
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Physiol. , 20 May 2022
Sec. Aquatic Physiology
Volume 13 - 2022 | https://doi.org/10.3389/fphys.2022.892550
This article is part of the Research Topic Nutritional Physiology of Aquacultured Species View all 12 articles
 Ilaria Biasato1
Ilaria Biasato1 Simona Rimoldi2
Simona Rimoldi2 Christian Caimi1
Christian Caimi1 Sara Bellezza Oddon1
Sara Bellezza Oddon1 Giulia Chemello3
Giulia Chemello3 Marino Prearo4
Marino Prearo4 Marco Saroglia2
Marco Saroglia2 Ronald Hardy5
Ronald Hardy5 Laura Gasco1*
Laura Gasco1* Genciana Terova2
Genciana Terova2The present study aimed to investigate the growth performance, whole-body proximate composition, and intestinal microbiome of rainbow trout strains when selected and non-selected for weight gain on all-plant protein diets. A 2x2 factorial design was applied, where a selected (United States) and a non-selected (ITA) rainbow trout strain were fed using either an all-plant protein (PP) or a commercial low-FM diet (C). Diets were fed to five replicates of 20 (PP) or 25 (C) fish for 105 days. At the end of the trial, growth parameters were assessed, and whole fish (15 pools of three fish/diet) and gut samples (six fish/diet) were collected for whole-body proximate composition and gut microbiome analyses, respectively. Independent of the administered diet, the United States strain showed higher survival, final body weight, weight gain, and specific growth rate when compared to the ITA fish (p < 0.001). Furthermore, decreased whole-body ether extract content was identified in the PP-fed United States rainbow trout when compared to the ITA strain fed the same diet (p < 0.001). Gut microbiome analysis revealed the Cetobacterium probiotic-like genus as clearly associated with the United States rainbow trout, along with the up-regulation of the pathway involved in starch and sucrose metabolism. In summary, the overall improvement in growth performance and, to a lesser extent, whole-body proximate composition observed in the selected rainbow trout strain was accompanied by specific, positive modulation of the intestinal microbiome.
Genetic selection has progressively gained a foothold in the aquaculture sector, as it allows obtaining individuals with desired phenotypic traits. Traditional selective breeding—where only fish that exhibit desirable characteristics for one or multiple traits are chosen and bred—represents the most commonly adopted genetic enhancement technique. It is mainly directed towards the maximization of growth potential, improvement in fillet quality attributes, or development of resistance to stress and pathogens (Xu et al., 2015). In particular, rainbow trout (Oncorhynchus mykiss) has frequently been the object of selective breeding, with current programs being predominantly based on mass selection for growth rates (Cleveland et al., 2020) and resistance to viral or bacterial diseases (Vallejo et al., 2020; Zuo et al., 2020). Among all the selective breeding programs carried out to improve the growth potential of rainbow trout, one of the most impactful is related to the efficiency of digesting all-plant protein feeds (Overturf et al., 2013; Lazzarotto et al., 2015; Callet et al., 2017). It is, indeed, well known that the aquaculture sector has been trying to overcome sustainability challenges for the past decade, as worldwide production from aquaculture has grown rapidly and overtaken capture fisheries, and per capita consumption of fish has doubled (FAO, 2020). Therefore, the unsustainability of a “fish-to-fish” feeding has progressively made it necessary to replace fishmeal (FM) by alternative protein sources of plant origin in diets for farming fish. However, all-plant protein feeds do not represent the optimal choice for salmonids, with reductions in feed intake, weight gain, feed efficiency, alteration of gut microflora and immune response, and intestinal dysfunction and inflammation (mainly affecting the distal gut) being commonly observed (Krogdahl et al., 2010). In the light of such negative outcomes, genetic selection has successfully proved to be an effective counteracting measure to create rainbow trout strains that outperform the commercial, parental lines in utilizing all-plant protein diets without showing worsened growth performance or developing distal enteritis (Burr et al., 2012; Abernathy et al., 2017; Callet et al., 2017).
So far, the efficacy of selective breeding for utilization of all-plant protein diets in rainbow trout has been investigated by comparing selected and non-selected strains when fed either an FM or an all-plant protein diet in terms of growth response, nutrient retention, plasma amino acid (AA) patterns, and gut, liver, and muscle transcriptomic profiles (Abernathy et al., 2017; Callet et al., 2017; Brezas and Hardy, 2020; Brezas et al., 2021; Callet et al., 2021). In particular, candidate genes for protein (i.e., intestinal AA transporters and ribosomal proteins), lipid (i.e., long-chain polyunsaturated fatty acids biosynthesis), and purine–thiamine metabolisms, as well as energy production and inflammation (i.e., interleukin receptors), have previously been linked to synchronization of AA absorption and protein digestion rates and, in turn, improved growth and protein retention in the selected strains (Abernathy et al., 2017; Brezas et al., 2021; Callet et al., 2021). However, as gut microbiome alterations can influence fish performance (Vargas-Albores et al., 2021), and the intestinal microbiome itself may be, in turn, modulated by dietary changes (Rimoldi et al., 2018 and, 2021; Gaudioso et al., 2021; Terova et al., 2021), the characterization of the gut microbiome in selected rainbow trout could provide novel insights into the adaptation mechanisms of the fish to all-plant protein feeds. Nowadays, it is widely accepted that there is a correlation between fish genotype and gut microbial communities, but to date, there is very little information on how genetic selection drives differences in intestinal microbiota composition and how this could affect diet plasticity, health, and disease resistance in both marine and freshwater fish species (Blaufuss et al., 2020; Piazzon et al., 2020).
Therefore, the present study was designed to investigate the growth performance, whole-body proximate composition, and intestinal microbiome of rainbow trout strains when selected and non-selected for weight gain on all-plant protein feeds in response to all-plant protein and commercial low-FM diets.
A feeding trial was conducted at the Experimental Facility of the Department of Agricultural, Forest and Food Sciences (DISAFA) of the University of Turin (Italy) using two different rainbow trout strains. The selected strain (United States) was developed by the United States Department of Agricultural–Agricultural Research Service (USDA-ARS) and the University of Idaho by introgression of nine domesticated commercial and conservation stocks (Overturf et al., 2013). The selection was based on growth performance when trout were fed an all-plant protein feed. Besides dietary utilization, the selected strain also displays an enhanced non-specific pathogen resistance and a concomitant resistance to the development of distal intestine enteritis (Venold et al., 2012; Abernathy et al., 2017). The non-selected strain (ITA) was selected for different performance traits when fed a commercial low-FM diet by the Italian hatchery Fratelli Leonardi (Trento, Italy). To understand the effects of both diet and genotype, a 2x2 factorial design was applied. In particular, each rainbow trout strain was fed using two different experimental diets: a diet containing only plant proteins (PP, provided by the University of Idaho), and a commercial low-FM diet (C) containing both the plant meals and FM (provided by Naturalleva, Cologna Veneta, Verona, Italy). No information was provided by the feed company about the C diet formulation as it was considered confidential. Both the diets were provided as extruded feeds.
