- 1Department of Family Medicine, Gangnam Severance Hospital, Yonsei University College of Medicine, Seoul, South Korea
- 2Department of Medical Biotechnology, Yeungnam University, Gyeongsan, South Korea
- 3Research Institute of Cell Culture, Yeungnam University, Gyeongsan, South Korea
- 4Department of Internal Medicine, College of Medicine, Yeungnam University, Daegu, South Korea
- 5Department of Physiology, College of Medicine, Yeungnam University, Daegu, South Korea
- 6School of Biosciences and Biotechnology, Baba Ghulam Shah Badshah University, Rajouri, India
Myostatin (MSTN) is a well-reported negative regulator of muscle growth and a member of the transforming growth factor (TGF) family. MSTN has important functions in skeletal muscle (SM), and its crucial involvement in several disorders has made it an important therapeutic target. Several strategies based on the use of natural compounds to inhibitory peptides are being used to inhibit the activity of MSTN. This review delivers an overview of the current state of knowledge about SM and myogenesis with particular emphasis on the structural characteristics and regulatory functions of MSTN during myogenesis and its involvements in various muscle related disorders. In addition, we review the diverse approaches used to inhibit the activity of MSTN, especially in silico approaches to the screening of natural compounds and the design of novel short peptides derived from proteins that typically interact with MSTN.
Introduction
Skeletal muscle (SM) is the largest organ, comprising ∼40% of total body weight, and one of the most dynamic and plastic tissues in the human body (Holmberg and Durbeej, 2013). These highly dynamic plastic tissues constitute 50–75% of body protein content and perform a large number of crucial body functions such as movement, temperature control, and maintaining glucose levels (Frontera and Ochala, 2015). Muscle satellite cells (MSCs) are multipotent precursor cells that are found between the sarcolemma and the basal lamina and provide anatomical and functional stability, preserving SM integrity (Lee et al., 2018). MSCs are capable of self-renewing and developing differentiated progeny. In particular, the proliferation and differentiation of MSCs to myotubes via the myogenic program relies on the coordinated regulations of paired box transcription factors (Pax3/Pax7) and the basic helix-loop-helix (bHLP) family of transcription factors (myogenic factor 5, Myf5; myogenic differentiation, MyoD; and myogenin, MYOG). During the gradual muscle loss associated with muscular dystrophy (MD) or aging, MSC activity is commonly impaired due to imprecise asymmetrical division or abnormal transcriptional control (Bianchi et al., 2020).
Aging is characterized by a progressive loss of muscle mass, leading to a loss of muscular strength (Cartee et al., 2016). Apart from aging, the loss of muscle mass is also associated with several other disease conditions such as cancer, chronic obstructive pulmonary disease (COPD), muscular dystrophies, acquired immune deficiency syndrome (AIDS), immune disorders, congestive heart failure, etc. (Fanzani et al., 2012). Furthermore, sarcopenia, a condition that impairs physical ability and metabolism, is also linked to age-related loss of SM mass and function (Curtis et al., 2015).
Myostatin; also known as growth differentiation factor 8 (GDF8) has been well reported to negatively regulate muscle growth and size (Carnac et al., 2007; Chen et al., 2021). The putative involvement of MSTN in muscle atrophy has been documented in several studies, prompting interest in MSTN as a therapeutic target to counteract muscle loss in patients with a range of muscle-wasting conditions (Baczek et al., 2020; Sartori et al., 2021). MSTN-deficient mice were found to have 2 to 3 times the SM mass of wild-type mice, which indicated MSTN acts as a negative regulator of muscle cells in vivo (McPherron et al., 1997). MSTN inhibition is also regarded as a crucial therapeutic target in the context of enhancing muscle strength and insulin sensitivity (Camporez et al., 2016).
MSTN inhibition is considered to be a potentially effective means of addressing the issue of muscle loss. Computational methods are widely used to discover novel inhibitors in a quick and cost-effective manner, typically through peptide design and compound screening. Usually, peptides are generated based on the 3D structures of protein complexes (Baig et al., 2018). Peptide fragments are often created from the interacting residues of protein-protein interactions (PPIs), which are central considerations in rational drug design (Baig et al., 2016). Computational screening of large compound collections against the binding sites of target proteins often results in the rapid identification of potential ligands. Virtual screening (VS.) is usually conducted using structure- and ligand-based approaches (Baig et al., 2016).
Other TGF-β superfamily members, in addition to MSTN, are documented to be effective negative muscle regulators, notably “activin A” being the second negative muscle regulator. Latres et al. (2017) found that MSTN and “activin A” adversely affect muscle development and function, and that blocking both of these ligands with antibodies resulted in a large increase in muscle and lean body mass in mice and monkeys, with a better therapeutic window than inhibiting all TGF-ligands.
The role played by MSTN in the development and growth of SM, and the mechanism by which it regulates the myogenic process are discussed in this review together with its role in different diseases and the strategies used to inhibit its activity [from natural compounds to myostatin inhibitory proteins (MIPs)]. In addition, we also review the state-of-the-art in silico approaches used to design MSTN inhibitors based on the structures of its interacting proteins.
Skeletal Muscle and Myogenesis
SM is composed of muscle fiber with a unique structure, which mainly consists of actin and myosin filaments that allow muscles to contract and relax. Each muscle fiber represents a muscle cell, which has a fundamental cellular unit known as the sarcomere. Fascicules are formed by bundles of myofibers, and muscle tissue is formed by bundles of fascicles, with each layer being contained by the ECM and maintained by cytoskeletal networks (Lieber and Friden, 2000). Thus, SM is responsible for body movement and posture. In addition, SM physically protects soft tissues, and internal organs, and maintains body temperature by producing heat using the energy generated during muscle contraction (Argilés et al., 2016).
The SMs present a strong capacity to regenerate, even after serious damage caused by heavy exercise, mechanical laceration, disease, or induced under experimental circumstances, e.g., by crushing or injecting cardiotoxin (CTX), begin a cascade of episodes leading to the muscle restoration (Collins and Morgan, 2003; Guardiola et al., 2017). MSCs divide symmetrically to increase their number, or asymmetrically to produce cohorts of committed satellite cells and consequently progenitors after they have been activated. Myogenic progenitors multiply and eventually differentiate by fusing with other myogenic progenitors or injured fibers in order to restore fiber integrity and function (Dumont et al., 2015; Dueweke et al., 2017).
Regulation of Myogenesis
Transcription Factors
MSCs present in SM originate from multipotent mesodermal cells. Like embryonic progenitors, the progression of MSCs along the myogenic lineage commences with the co-expressions of Pax3 and Pax7 and a family of bHLP transcription factors referred to as myogenic-regulatory factors (MRFs), such as Myf5, MyoD, Mrf4, and MYOG (Jan et al., 2016). These four MRFs are the fundamental constituents of the myogenic pathway. Myf5, the determining factor of myoblast, is expressed before commitment to myogenic fate. MyoD, which is induced by Myf5, drives cells to the myogenic lineage, and MYOG seems to work downstream of Myf5 and MyoD, and it is necessary for the establishment of the myogenic lineage as well as terminal differentiation of myoblasts (Lindon et al., 1998; Tapscott, 2005; Londhe and Davie, 2011; Ganassi et al., 2020).
When activated, MSCs undergo asymmetric division during muscle regeneration to give rise to two self-renewal daughter cells or emerge to form non-committed stem cells (Myf5-) for self-renewal or committed (Myf5+) cells (Kuang et al., 2008). The up-regulation of MyoD expression in activated MSCs (Pax7+/Myf5+) causes them to proliferate to generate myoblasts (Shefer et al., 2006). On the other hand, a decline in Pax7 expression in MyoD primed myoblasts marks their withdrawal from the cell cycle and entry into differentiation (Zammit et al., 2004). Collectively, the transcriptional network regulates the progression of the MSC lineage from origin to myogenic specification, differentiation, and fusion to produce myoblasts.
Hormones
The regulation of muscle growth also involves different hormones. A balance between the secretions of growth hormone (GH) and testosterone was found to be a prerequisite for optimizing muscle growth (Jan et al., 2016). Testosterone controls the sizes and numbers of muscle fibers by stimulating the longitudinal growth of muscle fibers. Though testosterone is associated with enhanced MSC proliferation and differentiation, estrogen influences their trans-differentiation, whereby lipids accumulate in differentiating myotubes (Wheeler and Koohmaraie, 1994). As a result of stimulation of insulin-like growth factor-1 (IGF-1) production via the liver JAK-STAT pathway, GH stimulates muscle fiber hypertrophy by enhancing the synthesis of proteins associated with MSC proliferation (Velloso, 2008). IGF-1 is an important hormone for muscle development and strength and helps in the proliferative efficiency of MSCs. Reportedly, TGF-β1 acts as a multifunctional cytokine that helps regulate muscle repair by activating MSCs (Delaney et al., 2017). Oxytocin injection rapidly enhanced muscle regeneration in young animals by aged MSCs and their proliferation by activating the MAPK/ERK pathway (Elabd et al., 2014).
GH-mediated conversion of thyroid hormone (TH) thyroxine (T4) to triiodothyronine (T3) helps their distribution to different tissues via binding to thyroxine-binding globulin, albumin, or transthyretin (TTR) (Alshehri et al., 2015). TTR-based T4 distribution was found to promote myoblast differentiation by regulating the expressions of myosin light chain 2 (MYL2) and the calcium channel genes Cav1.1 and Cav3.1 (Lee et al., 2013). We recently reported that during myoblast differentiation, TTR maintains muscle homeostasis via the unique TH shuttle mechanism. Furthermore, we postulated a unique mechanism for T4 and T3 absorption and release in myoblasts, as well as the role of TTR as an intracellular T4 sensor during myogenesis. (Lee et al., 2019).
Extracellular Matrix
The extracellular matrix (ECM) is a complex structure comprised of different structural molecules that provide mechanical support to cells and maintain biochemical signaling (Zhang et al., 2021). ECM proteins such as collagen IV and VI, laminins, and their receptors (e.g., integrin α7β1 and dystroglycan) have been reported to play crucial roles in SM development and to be responsible for SM homeostasis (Thorsteinsdóttir et al., 2011; Ahmad et al., 2020a; Ahmad et al., 2020b). ECM components interact with and regulate the MSC niche in muscle fibers, and alterations or inadequacies in the components of SM ECM can have dramatic effects on the characteristic functions of MSCs such as activation, self-renewal, proliferation, and differentiation (Thomas et al., 2015).
Some ECM proteins bind and modulate the function of MSTN, especially fibromodulin (FMOD), decorin, fibronectin, and laminins (Miura et al., 2010). Earlier, we investigated several ECM proteins, namely, FMOD (Lee et al., 2016; Lee et al., 2018), matrix gla protein (Ahmad et al., 2017), and dermatopontin (Kim et al., 2019), that play vital roles in the regulation of myogenesis. MSTN is known to inhibit the transcription factors Pax7, MYOD, and MYOG and thereby, regulate MSC proliferation and differentiation (Joulia-Ekaza and Cabello, 2006; McFarlane et al., 2008). Interestingly, it was observed FMOD bypassed the inhibitory effects of MSTN and maintained its transcriptional activity. We showed that FMOD directly bound with MSTN in myoblast differentiation by co-immunoprecipitation. Furthermore, PPIs between FMOD and MSTN and its receptor (Activin receptor type-IIB, ACVRIIB) showed that FMOD effectively reduced the interaction between MSTN and ACVRIIB (Lee et al., 2016).
Intracellular aggregation of methylglyoxal, a precursor of advanced glycation end-products (AGEs), and subsequent glycation of biomolecules impaired ECM remodeling, and curcumin and gingerol have been reported to reduce the impact of AGE on myoblasts (Baig et al., 2017). Moreover, enhanced AGE production and consequent RAGE (AGE receptor)-AGE interaction hinders the muscle development program. We also found by in silico analysis that the MSTN-ACVRIIB interaction is reduced by curcumin or gingerol. Protein-ligand (curcumin/gingerol and MSTN) and protein-protein interactions (MSTN and ACVRIIB) studies were carried out to explore the effect of curcumin and gingerol in the myogenesis processes. MSTN was found to have interacted with ACVRIIB with an energy score of −56.99. However, the free energy of MSTN to ACVRIIB binding fell to −46.55 and −47.26, correspondingly, for MSTN-curcumin and MSTN-gingerol complexes, showing that curcumin and gingerol interfere with MSTN-ACVRIIB interaction (Baig et al., 2017).
Myokines
SM produces several bioactive proteins, including cytokines, and numerous other peptides collectively called “myokines”. Skeletal myofibers produce a plethora of myokines, which exert auto-, para, and/or endocrine effects. Since myokine secretion is generally regulated by exercise, it has various advantageous effects on metabolic, cardiovascular, and mental health (Manole et al., 2018). Myokines are known to be involved in MSC activation and regulate their major functions, for example, they augment proliferation and differentiation rates (Mandai et al., 2017). The roles of myokines in the SM milieu appear to be directed toward MSC regulation at different stages, for instance, MSTN and growth differentiation factor 11 (GDF11) down-regulate MSC activation, proliferation, fusion, and differentiation, while IGF-1 promotes MSC fusion and differentiation (Mandai et al., 2017; Suh and Lee, 2020b).
