- Guangxi Key Laboratory of Agric-Environment and Agric-Products Safety, College of Agriculture, Guangxi University, Nanning, China
Olfactory and gustatory systems play an irreplaceable role in all cycles of growth of insects, such as host location, mating, and oviposition. Many chemosensory genes in many nocturnal moths have been identified via omics technology, but knowledge of these genes in diurnal moths is lacking. In our recent studies, we reported two sex pheromone compounds and three host plant volatiles that play a vital role in attracting the diurnal moth, Phauda flammans. The antennal full-length transcriptome sequence of P. flammans was obtained using the Pacbio sequencing to further explore the process of sex pheromone and host plant volatile recognition in P. flammans. Transcriptome analysis identified 166 candidate olfactory and gustatory genes, including 58 odorant-binding proteins (OBPs), 19 chemosensory proteins (CSPs), 59 olfactory receptors (ORs), 16 ionotropic receptors (IRs), 14 gustatory receptors (GRs), and 2 sensory neuron membrane proteins (SNMPs). Subsequently, a phylogenetic tree was established using P. flammans and other lepidopteran species to investigate orthologs. Among the 17 candidate pheromone receptor (PR) genes, the expression levels of PflaOR21, PflaOR25, PflaOR35, PflaOR40, PflaOR41, PflaOR42, PflaOR44, PflaOR49, PflaOR51, PflaOR61, and PflaOR63 in the antennae were significantly higher than those in other non-antennae tissues. Among these PR genes, PflaOR21, PflaOR27, PflaOR29, PflaOR35, PflaOR37, PflaOR40, PflaOR42, PflaOR44, PflaOR60, and PflaOR62 showed male-biased expression, whereas PflaOR49, PflaOR61, and PflaOR63 revealed female-biased expression. The functions of related OR genes were also discussed. This research filled the gap of the chemosensory genes of P. flammans and provided basic data for future functional molecular mechanisms studies on P. flammans olfaction.
Introduction
In long evolutionary histories, lepidopteran herbivores with abundant species have developed highly sensitive olfactory and gustatory systems, which are essential for survival and reproduction (Song et al., 2020). Host plant volatiles and sex pheromones are dominating odor compounds that usually function together and efficiently guide herbivores to locate mates, hosts, and oviposition sites and avoid natural enemies (Prestwich, 1996; Bruce et al., 2005; Xu and Turlings, 2018). The ability to identify intricate odors in lepidopteran herbivores specifically and extensively depends on their sophisticated sensilla and sensory genes. Among all the main chemosensory organs of adult insects, the antennae typically contain various types of sensilla that play important roles in a number of behaviors (Zacharuk and Shields, 1991; Hildebrand and Shepherd, 1997; Hallem et al., 2006; Gu et al., 2015; Liu et al., 2022). In the sensilla, many sensory genes, including chemosensory proteins, olfactory receptors, ionotropic receptors, gustatory receptors, odorant-degrading enzymes (ODEs), and sensory neuron membrane proteins, are supposed to be involved in host volatile and sex pheromone recognition (Vogt and Riddiford, 1981; Wu et al., 2022). The gene families’ odorant-binding proteins and chemosensory proteins are in charge of binding and transporting externally lipophilic odorant molecules through the sensilla lymph, which is the first step to identifying odorant molecules (Zhou et al., 2021). The first OBP and CSP in lepidopteron are identified from Antheraea polyphemus and Cactoblastis cactorum, respectively (Vogt and Riddiford, 1981; Maleszka and Stange, 1997). Extensive OBPs and CSPs of lepidopteran herbivores, such as Chilo suppressalis (Cao et al., 2014), Loxostege sticticalis (Wei et al., 2017), and Sesamia inferens (Zhang et al., 2013), have been identified, classified, and analyzed using transcriptomes. Normally, pheromone-binding proteins (PBPs) and general odorant–binding proteins (GOBPs) are the primary subfamilies in OBPs of lepidopteron and can recognize sex pheromone constituents and volatiles from host plants (Zhou et al., 2008; Xing et al., 2021). However, a recent study showed that CpinPBP2 and CpinGOBP1 have similar functions in the identification and transportation of sex pheromones and host plant volatiles in Conogethes pinicolalis (Jing et al., 2020).
Receptor protein families, that is, ORs, IRs, and GRs, are used to recognize semiochemicals in the fields (Clyne et al., 1999, 2000; Benton, 2009). The OBP–odorant complex or the OBP-released individual odorant in proximity are recognized by relevant receptor proteins on the dendritic membrane, resulting in chemical signals transforming into electrical signals and then guiding the central nervous system (CNS) to make corresponding commands about the ethological response of lepidopteran insects (Leal, 2013; Wicher and Miazzi, 2021; Wu et al., 2022). During the process of the odorant identification, ORs with odorant receptor co-receptor (Orco) form tetramer as the channels of odorant-gated ions to respond to odors and pheromones (Liu N et al., 2018; Sun et al., 2019). Orco is a unique and highly conserved ortholog considered as the necessary factor for the localization, stability, and correct protein folding of ORs (Larsson et al., 2004; Neuhaus et al., 2005; Martin and Alcorta, 2011). The knockout or mutation of the Orco gene can lead to the loss of odorant function in lepidopterons (Koutroumpa et al., 2016). PRs are highly conserved orthologs, and nearly all PR sequences in moth species are clustered into PR clades including the classical type I, novel, type II, and type 0 PR clades, which are separated from other odorant receptor clades (Zhang and Löfstedt, 2015; Yuvaraj et al., 2022). As a member of the ionotropic glutamate receptor (iGluR) gene family, IRs have a wide range of functions besides odor identification and can take part in the process of sensation of temperature, humidity, and taste (Chen et al., 2015; Zhang et al., 2019; Sun D. et al., 2020). IRs can be divided into antennal IRs (A-IRs), divergent IRs (D-IRs), and Lepidoptera-specific IRs (LS-IRs) (Croset et al., 2010; Zhou et al., 2021). GRs are necessary to sense gustatory substances (Knecht et al., 2016; Xing et al., 2021). GRs can also be classified as CO2, sugars, fructose, and bitter receptors, which share relatively high conservation (Ning et al., 2016; Xu, 2020). The ORs, IRs, and GRs of lepidopteran herbivores are first reported on Heliothis virescens (Krieger et al., 2002), Bombyx mori (Wanner and Robertson, 2008), and Spodoptera littoralis (Olivier et al., 2011). To date, a number of receptor protein genes, including Oraesia emarginata (Feng et al., 2017), Semiothisa cinerearia (Liu et al., 2020), Achelura yunnanensis (Li G.-C. et al., 2021), and Dioryctria abietella (Wang et al., 2021), have been identified. Among the receptor proteins, PRs are a special type of OR that participate in pheromone reception (Wu et al., 2022). Therefore, extensive studies have been conducted on its identification and function in insect pests for potential control (Tian et al., 2021), including BmOR1 and BmOR2 of B. mori (Sakurai et al., 2004; Nakagawa et al., 2005); HR13, HR14, and HR16 of H. virescens (Grosse-Wilde et al., 2007); and PxOR1, MsOR1, and DiOR1 of Plutella xylostella, Mythimna separata, and Diaphania indica, respectively (Mitsuno et al., 2008).
SNMP, a member of the CD36 protein family, plays an important role in the process of accepting sex pheromones, degrading the breakdown products of odors, and immunity (Pregitzer et al., 2014; Zhang et al., 2020; Li G.-C. et al., 2021; Cassau and Krieger, 2021). The latest studies indicated that SNMPs can be classified as SNMP1, SNMP2, and SNMP3 in lepidopterons (Liu J et al., 2015; Zhang et al., 2020). The SNMPs of lepidopteron insects are first identified in A. polyphemus (Rogers et al., 1997). Most SNMPs of moths, such as Carposina sasakii Matsumura (Tian et al., 2018), Galleria mellonella (Jiang et al., 2021), S. litura (Zhang et al., 2015), and S. exigua (Liu N et al., 2015), have been identified until now.
Phauda flammans (Walker) (Lepidoptera: Phaudidae) is a diurnal moth and oligophagous pest widely distributed in Southeast Asia and Southern China (Nageshchandra et al., 1972; Verma and Dogra, 1982; FOF, 2015; Liu J et al., 2015; Liu et al., 2016; Robinson et al., 2019). Larvae primarily consume leaves, and additionally, some of the phloem in Ficus, including Ficus microcarpa (Miq.) and F. benjamina L., resulting in leaf necrosis, and only the bare trunk remains (Liu N et al., 2015; Liu et al., 2016). Early studies found two sex pheromone components from P. flammans females, including Z-9-hexadecenal and (Z,Z,Z)-9,12,15-octadecatrienal, and their application in fields gain success (Zheng et al., 2019). In addition, β-rolene, β-caryophylene, and D-limonene in host plant volatiles show remarkable attraction to P. flammans adults (Guan et al., 2020). Two PflaPBPs are demonstrated to have high affinity with sex pheromones (unpublished data), as identified from the antennal unigene transcriptome. As the crucial next step of sex pheromone recognition, the identification and functional test of PflaPRs become urgent by comprehensive transcriptome sequencing tools.
Knowledge about olfactory- and gustatory-related genes that participate in the identification of sex pheromone and plant volatiles in P. flammans remains lacking. Therefore, the first antennal full-length transcriptome of P. flammans is generated using single-molecule real-time (SMRT) sequencing technology. Furthermore, 166 candidate olfactory and gustatory genes, including 58 OBPs, 19 CSPs, 59 ORs, 16 IRs, 14GRs, and 2 SNMPs, are identified by analyzing the transcriptome, constructing evolutionary trees of chemosensory genes, and evaluating the expression profiles of 17 candidate PflaPRs. The results on these chemosensory genes may provide fundamental data for understanding their roles in discerning odorants and identifying specific molecular targets in P. flammans.
Materials and Methods
Insect Rearing and Tissue Collection
Mature P. flammans larvae were collected from Lingli town (108° 81′ E, 22° 85′ N), Qingxiu District, Nanning City, Guangxi Zhuang Autonomous Region, P.R. China from June to July in 2020. Every 10 larvae were reared in a barrel-shaped transparent plastic box (2000 ml in volume) with enough pinholes (1 mm in diameter) under constant conditions of 25 ± 1°C, 80% ± 5% relative humidity, and a photoperiod of 14 L:10 D (light:dark). The larvae were fed with fresh leaves of F. benjamina until pupation. The pupae were sorted by sex and placed in different barrel-shaped transparent plastic boxes until eclosion. For real-time quantitative PCR (qRT-PCR), 45 male antennae, 45 female antennae; 30 heads (♀:♂ = 1:1), whose antennae were cut off; 30 legs (♀:♂ = 1:1), 30 wings (♀:♂ = 1:1), 30 thoraxes (♀:♂ = 1:1), and 30 abdomens (♀:♂ = 1:1) were collected in three replications from 1-day-old virgin adults. After the collection, samples were preserved in liquid nitrogen and stored at −80°C for later use.
RNA Extraction, cDNA Library Construction, Sequencing, and Functional Annotation of the Two Transcriptomes
Total RNA was extracted from one mixture sample including 50 male antennae and 50 female antennae of P. flammans using the RNAiso Plus (Takara, Japan) and in accordance with specifications. The degradation and integrity of the total RNA were tracked by 1% agarose gels. RNA concentration and quality were assessed using the NanoPhotometer® N60 spectrophotometer (Implen, Germany).
The cDNA library construction and SMRT (Single-Molecular, Real-Time) sequencing were conducted by the Gene Denovo Biotechnology Co., (Guangzhou, China) to acquire the antennal full-length transcriptome. The purification of mRNA from 3 μg total RNA was performed to produce cDNA libraries, mRNA was enriched by Oligo (dT) beads and reverse transcribed into cDNA using the Clontech SMARTer PCR cDNA Synthesis Kit, and the second-strand cDNA was synthesized. Fragments with size >4 kb were selected using the BluePippin™ Size-Selection System. The SMRTbell library was constructed with the large-scale PCR. DNA was damage repaired, end-repaired, and ligated to sequencing adapters. cDNAs were sequenced on the PacBio Sequel platform. High-quality cyclic consensus sequences were acquired from subreads BAM files. Full-length nonchimeric (FLNC) reads, including 5′ and 3′ primer and polyA structures, were obtained via removing primers, barcodes, polyA tail trimming, and concatemer of full passes. Similar FLNC reads were clustered using minimap2, and the consistency sequence was corrected by performing the quiver algorithm. High-quality isoforms (prediction accuracy ≥0.99) were collected. The final transcriptome isoform sequence was constructed after removing redundant sequences using CD-hit software (similarity threshold reached 0.99) (Fu et al., 2012). The quality and integrity of final transcriptome isoforms were evaluated by assembly quality statistics and BUSCO assessment. The BUSCO assessment of isoforms was performed using the BUSCO v5 ortholog search pipeline with “Insecta” as an ortholog set in the gVolante (Nishimura et al., 2017) (https://gvolante.riken.jp/).
