Corrigendum: Validation of the binding stoichiometry between HCN channels and their neuronal regulator TRIP8b by single molecule measurements
- 1Department of Biosciences, University of Milan, Milano, Italy
- 2Department of Physiology and Cellular Biophysics, Columbia University, New York, NY, United States
- 3Department of Anesthesiology, Columbia University Irving Medical Center, New York, NY, United States
- 4Irving Institute for Clinical and Translational Research, Columbia University, New York, NY, United States
Tetratricopeptide repeat–containing Rab8b-interacting (TRIP8b) protein is a brain-specific subunit of Hyperpolarization-activated Cyclic Nucleotide-gated (HCN) channels, a class of voltage-gated channels modulated by cyclic nucleotides. While the interaction between TRIP8b and the cytosolic C terminus of the channel has been structurally described, the HCN:TRIP8b stoichiometry is less characterized. We employed single molecule mass photometry (MP) to image HCN4 particles purified in complex with TRIP8b. Our data show that four TRIP8b subunits are bound to the tetrameric HCN4 particle, confirming a 1:1 stoichiometry.
Introduction
Hyperpolarization-activated cyclic nucleotide-gated (HCN1–4) channels conduct the h-current (Ih), which plays a critical role in regulating several neuronal properties, including membrane resting potential, dendritic excitability, and intrinsic rhythmicity (Robinson and Siegelbaum, 2003). HCNs belong to the superfamily of tetrameric voltage-gated K+ channels (Kv) and are uniquely regulated by the direct binding of cAMP to their cytoplasmic Cyclic Nucleotide Binding Domain (CNBD). In addition to cAMP, neuronal HCN channels are further modulated by TRIP8b, a brain-specific cytoplasmic subunit, which controls channel trafficking and gating (Santoro et al., 2011). Particularly, TRIP8b, by interacting with the CNBD and thus impairing cAMP binding, antagonizes the facilitatory effect of the cyclic nucleotide on the voltage-dependent gating of HCN channels (Hu et al., 2013).
The structural details of the mechanism of TRIP8b binding to the C terminus of HCN channels are well known (Bankston et al., 2017; Deberg et al., 2015; Hu et al., 2013; Porro et al., 2020; Saponaro et al., 2014). TRIP8b possesses at least two binding sites for HCN channels: 1) a core sequence of 40 amino acid, named TRIP8bnano, that binds to the CNBD in a cAMP-dependent manner (Saponaro et al., 2018); 2) the tetratricopepide (TPR) domain of TRIP8b that interacts with the last three amino acids (SNL) of the channel (Bankston et al., 2012). TRIP8bnano sequence is necessary and sufficient to inhibit the cAMP-dependent activation of HCN channels (Porro et al., 2020; Santoro et al., 2011; Saponaro et al., 2018; Hu et al., 2013).
The association stoichiometry between TRIP8b and the HCN tetramer is less characterized. By using single molecule photobleaching method, Bankston and co-workers have shown that HCN2 and EGFP-TRIP8b form a 4:4 complex in the membrane of Xenopus laevis oocytes (Bankston et al., 2012). Given that, often times, results obtained with this approach were further disproved by direct biochemical/structural data (Balasuriya et al., 2014; Hou et al., 2012; Ji et al., 2008; Yen et al., 2016), we decided to confirm the HCN:TRIP8b stoichiometry with a different approach.
Here, we present the results of the binding stoichiometry of HCN channels and TRIP8b performed by single molecule mass photometry (MP). Mass Photometry enables mass measurement of single molecules in solution by detecting light scattering as they non-specifically and transiently interact with a glass surface. Binding events change the refractive index at the water/glass interface. This alteration of the local reflectivity can be converted into the molecular mass of the molecule (Young et al., 2018).
By mass-imaging detergent-purified HCN4 - TRIP8b complexes, we detected four TRIP8b molecules bound to a tetrameric HCN4 channel particle. Addition of saturating concentrations of cAMP, which competes with TRIP8b for the binding to HCN channels (Hu et al., 2013), caused the disruption of the complex into the two single elements.