Interleukin (IL)-6 plays a pleiotropic function in multiple tissues and organs. SM produces and secretes IL-6 during prolonged exercise, and is thus reflected as myokines (Munoz-Canoves et al., 2013). Local IL-6 production increase MSCs activation and promote the regeneration of myotube (Munoz-Canoves et al., 2013). Besides, IL-6 treatment has been found to enhance MSCs proliferation by controlling the cyclin D1 and c-myc genes (Serrano et al., 2008). The importance of IL-6 in myogenic differentiation has been confirmed as myoblast obtained from IL-6 null mice exhibits reduced fusion ability in vitro (Hoene et al., 2013). Like IL-6, leukemia inhibitory factor (LIF) has also been identified as a myokine, released by SM in response to exercise (Broholm and Pedersen, 2010; Pedersen and Febbraio, 2012). LIF regulates MSCs proliferation both in mice and humans. Exogenous LIF promotes the proliferation of human myoblast by inducing the transcription factors JunB and c-Myc (Broholm et al., 2011). In addition, LIF has also been found to induce myoblast differentiation (Yang et al., 2009).
Interleukin-15 (IL-15) is highly expressed in SM and has anabolic effects on SM protein dynamics (Quinn et al., 2002). IL-15 mRNA expression is up-regulated during myoblast differentiation and its administration inhibits the white adipose tissue deposition in rodents (Quinn et al., 2005). Also, IL-15 treatment decreased muscle protein degradation and SM wasting in an in vivo rat model of cancer cachexia (Carbo et al., 2000). Furthermore, reduced exercise endurance has been reported in IL-15−/− mice, however, enhanced exercise induction has been found in SM-specific IL-15-transgenic mice (Quinn et al., 2013; Quinn et al., 2014). Altogether, the above studies indicate that IL-6, IL-15, and LIF appeared as a vital myogenesis controllers, functioning during both myoblast proliferation and differentiation.
Myostatin
The MSTN protein sequence includes a secretion signal sequence, a proteolytic processing site, and a carboxy-terminal region with a conserved pattern of nine cysteine residues, all of which are shared by TGF-superfamily members. MSTN activation requires proteolytic cleavages of the precursor protein by a furin family enzyme and BMP1/Tolloid matrix metalloproteinase (Huang et al., 2011). Natural MSTN mutations in increased SM mass in many species including humans and similar results have been observed in MSTN null experimental mice (Amthor et al., 2007). During embryogenesis, MSTN is produced by cells in the myotome and developing SM and regulates the overall amount of muscle fibers formed. In adults, MSTN is secreted by SM, circulates in the blood, and inhibits muscle fiber growth (Lee, 2012).
Structure of MSTN
MSTN is translated as a precursor protein, which undergoes several proteolytic processing events that result in the formation of active, mature MSTN (Qian et al., 2015). Initially, the amino(N)-terminal signal sequence is removed by a signal peptidase to form Pro-MSTN, and dimerization follows due to disulfide bond formation near carboxy(C)-termini. Subsequently, furin cleaves the dimer at its proteolytic processing site RXXR site. C-terminus cleavage results in an N-terminal propeptide with an N-linked glycosyl group and a receptor-binding domain at the C-terminal. Latent MSTN complex forms when the N-terminal propeptide binds the C-terminal region noncovalently through a crucial peptide sequence, which prevents MSTN from binding to its receptor. In the last stage, BMP-1/TLD cleaves the propeptide, which leads to the release of mature MSTN. Increased muscle growth in adult mice was attributed to an inability to cleave the latent complex (Wolfman et al., 2003).
Like other members of the pro-TGF-β superfamily, pro-MSTN is a homodimer comprised of two identical disulfide-linked subunits. Each chain consists of 109 amino acid residues containing a pro-domain (N-terminal) and a smaller growth factor (GF) domain (C-terminal). As found in other members of the TGF-β superfamily, the GF domain of MSTN contains a cystine-knot motif and four antiparallel β-strands referred to as “fingers”. The two identical GF domains of MSTN are connected by their concave “palms”, which are covalently linked to each other by disulfide bonds between C339 residues in the wrist region. The pro-domain contains N-terminal “forearm” helices, which grasp mature GF, and a globular “arm/shoulder” domain, which sits on top of the mature GF protomers (Cotton et al., 2018). Each MSTN monomer has four intermolecular disulfide bonds, three of which are involved in cysteine knot formation. When the two monomers of MSTN come together in an antiparallel direction they generate convex or concave surfaces. The cysteine knots and dimerization are the major determinants of MSTN stability. Due to its similarity with activin class members (∼40%) and its binding to activin receptors and inhibitors (follistatin; FST), MSTN has long been considered to be a member of the activin class, as it has been shown to interact with ACVRIIB and ACVRIIA, as well as FST (Lee and McPherron, 2001). However, later x-ray structural analysis demonstrated that it is a member of the TGF-β superfamily, though it exhibits remarkable differences in the N-terminal region and in the region preceding the wrist helix (Cotton et al., 2018).
Signaling Pathways
The mechanisms of MSTN-induced SM loss are mediated by reduced protein synthesis and/or enhanced protein catabolism (Elliott et al., 2012). MSTN decreases protein synthesis by inhibiting the Akt/mTOR signaling pathway and induces muscle atrophy by promoting the transcriptions of atrophy-related genes (atrogenes). MSTN signaling pathways can be divided into Smad and non-Smad mediated pathways (Figure 1).
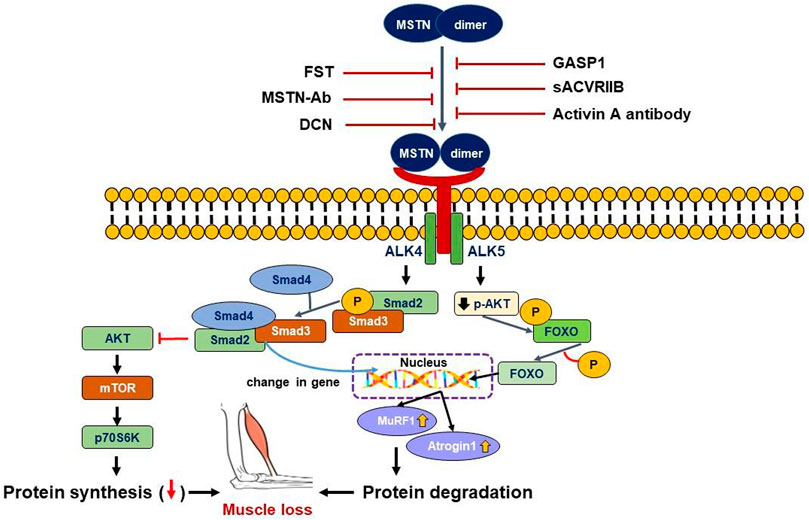
FIGURE 1. Smad and non-Smad mediated signaling pathway of MSTN. MSTN binds to the ACVRIIB and ALK4/5 complex resulting in successive phosphorylation of Smad2/3, leading to its binding with Smad4 and translocation of the complex to the nucleus. Non-Smad signaling, on the other hand, tends to suppress the AKT intracellular signaling pathways. Both Smad and non-Smad mediated signaling cause gene transcriptional alterations in the nucleus, as well as activation of muscle atrophy marker genes (MuRF1 and Atrogin1), resulting in muscle loss. Extracellularly, MSTN pathway inhibitors can bind MSTN directly or bind its receptor complex to prevent MSTN from interacting with its receptor complex and triggering downstream signals.
The Smad-Mediated Pathways
Mature MSTN contains a disulfide-linked dimer of the C-terminal domain and is 100% identical in humans, mice, rats, pigs, chickens, and dogs. The C-terminal dimer of mature MSTN binds to ACVRIIB and results in the recruitment and activation of activin type I receptors (Alk4 or Alk5), which in turn promote the activations of Smad 2 and Smad 3 to inhibit myoblast differentiation. Activation of MSTN signaling inhibits Akt phosphorylation by IGF-1 and the IGF-1-induced protein synthesis pathway possibly under Smad 3 mediation (Morissette et al., 2009). Inhibition of the MSTN-ACVRIIB-ALK4/ALK5-Smad2/3 pathway is strongly associated with muscle hypertrophy in adults, and Smad3 also increases E3 ligase atrogin-1 expression, which increases protein catabolism by the proteasome ubiquitination pathway (Goodman et al., 2013). Although Smad2/3 plays a predominant role in the protein catabolic pathway induced by MSTN, Smad3 null mice also exhibit MSTN-induced SM atrophy. Furthermore, MSTN overexpression was found to repress the differentiation of myoblasts by inhibiting MyoD and MyoD expression through MEK/Erk1/2 pathways (myogenic differentiation suppression pathways) (Yang et al., 2006). Thus, MSTN regulates muscle mass by acting through various signaling pathways that regulate muscle growth.
The Non-Smad Pathways
MSTN activates the JNK/Erk 1/2 (c-Jun N-terminal kinase/Erk 1/2) signaling pathway in proliferating and differentiating C2C12 cells (Huang et al., 2007). Philip et al. reported MSTN activates p38 MAPK through the TAK1-MKK6 cascade independently of Smad activation in proliferating A204 and C2C12 cells and that p38 MAPK plays an important role in the MSTN-regulated inhibition of myoblast proliferation (Philip et al., 2005). In another study, MSTN was found to act upstream of Wnt pathway components and suppress Wnt4 expression, which is capable of stimulating MSC proliferation. Therefore, inhibition of Wnt signaling downregulated MSC proliferation (Steelman et al., 2006). Altogether, these findings suggest the complexity of MSTN signal transduction is probably due to the involvement of different signaling pathways and that the precise integration of these pathways underlies the growth inhibitory effects of MSTN.
Pathologic Roles of MSTN
Muscle wasting is associated with cancer-related cachexia, age-related sarcopenia, and metabolic diseases such as obesity and diabetes, which are all directly related to morbidity and mortality. Associations between MSTN and muscle-wasting conditions have been investigated at the clinical level (Consitt and Clark, 2018). Plasma MSTN has been reported to increase with age, and its association with the prevalence of sarcopenia was found to be stronger in women (Bergen et al., 2015). In particular, plasma MSTN has been suggested to be a possible marker for the early diagnosis of cachexia in women with medullary thyroid carcinoma (Hedayati et al., 2016). Furthermore, the expression of MSTN in SM was significantly greater in gastric cancer patients before cachexia became clinically apparent, but this was not observed in lung cancer patients (Aversa et al., 2012). In another study, MSTN expression in the SMs of patients with gastric cancer was no different from that observed in healthy controls and was not associated with weight loss (D'Orlando et al., 2014). Increased protein degradation is probably a primary cause of pathologic muscle depletion, and it has been reported that protein degradation was enhanced in an experimental model of cancer cachexia and associated with muscle atrophy. In preclinical investigations of cancer cachexia, excessive proteolysis, particularly via autophagy and the ubiquitin-proteasome system, and low protein synthesis in SM have been documented (Sandri, 2016).
Diabetes and Obesity
Studies have explored the impact of MSTN on insulin resistance, and several studies conducted using mouse models have provided evidence that the absence of MSTN has significant effects on metabolism, that is, it improves insulin sensitivity and reduces obesity (Amor et al., 2019). Many studies have suggested that MSTN has a substantial impact on metabolism and may contribute to the development of obesity and diabetes (Allen et al., 2008; Allen et al., 2011). MSTN protein secretion was higher from the cultured myotubes of obese insulin-resistant subjects. High MSTN expression was first reported in the SMs of morbidly obese patients, whereas low expression was associated with subsequent fat loss (Hittel et al., 2009). In addition, MSTN expression and insulin sensitivity were found to be inversely proportional, and it was suggested a causal relationship exists between MSTN expression and insulin sensitivity (Hittel et al., 2010). In high-fat diet (HFD) mice, mutation-induced reductions in MSTN activity protected against obesity-induced insulin resistance (Wilkes et al., 2009). MSTN decreased insulin-induced GLUT4 membrane translocation and glucose absorption by inhibiting GLUT4 expression (Ahmad et al., 2018; Liu et al., 2018).
Cancer Cachexia and Sarcopenia
Because SM is largely composed of proteins, an imbalance between protein synthesis and degradation sensitively affects muscle mass, and reductions in muscle mass may lead to functional disability and an increase in the risk of injury and mortality. Cachexia and sarcopenia are two representative conditions that are closely related to gradual muscle loss and its inevitable consequences. Although the mechanisms of muscle loss are not clearly defined in either condition, the results obtained from experimental rodent models and clinical trials indicate that MSTN inhibition offers a promising means of controlling muscle loss and cancer-related cachexia (Sakuma and Yamaguchi, 2012).
Cachexia is a multifactorial syndrome associated with a chronic illness that causes involuntary weight loss due to reduced SM mass with or without fat mass loss. Cachexia is associated with chronic inflammatory disorders such as COPD, heart failure, chronic kidney disease, AIDS, sepsis, and most commonly cancer. The overall prevalence of cachexia due to any disease is around 1% among the patient population (i.e. approximately 9 million) (von Haehling and Anker, 2014). The most prominent clinical feature of cancer-related cachexia is SM loss due to anorexia and increased protein catabolism. Nutritional support does not reverse weight loss and the condition is treated using appetite-enhancing and/or anti-inflammatory drugs (Sadeghi et al., 2018).
