
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Physiol. , 18 July 2022
Sec. Reproduction
Volume 13 - 2022 | https://doi.org/10.3389/fphys.2022.926795
 Guohua Song1†
Guohua Song1† Yadong Wang1†
Yadong Wang1† Yaling Wang1
Yaling Wang1 Yixuan Jiang1
Yixuan Jiang1 Shuaijie Sun1
Shuaijie Sun1 Hanfang Cai1
Hanfang Cai1 Guirong Sun1
Guirong Sun1 Ming Li1*
Ming Li1* Massimo Bionaz2*
Massimo Bionaz2* Huifen Xu1*
Huifen Xu1*Coprophagy is an instinctive behavior in rabbit with important effects on growth and reproductive performance. The underlying mechanism of this effect in rabbit is unknown. Here, we used Elizabeth circle as a coprophagy preventing model in female rabbits and assess feed intake, growth, and reproductive performance. We found that preventing coprophagy did not affect feed intake but decreased body weight and weight of several organs and tissues and resulted in complete reproductive failure during the late pregnancy period, accompanied by reduced levels of plasma progesterone. RNA-seq analysis of rabbit ovarian tissues revealed that preventing coprophagy affected significantly 241 genes (DEGs), with the large majority being downregulated. Bioinformatic analyses revealed that those DEGs are mostly involved in apoptosis, immune response, and metabolic pathways. Among DEGs, the lysosomal cysteine protease cathepsin B (CTSB) was significantly downregulated in the coprophagy prevention group. Further studies using siRNA and adenovirus overexpression systems revealed that CTSB promotes the proliferation of rabbit granulosa cells (GCS) and prevents apoptosis. Measurement of transcripts coding for proteins related to apoptosis revealed a minor transcriptomic effect of CTSB, indicating that its effect is likely post-transcriptional. Overexpression of CTSB increased secretion of progesterone and estradiol, partly via upregulation of CYP19A1 while inhibition of CTSB decreased progesterone secretion partly via downregulation of the StAR gene. In conclusion, our study demonstrated the detrimental effect on reproduction by preventing coprophagy with a main role for this response played by CTSB on the granulosa cells of the ovary.
Coprophagy is a terminology that is used to describe the behavior of eating feces in small herbivores (Garcia et al., 2004; Ferreira et al., 2018). Two types of coprophagy behavior have been observed: the ingestion of own feces (autocoprophagy) or ingestion of excrements of another animal of the same species (allocoprophagy) (Galef and Bennett, 1979). This behavior allows hindgut-fermenting mammals to recover the nutrients that were liberated in the hindgut and aids the absorption of essential amino acids, vitamin B, vitamin K, and trace elements, thereby avoiding the loss of these nutrients (Savage, 1986). Due to the colonic separation mechanism, many small herbivores such as rabbits, golden hamsters, rodents, and koalas produce two types of feces (hard feces and soft feces), and they acquire the behavior of coprophagy during specific growing periods or under conditions of malnutrition (Sakaguchi, 2003).
The nutritional importance of coprophagy has been demonstrated, and it can increase the total intake of protein up to 38% in growing rabbits (Gidenne, 2000; Belenguer et al., 2005; Liu et al., 2007). Coprophagy can avoid the loss of endogenous nitrogen and increase absorption of essential amino acids, providing up to 23% of lysine for lactating rabbits (Abecia et al., 2008; García et al., 2004). In addition to nutritional benefits, coprophagy behavior is helpful for maintaining gut microflora homeostasis and this was supported by the evidence that fasting coprophagy resulted in marked changes in the diversity of caecal microorganisms (Klaasen et al., 2009; Kobayashi et al., 2019; Li et al., 2020), affecting nutrition and energy homeostasis (Tremaroli and Backhed, 2012; Zietak et al., 2016). The maintenance of the gut microbiota by coprophagy is also important for cognitive behavior, as observed in Brandt’s vole (Bo et al., 2020).
Coprophagy behavior in small herbivores is an adaptive strategy to the disadvantage of the small body size for the digestion of low-quality fiber (Pei et al., 2001; Liu and Wang, 2007; Sakamaki, 2010). Coprophagy has great impact on the growth of rabbits, as the body size and body weight are significantly decreased when coprophagy is prevented (Sukemori et al., 2006; Wang et al., 2019; Bo et al., 2020). Coprophagy is also important for pregnancy and lactation, as female mice exhibit coprophagy more frequently and ingest more feces during pregnancy and lactation than the non-pregnant period (Ebino et al., 1988). Prevention of coprophagy in mice increases abortion, likely due to deficiency of vitamin B12 and folic acid during pregnancy (Sukemori et al., 2006; Ebino et al., 1989).
The aforementioned data indicated that coprophagy is an important behavior during pregnancy in mice; however, the role of coprophagy on reproduction of rabbits has not been studied. Our hypothesis is that coprophagy positively affects reproduction in rabbits. To assess our hypothesis, we used Elizabeth circle to prevent coprophagy for 5 months in female rabbits and assessed its effects on growth and reproductive performance. Furthermore, via RNA-seq of ovary and molecular biology approaches using cumulus cells, we attempted to reveal the molecular mechanism responsible for the reproductive response mediated by coprophagy in rabbits.
The present study was approved by the Institutional Animals Care and Use Committee (IACUC) of the College of Animal Science and Technology of Henan Agricultural University, China (Permit Number: 11-0085; Data: 06-2011) and was designed and performed according to the institutional guidelines.
For the experiment we used 12 healthy, weaned 30-days old female New Zealand white rabbits with similar body weight (1.14 ± 0.12 kg) from the animal experimental center of Henan Agricultural University. The rabbits were maintained in individual cages and were randomly divided into two groups: control group and experimental group (i.e., coprophagy prevention group). The coprophagy prevention group was fitted with a 7-cm wide Elizabeth circle to prevent coprophagy (wang et al., 2019). Food intake of the rabbits was measured every day. Body weight of the rabbits was measured every week. After 22 weeks of feeding, rabbits received an intramuscular injection of chorionic gonadotrophin (SanSheng Biotechnology, NingBo, China), followed by artificial insemination (210 days). Pregnancy diagnosis was performed by using B-ultrasound (Mindary, DP-10) 15 days after artificial insemination (225 days).
After giving birth to pups and/or on the 30th day of pregnancy, rabbits were anesthetized followed by euthanization for the collection of samples. The abdominal fat, liver, stomach, kidney, uterus, lung, heart, and spleen were collected and weighed. All tissue samples, including ovaries, were flash-frozen in liquid nitrogen and preserved at -80°C. The blood samples were collected and placed in pro-coagulant tubes (Shandong Ao Sai Te Medical Devices Co’ LTD, Shandong, China) during sacrifice, the serum was obtained by centrifuging and stored at -80°C for analysis.
All kits used to measure serum biochemical parameters were purchased from the Nanjing Jiancheng Institute of Biological Engineering (Nanjing, China) and measured according to the manufacturer’s instructions using a microplate reader (Bio-Rad). The parameters examined included high-density lipoprotein cholesterol (Cat. No.F003-1-1), low-density lipoprotein cholesterol (Cat. No. A019-2-1), total cholesterol (Cat. No. F002-1-1), triglyceride (Cat. No. A110-1-1), albumin (Cat. No. A028-1-1), and progesterone (Cat. No. H089).
Total RNA of ovary samples was extracted by using TRIzol reagent (Invitrogen, CA, United States) and stored at -80°C. The quality and concentration of the total RNA was quantified by using the Nanodrop 2000 spectrophotometer (Thermo Scientific). RNA integrity was measured using the Nano 6000 LabChip kit (Agilent, Technologies, CA, United States). The RIN (RNA integrity number) of all RNA samples was >8.6. One aliquot of the RNA per each sample (n = 3 per group) was sent for the preparation of RNA-seq libraries (Biomarker Biotechnology Co., Ltd, Beijing, China), and the remaining aliquot of RNA samples was used for RT-qPCR analysis.
The library of each sample was sequenced using Illumina HiSeqTM 2500 (Biomarker Biotechnology Co., Ltd, Beijing, China). The Q20, Q30, GC-content, and sequence duplication level of the clean data were calculated, the clean data (clean reads) were obtained by removing reads containing adapter, reads containing ploy-N, and low-quality reads from raw data.
Clean reads were mapped to the reference genome of the rabbit (Orycun2.0) and annotated using TopHat 2 software (http://ccb.jhu.edu/software/tophat/index.shtml). Cufflinks software (Cufflinks, Berkeley, CA, United States) was used for splicing and expression quantification. The dataset is available in the public repository GEO NCBI Sequence Read Archive (SRA accession PRJNA767495).