In detail, the United States embryonated eggs were received from the University of Idaho and, after hatching in hatchery tanks, were fed commercial diets (crumbled extruded feeds of different sizes, with 55–62% crude protein [CP] and 11–18% ether extract [EE]; Skretting Italia Spa, Mozzecane, Verona, Italy) until the beginning of the experimental trial. In parallel, ITA fingerlings (mean weight of 2.0 ± 0.01 g) were provided by Fratelli Leonardi and fed the same commercial diets until the beginning of the feeding trial as well. After reaching the juvenile stage, 250 juvenile rainbow trout (125 from each strain) underwent light anesthesia (MS-222; PHARMAQ Ltd., United Kingdom; 60 mg/L) and were individually weighed (10.63 ± 0.01 g) by using electronic scales (KERN PLE-N v. 2.2; KERN and, Sohn GmbH, Balingen-Frommern, Germany; d: 0.1). Then, fish from each strain were randomly distributed into ten 100-L, rectangular-shaped tanks (five replicate tanks per strain, 25 fish per tank) connected to a flow-through open system supplied with artesian well water (constant temperature of 13 ± 1°C, 8 L min-1, dissolved oxygen ranged between 7.6 and 8.7 mg L), and fed the C diet (USA-C and ITA-C treatments). In parallel, at the same rearing conditions, 200 juvenile rainbow trout (100 from each strain; initial body weight of 13.47 ± 0.01 g) were randomly allotted to ten 100-L, rectangular-shaped tanks (five replicate tanks per strain, 20 fish per tank), and fed the PP diet (USA-PP and ITA-PP treatments). In order to update the daily amount of feed, the biomass in each tank was weighed in bulk every 14 days. Fish were fed by hand twice a day (08:00 and 15:00) and 6 days per week, up to a maximum of 2.5% of the tank biomass the first 6 weeks of the feeding trial. Then, according to the fish growth, the daily amount of feed per tank was progressively reduced to 1.7%. Feed intake was checked at each administration, as feed distribution was immediately suspended when fish stopped eating. Undistributed feed was weighed and this data was used for the correct calculation of the growth parameters. Mortality was daily recorded. The feeding trial lasted 105 days.
Feed samples were ground using a cutting mill (MLI 204; Bühler AG, Uzwil, Switzerland) and analyzed for dry matter (DM, AOAC #934.01), CP (AOAC #984.13), and ash (AOAC #942.05) contents according to AOAC International (2000). Feed samples were also analyzed for EE (AOAC #2003.05) content according to AOAC International (2003). The gross energy (GE) content was determined using an adiabatic calorimetric bomb (C7000; IKA, Staufen, Germany). All the chemical analyses of the feeds were performed in duplicate. The proximate composition of the experimental diets is shown in Table 1.
At the end of the feeding trial, the feed was withheld from the fish for 1 day, and then they were individually weighted after light anesthesia. The following performance indexes were calculated per tank:
Survival (%) = 100—[(number of dead fish/number of fish at start) × 100].
Individual weight gain (iWG, g) = individual mean final body weight (iFBW, g)—individual mean initial body weight (iIBW, g).
Feed conversion ratio (FCR) = total feed supplied (g, DM)/WG (g).
Protein efficiency ratio (PER) = WG (g)/total protein fed (g, DM).
Specific growth rate (SGR, % day-1) = [(lnFBW–lnIBW)/number of days] × 100.
At the end of the feeding trial, 3 fish per tank (15 fish per dietary treatment) were sacrificed by an overdose of anesthetic (MS-222, PHARMAQ Ltd., United Kingdom; 500 mg/L) and then frozen at -20°C. Frozen fish were finely ground with a knife mill (Grindomix 174 GM200; Retsch GmbH, Haan, Germany) to obtain one pool of three fish per tank (5 pools per dietary treatment). All the fish pools were freeze-dried and used for the determination of the final whole-body proximate composition. The proximate composition of fish whole body was determined following the same procedures used for feed analyses (AOAC International, 2000 and 2003).
At the end of the feeding trial, six fish per dietary treatment were sacrificed by an overdose of anesthetic, and the whole intestine was aseptically dissected out. The gut autochthonous microbiota was obtained by scraping the mucosa of the entire intestine (excluding pyloric caeca) with a sterile swab as reported in detail by Rimoldi et al. (2019). Briefly, each swab head was cut and immediately transferred to a sterile 1.5 ml microtube containing 200 μl of Xpedition Lysis/Stabilization Solution. The tube was then vortexed for shaking out the bacteria from the swab tip and stored at room temperature for up to 24 h until bacterial DNA extraction.
The DNA was extracted from 200 µl of bacterial suspension and from three aliquots (200 mg) of each experimental feed using the DNeasy PowerSoil® Kit (Qiagen, Milan, Italy), according to the manufacturer’s instructions. The samples were mechanically lysed in PowerBead Tubes by means of a TissueLyser II (Qiagen) for 2 min at 25 Hz. The extracted DNA was spectrophotometrically quantified using the NanoDropTM 2000 spectrophotometer (Thermo Scientific, Milan, Italy) and stored at -20°C until the PCR reaction was performed.
Preparation of the 16S metagenomic library and sequencing were performed by GalSeq srl (Italy), targeting the variable V4 region and applying Illumina protocol “16S Metagenomic Sequencing Library Preparation for Illumina MiSeq System” (#15044223 rev. B). Details of the methodology used have previously been described (Rimoldi et al., 2019; Terova et al., 2019). Briefly, primers are tailed with sequences to incorporate indexing barcodes, and samples are pooled into a single library for sequencing on the Illumina MiSeq device using the pair-ended sequencing (2 × 250) strategy. All FastQ sequence files were submitted to the European Nucleotide Archive (EBI ENA).
The raw sequence files were imported in QIIME™ 2 (v. 2018.4) (Bolyen et al., 2019) and processed at the default setting. Artefacts and primers were trimmed, the remaining sequences were filtered for base quality (Q > 30), and forward and reverse reads were merged. Filtered reads were then dereplicated and chimeras removed using the denoise-paired command of the DADA2 plug-in. The output was an amplicon sequence variant (ASV) table, analog to the OTU table, which records the number of times each exact ASV was observed in each sample. The representative sequences were classified using the Silva reference database (https://www.arb-silva.de/) down to the genus level. All sequences assigned to eukaryotes (chloroplasts and mitochondria) in gut samples were discarded. Alpha and beta diversity analyses were performed using QIIME scripts “alpha_rarefection.py” and “jackknifed_beta_diversity_.py”, respectively. Principal Coordinates Analysis (PCoA) based on both unweighted UniFrac and weighted UniFrac distance matrices (Lozupone and Knight, 2005; Lozupone et al., 2007) was conducted to visualize similarities or dissimilarities between bacterial communities.
The PICRUSt (Phylogenetic Investigation of Communities by Reconstruction of Unobserved States) software package was used to predict the functional profiling of microbial communities on the basis of 16S rRNA gene sequences (Langille et al., 2013) as previously described (Rimoldi et al., 2021). The inferred metagenomic functions were predicted using the Kyoto Encyclopedia of Genes and Genomes (KEGG database). The maximum allowed Nearest Sequenced Taxon Index (NSTI) value was set to two. Analysis of the output generated by PICRUSt was made using the Statistical Analysis of Metagenomic Profiles (STAMP) software package (Parks et al., 2014) applying a two-sided Welch t-test (p < 0.05) to identify differences in microbial metabolic pathways between two groups.
The experimental unit for statistical analyses was the tank for growth performance and whole-body proximate composition, and the fish for microbiome analysis.
The statistical analysis of growth performance and whole-body proximate composition was performed using IBM SPSS Statistics v. 28.0 (IBM, Armonk, NY, United States). The Shapiro–Wilk test assessed the normality distribution of the residuals, while the assumption of equal variances was assessed by Levene’s homogeneity of variance test. The growth performance and whole-body proximate composition data were analyzed by fitting a general linear model that allowed them to depend on two fixed factors (strain and diet) and their interaction. The interactions between the levels of the fixed factors were evaluated by pairwise contrasts. The results obtained were expressed as the least square mean and pooled standard error of the mean (SEM). p values ≤ 0.05 were considered statistically significant.