Inflammation probably contributes to muscle atrophy by regulating the NF-κB signaling pathway, and conversely, the suppression of inflammation reverses muscle atrophy (Yu et al., 2017). In a study conducted on animals with tumor-induced cachexia, the expression of MSTN was up-regulated (Samant et al., 2017), and another study showed that blockage/inhibition of MSTN in animals with cancer cachexia prevented muscular atrophy without affecting tumor growth (Aversa et al., 2017). Thus, it is believed that increased MSTN expression is responsible for the progression of cancer and cancer-associated cachexia. Furthermore, it was observed that genetic deletion of MSTN or inhibition of its expression using anti-sense oligonucleotide preserved muscle mass (Gallot et al., 2014). The addition of a soluble antagonist of ACVRIIB that antagonizes MSTN signaling also reduced cancer-associated cachexia and reversed muscle wasting (Zhou et al., 2010). In a recent review, Hulmi et al. reviewed the numerous anti-cachectic benefits of ACVR2 inhibition in preclinical cancer models and in combination with anticancer therapies (Hulmi et al., 2021). The administration of this antagonist to mice with Lewis lung carcinoma or transfected with colon-26 cells improved muscle strength and reversed muscle wasting and prolonged survival in mice with a C26 tumor (Zhou et al., 2010; Busquets et al., 2012; Hatakeyama et al., 2016). MSTN levels and MSTN-mediated signaling pathways were found to be upregulated in experimental cancer cachexia and cancer patients even before the development of cancer cachexia (Costelli et al., 2008). However, plasma levels of MSTN are not always correlated with muscle loss in human cancer patients (Loumaye et al., 2015), and genetic deletion of MSTN or its acute inhibition using trichostatin A or FST does not always have a preventive effect against cancer cachexia in experimental rodents (Bonetto et al., 2009; Benny Klimek et al., 2010). Antibody against MSTN failed to elicit any significant clinical benefit in muscle dystrophy or pancreatic cancer patients (Wagner et al., 2008; Golan et al., 2018).
Sarcopenia is the result of a decline in the number of motor units and muscle fiber atrophy and is more prevalent these days due to the increasing number of elderly (Stoever et al., 2017). Although it is well known that reduced protein synthesis and/or increased protein degradation induces SM atrophy, reports regarding the underlying molecular pathways are inconsistent. Currently, no approved therapy is available for treating sarcopenia. Nevertheless, numerous reports have demonstrated that MSTN is a potential therapeutic target (White and LeBrasseur, 2014).
Moreover, reported relations between MSTN and muscle mass and aging also vary. In healthy older men, lower serum MSTN levels were linked to lower SM mass, but not in women (Peng et al., 2018). It is also evident that serum MSTN does not differ in young and sarcopenic elderly men (Ratkevicius et al., 2011). On the other hand, serum MSTN levels are elevated in elderly people and inversely correlated with lean mass. This discordance may stem from the technical limitations of enzyme-linked immunosorbent assays or radioimmunoassays used to discriminate between active and inactive MSTN or between MSTN and the similar protein (GDF-11) (Yarasheski et al., 2002). In SM, although MSTN mRNA levels are not correlated with age, MSTN protein levels are elevated in elderly subjects. Furthermore, MSTN protein levels are higher in the muscle tissues of elderly men than in healthy young men after acute muscle exercise (McKay et al., 2012).
Drug development targeting MSTN or its signaling pathways is being actively pursued. The amount of nuclear FOXO1 was increased in myotubes after MSTN treatment (McFarlane et al., 2006). FOXO1 and Smad2 were found to synergistically increase the MSTN mRNA expression and its promoter activity in the myotube of the C2C12 cell (Allen and Unterman, 2007). MSTN induced cachexia has been reported to be triggered by the activation of the ubiquitin-proteasome system (UPS) via FOXO1-dependent signaling, and it has been shown that increased MSTN expression is involved in the production of COPD-related muscle atrophy (Testelmans et al., 2010).
Ghrelin is largely produced in gastric oxyntic mucosa (DeBoer, 2011), and ghrelin treatment reduces proinflammatory cytokine release in cachexia patients (Kishimoto et al., 2012). Furthermore, increases in anabolic activity by ghrelin enhance GH release and reduce the effects of inflammation, which offers promise for the treatment of cachexia (Yanagi et al., 2018). In addition, ghrelin has been shown to prevent muscle atrophy in rats by enhancing AKT phosphorylation, suppressing the MSTN pathway, and activating myogenin and MyoD (Chen et al., 2015). Ghrelin formulations for parenteral administration are being developed (Garin et al., 2013). However, administration by injection over extended periods can result in poor patient compliance and therapy failure, and ghrelin is prone to enzymatic breakdown in blood when delivered intravenously (Brimijoin et al., 2016). Liposomes are frequently utilized as drug carriers due to their ability to encapsulate hydrophilic, amphiphilic, and lipophilic compounds (Pashirova et al., 2020), and the protective phospholipid layers of these systems protect ghrelin from pH, free radical, and enzymatic degradation in vivo (Sessa and Weissmann, 1968) and metabolism in the mucosal layer of the nasal cavity (Vieira and Gamarra, 2016). Ghrelin liposomes coated with chitosan are being developed for nose-to-brain administration for the treatment of cachexia (Salade et al., 2017). Furthermore, it has been shown that anionic liposomes can protect ghrelin from enzymatic breakdown by trypsin and carboxylesterase. Ghrelin interacts with lipid bilayers electrostatically and hydrophobically. Coating ghrelin with N-(2-hydroxy) propyl-3-trimethyl ammonium chitosan chloride enhanced mucin adsorption capacity (22.9%), with improved permeability via Calu3 epithelial monolayers recovering 10.8% of ghrelin in the basal compartment versus nonloaded ghrelin was used. Ghrelin can also be protected from metabolic enzymes in nasal tissues. Anionic liposomes coated with chitosan in dry powder form exhibited better mucin adhesion, ghrelin loadings, and enzymatic protection against trypsin, and reduced ghrelin degradation during storage at room temperature (Howick et al., 2018).
MSTN Inhibition
Ever since its discovery, intensive research has been conducted to suppress the activity of MSTN using soluble activin type IIB (sACVRIIB) receptors, peptides or propeptides, small molecules, neutralizing antibodies, or MIPs (Table 1).
Antibodies
The injection of MSTN neutralizing antibodies is known to significantly increase muscle mass (by up to 17%) in aged mice. The administration of neutralizing antibodies targeting MSTN, such as LY-2495655, MYO-029, PF-06252616, ATA 842, and REGN1033/SAR391786, improves body metabolism as well as the increase of SM mass (Camporez et al., 2016; St Andre et al., 2017). Furthermore, these antibodies help increase muscle mass and strength while attenuating muscle atrophy and act by blocking the ability of MSTN to prevent the signaling of TGF-β family members (Latres et al., 2015).
MSTN Receptor Proteins
ACVRIIB is a widely reported signaling receptor for several members of the TGF-β superfamily. ACVRIIB is involved in the negative regulation of muscle mass and is extensively distributed in SM, adipose tissues, and other organs. ACE-031 is a soluble form of ACVRIIB, and various studies on an Amyotrophic Lateral Sclerosis mouse model have shown a single dose of ACE-031 increases muscle mass and strength. This fusion protein of ACVRIIB and IgG1-Fc acts by binding to MSTN, and thus, disrupts its inhibitory effect (Campbell et al., 2017). Experiments on ACE-031 were subsequently suspended due to possible safety issues of epistaxis and telangiectasia.
Peptides/Propeptides
The latent MSTN complex circulates in the blood and is subsequently converted into its mature form after the removal of associated propeptides (Lee and McPherron, 2001). The binding of MSTN by these propeptides prevents MSTN functioning, and this is considered potential means of increasing muscle mass (Yang et al., 2001). Soon after the removal of these propeptides by proteolytic cleavage and conversion of latent MSTN to its mature form, MSTN binds to its receptor (usually ACVRIIB) and initiates the signaling process that regulates muscle growth. At this stage, several MIPs inhibit the binding of MSTN to ACVRIIB.
Myostatin Inhibitory Proteins
Follistatin
Follistatin (FST) is an extracellular cysteine-rich glycoprotein, which is structurally dissimilar to TGF-β family members, and it has been established that FST interrupts the activity of MSTN by binding to it and preventing MSTN binding to its receptor (Lee and McPherron, 2001; Liu et al., 2021). In vivo studies have reported that the overexpression of this glycoprotein has hypertrophic effects on mouse muscles similar to those observed in MSTN null mice (Winbanks et al., 2012). Moreover, a homozygous mutation in the FST gene reduces muscle mass, which suggests it plays an important role in the regulation of myogenesis (Lee et al., 2010; Liu et al., 2021).
FST-Related Gene
FST-related gene (FLRG) also known as FSTL3 protein, exhibits high homology to a 10-cysteine repeat of FST. In vitro studies have shown that like FST, FLRG binds to activin and BMPs to inhibit their biological activities (Tsuchida et al., 2001). Furthermore, endogenous latent MSTN complex largely circulates in association with propeptide and FLRG, which both act independently as negative regulators of MSTN, probably by preventing MSTN binding to its receptor (Thies et al., 2001). Reports suggest that FLRG potently inhibits MSTN activity in a concentration-dependent manner (Hill et al., 2002). Monovalent FSTL3-Fc fusion protein (mono-FSTL3-Fc) generated with knobs-into-holes technology has recently been reported to overcome the limitations of existing anti-myostatin therapies, as systemic administration of mono-FSTL3-Fc in mice resulted in muscle fiber hypertrophy and improved muscle mass in vivo (Ozawa et al., 2021a; Ozawa et al., 2021b).
Growth and Differentiation Factor-Associated Serum Protein
Growth and differentiation factor-associated serum protein-1 and -2 (GASP-1 and GASP-2) also importantly regulate the biological activity of MSTN (Lee and Lee, 2013). These two proteins are mostly expressed in adult tissues and have been well reported to induce small but significant increases in muscle mass in mice (Monestier et al., 2012). One of the multiple domains of GASP-1 is homologous to the 10-cysteine repeat of FST, whereas, in GASP-2, the FSD domain is responsible for MSTN binding. Recombinant GASP-1 binds directly to mature MSTN and its propeptide, and the inhibitory potential of GASP-1 is reduced when its C-terminal domains are removed (Hill et al., 2003).
Decorin and FMOD
Decorin (DCN) is a component of the MSTN signaling pathway and has been reported to antagonize the effects of MSTN. This member of the small leucine-rich proteoglycan gene family has been found to suppress MSTN activity efficiently and to enhance the differentiation and proliferation rates of myogenic cells (Kishioka et al., 2008). Reportedly, MSTN and decorin are produced at the same time in muscle cells (Nishimura et al., 2002). FMOD is known to be actively involved in the assembly of ECM and was recently reported to be a novel regulator of MSTN during myoblast differentiation by regulating the transcriptional activities of MSTN and other myogenic marker genes, which include myogenin (MYOG) and myosin light chain 2 (MYL2) (Lee et al., 2016). FMOD suppresses muscle aging by negatively regulating the MSTN gene or reducing the action of MSTN protein, while MSTN promotes muscle aging by positively regulating the expressions of the Atrogin1, CD36, and PPAR genes in muscle tissues (Lee et al., 2021a).
Natural Compounds
Epicatechin
(-)-Epicatechin (EC) is a flavonol, anti-oxidant, and bioactive stereoisomer of catechin that is used as a food supplement and found in cocoa and green tea (Gadkari and Balaraman, 2015). EC treatment also reduced MSTN expression and significantly increased the levels of myogenic marker genes responsible for muscle growth in the quadriceps muscles of mice, and it has been well-established aging is associated with higher MSTN levels but reduced levels of several myogenic genes such as FST, MYOG, and MyoD (Mafi et al., 2019). These findings suggest EC promotes SM development by inhibiting MSTN.
Fructus Schisandrae
Fructus Schisandrae (FS; Schisandra chinensis) is a well-known traditional herb in Korea, China, and Japan. The dried fruit of this herb (called Baill) is used to enhance physical capacity and for its anti-inflammatory and anti-stress effects (Panossian and Wikman, 2008). It has been reported that FS extract helps increase SM mass. When administered to a mouse MD model that exhibited high MSTN mRNA levels, FS extract reduced these levels in a dose-dependent manner (Kim et al., 2015).
Sulforaphane
Sulforaphane (SFN), which is found in cruciferous vegetables, is a bioactive isothiocyanate that inhibits the activity of histone deacetylases (HDACs) (Myzak et al., 2006). SFN significantly reduces MSTN expression in porcine MSCs and can inhibit HDAC activities and DNA methyltransferase expression (Fan et al., 2012).
Astragalus Polysaccharide
Astragalus polysaccharide (APS; also known as Huang Qi) is a well-known extract of Astragalus membranaceus (Fisch.) Bge (AMB) (Fu et al., 2013). Injection of APS into the SMs of non-insulin-dependent type 2 diabetic KKAy mice ameliorated insulin resistance and hyperglycemia, and reduced MSTN levels in SM, which demonstrated APS might improve insulin sensitivity and reduce SM MSTN levels by downregulating the ROS-ERK-NF-κB pathway (Liu et al., 2013).
