
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Physiol., 29 September 2022
Sec. Striated Muscle Physiology
Volume 13 - 2022 | https://doi.org/10.3389/fphys.2022.969854
This article is part of the Research TopicMechanical and Genetic signaling in Striated Muscle Development, Aging and DiseaseView all 9 articles
Skeletal muscle, comprising approximately 40% of body mass, is a highly complex and heterogeneous tissue serving a multitude of functions in the organism. Non-coding RNAs (ncRNAs) are known to participate in skeletal muscle development as critical regulators. However, the regulatory mechanisms of ncRNAs on chicken muscle traits are not well understood. In the present study, we collected the leg muscle from male embryos of Tibetan chicken at embryonic (E) 10 and E18 for RNA sequencing. A total of 6,583 differentially expressed mRNAs (DEMs) including 3,055 down-regulated and 3,528 up-regulated were identified in E18. We identified 695 differentially expressed lncRNAs (DELs) (187 down-regulated and 508 up-regulated) and 1,906 differentially expressed circRNAs (DECs) (1,224 down-regulated and 682 up-regulated) in E18. Among the 130 differentially expressed miRNAs (DEMIs), 59 were up-regulated and 71 were down-regulated in E18. Numerous DEMs and target genes for miRNAs/lncRNAs were significantly enriched in the muscle system process and cell cycle. We constructed a miRNA-gene-pathway network by considering target relationships between genes related to skeletal muscle development and miRNAs. A competing endogenous RNA (ceRNA) network was also constructed by integrating competing relationships between DEMs, DELs, and DECs. Several DELs and DECs were predicted to regulate the ADRA1B, ATP2A2, ATP2B1, CACNA1S, CACNB4, MYLK2, and ROCK2 genes. We discovered the crosstalk between the ncRNAs and their competing mRNAs, which provides insights into ceRNA function and mechanisms in the skeletal muscle development of chicken.
The meat products developed by skeletal muscle have been one of the most important animal products for human consumption in livestock production. Mammalian skeletal muscle occupies approximately 40% of total body weight, which is essential for vital functions such as breathing, movement, and thermogenesis (Bentzinger et al., 2012; Dumont et al., 2015). Several levels of intrinsic complexity during myogenesis arise from hierarchical interactions between transcriptional regulators and regulatory RNAs in addition to other extrinsic regulators. The differentiation and formation of skeletal muscle entail activation of muscle-specific transcription network governed by muscle-specific regulatory factors (MRFs) (Delfini et al., 2000; Buckingham and Rigby, 2014). The impairment of skeletal muscle myogenesis is related in various muscle dysfunctions, including sarcopenia and muscular dystrophy (Chang et al., 2016; Cruz-Jentoft and Sayer, 2019). There are many studies show that skeletal muscle development is also significantly affected by post-transcriptional regulation (Cesana et al., 2011; Yue et al., 2019). However, the underlying molecular mechanism remains poorly understood.
Non-coding RNAs (ncRNAs) which are RNA molecules have little protein-coding capacity act as regulators in multifarious biological processes instead of transcriptional noise (Huarte, 2009; Mchugh et al., 2015). Recently, accumulating evidence has revealed that the ncRNAs including microRNA (miRNA), long non-coding RNA (lncRNA), and circular RNA (circRNA) regulated mRNA expression by functioning as a competing endogenous RNA (ceRNA) at the post-transcriptional level (Salmena et al., 2011). As non-protein-coding transcripts, lncRNAs range from 0.2 to 100 kb and have abundant binding sites for miRNAs (Phelps et al., 2016). For example, A newly identified lncRNA MAR1 promoted skeletal muscle differentiation and regeneration by acting as a miR-487b sponge (Zhang et al., 2018). LncIRS1 regulated muscle atrophy through sponging miR-15 family to activate IGF1-PI3K/AKT pathway in chicken (Li et al., 2019b). CircRNAs are a new type of endogenous ncRNAs that form closed continuous loops without 3′- and 5′- ends (Greene et al., 2017; Zeng et al., 2017). In recent years, there were many studies on the effect of circRNAs on the regulation of skeletal muscle myogenesis. For example, CircARID1A regulated mouse skeletal muscle regeneration by functioning as a sponge of miR-6368 (Liu et al., 2021). CircRNAs such as circTMTC1 (Shen et al., 2019), circFAM188B (Yin et al., 2020), circHIPK3 (Chen et al., 2019), and circSVIL (Ouyang et al., 2018) showed vital roles in myogenesis by acting as ceRNA in chicken. Only a few lncRNAs and circRNAs have been well annotated up to now. Our study aimed to establish an accurate regulatory mechanism based on the ceRNA theory in the skeletal muscle development of chicken.
The transcriptome is the set of all RNA molecules, including mRNA, lncRNA, circRNA, and miRNA produced in one or a population of cells (Liu et al., 2014). Unlike the genome, the transcriptome reflects the gene expression under certain physiological conditions or developmental stages (Liu et al., 2018). RNA-seq is a highly sensitive method for whole transcriptome analysis. In this study, we investigated the expression level of miRNAs, lncRNAs, circRNAs, and mRNAs in the skeletal muscle of Tibetan chicken by RNA-seq. Subsequently, we constructed the lncRNA/circRNA-miRNA-mRNA regulatory networks to identify the crucial factors involved in the development of chicken skeletal muscle.
Fertilized eggs were obtained from Tibetan chickens (TC) were incubated under humid conditions at 37.5°C until they reached appropriate stages. On the day of harvest, the chicken embryos were sacrificed and the gender was determined according to the fetus anatomical characteristics. The leg muscle from six male embryos (three randomly selected embryos from each period) at embryonic (E) 10 and E18 were obtained, divided into three parts. Two parts for RNA-seq and real time quantitative PCR detection (RT-qPCR) were immediately frozen in liquid nitrogen and stored at −80°C, whereas the other part was fixed in 4% paraformaldehyde and embedded in paraffin for histological observation. All treatment procedures were approved by the Institutional Animal Care and Use Committee of the Southwest Minzu University.
The histological characteristics in the skeletal muscle were evaluated by haematoxylin and eosin (H&E) staining. Paraffin sections were mounted on slides for H&E staining. Histological characteristics of the chicken skeletal muscle were observed using a BA210 Digital microscope (Motic) and Images Advanced Software (Motic). Two independent samples (E10 sample and E18 sample) were used in this experiment, and each of the samples had five biology repeats.
Total RNA was isolated from each sample using TRIzol reagent (Invitrogen™, Carlsbad, CA, United States) according to the manufacturer’s instructions. Integrity, purity, and quality of the isolated RNA were measured by agarose gel electrophoresis, Nano-Drop ND-2000 spectrophotometer (NanoDrop Products, Wilmington, DE, United States), and Agilent 2,100 Bioanalyzer (Agilent Technologies, Massy, France). The RNA integrity number (RIN) value of samples larger than eight were used for further analysis.
Approximately 9 μg of RNA per sample was used in the protocol to deplete ribosomal RNA (rRNA) by the Ribo-zero™ rRNA Removal kit (Epicentre, Madison, WI, United States). The rRNA-depleted RNAs were used for preparing the libraries following the manufacturer’s recommendations in the NEBNext® Ultra™ Directional RNA Library Prep Kit for Illumina (NEB, United States). The library fragments were purified with AMPure XP system (Beckman Coulter, Beverly, United States), and library quality was assessed on the Agilent Bioanalyzer 2,100 system. Paired-end sequencing with 125-bp reads was performed on the Illumina HiSeq 2,500 platform. A total of 3 μg of RNA per sample was used for the small RNA library. The libraries were also sequenced on an Illumina HiSeq 2,500 platform and 50-bp single-end reads were generated.
Clean data (clean reads) were obtained by removing the poor-quality bases and adapter sequences from the raw data using FastQC (v0.19.5), and all the downstream analyses were based on the clean data with high quality. The clean reads were aligned to the chicken genome sequence assembly (Genome Reference Consortium Chicken Build 6a, GRCg6a) by HISAT2 (v2.1.0) (Kim et al., 2019) and Bowtie (v2.2.9) (Langmead and Salzberg, 2012). The mapped reads of each sample were assembled by Cufflinks (v2.2.1) (Trapnell et al., 2010).
