
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Physiol., 17 August 2022
Sec. Aquatic Physiology
Volume 13 - 2022 | https://doi.org/10.3389/fphys.2022.961773
 Yingduo Huo1,2,3†
Yingduo Huo1,2,3† Yang Gu1,2,3†
Yang Gu1,2,3† Mulian Cao1,2,3
Mulian Cao1,2,3 Yingrui Mao1,2,3
Yingrui Mao1,2,3 Yayu Wang1,2,3
Yayu Wang1,2,3 Xiaoqiang Wang1,2,3
Xiaoqiang Wang1,2,3 Guiling Wang1,2,3*
Guiling Wang1,2,3* Jiale Li1,2,3
Jiale Li1,2,3Abstract: The process of spermatogenesis is complex and controlled by many genes. In mammals, Testis-expressed gene 11 (Tex11) and meiosis expressed gene 1 (Meig1) are typical spermatogenesis-related genes. In this study, we obtained the full length cDNAs for Tex11 (3143bp) and Meig1 (1649bp) in Hyriopsis cumingii by cloning. Among them, Hc-Tex11 contains 930 amino acids and Hc-Meig1 contains 91 amino acids. The protein molecular masses (MW) of Hc-Tex11 and Hc-Meig1 were 105.63 kDa and 10.95 kDa, respectively. Protein secondary structure analysis showed that Hc-TEX11 protein has three TPR domains. The expression of Hc-Tex11 and Hc-Meig1 in different tissues showed higher levels in testes. At different ages, the expression of Hc-Tex11 and Hc-Meig1 was higher levels in 3-year-old male mussels. During spermatogenesis, the mRNA levels of Hc-Tex11, Hc-Meig1 gradually increased with the development of spermatogonia and reached a peak during sperm maturation. Hc-Tex11 and Hc-Meig1 mRNA signals were detected on spermatogonia and spermatocytes by in situ hybridization. In addition, RNA interference (RNAi) experiments of Hc-Tex11 caused a down-regulated of Dmrt1, KinaseX, Tra-2 and Klhl10 genes and an up-regulated of β-catenin gene. Based on the above experimental results, it can be speculated that Hc-Tex11 and Hc-Meig1 are important in the development of the male gonadal and spermatogenesis in H. cumingii, which can provide important clues to better comprehend the molecular mechanism of Tex11 and Meig1 in regulating spermatogenesis of bivalves.
Reproduction is one of the most basic characteristics of living organisms, and most animals reproduce sexually. The process of spermatogenesis comprises mitosis, meiosis, and sperm deformation (Nishimura and L'Hernault, 2017). Many specific genes are involved in this process, and several genes involved in the spermatogenesis process have been identified in mammals, such as Stra8, Cabs1 and Spag6 (Liu Y. H. et al., 2019; Yue et al., 2019; Zhang et al., 2021), and in fish, Nanos1, Amh and Aqp1aa (Li et al., 2015; Guo et al., 2017; Liang et al., 2020). However, studies on specific genes regulating spermatogenesis in mollusks are limited. It has been shown that Klf4, Sox2, Sox17 in Chlamys farreri (Liang et al., 2017; Yang et al., 2017; Liang et al., 2019) and Tssk1 in Atrina pectinate (Li H. H. et al., 2016) play a significant role in spermatogenesis1. In China, Hyriopsis cumingii is the most dominant freshwater pearl-cultivating mussel, and it accounts for more than 80% of the pearl-cultivating volume in captive freshwater mussels (Wang et al., 2014)2. The study found marked difference in pearl production and morphology between female and male mussels, with males being better than females (Wang et al., 2020). The gonads of freshwater mussels are of the follicular type3, consisting of three parts: follicles, genital canal and gonoduct, with the follicles and genital canal being the main parts that form germ cells (Chen and Shi, 2002; Pan et al., 2010). Heteromorphic chromosomes have not been identified in current studies on H. cumingii, so studies of sex and gametogenesis in H. cumingii revolve around the regulation of related genes (Wang et al., 2021a)4. Understanding the molecular mechanism of gametogenesis in H. cumingii can provide a basis for artificial propagation and hybrid breeding and is of great importance for seedling breeding5.
Direct homologs of testis-expressed (Tex) have been identified in vertebrates (mammals6, birds and reptiles), invertebrates and yeast (Bellil et al., 2021). Testis expressed gene 11 (Tex11) was originally identified as a germ cell-specific7, X-linked gene in mice (Yu et al., 2021)8. Tex11, also known as Zip4h in mice, is a direct mammalian homolog and a meiosis-specific protein in Saccharomyces cerevisiae and Arabidopsis thaliana that regulates the level of meiotic crossover (Wang et al., 2001). In mammals, Tex11 is highly expressed specifically in the testis and localized to spermatocytes (Tang et al., 2011). Tex11 competes with the estrogen receptor (ER), and when Tex11 is overexpressed, it enhances the transcription of estrogen-response reporter genes but suppresses AKT and MAPK signaling pathways, resulting in reduced cell proliferation (Yu et al., 2012). In addition, some studies have shown that Tex11 may be a key factor in germ cell development and ovarian in Xenopus laevis toads and affects fertility (Haselman et al., 2015). During meiosis, homologous chromosome pairing and recombination are required, a process that is closely linked to the activity of the synaptonemal complex (SC) (Heyting, 1996). Defects in meiosis can lead to haploid and polyploid organisms (Hassold and Hunt, 2001). Tex11 is involved in the initiation of chromosomal synapses, and interacts with the central elements of SC, Tex12 and Sycp2, in the formation of meiotic crossovers, providing a physical link between meiotic processes (Yang et al., 2008). Tex11 genes have been studied more in mammals and less in aquatic animals. Tex11 is an interesting factor in the late developmental stages of male Anguilla (Rozenfeld et al., 2019). Meiosis expressed gene 1 (Meig1) is a critical gene for the control of spermiogenesis and manchette structure. The Meig1 defect destroys the head and tail of the sperm, impairing spermatogenesis and leading to infertility (Zhang et al., 2009). Parkinson’s co-regulatory gene (PACRG) interacts with Meig1 to form a complex in the sperm tail that is essential for the transport of sperm flagellin and construction of sperm flagellum (Li W. et al., 2016). Meig1 can also be used as a key gene for studying male sterility in Vulpes fulvus and Alopex lagopus hybrids (Yang et al., 2019). In Acanthopagrus schlegelii, Meig1 transcript levels were higher in the testis than in the ovary (Zhang et al., 2019).
Due to the low evolutionary status of mollusks, the regulatory mechanisms of gonadal development and spermatogenesis are not yet clear. Bivalve mollusks contain multiple reproductive modes, including dioecism, hermaphroditic, and sex-reversed (Yue et al., 2020). This suggests that bivalves are suitable animals for studying gametogenesis and sex development. Some sex-linked genes such as β-catenin, Dmrt1, Klhl10, KinaseX, Tra-2 have been identified in H. cumingii, and show expression characteristics of both sexes (Dong et al., 2020; Guo et al., 2020; Cui et al., 2021; Wang et al., 2021b). However, knowledge of the genes involved in spermatogenesis is still limited. Tex11 and Meig1 have been shown to be associated with mammalian spermatogenesis, while their role in invertebrate gametogenesis has not been reported. In this study, Hc-Tex11 and Hc-Meig1 were identified. The expression of Hc-Tex11 and Hc-Meig1 was analyzed in different tissues and at different periods. Additionally, the functions of Hc-Tex11 and Hc-Meig1 in gonads were analyzed by in situ hybridization and RNA interference (RNAi). Our data suggest that Hc-Tex11 and Hc-Meig1 are important for the spermatogenesis in H. cumingii. This provides a theoretical basis for the molecular mechanisms of spermatogenesis and sperm development in bivalves.
Zhejiang Weiming aquaculture farm provided all the samples for this study. The collection and handling of H. cumingii was approved by the Institutional Animal Care and Use Committee (IACUC) of the Shanghai Ocean University, China.
