Andreux P. A., Katsyuba E., Moullan N., Nicolet-Dit-Felix A. A., et al. (2016). Urolithin A induces mitophagy and prolongs lifespan in C. elegans and increases muscle function in rodents. Nat. Med. 22 (8), 879–888. doi:10.1038/nm.4132
Ryu W. I., Bormann M. K., Shen M., Kim D., Forester B., Park Y., et al. (2021). Brain cells derived from Alzheimer's disease patients have multiple specific innate abnormalities in energy metabolism. Mol. Psychiatry 26 (10), 5702–5714. doi:10.1038/s41380-021-01068-3
Saavedra L., Mohamed A., Ma V., Kar S., de Chaves E. P. (2007). Internalization of beta-amyloid peptide by primary neurons in the absence of apolipoprotein E. J. Biol. Chem. 282 (49), 35722–35732. doi:10.1074/jbc.M701823200
Saitoh T., Igura M., Miyazaki Y., Ose T., Maita N., Kohda D. (2011). Crystallographic snapshots of Tom20-mitochondrial presequence interactions with disulfide-stabilized peptides. Biochemistry 50 (24), 5487–5496. doi:10.1021/bi200470x
Saitoh T., Igura M., Obita T., Ose T., Kojima R., Maenaka K., et al. (2007). Tom20 recognizes mitochondrial presequences through dynamic equilibrium among multiple bound states. EMBO J. 26 (22), 4777–4787. doi:10.1038/sj.emboj.7601888
Sakaue H., Shiota T., Ishizaka N., Kawano S., Tamura Y., Tan K. S., et al. (2019). Porin associates with Tom22 to regulate the mitochondrial protein gate assembly. Mol. Cell. 73 (5), 1044–1055. doi:10.1016/j.molcel.2019.01.003
Samuel F., Flavin W. P., Iqbal S., Pacelli C., Sri Renganathan S. D., Trudeau L. E., et al. (2016). Effects of serine 129 phosphorylation on alpha-synuclein aggregation, membrane association, and internalization. J. Biol. Chem. 291 (9), 4374–4385. doi:10.1074/jbc.M115.705095
Sandbrink R., Masters C. L., Beyreuther K. (1996). APP gene family. Alternative splicing generates functionally related isoforms. Ann. N. Y. Acad. Sci. 777, 281–287. doi:10.1111/j.1749-6632.1996.tb34433.x
Sando S. B., Melquist S., Cannon A., Hutton M. L., Sletvold O., Saltvedt I., et al. (2008). APOE epsilon 4 lowers age at onset and is a high risk factor for Alzheimer's disease; a case control study from central Norway. BMC Neurol. 8, 9. doi:10.1186/1471-2377-8-9
Sarchione A., Marchand A., Taymans J. M., Chartier-Harlin M. C. (2021). Alpha-synuclein and lipids: the elephant in the room? Cells 10 (9), 2452. doi:10.3390/cells10092452
Sassano M. L., Felipe-Abrio B., Agostinis P. (2022). ER-mitochondria contact sites; a multifaceted factory for Ca(2+) signaling and lipid transport. Front. Cell. Dev. Biol. 10, 988014. doi:10.3389/fcell.2022.988014
Sathasivam K., Neueder A., Gipson T. A., Landles C., Benjamin A. C., Bondulich M. K., et al. (2013). Aberrant splicing of HTT generates the pathogenic exon 1 protein in Huntington disease. Proc. Natl. Acad. Sci. U. S. A. 110 (6), 2366–2370. doi:10.1073/pnas.1221891110
Saudou F., Humbert S. (2016). The biology of huntingtin. Neuron 89 (5), 910–926. doi:10.1016/j.neuron.2016.02.003
Sawa A., Wiegand G. W., Cooper J., Margolis R. L., Sharp A. H., Lawler J. F., et al. (1999). Increased apoptosis of Huntington disease lymphoblasts associated with repeat length-dependent mitochondrial depolarization. Nat. Med. 5 (10), 1194–1198. doi:10.1038/13518
Scheper W., Hoozemans J. J. (2015). The unfolded protein response in neurodegenerative diseases: a neuropathological perspective. Acta Neuropathol. 130 (3), 315–331. doi:10.1007/s00401-015-1462-8
Scherzinger E., Sittler A., Schweiger K., Heiser V., Lurz R., Hasenbank R., et al. (1999). Self-assembly of polyglutamine-containing huntingtin fragments into amyloid-like fibrils: implications for Huntington's disease pathology. Proc. Natl. Acad. Sci. U. S. A. 96 (8), 4604–4609. doi:10.1073/pnas.96.8.4604
Schilling G., Klevytska A., Tebbenkamp A. T., Juenemann K., Cooper J., Gonzales V., et al. (2007). Characterization of huntingtin pathologic fragments in human Huntington disease, transgenic mice, and cell models. J. Neuropathol. Exp. Neurol. 66 (4), 313–320. doi:10.1097/nen.0b013e318040b2c8
Schilling G., Sharp A. H., Loev S. J., Wagster M. V., Li S. H., Stine O. C., et al. (1995). Expression of the Huntington's disease (IT15) protein product in HD patients. Hum. Mol. Genet. 4 (8), 1365–1371. doi:10.1093/hmg/4.8.1365
Schmidt M. F., Gan Z. Y., Komander D., Dewson G. (2021). Ubiquitin signalling in neurodegeneration: mechanisms and therapeutic opportunities. Cell. Death Differ. 28 (2), 570–590. doi:10.1038/s41418-020-00706-7
Schneider A. (2018). Dihydrofolate reductase and membrane translocation: evolution of a classic experiment: classic landmark papers, irrespective of their age, can teach students how best science is practiced and inspire new experiments. EMBO Rep. 19 (3), e45692. doi:10.15252/embr.201745692
