
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Physiol., 26 June 2023
Sec. Cardiac Electrophysiology
Volume 14 - 2023 | https://doi.org/10.3389/fphys.2023.1183280
This article is part of the Research TopicMethods and Applications in Cardiac Electrophysiology - application to inherited arrhythmiasView all 8 articles
 Shijie Zhou1,2*
Shijie Zhou1,2* Raymond Wang3
Raymond Wang3 Avery Seagren1
Avery Seagren1 Noah Emmert2
Noah Emmert2 James W. Warren4
James W. Warren4 Paul J. MacInnis4
Paul J. MacInnis4 Amir AbdelWahab5
Amir AbdelWahab5 John L. Sapp5
John L. Sapp5Background: We previously developed a non-invasive approach to localize the site of early left ventricular activation origin in real time using 12-lead ECG, and to project the predicted site onto a generic LV endocardial surface using the smallest angle between two vectors algorithm (SA).
Objectives: To improve the localization accuracy of the non-invasive approach by utilizing the K-nearest neighbors algorithm (KNN) to reduce projection errors.
Methods: Two datasets were used. Dataset #1 had 1012 LV endocardial pacing sites with known coordinates on the generic LV surface and corresponding ECGs, while dataset #2 included 25 clinically-identified VT exit sites and corresponding ECGs. The non-invasive approach used “population” regression coefficients to predict the target coordinates of a pacing site or VT exit site from the initial 120-m QRS integrals of the pacing site/VT ECG. The predicted site coordinates were then projected onto the generic LV surface using either the KNN or SA projection algorithm.
Results: The non-invasive approach using the KNN had a significantly lower mean localization error than the SA in both dataset #1 (9.4 vs. 12.5 mm, p < 0.05) and dataset #2 (7.2 vs. 9.5 mm, p < 0.05). The bootstrap method with 1,000 trials confirmed that using KNN had significantly higher predictive accuracy than using the SA in the bootstrap assessment with the left-out sample (p < 0.05).
Conclusion: The KNN significantly reduces the projection error and improves the localization accuracy of the non-invasive approach, which shows promise as a tool to identify the site of origin of ventricular arrhythmia in non-invasive clinical modalities.
Sudden cardiac arrest is a major cause of death in developed countries, with approximately 350,000 deaths per year in the United States alone (Al-Khatib et al., 2017). The majority of those events are caused by ventricular arrhythmias (VAs) (Tang et al., 2017). Catheter ablation has emerged as an established therapeutic option for the treatment of VAs (Reddy et al., 2007; Kuck et al., 2010; Sapp et al., 2016). Accurate identification of the substrate responsible for the VA is key to the success of the modality and may be facilitated using 12-lead ECG to non-invasively localize the breakthrough site from which a focal ventricular tachycardia (VT) or premature ventricular contraction (PVC) arises, from an exit pathway of a transmural re-entry, or from which a re-entrant circuit exits the central isthmus to activate the “normal” myocardium. Computer-aided methods for rapidly and automatically localizing the site of early ventricular activation origin in real-time using 12-lead ECG can be particularly helpful for catheter ablation of VA (Cronin et al., 2020). Several non-invasive algorithms based on the 12-lead ECG have been proposed for localizing the site of early ventricular activation origin with varying degrees of accuracy (Josephson et al., 1981; Miller et al., 1988; Betensky et al., 2011; Yokokawa et al., 2012; Efimova et al., 2015; Misra et al., 2018; Alawad and Wang, 2019; He et al., 2020). Recent studies using deep learning—based VT exit/PVC origin localization have shown moderate localization accuracy (Yang et al., 2018; Gyawali et al., 2020), and the “black box” nature of these deep learning models makes it difficult to interpret how input variables interact to identify the VT exit/PVC origin site (Zhou et al., 2021).
Locating the site of origin within 10 mm is of great clinical significance. We have previously developed a non-invasive automated approach that combines information from 12-lead ECG recordings and a generic LV endocardial mesh surface consisting of 238 area elements to localize the site of early left-ventricular (LV) activation origin (Sapp et al., 2017; Zhou et al., 2019). We have shown that, using the non-invasive automated approach based on the 12-lead ECG (Sapp et al., 2017; Zhou et al., 2019), spatial localization of the site of early LV activation origin can be achieved without significant loss of accuracy in comparison with our 120-lead Electrocardiographic Imaging (ECGI) (Zhou et al., 2018a; Zhou et al., 2018b); ECGI uses CT/MRI volumes to delineate the cardiac anatomy and determine the relative location of body surface electrodes to reconstruct epicardial electrical events by a mathematical process known as calculation of inverse solution (Zhou et al., 2018a; Zhou et al., 2018b). The non-invasive automatic approach demonstrated a mean localization error of 12.2 ± 8.14 mm (Sapp et al., 2017; Zhou et al., 2019). Briefly, the non-invasive automated approach was based on the hypothesis of a linear relationship between coordinates of early left ventricular activation sites and ECG lead potentials (QRS integrals); a dataset comprising coordinates (exported from an electroanatomic mapping (EAM) system) of 1012 LV endocardial pacing sites and their corresponding ECGs was used to calculate population-derived regression coefficients; the non-invasive automated approach uses these population-derived regression coefficients to predict the target coordinates of a pacing site or VA from the initial 120-m QRS integrals of the pacing site ECG or the VA ECG. The predicted pacing-site/VA origin site coordinates are projected onto one of the 238 triangular area elements of the generic LV endocardial mesh surface using the smallest angle between two vectors algorithm (named SA algorithm), so that the projected site can be targeted for ablation. However, the prolate ellipsoid geometry of the normal LV shape, with a ratio of 2:1 from echocardiographic long axis dimension to minor axis dimension (Udelson, 2017), may lead to projection errors when using the SA algorithm.
