
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Physiol., 09 May 2022
Sec. Invertebrate Physiology
Volume 13 - 2022 | https://doi.org/10.3389/fphys.2022.896793
 Xiaoqian Ma1,2†
Xiaoqian Ma1,2† Xinming Lu1†
Xinming Lu1† Ping Zhang1
Ping Zhang1 Xun Deng2
Xun Deng2 Jianyang Bai1
Jianyang Bai1 Zhe Xu1
Zhe Xu1 Jian Diao1
Jian Diao1 Hongyang Pang3
Hongyang Pang3 Qi Wang2
Qi Wang2 Hongying Zhao2
Hongying Zhao2 Wei Ma4*
Wei Ma4* Ling Ma1*
Ling Ma1*The olfactory system plays a key role in regulating insect behaviors, such as locating host plants, spawning sites, and mating partners and avoiding predators. Chemosensory genes are required for olfactory recognition in insects. Curculio dieckmanni Faust. (Coleoptera: Curculionidae) damages hazelnuts and causes severe economic losses. There are no effective control measures, but understanding the olfaction mechanisms of this insect could lead to a new approach for population management. However, the genes that perform chemosensory functions in C. dieckmanni are still unclear. Using high-throughput sequencing, we assembled the antennal transcriptome of C. dieckmanni and annotated the major chemosensory gene families. Of the chemosensory gene families, we found 23 odorant-binding proteins, 15 chemosensory proteins, 2 sensory neuron membrane proteins, 15 odorant receptors, 23 ionotropic receptors, and nine gustatory receptors. Using Blast sequence alignment and phylogenetic analysis, the sequences of these proteins were identified. Male- and female-specific chemosensory genes involved in odorant detection and recognition were validated by qRT-PCR. Among the chemosensory genes, we found significant differences in the expression of CdieOBP8, CdieOBP9, CdieOBP19, CdieOBP20, CdieOBP21, CdieCSP15, CdieOR13, and CdieOR15 between adult male and female C. dieckmanni. A total of 87 expressed chemosensory proteins were found in C. dieckmanni. Investigating these proteins will help reveal the molecular mechanism of odorant recognition in C. dieckmanni and may aid the development of novel control strategies for this species.
Curculio dieckmanni Faust. is a pest beetle that infests hazelnut (Corylus heterophylla Fisch.) crops and is common in hazelnut orchards of China. The adults lay eggs in hazelnut trees, and the larvae feed on, and damage, the nut causing significant economic losses (Xiao, 1992; Zhang et al., 2021). Because of the damage caused by this pest, the State Forestry Administration of China listed C. dieckmanni as a national harmful pest for forestry in 2013 and initiated measures to reduce the economic impact that it causes in hazelnut orchards (Announcement of the State Forestry Administration of China, 2013). C. dieckmanni has overlapping generations and nonsynchronous life cycles, and it is presently controlled by pesticide applications. However, long-term use of broad-spectrum pesticides can result in toxic residues and lead to pest resistance (Ahmad et al., 2002; Chai and Lee, 2010; Downes, et al., 2016). In addition, pesticide residues in hazelnuts can be detrimental to human health and the environment. Therefore, it is crucial to develop novel control methods that are safer and less ecologically disruptive.
The olfactory chemosensory system performs important functions in the life stages of insects because it detects olfactory information carried by odorant molecules related to specific behaviors such as congregating, locating reproductive sites and mates, foraging, and avoiding predators (Field et al., 2000; Smadja and Butlin, 2009; Leal, 2013; Tian et al., 2018; Xiao et al., 2019). When lipophilic odorant molecules enter the lymph through micropores on chemosensory sensilla located in the antennal epidermis, they are detected by olfactory neurons (ORNs). The chemical information signals will be transformed into electrophysiological information signals. These physiological signals are transduced by ORNs to the central nervous system of the insect brain, where the information is processed to trigger specific physiological and behavioral responses through neurons (Vosshall et al., 1999; Vogt et al., 2009). In the process of odorant molecule detection, odorant molecules are first bound and transported by odorant-binding proteins (OBPs) and chemosensory proteins (CSPs) in sensory lymph fluid, which is the first step in the transport of hydrophobic odorants in olfactory recognition (Dippel, 2014; Keil and Steinbrecht, 1984; Wanner et al., 2005; Pelosi et al., 2006; Guo et al., 2011; Pelosi et al., 2014; Pelosi et al., 2018; Xiao et al., 2019; Liu et al., 2020). Then, these molecules are detected by odorant receptors protein (ORs) or ionotropic receptors (IRs) expressed on the olfactory neuron membrane (Benton et al., 2009; Croset et al., 2010; Abuin et al., 2011). ORs are divided into co-receptors (ORco) and common odorant receptors. ORco is highly conserved across species and can enhance the ability of specific common odorant receptors to bind to odorant molecules (Jacquin-Joly and Merlin, 2004; Benton et al., 2006; Sato and Touhara 2009). In coleopteran insects, ORs are divided into seven monophyletic subfamilies, called nine groups, according to their amino acid sequences (Mitchell and Andersson, 2020). Additionally, gustatory receptors (GRs) and sensory neuron membrane proteins (SNMPs) are also involved. GRs mainly detect sugars, bitter flavors, pheromones, CO2, or contact stimuli. They perform important functions in host-seeking and are not limited to gustatory functions (Clyne, et al., 2000; Park and Kwon, 2011; Mang et al., 2016). Sensory neuron membrane proteins (SNMPs) contain two transmembrane domains, which play an essential role in identifying and detecting pheromones (Clyne et al., 1999; Mang et al., 2016).
Early research on chemosensory genes in coleopteran insects has focused mainly on OBPs (Vogt and Riddiford, 1981). The use of OBPs to detect pheromones was subsequently recorded in scarabs (2001). In Coleoptera, the earliest study of receptors came from the wasp co-receptor ORco, followed by the first genome study of the full set of chemosensory genes in coleopteran model insects (Krieger et al., 2003). The most complete study of chemosensory genes was conducted in other species of Coleoptera (Engsontia et al., 2008; Nichols and Vogt, 2008; Richards et al., 2008). In recent years, most studies have focused on chemosensory gene sequences and gene expression, while studies on functional characterization are relatively rare. ORs have a key role in pheromone detection. The ItpyOR46 and ItpyOR49 of I. typographus were specifically reactive with pheromone compounds (S)-ipsenol and (R)-(-)-ipsdienol, respectively, and phylogenetic analysis was performed in the OR subfamily of coleopteran group 7, which is widely present in the weevil family (Yuvaraj et al., 2021). McOr3 and McOr20 of Megacyllene caryae are sensitive to the male-secreted pheromones (S)-2-methyl-1-butanol and (2S, 3R)-2,3-hexanediol, respectively (Mitchell et al., 2012). The red palm weevil is rendered incapable of recognizing the male-produced pheromones (4RS, 5RS)-4-methylnonan-5-ol and 4 (RS)-methylnonan-5-one by silencing RferOR1 (Antony et al., 2021).
With the development of next generation sequencing (NGS) in insects, many chemosensory genes have been sequenced and verified by genomic and transcriptomic data. NGS has improved our knowledge. Despite substantial progress in deciphering these chemical communication systems in insects, the study of chemosensory genes in Coleoptera is challenging because of the profound diversity of Coleoptera. There are nearly 200 families in Coleoptera, and the annotation of chemosensory genes mainly revolves around several genera in five families (Cerambycidae, Chrysomelidae, Curculionidae, Scarabaeidae, and Tenebrionidae) (Mckenna et al., 2019). In addition to the coleopteran model insect T. castaneum, the genomes of Leptinotarsa decemlineata, Anoplophora glabripennis, Agrilus planipennis, and Dendroctonus ponderosae have been established (Mckenna et al., 2016; Mitchell et al., 2017; Schoville et al., 2018; Andersson et al., 2019; Mitchell et al., 2020). These genomes help to establish the phylogenetic framework of coleopteran chemosensory genes. The annotations of the remaining chemosensory genes are derived from the transcriptome, which is currently a handy and economical sequencing method suitable for the annotation of insect chemosensory genes. However, there is little information regarding the molecular mechanism underlying olfactory recognition in Curculionoidea. Very little research is available on the molecular mechanisms of olfactory recognition of C. dieckmanni. The Curculionidae (weevils) is one of the largest coleopteran families with more than 80,000 species described, some of which are economically important pests. Only the chemosensory gene families of a dozen Curculionidae species have been studied (Andersson et al., 2013; Bin et al., 2017; Paula et al., 2018; Andersson et al., 2019; Zhang et al., 2019; Antony et al., 2021; Hou et al., 2021; Yuvaraj et al., 2021). The olfactory genes of C. dieckmanni have not been studied yet, and their identification will help to illustrate the molecular mechanisms of olfactory recognition in this family.
In this work, the antennal transcriptomes of female and male adults of C. dieckmanni were sequenced on an Illumina HiSeqTM 4000 platform. After the transcriptome data were assembled and analyzed, we identified 23 OBPs, 15 CSPs, 2 SNMPs, 15 ORs, 23 IRs and 9 GRs. The expression patterns of chemosensory genes in the antennae of male and female adults were analyzed by reads per kilobase per million (RPKM) and the expression profiles of these genes were verified by qRT-PCR. In addition, phylogenetic trees were constructed to predict the evolutionary relationship between the chemosensory genes of C. dieckmanni and other Curculionoidea species. These results help validate the functions of olfactory genes and provide a foundation for a more detailed study of the molecular mechanisms of olfactory recognition in C. dieckmanni.
