- 1School of Marine Sciences, Ningbo University, Ningbo, China
- 2Laboratory for Marine Biology and Biotechnology, Pilot Qingdao National Laboratory for Marine Science and Technology (Qingdao), Qingdao, China
DM domain genes are a group of transcription factors that are integral to sexual development and its evolution in metazoans. Their functions and regulatory mechanisms are not well understood in Malacostraca (crabs and crayfish) while these sex regulators have been widely identified in the past decade. In this study, the Dmrt family was investigated in the decapod crab, Eriocheir sinensis. We find that most members of the EsDmrt family begin to enrich around the juvenile 1 stage. In reproductive organs, EsDsx1, EsDsx2, EsiDMY and EsiDmrt1a highly express in the male-specific androgenic gland (AG), while EsDmrt-like, EsDsx-like, EsDmrt11E, and EsiDmrt1b show relatively high expression in testis. Also, we find the highly aberrant expression of EsiDMY and EsiDmrt1a in the chimeric AG, strongly indicating their function in AG development. Moreover, RNA interference of EsDsx1, EsiDMY, and EsiDmrt1a results in a significant decrease in transcription of the Insulin-like androgenic hormone (IAG), respectively. Our findings suggest that Dmrt genes in E. sinensis primarily function in male sexual differentiation, especially in AG development. Besides, this study identifies two unique groups of Dmrt genes in Malacostraca: Dsx and iDmrt1. In Malacostraca Dsx, we uncover a cryptic mutation in the eight zinc motif-specific residues, which were firmly believed to be invariant across the Dmrt family. This mutation sets the Malacostraca Dsx apart from all the other Dmrt genes and implies a different way of transcriptional regulation. Genes from the iDmrt1 group show phylogenetical limitation to the malacostracan species and underwent positive selection, suggesting their highly specialized gene function to this class. Based on these findings, we propose that Dsx and iDmrt1 in Malacostraca have developed unique transcriptional regulation mechanisms to facilitate AG development. We hope that this study would contribute to our understandings of sexual development in Malacostraca and provide new insights into the evolutionary history of the Dmrt family.
1 Introduction
Animals have evolved to become sexual dimorphism in the power of sexual selection (Darwin, 1888). Despite the various sexually dimorphic traits, the upstream regulators that contribute to these discrepancies are highly conserved (Zarkower, 2001). Recent years, the center of these regulators converge on the family of DM domain genes, also known as the Doublesex and male abnormal-3 related transcription factor (Dmrt) (Volff et al., 2003; Hong et al., 2007; Kopp, 2012). The essence of the Dmrt family is a group of transcription factors that contain the DM DNA binding domain, which was firstly identified in the Drosophila melanogaster Dsx (Hildreth, 1965) and Caenorhabditis elegans mab-3 (Shen and Hodgkin, 1988) genes. The discovery of the evolutionary conservation of these two genes in structure and function soon leads to numerous identifications in other species (Raymond et al., 1998).
The Dmrt family is widely distributed across the animal kingdom and includes various groups (Wexler et al., 2014; Picard et al., 2015). In vertebrates, the Dmrt1 gene is essential to sexual development: promoting postnatal testis differentiation in mammalian (Raymond et al., 2000; Matson et al., 2011), maintaining sexual bias in avian (Smith et al., 2009), and preventing sexual transition in fish (Wu et al., 2012). Sometimes, the mutated Dmrt1 homologue DM domain on Y (DMY) (Kobayashi et al., 2004) and DM domain on W (DMW) (Yoshimoto et al., 2008) could take over sex determination due to the amino acid substitution (Ogita et al., 2020).
In invertebrates, the genes of Dmrt family are broadly divided into four groups: Dsx, Dmrt11E, Dmrt93B and Dmrt99B (Panara et al., 2019). While the functions of some of these genes are not yet well understood, the Dsx gene has been found to have some similarities in expression pattern and functionality to the vertebrate Dmrt1 gene (Matson and Zarkower, 2012). Besides, a unique group of Dmrt genes with two DM domains was specifically found in the Malacostraca. Initial research of the group was conducted on Sagmariasus verreauxi (Chandler et al., 2017) and soon extended to other species, such as E. sinensis (Cui et al., 2021), Macrobrachium rosenbergii (Abayed et al., 2019) and Scylla paramamosain (Wan et al., 2021).
The Dmrt family is a group of transcription factors with a conserved DNA-binding motif known as the DM domain (Raymond et al., 1998). The DM domain consists of two intertwined structures that chelate two atoms of zinc and binds to the minor groove of target gene in transcriptional regulation (Zhu et al., 2000). In Pancrustacea, the Dmrt11E, Dmrt93B, and Dmrt99B genes have been shown to play less important roles in sexual development compared to the Dsx gene. (Kato et al., 2011; Panara et al., 2019). In Hexapoda, Dsx is alternatively spliced into sex-specific isoforms (Chikami et al., 2022), which control the transcription of the target gene yolk protein/vitellogenin (Burtis et al., 1991), contributing to the sexual differentiation of both genders. In Branchiopoda, the targeted gene of Dsx remains unclear, but a recent transcriptomic analysis suggests a vitellogenin receptor could be the downstream element of Dsx (Nong et al., 2020). In Branchiopoda and Malacostraca, Dsx transcribes male-specifically, controls the development of male-specific structures, and is dispensable for female sexual differentiation (Wexler et al., 2019). In Malacostraca, the common regulator of male sexual differentiation is IAG (Chandler et al., 2018), which is secreted by the class-specific endocrine organ androgenic gland (AG, Ventura et al., 2011). And putative binding sites of Dmrt have been found on the promoter of IAG in many malacostracan species (Li et al., 2018; Cui et al., 2021; Wan et al., 2022; Wei et al., 2022; Wang et al., 2023). Moreover, homologue of the vitellogenin was also found in the S. paramamosain, but no Dmrt binding site was found on the promoter (Wan et al., 2022).
In this study, we investigated the Dmrt family in E. sinensis and analyzed its function in AG development. Additionally, we conducted evolutionary analyses of the Dmrt genes, reconstructed their ancestral sequences, and proposed a hypothesis for the evolution of the family. The discovery of the Malacostraca-specific Dsx and iDmrt1 groups supports the idea that the Dmrt gene family in this class may have undergone a unique evolutionary event that differs from that of Hexapoda and Branchiopoda.
2 Materials and methods
2.1 Crab sample and RNA preparation
We reasoned that male reproductive tissues were likely to express Dmrt, including splicing isoforms of these genes that might exist. To obtain different stages of testis and androgenic gland, crabs were purchased separately in August and October from a local market in Ningbo city (Zhejiang, China). Crabs were anesthetized on ice for 15 min before dissection. Testis at the spermatid stage and AG at the synthesis stage were gently dissected from crabs bought in August, and testis at the sperm stage and AG at the secretion stage were gently dissected from crabs bought in October. The fresh reproductive organs were plunged into TRIzol reagent (Invitrogen, United States) and stored at −80°C for RNA extraction. Total RNA was extracted by grinding tissue in TRIzol reagent (Invitrogen, United States) and processed following the manufacturer’s instructions. RNA quality was guaranteed by NanoOne and 1.2% agarose gel. The integrity of the RNA was determined with the Agilent 2100 Bioanalyzer (Agilent Technologies, United States). Besides, chimeric crabs, collected in local crab farms, are genetically females whereas exhibit male characteristics such as AG and male gonopods (Zhu et al., 2022).
