- 1Henan International Laboratory for Green Pest Control, College of Plant Protection, Henan Agricultural University, Zhengzhou, China
- 2State Key Laboratory of Integrated Management of Pest Insects and Rodents, Institute of Zoology, Chinese Academy of Sciences, Beijing, China
Introduction: Insect cytochrome P450 (CYP450) genes play important roles in the detoxification and metabolism of xenobiotics, such as plant allelochemicals, mycotoxins and pesticides. The polyphagous Conogethes punctiferalis is a serious economic pest of fruit trees and agricultural crops, and it shows high adaptability to different living environments.
Methods: The two novel P450 genes CYP6CV1 and CYP6AB51 were identified and characterized. Quantitative real-time PCR (qRT-PCR) technology was used to study the expression patterns of the two target genes in different larval developmental stages and tissues of C. punctiferalis. Furthermore, RNA interference (RNAi) technology was used to study the potential functions of the two P450 genes by treating RNAi-silenced larvae with three commonly used pesticides.
Results: The CYP6CV1 and CYP6AB51 genes were expressed throughout various C. punctiferalis larval stages and in different tissues. Their expression levels increased along with larval development, and expression levels of the two target genes in the midgut were significantly higher than in other tissues. The toxicity bioassay results showed that the LC50 values of chlorantraniliprole, emamectin benzoate and lambda-cyhalothrin on C. punctiferalis larvae were 0.2028 μg/g, 0.0683 μg/g and 0.6110 mg/L, respectively. After treating with different concentrations of chlorantraniliprole, emamectin benzoate and lambda-cyhalothrin (LC10, LC30, LC50), independently, the relative expressions of the two genes CYP6CV1 and CYP6AB51 were significantly induced. After the dsRNA injection, the expression profiles of the two CYP genes were reduced 72.91% and 70.94%, respectively, and the mortality rates of the larvae significantly increased when treated with the three insecticides independently at LC10 values.
Discussion: In the summary, after interfering with the CYP6CV1 and CYP6AB51 in C. punctiferalis, respectively, the sensitivity of C. punctiferalis to chlorantraniliprole, emamectin benzoate and lambda-cyhalothrin was significantly increased, indicating that the two CYP6 genes were responsible for the adaptability of C. punctiferalis to the three chemical insecticides in C. punctiferalis. The results from this study demonstrated that CYP6CV1 and CYP6AB51 in C. punctiferalis play crucial roles in the detoxification of chlorantraniliprole, emamectin benzoate and lambda-cyhalothrin.
1 Introduction
The yellow peach moth, Conogethes punctiferalis (Guenée), is widely distributed in East Asia, South Asia, Australia and Papua New Guinea (Shwe et al., 2021a). It is one of the few polyphagous borer pests that not only damage woody plants, such as peach, apple, hawthorn and chestnut, but also feed on herbaceous plants, including maize, soybeans and cotton (Stanley et al., 2009; Chen et al., 2018). In recent years, C. punctiferalis has caused severe damage, and it is now considered a dominant economic lepidopteran pest of summer corn fields in the Huanghuaihai Region of China (Shwe et al., 2021b). Currently, chemical control is commonly used to control lepidopteran pests in corn fields, including the widely used pesticides chlorantraniliprole, emamectin benzoate and lambda-cyhalothrin. However, the excessive and frequent use of insecticides has caused a series of serious problems, including the evolution of pesticide resistance (Gao, 2010; Siegwart et al., 2017; Wang et al., 2020). Some lepidopteran pests have developed high resistance levels to pesticides commonly used in corn fields. For example, the field population of Plutella xylostella is more than 2,000 times resistant to chlorantraniliprole (Wang and Wu, 2012). Spodoptera frugiperda has a high risk of developing resistance to emamectin benzoate (Muraro et al., 2021), and field populations of Spodoptera exigua have developed a high level of resistance to lambda-cyhalothrin insecticides (Su and Sun, 2014). The increased metabolic activity levels of detoxification enzymes are main causes of insect resistance, including the cytochrome P450 monooxygenase (P450)-mediated detoxification of insecticides (Elzaki et al., 2015). The overexpression of P450 genes plays important roles in insecticide resistance. For example, overexpression of CYP6BG1 may contribute to chlorantraniliprole resistance in P. xylostella (Li et al., 2018), CYP337B3 in Helicoverpa armigera is involved in the resistance to cypermethrin (Rasool et al., 2014), and CYP6AY1 of Nilaparvata lugens is involved in resistance to imidacloprid (Ding et al., 2013).
P450 is a common and important detoxification enzyme in all living organisms, including animals, plants and microorganisms (Cui et al., 2016; Elfaki et al., 2018). Insect P450 can be divided into four main branches: CYP2, CYP3, CYP4 and mitochondrial P450s. The CYP3 family is a large group of insect P450s, which can be subdivided into CYP6, CYP9, CYP28 and several other families (Wang et al., 2018; Ullah et al., 2020). To date, the identified insect cytochrome P450s mainly have two functions. They catalyze the formation and decom position of endogenous substances (such as ecdysone, juvenile hormone and fatty acid) to maintain the normal functions of the organism (Helvig et al., 2004; Iga and Kataoka, 2012). In addition, they metabolize many exogenous substances (such as pesticides, plant secondary substances and other environmental chemicals), and this has detoxification and activation effects (Yang and Liu, 2011; Giraudo et al., 2015). Many insect P450 genes, especially in the families of CYP3 and CYP4, participate in the detoxification of, and metabolic resistance to, several insecticides (Amenya et al., 2008; Balabanidou et al., 2016; Wang et al., 2019; Jing et al., 2022).
Previous reports found that the mRNA expression levels of CYP6CV1 and CYP6AB51 in CYP6 family from C. punctiferalis were upregulated when larvae fed on resistant plant, and in other lepidopteran insects, the CYP6 family genes also played important role in the insect resistance to some pesticides (Chen et al., 2015; Jing et al., 2022). To examine whether CYP6CV1 and CYP6AB51 in C. punctiferalis also are involved in the detoxification of the commonly used pesticides, they were identified and characterized by molecular technologies. The results provide significant insights into the functions of CYP6CV1 and CYP6AB51 from C. punctiferalis in the detoxification of chlorantraniliprole, emamectin benzoate and lambda-cyhalothrin, which are three commonly used pesticides for the control of C. punctiferalis and other lepidopteran insects in corn fields (Siegwart et al., 2017; Wang et al., 2020).
2 Materials and methods
2.1 Insects and chemical insecticides
C. punctiferalis used in this study was collected from corn fields in 2019 at the Xuchang campus of Henan Agricultural University (Henan, China), and then, they were maintained in the laboratory under the follow conditions: 27°C ± 1°C and a 75% ± 5% relative humidity, with a 14-h light: 10-h dark photoperiod. The larvae were fed fresh maize ears, and the adults were provided a 10% sucrose solution without exposure to any insecticides.
Three chemical pesticides, 95% chlorantraniliprole, 91% emamectin benzoate and 96.9% lambda-cyhalothrin were kindly provided by the Insect Physiology, Biochemistry and Molecular Biology Group of Henan Agricultural University.
2.2 Molecular cloning of CYP6CV1 and CYP6AB51 from C. punctiferalis
The total RNA of fourth-instar larvae of C. punctiferalis was extracted using TRIzol reagent in accordance with the instructions (Invitrogen, Carlsbad, CA, United States). After the RNA quality and concentration were verified using 1.5% agarose gel electrophoresis and a NanoDrop 1,000 spectrophotometer (Thermo Scientific, Waltham, MA, United States), respectively. cDNA was synthesized using a FastKing RT Kit (with gDNase) (Tiangen Biotech, Beijing, China).