The statistical analysis of gut microbiome data was performed using Past3 software (Hammer et al., 2001). All the data were checked for normality and homogeneity of variance using Shapiro–Wilk and Levene’s test, respectively. Concerning gut microbiota composition, only those taxa with an overall abundance of more than 0.5% (up to order level) and 0.01% at family and genus level were considered for statistical analysis. Before being statistically analyzed, the percentage values were angular transformed (arcsine of the square root). To test the null hypothesis (p < 0.05), a two-way ANOVA was performed considering the influence of two main factors (strain and diet) and their interaction. Multivariate analysis of beta diversity was tested using a two-way non-parametric permutational multivariate analysis of variance (two-way PERMANOVA) with 999 permutations.
The growth performance of the rainbow trout in the present study is summarized in Table 2. Survival was high for all the dietary treatments (range: 95.00–100), with a significant influence of both the strain (p < 0.05) and the interaction between strain and diet (p < 0.001) being identified. In particular, the United States rainbow trout showed higher survival than the ITA group (p < 0.05), but greater survival was observed in the United States strain fed the PP diet when compared to the PP-fed ITA fish only (100.00 ± 0.51 vs. 95.00 ± 0.51; p < 0.001). In parallel, the ITA rainbow trout fed the C diet displayed higher survival than the C-fed United States group (99.20 ± 0.51 vs. 96.80 ± 0.51; p = 0.001). Higher survival was also observed in both the United States and the ITA strains when fed the PP and C diets, respectively (p < 0.001). Differently, the iFBW was influenced by both strain and diet (p < 0.001). The United States fish showed greater iFBW than the ITA group (p < 0.001), with higher values being also observed when the fish were fed the PP diet in comparison with the C one (p < 0.001). The iWG depended on all the considered variables (strain and diet: p < 0.001; strain × diet: p = 0.010). In particular, the United States rainbow trout showed higher iWG than the ITA group (p < 0.001), with greater iWG being also observed when fish were fed the PP diet in comparison with the C one (p < 0.001). However, the United States rainbow trout displayed higher iWG than the ITA strain when they were fed the C diet only (55.12 ± 1.25 vs. 47.43 ± 1.25; p < 0.001). The FCR and the PER were not affected by any of the considered variables (p > 0.05). Finally, the SGR depended on strain only (p < 0.001), with higher values being identified in the United States rainbow trout in comparison with the ITA fish (p < 0.001).
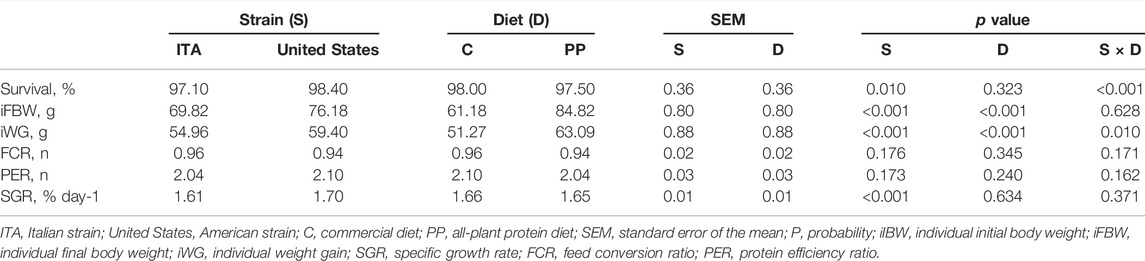
TABLE 2. Growth performance of the rainbow trout depending on strain, diet, and their interaction (n = 5).
Table 3 summarizes the whole-body proximate composition of the rainbow trout in the current research. The moisture depended on all the considered variables (strain and diet: p < 0.001; strain × diet: p = 0.042). In particular, the ITA rainbow trout showed lower moisture content than the United States group (p < 0.001), with a reduction in the whole-body moisture being also observed when fish were fed the PP diet in comparison with the C one (p < 0.001). However, the ITA strain displayed lower moisture content than the United States rainbow trout when they were fed the PP diet only (70.83 ± 0.25 vs. 72.43 ± 0.25; p < 0.001). The ash content was influenced by the strain × diet interaction only (p < 0.05), with the C-fed United States fish displaying increased whole-body ash in comparison with the USA-PP group (2.52 ± 0.07 vs. 2.26 ± 0.07; p < 0.05). Differently, the CP content uniquely depended on diet (p < 0.001). In detail, the PP diet led to higher whole-body CP than the C feed (p < 0.001). On the contrary, the EE content was affected by all the considered variables (p < 0.001). In particular, the ITA fish displayed higher whole-body EE than the United States strain (p < 0.001), with an increase in the EE content being also observed when fish were fed the PP diet in comparison with the C one (p < 0.001). However, greater whole-body EE was observed in the ITA rainbow trout fed the PP diet when compared to the PP-fed United States group only (9.84 ± 0.10 vs. 8.28 ± 0.10; p < 0.001). Furthermore, the ITA strain showed increased EE content when fed the PP diet (9.84 ± 0.10 vs. 8.71 ± 0.10 [C]; p < 0.001).
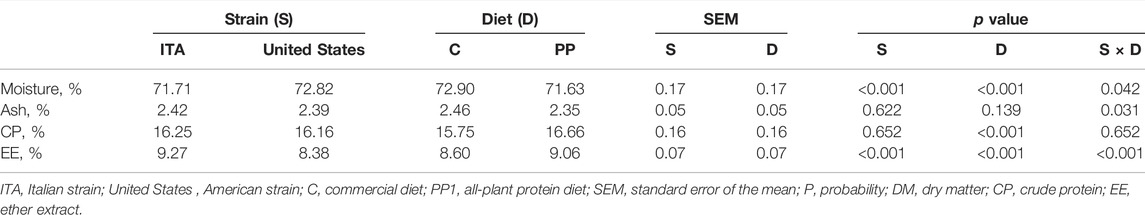
TABLE 3. Final whole-body proximate composition (as is) of the rainbow trout depending on strain, diet, and their interaction (n = 5).
Illumina sequencing of the 30 samples (24 mucosae + six feeds) yielded 1,015,591 high-quality reads, which were successfully taxonomically assigned according to the Silva database. The average number of reads (mean + SD) per sample was 24,838 ± 5,381 and 37,356 ± 4,810 for feed and gut mucosa samples, respectively (Supplementary data file S1). Rarefaction analysis of the chao1 index showed most of the curves approximating the saturation, with one intestinal sample from group United States_C failing only (Supplementary Figure S1). To ensure the adequateness of the remaining samples, the good coverage was also calculated, resulting in a good range (ranged 0.99–1) in all the samples.
When comparing the bacterial alpha diversity of gut mucosa samples, no significant differences were found in species richness and biodiversity indices (Table 4). Differently, the plant protein feed-related microbiota was characterized by higher species richness (Chao1 and Faith_PD) and lower diversity (Shannon and Simpson indices) than the C diet (Table 4).