2.2 PacBio sequencing
Four SMRT libraries were generated from collected tissues, each corresponding to a specific stage of testis or AG development: T_S2 (testis at the spermatid stage), T_S3 (testis at the sperm stage), AG_S2 (AG at the synthesis stage), and AG_S3 (AG at the secretion stage). The libraries were constructed using the SMRTbell Template Prep Kit from Pacbio (United States). The mRNA was first converted to cDNA using the SMARTer PCR cDNA Synthesis Kit from Takara (United States). The cDNA was then amplified by PCR and purified using AMPure PB beads from Pacbio (United States). Finally, the resulting templates were sequenced on a Sequel II instrument from Pacbio (United States).
2.3 Identification and characterization of the EsDmrt family
To get a comprehensive understanding of the EsDmrt family, we annotated a broad transcriptomic library of E. sinensis (annotation details are listed in Supplementary Table S1), spanning from embryonic and larva stages, normal tissues of adult crab, and reproductive organs after eyestalk ablation and IAG knockdown (accession numbers of transcriptomes used for annotation are listed in Supplementary Table S2). After annotation, we extracted all clusters that were annotated as DM domain genes in at least one database, serving as a query to conduct BLASTn against the newly published genome (GCA_013436485.1) of E. sinensis. The genomic locations of potential EsDmrt genes were marked on the general feature format (GFF) file. Then we aligned the merged SMRT library to the genome using Minimap2 v2.17 with the command “minimap2 -ax map-pb.” Corresponding full-length transcripts of EsDmrt genes were fetched (name of transcripts are listed in Supplementary Table S3). Gene structures and alternative splicing events were analyzed on IGV using the binary sequence alignment/map (BAM) file obtained from the alignment. ORFs and deduced amino acids were predicted by Expasy Translate (https://web.expasy.org/translate/). Functional domains were predicted by SMART program (http://smart.embl-heidelberg.de/). Genomic structure visualization was performed by GSDS v2.0 (http://gsds.gao-lab.org/). Visualization of chromosome location was conducted by MapChart (Voorrips, 2002). Multiple sequence alignment was conducted by MAFFT v7.407 (Katoh and Standley, 2013) and visualized by GeneDoc (Nicholas, 1997).
2.4 Molecular phylogenetic analysis
Phylogenetic analysis of DM domain genes was performed using amino acid sequences of the N-terminal DM domain (Dmrt genes used for the analysis are listed in Supplementary Table S4; amino acid sequences used for alignment can be found in Supplementary Material). We aligned the sequences using MAFFT with the L-INS-I option (most accurate model) and built a maximum-likelihood (ML) tree with 1,000 bootstrap values in the IQ-TREE software (Minh et al., 2020). The substitution model was automatically selected using “-MPF” and LG+I+G4 was the best-fit. Finally, the tree was visualized and annotated using GGTREE (Yu et al., 2017).
2.5 Gene expression analysis using transcriptomic data
To investigate the function of the EsDmrt family in sexual development, we conducted a wide-ranging analysis of the expression levels in gonads, early-stage transcriptomic library (Illumina data) and chimeric tissues (NanoPore data). Illumina data (SRA numbers are listed in Supplementary Table S2) were generated from our previous studies or downloaded from NCBI and then trimmed by Trimmomatic v0.39. The clean data were mapped to the genome with Hisat2 v2.1.0. Consequent BAM and GFF files were used to calculate the transcripts per million (TPM) values by Stringtie v2.1.3. NanoPore data (Unpublished data from our lab) were aligned to the genome using minimap2 v2.17 after quality control and consequent BAM files were used to calculate the TPM values as described above. All TPM values were presented as mean ± SD. Statistical analyses were performed by the one-way analysis of variance (ANOVA) or T-test using SPSS 28 software. p-values less than 0.05 and 0.01 were considered significant and extremely significant. Each group contained three biological repetitions. The results were visualized by ggplot2 (https://ggplot2.tidyverse.org).
2.6 RNAi and RT-qPCR
The siRNA of EsDsx1M1, EsDsx1M2, EsiDmrt1a, EsiDMYL and siControl were chemically synthesized by GenePharma (Shanghai, China) (sequences of the siRNAs and control siRNA are listed in Supplementary Table S5). Crab saline was synthesized in the lab (2.570 g NaCl, 0.084 g KCl, 0.148 g CaCl2, 0.248 g MgCl2, 0.327 g Na2SO4, 0.238 g HEPES, pH 7.4, total 100 mL). The siRNA was incubated with GP-transfect for 30 min before injection. The experiment included three treatments, namely, siRNAi, siControl and saline, with each treatment performed in six replicates (n = 6). A total of 40 μg/individual siRNA was injected into the fifth walking leg’s sinus. After 24 h of injection, only AG was collected in three treatments. Animal dissection and RNA extraction were conducted in the same way as described above. RT-qPCR was performed to investigate the expression level. Dmrt and IAG expression relative to the siControl and saline were determined by the 2−ΔΔCt method. The Shapiro-Wilk test and Levene’s test were used to check the normality and homogeneity of variance assumptions, respectively. Independent sample T-tests were used to analyze Dmrt and IAG expression between RNAi and siControl groups. p-values less than 0.05 and 0.01 were considered significant and extremely significant. Results of statistical tests are listed in Supplementary Table S6.
2.7 Consensus analysis of DM domain
To gain insights into the conservation and variation of the DM domain across different species and groups including Dmrt1 from various metazoans, four conserved groups from Pancrustacea and iDmrt1 from Malacostraca, we conducted amino acid sequence alignments and showed their conservation using Weblogo v3.7.4 (http://weblogo.threeplusone.com/). Accession numbers of sequences used for the analysis are listed in Supplementary Table S7.
2.8 Positive selection analysis and ancestral sequence reconstruction
The ratio of nonsynonymous (dN) and synonymous (dS) substitution rates (ω = dN/dS) were evaluated using codeML in PAML version 4.9 (Yang, 2007). For the iDmrt1 group, we used site models that assume different ω varies among sites but not branches, including model M0 (one ratio), M1a (neutral), M2a (selection), M3 (discrete), M7 (beta), and M8 (beta and ω > 1) (Yang et al., 2000). The reference tree was built first (tree diagram is shown in Supplementary Figure S1; Dmrt genes used for the analysis are listed in Supplementary Table S8; nucleotide sequences used for the alignment can be found in Supplementary Material), then pairwise comparisons of M0 versus M3, M1a versus M2a, and M7 versus M8 were used to perform likelihood ratio test (LRT). For the Dsx group, we used the branch-site model that assumes ω to vary both among sites and branches, to compare the model A against the model Null (fixed ω of 1). Also, we built a reference tree first (tree diagram is shown in Supplementary Figure S2; Dmrt genes used for the analysis are listed in Supplementary Table S9; nucleotide sequences used for the alignment can be found in Supplementary Material), then the comparison of model A versus model Null was used to perform LRT. Both significance tests were performed using the χ2 distribution with degrees of freedom of 2 and 1 for iDmrt1 and Dsx groups, respectively. If the results of LRT showed significance, Bayes empirical Bayes (BEB) analysis would be conducted to identify positively selected sites with a posterior probability greater than 0.95. Meanwhile, ancestral sequences of Dsx were reconstructed using the empirical Bayes approach implemented in PAML (Yang et al., 1995).
2.9 Protein structure prediction and motif prediction
The reconstructed ancestral sequences of Dsx were utilized as inferred protein sequences to model structural domains using the Alphafold2-based algorithm (ColabFold: Mirdita et al., 2022) with the default settings. The accuracy of predicted models was assessed using the predicted Local distance difference test (plDDT) score, which was automatically calculated on the ColabFold platform (diagram of plDDT score is shown in Supplementary Figure S3). The PyMOL Molecular Graphics System, Version 2.0 (Schrödinger, 2015) was used to visualize the 3D models of the predicted structures and we highlighted the zinc motif-specific residues cysteine and histidine with red and yellow, respectively. Then we used the online software ZincExplorer to detect the zinc binding ability of the reconstructed amino acids. Moreover, we modelled the Dsx protein from several malacostracan species using Swiss-Model.