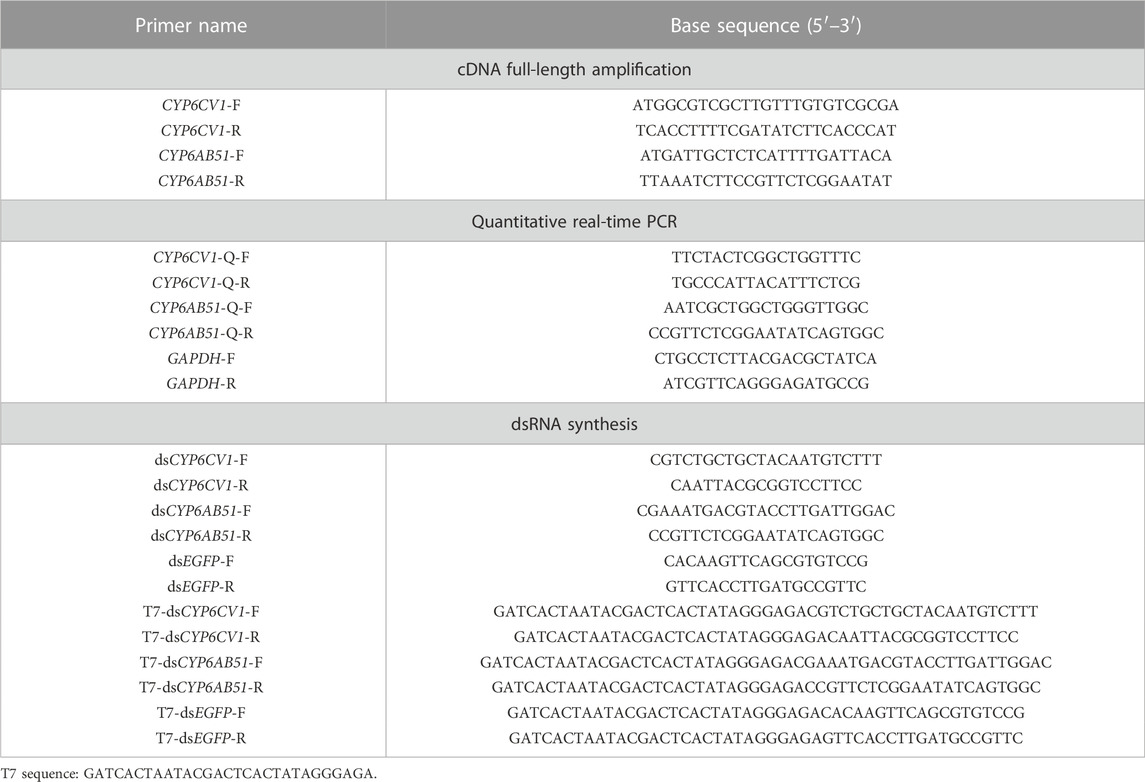
The gene-specific primers were designed to clone CYP6CV1 and CYP6AB51 (Table 1), and the above synthesized cDNA was used as the template for PCR amplification. The PCR reaction was performed in a total volume of 25 μL, containing 12.5 μL of PrimeSTAR® Max DNA Polymerase (TaKaRa, Dalian, China), 1 μL of each primer (10 μM), 1 μL of cDNA template and 9.5 μL of ddH2O, and the amplification conditions were as follows: 98°C for 5 min; 40 cycles of 98°C for 10 s, 55°C for 10 s and 72°C for 30 s; and a final extension step at 72°C for 7 min. After detecting the target band using 1.5% agarose gel electrophoresis, the gel was cut and then purified using a DNA gel extraction kit (Axygen Scientific, Union City, CA, United States). Afterwards, the product was inserted into the pClone 007 vector (Tsingke Biotech, Beijing, China) and then transformed into Escherichia coli DH5α competent cells (Sangon Biotech, Shanghai, China). Finally, the selected positive clones were cultured in LB liquid medium containing Amp (50 mg/ml) at 37°C and 180 r/min for 12 h. The positive clones were confirmed by PCR and sequencing (Sangon Biotech). The sequencing results were compared with the previously obtained transcriptome sequences.
2.3 Sequence analysis and phylogenetic analysis
ORF finder (https://www.ncbi.nlm.nih.gov/orffinder/) was used to determine the open reading frames (ORFs) of the genes. The online website ExPASy (http://web.expasy.org/compute_pi/) was used to predict the isoelectric points and molecular weights of the proteins encoded by the genes. The Neighbor-Joining method in MEGA 7.0 was used to analyze the phylogenetic relationships between the target genes and other amino acid sequences of the CYP6 family in lepidopteran insects. The ClustalX was used to align the sequences of C. punctiferalis CYP6CV1 and CYP6AB51 with the related lepidopteran cytochrome P450.
2.4 Analysis of the CYP6CV1 and CYP6AB51 expression patterns in C. punctiferalis
Larval samples of C. punctiferalis at different developmental stages were collected, including first- (80 individuals), second- (40 individuals), third- (10 individuals), fourth- (5 individuals) and fifth-instar (3 individuals) larvae, to analyze the expression levels of the two genes. Samples of different larval tissues, including head, salivary gland, midgut, fat body, cuticle and hemolymph from 20 fourth-instar larvae, were also dissected and collected. Each insect sample had three replicates. The total RNA extraction and synthesis of cDNA were the same as in section 2.2.
On the basis of the cloned cDNA sequences of CYP6CV1 and CYP6AB51, Primer 5.0 was used to design qRT-PCR primers (Table 1), and glyceraldehyde 3-phosphate dehydrogenase (GAPDH; GenBank accession no: KX668532.1) was used as the internal reference gene to normalize the qRT-PCR data. qRT-PCR experiments were performed using a QuantStudio™ three Real-Time PCR System (Thermo Scientific) in a total volume of 20 μL, which contained 10 μL of 2 × SuperReal PreMix Plus, 0.6 μL of each primer (10 μM), 0.4 μL of 50× ROX Reference Dye, 1 μL of cDNA template (200 ng) and 7.4 μL of RNase-Free ddH2O. The qRT-PCR program conditions were as follows: 95°C for 15 min; 40 cycles of 95°C for 10 s and 60°C for 32 s. To assess the specificity of each PCR amplification, a dissociation-curve analysis was performed at the end of the run. Each sample had three technical replicates.
2.5 The susceptibility of C. punctiferalis larvae to the three insecticides
The toxicity levels of 95% chlorantraniliprole and 91% emamectin benzoate to the third-instar larvae of C. punctiferalis were evaluated independently using the feed-mixing method (Yu et al., 2015). The two insecticides were dissolved and diluted with 0.1% TritionX-100 independently (Solarbio, Beijing, China) to obtain a series of different concentrations. Then, 1 ml of each different diluted solution was incorporated into 100 g of the artificial diet to obtain six concentration-gradient mixed insecticide feeds, and 0.1% TritionX-100 was used as a control. Then, the diets were cut into small pieces and provided to third-instar larvae of C. punctiferalis that had been starved for 12 h. The toxicity of 96.9% lambda-cyhalothrin to the third-instar larvae of C. punctiferalis was evaluated using the topical application method (Brewer and Trumble, 1989). The 96.9% lambda-cyhalothrin was diluted into a series of different concentrations using acetone (Fuyu Chemical, Tianjin, China), and 1 µL of the diluted solution was placed on the thoracic dorsum of the larvae using a microdropper (Envta Technology, Beijing, China). Three biological replicates were used for each concentration, and 20 larvae were treated per replicate. The number of dead insects was counted after 24 h or 48 h (24 h for 96.9% lambda-cyhalothrin and 48 h for both 95% chlorantraniliprole and 91% emamectin benzoate).
2.6 Effects of exposure to the three insecticides on the mRNA expression levels of the target genes
On the basis of the toxicity levels of the three chemical insecticides on the third-instar larvae, LC10, LC30 and LC50 were used to evaluate the effects of chlorantraniliprole, emamectin benzoate and lambda-cyhalothrin on the CYP6CV1 and CYP6AB51 expression levels in C. punctiferalis. The third-instar larvae were treated with the above concentrations of the three insecticides as described in section 2.5, and the surviving larvae were collected at 3, 6, 12, 24 and 48 h after treatment. Each replication included 10 larvae, with three biological replicates per concentration. The total RNA extraction, cDNA synthesis and qRT-PCR were performed as described above in section 2.2 and 2.4.