In this study, we hypothesized that K-Nearest Neighbors (KNN) algorithm can reduce the projection error and improve the localization performance of the non-invasive automated approach. The paper is organized as follows, Section 2 describes the methods in detail, Section 3 presents the results, and Section 4 concludes the study and discusses its limitations and future research.
This study utilized two datasets (Sapp et al., 2017; Zhou et al., 2019; Zhou et al., 2020). The first dataset (#1) comprised 1012 LV endocardial pacing sites pooled from 38 patients, which were exported from an EAM system (Carto 3, Biosense Webster, Inc., Irvine, CA, United States) and included corresponding ECGs (Sapp et al., 2017; Zhou et al., 2019). The second dataset (#2) included 25 clinically-identified LV endocardial VT exit sites with corresponding ECGs (Zhou et al., 2020). All participating patients gave written informed consent; the study protocol was approved by the Institutional Research Ethics Board (Nova Scotia Health Authority, Halifax, Canada).
In a previous study, we constructed a generic LV endocardial mesh surface consisting of 238 triangles, derived from the necropsy specimen of a normal human heart (Hren et al., 1998). The average distance between the centers of the 238 triangles was found to be 5.4 ± 1.4 mm (mean ± SD). In the current study, the Cartesian coordinates of each pacing site or clinically-identified VT exit site were manually registered from the patient-specific EAM geometry onto one of the 238-triangle centers of the generic LV endocardial surface by two independent observers (Drs Sapp and AbdelWahab) (Sapp et al., 2017; Zhou et al., 2019; Zhou et al., 2020). The registration process had an interobserver variability of approximately 3.7 mm in the surrounding area (Zhou et al., 2020). For each pacing site/clinical-identified VT exit site, the QRS integral was calculated over the initial 120 m of the QRS complex for the 8 independent leads (I, II, V1-V6) of the 12-lead ECG (∫QRS, in microvolt-seconds) (Sapp et al., 2017; Zhou et al., 2018a; Zhou et al., 2018b; Zhou et al., 2019; Zhou et al., 2020) by using summing up the area of the 120 m QRS window.
The non-invasive automated approach based on a training set (n samples) with a multiple linear regression (MLR) model can be applied to the 238-triangle generic LV endocardial mesh surface, provided the 8-variable set (P:i, i = 1, . . ., k = 8) can be generated from the 8 independent leads ECG (I, II, V1-V6) for the training-set pacing sites with known coordinates xj, yj, zj, (j = 1, . . . n.) (Sapp et al., 2017; Zhou et al., 2019). The MLR model with intercept is used to determine “population” regression coefficients in the regression equations linking each of the 3 coordinates of a known pacing site xj, yj, zj, with the values of the corresponding QRS integrals, Pji, where the subscript i indicates one of the 8 leads:
Here
The unknowns in this set of equations are the coordinates of the pacing-site origin or the clinically-induced VT exit site,
To project the predicted pacing/VT exit site
After calculating Eq. 3, the smallest angle,
The K-nearest neighbors (KNN) algorithm is a non-parametric approach for classification and regression tasks in supervised learning (Bzdok et al., 2018). In this study, the KNN is used to predict the class of the test data by calculating the distance between the test data and all the training points, then to select the ‘K’ number of points which is close to the test data. In this study, the test data is the predicted pacing/VT exit site (
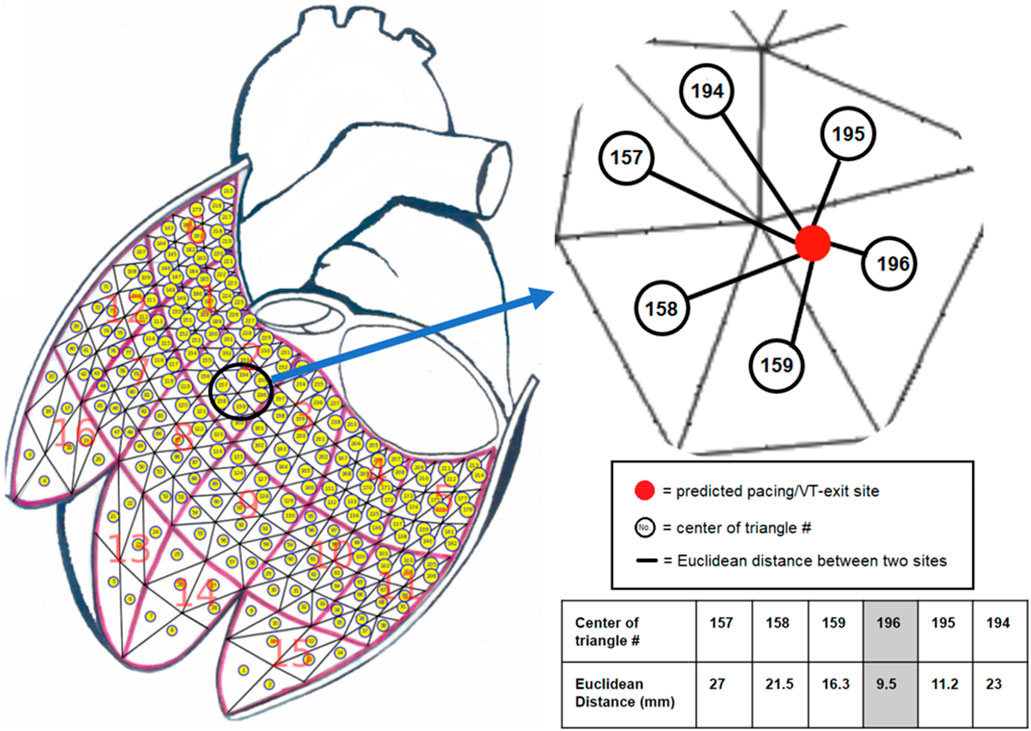
FIGURE 1. The K-nearest neighbors (KNN) algorithm with Euclidean distance measurement was used to project a predicted pacing/VT-exit site marked by the red ball onto one of the 238-triangle centers of the generic LV endocardial mesh surface. The example illustrates that a predicted pacing/VT-exit site marked by the red ball was projected onto the No. 196 triangle center by finding the shortest distance from all of the 238 Euclidean distances calculated by the predicted pacing/VT-exit site and all of the 238-triangle centers of the generic LV endocardial mesh surface.