Healthy 1, 2, and 3-year-old H. cumingii were brought back from the farm. The gonads were taken through a 1 ml microsyringe to differentiate gander under the microscope. The gonads, gills, kidneys, adductor, mantle, and liver were collected from 2-year-old H. cumingii. We collected gonadal tissues from the various stages of spermatogenesis (spermatogonia stage, spermatocyte stage, sperm maturation stage, sperm discharge stage, follicular atrophy stage). All samples were stored at 80°C for long-term storage, with three replicates per group.
The RNA was extracted by the Trizol, the purity of RNA was checked by NanoDrop 2000c (Thermo Scientific, US), followed by 1.0% agarose gel electrophoresis to check the integrity of RNA. cDNA synthesis according to PrimeScriptTM RT Reagent Kit with gDNA Eraser kit (TaKaRa, Japan), mixed in three parallel aliquots, and diluted 5-fold as a template for qRT-PCR.
The sequences of Hc-Tex11 and Hc-Meig1 were obtained from the gonadal transcriptome database and were found to be incomplete at the 3′ end. Based on the original sequences, 3′ RACE primers for Hc-Tex11 and Hc-Meig1 were designed (Table 1). Reverse conversion to cDNA as template according to 3′-Full RACE Core Set with PrimeScript RTase kit. Rapid amplification was performed by nested PCR and cDNA ends. After PCR, the PCR products were ligated into pMD19-T vector, transformed into receptor E. coli DH5α, and selected white strains after blue-white spot and sent for testing.
The NCBI ORF Finder program was used to obtain open reading frame predictions of amino acid sequences; BLAST program was used to perform nucleotide and amino acid sequence similarity analysis of the obtained gene sequences with homologous species; SMART program predicts the secondary structure of proteins; SWISS-MODEL program predicts the tertiary structure; SignalP 4.1 Server program was used to predict signal peptides; the TMHMM Server v2.0 program was used to predict its transmembrane structure, and the NetPhos 3.1 program was used to discover its phosphorylation sites. The cloned cDNAs and amino acid sequences were analyzed by DNAMAN software, GeneDoc software performed multiple sequence alignment, and phylogenetic trees were constructed by MEGA 7.0 software using the neighbor-joining (NJ) method. Bootstrap was repeated 1,000 times to calculate the confidence values among the species.
Real-time PCR primers for Hc-Tex11 and Hc-Meig1 were designed using Primer 5.0: Q-Tex11-F, Q-Tex11-R, Q-Meig1-F, Q-Meig1-R (Table 1). Using α subunit of elongation factor 1 (EF1-α) (GenBank no. GW694601) as the reference (Bai et al., 2014), the reaction system (20 μL) as follows, 2×TB GreenTM Premix Ex Taq TM (Takara) 10 μL, primers 0.8 μL each, cDNA 1.6 μL, ddH2O 6.8 μL, three replicates of each sample. The calculation of relative expressions by the 2−ΔΔCt method (Schmittgen and Livak, 2008). Significant differences were analyzed by SPSS18.0 software and plotted by SigmaPlot12.3.
In situ hybridization primers were designed in the ORF region (Table 1), and the T7 promoter (5′-TAATACGACTCACTATAGGG-3′) was added before the reverse primer. The T7 High Efficiency Transcription Kit (Transgen, China) was used for in vitro transcription. Gonadal tissues were fixed in 4% paraformaldehyde for 2 h, then paraffin embedded, cut to a thickness size of 6 μm and subjected to in situ hybridization using the Enhanced Sensitive ISH Assay Kit II (Boster, United States). Under a microscope, the hybridization signal was observed and photographed.
Two pairs of primers were designed using Primer 5.0 based on the Hc-Tex11 cDNA sequence. Name the two interference strands as G1, G2 respectively. The primers of G1 were T-RNAi-F1 (5′-TAATACGACTCACTATAGGGTGCGGAAGATGTGTGACTGT-3′), T-RNAi-R1 (5′-TAATACGACTCACTATAGGGGCTTGTCACATTGGAGAGCAA-3′), amplifying a fragment of 393 bp. The primers of G2 were T-RNAi-F2 (5′-TAATACGACTCACTATAGGGAGGGTGCCTTCCTGTCTATGTT-3′), T-RNAi-R2 (5′-TAATACGACTCACTATAGGGTTTCCGATGTCGGGAGAGTT-3′), amplifying a fragment of 469 bp. The negative control dsRNA was a green fluorescent protein (GFP) sequence with no homology to Hc-Tex11 (Table 1). The method for the synthesis step of dsRNA was referenced to that described by Wang (Wang et al., 2021b). Dilute to a certain concentration and store in the refrigerator at -80°C.
2-year-old male H. cumingii was divided into three groups (interference groups 1, 2, and negative control group), with six mussels in each group. Both dsGFP and two interference strands were diluted to 80 μg/μL, and 100 μL of Hc-Tex11 dsRNA was injected into the adductor with a 1 ml microinjector, and tissues were collected after 48 h. RNA was extracted from the gonads, and the interference efficiency was detected and calculated by qRT-PCR. The expression of Hc-Tra-2, Hc-Dmrt1, Hc-KinaseX, Hc-Klhl10 and Hc-β-catenin after dsRNA injection was also examined. The primer information is in Table 1.
The experimental data were statistically analyzed using SPSS 18.0 software, and the data obtained were expressed as mean ± standard deviation. Significant differences were calculated using the independent samples t-test; p < 0.05 was considered statistically significant.
The full-length cDNA of Hc-Tex11 gene was 3143 bp (GenBank no. ON804236), which includes 56 bp of 5′ untranslated region (5′-UTR) and 294 bp of 3′ untranslated region (3′-UTR) and 2793 bp of open reading frame (ORF), encoding 930 amino acids (Figure 1A). The protein predicted structure showed that it contains three tetrapeptide repeat proteins (TPR) (178-211aa, 414-447aa, and 455-488aa). The molecular weight (MW) of the protein was assessed to be approximately 105.63 kDa with a theoretical isoelectric point (PI) of 5.84. Phosphorylation site analysis revealed 38 serine (S) phosphorylation sites, 25 threonine (T) phosphorylation sites, and seven tyrosine (Y) phosphorylation sites. TMHMM predicted that the gene does not possess a transmembrane structure. SWISS-MODEL predicted the tertiary structure of the Hc-TEX11 protein (Figure 1B). The α-helix accounts for 70%, the irregular curl for 18% and no β-fold. Comparison of the amino acid sequence between Hc-Tex11 and other species, the results showed that the similarity of Hc-Tex11 among different species was high, including Mizuhopecten yessoensis (50.86%), Pecten maximus (50.23%), and Crassostrea gigas (45.57%). The homology in humans and mice was 35.03% and 29.28%. Multiplex sequence analysis using GeneDoc revealed high amino acid similarity of Hc-Tex11 with 10 other species (Figure 2). The phylogenetic tree was divided into two branches, vertebrate and invertebrate, in which the TEX11 protein of H. cumingii clusters into a branch with bivalve shellfish such as M. yessoensis and P. maximus (Figure 3). This result indicates that Hc-TEX11 is more closely related to mollusks and more distantly related to mammals, suggesting that the gene is relatively evolutionarily conserved.
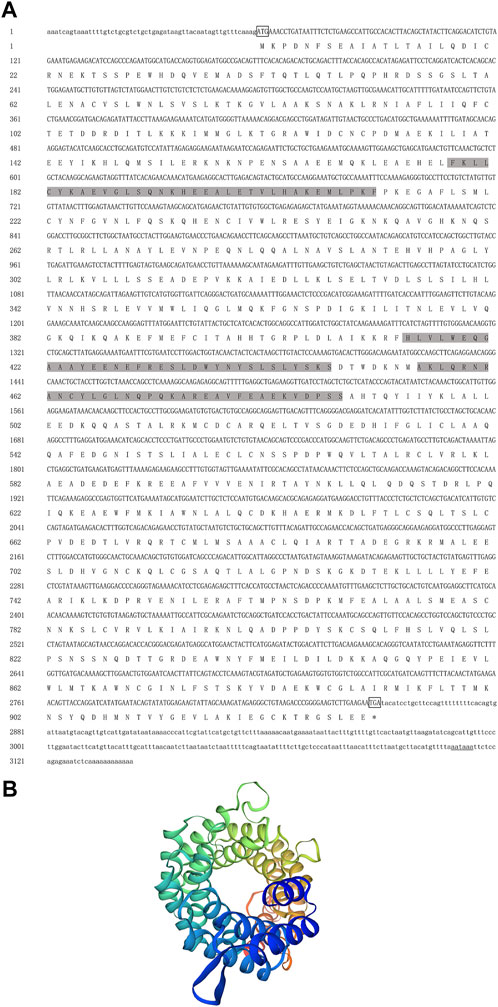
FIGURE 1. (A) Nucleotide and amino acid sequences of Hc-Tex11. Lowercase letters indicate 5′-UTR and 3′-UTR; start codon and stop codon are marked by boxes; plus-tail signals are underlined. The gray area is the TPR domain (B) SWISS-MODEL predicts the tertiary structure of Hc-TEX11.