2.7 RNAi silencing of CYP6CV1 and CYP6AB51 and interference efficiency detection
On the basis of the obtained gene sequences of CYP6CV1 and CYP6AB51 in C. punctiferalis, gene-specific primers containing the T7 promoter were designed, and the enhanced green fluorescent protein gene (EGFP) was used as a control (Table 1). DsCYP6CV1, dsCYP6AB51 and dsEGFP were synthesized using a T7 RiboMAX™ Express RNAi System kit (Promega, Madison, WI, United States) in accordance with the instructions. The dsRNA products were diluted with nuclease-free water to a final concentration of 3 μg/μL and then maintained at −80°C for later use.
The third-instar larvae of C. punctiferalis were injected with 2 μL (3 μg/μL) of gene-specific dsRNA (dsCYP6CV1 and dsCYP6AB51) at the second internode membrane of the abdomen using a manual microinjector. Larvae injected with same amount of dsEGFP (EGFP control) or nuclease-free water (DEPC control) were used as negative controls, and the non-injection treatment was used as a blank control (CK). At 24, 36 and 48 h after injection, 10 surviving larvae per treatment were collected, and qRT-PCR was used to detect the interference efficiency with the target gene. Three biological replicates were used per treatment, and each biological replicate had three technical replicates.
2.8 The susceptibility of C. punctiferalis to the three insecticides after target gene interference
The third-instar larvae of C. punctiferalis were injected with 2 μL (3 μg/μL) of the target gene dsRNA, dsEGFP or DEPC. At 36 h after injection, the surviving larvae were fed diets treated with chlorantraniliprole or emamectin benzoate at the LC10 dose, and another group of C. punctiferalis larvae were subjected to the topical application of lambda-cyhalothrin at the LC10 dose. After 6 h, the mortality of each treatment was recorded. Every treatment was performed in triplicate, and each replicate included 20 larvae.
2.9 Statistical analysis
The relative expression levels of the genes as assessed by qRT-PCR were analyzed using the 2−ΔΔCT method (Livak and Schmittgen, 2001). Excel was used to sort the data, and SPSS 20.0 was used for the statistical analysis. After a one-way ANOVA, Duncan’s multiple range tests (p < 0.05) were used to analyze the significant differences between different treatments. The bioassay used the Probit function of SPSS 20.0 for the statistical regression analysis.
3 Results
3.1 Identification of CYP6CV1 and CYP6AB51 in C. punctiferalis
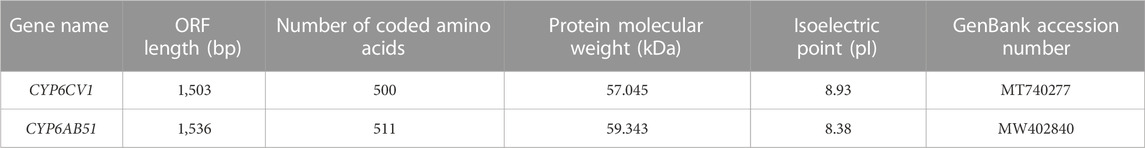
The cDNA sequences of CYP6CV1 and CYP6AB51 in C. punctiferalis were obtained through molecular cloning and sequencing. The ORF lengths, number of encoded amino acids, protein molecular weights and isoelectric points of CYP6CV1 and CYP6AB51 in C. punctiferalis were determined using the online tools of the NCBI and ExPASY, and then, the data were submitted to NCBI to obtain the GenBank accession numbers. The basic sequence characteristics of CYP6CV1 and CYP6AB51 from C. punctiferalis are shown in Table 2.
The constructed phylogenetic tree showed that C. punctiferalis CYP6CV1 was closely related to CYP6CV1 in Cnaphalocrocis medinalis, with a homology of 78.45% (Figure 1), whereas there was a close relationship between CYP6AB51 from C. punctiferalis and CYP6AB51 from Chilo suppressalis, with a homology of 61.36% (Figure 2). The predicted amino acid sequences encoded by the two genes contained five typical conserved structural regions of insect P450 proteins (Graham and Peterson, 1999; Deng et al., 2007), a C-helix region (WXXXR), I-helix region (AGXETS), K-helix region (EXXR), meander sequence (PXXFXPXXF) and heme-binding sequence (FXXGXRXCXG) (where X represents any amino acid) (Figure 3).
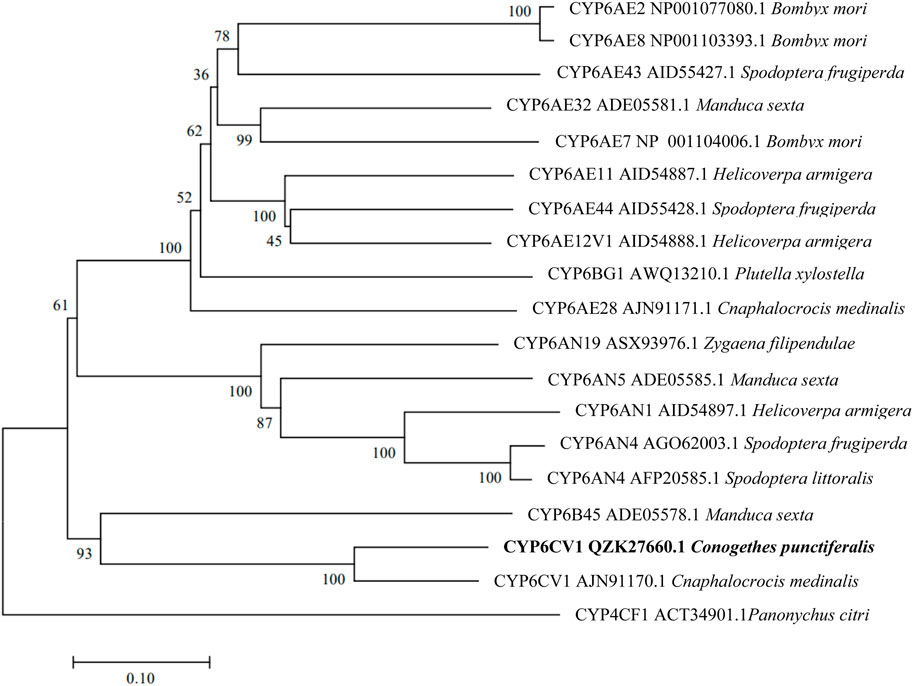
FIGURE 1. Phylogenetic relationships between CYP6CV1 from C. punctiferalis and homologs from other lepidopteran insects. The phylogram was generated by MEGA 7.0 using the neighbor-joining method, and the inferred phylogeny was tested using a bootstrap analysis with 1,000 pseudoreplicate datasets.
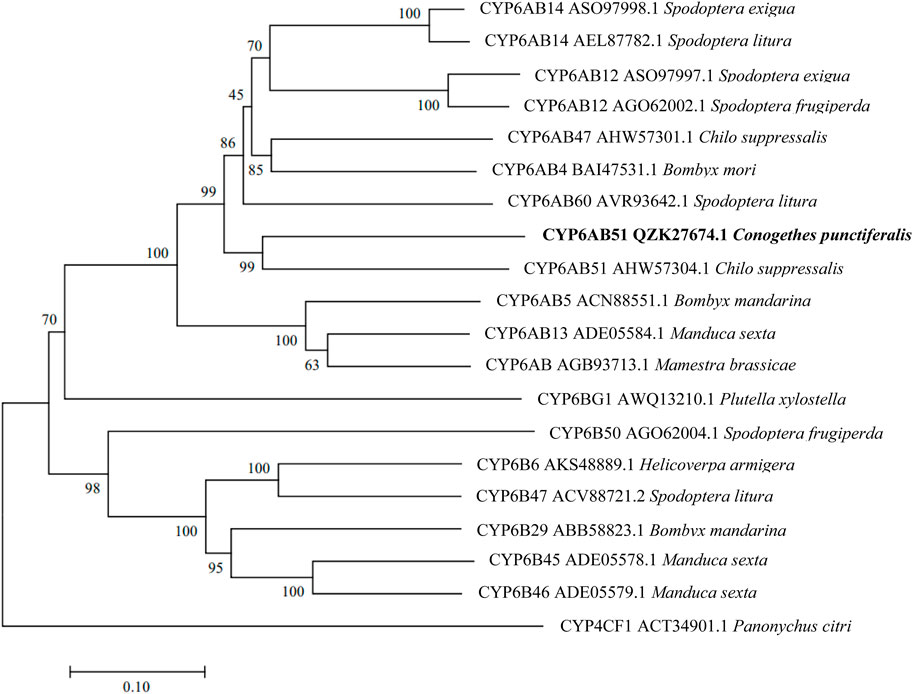
FIGURE 2. Phylogenetic relationships between CYP6AB51 from C. punctiferalis and homologs from other lepidopteran insects. The phylogram was generated by MEGA 7.0 using the neighbor-joining method, and the inferred phylogeny was tested using a bootstrap analysis with 1,000 pseudoreplicate datasets.
