
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Physiol., 01 March 2023
Sec. Aquatic Physiology
Volume 14 - 2023 | https://doi.org/10.3389/fphys.2023.1118341
This article is part of the Research TopicThe defense responses of aquatic animals to the environmentView all 7 articles
 Yun-Dong Li1,2,3,4†
Yun-Dong Li1,2,3,4† Meng-Ru Si2†
Meng-Ru Si2† Shi-Gui Jiang2
Shi-Gui Jiang2 Qi-Bin Yang3
Qi-Bin Yang3 Song Jiang2
Song Jiang2 Li-Shi Yang2
Li-Shi Yang2 Jian-Hua Huang2
Jian-Hua Huang2 Xu Chen3
Xu Chen3 Fa-Lin Zhou2,3,4*
Fa-Lin Zhou2,3,4* ErChao Li1,4*
ErChao Li1,4*Background: Salinity is one of the main influencing factors in the culture environment and is extremely important for the survival, growth, development and reproduction of aquatic animals.
Methods: In this study, a comparative transcriptome analysis (maintained for 45 days in three different salinities, 30 psu (HC group), 18 psu (MC group) and 3 psu (LC group)) was performed by high-throughput sequencing of economically cultured Penaeus monodon. P. monodon gill tissues from each treatment were collected for RNA-seq analysis to identify potential genes and pathways in response to low salinity stress.
Results: A total of 64,475 unigenes were annotated in this study. There were 1,140 upregulated genes and 1,531 downregulated genes observed in the LC vs. HC group and 1,000 upregulated genes and 1,062 downregulated genes observed in the MC vs. HC group. In the LC vs. HC group, 583 DEGs significantly mapped to 37 signaling pathways, such as the NOD-like receptor signaling pathway, Toll-like receptor signaling pathway, and PI3K-Akt signaling pathway; in the MC vs. HC group, 444 DEGs significantly mapped to 28 signaling pathways, such as the MAPK signaling pathway, Hippo signaling pathway and calcium signaling pathway. These pathways were significantly associated mainly with signal transduction, immunity and metabolism.
Conclusions: These results suggest that low salinity stress may affect regulatory mechanisms such as metabolism, immunity, and signal transduction in addition to osmolarity in P. monodon. The greater the difference in salinity, the more significant the difference in genes. This study provides some guidance for understanding the low-salt domestication culture of P. monodon.
The stress response is a complex physiological mechanism activated by stimuli that an organism perceives as potentially threatening (Vallejos-Vidal et al., 2022). It involves the sum of continuous and coordinated activation mechanisms at different functional levels, including molecular processes that have a direct impact on gene expression and protein synthesis, and will ultimately activate responses from the cellular to the systemic and performance levels (Parra et al., 2015). Salinity is one of the key factors regulating the distribution, abundance and diversity of aquatic animals and is one of the main environmental factors exerting selection pressure on aquatic animals (Sun et al., 2020). Changes in environmental salinity can directly affect the composition of aquatic animal fluids and the homeostasis of the internal environment (Chen et al., 2015). It has been shown that salinity stress can alter the diffusion of salts between the hemolymph and the environment, leading to altered cellular states. Within the critical point salinity range, significant changes in protein function and structure as well as in the physicochemical properties of NaCl solutions can occur (Normant et al., 2005). Low salinity alters the cell membrane potential, the optical density of solubilized proteins and the activity of various enzymes (Esparza-Leal et al., 2019). Prolonged hyposalinity exposure affects the physiology of the organism, reducing the immune response of aquatic animals to attack by external factors and leading to reduced survival (Chen and He, 2019). Under moderate hypoosmotic stress, aquatic animals typically meet the increased cost of osmoregulation by increasing their regular metabolic rate or reallocating energy to osmoregulatory processes (Rivera-Ingraham et al., 2016). Under extreme hypoosmotic stress, many aquatic animals are limited to maintaining basic functions to conserve energy reserves until the environment improves (Podbielski et al., 2022). Therefore, elucidating the adaptation mechanisms of aquatic animals to different salinities is crucial for the continued health of aquaculture.
Penaeus monodon (also known as black tiger shrimp) is one of the most commercially valuable species of aquaculture shrimp (Li et al., 2018; Li et al., 2020). P. monodon is popular with farmers and consumers due to its high economic and nutritional value (FAO, 2018). P. monodon has a wide range of salinity adaptations and can grow in a wide range of salinities from one psu ∼57 psu (Joseph and Philip, 2020). P. monodon has an isosmotic point of 750 mOsm·kg-1 (equivalent to 25 psu). In recent years, its culture has advanced significantly from coastal marine waters to inland low-salinity waters due to mariculture disease outbreaks and near-coastal pollution (Vezzulli, 2022). Therefore, understanding the mechanisms of P. monodon adaptation in long-term low-salt stress will be beneficial to its healthy culture. The regulatory mechanisms of various genes in salinity adaptation of P. monodon have been reported, including cold shock domain containing protein E1 (Si et al., 2022a), ecdysone-induced protein E74 (Si et al., 2022b), calreticulin, calnexin and endoplasmic reticulum protein 57 (Visudtiphole et al., 2017). However, systematic studies on the adaptation mechanisms of P. monodon in long-term low-salt stress environments are still lacking.
