- 1Department of Pediatrics, University of California, San Diego, San Diego, CA, United States
- 2Division of Gastroenterology, Hepatology and Nutrition, Rady Children’s Hospital San Diego, San Diego, CA, United States
Editorial on the Research Topic
The Microbiome in Hepatobiliary and Intestinal Disease
Introduction
This Editorial provides a brief overview of the changes in the intestinal microbiome with focus on the bacterial microbiome in a wide range of diseases affecting the human digestive system and then highlights the specific articles of this Research Topic.
Gut Microbiome Changes in Diseases Affecting the Human Digestive System
Essentially all diseases affecting the human digestive system are associated with significant increases and decreases of sub-populations of the gut microbiome compared with controls subjects; recurrent changes of the intestinal bacterial microbiome observed across 30 different conditions of the human digestive system are summarized in Figure 1. Briefly, Streptococcus (Llopis et al., 2016; Maccioni et al., 2020; Gao et al., 2021), Actinomyces (Ciocan et al., 2018; Maccioni et al., 2020; Gao et al., 2021), and Rothia (Ciocan et al., 2018; Maccioni et al., 2020) are increased, whereas Faecalibacterium (prausnitzii) (Gao et al., 2020a; Gao et al., 2020b; Maccioni et al., 2020) and Bacteroides (Puri et al., 2018; Gao et al., 2020a; Maccioni et al., 2020; Gao et al., 2021) are decreased in abundance in alcohol-associated liver disease (ALD). Faecalibacterium (prausnitzii) (Wong et al., 2013; Da Silva et al., 2018; Oh et al., 2020a) is also detected at diminished concentrations in non-alcoholic fatty liver disease (NAFLD) similar to Coprococcus (Zhu et al., 2013; Wang et al., 2016; Da Silva et al., 2018), whereas Escherichia (coli) (Zhu et al., 2013; Jiang et al., 2015; Oh et al., 2020a) and Lactobacillus (Raman et al., 2013; Jiang et al., 2015; Da Silva et al., 2018) are increased in NAFLD. Liver cirrhosis is associated with elevated intestinal levels of Enterococcus (faecalis) (Zhao et al., 2004; Chen et al., 2011; Bajaj et al., 2012), Prevotella (Qin et al., 2014; Chen et al., 2016; Shao et al., 2018; Ponziani et al., 2019; Zeng et al., 2020), Clostridium (Zhao et al., 2004; Chen et al., 2011; Bajaj et al., 2012; Qin et al., 2014; Heidrich et al., 2018; Shao et al., 2018), Veillonella (Qin et al., 2014; Chen et al., 2016; Shao et al., 2018; Oh et al., 2020a; Zeng et al., 2020), Lactobacillus (Qin et al., 2014; Heidrich et al., 2018; Shao et al., 2018; Ponziani et al., 2019; Zeng et al., 2020), Atopobium (Chen et al., 2016; Ponziani et al., 2019; Zeng et al., 2020), and Streptococcus (Qin et al., 2014; Shao et al., 2018; Ponziani et al., 2019; Oh et al., 2020a), and with reduced levels of Dorea (Bajaj et al., 2012; Oh et al., 2020a), Alistipes (Qin et al., 2014; Shao et al., 2018; Oh et al., 2020a), and Subdoligranulum (Bajaj et al., 2012; Qin et al., 2014; Shao et al., 2018). Gut microbiome changes in hepatocellular carcinoma (HCC) are similar to the ones identified in liver cirrhosis, indicating their relationship also on a microbial level. HCC is frequently linked to high intestinal amounts of Bacteroides (Ponziani et al., 2019; Huang et al., 2020; Zeng et al., 2020), Enterococcus (Ni et al., 2019; Ponziani et al., 2019; Xin et al., 2019), Veillonella (Ni et al., 2019; Zeng et al., 2020), and Atopobium (Ni et al., 2019; Zeng et al., 2020), and low amounts of Ruminococcus (Ren et al., 2019; Zeng et al., 2020), Alistipes (Ren et al., 2019; Huang et al., 2020), and Bifidobacterium (Ponziani et al., 2019; Xin et al., 2019). Hepatitis B is associated with increased Actinomyces (Wang et al., 2017; Yao et al., 2021), Megamonas (Wang et al., 2017; Joo et al., 2021), Enterococcus (faecalis) (Lu et al., 2011; Yao et al., 2021), Veillonella (Yang et al., 2020; Zeng et al., 2020; Yao et al., 2021), Streptococcus (Yang et al., 2020; Yao et al., 2021), and Atopobium (Zeng et al., 2020; Yao et al., 2021), and decreased Bifidobacterium spp. (Lu et al., 2011; Xu et al., 2012), Faecalibacterium (prausnitzii) (Lu et al., 2011; Yang et al., 2020), Parabacteroides (Wang et al., 2017; Yao et al., 2021), and Ruminococcus (Wang et al., 2017; Yao et al., 2021) in abundance, whereas patients with hepatitis C have been found enriched in Prevotella (Aly et al., 2016; Sultan et al., 2021a), Lactobacillus (Heidrich et al., 2018; Inoue et al., 2018), Streptococcus (Heidrich et al., 2018; Inoue et al., 2018), and Veillonella (Aly et al., 2016; Heidrich et al., 2018), and deplete of Ruminococcus (Aly et al., 2016; Mohieldeen et al., 2021) and Butyricimonas (Aly et al., 2016; Heidrich et al., 2018) compared with control subjects.
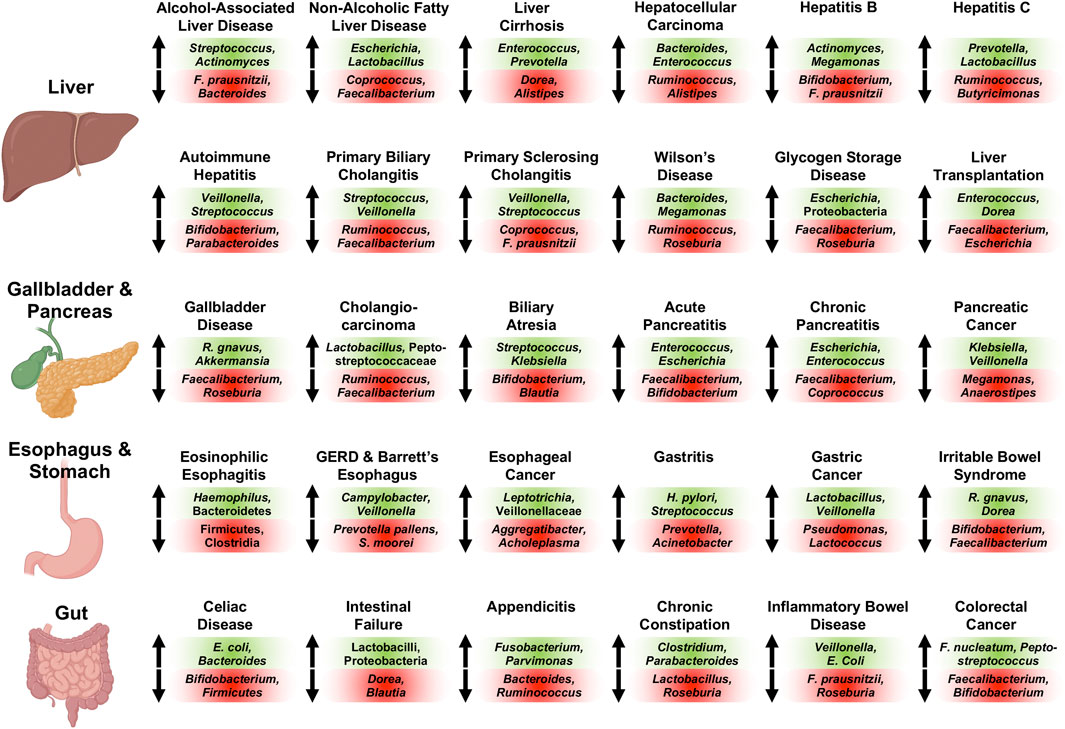
FIGURE 1. Representative associated intestinal bacterial microbiome changes across diseases of the human digestive system with focus on genera. Changes described here were observed in various publications for the respective conditions as detailed in the manuscript. Bacterial populations on green background are increased in abundance and populations on red background are decreased in abundance for the respective disease. Of note, Veillonella and Veillonellaceae, Streptococcus, Lactobacillus and Lactobacilli, Escherichia (coli), Enterococcus (faecalis and faecium), and Klebsiella are oftentimes found at increased concentrations, whereas Faecalibacterium (prausnitzii), Ruminococcus and Ruminococcaceae (except Ruminococcus gnavus), Bifidobacterium, Roseburia, and Coprococcus are frequently decreased in abundance in various digestive diseases. Created with a license from Biorender.com. E. coli, Escherichia coli; F. nucleatum, Fusobacterium nucleatum; F. prausnitzii, Faecalibacterium prausnitzii; GERD, gastroesophageal reflux disease; R. gnavus, Ruminococcus gnavus; S. moorei, Solobacterium moorei.
The various autoimmune liver diseases exhibit similar gut microbiome alterations: Stool samples of patients with autoimmune hepatitis are characterized by large quantities of Veillonella (Elsherbiny et al., 2020; Liwinski et al., 2020; Lou et al., 2020; Wei et al., 2020), Streptococcus (Elsherbiny et al., 2020; Liwinski et al., 2020; Wei et al., 2020), Haemophilus (Elsherbiny et al., 2020; Lou et al., 2020), and Klebsiella (Lou et al., 2020; Wei et al., 2020), and small quantities of Bifidobacterium (Lin et al., 2015; Liwinski et al., 2020), Parabacteroides (Elsherbiny et al., 2020; Lou et al., 2020; Wei et al., 2020), and Ruminococcaceae (Lou et al., 2020; Wei et al., 2020). Primary biliary cholangitis is associated with an elevated abundance of Streptococcus (Lv et al., 2016; Tang et al., 2018; Furukawa et al., 2020), Veillonella (Lv et al., 2016; Abe et al., 2018; Tang et al., 2018), Klebsiella (Lv et al., 2016; Tang et al., 2018), Haemophilus (Lv et al., 2016; Tang et al., 2018), and Lactobacillus (Tang et al., 2018; Furukawa et al., 2020), and diminished Ruminococcus/Ruminococcaceae (Lv et al., 2016; Furukawa et al., 2020) and Faecalibacterium (Tang et al., 2018; Furukawa et al., 2020), while primary sclerosing cholangitis is linked to enlarged proportions of Veillonella (Bajer et al., 2017; Kummen et al., 2017; Rühlemann et al., 2019; Cortez et al., 2020; Lapidot et al., 2021), Streptococcus (Sabino et al., 2016; Bajer et al., 2017; Rühlemann et al., 2019; Lapidot et al., 2021), and Enterococcus (Sabino et al., 2016; Bajer et al., 2017), as well as depressed proportions of Coprococcus (Bajer et al., 2017; Kummen et al., 2017; Rühlemann et al., 2019; Kummen et al., 2021), Faecalibacterium prausnitzii (Bajer et al., 2017; Lapidot et al., 2021), and Lachnospiraceae (Bajer et al., 2017; Kummen et al., 2017; Kummen et al., 2021; Lapidot et al., 2021). Enriched gut microbiota in Wilson’s Disease are Bacteroides (Geng et al., 2018; Cai et al., 2020) and Megamonas (Geng et al., 2018; Cai et al., 2020), whereas Ruminococcus (Cai et al., 2020) and Roseburia (Geng et al., 2018) are reduced. Glycogen storage disease shows overrepresented intestinal Escherichia (Colonetti et al., 2019; Ceccarani et al., 2020) and Proteobacteria (Colonetti et al., 2019; Ceccarani et al., 2020), and underrepresented Faecalibacterium (Colonetti et al., 2019; Ceccarani et al., 2020) and Roseburia (Colonetti et al., 2019; Ceccarani et al., 2020). Liver transplantation results in expansion of Enterococcus spp. (Wu et al., 2012; Annavajhala et al., 2019; Song et al., 2021a), Dorea (Bajaj et al., 2018; Annavajhala et al., 2019), Blautia (Sun et al., 2017; Bajaj et al., 2018; Song et al., 2021a), and Streptococcus (Bajaj et al., 2018; Annavajhala et al., 2019), and reduction of Faecalibacterium (prausnitzii) (Wu et al., 2012; Annavajhala et al., 2019; Lu et al., 2019; Song et al., 2021a), Escherichia (Bajaj et al., 2017; Sun et al., 2017; Bajaj et al., 2018), Shigella (Bajaj et al., 2017; Sun et al., 2017; Bajaj et al., 2018), and Bifidobacterium (Wu et al., 2012; Bajaj et al., 2018).