The pipelines for lncRNAs identification were as follows: 1) transcripts that were merged or were similar to the known gallinaceous mRNAs and other small RNAs (rRNA, snRNA, snoRNAs, tRNAs, pre-miRNA, pseudogenes) were removed using Cuffcompare software (Ghosh and Chan, 2016). 2) the transcripts, annotated as “i (a transfrag falling entirely within a reference intron),” “u (unknown, intergenic transcript),” “x (exonic overlap with reference on the opposite strand)” or “o (generic exonic overlap with a reference transcript)” by the Cuffcompare software, were left for the next filter. 3) the remaining transcripts that contained a single exon and were shorter than 200 bp were removed. 4) The remaining transcripts were analyzed by the Coding Potential Calculator (CPC, v2.0) (Kang et al., 2017), Coding-Non-Coding-Index (CNCI, v2.0) (Sun et al., 2013), Pfam and Coding Potential Assessment Tool (CPAT, V2.0.0) (Wang et al., 2013). The transcripts that passed through all these stages were considered to be lncRNAs. Transcripts per million reads (TPM) was used to reflect the lncRNA expressional levels.
After the clean reads were aligned to the gallinaceous reference genome, the junctions of the unmapped reads were identified using a back-splice algorithm. Then, the Findcirc software was used to predict the circRNAs (Memczak et al., 2013). MMapped back splicing junction reads per million mapped reads (RPM) was used to reflect the expressional levels of circRNAs.
The DESeq2 (v1.10.1) software (Love et al., 2014) was used to detect differentially expressed mRNAs (DEMs), miRNAs (DEMIs), lncRNAs (DELs), and circRNAs (DECs) with Padjust < 0.05 and |log2FoldChange| ≥ 1.
Gene Ontology (GO) analyses were performed for DEMs and target genes using Goatools (v0.6.5) software. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses for DEMs and target genes were carried out with the DAVID tool. The results with Padjust < 0.05 were considered to be significantly enriched.
To determine the possible interactions between miRNA with lncRNA, circRNA, and mRNA, the miRNA-lncRNA, miRNA-circRNA, and miRNA-mRNA interactions were predicted with miRanda software (John et al., 2004). The miRanda software was used to analyze the relationships between DEMIs and DELs, DECs, DEMs which predicted DEMIs targets based on sequence complementarity, conserved target sites, and free energy of formation. Only alignments with no mismatch in the positions, 2-8 in the 5′end and energies lower than −20 kcal/mol were retained. The potential lncRNA/circRNA-miRNA-mRNA network was constructed and visualized using Cytoscape software (v3.6.0) based on the ceRNA theory (Salmena et al., 2011).
Three DELs, DECs, DEMs, and DEMIs from the ceRNA network were selected to validate the results of RNA-seq by RT-qPCR, respectively. First-strand cDNA was synthesized with random hexamers (for mRNA, lncRNA, and circRNA) and stem-loop RT primers (for miRNA) by the PrimeScript RT reagent Kit (TaKaRa, Dalian, China). The glyceraldehyde-3-phosphate dehydrogenase (GAPDH, for mRNA, lncRNA, and circRNA) and 5S rRNA (for miRNA) were used as endogenous controls for normalization. All primers used in RT-qPCR are shown in Supplementary Table S1. All reactions were carried out in the CFX96 qPCR system (Bio-Rad, United States) with triplicate reactions for each sample. The quantification of relative expression of lncRNAs, circRNAs, mRNAs, and miRNAs was performed using the 2−ΔΔCt method.
To preferably understand the different performance of skeletal muscle fibers between E10 and E18, we used the muscle section following H&E staining to observe and analyze the morphology of the muscle fibers. The developmental muscle fibers are unclear and incomplete at the E10 stage which is in the mixed phase of myoblast proliferation and differentiation (Figure 1A). At the E18 stage, the muscle fibers have developed completely and the number of muscle fibers has been determined (Figure 1B).
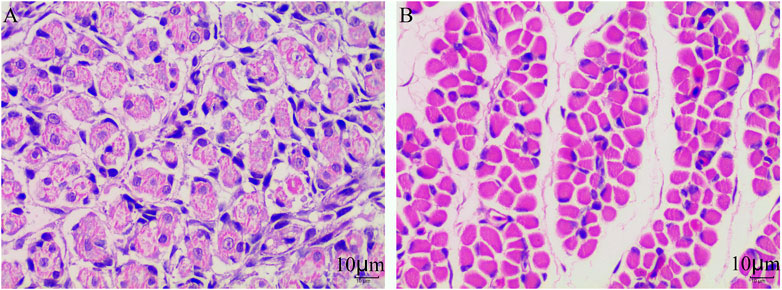
FIGURE 1. Muscle fiber characteristics of Tibetan chicken at E10 and E18. (A) H&E staining of the leg muscle cross-section from E10. (B) H&E staining of the leg muscle cross-section from E18.
To obtain a global view of the Tibetan chicken leg muscle transcriptome and identify the lncRNA, circRNA, and mRNA transcripts related to the two developmental stages, six libraries were constructed and then sequenced. A total of 710.03 M (million) raw reads were obtained and an average of 118.34 M of raw reads per library. 704.07 M (99.16%) of raw reads were filtered and preserved as clean reads for the following analyses, where an average of 117.35 M clean reads per library (Supplementary Tables S2, S3).
In all the sequencing libraries, a total of 27,457 protein-coding transcripts including 25,768 known and 1,689 novel protein-coding transcripts were identified. The transcripts abundances were quantified by TPM, the average expression level of 25,768 known protein-coding transcripts was 15.90 (Figure 2A). We detected 26,366 and 25,647 protein-coding transcripts in the whole transcriptome of E10 and E18 group, respectively. A total of 24,556 protein-coding transcripts were co-expressed in E10 and E18, while 1,810 and 1,091 protein-coding transcripts were specifically expressed in E10 and E18, respectively (Figure 2B).
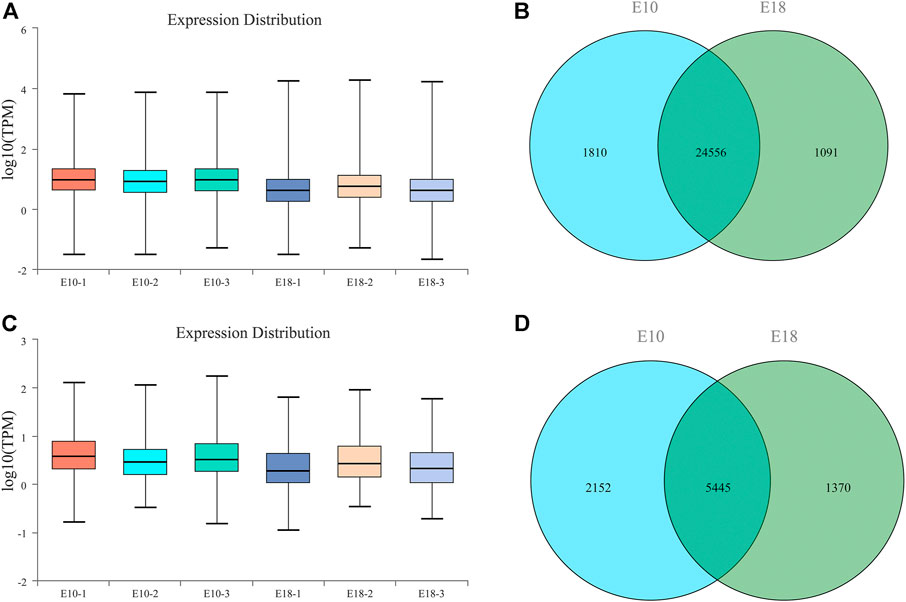
FIGURE 2. Overview of long RNA sequencing in the leg muscle of Tibetan chicken. (A) The boxplot of TPM (transcripts per million reads) for known protein-coding transcripts. (B) The Venn diagram of detected mRNAs identified in E10 and E18 group. (C) The boxplot of TPM (transcripts per million reads) for known lncRNA transcripts. (D) The Venn diagram of detected lncRNAs identified in E10 and E18 group.