The mRNA was enriched by Oligo (dT) beads and interrupted by ultrasound to acquire the antennal unigene transcriptome. The first strand of cDNA was synthesized in the M-MuLV reverse transcriptase system by using mRNAs as a template. Then, the second-strand cDNA was synthesized and purified. After the end repair, add a tail, and ligation of adapters, PCR was performed to amplify the cDNA. The cDNA was sequenced on the Illumina novaseq 6000 (Illumina, California, United States). After filtering the low-quality data and base quality analysis, clean reads were obtained. The final transcriptome unigene sequence was assembled and obtained using Trinity v2.8.4. The quality and integrity of final transcriptome unigenes were evaluated by assembly quality statistics and BUSCO assessment, respectively.
Isoforms and unigenes were annotated into different databases, including EuKaryotic Orthologous Groups/Clusters of Orthologous Groups (KOG/COG), NCBI nonredundant protein (Nr), Kyoto Encyclopedia of Genes and Genomes (KEGG), and Swiss-Prot database, to predict the assumed functional roles of isoforms and unigenes. Gene Ontology (GO) annotation was performed using Blast2GO software with Nr annotation results of isoforms (Conesa et al., 2005). The functional classification of isoforms and unigenes were analyzed using WEGO software (Ye et al., 2006).
Sequence Analysis and Evolutionary Tree Construction of P. flammans
Two antennal transcriptomes, that is, full-length transcriptome and unigene transcriptome, were used to identify candidate olfactory genes in P. flammans. “OBP and odorant-binding protein,” “CSP and chemosensory protein,” “OR and odorant receptor,” “IR and ionotropic receptor,” “GR and gustatory receptor,” and “SNMP and sensory neuron membrane protein” were used as keywords to screen the annotated chemosensory genes by the results of Nr annotation from two antennal transcriptome datasets. The similarity in candidate olfactory sequence was analyzed using the Blastx search in the NCBI database (Altschul et al., 1997) (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastx&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome). The open reading frames (ORFs) of candidate olfactory genes were estimated using the NCBI ORF finder (https://www.ncbi.nlm.nih.gov/orffinder/). The putative N-terminal signal peptides and transmembrane domains (TMDs) of candidate olfactory genes were predicted by running the SignalP 5.0 Server (Almagro Armenteros et al., 2019) (https://services.healthtech.dtu.dk/service.php?SignalP-5.0) and TOPCONS (Tsirigos et al., 2015) (https://topcons.net/pred/result/rst_651nhxzl/). The amino acid (aa) sequence alignment of candidate olfactory genes in P. flammans and the homologous sequence obtained from different lepidopteran families (Supplementary Tables S4, S6, S8, S10, S12, and S14) were analyzed using the Clustal W method (Ye et al., 2006) in the MEGA (version 7.0, Mega Limited, Auckland, New Zealand) software (Kumar et al., 2016). Then, a phylogenetic tree was constructed using the neighbor-joining method in the IQ-TREE web server (Trifinopoulos et al., 2016) (http://iqtree.cibiv.univie.ac.at/). The reliability of the tree structure was evaluated using the 1000-fold bootstrap replication. All phylogenetic trees were established utilizing the FigTree (version 1.4.3, Andrew Rambaut Institute of Evolutionary Biology, England) and Adobe Illustrator (CC 2019; Adobe, America). The olfactory sequence identified from an antennal unigene transcriptome in P. flammans (unpublished data) was also included in the phylogenetic analysis to achieve a comprehensive analysis.
Tissue Expression Profiles of PflaORs
The total RNA was extracted from 12 different tissues of P. flammans as mentioned earlier. The primers for PflaORs were designed using the Primer-BLAST (http://www.ncbi.nlm.nih.gov/tools/primer-blast). TUB1 (accession number: MN852477) and GADPH (accession number: MN852481) were used as reference genes in accordance with a previous study (Chen L et al., 2021). The primers used in this tissue expression are listed in Supplementary Table S1. The amplification efficiencies of primers for target genes were determined using the fivefold dilutions of male antennal cDNA. The specificity of products was evaluated by a melting curve analysis. Melting curves and primer amplification efficiencies are shown in Supplementary Presentation S1 and Supplementary Table S2. On the basis of the phylogeny of PflaORs, 17 candidate PflaPRs in P. flammans were selected, and their expression profiles in different tissues were further detected using qRT-PCR with the LightCycler 96 System Real-Time PCR System (Roche, Switzerland). qRT-PCR reactions were executed in 10 µl reaction mixtures containing 5 μl TB Green Premix Ex Taq II (Tli RNaseH Plus, Takara, China), 0.4 µl of each primer (forward and reverse primers, 10 µM), 2 µl sample cDNA, and 2.2 µl sterilized H2O. Thermocycling conditions are as follows: predenaturation (95°C for 30 s, one cycle), PCR reaction (95°C for 5 s and 60°C for 30 s, 40 cycles in total), and melt curve (95°C for 15 s, 60°C for 1 min, and 95°C for 15 s). Each qRT-PCR reaction was carried out with 3 technical and 3 biological replicates. The relative expression levels of PflaORs were measured using the 2−∆∆CT method, where the heads were used as the internal control (Livak and Schmittgen, 2001). All data were analyzed using SPSS Statistics 26.0 (IBM, Armonk, United States). The expression of PflaORs in different tissues was analyzed using a one-way analysis of variance followed by the Tukey’s honest significant difference test. In the same tissue, the expression of PflaORs in different sexes was analyzed using the independent sample t-test. Graphs were drawn using GraphPad Prism 5 (GraphPad Software, San Diego, California, United States).
Results
Identification and Phylogenetic Analysis of Candidate OBPs
The antennal full-length transcriptome of P. flammans was created using the PacBio Sequel platform. A total of 20,332 isoforms with a total length of 52,425,403 bp were acquired. The average length of isoforms was 2578.47 bp. The N50 value of isoforms was 2909 bp (Supplementary Figure S1). In the result of BUSCO assessment, the complete, fragmented, and missing BUSCOs accounted for 49.09%, 51.50%, and 48.50%, respectively (Supplementary Figure S2). The unigene transcriptome of P. flammans was also created using the Illumina novaseq 6000 platform. A total of 99 386 unigenes with a total length of 90 588 857 bp were acquired. The average length of unigenes was 911 bp. The N50 value of unigenes was 1862 bp (Supplementary Figure S3). The result of BUSCO assessment showed that the complete, fragmented, and missing BUSCOs accounted for 93.30%, 3.67%, and 1.94%, respectively (Supplementary Figures S4, S5).
A total of 58 candidate OBPs, including 43 PflaOBPs, 13 PflaGOBPs, and 2 PflaPBPs from the antennal full-length transcriptome and unigene transcriptome dataset of P. flammans were identified. This study showed that all candidate PflaOBPs had 312–999 bp, which could code 103–332 aa sequences. Among the candidate OBP genes, 44 PflaOBPs had complete ORFs, and 45 PflaOBPs were predicted that had signal peptides. In total, 16 PflaOBPs (i.e., PflaOBP1, PflaOBP3, PflaOBP4, PflaOBP6, PflaOBP12, PflaOBP17, PflaOBP28, PflaOBP35, PflaOBP41, PflaOBP42, PflaGOBP5, PflaGOBP6, PflaGOBP8, PflaGOBP15, PflaPflaPBP1, and PflaPBP2) contained six conserved cysteine residues, and this finding was the same as the classic structure of OBPs in other insects (Supplementary Table S3). The GOBP clade was next to the PBP clade, and both clades were phylogenetically far from other OBPs in accordance with phylogenetic analysis. In total, 13 PflaGOBPs could phylogenetically cluster into the GOBP1 and GOBP2 subtribe of other lepidopterans. PflaPBP1 and PflaPBP2 were also closely assembled with other PBPs. Most PflaOBPs were dispersedly distributed in the phylogenetic tree, building orthologous with other lepidopteran OBPs. PflaOBP8 and PflaOBP30 were clustered with SlituOBP10 in the same branch. PflaOBP3 and PflaOBP38 were clustered into an independent branch, as well as PflaOBP4, and PflaOBP23. PflaOBP16 formed separate branches by itself. All of them were considered to have a special function in P. flammans (Figure 1).
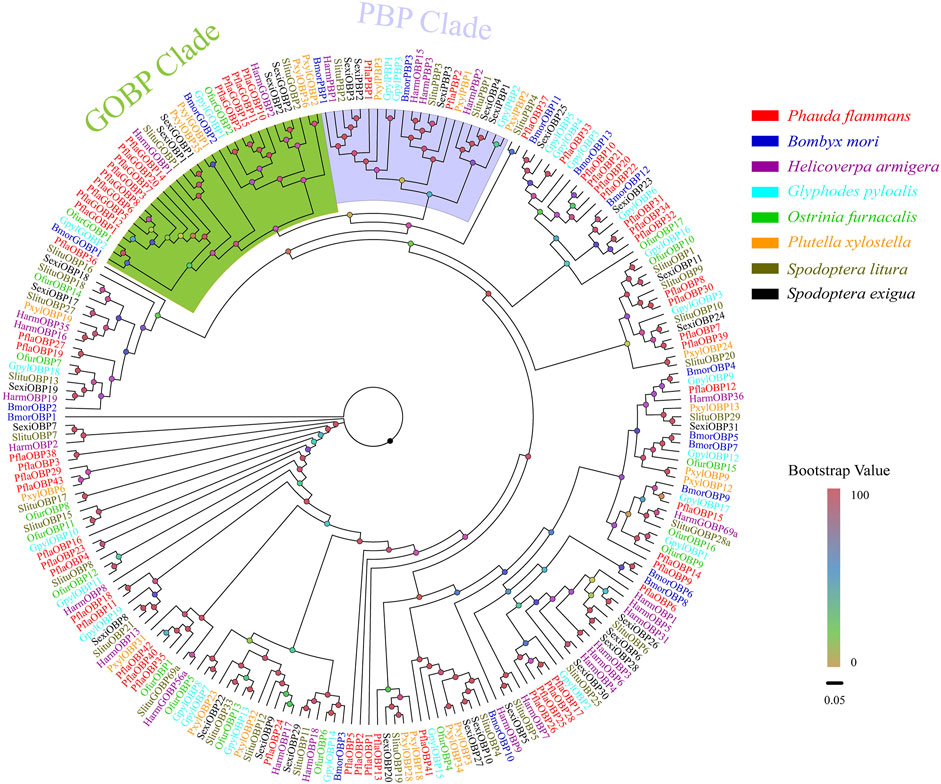
FIGURE 1. Phylogenetic tree of candidate odorant–binding proteins (OBPs) of P. flammans with other lepidopteran insects, including B. mori, H. armigera, Glyphodes pyloalis, O. furnacalis, P. xylostella, S. litura, and S. exigua. Bootstrap values are indicated by colors from yellow (0) to red (100). The protein names and gene accession of OBPs used in the phylogenetic tree are recorded in Supplementary Table S4.
Identification and Phylogenetic Analysis of Candidate CSPs
A total of 19 candidate CSPs, which had 312–546 bp, were also identified. Among these CSPs, 18 PflaCSPs were predicted to have complete ORF to code 103–181 aa sequences. All PflaCSPs had 15–19 aa signal peptides (Supplementary Table S5). The phylogeny of CSPs indicated that the amount of PflaCSPs except PflaCSP5, PflaCSP8, PflaCSP15, and PflaCSP17 was dispersed widespread in varied branches. In addition, PflaCSP2, PflaCSP6, and PflaCSP11 were clustered together with the homologous clade (Figure 2).