3 Results
3.1 PacBio sequencing of Eriocheir sinensis
We constructed four SMRT libraries (T_S2, T_S3, AG_S2 and AG_S3) using testis at spermatid and sperm stages, and AG at synthesis and secretion stages. We obtained a total of 171,436, 348,229, 187,041 and 204,937 circular consensus sequences from raw reads, of which 58,046,178,242,118,195 and 116,436 were identified as full length non-chimeric reads. After clustering, we identified 35,011, 103,219, 54,833 and 68,126 high-quality isoforms with an average length of 1,092, 1,633, 904 and 1,429 bp, respectively. Finally, four libraries were merged, resulting in a total of 197,160 nonredundant full-length transcript. The complete datasets were deposited in the NCBI Sequence Read Archive (SRA) under accession numbers SRR24067590, SRR24067589, SRR24067592 and SRR24067591.
3.2 Identification and characterization of EsDmrt genes
Our study identified nine members of the Dmrt family in E. sinensis, including EsDsx1, EsDsx2, EsDsx-like, EsiDmrt1a, EsiDmrt1b, EsiDMY, EsDmrt-like, EsDmrt11E and EsDmrt93B (Figure 1A). The nine genes exhibit highly conserved DM domains, except for EsDsx1 and EsDsx2, in which a point mutation was both detected in the zinc motif-specific residues (Figure 1B). EsDsx1 and EsiDMY are alternatively spliced (Figure 1A), and no sex-specific splicing isoforms were detected. While we found two EsDsx1 isoforms (EsDsx1M1 and EsDsx1M2) specifically expressed in AG and one isoform (EsDsx1C) that was commonly expressed (Supplementary Figure S4). Splicing isoforms of EsiDMY encoded three proteins of different lengths (EsiDMYS, EsiDMYM and EsiDMYL), and their presence was confirmed by RT-PCR (Supplementary Figure S5). Besides, most EsDmrt genes were located on a single chromosome, while EsDsx1 and EsDsx2 were adjacent to each other (Figure 1C).
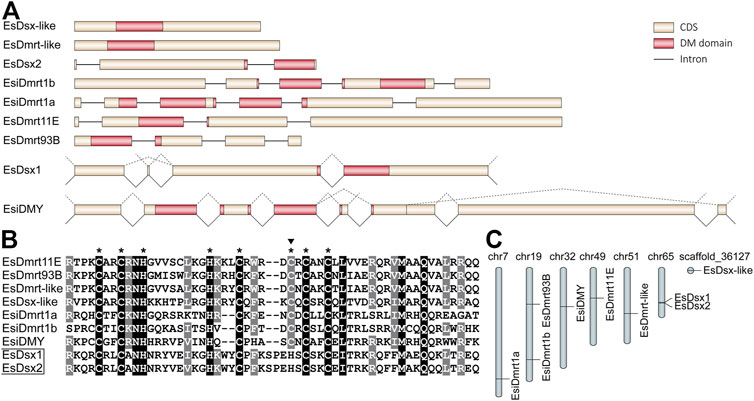
FIGURE 1. Characterization of Dmrt family genes in E. sinensis. (A) Structure features of nine EsDmrt genes. All elements are shown to scale except for introns. The alternative splicing events of EsDsx1 and EsiDMY are displayed above the genes with dotted lines. (B) Alignment of deduced amino acid sequences of DM domains (N-terminal domain of EsiDmrt1a, EsiDmrt1b and EsiDMY are used in this alignment). Identical amino acids are highlighted in black and similar ones in grey. Eight conserved motif-specific residues are indicated with asterisks and the point mutation in EsDsx1 and EsDsx2 is indicated with a top solid triangle. (C) Genome loci of nine EsDmrt genes. Except for the EsDsx-like, all genes have their clear chromosome locations.
3.3 Molecular phylogenetic analysis of DM domain genes
We built a ML tree based on 91 DM domain genes to reconstruct the phylogenetic relationship of the EsDmrt family (Figure 2). Our analysis revealed that five EsDmrt genes were distributed in four conserved clades of Pancrustacea: Dsx, Dmrt11E, Dmrt93B, and Dmrt99B. Notably, the Dsx clade was classified into three reliable subclades, including Hexapoda Dsx clade, Branchiopoda Dsx clade and Malacostraca Dsx clade. Besides, we found that three EsDmrt genes fall into a monophyletic branch consisting of iDmrt1 orthorlogues from malacostracan species. The phylogeny of EsDsx-like remained unclear in our study.
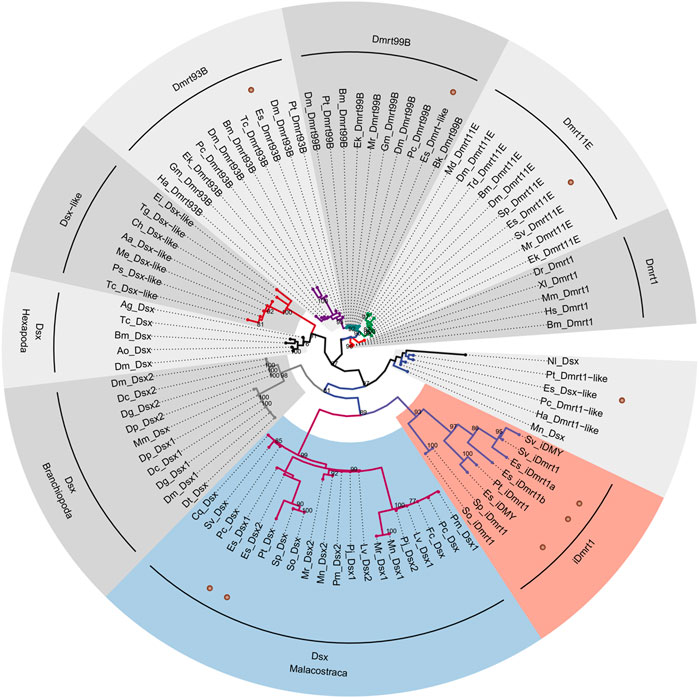
FIGURE 2. Molecular phylogeny of Dmrt family genes in E. sinensis. Phylogenetic analysis was based on amino acid sequences of DM domain and performed by IQ-TREE following multiple sequence alignment using MAFFT. The maximum-likelihood method was applied. The 91 taxonomic units conducted for this analysis are listed in Supplementary Table S4. Numerical value on each node indicates bootstrap value that is >70%. EsDmrt genes are indicated with brown dots. Malacostraca Dsx and iDmrt1 groups are colored in light steel blue and light coral.
3.4 Expression pattern of EsDmrt genes in three transcriptomic libraries
We analyzed expression patterns of the EsDmrt family using transcriptomic data from three libraries. The embryonic and larval transcriptomes show that most EsDmrt genes start to enrich around the juvenile 1 stage (Figure 3A), while EsDsx1M1, EsDsx1M2 and EsDsx2 show relatively high expression in gastrula and blastula stages. In reproductive organs, EsDsx1, EsDsx2, EsiDMY and EsiDmrt1a are co-expressed predominantly in male organs, particularly in two stages of AG, where their expression levels are significantly higher than testis and ovary (Figure 3B, one-way ANOVA, p < 0.05). Besides, the overall expression levels of EsiDmrt1b, EsDmrt11E, EsDmrt-like and EsDsx-like are relatively low, but they still show a significantly higher expression in testis (One-way ANOVA, p < 0.05). The expression level of EsDmrt93B is too low to show. In chimeric crabs, we find a significantly higher expression of EsiDMY in the chimeric AG compared to the normal ones (Figure 3C, independent sample T-test, *p < 0.05). Additionally, EsDsx1C, EsDsx2 and EsiDMYL show significantly higher expression levels in the chimeric eyestalk compared to the normal ones (independent sample T-test, *p < 0.05).