Glycyrrhiza Uralensis
G. uralensis is native to Asia and is used as a medicinal herb, sweetener, and in traditional Chinese medicine (Ji et al., 2016). Recently, we reported that G. uralensis inhibits MSTN expression and promotes myogenesis. In addition, liquiritigenin, tetrahydroxymethoxychalcone, and licochalcone B isolated from the EtOAc fraction of G. uralensis enhanced myoblast proliferation and differentiation, and liquiritigenin enhanced muscle regeneration in injured muscles (Lee et al., 2021b). These findings show that G. uralensis-derived compounds have therapeutic potential for the management of muscle-related disorders.
Developmental Approaches Used to Design MSTN Inhibitors
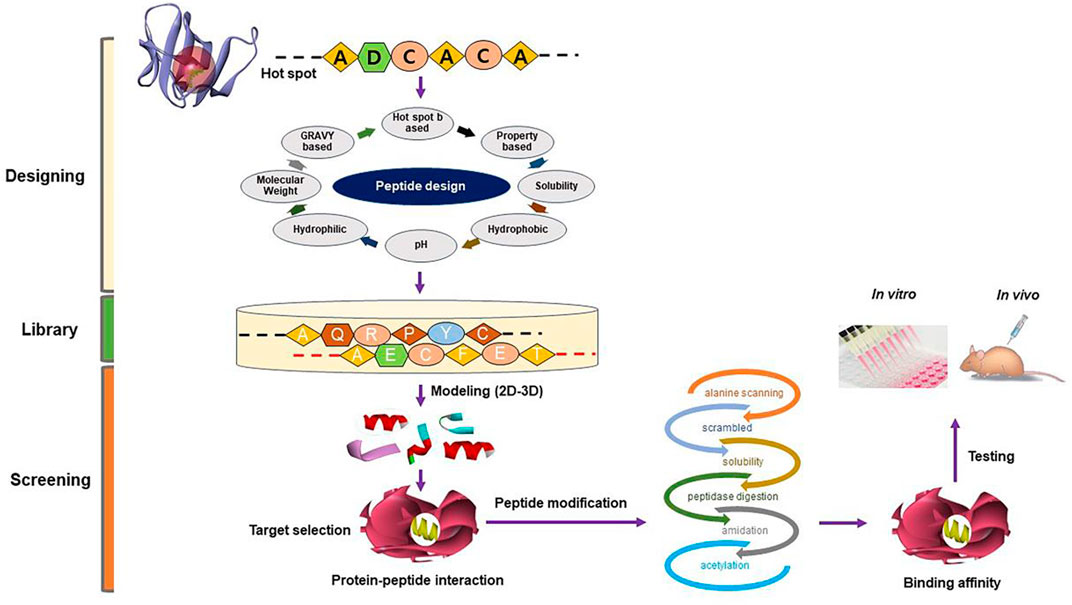
We have been working in the SM field using in silico, in vitro, and in vivo techniques with an emphasis on the mechanism responsible for SM development and regeneration, for more than a decade. Our quest for an efficacious natural MSTN inhibitor in the form of a small molecule or short peptide is ongoing. This section will give a brief understanding of screening natural compounds (Figure 2) and designing short peptides (Figure 3).
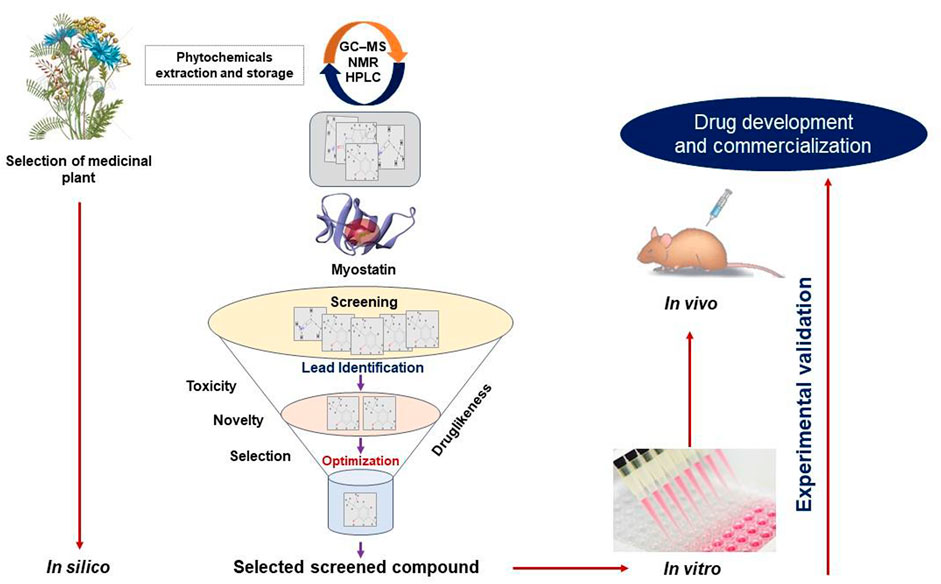
FIGURE 2. A typical approach for the conventional drug design and development strategy. An overview of the techniques for identifying MSTN inhibitors using in silico (virtual screening, molecular docking, ADMET, and so on), in vitro, and in vivo approaches.
Protein-Protein Interactions
PPIs play a vital role in mediating various cellular processes, and thus, have attracted research attention (Guo et al., 2014; Qiu et al., 2020). The advancement in the field of 3-dimensional structure predictions of proteins and PPI includes the recent discovery of AlphaFold (Jumper et al., 2021) and AlphaFold2 (Bryant et al., 2022). Studies have shown that PPI provides a means of effectively regulating various pathways and of developing therapeutic targets. However, all the interfaces of proteins do not contribute equally to PPI (Guo et al., 2014).
The mechanism of action of MSTN is based on its interaction with other proteins in the pathway leading to the transformation of latent MSTN to its mature form and further activation of the Smad pathway, which leads to the inhibition of myogenesis. Studies have shown PPIs are key mediators of various signaling and regulatory networks (Villoutreix et al., 2008). As discussed above, during the activations of different signaling processes, and thus, the activations of atrophic genes, MSTN interacts with a large number of different proteins, which provides clues for the design of peptide inhibitors of MSTN. Peptides that inhibit MSTN activity can be derived in two ways.
Derivation of Peptide Inhibitors From MIPs
MSTN is known to interact with FST, GASP-1, GASP-2, decorin, FMOD, and FLRG, which are collectively referred to as MIPs. These proteins inhibit the formation of mature MSTN complex and interfere with complex formation between MSTN and ACVRIIB. Designing short peptides based on the make-up of MIPs offers a potential means of effective MSTN inhibitors, and some studies indicate that these inhibitory peptides have therapeutic potential for the treatment of a range of muscular dystrophies (Tsuchida, 2008). FS I-I (MSTN-specific inhibitor derived from FST) provides an example of an FST-derived inhibitory peptide and increased SM mass in mdx/FS I-I mice and reduced cell infiltration into muscles (Tsuchida, 2008). DCN is another MIP protein with MSTN inhibitory potential. DCN48-71 and 42-65 are two short fragment peptides derived from members of the small leucine-rich proteoglycan family that demonstrated MSTN inhibitory activity in vitro (El Shafey et al., 2016). Similarly, other studies have successfully identified and tested short peptides capable of inhibiting MSTN activity. One such example is WRQNTRYSRIEAIKIQILSKLRL-amide, which was designed based on the mouse MSTN prodomain. Administration of this peptide to MDX mice (a model of DMD) significantly increased muscle mass. Subsequently, several peptides were designed by structure-activity relationship (SAR) analysis of this short peptide, and peptide ‘3d' (XRQNTRYSRIEWIKIQIISKLRL-amide) exhibited 11 times the potency of its parent and induced muscle growth in MDX and wild-type ICR mice (Takayama et al., 2017).
Detailed structural study of interactions between MIPs and MSTN helped identify residues largely responsible for inhibiting MSTN activity. These were later assembled in different combinations to produce candidate anti-MSTN peptides. Three-dimensional (3D) structures of MSTN in complex with various isomers of FST are available in the protein data bank (PDB) (e.g. PDB ID: 3HH2, 3SEK, and 5JHW) (Cash et al., 2012). Crystal structure analysis showed that the binding of two Fst-like 3 (FSTL3) molecules with MSTN dimer resulted in a highly compact structure and strong interactions in the docked complex.
Self-Inhibitory Peptides
Designing peptide inhibitors targeting PPIs is challenging due to the large sizes of PPIs. Nonetheless, substantial progress has been reported in the field of PPI inhibitor design during the last few years (Jones and Thornton, 1996; Lu et al., 2020; Valtonen et al., 2020). The use of self-derived peptide inhibitors has been one of the most successful PPI inhibitor design strategies. This strategy involves deriving inhibitory peptides from PPIs that act by inhibiting their cognate interactions. The use of self-inhibitory peptides has attracted much interest as a means of inhibiting PPIs that are considered important therapeutic targets (Vlieghe et al., 2010). MSTN activity can also be inhibited by disrupting the interaction between MSTN and its receptor. The approach of targeting protein-protein interfaces to block interactions between MSTN-ACVRIIB instead of enzyme active sites provides another way of reducing MSTN-mediated signaling activity without hampering the intrinsic catalytic functionality of these proteins (Chen et al., 2021).
Virtual Screening for Natural Antagonists of MSTN
Computer-aided drug design (CADD) and computer-assisted molecular design (CAMD) are used as drug discovery tools in the pharmaceutical science field (Baig et al., 2016). VS. is widely used for drug discovery and is complemented by High Throughput Screening (HTS). The VS./HTS approach is used to screen compound libraries quickly and cost-effectively using high-end computational approaches. Selected compounds are subsequently tested for biological activity. The activities of many identified natural compounds have yet to be determined, such as those detailed in the Chinese traditional medicine and Korean medicinal compound databases. Several groups have attempted to identify novel therapeutic candidates that target MSTN, but unfortunately, a large number of identified compounds were not MSTN specific and were also found to block activin A and TGF-β signaling (Suh and Lee, 2020a). The specific targeting of MSTN remains a significant research challenge as many TGF-β ligands exhibit considerable structural similarities. Recently, we performed a VS-based analysis on known muscle-enhancing natural compounds for MSTN inhibitory activity and identified curcumin and gingerol as candidates (Baig et al., 2017). Undoubtedly, there are limitations of the VS approach that should be taken into account. The prevalence of stereochemical and valence mistakes in biochemical compound libraries may potentially result in inviable molecules (Williams et al., 2012; Santana et al., 2021). There are a variety of open source and licensed virtual screening software/tools available, and each has its own constraints that must be overcome to prevent the production of erroneous conclusions or artifacts (Gimeno et al., 2019). For VS, licensed software such as Molecular Operating Environment (MOE) (Vilar et al., 2008), and the GLIDE module in Schrodinger (Bhachoo and Beuming, 2017) as well as open access tools such as Autodock Vina (Trott and Olson, 2010), are commonly used.
Conclusion
The inhibitory role played by MSTN in muscle development has made it an important therapeutic target for accelerating muscle mass. There are several ways of disrupting MSTN activity ranging from the use of MSTN antibodies to natural compounds. Detailed knowledge of these strategies and the use of in silico techniques should improve knowledge of the structural characteristics of MSTN and its bindings with inhibitory proteins, derived inhibitors, and other natural compounds. Structural insight of binding between different MIPs and MSTN should open new doors to the design of better therapeutic peptide candidates. Although no drugs have yet been developed to prevent muscle degeneration, we believe that research efforts targeting MSTN will result in treatments that attenuate muscle degeneration and improve the quality of life in the elderly and those suffering from MD.
Author Contributions
Conceptualization, IC, EL; writing—original draft preparation, MB, KA, JM, S-YP; writing—review and editing, JH, HC, AQ, YH, AJ, SA, SA, SS.