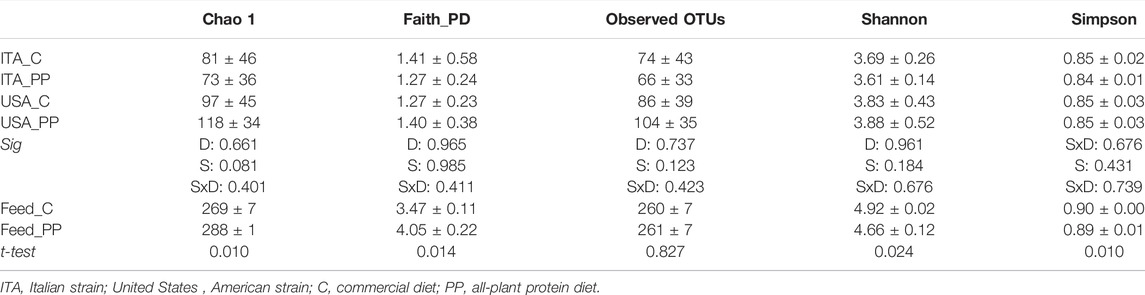
TABLE 4. Alpha diversity indices (mean ± SD, rarefied at 17,778 reads) of microbial communities of feed (n = 3) and gut mucosa (n = 6) samples. diet, D; strain, S.
The microbial community profiles of the feeds were outlined up to the genus level. Most of the sequences (81–82%) were of plant origin (p_Cyanobacteria and c_Chloroplast), deriving from vegetable ingredients of the diets. Considering only the most representative taxa, the overall feed-related microbial community comprised three phyla, four classes, five orders, 14 families, and 14 genera. The profiles of the feed-related microbial communities are shown at the phylum, family, and genus level in Figure 1. The list of the most abundant bacterial taxa, their relative abundance values, and statistical analysis are reported in Supplementary Table S1. Proteobacteria and Firmicutes were the dominant bacterial phyla in feeds, with a relative abundance of 13% and 4%, respectively (Figure 1A).
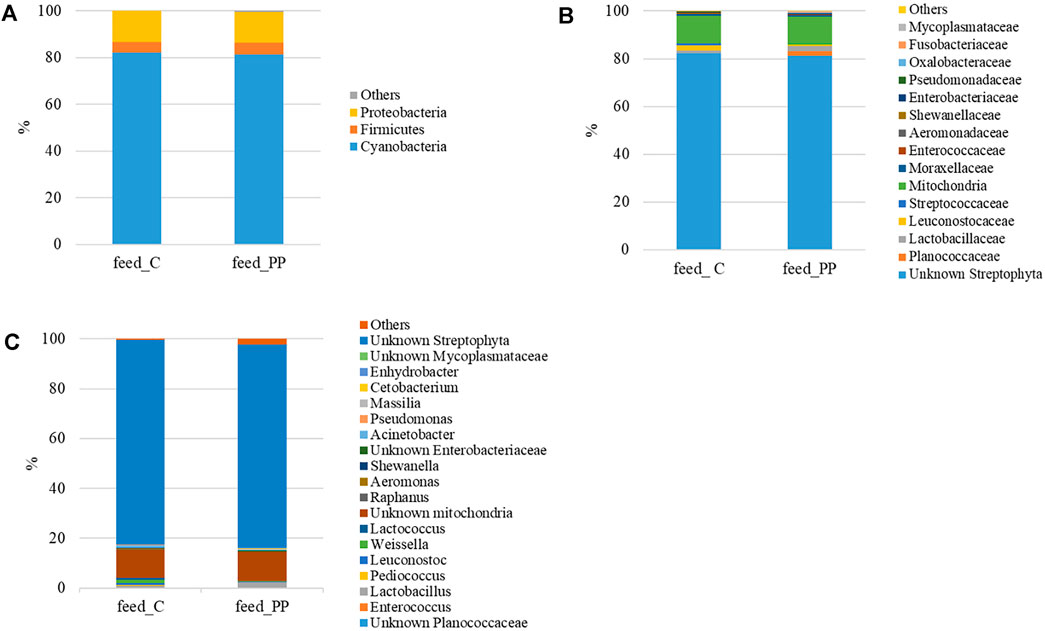
FIGURE 1. Mean relative abundance (%) of the most prevalent bacterial taxa in experimental feeds at (n = 3) phylum (A), family (B), and genus (C) levels.
Accordingly, Alphaproteobacteria and Bacilli were the predominant classes of bacteria in the feed samples. At the order level, Lactobacillales only differed between the two feeds, resulting in two times more abundant in the C diet than in the PP feed. Bacteria assigned to the Leuconostocaceae, Streptococcaceae, Aeromonadaceae, and Shewanellaceae families were preferentially associated with the C diet, while Fusobacteriaceae and Mycoplamataceae were found in the PP feed only (Figure 1B). Similarly, at the genus level, Pediococcus, Leuconostoc, Weissella, Lactococcus, Aeromonas, Shewanella, and Acinetobacter were significantly higher in the C diet when compared to the PP feed (Figure 1C). The relative abundance of the Lactobacillus genus only was increased in the PP diet, whereas Cetobacterium and Anhydrobacter genera were solely detected (Figure 1C).
The overall intestinal microbial community, considering only the most representative taxa, was mainly composed of three phyla, three classes, five orders, seven families, and six genera. The profiles of the intestinal microbial communities for each dietary treatment are shown at phylum, family, and genus level in Figure 2. The most abundant bacterial taxa with their relative abundance data and statistical analysis are listed in Supplementary Table S2.
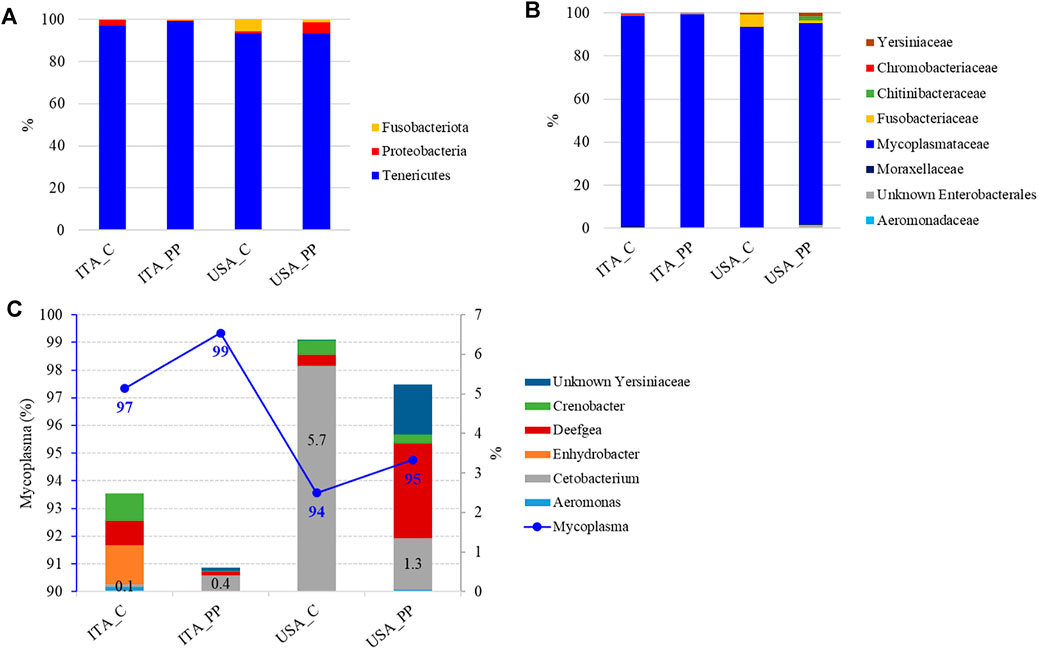
FIGURE 2. Mean relative abundance (%) of the most prevalent bacterial taxa in gut mucosa of two rainbow trout strains (the United States and ITA) fed experimental diets (C and PP) at phylum (A), and family (B), and genus (C) level. ITA_C (n = 6), ITA_PP (n = 6), United States_C (n = 5), and United States_PP (n = 6). Percentages of the Cetobacterium genus are indicated on histograms. Blueline and the corresponding values indicate the amount of Mycoplasma in each experimental feeding group.