With the development of high-throughput sequencing technologies, RNA-seq offers the opportunity to study the mechanisms of transcriptional variation in organisms under different environmental conditions (de Jong et al., 2019). Transcriptome sequencing not only allows the detection of almost all validated genes expressed in a specific cell or organ but also allows a comprehensive resolution of the regulatory mechanisms involved in a specific biological process based on the structure and function of the differential genes (Stupnikov et al., 2021). In addition, transcriptome sequencing allows the simultaneous analysis of all physiological processes, including metabolism (Xiao et al., 2020), proteostasis (Li et al., 2022), osmoregulation (Gao et al., 2021) and other cellular processes (Ge et al., 2022). Thus, transcriptome technologies address key gene expression patterns in non-model organisms in specific environments and enable the detection of unknown genes and the discovery of novel transcripts. In recent years, transcriptome technologies have been widely used for transcriptome assembly and annotation in many aquatic animals, such as Macrobrachium rosenbergii (Liu X. et al., 2021), Coilia nasus (Gao et al., 2021) and Penaeus vannamei (Yu et al., 2022). Thus, RNA-seq is expected to make an important contribution to the resolution of long-term low salinity adaptation mechanisms in P. monodon.
The gill is an important respiratory organ of P. monodon and is involved in its respiratory metabolism, acid-base balance, ammonia waste excretion, ion transport and osmotic pressure regulation (Henry et al., 2012). Therefore, in this study, the gene expression profiles of P. monodon gill tissues reared at three salinity levels were examined by RNA-seq. The results of this study will provide new insights into the mechanisms of osmotic pressure regulation in P. monodon under chronic low salinity stress.
Experimental shrimp were approved by the Animal Care and Use Committee of the Chinese Academy of Fisheries Sciences (CAFS) of the South China Sea Fisheries Research Institute of the Chinese Academy of Fisheries Science (NO: SCSFRI96-253). Our experiments were implemented in accordance with national and institutional guidelines for the protection and use of CAFS laboratory animals.
Healthy P. monodon were obtained from the Shenzhen base of the South China Sea Fisheries Research Institute of the Chinese Academy of Fisheries Science. The experimental P. monodon were 7.8 ± 0.8 cm in length and 7.3 ± 1.0 g in body mass. The experiments were carried out in concrete ponds at a salinity of approximately 30 psu, 27–28°C and pH 7.6–7.8 for 1 week prior to the start of the experiment. During this period, the ponds were aerated and fed commercial bait four times a day. One-third of the water volume was replaced daily using multistage filtered and disinfected seawater.
The experiment was divided into three groups: a pure seawater group with a salinity of 30 psu (high-salinity challenge, HC), a group with a salinity of 18 psu (moderate-salinity challenge, MC), and a group with a salinity of 3 psu (low-salinity challenge, LC). The different salinities required for the experiments were adjusted by mixing bay water disinfected by multistage sand filtration with tap water in proportions, and the salinity was measured using a high-precision salinometer (ATAGO, Guangzhou). The formal experiments were carried out in special buckets for shrimp culture with a volume of 500 L and 400 L of culture water. Four parallels were set up in each group, using one bucket per parallel. Each bucket contained 60 juvenile shrimp. The salinities used were all 18 psu at the beginning and then adjusted by 3 psu each day until each group reached the desired salinity of 30 psu, 18 psu and 3 psu, respectively, before the official start of the culture trials at the three salinities. For 45 days of culture, water quality was tested daily, and shrimp growth and feeding were observed. Feeding was performed 3 times a day, weighing the same bait and feeding regularly. Water was changed completely every 3 days to maintain the water quality. At the end of the culture, gill tissue samples were taken from each of the three groups for transcriptome sequencing. Every three shrimp from the same group were mixed into one tube. All tissues were immediately frozen in liquid nitrogen and stored at −80°C until RNA extraction.
Total RNA was extracted from each sample of P. monodon gill tissue according to the instructions of the mirVana™ miRNA ISOlation Kit (Ambion, America). The product (concentration ≥50 ng mL−1) was examined by 1.5% agarose gel electrophoresis for completeness, which showed clear 28S and 18S bands with no obvious dragging. RNA purity was measured by a NanoDrop 2000 (Thermo, America) with A260/280 ≈ 2.0. A total of 4 μg of total RNA was used to construct RNA-seq libraries according to the instructions of the TruSeq Stranded mRNA LTSample Prep Kit (Illumina, America) (Loureiro et al., 2019). The RNA libraries were tested by an Agilent 2100 microarray for length and quality. After passing quality control, all RNA libraries were sequenced using an Illumina HiSeqTM 2500 instrument (Illumina, United States). The raw reads were archived in the sequence read archive of the National Center for Biotechnology Information under accession number SRP262105.
Transcript sequences were obtained using the paired-end splicing method with Trinity trinityrnaseq_r20131110 software (Grabherr et al., 2011), and the final unigene was obtained using TGICL 2.1 software (Pertea et al., 2003) for clustering and deredundancy extension to obtain the final unigene.
Unigene sequences were aligned by blastx to non-redundant (NR), Swiss-Prot protein (Swiss-Prot), Clusters of orthologous groups for eukaryotic complete genomes (KOG), Kyoto encyclopedia of genes and genomes (KEGG) and gene ontology (GO) libraries for comparison. The annotations with e < 1e−5 were taken to obtain the protein with the highest sequence similarity to the given Unigene. The KEGG annotation information of UniGene was obtained by using KAAS (http://www.genome.jp/kaas-bin/kaas_main), and based on the annotation results of SWISSPROT, the GO term was mapped to obtain the GO protein function annotation information of the UniGene. The annotation summary table information was obtained by merging Unigene with the best one from each database.
