
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
HYPOTHESIS AND THEORY article
Front. Physiol. , 09 January 2024
Sec. Chronobiology
Volume 14 - 2023 | https://doi.org/10.3389/fphys.2023.1243455
This article is part of the Research Topic Re-Scoping Ultradian Biological Rhythms View all 5 articles
Environmental rhythms such as the daily light-dark cycle selected for endogenous clocks. These clocks predict regular environmental changes and provide the basis for well-timed adaptive homeostasis in physiology and behavior of organisms. Endogenous clocks are oscillators that are based on positive feedforward and negative feedback loops. They generate stable rhythms even under constant conditions. Since even weak interactions between oscillators allow for autonomous synchronization, coupling/synchronization of oscillators provides the basis of self-organized physiological timing. Amongst the most thoroughly researched clocks are the endogenous circadian clock neurons in mammals and insects. They comprise nuclear clockworks of transcriptional/translational feedback loops (TTFL) that generate ∼24 h rhythms in clock gene expression entrained to the environmental day-night cycle. It is generally assumed that this TTFL clockwork drives all circadian oscillations within and between clock cells, being the basis of any circadian rhythm in physiology and behavior of organisms. Instead of the current gene-based hierarchical clock model we provide here a systems view of timing. We suggest that a coupled system of autonomous TTFL and posttranslational feedback loop (PTFL) oscillators/clocks that run at multiple timescales governs adaptive, dynamic homeostasis of physiology and behavior. We focus on mammalian and insect neurons as endogenous oscillators at multiple timescales. We suggest that neuronal plasma membrane-associated signalosomes constitute specific autonomous PTFL clocks that generate localized but interlinked oscillations of membrane potential and intracellular messengers with specific endogenous frequencies. In each clock neuron multiscale interactions of TTFL and PTFL oscillators/clocks form a temporally structured oscillatory network with a common complex frequency-band comprising superimposed multiscale oscillations. Coupling between oscillator/clock neurons provides the next level of complexity of an oscillatory network. This systemic dynamic network of molecular and cellular oscillators/clocks is suggested to form the basis of any physiological homeostasis that cycles through dynamic homeostatic setpoints with a characteristic frequency-band as hallmark. We propose that mechanisms of homeostatic plasticity maintain the stability of these dynamic setpoints, whereas Hebbian plasticity enables switching between setpoints via coupling factors, like biogenic amines and/or neuropeptides. They reprogram the network to a new common frequency, a new dynamic setpoint. Our novel hypothesis is up for experimental challenge.
Life on Earth evolved in a highly rhythmic environment. Geophysical rhythms occur via cycles of Earth-Moon-Sun constellations and additional rhythms are generated by multiscale fast signaling between organisms. These environmental rhythms favored the evolution of multiscale endogenous clocks that allow organisms to predict relevant changes in environmental parameters, improving adaptation and survival (Pittendrigh, 1993; Franken et al., 1994; Kippert and Hunt, 2000; Tyson et al., 2003; Rodríguez-Sosa et al., 2008; Isomura and Kageyama, 2014; Takahashi, 2016; Aviram et al., 2021). Any rhythmically occurring external cue can function as a zeitgeber when it entrains an endogenous clock (Meijer and Schwartz, 2003; Yoshii et al., 2015). The entrained clock maintains the same frequency and a stable phase-relation to the zeitgeber’s rhythm. If an organism’s clock expresses specific receptors, like photo- or chemoreceptors, it can entrain to zeitgeber cues of different modalities (Meijer and Schwartz, 2003; Stengl and Schröder, 2021). Predominant zeitgebers are the daily rhythm of light and dark, circalunar monthly rhythms in the brightness of nocturnal light, and infradian (period >24 h) annual rhythms in the duration of light per day (photoperiod). Consequently, environmental rhythms of light, often associated with regular temperature rhythms, temporally structure life on our planet into days, months, and years.
Superimposed on slow rhythms in illumination are fast fluctuating environmental events that organisms need to detect and process. Ultradian rhythms of social signals, most of them chemosensory, are exchanged between individuals within and across species, with periods ranging from milliseconds to hours (Stengl and Schröder, 2021). Concentration changes in nutritious or hazardous chemicals provide spatio-temporal orientation to food sources or mates, or away from danger. Chemoreceptors detect fast fluctuations in chemicals (e.g., in turbulent water or air) that occur on the scale of milliseconds (Baker et al., 1985; Stengl, 2010; Stengl and Schröder, 2021). These fast ultradian odor fluctuations are superimposed on slow 24 h cycles of odor presence; for example, plants advertise their nectar to attract pollinating insects only at specific times during the day, in coordination with daily patterned release of pheromones by mate-calling insects (Itagaki and Conner, 1988; Fenske and Imaizumi, 2016; Fenske et al., 2018). These superimposed oscillations in the concentration of species-specific chemicals can act as multiscale zeitgebers coordinating multiscale endogenous clocks that orchestrate physiology and behavior. Thus, endogenous clocks allow for ultradian (period <24 h), infradian (>24 h), or circadian (∼24 h) orchestration of reproduction rates (Hunt and Sassone-Corsi, 2007; Johnson, 2010; Farshadi et al., 2020), coordinate circadian rest-activity (sleep-wake) rhythms, as well as circannual adaptations to seasons in single cell organisms, animals, and humans (Meijer and Schwartz, 2003; Yoshii et al., 2015; Helfrich-Förster, 2018; Häfker and Tessmar-Raible, 2020; Veedin Rajan et al., 2021).
In summary, endogenous clocks predict environmental rhythms at multiple timescales as imminent advantage for survival by providing cues for spatial-temporal orientation. The environmental niche of an organism determines the pace and phase of its clocks. Amongst different environmental zeitgebers the daily 24 h cycle of light and dark provides the most dominant timing cue for terrestrial life and was the driving force for the evolution of circadian clocks that control sleep-wake cycles (Aréchiga et al., 1993; Hall and Rosbash, 1993; Takahashi, 2016).
In insects and mammals, physiological and behavioral processes cycle at different periods, e.g., locomotion, feeding, breathing, or heartbeat. They are coupled at various strengths and are temporally orchestrated by the brain’s neuronal clocks (King and Sehgal, 2020; Patel and Rangan, 2021; Parviainen et al., 2022; Ijspeert and Daley, 2023; Luhmann, 2023; Nakamura et al., 2023; Park et al., 2023). Current research in chronobiology focuses on circadian clocks and transcriptional/translational feedback loop (TTFL)-based clock(work)s in circadian genetic model organisms such as fruit flies and mice. The predominant view of biological timing is hierarchical. It interprets the network of circadian clock neurons that are hubs of photic entrainment as master circadian clock centers: the suprachiasmatic nucleus (SCN) of mammals, the pineal of some avian species, and the accessory medulla (AME) of insects (Stetson and Watson-Whitmyre, 1976; Inouye and Kawamura, 1979; Takahashi and Menaker, 1979; Moore, 1983; Reischig and Stengl, 2003b; Reischig and Stengl, 2003a). Furthermore, the clock neuron’s TTFL clockwork is assumed to constitute the molecular master clock that drives all circadian oscillations of the clock cell as master clockwork outputs (Hardin, 2011; Harvey et al., 2020; Patton and Hastings, 2023).
Instead, here, we advocate a systems view of mutually interconnected endogenous TTFL and posttranslational feedback loop (PTFL) oscillators/clocks in single clock cells which run at multiple timescales. Furthermore, we suggest that clock neurons in the brain maintain a dynamically coupled and interconnected system of timing where single clock neurons can be recruited into different physiological/behavioral tasks, depending on coupling factors and zeitgebers. We do not attempt to provide a comprehensive review on timing at the levels of networks, transcription, translation, or metabolomics and refer to other reviews (e.g., Colwell, 2011; Hardin, 2011; Panda, 2016; Harvey et al., 2020; Parnell et al., 2021; Liu and Chiu, 2022; Adlanmerini and Lazar, 2023; Kahn et al., 2023; Patton and Hastings, 2023; Xiong and Garfinkel, 2023).
Our focus is the neuronal plasma membrane of mammalian and insect circadian clock neurons as a prominent endogenous PTFL clock ticking at multiple, superimposed timescales. First, we explain general properties of endogenous oscillators (Section 1.3). Then, we dive into details to elucidate also for the non-electrophysiologist how spontaneous ultradian membrane potential oscillations in clock neurons arise (Section 1.4) as mandatory prerequisite to circadian or any other frequency modulation. Briefly, important connections between membrane potential oscillations and second messenger oscillations at membrane associated signalosomes are pointed out before we sketch the neurophysiological concepts of neuronal homeostasis and plasticity, referring to more extensive reviews (Marder et al., 1996; Turrigiano, 1999; Turrigiano, 2008; Turrigiano, 2012; Turrigiano and Nelson, 2000; Caporale and Dan, 2008; Masquelier et al., 2009; Schulz and Lane, 2017; Lee and Kirkwood, 2019; McCormick et al., 2020; Debanne and Inglebert, 2023; Xiong and Garfinkel, 2023). In Section 2 we review the predominant hierarchical view on circadian clocks and contrast it with our novel systemic hypothesis of biological timing. In Section 3 we propose that mechanisms of homeostatic plasticity based on signalosomes maintain the stability of dynamic physiological setpoints in the system of interconnected PTFL and TTFL oscillators/clocks. Finally, in Section 4 we present our new systemic hypothesis of the neuronal plasma membrane as endogenous PTFL clock that is ticking at multiple timescales and interconnected with the endogenous TTFL clock as basis for a new concept of a dynamic homeostasis in physiology and behavior.
In organisms, endogenous oscillators generate self-sustained periodic events with stable cycle frequencies, even in the absence of rhythmic inputs (Figure 1). In the study of dynamical systems they are described mathematically as limit cycle oscillators with the limit cycle as the stable frequency (an attractor, here coined as “dynamic setpoint”) the oscillator returns to after perturbations (Leloup et al., 1999). Endogenous oscillators employ universal mechanisms despite their large variety, ranging from cycling conformations of molecular complexes to cells with oscillating membrane potentials to synchronously oscillating neural networks that orchestrate rhythmic behavior (Trujillo et al., 2019; Hille, 2022; Jabbur and Johnson, 2022; Fernandez-Ruiz et al., 2023; Kang et al., 2023). Oscillations are generated as soon as antagonistic elements/chemical reactions co-evolved (Gutekunst, 2018) and assembled to form a loop: positive feedforward pathways that are connected to delayed self-inhibitory feedback loops (Figure 1A). For example, genetic and molecular oscillators can be based on autoregulatory TTFLs (Figure 1B) (Hardin, 2011; Li et al., 2023b; Patton and Hastings, 2023). In contrast, not all feedback loops require transcription and translation. Post-translational feedback loops (PTFL, also known as post-translational oscillator: PTO; Figure 1B) work, for example, through cycles of autophosphorylation of molecular complexes. Positive feedforward elements promote autophosphorylation, while antagonistic negative feedback elements inhibit autophosphorylation (Bell-Pedersen et al., 2005; Johnson, 2010; Jabbur and Johnson, 2022; Li et al., 2023b).
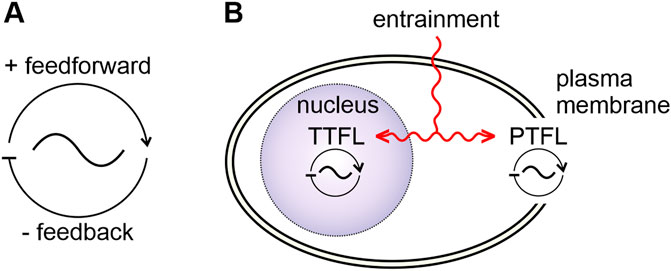
FIGURE 1. Basic properties of biological oscillators and clocks. (A) Biological oscillators consist of a positive feedforward- and a delayed negative feedback loop. (B) A clock is a specialized oscillator with entrainment mechanisms (red) that allow for synchronization to zeitgeber rhythms. Two general types of oscillators/clocks can be distinguished according to their “clockwork”: a transcription/translation feedback loop (TTFL)- based oscillator/clock, located in the nucleus (purple) and an oscillator/clock based on posttranslational feedback loops (PTFLs) located in other parts of the cell, such as the plasma membrane. Zeitgeber signals can entrain both the TTFL and the PTFL clockworks.
Interaction between endogenous oscillators with similar periods cause autonomous synchronization (Figure 2). This is the foundation for stable, self-organized timing, for autonomous assembly of a highly ordered sustainable biological system. Autonomous synchronization happens because inputs into an oscillator do not cause runaway acceleration or runaway braking but have phase-dependent antagonistic effects (Figures 2A, B). Self-organized, autonomous synchronization occurs because coupled oscillators either advance/accelerate or delay/decelerate each other until they maintain a common stable, intermediate frequency (a new dynamic setpoint) at stable phase relationships (Figure 2) (Tokuda et al., 2015; 2020; Ananthasubramaniam et al., 2018; Hahn et al., 2019; Lowet et al., 2022). Dependent on the context some oscillators become dominant and impose their frequency on other oscillators. While these pacemakers dictate the period of an oscillatory system, the coupled follower components can further pattern the system’s output (Marder and Bucher, 2001; Harris-Warrick, 2010; Daur et al., 2016; Marder et al., 2017; Jékely et al., 2018).
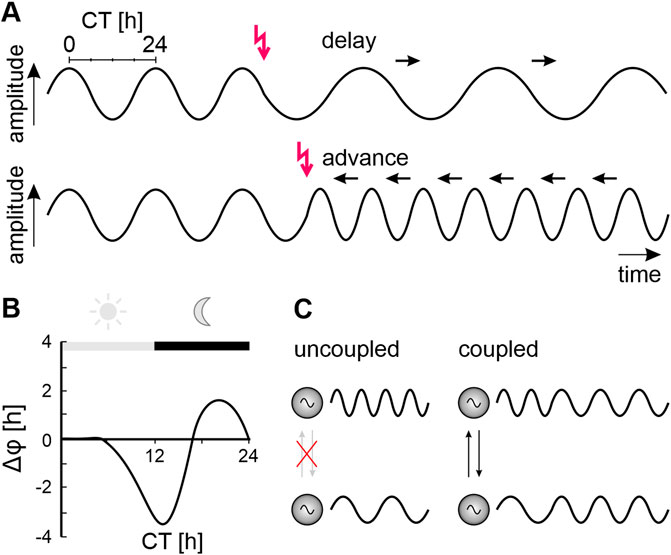
FIGURE 2. Phase-response curves characterize oscillators and clocks. (A) Inputs (red arrows) to an endogenous oscillator (sine waves) can either delay (top panel) or advance (bottom panel) the beginning of the next cycle, depending on the phase (relative timepoint within an endogeous cycle) at which they occur. The endogenous period (duration of the endogenous oscillator’s cycle) is measured in constant conditions. It is the shortest time difference between the respective same phases of the cycle divided into 24-time units (circadian time, CT). Repetitively delaying inputs (black arrows, top panel) prolong the period of the cycle (decrease oscillator frequency). Repetitively advancing inputs (black arrows, bottom panel) shorten the period of the cycle (increase oscillator frequency). (B) Plotting phase shifts against the circadian time (CT) of a subjective day-night cycle in constant conditions at which the phase-shifting input occurred yields a phase-response curve (PRC). Schematic PRC obtained from a nocturnal cockroach in a running wheel in constant darkness with light pulses applied at different CTs. Light pulses during the early subjective day (light grey bar, grey sun) have no effect. Light at the beginning of the subjective night (black bar, grey moon) delays the onset of locomotor activity rhythms, while light at the end of the night advances their onset. (C) Autonomous synchronization of two previously not interacting (crossed-out grey arrows) oscillators with endogenous fast (upper sine wave) or endogenous slow oscillations (lower sine wave). Mutual phase shifting through coupling/interactions (black arrows) results in a self-organized common period and stable phase relationship between both oscillators.
The term “clock” is less well defined than the term “oscillator” (Pittendrigh, 1993; Hall, 2003; 2005; Michel and Meijer, 2020). Generally, a clock is defined as a specialized endogenous oscillator that can be entrained by environmental zeitgebers. For example, a light-sensitive clock synchronizes (entrains) to the daily light-dark cycle. Entrainment by the zeitgeber via iterative phase-dependent phase-shifts eventually synchronizes and phase-locks the endogenous clock to the zeitgeber (Figures 2A, B). Because the entrained endogenous circadian clock has now, for example, the same 24 h period and a stable phase relationship to the rising and setting Sun, it can predict time of day. The endogenous period of clocks is species-specific and genetically determined and maintains stable oscillations even under constant conditions. In addition to unilateral entrainment by a zeitgeber the mutual coupling of endogenous clocks (Figure 2C) allows for autonomous multilateral synchronization which adds dynamics on a higher level than unilateral entrainment (Hardin, 2011; Hahn et al., 2019; Tokuda et al., 2020; Lowet et al., 2022; Jagannath et al., 2023; Kageyama et al., 2023; Kahn et al., 2023; Patton and Hastings, 2023; Wollmuth and Angert, 2023).
Endogenous clocks have a biochemical basis. While an increase in temperature usually speeds up biochemical processes, circadian clocks are temperature-compensated (being stable at different temperatures). Their endogenous cycle period remains the same over changing temperatures within a physiological range. Temperature compensation occurs automatically if both the positive feedforward and the negative feedback elements are symmetrically affected by temperature changes. Nevertheless, temperature changes can phase shift circadian clocks, for example, if they target only one of the clocks’ antagonistic elements (Rensing and Ruoff, 2002; Narasimamurthy and Virshup, 2017; Giesecke et al., 2023). So far, temperature-compensation is generally not considered a requirement for the definition of fast ultra- or slow infradian clocks, such as cell cycle clocks, metabolic clocks, photoperiodic clocks, or the clock that determines life span in a population of unicellular organisms (Rodríguez-Sosa et al., 2008; Zhu et al., 2017; Droin et al., 2019). Only few studies reported temperature-compensation for ultradian oscillators (Curras and Boulant, 1989; Roemschied et al., 2014; Städele et al., 2015; O’Leary and Marder, 2016).
In summary, endogenous oscillators evolved at different levels of complexity and timescales when antagonistic elements coupled to form robustly oscillating feedback loops. Respective endogenous oscillations are not an unwanted artefact but a process that can couple and reconcile antagonistic mechanisms into a stable system. Autonomous synchronization/entrainment of endogenous oscillators underlies the autonomously generated robust sustainable order and homeostasis of biological systems and embed organisms into their environmental niche (Schulz and Lane, 2017; Hahn et al., 2019; Liang et al., 2019; Rojas et al., 2019; Trujillo et al., 2019; Lowet et al., 2022; Jagannath et al., 2023; Kageyama et al., 2023; Kahn et al., 2023; Patton and Hastings, 2023; Wollmuth and Angert, 2023). The stable limit cycle oscillation frequency of a coupled system of endogenous oscillators can be viewed as dynamic setpoint of physiological homeostasis that the system bounces back to after perturbations.
So far, it is unknown how synchronization of endogenous oscillators/clocks can occur across largely different timescales. Here, we choose mammalian and insect brain neurons as an example to explain multiscale rhythms originating from the neuron’s excitable membrane.
