- 1Department of Hematology, the First Affiliated Hospital of China Medical University, Shenyang, LN, China
- 2Department of Hematology, General Hospital of PLA Northern Theater Command, Shenyang, LN, China
- 3Department of Cardiology, the First Affiliated Hospital of China Medical University, Shenyang, LN, China
E3 ubiquitin ligases, an important part of ubiquitin proteasome system, catalyze the covalent binding of ubiquitin to target substrates, which plays a role in protein ubiquitination and regulates different biological process. DNA damage response (DDR) is induced in response to DNA damage to maintain genome integrity and stability, and this process has crucial significance to a series of cell activities such as differentiation, apoptosis, cell cycle. The NEDD4 family, belonging to HECT E3 ubiquitin ligases, is reported as regulators that participate in the DDR process by recognizing different substrates. In this review, we summarize recent researches on NEDD4 family members in the DDR and discuss the roles of NEDD4 family members in the cascade reactions induced by DNA damage. This review may contribute to the further study of pathophysiology for certain diseases and pharmacology for targeted drugs.
Introduction
Ubiquitination is a reversible biological process in which proteins are post-translationally modified through a series of multiple catalytic steps, leading to protein degradation by the proteasome or lysosome. A wide range of proteins, including membrane proteins, cell cycle regulators, transcription factors, tumor suppressors and oncogenes, are ubiquitinated (Goel et al., 2015; Zhang et al., 2020a; Zhang et al., 2020b). Therefore, protein ubiquitination is involved in regulating a variety of biological processes (Chen and Matesic, 2007). The ubiquitin-proteasome system is composed of ubiquitin activating enzyme (E1), ubiquitin conjugation enzyme (E2) and ubiquitin ligase (E3). Ubiquitin is activated by E1 and transferred from E2 to substrate mediated by E3 (Xie et al., 2021). E3 ligases can be divided into three classes according to its structure and ubiquitin transfer mechanism: Really interesting new gene (RING)-type E3 ligases, Homologous to E6-AP COOH terminus (HECT)-type E3 ligases and Ring between Ring (RBR)-type E3 ligases (Qie, 2022). RING-type E3 ligases transfer ubiquitin from E2 to substrates directly, while HECT-type E3 ligases and RBR-type E3 ligases transfer ubiquitin in two steps: ubiquitin is transferred from E2 to E3 firstly and from E3 to substrates secondly (Cruz Walma et al., 2022). All seven lysine (K6, K11, K27, K29, K33, K48, and K63) and N-terminal M1 residues form linkage points during chain extension, among which K48 and K63 are mostly studied (Ikeda and Dikic, 2008; Zhang et al., 2020c). K63-linked Ub chains are involved in DDR pathways and NF-κB pathways as previously reported (Bennett and Harper, 2008) and K29/K33-linked Ub chains participate in the regulation of AMP-activated protein kinase (Al-Hakim et al., 2008). Most of E3 ubiquitin ligases belong to the RING family. The HECT-type E3 ligase in humans contains 28 members, and the neural precursor cell–expressed developmentally down-regulated 4 (NEDD4) family is the largest group (Manning and Kumar, 2018; Liu et al., 2021). Multiple studies have demonstrated that HECT E3 ubiquitin ligases are involved in various cellular activities such as protein transport (d'Azzo et al., 2005), subcellular localization (Laine and Ronai, 2007), immune response (Liu, 2004), viral infection (d'Azzo et al., 2005), DNA damage response (DDR) (Harvey and Kumar, 1999; Mohiuddin et al., 2016), oxidative stress (Zhang et al., 2021a) apoptosis (Li et al., 2008a; Kim et al., 2013; Li et al., 2013), cell cycle progression (Goto et al., 2017) and signal transduction (Michnov et al., 2012).
The NEDD4 family consists of nine members: NEDD4 (also known as NEDD4-1), NEDD4L (also known as NEDD4-2), ITCH (also known as AIP4), WWP1 (also known as AIP5), WWP2 (also known as AIP2), SMUEF1, SMURF2, NEDL1 (also known as HECW1) and NEDL2 (also known as HECW2) (Scheffner and Kumar, 2014). They work through binding to both E2-ub thioester and substrate and transferring ubiquitin to substrate finally (Mathieu et al., 2021) (Figure 1A). NEDD4 family proteins have three functional domains: the C2 domain in the amino terminus that binds the cell membrane; the WW domain in the central region that mediates protein-protein interaction; and the HECT domain in the carboxyl terminus that promotes transfer of ubiquitin to substrate proteins (Figure 1B) (Kumar et al., 1997). The C2 domain is a calcium ion–binding domain with a length of about 120 amino acids and binds to phospholipids, inositol, polyphosphates and some proteins (Plant et al., 1997). The WW domain contains 35–40 amino acids and interacts with the PY(PPXY) motif or phosphorylated serine/threonine residues in the substrate protein (Lu et al., 1999). The HECT domain consists of about 350 residues and is responsible for the transfer of ubiquitin to lysine residues in the substrate proteins (Ingham et al., 2004). Because of the diversity of WW domain (Ingham et al., 2004) and substrates (Dodson et al., 2015), the NEDD4 family proteins exert various cellular roles.
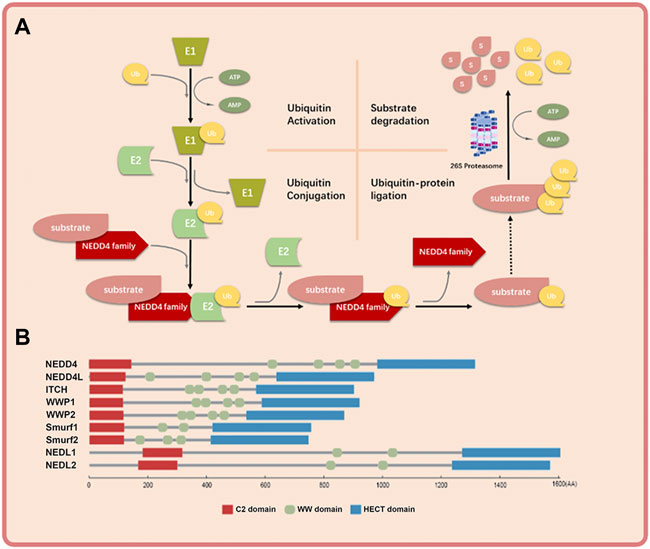
FIGURE 1. The basic mechanism of NEDD4 family in ubiquitin-proteosome system and the schematic structure of NEDD4 family members. (A) Ubiquitin is activated by ubiquitin activating enzyme (E1), transferred by ubiquitin conjugation enzyme (E2) to NEDD4 family (E3), and covalently bound to substrate protein by NEDD4 family. The polyubiquitinated substrate protein is recognized and degraded by 26s proteosome finally. (B) The NEDD4 family gene mainly has three functional domains: C2 domain, where the amino terminus can bind to cell membrane, WW domain, which is the central region of protein-protein interaction, and HECT domain, where the carboxyl terminus has ubiquitin-protein connection.
DNA damage is the alteration of DNA nucleotide sequence caused by endogenous factors (such as base changes or oxidation) or exogenous genotoxic factors (Molinaro et al., 2021). DNA damage is associated with aging (Vijg, 2021), cancer development (Perkhofer et al., 2021; Rivas-Domínguez et al., 2021) and some systemic diseases (Zhang et al., 2021b; Hu et al., 2021; Wang et al., 2021). To repair DNA damage and maintain the integrity of the genome, cells initiate a series of cascade reactions that detect DNA damage and transmit information, collectively known as the DNA damage response (DDR). The main outcomes of the DDR include cell cycle arrest, apoptosis, aging and DNA repair (Figure 2) (Visser and Thomas, 2021). DNA repair mechanisms vary depending on the type of DNA damage. Base excision repair (BER) repairs DNA single strand damage. Mismatch repair (MMR) detects and removes the insertion or deletion of mismatched nucleotides during DNA replication. Nucleotide excision repair (NER) repairs large or helically destabilizing DNA lesions. DNA double-strand breaks are repaired by homologous recombination (HR) and non-homologous end joining (NHEJ) (Rivas-Domínguez et al., 2021). It is reported that Ataxia-telangiectasia mutated (ATM), ATM- and RAD3-related (ATR) and DNA-dependent protein kinase (DNA-PK) play important roles in DDR (Blackford and Jackson, 2017). The ATM kinase phosphorylates proteins in response to recognition of DNA double strand breaks (DSBs) and ATR stops the DNA replication process and promotes DNA repair. DNA-PK mainly participates in NHEJ and activates a small subset of proteins (Kiss et al., 2021). The point of DDR is repairing DNA damage and activating cell death if the damage is irreparable, and thus preventing the transmission of potentially harmful DNA mutations (Goff et al., 2021).
