- Department of Cell Biology and Imaging, Faculty of Biology, Institute of Zoology and Biomedical Research, Jagiellonian University, Krakow, Poland
Autophagy is a self-degradative process which plays a role in removing misfolded or aggregated proteins, clearing damaged organelles, but also in changes of cell membrane size and shape. The aim of this phenomenon is to deliver cytoplasmic cargo to the lysosome through the intermediary of a double membrane-bound vesicle (autophagosome), that fuses with a lysosome to form autolysosome, where cargo is degraded by proteases. Products of degradation are transported back to the cytoplasm, where they can be re-used. In the present study we showed that autophagy is important for proper functioning of the glia and that it is involved in the regulation of circadian structural changes in processes of the pacemaker neurons. This effect is mainly observed in astrocyte-like glia, which play a role of peripheral circadian oscillators in the Drosophila brain.
Introduction
Autophagy is an evolutionary conserved process involved in cellular responses to starvation and other stress, and it also plays an important role in development, cell death, aging and immunity. Autophagy maintains cellular homeostasis under normal but also under stress conditions. In general, it is used for degradation of long-lived proteins. Three autophagy pathways have already been described: macroautophagy, microautophagy and chaperone-mediated autophagy. The most important for proper cell functioning seems to be macroautophagy, here referred as autophagy. It depends on the group of proteins called Autophagy-related (Atgs), which are involved in the formation of autophagosomes (Mizushima et al., 1998; Mizushima et al., 1999; Shintani et al., 1999; Tanida et al., 1999; Suzuki et al., 2007). The autophagy process starts when the kinase Atg1 forms complexes with other proteins, Unc76 and Atg13, and phosphorylates them (Reggiori et al., 2004). These complexes bind to fragments of endoplasmatic reticulum membrane and start to form an omegasome, developing cup-shape double-membrane structure. Phagophore formation is Atg9-dependent. Crucial role in this process plays also Atg8 protein. C-terminal amino acid following a glycine residue of Atg8 is cleaved by cystein protease belonging to Atg4 family. In the next step, the exposed glycine is conjugated to Atg7 (E1-like enzyme) and Atg3 (E2-like enzyme). In the meantime, Atg7 activates Atg12, and then Atg10 (E2-like enzyme) catalyzes Atg12 conjugation to Atg5. Atg5-Atg12 complex enhances the covalent binding of Atg8 to the membrane lipid of forming phagophore (Ichimura et al., 2000; Matsushita et al., 2007). This molecular mechanism allows sequestration of cytosolic components, and forming vesicles, called autophagosomes, which delivers the cargo to lysosomes for recycling (Kuma et al., 2002; Axe et al., 2008). Upon autophagosome formation, the inner autophagosome membrane and its contents are degraded by lysosomal hydrolases and cargo is subsequently released into the cytosol for recycling. Autophagy induced by structural remodeling of the cell increases the level of nutrients and energy, and removes damaged elements (Vabulas and Hartl, 2005).
The main pathway that regulates autophagy uses the protein kinase Target of rapamycin (TOR). When nutrients are available in the cell, TOR is activated through the Class Iphosphatidylinositol-3-kinase (PI3K) signaling pathway, and inhibits autophagy through direct phosphorylation and repression of Atg1 (Kamada et al., 2000; Scott et al., 2007). When nutrients are scarce, TOR becomes inactivated, its repression of Atg1 is relieved, and autophagy is induced. Another mechanism of the autophagy activation involves the engulfment receptor Draper (McPhee et al., 2010).
The knowledge about circadian regulation of autophagy and its impact on brain functioning is poorly recognized. It has been found, however, that TOR signaling and autophagy are involved in the regulation of circadian rhythms in behavior and neuronal plasticity in Drosophila melanogaster (Kijak and Pyza, 2017). Fruit flies provide an excellent animal model to study autophagy in vivo, as autophagy genes (Atg) and their regulators (like Tor) are conserved between insects and mammals (Meijer et al., 2007). The molecular mechanism of the circadian clock (reviewed by Özkaya and Rosato, 2012) and circadian neuronal plasticity have also been well described in this model (reviewed by Krzeptowski et al., 2018a). Rhythmic changes in behavior, metabolism and gene/protein expression are regulated by the main clock (pacemaker) as well as by peripheral oscillators located in many different tissues. The most important pacemaker neurons are the small ventral lateral neurons (sLNv) (Grima et al., 2004; Stoleru et al., 2004; Picot et al., 2007; Cusumano et al., 2009; Zhang et al., 2009; Yao and Shafer, 2014; Chatterjee et al., 2018; Díaz et al., 2019; Schlichting et al., 2019), which terminate in the dorsal brain. The proper functioning of clock neurons regulates rhythmic changes in behavior, like sleep/wake pattern and activity level. sLNv communicate with sleep-promoting dopaminergic cells (Potdar and Sheeba, 2018) and posterior lateral protocerebrum (PLP) AstA-expressing neurons (Chen et al., 2016) through PDF signaling, as well as with sleep centers located in mushroom bodies. One of the phenomena involved in the regulation of behavior is the daily structural plasticity of the sLNv terminals. The arborization complexity of these processes changes to have more complex branching at the beginning of the day than during the night (Fernández et al., 2008). This remodeling affects daily changes in synapse number and size within PDF processes (Fernández et al., 2008; Bushey et al., 2011) and causes changes of postsynaptic partners. In effect sLNvs communication with mushroom bodies is time-dependent (Gorostiza et al., 2014). sLNv daily structural remodeling is regulated by both, the pacemaker and glial clocks (Herrero et al., 2017). However, the mechanism by which glia can modulate clock neuron plasticity has not been described yet. In the present work, we showed that autophagy in glia plays a role in the regulation of sleep length during the night. This connection is observed for specific glia types, like astrocytes, which are involved in the maintenance of structural remodeling of the sLNv terminals.
Materials and Methods
Fly Strains
The following strains of Drosophila melanogaster were used: repo-Gal4 (Gal4 expressed in all glial cell types, kindly donated by Dr. J. Giebultowicz, Oregon State University), R29A12 (netB-Gal4 expressing Gal4 in the epithelial glia and medulla chandelier glia, BDSC no. 49478) (Edwards et al., 2012), alrm-Gal4 (astrocyte-like glia, BDSC no. 67032) (Doherty et al., 2009), moody-Gal4 (subperineurial and pseudocartridge glia, kindly donated by Dr. C. Klambt, Muenster University) (Edwards et al., 2012), Wnt4-Gal4 (chiasm giant glia, BDSC no. 49102) (Edwards et al., 2012), UAS-atg7RNAi (BDSC no. 27707), UAS-atg5RNAi (BDSC no. 34899) (strains with expression of dsRNA for specific gene under control of UAS sequence). Responder lines were crossed with Tub-Gal80ts to obtain strains with temperature dependent expression of Gal4.
Flies were maintained on a standard cornmeal medium under LD12:12 (12 h of light and 12 h of darkness) regime and at constant temperature 20°C. Two days old males of crosses were transferred to 29°C to induce adult specific gene silencing in the specific glia type.
