
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Physiol. , 29 March 2023
Sec. Avian Physiology
Volume 14 - 2023 | https://doi.org/10.3389/fphys.2023.1139321
This article is part of the Research Topic Avian Microbiome: from embryonic development to adulthood View all 10 articles
Microbes commonly administered to chickens facilitate development of a beneficial microbiome that improves gut function, feed conversion and reduces pathogen colonization. Competitive exclusion products, derived from the cecal contents of hens and shown to reduce Salmonella colonization in chicks, possess important pioneer-colonizing bacteria needed for proper intestinal development and animal growth. We hypothesized that inoculation of these pioneer-colonizing bacteria to day of hatch chicks would enhance the development of their intestinal anatomy and microbiome. A competitive exclusion product was administered to broiler chickens, in their drinking water, at day of hatch, and its impact on intestinal morphometrics, intestinal microbiome, and production parameters, was assessed relative to a control, no treatment group. 16S rRNA gene, terminal restriction fragment length polymorphism (T-RFLP) was used to assess ileal community composition. The competitive exclusion product, administered on day of hatch, increased villus height, villus height/width ratio and goblet cell production ∼1.25-fold and expression of enterocyte sugar transporters 1.25 to 1.5-fold in chickens at 3 days of age, compared to the control group. As a next step, chicks were inoculated with a defined formulation, containing Bacteroidia and Clostridia representing pioneer-colonizing bacteria of the two major bacterial phyla present in the competitive exclusion product. The defined formulation, containing both groups of bacteria, were shown, dependent on age, to improve villus height (jejunum: 1.14 to 1.46-fold; ileum: 1.17-fold), goblet cell numbers (ileum 1.32 to 2.51-fold), and feed efficiency (1.18-fold, day 1) while decreasing Lactobacillus ileal abundance by one-third to half in birds at 16 and 42 days of age, respectively; compared to the phosphate buffered saline treatment group. Therefore, specific probiotic formulations containing pioneer colonizing species can provide benefits in intestinal development, feed efficiency and body weight gain.
The microbiome has been shown to serve as an effective barrier to pathogen colonization or pathogenic behavior in numerous examples while the mechanisms underlying pathogen exclusion remains elusive (Nurmi and Rantala, 1973; Berg, 1980; Faure et al., 1984). Approximately 50 years ago, Nurmi demonstrated that chicks seeded with the cecal microbiome from adult birds were resistant to Salmonella colonization (Nurmi and Rantala, 1973) and termed the phenomena “competitive exclusion”. Since this discovery, numerous groups have investigated single or multiple microbial species as probiotics to replace the effectiveness of growth-promoting antibiotics (Vuong et al., 2016) or suppress other harmful microorganisms (Fukata et al., 1991; Hofacre et al., 1998a). However, no defined consortium has been quite as effective at pathogen exclusion as Nurmi’s approach using the cecal microbiome. Competitive exclusion has since been commercialized; amplifying cecal bacteria from an original seed stock and distributing lyophilized cultures with >5 log10 Salmonella reduction to customers, marketed as a Salmonella exclusion product for poultry (Lee et al., 2023). Hofacre et al. demonstrated that administration of this competitive exclusion product could also reduce the severity of necrotic enteritis in poultry (Hofacre et al., 1998a). The study, which was replicated in 2019 with conditions intended to increase the severity of disease (Hofacre et al., 2019), illustrated an important new concept in disease control. The chicks were given one dose at day of hatch, then challenged with three sequential high oral doses of an avian pathogenic C. perfringens isolate 3 weeks later. The findings suggested a paradigm shift because the principle of competitive exclusion was inadequate to explain the chicks’ resistance to repeated doses of a billion pathogenic, toxigenic Clostridium perfringens cells administered orally for 3 days in a row 3 weeks after receiving the intestinal bioproducts. In fact, a subsequent study showed that one dose of the intestinal bioproduct, Aviguard®, performed as well as continuously feeding bacitracin or virginiamycin to prevent necrotic enteritis (Hofacre et al., 1998b). These findings indicated that competitive exclusion products could alter the intestinal environment leading to greater resistance to enteropathogens.
Understanding the microbiome, a consortium of microbes found in and on animal and plant species, offers a new perspective on eukaryotic development. An understanding of the nutritional and physiological contributions of the monogastric vertebrate intestinal microbiome is still emerging. The role of the microbiome in disease, physiology or development is often inferred from studies using “germ-free” subjects versus “conventionally-raised” litter mates. Monogastrics can be raised “germ-free” (gnotobiotic) (Pleasants, 1959; Meyer et al., 1964) however, gnotobiotic mice, pigs and rats exhibit reduced growth and weight gain compared to conventionally raised litter mates (Dymsza et al., 1965; Waxler and Drees, 1972; Al-Asmakh and Zadjali, 2015). Gnotobiotic animals are also more susceptible to enteric infections which makes them an excellent model for studying enteric pathogens (Sprinz et al., 1961; Eaton et al., 2008; Reeves et al., 2012). Augmentation with certain bacterial species has been shown have a profound effect on the intestinal physiology, growth, and disease resistance of gnotobiotic animals (Dymsza et al., 1965; Shirkey et al., 2006; Mahowald et al., 2009; Cheled-Shoval et al., 2014; Greig et al., 2018; Yin et al., 2022). Furthermore, the microbiome composition can have a profound impact on animal weight gain as evident when gnotobiotic mice receive fecal transplants from obese mice (Turnbaugh et al., 2006).
In comparisons with other germ-free animal models, chickens possessing an intestinal microbiota were believed to be at a growth disadvantage except when grown on vitamin-deficient diets (Coates et al., 1968) or those with high fiber/low metabolizable energy (Muramatsu et al., 1991). Gnotobiotic-chickens produce fewer goblet cells, more sulfated mucins (Cheled-Shoval et al., 2014), shorter villi, and lower crypt depth, compared to conventionally-raised birds (Cowieson, 2022). However, weight gain and feed conversion improved when chickens were raised with antimicrobial-amended feed but this was not observed in germ free animals (Lev and Forbes, 1959). Furthermore, it was most pronounced in birds raised in heavily contaminated environments indicating that the growth disadvantage was likely the result of pathogens. Therefore, in commercial poultry production, antibiotics such as virginiamycin and bacitracin improved growth performance on farms with high stocking density or poor hygienic conditions or practices (Eyssen and de Somer, 1963). Because this improvement was believed to be due to the suppression of pathogenic intestinal bacterial species such as C. perfringens, it led to the widespread use of antibiotics, in the U.S., as a prophylactic to prevent necrotic enteritis in poultry. This practice has been in decline since the European ban of growth-promoting antibiotics (Casewell et al., 2003) and the movement in the U.S. towards antibiotic free production (Diaz-Sanchez et al., 2015).
The maternal intestinal microbiome is an important source of organisms for the progeny and many studies have shown that the initial microbiome seeding is crucial for health of the progeny (Neu and Rushing, 2011; Koleva et al., 2015; Chen et al., 2018; Kubasova et al., 2019a; Klein-Jöbstl et al., 2019; Treichel et al., 2019; Yu et al., 2019). The modern poultry production system, in order to increase productivity and reduce disease transmission, eliminated the physical presence of the progenitors during the incubation and hatching process thereby interrupting the transfer of bacteria from hen to chick. As a result, newly hatched chicks do not have access to a diverse maternal microbiome are easily seeded with environmental microbes (Pedroso et al., 2015) and these organisms would not be expected to provide beneficial effects in early intestinal development or pathogen exclusion. Yet, there is a consistent and predictable microbial succession within the chicken intestine, beginning with oxygen-consuming streptococci and γ-proteobacteria at 4 days of age, followed by their displacement with the obligate anaerobic Clostridia (Lu et al., 2003; Jurburg et al., 2019). However, if chicks are presented early with cecal microbiome, they develop a stable community resistant to pathogen colonization (Ramírez et al., 2020). Because the microbiome affects animal physiology (Cheled-Shoval et al., 2014; Heath-Heckman et al., 2014; Kremer et al., 2014; McFall-Ngai, 2014), this would be especially evident in early intestinal development as the host responds to early pioneer colonizers (Shao et al., 2014; De Maesschalck et al., 2015; Gourbeyre et al., 2015).
The ecological concept of pioneer colonizers is well established and has been shown to be crucial in augmenting development of a functionally diverse intestinal microbiome. In 2005, Backhed et al. conceptualized the process by which pioneer colonizers coevolve with their animal hosts and influence the intestinal environment from a nutritional and anatomical standpoint (Backhed et al., 2005). Using gnotobiotic mice, they demonstrated that the developmental deficiencies associated with the absence of an intestinal microbiome, could be fully mitigated by administering a single Bacteroides thetaiotaomicron species. Subsequent studies illustrated that stem cell differentiation was stimulated by bacterial metabolites from utilization of host intestinal mucin (Sommer and Backhed, 2013). These findings indicated that probiotic formulations, containing pioneer colonizers from the intestinal microbiota of mature chickens, may accelerate intestinal development and improve performance in newly hatched chicks.
In this study, we treated day-of-hatch broiler chickens with a competitive exclusion product and investigated the impact of the treatments on intestinal community structure, as measured by 16 S rRNA gene terminal restriction fragment length polymorphism (T-RFLP), intestinal morphometrics (villus height, villus height to width ratio, goblet cell numbers), body weight gain and feed conversion ratios; compared to the no treatment, control group. As a second step, we selected specific species from chicken ceca, that represent the two dominant phyla in the competitive exclusion product Aviguard® (Lee et al., 2023), and administered different formulations, consisting of these cecal organism, or PBS to newly hatched chicks in order to determine their effects on production performance, intestinal physiology and changes to the intestinal microbiome, relative to the PBS treatment group. Similar morphometric improvements to chicken gut function could be obtained with a two to five species, probiotic cocktail, consisting of obligate anaerobic pioneer colonizers, than a competitive exclusion product that consists of 20–50 distinct genera (Lee et al., 2023).