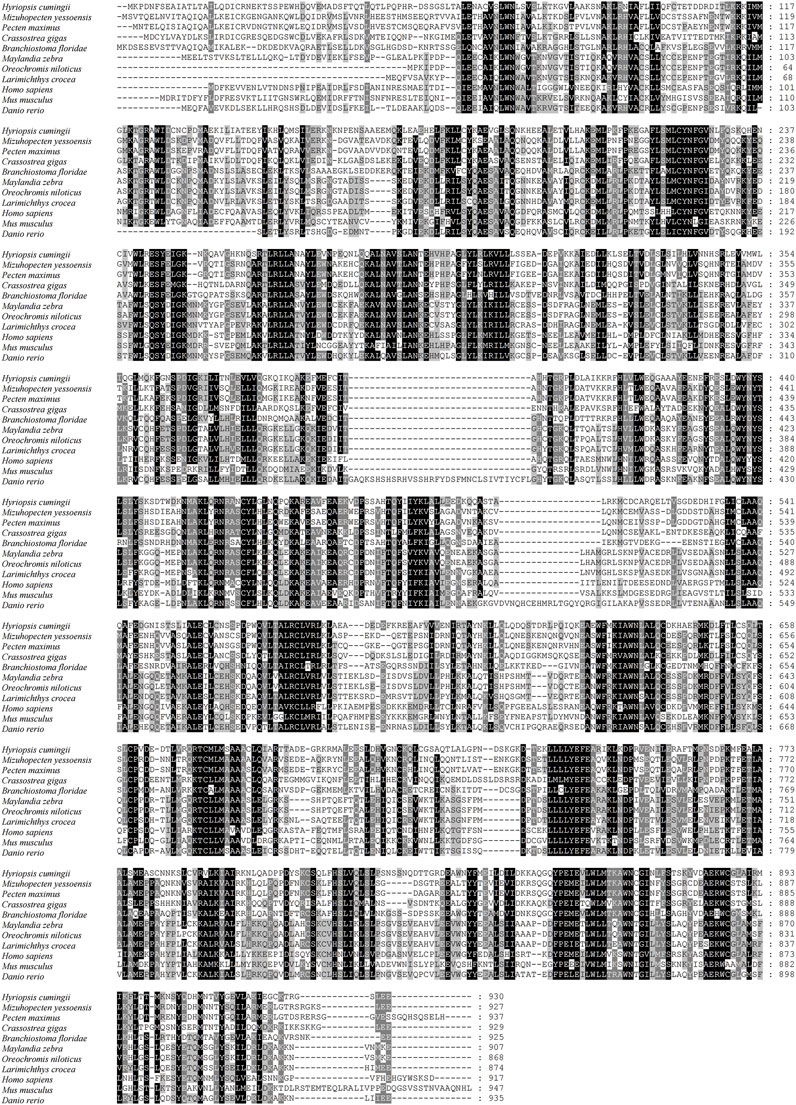
FIGURE 2. Multiple comparison of Hc-TEX11 amino acid sequence with other species. Black, same amino acid; grey, similar amino acid.
The full length of the Hc-Meig1 cDNA was 1649 bp (GenBank no. ON804237) with a 314 bp 5′-UTR, a 1059 bp 3′-UTR and a 276 bp ORF, encoding 91 amino acids (Figure 4A). Hc-Meig1 has no transmembrane structure and secondary structure. The Hc-Meig1 was 10.95 kDa and the PI was 8.94. Phosphorylation site analysis identified five serine (S) phosphorylation sites and two tyrosine (Y) phosphorylation sites. SWISS-MODEL predicted the tertiary structure of the Hc-MEIG1 protein (Figure 4B) with a QMEAN of -1.38, indicating a good match to the template protein. The α-helix accounted for 44%, the β-fold for 16%, and the irregular curl for 39%. Comparison of the amino acid sequence between Hc-Meig1 and other species, the results showed that the similarity of Hc-Meig1 among different species was high (66–74%), among which Crassostrea virginica (74.71%) was the highest. The homology in humans and mice was 52.94% and 56.47%. Multiple sequence analysis using GeneDoc revealed high amino acid similarity of Hc-Meig1 with 10 other species (Figure 5). The phylogenetic tree was produced by MAGE 7.0 software, in which the MEIG1 protein of H. cumingii clusters into a branch with bivalve shellfish such as C. virginica and C. gigas (Figure 6). This result indicates that Hc-MEIG1 is more closely related to mollusks and more distantly related to mammals, suggesting that the gene is relatively evolutionarily conserved.
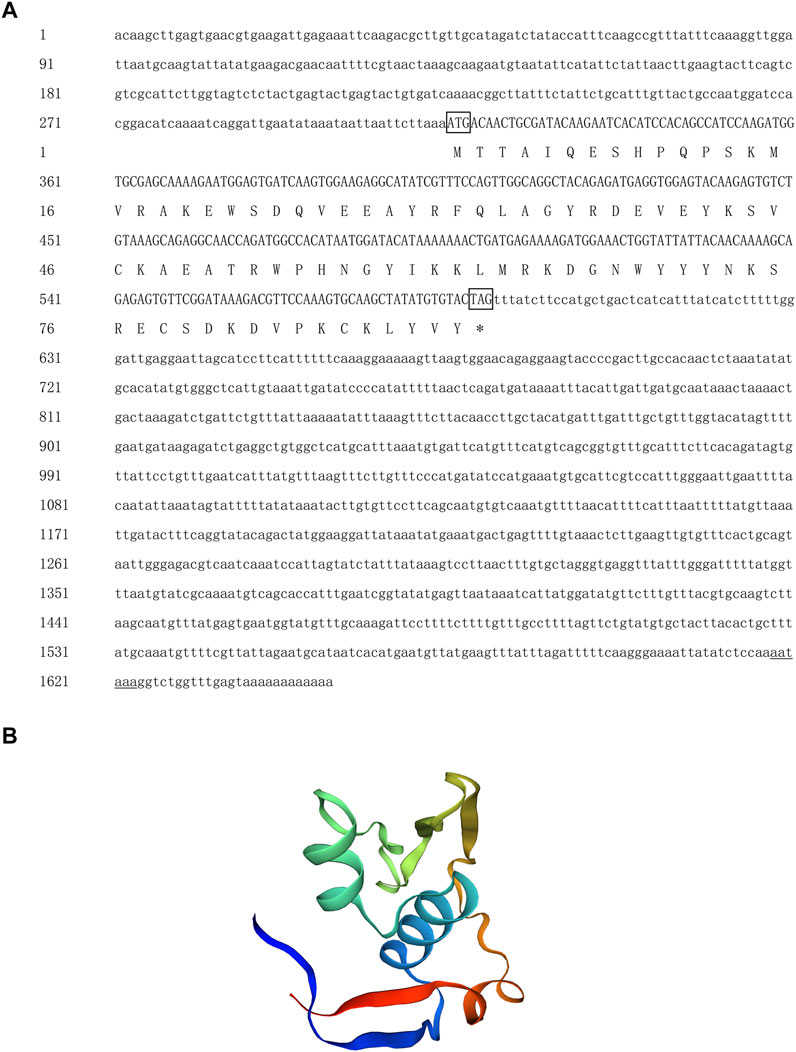
FIGURE 4. (A) Nucleotide and amino acid sequences of Hc-Meig1. Lowercase letters indicate 5′-UTR and 3′-UTR; start codon and stop codon are marked by boxes; plus-tail signals are underlined (B) SWISS-MODEL predicts the tertiary structure of Hc-MEIG1.