Neurons evolved as neurosecretory cells destined for orchestration of internal physiology and for communication between organisms and environment (Colgren and Burkhardt, 2022; Burkhardt et al., 2023). Across species, neuron-like cells and neuronal circuits employ oscillation-based mechanisms to autonomously regulate and stabilize physiology and behavior, providing for homeostasis (Schulz and Lane, 2017; Li et al., 2023c; Hürkey et al., 2023; Nikitin et al., 2023; Norekian and Moroz, 2023; Stöber et al., 2023). Homeostasis is defined as a dynamic equilibrium state of an open dynamical system, of the self-regulation of organisms to maintain stability while remaining adaptive (Encyclopedia Britannica, 2023). Despite being under intense investigation the functional basis of these different autonomous types of adaptive neuronal oscillations is still not understood but on the cellular level they all rely on membrane potential oscillations.
Neurons, as any other cell type, are enclosed by semi-permeable membranes separating water-based ionic solutions with different osmotic values (Hille, 2001). Thus, across semi-permeable neuronal membranes two opposing, counter-balancing driving forces build up: an electric gradient and an osmotic gradient. Usually, neurons face extracellular solutions with K+ concentrations being ∼10 times lower than the intracellular K+ concentration and extracellular Na+ concentrations being ∼10 times higher than intracellular (Hille, 2001; Cohen et al., 2009; Wells et al., 2012; Mulet et al., 2023). Excitable membranes of neurons of different species express highly conserved voltage-dependent, specific, or unspecific, cation and anion channels (Hille, 2022). In the plasma membrane of un-stimulated, resting neurons specific K+ channels (two-pore K+, K2P) are constitutively open and create “leak” currents (Goldstein et al., 2001; Patel and Honoré, 2001; Talley et al., 2001). Driven by their osmotic gradient, K+ ions diffuse through K2P channels out of the cell until osmotic and electrical gradients are at equilibrium. The resulting membrane potential at which no net flow of ions through the membrane’s ion channels occurs is termed equilibrium (or reversal) potential. The reversal potential for each type of ion channel depends on the intra- and extracellular concentrations of permeating ions (Hille, 2022). For K+ it is typically around −80 mV and for Na+ around +50 mV. The steepest chemical gradient and, thus, the strongest driving force across the neuronal membrane is formed by Ca2+ ions, with extracellular concentrations in the millimolar range and intracellular concentrations several orders of magnitude lower in the nano- to picomolar range. The ionic gradients across the membrane are maintained by energy-consuming electrogenic pumps (Läuger, 1991). Thus, neurons at rest remain stable at their negative K+ equilibrium potential because only leaky K2P channels are open. Nevertheless, although K2P channels usually do not show voltage dependence, they are tightly controlled by second messenger systems linking membrane potential to second messenger signaling as basis for homeostatic feedback control of neuronal activity (Goldstein et al., 2001; Patel and Honoré, 2001; Talley et al., 2001).
Characteristic for clock neurons is their ability to generate endogenous oscillations in membrane potential and action potential (spike) frequency at characteristic timescales. These endogenous oscillations occur even in the absence of electrical stimulation and are maintained at dynamic setpoints through mechanisms of homeostatic plasticity (Miller and Selverston, 1982; Adams and Benson, 1985; Marder et al., 1996; Turrigiano, 1999; Turrigiano and Nelson, 2000; Lee and Kirkwood, 2019; McCormick et al., 2020). Even though endogenous membrane potential oscillations can occur at various timescales, they all require the expression of pacemaker channels (Figure 3A). In general, pacemaker channels are cation channels that open at hyperpolarized voltages. They drive the membrane potential to more depolarized potentials (Lüthi and McCormick, 1998; Robinson and Siegelbaum, 2003; Bose et al., 2014; Cochet-Bissuel et al., 2014; Das et al., 2016; Golowasch et al., 2017; Ratliff et al., 2021; Sharma et al., 2023). The resulting depolarization both closes the pacemaker channels and typically activates voltage-gated K+ channels, which in turn hyperpolarize the membrane potential. The hyperpolarization causes pacemaker channels to open again, the cycle begins anew and results in membrane potential oscillations (Figure 3A). One prominent example of a pacemaker channel is the hyperpolarization-activated, cyclic nucleotide-gated (HCN) non-specific cation channel that gives rise to the Ih current (Atkinson et al., 2011; Combe and Gasparini, 2021; Crunelli et al., 2023). The HCN channels express reversed voltage dependence, i.e., they close with depolarization and open with hyperpolarization. In that way they resemble molecular oscillators (Lee and MacKinnon, 2019). The depolarization by Ih and other regenerative pacemaker currents counteract the K+ leak, increase the neuron’s excitability, and promote oscillations (Cymbalyuk et al., 2002; Blethyn et al., 2006; Golowasch et al., 2017; Schneider et al., 2021; Sharma et al., 2023). When the oscillating depolarizations reach the activation threshold (approximately −40 mV) of voltage-gated fast Na+ channels (Figure 3B) the neuron generates spikes. However, already the subthreshold membrane potential oscillations control response threshold and response kinetics of neurons. These oscillations are determined by the kinetics, permeabilities, and relative numbers of antagonistic (depolarizing vs. hyperpolarizing) ion channels with their respective posttranslational modifications (Figures 3B, C) (Hille, 2022). Furthermore, the HCN channel is a hub for interacting intracellular messenger cascades providing for tight homeostatic control of spontaneous neuronal activity (Atkinson et al., 2011; Puri, 2020; Combe and Gasparini, 2021; Crunelli et al., 2023). We will focus next on Ca2+ dependent intracellular signaling and its close coupling to the membrane potential in a multitude of negative feedback circuits that provide for homeostasis of neuronal functions.
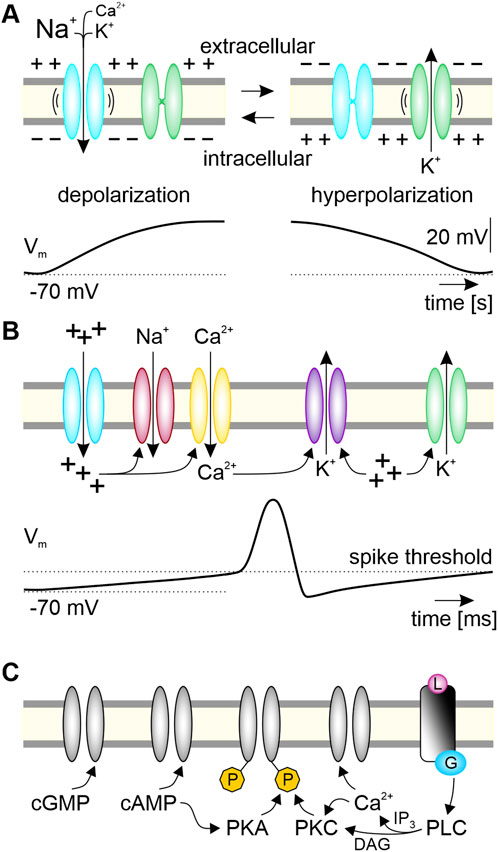
FIGURE 3. Ion channels contributing to endogenous oscillations of membrane potential and second messenger levels constitute a PTFL clock associated with a clock neuron’s plasma membrane. (A) Minimal requirement to generate spontaneous membrane potential oscillations are one pacemaker channel and one channel with antagonistic effects on the membrane potential. As example, the slow HCN-type pacemaker channels (blue) open at hyperpolarized voltages (“-”). The resulting influx of Na+, Ca2+, and K+ depolarizes the neuron from its negative membrane potential (−70 mV). This depolarization (“+”) opens voltage gated K+ channels (green), and the resulting efflux of K+ hyperpolarizes the neuron so that HCN channels can open again. Opening of hyperpolarization-activated HCN pacemaker channels constitute the positive feedforward element. The delayed negative feedback element, restarting the cycle of this posttranslational feedback loop (PTFL)-clock in the plasma membrane, is the slow HCN-dependent depolarization (“+”). It increases the open-time probability of the hyperpolarizing K+ channels and it closes the inverse-voltage-dependent HCN-pacemaker channels. (B) The respective complement of ion channels in a clock neuron leads to endogenous voltage oscillations at specific ultradian timescales. Depolarization via pacemaker channels (blue) triggers fast spikes via opening of voltage-gated fast Na+ channels (red) at the spike threshold. In addition, depolarization opens high voltage-activated (HVA-type) Ca2+ channels (yellow), increasing intracellular Ca2+ levels. Both depolarization and Ca2+ increase gate further channels, such as Ca2+-dependent (purple) and voltage-dependent (green) hyperpolarizing K+ channels. (C) Various interacting intracellular messenger cascades further gate and modulate antagonistic ion channels and enzymes via posttranslational modifications which leads to multiscale oscillations of the membrane potential interlinked with intracellular messenger oscillations. Cyclic nucleotides (cGMP, cAMP) modulate enzymes such as kinases (protein kinase A; PKA; protein kinase C; PKC) directly, and ion channels either directly, or indirectly via phosphorylating (P, yellow) kinases. Rising levels of intracellular messengers like Ca2+, as well as ligand (L, pink)-dependently activated G protein (blue)-coupled receptors activate and/or inhibit ion channels and enzymes. For example, activated phospholipase C (PLC) generates the second messengers diacyl glycerol (DAG) and inositol trisphosphate (IP3), orchestrating Ca2+-dependent signaling cascades. Positive feedforward elements of the PTFL clock cause depolarizations, delayed negative feedback elements hyperpolarize.
Any neuron expresses various sets of voltage-gated Ca2+ channels (Hille, 2022; Sharma et al., 2023). Therefore, both subthreshold membrane potential oscillations and regular spiking are accompanied by oscillatory changes in the intracellular Ca2+ concentration. Many enzymes, proteins, ion channels, and transcription factors are regulated by Ca2+ (Puri, 2020; Tokumitsu and Sakagami, 2022). Hence, the increase in intracellular Ca2+ concentration can both directly or indirectly activate and inactivate additional ion channels, which adds layers of complexity to the membrane potential oscillations (Figures 3B, C). Because prolonged high intracellular Ca2+ concentrations are highly toxic to neurons, intracellular Ca2+ levels are under tight autonomous homeostatic control (Duchen, 2000; Rizzuto et al., 2004; Sundararaj et al., 2021; Centeno et al., 2023; Nieto-Felipe et al., 2023).
Homeostatic mechanisms keep the intracellular Ca2+ concentrations tightly at nano- to picomolar levels as a dynamic setpoint via a multitude of interlinked negative feedback control circuits. When the intracellular Ca2+ concentrations rises, plasma membrane Ca2+ channels are either directly closed via the Ca2+-binding protein calmodulin, or their open time probability is decreased via protein kinase C-dependent phosphorylation (Hille, 2022; Sharma et al., 2023). Also, the number of ion channels and transporters located in the plasma membrane are controlled via a barrage of Ca2+-dependent homeostatic processes based on negative feedback (Lamothe and Zhang, 2016). Furthermore, elevated intracellular Ca2+ activates pumps that transport Ca2+ out of the neuron or into intracellular Ca2+ stores such as the endoplasmatic reticulum. This intracellular store contains Ca2+-conducting IP3 receptors and ryanodine-type ion channels which open in tight cooperation with plasma membrane Ca2+ channels when intracellular Ca2+ concentrations drop below the homeostatic setpoint and need to be restored (Duchen, 2000; Rizzuto et al., 2004; Sundararaj et al., 2021; Centeno et al., 2023; Nieto-Felipe et al., 2023).
Mechanisms of homeostatic plasticity constitute an interconnected network of feedforward/feedback elements. They are characteristic for endogenous oscillators and clocks that control neuronal activity at the single cell level as well as at the level of neuronal networks (Debanne and Inglebert, 2023; Xiong and Garfinkel, 2023). Traditionally, it is thought that homeostatic mechanisms evolved to maintain stationary setpoints of various physiological parameters such as fixed setpoints of electrical activity after stimulus-dependent activations (Marder et al., 1996; Turrigiano, 1999; Turrigiano and Nelson, 2000; Caporale and Dan, 2008; Masquelier et al., 2009; Schulz and Lane, 2017; Lee and Kirkwood, 2019; McCormick et al., 2020). Now, it is increasingly appreciated that homeostatic setpoints are not stationary but oscillating. In that way, they are “dynamic setpoints”. These observed physiological oscillations are not an undesired oversteering of feedback control circuits. Instead, oscillation-based coupling as basis for dynamic homeostatic setpoints allow for autonomously regulated plasticity. Oscillations interconnect and stabilize incompatible, antagonistic conditions for individual elements, such as oxidation and reduction, or phosphorylation and dephosphorylation (Xiong and Garfinkel, 2023).
On the single neuron level, mechanisms of homeostatic plasticity keep endogenous membrane potential oscillations at physiological dynamic setpoints, and maintain a characteristic spiking frequency via homeostatic control of ion channels (Xiong and Garfinkel, 2023). The negative feedback element of homeostatic control can be Ca2+ entry via the pacemaker channel that feeds back to decrease its open time probability, either directly or indirectly via phosphorylation by Ca2+-dependent protein kinase (He et al., 2014). Thereby, neuronal activity and responsiveness is oscillating but maintained within a stable physiological range.
Furthermore, at the brain’s network level, homeostatic plasticity maintains a common, synchronized ultradian spiking frequency of ensembles of synchronized neurons as dynamic homeostatic ensemble setpoint by orchestrating the gain of all synapses in the neuronal network without changing their respective weights (Marder et al., 1996; Turrigiano, 1999; Turrigiano and Nelson, 2000; Schulz and Lane, 2017; Lee and Kirkwood, 2019; McCormick et al., 2020; Valakh et al., 2023). During the course of each day (i.e., with circadian modulation) mammalian brains pass through different self-organized stable oscillatory states (i.e., dynamic setpoints) via autonomous sequential recruitment of neuronal ensembles (Figure 4). These ensembles spike synchronously at evolutionary conserved ultradian spike frequency bands that are connected to specific physiological functions (Laurent et al., 2016; Singer and Lazar, 2016; Hahn et al., 2019; McCormick et al., 2020; Tononi and Cirelli, 2020; Cirelli and Tononi, 2022). Ultradian oscillations in neuronal ensemble activity with superimposed circadian modulation are, for example, slow delta (0.5–4 Hz) waves predominating during sleep, or fast gamma (40 to >100 Hz) waves predominating during wakefulness (Figure 4). They correlate with different physiological states of sensory perception, of learning and memory, of sleep or wakefulness (Buzsáki and Draguhn, 2004; Colwell, 2011; Harvey et al., 2020; Stengl and Schröder, 2021; Brodt et al., 2023; Patton and Hastings, 2023). As foundation of robust, autonomous neuronal functions multiple mechanisms of homeostatic plasticity at the level of single neurons as well as at the level of neuronal ensembles maintain a specific dynamic setpoint measurable as stable spike frequency (McCormick et al., 2020).
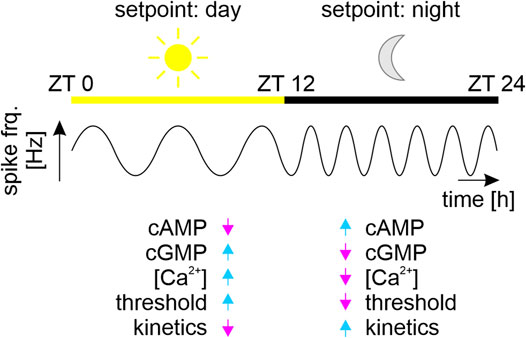
FIGURE 4. Different dynamic setpoints during the sleep-wake cycle. Schematic of synchronized physiological states of brain networks (ensembles) of clock neurons that regulate circadian sleep-wake cycles of a nocturnal animal. The multiscale clock neurons of the circadian ensembles fire at ultradian fast frequencies with superimposed circadian modulation. Ensembles fire with a common lower ultradian frequency (sine waves) during the day (sun, yellow bar, Zeitgebertime (ZT) 0–12 h), as compared to the night (moon, black bar, ZT 12–24 h). Each stable ultradian action potential frequency is the fingerprint (coined “setpoint”) of an ensemble of coupled oscillator neurons. The network’s “day” setpoint maintains the nocturnal animal’s physiological homeostasis of sleep, while the “night” setpoint maintains the physiological homeostasis of activity. Based on our work on peripheral insect circadian clock neurons we hypothesize that these antagonistic dynamic homeostatic setpoints correlate with antagonistic second messenger compositions (Stengl, 2010; Schendzielorz et al., 2012; Schendzielorz et al., 2015; Dolzer et al., 2021; Stengl and Schröder, 2021). The high sensory thresholds and slow response kinetics of sensory neurons during sleep correlate with increased intracellular Ca2+ and cGMP concentrations. In contrast, the low sensory thresholds and fast response kinetics during wakefulness correlate with increased cAMP levels and decreased concentrations in intracellular Ca2+ and cGMP levels (Stengl, 2010; Dolzer et al., 2021; Stengl and Schröder, 2021).
In contrast to homeostatic plasticity mechanisms that keep neuronal setpoints stable, mechanisms of Hebbian plasticity (including associative and non-associative forms of learning and memory), change neuronal setpoints. An example of non-associative learning is adaptation of olfactory sensory neurons. Olfactory adaptation employs, e.g., Ca2+-dependent negative feedback control mechanisms to prevent damage via overstimulation after a very strong or long odor/pheromone stimulus (Zufall and Leinders-Zufall, 2000; Dolzer et al., 2001; Spehr et al., 2009; Stengl, 2010). Hebbian plasticity mechanisms shift the current physiological dynamic setpoint to a new dynamic setpoint, for example, measurable as shifted dose-response curve of the sensory neurons. Thus, in an olfactory receptor neuron, after adapting to stimulation higher odor stimuli are necessary to further activate the sensory neuron which, thereby, preserved the memory of the adapting stimulus.
Hebbian plasticity at the network level allows for stimulus-dependent changes in brain function best described during synaptic processes of associative learning and memory (Hebb, 1949; Yee et al., 2017; Magee and Grienberger, 2020; McFarlan et al., 2023). In contrast to homeostatic plasticity Hebbian plasticity can push the neuronal network to a new physiological dynamic setpoint, detectable as different ensemble formation with different synchronous spiking frequency due to reconfiguration of the neuronal network. During processes of associative learning, such as long-term potentiation (LTP) or long-term depression (LTD), ultradian oscillatory processes in the pre- and postsynaptic cell are coupled at defined phase-differences to allow for spike time-dependent forms of plasticity causing synaptic weight changes (Debanne and Inglebert, 2023; Griffiths and Jensen, 2023; Yamakou et al., 2023). The autonomous synchronization of pre- and postsynaptic neurons to new synaptic weights, to a new physiological setpoint, depends on physiological conditions and specific coupling signals (neurotransmitters, neuropeptides) and can extend over several timescales (short, medium, long-term memory). Thus, mechanisms of homeostatic or Hebbian plasticity can also act on the circadian timescale and can target both the number of functional receptors as well as the gating and the number and types of voltage dependent ion channels expressed to regulate dynamic setpoints of electrical activity in neurons (Turrigiano et al., 1994; 1995; Marder et al., 1996; Golowasch et al., 1999; Turrigiano, 1999; Turrigiano and Nelson, 2000; Valakh et al., 2023).