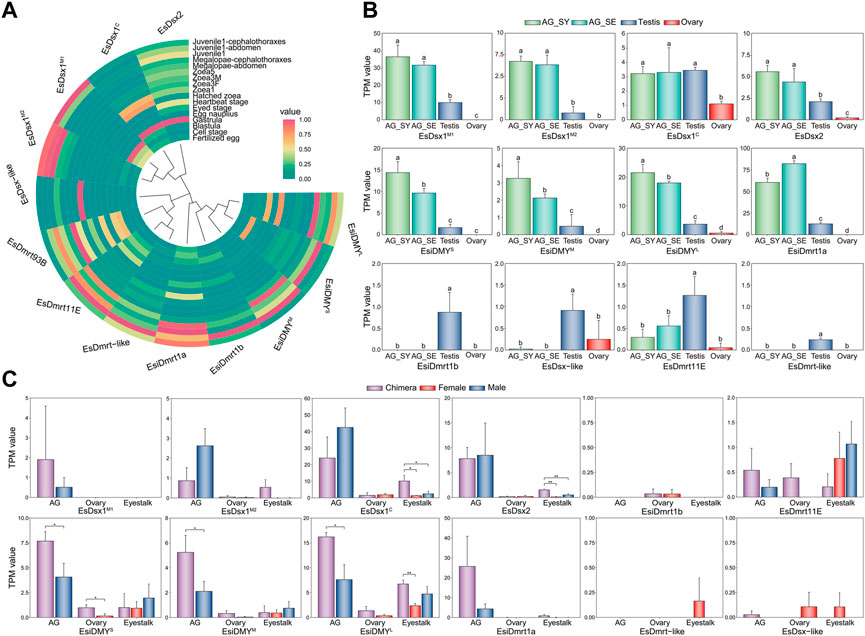
FIGURE 3. Expression level of the Dmrt family genes in three different transcriptomic libraries of E. sinensis. (A) Expression pattern of EsDmrt genes in embryonic, larval and juvenile stages. The heatmap was shown as normalized TPM value. (B) Expression pattern of EsDmrt genes (including splicing isoforms) in reproductive organs (AG, testis and ovary). The bar plot was shown as TPM value. Vertical bars represented mean ± SD, n = 3. Results were analyzed by one-way ANOVA and different letters (a-d) above the bars presented significant differences between groups (p < 0.05). AG_SY, androgenic gland at synthesis stage; AG_SE, androgenic gland at secretion stage. (C) Expression pattern of the EsDmrt genes in normal and chimeric E. sinensis. The bar plot was shown as TPM value. Vertical bars represented mean ± SD, n = 3. Tissues from left to right: AG-androgenic gland (chimeric, normal male); ovary (chimeric, normal female); eyestalk (chimeric, normal female and male). Tissue and organ from chimeric, normal female and male were colored with purple, red and blue. Vertical bars represented mean ± SD, n = 3. Results were analyzed by independent T-test comparing chimeric and normal tissue and organ (*p < 0.05, **p < 0.01).
3.5 Regulation of EsDmrt for EsIAG expression
We conducted siRNA interference to investigate the role of the EsDmrt family in AG development, with EsIAG serving as a molecular marker. The results prove successful silencing of EsDsx1M1, EsDsx1M2, EsiDmrt1a and EsiDMYL genes (Figure 4A, independent sample T-test, p = 4.11E-03 in siEsDsx1M1 group, 2.00E-03 in siEsDsx1M2 group, 4.97E-02 in siEsiDmrt1a group, and 3.80E-02 in siEsiDMYL group). Furthermore, all RNAi groups show a significant decrease in EsIAG expression (Figure 4B, independent sample T-test, p = 4.40E-02 in siEsDsx1M1 group, 3.00E-03 in siEsDsx1M2 group, 3.50E-02 in siEsiDmrt1a group and 3.00E-03 in siEsiDMYL group).
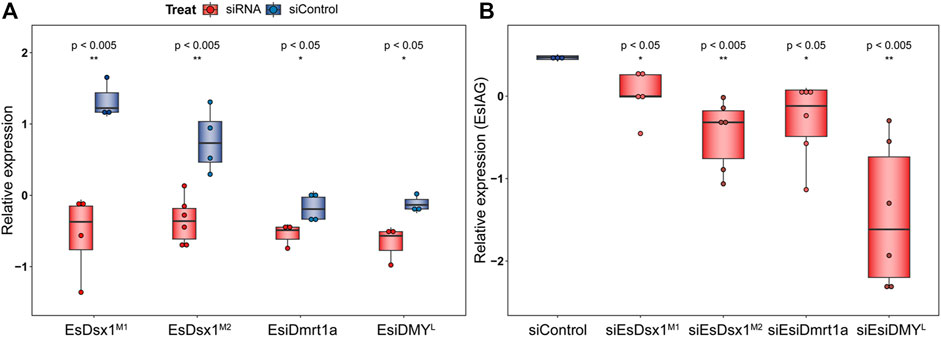
FIGURE 4. Function of Dmrt genes for IAG expression in E. sinensis. Transcript abundance was measured through RT-qPCR. Box plot is show as log-scale relative values of expression levels to the reference gene, Esβ-actin. Each dot represents the expression level of each individual. Independent T-test was performed to ascertain the difference between siRNA and siControl groups, with significance levels indicated (*p < 0.05, **p < 0.01) Statistical results are listed in the Supplementary Table S6. Expression levels of EsDmrt genes (A) and EsIAG gene (B) in RNAi and control groups are shown separately.
3.6 Consensus analysis of DM domain in different Dmrt genes
In the analysis of the conservation of DM domains (Figure 5). We find that Dsx and iDmrt1 groups exhibit great variability in the amino acids, which is different from the conservation observed in Dmrt1, Dmrt11E, Dmrt93B and Dmrt99B groups. The eight zinc motif-specific residues remain invariant in all tested Dmrt genes, except for the Malacostraca Dsx, where a cryptic mutation appears due to a substitution, from cysteine to histidine. Furthermore, there are some certain amino acids, including positions 9, 15, 16, 37, 38, and 53 that also distinguish the Malacostraca Dsx from other Dmrt genes.
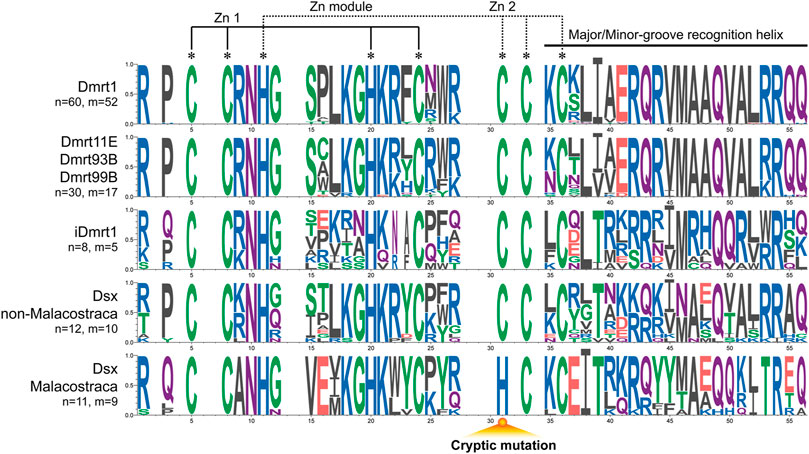
FIGURE 5. Consensus analysis of DM domains of the Dmrt gene family. Less conserved points are discarded. The cryptic mutation in Malacostraca Dsx is annotated below. Amino acids are colored according to their chemical properties: red (D, E) for acidic, green (C, G, S, T, Y) for polar, blue (H, K, R) for basic, black (A, F, I, L, M, V, W) for hydrophobic, and purple (Q, N) for neutral. n: number of unique sequences. m: number of different species.
