- 1Department of Biology, Institute for Molecular Evolution, Heinrich-Heine-Universität Düsseldorf, Düsseldorf, Germany
- 2Department of Life Sciences, Ben-Gurion University of the Negev and the National Institute for Biotechnology in the Negev, Marcus Family Campus, Be’er-Sheva, Israel
Oxygen sensing mechanisms are essential for metazoans, their origin and evolution in the context of oxygen in Earth history are of interest. To trace the evolution of a main oxygen sensing mechanism among metazoans, the hypoxia induced factor, HIF, we investigated the phylogenetic distribution and phylogeny of 11 of its components across 566 eukaryote genomes. The HIF based oxygen sensing machinery in eukaryotes can be traced as far back as 800 million years (Ma) ago, likely to the last metazoan common ancestor (LMCA), and arose at a time when the atmospheric oxygen content corresponded roughly to the Pasteur point, or roughly 1% of present atmospheric level (PAL). By the time of the Cambrian explosion (541–485 Ma) as oxygen levels started to approach those of the modern atmosphere, the HIF system with its key components HIF1α, HIF1β, PHD1, PHD4, FIH and VHL was well established across metazoan lineages. HIF1α is more widely distributed and therefore may have evolved earlier than HIF2α and HIF3α, and HIF1β and is more widely distributed than HIF2β in invertebrates. PHD1, PHD4, FIH, and VHL appear in all 13 metazoan phyla. The O2 consuming enzymes of the pathway, PHDs and FIH, have a lower substrate affinity, Km, for O2 than terminal oxidases in the mitochondrial respiratory chain, in line with their function as an environmental signal to switch to anaerobic energy metabolic pathways. The ancient HIF system has been conserved and widespread during the period when metazoans evolved and diversified together with O2 during Earth history.
Introduction
Oxygen on Earth stems from cyanobacterial photosynthesis. During Earth history, there were two main phases of atmospheric oxygen content change (Mills et al., 2022). The first was the great oxidation event, GOE, around 2.4 billion years (Ga) ago (Bekker et al., 2004), followed by almost 2 Ga of low oxygen levels and a second, late rise of oxygen corresponding to the origin of land plants and animals about 500 million years (Ma) ago (Lyons et al., 2014). The metazoan lineage arose roughly 700–1000 Ma ago, long before the origin of land plants, during a phase of Earth history when oxygen levels were much lower than today’s. The first metazoans were thus well adapted to low oxygen environments from the outset of their evolution, but adapted to rising oxygen levels during evolution, particularly with the transition to life on land in a highly oxic atmosphere starting about 450 Ma ago (Martin et al., 2021). Oxygen sensing pathways in animals are integral to their evolution during changing oxygen concentrations (Hammarlund et al., 2020) in Earth history. The main oxygen sensing pathway in animals is mediated by hypoxia-inducible factors, HIFs (Semenza 2001; Kaelin and Ratcliffe 2008; Zhang et al., 2014; Ivan and Kaelin 2017). The alpha subunit of HIF1, HIF1α is posttranslational modified by prolyl hydroxylases (PHD1—4), which catalyze the O2 dependent hydroxylation of prolyl residues, inducing ubiquitinylation of HIF by the von Hippel Lindau tumor suppressor (VHL) (Brihimi-Horn and Pouysségur 2009), leading to HIF1α degradation by proteasomes (Figure 1). The factor inhibiting HIF, FIH, hydroxylates asparaginyl residues in the alpha subunit of HIFs (HIF1—3α), in an O2 dependent reaction, which inhibits proline hydroxylation (Figure 1). HIF1α and PHDs are constitutively expressed. When O2 is lacking, the alpha subunit of HIFs is not hydroxylated by PHDs and therefore not degraded, but accumulates and is transported into the cell nucleus to create a heterodimer with the beta subunit of HIFs (HIF1β and 2β). The HIFαβ-heterodimer acts as the transcription factor, which binds to corresponding hypoxia response elements (HRE) of the promoters of the HIF-target genes to stimulate or repress the downstream gene transcription activities which can elicit a series of biological responses (Brihimi-Horn and Pouysségur 2009) (Figure 1). As it relates to energy metabolism, the main biological response governed by the HIF pathway is a shift from aerobic energy metabolism to anaerobic energy metabolism, which in land mammals, diverts pyruvate flux away from mitochondria and O2 dependent terminal oxidases towards cytosolic fermentations (Samanta and Semenza 2018).
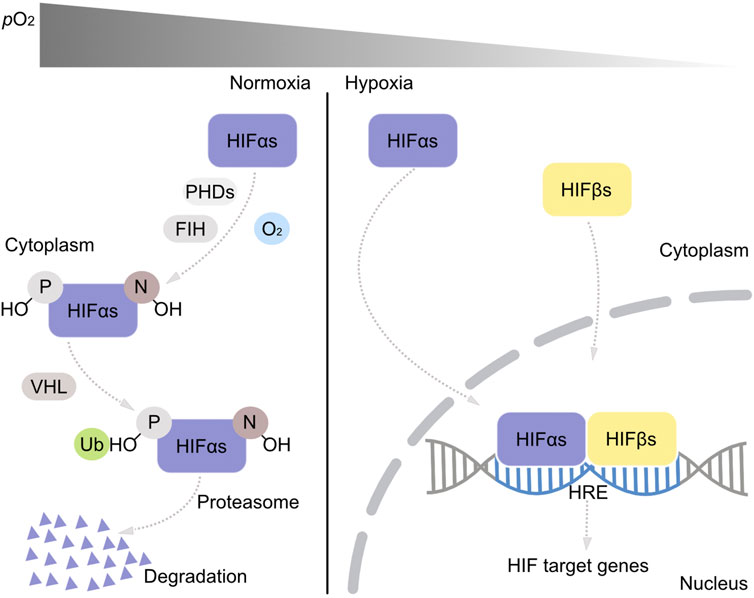
FIGURE 1. The HIF oxygen sensing pathway. Under normoxic conditions, proline (P) and asparaginyl (N) residues in HIFαs are first recognized and degraded by activated prolyl hydroxylases (PHDs) and asparaginyl hydroxylase (FIH). The hydroxylated proline residues (P-OH) bind on HIFαs and are ubiquitinated by the VHL and degraded by proteasomes within the cytoplasm. Under hypoxia, PHDs and FIH are inactivated and HIFαs are transported into the nucleus and form heterodimers with HIFβs. This heterodimer binds on the hypoxia responsive element (HRE) in the DNA structure and functions as a transcription factor activating or repressing the transcription activities of the downstream genes.