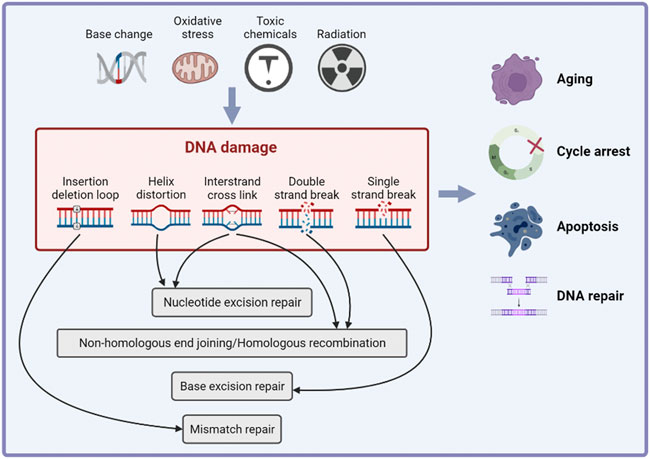
FIGURE 2. The inducements and effects of DNA damage response. Cells initiate a series of cascade reactions in response of endogenous of exogenous DNA damage inducements, which is known as DNA damage response (DDR). The main effects of DDR include cell cycle arrest, apoptosis, aging and DNA repair.
In recent years, several studies have shown that members of the NEDD4 family participate in and regulate multiple DDR pathways to maintain genome integrity. Therefore, this review aims to summarize the mechanistic role of NEDD4 family members in DDR and provide a general understanding of the roles of NEDD4 family members in the cascade reactions induced by DNA damage. This review may also contribute to the further study of pathophysiology for certain diseases and pharmacology for targeted drugs.
NEDD4
NEDD4 function
NEDD4, the first identified member of the NEDD4 family, was first isolated from neural progenitor cells in 1992 as a factor with mRNA levels down-regulated during mouse brain development (Kumar et al., 1992; Yang and Kumar, 2010). The human NEDD4 gene, located on chromosome 15Q21.3, contains 33 exons and encodes a protein with a molecular weight of 120 kDa. NEDD4 is expressed in heart, lung, brain, kidney, and other tissues (Fagerberg et al., 2014) and mainly localizes in the cytoplasm and specifically around the nucleus (Anan et al., 1998; Putz et al., 2008). NEDD4 was originally shown to regulate water and electrolyte balance by controlling the abundance of sodium channels in epithelial cells. Subsequent studies demonstrated that NEDD4 regulates embryonic and tumor development as an E3 ligase (Huang et al., 2019). In yeast and mammalian cells, NEDD4 regulates intracellular sorting and transport (Yalcin et al., 2019; Xie et al., 2020). NEDD4 also functions in protein degradation through the polyubiquitination of K48 and K63 sites (Xu et al., 2015; French et al., 2017; Sluimer and Distel, 2018) or monoubiquitin of K6 and K27 sites (Murillas et al., 2002; Fukushima et al., 2015).
NEDD4 and DDR
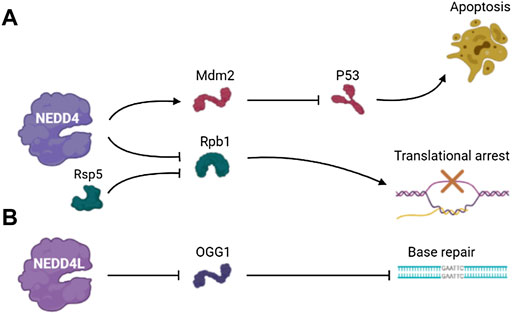
Mdm2 is an E3 ubiquitin ligase and important negative regulator of tumor suppressor protein p53 (Fang et al., 2000). Mdm2 overexpression leads to the inactivation of p53 (Lai et al., 2001). NEDD4 regulates the stability of Mdm2 in cells, thus contributing to regulation of p53 in the DNA damage response. NEDD4 physically interacts with Mdm2 via the RING domain of Mdm2 and promotes Mdm2 ubiquitination. In NEDD4 knockout cells, the expression level of p53, Mdm2-dependent behavior and DDR were increased. Therefore, NEDD4 is an important component of p53 pathway that influences Mdm2 stabilization in the DNA damage response (Xu et al., 2015).
RNA polymerase II (Rpb1) is the largest subunit of RNA polymerase II (Pol II), a substrate of Rsp5 that is an E3 ubiquitin-protein ligase and is essential for the synthesis of mRNA and transcript synthesis following repair (Somesh et al., 2005; Calvo, 2020). The ubiquitination and degradation of Rpb1 participates in the transcriptional arrest induced by DNA damage. The ubiquitination of Rpb1 mediated by Rsp5 is part of the DNA damage response, and Rpb1 is specifically identified to enhance DNA damage response. Beaudenon et al. demonstrated that NEDD4 mediated UV-induced degradation of Pol II by ubiquitination of Rpb1. This finding suggests that an irreversible disassembly of transcription complexes by the degradation of the major catalytic subunit of Pol II works in response to DNA damage (Beaudenon et al., 1999) (Figure 3A).
NEDD4L
NEDD4L function
NEDD4L is the closest homologue of NEDD4 in the NEDD4 family (Harvey and Kumar, 1999; Goel et al., 2015). The human NEDD4L gene, located on chromosome 18Q21.31, contains 38 exons and can produce multiple spliced mRNAs (Chen et al., 2001; Harvey et al., 2001). NEDD4L transcripts exist in many tissues, especially in liver, kidney, heart and lung (Dunn et al., 2002; Araki et al., 2008), and NEDD4L is down-regulated in melanoma (Cui et al., 2020), colorectal cancer (Yang et al., 2020), lung cancer (Wang et al., 2019) and other cancers, indicating NEDD4L is a tumor suppressor. NEDD4L regulates many membrane proteins, such as epithelial and voltage-gated sodium channels (Goel et al., 2015; Gao et al., 2021). It mediates the Wnt/β-catenin and TGF-β signaling pathways and plays a key role in preventing the progression of systemic chronic kidney disease (Manning et al., 2021). NEDD4L catalyzes K29-linked ubiquitination of cysteine residues in TRAF3, which is of vital importance against innate immunity to viruses (Gao et al., 2021). In addition, NEDD4L is also involved in virus budding by ubiquitination and activation of ESCRT-I (Chung et al., 2008).
NEDD4L and DDR
Eight-oxyguanine lesions are premutagenic lesions that lead to GC to TA translocation, and 8-oxyguanine DNA glycosylase (OGG1) is a major DNA glycosylase that removes 8-oxyguanine lesions from DNA (Boiteux and Radicella, 2000). OGG1 plays a regulatory role in controlling gene expression by the removal of 8-oxyguanine lesions in guanine-rich promoter sequences, leading to transcriptional activation of guanine quadruplet structures (Fleming et al., 2017; Wang et al., 2018). Hughes and Parsons showed that ubiquitination of OGG1 on lysine 341 by NEDD4L inhibited its DNA glycosylase/lyase activity, and the dysregulation of OGG1 increased DNA damage (Hughes and Parsons, 2020). This result suggests that the regulation of OGG1 protein level plays an important role in the repairing of bases and the maintenance of genomic stability, and this process involves the ubiquitination and degradation mediated by the NEDD4L E3 ubiquitin ligase (Figure 3B).
ITCH
ITCH function
ITCH was originally identified by genetic studies of mouse fur color changes and its deletion leads to the itchy phenotype of persistent skin scratching and multi-organ inflammation (WL et al., 1998). The human ITCH gene is located on chromosome 20q11.22 and encodes 864 amino acids (Infante et al., 2019; Li et al., 2020). It contains the typical domains of the NEDD4 family and four WW domains that interact with substrates. ITCH regulates a series of signaling pathways by promoting polyubiquitination of over 50 target proteins, including C-Jun, C-flip, LATS1, P63, P73, TCR-ζ and BRAF. ITCH not only catalyzes K48 polyubiquitination for proteolytic hydrolysis, but it also promotes the polyubiquitin chains linked to K63, K27 and K33 (Yin et al., 2020).
ITCH and DDR
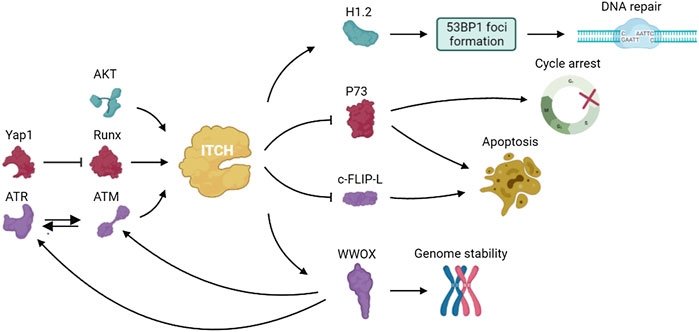
P73, a member of the p53 family of transcription factors, is up-regulated in response to DNA damage and induces cell cycle arrest and apoptosis. In 2005, Rossi found that ITCH bound to and ubiquitinated p73, leading to rapid proteasome-dependent degradation of p73 (Rossi et al., 2005). Studies have shown that YES-associated protein (Yap1) binds to p73 and enables p73 to escape ITCH-mediated ubiquitination by competing with ITCH (Levy et al., 2007). Under normal conditions, Yap1 enhances transcriptional activation of ITCH through Runx binding sites and causes degradation of p73. Yap1 is phosphorylated by c-abl at Tyr357 in response to DNA damage and failed to activate Runx and promote ITCH transcription (Levy et al., 2008a). The resulting decreased ITCH level leads to accumulation of p73 (Levy et al., 2008b). Hansen demonstrated that the ubiquitination of p73 mediated by ITCH played an important role in regulating DNA damage–induced apoptosis. ITCH knockout increased apoptosis in response to DNA damage agents used in chemotherapy, and reintroduction of ITCH into fibroblasts from ITCH-deficient mice reduced cell death after DNA damage (Hansen et al., 2007). These results indicated that inhibition of ITCH activity was important to modulate the chemosensitivity of DNA damage agents and targeting ITCH may help potentiate the effect of chemotherapeutic drugs on cancers.