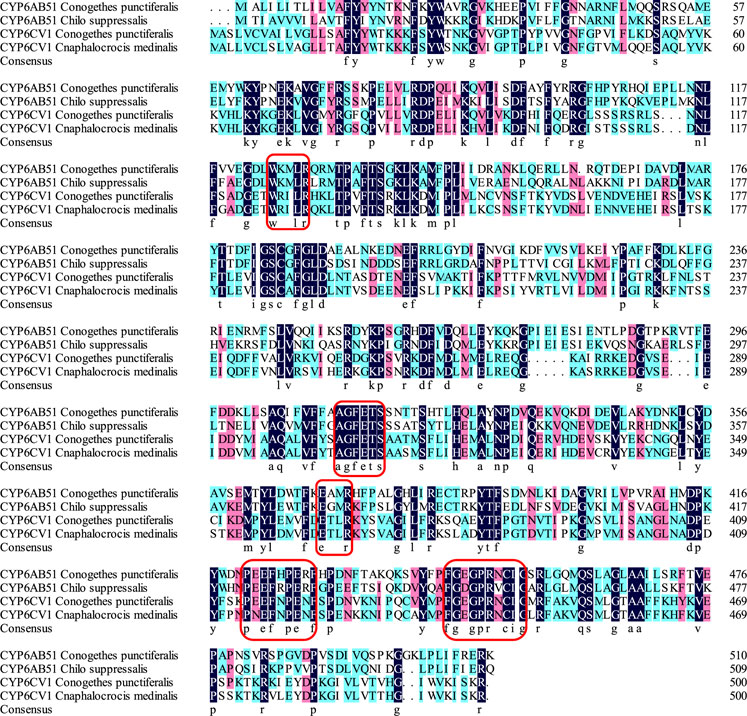
FIGURE 3. Alignment of C. punctiferalis CYP6CV1 and CYP6AB51 with related lepidopteran cytochrome P450s. The sequences of C. punctiferalis CYP6AB51, C. suppressalis CYP6AB51, C. punctiferalis CYP6CV1 and C. medinalis CYP6CV1 were aligned using ClustalX. The dark blue indicates a shared amino acid identity of 100%, the pink indicates a shared amino acid identity greater than or equal to 75%, and the green indicates a shared amino acid identity greater than or equal to 50%. The five conserved domains of the cytochrome P450 genes are marked with red boxes.
3.2 Expression profiles of CYP6CV1 and CYP6AB51 in different larval developmental stages and different tissues
qRT-PCR was used to detect the gene expression levels of CYP6CV1 and CYP6AB51 at different larval developmental stages and in different tissues of C. punctiferalis. The two target genes were expressed throughout the C. punctiferalis larval growth period and in different tissues. As the larvae developed, the expression levels of the two genes increased. The maximum expression levels of CYP6CV1 and CYP6AB51 genes were detected in the fifth-instar larvae stage, and they were 385.96 and 12.42 times higher than in the first-instar larvae, respectively. Among the different tissues, the expression levels of the two genes in the midgut were the highest, and they were significantly higher than in other larval tissues. The transcript levels in the head and cuticle were the lowest among the tested tissues (Figure 4).
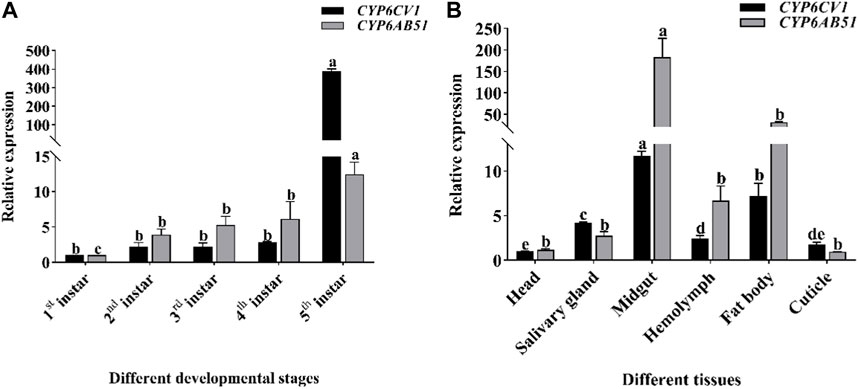
FIGURE 4. The transcription levels of CYP6CV1 and CYP6AB51 in different larval developmental stages (A) and tissues [(B), fourth-instar larvae] of C. punctiferalis. All the transcript levels were normalized using GAPDH as the internal reference gene. The values are represented as the means ± SEs. Significant differences among treatments are indicated by lowercase letters above each bar (p < 0.05).
3.3 Toxicity of the three insecticides on C. punctiferalis
The toxicity of the three chemical insecticides, chlorantraniliprole, emamectin benzoate and lambda-cyhalothrin, on C. punctiferalis larvae are shown in Table 3.
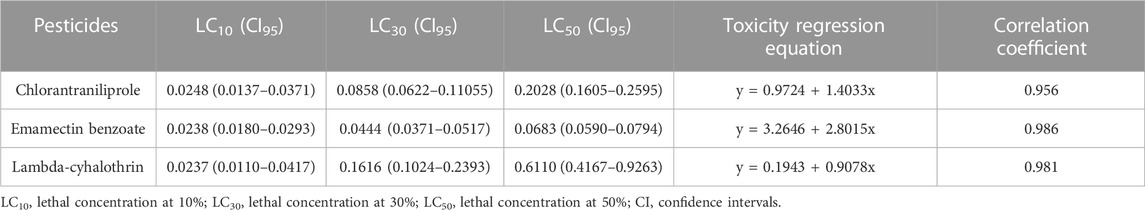
TABLE 3. Toxicity levels of the three tested chemical insecticides on the third-instar larvae of C. punctiferalis.
3.4 Effects of insecticide exposure on the mRNA expression levels of CYP6CV1 and CYP6AB51
Three pesticides commonly used in the control of C. punctiferalis were selected to evaluate their effects on the expression levels of C. punctiferalis CYP6CV1 and CYP6AB51 as assessed by qRT-PCR. When C. punctiferalis larvae were exposed to chlorantraniliprole at LC10, the expression of CYP6CV1 increased significantly and peaked at 3 h, whereas the expression of CYP6AB51 peaked at 6 h. When treating with chlorantraniliprole at LC30, the expression levels of CYP6CV1 in C. punctiferalis were obviously different from the control group at 3 h and 48 h, and the transcript levels of CYP6AB51 at different time points were all higher than in the control. Compared with the control group, the expression of CYP6CV1 in the LC50 treatment increased significantly at 3 h, and the expression of CYP6AB51 was induced notably at all the time points (Figures 5A,B).