The mechanism of the main oxygen sensing pathway is well known and its origin and distribution have also been widely studied from the phylogenetic perspective (Rytkönen et al., 2011; Rytkönen and Storz 2011; Mills et al., 2018; Rytkönen 2018; Graham and Barreto 2019; Graham and Barreto 2020) but less so from the geological perspective (Taylor and McElwain 2010; Hammarlund 2020). One hypothesis is that the main oxygen sensing pathway may have already occurred in the common ancestor of metazoans (Hammarlund et al., 2018; Hammarlund 2020). PHD like prolyl hydroxylases have been reported in unicellular eukaryotes such as Dictyostelium (Van der Wel et al., 2005) and Schizosaccharomyces pombe (Lee et al., 2009) and pathogenic proteobacteria like Vibrio cholerae (Aravind and Koonin 2001; McDonough et al., 2006) and Pseudomonas aeruginosa (Aravind and Koonin 2001; Scotti et al., 2014). The HIFα/PHD/VHL pathway is conserved in all animals but is not found in choanoflagellates (Monosiga brevicollis) or other protists (Loenarz et al., 2011), it is present in all eumetazoans, except Ctenophera (Mills et al., 2018). A recent study showed that choanoflagellates have PHD but not HIF and VHL (Rytkönen 2018). HIF1α is conserved among most metazoans while HIF2α appeared later (Graham and Presnell 2017). Some studies have suggested that the HIF pathway was lacking in the last common ancestor of animals and is not ubiquitous across metazoans (Mills et al., 2018; Graham and Barreto 2020). From the geological perspective, the HIF system requires the presence of O2, with HIF1α, HIF1β, PHD2, and VHL being the oldest components, HIF2α and PHD3 were suggested to have appeared around 460 to 421 Ma ago when the atmospheric oxygen level was lower than the present, whereby PHD1 and HIF3α are thought to have arisen most recently (about 312 Ma ago) with contemporary atmospheric oxygen levels (Taylor and McElwain 2010). FIH1 is missing in fruit flies and nematodes but not in intermediate beetles (Taylor and McElwain 2010).
Here, we focus on the origin and distribution of the HIF/PHD/FIH/VHL oxygen sensing pathway within metazoans from the perspective of oxygen in Earth history, addressing the two phases of the appearance of its essential components: First, the presence of HIF/PHD/FIH/VHL pathway related genes—HIFαs, HIFβs, PHDs, FIH, and VHL—across metazoan and non-metazoan phyla, and the oxygen affinity of oxygen sensing enzymes (PHDs and FIH) were compared to terminal oxidases in mitochondria by the measure of substrate affinity for O2, Kmapp (O2).
Methods
Identification and presence of hypoxia induced factor related genes
From the National Center for Biotechnology Information (NCBI) (Agarwala et al., 2018), Kyoto Encyclopedia of Genes and Genomes (KEGG) (Kanehisa et al., 2016), European Molecular Biology Laboratory (EMBL) (Nightingale et al., 2017), and Universal Protein (UniProt) KB database (Bateman 2019), 422 eukaryotic HIF related sequences from 11 gene categories were downloaded in September 2020 (Supplementary Table S1). In July and October 2020, protein sequences from 566 complete eukaryotic genomes were downloaded from the NCBI Reference Sequence database (O’Leary et al., 2016) with assembly levels ranging from contig to complete genome (Supplementary Table S2). The 566 genomes were classified into 15 phyla according to NCBI taxonomy. The proteins of the 566 eukaryotic genomes were blasted against the eukaryotic HIF related sequence database using DIAMOND v2.0.1.139 (Buchfink et al., 2015), to identify potential HIF related genes. DIAMOND blastp was executed with the options--very-sensitive, --evalue 0.0000001 and--id 25 to define the sensitivity as well as the e-value (10−7) and identity (25%) thresholds for the homology search and to determine the best corresponding HIF related genes in each genome. Duplicated sequences for the same identified HIF related gene were removed by only taking one of the best hits.
Presence and absence patterns of the selected hits were used to demonstrate the evolution of HIF related genes within metazoans over the last 3.0 Ga of Earth’s history (Figure 2). The reference phylogenetic tree in Figure 2 was taken from Telford et al., 2015 and the divergence time of the last common ancestor of each representative phylum on the phylogenetic tree was estimated from the free public database (Kumar et al., 2017) used for the estimation of time-scaled phylogenies (Dos Reis et al., 2015; Gold et al., 2015; Delsuc et al., 2018; Tedersoo et al., 2018). The presence of the genes in each phylum is based on the respective gene being present in at least one species within the indicated phylum and represented by filled colored circles, if the gene was not present in the respective phylum, the space was left blank. The time of the earliest eukaryote fossil record (Eme et al., 2014) is displayed in the time scale at the bottom in light brown and the earliest animal fossil record (Hoyal Cuthill and Han 2018) is indicated by a dark brown bar in the time scale.
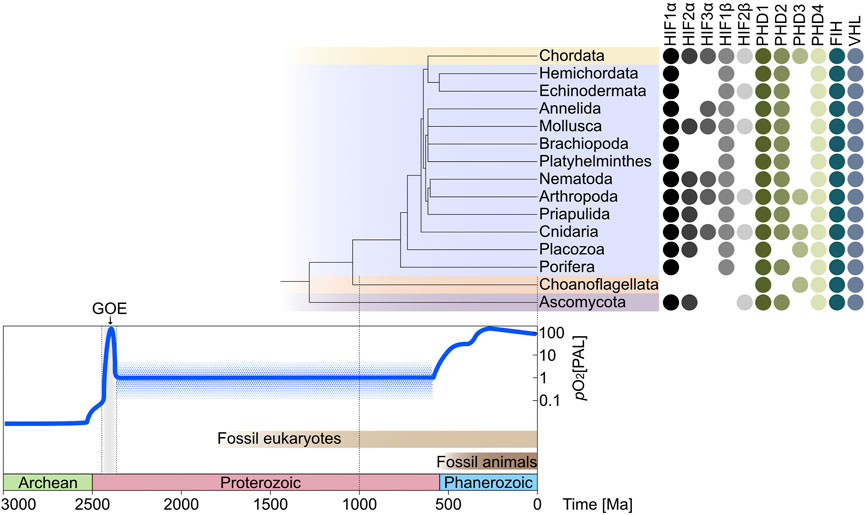
FIGURE 2. The evolution of the main oxygen sensing pathway across metazoans over the Earth’s oxygen history. The oxygen curve describes the Earth’s oxygen content during each geological time scale (Walker et al., 2018), the detailed information has been obtained from recent publications (Bekker et al., 2004; Lyons et al., 2014). More than 2500 million years (Ma) ago, the Earth oxygen content was almost at an anaerobic state (Catling and Zahnle 2020); during the Proterozoic period (2500–542 Ma), despite the great oxidation event (GOE) that happened at the beginning of this period, the oxygen content was still very low with around 1% present atmospheric levels (PAL) (Allen et al., 2019); during the Phanerozoic period (541–0 Ma), the atmospheric oxygen content increased constantly to the modern level (Lenton et al., 2016; Krause et al., 2018). The phylogenetic tree shown in the figure is scaled to the timeframe at the bottom of the figure. It is connected to the presence-absence pattern (PAP) of oxygen sensing genes. The PAP shows the distribution of the 11 gene categories across 13 metazoan phyla (Chordata, Hemichordata, Echinodermata, Annelida, Mollusca, Brachiopoda, Platyhelminthes, Nematoda, Arthropoda, Priapulida, Cnidaria, Placozoa, and Porifera) and two non-metazoan phyla (Choanoflagellata and Ascomycota) that include 566 species (Supplementary Table S2). The oxygen sensing regulators (HIF1α, HIF2α, HIF3α, HIF1β, and HIF2β) at the left of the PAP are the substrates of the oxygen sensors (PHD1, PHD2, PHD3, PHD4, and FIH) in oxygen-dependent reactions before the ubiquitination (Ub) of the tumor suppressor gene (VHL). (HIF—hypoxia-inducible factor; PHD—HIF prolyl hydroxylases; FIH—factor inhibiting HIF; VHL—von Hippel Lindau tumor suppressor).