Adult C. dieckmanni were collected in a hazelnut orchard in the Sandao Forestry Farm, Mudanjiang City, Heilongjiang Province, China (129°39′02″E, 44°07′04″N). The insects were fed with hydroponic C. heterophylla shoots in a cage (80 cm × 40 cm × 40 cm). Males and females were separated and dissected under a dissecting microscope to collect the antennae, proboscis, head, legs, wings, prothorax, and abdomen. Tissue samples were separately stored in Axygen tubes and frozen at −80°C. A total of 200 pairs of antennae were dissected from female and male adults for transcriptome sequencing, and there were three biological replicates for female and males, including six samples (DM1, DM2, DM3, DF1, DF2, and DF3).
Total RNA was isolated from the antennae of female and male adults of C. dieckmanni, using a Trizol reagent kit (Invitrogen, Carlsbad, CA, United States) based on the manufacturer’s instructions. The integrity of RNA quality was assessed using an Agilent 2100 bioanalyzer (Agilent Technologies, Palo Alto, CA, United States) and examined by RNase free agarose gel electrophoresis.
After the total RNA examination was qualified, the mRNA was enriched with magnetic beads with Oligo (dT). We added fragmentation buffer to break the mRNA into short fragments. We then used mRNA as a template for amplification and added random hexamers for reverse transcription to synthesize the first strand cDNA. We added buffer, dNTPs, and reagents, such as DNA polymerase I and RNase H, to synthesize the second-strand cDNA and purified the double-stranded cDNA with AMPure XP beads. The purified double-stranded cDNA was end repaired, poly (A) was added, and the cDNA was connected to Illumina sequencing adapters. We used AMPure XP beads to select the size of the fragments. PCR amplification was performed, and the PCR products were purified with AMPure XP beads to obtain the final cDNA library. After the library was constructed, Qubit 2.0 was used to determine the concentration, and the library was diluted to 1.5 ng/μl. Agilent 2100 was used to detect the insert size of the library. To ensure the quality of the library, the qPCR method was used to accurately quantify the effective concentration of the library. Qualified libraries of female and male adult antennae were sequenced using Illumina HiSeqTM 4000 by Gene Denovo Biotechnology Co. (Guangzhou, China). All the raw sequence data were saved in the NCBI Sequence Read Archive (SRA) database under BioProject accession No. PRJNA769532.
To ensure analysis quality, the original data obtained by sequencing were converted into sequence data by base calling, which contains joints, duplicates, and low sequencing quality. The reads were filtered to avoid affecting sequence assembly and subsequent analysis. We removed reads containing adaptors, reads with a proportion of unknown nucleotides (N) greater than 10%, and reads with low quality (q-value ≤ 20) over 40% bases. After obtaining the clean reads, we calculated the Q20, Q30 and GC content of the clean reads, and used Trinity to assemble the transcriptome from scratch. Trinity is a short reads assembly software (Grabherr et al., 2011).
The results of Trinity assembly were unigenes. To obtain the functional information of the unigenes, the unigene sequences were subjected to basic annotations, including protein function annotations, COG/KOG function annotations, pathway annotations and gene ontology (GO) annotations. For unigene annotation, at the E-value threshold of 1e-5, through BLASTx, all unigenes were compared in the NCBI non-redundant protein database (Nr), manually annotated, and reviewed in the Swiss-Prot protein database, the Kyoto Encyclopedia of Genes and Genomes (KEGG) database, and the Clusters of Orthologous Groups of proteins (COG/KOG) database. According to the best alignment results, unigene sequence directions were determined and protein function annotations were obtained. We used Blast2GO software for GO annotation and unigenes Nr result annotation (Conesa, et al., 2005). We selected the top 20 unigenes with the highest scores and no less than 33 High-scoring Segment Pair (HSPs) hits for Blast2GO analysis. WEGO software was used to classify the unigenes (Ye, et al., 2006).
The predicted conserved domains and open reading frames (ORFs) were manually validated for all candidate chemosensory genes (OBPs, CSPs, ORs, SNMPs, GRs, and IRs). Sequences with errors (insertions/gaps/deletions in homopolymer regions) were deleted. The putative N-terminal signal peptides of ligand-binding proteins (OBPs and CSPs) were searched using the SignalP v4.1 program. The transmembrane domains (TMDs) of ORs, IRs, and GRs were searched using TMHMM Server version 2.0. The translation of nucleotide sequences was performed using the Expasy program.
To validate the annotation of candidate chemosensory genes and the identification of homologous sequences, we conducted a phylogenetic analysis of C. dieckmanni including closely related Coleoptera species and other model insects. Insects selected for analysis were those whose chemosensory genes have been well studied (Supplementary Table S6). A multiple sequence alignment was performed using the MAFFT program, and the results were accessed using the Jalview software for editing and selecting the color scheme. Amino acid sequences of chemosensory genes of selected insects were obtained from the NCBI database, and Maximum likelihood (ML) phylogenetic trees were built based on the Jones-Taylor-Thornton (JTT) model in MEGA-X. Initial trees for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the JTT model, then selecting the topology with the superior log likelihood value. The tree was drawn to scale with branch lengths measured in the number of substitutions per site (Jones et al., 1992; Kumar et al., 2018). Bootstrap support values were based on 100 replicates.
To identify chemosensory DEGs, we first used RPKM to estimate the expression of chemosensory genes in the antennae of male and female adults (Mortazavi et al., 2008). These genes were selected as reference for the expression profile of genes in the antennae of male and female adults. Gene expression levels were determined, according to RPKM values. In addition, DEGs were identified by Benjamini-Hochberg-corrected p values (Guo and Rao, 2008). As a key indicator, the false discovery rate (FDR) was used to screen in multiple hypothesis testing for gene selection. In comparison, genes with multiple changes ≥ 2 and FDR < 0.05 were identified as significant DEGs (Ran et al., 2020).
The 31 genes in female and male transcriptomes were validated by qRT-PCR in different tissues and these genes, compared with all candidate genes, were highly expressed based on RPKM. Antennae (480) were collected from female and male adults. RNA was extracted using Trizol reagent and cDNA was synthesized using a PrimeScriptTM RT Reagent Kit with gDNA Eraser (TaKaRa, Shiga, Japan). We designed gene-specific primers using Primer3. The amplification efficiency was calculated for the genes and two candidate internal reference genes (actin and EF1-a). Based on the results, actin and EF1-a were selected as the reference genes for qRT-PCR. PCR was performed using Mx3000P QPCR (Agilent, United States) and ChamQ universal SYBR qPCR Master Mix (TaKaRa). The PCR program was as follows: 95°C for 30 s; 40 cycles of 95°C for 5 s and 60°C for 30 s; and 65–95°C at 0.5°C/5 s. A melting curve was generated. Each reaction had three independent biological replicates, and the relative gene expression level was obtained by the 2−∆∆Ct method (Guo and Rao, 2008). The relative gene expression in antennae was analyzed using SPSS v19.0 software. An independent sample t-test was performed to compare the gene expression levels between male and female antennae. Prism v6.0 was used to generate bar graphs.
A total of 42.5 Gb raw data were obtained after filtering. The transcriptomes of three female individuals yielded 49,095,002 (97.56%, DF1), 44,906,190 (97.60%, DF2), and 49,073,722 (97.47%, DF3) clean reads. The transcriptomes of three male individuals yielded 55,983,482 (97.52%, DM1), 49,406,426 (97.53%, DM2), and 41,989,468 (97.44%, DM3) clean reads. The Q20 value was >98%, and the Q30 value was >95% (Supplementary Table S1.1). The clean reads of six samples of females and males were assembled to yield a total of 43,060,197 unigenes. There were 44,024 unigenes longer than 200 bp (N50 = 1,861), including 42,971 unigenes from female samples (DF) and 43,190 unigenes from male samples (DM). The maximum length was 27,588 bp, the minimum length was 201 bp, and the average length was 978 bp (Supplementary Figure S1, Supplementary Table S1.2).
The unigene sequences were aligned using BLASTx against the protein databases NR, SwissProt, KEGG, and KOG (e-value < 0.00001). The protein with the highest similarity to a given unigene was selected for protein function annotation, and 21,803 of 44,024 (49.52%) unigenes were successfully annotated. The numbers of reference gene sequences annotated in NR, Swiss-Prot, KEGG, and KOG were 21,733, 12,094, 7,502, and 11,403, respectively, (Supplementary Figure S2, Supplementary Table S1.2). The sequence with the best alignment (lowest e-value) of a unigene, which was selected as homologous in the NR database (if there was a tie, the first sequence was selected). The species of the homologous sequence was determined, and the number of homologous sequences of each species was counted. The sequences with high similarity to those in transcripts were searched in the NR protein database by comparison with BLASTx. The analysis revealed a large number of homologous genes in Coleoptera. The insect species with the highest homology (40.93%) was Dendroctnous ponderosae, followed by Anoplophora glabripennis (14.13%), Tribolium castaneum (5.01%), and Lasius niger (4.68%), which had 8,895, 3,071, 1,088, and 1,018 matched transcripts, respectively (Supplementary Figure S3).
Based on the NR annotation, the Blast2GO (Conesa et al., 2005) software was used to obtain the GO annotations of unigenes. The WEGO software (Ye et al., 2006) was used for GO functional classification of all unigenes to draw a whole picture of the gene functions of this species. These transcripts were assigned to three categories: molecular function (1,808 unigenes), biological process (3,675 unigenes), and cellular component (2,544 unigenes; Supplementary Figure S4). To accurately evaluate the annotation information, the transcripts were aligned to the KOG database, and a total of 11,403 transcripts were assigned to 25 functional categories (Supplementary Figure S5).
Of the chemosensory gene families, a total of 87 expressed chemosensory proteins were found in C. dieckmanni: 23 odorant-binding proteins, 15 chemosensory proteins, 2 sensory neuron membrane proteins, 15 odorant receptors, 23 ionotropic receptors, and nine gustatory receptors (Information for genes in Supplementary Tables S2.1–S2.6). The sequences of chemosensory genes identified in C. dieckmanni transcriptomes are shown in Supplementary Table S5.