Behavioral Assays
Locomotor activity was recorded at 29°C using Drosophila Activity Monitoring System (DAMS, Trikinetics, Waltham) for 3 days in LD12:12 and 5 days in constant darkness (DD). Activity was counted every 1 min and analyzed in Excel by using “Befly!” software (Department of Genetics, Leicester University). Lomb–Scargle normalized periodogram was used to determine rhythmic flies; individuals with period value lower than 10 (confidence level 0.05) were regarded as arrhythmic. Flies, which did not survive until the end of experiments were removed from analyses. Every experiment was repeated three times, at least 60 flies in total were used, detailed data are presented in Supplementary Table S1.
Sleep analysis was performed on the third day of LD12:12, and sleep was recorded as at least 5 min of a fly immobility.
Immunohistochemistry
Flies were collected at ZT2 (two hours after lights-on in LD 12:12) and ZT14 (two hours after lights-off in LD12:12), their heads were fixed in 4% paraformaldehyde and brains were isolated. After washing in 0.2% phosphate buffer saline with Triton X-100 (PBST) and 30 min of blocking in Normal Goat Serum (NGS) they were incubated overnight with anti-PDF primary antibody (PDF C7 1:500, Developmental Studies Hybridoma Bank). Next, samples were washed in PBST and incubated with secondary antibodies (1:500 anti-mouse Cy3, Abcam). Whole brains were mounted in Vectashield medium (Vector) and examined with a Zeiss Meta 510 Laser Scanning Microscope.
Scholl Analysis
To visualize axon projections of the sLNvs whole brain confocal images were used. Pictures were taken using 40 × objective. Scholl’s analysis plugin in ImageJ software was used to quantify the axonal arbour in the dorsal protocerebrum. The point where the first dorsal ramification opens up was manually selected, and based on this software created concentric rings. The number of intersections of each projection with a particular ring was counted. The total number of intersections was calculated (according to Fernández et al., 2008).
Gene Expression Analysis by qPCR
repo > GFP flies were collected at specific time points (ZT1, ZT4, ZT13, and ZT16, where ZT0 marks the time of lights on, ZT12—lights off) and glia cells were obtained using FACS sorting as described in (Damulewicz et al., 2015). Expression of the selected genes was examined using a quantitative PCR (qPCR) technique. The total RNA was isolated using a TriReagent (Invitrogen), and the RNA quality was assessed by Nanodrop. To prepare cDNA, 1 μg of the total RNA was used with the High Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific) and random primers. cDNA (diluted 1:10) was used for SybrGreen qPCR (KapaBiosystems). The specific primers (the specificity was controlled with Primer BLAST and gel electrophoresis) used for the reaction are listed in Table 1. Expression level was calculated by ΔΔCT method. The reference gene used was rpl32.
Statistical Analysis
GraphPad Prism software was used for statistics and making graphs. Outliers were removed using Grubbs’ test (GraphPad online software). Shapiro-Wilk’s test was used to check normality in distribution. Data were analyzed using one-way ANOVA with post-hoc Tukey’s test or t test to detect statistically significant differences between groups. Detailed statistics are presented in Supplementary Table S2.
Results
Autophagy in Glia is a Rhythmic Process
Daily changes have been observed in many processes and they are regulated by circadian oscillators located in the brain and in other organs. One of these oscillators is located in glia (called glia clocks), but the mechanism of their functioning is not fully recognized yet. To check whether autophagy level in glia oscillates during the day we sorted out glial cells marked with GFP at specific time points, isolated mRNA and analyzed autophagy-related gene expression level. The obtained results showed that there are daily changes in the expression pattern of several genes. The atg5 gene had higher expression during the day (ZT1, ZT4) than during the night (ZT13, ZT16) (Figure 1A), atg7—in the middle of the day (ZT4) and at the beginning of the night (ZT13) (Figure 1B), while atg10 showed the highest mRNA level at early night (ZT13) (Figure 1C). The gene draper that is involved in the activation of autophagy had maximum of the transcript level during the night (ZT16) (Figure 1D), while Tor–encoding the repressor of autophagy exhibited no daily changes in the mRNA level (Figure 1E).
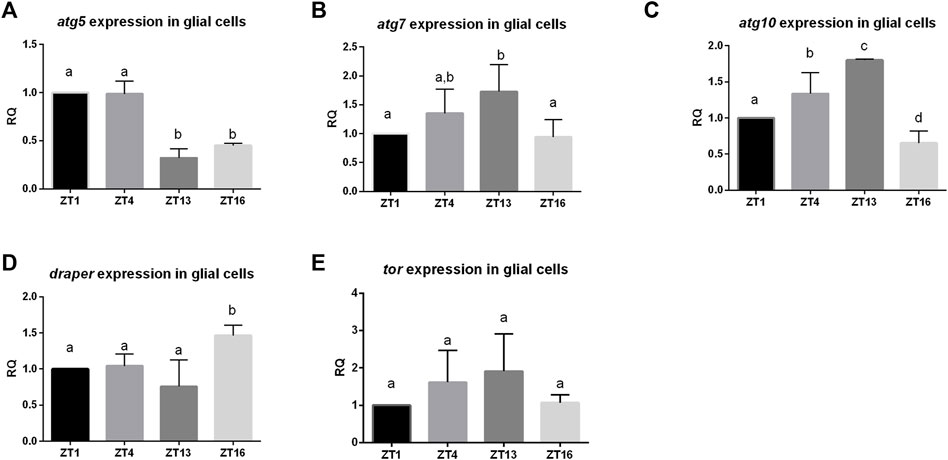
FIGURE 1. Daily pattern of expression of genes involved in autophagy in glia. Glia cells were sorted out at selected time points (ZT1,ZT4, ZT13, ZT16). qPCR data were calculated as ΔΔCT and normalized to ZT1 as 1. Statistically significant differences are marked as letters: different letters above two bars mean there is statistically significant difference. Genes analysed: (A)atg5 (B) atg7, (C) atg10 (D) draper, (E) tor.
Autophagy in Glia is Necessary for Sleep Regulation
We used transgenic flies to manipulate the selected gene expression level in all glia (repo-Gal4). To avoid developmental changes caused by this genetic manipulation, we forced adult-specific dsRNA production using TARGET system, in which higher temperature inhibits temperature-sensitive Gal80 and allows Gal4 activation. In effect we weakened autophagy process only in glia of adult flies. Silencing of atg5 and atg7 did not affect circadian rhythm of locomotor activity and flies were still rhythmic, with period of the rhythm similar to controls (Supplementary Table S1), but sleep amount, counted as sleep minutes per 12 h, was increased during the night compared to parental strains (Figures 2A–D, Supplementary Table S2). Because the previous data suggested that glia affects sleep through modulation of the rhythm in the sLNv terminal complexity (Herrero et al., 2017), we checked this phenomenon using flies with disrupted autophagy. After silencing of either atg5 or atg7 we observed that this complexity rhythm was inversed, with more intersections calculated during the night than during the day (Figures 2E,F) (p = 0.0002 for atg5RNAi and p = 0.0007 for atg7RNAi, respectively). The control flies, kept at a lower temperature, showed the normal pattern of these changes, with higher number of intersections in the morning (Figures 2E,F) (p = 0.0024 for atg5RNAi and p = 0.0345 for atg7RNAi, respectively).

FIGURE 2. Effect of pan-glial autophagy disruption. (A–D): Effect of silencing of atg5 or atg7 in glia on sleep level. Changes were observed only during the night. (E,F): Autophagy disruption in glia affects daily plasticity of sLNvs terminals arborisation complexity. Statistically significant differences marked as asterisks *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001.