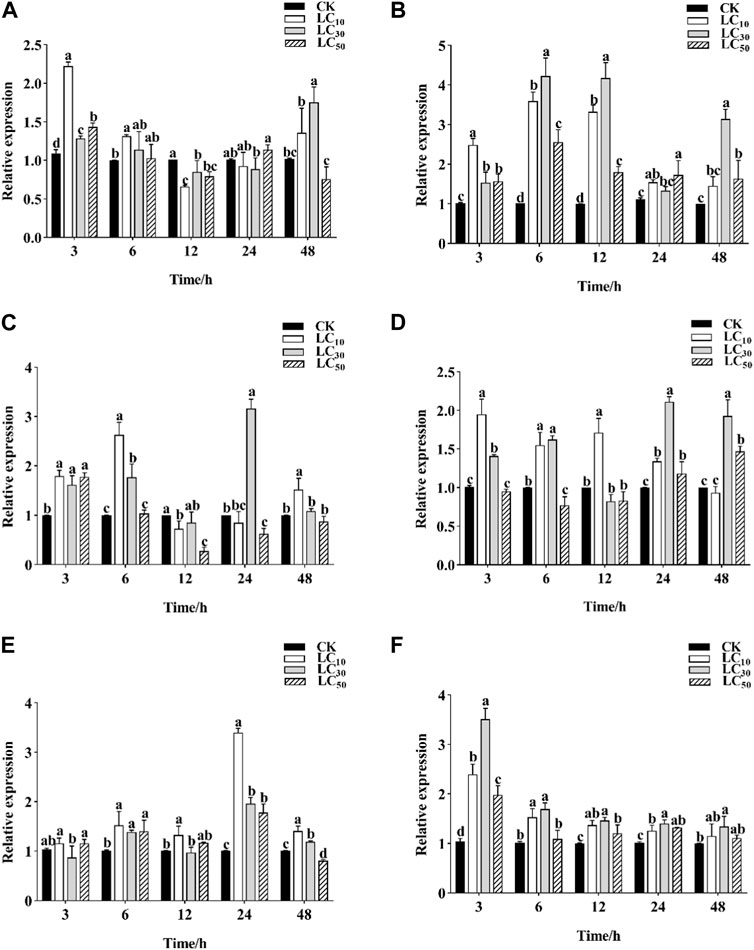
FIGURE 5. Effects of three different pesticides treatments on CYP6CV1 and CYP6AB51 expression level in C. punctiferalis. The relative transcription levels of (A) CYP6CV1 and (B) CYP6AB51 in the chlorantraniliprole treatment (C) CYP6CV1 and (D) CYP6AB51 in the emamectin benzoate treatment; and (E) CYP6CV1 and (F) CYP6AB51 in the lambda-cyhalothrin treatment. Values are represented by means ± SEs. Significant differences among groups are indicated by lowercase letters above each bar (p < 0.05).
Emamectin benzoate at LC10 obviously induced the expression of CYP6CV1 and CYP6AB51 at 3 h and 6 h. In addition, emamectin benzoate at LC30 also significantly increased the expression levels of the two genes relative to the control treatment. However, when C. punctiferalis larvae were exposed to emamectin benzoate at LC50, the expression of CYP6CV1 was significantly increased at 3 h and then decreased slowly, whereas the transcript levels of CYP6AB51 peaked at 48 h (Figures 5C,D).
In response to lambda-cyhalothrin exposure, the expression levels of CYP6CV1 at the three concentrations all peaked at 24 h, whereas the expression of CYP6AB51 was upregulated at all time points and peaked at 3 h compared with the control (Figures 5E,F).
3.5 The sensitivity of C. punctiferalis larvae to the three insecticides after target gene silencing
After the silencing of CYP6CV1 and CYP6AB51 independently in C. punctiferalis by injecting dsCYP6CV1 and dsCYP6AB51, respectively, the expression levels of CYP6CV1 and CYP6AB51 were both decreased most at 36 h, with reductions of 72.91% and 70.94%, respectively, when compared with the control groups (Figure 6).
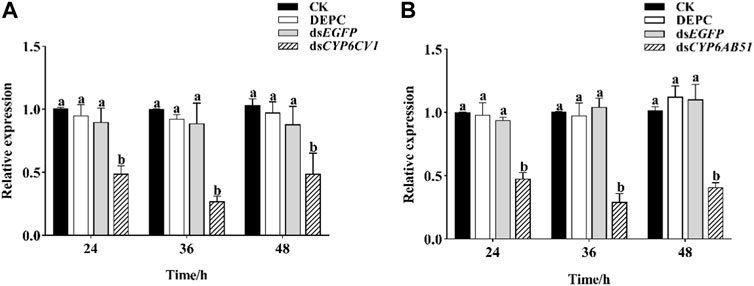
FIGURE 6. Effects of RNAi on the transcript levels of CYP6CV1 (A) and CYP6AB51 (B) in C. punctiferalis. Values represent means ± SEs for three independent replicates. Significant differences among groups are indicated by lowercase letters above each bar (p < 0.05).
After dsRNA injection, the mortality of CYP6CV1-silenced larvae and CYP6AB51-silenced larvae treated independently with LC10 values of chlorantraniliprole, emamectin benzoate and lambda-cyhalothrin were recorded. The results indicated that the delivery of dsCYP6CV1 increased the larval mortality caused by chlorantraniliprole (from 1.67% to 11.67%), emamectin benzoate (from 3.33% to 16.67%) and lambda-cyhalothrin (from 3.33% to 13.33%), and injection with dsCYP6AB51 significantly increased the larval mortality caused by chlorantraniliprole (from 1.67% to 8.33%), emamectin benzoate (from 3.33% to 11.67%) and lambda-cyhalothrin (from 3.33% to 11.67%) (Figure 7).
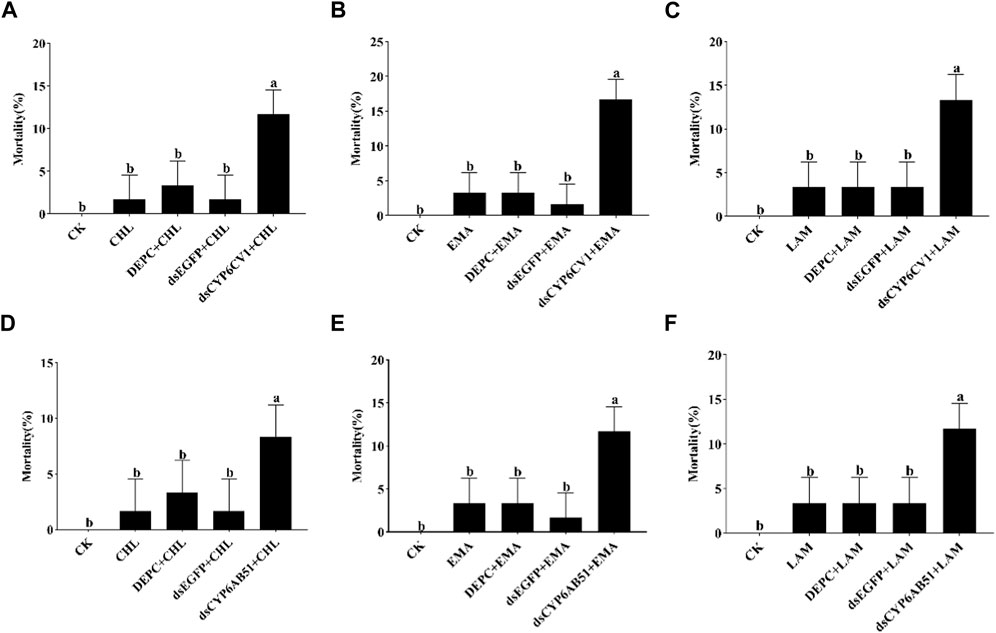
FIGURE 7. Silencing CYP6CV1 (A–C) and CYP6AB51 (D–F) to the sensitivity of C. punctiferalis larvae treat with three pesticides. Data shown are means ± SEs, and significant differences among groups are indicated by lowercase letters above each bar (p < 0.05). CK: uninjected larvae fed with normal artificial diets; CHL/EMA/LAM: uninjected larvae treated with chlorantraniliprole, emamectin benzoate or lambda-cyhalothrin at the LC10 value; DEPC + CHL/EMA/LAM: larvae injected with DEPC-treated water and then treated with chlorantraniliprole, emamectin benzoate or lambda-cyhalothrin at the LC10 value; dsEGFP + CHL/EMA/LAM: larvae injected with dsEGFP and then treated with chlorantraniliprole, emamectin benzoate or lambda-cyhalothrin at the LC10 value; dsCYP6CV1/dsCYP6AB51 + CHL/EMA/LAM: larvae injected with dsCYP6CV1 or dsCYP6AB51 and then treated with chlorantraniliprole, emamectin benzoate or lambda-cyhalothrin at the LC10 value.