Our results confirm the 4:4 HCN2:TRIP8b stoichiometry previously shown by single molecule photobleaching experiments (Bankston et al., 2012) and provides data on cAMP/TRIP8b competition in purified full length HCN proteins.
Results
For MP studies, HCN4 channels carrying an internal deletion (see Material and Methods for details) (Saponaro et al., 2021a) and EGFP- tagged TRIP8b (1a), one of the most abundant TRIP8b isoform in brain (Santoro et al., 2009), were transiently co-expressed in HEK293F cells by using a HCN4:TRIP8b DNA ratio of 1:1 to do not bias MP results by overexpressing TRIP8b. Electrophysiological analysis performed on HEK293T cells transiently expressing EGPF-TRIP8b (1a) together with the modified HCN4 channels (hereafter indicated as HCN4) using the same 1:1 DNA ratio (Figure 1) showed that TRIP8b fully antagonizes the cAMP effect as previously reported for full length HCN4 channels (Saponaro et al., 2018).
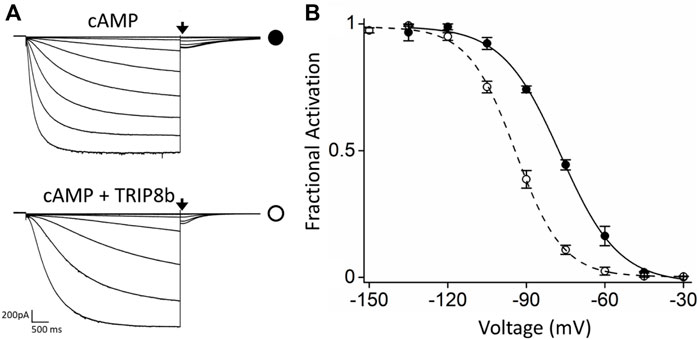
FIGURE 1. Functional characterization of the inhibitory effect of TRIP8b on the HCN4 construct employed for MP. (A) Representative whole-cell current traces of HCN4 channels recorded, with 0.25 µM cAMP in the patch pipette, in HEK293T cells transiently expressing the channel alone (top) or with GFP-TRIP8b (1a) (bottom). Black arrows indicate the current selected for analysis in (B). (B) Mean activation curves of HCN4 channels alone (black full circles) or co-expressed with GFP-TRIP8b (1a) (black open circles) with cAMP in the patch pipette obtained from tail currents collected at −40 mV (see arrows in panel (A). Dashed lines indicate Boltzmann fitting to the data (see Materials and methods) from which the half activation potential (V1/2) were derived. HCN4 + cAMP = −76.9 ± 0.6 mV; HCN4 + TRIP8b + cAMP = −94.2 ± 1.2 mV. Data are presented as mean ± SEM. Number of cells (N) ≥ 8. The two half activation potentials are statistically different. Statistical analysis performed with t-student test (p < 0.001).
After membrane solubilization with detergents, the HCN4—TRIP8b complex was affinity purified by using the polyhistidine tag at the N-terminus of EGFP-TRIP8b. The complex was further isolated by size-exclusion chromatography and the co-elution of HCN4 and EGFP-TRIP8b was confirmed by immunoblot (Figures 2A–C). Of note that HCN4 does not completely lose its quaternary structure when run into a denaturing gel (Figures 2B,C, left panel). This was already reported for detergent-purified HCN1 (Lee and MacKinnon, 2017) and HCN4 (Saponaro et al., 2021a; Saponaro et al., 2021b). The same occurs also for HCN4 - TRIP8b complex, although in the latter case the phenomenon is significantly reduced since most of TRIP8b molecules run in their monomeric state (Figures 2B,C, right panel). This is expected as TRIP8b is a soluble protein and thus it is more prone to be denatured and consequently to dissociate from HCN4.