In summary, evolutionarily conserved ultradian endogenous membrane potential oscillations at different timescales are a prerequisite to all endogenous (including circadian) clock neurons (Schneider and Stengl, 2005; Schneider and Stengl, 2006; Schneider and Stengl, 2007; Stengl and Schröder, 2021; Brodt et al., 2023). Spontaneously active neurons require the expression of pacemaker channels. Oscillations in membrane potential are tightly coupled to oscillations of intracellular Ca2+ levels. Both spike frequency and Ca2+ oscillations are controlled at dynamic setpoints via various interlinked mechanisms of homeostatic plasticity at multiple timescales. In contrast to mechanisms of homeostatic plasticity which preserve and maintain robust dynamic setpoints, mechanisms of Hebbian plasticity change the respective dynamic setpoint of the system of coupled oscillators/clocks (Turrigiano et al., 1994; Turrigiano et al., 1995; Marder et al., 1996; Golowasch et al., 1999; Turrigiano, 1999; Turrigiano and Nelson, 2000). It remains to be determined how concepts of endogenous clocks/oscillators are interconnected with, are the same as, or are different from concepts of homeostasis and plasticity in neurons.
In the following sections, we briefly state general principles of the master molecular TTFL clockwork in insect and mammalian clock neurons. Subsequently, we focus on plasma membrane-derived rhythms in mammalian and insect circadian clock neurons to summarize prevailing concepts. Since mammalian neurons and brains are much better analyzed with electrophysiological methods, we focus more on the mammalian than the insect circadian system. We challenge the current hierarchical view that membrane-dependent circadian rhythms are mere outputs of the master TTFL clockwork without denying strong mutual coupling between both. We highlight open questions, taking a systemic view of interlinked oscillators that use mechanisms of homeostatic and Hebbian plasticity. Finally, we propose a new hypothesis how circadian and ultradian oscillations could be linked by mechanisms of neuronal plasticity involving an endogenous multiscale PTFL membrane clock.
The search for the molecular clockwork which underlies circadian rhythms in behavior was initiated by the finding of a single gene that affected daily rhythms of both eclosion and locomotion in the fruit fly Drosophila melanogaster (Konopka and Benzer, 1971). Since then, it was generally believed that a molecular genetic clockwork in the nucleus rules circadian behavior such as daily sleep-wake cycles.
Best studied are the molecular circadian TTFL-based neuronal clocks in insects and mammals that generate oscillations in mRNA and protein levels with periods of ∼24 h (Hall, 2003; Hall, 2005; Hardin, 2011; Hastings et al., 2019). The core mechanism of the TTFL clockwork is astoundingly conserved between animal species and consists of various sets of homologous circadian clock genes. The transcription factors CLOCK and CYCLE (mammalian ortholog: BMAL1) are the positive feedforward elements that activate the transcription of the circadian clock genes period, timeless, and/or cryptochromes. Translation of mRNAs and posttranscriptional modifications of clock proteins (such as successive phosphorylations) generate delays before the clock proteins move back into the nucleus and inhibit their own transcription as negative feedback elements. This core circadian clockwork is interlinked with other oscillating TTFLs that increase stability and regularity of the resulting oscillations in mRNA and protein levels (Hardin, 2011; Mendoza-Viveros et al., 2017; Hastings et al., 2019; Park et al., 2020).
The prevailing assumption is that in mammalian or insect brains a single neuron becomes a circadian clock neuron because it contains this conserved TTFL-based molecular master clockwork that generates endogenous oscillations in gene transcription in the 24 h range. This TTFL master clockwork in master circadian clock neurons in the brain then drives all other circadian oscillations in this organism’s physiology and behavior, controlling and dominating peripheral clock cells in other organs (Green and Gillette, 1982; Groos and Hendriks, 1982; Welsh et al., 1995; Honma et al., 1998; Schaap et al., 2003; Colwell, 2011; Cox and Takahashi, 2019; Harvey et al., 2020; Patton and Hastings, 2023). Even though this review focuses on neuronal clocks in insects and mammals it should be noted that a wide variety of clock mechanisms, some of which are entirely independent of TTFLs, were reported in other organisms (Bell-Pedersen et al., 2005; Kitayama et al., 2008; Johnson, 2010; O’Neill and Reddy, 2011; Brown et al., 2012; Jabbur and Johnson, 2022; Li et al., 2023b). Furthermore, while different kinases, phosphatases, or O-GlcNAcylation processing enzymes regulate core elements of the neuron’s TTFL clockwork through posttranslational modifications (Tomita et al., 2005; Li et al., 2019; Anna and Kannan, 2021; Parnell et al., 2021; Liu and Chiu, 2022), it remains to be studied whether any of these signaling cascades constitute homeostatic control and/or are PTFL-based endogenous oscillators.
Circadian clock gene expressing neurons in the brains of both insects and mammals can generate spontaneous membrane potential oscillations at fast and slow frequencies that elicit ultradian and circadian spike rhythms (Pennartz et al., 2002; Jackson et al., 2004; Brown and Piggins, 2007; Schneider and Stengl, 2007; Belle et al., 2009; Colwell, 2011; Allen et al., 2017; Belle and Allen, 2018; Harvey et al., 2020). In mammals, the suprachiasmatic nucleus (SCN) houses the circadian pacemaker network that controls sleep-wake cycles, as well as any other physiological and behavioral circadian rhythms (Meijer and Schwartz, 2003; Michel and Meijer, 2020; Patton and Hastings, 2023). Under physiological conditions, SCN clock neurons spike spontaneously with circadian rhythms and, additionally, with higher ultradian frequency during the day (theta: ∼4 Hz) than during the night (delta: ∼1 Hz). In wild-type (WT) rodents, circadian and ultradian spike rhythms persist in SCN slices in constant conditions (Green and Gillette, 1982; Groos and Hendriks, 1982) and in individually dispersed clock neurons of both invertebrates and mammals (Michel et al., 1993; Welsh et al., 1995; Honma et al., 1998; Michel and Meijer, 2020). Therefore, the generation of electrical activity rhythms in the ultradian and circadian range is not an exclusive property of the SCN’s neuronal network but a property of single neurons with endogenous circadian clockworks. However, synchronized electrical activity of the SCN clock neuron network is a prerequisite for robust daily rhythms in behavioral activity: Application of the Na+ channel blocker TTX to the SCN of rats reversibly blocked both ultradian and circadian rhythms in electrical activity that correlated with a reversible block of daily locomotor and drinking behavioral rhythms (Schwartz et al., 1987; Earnest et al., 1991; Honma et al., 1998; Schaap et al., 2003; Colwell, 2011; Harvey et al., 2020).
It is generally assumed that the TTFL-based molecular master clockwork in individual neurons generates the ∼24 h rhythms in the neuron’s membrane potential via transcriptional control of key ion channels (Depetris-Chauvin et al., 2011; Allen et al., 2017; Harvey et al., 2020). Indeed, if core clock genes such as clock of the mammalian core TTFL clockwork are mutated or deleted, circadian sleep-wake cycles become arrhythmic (Albus et al., 2002). In addition, explants of the SCN network of TTFL clockwork mutants express arrhythmic electrical activity. However, improved clock mutants with targeted removal of the exons encoding the required dimerization region for BMAL1 that led to a loss of CLOCK immunoreactivity still express robust circadian rhythms in locomotor activity with only slightly shorter periods, with altered light-responses, and with some milder alterations in the TTFL clockwork (Collins and Blau, 2006; DeBruyne et al., 2006). Individually dispersed SCN clock neurons of clock mutants still display endogenous spiking rhythms in the 24 h range, albeit with a wider range of periods compared to WT SCN neurons (Herzog et al., 1998). Mutations of circadian clock genes in brain areas outside the SCN affected but did not delete all circadian rhythms in these neuronal circuits (Ray et al., 2020; Bering et al., 2023; Zheng et al., 2023). Similarly, in Drosophila, the core clock genes are not necessarily required for rhythmicity, since arrhythmicity in period01 mutants can be partially rescued by cryptochrome mutants (Collins et al., 2005). Therefore, the observed losses in rhythmicity in TTFL targeted mutants appear to be caused by loss of synchronization within and between neurons, rather than the loss of all endogenous circadian rhythmicity. Mutations in the core clock TTFL change the periodicity and phase of spiking and behavioral activity without abolishing rhythms (Wegner et al., 2017; Haque et al., 2019; Ray et al., 2020; Zheng et al., 2023). Thus, while there is convincing evidence for a tight coupling between membrane associated circadian oscillations and the circadian TTFL clockwork (Colwell, 2011; Harvey et al., 2020) it was not proven that all circadian membrane-associated rhythms are mere outputs of a master TTFL clock.
In summary, synchronized circadian rhythms in electrical activity of circadian clock neurons drive circadian rhythms in behavior. Mutations of core TTFL constituents are tightly coupled to, but do not delete all circadian membrane potential rhythms, possibly due to redundancy at the TTFL level, possibly due to other additional mechanisms such as endogenous PTFL oscillators/clocks. While nuclear clockwork and membrane potential rhythms clearly interact the respective elements and mechanisms are not yet known.
Circadian membrane potential oscillations feed back to the molecular TTFL clockwork (Mizrak et al., 2012; Harvey et al., 2020; Parnell et al., 2021; Matsuo et al., 2022). Electrically silencing or isolating circadian clock neurons of the SCN does not stop the molecular clockwork but renders its circadian rhythms less stable (Welsh et al., 1995; Herzog et al., 1998; Shirakawa et al., 2000; Yamaguchi et al., 2003; Aton et al., 2005; Lundkvist et al., 2005; Liu et al., 2007). In SCN clock neurons the blocking of Na+ channels and mutations of K+ ion channels (Kcnc1/Kcnc2) deleted circadian membrane potential rhythms without deleting circadian transcription of per2 of the core TTFL clockwork (Hermanstyne et al., 2023). Notably, in electrically silenced or isolated Drosophila neurons, transcription rhythms of the TTFL molecular clockwork desynchronized or dissipated altogether (Nitabach et al., 2002; Depetris-Chauvin et al., 2011; Schlichting et al., 2019; Tang et al., 2022). Apparently, the endogenous circadian molecular clockwork becomes more robust when it is tightly coupled to circadian spiking rhythms in the same clock neuron and/or the neuronal network.
In general, in both insect and mammalian circadian clock neurons, the ultradian membrane potential rhythms that are tightly interlinked with circadian activity rhythms are also tightly coupled with intracellular circadian rhythms in Ca2+ and cAMP levels (see Sections 1.4, 3.3). These membrane-associated oscillations at different timescales can be phase-shifted by neurotransmitters and neuropeptides that signal via G protein-coupled receptors (Ikeda et al., 2003b; Ikeda et al., 2003a; Schendzielorz et al., 2012; Schendzielorz et al., 2014; Schendzielorz et al., 2015; Wei and Stengl, 2012; Brancaccio et al., 2013; Brancaccio et al., 2017; Wei et al., 2014; Stengl et al., 2015; Stengl and Arendt, 2016; Harvey et al., 2020; Parnell et al., 2021; Stengl and Schröder, 2021; Matsuo et al., 2022). Several pathways have been identified in circadian clock neurons that demonstrate how these membrane-dependent rhythms in Ca2+ and cAMP levels couple to the molecular clockwork in the nucleus (Figure 5): Different Ca2+- and cAMP-dependent kinase pathways modify relevant transcription factors such as CREB, thereby phase-locking second messenger oscillations to the core TTFL clockwork oscillation (Harvey et al., 2020; Parnell et al., 2021). These signaling pathways connect plasma membrane-dependent activity with transcriptional control in the nucleus and are employed in many processes of homeostatic plasticity that stabilize ultradian spiking patterns of neurons (Turrigiano, 2012; Steven et al., 2020; Wu et al., 2021; Fitzpatrick and Kerschensteiner, 2023; Nieto-Felipe et al., 2023).
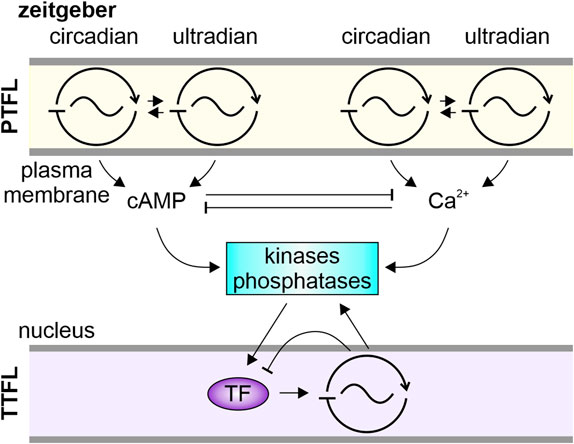
FIGURE 5. Schematic of our novel systemic hypothesis. Associated with the plasma membrane are multiple signalosomes comprising posttranslational feedback loop (PTFL)-oscillators. They anchor on scaffolding proteins pacemaker channels and specific receptors with their intracellular messenger cascades. Depending on the receptor, e.g., luminance- or chemoreceptor, the signalosome is susceptible to circadian or ultradian zeitgeber cues. Entrained to the zeitgeber frequency the signalosome’s transduction cascades generate oscillations in levels of cAMP and/or Ca2+. The intracellular messenger oscillations can interlink/couple by sharing common targets, like kinases and phosphatases. These can act as coupling factors synchronizing PTFL-oscillators as well as, via modulation of transcription factors (TF), transcriptional-translational feedback loop (TTFL) oscillators.
It is generally accepted that circadian membrane potential oscillations, which are tightly interconnected with circadian rhythms of second messenger cascades, do not drive but synchronize with the circadian TTFL clockwork. It remains to be studied whether these mechanisms of circadian synchronization/coupling are identical to general neuronal homeostatic plasticity mechanisms that work on fast and slow timescales (Turrigiano, 2008; Fox and Stryker, 2017; Liu et al., 2021; Parnell et al., 2021).
While circadian rhythms in electrical activity were observed in mammalian and insect circadian clock neurons, here, we focus mainly on the mammalian SCN (Cao and Nitabach, 2008; Colwell, 2011; Flourakis and Allada, 2015; Smith et al., 2019; Harvey et al., 2020; Barber et al., 2021; Fernandez-Chiappe et al., 2021; Stengl and Schröder, 2021; Schellinger et al., 2022). During the day, when nocturnal mice and rats sleep, most GABAergic SCN neurons express higher spike rates, higher intracellular Ca2+ levels, and higher input resistance as compared to the night (Pennartz et al., 2002; Colwell, 2011; Harvey et al., 2020; Patton and Hastings, 2023; Yang et al., 2023). This correlates with a multitude of ion channels in mammalian clock neurons which express daily rhythms in their current amplitudes and/or mRNA levels (Pitts et al., 2006; Colwell, 2011; CircaDB, 2013; Harvey et al., 2020; Hermanstyne et al., 2023).
During the night, the activity phase of rodents, the spontaneous spike frequency of most GABAergic SCN circadian clock neurons is lower as compared to the day. This correlates with a lower input resistance, apparently due to an increase in open time probability of hyperpolarizing ion channels. Thus, at night, hyperpolarizing ionic currents such as BK-type Ca2+ activated K+ currents dominate as compared to the day when depolarizing currents dominate (Flourakis et al., 2015; Harvey et al., 2020).
It is generally assumed that all membrane-dependent circadian rhythms, such as daily rhythms in ionic current amplitudes, are mere outputs of the molecular master TTFL clockwork (e.g., reviews: Harvey et al., 2020; Patton and Hastings, 2023). Thus, it is assumed that the TTFL clockwork activates daily expression of hyperpolarizing ion channels at night and activates daily expression of depolarizing ion channels during the day, connected to a strict circadian control of ion channel protein turnover. However, early models have shown that circadian changes in the electrical activity of neurons can be generated and sustained by the properties of ion channels and lipid membranes alone, without requiring transcriptional control connected to temporally regulated protein turnover (Njus et al., 1974).
To determine whether the TTFL clockwork in circadian clock neurons indeed controls membrane-derived circadian rhythms it would be necessary to show that the proteins of ion channels, such as pacemaker channels which are mandatory for expression of spontaneous endogenous circadian electrical rhythmicity, show circadian turnover and that respective ion channel genes require circadian transcriptional control by the TTFL clockwork. Current experiments mostly searched for correlations in maxima of ion channel mRNA levels with peaks in circadian spike rhythms in the SCN. In addition, pharmacological experiments using ion channel agonists and antagonists interfered with observed circadian firing rhythms of clock neurons, mostly at the network level (Colwell, 2011; Harvey et al., 2020; Patton and Hastings, 2023).
Two types of leak channels with antagonistic effects (K2P potassium leak and NALCN-type sodium leak channels) that control ultradian oscillations of spontaneous activity in circadian clock neurons of mammals would be ideal targets for the circadian modulation of spontaneous membrane potential oscillations (Brown and Piggins, 2007; Colwell, 2011; Flourakis et al., 2015; Harvey et al., 2020). While the current through the hyperpolarizing K2P channels does not express daily rhythms the currents through depolarizing NALCN-type (insect ortholog: NARROW ABDOMEN; NA) Na+ leak channels express higher amplitudes during the day. Therefore, it was suggested that under the control of the molecular TTFL clock NALCN channels are responsible for the stronger depolarization during the day (Talley et al., 2001; Cochet-Bissuel et al., 2014; Flourakis et al., 2015).
Next to the NALCN-type channels other pacemaker channels support spontaneous membrane potential oscillations in circadian clock neurons, such as T-type Ca2+ channels, and Ih -type cation channels (Akasu et al., 1993; Notomi and Shigemoto, 2004; Atkinson et al., 2011). In addition, subthreshold depolarization and oscillations are driven by TRPM4 cation channels, which are both voltage- and Ca2+ dependent (Li et al., 2021). Furthermore, L-type Ca2+ channels were found to control spontaneous activity of SCN neurons (Pennartz et al., 2002; Filosa and Putnam, 2003; Jackson et al., 2004; Imber and Putnam, 2012; Sanchez-Padilla et al., 2014). It remains to be examined which of the voltage-dependent ion channels, such as L-type Ca2+ channels or fast delayed rectifier (FDR) and A-type K+ channels, are driven secondarily by the stronger baseline depolarization in circadian clock neurons during the day, or primarily via transcriptional control (Colwell, 2011; Harvey et al., 2020).
As explained in previous sections membrane potential dynamics are connected to dynamics in the concentration of intracellular Ca2+, which is a central intracellular messenger in a multitude of signaling cascades connected to homeostatic control. Membrane-associated signaling cascades are often restricted to subcellular nanodomains and signalosomes to ensure specific responses (Musheshe et al., 2018; Tenner et al., 2020; Zaccolo et al., 2021; Anton et al., 2022; Lohse et al., 2023; Posner et al., 2023). Similarly, not all ion channel types are uniformly distributed throughout a neuron. Their spatial distribution influences the signaling properties of a neuron (Lai and Jan, 2006). Ion channels and their associated signaling molecules form channelosomes, which present a hub for interactions with other signalosomes to regulate ion channel activity. Since the activity of ion channels can be regulated by posttranslational modifications, such as trafficking to and localization in the plasma membrane or gating kinetics, it would be generally possible to achieve circadian and ultradian control independent of the molecular TTFL clockwork (Lamothe and Zhang, 2016).