Oxygen affinity of oxygen sensing enzymes and terminal oxidases
Kmapp (O2) values of oxygen sensing enzymes (PHD and FIH) and terminal oxidases were collected from literature (Supplementary Tables S3, S4). Km is the Michaelis constant, which represents the concentration of substrate needed to achieve half of the maximum reaction speed (Vmax) of the enzyme (Northrop 1998). Kmapp (O2) represents the concentration of oxygen required to achieve half of the Vmax of the examined enzyme; the lower the Km the higher the affinity of the enzyme for the substrate.
Results
The hypoxia induced factor oxygen sensing pathway traces to the last metazoan common ancestor
The two non-metazoan phyla in our dataset (Choanoflagellata and Ascomycota) possess the oxygen sensors PHD, FIH, and VHL. This result is based on the genome of Salpingoeca rosetta, as this species deals as a model organism of a group of single-celled eukaryotes that are considered to be the closest relatives of animals (Carr et al., 2008; Booth and King 2022) (Supplementary Table S2). Thus, S. rosetta was included to analyze the presence or absence of the HIF-pathway with its components in an organism that may give insight into oxygen sensing via hypoxia-inducible factors in simple premetazoan eukaryotes. It has been previously reported that choanoflagellates have PHD, but not HIF and VHL (Rytkönen 2018). Ascomycetes also possess HIF homologs, the result of Ascomycota is based on nine fungi genomes (Supplementary Table S2). Our results support those of Rytkönen et al. in that HIFα and HIFβ subunits are not present in S. rosetta, only homologs of PHD1/3/4 as well as FIH and VHL were found. Ascomycota are the largest phylum of fungi predating metazoans, within which Ofd1—the prolyl 4-hydroxylase-like 2-OG-Fe(II) dioxygenase—mediates the degradation and accumulation of the N-terminal transcription factor (Sre1N) (Hughes and Espenshade 2008). Porifera is the sister group of all other animals (Simion et al., 2017) and has been shown to contain HIF1α, HIF1β, PHD1, PHD2, PHD4, FIH, and VHL based on the sponge species Amphimedon queenslandica (Supplementary Table S2). Trichoplax adhaerens which belongs to the phylum Placozoa is discussed as the last common ancestor of all animals (Schierwater and DeSalle 2018) and was previously shown to possess the key components of the oxygen sensing mechanism—HIFα, PHD, and VHL (Loenarz et al., 2011). Our finding shows a more complete result as T. adhaerens not only contains HIFα and PHD homologs but also HIFβ, FIH, and VHL homologues. The main oxygen sensing pathway can thus be traced back to at least the last common ancestor of metazoans, which lived roughly 800 Ma ago, when the atmospheric oxygen content was around 1% PAL (Erwin et al., 2011) (Figure 2).
The hypoxia induced factor pathway is conserved and widely distributed across metazoans
The 13 metazoan phyla examined all possess HIF1α, HIF1β, PHD1, PHD4, FIH, and VHL. This shows that the core components of the main oxygen sensing pathway are relatively conserved and widespread across metazoans. Moreover, HIF1α is more widely distributed across all phyla and therefore probably evolved earlier than HIF2α and HIF3α (Figure 2). HIF1β is more widely distributed than HIF2β, whereas PHD1 and PHD4 may have arisen earlier than PHD2 and PHD3 as they are found in all phyla examined as shown in Figure 2. It was previously shown that HIF1α is more conserved among metazoans and probably arose before HIF2α (Graham and Presnell 2017), consistent with our findings. However, previous studies have shown that HIF homologs are ubiquitous across metazoans, with the exception of Porifera and Ctenophora, and HIF2α only appears in vertebrates (Loenarz et al., 2011; Rytkönen et al., 2011; Graham and Presnell 2017; Hammarlund et al., 2018; Rytkönen 2018). Accordingly, it has been suggested that the HIF/PHD/FIH/VHL oxygen sensing pathway might represent a recent lineage specific invention among recent animals (Hashimoto et al., 2016; Graham and Barreto 2019; Graham and Barreto 2020). The copepod Tigriopus californicus seems to have lost the HIF pathway but still tolerates nearly anoxic conditions for at least 24 h, the genes involved in cuticle reorganization and ion transport may act as the potential solution to low oxygen availability as a replacement to the HIF pathway (Graham and Barreto 2019). Ramazzottius varieornatus, one member of the most stress-tolerant tardigrade species, has lost HIF1α, PHD, and VHL selectively (Hashimoto et al., 2016). Three of the four orders of Copepoda were shown to have lost the use of the HIF pathway, but both barnacles and copepods have not fully lost VHL which suggests HIF-independent functions of VHL (Graham and Barreto 2020).
The oxygen affinity of terminal oxidases is far higher than that of oxygen sensing enzymes
The Kmapp (O2) of terminal oxidases were obtained from both prokaryotes and eukaryotes, the Kmapp (O2) values of oxygen sensing enzymes (PHDs and FIH) were obtained from the literature for eukaryotes (Supplementary Tables S3, S4). Figure 3 shows the distribution of the Kmapp (O2) values of terminal oxidases, PHDs and FIH, which are 0.0034–33 μM, 11.54–530 μM and 12–270 μM, respectively. Low Kmapp (O2) values indicate higher oxygen affinity, this shows that the oxygen affinity of terminal oxidases is far higher than that of PHD and FIH.