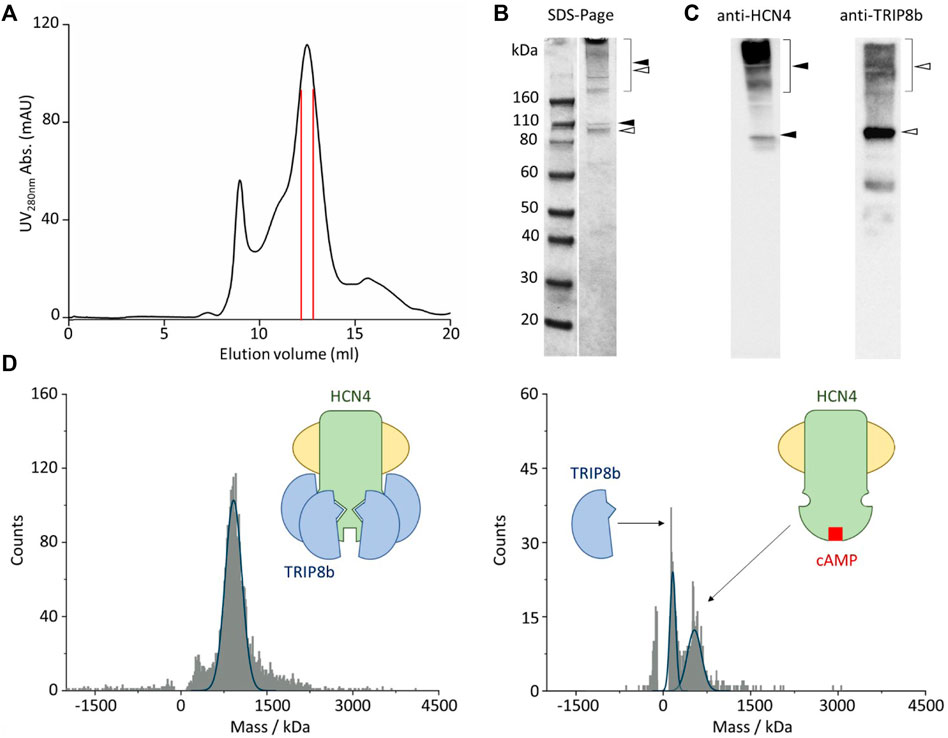
FIGURE 2. Single molecule mass photometry of HCN4—EGFP-TRIP8b complex. (A) SEC of HCN4—EGFP-TRIP8b complex following purification in LMNG/CHS. Peak fractions used for MP are delimited by the red lines. (B) SDS-PAGE gel of the pooled SEC fractions of HCN4—EGFP-TRIP8b complex, stained with Coomassie blue. Filled arrows indicate HCN4, while open arrows indicate EGFP-TRIP8b, in their oligomeric (square bracket), and monomeric forms. (C) Western blots of the pooled SEC fractions of HCN4—EGFP-TRIP8b complex performed by using anti-TRIP8b and anti-HCN4 antibodies, respectively. Open and filled arrows as in panel (B). Of note that the western blot anti-TRIP8b reveals the presence of a small number of molecules (faint band between 60 and 50 kDa markers) corresponding N terminally degraded TRIP8b. Indeed, they co-purified with HCN4 and thus have retained the HCN binding sites located in their C-terminal half (Santoro et al., 2004; Santoro et al., 2011). (D) Mass histogram of binding events for HCN4—EGFP-TRIP8b complex before (left) and after the addition of 2 mM cAMP (right). The blue lines represent the fitting of the data with Gaussian Distribution Function. Left, schematic representation of purified HCN4 (green) embedded into a detergent micelle (yellow) and bound to four TRIP8b molecules (blue). Right, purified HCN4 bound to cAMP (red) has lost TRIP8b.