Gallbladder disease is linked to enriched Ruminococcus gnavus (Wang et al., 2020a; Zhang et al., 2021a) and Akkermansia (Liu et al., 2015; Zhang et al., 2021a), and depleted Faecalibacterium (Wu et al., 2013; Wang et al., 2020a), Roseburia (Wu et al., 2013; Keren et al., 2015; Zhang et al., 2021a), and Prevotella 9 (Wang et al., 2020a; Zhang et al., 2021a). Cholangiocarcinoma is associated with enlarged fecal proportions of Lactobacillus (Jia et al., 2020; Zhang et al., 2021a) and Peptostreptococcaceae (Jia et al., 2020; Zhang et al., 2021a), and smaller proportions of Ruminococcus (Jia et al., 2020; Zhang et al., 2021a) and Faecalibacterium (Zhang et al., 2021a). The intestinal contributions of Streptococcus (Wang et al., 2020b; Song et al., 2021b) and Klebsiella (Wang et al., 2020b; Song et al., 2021b) are increased in biliary atresia, and those of Bifidobacterium (Wang et al., 2020b; Song et al., 2021b), Blautia (Wang et al., 2020b; Song et al., 2021b), and Faecalibacterium (Wang et al., 2020b; Song et al., 2021b) are decreased. The microbial changes occurring in acute and chronic pancreatitis compared with controls are similar: Acute pancreatitis is characterized by overrepresentation of Enterococcus (faecalis) (Zhu et al., 2019; Yu et al., 2020), Escherichia (coli) (Zhu et al., 2019; Yu et al., 2020), and Enterobacteriaceae (Tan et al., 2015; Zhu et al., 2019), and underrepresentation of Faecalibacterium (Zhu et al., 2019; Yu et al., 2020), Bifidobacterium (Zhu et al., 2019; Yu et al., 2020), and Blautia (Zhu et al., 2019; Yu et al., 2020), whereas chronic pancreatitis exhibits elevated fecal amounts of Escherichia (coli) (Savitskaia et al., 2002; Zhou et al., 2020a; Frost et al., 2020) and Enterococcus (faecalis and faecium) (Savitskaia et al., 2002; Frost et al., 2020), and lower amounts of Faecalibacterium (prausnitzii) (Jandhyala et al., 2017; Zhou et al., 2020a; Wang et al., 2020c; Frost et al., 2020), Coprococcus (Zhou et al., 2020a; Frost et al., 2020), Subdoligranulum (Zhou et al., 2020a; Wang et al., 2020c), and Collinsella (Zhou et al., 2020a; Wang et al., 2020c). Pancreatic cancer is associated with an expansion of Klebsiella (Ren et al., 2017; Pushalkar et al., 2018; Matsukawa et al., 2021), Veillonella/Veillonellaceae (Ren et al., 2017; Pushalkar et al., 2018; Half et al., 2019), Parabacteroides (Pushalkar et al., 2018; Matsukawa et al., 2021), and Lactobacillus (Ren et al., 2017; Matsukawa et al., 2021), as well as reduced Megamonas (Ren et al., 2017; Pushalkar et al., 2018), Anaerostipes (Ren et al., 2017; Pushalkar et al., 2018; Half et al., 2019), Dorea (Ren et al., 2017; Pushalkar et al., 2018), and Firmicutes (Ren et al., 2017; Matsukawa et al., 2021).
Eosinophilic esophagitis is linked to high abundances of Haemophilus (Harris et al., 2015; Hiremath et al., 2019) and Bacteroidetes (Benitez et al., 2015; Kashyap et al., 2019; Laserna-Mendieta et al., 2021), and low abundances of Firmicutes (Benitez et al., 2015; Kashyap et al., 2019) and Clostridia (Kashyap et al., 2019). Elevated concentrations of Campylobacter (concisus) (Macfarlane et al., 2007; Blackett et al., 2013; Deshpande et al., 2018; Snider et al., 2019) and Veillonella (Liu et al., 2013; Deshpande et al., 2018; Snider et al., 2018), and small quantities of Prevotella pallens (Snider et al., 2019; Kawar et al., 2021) and Solobacterium moorei (Zhou et al., 2020b; Kawar et al., 2021) can be detected in gastroesophageal reflux disease (GERD) and Barrett’s Esophagus. Esophageal adeno- and squamous cell carcinoma are associated with increased amounts of Leptotrichia (Lopetuso et al., 2020; Zhao et al., 2020), Veillonellaceae (Li et al., 2020a; Lopetuso et al., 2020; Zhao et al., 2020), and Bifidobacterium (Zhou et al., 2020b; Lopetuso et al., 2020; Zhao et al., 2020), and decreased amounts of Aggregatibacter (Chen et al., 2015; Zhao et al., 2020) and Acholeplasma (Chen et al., 2015; Zhao et al., 2020). Gastritis is characterized by enriched Helicobacter pylori (Parsons et al., 2017; Yang et al., 2019; Ndegwa et al., 2020) and Streptococcus (Li et al., 2009; Gao et al., 2018; Cui et al., 2019), and depleted Prevotella (Parsons et al., 2017; Cui et al., 2019; Ndegwa et al., 2020) and Acinetobacter (Parsons et al., 2017; Cui et al., 2019; Ndegwa et al., 2020). In gastric cancer, Lactobacillus (Qi et al., 2019; Wang et al., 2020d; Gantuya et al., 2020) and Veillonella (Castaño-Rodríguez et al., 2017; Qi et al., 2019; Wang et al., 2020d) are increased, whereas Pseudomonas (Wang et al., 2020d; Gantuya et al., 2020) and Lactococcus (Chen et al., 2019; Gunathilake et al., 2019; Wang et al., 2020d) are decreased. Irritable bowel syndrome is linked to overrepresented Ruminococcus gnavus (Rajilić–Stojanović et al., 2011; Rangel et al., 2015) and Dorea (formicigenerans) (Rajilić–Stojanović et al., 2011; Rangel et al., 2015; Maharshak et al., 2018), and underrepresented Bifidobacterium (catenulatum) (Malinen et al., 2005; Kerckhoffs et al., 2009; Rajilić–Stojanović et al., 2011) and Faecalibacterium (prausnitzii) (Carroll et al., 2012; Rangel et al., 2015; Maharshak et al., 2018).
Celiac disease is associated with enlarged proportions of Escherichia coli (Nadal et al., 2007; Collado et al., 2009; Schippa et al., 2010), Bacteroides (fragilis and vulgatus) (Nadal et al., 2007; Collado et al., 2009; De Palma et al., 2010; Schippa et al., 2010; Sánchez et al., 2012), and Staphylococcus (Collado et al., 2009; Sánchez et al., 2013), and contracted contributions of Bifidobacterium (Sanz et al., 2007; Collado et al., 2009; De Palma et al., 2010) and Firmicutes (Sánchez et al., 2013; Iaffaldano et al., 2018). The gut microbiome of patients with intestinal failure is enriched in Lactobacillus/Lactobacilli (Joly et al., 2010; Korpela et al., 2017) and Proteobacteria (Davidovics et al., 2016; Korpela et al., 2017), and diminished in Dorea (Huang et al., 2017; Piper et al., 2017) and Blautia (Huang et al., 2017; Piper et al., 2017). Acute appendicitis is characterized by elevated levels of Fusobacterium (Swidsinski et al., 2011; Guinane et al., 2013; Jackson et al., 2014; Zhong et al., 2014; Rogers et al., 2016), Parvimonas (Guinane et al., 2013; Jackson et al., 2014; Zhong et al., 2014; Rogers et al., 2016), Campylobacter jejuni (Campbell et al., 2006; Oh et al., 2020b), and Gemella (Guinane et al., 2013; Zhong et al., 2014), and reduced levels of Bacteroides (Swidsinski et al., 2011; Samuelsson et al., 2013; Zhong et al., 2014; Rogers et al., 2016), Ruminococcus (Samuelsson et al., 2013; Munakata et al., 2021), and Faecalibacterium (prausnitzii) (Swidsinski et al., 2011; Samuelsson et al., 2013). The gut microbiome signature of chronic constipation consists of large proportions of Clostridium (Zoppi et al., 1998; Zhu et al., 2014) and Parabacteroides (de Meij et al., 2016; Li et al., 2020b), and depressed amounts of Lactobacillus (Khalif et al., 2005; Moraes et al., 2016; Jomehzadeh et al., 2020) and Roseburia (Mancabelli et al., 2017; Li et al., 2020b). Stool analysis of patients with inflammatory bowel disease (IBD) frequently demonstrates overrepresentation of Veillonella (Gevers et al., 2014; Mottawea et al., 2016; Santoru et al., 2017; Schirmer et al., 2018) and Escherichia coli (Schwiertz et al., 2010; Sha et al., 2013; Gevers et al., 2014; Santoru et al., 2017), and underrepresentation of Faecalibacterium (prausnitzii) (Schwiertz et al., 2010; Joossens et al., 2011; Morgan et al., 2012; Kumari et al., 2013; Gevers et al., 2014; Machiels et al., 2014; Schirmer et al., 2018), and Roseburia (Morgan et al., 2012; Kumari et al., 2013; Rajilić-Stojanović et al., 2013; Gevers et al., 2014; Machiels et al., 2014). Fusobacterium (nucleatum) (Kostic et al., 2012; Ahn et al., 2013; Warren et al., 2013; Zeller et al., 2014; Gao et al., 2015; Mira-Pascual et al., 2015; Yu et al., 2017; Dai et al., 2018; Guo et al., 2018) is commonly enriched in colorectal cancer along with Peptostreptococcus (Wang et al., 2012; Ahn et al., 2013; Zeller et al., 2014; Gao et al., 2015; Yu et al., 2017), whereas Faecalibacterium (prausnitzii) (Balamurugan et al., 2008; Kostic et al., 2012; Guo et al., 2018) and Bifidobacterium (Mira-Pascual et al., 2015; Dai et al., 2018; Guo et al., 2018) are depressed in number.
Fungi, viruses, and other non-bacterial populations are also detected at aberrant proportions in disorders of the digestive system, e.g., the fungus Candida albicans is increased in ALD (Lang et al., 2020a; Hartmann et al., 2021), NAFLD (Demir et al., 2021), gastric cancer (Zhong et al., 2021), IBD (Sokol et al., 2017), and colorectal cancer. (Starý et al., 2020) Viruses have also been correlated with disease activity in alcoholic hepatitis (Jiang et al., 2020) and NAFLD (Lang et al., 2020b) among others. Archaea have been investigated as well, Methanosphaera stadtmaniae (Blais Lecours et al., 2014) has been found to be more abundant and Methanobrevibacter smithii (Ghavami et al., 2018) has been found to be depleted in the gut microbiome of patients with IBD.