Funding
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Korean Ministry of Education (2020R1A6A1A03044512) and the NRF funded by the Korean government (MSIP: Grant Nos. NRF-2021R1A2C2004177 and NRF-2019R1C1C1006542) and was supported by Korea Institute of Planning and Evaluation for Technology in Food, Agriculture, Forestry (IPET) through High Value-added Food Technology Development Program, funded by Ministry of Agriculture, Food and Rural Affairs (MAFRA) (321026-05).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ahmad K., Choi I., Lee Y. H. (2020a). Implications of Skeletal Muscle Extracellular Matrix Remodeling in Metabolic Disorders: Diabetes Perspective. Int. J. Mol. Sci. 21. doi:10.3390/ijms21113845
Ahmad K., Lee E., Moon J., Park S.-Y., Choi I. (2018). Multifaceted Interweaving between Extracellular Matrix, Insulin Resistance, and Skeletal Muscle. Cells 7, 148. doi:10.3390/cells7100148
Ahmad K., Shaikh S., Ahmad S. S., Lee E. J., Choi I. (2020b). Cross-Talk between Extracellular Matrix and Skeletal Muscle: Implications for Myopathies. Front. Pharmacol. 11, 142. doi:10.3389/fphar.2020.00142
Ahmad S., Jan A. T., Baig M. H., Lee E. J., Choi I. (2017). Matrix Gla Protein: An Extracellular Matrix Protein Regulates Myostatin Expression in the Muscle Developmental Program. Life Sci. 172, 55–63. doi:10.1016/j.lfs.2016.12.011
Allen D. L., Cleary A. S., Speaker K. J., Lindsay S. F., Uyenishi J., Reed J. M., et al. (2008). Myostatin, Activin Receptor IIb, and Follistatin-Like-3 Gene Expression Are Altered in Adipose Tissue and Skeletal Muscle of Obese Mice. Am. J. Physiology-Endocrinology Metabolism 294, E918–E927. doi:10.1152/ajpendo.00798.2007
Allen D. L., Hittel D. S., Mcpherron A. C. (2011). Expression and Function of Myostatin in Obesity, Diabetes, and Exercise Adaptation. Med. Sci. Sports Exerc 43, 1828–1835. doi:10.1249/mss.0b013e3182178bb4
Allen D. L., Unterman T. G. (2007). Regulation of Myostatin Expression and Myoblast Differentiation by FoxO and SMAD Transcription Factors. Am. J. Physiology-Cell Physiology 292, C188–C199. doi:10.1152/ajpcell.00542.2005
Alshehri B., D'souza D. G., Lee J. Y., Petratos S., Richardson S. J. (2015). The Diversity of Mechanisms Influenced by Transthyretin in Neurobiology: Development, Disease and Endocrine Disruption. J. Neuroendocrinol. 27, 303–323. doi:10.1111/jne.12271
Amor M., Itariu B. K., Moreno-Viedma V., Keindl M., Jürets A., Prager G., et al. (2019). Serum Myostatin Is Upregulated in Obesity and Correlates with Insulin Resistance in Humans. Exp. Clin. Endocrinol. Diabetes 127, 550–556. doi:10.1055/a-0641-5546
Amthor H., Macharia R., Navarrete R., Schuelke M., Brown S. C., Otto A., et al. (2007). Lack of Myostatin Results in Excessive Muscle Growth but Impaired Force Generation. Proc. Natl. Acad. Sci. U.S.A. 104, 1835–1840. doi:10.1073/pnas.0604893104
Argilés J. M., Campos N., Lopez-Pedrosa J. M., Rueda R., Rodriguez-Mañas L. (2016). Skeletal Muscle Regulates Metabolism via Interorgan Crosstalk: Roles in Health and Disease. J. Am. Med. Dir. Assoc. 17, 789–796. doi:10.1016/j.jamda.2016.04.019
Aversa Z., Bonetto A., Penna F., Costelli P., Di Rienzo G., Lacitignola A., et al. (2012). Changes in Myostatin Signaling in Non-weight-losing Cancer Patients. Ann. Surg. Oncol. 19, 1350–1356. doi:10.1245/s10434-011-1720-5
Aversa Z., Costelli P., Muscaritoli M. (2017). Cancer-induced Muscle Wasting: Latest Findings in Prevention and Treatment. Ther. Adv. Med. Oncol. 9, 369–382. doi:10.1177/1758834017698643
Baczek J., Silkiewicz M., Wojszel Z. B. (2020). Myostatin as a Biomarker of Muscle Wasting and Other Pathologies-State of the Art and Knowledge Gaps. Nutrients 12, 2401. doi:10.3390/nu12082401
Baig M. H., Ahmad K., Roy S., Ashraf J. M., Adil M., Siddiqui M. H., et al. (2016). Computer Aided Drug Design: Success and Limitations. Curr. Pharm. Des. 22, 572–581. doi:10.2174/1381612822666151125000550
Baig M. H., Jan A. T., Rabbani G., Ahmad K., Ashraf J. M., Kim T., et al. (2017). Methylglyoxal and Advanced Glycation End Products: Insight of the Regulatory Machinery Affecting the Myogenic Program and of its Modulation by Natural Compounds. Sci. Rep. 7, 5916. doi:10.1038/s41598-017-06067-5
Baig M. H., Rabbani G., Ahmad K., Choi I. (2018). Use of Peptides for the Management of Alzheimer's Disease: Diagnosis and Inhibition. Front. Aging Neurosci. 10, 21. doi:10.3389/fnagi.2018.00021
Becker C., Lord S. R., Studenski S. A., Warden S. J., Fielding R. A., Recknor C. P., et al. (2015). Myostatin Antibody (LY2495655) in Older Weak Fallers: a Proof-Of-Concept, Randomised, Phase 2 Trial. Lancet Diabetes & Endocrinol. 3, 948–957. doi:10.1016/s2213-8587(15)00298-3
Benny Klimek M. E., Aydogdu T., Link M. J., Pons M., Koniaris L. G., Zimmers T. A. (2010). Acute Inhibition of Myostatin-Family Proteins Preserves Skeletal Muscle in Mouse Models of Cancer Cachexia. Biochem. Biophysical Res. Commun. 391, 1548–1554. doi:10.1016/j.bbrc.2009.12.123
Bergen H. R., Farr J. N., Vanderboom P. M., Atkinson E. J., White T. A., Singh R. J., et al. (2015). Myostatin as a Mediator of Sarcopenia versus Homeostatic Regulator of Muscle Mass: Insights Using a New Mass Spectrometry-Based Assay. Skelet. Muscle 5, 21. doi:10.1186/s13395-015-0047-5
Bhachoo J., Beuming T. (2017). Investigating Protein–Peptide Interactions Using the Schrödinger Computational Suite. Humana New York, NY: Modeling peptide-protein interactions, 235–254. doi:10.1007/978-1-4939-6798-8_14
Bianchi A., Mozzetta C., Pegoli G., Lucini F., Valsoni S., Rosti V., et al. (2020). Dysfunctional Polycomb Transcriptional Repression Contributes to Lamin A/C-dependent Muscular Dystrophy. J. Clin. Invest. 130, 2408–2421. doi:10.1172/jci128161
Bonetto A., Penna F., Minero V., Reffo P., Bonelli G., Baccino F., et al. (2009). Deacetylase Inhibitors Modulate the Myostatin/follistatin axis without Improving Cachexia in Tumor-Bearing Mice. Curr. Cancer Drug Targets 9, 608–616. doi:10.2174/156800909789057015
Bose C., Alves I., Singh P., Palade P. T., Carvalho E., Børsheim E., et al. (2020). Sulforaphane Prevents Age-Associated Cardiac and Muscular Dysfunction through Nrf2 Signaling. Aging Cell 19, e13261. doi:10.1111/acel.13261
Brimijoin S., Chen V. P., Pang Y.-P., Geng L., Gao Y. (2016). Physiological Roles for Butyrylcholinesterase: A BChE-Ghrelin axis. Chemico-Biological Interact. 259, 271–275. doi:10.1016/j.cbi.2016.02.013
Broholm C., Pedersen B. K. (2010). Leukaemia Inhibitory Factor-Aan Exercise-Induced Myokine. Exerc Immunol. Rev. 16, 77–85.
Broholm C., Laye M. J., Brandt C., Vadalasetty R., Pilegaard H., Pedersen B. K., et al. (2011). LIF Is a Contraction-Induced Myokine Stimulating Human Myocyte Proliferation. J. Appl. Physiology 111, 251–259. doi:10.1152/japplphysiol.01399.2010
Bryant P., Pozzati G., Elofsson A. (2022). Improved Prediction of Protein-Protein Interactions Using AlphaFold2. Nat. Commun. 13, 1–11. doi:10.1038/s41467-022-28865-w
Busquets S., Toledo M., Orpí M., Massa D., Porta M., Capdevila E., et al. (2012). Myostatin Blockage Using actRIIB Antagonism in Mice Bearing the Lewis Lung Carcinoma Results in the Improvement of Muscle Wasting and Physical Performance. J. Cachexia Sarcopenia Muscle 3, 37–43. doi:10.1007/s13539-011-0049-z
Campbell C., Mcmillan H. J., Mah J. K., Tarnopolsky M., Selby K., Mcclure T., et al. (2017). Myostatin Inhibitor ACE-031 Treatment of Ambulatory Boys with Duchenne Muscular Dystrophy: Results of a Randomized, Placebo-Controlled Clinical Trial. Muscle Nerve 55, 458–464. doi:10.1002/mus.25268
Camporez J.-P. G., Petersen M. C., Abudukadier A., Moreira G. V., Jurczak M. J., Friedman G., et al. (2016). Anti-myostatin Antibody Increases Muscle Mass and Strength and Improves Insulin Sensitivity in Old Mice. Proc. Natl. Acad. Sci. U.S.A. 113, 2212–2217. doi:10.1073/pnas.1525795113
Carbó N., López-Soriano J., Costelli P., Busquets S., Alvarez B., Baccino F. M., et al. (2000). Interleukin-15 Antagonizes Muscle Protein Waste in Tumour-Bearing Rats. Br. J. Cancer 83, 526–531. doi:10.1054/bjoc.2000.1299
Carnac G., Vernus B., Bonnieu A. (2007). Myostatin in the Pathophysiology of Skeletal Muscle. Curr. Genomics 8, 415–422. doi:10.2174/138920207783591672
Cartee G. D., Hepple R. T., Bamman M. M., Zierath J. R. (2016). Exercise Promotes Healthy Aging of Skeletal Muscle. Cell Metab. 23, 1034–1047. doi:10.1016/j.cmet.2016.05.007
Cash J. N., Angerman E. B., Kattamuri C., Nolan K., Zhao H., Sidis Y., et al. (2012). Structure of Myostatin·Follistatin-like 3. J. Biol. Chem. 287, 1043–1053. doi:10.1074/jbc.m111.270801
Chen J.-a., Splenser A., Guillory B., Luo J., Mendiratta M., Belinova B., et al. (2015). Ghrelin Prevents Tumour- and Cisplatin-Induced Muscle Wasting: Characterization of Multiple Mechanisms Involved. J. Cachexia Sarcopenia Muscle 6, 132–143. doi:10.1002/jcsm.12023
Chen M.-M., Zhao Y.-P., Zhao Y., Deng S.-L., Yu K. (2021). Regulation of Myostatin on the Growth and Development of Skeletal Muscle. Front. Cell Dev. Biol. 9, 785712. doi:10.3389/fcell.2021.785712
Collins C. A., Morgan J. E. (2003). Duchenne's Muscular Dystrophy: Animal Models Used to Investigate Pathogenesis and Develop Therapeutic Strategies. Int. J. Exp. Pathol. 84, 165–172. doi:10.1046/j.1365-2613.2003.00354.x
Consitt L. A., Clark B. C. (2018). The Vicious Cycle of Myostatin Signaling in Sarcopenic Obesity: Myostatin Role in Skeletal Muscle Growth, Insulin Signaling and Implications for Clinical Trials. J. Frailty Aging 7, 21–27. doi:10.14283/jfa.2017.33
Costelli P., Muscaritoli M., Bonetto A., Penna F., Reffo P., Bossola M., et al. (2008). Muscle Myostatin Signalling Is Enhanced in Experimental Cancer Cachexia. Eur. J. Clin. Invest. 38, 531–538. doi:10.1111/j.1365-2362.2008.01970.x
Cotton T. R., Fischer G., Wang X., Mccoy J. C., Czepnik M., Thompson T. B., et al. (2018). Structure of the Human Myostatin Precursor and Determinants of Growth Factor Latency. EMBO J. 37, 367–383. doi:10.15252/embj.201797883
Curtis E., Litwic A., Cooper C., Dennison E. (2015). Determinants of Muscle and Bone Aging. J. Cell. Physiol. 230, 2618–2625. doi:10.1002/jcp.25001
D'Orlando C., Marzetti E., François S., Lorenzi M., Conti V., Di Stasio E., et al. (2014). Gastric Cancer Does Not Affect the Expression of Atrophy-Related Genes in Human Skeletal Muscle. Muscle Nerve 49, 528–533. doi:10.1002/mus.23945
DeBoer M. D. (2011). Ghrelin and Cachexia: Will Treatment with GHSR-1a Agonists Make a Difference for Patients Suffering from Chronic Wasting Syndromes? Mol. Cell. Endocrinol. 340, 97–105. doi:10.1016/j.mce.2011.02.012