The complex was then imaged with MP. Figure 2D shows the mass distribution histograms of the purified HCN4 - EGFP-TRIP8b complex. Each histogram represents a single molecule binding signal arising from 1-min recording of binding events to the glass surface. HCN4—TRIP8b complex produced a mostly homogeneous mass distribution with a main peak of 935 kDa. The theoretical molecular weight (MW) of the complex with 4:4 stoichiometry is 768 kDa since the expected MW for the monomeric HCN4 is 98.4 kDa and that of EGFP-TRIP8b is 93.6 kDa. Given the absence of protein contaminants (Figures 2B,C), The about 167 kDa difference between the calculated and the theoretical MW of HCN4 - EGFP-TRIP8b complex may be ascribed to the mass of the LMNG-CHS detergent micelle surrounding HCN4. It is worth noting that the micelles are dynamic structures in equilibrium with the surrounding environment. This introduces a certain degree of variability in the mass of a detergent-purified membrane protein.
We further tested the complex by adding an excess of cAMP (2 mM) that should disrupt the interaction, leading to the appearance of two peaks: 1) the homotetrameric HCN4 channel embedded into the detergent micelle (expected mass of about 500kDa); 2) the monomeric EGFP-TRIP8b (expected mass of 93.6 kDa). In line with our prediction, in the presence of cAMP, the 935 kDa peak disappeared and was substituted by two peaks of 498 and 100 kDa respectively (Figure 2D). Of note that the binding events recorded from the sample with cAMP (Figure 2D, right panel) are less than the ones recorded from the sample without the ligand (Figure 2D, left panel). This is due to the dilution of HCN4—EGFP-TRIP8b complex caused by the addition of 2 mM cAMP.
Discussion
Here we present the measurement of the association stoichiometry of HCN:TRIP8b complex based on a highly accurate determination of the mass of their complex. Such high resolution was achieved by mass photometry (MP). Currently used methods for assessing the mass of macromolecules are solution-based ensemble techniques, with limited mass accuracy and resolution. Instead, MP determines the mass of biomolecules with single molecule sensitivity. The latter feature provides the powerful advantage of an unprecedented resolution/precision, and thus mass accuracy (∼2% mass error) (Young et al., 2018). Therefore, MP allowed us to conclusively establish that TRIP8b forms a tetrameric assembly with HCN4 channels. This finding validates the 4:4 stoichiometry of HCN2:TRIP8b previously determined by Bankston et al. (2012) with fluorescence bleaching.
The finding that TRIP8b shows 4:4 stoichiometry with HCN2 and HCN4 allows to speculate that this will be the case for HCN1 as well, given the high degree of conservation between the three isotypes.
What is the significance of the 4:4 stoichiometry?
HCN channels play a crucial role in regulating dendritic excitability by filtering excitatory inputs (Magee, 1999; Williams and Stuart, 2000; Tsay et al., 2007; George et al., 2009). Therefore, HCN channels need to be finely controlled, in particular in response to the complexity of cAMP dynamics. TRIP8b is expressed in the brain to this end. In hippocampal CA1 pyramidal neurons, for instance, the sensitivity of Ih current to cAMP decreases symmetrically along the dorsoventral axis in well agreement with the dorsoventral gradient of TRIP8b (Marcelin, 2012). Given the pronounced cooperativity of HCN channels in cAMP binding and activation (Ulens and Siegelbaum, 2003; Kusch et al., 2010; Lolicato et al., 2011; Wu et al., 2011; Chow et al., 2012; Kusch et al., 2012; Thon et al., 2015) it is likely to conclude that the obligate saturation of TRIP8b molecules to the four subunits forming the channel appears to be the fastest way possible to strictly and finely control the response of Ih to cAMP fluctuations.
A major advantage of MP measurement is the small amount and the low concentration of the protein sample employed (1 μL at 200 nM concentration). This is extremely relevant for eukaryotic membrane proteins as they are difficult to purify in large amounts. In the case of HCN proteins, such limitation has long restricted the biochemical studies of HCN-TRIP8b interaction to their isolated soluble domains (Bankston et al., 2012; Bankston et al., 2017; Deberg et al., 2015; Porro et al., 2020; Saponaro et al., 2014; Saponaro et al., 2018) and thus prevented the study of the complex in full length proteins.