4 Discussion
The number of CYP genes carried by insect genomes range from 36 for Pediculus humanus to 170 for Culex quinquefasciatus (Arensburger et al., 2010; Lee et al., 2010). The first identified insect P450 gene was CYP6A1 cloned from Musca domestica (Feyereisen et al., 1989), and since then, more and more novel insect P450 genes have been identified with the development of molecular biology-related technology (Li F. et al., 2019). Some P450 genes, especially CYP3 (including CYP6 and CYP9) and CYP4 are involved in the detoxification and metabolism of exogenous compounds (Feyereisen, 2011). In Mamestra brassicae, the CYP4M51 and CYP6AB56 genes have potential roles in the detoxification of deltamethrin (Zhou et al., 2017). In addition, the CYP9A105 gene may play an important role in the detoxification of α-cypermethrin, deltamethrin and fenvalerate in Spodoptera exigua (Wang et al., 2018). In this study, the obtained P450 CYP6 genes, CYP6CV1 and CYP6AB51 in C. punctiferalis, also contained the five conserved domains of insect P450s. The phylogenetic analysis showed that CYP6CV1 and CYP6AB51 in C. punctiferalis were closely related to C. medinalis CYP6CV1 and C. suppressalis CYP6AB51, respectively.
Analyses of P450 gene expression patterns provide useful information on the potential roles of these genes in biological functions (Lyu et al., 2020). Insect P450 genes have a diverse distribution. Some genes are expressed throughout all the developmental stages and distributed widely in all the tissues, whereas some genes are only expressed in specific developmental stages and specific tissues (Scott et al., 1999; Feyereisen, 2015; Xu et al., 2009). Larval midguts and fat bodies play important roles in the metabolism of xenobiotics; therefore, the P450 genes associated with the detoxification of xenobiotics may be highly expressed in these tissues (Hou et al., 2021). Consequently, the expression profiles of CYP6CV1 and CYP6AB51 in C. punctiferalis at different developmental stages and in different larval tissues were assessed by qRT-PCT. As the larvae developed, the gene expression levels increased. The expression levels of CYP6CV1 and CYP6AB51 in fifth-instar larvae were 385.96 and 12.42 times higher than in first-instar larvae, respectively. This expression pattern is similar to the larval expression pattern of CYP6CV1 in C. medinalis (Chen et al., 2015). A tissue-specific expression analysis showed that CYP6CV1 and CYP6AB51 are highly expressed in the larval midguts of C. punctiferalis, with expression levels being approximately 11.60-fold and 156.96-fold higher than in the head, respectively, and a similar result was also found for the CYP6AB60 expression levels in Spodoptera litura (Sun et al., 2019). The detoxification system of the midgut responds quickly to the ingestion of xenobiotics (plant secondary substances and pesticides), indicating that the detoxification enzymes of midguts play important roles in the metabolism of xenobiotics (Li Q. et al., 2019; Huang et al., 2021).
An important feature of the P450 enzyme system is its inducibility (Yang et al., 2013). The expression of insect P450 can be induced by a broad range of xenobiotics, thereby enhancing their ability to metabolize the xenobiotics after exposure (Feyereisen, 2015). CYP6B8 in Helicoverpa zea metabolizes plant allelochemicals, such as xanthotoxin, quercetin and flavone, as well as three insecticides, diazinon, cypermethrin and aldrin (Li et al., 2004). The upregulation of CYP6CM1 in Bemisia tabaci is related to its resistance to neonicotinoid insecticides (Jones et al., 2011). In Leptinotarsa decemlineata, six CYP6 genes (CYP6BH2, CYP6BJ1, CYP6BQ17, CYP6EG1, CYP6EH1 and CYP6EJ1) are involved in cyhalothrin detoxification (Wan et al., 2013). In this study, the expression of CYP6CV1 and CYP6AB51 was significantly induced by chlorantraniliprole, emamectin benzoate and lambda-cyhalothrin treatments, which indicated that CYP6CV1 and CYP6AB51 may participate in the detoxification and metabolism of the three pesticides in C. punctiferalis.
RNAi is a commonly used gene silencing technology that inhibits the expression of target genes in the test organisms. It has been widely used to study the functional roles of insect P450s (Bautista et al., 2009; Shi et al., 2016). Furthermore, it is a promising bioengineering technology that can be applied to pest control (Zhu and Palli, 2020). It has been successfully used in the functional studies of a variety of insect P450 genes (Zhao et al., 2021). In the current study, independent injections with dsRNA targeting CYP6CV1 and CYP6AB51 significantly increased the mortality of C. punctiferalis larvae treated with chlorantraniliprole, emamectin benzoate or lambda-cyhalothrin, which suggested the potential roles of CYP6CV1 and CYP6AB51 in the detoxification of the three pesticides. Similarly, after the injection of dsRNA targeting CYP6B7, the sensitivity of H. armigera larvae significantly increases after fenvalerate exposure (Tang et al., 2012). In addition, the gene silencing of CYP6B6 in H. armigera significantly reduces the survival rate and development of larvae (Zhang et al., 2013). After injecting the dsRNA of CYP6AB14, the transcription level of CYP6AB14 in S. litura is significantly reduced, and the mortality of larvae significantly increases (Wang et al., 2015).
5 Conclusion
In this study, it was found that after interfering with the CYP6CV1 and CYP6AB51 in C. punctiferalis, respectively, the sensitivity of C. punctiferalis to chlorantraniliprole, emamectin benzoate and lambda-cyhalothrin was significantly increased, indicating that the two CYP6 genes were responsible for the adaptability of C. punctiferalis to the three chemical insecticides in C. punctiferalis. The results from this study demonstrated that CYP6CV1 and CYP6AB51 in C. punctiferalis play crucial roles in the detoxification of chlorantraniliprole, emamectin benzoate and lambda-cyhalothrin.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Author contributions
Formal analysis, QZ; Investigation, XY and HL; Supervision, XG and HJ; Writing—original draft, LZ, GW, WL, and MZ. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the National Natural Science Foundation of China (Grant No. 31801735) and the State Key Laboratory of Integrated Management of Pest Insects and Rodents (Grant No. IPM2117).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Amenya, D. A., Naguran, R., Lo, T. C., Ranson, H., Spillings, B. L., Wood, O. R., et al. (2008). Over expression of a cytochrome P450 (CYP6P9) in a major African malaria vector, Anopheles funestus, resistant to pyrethroids. Insect Mol. Biol. 17, 19–25. doi:10.1111/j.1365-2583.2008.00776.x
Arensburger, P., Megy, K., Waterhouse, R. M., Abrudan, J., Amedeo, P., Antelo, B., et al. (2010). Sequencing of Culex quinquefasciatus establishes a platform for mosquito comparative genomics. Science 330, 86–88. doi:10.1126/science.1191864
Balabanidou, V., Kampouraki, A., MacLean, M., Blomquist, G. J., Tittiger, C., Juárez, M. P., et al. (2016). Cytochrome P450 associated with insecticide resistance catalyzes cuticular hydrocarbon production in Anopheles gambiae. Proc. Natl. Acad. Sci. U. S. A. 113, 9268–9273. doi:10.1073/pnas.1608295113
Bautista, M. A. M., Miyata, T., Miura, K., and Tanaka, T. (2009). RNA interference-mediated knockdown of a cytochrome P450, CYP6BG1, from the diamondback moth, Plutella xylostella, reduces larval resistance to permethrin. Insect Biochem. Mol. Biol. 39, 38–46. doi:10.1016/j.ibmb.2008.09.005
Brewer, M. J., and Trumble, J. T. (1989). Field monitoring for insecticide resistance in beet armyworm (Lepidoptera: Noctuidae). J. Econ. Entomol. 82, 1520–1526. doi:10.1093/jee/82.6.1520
Chen, J., Li, C., and Yang, Z. (2015). Identification and expression of two novel cytochrome P450 genes, CYP6CV1 and CYP9A38, in Cnaphalocrocis medinalis (Lepidoptera: Pyralidae). J. Insect Sci. 15, 50. doi:10.1093/jisesa/ieu174
Chen, G. M., Chi, H., Wang, R. C., Wang, Y. P., Xu, Y. Y., Li, X. D., et al. (2018). Demography and uncertainty of population growth of Conogethes punctiferalis (Lepidoptera: Crambidae) reared on five host plants with discussion on some life history statistics. J. Econ. Entomol. 111, 2143–2152. doi:10.1093/jee/toy202
Cui, S., Wang, L., Ma, L., and Geng, X. (2016). P450-mediated detoxification of botanicals in insects. Phytoparasitica 44, 585–599. doi:10.1007/s12600-016-0550-1
Deng, J., Carbone, I., and Dean, R. A. (2007). The evolutionary history of cytochrome P450 genes in four filamentous Ascomycetes. BMC Evol. Biol. 7, 30–22. doi:10.1186/1471-2148-7-30
Ding, Z., Wen, Y., Yang, B., Zhang, Y., Liu, S., Liu, Z., et al. (2013). Biochemical mechanisms of imidacloprid resistance in Nilaparvata lugens: Over-expression of cytochrome P450 CYP6AY1. Insect biochem. Mol. Biol. 43, 1021–1027. doi:10.1016/j.ibmb.2013.08.005
Elfaki, I., Mir, R., Almutairi, F. M., and Duhier, F. M. A. (2018). Cytochrome P450: Polymorphisms and roles in cancer, diabetes and atherosclerosis. Asian pac. J. Cancer Prev. 19, 2057–2070. doi:10.22034/APJCP.2018.19.8.2057
Elzaki, M. E. A., Zhang, W., and Han, Z. (2015). Cytochrome P450 CYP4DE1 and CYP6CW3v2 contribute to ethiprole resistance in Laodelphax striatellus (Fallén). Insect Mol. Biol. 24, 368–376. doi:10.1111/imb.12164
Feyereisen, R., Koener, J. F., Farnsworth, D. E., and Nebert, D. W. (1989). Isolation and sequence of cDNA encoding a cytochrome P450 from an insecticide-resistant strain of the house fly, Musca domestica. Proc. Natl. Acad. Sci. U. S. A. 86, 1465–1469. doi:10.1073/pnas.86.5.1465
Feyereisen, R. (2011). Arthropod CYPomes illustrate the tempo and mode in P450 evolution. Biochim. Biophys. Acta, Proteins Proteomics 1814, 19–28. doi:10.1016/j.bbapap.2010.06.012
Feyereisen, R. (2015). Insect P450 inhibitors and insecticides: Challenges and opportunities. Pest Manage. Sci. 71, 793–800. doi:10.1002/ps.3895
Gao, X. W. (2010). Current status and development strategy for chemical control in China. Plant Prot. 36, 19–22.