The depolarizing NALCN is linked to many cellular rhythms and signaling cascades (Flourakis et al., 2015; Lee et al., 2019; Harvey et al., 2020; Impheng et al., 2021; Kschonsak et al., 2022). It is part of a large channelosome complex and functions as a hub for signaling cascades, with links to extracellular Ca2+ concentrations (Cochet-Bissuel et al., 2014; Lee et al., 2019). A multitude of signals, including G protein-coupled receptors, alternative splicing, long non-coding RNA interactions, and several posttranslational mechanisms such as methylations control NALCN functions (Flourakis et al., 2015; Lee et al., 2019). Thus, it is possible that coupling of the TTFL circadian clockwork with PTFLs that maintain homeostatic control of NALCN could be required for stable rhythmicity on the circadian timescale (Lear et al., 2013; Kang and Chen, 2022). The NALCN subunit expression is not directly under transcriptional control. However, localization of NALCN to membranes via the endoplasmatic reticulum resident protein NFL-1 is controlled by the TTFL clockwork because nlf-1 transcripts express circadian rhythms. Knockdown of nlf-1 transcripts in different populations of clock neurons reduced NALCN protein levels, circadian spiking rhythms, as well as morning and evening anticipation in circadian locomotor behavior (Flourakis et al., 2015).
Concerning the above-mentioned K+ channels, circadian expression has only been observed for the subunit that determines the fast inactivation kinetics (Kcnma1) of the BK-type K+ channels during the day. On the other hand, L-type Ca2+ channels and FDR K+ channels (Kcnc1,2) show circadian expression levels (Colwell, 2011; CircaDB, 2013; Harvey et al., 2020). However, when L-type Ca2+ channels or BK-type K+ channels are mutated, or the TTFL clockwork was disrupted, circadian membrane potential rhythms persist, albeit with reduced stability and amplitude (Colwell, 2011; Harvey et al., 2020). Thus, only a small fraction of ion channel mRNA level oscillations depends on the molecular circadian TTFL clockwork, and they do not appear to drive all the circadian rhythms of electrical activity in clock neurons.
The current hypothesis is that circadian rhythms in SCN spike rates are caused by an increased expression of hyperpolarizing ion channels during the night. While different K+ channels express higher current amplitudes at night, only deletions or knock-down of Kv12.1 and Kv12.2-encoded K+ channels (Kcnh8, Kcnh2 locus from the ether-á-go-go (EAG) family of voltage-gated K+ channels (Bauer and Schwarz, 2018)) deleted the circadian rhythm in electrical activity (Hermanstyne et al., 2023). Interestingly, mRNA levels of both Kv12 channels do not show any circadian rhythm, and deletions of Kv12.1, Kv12.2 leaves behavioral circadian rhythms intact (Hermanstyne et al., 2023). Thus, their circadian rhythm is not directly controlled by the molecular circadian TTFL clockwork. Other, so far not identified, mechanisms must drive the daily rhythms in those Kv12-dependent current levels. Members of the EAG family are hubs for several homeostatic processes and are linked to the control of membrane excitability, intracellular pH, Ca2+, and cyclic nucleotide levels (Bauer and Schwarz, 2018). Therefore, it is likely that they are controlled by PTFL-based circadian oscillators/clocks that are employed in mechanisms of homeostatic plasticity (O’Leary et al., 2014). Oscillating spike rates that are based on voltage and Ca2+ (-dependent) oscillations are maintained via mechanisms of homeostatic plasticity that comprise, amongst others, the activity-dependent control of ion channel expression levels (e.g., Colwell, 2011; Turrigiano, 2012; Lee and Kirkwood, 2019; Harvey et al., 2020; Hickey et al., 2020; McCormick et al., 2020; Wu et al., 2021; Kschonsak et al., 2022; Brodt et al., 2023; Fitzpatrick and Kerschensteiner, 2023; Valakh et al., 2023).
In summary, more than half of all ion channels found in circadian clock neurons express daily oscillations in current amplitudes while only few of these daily rhythms are directly controlled at the level of transcription via the molecular circadian TTFL clockwork. Biochemical data concerning timing of ion channel protein turnover are missing. None of the ion channels that are shown to be directly controlled by the TTFL clockwork were proven to be a mandatory prerequisite for the circadian rhythms in spike frequency or behavioral activity. However, circadian changes in the electrical activity stabilizes the output of the molecular clockwork, and circadian output of the molecular clockwork stabilizes the circadian changes in electrical activity (Colwell, 2011; Harvey et al., 2020). Furthermore, published data are neither consistent nor conclusive because the small, abundant SCN clock neurons could not be identified individually and appear to express different combinations of ion channels. It becomes increasingly apparent that a multitude of posttranslational feedback loops (PTFL oscillators) intertwine with the TTFL clockwork which are coupled to manifold receptor-dependent signaling cascades, driven by homeostatic control mechanisms that work at different timescales (Turrigiano, 2012; Li et al., 2014; Lee and Kirkwood, 2019; Hickey et al., 2020; McCormick et al., 2020; Parnell et al., 2021; Wu et al., 2021; Kschonsak et al., 2022; Tang et al., 2022; Yang et al., 2022; Brodt et al., 2023; Fitzpatrick and Kerschensteiner, 2023; Ma et al., 2023; Valakh et al., 2023).
The mechanisms of homeostatic plasticity are intimately connected to PTFL oscillators that constitute localized signaling cascades, termed signalosomes. Signalosomes are multimolecular complexes that anchor complete signal transduction cascades from the receptor to effectors at distinct cellular locations, such as to the plasma membrane, through scaffolding molecules (Dunn and Ferguson, 2015; Zaccolo et al., 2021; Excoffon et al., 2022; Ma et al., 2023; Paolocci and Zaccolo, 2023). They allow for compartmentalization of intracellular (second) messengers in subcellular nanodomains (Zaccolo et al., 2021). Furthermore, signalosomes can constitute oscillators controlling second messenger oscillations via homeostatic mechanisms.
Especially well studied is the cAMP-dependent signal transduction cascade (Blair and Baillie, 2019; Zaccolo et al., 2021; Paolocci and Zaccolo, 2023). Extracellular signal-dependent G-protein coupled receptors activate adenylyl cyclases via Gαs causing rises in cAMP levels, or inhibit adenylyl cyclases via Gαi (Rodbell et al., 1971; Tulsian et al., 2020; Boczek et al., 2021). The changing cAMP concentrations can modulate ion channels (Fesenko et al., 1985; Kar et al., 2021; Saponaro et al., 2021; Lohse et al., 2023), as well as cAMP-dependent protein kinase A (PKA) (Walsh et al., 1968; Corbin and Keely, 1977; Taylor et al., 1993; Taylor et al., 2013; Diskar et al., 2010; Wiggins et al., 2018; Paolocci and Zaccolo, 2023), cAMP exchange protein (EPAC) (de Rooij et al., 1998), or Popeye domain-containing proteins (POPDC) (Brand, 2005). Via different mechanisms PKA actions can remain localized (Smith et al., 2017; Walker-Gray et al., 2017; Chao et al., 2019). For example, PKA anchoring proteins (AKAPs) can keep activated PKA compartmentalized, as opposed to the previous assumption that the catalytic PKA subunit may diffuse freely in the cytoplasm (Colledge and Scott, 1999; Wong and Scott, 2004; Bachmann et al., 2016).
Next to the positive feedforward cascades connecting rising cAMP concentrations to the respective effectors, cAMP levels are restricted via negative feedback mechanisms such as hydrolysis via differently regulated phosphodiesterase that provide further links between different signaling cascades. Different isoforms of phosphodiesterase can be regulated directly or indirectly (e.g., via respective phosphorylations) via cAMP, Ca2+, and cGMP levels confined to specific subcellular locations (Marchmont and Houslay, 1980; Conti et al., 1984; Jurevicius and Fischmeister, 1996; Mongillo et al., 2004; Bender and Beavo, 2006; Lugnier, 2006; Di Benedetto et al., 2008; Weber et al., 2017; Vinogradova and Lakatta, 2021; Paolocci and Zaccolo, 2023). Furthermore, cAMP signaling can be curtailed via phosphatases that antagonize PKA-dependent phosphorylations (Ingebritsen and Cohen, 1983; Francis et al., 2011; Mehta and Zhang, 2021).
In summary, we hypothesize that plasma membrane associated PTFL clocks constitute localized signalosomes maintained via mechanisms of homeostatic plasticity, interlinked with other PTFL and TTFL clocks/oscillators via their signaling cascades as coupling factors.
Our opinion paper focuses on membrane-associated ultradian and circadian rhythms in insect and mammalian clock neurons, offering a new perspective by taking a systemic view on biological timing. We suggest bridges between concepts of chronobiology and homeostatic or Hebbian plasticity in the neurosciences (for details please see previous sections with respective citations). We present a fresh view of the plasma membrane as an endogenous plastic multiscale PTFL clock, tightly coupled to multiscale molecular TTFL clocks. Interlinked posttranslational feedback loops as part of PTFL clocks guarantee a balance of physiological homeostasis and plasticity at all timescales that are relevant for dynamic brain function (Figure 5). The various elements of these PTFL and TTFL oscillators/clocks are suggested to have multiple systemic functions as in- and outputs, as gates and coupling signals, as functional units in mechanisms of homeostatic or Hebbian plasticity.
How dynamic setpoints of homeostasis in brain function are controlled is one of the most important tasks to be resolved in brain research with imminent relevance in medicine and healthcare. Based on our own work as electrophysiologists experienced with individually identifiable invertebrate clock neurons (Stengl et al., 2015; Stengl and Arendt, 2016; Stengl and Schröder, 2021) we suggest a paradigm switch, presenting our novel hypothesis together with an outlook comprising suggestions how to challenge our hypothesis experimentally.
Based on our electrophysiological analysis of insect circadian clock neurons (reviews: Stengl, 2010; Stengl and Arendt, 2016; Stengl and Schröder, 2021) we propose that the excitable plasma membrane of spontaneously active clock neurons contains a system of coupled, endogenous, adaptive PTFL oscillators/clocks, each generating membrane potential oscillations at a characteristic infradian, ultradian, or circadian oscillation frequency (Figure 5). At the core of each neuronal membrane oscillator/clock, associated with signalosomes, specific pacemaker channels drive specific frequencies of membrane potential oscillations. The channels’ activity is tightly coupled to oscillations in concentrations of specific intracellular messengers like Ca2+ or cAMP (Craven and Zagotta, 2006; Biel and Michalakis, 2009; Johnstone et al., 2018; Stengl and Schröder, 2021). The Ca2+ oscillations in turn interlink with endogenous cAMP oscillations, e.g., via Ca2+-dependent adenylyl cyclases, or vice versa via cAMP-dependent Ca2+ permeable ion channels (Biel and Michalakis, 2009; Islam, 2020; Schultz, 2022; Li et al., 2023a). Via different intracellular messenger cascades multiscale membrane-associated oscillations link to multiscale TTFL oscillators/clocks engaged in gene regulation networks (Colwell, 2011; Patton et al., 2016; Stengl and Arendt, 2016; Hastings et al., 2019; Perfitt et al., 2020; Steven et al., 2020; Stengl and Schröder, 2021). The endogenous oscillations in membrane potential and second messenger concentrations are coupled/entrained via various receptors which repetitively phase-shift the respective endogenous oscillations until they remain synchronized with the respective internal oscillator or external zeitgeber.
We suggest that signalosomes associated with the plasma membrane of sensory clock neurons in different parts of the body comprise receptors specialized for the detection of specific environmental rhythms/zeitgebers such as the daily light dark cycle or fast chemical fluctuations (Calebiro and Grimes, 2020; Erofeeva et al., 2023; Takeuchi and Kurahashi, 2023). Signalosome receptors of central clock neurons in the brain link to other physiological oscillators/clocks via neurotransmitters, neurohormones, and neuropeptides as coupling factors (Choi et al., 2012; Wei et al., 2014; Gestrich et al., 2018; Ono et al., 2021). Depending on the signalosome-specific receptors and signal transduction cascades, entrained rhythms of specific intracellular messenger concentrations are generated that oscillate at the same frequency as their respective zeitgeber or coupled oscillator/clock forming an interlinked system of physiological/behavioral timing (Figure 5).
We hypothesize that the negative feedback elements of the membrane associated PTFL oscillators are identical to mechanisms of homeostatic plasticity that act as gain control mechanisms preventing runaway activations. For example, in hawkmoth pheromone-receptor neurons, which are both ultradian and circadian oscillators, Ca2+ activates non-specific cation channels that also permeate Ca2+. The influx of Ca2+ via the cation channel then decreases their open-time probability if intracellular Ca2+-concentrations rise above a certain level (Stengl, 1993; Stengl and Schröder, 2021). Negative feedback mechanisms are suggested to maintain respective dynamic setpoints (i.e., stable oscillation frequency). Alternatively, via different forms of non-associative learning (adaptation, sensitization) or associative forms of learning (Hebbian learning) new dynamic setpoints at different frequencies can be obtained (as described in previous sections).
In contrast to a hierarchical view of TTFL clocks that generate all rhythms via transcriptional control we suggest that specific TTFL- or PTFL-based clocks/oscillators can become dominant or dormant, dependent on the respectively impacting zeitgeber/coupling factor, or respective physiological or behavioral circumstances. Furthermore, we suggest that zeitgeber-dependent oscillations in intracellular concentrations of Ca2+ and cAMP, and the type and relative amounts of activated kinases and phosphatases, play a key role as coupling factors. They couple oscillations at different timescales, couple PTFL- and TTFL clockworks in single clock neurons, and connect various nanodomains via controlled diffusion (Wei et al., 2014). For example, PKC and/or PKA can act as coupling factors between PTFL- and TTFL clockworks by modification of CREB or other transcription factors that are positive feedforward elements of TTFLs (Colwell, 2011; Patton et al., 2016; Stengl and Arendt, 2016; Hastings et al., 2019; Perfitt et al., 2020; Steven et al., 2020; Stengl and Schröder, 2021).
Furthermore, at the network level signalosomes may comprise various types of neuropeptide- and neurotransmitter receptors to mediate coupling between clock neurons. Neuropeptides or neurotransmitters could act as coupling factors or as gates that reprogram a neuronal circuit via phase-dependent synchronization of receptor expressing neurons in the network (Schneider and Stengl, 2005). Thus, neuropeptides cause neuronal ensemble formation and thereby change homeostatic dynamic setpoints in neuronal networks, guiding the brain to a new physiological and behavioral context (Marder, 2012; Stengl et al., 2015; Stengl and Arendt, 2016).
We suggest that a stably coupled, interconnected system of endogenous oscillators and clocks oscillating at defined frequency bands defines dynamic homeostatic setpoints of physiology and behavior. This systems-based concept comprising different time scales and different levels of complexity differs from the currently dominating hierarchical concepts of chronobiology (e.g., Harvey et al., 2020; Rosbash, 2021; Patton and Hastings, 2023) but appears to find increasing support (Hall, 2005; Stengl et al., 2015; Stengl and Arendt, 2016; Rojas et al., 2019; Jaumouillé et al., 2021; Stengl and Schröder, 2021; Yang et al., 2022; Heigwer et al., 2023). Furthermore, contrasting the current hypothesis (Hughes et al., 2009; Zhu et al., 2017; Ballance and Zhu, 2021) we propose an equally important role for plasma membrane associated PTFL clocks as compared to TTFL clocks associated with nuclear gene regulatory networks. The PTFL membrane clocks generate superimposed endogenous multiscale oscillations in membrane potential and intracellular messenger levels connecting to multiscale TTFL oscillations. Like waves on the surface of a lake, endogenous ultradian, circadian, and infradian oscillations of the membrane potential spread from their locally structured signalosomes across the plasma membrane to other compartments of the clock cells. The stable systemic superposition of oscillations at different timescales, originating at diverse signalosomes are suggested to allow equally for stability and plasticity, being hallmarks of the neuron’s dynamic homeostatic setpoints (Figure 5) (Hutcheon and Yarom, 2000; Hughes et al., 2009; Albert, 2011; Cochet-Bissuel et al., 2014; Ananthasubramaniam et al., 2018; Bauer and Schwarz, 2018; Alza et al., 2022; Hille, 2022; Bronson and Kalluri, 2023).
To challenge our hypothesis and to reveal mechanisms of timing that span multiple timescales, it would be necessary to perform long-term physiological experiments with endogenous clock neurons over several days. With improved data analysis and modelling a careful search for periodicities at different timescales, that can be detected as stably oscillating frequency band, needs to be performed, with/without compromising the TTFL clockwork. Furthermore, since clock neurons in the SCN or the insect AME clock are very heterogeneous it is necessary to work with individually identified and characterized endogenous clock neurons over the course of the respectively expressed endogenous ultradian and circadian cycles. Thus, long-term physiological recordings of primary cell cultures in vitro and of intact clock networks in situ need to be accomplished, such as cell-attached patch clamp recordings in search for oscillations in electrical activity at specific interlinked frequency bands, as candidates for “physiological fingerprints”. In search of interlinked intracellular messenger oscillations long-term imaging of Ca2+and/or cAMP levels in individual clock cells needs to be performed together with electrophysiological analysis. These experimentally very challenging experiments will reveal and characterize specific physiological clock types. Single-cell transcriptomics and single cell mass spectrometry at different times of the circadian and ultradian cycles will reveal different types of clock neurons at the transcriptional level and will identify candidates of coupling factors. With refined data analysis and modelling the different types of clock cells need to be matched and their interdependence in the network needs to be calculated. Furthermore, most interesting is to determine which modification of predicted coupling factors affect physiological oscillations at different timescales as predicted hallmarks of dynamic setpoints of homeostasis and plasticity. Finally, combined physiological and behavioral assays need to determine whether changes of the respective oscillation frequencies (“physiological fingerprints”) correlate with/are functionally connected to changes in dynamic homeostatic setpoints. Currently, we are testing our hypothesis in respective long-term assays of peripheral and central insect circadian clock cells in the hawkmoth Manduca sexta and the cockroaches Rhyparobia maderae and Periplaneta americana. We encourage the interested scientists to try to falsify our new hypothesis experimentally in their respective model systems.
The original contributions presented in the study are included in the article, further inquiries can be directed to the corresponding author.
MS: development of the novel hypothesis, conceptualization, writing of the first draft, manuscript editing. ACS: critical discussion of concepts, conceptualization, writing of the first draft, manuscript editing, visualization. All authors contributed to the article and approved the submitted version.
This research was supported by the DFG Research Training Group 2749: Biological Clocks on Multiple Time Scales [DFG GRK 2749/1] and DFG grants STE 531/25-1, STE 531/26-1 to MS.