Schneider H. C., Berthold J., Bauer M. F., Dietmeier K., Guiard B., Brunner M., et al. (1994). Mitochondrial Hsp70/MIM44 complex facilitates protein import. Nature 371 (6500), 768–774. doi:10.1038/371768a0
Schneider H. C., Westermann B., Neupert W., Brunner M. (1996). The nucleotide exchange factor MGE exerts a key function in the ATP-dependent cycle of mt-Hsp70-Tim44 interaction driving mitochondrial protein import. EMBO J. 15 (21), 5796–5803. doi:10.1002/j.1460-2075.1996.tb00966.x
Schulz C., Lytovchenko O., Melin J., Chacinska A., Guiard B., Neumann P., et al. (2011). Tim50's presequence receptor domain is essential for signal driven transport across the TIM23 complex. J. Cell. Biol. 195 (4), 643–656. doi:10.1083/jcb.201105098
Selkoe D. J., Hardy J. (2016). The amyloid hypothesis of Alzheimer's disease at 25 years. EMBO Mol. Med. 8 (6), 595–608. doi:10.15252/emmm.201606210
Seong I. S., Ivanova E., Lee J. M., Choo Y. S., Fossale E., Anderson M., et al. (2005). HD CAG repeat implicates a dominant property of huntingtin in mitochondrial energy metabolism. Hum. Mol. Genet. 14 (19), 2871–2880. doi:10.1093/hmg/ddi319
Serpell L. C. (2000). Alzheimer's amyloid fibrils: structure and assembly. Biochim. Biophys. Acta 1502 (1), 16–30. doi:10.1016/s0925-4439(00)00029-6
Seubert P., Vigo-Pelfrey C., Esch F., Lee M., Dovey H., Davis D., et al. (1992). Isolation and quantification of soluble Alzheimer's beta-peptide from biological fluids. Nature 359 (6393), 325–327. doi:10.1038/359325a0
Shahmoradian S. H., Lewis A. J., Genoud C., Hench J., Moors T. E., Navarro P. P., et al. (2019). Lewy pathology in Parkinson's disease consists of crowded organelles and lipid membranes. Nat. Neurosci. 22 (7), 1099–1109. doi:10.1038/s41593-019-0423-2
Sharon R., Bar-Joseph I., Frosch M. P., Walsh D. M., Hamilton J. A., Selkoe D. J. (2003). The formation of highly soluble oligomers of alpha-synuclein is regulated by fatty acids and enhanced in Parkinson's disease. Neuron 37 (4), 583–595. doi:10.1016/s0896-6273(03)00024-2
Shen K., Gamerdinger M., Chan R., Gense K., Martin E. M., Sachs N., et al. (2019a). Dual role of ribosome-binding domain of NAC as a potent suppressor of protein aggregation and aging-related proteinopathies. Mol. Cell. 74 (4), 729–741. doi:10.1016/j.molcel.2019.03.012
Shen Y., Ding M., Xie Z., Liu X., Yang H., Jin S., et al. (2019b). Activation of mitochondrial unfolded protein response in SHSY5Y expressing APP cells and APP/PS1 mice. Front. Cell. Neurosci. 13, 568. doi:10.3389/fncel.2019.00568
Shi Q., Chang C., Saliba A., Bhat M. A. (2022). Microglial mTOR activation upregulates Trem2 and enhances beta-amyloid plaque clearance in the 5XFAD alzheimer's disease model. J. Neurosci. 42 (27), 5294–5313. doi:10.1523/JNEUROSCI.2427-21.2022
Shin C. S., Meng S., Garbis S. D., Moradian A., Taylor R. W., Sweredoski M. J., et al. (2021). LONP1 and mtHSP70 cooperate to promote mitochondrial protein folding. Nat. Commun. 12 (1), 265. doi:10.1038/s41467-020-20597-z
Shindyapina A. V., Cho Y., Kaya A., Tyshkovskiy A., Castro J. P., Deik A., et al. (2022). Rapamycin treatment during development extends life span and health span of male mice and Daphnia magna. Sci. Adv. 8 (37), eabo5482. doi:10.1126/sciadv.abo5482
Shiota T., Imai K., Qiu J., Hewitt V. L., Tan K., Shen H. H., et al. (2015). Molecular architecture of the active mitochondrial protein gate. Science 349 (6255), 1544–1548. doi:10.1126/science.aac6428
Shiota T., Mabuchi H., Tanaka-Yamano S., Yamano K., Endo T. (2011). In vivo protein-interaction mapping of a mitochondrial translocator protein Tom22 at work. Proc. Natl. Acad. Sci. U. S. A. 108 (37), 15179–15183. doi:10.1073/pnas.1105921108
Shirasaki D. I., Greiner E. R., Al-Ramahi I., Gray M., Boontheung P., Geschwind D. H., et al. (2012). Network organization of the huntingtin proteomic interactome in mammalian brain. Neuron 75 (1), 41–57. doi:10.1016/j.neuron.2012.05.024
Shirendeb U. P., Calkins M. J., Manczak M., Anekonda V., Dufour B., McBride J. L., et al. (2012). Mutant huntingtin's interaction with mitochondrial protein Drp1 impairs mitochondrial biogenesis and causes defective axonal transport and synaptic degeneration in Huntington's disease. Hum. Mol. Genet. 21 (2), 406–420. doi:10.1093/hmg/ddr475
Sichting M., Mokranjac D., Azem A., Neupert W., Hell K. (2005). Maintenance of structure and function of mitochondrial Hsp70 chaperones requires the chaperone Hep1. EMBO J. 24 (5), 1046–1056. doi:10.1038/sj.emboj.7600580
Silva A. C., Almeida S., Laco M., Duarte A. I., Domingues J., Oliveira C. R., et al. (2013). Mitochondrial respiratory chain complex activity and bioenergetic alterations in human platelets derived from pre-symptomatic and symptomatic Huntington's disease carriers. Mitochondrion 13 (6), 801–809. doi:10.1016/j.mito.2013.05.006