Statistical analysis of data normalized by the RPKM method (reads per kilo base transcriptome per million mapped reads) was performed using the DESeq R package (1.10.1) (Anders and Huber, 2010), with negative binomial distribution. The resulting p-value was adjusted using Benjamin and Hochberg’s multiple testing correction for controlling the false discovery rate (FDR). An FDR <0.05 was considered as the thresholds for screening significantly differentially expressed mRNA (DEGs).
Bioinformatics analysis was performed using three different tools. The KEGG database was used for the DEGs pathway analysis using KOBAS (http://kobas.cbi.pku.edu.cn/) (Mao et al., 2005). Pathways with a Q value ≤0.05 were defined as significantly enriched. The data were analyzed also using the Dynamic Impact Approach (DIA) (Bionaz et al., 2012) and Database for Annotation, Visualization, and Integrated Discovery (DAVID) (Sherman et al., 2022). For the DIA, the orthologous mouse gene IDs were used after annotation of the rabbit gene IDs were performed using the biological DataBase network (bioDBnet) (Mudunuri et al., 2009). The whole annotated gene IDs were used as background for both DAVID (where the Ensembl gene IDs were used) and DIA (where the Entrez Gene IDs were used). For DAVID, an EASE score ≤0.05 was used to identify significant enriched terms, and results were downloaded as a functional annotation chart, using the default DAVID annotation categories. The Ensembl gene IDs with an FDR<0.05 were uploaded into DAVID as all DEGs, upregulated DEGs, and downregulated DEGs.
The rabbits’ granulosa cells (GCS) were isolated and cultured as previously described (Song et al., 2021) from the ovaries of three rabbits purchased from the Laboratory Animal Center (Zhengzhou, Henan, China). Briefly, ovaries from 6 months female rabbits were collected and washed in PBS (with 1% penicillin/streptomycin, 37°C). These rabbits were first intramuscularly injected with 50 IU pregnant mare serum gonadotropin (Solarbio, Beijing, China) to induce estrus. The follicles were punctured with 1 ml syringe needle to collect GCS. Those were cultured in a basal culture medium which consisted of DMEM/F12 medium (Gibco, United States) supplemented with 15% fetal bovine serum (Gibco, United States) and 1% penicillin/streptomycin (Hyclone, United States). The GCS were collected by centrifugation (5 min, 1,000 rpm/min) and incubated in basal culture medium at 37°C in 5% CO2, and cell medium was changed every 24 h.
Triplicate for each rabbit of sections of ovarian tissue and GCS-crawling film were used for immunofluorescence staining. Ovarian tissue sections and GCS-crawling film were fixed with 4% paraformaldehyde for 15 min and then permeabilized with 0.1% Triton X-100 for 10 min. Then cell-climbing slices were incubated with an antifollicle-stimulating hormone receptor (MAB65591, 1: 200, R&D Systems, Minneapolis, United States) and ovarian sections were incubated with anti-CTSB (48118-1, 1:300, Signalway Antibody, Maryland, United States) rabbit polyclonal antibody for 1 h at 37°C. The sections were then incubated with allophycoerythrin (APC)-goat antirabbit Ig G (GB25303, 1:400, Servicebio, Wuhan, China) at 37°C for 45 min. Finally, the ovarian section and cell-climbing slices were taken out and mounted with DAPI medium (Solarbio). The results were observed under a laser scanning confocal microscope (Nikon Eclipse C1).
The ovaries were made into paraffin sections, deparaffinized, and rehydrated. The ovaries were incubated with proteinase K working solution at 37°C for 25 min, and covered with permeabilization working solution (Servicebio; Hubei, China) for 20 min. According to the instructions of the TUNEL assay kit (Roche; Basel, Switzerland), the ovaries were incubated with mix reagent 1 (TdT) and reagent 2 (dUTP) at 37°C for 2 h. Cell nuclei were stained with DAPI (Beyotime). The cells were observed under a fluorescence microscope (Nikon, Tokyo, Japan).
GCS were seeded in 6-well culture plates with cell-climbing slices. The cells were fixed with 95% ethanol for 20 min and stained with hematoxylin dye solution for 3 min. After dehydration with gradient alcohol, the cells were dyed with eosin dye solution for 5 min. Finally, the cells were mounted with neutral gum and observed under a microscope (Nikon Eclipse E100).
The specific siRNA sequence targeting CTSB was designed and synthesized by GenePharma (Shanghai, China). CTSB-siRNA, sense: 5′-CCGGGCACAACUUCUUCAATT-3′ and antisense: 5′-UUGAAGAAGUUGUGCCCGGTT-3′. NC-siRNA, sense: 5′-UUCUCCGAACGUGUCACGUTT-3′ and antisense: 5′-ACGUGACACGUUCGGAGAATT-3′. siRNA was transfected into GCS by using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, United States), according to the manufacturer’s instructions.
The production of recombinant adenovirus was previously described (Song et al., 2021). Briefly, CTSB gene was amplified and subcloned into the shuttle vector pAdTrack-CMV. The linearized product of pAdTrack-CMV-CTSB was transformed into Escherichia coli BJ 5183 competent cells containing the backbone vector pAdEasy-1 to obtain the positive recombinants of pAd-CTSB. Linearized pAd-CTSB fragment was transfected into HEK293 cells for adenovirus packaging and amplification. GCS were infected with pAd-CTSB at an MOI value of 40.
Total RNA of ovary tissues and rabbit GCS was extracted with the TRIzol reagent (Invitrogen, Carlsbad, CA, United States) and RNAprep pure cell kit (Tiangen Biotech Co. Ltd., Beijing, China), respectively. cDNA was synthesized using PrimeScript RT kit (Takara, Tokyo, Japan). RT-qPCR was performed using SYBR®Premix Ex TaqTM ⅡKit (Takara, Tokyo, Japan) on a CFX96TM real-time PCR detection system (Bio-Rad Laboratories Inc, Hercules, CA). All the primer sequences were designed by using NCBI (https://www.ncbi.nlm.nih.gov/) and Oligo 7 software (Molecular Biology Insights, Cascade, CO) and are listed in Supplementary Table S1. LinRegPCR was used to calculate the final RT-qPCR data (Ruijter et al., 2009). Four internal control genes were tested (GAPDH, YWHAZ, ACTB, and HPRT1). According to geNorm results, the geometrical mean of three reference genes (YWHAZ, ACTB, and HPRT1) was used to normalize RT-qPCR for the ovary transcriptome and the geometrical mean of ACTB and HPRT1 for the CTSB overexpression or inhibition experiments (see Supplementary Figure S3). Final data for each sample were calculated as expression ratio relative to the mean of the control group in all experiments.
After 48 h treatment with recombinant CTSB adenovirus or siRNA, rabbit GCS cultured in 6-well culture plates were washed with PBS and lysed using RIPA lysis buffer (Epizyme, Shanghai, China). Protein concentration was quantified with a BCA kit (Beijing, China), processed with 12.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and translocated to nitrocellulose membranes. The membrane was sealed with 5% skimmed milk for 2 h and incubated with anti-CTSB (48118-1, 1:5000, Signalway Antibody, Maryland, United States) and β-actin (GB12001, 1:1000, Servicebio, Wuhan, China) at 4°C overnight followed by incubation with goat antirabbit IgG antibody (SA00001-1, 1:1000, Proteintech, Wuhan, China) at room temperature for 2 h. Blots were visualized using GE Amersham Imager 600 (General Electric Company, Bosto, MA, United States) and analyzed with ImageJ.
Rabbits GCS were cultured in a 6-well culture plate, after 48 h treatment with recombinant CTSB adenovirus and siRNA, and the culture medium was collected for hormone detection. Progesterone and estradiol were quantified with commercial kits, according to the manufacturer’s instructions (Ruixin, Quanzhou, China). The absorbance was measured using a microplate reader (Bio-Rad) at 450 nm.
GCS (1×104/well) were seeded in 96-well plates. After treatment with recombinant CTSB adenovirus and siRNA for 12, 24, 36, and 48 h, GCS were collected for proliferation assay by Cell Counting Kit-8 (CCK8) (US Everbright® Inc., Jiangsu, China). Ten μL of CCK8 solution was added to each well. After 2 h incubation, the absorbance was measured by a microplate reader (Bio-Rad) at 450 nm.
Rabbits GCS were cultured in a 12-well culture plate, after treatment of GCS for 48 h treatment with recombinant CTSB adenovirus or siRNA, cell apoptosis was detected with the Annexin V-Alexa Fluor 647/7-AAD apoptosis kit (4A Biotech., Beijing, China). In short, GCS were digested with 0.05% trypsin (without EDTA) and resuspended with cold PBS, and then mixed with 5 μl Annexin V/Alexa Fluor 647 at room temperature for 5 min. Finally, 10 μl 7-AAD and 400 μl PBS were added. The apoptosis rate of GCS was measured with a CytoFLEX flow cytometer (Beckman CytoFlex, CA, United States).