Our study further confirms that the interaction of the two partners can be disrupted by increasing amount of cAMP (Bankston et al., 2017; Hu et al., 2013; Saponaro et al., 2014). Such a competitive action of cAMP on TRIP8b was so far never reproduced in isolated full length HCN proteins. This is particularly relevant in the light of recent functional studies (Gross et al., 2018; Porro et al., 2019), based on the cryo-EM structures of HCN1 (Lee and MacKinnon, 2017), which assigned a key role in the propagation of the cAMP-pathway to an unknown cytoplasmic N terminal domain (HCND) that wedges in between the voltage sensor module and C terminal cytoplasmic regulatory region. This implies that the HCND, as well as other intracellular modules, may contribute to regulate the association between HCN and TRIP8b, and that the interaction between the two proteins is considerably more complex than the mechanism so far described.
Methods
Constructs
The cDNA encoding full-length mouse TRIP8b (splice variant 1a4) was cloned into a modified pEG BacMam vector (Goehring et al., 2014) (hereafter pEGA). The cDNA encoding rabbit HCN4 carrying an internal deletion (from residues 783 to 1064, hereafter HCN4) was cloned into pCI vector (Promega Corporation). Is it worth noting that the internal deletion eliminates a poorly conserved region in the C-terminal portion of the HCN channel protein, but preserves the extreme C-terminal SNL tripeptide sequence responsible for binding to TRIP8b (Santoro et al., 2004; Santoro et al., 2011).
Electrophysiology
HEK293-T cells (ATCC) were cultured, transiently transfected, and measured by patch-clamp technique using a ePatch amplifier (Elements srl) as described in Porro et al. (2019), Porro et al. (2020), Saponaro et al. (2018).
Protein complex expression and purification
Freestyle HEK293-F cell cultures (Thermo Fisher) were transiently co-transfected with pCI: HCN4ΔC (0.75 μg per ml) and pEGA: TRIP8b (0.75 µg per ml) according to the procedure detailed in Saponaro et al. (2021a).
Membrane isolation and HCN4—TRIP8b complex purification were pursed according to the protocols details in Saponaro et al. (2021a), with the following modification: HCN4—TRIP8b complex eluted form Ni2+-NTA resin (Qiagen) was loaded on a Superose 6 increase 10/300 GL SEC column (GE Healthcare Life Sciences) pre-equilibrated with buffer containing 200 mM NaCl, 20 mM HEPES pH 7.0 and detergent (LMNG-CHS) at the concentration of 0.002% (w/v). Peak fractions (see Figure 2A) were used for mass photometry. For the protein complex analyzed in the presence of cAMP, 2 mM of ligand was added to the sample prior mass photometry.
SDS-page denaturing gel and western blots
Protein samples were resolved by precast 4–12% SDS-PAGE gels (Thermo Fisher Scientific) and either stained with Coomassie Brilliant Blue R (Merck) or transferred to PVDF membranes (Thermo Fisher Scientific) for Western blotting. Primary antibody dilutions were as follows: anti-HCN4 (rabbit polyclonal, Alomone) 1:1000; anti-TRIP8b (mouse monoclonal, NeuroMab) 1:1000.
Anti-mouse alkaline phosphatase conjugated antibody (Merck) or anti-rabbit alkaline phosphatase conjugated antibody (Merck) diluted 1:1000 were used as secondary antibodies. The protein bands were visualized using SIGMAFAST BCIP®/NBT reagent (Merck).