Twenty-three OBPs were obtained using BLASTx (Table S2.1). In the NCBI database, the homology of OBP sequences from C. dieckmanni and other Coleoptera species ranged from 37.40 to 78.05% (58.26% on average). In a further sequence analysis based on the number and location of cysteine (Figure 1), 11 OBPs (CdieOBP1, CdieOBP3, CdieOBP5, CdieOBP6, CdieOBP7, CdieOBP11, CdieOBP13, CdieOBP14, CdieOBP15, CdieOBP16, and CdieOBP21) were identified as typical “Classic OBP” subfamily with the sequence C1-X26-29-C2-X3-C3-X35-40-C4-X7-10-C5-X8-C6 (X is any amino acid), and 12 OBPs (CdieOBP2, CdieOBP4, CdieOBP8, CdieOBP9, CdieOBP10, CdieOBP12, CdieOBP17, CdieOBP18, CdieOBP19, CdieOBP20, CdieOBP22, and CdieOBP23) belonged to the “Minus-C OBP” subfamily with the sequence C1-X30-33-C2-X38-40-C3- X16-17-C4. The ORFs of 23 OBP genes ranged from 330 to 474 bp and encoded 110–157 amino acids. All genes except CdieOBP23 had complete ORFs, and six OBPs (CdieOBP9, CdieOBP11, CdieOBP18, CdieOBP19, CdieOBP21, and CdieOBP22) lacked signal peptides.

FIGURE 1. Alignment of candidate OBPs of C. dieckmanni. The highly conserved cysteine residues are marked with red triangles.
A phylogenetic tree was built using the 23 OBPs obtained from the transcriptomes and the OBPs from five Curculionoidea species (Dendroctonus adjunctus, D. armandi, Lissorhoptrus oryzophilus, Sitophilus zeamais, and Tomicus yunnanensis; Figure 2). These OBPs were highly conserved and were classified into “Minus-C OBP” or “Classic OBP” according to their functions. OBPs showed distinct patterns and did not cluster together. A homology matrix showed that adjacent branches had higher homology.
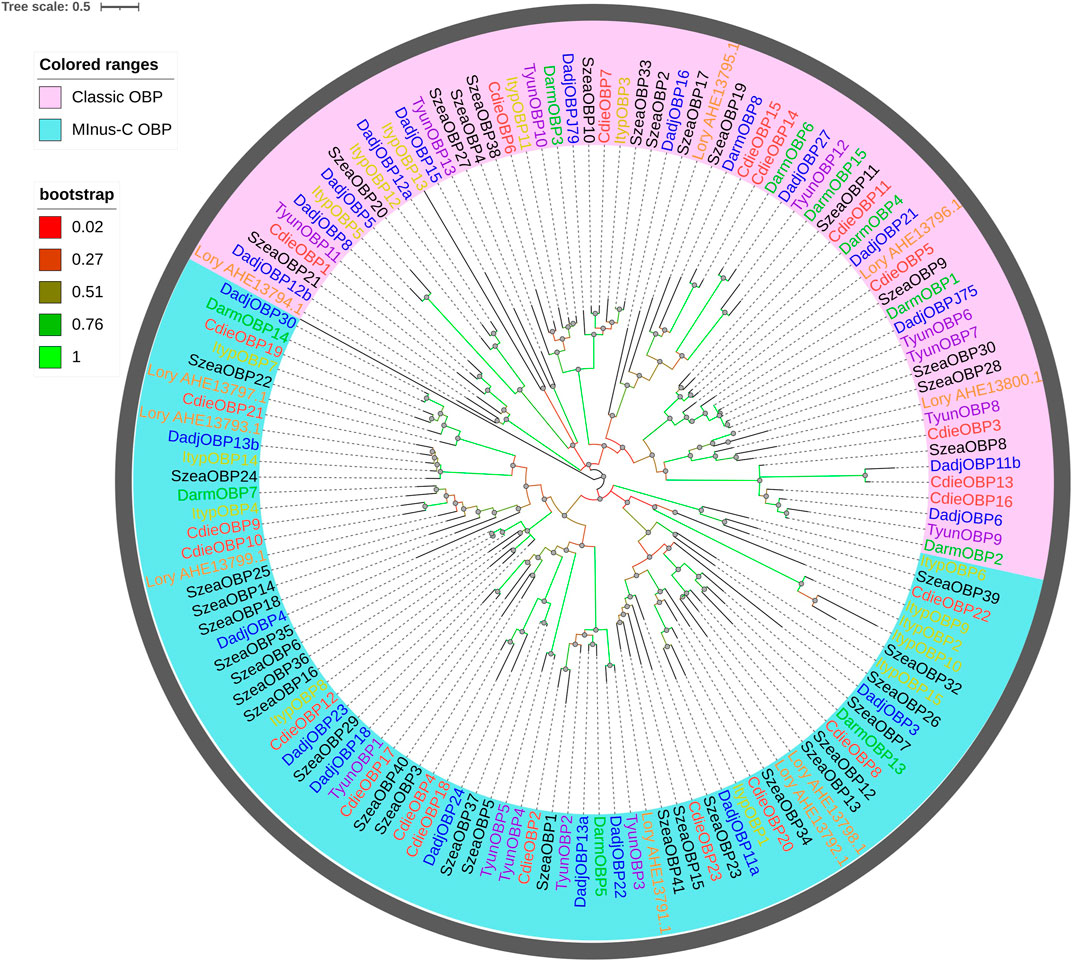
FIGURE 2. Maximum likelihood tree of OBPs in C. dieckmanni (Cdie, red), D. adjunctus (Dadj, blue), D. armandi (Darm, green), Ips typographus (Ityp, yellow), Lissorhoptrus oryzophilus (Lory, orange), S. zeamais (Szea, black), and T. yunnanensis (Tyun, purple). The scale bar represents 0.5 amino acid substitutions per site.
The analysis of transcriptome data revealed 15 CSPs, which had four conserved cysteine residues and the featured sequence C1-X6-7-C2-X18-C3-X2-C4 (X is any amino acid) of typical insect CSPs (Figure 3). The genes of 15 CSPs had ORFs ranging from 231 to 963 bp and encoding 76–320 amino acids, of which the genes of seven CSPs (CdieCSP1, CdieCSP2, CdieCSP4, CdieCSP5, CdieCSP6, CdieCSP7, and CdieCSP8) had incomplete ORFs, and eight CSPs (CdieCSP1, CdieCSP2, CdieCSP4, CdieCSP5, CdieCSP8, CdieCSP10, CdieCSP12, and CdieCSP13) lacked signal peptides (Supplementary Table S2.2). A phylogenetic tree was built using the 15 CSPs obtained from the transcriptomes and CSPs from five Curculionoidea species (Dendroctonus adjunctus, D. armandi, Lissorhoptrus oryzophilus, Dendroctonus ponderosae, and Tomicus yunnanensis) (Figure 4).

FIGURE 3. Alignment of candidate CSPs of C. dieckmanni. Each highly conserved cysteine residue is marked with a red triangle above it.
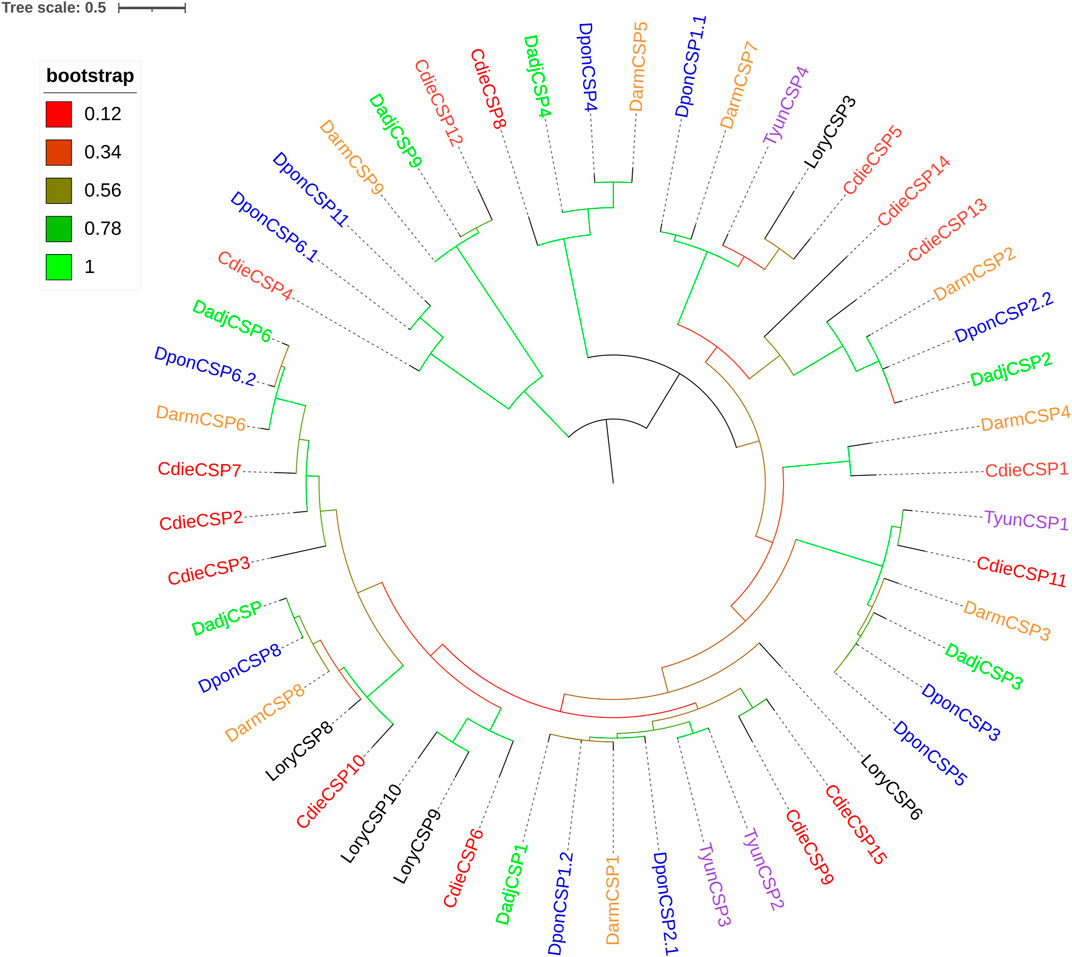
FIGURE 4. Maximum likelihood tree of CSPs in C. dieckmanni (Cdie, red), Dendroctonus adjunctus (Dadj, green), Dendroctonus armandi (Darm, orange), Lissorhoptrus oryzophilus (Lory, black), Dendroctonus ponderosae (Dpon, blue), and Tomicus yunnanensis (Tyun, purple). The scale bar represents 0.5 amino acid substitutions per site.
