
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Physiol. , 20 May 2022
Sec. Craniofacial Biology and Dental Research
Volume 13 - 2022 | https://doi.org/10.3389/fphys.2022.899699
 Lay Thant1,2,3
Lay Thant1,2,3 Masaru Kaku4*
Masaru Kaku4* Yoshito Kakihara1
Yoshito Kakihara1 Masaru Mizukoshi2
Masaru Mizukoshi2 Megumi Kitami1,3
Megumi Kitami1,3 Moe Arai2
Moe Arai2 Kohei Kitami2
Kohei Kitami2 Daiki Kobayashi5
Daiki Kobayashi5 Yutaka Yoshida6
Yutaka Yoshida6 Takeyasu Maeda3
Takeyasu Maeda3 Isao Saito2
Isao Saito2 Katsumi Uoshima4
Katsumi Uoshima4 Makio Saeki1
Makio Saeki1The periodontal ligament (PDL) is a specialized connective tissue that provides structural support to the tooth and is crucial for oral functions. The mechanical properties of the PDL are mainly derived from the tissue-specific composition and structural characteristics of the extracellular matrix (ECM). The ECM also plays key roles in determining cell fate in the cellular microenvironment thus crucial in the PDL tissue homeostasis. In the present study, we determined the comprehensive ECM profile of mouse molar PDL using laser microdissection and mass spectrometry-based proteomic analysis with ECM-oriented data curation. Additionally, we evaluated changes in the ECM proteome under mechanical loading using a mouse orthodontic tooth movement (OTM) model and analyzed potential regulatory networks using a bioinformatics approach. Proteomic changes were evaluated in reference to the novel second harmonic generation (SHG)-based fiber characterization. Our ECM-oriented proteomics approach succeeded in illustrating the comprehensive ECM profile of the mouse molar PDL. We revealed the presence of type II collagen in PDL, possibly associated with the load-bearing function upon occlusal force. Mechanical loading induced unique architectural changes in collagen fibers along with dynamic compositional changes in the matrisome profile, particularly involving ECM glycoproteins and matrisome-associated proteins. We identified several unique matrisome proteins which responded to the different modes of mechanical loading in PDL. Notably, the proportion of type VI collagen significantly increased at the mesial side, contributing to collagen fibrogenesis. On the other hand, type XII collagen increased at the PDL-cementum boundary of the distal side. Furthermore, a multifaceted bioinformatics approach illustrated the potential molecular cues, including PDGF signaling, that maintain ECM homeostasis under mechanical loading. Our findings provide fundamental insights into the molecular network underlying ECM homeostasis in PDL, which is vital for clinical diagnosis and development of biomimetic tissue-regeneration strategies.
The extracellular matrix (ECM) is a principal tissue component that primarily provides a structural framework for tissues and plays key roles in determining cell fate in the cellular microenvironment (Frantz et al., 2010). Physical cues, such as externally applied deformation forces, biomaterial-induced forces, and cell-induced forces, evoke various cellular reactions that determine cell fate, with these broadly modified by the ECM (Kumar et al., 2017). Tight ECM-cell communication provides crucial biochemical and biomechanical cues that regulate cell behaviors; therefore, any physiological changes in the tissue can be traced back to the ECM (Iozzo and Gubbiotti, 2018). Hence, a dysfunctional ECM and its associated regulators affect tissue homeostasis, leading to a wide spectrum of pathological conditions, including skeletal disease, fibrosis, and cancer (Bonnans et al., 2014).
Proteomics involves the identification and quantification of the entire protein complement of a cell, tissue, or organism (Wilkins et al., 1996). Laser microdissection (LMD) combined with mass spectrometry-based proteomics such as liquid chromatography-tandem mass spectrometry (LC-MS/MS) enables the characterization of the protein composition of anatomically complex tissues in a sensitive and unbiased manner (Aebersold and Mann, 2016). Although the MS-based method offers the acquisition of comprehensive proteome, characterization of the ECM has been technically challenging due to the lack of basic knowledge of their protein constituents (Krasny and Huang, 2021). To overcome such difficulties, matrisome, a consensus catalog of proteins that are either part of or are associated with the ECM, was developed based on a domain-based in silico prediction algorithm (Naba et al., 2012). The matrisome comprised of two classes: the core matrisome which includes collagens, proteoglycans, and ECM glycoproteins; matrisome-associated proteins encompassing ECM-affiliated proteins, ECM regulators, and secreted factors (Naba et al., 2012; Naba et al., 2016).
The periodontal ligament (PDL) is a highly specialized fibrous connective tissue that anchors the tooth to the alveolar bone socket and is important for oral function, including the dissipation of masticatory force, neuronal feedback, and tooth eruption (Beertsen et al., 1997). The major ECM components of the PDL are type I and III collagens, with minor collagens (e.g., types V, VI, and XII), proteoglycans, and ECM glycoproteins (Butler et al., 1975; Beertsen et al., 1997; Nanci and Bosshardt, 2006). Mechanoregulation of the ECM in the PDL is of clinical interest because controlling PDL tissue homeostasis under mechanical loading is crucial in dental practice, such as in occlusal adjustment and managing orthodontic tooth movement (OTM). Moreover, the PDL is an adequate in vivo experimental model for studying ECM organization and remodeling under mechanical loading (Kaku and Yamauchi, 2014) because of its well-organized fiber alignment, rapid tissue-turnover rate (Sodek, 1977), good responsiveness, and ease of controlling the mode of mechanical loading (Nozaki et al., 2010; Kaku et al., 2016; Mizukoshi et al., 2021).
Although previous studies have attempted to identify the unique genes/proteins in the PDL (Nishida et al., 2007; Yamada et al., 2007) the global landscape of the ECM proteome in the PDL, even in the physiological state, remains unclear. Additionally, understanding changes in the ECM proteome under mechanical loading may accelerate its etiological consideration in various clinical conditions. Thus, studying the detailed ECM proteome of the PDL may enable a more precise diagnosis of the clinical state and its variations in individuals (Taha and Naba, 2019). Moreover, understanding the ECM proteome is indispensable for developing biomimetic tissue-regeneration protocols (Fernandes and Yang, 2016).
In this study, we analyzed the ECM proteome of the mouse molar PDL using LMD and LC-MS/MS with ECM-oriented data curation. Additionally, we evaluated changes in the ECM proteome under mechanical loading using a mouse OTM model and analyzed potential regulatory networks using bioinformatics approaches. Proteomic changes were evaluated in reference to the novel second harmonic generation (SHG)-based fiber characterization to better illustrate the homeostatic balance of the ECM network under different modes of mechanical loading.
The Niigata University Animal Experiment Ethics Committee reviewed and approved all animal procedures (SA00532). All animal handling and experiments strictly followed the ARRIVE guidelines for animal research reporting for in vivo experiments.
Eight-week-old male mice (C57BL/6J) were obtained from Charles River Laboratories Japan (Yokohama, Japan) and maintained in the animal facility of Niigata University. Mice were anesthetized with intraperitoneal injections of chloral hydrate (400 mg/kg body weight). Ni-Ti closed-coil springs (25 gf; TOMY International Inc., Tokyo, Japan) were inserted between the left maxillary 1st molars and incisors to induce mesial movement of the 1st molars (Mizukoshi et al., 2021). The ends of the Ni-Ti springs were tied to the molars and incisors with stainless steel wire (φ 0.09 mm) and secured with light-cure composite resin (BEAUTIFIL Flow; Shofu, Kyoto, Japan). At 14 days after coil application, the mice were sacrificed, and both sides of the maxilla were harvested and fixed with 4% w/v paraformaldehyde (PFA) for 24 h. The contralateral 1st molars served as the controls.
Micro-CT images of the maxilla were obtained with a CosmoScan GX high-resolution X-ray tomographic system (Rigaku Corporation, Tokyo, Japan) at 90 kV and 88-μA irradiation and structurally examined using Analyze software (v.12.0; AnalyzeDirect, Stilwell, KS, United States). The distance between the closest points of the 1st and 2nd molars was measured after the outline of the teeth was defined with a threshold intensity of between 6,000 and 12,000 Hounsfield units (n = 5) (Mizukoshi et al., 2021).
Upper 1st molars with surrounding tissue were dissected, decalcified with 10% (w/v) EDTA for 3 weeks at 4°C, and embedded in Tissue-Tek O.C.T. compound (Sakura Finetek, Tokyo, Japan) or paraffin according to a standard protocol (Kaku et al., 2016; Mizukoshi et al., 2021). Sagittal cryosections (10-µm thick) were prepared on membrane-coated slides (Leica Biosystems, Wetzlar, Germany), and 5-µm thick sagittal paraffin sections were prepared using a microtome (REM-710+MC-802C; Yamato Kohki, Saitama, Japan). Paraffin sections were stained with hematoxylin and eosin and a tartrate-resistant acid phosphatase/alkaline phosphatase (TRAP/ALP)-staining kit (FUJIFILM Wako Pure Chemical Corporation, Osaka, Japan) according to the manufacturer’s instructions.
Images were developed using a custom-built multiphoton microscope at Niigata University (Zeiss, Oberkochen, Germany). The second harmonic generation (SHG) signal was generated by a tunable Ti-sapphire laser with an excitation wavelength of 800 nm. Morphological analysis of the SHG images was performed using CT-FIRE software with default settings (Bredfeldt et al., 2014) to measure the individual collagen-fiber metrics of number, length, width, and angulation. The parameters were expressed in pixels.