Disease Association Index
When evaluating the microbiome findings of these 30 disorders of the digestive system above, striking observations can be made: Faecalibacterium (prausnitzii) is associated with 16 of these conditions, and this bacterium is decreased in all of these 16 conditions. In contrast, the fecal abundance of Veillonella and Veillonellaceae is increased in 13 out of 13 diseases and that of Streptococcus is increased in 10 out of 10 conditions in which a robust association has been demonstrated. To evaluate how likely a microbial population is increased or decreased across diseases that it is associated with, a Disease Association Index (DAI) can be calculated by dividing the increased-decreased net value (= the number of diseases in which the microbial population is increased minus the number of diseases in which the microbial population is decreased) by the total number of conditions that the population has been associated with. E.g. the DAI for Faecalibacterium (prausnitzii) among the digestive diseases discussed above is −1 (= (0–16)/16); the DAI for Veillonella and Veillonellaceae as well as for Streptococcus is +1 (= (13–0)/13) and +1 (= (10–0)/10), respectively. The DAI ranges from −1 to +1; the higher the value the more likely the microbial population to be increased in the evaluated diseases, and the lower the more likely that the population is decreased in the analyzed conditions. The closer the DAI is to 0, the more ambivalent is the microbial population. A DAI of +0.6 or higher, and a DAI of −0.6 or lower indicates that the abundance of a microbe can be considered highly positively or negatively correlated with disease, respectively. Ruminococcus has a DAI of −0.6 (2 increases/8 decreases), indicating that it is predominantly decreased in digestive diseases (of note, the species Ruminococcus gnavus is responsible for both increased abundances of Ruminococcus in these diseases, see above). Additional notable DAIs: Bifidobacterium −0.8 (1 increase/9 decreases), Lactobacillus/Lactobacilli +0.78 (8/1), Escherichia (coli) +0.71 (6/1), Enterococcus (faecalis/faecium) +1 (7/0), Dorea −0.2 (2/3), Prevotella −0.2 (2/3), Bacteroides (fragilis/vulgatus) +0.2 (3/2), Roseburia −1 (0/5), and Klebsiella +1 (4/0).
Targeted repletion trials for bacterial populations that are predominantly decreased (such as repletion of Faecalibacterium (prausnitzii) in a murine colitis model), (Martín et al., 2014) or targeted elimination trials for populations that are predominantly increased (such as elimination of cytolytic Enterococcus faecalis via targeted bacteriophages in a murine model of alcohol-induced liver disease) (Duan et al., 2019) could be attempted in the future.
Articles in this Research Topic
The articles in this Research Topic are very diverse and wide-ranging. Zheng et al. describe microbiome and metabolite differences in various autoimmune liver diseases (Zheng et al.). Warner et al. characterize the role of human beta defensin-2 in alcohol-induced liver injury in mice (Warner et al.). Chen et al. discuss the role of the microbiota in the pathogenesis of chemical-induced acute liver injury models in rodents and the protective use of probiotics herein (Chen et al.). Zhang et al. demonstrate that hepatic branch vagotomy results in decreased dysbiosis but increased hepatic steatosis and continued neuro-inflammation in murine cirrhosis secondary to carbon tetrachloride injections (Zhang et al.). Song et al. analyzed changes of the gut microbiome in patients with biliary atresia after liver transplantation (Song et al.). Chen et al. report microbiome and metabolite shifts in a mouse model of gallstone disease (Chen et al.). Rao et al. discuss microbiome changes in cholangiocarcinoma and related precancerous conditions (Rao et al.). Jihan et al. identified specific microbiome signatures in cancers affecting the esophagus, stomach, colon, and rectum (Wang et al.). Busing et al. review various changes in the microbiota and metabolism in eosinophilic esophagitis (Busing et al.). Wang et al. associate intratumor microbiome signatures with subtype, tumor stage, and survival in patients with esophageal carcinoma (Wang et al.). Hu et al. discuss alterations seen in the gut microbiota in food allergies and other allergic conditions (Hu et al.). Chu et al. use a variety of mouse models to induce gastritis and analyze the associated modulations of the intestinal microflora (Chu et al.). Sultan et al. discuss metabolite alterations associated with intestinal dysbiosis in IBD (Sultan et al.). Houshyar et al. review what is known about the role of fungi and archaea in IBD (Houshyar et al.). Zhao et al. evaluate the role of gut bacteria in a rat model of intra-abdominal hypertension (Zhao et al.). Montanari et al. detail the relationship of an pro-inflammatory state and gut dysbiosis, and the effects of diet and medications on the gut microbiota observed in disorders of inborn errors of metabolism (Montanari et al.). Lastly, Li et al. introduce Amadis, a comprehensive, manually curated database that documents experimentally supported microbiota-disease associations (Li et al.).
Author Contributions
PH was responsible for the preparation of the manuscript.
Funding
The manuscript was supported by NIH grant K12 HD000850 to PH.
Conflict of Interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The author wants to thank Bernd Schnabl for helpful comments in preparing the manuscript.
References
Abe K., Takahashi A., Fujita M., Imaizumi H., Hayashi M., Okai K., et al. (2018). Dysbiosis of Oral Microbiota and its Association with Salivary Immunological Biomarkers in Autoimmune Liver Disease. PLoS One 13 (7), e0198757. Epub 20180703PubMed PMID: 29969462. doi:10.1371/journal.pone.0198757
Ahn J., Sinha R., Pei Z., Dominianni C., Wu J., Shi J., et al. (2013). Human Gut Microbiome and Risk for Colorectal Cancer. J. Natl. Cancer Inst. 105 (24), 1907–1911. Epub 20131206PubMed PMID: 24316595 ; PMC3866154. doi:10.1093/jnci/djt300
Aly A. M., Adel A., El-Gendy A. O., Essam T. M., Aziz R. K. (2016). Gut Microbiome Alterations in Patients with Stage 4 Hepatitis C. Gut Pathog. 8 (1), 42. Epub 20160913PubMed PMID: 27625705. doi:10.1186/s13099-016-0124-2
Annavajhala M. K., Gomez-Simmonds A., Macesic N., Sullivan S. B., Kress A., Khan S. D., et al. (2019). Colonizing Multidrug-Resistant Bacteria and the Longitudinal Evolution of the Intestinal Microbiome after Liver Transplantation. Nat. Commun. 10 (1), 4715. Epub 20191017PubMed PMID: 31624266 ; PMC6797753. doi:10.1038/s41467-019-12633-4
Bajaj J. S., Fagan A., Sikaroodi M., White M. B., Sterling R. K., Gilles H., et al. (2017). Liver Transplant Modulates Gut Microbial Dysbiosis and Cognitive Function in Cirrhosis. Liver Transpl. 23 (7), 907–914. PubMed PMID: 28240840. doi:10.1002/lt.24754
Bajaj J. S., Hylemon P. B., Ridlon J. M., Heuman D. M., Daita K., White M. B., et al. (2012). Colonic Mucosal Microbiome Differs from Stool Microbiome in Cirrhosis and Hepatic Encephalopathy and Is Linked to Cognition and Inflammation. Am. J. Physiology-Gastrointestinal Liver Physiol. 303 (6), G675–G685. Epub 20120719PubMed PMID: 22821944. doi:10.1152/ajpgi.00152.2012
Bajaj J. S., Kakiyama G., Cox I. J., Nittono H., Takei H., White M., et al. (2018). Alterations in Gut Microbial Function Following Liver Transplant. Liver Transpl. 24, 752–761. Epub 20180513PubMed PMID: 29500907. doi:10.1002/lt.25046
Bajer L., Kverka M., Kostovcik M., Macinga P., Dvorak J., Stehlikova Z., et al. (2017). Distinct Gut Microbiota Profiles in Patients with Primary Sclerosing Cholangitis and Ulcerative Colitis. Wjg 23 (25), 4548–4558. PubMed PMID: 28740343. doi:10.3748/wjg.v23.i25.4548
Balamurugan R., Rajendiran E., George S., Samuel G. V., Ramakrishna B. S. (2008). Real-time Polymerase Chain Reaction Quantification of Specific Butyrate-Producing bacteria,DesulfovibrioandEnterococcus Faecalisin the Feces of Patients with Colorectal Cancer. J. Gastroenterol. Hepatol. 23 (8 Pt 1), 1298–1303. Epub 20080708PubMed PMID: 18624900. doi:10.1111/j.1440-1746.2008.05490.x
Benitez A. J., Hoffmann C., Muir A. B., Dods K. K., Spergel J. M., Bushman F. D., et al. (2015). Inflammation-associated Microbiota in Pediatric Eosinophilic Esophagitis. Microbiome 3, 23. Epub 20150601PubMed PMID: 26034601 ; PMC4450515. doi:10.1186/s40168-015-0085-6
Blackett K. L., Siddhi S. S., Cleary S., Steed H., Miller M. H., Macfarlane S., et al. (2013). Oesophageal Bacterial Biofilm Changes in Gastro-Oesophageal Reflux Disease, Barrett's and Oesophageal Carcinoma: Association or Causality? Aliment. Pharmacol. Ther. 37 (11), 1084–1092. Epub 20130422PubMed PMID: 23600758. doi:10.1111/apt.12317
Blais Lecours P., Marsolais D., Cormier Y., Berberi M., Haché C., Bourdages R., et al. (2014). Increased Prevalence of Methanosphaera Stadtmanae in Inflammatory Bowel Diseases. PLoS One 9 (2), e87734. Epub 20140203PubMed PMID: 24498365. doi:10.1371/journal.pone.0087734
Cai X., Deng L., Ma X., Guo Y., Feng Z., Liu M., et al. (2020). Altered Diversity and Composition of Gut Microbiota in Wilson's Disease. Sci. Rep. 10 (1), 21825. Epub 20201211PubMed PMID: 33311635 ; PMC7732847. doi:10.1038/s41598-020-78988-7
Campbell L. K., Havens J. M., Scott M. A., Lamps L. W. (2006). Molecular Detection of Campylobacter Jejuni in Archival Cases of Acute Appendicitis. Mod. Pathol. 19 (8), 1042–1046. Epub 20060519PubMed PMID: 16715071. doi:10.1038/modpathol.3800640
Carroll I. M., Ringel-Kulka T., Siddle J. P., Ringel Y. (2012). Alterations in Composition and Diversity of the Intestinal Microbiota in Patients with Diarrhea-Predominant Irritable Bowel Syndrome. Neurogastroenterol Motil. 24 (6), 521–e248. e248Epub 20120220PubMed PMID: 22339879. doi:10.1111/j.1365-2982.2012.01891.x
