
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Physiol., 27 June 2023
Sec. Environmental, Aviation and Space Physiology
Volume 14 - 2023 | https://doi.org/10.3389/fphys.2023.1215535
This article is part of the Research TopicMulti-omics Approaches for Understanding Space Flight ImpactView all 5 articles
 Soren Impey1,2*
Soren Impey1,2* Carl Pelz1
Carl Pelz1 Lara-Kirstie Riparip3
Lara-Kirstie Riparip3 Amanuel Tafessu1
Amanuel Tafessu1 Fatema Fareh1
Fatema Fareh1 Damian G. Zuloaga4
Damian G. Zuloaga4 Tessa Marzulla4
Tessa Marzulla4 Blair Stewart4
Blair Stewart4 Susanna Rosi3
Susanna Rosi3 Mitchell S. Turker5
Mitchell S. Turker5 Jacob Raber4,6*
Jacob Raber4,6*Introduction: The response of the brain to space radiation is an important concern for astronauts during space missions. Therefore, we assessed the response of the brain to 28Si ion irradiation (600 MeV/n), a heavy ion present in the space environment, on cognitive performance and whether the response is associated with altered DNA methylation in the hippocampus, a brain area important for cognitive performance.
Methods: We determined the effects of 28Si ion irradiation on object recognition, 6-month-old mice irradiated with 28Si ions (600 MeV/n, 0.3, 0.6, and 0.9 Gy) and cognitively tested two weeks later. In addition, we determined if those effects were associated with alterations in hippocampal networks and/or hippocampal DNA methylation.
Results: At 0.3 Gy, but not at 0.6 Gy or 0.9 Gy, 28Si ion irradiation impaired cognition that correlated with altered gene expression and 5 hmC profiles that mapped to specific gene ontology pathways. Comparing hippocampal DNA hydroxymethylation following proton, 56Fe ion, and 28Si ion irradiation revealed a general space radiation synaptic signature with 45 genes that are associated with profound phenotypes. The most significant categories were glutamatergic synapse and postsynaptic density.
Discussion: The brain’s response to space irradiation involves novel excitatory synapse and postsynaptic remodeling.
A unique feature of the space radiation environment is the presence of galactic cosmic rays (GCR) and solar particle events (SPE) (Kronenberg and Cucinotta, 2012). The former involves protons and fully ionized atomic nuclei such as 28Si, while the latter includes predominantly low to medium energy protons with a small heavy ion component. Space irradiation exposures pose a significant health hazard to space flight crews during missions (Ferlazzo and Foray, 2017) but uncertainty exists with respect to the level of risk to develop cognitive changes following irradiation doses relevant to those encountered during space missions and there are individual difference of risk as well to consider (Foray et al., 2016; Restier-Verlet et al., 2021). In the central nervous system (CNS), radiation exposure affects the hippocampus (for a review, see Kiffer et al., 2019), a structure critical for memory function. We reported that object recognition memory (Ennaceur, 2010), which uses a 24-h interval between learning and memory assessment to test hippocampal function (Raber, 2015), is impaired 2 weeks following irradiation of 6-month-old mice with 56Fe ions (600 MeV/n, 0.1 Gy) (Impey et al., 2016a) and protons (150 MeV, 1 Gy) and associated with alterations in hippocampal DNA methylation (Impey et al., 2016b).
The mechanisms mediating the effects of simulated space irradiation on hippocampus-dependent cognitive function might be associated with changes in hippocampal networks. We earlier reported 56Fe ion- and proton-irradiation related changes in immediate early gene Activity-Regulated Cytoskeleton-Associated Protein (Arc) in the hippocampus (Penner et al., 2010; Impey et al., 2016a; Impey et al., 2016b).
The mechanisms mediating the effects of simulated space irradiation on hippocampus-dependent cognitive function might be associated with alteration in hippocampal DNA methylation as well. Changes in cytosine methylation involving the addition of a methyl group to cytosine (5 mC and especially those involving addition of a hydroxy group to 5 mC (hydroxymethylcytosine or 5 hmC) (Veron and Peters, 2011) play a key role in regulating expression of genes required for learning and memory (Miller and Sweatt, 2007; Lubin et al., 2008). Hippocampal DNA methylation is affected following proton (Impey et al., 2016b) and 56Fe irradiation (Impey et al., 2016a).
In the current study, to determine the effects of 28Si ion irradiation on object recognition, 6-month-old mice were irradiated with 28Si ions (600 MeV/n, 0.3, 0.6, an 0.9 Gy) and cognitively tested 2 weeks later. In addition, we determined if those effects were associated with alterations in hippocampal networks and/or hippocampal DNA methylation.
Six-month-old C57BL/6J male mice (n = 84 mice in total) were obtained from Jackson Laboratories, Bar Harbor Maine. The biological age of the mice was selected to be relevant to the biological age of astronauts during space missions. The mice were shipped from Jackson Laboratories to Brookhaven National Laboratory (BNL), Upton, New York, and allowed to accommodate to the housing facility there for 1 week. Subsequently, the mice were irradiated with 0.3, 0.6, or 0.9 Gy of 600 MeV 28Si ion or sham-irradiated (n = 42 mice/radiation dose). The duration of the radiation exposures was 0.99 ± 0.08 min, 1.53 ± 0.08 min, and 2.10 ± 0.07 min, for the 0.3, 0.6, and 0.9 Gy dose respectively. For irradiation, mice were individually loaded into 8 × 3 × 3 cm plastic square enclosures with air holes and placed in a foam fixture in the beam line of the NASA Space Radiation Laboratory (NSRL). They were exposed to a rectangular beam of approximately 20 × 20 cm. The focused beam of high-energy was generated by the Booster accelerator at BNL and transferred to the experimental beam line at the NSRL facility. Dose calibration was performed so that the desired dose could be delivered. Sham-irradiated mice were placed into the plastic enclosures for the same time as the irradiated mice. Mice were randomly assigned to the experimental groups. The week after the irradiation or sham-irradiation, the mice were shipped to Oregon Health & Science University (OHSU) and cognitive testing started 2 weeks following irradiation. Mice were tested in the open field and for novel object recognition in week 1. In week 3, mice were tested for hippocampal network stability and euthanized immediately after by cervical dislocation. The hippocampus of one hemibrain was dissected for DNA methylation and RNAseq analyses. The other hemibrain was frozen in −70°C isopentane (2-methyl butane, Sigma) and processed for Arc mRNA and TET2 immunohistochemical analyses. The hemibrains were blocked together such that each slide contained sections from different experimental groups. All slides were cryosectioned and stored at −70°C until processed for immunocytochemistry or fluorescence in situ hybridization (FISH). All protocols were reviewed and approved by the Institutional Animal Care and Use Committees (IACUC) of OHSU and BNL and were in compliance with all Federal regulations.
The novel object recognition test was performed as described (Raber et al., 2002). The mice were habituated to an open field (16 × 16 inches, Kinder Scientific, Poway, CA) for 3 times for 10 min each over three subsequent days. On day 4, the mice were placed in the open field containing two identical objects and they were allowed to freely explore for 15 min. On day 5, the mice were placed again in the open field, but one familiar object was replaced with a novel object. The mice were allowed to explore for 15 min. Movement and time spent exploring each object was recorded using Ethovision XT video tracking system (Noldus Information Technology, Sterling, VA) and hand scored by a researcher blinded to the treatment of the mice. The percent time exploring the novel object, out of the total time exploring the novel and familiar objects on day 5, was used to assess novel object recognition. For each group, the preference for the novel versus the familiar object was assessed. The open field arena and objects were cleaned with 5% acetic acid between mice and trials.
Exploration of identical or different environments was used to study the stability of hippocampal networks (Guzowski et al., 1999), using a method called catFISH (cellular compartment analysis of temporal activity using fluorescence in situ hybridization) that relies on the precise temporal kinetics of the IEG and neuronal gene Arc, which is involved in synaptic plasticity and memory, as described previously (Impey et al., 2017). When neurons are engaged in information processing, Arc is rapidly transcribed and can be visualized and quantified after ∼5 min. Subsequently, the mRNA is translocated to the cytoplasm where it remains detectable for ∼20–30 min after the initial transcription. The mRNA is translocated to tagged synapses for protein synthesis. Two different cellular compartments (nuclear and cytoplasmic) can be clearly distinguished, allowing identification of neurons active during distinct behavioral experiences (Guzowski et al., 1999).
Eight mice from each experimental radiation condition were placed individually into a novel environment (A; a square open field; 61 × 61 cm box with 20-cm high walls) and allowed to explore for 5 minutes. Mice were returned to their cage for 25 min, returned to the same environment for an additional 5 min (AA Paradigm). Another 8 mice per radiation dose were allowed to explore environment A for 5 min, and 25 min later they were placed in a different environment (B; a circular arena 45 cm in diameter, and allowed to explore for 5 min; AB Paradigm). Following the last environmental exposure, the mice were killed by cervical dislocation and the brains quickly removed, as described above.
Four to six slides from each treatment group, each containing sections from all the mice in that group, were prepared for fluorescence in situ hybridization. Arc mRNA were detected as previously reported in detail (Rosi et al., 2009; Ramirez-Amaya et al., 2013). Briefly, hapten-labeled antisense riboprobes were hybridized together with the tissues overnight. The digoxigenin-labeled Arc full-probe riboprobe was detected with anti–digoxigenin-HRP conjugate (Roche) and revealed with a cyanine-3 (CY3) substrate kit. Nuclei were counterstained with sytox-green (Molecular Probes).
Microscopic imaging for Arc mRNA, was performed using a Zeiss AXIO IMAGER Z1 microscope with motorized Z-drive for transmitted light and epifluorescence (Rosi et al., 2009). For each end point, the four to six coronal sections per mouse were used to image CA1 and CA3.
Manual cell counts of cells expressing Arc mRNA were performed by an experimenter blind to the relationship between the experimental conditions they represented. Cytoplasmic Arc mRNA–positive neurons were identified when the staining constituted at least 60% of the cell body (Rosi et al., 2009) and was detectable throughout three planes across the Z-stack. To avoid classification errors, we carefully verified that the staining belonged to the cell of interest by checking the nuclear counterstaining (Rosi et al., 2009). The neuronal nuclei were classified as follows: negative (no staining), Arc mRNA foci positive (containing only Arc intranuclear foci staining detected with the Arc intron probe), Arc mRNA cytoplasmic positive (containing only cytoplasmic staining detected with the full-length probe) (Arc-cyto), or as double-labeled for Arc pre-mRNA foci and Arc mRNA cytoplasmic (containing both intranuclear Arc pre-mRNA foci and cytoplasmic Arc mRNA transcripts in two different colors). When a count included all classifications we refer to it as an Arc-positive neuron (this includes the total number of the three above mentioned classifications). The percentages of Arc-foci, Arc-cyto, or Arc-double were calculated relative to the total number of pyramidal neurons included in the analysis as previously reported (Rosi et al., 2009).