Silva P. D., Schilke B., Walter W., Andrew A., Craig E. A. (2003). J protein cochaperone of the mitochondrial inner membrane required for protein import into the mitochondrial matrix. Proc. Natl. Acad. Sci. U. S. A. 100 (24), 13839–13844. doi:10.1073/pnas.1936150100
Silva P., Liu Q., Walter W., Craig E. A. (2004). Regulated interactions of mtHsp70 with Tim44 at the translocon in the mitochondrial inner membrane. Nat. Struct. Mol. Biol. 11 (11), 1084–1091. doi:10.1038/nsmb846
Sim S. I., Chen Y., Lynch D. L., Gumbart J. C., Park E. (2023). Structural basis of mitochondrial protein import by the TIM23 complex. Nature 621, 620–626. doi:10.1038/s41586-023-06239-6
Singh R., Jamdar S. N., Goyal V. D., Kumar A., Ghosh B., Makde R. D. (2017). Structure of the human aminopeptidase XPNPEP3 and comparison of its in vitro activity with Icp55 orthologs: insights into diverse cellular processes. J. Biol. Chem. 292 (24), 10035–10047. doi:10.1074/jbc.M117.783357
Sinha D., Joshi N., Chittoor B., Samji P., D'Silva P. (2010). Role of Magmas in protein transport and human mitochondria biogenesis. Hum. Mol. Genet. 19 (7), 1248–1262. doi:10.1093/hmg/ddq002
Sinha D., Srivastava S., Krishna L., D'Silva P. (2014). Unraveling the intricate organization of mammalian mitochondrial presequence translocases: existence of multiple translocases for maintenance of mitochondrial function. Mol. Cell. Biol. 34 (10), 1757–1775. doi:10.1128/MCB.01527-13
Sirk D., Zhu Z., Wadia J. S., Shulyakova N., Phan N., Fong J., et al. (2007). Chronic exposure to sub-lethal beta-amyloid (Abeta) inhibits the import of nuclear-encoded proteins to mitochondria in differentiated PC12 cells. J. Neurochem. 103 (5), 1989–2003. doi:10.1111/j.1471-4159.2007.04907.x
Sivanandam V. N., Jayaraman M., Hoop C. L., Kodali R., Wetzel R., van der Wel P. C. (2011). The aggregation-enhancing huntingtin N-terminus is helical in amyloid fibrils. J. Am. Chem. Soc. 133 (12), 4558–4566. doi:10.1021/ja110715f
Skerjanc I. S., Shore G. C., Silvius J. R. (1987). The interaction of a synthetic mitochondrial signal peptide with lipid membranes is independent of transbilayer potential. EMBO J. 6 (10), 3117–3123. doi:10.1002/j.1460-2075.1987.tb02621.x
Somayaji M., Lanseur Z., Choi S. J., Sulzer D., Mosharov E. V. (2021). Roles for α-synuclein in gene expression. Genes. (Basel) 12 (8), 1166. doi:10.3390/genes12081166
Song W., Chen J., Petrilli A., Liot G., Klinglmayr E., Zhou Y., et al. (2011). Mutant huntingtin binds the mitochondrial fission GTPase dynamin-related protein-1 and increases its enzymatic activity. Nat. Med. 17 (3), 377–382. doi:10.1038/nm.2313
Sonsky I., Vodicka P., Vodickova Kepkova K., Hansikova H. (2021). Mitophagy in Huntington's disease. Neurochem. Int. 149, 105147. doi:10.1016/j.neuint.2021.105147
Sorolla M. A., Reverter-Branchat G., Tamarit J., Ferrer I., Ros J., Cabiscol E. (2008). Proteomic and oxidative stress analysis in human brain samples of Huntington disease. Free Radic. Biol. Med. 45 (5), 667–678. doi:10.1016/j.freeradbiomed.2008.05.014
Sorrentino V., Romani M., Mouchiroud L., Beck J. S., Zhang H., D'Amico D., et al. (2017). Enhancing mitochondrial proteostasis reduces amyloid-beta proteotoxicity. Nature 552 (7684), 187–193. doi:10.1038/nature25143
Spillantini M. G., Crowther R. A., Jakes R., Hasegawa M., Goedert M. (1998). alpha-Synuclein in filamentous inclusions of Lewy bodies from Parkinson's disease and dementia with lewy bodies. Proc. Natl. Acad. Sci. U. S. A. 95 (11), 6469–6473. doi:10.1073/pnas.95.11.6469
Srinivasan E., Chandrasekhar G., Chandrasekar P., Anbarasu K., Vickram A. S., Karunakaran R., et al. (2021). Alpha-synuclein aggregation in Parkinson's disease. Front. Med. (Lausanne) 8, 736978. doi:10.3389/fmed.2021.736978
Srivastava S., Savanur M. A., Sinha D., Birje A., Saha P. P. (2017). Regulation of mitochondrial protein import by the nucleotide exchange factors GrpEL1 and GrpEL2 in human cells. J. Biol. Chem. 292 (44), 18075–18090. doi:10.1074/jbc.M117.788463
Stefanis L. (2012). α-Synuclein in Parkinson's disease. Cold Spring Harb. Perspect. Med. 2 (2), a009399. doi:10.1101/cshperspect.a009399
Steffan J. S., Kazantsev A., Spasic-Boskovic O., Greenwald M., Zhu Y. Z., Gohler H., et al. (2000). The Huntington's disease protein interacts with p53 and CREB-binding protein and represses transcription. Proc. Natl. Acad. Sci. U. S. A. 97 (12), 6763–6768. doi:10.1073/pnas.100110097
Stockl M., Fischer P., Wanker E., Herrmann A. (2008). Alpha-synuclein selectively binds to anionic phospholipids embedded in liquid-disordered domains. J. Mol. Biol. 375 (5), 1394–1404. doi:10.1016/j.jmb.2007.11.051
Su J., Liu D., Yang F., Zuo M. Q., Li C., Dong M. Q., et al. (2022). Structural basis of Tom20 and Tom22 cytosolic domains as the human TOM complex receptors. Proc. Natl. Acad. Sci. U. S. A. 119 (26), e2200158119. doi:10.1073/pnas.2200158119