The expression levels of all DEGs were quantified based on the negative binomial distribution test in DESeq software (http://bioconductor.org/packages/release/bioc/html/DESeq.html). The NB (negative binomial distribution test) was used to test the significance of differences in the number of reads, and baseMean values were used to estimate the amount of gene expression.
The number of differential genes included in each GO entry was counted, and the significance of differential gene enrichment in each GO entry was calculated using the hypergeometric distribution test. A p-value <0.05 and log2-fold change value >2 indicated that differential genes were enriched in that GO entry.
To investigate the dynamic mRNA expression levels of P. monodon under different salinity stresses, mRNA-seq libraries were constructed from nine samples and sequenced by the Illumina NovaSeq platform. These libraries were HC1, HC2, HC3, MC1, MC2, MC3, LC1, LC2 and LC3. Nine libraries were obtained with approximately 49,236,624, 48,478,994, 48,743,030, 47,468,446, 48,525,278, 48,485,970, 48,220,504, 47,663,910 and 47,972,088 clean reads with average GC contents of 45.5%, 45%, 44%, 45.5%, 45.5%, 45%, 45%, 46%, and 45%, respectively. The samples were tested with Q30 values higher than 94.00%, indicating a good quality of transcriptome sequencing (Table 1). Due to the lack of a reference genome for P. monodon transcriptome analysis, the Trinity method was used to assemble high-quality reads. A total of 64,475 unigenes were obtained from transcriptome sequencing. The maximum length was 38,289 bp, the average length was 1,275.72 bp, and the N50 was 2,158 bp. Among these unigenes, 37,266 (36.88%) were longer than 1,000 bp, and the 300–500 bp and 500–1,000 bp unigenes were 21,796 (33.81%) and 18,898 (29.31%), respectively.
A total of 64,475 unigenes were annotated in five commonly used databases, including NR, Swiss-Prot, KOG, KEGG and GO. E-values less than 1e−60 were present in 55.02% of the 20,990 unigenes (32.56%) annotated to the NR database. When mapped to other species, 10,780 unigenes (27.75%) showed a preference for sequence similarity to Hyalella azteca species. Sequences from 472 unigenes (2.25%) showed similarities to P. monodon species. Due to the shortage of decapoda genomic resources, up to 51.36% and 26.65% of unigenes could not be annotated in the NR and Swiss-Prot databases, respectively, and may represent the genetic specificity of P. monodon.
The 64,475 unigenes (total sequence length 82,251,807bp) were examined using MISA and Primmer 3 software, and their SSR loci were analyzed. We identified 51,544 SSR loci (31.70% of the total unigenes), of which 12,634 sequences contained more than one SSR. Among all SSRs, 5,387 SSRs were present in compound formation. Among them, the most represented repeat type was mononucleotide (46.04%), and the least represented repeat types were pentanucleotide and hexanucleotide (0.17%) (Table 2).
The genes responsive to low salinity stress and the preliminary mechanism of P. monodon adaptation to low salinity stress were screened. The expression levels were calculated and compared between the three groups (HC, MC and LC groups), and the DEGs of the MC and LC groups compared to the HC group were determined (the conditions for significant differences were p-value <0.05 and difference multiplicity >2). The DEG results showed that compared to the HC group, 1,000 upregulated genes and 1,062 downregulated genes were observed in the MC group, and 1,140 upregulated genes and 1,531 downregulated genes were observed in the LC group compared to the HC group (Figure 1).
Figure 2 shows the grouped clustering analysis of differentially expressed genes in three salinity groups, HC, MC and LC. The results showed that the differentially expressed genes basically had a clear up- and downregulation relationship in the two samples. The genes thus classified are good references for conducting gene enrichment analysis and the resolution of salinity adaptation mechanisms.
Functional annotation analysis of DEGs was used to further compare and explore the transcriptome regulatory patterns under different salt concentration stresses. Figure 3 shows the GO enrichment analysis of all differentially expressed genes. The GO enrichment results showed the highest enrichment (p-value <0.05) in the three clusters of biological processes, molecular functions and cellular components. Figure 4 shows the integration of GO Level 2 classification of upregulated genes and downregulated genes in the differential grouping. The results showed that the differentially expressed genes were more annotated in the biological process category in cellular process, single-organism process, metabolic process, biological regulation, regulation of biological process and other functional localizations. The enriched cellular components were mainly functions such as cell, cell part, organelle, organelle part and membrane. The enrichment in molecular function was mainly in the functional classification of binding, catalytic activity, structural molecule activity, and transporter activity.
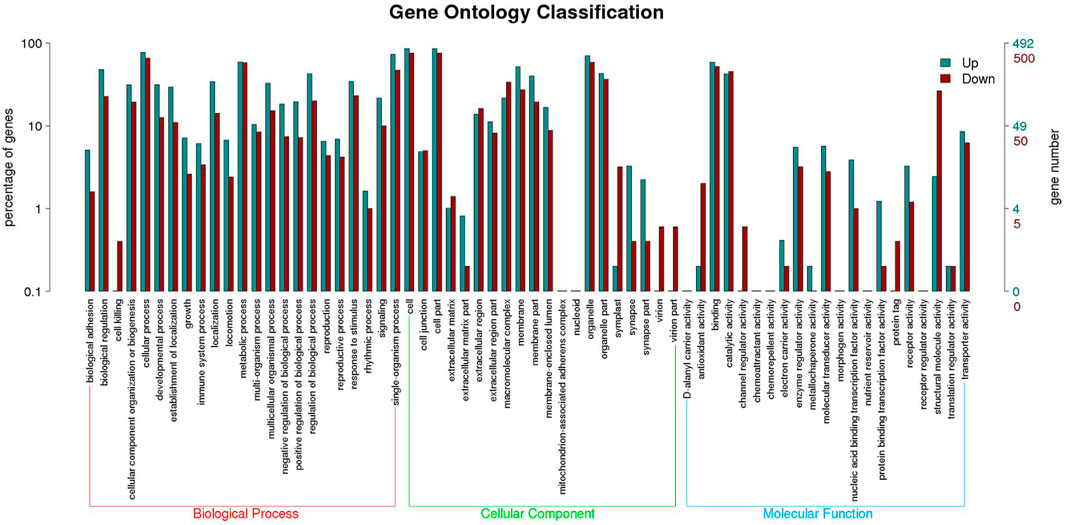
FIGURE 4. GO Level 2 classification statistics results of up- and downregulated differentially expressed genes.