To evaluate the localization performance of the non-invasive automated approach, the entire dataset #1 (n = 1,012) was randomly partitioned into a training set with 80% of the entire set (n = 810) and a test set with the remaining 20% (n = 202). The “population” regression coefficients were calculated from the training set (n = 810) using Eq. 1, and then applied as constants with the ECG variables extracted from the 8 independent leads ECG of any activation sequence of interest initiated at the unknown site obtained from the test set (n = 202)., The resulting coordinates
In addition, the bootstrap method with replacement was used to assess the statistical inference (Efron and Tibshirani, 1993). To estimate the optimistic bias of the proposed projection algorithm, the mean error with standard deviation was calculated and applied to the left-out sample (n = 1,012/e≃371), which served as a test set. The mean and standard deviation of 1,000 bootstrap trials was calculated. The standard error was used to construct the 95%-confidence interval of the localization performance. The Minitab’s boxplot was used to visualize the distribution of data and identify any potential outliers. To determine the outliers, the Minitab involved identifying data points that are more than 1.5 times the interquartile range (IQR) below the first quartile or above the third quartile.
The actual pacing site is known and specified on the 238-triangle generic LV endocardial mesh surface as triangle centroid (
We evaluated the localization performance of the non-invasive automated approach using the KNN projection algorithm on dataset #2, which consists of 25 clinically-identified VT-exit sites. We calculated the “population” regression coefficients using the entire dataset #1 (n = 1,012), and compared the results to those of the non-invasive automated approach using the SA projection algorithm. In the Dataset #2, we used the Euclidean distance to assess the localization accuracy of the non-invasive automated approach using the SA projection algorithm for predicting the 25 clinically-identified VT-exit sites in our previous study (Zhou et al., 2020). To ensure consistency in this study, the localization error of the VT exit site was assessed by measuring the Euclidean distance between the clinically-identified site and the estimated site both on the generic LV endocardial mesh surface.
The study evaluated the localization error of a non-invasive automated approach using two projection algorithms. Geodesic distance was calculated from the centroid of the predicted triangle to the centroid of the known reference triangle on the 238-triangle generic LV endocardial mesh surface. After deriving “population” regression coefficients from a training set (n = 810), the accuracy of the approach was assessed on a test set (n = 202). The mean accuracy of the non-invasive automated approach was 9.4 mm and 12.5 mm for using the KNN projection and the SA projection algorithms, respectively. Figure 2 (left panel) shows that the mean localization error using the KNN projection algorithm was significantly lower than that of the SA projection algorithm (9.4 vs. 12.5 mm, p < 0.05).
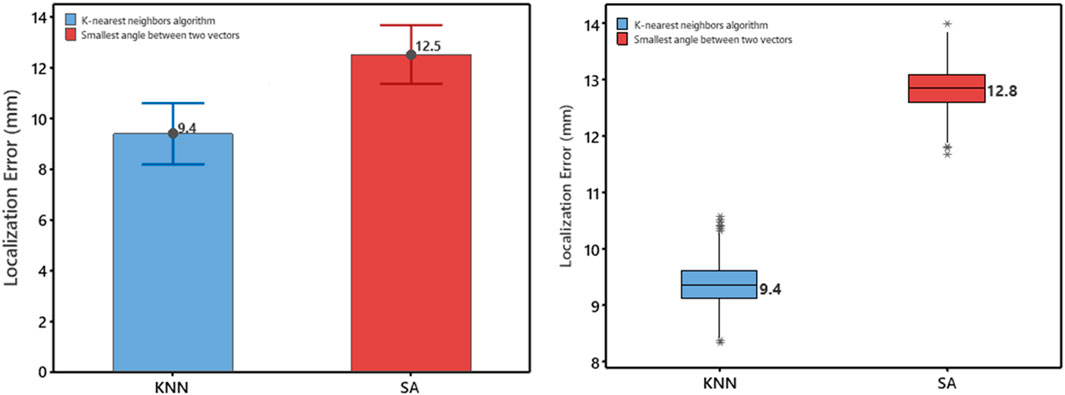
FIGURE 2. Left Panel, Comparison based on the test set (n = 202) for the error measured as geodesic distance using the two projection algorithms (KNN, SA). Mean values are shown numerically. Right Panel: Box plot of localization error for using the two projection algorithms (KNN, SA). Plots represent data for mean localization error in terms of geodesic distance on the generic LV-endocardial surface for the left-out sample (n = 1,012/e≃371). Boxes represent interquartile ranges; a line inside the box marks the median, “whiskers” above and below the box indicate range, ∗∗ represent outliers.