We thank Dr. Uwe Homberg, University of Marburg, for critical comments on our manuscript. Furthermore, we thank our unknown referees that helped to considerably improve our manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Adams W. B., Benson J. A. (1985). The generation and modulation of endogenous rhythmicity in the Aplysia bursting pacemaker neurone R15. Prog. Biophys. Mol. Biol. 46, 1–49. doi:10.1016/0079-6107(85)90011-2
Adlanmerini M., Lazar M. A. (2023). The REV-ERB nuclear receptors: timekeepers for the core clock period and metabolism. Endocrinology 164, bqad069. doi:10.1210/endocr/bqad069
Akasu T., Shoji S., Hasuo H. (1993). Inward rectifier and low-threshold calcium currents contribute to the spontaneous firing mechanism in neurons of the rat suprachiasmatic nucleus. Pflüg. Arch. 425, 109–116. doi:10.1007/BF00374510
Albert A. P. (2011). Gating mechanisms of canonical transient receptor potential channel proteins: role of phosphoinositols and diacylglycerol. Adv. Exp. Med. Biol. 704, 391–411. doi:10.1007/978-94-007-0265-3_22
Albus H., Bonnefont X., Chaves I., Yasui A., Doczy J., van der Horst G. T. J., et al. (2002). Cryptochrome-deficient mice lack circadian electrical activity in the suprachiasmatic nuclei. Curr. Biol. CB 12, 1130–1133. doi:10.1016/s0960-9822(02)00923-5
Allen C. N., Nitabach M. N., Colwell C. S. (2017). Membrane currents, gene expression, and circadian clocks. Cold Spring Harb. Perspect. Biol. 9, a027714. doi:10.1101/cshperspect.a027714
Alza L., Visa A., Herreros J., Cantí C. (2022). T-type channels in cancer cells: driving in reverse. Cell Calcium 105, 102610. doi:10.1016/j.ceca.2022.102610
Ananthasubramaniam B., Diernfellner A., Brunner M., Herzel H. (2018). Ultradian rhythms in the transcriptome of Neurospora crassa. iScience 9, 475–486. doi:10.1016/j.isci.2018.11.012
Anna G., Kannan N. N. (2021). Post-transcriptional modulators and mediators of the circadian clock. Chronobiol. Int. 38, 1244–1261. doi:10.1080/07420528.2021.1928159
Anton S. E., Kayser C., Maiellaro I., Nemec K., Möller J., Koschinski A., et al. (2022). Receptor-associated independent cAMP nanodomains mediate spatiotemporal specificity of GPCR signaling. Cell 185, 1130–1142. doi:10.1016/j.cell.2022.02.011
Aréchiga H., Fernéndez-Quiróz F., de Miguel F. F., Rodríguez-Sosa L. (1993). The circadian system of Crustaceans. Chronobiol. Int. 10, 1–19. doi:10.3109/07420529309064477
Atkinson S. E., Maywood E. S., Chesham J. E., Wozny C., Colwell C. S., Hastings M. H., et al. (2011). Cyclic AMP signaling control of action potential firing rate and molecular circadian pacemaking in the suprachiasmatic nucleus. J. Biol. Rhythms 26, 210–220. doi:10.1177/0748730411402810
Aton S. J., Colwell C. S., Harmar A. J., Waschek J., Herzog E. D. (2005). Vasoactive intestinal polypeptide mediates circadian rhythmicity and synchrony in mammalian clock neurons. Nat. Neurosci. 8, 476–483. doi:10.1038/nn1419
Aviram R., Dandavate V., Manella G., Golik M., Asher G. (2021). Ultradian rhythms of AKT phosphorylation and gene expression emerge in the absence of the circadian clock components Per1 and Per2. PLOS Biol. 19, e3001492. doi:10.1371/journal.pbio.3001492
Bachmann V. A., Mayrhofer J. E., Ilouz R., Tschaikner P., Raffeiner P., Röck R., et al. (2016). Gpr161 anchoring of PKA consolidates GPCR and cAMP signaling. Proc. Natl. Acad. Sci. 113, 7786–7791. doi:10.1073/pnas.1608061113
Baker T. C., Willis M. A., Haynes K. F., Phelan P. L. (1985). A pulsed cloud of sex pheromone elicits upwind flight in male moths. Physiol. Entomol. 10, 257–265. doi:10.1111/j.1365-3032.1985.tb00045.x
Ballance H., Zhu B. (2021). Revealing the hidden reality of the mammalian 12-h ultradian rhythms. Cell. Mol. Life Sci. CMLS 78, 3127–3140. doi:10.1007/s00018-020-03730-5
Barber A. F., Fong S. Y., Kolesnik A., Fetchko M., Sehgal A. (2021). Drosophila clock cells use multiple mechanisms to transmit time-of-day signals in the brain. Proc. Natl. Acad. Sci. 118, e2019826118. doi:10.1073/pnas.2019826118
Bauer C. K., Schwarz J. R. (2018). Ether-à-go-go K+ channels: effective modulators of neuronal excitability. J. Physiol. 596, 769–783. doi:10.1113/JP275477
Belle M. D. C., Allen C. N. (2018). The circadian clock: a tale of genetic-electrical interplay and synaptic integration. Curr. Opin. Physiol. 5, 75–79. doi:10.1016/j.cophys.2018.08.002
Belle M. D. C., Diekman C. O., Forger D. B., Piggins H. D. (2009). Daily electrical silencing in the mammalian circadian clock. Science 326, 281–284. doi:10.1126/science.1169657
Bell-Pedersen D., Cassone V. M., Earnest D. J., Golden S. S., Hardin P. E., Thomas T. L., et al. (2005). Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nat. Rev. Genet. 6, 544–556. doi:10.1038/nrg1633
Bender A. T., Beavo J. A. (2006). Cyclic nucleotide phosphodiesterases: molecular regulation to clinical use. Pharmacol. Rev. 58, 488–520. doi:10.1124/pr.58.3.5
Bering T., Gadgaard C., Vorum H., Honoré B., Rath M. F. (2023). Diurnal proteome profile of the mouse cerebral cortex: conditional deletion of the Bmal1 circadian clock gene elevates astrocyte protein levels and cell abundance in the neocortex and hippocampus. Glia 71, 2623–2641. doi:10.1002/glia.24443
Biel M., Michalakis S. (2009). “Cyclic nucleotide-gated channels,” in cGMP: generators, Effectors and therapeutic implications handbook of experimental pharmacology. Editors H. H. H. W. Schmidt, F. Hofmann, and J.-P. Stasch (Berlin, Heidelberg: Springer), 111–136. doi:10.1007/978-3-540-68964-5_7
Blair C. M., Baillie G. S. (2019). Reshaping cAMP nanodomains through targeted disruption of compartmentalised phosphodiesterase signalosomes. Biochem. Soc. Trans. 47, 1405–1414. doi:10.1042/BST20190252
Blethyn K. L., Hughes S. W., Tóth T. I., Cope D. W., Crunelli V. (2006). Neuronal basis of the slow (∼1 Hz) oscillation in neurons of the nucleus reticularis thalami in vitro. J. Neurosci. 26, 2474–2486. doi:10.1523/JNEUROSCI.3607-05.2006
Boczek T., Yu Q., Zhu Y., Dodge-Kafka K. L., Goldberg J. L., Kapiloff M. S. (2021). cAMP at perinuclear mAKAPα signalosomes is regulated by local Ca2+ signaling in primary hippocampal neurons. eNeuro 8, ENEURO.0298, 20.2021. doi:10.1523/ENEURO.0298-20.2021
Bose A., Golowasch J., Guan Y., Nadim F. (2014). The role of linear and voltage-dependent ionic currents in the generation of slow wave oscillations. J. Comput. Neurosci. 37, 229–242. doi:10.1007/s10827-014-0498-4
Brancaccio M., Maywood E. S., Chesham J. E., Loudon A. S. I., Hastings M. H. (2013). A Gq-Ca2+ axis controls circuit-level encoding of circadian time in the suprachiasmatic nucleus. Neuron 78, 714–728. doi:10.1016/j.neuron.2013.03.011
Brancaccio M., Patton A. P., Chesham J. E., Maywood E. S., Hastings M. H. (2017). Astrocytes control circadian timekeeping in the suprachiasmatic nucleus via glutamatergic signaling. Neuron 93, 1420–1435. doi:10.1016/j.neuron.2017.02.030
Brand T. (2005). The Popeye domain-containing gene family. Cell biochem. Biophys. 43, 95–103. doi:10.1385/CBB:43:1:095
Brodt S., Inostroza M., Niethard N., Born J. (2023). Sleep—a brain-state serving systems memory consolidation. Neuron 111, 1050–1075. doi:10.1016/j.neuron.2023.03.005
Bronson D., Kalluri R. (2023). Muscarinic acetylcholine receptors modulate HCN channel properties in vestibular ganglion neurons. J. Neurosci. Off. J. Soc. Neurosci. 43, 902–917. doi:10.1523/JNEUROSCI.2552-21.2022
Brown S. A., Kowalska E., Dallmann R. (2012). (Re)inventing the circadian feedback loop. Dev. Cell 22, 477–487. doi:10.1016/j.devcel.2012.02.007
Brown T. M., Piggins H. D. (2007). Electrophysiology of the suprachiasmatic circadian clock. Prog. Neurobiol. 82, 229–255. doi:10.1016/j.pneurobio.2007.05.002
Burkhardt P., Colgren J., Medhus A., Digel L., Naumann B., Soto-Angel J. J., et al. (2023). Syncytial nerve net in a ctenophore adds insights on the evolution of nervous systems. Science 380, 293–297. doi:10.1126/science.ade5645
Buzsáki G., Draguhn A. (2004). Neuronal oscillations in cortical networks. Science 304, 1926–1929. doi:10.1126/science.1099745
Calebiro D., Grimes J. (2020). G protein-coupled receptor pharmacology at the single-molecule level. Annu. Rev. Pharmacol. Toxicol. 60, 73–87. doi:10.1146/annurev-pharmtox-010919-023348
Cao G., Nitabach M. N. (2008). Circadian control of membrane excitability in Drosophila melanogaster lateral ventral clock neurons. J. Neurosci. Off. J. Soc. Neurosci. 28, 6493–6501. doi:10.1523/JNEUROSCI.1503-08.2008
Caporale N., Dan Y. (2008). Spike timing-dependent plasticity: a Hebbian learning rule. Annu. Rev. Neurosci. 31, 25–46. doi:10.1146/annurev.neuro.31.060407.125639
Centeno P. P., Binmahfouz L. S., Alghamdi K., Ward D. T. (2023). Inhibition of the calcium-sensing receptor by extracellular phosphate ions and by intracellular phosphorylation. Front. Physiol. 14, 1154374. doi:10.3389/fphys.2023.1154374
Chao Y.-C., Surdo N. C., Pantano S., Zaccolo M. (2019). Imaging cAMP nanodomains in the heart. Biochem. Soc. Trans. 47, 1383–1392. doi:10.1042/BST20190245
Choi C., Cao G., Tanenhaus A. K., McCarthy E. V., Jung M., Schleyer W., et al. (2012). Autoreceptor control of peptide/neurotransmitter corelease from PDF neurons determines allocation of circadian activity in Drosophila. Cell Rep. 2, 332–344. doi:10.1016/j.celrep.2012.06.021
CircaDB (2013). CIRCA: circadian gene expression profiles. Available at: http://circadb.hogeneschlab.org/ (Accessed May 19, 2023).
Cirelli C., Tononi G. (2022). The why and how of sleep-dependent synaptic down-selection. Semin. Cell Dev. Biol. 125, 91–100. doi:10.1016/j.semcdb.2021.02.007
Cochet-Bissuel M., Lory P., Monteil A. (2014). The sodium leak channel, NALCN, in health and disease. Front. Cell. Neurosci. 8, 132. doi:10.3389/fncel.2014.00132
Cohen A., Ben-Abu Y., Zilberberg N. (2009). Gating the pore of potassium leak channels. Eur. Biophys. J. 39, 61–73. doi:10.1007/s00249-009-0457-6
Colgren J., Burkhardt P. (2022). The premetazoan ancestry of the synaptic toolkit and appearance of first neurons. Essays Biochem. 66, 781–795. doi:10.1042/EBC20220042
Colledge M., Scott J. D. (1999). AKAPs: from structure to function. Trends Cell Biol. 9, 216–221. doi:10.1016/S0962-8924(99)01558-5
Collins B., Blau J. (2006). Keeping time without a clock. Neuron 50, 348–350. doi:10.1016/j.neuron.2006.04.022
Collins B. H., Dissel S., Gaten E., Rosato E., Kyriacou C. P. (2005). Disruption of Cryptochrome partially restores circadian rhythmicity to the arrhythmic period mutant of Drosophila. Proc. Natl. Acad. Sci. 102, 19021–19026. doi:10.1073/pnas.0505392102
Colwell C. S. (2011). Linking neural activity and molecular oscillations in the SCN. Nat. Rev. Neurosci. 12, 553–569. doi:10.1038/nrn3086
Combe C. L., Gasparini S. (2021). Ih from synapses to networks: HCN channel functions and modulation in neurons. Prog. Biophys. Mol. Biol. 166, 119–132. doi:10.1016/j.pbiomolbio.2021.06.002
Conti M., Kasson B. G., Hsueh A. J. W. (1984). Hormonal regulation of 3′,5′-adenosine monophosphate phosphodiesterases in cultured rat granulosa cells. Endocrinology 114, 2361–2368. doi:10.1210/endo-114-6-2361
Corbin J. D., Keely S. L. (1977). Characterization and regulation of heart adenosine 3’:5’-monophosphate-dependent protein kinase isozymes. J. Biol. Chem. 252, 910–918. doi:10.1016/S0021-9258(19)75184-9
Cox K. H., Takahashi J. S. (2019). Circadian clock genes and the transcriptional architecture of the clock mechanism. J. Mol. Endocrinol. 63, R93–R102. doi:10.1530/JME-19-0153
Craven K. B., Zagotta W. N. (2006). CNG and HCN channels: two peas, one pod. Annu. Rev. Physiol. 68, 375–401. doi:10.1146/annurev.physiol.68.040104.134728
Crunelli V., David F., Morais T. P., Lorincz M. L. (2023). HCN channels and absence seizures. Neurobiol. Dis. 181, 106107. doi:10.1016/j.nbd.2023.106107
Curras M. C., Boulant J. A. (1989). Effects of ouabain on neuronal thermosensitivity in hypothalamic tissue slices. Am. J. Physiol. 257, R21–R28. doi:10.1152/ajpregu.1989.257.1.R21
Cymbalyuk G. S., Gaudry Q., Masino M. A., Calabrese R. L. (2002). Bursting in leech heart interneurons: cell-autonomous and network-based mechanisms. J. Neurosci. 22, 10580–10592. doi:10.1523/JNEUROSCI.22-24-10580.2002
Das A., Holmes T. C., Sheeba V. (2016). dTRPA1 in non-circadian neurons modulates temperature-dependent rhythmic activity in Drosophila melanogaster. J. Biol. Rhythms 31, 272–288. doi:10.1177/0748730415627037
Daur N., Nadim F., Bucher D. (2016). The complexity of small circuits: the stomatogastric nervous system. Curr. Opin. Neurobiol. 41, 1–7. doi:10.1016/j.conb.2016.07.005
Debanne D., Inglebert Y. (2023). Spike timing-dependent plasticity and memory. Curr. Opin. Neurobiol. 80, 102707. doi:10.1016/j.conb.2023.102707
DeBruyne J. P., Noton E., Lambert C. M., Maywood E. S., Weaver D. R., Reppert S. M. (2006). A clock shock: mouse CLOCK is not required for circadian oscillator function. Neuron 50, 465–477. doi:10.1016/j.neuron.2006.03.041
Depetris-Chauvin A., Berni J., Aranovich E. J., Muraro N. I., Beckwith E. J., Ceriani M. F. (2011). Adult-specific electrical silencing of pacemaker neurons uncouples molecular clock from circadian outputs. Curr. Biol. 21, 1783–1793. doi:10.1016/j.cub.2011.09.027
de Rooij J., Zwartkruis F. J. T., Verheijen M. H. G., Cool R. H., Nijman S. M. B., Wittinghofer A., et al. (1998). Epac is a Rap1 guanine-nucleotide-exchange factor directly activated by cyclic AMP. Nature 396, 474–477. doi:10.1038/24884
Di Benedetto G., Zoccarato A., Lissandron V., Terrin A., Li X., Houslay M. D., et al. (2008). Protein kinase A type I and type II define distinct intracellular signaling compartments. Circ. Res. 103, 836–844. doi:10.1161/CIRCRESAHA.108.174813
Diskar M., Zenn H.-M., Kaupisch A., Kaufholz M., Brockmeyer S., Sohmen D., et al. (2010). Regulation of cAMP-dependent protein kinases: the human protein kinase X (PrKX) reveals the role of the catalytic subunit alphaH-alphaI loop. J. Biol. Chem. 285, 35910–35918. doi:10.1074/jbc.M110.155150
Dolzer J., Krannich S., Fischer K., Stengl M. (2001). Oscillations of the transepithelial potential of moth olfactory sensilla are influenced by octopamine and serotonin. J. Exp. Biol. 204, 2781–2794. doi:10.1242/jeb.204.16.2781
Dolzer J., Schröder K., Stengl M. (2021). Cyclic nucleotide-dependent ionic currents in olfactory receptor neurons of the hawkmoth Manduca sexta suggest pull–push sensitivity modulation. Eur. J. Neurosci. 54, 4804–4826. doi:10.1111/ejn.15346
Droin C., Paquet E. R., Naef F. (2019). Low-dimensional dynamics of two coupled biological oscillators. Nat. Phys. 15, 1086–1094. doi:10.1038/s41567-019-0598-1
Duchen M. R. (2000). Mitochondria and calcium: from cell signalling to cell death. J. Physiol. 529 (1), 57–68. doi:10.1111/j.1469-7793.2000.00057.x
Dunn H. A., Ferguson S. S. (2015). PDZ protein regulation of G protein-coupled receptor trafficking and signaling pathways. Mol. Pharmacol. 88, 624–639. doi:10.1124/mol.115.098509
Earnest D. J., Digiorgio S. M., Sladek C. D. (1991). Effects of tetrodotoxin on the circadian pacemaker mechanism in suprachiasmatic explants in vitro. Brain Res. Bull. 26, 677–682. doi:10.1016/0361-9230(91)90160-L
Encyclopedia Britannica (2023). Homeostasis | definition, function, examples, & facts | Britannica. Available at: https://www.britannica.com/science/homeostasis (Accessed December 1, 2023).