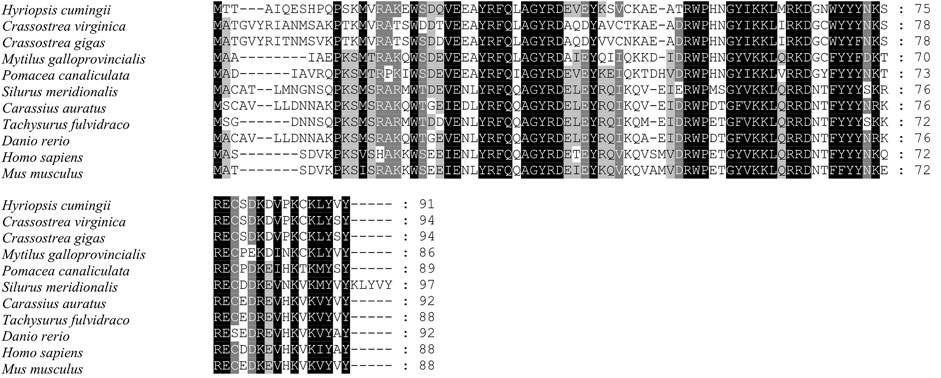
FIGURE 5. Multiple comparison of the Hc-MEIG1 amino acid sequence with other species. Black, same amino acid; grey, similar amino acid.
The expression of Hc-Tex11 and Hc-Meig1 genes was detected in gonads, liver, gill, kidney, adductor, mantle. As well as the expression of male and female gonads at ages 1, two and three and spermatogenesis at each stage. The results showed that Hc-Tex11 expression was highest in the testis and significantly higher than in other tissues (p < 0.01) (Figure 7A). The expression of Hc-Tex11 increased with age in males, with the highest expression at 3 years of age and no significant change in expression in females (p < 0.01) (Figure 7C). During all stages of spermatogenesis, the expression level of Hc-Tex11 gradually increases as spermatogonia continue to develop into sperm, reaching a maximum during the sperm maturation stage. A decreasing trend was observed from the sperm maturation stage to the follicular atrophy stage (p < 0.05) (Figure 7E).
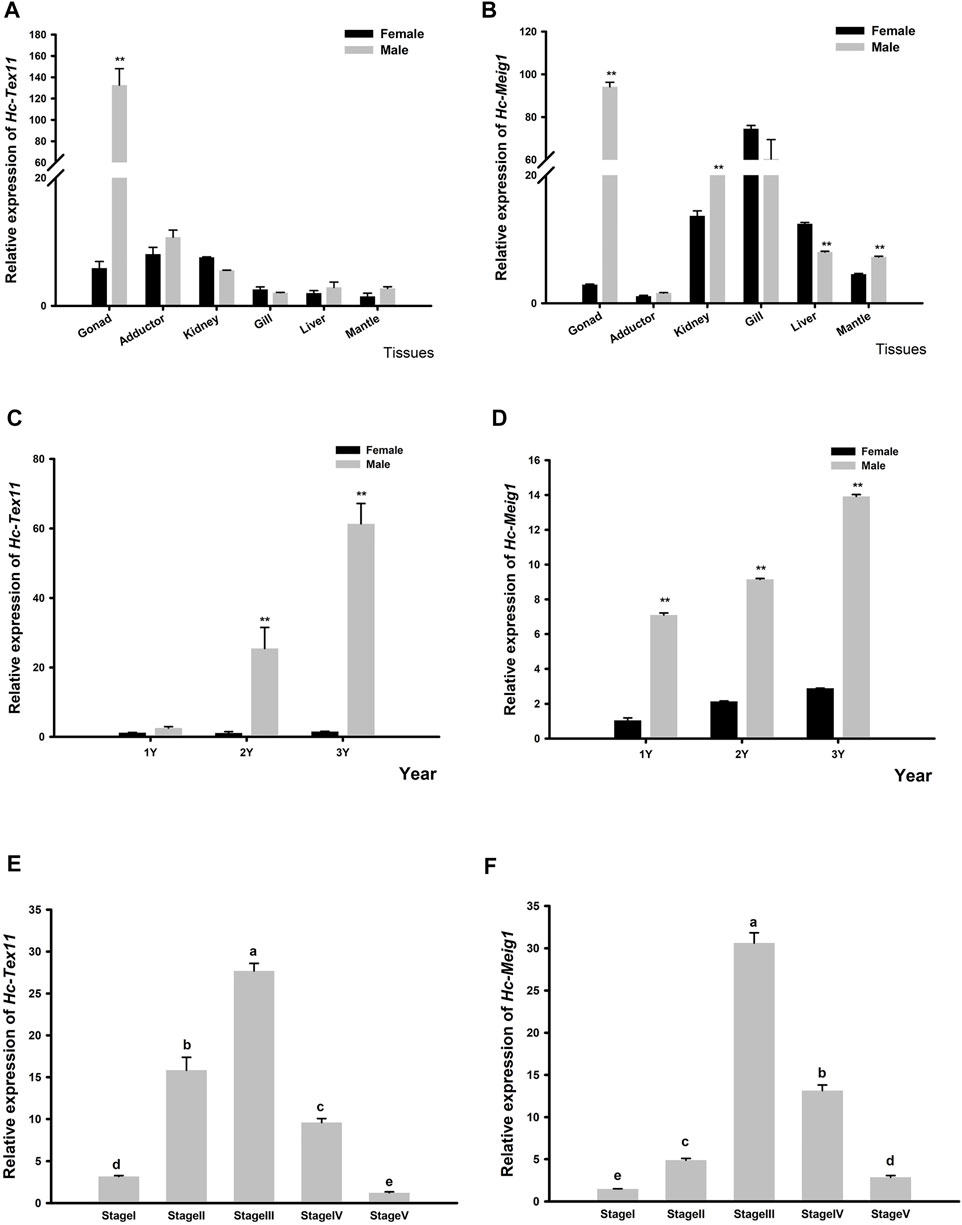
FIGURE 7. (A) Relative expression of Hc-Tex11 gene in male and female tissue (B) Relative expression of Hc-Meig1 gene in male and female tissue (C) Relative expression of the Hc-Tex11 gene in 1-3-year-old male and female gonads (D) Relative expression of the Hc-Meig1 gene in 1-3-year-old male and female gonads. Significant differences, * indicates p < 0.05, ** indicates p < 0.01 (E) Relative expression of Hc-Tex11 gene during spermatogenesis (F) Relative expression of Hc-Meig1 gene during spermatogenesis. Results are expressed as mean ± SD, and different letters (A–E) indicate statistically significant differences (p < 0.05). Stage1, spermatogonia stage; stage2, spermatocyte stage; stage3, sperm maturation stage; stage4, sperm discharge period; stage5, follicular atrophy stage.
The expression of Hc-Meig1 was highest in the testes, followed by the gills, and the expression of the testes was significantly higher than that of the ovaries (p < 0.01) (Figure 7B). Hc-Meig1 expression increases with age in both gonads, with higher expression in the testis than in the ovary at the same age (p < 0.01) (Figure 7D). In all stages of spermatogenesis, Hc-Meig1 expression was significantly higher in sperm maturation than in other stages (p < 0.05) (Figure 7F). The expression pattern of Hc-Meig1 in spermatogenesis is similar to that of Hc-Tex11.
The localization of Hc-Tex11 and Hc-Meig1 in the testis was detected by in situ hybridization, and male germ cells at different developmental stages (from spermatogonia to mature sperm) can be seen in the figure. Hc-Tex11 (Figure 8B) and Hc-Meig1 (Figure 8E) have distinct signals compared with the control group (Figures 8A,D). The results showed that the signals were located on spermatogonia and spermatocytes, whereas no signal was evident on spermatids and sperms.