There were 8,967 lncRNA transcripts including 7,883 known and 1,084 novel protein-coding transcripts in total. 7,597 lncRNA transcripts were detected in the whole transcriptome of E10, and 6,815 lncRNA transcripts in E18. The average expression level of known lncRNA transcripts (1.93) was lower than known protein-coding transcripts (15.90) (Figure 2C). A total of 5,445 lncRNA transcripts were co-expressed in E10 and E18, while 2,152 and 1,370 lncRNA transcripts were specifically expressed in E10 andE18, respectively (Figure 2D).
Overall, 6,583 DEMs including 3,055 down-regulated (46.41%) and 3,528 up-regulated (53.59%) were discovered between the two developmental stages (Figure 3A). The top 20 up-regulated and down-regulated mRNAs were summarized in Table 1. GO and KEGG pathway enrichment analysis were performed to identify biological functions of DEMs. GO analysis of the DEMs showed that respiratory electron transport chain, muscle system process, striated muscle contraction, generation of precursor metabolites and energy, ATP metabolic process, regulation of cellular component movement, and cell cycle process were the most abundant terms in the biological process category. In the cellular component category, respirasome, mitochondrial respiratory chain complex I, and NADH dehydrogenase complex were the top three terms, while NADH dehydrogenase activity, actin binding, cytoskeletal protein binding, and NADH dehydrogenase (quinone) activity were the most abundant terms in the molecular function category (Figure 3B; Supplementary Table S4). Pathway analysis indicated that the DEMs were significantly enriched in 28 KEGG pathways, in which several pathways were related to skeletal muscle development, such as cell cycle, DNA replication, MAPK signaling pathway, cGMP-PKG signaling pathway, and Arginine biosynthesis (Figure 3C; Supplementary Table S5).
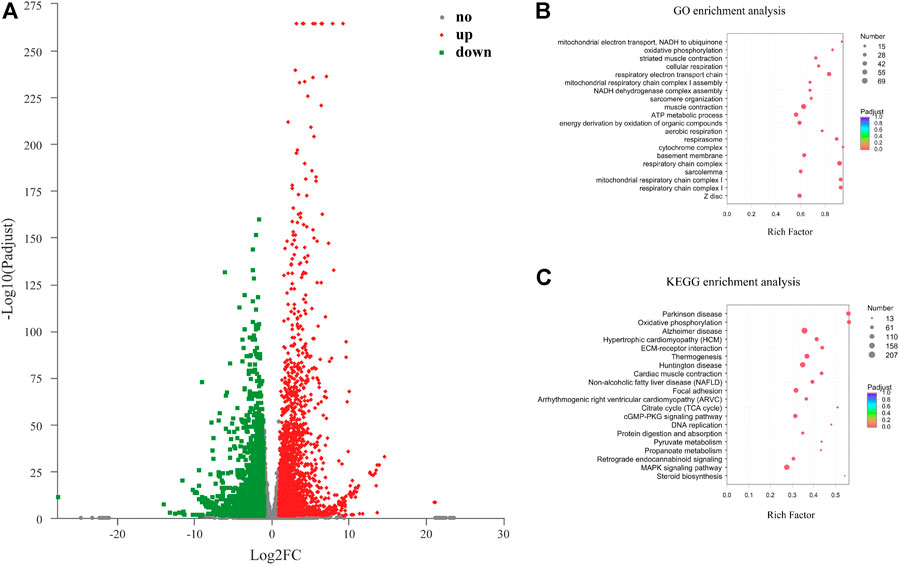
FIGURE 3. GO and KEGG pathway analysis of the differentially expressed mRNAs (DEMs) in the chicken leg muscle. (A) Volcano plot of 6,583 DEMs. (B) Top 20 significantly enriched GO terms for DEMs. (C) Top 20 significantly enriched pathways for DEMs.
There are 695 lncRNA transcripts that were differentially expressed between the two developmental stages, including 187 that were down-regulated (26.91%) and 508 that were up-regulated (73.09%) in E18 (Supplementary Figure S1A). The top 20 up-regulated and down-regulated lncRNAs were summarized in Table 2. We predicted the potential cis and trans target DEMs of lncRNAs for investigating the function of DELs. For the cis action of DELs, protein-coding genes 10 and 100 kb upstream and downstream of the DELs were searched, respectively. The results showed 695 DELs corresponding to 133 DEMs within a range of 10 kb, as well as 272 DEMs within a range of 100 kb (Supplementary Table S6). On the other hand, the trans role of 695 DELs in protein-coding genes was examined based on their expression correlation coefficient (Pearson correlation ≥0.95 or ≤ −0.95). A total of 25,290 interaction relationships were detected in the trans form between 695 DELs and 6,583 DEMs in the chicken genome (Supplementary Table S7).
Functional analysis showed that these target genes were significantly enriched in 504 GO terms (350 biological process terms, 91 molecular function terms, and 63 cellular component terms). Many GO terms were related to regulation of muscle system process, muscle fiber development, muscle cell development, muscle structure development, and muscle tissue development (Supplementary Figure S1B and Supplementary Table S8). In addition, the target genes were enriched in 23 pathways, some of which were associated with muscle development including DNA replication, ECM-receptor interaction, cell cycle, p53 signaling, glycine, serine, and threonine metabolism pathways (Supplementary Figure S1C and Supplementary Table S9). These results indicated that lncRNAs took part in several biological processes in the development of chicken muscle.
There were 1,906 DECs including 1,224 down-regulated (64.22%) and 682 up-regulated (35.78%) were discovered between the two developmental stages (Supplementary Figure S2A). The top 20 up-regulated and down-regulated circRNAs were summarized in Table 3. We performed GO and pathway enrichment analysis of host genes to identify biological functions of DECs. These host genes were mainly associated with GO terms including regulation of cell differentiation, developmental process, regulation of cell morphogenesis, and ncRNA-mediated regulation of translation (Supplementary Figure S2B and Supplementary Table S10). The KEGG results revealed that the enriched pathways involved Wnt signaling pathway and MAPK signaling pathway (Supplementary Figure S2C and Supplementary Table S11).
As shown in Supplementary Tables S11, S12, six libraries were constructed and sequenced, resulting in a total of 67.21 M raw reads, and an average of 11.20 M of raw reads was obtained per library. 63.46 M (94.43%) of raw reads were filtered and preserved as useful reads for the subsequent analyses, where an average of 10.58 M useful reads per library (Supplementary Tables S12, S13). After alignment with small RNAs in the reference genome, miRBase and Rfam, we identified 90.34% miRNAs, and other small RNAs (9.66%) including rRNA, tRNA, snRNA, snoRNA as well as repbase (Figure 4A). We identified a total of 1,143 miRNAs in six small RNA libraries, and the length of most miRNAs were 18–24 nucleotides (Figure 4B). Total 1,094 and 795 miRNAs were detected in E10 and E18, respectively. A total of 746 miRNAs were co-expressed in E10 and E18, while 348 and 49 miRNAs were specifically expressed in E10 and E18, respectively (Figure 4C). Twenty mature miRNAs with the highest expression comprised 71.88 and 91.02% of all miRNAs in E10 and E18 (Figure 4D), of which gga-miR-1a-3p, gga-miR-143-3p, gga-miR-199-3p, gga-miR-148a-3p, and gga-miR-21-5p accounting for 56% of the total distribution, were the most abundant miRNAs.