In order to recover bacteria from the commercial competitive exclusion product, the bacterial cells were rehydrated by incubation for 10 min in saline solution and recovered by centrifugation. Chicken intestines were collected from the various treatments and time points outlined below. The bacterial fraction was recovered from the intestinal contents through multiple rounds of differential centrifugation as described previously (Apajalahti et al., 1998; Lu et al., 2003). DNA was extracted using Mo Bio kit (Mo Bio Laboratories Inc., Solana Beach, CA), beating cell suspensions at 6,000 rpm for 20 min (Lu et al., 2008). The bacterial communities were assayed by 16 S rRNA terminal restriction fragment length polymorphism (T-RFLP) analysis, using a sequence-based database, as previously described (Lu et al., 2006; Lu et al., 2008). universal 16S rRNA primers 8F labeled with 5′FAM (carboxyfluorescein-N-hydroxysuccinimide ester-dimethyl sulfoxide) and unlabeled 1429R were used to amplify community DNA (Lu et al., 2008). Three separate 18-cycle PCR reactions were performed for each DNA sample and pooled for T-RFLP analysis. No DNA template was included with PCR, as a negative control. No amplicons were ever observed for this negative control. Amplicons were digested with restriction enzyme HaeIII (New England BioLabs; Ipswich, MA) and analyzed by electrophoresis on ABI PRISM 310 DNA sequencer (PE Biosystems; Foster City, CA). For each sample, only peak areas and peak heights over a threshold of 50 units, above background were analyzed by manually aligning fragments to size standards; and only DNA fragments between 35 and 525 bp were examined. T-RFL peaks were identified by comparison to a 16S rRNA gene database, of Insilco HaeIII patterns, from previously published clone libraries (Lu et al., 2003).
The relative abundance of bacterial species or phylotypes detected by T-RFLP was determined by calculating the ratio between the areas of each peak and the total areas of all peaks within one sample (Lukow et al., 2000); mean ratios of three analyses were converted to percentages. The Shannon diversity information index (Shannon and Wiener, 1963) was used to evaluate the diversity of the bacterial communities. The diversity indices were analyzed using analysis of variance (SAS, 2008) to determine differences between the intestinal communities from birds given Aviguard® or nothing (no treatment control group).
Aviguard®, consists predominantly of obligate anaerobes (Pedroso et al., 2015; Lee et al., 2023), belonging to Clostridia and Bacteroidia orders and because it improved intestinal morphometrics in young birds, we sought to isolate and identify pioneer colonizing species that could supplant this competitive exclusion product. Anaerobic, pioneer colonizing bacterial were isolated from the ceca of commercial broiler chicken carcasses obtained from a local processing plant. The cecal contents of three chickens were squeezed into pre-reduced serum bottles and serially diluted with 20 ml of phosphate-buffered saline (PBS) in within an anaerobic chamber containing 90% N2 and 10% H2. The suspensions were plated on rumen fluid-glucose-cellobiose plus peptone (RGCAP)-10, RGCAP-30, 10% modified rumen fluid medium (M98-5), and rich medium (RM) (ATCC Medium 1,341; 20 g glucose, 10 g yeast extract, 2 g K2HPO4, 15 g agar per 1L dH2O) agar (Kelley, 1983), and incubated under 95% N2 and 5% H2 for 5 days at 41°C. Isolated colonies were characterized by 16 S rDNA sequencing as previously described (Lu et al., 2003). Selected isolates of Escherichia coli, Parabacteroides distasonis, Bacteroides salyersiae, Phocaeicola dorei and Romboutsia lituseburensis ATCC 25759 were grown on RGCAP-10 agar under anaerobic conditions (80% N2, 10% CO2 and 10% H2) for 5 days. Colonies were harvested and resuspended in pre-reduced saline solution to reach the concentration of 109 CFU/ml.
Obligate anaerobes were isolated by culture and identified by 16S rRNA amplicon sequencing. Subculture yielded obligate anaerobes belonging to the order Bacteroidia which were identified as P. distasonis, B. salyersiae, and P. dorei (formerly, Bacteroides dorei). Partial sequence of their genomes revealed polysaccharide utilization loci and associated glycosyl hydrolases (Grondin et al., 2017) characteristic of the Bacteroidia. DNA sequence of these genes, 16S rRNA, and other housekeeping genes confirmed their identity to the species level (BLAST scores: ≥98% nucleotide identity; 100% coverage) (Table 1). The Bacteroidia genomes exhibited several annotated genes for acetate and propionate metabolism. Bacteroides salyersiae and P. dorei also possessed genes annotated as phosphotransbutyrylase and butyrate kinase, responsible for butyrate production. These genes were absent in a search of the isolated P. distasonis’ genome as well as a search of published, annotated P. distasonis genomes, including a specified BLAST search, at the amino acid level. While these Bacteroidia contained core carbohydrate-active enzymes (CAZymes), there were differences in the distribution of other CAZymes among these isolates. Because of the variances in carbohydrate and fermentation metabolism, it was decided to include multiple species as part of a Bacteroidia cocktail to administer to birds.
As R. lituseburensis was an abundant phylotype in birds fed the competitive exclusion product Aviguard® or other feed additives (Lu et al., 2008), an R. lituseburensis isolate was purchased from the American Type Culture Collection (ATCC 25759) to be included in this study. In addition, an E. coli isolated from the chicken intestinal samples, served as a γ-proteobacteria pioneer for establishing the anaerobic environment needed for seeding chicks with obligate anaerobes (Espey, 2013).
Pools of isolates were created by mixing equal volumes of suspensions. Three pools of probiotic cultures were prepared for administration to day of hatch chicks and consisted of the following formulations; probiotic cocktail 1: P. distasonis, B. salyersiae, and P. dorei, and E. coli; probiotic cocktail 2: R. lituseburensis and E. coli; and probiotic cocktail three containing P. distasonis, B. salyersiae, P. dorei, R. lituseburensis and E. coli. Glycerol (15%) was added to aliquots of each probiotic formulation and stored at −80°C.
For assessment of the effects of the commercial competitive exclusion product (Aviguard®, Lallemand, Montreal Canada), 120 one-day-old commercial Ross-Cobb hybrid broiler chicks were raised in two groups of 60 on sawdust bedding. Both groups were fed a commercial corn-soy bean meal diet devoid of antimicrobials. Chicks in one group were administered the commercial competitive exclusion product, Aviguard® in their drinking water on the day of hatch, as per the manufacturer’s instructions, while the other group just received standard drinking water, no product. Birds were sacrificed, by cervical dislocation, at 3, 7, 14, 21, 28, and 49 days of age and intestines were collected. The ileal contents were collected and processed as previously described for 16S rRNA gene TRFLP analysis. Intestinal morphometrics and glucose transporter gene expression were performed on intestines collected from birds at 3 days of age, as described below.
Eight hundred and 40 day of hatch chicks (Cobb 500) were divided into four treatments of three replications each containing 70 chicks. Chicks were orally inoculated with 50 µl of 1 × 108 Bacteroidia cocktail (P. distasonis, B. salyersiae, and P. dorei) with E. coli, R. lituseburensis with E. coli, and Bacteroidia cocktail with R. lituseburensis and E. coli. The control group received 50 μl sterile PBS. Chickens were fed a corn-soy bean meal diet free of antimicrobials (Table 1). Birds were sacrificed by cervical dislocation 3 hours following oral administration with probiotic formulation or PBS and at days 1, 2, 3, 7, 16 and 42 days and intestines were collected.
Following inoculation with the competitive exclusion product Aviguard® (n = 60), probiotic formulation (3 different formulations, 210 birds per treatment) or PBS (n = 210), chickens were sacrificed at 3 hours after inoculation, or at time points described above and the small intestines were collected. A no treatment group (n = 60) was included with the competitive exclusion trial. The middle portion of the jejunum and ileum from 4 birds per experimental unit were excised, fixed in 10% formalin, embedded in paraffin and cut in five um thick sections. Three intact, well-oriented villi were selected in eight replicates for each intestinal cross section, totaling 24 villus height and width measurements for each intestinal sample and 288 measurements per treatment. In addition, intestinal sections were stained using Mayer’s Mucicarmine (Val-Bernal et al., 1999) and the number of goblet cells counted. Morphological indices were determined using a light microscope and a ×16 magnification lens. Images were analysis using the Image-Pro Plus Version 3.0 software (Media Cybernetics, Silver spring, MD). Expression of glucose transporters were measured by reverse-transcriptase (RT) qPCR according to method described by Gilbert et al. (2007).
The jejunum and ileum from four animals per treatment group (three probiotic cocktails and PBS control) were collected, measured, flushed using deionized water, and the empty weight recorded. Relative intestinal weight (grams/kg of body weight) and relative intestinal lengths (mm/kg of body weight) were determined.
Body weight and feed intake were recorded and body weight gain and gain: feed were calculated. At the occurrence of mortality, feed intake was adjusted based on bird days on feed. At 42 days of age, fifteen chickens per pen were randomly selected and wing-banded and fasted overnight. Birds were weighed individually, slaughtered, eviscerated, and carcasses were chilled for 12 h. The yield was obtained for the entire carcass, and parts.
Two samples were thawed on ice, centrifuged at 10,000xg at 4°C for 15 min, and DNA was extracted from the bacterial pellet using Promega Wizard® Genomic DNA extraction kit (Madison, WI), with an added lysozyme treatment, as described by the manufacturer. DNA was submitted to Georgia Genomics and Bioinformatics Core for sequencing using Illumina sequencing (San Diego, CA). FastQC and FastQ/A were used to clean raw sequence reads of adapters and low-quality sequences (Patel and Jain, 2012; Afgan et al., 2018). SPADES sequence alignment tool was used to assemble processed pair-end Illumina reads (Bankevich et al., 2012). Assembled sequence files were uploaded and annotated in the Rapid Annotation using Subsystem Technology (RAST) (Aziz et al., 2008). Species identity of individual contigs (≥17 kb) was determined by Basic Local Alignment Search Tool (BLAST) (Altschul et al., 1990) at nucleotide level (≥98% identity). Identity of species, within this probiotic formulation, was confirmed by BLAST for sequences annotated as: “small ribosomal subunit RNA” (16 S rRNA); “large ribosomal subunit RNA (23S rRNA); gene annotated as “SusC”, “SusD”, polysaccharide utilization genes commonly present in the Bacteroidia (Grondin et al., 2017); or housekeeping genes listed in Table 1 (≥98% identity, 100% coverage). The Bacteroidia are adept at metabolizing complex carbohydrates, whether its indigestible fiber from the animal’s diet or mucin, and producing volatile fatty acid from said metabolism for its host. As multiple Bacteroidia species were identified, a more detailed genomic analysis was performed to determine which isolates to include in the probiotic formulation that had the broadest repertoire of carbohydrate metabolism. Carbohydrate-active enzymes (CAZymes) were identified among the annotated sequences through a word search for genes annotated as “Sus”, “glycosyl hydrolases”, “amylase”, “pullanase” or “idase”; and species identity and enzyme confirmation was determined by BLAST at the nucleotide and amino acid level, respectively. CAZymes identified had motifs consistent with these enzymes at the amino acid level. In Bacteroidia, CAZymes are often associated with Polysaccharide Utilization Loci (PUL) denoted by polysaccharide transporters Sus (Grondin et al., 2017). Several loci were identified with Sus minus any genes annotated as some CAZyme. Genes annotated as “hypothetical protein” were identified as CAZyme via BLASTX search of annotated bacterial genomes. Fermentation enzymes were identified by similar word search of gene annotations for enzymes listed in Table 1. Their identity and species assignment were determined by BLAST search at the amino acid and nucleotide level, respectively.