To determine the impact of strain and diet on intestinal phylotypic diversity, UniFrac analysis was performed. Principal Coordinates Analysis (PCoA) of unweighted UniFrac distances revealed an effect of both the strain and the diet on gut microbial community profiles, with the first principal coordinate PC1 explaining up to 48.9% of the variation among the individuals (Figure 3).
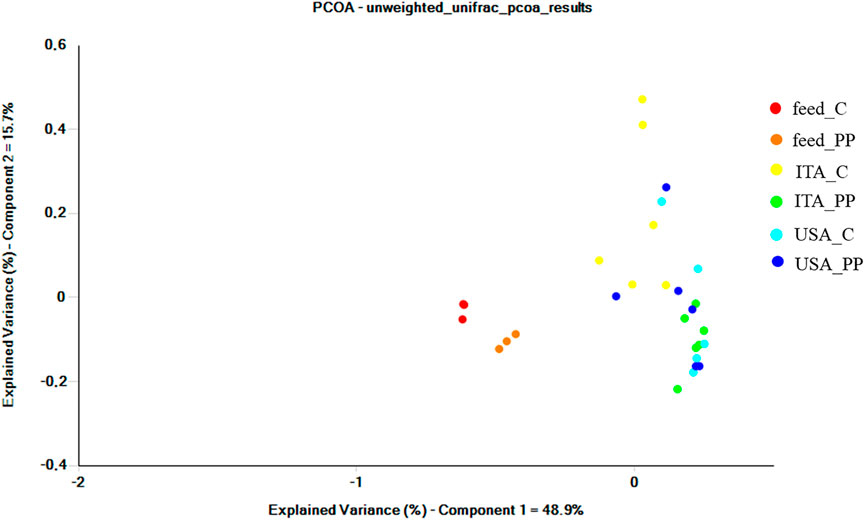
FIGURE 3. Beta diversity metrics. Principal coordinate analysis (PCoA) of unweighted UniFrac distances of microbial communities associated with gut mucosa and feeds. The figure shows the bi-dimensional plot of individual fish and feeds samples according to their microbiome profile.
Intestinal samples appeared clearly separated from the feed ones, thus indicating that observed differences between intestinal bacterial communities were not simply a consequence of undigested feed-related bacteria. Gut communities from the ITA strain clustered separately according to the diet. Similarly, distinct clusters were observed between the trout strains fed the C diet. Results of unweighted UniFrac PCoA were wholly confirmed by the non-parametric multivariate statistical test PERMANOVA (Table 5). The unweighted data showed a significant interaction between strain and diet. Results of pairwise comparisons showed differences between strains fed the same diet, while an overall effect of the diet was evident for the ITA strain only (p = 0.004). The weighted UniFrac analysis was also performed, but no significant differences were found among the groups (data not shown).
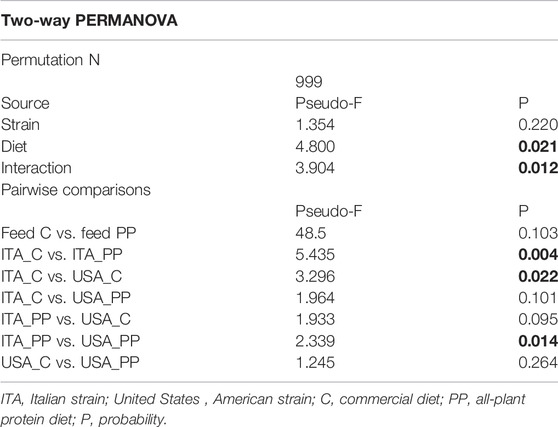
TABLE 5. Results of non-parametric multivariate analysis PERMANOVA on the unweighted UniFrac data of intestinal and feed samples. Significant p values (p ≤ 0.05) are in bold.
The autochthonous gut microbiome of rainbow trout was mostly dominated, regardless of the diet, by the Tenericutes phylum, mainly represented by the Mycoplasma genus, with a relative abundance ranging between 93 and 99% in all the samples (Figure 2). Proteobacteria and Fusobacteria completed the microbiota profile, while Firmicutes were unexpectedly not detected. The difference in gut microbiota composition between the two strains was limited to the Cetobacterium genus, belonging to the Fusobaceriaceae family (Figure 2). Two-way ANOVA indicated that the relative abundance of this genus was influenced by genotype only, with results being higher in the United States strain when compared to the ITA fish (Figure 2, Supplementary Table S2).
To attain a comprehensive analysis of the functional composition of the metagenome of each gut microbial community based on 16S rRNA sequencing, the PICRUSt method was used at Level 3 KEGG. Comparative analysis of PICRUSt functional inferences revealed an increased abundance of genes responsible for replication and repair pathway in both the ITA and the United States strains fed the all-plant protein diet (Figure 4). A total of six pathways were, instead, significantly different between the two strains when fed the C feed. Specifically, the pathway involved in starch and sucrose metabolism was more abundant in the United States trout in comparison with the ITA strain. On the contrary, the ITA trout showed an over-representation of the ubiquinone biosynthesis pathway (Figure 4).

FIGURE 4. Extended error bar plots showing PICRUSt-predicted bacterial KEGG pathways statistically different between experimental groups.
Independent of the administered diet, the United States rainbow trout strain in the present study overall showed better growth performance (in terms of increased FBW, iWGm, and SGR) when compared to the ITA one. Furthermore, a clear strain×diet interaction indicating an improvement in the iWG in the United States fish was observed when the C diet only was provided. This represents an interesting finding, as the selected strains usually outperform the non-selected ones on the plant-based selection diet (Overturf et al., 2013). Therefore, the ability of the United States rainbow trout to grow more efficiently when fed either the low-FM or the all-plant protein diets–with even more pronounced effects when using the former–reflects the genetic selection directed towards the utilization of the vegetable proteins, which efficiently integrates the physiological, carnivorous feeding behavior of this fish species. Analogously to what was reported by Overturf et al. (2013), the selected strain of the current research displayed similar FCR to that of the non-selected one, thus still not excluding a primary role of the feed intake–rather than the feed efficiency–in the improvement of the growth performance (Callet et al., 2017). Differently, the percent survival of the rainbow trout of the present study–while remaining high (≥ 95%)—displayed a clear strain × diet interaction, with the United States strain surviving more when fed the all-plant protein diet, and the ITA fish behaving similarly when fed the low-FM diet. This is in partial agreement with Callet et al. (2017) that observed higher survival in the selected strain fed the plant-based diets in comparison with the non-selected fish, thus reasonably reflecting genetic selection. As a final aspect to consider, independently of the strain, the rainbow trout used in the current research showed greater FBW and iWG when fed the PP diet. This may be related to the high protein digestibility of all-plant protein diets that have previously been highlighted, as such diets are usually formulated with highly digestible protein concentrates (Gaylord et al., 2008; Callet et al., 2017; Brezas and Hardy, 2020). Furthermore, the ability of rainbow trout to grow efficiently on a diet containing soy protein concentrate-based blend as a replacement of high percentages (until 87%) of the FM in the formulation has already been reported (Burr et al., 2012).