ATM, which is induced by DNA damage, regulates cellular responses mainly through the phosphorylation of downstream target proteins (Shiloh, 2003; Bhatti et al., 2011). Santini et al. found that the S161 residue in ITCH was necessary for ATM-dependent ITCH activation. ATM enhanced ITCH enzyme activity and promoted the ubiquitination and degradation of c-FLIP-L (Gao et al., 2004; Chang et al., 2006), which participates in DDR and the modulation of death receptor signaling (Santini et al., 2014).
WW domain oxidoreductase (WWOX) is a tumor suppressor that spans the common fragile site FRA16D on chromosomes and plays an important role in DDR (Bednarek et al., 2000). WWOX deletion results in impaired DNA repair. WWOX is ubiquitinated at lysine 274 by ITCH and interacts with ATM when DNA single strand breaks occurs, thereby regulating ATR checkpoint and leading to cell cycle arrest to repair damaged DNA (Abu-Odeh et al., 2014; Abu-Odeh et al., 2015). These data indicated that regulation of WWOX by ITCH plays an important role in genome stability and clonal expansion of neoplastic cells.
Down-regulation of the DDR can enable the uncontrolled proliferation of invasive tumors. Studies showed that ITCH is highly expressed in triple negative breast cancer (TNBC) and modulated DDR in TNBC (Chang et al., 2019). Phosphorylation of ITCH at Ser257 by AKT leads to the nuclear localization of ITCH and the ubiquitination of H1.2. ITCH mediates H1.2 polyubiquitination and inhibits the formation of 53BP1 foci, which play a part in cell cycle checkpoints and DNA damage repair (Thorslund et al., 2015). These results demonstrated that the AKT-activated ITCH-H1.2 axis may lead to DDR inhibition in TNBC cells, which can offset replication stress and improve the survival and growth potential of tumor cells (Figure 4).
WWP1
WWP1 function
The gene encoding human E3 ubiquitin ligase WWP1 is located at 8q21 (Chen et al., 2007). WWP1 is a 922 amino acid protein that contains one C2 domain, four WW domains and one HECT domain (Zhi and Chen, 2012). The C2 domain determines the subcellular localization of the molecule, while the WW domain binds to the proline-rich sequence (PY motif) of the substrate protein (Hoshino et al., 2020). Studies have shown that WWP1 regulates a variety of cellular biological processes such as protein transport and degradation, signal transduction and transcription (Jia et al., 2021).
WWP1 and DDR
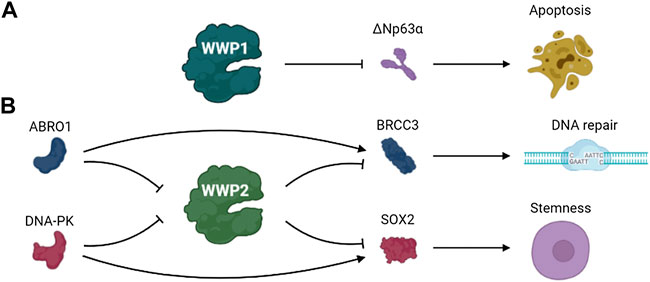
ΔNp63α, an isomer of the protein encoded by p63 gene, is homologous to p53 and plays a role in cell survival and proliferation (Mills et al., 1999; Kurinna et al., 2021; Xu et al., 2021). Previous studies identified ΔNp63α as a ubiquitination substrate of WWP1 (Li and Xiao, 2014). DNA damage stimulates WWP1 transcription, and WWP1 knockout eliminated the down-regulation of ΔNp63α expression induced by DNA damage and salvaged the apoptosis caused by DNA damage (Chen et al., 2017). The expression of WWP1 is up-regulated under the stimulation of DNA damage chemotherapeutic drugs. These results indicate that WWP1 may be involved in the effects of chemotherapeutic drugs (Li et al., 2008b) (Figure 5A).
WWP2
WWP2 function
In 1997, Wood and others identified WWP2 as binding to atrophin-1 by yeast two-hybrid and vitro binding analysis and named WWP2 as atrophin-1 interacting protein 2 (AIP2) (Zhang et al., 2019). The same gene locus produces three protein subtypes: full-length WWP2 (WWP2-FL, 870 AA), N-terminal WWP2-N (WWP2-N, 336 AA), and C-terminal WWP2-C (WWP2-C, 440 AA) (Soond and Chantry, 2011; Chen et al., 2014). WWP2 is widely expressed in the heart, placenta, lungs, liver, muscles, kidneys, pancreas and brain (Xu et al., 2009). Several substrates of WWP2 have been identified such as Oct4 (Liao and Jin, 2010), PTEN (Fang et al., 2020), TIRF (Yang et al., 2013), Septin4 (Zhang et al., 2020b) and PARP1 (Zhang et al., 2020a). WWP2 regulates various cellular physiological processes by regulating ubiquitin-dependent degradation of these proteins (Shao et al., 2016; Mokuda et al., 2019; Zhang et al., 2019). Identification of new substrates for WWP2 will expand the understanding of its physiological functions (Zhang et al., 2021c).
WWP2 and DDR
BRCA1/BRCA2-containing complex subunit 3 (BRCC3) is a lysine 63–specific deubiquitinating enzyme that is involved in various biological processes such as DNA damage repair (Ng et al., 2016; Rabl et al., 2019). WWP2 regulates the ubiquitination and degradation of BRCC3, thus regulating the functions of BRCC3. ABRO1, a subunit of the BRCC36 isopeptidase complex (BRISC), prevents WWP2-mediated BRCC3 ubiquitination and enhances BRCC3 stability by competing with WWP2 to bind to BRCC3. Thus, the stability of BRCC3 regulated by WWP2 and ABRO1 may play a role in DDR (Zhang et al., 2021c).
Glioblastoma, a lethal primary brain tumor, contains glioma stem cells that promote malignant progression and therapeutic resistance. SOX2 plays crucial roles in maintaining the self-renewal potential of normal stem cells in glioblastoma (Masui et al., 2007; Kim et al., 2008; Boumahdi et al., 2014; Zhang et al., 2020d). S251 of SOX2 is phosphorylated by DNA-PK under normal conditions, preventing the ubiquitination and degradation of SOX2 mediated by WWP2 and maintaining the stem status of glioma stem cells. The separation of DNA-PK from SOX2 because of DNA double-strand breaks promoted WWP2 binding with SOX2 and failed to stabilize SOX by preventing the ubiquitination and degradation of SOX2 mediated by WWP2, thus promoting the differentiation of glioma stem cells (Fang et al., 2021). These data indicate that DNA damage triggers glioma stem cell differentiation through precise regulation of SOX2 stability by DNA-PK and WWP2, suggesting this pathway may be a possible therapeutic target of glioblastoma (Figure 5B).
Smurf1
Smurf1 function
The human SMURF1 gene is located on chromosome 7q22.1 (Xia et al., 2021). Smurf1 is usually found in the cytoplasm, but sometimes localizes in the plasma membrane or nucleus by interacting with other proteins (Suzuki et al., 2002; Horiki et al., 2004). Smurf1 contains one C2 domain, two WW domains (WW1 and WW2) and one HECT domain. Smurf1 substrates are ubiquitinated either through direct interactions between the PY motifs in substrates and the WW domain or through indirect interactions mediated by adaptor proteins (Cao and Zhang, 2013). Nearly 40 substrate proteins of Smurf1 have been discovered. Smurf1 participates in many processes including tumorigenesis, embryogenesis, regulation of cell localization and maintenance of homeostasis through the ubiquitin proteasome pathway by targeting substrate proteins (Shimazu et al., 2016; Feng et al., 2019; Scott et al., 2020).
Smurf1 and DDR
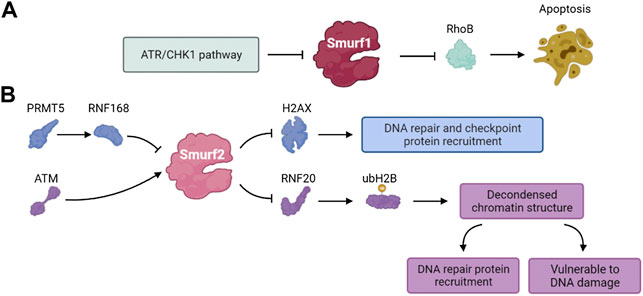
The Rho GTPase family of proteins are key regulators of actin cytoskeletal dynamics and participate in regulating the cell cycle, gene expression, vesicle transport and cell polarity (Kaibuchi et al., 1999; Bustelo et al., 2007). RhoB is a member of the Rho GTPase family (Gutierrez et al., 2019). Some studies have shown that RhoB deficiency significantly reduced the apoptosis of transformed cells induced by DNA damage (Liu et al., 2001). The ATR/Chk1 signaling pathway, activated by DNA damage, mediates the phosphorylation and self-degradation of Smurf1, leading to the accumulation of RhoB and promoting cell apoptosis (Wang et al., 2014). These results suggest that Smurf1 is involved in the DNA damage response and regulates various cell functions (Figure 6A).