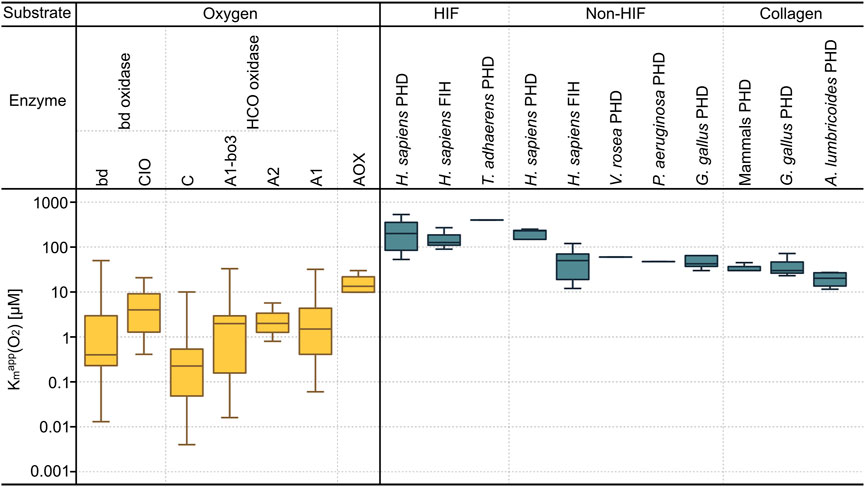
FIGURE 3. The Kmapp(O2) values of terminal oxidases and oxygen sensing enzymes. On the left (yellow) Kmapp (O2) values of the three main families of terminal oxidases are shown and indicated on the right (dark cyan) are Kmapp (O2) values of oxygen sensing enzymes (PHDs and FIH). For cytochrome bd oxidases, two subtypes (bd and CIO) are shown; for HCOs, four subtypes (C, A1-bo3, A2, and A1) are shown. On the top the substrates of the enzymes are indicated; oxygen as substrate for terminal oxidases, as their main function is to reduce oxygen within organism organelles. The Kmapp (O2) values of oxygen sensors (PHDs and FIH) are combined based on their substrates, which are sorted into three groups—HIF protein peptides, non-HIF protein peptides, and collagen. All underlying Kmapp (O2) values are detailed in Supplementary Tables S3, S4 (AOX: Alternative Oxidase, CIO: Cyanide Insensitive Oxidase, HCO: Heme Copper Oxidase).
Discussion
In previous studies, the characteristic domains of HIF, PHD, FIH, and VHL were used to identify HIF related homologs as the characteristic domains are usually the most conserved and representative part of one protein family. Here, complete HIF related protein sequences were used to find possible HIF pathway homologues from sequenced genomes. This identified HIF pathway homologues that were not detected in previous studies. We used Diamond (Buchfink et al., 2015) for searching, which is generally faster and often more accurate than the traditional Basic Local Alignment Search Tool (BLAST; Altschul et al., 1990), which helped to identify previously missing HIF pathway genes.
The HIF/PHD/FIH/VHL pathway was present during the Cambrian explosion
The largest fungal phylum Ascomycota, the metazoan’s closest sister lineage Choanoflagellata as well as the earliest invertebrate phyla Porifera and Placozoa originated in a low oxygen environment, when the oxygen level was roughly 1%–10% of the present atmospheric level. They are regarded as the low oxygen species group here. According to Figure 2, this group could have the evolved repression machinery - PHD, FIH, and VHL, followed by the first appearance of the HIF1α and HIF1β subunits and gene duplications leading to HIF2α, HIF3α, and HIF2β. The other invertebrate phyla (Cnidaria to Hemichordata in Figure 2) and the vertebrate phylum Chordata appeared before or after the Cambrian period, when the oxygen content increased to the around 10%–100% PAL. Thus, we classify these phyla as the high oxygen group. The HIF oxygen sensing mechanism may have been fully adapted across this group and is responsible for the adaptive transcriptional responses to hypoxia. In this view, the pathway originated in low oxygen environments and evolved vertically in different animal lineages as atmospheric oxygen content rose.
Genes for enzymes of the HIF oxygen sensing pathway—PHD1, PHD4, FIH, and VHL—are distributed across all examined phyla (Figure 2), tracing their first appearance to lineages during the course of metazoan and non-metazoan phylum diversification. HIF1α was the progenitor from which HIF2α and HIF3α arose, and HIF1β may have emerged before HIF2β as it is more widely distributed, giving rise to the hypoxia-inducible transcription factor (HIF1), which is composed of HIF1α and HIF1β, and is the major regulator of oxygen homeostasis (Wang et al., 1995; Semenza 2007). HIFα and HIFβ are only absent in the phylum of Choanoflagellata. As S. rosetta is capable of aerotaxis (movement in the direction of oxygen) this species might possess an oxygen sensing mechanism other than HIF because this process requires the ability to detect changes in oxygen levels (Kirkegaard et al., 2016). This alternative oxygen sensing mechanism might also play a role in hypoxia-related responses which obviate a need for retention of HIFα and -β subunits.
Purifying selection is a dominant factor underlying functional diversification and structural conservation among HIFα genes (Brihimi-Horn and Pouysségur 2009; Schödel et al., 2013; Tarade et al., 2019). HIF1α has a high affinity to VHL through the close contact between HIF1α Met561 and VHL Phe91. It is furthermore bound in closer proximity to promoters compared with HIF2α. HIF1β and HIF2β have the same domain architectures and can dimerize with HIF-α subunits, but HIF1β is more conserved across animal lineages than HIF2β. These evolutionary changes accompanied the strict retention of HIF1α and HIF1β across animal lineages.
PHD3 only appears in five of the 13 phyla analyzed but was present in choanoflagellates. Previous work reported that PHD3 may function as an ancient signaling protein due to its involvement in several cell signaling mechanisms (Place and Domann 2013), which enables large animals to sense and deliver oxygen for development and metabolism. Therefore, the retention of PHD3 in choanoflagellates might not reflect the oxygen sensing pathway but other functions instead.