Erofeeva N., Meshalkina D., Firsov M. (2023). Multiple roles of cAMP in vertebrate retina. Cells 12, 1157. doi:10.3390/cells12081157
Excoffon K. J. D. A., Avila C. L., Alghamri M. S., Kolawole A. O. (2022). The magic of MAGI-1: a scaffolding protein with multi signalosomes and functional plasticity. Biol. Cell 114, 185–198. doi:10.1111/boc.202200014
Farshadi E., van der Horst G. T. J., Chaves I. (2020). Molecular links between the circadian clock and the cell cycle. J. Mol. Biol. 432, 3515–3524. doi:10.1016/j.jmb.2020.04.003
Fenske M. P., Imaizumi T. (2016). Circadian rhythms in floral scent emission. Front. Plant Sci. 7, 462. doi:10.3389/fpls.2016.00462
Fenske M. P., Nguyen L. P., Horn E. K., Riffell J. A., Imaizumi T. (2018). Circadian clocks of both plants and pollinators influence flower seeking behavior of the pollinator hawkmoth Manduca sexta. Sci. Rep. 8, 2842. doi:10.1038/s41598-018-21251-x
Fernandez-Chiappe F., Frenkel L., Colque C. C., Ricciuti A., Hahm B., Cerredo K., et al. (2021). High-frequency neuronal bursting is essential for circadian and sleep behaviors in Drosophila. J. Neurosci. Off. J. Soc. Neurosci. 41, 689–710. doi:10.1523/JNEUROSCI.2322-20.2020
Fernandez-Ruiz A., Sirota A., Lopes-Dos-Santos V., Dupret D. (2023). Over and above frequency: gamma oscillations as units of neural circuit operations. Neuron 111, 936–953. doi:10.1016/j.neuron.2023.02.026
Fesenko E. E., Kolesnikov S. S., Lyubarsky A. L. (1985). Induction by cyclic GMP of cationic conductance in plasma membrane of retinal rod outer segment. Nature 313, 310–313. doi:10.1038/313310a0
Filosa J. A., Putnam R. W. (2003). Multiple targets of chemosensitive signaling in locus coeruleus neurons: role of K+ and Ca2+channels. Am. J. Physiol.-Cell Physiol. 284, C145–C155. doi:10.1152/ajpcell.00346.2002
Fitzpatrick M. J., Kerschensteiner D. (2023). Homeostatic plasticity in the retina. Prog. Retin. Eye Res. 94, 101131. doi:10.1016/j.preteyeres.2022.101131
Flourakis M., Allada R. (2015). Patch-clamp electrophysiology in Drosophila circadian pacemaker neurons. Methods Enzymol. 552, 23–44. doi:10.1016/bs.mie.2014.10.005
Flourakis M., Kula-Eversole E., Hutchison A. L., Han T. H., Aranda K., Moose D. L., et al. (2015). A conserved bicycle model for circadian clock control of membrane excitability. Cell 162, 836–848. doi:10.1016/j.cell.2015.07.036
Fox K., Stryker M. (2017). Integrating Hebbian and homeostatic plasticity: introduction. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 372, 20160413. doi:10.1098/rstb.2016.0413
Francis S. H., Blount M. A., Corbin J. D. (2011). Mammalian cyclic nucleotide phosphodiesterases: molecular mechanisms and physiological functions. Physiol. Rev. 91, 651–690. doi:10.1152/physrev.00030.2010
Franken P., Dijk D.-J., Tobler I., Borbély A. A. (1994). High-frequency components of the rat electrocorticogram are modulated by the vigilance states. Neurosci. Lett. 167, 89–92. doi:10.1016/0304-3940(94)91034-0
Gestrich J., Giese M., Shen W., Zhang Y., Voss A., Popov C., et al. (2018). Sensitivity to pigment-dispersing factor (PDF) is cell-type specific among PDF-expressing circadian clock neurons in the madeira cockroach. J. Biol. Rhythms 33, 35–51. doi:10.1177/0748730417739471
Giesecke A., Johnstone P. S., Lamaze A., Landskron J., Atay E., Chen K.-F., et al. (2023). A novel period mutation implicating nuclear export in temperature compensation of the Drosophila circadian clock. Curr. Biol. CB 33, 336–350.e5. doi:10.1016/j.cub.2022.12.011
Goldstein S. A. N., Bockenhauer D., O’Kelly I., Zilberberg N. (2001). Potassium leak channels and the KCNK family of two-p-domain subunits. Nat. Rev. Neurosci. 2, 175–184. doi:10.1038/35058574
Golowasch J., Bose A., Guan Y., Salloum D., Roeser A., Nadim F. (2017). A balance of outward and linear inward ionic currents is required for generation of slow-wave oscillations. J. Neurophysiol. 118, 1092–1104. doi:10.1152/jn.00240.2017
Golowasch J., Casey M., Abbott L. F., Marder E. (1999). Network stability from activity-dependent regulation of neuronal conductances. Neural Comput. 11, 1079–1096. doi:10.1162/089976699300016359
Green D. J., Gillette R. (1982). Circadian rhythm of firing rate recorded from single cells in the rat suprachiasmatic brain slice. Brain Res. 245, 198–200. doi:10.1016/0006-8993(82)90361-4
Griffiths B. J., Jensen O. (2023). Gamma oscillations and episodic memory. Trends Neurosci. 46, 832–846. doi:10.1016/j.tins.2023.07.003
Groos G., Hendriks J. (1982). Circadian rhythms in electrical discharge of rat suprachiasmatic neurones recorded in vitro. Neurosci. Lett. 34, 283–288. doi:10.1016/0304-3940(82)90189-6
Gutekunst K. (2018). Hypothesis on the synchronistic evolution of autotrophy and heterotrophy. Trends biochem. Sci. 43, 402–411. doi:10.1016/j.tibs.2018.03.008
Häfker N. S., Tessmar-Raible K. (2020). Rhythms of behavior: are the times changin’? Curr. Opin. Neurobiol. 60, 55–66. doi:10.1016/j.conb.2019.10.005
Hahn G., Ponce-Alvarez A., Deco G., Aertsen A., Kumar A. (2019). Portraits of communication in neuronal networks. Nat. Rev. Neurosci. 20, 117–127. doi:10.1038/s41583-018-0094-0
Hall J. C. (2003). Genetics and molecular biology of rhythms in Drosophila and other insects. Adv. Genet. 48, 1–280. doi:10.1016/s0065-2660(03)48000-0
Hall J. C. (2005). “Systems approaches to biological rhythms in Drosophila,” in Methods in enzymology circadian rhythms. Editor M. W Young (Cambridge: Academic Press), 61–185. doi:10.1016/S0076-6879(05)93004-8
Hall J. C., Rosbash M. (1993). Oscillating molecules and how they move circadian clocks across evolutionary boundaries. Proc. Natl. Acad. Sci. U. S. A. 90, 5382–5383. doi:10.1073/pnas.90.12.5382
Haque S. N., Booreddy S. R., Welsh D. K. (2019). Effects of BMAL1 manipulation on the brain’s master circadian clock and behavior. Yale J. Biol. Med. 92, 251–258.
Hardin P. E. (2011). Molecular genetic analysis of circadian timekeeping in Drosophila. Adv. Genet. 74, 141–173. doi:10.1016/B978-0-12-387690-4.00005-2
Harris-Warrick R. M. (2010). General principles of rhythmogenesis in central pattern generator networks. Prog. Brain Res. 187, 213–222. doi:10.1016/B978-0-444-53613-6.00014-9
Harvey J. R. M., Plante A. E., Meredith A. L. (2020). Ion channels controlling circadian rhythms in suprachiasmatic nucleus excitability. Physiol. Rev. 100, 1415–1454. doi:10.1152/physrev.00027.2019
Hastings M. H., Maywood E. S., Brancaccio M. (2019). The mammalian circadian timing system and the suprachiasmatic nucleus as its pacemaker. Biology 8, 13. doi:10.3390/biology8010013
He C., Chen F., Li B., Hu Z. (2014). Neurophysiology of HCN channels: from cellular functions to multiple regulations. Prog. Neurobiol. 112, 1–23. doi:10.1016/j.pneurobio.2013.10.001
Hebb D. O. (1949). The organization of behavior; a neuropsychological theory. Oxford, England: Wiley.
Heigwer F., Scheeder C., Bageritz J., Yousefian S., Rauscher B., Laufer C., et al. (2023). A global genetic interaction network by single-cell imaging and machine learning. Cell Syst. 14, 346–362.e6. doi:10.1016/j.cels.2023.03.003
Helfrich-Förster C. (2018). Sleep in insects. Annu. Rev. Entomol. 63, 69–86. doi:10.1146/annurev-ento-020117-043201
Hermanstyne T. O., Yang N.-D., Granados-Fuentes D., Li X., Mellor R. L., Jegla T., et al. (2023). Kv12-encoded K+ channels drive the day–night switch in the repetitive firing rates of SCN neurons. J. Gen. Physiol. 155, e202213310. doi:10.1085/jgp.202213310
Herzog E. D., Takahashi J. S., Block G. D. (1998). Clock controls circadian period in isolated suprachiasmatic nucleus neurons. Nat. Neurosci. 1, 708–713. doi:10.1038/3708
Hickey P., Merseal H., Patel A. D., Race E. (2020). Memory in time: neural tracking of low-frequency rhythm dynamically modulates memory formation. NeuroImage 213, 116693. doi:10.1016/j.neuroimage.2020.116693
Hille B. (2001). Ion channels of excitable membranes. Third Edition. Oxford, New York: Oxford University Press.
Hille B. (2022). Ionic channels in nerve membranes, 50 years on. Prog. Biophys. Mol. Biol. 169–170, 12–20. doi:10.1016/j.pbiomolbio.2021.11.003
Honma S., Shirakawa T., Katsuno Y., Namihira M., Honma K. (1998). Circadian periods of single suprachiasmatic neurons in rats. Neurosci. Lett. 250, 157–160. doi:10.1016/S0304-3940(98)00464-9
Hughes M. E., DiTacchio L., Hayes K. R., Vollmers C., Pulivarthy S., Baggs J. E., et al. (2009). Harmonics of circadian gene transcription in mammals. PLoS Genet. 5, e1000442. doi:10.1371/journal.pgen.1000442
Hunt T., Sassone-Corsi P. (2007). Riding tandem: circadian clocks and the cell cycle. Cell 129, 461–464. doi:10.1016/j.cell.2007.04.015
Hürkey S., Niemeyer N., Schleimer J.-H., Ryglewski S., Schreiber S., Duch C. (2023). Gap junctions desynchronize a neural circuit to stabilize insect flight. Nature 618, 118–125. doi:10.1038/s41586-023-06099-0
Hutcheon B., Yarom Y. (2000). Resonance, oscillation and the intrinsic frequency preferences of neurons. Trends Neurosci. 23, 216–222. doi:10.1016/S0166-2236(00)01547-2
Ijspeert A. J., Daley M. A. (2023). Integration of feedforward and feedback control in the neuromechanics of vertebrate locomotion: a review of experimental, simulation and robotic studies. J. Exp. Biol. 226, jeb245784. doi:10.1242/jeb.245784
Ikeda M., Sugiyama T., Wallace C. S., Gompf H. S., Yoshioka T., Miyawaki A., et al. (2003a). Circadian dynamics of cytosolic and nuclear Ca2+ in single suprachiasmatic nucleus neurons. Neuron 38, 253–263. doi:10.1016/s0896-6273(03)00164-8
Ikeda M., Yoshioka T., Allen C. N. (2003b). Developmental and circadian changes in Ca2+ mobilization mediated by GABAA and NMDA receptors in the suprachiasmatic nucleus. Eur. J. Neurosci. 17, 58–70. doi:10.1046/j.1460-9568.2003.02427.x
Imber A. N., Putnam R. W. (2012). Postnatal development and activation of L-type Ca2+ currents in locus ceruleus neurons: implications for a role for Ca2+ in central chemosensitivity. J. Appl. Physiol. 112, 1715–1726. doi:10.1152/japplphysiol.01585.2011
Impheng H., Lemmers C., Bouasse M., Legros C., Pakaprot N., Guérineau N. C., et al. (2021). The sodium leak channel NALCN regulates cell excitability of pituitary endocrine cells. FASEB J. 35, e21400. doi:10.1096/fj.202000841RR
Ingebritsen T. S., Cohen P. (1983). Protein phosphatases: properties and role in cellular regulation. Science 221, 331–338. doi:10.1126/science.6306765
Inouye S. T., Kawamura H. (1979). Persistence of circadian rhythmicity in a mammalian hypothalamic “island” containing the suprachiasmatic nucleus. Proc. Natl. Acad. Sci. 76, 5962–5966. doi:10.1073/pnas.76.11.5962
Islam M. S. (2020). Calcium signaling: from basic to bedside. Adv. Exp. Med. Biol. 1131, 1–6. doi:10.1007/978-3-030-12457-1_1
Isomura A., Kageyama R. (2014). Ultradian oscillations and pulses: coordinating cellular responses and cell fate decisions. Dev. Camb. Engl. 141, 3627–3636. doi:10.1242/dev.104497
Itagaki H., Conner W. E. (1988). Calling behavior of Manduca sexta (L.) (Lepidoptera: sphingidae) with notes on the morphology of the female sex pheromone gland. Ann. Entomol. Soc. Am. 81, 798–807. doi:10.1093/aesa/81.5.798
Jabbur M. L., Johnson C. H. (2022). Spectres of clock evolution: past, present, and yet to come. Front. Physiol. 12, 815847. doi:10.3389/fphys.2021.815847
Jackson A. C., Yao G. L., Bean B. P. (2004). Mechanism of spontaneous firing in dorsomedial suprachiasmatic nucleus neurons. J. Neurosci. Off. J. Soc. Neurosci. 24, 7985–7998. doi:10.1523/JNEUROSCI.2146-04.2004
Jagannath A., Taylor L., Ru Y., Wakaf Z., Akpobaro K., Vasudevan S., et al. (2023). The multiple roles of salt-inducible kinases in regulating physiology. Physiol. Rev. 103, 2231–2269. doi:10.1152/physrev.00023.2022
Jaumouillé E., Koch R., Nagoshi E. (2021). Uncovering the roles of clocks and neural transmission in the resilience of Drosophila circadian network. Front. Physiol. 12, 663339. doi:10.3389/fphys.2021.663339
Jékely G., Melzer S., Beets I., Kadow I. C. G., Koene J., Haddad S., et al. (2018). The long and the short of it – a perspective on peptidergic regulation of circuits and behaviour. J. Exp. Biol. 221, jeb166710. doi:10.1242/jeb.166710
Johnson C. H. (2010). Circadian clocks and cell division. Cell Cycle 9, 3864–3873. doi:10.4161/cc.9.19.13205
Johnstone T. B., Agarwal S. R., Harvey R. D., Ostrom R. S. (2018). cAMP signaling compartmentation: adenylyl cyclases as anchors of dynamic signaling complexes. Mol. Pharmacol. 93, 270–276. doi:10.1124/mol.117.110825
Jurevicius J., Fischmeister R. (1996). cAMP compartmentation is responsible for a local activation of cardiac Ca2+ channels by beta-adrenergic agonists. Proc. Natl. Acad. Sci. 93, 295–299. doi:10.1073/pnas.93.1.295
Kageyama R., Isomura A., Shimojo H. (2023). Biological significance of the coupling delay in synchronized oscillations. Physiology 38, 0–72. doi:10.1152/physiol.00023.2022
Kahn R. E., Dayanidhi S., Lacham-Kaplan O., Hawley J. A. (2023). Molecular clocks, satellite cells, and skeletal muscle regeneration. Am. J. Physiol.-Cell Physiol. 324, C1332–C1340. doi:10.1152/ajpcell.00073.2023
Kang X., Wang J., Yan L. (2023). Endoplasmic reticulum in oocytes: spatiotemporal distribution and function. J. Assist. Reprod. Genet. 40, 1255–1263. doi:10.1007/s10815-023-02782-3
Kang Y., Chen L. (2022). Structure and mechanism of NALCN-FAM155A-UNC79-UNC80 channel complex. Nat. Commun. 13, 2639. doi:10.1038/s41467-022-30403-7
Kar P., Barak P., Zerio A., Lin Y.-P., Parekh A. J., Watts V. J., et al. (2021). AKAP79 orchestrates a cyclic AMP signalosome adjacent to Orai1 Ca2+ channels. Function 2, zqab036. doi:10.1093/function/zqab036
King A. N., Sehgal A. (2020). Molecular and circuit mechanisms mediating circadian clock output in the Drosophila brain. Eur. J. Neurosci. 51, 268–281. doi:10.1111/ejn.14092
Kippert F., Hunt P. (2000). Ultradian clocks in eukaryotic microbes: from behavioural observation to functional genomics. BioEssays News Rev. Mol. Cell. Dev. Biol. 22, 16–22. doi:10.1002/(SICI)1521-1878(200001)22:1<16::AID-BIES5>3.0.CO;2-1
Kitayama Y., Nishiwaki T., Terauchi K., Kondo T. (2008). Dual KaiC-based oscillations constitute the circadian system of cyanobacteria. Genes Dev. 22, 1513–1521. doi:10.1101/gad.1661808
Konopka R. J., Benzer S. (1971). Clock mutants of Drosophila melanogaster. Proc. Natl. Acad. Sci. U. S. A. 68, 2112–2116. doi:10.1073/pnas.68.9.2112
Kschonsak M., Chua H. C., Weidling C., Chakouri N., Noland C. L., Schott K., et al. (2022). Structural architecture of the human NALCN channelosome. Nature 603, 180–186. doi:10.1038/s41586-021-04313-5
Lai H. C., Jan L. Y. (2006). The distribution and targeting of neuronal voltage-gated ion channels. Nat. Rev. Neurosci. 7, 548–562. doi:10.1038/nrn1938
Lamothe S. M., Zhang S. (2016). Chapter five - ubiquitination of ion channels and transporters. Prog. Mol. Biol. Transl. Sci. 141, 161–223. doi:10.1016/bs.pmbts.2016.02.005
Laurent G., Fournier J., Hemberger M., Müller C., Naumann R., Ondracek J. M., et al. (2016). “Cortical evolution: introduction to the reptilian cortex,” in Micro-, meso- and macro-Dynamics of the brain research and perspectives in neurosciences. Editors G. Buzsáki,, and Y. Christen (Cham: Springer International Publishing), 23–33. doi:10.1007/978-3-319-28802-4_2
Lear B. C., Darrah E. J., Aldrich B. T., Gebre S., Scott R. L., Nash H. A., et al. (2013). UNC79 and UNC80, putative auxiliary subunits of the NARROW ABDOMEN ion channel, are indispensable for robust circadian locomotor rhythms in Drosophila. PloS One 8, e78147. doi:10.1371/journal.pone.0078147
Lee C.-H., MacKinnon R. (2019). Voltage sensor movements during hyperpolarization in the HCN channel. Cell 179, 1582–1589. doi:10.1016/j.cell.2019.11.006
Lee H.-K., Kirkwood A. (2019). Mechanisms of homeostatic synaptic plasticity in vivo. Front. Cell. Neurosci. 13, 520. doi:10.3389/fncel.2019.00520
Lee S.-Y., Vuong T. A., Wen X., Jeong H.-J., So H.-K., Kwon I., et al. (2019). Methylation determines the extracellular calcium sensitivity of the leak channel NALCN in hippocampal dentate granule cells. Exp. Mol. Med. 51, 1–14. doi:10.1038/s12276-019-0325-0
Leloup J.-C., Gonze D., Goldbeter A. (1999). Limit cycle models for circadian rhythms based on transcriptional regulation in Drosophila and neurospora. J. Biol. Rhythms 14, 433–448. doi:10.1177/074873099129000948