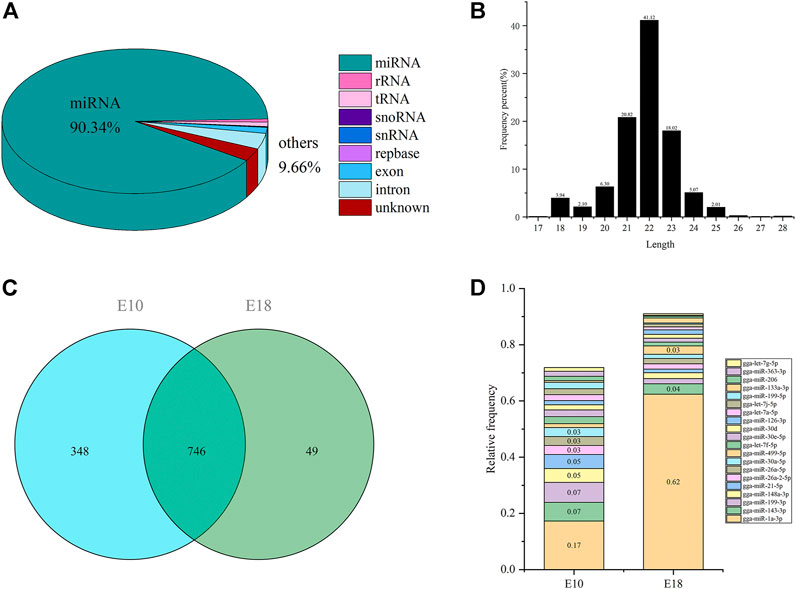
FIGURE 4. Overview of small RNA sequencing in the leg muscle of Tibetan chicken. (A) Portions of small RNA types in the useful reads. The percent of miRNA is 90.34%, and the other 9.66% included rRNA, tRNA, snRNA, snoRNA, repbase, exon, and intron. (B) Length distribution of all miRNAs. (C) The Venn diagram of detected miRNAs identified in E10 and E18 group. (D) The relative proportion of the top 20 miRNAs in the total amount of miRNAs.
There were 130 DEMIs including 59 that were up-regulated (45.38%) and 71 that were down-regulated (54.61%) between the two developmental stages (Supplementary Figure S3A). The top 20 up-regulated and down-regulated miRNAs were summarized in Table 4. A total of 3,834 target DEMs (Supplementary Table S14), 440 target DELs (Supplementary Table S15), and 1,581 target DECs (Supplementary Table S16) for DEMIs were identified. GO and pathway enrichment analysis were performed on target DEMs to identify biological functions of DEMIs. These genes were mainly associated with muscle system process, myofilament, striated muscle cell development, structural constituent of muscle, skeletal muscle thin filament assembly, muscle cell development, skeletal muscle myosin thick filament assembly, and muscle fiber development (Supplementary Figure S3B and Supplementary Table S17). The result of KEGG pathway analysis indicated the enriched pathways involved DNA replication, ECM-receptor interaction, valine, leucine and isoleucine degradation, and arginine biosynthesis (Supplementary Figure S3C and Supplementary Table S18).
Among the DEMs, 19 of them involved in muscle system process, ECM-receptor interaction, Focal adhesion, cGMP-PKG signaling pathway, MAPK signaling pathway, and cell cycle corresponded with 39 DEMIs (Table 5). Rho associated coiled-coil containing protein kinase 2 (ROCK2), calcium voltage-gated channel subunit alpha1 S (CACNA1S), filamin B (FLNB), myosin light chain kinase 2 (MYLK2), and ras homolog family member A (RHOA) genes participate in multiple pathways. To further understand and visualize the interactions and investigate the function of corresponding DEMIs, a miRNA-gene-pathway network was constructed (Figure 5) using the data from Table 5. The potential functions of a few miRNAs were identified by the interaction analysis in the skeletal muscle, for example, gga-miR-338-3p may play important roles in cell proliferation, gga-let-7b, gga-miR-1625-5p, gga-miR-1635, gga-miR-1677-3p, gga-miR-205b, gga-miR-214b-3p, gga-miR-338-3p, gga-miR-6549-5p, novel-1-4,798, and novel-26-28681were closely associated with multiple pathways including cGMP-PKG signaling pathway, focal adhesion and MAPK signaling pathway.
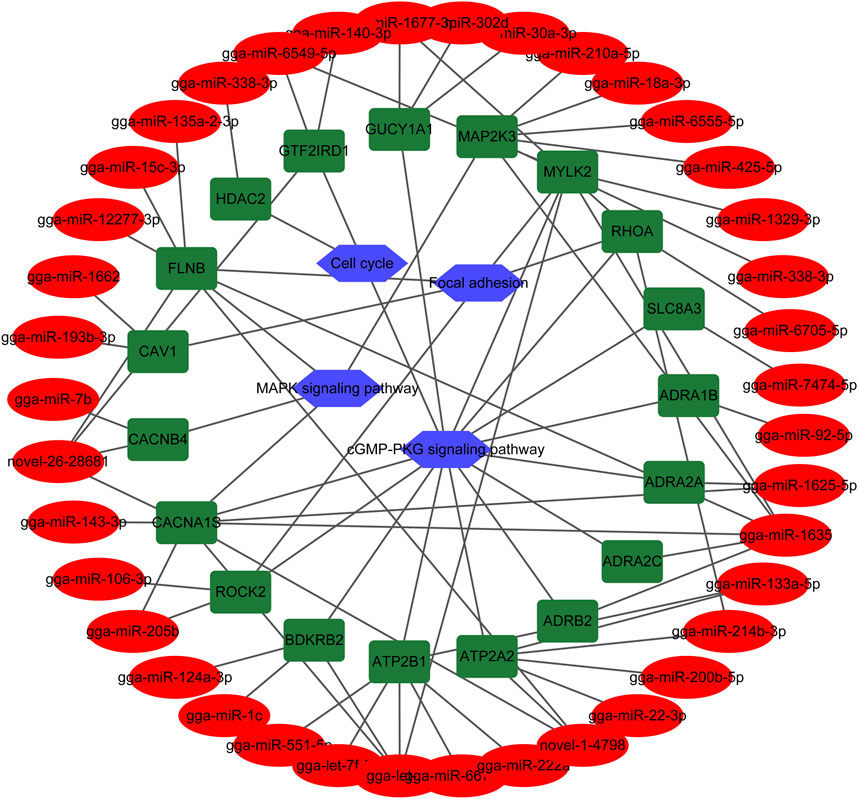
FIGURE 5. The miRNA-gene-pathway network between 19 DEMs involved in muscle system process, and their corresponding pathways and DEMIs. Blue hexagon, green round rectangle and red ellipse indicate pathway, gene and miRNA, respectively.
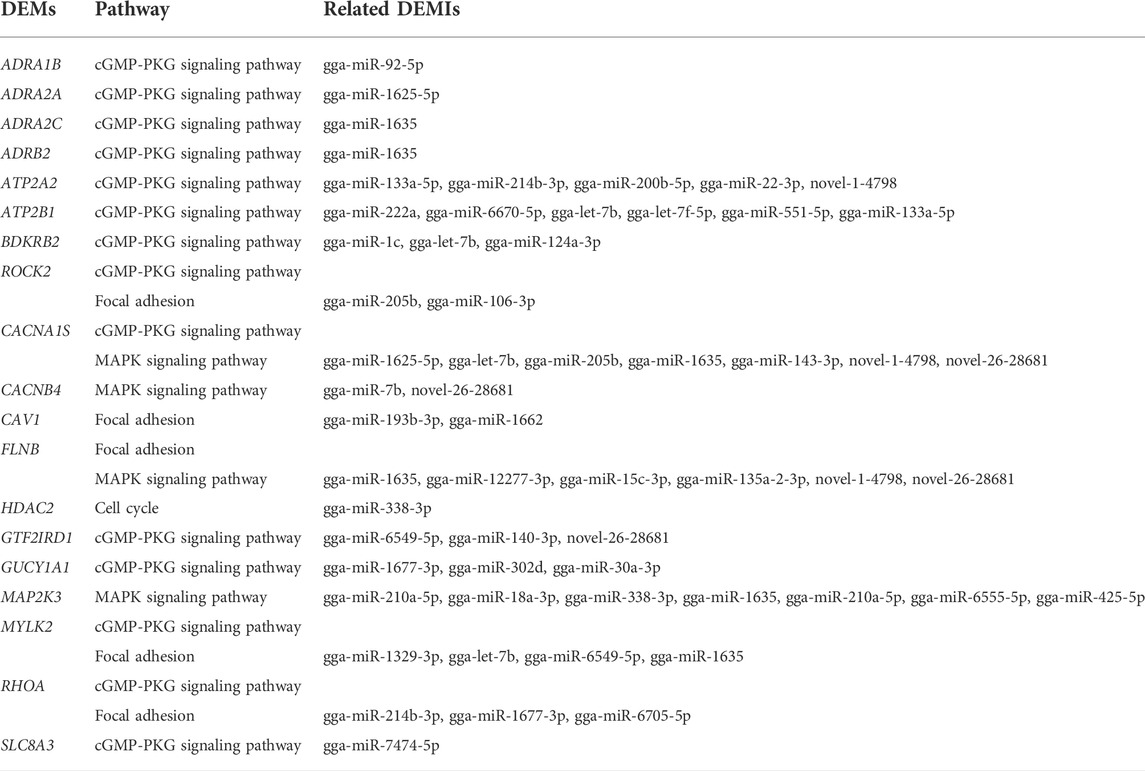
Table 5. 19 DEMs involved in muscle system process and their corresponding pathways and related DEMIs.