3.7 Selection analysis of iDmrt1 group
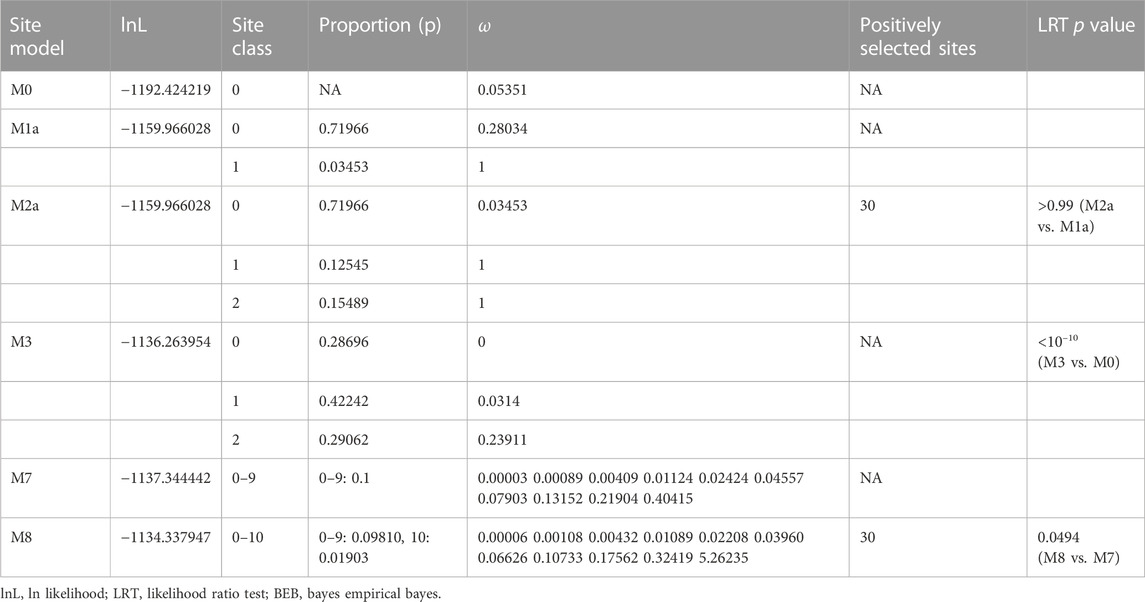
We used the site codon model in PAML to investigate ongoing selection pressure on the iDmrt1 group. The selection test between M8 and M7 site models shows evidence of positive selection for the iDmrt1 group (LRT, p = 0.0494, Table 1). The BEB analysis demonstrates that position 30 on the DM domain is under positive selection with a posterior probability greater than 0.95 (Table 1). Additionally, the comparison between M3 and M0 site models suggests variable ω values among different positions (LRT, p < 10^−10, Table 1).
3.8 Evolutionary history of Dsx
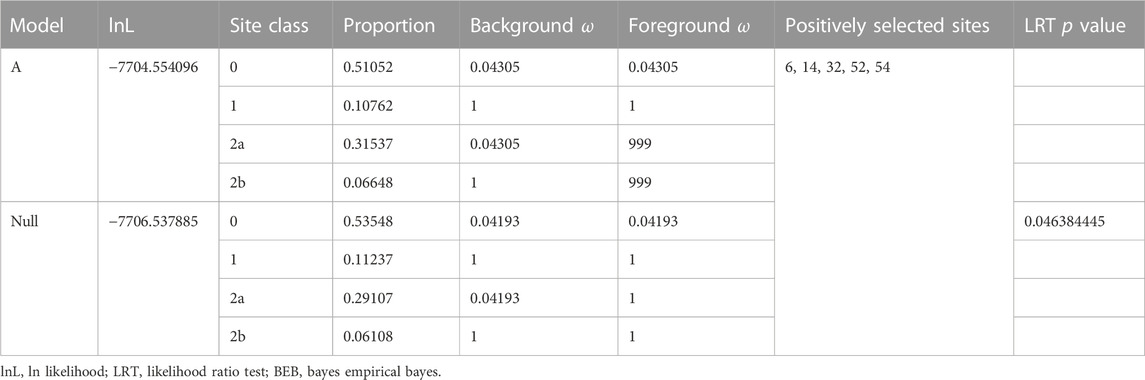
The LRT result shows that Model A fits the reference tree significantly better than the Model Null (LRT, p = 0.046384445, Table 2), indicating selection pressure acting on the foreground branch. The BEB analysis demonstrates that five positions on the DM domain, including the cryptic mutation shown in Figure 5, are under positive selection. Ancestral sequence reconstruction reveals that the cryptic mutation is also present in the common ancestor of Pancrustacea, but not in the common ancestor of Hexapoda and Branchiopoda (Figure 6A). According to the prediction of ZincExplorer (Supplementary Figure S6), the Dsx protein in the common ancestor of Pancrustacea could bind only one zinc atom (Figure 6B), which is consistent with the three-dimension structures of Malacostraca Dsx modelled using Swiss-Model (Supplementary Figure S7).
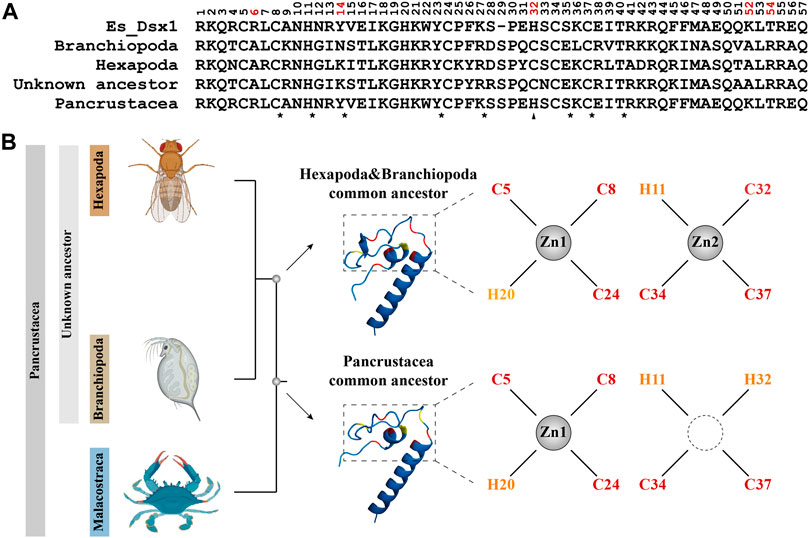
FIGURE 6. Ancestral sequence reconstruction of the DM domain from Dsx in Pancrustacea. (A) Alignment of the ancestral amino acid sequences of the DM domain from different common ancestor. Positions assumed to be positively selected are colored in red. Conserved residues are indicated with asterisks and mutation is indicated with a triangle. Es_Dsx1, Dsx1 of E. sinensis, is shown as a reference. (B) Predicted protein structures of Dsx in different common ancestors. The phylogenetic relationship is based on topology from Rota-Stabelli et al. (2013). 3D images in the midst indicate modelled proteins of DM domains with the motif-specific residues highlighted in red (cysteine) and yellow (histidine). Chelation of zinc atoms is visualized as a result of ZincExplore.