The localization performance of the non-invasive automated approach using the two projection algorithms was further evaluated in terms of geodesic distance between the centroid of the projected and actual pacing site, measured on the generic LV endocardial mesh surface. The bootstrap method with replacement, using 1,000 trials (Efron and Tibshirani, 1993), was employed to summarize the results. Table 1 and in Figure 2 (right panel) provide an overview of the results. As shown in Figure 2 (right panel), the KNN projection algorithm yielded a significantly lower mean localization errors compared to the SA projection algorithm (KNN vs SA, p < 0.05) in the left-out sample (n≃371). The distributions of the left-out-sample errors are depicted in Figure 3, based on the pacing-site localization error measured as a geodesic distance in the generic LV endocardial mesh surface, for the 1,000 bootstrap trials using the non-invasive automated approach with the two projection algorithms.
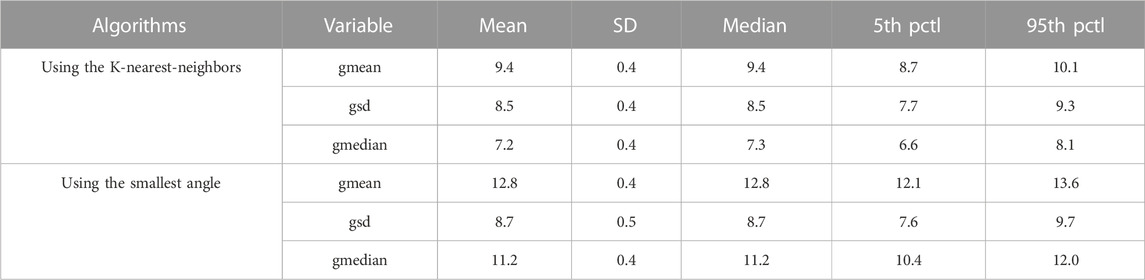
TABLE 1. Bootstrap method with replacement (Efron and Tibshirani, 1993), using 1,000 trials, was used; the left-out sample had n = 1,012/e≃371 pacing sites. Three g-variables quantify accuracy of localization by eight-predictor regression in terms of geodesic distance (in mm) from the centroid of the predicted triangle to the centroid of the pacing-site triangle on the 238-triangle generic LV endocardial mesh surface; gmean, mean value; gsd, standard deviation; gmedian, median value; Pctl, percentile.
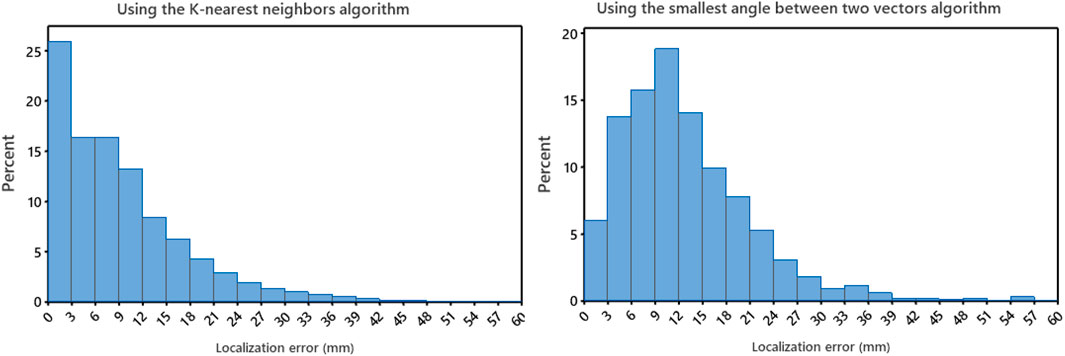
FIGURE 3. Left panel: the distributions for the non-invasive automated approach using the K-nearest neighbors (KNN) projection algorithm of the bootstrap left-out-sample errors, based on localization errors on the LV endocardial surface for 1,000 bootstrap trials. Right panel: the distributions for the non-invasive automated approach using the smallest angle between two vectors algorithm of the bootstrap left-out-sample errors, based on localization errors on the LV endocardial surface for 1,000 bootstrap trials. Error was measured as geodesic distance (approximated by the arc length) between the centroid of the projected triangle and the centroid of the pacing-site triangle on the 238-triangle generic LV endocardial mesh surface.