Effect of Autophagy Modification in Specific Glia Types on Sleep
To study the role of autophagy in glia in details, we proceeded with the sleep analysis using driver lines which allowed us to modify autophagy level in the specific glia types. We showed that autophagy in the epithelial glia (netB driver) (Figures 3A,B, Supplementary Figures S1A,B), ensheathing glia (sws driver) (Figures 3C,D, Supplementary Figures S1C,D), marginal glia (ds driver) (Figures 3E,F, Supplementary Figures S1E,F) and optic chiasm glia (Wnt4 driver) (Figures 3G,H, Supplementary Figures S1G,H) is not involved in sleep regulation, since atg5 and atg7 silencing in these glia types did not change sleep amount (Supplementary Table S2).
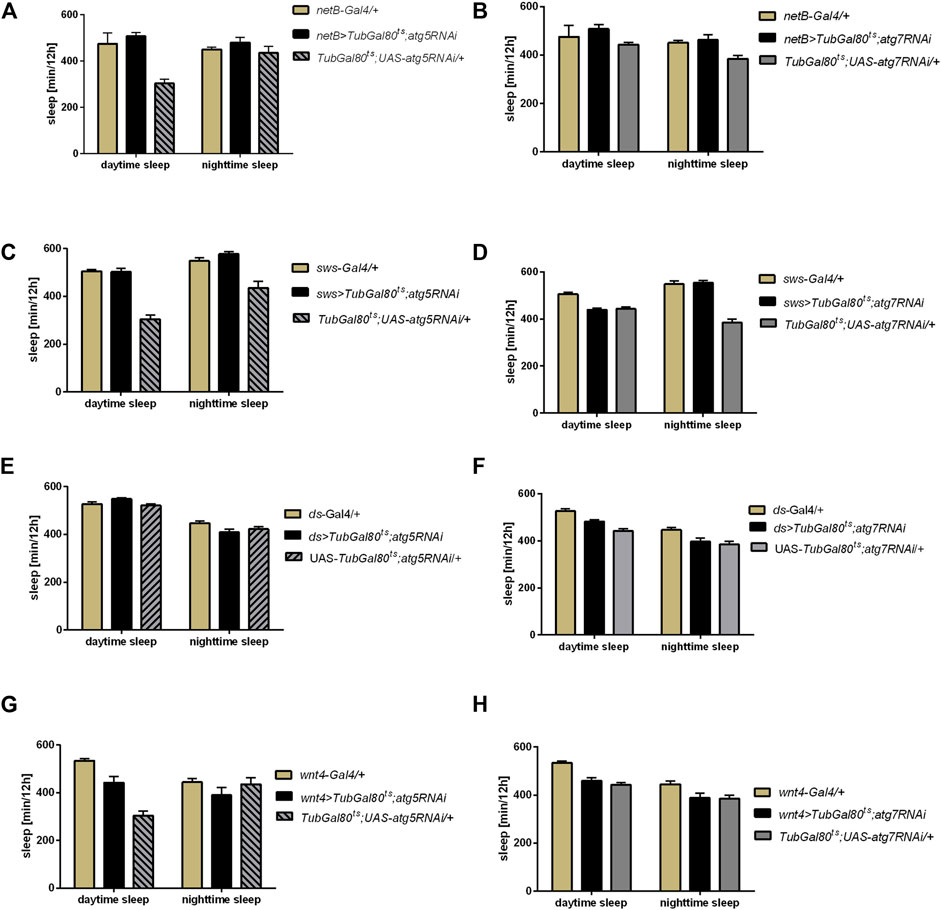
FIGURE 3. Effect of autophagy disruption in specific glia types on sleep level. Expression of atg5RNAi and atg7RNAi in epithelial glia (netB marker) (A,B), ensheathing glia (sws driver) (C,D), marginal glia (ds driver) (E,F) and optic chiasm glia (wnt marker) (G,H) did not affect sleep time.
Subperineurial glia and/or pseudocartridge glia seem to use some of the Atg proteins in the glia-clock neuron communication, because atg7 silencing with the moody driver caused increased sleep level during the night (Figures 4B,D, Supplementary Table S2) and decreased night offset (Supplementary Table S3). Surprisingly, atg5 silencing in this type of glia did not affect the sleep time (Figures 4A,C, Supplementary Table S2). The sLNv complexity of processes after atg7 silencing was similar at the beginning of the day and night (Figure 4E) (p = 0.502 at 29°C and p < 0.0001 at 20°C, respectively), which means that oscillation in remodeling of sLNv processes was interrupted once autophagy in this type of glia was reduced.
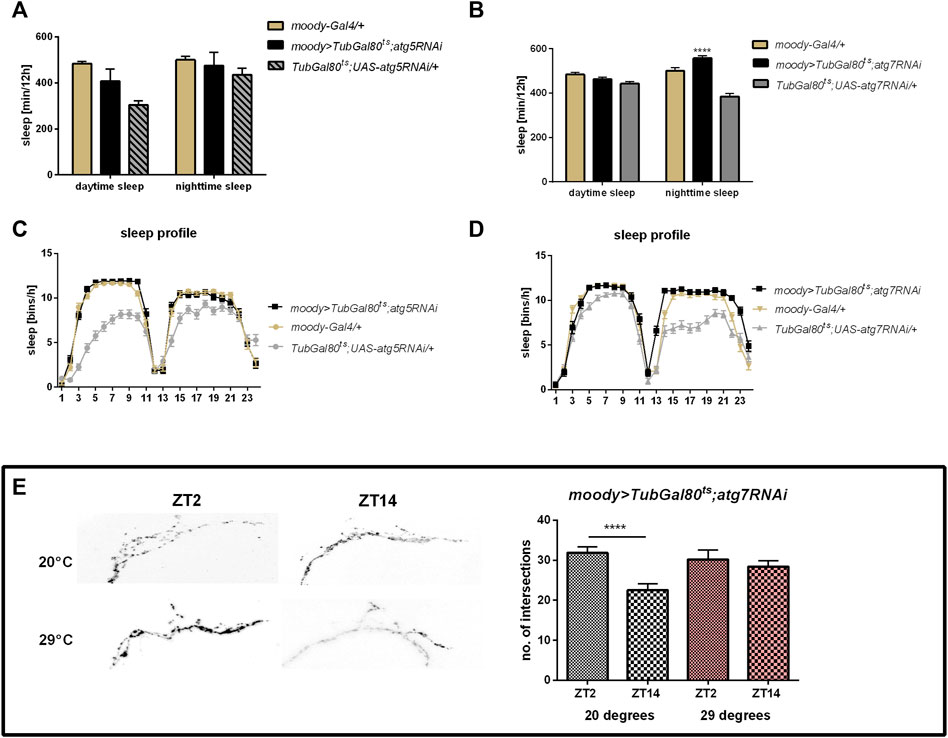
FIGURE 4. Effect of autophagy disruption in subperineurial and pseudocartridge glia (moody marker) Adult specific atg5 silencing did not affect sleep time regulates (A,C), however atg7RNAi expression increased sleep level during the night (B,D) and detained daily changes in sLNv terminal arborisation complexity (E). Statistically significant changes marked as asterisks ***p ≤ 0.001, ****p ≤ 0.0001.