Li B., Jie W., Huang L., Wei P., Li S., Luo Z., et al. (2014). Nuclear BK channels regulate gene expression via the control of nuclear calcium signaling. Nat. Neurosci. 17, 1055–1063. doi:10.1038/nn.3744
Li H. L., Go S., Chang J.-C., Verhoeven A., Elferink R. O. (2023a). Soluble adenylyl cyclase, the cell-autonomous member of the family. Biochim. Biophys. Acta Mol. Basis Dis. 1870, 166936. doi:10.1016/j.bbadis.2023.166936
Li K., Shi Y., Gonye E. C., Bayliss D. A. (2021). TRPM4 contributes to subthreshold membrane potential oscillations in multiple mouse pacemaker neurons. eNeuro 8, ENEURO.0212–21.2021. doi:10.1523/ENEURO.0212-21.2021
Li W., Wang Z., Cao J., Dong Y., Chen Y. (2023b). Perfecting the life clock: the journey from PTO to TTFL. Int. J. Mol. Sci. 24, 2402. doi:10.3390/ijms24032402
Li X., Itani O., Bucher D. M., Rotstein H. G., Nadim F. (2023c). Distinct mechanisms underlie electrical coupling resonance and its interaction with membrane potential resonance. Front. Syst. Biol. 3. doi:10.3389/fsysb.2023.1122433
Li Y. H., Liu X., Vanselow J. T., Zheng H., Schlosser A., Chiu J. C. (2019). O-GlcNAcylation of PERIOD regulates its interaction with CLOCK and timing of circadian transcriptional repression. PLOS Genet. 15, e1007953. doi:10.1371/journal.pgen.1007953
Liang X., Ho M. C. W., Zhang Y., Li Y., Wu M. N., Holy T. E., et al. (2019). Morning and evening circadian pacemakers independently drive premotor centers via a specific dopamine relay. Neuron 102, 843–857. doi:10.1016/j.neuron.2019.03.028
Liu A. C., Welsh D. K., Ko C. H., Tran H. G., Zhang E. E., Priest A. A., et al. (2007). Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell 129, 605–616. doi:10.1016/j.cell.2007.02.047
Liu N., Wang Y., Li T., Feng X. (2021). G-protein coupled receptors (GPCRs): signaling pathways, characterization, and functions in insect physiology and toxicology. Int. J. Mol. Sci. 22, 5260. doi:10.3390/ijms22105260
Liu X., Chiu J. C. (2022). Nutrient-sensitive protein O-GlcNAcylation shapes daily biological rhythms. Open Biol. 12, 220215. doi:10.1098/rsob.220215
Lohse M. J., Bock A., Zaccolo M. (2023). G protein–coupled receptor signaling: new insights define cellular nanodomains. Annu. Rev. Pharmacol. Toxicol. 64. doi:10.1146/annurev-pharmtox-040623-115054
Lowet E., De Weerd P., Roberts M. J., Hadjipapas A. (2022). Tuning neural synchronization: the role of variable oscillation frequencies in neural circuits. Front. Syst. Neurosci. 16, 908665. doi:10.3389/fnsys.2022.908665
Lugnier C. (2006). Cyclic nucleotide phosphodiesterase (PDE) superfamily: a new target for the development of specific therapeutic agents. Pharmacol. Ther. 109, 366–398. doi:10.1016/j.pharmthera.2005.07.003
Luhmann H. J. (2023). Dynamics of neocortical networks: connectivity beyond the canonical microcircuit. Pflüg. Arch. - Eur. J. Physiol. 475, 1027–1033. doi:10.1007/s00424-023-02830-y
Lundkvist G. B., Kwak Y., Davis E. K., Tei H., Block G. D. (2005). A calcium flux is required for circadian rhythm generation in mammalian pacemaker neurons. J. Neurosci. Off. J. Soc. Neurosci. 25, 7682–7686. doi:10.1523/JNEUROSCI.2211-05.2005
Lüthi A., McCormick D. A. (1998). H-current: properties of a neuronal and network pacemaker. Neuron 21, 9–12. doi:10.1016/S0896-6273(00)80509-7
Ma D., Herndon N., Le J. Q., Abruzzi K. C., Zinn K., Rosbash M. (2023). Neural connectivity molecules best identify the heterogeneous clock and dopaminergic cell types in the Drosophila adult brain. Sci. Adv. 9, eade8500. doi:10.1126/sciadv.ade8500
Magee J. C., Grienberger C. (2020). Synaptic plasticity forms and functions. Annu. Rev. Neurosci. 43, 95–117. doi:10.1146/annurev-neuro-090919-022842
Marchmont R. J., Houslay M. D. (1980). Insulin trigger, cyclic AMP-dependent activation and phosphorylation of a plasma membrane cyclic AMP phosphodiesterase. Nature 286, 904–906. doi:10.1038/286904a0
Marder E. (2012). Neuromodulation of neuronal circuits: back to the future. Neuron 76, 1–11. doi:10.1016/j.neuron.2012.09.010
Marder E., Abbott L. F., Turrigiano G. G., Liu Z., Golowasch J. (1996). Memory from the dynamics of intrinsic membrane currents. Proc. Natl. Acad. Sci. U A 93, 13481–13486. doi:10.1073/pnas.93.24.13481
Marder E., Bucher D. (2001). Central pattern generators and the control of rhythmic movements. Curr. Biol. 11, R986–R996. doi:10.1016/S0960-9822(01)00581-4
Marder E., Gutierrez G. J., Nusbaum M. P. (2017). Complicating connectomes: electrical coupling creates parallel pathways and degenerate circuit mechanisms. Dev. Neurobiol. 77, 597–609. doi:10.1002/dneu.22410
Masquelier T., Hugues E., Deco G., Thorpe S. J. (2009). Oscillations, phase-of-firing coding, and spike timing-dependent plasticity: an efficient learning scheme. J. Neurosci. 29, 13484–13493. doi:10.1523/JNEUROSCI.2207-09.2009
Matsuo M., Seo K., Taruno A., Mizoro Y., Yamaguchi Y., Doi M., et al. (2022). A light-induced small G-protein gem limits the circadian clock phase-shift magnitude by inhibiting voltage-dependent calcium channels. Cell Rep. 39, 110844. doi:10.1016/j.celrep.2022.110844
McCormick D. A., Nestvogel D. B., He B. J. (2020). Neuromodulation of brain state and behavior. Annu. Rev. Neurosci. 43, 391–415. doi:10.1146/annurev-neuro-100219-105424
McFarlan A. R., Chou C. Y. C., Watanabe A., Cherepacha N., Haddad M., Owens H., et al. (2023). The plasticitome of cortical interneurons. Nat. Rev. Neurosci. 24, 80–97. doi:10.1038/s41583-022-00663-9
Mehta S., Zhang J. (2021). Biochemical activity architectures visualized–using genetically encoded fluorescent biosensors to map the spatial boundaries of signaling compartments. Acc. Chem. Res. 54, 2409–2420. doi:10.1021/acs.accounts.1c00056
Meijer J. H., Schwartz W. J. (2003). In search of the pathways for light-induced pacemaker resetting in the suprachiasmatic nucleus. J. Biol. Rhythms 18, 235–249. doi:10.1177/0748730403018003006
Mendoza-Viveros L., Bouchard-Cannon P., Hegazi S., Cheng A. H., Pastore S., Cheng H.-Y. M. (2017). Molecular modulators of the circadian clock: lessons from flies and mice. Cell. Mol. Life Sci. 74, 1035–1059. doi:10.1007/s00018-016-2378-8
Michel S., Geusz M. E., Zaritsky J. J., Block G. D. (1993). Circadian rhythm in membrane conductance expressed in isolated neurons. Science 259, 239–241. doi:10.1126/science.8421785
Michel S., Meijer J. H. (2020). From clock to functional pacemaker. Eur. J. Neurosci. 51, 482–493. doi:10.1111/ejn.14388
Miller J. P., Selverston A. I. (1982). Mechanisms underlying pattern generation in lobster stomatogastric ganglion as determined by selective inactivation of identified neurons. II. Oscillatory properties of pyloric neurons. J. Neurophysiol. 48, 1378–1391. doi:10.1152/jn.1982.48.6.1378
Mizrak D., Ruben M., Myers G. N., Rhrissorrakrai K., Gunsalus K. C., Blau J. (2012). Electrical activity can impose time of day on the circadian transcriptome of pacemaker neurons. Curr. Biol. CB 22, 1871–1880. doi:10.1016/j.cub.2012.07.070
Mongillo M., McSorley T., Evellin S., Sood A., Lissandron V., Terrin A., et al. (2004). Fluorescence resonance energy transfer–based analysis of cAMP dynamics in live neonatal rat cardiac myocytes reveals distinct functions of compartmentalized phosphodiesterases. Circ. Res. 95, 67–75. doi:10.1161/01.RES.0000134629.84732.11
Moore R. Y. (1983). Organization and function of a central nervous system circadian oscillator: the suprachiasmatic hypothalamic nucleus. Fed. Proc. 42, 2783–2789.
Mulet J. M., Porcel R., Yenush L. (2023). Modulation of potassium transport to increase abiotic stress tolerance in plants. J. Exp. Bot. 74, 5989–6005. doi:10.1093/jxb/erad333
Musheshe N., Schmidt M., Zaccolo M. (2018). cAMP: from long-range second messenger to nanodomain signalling. Trends Pharmacol. Sci. 39, 209–222. doi:10.1016/j.tips.2017.11.006
Nakamura N. H., Oku Y., Fukunaga M. (2023). “Brain–breath” interactions: respiration-timing–dependent impact on functional brain networks and beyond. Rev. Neurosci., 1–16. doi:10.60124/j.pneuro.2023.10.01
Narasimamurthy R., Virshup D. M. (2017). Molecular mechanisms regulating temperature compensation of the circadian clock. Front. Neurol. 8, 161. doi:10.3389/fneur.2017.00161
Nieto-Felipe J., Macias-Diaz A., Sanchez-Collado J., Berna-Erro A., Jardin I., Salido G. M., et al. (2023). Role of Orai-family channels in the activation and regulation of transcriptional activity. J. Cell. Physiol. 238, 714–726. doi:10.1002/jcp.30971
Nikitin M. A., Romanova D. Y., Borman S. I., Moroz L. L. (2023). Amino acids integrate behaviors in nerveless placozoans. Front. Neurosci. 17, 1125624. doi:10.3389/fnins.2023.1125624
Nitabach M. N., Blau J., Holmes T. C. (2002). Electrical silencing of Drosophila pacemaker neurons stops the free-running circadian clock. Cell 109, 485–495. doi:10.1016/s0092-8674(02)00737-7
Njus D., Sulzman F. M., Hastings J. W. (1974). Membrane model for the circadian clock. Nature 248, 116–120. doi:10.1038/248116a0
Norekian T. P., Moroz L. L. (2023). Recording cilia activity in ctenophores: effects of nitric oxide and low molecular weight transmitters. Front. Neurosci. 17, 1125476. doi:10.3389/fnins.2023.1125476
Notomi T., Shigemoto R. (2004). Immunohistochemical localization of Ih channel subunits, HCN1–4, in the rat brain. J. Comp. Neurol. 471, 241–276. doi:10.1002/cne.11039
O’Leary T., Marder E. (2016). Temperature-robust neural function from activity-dependent ion channel regulation. Curr. Biol. 26, 2935–2941. doi:10.1016/j.cub.2016.08.061
O’Leary T., Williams A. H., Franci A., Marder E. (2014). Cell types, network homeostasis, and pathological compensation from a biologically plausible ion channel expression model. Neuron 82, 809–821. doi:10.1016/j.neuron.2014.04.002
O’Neill J. S., Reddy A. B. (2011). Circadian clocks in human red blood cells. Nature 469, 498–503. doi:10.1038/nature09702
Ono D., Honma K., Honma S. (2021). GABAergic mechanisms in the suprachiasmatic nucleus that influence circadian rhythm. J. Neurochem. 157, 31–41. doi:10.1111/jnc.15012
Panda S. (2016). Circadian physiology of metabolism. Science 354, 1008–1015. doi:10.1126/science.aah4967
Paolocci E., Zaccolo M. (2023). Compartmentalised cAMP signalling in the primary cilium. Front. Physiol. 14, 1187134. doi:10.3389/fphys.2023.1187134
Park I., Kim D., Kim J., Jang S., Choi M., Choe H. K., et al. (2020). microRNA-25 as a novel modulator of circadian Period2 gene oscillation. Exp. Mol. Med. 52, 1614–1626. doi:10.1038/s12276-020-00496-5
Park K. S., Choi S. H., Yoon H. (2023). Modulation of sleep using noninvasive stimulations during sleep. Biomed. Eng. Lett. 13, 329–341. doi:10.1007/s13534-023-00298-4
Parnell A. A., De Nobrega A. K., Lyons L. C. (2021). Translating around the clock: multi-level regulation of post-transcriptional processes by the circadian clock. Cell. Signal. 80, 109904. doi:10.1016/j.cellsig.2020.109904
Parviainen T., Lyyra P., Nokia M. S. (2022). Cardiorespiratory rhythms, brain oscillatory activity and cognition: review of evidence and proposal for significance. Neurosci. Biobehav. Rev. 142, 104908. doi:10.1016/j.neubiorev.2022.104908
Patel A. J., Honoré E. (2001). Properties and modulation of mammalian 2P domain K+ channels. Trends Neurosci. 24, 339–346. doi:10.1016/S0166-2236(00)01810-5
Patel M., Rangan A. (2021). Olfactory encoding within the insect antennal lobe: the emergence and role of higher order temporal correlations in the dynamics of antennal lobe spiking activity. J. Theor. Biol. 522, 110700. doi:10.1016/j.jtbi.2021.110700
Patton A. P., Chesham J. E., Hastings M. H. (2016). Combined pharmacological and genetic manipulations unlock unprecedented temporal elasticity and reveal phase-specific modulation of the molecular circadian clock of the mouse suprachiasmatic nucleus. J. Neurosci. Off. J. Soc. Neurosci. 36, 9326–9341. doi:10.1523/JNEUROSCI.0958-16.2016
Patton A. P., Hastings M. H. (2023). The mammalian circadian time-keeping system. J. Huntingt. Dis. 12, 91–104. doi:10.3233/JHD-230571
Pennartz C. M. A., de Jeu M. T. G., Bos N. P. A., Schaap J., Geurtsen A. M. S. (2002). Diurnal modulation of pacemaker potentials and calcium current in the mammalian circadian clock. Nature 416, 286–290. doi:10.1038/nature728
Perfitt T. L., Wang X., Dickerson M. T., Stephenson J. R., Nakagawa T., Jacobson D. A., et al. (2020). Neuronal L-type calcium channel signaling to the nucleus requires a novel CaMKIIα-shank3 interaction. J. Neurosci. Off. J. Soc. Neurosci. 40, 2000–2014. doi:10.1523/JNEUROSCI.0893-19.2020
Pittendrigh C. S. (1993). Temporal organization: reflections of a darwinian clock-watcher. Annu. Rev. Physiol. 55, 16–54. doi:10.1146/annurev.ph.55.030193.000313
Pitts G. R., Ohta H., McMahon D. G. (2006). Daily rhythmicity of large-conductance Ca2+ -activated K+ currents in suprachiasmatic nucleus neurons. Brain Res. 1071, 54–62. doi:10.1016/j.brainres.2005.11.078
Posner C., Mehta S., Zhang J. (2023). Fluorescent biosensor imaging meets deterministic mathematical modelling: quantitative investigation of signalling compartmentalization. J. Physiol. 601, 4227–4241. doi:10.1113/JP282696
Puri B. K. (2020). “Calcium signaling and gene expression,” in Calcium signaling advances in experimental medicine and biology. Editor Md. S. Islam (Cham: Springer International Publishing), 537–545. doi:10.1007/978-3-030-12457-1_22
Ratliff J., Franci A., Marder E., O’Leary T. (2021). Neuronal oscillator robustness to multiple global perturbations. Biophys. J. 120, 1454–1468. doi:10.1016/j.bpj.2021.01.038
Ray S., Valekunja U. K., Stangherlin A., Howell S. A., Snijders A. P., Damodaran G., et al. (2020). Circadian rhythms in the absence of the clock gene Bmal1. Science 367, 800–806. doi:10.1126/science.aaw7365
Reischig T., Stengl M. (2003a). Ectopic transplantation of the accessory medulla restores circadian locomotor rhythms in arrhythmic cockroaches (Leucophaea maderae). J. Exp. Biol. 206, 1877–1886. doi:10.1242/jeb.00373
Reischig T., Stengl M. (2003b). Ultrastructure of pigment-dispersing hormone-immunoreactive neurons in a three-dimensional model of the accessory medulla of the cockroach Leucophaea maderae. Cell Tissue Res. 314, 421–435. doi:10.1007/s00441-003-0772-7
Rensing L., Ruoff P. (2002). Temperature effect on entrainment, phase shifting, and amplitude of circadian clocks and its molecular bases. Chronobiol. Int. 19, 807–864. doi:10.1081/CBI-120014569
Rizzuto R., Duchen M. R., Pozzan T. (2004). Flirting in little space: the ER/mitochondria Ca2+ liaison. Sci. STKE 2004, re1. doi:10.1126/stke.2152004re1
Robinson R. B., Siegelbaum S. A. (2003). Hyperpolarization-activated cation currents: from molecules to physiological function. Annu. Rev. Physiol. 65, 453–480. doi:10.1146/annurev.physiol.65.092101.142734
Rodbell M., Birnbaumer L., Pohl S. L., Krans H. M. J. (1971). The glucagon-sensitive adenyl cyclase system in plasma membranes of rat liver: V. An obligatory role of guanyl nucleotides in glucagon action. J. Biol. Chem. 246, 1877–1882. doi:10.1016/S0021-9258(18)62390-7
Rodríguez-Sosa L., Calderón-Rosete G., Flores G. (2008). Circadian and ultradian rhythms in the crayfish caudal photoreceptor. Synap. N. Y. N. 62, 643–652. doi:10.1002/syn.20540
Roemschied F. A., Eberhard M. J., Schleimer J.-H., Ronacher B., Schreiber S. (2014). Cell-intrinsic mechanisms of temperature compensation in a grasshopper sensory receptor neuron. eLife 3, e02078. doi:10.7554/eLife.02078
Rojas P., Plath J. A., Gestrich J., Ananthasubramaniam B., Garcia M. E., Herzel H., et al. (2019). Beyond spikes: multiscale computational analysis of in vivo long-term recordings in the cockroach circadian clock. Netw. Neurosci. 3, 944–968. doi:10.1162/netn_a_00106
Rosbash M. (2021). Circadian rhythms and the transcriptional feedback loop (nobel lecture). Angew. Chem. Int. Ed. 60, 8650–8666. doi:10.1002/anie.202015199
Sanchez-Padilla J., Guzman J. N., Ilijic E., Kondapalli J., Galtieri D. J., Yang B., et al. (2014). Mitochondrial oxidant stress in locus coeruleus is regulated by activity and nitric oxide synthase. Nat. Neurosci. 17, 832–840. doi:10.1038/nn.3717
Saponaro A., Thiel G., Moroni A. (2021). Structural and functional approaches to studying cAMP regulation of HCN channels. Biochem. Soc. Trans. 49, 2573–2579. doi:10.1042/BST20210290
Schaap J., Albus H., vanderLeest H. T., Eilers P. H. C., Détári L., Meijer J. H. (2003). Heterogeneity of rhythmic suprachiasmatic nucleus neurons: implications for circadian waveform and photoperiodic encoding. Proc. Natl. Acad. Sci. 100, 15994–15999. doi:10.1073/pnas.2436298100