Smurf2
Smurf2 function
The human Smurf2 gene is located on chromosome 17. Similar to other members of the NEDD4 family, Smurf2 contains one C2 domain, three WW domains with two conserved tryptophan residues each and one HECT domain (Bai and Ying, 2020). The WW domain is responsible for substrate recognition through specific binding to the PPXY motif (Lin et al., 2000). Smurf2 was initially found to negatively regulate TGF-β/BMP signaling by ubiquitination and degradation of substrates, preventing overactivation of TGF-β/BMP signaling (Kushioka et al., 2020). Smurf2 also contributes to genomic stability, cell polarity, tissue homeostasis, embryogenesis and tumorigenesis (Koganti et al., 2018; Bai and Ying, 2020; Shepley-McTaggart et al., 2021).
Smurf2 and DDR
Phosphorylation of histone H2AX (γH2AX) and localization of γH2AX to DSB sites play a vital role in the DDR. The γH2AX foci formation functions as recruiting other DDR effectors and is a mark of early step of DDR (Celeste et al., 2003; Yuan et al., 2010; Machitani et al., 2020). Du et al. found that the PRMT5-RNF168-Smurf2 cascade regulated the protein stability of H2AX and confirmed Smurf2 was essential to DDR. RNF168 and SMURF2 regulate the expression of H2AX as stabilizers and destabilizers of H2AX, respectively. Inhibition of PRMT5 attenuates the expression of RNF168 and causes Smurf2 to destabilize H2AX by ubiquitination and degradation. Thus, Smurf2 regulates the levels of H2AX in the PRMT5- RNF168-SmurF2 cascade and the related effect in the process of DDR (Du et al., 2019).
Genomic ablation of Smurf2 results in dysregulation of the DNA damage response and genomic stability. Smurf2 is phosphorylated by ATM at S384 when DNA damage occurs, and its phosphorylation is required for its interaction with ring finger protein 20 (RNF20) (Blank et al., 2012). Smurf2-mediated ubiquitination and degradation of RNF20 contributes to a negative feedback loop that regulates DSB repair. The mono-ubiquitination of H2B mediated by RNF20 relaxes chromatin, which plays dual roles in DDR. The SMURF2-mediated RNF20 ubiquitination leads to down-regulation of H2B, and the chromatin compaction protects cells from DNA damage but disturbs the recruitment of DNA repair proteins (Tang et al., 2020). Ayyathan et al. also demonstrated that the inactivation of Smurf2 triggered a variety of changes in cellular activities including cell mobility, self-replication and DNA damage repair (Manikoth Ayyathan et al., 2020). Thus, Smurf2 is a key regulator of the DDR, chromatin structure regulation and genome integrity maintenance (Figure 6B).
Conclusion and prospects
The DDR functions to maintain the integrity of the genome is necessary for the growth and survival of cells and organisms. Activating DDR signaling pathways promote cellular homeostasis and survival in health and disease (Chatzidoukaki et al., 2020). While, an impaired DDR is related to some disease such as autosomal dominant polycystic kidney disease and Alzheimer’s disease (Farmer et al., 2020; Zhang et al., 2021b). An abnormal DDR process leads to a change of cellular activities facing to stimulus, which provides a clue for treating some diseases. In this review, we summarize the recent literature on the function and regulation of NEDD4 family members in DDR (Table 1). It showed NEDD4 family play important roles in maintaining genome integrity and stability by regulating different substrate proteins and participating in various pathways involved in DDR. Therefore, identification of new target proteins of the NEDD4 family may help elucidate more mechanisms of DDR, which may provide an idea to the study of cellular homeostasis and the balance between health and disease, and contribute to the further study of pathophysiology for certain diseases and pharmacology for targeted drugs.
Author contributions
Conceptualization, XL, NZ, and LZ; writing—original draft preparation, XL; writing—review and editing, XL, HX, JX, SL, SY, XH, NZ, and LZ. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by National Natural Science Foundation of China (Grant No. 82000753, No. 82171572, 82171571, 81900355, 81900372, 81970211), National Postdoctoral Innovative Talents Support Program (BX2021376).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abu-Odeh M., Hereema N. A., Aqeilan R. I. (2015). WWOX modulates the ATR-mediated DNA damage checkpoint response. Oncotarget 7 (4), 4344–4355. doi:10.18632/oncotarget.6571
Abu-Odeh M., Salah Z., Herbel C., Hofmann T. G., Aqeilan R. I. (2014). WWOX, the common fragile site FRA16D gene product, regulates ATM activation and the DNA damage response. Proc. Natl. Acad. Sci. U. S. A. 111 (44), E4716–E4725. doi:10.1073/pnas.1409252111
Al-Hakim A. K., Zagorska A., Chapman L., Deak M., Peggie M., Alessi D. R. (2008). Control of AMPK-related kinases by USP9X and atypical Lys29/Lys33-linked polyubiquitin chains. Biochem. J. 411 (2), 249–260. doi:10.1042/bj20080067
Anan T., Nagata Y., Koga H., Honda Y., Yabuki N., Miyamoto C., et al. (1998). Human ubiquitin-protein ligase Nedd4: Expression, subcellular localization and selective interaction with ubiquitin-conjugating enzymes. Genes Cells 3, 751–763. doi:10.1046/j.1365-2443.1998.00227.x
Araki N., Umemura M., Miyagi Y., Yabana M., Miki Y., Tamura K., et al. (2008). Expression, transcription, and possible antagonistic interaction of the human Nedd4L gene variant. Hypertension 51 (3), 773–777. doi:10.1161/hypertensionaha.107.102061
Bai Y., Ying Y. (2020). The post-translational modifications of Smurf2 in TGF-β signaling. Front. Mol. Biosci. 7, 128. doi:10.3389/fmolb.2020.00128
Beaudenon S. L., Huacani M. R., Wang G., McDonnell D. P., Huibregtse J. M. (1999). Rsp5 ubiquitin-protein ligase mediates DNA damage-induced degradation of the large subunit of RNA polymerase II in Saccharomyces cerevisiae. Mol. Cell Biol. 19 (10), 6972–6979. doi:10.1128/mcb.19.10.6972
Bednarek A., Laflin K., Daniel R., Liao Q., Hawkins K., Aldaz C. M. (2000). WWOX, a novel WW domain-containing protein mapping to human chromosome 16q23.3-24.1, a region frequently affected in breast cancer. Cancer Res. 60 (8), 2140–2145.