Castaño-Rodríguez N., Goh K.-L., Fock K. M., Mitchell H. M., Kaakoush N. O. (2017). Dysbiosis of the Microbiome in Gastric Carcinogenesis. Sci. Rep. 7 (1), 15957. Epub 20171121PubMed PMID: 29162924 ; PMC5698432. doi:10.1038/s41598-017-16289-2
Ceccarani C., Bassanini G., Montanari C., Casiraghi M. C., Ottaviano E., Morace G., et al. (2020). Proteobacteria Overgrowth and Butyrate-Producing Taxa Depletion in the Gut Microbiota of Glycogen Storage Disease Type 1 Patients. Metabolites 10 (4), 133. Epub 20200330PubMed PMID: 32235604. doi:10.3390/metabo10040133
Chen X.-H., Wang A., Chu A.-N., Gong Y.-H., Yuan Y. (2019). Mucosa-Associated Microbiota in Gastric Cancer Tissues Compared with Non-cancer Tissues. Front. Microbiol. 10, 1261. Epub 20190605PubMed PMID: 31231345. doi:10.3389/fmicb.2019.01261
Chen X., Winckler B., Lu M., Cheng H., Yuan Z., Yang Y., et al. (2015). Oral Microbiota and Risk for Esophageal Squamous Cell Carcinoma in a High-Risk Area of China. PLoS One 10 (12), e0143603. Epub 20151207PubMed PMID: 26641451. doi:10.1371/journal.pone.0143603
Chen Y., Ji F., Guo J., Shi D., Fang D., Li L. (2016). Dysbiosis of Small Intestinal Microbiota in Liver Cirrhosis and its Association with Etiology. Sci. Rep. 6, 34055. Epub 20160930PubMed PMID: 27687977. doi:10.1038/srep34055
Chen Y., Yang F., Lu H., Wang B., Chen Y., Lei D., et al. (2011). Characterization of Fecal Microbial Communities in Patients with Liver Cirrhosis. Hepatology 54 (2), 562–572. Epub 20110626PubMed PMID: 21574172. doi:10.1002/hep.24423
Ciocan D., Voican C. S., Wrzosek L., Hugot C., Rainteau D., Humbert L., et al. (2018). Bile Acid Homeostasis and Intestinal Dysbiosis in Alcoholic Hepatitis. Aliment. Pharmacol. Ther. 48 (9), 961–974. Epub 20180824PubMed PMID: 30144108. doi:10.1111/apt.14949
Collado M. C., Donat E., Ribes-Koninckx C., Calabuig M., Sanz Y. (2009). Specific Duodenal and Faecal Bacterial Groups Associated with Paediatric Coeliac Disease. J. Clin. Pathol. 62 (3), 264–269. Epub 20081107PubMed PMID: 18996905. doi:10.1136/jcp.2008.061366
Colonetti K., Bento Dos Santos B., Nalin T., Moura de Souza C. F., Triplett E. W., Dobbler P. T., et al. (2019). Hepatic Glycogen Storage Diseases Are Associated to Microbial Dysbiosis. PLoS One 14 (4), e0214582. Epub 20190402PubMed PMID: 30939160. doi:10.1371/journal.pone.0214582
Cortez R. V., Moreira L. N., Padilha M., Bibas M. D., Toma R. K., Porta G., et al. (2020). Gut Microbiome of Children and Adolescents with Primary Sclerosing Cholangitis in Association with Ulcerative Colitis. Front. Immunol. 11, 598152. Epub 20210205PubMed PMID: 33613519 ; PMC7893080. doi:10.3389/fimmu.2020.598152
Cui J., Cui H., Yang M., Du S., Li J., Li Y., et al. (2019). Tongue Coating Microbiome as a Potential Biomarker for Gastritis Including Precancerous cascade. Protein Cell 10 (7), 496–509. Epub 20181126PubMed PMID: 30478535 ; PMC6588651. doi:10.1007/s13238-018-0596-6
Da Silva H. E., Teterina A., Comelli E. M., Taibi A., Arendt B. M., Fischer S. E., et al. (2018). Nonalcoholic Fatty Liver Disease Is Associated with Dysbiosis Independent of Body Mass index and Insulin Resistance. Sci. Rep. 8 (1), 1466. Epub 20180123PubMed PMID: 29362454 ; PMC5780381. doi:10.1038/s41598-018-19753-9
Dai Z., Coker O. O., Nakatsu G., Wu W. K. K., Zhao L., Chen Z., et al. (2018). Multi-cohort Analysis of Colorectal Cancer Metagenome Identified Altered Bacteria across Populations and Universal Bacterial Markers. Microbiome 6 (1), 70. Epub 20180411PubMed PMID: 29642940 ; PMC5896039. doi:10.1186/s40168-018-0451-2
Davidovics Z. H., Carter B. A., Luna R. A., Hollister E. B., Shulman R. J., Versalovic J. (2016). The Fecal Microbiome in Pediatric Patients with Short Bowel Syndrome. J. Parenter. Enteral Nutr. 40 (8), 1106–1113. Epub 20150609PubMed PMID: 26059898 ; PMC4874906. doi:10.1177/0148607115591216
de Meij T. G. J., de Groot E. F. J., Eck A., Budding A. E., Kneepkens C. M. F., Benninga M. A., et al. (2016). Characterization of Microbiota in Children with Chronic Functional Constipation. PLoS One 11 (10), e0164731. Epub 20161019PubMed PMID: 27760208. doi:10.1371/journal.pone.0164731
De Palma G., Nadal I., Medina M., Donat E., Ribes-Koninckx C., Calabuig M., et al. (2010). Intestinal Dysbiosis and Reduced Immunoglobulin-Coated Bacteria Associated with Coeliac Disease in Children. BMC Microbiol. 10, 63. Epub 20100224PubMed PMID: 20181275 ; PMC2843610. doi:10.1186/1471-2180-10-63
Demir M., Lang S., Hartmann P., Duan Y., Martin A., Miyamoto Y., et al. (2021). The Fecal Mycobiome in Non-alcoholic Fatty Liver Disease. J. Hepatol. Epub 20211209PubMed PMID: 34896404. doi:10.1016/j.jhep.2021.11.029
Deshpande N. P., Riordan S. M., Castaño-Rodríguez N., Wilkins M. R., Kaakoush N. O. (2018). Signatures within the Esophageal Microbiome Are Associated with Host Genetics, Age, and Disease. Microbiome 6 (1), 227. Epub 20181217PubMed PMID: 30558669 ; PMC6297961. doi:10.1186/s40168-018-0611-4
Duan Y., Llorente C., Lang S., Brandl K., Chu H., Jiang L., et al. (2019). Bacteriophage Targeting of Gut Bacterium Attenuates Alcoholic Liver Disease. Nature 575 (7783), 505–511. Epub 20191113PubMed PMID: 31723265 ; PMC6872939. doi:10.1038/s41586-019-1742-x
Elsherbiny N. M., Rammadan M., Hassan E. A., Ali M. E., El-Rehim A. S. A., Abbas W. A., et al. (2020). Autoimmune Hepatitis: Shifts in Gut Microbiota and Metabolic Pathways Among Egyptian Patients. Microorganisms 8 (7), 1011. Epub 20200706PubMed PMID: 32640728 ; PMC7409351. doi:10.3390/microorganisms8071011
Frost F., Weiss F. U., Sendler M., Kacprowski T., Rühlemann M., Bang C., et al. (2020). The Gut Microbiome in Patients with Chronic Pancreatitis Is Characterized by Significant Dysbiosis and Overgrowth by Opportunistic Pathogens. Clin. Translational Gastroenterol. 11 (9), e00232. PubMed PMID: 33094959 ; PMC7494146. doi:10.14309/ctg.0000000000000232
Furukawa M., Moriya K., Nakayama J., Inoue T., Momoda R., Kawaratani H., et al. (2020). Gut Dysbiosis Associated with Clinical Prognosis of Patients with Primary Biliary Cholangitis. Hepatol. Res. 50 (7), 840–852. Epub 20200610PubMed PMID: 32346970. doi:10.1111/hepr.13509
Gantuya B., El Serag H. B., Matsumoto T., Ajami N. J., Uchida T., Oyuntsetseg K., et al. (2020). Gastric Mucosal Microbiota in a Mongolian Population with Gastric Cancer and Precursor Conditions. Aliment. Pharmacol. Ther. 51 (8), 770–780. Epub 20200304PubMed PMID: 32133670 ; PMC8761497. doi:10.1111/apt.15675
Gao B., Duan Y., Lang S., Barupal D., Wu T. C., Valdiviez L., et al. (2020). Functional Microbiomics Reveals Alterations of the Gut Microbiome and Host Co‐Metabolism in Patients with Alcoholic Hepatitis. Hepatol. Commun. 4 (8), 1168–1182. Epub 20200619PubMed PMID: 32766476 ; PMC7395072. doi:10.1002/hep4.1537
Gao B., Emami A., Zhou R., Lang S., Duan Y., Wang Y., et al. (2020). Functional Microbial Responses to Alcohol Abstinence in Patients with Alcohol Use Disorder. Front. Physiol. 11, 370. Epub 20200424PubMed PMID: 32390870. doi:10.3389/fphys.2020.00370
Gao B., Zhu Y., Gao N., Shen W., Stärkel P., Schnabl B. (2021). Integrative Analysis of Metabolome and Microbiome in Patients with Progressive Alcohol-Associated Liver Disease. Metabolites 11 (11), 766. Epub 20211110PubMed PMID: 34822424. doi:10.3390/metabo11110766
Gao J.-J., Zhang Y., Gerhard M., Mejias-Luque R., Zhang L., Vieth M., et al. (2018). Association between Gut Microbiota and Helicobacter Pylori-Related Gastric Lesions in a High-Risk Population of Gastric Cancer. Front. Cel. Infect. Microbiol. 8, 202. Epub 20180619PubMed PMID: 29971220. doi:10.3389/fcimb.2018.00202
Gao Z., Guo B., Gao R., Zhu Q., Qin H. (2015). Microbiota Disbiosis Is Associated with Colorectal Cancer. Front. Microbiol. 6, 20. Epub 20150202PubMed PMID: 25699023. doi:10.3389/fmicb.2015.00020
Geng H., Shu S., Dong J., Li H., Xu C., Han Y., et al. (2018). Association Study of Gut flora in Wilson's Disease through High-Throughput Sequencing. Medicine (Baltimore) 97 (31), e11743. PubMed PMID: 30075590. doi:10.1097/MD.0000000000011743
Gevers D., Kugathasan S., Denson L. A., Vázquez-Baeza Y., Van Treuren W., Ren B., et al. (2014). The Treatment-Naive Microbiome in New-Onset Crohn's Disease. Cell Host & Microbe 15 (3), 382–392. PubMed PMID: 24629344. doi:10.1016/j.chom.2014.02.005
Ghavami S. B., Rostami E., Sephay A. A., Shahrokh S., Balaii H., Aghdaei H. A., et al. (2018). Alterations of the Human Gut Methanobrevibacter Smithii as a Biomarker for Inflammatory Bowel Diseases. Microb. Pathogenesis 117, 285–289. Epub 20180222PubMed PMID: 29477743. doi:10.1016/j.micpath.2018.01.029
Guinane C. M., Tadrous A., Fouhy F., Ryan C. A., Dempsey E. M., Murphy B., et al. (2013). Microbial Composition of Human Appendices from Patients Following Appendectomy. mBio 4 (1). Epub 20130115PubMed PMID: 23322636 ; PMC3551545. doi:10.1128/mBio.00366-12
Gunathilake M. N., Lee J., Choi I. J., Kim Y.-I., Ahn Y., Park C., et al. (2019). Association between the Relative Abundance of Gastric Microbiota and the Risk of Gastric Cancer: a Case-Control Study. Sci. Rep. 9 (1), 13589. Epub 20190919PubMed PMID: 31537876 ; PMC6753194. doi:10.1038/s41598-019-50054-x
Guo S., Li L., Xu B., Li M., Zeng Q., Xiao H., et al. (2018). A Simple and Novel Fecal Biomarker for Colorectal Cancer: Ratio of Fusobacterium Nucleatum to Probiotics Populations, Based on Their Antagonistic Effect. Clin. Chem. 64 (9), 1327–1337. Epub 20180618PubMed PMID: 29914865. doi:10.1373/clinchem.2018.289728
Half E., Keren N., Reshef L., Dorfman T., Lachter I., Kluger Y., et al. (2019). Fecal Microbiome Signatures of Pancreatic Cancer Patients. Sci. Rep. 9 (1), 16801. Epub 20191114PubMed PMID: 31727922 ; PMC6856127. doi:10.1038/s41598-019-53041-4