Cryosections were stained with 0.05% (w/v) Toluidine Blue (Merck), and PDL samples from the mesial and distal sides of the distal root PDL were separately dissected using an LMD 7000 microdissection system (Leica, Wetzlar, Germany) with a cutting speed of 10 mm/s. The LMD settings were optimized for capture under a 10 × objective lens with a laser power of 20 mW. Approximately 20 cryosections were prepared for each mouse, and a total of eight mice (∼ 160 cryosections) were used to obtain enough protein for proteomic analysis. The proteomic experiment was repeated three times (total of 24 mice) for getting data from three biological replicates (n = 3). Samples were kept at −80°C for subsequent analysis.
Micro-dissected tissue samples were dispersed and equilibrated with 50 µL of SDS extraction buffer [2% (w/v) SDS, 50 mM Tris-HCl (pH 8.8), 2% (v/v) 2-mercaptoethanol, and 10% (v/v) glycerol] for 30 min, followed by incubation at 95°C for 20 min and 60°C for 2 h. The mixtures were centrifuged at 10,000 × g for 5 min to obtain the supernatants as protein extracts. The extracts were co-polymerized with acrylamide (Lu and Zhu, 2005) and in-gel digested with trypsin (T6567; Sigma-Aldrich, St. Louis, MO, United States) to generate dithiothreitol-reduced iodoacetamide-alkylated tryptic peptides (Katayama et al., 2001).
Trypsin-digested peptides were dissolved in 0.3% formic acid and filtered through a 0.45-µm Ultrafree-MC membrane filter (Merck-Millipore, Billerica, MA, United States). Peptide concentration of samples was estimated using a modified bicinchoninic acid assay (Kappor et al., 2009). Samples containing 0.13 µg peptide were analyzed in triplicate using direct-injection mode on an Eksigent NanoLC 415 nano-flow liquid chromatography system (Sciex, Framingham, MA, United States) using a 75 μm × 150 mm C18 spray-tip column (3 μm, 120 Å; Nikkyo Technos, Tokyo, Japan) coupled with a TripleTOF 5600 + tandem mass spectrometer (Sciex). Separation involved a 40-min gradient elution using 98% A, 2% B to 68% A, and 32% B at 300 nL/min (A and B refer to mobile phases comprising 0.1% aqueous formic acid and 0.1% formic acid in acetonitrile, respectively). The MS spectrum (250 ms), followed by 10 MS/MS spectra (100-ms each) were acquired in data-dependent mode. Auto-calibration using 50 fmol of bovine serum albumin tryptic digests (KYA Technology, Tokyo, Japan) was performed for every four to five samples.
Raw data generated using Analysis TF 1.6 software (Sciex) were converted to mascot generic files by MS Data Converter (Sciex) and searched against an in-house mouse reference protein sequence database (Swiss-Prot; downloaded on July 7, 2020) using a Mascot search engine (v.2.6; Matrix Science, Boston, MA, United States). Peptide and MS/MS tolerances were set at ± 20 ppm and ± 0.1 Da, respectively. A maximum of two missed cleavages was allowed. Search settings involved cysteine carbamidomethylation as a fixed modification and the following variable modifications: deamidation of asparagine and/or glutamine, N-terminal glutamine to pyroglutamate, oxidation of methionine, oxidation of proline, and oxidation of lysine. The target false discovery rate (FDR) was set to < 1%. Proteins with more than three spectral matches were considered as identified proteins.
Matrisome proteins were selected from all identified proteins using the Matrisome Annotator (http://matrisomeproject.mit.edu/analytical-tools/matrisome-annotator/) (Naba et al., 2016). The occupancy of each matrisome protein in controls was calculated based on the number of spectral counts (Lundgren et al., 2010), and the relative abundance of each protein was determined using the normalized spectral abundance factor (NSAF) (Zybailov et al., 2006). Statistical significance between the control and experimental groups was calculated using the Benjamini–Hochberg procedure (Benjamini and Hochberg, 1995) with log (fold change) > |0.2| and log (false discovery rate) > 2 as the threshold. The statistically significant proteins and proteins exclusively detected either in control or at day 14 were considered differentially expressed proteins (DEPs). Pathway- and process-enrichment analyses were performed using Metascape (Zhou et al., 2019) based on the following ontology sources: KEGG Pathway, GO Biological Processes, GO Cellular Components, GO Molecular Functions, Reactome Gene Sets, CORUM (comprehensive resource of mammalian protein complexes) WikiPathways, and Protein Analysis Through Evolutionary Relationships (PANTHER) Pathway. Terms with a p-value < 0.01, a minimum count of three, and an enrichment factor > 1.5 were grouped into clusters based on their similarities, with the most statistically significant term representing the cluster. PPI enrichment analysis was performed using STRING v11.5 (Szklarczyk et al., 2019). Local STRING network clusters were identified by hierarchical clustering of the full STRING network using an average linkage algorithm.
Histological sections were deparaffinized in xylene and rehydrated with ethanol. Endogenous peroxidase activity was quenched with 3% hydrogen peroxide, blocked with 5% sheep or goat serum, and incubated with the primary antibody at 4°C overnight. Samples were washed three times with PBS-T and treated with an avidin-biotin complex kit (Vector Laboratories, Burlingame, CA, United States) according to manufacturer instructions, followed by visualization by incubation with 3,3′-diaminobenzidine tetrahydrochloride (DAB) (Vector Laboratories) and counter-staining with hematoxylin. The following primary antibodies were used: rabbit anti-collagen type II (1:100; 15943-1-AP), rabbit anti-collagen type III (1:200; 22734-1-AP), and rabbit anti-collagen type VI (1:100; 17023-1-AP) from ProteinTech (Manchester, United Kingdom); and rabbit anti-collagen type XII alpha 1 chain (1:100; HPA009143) from Merck (Darmstadt, Germany). Normal rabbit serum was used as a negative control. Histological sections were imaged using a BX53 microscope (Olympus, Tokyo, Japan) and processed using Fiji/ImageJ (NIH, Bethesda, MD, United States) and cellSens (Olympus) software.
Picrosirius Red staining was performed, as previously described (Kaku et al., 2016). Histological sections were deparaffinized in xylene, rehydrated, and incubated with Picrosirius Red staining solution [1% (w/v) Sirius Red S in saturated aqueous picric acid] for 30 min, washed thoroughly with deionized water, dehydrated, and analyzed with a BX53 light microscope (Olympus) equipped with polarized filters.
Results are expressed as the means ± SD from three or more independent experiments. Statistical analysis for differences between the groups was performed using a two-tailed unpaired t-test with Welch’s correction using Prism 9 (GraphPad Software, San Diego, CA, United States) and a p value < 0.05 was considered significant.
To apply static mechanical loading to the PDL, we placed a coil spring between the upper left 1st molar and the upper incisors (Figure 1A). The mesial and distal sides of the PDL of the 1st molar distal root were selected as the regions of interest. Micro-CT images and H&E staining showed the widening of the PDL space of the 1st molar on day 14 (Figures 1B,C). An increased distance between the proximal surfaces of the 1st and 2nd molar confirmed mesial drift of the 1st molar on day 14 (Figure 1B). Additionally, the number of TRAP-positive cells increased on the mesial side, whereas the ALP activity increased on the distal side on day 14 (Figure 1D), with high TRAP and ALP activities in the PDL, indicating persistent tooth movement.
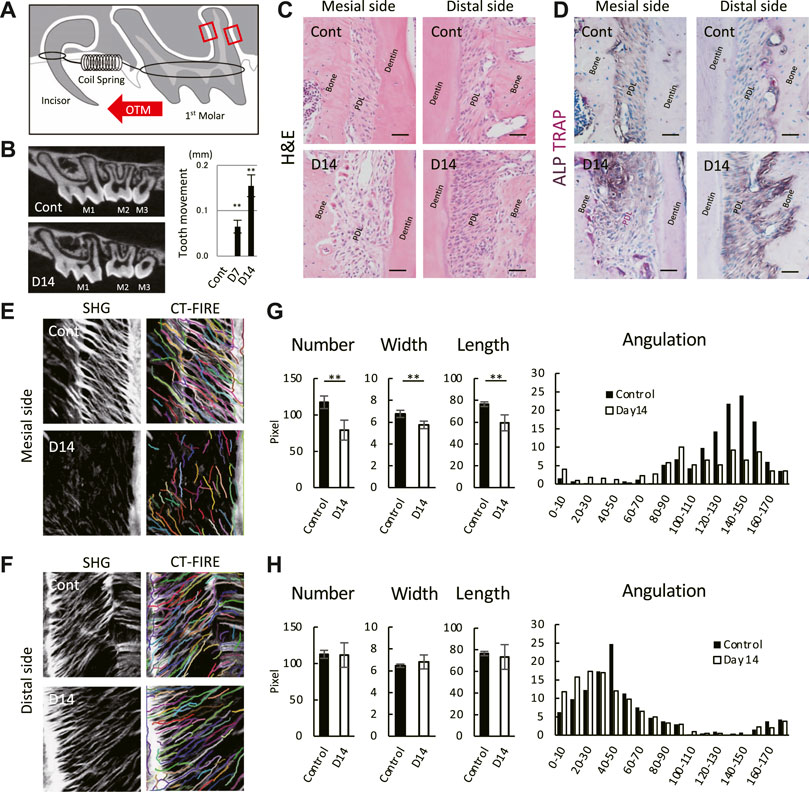
FIGURE 1. Mouse molar orthodontic tooth movement (OTM) and macroscale characteristics of collagenous fibers in the PDL. (A) Schematic of the OTM. Region of interests are indicated by red rectangles. (B) Representative micro-CT images of controls (Cont) and at day 14 (D14) of the OTM. The distance between the proximal surfaces of the 1st and 2nd molar. **p < 0.01 (n = 8, mean ± SD) (C,D) Staining at the mesial and distal sides of the PDL by hematoxylin and eosin (H,E) (n = 3) (C) and TRAP/ALP (n = 3) (D). Scale bar: 50 μm (E,F) SHG microscopy image and collagenous fibers at the mesial (E) and distal (F) sides of the PDL. Individual fibers were detected using CT-FIRE. Quantification of detected fibers at the mesial (G) and distal (H) sides of the PDL. **p < 0.01 (n = 3, mean ± SD).