Bennett E. J., Harper J. W. (2008). DNA damage: Ubiquitin marks the spot. Nat. Struct. Mol. Biol. 15 (1), 20–22. doi:10.1038/nsmb0108-20
Bhatti S., Kozlov S., Farooqi A. A., Naqi A., Lavin M., Khanna K. K. (2011). ATM protein kinase: The linchpin of cellular defenses to stress. Cell. Mol. Life Sci. 68 (18), 2977–3006. doi:10.1007/s00018-011-0683-9
Blackford A. N., Jackson S. P. (2017). ATM, ATR, and DNA-PK: The trinity at the heart of the DNA damage response. Mol. Cell 66 (6), 801–817. doi:10.1016/j.molcel.2017.05.015
Blank M., Tang Y., Yamashita M., Burkett S. S., Cheng S. Y., Zhang Y. E. (2012). A tumor suppressor function of Smurf2 associated with controlling chromatin landscape and genome stability through RNF20. Nat. Med. 18 (2), 227–234. doi:10.1038/nm.2596
Boiteux S., Radicella J. P. (2000). The human OGG1 gene: Structure, functions, and its implication in the process of carcinogenesis. Archives Biochem. Biophysics 377 (1), 1–8. doi:10.1006/abbi.2000.1773
Boumahdi S., Driessens G., Lapouge G., Rorive S., Nassar D., Le Mercier M., et al. (2014). SOX2 controls tumour initiation and cancer stem-cell functions in squamous-cell carcinoma. Nature 511 (7508), 246–250. doi:10.1038/nature13305
Bustelo X. R., Sauzeau V., Berenjeno I. M. (2007). GTP-Binding proteins of the rho/rac family: Regulation, effectors and functions in vivo. BioEssays 29 (4), 356–370. doi:10.1002/bies.20558
Calvo O. (2020). RNA polymerase II phosphorylation and gene looping: New roles for the rpb4/7 heterodimer in regulating gene expression. Curr. Genet. 66 (5), 927–937. doi:10.1007/s00294-020-01084-w
Cao Y., Zhang L. (2013). Pharmaceutical perspectives of HECT-type ubiquitin ligase Smurf1. Cpd 19 (18), 3226–3233. doi:10.2174/1381612811319180007
Celeste A., Difilippantonio S., Difilippantonio M. J., Fernandez-Capetillo O., Pilch D. R., Sedelnikova O. A., et al. (2003). H2AX haploinsufficiency modifies genomic stability and tumor susceptibility. Cell 114 (3), 371–383. doi:10.1016/s0092-8674(03)00567-1
Chang L., Kamata H., Solinas G., Luo J. L., Maeda S., Venuprasad K., et al. (2006). The E3 ubiquitin ligase itch couples JNK activation to tnfα-induced cell death by inducing c-FLIPL turnover. Cell 124 (3), 601–613. doi:10.1016/j.cell.2006.01.021
Chang L., Shen L., Zhou H., Gao J., Pan H., Zheng L., et al. (2019). ITCH nuclear translocation and H1.2 polyubiquitination negatively regulate the DNA damage response. Nucleic Acids Res. 47 (2), 824–842. doi:10.1093/nar/gky1199
Chatzidoukaki O., Goulielmaki E., Schumacher B., Garinis G. A. (2020). DNA damage response and metabolic reprogramming in health and disease. Trends Genet. 36 (10), 777–791. doi:10.1016/j.tig.2020.06.018
Chen C., Matesic L. E. (2007). The Nedd4-like family of E3 ubiquitin ligases and cancer. Cancer Metastasis Rev. 26 (3-4), 587–604. doi:10.1007/s10555-007-9091-x
Chen C., Sun X., Guo P., Dong X. Y., Sethi P., Zhou W., et al. (2007). Ubiquitin E3 ligase WWP1 as an oncogenic factor in human prostate cancer. Oncogene 26 (16), 2386–2394. doi:10.1038/sj.onc.1210021
Chen H., Ross C. A., Wang N., Huo Y., MacKinnon D. F., Potash J. B., et al. (2001). NEDD4L on human chromosome 18q21 has multiple forms of transcripts and is a homologue of the mouse Nedd4-2 gene. Eur. J. Hum. Genet. 9 (9), 922–930. doi:10.1038/sj.ejhg.5200747
Chen J., Shi H., Chen Y., Fan S., Liu D., Li C. (2017). DNA damage induces expression of WWP1 to target ΔNp63α to degradation. PLoS One 12 (4), e0176142. doi:10.1371/journal.pone.0176142
Chen W., Jiang X., Luo Z. (2014). WWP2: A multifunctional ubiquitin ligase gene. Pathol. Oncol. Res. 20 (4), 799–803. doi:10.1007/s12253-014-9838-y
Chung H. Y., Morita E., von Schwedler U., Müller B., Kräusslich H. G., Sundquist W. I. (2008). NEDD4L overexpression rescues the release and infectivity of human immunodeficiency virus type 1 constructs lacking PTAP and YPXL late domains. J. Virol. 82 (10), 4884–4897. doi:10.1128/jvi.02667-07
Cruz Walma D. A., Chen Z., Bullock A. N., Yamada K. M. (2022). Ubiquitin ligases: Guardians of mammalian development. Nat. Rev. Mol. Cell Biol. 23 (5), 350–367. doi:10.1038/s41580-021-00448-5
Cui J., Shu C., Xu J., Chen D., Li J., Ding K., et al. (2020). JP1 suppresses proliferation and metastasis of melanoma through MEK1/2 mediated NEDD4L-SP1-Integrin αvβ3 signaling. Theranostics 10 (18), 8036–8050. doi:10.7150/thno.45843
d'Azzo A., Bongiovanni A., Nastasi T. (2005). E3 ubiquitin ligases as regulators of membrane protein trafficking and degradation. Traffic 6 (6), 429–441. doi:10.1111/j.1600-0854.2005.00294.x
Dodson E. J., Fishbain-Yoskovitz V., Rotem-Bamberger S., Schueler-Furman O. (2015). Versatile communication strategies among tandem WW domain repeats. Exp. Biol. Med. (Maywood) 240 (3), 351–360. doi:10.1177/1535370214566558
Du C., Hansen L. J., Singh S. X., Wang F., Sun R., Moure C. J., et al. (2019). A PRMT5-rnf168-SMURF2 Axis controls H2AX proteostasis. Cell Rep. 28 (12), 3199–3211. doi:10.1016/j.celrep.2019.08.031
Dunn D. M., Ishigami T., Pankow J., Niederhausern Av, Alder J., Hunt S. C., et al. (2002). Common variant of human NEDD4L activates a cryptic splice site to form a frameshifted transcript. J. Hum. Genet. 47, 665–676. doi:10.1007/s100380200102
Fagerberg L., Hallström B. M., Oksvold P., Kampf C., Djureinovic D., Odeberg J., et al. (2014). Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell. Proteomics 13 (2), 397–406. doi:10.1074/mcp.m113.035600
Fang S., Jensen J. P., Ludwig R. L., Vousden K. H., Weissman A. M. (2000). Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J. Biol. Chem. 275 (12), 8945–8951. doi:10.1074/jbc.275.12.8945
Fang S., Zhang D., Weng W., Lv X., Zheng L., Chen M., et al. (2020). CPSF7 regulates liver cancer growth and metastasis by facilitating WWP2-FL and targeting the WWP2/PTEN/AKT signaling pathway. Biochimica Biophysica Acta (BBA) - Mol. Cell Res. 1867 (2), 118624. doi:10.1016/j.bbamcr.2019.118624
Fang X., Huang Z., Zhai K., Huang Q., Tao W., Kim L., et al. (2021). Inhibiting DNA-PK induces glioma stem cell differentiation and sensitizes glioblastoma to radiation in mice. Sci. Transl. Med. 13 (600), 1–14. doi:10.1126/scitranslmed.abc7275
Farmer K. M., Ghag G., Puangmalai N., Montalbano M., Bhatt N., Kayed R. (2020). P53 aggregation, interactions with tau, and impaired DNA damage response in Alzheimer's disease. Acta Neuropathol. Commun. 8 (1), 132. doi:10.1186/s40478-020-01012-6
Feng X., Jia Y., Zhang Y., Ma F., Zhu Y., Hong X., et al. (2019). Ubiquitination of UVRAG by SMURF1 promotes autophagosome maturation and inhibits hepatocellular carcinoma growth. Autophagy 15 (7), 1130–1149. doi:10.1080/15548627.2019.1570063
Fleming A. M., Ding Y., Burrows C. J. (2017). Oxidative DNA damage is epigenetic by regulating gene transcription via base excision repair. Proc. Natl. Acad. Sci. U.S.A. 114 (10), 2604–2609. doi:10.1073/pnas.1619809114
French M. E., Klosowiak J. L., Aslanian A., Reed S. I., Yates J. R., Hunter T. (2017). Mechanism of ubiquitin chain synthesis employed by a HECT domain ubiquitin ligase. J. Biol. Chem. 292 (25), 10398–10413. doi:10.1074/jbc.m117.789479
Fukushima T., Yoshihara H., Furuta H., Kamei H., Hakuno F., Luan J., et al. (2015). Nedd4-induced monoubiquitination of IRS-2 enhances IGF signalling and mitogenic activity. Nat. Commun. 6, 6780. doi:10.1038/ncomms7780
Gao M., Labuda T., Xia Y., Gallagher E., Fang D., Liu Y-C., et al. (2004). Jun turnover is controlled through JNK-dependent phosphorylation of the E3 ligase Itch. Science 306, 271–275. doi:10.1126/science.1099414
Gao P., Ma X., Yuan M., Yi Y., Liu G., Wen M., et al. (2021). E3 ligase Nedd4l promotes antiviral innate immunity by catalyzing K29-linked cysteine ubiquitination of TRAF3. Nat. Commun. 12 (1), 1194. doi:10.1038/s41467-021-21456-1
Goel P., Manning J. A., Kumar S. (2015). NEDD4-2 (NEDD4L): The ubiquitin ligase for multiple membrane proteins. Gene 557 (1), 1–10. doi:10.1016/j.gene.2014.11.051
Goff P. H., Bhakuni R., Pulliam T., Lee J. H., Hall E. T., Nghiem P. (2021). Intersection of two checkpoints: Could inhibiting the DNA damage response checkpoint rescue immune checkpoint-refractory cancer? Cancers (Basel) 13 (14). doi:10.3390/cancers13143415
Goto Y., Koyasu S., Kobayashi M., Harada H. (2017). The emerging roles of the ubiquitination/deubiquitination system in tumor radioresistance regarding DNA damage responses, cell cycle regulation, hypoxic responses, and antioxidant properties: Insight into the development of novel radiosensitizing strategies. Mutat. Research/Fundamental Mol. Mech. Mutagen. 803-805, 76–81. doi:10.1016/j.mrfmmm.2017.07.007