Suarez-Rivero J. M., Pastor-Maldonado C. J., Povea-Cabello S., Alvarez-Cordoba M., Villalon-Garcia I., Talaveron-Rey M., et al. (2022). Activation of the mitochondrial unfolded protein response: a new therapeutic target? Biomedicines 10 (7), 1611. doi:10.3390/biomedicines10071611
Sugrue M. M., Tatton W. G. (2001). Mitochondrial membrane potential in aging cells. Biol. Signals Recept 10 (3-4), 176–188. doi:10.1159/000046886
Suzuki H., Okazawa Y., Komiya T., Saeki K., Mekada E., Kitada S., et al. (2000). Characterization of rat TOM40, a central component of the preprotein translocase of the mitochondrial outer membrane. J. Biol. Chem. 275 (48), 37930–37936. doi:10.1074/jbc.M006558200
Swerdlow R. H., Burns J. M., Khan S. M. (2014). The Alzheimer's disease mitochondrial cascade hypothesis: progress and perspectives. Biochim. Biophys. Acta 1842 (8), 1219–1231. doi:10.1016/j.bbadis.2013.09.010
Szczepanowska K., Trifunovic A. (2022). Mitochondrial matrix proteases: quality control and beyond. FEBS J. 289 (22), 7128–7146. doi:10.1111/febs.15964
Szeto H. H. (2014). First-in-class cardiolipin-protective compound as a therapeutic agent to restore mitochondrial bioenergetics. Br. J. Pharmacol. 171 (8), 2029–2050. doi:10.1111/bph.12461
Tam S., Spiess C., Auyeung W., Joachimiak L., Chen B., Poirier M. A., et al. (2009). The chaperonin TRiC blocks a huntingtin sequence element that promotes the conformational switch to aggregation. Nat. Struct. Mol. Biol. 16 (12), 1279–1285. doi:10.1038/nsmb.1700
Tambini M. D., Pera M., Kanter E., Yang H., Guardia-Laguarta C., Holtzman D., et al. (2016). ApoE4 upregulates the activity of mitochondria-associated ER membranes. EMBO Rep. 17 (1), 27–36. doi:10.15252/embr.201540614
Tamura Y., Harada Y., Shiota T., Yamano K., Watanabe K., Yokota M., et al. (2009). Tim23-Tim50 pair coordinates functions of translocators and motor proteins in mitochondrial protein import. J. Cell. Biol. 184 (1), 129–141. doi:10.1083/jcb.200808068
Tanaka M., Morishima I., Akagi T., Hashikawa T., Nukina N. (2001). Intra- and intermolecular beta-pleated sheet formation in glutamine-repeat inserted myoglobin as a model for polyglutamine diseases. J. Biol. Chem. 276 (48), 45470–45475. doi:10.1074/jbc.M107502200
Taylor A. B., Smith B. S., Kitada S., Kojima K., Miyaura H., Otwinowski Z., et al. (2001). Crystal structures of mitochondrial processing peptidase reveal the mode for specific cleavage of import signal sequences. Structure 9 (7), 615–625. doi:10.1016/s0969-2126(01)00621-9
Taylor R. C., Dillin A. (2011). Aging as an event of proteostasis collapse. Cold Spring Harb. Perspect. Biol. 3 (5), a004440. doi:10.1101/cshperspect.a004440
Tcw J., Goate A. M. (2017). Genetics of beta-amyloid precursor protein in alzheimer's disease. Cold Spring Harb. Perspect. Med. 7 (6), a024539. doi:10.1101/cshperspect.a024539
Tebbenkamp A. T., Crosby K. W., Siemienski Z. B., Brown H. H., Golde T. E., Borchelt D. R. (2012). Analysis of proteolytic processes and enzymatic activities in the generation of huntingtin n-terminal fragments in an HEK293 cell model. PLoS One 7 (12), e50750. doi:10.1371/journal.pone.0050750
Teixeira P. F., Glaser E. (2013). Processing peptidases in mitochondria and chloroplasts. Biochim. Biophys. Acta 1833 (2), 360–370. doi:10.1016/j.bbamcr.2012.03.012
Terada K., Ohtsuka K., Imamoto N., Yoneda Y., Mori M. (1995). Role of heat shock cognate 70 protein in import of ornithine transcarbamylase precursor into mammalian mitochondria. Mol. Cell. Biol. 15 (7), 3708–3713. doi:10.1128/mcb.15.7.3708
Terni B., Boada J., Portero-Otin M., Pamplona R., Ferrer I. (2010). Mitochondrial ATP-synthase in the entorhinal cortex is a target of oxidative stress at stages I/II of Alzheimer's disease pathology. Brain Pathol. 20 (1), 222–233. doi:10.1111/j.1750-3639.2009.00266.x
Thakur A. K., Jayaraman M., Mishra R., Thakur M., Chellgren V. M., Byeon I. J., et al. (2009). Polyglutamine disruption of the huntingtin exon 1 N terminus triggers a complex aggregation mechanism. Nat. Struct. Mol. Biol. 16 (4), 380–389. doi:10.1038/nsmb.1570
Theillet F. X., Binolfi A., Bekei B., Martorana A., Rose H. M., Stuiver M., et al. (2016). Structural disorder of monomeric alpha-synuclein persists in mammalian cells. Nature 530 (7588), 45–50. doi:10.1038/nature16531
Thorne N. J., Tumbarello D. A. (2022). The relationship of alpha-synuclein to mitochondrial dynamics and quality control. Front. Mol. Neurosci. 15, 947191. doi:10.3389/fnmol.2022.947191
Tornquist M., Michaels T. C. T., Sanagavarapu K., Yang X., Meisl G., Cohen S. I. A., et al. (2018). Secondary nucleation in amyloid formation. Chem. Commun. (Camb). 54 (63), 8667–8684. doi:10.1039/c8cc02204f