It has been suggested that rudimentary HIF1α-dominant stemness control can function in such a way as to generate oxygen gradients within a tissue in the presence of 1% oxygen and thus maintain the inner tissue’s hypoxic cell stemness, with HIF2α-driven pseudohypoxia maintaining stemness of cells in nearby well-vascularized and oxygenated tissue with more than 1%–3% oxygen (Hammarlund et al., 2018). Under pseudohypoxic conditions, cells can activate hypoxia-related metabolic pathways even though sufficient oxygen supply is provided. Pseudohypoxia conditions can include the accumulation of hypoxia-inducible factor subunits and utilization of glycolysis under normal oxygen conditions in cancer cells (Masoud and Li 2015; Hayashi et al., 2019).
During early animal evolution, refined HIF stemness control might have played an essential role in promoting the larger size of animals and in energy metabolism. Before the Cambrian, low oxygen levels were sufficient to meet the physiological requirements of the simple invertebrates, but after the Cambrian, higher oxygen levels would have interfered with the stemness of animals, which would implicate a role of HIF in the evolution of animal size at the Cambrian explosion. At the same time, collagen hydroxylation at proline residues would have led to more rigid invertebrate body structures and better fossilization properties (Towe 1970).
The functional ranges of both HIF1α and HIF2α (5%–24% PAL or 1%–5% O2) hydroxylation (Holmquist-Mengelbier et al., 2006) suggest that they are a relic from the time of O2 sensing during low O2 concentrations in the Cambrian (15%–20% PAL or 3%–4% O2), marking a role for the HIF pathway during the adaptation of larger animals to the rising atmospheric oxygen concentrations. Under low oxygen concentrations of around 1%, both HIF1α and HIF2α are stabilized with HIF1α driving the hypoxia induced reactions which can create oxygen gradients in tissue and thus result in hypoxia and cell stemness within invertebrate tissue. HIF2α may have gained a role for creating the pseudohypoxic phenotype in order to facilitate the activation of genes related with promoting stemness within vertebrate tissue, while HIF2α maintained HIF2α-driven pseudohypoxia, promoting stemness even at oxygen levels of roughly 5% (Hammarlund et al., 2018). Clearly HIF dependent oxygen sensing was integral to the Cambrian explosion (541–485 Ma) and likely facilitated an increase in animal diversity (Knoll and Carroll 1999).
Low O2 affinity of hypoxia induced factor and high O2 affinity of terminal oxidases
The Kmapp (O2) values in Figure 3 show that the O2 affinity of O2 sensing enzymes is lower than that of terminal oxidases by one to two orders of magnitude. The O2 affinity of A1 type terminal oxidases in mitochondria is on the order of 10 μM, corresponding to 1% [v/v] O2 in air reflecting the environmental concentration of O2 at the time of mitochondrial origin (Zimorski et al., 2019) which is, in turn, very close to the O2 concentration in functioning mitochondria of human tissues because of the oxygen cascade from air to blood to capillaries (Martin et al., 2021). This affinity seems to have changed little during evolution. The situation is different with HIF however, as different animals respond to hypoxia at different levels. For example, Caenorhabditis elegans prefers O2 levels around 7%, and responds to hypoxia only at about 1% O2 (Branicky and Schafer 2008), reflecting diversification of oxygen sensing physiology during metazoan evolution (Kaelin and Ratcliffe 2008; Hampton-Smith and Peet 2009).
PHDs and FIH can target alternative substrates other than HIF, due to their involvement in both HIF and non-HIF related pathways. Their protein hydroxylation is neither unique nor ubiquitous. The substrates of PHDs and FIH can be sorted into three groups (Strowitzki et al., 2019) (Figure 3). Within the oxygen metabolizing enzymes, FIH has a relatively high oxygen affinity compared to the PHDs suggesting that FIH is more hypoxia-tolerant than the PHDs (Tarhonskaya et al., 2015). The Kmapp (O2) values of FIH are normally between 90 up to 270 μM (Koivunen et al., 2004; Hangasky et al., 2014; Wilson et al., 2020) while that of PHDs are 30–250 μM (Hirsilä et al., 2003). One study also reported an outlier Kmapp (O2) value of PHD around 1700 μM, which is not considered here (Dao et al., 2009). PHD2 was identified as the main regulator of the normal development of growth plate chondrocytes in the avascular environment, as it can inactivate HIF1α to avoid prolonged HIF1α-induced skeletal dysplasia (Stegen et al., 2019). Overexpressed HIF1α can reprogram cellular metabolism from respiration to fermentation (Stegen et al., 2019). Appropriate control of PHD2 on HIF1α activation is also important for collagen synthesis. For the comparison of the same oxygen sensing enzyme type from the same substrate group, it can be seen that larger sized vertebrates have higher oxygen affinity than smaller sized vertebrates and invertebrates on HIF-based substrates and collagen but not non-HIF substrates. This might reflect selection for a greater O2 demand for structural rigidity among larger vertebrates that are adapted to life above the soil line.
Within the terminal oxidases, the Kmapp (O2) values of other bacteria except for Alphaproteobacteria, archaea, and eukaryotes have also been added here. The mean Kmapp (O2) values of these seven of terminal oxidase subtypes, from high to low, are AOX (14.91 μM) > CIO (8.40 μM) > A1-bo3 (4.38 μM) > A1 (4.32 μM) > bd (4.18 μM) > A2 (2.83 μM) > C (1.62 μM) respectively (Degli Esposti et al., 2019). Cytochrome c oxidase was reported to exhibit a much smaller Kmapp (O2) than the mean physiological oxygen concentration compared to other oxygen utilizing enzymes in mammals, while the reaction rates of other oxygen-consuming enzymes could be limited by the physiological oxygen tensions commonly present in organs (Vanderkooi et al., 1991). The lower affinity for O2 of HIF dependent O2 sensing oxygenases relative to terminal oxidases suggests that the former have undergone evolutionary adaptations in response to changing O2 environments that reflect the physiological needs of the whole animal as opposed to its mitochondria, which are supplied via circulatory and respiratory systems with roughly the same concentration of O2 as the mitochondria of the first unicellular eukaryotes more than 1.5 billion years ago.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Materials, further inquiries can be directed to the corresponding author.
Author contributions
Conceptualization, NK, IM, and WFM; Data curation, BS and LDM; Formal analysis, BS and LDM; Funding acquisition, IM and WFM; Supervision, WFM; Visualization, BS and LDM; Writing—original draft, BS and NK; Writing—review and editing, BS, LDM, NK, IM, and WFM.
Funding
This work received financial support from the Deutsch-Israelische Projektkooperation (DIP) der Deutschen Forschungsgemeinschaft (MA 1426/23-1), from the Moore Simons Initiative on the Origin of the Eukaryotic Cell (grant 9743) and from the Volkswagen Foundation (grant 93 046). The stipend of Bing Song was funded by the China Scholarship Council (Bing Song’s application number 201706300100). IM was funded by the DIP (MI 2476/2-1) and ERC (866530). Funding was also provided by ISF (1947/19) to IM.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2022.977391/full#supplementary-material
References
Agarwala R., Barrett T., Beck J., Benson D. A., Bollin C., Bolton E., et al. (2018). Database resources of the national center for biotechnology information. Nucleic Acids Res. 46, 8–13.