Hemibrains were processed for Tet2 immunoreactivity using a specific primary antibody from Santa Cruz Biotechnology (Tet2 S-13, 1:250, catalog number sc-136926) and donkey anti-rabbit Alexa 488 (1:200) as secondary antibody and n = 3 sections per hemibrain, approximately 200 µm apart, per mouse, as described (Impey et al., 2017). Background threshold levels were set and applied to all images. Pixel intensities above this threshold were used for quantification measures (area occupied by pixels and intensities of pixels). The total intensity was also quantified as a measure of overall pixel intensity within a specific brain region.
DNA was isolated from the hippocampus. Antibodies against 5 mC and 5 hmC were used to immunoprecipitate sonicated DNA preparations for methyl-DNA immunoprecipitation (meDIP--anti-5mC mouse mAb, EMD-Millipore NA8; catalog number 162 33 D3) and hydroxymethyl-DNA immunoprecipitation (hmeDIP; anti-5hmC rabbit polyclonal; catalog number 39769; 2 ul, Active Motif)), respectively, and Dynal anti-mouse IgG beads from pools of tissues. Beads were rinsed 7 times with IP buffer, eluted with 1% SDS at room temperature and the eluted DNA purified and subjected to limited amplification (∼18 cycles). Libraries were sequenced on the HiSeq2000 platform at the OHSU Massively Parallel Sequencing Shared Resource or the Oregon State University Center for Genome ReseArch. DIP-Seq regions methylated above “background” were identified using a sliding window method and enriched regions selected via a Monte Carlo-permutation test (Fejes et al., 2008).
To facilitate direct comparison of DIP-Seq data with gene expression data, RNA-Seq was used to profile transcription from the same animals used for the DIP-Seq experiments. RNA was isolated using the NEBnext poly A selection kit (New England Biolabs). Illumina high-throughput sequencing technology was used to profile RNA levels in an unbiased manner. For Illumina RNA-Seq library preparation, NEBnext Ultra kit was used according to the manufacturers specifications (New England Biolabs). Libraries were sequenced on the HiSeq2000 platform at the OHSU Massively Parallel Sequencing Shared Resource. Illumina data were mapped to the UC Santa Cruz assembly using Bowtie (Langmead et al., 2009). For RNA-Seq analyses, tags that overlap with known RefSeq gene models (UCSC RefSeq annotation) were counted using R scripts (Gentleman et al., 2004). Significance was assessed using the DESeq2 package (Love et al., 2014). The Storey Q-test was used to adjust for multiple comparisons (Storey and Tibshirani, 2003).
The cognitive and Arc data are shown as mean ± SEM. The statistical analyses of the data were performed using SPSS™ (Chicago, IL) and GraphPad Prism™ (San Diego, CA) software packages. To analyze locomotor activity over 3 days, ability to locate a visible platform over 2 days, and ability to locate a hidden platform over 3 days, repeated measures ANOVA was used. To compare exploration of the objects and the percentage of Arc-positive cells and total number of Arc cells following exposure to the two different environmental conditions, 2-tailed t-tests were used. The cognitive and Arc figures were generated using GraphPad Prism software. We considered p < 0.05 as statistically significant.
Single read sequence data was mapped to the mouse reference genome (UCSC mm9) using the Bowtie algorithm using standard flags and allowing 2 mismatches (Langmead et al., 2009). Sequences that map to a single location were selected and domains enriched for 5 mC or 5 hmC were selected using a parameter-optimized Monte-Carlo-based segmentation algorithm (Fejes et al., 2008). A 1000 bp sliding-window was used based on iterative analyses that maximized the number of enriched regions. A comparison of different high-throughput sequencing based methods to study DNA methylation concluded that MeDIP-Seq covers ∼67% of genomic CpGs (Harris et al., 2010).
For statistical comparisons of biological samples, regions of methylation enrichment were merged and differences in methylation interrogated with FDR-adjusted negative binomial statistics (Anders and Huber, 2010). Statistical and visualization studies involved the R programming language and Bioconductor packages (Gentleman et al., 2004). Gene ontology analyses utilized the Bioconductor Goseq package, which adjusts for sequence-length bias artifacts in genomic data (Young et al., 2010). KEGG analyses were conducted using the DAVID-EASE site (Dennis et al., 2003). For gene ontology analyses the top DMRs (differentially methylation regions) or DHRs (differentially hydroxymethylated regions) (p < 0.01) within a 50 kb window centered on the RefSeq transcriptional start site or within the RefSeq gene body were non-redundantly annotated. Unless otherwise stated, overlap between DMRs and RNA-Seq data was analyzed using a similar windowing approach. For statistical comparisons of gene-annotated DHRs with DHRs or RNA-Seq gene data we used the Fisher Exact test (“stats” R core library) or for multi-set comparisons (>2 sets) the SuperExactTest R package (Wang et al., 2015). For Fisher Exact tests FDR-adjusted p < 1 × 10−3 was considered significant.
DIP sequence-tag heatmaps were generated in R by plotting median-normalized DIP-Seq tag density in gene bodies and indicated flanking regions with color-maps scaled to the 80% quantile. Statistical analyses of pathway data were conducted using FDR-adjusted Fisher exact test. Statistical analyses of DHR density in genomic regions were conducted using a Monte-Carlo-based permutation statistic (“coin” R package). Unless otherwise stated, FDR-adjusted p < 0.01 was considered statistically significant.
The mice were first habituated to an open field for three subsequent days. 28Si ion irradiation did not affect activity levels in the open field over these 3 days (Figure 1A). On day 4, the mice were exposed to the open field containing two objects for 15 min. On day 5, the mice were exposed to the open field with two objects for 15 min, but one familiar object (present on day 4) was replaced with a novel one. There was no effects of radiation on the time the mice spent exploring the objects (Figure 1B). The percent time exploring the novel object was used to assess novel object recognition. Following 28Si (600 MeV) ion irradiation, mice irradiated with 0.3 Gy were impaired, as revealed by no discrimination between the familiar and novel object, while those irradiated with higher doses were not (Figure 1C).
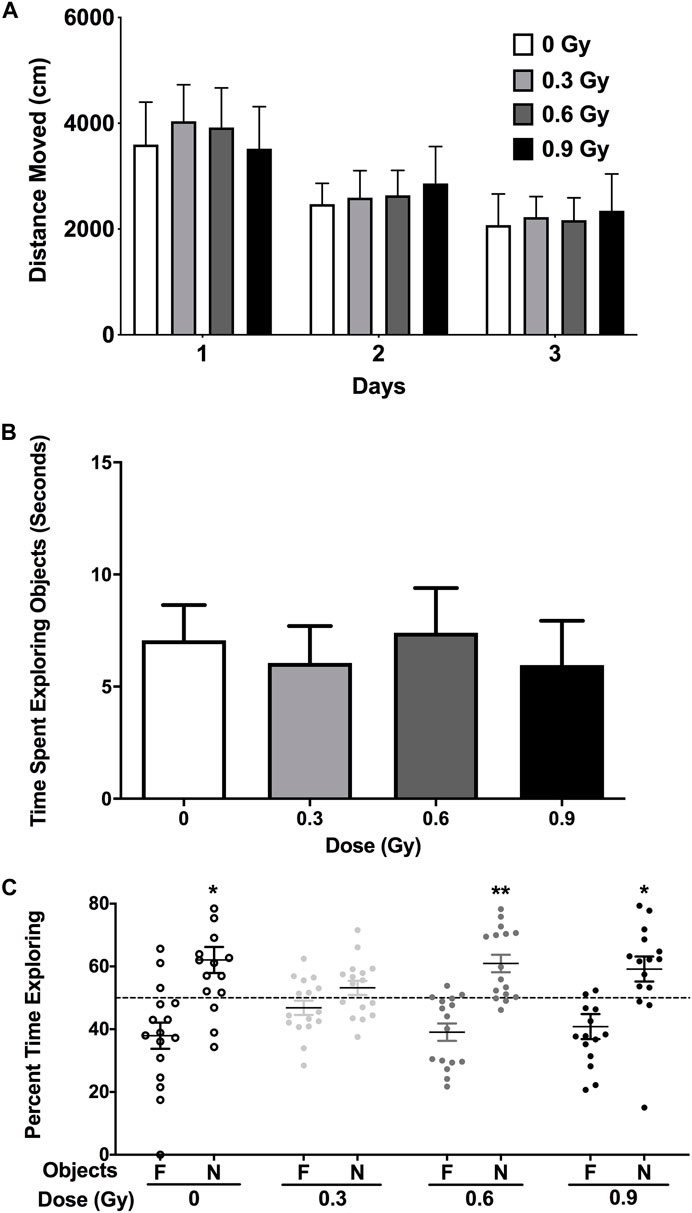
FIGURE 1. (A) 28Si ion irradiation did not affect activity levels in the open field on three subsequent day. (B) Object recognition. There was no effects of radiation on the time the mice spent exploring the objects. (C) Sham-irradiated mice and mice irradiated with a dose of 0.6 or 0.9 Gy showed object recognition and spent significantly more time exploring the novel than the familiar object. In contrast, mice irradiation with a dose of 0.3 Gy did not. N = 16 mice/dose. *p < 0.05, **p < 0.01 versus familiar object. N: novel object; F: familiar object.
We next assessed whether the mechanisms mediating the cognitive effects of 28Si ion irradiation might be associated with reduced ability of hippocampal neurons to recognize similar environments and to discriminate distinct environments using the temporal kinetics of mRNA encoding Arc. Exploratory behavior activated a comparable total number of neurons in sham-irradiated and irradiated mice in the CA1 (Figures 2, 4A) and CA3 (Figures 3, 4B) regions of the hippocampus. However, there was an effect of 28Si ion irradiation on the percent of neurons activated by both experiences. The percentage of Arc-positive neurons expressing Arc mRNA in the nucleus and cytoplasm in the CA3 region of the hippocampus of sham-irradiated mice was significantly higher following exposure twice to the same environment, as opposed to exposure to two different environments (t = 3.191, p = 0.0110, Figure 4D) and there was a trend towards a difference in the CA1 region of the hippocampus of sham-irradiated mice (t = 2.108, p = 0.0567, Figure 4C). This was not seen in mice irradiated with 0.3 Gy that showed impaired cognitive performance (CA1: t = 1.286, p = 0.2273, Figure 3C; CA3: t = 1.051, p = 0.3280; Figure 2D). In mice irradiated with 0.9 Gy, who showed no impaired cognitive performance (Figure 1B), there was a trend towards a difference in the CA3 region (t = 2.237, p = 0.00557, Figure 4D), but not the CA1 region (t = 0.5842, p = 0.5752, Figure 4C).