Genes with complete ORFs encoding CdieSNMP1 (SNMP1) and CdieSNMP2 (SNMP2) were identified based on BLASTx and cluster analysis. The ORF was 1,695 bp for CdieSNMP1 and 1,575 bp for CdieSNMP2, indicating that both were encoded by full-length genes. CdieSNMP1 and CdieSNMP1 had six and three transmembrane regions as well as 74.04 and 56.84% homology, respectively, (Supplementary Table S2.3). Phylogenetic analysis showed that CdieSNMP1 and CdieSNMP2 clustered with the SNMP1 and SNMP2 of other Curculionoidea species, respectively, and were most closely related to D. ponderosae, which confirmed the above results (Figure 5).
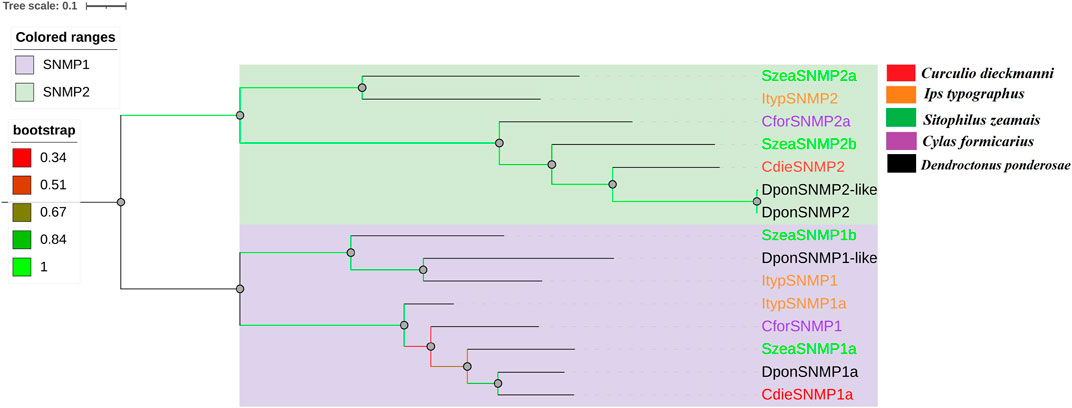
FIGURE 5. Maximum likelihood tree of SNMPs in C. dieckmanni (Cdie, red), S. zeamais (Szea, green), Ips typographus (Ityp, orange), Cylas formicarius (Cfor, purple), and Dendroctonus ponderosae (Dpon, black). The scale bar represents the 0.1 amino acid substitutions per site.
A total of 15 ORs were identified in the analysis of transcriptome data, including one odorant receptor coreceptor (Orco). The alignment of ORs against the NR database showed 36.02–94.40% homology and 78–1,449 bp ORFs encoding 25–482 amino acids. Given the sequence characteristics of ORs, 1–9 transmembrane domains were predicted (Supplementary Table S2.4). A phylogenetic tree was established along with the ORs identified in other Curculionoidea species (D. ponderosae, Ips typographus, and Sitophilus oryzae; Figure 6). Based on the amino acid sequence analysis, the ORs from C. dieckmanni was contained in six groups of beetles (Bin et al., 2017; Mitchell et al., 2020). Among them, CdieOR15 clustered with the highly conserved ORco in a branch. CdieOR14 clustered in group 1, CdieOR12 clustered in group 3, CdieOR10 clustered in group 5, three candidate ORs (CdieOR2, CdieOR7, and CdieOR9) clustered in group 7, and four candidate ORs (CdieOR5, CdieOR6, CdieOR8, and CdieOR11) clustered in group 2.
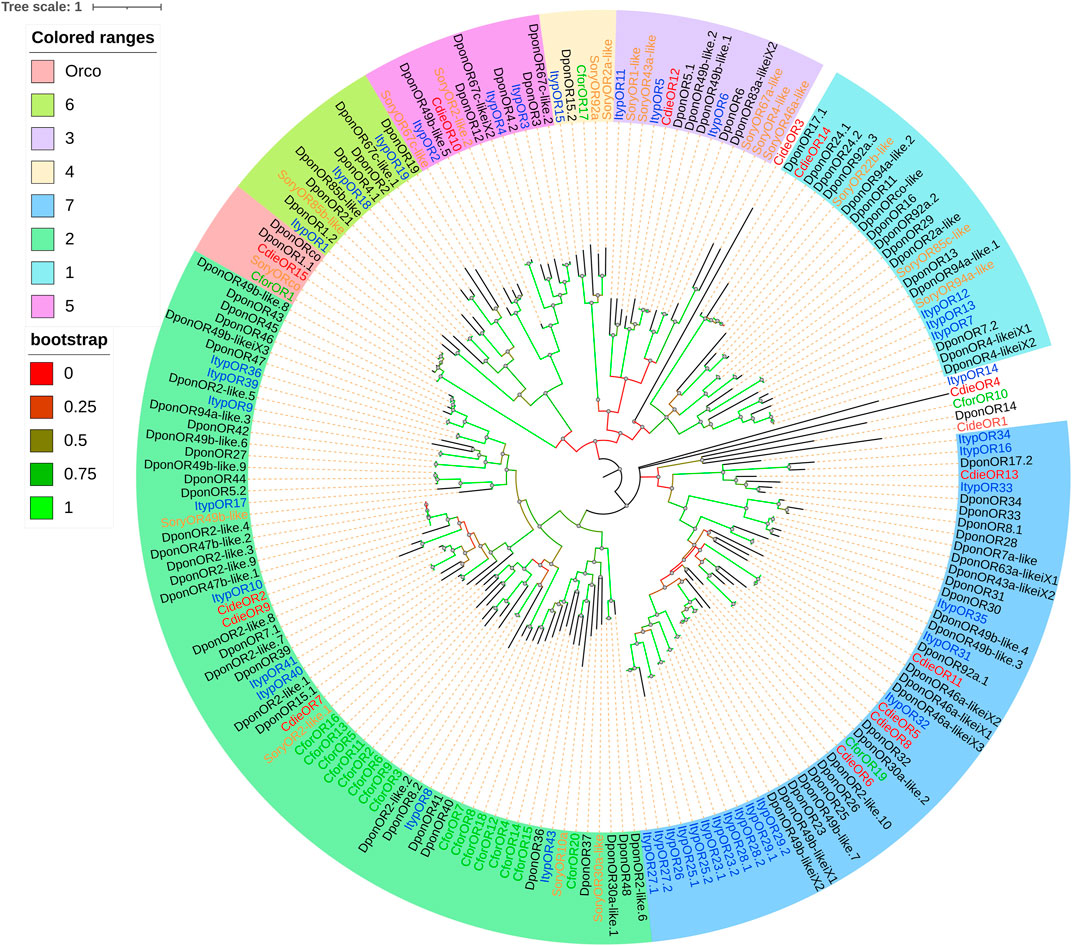
FIGURE 6. Maximum likelihood phylogenetic tree of candidate ORs in C. dieckmanni (Cdie, red), Dendroctonus ponderosae (Dpon, black), Ips typographus (Ityp, blue), Cylas formicarius (Cfor, green), and Sitophilus oryzae (Sory, orange). The scale bar represents 1 amino acid substitution per site.
We identified 23 IRs from C. dieckmanni antennae transcriptomes. A phylogenetic tree was built along with the IRs identified in other Curculionoidea species (Cylas formicarius, D. ponderosae, Ips typographus, L. oryzophilus, and S. oryzae; Figure 7). Phylogenetic analysis showed that CdieIR2, CdieIR4, CdieIR15, and CdieIR17 were clustered in IR75; CdieIR10 clustered in IR8a; CdieIR11 clustered in IR25a; CdieIR13 clustered in IR76b; and CdieIR21 clustered in IR41a. These were highly conserved. Related genes were identified based on other Curculionoidea species. The genes encoding nine IRs (CdieIR1, CdieIR8, CdieIR11, CdieIR12, CdieIR13, CdieIR19, CdieIR20, CdieIR22, and CdieIR23) had ORFs similar to or longer than 1,600 bp, and the ORF of CdieIR7 gene was 1,431 bp. The lengths of these nine IRs were comparable to the full lengths of such genes, and the genes of the remaining 14 IRs (CdieIR2, CdieIR3, CdieIR4, CdieIR5, CdieIR6, CdieIR7, CdieIR9, CdieIR10, CdieIR14, CdieIR15, CdieIR16, CdieIR17, CdieIR18, and CdieIR21) had partial ORFs of such genes. The number of transmembrane domains of the 23 IRs ranged from one to six (Supplementary Table S2.5).