Figure 4 shows the localization errors of 25 clinically-identified VT exit sites in the dataset #2, based on the non-invasive automated approach using the two different projection algorithms. The mean localization error of the non-invasive automated approach, using the KNN projection algorithm (named proposed approach), was significantly lower than that of the non-invasive automated approach using the SA projection algorithm (published in reference 23). (7.2 vs. 9.5 mm, p < 0.05). Figure 5 illustrates the non-invasive automated approach for localizing the exit site of a ventricular tachycardia (VT) using two projection algorithms. This VT had a cycle length of 315 m, with right bundle branch block-type morphology in lead V1, and a rightward axis (Figure 5A). The site of exit was identified at the mid-apical anterolateral wall inferior to the anterolateral papillary muscle. The VT exit site was localized to the more apical portion of the mid-anterolateral segment identified by the non-invasive automated approach using the two projection algorithms, respectively (Figure 5B for using the SA projection algorithm; Figure 5C for using the KNN projection algorithm). The electroanatomic substrate map is shown in Figure 5D, with the site of VT exit identified (yellow arrow, yellow star and gold ball).
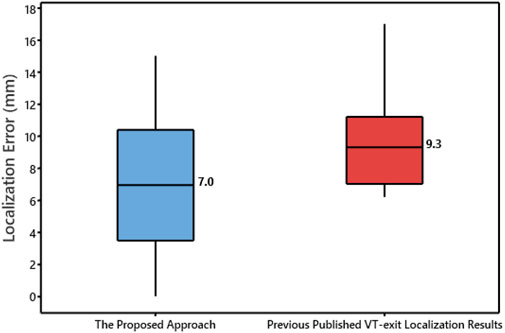
FIGURE 4. Box plot of localization error of the 25 VT exit sites for using the non-invasive automated approach based the two projection algorithms. Plots represent data for mean localization error in terms of Euclidean distance.
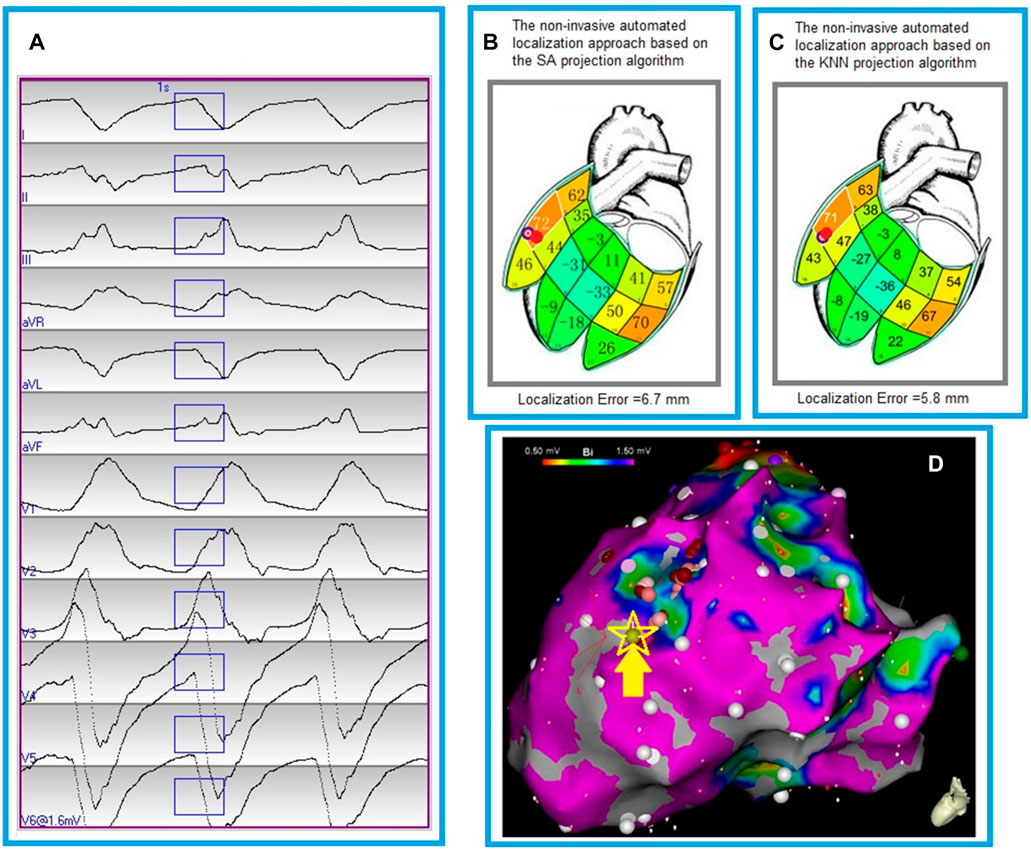
FIGURE 5. Localization of a ventricular tachycardia (VT) exit by the non-invasive automated localization using the two projection algorithms. (A), The recorded 12-lead ECG of an induced monomorphic VT during the procedure. The onset of one VT beat was automatically detected (Kemmelings et al., 1994); the user can edit the onset of the 120 m window (rectangle box) if correction is necessary. (B), Bull’s eye icon that indicates the estimated VT-exit locations using the non-invasive automated approach based on the smallest angle between two vectors (SA algorithm). The red ball indicates the VT reference site on the 238-triangle generic LV endocardial mesh surface, which was registered manually from an endocardial electroanatomic mapping map (panel D). Localization error of the VT exit site is 6.7 mm between the bull’s eye icon and the red ball. The large number within each segment is the correlation coefficient (%) for match by the 12-lead ECG VT pattern with population based 12-lead ECG templates; the small number identifies the segment. (C), Bull’s eye icon that indicates the estimated VT-exit locations using the non-invasive automated approach based on the K-nearest neighbors algorithm (KNN). The red ball registered manually from an endocardial electroanatomic mapping map (panel D) indicates the VT reference site on the 238-triangle generic LV endocardial mesh surface. Localization error of the VT exit site is 5.8 mm between the bull’s eye icon and the red ball. The large number within each segment is the correlation coefficient (%) for match by the 12-lead ECG VT pattern with population based 12-lead ECG templates; the small number identifies the segment. (D), an endocardial electroanatomic substrate map, with areas featuring bipolar signal amplitude ≥1.50 mV in purple, and the site of VT exit (identified by contact mapping) depicted by the yellow arrow, yellow starand gold ball.