All treatments were performed in biological triplicate, and data are presented as means ± SE. Statistical analysis for all parameters except RT-qPCR data were performed with one-way ANOVA to compare the effects of different treatments by SPSS statistics 22.0 (SPSS, Chicago, IL, United States). For RT-qPCR data, a general linear model (SAS v9; SAS Institute, Cary, NC, United States) with treatment (coprophagy prevention or control; overexpression of CTSB or wild-type; and interference CTSB or wild-type) as main effect was used. Differences were considered statistically significant when p < 0.05.
Prevention of coprophagy did not affect feed intake (Figure 1A, p = 0.53) but decreased body weight of the rabbits from the fifth week to the end of the experiment (Figure 1B, p < 0.05). The weight of the liver, perirenal fat, stomach, and kidney in the coprophagy prevention group was significantly lower than the control group (Figure 1C, p < 0.05). No differences were observed in the levels of serum albumin, triglycerides, total cholesterol, HDL-cholesterol, and LDL-cholesterol between the control group and the coprophagy prevention group (Figure 1D, p > 0.05). However, serum progesterone was significantly decreased by preventing coprophagy (Figure 1D, p < 0.05).
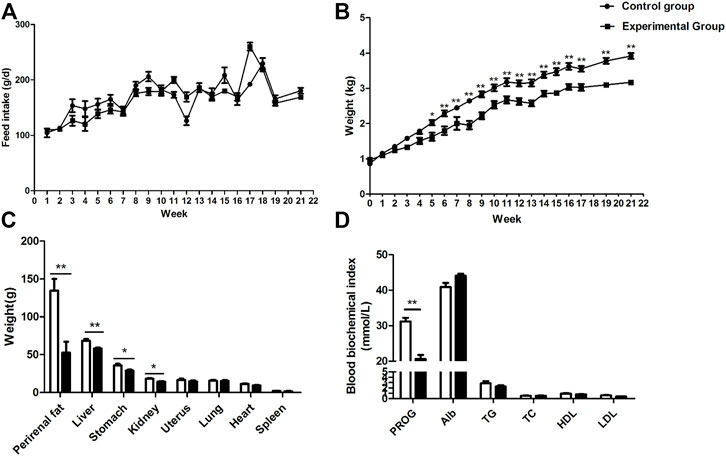
FIGURE 1. Effects of coprophagy prevention on (A) feed intakes; (B) body weight; (C) weight of different tissues and internal organs; and (D) serum biochemical parameters in rabbits [progesterone (PROG), albumin (Alb), triglycerides (TG), total cholesterol (TC), HDL-cholesterol (HDL), and LDL-cholesterol (LDL)]. Statistical differences between groups were indicated as * for p < 0.05, ** for p < 0.01, and *** for p < 0.001.
Fifteen days after artificial insemination, rabbits of the two groups were subjected to B-mode ultrasound examination to check the status of pregnancy of the animals. As shown in Supplementary Figure S1, compared with non-pregnant rabbits, embryos in the rabbits of the control group and coprophagy prevention group were clearly detected, indicating that rabbits in both groups were all pregnant. However, after 30 days of pregnancy, only the rabbits in the control group gave birth to newborn rabbits, while rabbits in the coprophagy prevention group did not gave birth to any newborn rabbits (Table 1). Despite the lack of any litter, we did not observe dead fetuses in the uterus after slaughter that happened just after the due date.
In order to investigate the possible mechanism of coprophagy prevention on reproduction, TUNEL assay was performed to measure the apoptosis of ovarian tissue cells. As shown in Figure 2, the proportion of apoptotic cells was significantly higher in the ovarian tissue of rabbits where coprophagy was prevented compared to the control group.
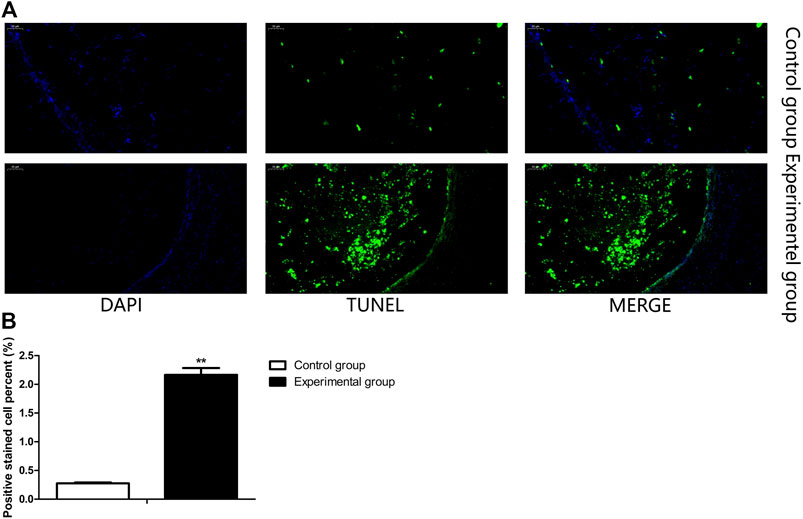
FIGURE 2. Coprophagy prevention increased cell apoptosis in ovary of rabbits. (A) Representative images of ovarian stained by TUNEL assay, cell nuclei were stained by DAPI (blue), apoptotic cells were stained with TUNEL (green). (B) Quantification of the proportion of ovarian apoptotic cells as detected by TUNEL assay in each group of rabbits. Scale bar = 50 μm **p < 0.01.
A total of 14,378 transcripts with a minimum count of four in at the least two samples were expressed in ovaries of the two groups of rabbits, 241 DEGs were present in these two groups, in which 210 were downregulated and 31 were upregulated DEGs (Figure 3A, FDR ≤0.05; complete dataset is available in Supplementary File S1).
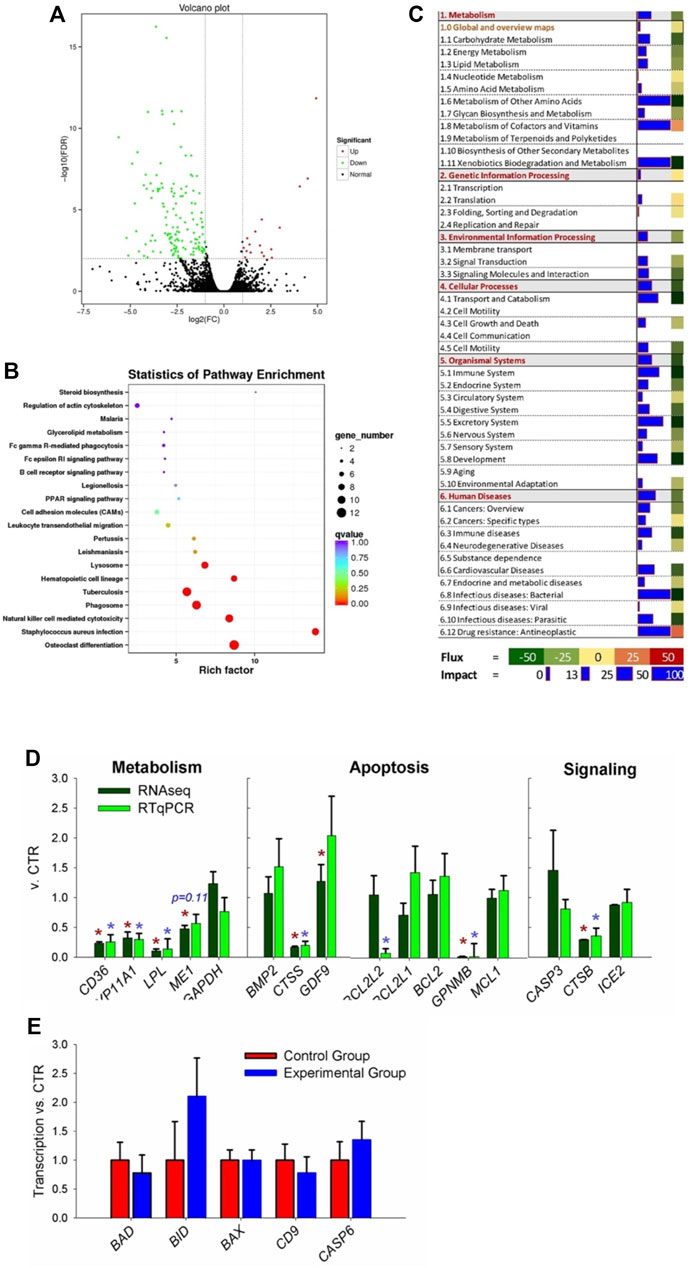
FIGURE 3. Transcriptomic analysis of ovary tissues in rabbit where coprophagy was prevented (i.e., experimental group) and a control group (n = 3 per group). (A) Volcano plot of differentially expressed genes (expression ratio experimental/control group). (B) KEGG pathways enriched (p < 0.05) in DEGs by using KOBAS. (C) Summary of main categories of KEGG pathways generated by using the Dynamic Impact Approach. Blue horizontal bars denote impact (larger the bar, larger the impact) and squares besides the blue horizontal bars denote flux of the pathway that can be induced or inhibited as indicated in the bottom legend. (D) RNA-seq vs. RT-qPCR for transcripts selected among the ones related to metabolism, apoptosis, and signaling pathways. (E) Additional transcripts measured by RT-qPCR involved in apoptosis and related to cathepsin B activity.