KEGG pathway analysis can provide detailed information on the pathways in which DEGs are involved. In this study, 1,223 DEGs were annotated into 36 pathways. These pathways were divided into six main groups at Level 1: Metabolism, genetic information processing, environmental information processing, cellular processes, biological systems, and human diseases (Figure 5). There were significant differences in the enriched pathways between the experimental groups. In the HC vs. LC group, 583 DEGs significantly mapped to 37 signaling pathways. In the HC vs. MC group, 444 DEGs were significantly mapped to 28 signaling pathways. These pathways were mainly significantly associated with infectious diseases, signal transduction, immunity and metabolism. The top twenty pathways enriched in each group are shown in Figure 6.
To investigate the effects of low salinity on the metabolic regulation of P. monodon, gill tissues were screened for metabolism-related DEGs after 45 days of stress with different salinities. Twelve metabolic pathways were identified, involving processes in amino acid metabolism, glucose metabolism and lipid metabolism. Five significantly enriched metabolic pathways were detected in the LC vs. HC group, including pentose and glucuronate interconversions, ascorbate and aldarate metabolism, cutin, suberin and wax biosynthesis, and sulfur metabolism. Three significantly enriched metabolic pathways were detected in the MC vs. HC group, including cutin, suberin and wax biosynthesis, and butanoate. Some representative differentially expressed genes are shown in Table 3.
To investigate the effect of chronic low salinity on immune regulation in P. monodon, the study analyzed immune system pathways. A total of 17 immune system regulatory signaling pathways were detected, including the NOD-like receptor signaling pathway, Toll-like receptor signaling pathway, and Toll and IMD signaling pathways. The study detected more differentially expressed genes in the NOD-like receptor signaling pathway and Toll and IMD signaling pathways. In addition, antigen processing and presentation were significantly different only in the LC vs. HC group, and cell differentiation was only different in the MC vs. HC group. Some representative differentially expressed genes are shown in Table 4.
Understanding signal transduction pathways is an important guide for resolving the adaptation mechanisms of organisms to environmental changes. Therefore, the signal transduction pathways of P. monodon under chronic low salinity stress were analyzed during this study. A total of 31 signal transduction pathways were identified, including the PI3K-Akt signaling pathway, MAPK signaling pathway, AMPK signaling pathway, Hippo signaling pathway, calcium signaling pathway, sphingolipid signaling pathway, cGMP-PKG signaling pathway, HIF-1 signaling pathway, cAMP signaling pathway and Ras signaling pathway. Among them, the MAPK signaling pathway was significantly different in both the LC vs. HC and MC vs. HC groups, while the Hippo signaling pathway was only significantly different in the MC vs. HC groups. Some representative differentially expressed genes are shown in Table 5.
Although previous studies have shown that P. monodon can tolerate a wide range of salinities, studies on its osmotic adaptation mechanisms under chronic low salinity stress are still lacking. The mechanisms of adaptation of P. monodon to long-term varying salinity were assessed by full-length transcriptome amplification in our study. This study identified 2,062 DEGs from the HC vs. MC group, including 1,000 upregulated and 1,062 downregulated genes, and identified 2,671 DEGs from the HC vs. LC group, including 1,140 upregulated and 1,531 downregulated genes. This difference in gene regulation suggests that although P. monodon can cope with this stress, 18 psu and 3 psu may cause subtle stress. The GO analysis results are consistent with previous studies on aquatic animals that classified DEGs into three categories of biological processes, molecular functions and cellular components, such as P. vannamei (Yu et al., 2022), Pelteobagrus fulvidraco (Wang et al., 2022) and Oratosquilla oratoria (Lou et al., 2019). Furthermore, KEGG analysis showed that changes in salinity significantly affected metabolic, immune and signaling pathways in P. monodon, consistent with the results of transcriptome analyses of P. vannamei (Chen et al., 2015), Acanthogobius ommaturus (Sun et al., 2020) and Eriocheir sinensis (Yang et al., 2019) under low salinity stress. Therefore, the present study focused on the adaptation of metabolic, immune and signal transduction mechanisms in relation to salinity.