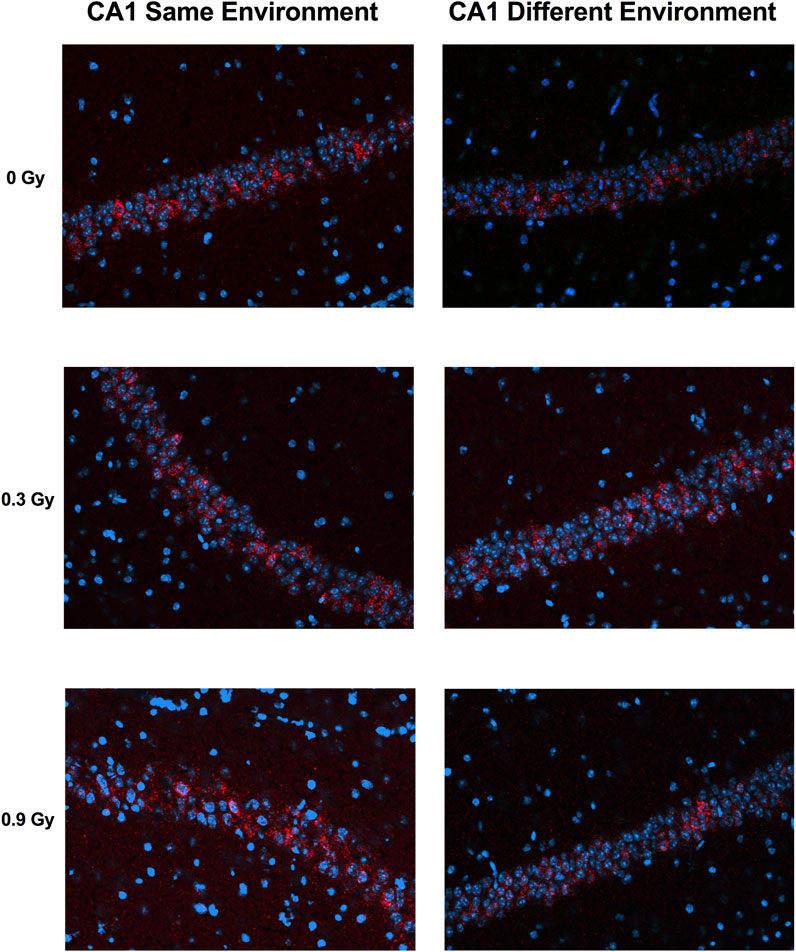
FIGURE 2. Representative images for Arc catFISH data in the CA1 region of the hippocampus. Representative fluorescence images showing Arc mRNA expression following exploration of the same or different environments (images taken at 20 Å∼ 1 z stack). Scale bar: 100 μm. The Arc mRNA is illustrated in red and cell nuclei are indicated in blue (DAPI).
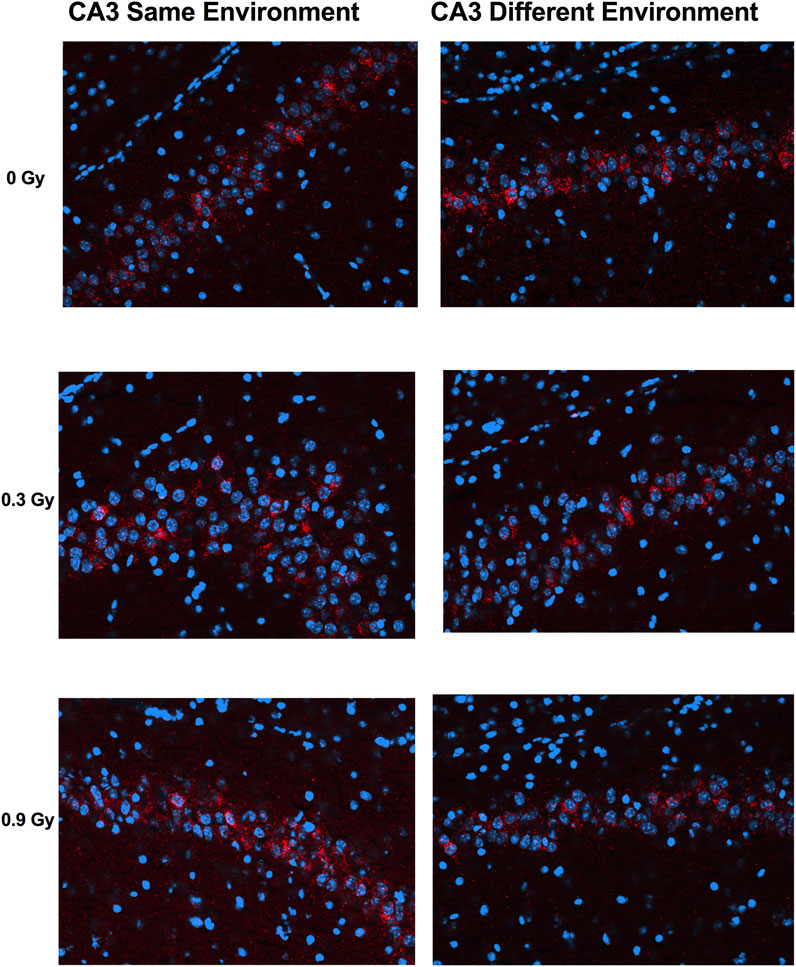
FIGURE 3. Representative images for Arc catFISH data in the CA3 region of the hippocampus. Representative fluorescence images showing Arc mRNA expression following exploration of the same or different environments (images taken at 20 Å∼ 1 z stack). Scale bar: 100 μm. The Arc mRNA is illustrated in red and cell nuclei are indicated in blue (DAPI).
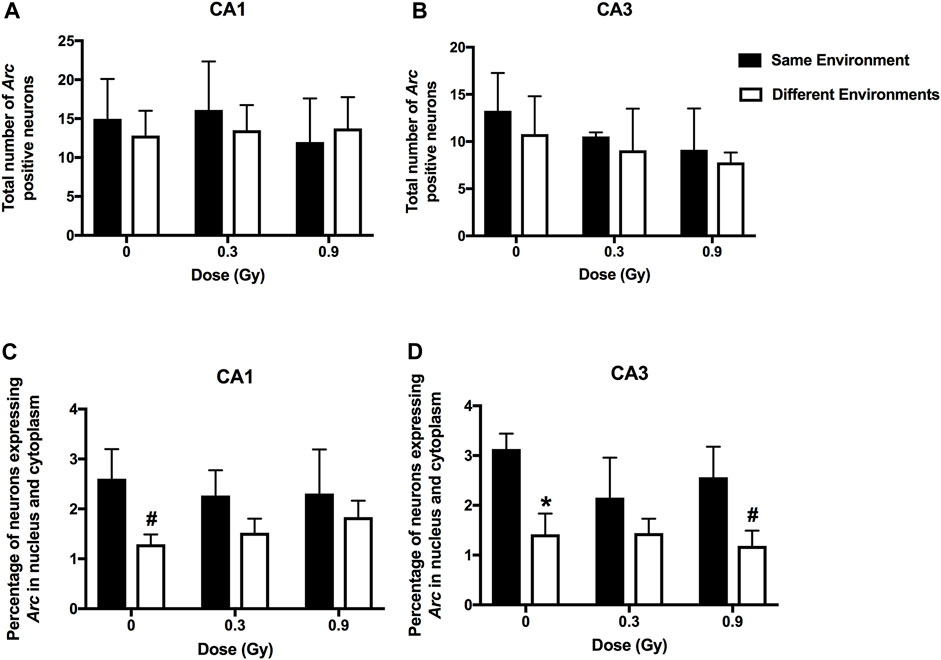
FIGURE 4. A comparable total number of Arc-positive neurons in the CA1 (A) and CA3 (B) regions of the hippocampus of sham-irradiated mice and mice irradiated with 0.3 or 0.9 Gy of 28Si ions. (C) There was a trend towards a higher percentage of Arc-positive neurons in the CA1 region of the hippocampus of sham-irradiated mice following exposure to twice the same environment than two different environments. This was not seen in mice irradiated with 0.3 or 0.9 Gy. (D) In sham-irradiated mice, the percentage of Arc-positive neurons expressing Arc mRNA in the nucleus and cytoplasm in the CA3 region of the hippocampus following exposure twice to the same environment was higher than following exposure to two different environments and there was a trend towards a higher percentage of Arc-positive neurons in the CA3 region of the hippocampus of mice irradiated with a dose of 0.9 Gy following exposure to twice the same environment than two different environments. *p = 0.011 versus same environment; #p = 0.05. CA1; N = 4–7 mice/dose/environment; CA3: N = 3–6 mice/dose/environment. Due to technical problems, tissues from mice irradiated with a dose of 0.6 Gy could not be analyzed.
We next determined whether 28Si ion irradiation-induced cognitive injury is associated with pathway changes in the hippocampus by comparing hippocampal pathway changes in mice irradiated at a lower dose that caused cognitive injury with hippocampal pathway changes in mice irradiated at a higher dose that did not cause cognitive injury. We did not see effects of 28Si ion irradiation on TET2 immunoreactivity in the dentate gyrus, CA1 or CA3 region of the hippocampus, or cortex. The levels of 5 mC and 5 hmC levels are high and exceptionally dynamic during brain development and aging (Jin et al., 2011; Szulwach et al., 2011), suggesting that they play critical roles.
Recently, we reported alterations in hippocampal DNA methylation following proton (Impey et al., 2016b; Impey et al., 2017) and 56Fe ion (Impey et al., 2016a) irradiation. To determine the effects of 28Si ion irradiation on hippocampal DNA methylation, we generated cytosine methylation (5 mC) and/or cytosine hydroxymethylation (5 hmC)-DIP-Seq libraries from the hippocampi of sham irradiated mice and mice exposed to 0.3 or 0.6 Gy of 28Si ion radiation using highly specific antibodies (Impey et al., 2016a; Impey et al., 2016b). These libraries were sequenced to an average depth of ∼14 million tags of which ∼61% mapped to a unique genomic location (Supplementary Table S1). Because 5mc-DIP-Seq tends to map to repetitive areas the lower percentage of unique alignments relative to 5 hmc-DIP-Seq is expected. Genomic regions enriched for 5 mC or 5 hmC were segmented using a previously published Monte Carlo-based algorithm (Impey et al., 2016a; Impey et al., 2017; Johnson et al., 2017) and the union of these regions was tested for significant differences based on a negative binomial distribution (Love et al., 2014). This pipeline identified thousands of differentially methylated and hydroxymethylated regions (DMRs and DHRs, respectively), of which the majority were within 25 kb of a transcription start site (Figure 5A).