4 Discussion
In our previous studies, we have confirmed the names of EsDmrt11E (Du et al., 2019) and EsiDMY (Cui et al., 2021). EsDmrt-like and EsDsx-like were reported in Zhang and Qiu (2010) and Wang et al. (2023), respectively. In this study, we rename three genes previously identified in Du et al. (2019) based on their phylogenetic relationships, which are EsDmrt93B, EsiDmrt1a and EsiDmrt1b (formerly known as EsDmrt3, EsIdmrt1-1 and EsDsx2). Furthermore, we find the duplicated Dsx genes, EsDsx1 and EsDsx2, which form a tandem gene cluster on the genome. As a result, we identify a total of nine members of the Dmrt family in the crab.
In invertebrates, Dmrt genes are known to play a crucial role in sexual differentiation, driving sex-specific development in varied tissues and cell types (Matson and Zarkower, 2012). In crustaceans, Dmrt genes have been found to transcribe in a strongly male-biased way, indicating their specific involvement in male morphogenesis (Wexler et al., 2019). Our transcriptomic analyses of the Dmrt family in E. sinensis support and expand on previous findings, suggesting that the EsDmrt family primarily functions in male sexual differentiation, specifically promoting the development of testis and AG. Previous studies have meticulously described the function of EsDmrt-like (Zhang and Qiu, 2010) and EsDsx-like (Wang et al., 2023) during spermatogenesis. In our research, we find the abundant expression of EsDsx1, EsDsx2, EsiDmrt1a and EsiDMY in AG, as well as the aberrant transcription of EsiDmrt1a and EsiDMY in the chimeric AG. Furthermore, RNA knockdown on the transcripts from EsDsx1, EsiDmrt1a and EsiDMY could result in a significant decrease in EsIAG transcription. Taken together, our findings propose that in E. sinensis, both genes from Dsx and iDmrt1 groups may function in AG development.
In Malacostraca, Dsx genes have been extensively studied and are known to upregulate IAG in various species, such as F. chinensis (Li et al., 2018), S. paramamosain (Wan et al., 2022) and P. monodon (Wei et al., 2022). And iDmrt1 genes has also showed closed relationship with AG in species such as M. rosenbergii (Abayed et al., 2019), where the MroiDmrt1b and MroiDmrt1c genes are highly enriched in AG, and Portunus trituberculatus, where the PtiDmrt1 gene upregulates the PtIAG expression (Wang et al., 2020). These findings suggest that the involvement of both Dsx and iDmrt1 genes in AG development might be a common feature of Malacostraca.
Our phylogenetic tree showed that the Dsx and iDmrt1 genes in Malacostraca form two distinct monophyletic clades. On the one hand, the iDmrt1 gene was first identified in the S. verreauxi (Ventura et al., 2020), and subsequently identified solely in malacostracan species. Our selection analysis (Table 1) reveals that the iDmrt1 group is subjected to positive selection, suggesting a unique function that is specific to the Malacostraca, i.e., promoting AG development. On the other hand, the Malacostraca Dsx shows unique amino acid residues at position 9, 38 and 53 (Figure 5). These positions have been confirmed to play critical roles in DNA binding and homodimerization (Murphy et al., 2015). Moreover, and perhaps more significantly, an invariant cysteine residue in the zinc binding module, known to coordinate with zinc atom (Zhu et al., 2000), is replaced with a histidine only in the Malacostraca Dsx (Figure 5). Therefore, we suggest that the Malacostraca Dsx may not bind to DNA directly but instead form heterodimers with the iDmrt1. Such binding mode also rationalizes their co-expression in AG (Figure 2B), which is similar to the situation observed in vertebrates, where Dmrt3 heterodimerizes with the co-expressed Dmrt1 in testis (Murphy et al., 2007).
Selection analysis (Table 2) and ancestral sequence reconstruction (Figure 6) place Malacostraca Dsx as an outgroup to Hexapoda and Branchiopoda, which is consistent with previous studies on Hox (Averof and Akam, 1995) and elongation factor-1 alpha (Regier and shultz, 1997) genes. Based on these findings, we incorporated Malacostraca into the stepwise evolutionary history proposed by Wexler et al. (2019) and Chikami et al. (2022) (Figure 7) that Dsx has become essential for male development in the common ancestor of Pancrustacea; then during the split of Malacostraca and the common ancestor of Branchiopoda and Hexapoda in the late Cambrian, a selective event, which might result from ecological or physiological differences (Glenner et al., 2006), either builds or reserves the unique regulatory mode of Dmrt genes in Malacostraca.
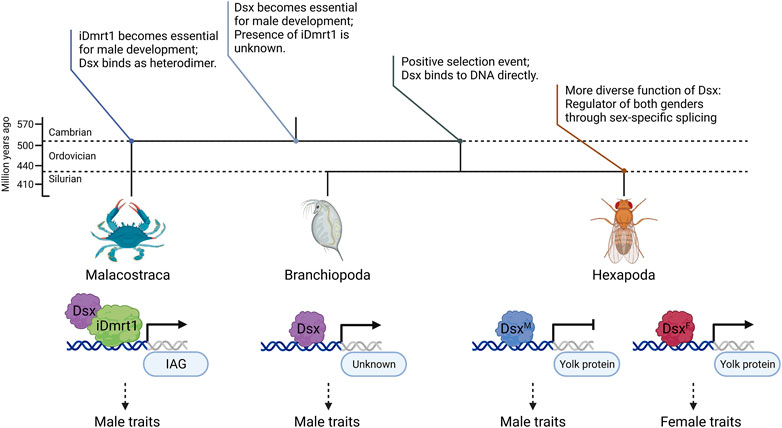
FIGURE 7. Diagram of the evolutionary history of Dsx and iDmrt1 proposed in this study. The upper topological structure and annotations describe possible statuses of Dmrt in different common ancestors. The lower part compares the transcriptional regulation of Dsx in Malacostraca, Hexapoda and Branchiopoda. Previous research has suggested that heterodimer formation of Dmrt proteins may contribute to transcriptional regulation (Murphy et al., 2007). Mutations in the DM domain and the lack of an oligomerization domain (Zheng et al., 2020) suggest that Malacostraca Dsx may not directly bind to, but rather heterodimerize on DNA with another Dmrt protein, which, according to our evolutionary analysis, could be iDmrt1. Figure created using BioRender.
In this study, we investigated the Dmrt family in a decapod crab and explored its function in AG development, which extend our knowledge of sexual development mechanisms in this species. Besides, we proposed a hypothesis of transcriptional regulation based on the finding of the intriguing Dsx and iDmrt1 groups in Malacostraca. Further investigation would be necessary to test our hypothesis and unravel the evolutionary history of DM domain genes.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
PZ: conceptualization, investigation, methodology, drawing, writing—original draft. YY: supervision, writing—review and editing. YX: methodology and software supporting. ZC: supervision, funding, writing—review and editing. All authors contributed to the article and approved the submitted version.
Funding
The work was supported by the National Natural Science Foundation of China (32072964), the National Key R and D Program of China (2018YFD0900303), and the Ten Thousand Talents Program.