Performance and intestinal measurements were subjected to Analysis of Variance (ANOVA) procedure for completely randomized design using the general linear model procedure of SAS (SAS, 2008). Statistical significance of differences among treatments was assessed using the least significant difference test (Steel and Torrie, 1980). A probability level of p < 0.05 was used to determine statistical significance. The Standard Error of Measure (SEM) was calculated from the standard deviation of all values divided by the square-root of the sample size.
While competitive exclusion products have been shown effective at pathogen exclusion, can this microbial consortium, of chicken intestinal origin, also effectively modulate intestinal morphology and function? To address this question, birds were either administered the competitive exclusion product, Aviguard® at day of hatch, in their drinking water or not (untreated, control group). Administration of Aviguard® improved intestinal morphology, increasing villus height, height/width ratio; and percentage of goblet cells per villus 1.26 to 1.36-fold, p < 0.05 (Table 2), compared to untreated birds. Furthermore, increased expression 1.25 to 1.5-fold of the enterocyte transporters, GLUT2, GLUT5, and SGLT1 was exhibited in the ileum compared to control group in 3-day-old broiler chickens (Figure 1; p < 0.05).
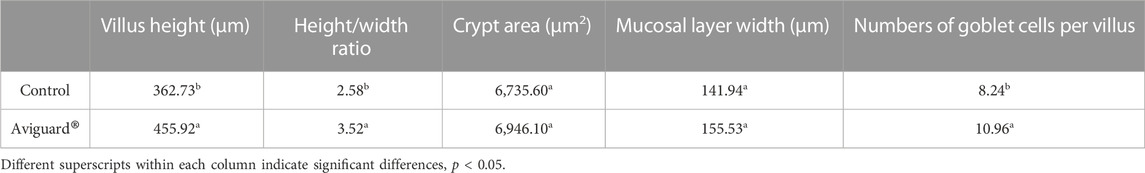
TABLE 2. Ileal morphology of broiler chicks at 3 days of age that were administered an intestinal bioproduct Aviguard®.
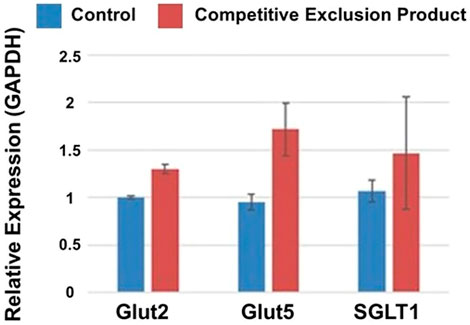
FIGURE 1. Relative expression of glucose transporters Glut2, Glut5 and SGLT1 (p < 0.05) in the small intestine of 3 d old broiler chickens receiving a competitive exclusion product (Aviguard®) or no treatment (control) on the day of hatch. Relative gene expression was determined using the 2−ΔΔCT method. GAPDH was used to normalize gene expression for targeted genes.
Figure 2 and Figure 3 illustrate the composition and successions of bacteria in the microbiome in response to administration of Aviguard® to birds at day of hatch. There were differences in the succession of bacterial phylotypes over the 49-day period between the control and birds administered the competitive exclusion product, Aviguard® (Supplementary Figure S1). There were also significant differences in the distribution of phylotypes between the control and birds administered the competitive exclusion product, especially evident were differences in Lactobacillus crispatus and R. lituseburensis (Clostridia) abundance. This was most pronounced in birds at 21 days of age and older (Figure 2). Lactobacillus species were the most abundant group in untreated birds (Figure 2, Supplementary Figure S1), while the abundance of other species varied. However, administration of Aviguard® produced an ileal bacterial community in which the Clostridia were abundant at 3 days of age while Enterococcus phylotypes represented 60% of total phylotypes on day 7 (Figure 2, Supplementary Figure S1). But SFB/Bacteroides phylotypes emerged with R. lituseburensis day 7 with Romboutsia becoming the most abundant ileal species by day 28 representing 70% of the total phylotypes for the treatment group. This observation suggested that Bacteroides may act as a pioneer colonizer in chicks enabling successional colonization of poultry anaerobic bacteria.
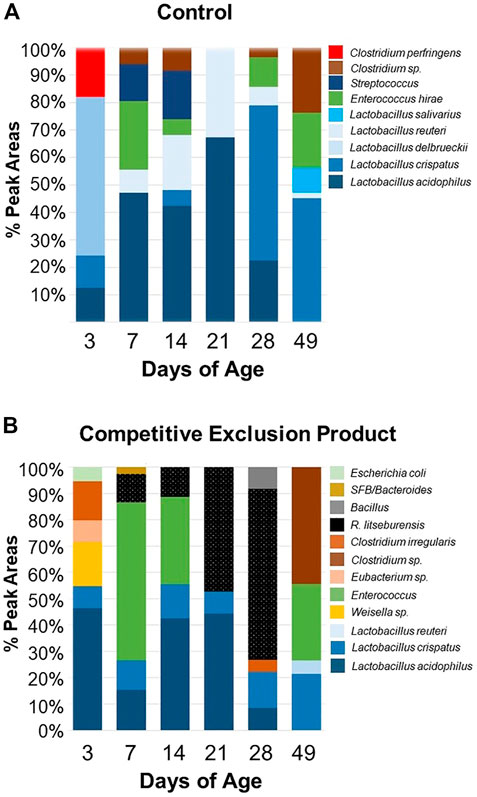
FIGURE 2. Composition of the ileal bacterial community of chicks administered Aviguard® (panel (B)) or no treatment (control, Panel (A) on day of hatch as determined by 16S rRNA T-RFLP at 3, 7, 14, 21, 28 and 49 days of age.
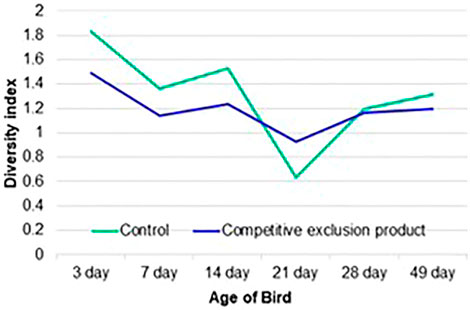
FIGURE 3. Shannon’s H diversity index of samples collected from the ileal bacterial community of chicken from the control (no treatment) or birds administered a competitive exclusion product (Aviguard®) at day of hatch. Samples were collected at 3, 7, 14, 21, 28 and 49 days of age.
The Shannon diversity index indicated that age-related instability in the diversity of the ileal communities could be reduced by Aviguard® (Figure 3). There were significant differences between the control and treatment groups at all ages analyzed (p < 0.05). At 21 days of age, there was a distinct reduction in diversity which was most pronounced for the control group. Aviguard® administration lowered diversity but provided stability compared to the dramatic shifts in control birds.
Newly hatched chicks inoculated with R. lituseburensis and E. coli cocktail had the greatest (15.8 vs 14.7 g; p < 0.05), body weight gain 24 h following its administration (Table 3), relative to the PBS control. Similarly, the feed: gain ratio was improved 1.18-fold relative to PBS control (1,408 vs. 1,198 kg/g; p < 0.05) in chicks receiving the P. distasonis, B. salyersiae, P. dorei, E. coli and R. lituseburensis cocktail. In addition, R. lituseburensis and E. coli cocktail or the P. distasonis, B. salyersiae, P. dorei, E. coli and R. lituseburensis cocktail produced a higher body weight gain at 16 days of age (438 or 421 vs. 411; p < 0.05). Probiotic cocktails consisting of R. lituseburensis and E. coli or P. distasonis, B. salyersiae, P. dorei, E. coli and R. lituseburensis reduced body weight gains by 4% at the end of the rearing period, compared to birds administered PBS or the P. distasonis, B. salyersiae, P. dorei, and E. coli cocktail. However, birds administered R. lituseburensis and E. coli cocktail had higher carcass yield (Table 4; 76.1% vs 73.7%, p < 0.05). No differences were observed on the legs, thighs, wings and breast yield for either probiotic formulations or PBS control therefore the weight gains were likely tied to changes in intestinal development.
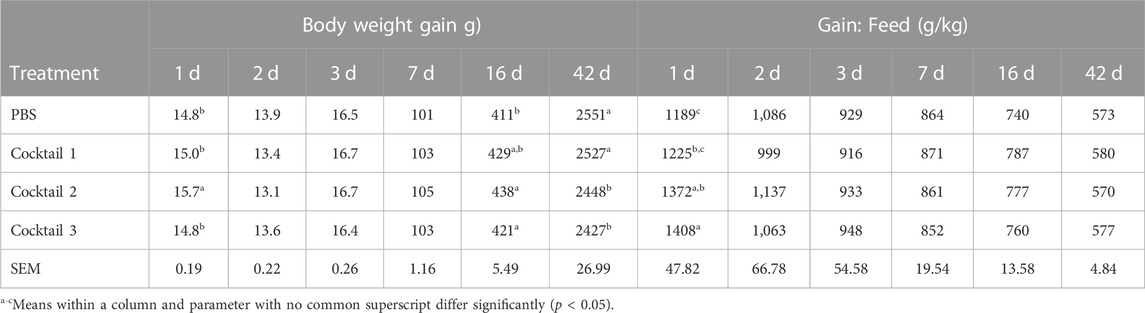
TABLE 3. Body weight gain and feed efficiency of birds inoculated with Parabacteroides distasonis, Bacteroides salyersiae, Phocaeicola dorei, and Escherichia coli (Cocktail 1); Romboutsia lituseburensis and E. coli (Cocktail 2); P. distasonis, B. salyersiae, P. dorei, E. coli and R. lituseburensis (Cocktail 3); or PBS.
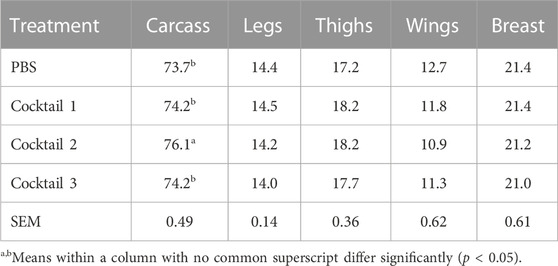
TABLE 4. Carcass yield (%) of chickens at 42 d old that were administered Parabacteroides distasonis, Bacteroides salyersiae, Phocaeicola dorei, and Escherichia coli (Cocktail 1); Romboutsia lituseburensis and E. coli (Cocktail 2); P. distasonis, B. salyersiae, P. dorei, E. coli and R. lituseburensis (Cocktail 3); or PBS.