Giraudo, M., Hilliou, F., Fricaux, T., Audant, P., Feyereisen, R., and Le Goff, G. (2015). Cytochrome P450s from the fall armyworm (Spodoptera frugiperda): Responses to plant allelochemicals and pesticides. Insect Mol. Biol. 24, 115–128. doi:10.1111/imb.12140
Graham, S. E., and Peterson, J. A. (1999). How similar are P450s and what can their differences teach us? Arch. Biochem. Biophys. 369, 24–29. doi:10.1006/abbi.1999.1350
Helvig, C., Tijet, N., Feyereisen, R., Walker, F. A., and Restifo, L. L. (2004). Drosophila melanogaster CYP6A8, an insect P450 that catalyzes lauric acid (ω-1)-hydroxylation. Biochem. Biophys. Res. Commun. 325, 1495–1502. doi:10.1016/j.bbrc.2004.10.194
Hou, W. T., Staehelin, C., Elzaki, M. E. A., Hafeez, M., Luo, Y. S., and Wang, R. L. (2021). Functional analysis of CYP6AE68, a cytochrome P450 gene associated with indoxacarb resistance in Spodoptera litura (Lepidoptera: Noctuidae). Pestic. Biochem. Physiol. 178, 104946. doi:10.1016/j.pestbp.2021.104946
Huang, Y., Luo, Y., Wu, P., Zheng, J., Qiu, L., and Hou, L. (2021). Effects of three insecticides on the expression of cytochrome P450 CYP6B7 in Helicoverpa armigera. J. Appl. Entomol. 145, 440–447. doi:10.1111/iwj.13544
Iga, M., and Kataoka, H. (2012). Recent studies on insect hormone metabolic pathways mediated by cytochromeP450 enzymes. Biol. Pharm. Bull. 35, 838–843. doi:10.1248/bpb.35.838
Jing, D. P., Huang, X. D., Zhang, T. T., and Wang, Z. Z. (2022). Involvement of CYP6AE76 gene in detoxifying the Cry1Ab protein in Conogethes punctiferalis (Lepidoptera: Crambidae). Acta Entomol. Sin. 65, 1136–1143.
Jones, C. M., Daniels, M., Andrews, M., Slater, R., Lind, R. J., Gorman, K., et al. (2011). Age-specific expression of a P450 monooxygenase (CYP6CM1) correlates with neonicotinoid resistance in Bemisia tabaci. Pestic. Biochem. Physiol. 101, 53–58. doi:10.1016/j.pestbp.2011.07.004
Lee, S. H., Kang, J. S., Min, J. S., Yoon, K. S., Strycharz, J. P., Johnson, R., et al. (2010). Decreased detoxification genes and genome size make the human body louse an efficient model to study xenobiotic metabolism. Insect Mol. Biol. 19, 599–615. doi:10.1111/j.1365-2583.2010.01024.x
Li, X., Baudry, J., Berenbaum, M. R., and Schuler, M. A. (2004). Structural and functional divergence of insect CYP6B proteins: From specialist to generalist cytochrome P450. Proc. Natl. Acad. Sci. U. S. A. 101, 2939–2944. doi:10.1073/pnas.0308691101
Li, X., Li, R., Zhu, B., Gao, X., and Liang, P. (2018). Overexpression of cytochrome P450 CYP6BG1 may contribute to chlorantraniliprole resistance in Plutella xylostella (L). Pest Manag. Sci. 74, 1386–1393. doi:10.1002/ps.4816
Li, F., Zhao, X., Li, M., He, K., Huang, C., Zhou, Y., et al. (2019). Insect genomes: Progress and challenges. Insect Mol. Biol. 28, 739–758. doi:10.1111/imb.12599
Li, Q., Sun, Z., Shi, Q., Wang, R., Xu, C. C., Wang, H., et al. (2019). RNA-seq analyses of midgut and fat body tissues reveal the molecular mechanism underlying Spodoptera litura resistance to tomatine. Front. Physiol. 10, 8. doi:10.3389/fphys.2019.00008
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 25, 402–408. doi:10.1006/meth.2001.1262
Lyu, Z., Kong, D., Cheng, J., and Lin, T. (2020). Identification and expression analysis of cytochrome P450 genes in Plecoptera oculata (Lepidoptera: Noctuidae). Entomol. Res. 50, 90–99. doi:10.1111/1748-5967.12412
Maïbèche-Coisne, M., Jacquin-Joly, E., François, M. C., and Nagnan-Le Meillour, P. (2002). cDNA cloning of biotransformation enzymes belonging to the cytochrome P450 family in the antennae of the noctuid moth Mamestra brassicae. Insect Mol. Biol. 11, 273–281. doi:10.1046/j.1365-2583.2002.00335.x
Muraro, D. S., de Oliveira Abbade Neto, D., Kanno, R. H., Kaiser, I. S., Bernardi, O., and Omoto, C. (2021). Inheritance patterns, cross-resistance and synergism in Spodoptera frugiperda (Lepidoptera: Noctuidae) resistant to emamectin benzoate. Pest Manage. Sci. 77, 5049–5057. doi:10.1002/ps.6545
Rasool, A., Joußen, N., Lorenz, S., Ellinger, R., Schneider, B., Khan, S. A., et al. (2014). An independent occurrence of the chimeric P450 enzyme CYP337B3 of Helicoverpa armigera confers cypermethrin resistance in Pakistan. Insect Biochem. Mol. Biol. 53, 54–65. doi:10.1016/j.ibmb.2014.07.006
Scott, J. G., Liu, N., Wen, Z., Smith, F. F., Kasai, S., and Horak, C. E. (1999). House-fly cytochrome P450 CYP6D1: 5′ flanking sequences and comparison of alleles. Gene 226, 347–353. doi:10.1016/s0378-1119(98)00545-9
Shi, L., Zhang, J., Shen, G., Xu, Z., Xu, Q., and He, L. (2016). Collaborative contribution of six cytochrome P450 monooxygenase genes to fenpropathrin resistance in Tetranychus cinnabarinus (Boisduval). Insect Mol. Biol. 25, 653–665. doi:10.1111/imb.12251
Shwe, S. M., Wang, Y., Gao, Z., Li, X., Liu, S., Bai, S., et al. (2021a). Toxicity of Cry1-Class, Cry2Aa, and Vip3Aa19 Bt proteins and their interactions against yellow peach moth, Conogethes punctiferalis (Guenée) (Lepidoptera: Crambidae). J. Invertebr. Pathol. 178, 107507. doi:10.1016/j.jip.2020.107507
Shwe, S. M., Prabu, S., Chen, Y., Li, Q., Jing, D., Bai, S., et al. (2021b). Baseline susceptibility and laboratory selection of resistance to Bt Cry1Ab protein of Chinese populations of yellow peach moth, Conogethes punctiferalis (Guenée). Toxins 13, 335. doi:10.3390/toxins13050335
Siegwart, M., Thibord, J. B., Olivares, J., Hirn, C., Elias, J., Maugin, S., et al. (2017). Biochemical and molecular mechanisms associated with the resistance of the European corn borer (Lepidoptera: Crambidae) to lambda-cyhalothrin and first monitoring tool. J. Econ. Entomol. 110, 598–606.