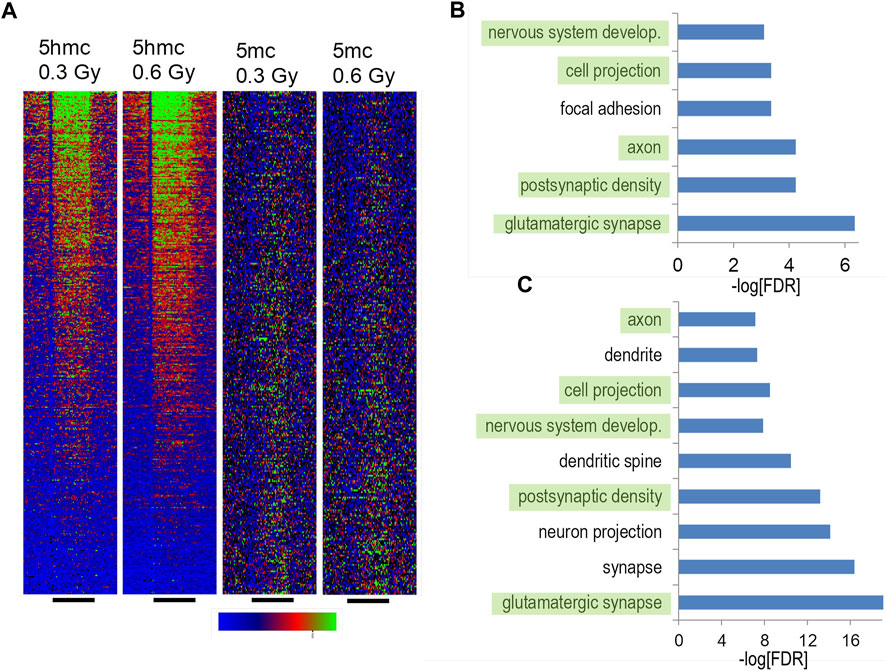
FIGURE 5. Hippocampal DNA methylation and hydroxymethylation pathways regulated by silicon irradiation. (A) DIP density histograms illustrating 5 mC and 5 hmC signals at RefSeq genes sorted by RNA-Seq gene expression levels. The antibodies used to pull down 5mC and 5 hmC regions do not cross react. (B) Gene ontology analyses of upregulated 5 hmC DHRs irradiated with 0.3 Gy. DHRs were annotated with the closest RefSeq gene start site within 50 kb. (C) Gene ontology analyses of upregulated 5 hmC DHRs irradiated with 0.1 56Fe Gy and reanalyzed using the same pipeline as the silicon data. DHRs were annotated with the closest RefSeq gene start site within 50 kb.
Consistent with our and other labs’ DIP-Seq data, DMRs and DHRs distribution showed an expected bias towards genes with high levels of expression or low levels of expression respectively (Figure 5A). Also consistent with our analyses of 56Fe irradiation and literature indicating that 5 hmC is a stable epigenetic mark associated with gene expression in brain (Hahn et al., 2014), there was a highly significant enrichment for gene ontology annotation for increased DHRs at the 0.3 Gy dose (Figure 5B). There was weak enrichment for one gene ontology category (“glutamatergic synapse”, FDR-adjusted p < 5 × 10−4) for decreased DHRs at the 0.3 Gy while all other DHR or DMR conditions showed no significant enrichment for gene ontology categories. Therefore, in this study we focused further analyses on DHRs that showed a significant increase. There was a striking overlap between gene ontology pathways linked to synaptic function and development between the 0.3 Gy 28Si dose and our previous 56Fe 0.1 Gy data (Figures 5B, C) (Impey et al., 2016a), suggesting that these two forms of space radiation regulate similar DNA methylation responses. Consistent with this, there was only a ∼14% overlap between 28Si DHR-associated gene ontology categories and our previously-reported proton-irradiation DHR-associated gene ontology analysis (Impey et al., 2017).
We next sought to compare 0.3 Gy 28Si DHRs (600 MeV/n) with previously published proton (150 MeV; 1 Gy) (Impey et al., 2016b) (Impey et al., 2016b), and 56Fe data (600 MeV/n, 0.1 Gy) (Impey et al., 2016a) by selecting the most-significant gene-associated DHRs from all data sets (top 3,000 DHRs ranked by p-value; p < 0.01 cut off; duplicate genes removed) and testing for overlap of resulting RefSeq genes. These comparisons revealed highly significant overlap between 28Si DHR-associated genes and analogous 56Fe and proton DHR-associated genes (Figure 6A). Interestingly, the overlap between the 0.1 Gy 56Fe and 0.3 Gy 28Si sets was markedly more significant (0.3 Gy 28Si vs. 0.1 Gy 56Fe, Fisher exact p < 2 × 10−69; 0.3 Gy 28Si vs. 0.2 Gy 56Fe, Fisher exact p < 4 × 10−37) than the overlap between the 0.3 Gy 28Si and proton 1 Gy data (Fisher exact p < 5 × 10−48) (Figure 6A). The overlapping set of DHRs led us to determine the subset of DHR-associated genes that were in common to all three forms of radiation exposure. We identified overlapping genes that were associated with increased hydroxymethylation in response to all three forms of radiation (Superexact test: Figure 6B, p < 3 × 10−6; Figure 6C p < 3 × 10−4). The intersection of all 4 data sets was very highly significant (Superexact test: Figure 6D p < 3 × 10−20) and this 45 DHR-associated gene “signature” was significantly enriched for “glutamatergic synapse”, “learning”, and “neuron projection” gene-ontology categories which suggest that this set of DHR-associated genes denotes a pathway linked to changes in synapse function and synaptic plasticity (Figure 6E).
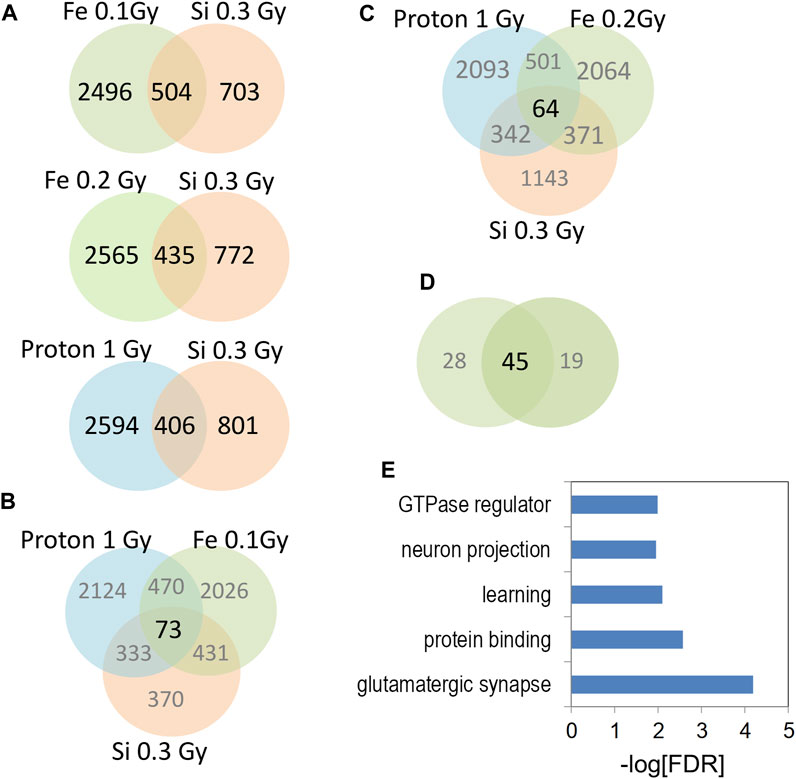
FIGURE 6. (A) Venn diagrams depict the overlap of gene-associated DHRs significantly increase by the indicated radiation treatment. DHRs were selected by p-value rank (top 1,500) and absolute tag difference (>10). DHRs were annotated with the closest RefSeq gene start site within 50 kb. (B,C) Venn diagrams depict the three way overlap of gene-associated DHRs significantly increased by the indicated radiation treatments. (D) Venn diagram illustrates the overlap between the 3-way intersections in panels (B,C). (E) Gene-ontology analyses of 45 genes from (D).
A similar set of analyses that compared 0.6 Gy 28Si DHR-associated genes with 56Fe and proton DHR-associated genes also found more significant overlap for the 0.1 Gy 56Fe data (Figure 7A: 0.6 Gy 28Si vs. 0.1 Gy 56Fe, Fisher exact p < 5 × 10−68) than for the proton 1Gy (Fisher exact, p < 9 × 1049) or 0.2 Gy 56Fe comparisons (0.6 Gy 28Si vs. 0.2 Gy 56Fe, Fisher exact p < 4 × 10−30). The DHR-associated gene overlap between all three forms of radiation was again highly significant (Figure 7B, p < 2x−100; Figure 7C, p < 7 × 10−70) as was the overlap between the proton, 28Si, and two doses of 56Fe radiation (Figure 7D, SuperExact test, 9 × 10−200).

FIGURE 7. (A) Venn diagrams depict the overlap of gene-associated DHRs significantly increase by the indicated radiation treatment. DHRs were selected by p-value rank (top 1,500) and absolute tag difference (>10). DHRs were annotated with the closest RefSeq gene start site within 50 kb. (B,C) Venn diagrams depict the three way overlap of gene-associated DHRs significantly increased by the indicated radiation treatment. (D) Venn diagram illustrates the overlap between the 3-way intersections in panels (B,C). (E) Gene-ontology analyses of 133 genes from (D).