ECM organization at the macroscale showed individual collagenous fibers detected using SHG imaging and an automated tracking algorithm by CT-FIRE (Figures 1E,F). Controls showed well-aligned fibers on the mesial and distal sides of the PDL; however, the SHG signal for the mesial side of the PDL was difficult to detect on day 14 (Figure 1E bottom), and the fiber number, width, and length were significantly decreased (Figure 1G). The tendency for fiber angulation with respect to the cementum surface on the controls was not observed at day 14 on the mesial side. In contrast, the fiber number, width, length, and angulation remained unchanged at day 14 on the distal side (Figure 1H). These results indicate that the OTM used in this study induced destructive changes in macroscale fiber organization on the mesial side, whereas these changes were within the homeostatic balance on the distal side of the PDL.
Next, we performed a comprehensive view of the ECM proteome in the mouse molar PDL. The mesial and distal sides of the PDL were precisely dissected from the surrounding tissue to obtain PDL tissue-specific proteome (Supplementary Figure S1). On the mesial side, the number of identified proteins increased from 443 to 691 on day 14 (Figure 2A). Among the 726 identified proteins on the mesial side, 84 were matrisome proteins. On the distal side, the number of identified proteins increased from 518 to 714 on day 14 (Figure 2B), 83 of which were matrisome proteins. Additional unique proteins were detected at day 14 compared to the control on both mesial and distal sides. In controls, matrisome proteins accounted for ∼30% of the total spectral count (Figure 2C). Among matrisome proteins, collagens were the major sub-class, occupying ∼20% of the total spectral counts. The proportion of matrisome proteins decreased on day 14 at mesial and distal sides. The number of detected ECM regulators and ECM-affiliated proteins was increased, whereas that of collagens and proteoglycans was decreased on day 14 (Figure 2D). A complete list of core matrisome proteins in the controls comprised 13 collagen types with 16 chains, 9 proteoglycans, and 20 ECM glycoproteins (Figure 3A). Type I collagen (COL1A1 and COL1A2) was the most abundant matrisome protein in the PDL. accounting for 86.5% of the spectral count among collagens (Figure 3B). Type III and XII collagen (COL3A1 and COL12A1) each accounted for 4.5% of collagens, representing the 2nd and 3rd most abundant collagens in mouse molar PDL. Unexpectedly, type II collagen (COL2A1) was detected to account for 0.8% of collagens. Additionally, type V, VI, VII, VIII, XI, XIV, XVI, XX, and XXVIII collagens were identified (Figure 3A). Our proteome data revealed that lumican (LUM), asporin (ASPN), biglycan (BGN), and matrix gla protein (MGP) were the major proteoglycans, and periostin (POSTN) and tenascin-N (TNN) were the dominant ECM glycoproteins in the mouse molar PDL (Figure 3C).
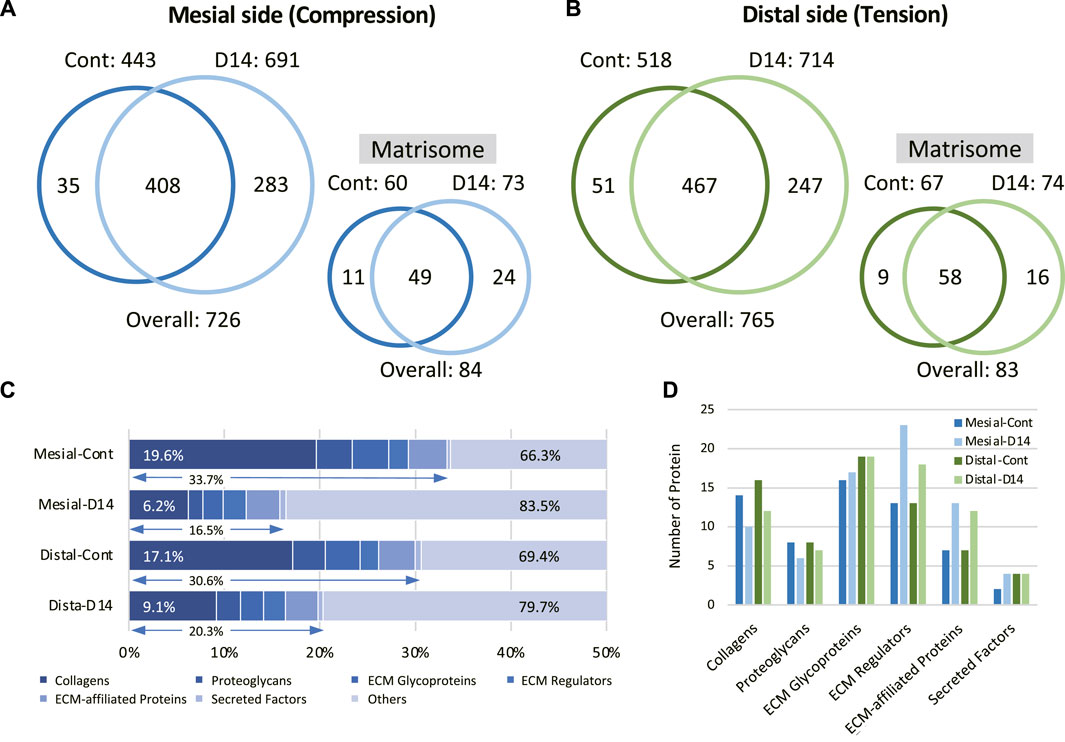
FIGURE 2. Identification and quantification of the mouse molar PDL matrisome proteins. Venn diagram of entire and matrisome proteins detected in control (Cont) and at day 14 (D14) of OTM on the mesial (A) and distal (B) sides of the PDL (n = 3) (C) Matrisome profile of the mouse molar PDL in each condition. Proportions of matrisome and collagens in reference to the entire proteome are shown. (D) Number of identified proteins in each matrisome subclass for each condition.
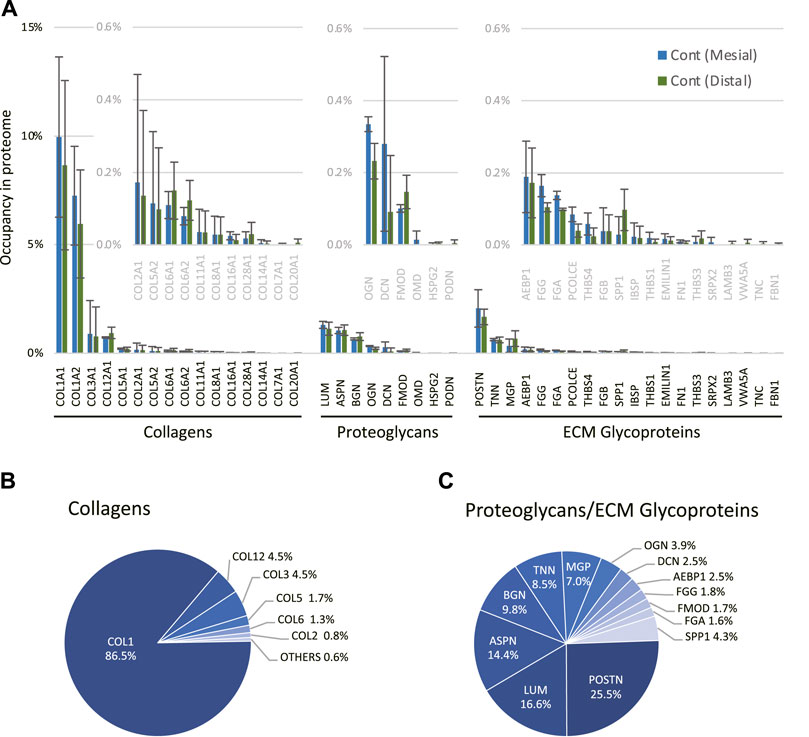
FIGURE 3. Profile of core matrisome proteins in the mouse molar PDL. (A) Profile of core matrisome (collagens, proteoglycans, and ECM glycoproteins) in the mouse molar PDL in controls. Insets are magnified graphs of the corresponding regions (n = 3, mean ± SD) (B) Profile of collagens in the mouse molar PDL in controls. (C) Profile of proteoglycans and ECM glycoproteins in the mouse molar PDL in controls.
We then analyzed changes in the matrisome profile under mechanical loading at the mesial and distal sides. The proportion of collagens and proteoglycans in the mesial side of the PDL decreased on day 14, whereas that of matrisome-associated proteins increased (Figure 4A). A complete list of matrisome proteins on the mesial side showed that, on day 14, many ECM glycoproteins and ECM-associated proteins were detected that were not detected in controls (Supplementary Figure S2). The matrisome profile of the distal side of the PDL showed that the proportion of collagens decreased, while that of ECM regulators increased on day 14 (Figure 4B). A complete list of matrisome proteins on the distal side also showed that many ECM glycoproteins and ECM-associated proteins were detected on day 14 but were not found in controls (Supplementary Figure S3). We observed minor changes in the matrisome profile on the distal side compared with that of the mesial side of the PDL, which is consistent with the macroscale fiber characterization (Figures 1E–H).