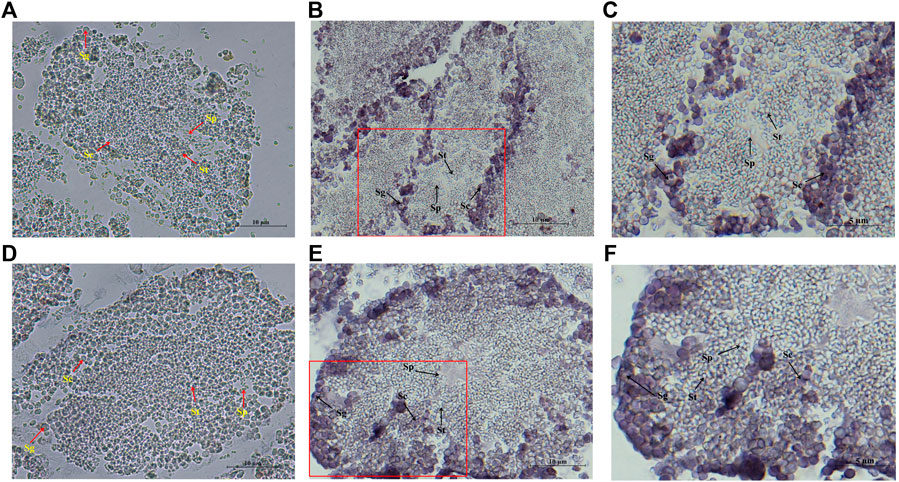
FIGURE 8. In situ hybridization of Hc-Tex11 and Hc-Meig1 genes in testes, (A) Hc-Tex11 control group, (B) Hc-Tex11 experimental group, (C) magnified image of Hc-Te×11 experimental group, (D) Hc-Meig1 control group, (E) Hc-Meig1 experimental group, (F) magnified image of Hc-Meig1 experimental group. Sg, spermatogonia; Sc, spermatocyte; St, spermatid; Sp, sperm.
The expression of Tex11 was measured in the male gonads of the H. cumingii after 48 h of interference. The results showed that the two interference strands synthesized in this experiment all played a role in the interference. The results of SPSS difference significance analysis showed that G1 was significantly different from the control group (p < 0.05) and G2 was highly different from the control group (p < 0.01). The silencing efficiency of the two interference strands was 26.74% and 90.13% (Figure 9A). And G2 had been selected as the interference strand for the subsequent experiments.
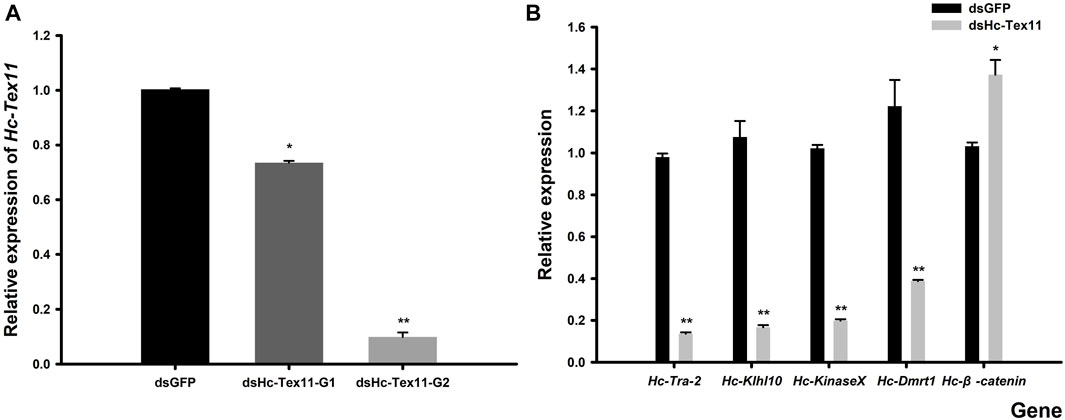
FIGURE 9. (A) Relative expression of Hc-Tex11 gene in male gonadal tissue after RNAi (B) Effect of dsHc-Tex11-2 injection on the expression of Hc-Tra-2, Hc-Klhl10, Hc-KinaseX and Hc-Dmrt1 and Hc-β-catenin genes in the testes of the H. cumingii. Control group, dsGFP; experimental group, dsHc-Tex11.
The silencing effect of dsHc-Tex11-G2 was 90.13%, which indicated that this interference strand was effective. Detect the effect of Hc-Tex11 gene on other sex genes. Compared with the negative control group, the expression levels of Hc-Tra-2, Hc-Klhl10, Hc-KinaseX and Hc-Dmrt1 were decreased, with interference efficiencies of 86.11%, 84.46%, 80.60% and 68.27% (p < 0.01), respectively, and the Hc-β-catenin expression level was significantly increased, 33.04% higher than that of the control group (p < 0.05) (Figure 9B).
In this study, the full length of Hc-Tex11 was 3143bp, encoding 930 amino acids and containing three TPR domains (Figure 1A). The TPR domain usually forms a helix-turn-helix structure with internal gaps and contains 34 amino acids (Blatch and Lassle, 1999; Perez-Riba and Itzhaki, 2019). Proteins containing TPR domains are usually arranged in tandem 3–16 TPRs (Allan and Ratajczak, 2011). TPR structures are important in transcriptional regulation, protein kinase inhibition, peroxisomal protein and mitochondrial transport, cell cycle, immunity and viral replication, protein folding (Zhang et al., 2004; Liu et al., 2012; Rozenfeld et al., 2019). Hsd-3.8, Klc3, Pp2ac and Ttc21a genes possessing TPR domains were shown to be involved in mammalian spermatogenesis (Lin et al., 2002; Zhang et al., 2012; Pan et al., 2015; Liu W. J. et al., 2019). In aquatic animals, genes possessing the TPR domain also play an important role in the gonads. Fem-1c is involved in ovarian differentiation in Danio rerio (Shi et al., 2015), and CypD is highly expressed in the gonads of H. cumingii (Liu et al., 2017). Multiple sequence comparisons revealed that the structural domain of Hc-Tex11 was more similar to that of mollusks compared to vertebrates, indicating that the gene has been relatively conserved during evolution. The full length of Hc-Meig1 was 1649 bp (Figure 4A). The MEIG1 protein sequence of the H. cumingii was not found to have any known functional structure. However, Hc-MEIG1 has a high degree of amino acid sequence similarity to other species (Figure 5). It has been shown that the MEIG1 protein was phosphorylated in vivo and forms a dimer that enters the nucleus and binds to meiotic chromatin during the first meiotic division (ChenMoses et al., 1997; Steiner et al., 1999). Hc-MEIG1 contains seven phosphorylation sites, including serine and tyrosine. Multiple sequence comparisons showed that Meig1 was highly conserved across species, implying that Hc-Meig1 may function similarly to mammals in mollusks.
Up to now, the role of Tex11 and Meig1 genes in the testis and ovary has only been studied in mammals. Tex11 is a testis-specific transcript that is required for chromosomal synapses, meiotic crossover and recombination, DNA double-strand break repair, and has been shown to be detected only in mammalian testes (Yu et al., 2021). In mature mice, Meig1 is differentially expressed in females and males. Transcripts of Meig1 are detected in pre-meiotic oocytes and mature testes, but not in mature ovaries (Don et al., 1994; Teves et al., 2013). In this study, Hc-Tex11 was expressed in trace amounts in tissues other than the gonads, showing sexual dimorphism in the gonads and specific high expression in the testes (Figure 7A). This result is similar to the results of Tex11 expression in pigs (Lin et al., 2002). Hc-Meig1 was expressed in all the tissues examined, with the highest expression in the testis (Figure 7B). This is similar to the Meig1_v1 results in the three Meig1 transcripts in mice (Zhang et al., 2009). Expression in tissues other than gonads suggests that Hc-Meig1 gene may also be involved in other functions in shellfish. Hc-Tex11 and Hc-Meig1 were most highly expressed in testes of 3 years old (Figures 7C,D). Based on the expression pattern, it is postulated that Hc-Tex11 and Hc-Meig1 are involved in the gonadal development of H. cumingii. All stages of the spermatogenesis were observed in male mussels above 2 years of age (Mi et al., 2002). Spermatogenesis is a cyclic process in which primordial germ cells undergo mitosis and meiosis to continuously form sperm, and mature sperm are gradually discharged until the follicle atrophy (Yu et al., 2008; Wang, 2010). In the spermatogenesis process of H. cumingii, the expression of Hc-Tex11 and Hc-Meig1 gradually increased during the development of spermatogonia to sperm and was highest during the sperm maturation stage (Figures 7E,F). This expression pattern suggests that Hc-Tex11 and Hc-Meig1 may be involved in spermatocyte meiosis like mammals. It is speculated that they play a potential role in the regulation of spermatogenesis in H. cumingii.