Allen J. F., Thake B., Martin W. F. (2019). Nitrogenase inhibition limited oxygenation of Earth’s proterozoic atmosphere. Trends Plant Sci. 24, 1022–1031. doi:10.1016/j.tplants.2019.07.007
Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi:10.1016/S0022-2836(05)80360-2
Aravind L., Koonin E. V. (2001). The DNA-repair protein AlkB, EGL-9, and leprecan define new families of 2-oxoglutarate- and iron-dependent dioxygenases. Genome Biol. 2, RESEARCH0007–8. doi:10.1186/gb-2001-2-3-research0007
Bateman A. (2019). UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 47, D506–D515. doi:10.1093/nar/gky1049
Bekker A., Holland H. D., Wang P. L., Rumble D. I. I. I., Stein H. J., Hannah J. L., et al. (2004). Dating the rise of atmospheric oxygen. Nature 427, 117–120. doi:10.1038/nature02260
Booth D. S., King N. (2022). The history of Salpingoeca rosetta as a model for reconstructing animal origins. Curr. Top. Dev. Biol. 147, 73–91. doi:10.1016/bs.ctdb.2022.01.001
Brahimi-Horn M. C., Pouysségur J. (2009). HIF at a glance. J. Cell Sci. 122, 1055–1057. doi:10.1242/jcs.035022
Branicky R. S., Schafer W. R. (2008). Oxygen homeostasis: How the worm adapts to variable oxygen levels. Curr. Biol. 18, 559–560. doi:10.1016/j.cub.2008.05.023
Buchfink B., Xie C., Huson D. H. (2015). Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60. doi:10.1038/nmeth.3176
Carr M., Leadbeater B. S., Hassan R., Nelson M., Baldauf S. L. (2008). Molecular phylogeny of choanoflagellates, the sister group to Metazoa. Proc. Natl. Acad. Sci. U. S. A. 105, 16641–16646. doi:10.1073/pnas.0801667105
Catling D. C., Zahnle K. J. (2020). The archean atmosphere. Sci. Adv. 6, 1420. doi:10.1126/sciadv.aax1420
Dao J. H., Kurzeja R. J., Morachis J. M., Veith H., Lewis J., Yu V., et al. (2009). Kinetic characterization and identification of a novel inhibitor of hypoxia-inducible factor prolyl hydroxylase 2 using a time-resolved fluorescence resonance energy transfer-based assay technology. Anal. Biochem. 384, 213–223. doi:10.1016/j.ab.2008.09.052
Degli Esposti M., Mentel M., Martin W., Sousa F. L. (2019). Oxygen reductases in alphaproteobacterial genomes: Physiological evolution from low to high oxygen environments. Front. Microbiol. 10, 499. doi:10.3389/fmicb.2019.00499
Delsuc F., Philippe H., Tsagkogeorga G., Simion P., Tilak M. K., Turon X., et al. (2018). A phylogenomic framework and timescale for comparative studies of tunicates. BMC Biol. 16, 39–14. doi:10.1186/s12915-018-0499-2
Dos Reis M., Thawornwattana Y., Angelis K., Telford M. J., Donoghue P. C., Yang Z. (2015). Uncertainty in the timing of origin of animals and the limits of precision in molecular timescales. Curr. Biol. 25, 2939–2950. doi:10.1016/j.cub.2015.09.066
Eme L., Sharpe S. C., Brown M. W., Roger A. J. (2014). On the age of eukaryotes: Evaluating evidence from fossils and molecular clocks. Cold Spring Harb. Perspect. Biol. 6, a016139. doi:10.1101/cshperspect.a016139
Erwin D. H., Laflamme M., Tweedt S. M., Sperling E. A., Pisani D., Peterson K. J. (2011). The cambrian conundrum: Early divergence and later ecological success in the early history of animals. Science 334, 1091–1097. doi:10.1126/science.1206375
Gold D. A., Runnegar B., Gehling J. G., Jacobs D. K. (2015). Ancestral state reconstruction of ontogeny supports a bilaterian affinity for Dickinsonia. Evol. Dev. 17, 315–324. doi:10.1111/ede.12168
Graham A. M., Barreto F. S. (2020). Independent losses of the hypoxia-inducible factor (HIF) pathway within Crustacea. Mol. Biol. Evol. 37, 1342–1349. doi:10.1093/molbev/msaa008
Graham A. M., Barreto F. S. (2019). Loss of the HIF pathway in a widely distributed intertidal crustacean, the copepod Tigriopus californicus. Proc. Natl. Acad. Sci. U. S. A. 116, 12913–12918. doi:10.1073/pnas.1819874116
Graham A. M., Presnell J. S. (2017). Hypoxia Inducible Factor (HIF) transcription factor family expansion, diversification, divergence and selection in eukaryotes. PLoS One 12, e0179545. doi:10.1371/journal.pone.0179545
Hammarlund E. U., Flashman E., Mohlin S., Licausi F. (2020). Oxygen-sensing mechanisms across eukaryotic kingdoms and their roles in complex multicellularity. Science 370, eaba3512. doi:10.1126/science.aba3512
Hammarlund E. U. (2020). Harnessing hypoxia as an evolutionary driver of complex multicellularity. Interface Focus 10, 20190101. doi:10.1098/rsfs.2019.0101