Delaney K., Kasprzycka P., Ciemerych M. A., Zimowska M. (2017). The Role of TGF-Β1 during Skeletal Muscle Regeneration. Cell Biol. Int. 41, 706–715. doi:10.1002/cbin.10725
Dueweke J. J., Awan T. M., Mendias C. L. (2017). Regeneration of Skeletal Muscle after Eccentric Injury. J. Sport Rehabil. 26, 171–179. doi:10.1123/jsr.2016-0107
Dumont N. A., Bentzinger C. F., Sincennes M. C., Rudnicki M. A. (2015). Satellite Cells and Skeletal Muscle Regeneration. Compr. Physiol. 5, 1027–1059. doi:10.1002/cphy.c140068
Dutt V., Gupta S., Dabur R., Injeti E., Mittal A. (2015). Skeletal Muscle Atrophy: Potential Therapeutic Agents and Their Mechanisms of Action. Pharmacol. Res. 99, 86–100. doi:10.1016/j.phrs.2015.05.010
El Shafey N., Guesnon M., Simon F., Deprez E., Cosette J., Stockholm D., et al. (2016). Inhibition of the myostatin/Smad Signaling Pathway by Short Decorin-Derived Peptides. Exp. Cell Res. 341, 187–195. doi:10.1016/j.yexcr.2016.01.019
Elabd C., Cousin W., Upadhyayula P., Chen R. Y., Chooljian M. S., Li J., et al. (2014). Oxytocin Is an Age-specific Circulating Hormone that Is Necessary for Muscle Maintenance and Regeneration. Nat. Commun. 5, 4082. doi:10.1038/ncomms5082
Elliott B., Renshaw D., Getting S., Mackenzie R. (2012). The Central Role of Myostatin in Skeletal Muscle and Whole Body Homeostasis. Acta Physiol. 205, 324–340. doi:10.1111/j.1748-1716.2012.02423.x
Fan H., Zhang R., Tesfaye D., Tholen E., Looft C., Hölker M., et al. (2012). Sulforaphane Causes a Major Epigenetic Repression of Myostatin in Porcine Satellite Cells. Epigenetics 7, 1379–1390. doi:10.4161/epi.22609
Fanzani A., Conraads V. M., Penna F., Martinet W. (2012). Molecular and Cellular Mechanisms of Skeletal Muscle Atrophy: an Update. J. Cachexia Sarcopenia Muscle 3, 163–179. doi:10.1007/s13539-012-0074-6
Frontera W. R., Ochala J. (2015). Skeletal Muscle: a Brief Review of Structure and Function. Calcif. Tissue Int. 96, 183–195. doi:10.1007/s00223-014-9915-y
Fu J., Huang L., Zhang H., Yang S., Chen S. (2013). Structural Features of a Polysaccharide fromAstragalus membranaceus(Fisch.) Bge. var.mongholicus(Bge.) Hsiao. J. Asian Nat. Prod. Res. 15, 687–692. doi:10.1080/10286020.2013.778832
Gadkari P. V., Balaraman M. (2015). Catechins: Sources, Extraction and Encapsulation: A Review. Food Bioprod. Process. 93, 122–138. doi:10.1016/j.fbp.2013.12.004
Gallot Y. S., Durieux A.-C., Castells J., Desgeorges M. M., Vernus B., Plantureux L., et al. (2014). Myostatin Gene Inactivation Prevents Skeletal Muscle Wasting in Cancer. Cancer Res. 74, 7344–7356. doi:10.1158/0008-5472.can-14-0057
Ganassi M., Badodi S., Wanders K., Zammit P. S., Hughes S. M. (2020). Myogenin Is an Essential Regulator of Adult Myofibre Growth and Muscle Stem Cell Homeostasis. Elife 9, e60445. doi:10.7554/eLife.60445
Garin M. C., Burns C. M., Kaul S., Cappola A. R. (2013). The Human Experience with Ghrelin Administration. J. Clin. Endocrinol. Metab. 98, 1826–1837. doi:10.1210/jc.2012-4247
Gimeno A., Ojeda-Montes M., Tomás-Hernández S., Cereto-Massagué A., Beltrán-Debón R., Mulero M., et al. (2019). The Light and Dark Sides of Virtual Screening: what Is There to Know? Int. J. Mol. Sci. 20, 1375. doi:10.3390/ijms20061375
Glasser C. E., Gartner M. R., Wilson D., Miller B., Sherman M. L., Attie K. M. (2018). Locally Acting ACE-083 Increases Muscle Volume in Healthy Volunteers. Muscle Nerve 57, 921–926. doi:10.1002/mus.26113
Golan T., Geva R., Richards D., Madhusudan S., Lin B. K., Wang H. T., et al. (2018). LY2495655, an Antimyostatin Antibody, in Pancreatic Cancer: a Randomized, Phase 2 Trial. J. Cachexia, Sarcopenia Muscle 9, 871–879. doi:10.1002/jcsm.12331
Goodman C. A., Mcnally R. M., Hoffmann F. M., Hornberger T. A. (2013). Smad3 Induces Atrogin-1, Inhibits mTOR and Protein Synthesis, and Promotes Muscle Atrophy In Vivo. Mol. Endocrinol. 27, 1946–1957. doi:10.1210/me.2013-1194
Guardiola O., Andolfi G., Tirone M., Iavarone F., Brunelli S., Minchiotti G. (2017). Induction of Acute Skeletal Muscle Regeneration by Cardiotoxin Injection. J. Vis. Exp. 119, 54515. doi:10.3791/54515
Guo W., Wisniewski J. A., Ji H. (2014). Hot Spot-Based Design of Small-Molecule Inhibitors for Protein-Protein Interactions. Bioorg. Med. Chem. Lett. 24, 2546–2554. doi:10.1016/j.bmcl.2014.03.095
Gutierrez-Salmean G., Ciaraldi T. P., Nogueira L., Barboza J., Taub P. R., Hogan M. C., et al. (2014). Effects of (−)-epicatechin on Molecular Modulators of Skeletal Muscle Growth and Differentiation. J. Nutr. Biochem. 25, 91–94. doi:10.1016/j.jnutbio.2013.09.007
Hatakeyama S., Summermatter S., Jourdain M., Melly S., Minetti G. C., Lach-Trifilieff E. (2016). ActRII Blockade Protects Mice from Cancer Cachexia and Prolongs Survival in the Presence of Anti-cancer Treatments. Skelet. Muscle 6, 26–12. doi:10.1186/s13395-016-0098-2
Hedayati M., Nozhat Z., Hannani M. (2016). Can the Serum Level of Myostatin Be Considered as an Informative Factor for Cachexia Prevention in Patients with Medullary Thyroid Cancer? Asian Pac. J. Cancer Prev. 17, 119–123. doi:10.7314/apjcp.2016.17.s3.119
Hill J. J., Davies M. V., Pearson A. A., Wang J. H., Hewick R. M., Wolfman N. M., et al. (2002). The Myostatin Propeptide and the Follistatin-Related Gene Are Inhibitory Binding Proteins of Myostatin in Normal Serum. J. Biol. Chem. 277, 40735–40741. doi:10.1074/jbc.m206379200
Hill J. J., Qiu Y., Hewick R. M., Wolfman N. M. (2003). Regulation of Myostatin In Vivo by Growth and Differentiation Factor-Associated Serum Protein-1: a Novel Protein with Protease Inhibitor and Follistatin Domains. Mol. Endocrinol. 17, 1144–1154. doi:10.1210/me.2002-0366
Hittel D. S., Axelson M., Sarna N., Shearer J., Huffman K. M., Kraus W. E. (2010). Myostatin Decreases with Aerobic Exercise and Associates with Insulin Resistance. Med. Sci. Sports Exerc 42, 2023–2029. doi:10.1249/mss.0b013e3181e0b9a8
Hittel D. S., Berggren J. R., Shearer J., Boyle K., Houmard J. A. (2009). Increased Secretion and Expression of Myostatin in Skeletal Muscle from Extremely Obese Women. Diabetes 58, 30–38. doi:10.2337/db08-0943
Hoene M., Runge H., Häring H. U., Schleicher E. D., Weigert C. (2013). Interleukin-6 Promotes Myogenic Differentiation of Mouse Skeletal Muscle Cells: Role of the STAT3 Pathway. Am. J. Physiology-Cell Physiology 304, C128–C136. doi:10.1152/ajpcell.00025.2012
Holmberg J., Durbeej M. (2013). Laminin-211 in Skeletal Muscle Function. Cell Adhesion Migr. 7, 111–121. doi:10.4161/cam.22618
Howick K., Alam R., Chruscicka B., Kandil D., Fitzpatrick D., Ryan A. M., et al. (2018). Sustained-release Multiparticulates for Oral Delivery of a Novel Peptidic Ghrelin Agonist: Formulation Design and In Vitro Characterization. Int. J. Pharm. 536, 63–72. doi:10.1016/j.ijpharm.2017.11.051
Huang Z., Chen D., Zhang K., Yu B., Chen X., Meng J. (2007). Regulation of Myostatin Signaling by C-Jun N-Terminal Kinase in C2C12 Cells. Cell. Signal. 19, 2286–2295. doi:10.1016/j.cellsig.2007.07.002
Huang Z., Chen X., Chen D. (2011). Myostatin: a Novel Insight into its Role in Metabolism, Signal Pathways, and Expression Regulation. Cell. Signal. 23, 1441–1446. doi:10.1016/j.cellsig.2011.05.003
Hulmi J. J., Nissinen T. A., Penna F., Bonetto A. (2021). Targeting the Activin Receptor Signaling to Counteract the Multi-Systemic Complications of Cancer and its Treatments. Cells 10, 516. doi:10.3390/cells10030516
Jan A. T., Lee E. J., Ahmad S., Choi I. (2016). Meeting the Meat: Delineating the Molecular Machinery of Muscle Development. J. Anim. Sci. Technol. 58, 18–10. doi:10.1186/s40781-016-0100-x
Ji S., Li Z., Song W., Wang Y., Liang W., Li K., et al. (2016). Bioactive Constituents of Glycyrrhiza Uralensis (Licorice): Discovery of the Effective Components of a Traditional Herbal Medicine. J. Nat. Prod. 79, 281–292. doi:10.1021/acs.jnatprod.5b00877
Jones S., Thornton J. M. (1996). Principles of Protein-Protein Interactions. Proc. Natl. Acad. Sci. U.S.A. 93, 13–20. doi:10.1073/pnas.93.1.13
Joulia-Ekaza D., Dominique J. E., Cabello G., Gérard C. (2006). Myostatin Regulation of Muscle Development: Molecular Basis, Natural Mutations, Physiopathological Aspects. Exp. Cell Res. 312, 2401–2414. doi:10.1016/j.yexcr.2006.04.012
Lee E. J., Tasleem Jan A., Hassan Baig M., Ahmad K., Malik A., Rabbani G., et al. (2018). Fibromodulin and Regulation of the Intricate Balance between Myoblast Differentiation to Myocytes or Adipocyte‐like Cells. FASEB J. 32, 768–781. doi:10.1096/fj.201700665r
Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., et al. (2021). Highly Accurate Protein Structure Prediction with AlphaFold. Nature 596, 583–589. doi:10.1038/s41586-021-03819-2
Kim J. W., Ku S.-K., Han M. H., Kim K. Y., Kim S. G., Kim G.-Y., et al. (2015). The Administration of Fructus Schisandrae Attenuates Dexamethasone-Induced Muscle Atrophy in Mice. Int. J. Mol. Med. 36, 29–42. doi:10.3892/ijmm.2015.2200
Kim T., Ahmad K., Shaikh S., Jan A. T., Seo M. G., Lee E. J., et al. (2019). Dermatopontin in Skeletal Muscle Extracellular Matrix Regulates Myogenesis. Cells 8. doi:10.3390/cells8040332
Kishimoto I., Tokudome T., Hosoda H., Miyazato M., Kangawa K. (2012). Ghrelin and Cardiovascular Diseases. J. Cardiol. 59, 8–13. doi:10.1016/j.jjcc.2011.11.002
Kishioka Y., Thomas M., Wakamatsu J.-i., Hattori A., Sharma M., Kambadur R., et al. (2008). Decorin Enhances the Proliferation and Differentiation of Myogenic Cells through Suppressing Myostatin Activity. J. Cell. Physiol. 215, 856–867. doi:10.1002/jcp.21371
Kuang S., Gillespie M. A., Rudnicki M. A. (2008). Niche Regulation of Muscle Satellite Cell Self-Renewal and Differentiation. Cell stem Cell 2, 22–31. doi:10.1016/j.stem.2007.12.012
Latres E., Mastaitis J., Fury W., Miloscio L., Trejos J., Pangilinan J., et al. (2017). Activin A More Prominently Regulates Muscle Mass in Primates Than Does GDF8. Nat. Commun. 8, 15153. doi:10.1038/ncomms15153
Latres E., Pangilinan J., Miloscio L., Bauerlein R., Na E., Potocky T. B., et al. (2015). Myostatin Blockade with a Fully Human Monoclonal Antibody Induces Muscle Hypertrophy and Reverses Muscle Atrophy in Young and Aged Mice. Skelet. Muscle 5, 34. doi:10.1186/s13395-015-0060-8
Lee E. J., Shaikh S., Choi D., Ahmad K., Baig M. H., Lim J. H., et al. (2019). Transthyretin Maintains Muscle Homeostasis through the Novel Shuttle Pathway of Thyroid Hormones during Myoblast Differentiation. Cells 8. doi:10.3390/cells8121565