Truscott K. N., Kovermann P., Geissler A., Merlin A., Meijer M., Driessen A. J., et al. (2001). A presequence- and voltage-sensitive channel of the mitochondrial preprotein translocase formed by Tim23. Nat. Struct. Biol. 8 (12), 1074–1082. doi:10.1038/nsb726
Truscott K. N., Voos W., Frazier A. E., Lind M., Li Y., Geissler A., et al. (2003). A J-protein is an essential subunit of the presequence translocase-associated protein import motor of mitochondria. J. Cell. Biol. 163 (4), 707–713. doi:10.1083/jcb.200308004
Trushina E., Dyer R. B., Badger J. D., Ure D., Eide L., Tran D. D., et al. (2004). Mutant huntingtin impairs axonal trafficking in mammalian neurons in vivo and in vitro. Mol. Cell. Biol. 24 (18), 8195–8209. doi:10.1128/MCB.24.18.8195-8209.2004
Tsacopoulos M., Magistretti P. J. (1996). Metabolic coupling between glia and neurons. J. Neurosci. 16 (3), 877–885. doi:10.1523/JNEUROSCI.16-03-00877.1996
Tucker K., Park E. (2019). Cryo-EM structure of the mitochondrial protein-import channel TOM complex at near-atomic resolution. Nat. Struct. Mol. Biol. 26 (12), 1158–1166. doi:10.1038/s41594-019-0339-2
Uddin M. S., Kabir M. T., Rahman M. S., Behl T., Jeandet P., Ashraf G. M., et al. (2020). Revisiting the amyloid cascade hypothesis: from anti-aβ therapeutics to auspicious new ways for alzheimer's disease. Int. J. Mol. Sci. 21 (16), 5858. doi:10.3390/ijms21165858
Ullman O., Fisher C. K., Stultz C. M. (2011). Explaining the structural plasticity of alpha-synuclein. J. Am. Chem. Soc. 133 (48), 19536–19546. doi:10.1021/ja208657z
Ulmer T. S., Bax A., Cole N. B., Nussbaum R. L. (2005). Structure and dynamics of micelle-bound human alpha-synuclein. J. Biol. Chem. 280 (10), 9595–9603. doi:10.1074/jbc.M411805200
Uversky V. N. (2002). Natively unfolded proteins: a point where biology waits for physics. Protein Sci. 11 (4), 739–756. doi:10.1110/ps.4210102
van der Laan M., Wiedemann N., Mick D. U., Guiard B., Rehling P., Pfanner N. (2006). A role for Tim21 in membrane-potential-dependent preprotein sorting in mitochondria. Curr. Biol. 16 (22), 2271–2276. doi:10.1016/j.cub.2006.10.025
van Wilpe S., Ryan M. T., Hill K., Maarse A. C., Meisinger C., Brix J., et al. (1999). Tom22 is a multifunctional organizer of the mitochondrial preprotein translocase. Nature 401 (6752), 485–489. doi:10.1038/46802
Vande Walle L., Lamkanfi M., Vandenabeele P. (2008). The mitochondrial serine protease HtrA2/Omi: an overview. Cell. Death Differ. 15 (3), 453–460. doi:10.1038/sj.cdd.4402291
Vangavaragu J. R., Valasani K. R., Gan X., Yan S. S. (2014). Identification of human presequence protease (hPreP) agonists for the treatment of Alzheimer's disease. Eur. J. Med. Chem. 76, 506–516. doi:10.1016/j.ejmech.2014.02.046
Varshavsky A. (2011). The N-end rule pathway and regulation by proteolysis. Protein Sci. 20 (8), 1298–1345. doi:10.1002/pro.666
Ventura A., Maccarana M., Raker V. A., Pelicci P. G. (2004). A cryptic targeting signal induces isoform-specific localization of p46Shc to mitochondria. J. Biol. Chem. 279 (3), 2299–2306. doi:10.1074/jbc.M307655200
Vicario M., Cieri D., Brini M., Cali T. (2018). The close encounter between alpha-synuclein and mitochondria. Front. Neurosci. 12, 388. doi:10.3389/fnins.2018.00388
Vijayvargia R., Epand R., Leitner A., Jung T. Y., Shin B., Jung R., et al. (2016). Huntingtin's spherical solenoid structure enables polyglutamine tract-dependent modulation of its structure and function. Elife 5, e11184. doi:10.7554/eLife.11184
Vogtle F. N., Prinz C., Kellermann J., Lottspeich F., Pfanner N., Meisinger C. (2011). Mitochondrial protein turnover: role of the precursor intermediate peptidase Oct1 in protein stabilization. Mol. Biol. Cell. 22 (13), 2135–2143. doi:10.1091/mbc.E11-02-0169
Vogtle F. N., Wortelkamp S., Zahedi R. P., Becker D., Leidhold C., Gevaert K., et al. (2009). Global analysis of the mitochondrial N-proteome identifies a processing peptidase critical for protein stability. Cell. 139 (2), 428–439. doi:10.1016/j.cell.2009.07.045
Waegemann K., Popov-Celeketic D., Neupert W., Azem A., Mokranjac D. (2015). Cooperation of TOM and TIM23 complexes during translocation of proteins into mitochondria. J. Mol. Biol. 427 (5), 1075–1084. doi:10.1016/j.jmb.2014.07.015
Waingankar T. P., Silva P. (2021). Multiple variants of the human presequence translocase motor subunit Magmas govern the mitochondrial import. J. Biol. Chem. 297 (6), 101349. doi:10.1016/j.jbc.2021.101349
Wang M., Oge L., Perez-Garcia M. D., Hamama L., Sakr S. (2018). The PUF protein family: overview on PUF RNA targets, biological functions, and post transcriptional regulation. Int. J. Mol. Sci. 19 (2), 410. doi:10.3390/ijms19020410
Wang W., Chen X., Zhang L., Yi J., Ma Q., Yin J., et al. (2020a). Atomic structure of human TOM core complex. Cell. Discov. 6, 67. doi:10.1038/s41421-020-00198-2