FIGURE 7. Maximum likelihood phylogenetic tree of candidate IRs in C. dieckmanni (Cdie, red), Dendroctonus ponderosae (Dpon, black), Ips typographus (Ityp, blue), Lissorhoptrus oryzophilus (Lory, green), Sitophilus oryzae (Sory, orange), and Cylas formicarius (Cfor, purple). The scale bar represents 1 amino acid substitution per site.
From the transcriptome data, we identified 9 GR candidate transcripts with 1–11 transmembrane domains and homology ranging from 29.16 to 61.07%. The genes of 5 GRs (CdieGR1, CdieGR5, CdieGR6, CdieGR8, and CdieGR9) had ORFs longer than 1,000 bp (Supplementary Table S2.6). A phylogenetic tree was constructed to analyze the functional classification of GRs in C. dieckmanni transcriptomes based on the GRs identified in other Curculionoidea species (C. formicarius, D. ponderosae, I. typographus, L. oryzophilus, and S. oryzae; Figure 8). Phylogenetic analysis showed that three GRs (CdieGR4, CdieGR7, and CdieGR8) clustered in the gustatory receptor for sugar receptors, while CdieGR1 clustered in the gustatory receptor for CO2 receptors.
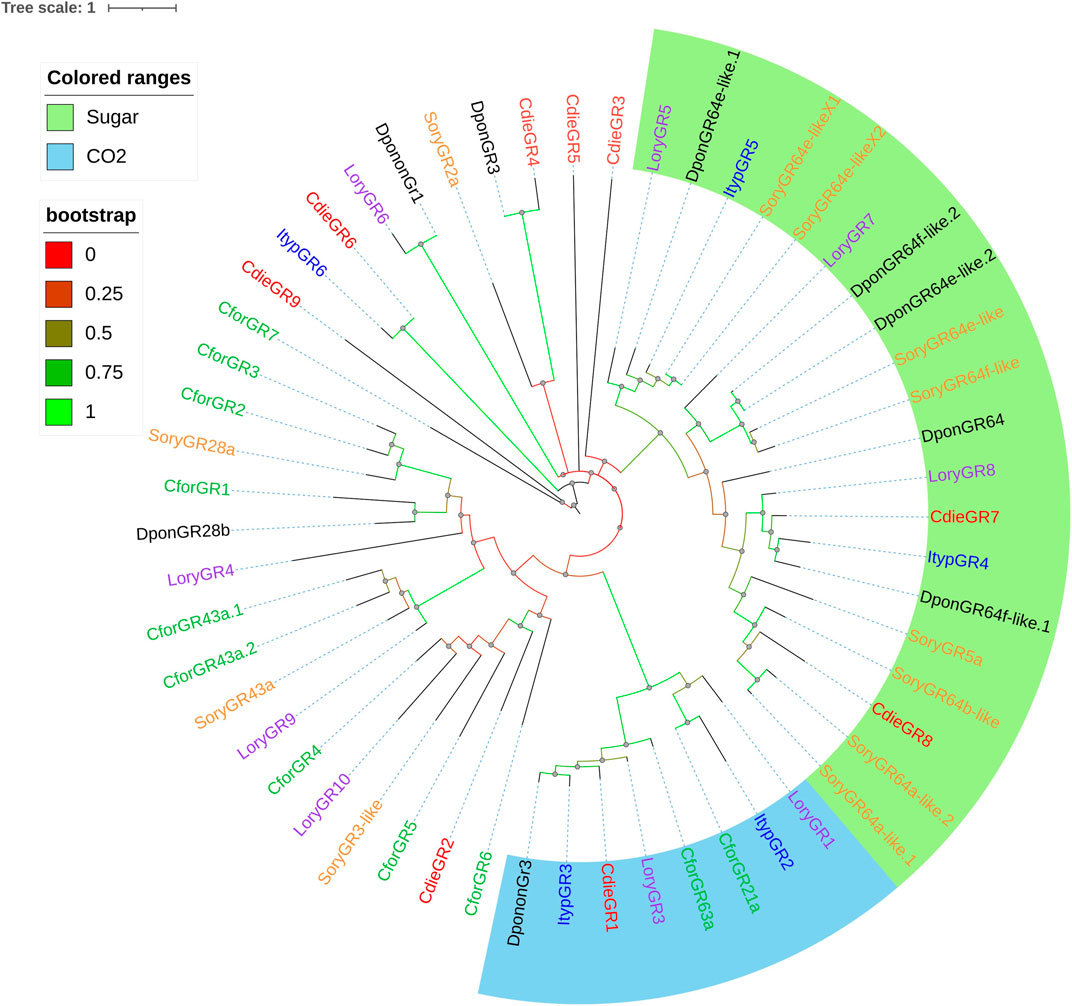
FIGURE 8. Maximum likelihood phylogenetic tree of candidate GRs in C. dieckmanni (Cdie, red), Dendroctonus ponderosae (Dpon, black), Lissorhoptrus oryzophilus (Lory, purple), Ips typographus (Ityp, blue), and Cylas formicarius (Cfor, green). The scale bar represents the 1 amino acid substitutions per site.
The RPKM analysis showed that a total of 355 genes were differentially expressed in male and female antennae, among which, 69 were significantly male-biased and 286 were significantly female-biased (Figure 9, Supplementary Tables S3, S4), including only three chemosensory genes. Two candidate odorant-binding proteins (CdieOBP19 and CdieOBP20) were expressed at significantly higher levels in female antennae than in male antennae (FDR < 0.05) (Supplementary Table S4). One candidate odorant receptor protein (CdieOR13) was expressed at significantly higher levels in female antennae than in male antennae (FDR < 0.05) (Supplementary Table S4).

FIGURE 9. Heatmap of significant differentially expressed genes in C. dieckmanni. Cluster analyses were based on log2RPKM. Each column represents a sample and each row represents a gene. Red boxes represent highly expressed genes. Blue boxes represent lowly expressed genes. DM1, DM2, and DM3 represent three replicate samples of males. DF1, DF2, and DF3 represent three replicate samples of females.
To validate and further understand the functions of the chemosensory genes in adult female and male C. dieckmanni antennae, we studied the expression profiles of these genes using qRT-PCR. The primers are listed in Supplementary Table S7. The qRT-PCR results showed that all chemosensory genes were expressed in both male and female antennae, except CdieOBP19, which was only expressed in female antennae. Three OBPs (CdieOBP9, CdieOBP20, and CdieOBP21) were significantly female-biased, one OBP (CdieOBP8) was significantly male-biased, one CSP (CdieCSP15) was significantly female-biased, two ORs (CdieOR13 and CdieOR15) were significantly female-biased, and no SNMPs, IRs, or GRs showed differential expression between males and females (Figure 10).
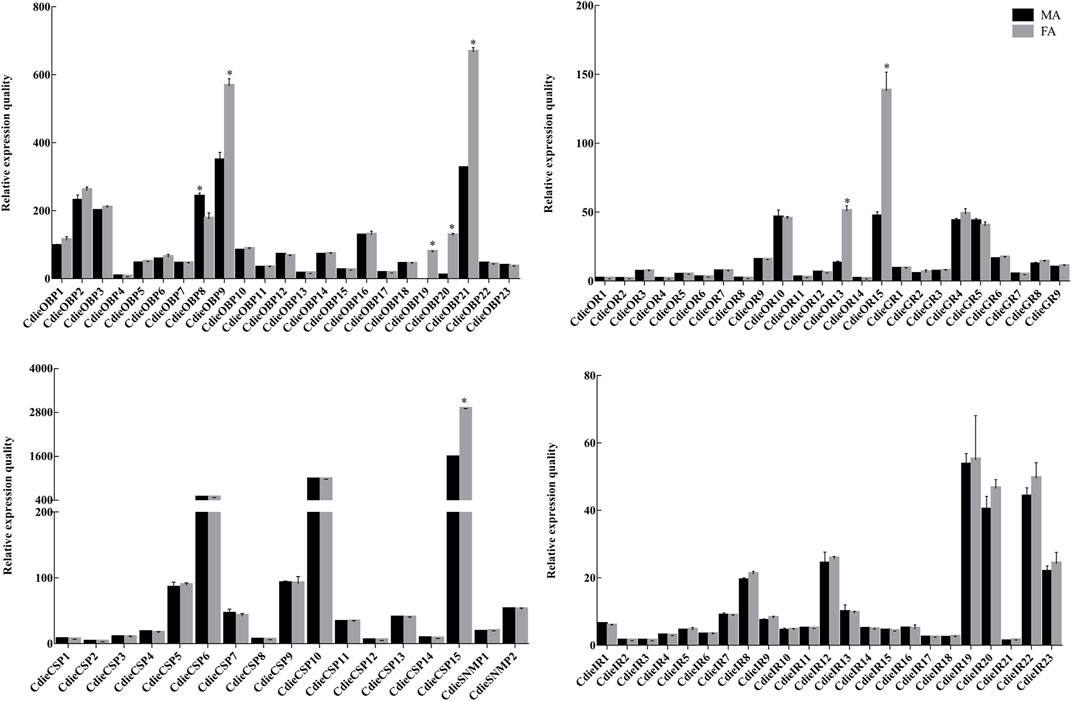
FIGURE 10. Relative expression levels of chemosensory genes in C. dieckmanni OBPs. FA: Female antennae; MA: Male antennae. The asterisks above each bar represent significant differences (p < 0.05).