Based on the data of 6,583 DEMs, 695 DELs, 1,906 DECs, and 130 DEMIs, we identified putative miRNA-mRNA, miRNA-lncRNA, and miRNA-circRNA interactions. In total, we obtained 11,326 miRNA-mRNA, 1,266 miRNA-lncRNA, and 11,905 miRNA-circRNA interaction pairs. We next extracted miRNAs that paired with both lncRNA/circRNAs and mRNAs to construct the ceRNA network which included 115 miRNAs, 788 mRNAs, 56 lncRNAs, 398 circRNAs, and 2,161 interaction pairs (Figure 6).
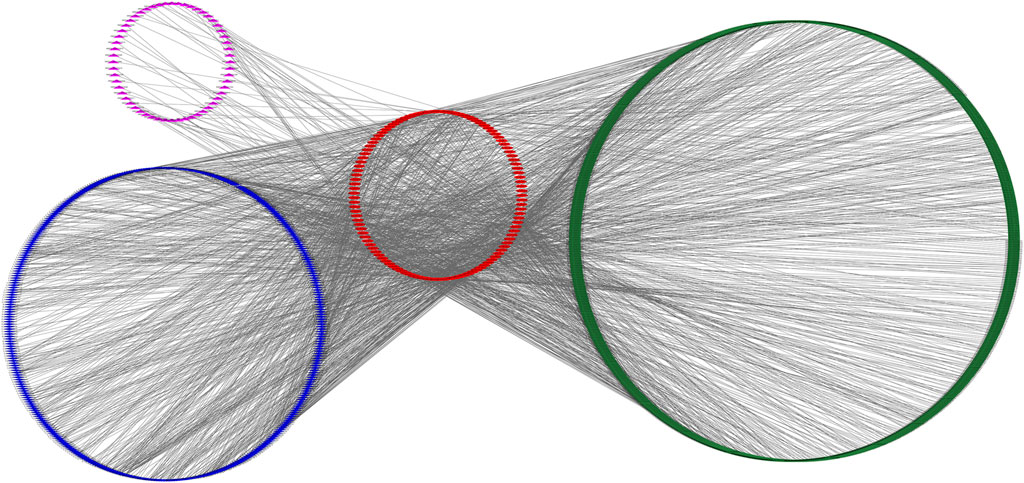
FIGURE 6. An overview of the competing endogenous RNA (ceRNA) network. Pink triangle, blue rhombus, red ellipse, and green round rectangle indicate circRNA transcript, lncRNA transcript, miRNA and mRNA transcript, respectively.
The interactions between lncRNA/circRNAs and mRNAs were predicted through combining analysis of the ceRNA and miRNA-gene-pathway networks (Figure 7). These interactions referred to 123 circRNAs, 27 lncRNAs, 11miRNAs (gga-let-7f-5b, gga-miR-106-3p, gga-miR-143-3p, gga-miR-1625-5p, gga-miR-1635, gga-miR-193b-3p, gga-miR-205b, gga-miR-22-3p, gga-miR-7b, gga-miR-92-5p, and novel-26-28681), and 8 mRNAs (ADRA1B, ATP2A2, ATP2B1, CACNA1S, CACNB4, MYLK2, and ROCK2).
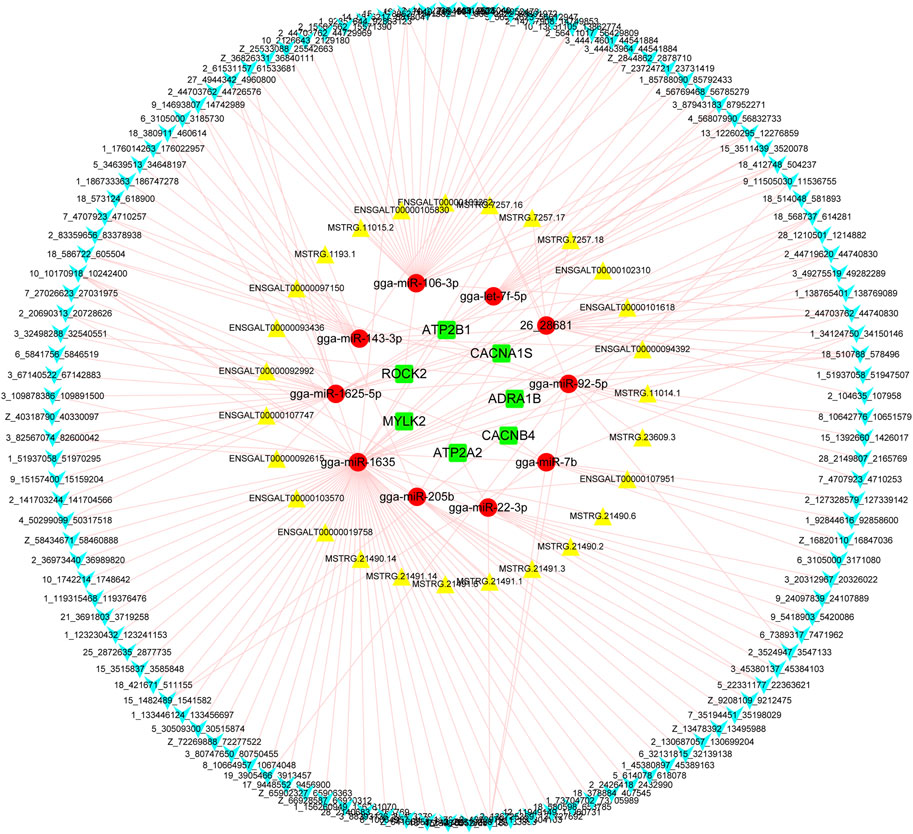
FIGURE 7. The predicted interaction between circRNA transcript, lncRNA transcripts and ADRA1B, ATP2A2, ATP2B1, CACNA1S, CACNB4, MYLK2, and ROCK2 genes. Blue polygon, yellow triangle, red ellipse, and green round rectangle indicate circRNA transcript, lncRNA transcript, miRNA, and mRNA, respectively.
Three DEMs, DELs, DECs, and DEMIs from the ceRNA network were selected to validate the RNA-seq results using RT-qPCR, respectively. The results were consistent with the RNA-seq data (Figure 8A) and a high correlation was detected, with a Pearson’s correlation coefficient of 0.8193 (p < 0.01) (Figure 8B).