Wang W., Perovic I., Chittuluru J., Kaganovich A., Nguyen L. T., Liao J., et al. (2011). A soluble alpha-synuclein construct forms a dynamic tetramer. Proc. Natl. Acad. Sci. U. S. A. 108 (43), 17797–17802. doi:10.1073/pnas.1113260108
Wang W., Zhao F., Ma X., Perry G., Zhu X. (2020c). Mitochondria dysfunction in the pathogenesis of Alzheimer's disease: recent advances. Mol. Neurodegener. 15 (1), 30. doi:10.1186/s13024-020-00376-6
Wang W., Zhao F., Ma X., Perry G., Zhu X. (2020b). Mitochondria dysfunction in the pathogenesis of Alzheimer’s disease: recent advances. Mol. Neurodegener. 15 (1), 30. doi:10.1186/s13024-020-00376-6
Wang X., Chen X. J. (2015). A cytosolic network suppressing mitochondria-mediated proteostatic stress and cell death. Nature 524 (7566), 481–484. doi:10.1038/nature14859
Wang X., Su B., Lee H. G., Li X., Perry G., Smith M. A., et al. (2009). Impaired balance of mitochondrial fission and fusion in Alzheimer's disease. J. Neurosci. 29 (28), 9090–9103. doi:10.1523/JNEUROSCI.1357-09.2009
Wang X., Su B., Siedlak S. L., Moreira P. I., Fujioka H., Wang Y., et al. (2008a). Amyloid-beta overproduction causes abnormal mitochondrial dynamics via differential modulation of mitochondrial fission/fusion proteins. Proc. Natl. Acad. Sci. U. S. A. 105 (49), 19318–19323. doi:10.1073/pnas.0804871105
Wang X., Zhu S., Pei Z., Drozda M., Stavrovskaya I. G., Del Signore S. J., et al. (2008b). Inhibitors of cytochrome c release with therapeutic potential for Huntington's disease. J. Neurosci. 28 (38), 9473–9485. doi:10.1523/JNEUROSCI.1867-08.2008
Wang Y., Guo X., Ye K., Orth M., Gu Z. (2021). Accelerated expansion of pathogenic mitochondrial DNA heteroplasmies in Huntington's disease. Proc. Natl. Acad. Sci. U. S. A. 118 (30), e2014610118. doi:10.1073/pnas.2014610118
Weidberg H., Amon A. (2018). MitoCPR-A surveillance pathway that protects mitochondria in response to protein import stress. Science 360 (6385), eaan4146. doi:10.1126/science.aan4146
Weill U., Yofe I., Sass E., Stynen B., Davidi D., Natarajan J., et al. (2018). Genome-wide SWAp-Tag yeast libraries for proteome exploration. Nat. Methods 15 (8), 617–622. doi:10.1038/s41592-018-0044-9
Weinreb P. H., Zhen W., Poon A. W., Conway K. A., Lansbury P. T. (1996). NACP, a protein implicated in Alzheimer's disease and learning, is natively unfolded. Biochemistry 35 (43), 13709–13715. doi:10.1021/bi961799n
Wellington C. L., Ellerby L. M., Hackam A. S., Margolis R. L., Trifiro M. A., Singaraja R., et al. (1998). Caspase cleavage of gene products associated with triplet expansion disorders generates truncated fragments containing the polyglutamine tract. J. Biol. Chem. 273 (15), 9158–9167. doi:10.1074/jbc.273.15.9158
Wells C., Brennan S., Keon M., Ooi L. (2021). The role of amyloid oligomers in neurodegenerative pathologies. Int. J. Biol. Macromol. 181, 582–604. doi:10.1016/j.ijbiomac.2021.03.113
Wentink A. S., Nillegoda N. B., Feufel J., Ubartaite G., Schneider C. P., De Los Rios P., et al. (2020). Molecular dissection of amyloid disaggregation by human HSP70. Nature 587 (7834), 483–488. doi:10.1038/s41586-020-2904-6
Weydt P., Pineda V. V., Torrence A. E., Libby R. T., Satterfield T. F., Lazarowski E. R., et al. (2006). Thermoregulatory and metabolic defects in Huntington's disease transgenic mice implicate PGC-1alpha in Huntington's disease neurodegeneration. Cell. Metab. 4 (5), 349–362. doi:10.1016/j.cmet.2006.10.004
Wiedemann N., Pfanner N., Ryan M. T. (2001). The three modules of ADP/ATP carrier cooperate in receptor recruitment and translocation into mitochondria. EMBO J. 20 (5), 951–960. doi:10.1093/emboj/20.5.951
Wiedmann B., Sakai H., Davis T. A., Wiedmann M. (1994). A protein complex required for signal-sequence-specific sorting and translocation. Nature 370 (6489), 434–440. doi:10.1038/370434a0
Wieprecht T., Apostolov O., Beyermann M., Seelig J. (2000). Interaction of a mitochondrial presequence with lipid membranes: role of helix formation for membrane binding and perturbation. Biochemistry 39 (50), 15297–15305. doi:10.1021/bi001774v
Williams C. C., Jan C. H., Weissman J. S. (2014). Targeting and plasticity of mitochondrial proteins revealed by proximity-specific ribosome profiling. Science 346 (6210), 748–751. doi:10.1126/science.1257522
Wodrich A. P. K., Scott A. W., Shukla A. K., Harris B. T., Giniger E. (2022). The unfolded protein responses in health, aging, and neurodegeneration: recent advances and future considerations. Front. Mol. Neurosci. 15, 831116. doi:10.3389/fnmol.2022.831116
Woellhaf M. W., Hansen K. G., Garth C., Herrmann J. M. (2014). Import of ribosomal proteins into yeast mitochondria. Biochem. Cell. Biol. 92 (6), 489–498. doi:10.1139/bcb-2014-0029
Wong Y. C., Holzbaur E. L. (2014). The regulation of autophagosome dynamics by huntingtin and HAP1 is disrupted by expression of mutant huntingtin, leading to defective cargo degradation. J. Neurosci. 34 (4), 1293–1305. doi:10.1523/JNEUROSCI.1870-13.2014