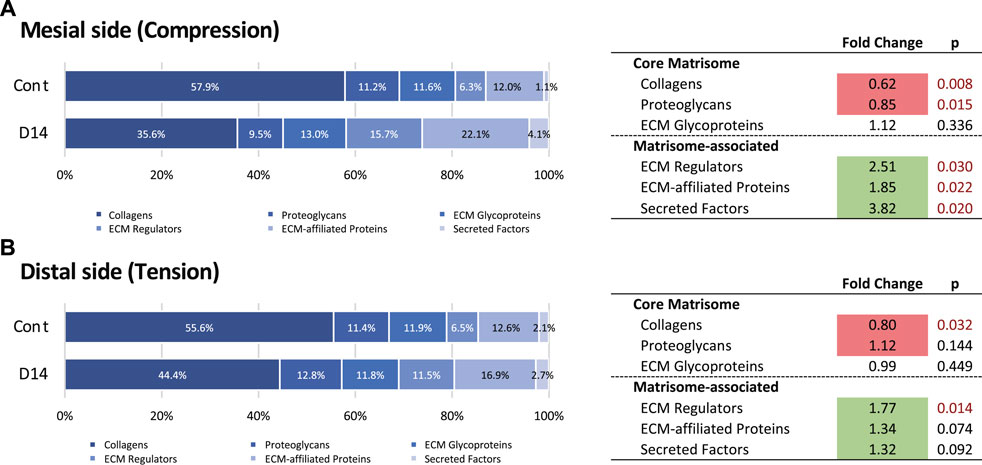
FIGURE 4. Changes in the matrisome profile at the mesial and distal sides of the PDL. Proportions of matrisome proteins and fold change of matrisome classes during OTM at mesial (A) and distal (B) sides of the PDL. Matrisome classes that significantly increased and decreased in proportion on day 14 (D14) compared with the control (Cont) are highlighted in green and red, respectively.
We next analyzed changes in individual matrisome proteins on day 14 and presented these using volcano plots (Figures 5A,B). On the mesial side, 53 DEPs (32 increased and 21 decreased in proportion) were identified (Figure 4A). The proportion of type VI collagen (COL6A1 and COL6A2) increased, but that of other collagens tended to decrease. The increased proportion of proteases (CTSK, CTSZ, and CTSD) and a matrix metalloproteinase (MMP13) indicated matrix degradation and bone resorption at the mesial side of the PDL. Proteins with increased proportion also included S100 proteins (A100A4, S100A9) and annexins (ANXA4, ANXA7, ANXA8, ANXA11). Proteins with decreased proportion include proteoglycans (ASPN, LUM, OGN, FMOD, OMD) and an ECM glycoprotein (TNN).
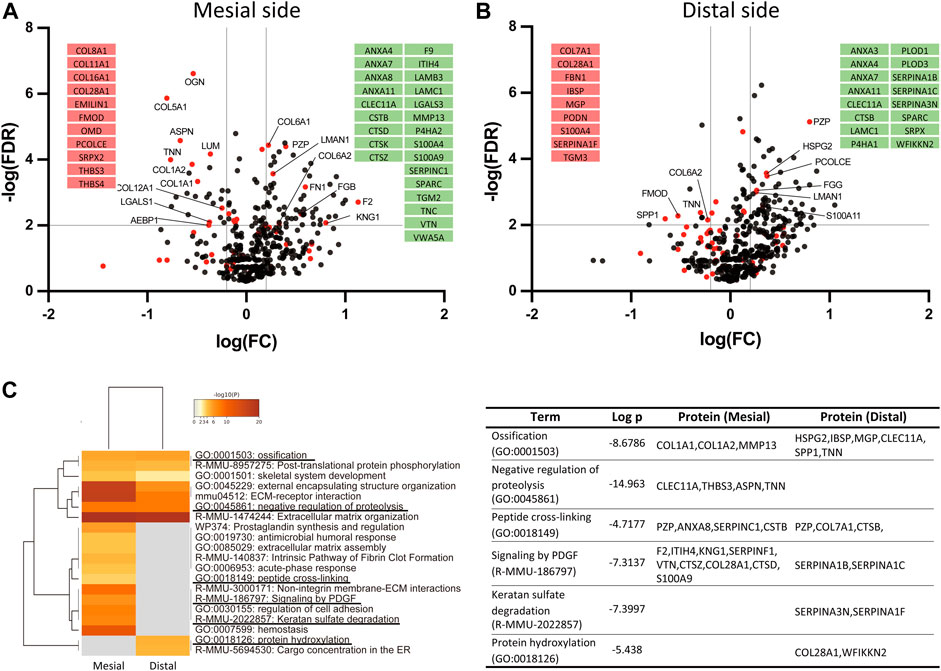
FIGURE 5. Identification of ECM-oriented differentially expressed proteins (DEPs) and enrichment analysis. Volcano plot of the DEPs at the mesial (A) and distal (B) sides of the PDL. Matrisome proteins are shown in red dots and other proteins are shown in black dots. The matrisome proteins exclusively detected only in control (red) or at day 14 (green) are shown in insets. (C) Pathway- and process-enrichment analyses of ECM-oriented DEPs in the mesial and distal sides of the PDL.
On the distal side, 35 DEPs (22 increased and 13 decreased in proportion) were identified (Figure 4B). The proportion of proteins associated with mineralization, such as matrix gla protein (MGP), osteopontin (SPP1), and bone sialoprotein (IBSP) was decreased. Notably, the proportion of collagen-modifying enzymes, including prolyl 4-hydroxylase (P4HA1) and lysyl hydroxylases (PLOD1 and PLOD3) was increased at the distal side of the PDL.
Among matrisome-oriented DEPs in the PDL, the expression of 9 proteins (ANXA11, ANXA4, ANXA7, CLEC11A, FGG, LAMC1, LMAN1, PZP, and SPARC) was increased and that of 2 proteins (FMOD and TNN) was decreased at both mesial and distal sides of the PDL.
To better understand the molecular network regulating mechanical loading-induced ECM alterations in the PDL, we performed a matrisome-oriented protein-enrichment analysis (Figure 5C). Pathway-enrichment and process-enrichment analyses revealed that ossification (GO:0001503) and negative regulation of proteolysis (GO:0045861) were enriched at both mesial and distal sides of the PDL. PDGF signaling (R-MMU-186797) and keratan sulfate degradation (R-MMU-2022857) were enriched at the mesial side, and protein hydroxylation (GO:0018126) was enriched at the distal side.
We further performed protein-protein interaction (PPI) enrichment analysis of matrisome-oriented DEPs using the STRING database (Supplementary Figure S4). The selected PPI networks contributing to “ECM organization (R-MMU-1474244)” at the mesial side of the PDL showed that type I collagen (COL1A1 and COL1A2) was present at the center of the network and in close association with type VI collagen (COL6A1 and COL6A2) (Supplementary Figure S4A). Fibronectin1 (FN1) and SPARC were present at the interface of the “ECM organization” and “ECM-receptor interaction (R-MMU04512)” networks, likely playing a role as hub proteins. The selected PPI networks on the distal side of the PDL showed no strong relations at the network interface (Supplementary Figure S4B).
Enrichment analysis was also performed for all identified proteins, using 431 DEPs of 726 proteins on the mesial side and 407 DEPs of 765 proteins on the distal side as inputs. The analytical method applied was identical to that used in the matrisome-oriented enrichment analysis. Although ECM-associated gene ontology terms were enriched at both the mesial and distal sides of the PDL, most enriched terms were associated with general cell metabolic processes (Supplementary Figure S5). PPI enrichment analysis using all DEPs showed no indicative results, although a matrisome protein-enriched cluster was clearly detected at the mesial and distal sides of the PDL (Supplementary Figures S6, S7).
Our proteomics data revealed a considerable amount of type II collagen in the PDL, which has not attracted attention so far. Our immunohistochemistry (IHC) data clearly showed that type II collagen was distributed throughout the PDL with intense signals at the mineralized-tissue surface, particularly at the bone surface of the furcation area and cementum surface of the apical area (Figure 6A). There were no notable changes in the staining intensity or distribution pattern of type II collagen on day 14, which is consistent with the results of the proteomic analysis (Supplementary Figures S2, S3).