Bioinformatics analysis of DEGs performed using KOBAS not only revealed an enrichment of KEGG pathways related to differentiation (specifically of osteoclasts and hematopoietic cells), infection, and immune response (e.g., response to Staphylococcus infection, natural killer cell-mediated cytotoxicity, leukocytes migration, and several human-related diseases) but also pathways related to apoptosis, such as ‘Lysosome’ and ‘Phagosome’ (Figure 3B). Few KEGG pathways related to metabolism were significantly enriched, including PPAR signaling, glycerol-lipid metabolism, and steroid biosynthesis.
Summary of KEGG pathway as performed by DIA (Figure 3C), revealed an overall inhibition on all categories of pathways except ‘genetic information processing.’ Metabolism of other amino acids, cofactors, vitamins, and xenobiotic were among the most impacted and inhibited metabolic sub-category of pathways. When looking at the detail of each metabolic pathway, ‘nicotinate and nicotinamide metabolism’ was highly activated and ‘folate biosynthesis,’ ‘drug metabolism–cytochrome P450,’ and ‘glycolipid metabolism’ were highly inhibited (Supplementary File S2). Several signaling pathways were impacted by the prevention of coprophagy on ovaries, such as ‘PPAR signaling’ and ‘cytokine–cytokine receptor interaction’ that were inhibited and ‘Wnt signaling pathway’ that was activated (Supplementary File S2). The pathways associated with apoptosis were overall inhibited as well pathways associated with the immune system, such as ‘natural killer cell mediated cytotoxicity,’ ‘antigen processing and presentation,’ and ‘Toll-like receptor signaling pathway.’ Highly inhibited pathways were also associated with transport, catabolism, and apoptosis, such as ‘lysosome,’ ‘endocytosis,’ ‘phagosome,’ and ‘apoptosis’ (Supplementary File S2). Among pathways associated with diseases, the most impacted and inhibited was ‘Staphylococcus aureus infection,’ ‘tuberculosis,’ and ‘chemical carcinogenesis’ (Supplementary File S2).
Results from DAVID were somewhat consistent with the other bioinformatics tools. Highly enriched were pathways associated with disease and immune response (e.g., ‘Staphylococcus aureus infection’ and ‘natural killer cell mediated cytotoxicity’), lipid metabolism (e.g., ‘glycerolipid metabolism’), signaling (e.g., ‘JAK-STAT signaling pathway’), and organelles involved with apoptosis (e.g., ‘lysosome’ and ‘phagosome’) (Supplementary File S2). When considering additional annotations, DAVID results revealed significant enrichment of membrane-associated components, immunoglobulins, diseases-associated pathways, glycoproteins, peptidases associated with the lysosome, and lipid metabolism-related terms. All the enriched terms in DAVID were downregulated DEGs (Supplementary File S2).
We selected 16 transcripts coding for proteins related to metabolism, apoptosis, and signaling to validate the RNA-seq data (Figure 3D). Except for 1 gene (BCL2L2), all of the other transcripts had the same pattern and for 14 out 16 transcripts the statistical effect was conserved.
Among the transcripts selected for validation, the cathepsin B (CTSB) was significantly decreased by the prevention of coprophagy (Figure 3D). The reasons for focusing on this transcript are: 1) the CTSB transcript codes for a member of the cathepsin family, which is mainly expressed in lysosomes and plays a variety of roles in antigen presentation (Brix et al., 2008), apoptosis (Conus and Simon, 2010), autophagy (Tardy et al., 2006), and cell homeostasis (Li and Albertini, 2013), all functions and terms that were revealed to be important by bioinformatics results of the transcriptomic data in the ovary; 2) CTSB plays an important role in cell autophagy and apoptosis (Morchang et al., 2013; Tatti et al., 2013), which could help explaining the high apoptosis we observed in ovaries when rabbit were prevented to perform coprophagy. It has been reported that CTSB plays an important role in regulating the formation of cumulus-oocyte complex and the maturation of oocytes, which will subsequently affect reproductive performance (Balboula et al., 2010; Balboula et al., 2013). However, the role of the CTSB gene in rabbit reproductive performance has not been previously studied.
CTSB in the cytoplasm initiates the lysosomal pathway of apoptosis, mainly through cleavage of BH3-interacting domain death agonist (BID) and anti-apoptotic degradation of BCL-2 homologues (Repnik et al., 2012). Thus, to further evaluate if CTSB is implied in regulating apoptosis in the ovaries of rabbit via the BCL-2 pathway, we performed RT-qPCR analysis of additional transcripts that were not in the RNA-seq dataset but are known to be important in apoptosis, such as BCL2-associated X (BAX), BID and BCL2-associated agonist of cell death (BAD) (Figure 3E). We were able to detect all those transcripts but none resulted to be differentially expressed in the ovary of the two groups.
In order to investigate the role of CTSB in reproduction and the association between coprophagy prevention and reproductive failure, GCS of rabbits were isolated from three female rabbits and stained with hematoxylin and eosin (H&E) (Figure 4A). The cells exhibited a typical morphology expected for GCS, with obvious nuclei (blue) and cytoplasm (red) and with pseudopodia between the cells closely connected. Identification of rabbits GCS was performed by using immunofluorescence staining with the marker protein follicle-stimulating hormone receptor (FSHR). As shown in Figure 4B, FSHR protein was abundantly present in the cytoplasm of the cells, indicating that the isolated cells were indeed GCS.
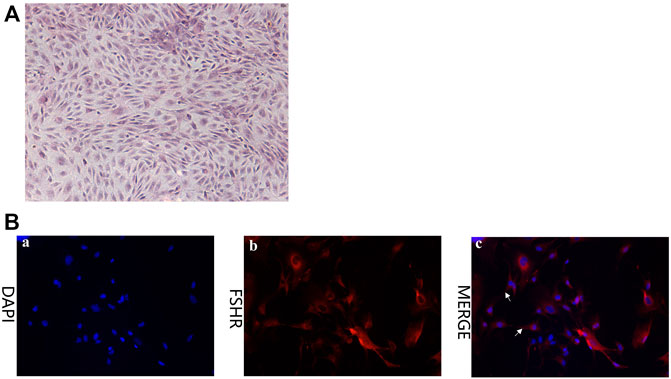
FIGURE 4. Characterization of granulosa cells (GCS) isolate from ovaries of healthy female rabbits (5 months of age). (A) H&E staining (40×). (B) Expression of follicle-stimulating hormone receptor (FSHR) in GCS isolated from ovaries of rabbits as detected via immunofluorescence staining (red = FSHR; blue = nuclei) (200×). Pseudopodia between cells are indicated with white arrows.
To study the role of CTSB in GCS, we overexpressed and silenced the gene. The transcription and protein expression of CTSB were significantly affected by overexpression (Ad-CTSB) and silencing (siRNA) (Figure 5).
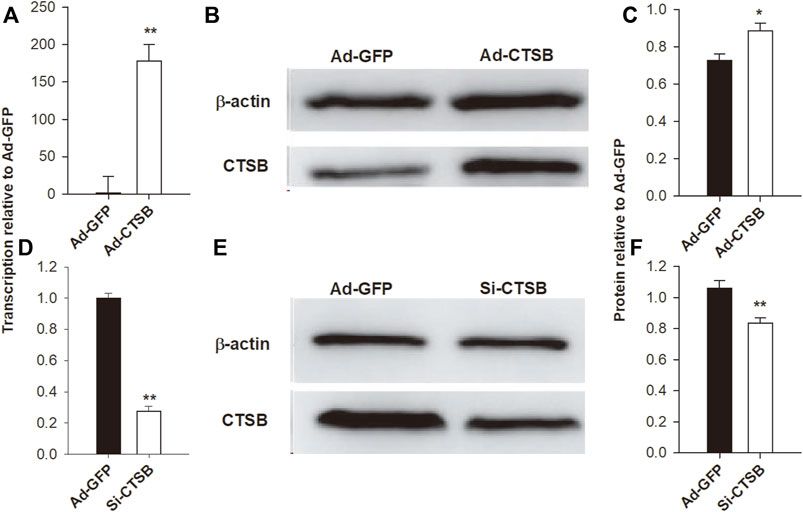
FIGURE 5. Adenovirus-mediated overexpression and siRNA-mediated interference of CTSB in rabbit GCS. The transcription of mRNA via RT-qPCR (A,D) and the protein expression via Western blot (B,C,E,F) of CTSB were highly affected by overexpression and interference treatments. Each experiment was performed with three biological replicates, and the results are expressed as the mean ± SEM. *p < 0.05, **p < 0.01.