Mass photometry (MP, iSCAMS)
Mass photometry experiments were performed with a Refeyn OneMP (Refeyn Ltd.). Data acquisition was performed using AcquireMP (Refeyn Ltd. 172 v2.3). Samples were evaluated with microscope coverslips (70 × 26 174 mm). The coverslips were washed with ddH2O and isopropanol. A silicone template was placed on top of the coverslip to form reaction chambers immediately prior to measurement. The instrument was calibrated using NativeMark Protein Standard (Thermo Fisher). 10 μL of fresh room temperature buffer was pipetted into a well, the focal position was identified and locked. For each acquisition 1 μL of the protein (at a concentration of 200 nM) was added to the well and thoroughly mixed. MP signals were recorded for 60 s to allow detection of at least 2 × 103 individual protein molecules. The data were analyzed using the Refeyn AcquireMP 2.3.0 software.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author contributions
AS: Investigation, Formal analysis, Conceptualization, Resources, Writin—Original Draft, Writing—Review and Editing, Visualization, Supervision, Project administration, Funding acquisition. FV: Investigation, Formal analysis; AP: Investigation, Formal analysis; OC: Resources, Data Curation.
Funding
This work was supported by NINDS R01-NS109366 to OC, and the authors acknowledge the support of COMPPÅ, funded by P41-GM107462, for the use of the Refeyn mass photometry instrument.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Balasuriya, D., Srivats, S., Murrell-Lagnado, R. D., and Edwardson, J. M. (2014). Atomic force microscopy (AFM) imaging suggests that stromal interaction molecule 1 (STIM1) binds to Orai1 with sixfold symmetry. FEBS Lett. 588, 2874–2880. doi:10.1016/j.febslet.2014.06.054
Bankston, J. R., Camp, S. S., DiMaio, F., Lewis, A. S., Chetkovich, D. M., and Zagotta, W. N. (2012). Structure and stoichiometry of an accessory subunit TRIP8b interaction with hyperpolarization-activated cyclic nucleotide-gated channels. Proc. Natl. Acad. Sci. U. S. A. 109, 7899–7904. doi:10.1073/pnas.1201997109
Bankston, J. R., DeBerg, H. A., Stoll, S., and Zagotta, W. N. (2017). Mechanism for the inhibition of the cAMP dependence of HCN ion channels by the auxiliary subunit TRIP8b. J. Biol. Chem. 292, 17794–17803. doi:10.1074/jbc.M117.800722
Chow, S. S., Van Petegem, F., and Accili, E. A. (2012). Energetics of cyclic AMP binding to HCN channel C terminus reveal negative cooperativity. J. Biol. Chem. 287, 600–606. doi:10.1074/jbc.M111.269563
Deberg, H. A., Bankston, J. R., Rosenbaum, J. C., Brzovic, P. S., Zagotta, W. N., and Stoll, S. (2015). Structural mechanism for the regulation of HCN ion channels by the accessory protein TRIP8b. Structure 23, 734–744. doi:10.1016/j.str.2015.02.007
George, M. S., Abbott, L. F., and Siegelbaum, S. A. (2009). HCN hyperpolarization-activated cation channels inhibit EPSPs by interactions with M-type K(+) channels. Nat. Neurosci. 12, 577–584. doi:10.1038/nn.2307
Goehring, A., Lee, C.-H., Wang, K. H., Michel, J. C., Claxton, D. P., Baconguis, I., et al. (2014). Screening and large-scale expression of membrane proteins in mammalian cells for structural studies. Nat. Protoc. 9, 2574–2585. doi:10.1038/nprot.2014.173