Gutierrez E., Cahatol I., Bailey C. A. R., Lafargue A., Zhang N., Song Y., et al. (2019). Regulation of RhoB gene expression during tumorigenesis and aging process and its potential applications in these processes. Cancers (Basel) 11 (6). doi:10.3390/cancers11060818
Hansen T. M., Rossi M., Roperch J. P., Ansell K., Simpson K., Taylor D., et al. (2007). Itch inhibition regulates chemosensitivity in vitro. Biochem. Biophysical Res. Commun. 361 (1), 33–36. doi:10.1016/j.bbrc.2007.06.104
Harvey K. F., Dinudom A., Cook D. I., Kumar S. (2001). The Nedd4-like protein KIAA0439 is a potential regulator of the epithelial sodium channel. J. Biol. Chem. 276 (11), 8597–8601. doi:10.1074/jbc.c000906200
Harvey K. F., Kumar S. (1999). Nedd4-like proteins: An emerging family of ubiquitin-protein ligases implicated in diverse cellular functions. Trends Cell Biol. 9 (5), 166–169. doi:10.1016/s0962-8924(99)01541-x
Horiki M., Imamura T., Okamoto M., Hayashi M., Murai J., Myoui A., et al. (2004). Smad6/Smurf1 overexpression in cartilage delays chondrocyte hypertrophy and causes dwarfism with osteopenia. J. Cell Biol. 165 (3), 433–445. doi:10.1083/jcb.200311015
Hoshino S., Kobayashi M., Tagawa R., Konno R., Abe T., Furuya K., et al. (2020). WWP1 knockout in mice exacerbates obesity‐related phenotypes in white adipose tissue but improves whole‐body glucose metabolism. FEBS Open Bio 10 (3), 306–315. doi:10.1002/2211-5463.12795
Hu L., Wang Z., Carmone C., Keijer J., Zhang D. (2021). Role of oxidative DNA damage and repair in atrial fibrillation and ischemic heart disease. Int. J. Mol. Sci. 22 (8). doi:10.3390/ijms22083838
Huang X., Chen J., Cao W., Yang L., Chen Q., He J., et al. (2019). The many substrates and functions of NEDD4-1. Cell Death Dis. 10 (12). doi:10.1038/s41419-019-2142-8
Hughes J. R., Parsons J. L. (2020). The E3 ubiquitin ligase NEDD4L targets OGG1 for ubiquitylation and modulates the cellular DNA damage response. Front. Cell Dev. Biol. 8, 607060. doi:10.3389/fcell.2020.607060
Ikeda F., Dikic I. (2008). Atypical ubiquitin chains: New molecular signals. 'Protein modifications: Beyond the usual suspects' review series. EMBO Rep. 9 (6), 536–542. doi:10.1038/embor.2008.93
Infante P., Lospinoso Severini L., Bernardi F., Bufalieri F., Di Marcotullio L. (2019). Targeting hedgehog signalling through the ubiquitylation process: The multiple roles of the HECT-E3 ligase itch. Cells 8 (2). doi:10.3390/cells8020098
Ingham R. J., Gish G., Pawson T. (2004). The Nedd4 family of E3 ubiquitin ligases: Functional diversity within a common modular architecture. Oncogene 23 (11), 1972–1984. doi:10.1038/sj.onc.1207436
Jia X., Chen H., Ren Y., Dejizhuoga G., Gesangyuzhen N., Gao N., et al. (2021). BAP1 antagonizes WWP1-mediated transcription factor KLF5 ubiquitination and inhibits autophagy to promote melanoma progression. Exp. Cell Res. 402 (1), 112506. doi:10.1016/j.yexcr.2021.112506
Kaibuchi K., Kuroda S., Amano M. (1999). Regulation of the cytoskeleton and cell adhesion by the Rho family GTPases in mammalian cells. Annu. Rev. Biochem. 68 (68), 459–486. doi:10.1146/annurev.biochem.68.1.459
Kim J. B., Kim S. Y., Kim B. M., Lee H., Kim I., Yun J., et al. (2013). Identification of a novel anti-apoptotic E3 ubiquitin ligase that ubiquitinates antagonists of inhibitor of apoptosis proteins SMAC, HtrA2, and ARTS. J. Biol. Chem. 288 (17), 12014–12021. doi:10.1074/jbc.m112.436113
Kim J. B., Zaehres H., Wu G., Gentile L., Ko K., Sebastiano V., et al. (2008). Pluripotent stem cells induced from adult neural stem cells by reprogramming with two factors. Nature 454 (7204), 646–650. doi:10.1038/nature07061
Kiss R. C., Xia F., Acklin S. (2021). Targeting DNA damage response and repair to enhance therapeutic index in cisplatin-based cancer treatment. Int. J. Mol. Sci. 22 (15). doi:10.3390/ijms22158199
Koganti P., Levy-Cohen G., Blank M. (2018). Smurfs in protein homeostasis, signaling, and cancer. Front. Oncol. 8, 295. doi:10.3389/fonc.2018.00295
Kumar S., Harvey K. F., Kinoshita M., Copeland N. G., Noda M., Jenkins N. A. (1997). cDNA cloning, expression analysis, and mapping of the MouseNedd4Gene. GENOMICS 40, 435–443. doi:10.1006/geno.1996.4582
Kumar S., Tomooka Y., Noda M. (1992). Identification of a set of genes with developmentally down-regulated expression in the mouse brain. Biochem. BIOPHYSICAL Res. Commun. 185 (3), 1155–1161. doi:10.1016/0006-291x(92)91747-e
Kurinna S., Seltmann K., Bachmann A. L., Schwendimann A., Thiagarajan L., Hennig P., et al. (2021). Interaction of the NRF2 and p63 transcription factors promotes keratinocyte proliferation in the epidermis. Nucleic Acids Res. 49 (7), 3748–3763. doi:10.1093/nar/gkab167
Kushioka J., Kaito T., Okada R., Ishiguro H., Bal Z., Kodama J., et al. (2020). A novel negative regulatory mechanism of Smurf2 in BMP/Smad signaling in bone. Bone Res. 8 (1), 41. doi:10.1038/s41413-020-00115-z
Lai Z., Ferry K. V., Diamond M. A., Wee K. E., Kim Y. B., Ma J., et al. (2001). Human mdm2 mediates multiple mono-ubiquitination of p53 by a mechanism requiring enzyme isomerization. J. Biol. Chem. 276 (33), 31357–31367. doi:10.1074/jbc.m011517200
Laine A., Ronai Z. (2007). Regulation of p53 localization and transcription by the HECT domain E3 ligase WWP1. Oncogene 26 (10), 1477–1483. doi:10.1038/sj.onc.1209924
Levy D., Adamovich Y., Reuven N., Shaul Y. (2007). The Yes-associated protein 1 stabilizes p73 by preventing Itch-mediated ubiquitination of p73. Cell Death Differ. 14 (4), 743–751. doi:10.1038/sj.cdd.4402063
Levy D., Adamovich Y., Reuven N., Shaul Y. (2008a). Yap1 phosphorylation by c-Abl is a critical step in selective activation of proapoptotic genes in response to DNA damage. Mol. Cell 29 (3), 350–361. doi:10.1016/j.molcel.2007.12.022
Levy D., Reuven N., Shaul Y. (2008b). A regulatory circuit controlling Itch-mediated p73 degradation by Runx. J. Biol. Chem. 283 (41), 27462–27468. doi:10.1074/jbc.m803941200
Li C., Xiao Z. X. (2014). Regulation of p63 protein stability via ubiquitin-proteasome pathway. Biomed. Res. Int. 2014, 175721. doi:10.1155/2014/175721
Li Y., Chen X., Wang Z., Zhao D., Chen H., Chen W., et al. (2013). The HECTD3 E3 ubiquitin ligase suppresses cisplatin-induced apoptosis via stabilizing MALT1. Neoplasia 15 (1), 39–IN15. doi:10.1593/neo.121362
Li Y., Ge Y. Z., Xu L., Jia R. (2020). Circular RNA ITCH: A novel tumor suppressor in multiple cancers. Life Sci. 254, 117176. doi:10.1016/j.lfs.2019.117176
Li Y., Ozaki T., Kikuchi H., Yamamoto H., Ohira M., Nakagawara A. (2008). A novel HECT-type E3 ubiquitin protein ligase NEDL1 enhances the p53-mediated apoptotic cell death in its catalytic activity-independent manner. Oncogene 27 (26), 3700–3709. doi:10.1038/sj.onc.1211032
Li Y., Zhou Z., Chen C. (2008). WW domain-containing E3 ubiquitin protein ligase 1 targets p63 transcription factor for ubiquitin-mediated proteasomal degradation and regulates apoptosis. Cell Death Differ. 15 (12), 1941–1951. doi:10.1038/cdd.2008.134
Liao B., Jin Y. (2010). Wwp2 mediates Oct4 ubiquitination and its own auto-ubiquitination in a dosage-dependent manner. Cell Res. 20 (3), 332–344. doi:10.1038/cr.2009.136
Lin X., Liang M., Feng X. H. (2000). Smurf2 is a ubiquitin E3 ligase mediating proteasome-dependent degradation of Smad2 in transforming growth factor-β signaling. J. Biol. Chem. 275 (47), 36818–36822. doi:10.1074/jbc.c000580200
Liu A-x., Cerniglia G. J., Bernhard E. J., Prendergast G. C. (2001). RhoB is required to mediate apoptosis in neoplastically transformed cells after DNA damage. Proc. Natl. Acad. Sci. U.S.A. 98 (11), 6192–6197. doi:10.1073/pnas.111137198
Liu Y., Duan C., Zhang C. (2021). E3 ubiquitin ligase in anticancer drugdsla resistance: Recent advances and future potential. Front. Pharmacol. 12, 645864. doi:10.3389/fphar.2021.645864
Liu Y. C. (2004). Ubiquitin ligases and the immune response. Annu. Rev. Immunol. 22, 81–127. doi:10.1146/annurev.immunol.22.012703.104813
Lu P. J., Zhou X. Z., Shen M., Lu K. P. (1999). Function of WW domains as phosphoserine- or phosphothreonine-binding modules. Science 283 (5406), 1325–1328. doi:10.1126/science.283.5406.1325