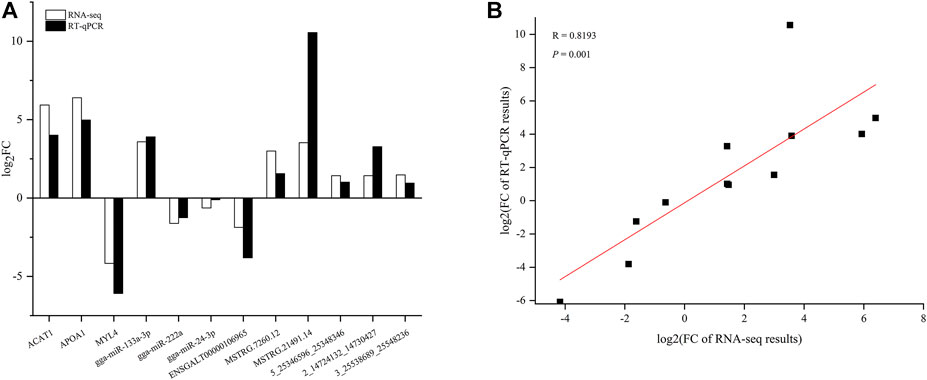
FIGURE 8. Validation of RNA-seq data using real time quantitative PCR (RT-qPCR) (A) and correlation between RNA-seq and RT-qPCR results (B).
Chicken is an important agricultural animal that has become the largest consumer meat worldwide over the past 50 years (Petracci and Cavani, 2012). The production and quality of poultry meat are closely related to the development of skeletal muscle (Dransfield and Sosnicki, 1999). Leg muscle is a primary contributor to skeletal muscle and is directly related with the quantity and quality of meat. The exploration of molecular mechanisms underlying skeletal muscle development has been a focus in the field of poultry genetic breeding. Muscle growth is characterized by an increase in myoblast cell number through hyperplasia during the embryonic period of development, and the process is nearly complete at the time of hatch (Velleman, 2007). To obtain a full-scale picture of the transcriptome level changes that occur within the increase of myoblast cell number, whole transcriptome analysis was used to detect candidate gene function and their regulatory effectors. In total, 6,583 DEMs, 695 DELs, 1,906 DECs, and 130 DEMIs were detected between the two developmental stages of Tibetan chicken.
It is clear that RNA has a diverse set of functions and is more than just a messenger between gene and protein. Considering the differential expression of RNAs from these two stages, these RNAs might be associated with myogenesis, particularly thousands of ncRNAs are well-expressed with exquisite cell-type and tissue specificity (Mercer et al., 2008; Guttman et al., 2010). In the present study, we detected DEMs relevant to the development of skeletal muscle in the leg muscle of chicken at E18. For example, myogenin (MYOG), as an MRF, which has a unique role in fetal myogenesis, was decreased in chickens at E18. MYOG is indispensable for myogenic differentiation, and fetal myogenesis essentially fails in MYOG-null mice, with few differentiated myofibers present (Hasty et al., 1993; Nabeshima et al., 1993). Primary myogenesis is complete by E14.5, with all muscle groups largely established, and secondary myogenesis begins at approximately E16.5, with fetal myoblasts proliferating (Zammit, 2017). The differential expression of MYOG indicates that E10 and E18 were two different phases of developmental myogenesis. Notably, several detected DEMs were well-known related to muscle development. For example, caveolin 3 (CAV3) impairs skeletal muscle mitochondrial form and function (Shah et al., 2020). Myopalladin (MYPN) promotes muscle growth through modulation of the serum response factor pathway (Filomena et al., 2020). The myocyte enhancer binding factor 2 (MEF2) family including MEF2A, MEF2B, and MEF2C act synergistically with MyoD and myogenin to promote muscle differentiation and activation of muscle genes (Taylor and Hughes, 2017).
Muscle development is a complex and multi-stage process with many genes cooperatively involved in the regulation of each stage. It is also very conservative among vertebrates. The process can be divided into four major stages: somite differentiation into dermomyotome-containing myogenic precursors, myogenic precursor proliferation and differentiation into myoblasts, myoblast proliferation, determination, differentiation and fusion into myotube, and finally maturation (or differentiation) of myotubes into myofibers (Luo et al., 2013). Compared to the unclear and incomplete muscle fibers at E10, the complete and determined muscle fibers at E18 were closely related to the DEMs significantly enriched in many signal transduction pathways including focal adhesion, cGMP-PKG signaling, MAPK signaling, and cell cycle pathways, resulting in skeletal muscle development. In our study, we found increased expression of genes related to the above-mentioned pathways at E18, indicating that myoblast proliferation, differentiation, fusion into myotube, and finally maturation of myotubes into myofibers were mediated by the above pathways. Some of the pathways related to muscle development involved in DEMs such as MAPK signaling pathway were also found in the embryonic muscle development of the Chengkou Mountain Chicken (Ren et al., 2021). In addition, the target genes of miRNAs and lncRNAs were also found to be enriched in the mentioned pathways.
Expression of ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 2 (ATP2A2) and CACNA1S was increased. Intracellular Ca2+ is a critical coordinator of various aspects of cellular physiology. The Ca2+ adenosine triphosphatase SERCA encoded by ATP2A2 controls of cell death and survival in various cell types (Nelson et al., 2016; Chemaly et al., 2018). The α1s subunit of the dihydropyridine receptor (DHPR α1s) encoded by CACNA1S controls skeletal muscle mass and morphogenesis (Piétri-Rouxel et al., 2010). We also found that the level of MYLK2 and ROCK2 were increased. The MYLK2 gene which encodes skeletal muscle myosin light chain kinase regulates skeletal myogenesis by phosphorylation of MEF2C (Al Madhoun et al., 2011; Stull et al., 2011). Goetsch et al. (2014) found that ROCK2 regulates directional myoblast migration through focal adhesion formation and maturation. Altogether, these results suggest that E10 to E18 are critical stages for myoblast proliferation, differentiation and finally maturation (or differentiation) of myotubes into myofibers.
In recent years, lncRNA and circRNA ceRNAs have received increased attention for their involvement in the development of skeletal muscle. For example, lncRNA 2310043L19Rik inhibits differentiation and promotes proliferation of myoblast by sponging miR-125a-5p (Li et al., 2020). As a ceRNA for miR-351-5p and miR-125b, lnc-mg mediates skeletal myogenesis by indirectly targeting lactamase-β (Zhu et al., 2017; Du et al., 2019). It was reported that circ-FoxO3 is a sponge of miR-138-5p which inhibits myoblast cells differentiation (Li et al., 2019a). CircMYBPC1 was identified to promote myoblast differentiation by directly binding miR-23a to relieve its inhibition on MyHC (Chen et al., 2021). Integrated analysis of lncRNA and mRNA expression profiles revealed that lncRNAs and target genes that might contribute to the regulation of different embryonic stages of skeletal muscle development in chicken (Li et al., 2016; Wu et al., 2022). Many miRNAs related to embryonic muscle development identified in Chengkou mountain chicken were also found in Tibetan chicken (Shi et al., 2022). We have not found research on circRNA sequencing related to embryonic muscle development in chicken. Our study showed expression profiles of lncRNA and circRNA transcripts in the chicken skeletal muscle at E10 and E18 by whole transcriptome sequencing. Through interaction analysis of DEMs, DELs, DECs, and DEMIs, we also discovered that lncRNA and circRNA could compete for miRNAs binding sites with mRNA and subsequently influence their expression. The genes associated with skeletal muscle development such as ATP2A2, CACNA1S, MYLK2, and ROCK2 could be modulated by lncRNAs and circRNAs. These findings indicate that these lncRNAs and circRNAs could modulate multiple subsystems involved in the development skeletal muscle as regulators.
Integration of multi-omics can generate new knowledge that is not accessible by analysis of single datasets alone (Stanberry et al., 2013). Recent reports have described an intricate interplay among diverse RNA species, including protein-coding messenger RNAs and non-coding RNAs such as lncRNAs, pseudogenes and circRNAs (Tay et al., 2014). These RNA transcripts containing miRNA response elements (MREs) act as ceRNAs, communicate with and co-regulate each other by competing for binding to shared miRNAs. Our study constructed a ceRNA network by integrating multiple omics analyses, and subsequently detected lncRNA, circRNA, and miRNA highly relevant to skeletal muscle development.
In summary, we characterized and compared the expressional features of mRNAs, lncRNAs, circRNAs, and miRNAs in the skeletal muscle of the Tibetan chicken at E10 and E18 by RNA-seq. The DEMs, DELs, DECs, and DEMIs between the two stages involved in skeletal muscle development were identified and characterized. We conducted lncRNAs/circRNAs-miRNA-mRNA regulatory networks of the molecular mechanism for skeletal muscle development. The present study provided a new insight of the molecular mechanism underlying skeletal muscle development in chicken.