During the first week, changes in the intestinal development were observed in response to the probiotics administered to day of hatch chicks. Birds administered the P. distasonis, B. salyersiae, P. dorei, and E. coli cocktail had a higher relative jejunal weight, 1.28 to 1.44-fold increase, just 3 h following administration compared to chicks receiving R. lituseburensis and E. coli cocktail or the PBS control, respectively (Table 5). At 2 days of age, the group that received P. distasonis, B. salyersiae, P. dorei, and E. coli cocktail had a relative jejunal weight, ∼1.2-fold greater than the control or the other probiotic formulations. The relative weight of the jejunum was significantly decreased by 18% for birds administered R. lituseburensis and E. coli cocktail in comparison to the PBS control at 42 days (p < 0.05). There were no significant differences in jejunal length for either probiotic administration compared to the control. The probiotics also did not impact the relative weight or length of the ileum (Supplementary Table S2).
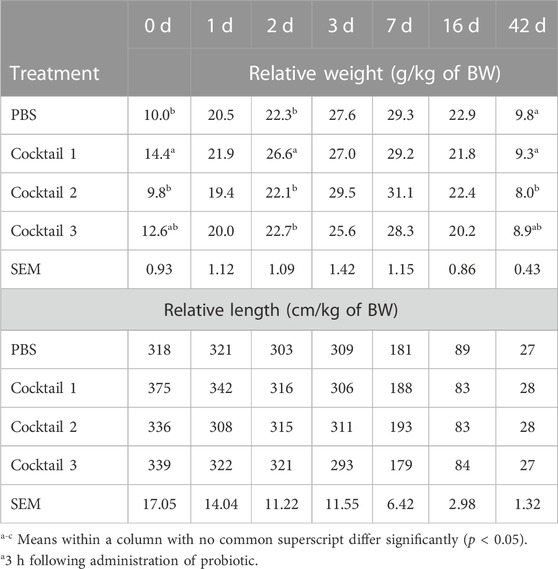
TABLE 5. Relative intestinal weight and length of the jejunum of chickens administered Parabacteroides distasonis, Bacteroides salyersiae, Phocaeicola dorei, and Escherichia coli (Cocktail 1); Romboutsia lituseburensis and E. coli (Cocktail 2); P. distasonis, B. salyersiae, P. dorei, E. coli and R. lituseburensis (Cocktail 3); or PBS.
The P. distasonis, B. salyersiae, P. dorei, E. coli and R. lituseburensis cocktail induced longer jejunal villi just 3 h following administration (Table 6; Supplementary Figure S2), and continued to increase villus height at 7 and 16 days of age, compared to the PBS control (1.46, 1.14, and 1.15-fold increase, respectively; p < 0.05). However, the villus height was shorter in birds at 2 and 3 days of age, for R. lituseburensis and E. coli cocktail or P. distasonis, B. salyersiae, P. dorei, and E. coli cocktail compared to the PBS control (∼20% decrease; p < 0.05). By 42 days of age, there were no significant differences in jejunal villus height for either group. The probiotic formulations did not seem to elicit enhancement of villi height in the ileum as seen in the jejunum until birds were 42 days of age. At this time point, all three formulations increased villi height compared to the PBS control with R. lituseburensis and E. coli cocktail or P. distasonis, B. salyersiae, P. dorei, E. coli and R. lituseburensis having the most pronounced effect on villus height (1.39 and 1.16-fold increase, respectively; p < 0.05). At earlier time points, the probiotics appeared to reduce ileal villus height, compared to the control group, 3 h (P. distasonis, B. salyersiae, P. dorei, and E. coli cocktail; 40% decrease; p < 0.05) following probiotic administration; and at day 7, all three probiotic formulations reduced villus height ∼20% relative to the PBS control (p < 0.05).
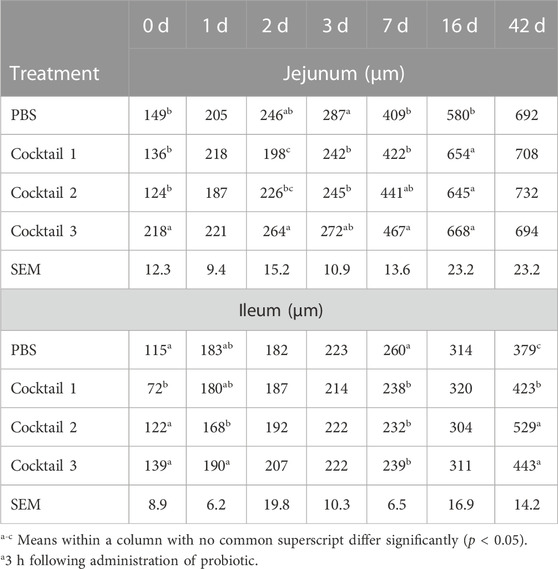
TABLE 6. Jejunal and ileal villi height of chickens administered Parabacteroides distasonis, Bacteroides salyersiae, Phocaeicola dorei, and Escherichia coli (Cocktail 1); Romboutsia lituseburensis and E. coli (Cocktail 2); P. distasonis, B. salyersiae, P. dorei, E. coli and R. lituseburensis (Cocktail 3); or PBS.
The P. distasonis, B. salyersiae, P. dorei, R. lituseburensis and E. coli cocktail significantly increased 1.3 to 2.5-fold the number of goblet cells in the ileum in newly hatched chicks, just 3 h following its administration, and at day 2 in the ileum, respectively (Table 7; p < 0.05). The P. distasonis, B. salyersiae, P. dorei, and E. coli cocktail increased goblet cell numbers 1.5-fold at day 3 and the R. lituseburensis and E. coli cocktail improved goblet cells numbers at day 7 relative to the PBS control (p < 0.05). All probiotic formulations increased ∼1.5-fold goblet cells per villus at 42 days (Table 7), in the ileum, however, a significant decrease was observed in the proportion of goblet cells in the jejunum with probiotic formulations R. lituseburensis and E. coli (30% reduction, day 7), or P. distasonis, B. salyersiae, P. dorei, R. lituseburensis and E. coli cocktail (28% and 40% reductions on days 3 and 42, respectively) relative to PBS control (Table 7; p < 0.05).
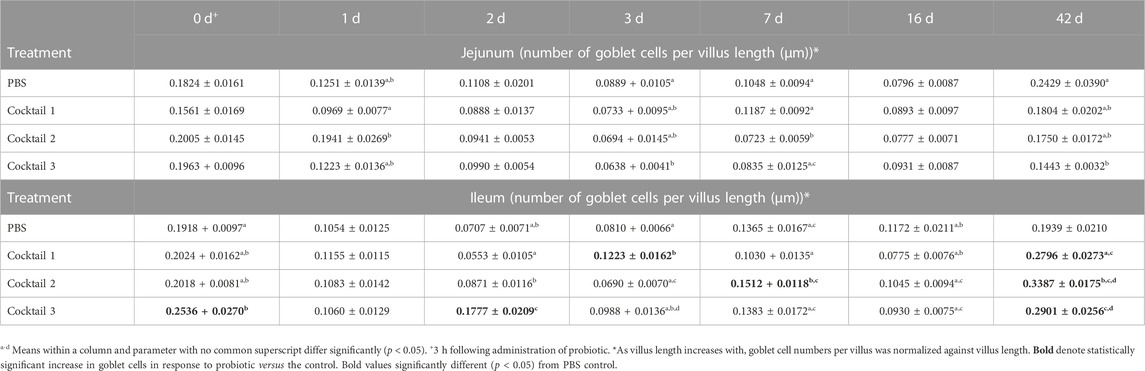
TABLE 7. Goblet cells number per jejunum and ileal villus length of chickens inoculated Parabacteroides distasonis, Bacteroides salyersiae, Phocaeicola dorei, and Escherichia coli (Cocktail 1); Romboutsia lituseburensis and E. coli (Cocktail 2); P. distasonis, B. salyersiae, P. dorei, E. coli and R. lituseburensis (Cocktail 3); or PBS.
The probiotic cocktails were shown to modify the intestinal microbiota of birds compared to PBS control (Figure 4). Similar to Aviguard® treatment, the probiotic formulations affected the Lactobacillus population in the intestine. With the exception of day 3, P. distasonis, B. salyersiae, P. dorei, R. lituseburensis and E. coli cocktail reduced ileal Lactobacillus abundance 23%–60% compared to the PBS control. However, other probiotic formulations increased Lactobacillus abundance, depending on the intestinal segment (jejunum vs. ileum) or day of age.
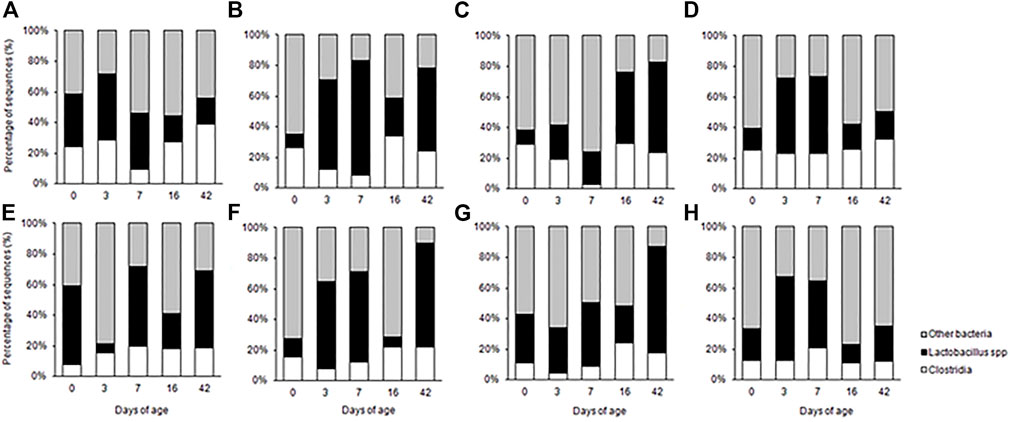
FIGURE 4. Composition of the small intestine bacterial community of chicks administered pioneer colonizers on day of hatch as determined by 16 S rRNA T-RFLP from samples collected from the jejunum (A–D) and ileum (E–H) of chickens from the phosphate buffered saline control group (A, E), Romboutsia lituseburense and Escherichia coli cocktail (B, F), Parabacteroides distasonis, Bacteroides salyersiae, Phocaeicola dorei, and Escherichia coli cocktail (C, G), P. distasonis, B. salyersiae, P. dorei, R. lituseburense and E. coli cocktail (D, H).