TEX11 protein was observed in mice from late spermatogonia onwards, with the highest levels in zygotene spermatocytes and the lowest levels in pachytene spermatocytes (Adelman and Petrini, 2008). Meig1 was most abundantly expressed in pachytene spermatocytes (Don and Wolgemuth, 1992). In this study, mRNA signals of Hc-Tex11, Hc-Meig1 were detected on both spermatogonia and spermatocytes of the testes (Figure 8). This result was similar to the results of mouse cell localization, further confirming the potential role of Hc-Tex11 and Hc-Meig1 in the spermatogenesis of the H. cumingii.
Several studies have shown that Tex11 is strongly associated with azoospermia in human males (Sha et al., 2018; Yu et al., 2021). Tex11 gene affects the quality of bull sperm and thus the early embryo, and deletion or mutation of the Tex11 gene has been shown to cause meiotic arrest in animals (Yatsenko et al., 2015; Wu et al., 2020). In recent years, siRNA-mediated RNA interference has been widely used in the study of genes affecting spermatogenesis (Yang et al., 2020). In this study, Hc-Tex11 was silenced by RNAi to further explore the role of Hc-Tex11 in the spermatogenesis of H. cumingii (Figure 9A). To further investigate the effect of Hc-Tex11 deficiency on gonadal development, we examined the expression levels of several genes that have been shown to play evolutionarily conserved roles in gonadal development in the H. cumingii, including Hc-Dmrt1, Hc-β-catenin, Hc-KinaseX, Hc-Tra-2, and Hc-Klhl10. The expression of Hc-Dmrt1 and Hc-Tra-2 genes, which are important in sex determination and differentiation, and early gonadal development (Kim et al., 2003; Erickson and Quintero, 2007; Wang et al., 2019), was down-regulated following knockdown of Hc-Tex11. Dmrt1 maintains male germ cell differentiation and low expression results in impaired gonadal differentiation (Guo et al., 2020). As a result, it is speculated that Hc-Tex11 may be involved in gonadal development and germ cell differentiation in H. cumingii, by regulating the expression of Hc-Dmrt1 and Hc-Tra-2. KinaseX and Klhl10 genes are involved in spermatogenesis, sperm capacitation and fertilization (Stogios et al., 2005; Dong et al., 2020; Cui et al., 2021). The expression of Hc-KinaseX and Hc-Klhl10 was down-regulated after Hc-Tex11 knockdown. It is speculated that Hc-Tex11 may be involved in the spermatogenesis of H. cumingii by regulating the Hc-KinaseX and Hc-Klhl10. β-catenin is a key factor for pro-ovarian and anti-testicular whose activation prevents testis development (Li et al., 2014; Wang et al., 2019). The female-associated gene Hc-β-catenin was up-regulated after knockdown of Hc-Tex11, suggesting a possible negative regulatory relationship between Hc-Tex11 and Hc-β-catenin (Figure 9B). In summary, there is a link between the Hc-Tex11 and sex-related genes, but their interrelationships and the mechanisms regulating spermatogenesis need further research to be explored.
In conclusion, in this study, we obtained the full length cDNAs of Hc-Tex11 and Hc-Meig1 by cloning. Hc-Tex11 and Hc-Meig1 play a significant role in spermatogenesis and male gonad development in H. cumingii, as shown by qRT-PCR and in situ hybridization experiments, and revealed that their cellular localization was on spermatogonia and spermatocytes. The RNAi results illustrate some association between Hc-Tex11 and sex-related genes in H. cumingii. Among them, Hc-Dmrt1, Hc-KinaseX, HcTra-2 and Hc-Klhl10 were down-regulated and Hc-β-catenin was up-regulated. It shows that Hc-Tex11 regulates gonadal development, but this regulation needs to be further investigated.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
The animal study was reviewed and approved by Institutional Animal Care and Use Committee (IACUC) of the Shanghai Ocean University, China.
XW and YG Participate in sample collection, YH, YG, MC, YM, and YW participated in the experimental design and analyzed the data. YH was responsible for the writing of the manuscript. GW and JL presented ideas for the study and provided feedback on the experiments. All the authors agreed to the final version of the manuscript.
This work was supported by the National Key Research and Development Program of China (grant number 2018YFD0901406), the National Natural Science Foundation of China (grant number 31772835), the Ministry of Finance, and the China Agricultural Research System of MOF and MARA.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Tex11 Testis-expressed gene 11; Meig1 Meiosis expressed gene one; β-catenin beta-catenin; Dmrt1 Doublesex and Mab-3 Related Transcription Factor 1; Klhl10 Kelch-like protein 10; KinaseX protein kinaseX gene Tra-2 Transformer-2; TPR tetrapeptide repeat proteins.
1https://www.ncbi.nlm.nih.gov/orffinder/
2https://blast.ncbi.nlm.nih.gov/Blast.cgi.
3http://smart.embl-heidelberg.de/
4https://swissmodel.expasy.org/
5http://www.cbs.dtu.dk/services/TMHMM/
6http://www.cbs.dtu.dk/services/SignalP/
7https://web.expasy.org/protparam/
8http://www.cbs.dtu.dk/services/NetPhos/
Adelman C. A., Petrini J. H. J. (2008). ZIP4H (TEX11) deficiency in the mouse impairs meiotic double strand break repair and the regulation of crossing over. PLoS Genet. 4, e1000042. doi:10.1371/journal.pgen.1000042
Allan R. K., Ratajczak T. (2011). Versatile TPR domains accommodate different modes of target protein recognition and function. Cell. Stress Chaperones 16, 353–367. doi:10.1007/s12192-010-0248-0
Bai Z., Lin J., Ma K., Wang G., Niu D., Li J., et al. (2014). Identification of housekeeping genes suitable for gene expression analysis in the pearl mussel, Hyriopsis cumingii, during biomineralization. Mol. Genet. Genomics 289, 717–725. doi:10.1007/s00438-014-0837-1
Bellil H., Ghieh F., Hermel E., Mandon-Pepin B., Vialard F. (2021). Human testis-expressed (TEX) genes: A review focused on spermatogenesis and male fertility. Basic Clin. Androl. 31, 9. doi:10.1186/s12610-021-00127-7
Blatch G. L., Lassle M. (1999). The tetratricopeptide repeat: A structural motif mediating protein-protein interactions. Bioessays 21, 932–939. doi:10.1002/(SICI)1521-1878(199911)21:11<932:AID-BIES5>3.0.CO;2-N
Chen J. C., Shi A. J. (2002). Studies of oogenesis of Anodonta woodiana elliptica (Heude). Acta Sci. Nat. Univ. Szechuan. 3, 546–551. doi:10.3969/j.issn.0490-6756.2002.03.037
ChenMoses A., Malkov M., Shalom S., Ever L., Don J., Chen-Moses A. (1997). A switch in the phosphorylation state of the dimeric form of the meg1 protein correlates with progression through meiosis in the mouse. Cell. Growth Differ. 8, 711–719.
Cui X. Y., Dong S. S., Duan S. H., Wang G. L., Li J. L. (2021). Cloning and functional investigation of the kinase X gene of Hyriopsis cumingii. J. Fish. China. 46, 537–545. doi:10.11964/jfc.20201012453
Don J., Winer M. A., Wolgemuth D. J. (1994). Developmentally regulated expression during gametogenesis of the murine gene meg1 suggests a role in meiosis. Mol. Reprod. Dev. 38, 16–23. doi:10.1002/mrd.1080380104
Don J., Wolgemuth D. J. (1992). Identification and characterization of the regulated pattern of expression of a novel mouse gene, meg1, during the meiotic cell cycle. Cell. Growth Differ. 3, 495–505.