Acknowledgments
We would like to express our sincere gratitude to Prof. David Zarkower for providing valuable insights on the transcriptional regulation of DM domain genes. Our gratitude also extends to Prof. Yilei Wang for generously providing the amino acid sequences of Malacostraca Dsx.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2023.1201846/full#supplementary-material
References
Abayed, F. A. A., Manor, R., Aflalo, E. D., and Sagi, A. (2019). Screening for Dmrt genes from embryo to mature Macrobrachium rosenbergii prawns. Gen. Comp. Endocrinol. 282, 113205. doi:10.1016/j.ygcen.2019.06.009
Averof, M., and Akam, M. (1995). Hox genes and the diversification of insect and crustacean body plans. Nature 376 (6539), 420–423. doi:10.1038/376420a0
Burtis, K. C., Coschigano, K. T., Baker, B. S., and Wensink, P. C. (1991). The doublesex proteins of Drosophila melanogaster bind directly to a sex-specific yolk protein gene enhancer. EMBO J. 10 (9), 2577–2582. doi:10.1002/j.1460-2075.1991.tb07798.x
Chandler, J. C., Elizur, A., and Ventura, T. (2018). The decapod researcher’s guide to the galaxy of sex determination. Hydrobiologia 825, 61–80. doi:10.1007/s10750-017-3452-4
Chandler, J. C., Fitzgibbon, Q. P., Smith, G., Elizur, A., and Ventura, T. (2017). Y-linked iDmrt1 paralogue (iDMY) in the Eastern spiny lobster, Sagmariasus verreauxi: The first invertebrate sex-linked Dmrt. Dev. Biol. 430 (2), 337–345. doi:10.1016/j.ydbio.2017.08.031
Chikami, Y., Okuno, M., Toyoda, A., Itoh, T., and Niimi, T. (2022). Evolutionary history of sexual differentiation mechanism in insects. Mol. Biol. Evol. 39, msac145. doi:10.1101/2021.11.30.470672
Cui, Z. X., Liu, Y., Yuan, J. B., Zhang, X. J., Ventura, T., Ma, K. Y., et al. (2021). The Chinese mitten crab genome provides insights into adaptive plasticity and developmental regulation. Nat. Commun. 12, 1–13. doi:10.1038/s41467-021-22604-3
Darwin, C. (1888). The descent of man, and selection in relation to sex. London: Published material.
Du, J., Liu, Y., Song, C. W., and Cui, Z. X. (2019). Discovery of sex-related genes from embryonic development stage based on transcriptome analysis in Eriocheir sinensis. Gene 710, 1–8. doi:10.1016/j.gene.2019.05.021
Glenner, H., Thomsen, P. F., Hebsgaard, M. B., Sørensen, M. V., and Willerslev, E. (2006). Evolution. The origin of insects. Science 314 (5807), 1883–1884. doi:10.1126/science.1129844
Hildreth, P. E. (1965). Doublesex, a recessive gene that tranforms both males and females of Drosophila into intersexes. Genetics 51 (4), 659–678. doi:10.1093/genetics/51.4.659
Hong, C. S., Park, B. Y., and Saint-Jeannet, J. P. (2007). The function of dmrt genes in vertebrate development: It is not just about sex. Dev. Biol. 310 (1), 1–9. doi:10.1016/j.ydbio.2007.07.035
Kato, Y., Kobayashi, K., Watanabe, H., and Iguchi, T. (2011). Environmental sex determination in the branchiopod crustacean Daphnia magna: Deep conservation of a doublesex gene in the sex-determining pathway. PLoS Genet. 7 (3), e1001345. doi:10.1371/journal.pgen.1001345
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30 (4), 772–780. doi:10.1093/molbev/mst010
Kobayashi, T., Matsuda, M., Kajiura-Kobayashi, H., Suzuki, A., Saito, N., Nakamoto, M., et al. (2004). Two DM domain genes, DMY and DMRT1, involved in testicular differentiation and development in the medaka. Oryzias Latipes. Dev. Dyn. 231 (3), 518–526. doi:10.1002/dvdy.20158
Kopp, A. (2012). Dmrt genes in the development and evolution of sexual dimorphism. Trends Genet. 28 (4), 175–184. doi:10.1016/j.tig.2012.02.002
Li, S., Li, F., Yu, K., and Xiang, J. (2018). Identification and characterization of a doublesex gene which regulates the expression of insulin-like androgenic gland hormone in Fenneropenaeus chinensis. Gene 649, 1–7. doi:10.1016/j.gene.2018.01.043
Matson, C. K., Murphy, M. W., Sarver, A. L., Griswold, M. D., Bardwell, V. J., and Zarkower, D. (2011). DMRT1 prevents female reprogramming in the postnatal mammalian testis. Nature 476 (7358), 101–104. doi:10.1038/nature10239
Matson, C. K., and Zarkower, D. (2012). Sex and the singular DM domain: Insights into sexual regulation, evolution and plasticity. Nat. Rev. Genet. 13 (3), 163–174. doi:10.1038/nrg3161
Minh, B. Q., Schmidt, H. A., Chernomor, O., Schrempf, D., Woodhams, M. D., Von Haeseler, A., et al. (2020). IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37 (5), 1530–1534. doi:10.1093/molbev/msaa015
Mirdita, M., Schütze, K., Moriwaki, Y., Heo, L., Ovchinnikov, S., and Steinegger, M. (2022). ColabFold: Making protein folding accessible to all. Nat. Methods 19 (6), 679–682. doi:10.1038/s41592-022-01488-1
Murphy, M. W., Lee, J. K., Rojo, S., Gearhart, M. D., Kurahashi, K., Banerjee, S., et al. (2015). An ancient protein-DNA interaction underlying metazoan sex determination. Nat. Struct. Mol. Biol. 22 (6), 442–451. doi:10.1038/nsmb.3032
Murphy, M. W., Zarkower, D., and Bardwell, V. J. (2007). Vertebrate DM domain proteins bind similar DNA sequences and can heterodimerize on DNA. BMC Mol. Biol. 8 (1), 58–14. doi:10.1186/1471-2199-8-58
Nicholas, K. B. (1997). GeneDoc: A tool for editing and annotating multiple sequence alignments. doi:10.1186/1471-2105-8-381
Nong, Q. D., Matsuura, T., Kato, Y., and Watanabe, H. (2020). Two Doublesex1 mutants revealed a tunable gene network underlying intersexuality in Daphnia magna. PLoS One 15 (8), e0238256. doi:10.1371/journal.pone.0238256
Ogita, Y., Mawaribuchi, S., Nakasako, K., Tamura, K., Matsuda, M., Katsumura, T., et al. (2020). Parallel evolution of two dmrt1-derived genes, dmy and dm-W, for vertebrate sex determination. iScience 23 (1), 100757. doi:10.1016/j.isci.2019.100757
Panara, V., Budd, G. E., and Janssen, R. (2019). Phylogenetic analysis and embryonic expression of panarthropod Dmrt genes. Front. Zool. 16 (1), 23–18. doi:10.1186/s12983-019-0322-0
Picard, M. A. L., Cosseau, C., Mouahid, G., Duval, D., Grunau, C., Toulza, E., et al. (2015). The roles of Dmrt (Double sex/Male-abnormal-3 Related Transcription factor) genes in sex determination and differentiation mechanisms: Ubiquity and diversity across the animal kingdom. C. R. Biol. 338 (7), 451–462. doi:10.1016/j.crvi.2015.04.010
Raymond, C. S., Murphy, M. W., O'Sullivan, M. G., Bardwell, V. J., and Zarkower, D. (2000). Dmrt1, a gene related to worm and fly sexual regulators, is required for mammalian testis differentiation. Genes Dev. 14 (20), 2587–2595. doi:10.1101/gad.834100
Raymond, C. S., Shamu, C. E., Shen, M. M., Seifert, K. J., Hirsch, B., Hodgkin, J., et al. (1998). Evidence for evolutionary conservation of sex-determining genes. Nature 391 (6668), 691–695. doi:10.1038/35618