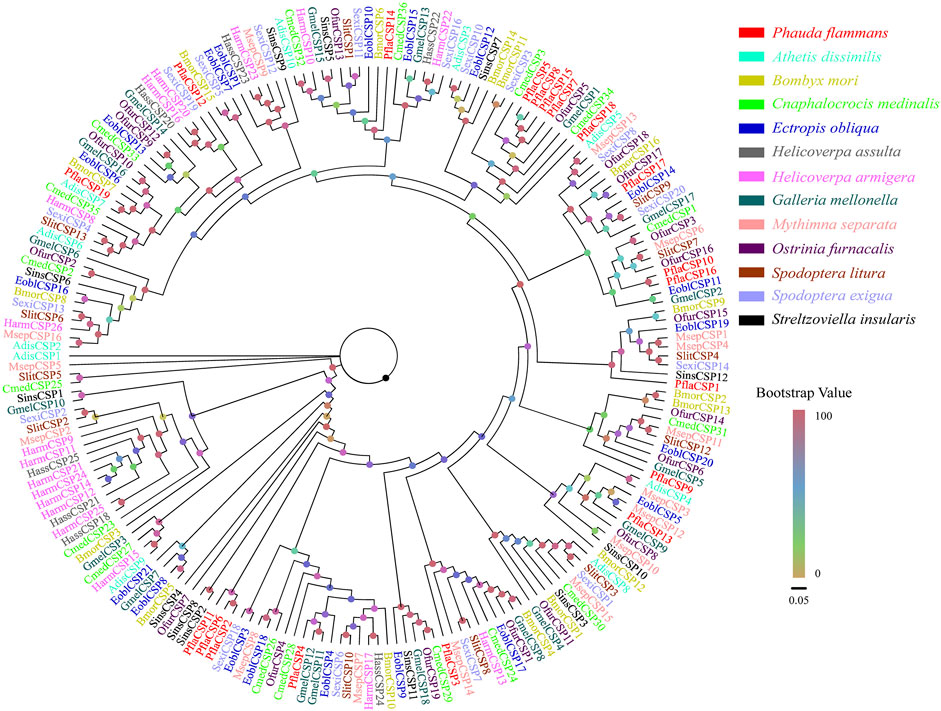
FIGURE 2. Phylogenetic tree of candidate chemosensory proteins (CSPs) of P. flammans with other lepidopteran insects, including Athetis dissimilis, B. mori, Cnaphalocrocis medinalis, Ectropis obliqua, H. assulta, H. armigera, G. mellonella, M. separata, O. furnacalis, S. litura, S. exigua, and Streltzoviella insularis. Bootstrap values are indicated by colors from yellow (0) to red (100). The protein names and gene accession of CSPs used in the phylogenetic tree are recorded in Supplementary Table S6.
Identification and Phylogenetic Analysis of Candidate ORs
A total of 59 candidate PflaORs, which were identified by bioinformatics analysis from the two transcriptomes of P. flammans, consisted of 687–1440 bp nucleotide sequences and could code 228–479 aa sequences. Among these PflaORs, 51 were forecasted to own full-length ORFs containing 3–7 TMDs (Supplementary Table S7). The phylogenetic tree of ORs clearly showed that PflaORs exhibited high differentiation. Most PflaORs split into different clades. By contrast, the Orco clade and three PR clades were comparatively highly conserved. One PflaOR (PflaOrco) was clustered in the Orco clade. A total of 9 PflaORs (i.e., PflaOR27, PflaOR29, PflaOR44, PflaOR49, PflaOR51, and PflaOR60-63), 5 PflaORs (i.e., PflaOR21, PflaOR35, PflaOR37, PflaOR40, and PflaOR42), and 1 PflaOR (i.e., PflaOR25) were clustered in the classical PR clade, novel clade of type I PR clade, and type 0 PR clade, respectively. Moreover, compared with the ORs from the six other lepidopteran insects, two expansions in the number of ORs in P. flammans were found. These expansions were branches of PflaOR14, PflaOR19, and PflaOR24 and branches of PflaOR33, PflaOR38, and PflaOR64. A unique gene, namely, PflaOR53, which had no homologous gene in other species, was also found (Figure 3).
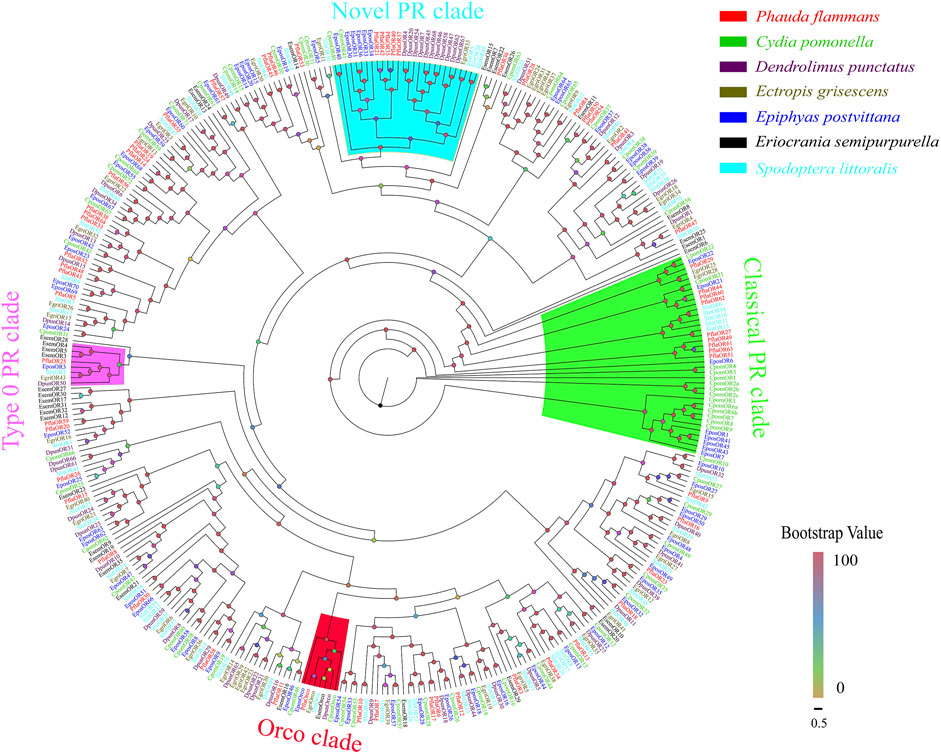
FIGURE 3. Phylogenetic tree of candidate olfactory receptors (ORs) of P. flammans with other lepidopteran insects, including Cydia pomonella, Dendrolimus punctatus, E. grisescens, E. postvittana, Eriocrania semipurpurella, and S. littoralis. Bootstrap values are indicated by colors from yellow (0) to red (100). The protein names and gene accession of ORs used in the phylogenetic tree are recorded in Supplementary Table S8.
Identification and Phylogenetic Analysis of Candidate IRs
A total of 16 isoforms were recognized as candidate PflaIRs. The lengths of isoforms were all 1116–2700 bp, which could code 371–899 aa sequences. Then, 16 PflaIRs were predicted to possess complete ORFs containing 1–5 TMDs (Supplementary Table S9). The phylogenetic tree of IRs showed that these PflaIRs were grouped into 15 highly conserved IR subtribes, including IR7d, IR8a, IR21a, IR25a, IR40a, IR41a, IR60a, IR64a, IR68a, IR75, IR76b, IR85a, IR87a, IR93a, and iGluRs (Figure 4). Interestingly, the aa of PflaIR25a was found to share 90.55% identity with HnubIR25a using the best blast-x match. Equally, the aa of PflaIR8a shared 74.6% similarity with EhipIR8a (Supplementary Table S9). In the phylogenetic tree, PflaIR25a and PflaIR8a, which belonged to co-receptor groups IR25a and IR8a, respectively, were clustered with conserved branches. Several PflaIRs (i.e., PflaIR75d, PflaIR75c, PflaIR75p, PflaIR75q, and PflaIR2) were clustered into the IR75 subfamily. In addition, PflaIR76b showed homology with SinsIR76b in the evolutionary tree. PflaIR85a was clustered with PsauIR85a in the same branch (Figure 4).
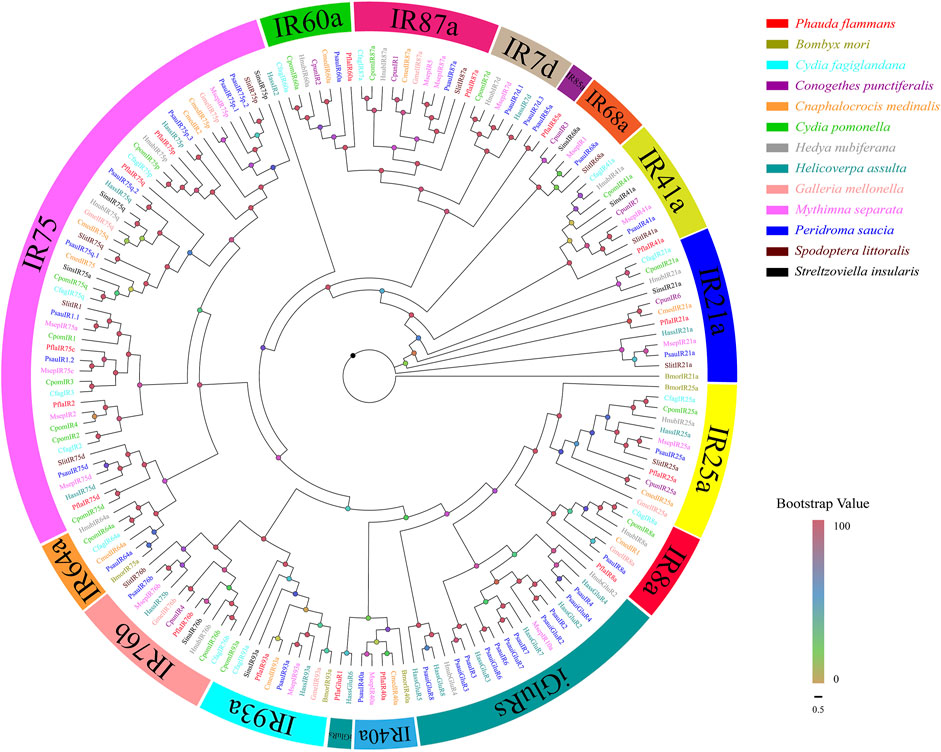
FIGURE 4. Phylogenetic tree of candidate ionotropic receptors (IRs) of P. flammans with other lepidopteran insects, including B. mori, Cydia fagiglandana, Conogethes punctiferalis, C. medinalis, C. pomonella, H. nubiferana, H. assulta, G. mellonella, M. separata, Peridroma saucia, S. littoralis, and S. insularis. Bootstrap values are indicated by colors from yellow (0) to red (100). The protein names and gene accession of IRs used in the phylogenetic tree are recorded in Supplementary Table S10.
Identification and Phylogenetic Analysis of Candidate GRs
A total of 14 candidate PflaGRs were singled out from the antennal transcriptomes of P. flammans. They had 621–1275 bp and could code 206–424 aa sequences. Subsequently, five PflaGRs were predicted to retain complete ORFs containing 6–7 TMDs (Supplementary Table S11). The phylogenetic tree of GRs manifested that PflaGRs were classified as four GR subfamilies of lepidopteran, including CO2, sugar, fructose, and bitter receptors. Among these PflaGRs, 2 (PflaGR8 and PflaGR12) showed homology and were clustered in the fructose receptor branch, and 2 (i.e., PflaGR9 and PflaGR13) showed homology and were clustered in the CO2 receptor branch (Figure 5).
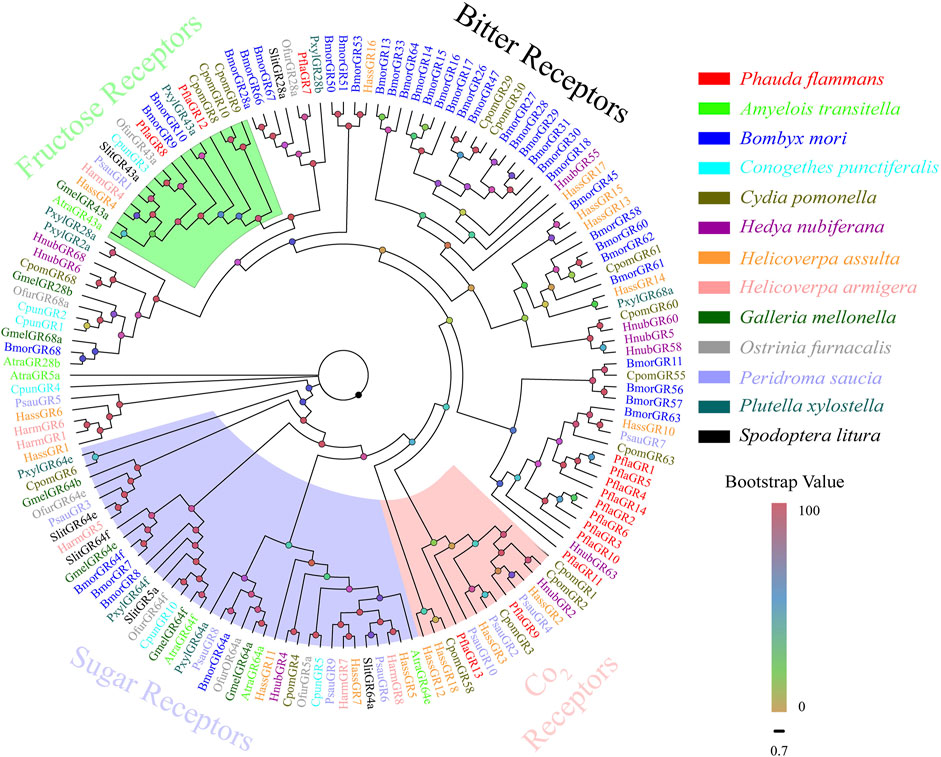
FIGURE 5. Phylogenetic tree of candidate gustatory receptors (GRs) of P. flammans with other lepidopteran insects, including A. transitella, B. mori, C. punctiferalis, C. pomonella, H. nubiferana, H. assulta, H. armigera, G. mellonella, O. furnacalis, P. saucia, P. xylostella, and S. litura. Bootstrap values are indicated by colors from yellow (0) to red (100). The protein names and gene accession of GRs used in the phylogenetic tree are recorded in Supplementary Table S12.