Chemosensory proteins play essential roles in insect survival and reproduction and, thus, have been studied in several insect species (Guo et al., 2011). The present study contributes to build up our knowledge on the functioning of olfactory genes in the Curculionidae family and, specifically, constitutes a foundation for understanding the olfactory mechanisms of C. dieckmanni, a pest that affect hazelnut crops. Our results provide new directions for safer and less toxic methods to control C. dieckmanni populations.
Transcriptome analysis using BLASTx identified 87 chemosensory genes, including OBPs, CSPs, SNMPs, ORs, IRs, and GRs, from the antennae of male and female C. dieckmanni. These results enriched the chemosensory genes of Curculionidae. OBPs and CSPs are involved in the first step of hydrophobic odorant recognition, and their identification is a prerequisite for exploring the olfactory mechanisms of C. dieckmanni.
The numbers of OBPs and CSPs identified in C. dieckmanni were 23 and 15, respectively, which were lower than the OBPs (49) and CSPs (20) of T. castaneum but were similar to the OBPs (24) and CSPs (12) of Anthonomus grandis (Paula et al., 2018). We found that 11 were typical OBPs, 12 were Minus-C OBPs, with the OBPs from five Curculionoidea species (D. adjunctus, D. armandi, L. oryzophilus, S. zeamais, and T. yunnanensis), which indicated that they may serve the same functions and have the same ancestor gene. The qRT-PCR results showed that the expression levels of CdieOBP9, CdieOBP20, and CdieOBP21 in female antennae were significantly higher than in male antennae, and CdieOBP19 was only expressed in female antennae, indicating that these genes may play the role of binding sex pheromones. Odorant-binding proteins are the main carriers of chemical signals as aggregation, alarm, and sex pheromones. ApisOBP3 and ApisOBP7 in adults and nymphs of A. pisum carry the alarm pheromone (E)-β-farnesene (Zhang et al., 2017). Most weevil sex pheromones are released by males, such as boll weevils and pecan weevils, the sex pheromones of boll weevils are alpha-pinene, limonene, beta-caryophyllene, and beta-bisabolol (Tumlinson et al., 1969). The sex pheromones of pecan weevils are delta3-21Hy, deta1,3-22Hy, delta1-23Hy, and delta1-24Hy (Hedin et al., 1979). Because CdieOBP9, CdieOBP20, and CdieOBP21 are also expressed in males, it is speculated that they may function as pheromone carriers for male sexual behavior. OBPs showed an average of 58.26% homology with other insects, while the homology value for CSPs reached 70.83%. The reason for this discrepancy is that, as compared with OBPs, CSPs have four highly conserved cysteines and lower binding specificity but were more flexible in recognizing different ligands (Sánchez-Gracia et al., 2009; Pelosi et al., 2018). We found that one CSP (CdieCSP15) was highly expressed in female antennae; The function of this CSP requires further investigation.
Moreover, because SNMPs were discovered in ORNs, they may play important roles in insect olfactory recognition (Nichols and Vogt, 2008). SNMP1 and SNMP2 were identified in C. dieckmanni transcriptomes and in other insect species (Hildebbrand, 1996). Hylogenetic analysis showed that CdieSNMP1 and CdieSNMP2 clustered with the SNMP1 and SNMP2 of other Curculionoidea species (S. zeamais, I. typographus, C. formicarius, and D. ponderosae) and were most closely related to D. ponderosae. The RPKM and qRT-PCR results showed that no SNMPs were differentially expressed between males and females.
Olfactory sensilla in insect antennae are controlled by two or more ORNs, which express a range of chemosensory receptor protein families and are involved in the detection and transduction of odorant signals (Benton et al., 2006; Sato and Touhara, 2009). These proteins mainly include ORs, IRs, and GRs (Benton et al., 2009; Koenig et al., 2015). Our study identified a total of 15 ORs in C. dieckmanni transcriptomes, which was lower than that of D. ponderosae and I. typographus (49 and 43) (Andersson et al., 2013). The difference in the number of OR genes may be a result of the method and depth of sequencing. By comparing the complete amino acid sequences, we identified the main OR subfamily and ORco genes from D. ponderosae, I. typographus, C. formicarius, and S. oryzae. Phylogenetic analyses showed that 11 ORs of C. dieckmanni (except CdieOR1, CdieOR3, and CdieOR4) were contained in six groups (1, 2, 3, 4, 5, and 7, with no CdieORs in group 6 of the nine beetle groups) (Mitchell et al., 2020), which is consistent with other Curculionoidea species, including D. ponderosae, I. typographus, C. formicarius, and S. oryzae (Andersson et al., 2013; Bin et al., 2017). One OR (CdieOR15) clustered in a branch with the highly conserved Orco of D. Ponderosae, C. formicarius, and S. oryzae. They may serve the same role in odorant detection. The qRT-PCR results showed that two ORs (CdieOR13 and CdieOR15) were significantly female-biased. CdieOR15 may serve a different function in males and females when detecting specific odorants, for example when a female is looking for a male or for a place to lay eggs (Jacquin-Joly and Merlin, 2004; Benton et al., 2006; Sato and Touhara 2009). CdieOR13 clustered on the branch of group 7 together with DponOR33, DponOR34, and ItypOR33. It may play a role in the female detection of sex pheromones released from males (Antony et al., 2021; Yuvaraj et al., 2021). This needs to be verified by further ligand binding studies.
IRs are involved in odorant detection (Benton et al., 2009; Croset et al., 2010; Abuin et al., 2011). CdieIR10 clustered in the IR8a branch, CdieIR11 clustered in the IR25a branch, and IR8a and IR25a are co-expressed with IRs and essential for the functioning of IRs (Benton et al., 2009; Rytz et al., 2013; Joseph and Carlson, 2015; Fleischer et al., 2018). CdieIR2 clustered in IR75p, CdieIR4 clustered in IR75s, CdieIR13 clustered in IR76b, CdieIR15 clustered in IR76q, CdieIR17 clustered in IR75c, and CdieIR21 clustered in IR41a. These results may suggest that IRs of the same branch may play the same role in the antennae. GRs play key roles in detecting gustatory chemicals that govern insect feeding, courtship, mating, and the location of spawning sites (Freeman et al., 2014; Sollai et al., 2018). We identified 9 GRs in C. dieckmanni antennae transcriptomes. This number was lower than the number of ORs, presumably because antennae are not major gustatory organs. The phylogenetic tree showed that CdieGR1 clustered in the CO2 branch and may be involved in CO2 detection. Interestingly, CdieGR4, CdieGR7, and CdieGR8 clustered in the sugar branch, and presumably play a role in sugar detection. CdieGR4 and DponGR3 clustered in the same branch, CdieGR6 and ItypGR6 clustered in the same branch, CdieGR9 and CforGR7 clustered in the same branch, and CforGR4, CforGR5, CforGR6, LoryGR10, SoryGR3-like, and CdieGR2 clustered in the same branch. GRs on the same branch may have the same function.
In this study, a total of 44,024 unigenes were obtained from the antennae of female and male adults of C. dieckmanni using high-throughput sequencing. Our transcriptome analysis identified 87 olfactory genes in antennae, including 23 OBPs, 15 CSPs, 2 SNMPs, 15 ORs, 23 IRs, and 9 GRs. Male- and female-specific chemosensory genes are possibly involved in odorant detection and recognition, and their expression was validated by qRT-PCR. Among the chemosensory genes, we found significant differences in the expression of CdieOBP8, CdieOBP9, CdieOBP19, CdieOBP20, CdieOBP21, CdieCSP15, CdieOR13, and CdieOR15 between adult male and female C. dieckmanni. These results add to the known chemosensory genes of Curculionidae and contribute to further understand the olfactory and gustatory mechanisms of C. dieckmanni. Our results may provide new insights for the control of this pest.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA769532.
Ethical review and approval was not required for the animal study because Adult C. dieckmanni were collected in a hazelnut orchard in the Sandao Forestry Farm, Mudanjiang City, Heilongjiang Province, China (129°39′02″E, 44°07′04"N).The breeding and experimentation of these weevils are faithfully recorded in the manuscript. It was collected with the permission of the Hazel Garden owner. The collected insects did not come from national parks or protected nature reserves. Also, these weevils are definitely not an endangered species.
Insect rearing and antenna collection, XM, PZ, XD, HP, and HZ. Conceived and designed the research, XM and LM. Conducted the experiments, XM and XL. Collected and analyzed the data, XM, XL, and QW. Wrote the manuscript, XM. Reviewed and edited the manuscript, XM, LM, and WM. All authors have read and agreed to the published version of the manuscript.
This research was supported and funded by the National key Research and Development project of China (2016YFC0500308), Heilongjiang Provincial Natural Science Foundation of China (C2018055), the Scientific research fund project of Heilongjiang Provincial-administrated Scientific research institutes, China (CZKYF 2021C03) and the Application Research Project of Heilongjiang Provincial Forest Industry Administration, China (sgzjY2014004, sgzjY2016015).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2022.896793/full#supplementary-material
Abuin L., Bargeton B., Ulbrich M. H., Isacoff E. Y., Kellenberger S., Benton R. (2011). Functional Architecture of Olfactory Ionotropic Glutamate Receptors. Neuron 69 (1), 44–60. doi:10.1016/j.neuron.2010.11.042
Ahmad M., Hollingworth R. M., Wise J. C. (2002). Broad-Spectrum Insecticide Resistance in Obliquebanded LeafrollerChoristoneura Rosaceana (Lepidoptera: Tortricidae) from Michigan. Pest Manag. Sci. 58 (8), 834–838. doi:10.1002/ps.531
Andersson M. N., Grosse-Wilde E., Keeling C. I., Bengtsson J. M., Yuen M. M., Li M., et al. (2013). Antennal Transcriptome Analysis of the Chemosensory Gene Families in the Tree Killing Bark Beetles, Ips Typographus and Dendroctonus Ponderosae (Coleoptera: Curculionidae: Scolytinae). BMC Genomics 14 (14), 198–2164. doi:10.1186/1471-2164-14-198
Andersson M. N., Keeling C. I., Mitchell R. F. (2019). Genomic Content of Chemosensory Genes Correlates with Host Range in wood-boring Beetles (Dendroctonus Ponderosae, Agrilus Planipennis, and Anoplophora Glabripennis). BMC Genomics 20, 690. doi:10.1186/s12864-019-6054-x
Announcement of the State Forestry Administration of China (2013). National Forestry Dangerous Pests List. (No.4 of 2013). Available at: http://www.forestry.gov.cn/portal/main/s/72/content-580486.html.