Machitani M., Taniguchi I., McCloskey A., Suzuki T., Ohno M. (2020). The RNA transport factor PHAX is required for proper histone H2AX expression and DNA damage response. RNA 26 (26), 1716–1725. doi:10.1261/rna.074625.120
Manikoth Ayyathan D., Koganti P., Marcu-Malina V., Litmanovitch T., Trakhtenbrot L., Emanuelli A., et al. (2020). SMURF2 prevents detrimental changes to chromatin, protecting human dermal fibroblasts from chromosomal instability and tumorigenesis. Oncogene 39 (16), 3396–3410. doi:10.1038/s41388-020-1226-3
Manning J. A., Kumar S. (2018). Physiological functions of nedd4-2: Lessons from knockout mouse models. Trends Biochem. Sci. 43 (8), 635–647. doi:10.1016/j.tibs.2018.06.004
Manning J. A., Shah S. S., Nikolic A., Henshall T. L., Khew-Goodall Y., Kumar S. (2021). The ubiquitin ligase NEDD4-2/NEDD4L regulates both sodium homeostasis and fibrotic signaling to prevent end-stage renal disease. Cell Death Dis. 12 (4), 398. doi:10.1038/s41419-021-03688-7
Masui S., Nakatake Y., Toyooka Y., Shimosato D., Yagi R., Takahashi K., et al. (2007). Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells. Nat. Cell Biol. 9 (6), 625–635. doi:10.1038/ncb1589
Mathieu N. A., Levin R. H., Spratt D. E. (2021). Exploring the roles of HERC2 and the NEDD4L HECT E3 ubiquitin ligase subfamily in p53 signaling and the DNA damage response. Front. Oncol. 11, 659049. doi:10.3389/fonc.2021.659049
Michnov O., Solomayer E., Fehm T., Stubenrauch F., Iftner T. (2012). Knock down of p53 or its ubiquitin ligase E6AP does not affect the sensitivity of human papillomavirus-positive cervical cancer cells to cisplatin. Am. J. Cancer Res. 2 (3), 309–321.
Mills A. A., Zheng B., Wang X-J., Vogel H., Roop D. R., Bradley A. (1999). p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature 398, 708–713. doi:10.1038/19531
Mohiuddin , Kobayashi S., Keka I. S., Guilbaud G., Sale J., Narita T., et al. (2016). The role of HERC2 and RNF8 ubiquitin E3 ligases in the promotion of translesion DNA synthesis in the chicken DT40 cell line. DNA Repair 40, 67–76. doi:10.1016/j.dnarep.2016.02.002
Mokuda S., Nakamichi R., Matsuzaki T., Ito Y., Sato T., Miyata K., et al. (2019). Wwp2 maintains cartilage homeostasis through regulation of Adamts5. Nat. Commun. 10 (1), 2429. doi:10.1038/s41467-019-10177-1
Molinaro C., Martoriati A., Cailliau K. (2021). Proteins from the DNA damage response: Regulation, dysfunction, and anticancer strategies. Cancers (Basel) 13 (15). doi:10.3390/cancers13153819
Murillas R., Simms K. S., Hatakeyama S., Weissman A. M., Kuehn M. R. (2002). Identification of developmentally expressed proteins that functionally interact with Nedd4 ubiquitin ligase. J. Biol. Chem. 277 (4), 2897–2907. doi:10.1074/jbc.m110047200
Ng H. M., Wei L., Lan L., Huen M. S. (2016). The lys63-deubiquitylating enzyme BRCC36 limits DNA break processing and repair. J. Biol. Chem. 291 (31), 16197–16207. doi:10.1074/jbc.m116.731927
Perkhofer L., Golan T., Cuyle P. J., Matysiak-Budnik T., Van Laethem J. L., Macarulla T., et al. (2021). Targeting DNA damage repair mechanisms in pancreas cancer. Cancers (Basel) 13 (17). doi:10.3390/cancers13174259
Plant P. J., Yeger H., Staub O., Howard P., Rotin D. (1997). The C2 domain of the ubiquitin protein ligase Nedd4 mediates Ca2+-dependent plasma membrane localization. J. Biol. Chem. 272 (51), 32329–32336. doi:10.1074/jbc.272.51.32329
Putz U., Howitt J., Lackovic J., Foot N., Kumar S., Silke J., et al. (2008). Nedd4 family-interacting protein 1 (Ndfip1) is required for the exosomal secretion of Nedd4 family proteins. J. Biol. Chem. 283 (47), 32621–32627. doi:10.1074/jbc.m804120200
Qie S. (2022). The E3 ubiquitin ligase Fbxo4 functions as a tumor suppressor: Its biological importance and therapeutic perspectives. Cancers (Basel) 14 (9). doi:10.3390/cancers14092133
Rabl J., Bunker R. D., Schenk A. D., Cavadini S., Gill M. E., Abdulrahman W., et al. (2019). Structural basis of BRCC36 function in DNA repair and immune regulation. Mol. Cell 75 (3), 483–497. doi:10.1016/j.molcel.2019.06.002
Rivas-Domínguez A., Pastor N., Martínez-López L., Colón-Pérez J., Bermúdez B., Orta M. L. (2021). The role of DNA damage response in dysbiosis-induced colorectal cancer. Cells 10 (8). doi:10.3390/cells10081934
Rossi M., De Laurenzi V., Munarriz E., Green D. R., Liu Y. C., Vousden K. H., et al. (2005). The ubiquitin-protein ligase Itch regulates p73 stability. EMBO J. 24 (4), 836–848. doi:10.1038/sj.emboj.7600444
Santini S., Stagni V., Giambruno R., Fianco G., Di Benedetto A., Mottolese M., et al. (2014). ATM kinase activity modulates ITCH E3-ubiquitin ligase activity. Oncogene 33 (9), 1113–1123. doi:10.1038/onc.2013.52
Scheffner M., Kumar S. (2014). Mammalian HECT ubiquitin-protein ligases: Biological and pathophysiological aspects. Biochimica Biophysica Acta (BBA) - Mol. Cell Res. 1843 (1), 61–74. doi:10.1016/j.bbamcr.2013.03.024
Scott J. L., Frick C. T., Johnson K. A., Liu H., Yong S. S., Varney A. G., et al. (2020). Molecular analysis of membrane targeting by the C2 domain of the E3 ubiquitin ligase Smurf1. Biomolecules 10 (2). doi:10.3390/biom10020229
Shao R., Liu J., Yan G., Zhang J., Han Y., Guo J., et al. (2016). Cdh1 regulates craniofacial development via APC-dependent ubiquitination and activation of Goosecoid. Cell Res. 26 (6), 699–712. doi:10.1038/cr.2016.51
Shepley-McTaggart A., Schwoerer M. P., Sagum C. A., Bedford M. T., Jaladanki C. K., Fan H., et al. (2021). Ubiquitin ligase SMURF2 interacts with filovirus VP40 and promotes egress of VP40 VLPs. Viruses 13 (2). doi:10.3390/v13020288
Shiloh Y. (2003). ATM and related protein kinases: Safeguarding genome integrity. Nat. Rev. Cancer 3 (3), 155–168. doi:10.1038/nrc1011
Shimazu J., Wei J., Karsenty G. (2016). Smurf1 inhibits osteoblast differentiation, bone formation, and glucose homeostasis through serine 148. Cell Rep. 15 (1), 27–35. doi:10.1016/j.celrep.2016.03.003
Sluimer J., Distel B. (2018). Regulating the human HECT E3 ligases. Cell. Mol. Life Sci. 75 (17), 3121–3141. doi:10.1007/s00018-018-2848-2
Somesh B. P., Reid J., Liu W. F., Søgaard T. M., Erdjument-Bromage H., Tempst P., et al. (2005). Multiple mechanisms confining RNA polymerase II ubiquitylation to polymerases undergoing transcriptional arrest. Cell 121 (6), 913–923. doi:10.1016/j.cell.2005.04.010
Soond S. M., Chantry A. (2011). Selective targeting of activating and inhibitory Smads by distinct WWP2 ubiquitin ligase isoforms differentially modulates TGFβ signalling and EMT. Oncogene 30 (21), 2451–2462. doi:10.1038/onc.2010.617
Suzuki C., Murakami G., Fukuchi M., Shimanuki T., Shikauchi Y., Imamura T., et al. (2002). Smurf1 regulates the inhibitory activity of Smad7 by targeting Smad7 to the plasma membrane. J. Biol. Chem. 277 (42), 39919–39925. doi:10.1074/jbc.m201901200
Tang L. Y., Thomas A., Zhou M., Zhang Y. E. (2020). Phosphorylation of SMURF2 by ATM exerts a negative feedback control of DNA damage response. J. Biol. Chem. 295 (52), 18485–18493. doi:10.1074/jbc.ra120.014179
Thorslund T., Ripplinger A., Hoffmann S., Wild T., Uckelmann M., Villumsen B., et al. (2015). Histone H1 couples initiation and amplification of ubiquitin signalling after DNA damage. Nature 527 (7578), 389–393. doi:10.1038/nature15401
Vijg J. (2021). From DNA damage to mutations: All roads lead to aging. Ageing Res. Rev. 68, 101316. doi:10.1016/j.arr.2021.101316
Visser H., Thomas T. (2021). MicroRNAs and the DNA damage response: How is cell fate determined? DNA Repair 108, 103245. doi:10.1016/j.dnarep.2021.103245
Wang H., Lautrup S., Caponio D., Zhang J., Fang E. F. (2021). DNA damage-induced neurodegeneration in accelerated ageing and alzheimer's disease. Int. J. Mol. Sci. 22 (13). doi:10.3390/ijms22136748
Wang M., Guo L., Wu Q., Zeng T., Lin Q., Qiao Y., et al. (2014). ATR/Chk1/Smurf1 pathway determines cell fate after DNA damage by controlling RhoB abundance. Nat. Commun. 5, 4901. doi:10.1038/ncomms5901
Wang R., Hao W., Pan L., Boldogh I., Ba X. (2018). The roles of base excision repair enzyme OGG1 in gene expression. Cell. Mol. Life Sci. 75 (20), 3741–3750. doi:10.1007/s00018-018-2887-8
Wang X., Duan J., Fu W., Yin Z., Sheng J., Lei Z., et al. (2019). Decreased expression of NEDD4L contributes to NSCLC progression and metastasis. Biochem. Biophysical Res. Commun. 513 (2), 398–404. doi:10.1016/j.bbrc.2019.04.001
Wl P., Cm H., Da S., Tn O. S., Na J., Copeland N. G. (1998). The itchy locus encodes a novel ubiquitin protein ligase that is disrupted in a18H mice. Nat. Genet. 18 (2), 143–146.