In this study, we introduced the K-nearest neighbors (KNN) algorithm to improve the localization performance of a non-invasive automated approach by reducing the projection error. The KNN algorithm was used to project the predicted pacing/VT-exit site onto a generic LV endocardial mesh surface, and compare its accuracy to the smallest angle between two vectors projection algorithm. The non-invasive automated approach utilizing the KNN projection algorithm achieved a mean localization accuracy of < 10 mm in both datasets, highlighting its clinical significance.
Based on the comprehensive assessments, we conclude that the KNN projection algorithm can enhance the localization accuracy of the non-invasive automated approach in clinical cardiac electrophysiology. The KNN algorithm does not rely on any machine learning model that requires a pre-existing training on a dataset to make predictions. In other words, the KNN does not require any training, which saves the training dataset and uses it only when making real-time predictions to learn. This makes the KNN algorithm much faster than other training-based algorithms, such as, random forest or support vector machine. In addition, the KNN requires knowing the number of categories (one or more), which means that the K value has a powerful effect on the KNN performance. In our specific situation, the ‘K’ value was required to be 1, which completely solved the most prominent issue—the optimal K number for determining the KNN performance. Therefore, the 1-NN (‘K’ NN) algorithm was directly used to calculate the Euclidean distances between the predicted pacing/VT-exit site and all of the 238-triangle centers of the generic LV endocardial mesh surface, finding the shortest distance for the calculated 238 Euclidean distances.
Our study represents a significant step towards improving the localization accuracy of the non-invasive automated approach for real-time localization of early LV activation origin, which has potential applications for catheter ablation of VA (Cronin et al., 2020) and targeting VT locations for substrate modification using cardiac stereotactic body radiotherapy (cSBRT) (Qian et al., 2021). Recently cardiac SBRT as a non-invasive alternative has been shown to provide a viable option for VT which is refractory to ablation and medication (Cuculich et al., 2017). While ECGI has been proposed as a means to identify VT substrate for guiding cardiac SBRT (Graham et al., 2019; Samson et al., 2020), its spatial accuracy is limited and depends on several factors (Hohmann et al., 2019). Septal activation, for example, is inherently difficult to represent on the epicardial surface (Duchateau et al., 2019). Duchateau et al. compared ECGI with invasive epicardial mapping, and found an inadequate correlation between the two modalities, with an average distance of 75 mm from the invasively mapped focal breakthrough locations to the predicted origin sites (Bhaskaran et al., 2019). Our proposed approach provides a promising foundation for future studies in the non-invasive cSBRT. However, there are several limitations of this study. Specifically, the proposed study has limited applicability in the right ventricle (RV) endocardium and epicardium. To overcome this limitation, future studies could utilize the same method to identify the site of early RV/epicardial activation origin. Additionally, the proposed study is based on a generic LV endocardial mesh surface and does not account for the patient-specific LV endocardial surface. Future studies would explore a non-invasive ECG-image-based mapping approach that relies on personalized ventricular surfaces from CT/MRI scans and the proposed approach for identifying the site of early ventricular activation origin in both ventricles.
The K-nearest neighbors (KNN) algorithm can greatly reduce the projection error, improve the localization accuracy of the non-invasive automated approach. By utilizing the KNN projection algorithm, the non-invasive automated approach outperforms any prior published approached, which may potentially facilitate the use of non-invasive clinical modalities in identifying the site of origin of ventricular arrhythmia.
The original contributions presented in the study are included in the article/supplementary materials, further inquiries can be directed to the corresponding author.
The studies involving human participants were reviewed and approved by Nova Scotia Health Authority. The patients/participants provided their written informed consent to participate in this study. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
SZ processed all clinical data, ran all algorithms and analyses and wrote the manuscript. RW implemented the KNN algorithm, AS and NE helped data processing. PM developed the localization software. AA and JS helped obtain and provide the clinical data. JW, AA, and JS contributed to the preparation of the manuscript. All authors contributed to the article and approved the submitted version.
American Heart Association (AIREA grant # 949812).