Poultry feed has diversified to vegetarian options and use of non-traditional ingredients that result in additional supplementation with amino acids and vitamins that enhance animal growth, physiology and performance (Alagawany et al., 2020). Gone are antibiotics once used to promote animal growth and prevent disease; replaced by probiotics, prebiotics, organic acids or essential oils. Some of these same feed additives have been shown to be comparable to growth-promoting antibiotics in improving intestinal development, animal growth, and pathogen exclusion or control (Gadde et al., 2017; Ricke, 2021; Abd El-Hack et al., 2022). These additives have been shown to alter the chicken gastrointestinal microbiome (Dittoe et al., 2018; Khan et al., 2020; Ali et al., 2021). The challenge now is piecing out their mechanism of action.
Poultry producers seek to imprint desirable attributes such as optimal feed conversion, disease and pathogen resistance, onto recipient hatchlings. Many studies have shown that a complex microbiota prohibits the establishment of harmful pathogens and fosters beneficial microbes that reduce inflammation, promote healing, improve feed efficiency and promote growth (van der Waaij et al., 1972; Candela et al., 2008; Fukuda et al., 2011; McNulty et al., 2011). Based on this concept, early intestinal colonization is essential to intestinal development, feed conversion and animal growth. Pioneer colonizers, as probiotics, offer an approach to ensure a mature and stable microbiome for newly hatched chicks.
Aviguard®, a commercially available competitive exclusion product, has been shown in multiple studies to improve disease resistance in broilers (Hofacre et al., 1998a; Hofacre et al., 1998b; Hofacre et al., 2019). In our current study it also altered the microbiome of chicks and improved development of the small intestine. The Lactobacillus population of the jejunum and ileum was more quickly replaced with intestinal anaerobes and the diversity of the ileal community was more stable indicating that the previously reported community successions could be altered (Lu et al., 2003). A more stable intestinal community structure in chicks at 3 weeks of age is important because this is a critical time of vulnerability for intestinal health (Hofacre et al., 1998a). Compositionally, the competitive exclusion product contained abundant intestinal member species, as potential pioneer colonizers, with sufficient diversity to induce intestinal development and animal growth (Flint et al., 2015; Kettle et al., 2015; Pedroso et al., 2015).
The most abundant bacterial phyla in the small intestine, following administration of the competitive exclusion product Aviguard®, were the phyla Bacteroidetes and Firmicutes, and specifically, with regard to the latter phyla, Clostridia was the dominant order. The impact of the intestinal microbiota on host physiology is being intensively studied and it is becoming increasingly clear that the intestine does not function or develop properly in the absence of its resident microbiota (Dubos et al., 1968; Smith et al., 2007; Sommer and Backhed, 2013). While Clostridia and Bacteroidia have fundamental differences in polysaccharide utilization and feeding strategies, they are similar in their reliance on carbohydrates for metabolism. Bacteroidetes harvest mucus glycans, a nutrient generated by its animal host (Koropatkin et al., 2012) but Clostridia are also known for their ability to harvest energy from indigestible fiber. The Bacteroidia and Clostridia species, examined in this study possessed many CAZymes for liberating sugars from mucin and indigestible fibers. In addition, the Bacteroidia have been shown to influence the carbohydrate composition of the intestinal glycome by liberating fucose by hydrolysis of mucin and the byproducts of fucose fermentation stimulate stem cell development (Bry et al., 1996). Fucose has been shown to be a terminal carbohydrate in the chicken’s intestinal glycome indicating that Bacteroides may also function as a pioneer colonizer in birds (Alroy et al., 1989; Madrid et al., 1989; Bryk et al., 1999). In addition, the species used in this study, B. salyersiae, P. dorei, and R. lituseburensis, possess fermentation enzymes and pathways for producing butyrate. While P. distasonis lacked these enzymes, it did possess enzymes necessary for producing propionate and several of the isolates also had acetyl-CoA hydrolases involved in acetate production.
Therefore, these probiotic isolates produce volatile fatty acids (VFA) that can be metabolized by the host animal. Metabolically, members of the order Clostridia and Bacteroidia cooperate rather than compete for the same nutrients in the intestine (Mahowald et al., 2009). This cooperation has the added benefit of increasing the VFA butyrate, which benefits their animal host in a number of ways including stimulating stem cell differentiation and reducing expression of inflammatory cytokines (Mahowald et al., 2009). Both Clostridia and Bacteroidia produce a variety of VFA, as end-products of fermentation, that can alter the composition of the microbiome and affect intestinal physiology (Segain et al., 2000; Pryde et al., 2002; Atarashi et al., 2013; Cockburn et al., 2015). Butyrate stimulates butyrate transporters in the host intestinal cells (Mahowald et al., 2009), dampens inflammation (Vieira et al., 2012), promotes intestinal integrity (Peng et al., 2009) and healing of intestinal damage (Butzner et al., 1996). In contrast, use of proteobacteria such as E. coli and Citrobacter, as pioneer colonizers in chicks elicited an intestinal inflammatory state that may lead to reduced intestinal health (Wilson et al., 2020; Chasser et al., 2021a; Chasser et al., 2021b).
The P. distasonis isolate used in this study also possessed a Vitamin B12 dependent ethanolamine utilization locus and vitamin B12 transporters that would allow it to compete with proteobacteria such as Salmonella and other intestinal bacteria for ethanolamine (Thiennimitr et al., 2011; Anderson et al., 2015; Kaval and Garsin, 2018). Furthermore, these Clostridia and Bacteroidia species may have elicited an indirect improvement of feed conversion by suppression of the Lactobacillus population. The lactobacilli are auxotrophs, deficient in their ability to synthesize up to eight different amino acids, vitamins and important co-factors (Makarova et al., 2006; Cai et al., 2009). While they are capable of fermenting a large repertoire of carbohydrates, they do not possess the enzymes to acquire these sugars from mucin (Makarova et al., 2006; Cai et al., 2009). Therefore, Lactobacillus is in competition with its host for free sugars, peptides and amino acids while the strict anaerobes such as Clostridia and Bacteroidia focus on utilizing mucin. Under feed restriction or a diet with low digestibility such as a wheat vs corn-soy diet, the composition of the small intestinal microbiome may have a significant impact on feed conversion and weight gain because of this competition (Torok et al., 2008; Metzler-Zebeli et al., 2019). In fact, a negative correlation between Lactobacillus abundance in the ileum and total body weight gain has been shown under feed restriction (Metzler-Zebeli et al., 2019). Low body weight birds tend to also have microbiome dominated by lactic acid bacteria (Zhao et al., 2013).
This is not to say the lactobacilli do not perform important functions for its animal host including dampening inflammation (Chen et al., 2012; Gong et al., 2020; Šefcová et al., 2020) or pathogen exclusion (Chen et al., 2012; Gong et al., 2020). However the mechanism of action of growth-promoting antibiotics may not only be due to suppression of pathogens (Arakawa and Oe, 1975), but the streptogramin (Lamb et al., 1999), glycopeptide (Chow and Cheng, 1988) and bacitracin (Toscano and Storm, 1982) antibiotics have broad activity against lactobacilli. In fact, antibiotic growth promoters suppress Lactobacillus, reduce community diversity and favor Clostridia in the ileum, similar to results observed with the competitive exclusion product used in this study (Lu et al., 2008). Therefore it is not surprising that growth promoting antibiotics profoundly affect microbiome composition and diversity (Lu et al., 2008). And while the growth-promoting properties of antibiotics and probiotics might be attributed to control of intestinal pathogens such as C. perfringens, it is also likely that their true impact is from enhancing intestinal development, modulating metabolism of the microbiome, and allowing the animal to better compete for nutrients liberated in the small intestine.
The Bacteroidia contain foundational genera, Bacteroides and Parabacteroides transmitted from the adult hen to its progeny, when hens are reared with their chicks (Kubasova et al., 2019a). The Bacteroidetes become the dominant phyla by day 18, for gnotobiotic chicks seeded with the intestinal microbiome from feral chickens (Thomas et al., 2019) and members of this phyla can stably colonize the cecum of chicks administered a complex cocktail containing this phyla, Firmicutes and Proteobacteria (Kubasova et al., 2019b). Bacteroidia member species have been shown to exclude certain pathogens from the chicken gastrointestinal tract (Kubasova et al., 2019b; Papouskova et al., 2023).
While we observed significant improvement to intestinal development and animal performance with our five-member probiotic formulation, this does not imply that this probiotic performs all the same functions as the competitive exclusion product examined in this study. While variable in composition, this product is consistent at reducing Salmonella colonization in chicks (Lee et al., 2023) and has been shown to be effective at controlling other enteropathogens (Hofacre et al., 1998a; Hofacre et al., 2019). This competitive exclusion product is a complex consortium, of chicken cecal origin, that consists of 20–50 distinct genera. While Kubasova et al. demonstrated significant Salmonella exclusion with an eight-member probiotic formulation, including P. distasonis (Kubasova et al., 2019b), it is not known whether this same formulation can exclude other enteropathogens or has growth promoting properties. Perhaps, it requires sufficient community diversity to outcompete pathogens, promote intestinal development and function, and repair any perturbation to gut function brought about by disease. Microbiome diversity is important in pathogen exclusion (Pedroso et al., 2021) and restoring homeostasis following any perturbation to the gastrointestinal tract (Weimer, 2015).
In addition to excluding pathogens, competitive exclusion product contains foundational bacterial species to promote intestinal function, development and animal growth. Intestinal pioneering colonizers selected from chicken ceca, based on their prominence in competitive exclusion product and consisting of five-member intestinal species, was comparable to a competitive exclusion product in improving intestinal morphology and animal performance. The balance of proteobacter, lactobacilli and anaerobic intestinal member species is critical to a healthy microbiome that promotes intestinal development, feed efficiency and animal growth (Foo et al., 2017). In the past, growth-promoting antibiotics provided this balance. Now, as the poultry industry has moved towards antibiotic free production, defined intestinal bioproducts are needed to stimulate intestinal development and function, support lower feed conversion rates and improved body weight gains, and maintain a healthy balance in the intestinal microbiota.
The original contributions presented in the study are included in the article/Supplementary Materials, further inquiries can be directed to the corresponding author.
The animal study was reviewed and approved by University of Georgia Institutional Animal Care and Use Committee.
ML was responsible for funding acquisition, conceptualization, writing, review and editing. AP, BL, and YC performed the experiments and formal analysis described in this article. JM performed metagenome analyses and data curation. ML and JM were involved in the writing, review and editing.
This work was supported by United States Department of Agriculture formula funds and the State of Georgia’s Veterinary Medicine Agricultural Research Grant. JM was supported by USDA HATCH fund: VA-160130.