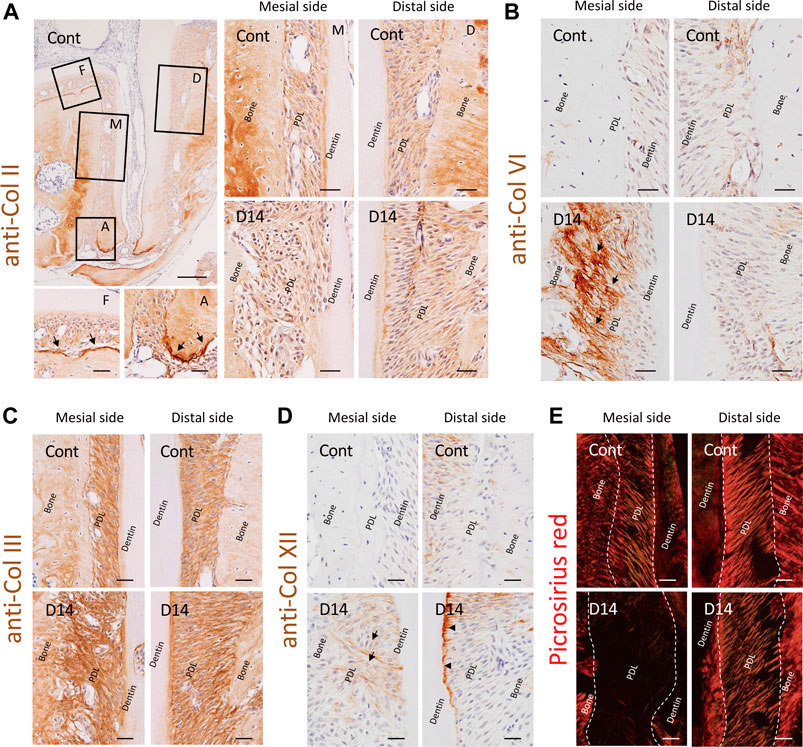
FIGURE 6. Effect of mechanical loading on collagen distribution in the mouse molar PDL. (A) Distribution of type II collagen in the PDL. Type II collagen was distributed throughout the PDL space, and intense staining was observed at the mineralized tissue surface, particularly at the bone surface of the furcation area and cementum surface of the apical area (arrows). F, furcation area; A, apical area; M, mesial side; D, distal side. (B) Distribution of type VI collagen in the PDL. Staining intensity of type VI collagen markedly increased along with that of the collagen fibers at the mesial side of the PDL, specifically at the bone side (arrows). (C) Distribution of type III collagen in the PDL. (D) Distribution of type XII collagen in the PDL. Staining intensity of type XII collagen was increased at the newly formed fibers on the mesial side of the PDL (arrows) and at the cementum surface on the distal side of the PDL (arrowheads) on day 14. (E) Polarized observation of picrosirius red staining, detecting matured collagen fibers. Scale bars: 200 μm at low magnification (a, top left) and 50 μm at high magnification (all other images).
In contrast, the staining intensity of type VI collagen was markedly increased at the mesial side of the PDL on day 14, whereas there was no noticeable change on the distal side of the PDL (Figure 6B). This correlated with the observed increase in abundance of type VI collagen at the mesial side of the PDL in response to mechanical loading according to proteomic analysis results (Figure 4A).
We also analyzed the 2nd and 3rd most abundant collagens, type III and XII collagens, using IHC, and analyzed type I collagen with picrosirius red staining. Type III collagen was distributed ubiquitously in the PDL space along without a noticeable change in the staining intensity on day 14 (Figure 6C). Type XII collagen was more abundant on the distal side of the PDL than on the mesial side of the PDL in controls (Figure 3A, Figure 6D), whereas the staining intensity increased on the newly formed fibers at the mesial side of the PDL and on the cementum surface at the distal side of the PDL on day 14. Picrosirius red staining showed trace signals at the mesial side of the PDL on day 14, indicative of the absence of matured type I collagen (Fig. 7e), despite the abundance of type III, VI, and XII collagens detected using IHC.
ECM is a principal component of tissues involving various functions; however, the composition of ECM in tissues is still largely unknown. One reason for this is that proteomic analysis in structurally complex tissue, such as the PDL, has been technically challenging (Aebersold and Mann, 2016). In the present study, we performed LMD and LC-MS/MS to comprehensively analyze the PDL tissue-specific proteome. Furthermore, matrisome-oriented data curation enabled the acquisition of a global ECM profile in the mouse molar PDL, to the best of our knowledge, for the first time. The detailed ECM profile of the PDL observed in this study provides fundamental insights that would be useful in understanding the role of the PDL tissue in health and diseases. Additionally, if the ECM composition of the PDL can be mimicked, it is possible to regulate cell behavior to enable tissue regeneration in vivo (Hussey et al., 2018). In fact, the ECM of the cultured PDL-derived cells was reported to produce PDL stem cell-like cells from iPS cells (Hamano et al., 2018). Therefore, the ECM profile of the PDL obtained in this study provides a foundation for successful biomimetic tissue regeneration.
Our proteome analysis revealed that the ECM of the PDL primarily comprises collagens; major collagen types are type I (86.5%), type III (4.5%), type XII (4.5%), and type V (1.7%) based on the spectral count (Figure 3B). Although we successfully illustrated the global ECM proteome of the PDL, there is a discrepancy in collagen proportion among previous reports, which indicate that type I, III, and V collagens occupied 75, 20, and 5% of the collagen in PDL, respectively (Butler et al., 1975; Yamauchi et al., 1986). This discrepancy is possibly due to the difference in the protein extraction method and identification strategies. The ECM proteins, especially collagens, are heavily cross-linked and have large molecular weight, which results in the insolubility during extraction (Byron et al., 2013). The earlier studies used acid extraction combined with chemical and enzymatic digestion and protein characterized by liquid chromatography (Butler et al., 1975; Yamauchi et al., 1986), while we employed the SDS extraction method with in-gel trypsin digestion for LC-MS/MS. Our method covered both core matrisome and matrisome-associated proteins, but the collagen profile of the tissue could be under-represented because of the incomplete extraction of heavily cross-linked proteins. Besides, matrisome analysis showed that the proportion of collagen decreased significantly (0.8-fold, p<0.05) at the distal side (Figure 4B), whereas characterization of the macroscale fiber did not indicate an alteration in this region (Figure 1H). One possible explanation is that the mechanical stress accelerates the collagen crosslinking in the PDL (Kaku et al., 2016) resulting in the decreased solubility of collagen (Taha and Naba, 2019). This may lead to the decreased proportion of collagen in the analytical sample from the distal side of PDL. To overcome technical limitations attributable to the insolubility of the ECM, several ECM-enrichment protocols including decellularization, deglycosylation, and chemical digestion have been developed (de Castro Bras et al., 2013; McCabe et al., 2021). Although these methods were shown to effectively enrich the core matrisome proteins, they also present a significant loss in the number of detected matrisome-associated proteins (Krasny and Huang, 2021). Therefore, an appropriate protein extraction protocol needs to be established on the basis of the biochemical nature of the target proteins. Further studies are needed to acquire a quantitatively accurate ECM profile of the PDL.
One of the key findings of this study is the presence of type II collagen in the PDL, which is in contrast to the results of previous studies citing this protein as a predominant collagen type in the cartilage (Ricard-Blum, 2011). Recent proteomic studies have identified type II collagens in mouse PDL (Connizzo et al., 2021) and rat PDL (Denes et al., 2020); however, these observations have not received attention. The increased type II collagen expression reported in cultured PDL-derived cells under cyclic tension force (Wescott et al., 2007) and hypoxic conditions (Li Q. et al., 2019) has been considered a result of their multipotent nature and chondrogenic differentiation potential (Seo et al., 2004). The distribution pattern of type II collagen, analyzed using IHC, indicated an association with bone/cementum apposition, similar to the fibrocartilage at enthesis, where type II collagen is present (Benjamin and Ralphs, 1998). Owing to the abundance of glycosylated hydroxylysine residues, type II collagen has larger molecular spacing than type I collagen (Grynpas et al., 1980), enabling it to hold more water and contributing to the dissipation of compressive forces (Bielajew et al., 2020). Collectively, type II collagen may play important roles in maintaining the tissue integrity of the PDL, and thus, further detailed investigation is required.
Our proteomic data showed that the proportion of type VI collagen increased at the mesial side of the PDL, but that of other collagens tended to decrease. PPI network analysis indicated a close association of type VI collagen with collagen fibrogenesis, particularly posttranslational modifications. Reportedly, type VI collagen interacts with the surrounding ECM and organizes the collagen fibril assembly and three-dimensional tissue architecture (Cescon et al., 2015). In periodontal tissue, type VI collagen localizes to the pericellular region surrounding fibroblasts and establishes extracellular microdomains that are associated with microfibrils and oxytalan fibers (Everts et al., 1998). Although details are still unclear, our results indicate that type VI collagen plays a crucial role in ECM alteration under mechanical loading in the PDL.
We noted the increased staining intensity of type XII collagen on the distal side of the PDL by IHC (Figure 6D); however, this was not detected in the proteome datasets (Supplementary Figure S3). The increased production of type XII collagen in response to the mechanical loading was reported in PDL tissue (Karimbux and Nishimura, 1995; Tsuzuki et al., 2016) and PDL-derived cells (Nemoto et al., 2010). In line with our observation, the fibril-associated type XII collagen has been shown to regulate collagen fibrillogenesis (Chiquet et al., 2014). Therefore, the increased staining intensity of type XII collagen at the cementum surface on the distal side of the PDL suggests enhanced fibril stabilization in this region. On the other hand, the discrepancy between our proteomic and IHC data on type XII collagen may be explained by the destruction of the tissue boundary during LMD, because the changes in the production of type XII collagen at the distal side of the PDL specifically occurred at the PDL-cementum boundary. Although the tissue-level proteome is a powerful tool to illustrate the tissue-specific ECM proteome, attention is required when the region-specific proteomic changes are of particular interest.
Our proteome analysis identified 2 calcium-binding protein families, S100 proteins and annexins; these proteins interact with each other and form complexes that exhibit biological activities (Miwa et al., 2008). S100A4 is highly expressed in PDL (Duarte et al., 1998) and is responsive to mechanical stress (Duarte et al., 1999). Furthermore, S100A4 downregulates osteogenic gene expression and upregulates matrix degradation-related gene expression in PDL cells (Zhou et al., 2015) as well as enhances osteoclast formation (Mah et al., 2015). We found that the proportion of S100A4 was increased on the mesial side, where the osteoclast recruitment was evident. In addition, annexin a2, an S100A4-binding partner, was also detected in PDL cells in high abundance (Xiong et al., 2016). Further, many S100 proteins (A100A4, S100A9) and annexins (ANXA4, ANXA7, ANXA8, ANXA11) were increased on the mesial side, and three annexins (ANXA4, ANXA7, ANXA11) were increased at both mesial and distal sides of the PDL. Although further investigations are needed, the S100-annexin complex may have reciprocal roles in PDL tissue maintenance under mechanical loading.