Finally, using a driver line specific for astrocyte-like glia we obtained results corresponding to the pan-glial autophagy changes, meaning that sleep time was increased after silencing of both atg5 and atg7 (Figures 5A–D, Supplementary Table S2) and night offset was decreased (Supplementary Table S3). We also observed the effect on sLNv terminals complexity, since atg5 silencing caused a similar pattern to pan-glial autophagy disruption (p = 0.0265 at 29°C and p = 0.0125 at 20°C, respectively), while atg7 down-regulation disrupted the daily changes in sLNv plasticity (Figures 5E,F) (p = 0.4557 at 29°C and p < 0.0001 at 20°C, respectively).
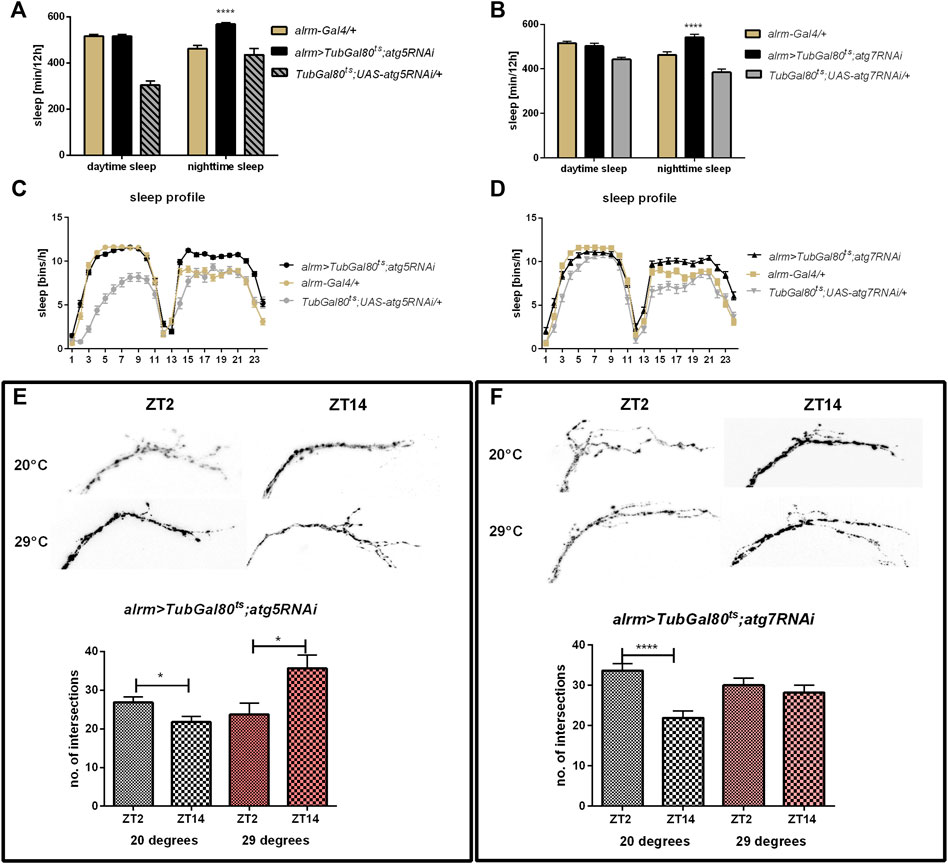
FIGURE 5. Effect of autophagy disruption in astrocytes-like glia (alrm marker). Adult specific atg5 (A,C) and atg7 silencing (B,D) increased sleep time during the night. atg5RNAi expression inversed pattern of sLNv arborisations (E), while atg7RNAi stopped daily changes in intersections number (F). Statistically significant differences marked as asterisks *p ≤ 0.05, ****p ≤ 0.0001.
Discussion
Autophagy is an important mechanism, which maintains normal metabolism of the cell, and changes in this process pathway may lead to pathological processes. Autophagy has been described in most tissues, however, in the nervous system it has been studied mostly in neurons but unexplored in glia. Because autophagy level and its regulation seem to differ in neurons and in other cells (Komatsu et al., 2007; Boland et al., 2008), they can differently respond to stress, which in effect may be a key factor to understand the basis of cognition disruption and development of neurodegenerative diseases.
Daily changes in the number of autophagosomes were described in many mammalian tissues, including the inner segment of retina rod cells, cardiomyocytes, hepatocytes, pancreatic cells, skeletal muscles and kidneys (Pfeifer et al., 1975; Pfeifer and Strauss, 1981; Ma et al., 2011). The other parameter showing the autophagy level is autophagy flux, which is the rate of autophagosome formation, its conversion to autolysosome and degradation in lysosome. It has been shown that autophagy flux in mice liver is under circadian control with the highest level in the afternoon and low level during the night (Ma et al., 2011).
Although circadian autophagy was described in many tissues, little is known about this process in the brain. Neuronal and glial cells are highly rhythmic, they show daily changes in size and shape, in the level of protein and in the number of synaptic contacts (Pyza and Meinertzhagen, 1999; Pyza and Górska-Andrzejak, 2004a; Weber et al., 2009; Górska-Andrzejak, 2013). They also have high metabolism, and are under the risk of oxidative stress, which is also regulated in clock-dependent manner (Damulewicz et al., 2016). However, we still do not know whether degradative processes in these cells, like autophagy, are rhythmic, and whether these changes play a role in the brain daily plasticity. Rhythmic autophagy-related gene expression was not observed in the whole head (Claridge-Chang et al., 2001; McDonald and Rosbash, 2001; Ceriani et al., 2002), however it was detected in the brain (Kijak and Pyza, 2017). These results suggest that daily profile of autophagy level may differ according to cell type. This was supported by analysis of gene expression of isolated PDF-producing cells, which showed significant daily changes in atg5 and atg7 mRNA level (Kula-Eversole et al., 2010). In our work we focused on glial cells as they are known to be involved in the regulation of brain function, through their impact on neuronal metabolism, survival and neurotransmitter turnover (reviewed by Freeman, 2015). Additionally, this regulation seems to be more complex since the function of the glia is under circadian clock control and glia possess their own oscillators (Prolo et al., 2005; Marpegan et al., 2011; Hayashi et al., 2013; Krzeptowski et al., 2018b; Long and Giebultowicz, 2018).
Our results showed that expression of autophagy-related genes in glia oscillates during the day. We observed the sequence of the expression peaks congruent with their temporal role in the autophagy machinery. The atg5 coding Atg5, which is involved in autophagosome formation, reaches its maximum expression during the day. Next, atg7 and atg10 encoding proteins, which conjugate to elongating membranes of a vesicle, have peaks of mRNA at the end of the day. In addition, draper, which protein is known to increase autophagy, is highly expressed during the night. These data suggest that autophagy in glia is enhanced during the night. It is consistent with the previously published data which have shown that also Drosophila neuronal cells produce more autophagosomes early in the night than at the beginning of the day (Bedont et al., 2021). Atg proteins play also additional functions, not connected with autophagy, which may explain why there are differences in the expression pattern of specific genes. However, mRNA of specific Atg gene is accumulated and transported to the destination site, and once all components are present autophagy process can be activated, which seems to be enhanced during the night.