Wrobel L., Topf U., Bragoszewski P., Wiese S., Sztolsztener M. E., Oeljeklaus S., et al. (2015). Mistargeted mitochondrial proteins activate a proteostatic response in the cytosol. Nature 524 (7566), 485–488. doi:10.1038/nature14951
Wu Y., Sha B. (2006). Crystal structure of yeast mitochondrial outer membrane translocon member Tom70p. Nat. Struct. Mol. Biol. 13 (7), 589–593. doi:10.1038/nsmb1106
Wynne M. E., Ogunbona O., Lane A. R., Gokhale A., Zlatic S. A., Xu C., et al. (2023). APOE expression and secretion are modulated by mitochondrial dysfunction. Elife 12, e85779. doi:10.7554/eLife.85779
Xie W., Chung K. K. (2012). Alpha-synuclein impairs normal dynamics of mitochondria in cell and animal models of Parkinson's disease. J. Neurochem. 122 (2), 404–414. doi:10.1111/j.1471-4159.2012.07769.x
Xin N., Durieux J., Yang C., Wolff S., Kim H. E., Dillin A. (2022). The UPRmt preserves mitochondrial import to extend lifespan. J. Cell. Biol. 221 (7), e202201071. doi:10.1083/jcb.202201071
Xu J., Du W., Zhao Y., Lim K., Lu L., Zhang C., et al. (2022b). Mitochondria targeting drugs for neurodegenerative diseases-Design, mechanism and application. Acta Pharm. Sin. B 12 (6), 2778–2789. doi:10.1016/j.apsb.2022.03.001
Xu J., Minobe E., Kameyama M. (2022a). Ca(2+) dyshomeostasis links risk factors to neurodegeneration in Parkinson's disease. Front. Cell. Neurosci. 16, 867385. doi:10.3389/fncel.2022.867385
Xu Y. J., Mei Y., Qu Z. L., Zhang S. J., Zhao W., Fang J. S., et al. (2018). Ligustilide ameliorates memory deficiency in APP/PS1 transgenic mice via restoring mitochondrial dysfunction. Biomed. Res. Int. 2018, 4606752. doi:10.1155/2018/4606752
Yablonska S., Ganesan V., Ferrando L. M., Kim J., Pyzel A., Baranova O. V., et al. (2019). Mutant huntingtin disrupts mitochondrial proteostasis by interacting with TIM23. Proc. Natl. Acad. Sci. U. S. A. 116 (33), 16593–16602. doi:10.1073/pnas.1904101116
Yamamoto A., Lucas J. J., Hen R. (2000). Reversal of neuropathology and motor dysfunction in a conditional model of huntington's disease. Cell. 101 (1), 57–66. doi:10.1016/S0092-8674(00)80623-6
Yamamoto H., Esaki M., Kanamori T., Tamura Y., Nishikawa S., Endo T. (2002). Tim50 is a subunit of the TIM23 complex that links protein translocation across the outer and inner mitochondrial membranes. Cell. 111 (4), 519–528. doi:10.1016/s0092-8674(02)01053-x
Yamamoto H., Fukui K., Takahashi H., Kitamura S., Shiota T., Terao K., et al. (2009). Roles of Tom70 in import of presequence-containing mitochondrial proteins. J. Biol. Chem. 284 (46), 31635–31646. doi:10.1074/jbc.M109.041756
Yamano K., Yatsukawa Y., Esaki M., Hobbs A. E., Jensen R. E., Endo T. (2008). Tom20 and Tom22 share the common signal recognition pathway in mitochondrial protein import. J. Biol. Chem. 283 (7), 3799–3807. doi:10.1074/jbc.M708339200
Yang L., Zhao K., Calingasan N. Y., Luo G., Szeto H. H., Beal M. F. (2009). Mitochondria targeted peptides protect against 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine neurotoxicity. Antioxid. Redox Signal 11 (9), 2095–2104. doi:10.1089/ars.2009.2445
Yang L. G., March Z. M., Stephenson R. A., Narayan P. S. (2023). Apolipoprotein E in lipid metabolism and neurodegenerative disease. Trends Endocrinol. Metab. 34 (8), 430–445. doi:10.1016/j.tem.2023.05.002
Yang Y., Shi Y., Schweighauser M., Zhang X., Kotecha A., Murzin A. G., et al. (2022). Structures of α-synuclein filaments from human brains with Lewy pathology. Nature 610 (7933), 791–795. doi:10.1038/s41586-022-05319-3
Yano H., Baranov S. V., Baranova O. V., Kim J., Pan Y., Yablonska S., et al. (2014). Inhibition of mitochondrial protein import by mutant huntingtin. Nat. Neurosci. 17 (6), 822–831. doi:10.1038/nn.3721
Yano M., Terada K., Mori M. (2004). Mitochondrial import receptors Tom20 and Tom22 have chaperone-like activity. J. Biol. Chem. 279 (11), 10808–10813. doi:10.1074/jbc.M311710200
Yapa N. M. B., Lisnyak V., Reljic B., Ryan M. T. (2021). Mitochondrial dynamics in health and disease. FEBS Lett. 595 (8), 1184–1204. doi:10.1002/1873-3468.14077
Yiannopoulou K. G., Papageorgiou S. G. (2020). Current and future treatments in alzheimer disease: an update. J. Cent. Nerv. Syst. Dis. 12, 1179573520907397. doi:10.1177/1179573520907397
Yin F. (2023). Lipid metabolism and Alzheimer's disease: clinical evidence, mechanistic link and therapeutic promise. FEBS J. 290 (6), 1420–1453. doi:10.1111/febs.16344
Yin X., Manczak M., Reddy P. H. (2016). Mitochondria-targeted molecules MitoQ and SS31 reduce mutant huntingtin-induced mitochondrial toxicity and synaptic damage in Huntington's disease. Hum. Mol. Genet. 25 (9), 1739–1753. doi:10.1093/hmg/ddw045