JS: a co-holder of a patent for automated VT localization; no licensing, royalties or income currently or anticipated. Research funding from Biosense-Webster and Abbott (for clinical trial of catheter ablation of VT); modest speaker honoraria Medtronic, Biosense Webster, Abbott. AA: speaker honoraria Abbott, Medtronic.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Alawad, M., and Wang, L. (2019). Learning domain shift in simulated and clinical data: Localizing the origin of ventricular activation from 12-lead electrocardiograms. IEEE Trans. Med. Imaging 38, 1172–1184. doi:10.1109/TMI.2018.2880092
Al-Khatib, S. M., Stevenson, W. G., Ackerman, M. J., Bryant, W. J., Callans, D. J., Curtis, A. B., et al. (2017). 2017 AHA/ACC/HRS guideline for management of patients with ventricular arrhythmias and the prevention of sudden cardiac death: Executive summary: A report of the American college of cardiology/American heart association task force on clinical practice guidelines and the heart rhythm society. Circulation 138, e210–e271. doi:10.1161/CIR.0000000000000548
Betensky, B. P., Park, R. E., Marchlinski, F. E., Hutchinson, M. D., Garcia, F. C., Dixit, S., et al. (2011). The V(2) transition ratio: A new electrocardiographic criterion for distinguishing left from right ventricular outflow tract tachycardia origin. J. Am. Coll. Cardiol. 57, 2255–2262. doi:10.1016/j.jacc.2011.01.035
Bhaskaran, A., Nayyar, S., Porta-Sanchez, A., Haldar, S., Bokhari, M., Massé, S., et al. (2019). Exit sites on the epicardium rarely subtend critical diastolic path of ischemic VT on the endocardium: Implications for noninvasive ablation. J. Cardiovasc Electrophysiol. 30, 520–527. doi:10.1111/jce.13843
Bzdok, D., Krzywinski, M., and Altman, N. (2018). Machine learning: Supervised methods. Nat. Methods 15, 5–6. doi:10.1038/nmeth.4551
Cronin, E. M., Bogun, F. M., Maury, P., Peichl, P., Chen, M., Namboodiri, N., et al. (2020). 2019 HRS/EHRA/APHRS/LAHRS expert consensus statement on catheter ablation of ventricular arrhythmias: Executive summary. Heart rhythm. 17, e155–e205. doi:10.1016/j.hrthm.2019.03.014
Cuculich, P. S., Schill, M. R., Kashani, R., Mutic, S., Lang, A., Cooper, D., et al. (2017). Noninvasive cardiac radiation for ablation of ventricular tachycardia. N. Engl. J. Med. 377, 2325–2336. doi:10.1056/NEJMoa1613773
Duchateau, J., Sacher, F., Pambrun, T., Derval, N., Chamorro-Servent, J., Denis, A., et al. (2019). Performance and limitations of noninvasive cardiac activation mapping. Heart rhythm. 16, 435–442. doi:10.1016/j.hrthm.2018.10.010
Efimova, E., Dinov, B., Acou, W. J., Schirripa, V., Kornej, J., Kosiuk, J., et al. (2015). Differentiating the origin of outflow tract ventricular arrhythmia using a simple, novel approach. Heart rhythm. 12, 1534–1540. doi:10.1016/j.hrthm.2015.04.004
Efron, B., and Tibshirani, R. J. (1993). An introduction to the bootstrap. New York: Chapman and Hall.
Graham, A. J., Orini, M., Zacur, E., Dhillon, G., Daw, H., Srinivasan, N. T., et al. (2019). Simultaneous comparison of electrocardiographic imaging and epicardial contact mapping in structural heart disease. Circ. Arrhythm. Electrophysiol. 12, e007120. doi:10.1161/CIRCEP.118.007120
Gyawali, P. K., Horacek, B. M., Sapp, J. L., and Wang, L. (2020). Sequential factorized autoencoder for localizing the origin of ventricular activation from 12-lead electrocardiograms. IEEE Trans. Biomed. Eng. 67, 1505–1516. doi:10.1109/TBME.2019.2939138
He, K., Nie, Z., Zhong, G., Yang, C., and Sun, J. (2020). Localization of origins of premature ventricular contraction in the whole ventricle based on machine learning and automatic beat recognition from 12-lead ECG. Physiol. Meas. 41, 055007. doi:10.1088/1361-6579/ab86d7
Hohmann, S., Rettmann, M. E., Konishi, H., Borenstein, A., Wang, S., Suzuki, A., et al. (2019). Spatial accuracy of a clinically established noninvasive electrocardiographic imaging system for the detection of focal activation in an intact porcine model. Circ. Arrhythm. Electrophysiol. 12, e007570. doi:10.1161/CIRCEP.119.007570
Hren, R., Nenonen, J., and Horáček, B. M. (1998). Simulated epicardial potential maps during paced activation reflect myocardial fibrous structure. Ann. Biomed. Eng. 26, 1022–1035. doi:10.1114/1.73
Josephson, M. E., Horowitz, L. N., Waxman, H. L., Cain, M. E., Spielman, S. R., Greenspan, A. M., et al. (1981). Sustained ventricular tachycardia: Role of the 12-lead electrocardiogram in localizing site of origin. Circulation 64, 257–272. doi:10.1161/01.cir.64.2.257
Kemmelings, J. G., Linnenbank, A. C., Muilwijk, S. L., SippensGroenewegen, A., Peper, A., and Grimbergen, C. A. (1994). Automatic QRS onset and offset detection for body surface QRS integral mapping of ventricular tachycardia. IEEE Trans. Biomed. Eng. 41, 830–836. doi:10.1109/10.312090
Kuck, K. H., Schaumann, A., Eckardt, L., Willems, S., Ventura, R., Delacrétaz, E., et al. (2010). Catheter ablation of stable ventricular tachycardia before defibrillator implantation in patients with coronary heart disease (VTACH): A multicentre randomised controlled trial. Lancet 375, 31–40. doi:10.1016/S0140-6736(09)61755-4
Miller, J. M., Marchlinski, F. E., Buxton, A. E., and Josephson, M. E. (1988). Relationship between the 12-lead electrocardiogram during ventricular tachycardia and endocardial site of origin in patients with coronary artery disease. Circulation 74, 759–766. doi:10.1161/01.cir.77.4.759
Misra, S., van Dam, P., Chrispin, J., Assis, F., Keramati, A., Kolandaivelu, A., et al. (2018). Initial validation of a novel ECGI system for localization of premature ventricular contractions and ventricular tachycardia in structurally normal and abnormal hearts. J. Electrocardiol. 51, 801–808. doi:10.1016/j.jelectrocard.2018.05.018
Qian, P. C., Quadros, K., Aguilar, M., Wei, C., Boeck, M., Bredfeldt, J., et al. (2021). Substrate modification using stereotactic radioablation to treat refractory ventricular tachycardia in patients with ischemic cardiomyopathy. JACC Clin. Electrophysiol. 8, 49–58. doi:10.1016/j.jacep.2021.06.016
Reddy, V. Y., Reynolds, M. R., Neuzil, P., Richardson, A. W., Taborsky, M., Jongnarangsin, K., et al. (2007). Prophylactic catheter ablation for the prevention of defibrillator therapy. N. Engl. J. Med. 357, 2657–2665. doi:10.1056/NEJMoa065457
Samson, P., Hugo, G., Moore, K., Knutson, N., Cuculich, P., Robinson, C., et al. (2020). Noninvasive cardiac radioablation at Washington university: Past, present, future directions for the treatment of VT. Appl. Rad. Oncol. 9, 10–14.