Independent of the considered strain, the PP diet led to the lower whole-body moisture content in comparison with the C one, as previously reported (Overturf et al., 2013). Furthermore, a clear strain × diet interaction indicating a reduction in the moisture content of the ITA fish was observed when the PP diet only was provided. This may represent the logical consequence of the increased whole-body EE identified in the PP-fed ITA rainbow trout when compared to the United States fish fed the same diet. As far as ash content is concerned, the C diet determined an overall increase in the United States strain only. Overturf et al. (2013) previously reported that whole-body ash was unaffected in either the selected or the non-selected rainbow trout in response to both a FM- or PP-based diet, but no information about selection-related ash digestibility changes is currently available. Therefore, the increased ash content herein highlighted may result from a better ash digestibility, thus potentially strengthening the effect of the higher ash content of the C diet than the PP feed. Differently, the United States strain showed that the whole-body CP content was similar to the non-selected one, as previously reported (Overturf et al., 2013). This also confirms that CP digestibility is not different in the selected rainbow trout as previously observed (Callet et al., 2017; Brezas and Hardy, 2020). However, independently of the considered strain, the PP diet led to an increased CP content in comparison with the C feed, thus reflecting the above-mentioned high digestibility of the vegetable proteins (Gaylord et al., 2008; Callet et al., 2017; Brezas and Hardy, 2020). As a final aspect to consider, the whole-body EE content of the rainbow trout of the present study displayed a clear strain×diet interaction, with the ITA strain body containing more EE than the United States fish when fed the all-plant protein diet. Genetic selection towards the utilization of vegetable proteins has been reported to not influence the whole-body EE content of rainbow trout (Overturf et al., 2013; Callet et al., 2017; Brezas and Hardy, 2020). However, the higher whole-body EE content of the PP-fed ITA rainbow trout when compared to the United States fish may result from a synergic effect of both the physiological response to all-plant protein diets (Burel et al., 1998; Kaushik et al., 2004; Lazzarotto et al., 2015) and the use of a C diet (characterized by higher EE content) across the generations.
The high-throughput sequencing on the Illumina MiSeq platform has been used in the present research for intestinal microbiome characterization. In line with the previous studies performed on rainbow trout, results of the metabarcoding analysis showed that the most abundant phylum of autochthonous gut microbiota in rainbow trout, regardless of the diet and genotype, was Tenericutes, mainly represented by the Mycoplasma genus (Rimoldi et al., 2019). Accordingly, it has recently been suggested that Mycoplasma, due to its dominance in the distal intestine of rainbow trout and other farmed salmonids, could have a mutualistic relationship with its host (Rasmussen et al., 2021). In Atlantic salmon (Salmo salar), the abundance of Mycoplasma has positively been associated with improved growth, carotenoid utilization, and disease resilience of the host (Bozzi et al., 2021). In addition, this genus harbors genes for long-chain polymer degradation, such as chitin, which is particularly abundant in insects, that are part of the natural diet of juvenile salmonids. This ability of Mycoplasma could be beneficial for its host, which can utilize all the nutritional value of a chitin-rich diet. Indeed, an increased presence of Mycoplasma was also shown in the gut of rainbow trout fed an insect-based diet (Rimoldi et al., 2019). An unexpected result was the absence of bacteria belonging to the Firmicutes phylum in all the gut samples, even in those fish fed the plant-based diet. Members of this phylum are known to play a key role in the fermentation of dietary carbohydrates (Corrigan et al., 2015), and are usually associated with plant ingredients, while low-FM diet generally favors the presence of Proteobacteria (Desai et al., 2012; Ingerslev et al., 2014a and b; Rimoldi et al., 2018). An increase in Firmicutes to Proteobacteria ratio was, however, observed by Blaufuss et al. (2020) in analogously selected trout for growth on an all-plant protein diet when fed all-plant protein feeds.
In terms of alpha diversity, no differences were detected between ITA and United States strains in microbial richness and biodiversity, regardless of the administered diet. Comparison of the overall microbial composition among all the samples according to weighted and unweighted UniFrac showed an overlap among all the gut samples, with the feed samples clustering separately. The composition of feed-related microbial communities was, indeed, significantly different from gut microbiota, resulting mainly constituted of cyanobacteria, Proteobacteria, and Firmicutes phyla. According to PERMANOVA analysis, unweighted UniFrac indicated that either the diet or the host genotype plays a crucial role in shaping the gut microbiome, showing differences between the two strains fed the same diet and an overall effect of diet for ITA strain only. This result could explain the differences observed in growth performance between the two strains, with the United States trout growing more efficiently than the ITA fish when fed either the low-FM or the all-plant protein diets.
The current results seemed to indicate an existence of a relationship between the selected United States strain and the Cetobacterium genus, belonging to the Fusobacteriaceae family, whose relative abundance was an order of magnitude larger than that detected in the ITA strain. Similarly, higher abundances of the phyla Bacteroidetes and Fusobacteria phyla are associated with trout selected for high-muscle yield (Chapagain et al., 2020). Fusobacteria is commonly found in the freshwater fish guts, and Cetobacterium is an important commensal bacterium (Navarrete et al., 2012). Metabolites produced by Cetobacterium include, indeed, short-chain fatty acids (SCFAs), such as acetate, propionate, and butyrate, and vitamin B12, which can enhance fish health (Xie et al., 2021).
The in silico prediction of the metabolic capability of the bacterial populations revealed an up-regulation of the pathway related to starch and sucrose metabolism in the United States strain fed the C diet, thus indicating a better ability to utilize carbohydrates. This is not surprising, as the United States strain has been selected for its growth performance when fed all-plant protein feeds (rich in fibers and non-digestible carbohydrates). The fatty acid metabolism pathway has previously been reported to be enriched in high-muscle yield selected trout (Chapagain et al., 2020). However, despite the different pathways being activated, an up-regulation of bacterial pathways involved in energy supply–which is essential for host growth–was herein analogously observed.
In conclusion, the selected strain of rainbow trout overall displayed improved growth performance and (to a lesser extent) whole-body proximate composition when compared to the non-selected one. However, the absence of negative changes in the non-selected strain in response to all-plant protein feed is indicative of a proper adaptation of the fish to such protein sources over time, thus suggesting that a complete FM replacement in diets for rainbow trout is technically feasible. Furthermore, differences between microbial communities were mainly driven by host genetic selection, with the Cetobacterium probiotic-like genus being associated with the gut of the selected rainbow trout. Such microbiota signature, along with the up-regulation of the pathway involved in starch and sucrose metabolism, may reasonably predispose the selected strain to efficiently utilize all-plant protein feeds. Future research assessing additional parameters related to the gut health of the fish (i.e., gut mucosa morphology, mucin dynamics, inflammatory response, and metabolomics) are strongly recommended to further investigate the role of genetic selection in the digestion of all-plant protein diets.
All fastq sequencing files were deposited in the EBI ENA public database under the accession project code PRJEB51166.
The animal study was reviewed and approved by Ethical Committee of the University of Turin.
LG, GT, MS, and RH designed the study. IB, CC, GC, MP, and SBO carried out the rearing work. CC and GC performed the whole-body proximate composition analysis. SR and GT performed the intestinal microbiome analysis. IB wrote the first draft of the manuscript. All the authors contributed to the article and approved the submitted version.