Harris J. K., Fang R., Wagner B. D., Choe H. N., Kelly C. J., Schroeder S., et al. (2015). Esophageal Microbiome in Eosinophilic Esophagitis. PLoS One 10 (5), e0128346. Epub 20150528PubMed PMID: 26020633. doi:10.1371/journal.pone.0128346
Hartmann P., Lang S., Zeng S., Duan Y., Zhang X., Wang Y., et al. (2021). Dynamic Changes of the Fungal Microbiome in Alcohol Use Disorder. Front. Physiol. 12, 699253. Epub 20210719PubMed PMID: 34349667. doi:10.3389/fphys.2021.699253
Heidrich B., Vital M., Plumeier I., Döscher N., Kahl S., Kirschner J., et al. (2018). Intestinal Microbiota in Patients with Chronic Hepatitis C with and without Cirrhosis Compared with Healthy Controls. Liver Int. 38 (1), 50–58. Epub 20170620PubMed PMID: 28561276. doi:10.1111/liv.13485
Hiremath G., Shilts M. H., Boone H. H., Correa H., Acra S., Tovchigrechko A., et al. (2019). The Salivary Microbiome Is Altered in Children with Eosinophilic Esophagitis and Correlates with Disease Activity. Clin. Translational Gastroenterol. 10 (6), e00039. PubMed PMID: 31107724. doi:10.14309/ctg.0000000000000039
Huang H., Ren Z., Gao X., Hu X., Zhou Y., Jiang J., et al. (2020). Integrated Analysis of Microbiome and Host Transcriptome Reveals Correlations between Gut Microbiota and Clinical Outcomes in HBV-Related Hepatocellular Carcinoma. Genome Med. 12 (1), 102. Epub 20201123PubMed PMID: 33225985 ; PMC7682083. doi:10.1186/s13073-020-00796-5
Huang Y., Guo F., Li Y., Wang J., Li J. (2017). Fecal Microbiota Signatures of Adult Patients with Different Types of Short Bowel Syndrome. J. Gastroenterol. Hepatol. 32 (12), 1949–1957. PubMed PMID: 28425133. doi:10.1111/jgh.13806
Iaffaldano L., Granata I., Pagliuca C., Esposito M. V., Casaburi G., Salerno G., et al. (2018). Oropharyngeal Microbiome Evaluation Highlights Neisseria Abundance in Active Celiac Patients. Sci. Rep. 8 (1), 11047. Epub 20180723PubMed PMID: 30038321 ; PMC6056421. doi:10.1038/s41598-018-29443-1
Inoue T., Nakayama J., Moriya K., Kawaratani H., Momoda R., Ito K., et al. (2018). Gut Dysbiosis Associated with Hepatitis C Virus Infection. Clin. Infect. Dis. 67 (6), 869–877. PubMed PMID: 29718124. doi:10.1093/cid/ciy205
Jackson H. T., Mongodin E. F., Davenport K. P., Fraser C. M., Sandler A. D., Zeichner S. L. (2014). Culture-independent Evaluation of the Appendix and Rectum Microbiomes in Children with and without Appendicitis. PLoS One 9 (4), e95414. Epub 20140423PubMed PMID: 24759879. doi:10.1371/journal.pone.0095414
Jandhyala S. M., Madhulika A., Deepika G., Rao G. V., Reddy D. N., Subramanyam C., et al. (2017). Altered Intestinal Microbiota in Patients with Chronic Pancreatitis: Implications in Diabetes and Metabolic Abnormalities. Sci. Rep. 7, 43640. Epub 20170303PubMed PMID: 28255158. doi:10.1038/srep43640
Jia X., Lu S., Zeng Z., Liu Q., Dong Z., Chen Y., et al. (2020). Characterization of Gut Microbiota, Bile Acid Metabolism, and Cytokines in Intrahepatic Cholangiocarcinoma. Hepatology 71 (3), 893–906. Epub 20190819PubMed PMID: 31298745. doi:10.1002/hep.30852
Jiang L., Lang S., Duan Y., Zhang X., Gao B., Chopyk J., et al. (2020). Intestinal Virome in Patients with Alcoholic Hepatitis. Hepatology 72 (6), 2182–2196. Epub 20201010PubMed PMID: 32654263. doi:10.1002/hep.31459
Jiang W., Wu N., Wang X., Chi Y., Zhang Y., Qiu X., et al. (2015). Dysbiosis Gut Microbiota Associated with Inflammation and Impaired Mucosal Immune Function in Intestine of Humans with Non-alcoholic Fatty Liver Disease. Sci. Rep. 5, 8096. Epub 20150203PubMed PMID: 25644696. doi:10.1038/srep08096
Joly F., Mayeur C., Bruneau A., Noordine M.-L., Meylheuc T., Langella P., et al. (2010). Drastic Changes in Fecal and Mucosa-Associated Microbiota in Adult Patients with Short Bowel Syndrome. Biochimie 92 (7), 753–761. Epub 20100219PubMed PMID: 20172013. doi:10.1016/j.biochi.2010.02.015
Jomehzadeh N., Javaherizadeh H., Amin M., Rashno M., Teimoori A. (2020). Quantification of Intestinal Lactobacillus Species in Children with Functional Constipation by Quantitative Real-Time PCR. Ceg Vol. 13, 141–150. Epub 20200505PubMed PMID: 32440191. doi:10.2147/CEG.S250755
Joo E.-J., Cheong H. S., Kwon M.-J., Sohn W., Kim H.-N., Cho Y. K. (2021). Relationship between Gut Microbiome Diversity and Hepatitis B Viral Load in Patients with Chronic Hepatitis B. Gut Pathog. 13 (1), 65. Epub 20211030PubMed PMID: 34717727 ; PMC8557478. doi:10.1186/s13099-021-00461-1
Joossens M., Huys G., Cnockaert M., De Preter V., Verbeke K., Rutgeerts P., et al. (2011). Dysbiosis of the Faecal Microbiota in Patients with Crohn's Disease and Their Unaffected Relatives. Gut 60 (5), 631–637. Epub 20110105PubMed PMID: 21209126. doi:10.1136/gut.2010.223263
Kashyap P. C., Johnson S., Geno D. M., Lekatz H. R., Lavey C., Alexander J. A., et al. (2019). A Decreased Abundance of Clostridia Characterizes the Gut Microbiota in Eosinophilic Esophagitis. Physiol. Rep. 7 (20), e14261. PubMed PMID: 31650712. doi:10.14814/phy2.14261
Kawar N., Park S. G., Schwartz J. L., Callahan N., Obrez A., Yang B., et al. (2021). Salivary Microbiome with Gastroesophageal Reflux Disease and Treatment. Sci. Rep. 11 (1), 188. Epub 20210108PubMed PMID: 33420219 ; PMC7794605. doi:10.1038/s41598-020-80170-y
Kerckhoffs A. P., Samsom M., Rest M. E. v. d., Vogel J. d., Knol J., Ben-Amor K., et al. (2009). Lower Bifidobacteria Counts in Both Duodenal Mucosa-Associated and Fecal Microbiota in Irritable Bowel Syndrome Patients. Wjg 15, 2887–2892. PubMed PMID: 19533811. doi:10.3748/wjg.15.2887
Keren N., Konikoff F. M., Paitan Y., Gabay G., Reshef L., Naftali T., et al. (2015). Interactions between the Intestinal Microbiota and Bile Acids in Gallstones Patients. Environ. Microbiol. Rep. 7 (6), 874–880. Epub 20150807PubMed PMID: 26149537. doi:10.1111/1758-2229.12319
Khalif I., Quigley E., Konovitch E., Maximova I. (2005). Alterations in the Colonic flora and Intestinal Permeability and Evidence of Immune Activation in Chronic Constipation. Dig. Liver Dis. 37 (11), 838–849. Epub 20051005PubMed PMID: 16169298. doi:10.1016/j.dld.2005.06.008
Korpela K., Mutanen A., Salonen A., Savilahti E., de Vos W. M., Pakarinen M. P. (2017). Intestinal Microbiota Signatures Associated with Histological Liver Steatosis in Pediatric-Onset Intestinal Failure. JPEN J. Parenter. Enteral Nutr. 41 (2), 238–248. Epub 20160930PubMed PMID: 25934046. doi:10.1177/0148607115584388
Kostic A. D., Gevers D., Pedamallu C. S., Michaud M., Duke F., Earl A. M., et al. (2012). Genomic Analysis Identifies Association of Fusobacterium with Colorectal Carcinoma. Genome Res. 22 (2), 292–298. Epub 20111018PubMed PMID: 22009990. doi:10.1101/gr.126573.111
Kumari R., Ahuja V., Paul J. (2013). Fluctuations in Butyrate-Producing Bacteria in Ulcerative Colitis Patients of North India. Wjg 19 (22), 3404–3414. PubMed PMID: 23801832. doi:10.3748/wjg.v19.i22.3404
Kummen M., Holm K., Anmarkrud J. A., Nygård S., Vesterhus M., Høivik M. L., et al. (2017). The Gut Microbial Profile in Patients with Primary Sclerosing Cholangitis Is Distinct from Patients with Ulcerative Colitis without Biliary Disease and Healthy Controls. Gut 66 (4), 611–619. Epub 20160217PubMed PMID: 26887816. doi:10.1136/gutjnl-2015-310500
Kummen M., Thingholm L. B., Rühlemann M. C., Holm K., Hansen S. H., Moitinho-Silva L., et al. (2021). Altered Gut Microbial Metabolism of Essential Nutrients in Primary Sclerosing Cholangitis. Gastroenterology 160 (5), 1784–1798. e0. Epub 20201231PubMed PMID: 33387530. doi:10.1053/j.gastro.2020.12.058
Lang S., Demir M., Martin A., Jiang L., Zhang X., Duan Y., et al. (2020). Intestinal Virome Signature Associated with Severity of Nonalcoholic Fatty Liver Disease. Gastroenterology 159 (5), 1839–1852. Epub 20200709PubMed PMID: 32652145. doi:10.1053/j.gastro.2020.07.005
Lang S., Duan Y., Liu J., Torralba M. G., Kuelbs C., Ventura‐Cots M., et al. (2020). Intestinal Fungal Dysbiosis and Systemic Immune Response to Fungi in Patients with Alcoholic Hepatitis. Hepatology 71 (2), 522–538. Epub 2019/08/20PubMed PMID: 31228214 ; PMC6925657. doi:10.1002/hep.30832
Lapidot Y., Amir A., Ben-Simon S., Veitsman E., Cohen-Ezra O., Davidov Y., et al. (2021). Alterations of the Salivary and Fecal Microbiome in Patients with Primary Sclerosing Cholangitis. Hepatol. Int. 15 (1), 191–201. Epub 20200919PubMed PMID: 32949377. doi:10.1007/s12072-020-10089-z
Laserna-Mendieta E. J., FitzGerald J. A., Arias-Gonzalez L., Ollala J. M., Bernardo D., Claesson M. J., et al. (2021). Esophageal Microbiome in Active Eosinophilic Esophagitis and Changes Induced by Different Therapies. Sci. Rep. 11 (1), 7113. Epub 20210329PubMed PMID: 33782490. doi:10.1038/s41598-021-86464-z
Li D., He R., Hou G., Ming W., Fan T., Chen L., et al. (2020). Characterization of the Esophageal Microbiota and Prediction of the Metabolic Pathways Involved in Esophageal Cancer. Front. Cel. Infect. Microbiol. 10, 268. Epub 20200626PubMed PMID: 32676460. doi:10.3389/fcimb.2020.00268
Li H., Chen J., Ren X., Yang C., Liu S., Bai X., et al. (2020). Gut Microbiota Composition Changes in Constipated Women of Reproductive Age. Front. Cel. Infect. Microbiol. 10, 557515. Epub 20210121PubMed PMID: 33552996. doi:10.3389/fcimb.2020.557515
Li X.-X., Wong G. L.-H., To K.-F., Wong V. W.-S., Lai L. H., Chow D. K.-L., et al. (2009). Bacterial Microbiota Profiling in Gastritis without Helicobacter pylori Infection or Non-steroidal Anti-inflammatory Drug Use. PLoS One 4 (11), e7985. Epub 20091124PubMed PMID: 19956741. doi:10.1371/journal.pone.0007985
Lin R., Zhou L., Zhang J., Wang B. (2015). Abnormal Intestinal Permeability and Microbiota in Patients with Autoimmune Hepatitis. Int. J. Clin. Exp. Pathol. 8 (5), 5153–5160. Epub 20150501. PubMed PMID: 26191211 ; PMC4503083.