Sapp, J. L., Wells, G. A., Parkash, R., Stevenson, W. G., Blier, L., Sarrazin, J. F., et al. (2016). Ventricular tachycardia ablation versus escalation of antiarrhythmic drugs. N. Engl. J. Med. 375, 111–121. doi:10.1056/NEJMoa1513614
Sapp, J. L., Bar-Tal, M., Howes, A. J., Toma, J. E., El-Damaty, A., Warren, J. W., et al. (2017). Real-time localization of Ventricular Tachycardia origin from the 12-lead electrocardiogram. JACC Clin. Electrophysiol. 3, 687–699. doi:10.1016/j.jacep.2017.02.024
Tang, P. T., Shenasa, M., and Boyle, N. G. (2017). Ventricular arrhythmias and sudden cardiac death. Card. Electrophysiol. Clin. 9, 693–708. doi:10.1016/j.ccep.2017.08.004
Udelson, J. E. (2017). Left ventricular shape: The forgotten stepchild of remodeling parameters. JACC Heart Fail. 5, 179–181. doi:10.1016/j.jchf.2017.01.005
Yang, T., Yu, L., Jin, Q., Wu, L., and He, B. (2018). Localization of origins of premature ventricular contraction by means of convolutional neural network from 12-lead ECG. IEEE Trans. Biomed. Eng. 65, 1662–1671. doi:10.1109/TBME.2017.2756869
Yokokawa, M., Liu, T., Yoshida, K., Scott, C., Hero, A., Good, E., et al. (2012). Automated analysis of the 12-lead electrocardiogram to identify the exit site of postinfarction ventricular tachycardia. Heart rhythm. 9, 330–334. doi:10.1016/j.hrthm.2011.10.014
Zhou, S., Horáček, B. M., Warren, J. W., AbdelWahab, A., and Sapp, J. L. (2018a). Rapid 12-lead automated localization method: Comparison to electrocardiographic imaging (ECGI) in patient-specific geometry. J. Electrocardiol. 51 (6), S92–S97. doi:10.1016/j.jelectrocard.2018.07.022
Zhou, S., Sapp, J. L., AbdelWahab, A., Stovicek, P., and Horáček, B. M. (2018b). Localization of ventricular activation origin using patient-specific geometry: Preliminary results. J. Cardiovasc Electrophysiol. 29 (7), 979–986. doi:10.1111/jce.13622
Zhou, S., AbdelWahab, A., Sapp, J. L., Warren, J. W., and Horáček, B. M. (2019). Localization of ventricular activation origin from the 12-lead ECG: A comparison of linear regression with non-linear methods of machine learning. Ann. Biomed. Eng. 47, 403–412. doi:10.1007/s10439-018-02168-y
Zhou, S., AbdelWahab, A., Horacek, B. M., MacInnis, P. J., Warren, J. W., Davis, J. S., et al. (2020). Prospective assessment of an automated intraprocedural 12-lead ECG-based system for localization of early left ventricular activation. Circ. Arrhythm. Electrophysiol. 13, e008262. doi:10.1161/CIRCEP.119.008262
Keywords: k-nearest neighbors (KNN) algorithm, ventricular tachycardia, pace-mapping, radiofrequency ablation, ECG
Citation: Zhou S, Wang R, Seagren A, Emmert N, Warren JW, MacInnis PJ, AbdelWahab A and Sapp JL (2023) Improving localization accuracy for non-invasive automated early left ventricular origin localization approach. Front. Physiol. 14:1183280. doi: 10.3389/fphys.2023.1183280
Received: 09 March 2023; Accepted: 02 June 2023;
Published: 26 June 2023.
Edited by:
Ange Maguy, University of Bern, SwitzerlandReviewed by:
Peter Michael Van Dam, University Medical Center Utrecht, NetherlandsCopyright © 2023 Zhou, Wang, Seagren, Emmert, Warren, MacInnis, AbdelWahab and Sapp. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shijie Zhou, c2hpamllLnpob3VAbWlhbWlvaC5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.