We observed increased production of collagen-modifying enzymes in response to mechanical loading, including that of prolyl hydroxylases (P4HA1/2), lysyl hydroxylases (PLOD1/3), and transglutaminases (TGM2/3). Type I collagen in the mouse molar PDL undergo distinct posttranslational modifications that significantly affect the formation of intermolecular cross-links and determine the structural and mechanical properties of the tissue (Hudson et al., 2017). We previously reported that the occlusal loading-induced mechanical stress enhances ECM maturation through the posttranslational modifications of collagen (Kaku et al., 2016). Our current data further support the notion that the posttranslational modifications of collagen play important roles in ECM organization in a mechanically challenging environment in the PDL.
This study reports putative mechanoresponsive proteins in PDL, whose roles regarding the ECM and/or PDL have not received attention. One example is the pregnancy zone protein (PZP), which is highly expressed in late-pregnancy serum and inhibits the activity of all four classes of proteinases (Wyatt et al., 2016). A recent study proposed that the PZP inhibits the aggregation of misfolded proteins that helps to maintain protein homeostasis during pregnancy (Cater et al., 2019). Our proteome data showed that the PZP increased 2.53-fold (p < 0.01) at the mesial side of the PDL and 6.22-fold (p < 0.01) at the distal side of the PDL. Other DEPs found on both sides of the PDL may also have undiscovered functions that involve mechano-transduction and ECM organization.
Although we identified numerous proteins via proteomic analysis, the number of detected proteins differed between the mesial and distal sides of the PDL in controls. Additionally, spectral counting values presented large standard deviations among biological replicates. These differences are likely because of the heterogeneity of the PDL tissue, given that the cell characteristics (Takimoto et al., 2015), fibrous network structure (Naveh et al., 2018), ECM composition (Denes et al., 2020; Connizzo et al., 2021), microvascular distribution (Sims et al., 1996), and response to mechanical stress (Takimoto et al., 2015; Kaku et al., 2016) differ between the respective regions of the PDL. The IHC data similarly showed that each protein had a distinct distribution pattern within the PDL. Collectively, it is necessary to consider the biomechanical and biochemical non-uniformity of the PDL (Connizzo et al., 2021). Indeed, non-uniformity of the PDL tissue is a critical factor that makes the clinical control and regeneration of this tissue extremely difficult.
In the ideal OTM theory, when the tooth is moved in the mesial direction, symmetric compression and tension forces are present at the mesial and distal sides of the PDL, respectively. However, the mesial movement of the tooth generates the compression force at the cervical region of the mesial and apical PDL, whereas the tension force is applied at the coronal two-thirds of the distal PDL (Li Z. et al., 2019). Additionally, the intrusive and/or tilting movement also occurs throughout the OTM process; therefore, the force is not completely uniform. This is one of the possible explanations for the overlapping of some of the DEPs, both at the mesial and distal sides of the PDL, while these may be involved simultaneously, maintaining PDL homeostasis in both compression and tension regions. As mentioned above, region-specific analysis of the ECM proteome and targeted analysis of specific proteins should be considered in future studies.
To enable the LMD of mineralized tissue-containing samples, we used PFA fixation for 24 h, followed by decalcification with 10% EDTA for 3 weeks at 4°C. Although fixation may affect the protein extraction efficiency, in theory, previous reports have shown that the number of identified proteins from formalin-fixed samples in the bottom-up proteomics is in concurrence with that obtained with fresh frozen tissue samples (Thompson et al., 2013). The decalcification may result in the loss of the mineral binding proteins (Chun et al., 2011; Salmon et al., 2017); however, this process is necessary for mineralized tissues, that is, enamel, dentin, cementum, and bone, and therefore, the effect on PDL is minimal. Special attention is required when interpreting the proteome data of the mineralized tissue using decalcified samples. Furthermore, we analyzed potential molecular networks involving ECM organization in the PDL using bioinformatics approaches. The use of ECM-oriented DEPs resulted in better resolution of the enriched terms, which included secretion, posttranslational modifications, and organization of the ECM; using the entire PDL proteome as an input resulted in enriched terms mainly related to general cellular metabolic processes. The process and pathway enrichment analysis showed that ossification (GO:0001503) and negative regulation of proteolysis (GO:0045861) were enriched at mesial and distal sides of the PDL. It has been well accepted that the cells in PDL maintain the adjacent alveolar bone by resorption and apposition of the bone matrix (Jiang et al., 2016; Moon et al., 2019; Lee et al., 2021). The enrichment of the “negative regulation of proteolysis” indicated that antagonizing mechanisms against ECM degradation were enhanced in response to the mechanical loading. We also showed that PDGF signaling (R-MMU-186797) involved ECM alteration on the mesial side of the PDL. PDGF is a widely studied growth factor (Giannobile, 1996) based on its abundance in platelet-rich plasma, demonstrating its capacity for periodontal-tissue regeneration (Fernandes and Yang, 2016). Moreover, PDGF stimulates proliferation, chemotaxis, and ECM production of human PDL-derived cells (Ojima et al., 2003) and acts synergistically with other growth factors to modulate the wound-healing process (Lynch et al., 1987). Our data suggest that the PDGF could be a potential target to achieve effective tissue regeneration of the PDL.
Our ECM-oriented proteomics approach succeeded in illustrating the comprehensive ECM profile of the mouse molar PDL and its compositional changes following mechanical loading. Mechanical loading induced unique architectural changes in collagen fibers along with dynamic compositional changes in the matrisome profile, particularly involving ECM glycoproteins and matrisome-associated proteins. We identified several unique matrisome proteins that responded to different modes of mechanical loading in the PDL. Furthermore, the bioinformatics approach revealed potential molecular cues that maintain ECM homeostasis in a mechanically challenging environment. As a future prospect, the specific DEPs identified in this study need to be characterized extensively. Furthermore, the human PDL proteome needs to be clarified for the translation of the scientific findings to clinical practice. Our findings provide fundamental insights into the ECM network in the PDL, which are vital for precise clinical diagnosis and development of biomimetic tissue-regeneration strategies.
The data that support the findings of this study are available within the article/Supplementary Material. The mass spectrometry proteomics data have been deposited to the jPOST (Japan ProteOme STandard Repository/Database, https://jpostdb.org) with the data set identifier JPST001418/PXD030430.
The animal study was reviewed and approved by The Niigata University Animal Experiment Ethics Committee.
LT contributed to data acquisition, analysis, and interpretation, drafted, and critically revised the manuscript; Ma K contributed to conception, design, data analysis, and interpretation, drafted, and critically revised the manuscript; YK contributed to conception, design, data acquisition, analysis, and interpretation, and critically revised the manuscript; MM, Me K, MA, KK, DK, and YY contributed to data acquisition, analysis, and interpretation, and critically revised the manuscript; TM, IS, KU, and MS. contributed to data analysis and interpretation, and critically revised the manuscript. All authors gave final approval and agreed to be accountable for all aspects of the work.