The 133 gene “4-way” intersection (Figure 7D) was significantly enriched for gene ontology categories linked to synaptic function and neuron projection (Figure 7E) suggesting that 0.6 Gy 28Si irradiation induced a response more closely correlated with our published 56Fe and proton radiation responses (Impey et al., 2016a; Impey et al., 2016b). We next tested whether this 133 gene space radiation “signature” contained genes previously linked to radiation in rodent and human models (Xu et al., 2020) and found a non-significant intersection (Table 1) that included genes involved in growth-factor and small GTPase signaling (WIPF1, PIAS1, and IGFR1). Remarkably, the 45 genes in the 0.3 Gy 28Si “4-way” intersection (Figure 6D) were all present in the 133 gene 0.6 Gy 28Si “4-way” intersection (Figure 8A, SuperExact test, p < 7 × 10−68) indicating that there is a DHR-associated gene signature shared between the two 28Si doses (0.3 Gy, 0.6 Gy), the two 56Fe doses (0.1 Gy, 0.2 Gy), and the 1 Gy proton dose (Figure 8B; Table 2) (Cekanaviciute et al., 2023). This space radiation signature of 45 genes contained only one gene, PIAS1, previously linked to radiation response in a database largely limited to “photon” radiation studies (Xu et al., 2020). To provide better context for this space-radiation hydroxymethylation-related gene signature we annotated each gene using PubMed and Google Scholar searches for “radiation”, “ionizing radiation,” or “space radiation;” 23 of 45 genes were associated with studies indicating a radiation response ranging from “radiation response” gene ontology category membership (3), changes in gene expression (19), differential methylation of genetic loci (2), and radiation-associated mutagenesis or crosslinking (3) (Table 2). Interestingly, Npas3 was identified as a significant space-radiation decreased gene (12C ion beam) (Iwakawa et al., 2008) while Ncald and Parkn were identified as genes that are preferentially mutated in response to space radiation-associated DNA damage (40Ar and 65Fe ion beam) (Cekanaviciute et al., 2023).
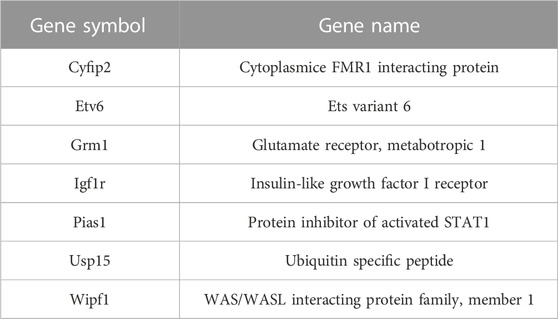
TABLE 1. Intersection between 133 gene space radiation signature and Radatlas ionizing-radiation-regulated genes (MESH terms).
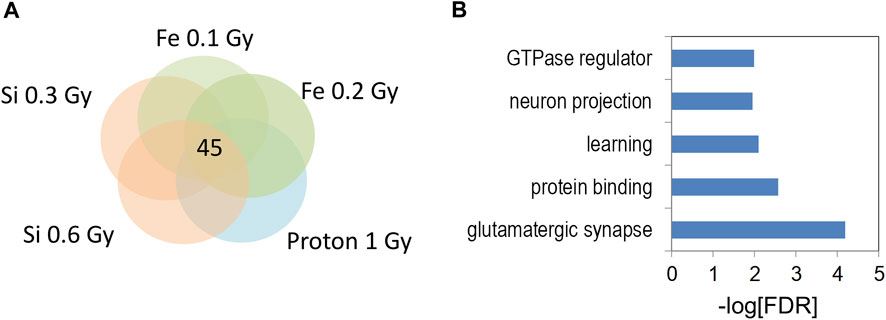
FIGURE 8. (A) Venn diagram illustrates the five way overlap of gene-associated DHRs significantly increased by indicated radiation treatments in Figures 5, 6. (B) Gene ontology analyses of 5 hmC DHRs from 5-way intersection in (A).

TABLE 2. Summary of the novel object recognition data of the 56Fe ion (600 MeV) and proton (150 MeV) studiesa.
We next utilized RNA-Seq to profile the transcriptional response to 28Si irradiation using hippocampal RNA co-extracted from the same tissue used for DIP-Seq experiments. Although our 56Fe irradiation study (Impey et al., 2016a) identified highly significant changes in gene expression that correlated with DHRs (but not DMRs), we did not observe a significant transcriptional response to either 0.3 Gy or 0.6 Gy 28Si irradiation at the 2 weeks time point (no genes reached the FDR-adjusted p < 0.01 cut off, Supplementary Tables S1, S2). This result is consistent with our observation that 28Si irradiation at the 2 weeks time point only demonstrated significant gene ontologies at the 0.3 Gy dose whereas we saw highly significant gene ontology and KEGG pathway enrichment for both down and upregulated DHRs following 0.1 Gy and 0.2 Gy 56Fe irradiation (Impey et al., 2016a). Because gene expression pathway data is often regulated in a biologically meaningful manner even when individual genes do not achieve FDR-adjusted significance, we selected differentially-expressed genes at an unadjusted p-value cut off of p < 0.01 and assessed gene-ontology category and KEGG pathway enrichment via the Fisher-exact test. For genes increased by the 0.3 Gy 28Si dose, gene ontology categories associated with mitochondrial oxidative phosphorylation were most-significantly enriched as was the KEGG pathway for oxidative phosphorylation (Figure 9A; Supplementary Figure S1). Interestingly, we also observed significant enrichment for KEGG pathways associated with Huntington’s, Alzheimer’s, and Parkinson’s diseases (Supplementary Figures S2–S4). We also saw enrichment for Huntington’s, Alzheimer’s, and Parkinson’s disease KEGG pathways for genes significantly decreased by 56Fe irradiation in a previous study (Impey et al., 2016a), suggesting that these pathways may represent a more generalized space-radiation response. Because oxidative stress has been linked to the neuropathology of these neurodegenerative disorders (Kim et al., 2015), our data suggest that a 0.3 Gy 28Si radiation dose may trigger persistent oxidative stress that adversely impacts neuronal homeostasis/metabolism. Genes decreased by 0.3 Gy 28Si irradiation were significantly enriched for synaptic components and neuronal proteins (Figure 9B) which is consistent with the idea that the radiation-induced oxidative stress adversely impacts synaptic function/homeostasis.

FIGURE 9. (A–D) Bar graphs depicts gene ontology analyses of RefSeq genes significantly up- and downregulated (p < 0.01) for indicated RNA-Seq comparisons.
Genes increased by 0.6 Gy 28Si irradiation were enriched for gene ontology categories associated with synaptic and neuronal proteins (Figure 9C) while genes decreased by 0.6 Gy 28Si irradiation showed less-significant enrichment for neuron-associated gene ontology categories (Figure 9D) and KEGG pathways (Supplementary Figure S5). The Alzheimer’s disease KEGG pathway was significantly enriched for genes decreased by 0.6 Gy 28Si irradiation which is the opposite effect seen for 0.3 Gy 28Si. It is conceivable that the 0.6 Gy 28Si dose triggered compensatory neuroprotective gene expression changes. Gene ontology analyses of indicated intersections between gene-annotated DHRs and differentially-expressed genes are illustrated in Figure 10.
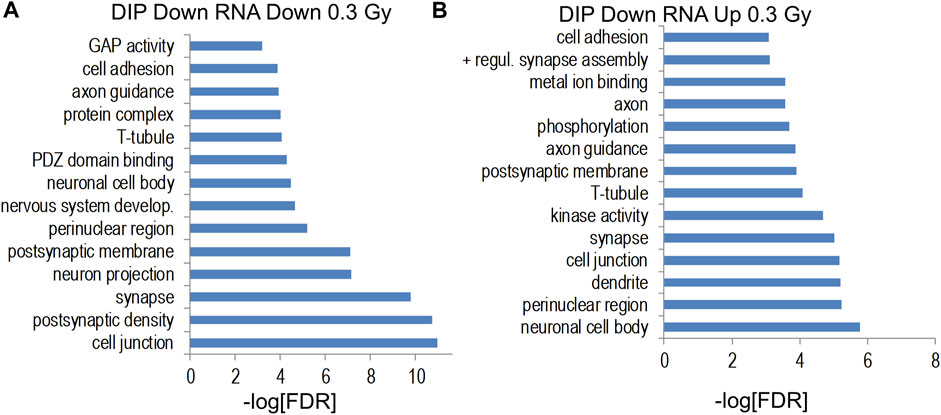
FIGURE 10. (A,B) Gene ontology analyses of indicated intersections between gene-annotated DHRs and differentially-expressed genes (unadjusted p < 0.011).
We next assessed the correlation between gene-associated differential DNA hydroxymethylation and gene expression. Although we did not detect the highly-correlated spatial correlation of DHRs and gene expression seen for 56Fe irradiation (Impey et al., 2016a), we found significant overlap between downregulated genes and both increased and decreased DHRs at the 0.3 Gy and 0.6 Gy 28Si radiation doses (Supplementary Figure S6). These data suggest that by intersecting DIP-Seq data with RNA-Seq data, we increased statistical power allowing us to detect biologically relevant epigenetic remodeling. Consistent with this idea, the highly significant intersection between decreased 0.3 Gy 28Si DHRs and both up and downregulated genes were significantly enriched for gene ontology categories linked to synapses, neuronal connectivity, and neuronal development (Supplementary Figure S7). We also identified the “mitochondrion” gene ontology category when comparing increased 0.3 Gy 28Si DHRs with increased 0.3 Gy 28Si gene expression and the “nervous system development” gene ontology category for the intersection of decreased 0.6 Gy 28Si DHRs with decreased 0.6 Gy 28Si gene expression.