Xia Q., Li Y., Han D., Dong L. (2021). SMURF1, a promoter of tumor cell progression? Cancer Gene Ther. 28 (6), 551–565. doi:10.1038/s41417-020-00255-8
Xie S., Xia L., Song Y., Liu H., Wang Z. W., Zhu X. (2021). Insights into the biological role of NEDD4L E3 ubiquitin ligase in human cancers. Front. Oncol. 11, 774648. doi:10.3389/fonc.2021.774648
Xie W., Jin S., Wu Y., Xian H., Tian S., Liu D. A., et al. (2020). Auto-ubiquitination of NEDD4-1 recruits USP13 to facilitate autophagy through deubiquitinating VPS34. Cell Rep. 30 (8), 2807–2819. doi:10.1016/j.celrep.2020.01.088
Xu C., Fan C. D., Wang X. (2015). Regulation of Mdm2 protein stability and the p53 response by NEDD4-1 E3 ligase. Oncogene 34 (3), 281–289. doi:10.1038/onc.2013.557
Xu H., Wang W., Li C., Yu H., Yang A., Wang B., et al. (2009). WWP2 promotes degradation of transcription factor OCT4 in human embryonic stem cells. Cell Res. 19 (5), 561–573. doi:10.1038/cr.2009.31
Xu J., Li F., Gao Y., Guo R., Ding L., Fu M., et al. (2021). E47 upregulates ΔNp63α to promote growth of squamous cell carcinoma. Cell Death Dis. 12 (4), 381. doi:10.1038/s41419-021-03662-3
Yalcin E., Beker M. C., Turkseven S., Caglayan B., Gurel B., Kilic U., et al. (2019). Evidence that melatonin downregulates Nedd4-1 E3 ligase and its role in cellular survival. Toxicol. Appl. Pharmacol. 379, 114686. doi:10.1016/j.taap.2019.114686
Yang B., Kumar S. (2010). Nedd4 and nedd4-2: Closely related ubiquitin-protein ligases with distinct physiological functions. Cell Death Differ. 17 (1), 68–77. doi:10.1038/cdd.2009.84
Yang H., Zhu J., Wang G., Liu H., Zhou Y., Qian J. (2020). STK35 is ubiquitinated by NEDD4L and promotes glycolysis and inhibits apoptosis through regulating the AKT signaling pathway, influencing chemoresistance of colorectal cancer. Front. Cell Dev. Biol. 8, 582695. doi:10.3389/fcell.2020.582695
Yang Y., Liao B., Wang S., Yan B., Jin Y., Shu H. B., et al. (2013). E3 ligase WWP2 negatively regulates TLR3-mediated innate immune response by targeting TRIF for ubiquitination and degradation. Proc. Natl. Acad. Sci. U.S.A. 110 (13), 5115–5120. doi:10.1073/pnas.1220271110
Yin Q., Wyatt C. J., Han T., Smalley K. S. M., Wan L. (2020). ITCH as a potential therapeutic target in human cancers. Seminars Cancer Biol. 67 (2), 117–130. doi:10.1016/j.semcancer.2020.03.003
Yuan J., Adamski R., Chen J. (2010). Focus on histone variant H2AX: To be or not to be. FEBS Lett. 584 (17), 3717–3724. doi:10.1016/j.febslet.2010.05.021
Zhang J. Q. J., Saravanabavan S., Chandra A. N., Munt A., Wong A. T. Y., Harris P. C., et al. (2021). Up-regulation of DNA damage response signaling in autosomal dominant polycystic kidney disease. Am. J. Pathology 191 (5), 902–920. doi:10.1016/j.ajpath.2021.01.011
Zhang N., Zhang Y., Qian H., Wu S., Cao L., Sun Y. (2020). Selective targeting of ubiquitination and degradation of PARP1 by E3 ubiquitin ligase WWP2 regulates isoproterenol-induced cardiac remodeling. Cell Death Differ. 27 (9), 2605–2619. doi:10.1038/s41418-020-0523-2
Zhang N., Zhang Y., Wu B., Wu S., You S., Lu S., et al. (2021). Deacetylation-dependent regulation of PARP1 by SIRT2 dictates ubiquitination of PARP1 in oxidative stress-induced vascular injury. Redox Biol. 47, 102141. doi:10.1016/j.redox.2021.102141
Zhang N., Zhang Y., Wu B., You S., Sun Y. (2020). Role of WW domain E3 ubiquitin protein ligase 2 in modulating ubiquitination and Degradation of Septin4 in oxidative stress endothelial injury. Redox Biol. 30, 101419. doi:10.1016/j.redox.2019.101419
Zhang R., Zhang J., Luo W., Luo Z., Shi S. (2019). WWP2 is one promising novel oncogene. Pathol. Oncol. Res. 25 (2), 443–446. doi:10.1007/s12253-018-0506-5
Zhang S., Xiong X., Sun Y. (2020). Functional characterization of SOX2 as an anticancer target. Sig Transduct. Target Ther. 5 (1), 135. doi:10.1038/s41392-020-00242-3
Zhang W., Tao S. S., Wang T., Zhang J., Liu X., Li Y. T., et al. (2021). ABRO1 stabilizes the deubiquitinase BRCC3 through inhibiting its degradation mediated by the E3 ubiquitin ligase WWP2. FEBS Lett. 595 (2), 169–182. doi:10.1002/1873-3468.13970
Zhang Y., Qian H., Wu B., You S., Wu S., Lu S., et al. (2020). E3 Ubiquitin ligase NEDD4 family‑regulatory network in cardiovascular disease. Int. J. Biol. Sci. 16 (14), 2727–2740. doi:10.7150/ijbs.48437
Keywords: E3 ubiquitin ligase, NEDD4 family, DNA damage response (DDR), DNA damage, ubiquitin-proteasome system
Citation: Lu X, Xu H, Xu J, Lu S, You S, Huang X, Zhang N and Zhang L (2022) The regulatory roles of the E3 ubiquitin ligase NEDD4 family in DNA damage response. Front. Physiol. 13:968927. doi: 10.3389/fphys.2022.968927
Received: 14 June 2022; Accepted: 21 July 2022;
Published: 26 August 2022.
Edited by:
Mustafa Ark, Gazi University, TurkeyReviewed by:
Arun Upadhyay, Northwestern University, United StatesVibhuti Joshi, Bennett University, India
Copyright © 2022 Lu, Xu, Xu, Lu, You, Huang, Zhang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lijun Zhang, bHpoYW5nMjAyMDAzQDE2My5jb20=; Naijin Zhang, bmp6aGFuZ0BjbXUuZWR1LmNu
 Xinxin Lu1
Xinxin Lu1 Shilong You
Shilong You Naijin Zhang
Naijin Zhang Lijun Zhang
Lijun Zhang