Hammarlund E. U., Von Stedingk K., Påhlman S. (2018). Refined control of cell stemness allowed animal evolution in the oxic realm. Nat. Ecol. Evol. 2, 220–228. doi:10.1038/s41559-017-0410-5
Hampton-Smith R. J., Peet D. J. (2009). From polyps to people: A highly familiar response to hypoxia. Ann. N. Y. Acad. Sci. 1177, 19–29. doi:10.1111/j.1749-6632.2009.05035.x
Hangasky J. A., Gandhi H., Valliere M. A., Ostrom N. E., Knapp M. J. (2014). The rate-limiting step of O2 activation in the α-ketoglutarate oxygenase factor inhibiting hypoxia inducible factor. Biochemistry 53, 8077–8084. doi:10.1021/bi501246v
Hashimoto T., Horikawa D. D., Saito Y., Kuwahara H., Kozuka-Hata H., Shin T., et al. (2016). Extremotolerant tardigrade genome and improved radiotolerance of human cultured cells by tardigrade-unique protein. Nat. Commun. 7, 12808–12814. doi:10.1038/ncomms12808
Hayashi Y., Yokota A., Harada H., Huang G. (2019). Hypoxia/pseudohypoxia-mediated activation of hypoxia-inducible factor-1α in cancer. Cancer Sci. 110 (5), 1510–1517. doi:10.1111/cas.13990
Hirsilä M., Koivunen P., Günzler V., Kivirikko K. I., Myllyharju J. (2003). Characterization of the human prolyl 4-hydroxylases that modify the hypoxia-inducible factor. J. Biol. Chem. 278, 30772–30780. doi:10.1074/jbc.M304982200
Holmquist-Mengelbier L., Fredlund E., Löfstedt T., Noguera R., Navarro S., Nilsson H., et al. (2006). Recruitment of HIF-1alpha and HIF-2alpha to common target genes is differentially regulated in neuroblastoma: HIF-2alpha promotes an aggressive phenotype. Cancer Cell 10, 413–423. doi:10.1016/j.ccr.2006.08.026
Hoyal Cuthill J. F., Han J. (2018). Cambrian petalonamid Stromatoveris phylogenetically links Ediacaran biota to later animals. Palaeontology 61, 813–823. doi:10.1111/pala.12393
Hughes B. T., Espenshade P. J. (2008). Oxygen-regulated degradation of fission yeast SREBP by Ofd1, a prolyl hydroxylase family member. EMBO J. 27, 1491–1501. doi:10.1038/emboj.2008.83
Ivan M., Kaelin W. G. (2017). The EGLN-HIF O2-sensing system: Multiple inputs and feedbacks. Mol. Cell 66, 772–779. doi:10.1016/j.molcel.2017.06.002
Kaelin W. G., Ratcliffe P. J. (2008). Oxygen sensing by metazoans: The central role of the HIF hydroxylase pathway. Mol. Cell 30, 393–402. doi:10.1016/j.molcel.2008.04.009
Kanehisa M., Sato Y., Kawashima M., Furumichi M., Tanabe M. (2016). KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 44, 457–462. doi:10.1093/nar/gkv1070
Kirkegaard J. B., Bouillant A., Marron A. O., Leptos K. C., Goldstein R. E. (2016). Aerotaxis in the closest relatives of animals. eLife 5, e18109. doi:10.7554/eLife.18109
Knoll A. H., Carroll S. B. (1999). Early animal evolution: Emerging views from comparative biology and geology. Science 284, 2129–2137. doi:10.1126/science.284.5423.2129
Koivunen P., Hirsilä M., Günzler V., Kivirikko K. I., Myllyharju J. (2004). Catalytic properties of the asparaginyl hydroxylase (FIH) in the oxygen sensing pathway are distinct from those of its prolyl 4-hydroxylases. J. Biol. Chem. 279, 9899–9904. doi:10.1074/jbc.M312254200
Krause A. J., Mills B. J., Zhang S., Planavsky N. J., Lenton T. M., Poulton S. W. (2018). Stepwise oxygenation of the Paleozoic atmosphere. Nat. Commun. 9, 4081. doi:10.1038/s41467-018-06383-y
Kumar S., Stecher G., Suleski M., Hedges S. B. (2017). TimeTree: A resource for timelines, timetrees, and divergence times. Mol. Biol. Evol. 34, 1812–1819. doi:10.1093/molbev/msx116
Lee C. Y. S., Stewart E. V., Hughes B. T., Espenshade P. J. (2009). Oxygen-dependent binding of Nro1 to the prolyl hydroxylase Ofd1 regulates SREBP degradation in yeast. EMBO J. 28, 135–143. doi:10.1038/emboj.2008.271
Lenton T. M., Dahl T. W., Daines S. J., Mills B. J., Ozaki K., Saltzman M. R., et al. (2016). Earliest land plants created modern levels of atmospheric oxygen. Proc. Natl. Acad. Sci. U. S. A. 113, 9704–9709. doi:10.1073/pnas.1604787113
Loenarz C., Coleman M. L., Boleininger A., Schierwater B., Holland P. W., Ratcliffe P. J., et al. (2011). The hypoxia-inducible transcription factor pathway regulates oxygen sensing in the simplest animal, Trichoplax adhaerens. EMBO Rep. 12, 63–70. doi:10.1038/embor.2010.170
Lyons T. W., Reinhard C. T., Planavsky N. J. (2014). The rise of oxygen in Earth’s early ocean and atmosphere. Nature 506, 307–315. doi:10.1038/nature13068
Martin W. F., Tielens A. G., Mentel M. (2021). Mitochondria and anaerobic energy metabolism in eukaryotes. Berlin: De Gruyter.