The results of the current study show that at 0.3 Gy, but not at 0.6 Gy or 0.9 Gy, 28Si ion irradiation impaired cognition and network activity in the CA3 region of the hippocampus. Consistent with this pattern of the dose-response curve for cognitive performance, sham-irradiated mice and those irradiated with 56Fe (600 MeV) at 0.2 Gy showed novel object recognition but mice irradiated with 0.1 Gy were impaired (Impey et al., 2016a) (Table 2 for Summary of the cognitive data of the 56Fe (600 MeV) and proton (150 MeV) studies). This pattern is especially concerning as the lower doses would be expected to be more pertinent to doses astronauts will be exposed to during space missions. While the percentage of Arc-positive neurons expressing Arc mRNA in the nucleus and cytoplasm in the CA3 region of the hippocampus of sham-irradiated mice was higher following exposure twice to the same environment, as opposed to exposure to two different environments, this was not seen in mice irradiated with 0.3 Gy that showed impaired cognitive performance. In mice irradiated with 0.9 Gy, who showed no impaired cognitive performance, there was a trend towards a difference in the CA3 but not the CA1 region. The CA3 region is involved in rapid encoding of new detailed memories and bound to a large degree to a spatial coordinate system and more important for distinguishing different environments, while the downstream CA1 region can interpret this information from the CA3 region in the context of less spatially restricted information and is more important for recognizing the same environment (Leutgeb et al., 2004; Leutgeb and Leutgeb, 2007). These data suggest that in mice irradiated with 0.3 Gy, the CA3 region might be especially affected, while in mice irradiated with 0.9 Gy, the CA3 region might be less affected than the CA1 region and sufficient for detecting a novel object in the environment. We detected significant enrichment of gene ontology categories only for DHRs at the 0.3 Gy 28Si dose but no significant enrichment of gene ontology categories for DMRs. As we observed following 65Fe and proton irradiation, DHRs were significantly associated with gene ontology pathways for synapses, postsynaptic specialization, and axons. In previous studies, when we detected gene-ontology category enrichment, it was consistently more significant for 5 hmC data than for 5 mC data. Moreover, for multiple conditions (65Fe 20 weeks, proton radiation 2 weeks), there was a significant gene ontology enrichment only for DHRs. Therefore, our results are consistent with our general observation that space-radiation-associated 5 hmC remodeling is more tightly associated with genes and gene pathways than 5 mC remodeling. It should be noted that because 5 hmC is generated by enzymatic hydroxylation of 5mC, remodeling of 5 mC is also captured by methodologies that measure changes in 5 hmc. Interestingly, the 0.3 Gy 28Si dose was also the only dose that caused a significant deficit in novel object recognition learning, suggesting that 5 hmC remodeling of synapse-associated gene ontology category may be linked to cognitive dysfunction. These data show that 28Si ion radiation-induced cognitive injury is associated with hippocampal changes in DNA hydroxymethylation of cytosine, a major epigenetic modification involving dynamic changes methyl groups, and specific gene ontology categories linked to synaptic pathways and the post-synaptic density. Consistent with these data, hippocampal changes in DNA methylation are involved in the regulation of expression of genes required for cognitive performance (Miller and Sweatt, 2007; Lubin et al., 2008). 5 hmC DNA methylation is derived from 5 mC by the action of three TET enzymes (TET 1–3), is hydroxymethylcytosine (5 hmC) (Veron and Peters, 2011). While in brain, levels of TET2 are higher than those of TET1 or TET3 in the brain, and therefore TET2 is believed most important for brain function (Chen et al., 2012; Dzitoyeva et al., 2012); however, we did not see effects of radiation on TET2 levels.
While altered hippocampal DNA methylation was seen following 0.6 Gy 28Si ion irradiation, there was no impairment in novel object recognition. Genes increased by 0.6 Gy 28Si irradiation were enriched for gene ontology categories associated with synaptic and neuronal proteins. In addition, the Alzheimer’s disease KEGG pathway was significantly enriched for genes decreased by 0.6 Gy, but increased by 0.3 Gy 28Si irradiation. Based on these results, we hypothesize that the 0.6 Gy 28Si dose triggered compensatory neuroprotective gene expression changes that prevented impairments in novel object recognition seen following 0.3 Gy.
We next assessed whether there is a general space radiation synaptic signature and/or distinct changes in hippocampal DNA methylation comparing effects of proton, 56Fe ion, and 28Si ion irradiation. Analyzing the shared hippocampal pathways affected 2 weeks following proton (150 MeV; 1 Gy), 56Fe (600 MeV/n, 0.1 Gy and 0.2 Gy) and 28Si (600 MeV/n, 0.3 Gy and 0.6 Gy) ion irradiation revealed 45 genes (Table 2). Although this set should not be taken as a total number of radiation response genes in the hippocampus because we only included those that were observed in five independent experiments with three different types of particles, their reproducible presence in these experiments represents a remarkable degree of specificity, and thus importance. This set of 45 genes was significantly enriched for gene ontology categories including “glutamatergic synapse”, “learning”, and “neuron projection”. The “glutamatergic synapse” gene ontology category included genes such as Neuron-glial related cell adhesion molecule (Nrcam), Parkin 2 (Prkn), Amphiphysin (Amph), Contactin associated protein-like 2 (Cntnap2)¸ and Sortilin-Related Receptor3 (Sorcs3). Interestingly, Nrcam, Amph, Cntnap2, and Sorcs3 regulate presynaptic glutamatergic function suggesting remodeling of genes involved in glutamate release. The “learning” gene ontology category was comprised of an overlapping set of enriched genes including Parkin 2 (Prkn), Amphiphysin (Amph), Contactin associated protein-like 2(Cntnap2)¸ Sortilin-Related Receptor3 (Sorcs3), and phospholipase C, beta 1 (Plcb1). The biological relevance of these associations is supported by a robust literature linking space radiation to changes in glutamatergic synapse gene expression, glutamatergic synapse function, and regulation of glutamatergic postsynaptic density genes/proteins (Machida et al., 2010; Allen et al., 2015; Parihar et al., 2015; Sokolova et al., 2015; Parihar et al., 2016; Krukowski et al., 2018; Parihar et al., 2018). Our results indicate that, in addition, to a microglial stress response (Parihar et al., 2016; Krukowski et al., 2018; Parihar et al., 2018) space radiation targets long-lasting gene expression and epigenetic remodeling responses to glutamatergic synapse genes.
Table 3 annotates these 45 genes by their association with radiation response in the scientific literature as well as their association with neurological disorders. The majority of these genes were associated with a radiation response and two (Asic2 and Celf4) were associated with both radiation-induced differential methylation and gene expression. Although only three genes (Asic2, Cntnap, and Pias1) were annotated in the “radiation response” gene ontology category another 20 were identified as radiation-regulated genes in the literature. Notably, one gene (Npas3) was identified as being decreased by space radiation (Iwakawa et al., 2008) and two genes (Nfia and Prkn) were identified as prone to space-radiation-mediated DNA damage (Cekanaviciute et al., 2023). The high degree of overlap between our 45 gene space radiation signature and the previous literature linking these genes to potential radiation response supports the idea that these sites of 5 hmC remodeling are biologically relevant to radiation injury The profound phenotype seen with those genes confirms the potential of this approach to identify genes and pathways involved in the CNS radiation response that are critical for CNS function.
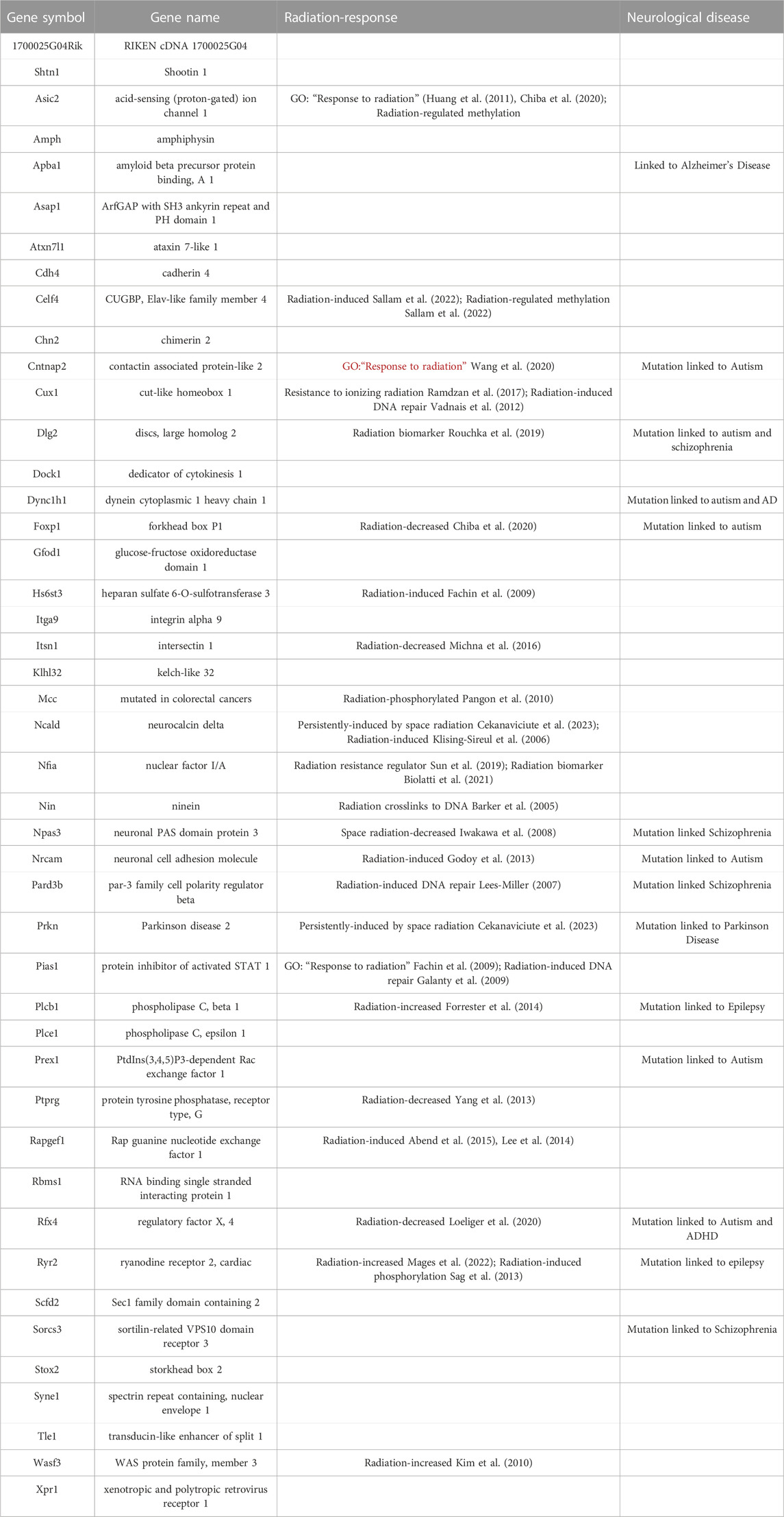
TABLE 3. Comparing hippocampal DNA hydroxymethylation following proton, 56Fe ion, and 28Si ion irradiation revealed a general space radiation synaptic signature with 45 genes that are associated with profound phenotypes.
In summary, 0.3 Gy of 28Si ion irradiation (600 MeV/n) affects hippocampus-dependent cognitive performance, network activity in the CA3 region of the hippocampus, with impaired cognition correlating with altered gene expression and 5 hmC profiles that mapped to specific gene ontology pathways. The general space radiation synaptic signature with 45 genes that are associated with profound phenotypes following proton, 56Fe ion, and 28Si ion irradiation revealed that the most significant categories are glutamatergic synapse and postsynaptic density. Thus, the brain’s response to space irradiation involves novel excitatory synapse and postsynaptic remodeling. Future efforts are warranted to determine how this general radiation signature might be targeted to reduce detrimental effects of space radiation on the brain during and following missions.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
The animal study was reviewed and approved by the OHSU IACUC and the BNL IACUC.