Regier, J. C., and Shultz, J. W. (1997). Molecular phylogeny of the major arthropod groups indicates polyphyly of crustaceans and a new hypothesis for the origin of hexapods. Mol. Biol. Evol. 14 (9), 902–913. doi:10.1093/oxfordjournals.molbev.a025833
Rota-Stabelli, O., Daley, A. C., and Pisani, D. (2013). Molecular timetrees reveal a Cambrian colonization of land and a new scenario for ecdysozoan evolution. Curr. Biol. 23 (5), 392–398. doi:10.1016/j.cub.2013.01.026
Schrödinger, L. L. C. (2015). The PyMOL molecular graphics system. Version 1, 8. doi:10.18356/0cd8e28e-zh
Shen, M. M., and Hodgkin, J. (1988). mab-3, a gene required for sex-specific yolk protein expression and a male-specific lineage in C. elegans. Cell 54 (7), 1019–1031. doi:10.1016/0092-8674(88)90117-1
Smith, C. A., Roeszler, K. N., Ohnesorg, T., Cummins, D. M., Farlie, P. G., Doran, T. J., et al. (2009). The avian Z-linked gene DMRT1 is required for male sex determination in the chicken. Nature 461 (7261), 267–271. doi:10.1038/nature08298
Ventura, T., Chandler, J. C., Nguyen, T. V., Hyde, C. J., Elizur, A., Fitzgibbon, Q. P., et al. (2020). Multi-tissue transcriptome analysis identifies key sexual development-related genes of the ornate spiny lobster (Panulirus ornatus). Genes 11 (10), 1150. doi:10.3390/genes11101150
Ventura, T., Rosen, O., and Sagi, A. (2011). From the discovery of the crustacean androgenic gland to the insulin-like hormone in six decades. Gen. Comp. Endocrinol. 173 (3), 381–388. doi:10.1016/j.ygcen.2011.05.018
Volff, J. N., Zarkower, D., Bardwell, V. J., and Schartl, M. (2003). Evolutionary dynamics of the DM domain gene family in metazoans. J. Mol. Evol. 57, S241–S249. doi:10.1007/s00239-003-0033-0
Voorrips, R. (2002). MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 93 (1), 77–78. doi:10.1093/jhered/93.1.77
Wan, H. F., Zhong, J. Y., Zhang, Z. P., Zou, P. F., Zeng, X. Y, and Wang, Y. L. (2021). Discovery of the Dmrt gene family members based on transcriptome analysis in mud crab Scylla paramamosain. Gene 784, 145576. doi:10.1016/j.gene.2021.145576
Wan, H., Zhong, J., Zhang, Z., Sheng, Y., and Wang, Y. (2022). Identification and functional analysis of the doublesex gene in the mud crab Scylla paramamosain. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 266, 111150. doi:10.1016/j.cbpa.2022.111150
Wang, L., Liu, Z. Q., Ma, K. Y., Tao, J. R., Fang, X., and Qiu, G. F. (2023). A testis-specific gene doublesex is involved in spermatogenesis and early sex differentiation of the Chinese mitten crab Eriocheir sinensis. Aquaculture 569, 739401. doi:10.1016/j.aquaculture.2023.739401
Wang, Y., Xie, X., Wang, M., Zheng, H., Zheng, L., and Zhu, D. (2020). Molecular characterization and expression analysis of the inverbrate Dmrt1 homologs in the swimming crab, Portunus trituberculatus (Miers, 1876) (Decapoda, Portunidae). Crustaceana 93 (8), 851–866. doi:10.1163/15685403-bja10025
Wei, W. Y., Huang, J. H., Zhou, F. L., Yang, Q. B., Li, Y. D., Jiang, S., et al. (2022). Identification and expression analysis of Dsx and its positive transcriptional regulation of IAG in black tiger shrimp (Penaeus monodon). Int. J. Mol. Sci. 23 (20), 12701. doi:10.3390/ijms232012701
Wexler, J., Delaney, E. K., Belles, X., Schal, C., Wada-Katsumata, A., Amicucci, M. J., et al. (2019). Hemimetabolous insects elucidate the origin of sexual development via alternative splicing. eLife 8, e47490. doi:10.7554/eLife.47490
Wexler, J. R., Plachetzki, D. C., and Kopp, A. (2014). Pan-metazoan phylogeny of the DMRT gene family: A framework for functional studies. Dev. Genes Evol. 224, 175–181. doi:10.1007/s00427-014-0473-0
Wu, G. C., Chiu, P. C., Lin, C. J., Lyu, Y. S., Lan, D. S., and Chang, C. F. (2012). Testicular dmrt1 is involved in the sexual fate of the ovotestis in the protandrous black porgy. Biol. Reprod. 86 (2), 41–1. doi:10.1095/biolreprod.111.095695
Yang, Z., Kumar, S., and Nei, M. (1995). A new method of inference of ancestral nucleotide and amino acid sequences. Genetics 141 (4), 1641–1650. doi:10.1093/genetics/141.4.1641
Yang, Z., Nielsen, R., Goldman, N., and Pedersen, A. M. K. (2000). Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics 155 (1), 431–449. doi:10.1093/genetics/155.1.431
Yang, Z. (2007). Paml 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24 (8), 1586–1591. doi:10.1093/molbev/msm088
Yoshimoto, S., Okada, E., Umemoto, H., Tamura, K., Uno, Y., Nishida-Umehara, C., et al. (2008). A W-linked DM-domain gene, DM-W, participates in primary ovary development in Xenopus laevis. Proc. Natl. Acad. Sci. U.S.A. 105 (7), 2469–2474. doi:10.1073/pnas.0712244105
Yu, G., Smith, D. K., Zhu, H., Guan, Y., and Lam, T. T. Y. (2017). ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 8 (1), 28–36. doi:10.1111/2041-210x.12628
Zarkower, D. (2001). Establishing sexual dimorphism: Conservation amidst diversity? Nat. Rev. Genet. 2 (3), 175–185. doi:10.1038/35056032
Zhang, E. F., and Qiu, G. F. (2010). A novel Dmrt gene is specifically expressed in the testis of Chinese mitten crab, Eriocheir sinensis. Dev. Genes Evol. 220 (5), 151–159. doi:10.1007/s00427-010-0336-2
Zheng, J., Cai, L., Jia, Y., Chi, M., Cheng, S., Liu, S., et al. (2020). Identification and functional analysis of the doublesex gene in the redclaw crayfish, Cherax quadricarinatus. Gene Expr. Patterns 37, 119129. doi:10.1016/j.gep.2020.119129
Zhu, D., Feng, T., Mo, N., Han, R., Lu, W., Shao, S., et al. (2022). New insights for the regulatory feedback loop between type 1 crustacean female sex hormone (CFSH-1) and insulin-like androgenic gland hormone (IAG) in the Chinese mitten crab (Eriocheir sinensis). Front. Physiol. 13, 1054773. doi:10.3389/fphys.2022.1054773
Keywords: sexual development, Dmrt, evolution, malacostraca, Eriocheir sinensis
Citation: Zhang P, Yang Y, Xu Y and Cui Z (2023) Analyses of the Dmrt family in a decapod crab, Eriocheir sinensis uncover new facets on the evolution of DM domain genes. Front. Physiol. 14:1201846. doi: 10.3389/fphys.2023.1201846
Received: 07 April 2023; Accepted: 16 May 2023;
Published: 26 May 2023.
Edited by:
Yafei Duan, South China Sea Fisheries Research Institute, ChinaReviewed by:
Yilei Wang, Jimei University, ChinaJianjian Lv, Chinese Academy of Fishery Sciences (CAFS), China
Copyright © 2023 Zhang, Yang, Xu and Cui. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhaoxia Cui, Y3Vpemhhb3hpYUBuYnUuZWR1LmNu
 Peng Zhang
Peng Zhang Yanan Yang
Yanan Yang Yuanfeng Xu
Yuanfeng Xu Zhaoxia Cui
Zhaoxia Cui