Dong S. S., Cui X. Y., Duan S. H., Xia S. Y., Liu F. F., Ge J. Y., et al. (2020). Characterization and expression analysis of KLHL10 gene in freshwater mussel Hyriopsis cumingii. J. Shanghai Ocean. Univ. 30, 389–398. doi:10.12024/jsou.20200402983
Erickson J. W., Quintero J. J. (2007). Indirect effects of pl.oidy suggest X chromosome dose, not the X : A ratio, signals sex in Drosophila. PLoS Biol. 5, e332. doi:10.1371/journal.pbio.0050332
Guo H., Wei M., Liu Y., Zhu Y., Xu W. T., Meng L., et al. (2017). Molecular cloning and expression analysis of the aqp1aa gene in half-smooth tongue sole (Cynoglossus semilaevis). Plos One 12, e0175033. doi:10.1371/journal.pone.0175033
Guo P. F., Duan S. H., Dong S. S., Wu C. D., Wang G. L. (2020). Molecular characterization and expression analysis of Dmrt1 gene in Hyriopsis cumingii. Genomics Appl. Biol. 39, 2033–2041. doi:10.13417/j.gab.039.002033
Haselman J. T., Olmstead A. W., Degitz S. J. (2015). Global gene expression during early differentiation of Xenopus (Silurana) tropicalis gonad tissues. Gen. Comp. Endocrinol. 214, 103–113. doi:10.1016/j.ygcen.2014.06.009
Hassold T., Hunt P. (2001). To err (meiotliuically) is human: the genesis of human aneuploidy. Nat. Rev. Genet. 2, 280–291. doi:10.1038/35066065
Heyting C. (1996). Synaptonemal complexes: Structure and function. Curr. Opin. Cell. Biol. 8, 389–396. doi:10.1016/s0955-0674(96)80015-9
Kim S. S., Kettlewell J. R., Anderson R. C., Bardwell V. J., Zarkower D. (2003). Sexually dimorphic expression of multiple doublesex-related genes in the embryonic mouse gonad. Gene Expr. Patterns 3, 77–82. doi:10.1016/s1567-133x(02)00071-6
Li H. H., Kong L. F., Yu R. H., Li Q. (2016a). Characterization, expression, and functional analysis of testis-specific serine/threonine kinase 1 (Tssk1) in the pen shell Atrina pectinata. Invertebr. Reprod. Dev. 60, 118–125. doi:10.1080/07924259.2016.1161667
Li H. L., Zhang Z. F., Bi Y., Yang D. D., Zhang L. T., Liu J. G., et al. (2014). Expression characteristics of beta-catenin in scallop Chlamys farreri gonads and its role as a potential upstream gene of Dax1 through canonical wnt signalling pathway regulating the spermatogenesis. Plos One 9, e115917. doi:10.1371/journal.pone.0115917
Li M. H., Sun Y. L., Zhao J. E., Shi H. J., Zeng S., Ye K., et al. (2015). A tandem duplicate of anti-mullerian hormone with a missense SNP on the Y chromosome is essential for male sex determination in nile Tilapia, Oreochromis niloticus. PLoS Genet. 11, e1005678. doi:10.1371/journal.pgen.1005678
Li W., Walavalkar N. M., Buchwald W. A., Teves M. E., Zhang L., Liu H., et al. (2016b). Dissecting the structural basis of MEIG1 interaction with PACRG. Sci. Rep. 6, 18278. doi:10.1038/srep18278
Liang S. S., Liu D. W., Li X. X., Wei M. K., Yu X. H., Li Q., et al. (2019). SOX2 participates in spermatogenesis of Zhikong scallop Chlamysfarreri. Sci. Rep. 9, 76. doi:10.1038/s41598-018-35983-3
Liang S. S., Zhang Z. F., Yang D. D., Chen Y. Y., Qin Z. K. (2017). Different expression of sox17 gene during gametogenesis between scallop Chlamys farreri and vertebrates. Gene Expr. Patterns 25-26, 102–108. doi:10.1016/j.gep.2017.06.009
Liang Y. Q., Jing Z. X., Pan C. G., Lin Z., Zhen Z., Hou L. P., et al. (2020). The progestin norethindrone alters growth, reproductive histology and gene expression in zebrafish (Danio rerio). Chemosphere 242, 125285. doi:10.1016/j.chemosphere.2019.125285
Lin W., Mu S. Y., Zhang L., Wang L. F. (2002). Study on the function of HSD-3.8 gene encoding testis-specific protein with yeast two-hybrid system. Acta Acad. Med. Sin. 6, 582–587. doi:10.1038/sj.cr.7290128
Liu D. K., Li L., Chen X., Tang C., Li J. M. (2012). Interaction of the TPR structural domains of Ppp5c and TTC16 genes with Hsp70 and Hsp90 proteins and effects on the cell cycle. Prog. Mod. Biomed. 12, 2005–2009. doi:10.13241/j.cnki.pmb.2012.11.014
Liu W. J., He X. J., Yang S. M., Zouari R., Wang J. X., Wu H., et al. (2019a). Bi-Allelic mutations in TTC21A induce asthenoteratospermia in humans and mice. Am. J. Hum. Genet. 104, 738–748. doi:10.1016/j.ajhg.2019.02.020
Liu X. X., Wang C. Y., Luo C., Sheng J. Q., Wu D., Hu B. J., et al. (2017). Characterization of cyclophilin D in freshwater pearl mussel (Hyriopsis schlegelii). Zool. Res. 38, 103–109. doi:10.24272/j.issn.2095-8137.2017.018
Liu Y. H., Zhang L., Li W., Huang Q., Yuan S., Li Y. H., et al. (2019b). The sperm-associated antigen 6 interactome and its role in spermatogenesis. Reproduction 158, 181–197. doi:10.1530/rep-18-0522
Mi Z. X., Wang D. W., Wang G. F., Li J. P. (2002). Observation of ultrastructure of spermatozoa of Hyriopsis cumingii. J. Chin. Electron Microsc. Soc. 21, 578–579.
Nishimura H., L'Hernault S. W. (2017). Spermatogenesis. Curr. Biol. 27, R988–R994. doi:10.1016/j.cub.2017.07.067
Pan B. B., Li J. L., Bai Z. Y. (2010). Histological study on ovarian development and oogenesis of Hyriopsis cumingii in the pond. J. Shanghai Ocean. Univ. 19, 452–456.
Pan X. Y., Chen X., Tong X., Tang C., Li J. M. (2015). Ppp2ca knockout in mice spermatogenesis. Reproduction 149, 385–391. doi:10.1530/rep-14-0231
Perez-Riba A., Itzhaki L. S. (2019). The tetratricopeptide-repeat motif is a versatile platform that enables diverse modes of molecular recognition. Curr. Opin. Struct. Biol. 54, 43–49. doi:10.1016/j.sbi.2018.12.004
Rozenfeld C., Garcia-Carpintero V., Perez L., Gallego V., Herranz-Jusdado J. G., Tveiten H., et al. (2019). Cold seawater induces early sexual developmental stages in the BPG axis of European eel males. BMC Genomics 20, 597. doi:10.1186/s12864-019-5969-6
Schmittgen T. D., Livak K. J. (2008). Analyzing real-time PCR data by the comparative C-T method. Nat. Protoc. 3, 1101–1108. doi:10.1038/nprot.2008.73
Sha Y. W., Zheng L. K., Ji Z. Y., Mei L. B., Ding L., Lin S. B., et al. (2018). A novel TEX11 mutation induces azoospermia: A case report of infertile brothers and literature review. BMC Med. Genet. 19, 63. doi:10.1186/s12881-018-0570-4
Shi J. T., Li Z., Gui J. F., Zhou L. (2015). The cloning and experssion analysis of zebrafish fem-1c, a member of fem-1 famly. Acta Hydrobiol. Sin. 39, 459–467. doi:10.7541/2015.61
Steiner R., Ever L., Don J. (1999). MEIG1 localizes to the nucleus and binds to meiotic chromosomes of spermatocytes as they initiate meiosis. Dev. Biol. 216, 635–645. doi:10.1006/dbio.1999.9520
Stogios P. J., Downs G. S., Jauhal J. J. S., Nandra S. K., Prive G. G. (2005). Sequence and structural analysis of BTB domain proteins. Genome Biol. 6, R82. doi:10.1186/gb-2005-6-10-r82
Tang L., Zeng W., Clark R. K., Dobrinski I. (2011). Characterization of the porcine testis-expressed gene 11 (Tex11). Spermatogenesis 1, 147–151. doi:10.4161/spmg.1.2.16680
Teves M. E., Jha K. N., Song J., Nagarkatti-Gude D. R., Herr J. C., Foster J. A., et al. (2013). Germ cell-specific disruption of the Meig1 gene causes impaired spermiogenesis in mice. Andrology 1, 37–46. doi:10.1111/j.2047-2927.2012.00001.x
Wang B. H. (2010). Study on Development of the Gonad of Hyriopsis schlegeli. master’s thesis. China: Nangchang University.