Antony B., Johny J., Montagné N., Jacquin‐Joly E., Capoduro R., Cali K., et al. (2021). Pheromone Receptor of the Globally Invasive Quarantine Pest of the palm Tree, the Red Palm Weevil ( Rhynchophorus Ferrugineus ). Mol. Ecol. 30 (9), 2025–2039. doi:10.1111/mec.15874
Benton R., Sachse S., Michnick S. W., Vosshall L. B. (2006). Atypical Membrane Topology and Heteromeric Function of Drosophila Odorant Receptors In Vivo. Plos Biol. 4 (2), e20–57. doi:10.1371/journal.pbio.0040020
Benton R., Vannice K. S., Gomez-Diaz C., Vosshall L. B. (2009). Variant Ionotropic Glutamate Receptors as Chemosensory Receptors in Drosophila. Cell 136 (1), 149–162. doi:10.1016/j.cell.2008.12.001
Bin S.-Y., Qu M.-Q., Pu X.-H., Wu Z.-Z., Lin J.-T. (2017). Antennal Transcriptome and Expression Analyses of Olfactory Genes in the Sweetpotato Weevil Cylas Formicarius. Sci. Rep. 7 (1), 11073. doi:10.1038/s41598-017-11456-x
Chai R.-Y., Lee C.-Y. (2010). Insecticide Resistance Profiles and Synergism in Field Populations of the German Cockroach (Dictyoptera: Blattellidae) From Singapore. J. Econo Entomol. 103 (2), 460–471. doi:10.1603/ec09284
Clyne P. J., Warr C. G., Carlson J. R. (2000). Candidate Taste Receptors in Drosophila. Science 287 (5459), 1830–1834. doi:10.1126/science.287.5459.1830
Clyne P. J., Warr C. G., Freeman M. R., Lessing D., Kim J., Carlson J. R. (1999). A Novel Family of Divergent Seven-Transmembrane Proteins. Neuron 22, 327–338. doi:10.1016/S0896-6273(00)81093-4
Conesa A., Gotz S., Garcia-Gomez J. M., Terol J., Talon M., Robles M. (2005). Blast2GO: a Universal Tool for Annotation, Visualization and Analysis in Functional Genomics Research. Bioinformatics 21 (18), 3674–3676. doi:10.1093/bioinformatics/bti610
Croset V., Rytz R., Cummins S. F., Budd A., Brawand D., Kaessmann H., et al. (2010). Ancient Protostome Origin of Chemosensory Ionotropic Glutamate Receptors and the Evolution of Insect Taste and Olfaction. Plos Genet. 6 (8), e1001064. doi:10.1371/journal.pgen.1001064
Dippel S., Oberhofer G., Kahnt J., Gerischer L., Opitz L., Schachtner J., et al. (2014). Tissue-specific Transcriptomics, Chromosomal Localization, and Phylogeny of Chemosensory and Odorant Binding Proteins from the Red Flour Beetle Tribolium castaneum Reveal Subgroup Specificities for Olfaction or More General Functions. BMC Genomics 15, 1141. doi:10.1186/1471-2164-15-1141
Downes S., Kriticos D., Parry H., Paull C., Schellhorn N., Zalucki M. P. (2016). A Perspective on Management ofHelicoverpa Armigera: Transgenic Bt Cotton, IPM, and Landscapes. Pest Manag. Sci. 73 (3), 485–492. doi:10.1002/ps.4461
Engsontia P., Sanderson A. P., Cobb M., Walden K. K. O., Robertson H. M., Brown S. (2008). The Red Flour Beetle's Large Nose: An Expanded Odorant Receptor Gene Family in Tribolium castaneum. Insect Biochem. Mol. Biol. 38 (4), 387–397. doi:10.1016/j.ibmb.2007.10.005
Field L. M., Pickett J. A., Wadhams L. J. (2000). Molecular Studies in Insect Olfaction. Insect Mol. Biol. 9 (6), 545–551. doi:10.1046/j.1365-2583.2000.00221.x
Fleischer J., Pregitzer P., Breer H., Krieger J. (2018). Access to the Odor World: Olfactory Receptors and Their Role for Signal Transduction in Insects. Cell. Mol. Life Sci. 75, 485–508. doi:10.1007/s00018-017-2627-5
Freeman E. G., Wisotsky Z., Dahanukar A. (2014). Detection of Sweet Tastants by a Conserved Group of Insect Gustatory Receptors. Proc. Natl. Acad. Sci. U.S.A. 111 (4), 1598–1603. doi:10.1073/pnas.1311724111
Grabherr M. G., Haas B. J., Yassour M., Levin J. Z., Thompson D. A., Amit I., et al. (2011). Full-Length Transcriptome Assembly from RNA-Seq Data without a Reference Genome. Nat. Biotechnol. 29, 644–652. doi:10.1038/nbt.1883
Guo W., Bhaskara Rao M. (2008). On Optimality of the Benjamini-Hochberg Procedure for the False Discovery Rate. Stat. Probab. Lett. 78 (14), 2024–2030. doi:10.1016/j.spl.2008.01.069
Guo W., Wang X., Ma Z., Xue L., Han J., Yu D., et al. (2011). CSP and Takeout Genes Modulate the Switch between Attraction and Repulsion during Behavioral Phase Change in the Migratory Locust. Plos Genet. 7 (2), e1001291. doi:10.1371/journal.pgen.1001291
Hedin P. A., Payne J. A., Carpenter T. L., Neel W. (1979). Sex Pheromones of the Male and Female Pecan Weevil, Curculio Caryae1 : Behavioral and Chemical Studies 23. Environ. Entomol. 8 (3), 521–523. doi:10.1093/ee/8.3.521
Hildebrand J. G. (1996). Olfactory Control of Behavior in Moths: central Processing of Odor Information and the Functional Significance of Olfactory Glomeruli. J. Comp. Physiol. A. 178 (1), 5–19. doi:10.1007/BF00189586
Hou X.-Q., Yuvaraj J. K., Roberts R. E., Zhang D.-D., Unelius C. R., Löfstedt C., et al. (2021). Functional Evolution of a Bark Beetle Odorant Receptor Clade Detecting Monoterpenoids of Different Ecological Origins. Mol. Biol. Evol. 38 (11), 4934–4947. doi:10.1093/molbev/msab218
Jacquin-Joly E., Merlin C. (2004). Insect Olfactory Receptors: Contributions of Molecular Biology to Chemical Ecology. J. Chem. Ecol. 30, 2359–2397. doi:10.1007/s10886-004-7941-3
Jones D. T., Taylor W. R., Thornton J. M. (1992). The Rapid Generation of Mutation Data Matrices from Protein Sequences. Bioinformatics 8, 275–282. doi:10.1093/bioinformatics/8.3.275
Joseph R. M., Carlson J. R. (2015). Drosophila Chemoreceptors: A Molecular Interface Between the Chemical World and the Brain. Trends Genet. 31 (12), 683–695. doi:10.1016/j.tig.2015.09.005
Keil T. A., Steinbrecht R. A. (1984). Mechanosensitive and Olfactory Sensilla of InsectsInsect Ultrastructure. New York: Plenum Press, 477–516.