3.6 Identification and Phylogenetic Analysis of Candidate SNMPs
Two PflaSNMPs containing complete ORFs were recognized in the two transcriptomes of P. flammans. Both of their nucleotide sequences were 1563 bp and could code 520 aa sequences. PflaSNMPs (i.e., PflaSNMP1 and PflaSNMP2) were predicted to have two conserved TMDs, belonging to the CD36 protein family. PflaSNMP1 shared 77.78% aa identity with SNMP1 in H. kahamanoa, whereas PflaSNMP2 was more analogous with OnubSNMP2 (identity: 66.48%, Supplementary Table S13). Phylogenetic analysis showed that PflaSNMP1 and PflaSNMP2 were closely clustered into lepidopteran SNMP1 and SNMP2 branches, respectively (Figure 6).
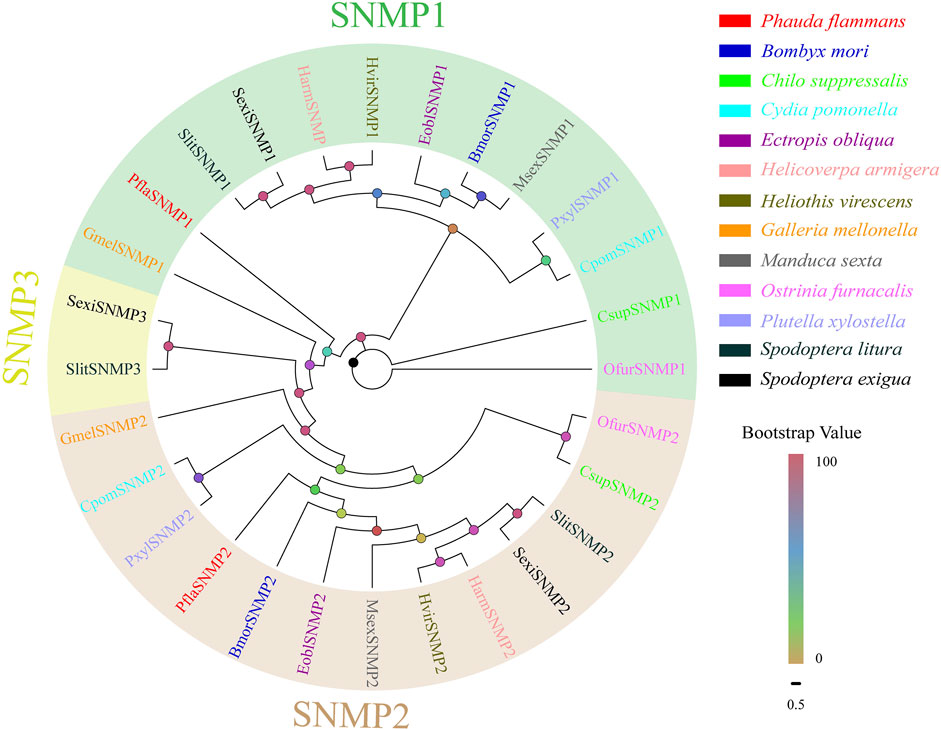
FIGURE 6. Phylogenetic tree of candidate sensory neuron membrane proteins (SNMPs) of P. flammans with other lepidopteran insects, including B. mori, C. suppressalis, C. pomonella, E. obliqua, H. armigera, H. virescens, G. mellonella, M. sexta, O. furnacalis, P. xylostella, S. litura, and S. exigua. Bootstrap values are indicated by colors from yellow (0) to red (100). The protein names and gene accession of SNMPs used in the phylogenetic tree are recorded in Supplementary Table S14.
Expression Profile Analysis of Candidate PRs of P. flammans
A total of 17 candidate PR genes (i.e., PflaOR21, PflaOR25, PflaOR27, PflaOR29, PflaOR35, PflaOR36, PflaOR37, PflaOR40, PflaOR41, PflaOR42, PflaOR44, PflaOR49, PflaOR51, PflaOR60, PflaOR61, PflaOR62, and PflaOR63) were used to compare tissue- and sex-specific expression in P. flammans. RT-qPCR results discovered that the expression levels of 11 candidate PRs (i.e., PflaOR21, PflaOR25, PflaOR35, PflaOR40, PflaOR41, PflaOR42, PflaOR44, PflaOR49, PflaOR51, PflaOR61, and PflaOR63) in the antennae were significantly higher than those in five other tissues of males and females. However, PflaOR27, PflaOR29, PflaOR60, and PflaOR62 were only significantly expressed in the antennae of males. PflaOR36 was significantly expressed in the abdomens of females. PflaOR37 was significantly expressed in the wings of females. Clearly, 13 candidate PRs (i.e., PflaOR21, PflaOR27, PflaOR29, PflaOR35, PflaOR37, PflaOR40, PflaOR42, PflaOR44, PflaOR49, PflaOR60, PflaOR61, PflaOR62, and PflaOR63) exhibited significantly sex-biased expression in the antennae. Among them, PflaOR27 and PflaOR40 showed sex-biased expression in the male antennae, whereas PflaOR21, PflaOR29, PflaOR35, PflaOR37, PflaOR42, PflaOR44, PflaOR60, and PflaOR62 showed highly expressed significant differences in the male antennae compared with those in their female counterpart. By contrast, PflaOR49, PflaOR61, and PflaOR63 revealed the female-biased expression in the antennae. PflaOR36 also displayed the female-biased expression in the abdomens. Other candidate PRs (i.e., PflaOR25, PflaOR41, and PflaOR51) had almost similar expression between female and male antennae (Figure 7).
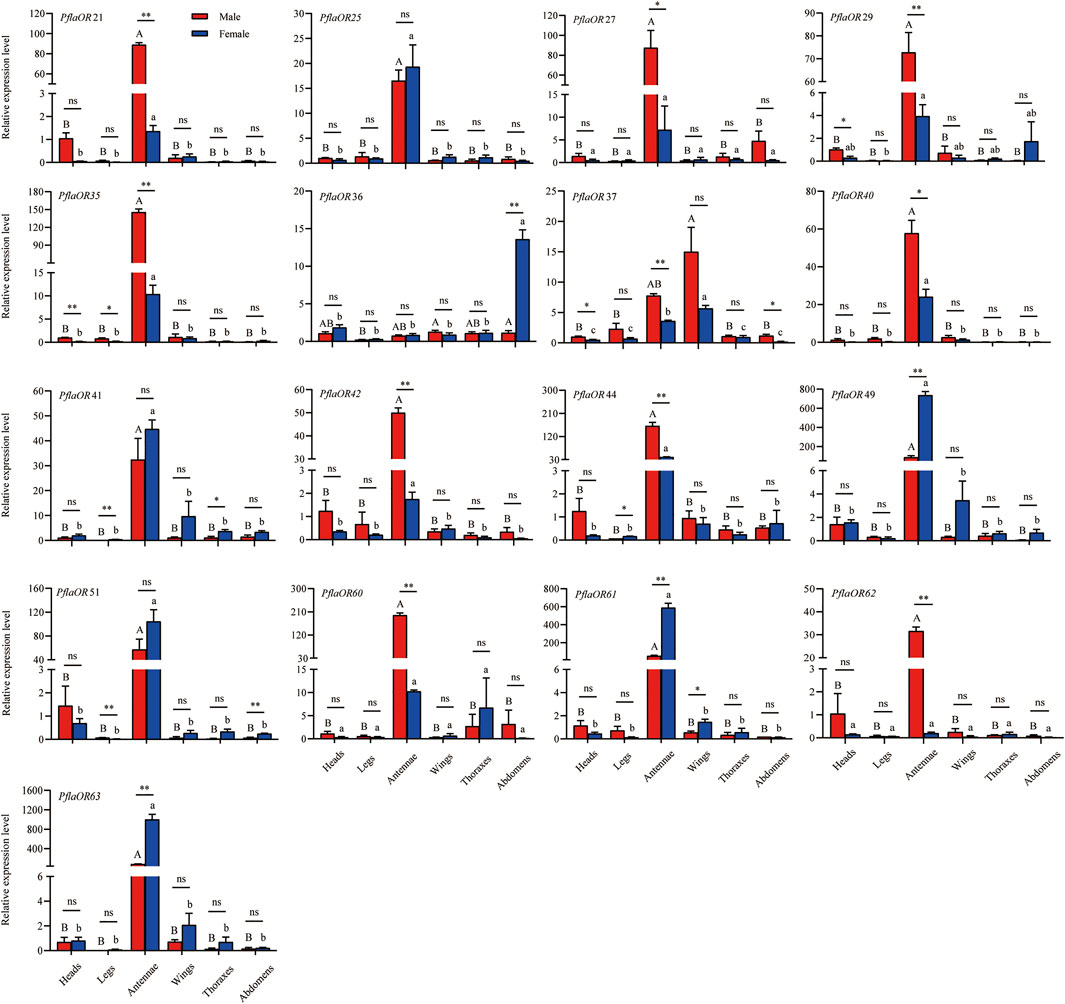
FIGURE 7. Expression profile of candidate pheromone receptors (PRs) in different tissues of male and female adults of P. flammans. Note: Data are presented as mean ± SE. Different uppercase and lowercase letters on the column indicated significant differences in the OR expression levels among different tissues of male and female, respectively (ANOVA and Tukey tests, p < 0.05). ns, *, and ** on the column indicated no significant difference (p > 0.05), significant difference (p < 0.05), and highly significant difference (p < 0.01), respectively, in the gene expression between different sexes in the same tissue (independent sample t-test).
Discussion
Phauda flammans is a serious pest of Ficus trees (Liu J et al., 2015; Liu et al., 2016). Although the sex pheromone components of P. flammans females and the main components of host plant volatiles have been identified (Zheng et al., 2019; Guan et al., 2020), the mechanisms of how P. flammans recognizes these odors through their olfactory and gustatory sensory systems remain unclear. To date, many chemosensory receptor genes of lepidopteran herbivores, e.g., at least 11 families of nocturnal moths (such as Noctuidae, Crambidae, and Cossidae) and some diurnal moths (such as Zygaenidae) (Hu et al., 2016; Jia et al., 2016; Sun et al., 2016; Yang et al., 2020), have been identified by increasingly mature transcriptome sequencing technologies. Most of these studies are focused on nocturnal moths that larvae can cause loss to the agricultural economy, and only a few diurnal moths are paid attention. Therefore, the identification and analysis of candidate olfactory and gustatory genes of P. flammans can provide essential information to further understand the chemosensory mechanism of mate and host location in diurnal moths.
In this research, 166 candidate olfactory and gustatory genes, including 58 OBPs, 19 CSPs, 59 ORs, 16 IRs, 14 GRs, and 2 SNMPs, are identified by analyzing the antennal full-length transcriptome and RNA-seq transcriptome of P. flammans (the sequences of the 166 genes are provided in Supplementary Material S1). This number is less than that in D. abietella (42 OBPs, 23 CSPs, 75 ORs, 30 IRs, 62 GRs, and 3 SNMPs), an oligophagous pest (Wang et al., 2021), and more than that in O. emarginata (41 OBPs, 20 CSPs, 35 ORs, six IRs, and 2 SNMPs); M. separata (50 OBPs, 20 CSPs, 35 ORs, six IRs, and 2 SNMPs); and C. sasakii (29 OBPs, 13 CSPs, 52 ORs, eight IRs, 11 GRs, and 1 SNMP) (Feng et al., 2017; Liu et al., 2017; Tian et al., 2018), an omnivorous pest; and P. xyllostella (24 OBPs, 15 CSPs, 54 ORs, 16 IRs, 7 GRs, and 2 SNMPs), an oligophagous pest (Yang, et al., 2017). These findings show that the transcriptome database owns high-quality gene numbers belonging to a normal range of moth species.