This research was funded by the AGER project “Fine Feed for Fish (4F),” Rif. nr. 2016-01-01. Dr. Giulia Chemello was supported by a research grant of the University of Turin.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors are grateful to Mr. Dario Sola for fish care and technical support.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2022.892550/full#supplementary-material
Abernathy J., Brezas A., Snekvik K. R., Hardy R. W., Overturf K. (2017). Integrative Functional Analyses Using Rainbow trout Selected for Tolerance to Plant Diets Reveal Nutrigenomic Signatures for Soy Utilization without the Concurrence of Enteritis. PLoS ONE 12, e0180972. doi:10.1371/journal.pone.0180972
Blaufuss P. C., Bledsoe J. W., Gaylord T. G., Sealey W. M., Overturf K. E., Powell M. S. (2020). Selection on a Plant-Based Diet Reveals Changes in Oral Tolerance, Microbiota and Growth in Rainbow trout (Oncorhynchus mykiss) when Fed a High Soy Diet. Aquaculture 525, 735287. doi:10.1016/j.aquaculture.2020.735287
Bolyen E., Rideout J. R., Dillon M. R., Bokulich N. A., Abnet C. C., Ghalith G. A., et al. (2019). Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol 37, 852–857. doi:10.1038/s41587-019-0209-9
Bozzi D., Rasmussen J. A., Carøe C., Sveier H., Nordøy K., Gilbert M. T. P., et al. (2021). Salmon Gut Microbiota Correlates with Disease Infection Status: Potential for Monitoring Health in Farmed Animals. Anim. Microbiome 3, 1–17. doi:10.1186/s42523-021-00096-2
Brezas A., Hardy R. W. (2020). Improved Performance of a Rainbow trout Selected Strain Is Associated with Protein Digestion Rates and Synchronization of Amino Acid Absorption. Sci. Rep. 10, 4678. doi:10.1038/s41598-020-61360-0
Brezas A., Kumar V., Overturf K., Hardy R. W. (2021). Dietary Amino Acid Supplementation Affects Temporal Expression of Amino Acid Transporters and Metabolic Genes in Selected and Commercial Strains of Rainbow trout (Oncorhynchus mykiss). Comp. Biochem. Physiol. B: Biochem. Mol. Biol. 255, 110589. doi:10.1016/j.cbpb.2021.110589
Burel C., Boujard T., Corraze G., Kaushik S. J., Boeuf G., Mol K. A., et al. (1998). Incorporation of High Levels of Extruded Lupin in Diets for Rainbow trout (Oncorhynchus mykiss): Nutritional Value and Effect on Thyroid Status. Aquaculture 163, 325–345. doi:10.1016/S0044-8486(98)00241-5
Burr G. S., Wolters W. R., Barrows F. T., Hardy R. W. (2012). Replacing Fishmeal with Blends of Alternative Proteins on Growth Performance of Rainbow trout (Oncorhynchus mykiss), and Early or Late Stage Juvenile Atlantic salmon (Salmo salar). Aquaculture 334-337, 110–116. doi:10.1016/j.aquaculture.2011.12.044
Callet T., Dupont-Nivet M., Danion M., Burel C., Cluzeaud M., Surget A., et al. (2021). Why Do Some Rainbow Trout Genotypes Grow Better with a Complete Plant-Based Diet? Transcriptomic and Physiological Analyses on Three Isogenic Lines. Front. Physiol. 12, 732321. doi:10.3389/fphys.2021.732321
Callet T., Médale F., Larroquet L., Surget A., Aguirre P., Kerneis T., et al. (2017). Successful Selection of Rainbow trout (Oncorhynchus mykiss) on Their Ability to Grow with a Diet Completely Devoid of Fishmeal and Fish Oil, and Correlated Changes in Nutritional Traits. PLoS One 12, e0186705. doi:10.1371/journal.pone.0186705
Chapagain P., Walker D., Leeds T., Cleveland B. M., Salem M. (2020). Distinct Microbial Assemblages Associated with Genetic Selection for High- and Low- Muscle Yield in Rainbow trout. BMC Genomics 21, 820. doi:10.1186/s12864-020-07204-7
Cleveland B. M., Gao G., Leeds T. D. (2020). Transcriptomic Response to Selective Breeding for Fast Growth in Rainbow Trout (Oncorhynchus mykiss). Mar. Biotechnol. 22, 539–550. doi:10.1007/s10126-020-09974-3
Corrigan A., De Leeuw M., Penaud-Frézet S., Dimova D., Murphy R. A. (2015). Phylogenetic and Functional Alterations in Bacterial Community Compositions in Broiler Ceca as a Result of Mannan Oligosaccharide Supplementation. Appl. Environ. Microbiol. 81, 3460–3470. doi:10.1128/AEM.04194-14
Desai A. R., Links M. G., Collins S. A., Mansfield G. S., Drew M. D., Van Kessel A. G., et al. (2012). Effects of Plant-Based Diets on the Distal Gut Microbiome of Rainbow trout (Oncorhynchus mykiss). Aquaculture 350-353, 134–142. doi:10.1016/j.aquaculture.2012.04.005
FAO (2020). The State of World Fisheries and Aquaculture 2020.” in Sustainability in Action. Rome: FAO. doi:10.4060/ca9229en
Gaudioso G., Marzorati G., Faccenda F., Weil T., Lunelli F., Cardinaletti G., et al. (2021). Processed Animal Proteins from Insect and Poultry By-Products in a Fish Meal-Free Diet for Rainbow Trout: Impact on Intestinal Microbiota and Inflammatory Markers. Int. J. Mol. Sci 22, 5454. doi:10.3390/ijms22115454
Gaylord T. G., Barrows F. T., Rawles S. D. (2008). Apparent Digestibility of Gross Nutrients from Feedstuffs in Extruded Feeds for Rainbow Trout,Oncorhynchus mykiss. J. World Aquac. Soc. 39, 827–834. doi:10.1111/j.1749-7345.2008.00220.x
Hammer Ø., Harper D. A. T., Paul D. R. (2001). Past: Paleontological Statistics Software Package for Education and Data Analysis. Palaeontol. Electron 4 (1), 9.