Autophagy-related proteins (Atgs) are involved in many processes. The most common role is protein or cell compartment degradation, however their functions in many non-autophagic processes were described (Subramani and Malhotra, 2013; Galluzzi and Green, 2019). Here, we showed that autophagy in glia, especially in astrocytes and possibly also in subperineurial and/or pseudocartridge glia, is involved in the sleep regulation through modification of the sLNv terminal plasticity. First, we examined the effect of adult-specific atg5 and atg7 gene silencing in selected glia types on sleep level and its pattern. Because Atg5 and Atg7 are necessary for autophagosome formation, in effect, their lower level significantly decreases autophagic effectiveness. In our research, we showed that pan-glial atg5 or atg7 silencing do not change period of locomotor activity but it affects sleep level during the night. Similar effect was observed after autophagy gene silencing in per-expressing cells (Kijak and Pyza, 2017) and when we used alrm or moody drivers, which are markers for astrocytes, and subperineurial and/or pseudocartridge glia, respectively. The other types of glia seem to not be involved in this regulation, as flies with autophagy disruption in giant optic chiasm glia, epithelial, ensheathing and marginal glia did not show differences in the sleep time. The weakening of autophagy in the specific types of glia increases sleep time during the night suggesting that autophagy in these cells promotes awakeness during the resting time. The involvement of autophagy in the regulation of brain functioning is not surprising. It was previously shown that autophagy in the brain is involved in the memory formation, as flies with atg5 or atg9 silencing in the mushroom bodies attenuate associative olfactory memory and neuropeptide Y level in their brains is changed (Bhukel et al., 2019). In turn atg7 mutants are hypersensitive to starvation and oxidative stress, and they exhibit degenerative neuronal defects and accumulate protein aggregates in neurons (Juhasz et al., 2007; Simonsen et al., 2008).
Glia plays a role in the regulation of rhythmic behavior, which is modulated by releasing gliotransmitters (Ng et al., 2011) or affecting clock neuron activity by changes in PDF transport or release (Ng et al., 2011). The impact of astrocytes on behavior has already been shown (You et al., 2018). In flies, astrocytes are involved in regulating metabolism, neurotransmitter turnover and transport, proper vesicle trafficking (Ng et al., 2011; Jackson et al., 2015; Ng and Jackson, 2015; Ng et al., 2016) and modulation of the fast synaptic transmission (Macnamee et al., 2016), all of them eventually affect behavior (Walkowicz et al., 2021). The role of astrocytes on sleep regulation in fruit flies was already described. Several factors secreted from astrocytes activate different pathways and regulate particular sleep phase or parameter. Noktochor (NKT), which belongs to small secreted immunoglobulin promotes sleep during the night. Knockdown of nkt gene expression causes night sleep fragmentation and reduction but it does not affect daytime siesta (Ng et al., 2016; Sengupta et al., 2019). The other astrocytes-secreted factor Eiger (EGR) promotes baseline sleep both during the day and night, while its neuronal receptor affects recovery sleep after deprivation (Ng et al., 2016; Vanderheyden et al., 2018). Moreover, homeostatic sleep is also regulated by glia. It is known that calcium signaling in astrocytes increases sleep time, and similar effect is observed after sleep deprivation. Mechanism of glia-dependent sleep homeostasis requires L-type Ca2+ channel, and monoaminergic receptor, TyrRII, which level is increased with higher sleep need. In effect, astrocytes release Spätzle, interleukin analog, acting on Toll receptors on R5 ellipsoid body neurons to promote sleep. Additionally, activated astrocytes reduce spiking frequency and resting membrane potential of l-LNvs, in effect inhibiting an arousal circuit (Blum et al., 2021).
The subperineurial glia maintains the hemolymph-brain barrier, which protects neurons from unregulated exchange with humoral fluids. The permeability of this barrier changes during the day and is clock-dependent (Zhang et al., 2018). It has been shown that disruption of endocytosis in moody-expressing cells enhances total and daytime sleep (Artiushin et al., 2018).
Glial oscillators mediate the daily structural plasticity of clock neuron terminals in the dorsal brain (Herrero et al., 2017) and morphological changes in the visual system (Pyza and Górska-Andrzejak, 2004b). Here, we showed that the daily changes in the complexity of arborization depends not only on the clock but also on autophagy in glial cells. The mechanism of this process needs further investigations, however, our results suggest that observed changes in sleep length during the night and night offset are connected with changes in sLNv terminals plasticity. How is it possible that autophagy in glia affects neuronal remodeling? First of all, rhythmic autophagy may affect temporal changes of specific cell proteomes. There are many proteins whose levels oscillate while they lack corresponding mRNA cycling. In mouse liver many proteins have expression peaks during the time when autophagy activity is lower, which suggests that their level might be regulated by this process (Pfeifer and Strauss, 1981; Reddy et al., 2006; Ma et al., 2011). There are many evidences that glia produce transmitters which affects the physiology of clock neurons. It was previously shown that blocking of vesicle trafficking from glia decreases PDF transport or release from sLNv terminals without changing its expression (Ng et al., 2011). In turn, PDF level regulates sLNv terminals branching (Herrero et al., 2020). The other possibility is the autophagy-dependent daily remodeling of organelles. It is known that the number and morphology of mitochondria, peroxisomes and endoplasmic reticulum vary during the day (Doktór et al., 2018), and there is evidence that these changes are mediated by autophagy, at least in specific tissues, like liver and heart (Pfeifer and Strauss, 1981; Youle and Narendra, 2011). Mitophagy is also observed in the brain and is involved in proper functioning of dopaminergic neurons (Martinez-Vicente, 2017). Autophagy seems to be also involved in recycling of cell membranes, which is particularly important in neuronal plasticity. In the fly’s first optic neuropil (lamina) daily changes in size of interneurons are autophagy-dependent (Weber et al., 2009; Kijak and Pyza, 2017) and they seem to be compensated by changes in the epithelial glial cells (Pyza and Górska-Andrzejak, 2004b). It is possible that similar cross-talk exists between astrocytes and PDF-expressing neurons, as it was shown that this kind of glia affects LNv morphology through mesencephalic astrocyte-derived neurotrophic factor (MANF) (Walkowicz et al., 2017; Walkowicz et al., 2021). The last possibility is that the cyclic autophagy maintains nutrient and energy homeostasis in glial cells (Ezaki et al., 2011). All these autophagy-dependent processes may affect glia physiology in rhythmic manner and provide different glia - neurons communication throughout the day. Interestingly, sleep length also regulates glia plasticity. It has been shown that sleep promotes engulfment of damaged neurites, and in effect their clearance. This process requires enhanced expression of Draper (Stanhope et al., 2020), which is known as autophagy activator. It is possible that this mechanism works as feedback loop–higher sleep level activate autophagy in glia, which in turn inhibits sleep during the night to maintain proper wake/sleep proportion.
Data Availability Statement
The raw data supporting the conclusion of this article will be made available by the authors, without undue reservation.