Oligophagous pests generally require relatively few binding proteins to sense external compounds due to their specificity to host plants (Gouin et al., 2017). In the current study, 58 candidates OBPs (43 PflaOBPs, 13 PflaGOBPs, and 2 PflaPBPs) are found from the transcriptome of P. flammans. This number is more than most of the identified OBPs from transcriptomes in lepidopteran herbivores, such as C. punctiferalis (29 OBPs), M. separata (50 OBPs), P. xyllostella (24 OBPs), and Athetis lepigone (28 OBPs) (Xiao et al., 2016; Liu et al., 2017; Yang et al., 2017; Zhang et al., 2017). This finding may be because two transcriptome databases are integrated, and a mixture of male and female antennae is used as samples. The phylogenetic tree indicates that PflaPBP1 and PflaPBP2 can cluster in the PBP branch of lepidopterons. However, a third PflaPBP is not found. Three or more PBPs are commonly found in moth, such as C. punctiferalis (5 PBPs), O. emarginata, and Clostera restitura (3 PBPs) (Xiao et al., 2016; Feng et al., 2017; Gu et al., 2019). However, the result of the present study is similar to that of another study, which reports that only two PBPs are found in the diurnal moth Histia rhodope. A total of 13 PflaGOBPs are identified in the transcriptome of P. flammans. This number is substantially more than those of other moths, such as S. littoralis (2 GOBPs) and G. mellonella (2 GOBPs) (Jiang et al., 2021; Koutroumpa et al., 2021). PflaGOBP and PflaPBP subfamilies cluster together adjacently in the phylogenetic tree, indicating that they may evolve from the same ancestral gene and produce differentiation along with the evolution of species. Many studies showed that some GOBPs have similar functions with PBPs, which cannot only bind plant volatile but also sex pheromones. For example, AtraGOBPs have high affinities with their sex pheromone components in A. transitella (Liu et al., 2010). Han et al. (2022) found that SlitGOBP2 also takes part in sensing sex pheromone components using in vivo CRISPR/Cas9 technique. P. flammans is an oligophagous moth with a narrow host range. Thus, males are likely to find females on host plants. Therefore, sex pheromones and host plant volatiles may co-regulate the mate finding of P. flammans males. In this process, PflaGOBPs and PflaPBPs are possibly sensing sex pheromones and host plant volatiles at the same time.
As a binding protein family, CSPs are considered to have similar binding capacities with OBPs, same with host plant volatiles and sex pheromones (Pelosi et al., 2014; Li et al., 2020). However, increasing studies suggested that CSPs have a wider range of functions than OBPs, including non-olfactory functions (Benton et al., 2007; Yang et al., 2019). For example, in S. exigua, SexiCSP3 is demonstrated to be involved in the survival and reproduction by the RNA interference technology (Gong et al., 2012). PrapCSPs are conjectured to be related to the reproduction of Pieris rapae because PrapCSP7, PrapCSP18, and PrapCSP20 are evidently expressed in the reproductive organs (Li et al., 2020). Thus, CSPs are considered to take part in releasing some chemical substances in male glands, such as secreting vaccenyl acetate (Dyanov and Dzitoeva, 1995). Furthermore, CSPs may be associated with detecting nonvolatile chemicals. For example, CSPs are detected to have a high expression in the legs of C. sacchariphagus (Liu et al., 2021). In the present study, PflaCSPs are widely distributed in the whole branches of the CSP phylogenetic tree, and this finding is consistent with the hypothesis that CSP has a broad function in chemical communication (Benton et al., 2007; Yang et al., 2019). Among PflaCSPs, PflaCSP5/7/8/15/18, PflaCSP10/16/17, and PflaCSP2/6/11 are clustered in the same branch, indicating that they have recent gene differentiation.
A total of 16 candidate PflaIRs are recognized through the antennal transcriptome of P. flammans and are divided into 13 A-IRs (i.e., PflaIR2, PflaIR8a, PflaIR21a, PflaIR25a, PflaIR40a, PflaIR41a, PflaIR60a, PflaIR75c, PflaIR75d, PflaIR75p, PflaIR75q, PflaIR76b, and PflaIR93a), 1 D-IR (i.e., PflaIR85a), 1 LS-IR (i.e., PflaIR87a), and 1 iGluR (i.e., PflaGluR1). A-IRs and LS-IRs ordinarily belong to a conservative subfamily among different lepidopteran species. By contrast, D-IRs have variable gene numbers that contribute to the difference in IR numbers (Liu Y et al., 2018; Zhu et al., 2018; Wang et al., 2021; Yin et al., 2021). The phylogenetic tree analysis shows that all PflaIRs can be separated into 15 IR orthologous clades, including IR7d, IR8a, IR21a, IR25a, IR40a, IR41a, IR60a, IR64a, IR68a, IR75, IR76b, IR85a, IR87a, IR93a, and iGluRs. PflaIR8a and PflaIR25a are clustered into the IR8a/25a subfamily, and PflaIR76b is clustered into the IR76b subfamily, which is composed of IR co-receptors. Previous studies showed that IR co-receptors are highly expressed in antennae and other tissues and can sense carboxylic acids, that is, 3-methylpentanoicand hexanoic acids, and temperature variation (Ni et al., 2016; Zhang et al., 2019; Koutroumpa et al., 2021). Moreover, the IR75 gene family exhibits the substantially female-biased expression in the antennae of A. yunnanensis, and the expression is upregulated extensively after mating in H. armigera. The IR75 may be related to mating and spawning behavior (Liu N et al., 2018; Li H. et al., 2021). Therefore, PflaIR75d, PflaIR75c, PflaIR75p, PflaIR75q, and PflaIR2 may have the same function, but validation is needed.
A total of 14 candidate PflaGRs are identified in the P. flammans transcriptome. GRs can be categorized into fructose, sugar, CO2, and bitter receptors by differentiation in function (Tian et al., 2018). Some studies found that sugars and sugar alcohols can influence the host plant search and the oviposition behavior of female C. pomonella (Lombarkia and Derridj, 2008). In the present study, PflaGR8 and PflaGR12 are clustered into fructose receptors that hint the two PflaGRs may have a reaction for fructose the same as BmorGR9 in B. mori (Sato et al., 2011) and SlitGR9 in S. littoralis (Walker et al., 2019) on fructose detection. Two PflaGRs could mediate internal nutrient sensing in the brain, such as DmelGR43a of Drosophila melanogaster (Miyamoto et al., 2012). Moths usually use CO2 as cues to detect the floral food source, and CO2 receptors play an important role in this process (Thom et al., 2004). Most moths have 2–3 CO2 receptors, such as A. yunnanensis (Li G.-C. et al., 2021), S. littoralis (Koutroumpa et al., 2021), and Plodia interpunctella (Jia et al., 2018). PflaGR9 and PflaGR13 are clustered into the branch of the CO2 receptor subfamily. CO2 receptors are usually highly expressed in gustatory organs, including proboscis, tarsi, mouthparts, ovipositors, and legs but have a low expression in the antenna (Guerenstein and Hildebrand, 2008; Koutroumpa et al., 2021). The low number of GRs identified in the current study may be explained in this respect.
In S. insularis, H. assulta, and D. abietella, SNMP1 and SNMP2 are highly expressed in the antenna of male and female adults and are speculated to participate in pheromone detection (Yang et al., 2019; Li H. et al., 2021; Wang et al., 2021). In P. flammans, PflaSNMP1 and PflaSNMP2 are closely clustered into lepidopteran SNMP1 and SNMP2 branches by the evolutionary tree. Many expression studies showed that SNMP1 has a wide association with pheromone sensing (Nichols and Vogt, 2008; Gomez-Diaz et al., 2016; Liu et al., 2021). However, some studies considered that SNMP1 is not necessary for pheromone sensing (Cassau and Krieger, 2021). Therefore, the function of PflaSNMP1 in the sexual communication of P. flammans needs to be further studied. In addition, two PflaSNMPs are recognized in P. flammans, whereas SNMPs are phylogenetically partitioned into three groups in most lepidopteran insects in a recent study (Zhang et al., 2020). Therefore, subsequent verification is needed to prove whether SNMP3 exists in P. flammans.
ORs are the most essential part of insect olfactory recognition (Dani et al., 2011; Leal, 2013). The phylogenetic tree analysis revealed that PflaOrco is clustered into the conservative Orco branch, indicating that it may be Orco in P. flammans. PflaOrco may also become the most highly expressed OR in P. flammans antennae, such as in P. saucia (Sun Y.-L. et al., 2020), S. litura (Wu et al., 2013), and Lymantria dispar (Lin et al., 2015), in which Orco is highly expressed in male antennae. By contrast, the Orco of some moths, such as M. sexta (Howlett et al., 2012), S. cinerearia (Liu et al., 2020), and E. grisescens (Li et al., 2017), shows high expression in female antennae. The olfactory function of Orco can be further confirmed using the RNAi technology and EAG response assays. For example, the expression quantity of GmolOrco and responses to sex pheromones show significant reduction in male Grapholita molesta after injecting dsRNA (Chen X et al., 2021). Fandino et al. (2019) used CRISPR-Cas9-targeted mutagenesis to knockout MsexOrco and showed that the foraging and oviposition behavior of M. sexta are affected. The aforementioned research can provide a reference for the functional verification of PflaOrco.
In the phylogenetic tree, 15 PflaORs (i.e., PflaOR21, PflaOR25, PflaOR27, PflaOR29, PflaOR35, PflaOR37, PflaOR40, PflaOR42, PflaOR44, PflaOR49, PflaOR51, PflaOR60, PflaOR61, PflaOR62, and PflaOR63) are found to be clustered together with three PR clades. Thus, these 15 PflaORs are screened, and two PflaORs (i.e., PflaOR36 and PflaOR41), which are close to the PR clade, are added as candidate PRs to conduct the tissue expression. The results of RTq-PCR analysis show that PflaOR21, PflaOR27, PflaOR29, PflaOR35, PflaOR37, PflaOR40, PflaOR42, PflaOR44, PflaOR60, and PflaOR62 have sex-biased expression in male antennae and may be involved as specificity-sensing sex pheromone compound. These results are similar to those of CpomPRs (i.e., CpomOR1, CpomOR2a, CpomOR5, and CpomOR7) in C. pomonella, in which CpomOR2a and CpomOR5 respond to an important component of C. pomonella sex pheromone (Tian et al., 2021). Classical (i.e., PflaOR27, PflaOR29, PflaOR44, PflaOR60, and PflaOR62) and novel (i.e., PflaOR21, PflaOR35, PflaOR37, PflaOR40, and PflaOR42) PR clades show male sex-biased expression in the antennae. Yuvaraj et al. (2022) found that the PRs of both clades of light brown apple moth show no difference in acetates and both isomers, behavioral attractants, and behavioral antagonists. Two clades have evolved convergently on the ability to detect similar compounds. By contrast, PflaOR36, PflaOR49, PflaOR61, and PflaOR63 reveal female-biased expression in the abdomens and antennae, and this finding is similar to HillOR32, HillOR44, HillOR56, HillOR57, HillOR60, and HillOR66 in Hermetia illucens (Xu et al., 2020). This finding suggested that the four PflaORs act on the male-released pheromone detection, localization of oviposition sites, and oviposition for females. PflaOR25, PflaOR41, and PflaOR51 have almost similar expressions between female and male antennae, such as in G. mellonella (GmelOR13 and GmelOR50) (Wang et al., 2021) and P. saucia (e.g., PsauOR43 and PsauOR54) (Sun Y.-L. et al., 2020), and are likely to function in food source odor perception. Some PRs, such as BmorOR9 of B. mori and HvirOR6 of H. virescens (Krieger et al., 2004; Wanner et al., 2007), also display the no sex-biased expression pattern. PflaOR25, PflaOR41, and PflaOR51 may be similar to these PRs. PflaOR29, PflaOR36, and PflaOR37 can be expressed in nonantennal tissue, indicating wide functions not limited to odor recognition, and odor recognition may be happening in other tissues besides the antennae such as proboscis, palpae, and ovipositor.
In recent years, many sex pheromones of lepidopteron insects show practical application in the monitoring of pest populations, mass trapping, mating disruption, and push–pull strategies (Rizvi et al., 2021). In accordance with the response spectrum of PR and combined with behavior experiments, many sexual behavior antagonists can be screened to interfere with the courtship behavior of target lepidopteran pests, including S. litura (Zhang et al., 2014), Helicoverpa armigera (Chang et al., 2017), and Epiphyas postvittana (Yuvaraj et al., 2022), and have potential applications in developing new control methods (You and Zhang, 2021).
Conclusion
In this study, a full-length transcriptome and an antennal unigene transcriptome of P. flammans antennae were formed. A total of 166 candidate olfactory and gustatory genes, including 58 OBPs, 19 CSPs, 59 ORs, 16 IRs, 14 GRs, and 2 SNMPs, were identified. Their evolution and phylogenetic relationships with those of other lepidopteran insects were elaborated. The expression profile analysis of candidate PflaPRs revealed tissue-specific and sex-biased expression in P. flammans, indicating their probable important role in olfactory recognition processes, such as sexual communication and host recognition. This research filled the gap in olfactory and gustatory genes of P. flammans and provided basic data for future functional molecular mechanism studies on P. flammans olfaction.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The data presented in the study are deposited in the repository: https://www.ncbi.nlm.nih.gov/, accession number:PRJNA806455.