Yogev O., Karniely S., Pines O. (2007). Translation-coupled translocation of yeast fumarase into mitochondria in vivo. J. Biol. Chem. 282 (40), 29222–29229. doi:10.1074/jbc.M704201200
Young J. C., Hoogenraad N. J., Hartl F. U. (2003). Molecular chaperones Hsp90 and Hsp70 deliver preproteins to the mitochondrial import receptor Tom70. Cell. 112 (1), 41–50. doi:10.1016/s0092-8674(02)01250-3
Zeytuni N., Zarivach R. (2012). Structural and functional discussion of the tetra-trico-peptide repeat, a protein interaction module. Structure 20 (3), 397–405. doi:10.1016/j.str.2012.01.006
Zhai P., Stanworth C., Liu S., Silberg J. J. (2008). The human escort protein Hep binds to the ATPase domain of mitochondrial hsp70 and regulates ATP hydrolysis. J. Biol. Chem. 283 (38), 26098–26106. doi:10.1074/jbc.M803475200
Zhai S., Tanimura A., Graves S. M., Shen W., Surmeier D. J. (2018). Striatal synapses, circuits, and Parkinson's disease. Curr. Opin. Neurobiol. 48, 9–16. doi:10.1016/j.conb.2017.08.004
Zhang H., Alder N. N., Wang W., Szeto H., Marcinek D. J., Rabinovitch P. S. (2020). Reduction of elevated proton leak rejuvenates mitochondria in the aged cardiomyocyte. Elife 9, e60827. doi:10.7554/eLife.60827
Zhang L., Guo X. Q., Chu J. F., Zhang X., Yan Z. R., Li Y. Z. (2015). Potential hippocampal genes and pathways involved in Alzheimer's disease: a bioinformatic analysis. Genet. Mol. Res. 14 (2), 7218–7232. doi:10.4238/2015.June.29.15
Zhang L., Zhang C., Zhu Y., Cai Q., Chan P., Ueda K., et al. (2008). Semi-quantitative analysis of alpha-synuclein in subcellular pools of rat brain neurons: an immunogold electron microscopic study using a C-terminal specific monoclonal antibody. Brain Res. 1244, 40–52. doi:10.1016/j.brainres.2008.08.067
Zhang Y., Chen Y., Gucek M., Xu H. (2016). The mitochondrial outer membrane protein MDI promotes local protein synthesis and mtDNA replication. EMBO J. 35 (10), 1045–1057. doi:10.15252/embj.201592994
Zhang Y., Leavitt B. R., van Raamsdonk J. M., Dragatsis I., Goldowitz D., MacDonald M. E., et al. (2006). Huntingtin inhibits caspase-3 activation. EMBO J. 25 (24), 5896–5906. doi:10.1038/sj.emboj.7601445
Zhao J., Liu X., Xia W., Zhang Y., Wang C. (2020). Targeting amyloidogenic processing of APP in alzheimer's disease. Front. Mol. Neurosci. 13, 137. doi:10.3389/fnmol.2020.00137
Zhao W., Xu Z., Cao J., Fu Q., Wu Y., Zhang X., et al. (2019). Elamipretide (SS-31) improves mitochondrial dysfunction, synaptic and memory impairment induced by lipopolysaccharide in mice. J. Neuroinflammation 16 (1), 230. doi:10.1186/s12974-019-1627-9
Zheng H., Koo E. H. (2011). Biology and pathophysiology of the amyloid precursor protein. Mol. Neurodegener. 6 (1), 27. doi:10.1186/1750-1326-6-27
Zhou Z. D., Chan C. H., Ma Q. H., Xu X. H., Xiao Z. C., Tan E. K. (2011). The roles of amyloid precursor protein (APP) in neurogenesis: implications to pathogenesis and therapy of Alzheimer disease. Cell. Adh Migr. 5 (4), 280–292. doi:10.4161/cam.5.4.16986
Zhu L., Zhou Q., He L., Chen L. (2021). Mitochondrial unfolded protein response: an emerging pathway in human diseases. Free Radic. Biol. Med. 163, 125–134. doi:10.1016/j.freeradbiomed.2020.12.013
Zhu Y., Duan C., Lu L., Gao H., Zhao C., Yu S., et al. (2011). α-Synuclein overexpression impairs mitochondrial function by associating with adenylate translocator. Int. J. Biochem. Cell. Biol. 43 (5), 732–741. doi:10.1016/j.biocel.2011.01.014
Zhu Y., Wang H., Fang J., Dai W., Zhou J., Wang X., et al. (2018). SS-31 provides neuroprotection by reversing mitochondrial dysfunction after traumatic brain injury. Oxid. Med. Cell. Longev. 2018, 4783602. doi:10.1155/2018/4783602
Zigoneanu I. G., Yang Y. J., Krois A. S., Haque E., Pielak G. J. (2012). Interaction of alpha-synuclein with vesicles that mimic mitochondrial membranes. Biochim. Biophys. Acta 1818 (3), 512–519. doi:10.1016/j.bbamem.2011.11.024
Keywords: mitochondria, amyloids, neurodegeneration, protein import, cryptic targeting, targeting signals
Citation: Reed AL, Mitchell W, Alexandrescu AT and Alder NN (2023) Interactions of amyloidogenic proteins with mitochondrial protein import machinery in aging-related neurodegenerative diseases. Front. Physiol. 14:1263420. doi: 10.3389/fphys.2023.1263420
Received: 19 July 2023; Accepted: 02 October 2023;
Published: 02 November 2023.
Edited by:
Martin Van Der Laan, Saarland University, GermanyReviewed by:
Julia C. Fitzgerald, University of Tübingen, GermanyDejana Mokranjac, Ludwig Maximilian University of Munich, Germany
Copyright © 2023 Reed, Mitchell, Alexandrescu and Alder. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nathan N. Alder, bmF0aGFuLmFsZGVyQHVjb25uLmVkdQ==
† These authors have contributed equally to this work
 Ashley L. Reed
Ashley L. Reed Wayne Mitchell
Wayne Mitchell Andrei T. Alexandrescu
Andrei T. Alexandrescu Nathan N. Alder
Nathan N. Alder