Masoud G. N., Li W. (2015). HIF-1α pathway: Role, regulation and intervention for cancer therapy. Acta Pharm. Sin. B 5, 378–389. doi:10.1016/j.apsb.2015.05.007
McDonough M. A., Li V., Flashman E., Chowdhury R., Mohr C., Liénard B. M., et al. (2006). Cellular oxygen sensing: Crystal structure of hypoxia-inducible factor prolyl hydroxylase (PHD2). Proc. Natl. Acad. Sci. U. S. A. 103, 9814–9819. doi:10.1073/pnas.0601283103
Mills D. B., Boyle R. A., Dainesm S. J., Sperling E. A., Pisani D., Donoghue P. C. J., et al. (2022). Eukaryogenesis and oxygen in Earth history. Nat. Ecol. Evol. 6, 520–532. doi:10.1038/s41559-022-01733-y
Mills D. B., Francis W. R., Vargas S., Larsen M., Elemans C. P., Canfield D. E., et al. (2018). The last common ancestor of animals lacked the HIF pathway and respired in low-oxygen environments. Elife 7, e31176. doi:10.7554/eLife.31176
Nightingale A., Antunes R., Alpi E., Bursteinas B., Gonzales L., Liu W., et al. (2017). The proteins API: Accessing key integrated protein and genome information. Nucleic Acids Res. 45, W539–W544. doi:10.1093/nar/gkx237
Northrop D. B. (1998). On the meaning of Km and V/K in enzyme kinetics. J. Chem. Educ. 75, 1153. doi:10.1021/ed075p1153
O'Leary N. A., Wright M. W., Brister J. R., Ciufo S., Haddad D., McVeigh R., et al. (2016). Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 44, 733–745. doi:10.1093/nar/gkv1189
Place T. L., Domann F. E. (2013). Prolyl-hydroxylase 3: Evolving roles for an ancient signaling protein. Hypoxia 1, 13–17. doi:10.2147/HP.S50091
Rytkönen K. T. (2018). Evolution: Oxygen and early animals. Elife 7, e34756. doi:10.7554/eLife.34756
Rytkönen K. T., Storz J. F. (2011). Evolutionary origins of oxygen sensing in animals. EMBO Rep. 12, 3–4. doi:10.1038/embor.2010.192
Rytkönen K. T., Williams T. A., Renshaw G. M., Primmer C. R., Nikinmaa M. (2011). Molecular evolution of the metazoan PHD-HIF oxygen-sensing system. Mol. Biol. Evol. 28, 1913–1926. doi:10.1093/molbev/msr012
Samanta D., Semenza G. L. (2018). Metabolic adaptation of cancer and immune cells mediated by hypoxia-inducible factors. Biochim. Biophys. Acta. Rev. Cancer 1870, 15–22. doi:10.1016/j.bbcan.2018.07.002
Schödel J., Mole D. R., Ratcliffe P. J. (2013). Pan-genomic binding of hypoxia-inducible transcription factors. Biol. Chem. 394, 507–517. doi:10.1515/hsz-2012-0351
Scotti J. S., Leung I. K., Ge W., Bentley M. A., Paps J., Kramer H. B., et al. (2014). Human oxygen sensing may have origins in prokaryotic elongation factor Tu prolyl-hydroxylation. Proc. Natl. Acad. Sci. U. S. A. 111, 13331–13336. doi:10.1073/pnas.1409916111
Semenza G. L. (2001). HIF-1, O2, and the 3 PHDs: How animal cells signal hypoxia to the nucleus. Cell 107, 1–3. doi:10.1016/s0092-8674(01)00518-9
Semenza G. L. (2007). Hypoxia-inducible factor 1 (HIF-1) pathway. Sci. STKE 407, 8. doi:10.1126/stke.4072007cm8
Simion P., Philippe H., Baurain D., Jager M., Richter D. J., Di Franco A., et al. (2017). A large and consistent phylogenomic dataset supports sponges as the sister group to all other animals. Curr. Biol. 27, 958–967. doi:10.1016/j.cub.2017.02.031
Stegen S., Laperre K., Eelen G., Rinaldi G., Fraisl P., Torrekens S., et al. (2019). HIF-1α metabolically controls collagen synthesis and modification in chondrocytes. Nature 565, 511–515. doi:10.1038/s41586-019-0874-3
Strowitzki M. J., Cummins E. P., Taylor C. T. (2019). Protein hydroxylation by hypoxia-inducible factor (HIF) hydroxylases: Unique or ubiquitous? Cells 8, 384. doi:10.3390/cells8050384
Tarade D., Lee J. E., Ohh M. (2019). Evolution of metazoan oxygen-sensing involved a conserved divergence of VHL affinity for HIF1α and HIF2α. Nat. Commun. 10, 3293. doi:10.1038/s41467-019-11149-1
Tarhonskaya H., Hardy A. P., Howe E. A., Loik N. D., Kramer H. B., McCullagh J. S., et al. (2015). Kinetic investigations of the role of factor inhibiting hypoxia-inducible factor (FIH) as an oxygen sensor. J. Biol. Chem. 290, 19726–19742. doi:10.1074/jbc.M115.653014
Taylor C. T., McElwain J. C. (2010). Ancient atmospheres and the evolution of oxygen sensing via the hypoxia-inducible factor in metazoans. Physiology 25, 272–279. doi:10.1152/physiol.00029.2010
Tedersoo L., Sánchez-Ramírez S., Koljalg U., Bahram M., Döring M., Schigel D., et al. (2018). High-level classification of the fungi and a tool for evolutionary ecological analyses. Fungal Divers. 90, 135–159. doi:10.1007/s13225-018-0401-0
Telford M. J., Budd G. E., Philippe H. (2015). Phylogenomic insights into animal evolution. Curr. Biol. 25, 876–887. doi:10.1016/j.cub.2015.07.060
Towe K. M. (1970). Oxygen-collagen priority and the early metazoan fossil record. Proc. Natl. Acad. Sci. U. S. A. 65, 781–788. doi:10.1073/pnas.65.4.781
Van der Wel H., Ercan A., West C. M. (2005). The Skp1 prolyl hydroxylase from Dictyostelium is related to the hypoxia-inducible factor-α class of animal prolyl 4-hydroxylases. J. Biol. Chem. 280, 14645–14655. doi:10.1074/jbc.M500600200
Vanderkooi J. M., Erecinska M., Silver I. A. (1991). Oxygen in mammalian tissue: Methods of measurement and affinities of various reactions. Am. J. Physiol. 260, 1131–1150. doi:10.1152/ajpcell.1991.260.6.C1131
Walker J. D., Geissman J. W., Bowring S. A., Babcock L. E. (2018). Geologic Time Scale v. 5.0: Geological Society of America. Available at: https://www.geosociety.org/documents/gsa/timescale/timescl.pdf.
Wang G. L., Jiang B. H., Rue E. A., Semenza G. L. (1995). Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc. Natl. Acad. Sci. U. S. A. 92, 5510–5514. doi:10.1073/pnas.92.12.5510
Wilson J. W., Shakir D., Batie M., Frost M., Rocha S. (2020). Oxygen-sensing mechanisms in cells. FEBS J. 287, 3888–3906. doi:10.1111/febs.15374
Zhang P., Yao Q., Lu L., Li Y., Chen P. J., Duan C. (2014). Hypoxia-inducible factor 3 is an oxygen-dependent transcription activator and regulates a distinct transcriptional response to hypoxia. Cell Rep. 6, 1110–1121. doi:10.1016/j.celrep.2014.02.011
Keywords: anaerobes, metazoan, origin, oxygen sensing mechanism, terminal oxidase
Citation: Song B, Modjewski LD, Kapust N, Mizrahi I and Martin WF (2022) The origin and distribution of the main oxygen sensing mechanism across metazoans. Front. Physiol. 13:977391. doi: 10.3389/fphys.2022.977391
Received: 24 June 2022; Accepted: 07 October 2022;
Published: 17 October 2022.
Edited by:
David Wright, University of Guelph, CanadaReviewed by:
Chandan Kumar Maurya, The Ohio State University, United StatesLei Wang, University of Miami, United States
Copyright © 2022 Song, Modjewski, Kapust, Mizrahi and Martin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: William F. Martin, YmlsbEBoaHUuZGU=
†These authors have contributed equally to this work
 Bing Song
Bing Song Luca David Modjewski
Luca David Modjewski Nils Kapust1
Nils Kapust1 Itzhak Mizrahi
Itzhak Mizrahi William F. Martin
William F. Martin