Author Contributions
The conception and the design of the work were formulated by JH, X-YW, L-ST, WL, and X-LZ. Bioinformatics analyses were performed by JH, X-YW, and L-ST. The laboratory experiments were performed by JH and L-ST. The statistics analysis was carried out by JH, X-YW and L-ST. The analyses of experimental data and interpretation of data were performed by JH, X-YW, L-ST, WL, and X-LZ. The manuscript was written, validated, and revised by JH, X-YW, L-ST, WL, and X-LZ.
Funding
This work was supported by the National Natural Science Foundation of China (31660206) and the Guangxi Scholarship Fund of Guangxi Education Department.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2022.907694/full#supplementary-material
References
Almagro Armenteros J. J., Tsirigos K. D., Sønderby C. K., Petersen T. N., Winther O., Brunak S., et al. (2019). SignalP 5.0 Improves Signal Peptide Predictions Using Deep Neural Networks. Nat. Biotechnol. 37, 420–423. doi:10.1038/s41587-019-0036-z
Altschul S., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., et al. (1997). Gapped BLAST and PSI-BLAST: a New Generation of Protein Database Search Programs. Nucleic Acids Res. 25, 3389–3402. doi:10.1093/nar/25.17.3389
Benton R. (2009). Evolution and Revolution in Odor Detection. Science 326, 382–383. doi:10.1126/science.1181998
Benton R., Vannice K. S., Vosshall L. B. (2007). An Essential Role for a CD36-Related Receptor in Pheromone Detection in Drosophila. Nature 450, 289–293. doi:10.1038/nature06328
Bruce T. J. A., Wadhams L. J., Woodcock C. M. (2005). Insect Host Location: a Volatile Situation. Trends Plant Sci. 10, 269–274. doi:10.1016/j.tplants.2005.04.003
Cao D., Liu Y., Wei J., Liao X., Walker W. B., Li J., et al. (2014). Identification of Candidate Olfactory Genes in Chilo Suppressalis by Antennal Transcriptome Analysis. Int. J. Biol. Sci. 10, 846–860. doi:10.7150/ijbs.9297
Cassau S., Krieger J. (2021). The Role of SNMPs in Insect Olfaction. Cell Tissue Res. 383, 21–33. doi:10.1007/s00441-020-03336-0
Chang H., Liu Y., Ai D., Jiang X., Dong S., Wang G. (2017). A Pheromone Antagonist Regulates Optimal Mating Time in the Moth Helicoverpa Armigera. Curr. Biol. 27, 1610–1615. doi:10.1016/j.cub.2017.04.035
Chen C., Buhl E., Xu M., Croset V., Rees J. S., Lilley K. S., et al. (2015). Drosophila Ionotropic Receptor 25a Mediates Circadian Clock Resetting by Temperature. Nature 527, 516–520. doi:10.1038/nature16148
Chen L., Tian Z., Wang X. Y., Chen X. M., Lu W., Wang X. P., et al. (2021). Screening of Reference Genes for Quantitative Real-Time PCR in Phauda Flammans (Walker) (Lepidoptera: Phaudidae). J. Environ. Entomol. 43, 15–24.
Chen X. L., Li B. L., Chen Y. X., Li G. W., Wu J. X. (2021). Functional Analysis of the Odorant Receptor Coreceptor in Odor Detection in Grapholita Molesta (Lepidoptera: Tortricidae). Arch. Insect Biochem. Physiol. 108, e21837. doi:10.1002/arch.21837
Clyne P. J., Warr C. G., Carlson J. R. (2000). Candidate Taste Receptors in Drosophila. Science 287, 1830–1834. doi:10.1126/science.287.5459.1830
Clyne P. J., Warr C. G., Freeman M. R., Lessing D., Kim J., Carlson J. R. (1999). A Novel Family of Divergent Seven-Transmembrane Proteins. Neuron 22, 327–338. doi:10.1016/s0896-6273(00)81093-4
Conesa A., Gotz S., Garcia-Gomez J. M., Terol J., Talon M., Robles M. (2005). Blast2GO: a Universal Tool for Annotation, Visualization and Analysis in Functional Genomics Research. Bioinformatics 21, 3674–3676. doi:10.1093/bioinformatics/bti610
Croset V., Rytz R., Cummins S. F., Budd A., Brawand D., Kaessmann H., et al. (2010). Ancient Protostome Origin of Chemosensory Ionotropic Glutamate Receptors and the Evolution of Insect Taste and Olfaction. PLoS Genet. 6, e1001064. doi:10.1371/journal.pgen.1001064
Dani F. R., Michelucci E., Francese S., Mastrobuoni G., Cappellozza S., La Marca G., et al. (2011). Odorant-binding Proteins and Chemosensory Proteins in Pheromone Detection and Release in the Silkmoth Bombyx mori. Chem. Senses 36, 335–344. doi:10.1093/chemse/bjq137
Dyanov H. M., Dzitoeva S. G. (1995). Method for Attachment of Microscopic Preparations on Glass for In Situ Hybridization, PRINS and In Situ PCR Studies. Biotechniques 18, 822–826.
Fandino R. A., Haverkamp A., Bisch-Knaden S., Zhang J., Bucks S., Nguyen T. A. T., et al. (2019). Mutagenesis of Odorant Coreceptor Orco Fully Disrupts Foraging but Not Oviposition Behaviors in the Hawkmoth Manduca Sexta. Proc. Natl. Acad. Sci. U.S.A. 116, 15677–15685. doi:10.1073/pnas.1902089116
Feng B., Guo Q., Zheng K., Qin Y., Du Y. (2017). Antennal Transcriptome Analysis of the Piercing Moth Oraesia Emarginata (Lepidoptera: Noctuidae). PLoS One 12, e0179433. doi:10.1371/journal.pone.0179433
Fof (2015). Diurnal Leaf Skeletonizing Moth from Vietnam: Phauda Flammans. Available online at: https://www.whatsthatbug.com/2015/06/08/diurnal-leaf-skeletonizing-moth-from-vietnam-phauda-flammans/ (accessed March 14, 2022).
Fu L., Niu B., Zhu Z., Wu S., Li W. (2012). CD-HIT: Accelerated for Clustering the Next-Generation Sequencing Data. Bioinformatics 28, 3150–3152. doi:10.1093/bioinformatics/bts565
Gomez-Diaz C., Bargeton B., Abuin L., Bukar N., Reina J. H., Bartoi T., et al. (2016). A CD36 Ectodomain Mediates Insect Pheromone Detection via a Putative Tunnelling Mechanism. Nat. Commun. 7, 11866. doi:10.1038/ncomms11866
Gong L., Luo Q., Rizwan-Ul-Haq M., Hu M.-Y. (2012). Cloning and Characterization of Three Chemosensory Proteins from Spodoptera Exigua and Effects of Gene Silencing on Female Survival and Reproduction. Bull. Entomol. Res. 102, 600–609. doi:10.1017/S0007485312000168
Gouin A., Bretaudeau A., Nam K., Gimenez S., Aury J.-M., Duvic B., et al. (2017). Two Genomes of Highly Polyphagous Lepidopteran Pests (Spodoptera Frugiperda, Noctuidae) with Different Host-Plant Ranges. Sci. Rep. 7, 1186. doi:10.1038/s41598-017-10461-4
Grosse-Wilde E., Gohl T., Bouché E., Breer H., Krieger J. (2007). Candidate Pheromone Receptors Provide the Basis for the Response of Distinct Antennal Neurons to Pheromonal Compounds. Eur. J. Neurosci. 25, 2364–2373. doi:10.1111/j.1460-9568.2007.05512.x
Gu T., Huang K., Tian S., Sun Y., Li H., Chen C., et al. (2019). Antennal Transcriptome Analysis and Expression Profiles of Odorant Binding Proteins in Clostera Restitura. Comp. Biochem. Physiology Part D Genomics Proteomics 29, 211–220. doi:10.1016/j.cbd.2018.12.002
Gu X.-C., Zhang Y.-N., Kang K., Dong S.-L., Zhang L.-W. (2015). Antennal Transcriptome Analysis of Odorant Reception Genes in the Red Turpentine Beetle (RTB), Dendroctonus Valens. PLoS One 10, e0125159. doi:10.1371/journal.pone.0125159
Guan W. K., Wang X. Y., Lu W., Zhang Z. L., Zheng X. L. (2020). Electrophysiological and Behavioral Responses of Phauda Flammans to Plant Volatiles Isolated from Two Ficus Species. J. For. Environ. 40, 442–448. doi:10.13324/j.cnki.jfcf.2020.04.015
Guerenstein P. G., Hildebrand J. G. (2008). Roles and Effects of Environmental Carbon Dioxide in Insect Life. Annu. Rev. Entomol. 53, 161–178. doi:10.1146/annurev.ento.53.103106.093402
Hallem E. A., Dahanukar A., Carlson J. R. (2006). Insect Odor and Taste Receptors. Annu. Rev. Entomol. 51, 113–135. doi:10.1146/annurev.ento.51.051705.113646
Han W.-K., Yang Y.-L., Si Y.-X., Wei Z.-Q., Liu S.-R., Liu X.-L., et al. (2022). Involvement of GOBP2 in the Perception of a Sex Pheromone Component in Both Larval and Adult Spodoptera Litura Revealed Using CRISPR/Cas9 Mutagenesis. Insect Biochem. Mol. Biol. 141, 103719. doi:10.1016/j.ibmb.2022.103719
Hildebrand J. G., Shepherd G. M. (1997). Mechanisms of Olfactory Discrimination: Converging Evidence for Common Principles across Phyla. Annu. Rev. Neurosci. 20, 595–631. doi:10.1146/annurev.neuro.20.1.595
Howlett N., Dauber K. L., Shukla A., Morton B., Glendinning J. I., Brent E., et al. (2012). Identification of Chemosensory Receptor Genes in Manduca Sexta and Knockdown by RNA Interference. BMC Genomics 13, 211. doi:10.1186/1471-2164-13-211
Hu P., Tao J., Cui M., Gao C., Lu P., Luo Y. (2016). Antennal Transcriptome Analysis and Expression Profiles of Odorant Binding Proteins in Eogystia Hippophaecolus (Lepidoptera: Cossidae). BMC Genomics 17, 651. doi:10.1186/s12864-016-3008-4
Jia X.-J., Wang H.-X., Yan Z.-G., Zhang M.-Z., Wei C.-H., Qin X.-C., et al. (2016). Antennal Transcriptome and Differential Expression of Olfactory Genes in the Yellow Peach Moth, Conogethes Punctiferalis (Lepidoptera: Crambidae). Sci. Rep. 6, 29067. doi:10.1038/srep29067
Jia X., Zhang X., Liu H., Wang R., Zhang T. (2018). Identification of Chemosensory Genes from the Antennal Transcriptome of Indian Meal Moth Plodia Interpunctella. PLoS One 13, e0189889. doi:10.1371/journal.pone.0189889
Jiang X.-C., Liu S., Jiang X.-Y., Wang Z.-W., Xiao J.-J., Gao Q., et al. (2021). Identification of Olfactory Genes from the Greater Wax Moth by Antennal Transcriptome Analysis. Front. Physiol. 12, 663040. doi:10.3389/fphys.2021.663040
Jing D., Zhang T., Prabu S., Bai S., He K., Wang Z. (2020). Molecular Characterization and Volatile Binding Properties of Pheromone Binding Proteins and General Odorant Binding Proteins in Conogethes Pinicolalis (Lepidoptera: Crambidae). Int. J. Biol. Macromol. 146, 263–272. doi:10.1016/j.ijbiomac.2019.12.248
Knecht Z. A., Silbering A. F., Ni L., Klein M., Budelli G., Bell R., et al. (2016). Distinct Combinations of Variant Ionotropic Glutamate Receptors Mediate Thermosensation and Hygrosensation in Drosophila. eLife 5, e17879. doi:10.7554/eLife.17879.001
Koutroumpa F. A., Monsempes C., François M.-C., de Cian A., Royer C., Concordet J.-P., et al. (2016). Heritable Genome Editing with CRISPR/Cas9 Induces Anosmia in a Crop Pest Moth. Sci. Rep. 6, 29620. doi:10.1038/srep29620
Koutroumpa F. A., Monsempes C., François M.-C., Severac D., Montagné N., Meslin C., et al. (2021). Description of Chemosensory Genes in Unexplored Tissues of the Moth Spodoptera Littoralis. Front. Ecol. Evol. 9, 678277. doi:10.3389/fevo.2021.678277
Krieger J., Grosse-Wilde E., Gohl T., Dewer Y. M. E., Raming K., Breer H. (2004). Genes Encoding Candidate Pheromone Receptors in a Moth ( Heliothis virescens ). Proc. Natl. Acad. Sci. U.S.A. 101, 11845–11850. doi:10.1073/pnas.0403052101
Krieger J., Raming K., Dewer Y. M. E., Bette S., Conzelmann S., Breer H. (2002). A Divergent Gene Family Encoding Candidate Olfactory Receptors of the mothHeliothis Virescens. Eur. J. Neurosci. 16, 619–628. doi:10.1046/j.1460-9568.2002.02109.x
Kumar S., Stecher G., Tamura K. (2016). MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 33, 1870–1874. doi:10.1093/molbev/msw054
Larsson M. C., Domingos A. I., Jones W. D., Chiappe M. E., Amrein H., Vosshall L. B. (2004). Or83b Encodes a Broadly Expressed Odorant Receptor Essential for Drosophila Olfaction. Neuron 43, 703–714. doi:10.1016/j.neuron.2004.08.019
Leal W. S. (2013). Odorant Reception in Insects: Roles of Receptors, Binding Proteins, and Degrading Enzymes. Annu. Rev. Entomol. 58, 373–391. doi:10.1146/annurev-ento-120811-153635
Li G.-C., Nuo S.-M., Wang Z.-Q., Yang A.-J., Liu N.-Y. (2021a). Identification and Expression Profiling of Chemosensory Membrane Protein Genes in Achelura Yunnanensis (Lepidoptera: Zygaenidae). Comp. Biochem. Physiology Part D Genomics Proteomics 40, 100876. doi:10.1016/j.cbd.2021.100876
Li H., Li W., Miao C., Wang G., Zhao M., Yuan G., et al. (2021b). Identification of the Differences in Olfactory System between Male and Female Oriental Tobacco Budworm Helicoverpa Assulta. Arch. Insect Biochem. Physiol. 107, e21829. doi:10.1002/arch.21829
Li M.-Y., Jiang X.-Y., Liu X.-Y., Huang Y.-J., Li S.-G., Liu S. (2020). Genome-wide Analysis of Chemosensory Protein Genes in the Small White Butterfly Pieris Rapae (Lepidoptera: Pieridae). J. Asia-Pacific Entomology 23, 772–780. doi:10.1016/j.aspen.2020.07.005
Li Z.-Q., Luo Z.-X., Cai X.-M., Bian L., Xin Z.-J., Liu Y., et al. (2017). Chemosensory Gene Families in Ectropis Grisescens and Candidates for Detection of Type-II Sex Pheromones. Front. Physiol. 8, 953. doi:10.3389/fphys.2017.00953
Lin W., Yu Y., Zhou P., Zhang J., Dou L., Hao Q., et al. (2015). Identification and Knockdown of the Olfactory Receptor (OrCo) in Gypsy Moth, Lymantria dispar. Int. J. Biol. Sci. 11, 772–780. doi:10.7150/ijbs.11898
Liu J., Liu H., Yi J., Mao Y., Li J., Sun D., et al. (2021). Transcriptome Characterization and Expression Analysis of Chemosensory Genes in Chilo Sacchariphagus (Lepidoptera Crambidae), a Key Pest of Sugarcane. Front. Physiol. 12, 636353. doi:10.3389/fphys.2021.636353
Liu J. Y., He Q. L., Wei H., Yang J., Li J., Lu W., et al. (2015). Biological Characteristics of Phauda Flammans (Lepidoptera: Zygaenidae). Plant Prot. 41, 188–192.