We wish to thank Jingrang Lu advising us on T-RFLP methodology, analysis and interpretation.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2023.1139321/full#supplementary-material
Abd El-Hack, M. E., El-Saadony, M. T., Saad, A. M., Salem, H. M., Ashry, N. M., Abo Ghanima, M. M., et al. (2022). Essential oils and their nanoemulsions as green alternatives to antibiotics in poultry nutrition: A comprehensive review. Poult. Sci. 101, 101584. doi:10.1016/j.psj.2021.101584
Afgan, E., Baker, D., Batut, B., Van Den Beek, M., Bouvier, D., Cech, M., et al. (2018). The galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 46, W537–W544. doi:10.1093/nar/gky379
Alagawany, M., Elnesr, S. S., Farag, M. R., Tiwari, R., Yatoo, M. I., Karthik, K., et al. (2020). Nutritional significance of amino acids, vitamins and minerals as nutraceuticals in poultry production and health - a comprehensive review. Vet. Q. 41, 1–29. doi:10.1080/01652176.2020.1857887
Al-Asmakh, M., and Zadjali, F. (2015). Use of germ-free animal models in microbiota-related research. J. Microbiol. Biotechnol. 25, 1583–1588. doi:10.4014/jmb.1501.01039
Ali, A., Ponnampalam, E. N., Pushpakumara, G., Cottrell, J. J., Suleria, H. a. R., and Dunshea, F. R. (2021). Cinnamon: A natural feed additive for poultry health and production-A review. Anim. (Basel) 11, 2026. doi:10.3390/ani11072026
Alroy, J., Goyal, V., Lukacs, N. W., Taylor, R. L., Strout, R. G., Ward, H. D., et al. (1989). Glycoconjugates of the intestinal epithelium of the domestic fowl (Gallus domesticus): A lectin histochemistry study. Histochem. J. 21, 187–193. doi:10.1007/BF01747519
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi:10.1016/S0022-2836(05)80360-2
Anderson, C. J., Clark, D. E., Adli, M., and Kendall, M. M. (2015). Ethanolamine signaling promotes Salmonella niche recognition and adaptation during infection. PLoS Pathog. 11, e1005278. doi:10.1371/journal.ppat.1005278
Apajalahti, J. H., Sarkilahti, L. K., Maki, B. R., Heikkinen, J. P., Nurminen, P. H., and Holben, W. E. (1998). Effective recovery of bacterial DNA and percent-guanine-plus-cytosine-based analysis of community structure in the gastrointestinal tract of broiler chickens. Appl. Environ. Microbiol. 64, 4084–4088. doi:10.1128/AEM.64.10.4084-4088.1998
Arakawa, A., and Oe, O. (1975). Reduction of Clostridium perfringens by feed additive antibiotics in the ceca of chickens infected with Eimeria tenella. Poult. Sci. 54, 1000–1007. doi:10.3382/ps.0541000
Atarashi, K., Tanoue, T., Oshima, K., Suda, W., Nagano, Y., Nishikawa, H., et al. (2013). Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 500, 232–236. doi:10.1038/nature12331
Aziz, R. K., Bartels, D., Best, A. A., Dejongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST server: Rapid annotations using subsystems technology. Bmc Genomics 9, 75. doi:10.1186/1471-2164-9-75
Backhed, F., Ley, R. E., Sonnenburg, J. L., Peterson, D. A., and Gordon, J. I. (2005). Host-bacterial mutualism in the human intestine. science 307, 1915–1920. doi:10.1126/science.1104816
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi:10.1089/cmb.2012.0021
Berg, R. D. (1980). Inhibition of Escherichia coli translocation from the gastrointestinal tract by normal cecal flora in gnotobiotic or antibiotic-decontaminated mice. Infect. Immun. 29, 1073–1081. doi:10.1128/iai.29.3.1073-1081.1980
Bry, L., Falk, P. G., Midtvedt, T., and Gordon, J. I. (1996). A model of host-microbial interactions in an open mammalian ecosystem. Science 273, 1380–1383. doi:10.1126/science.273.5280.1380
Bryk, S. G., Sgambati, E., and Gheri Bryk, G. (1999). Lectin histochemistry of goblet cell sugar residues in the gut of the chick embryo and of the newborn. Tissue Cell. 31, 170–175. doi:10.1054/tice.1999.0027
Butzner, J. D., Parmar, R., Bell, C. J., and Dalal, V. (1996). Butyrate enema therapy stimulates mucosal repair in experimental colitis in the rat. Gut 38, 568–573. doi:10.1136/gut.38.4.568
Cai, H., Thompson, R., Budinich, M. F., Broadbent, J. R., and Steele, J. L. (2009). Genome sequence and comparative genome analysis of Lactobacillus casei: Insights into their niche-associated evolution. Genome Biol. Evol. 1, 239–257. doi:10.1093/gbe/evp019
Candela, M., Perna, F., Carnevali, P., Vitali, B., Ciati, R., Gionchetti, P., et al. (2008). Interaction of probiotic Lactobacillus and bifidobacterium strains with human intestinal epithelial cells: Adhesion properties, competition against enteropathogens and modulation of IL-8 production. Int. J. Food Microbiol. 125, 286–292. doi:10.1016/j.ijfoodmicro.2008.04.012
Casewell, M., Friis, C., Marco, E., Mcmullin, P., and Phillips, I. (2003). The European ban on growth-promoting antibiotics and emerging consequences for human and animal health. J. Antimicrob. Chemother. 52, 159–161. doi:10.1093/jac/dkg313
Chasser, K., Mcgovern, K., Duff, A., Graham, B., Briggs, W., Rodrigues, D., et al. (2021a). Evaluation of day of hatch exposure to various Enterobacteriaceae on inducing gastrointestinal inflammation in chicks through two weeks of age. Poult. Sci. 100, 101193. doi:10.1016/j.psj.2021.101193
Chasser, K., Mcgovern, K., Duff, A., Trombetta, M., Graham, B., Graham, L., et al. (2021b). Enteric permeability and inflammation associated with day of hatch Enterobacteriaceae inoculation. Poult. Sci. 100, 101298. doi:10.1016/j.psj.2021.101298
Cheled-Shoval, S. L., Gamage, N. S., Amit-Romach, E., Forder, R., Marshal, J., Van Kessel, A., et al. (2014). Differences in intestinal mucin dynamics between germ-free and conventionally reared chickens after mannan-oligosaccharide supplementation. Poult. Sci. 93, 636–644. doi:10.3382/ps.2013-03362
Chen, C. Y., Tsen, H. Y., Lin, C. L., Yu, B., and Chen, C. S. (2012). Oral administration of a combination of select lactic acid bacteria strains to reduce the Salmonella invasion and inflammation of broiler chicks. Poult. Sci. 91, 2139–2147. doi:10.3382/ps.2012-02237
Chen, X., Xu, J., Ren, E., Su, Y., and Zhu, W. (2018). Co-occurrence of early gut colonization in neonatal piglets with microbiota in the maternal and surrounding delivery environments. Anaerobe 49, 30–40. doi:10.1016/j.anaerobe.2017.12.002
Chow, A. W., and Cheng, N. (1988). In vitro activities of daptomycin (LY146032) and paldimycin (U-70,138F) against anaerobic gram-positive bacteria. Antimicrob. Agents Chemother. 32, 788–790. doi:10.1128/AAC.32.5.788
Coates, M. E., Ford, J. E., and Harrison, G. F. (1968). Intestinal synthesis of vitamins of the B complex in chicks. Br. J. Nutr. 22, 493–500. doi:10.1079/bjn19680057
Cockburn, D. W., Orlovsky, N. I., Foley, M. H., Kwiatkowski, K. J., Bahr, C. M., Maynard, M., et al. (2015). Molecular details of a starch utilization pathway in the human gut symbiont Eubacterium rectale. Mol. Microbiol. 95, 209–230. doi:10.1111/mmi.12859
Cowieson, A. J. (2022). Comparative biology of germ-free and conventional poultry. Poult. Sci. 101, 102105. doi:10.1016/j.psj.2022.102105
De Maesschalck, C., Eeckhaut, V., Maertens, L., De Lange, L., Marchal, L., Nezer, C., et al. (2015). Effects of xylo-oligosaccharides on broiler chicken performance and microbiota. Appl. Environ. Microbiol. 81, 5880–5888. doi:10.1128/AEM.01616-15
Diaz-Sanchez, S., Moscoso, S., Solis De Los Santos, F., Andino, A., and Hanning, I. (2015). Antibiotic use in poultry: A driving force for organic poultry production. Food Prot. Trends 35, 440–447.