Ingerslev H.-C., Strube M. L., Jørgensen L. v. G., Dalsgaard I., Boye M., Madsen L. (2014a). Diet Type Dictates the Gut Microbiota and the Immune Response against Yersinia ruckeri in Rainbow trout (Oncorhynchus mykiss). Fish Shellfish Immunol. 40, 624–633. doi:10.1016/j.fsi.2014.08.021
Ingerslev H.-C., von Gersdorff Jørgensen L., Lenz Strube M., Larsen N., Dalsgaard I., Boye M., et al. (2014b). The Development of the Gut Microbiota in Rainbow trout (Oncorhynchus mykiss) Is Affected by First Feeding and Diet Type. Aquaculture 424-425, 24–34. doi:10.1016/j.aquaculture.2013.12.032
Kaushik S. J., Covès D., Dutto G., Blanc D. (2004). Almost Total Replacement of Fish Meal by Plant Protein Sources in the Diet of a marine Teleost, the European Seabass, Dicentrarchus labrax. Aquaculture 230, 391–404. doi:10.1016/S0044-8486(03)00422-8
Krogdahl Å., Penn M., Thorsen J., Refstie S., Bakke A. M. (2010). Important Antinutrients in Plant Feedstuffs for Aquaculture: an Update on Recent Findings Regarding Responses in Salmonids. Aquac. Res. 41, 333–344. doi:10.1111/j.1365-2109.2009.02426.x
Langille M. G. I., Zaneveld J., Caporaso J. G., McDonald D., Knights D., Reyes J. A., et al. (2013). Predictive Functional Profiling of Microbial Communities Using 16S rRNA Marker Gene Sequences. Nat. Biotechnol. 31, 814–821. doi:10.1038/nbt.2676
Lazzarotto V., Corraze G., Leprevost A., Quillet E., Dupont-Nivet M., Médale F. (2015). Three-year Breeding Cycle of Rainbow trout (Oncorhynchus mykiss) Fed a Plant-Based Diet, Totally Free of marine Resources: Consequences for Reproduction, Fatty Acid Composition and Progeny Survival. PLoS One 10, e0117609. doi:10.1371/journal.pone.0117609
Lozupone C. A., Hamady M., Kelley S. T., Knight R. (2007). Quantitative and Qualitative β Diversity Measures Lead to Different Insights into Factors that Structure Microbial Communities. Appl. Environ. Microbiol. 73, 1576–1585. doi:10.1128/AEM.01996-06
Lozupone C., Knight R. (2005). UniFrac: a New Phylogenetic Method for Comparing Microbial Communities. Appl. Environ. Microbiol. 71, 8228–8235. doi:10.1128/AEM.71.12.8228-8235.2005
Navarrete P., Magne F., Araneda C., Fuentes P., Barros L., Opazo R., et al. (2012). PCR-TTGE Analysis of 16S rRNA from Rainbow trout (Oncorhynchus mykiss) Gut Microbiota Reveals Host-specific Communities of Active Bacteria. PLoS One 7, e31335. doi:10.1371/journal.pone.0031335
Overturf K., Barrows F. T., Hardy R. W. (2013). Effect and Interaction of Rainbow trout Strain (Oncorhynchus mykiss) and Diet Type on Growth and Nutrient Retention. Aquac. Res. 44, 604–611. doi:10.1111/j.1365-2109.2011.03065.x
Parks D. H., Tyson G. W., Hugenholtz P., Beiko R. G. (2014). STAMP: Statistical Analysis of Taxonomic and Functional Profiles. Bioinformatics 30, 3123–3124. doi:10.1093/bioinformatics/btu494
Piazzon M. C., Naya-Català F., Perera E., Palenzuela O., Sitjà-Bobadilla A., Pérez-Sánchez J. (2020). Genetic Selection for Growth Drives Differences in Intestinal Microbiota Composition and Parasite Disease Resistance in Gilthead Sea Bream. Microbiome 8, 168. doi:10.1186/s40168-020-00922-w
Rasmussen J. A., Villumsen K. R., Duchêne D. A., Puetz L. C., Delmont T. O., Sveier H., et al. (2021). Genome-resolved Metagenomics Suggests a Mutualistic Relationship between Mycoplasma and Salmonid Hosts. Commun. Biol. 4, 579. doi:10.1038/s42003-021-02105-1
Rimoldi S., Antonini M., Gasco L., Moroni F., Terova G. (2021). Intestinal Microbial Communities of Rainbow trout (Oncorhynchus mykiss) May Be Improved by Feeding a Hermetia Illucens Meal/low-Fishmeal Diet. Fish. Physiol. Biochem. 47, 365–380. doi:10.1007/s10695-020-00918-1
Rimoldi S., Gini E., Iannini F., Gasco L., Terova G. (2019). The Effects of Dietary Insect Meal from Hermetia Illucens Prepupae on Autochthonous Gut Microbiota of Rainbow trout (Oncorhynchus mykiss). Animals 9, 143. doi:10.3390/ani9040143
Rimoldi S., Terova G., Ascione C., Giannico R., Brambilla F. (2018). Next Generation Sequencing for Gut Microbiome Characterization in Rainbow trout (Oncorhynchus mykiss) Fed Animal By-Product Meals as an Alternative to Fishmeal Protein Sources. PLoS One 13, e0193652–29. doi:10.1371/journal.pone.0193652
Terova G., Gini E., Gasco L., Moroni F., Antonini M., Rimoldi S. (2021). Effects of Full Replacement of Dietary Fishmeal with Insect Meal from Tenebrio molitor on Rainbow trout Gut and Skin Microbiota. J. Anim. Sci Biotechnol 12, 30. doi:10.1186/s40104-021-00551-9
Terova G., Rimoldi S., Ascione C., Gini E., Ceccotti C., Gasco L. (2019). Rainbow trout (Oncorhynchus mykiss) Gut Microbiota Is Modulated by Insect Meal from Hermetia Illucens Prepupae in the Diet. Rev. Fish. Biol. Fish. 29, 465–486. doi:10.1007/s11160-019-09558-y
Vallejo R. L., Fragomeni B. O., Cheng H., Gao G., Long R. L., Shewbridge K. L., et al. (2020). Assessing Accuracy of Genomic Predictions for Resistance to Infectious Hematopoietic Necrosis Virus with Progeny Testing of Selection Candidates in a Commercial Rainbow Trout Breeding Population. Front. Vet. Sci. 7, 590048. doi:10.3389/fvets.2020.590048
Vargas-Albores F., Martínez-Córdova L. R., Hernández-Mendoza A., Cicala F., Lago-Lestón A., Martínez-Porchas M. (2021). Therapeutic Modulation of Fish Gut Microbiota, a Feasible Strategy for Aquaculture? Aquaculture 544, 737050. doi:10.1016/j.aquaculture.2021.737050
Venold F. F., Penn M. H., Krogdahl Å., Overturf K. (2012). Severity of Soybean Meal Induced Distal Intestinal Inflammation, Enterocyte Proliferation Rate, and Fatty Acid Binding Protein (Fabp2) Level Differ between Strains of Rainbow trout (Oncorhynchus mykiss). Aquaculture 364-365, 281–292. doi:10.1016/j.aquaculture.2012.08.035
Xie M., Zhou W., Xie Y., Li Y., Zhang Z., Yang Y., et al. (2021). Effects of Cetobacterium Somerae Fermentation Product on Gut and Liver Health of Common Carp (Cyprinus carpio) Fed Diet Supplemented with Ultra-micro Ground Mixed Plant Proteins. Aquaculture 543, 736943. doi:10.1016/j.aquaculture.2021.736943
Xu K., Duan W., Xiao J., Tao M., Zhang C., Liu Y., et al. (2015). Development and Application of Biological Technologies in Fish Genetic Breeding. Sci. China Life Sci. 58, 187–201. doi:10.1007/s11427-015-4798-3
Keywords: rainbow trout, selective breeding, performance, vegetable proteins, gut microbiome
Citation: Biasato I, Rimoldi S, Caimi C, Bellezza Oddon S, Chemello G, Prearo M, Saroglia M, Hardy R, Gasco L and Terova G (2022) Efficacy of Utilization of All-Plant-Based and Commercial Low-Fishmeal Feeds in Two Divergently Selected Strains of Rainbow Trout (Oncorhynchus mykiss): Focus on Growth Performance, Whole-Body Proximate Composition, and Intestinal Microbiome. Front. Physiol. 13:892550. doi: 10.3389/fphys.2022.892550
Received: 09 March 2022; Accepted: 11 April 2022;
Published: 20 May 2022.
Edited by:
Mohamed Salah Azaza, National Institute of Marine Science and Technology, TunisiaReviewed by:
Ines Ben Khemis, Institut National des Sciences et Technologies de la Mer (Tunisia), TunisiaCopyright © 2022 Biasato, Rimoldi, Caimi, Bellezza Oddon, Chemello, Prearo, Saroglia, Hardy, Gasco and Terova. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Laura Gasco, bGF1cmEuZ2FzY29AdW5pdG8uaXQ=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.