Liu J. Y., Ma Z. H., Wu S. Y., Liu M. Y., Zhang J., Lu W., et al. (2016). Studies on the Feeding Preferences of Phauda Flammans Walker (Lepidoptera: Zygaenidae). J. Environ. Entomol. 38, 924–930.
Liu N.-Y., Xu W., Dong S.-L., Zhu J.-Y., Xu Y.-X., Anderson A. (2018). Genome-wide Analysis of Ionotropic Receptor Gene Repertoire in Lepidoptera with an Emphasis on its Functions of Helicoverpa Armigera. Insect Biochem. Mol. Biol. 99, 37–53. doi:10.1016/j.ibmb.2018.05.005
Liu N.-Y., Zhang T., Ye Z.-F., Li F., Dong S.-L. (2015). Identification and Characterization of Candidate Chemosensory Gene Families from Spodoptera Exigua Developmental Transcriptomes. Int. J. Biol. Sci. 11, 1036–1048. doi:10.7150/ijbs.12020
Liu P., Zhang X., Meng R., Liu C., Li M., Zhang T. (2020). Identification of Chemosensory Genes from the Antennal Transcriptome of Semiothisa Cinerearia. PLoS One 15, e0237134. doi:10.1371/journal.pone.0237134
Liu X., Tong N., Wu Z., Li Y., Ma M., Liu P., et al. (2022). Identification of Chemosensory Genes Based on the Antennal Transcriptomic Analysis of Plagiodera Versicolora. Insects 13, 36. doi:10.3390/insects13010036
Liu Y., Liu Y., Jiang X., Wang G. (2018). Cloning and Functional Characterization of Three New Pheromone Receptors from the Diamondback Moth, Plutella Xylostella. J. Insect Physiology 107, 14–22. doi:10.1016/j.jinsphys.2018.02.005
Liu Z., Vidal D. M., Syed Z., Ishida Y., Leal W. S. (2010). Pheromone Binding to General Odorant-Binding Proteins from the Navel Orangeworm. J. Chem. Ecol. 36, 787–794. doi:10.1007/s10886-010-9811-5
Liu Z., Wang X., Lei C., Zhu F. (2017). Sensory Genes Identification with Head Transcriptome of the Migratory Armyworm, Mythimna Separata. Sci. Rep. 7, 46033. doi:10.1038/srep46033
Livak K. J., Schmittgen T. D. (2001). Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 25, 402–408. doi:10.1006/meth.2001.1262
Lombarkia N., Derridj S. (2008). Resistance of Apple Trees to Cydia Pomonella Egg‐laying Due to Leaf Surface Metabolites. Entomologia Exp. Appl. 128, 57–65. doi:10.1111/j.1570-7458.2008.00741.x
Maleszka R., Stange G. (1997). Molecular Cloning, by a Novel Approach, of a cDNA Encoding a Putative Olfactory Protein in the Labial Palps of the Moth Cactoblastis cactorum. Gene 202, 39–43. doi:10.1016/S0378-1119(97)00448-4
Martin F., Alcorta E. (2011). Regulation of Olfactory Transduction in the Orco Channel. Front. Cell. Neurosci. 5, 21. doi:10.3389/fncel.2011.00021
Mitsuno H., Sakurai T., Murai M., Yasuda T., Kugimiya S., Ozawa R., et al. (2008). Identification of Receptors of Main Sex-Pheromone Components of Three Lepidopteran Species. Eur. J. Neurosci. 28, 893–902. doi:10.1111/j.1460-9568.2008.06429.x
Miyamoto T., Slone J., Song X., Amrein H. (2012). A Fructose Receptor Functions as a Nutrient Sensor in the Drosophila Brain. Cell 151, 1113–1125. doi:10.1016/j.cell.2012.10.024
Nageshchandra B. K., Rajagopal B. K., Balasubramanian R. (1972). Occurrence of Slug Caterpillar Phauda Flammans Wlk. (Lepidoptera: Zygaenidae) on Ficus Racemosa L. In South India. Mysore J. Agr. Sci. 6, 186–189.
Nakagawa T., Sakurai T., Nishioka T., Touhara K. (2005). Insect Sex-Pheromone Signals Mediated by Specific Combinations of Olfactory Receptors. Science 307, 1638–1642. doi:10.1126/science.1106267
Neuhaus E. M., Gisselmann G., Zhang W., Dooley R., Störtkuhl K., Hatt H. (2005). Odorant Receptor Heterodimerization in the Olfactory System of Drosophila melanogaster. Nat. Neurosci. 8, 15–17. doi:10.1038/nn1371
Ni L., Klein M., Svec K. V., Budelli G., Chang E. C., Ferrer A. J., et al. (2016). The Ionotropic Receptors IR21a and IR25a Mediate Cool Sensing in Drosophila. eLife 5, e13254. doi:10.7554/eLife.13254
Nichols Z., Vogt R. G. (2008). The SNMP/CD36 Gene Family in Diptera, Hymenoptera and Coleoptera: Drosophila melanogaster, D. pseudoobscura, Anopheles gambiae, Aedes aegypti, Apis mellifera, and Tribolium castaneum. Insect Biochem. Mol. Biol. 38, 398–415. doi:10.1016/j.ibmb.2007.11.003
Ning C., Yang K., Xu M., Huang L.-Q., Wang C.-Z. (2016). Functional Validation of the Carbon Dioxide Receptor in Labial Palps of Helicoverpa Armigera Moths. Insect Biochem. Mol. Biol. 73, 12–19. doi:10.1016/j.ibmb.2016.04.002
Nishimura O., Hara Y., Kuraku S. (2017). gVolante for Standardizing Completeness Assessment of Genome and Transcriptome Assemblies. Bioinformatics 33, 3635–3637. doi:10.1093/bioinformatics/btx445
Olivier V., Monsempes C., François M.-C., Poivet E., Jacquin-Joly E. (2011). Candidate Chemosensory Ionotropic Receptors in a Lepidoptera. Insect Mol. Biol. 20, 189–199. doi:10.1111/j.1365-2583.2010.01057.x
Pelosi P., Iovinella I., Felicioli A., Dani F. R. (2014). Soluble Proteins of Chemical Communication: an Overview across Arthropods. Front. Physiol. 5, 320. doi:10.3389/fphys.2014.00320
Pregitzer P., Greschista M., Breer H., Krieger J. (2014). The Sensory Neurone Membrane Protein SNMP1 Contributes to the Sensitivity of a Pheromone Detection System. Insect Mol. Biol. 23, 733–742. doi:10.1111/imb.12119
Prestwich G. D. (1996). Proteins that Smell: Pheromone Recognition and Signal Transduction. Bioorg. Med. Chem. 4, 505–513. doi:10.1016/0968-0896(96)00033-8
Rizvi S. A. H., George J., Reddy G. V. P., Zeng X., Guerrero A. (2021). Latest Developments in Insect Sex Pheromone Research and its Application in Agricultural Pest Management. Insects 12, 484. doi:10.3390/insects12060484
Robinson G. S., Ackery P. R., Kitching I. J., Beccaloni G. W., Hernández L. M. (2019). Hosts–A Database of the World’s Lepidopteran Hostplants. (accessed March 14, 2022).
Rogers M. E., Sun M., Lerner M. R., Vogt R. G. (1997). Snmp-1, a Novel Membrane Protein of Olfactory Neurons of the Silk Moth Antheraea polyphemus with Homology to the CD36 Family of Membrane Proteins. J. Biol. Chem. 272, 14792–14799. doi:10.1074/jbc.272.23.14792
Sakurai T., Nakagawa T., Mitsuno H., Mori H., Endo Y., Tanoue S., et al. (2004). Identification and Functional Characterization of a Sex Pheromone Receptor in the Silkmoth Bombyx mori. Proc. Natl. Acad. Sci. U.S.A. 101, 16653–16658. doi:10.1073/pnas.0407596101
Sato K., Tanaka K., Touhara K. (2011). Sugar-regulated Cation Channel Formed by an Insect Gustatory Receptor. Proc. Natl. Acad. Sci. U.S.A. 108, 11680–11685. doi:10.1073/pnas.1019622108
Song Y. Q., Sun H. Z., Dong J. F. (2020). Identification and Expression Analysis of an Olfactory Receptor Gene Family in the Yemma Signatus Hsiao (Hemiptera: Berytidae). Pjz 52, 1911–1919. doi:10.17582/journal.pjz/20180616040644
Sun D., Huang Y., Qin Z., Zhan H., Zhang J., Liu Y., et al. (2020a). Identification of Candidate Olfactory Genes in the Antennal Transcriptome of the Stink Bug Halyomorpha Halys. Front. Physiol. 11, 876. doi:10.3389/fphys.2020.00876
Sun H., Song Y., Du J., Wang X., Cheng Z. (2016). Identification and Tissue Distribution of Chemosensory Protein and Odorant Binding Protein Genes in Athetis Dissimilis (Lepidoptera: Noctuidae). Appl. Entomol. Zool. 51, 409–420. doi:10.1007/s13355-016-0413-8
Sun Y.-L., Dong J.-F., Gu N., Wang S.-L. (2020b). Identification of Candidate Chemosensory Receptors in the Antennae of the Variegated Cutworm, Peridroma Saucia Hübner, Based on a Transcriptome Analysis. Front. Physiol. 11, 39. doi:10.3389/fphys.2020.00039
Sun Y.-L., Dong J.-F., Ning C., Ding P.-P., Huang L.-Q., Sun J.-G., et al. (2019). An Odorant Receptor Mediates the Attractiveness of Cis -jasmone to Campoletis Chlorideae, the Endoparasitoid of Helicoverpa Armigera. Insect Mol. Biol. 28, 23–34. doi:10.1111/imb.12523
Thom C., Guerenstein P. G., Mechaber W. L., Hildebrand J. G. (2004). Floral CO2Reveals Flower Profitability to Moths. J. Chem. Ecol. 30, 1285–1288. doi:10.1023/B:JOEC.0000030298.77377.7d
Tian K., Liu W., Feng L. K., Huang T. Y., Wang G. R., Lin K. J. (2021). Functional Characterization of Pheromone Receptor Candidates in Codling Moth Cydia Pomonella (Lepidoptera: Tortricidae). Insect Sci. 28, 445–456. doi:10.1111/1744-7917.12775
Tian Z., Sun L., Li Y., Quan L., Zhang H., Yan W., et al. (2018). Antennal Transcriptome Analysis of the Chemosensory Gene Families in Carposina Sasakii (Lepidoptera: Carposinidae). BMC Genomics 19, 544. doi:10.1186/s12864-018-4900-x
Trifinopoulos J., Nguyen L.-T., von Haeseler A., Minh B. Q. (2016). W-IQ-TREE: a Fast Online Phylogenetic Tool for Maximum Likelihood Analysis. Nucleic Acids Res. 44, W232–W235. doi:10.1093/nar/gkw256
Tsirigos K. D., Peters C., Shu N., Käll L., Elofsson A. (2015). The TOPCONS Web Server for Consensus Prediction of Membrane Protein Topology and Signal Peptides. Nucleic Acids Res. 43, W401–W407. doi:10.1093/nar/gkv485
Verma T. D., Dogra G. S. (1982). Occurrence of Phauda Flammans Wlk. (Lepidoptera: Zygaenidae) on ficus Species in Himachal Pradesh. J. Tree. Sci. 1, 130–132.