Dittoe, D. K., Ricke, S. C., and Kiess, A. S. (2018). Organic acids and potential for modifying the avian gastrointestinal tract and reducing pathogens and disease. Front. Vet. Sci. 5, 216. doi:10.3389/fvets.2018.00216
Dubos, R., Schaedler, R. W., and Costello, R. (1968). Lasting biological effects of early environmental influences. I. Conditioning of adult size by prenatal and postnatal nutrition. J. Exp. Med. 127, 783–799. doi:10.1084/jem.127.4.783
Dymsza, H. A., Stoewsand, G. S., Enright, J. J., Trexler, P. C., and Gall, L. C. (1965). Human indigenous microflora in gnotobiotic rats. Nature 208, 1236–1237. doi:10.1038/2081236a0
Eaton, K. A., Friedman, D. I., Francis, G. J., Tyler, J. S., Young, V. B., Haeger, J., et al. (2008). Pathogenesis of renal disease due to enterohemorrhagic Escherichia coli in germ-free mice. Infect. Immun. 76, 3054–3063. doi:10.1128/IAI.01626-07
Espey, M. G. (2013). Role of oxygen gradients in shaping redox relationships between the human intestine and its microbiota. Free Radic. Biol. Med. 55, 130–140. doi:10.1016/j.freeradbiomed.2012.10.554
Eyssen, H., and De Somer, P. (1963). The mode of action of antibiotics in stimulating growth of chicks. J. Exp. Med. 117, 127–138. doi:10.1084/jem.117.1.127
Faure, J. C., Schellenberg, D. A., Bexter, A., and Wuerzner, H. P. (1984). Barrier effect of Bifidobacterium longum on a pathogenic Escherichia coli strain by gut colonization in the germ-free rat. Z Ernahrungswiss 23, 41–51. doi:10.1007/BF02020895
Flint, H. J., Duncan, S. H., Scott, K. P., and Louis, P. (2015). Links between diet, gut microbiota composition and gut metabolism. Proc. Nutr. Soc. 74, 13–22. doi:10.1017/S0029665114001463
Foo, J. L., Ling, H., Lee, Y. S., and Chang, M. W. (2017). Microbiome engineering: Current applications and its future. Biotechnol. J. 12, 1600099. doi:10.1002/biot.201600099
Fukata, T., Hadate, Y., Baba, E., and Arakawa, A. (1991). Influence of bacteria on Clostridium perfringens infections in young chickens. Avian Dis. 35, 224–227. doi:10.2307/1591319
Fukuda, S., Toh, H., Hase, K., Oshima, K., Nakanishi, Y., Yoshimura, K., et al. (2011). Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature 469, 543–547. doi:10.1038/nature09646
Gadde, U., Kim, W. H., Oh, S. T., and Lillehoj, H. S. (2017). Alternatives to antibiotics for maximizing growth performance and feed efficiency in poultry: A review. Anim. Health Res. Rev. 18, 26–45. doi:10.1017/S1466252316000207
Gilbert, E. R., Li, H., Emmerson, D. A., Webb, K. E., and Wong, E. A. (2007). Developmental regulation of nutrient transporter and enzyme mRNA abundance in the small intestine of broilers. Poult. Sci. 86, 1739–1753. doi:10.1093/ps/86.8.1739
Gong, L., Wang, B., Zhou, Y., Tang, L., Zeng, Z., Zhang, H., et al. (2020). Protective effects of Lactobacillus plantarum 16 and paenibacillus polymyxa 10 against Clostridium perfringens infection in broilers. Front. Immunol. 11, 628374. doi:10.3389/fimmu.2020.628374
Gourbeyre, P., Berri, M., Lippi, Y., Meurens, F., Vincent-Naulleau, S., Laffitte, J., et al. (2015). Pattern recognition receptors in the gut: Analysis of their expression along the intestinal tract and the crypt/villus axis. Physiol. Rep. 3, e12225. doi:10.14814/phy2.12225
Greig, C. J., Alper, A., Goodman, A. L., and Cowles, R. A. (2018). Mucosal homeostasis is altered in the ileum of gnotobiotic mice. J. Surg. Res. 231, 331–337. doi:10.1016/j.jss.2018.05.055
Grondin, J. M., Tamura, K., Déjean, G., Abbott, D. W., and Brumer, H. (2017). Polysaccharide utilization loci: Fueling microbial communities. J. Bacteriol. 199, 1–15. doi:10.1128/JB.00860-16
Heath-Heckman, E. A., Gillette, A. A., Augustin, R., Gillette, M. X., Goldman, W. E., and Mcfall-Ngai, M. J. (2014). Shaping the microenvironment: Evidence for the influence of a host galaxin on symbiont acquisition and maintenance in the squid-Vibrio symbiosis. Environ. Microbiol. 16, 3669–3682. doi:10.1111/1462-2920.12496
Hofacre, C. L., Froyman, R., Gautrias, B., George, B., Goodwin, M. A., and Brown, J. (1998a). Use of Aviguard and other intestinal bioproducts in experimental Clostridium perfringens-associated necrotizing enteritis in broiler chickens. Avian Dis. 42, 579–584. doi:10.2307/1592685
Hofacre, C. L., Froyman, R., George, B., Goodwin, M. A., and Brown, J. (1998b). Use of Aviguard, virginiamycin, or bacitracin MD against Clostridium perfringens-associated necrotizing enteritis. J. Appl. Poult. Res. 7, 412–418. doi:10.1093/japr/7.4.412
Hofacre, C., Reynolds, D., Mathis, G., Lumpkins, B., Ollis, N., Smith, J., et al. (2019). Effect of a competitive exclusion culture in a necrotic enteritis challenge model in broilers. J. Appl. Poult. Res. 28, 350–355. doi:10.3382/japr/pfy078
Jurburg, S. D., Brouwer, M. S. M., Ceccarelli, D., Van Der Goot, J., Jansman, A. J. M., and Bossers, A. (2019). Patterns of community assembly in the developing chicken microbiome reveal rapid primary succession. Microbiologyopen 8, e00821. doi:10.1002/mbo3.821
Kaval, K. G., and Garsin, D. A. (2018). Ethanolamine utilization in bacteria. mBio 9, 1–13. doi:10.1128/mBio.00066-18
Kelley, R. W. (1983). Comparison of media for isolation of poultry intestinal bacteria. Appl. Environ. Microbiol. 46, 421–424. doi:10.1128/aem.46.2.421-424.1983
Kettle, H., Louis, P., Holtrop, G., Duncan, S. H., and Flint, H. J. (2015). Modelling the emergent dynamics and major metabolites of the human colonic microbiota. Environ. Microbiol. 17, 1615–1630. doi:10.1111/1462-2920.12599
Khan, S., Moore, R. J., Stanley, D., and Chousalkar, K. K. (2020). The gut microbiota of laying hens and its manipulation with prebiotics and probiotics to enhance gut health and food safety. Appl. Environ. Microbiol. 86, e00600–e00620. doi:10.1128/AEM.00600-20
Klein-Jöbstl, D., Quijada, N. M., Dzieciol, M., Feldbacher, B., Wagner, M., Drillich, M., et al. (2019). Microbiota of newborn calves and their mothers reveals possible transfer routes for newborn calves' gastrointestinal microbiota. PLoS One 14, e0220554. doi:10.1371/journal.pone.0220554
Koleva, P. T., Kim, J. S., Scott, J. A., and Kozyrskyj, A. L. (2015). Microbial programming of health and disease starts during fetal life. Birth Defects Res. Part C Embryo Today Rev. 105, 265–277. doi:10.1002/bdrc.21117
Koropatkin, N. M., Cameron, E. A., and Martens, E. C. (2012). How glycan metabolism shapes the human gut microbiota. Nat. Rev. Microbiol. 10, 323–335. doi:10.1038/nrmicro2746
Kremer, N., Schwartzman, J., Augustin, R., Zhou, L., Ruby, E. G., Hourdez, S., et al. (2014). The dual nature of haemocyanin in the establishment and persistence of the squid-vibrio symbiosis. Proc. Biol. Sci. 281, 20140504. doi:10.1098/rspb.2014.0504
Kubasova, T., Kollarcikova, M., Crhanova, M., Karasova, D., Cejkova, D., Sebkova, A., et al. (2019a). Contact with adult hen affects development of caecal microbiota in newly hatched chicks. PLoS One 14, e0212446. doi:10.1371/journal.pone.0212446
Kubasova, T., Kollarcikova, M., Crhanova, M., Karasova, D., Cejkova, D., Sebkova, A., et al. (2019b). Gut anaerobes capable of chicken caecum colonisation. Microorganisms 7, 597. doi:10.3390/microorganisms7120597
Lamb, H. M., Figgitt, D. P., and Faulds, D. (1999). Quinupristin/dalfopristin: A review of its use in the management of serious gram-positive infections, quinupristin/dalfopristin. Drugs 58, 1061–1097. doi:10.2165/00003495-199958060-00008
Lee, M. D., Pedroso, A. A., and Maurer, J. J. (2023). Bacterial composition of a competitive exclusion product and its correlation with product efficacy at reducing Salmonella in poultry. Front. Physiol. 13, 1043383. doi:10.3389/fphys.2022.1043383
Lev, M., and Forbes, M. (1959). Growth response to dietary penicillin of germ-free chicks and of chicks with a defined intestinal flora. Br. J. Nutr. 13, 78–84. doi:10.1079/bjn19590012
Lu, J., Idris, U., Harmon, B., Hofacre, C., Maurer, J. J., and Lee, M. D. (2003). Diversity and succession of the intestinal bacterial community of the maturing broiler chicken. Appl. Environ. Microbiol. 69, 6816–6824. doi:10.1128/AEM.69.11.6816-6824.2003
Lu, J., Hofacre, C. L., and Lee, M. D. (2006). Emerging technologies in microbial ecology aid in understanding the effect of monensin in the diets of broilers in regard to the complex disease necrotic enteritis. J. Appl. Poult. Res. 15, 145–153. doi:10.1093/japr/15.1.145
Lu, J., Hofacre, C., Smith, F., and Lee, M. D. (2008). Effects of feed additives on the development on the ileal bacterial community of the broiler chicken. Animal 2, 669–676. doi:10.1017/S1751731108001894
Lukow, T., Dunfield, P. F., and Liesack, W. (2000). Use of the T-RFLP technique to assess spatial and temporal changes in the bacterial community structure within an agricultural soil planted with transgenic and non-transgenic potato plants. FEMS Microbiol. Ecol. 32, 241–247. doi:10.1111/j.1574-6941.2000.tb00717.x
Madrid, J., Ballesta, J., Castells, M., Marin, J., and Pastor, L. (1989). Characterization of glycoconjugates in the intestinal mucosa of vertebrates by means of lectin histochemistry. Acta Histochem. Cytochem. 22, 1–14. doi:10.1267/ahc.22.1
Mahowald, M. A., Rey, F. E., Seedorf, H., Turnbaugh, P. J., Fulton, R. S., Wollam, A., et al. (2009). Characterizing a model human gut microbiota composed of members of its two dominant bacterial phyla. Proc. Natl. Acad. Sci. U. S. A. 106, 5859–5864. doi:10.1073/pnas.0901529106
Makarova, K., Slesarev, A., Wolf, Y., Sorokin, A., Mirkin, B., Koonin, E., et al. (2006). Comparative genomics of the lactic acid bacteria. Proc. Natl. Acad. Sci. U. S. A. 103, 15611–15616. doi:10.1073/pnas.0607117103
Mcfall-Ngai, M. J. (2014). The importance of microbes in animal development: Lessons from the squid-vibrio symbiosis. Annu. Rev. Microbiol. 68, 177–194. doi:10.1146/annurev-micro-091313-103654
Mcnulty, N. P., Yatsunenko, T., Hsiao, A., Faith, J. J., Muegge, B. D., Goodman, A. L., et al. (2011). The impact of a consortium of fermented milk strains on the gut microbiome of gnotobiotic mice and monozygotic twins. Sci. Transl. Med. 3, 106ra106. doi:10.1126/scitranslmed.3002701
Metzler-Zebeli, B. U., Siegerstetter, S. C., Magowan, E., Lawlor, P. G., Petri, R. M., Ne, O. C., et al. (2019). Feed restriction modifies intestinal microbiota-host mucosal networking in chickens divergent in residual feed intake. mSystems 4, 1–15. doi:10.1128/mSystems.00261-18
Meyer, R. C., Bohl, E. H., and Kohler, E. M. (1964). Procurement and maintenance of germ-free seine for microbiological investigations. Appl. Microbiol. 12, 295–300. doi:10.1128/am.12.4.295-300.1964
Muramatsu, T., Kodama, H., Morishita, T., Furuse, M., and Okumura, J. (1991). Effect of intestinal microflora on digestible energy and fiber digestion in chickens fed a high-fiber diet. Am. J. Vet. Res. 52, 1178–1181.