Wang G. L., Bai Z. Y., Liu X. J., Li J. L. (2014). Research progress on germplasm resources of Hyriopsis cumingii. J. Fish. China. 38, 1618–1627. doi:10.3724/SP.J.1231.2014.49308
Wang G. L., Dong S. S., Guo P. F., Cui X. Y., Duan S. H., Li J. L., et al. (2020). Identification of Foxl2 in freshwater mussel Hyriopsis cumingii and its involvement in sex differentiation. Gene 754, 144853. doi:10.1016/j.gene.2020.144853
Wang G. L., Liu F. F., Xu Z. C., Ge J. Y., Li J. L. (2019). Identification of Hc-beta-catenin in freshwater mussel Hyriopsis cumingii and its involvement in innate immunity and sex determination. Fish. Shellfish Immunol. 91, 99–107. doi:10.1016/j.fsi.2019.05.009
Wang P. J., McCarrey J. R., Yang F., Page D. C. (2001). An abundance of X-linked genes expressed in spermatogonia. Nat. Genet. 27, 422–426. doi:10.1038/86927
Wang Y. Y., Duan S. H., Wang G. L., Li J. L. (2021a). Integrated mRNA and miRNA expression profile analysis of female and male gonads in Hyriopsis cumingii. Sci. Rep. 11, 665. doi:10.1038/s41598-020-80264-7
Wang Y. Y., Wang X. Q., Ge J. Y., Wang G. L., Li J. L. (2021b). Identification and functional analysis of the sex-determiner transformer-2 homologue in the freshwater pearl mussel, Hyriopsis cumingii. Front. Physiol. 12, 704548. doi:10.3389/fphys.2021.704548
Wu C. Y., Blondin P., Vigneault C., Labrecque R., Sirard M. A. (2020). The age of the bull influences the transcriptome and epigenome of blastocysts produced by IVF. Theriogenology 144, 122–131. doi:10.1016/j.theriogenology.2019.12.020
Yang D. D., Zhang Z. F., Liang S. S., Yang Q. K., Wang Y. R., Qin Z. K., et al. (2017). A novel role of Kruppel-like factor 4 in Zhikong scallop Chlamys farreri during spermatogenesis. Plos One 12, e0180351. doi:10.1371/journal.pone.0180351
Yang F., Gell K., van der Heijden G. W., Eckardt S., Leu N. A., Page D. C., et al. (2008). Meiotic failure in male mice lacking an X-linked factor. Genes Dev. 22, 682–691. doi:10.1101/gad.1613608
Yang T. A., Yang Y. H., Liu H. M., Song X. C., Liu L. L., Yang F. H., et al. (2019). MEIG1 gene Expression in foxes and their Hybrids during Preparative mating period and breeding period. J. Jilin Agric. Univ. 41, 621–624. doi:10.13327/j.jjlau.2019.4993
Yang T., Wei B. H., Hao S. L., Wei Y. L., Yang W. X. (2020). Bone morphogenetic protein 2 (BMP2) mediates spermatogenesis in Chinese mitten crab Eriocheir sinensis by regulating kinesin motor KIFC1 expression. Gene 754, 144848. doi:10.1016/j.gene.2020.144848
Yatsenko A. N., Georgiadis A. P., Ropke A., Berman A. J., Jaffe T., Olszewska M., et al. (2015). X-linked TEX11 mutations, meiotic arrest, and azoospermia in infertile men. N. Engl. J. Med. 372, 2097–2107. doi:10.1056/NEJMoa1406192
Yu X. C., Li M. J., Cai F. F., Yang S. J., Liu H. B., Zhang H. B., et al. (2021). A new TEX11 mutation causes azoospermia and testicular meiotic arrest. Asian J. Androl. 23, 510–515. doi:10.4103/aja.aja_8_21
Yu Y. H., Siao F. P., Hsu L. C. L., Yen P. H. (2012). TEX11 modulates germ cell proliferation by competing with estrogen receptor beta for the binding to HPIP. Mol. Endocrinol. 26, 630–642. doi:10.1210/me.2011-1263
Yu Y., Jiang Y. H., Qiu Q. J., Wang J. H., Xu M. X., Li Y. J. (2008). Embryonic developmene and breading-season gonad in Hyriopsis schlegelii. Chin. J. Zool. 3, 102–107. doi:10.13859/j.cjz.2008.03.030
Yue C. Y., Li Q., Yu H., Liu S. K., Kong L. F. (2020). Restriction site-associated DNA sequencing (RAD-seq) analysis in Pacific oyster Crassostrea gigas based on observation of individual sex changes. Sci. Rep. 10, 9873. doi:10.1038/s41598-020-67007-4
Yue M. S., Fan X. R., Liu Y. H., Yue W. D., Ren G. Y., Zhang J. W., et al. (2019). Effects of body temperature on the expression and localization of meiosis-related proteins STRA8 and SCP3 in boar testes. Acta Histochem. 121, 718–723. doi:10.1016/j.acthis.2019.06.007
Zhang K., Xu J., Zhang Z. W., Huang Y., Ruan Z. Q., Chen S. Y., et al. (2019). A comparative transcriptomic study on developmental gonads provides novel insights into sex change in the protandrous black porgy (Acanthopagrus schlegelii). Genomics 111, 277–283. doi:10.1016/j.ygeno.2018.11.006
Zhang X. N., Zhou W. W., Zhang P., Gao F. X., Zhao X. L., Shum W. W., et al. (2021). Cabs1 maintains structural integrity of mouse sperm flagella during epididymal transit of sperm. Int. J. Mol. Sci. 22, 652. doi:10.3390/ijms22020652
Zhang Y., Oko R., van der Hoorn F. A. (2004). Rat kinesin light chain 3 associates with spermatid mitochondria. Dev. Biol. 275, 23–33. doi:10.1016/j.ydbio.2004.07.014
Zhang Y., Ou Y., Cheng M., Saadi H. S., Thundathil J. C., van der Hoorn F. A., et al. (2012). KLC3 is involved in sperm tail midpiece formation and sperm function. Dev. Biol. 366, 101–110. doi:10.1016/j.ydbio.2012.04.026
Keywords: Hyriopsis cumingii, Tex11, Meig1, spermatogenesis, gonadal development, RNAi
Citation: Huo Y, Gu Y, Cao M, Mao Y, Wang Y, Wang X, Wang G and Li J (2022) Identification and functional analysis of Tex11 and Meig1 in spermatogenesis of Hyriopsis cumingii. Front. Physiol. 13:961773. doi: 10.3389/fphys.2022.961773
Received: 05 June 2022; Accepted: 04 July 2022;
Published: 17 August 2022.
Edited by:
Rui Jia, Freshwater Fisheries Research Center (CAFS), ChinaReviewed by:
Shuang Zhang, Guangdong Ocean University, ChinaCopyright © 2022 Huo, Gu, Cao, Mao, Wang, Wang, Wang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guiling Wang, eWh3dXdhbmcyMDA4QDEyNi5jb20=
†These authors share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.