Author Contributions
MD designed the study, performed the experiments, analysed data and wrote the manuscript. KS performed experiments, EP contributed to the writing of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by grant from the Polish National Science Centre (Narodowe Centrum Nauki, NCN) no. UMO-2017/27/B/NZ3/00859 to MD.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2022.886273/full#supplementary-material
References
Artiushin G., Zhang S. L., Tricoire H., Sehgal A. (2018). Endocytosis at the Drosophila Blood-Brain Barrier as a Function for Sleep. eLife 7, 1–18. doi:10.7554/eLife.43326
Axe E. L., Walker S. A., Manifava M., Chandra P., Roderick H. L., Habermann A., et al. (2008). Autophagosome Formation from Membrane Compartments Enriched in Phosphatidylinositol 3-phosphate and Dynamically Connected to the Endoplasmic Reticulum. J. Cel Biol. 182, 685–701. doi:10.1083/jcb.200803137
Bedont J. L., Toda H., Shi M., Park C. H., Quake C., Stein C., et al. (2021). Short and Long Sleeping Mutants Reveal Links between Sleep and Macroautophagy. eLife 10, 1–27. doi:10.7554/eLife.64140
Bhukel A., Beuschel C. B., Maglione M., Lehmann M., Juhász G., Madeo F., et al. (2019). Autophagy within the Mushroom Body Protects from Synapse Aging in a Non-cell Autonomous Manner. Nat. Commun. 10, 1–13. doi:10.1038/s41467-019-09262-2
Blum I. D., Keleş M. F., Baz E.-S., Han E., Park K., Luu S., et al. (2021). Astroglial Calcium Signaling Encodes Sleep Need in Drosophila. Curr. Biol. 31, 150–162. e7. doi:10.1016/j.cub.2020.10.012
Boland B., Kumar A., Lee S., Platt F. M., Wegiel J., Yu W. H., et al. (2008). Autophagy Induction and Autophagosome Clearance in Neurons: Relationship to Autophagic Pathology in Alzheimer's Disease. J. Neurosci. 28, 6926–6937. doi:10.1523/JNEUROSCI.0800-08.2008
Bushey D., Tononi G., Cirelli C. (2011). Sleep and Synaptic Homeostasis: Structural Evidence in Drosophila. Science 332, 1576–1581. doi:10.1126/science.1202839.Sleep
Ceriani M. F., Hogenesch J. B., Yanovsky M., Panda S., Straume M., Kay S. A. (2002). Genome-Wide Expression Analysis inDrosophilaReveals Genes Controlling Circadian Behavior. J. Neurosci. 22, 9305–9319. doi:10.1523/jneurosci.22-21-09305.2002
Chatterjee A., Lamaze A., De J., Mena W., Chélot E., Martin B., et al. (2018). Reconfiguration of a Multi-Oscillator Network by Light in the Drosophila Circadian Clock. Curr. Biol. 28, 2007–2017. doi:10.1016/j.cub.2018.04.064
Chen J., Reiher W., Hermann-Luibl C., Sellami A., Cognigni P., Kondo S., et al. (2016). Allatostatin A Signalling in Drosophila Regulates Feeding and Sleep and Is Modulated by PDF. Plos Genet. 12, e1006346. doi:10.1371/journal.pgen.1006346
Claridge-Chang A., Wijnen H., Naef F., Boothroyd C., Rajewsky N., Young M. W. (2001). Circadian Regulation of Gene Expression Systems in the Drosophila Head. Neuron 32, 657–671. doi:10.1016/s0896-6273(01)00515-3
Cusumano P., Klarsfeld A., Chélot E., Picot M., Richier B., Rouyer F. (2009). PDF-modulated Visual Inputs and Cryptochrome Define Diurnal Behavior in Drosophila. Nat. Neurosci. 12, 1431–1437. doi:10.1038/nn.2429
Damulewicz M., Loboda A., Bukowska-Strakova K., Jozkowicz A., Dulak J., Pyza E. (2015). Clock and Clock-Controlled Genes Are Differently Expressed in the Retina, Lamina and in Selected Cells of the Visual System of Drosophila melanogaster. Front. Cel. Neurosci. 9, 353. doi:10.3389/fncel.2015.00353
Damulewicz M., Loboda A., Jozkowicz A., Dulak J., Pyza E. (2016). Interactions between the Circadian Clock and Heme Oxygenase in the Retina of Drosophila melanogaster. Mol. Neurobiol. 54, 4953–4962. doi:10.1007/s12035-016-0026-9
Díaz M. M., Schlichting M., Abruzzi K. C., Long X., Rosbash M. (2019). Allatostatin-C/AstC-R2 Is a Novel Pathway to Modulate the Circadian Activity Pattern in Drosophila. Curr. Biol. 29, 13–22. doi:10.1016/j.cub.2018.11.005
Doherty J., Logan M. A., Tasdemir O. E., Freeman M. R. (2009). Ensheathing Glia Function as Phagocytes in the Adult Drosophila Brain. J. Neurosci. 29, 4768–4781. doi:10.1523/JNEUROSCI.5951-08.2009
Doktór B., Damulewicz M., Krzeptowski W., Bednarczyk B., Pyza E. (2018). Effects of Pink1 Mutation on Synapses and Behavior in the Brain of drosophila Melanogaster. Acta Neurobiologiae Experimentalis 78, 231–241. doi:10.21307/ane-2018-021
Edwards T. N., Nuschke A. C., Nern A., Meinertzhagen I. A. (2012). Organization and Metamorphosis of Glia in the Drosophila Visual System. J. Comp. Neurol. 520, 2067–2085. doi:10.1002/cne.23071
Ezaki J., Matsumoto N., Takeda-Ezaki M., Komatsu M., Takahashi K., Hiraoka Y., et al. (2011). Liver Autophagy Contributes to the Maintenance of Blood Glucose and Amino Acid Levels. Autophagy 7, 727–736. doi:10.4161/auto.7.7.15371
Fernández M. P., Berni J., Ceriani M. F. (2008). Circadian Remodeling of Neuronal Circuits Involved in Rhythmic Behavior. Plos Biol. 6, e69–0524. doi:10.1371/journal.pbio.0060069
Freeman M. R. (2015). DrosophilaCentral Nervous System Glia. Cold Spring Harb Perspect. Biol. 7, a020552–14. doi:10.1101/cshperspect.a020552
Galluzzi L., Green D. R. (2019). Autophagy-Independent Functions of the Autophagy Machinery. Cell 177, 1682–1699. doi:10.1016/j.cell.2019.05.026
Gorostiza E. A., Depetris-Chauvin A., Frenkel L., Pírez N., Ceriani M. F. (2014). Circadian Pacemaker Neurons Change Synaptic Contacts across the Day. Curr. Biol. 24, 2161–2167. doi:10.1016/j.cub.2014.07.063
Górska-Andrzejak J. (2013). Glia-related Circadian Plasticity in the Visual System of Diptera. Front. Physiol. 4, 1–8. doi:10.3389/fphys.2013.00036
Grima B., Chélot E., Xia R., Rouyer F. (2004). Morning and Evening Peaks of Activity Rely on Different Clock Neurons of the Drosophila Brain. Nature 431, 869–873. doi:10.1038/nature02935
Hayashi Y., Koyanagi S., Kusunose N., Okada R., Wu Z., Tozaki-Saitoh H., et al. (2013). The Intrinsic Microglial Molecular Clock Controls Synaptic Strength via the Circadian Expression of Cathepsin S. Sci. Rep. 3, 1–7. doi:10.1038/srep02744