Transcription of proliferating cell nuclear antigen (PCNA), a marker of cell proliferation, was significantly decreased (p < 0.05) by CTSB overexpression (Figure 6A) and numerically decreased with CTSB knockdown (Figure 6C). While transcription of the other two proliferation related genes, CCND1 and NOTCH2, was not affected by CTSB overexpression (p > 0.05) but NOTCH2 tended (p = 0.06) to be reduced by CTSB knockdown.
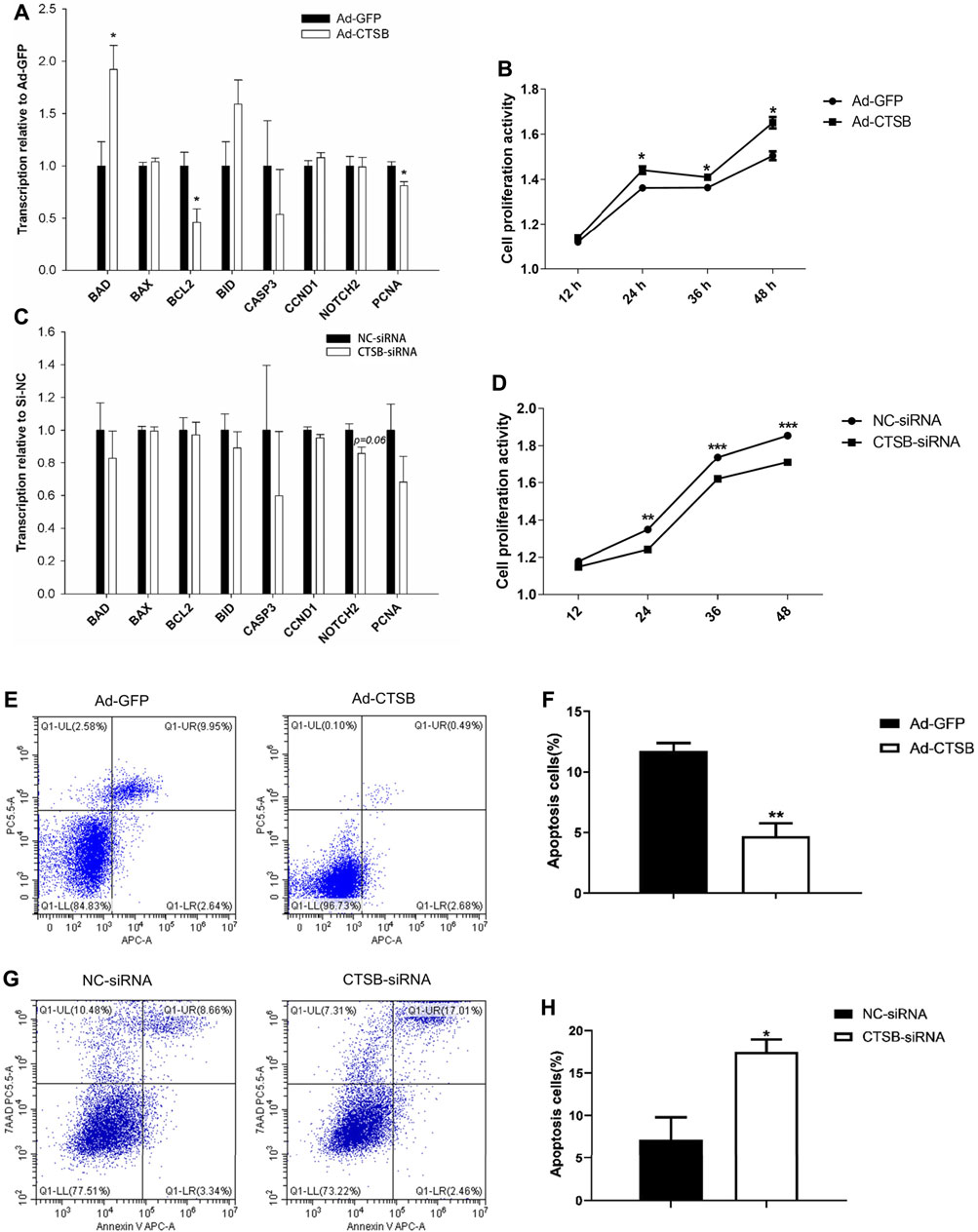
FIGURE 6. CTSB overexpression and interference on the proliferation and apoptosis of rabbit GCS. Transcription of genes associated with cell proliferation and apoptosis after CTSB overexpression (A) and knockdown (C). Rabbit GCS proliferation after CTSB overexpression (B) and knockdown (D). CTSB overexpression decreased the apoptosis in GCS as assessed by flow cytometer (E,F). CTSB knockdown increased apoptosis of GCS as assessed by using a flow cytometer (G,H). *p < 0.05; **p < 0.01.
In spite of no important changes in genes associated with cell proliferation, overexpression or inhibition of CTSB strongly affected proliferation when measured at 12, 24, 36, and 48 h after insertion of Ad-CTSB or CTSB-siRNA (Figure 6B), with greater cell proliferation activity by CTSB overexpression (p < 0.05), while CTSB interference decreased cell proliferation activity (Figure 6D, p < 0.05).
Compared with the Ad-GFP group, the mRNA level of the main anti-apoptotic gene, BCL2, was decreased, while the pro-apoptotic gene, BAD, was increased by CTSB overexpression (Figure 6B, p < 0.05). Transcription of genes involved in apoptosis was not affected by silencing CTSB. Despite the transcription of apoptosis-related genes indicated no effect by silencing and minor effect by the overexpression of CTSB; flow cytometry analysis revealed that CTSB overexpression inhibits apoptosis of rabbit GCS (Figures 6E,F, p < 0.01). As shown in Figures 6G,H, CTSB knockdown led to a remarkable increase of apoptosis in rabbit GCS (p < 0.05).
In order to further explore the function of the CTSB gene in reproduction and the potential link between this gene and the reduced fertility when coprophagy was prevented, we measured the effects of CTSB on the mRNA expression of genes related to progesterone and estradiol secretion. Transcription of CYP19A1 was significantly upregulated by CTSB overexpression (Figure 7A, p < 0.05), together with the significant increase of progesterone and estradiol secretion (Figures 7B,C, p < 0.05). CTSB knockdown decreased the mRNA expression of steroidogenic acute regulatory protein (STAR) and progesterone level (Figures 7D,E, p < 0.05), while the estradiol level was not affected (Figure 7F, p > 0.05).
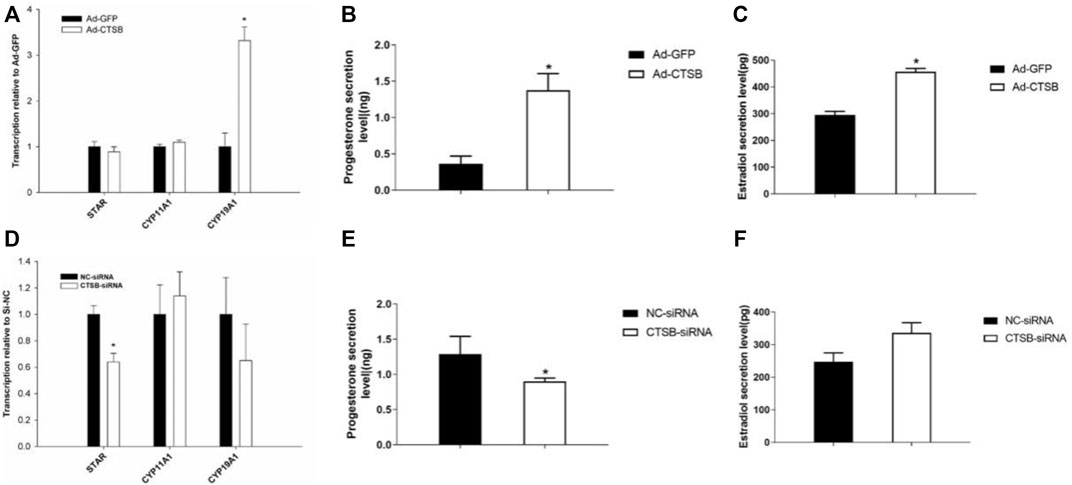
FIGURE 7. Effects of CTSB overexpression and interference on progesterone and estradiol secretion in rabbit GCS. Transcription of key genes related to hormone secretion in GCS with CTSB overexpression (A) or interference (D). Level of progesterone and estradiol secretion in rabbit GCS after CTSB overexpression (B,C) and interference (E,F). Each experiment had three biological replicates and the results are expressed as the mean ± SE. * p < 0.05, ** p < 0.01.