Koenig C., Hirsh A., Bucks S., Klinner C., Vogel H., Shukla A., et al. (2015). A Reference Gene Set for Chemosensory Receptor Genes of Manduca Sexta. Insect Biochem. Mol. Biol. 66, 51–63. doi:10.1016/j.ibmb.2015.09.007
Kumar S., Stecher G., Li M., Knyaz C., Tamura K. (2018). MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 35, 1547–1549. doi:10.1093/molbev/msy096
Leal W. S. (2013). Odorant Reception in Insects: Roles of Receptors, Binding Proteins, and Degrading Enzymes. Annu. Rev. Entomol. 58, 373–391. doi:10.1146/annurev-ento-120811-153635
Liu Z., Liang X.-F., Xu L., Keesey I. W., Lei Z.-R., Smagghe G., et al. (2020). An Antennae-specific Odorant-Binding Protein Is Involved in Bactrocera Dorsalis Olfaction. Front. Ecol. Evol. 8, 63. doi:10.3389/fevo.2020.00063
Mang D., Shu M., Endo H., Yoshizawa Y., Nagata S., Kikuta S., et al. (2016). Expression of a Sugar Clade Gustatory Receptor, BmGr6, in the Oral Sensory Organs, Midgut, and central Nervous System of Larvae of the Silkworm Bombyx mori. Insect Biochem. Mol. Biol. 70, 85–98. doi:10.1016/j.ibmb.2015.12.008
Mckenna D. D., Scully E. D., Pauchet Y., Hoover K., Kirsch R., Geib S. M., et al. (2016). Genome of the Asian Longhorned Beetle (Anoplophora Glabripennis), a Globally Significant Invasive Species, Reveals key Functional and Evolutionary Innovations at the Beetle–Plant Interface. Genome Biol. 17 (1), 227. doi:10.1186/s13059-016-1088-8
Mckenna D. D., Shin S., Ahrens D., Balke M., Beza-Beza C., Clarke D. J., et al. (2019). The Evolution and Genomic Basis of Beetle Diversity. Proc. Natl. Acad. Sci. U.S.A. 116 (49), 24729–24737. doi:10.1073/pnas.1909655116
Mitchell R. F., Andersson M. N. (2021). Olfactory Genomics of the Coleoptera. Insect Pheromone Biochem. Mol. Biol. 2021, 547–590. doi:10.1016/B978-0-12-819628-1.00017-1
Mitchell R. F., Hall L. P., Reagel P. F., McKenna D. D., Baker T. C., Hildebrand J. G. (2017). Odorant Receptors and Antennal Lobe Morphology Offer a New Approach to Understanding Olfaction in the Asian Longhorned Beetle. J. Comp. Physiol. A. 203 (2), 99–109. doi:10.1007/s00359-016-1138-4
Mitchell R. F., Hughes D. T., Luetje C. W., Millar J. G., Soriano-Agatón F., Hanks L. M., et al. (2012). Sequencing and Characterizing Odorant Receptors of the Cerambycid Beetle Megacyllene Caryae. Insect Biochem. Mol. Biol. 42 (7), 499–505. doi:10.1016/j.ibmb.2012.03.007
Mitchell R. F., Schneider T. M., Schwartz A. M., Andersson M. N., McKenna D. D. (2020). The Diversity and Evolution of Odorant Receptors in Beetles (Coleoptera). Insect Mol. Biol. 29, 77–91. doi:10.1111/imb.12611
Mortazavi A., Williams B. A., Mccue K., Schaeffer L., Wold B. (2008). Mapping and Quantifying Mammalian Transcriptomes by RNA-Seq. Nat. Methods 5, 621–628. doi:10.1038/nmeth.1226
Nichols Z., Vogt R. G. (2008). The SNMP/CD36 Gene Family in Diptera, Hymenoptera and Coleoptera: Drosophila melanogaster, D. pseudoobscura, Anopheles gambiae, Aedes aegypti, Apis mellifera, and Tribolium castaneum. Insect Biochem. Mol. Biol. 38 (4), 398–415. doi:10.1016/j.ibmb.2007.11.003
Park J.-H., Kwon J. Y. (2011). Heterogeneous Expression of Drosophila Gustatory Receptors in Enteroendocrine Cells. Plos one 6, e29022. doi:10.1371/journal.pone.0029022
Paula D. P., Togawa R. C., do Carmo Costa M. M., Grynberg P., Martins N. F., Andow D. A. (2018). Systemic and Sex-Biased Regulation of OBP Expression under Semiochemical Stimuli. Sci. Rep. 8, 6035. doi:10.1038/s41598-018-24297-z
Pelosi P., Iovinella I., Felicioli A., Dani F. R. (2014). Soluble Proteins of Chemical Communication: an Overview across Arthropods. Front. Physiol. 5 (320), 320. doi:10.3389/fphys.2014.00320
Pelosi P., Iovinella I., Zhu J., Wang G., Dani F. R. (2018). Beyond Chemoreception: Diverse Tasks of Soluble Olfactory Proteins in Insects. Biol. Rev. 93, 184–200. doi:10.1111/brv.12339
Pelosi P., Zhou J.-J., Ban L. P., Calvello M. (2006). Soluble Proteins in Insect Chemical Communication. Cell. Mol. Life Sci. 63, 1658–1676. doi:10.1007/s00018-005-5607-0
Ran L. y., Jiang C. x., Yang Q. f., Wang H. j., Bai ma T. z., Chen L., et al. (2020). Comparative Transcriptome Analysis of Locusta migratoria Tibetensis Chen (Orthoptera: Oedipodidae) Under High‐ and Low‐temperature Stress. J. Appl. Entomol. 144 (3), 181–190. doi:10.1111/jen.12728
Richards S., Richards S., Gibbs R. A., Weinstock G. M., Brown S. J., Denell R., et al. (2008). The Genome of the Model Beetle and Pest Tribolium castaneum. Nature 452, 949–955. doi:10.1038/nature06784
Rytz R., Croset V., Benton R. (2013). Ionotropic Receptors (IRs): Chemosensory Ionotropic Glutamate Receptors in Drosophila and beyond. Insect Biochem. Mol. Biol. 43 (9), 888–897. doi:10.1016/j.ibmb.2013.02.007
Sánchez-Gracia A., Vieira F. G., Rozas J. (2009). Molecular Evolution of the Major Chemosensory Gene Families in Insects. Heredity 103 (3), 208–216. doi:10.1038/hdy.2009.55
Sato K., Touhara K. (2008). Insect Olfaction: Receptors, Signal Transduction, and Behavior. Results Probl. Cell Differ 7, 203–220. doi:10.1007/400_2008_10
Schoville S. D., Chen Y. H., Andersson M. N., Benoit J. B., Bhandari A., Bowsher J. H., et al. (2018). A Model Species for Agricultural Pest Genomics: the Genome of the Colorado Potato Beetle, Leptinotarsa decemlineata (Coleoptera: Chrysomelidae). Sci. Rep. 8 (1), 1931. doi:10.1101/19264110.1038/s41598-018-20154-1
Smadja C., Butlin R. K. (2009). On the Scent of Speciation: the Chemosensory System and its Role in Premating Isolation. Heredity 102 (1), 77–97. doi:10.1038/hdy.2008.55
Sollai G., Biolchini M., Crnjar R. (2018). Taste Receptor Plasticity in Relation to Feeding History in Two Congeneric Species of Papilionidae (Lepidoptera). J. Insect Physiol. 107, 41–56. doi:10.1016/j.jinsphys.2018.02.007
Tian Z., Sun L., Li Y., Quan L., Zhang H., Yan W., et al. (2018). Antennal Transcriptome Analysis of the Chemosensory Gene Families in Carposina Sasakii (Lepidoptera: Carposinidae). BMC Genomics 19 (19), 3–16. doi:10.1186/s12864-018-4900-x
Tumlinson J. H., Hardee D. D., Gueldner R. C., Thompson A. C., Hedin P. A., Minyard J. P. (1969). Sex Pheromones Produced by Male Boll Weevil: Isolation, Identification, and Synthesis. Science 166 (21), 1010–1012. doi:10.1126/science.166.3908.1010
Vogt R. G., Miller N. E., Litvack R., Fandino R. A., Sparks J., Staples J., et al. (2009). The Insect SNMP Gene Family. Insect Biochem. Mol. Biol. 39, 448–456. doi:10.1016/j.ibmb.2009.03.007
Vosshall L. B., Amrein H., Morozov P. S., Rzhetsky A., Axel R. (1999). A Spatial Map of Olfactory Receptor Expression in the Drosophila Antenna. Cell 96, 725–736. doi:10.1016/S0092-8674(00)80582-6
Wanner K. W., Isman M. B., Feng Q., Plettner E., Theilmann D. A. (2005). Developmental Expression Patterns of Four Chemosensory Protein Genes from the Eastern spruce Budworm, Chroistoneura Fumiferana. Insect Mol. Biol. 14 (3), 289–300. doi:10.1111/j.1365-2583.2005.00559.x
Xiao S., Sun J. S., Carlson J. R. (2019). Robust Olfactory Responses in the Absence of Odorant Binding Proteins. eLife 8, e51040. doi:10.7554/eLife.51040
Ye J., Fang L., Zheng H., Zhang Y., Chen J., Zhang Z., et al. (2006). WEGO: a Web Tool for Plotting GO Annotations. Nucleic Acids Res. 34, W293–W297. doi:10.1093/nar/gkl031
Yuvaraj J. K., Roberts R. E., Sonntag Y., Hou X.-Q., Grosse-Wilde E., Machara A., et al. (2021). Putative Ligand Binding Sites of Two Functionally Characterized Bark Beetle Odorant Receptors. BMC Biol. 19, 16. doi:10.1186/s12915-020-00946-6
Zhang P., Zhao Q., Ma X., Ma L. (2021). Pathogenicity of Serratia marcescens to Hazelnut Weevil (Curculio Dieckmanni). J. For. Res. 32, 409–417. doi:10.1007/s11676-020-01096-9
Zhang R., Wang B., Grossi G., Falabella P., Liu Y., Yan S., et al. (2017). Molecular Basis of Alarm Pheromone Detection in Aphids. Curr. Biol. 27, 55–61. doi:10.1016/j.cub.2016.10.013
Zhang X., Yang S., Zhang J., Wang X., Wang S., Liu M., et al. (2019). Identification and Expression Analysis of Candidate Chemosensory Receptors Based on the Antennal Transcriptome of Lissorhoptrus Oryzophilus. Comp. Biochem. Physiol. D: Genomics Proteomics 30, 133–142. doi:10.1016/j.cbd.2019.02.007
Keywords: curculionoidea, antennal transcriptome, chemosensory genes, control strategies, curculio dieckmanni
Citation: Ma X, Lu X, Zhang P, Deng X, Bai J, Xu Z, Diao J, Pang H, Wang Q, Zhao H, Ma W and Ma L (2022) Transcriptome Analysis of Antennal Chemosensory Genes in Curculio Dieckmanni Faust. (Coleoptera: Curculionidae). Front. Physiol. 13:896793. doi: 10.3389/fphys.2022.896793
Received: 18 March 2022; Accepted: 18 April 2022;
Published: 09 May 2022.
Edited by:
Peng He, Guizhou University, ChinaReviewed by:
Sun Ya-Lan, Henan University of Science and Technology, ChinaCopyright © 2022 Ma, Lu, Zhang, Deng, Bai, Xu, Diao, Pang, Wang, Zhao, Ma and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Ma, bWF3ZWlAaGxqdWNtLm5ldA==; Ling Ma, bWFsaW5nNjNAMTYzLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.