Liu J., Yan Q., Luo F., Shang D., Wu D., Zhang H., et al. (2015). Acute Cholecystitis Associated with Infection of Enterobacteriaceae from Gut Microbiota. Clin. Microbiol. Infect. 21 (9), e1–8519. Epub 20150527PubMed PMID: 26025761. doi:10.1016/j.cmi.2015.05.017
Liu N., Ando T., Ishiguro K., Maeda O., Watanabe O., Funasaka K., et al. (2013). Characterization of Bacterial Biota in the Distal Esophagus of Japanese Patients with Reflux Esophagitis and Barrett's Esophagus. BMC Infect. Dis. 13, 130. Epub 20130311PubMed PMID: 23496929 ; PMC3599685. doi:10.1186/1471-2334-13-130
Liwinski T., Casar C., Ruehlemann M. C., Bang C., Sebode M., Hohenester S., et al. (2020). A Disease-specific Decline of the Relative Abundance of Bifidobacterium in Patients with Autoimmune Hepatitis. Aliment. Pharmacol. Ther. 51 (12), 1417–1428. Epub 20200507PubMed PMID: 32383181. doi:10.1111/apt.15754
Llopis M., Cassard A. M., Wrzosek L., Boschat L., Bruneau A., Ferrere G., et al. (2016). Intestinal Microbiota Contributes to Individual Susceptibility to Alcoholic Liver Disease. Gut 65 (5), 830–839. Epub 20151207PubMed PMID: 26642859. doi:10.1136/gutjnl-2015-310585
Lopetuso L. R., Severgnini M., Pecere S., Ponziani F. R., Boskoski I., Larghi A., et al. (2020). Esophageal Microbiome Signature in Patients with Barrett's Esophagus and Esophageal Adenocarcinoma. PLoS One 15 (5), e0231789. Epub 20200505PubMed PMID: 32369505. doi:10.1371/journal.pone.0231789
Lou J., Jiang Y., Rao B., Li A., Ding S., Yan H., et al. (2020). Fecal Microbiomes Distinguish Patients with Autoimmune Hepatitis from Healthy Individuals. Front. Cel. Infect. Microbiol. 10, 342. Epub 20200803PubMed PMID: 32850468. doi:10.3389/fcimb.2020.00342
Lu H.-F., Ren Z.-G., Li A., Zhang H., Xu S.-Y., Jiang J.-W., et al. (2019). Fecal Microbiome Data Distinguish Liver Recipients with Normal and Abnormal Liver Function from Healthy Controls. Front. Microbiol. 10, 1518. Epub 20190703PubMed PMID: 31333622. doi:10.3389/fmicb.2019.01518
Lu H., Wu Z., Xu W., Yang J., Chen Y., Li L. (2011). Intestinal Microbiota Was Assessed in Cirrhotic Patients with Hepatitis B Virus Infection. Microb. Ecol. 61 (3), 693–703. Epub 20110201PubMed PMID: 21286703. doi:10.1007/s00248-010-9801-8
Lv L.-X., Fang D.-Q., Shi D., Chen D.-Y., Yan R., Zhu Y.-X., et al. (2016). Alterations and Correlations of the Gut Microbiome, Metabolism and Immunity in Patients with Primary Biliary Cirrhosis. Environ. Microbiol. 18 (7), 2272–2286. PubMed PMID: 27243236. doi:10.1111/1462-2920.13401
Maccioni L., Gao B., Leclercq S., Pirlot B., Horsmans Y., De Timary P., et al. (2020). Intestinal Permeability, Microbial Translocation, Changes in Duodenal and Fecal Microbiota, and Their Associations with Alcoholic Liver Disease Progression in Humans. Gut Microbes 12 (1), 1782157. Epub 2020/06/26PubMed PMID: 32588725. doi:10.1080/19490976.2020.1782157
Macfarlane S., Furrie E., Macfarlane G. T., Dillon J. F. (2007). Microbial Colonization of the Upper Gastrointestinal Tract in Patients with Barrett's Esophagus. Clin. Infect. Dis. 45 (1), 29–38. Epub 20070522PubMed PMID: 17554697. doi:10.1086/518578
Machiels K., Joossens M., Sabino J., De Preter V., Arijs I., Eeckhaut V., et al. (2014). A Decrease of the Butyrate-Producing speciesRoseburia hominisandFaecalibacterium Prausnitziidefines Dysbiosis in Patients with Ulcerative Colitis. Gut 63 (8), 1275–1283. Epub 20130910PubMed PMID: 24021287. doi:10.1136/gutjnl-2013-304833
Maharshak N., Ringel Y., Katibian D., Lundqvist A., Sartor R. B., Carroll I. M., et al. (2018). Fecal and Mucosa-Associated Intestinal Microbiota in Patients with Diarrhea-Predominant Irritable Bowel Syndrome. Dig. Dis. Sci. 63 (7), 1890–1899. Epub 20180517PubMed PMID: 29777439. doi:10.1007/s10620-018-5086-4
Malinen E., Rinttila T., Kajander K., Matto J., Kassinen A., Krogius L., et al. (2005). Analysis of the Fecal Microbiota of Irritable Bowel Syndrome Patients and Healthy Controls with Real-Time PCR. Am. J. Gastroenterol. 100 (2), 373–382. PubMed PMID: 15667495. doi:10.1111/j.1572-0241.2005.40312.x
Mancabelli L., Milani C., Lugli G. A., Turroni F., Mangifesta M., Viappiani A., et al. (2017). Unveiling the Gut Microbiota Composition and Functionality Associated with Constipation through Metagenomic Analyses. Sci. Rep. 7 (1), 9879. Epub 20170829PubMed PMID: 28852182 ; PMC5575163. doi:10.1038/s41598-017-10663-w
Martín R., Chain F., Miquel S., Lu J., Gratadoux J.-J., Sokol H., et al. (2014). The Commensal Bacterium Faecalibacterium Prausnitzii Is Protective in DNBS-Induced Chronic Moderate and Severe Colitis Models. Inflamm. Bowel Dis. 20 (3), 417–430. PubMed PMID: 24418903. doi:10.1097/01.MIB.0000440815.76627.64
Matsukawa H., Iida N., Kitamura K., Terashima T., Seishima J., Makino I., et al. (2021). Dysbiotic Gut Microbiota in Pancreatic Cancer Patients Form Correlation Networks with the Oral Microbiota and Prognostic Factors. Am. J. Cancer Res. 11 (6), 3163–3175. Epub 20210615. PubMed PMID: 34249452 ; PMC8263681.
Mira-Pascual L., Cabrera-Rubio R., Ocon S., Costales P., Parra A., Suarez A., et al. (2015). Microbial Mucosal Colonic Shifts Associated with the Development of Colorectal Cancer Reveal the Presence of Different Bacterial and Archaeal Biomarkers. J. Gastroenterol. 50 (2), 167–179. Epub 20140509PubMed PMID: 24811328. doi:10.1007/s00535-014-0963-x
Mohieldeen K., Hamoda S. A. F., Ahmed S. M., Najeeb A., Ellakany W. I. (2021). Gut Microbiome in Cirrhotic Hepatitis C Virus Patients with and without Hepatocellular Carcinoma. Egypt. Liver J. 11, 79. doi:10.1186/s43066-021-00147-y
Moraes J. G. d., Motta M. E. F. d. A., Beltrão M. F. d. S., Salviano T. L., Silva G. A. P. d. (2016). Fecal Microbiota and Diet of Children with Chronic Constipation. Int. J. Pediatr. 2016, 1–8. Epub 20160623PubMed PMID: 27418934 ; PMC4935906. doi:10.1155/2016/6787269
Morgan X. C., Tickle T. L., Sokol H., Gevers D., Devaney K. L., Ward D. V., et al. (2012). Dysfunction of the Intestinal Microbiome in Inflammatory Bowel Disease and Treatment. Genome Biol. 13 (9), R79. Epub 20120416PubMed PMID: 23013615 ; PMC3506950. doi:10.1186/gb-2012-13-9-r79
Mottawea W., Chiang C.-K., Mühlbauer M., Starr A. E., Butcher J., Abujamel T., et al. (2016). Altered Intestinal Microbiota-Host Mitochondria Crosstalk in New Onset Crohn's Disease. Nat. Commun. 7, 13419. Epub 20161123PubMed PMID: 27876802. doi:10.1038/ncomms13419
Munakata S., Tohya M., Matsuzawa H., Tsuchiya Y., Amemiya K., Hagiwara T., et al. (2021). Analysis of Appendectomy Samples Identified Dysbiosis in Acute Appendicitis. Biosci. Microbiota Food Health 40 (2), 92–97. Epub 20201114PubMed PMID: 33996365. doi:10.12938/bmfh.2020-051
Nadal I., Donant E., Ribes-Koninckx C., Calabuig M., Sanz Y. (2007). Imbalance in the Composition of the Duodenal Microbiota of Children with Coeliac Disease. J. Med. Microbiol. 56 (Pt 12), 1669–1674. PubMed PMID: 18033837. doi:10.1099/jmm.0.47410-0
Ndegwa N., Ploner A., Andersson A. F., Zagai U., Andreasson A., Vieth M., et al. (2020). Gastric Microbiota in a Low-Helicobacter pylori Prevalence General Population and Their Associations with Gastric Lesions. Clin. Translational Gastroenterol. 11 (7), e00191. PubMed PMID: 32764211. doi:10.14309/ctg.0000000000000191
Ni J., Huang R., Zhou H., Xu X., Li Y., Cao P., et al. (2019). Analysis of the Relationship between the Degree of Dysbiosis in Gut Microbiota and Prognosis at Different Stages of Primary Hepatocellular Carcinoma. Front. Microbiol. 10, 1458. Epub 20190625PubMed PMID: 31293562. doi:10.3389/fmicb.2019.01458
Oh S. J., Pimentel M., Leite G. G. S., Celly S., Villanueva-Millan M. J., Lacsina I., et al. (2020). Acute Appendicitis Is Associated with Appendiceal Microbiome Changes Including elevatedCampylobacter Jejunilevels. BMJ Open Gastroenterol. 7 (1), e000412. PubMed PMID: 32499276 ; PMC7279619. doi:10.1136/bmjgast-2020-000412
Oh T. G., Kim S. M., Caussy C., Fu T., Guo J., Bassirian S., et al. (2020). A Universal Gut-Microbiome-Derived Signature Predicts Cirrhosis. Cel Metab. 32 (5), 878–888. e6. Epub 20200630PubMed PMID: 32610095. doi:10.1016/j.cmet.2020.06.005
Parsons B. N., Ijaz U. Z., D’Amore R., Burkitt M. D., Eccles R., Lenzi L., et al. (2017). Comparison of the Human Gastric Microbiota in Hypochlorhydric States Arising as a Result of Helicobacter Pylori-Induced Atrophic Gastritis, Autoimmune Atrophic Gastritis and Proton Pump Inhibitor Use. Plos Pathog. 13 (11), e1006653. Epub 20171102PubMed PMID: 29095917. doi:10.1371/journal.ppat.1006653
Piper H. G., Fan D., Coughlin L. A., Ho E. X., McDaniel M. M., Channabasappa N., et al. (2017). Severe Gut Microbiota Dysbiosis Is Associated with Poor Growth in Patients with Short Bowel Syndrome. JPEN J. Parenter. Enteral Nutr. 41 (7), 1202–1212. Epub 20160712PubMed PMID: 27406942. doi:10.1177/0148607116658762
Ponziani F. R., Bhoori S., Castelli C., Putignani L., Rivoltini L., Del Chierico F., et al. (2019). Hepatocellular Carcinoma Is Associated with Gut Microbiota Profile and Inflammation in Nonalcoholic Fatty Liver Disease. Hepatology 69 (1), 107–120. Epub 20180710PubMed PMID: 29665135. doi:10.1002/hep.30036