Lee E. J., Ahmad S. S., Lim J. H., Ahmad K., Shaikh S., Lee Y.-S., et al. (2021a). Interaction of Fibromodulin and Myostatin to Regulate Skeletal Muscle Aging: An Opposite Regulation in Muscle Aging, Diabetes, and Intracellular Lipid Accumulation. Cells 10, 2083. doi:10.3390/cells10082083
Lee E. J., Jan A. T., Baig M. H., Ashraf J. M., Nahm S. S., Kim Y. W., et al. (2016). Fibromodulin: a Master Regulator of Myostatin Controlling Progression of Satellite Cells through a Myogenic Program. FASEB J. 30, 2708–2719. doi:10.1096/fj.201500133r
Lee E. J., Kamli M. R., Pokharel S., Malik A., Tareq K. M. A., Roouf Bhat A., et al. (2013). Expressed Sequence Tags for Bovine Muscle Satellite Cells, Myotube Formed-Cells and Adipocyte-like Cells. PLoS One 8, e79780. doi:10.1371/journal.pone.0079780
Lee E. J., Shaikh S., Ahmad K., Ahmad S. S., Lim J. H., Park S., et al. (2021b). Isolation and Characterization of Compounds from Glycyrrhiza Uralensis as Therapeutic Agents for the Muscle Disorders. Int. J. Mol. Sci. 22, 876. doi:10.3390/ijms22020876
Lee S.-J., Lee Y.-S., Zimmers T. A., Soleimani A., Matzuk M. M., Tsuchida K., et al. (2010). Regulation of Muscle Mass by Follistatin and Activins. Mol. Endocrinol. 24, 1998–2008. doi:10.1210/me.2010-0127
Lee S.-J., McPherron A. C. (2001). Regulation of Myostatin Activity and Muscle Growth. Proc. Natl. Acad. Sci. U.S.A. 98, 9306–9311. doi:10.1073/pnas.151270098
Lee S.-J. (2012). “Myostatin: Regulation, Function, and Therapeutic Applications,” in Muscle (Elsevier), 1077–1084. doi:10.1016/b978-0-12-381510-1.00079-x
Lee Y. S., Lee S. J. (2013). Regulation of GDF-11 and Myostatin Activity by GASP-1 and GASP-2. Proc. Natl. Acad. Sci. U. S. A. 110, E3713–E3722. doi:10.1073/pnas.1309907110
Lieber R. L., Friden J. (2000). Functional and Clinical Significance of Skeletal Muscle Architecture. Muscle Nerve 23, 1647–1666. doi:10.1002/1097-4598(200011)23:11<1647::aid-mus1>3.0.co;2-m
Lindon C., Montarras D., Pinset C. (1998). Cell Cycle-Regulated Expression of the Muscle Determination Factor Myf5 in Proliferating Myoblasts. J. Cell Biol. 140, 111–118. doi:10.1083/jcb.140.1.111
Liu M., Qin J., Hao Y., Liu M., Luo J., Luo T., et al. (2013). Astragalus Polysaccharide Suppresses Skeletal Muscle Myostatin Expression in Diabetes: Involvement of ROS-ERK and NF-Κb Pathways. Oxid. Med. Cell Longev. 2013, 782497. doi:10.1155/2013/782497
Liu X.-H., Bauman W. A., Cardozo C. P. (2018). Myostatin Inhibits Glucose Uptake via Suppression of Insulin-dependent and -independent Signaling Pathways in Myoblasts. Physiol. Rep. 6, e13837. doi:10.14814/phy2.13837
Liu Y., Lehar A., Rydzik R., Chandok H., Lee Y.-S., Youngstrom D. W., et al. (2021). Local versus Systemic Control of Bone and Skeletal Muscle Mass by Components of the Transforming Growth Factor-β Signaling Pathway. Proc. Natl. Acad. Sci. 118. doi:10.1073/pnas.2111401118
Londhe P., Davie J. K. (2011). Sequential Association of Myogenic Regulatory Factors and E Proteins at Muscle-specific Genes. Skelet. Muscle 1, 14–18. doi:10.1186/2044-5040-1-14
Loumaye A., De Barsy M., Nachit M., Lause P., Frateur L., Van Maanen A., et al. (2015). Role of Activin A and Myostatin in Human Cancer Cachexia. J. Clin. Endocrinol. Metabolism 100, 2030–2038. doi:10.1210/jc.2014-4318
Lu H., Zhou Q., He J., Jiang Z., Peng C., Tong R., et al. (2020). Recent Advances in the Development of Protein-Protein Interactions Modulators: Mechanisms and Clinical Trials. Sig Transduct. Target Ther. 5, 213. doi:10.1038/s41392-020-00315-3
Mafi F., Biglari S., Ghardashi Afousi A., Gaeini A. A. (2019). Improvement in Skeletal Muscle Strength and Plasma Levels of Follistatin and Myostatin Induced by an 8-Week Resistance Training and Epicatechin Supplementation in Sarcopenic Older Adults. J. Aging Phys. Activity 27, 384–391. doi:10.1123/japa.2017-0389
Mandai S., Furukawa S., Kodaka M., Hata Y., Mori T., Nomura N., et al. (2017). Loop Diuretics Affect Skeletal Myoblast Differentiation and Exercise-Induced Muscle Hypertrophy. Sci. Rep. 7, 46369. doi:10.1038/srep46369
Manole E., Ceafalan L. C., Popescu B. O., Dumitru C., Bastian A. E. (2018). Myokines as Possible Therapeutic Targets in Cancer Cachexia. J. Immunol. Res. 2018, 8260742. doi:10.1155/2018/8260742
McFarlane C., Hennebry A., Thomas M., Plummer E., Ling N., Sharma M., et al. (2008). Myostatin Signals through Pax7 to Regulate Satellite Cell Self-Renewal. Exp. Cell Res. 314, 317–329. doi:10.1016/j.yexcr.2007.09.012
McFarlane C., Plummer E., Thomas M., Hennebry A., Ashby M., Ling N., et al. (2006). Myostatin Induces Cachexia by Activating the Ubiquitin Proteolytic System through an NF-κb-independent, FoxO1-dependent Mechanism. J. Cell. Physiol. 209, 501–514. doi:10.1002/jcp.20757
McKay B. R., Ogborn D. I., Bellamy L. M., Tarnopolsky M. A., Parise G. (2012). Myostatin Is Associated with Age‐related Human Muscle Stem Cell Dysfunction. FASEB J. 26, 2509–2521. doi:10.1096/fj.11-198663
McPherron A. C., Lawler A. M., Lee S.-J. (1997). Regulation of Skeletal Muscle Mass in Mice by a New TGF-P Superfamily Member. Nature 387, 83–90. doi:10.1038/387083a0
Miura T., Kishioka Y., Wakamatsu J.-i., Hattori A., Nishimura T. (2010). Interaction between Myostatin and Extracellular Matrix Components. Anim. Sci. J. 81, 102–107. doi:10.1111/j.1740-0929.2009.00700.x
Monestier O., Brun C., Heu K., Passet B., Malhouroux M., Magnol L., et al. (2012). Ubiquitous Gasp1 Overexpression in Mice Leads Mainly to a Hypermuscular Phenotype. BMC Genomics 13, 541. doi:10.1186/1471-2164-13-541
Morissette M. R., Cook S. A., Buranasombati C., Rosenberg M. A., Rosenzweig A. (2009). Myostatin Inhibits IGF-I-Induced Myotube Hypertrophy through Akt. Am. J. Physiol. Cell Physiol. 297, C1124–C1132. doi:10.1152/ajpcell.00043.2009
Muñoz-Cánoves P., Scheele C., Pedersen B. K., Serrano A. L. (2013). Interleukin-6 Myokine Signaling in Skeletal Muscle: a Double-Edged Sword? FEBS J. 280, 4131–4148. doi:10.1111/febs.12338
Myzak M. C., Dashwood W. M., Orner G. A., Ho E., Dashwood R. H. (2006). Sulforaphane Inhibits Histone Deacetylase In Vivo and Suppresses Tumorigenesis in Apc Min Mice. FASEB J. 20, 506–508. doi:10.1096/fj.05-4785fje
Nishimura T., Futami E., Taneichi A., Mori T., Hattori A. (2002). Decorin Expression during Development of Bovine Skeletal Muscle and its Role in Morphogenesis of the Intramuscular Connective Tissue. Cells Tissues Organs 171, 199–214. doi:10.1159/000063713
Ozawa T., Miyazono K., Morikawa M. (2021a). Preparation of Monovalent Follistatin-like 3-Fc-Fusion Protein and Evaluation of its Effects on Muscle Mass in Mice. Star. Protoc. 2, 100839. doi:10.1016/j.xpro.2021.100839
Ozawa T., Morikawa M., Morishita Y., Ogikubo K., Itoh F., Koinuma D., et al. (2021b). Systemic Administration of Monovalent Follistatin-like 3-Fc-Fusion Protein Increases Muscle Mass in Mice. Iscience 24, 102488. doi:10.1016/j.isci.2021.102488
Panossian A., Wikman G. (2008). Pharmacology of Schisandra Chinensis Bail.: an Overview of Russian Research and Uses in Medicine. J. Ethnopharmacol. 118, 183–212. doi:10.1016/j.jep.2008.04.020
Pashirova T. N., Sapunova A. S., Lukashenko S. S., Burilova E. A., Lubina A. P., Shaihutdinova Z. M., et al. (2020). Synthesis, Structure-Activity Relationship and Biological Evaluation of Tetracationic Gemini Dabco-Surfactants for Transdermal Liposomal Formulations. Int. J. Pharm. 575, 118953. doi:10.1016/j.ijpharm.2019.118953
Pedersen B. K., Febbraio M. A. (2012). Muscles, Exercise and Obesity: Skeletal Muscle as a Secretory Organ. Nat. Rev. Endocrinol. 8, 457–465. doi:10.1038/nrendo.2012.49
Peng L.-N., Lee W.-J., Liu L.-K., Lin M.-H., Chen L.-K. (2018). Healthy Community-Living Older Men Differ from Women in Associations between Myostatin Levels and Skeletal Muscle Mass. J. cachexia, sarcopenia muscle 9, 635–642. doi:10.1002/jcsm.12302
Philip B., Lu Z., Gao Y. (2005). Regulation of GDF-8 Signaling by the P38 MAPK. Cell. Signal. 17, 365–375. doi:10.1016/j.cellsig.2004.08.003
Qian L., Tang M., Yang J., Wang Q., Cai C., Jiang S., et al. (2015). Targeted Mutations in Myostatin by Zinc-Finger Nucleases Result in Double-Muscled Phenotype in Meishan Pigs. Sci. Rep. 5, 14435. doi:10.1038/srep14435
Qiu Y., Li X., He X., Pu J., Zhang J., Lu S. (2020). Computational Methods-Guided Design of Modulators Targeting Protein-Protein Interactions (PPIs). Eur. J. Med. Chem. 207, 112764. doi:10.1016/j.ejmech.2020.112764
Quinn L. S., Anderson B. G., Conner J. D., Wolden-Hanson T. (2013). IL-15 Overexpression Promotes Endurance, Oxidative Energy Metabolism, and Muscle PPARδ, SIRT1, PGC-1α, and PGC-1β Expression in Male Mice. Endocrinology 154, 232–245. doi:10.1210/en.2012-1773
Quinn L. S., Anderson B. G., Conner J. D., Wolden-Hanson T., Marcell T. J. (2014). IL-15 Is Required for Postexercise Induction of the Pro-oxidative Mediators PPARδ and SIRT1 in Male Mice. Endocrinology 155, 143–155. doi:10.1210/en.2013-1645
Quinn L. S., Anderson B. G., Drivdahl R. H., Alvarez B., Argilés J. M. (2002). Overexpression of Interleukin-15 Induces Skeletal Muscle Hypertrophy In Vitro: Implications for Treatment of Muscle Wasting Disorders. Exp. Cell Res. 280, 55–63. doi:10.1006/excr.2002.5624
Quinn L., Straitbodey L., Anderson B., Argiles J., Havel P. (2005). Interleukin-15 Stimulates Adiponectin Secretion by 3T3-L1 Adipocytes: Evidence for a Skeletal Muscle-To-Fat Signaling Pathway. Cell Biol. Int. 29, 449–457. doi:10.1016/j.cellbi.2005.02.005
Ratkevicius A., Joyson A., Selmer I., Dhanani T., Grierson C., Tommasi A. M., et al. (2011). Serum Concentrations of Myostatin and Myostatin-Interacting Proteins Do Not Differ between Young and Sarcopenic Elderly Men. Journals Gerontology Ser. A Biol. Sci. Med. Sci. 66A, 620–626. doi:10.1093/gerona/glr025
Sadeghi M., Keshavarz-Fathi M., Baracos V., Arends J., Mahmoudi M., Rezaei N. (2018). Cancer Cachexia: Diagnosis, Assessment, and Treatment. Crit. Rev. Oncology/Hematology 127, 91–104. doi:10.1016/j.critrevonc.2018.05.006
Sakuma K., Yamaguchi A. (2012). Sarcopenia and Cachexia: the Adaptations of Negative Regulators of Skeletal Muscle Mass. J. Cachexia Sarcopenia Muscle 3, 77–94. doi:10.1007/s13539-011-0052-4
Salade L., Wauthoz N., Deleu M., Vermeersch M., De Vriese C., Amighi K., et al. (2017). Development of Coated Liposomes Loaded with Ghrelin for Nose-To-Brain Delivery for the Treatment of Cachexia. Int. J. Nanomedicine 12, 8531–8543. doi:10.2147/ijn.s147650