Gross, C., Saponaro, A., Santoro, B., Moroni, A., Thiel, G., and Hamacher, K. (2018). Mechanical transduction of cytoplasmic-to-transmembrane-domain movements in a hyperpolarization-activated cyclic nucleotide-gated cation channel. J. Biol. Chem. 293, 12908–12918. doi:10.1074/jbc.RA118.002139
Hou, X., Pedi, L., Diver, M. M., and Long, S. B. (2012). Crystal structure of the calcium release-activated calcium channel Orai. Science 338, 1308–1313. doi:10.1126/science.1228757
Hu, L., Santoro, B., Saponaro, A., Liu, H., Moroni, A., and Siegelbaum, S. (2013). Binding of the auxiliary subunit TRIP8b to HCN channels shifts the mode of action of cAMP. J. Gen. Physiol. 142, 599–612. doi:10.1085/jgp.201311013
Ji, W., Xu, P., Li, Z., Lu, J., Liu, L., Zhan, Y., et al. (2008). Functional stoichiometry of the unitary calcium-release-activated calcium channel. Proc. Natl. Acad. Sci. U. S. A. 105, 13668–13673. doi:10.1073/pnas.0806499105
Kusch, J., Biskup, C., Thon, S., Schulz, E., Nache, V., Zimmer, T., et al. (2010). Interdependence of receptor activation and ligand binding in hcn2 pacemaker channels. Neuron 67, 75–85. doi:10.1016/j.neuron.2010.05.022
Kusch, J., Thon, S., Schulz, E., Biskup, C., Nache, V., Zimmer, T., et al. (2012). How subunits cooperate in cAMP-induced activation of homotetrameric HCN2 channels. Nat. Chem. Biol. 8, 162–169. doi:10.1038/nchembio.747
Lee, C.-H., and MacKinnon, R. (2017). Structures of the human HCN1 hyperpolarization-activated channel. Cell. 168, 111–120. e11. doi:10.1016/j.cell.2016.12.023
Lolicato, M., Nardini, M., Gazzarrini, S., Moller, S., Bertinetti, D., Herberg, F. W., et al. (2011). Tetramerization dynamics of C-terminal domain underlies isoform-specific cAMP gating in hyperpolarization-activated cyclic nucleotide-gated channels. J. Biol. Chem. 286, 44811–44820. doi:10.1074/jbc.M111.297606
Magee, J. C. (1999). Dendritic Ih normalizes temporal summation in hippocampal CA1 neurons. Nat. Neurosci. 2, 848. doi:10.1038/12229
Marcelin, B., Liu, Z., Chen, Y., Lewis, A. S., Becker, A., McClelland, S., et al. (2012). Dorsoventral differences in intrinsic properties in developing CA1 pyramidal cells. J. Neurosci. 32, 3736–3747. doi:10.1523/JNEUROSCI.5870-11.2012
Porro, A., Binda, A., Pisoni, M., Donadoni, C., Rivolta, I., and Saponaro, A. (2020). Rational design of a mutation to investigate the role of the brain protein TRIP8b in limiting the cAMP response of HCN channels in neurons. J. Gen. Physiol. 152, e202012596. doi:10.1085/jgp.202012596
Porro, A., Saponaro, A., Gasparri, F., Bauer, D., Gross, C., Pisoni, M., et al. (2019). The HCN domain couples voltage gating and cAMP response in hyperpolarization-activated cyclic nucleotide-gated channels. Elife 8, e49672. doi:10.7554/eLife.49672
Robinson, R. B., and Siegelbaum, S. A. (2003). Hyperpolarization-activated cation currents: From molecules to physiological function. Annu. Rev. Physiol. 65, 453–480. doi:10.1146/annurev.physiol.65.092101.142734
Santoro, B., Hu, L., Liu, H., Saponaro, A., Pian, P., Piskorowski, R. A., et al. (2011). TRIP8b regulates HCN1 channel trafficking and gating through two distinct C-terminal interaction sites. J. Neurosci. 31, 4074–4086. doi:10.1523/JNEUROSCI.5707-10.2011
Santoro, B., Piskorowski, R. A., Pian, P., Hu, L., Liu, H., and Siegelbaum, S. A. (2009). TRIP8b splice variants form a family of auxiliary subunits that regulate gating and trafficking of HCN channels in the brain. Neuron 62, 802–813. doi:10.1016/j.neuron.2009.05.009