Puri P., Liangpunsakul S., Christensen J. E., Shah V. H., Kamath P. S., Gores G. J., et al. (2018). The Circulating Microbiome Signature and Inferred Functional Metagenomics in Alcoholic Hepatitis. Hepatology 67 (4), 1284–1302. Epub 20180222PubMed PMID: 29083504 ; PMC5867221. doi:10.1002/hep.29623
Pushalkar S., Hundeyin M., Daley D., Zambirinis C. P., Kurz E., Mishra A., et al. (2018). The Pancreatic Cancer Microbiome Promotes Oncogenesis by Induction of Innate and Adaptive Immune Suppression. Cancer Discov. 8 (4), 403–416. Epub 20180322PubMed PMID: 29567829 ; PMC6225783. doi:10.1158/2159-8290.CD-17-1134
Qi Y.-f., Sun J.-n., Ren L.-f., Cao X.-l., Dong J.-h., Tao K., et al. (2019). Intestinal Microbiota Is Altered in Patients with Gastric Cancer from Shanxi Province, China. Dig. Dis. Sci. 64 (5), 1193–1203. Epub 20181207PubMed PMID: 30535886. doi:10.1007/s10620-018-5411-y
Qin N., Yang F., Li A., Prifti E., Chen Y., Shao L., et al. (2014). Alterations of the Human Gut Microbiome in Liver Cirrhosis. Nature 513 (7516), 59–64. Epub 20140723PubMed PMID: 25079328. doi:10.1038/nature13568
Rajilić-Stojanović M., Shanahan F., Guarner F., de Vos W. M. (2013). Phylogenetic Analysis of Dysbiosis in Ulcerative Colitis during Remission. Inflamm. Bowel Dis. 19 (3), 481–488. PubMed PMID: 23385241. doi:10.1097/MIB.0b013e31827fec6d
Rajilić–Stojanović M., Biagi E., Heilig H. G. H. J., Kajander K., Kekkonen R. A., Tims S., et al. (2011). Global and Deep Molecular Analysis of Microbiota Signatures in Fecal Samples from Patients with Irritable Bowel Syndrome. Gastroenterology 141 (5), 1792–1801. Epub 20110805PubMed PMID: 21820992. doi:10.1053/j.gastro.2011.07.043
Raman M., Ahmed I., Gillevet P. M., Probert C. S., Ratcliffe N. M., Smith S., et al. (2013). Fecal Microbiome and Volatile Organic Compound Metabolome in Obese Humans with Nonalcoholic Fatty Liver Disease. Clin. Gastroenterol. Hepatol. 11 (7), 868–875. e1-3. Epub 20130227PubMed PMID: 23454028. doi:10.1016/j.cgh.2013.02.015
Rangel I., Sundin J., Fuentes S., Repsilber D., de Vos W. M., Brummer R. J. (2015). The Relationship between Faecal-Associated and Mucosal-Associated Microbiota in Irritable Bowel Syndrome Patients and Healthy Subjects. Aliment. Pharmacol. Ther. 42 (10), 1211–1221. Epub 20150917PubMed PMID: 26376728. doi:10.1111/apt.13399
Ren Z., Jiang J., Xie H., Li A., Lu H., Xu S., et al. (2017). Gut Microbial Profile Analysis by MiSeq Sequencing of Pancreatic Carcinoma Patients in China. Oncotarget 8 (56), 95176–95191. Epub 20170629PubMed PMID: 29221120. doi:10.18632/oncotarget.18820
Ren Z., Li A., Jiang J., Zhou L., Yu Z., Lu H., et al. (2019). Gut Microbiome Analysis as a Tool towards Targeted Non-invasive Biomarkers for Early Hepatocellular Carcinoma. Gut 68 (6), 1014–1023. Epub 20180725PubMed PMID: 30045880 ; PMC6580753. doi:10.1136/gutjnl-2017-315084
Rogers M. B., Brower-Sinning R., Firek B., Zhong D., Morowitz M. J. (2016). Acute Appendicitis in Children Is Associated with a Local Expansion of Fusobacteria. Clin. Infect. Dis. 63 (1), 71–78. Epub 20160407PubMed PMID: 27056397. doi:10.1093/cid/ciw208
Rühlemann M., Liwinski T., Heinsen F.-A., Bang C., Zenouzi R., Kummen M., et al. (2019). Consistent Alterations in Faecal Microbiomes of Patients with Primary Sclerosing Cholangitis Independent of Associated Colitis. Aliment. Pharmacol. Ther. 50 (5), 580–589. Epub 20190628PubMed PMID: 31250469 ; PMC6899739. doi:10.1111/apt.15375
Sabino J., Vieira-Silva S., Machiels K., Joossens M., Falony G., Ballet V., et al. (2016). Primary Sclerosing Cholangitis Is Characterised by Intestinal Dysbiosis Independent from IBD. Gut 65 (10), 1681–1689. Epub 20160520PubMed PMID: 27207975 ; PMC5036217. doi:10.1136/gutjnl-2015-311004
Samuelsson A., Wefer H., Fahlén A., Agréus L. (2013). Disturbed Intestinal Microbiota (Dysbiosis) and Micro Dynamics in Patients Treated for Appendicitis and Diverticulitis. Linköping: Linköping University.
Sánchez E., Donat E., Ribes-Koninckx C., Fernández-Murga M. L., Sanz Y. (2013). Duodenal-mucosal Bacteria Associated with Celiac Disease in Children. Appl. Environ. Microbiol. 79 (18), 5472–5479. Epub 20130708PubMed PMID: 23835180 ; PMC3754165. doi:10.1128/AEM.00869-13
Sánchez E., Laparra J. M., Sanz Y. (2012). Discerning the Role of Bacteroides Fragilis in Celiac Disease Pathogenesis. Appl. Environ. Microbiol. 78 (18), 6507–6515. Epub 20120706PubMed PMID: 22773639 ; PMC3426693. doi:10.1128/AEM.00563-12
Santoru M. L., Piras C., Murgia A., Palmas V., Camboni T., Liggi S., et al. (2017). Cross Sectional Evaluation of the Gut-Microbiome Metabolome axis in an Italian Cohort of IBD Patients. Sci. Rep. 7 (1), 9523. Epub 20170825PubMed PMID: 28842640 ; PMC5573342. doi:10.1038/s41598-017-10034-5
Sanz Y., Sánchez E., Marzotto M., Calabuig M., Torriani S., Dellaglio F. (2007). Differences in Faecal Bacterial Communities in Coeliac and Healthy Children as Detected by PCR and Denaturing Gradient Gel Electrophoresis. FEMS Immunol. Med. Microbiol. 51 (3), 562–568. Epub 20071004PubMed PMID: 17919298. doi:10.1111/j.1574-695X.2007.00337.x
Savitskaia K. I., Mel'nikova E. F., Vorob'ev A. A., Zagal'skaia N. V. (2002). Evaluation of Microecology of Colonic Contents in Patients with Chronic Pancreatitis. Vestn Ross Akad Med. Nauk (4), 20–23. PubMed PMID: 12046331.
Schippa S., Iebba V., Barbato M., Di Nardo G., Totino V., Checchi M., et al. (2010). A Distinctive 'microbial Signature' in Celiac Pediatric Patients. BMC Microbiol. 10, 175. Epub 20100617PubMed PMID: 20565734 ; PMC2906462. doi:10.1186/1471-2180-10-175
Schirmer M., Denson L., Vlamakis H., Franzosa E. A., Thomas S., Gotman N. M., et al. (2018). Compositional and Temporal Changes in the Gut Microbiome of Pediatric Ulcerative Colitis Patients Are Linked to Disease Course. Cell Host & Microbe 24 (4), 600–610. e4PubMed PMID: 30308161. doi:10.1016/j.chom.2018.09.009
Schwiertz A., Jacobi M., Frick J.-S., Richter M., Rusch K., Köhler H. (2010). Microbiota in Pediatric Inflammatory Bowel Disease. J. Pediatr. 157 (2), 240–244. e1. Epub 20100418PubMed PMID: 20400104. doi:10.1016/j.jpeds.2010.02.046
Sha S., Xu B., Wang X., Zhang Y., Wang H., Kong X., et al. (2013). The Biodiversity and Composition of the Dominant Fecal Microbiota in Patients with Inflammatory Bowel Disease. Diagn. Microbiol. Infect. Dis. 75 (3), 245–251. Epub 20121228PubMed PMID: 23276768. doi:10.1016/j.diagmicrobio.2012.11.022
Shao L., Ling Z., Chen D., Liu Y., Yang F., Li L. (2018). Disorganized Gut Microbiome Contributed to Liver Cirrhosis Progression: A Meta-Omics-Based Study. Front. Microbiol. 9, 3166. Epub 20181218PubMed PMID: 30631318. doi:10.3389/fmicb.2018.03166
Snider E. J., Compres G., Freedberg D. E., Giddins M. J., Khiabanian H., Lightdale C. J., et al. (2018). Barrett's Esophagus Is Associated with a Distinct Oral Microbiome. Clin. Transl Gastroenterol. 9 (3), e135. Epub 20180220PubMed PMID: 29491399 ; PMC5862155. doi:10.1038/s41424-018-0005-8
Snider E. J., Compres G., Freedberg D. E., Khiabanian H., Nobel Y. R., Stump S., et al. (2019). Alterations to the Esophageal Microbiome Associated with Progression from Barrett's Esophagus to Esophageal Adenocarcinoma. Cancer Epidemiol. Biomarkers Prev. 28 (10), 1687–1693. Epub 20190829PubMed PMID: 31466948 ; PMC6774849. doi:10.1158/1055-9965.EPI-19-0008
Sokol H., Leducq V., Aschard H., Pham H.-P., Jegou S., Landman C., et al. (2017). Fungal Microbiota Dysbiosis in IBD. Gut 66 (6), 1039–1048. Epub 20160203PubMed PMID: 26843508. doi:10.1136/gutjnl-2015-310746
Song W., Sun L.-Y., Zhu Z.-J., Wei L., Qu W., Zeng Z.-G., et al. (2021). Association of Gut Microbiota and Metabolites with Disease Progression in Children with Biliary Atresia. Front. Immunol. 12, 698900. Epub 20210923PubMed PMID: 34630385. doi:10.3389/fimmu.2021.698900
Starý L., Mezerová K., Vysloužil K., Zbořil P., Skalický P., Stašek M., et al. (2020). Candida Albicans Culture from a Rectal Swab Can Be Associated with Newly Diagnosed Colorectal Cancer. Folia Microbiol. 65 (6), 989–994. Epub 20200629PubMed PMID: 32602070. doi:10.1007/s12223-020-00807-3
Sultan S., El-Mowafy M., Elgaml A., El-Mesery M., El Shabrawi A., Elegezy M., et al. (2021). Alterations of the Treatment-Naive Gut Microbiome in Newly Diagnosed Hepatitis C Virus Infection. ACS Infect. Dis. 7 (5), 1059–1068. Epub 20201029PubMed PMID: 33119247. doi:10.1021/acsinfecdis.0c00432
Sun L.-Y., Yang Y.-S., Qu W., Zhu Z.-J., Wei L., Ye Z.-S., et al. (2017). Gut Microbiota of Liver Transplantation Recipients. Sci. Rep. 7 (1), 3762. Epub 20170619PubMed PMID: 28630433 ; PMC5476624. doi:10.1038/s41598-017-03476-4
Swidsinski A., Dorffel Y., Loening-Baucke V., Theissig F., Ruckert J. C., Ismail M., et al. (2011). Acute Appendicitis Is Characterised by Local Invasion with Fusobacterium Nucleatum/necrophorum. Gut 60 (1), 34–40. Epub 20091118PubMed PMID: 19926616. doi:10.1136/gut.2009.191320
Tan C., Ling Z., Huang Y., Cao Y., Liu Q., Cai T., et al. (2015). Dysbiosis of Intestinal Microbiota Associated with Inflammation Involved in the Progression of Acute Pancreatitis. Pancreas 44 (6), 868–875. PubMed PMID: 25931253. doi:10.1097/MPA.0000000000000355