Herrero A., Duhart J. M., Ceriani M. F. (2017). Neuronal and Glial Clocks Underlying Structural Remodeling of Pacemaker Neurons in Drosophila. Front. Physiol. 8. doi:10.3389/fphys.2017.00918
Herrero A., Yoshii T., Ispizua J. I., Colque C., Veenstra J. A., Muraro N. I., et al. (2020). Coupling Neuropeptide Levels to Structural Plasticity in Drosophila Clock Neurons. Curr. Biol. 30, 3154–3166. e4. doi:10.1016/j.cub.2020.06.009
Ichimura Y., Kirisako T., Takao T., Satomi Y., Shimonishi Y., Ishihara N., et al. (2000). A Ubiquitin-like System Mediates Protein Lipidation. Nature 408, 488–492. doi:10.1038/35044114
Jackson F. R., Ng F. S., Sengupta S., You S., Huang Y. (2015). Glial Cell Regulation of Rhythmic Behavior. Methods Enzymol. 552, 45–73. doi:10.1016/bs.mie.2014.10.016
Kamada Y., Funakoshi T., Shintani T., Nagano K., Ohsumi M., Ohsumi Y. (2000). Tor-mediated Induction of Autophagy via an Apg1 Protein Kinase Complex. J. Cel Biol. 150, 1507–1513. doi:10.1083/jcb.150.6.1507
Kijak E., Pyza E. (2017). TOR Signaling Pathway and Autophagy Are Involved in the Regulation of Circadian Rhythms in Behavior and Plasticity of L2 Interneurons in the Brain of Drosophila melanogaster. PLoS ONE 12, e0171848. doi:10.1371/journal.pone.0171848
Komatsu M., Ueno T., Waguri S., Uchiyama Y., Kominami E., Tanaka K. (2007). Constitutive Autophagy: Vital Role in Clearance of Unfavorable Proteins in Neurons. Cell Death Differ 14, 887–894. doi:10.1038/sj.cdd.4402120
Krzeptowski W., Hess G., Pyza E. (2018a). Circadian Plasticity in the Brain of Insects and Rodents. Front. Neural Circuits 12, 1–14. doi:10.3389/fncir.2018.00032
Krzeptowski W., Walkowicz L., Płonczyńska A., Górska-Andrzejak J. (2018b). Different Levels of Expression of the Clock Protein PER and the Glial Marker REPO in Ensheathing and Astrocyte-like Glia of the Distal Medulla of drosophila Optic Lobe. Front. Physiol. 9. doi:10.3389/fphys.2018.00361
Kula-Eversole E., Nagoshi E., Shang Y., Rodriguez J., Allada R., Rosbash M. (2010)., Surprising Gene Expression Patterns within and between PDF-Containing Circadian Neurons in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 107, 13497–13502. doi:10.1073/pnas.1002081107
Kuma A., Mizushima N., Ishihara N., Ohsumi Y. (2002). Formation of the ∼350-kDa Apg12-Apg5·Apg16 Multimeric Complex, Mediated by Apg16 Oligomerization, Is Essential for Autophagy in Yeast. J. Biol. Chem. 277, 18619–18625. doi:10.1074/jbc.M111889200
Long D. M., Giebultowicz J. M. (2018). Age-related Changes in the Expression of the Circadian Clock Protein PERIOD in Drosophila Glial Cells. Front. Physiol. 8. doi:10.3389/fphys.2017.01131
Ma D., Panda S., Lin J. D. (2011). Temporal Orchestration of Circadian Autophagy Rhythm by C/EBPβ. EMBO J. 30, 4642–4651. doi:10.1038/emboj.2011.322
Macnamee S. E., Liu K. E., Gerhard S., Tran C. T., Fetter R. D., Cardona A., et al. (2016). Astrocytic Glutamate Transport Regulates a Drosophila CNS Synapse that Lacks Astrocyte Ensheathment. J. Comp. Neurol. 524, 1979–1998. doi:10.1002/cne.24016
Marpegan L., Swanstrom A. E., Chung K., Simon T., Haydon P. G., Khan S. K., et al. (2011). Circadian Regulation of ATP Release in Astrocytes. J. Neurosci. 31, 8342–8350. doi:10.1523/JNEUROSCI.6537-10.2011
Martinez-Vicente M. (2017). Neuronal Mitophagy in Neurodegenerative Diseases. Front. Mol. Neurosci. 10, 1–13. doi:10.3389/fnmol.2017.00064
Matsushita M., Suzuki N. N., Obara K., Fujioka Y., Ohsumi Y., Inagaki F. (2007). Structure of Atg5·Atg16, a Complex Essential for Autophagy. J. Biol. Chem. 282, 6763–6772. doi:10.1074/jbc.M609876200
McDonald M. J., Rosbash M. (2001). Microarray Analysis and Organization of Circadian Gene Expression in Drosophila. Cell 107, 567–578. doi:10.1016/S0092-8674(01)00545-1
McPhee C. K., Logan M. A., Freeman M. R., Baehrecke E. H. (2010). Activation of Autophagy during Cell Death Requires the Engulfment Receptor Draper. Nature 465, 1093–1096. doi:10.1038/nature09127
Meijer W. H., Van Der Klei I. J., Veenhuis M., Kiel J. A. K. W. (2007). ATGGenes Involved in Non-selective Autophagy Are Conserved from Yeast to Man, but the Selective Cvt and Pexophagy Pathways Also Require Organism-specific Genes. Autophagy 3, 106–116. doi:10.4161/auto.3595
Mizushima N., Noda T., Ohsumi Y. (1999). Apg16p Is Required for the Function of the Apg12p-Apg5p Conjugate in the Yeast Autophagy Pathway. Embo J. 18, 3888–3896. doi:10.1093/emboj/18.14.3888
Mizushima N., Sugita H., Yoshimori T., Ohsumi Y. (1998). A New Protein Conjugation System in Human. J. Biol. Chem. 273, 33889–33892. doi:10.1074/jbc.273.51.33889
Ng F. S., Jackson F. R. (2015). The ROP Vesicle Release Factor Is Required in Adult Drosophila Glia for normal Circadian Behavior. Front. Cel. Neurosci. 9. doi:10.3389/fncel.2015.00256
Ng F. S., Sengupta S., Huang Y., Yu A. M., You S., Roberts M. A., et al. (2016). TRAP-seq Profiling and RNAi-Based Genetic Screens Identify Conserved Glial Genes Required for Adult Drosophila Behavior. Front. Mol. Neurosci. 9, 1–15. doi:10.3389/fnmol.2016.00146
Ng F. S., Tangredi M. M., Jackson F. R. (2011). Glial Cells Physiologically Modulate Clock Neurons and Circadian Behavior in a Calcium-dependent Manner. Curr. Biol. 21, 625–634. doi:10.1016/j.cub.2011.03.027
Özkaya Ö., Rosato E. (2012). The Circadian Clock of the Fly: A Neurogenetics Journey through Time. Adv. Genet. 77, 79–123. doi:10.1016/B978-0-12-387687-4.00004-0
Pfeifer U., Scheller H., Germany W. (1975). A Morphometric Study of Cellular Autophagy Including Diurnal Variations in Kidney Tubules of normal Rats. J. Cel Biol. 64, 608–621. doi:10.1083/jcb.64.3.608
Pfeifer U., Strauss P. (1981). Autophagic Vacuoles in Heart Muscle and Liver. A Comparative Morphometric Study Including Circadian Variations in Meal-Fed Rats. J. Mol. Cell Cardiol. 13, 37–49. doi:10.1016/0022-2828(81)90227-3
Picot M., Cusumano P., Klarsfeld A., Ueda R., Rouyer F. (2007). Light Activates Output from Evening Neurons and Inhibits Output from Morning Neurons in the Drosophila Circadian Clock. Plos Biol. 5, e315–2521. doi:10.1371/journal.pbio.0050315