Samant S. A., Kanwal A., Pillai V. B., Bao R., Gupta M. P. (2017). The Histone Deacetylase SIRT6 Blocks Myostatin Expression and Development of Muscle Atrophy. Sci. Rep. 7, 11877. doi:10.1038/s41598-017-10838-5
Sandri M. (2016). Protein Breakdown in Cancer Cachexia. Seminars Cell & Dev. Biol. 54, 11–19. doi:10.1016/j.semcdb.2015.11.002
Santana K., Do Nascimento L. D., Lima E Lima A., Damasceno V., Nahum C., Braga R. C., et al. (2021). Applications of Virtual Screening in Bioprospecting: Facts, Shifts, and Perspectives to Explore the Chemo-Structural Diversity of Natural Products. Front. Chem. 9, 155. doi:10.3389/fchem.2021.662688
Sartori R., Romanello V., Sandri M. (2021). Mechanisms of Muscle Atrophy and Hypertrophy: Implications in Health and Disease. Nat. Commun. 12, 330. doi:10.1038/s41467-020-20123-1
Serrano A. L., Baeza-Raja B., Perdiguero E., Jardí M., Muñoz-Cánoves P. (2008). Interleukin-6 Is an Essential Regulator of Satellite Cell-Mediated Skeletal Muscle Hypertrophy. Cell Metab. 7, 33–44. doi:10.1016/j.cmet.2007.11.011
Sessa G., Weissmann G. (1968). Phospholipid Spherules (Liposomes) as a Model for Biological Membranes. J. Lipid Res. 9, 310–318. doi:10.1016/s0022-2275(20)43097-4
Shefer G., Van De Mark D. P., Richardson J. B., Yablonka-Reuveni Z. (2006). Satellite-cell Pool Size Does Matter: Defining the Myogenic Potency of Aging Skeletal Muscle. Dev. Biol. 294, 50–66. doi:10.1016/j.ydbio.2006.02.022
St. Andre M., Johnson M., Bansal P. N., Wellen J., Robertson A., Opsahl A., et al. (2017). A Mouse Anti-myostatin Antibody Increases Muscle Mass and Improves Muscle Strength and Contractility in the Mdx Mouse Model of Duchenne Muscular Dystrophy and its Humanized Equivalent, Domagrozumab (PF-06252616), Increases Muscle Volume in Cynomolgus Monkeys. Skelet. Muscle 7, 25. doi:10.1186/s13395-017-0141-y
Steelman C. A., Recknor J. C., Nettleton D., Reecy J. M. (2006). Transcriptional Profiling of Myostatin‐knockout Mice Implicates Wnt Signaling in Postnatal Skeletal Muscle Growth and Hypertrophy. FASEB J. 20, 580–582. doi:10.1096/fj.05-5125fje
Stoever K., Heber A., Eichberg S., Brixius K. (2017). Sarcopenia and Predictors of Skeletal Muscle Mass in Elderly Men with and without Obesity. Gerontol. Geriatr. Med. 3, 2333721417713637. doi:10.1177/2333721417713637
Suh J., Lee Y.-S. (2020a). Myostatin Inhibitors: Panacea or Predicament for Musculoskeletal Disorders? J. Bone Metab. 27, 151–165. doi:10.11005/jbm.2020.27.3.151
Suh J., Lee Y.-S. (2020b). Similar Sequences but Dissimilar Biological Functions of GDF11 and Myostatin. Exp. Mol. Med. 52, 1673–1693. doi:10.1038/s12276-020-00516-4
Takayama K., Rentier C., Asari T., Nakamura A., Saga Y., Shimada T., et al. (2017). Development of Potent Myostatin Inhibitory Peptides through Hydrophobic Residue-Directed Structural Modification. ACS Med. Chem. Lett. 8, 751–756. doi:10.1021/acsmedchemlett.7b00168
Tapscott S. J. (2005). The Circuitry of a Master Switch: Myod and the Regulation of Skeletal Muscle Gene Transcription. Development 132, 2685–2695. doi:10.1242/dev.01874
Tauer J. T., Rauch F. (2019). Novel ActRIIB Ligand Trap Increases Muscle Mass and Improves Bone Geometry in a Mouse Model of Severe Osteogenesis Imperfecta. Bone 128, 115036. doi:10.1016/j.bone.2019.115036
Testelmans D., Crul T., Maes K., Agten A., Crombach M., Decramer M., et al. (2010). Atrophy and Hypertrophy Signalling in the Diaphragm of Patients with COPD. Eur. Respir. J. 35, 549–556. doi:10.1183/09031936.00091108
Thies R. S., Chen T., Davies M. V., Tomkinson K. N., Pearson A. A., Shakey Q. A., et al. (2001). GDF-8 Propeptide Binds to GDF-8 and Antagonizes Biological Activity by Inhibiting GDF-8 Receptor Binding. Growth factors. 18, 251–259. doi:10.3109/08977190109029114
Thomas K., Engler A. J., Meyer G. A. (2015). Extracellular Matrix Regulation in the Muscle Satellite Cell Niche. Connect. tissue Res. 56, 1–8. doi:10.3109/03008207.2014.947369
Thorsteinsdóttir S., Deries M., Cachaço A. S., Bajanca F. (2011). The Extracellular Matrix Dimension of Skeletal Muscle Development. Dev. Biol. 354, 191–207. doi:10.1016/j.ydbio.2011.03.015
Trott O., Olson A. J. (2010). AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 31, 455–461. doi:10.1002/jcc.21334
Tsuchida K. (2008). Myostatin Inhibition by a Follistatin-Derived Peptide Ameliorates the Pathophysiology of Muscular Dystrophy Model Mice. Acta Myol. 27, 14–18.
Tsuchida K., Matsuzaki T., Yamakawa N., Liu Z., Sugino H. (2001). Intracellular and Extracellular Control of Activin Function by Novel Regulatory Molecules. Mol. Cell. Endocrinol. 180, 25–31. doi:10.1016/s0303-7207(01)00522-6
Valtonen S., Vuorinen E., Kariniemi T., Eskonen V., Le Quesne J., Bushell M., et al. (2020). Nanomolar Protein-Protein Interaction Monitoring with a Label-free Protein-Probe Technique. Anal. Chem. 92, 15781–15788. doi:10.1021/acs.analchem.0c02823
Velloso C. P. (2008). Regulation of Muscle Mass by Growth Hormone and IGF-I. Br. J. Pharmacol. 154, 557–568. doi:10.1038/bjp.2008.153
Vieira D., Gamarra L. (2016). Getting into the Brain: Liposome-Based Strategies for Effective Drug Delivery across the Blood–brain Barrier. Int. J. Nanomedicine 11, 5381–5414. doi:10.2147/ijn.s117210
Vilar S., Cozza G., Moro S. (2008). Medicinal Chemistry and the Molecular Operating Environment (MOE): Application of QSAR and Molecular Docking to Drug Discovery. Curr. Top. Med. Chem. 8, 1555–1572. doi:10.2174/156802608786786624
Villoutreix B., Bastard K., Sperandio O., Fahraeus R., Poyet J.-L., Calvo F., et al. (2008). In Silico-In Vitro Screening of Protein-Protein Interactions: towards the Next Generation of Therapeutics. Curr. Pharm. Biotechnol. 9, 103–122. doi:10.2174/138920108783955218
Vlieghe P., Lisowski V., Martinez J., Khrestchatisky M. (2010). Synthetic Therapeutic Peptides: Science and Market. Drug Discov. today 15, 40–56. doi:10.1016/j.drudis.2009.10.009
von Haehling S., Anker S. D. (2014). Prevalence, Incidence and Clinical Impact of Cachexia: Facts and Numbers-Update 2014. J. Cachexia Sarcopenia Muscle 5, 261–263. doi:10.1007/s13539-014-0164-8
Wagner K. R., Fleckenstein J. L., Amato A. A., Barohn R. J., Bushby K., Escolar D. M., et al. (2008). A Phase I/IItrial of MYO-029 in Adult Subjects with Muscular Dystrophy. Ann. Neurol. 63, 561–571. doi:10.1002/ana.21338
Wheeler T. L., Koohmaraie M. (1994). Prerigor and Postrigor Changes in Tenderness of Ovine Longissimus Muscle2. J. Anim. Sci. 72, 1232–1238. doi:10.2527/1994.7251232x
White T. A., LeBrasseur N. K. (2014). Myostatin and Sarcopenia: Opportunities and Challenges - a Mini-Review. Gerontology 60, 289–293. doi:10.1159/000356740
Wilkes J. J., Lloyd D. J., Gekakis N. (2009). Loss-of-Function Mutation in Myostatin Reduces Tumor Necrosis Factor α Production and Protects Liver against Obesity-Induced Insulin Resistance. Diabetes 58, 1133–1143. doi:10.2337/db08-0245
Williams A. J., Ekins S., Tkachenko V. (2012). Towards a Gold Standard: Regarding Quality in Public Domain Chemistry Databases and Approaches to Improving the Situation. Drug Discov. today 17, 685–701. doi:10.1016/j.drudis.2012.02.013
Winbanks C. E., Weeks K. L., Thomson R. E., Sepulveda P. V., Beyer C., Qian H., et al. (2012). Follistatin-mediated Skeletal Muscle Hypertrophy Is Regulated by Smad3 and mTOR Independently of Myostatin. J. Cell Biol. 197, 997–1008. doi:10.1083/jcb.201109091
Wolfman N. M., Mcpherron A. C., Pappano W. N., Davies M. V., Song K., Tomkinson K. N., et al. (2003). Activation of Latent Myostatin by the BMP-1/tolloid Family of Metalloproteinases. Proc. Natl. Acad. Sci. U.S.A. 100, 15842–15846. doi:10.1073/pnas.2534946100
Yanagi S., Sato T., Kangawa K., Nakazato M. (2018). The Homeostatic Force of Ghrelin. Cell Metab. 27, 786–804. doi:10.1016/j.cmet.2018.02.008
Yang J., Ratovitski T., Brady J. P., Solomon M. B., Wells K. D., Wall R. J. (2001). Expression of Myostatin Pro Domain Results in Muscular Transgenic Mice. Mol. Reprod. Dev. 60, 351–361. doi:10.1002/mrd.1097
Yang W., Chen Y., Zhang Y., Wang X., Yang N., Zhu D. (2006). Extracellular Signal-Regulated Kinase 1/2 Mitogen-Activated Protein Kinase Pathway Is Involved in Myostatin-Regulated Differentiation Repression. Cancer Res. 66, 1320–1326. doi:10.1158/0008-5472.can-05-3060
Yang Y., Xu Y., Li W., Wang G., Song Y., Yang G., et al. (2009). STAT3 Induces Muscle Stem Cell Differentiation by Interaction with myoD. Cytokine 46, 137–141. doi:10.1016/j.cyto.2008.12.015
Yarasheski K. E., Bhasin S., Sinha-Hikim I., Pak-Loduca J., Gonzalez-Cadavid N. F. (2002). Serum Myostatin-Immunoreactive Protein Is Increased in 60-92 Year Old Women and Men with Muscle Wasting. J. Nutr. Health Aging 6, 343–348.
Yu R., Chen J.-a., Xu J., Cao J., Wang Y., Thomas S. S., et al. (2017). Suppression of Muscle Wasting by the Plant-Derived Compound Ursolic Acid in a Model of Chronic Kidney Disease. J. Cachexia, Sarcopenia Muscle 8, 327–341. doi:10.1002/jcsm.12162
Zammit P. S., Golding J. P., Nagata Y., Hudon V., Partridge T. A., Beauchamp J. R. (2004). Muscle Satellite Cells Adopt Divergent Fates. J. Cell Biol. 166, 347–357. doi:10.1083/jcb.200312007
Zhang W., Liu Y., Zhang H. (2021). Extracellular Matrix: an Important Regulator of Cell Functions and Skeletal Muscle Development. Cell & Biosci. 11, 1–13. doi:10.1186/s13578-021-00579-4
Keywords: myostatin, skeletal muscle, MSTN inhibitors, natural compounds, peptides
Citation: Baig MH, Ahmad K, Moon JS, Park S-Y, Ho Lim J, Chun HJ, Qadri AF, Hwang YC, Jan AT, Ahmad SS, Ali S, Shaikh S, Lee EJ and Choi I (2022) Myostatin and its Regulation: A Comprehensive Review of Myostatin Inhibiting Strategies. Front. Physiol. 13:876078. doi: 10.3389/fphys.2022.876078
Received: 16 February 2022; Accepted: 06 June 2022;
Published: 23 June 2022.
Edited by:
Jianjie Ma, The Ohio State University, United StatesReviewed by:
Ahlke Heydemann, University of Illinois at Chicago, United StatesHirofumi Zempo, Tokyo Seiei College, Japan
Narendra Bharathy, University of Pennsylvania, United States
Copyright © 2022 Baig, Ahmad, Moon, Park, Ho Lim, Chun, Qadri, Hwang, Jan, Ahmad, Ali, Shaikh, Lee and Choi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Eun Ju Lee, Z29yYXBhZG9jMDMxNUBoYW5tYWlsLm5ldA==; Inho Choi, aW5ob2Nob2lAeW51LmFjLmty
†These authors have contributed equally to this work
 Mohammad Hassan Baig
Mohammad Hassan Baig Khurshid Ahmad
Khurshid Ahmad Jun Sung Moon
Jun Sung Moon So-Young Park5
So-Young Park5 Arif Tasleem Jan
Arif Tasleem Jan Syed Sayeed Ahmad
Syed Sayeed Ahmad Shahid Ali
Shahid Ali Sibhghatulla Shaikh
Sibhghatulla Shaikh Eun Ju Lee
Eun Ju Lee Inho Choi
Inho Choi