Neu, J., and Rushing, J. (2011). Cesarean versus vaginal delivery: Long-term infant outcomes and the hygiene hypothesis. Clin. Perinatol. 38, 321–331. doi:10.1016/j.clp.2011.03.008
Nurmi, E., and Rantala, M. (1973). New aspects of Salmonella infection in broiler production. Nature 241, 210–211. doi:10.1038/241210a0
Papouskova, A., Rychlik, I., Harustiakova, D., and Cizek, A. (2023). Research note: A mixture of Bacteroides spp. and other probiotic intestinal anaerobes reduces colonization by pathogenic E. coli strain O78:H4-st117 in newly hatched chickens. Poult. Sci. 102, 102529. doi:10.1016/j.psj.2023.102529
Patel, R. K., and Jain, M. (2012). NGS QC toolkit: A toolkit for quality control of next generation sequencing data. PLoS One 7, e30619. doi:10.1371/journal.pone.0030619
Pedroso, A. A., Batal, A., and Lee, M. D. (2015). Could in ovo administration of an adult derived microbiota augment earlier establishment of a beneficial chicken microbiome? Am. J. Veterinary Res. 76. doi:10.2460/ajvr.77.5.514
Pedroso, A. A., Lee, M. D., and Maurer, J. J. (2021). Strength lies in diversity: How community diversity limits Salmonella abundance in the chicken intestine. Front. Microbiol. 12, 694215. doi:10.3389/fmicb.2021.694215
Peng, L., Li, Z. R., Green, R. S., Holzman, I. R., and Lin, J. (2009). Butyrate enhances the intestinal barrier by facilitating tight junction assembly via activation of AMP-activated protein kinase in Caco-2 cell monolayers. J. Nutr. 139, 1619–1625. doi:10.3945/jn.109.104638
Pleasants, J. R. (1959). Rearing germfree cesarean-born rats, mice, and rabbits through weaning. Ann. N. Y. Acad. Sci. 78, 116–126. doi:10.1111/j.1749-6632.1959.tb53099.x
Pryde, S. E., Duncan, S. H., Hold, G. L., Stewart, C. S., and Flint, H. J. (2002). The microbiology of butyrate formation in the human colon. FEMS Microbiol. Lett. 217, 133–139. doi:10.1111/j.1574-6968.2002.tb11467.x
Ramírez, G. A., Richardson, E., Clark, J., Keshri, J., Drechsler, Y., Berrang, M. E., et al. (2020). Broiler chickens and early life programming: Microbiome transplant-induced cecal community dynamics and phenotypic effects. PLoS One 15, e0242108. doi:10.1371/journal.pone.0242108
Reeves, A. E., Koenigsknecht, M. J., Bergin, I. L., and Young, V. B. (2012). Suppression of Clostridium difficile in the gastrointestinal tracts of germfree mice inoculated with a murine isolate from the family Lachnospiraceae. Infect. Immun. 80, 3786–3794. doi:10.1128/IAI.00647-12
Ricke, S. C. (2021). Prebiotics and alternative poultry production. Poult. Sci. 100, 101174. doi:10.1016/j.psj.2021.101174
Šefcová, M., Larrea-Álvarez, M., Larrea-Álvarez, C., Karaffová, V., Revajová, V., Gancarčíková, S., et al. (2020). Lactobacillus fermentum administration modulates cytokine expression and lymphocyte subpopulation levels in broiler chickens challenged with Campylobacter coli. Foodborne Pathog. Dis. 17, 485–493. doi:10.1089/fpd.2019.2739
Segain, J. P., Raingeard De La Bletiere, D., Bourreille, A., Leray, V., Gervois, N., Rosales, C., et al. (2000). Butyrate inhibits inflammatory responses through NFkappaB inhibition: Implications for crohn's disease. Gut 47, 397–403. doi:10.1136/gut.47.3.397
Shannon, C. E., and Wiener, W. (1963). The mathematical theory of communication. Urbana: University of Juionis Press.
Shao, Y., Lei, Z., Yuan, J., Yang, Y., Guo, Y., and Zhang, B. (2014). Effect of zinc on growth performance, gut morphometry, and cecal microbial community in broilers challenged with Salmonella enterica serovar typhimurium. J. Microbiol. 52, 1002–1011. doi:10.1007/s12275-014-4347-y
Shirkey, T. W., Siggers, R. H., Goldade, B. G., Marshall, J. K., Drew, M. D., Laarveld, B., et al. (2006). Effects of commensal bacteria on intestinal morphology and expression of proinflammatory cytokines in the gnotobiotic pig. Exp. Biol. Med. (Maywood) 231, 1333–1345. doi:10.1177/153537020623100807
Smith, K., Mccoy, K. D., and Macpherson, A. J. (2007). Use of axenic animals in studying the adaptation of mammals to their commensal intestinal microbiota. Semin. Immunol. 19, 59–69. doi:10.1016/j.smim.2006.10.002
Sommer, F., and Backhed, F. (2013). The gut microbiota-masters of host development and physiology. Nat. Rev. Microbiol. 11, 227–238. doi:10.1038/nrmicro2974
Sprinz, H., Kundel, D. W., Dammin, G. J., Horowitz, R. E., Schneider, H., and Formal, S. B. (1961). The response of the germfree Guinea pig to oral bacterial challenge with Escherichia coli and Shigella flexneri. Am. J. Pathol. 39, 681–695.
Steel, R. G. D., and Torrie, J. H. (1980). Principles and procedures of statistics: A biometrical approch. New York: Mc Graw-Hill Book.
Thiennimitr, P., Winter, S. E., Winter, M. G., Xavier, M. N., Tolstikov, V., Huseby, D. L., et al. (2011). Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc. Natl. Acad. Sci. U. S. A. 108, 17480–17485. doi:10.1073/pnas.1107857108
Thomas, M., Wongkuna, S., Ghimire, S., Kumar, R., Antony, L., Doerner, K. C., et al. (2019). Gut microbial dynamics during conventionalization of germfree chicken. mSphere 4, e00035. doi:10.1128/mSphere.00035-19
Torok, V. A., Ophel-Keller, K., Loo, M., and Hughes, R. J. (2008). Application of methods for identifying broiler chicken gut bacterial species linked with increased energy metabolism. Appl. Environ. Microbiol. 74, 783–791. doi:10.1128/AEM.01384-07
Toscano, W. A., and Storm, D. R. (1982). Bacitracin. Pharmacol. Ther. 16, 199–210. doi:10.1016/0163-7258(82)90054-7
Treichel, N. S., Prevoršek, Z., Mrak, V., Kostrić, M., Vestergaard, G., Foesel, B., et al. (2019). Effect of the nursing mother on the gut microbiome of the offspring during early mouse development. Microb. Ecol. 78, 517–527. doi:10.1007/s00248-019-01317-7
Turnbaugh, P. J., Ley, R. E., Mahowald, M. A., Magrini, V., Mardis, E. R., and Gordon, J. I. (2006). An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444, 1027–1031. doi:10.1038/nature05414
Val-Bernal, J. F., Gomez-Roman, J. J., Vallina, T., Villoria, F., Mayorga, M., and Garcia-Arranz, P. (1999). Papillary (chromophil) renal cell carcinoma with mucinous secretion. Pathol. Res. Pract. 195, 11–17. doi:10.1016/s0344-0338(99)80088-0
Van Der Waaij, D., Berghuis, J. M., and Lekkerkerk, J. E. (1972). Colonization resistance of the digestive tract of mice during systemic antibiotic treatment. J. Hyg. (Lond) 70, 605–610. doi:10.1017/s0022172400022464
Vieira, E. L., Leonel, A. J., Sad, A. P., Beltrão, N. R., Costa, T. F., Ferreira, T. M., et al. (2012). Oral administration of sodium butyrate attenuates inflammation and mucosal lesion in experimental acute ulcerative colitis. J. Nutr. Biochem. 23, 430–436. doi:10.1016/j.jnutbio.2011.01.007
Vuong, C. N., Chou, W.-K., Hargis, B. M., Berghman, L. R., and Bielke, L. R. (2016). Role of probiotics on immune function and their relationship to antibiotic growth promoters in poultry, a brief review. Int. J. Probiotics Prebiotics 11, 1–7.
Waxler, G. L., and Drees, D. T. (1972). Comparison of body weights, organ weights and histological features of selected organs of gnotobiotic, conventional and isolator-reared contaminated pigs. Can. J. Comp. Med. 36, 265–274.
Weimer, P. J. (2015). Redundancy, resilience, and host specificity of the ruminal microbiota: Implications for engineering improved ruminal fermentations. Front. Microbiol. 6, 296. doi:10.3389/fmicb.2015.00296
Wilson, K., Rodrigues, D., Briggs, W., Duff, A., Chasser, K., Bottje, W., et al. (2020). Impact of in ovo administered pioneer colonizers on intestinal proteome on day of hatch. Poult. Sci. 99, 1254–1266. doi:10.1016/j.psj.2019.10.017
Yin, H. C., Liu, Z. D., Zhang, W. W., Yang, Q. Z., Yu, T. F., and Jiang, X. J. (2022). Chicken intestinal microbiota modulation of resistance to nephropathogenic infectious bronchitis virus infection through IFN-I. Microbiome 10, 162. doi:10.1186/s40168-022-01348-2
Yu, K., Rodriguez, M. D., Paul, Z., Gordon, E., Rice, K., Triplett, E. W., et al. (2019). Proof of principle: Physiological transfer of small numbers of bacteria from mother to fetus in late-gestation pregnant sheep. PLoS One 14, e0217211. doi:10.1371/journal.pone.0217211
Keywords: microbiome, performance, feed efficiency, intestinal development, anaerobes
Citation: Lee MD, Pedroso AA, Lumpkins B, Cho Y and Maurer JJ (2023) Pioneer colonizers: Bacteria that alter the chicken intestinal morphology and development of the microbiota. Front. Physiol. 14:1139321. doi: 10.3389/fphys.2023.1139321
Received: 06 January 2023; Accepted: 14 March 2023;
Published: 29 March 2023.
Edited by:
Sundus Javed, COMSATS University, PakistanReviewed by:
Ciara Keating, University of Glasgow, United KingdomCopyright © 2023 Lee, Pedroso, Lumpkins, Cho and Maurer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Margie D. Lee, bWxlZTJAdnQuZWR1
†Present address: Adriana A. Pedroso, Ventura Coastal, LLC, Tipton, CA, United States; Brett Lumpkin, Southern Poultry Feed and Research, Inc., Athens, GA, United States; Youngjae Cho, Pusan National University, Busan, Republic of Korea
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.