In our study we used the Elizabeth circle to establish a coprophagy prevention model of rabbits (Hughes and Gorospe, 1991). We found that preventing coprophagy did not cause any difference in food intake, but the body weight was decreased. This phenomenon may be caused by change in the intestinal microbiota and food digestion (McBee, 1971; Naumova et al., 2015).
As hypothesized, preventing coprophagy drastically affected reproduction by both decreasing progesterone secretion and inducing 100% fetal death in pregnant rabbits. Coprophagy is important for maintaining the normal reproductive performance, mainly because of the nutritional value of soft feces. However, the present study demonstrated that the failure to give birth to live pups was partly due to an increased apoptosis of ovaries, especially in granulosa cells. This is an important finding, since granulosa cells play an important role in the reproductive process and are mainly involved in the secretion of progesterone and estrogen (Gore-Langton and Daniel, 1990; Cai et al., 2015). Estradiol and progesterone are important hormones that play a critical regulatory role in maintaining normal reproduction in mammals (Cerri et al., 2011; Martins et al., 2014), including development of follicles (Chang et al., 2016). Granulosa cells maintain healthy oocytes via transzonal projections (Li and Albertini, 2013) and apoptosis of granulosa cells affects oocytes directly (Emanuelli et al., 2019), also causing atresia of the follicles (Hughes and Gorospe, 1991; Tilly et al., 1991; Manabe et al., 2004). Overall, our data indicated an important role of coprophagy in maintaining normal reproduction performance of rabbits, but it is unclear the reason for such an effect.
In order to further reveal the mechanism of coprophagy on reproduction and specifically on the ovaries, we performed a whole transcriptome sequencing of RNA isolated from the ovaries. The results indicate a very modest effect of coprophagy on ovary transcriptome but revealed a downregulation of pathways involved in lipid metabolism, immune response, cell proliferation, and organelles involved with apoptosis, such as lysosomes. Based on this, the CTSB gene which was significantly decreased in expression by coprophagy appeared to be an important hub, suggesting a possible role in connecting the prevention of coprophagy with reproductive failure.
As one member of the cathepsin family, CTSB is the most abundant lysosomal cysteine protease, which is generally considered as intracellular protease (Aggarwal and Sloane, 2014). It plays an important role in the catabolism of proteins in cells and participates in various physiological processes, including oxidative stress and inflammatory response (Bai et al., 2018; Lian et al., 2018). Moreover, CTSB is related to the development of the cumulus cells-oocyte complex, thereby affecting reproductive performance (Oksjoki et al., 2001; Wei et al., 2019). In our study, we detected the good expression of CTSB in rabbit ovarian tissue.
In order to identify the function of CTSB in the apoptosis of cumulus cells in rabbits, we manipulated the expression of the CTSB gene in primary cumulus cells. We found that the overexpression or underexpression of the CTSB gene promotes or inhibits the growth of GCS, respectively. Reduced cell proliferation was observed when the CTSB gene was inhibited in cholangiocarcinoma cells (Li et al., 2018). Among all genes associated with cell growth, overexpression of CTSB increased the expression of the pro-apoptotic gene BAD (Basu, 2021), while decreased the expression of anti-apoptotic BCL2 (Basu, 2021) and the PCNA which is important for cell proliferation (Zhang, 2021). Interference of CTSB expression did not affect transcription of any apoptosis-related genes. Those findings indicate that the effect of CTSB on apoptosis is not via change in the transcription of apoptosis-related genes. The effect on apoptosis by CTSB might be by protease activity toward pro-apoptotic proteins, as previously observed in human cells (Seng et al., 2016). Taken together, our results indicated that the expression of CTSB was decreased by preventing coprophagy in rabbits. The decrease of CTSB appears to be related to the increased apoptosis of GCS likely via reduced proteolysis of pro-apoptotic proteins affecting the reproductive performance of rabbits. The negative role of apoptosis of granulosa cells on reproduction is supported by a recent study, indicating that the apoptosis of granulosa cells can cause follicular atresia and decrease reproductive performance, as observed in pigs (Zhao et al., 2014; He et al., 2016). This effect was also confirmed during the process of cancer progression, in which CTSB deletion significantly increases tumor cell death (Gocheva et al., 2006).
`Interestingly, we also found that modulation of the CTSB gene had a great influence on the secretion of progesterone and estradiol in rabbit GCS. In this study, overexpression of the CTSB gene increased the secretion of sterol hormones (progesterone and estradiol) and the expression of CYP19A1, coding for the estrogen synthetase. The central role of CTSB on the decreased secretion of progesterone in rabbits that were not allowed to do coprophagy was demonstrated in our study by the manipulation of CTSB expression. Although CTSB gene interference resulted in decreased progesterone secretion, it did not affect estradiol secretion. However, the interference of CTSB expression did not affect estrogen synthetase but decreased the expression of the steroidogenic acute regulatory protein (STAR), a key gene for the transport of cholesterol into the mitochondria for the synthesis of pregnenolone, the main steroidogenic precursor (Schumacher et al., 2017).
In our study, we only evaluated the role of one of the 241 DEGs in ovary as consequence of preventing coprophagy. Thus, we cannot exclude that change in those other DEGs played a critical role; however, this need to be evaluated via molecular approaches as performed in the present manuscript. In addition, the link between CTSB and the change in expression of the few affected genes is still unclear, since CTSB is a protease.
In conclusion, our study demonstrated an important role of coprophagy on growth and reproductive performance of rabbits. Our data indicated an increased apoptosis of granulosa and a decrease secretion of progesterone when coprophagy was prevented. Our data revealed some effects on the transcriptome of the ovaries by preventing coprophagy with downregulation of genes associated with pathways related to cell growth, immune response, and metabolism. The transcriptome data together with the bioinformatic analyses allowed to identify CTSB as a potential central hub in the observed effects. The subsequent molecular studies on granulosa cells demonstrated a role of CTSB in controlling apoptosis and secretion of progesterone. The role of CTSB is not via change in expression of genes but likely via protease activity on apoptosis-related proteins. In Figure 8 we summarized our findings.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the following link: https://www.ncbi.nlm.nih.gov/, PRJNA767495.
The animal study was reviewed and approved. The present study was approved by the Institutional Animals Care and Use Committee (IACUC) of College of Animal Science and Technology of Henan Agricultural University, China (Permit Number: 11-0085; Data: 06-2011) and was designed and performed according to the institutional guidelines. Written informed consent was obtained from the owners for the participation of their animals in this study.
The work was mainly conceived and designed by HX, MB, and ML. Experiments were performed by GHS, YDW, YLW, YJ, and SS. Experimental data were collected and analyzed by GHS, MB, and GRS. The manuscript was written, revised, and edited by GHS, HX, and MB. All the authors contributed to, read, and approved the final manuscript.