Salt stress is largely responsible for altering the metabolic state of organisms (Sun et al., 2020; Liu Z. et al., 2021). Our study found that the expression of metabolism-related genes was significantly affected and showed metabolic imbalance in P. monodon during prolonged low salinity stress. Many studies have reported similar results, but most of the studies on metabolism in osmoregulation have focused on osmotic physiological responses (Guo et al., 2020). In the present study, three significantly enriched metabolic pathways were detected in the MC vs. HC group, including Cutin, suberin and wax biosynthesis, Butanoate metabolism and Valine, leucine and isoleucine biosynthesis; five significantly enriched metabolic pathways were detected in the LC vs. HC group, including Pentose and glucuronate interconversions, Ascorbate and aldarate metabolism, Cutin, suberin and wax biosynthesis, Sulfur metabolism, Arginine biosynthesis and Phenylalanine metabolism. These pathways are mainly related to the regulation of glucose, lipid and amino acid metabolism. It has been found that crustaceans exposed to salinity fluctuations require additional energy to ensure osmotic pressure regulation and ion exchange (Chen et al., 2014). Lipids may provide sufficient energy to maintain ion homeostasis and regulate the permeability of membrane structures and therefore may play an important role in osmotic pressure regulation (Palacios et al., 2004; Tseng and Hwang, 2008). Free amino acids are osmotic effectors in crustaceans, and the metabolism of free amino acids may provide energy and eventually lead to cell growth in crustaceans (Lemos et al., 2001). Thus, metabolic pathways play an important role in the regulation of osmotic pressure in crustaceans.
Chronic low salinity stress can damage the immune system of aquatic animals. Antigen processing and presentation and the NOD-like receptor signaling pathway are the immune pathways that are more affected by chronic low salinity stress in P. monodon. Antigen processing and presentation are the cornerstones of adaptive immunity, through which organisms immunologically recognize some bacteria, viruses and parasites (Pishesha et al., 2022). Therefore, changes in genes associated with this pathway suggest that chronic low salinity stress may disrupt the innate immunity of shrimp and increase their susceptibility to bacteria, viruses, and parasites, among others (Kelly and Trowsdale, 2019). Complement and coagulation cascades are evolutionarily relevant blood enzyme cascade reactions in the blood. The complement system has immunoprotective and regulatory functions, while the coagulation cascades are responsible for ensuring hemostasis, and together, they limit pathogen infection through innate immune mechanisms. Thus, both protein C (PROC) and plasminogen (PLG), important regulators in the complement and coagulation cascades in this study, were significantly upregulated, suggesting that prolonged hyposalinity stress may have some effect on coagulation function in P. monodon (Ayón-Núñez et al., 2018). In addition, the NOD-like receptor signaling pathway (NLR) immune recognition factor was significantly altered in both groups. This regulation has important implications for NLR and complement components, which are pattern recognition receptors that recognize molecular patterns associated with viral, bacterial and eukaryotic pathogens (Ward and Rosenthal, 2014). For crustaceans, the innate immune system plays a crucial role in defense against microbial infections and environmental stress (Tran et al., 2022). These results suggest that P. monodon may have an increased susceptibility to pathogen attack in low salinity environments.
The adaptive and stimulatory responses of aquatic animals to salt stress depend on effective mechanisms of osmotic sensing and osmotic stress signaling (Nie et al., 2017). Furthermore, osmotic sensing signals are not only rapidly transduced intracellularly but may also be amplified and distributed to multiple downstream osmotic effects (Sun et al., 2020). cAMP, as a signaling factor, is involved in the regulation of osmotic pressure by mediating the increased uptake of Na+ by cells in the internal environment (Liang et al., 2022). At the same time, cAMP stimulates the production of calmodulin (CaM) and ATP, thereby regulating stress, apoptosis and calcium homeostasis (Huang et al., 2020). Thus, as important ion transport regulatory proteins, CaM and NKA are significantly upregulated at low salinity. The PI3K-AKT pathway is one of the most actively studied kinase pathways, mediating cell growth, survival, cell cycle progression, differentiation, transcription, translation, and glucose metabolism in animals (Liu et al., 2018). Therefore, downregulation of this pathway may be responsible for the slow growth of P. monodon under prolonged hyposaline conditions, as shrimp require more energy for osmoregulation and ion homeostasis during normal growth (Cui et al., 2020). The MAPK pathway is important for osmotic pressure regulation, and chilling and hypoxia are important. In the present study, DEGs in the gill tissue of P. monodon under salt stress were enriched in the MAPK signaling pathway, indicating that the pathway was induced. Similar results were found in P. vannamei hemocytes under salt stress (Zhao et al., 2015). Furthermore, AMPK is suitable as an intracellular energy and pressure sensor to reveal the molecular mechanisms underlying temperature, hypoxia and salinity fluctuations in animals (Hardie, 2011). Salinity stress can temporarily activate AMPK, while glucose depletion leads to sustained AMPK activation. Moreover, in both cases, AMPK activation levels depend on the degree of stress (Schutt and Moseley, 2017). The activation of the AMPK signaling pathway and the expression of related genes in P. monodon under low salinity stress may be due to its continuous activation by glucose depletion. The differences in the expression of genes related to the AMPK signaling pathway in gill tissues of P. monodon at different salinities may be due to different salt stress stresses, resulting in different energy regulation mechanisms.
In this study, RNA-seq analysis of P. monodon exposed to three salinity gradients (namely, HC, MC and LC) was carried out, and the high-quality data obtained are the best guarantee of experimental reliability. These changes were verified by this experiment to be related to metabolic, immune system and signal transduction properties. Thus, P. monodon may adapt to changes in environmental salinity by regulating mechanisms such as metabolism, immunity and signal transduction. These data will provide basic data for future selection and breeding for resistance in the P. monodon industry, which will contribute to the sustainable and healthy development of shrimp farming.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, SRR11805651, SRR11805650, SRR11805649, SRR11805654, SRR11805653, SRR11805652, SRR11805665, SRR11805664, and SRR11805655.
The animal study was reviewed and approved by Animal Care and Use Committee of South China Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences (No. SCSFRI96-253).