This work was supported by JSPS KAKENHI (grant nos. JP18H02989, JP21H03127, and JP21K19895 to Ma K).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
This work was supported by research equipment shared in MEXT Project for promoting public utilization of advanced research infrastructure (Program for supporting introduction of the new sharing system) at Niigata University.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2022.899699/full#supplementary-material
Aebersold R., Mann M. (2016). Mass-Spectrometric Exploration of Proteome Structure and Function. Nature 537, 347–355. doi:10.1038/nature19949
Beertsen W., McCulloch C. A. G., Sodek J. (1997). The Periodontal Ligament: A Unique, Multifunctional Connective Tissue. Periodontol. 13, 20–40. doi:10.1111/j.1600-0757.1997.tb00094.x
Benjamin M., Ralphs J. R. (1998). Fibrocartilage in Tendons and Ligaments-Aan Adaptation to Compressive Load. J. Anat. 193 ( Pt 4) (Pt 4), 481–494. doi:10.1046/j.1469-7580.1998.19340481.x
Benjamini Y., Hochberg Y. (1995). Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B (Methodological) 57, 289–300. doi:10.1111/j.2517-6161.1995.tb02031.x
Bielajew B. J., Hu J. C., Athanasiou K. A. (2020). Collagen: Quantification, Biomechanics and Role of Minor Subtypes in Cartilage. Nat. Rev. Mater. 5, 730–747. doi:10.1038/s41578-020-0213-1
Bonnans C., Chou J., Werb Z. (2014). Remodelling the Extracellular Matrix in Development and Disease. Nat. Rev. Mol. Cel. Biol. 15, 786–801. doi:10.1038/nrm3904
Bredfeldt J. S., Liu Y., Pehlke C. A., Conklin M. W., Szulczewski J. M., Inman D. R., et al. (2014). Computational Segmentation of Collagen Fibers from Second-Harmonic Generation Images of Breast Cancer. J. Biomed. Opt. 19, 16007. doi:10.1117/1.JBO.19.1.016007
Butler W., Birkedal-Hansen H., Beegle W., Taylor R., Chung E. (1975). Proteins of the Periodontium. Identification of Collagens with the [alpha1(i)]2alpha2 and [alpha1(iii)]3 Structures in Bovine Periodontal Ligament. J. Biol. Chem. 250, 8907–8912. doi:10.1016/s0021-9258(19)40671-6
Byron A., Humphries J. D., Humphries M. J. (2013). Defining the Extracellular Matrix Using Proteomics. Int. J. Exp. Path. 94, 75–92. doi:10.1111/iep.12011
Cater J. H., Kumita J. R., Zeineddine Abdallah R., Zhao G., Bernardo-Gancedo A., Henry A., et al. (2019). Human Pregnancy Zone Protein Stabilizes Misfolded Proteins Including Preeclampsia- and Alzheimer's-Associated Amyloid Beta Peptide. Proc. Natl. Acad. Sci. U.S.A. 116, 6101–6110. doi:10.1073/pnas.1817298116
Cescon M., Gattazzo F., Chen P., Bonaldo P. (2015). Collagen Vi at a Glance. J. Cel. Sci. 128, 3525–3531. doi:10.1242/jcs.169748
Chiquet M., Birk D. E., Bönnemann C. G., Koch M. (2014). Collagen Xii: Protecting Bone and Muscle Integrity by Organizing Collagen Fibrils. Int. J. Biochem. Cel. Biol. 53, 51–54. doi:10.1016/j.biocel.2014.04.020
Chun S. Y., Lee H. J., Choi Y. A., Kim K. M., Baek S. H., Park H. S., et al. (2011). Analysis of the Soluble Human Tooth Proteome and its Ability to Induce Dentin/Tooth Regeneration. Tissue Eng. A 17, 181–191. doi:10.1089/ten.tea.2010.0121
Connizzo B. K., Sun L., Lacin N., Gendelman A., Solomonov I., Sagi I., et al. (2021). Nonuniformity in Periodontal Ligament: Mechanics and Matrix Composition. J. Dent. Res. 100, 179–186. doi:10.1177/0022034520962455
de Castro Brás L. E., Ramirez T. A., DeLeon-Pennell K. Y., Chiao Y. A., Ma Y., Dai Q., et al. (2013). Texas 3-Step Decellularization Protocol: Looking at the Cardiac Extracellular Matrix. J. Proteomics 86, 43–52. doi:10.1016/j.jprot.2013.05.004
Denes B. J., Ait-Lounis A., Wehrle-Haller B., Kiliaridis S. (2020). Core Matrisome Protein Signature During Periodontal Ligament Maturation from Pre-Occlusal Eruption to Occlusal Function. Front. Physiol. 11, 174. doi:10.3389/fphys.2020.00174
Duarte W. R., Mikuni-Takagaki Y., Kawase T., Limura T., Oida S., Ohya K., et al. (1999). Effects of Mechanical Stress on the Mrna Expression of S100a4 and Cytoskeletal Components by Periodontal Ligament Cells. J. Med. Dent. Sci. 46, 117.
Duarte W. R., Kasugai S., Iimura T., Oida S., Takenaga K., Ohya K., et al. (1998). Cdna Cloning of S100 Calcium-Binding Proteins from Bovine Periodontal Ligament and Their Expression in Oral Tissues. J. Dent. Res. 77, 1694–1699. doi:10.1177/00220345980770090501
Everts V., Niehof A., Jansen D., Beertsen W. (1998). Type Vi Collagen Is Associated with Microfibrils and Oxytalan Fibers in the Extracellular Matrix of Periodontium, Mesenterium and Periosteum. J. Periodontal Res. 33, 118–125. doi:10.1111/j.1600-0765.1998.tb02300.x
Fernandes G., Yang S. (2016). Application of Platelet-Rich Plasma with Stem Cells in Bone and Periodontal Tissue Engineering. Bone Res. 4, 16036. doi:10.1038/boneres.2016.36
Frantz C., Stewart K. M., Weaver V. M. (2010). The Extracellular Matrix at a Glance. J. Cel. Sci. 123, 4195–4200. doi:10.1242/jcs.023820
Giannobile W. V. (1996). Periodontal Tissue Engineering by Growth Factors. Bone 19, 23S–37S. doi:10.1016/s8756-3282(96)00127-5
Grynpas M. D., Eyre D. R., Kirschner D. A. (1980). Collagen Type Ii Differs from Type I in Native Molecular Packing. Biochim. Biophys. Acta (Bba) - Protein Struct. 626, 346–355. doi:10.1016/0005-2795(80)90129-4
Hamano S., Tomokiyo A., Hasegawa D., Yoshida S., Sugii H., Mitarai H., et al. (2018). Extracellular Matrix from Periodontal Ligament Cells Could Induce the Differentiation of Induced Pluripotent Stem Cells to Periodontal Ligament Stem Cell-Like Cells. Stem Cell Develop. 27, 100–111. doi:10.1089/scd.2017.0077
Hudson D. M., Garibov M., Dixon D. R., Popowics T., Eyre D. R. (2017). Distinct Post-Translational Features of Type I Collagen Are Conserved in Mouse and Human Periodontal Ligament. J. Periodont. Res. 52, 1042–1049. doi:10.1111/jre.12475
Hussey G. S., Dziki J. L., Badylak S. F. (2018). Extracellular Matrix-Based Materials for Regenerative Medicine. Nat. Rev. Mater. 3, 159–173. doi:10.1038/s41578-018-0023-x
Iozzo R. V., Gubbiotti M. A. (2018). Extracellular Matrix: The Driving Force of Mammalian Diseases. Matrix Biol. 71-72, 1–9. doi:10.1016/j.matbio.2018.03.023
Jiang N., Guo W., Chen M., Zheng Y., Zhou J., Kim S. G., et al. (2016). Periodontal Ligament and Alveolar Bone in Health and Adaptation: Tooth Movement. Front. Oral Biol. 18, 1–8. doi:10.1159/000351894
Kaku M., Rosales Rocabado J. M., Kitami M., Ida T., Akiba Y., Yamauchi M., et al. (2016). Mechanical Loading Stimulates Expression of Collagen Cross-Linking Associated Enzymes in Periodontal Ligament. J. Cel. Physiol. 231, 926–933. doi:10.1002/jcp.25184
Kaku M., Yamauchi M. (2014). Mechano-Regulation of Collagen Biosynthesis in Periodontal Ligament. J. Prosthodontic Res. 58, 193–207. doi:10.1016/j.jpor.2014.08.003
Karimbux N. Y., Nishimura I. (1995). Temporal and Spatial Expressions of Type Xii Collagen in the Remodeling Periodontal Ligament During Experimental Tooth Movement. J. Dent. Res. 74, 313–318. doi:10.1177/00220345950740010501
Katayama H., Nagasu T., Oda Y. (2001). Improvement of In-Gel Digestion Protocol for Peptide Mass Fingerprinting by Matrix-Assisted Laser Desorption/Ionization Time-Of-Flight Mass Spectrometry. Rapid Commun. Mass. Spectrom. 15, 1416–1421. doi:10.1002/rcm.379
Krasny L., Huang P. H. (2021). Advances in the Proteomic Profiling of the Matrisome and Adhesome. Expert Rev. Proteomics 18, 781–794. doi:10.1080/14789450.2021.1984885
Kumar A., Placone J. K., Engler A. J. (2017). Understanding the Extracellular Forces that Determine Cell Fate and Maintenance. Development 144, 4261–4270. doi:10.1242/dev.158469
Lee S. Y., Moon J. S., Yang D. W., Yoo H. I., Jung J. Y., Kim O. S., et al. (2021). SLPI in Periodontal Ligament Is Not Sleepy During Biophysical Force‐Induced Tooth Movement. J. Clin. Periodontol. 48, 528–540. doi:10.1111/jcpe.13416
Li Q., Luo T., Lu W., Yi X., Zhao Z., Liu J. (2019a). Proteomic Analysis of Human Periodontal Ligament Cells Under Hypoxia. Proteome Sci. 17, 3. doi:10.1186/s12953-019-0151-2
Li Z., Yu M., Jin S., Wang Y., Luo R., Huo B., et al. (2019b). Stress Distribution and Collagen Remodeling of Periodontal Ligament during Orthodontic Tooth Movement. Front. Pharmacol. 10, 1263. doi:10.3389/fphar.2019.01263
Lu X., Zhu H. (2005). Tube-Gel Digestion. Mol. Cell Proteomics 4, 1948–1958. doi:10.1074/mcp.m500138-mcp200