Vogt R. G., Riddiford L. M. (1981). Pheromone Binding and Inactivation by Moth Antennae. Nature 293, 161–163. doi:10.1038/293161a0
Walker W. B., Roy A., Anderson P., Schlyter F., Hansson B. S., Larsson M. C. (2019). Transcriptome Analysis of Gene Families Involved in Chemosensory Function in Spodoptera Littoralis (Lepidoptera: Noctuidae). BMC genomics 20, 428. doi:10.1186/s12864-019-5815-x
Wang Z.-Q., Wu C., Li G.-C., Nuo S.-M., Yin N.-N., Liu N.-Y. (2021). Transcriptome Analysis and Characterization of Chemosensory Genes in the Forest Pest, Dioryctria Abietella (Lepidoptera: Pyralidae). Front. Ecol. Evol. 9, 748199. doi:10.3389/fevo.2021.748199
Wanner K. W., Anderson A. R., Trowell S. C., Theilmann D. A., Robertson H. M., Newcomb R. D. (2007). Female-biased Expression of Odourant Receptor Genes in the Adult Antennae of the Silkworm, Bombyx mori. Insect Mol. Biol. 16, 107–119. doi:10.1111/j.1365-2583.2007.00708.x
Wanner K. W., Robertson H. M. (2008). The Gustatory Receptor Family in the Silkworm mothBombyx Moriis Characterized by a Large Expansion of a Single Lineage of Putative Bitter Receptors. Insect Mol. Biol. 17, 621–629. doi:10.1111/j.1365-2583.2008.00836.x
Wei H.-S., Li K.-B., Zhang S., Cao Y.-Z., Yin J. (2017). Identification of Candidate Chemosensory Genes by Transcriptome Analysis in Loxostege Sticticalis Linnaeus. PLoS One 12, e0174036. doi:10.1371/journal.pone.0174036
Wicher D., Miazzi F. (2021). Functional Properties of Insect Olfactory Receptors: Ionotropic Receptors and Odorant Receptors. Cell Tissue Res. 383, 7–19. doi:10.1007/s00441-020-03363-x
Wu Z.-N., Chen X., Du Y.-J., Zhou J.-J., ZhuGe Q.-C. (2013). Molecular Identification and Characterization of the Orco Orthologue ofSpodoptera Litura. Insect Sci. 20, 175–182. doi:10.1111/j.1744-7917.2011.01483.x
Wu Z., Ye J., Qian J., Purba E., Zhang Q., Zhang L., et al. (2022). Identification and Expression Profile of Chemosensory Receptor Genes in Aromia Bungii (Faldermann) Antennal Transcriptome. Insects 13, 96. doi:10.3390/insects13010096
Xiao W., Yang L., Xu Z., He L. (2016). Transcriptomics and Identification of Candidate Chemosensory Genes in Antennae of Conogethes Punctiferalis (Lepidoptera: Crambidae). J. Asia-Pacific Entomology 19, 911–920. doi:10.1016/j.aspen.2016.08.007
Xing Y., Thanasirungkul W., Adeel M. M., Yu J., Aslam A., Chi D.-F. (2021). Identification and Analysis of Olfactory Genes in Dioryctria Abietella Based on the Antennal Transcriptome. Comp. Biochem. Physiology Part D Genomics Proteomics 38, 100814. doi:10.1016/j.cbd.2021.100814
Xu H., Turlings T. C. J. (2018). Plant Volatiles as Mate-Finding Cues for Insects. Trends Plant Sci. 23, 100–111. doi:10.1016/j.tplants.2017.11.004
Xu Q., Wu Z., Zeng X., An X. (2020). Identification and Expression Profiling of Chemosensory Genes in Hermetia Illucens via a Transcriptomic Analysis. Front. Physiol. 11, 720. doi:10.3389/fphys.2020.00720
Xu W. (2020). How Do Moth and Butterfly Taste?-Molecular Basis of Gustatory Receptors in Lepidoptera. Insect Sci. 27, 1148–1157. doi:10.1111/1744-7917.12718
Yang H., Dong J., Sun Y., Hu Z., Lv Q., Li D. (2020). Antennal Transcriptome Analysis and Expression Profiles of Putative Chemosensory Soluble Proteins in Histia Rhodope Cramer (Lepidoptera: Zygaenidae). Comp. Biochem. Physiology Part D Genomics Proteomics 33, 100654. doi:10.1016/j.cbd.2020.100654
Yang S., Cao D., Wang G., Liu Y. (2017). Identification of Genes Involved in Chemoreception in Plutella Xyllostella by Antennal Transcriptome Analysis. Sci. Rep. 7, 11941. doi:10.1038/s41598-017-11646-7
Yang Y., Li W., Tao J., Zong S. (2019). Antennal Transcriptome Analyses and Olfactory Protein Identification in an Important Wood-Boring Moth Pest, Streltzoviella Insularis (Lepidoptera: Cossidae). Sci. Rep. 9, 17951. doi:10.1038/s41598-017-11646-710.1038/s41598-019-54455-w
Ye J., Fang L., Zheng H., Zhang Y., Chen J., Zhang Z., et al. (2006). WEGO: a Web Tool for Plotting GO Annotations. Nucleic Acids Res. 34, W293–W297. doi:10.1093/nar/gkl031
Yin N.-N., Nuo S.-M., Xiao H.-Y., Zhao Y.-J., Zhu J.-Y., Liu N.-Y. (2021). The Ionotropic Receptor Gene Family in Lepidoptera and Trichoptera: Annotation, Evolutionary and Functional Perspectives. Genomics 113, 601–612. doi:10.1016/j.ygeno.2020.09.056,
You Y. W., Zhang L. (2021). Progress in the Functional Research of Odorant Receptors of Agricultural Insects. Acta Entomol. Sin. 64, 627–644. doi:10.16380/j.kcxb.2021.05.010
Yuvaraj J. K., Jordan M. D., Zhang D.-D., Andersson M. N., Löfstedt C., Newcomb R. D., et al. (2022). Sex Pheromone Receptors of the Light Brown Apple Moth, Epiphyas Postvittana, Support a Second Major Pheromone Receptor Clade within the Lepidoptera. Insect Biochem. Mol. Biol. 141, 103708. doi:10.1016/j.ibmb.2021.103708
Zacharuk R. Y., Shields V. D. (1991). Sensilla of Immature Insects. Annu. Rev. Entomol. 36, 331–354. doi:10.1146/annurev.en.36.010191.001555
Zhang H.-J., Xu W., Chen Q.-m., Sun L.-N., Anderson A., Xia Q.-Y., et al. (2020). A Phylogenomics Approach to Characterizing Sensory Neuron Membrane Proteins (SNMPs) in Lepidoptera. Insect Biochem. Mol. Biol. 118, 103313. doi:10.1016/j.ibmb.2020.103313
Zhang J., Bisch-Knaden S., Fandino R. A., Yan S., Obiero G. F., Grosse-Wilde E., et al. (2019). The Olfactory Coreceptor IR8a Governs Larval Feces-Mediated Competition Avoidance in a Hawkmoth. Proc. Natl. Acad. Sci. U.S.A. 116, 21828–21833. doi:10.1073/pnas.1913485116
Zhang J., Liu Y., Walker W. B., Dong S.-L., Wang G.-R. (2015). Identification and Localization of Two Sensory Neuron Membrane Proteins fromSpodoptera litura(Lepidoptera: Noctuidae). Insect Sci. 22, 399–408. doi:10.1111/1744-7917.12131
Zhang D. D., Löfstedt C. (2015). Moth Pheromone Receptors: Gene Sequences, Function, and Evolution. Front. Ecol. Evol. 3, 105. doi:10.3389/fevo.2015.00105
Zhang J., Yan S., Liu Y., Jacquin-Joly E., Dong S., Wang G. (2014). Identification and Functional Characterization of Sex Pheromone Receptors in the Common Cutworm (Spodoptera Litura). Chem. Senses 40, 7–16. doi:10.1093/chemse/bju052
Zhang Y.-N., Jin J.-Y., Jin R., Xia Y.-H., Zhou J.-J., Deng J.-Y., et al. (2013). Differential Expression Patterns in Chemosensory and Non-chemosensory Tissues of Putative Chemosensory Genes Identified by Transcriptome Analysis of Insect Pest the Purple Stem Borer Sesamia Inferens (Walker). PLoS One 8, e69715. doi:10.1371/journal.pone.0069715
Zhang Y.-N., Zhu X.-Y., Ma J.-F., Dong Z.-P., Xu J.-W., Kang K., et al. (2017). Molecular Identification and Expression Patterns of Odorant Binding Protein and Chemosensory Protein Genes inAthetis lepigone(Lepidoptera: Noctuidae). PeerJ 5, e3157. doi:10.7717/peerj.3157
Zheng X. L., Liu J. Y., Zhang Z. L., Wang P., Lu W. (2019). Diel Rhythms of Sexual Behavior and Pheromone Responses in Phauda Flammans Walker (Lepidoptera: Zygaenidae). Pest. Manag. Sci. 75, 3070–3075. doi:10.1002/ps.5423
Zhou J.-J., He X.-L., Pickett J. A., Field L. M. (2008). Identification of Odorant-Binding Proteins of the Yellow Fever Mosquito Aedes aegypti: Genome Annotation and Comparative Analyses. Insect Mol. Biol. 17, 147–163. doi:10.1111/j.1365-2583.2007.00789.x
Zhou X., Guo J., Zhang M., Bai C., Wang Z., Li Z. (2021). Antennal Transcriptome Analysis and Candidate Olfactory Genes in Crematogaster Rogenhoferi. Bull. Entomol. Res. 111, 464–475. doi:10.1017/S0007485321000134
Keywords: transcriptome, chemosensory genes, Phauda flammans, gene expression, pheromone receptors
Citation: Hu J, Wang X-Y, Tan L-S, Lu W and Zheng X-L (2022) Identification of Chemosensory Genes, Including Candidate Pheromone Receptors, in Phauda flammans (Walker) (Lepidoptera: Phaudidae) Through Transcriptomic Analyses. Front. Physiol. 13:907694. doi: 10.3389/fphys.2022.907694
Received: 30 March 2022; Accepted: 24 May 2022;
Published: 01 July 2022.
Edited by:
Kayvan Etebari, The University of Queensland, AustraliaReviewed by:
Youssef Dewer, Agricultural Research Center, EgyptJacob Corcoran, Biological Control of Insects Research Laboratory (USDA-ARS), United States
William Benjamin Walker III, United States Department of Agriculture (USDA-ARS), United States
Copyright © 2022 Hu, Wang, Tan, Lu and Zheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xia-Lin Zheng, emhlbmcteGlhLWxpbkAxNjMuY29t
 Jin Hu
Jin Hu Xiao-Yun Wang
Xiao-Yun Wang Liu-Su Tan
Liu-Su Tan Xia-Lin Zheng
Xia-Lin Zheng