Y-DL Writing—original draft, F-LZ project administration EL Supervision. S-GJ Writing—review and editing. M-RS Writing—original draft Q-BY Supervision. SJ Data curation. L-SY Visualization, Investigation. J-HH Methodology, Software. XC Software, Validation.
This research was supported by National Key R & D Program of China (2022YFD2401900); China Agriculture Research System (CARS-48); Hainan Yazhou Bay Seed Laboratory (Project of B21Y10701, B21HJ0701); Central Public Interest Scientific Institution Basal Research Fund, South China Sea Fisheries Research Institute, CAFS (2020ZD01, 2021SD13); Hainan Provincial Natural Science Foundation of China (320QN359, 322RC806); Guangdong Basic and Applied Basic Research Foundation (2020A1515110200); Guangzhou Science and Technology Planning Project (202102020208); and Hainan Provincial Association for Science and Technology of Young Science and Technology Talents Innovation Plan Project (QCQTXM202206).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Ayón-Núñez, D. A., Fragoso, G., Bobes, R. J., and Laclette, J. P. (2018). Plasminogen-binding proteins as an evasion mechanism of the host’s innate immunity in infectious diseases. Biosci. Rep. 5, BSR20180705. doi:10.1042/bsr20180705
Chen, K., Li, E. C., Li, T. Y., Xu, C., Wang, X. D., Lin, H. Z., et al. (2015). Transcriptome and molecular pathway analysis of the hepatopancreas in the pacific white shrimp Litopenaeus vannamei under chronic low-salinity stress. Plos One 7, e0131503. doi:10.1371/journal.pone.0131503
Chen, K., Li, E., Gan, L., Wang, X., Xu, C., Lin, H., et al. (2014). Growth and lipid metabolism of the pacific white shrimp Litopenaeus vannamei at different salinities. J. Shellfish Res. 3, 825–832. doi:10.2983/035.033.0317
Chen, Y.-H., and He, J.-G. (2019). Effects of environmental stress on shrimp innate immunity and white spot syndrome virus infection. Fish. Shellfish Immun. 84, 744–755. doi:10.1016/j.fsi.2018.10.069
Cui, W., Ma, A., and Wang, X. (2020). Response of the PI3K-AKT signaling pathway to low salinity and the effect of its inhibition mediated by wortmannin on ion channels in turbot Scophthalmus maximus. Aquac. Res. 51 (7), 2676–2686. doi:10.1111/are.14607
de Jong, T. V., Moshkin, Y. M., and Guryev, V. (2019). Gene expression variability: The other dimension in transcriptome analysis. Physiol. Genomics 51 (5), 145–158. doi:10.1152/physiolgenomics.00128.2018
Esparza-Leal, H. M., Ponce-Palafox, J. T., Cervantes-Cervantes, C. M., Valenzuela-Quinonez, W., Luna-Gonzalez, A., Lopez-Alvarez, E. S., et al. (2019). Effects of low salinity exposure on immunological, physiological and growth performance in Litopenaeus vannamei. Aquac. Res. 50 (3), 944–950. doi:10.1111/are.13969
Gao, J., Xu, G., and Xu, P. (2021). Gills full-length transcriptomic analysis of osmoregulatory adaptive responses to salinity stress in Coilia nasus. Ecotox Environ. Safe 226, 112848. doi:10.1016/j.ecoenv.2021.112848
Ge, J., Tao, J., Zhao, J., Wu, Z., Zhang, H., Gao, Y., et al. (2022). Transcriptome analysis reveals candidate genes involved in multiple heavy metal tolerance in hyperaccumulator Sedum alfredii. Ecotox Environ. Safe 241, 113795. doi:10.1016/j.ecoenv.2022.113795
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29 (7), 644–652. doi:10.1038/nbt.1883
Guo, T., Yang, Y., Meng, F., Wang, S., Xia, S., Qian, Y., et al. (2020). Effects of low salinity on gill and liver glycogen metabolism of great blue-spotted mudskippers (Boleophthalmus pectinirostris). Comp. Biochem. Phys. C 230, 108709. doi:10.1016/j.cbpc.2020.108709
Hardie, D. G. (2011). AMP-activated protein kinase-an energy sensor that regulates all aspects of cell function. Gene Dev. 25 (18), 1895–1908. doi:10.1101/gad.17420111
Henry, R., Lucu, C., Onken, H., and Weihrauch, D. (2012). Multiple functions of the crustacean gill: Osmotic/ionic regulation, acid-base balance, ammonia excretion, and bioaccumulation of toxic metals. Front. Physiol. 3, 431. doi:10.3389/fphys.2012.00431
Huang, S., Jiang, S., and Xu, Q. (2020). Acute salinity stress in gill Na(+)/K+-atpase in the swimming crab, Portunus tritubercutatus (brachyura, portunidae). Crustaceana 93 (6), 587–609. doi:10.1163/15685403-bja10012
Joseph, A., and Philip, R. (2020). Immunocompetence of Penaeus monodon under acute salinity stress and pathogenicity of Vibrio harveyi with respect to ambient salinity. Fish. Shellfish Immun. 106, 555–562. doi:10.1016/j.fsi.2020.07.067
Kelly, A., and Trowsdale, J. (2019). Genetics of antigen processing and presentation. Immunogenetics 71 (3), 161–170. doi:10.1007/s00251-018-1082-2