The data presented in the study are deposited in the SRA repository, accession number "PRJNA758717".
The animal study was reviewed and approved by the Institutional Animal Care and Use Committee of the Southwest Minzu University.
ZP and ZL conceived the research; ZP and CY performed the experiments; XJ and CY shared regents; ZP, RZ, and ZL analyzed the data; ZL wrote the paper. All authors read and approved the final manuscript.
This work was funded by the Science and Technology Support Program of Sichuan Province (2021YFYZ0031), the earmarked fund for China Agriculture Research System (CARS-41-G07), and the Fundamental Research Funds for the Central Universities (2021PTJS20).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2022.969854/full#supplementary-material
Al Madhoun A. S., Mehta V., Li G., Figeys D., Wiper-Bergeron N., Skerjanc I. S. (2011). Skeletal myosin light chain kinase regulates skeletal myogenesis by phosphorylation of MEF2C. EMBO J. 30, 2477–2489. doi:10.1038/emboj.2011.153
Bentzinger C. F., Wang Y. X., Rudnicki M. A. (2012). Building muscle: Molecular regulation of myogenesis. Cold Spring Harb. Perspect. Biol. 4, a008342. doi:10.1101/cshperspect.a008342
Buckingham M., Rigby P. W. (2014). Gene regulatory networks and transcriptional mechanisms that control myogenesis. Dev. Cell 28, 225–238. doi:10.1016/j.devcel.2013.12.020
Cesana M., Cacchiarelli D., Legnini I., Santini T., Sthandier O., Chinappi M., et al. (2011). A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 147, 358–369. doi:10.1016/j.cell.2011.09.028
Chang N. C., Chevalier F. P., Rudnicki M. A. (2016). Satellite cells in muscular dystrophy - lost in polarity. Trends Mol. Med. 22, 479–496. doi:10.1016/j.molmed.2016.04.002
Chemaly E. R., Troncone L., Lebeche D. (2018). SERCA control of cell death and survival. Cell calcium 69, 46–61. doi:10.1016/j.ceca.2017.07.001
Chen B., Yu J., Guo L., Byers M. S., Wang Z., Chen X., et al. (2019). Circular RNA circHIPK3 promotes the proliferation and differentiation of chicken myoblast cells by sponging miR-30a-3p. Cells 8, 177. doi:10.3390/cells8020177
Chen M., Wei X., Song M., Jiang R., Huang K., Deng Y., et al. (2021). Circular RNA circMYBPC1 promotes skeletal muscle differentiation by targeting MyHC. Mol. Ther. Nucleic Acids 24, 352–368. doi:10.1016/j.omtn.2021.03.004
Cruz-Jentoft A. J., Sayer A. A. (2019). Sarcopenia. Lancet 393, 2636–2646. doi:10.1016/S0140-6736(19)31138-9
Delfini M. C., Hirsinger E., Pourquié O., Duprez D. (2000). Delta 1-activated notch inhibits muscle differentiation without affecting Myf5 and Pax3 expression in chick limb myogenesis. Development 127, 5213–5224. doi:10.1242/dev.127.23.5213
Dransfield E., Sosnicki A. A. (1999). Relationship between muscle growth and poultry meat quality. Poult. Sci. 78, 743–746. doi:10.1093/ps/78.5.743
Du J., Zhang P., Zhao X., He J., Xu Y., Zou Q., et al. (2019). MicroRNA-351-5p mediates skeletal myogenesis by directly targeting lactamase-β and is regulated by lnc-mg. FASEB J. official Publ. Fed. Am. Soc. Exp. Biol. 33, 1911–1926. doi:10.1096/fj.201701394RRR
Dumont N. A., Bentzinger C. F., Sincennes M.-C., Rudnicki M. A. (2015). Satellite cells and skeletal muscle regeneration. Compr. Physiol. 5, 1027–1059. doi:10.1002/cphy.c140068
Filomena M. C., Yamamoto D. L., Caremani M., Kadarla V. K., Mastrototaro G., Serio S., et al. (2020). Myopalladin promotes muscle growth through modulation of the serum response factor pathway. J. Cachexia Sarcopenia Muscle 11, 169–194. doi:10.1002/jcsm.12486
Ghosh S., Chan C.-K. K. (2016). Analysis of RNA-seq data using TopHat and Cufflinks. Methods Mol. Biol. 1374, 339–361. doi:10.1007/978-1-4939-3167-5_18
Goetsch K. P., Snyman C., Myburgh K. H., Niesler C. U. (2014). ROCK-2 is associated with focal adhesion maturation during myoblast migration. J. Cell. Biochem. 115, 1299–1307. doi:10.1002/jcb.24784
Greene J., Baird A.-M., Brady L., Lim M., Gray S. G., Mcdermott R., et al. (2017). Circular RNAs: Biogenesis, function and role in human diseases. Front. Mol. Biosci. 4, 38. doi:10.3389/fmolb.2017.00038
Guttman M., Garber M., Levin J. Z., Donaghey J., Robinson J., Adiconis X., et al. (2010). Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat. Biotechnol. 28, 503–510. doi:10.1038/nbt.1633
Hasty P., Bradley A., Morris J. H., Edmondson D. G., Venuti J. M., Olson E. N., et al. (1993). Muscle deficiency and neonatal death in mice with a targeted mutation in the myogenin gene. Nature 364, 501–506. doi:10.1038/364501a0
Huarte M. (2009). Journal club. A biologist looks at new functions for non-coding RNAs. Nature 459, 487. doi:10.1038/459487e
John B., Enright A. J., Aravin A., Tuschl T., Sander C., Marks D. S. (2004). Human MicroRNA targets. PLoS Biol. 2, e363. doi:10.1371/journal.pbio.0020363
Kang Y.-J., Yang D.-C., Kong L., Hou M., Meng Y.-Q., Wei L., et al. (2017). CPC2: A fast and accurate coding potential calculator based on sequence intrinsic features. Nucleic Acids Res. 45, W12–W16. doi:10.1093/nar/gkx428
Kim D., Paggi J. M., Park C., Bennett C., Salzberg S. L. (2019). Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915. doi:10.1038/s41587-019-0201-4
Langmead B., Salzberg S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi:10.1038/nmeth.1923
Li R., Li B., Shen M., Cao Y., Zhang X., Li W., et al. (2020). LncRNA 2310043L19Rik inhibits differentiation and promotes proliferation of myoblast by sponging miR-125a-5p. Aging 12, 5625–5639. doi:10.18632/aging.102905
Li X., Li C., Liu Z., Ni W., Yao R., Xu Y., et al. (2019a). Circular RNA circ-FoxO3 inhibits myoblast cells differentiation. Cells 8, E616. doi:10.3390/cells8060616
Li Z., Cai B., Abdalla B. A., Zhu X., Zheng M., Han P., et al. (2019b). LncIRS1 controls muscle atrophy via sponging miR-15 family to activate IGF1-PI3K/AKT pathway. J. Cachexia Sarcopenia Muscle 10, 391–410. doi:10.1002/jcsm.12374
Li Z., Ouyang H., Zheng M., Cai B., Han P., Abdalla B. A., et al. (2016). Integrated analysis of long non-coding RNAs (LncRNAs) and mRNA expression profiles reveals the potential role of LncRNAs in skeletal muscle development of the chicken. Front. Physiol. 7, 687. doi:10.3389/fphys.2016.00687
Liu H., Pecka J. L., Zhang Q., Soukup G. A., Beisel K. W., He D. Z. Z. (2014). Characterization of transcriptomes of cochlear inner and outer hair cells. J. Neurosci. 34, 11085–11095. doi:10.1523/JNEUROSCI.1690-14.2014
Liu J., Li M., Kong L., Cao M., Zhang M., Wang Y., et al. (2021). CircARID1A regulates mouse skeletal muscle regeneration by functioning as a sponge of miR-6368. FASEB J. official Publ. Fed. Am. Soc. Exp. Biol. 35, e21324. doi:10.1096/fj.202001992R