Schellinger J. N., Sun Q., Pleinis J. M., An S.-W., Hu J., Mercenne G., et al. (2022). Chloride oscillation in pacemaker neurons regulates circadian rhythms through a chloride-sensing WNK kinase signaling cascade. Curr. Biol. 32, 1429–1438.e6. doi:10.1016/j.cub.2022.03.017
Schendzielorz J., Schendzielorz T., Arendt A., Stengl M. (2014). Bimodal oscillations of cyclic nucleotide concentrations in the circadian system of the madeira cockroach Rhyparobia maderae. J. Biol. Rhythms 29, 318–331. doi:10.1177/0748730414546133
Schendzielorz T., Peters W., Boekhoff I., Stengl M. (2012). Time of day changes in cyclic nucleotides are modified via octopamine and pheromone in antennae of the madeira cockroach. J. Biol. Rhythms 27, 388–397. doi:10.1177/0748730412456265
Schendzielorz T., Schirmer K., Stolte P., Stengl M. (2015). Octopamine regulates antennal sensory neurons via daytime-dependent changes in cAMP and IP3 levels in the hawkmoth Manduca sexta. PLOS ONE 10, e0121230. doi:10.1371/journal.pone.0121230
Schlichting M., Díaz M. M., Xin J., Rosbash M. (2019). Neuron-specific knockouts indicate the importance of network communication to Drosophila rhythmicity. eLife 8, e48301. doi:10.7554/eLife.48301
Schneider A. C., Fox D., Itani O., Golowasch J., Bucher D., Nadim F. (2021). Frequency-dependent action of neuromodulation. eNeuro 8, ENEURO.0338, 21.2021. doi:10.1523/ENEURO.0338-21.2021
Schneider N. L., Stengl M. (2005). Pigment-Dispersing factor and GABA synchronize cells of the isolated circadian clock of the cockroach Leucophaea maderae. J. Neurosci. 25, 5138–5147. doi:10.1523/JNEUROSCI.5138-A-04.2005
Schneider N. L., Stengl M. (2006). Gap junctions between accessory medulla neurons appear to synchronize circadian clock cells of the cockroach Leucophaea maderae. J. Neurophysiol. 95, 1996–2002. doi:10.1152/jn.00835.2005
Schneider N. L., Stengl M. (2007). Extracellular long-term recordings of the isolated accessory medulla, the circadian pacemaker center of the cockroach Leucophaea maderae, reveal ultradian and hint circadian rhythms. J. Comp. Physiol. A 193, 35–42. doi:10.1007/s00359-006-0169-7
Schultz J. E. (2022). The evolutionary conservation of eukaryotic membrane-bound adenylyl cyclase isoforms. Front. Pharmacol. 13, 1009797. doi:10.3389/fphar.2022.1009797
Schulz D. J., Lane B. J. (2017). Homeostatic plasticity of excitability in crustacean central pattern generator networks. Curr. Opin. Neurobiol. 43, 7–14. doi:10.1016/j.conb.2016.09.015
Schwartz W. J., Gross R. A., Morton M. T. (1987). The suprachiasmatic nuclei contain a tetrodotoxin-resistant circadian pacemaker. Proc. Natl. Acad. Sci. 84, 1694–1698. doi:10.1073/pnas.84.6.1694
Sharma A., Rahman G., Gorelik J., Bhargava A. (2023). Voltage-gated T-type calcium channel modulation by kinases and phosphatases: the old ones, the new ones, and the missing ones. Cells 12, 461. doi:10.3390/cells12030461
Shirakawa T., Honma S., Katsuno Y., Oguchi H., Honma K. I. (2000). Synchronization of circadian firing rhythms in cultured rat suprachiasmatic neurons. Eur. J. Neurosci. 12, 2833–2838. doi:10.1046/j.1460-9568.2000.00170.x
Singer W., Lazar A. (2016). Does the cerebral cortex exploit high-dimensional, non-linear dynamics for information processing? Front. Comput. Neurosci. 10, 99. doi:10.3389/fncom.2016.00099
Smith F. D., Esseltine J. L., Nygren P. J., Veesler D., Byrne D. P., Vonderach M., et al. (2017). Local protein kinase A action proceeds through intact holoenzymes. Science 356, 1288–1293. doi:10.1126/science.aaj1669
Smith P., Buhl E., Tsaneva-Atanasova K., Hodge J. J. L. (2019). Shaw and Shal voltage-gated potassium channels mediate circadian changes in Drosophila clock neuron excitability. J. Physiol. 597, 5707–5722. doi:10.1113/JP278826
Spehr J., Hagendorf S., Weiss J., Spehr M., Leinders-Zufall T., Zufall F. (2009). Ca2+–Calmodulin feedback mediates sensory adaptation and inhibits pheromone-sensitive ion channels in the vomeronasal organ. J. Neurosci. 29, 2125–2135. doi:10.1523/JNEUROSCI.5416-08.2009
Städele C., Heigele S., Stein W. (2015). Neuromodulation to the rescue: compensation of temperature-induced breakdown of rhythmic motor patterns via extrinsic neuromodulatory input. PLOS Biol. 13, e1002265. doi:10.1371/journal.pbio.1002265
Stengl M. (1993). Intracellular-messenger-Mediated cation channels in cultured olfactory receptor neurons. J. Exp. Biol. 178, 125–147. doi:10.1242/jeb.178.1.125
Stengl M. (2010). Pheromone transduction in moths. Front. Cell. Neurosci. 4, 133. doi:10.3389/fncel.2010.00133
Stengl M., Arendt A. (2016). Peptidergic circadian clock circuits in the Madeira cockroach. Curr. Opin. Neurobiol. 41, 44–52. doi:10.1016/j.conb.2016.07.010
Stengl M., Schröder K. (2021). “14 - multiscale timing of pheromone transduction in hawkmoth olfactory receptor neurons,” in Insect pheromone biochemistry and molecular biology. Editors G. J Blomquist,, and R. G Vogt. Second Edition (London: Academic Press), 435–468. doi:10.1016/B978-0-12-819628-1.00014-6
Stengl M., Werckenthin A., Wei H. (2015). How does the circadian clock tick in the Madeira cockroach? Curr. Opin. Insect Sci. 12, 38–45. doi:10.1016/j.cois.2015.09.007
Stetson M. H., Watson-Whitmyre M. (1976). Nucleus suprachiasmaticus: the biological clock in the hamster? Science 191, 197–199. doi:10.1126/science.942799
Steven A., Friedrich M., Jank P., Heimer N., Budczies J., Denkert C., et al. (2020). What turns CREB on? And off? And why does it matter? Cell. Mol. Life Sci. 77, 4049–4067. doi:10.1007/s00018-020-03525-8
Stöber T. M., Batulin D., Triesch J., Narayanan R., Jedlicka P. (2023). Degeneracy in epilepsy: multiple routes to hyperexcitable brain circuits and their repair. Commun. Biol. 6, 479. doi:10.1038/s42003-023-04823-0
Sundararaj S., Ravindran A., Casarotto M. G. (2021). AHNAK: the quiet giant in calcium homeostasis. Cell Calcium 96, 102403. doi:10.1016/j.ceca.2021.102403
Takahashi J. S. (2016). “Molecular architecture of the circadian clock in mammals,” in A Time for Metabolism and hormones research and perspectives in endocrine interactions. Editors P. Sassone-Corsi,, and Y. Christen (Cham: Springer International Publishing), 13–24. doi:10.1007/978-3-319-27069-2_2
Takahashi J. S., Menaker M. (1979). Physiology of avian circadian pacemakers. Fed. Proc. 38, 2583–2588.
Takeuchi H., Kurahashi T. (2023). Segregation of Ca2+ signaling in olfactory signal transduction. J. Gen. Physiol. 155, e202213165. doi:10.1085/jgp.202213165
Talley E. M., Solórzano G., Lei Q., Kim D., Bayliss D. A. (2001). CNS distribution of members of the two-pore-domain (KCNK) potassium channel family. J. Neurosci. 21, 7491–7505. doi:10.1523/JNEUROSCI.21-19-07491.2001
Tang M., Cao L.-H., Yang T., Ma S.-X., Jing B.-Y., Xiao N., et al. (2022). An extra-clock ultradian brain oscillator sustains circadian timekeeping. Sci. Adv. 8, eabo5506. doi:10.1126/sciadv.abo5506
Taylor S. S., Radzio-Andzelm E., Knighton D. R., Ten Eyck L. F., Sowadski J. M., Herberg F. W., et al. (1993). Crystal structures of the catalytic subunit of cAMP-dependent protein kinase reveal general features of the protein kinase family. Receptor 3, 165–172.
Taylor S. S., Zhang P., Steichen J. M., Keshwani M. M., Kornev A. P. (2013). PKA: lessons learned after twenty years. Biochim. Biophys. Acta BBA - Proteins Proteomics 1834, 1271–1278. doi:10.1016/j.bbapap.2013.03.007
Tenner B., Getz M., Ross B., Ohadi D., Bohrer C. H., Greenwald E., et al. (2020). Spatially compartmentalized phase regulation of a Ca2+-cAMP-PKA oscillatory circuit. eLife 9, e55013. doi:10.7554/eLife.55013
Tokuda I. T., Ono D., Ananthasubramaniam B., Honma S., Honma K. I., Herzel H. (2015). Coupling controls the synchrony of clock cells in development and knockouts. Biophys. J. 109, 2159–2170. doi:10.1016/j.bpj.2015.09.024
Tokuda I. T., Schmal C., Ananthasubramaniam B., Herzel H. (2020). Conceptual models of entrainment, jet lag, and seasonality. Front. Physiol. 11, 334. doi:10.3389/fphys.2020.00334
Tokumitsu H., Sakagami H. (2022). Molecular mechanisms underlying Ca2+/calmodulin-dependent protein kinase kinase signal transduction. Int. J. Mol. Sci. 23, 11025. doi:10.3390/ijms231911025
Tomita J., Nakajima M., Kondo T., Iwasaki H. (2005). No transcription-translation feedback in circadian rhythm of KaiC phosphorylation. Science 307, 251–254. doi:10.1126/science.1102540
Tononi G., Cirelli C. (2020). Sleep and synaptic down-selection. Eur. J. Neurosci. 51, 413–421. doi:10.1111/ejn.14335
Trujillo C. A., Gao R., Negraes P. D., Gu J., Buchanan J., Preissl S., et al. (2019). Complex oscillatory waves emerging from cortical organoids model early human brain network development. Cell Stem Cell 25, 558–569. doi:10.1016/j.stem.2019.08.002
Tulsian N. K., Ghode A., Anand G. S. (2020). Adenylate control in cAMP signaling: implications for adaptation in signalosomes. Biochem. J. 477, 2981–2998. doi:10.1042/BCJ20200435
Turrigiano G. (2012). Homeostatic synaptic plasticity: local and global mechanisms for stabilizing neuronal function. Cold Spring Harb. Perspect. Biol. 4, a005736. doi:10.1101/cshperspect.a005736
Turrigiano G., Abbott L. F., Marder E. (1994). Activity-dependent changes in the intrinsic properties of cultured neurons. Science 264, 974–977. doi:10.1126/science.8178157
Turrigiano G., LeMasson G., Marder E. (1995). Selective regulation of current densities underlies spontaneous changes in the activity of cultured neurons. J. Neurosci. 15, 3640–3652. doi:10.1523/JNEUROSCI.15-05-03640.1995
Turrigiano G. G. (1999). Homeostatic plasticity in neuronal networks: the more things change, the more they stay the same. Trends Neurosci. 22, 221–227. doi:10.1016/s0166-2236(98)01341-1
Turrigiano G. G. (2008). The self-tuning neuron: synaptic scaling of excitatory synapses. Cell 135, 422–435. doi:10.1016/j.cell.2008.10.008
Turrigiano G. G., Nelson S. B. (2000). Hebb and homeostasis in neuronal plasticity. Curr. Opin. Neurobiol. 10, 358–364. doi:10.1016/s0959-4388(00)00091-x
Tyson J. J., Chen K. C., Novak B. (2003). Sniffers, buzzers, toggles and blinkers: dynamics of regulatory and signaling pathways in the cell. Curr. Opin. Cell Biol. 15, 221–231. doi:10.1016/S0955-0674(03)00017-6
Valakh V., Wise D., Zhu X. A., Sha M., Fok J., Van Hooser S. D., et al. (2023). A transcriptional constraint mechanism limits the homeostatic response to activity deprivation in mammalian neocortex. eLife 12, e74899. doi:10.7554/eLife.74899
Veedin Rajan V. B., Häfker N. S., Arboleda E., Poehn B., Gossenreiter T., Gerrard E., et al. (2021). Seasonal variation in UVA light drives hormonal and behavioural changes in a marine annelid via a ciliary opsin. Nat. Ecol. Evol. 5, 204–218. doi:10.1038/s41559-020-01356-1
Vinogradova T. M., Lakatta E. G. (2021). Dual activation of phosphodiesterase 3 and 4 regulates basal cardiac pacemaker function and beyond. Int. J. Mol. Sci. 22, 8414. doi:10.3390/ijms22168414
Walker-Gray R., Stengel F., Gold M. G. (2017). Mechanisms for restraining cAMP-dependent protein kinase revealed by subunit quantitation and cross-linking approaches. Proc. Natl. Acad. Sci. 114, 10414–10419. doi:10.1073/pnas.1701782114
Walsh D. A., Perkins J. P., Krebs E. G. (1968). An adenosine 3′,5′-Monophosphate-dependant protein kinase from rabbit skeletal muscle. J. Biol. Chem. 243, 3763–3765. doi:10.1016/S0021-9258(19)34204-8
Weber S., Zeller M., Guan K., Wunder F., Wagner M., El-Armouche A. (2017). PDE2 at the crossway between cAMP and cGMP signalling in the heart. Cell. Signal. 38, 76–84. doi:10.1016/j.cellsig.2017.06.020
Wegner S., Belle M. D. C., Hughes A. T. L., Diekman C. O., Piggins H. D. (2017). Delayed cryptochrome degradation asymmetrically alters the daily rhythm in suprachiasmatic clock neuron excitability. J. Neurosci. Off. J. Soc. Neurosci. 37, 7824–7836. doi:10.1523/JNEUROSCI.0691-17.2017
Wei H., Stengl M. (2012). Ca2+-dependent ion channels underlying spontaneous activity in insect circadian pacemaker neurons. Eur. J. Neurosci. 36, 3021–3029. doi:10.1111/j.1460-9568.2012.08227.x
Wei H., Yasar H., Funk N. W., Giese M., Baz E.-S., Stengl M. (2014). Signaling of pigment-dispersing factor (PDF) in the madeira cockroach Rhyparobia maderae. PLOS ONE 9, e108757. doi:10.1371/journal.pone.0108757
Wells G. D., Tang Q.-Y., Heler R., Tompkins-MacDonald G. J., Pritchard E. N., Leys S. P., et al. (2012). A unique alkaline pH-regulated and fatty acid-activated tandem pore domain potassium channel (K2P) from a marine sponge. J. Exp. Biol. 215, 2435–2444. doi:10.1242/jeb.066233
Welsh D. K., Logothetis D. E., Meister M., Reppert S. M. (1995). Individual neurons dissociated from rat suprachiasmatic nucleus express independently phased circadian firing rhythms. Neuron 14, 697–706. doi:10.1016/0896-6273(95)90214-7
Wiggins S. V., Steegborn C., Levin L. R., Buck J. (2018). Pharmacological modulation of the CO2/HCO3−/pH-calcium-and ATP-sensing soluble adenylyl cyclase. Pharmacol. Ther. 190, 173–186. doi:10.1016/j.pharmthera.2018.05.008
Wollmuth E. M., Angert E. R. (2023). Microbial circadian clocks: host-microbe interplay in diel cycles. BMC Microbiol. 23, 124. doi:10.1186/s12866-023-02839-4
Wong W., Scott J. D. (2004). AKAP signalling complexes: focal points in space and time. Nat. Rev. Mol. Cell Biol. 5, 959–970. doi:10.1038/nrm1527
Wu C.-H., Ramos R., Katz D. B., Turrigiano G. G. (2021). Homeostatic synaptic scaling establishes the specificity of an associative memory. Curr. Biol. CB 31, 2274–2285.e5. doi:10.1016/j.cub.2021.03.024
Xiong L. I., Garfinkel A. (2023). Are physiological oscillations physiological? J. Physiol. doi:10.1113/JP285015
Yamaguchi S., Isejima H., Matsuo T., Okura R., Yagita K., Kobayashi M., et al. (2003). Synchronization of cellular clocks in the suprachiasmatic nucleus. Science 302, 1408–1412. doi:10.1126/science.1089287
Yamakou M. E., Desroches M., Rodrigues S. (2023). Synchronization in STDP-driven memristive neural networks with time-varying topology. J. Biol. Phys. 49, 483–507. doi:10.1007/s10867-023-09642-2
Yang N.-D., Mellor R. L., Hermanstyne T. O., Nerbonne J. M. (2023). Effects of NALCN-encoded Na+ leak currents on the repetitive firing properties of SCN neurons depend on K+-Driven rhythmic changes in input resistance. J. Neurosci. 43, 5132–5141. doi:10.1523/JNEUROSCI.0182-23.2023
Yang S., Yamazaki S., Cox K. H., Huang Y.-L., Miller E. W., Takahashi J. S. (2022). Coupling-dependent metabolic ultradian rhythms in confluent cells. Proc. Natl. Acad. Sci. 119, e2211142119. doi:10.1073/pnas.2211142119
Yee A. X., Hsu Y.-T., Chen L. (2017). A metaplasticity view of the interaction between homeostatic and Hebbian plasticity. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 372, 20160155. doi:10.1098/rstb.2016.0155
Yoshii T., Hermann-Luibl C., Kistenpfennig C., Schmid B., Tomioka K., Helfrich-Förster C. (2015). Cryptochrome-dependent and -independent circadian entrainment circuits in Drosophila. J. Neurosci. 35, 6131–6141. doi:10.1523/JNEUROSCI.0070-15.2015
Zaccolo M., Zerio A., Lobo M. J. (2021). Subcellular organization of the cAMP signaling pathway. Pharmacol. Rev. 73, 278–309. doi:10.1124/pharmrev.120.000086
Zheng Y., Pan L., Wang F., Yan J., Wang T., Xia Y., et al. (2023). Neural function of Bmal1: an overview. Cell Biosci. 13, 1. doi:10.1186/s13578-022-00947-8
Zhu B., Zhang Q., Pan Y., Mace E. M., York B., Antoulas A. C., et al. (2017). A cell-autonomous mammalian 12 hr clock coordinates metabolic and stress rhythms. Cell Metab. 25, 1305–1319. doi:10.1016/j.cmet.2017.05.004
Keywords: endogenous clocks, coupled oscillators, homeostasis, plasticity, circadian rhythms, ultradian rhythms, TTFL, PTFL
Citation: Stengl M and Schneider AC (2024) Contribution of membrane-associated oscillators to biological timing at different timescales. Front. Physiol. 14:1243455. doi: 10.3389/fphys.2023.1243455
Received: 20 June 2023; Accepted: 12 December 2023;
Published: 09 January 2024.
Edited by:
Daan R. van der Veen, University of Surrey, United KingdomReviewed by:
Jerry Yin, University of Wisconsin-Madison, United StatesCopyright © 2024 Stengl and Schneider. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Monika Stengl, c3RlbmdsQHVuaS1rYXNzZWwuZGU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.