Tang R., Wei Y., Li Y., Chen W., Chen H., Wang Q., et al. (2018). Gut Microbial Profile Is Altered in Primary Biliary Cholangitis and Partially Restored after UDCA Therapy. Gut 67 (3), 534–541. Epub 20170217PubMed PMID: 28213609. doi:10.1136/gutjnl-2016-313332
Wang B., Jiang X., Cao M., Ge J., Bao Q., Tang L., et al. (2016). Altered Fecal Microbiota Correlates with Liver Biochemistry in Nonobese Patients with Non-alcoholic Fatty Liver Disease. Sci. Rep. 6, 32002. Epub 20160823PubMed PMID: 27550547 ; PMC4994089. doi:10.1038/srep32002
Wang J., Qian T., Jiang J., Yang Y., Shen Z., Huang Y., et al. (2020). Gut Microbial Profile in Biliary Atresia: a Case‐control Study. J. Gastroenterol. Hepatol. 35 (2), 334–342. Epub 20190801PubMed PMID: 31271681. doi:10.1111/jgh.14777
Wang J., Wang Y., Zhang X., Liu J., Zhang Q., Zhao Y., et al. (2017). Gut Microbial Dysbiosis Is Associated with Altered Hepatic Functions and Serum Metabolites in Chronic Hepatitis B Patients. Front. Microbiol. 8, 2222. Epub 20171113PubMed PMID: 29180991. doi:10.3389/fmicb.2017.02222
Wang Q., Hao C., Yao W., Zhu D., Lu H., Li L., et al. (2020). Intestinal flora Imbalance Affects Bile Acid Metabolism and Is Associated with Gallstone Formation. BMC Gastroenterol., 20, 59. Epub 20200306PubMed PMID: 32143645. doi:10.1186/s12876-020-01195-1
Wang T., Cai G., Qiu Y., Fei N., Zhang M., Pang X., et al. (2012). Structural Segregation of Gut Microbiota between Colorectal Cancer Patients and Healthy Volunteers. ISME J. 6 (2), 320–329. Epub 20110818PubMed PMID: 21850056 ; PMC3260502. doi:10.1038/ismej.2011.109
Wang W., Xiao Y., Wang X., Zhou Y., Wang T., Xv C., et al. (2020). Disordered Gut Microbiota in Children Who Have Chronic Pancreatitis and Different Functional Gene Mutations. Clin. Translational Gastroenterol. 11 (3), e00150. PubMed PMID: 32352720. doi:10.14309/ctg.0000000000000150
Wang Z., Gao X., Zeng R., Wu Q., Sun H., Wu W., et al. (2020). Changes of the Gastric Mucosal Microbiome Associated with Histological Stages of Gastric Carcinogenesis. Front. Microbiol. 11, 997. Epub 20200529PubMed PMID: 32547510. doi:10.3389/fmicb.2020.00997
Warren R. L., Freeman D. J., Pleasance S., Watson P., Moore R. A., Cochrane K., et al. (2013). Co-occurrence of Anaerobic Bacteria in Colorectal Carcinomas. Microbiome 1 (1), 16. Epub 20130515PubMed PMID: 24450771 ; PMC3971631. doi:10.1186/2049-2618-1-16
Wei Y., Li Y., Yan L., Sun C., Miao Q., Wang Q., et al. (2020). Alterations of Gut Microbiome in Autoimmune Hepatitis. Gut 69 (3), 569–577. Epub 20190614PubMed PMID: 31201284. doi:10.1136/gutjnl-2018-317836
Wong V. W.-S., Tse C.-H., Lam T. T.-Y., Wong G. L.-H., Chim A. M.-L., Chu W. C.-W., et al. (2013). Molecular Characterization of the Fecal Microbiota in Patients with Nonalcoholic Steatohepatitis - A Longitudinal Study. PLoS One 8 (4), e62885. Epub 20130425PubMed PMID: 23638162. doi:10.1371/journal.pone.0062885
Wu T., Zhang Z., Liu B., Hou D., Liang Y., Zhang J., et al. (2013). Gut Microbiota Dysbiosis and Bacterial Community Assembly Associated with Cholesterol Gallstones in Large-Scale Study. BMC Genomics 14, 669. Epub 20131001PubMed PMID: 24083370 ; PMC3851472. doi:10.1186/1471-2164-14-669
Wu Z.-W., Ling Z.-X., Lu H.-F., Zuo J., Sheng J.-F., Zheng S.-S., et al. (2012). Changes of Gut Bacteria and Immune Parameters in Liver Transplant Recipients. Hepatobiliary Pancreat. Dis. Int. 11 (1), 40–50. PubMed PMID: 22251469. doi:10.1016/s1499-3872(11)60124-0
Xin H., Li X., Sun R., Meng Y., Yu Q., Hao Y. (2019). Endotoxin and Intestinal Microflora in Patients with Hepatocellular Carcinoma. Chin. J. Gen. Surg. 12, 686–688.
Xu M., Wang B., Fu Y., Chen Y., Yang F., Lu H., et al. (2012). Changes of Fecal Bifidobacterium Species in Adult Patients with Hepatitis B Virus-Induced Chronic Liver Disease. Microb. Ecol. 63 (2), 304–313. Epub 20110804PubMed PMID: 21814872. doi:10.1007/s00248-011-9925-5
Yang L., Zhang J., Xu J., Wei X., Yang J., Liu Y., et al. (2019). Helicobacter pylori Infection Aggravates Dysbiosis of Gut Microbiome in Children with Gastritis. Front. Cel. Infect. Microbiol. 9, 375. Epub 20191107PubMed PMID: 31781514. doi:10.3389/fcimb.2019.00375
Yang X.-A., Lv F., Wang R., Chang Y., Zhao Y., Cui X., et al. (2020). Potential Role of Intestinal Microflora in Disease Progression Among Patients with Different Stages of Hepatitis B. Gut Pathog. 12, 50. Epub 20201027PubMed PMID: 33117435 ; PMC7590496. doi:10.1186/s13099-020-00391-4
Yao X., Yu H., Fan G., Xiang H., Long L., Xu H., et al. (2021). Impact of the Gut Microbiome on the Progression of Hepatitis B Virus Related Acute-On-Chronic Liver Failure. Front. Cel. Infect. Microbiol. 11, 573923. Epub 20210406PubMed PMID: 33889550. doi:10.3389/fcimb.2021.573923
Yu J., Feng Q., Wong S. H., Zhang D., Liang Q. y., Qin Y., et al. (2017). Metagenomic Analysis of Faecal Microbiome as a Tool towards Targeted Non-invasive Biomarkers for Colorectal Cancer. Gut 66 (1), 70–78. Epub 20150925PubMed PMID: 26408641. doi:10.1136/gutjnl-2015-309800
Yu S., Xiong Y., Xu J., Liang X., Fu Y., Liu D., et al. (2020). Identification of Dysfunctional Gut Microbiota through Rectal Swab in Patients with Different Severity of Acute Pancreatitis. Dig. Dis. Sci. 65 (11), 3223–3237. Epub 20200219PubMed PMID: 32076933. doi:10.1007/s10620-020-06061-4
Zeller G., Tap J., Voigt A. Y., Sunagawa S., Kultima J. R., Costea P. I., et al. (2014). Potential of Fecal Microbiota for Early‐stage Detection of Colorectal Cancer. Mol. Syst. Biol. 10, 766. Epub 20141128PubMed PMID: 25432777 ; PMC4299606. doi:10.15252/msb.20145645
Zeng Y., Chen S., Fu Y., Wu W., Chen T., Chen J., et al. (2020). Gut Microbiota Dysbiosis in Patients with Hepatitis B Virus-Induced Chronic Liver Disease Covering Chronic Hepatitis, Liver Cirrhosis and Hepatocellular Carcinoma. J. Viral Hepat. 27 (2), 143–155. Epub 20191029PubMed PMID: 31600845. doi:10.1111/jvh.13216
Zhang T., Zhang S., Jin C., Lin Z., Deng T., Xie X., et al. (2021). A Predictive Model Based on the Gut Microbiota Improves the Diagnostic Effect in Patients with Cholangiocarcinoma. Front. Cel. Infect. Microbiol. 11, 751795. Epub 20211123PubMed PMID: 34888258. doi:10.3389/fcimb.2021.751795
Zhao H. Y., Wang H. J., Lu Z., Xu S. Z. (2004). Intestinal Microflora in Patients with Liver Cirrhosis. Chin. Dig. Dis. 5 (2), 64–67. PubMed PMID: 15612659. doi:10.1111/j.1443-9573.2004.00157.x
Zhao Q., Yang T., Yan Y., Zhang Y., Li Z., Wang Y., et al. (2020). Alterations of Oral Microbiota in Chinese Patients with Esophageal Cancer. Front. Cel. Infect. Microbiol. 10, 541144. Epub 20201021PubMed PMID: 33194789 ; PMC7609410. doi:10.3389/fcimb.2020.541144
Zhong D., Brower-Sinning R., Firek B., Morowitz M. J. (2014). Acute Appendicitis in Children Is Associated with an Abundance of Bacteria from the Phylum Fusobacteria. J. Pediatr. Surg. 49 (3), 441–446. PubMed PMID: 24650474. doi:10.1016/j.jpedsurg.2013.06.026
Zhong M., Xiong Y., Zhao J., Gao Z., Ma J., Wu Z., et al. (2021). Candida Albicans Disorder Is Associated with Gastric Carcinogenesis. Theranostics 11 (10), 4945–4956. Epub 20210305PubMed PMID: 33754037 ; PMC7978306. doi:10.7150/thno.55209
Zhou C.-H., Meng Y.-T., Xu J.-J., Fang X., Zhao J.-L., Zhou W., et al. (2020). Altered Diversity and Composition of Gut Microbiota in Chinese Patients with Chronic Pancreatitis. Pancreatology 20 (1), 16–24. Epub 20191127PubMed PMID: 31806503. doi:10.1016/j.pan.2019.11.013
Zhou J., Shrestha P., Qiu Z., Harman D. G., Teoh W.-C., Al-Sohaily S., et al. (2020). Distinct Microbiota Dysbiosis in Patients with Non-erosive Reflux Disease and Esophageal Adenocarcinoma. Jcm 9 (7), 2162. Epub 20200708PubMed PMID: 32650561 ; PMC7408827. doi:10.3390/jcm9072162
Zhu L., Baker S. S., Gill C., Liu W., Alkhouri R., Baker R. D., et al. (2013). Characterization of Gut Microbiomes in Nonalcoholic Steatohepatitis (NASH) Patients: a Connection between Endogenous Alcohol and NASH. Hepatology 57 (2), 601–609. Epub 20130108PubMed PMID: 23055155. doi:10.1002/hep.26093
Zhu L., Liu W., Alkhouri R., Baker R. D., Bard J. E., Quigley E. M., et al. (2014). Structural Changes in the Gut Microbiome of Constipated Patients. Physiol. Genomics 46 (18), 679–686. Epub 20140729PubMed PMID: 25073603. doi:10.1152/physiolgenomics.00082.2014
Zhu Y., He C., Li X., Cai Y., Hu J., Liao Y., et al. (2019). Gut Microbiota Dysbiosis Worsens the Severity of Acute Pancreatitis in Patients and Mice. J. Gastroenterol. 54 (4), 347–358. Epub 20181205PubMed PMID: 30519748. doi:10.1007/s00535-018-1529-0
Keywords: Microbiome and dysbiosis, digestive diseases, Liver disease, Intestinal disease, microbiota
Citation: Hartmann P (2022) Editorial: The Microbiome in Hepatobiliary and Intestinal Disease. Front. Physiol. 13:893074. doi: 10.3389/fphys.2022.893074
Received: 09 March 2022; Accepted: 11 March 2022;
Published: 13 April 2022.
Edited and reviewed by:
Stephen J. Pandol, Cedars Sinai Medical Center, United StatesCopyright © 2022 Hartmann. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Phillipp Hartmann, cGhoYXJ0bWFubkB1Y3NkLmVkdQ==
 Phillipp Hartmann
Phillipp Hartmann