Liu L., Xiao Q., Gilbert E. R., Cui Z., Zhao X., Wang Y., et al. (2018). Whole-transcriptome analysis of atrophic ovaries in broody chickens reveals regulatory pathways associated with proliferation and apoptosis. Sci. Rep. 8, 7231. doi:10.1038/s41598-018-25103-6
Love M. I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. doi:10.1186/s13059-014-0550-8
Luo W., Nie Q., Zhang X. (2013). MicroRNAs involved in skeletal muscle differentiation. J. Genet. genomics = Yi chuan xue bao 40, 107–116. doi:10.1016/j.jgg.2013.02.002
Mchugh C. A., Chen C.-K., Chow A., Surka C. F., Tran C., Mcdonel P., et al. (2015). The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature 521, 232–236. doi:10.1038/nature14443
Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., et al. (2013). Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495, 333–338. doi:10.1038/nature11928
Mercer T. R., Dinger M. E., Sunkin S. M., Mehler M. F., Mattick J. S. (2008). Specific expression of long noncoding RNAs in the mouse brain. Proc. Natl. Acad. Sci. U. S. A. 105, 716–721. doi:10.1073/pnas.0706729105
Nabeshima Y., Hanaoka K., Hayasaka M., Esumi E., Li S., Nonaka I., et al. (1993). Myogenin gene disruption results in perinatal lethality because of severe muscle defect. Nature 364, 532–535. doi:10.1038/364532a0
Nelson B. R., Makarewich C. A., Anderson D. M., Winders B. R., Troupes C. D., Wu F., et al. (2016). A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Sci. (New York, N.Y.) 351, 271–275. doi:10.1126/science.aad4076
Ouyang H., Chen X., Li W., Li Z., Nie Q., Zhang X. (2018). Circular RNA circSVIL promotes myoblast proliferation and differentiation by sponging miR-203 in chicken. Front. Genet. 9, 172. doi:10.3389/fgene.2018.00172
Petracci M., Cavani C. (2012). Muscle growth and poultry meat quality issues. Nutrients 4, 1–12. doi:10.3390/nu4010001
Phelps M., Coss C., Wang H., Cook M. (2016). Registered report: Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. eLife 5, e12470. doi:10.7554/eLife.12470
Piétri-Rouxel F., Gentil C., Vassilopoulos S., Baas D., Mouisel E., Ferry A., et al. (2010). DHPR alpha1S subunit controls skeletal muscle mass and morphogenesis. EMBO J. 29, 643–654. doi:10.1038/emboj.2009.366
Ren L., Liu A., Wang Q., Wang H., Dong D., Liu L. (2021). Transcriptome analysis of embryonic muscle development in Chengkou Mountain Chicken. BMC Genomics 22, 431. doi:10.1186/s12864-021-07740-w
Salmena L., Poliseno L., Tay Y., Kats L., Pandolfi P. P. (2011). A ceRNA hypothesis: The rosetta stone of a hidden RNA language? Cell 146, 353–358. doi:10.1016/j.cell.2011.07.014
Shah D. S., Nisr R. B., Stretton C., Krasteva-Christ G., Hundal H. S. (2020). Caveolin-3 deficiency associated with the dystrophy P104L mutation impairs skeletal muscle mitochondrial form and function. J. Cachexia Sarcopenia Muscle 11, 838–858. doi:10.1002/jcsm.12541
Shen X., Liu Z., Cao X., He H., Han S., Chen Y., et al. (2019). Circular RNA profiling identified an abundant circular RNA circTMTC1 that inhibits chicken skeletal muscle satellite cell differentiation by sponging miR-128-3p. Int. J. Biol. Sci. 15, 2265–2281. doi:10.7150/ijbs.36412
Shi J. A., Li W., Liu A., Ren L., Zhang P., Jiang T., et al. (2022). MiRNA sequencing of embryonic myogenesis in Chengkou mountain chicken. BMC Genomics 23, 571. doi:10.1186/s12864-022-08795-z
Stanberry L., Mias G. I., Haynes W., Higdon R., Snyder M., Kolker E. (2013). Integrative analysis of longitudinal metabolomics data from a personal multi-omics profile. Metabolites 3, 741–760. doi:10.3390/metabo3030741
Stull J. T., Kamm K. E., Vandenboom R. (2011). Myosin light chain kinase and the role of myosin light chain phosphorylation in skeletal muscle. Arch. Biochem. Biophys. 510, 120–128. doi:10.1016/j.abb.2011.01.017
Sun L., Luo H., Bu D., Zhao G., Yu K., Zhang C., et al. (2013). Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res. 41, e166. doi:10.1093/nar/gkt646
Tay Y., Rinn J., Pandolfi P. P. (2014). The multilayered complexity of ceRNA crosstalk and competition. Nature 505, 344–352. doi:10.1038/nature12986
Taylor M. V., Hughes S. M. (2017). Mef2 and the skeletal muscle differentiation program. Semin. Cell Dev. Biol. 72, 33–44. doi:10.1016/j.semcdb.2017.11.020
Trapnell C., Williams B. A., Pertea G., Mortazavi A., Kwan G., Van Baren M. J., et al. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515. doi:10.1038/nbt.1621
Velleman S. G. (2007). Muscle development in the embryo and hatchling. Poult. Sci. 86, 1050–1054. doi:10.1093/ps/86.5.1050
Wang L., Park H. J., Dasari S., Wang S., Kocher J.-P., Li W. (2013). Cpat: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res. 41, e74. doi:10.1093/nar/gkt006
Wu P., Zhou K., Zhang J., Ling X., Zhang X., Li P., et al. (2022). Transcriptome integration analysis at different embryonic ages reveals key lncRNAs and mRNAs for chicken skeletal muscle. Front. Vet. Sci. 9, 908255. doi:10.3389/fvets.2022.908255
Yin H., Shen X., Zhao J., Cao X., He H., Han S., et al. (2020). Circular RNA CircFAM188B encodes a protein that regulates proliferation and differentiation of chicken skeletal muscle satellite cells. Front. Cell Dev. Biol. 8, 522588. doi:10.3389/fcell.2020.522588
Yue B., Li H., Liu M., Wu J., Li M., Lei C., et al. (2019). Characterization of lncRNA-miRNA-mRNA network to reveal potential functional ceRNAs in bovine skeletal muscle. Front. Genet. 10, 91. doi:10.3389/fgene.2019.00091
Zammit P. S. (2017). Function of the myogenic regulatory factors Myf5, MyoD, Myogenin and MRF4 in skeletal muscle, satellite cells and regenerative myogenesis. Semin. Cell Dev. Biol. 72, 19–32. doi:10.1016/j.semcdb.2017.11.011
Zeng X., Lin W., Guo M., Zou Q. (2017). A comprehensive overview and evaluation of circular RNA detection tools. PLoS Comput. Biol. 13, e1005420. doi:10.1371/journal.pcbi.1005420
Zhang Z.-K., Li J., Guan D., Liang C., Zhuo Z., Liu J., et al. (2018). A newly identified lncRNA MAR1 acts as a miR-487b sponge to promote skeletal muscle differentiation and regeneration. J. Cachexia Sarcopenia Muscle 9, 613–626. doi:10.1002/jcsm.12281
Keywords: chicken, ceRNA, lncRNA/circRNA, miRNA, skeletal muscle
Citation: Pan Z, Yang C, Zhao R, Jiang X, Yu C and Li Z (2022) Characterization of lncRNA/circRNA-miRNA-mRNA network to reveal potential functional ceRNAs in the skeletal muscle of chicken. Front. Physiol. 13:969854. doi: 10.3389/fphys.2022.969854
Received: 15 June 2022; Accepted: 05 September 2022;
Published: 29 September 2022.
Edited by:
Emma Louise Robinson, University of Colorado, Denver, United StatesReviewed by:
Genxi Zhang, Yangzhou University, ChinaCopyright © 2022 Pan, Yang, Zhao, Jiang, Yu and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhixiong Li, bGl6aGl4aW9uZ0Bzd3VuLmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.