Lemos, D., Phan, V. N., and Alvarez, G. (2001). Growth, oxygen consumption, ammonia-N excretion, biochemical composition and energy content of Farfantepenaeus paulensis Perez-Farfante (Crustacea, Decapoda, Penaeidae) early postlarvae in different salinities. J. Exp. Mar. Biol. Ecol. 261 (1), 55–74. doi:10.1016/s0022-0981(01)00260-x
Li, B.-B., Fan, J.-Q., Hong, Q.-M., Yan, Z.-Y., Yang, X.-J., Lu, K.-C., et al. (2022). Transcriptome analysis endoplasmic reticulum-stress response in Litopenaeus vannamei hemocytes. Fish. Shellfish Immun. 124, 421–429. doi:10.1016/j.fsi.2022.04.008
Li, L., Wang, P., Zhao, C., and Qiu, L. (2018). The anti-stresses capability of GRP78 in Penaeus monodon: Evidence from in vitro and in vivo studies. Fish. Shellfish Immun. 72, 132–142. doi:10.1016/j.fsi.2017.10.052
Li, Y., Zhou, F., Huang, J., Yang, L., Jiang, S., Yang, Q., et al. (2020). Transcriptome and miRNA profiles of black tiger shrimp, Penaeus monodon, under different salinity conditions. Front. Mar. Sci. 7, 579381. doi:10.3389/fmars.2020.579381
Liang, G., Qin, K., Chen, Y., Niu, M., Wang, H., Wang, C., et al. (2022). Transcriptomic analysis of adaptive mechanisms in response to inland saline-alkaline water in the mud crab, Scylla paramamosain. Front. Mar. Sci. 9, 974501. doi:10.3389/fmars.2022.974501
Liu, X., Jiang, H., Ye, B., Qian, H., Guo, Z., Bai, H., et al. (2021a). Comparative transcriptome analysis of the gills and hepatopancreas from Macrobrachium rosenbergii exposed to the heavy metal Cadmium (Cd2+). Sci. Rep-Uk 11 (1), 16140. doi:10.1038/s41598-021-95709-w
Liu, Y., Li, E., Xu, C., Su, Y., Qin, J. G., Chen, L., et al. (2018). Brain transcriptome profiling analysis of nile Tilapia (Oreochromis niloticus) under long-term hypersaline stress. Front. Physiol. 9, 219. doi:10.3389/fphys.2018.00219
Liu, Z., Ma, A., Yuan, C., Zhao, T., Chang, H., and Zhang, J. (2021b). Transcriptome analysis of liver lipid metabolism disorders of the turbot Scophthalmus maximus in response to low salinity stress. Aquaculture 534, 736273. doi:10.1016/j.aquaculture.2020.736273
Lou, F., Gao, T., and Han, Z. (2019). Effect of salinity fluctuation on the transcriptome of the Japanese mantis shrimp Oratosquilla oratoria. Int. J. Biol. Macromol. 140, 1202–1213. doi:10.1016/j.ijbiomac.2019.08.223
Loureiro, S., Amorim, A., Cainé, L., Silva, B., and Gomes, I. (2019). Evaluation of two DNA/RNA co-extraction methods for body fluid identification in forensics. Forensic Sci. Int. Genet. Suppl. Ser. 7, 250–252. doi:10.1016/j.fsigss.2019.09.096
Nie, H., Jiang, L., Chen, P., Huo, Z., Yang, F., and Yan, X. (2017). High throughput sequencing of RNA transcriptomes in Ruditapes philippinarum identifies genes involved in osmotic stress response. Sci. Rep-Uk 7 (1), 4953. doi:10.1038/s41598-017-05397-8
Normant, M., Kubicka, M., Lapucki, T., Czarnowski, W., and Michalowska, M. (2005). Osmotic and ionic hemolymph concentrations in the Baltic Sea amphipod Gammarus oceanicus in relation to water salinity. Comp. Biochem. Phys. A 141 (1), 94–99. doi:10.1016/j.cbpb.2005.04.007
Palacios, E., Bonilla, A., Luna, D., and Racotta, I. S. (2004). Survival, Na+/K+-ATPase and lipid responses to salinity challenge in fed and starved white Pacific shrimp (Litopenaeus vannamei) postlarvae. Aquaculture 234 (1-4), 497–511. doi:10.1016/j.aquaculture.2003.12.001
Parra, D., Reyes-Lopez, F. E., and Tort, L. (2015). Mucosal immunity and B cells in teleosts: Effect of vaccination and stress. Front. Immunol. 6, 354. doi:10.3389/fimmu.2015.00354
Pertea, G., Huang, X., Liang, F., Antonescu, V., Sultana, R., Karamycheva, S., et al. (2003). TIGR gene indices clustering tools (TGICL): A software system for fast clustering of large EST datasets. Bioinformatics 19 (5), 651–652. doi:10.1093/bioinformatics/btg034
Pishesha, N., Harmand, T. J., and Ploegh, H. L. (2022). A guide to antigen processing and presentation. Nat. Rev. Immunol. 22, 751–764. doi:10.1038/s41577-022-00707-2
Podbielski, I., Hiebenthal, C., Hajati, M.-C., Bock, C., Bleich, M., and Melzner, F. (2022). Capacity for cellular osmoregulation defines critical salinity of marine invertebrates at low salinity. Front. Mar. Sci. 9, 898364. doi:10.3389/fmars.2022.898364
Rivera-Ingraham, G. A., Nommick, A., Blondeau-Bidet, E., Ladurner, P., and Lignot, J.-H. (2016). Salinity stress from the perspective of the energy-redox axis: Lessons from a marine intertidal flatworm. Redox Biol. 10, 53–64. doi:10.1016/j.redox.2016.09.012