This work was supported by the National Key Research and Development Program of China (2018YFD0502203) and the Special Fund for the Henan Agriculture Research System (HARS-22-13-G1).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2022.926795/full#supplementary-material
Abecia L., Balcells J., Fondevila M., Belenguer A., Holtrop G., Lobley G. E. (2008). Contribution of Gut Microbial Lysine to Liver and Milk Amino Acids in Lactating Does. Br. J. Nutr. 100, 977–983. doi:10.1017/S0007114508957986
Aggarwal N., Sloane B. F. (2014). Cathepsin B: Multiple Roles in Cancer. Prot. Clin. Appl. 8, 427–437. doi:10.1002/prca.201300105
Anders S., Huber W. (2010). Differential Expression Analysis for Sequence Count Data. Genome Biol. 11, R106. doi:10.1186/gb-2010-11-10-r106
Bai H., Yang. B., Yu. W., Xiao Y., Yu. D., Zhang Q. (2018). Cathepsin B Links Oxidative Stress to the Activation of NLRP3 Inflammasome. Exp. Cell Res. 362, 180–187. doi:10.1016/j.yexcr.2017.11.015
Balboula A. Z., Yamanaka. K., Sakatani M., Hegab A. O., Zaabel S. M., Takahashi M. (2010). Intracellular Cathepsin B Activity Is Inversely Correlated with the Quality and Developmental Competence of Bovine Preimplantation Embryos. Mol. Reprod. Dev. 77, 1031–1039. doi:10.1002/mrd.21250
Balboula A. Z., Yamanaka. K., Sakatani M., Kawahara M., Hegab A. O., Zaabel S. M., et al. (2013). Cathepsin B Activity Has a Crucial Role in the Developmental Competence of Bovine Cumulus-Oocyte Complexes Exposed to Heat Shock during In Vitro Maturation. Reproduction 146, 407–417. doi:10.1530/REP-13-0179
Basu A. (2022). The Interplay between Apoptosis and Cellular Senescence: Bcl-2 Family Proteins as Targets for Cancer Therapy. Pharmacol. Ther. 230, 107943. doi:10.1016/j.pharmthera.2021.107943
Belenguer Á., Balcells J., Guada J. A., Decoux M., Milne E. (2005). Protein Recycling in Growing Rabbits: Contribution of Microbial Lysine to Amino Acid Metabolism. Br. J. Nutr. 94, 763–770. doi:10.1079/bjn20051508
Bionaz M., Periasamy K., Rodriguez-Zas S. L., Hurley W. L., Loor J. J. (2012). A Novel Dynamic Impact Approach (DIA) for Functional Analysis of Time-Course Omics Studies: Validation Using the Bovine Mammary Transcriptome. PLoS One 7, e32455. doi:10.1371/journal.pone.0032455
Bo T.-B., Zhang X.-Y., Kohl K. D., Wen J., Tian S.-J., Wang D.-H. (2020). Coprophagy Prevention Alters Microbiome, Metabolism, Neurochemistry, and Cognitive Behavior in a Small Mammal. ISME J. 14, 2625–2645. doi:10.1038/s41396-020-0711-6
Brix K., Dunkhorst A., Mayer. K., Jordans S. (2008). Cysteine Cathepsins: Cellular Roadmap to Different Functions. Biochimie 90, 194–207. doi:10.1016/j.biochi.2007.07.024
Cai L., Sun. A., Li H., Tsinkgou A., Yu. J., Ying S., et al. (2015). Molecular Mechanisms of Enhancing Porcine Granulosa Cell Proliferation and Function by Treatment In Vitro with Anti-inhibin Alpha Subunit Antibody. Reprod. Biol. Endocrinol. 13, 1–10. doi:10.1186/s12958-015-0022-3
Cerri R. L. A., Chebel R. C., Rivera. F., Narciso C. D., Oliveira. R. A., Thatcher W. W., et al. (2011). Concentration of Progesterone during the Development of the Ovulatory Follicle: I. Ovarian and Embryonic Responses. J. Dairy Sci. 94, 3342–3351. doi:10.3168/jds.2010-3734
Chang H.-M., Qiao. J., Leung P. C. K. (2016). Oocyte-somatic Cell Interactions in the Human Ovary-Novel Role of Bone Morphogenetic Proteins and Growth Differentiation Factors. Hum. Reprod. Update 23, 1–18. doi:10.1093/humupd/dmw039
Conus S., Simon H. (2010). Cathepsins and Their Involvement in Immune Responses. Swiss Med. Wkly. 140, w13042. doi:10.4414/smw.2010.13042
Ebino K. Y., Suwa T., Kuwabara. Y., Saito T. R., Takahashi K. W. (1988). Coprophagy in Female Mice during Pregnancy and Lactation. Exp. Anim. 37, 101–104. doi:10.1538/expanim1978.37.1_101
Ebino K. Y., Yoshinaga K., Suwa T., Kuwabara. Y., Takahashi K. W. (1989). Effects of Prevention of Coprophagy on Pregnant Mice. Exp. Anim. 38, 245–252. doi:10.1538/expanim1978.38.3_245
Emanuelli I. P., Costa C. B., Rafagnin Marinho. L. S., Seneda M. M., Meirelles F. V. (2019). Cumulus-oocyte Interactions and Programmed Cell Death in Bovine Embryos Produced In Vitro. Theriogenology 126, 81–87. doi:10.1016/j.theriogenology.2018.11.028
Ferreira W. M., Ferreira F. N. A., Inácio D. F. d. S., Mota K. C. d. N., Costa Júnior M. B. d., Silva Neta C. S., et al. (2018). Effects of Dietary Inclusion of Macaúba Seed Cake Meal on Performance, Caecotrophy Traits and In Vitro Evaluations for Growing Rabbits. Archives Animal Nutr. 72, 138–152. doi:10.1080/1745039X.2018.1435480
Galef B. G., Bennett G. (1979). Investigation of the Functions of Coprophagy in Juvenile Rats. J. Comp. Physiological Psychol. 93, 295–305. doi:10.1037/h0077551
García A. I., De Bias J. C., Carabaño R. (2004). Effect of Type of Diet (Casein-based or Protein-free) and Caecotrophy on Ileal Endogenous Nitrogen and Amino Acid Flow in Rabbits. Anim. Sci. 79, 231–240. doi:10.1111/j.1365-2052.2004.01185.x10.1017/s1357729800090093
GlDENNE T. (2000). Recent Advances in Rabbit Nutrition : Emphasis on Fibre Requirements. A Review. World rabbit Sci. 8, 23–32. doi:10.4995/wrs.2000.414
Gocheva V., Zeng W., Ke. D., Klimstra D., Reinheckel T., Peters C., et al. (2006). Distinct Roles for Cysteine Cathepsin Genes in Multistage Tumorigenesis. Genes Dev. 20, 543–556. doi:10.1101/gad.1407406
Gore-Langton R. E., Daniel S. A. J. (1990). Follicle-Stimulating Hormone and Estradiol Regulate Antrum-like Reorganization of Granulosa Cells in Rat Preantral Follicle Cultures1. Biol. Reprod. 43, 65–72. doi:10.1095/biolreprod43.1.65
He Y., Deng H., Jiang Z., Li Q., Shi M., Chen H., et al. (2016). Effects of Melatonin on Follicular Atresia and Granulosa Cell Apoptosis in the Porcine. Mol. Reprod. Dev. 83, 692–700. doi:10.1002/mrd.22676
Hughes F. M., Gorospe W. C. (1991). Biochemical Identification of Apoptosis (Programmed Cell Death) in Granulosa Cells: Evidence for a Potential Mechanism Underlying Follicular Atresia*. Endocrinology 129, 2415–2422. doi:10.1210/endo-129-5-2415
Klaasen H. L. B. M., Koopman J. P., Scholten P. M., Van Den Brink M. E., Theeuwes A. G. M. (2009). Effect of Preventing Coprophagy on Colonisation by Segmented Filamentous Bacteria in the Small Bowel of Mice. Microb. Ecol. Health Dis. 3, 99–103. doi:10.3109/08910609009140123
Kobayashi A., Tsuchida S., Ueda A., Yamada T., Murata K., Nakamura H., et al. (2019). Role of Coprophagy in the Cecal Microbiome Development of an Herbivorous Bird Japanese Rock Ptarmigan. J. Veterinary Med. Sci. 81, 1389–1399. doi:10.1292/jvms.19-0014
Li J. X., Ding X. M., Han S., Wang. K., Jiao C. Y., Li X. C. (2018). mir-637 Inhibits the Proliferation of Cholangiocarcinoma Cell QBC939 through Interfering CTSB Expression. Eur. Rev. Med. Pharmacol. Sci. 22, 1265–1276. doi:10.26355/eurrev_201803_14467
Li R., Albertini D. F. (2013). The Road to Maturation: Somatic Cell Interaction and Self-Organization of the Mammalian Oocyte. Nat. Rev. Mol. Cell Biol. 14, 141–152. doi:10.1038/nrm3531
Li R., Li X., Huang T., Wang. Y., Xue M., Sun. S., et al. (2020). Influence of Cecotrophy on Fat Metabolism Mediated by Caecal Microorganisms in New Zealand White Rabbits. J. Anim. Physiol. Anim. Nutr. 104, 749–757. doi:10.1111/jpn.13309