Stanley, J., Chandrasekaran, S., and Preetha, G. (2009). Conogethes punctiferalis (Lepidoptera: Pyralidae) its biology and field parasitization. Indian J. Agric. Sci. 79, 906–909. doi:10.1093/jee/tow267
Su, J., and Sun, X. X. (2014). High level of metaflumizone resistance and multiple insecticide resistance in field populations of Spodoptera exigua (Lepidoptera: Noctuidae) in Guangdong Province, China. Crop Prot. 61, 58–63. doi:10.1016/j.cropro.2014.03.013
Sun, Z., Shi, Q., Li, Q., Wang, R., Xu, C., Wang, H., et al. (2019). Identification of a cytochrome P450 CYP6AB60 gene associated with tolerance to multi-plant allelochemicals from a polyphagous caterpillar tobacco cutworm (Spodoptera litura). Pestic. Biochem. Physiol. 154, 60–66. doi:10.1016/j.pestbp.2018.12.006
Tang, T., Zhao, C., Feng, X., Liu, X., and Qiu, L. (2012). Knockdown of several components of cytochrome P450 enzyme systems by RNA interference enhances the susceptibility of Helicoverpa armigera to fenvalerate. Pest Manage. Sci. 68, 1501–1511. doi:10.1002/ps.3336
Ullah, F., Gul, H., Tariq, K., Desneux, N., Gao, X., and Song, D. (2020). Functional analysis of cytochrome P450 genes linked with acetamiprid resistance in melon aphid, Aphis gossypii. Aphis Gossypii. Pestic. Biochem. Physiol. 170, 104687. doi:10.1016/j.pestbp.2020.104687
Wan, P. J., Shi, X. Q., Kong, Y., Zhou, L. T., Guo, W. C., Ahmat, T., et al. (2013). Identification of cytochrome P450 monooxygenase genes and their expression profiles in cyhalothrin-treated 549 Colorado potato beetle, Leptinotarsa decemlineata. Pestic. Biochem. Physiol. 107, 360–368. doi:10.1016/j.pestbp.2013.10.004
Wang, X., and Wu, Y. (2012). High levels of resistance to chlorantraniliprole evolved infield populations of Plutella xylostella. J. Econ. Entomol. 105, 1019–1023. doi:10.1603/ec12059
Wang, R. L., Xia, Q. Q., Baerson, S. R., Ren, Y., Wang, J., Su, Y. J., et al. (2015). A novel cytochrome P450 CYP6AB14 gene in Spodoptera litura (Lepidoptera: Noctuidae) and its potential role in plant allelochemical detoxification. J. Insect Physiol. 75, 54–62. doi:10.1016/j.jinsphys.2015.02.013
Wang, R. L., Liu, S. W., Baerson, S. R., Qin, Z., Ma, Z. H., Su, Y. J., et al. (2018). Identification and functional analysis of a novel cytochrome P450 gene CYP9A105 associated with pyrethroid detoxification in Spodoptera exigua Hübner. Int. J. Mol. Sci. 19, 737. doi:10.3390/ijms19030737
Wang, Z. G., Jiang, S. S., Mota-Sanchez, D., Wang, W., Li, X. R., Gao, Y. L., et al. (2019). Cytochrome P450-mediated λ-cyhalothrin-resistance in a field strain of Helicoverpa armigera from Northeast China. J. Agric. Food Chem. 67, 3546–3553. doi:10.1021/acs.jafc.8b07308
Wang, R., Liu, J., Zhu, X., Pang, Y., Wang, Y., Liu, X., et al. (2020). Screening of insecticides for controlling yellow peach moth, Conogethes Punctiferalis (Guenée) on maize. Pestic. Sci. Adm. 41, 42–49. doi:10.1093/gastro/goz055
Xu, Y. Q., Wang, J. J., Jiang, H. B., Wei, D., Tang, P. A., and An, F. M. (2009). Identification, characterization, and expression of P450 gene encoding CYP6BQ13v2 from the red flour beetle, Tribolium castaneum (Herbst) (Coleoptera: Tenebrionidae). Agric. Sci. China. 8, 1210–1218. doi:10.1016/s1671-2927(08)60331-4
Yang, T., and Liu, N. (2011). Genome analysis of cytochrome P450s and their expression profiles in insecticide resistant mosquitoes, Culex quinquefasciatus. PloS One 6, e29418. doi:10.1371/journal.pone.0029418
Yang, X., Li, X., and Zhang, Y. (2013). Molecular cloning and expression of CYP9A61: A chlorpyrifos-ethyl and lambda-cyhalothrin-inducible cytochrome P450 cDNA from Cydia pomonella. Int. J. Mol. Sci. 14, 24211–24229. doi:10.3390/ijms141224211
Yu, H. L., Xiang, X., Yuan, G. X., Chen, Y. Q., and Wang, X. G. (2015). Effects of sublethal doses of cyantraniliprole on the growth and development and the activities of detoxifying enzymes in Spodoptera exigua (Lepidoptera: Noctuidae). Acta Entomol. Sin. 58, 634–641.
Zhang, X., Liu, X., Ma, J., and Zhao, J. (2013). Silencing of cytochrome P450 CYP6B6 gene of cotton bollworm (Helicoverpa armigera) by RNAi. B. Entomol. Res. 103, 584–591. doi:10.1017/S0007485313000151
Zhao, P., Xue, H., Zhu, X., Wang, L., Zhang, K., Li, D., et al. (2021). Silencing of cytochrome P450 gene CYP321A1 effects tannin detoxification and metabolism in Spodoptera litura. Int. J. Biol. Macromol. 194, 895–902. doi:10.1016/j.ijbiomac.2021.11.144
Zhou, X., Fan, X., Gao, Y., Yang, J., Qian, J., and Fan, D. (2017). Identification of two novel P450 genes and their responses to deltamethrin in the cabbage moth, Mamestra brassicae Linnaeus. Pestic. Biochem. Physiol. 141, 76–83. doi:10.1016/j.pestbp.2016.12.001
Keywords: Conogethes punctiferalis, cytochrome P450, adaptability, chemical pesticides, RNA interference
Citation: Yuan X, Li H, Guo X, Jiang H, Zhang Q, Zhang L, Wang G, Li W and Zhao M (2023) Functional roles of two novel P450 genes in the adaptability of Conogethes punctiferalis to three commonly used pesticides. Front. Physiol. 14:1186804. doi: 10.3389/fphys.2023.1186804
Received: 15 March 2023; Accepted: 05 June 2023;
Published: 28 June 2023.
Edited by:
Jia Fan, Institute of Plant Protection (CAAS), ChinaCopyright © 2023 Yuan, Li, Guo, Jiang, Zhang, Zhang, Wang, Li and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Man Zhao, emhhb21hbjgyMUBoZW5hdS5lZHUuY24=
†These authors have contributed equally to this work
 Xingxing Yuan1†
Xingxing Yuan1† Xianru Guo
Xianru Guo He Jiang
He Jiang Man Zhao
Man Zhao