SI, SR, MT, and JR: conception and design of the study, analysis, and interpretation of data, drafting, revising, and final approval of the manuscript. CP, AT, FF, DZ, TM, L-KR, and BS: acquisition of data and analysis of data, and revising and final approval of the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by NASA grants NNJ12ZSA001N, 80NSSC19K0498–P00001, NIDA T32DA007262, the Legacy Good Samaritan Foundation, and the development accounts of JR and SI.
The authors wish to thank Peter Guida, Adam Rusek, and other NSRL physics staffs for their invaluable help. We also thank Dr. Searles, the Director of the Massively Parallel Sequencing Shared Resource and Associate Director Integrated Genomics Laboratory at OHSU, for providing his expertise for the design and interpretation of the DNA methylation and RNAseq experiments in this study.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2023.1215535/full#supplementary-material
Abend, M., Azizova, T., Muller, K., Dorr, H., Doucha-Senf, S., Kreppel, H., et al. (2015). Association of radiation-induced genes with noncancer chronic diseases in Mayak workers occupationally exposed to prolonged radiation. Radiat. Res. 183, 249–261. doi:10.1667/RR13758.1
Allen, A. R., Raber, J., Chakraborti, A., Sharma, S., and Fike, J. R. (2015). (56 Fe irradiation alters spine density and dendritic complexity in the mouse Hippocampus. Radiat. Res. 184, 586–594. doi:10.1667/RR14103.1
Anders, S., and Huber, W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11, R106. doi:10.1186/gb-2010-11-10-r106
Barker, S., Weinfeld, M., Zheng, J., Li, L., and Murray, D. (2005). Identification of mammalian proteins cross-linked to DNA by ionizing radiation. J. Biol. Chem. 280, 33826–33838. doi:10.1074/jbc.M502477200
Biolatti, V., Negrin, L., Bellora, N., and Ibanez, I. L. (2021). High-throughput meta-analysis and validation of differentially expressed genes as potential biomarkers of ionizing radiation-response. Radiother. Oncol. 154, 21–28. doi:10.1016/j.radonc.2020.09.010
Cekanaviciute, E., Tran, D., Nguyen, H., Lopez Macha, A., Pariset, E., Langley, S., et al. (2023). Mouse genomic associations with in vitro sensitivity to simulated space radiation. Life Sci. Space Res. (Amst) 36, 47–58. doi:10.1016/j.lssr.2022.07.006
Chen, H., Dzitoyeva, S., and Manev, H. (2012). Effect of aging on 5-hydroxymethylcytosine in the mouse hippocampus. Restor. Neurol. Neurosci. 30, 237–245. doi:10.3233/RNN-2012-110223
Chiba, M., Uehara, H., Niiyama, I., Kuwata, H., and Monzen, S. (2020). Changes in miRNA expressions in the injured small intestine of mice following high-dose radiation exposure. Mol. Med. Rep. 21, 2452–2458. doi:10.3892/mmr.2020.11054
Dennis, G., Sherman, B. T., Hosack, D. A., Yang, J., Gao, W., Lane, H. C., et al. (2003). David: Database for annotation, visualization, and integrated discovery. Genome Biol. 4, P3. doi:10.1186/gb-2003-4-5-p3
Dzitoyeva, S., Chen, H., and Manev, H. (2012). Effect of aging on 5-hydroxymethylcytosine in brain mitochondria. Neurobiol. Aging 33, 2881–2891. doi:10.1016/j.neurobiolaging.2012.02.006
Ennaceur, A. (2010). One-trial object recognition in rats and mice: Methodological and theoretical issues. Behav. Brain Res. 215, 244–254. doi:10.1016/j.bbr.2009.12.036
Fachin, A. L., Mello, S. S., Sandrin-Garcia, P., Junta, C. M., Ghilardi-Netto, T., Donadi, E. A., et al. (2009). Gene expression profiles in radiation workers occupationally exposed to ionizing radiation. J. Radiat. Res. 50, 61–71. doi:10.1269/jrr.08034
Fejes, A. P., Robertson, G., Bilenky, M., Varhol, R., Bainbridge, M., and Jones, S. J. (2008). FindPeaks 3.1: A tool for identifying areas of enrichment from massively parallel short-read sequencing technology. Bioinformatics 24, 1729–1730. doi:10.1093/bioinformatics/btn305
Ferlazzo, M., and Foray, N. (2017). Space radiobiology needs realistic hypotheses and relevant methodology. Proc. Nath Acad. Sci. U. S. A. 114, E6733. doi:10.1073/pnas.1710545114
Foray, N., Bourguignon, M., and Hamada, N. (2016). Individual response to ionizing radiation. Mut Res. 770, 369–386. doi:10.1016/j.mrrev.2016.09.001
Forrester, H. B., Li, J., Leong, T., Mckay, M. J., and Sprung, C. N. (2014). Identification of a radiation sensitivity gene expression profile in primary fibroblasts derived from patients who developed radiotherapy-induced fibrosis. Radiother. Oncol. 111, 186–193. doi:10.1016/j.radonc.2014.03.007
Galanty, Y., Belotserkovskaya, R., Coates, J., Polo, S., Miller, K. M., and Jackson, S. P. (2009). Mammalian SUMO E3-ligases PIAS1 and PIAS4 promote responses to DNA double-strand breaks. Nature 462, 935–939. doi:10.1038/nature08657
Gentleman, R. C., Carey, V. J., Bates, D. M., Bolstad, B., Dettling, M., Dudoit, S., et al. (2004). Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 5, R80. doi:10.1186/gb-2004-5-10-r80
Godoy, P., Mello, S., Magalhaes, D., Donaires, F., Nicolucci, P., Donadi, E., et al. (2013). Ionizing radiation-induced gene expression changes in TP53 proficient and deficient glioblastoma cell lines. Mutat. Res. 756, 46–55. doi:10.1016/j.mrgentox.2013.06.010
Guzowski, J., Mcnaughton, B. L., Barnes, C. A., and Worley, P. F. (1999). Environment-specific expression of the immediate-early gene Arc in hippocampal neuronal ensembles. Nat. Neurosci. 2, 1120–1124. doi:10.1038/16046
Hahn, M. A., Szabo, P. E., and Pfeifer, G. P. (2014). 5-Hydroxymethylcytosine: A stable or transient DNA modification? Genomics 104, 314–323. doi:10.1016/j.ygeno.2014.08.015
Harris, R. A., Wang, T., Coarfa, C., Nagarajan, R. P., Hong, C., Downey, S. L., et al. (2010). Comparison of sequencing-based methods to profile DNA methylation and identification of monoallelic epigenetic modifications. Nat. Biotechnol. 28, 1097–1105. doi:10.1038/nbt.1682
Huang, C.-C., Sun, X.-J., and Hu, M.-J. (2011). DNA methylation changes induced by 60Co γ-ray irradiation. J. Chin. Public Health 27, 1263–1266. doi:10.11847/zgggws-2011-27-10-19
Impey, S., Jopson, T., Pelz, C., Tafessu, A., Fareh, F., Zuloaga, D., et al. (2017). Bi-directional and shared epigenomic signatures following proton and 56Fe irradiation. Sci. Rep. 7, 10227. doi:10.1038/s41598-017-09191-4
Impey, S., Jopson, T., Pelz, C., Tafessu, A., Fareh, F., Zuloaga, D., et al. (2016a). Short- and long-term effects of 56Fe irradiation on cognition and hippocampal DNA methylation and gene expression. BMC Genomics 17, 825. doi:10.1186/s12864-016-3110-7
Impey, S., Pelz, C., Tafessu, A., Marzulla, T., Turker, M., and J, R. (2016b). Proton irradiation induces persistent and tissue-specific DNA methylation changes in the left ventricle and hippocampus. BMC genomics 17, 273. doi:10.1186/s12864-016-2581-x
Iwakawa, M., Hamada, N., Imadome, K., Funayama, T., Sakashita, T., Kobayashi, Y., et al. (2008). Expression profiles are different in carbon ion-irradiated normal human fibroblasts and their bystander cells. Mutat. Res. 642, 57–67. doi:10.1016/j.mrfmmm.2008.04.007
Jin, S. G., Wu, X., Li, A., and Pfeifer, G. (2011). Genomic mapping of 5-hydroxymethylcytosine in the human brain. Nucleic Acids Res. 39, 5015–5024. doi:10.1093/nar/gkr120
Johnson, L. A., Torres, E. R., Impey, S., Stevens, J. F., and Raber, J. (2017). Apolipoprotein E4 and insulin resistance interact to impair cognition and alter the epigenome and metabolome. Sci. Rep. 7, 43701. doi:10.1038/srep43701
Kiffer, F., Boerma, M., and Allen, A. (2019). Behavioral effects of space radiation: A comprehensive review of animal studies. Life Sci. Space Res. 21, 1–21. doi:10.1016/j.lssr.2019.02.004
Kim, B. C., Han, N. K., Byun, H. O., Kim, S. S., Ahn, E. K., Chu, I. S., et al. (2010). Time-dependently expressed markers and the characterization for premature senescence induced by ionizing radiation in MCF7. Oncol. Rep. 24, 395–403. doi:10.3892/or_00000872
Kim, G. H., Kim, J. E., Rhie, S. J., and Yoon, S. (2015). The role of oxidative stress in neurodegenerative diseases. Exp. Neurobiol. 24, 325–340. doi:10.5607/en.2015.24.4.325
Klising-Sireul, E., Rigaud, O., Ory, K., Ugolin, N., Lebeau, J., Levalois, C., et al. (2006). Transcriptional response of wild-type and ataxia telangiectasia lymphoblasts following exposure to equitoxic doses of ionizing radiation. J. Radiat. Res. 47, 259–272. doi:10.1269/jrr.0594
Kronenberg, A., and Cucinotta, F. A. (2012). Space radiation protection issues. Health Phys. 103, 556–567. doi:10.1097/HP.0b013e3182690caf
Krukowski, K., Grue, K., Frias, E. S., Pietrykowski, J., Jones, T., Nelson, G., et al. (2018). Female mice are protected from space radiation-induced maladaptive responses. Brain Behav. Immun. 74, 106–120. doi:10.1016/j.bbi.2018.08.008