Lundgren D. H., Hwang S.-I., Wu L., Han D. K. (2010). Role of Spectral Counting in Quantitative Proteomics. Expert Rev. Proteomics 7, 39–53. doi:10.1586/epr.09.69
Lynch S. E., Nixon J. C., Colvin R. B., Antoniades H. N. (1987). Role of Platelet-Derived Growth Factor in Wound Healing: Synergistic Effects with Other Growth Factors. Proc. Natl. Acad. Sci. U.S.A. 84, 7696–7700. doi:10.1073/pnas.84.21.7696
Mah S.-j., Lee J., Kim H., Kang Y.-G., Baek S.-H., Kim H.-H., et al. (2015). Induction of S100a4 in Periodontal Ligament Cells Enhances Osteoclast Formation. Arch. Oral Biol. 60, 1215–1221. doi:10.1016/j.archoralbio.2015.05.014
McCabe M. C., Schmitt L. R., Hill R. C., Dzieciatkowska M., Maslanka M., Daamen W. F., et al. (2021). Evaluation and Refinement of Sample Preparation Methods for Extracellular Matrix Proteome Coverage. Mol. Cell Proteomics 20, 100079. doi:10.1016/j.mcpro.2021.100079
Miwa N., Uebi T., Kawamura S. (2008). S100-Annexin Complexes - Biology of Conditional Association. FEBS J. 275, 4945–4955. doi:10.1111/j.1742-4658.2008.06653.x
Mizukoshi M., Kaku M., Thant L., Kitami K., Arai M., Saito I., et al. (2021). In Vivo Cell Proliferation Analysis and Cell-Tracing Reveal the Global Cellular Dynamics of Periodontal Ligament Cells Under Mechanical-Loading. Sci. Rep. 11, 9813. doi:10.1038/s41598-021-89156-w
Moon J. S., Lee S. Y., Kim J. H., Choi Y. H., Yang D. W., Kang J. H., et al. (2019). Synergistic Alveolar Bone Resorption by Diabetic Advanced Glycation End Products and Mechanical Forces. J. Periodontol. 90, 1457–1469. doi:10.1002/jper.18-0453
Naba A., Clauser K. R., Hoersch S., Liu H., Carr S. A., Hynes R. O. (2012). The Matrisome: In Silico Definition and In Vivo Characterization by Proteomics of normal and Tumor Extracellular Matrices. Mol. Cel. Proteomics 11, M111014647–014647. doi:10.1074/mcp.M111.014647
Naba A., Clauser K. R., Ding H., Whittaker C. A., Carr S. A., Hynes R. O. (2016). The Extracellular Matrix: Tools and Insights for the “omics” Era. Matrix Biol. 49, 10–24. doi:10.1016/j.matbio.2015.06.003
Nanci A., Bosshardt D. D. (2006). Structure of Periodontal Tissues in Health and Disease*. Periodontol 40, 11–28. doi:10.1111/j.1600-0757.2005.00141.x
Naveh G. R. S., Foster J. E., Silva Santisteban T. M., Yang X., Olsen B. R. (2018). Nonuniformity in Ligaments Is a Structural Strategy for Optimizing Functionality. Proc. Natl. Acad. Sci. U.S.A. 115, 9008–9013. doi:10.1073/pnas.1807324115
Nemoto T., Kajiya H., Tsuzuki T., Takahashi Y., Okabe K. (2010). Differential Induction of Collagens by Mechanical Stress in Human Periodontal Ligament Cells. Arch. Oral Biol. 55, 981–987. doi:10.1016/j.archoralbio.2010.08.004
Nishida E., Sasaki T., Ishikawa S. K., Kosaka K., Aino M., Noguchi T., et al. (2007). Transcriptome Database Kk-Periome for Periodontal Ligament Development: Expression Profiles of the Extracellular Matrix Genes. Gene 404, 70–79. doi:10.1016/j.gene.2007.09.009
Nozaki K., Kaku M., Yamashita Y., Yamauchi M., Miura H. (2010). Effect of Cyclic Mechanical Loading on Osteoclast Recruitment in Periodontal Tissue. J. Periodontal Res. 45, 8–15. doi:10.1111/j.1600-0765.2008.01193.x
Ojima Y., Mizuno M., Kuboki Y., Komori T. (2003). In Vitro effect of Platelet-Derived Growth Factor-BB on Collagen Synthesis and Proliferation of Human Periodontal Ligament Cells. Oral Dis. 9, 144–151. doi:10.1034/j.1601-0825.2003.02906.x
Ricard-Blum S. (2011). The Collagen Family. Cold Spring Harbor Perspect. Biol. 3, a004978. doi:10.1101/cshperspect.a004978
Salmon C. R., Giorgetti A. P. O., Paes Leme A. F., Domingues R. R., Kolli T. N., Foster B. L., et al. (2017). Microproteome of Dentoalveolar Tissues. Bone 101, 219–229. doi:10.1016/j.bone.2017.05.014
Seo B.-M., Miura M., Gronthos S., Mark Bartold P., Batouli S., Brahim J., et al. (2004). Investigation of Multipotent Postnatal Stem Cells from Human Periodontal Ligament. The Lancet 364, 149–155. doi:10.1016/s0140-6736(04)16627-0
Sims M. R., Leppard P. I., Sampson W. J., Dreyer C. W. (1996). Microvascular Luminal Volume Changes in Aged Mouse Periodontal Ligament. J. Dent. Res. 75, 1503–1511. doi:10.1177/00220345960750071101
Sodek J. (1977). A Comparison of the Rates of Synthesis and Turnover of Collagen and Non-Collagen Proteins in Adult Rat Periodontal Tissues and Skin Using a Microassay. Arch. Oral Biol. 22, 655–665. doi:10.1016/0003-9969(77)90095-4
Szklarczyk D., Gable A. L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., et al. (2019). String V11: Protein-Protein Association Networks with Increased Coverage, Supporting Functional Discovery in Genome-Wide Experimental Datasets. Nucleic Acids Res. 47, D607–D613. doi:10.1093/nar/gky1131
Taha I. N., Naba A. (2019). Exploring the Extracellular Matrix in Health and Disease Using Proteomics. Essays Biochem. 63, 417–432. doi:10.1042/ebc20190001
Takimoto A., Kawatsu M., Yoshimoto Y., Kawamoto T., Seiryu M., Takano-Yamamoto T., et al. (2015). Scleraxis and Osterix Antagonistically Regulate Tensile Force-Responsive Remodeling of the Periodontal Ligament and Alveolar Bone. Development 142, 787–796. doi:10.1242/dev.116228
Thompson S. M., Craven R. A., Nirmalan N. J., Harnden P., Selby P. J., Banks R. E. (2013). Impact of Pre‐Analytical Factors on the Proteomic Analysis of Formalin‐Fixed Paraffin‐Embedded Tissue. Prot. Clin. Appl. 7, 241–251. doi:10.1002/prca.201200086
Tsuzuki T., Kajiya H., T-Goto K., Tsutsumi T., Nemoto T., Okabe K., et al. (2016). Hyperocclusion Stimulates the Expression of Collagen Type Xii in Periodontal Ligament. Arch. Oral Biol. 66, 86–91. doi:10.1016/j.archoralbio.2016.02.009
Wescott D. C., Pinkerton M. N., Gaffey B. J., Beggs K. T., Milne T. J., Meikle M. C. (2007). Osteogenic Gene Expression by Human Periodontal Ligament Cells Under Cyclic Tension. J. Dent. Res. 86, 1212–1216. doi:10.1177/154405910708601214
Wilkins M. R., Sanchez J.-C., Gooley A. A., Appel R. D., Humphery-Smith I., Hochstrasser D. F., et al. (1996). Progress with Proteome Projects: Why All Proteins Expressed by a Genome Should Be Identified and How to Do it. Biotechnol. Genet. Eng. Rev. 13, 19–50. doi:10.1080/02648725.1996.10647923
Wyatt A. R., Cater J. H., Ranson M. (2016). Pzp and Pai-2: Structurally-Diverse, Functionally Similar Pregnancy Proteins? Int. J. Biochem. Cell Biol. 79, 113–117. doi:10.1016/j.biocel.2016.08.018
Xiong J., Menicanin D., Zilm P. S., Marino V., Bartold P. M., Gronthos S. (2016). Investigation of the Cell Surface Proteome of Human Periodontal Ligament Stem Cells. Stem. Cells. Int. 2016, 1947157. doi:10.1155/2016/1947157
Yamada S., Tomoeda M., Ozawa Y., Yoneda S., Terashima Y., Ikezawa K., et al. (2007). Plap-1/Asporin, a Novel Negative Regulator of Periodontal Ligament Mineralization. J. Biol. Chem. 282, 23070–23080. doi:10.1074/jbc.m611181200
Yamauchi M., Katz E. P., Mechanic G. L. (1986). Intermolecular Crosslinking and Stereospecific Molecular Packing in Type I Collagen Fibrils of the Periodontal Ligament. Biochemistry 25, 4907–4913. doi:10.1021/bi00365a027
Zhou M., Li Z.-q., Wang Z.-l. (2015). S100a4 Upregulation Suppresses Tissue Ossification and Enhances Matrix Degradation in Experimental Periodontitis Models. Acta Pharmacol. Sin. 36, 1388–1394. doi:10.1038/aps.2015.77
Zhou Y., Zhou B., Pache L., Chang M., Khodabakhshi A. H., Tanaseichuk O., et al. (2019). Metascape Provides a Biologist-Oriented Resource for the Analysis of Systems-Level Datasets. Nat. Commun. 10, 1523. doi:10.1038/s41467-019-09234-6
Keywords: periodontal ligament, extracellular matrix, collagen, matrisome, mechanical stress, proteomics, bioinformatics
Citation: Thant L, Kaku M, Kakihara Y, Mizukoshi M, Kitami M, Arai M, Kitami K, Kobayashi D, Yoshida Y, Maeda T, Saito I, Uoshima K and Saeki M (2022) Extracellular Matrix-Oriented Proteomic Analysis of Periodontal Ligament Under Mechanical Stress. Front. Physiol. 13:899699. doi: 10.3389/fphys.2022.899699
Received: 19 March 2022; Accepted: 11 April 2022;
Published: 20 May 2022.
Edited by:
Joo Cheol Park, Seoul National University, South KoreaReviewed by:
Sun-Hun Kim, Chonnam National University, South KoreaCopyright © 2022 Thant, Kaku, Kakihara, Mizukoshi, Kitami, Arai, Kitami, Kobayashi, Yoshida, Maeda, Saito, Uoshima and Saeki. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Masaru Kaku, a2FrdW1AZGVudC5uaWlnYXRhLXUuYWMuanA=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.