Santoro, B., Wainger, B. J., and Siegelbaum, S. A. (2004). Regulation of HCN channel surface expression by a novel C-terminal protein-protein interaction. J. Neurosci. 24, 10750–10762. doi:10.1523/JNEUROSCI.3300-04.2004
Saponaro, A., Bauer, D., Giese, M. H., Swuec, P., Porro, A., Gasparri, F., et al. (2021a). Gating movements and ion permeation in HCN4 pacemaker channels. Mol. Cell. 81, 2929–2943. e6. doi:10.1016/j.molcel.2021.05.033
Saponaro, A., Cantini, F., Porro, A., Bucchi, A., DiFrancesco, D., Maione, V., et al. (2018). A synthetic peptide that prevents cAMP regulation in mammalian hyperpolarization-activated cyclic nucleotide-gated (HCN) channels. Elife 7, e35753. doi:10.7554/eLife.35753
Saponaro, A., Pauleta, S. R., Cantini, F., Matzapetakis, M., Hammann, C., Donadoni, C., et al. (2014). Structural basis for the mutual antagonism of cAMP and TRIP8b in regulating HCN channel function. Proc. Natl. Acad. Sci. U. S. A. 111, 14577–14582. doi:10.1073/pnas.1410389111
Saponaro, A., Sharifzadeh, A. S., and Moroni, A. (2021b). Detection of ligand binding to purified HCN channels using fluorescence-based size exclusion chromatography. Methods Enzymol. 652, 105–123. doi:10.1016/bs.mie.2021.01.043
Thon, S., Schulz, E., Kusch, J., and Benndorf, K. (2015). Conformational flip of nonactivated HCN2 channel subunits evoked by cyclic nucleotides. Biophys. J. 109, 2268–2276. doi:10.1016/j.bpj.2015.08.054
Tsay, D., Dudman, J. T., and Siegelbaum, S. A. (2007). HCN1 channels constrain synaptically evoked Ca2+ spikes in distal dendrites of CA1 pyramidal neurons. Neuron 56, 1076–1089. doi:10.1016/j.neuron.2007.11.015
Ulens, C., and Siegelbaum, S. A. (2003). Regulation of hyperpolarization-activated HCN channels by cAMP through a gating switch in binding domain symmetry. Neuron 40, 959–970. doi:10.1016/s0896-6273(03)00753-0
Williams, S. R., and Stuart, G. J. (2000). Site independence of EPSP time course is mediated by dendritic I(h) in neocortical pyramidal neurons. J. Neurophysiol. 83, 3177–3182. doi:10.1152/jn.2000.83.5.3177
Wu, S., Vysotskaya, Z. V., Xu, X., Xie, C., Liu, Q., and Zhou, L. (2011). State-dependent cAMP binding to functioning HCN channels studied by patch-clamp fluorometry. Biophys. J. 100, 1226–1232. doi:10.1016/j.bpj.2011.01.034
Yen, M., Lokteva, L. A., and Lewis, R. S. (2016). Functional analysis of Orai1 concatemers supports a hexameric stoichiometry for the CRAC channel. Biophys. J. 111, 1897–1907. doi:10.1016/j.bpj.2016.09.020
Keywords: HCN channels, Ih current, TRIP8b, cAMP, mass photometry, stoichiometry
Citation: Saponaro A, Vallese F, Porro A and Clarke OB (2022) Validation of the binding stoichiometry between HCN channels and their neuronal regulator TRIP8b by single molecule measurements. Front. Physiol. 13:998176. doi: 10.3389/fphys.2022.998176
Received: 19 July 2022; Accepted: 05 September 2022;
Published: 26 September 2022.
Edited by:
Padmaja Prasad Mishra, Saha Institute of Nuclear Physics (SINP), IndiaReviewed by:
Tapas Paul, Johns Hopkins University, United StatesElisabetta Cerbai, University of Florence, Italy
Copyright © 2022 Saponaro, Vallese, Porro and Clarke. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Andrea Saponaro, andrea.saponaro@unimi.it
 Andrea Saponaro
Andrea Saponaro Francesca Vallese2,3,4
Francesca Vallese2,3,4