Langmead, B., Trapnell, C., Pop, M., and Salzberg, S. L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25. doi:10.1186/gb-2009-10-3-r25
Lee, K. F., Weng, J. T., Hsu, P. W., Chi, Y. H., Chen, C. K., Liu, I. Y., et al. (2014). Gene expression profiling of biological pathway alterations by radiation exposure. Biomed. Res. Int. 2014, 834087. doi:10.1155/2014/834087
Lees-Miller, S. P. (2007). The double (strand break) life of Par-3. Nat. Cell. Biol. 9, 363–365. doi:10.1038/ncb0407-363
Leutgeb, S., and Leutgeb, J. (2007). Pattern separation, pattern completion, and new neuronal codes within a continuous CA3 map. Learn Mem. 14, 745–757. doi:10.1101/lm.703907
Leutgeb, S., Leutgeb, J., Treves, A., Moser, M.-B., and Moser, E. (2004). Distinct ensemble codes in hippocampal areas CA3 and CA1. Science 305, 1295–1298. doi:10.1126/science.1100265
Loeliger, B. W., Hanu, C., Panyutin, I. V., Maass-Moreno, R., Wakim, P., Pritchard, W. F., et al. (2020). Effect of ionizing radiation on transcriptome during neural differentiation of human embryonic stem cells. Radiat. Res. 193, 460–470. doi:10.1667/RR15535.1
Love, M., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. doi:10.1186/s13059-014-0550-8
Lubin, F., Roth, T., and Sweatt, J. (2008). Epigenetic regulation of BDNF gene transcription in the consolidation of fear memory. J. Neurosci. 28, 10576–10586. doi:10.1523/JNEUROSCI.1786-08.2008
Machida, M., Lonart, G., and Britten, R. A. (2010). Low (60 cGy) doses of (56)Fe HZE-particle radiation lead to a persistent reduction in the glutamatergic readily releasable pool in rat hippocampal synaptosomes. Radiat. Res. 174, 618–623. doi:10.1667/RR1988.1
Mages, C., Gampp, H., Rahm, A. K., Ullrich, N. D., Seidensaal, K., Rhein, B., et al. (2022). Radiation-induced electrophysiological remodelling in neonatal rat ventricular cardiomyocytes. Eur. Heart J. 43, ehac544.2975. doi:10.1093/eurheartj/ehac544.2975
Michna, A., Schotz, U., Selmansberger, M., Zitzelsberger, H., Lauber, K., Unger, K., et al. (2016). Transcriptomic analyses of the radiation response in head and neck squamous cell carcinoma subclones with different radiation sensitivity: Time-course gene expression profiles and gene association networks. Radiat. Oncol. 11, 94. doi:10.1186/s13014-016-0672-0
Miller, C., and Sweatt, J. (2007). Covalent modification of DNA regulates memory formation. Neuron 53, 857–869. doi:10.1016/j.neuron.2007.02.022
Pangon, L., Sigglekow, N. D., Larance, M., Al-Sohaily, S., Mladenova, D. N., Selinger, C. I., et al. (2010). The "mutated in colorectal cancer" protein is a novel target of the UV-induced DNA damage checkpoint. Genes. Cancer 1, 917–926. doi:10.1177/1947601910388937
Parihar, V. K., Allen, B. D., Caressi, C., Kwok, S., Chu, E., Tran, K. K., et al. (2016). Cosmic radiation exposure and persistent cognitive dysfunction. Sci. Rep. 6, 34774. doi:10.1038/srep34774
Parihar, V. K., Allen, B., Tran, K. K., Macaraeg, T. G., Chu, E. M., Kwok, S. F., et al. (2015). What happens to your brain on the way to Mars. Sci. Adv. 1, e1400256. doi:10.1126/sciadv.1400256
Parihar, V. K., Maroso, M., Syage, A., Allen, B. D., Angulo, M. C., Soltesz, I., et al. (2018). Persistent nature of alterations in cognition and neuronal circuit excitability after exposure to simulated cosmic radiation in mice. Exp. Neurol. 305, 44–55. doi:10.1016/j.expneurol.2018.03.009
Penner, M. R., Roth, T. L., Chawla, M. K., Hoang, L. T., Roth, E. D., Lubin, F. D., et al. (2010). Age-related changes in Arc transcription and DNA methylation within the hippocampus. Neurobiol. Aging 32, 2198–2210. doi:10.1016/j.neurobiolaging.2010.01.009
Raber, J., Bongers, G., Lefevour, A., Buttini, M., and Mucke, L. (2002). Androgens protect against apolipoprotein E4-induced cognitive deficits. J. Neurosci. 22, 5204–5209. doi:10.1523/JNEUROSCI.22-12-05204.2002
Raber, J. (2015). Novel images and novel locations of familiar images as sensitive translational cognitive tests in humans. Beh Brain Res. 285, 53–59. doi:10.1016/j.bbr.2015.01.046
Ramdzan, Z. M., Ginjala, V., Pinder, J. B., Chung, D., Donovan, C. M., Kaur, S., et al. (2017). The DNA repair function of CUX1 contributes to radioresistance. Oncotarget 8, 19021–19038. doi:10.18632/oncotarget.14875
Ramirez-Amaya, V., Angulo-Perkins, A., Chawla, M., Barnes, C., and Rosi, S. (2013). Sustained transcription of the immediate early gene Arc in the dentate gyrus after spatial exploration. J. Neurosci. 33, 1631–1639. doi:10.1523/JNEUROSCI.2916-12.2013
Restier-Verlet, J., El-Nachef, L., Ferlazzo, M., Al-Choboq, J., Granzotto, A., Bouchet, A., et al. (2021). Radiation on earth or in space: What does it change? Int. J. Mol. Sci. 22, 3739. doi:10.3390/ijms22073739
Rosi, S., Ramirez-Amaya, V., Vazdarjanova, A., Esparza, E., Larkin, P., Fike, J., et al. (2009). Accuracy of hippocampal network activity is disrupted by neuroinflammation: Rescue by memantine. Brain 1, 2464–2477. doi:10.1093/brain/awp148
Rouchka, E. C., Flight, R. M., Fasciotto, B. H., Estrada, R., Eaton, J. W., Patibandla, P. K., et al. (2019). Dataset for dose and time-dependent transcriptional response to ionizing radiation exposure. Data Brief. 27, 104624. doi:10.1016/j.dib.2019.104624
Sag, C. M., Wolff, H. A., Neumann, K., Opiela, M. K., Zhang, J., Steuer, F., et al. (2013). Ionizing radiation regulates cardiac Ca handling via increased ROS and activated CaMKII. Basic Res. Cardiol. 108, 385. doi:10.1007/s00395-013-0385-6
Sallam, M., Mysara, M., Benotmane, M. A., Crijns, A. P. G., Spoor, D., Van Nieuwerburgh, F., et al. (2022). DNA methylation alterations in fractionally irradiated rats and breast cancer patients receiving radiotherapy. Int. J. Mol. Sci. 23, 16214. doi:10.3390/ijms232416214
Sokolova, I. V., Schneider, C. J., Bezaire, M., Soltesz, I., Vlkolinsky, R., and Nelson, G. A. (2015). Proton radiation alters intrinsic and synaptic properties of CA1 pyramidal neurons of the mouse hippocampus. Radiat. Res. 183, 208–218. doi:10.1667/RR13785.1
Storey, J. D., and Tibshirani, R. (2003). Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. U. S. A. 100, 9440–9445. doi:10.1073/pnas.1530509100
Sun, C., Li, Y., Tan, Y., Zhang, H., Liang, Y., Zeng, J., et al. (2019). A novel role for NFIA in restoring radiosensitivity in radioresistant NSCLC cells by downregulating the AKT and ERK pathways. Biochem. Biophys. Res. Commun. 515, 558–564. doi:10.1016/j.bbrc.2019.06.011
Szulwach, K., Li, X., Li, Y., Song, C.-X., Wu, H., Dai, Q., et al. (2011). 5-hmC–mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat. Neurosci. 14, 1607–1616. doi:10.1038/nn.2959
Vadnais, C., Davoudi, S., Afshin, M., Harada, R., Dudley, R., Clermont, P. L., et al. (2012). CUX1 transcription factor is required for optimal ATM/ATR-mediated responses to DNA damage. Nucleic Acids Res. 40, 4483–4495. doi:10.1093/nar/gks041
Veron, N., and Peters, A. (2011). Epigenetics: Tet proteins in the limelight. Nature 273, 293–294. doi:10.1038/473293a
Wang, C., Kao, W.-H., and Hsiao, C. K. (2015). Using hamming distance as information for SNP-sets clustering and testing in disease association studies. Plos One 10, e0135918. doi:10.1371/journal.pone.0135918
Wang, H. B., Tahara, Y., Luk, S. H. C., Kim, Y. S., Hitchcock, O. N., Macdowell Kaswan, Z. A., et al. (2020). Melatonin treatment of repetitive behavioral deficits in the Cntnap2 mouse model of autism spectrum disorder. Neurobiol. Dis. 145, 105064. doi:10.1016/j.nbd.2020.105064
Xu, H., Liu, Y., Li, Y., Diao, L., Xun, Z., Zhang, Y., et al. (2020). RadAtlas 1.0: A knowledgebase focusing on radiation-associated genes. Int. J. Radiat. Biol. 96, 980–987. doi:10.1080/09553002.2020.1761567
Yang, H. J., Kim, N., Seong, K. M., Youn, H., and Youn, B. (2013). Investigation of radiation-induced transcriptome profile of radioresistant non-small cell lung cancer A549 cells using RNA-seq. Plos One 8, e59319. doi:10.1371/journal.pone.0059319
Keywords: postsynaptic, DNA methylation, hippocampus, object recognition, space radiation
Citation: Impey S, Pelz C, Riparip L-K, Tafessu A, Fareh F, Zuloaga DG, Marzulla T, Stewart B, Rosi S, Turker MS and Raber J (2023) Postsynaptic density radiation signature following space irradiation. Front. Physiol. 14:1215535. doi: 10.3389/fphys.2023.1215535
Received: 02 May 2023; Accepted: 14 June 2023;
Published: 27 June 2023.
Edited by:
Joseph Borg, University of Malta, MaltaReviewed by:
Jean-luc Morel, Centre National de la Recherche Scientifique (CNRS), FranceCopyright © 2023 Impey, Pelz, Riparip, Tafessu, Fareh, Zuloaga, Marzulla, Stewart, Rosi, Turker and Raber. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Soren Impey, c2ltcGV5QExIUy5vcmc=; Jacob Raber, cmFiZXJqQG9oc3UuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.