Lian D., Lai. J., Wu. Y., Wang. L., Chen Y., Zhang Y., et al. (2018). Cathepsin B-Mediated NLRP3 Inflammasome Formation and Activation in Angiotensin II -Induced Hypertensive Mice: Role of Macrophage Digestion Dysfunction. Cell Physiol. Biochem. 50, 1585–1600. doi:10.1159/000494656
Liu Q.-S., Li J.-Y., Wang D.-H. (2007). Ultradian Rhythms and the Nutritional Importance of Caecotrophy in Captive Brandt's Voles (Lasiopodomys Brandtii). J. Comp. Physiol. B 177, 423–432. doi:10.1007/s00360-006-0141-4
Liu Q.-S., Wang D.-H. (2007). Effects of Diet Quality on Phenotypic Flexibility of Organ Size and Digestive Function in Mongolian Gerbils (Meriones unguiculatus). J. Comp. Physiol. B 177, 509–518. doi:10.1007/s00360-007-0149-4
Manabe N., Goto Y., Matsuda-Minehata F., Inoue N., Maeda A., Sakamaki K., et al. (2004). Regulation Mechanism of Selective Atresia in Porcine Follicles: Regulation of Granulosa Cell Apoptosis during Atresia. J. Reproduction Dev. 50, 493–514. doi:10.1262/jrd.50.493
Mao X., Cai T., Olyarchuk J. G., Wei L. (2005). Automated Genome Annotation and Pathway Identification Using the KEGG Orthology (KO) as a Controlled Vocabulary. Bioinformatics 21, 3787–3793. doi:10.1093/bioinformatics/bti430
Martins T., Peres R. F. G., Rodrigues A. D. P., Pohler K. G., Pereira M. H. C., Day M. L., et al. (2014). Effect of Progesterone Concentrations, Follicle Diameter, Timing of Artificial Insemination, and Ovulatory Stimulus on Pregnancy Rate to Synchronized Artificial Insemination in Postpubertal Nellore Heifers. Theriogenology 81, 446–453. doi:10.1016/j.theriogenology.2013.10.020
McBee R. H. (1971). Significance of Intestinal Microflora in Herbivory. Annu. Rev. Ecol. Syst. 2, 165–176. doi:10.1146/annurev.es.02.110171.001121
Morchang A., Panaampon J., Suttitheptumrong A., Yasamut U., Noisakran S., Yenchitsomanus P.-t., et al. (2013). Role of Cathepsin B in Dengue Virus-Mediated Apoptosis. Biochem. Biophysical Res. Commun. 438, 20–25. doi:10.1016/j.bbrc.2013.07.009
Mudunuri U., Che A., Yi M., Stephens R. M. (2009). bioDBnet: the Biological Database Network. Bioinformatics 25, 555–556. doi:10.1093/bioinformatics/btn654
Naumova E. I., Zharova G. K., Chistova T. Y., Kuznetsova T. A. (2015). The Effect of Coprophagy on the Size of Plant Fibers in the Digestive Tract of Hares Lepus Europaeus and L. timidus (Lagomorpha, Leporidae). Biol. Bull. Russ. Acad. Sci. 42, 426–431. doi:10.1134/s106235901505009x
Oksjoki S., Söderström M., Vuorio E., Anttila L. (2001). Differential Expression Patterns of Cathepsins B, H, K, L and S in the Mouse Ovary. Mol. Hum. Reprod. 7, 27–34. doi:10.1093/molehr/7.1.27
Repnik U., Stoka V., Turk V., Turk B. (2012). Lysosomes and Lysosomal Cathepsins in Cell Death. Biochimica Biophysica Acta (BBA) - Proteins Proteomics 1824, 22–33. doi:10.1016/j.bbapap.2011.08.016
Ruijter J. M., Ramakers C., Hoogaars W. M. H., Karlen Y., Bakker O., van den Hoff M. J. B., et al. (2009). Amplification Efficiency: Linking Baseline and Bias in the Analysis of Quantitative PCR Data. Nucleic Acids Res. 37, e45. doi:10.1093/nar/gkp045
Sakaguchi E. (2003). Digestive Strategies of Small Hindgut Fermenters. Animal Sci. J. 74, 327–337. doi:10.1046/j.1344-3941.2003.00124.x
Sakamaki T. (2010). Coprophagy in Wild Bonobos (Pan paniscus) at Wamba in the Democratic Republic of the Congo: a Possibly Adaptive Strategy? Primates 51, 87–90. doi:10.1007/s10329-009-0167-9
Savage D. C. (1986). Gastrointestinal Microflora in Mammalian Nutrition. Annu. Rev. Nutr. 6, 155–178. doi:10.1146/annurev.nu.06.070186.001103
Schumacher M., Zhu X., Guennoun R. (2017). Progesterone: Synthesis, Metabolism, Mechanism of Action, and Effects in the Nervous System. Hormones, Brain Behav. 3, 215–244. doi:10.1016/B978-0-12-803592-4.00054-7Third Edition
Seng N., Megyesi J., Tarcsafalvi A., Price P. (2016). Mimicking Cdk2 Phosphorylation of Bcl-xL at Ser73 Results in Caspase Activation and Bcl-xL Cleavage. Cell Death Discov. 2, 16001. doi:10.1038/cddiscovery.2016.1
Sherman B. T., Hao M., Qiu J., Jiao X., Baseler M. W., Lane H. C., et al. (2022). DAVID: a Web Server for Functional Enrichment Analysis and Functional Annotation of Gene Lists (2021 Update). Nucleic Acids Res., gkac194. doi:10.1093/nar/gkac194
Song G., Jiang Y., Wang. Y., Song M., Niu X., Xu H., et al. (2021). Modulation of Cathepsin S (CTSS) Regulates the Secretion of Progesterone and Estradiol, Proliferation, and Apoptosis of Ovarian Granulosa Cells in Rabbits. Animals 11, 1770. doi:10.3390/ani11061770
Sukemori S., Kurosawa. A., Ikeda S., Kurihara Y. (2006). Investigation on the Growth of Coprophagy-Prevented Rats with Supplemented Vitamin B12. J. Anim. Physiol. Anim. Nutr. 90, 402–406. doi:10.1111/j.1439-0396.2006.00619.x
Tardy C., Codogno P., Autefage H., Levade T., Andrieu-Abadie N. (2006). Lysosomes and Lysosomal Proteins in Cancer Cell Death (New Players of an Old Struggle). Biochimica Biophysica Acta (BBA) - Rev. Cancer 1765, 101–125. doi:10.1016/j.bbcan.2005.11.003
Tatti M., Motta M., Di Bartolomeo S., Cianfanelli V., Salvioli R. (2013). Cathepsin-mediated Regulation of Autophagy in Saposin C Deficiency. Autophagy 9, 241–243. doi:10.4161/auto.22557
Tilly J. L., Kowalski K. I., Johnson A. L., Hsueh A. J. W. (1991). Involvement of Apoptosis in Ovarian Follicular Atresia and Postovulatory Regression. Endocrinology 129, 2799–2801. doi:10.1210/endo-129-5-2799
Tremaroli V., Bäckhed F. (2012). Functional Interactions between the Gut Microbiota and Host Metabolism. Nature 489, 242–249. doi:10.1038/nature11552
Wang Y., Xu H., Sun. G., Xue M., Sun. S., Huang T., et al. (2019). Transcriptome Analysis of the Effects of Fasting Caecotrophy on Hepatic Lipid Metabolism in New Zealand Rabbits. Animals 9, 648. doi:10.3390/ani9090648
Wei Z., Li P., Huang S., Lkhagvagarav P., Zhu M., Liang C., et al. (2019). Identification of Key Genes and Molecular Mechanisms Associated with Low Egg Production of Broiler Breeder Hens in Ad Libitum. BMC Genomics 20, 408. doi:10.1186/s12864-019-5801-3
Y.-X. P., D.-H. W., I. H. (2001). Selective Digesta Retention and Coprophagy in Brandt's Vole ( Microtus Brandti ). J. Comp. Physiology B Biochem. Syst. Environ. Physiology 171, 457–464. doi:10.1007/s003600100195
Zhang H. (2021). Regulation of DNA Replication Licensing and Re-replication by Cdt1. Ijms 22, 5195. doi:10.3390/ijms22105195
Zhao F., Zhao W., Ren. S., Fu Y., Fang X., Wang. X., et al. (2014). Roles of SIRT1 in Granulosa Cell Apoptosis during the Process of Follicular Atresia in Porcine Ovary. Animal Reproduction Sci. 151, 34–41. doi:10.1016/j.anireprosci.2014.10.002
Keywords: rabbits, coprophagy, reproductive performance, granulosa cells, CTSB
Citation: Song G, Wang Y, Wang Y, Jiang Y, Sun S, Cai H, Sun G, Li M, Bionaz M and Xu H (2022) Coprophagy Prevention Decreases the Reproductive Performance and Granulosa Cell Apoptosis via Regulation of CTSB Gene in Rabbits. Front. Physiol. 13:926795. doi: 10.3389/fphys.2022.926795
Received: 23 April 2022; Accepted: 23 June 2022;
Published: 18 July 2022.
Edited by:
Vikas Kumar Roy, Mizoram University, IndiaReviewed by:
Xiaopeng An, Northwest A&F University, ChinaCopyright © 2022 Song, Wang, Wang, Jiang, Sun, Cai, Sun, Li, Bionaz and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huifen Xu, aHVpZmVuMjIxQDEyNi5jb20=; Massimo Bionaz, bWFzc2ltby5iaW9uYXpAb3JlZ29uc3RhdGUuZWR1; Ming Li, bGltaW5nQGhlbmF1LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.