Potdar S., Sheeba V. (2018). Wakefulness Is Promoted during Day Time by PDFR Signalling to Dopaminergic Neurons in Drosophila melanogaster. eNeuro 5, 0129–218. doi:10.1523/ENEURO.0129-18.2018
Prolo L. M., Takahashi J. S., Herzog E. D. (2005). Circadian Rhythm Generation and Entrainment in Astrocytes. J. Neurosci. 25, 404–408. doi:10.1523/JNEUROSCI.4133-04.2005
Pyza E., Górska-Andrzejak J. (2004a). Involvement of Glial Cells in Rhythmic Size Changes in Neurons of the Housefly's Visual System. J. Neurobiol. 59, 205–215. doi:10.1002/neu.10307
Pyza E., Górska-Andrzejak J. (2004b). Involvement of Glial Cells in Rhythmic Size Changes in Neurons of the Housefly's Visual System. J. Neurobiol. 59, 205–215. doi:10.1002/neu.10307
Pyza E., Meinertzhagen I. A. (19991999). Daily Rhythmic Changes of Cell Size and Shape in the First Optic Neuropil inDrosophila Melanogaster. J. Neurobiol. 40, 77–88. doi:10.1002/(SICI)1097-4695
Reddy A. B., Karp N. A., Maywood E. S., Sage E. A., Deery M., O'Neill J. S., et al. (2006). Circadian Orchestration of the Hepatic Proteome. Curr. Biol. 16, 1107–1115. doi:10.1016/j.cub.2006.04.026
Reggiori F., Tucker K. A., Stromhaug P. E., Klionsky D. J. (2004). The Atg1-Atg13 Complex Regulates Atg9 and Atg23 Retrieval Transport from the Pre-autophagosomal Structure. Develop. Cel 6, 79–90. doi:10.1016/S1534-5807(03)00402-7
Schlichting M., Weidner P., Diaz M., Menegazzi P., Dalla Benetta E., Helfrich-Förster C., et al. (2019). Light-Mediated Circuit Switching in the Drosophila Neuronal Clock Network. Curr. Biol. 29, 3266–3276. doi:10.1016/j.cub.2019.08.033
Scott R. C., Juhász G., Neufeld T. P. (2007). Direct Induction of Autophagy by Atg1 Inhibits Cell Growth and Induces Apoptotic Cell Death. Curr. Biol. 17, 1–11. doi:10.1016/j.cub.2006.10.053
Sengupta S., Crowe L. B., You S., Roberts M. A., Jackson F. R. (2019). A Secreted Ig-Domain Protein Required in Both Astrocytes and Neurons for Regulation of Drosophila Night Sleep. Curr. Biol. 29, 2547–2554. doi:10.1016/j.cub.2019.06.055
Shintani T., Mizushima N., Ogawa Y., Matsuura A., Noda T., Ohsumi Y. (1999). Apg10p, a Novel Protein-Conjugating Enzyme Essential for Autophagy in Yeast. EMBO J. 18, 5234–5241. doi:10.1093/emboj/18.19.5234
Stanhope B. A., Jaggard J. B., Gratton M., Brown E. B., Keene A. C. (2020). Sleep Regulates Glial Plasticity and Expression of the Engulfment Receptor Draper Following Neural Injury. Curr. Biol. 30, 1092–1101. e3. doi:10.1016/j.cub.2020.02.057
Stoleru D., Peng Y., Agosto J., Rosbash M. (2004). Coupled Oscillators Control Morning and Evening Locomotor Behaviour of Drosophila. Nature 431, 862–868. doi:10.1038/nature02926
Subramani S., Malhotra V. (2013). Non‐autophagic Roles of Autophagy‐related Proteins. EMBO Rep. 14, 143–151. doi:10.1038/embor.2012.220
Suzuki K., Kubota Y., Sekito T., Ohsumi Y. (2007). Hierarchy of Atg Proteins in Pre-autophagosomal Structure Organization. Genes Cells 12, 209–218. doi:10.1111/j.1365-2443.2007.01050.x
Tanida I., Mizushima N., Kiyooka M., Ohsumi M., Ueno T., Ohsumi Y., et al. (1999). Apg7p/Cvt2p: A Novel Protein-Activating Enzyme Essential for Autophagy. MBoC 10, 1367–1379. doi:10.1091/mbc.10.5.1367
Vabulas R. M., Hartl F. U. (2005). Protein Synthesis upon Acute Nutrient Restriction Relies on Proteasome Function. Science 310, 1960–1963. doi:10.1126/science.1121925
Vanderheyden W. M., Goodman A. G., Taylor R. H., Frank M. G., Van Dongen H. P. A., Gerstner J. R. (2018). Astrocyte Expression of the Drosophila TNF-Alpha Homologue, Eiger, Regulates Sleep in Flies. Plos Genet. 14, e1007724. doi:10.1371/journal.pgen.1007724
Walkowicz L., Kijak E., Krzeptowski W., Górska-Andrzejak J., Stratoulias V., Woznicka O., et al. (2017). Downregulation of DmMANF in Glial Cells Results in Neurodegeneration and Affects Sleep and Lifespan in Drosophila melanogaster. Front. Neurosci. 11. doi:10.3389/fnins.2017.00610
Walkowicz L., Krzeptowski W., Krzeptowska E., Warzecha K., Sałek J., Górska‐Andrzejak J., et al. (2021). Glial Expression of DmMANF Is Required for the Regulation of Activity, Sleep and Circadian Rhythms in the Visual System of Drosophila melanogaster. Eur. J. Neurosci. 54, 5785–5797. doi:10.1111/ejn.15171
Weber P., Kula-Eversole E., Pyza E. (2009). Circadian Control of Dendrite Morphology in the Visual System of Drosophila melanogaster. PLoS ONE 4, e4290–12. doi:10.1371/journal.pone.0004290
Yao Z., Shafer O. T. (2014). The Drosophila Circadian Clock Is a Variably Coupled Network of Multiple Peptidergic Units. Science 343, 1516–1520. doi:10.1126/science.1251285
You S., Fulga T. A., Van Vactor D., Jackson F. R. (2018). Regulation of Circadian Behavior by Astroglial microRNAs in Drosophila. Genetics 208, 1195–1207. doi:10.1534/genetics.117.300342
Youle R. J., Narendra D. P. (2011). Mechanisms of Mitophagy. Nat. Rev. Mol. Cel Biol 12, 9–14. doi:10.1038/nrm3028
Zhang L., Lear B. C., Seluzicki A., Allada R. (2009). The CRYPTOCHROME Photoreceptor Gates PDF Neuropeptide Signaling to Set Circadian Network Hierarchy in Drosophila. Curr. Biol. 19, 2050–2055. doi:10.1016/j.cub.2009.10.058
Keywords: neuronal plasticity, circadian clock, sleep, Drosophila, autophagy
Citation: Damulewicz M, Szypulski K and Pyza E (2022) Glia-Neurons Cross-Talk Regulated Through Autophagy. Front. Physiol. 13:886273. doi: 10.3389/fphys.2022.886273
Received: 28 February 2022; Accepted: 11 April 2022;
Published: 29 April 2022.
Edited by:
Emi Nagoshi, Université de Genève, SwitzerlandReviewed by:
Sheeba Vasu, Jawaharlal Nehru Centre for Advanced Scientific Research, IndiaMasashi Tabuchi, Case Western Reserve University, United States
Copyright © 2022 Damulewicz, Szypulski and Pyza. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Milena Damulewicz, bWlsZW5hLmRhbXVsZXdpY3pAdWouZWR1LnBs
 Milena Damulewicz
Milena Damulewicz Kornel Szypulski
Kornel Szypulski Elzbieta Pyza
Elzbieta Pyza