Schutt, K. L., and Moseley, J. B. (2017). Transient activation of fission yeast AMPK is required for cell proliferation during osmotic stress. Mol. Biol. Cell 28 (13), 1804–1814. doi:10.1091/mbc.e17-04-0235
Si, M.-R., Li, Y.-D., Jiang, S.-G., Yang, Q.-B., Jiang, S., Yang, L.-S., et al. (2022a). A CSDE1/Unr gene from Penaeus monodon: Molecular characterization, expression and association with tolerance to low salt stress. Aquaculture 561, 738660. doi:10.1016/j.aquaculture.2022.738660
Si, M.-R., Li, Y.-D., Jiang, S.-G., Yang, Q.-B., Jiang, S., Yang, L.-S., et al. (2022b). Identification of multifunctionality of the PmE74 gene and development of SNPs associated with low salt tolerance in Penaeus monodon. Fish. Shellfish Immun. 128, 7–18. doi:10.1016/j.fsi.2022.07.010
Stupnikov, A., McInerney, C. E., Savage, K. I., McIntosh, S. A., Emmert-Streib, F., Kennedy, R., et al. (2021). Robustness of differential gene expression analysis of RNA-seq. Comput. Struct. Biotec 19, 3470–3481. doi:10.1016/j.csbj.2021.05.040
Sun, Z., Lou, F., Zhang, Y., and Song, N. (2020). Gill transcriptome sequencing and de novo annotation of Acanthogobius ommaturus in response to salinity stress. Genes 6 (11), 631. doi:10.3390/genes11060631
Tran, N. T., Liang, H., Zhang, M., Bakky, M. A. H., Zhang, Y., and Li, S. (2022). Role of cellular receptors in the innate immune system of Crustaceans in response to white spot syndrome virus. Viruses 14 (4), 743. doi:10.3390/v14040743
Tseng, Y.-C., and Hwang, P.-P. (2008). Some insights into energy metabolism for osmoregulation in fish. Comp. Biochem. Phys. C 148 (4), 419–429. doi:10.1016/j.cbpc.2008.04.009
Vallejos-Vidal, E., Khansari, A. R., Soliva-Dueso, L., Balasch, J. C., Tort, L., and Reyes-López, F. E. (2022). The direct exposure of cortisol does not modulate the expression of immune-related genes on tissue explants of mucosal surfaces in rainbow trout (Oncorhynchus mykiss) nor in gilthead Sea bream (Sparus aurata). Front. Mar. Sci. 9, 828050. doi:10.3389/fmars.2022.828050
Vezzulli, L. (2022). Global expansion of Vibrio spp. in hot water. Env. Microbiol. Rep., 1–3. doi:10.1111/1758-2229.13135
Visudtiphole, V., Petkon, S., Sittikankaew, K., Sakayaroj, J., and Klinbunga, S. (2017). Calcium signaling-related genes in Penaeus monodon respond to abiotic stress and pathogenic bacteria common in aquaculture. Aquaculture 479, 626–637. doi:10.1016/j.aquaculture.2017.07.010
Wang, M., Li, B., Wang, J., Xie, S., and Zhang, L. (2022). Skin transcriptome and physiological analyses reveal the metabolic and immune responses of yellow catfish (Pelteobagrus fulvidraco) to acute hypoxia. Aquaculture 546, 737277. doi:10.1016/j.aquaculture.2021.737277
Ward, A. E., and Rosenthal, B. M. (2014). Evolutionary responses of innate immunity to adaptive immunity. Infect. Genet. Evol. 21, 492–496. doi:10.1016/j.meegid.2013.12.021
Xiao, J., Liu, Q.-Y., Du, J.-H., Zhu, W.-L., Li, Q.-Y., Chen, X.-L., et al. (2020). Integrated analysis of physiological, transcriptomic and metabolomic responses and tolerance mechanism of nitrite exposure in Litopenaeus vannamei. Sci. Total Environ. 711, 134416. doi:10.1016/j.scitotenv.2019.134416
Yang, Z., Zhou, J., Wei, B., Cheng, Y., Zhang, L., and Zhen, X. (2019). Comparative transcriptome analysis reveals osmotic-regulated genes in the gill of Chinese mitten crab (Eriocheir sinensis). Plos One 1, e0210469. doi:10.1371/journal.pone.0210469
Yu, D., Zhai, Y., He, P., and Jia, R. (2022). Comprehensive transcriptomic and metabolomic analysis of the Litopenaeus vannamei hepatopancreas after WSSV challenge. Front. Immunol. 13, 826794. doi:10.3389/fimmu.2022.826794
Keywords: Penaeus Monodon, Chronic low-salinity stress, transcriptome, osmoregulation, adaptation mechanisms
Citation: Li Y-D, Si M-R, Jiang S-G, Yang Q-B, Jiang S, Yang L-S, Huang J-H, Chen X, Zhou F-L and Li E (2023) Transcriptome and molecular regulatory mechanisms analysis of gills in the black tiger shrimp Penaeus monodon under chronic low-salinity stress. Front. Physiol. 14:1118341. doi: 10.3389/fphys.2023.1118341
Received: 07 December 2022; Accepted: 14 February 2023;
Published: 01 March 2023.
Edited by:
Daniel Carneiro Moreira, University of Brasilia, BrazilReviewed by:
Ngoc Tuan Tran, Shantou University, ChinaCopyright © 2023 Li, Si, Jiang, Yang, Jiang, Yang, Huang, Chen, Zhou and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fa-Lin Zhou, emhvdWZhbGluQGFsaXl1bi5jb20=; ErChao Li, ZWNsaUBiaW8uZWNudS5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.